Abstract
Background: Antigen detection assays and viral isolation techniques are routinely used to detect adenoviruses (Ad) associated with respiratory infections, and the value of the polymerase chain reaction (PCR) has recently been assessed. Objectives: This paper describes a PCR-hybridization-immunoenzymatic assay (PCR Adenovirus consensus®) used to detect Ad and identify Ad species in respiratory specimens. Results: On seven representative serotypes Ad 12, Ad 3, Ad 7, Ad 11, Ad 1, Ad 8, Ad 4, the mean genome equivalents per ml and the mean 50% infectious doses per ml were 106.3and 104, respectively. Using 362 nasal aspirates from children, Ad were detected by immunofluorescence (IF) and culture in 97 cases (27%), by the PCR-Ad hexon method in 107 cases (29.5%) and by the PCR Adenovirus Consensus® method in 113 cases (31.2%); 13 samples were found positive by both PCR and negative by the IF and culture methods; five samples were only positive according to the PCR Adenovirus Consensus® method. The sensitivity, specificity, predictive positive value and predictive negative value of the PCR Adenovirus Consensus® method were 97.9%, 93.2%, 84%, 99.1%, respectively. The method identified the species (sp) from 91 positive amplicons: 1 Ad sp A, 44 Ad sp B, 42 Ad sp C, 3 Ad sp E, and 1 Ad sp F; 85 isolates were identified by IF or the neutralisation in culture, and 86 by a PCR-RE digestion method. The PCR Adenovirus Consensus® detected six positive samples that were negative according to the IF and culture methods, and it identified the precise species of nine IF-positive and culture-negative nasal aspirates. Conclusion: The PCR Adenovirus Consensus® technique is more efficient than the classical IF or culture techniques for the detection of Ad in respiratory samples. An internal control is included to validate the screening results, and specific probes are used to identify the Ad species.
Keywords: Adenovirus, PCR, Species
1. Introduction
Immunofluorescence assays, enzyme-linked immunosorbent assays (ELISA), and viral isolation techniques are routinely used to detect adenoviruses (Ad) associated with respiratory infections. However, Ad can take several days to grow in cell culture conditions and are susceptible to specimen toxicity or to be out-competed by bacteria or fungi. Ad isolates can be identified to the species level by the haemagglutination inhibition test and the neutralisation tests (Hierholzer, 1995). The limitations of the neutralisation test are the need for the availability of type-specific antisera and the need to test multiple antisera against a single isolate. For that reason most laboratories choose to limit the test to the Ad that are most frequently isolated from patients with respiratory tract diseases: Ad serotypes 1, 2, 5, 6 from species C, Ad serotypes 3, 7 from species B1, and Ad serotype 4 from species E (Schmidtz et al., 1983) according to the recently amended classification system (Benkö et al., 1999) (formerly subgenera). However, other Ad serotypes are occasionally detected in respiratory infections, especially in immunocompromised patients (Hierholzer, 1992). The use of IFA and ELISA to detect antigens provides a rapid diagnosis, but requires significant levels of antigens for detection, and is generally less sensitive than the culture method. The value of the polymerase chain reaction (PCR) for the diagnosis of Ad infections has recently been assessed. Most of the PCR assays used rely on the detection of sequences in the hexon gene region (Allard et al., 1990, Hierholzer et al., 1993, Morris et al., 1996, Raty et al., 1999), whereas others use sequences in theVA RNA gene (Kidd et al., 1996), the pIX gene (Akalu et al., 1998) or in the fiber gene (Xu et al., 2000). Non-nested and nested PCRs have also been developed for the detection of the six species of Ad. These methods are based on the comparison of the migration of amplimers (Kidd et al., 1996), the analysis of restriction enzyme profiles (Elnifro et al., 2000) and the use of multiplex PCR (Na et al., 2002, Pring-Akerblom et al., 1999, Xu et al., 2000). Finally, a method based on a combination of PCR and restriction endonuclease digestion has been developed for the rapid identification of species and serotypes of Ad (Allard et al., 2001).
In this paper, we describe a PCR-hybridization-immunoenzymatic assay (PCR-HYB-EIA), PCR Adenovirus consensus®, which uses Ad genus-specific primers and one generic and six species-specific probes defined in the VA RNA gene. We compare this method to another PCR method that detects specific sequences in the hexon gene and to the conventional methods (immunofluorescence (IF) and culture) used to detect Ad in respiratory specimens.
2. Methods
2.1. Sensitivity and specificity of the primers and probes
The analytical sensitivity of the PCR-HYB-EIA (PCR Adenovirus consensus®) was assessed by analysing serial 10-fold dilutions of MRC5 cells infected with Ad representative of each species A (Ad 12), B1 (Ad 3, Ad 7), B2 (Ad 11), C (Ad 1), D (Ad 8) and E (Ad 4). The specificity was demonstrated by using samples infected with infectious agents other than Ad and commonly found in the respiratory tract.
2.2. Clinical samples
The clinical sensitivity of the technique was assessed on 362 nasal aspirates collected from children hospitalised in the University Hospital of Caen and in the Flers hospital with acute respiratory tract disease during the winters of 1996–2000. Series of specimens were randomly selected from each winter period. They were classified as having or not having an Ad infection according to the results of the immunofluorescence assay or the viral isolation technique or the PCR Adenovirus Consensus®.
2.3. Ad detection and identification
A monoclonal antibody directed against Ad (Imagen®, Dako, UK) was used for the immunofluorescence assay on nasal smears or cell cultures. NCI-H292 and MRC5 cell cultures were used for the viral isolation technique. The immunofluorescence assay was systematically carried out in cultures exhibiting cytopathogenic effects and on NCI-H292 cells that had been incubated for 5 days. Ad serotypes were then identified using the classical neutralisation test in cell cultures and ATCC neutralising antibodies (Hierholzer, 1995).
2.4. PCR assays
-
(i)
DNA extraction: Nucleic acids were extracted from 200 μl of nasal aspirates by use of QIAamp® DNA Blood Mini Kit (QIAgen, France).
-
(ii)
PCR Adenovirus Consensus ®: The amplification uses a consensus set of two primers followed by a generic or specific hybridization. Two Ad group-specific primers were designed according to the reported sequences of the VA RNA gene between positions 10,065 and 10,235 for the Ad 40 sequence and between positions 10,607 and 11,023 for the Ad 2 sequence. DNA was amplified in a 50 μl volumes containing 5 μl of DNA and 45 μl of reaction mixture (1× PCR buffer, 200 μM each deoxynucleoside triphosphate, 0.4 mM each primer, 1.5 U of Taq DNA polymerase [HotStarTaq, Qiagen, France]). The amplification conditions were as follows: (i) initial denaturation for 15 min at 95 °C to activate the enzyme, followed by: (ii) 10 cycles of denaturation at 94 °C for 10 s, touch down annealing temperature from 65 to 55 °C for 30 s, and primer extension at 72 °C for 30 s, then 30 cycles of 94 °C, for 10 s; 55 °C, for 30 s and 72 °C, for 30 s and (iii) a final extension at 72 °C for 2 min. Amplified products were analysed by hybridisation on microtiter plates with biotinylated probes. Hybrids were detected with a streptavidin peroxidase conjugate. A tetramethylbenzidine-hydrogen peroxide solution was used as a substrate for color development (Hybridowell™, Argène, France). Adenovirus Consensus® kit, is now commercialy available (Argene, ref 67-065). One generic probe characterizing the Ad group and seven specific probes corresponding to Ad A, Ad B1, Ad B2, Ad C, Ad D, Ad E, Ad F are provided in the kit. However, an internal control, a composite plasmid containing sequences recognized by the primers flanking a plasmidic sequence, with thermodynamics similar to those of the viral sequence has been added to follow the inhibition of the amplification. This control is hybridized with a specific probe. A negative control of the amplification is performed for each experience with the premix amplication using sterile water. A negative detection control, human amplified product, is provided in order to determine the cut-off value. After detection of the positive samples with the generic probe, the species of the virus involved is determined using the specific probes on the same amplified product.
-
(iii)
PCR Ad hexon: We used the two hexon gene primers described by Hierholzer et al. (1993) (positions 2728–2750 and 2888–2866), and a 5′-biotinylated probe B (positions 2833–2858). The PCR and the detection of the amplified products by agarose gel electrophoresis (band at 161 bp) and by a DNA Enzyme Immunoassay (GEN-ETI-K® DEIA, Sorin, France) were carried out as previously reported (Freymuth et al., 1997).
-
(iv)
Serotyping: The results obtained with PCR Adenovirus Consensus® were compared with those obtained with the neutralisation test and the test described by Allard et al. (2001) using PCR to amplify sequences in the hexon gene combined with restriction endonuclease analysis with the aim of identifying serotypes that were identified by all three methods.
3. Results
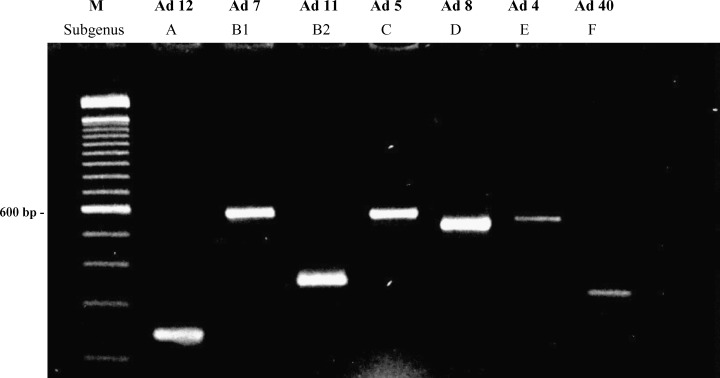
Primers were not tested against all 51 Ad serotypes, but were designed as broadly reactive as possible to detect all these serotypes. These primers were chosen by comparing the sequences of VA RNA regions in the Genbank database. These sequences included Ad 1–11 plus Ad 16, 18, 21, 30, 40, 41 and 46, which are representative of the seven species. One Ad from each species was amplified and the resulting products were subjected to agarose gel electrophoresis. The seven species had been correctly amplified, and the expected sizes of the bands, corresponding to one VA RNA gene (Ad from species A, B2, F) or to two VA RNA genes (Ad from B1, C, D, E), was as follows:: Ad 12 (sp A): 306 bp, Ad 7 (Sp B1): 584 bp, Ad 11 (sp B2): 354 bp, Ad 1 (sp C): 601 bp, Ad 8 (sp D): 520 bp, Ad 4 (sp E): 548 bp, Ad 40 (sp F): 323 bp (Fig. 1 ). However, as large bands (>500 bp) migrate slowly, Ad species B1, C, D and E cannot be reliably differentiated on this basis, thus species-specific probes were also designed as shown below.
Fig. 1.
Ethidium bromide-stained agarose gel showing VA RNA gene region PCR products derived from Ad representative of the indicated species. Bands of two size ranges were observed: 290–340 bp for one gene VA RNA Ad 12, Ad 11, Ad 40; and 460–560 bp for two genes VA RNA Ad 7, Ad 5, Ad 8, Ad 4. Lane M: 100 bp molecular weight marker
The specificity of PCR Adenovirus Consensus® was demonstrated by the fact that no amplified products were detected from nucleic acids extracted of cell cultures infected with other viruses and of bacterias that cause respiratory infections or are commonly found in the upper respiratory tract: influenza virus A and B, respiratory syncytial virus A and B, parainfluenza virus 1, 2 and 3, coronavirus 229E and OC 43, Mycoplasma pneumoniae, Chlamydia pneumoniae, herpes simplex virus type 1, herpesvirus varicellae, cytomegalovirus, Epstein Barr virus, herpes virus type 6, parvovirus B19, BK and JC viruses, measles virus, mumps virus. The analytical sensitivity of PCR Adenovirus Consensus® was further delineated by testing the DNA extracted from serial dilutions of MRC-5-infected cells. We used Ad prototype strains belonging to the main species and representative of those isolated from patients with respiratory Ad infections: Ad 12 (sp A), Ad 3 and Ad 7 (sp B1), Ad 11 (sp B2), Ad 1 (sp C), Ad 8 (sp D), Ad 4 (sp E). The numbers of genome equivalents per ml (GE/ml) were calculated using the end point dilution method and were compared to the 50% infectious dose per ml (ID50/ml) for each strain (Table 1 ). The mean GE/ml and ID50/ml for the seven serotypes, were 106.3 and 104, respectively. The large difference between the titres of the Ad strains according to their ID/ml or GE/ml, is probably to be due to the fact that MRC cells are not the best culture system for the isolation of Ad and that higher GE values reflects the proportion of nonviable virus unable to initiate productive infection. As usual, the result was slightly higher with the hybridisation method than with the gel electrophoresis method (mean titres of Ad strains: 106.2versus 105, respectively). However, the titres obtained after hybridisation in the PCR Adenovirus Consensus® technique did not differ according to whether the consensus probe or the sugbroup-specific probes were used.
Table 1.
Analytical sensitivity of the PCR Ad consensus method
| Strain | Species | GE/ml |
|||
|---|---|---|---|---|---|
| ID5O/ml | Gel electrophoresis | Group-specific hybridization | Species-specific hybridization | ||
| Ad 12 | A | 102.0 | 103.5 | 104.5 | 105.5 |
| Ad 3 | B1 | 104.0 | 104.5 | 106.5 | 106.5 |
| Ad 7 | B1 | 103.2 | 106.5 | 106.5 | 107.5 |
| Ad 11 | B2 | 103.4 | 106.5 | 108.5 | 108.5 |
| Ad 1 | C | 104.9 | 104.5 | 106.5 | 106.5 |
| Ad 8 | D | 102.9 | 104.5 | 105.5 | 105.5 |
| Ad 4 | E | 103.2 | 105.5 | 105.5 | 106.5 |
| Mean | 104.0 | 105.0 | 106.35 | 106.35 | |
Serial dilutions of MRC5 cells infected with Ad prototype strains from the main species and representative of respiratory Ad infections: Ad 12 (sg A), Ad 3 and Ad 7 (sg B1), Ad 11 (sg B2), Ad 1 (sg C), Ad 8 (sg D), Ad 4 (sg E) were used to count the number of genome equivalents per ml (GE/ml) and the 50% infectious doses per ml (ID50/ml).
The clinical sensitivities of the PCR Adenovirus Consensus® and PCR Ad hexon methods were compared to those of the immunofluorescence and culture methods, using 362 nasal aspirates. Ad were detected by immunofluorescence and culture in 97 cases (27%), by the PCR-Ad hexon method in 107 cases (29.5%) and by the PCR Adenovirus Consensus® method in 113 cases (31.2%) (Table 2 ). Thirteen of the samples found to be positive by both PCR were found to be negative by the immunofluorescence and culture methods. Five samples were only positive according to the PCR Adenovirus Consensus® method; these samples were re-extracted and still found to be Ad positive. Two of the samples found to be negative according to the PCR Adenovirus Consensus® method were found positive by PCR in one case and by culture in the other case and the isolate was not tested by the PCR assay. The sensitivity, specificity, predictive positive value and predictive negative value of the PCR Adenovirus Consensus® method and of the PCR-Ad hexon method were given in Table 2.
Table 2.
Comparison of the diagnostic efficacies of the Ad-specific PCR methods with that of virus culture or immunofluorescence in 362 nasal aspirates samples
| Diagnostic Method | PCR Ad consensus |
PCR Ad hexon |
||
|---|---|---|---|---|
| Positive | Negative | Positive | Negative | |
| Virus culture/Immunofluorescence | ||||
| Positive | 95 | 2 | 94 | 3 |
| Negative | 18a | 247 | 13 | 252 |
| Total | 362 | 362 | ||
| Sensitivity (%) | 97.9 | 96.9 | ||
| Specificity (%) | 93.2 | 95 | ||
| PPV (%) | 84 | 87.8 | ||
| NPV (%) | 99.1 | 98.8 | ||
Thirteen specimens were positive in both PCR Ad consensus and PCR Ad hexon but not in according to the culture/immunofluorescence method. Five additional samples were only positive according to PCR Ad consensus, these samples were re-extracted and controlled as positive.
The PCR Adenovirus Consensus® kit included probes specific for each species. The ability of this method to determine the species was assessed on 160 specimens which were randomly selected from November to February during winters 1998–1999 and 1999–2000. All samples were systematically tested by immunofluorescence, culture methods and by the PCR-RE digestion method proposed by Allard et al. (2001). Overall, 91 Ad-positive samples were identified (Table 3 ). Their distribution was as follows: 1 Ad sp A, 44 Ad sp B, 42 Ad sp C, 3 Ad sp E, and 1 Ad sp F. In all cases, the PCR Adenovirus Consensus® method gave positive results. Eighty five isolates were identified by the immunofluorescence or the neutralisation in culture method, and 86 by the PCR-RE digestion method. The PCR Adenovirus Consensus® detected six positive samples that were negative according to the immunofluorescence and culture methods: two Ad sp B1, three Ad sp C and one Ad species F. It also, identified the precise species of nine immunofluorescence-positive and culture-negative nasal aspirates. The PCR Adenovirus Consensus® method, was slightly more efficient than the PCR-RE digestion method, identifying five new positive samples: three Ad sp B1, one Ad sp C and one Ad sp F. All the other serotypes were identified as belonging to the same species by the two techniques.
Table 3.
Comparison of Ad identification by the species-specific PCR Adenovirus consensus®, by type-specific neutralisation in culture and by a combination of PCR and restriction endonuclease digestion
| Number of strains | Detection by IF or culturea | PCR AdV Consensus® | PCR-RE digestionb |
|---|---|---|---|
| 1 Ad species A | 1 Ad (IF) | 1 sp A | 1 Ad 31 |
| 44 Ad species B | 36 Ad 3 | 36 sp B1 | 36 Ad 3 |
| 1 Ad 3 | 1 sp B1 | 1 negative | |
| 2 Ad 7 | 2 sp B1 | 2 Ad 7 | |
| 2 Ad (IF) | 2 sp B1 | 2 Ad 3 | |
| 1 Ad 11 | 1 sp B2 | 1 sp B2 | |
| 2 negative | 2 sp B1 | 2 negative | |
| 42 Ad species C | 9 Ad 1 | 9 sp C | 9 Ad 1 |
| 13 Ad 2 | 13 sp C | 13 Ad (2.5.6) | |
| 3 Ad 5 | 3 sp C | 3 Ad (2.5.6) | |
| 1 Ad 5 | 1 sp C | 1 negative | |
| 10 Ad 6 | 10 sp C | 10 Ad (2.5.6) | |
| 3 Ad (IF) | 3 sp C | 2 Ad 1;1 Ad (2.5.6) | |
| 3 Negative | 3 sp C | 3 Ad (2.5.6) | |
| 3 Ad species E | 3 Ad (IF) | 3 sp E | 3 sp E |
| 1 Ad species F | 1 negative | 1 sp F | Negative |
| 69 Ad negative | Negative | Negative | Negative |
Detection by immunofluorescence (IF), or by culture followed by a type-specific neutralisation assay adapted from Hierholzer (1995).
PCR and restriction endonuclease digestion, adapted from Allard et al. (2001).
4. Discussion
The direct immunofluorescence method is less sensitive than the cell culture method to diagnose Ad respiratory infections (Lehtomaki et al., 1986, Takimoto et al., 1991). The variation is between 28% and 75% depending on the author (Ruuskanen et al., 1997). Nevertheless, the IF method is rapid which is especially useful in infants, for which symptoms may be misleading and be reminiscent to those of a bacterial infection (Brouard et al., 2001). Immunoenzymatic techniques are more sensitive than IF, 43–89% depending on the authors (Ruuskanen et al., 1997).
The cell culture method is less suitable for the identification of Ad infections because Ad can take up to 2 weeks to grow. In this study, the direct IF method and/or the cell culture method on MRC5 cells and a NCI-H292 cells were used as a reference. Ninety-seven Ad infections were identified in our study on 362 nasal aspirates.
A number of different PCR techniques, based on the amplification of fragments located in the most conserved areas of the genome, have been designed to identify Ad infections. The technique described by Allard et al. (1990) amplifies a 308 bp fragment partially located in the P1 domain of the hexon gene. In comparison with the culture method, the sensitivity of the Allard method was 93% for species C (types 1, 2, 5, 6) and only 50% for Ad 3 and Ad 7 belonging to B1 species, which are particularly involved in the respiratory diseases (Morris et al., 1996). Hierholzer et al. (1993) described a PCR method, which amplifies a 161 bp fragment located in the P2 region of the hexon gene associated with hybridisation to a fluorescent probe. In a study on 103 samples including 68 respiratory samples, the same number of positive samples were obtained with the culture method, the direct IF method and with the new PCR method. The PCR detection of Ad sequences may not reflect an active viral infection because some Ad (especially Ad 1, 2, 5, 6 from species C) can induce a latent infection. Some authors have suggested combining a PCR method with in situ hybridisation for positive samples (Matsuse et al., 1994). Raty et al. (1999) compared techniques for the detection of Ad on 268 nasopharyngeal specimens obtained from soldiers affected by acute respiratory infections. According to the culture method, 59.1% of the samples were Ad positive compared to 31.9% for the antigen detection method and 62.9% for the PCR method using a crude isolate extracted by “boiling.” In this PCR method, the primers were located in the hexon gene (Allard et al., 1990) and an internal control was included for each sample, to avoid false negative results. Two samples (0.7%) were shown to contain inhibitors. Compared to the culture method, the sensitivity and specificity of the PCR method were 94% and 85%, respectively; 15 samples were positive according to the culture method and negative according to the PCR method. In our study on 362 nasal aspirates, the sensitivity and specificity of the PCR methods were higher: 97.9% and 93.2% respectively for the Adenovirus Consensus® PCR, and higher: 96.9% and 95% for the PCR hexon method. Eighteen samples were positive according to the Adenovirus Consensus® PCR and negative according to the IF and culture methods. These cannot be false positives due to contamination because: (i) 13 were confirmed as positive with a second PCR method and (ii) the five others were confirmed on a second extract. These samples probably contained a low viral yield, below the detection limits of the IF and culture methods. Alternatively, these Ad strains may have been uncultivable or virtually uncultivable (Hierholzer, 1992). Finally, the Ad Consensus® PCR, method appeared to be more sensitive than the hexon PCR method with the screening probe, and was even more convenient following the introduction of an internal control for each sample, which reduced the number of false negative results by displaying inhibitors.
Ad can be identified by seroneutralisation on cell culture (Hierholzer, 1995), enzymatic restriction of DNA extracted from infected cells (Wadell et al., 1980) or sometimes by haemagglutination inhibition (Hierholzer, 1995). These techniques are time-consuming and are not suitable for clinical diagnosis. Kidd et al. (1996) proposed a one-step PCR method able to screen and to identify Ad species associated with the study of the enzymatic restriction profile (Kidd et al., 1996). The size of the amplified fragment varies in the different species and according to the number of copies of the VA RNA gene. Indeed, the A, B2 and F species, have one copy of the VA RNA gene, whereas the B1, C, D and E species have two copies, leading to 240–290 bp amplified fragments for the first group and 490–520 bp fragments for the second. To discriminate between members of each group (A, B2, F and B1, C, D, E) they analysed the enzymatic restriction profiles of the amplified products. On 200 Ad culture isolates, only 3% of the results were discordant with the seroneutralisation results. Our PCR, AdV Consensus® method, uses different primers to Kidd et al. (1996), located in the same VA RNA region, and specific probes to identify each Ad species. Seventy-six of the species identified by culture seroneutralisation were confirmed by the AdV Consensus® method. In addition, six Ad detected by direct IF and not isolated by culture were identified as being B1 and C species, and six others, negative according to IF and culture, were detected and identified as being B1, C, and F species.
Several teams have designed other approaches to identify the Ad species involving enzyme restriction analysis (Elnifro et al., 2000) or multiplex PCR methods (Na et al., 2002, Pring-Akerblom et al., 1999, Xu et al., 2000). These techniques seemed to be very sensitive on isolates, but they were not evaluated on respiratory samples.
The AdV Consensus® technique is a one-step test that can screen and identify Ad in respiratory samples, but cannot identify the serotype. The technique described by Allard et al. (2001), using degenerate primers in the conserved hexon region followed by restriction enzyme digestion of the amplified product, can identify the Ad species and the serotype of the most Ad involved in disease. This method gave good results on 51 Ad prototype strains and 44 variant strains. Thirty-four of the 40 clinical samples (including eight respiratory samples) were positive according to both the PCR and the culture methods. Six were not amplified and had to be further tested by nested PCR, but the amplimer was too short to be analysed by restriction endonucleases. We compared this identification technique with the conventional seroneutralisation technique and the AdV Consensus® technique on the 160 nasal aspirates of our study. Five of the 91 Ad strains, amplified and identified by the AdV Consensus® technique, were not detected by the Allard method: one Ad 3 and two Ad sp B1 (not detected by culture), one Ad sp C and one Ad sp F (not detected by culture).
In conclusion, the AdV Consensus® technique is an useful method for the detection of Ad in respiratory samples. It is more efficient than the classical IF or culture techniques. It consists of one-step PCR followed by hybridisation. An internal control is included to validate the screening results, and specific probes are used to identify the Ad species. This technique is easier and quicker than the enzymatic digestion methods described in the literature, which have not generally been evaluated on clinical samples.
References
- Akalu A., Seidel W., Lieberman H., Bauer U., Dohner L. Rapid identification of subgenera of human adenovirus by serological and PCR assays. J. Virol. Methods. 1998;71:187–196. doi: 10.1016/s0166-0934(97)00213-9. [DOI] [PubMed] [Google Scholar]
- Allard A., Girones R., Juoto P., Wadell G. Polymerase chain reaction for the detection of adenoviruses in stools samples. J. Clin. Microbiol. 1990;28:2859–2867. doi: 10.1128/jcm.28.12.2659-2667.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allard A., Albinsson B., Wadell G. Rapid typing of human adenoviruses by a general PCR combined with restriction endonuclease analysis. J. Clin. Microbiol. 2001;39:498–505. doi: 10.1128/JCM.39.2.498-505.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkö M, Harrach B, Russell WC, Family Adenoviridae. In: Van Regenmortel MHV, Fauquet CM, Bishop DKL, Carstens EB, Estes MK, Lemon SM, et al., editors. Virus taxonomy. Seventh report of the International Committee on Taxonomy of Viruses. New York, NY: Academic Press Inc.; 1999. p. 227–38.
- Brouard J., Toutain F., Vabret A., Gouarin S., Freymuth F., Duhamel J.F. Etude clinique et virologique de 116 enfants immunocompétents hospitalisés pour une infection respiratoire adenovirale. Arch. Pediatr. 2001;8:538s. [Google Scholar]
- Elnifro E.M., Cooper R.J., Klapper P.E., Bailey A.S. PCR and restriction endonuclease analysis for rapid identification of human adenovirus subgenera. J. Clin. Microbiol. 2000;38:2055–2061. doi: 10.1128/jcm.38.6.2055-2061.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freymuth F., Vabret A., Galateau-Salle F., Ferey J., Eugène G., Petitjean J. Detection of respiratory syncytial virus, parainfluenza 3, adenovirus and rhinovirus sequences in respiratory tract of infants by polymerase chain reaction and hybridization. Clin. Diagn. Virol. 1997;8:31–40. doi: 10.1016/s0928-0197(97)00060-3. [DOI] [PubMed] [Google Scholar]
- Hierholzer J.C. Adenoviruses in the immunocompromised host. J. Clin. Microbiol. 1992;5:262–274. doi: 10.1128/cmr.5.3.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hierholzer J.C., Halonen P.E., Dahlen P.O., Bingham P.G., McDonough M.M. Detection of adenovirus in clinical specimens by polymerase chain reaction and liquid-phase hybridization quantitated by time-resolved fluorimetry. J. Clin. Microbiol. 1993;31:1886–1891. doi: 10.1128/jcm.31.7.1886-1891.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hierholzer JC, Adenoviruses. In: Lennette EH, Lennette AD, Lennette TE, editors. Diagnostic procedures for viral, rickettsial and chlamydial infections. 7th ed. Washington, DC: American Public Health Association Inc.; 1995. p. 169–88.
- Kidd A.H., Jonsson M., Garwicz D., Kajon A.E., Wermenbol A.G., Verweij M.W. Rapid subgenus identification of human adenovirus isolates by a general PCR. J. Clin. Microbiol. 1996;34:622–627. doi: 10.1128/jcm.34.3.622-627.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtomaki K., Julkunen I., Sandelin K., Salonen J., Virtanen M., Ranki M. Rapid diagnosis of respiratory adenovirus infections in young adult men. J. Clin. Microbiol. 1986;24:108–111. doi: 10.1128/jcm.24.1.108-111.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuse T., Matsui H., Shu C.Y., Nagase T., Wakabayashi T., Mori S. Adenovirus pulmonary infections identified by PCR and in situ hybridization on bone marrow transplant recipients. J. Clin. Pathol. 1994;47:973–977. doi: 10.1136/jcp.47.11.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris D.J., Cooper R.J., Barr T., Bailey A.S. Polymerase chain reaction for rapid diagnosis of respiratory adenovirus infection. J. Infect. 1996;32:113–117. doi: 10.1016/s0163-4453(96)91250-5. [DOI] [PubMed] [Google Scholar]
- Na B.K., Kim J.H., Shin G.C., Lee J.S., Kang C., Kim W.J. Detection and typing of respiratory adenoviruses in a single-tube multiplex polymerase chain reaction. J. Med. Virol. 2002;66:512–517. doi: 10.1002/jmv.2174. [DOI] [PubMed] [Google Scholar]
- Pring-Akerblom P., Trijssenaar F.E., Adrian T., Hoyer H. Multiplex polymerase chain reaction for subgenus-specific detection of human adenoviruses in clinical samples. J. Med. Virol. 1999;58:87–92. [PubMed] [Google Scholar]
- Raty R., Kleemola M., Melen K., Stenvik M., Julkunen I. Efficacy of PCR and other diagnostic methods for the detection of respiratory adenoviral infections. J. Med. Virol. 1999;59:66–72. [PubMed] [Google Scholar]
- Ruuskanen O, Meurmann O, Akusjarvi G, Adenoviruses. In: Richman DG, Whitley RJ, Hayden FG, editors. Clinical virology. 1st ed. New York: Churchill Livingstone; 1997. p. 525–47.
- Schmidtz H.R., Wigand R., Heinrich W. Worldwide epidemiology of human adenovirus infections. Am J. Epidemiol. 1983;117:455–466. doi: 10.1093/oxfordjournals.aje.a113563. [DOI] [PubMed] [Google Scholar]
- Takimoto T., Grandien M., Ishida M.A., Pereira M.S., Paiva T.M., Ishimaru T. Comparison of enzyme-linked immunosorbent assay, indirect immunofluorescence assay, and virus isolation for detection of respiratory viruses in nasopharyngeal secretions. J. Clin. Microbiol. 1991;29:470–474. doi: 10.1128/jcm.29.3.470-474.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadell G., Hammarskjold M.L., Winberg G., Varsanyi T.W., Sundell G. The genetic variability of adenoviruses. Ann N. Y. Acad. Sci. 1980;354:16–42. doi: 10.1111/j.1749-6632.1980.tb27955.x. [DOI] [PubMed] [Google Scholar]
- Xu W., Mc Donough M.C., Erdman D.D. Species-specific identification of human adenoviruses by a multiplex PCR assay. J. Clin. Microbiol. 2000;38:4114–4120. doi: 10.1128/jcm.38.11.4114-4120.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]