Abstract
Coronavirus disease 2019 SARS-CoV-2 (COVID-19) is a zoonotic virus causing a variety of severe respiratory diseases. SARS-CoV-2 is closest to SARS-CoV and MERS-CoV in structure. The high prevalence of COVID-19 is a result of a lack of symptoms at onset. Our study aimed to present an overview of the virus in terms of structure, epidemiology, symptoms, treatment and prevention. Whole genome sequences and some viral proteins were investigated to determine gaps and changes in alternation of nucleotides and amino acid sequences. We evaluate 11 complete genome sequences of different coronaviruses using BAST and MAFFT software. We also selected seven types of structural proteins. We conclude that COVID-19 might produce new mutations, specifically in glycoproteins, so caution and complete preparation by health authorities is required.
Keywords: COVID-19, Middle-East respiratory syndrome, outbreaks, severe acute respiratory syndrome, zoonotic
Introduction
Novel coronavirus disease 19 (COVID-19) first emerged on 31 December 2019 in Wuhan city, China. COVID-19 is classified as the seventh member of the subfamily Orthocoronavirinae under the family Coronaviridae. Most members of this family are zoonotic viruses transmitted to humans through contact with infected animals. Although bats and snakes are the natural reservoir of most coronaviruses, there is no evidence so far that COVID-19 originated in or was transmitted from a seafood market [1]. Comparison of the lipid rafts of coronaviruses has indicated that the new strain COVID-19 has 80% identity with severe acute respiratory syndrome coronavirus (SARS-CoV). Lipid molecules such as caveolins, clathrins and dynamin have a fundamental role in the internalization of viruses. These molecules are involved in the entry of viruses into host cells, and targeting host lipids is being studied as an antiviral strategy and could have various applications [2]. COVID-19 seems to need to bind to the angiotensin-converting enzyme-2 receptor on the membrane host cell to enable it to infect the host cell upon coupled with a reliance of serine protease TMPRSS2. This intracellular protein seems to be a determinant of the virus ability to infect the cell [3].
Prevalence and epidemiology
Over the past two decades, outbreaks of coronavirus have been observed—SARS-CoV in 2003 and Middle-East respiratory syndrome (MERS) -CoV— and have been described as major public health threats. The WHO considers COVID-19 to be a more serious and widespread epidemic disease [4]. To date, it seems that the mortality rate of COVID-19 is lower than those of SARS or MERS. A significant increase in number of COVID-19 cases was observed as the result of the absence of emerging pathological symptoms in virus carriers. For this reason, it may lead to the collapse of local health-care systems [4]. Some countries face an outbreak crisis and are trying to prevent the spread of COVID-19 through preventing human gatherings, imposing a curfew in cities, preventing travel between countries and closing land borders, all of which may reduce the outbreaks.
The main transmission of COVID-19 starts with human-to-human contact, including relatives and friends who have intimate contact with patients or incubating carriers. Many studies have reported that coughing and sneezing are quicker routes of virus dispersion, indicating the need for droplet and airborne precautions when encountering an infected person [5].
Virus structure
COVID-19 is related to the beta-coronavirus that infects humans and probably developed from bat coronaviruses. Structural analysis shows that COVID-19 probably derives from a bat SARS-like coronavirus, which has mutated in the spike glycoprotein (protein S) and nucleocapsid N protein The positive-sense RNA genomes of COVID-19 differ from SARS-CoV and MERS-CoV, being approximately 29.9 kb, 27.9 kb and 30.1 kb, respectively [6]. The COVID-19 complete genome was annotated to possess 14 open reading frames (ORFs) encoding 27 proteins. Sequence analysis revealed that COVID-19 has > 80% identity with SARS-CoV and 50% with MERS-CoV, which originated in bats [7,8]. In addition, the spherical external spike protein displays a characteristic crown shape with electron microscopy [9]. In the current study, we have compared the novel COVID-19 complete genome with other related coronaviruses to identify mutations and gaps. We selected data from NCBI and we performed the FASTA and BLAST. The comparison between genomes with alignment used MAAFT-7 software. COVID-19 (GenBank MT188341.1) and COVID-19 (MT066175.1), bat-SL-CoVZC45 (MG772933.1) and SARS-CoV BJ182b (EU371561.1) showed alignment identities of 99%, 89% and 82%, respectively. Fig. 1 shows the differences between the four complete genomes.
Fig. 1.
Final alignment of four complete genomes.
According to the alignment analysis, the closest similarity was between the two COVID-19 genomes (99%) compared with the two bat CoV. Seven complete genomes have been aligned for different coronavirus strains (Fig. 2).
Fig. 2.
MAFFT considers similarities in forward strands (red) only, but ignores similarities in reverse strands (blue), Plot 1/2: alignment MT066175.1 + MT039873.1 99.7%, plot 1/3: MT066175.1+ MN997409.1 99.7%, plot 1/4: MT066175.1 + MN996532.1 89%, plot 1/5: MT066175.1 + EU371564.1 82.4%, plot 1/6: MT066175.1 + KY938558.1 = 68%, plot 1/7: MT066175.1 + MN985325.1 95%.
Results revealed that all COVID-19 strains were similar compared with other strains related to the same family. We believe, therefore, that COVID-19 came from several mutations occurring in other coronaviruses related to the same infection. Genomic analysis does not support the belief that COVID-19 is a laboratory construct, but it is impossible to disprove or prove the theories of its origin. To identify COVID-19's origin, virus sequences should be obtained from immediate animal sources.
The first ORF (ORF1a/b) translates two polyproteins, pp1a and pp1ab, and encodes 16 non-structural proteins (NSP); this takes up two-thirds of the viral RNA. The remaining ORFs encode structural proteins including spike (S) glycoprotein, small envelope (E) protein, matrix (M) protein and nucleocapsid (N) protein. COVID-19 also possesses accessory proteins that interfere with the host's innate immune response [8].
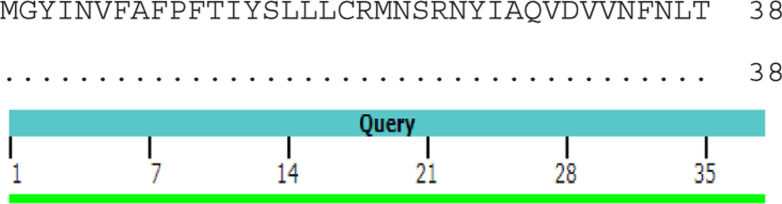
We performed alignment of ORF10 for COVID-19 (GenBank QIK50446.1) and COVID-19 (YP_009725255.1). The result showed similarity of 100% without any mutation in amino acids (Fig. 3).
Fig. 3.
100% similarity between open-reading frame 10 of QIK50446.1 and YP_009725255.1.
Nucleocapsid phosphoprotein COVID-19 (GenBank QIK50445.1) was aligned with NP COVID-19 (QIC50514.1) and showed similarity of 100%; against NP SARS CoV CUHK-L2 (AAS01074.1) and bat COV HKU5 (QHA24694.1) similarities were 85% and 61%, respectively (Fig. 4). The crystal structure of NP COVID-19 (6VYO) was released on 11 March 2020 from RCSB PDB (Fig. 5).
Fig. 4.
Alignment of QIK50445.1, QIC50514.1, AAS01074.1 and QHA24694.1.
Fig. 5.
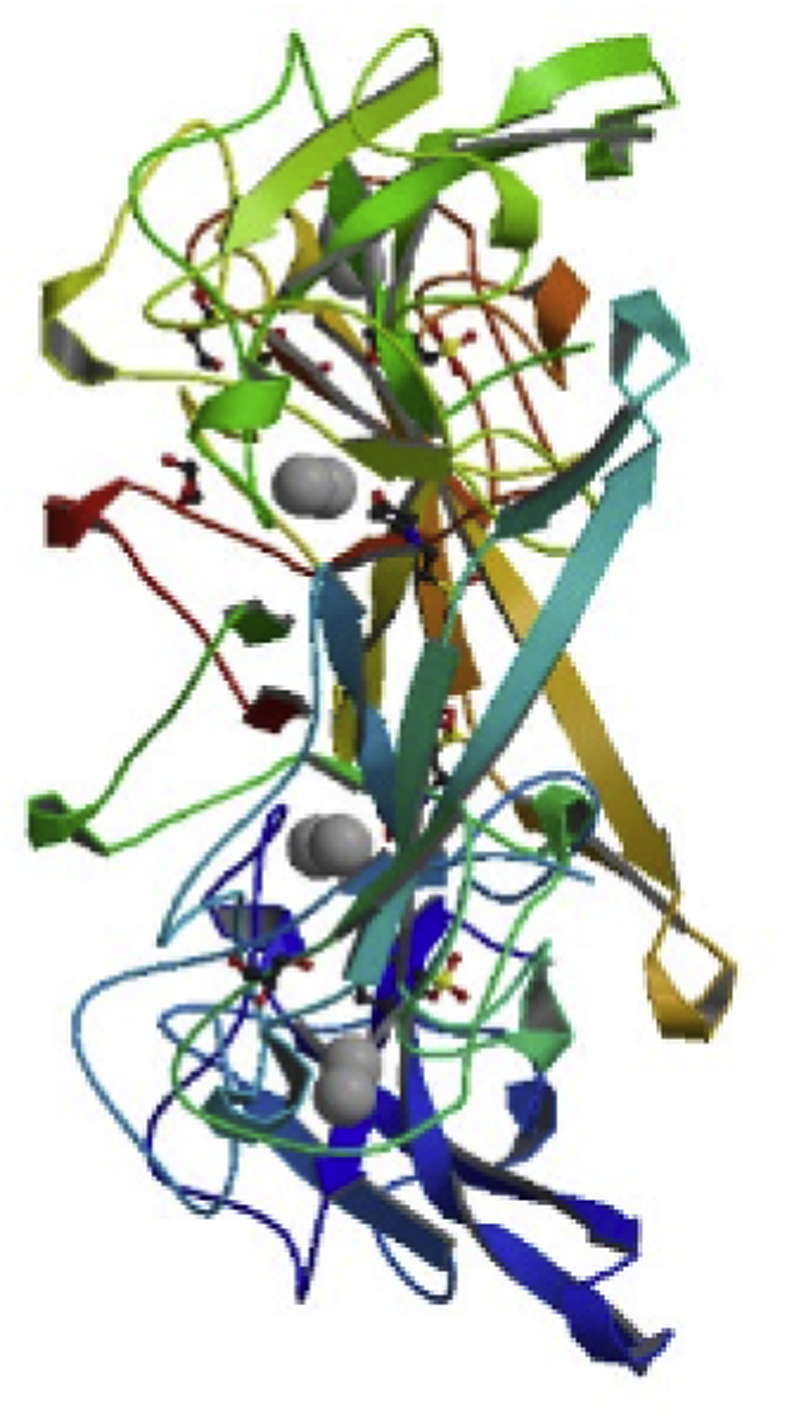
Crystal structure of NP COVID-19 (6VYO) from RCSB PDB.
ORF7a protein COVID-19 (GenBank QIK50443.1) aligned with NS7a Bat CoV RaTG13 (QHR63305.1) showed 99% similarity, aligned with hypothetical protein SARS 7 CoV (AFR58706.1) it showed 89% similarity and aligned with putative uncharacterized protein 4 SARS CoV (AAX16199.1) it showed 68% similarity (Fig. 6).
Fig. 6.
Alignment of QIK50443.1, QHR63305.1, AFR58706.1 and AAX16199.1.
Alignment of ORF8 protein COVID-19 (GenBank QIK50444.1) with ORF8 protein COVID-19 (QHN73801.1), hypothetical protein Bat SARS CoV Rs806/2006 (ACU31050.1) and hypothetical protein Bat SARS CoV HKU3-8 (ADE34775.1) showed similarities of 99%, 76% and 84%, respectively (Fig. 7).
Fig. 7.
Alignment of QIK50444.1, QHN73801.1, ACU31050.1 and ADE34775.1.
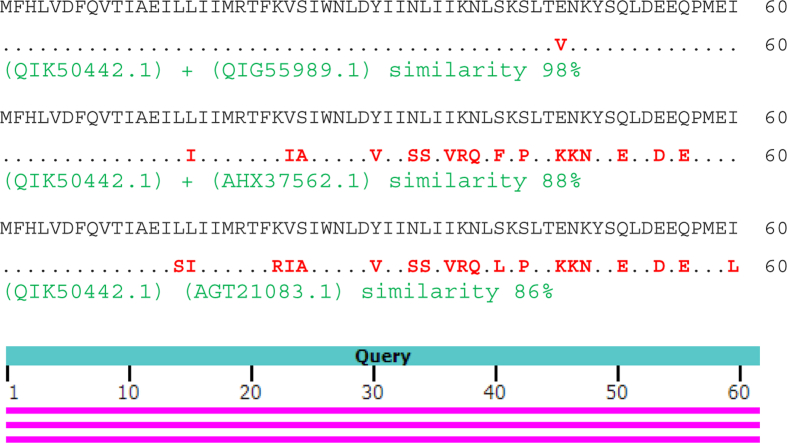
ORF6 protein COVID-19 (GenBank QIK50442.1) was aligned with ORF6 protein COVID-19 (QIG55989.1), protein 7 Rhinolophus affinis CoV (AHX37562.1) and NSP 6 SARS CoV ExoN1 (AGT21083.1)and showed similarities of 98%, 88% and 86%, respectively (Fig. 8).
Fig. 8.
Alignment of QIK50442.1, QIG55989.1, AHX37562.1 and AGT21083.1.
Membrane glycoprotein COVID-19 (GenBank QIK50441.1) aligned with MG COVID-19 (QIG55988.1), M protein COVID-19 (APO40582.1) and membrane glycoprotein Rousettus Bat CoV HKU9 (YP_001039974.1) gave similarities of 99%, 93% and 61%, respectively (Fig. 9).
Fig. 9.
Alignment of QIK50441.1, QIG55988.1, APO40582.1 and YP_001039974.1.
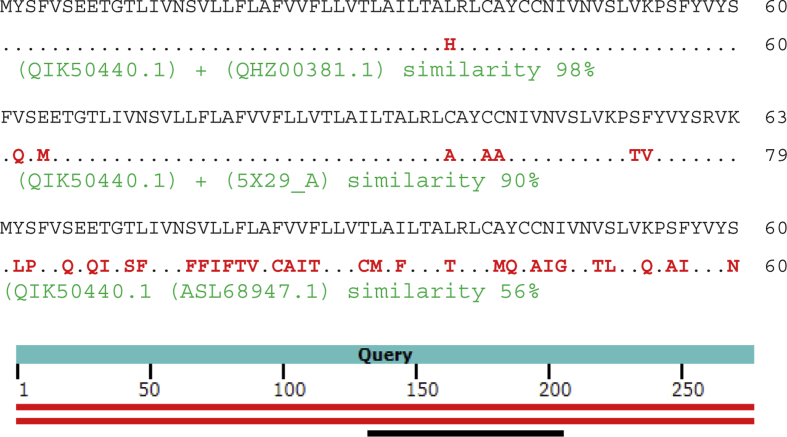
Alignment of envelope protein COVID-19 (GenBank QIK50440.1) with EP COVID-19 (QHZ00381.1) showed 98% similarity, with Chain A Envelope small membrane protein SARS CoV (5X29_A) showed 90% similarity and with envelope protein Hypsugo Bat CoV HKU25 (ASL68947.1) showed 56% similarity (Fig. 10).
Fig. 10.
Alignment of QIK50440.1, QHZ00381.1, 5X29_A and ASL68947.1.
By analysing the series compatibility of the protein sequences under study, we confirm that there is a match between strains of COVID-19. There are obvious differences compared with other species in the coronavirus family. This may indicate that COVID-19 originated from mutations within the coronavirus family. In clearer terms, new mutations may be created as there is a high probability, specifically in glycoproteins.
We are unable to give reasonable explanations for the significant number of amino acid substitutions between COVID-19 and SARS-CoV or MERS-CoV due to very limited knowledge of this novel virus.
Clinical manifestation and symptoms
The incubation period of the virus may vary with age and immune status. In general, it has been assumed that the incubation period is between 2 and 14 days, although cases have been observed up to 23 days after exposure. The main symptoms are easily seen in those aged over 70 years and in immunocompromised and diabetic individuals. Symptoms start with fever, dry cough and dyspnoea, as well as sore throat, nasal congestion, malaise; bilateral infiltrates may be seen on chest X-ray. However some cases are detected in the absence of fever. Clinical features of COVID-19 include the targeting of the lower airway, as well as upper respiratory tract symptoms like rhinorrhoea, sneezing, and sore throat, developed into gastrointestinal symptoms like diarrhoea [10]. Severe cases may present with sepsis, heart attack or even shock. Conversely, some cases may show mild illness or be asymptomatic.
From WHO records, the period from symptom onset to death ranges from 6 to 41 days with a median of 14 days. This period depends on the age and immune status of the individual and is shorter in those <70 years old [11].
Preventions
To prevent spreading virus, managed care of patients is required with early identification, rapid isolation, timely establishment of infection prevention and control measures, together with symptomatic care for patients with mild disease. Supportive treatment is needed for those with severe COVID-19. Specific attention should be given to and more efforts made to reduce transmission to susceptible populations, including health-care providers, immunocompromised patients, children and the elderly [5]. Health-care systems around the world must operate with more than one maximum capacity. Cooperation between health-care systems and the WHO is required to reduce infection. International media, social media and societal culture should be used to maintain personal cleaning, minimize risk of exposure, avoid gatherings and prevent all phenomena that lead to contact between persons [12]. COVID-19 vaccines is under accelerated development.
The global public health community must consider the effects of mass gathering cancellations on the future well-being of communities through economic recession as well as through the spread, or otherwise, of COVID-19 [13].
Diagnosis
Quantitative RT-PCR is the most specific and sensitive assay approved and straightforwardly used by many reference laboratories worldwide. Other laboratory tests may help in assessing disease severity and predicting the risk of evolution such as acute respiratory distress syndrome, disseminated intravascular coagulation and multiorgan failure. Moreover, C-reactive protein, lactate dehydrogenase, erythrocyte sedimentation rate and D-dimer, along with diminished concentration of serum albumin, increased values of lactate dehydrogenase, aspartate aminotransferase, alanine aminotransferase, total bilirubin, creatinine and cardiac troponins, are used as helpful tests for organ function. Notably, a combined IgM–IgG rapid immunoassay has also been recently developed, as well as investigation of elevation of pro-inflammatory cytokine detection kits, such as those for interleukin-1β (IL-1β), IL-1RA, IL-7, IL-8, IL-9, IL-10, basic granulocyte colony-stimulating factor, interferon-γ, tumour necrosis factor-α, and interferon-γ inducible protein 10 [14]. A study revealed that gastrointestinal symptoms with COVID-19 was not associated with viral RNA in the faecal sample, nor with extended duration of the viral RNA positivity in the faeces.
COVID-19 RNA has been isolated from human saliva, nasopharynx and lower respiratory tract. Computed tomography findings of lung abnormalities increased quickly after the onset of symptoms, and persisted for long durations. Computed tomography manifestations are important when inspected for patterns over time [15]. A study showed no evidence that tumour necrosis factor-α inhibition increased the risk of COVID-19 outbreaks [16].
Treatment
There is currently no approved antiviral treatment for COVID-19. The implementation of antiviral treatment and prophylaxis has several requirements to reduce risk. Drugs can be administered shortly after symptom onset to reduce infectiousness and shedding of virus in respiratory secretions. Some studies have indicated that hydroxychloroquine has antiviral activity in vitro against coronaviruses, and specifically, COVID-19. Remarkably, this drug was licensed for the chemoprophylaxis and treatment of malaria. Furthermore, drug testing suggest that prophylaxis with hydroxychloroquine at approved doses may prevent COVID-19 infection and amend viral shedding [17]. Clinical trials of hydroxychloroquine treatment for COVID-19 pneumonia have shown positive preliminary outcomes in China.
Unfortunately, the corticosteroid treatments commonly used in clinical practice for influenza virus such as acyclovir, ganciclovir, ribavirin and methylprednisolone, as well as neuraminidase inhibitors including peramivir, oseltamivir and zanamivir, are invalid against COVID-19 and are not recommended [18].
Conclusions
Scientists have made progress in characterizing the new coronavirus but there are still many questions to be answered. COVID-19 is a great biological hazard and is a worldwide threat. COVID-19 is highly contagious during the latency period. It is may be necessary to adopt and invest in more modern technologies both to facilitate notification, and to allow speedier data dissemination and analysis in keeping with the principles of precision epidemiology.
We suggest that close contact with an infected person is the major factor in disease transmission. Health-care workers must also follow CDC guidelines and should not attempt to perform any virus isolation or characterization. The effect of mass gathering cancellations on reducing the spread of COVID-19 needs to be determined. Any mutation occurring will be especially important. There is no evidence that part of COVID-19 is synthetic.
Transparency declaration
The authors declare no conflicts of interest.
Acknowledgement
We would like to state that all research budgets were self-supporting without any institute donation.
References
- 1.Lana R., Coellaho F.C., Gomes M.F., Cruz O.G., Bastos L.S., Villela D.A. The novel coronavirus (SARS-CoV-2) emergency and the role of timely and effective national health surveillance. Rep Public Health. 2020 doi: 10.1590/0102-311x00019620. http://doi:10.1590/0102-311X00019620 Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 2.Baglivo M., Baronio M., Natalini G., Beccari T., Fuulcheri P.C., Petralia P. Natural small molecules as inhibitors of coronavirus lipid-dependent attachment to host cells: a possible strategy for reducing SARS-COV-2 infectivity? Acta Biomed. 2020;91:161–164. doi: 10.23750/abm.v91i1.9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thomso G. COVID-19: social distancing, ACE 2 receptors, protease inhibitors and beyond? Int J Clin Prac. 2020 doi: 10.1111/ijcp.13503. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lippi G., Plebani M. The critical role of laboratory medicine during coronavirus disease 2019 (COVID-19) and other viral outbreaks. Clin Chem Lab Med. 2020 doi: 10.1515/cclm-2020-0240. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 5.Karako K., Song P., Chen Y., Tang W. Analysis of COVID-19 infection spread in Japan based on stochastic transition model. BioSci Tre. 2020 doi: 10.5582/bst.2020.01482. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 6.Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020 doi: 10.1016/j.chom.2020.02.001. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheepsattayakorn A., Cheepsattayakorn R. Proximal origin and phylogenetic analysis of COVID-19 (2019-nCoV or SARS-CoV-2) EC Microbiol. 2020 Epub ahead of print. [Google Scholar]
- 8.Guo Y.-R., Cao Q.-D., Hong Z.-S., Tan Y.-Y., Chen S.-D., Jin H.-J. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak—an update on the status. Milit Med Res. 2020;7:11. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim J.-M., Chung Y.-S., Jo H.J., Lee N.-J., Kim M.S., Woo S.H. Identification of coronavirus isolated from a patient in Korea with COVID-19. Osped Pub Health Res Per. 2020;11:3–7. doi: 10.24171/j.phrp.2020.11.1.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rothan H.A., Byrareddy S.N. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J Autoimm. 2020 doi: 10.1016/j.jaut.2020.102433. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koenig K.L., Beÿ C.K., McDonald E.C. 019-nCoV: the Identify-Isolate-Inform (3I) tool applied to a novel emerging coronavirus. West J Emerg Med. 2020;21:184–190. doi: 10.5811/westjem.2020.1.46760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dany L. COVID-19: protecting health-care workers. Lancet. 2020;395 doi: 10.1016/S0140-6736(20)30627-9. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mass gathering events and reducing further global spread of COVID-19: a political and public health dilemma. Lancet. 2020 doi: 10.1016/S0140-6736(20)30681-4. Letter to the Editor. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y. Structure-based drug design, virtual screening and high-throughput screening rapidly identify antiviral leads targeting COVID-19. BioRxiv. 2020 doi: 10.1101/2020.02.26.964882. Epub ahead of print. [DOI] [Google Scholar]
- 15.Wang Y., Dong C., Hu Y., Li C., Ren Q., Zhang X. Temporal changes of CT findings in 90 patients with COVID-19 pneumonia: a longitudinal study. Radiology. 2020 doi: 10.1148/radiol.2020200843. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu Y., Guo C., Tang L., Hong Z., Zhou J., Dong X. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol Hepatol. 2020 doi: 10.1016/S2468-1253(20)30083-2. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mitjà O., Clotet B. Use of antiviral drugs to reduce COVID-19 transmission. Lancet Glob Health. 2020 doi: 10.1016/S2214-109X(20)30114-5. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bashyam A.M., Feldman S.R. Should patients stop their biologic treatment during the COVID-19 pandemic. J Dermatol Treatm. 2020 doi: 10.1080/09546634.2020.1742438. Epub ahead of print. [DOI] [PubMed] [Google Scholar]