Abstract
Objectives
To provide a summary of evidence for the diagnostic accuracies of three multiplex PCR systems (mPCRs)—BioFire FilmArray RP (FilmArray), Nanosphere Verigene RV+ test (Verigene RV+) and Hologic Gen-Probe Prodesse assays—on the detection of viral respiratory infections.
Methods
A comprehensive search up to 1 July 2017 was conducted on Medline and Embase for studies that utilized FilmArray, Verigene RV+ and Prodesse for diagnosis of viral respiratory infections. A summary of diagnostic accuracies for the following five viruses were calculated: influenza A virus (FluA), influenza B virus, respiratory syncytial virus, human metapneumovirus and adenovirus. Hierarchical summary receiver operating curves were used for estimating the viral detection performance per assay.
Results
Twenty studies of 5510 patient samples were eligible for analysis. Multiplex PCRs demonstrated high diagnostic accuracy, with area under the receiver operating characteristic curve (AUROC) equal to or more than 0.98 for all the above viruses except for adenovirus (AUROC 0.89). FilmArray, Verigene RV+ and ProFlu+ (the only Prodesse assay with enough data) demonstrated a summary sensitivity for FluA of 0.911 (95% confidence interval, 0.848–0.949), 0.949 (95% confidence interval, 0.882–0.979) and 0.954 (95% confidence interval, 0.871–0.985), respectively. The three mPCRs were comparable in terms of detection of FluA.
Conclusions
Point estimates calculated from eligible studies showed that the three mPCRs (FilmArray, Verigene RV+ and ProFlu+) are highly accurate and may provide important diagnostic information for early identification of respiratory virus infections. In patients with low pretest probability for FluA, these three mPCRs can predict a low possibility of infection and may justify withholding empirical antiviral treatments.
Keywords: Multiplex PCR, Point-of-care test, Respiratory virus infection
Introduction
Acute respiratory tract infections (ARI) cause high morbidity and mortality [1]. Among them, viral ARIs are one of the leading causes for paediatric and geriatric hospitalization and clinic visits [2], [3]. Each year, seasonal influenza causes >200 000 hospitalizations and more than $10 billion direct medical costs in the United States. In specific populations (e.g. immunocompromised patients, neonates, and chronic pulmonary disease patients), the high complication and mortality rates from viral ARIs is a major concern [4]. Moreover, empirical antibiotics are commonly prescribed to patients with viral ARIs because of the lack of rapid and sensitive diagnostic methods and nonspecific symptoms, which delay proper treatments and precipitate antibiotic resistance [4], [5], [6].
Traditional diagnostic techniques (e.g. virus culture, haemagglutination inhibition assay, enzyme immunoassay and direct fluorescent antibody) were once the mainstays for pathogen detection. However, these methods were either insensitive, time consuming, labor intensive or operator dependent [7], [8], [9]. New technologies have emerged as a result of massive clinical demands, such as melting curve analysis, microfluidic device and nucleic acid amplification technologies [5], [6], [10], [11], [12], [13]. These molecular diagnostic tools have shorter turnaround times and higher sensitivity for viral pathogens [14], [15]. In addition, they allow for detection of a broader panel of viruses and coinfection [15], [16], and they thus have become more widely used than the conventional virologic assays [8], [17], [18], [19]. In particular, multiplex PCR (mPCR) is a validated strategy for the rapid detection and precise identification of a large number of respiratory viruses [19], [20], [21], [22] by incorporating several primers within one reaction tube to amplify genomic fragments of many pathogens [22], [23]. With the use of a mPCR panel, one study demonstrated a 30% to 50% increase in the diagnostic yield of respiratory viruses compared to direct fluorescent antibody and culture [24].
There are a number of US Food and Drug Association (FDA)-cleared mPCRs available today for detecting respiratory pathogens, each with pros and cons. The characteristics of the three FDA-approved mPCR systems included in our study are listed in Table 1 . The BioFire FilmArray RP (FilmArray) respiratory panel [24], which utilizes melting curve analysis, is a random-access molecular test using principles of real-time PCR. The Verigene RV+ test is based on gold nanoparticle technology and silver signal amplification. Lastly, Hologic Gen-Probe Prodesse launches several assays with variable run sizes that also utilize melting curve analysis but with limited multiplexing ability. Although each Prodesse assay can only detect two to three viruses at a time, the Prodesse assays are still viewed as mPCR [25]. These three mPCRs were chosen because they have shorter turnaround times and have more available studies for analysis among a number of FDA-approved mPCRs. There is also one original study that provided direct comparison of these three mPCRs [25].
Table 1.
Characteristics of BioFire FilmArray RP, Nanosphere Verigene RV+ Test and Hologic Gen-Probe Prodesse assays
| Name | BioFire FilmArray | Verigene | GenProbe Prodesse |
|---|---|---|---|
| Technology | Melting curve analysis | Gold nanoparticles with silver signal amplification | Melting curve analysis |
| Assays | Respiratory panel | Respiratory virus plus test | ProFlu+, ProFAST+, ProAdeno+, ProParaflu+, Pro hMPV+ |
| Targets |
|
|
|
| Throughput | 1 sample per instrument | 1 sample per processor | 14 samples per run |
| Run time (hours) | 1 | <2.5 | 4–5 |
| Hands-on time | 2 minutes | 5 minutes | 1.5 hours |
| Sample preparation included? | Yes | Yes | No |
| Reagent storage conditions | Room temperature | 2–8°C and −20°C | −70°C |
FluA, influenza A virus; hMPV, human metapneumovirus; RSV, respiratory syncytial virus.
To gain insight into the optimal diagnostic tool for routine clinical use, we here provide a summary of evidence comparing the diagnostic accuracies of FilmArray, Verigene RV+ and Hologic Gen-Probe Prodesse assays for the detection of viral respiratory infections.
Methods
The protocol of our study was based on the PRISMA (Preferred Reporting Items for Systematic Review and Meta-analysis) statement [26] and the standard guideline for systematic reviews of diagnostic tests by the Cochrane Collaboration [27].
Search strategy
A comprehensive search of literature was conducted using two databases: PubMed (from inception to April 2015) and Embase (from inception to April 2015). The search term combination was: (multiplex AND pcr OR (multiplex AND polymerase AND chain AND reaction) OR filmarray OR verigene OR prodesse OR proflu OR profast OR proadeno OR proparaflu OR (pro hmpv)) AND ((respiratory AND tract AND infection) OR (respiratory AND infection) OR (respiratory AND virus) OR (respiratory AND tract AND disease) OR (respiratory AND disease) OR (common AND cold) OR influenza OR pneumonia OR bronchitis OR bronchiolitis OR rhinosinusitis OR pharyngitis OR laryngitis OR (otitis AND media) OR tonsillitis OR asthma OR copd OR (chronic AND obstructive AND lung AND disease)). The detailed search strategy is provided in Supplementary Materials S1. No language restrictions were applied to the search. The search was then supplemented by bibliographies of retrieved full-text articles and the latest narrative reviews. We also contacted the authors of publications that did not provide required data. An updated search to 1 July 2017 was performed before starting the statistical analysis.
Study selection
Studies that evaluated the performance of FDA-approved mPCR systems for the detection of viral respiratory infection were included, as follow: (a) they assessed the accuracy of one or more the following systems: FilmArray, Nanosphere Verigene RV+ and Hologic Gen-Probe Prodesse assays (ProFlu+, ProFAST, ProParaflu+, ProAdeno+ and Pro hMPV+) against reference standards and (b) they provided sufficient information to calculate sensitivity and specificity. Studies outside of the United States that used non–US FDA–approved versions of the mPCRs were also included. We included conference abstracts, posters and oral presentations only if they provided sufficient information to calculate the sensitivities and specificities of the FDA-approved mPCR systems. Studies were excluded for the following reasons: (a) they only analysed clinical specimens previously determined to be positive (without negative specimens), (b) they used our target mPCR systems as the sole standard to validate other mPCR systems and (c) they used an mPCR assay not approved by FDA as reference. In addition, we excluded reviews, guidelines, case reports, editorials, panel discussions, letters, notes and comments. For multiple publications, only the latest available publications with complete data of the same patient group were included. Studies that compared more than one FDA-approved mPCR systems with the same reference standard, used different reference methods for different viruses, used different samples for different viruses or reported prospective and retrospective samples independently have separate data sets for each comparison. Three reviewers independently screened the titles and abstracts from PubMed and Embase. Disagreements or uncertainties were resolved by discussion.
Data extraction
A data extraction form was used in 20 included studies by two reviewers before being finalized. Each data set was extracted by two authors independently to avoid bias. The form consisted of the following characteristics: study type, study design, patient age, patient inclusion criteria, specimen type, mPCR systems used, reference standard and 2 × 2 tables. The 2 × 2 tables were further used to calculate sensitivities and specificities of the target assays. For the reference methods, virus culture and direct fluorescent antibody were grouped together because they are universally recognized as the reference standard [28], [29]. Reverse transcription (RT) PCR referred to both commercial RT-PCRs and in-house RT-PCRs. Composite reference standard was defined as a standard that used more than one comparator assays. Discrepant analyses encompassed studies that resolved discrepancies between target assay and comparator assay by bidirectional sequencing, RT-PCR, repeated testing or other methods. Studies were also grouped on the basis of whether they incorporated FilmArray, Verigene RV+ and Hologic Gen-Probe Prodesse assays as part of a composite reference standard. Studies that had <5% of the samples taken from the lower respiratory tract were categorized as using upper respiratory specimens. Children were defined as patients younger than 18 years. Studies that included both children and adult population were categorized as mixed.
Quality assessment
The quality of the eligible studies was independently assessed by two reviewers using the Quality Assessment of Diagnostic Accuracy Studies 2 tool (QUADAS-2) [30]. For each diagnostic study, we determined the risk for bias and general applicability in all four domains of QUADAS-2 and reported them separately. Those with low risk of bias or low concern regarding applicability were judged as low. A study would be judged as unclear if there were insufficient data for interpretation.
Data synthesis and statistical analysis
A bivariate model was applied to estimate summary sensitivity and specificity. The positive likelihood ratios (LR+) and negative likelihood ratios (LR−) were then calculated from summary sensitivity and specificity. The bivariate model approach modelled the logit-transformed sensitivity and specificity simultaneously to account for the inherent negative correlation between sensitivity and specificity that may arise due to different thresholds in different studies [30]. In addition, the bivariate model could also account for between-study heterogeneity. All analyses except for the summary receiver operating characteristic curve (ROC) were performed by the ‘mada’ package in R software (R Foundation for Statistical Computing, Vienna, Austria; http://www.r-project.org/). The summary ROC and area under the ROC was calculated by the ‘midas’ package in STATA (STATA Inc, College Station, Texas). A two-sided p value of <0.05 indicated statistical significance for all tests.
Results
Identification of studies
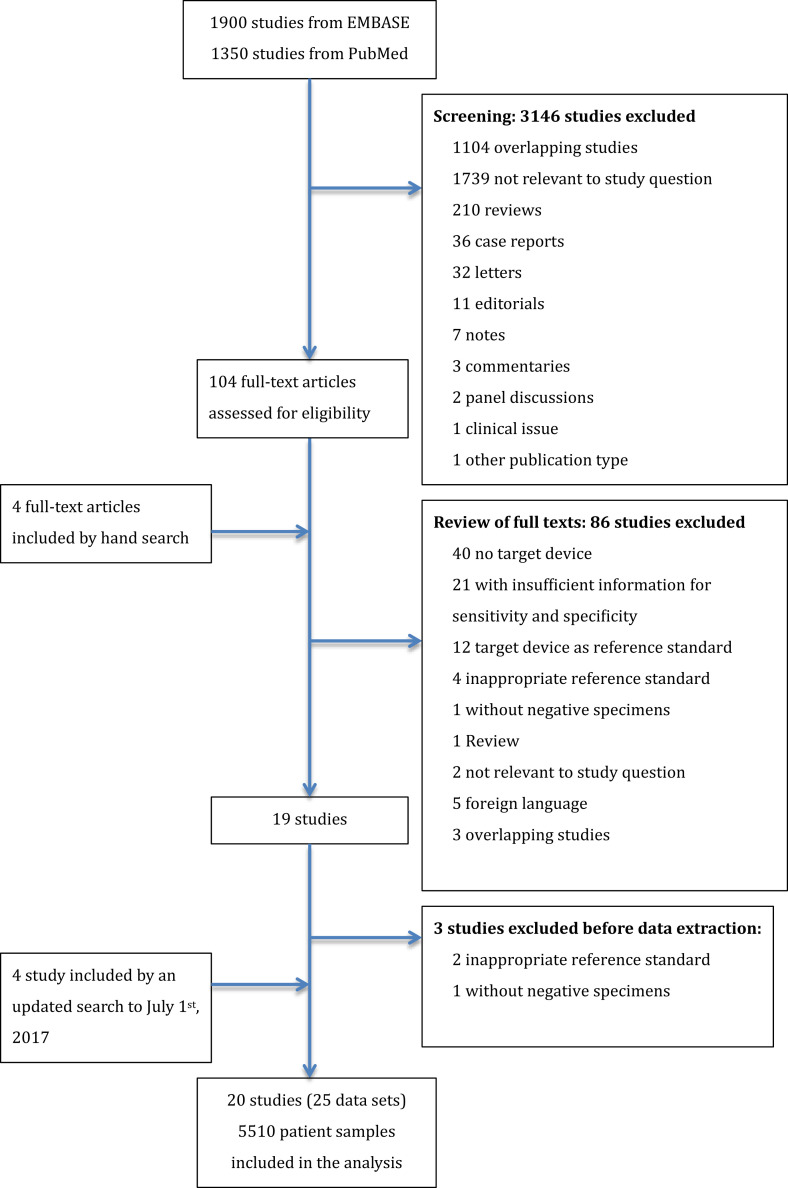
Our comprehensive search yielded 1900 studies from Embase and 1350 studies from PubMed. After inclusion and exclusion following the protocol and an updated search to 1 July 2017 (Fig. 1 ), a total of 20 studies (25 data sets) were eligible for analysis, encompassing a total of 5510 patient samples. ProFlu+ was the only Prodesse assay with enough data sets for quantitative analysis. Parainfluenza virus was not analysed because of insufficient data.
Fig. 1.
PRISMA flow diagram for selection of articles for meta-analysis.
Characteristics of included studies
The laboratory characteristics of FilmArray RP, Verigene RV+ and Prodesse ProFlu+ are illustrated in Table 1. A simplified summary of the characteristics of the included studies is provided in Table 2 . Details of the characteristics and key results of each individual study are provided in Supplementary Materials S2. None of the studies specifically recruited adults only, and approximately 30% of the studies obtained their samples from children. Discrepant analysis and composite reference standard were the two most commonly applied reference standards.
Table 2.
Characteristics of the 20 included studies (25 data sets)
| Characteristic | n (data sets) (%) |
|---|---|
| Age | |
| Children | 8 (32%) |
| Mixed | 10 (40%) |
| Unclear | 7 (28%) |
| Index test | |
| FilmArray RP | 10 (40%) |
| Verigene RV+ | 6 (24%) |
| Prodesse ProFlu+ | 9 (36%) |
| Reference standard | |
| Virus culture/direct fluorescent antibody | 4 (16%) |
| RT-PCR | 4 (16%) |
| Composite | 7 (28%) |
| Discrepant analysis | 10 (40%) |
| Specimen type | |
| Upper respiratory specimen | 22 (88%) |
| Mixed upper and lower respiratory specimen | 2 (8%) |
| Unclear | 1 (4%) |
| Study design | |
| Prospective | 11 (44%) |
| Retrospective | 12 (48%) |
| Mixed | 2 (8%) |
| Country | |
| United States | 18 (72%) |
| Belgium | 3 (12%) |
| Korea | 1 (4%) |
| Japan | 2 (8%) |
| Taiwan | 1 (4%) |
RT-PCR, reverse transcription–PCR.
Quality assessment
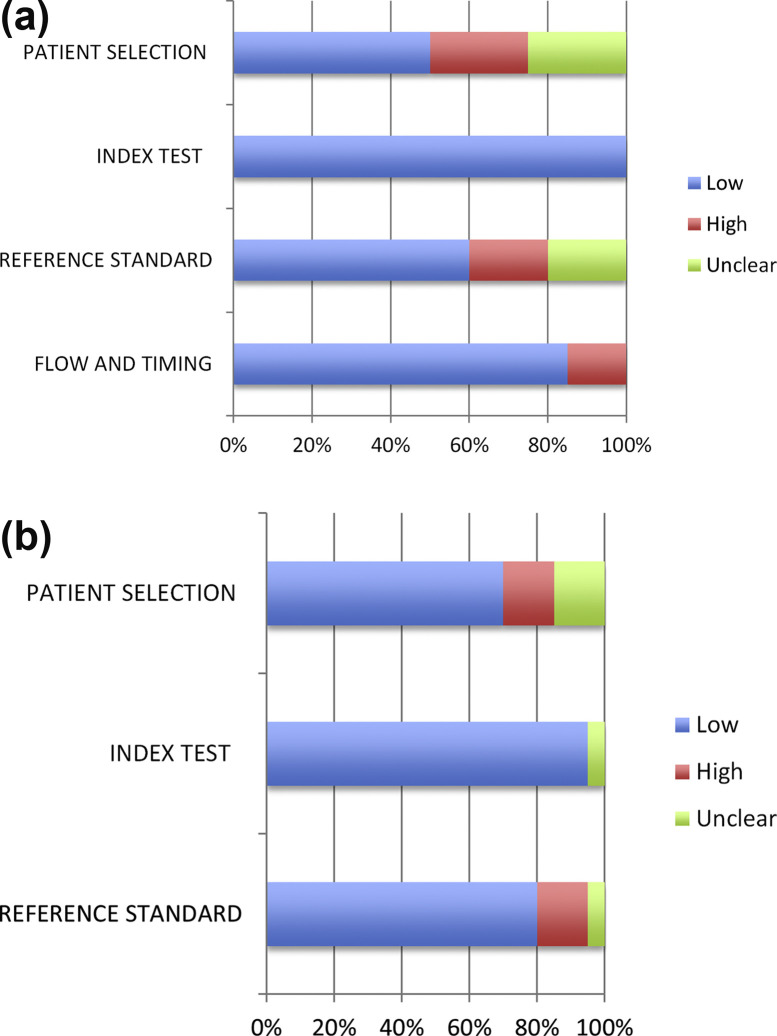
The studies varied in quality. Quality assessment by the QUADAS-2 tool is demonstrated in Fig. 2 . For the ‘Patient Selection’ domain, some studies were not clear about how the patients were recruited. Some studies used refrigerated samples that were precollected without specifying their original sources; others did not avoid a case–control design. For the ‘Reference Standard’ domain, most studies did not specify the use of blinding for reference standard. Some studies that were of low quality in these regards used the index test as part of the reference method. Others did not use the same reference method on all their samples. However, most studies were identified as high quality for flow and timing. Overall, the majority of the studies had low concern regarding applicability.
Fig. 2.
QUADAS-2 for included studies. (a) QUADAS-2 bias assessment. (b) QUADAS-2 applicability assessment. QUADAS-2, Quality Assessment of Diagnostic Accuracy Studies 2.
Diagnostic accuracy of mPCRs
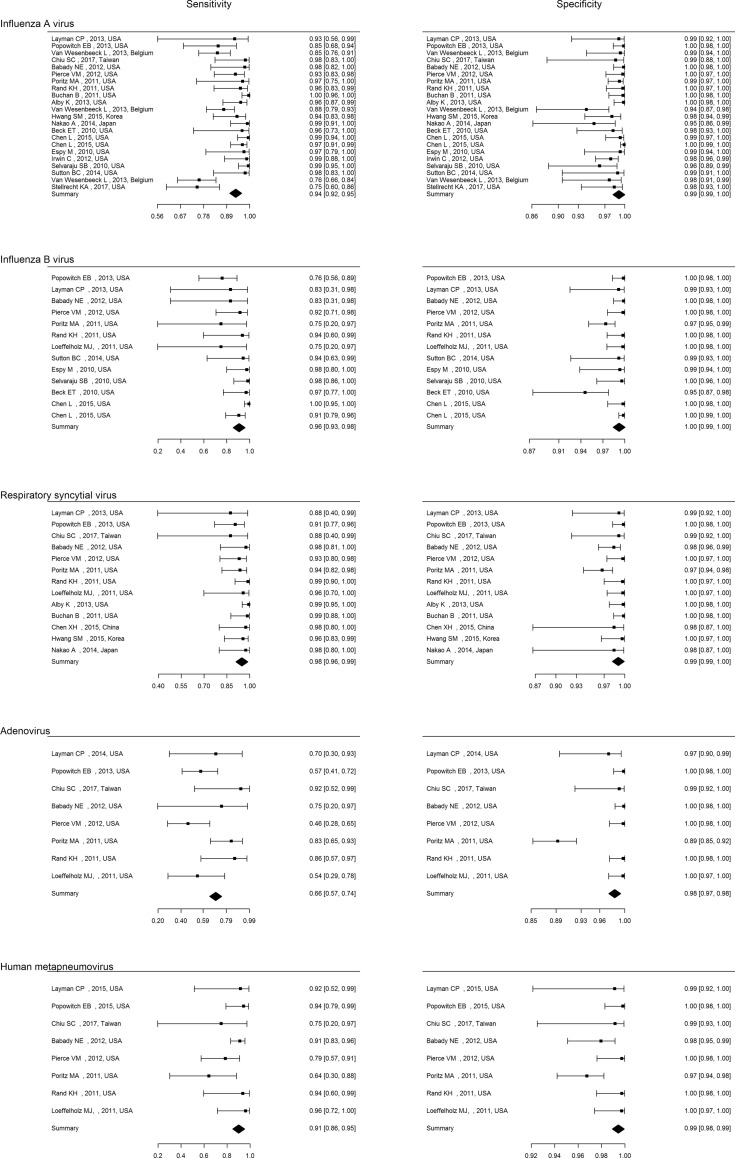
Table 3 lists the point estimates of sensitivity, specificity, LR+, LR− and area under the plasma concentration vs. time curve value of each mPCR assay for each virus. Multiplex PCRs demonstrated very high area under the receiver operating characteristic curve (AUROC) (≥0.98) for influenza A virus (FluA), influenza B virus (FluB), respiratory syncytial virus (RSV) and human metapneumovirus (hMPV) and high to moderate AUROC for adenovirus. For FluA, sensitivity estimates were acceptable (>0.9) for FilmArray RP and Verigene RV+, and high (>0.95) for Prodesse ProFlu+; specificity estimates were high (>0.98) for all three mPCRs. The overall LR+ were well beyond 10, indicating a high rule-in value. The overall LR− were all below 0.1, which suggested a high rule-out value. Table 3 provides point estimates for FluB, RSV, hMPV and adenovirus. Fig. 3 shows the forest plot of sensitivity and specificity of the mPCRs for each virus. Fig. 4 illustrates the receiver operating characteristic curve and area under the receiver operating characteristic curve for the detection of each virus.
Table 3.
Accuracy estimates of included studies
| Test | Sensitivity (95% confidence interval) | Specificity (95% confidence interval) | LR+ (95% confidence interval) | LR− (95% confidence interval) | AUC (95% confidence interval) |
|---|---|---|---|---|---|
| Influenza A virus | |||||
| FilmArray | 0.911 (0.848, 0.949) | 0.995 (0.988, 0.998) | 186 (74.9, 368) | 0.0928 (0.052, 0.153) | 0.99 (0.98, 1) |
| Verigene | 0.949 (0.882, 0.979) | 0.982 (0.944, 0.995) | 65.2 (15.9, 185) | 0.058 (0.0206, 0.122) | 0.99 (0.98, 1) |
| Prodesse | 0.954 (0.871, 0.985) | 0.983 (0.973, 0.989) | 57.65 (40.48, 76.94) | 0.053 (0.022, 0.0896) | 0.99 (0.99, 1) |
| Summary | 0.940 (0.902, 0.964) | 0.987 (0.979, 0.992) | 76.9 (42.4, 126) | 0.06 (0.03, 0.101) | 0.99 (0.98, 1) |
| Influenza B virus | |||||
| FilmArray | 0.822 (0.689, 0.905) | 0.994 (0.980, 0.998) | 167.50 (40.9, 503.00) | 0.188 (0.093, 0.313) | 0.98 (0.94, 1) |
| Prodesse | 0.963 (0.907, 0.986) | 0.992 (0.969, 0.998) | 136.73 (30.5, 385) | 0.04 (0.014, 0.097) | 0.99 (0.99, 1) |
| Summary | 0.932 (0.877, 0.963) | 0.993 (0.986, 0.997) | 154.4 (66.5, 304) | 0.072 (0.034, 0.124) | 0.99 (0.99, 1) |
| RSV | |||||
| FilmArray | 0.911 (0.821, 0.958) | 0.987 (0.971, 0.994) | 73.1 (29.4, 150) | 0.09 (0.0412, 0.172) | 0.98 (0.98, 0.99) |
| Verigene | 0.977 (0.929, 0.993) | 0.993 (0.962, 0.999) | 219.30 (23.5, 868) | 0.027 (0.0076, 0.072) | 0.99 (0.98, 1) |
| Summary | 0.942 (0.84, 0.972) | 0.991 (0.980, 0.996) | 109.45 (47.5, 226) | 0.06 (0.03, 0.118) | 0.99 (0.99, 1) |
| Adenovirus | |||||
| FilmArray | 0.670 (0.516, 0.794) | 0.991 (0.961, 0.998) | 86.9 (20.3, 273) | 0.337 (0.212, 0.494) | 0.89 (0.85, 0.91) |
| hMPV | |||||
| FilmArray | 0.914 (0.835, 0.956) | 0.999 (0.854, 1) | 74.5 (29.4, 188.98) | 0.167 (0.093, 0.301) | 0.98 (0.97, 0.99) |
AUC, area under the curve; hMPV, human metapneumovirus; LR−, negative likelihood ratio; LR+, positive likelihood ratio; RSV, respiratory syncytial virus.
Fig. 3.
Forest plot of sensitivity and specificity of multiplex PCRs for diagnosis of each virus.
Fig. 4.
Receiver operating curve analysis of multiplex PCRs for detection of different virus infections. Solid lines indicate the 95% confidence region, and the dashed lines indicate the 95% prediction region. AUC, area under the curve.
Discussion
Overall, in our analyses including 5510 patient samples, mPCR systems demonstrated high diagnostic accuracy (AUROC ≥0.98) for FluA, FluB, RSV and hMPV, with the exception of adenovirus (AUROC 0.89). FilmArray RP, Verigene RV+ and Prodesse ProFlu+ demonstrated a summary sensitivity for FluA of 0.911 (95% confidence interval (CI), 0.848–0.949), 0.949 (95% CI, 0.882–0.979) and 0.954 (95% CI, 0.871–0.985), respectively. The three mPCRs were comparable in terms of detection of FluA, and FilmArray RP and Verigene RV+ were comparable for RSV detection. Although Prodesse ProFlu+ was found to have a statistically significant higher sensitivity than FilmArray RP for FluB detection, they demonstrated comparable AUROC and thus comparable accuracy.
Of the five respiratory viruses that these systems detect, the diagnosis of influenza virus infection may have the greatest clinical impact [31]. On the basis of our study, overall, the mPCRs exhibited reasonable sensitivity (0.940; 95% CI, 0.902–0.964) and high specificity (0.987; 95% CI, 0.979,0.992) for FluA. In current clinical practice, the immunoassay-based rapid influenza diagnostic test is most widely used for screening influenza infections [8]. Although the rapid influenza diagnostic test can detect FluA and FluB in respiratory specimens in approximately 15 to 30 minutes, its sensitivity is limited, as reported by a previous meta-analysis [32]. This may give false-negative results, which prevents its use as a reliable excluding diagnostic tool in clinical practice. Furthermore, commonly used reference standards such as RT-PCR or virus culture may take several days to yield results and have little value for treatment decision. Our meta-analysis showed that mPCRs provide highly accurate results in a clinically relevant time frame and may potentially change the current diagnostic and treatment practice for FluA infection.
As a result of the low sensitivity of the rapid influenza diagnostic tests, the decision of antiviral treatment for FluA infection is largely based on clinical grounds alone [5], which may lead to overprescription of anti-influenza drugs and increasing drug resistance [33], [34]. The high sensitivity and specificity of the mPCRs, as demonstrated, could thus guide the precise use of the antiviral treatment. In a virtual population with a 20% prevalence (pretest probability) of FluA infection, a LR+ of 186 (FilmArray RP), 65.2 (Verigene RV+) and 57.65 (Prodesse ProFlu+) translates into a posttest probability of 97.89%, 94.22% and 93.51%. Similarly, an overall LR+ for FluA of 76.9 translates into a posttest probability of 95.06%. In other words, approximately 19 of 20 patients who tested positive for FluA truly have FluA infection. In the same population with a 20% prevalence of FluA infection, a LR− of 0.0928 (FilmArray RP), 0.058 (Verigene RV+) and 0.053 (Prodesse ProFlu+) translates into a posttest probability of 2.27%, 1.43% and 1.31%, while an overall LR− for FluA of 0.06 translates into a negative predictive value of 1.48%. That is, among 200 patients who tested negative for FluA, only three with FluA infection may be missed. Although influenza virus infection may have dire consequences in certain vulnerable population, such as the elderly or those who are immunocompromised, a negative test result may justify withholding empiric antiviral treatment in apparently healthy young men, pregnant woman or children.
A study by Van Wesenbeeck et al. [25] of 171 clinical samples concluded that FilmArray RP and Prodesse ProFlu+ have better sensitivity for FluA than Verigene RV+. This is contrary to our results, which found the three mPCRs to be comparable for FluA detection. Our meta-analysis synthesized data across different studies encompassing 5510 patient samples and may present a more accurate estimate of the real situation. Although these three systems have comparable accuracy for FluA, FilmArray RP has the shortest hands-on time (2 minutes) and run time (1 hour) among the three mPCRs and includes sample preparation in its panel. Furthermore, its reagents can be stored at room temperature, and its assay detects the largest number of targets. Therefore, FilmArray RP may be the best choice in emergency rooms or during the influenza season.
There is less interest in rapid diagnostic tests for RSV, hMPV and adenovirus as a result of a lack of specific treatments available against these viruses and the common practice of supportive care in clinical settings. Nevertheless, RSV is one of the leading causes of ARIs in children [35] and immunocompromised patients, and it may cause severe complications; it therefore requires definitive diagnosis. Our results revealed that mPCRs are highly sensitive and specific for RSV and hMPV, and they offer a more rapid and accurate alternative to traditional methods [7], [36]. The current treatment is ribavirin for high-risk infants and young children [37], with several clinical trials for novel RSV treatments underway [38], [39], [40], [41]. In contrast, for adenovirus, FilmArray RP had a moderate to low sensitivity (<70%), but its superior specificity (>0.99) makes it a reliable rule-in tool. The literature has demonstrated that commercial FilmArray RP (v1.6) has low sensitivity for adenovirus [24], [42], [43], [44], but its sensitivity greatly improved in the commercial FilmArray 1.7 [42], developed later. Of note, none of our included studies specifically reported using FilmArray 1.7.
To our knowledge, this is the first report to perform a comparative meta-analysis on different mPCR platforms and different viruses. Not only did we provide detailed comparison on the characteristics of the three systems but we also provided quantitative accuracy measure for the different viruses by different systems. There are several limitations to our study, however. Firstly, some included studies did not provide sufficient details regarding the version of FilmArray RP used (premarket or commercial). We traced their publication dates and contacted the FilmArray RP manufacturers as well as the authors of the original study at an attempt to confirm the version of FilmArray RP. However, sensitivity analysis showed no clinically significant difference between the sensitivities and specificities of precommercial and FDA-approved commercial assays (data not published). Secondly, some included studies have a retrospective study design; they failed to specify their inclusion criteria, and they used only previously stored samples from patients with unknown characteristics. There is also concern regarding diagnostic review bias, in which the interpretation of the result of reference test is made with knowledge of the index test result [45]. However, a test review bias is highly unlikely because the interpretation of the index tests (mPCRs) was objective. Of note, five of the included studies adopted the case–control design, which tends to give an overestimation of the accuracy of the index test [45]. In addition, results of this study only showed the accuracy of three commercial mPCR systems for diagnosis of respiratory virus infection. Whether they have equal strength in therapeutic guidance remains to be validated. Lastly, our study does not provide an answer to whether mPCR systems improve the outcome of patients with respiratory virus infection compared to clinical diagnosis alone.
Conclusions
Analysis of eligible studies showed that three commercial mPCR systems—FilmArray, Verigene RV+ and GenProbe Prodesse ProFlu+—may provide important diagnostic information to help early identification of influenza virus, respiratory syncytial virus, adenovirus and hMPV. The great improvement of the sensitivity of the rapid diagnosis of influenza may have the potential to change current modes of diagnosis and management. However, not all of these systems are equally useful in terms of inclusion or exclusion diagnosis. Clinicians should interpret the results on the basis of likelihood ratio and pretest probability. The next step would be to assess whether provision of information from these mPCR tests can modify clinical outcomes.
Acknowledgements
We thank the staff of Core Labs, Department of Medical Research, National Taiwan University Hospital, for technical support; and we thank our medical librarian, H.-P. Chiu, for consultation in formulating the search strategy.
Editor: M. Leeflang
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.cmi.2017.11.018.
Transparency declaration
Supported in part by National Taiwan University Hospital Research Grant NTUH106-P04. All authors report no conflicts of interest relevant to this article.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
References
- 1.Williams B.G., Gouws E., Boschi-Pinto C., Bryce J., Dye C. Estimates of world-wide distribution of child deaths from acute respiratory infections. Lancet Infect Dis. 2002;2:25–32. doi: 10.1016/s1473-3099(01)00170-0. [DOI] [PubMed] [Google Scholar]
- 2.Liu J., Ai H., Xiong Y., Li F., Wen Z., Liu W. Prevalence and correlation of infectious agents in hospitalized children with acute respiratory tract infections in Central China. PLoS One. 2015;10 doi: 10.1371/journal.pone.0119170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marrie T.J., Durant H., Yates L. Community-acquired pneumonia requiring hospitalization: 5-year prospective study. Rev Infect Dis. 1989;11:586–599. doi: 10.1093/clinids/11.4.586. [DOI] [PubMed] [Google Scholar]
- 4.Byington C.L., Castillo H., Gerber K., Daly J.A., Brimley L.A., Adams S. The effect of rapid respiratory viral diagnostic testing on antibiotic use in a children’s hospital. Arch Pediatr Adolesc Med. 2002;156:1230–1234. doi: 10.1001/archpedi.156.12.1230. [DOI] [PubMed] [Google Scholar]
- 5.Henrickson K.J. Cost-effective use of rapid diagnostic techniques in the treatment and prevention of viral respiratory infections. Pediatr Ann. 2005;34:24–31. doi: 10.3928/0090-4481-20050101-08. [DOI] [PubMed] [Google Scholar]
- 6.Gonzales R., Malone D.C., Maselli J.H., Sande M.A. Excessive antibiotic use for acute respiratory infections in the United States. Clin Infect Dis. 2001;33:757–762. doi: 10.1086/322627. [DOI] [PubMed] [Google Scholar]
- 7.Mahony J.B. Detection of respiratory viruses by molecular methods. Clin Microbiol Rev. 2008;21:716–747. doi: 10.1128/CMR.00037-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vemula S.V., Zhao J., Liu J., Wang X., Biswas S., Hewlett I. Current approaches for diagnosis of influenza virus infections in humans. Viruses. 2016;8:96. doi: 10.3390/v8040096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ginocchio C.C., Alexander J.M. Current best practices for respiratory virus testing. J Clin Microbiol. 2011;49:S44–S48. [Google Scholar]
- 10.Woo P.C., Chiu S.S., Seto W.H., Peiris M. Cost-effectiveness of rapid diagnosis of viral respiratory tract infections in pediatric patients. J Clin Microbiol. 1997;35:1579–1581. doi: 10.1128/jcm.35.6.1579-1581.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mahony J.B., Blackhouse G., Babwah J., Smieja M., Buracond S., Chong S. Cost analysis of multiplex PCR testing for diagnosing respiratory virus infections. J Clin Microbiol. 2009;47:2812–2817. doi: 10.1128/JCM.00556-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Phaneuf C.R., Pak N., Saunders D.C., Holst G.L., Birjiniuk J., Nagpal N. Thermally multiplexed polymerase chain reaction. Biomicrofluidics. 2015;9 doi: 10.1063/1.4928486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rafiefard F., Orvell C., Bondeson K. Genotyping of respiratory syncytial virus (RSV) group A in Stockholm, Sweden, using PCR and two-dimensional melting curve analysis. APMIS. 2008;116:317–322. doi: 10.1111/j.1600-0463.2008.00758.x. [DOI] [PubMed] [Google Scholar]
- 14.Mahony J.B. Nucleic acid amplification–based diagnosis of respiratory virus infections. Expert Rev Anti Infect Ther. 2010;8:1273–1292. doi: 10.1586/eri.10.121. [DOI] [PubMed] [Google Scholar]
- 15.Ginocchio C.C., Zhang F., Manji R., Arora S., Bornfreund M., Falk L. Evaluation of multiple test methods for the detection of the novel 2009 influenza A (H1N1) during the New York City outbreak. J Clin Virol. 2009;45:191–195. doi: 10.1016/j.jcv.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu W., Tang Y.W. Emerging molecular assays for detection and characterization of respiratory viruses. Clin Lab Med. 2009;29:673–693. doi: 10.1016/j.cll.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mahony J., Chong S., Merante F., Yaghoubian S., Sinha T., Lisle C. Development of a respiratory virus panel test for detection of twenty human respiratory viruses by use of multiplex PCR and a fluid microbead-based assay. J Clin Microbiol. 2007;45:2965–2970. doi: 10.1128/JCM.02436-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marshall D.J., Reisdorf E., Harms G., Beaty E., Moser M.J., Lee W.M. Evaluation of a multiplexed PCR assay for detection of respiratory viral pathogens in a public health laboratory setting. J Clin Microbiol. 2007;45:3875–3882. doi: 10.1128/JCM.00838-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reijans M., Dingemans G., Klaassen C.H., Meis J.F., Keijdener J., Mulders B. RespiFinder: a new multiparameter test to differentially identify fifteen respiratory viruses. J Clin Microbiol. 2008;46:1232–1240. doi: 10.1128/JCM.02294-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frobert E., Escuret V., Javouhey E., Casalegno J.S., Bouscambert-Duchamp M., Moulinier C. Respiratory viruses in children admitted to hospital intensive care units: evaluating the CLART® Pneumovir DNA array. J Med Virol. 2011;83:150–155. doi: 10.1002/jmv.21932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huguenin A., Moutte L., Renois F., Leveque N., Talmud D., Abely M. Broad respiratory virus detection in infants hospitalized for bronchiolitis by use of a multiplex RT-PCR DNA microarray system. J Med Virol. 2012;84:979–985. doi: 10.1002/jmv.23272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H., McCormac M.A., Estes R.W., Sefers S.E., Dare R.K., Chappell J.D. Simultaneous detection and high-throughput identification of a panel of RNA viruses causing respiratory tract infections. J Clin Microbiol. 2007;45:2105–2109. doi: 10.1128/JCM.00210-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Renois F., Talmud D., Huguenin A., Moutte L., Strady C., Cousson J. Rapid detection of respiratory tract viral infections and coinfections in patients with influenza-like illnesses by use of reverse transcription–PCR DNA microarray systems. J Clin Microbiol. 2010;48:3836–3842. doi: 10.1128/JCM.00733-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Babady N.E. The FilmArray® respiratory panel: an automated, broadly multiplexed molecular test for the rapid and accurate detection of respiratory pathogens. Expert Rev Mol Diagn. 2013;13:779–788. doi: 10.1586/14737159.2013.848794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Wesenbeeck L., Meeuws H., Van Immerseel A., Ispas G., Schmidt K., Houspie L. Comparison of the FilmArray RP, Verigene RV+, and Prodesse ProFLU+/FAST+ multiplex platforms for detection of influenza viruses in clinical samples from the 2011–2012 influenza season in Belgium. J Clin Microbiol. 2013;51:2977–2985. doi: 10.1128/JCM.00911-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liberati A., Altman D.G., Tetzlaff J., Mulrow C., Gøtzsche P.C., Ioannidis J.P.A. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate healthcare interventions: explanation and elaboration. BMJ. 2009;339 doi: 10.1136/bmj.b2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leeflang M.M., Deeks J.J., Gatsonis C., Bossuyt P.M., Cochrane Diagnostic Test Accuracy Working Group Systematic reviews of diagnostic test accuracy. Ann Intern Med. 2008;149:889–897. doi: 10.7326/0003-4819-149-12-200812160-00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsiung G. Diagnostic virology: from animals to automation. Yale J Biol Med. 1984;57:727. [PMC free article] [PubMed] [Google Scholar]
- 29.Leland D.S., Ginocchio C.C. Role of cell culture for virus detection in the age of technology. Clin Microbiol Rev. 2007;20:49–78. doi: 10.1128/CMR.00002-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leeflang M.M., Deeks J.J., Takwoingi Y., Macaskill P. Cochrane diagnostic test accuracy reviews. Syst Rev. 2013;2:82. doi: 10.1186/2046-4053-2-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hsu J., Santesso N., Mustafa R., Brozek J., Chen Y.L., Hopkins J.P. Antivirals for treatment of influenza—a systematic review and meta-analysis of observational studies. Ann Intern Med. 2012;156:512–524. doi: 10.7326/0003-4819-156-7-201204030-00411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chartrand C., Leeflang M.M., Minion J., Brewer T., Pai M. Accuracy of rapid influenza diagnostic tests: a meta-analysis. Ann Intern Med. 2012;156:500–511. doi: 10.7326/0003-4819-156-7-201204030-00403. [DOI] [PubMed] [Google Scholar]
- 33.Hayden F.G., de Jong M.D. Emerging influenza antiviral resistance threats. J Infect Dis. 2011;203:6–10. doi: 10.1093/infdis/jiq012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pizzorno A., Abed Y., Boivin G. Influenza drug resistance. Seminars in respiratory and critical care medicine. Semin Respir Crit Care Med. 2011;32:409–422. doi: 10.1055/s-0031-1283281. [DOI] [PubMed] [Google Scholar]
- 35.Glezen W.P., Taber L.H., Frank A.L., Kasel J.A. Risk of primary infection and reinfection with respiratory syncytial virus. Am J Dis Child. 1986;140:543–546. doi: 10.1001/archpedi.1986.02140200053026. [DOI] [PubMed] [Google Scholar]
- 36.Simoes E.A. Respiratory syncytial virus infection. Lancet. 1999;354(9181):847–852. doi: 10.1016/S0140-6736(99)80040-3. [DOI] [PubMed] [Google Scholar]
- 37.Ventre K., Randolph A.G. Ribavirin for respiratory syncytial virus infection of the lower respiratory tract in infants and young children. Cochrane Database Syst Rev. 2007;(1) doi: 10.1002/14651858.CD000181.pub3. [DOI] [PubMed] [Google Scholar]
- 38.DeVincenzo J.P., Whitley R.J., Mackman R.L., Scaglioni-Weinlich C., Harrison L., Farrell E. Oral GS-5806 activity in a respiratory syncytial virus challenge study. N Engl J Med. 2014;371:711–722. doi: 10.1056/NEJMoa1401184. [DOI] [PubMed] [Google Scholar]
- 39.Mackman R.L., Sangi M., Sperandio D., Parrish J.P., Eisenberg E., Perron M. Discovery of an oral respiratory syncytial virus (RSV) fusion inhibitor (GS-5806) and clinical proof of concept in a human RSV challenge study. J Med Chem. 2015;58:1630–1643. doi: 10.1021/jm5017768. [DOI] [PubMed] [Google Scholar]
- 40.DeVincenzo J.P., McClure M.W., Symons J.A., Fathi H., Westland C., Chanda S. Activity of oral ALS-008176 in a respiratory syncytial virus challenge study. N Engl J Med. 2015;373:2048–2058. doi: 10.1056/NEJMoa1413275. [DOI] [PubMed] [Google Scholar]
- 41.Parker S., Crump R., Foster S., Hartzler H., Hembrador E., Lanier E.R. Co-administration of the broad-spectrum antiviral, brincidofovir (CMX001), with smallpox vaccine does not compromise vaccine protection in mice challenged with ectromelia virus. Antivir Res. 2014;111:42–52. doi: 10.1016/j.antiviral.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Doern C.D., Lacey D., Huang R., Haag C. Evaluation and implementation of FilmArray version 1.7 for improved detection of adenovirus respiratory tract infection. J Clin Microbiol. 2013;51:4036–4039. doi: 10.1128/JCM.02546-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pierce V.M., Elkan M., Leet M., McGowan K.L., Hodinka R.L. Comparison of the Idaho Technology FilmArray system to real-time PCR for detection of respiratory pathogens in children. J Clin Microbiol. 2012;50:364–371. doi: 10.1128/JCM.05996-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Loeffelholz M.J., Pong D.L., Pyles R.B., Xiong Y., Miller A.L., Bufton K.K. Comparison of the FilmArray Respiratory Panel and Prodesse real-time PCR assays for detection of respiratory pathogens. J Clin Microbiol. 2011;49:4083–4088. doi: 10.1128/JCM.05010-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mower W.R. Evaluating bias and variability in diagnostic test reports. Ann Emerg Med. 1999;33:85–91. doi: 10.1016/s0196-0644(99)70422-1. [DOI] [PubMed] [Google Scholar]
Further reading
- 46.Sutton B.C., Maggert K., Rowell B., Etter R. Comparison of the focus diagnostics simplexa Flu A/B & RSV direct assay with the prodesse ProFlu+ assay for detection of influenza A virus (IAV), influenza B virus (IBV), and respiratory syncytial virus (RSV) in clinical specimens. J Mol Diagn. 2014;16:727–728. [Google Scholar]
- 47.Espy M., Schneider S.K., Sloan L.M., Pritt B.S. Detection of influenza A, B and RSV in respiratory specimens by the proFlu+TM Kit and roche LC 480. J Mol Diagn. 2010;12:875. [Google Scholar]
- 48.Irwin C., Matthews-Greer J., McRae K. Performance analysis of pandemic influenza detection methods. Am J Clin Pathol. 2012;138 [Google Scholar]
- 49.Buchan B., Anderson N., Jannetto P., Ledeboer N. Simultaneous detection of influenza A and its subtypes (H1, H3, 2009 H1N1), influenza B, and RSV A and B in respiratory specimens on an automated, random access, molecular platform. Clin Microbiol Infect. 2011;17:S3–S4. [Google Scholar]
- 50.Alby K., Popowitch E.B., Miller M.B. Comparative evaluation of the Nanosphere Verigene RV+ assay and the Simplexa Flu A/B & RSV kit for detection of influenza and respiratory syncytial viruses. J Clin Microbiol. 2013;51:352–353. doi: 10.1128/JCM.02504-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Popowitch E.B., O’Neill S.S., Miller M.B. Comparison of the Biofire FilmArray RP, GenMark eSensor RVP, Luminex xTAG RVPv1, and Luminex xTAG RVP fast multiplex assays for detection of respiratory viruses. J Clin Microbiol. 2013;51:1528–1533. doi: 10.1128/JCM.03368-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Babady N.E., Mead P., Stiles J., Brennan C., Li H., Shuptar S. Comparison of the Luminex xTAG RVP Fast assay and the Idaho Technology FilmArray RP assay for detection of respiratory viruses in pediatric patients at a cancer hospital. J Clin Microbiol. 2012;50:2282–2288. doi: 10.1128/JCM.06186-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rand K.H., Rampersaud H., Houck H.J. Comparison of two multiplex methods for detection of respiratory viruses: FilmArray RP and xTAG RVP. J Clin Microbiol. 2011;49:2449–2453. doi: 10.1128/JCM.02582-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hwang S.M., Lim M.S., Han M., Hong Y.J., Kim T.S., Lee H.R. Comparison of xTAG respiratory virus panel and verigene respiratory virus plus for detecting influenza virus and respiratory syncytial virus. J Clin Lab Anal. 2015;29:116–121. doi: 10.1002/jcla.21738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Poritz M.A., Blaschke A.J., Byington C.L., Meyers L., Nilsson K., Jones D.E. FilmArray, an automated nested multiplex PCR system for multi-pathogen detection: development and application to respiratory tract infection. PLoS One. 2011;6 doi: 10.1371/journal.pone.0026047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Layman C.P., Gordon S.M., Elegino-Steffens D.U., Agee W., Barnhill J., Hsue G. Rapid multiplex PCR assay to identify respiratory viral pathogens: moving forward diagnosing the common cold. Hawaii J Public Health. 2013;72(9 Suppl. 4):24–26. [PMC free article] [PubMed] [Google Scholar]
- 57.Nakao A., Hisata K., Matsunaga N., Fujimori M., Yoshikawa N., Komatsu M. The clinical utility of a near patient care rapid microarray-based diagnostic test for influenza and respiratory syncytial virus infections in the pediatric setting. Diagn Microbiol Infect Dis. 2014;78:363–367. doi: 10.1016/j.diagmicrobio.2013.11.005. [DOI] [PubMed] [Google Scholar]
- 58.Selvaraju S.B., Selvarangan R. Evaluation of three influenza A and B real-time reverse transcription–PCR assays and a new 2009 H1N1 assay for detection of influenza viruses. J Clin Microbiol. 2010;48:3870–3875. doi: 10.1128/JCM.02464-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Beck E.T., Jurgens L.A., Kehl S.C., Bose M.E., Patitucci T., LaGue E. Development of a rapid automated influenza A, influenza B, and respiratory syncytial virus A/B multiplex real-time RT-PCR assay and its use during the 2009 H1N1 swine-origin influenza virus epidemic in Milwaukee, Wisconsin. J Mol Diagn. 2010;12:74–81. doi: 10.2353/jmoldx.2010.090095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen L., Tian Y., Chen S., Liesenfeld O. Performance of the Cobas® Influenza A/B assay for rapid PCR-based detection of influenza compared to Prodesse ProFlu+ and viral culture. Eur J Microbiol Immunol. 2015;5:236–245. doi: 10.1556/1886.2015.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chiu S.C., Lin Y.C., Wang H.C., Hsu J.J., Yeh T.K., Liu H.F. Surveillance of upper respiratory infections using a new multiplex PCR assay compared to conventional methods during the influenza season in Taiwan. Int J Infect Dis. 2017;61:97–102. doi: 10.1016/j.ijid.2017.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stellrecht K.A., Nattanmai S.M., Butt J., Maceira V.P., Espino A.A., Castro A.J. Effect of genomic drift of influenza PCR tests. J Clin Virol. 2017;93:25–29. doi: 10.1016/j.jcv.2017.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.