Abstract
Rhinoviruses (RVs) are frequently detected respiratory viruses that cause mild common cold symptoms, but may also lead to more severe respiratory tract infections. The large number of RV types, classified into species A, B and C, hampers clear insights into the epidemiology and clinical significance of each RV type. The aim of this study was to map the circulation of RV types in the Amsterdam area. RV-positive nasopharyngeal and oropharyngeal samples, collected from 2007 to 2012 in the Academic Medical Centre (Amsterdam, the Netherlands), were typed based on the sequence of the region coding for capsid proteins VP4 and VP2. RV-A, RV-B and RV-C were found in proportions of of 52.4% (334/637), 11.3% (72/637), and 36.2% (231/637), respectively. We detected 129 of the 167 currently classified types. RVs circulated throughout the entire year with a peak in the autumn and a decline in the summer. Some RV types were observed throughout the entire sampling period and others had a more seasonal pattern. Nine RV-A and four RV-B novel provisionally assigned types were identified. This study provides an insight into the molecular epidemiology of RVs in the Amsterdam area. The RVs circulating are diverse and include several provisionally new types.
Keywords: Epidemiology, Genotype, Respiratory tract infection, Rhinovirus, Seasonality
Introduction
Rhinoviruses (RVs) are causative agents of upper and lower respiratory tract infections [1]. Symptoms range from a common cold to more serious infections such as bronchiolitis, and pneumonia. Also, RVs can cause acute exacerbations of asthma, chronic obstructive pulmonary disease and cystic fibrosis and have been implicated in the pathogenesis of asthma [2].
Rhinoviruses belong to the enterovirus (EV) genus of the picornavirus family, and currently 167 RV types have been classified into three species, named A, B and C [3]. Species RV-A and RV-B have been known since the 1950s [1], but species RV-C was only discovered in 2006 upon the introduction of molecular techniques because these viruses cannot be cultured on standard immortalized cell lines [4], [5], [6], [7].
Studies suggested that RV-C infections may elicit more severe disease compared with infections with RV-A or RV-B types [8]. However, this has been refuted by several studies that did not observe a difference in outcome between RV-C and RV-A infections [8]. Due to the large number of RV types, the severity of infections caused by individual RV types is even more elusive.
As a result of small sample sizes, short observation periods and the large number of RV types there is a lack of insight into the prevalence and circulation patterns of RV types. Compared with the other EVs, RVs co-circulate to a much larger extent, and seasonal patterns are less prominent [1], [9], [10]. Increased insight into prevalence, circulation patterns and clinical significance is not only of importance for surveillance purposes, but also for the future development of antiviral therapy and vaccines. The aim of our study was therefore to investigate the prevalence of RV types in the hospital population of the Academic Medical Centre (Amsterdam, the Netherlands), by genotyping all RV-positive samples submitted for respiratory viral diagnostics from 2007 to 2012.
Materials and methods
Study design
This study was conducted at the Academic Medical Centre in Amsterdam, the Netherlands. From 2007 to 2012, a total of 6258 nasopharyngeal and oropharyngeal samples were submitted to the Laboratory of Clinical Virology, Department of Medical Microbiology of the Academic Medical Centre in Amsterdam for diagnostic evaluation. The Academic Medical Centre receives mainly samples from the southeast area of Amsterdam. The samples were acquired from hospitalized patients and non-hospitalized patients visiting the outpatient clinics or emergency room. The reason for sampling was not systematically documented on the accompanying form for the laboratory and so could not be adequately monitored or analysed. Respiratory samples collected for research purposes and those that were not tested for RV were excluded from the analyses. A total of 1102 (17.6%) respiratory samples were positive for RV. There was no material available from five samples for additional gene sequencing. All available RV-positive samples were further characterized by sequencing the VP4/VP2 region. The sampling and virological testing were part of routine care and were executed according to hospital ethical guidelines and the Dutch code of conduct for responsible use of human tissue and medical research 2011.
Virological assessments
RNA was extracted from nasopharyngeal and/or oropharyngeal samples with the MagnaPure LC instrument using the total nucleic acid isolation kit (Roche Diagnostics, Basel, Switzerland). Samples were tested for the presence of RV, EV, human parechovirus (HPeV), influenza viruses A and B, parainfluenzavirus 1 to 4, human bocavirus, human coronavirus (HKU1, NL63, 229E and OC43), respiratory syncytial virus, adenovirus and human metapneumovirus, with a multiplex real-time PCR as described previously [11]. Primers used for the detection of RV were reported in Jaramillo-Gutierrez et al. [12]. Ct values ≥40 were considered negative.
RV genotyping
RV-positive samples were genotyped based on a 540-bp fragment of the VP4/VP2 region as described previously [13] using a two-step semi-nested protocol with primers in the 5′-untranslated region and in VP2 (Table 1 ). The VP4/VP2 sequences were phylogenetically compared with published reference sequences as proposed and provided by McIntyre et al. [14]. Sequences were analysed using CodonCode Aligner version 3.7.1, aligned using Clustal X version 3.0.11 and edited using Genedoc version 2.7 software [15], [16]. Phylogenetic trees were constructed using neighbour-joining trees under a p-distance model as implemented in MEGA version 5.10 [17]. Trees were unrooted and bootstrap values were determined from 100 bootstrap resamplings of the original data.
Table 1.
Primers
| Orientation | Name | Sequence |
|---|---|---|
| Step 1 | ||
| Sense | HRV-VP4-1 | GGG ACC AAC TAC TTT GGG TGT |
| Antisense | 9565-reverse | GCA TCI GGY ARY TTC CAC CAC CAN CC |
| Step 2 | ||
| Sense | HRV-VP4-2-forward | GGG GAC CAA CTA CTT TGG GTG TCC GTG T |
Sequences for which the genetic distance, i.e. nucleotide divergence, was above the threshold of 10%, 9.5% and 10% for RV-A, RV-B and RV-C, respectively [14], were submitted to GenBank (nos KP003842-KP003896 and KT272022-KT272030) and to the Picornavirus Study Group to be designated a provisionally assigned type.
Definitions
If multiple samples were available from the same patient, infections were defined as a new infection when the sample was the first RV-positive sample from that patient, when the sample yielded a different type than the previous sample from the same patient, and/or when the interval period between the two samples was >3.0 months.
As a result of the close genetic relationship between RVs and non-RV EVs, diagnostic detection assays may be cross-reactive and result in a false-positive outcome. An RV false-positive sample was defined as a sample that was RV-positive by real-time PCR and resulted in a non-RV EV sequence after phylogenetic analysis. A sample was only classified as an EV co-infection if the RV infection was verified by RV typing and if either of two EV typing protocols [18] yielded a non-RV EV sequence. The EV-A/EV-B protocol was adapted to a semi-nested PCR and used the same forward outer primer for the first and the second PCR for both EV-A and EV-B.
Statistical analysis
For data analysis, all new infections were considered statistically independent. Data were analysed with the IBM SPSS Statistics software (version 22.0, IBM Corporation, Armonk, NY, USA) and GraphPad Prism (version 5.01, GraphPad Software, La Jolla, CA, USA). Categorical variables were compared by means of a chi-square test. Differences between continuous variables were determined using Student's t-test or analysis of variance if normally distributed and non-parametric tests if not normally distributed. A two-sided p-value of <0.05 was considered statistically significant.
Results
RV genotyping
We analysed 1098 RV-positive nasopharyngeal and oropharyngeal samples. VP4/VP2 sequences were obtained for 745 samples (67.9%), yielding 709 samples with an RV sequence (64.6%, 64/1098) and 36 samples with a non-RV EV sequence (3.3%, 36/1098) (see Supplementary material, Table S1).
A total of 637 new RV infections were observed in 557 patients of whom 310 (55.7%) were male and 247 (44.3%) were female. Overall, the patients were young children, with a median age at time of infection of 1.6 years (interquartile range (IQR) 0.5–17.6).
Distribution of RV species
Typing of the available samples revealed that 334 of 637 infections (52.4%) typed were caused by RV species A, 72 (11.3%) by species RV-B, and 231 (36.2%) by RV-C. RV-B contains the smallest number of RV types (32 types, compared with 80 RV-A types and 55 RV-C types) and so one would expect a lower prevalence of RV-B infections if all RV types were equally prevalent. After correction for the skewed proportions of the different species, RV-B types were less prevalent than RV-A and RV-C types (chi-square test, p <0.0001).
Patients infected with RV-B types were significantly older (median 10.4 years, IQR 0.7–35.3) than those infected with RV-A types (median 1.8 years, IQR 0.5–26.5) or RV-C (median 1.2 years, IQR 0.5–4.3) (Kruskal–Wallis test, p 0.007) (Fig. 1 ). An analysis including only the first infection of each patient resulted in a similar outcome (Kruskal–Wallis test, p 0.002). To investigate whether the low median age of our population could explain the under-representation of RV-B infections, we calculated the ratio of RV species in patients over 10 years of age. In this selected patient population the under-representation of RV-B in our study was retained (chi-square test, p 0.005), indicating that the lower proportion of RV-B infections was not (solely) due to the lower detection of RV-B in the young children.
Fig. 1.
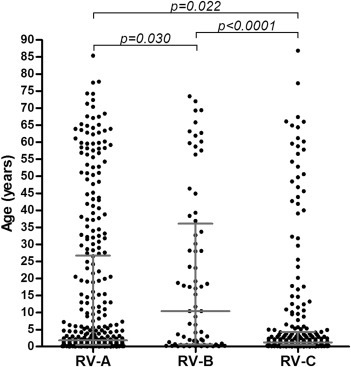
Age distribution of patients infected with rhinovirus type A (RV-A), RV-B or RV-C. The median and the interquartile range are depicted as well as p-values calculated with Dunn–Bonferroni post hoc test.
The detection of co-infections
In 130 RV infections (20.4%, 130/637) there was a viral co-infection (Fig. 2 ). In the majority of these viral co-infections, there was one co-infecting viral agent (n = 104), but up to four co-infecting viruses were detected, of which human bocavirus (n = 42) and adenovirus (n = 33) were the most prevalent.
Fig. 2.
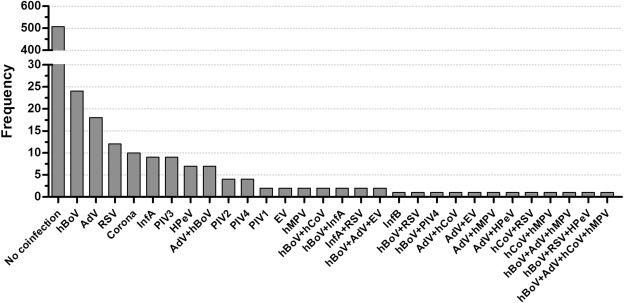
Observed rhinovirus (RV) co-infections. Patient samples were tested with a multiplex real-time PCR for other respiratory viruses. The viral co-infections detected in RV-positive samples are depicted. Enterovirus (EV) infections were only included if EV genotyping yielded a non-RV EV sequence.
Co-infection rates were similar between the different infecting RV species (22.4% (74/334) RV-A, 20.8% (15/72) RV-B and 17.7% (41/231) RV-C infected patients, chi-square test, p 0.44).
Circulation of RV types
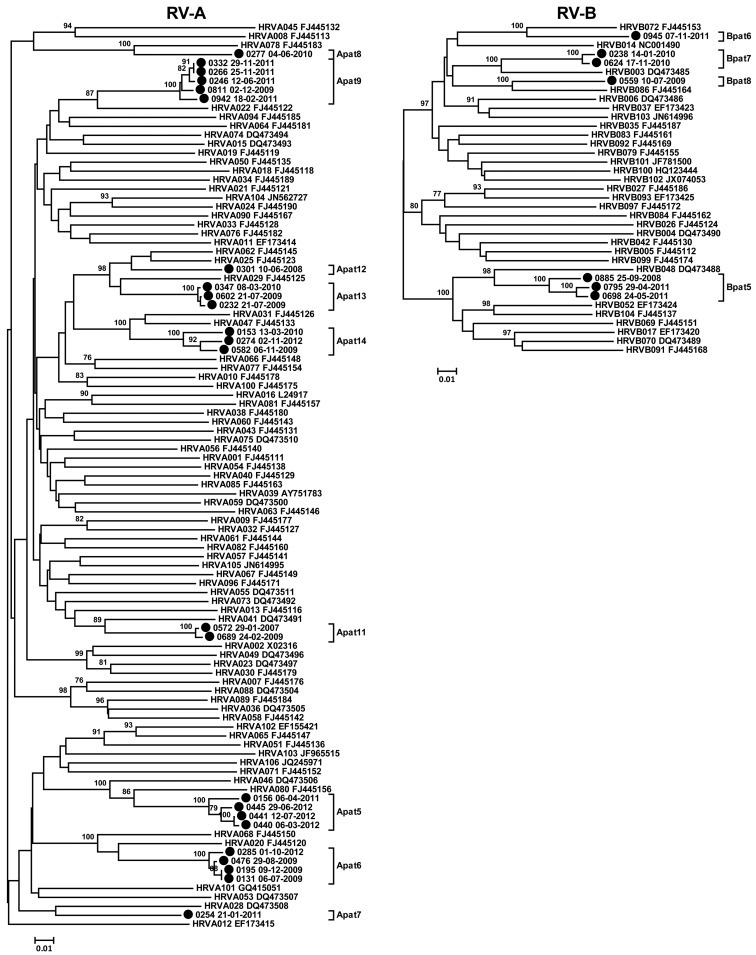
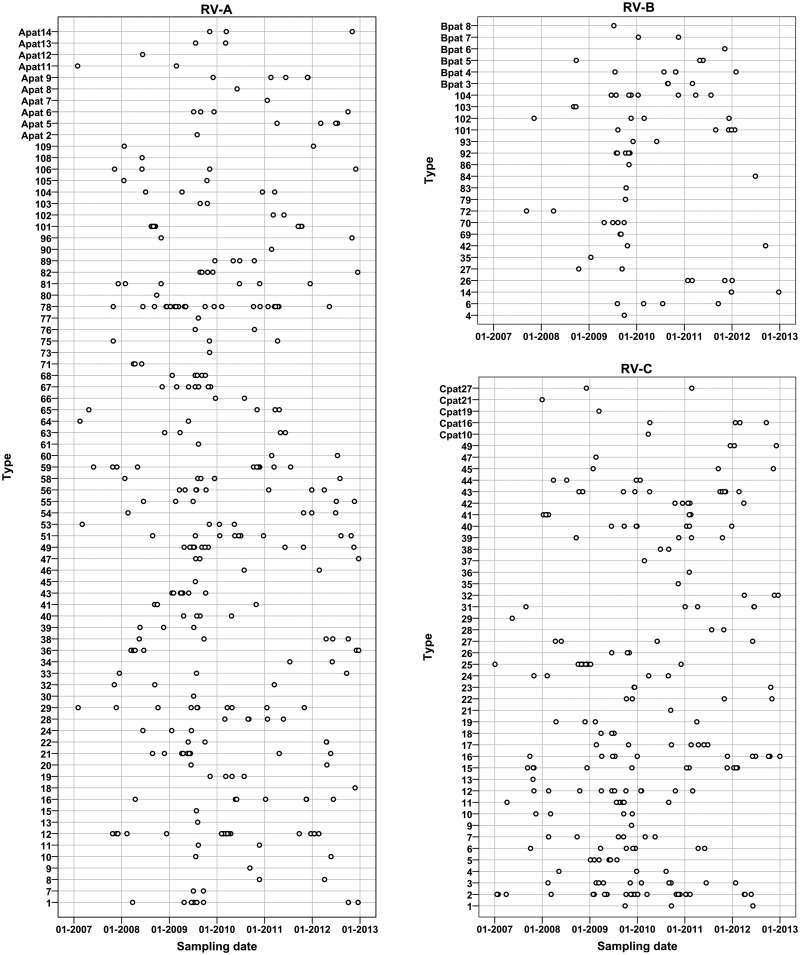
In our population we could detect the majority of the currently classified 167 RV types (77.2% (128/167) of all RV types, 83.8% (67/80) of all RV-A types, 62.5% (20/32) of all RV-B types, and 74.5% (41/55) of all RV-C types) (Fig. 3 ), suggesting that most types circulate ubiquitously. In addition, we detected eight previously described provisionally assigned types (PATs). These are virus strains predicted to be new types based on the VP4/VP2 sequence, but for which the VP1 sequence is not yet available [14]. VP1 is regarded as the reference standard for type identification because it shows more sequence variation than VP4/VP2. Furthermore, we detected 31 strains that were classified as 13 novel PATs (nine RV-A and four RV-B). Phylogenetic trees including these PATs are provided in the Supplementary material (see Fig. S1).
Fig. 3.
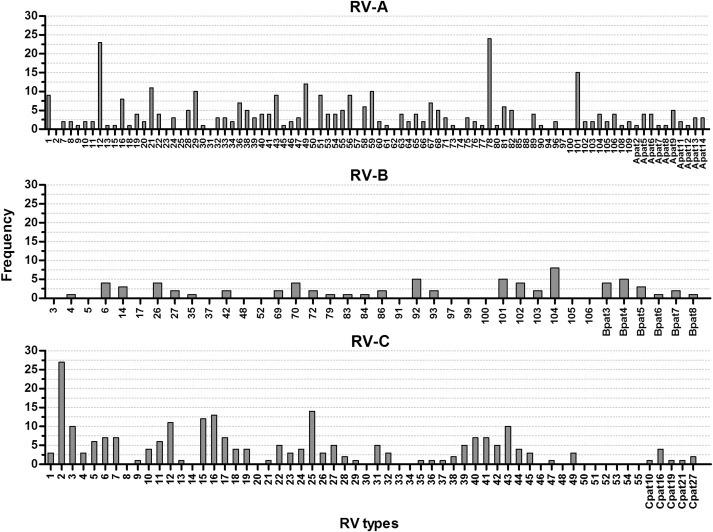
Detection of rhinovirus (RV) types. The frequency of all currently known RV types and all detected provisionally assigned types are indicated.
Most types were detected at low frequency, but some types, such as RV-A12 (n = 23), RV-A78 (n = 24) and RV-C2 (n = 27), were present at a higher rate.
Time of circulating species and types
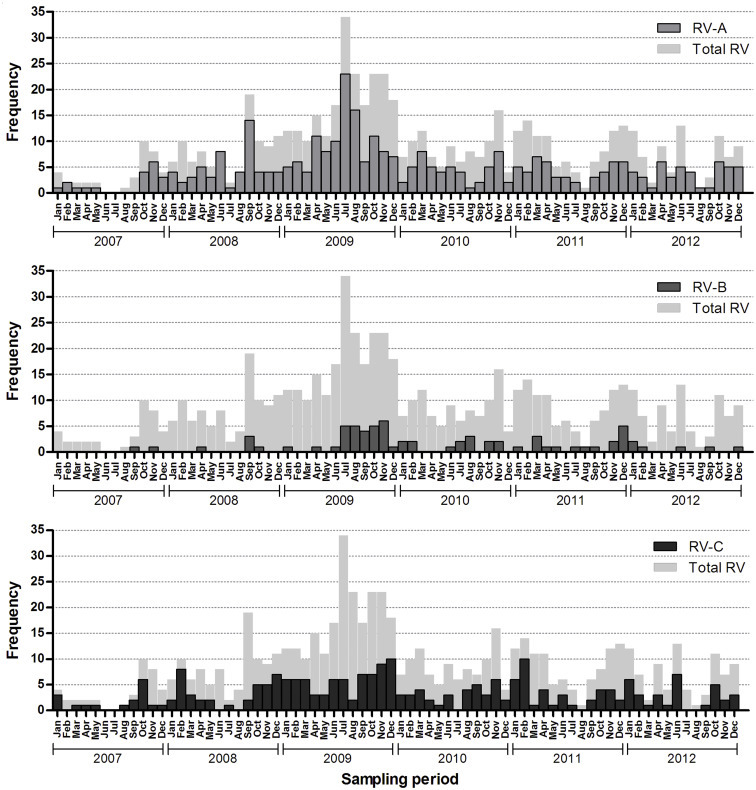
Rhinoviruses circulated throughout the year, with a slightly higher number of infections occurring in the autumn and a decline in the summer (Fig. 4 , and see Supplementary material, Fig. S2). In the second half of 2009 there was an exceptional large increase in RV-positive samples compared with other years (see Supplementary material, Fig. S2).
Fig. 4.
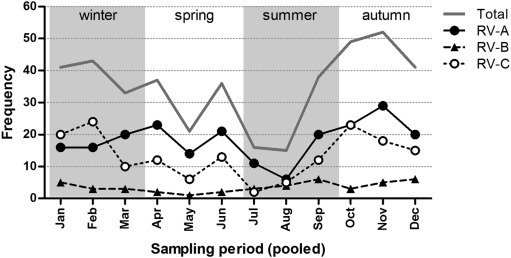
Frequency of rhinovirus type A (RV-A), RV-B and RV-C infections per month. The amount of infections with RV species A, B and C observed in each month, pooling the years 2007–2012. The year 2009 was excluded, as RV circulation in that year was atypical.
Over the year, the species distribution fluctuated slightly (Fig. 4). RV-C tended to be more dominant in the winter months, whereas the rest of the year RV-A infections were more prevalent. RV-B showed low-level circulation throughout the year with a moderate increase of infections in the second half of the year.
Specific RV types (e.g. RV-A29) were consistently detected throughout the study period, whereas other RV types were mainly detected in peak seasons (see Supplementary material, Fig. S3). For example, RV-A12 was detected in clusters in the winters of 2007/08, 2009/10 and 2011/12. Furthermore, RV-A101 was only detected during a small outbreak in August/September 2008 (n = 11) and a few times in September/October 2011 (n = 4). Part of these RV-A101 infections may have been hospital-acquired, as deduced from the reason for admission and the time between the admittance of the patient and the sampling, but other RV infections were clearly contracted outside the hospital (sample taken within 1 day of admittance to the hospital for respiratory illness). RV-C2 and RV-A78, the most commonly detected RV types in our population, were detected every year and followed a similar pattern as RVs in general: a higher detection rate in the autumn/winter and fewer infections in the summer.
Discussion
In our study we described the prevalence of RV types in Amsterdam by typing RV-positive samples submitted for diagnostics in 2007–2012 to the Academic Medical Centre. In our population we found a large number of different RV types, of which most belonged to species RV-A, and the least belonged to species RV-B. Relatively many RV strains were classified as novel PATs.
Among the 637 typed RV infections, 52.4% (334/637) belonged to RV-A, 11.3% (72/637) belonged to RV-B, and 36.2% (231/637) of the typed RV samples belonged to RV-C. The detection rates in our study are in line with previous studies that reported ratios of 47%–-64%, 2%–13% and 21%–45% for respectively RV-A, RV-B and RV-C [19], [20], [21], [22], [23]. RV-A and RV-C infections are consistently reported to be more frequent than RV-B infections.
We could detect most of the currently classified types even though our samples were obtained in a geographically small area. These results suggest that most RVs are widespread, which is in agreement with previous reports that also describe the simultaneous circulation of a high number of types [20], [24], [25]. Nevertheless, some types have repeatedly been detected at higher frequencies than other types over the world [14], [22], [26], [27]. Most of these types were also present at intermediate-to-high frequency in our population, such as RV-A12, RV-A78, RV-A101, RV-B104, RV-C2, RV-C16 and RV-C43. A limitation of our study was that we used population sequencing, which in most cases only detects the major type present and misses most RV–RV co-infections. However, there were no indications for RV–RV co-infections, such as sequence ambiguities.
Of all the sequences obtained from RV-positive samples, 4.8% (36/745) were characterized as an EV sequence. The finding of EV sequences reflects the well-reported cross-reactivity of the RV detection PCR for non-RV EVs, due to their close relationship and the conservation of the 5′-untranslated region [12], [28]. Almost half of the EVs (15/36) were EV-D68, an EV that is associated with respiratory diseases. The same holds true for EV-C104, which was detected in five samples (from four infections). The other EVs detected are found only occasionally in respiratory samples [8], [29].
Our study monitored circulation of RVs over a relatively long time period, which allowed us to examine the circulation of RV species and types over 6 years, thereby limiting the influence of single outbreaks. Our observation that RVs circulate during the whole year has been found repeatedly [1], [8]. Interestingly, Linder et al. also reported a dominance of RV-C in the winter months [30]. We found that some types circulate persistently, but for other types we saw a different circulation pattern where the type was detected mainly in the winter months and occasionally early spring but not throughout the rest of the year. Further studies are required to determine if this circulation pattern is type-specific or due to the small sample size of specific RV types in our study.
We were able to detect 13 putative new RV types. The finding of this many possible new types was unexpected compared with the 20 new RV types/PATs reported since 2010 (complete genome, VP1 or VP4/VP2 sequence published in 2010 or later, or sequence made available to the GenBank in 2010 or later if unpublished, www.picornastudygroup.com), though VP1 sequencing is in progress for our strains to confirm that these are new types. Theoretically, the large amount of new PATs could be the result of local circulation of specific RV types, but as widespread occurrence has been reported for most RV types [14], we hypothesize that there may be many more RV types to be discovered and classified.
In July and August 2009 an atypically high number of RV infections was detected. This may be due to increased sampling as a result of the start of the influenza H1N1 epidemic [31]. This emphasizes that one should be aware that the number of incoming samples can confound the estimated incidence when studying epidemiology by typing samples submitted for diagnostics. This is especially the case for a virus like RV, of which infections occur at high frequency and often asymptomatically.
Our study has some limitations. We were not able to perform associations between the clinical symptoms of the patients and the different RV types due to the retrospective character of the study and the limited information provided by the clinicians on the accompanying form to the laboratory. Because of the limited clinical information and heterogeneous composition of our study group, we were not able to evaluate the prevalence of various RV types in specific populations. A selection bias is that our study is composed of individuals attending the hospital and therefore might not completely reflect the prevalence of RV types in the general population. Nevertheless, the large variety in RV types in our population points towards continuous variations in RV types co-circulating in the community.
In summary, in this study we present an overview of the RV types circulating in the Amsterdam area in the Netherlands in the years 2007 to 2012. RV epidemiology is complex and many RV types are co-circulating simultaneously. RV-targeted vaccination and antiviral strategies should therefore aim for broad-spectrum activity and can probably not afford to focus on a selection of dominant RV types.
Transparency declaration
L.L. and X.T. were completely and A.B. was partially funded by a grant from the Crucell Vaccine Institute. K.W. and A.B. are partially funded by the Seventh Framework Programme of the European Union IAPP (AIROPico, PIAPP-GA-2013-612308). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Acknowledgements
The authors wish to thank Victoria A. Janes for critical reading of the manuscript. Part of the data in this manuscript have previously been presented at the ASV 2015, London, Canada, 14 July 2015.
Editor: L. Kaiser
Footnotes
Additional Supporting Information may be found in the online version of this article at http://dx.doi.org/10.1016/j.cmi.2016.08.007.
Appendix A. Supplementary data
The following supplementary materials are available for this article:
Enterovirus types detected
References
- 1.Jacobs S.E., Lamson D.M., Kirsten S., Walsh T.J. Human rhinoviruses. Clin Microbiol Rev. 2013;26:135–162. doi: 10.1128/CMR.00077-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leigh R., Proud D. Virus-induced modulation of lower airway diseases: pathogenesis and pharmacologic approaches to treatment. Pharmacol Ther. 2015;148:185–198. doi: 10.1016/j.pharmthera.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Knowles N.J., Hovi T., Hyypiä T., King A.M.Q., Lindberg A.M., Pallansch M.A. Picornaviridae. In: King A.M.Q., Adams M.J., Carstens E.B., Lefkowitz E.J., editors. Virus taxonomy: classification and nomenclature of viruses: Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier; San Diego, CA: 2012. pp. 855–880. [Google Scholar]
- 4.Lau S.K.P., Yip C.C.Y., Tsoi H.-W., Lee R.A., So L.-Y., Lau Y.-L. Clinical features and complete genome characterization of a distinct human rhinovirus (HRV) genetic cluster, probably representing a previously undetected HRV species, HRV-C, associated with acute respiratory illness in children. J Clin Microbiol. 2007;45:3655–3664. doi: 10.1128/JCM.01254-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lamson D., Renwick N., Kapoor V., Liu Z., Palacios G., Ju J. MassTag polymerase-chain-reaction detection of respiratory pathogens, including a new rhinovirus genotype, that caused influenza-like illness in New York State during 2004–2005. J Infect Dis. 2006;194:1398–1402. doi: 10.1086/508551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arden K.E., McErlean P., Nissen M.D., Sloots T.P., Mackay I.M. Frequent detection of human rhinoviruses, paramyxoviruses, coronaviruses, and bocavirus during acute respiratory tract infections. J Med Virol. 2006;78:1232–1240. doi: 10.1002/jmv.20689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bochkov Y a, Palmenberg A.C., Lee W.-M., Rathe J.A., Amineva S.P., Sun X. Molecular modeling, organ culture and reverse genetics for a newly identified human rhinovirus C. Nat Med. 2011;17:627–632. doi: 10.1038/nm.2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Royston L., Tapparel C. Rhinoviruses and respiratory enteroviruses: not as simple as ABC. Viruses. 2016;8:16. doi: 10.3390/v8010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.L'Huillier A.G., Kaiser L., Petty T.J., Kilowoko M., Kyungu E., Hongoa P. Molecular epidemiology of human rhinoviruses and enteroviruses highlights their diversity in sub-Saharan Africa. Viruses. 2015;7:6412–6423. doi: 10.3390/v7122948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khetsuriani N., Lamonte-Fowlkes A., Oberst S., Pallansch M.A. Enterovirus surveillance—United States, 1970–2005. MMWR Surveill Summ. 2006;55:1–20. [PubMed] [Google Scholar]
- 11.Jansen R.R., Schinkel J., Koekkoek S., Pajkrt D., Beld M., de Jong M.D. Development and evaluation of a four-tube real time multiplex PCR assay covering fourteen respiratory viruses, and comparison to its corresponding single target counterparts. J Clin Virol. 2011;51:179–185. doi: 10.1016/j.jcv.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jaramillo-Gutierrez G., Benschop K.S.M., Claas E.C.J., de Jong A.S., van Loon A.M., Pas S.D. September through October 2010 multi-centre study in the Netherlands examining laboratory ability to detect enterovirus 68, an emerging respiratory pathogen. J Virol Methods. 2013;190:53–62. doi: 10.1016/j.jviromet.2013.02.010. [DOI] [PubMed] [Google Scholar]
- 13.Bruning A.H.L., Thomas X.V., van der Linden L., Wildenbeest J.G., Minnaar R.P., Jansen R.R. Clinical, virological and epidemiological characteristics of rhinovirus infections in early childhood: a comparison between non-hospitalised and hospitalised children. J Clin Virol. 2015;73:120–126. doi: 10.1016/j.jcv.2015.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McIntyre C.L., Knowles N.J., Simmonds P. Proposals for the classification of human rhinovirus species A, B and C into genotypically assigned types. J Gen Virol. 2013;94:1791–1806. doi: 10.1099/vir.0.053686-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nicholas K., Nicholas H., Deerfield D. GeneDoc: analysis and visualization of genetic variation. EMBNEW NEWS. 1997;4 [Google Scholar]
- 16.Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 17.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Minnaar R.P., Koen G., de Haan K., Wolthers K.C., Benschop K.S.M. Evaluation of two (semi-)nested VP1 based-PCRs for typing enteroviruses directly from cerebral spinal fluid samples. J Virol Methods. 2012;185:228–233. doi: 10.1016/j.jviromet.2012.07.009. [DOI] [PubMed] [Google Scholar]
- 19.Rahamat-Langendoen J.C., Riezebos-Brilman A., Hak E., Schölvinck E.H., Niesters H.G.M. The significance of rhinovirus detection in hospitalized children: clinical, epidemiological and virological features. Clin Microbiol Infect. 2013;19:E435–E442. doi: 10.1111/1469-0691.12242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marcone D.N., Culasso A., Carballal G., Campos R., Echavarría M. Genetic diversity and clinical impact of human rhinoviruses in hospitalized and outpatient children with acute respiratory infection, Argentina. J Clin Virol. 2014;61:558–564. doi: 10.1016/j.jcv.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen W.-J., Arnold J.C., Fairchok M.P., Danaher P.J., McDonough E.A., Blair P.J. Epidemiologic, clinical, and virologic characteristics of human rhinovirus infection among otherwise healthy children and adults: rhinovirus among adults and children. J Clin Virol. 2015;64:74–82. doi: 10.1016/j.jcv.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsatsral S., Xiang Z., Fuji N., Maitsetseg C., Khulan J., Oshitani H. Molecular epidemiology of the human rhinovirus infection in Mongolia during 2008–2013. Jpn J Infect Dis. 2015;68:280–287. doi: 10.7883/yoken.JJID.2014.090. [DOI] [PubMed] [Google Scholar]
- 23.Jacobs S.E., Lamson D.M., Soave R., Guzman B.H., Shore T.B., Ritchie E.K. Clinical and molecular epidemiology of human rhinovirus infections in patients with hematologic malignancy. J Clin Virol. 2015;71:51–58. doi: 10.1016/j.jcv.2015.07.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wisdom A., Leitch E.C.M., Gaunt E., Harvala H., Simmonds P. Screening respiratory samples for detection of human rhinoviruses (HRVs) and enteroviruses: comprehensive VP4-VP2 typing reveals high incidence and genetic diversity of HRV species C. J Clin Microbiol. 2009;47(12):3958–3967. doi: 10.1128/JCM.00993-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang T., Wang W., Bessaud M., Ren P., Sheng J., Yan H. Evidence of recombination and genetic diversity in human rhinoviruses in children with acute respiratory infection. PLoS One. 2009;4:e6355. doi: 10.1371/journal.pone.0006355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tran D.N., Trinh Q.D., Pham N.T.K., Pham T.M.H., Ha M.T., Nguyen T.Q.N. Human rhinovirus infections in hospitalized children: clinical, epidemiological and virological features. Epidemiol Infect. 2015;144:1–9. doi: 10.1017/S0950268815000953. [DOI] [PubMed] [Google Scholar]
- 27.Lu Q.-B., Wo Y., Wang L.-Y., Wang H.-Y., Huang D.-D., Zhang X.-A. Molecular epidemiology of human rhinovirus in children with acute respiratory diseases in Chongqing, China. Sci Rep. 2014;4:6686. doi: 10.1038/srep06686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McAllister S.C., Schleiss M.R., Arbefeville S., Steiner M.E., Hanson R.S., Pollock C. Epidemic 2014 enterovirus D68 cross-reacts with human rhinovirus on a respiratory molecular diagnostic platform. PLoS One. 2015;10:e0118529. doi: 10.1371/journal.pone.0118529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yip C.C.Y., Lau S.K.P., Woo P.C.Y., Wong S.S.Y., Tsang T.H.F., Lo J.Y.C. Recombinant coxsackievirus A2 and deaths of children, Hong Kong, 2012. Emerg Infect Dis. 2013;19(8):1285–1288. doi: 10.3201/eid1908.121498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Linder J.E., Kraft D.C., Mohamed Y., Lu Z., Heil L., Tollefson S. Human rhinovirus C: age, season, and lower respiratory illness over the past 3 decades. J Allergy Clin Immunol. 2013;131:69–77. doi: 10.1016/j.jaci.2012.09.033. e1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hahné S, Donker T, Meijer A, Timen A, Steenbergen J van, Osterhaus AD, et al. Epidemiology and control of influenza A(H1N1)v in the Netherlands: the first 115 cases European Centre for Disease Prevention and Control (ECDC)—Health Communication Unit; 2009 [cited 2016 Feb 15]. Available at: http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19267. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Enterovirus types detected


