Paediatric infections
P1231: Acute bacterial conjuctivitis in children
A. Makri, X. Agathokleous, H. Papavasileiou, F. Lykou, N. Manios, A. Voyatzi (Athens, GR)
Introduction: Acute bacterial conjunctivitis (ABC) is one of the most commonly ocular diseases in children examined by pediatricians at the outpatients Department. Since the treatment is usually empirical and prior cultures are not normally taken, the virulent factors involved in the process are often unidentified.
Objectives: 1. To determine the incidence of the isolated pathogens from conjunctival exudates cultures and 2. To examine the antibiotic susceptibility for the efficacy of the therapy in the management of confirmed ABC in children.
Methods: Conjunctival swabs were collected from 780 children, aged 1 month to 14 years old, who were attended with the diagnosis of ABC in our Children Hospital, during a 5-years period (2000–2004). All bacteria were identified by classical microbiological methods and the antimicrobial susceptibility was performed according to NCCLS guidelines.
Results: 453 cultures out of 780 ocular specimens were positive(58%). A total of 501 bacteria were isolated. The most frequent isolated bacteria were: Haemophilus influenzae (37.5%), Streptococcus pneumoniae (20%), Staphylococcus coagulase negative (CNS) (15.2%), and Staphylococcus aureus (14.8%) followed by Streptococcus viridans (4%), various Gram negative bacteria (3%), Moraxella catarrhalis (2.5%) and Haemophilus spp (1.6%).42 patients developed mixed cultures with two or more types of bacteria. Among 99 isolates of S. pneumoniae tested, were resistant for penicillin 14.1% and for gentamicin (Gm) and tobramycin (Tob) 45.4% and 49.5% respectively. Resistance to ampicillin was recovered in 9.5% of H. influenzae, of 66.6% of S. aureus and in 79.5% of CNS strains. Aminoglycosides showed higher resistance (Gm 17.9%, Tob 25.6%) against CNS isolates, in comparison to S. aureus (Gm 12%, Tob 14.6%) and H. influenzae strains (Gm 1.6%, Tob 2.6%) respectively. The most isolates were sensitive to chloramphenicol and ciprofloxacin.
Conclusions: 1. The main pathogens causing ABC were H. influenzae and S. pneumoniae. 2. Chloramphenicol and ciprofloxacin showed favorable in vitro activity against the majority of pathogens. 3. Resistance rates indicate the need for continuous surveillance and for motoring studies.
P1232: Pertussis of adults and infants in Bulgarian population: role of PCR diagnosis
S. Panaiotov, V. Levterova, I. Ivanov, T. Kantardjiev, N. Vladimirova, V. Voinova, E. Kazarova (Sofia, BG)
Pertussis continues to be an important vaccine preventable disease. There is a long lasting tradition in Bulgaria in pertussis prevention. More than 40 years a whole-cell pertussis vaccine is administered to infants and a high vaccine coverage is achieved and maintained. Pertussis morbidity decreased dramatically but pertussis continues to occur among different ages. Making a specific diagnosis of pertussis in patients with clinical evidence of infection is one of the many challenges presented by Bordetella pertussis. For more than 20 years, pertussis diagnosis in Bulgaria routinely is done using two classic lab methods: the direct fluorescent antibody test and the pertussis-agglutinating antibody test. Recently ELISA method is introduced too but only for research purposes.
Objectives: 1. To describe the PCR diagnosis of B. pertussis in our country and to present first results of this assay in pertussis suspect patients
2. To compare results between PCR diagnosis and direct fluorescent antibody test in the same patients.
3. To assess the role of Mycoplasma pneumoniae and Chlamidia pneumoniae as causatives of infants' respiratory infections mis-diagnosed by the general practitioners as pertussis infection.
Results: 334 nasopharyngeal swabs were collected from infants with pertussis symptoms and contact adults. All samples were tested by PCR. PCR was performed for insertion element IS481 of B. pertussis. 69 samples out of them were found positive for B. pertussis by PCR. Two were positive for M. pneumoniae by PCR.100 samples were tested comparatively by two methods :PCR and direct fluorescent antibody test. Vaccination status of patients and clinical data were analysed too.
| Method | Age-group 0-3 | Age-group 3–14 | Age-group >14 years | Total |
|---|---|---|---|---|
| PCR positive | 34 | 30 | 5 | 69 |
| PCR negative | 118 | 110 | 47 | 275 |
Conclusions: First results of B. pertussis PCR assay are presented. Pertussis continues to circulate in Bulgaria even the achieved high vaccine coverage of infants and children. PCR is rapid and simplifies the laboratory diagnosis of pertussis. Further serological, immunofluorescence or culture comparative studies are needed with the aim to better evaluate the role of PCR diagnosis.
P1233: Molecular epidemiology of respiratory syncytial virus infection in a university hospital in Kuala Lumpur, Malaysia
Z. Sekawi, G. Vinomarlini, N.S. Mariana, D.S. Shamala, K. Thilakavathy (Kuala Lumpur, MYS)
Respiratory syncytial virus (RSV) is an important cause of acute lower respiratory infections in babies and young children worldwide. There are two major distinct RSV groups, RSV Group A and RSV Group B. To add to its complexity, additional genetic variability occurs within the groups. RSV infection is endemic in Malaysia with peak pattern reported to be related to the rainy season which is usually at the end of the year. However, the molecular epidemiology of RSV in Malaysia is unknown.
Objective: To determine the molecular epidemiology of RSV infection in a University Hospital, Kuala Lumpur, Malaysia.
Method: Paediatric patients who were hospitalized with acute lower respiratory infections from September 2002 to March 2004 were screened for RSV infection. Nasopharyngeal aspirate (NPA) samples were collected and subjected to direct immunofluorescence (DIF) assay and isolation by tissue culture. The total viral RNA was extracted and subjected to reverse transcriptase – polymerase chain reaction (RT-PCR). Seminested PCR was done by using specific primers for each RSV group. Random Amplification of Polymorphic DNA (RAPD) technique was performed as a simple method to observe the genetic diversity of the isolated RSV.
Results: Thirty NPA samples were positive for RSV using the DIF assay but only 20 samples were positive by tissue culture. The peak months are from the months of October to January for both years. Seminested PCR on RSV isolates revealed 17 cases of RSV Group A while the remaining three cases, RSV Group B. RAPD result suggested that there may be five main subgroups during the study period.
Conclusion: This study confirmed RSV Group A as the more dominant group and may be more virulent in the Malaysian context. This is similar to most studies in other parts of the world. Intragroup variation occurs throughout the duration of the study. This variation may have a considerable effect of the efficiency of transmission and virulence. Restriction analysis study will be done in order to compare the isolates objectively with other studies. These findings can be used as part of the globally initiative towards vaccine development.
P1234: Respiratory syncytial virus infections in Croatia, 1994–99
A. Lukic-Grlic, G. Mlinaric-Galinovic, V. Drazenovic, A. Barisin, A. Bace, V. Hresic-Krsulovic, R. Sim, B. Berberovic, E. Berberovic (Zagreb, HR)
Objective: To determine epidemiological characteristics, i.e. the occurrence of respiratory syncytial virus (RSV) infection in Croatian children with acute respiratory tract infections.
Methods: At Virology Department, Croatian National Institute of Public Health (CNIPH), we tested nasopharyngeal secretions obtained from 1232 patients most of whom were hospitalized in two Zagreb hospitals for acute respiratory infections. Demonstration of the virus was by isolating it in cell culture and/or by detecting it with monoclonal antibodies in the direct immunofluorescence assay.
Results: Most often, the virus demonstrated was RSV (43.8%; 540/1232). Other respiratory viruses (adeno, parainfluenza, influenza) were shown considerably less commonly (5.1%). Viral infection could not be demonstrated in 629 (51.1%) patients. As to bronchiolitis, RSV was demonstrated to be its most common cause (60.77%; 251/413). It was also proven to be the most common causative agent of infections in children aged 0–6 months (55.6%; 300/540). Bronchiolitis (63%; 190/300) and, less commonly, pneumonia (9.7%; 29/300) were the diagnoses linked with RSV in this age group. On the other hand, RSV was demonstrated in 21% (63/300) of the children diagnosed with upper respiratory tract infection (URTI). We showed the presence of the majority of RSV infections in winter months, i.e. between November and June.
Conclusion: RSV is a common cause of lower respiratory tract infections in Croatian infants and young children with its annual outbreaks occurring in winter season. Their onset is mostly in November.
P1235: Human metapneumovirus infection among children hospitalised with acute respiratory illness in Taiwan
C.Y. Lin, J.S. Lin (Changhua, TW)
Objectives: To understand the association of human metapneumovirus (hMPV) with acute respiratory infection (RI) among hospitalized children in Taiwan, we conducted this study.
Methods: From February to August 2004, consecutive nasopharyngeal aspirates (NPAs) collected from children hospitalized with acute RI and submitted to a virology laboratory for rapid detection of respiratory syncytial virus (RSV) antigens were retrospectively analysed for hMPV. After extraction of nucleic acids from these frozen NPAs, RSV was first detected by amplifying RSV-specific fusion (F) and nucleocapsid (N) genes, respectively, using reverse transcription-polymerase chain reaction (RT-PCR). The same extracts were subsequently detected for hMPV by amplifying hMPV-specific F and N genes, respectively, using nested RT-PCR. If multiple samples during the same course of illness were submitted, either one of the virus-positive samples (first choice) or one of the virus-negative samples (second choice) was included for analysis to decrease a statistical bias.
Results: 667 NPAs collected from 623 hospitalized children with acute RI were examined, however, only 580 NPAs of them had been successfully detected for both RSV and hMPV by RT-PCR. The age of patients ranged from 0 to 9 years. A positive rate of RSV or hMPV in NPAs was 39.1% (n = 227) and 8.6 (n = 50) respectively. In 353 NPAs negative for RSV, hMPV was found in 13.3% (n = 47) of samples. Only 2 NPAs (0.3%) were positive for both viruses. The infection rates of hMPV were significantly related to the month of recruitment (p = 0.001, Pearson Chi-squared test), with peak rates of 17.0% and 16.9% in May and June, respectively. According to the clinical diagnosis, 486 samples (83.8%) with pneumonia, bronchiolitis, bronchitis or brochopneumonia were arbitrarily defined to have a lower RI, otherwise, upper RI was recognized (16.2%, n = 94). Among NPAs negative for RSV, lower RI was significantly associated with hMPV when compared with upper RI (p = 0.034, Fisher's exact test).
Conclusions: This preliminary study discloses that hMPV plays a role in Taiwanese children with acute RI with a comparable rate of infection as reported in western countries. hMPV infection in children may have seasonal preference as shown in this study, though a longer period of evaluation remains necessary. The rare co-infection of RSV and hMPV in children with acute RI implies the importance of hMPV detection especially in NPAs negative for RSV.
P1236: Acute CMV infection in paediatric population
P. Koudounis, M. Pagkalou, G. Triantafillou, P. Rozi (Serres, GR)
Objectives: The aim of this study is the laboratory confirmation of acute CMV infection in children who have been hospitalized or have attended the Outpatient's Department with signs and symptoms that suggest recent CMV infection.
Methods: Blood samples were taken from 66 children (40 boys and 26 girls), aged 9 months to 13 years, with symptoms suggesting recent CMV infection, from July 2002 to December 2003. They were tested for detection of CMV antibodies (IgM and IgG) by Microparticle Enzyme Immunoassay (AXSYM, ABBOTT) Enzyme linked Fluorescent Assay was used as a method of confirmation of the IgM positive tests. Also, the test of IgG-avidity was used as a supplementary means for the exclusion of a recent primary infection of less than 3 months (Vidas, bioMerieux).
Results: Out of the total of 66 children examined, the 17 (group I) were found to be positive both for IgM and IgG antibodies, 3 (group II) were found to be positive only for IgM antibodies and 24 (group III) were found to be positive only for IgG antibodies. 22 children haven't been exposed to the disease (IgM-/IgG-). In group I children, with the use of the CMV-avidity test, the acute infection was excluded in 8 out of 17 children, a fact that was also confirmed with the second antibody detection test. In group II, 2 in 3 children were excluded in the same way, while in the other child acute infection was confirmed in the tests of confirmation. In group III, old infection was found with the CMV avidity test.
Conclusion: Those results give us indicative information about the incidence of CMV infections in our hospital in the chronological period that is referred above. This study showed that the use of confirmation tests are necessary in several cases, especially those with unclear clinical and laboratory findings.
P1237: The patterns of nasopharyngeal microflora in pre-school children with recurrent respiratory tract infections
U. Kosikowska, I. Korona-Glowniak, R. Los, A. Biernasiuk, A. Malm (Lublin, PL)
Objectives: Respiratory tract infections, predominantly of viral etiology, are the most common, community-acquired infections in young children. Some of them are complicated by bacterial infections usually of endogenous origin. The mucous membranes of nasopharynx are known to be the important reservoir of some opportunistic or potentially pathogenic bacteria. The aim of the present study was to compare the nasopharyngeal microflora in two groups of pre-school children – without or with the recurrent respiratory tract infections.
Methods: Nasal and throat specimens were obtained from 225 children aged 3–5 years. The cotton swabs were immediately placed onto appropriate nonselective (blood agar) or selective media (Haemophilus chocolate agar, Chapman agar, McConkey agar or Sabouraud agar). Plates were incubated in an appropriate atmosphere – with or without increased CO2 concentration) for 18–48 hrs at 35°C. The isolated microorganisms were identified on the basis of routinely methods (macroscopic, microscopic or biochemical assays) or by rapid commercial latex tests – Slidex Staph-Kit and Slidex Pneumo-Kit (bioMerieux).
Results: The prevalence of potentially pathogenic bacteria typical for nasopharynx such as Staphylococcus aureus, Streptococcus pneumoniae, Moraxella catarrhalis or Haemophilus influenzae and also of yeast-like fungi (Candida sp., mostly C. albicans) was similar in both groups of children. However, in nasopharynx of children with the recurrent respiratory tract infections several opportunistic bacteria belonging to Enterobacteriaceae (Escherichia coli, Citrobacter freundii) or non-fermentative rods (Pseudomonas putida, Agrobacterium radiobacter, Acinetobacter lwoffii) were found. Also other bacteria such as slime-producing Bacillus sp. or streptomycetes were isolated from this group of children.
Conclusion: The obtained data suggest that young children with the recurrent respiratory tract infections are predisposed for nasopharynx colonization by several opportunistic bacterial species of Gram-negative rods or even by some unusual microorganisms, e.g. slime-producing Bacillus sp. or streptomycetes.
P1238: Correlation of S. pneumoniae and H. influenzae antimicrobial resistance and history of antimicrobial use or vaccination in children with upper respiratory tract infections – Brazil 2002 to 2004
C.M. Mendes, C. Oplustil, J.L. Sampaio, P. Garbes, P. Lima, L. Pedneault, C.R. Kiffer (Sao Paulo, Rio de Janeiro, BR; Brussels, B)
Objective: Establish a correlation between antimicrobial resistance in S. pneumoniae and H. influenzae, isolated from children with upper respiratory tract infection (URTI) in São Paulo, Brazil, and their history of antimicrobial use and/or vaccination.
Methods: Samples (one per patient) were selected from patients under 7 years old between 2002 and 2004. All subjects were diagnosed with URTI and had a positive culture result for at least one of the selected pathogens (S. pneumoniae and H. influenzae). Clinical data related to age, gender, diagnosis and samples are described. S. pneumoniae isolates were tested against penicillin and another five antimicrobials for minimum inhibitory concentrations (MICs) by E-test methodology. Interpretative criteria used were those described by NCCLS document M100-S14. H. influenzae isolates were tested for betalactamase production by a chromogenic cephalosporin method. Logistic regression was performed to investigate the correlation between S. pneumoniae (intermediate and high) penicillin resistance and previous antimicrobial use and pneumococcal vaccination. The same procedure was done for beta-lactamase-positive H. influenzae and previous antimicrobial use and vaccination against H. influenzae type b (Hib).
Results: Of patients with S. pneumoniae isolation, 62 had information on previous antibiotic use and 53 on pneumococcal vaccination. Of patients with H. influenzae isolation, 121 had information on previous antibiotic use and 101 on vaccination to H. influenzae. There was no correlation between presence of penicillin-resistant S. pneumoniae and previous antibiotic use (p = 0.16; OR = 2.28; IC95% = 0.73–7.16) or vaccination against pneumococci (p = 0.61; OR = 0.72; IC95% = 0.20–2.54). Similarly, no correlation was found between presence of betalactamase-producing H. influenzae and previous antibiotic use (p = 0.71; OR = 0.78; IC95% = 0.21–2.96) or Hib vaccination (p = 0.78; OR = 0.72; IC95% = 0.08–6.72).
Conclusions: No correlation was found between antibiotic use and respective vaccinations against S. pneumoniae and Hib and their related resistance characteristics, penicillin resistance and beta-lactamase production respectively. However, among possible bias influencing the results, vaccination against both pathogens is still low in the study population and it was not possible to compare resistance and previous antimicrobial use by drug class.
P1239: Staphylococcus aureus small colony variants in patients with cystic fibrosis in Franfurt/Main, Germany
S. Besier, A. Ludwig, C. von Mallinckrodt, H.-G. Posselt, T.A. Wichelhaus (Frankfurt/ Main, D)
Objectives: In spite of intense and modern antibiotic treatment, mortality and morbidity of patients with cystic fibrosis (CF) are dominated by chronic bronchopulmonary infections. Staphylococcus aureus is one of the most important pathogens isolated from the chronically infected airways of CF patients. Interestingly, the persistence of S. aureus in the bronchial system is associated with the isolation of a subpopulation of S. aureus -- the small colony variants (SCVs). In contrast to normal S. aureus, SCVs are often affected in their electron transport activity or thymidine synthesis, thus growing as nonhemolytic, nonpigmented small colonies exhibiting auxotrophism for distinct growth factors.
Methods: Sputa and deep throat swabs of CF patients attending the ambulance of the university hospital in Frankfurt/Main, Germany, between January 2004 and May 2004 were screened for the prevalence of S. aureus SCVs.
Results: Of 113 patients, 34 (30.1%) harboured S. aureus in their respiratory specimens. Of these 34, 23 (67.6%) had normal S. aureus (NCVs) and 11 (32.4%) had NCVs plus SCVs or SCVs alone. The median age of patients with SCVs differed distinctly from the median age of patients with only NCVs [28.6 years (range, 2–45) vs. 19 years (range, 1–39)]. Persistence of the SCVs could be demonstrated for all SCV-positive patients with subsequent microbiological investigation during the study period. An antibiotic prophylaxis with trimethoprim-sulfamethoxazol could be elicited for 7 (63.6%) of the 11 SCV-positive patients. All SCV isolates were resistant to trimethoprimsulfamethoxazol and high resistance rates were also documented for ciprofloxacin (53.8%) and gentamicin (46.2%). Analysis of the underlying auxotrophism revealed a predominance of thymidine dependence in 69.2% of the SCV isolates. Most of the SCVs showed the known fried-egg or pinpoint morphology, while 4 strains exhibited a mucous phenotype not previously observed.
Conclusions: The presented data show that S. aureus SCVs can be isolated frequently and repeatedly from the airways of CF patients. Further investigations are required to illuminate the genetic background and pathogenic role of this S. aureus phenotype in persistent infections.
P1240: Epidemiological features of Stenotrophomonas maltophilia isolation from the sputum of children with cystic fibrosis
H. Alexandrou-Athanasoulis, S. Doudounakis, A. Sergounioti, I. Loucou, A. Pangalis (Athens, GR)
Stenotrophomonas maltophilia (Sm) has been isolated from the airway secretions of children with Cystic Fibrosis (CF) with increasing prevalence last years. The aim of our study is to determine trends in prevalence and in resistance to the 1st choice antibiotics for these patients.
Material and method: We reviewed retrospectively the clinical, demographic and laboratory data of the 388 patients who have been attended our hospital, between 01/2000 and 11/2004 for at least one year. During this period 5010 sputum or deep throat specimens were cultured. The identification of the isolates was made by the API 20NE identification system (Biomerieux, France) and the antibiotic resistance was determined with the E-test strips (AB Biodisk, Sweden).
Results: (1) Sm has been harboured by 62 (16.0 %) of our patients (38 were female and 24 male). Chronically infected remained 11.3 % of them, while 59.7 % had only one positive culture for Sm. (2) The mean age of the 1st isolation of Sm was 8.3 ± 4.9 years. (3) The yearly incidence rate of Sm acquisition and the yearly prevalence were 2.8–3.6% and 4.7–9.2% respectively. (4) The sensitivity to ticarcillin/clavulanic acid as well as to co-trimoxazole were 59.3% and 62.6% respectively.
Conclusions: (1)Yearly prevalence and incidence rates for Sm in CF patients showed no trends. (2)the resistance to ticarcillin/ clavulanic acid is gradually increasing.
P1241: Subclonal variation of a cystic fibrosis epidemic strain of Pseudomonas aeruginosa
S. Panagea, J.E. Corkill, C. Winstanley, A.C. Hart (Liverpool, UK)
Objectives: A highly transmissible, epidemic strain of Pseudomonas aeruginosa is widespread among cystic fibrosis patients attending clinics in Liverpool, United Kingdom. We studied the emergence of clonal variants of this Liverpool Epidemic strain (LES) over time in chronically infected patients.
Methods: Forty LES isolates from seven patients were typed by macrorestriction analysis of SpeI and XbaI-digested DNA by pulsed field gel electrophoresis (PFGE). A total of 4 to 8 isolates from each patient were studied, collected at three different times during their colonisation (1995–2003). Macrorestriction fragment patterns were compared with the first LES in our collection isolated in 1988 and clonal variants had 1 to 3 band differences from this control strain. All isolates were tested positive with a PCR assay specific for LES.
Results: PFGE analysis of SpeI and XbaI digests revealed 9 and 7 clonal variants of LES respectively. There was overall good correlation between SpeI and XbaI fragment patterns, however digestion with SpeI was more discriminatory in this study. All patients had clonal variants of LES (range 2–4). The variant with identical fragment pattern to the 1988 isolate was the most common and was isolated from all but one patient at least once. Two other variants were also common and were detected in three or more patients. Emergence of clonal variants in individual patients appeared to occur at random and did not persist throughout the study period in most of them. There was no correlation between clonal variants and phenotype or antibiotic sensitivity pattern of the isolates.
Conclusion: Clonal variants of LES are common and were detected in all the patients. This study confirms that genomic diversity and evolution of a clonal lineage as indicated by subtle band shifts of the macrorestriction fragment pattern is common in chronically infected cystic fibrosis patients.
P1242: In vitro effect of subMICs of antibiotics on the antigenic structure of Pseudomonas aeruginosa strains isolated from patients with cystic fibrosis
J. Vranes, B. Bedenic, D. Tjesic-Drinkovic, N. Jarza-Davila (Zagreb, HR)
Objectives: Pseudomonas aeruginosa is the leading pathogen responsible for morbidity and mortality in patients with cystic fibrosis (CF), and is the primary reason for the reduced survival age. Different molecules are responsible for P. aeruginosa virulence, including lipopolysaccharide (LPS) as a cellular component. Suppression or modulation of P. aeruginosa virulence factors may be an alternative strategy for treatment of lung infections in CF. The aim of this study was to examine the influence of subminimal inhibitory concentrations (subMICs) of ceftazidime and ciprofloxacin on the LPS structure of P. aeruginosa strains isolated from patients with CF.
Methods: A total of 20 strains isolated from sputa of patients with CF were used. Serotyping based on the detection of heat stable LPS bacterial surface antigens (O-antigens) was performed as slide agglutination tests using O-specific polyclonal sera and a suspension of live bacterial isolate to be examined, according to the International Antigenic Typing System. Serotyping was performed before and after exposure of strains to 1/ 2, 1/4, 1/8, 1/16, and 1/32 MIC of antibiotics.
Results: The slide agglutination test with polyclonal diagnostic sera showed that only nine strains were typable, while six strains were polyagglutinable, three strains were non-typable, and two strains were auto-agglutinable. After the exposure of strains in exponential phase of their growth to subMICs of antibiotics, a significant difference in typeability of the strains were observed. Treatment with subMICs of ceftazidime and ciprofloxacin resulted in the alteration of polysaccharide structure of typable strains. Out of nine monoagglutinable strains, seven strains have become auto-agglutinable after exposure to 1/2, 1/4, 1/8 and 1/16 MIC of ceftazidime, while two strains have become polyagglutinable, suggesting the loss of most of the O-antigenic determinants of the LPS. The polyagglutinability was observed more frequently after exposure of those strains to 1/2, 1/4 and 1/8 MIC of ciprofloxacin (five out of nine strains). The non-typable strains have become auto-agglutinable or polyagglutinable, depending on the concentration of antibiotic.
Conclusion: The results of this study have shown that the antigenic structure of P. aeruginosa strains isolated from sputa of patients with CF was significantly changed after in vitro exposure to subMICs of ceftazidime and ciprofloxacin, due to the loss of O-antigenic determinants.
P1243: Dynamics of antibody response to Pneumocystis jirovecii in patients with cystic fibrosis
N. Respaldiza, J. Dapena, M. Montes-Cano, C. de la Horra, I. Mateos, V. Friaza, I. Wichmann, J.M. Varela, E. Calderon, F.J. Medrano (Seville, E)
Objectives: The asymptomatic colonisation by Pneumocystis jirovecii (Pc) has been recently demonstrated in immunocompetent patients, mainly in subjects with different pulmonary diseases. Cross-sectional studies have shown that healthy adults have frequently serum antibodies to Pneumocystis, and the primary contact usually occurs at an early age. However, the dynamic of the humoral immune response against this pathogen still remains unknown. The objective of this study was to evaluate the dynamic of antibody response to P. jirovecii in patients with cystic fibrosis (CF).
Methods: (1) Population: Cohort study in which were included a total of 75 consecutive patients with CF, attended in a specialized unit between May 2001 and July 2002. Patients were followed every six months during one year. At each visit respiratory and serum samples were collected for Pc evaluation. (2)Methods: Nested PCR was performed in respiratory samples to detect the presence of Pc colonization. Serum specimens were assayed for total IgA/IgM/IgG antibodies to Pneumocystis antigens by western blot (WB). A commercial monoclonal antibody (MoAb) to a human Pc component (DAKO, Glostrup, Denmark) was used as a positive control. The presence of an immunoreactive band of 120 kDa (molecular weight of MoAb) was interpreted as a positive result.
Results: The mean age of the CF patients was 16.4 ± 6.7 years (range: 1–35), 31 (41%) of whom were male. Pc carriage was found in 14/75 (18.7%) of the CF patients at baseline. During the follow-up Pc colonisation was observed in 36 (48%) of the 61 initially PCR-negative subjects. WB was positive at baseline in 37/75 (49%) of the patients with CF. During the follow-up serologic dynamic was as follows: among the 37 initially WB-positive subjects 32 (86%) remained positive, but in 5 (14%) cases a seroreversion was observed (WB became negative); among the 38 initially WB-negative subjects a seroconversion was observed in 20 (53%) cases and the remaining 18 (47%) were persistently negative.
Conclusions: There is a high rate of Pneumocystis exposition in patients with cystic fibrosis. Contrary to previous belief, our results show for the first time that serologic response to P. jirovecii is not permanent, and that the presence of specific antibodies could be a result of repeated exposure to the pathogen. This study was supported by the Grants FIS 03/1743 and Consejería de Salud 32/02 and by Thematic Network for Joint Research (G03/90.
P1244: Nosocomial infections in paediatric patients
M. Renko, S. Kinnula, M. Uhari (Oulu, FIN)
Background and aim: We evaluated the incidence of nosocomial infections occurring within 14 days after care at the paediatric infection ward of Oulu University Hospital between VI/2001 and V/2003.
Methods: At discharge from the hospital data on clinical diagnosis, aetiology of the infection, number of patients in the same room and whether or not the child got a nosocomial infection during the hospital stay were registered. Two weeks after the child was discharged the parents filled in a questionnaire concerning whether the child became ill after the discharge.
Results: We have data from 1922 patients. Most of the patients (90%) were treated in a single patient room. The most common diagnoses were obstructive bronchitis in 437 (23%) and gastroenteritis in 566 (29%) cases. Twenty-three out of 1922 (1.3%) got a nosocomial infection during their stay at the ward, most of them gastroenteritis. Parents of 1136 patients (59%) returned the questionnaires. 313 children out of 1136 (28%) were taken ill after discharge, 88 (8%) of these within 72 hours. Cough was the most common symptom on those taken ill 4–14 days after discharge (53%), and diarrhoea on those taken ill within the first 72 hours (49%) (Figure). The children who got nosocomial infections were younger than the other patients (mean 2.3 years, vs. others 3.0, P = 0.03) and the duration of the hospital care was longer in them (mean 3.6 vs 2.9 days, P = 0.005).
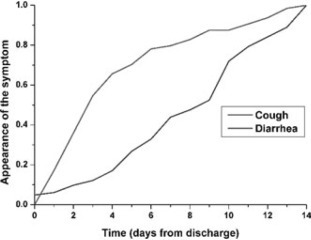
Conclusions: Due to the short hospital care only a few children were reported to get a nosocomial infection during their stay. Yet, 8% of the children discharged from the ward were taken ill within 72 hours. Viruses causing gastroenteritis seem to be more difficult to prevent from spreading than respiratory viruses.
P1245: Procalcitonin controlled administration of IgM-enriched immunoglobulins in early post-op can prevent postoperative infectious complications in high-risk children with congenital heart diseases
N.V. Beloborodova, D.A. Popov, A.I. Melko (Moscow, RUS)
Objectives: To estimate the effectiveness of procalcitonin (PCT) controlled administration of IgM-enriched immunoglobulins in high-risk children with congenital heart diseases after cardiac surgery with cardiopulmonary bypass (CPB).
Methods: Thirty-one consecutive patients (mean age 25 ± 8 months) were studied. Patients who had PCT blood plasma levels above 2 ng/ml on the 1st day after surgery (n = 28) were randomized into two groups. IgM-enriched immunoglobulin preparation (Pentaglobin, Biotest Pharma GmbH, Germany) was administrated to the patients in the study group (n = 15) in addition to standard treatment (1–3 days after surgery, 5 ml/kg each day). Patients in the control group (n = 13) received only standard treatment. Patients in both groups were comparable by severity of initial condition, age and CPB time. PCT blood plasma concentrations were measured on the 1st, 2nd, 3rd and 6th days after surgery by immunoluminometric method (PCT LIA, B.R.A.H.M.S Aktiengesellschaft GmbH, Germany). Post-op infections rates were analysed in these groups. The data were compared by Mann–Whitney U-test and p-value of (0.05 was considered statistically significant.
Results: None of the patients had exhibited any signs of infection before surgery. Patients with PCT blood plasma levels less than 2 ng/ml on the 1st day after surgery (n = 3) had smooth post-op period. The rate of post-op infectious complications was significantly lower in the study group (1/15 (6.7%) vs. 5/13 (38.5%), p = 0.03). Two deaths in the control group occurred due to sepsis and (n = 1) and peritonitis (n = 1). Postoperative PCT levels were significantly higher in the control group during all observation period (see the table). The data are expressed as median (ng/ml) and 25th and 75th percentiles.
| Group | PCT levels, ng/ml |
|||
|---|---|---|---|---|
| 1 day | 2 day | 3 day | 6 day | |
| Control (n=13) | 7.77 (5.95-10.72) | 4.24 (3.37-5.21) | 2.12 (1.67-3.50) | 0.52 (0.45-0.59) |
| Study (n=15) | 3.60 (2.98-6.54) | 1.91 (1.32-3.90) | 0.96 (0.72-1.99) | 0.41 (0.32-0.46) |
| p | 0.009 | 0.01 | 0.02 | 0.045 |
Conclusions: High PCT levels on the 1st day after surgery are associated with infectious complications. PCT monitoring allows to select patients with systemic bacterial inflammation after CPB. Early post-op administration of IgM-enriched immunoglobulins effectively prevents infectious complications in these patients after cardiac surgery.
P1246: Clinical characteristics of rotavirus, enteric adenovirus and astrovirus infections among hospitalised children in Hungary
F. Jakab, L. Varga, Z. Nyul, D. Mitchell, J. Walter, T. Berke, D. Matson, G. Szucs (Pecs, HUN; Norfolk, USA)
Objectives: The aim of this study was to determine the prevalence, severity and clinical characteristics of viral gastroenteritis caused by astrovirus (HAstV), rotavirus (RV) or enteric adenovirus (EAd) resulting in hospitalization among children in Baranya County, Hungary.
Methods: Stool specimens were collected between May 2003 and May 2004 from children hospitalized for gastroenteritis. Samples were tested for HAstV, RV, and EAd. HAstVs were detected by reverse transcriptase-polymerase chain reaction while RV and EAd were identified by latex agglutination test. Demographic and clinical data were collected for all enrolled children. Clinical symptoms and severity of illness were determined.
Results: During this 1-year period, 227 children hospitalized for acute gastroenteritis were enrolled. The mean age was 37 months (range: 21 days to 213 months). HAstV, RV or EAd were detected in 51% of enrolled children. RV was detected in 94 (41%), EAd in 13 (6%) and HAstV in 12 (5%) from the collected stool samples. The most common clinical presentation of RV infected children were the combination of diarrhoea, vomiting and fever (34%), while in children with HAstV or EAd infection diarrhoea alone was more characteristic (50% and 62%, respectively). Children infected with either virus had diarrhoea lasting 1–4 days with 1–3 episodes per day, vomiting episodes lasted 1–2 days and fever ≥38.0°C. Based on the 20-point Vesikari severity score system; 58% of RV infected children had moderate or severe infection (score: ≥8) while 77% of children with EAd or 67% with HAstV had rather mild disease (score: ≤ 7); (p < 0.05).
Conclusion: Clinical presentations of gastroenteritis by RV, HAstV or EAd are similar with diarrhoea the most common manifestation followed by vomiting and/or fever. RV infected patients had a more severe illness than those children infected with EAd or HAstV. Based on the epidemiological and clinical data we can conclude that these enteric viruses are a significant burden of disease based upon hospitalizations for childhood gastroenteritis in Baranya County, Hungary.
P1247: Investigation of the modulatory effect of pre- and probiotics on the gastrointestinal flora in post surgical neonates
N. Mackay, C. Lucas, G. Mackay, C. Davies, C. Wiliams (Glasgow, UK)
Background: The neonatal gut normally becomes colonised with a variety of bacterial species. However, surgical intervention, nutritional deprivation and antibiotic therapy devastate the normal gastro-intestinal (GI) ecosystem and may contribute to the risk of bacterial translocation (BT). Intervention using probiotic bacteria or prebiotic carbohydrates has been proposed to reduce this problem but few studies have looked at the ability of probiotic bacteria to colonise the abnormal gut.
Objectives: To demonstrate the persistence of probiotic bacteria demonstrate a biological effect using short chain fatty acid profiles.
Methods: Conventional culture of stools and stoma fluid was performed on 5% Horse blood, neomycin blood agar and MacConkey agar Extended culture was performed using Rogosa agar for Lactobacillus spp. and Beeren's Agar for Bifidobacterium spp. Short Chain Fatty Acid (SCFA) Analysis was performed by Gas Liquid Chromatography Chromatography and Isolates of lactobacilli were genotyped using. Random Amplification of Polymorphic DNA (RAPD) using primer 272 a eubacterial primer followed by gel electrophoresis.
Results: Routine bacterial culture on 320 specimens yielded Coagulase Negative Staphylococci (CNS), Enterococci and coliforms with Gram positive bacilli being isolated from only 7 specimens. On extended culture of 170 specimens Gram positive bacilli were isolated on 39 occasions. Short Chain Fatty Acid (SCFA) analysis showed that no patient samples demonstrated a SCFA profile similar to a healthy breastfed infants. In one patient studied longitudinally a change of diet along with administration of a probiotic resulted in a change in the fatty acid profile from being predominantly octanoic acid to proprionic acid. RAPD analysis showed that a lactobacillus indistinguishable from the probiotic strain could be isolated from the faeces of one patient throughout the period of probiotic administration.
Conclusions: This pilot study confirms the potential for SCFA analysis to detect changes as a result of pre and probiotic dietary supplements and the RAPD method used successfully discriminated between the Lactobacillus spp investigated. Further work needs to be done in this area in particular the on resolution of RAPD and whether other DNA-based methodologies e.g. FISH may further improve the detection of specific probiotic strains and allow greater understanding of the effects detected by SCFA analysis.
P1248: Children's urinary pathogens: one-year survey in a Greek hospital
L. Zachariadou, M. Chondrogianni, M. Nikolaou, M. Liakata, E. Petridou, A. Pangalis (Athens, GR)
Objectives: To present the frequency of pathogens of urinary tract infections in children and the possible differentiation in isolation rates of species among hospitalized and non hospitalized patients.
Methods: 12819 urine specimens were examined from August 2003 to July 2004. 7583 samples were taken from hospitalized children and 5236 from non hospitalized ones either visiting the emergency unit or on a monthly follow up due to urinary tract congenital anatomic or functional abnormalities. The percentage of boys and girls was 50.3 and 49.7 respectively. Samples were collected by suprapubic aspiration, urethral catheterization, clean-catch midstream or ‘bagged’ specimen, depending on the age of the little patients. The diagnosis of urinary tract infection was based on precise clinical and laboratory criteria according to the European guidelines and in case of doubt the culture was repeated.
Results: 1062(8.3%) cultures were positive for one causative pathogen and only 11(0.08%) for two. As expected, the dominant pathogen in all isolates was E. coli (73.6%), followed by K. pneumoniae (6.8%), P. mirabilis (6.7%), P. aeruginosa (3.5%) and Enterococcus spp (3.3%). The remaining 6.1% of isolates belonged to C. albicans, E. cloacae, K. oxytoca, Coagulase Negative Staphylococci (CNS), P. vulgaris, C. freundii, S. marcescens, S. aureus, S. bovis, E. vulneris, Y. enterocolitica, A. calco. lwoffi, M. morganii and K. terrigena. The incidence of E. coli in non hospitalized children reached 79.6%, followed by P. mirabilis (10.1%) and K. pneumoniae (4.0%). In hospitalized children, the most frequent isolate was E. coli (69.4%), followed by K. pneumoniae (8.8%) and P. aeruginosa (4.9%). P. mirabilis was more frequent in girls(55.6%) than in boys. The vast majority of P. aeruginosa, P. vulgaris and E. cloacae strains were isolated from children with vesicoureteral reflux(3rd grade or more), surgery in the urinary tract or immunosuppression. C. albicans, CNS, S. aureus, S. bovis and A. calco.lwoffi were isolated only from hospitalized patients. 5/6 CNS strains were isolated from neonates (one with concomitant isolation of C. albicans) and 1/ 6 from a child with malignancy.
Conclusions: 1. E. coli is the predominant causative pathogen of urinary tract in children, with an increased prevalence in non hospitalized patients. 2. The higher isolation of P. mirabilis in girls(55.6%) shows that boys are not the great preference of this uropathogen. 3. There is a low frequency of mixed urinary tract infections in children.
P1249: Antimicrobial susceptibility of urinary pathogens: comparison between paediatric and paediatric surgical units
E. Iosifidis, E. Farmaki, C. Antachopoulos, D. Sofianou, E. Roilides (Thessaloniki, GR)
Objectives: Urinary tract infections (UTI) are usually treated empirically before results of urine cultures are available. Periodic evaluation of antimicrobial susceptibility of the causative microorganisms is warranted due to the emerging bacterial resistance. Purpose of this study was to review current antimicrobial susceptibility patterns of pathogens causing UTI and compare them between paediatric (PU) and paediatric surgical (PSU) in a tertiary care hospital.
Methods: Retrospective analysis of microbiology records on bacterial isolates from urine cultures of children with UTI admitted in two PU and one PSU in Hippokration Hospital during 2002-03. Identification and susceptibility testing were performed using the Vitek 2 automated system (bioMerieux, France).
Results: A total of 369 isolates were reviewed from 270 children. The distribution of Escherichia coli (E. coli) varied significantly between PU and PSU (69.4% vs 29.6%, p <0.0001). Susceptibility rates of E. coli varied (p (0.006) among isolates from PU and PSU in amikacin (AM, 99 vs 62%), ampicillin (54 vs 15%), trimethoprim/sulfamethoxazole (S/T, 71 vs 35%), cefalothin (66 vs 19%) and ceftazidime (TAZ, 95 vs 39%), respectively. On the other hand, there were no significant differences between PU and PSU for cefoxitin (96% each), amoxicillin/clavulanate (84 vs 73%), nitrofurantoin (96 vs 92%) and ciprofloxacin (CIP, 100 vs 96%). In addition, 4% of E. coli isolates in PU and 54% in PSU had the phenotype of extended-spectrum a-lactamase (ESBL) producers. Other common pathogens were Pseudomonas aeruginosa (PSA), Klebsiella pneumoniae (KPN) and Enterobacter cloacae (ECL). Most PSA isolates were susceptible to AM, TAZ, piperacillin/tazobactam, ticarcillin and imipenem (IMI) in both departments (74-100%). Susceptibility rates of KPN and ECL in patients from PU were significantly higher than those in PSU (p < 0.05) for AM, TAZ and S/T, but there was no difference for IMI and CIP ((95%).
Conclusions: Resistance of E. coli isolates from PU and particularly PSU to common antimicrobials and the presence of ESBL-producing strains are of concern. The increased resistance rates of all common isolates in PSU patients should be highlighted. Continuous evaluation of antimicrobial susceptibility patterns of uropathogens is necessary to ascertain optimal empiric therapy.
P1250: Prevalence of vaginal pathogens in childhood and adolescence
R. Kourkouli, S. Baka, E. Kouskouni (Athens, GR)
Objective: The microbiology of the female genital tract is a complex entity always found in a dynamic situation. In children and adolescents normal flora cannot be easily defined. The purpose of this study was to assess the prevalence of pathogens in vaginal cultures obtained from children presenting to our hospital.
Methods: Between January 2003 and October 2004 we examined 227 vaginal cultures from an equal number of symptomatic non-sexually active young girls (mean age 8.5 years, range 5-14 years). Vaginal secretions were inoculated onto culture plates and were incubated in aerobic and anaerobic conditions for 24 and 48 h, respectively. Isolation, identification and susceptibility to antibiotics were carried out under standard conditions using the VITEK System ATB Expression (BioMerieux, France). For the detection of Mycoplasma hominis each specimen was inoculated on DNA agar and then incubated in anaerobic conditions for 48 h whereas Ureaplasma urealyticum was identified by urease production using urea broth.
Results: Our results show that out of 227 young girls 146 (64.3%) had positive cultures, 69 (30.4%) had negative cultures for pathogens and in 12 cases (5.2%) there was no growth on the plates whatsoever. Of the 146 positive cultures we isolated 179 pathogens: 29.6% anaerobic bacteria, 22.9% Streptococci, 16.8% Ureaplasma urealyticum, 13.4% Gram-negative rods, 13.4% Gard-nerella vaginalis, 3.3% Staphylococci and 0.6% Mycoplasma hominis.
Conclusions: Distinguishing pathogenic isolates from nonpathogenic ones is not a simple task in children and adolescents. The most commonly isolated pathogens were anaerobic bacteria and streptococci. The prevalence of Ureaplasma urealyticum in our study group was high. Since our results indicate the involvement of Ureaplasma urealyticum in young girls’ vulvovaginitis a routine examination for this pathogen is suggested.
P1251: Fever in infants less than three months of age
M. Christodoulaki, F. Barabouti, V. Makri, D. Athanasopoulos, E. Kataki, E. Balomenaki (Chania, GR)
Objective: Management of febrile infants younger than 3 months of age is a difficult challenge for pediatricians because of the relatively high prevalence of serious bacterial infection. The purpose of this study was to evaluate the clinical characteristics, management and infectious outcomes of febrile infants aged 1 to 3 months.
Methods: We reviewed 153 cases of febrile infants (rectal temperature ≥ 38°C) 29–90 days old who admitted to our hospital during the period 2002–2003. Infants were considered as low risk for serious bacterial infection if they met the following (Rochester) criteria: (1) appear well; (2) were previously healthy; (3) have no focal infection; (4) have white blood cell (WBC) count 5000–15,000/mm3, band form count ≤1500/mm3, ≤ 10 WBC per high power field on microscopic examination of urine sediment, and ≤ 5 WBC per high power field on microscopic examination of a stool smear (if diarrhoea). The remaining infants were classified as being at high risk group. Serious bacterial infection was defined as a positive culture of blood, urine, cerebrospinal fluid, stool or any aspirated specimen.
Results: Of 153 infants that were studied, 89 (58%) were boys and 64 (42%) were girls. 71 (46.4%) of all infants classified as being at low risk group and 82 (53.6%) as being at high risk group. The overall incidence of serious bacterial infection was 34% (53 patients): 32 infants (60.3%) had a urinary tract infection; 14 (26.4%) bacteraemia; 5 (9.4%) meningitis; 1 (1.8%) bacterial enteritis and 1 (1.8%) pneumonia. Antibiotics administered in 85 infants (55.5%). The mean duration of hospitalization was 7.8 (4–14) days. All infants had good outcome except of one infant with meningitis who was transferred to intensive care unit because of refractory seizures. Only 4 (5.6%) of the infants in the low risk group had a serious infection, compared with 49 (55.5%) of the infants in the high risk group (p < 0.0001). None of the infants in the low risk group had bacteraemia.
Conclusion: Infants younger than three months of age with fever who meet the low risk criteria are unlikely to have serious bacterial infection. This probability forms a basis for well founded decisions in the management of the individual febrile infants.
P1252: Evaluation in new born child of the structures with prolonged post partum morphogenesis. Implications for paediatric infections
G.S. Dragoi, E. Trasca, L. Rizea, F.A. Vreju, R. Dogaru (Craiova, RO)
Objective: The frequency, relative enlarged, in the new born and toddler, of the respiratory system's infections, accompanied by morpho-functional alterations of the excretory renal system and/or of the suprarenal glands, draw attention on the knowledge of the structural particularities of those systems that can become a risk factor in the prognostic of the infections’ evolutions. It is known that the morphogenesis of those systems is incomplete in the moment of the birth and that it continues long time post partum. We proposed to ourselves to evaluate the morpho-differentiation of the functional structures from the respiratory, renal excretory in order to know the potentialities of evolution and/or extension of the inflammatory processes.
Methods: The lung and kidney fragments, harvested from 15 new born and 10 toddlers deceased by cardio-respiratory insufficiency, were fixed using formaldehyde 1% at the pH 7.5. The sections made after the paraffin inclusion, were colored with HE, Van Gieson, Gömöri and PAS stains.
Results: In the serial sections’ analysis of the new born's lung, we noticed the presence of the “terminal bags” (synonym with “pulmonary alveoli”), with entoblastic origin, lined by a cylindrical epithelium and here and there cubical. On the sections realized on the new born kidney we noticed a structural heterogeneity of the renal cortex, due to the presence pf the intermediary stages in genesis of the ‘excretory unities of the metanephros’. In the extracellular matrix, surrounding the renal corpuscle partially differentiated, we found an enlargement of the heparin-sulfate proteoglycans’ quantities.
Conclusions: The identification in new born's lungs and kidneys of the functional structures, still in differentiation shows that those organs are reactional instability sources to the infectious aggressions.
P1253: The influence of severe early-onset infections on dynamic changes of serum gastrin concentration in full-term and pre-term neonates
J. Behrendt, M. Stojewska, J. Karpe, B. Sadownik, U. Godula-Stuglik (Zabrze, PL)
Objectives: To evaluate the influence of early-onset sepsis and pneumonia on serum gastrin concentration (s.g.c.) in the neonates according to their gestational age and to find out the relation between the functional disorders of the alimentary tract and s.g.c.
Methods: The study population consisted of 74 neonates (45 boys, 29 girls), among them 42 full-term (25 infected, 17 healthy) and 32 preterm (20 infected, 12 healthy) Sepsis in 23 cases (11 due to staphyloccocal strains: Staph. haemolyticus, hominis, epidermidis and aureus, 12 due to gram-negative bacteria: Klebsiella pn. 5 cases, Pseudomonas aerug.-3 cases, Enterobacter−2 cases, Haemophilus latens-1 case, Acinetobacter baumanii−1 case) and pneumonia in 22 cases were diagnosed. Main clinical signs of sepsis: gastrointestinal disorders and pneumonia were observed in all cases. Septic shock and DIC syndrome in 63%, intensive jaundice with hepatosplenomegaly in 75% and purulent meningitis in 25% of septic neonates was noted. The functional disorders were noted in 34 infected neonates (in 20 with sepsis, in 14 with pneumonia). S.g.c. (pg/ml) was determined in peripheral vein blood twice (first between 3rd and 4th day of life and second between 14th and 21st day of life) using radioimmunologic method with ready DCT sets.
Results: In the first week of life infected full-term neonates had significantly (p < 0.001) higher mean s.g.c. (29.38 ± 10.27, ranged from 11.90 to 67.90) than healthy (14.76 ± 3.24, ranged from 11.46 to 19.96) and infected prematures (19.29 ± 19.26, ranged from 11.45 to 21.32). The infected prematures in 3rd week of life had significantly higher mean s.g.c. (120.03 ± 32.03, ranged from 87.84 to 167.71) than the healthy ones (61.72 ± 23.51, ranged from 46.39 to 113.38) and infected full-term neonates (88.99 ± 29.44, ranged from 35.91 to 133.72). The mean value of s.g.c. in first determination in infected premature neonates with functional disorders of alimentary tract was significantly higher than in infected prematures without these symptoms. In second determination, the mean s.g.c. in full-term infected neonates with alimentary tract disorders (79.09 ± 22.37) was lower (p < 0.005) than in full-term neonates without such disorders (114.47 ± 31.56).
Conclusion: The dynamic changes in serum gastrin concentration in course of severe early-onset neonatal infections, observed in first month of life, both in full-term and preterm newborns, indicate its function on alimentary tract disorders.
P1254: An abrupt onset of tularaemia in children in Kosovo
M. Djordjevic, J. Garbino, I. Uckay, V. Kostic, D. Tasic (Nis, CS; Geneve, CH)
Objective: An unexpected, suddenly outbreak of fever, sore throat and enlargement of cervical lymph node happened among children from the same school, in village Donja Budriga around Gnjilane, in Kosovo, during winter months, from December 2001 to March 2002. There were 8 children, age from 6 to 14, complained about the similar difficulties. All of them had abrupt onset of the disease with high fever from 38.8 to 39.9°C in duration of 3–7 days, sore throat and unilateral enlargement of neck mass. Until then, it was the first epidemic of unknown disease in that region.
Methods and Results: Epidemiologic questionnaire revealed the presumptive water-borne disease: children drunk the same water from the common school's well where the dead cat was found from. Before hospitalization the children were treated by lactam antibiotics unsuccessfully. After the hospitalization, presumptive clinical diagnosis of tonsiloglandular tularemia and glandular tularemia were confirmed by serological micro-agglutination test: MAT was from 1:160 to 1: 640. In three children out of eight, pathohistological diagnosis of lymphadenitis granulomatosa was obtained. In one of them PH diagnosis of Lymphadenitis tuberculosa caseoproductiva was excluded after serological confirmance by MAT (1:640). All were treated by gentamycin or amikacin in duration from 7 to 14 days and four of them were undergone by surgical incision and biopsy. In all cases, a large amount of pus was obtained. Microbiological isolation F. tularensis from culture was negative. All of children were completely recovered. Relapse happened at one patient after l9 months of byopsy of cervical lymph node with new enlargement of adjacent lymph node which did not respond to antibiotic therapy. Byopsy was recommended again. Tularemia was serological confirmed again. MAT 1:320, ELISA IgM positive.
Conclusion: The most probably mode of transmission tularaemia in children in Kososvo, was water-borne. That is why the most common clinical form was tosiloglandular and glandular. The high per cent of persisted lymphadenopathy, recurrence of lymphadenitis and surgical approach (biopsy and incision), pointed out to greater resistance of F. tularesis on antibiotics.
P1255: Efficacy of cotrimoxasol plus rifampin for six and eight weeks of therapy in childhood brucellosis
M.R Hasanjani Roushan, S.M Smailnejad Gangi, N. Janmohammadi, M.J Soleimani Amiri (Babol, IR)
Objectives: In endemic regions of brucellosis, the disease frequently occurs in children. The purpose of this study was to evaluate the efficacy of cotrimoxasol plus rifampin for six and eight weeks of therapy.
Methods: From March 1998 to July 2003, 41 patients (group 1) and 38 patients (group 2) were treated randomly with cotrimoxasol plus rifampin for six and eight weeks, respectively. All patients were followed-up for one year after completion of therapy. Clinical features and outcome of treatment for all cases were recorded.
Results: In group1, twenty five males and 16 females, with the mean age of 10.17 ± 3.58 years and in group two,16 males and 22 females, with the mean age of 8.42 ± 2.81 years were treated (P = 0.073). Clinical manifestations in two groups of patients were similar (P > 0.05%). Failure of therapy was seen in 3 (7.3%) cases in group 1, and in no case in group 2. Relapse was seen in 3 (7.3%) cases in group 1, but 1 (2.7%) case in group 2. Cure rate with eight weeks of therapy was 97.4% and for six weeks was 85.4% (P = 0.067).
Conclusion: Eight weeks of therapy with cotrimoxasol plus rifampin is better than six weeks of therapy in children with brucellosis. Follow-up of these patients for finding of relapse cases are necessary.
P1256: Linezolid efficacy and tolerability in the treatment of infectious complications in children
M. Shneider, E. Kurnikova, L. Zharikova, E. Boichenko, A. Kolbin, A. Maschan (Moscow, St. Petersburg, RUS)
Objectives: Infections caused by Gram(+) flora are a major cause of morbidity in patients with haematological conditions associated with neutropenia. Although vancomycin is often indicated for these infections, it is associated with infusion-related reactions and renal toxicity. This study was to evaluate the efficacy and tolerability of linezolid, the first member of a new class of antibacterial agents, oxazolidinones, in the treatment of infectious complications in oncohematological patients.
Methods: This was a retrospective analysis based on data from 46 paediatric patients (age 8 mo to 16 y) with haematological conditions. Linezolid 10 mg/kg bid was administered by 1-h intravenous infusion, always as a component of combined antibacterial therapy (ceftazidime, cefepim or carbapenem with or without amikacin). Indications for antibacterial therapy included fever of unknown origin in neutropenic patients, soft tissue infection, mucositis, enterocolitis, catheter related infections, pneumonia, osteomyelitis, urinary tract infection and subhepatic abscesses. Microbiological samples were taken before the start of therapy and repeated on days 7 and 14 of treatment if the results were positive.
Results: A total of 50 courses of linezolid were delivered. The mean duration of therapy was 9 days (5–106 days). Clinical response to therapy was observed in 76% (38/50) of courses, including complete resolution of the symptoms in 68% (34/50) of courses and eradication of Gram(+) organisms in patients with mixed fungal and bacterial infection in 8% (4/50) of courses. Clinical response was achieved in 70% (26/37) of empirical therapy courses. In courses with documented Gram(+) infection, eradication was 100% (13/13). Complete resolution of symptoms was achieved in 50% (8/16) and improvement in 19% (3/13) of courses in patients who were switched to linezolid following ineffective vancomycin therapy. Only 1 (2%) course of linezolid therapy had to be discontinued due to exacerbation of cytopenia in a child with acquired severe aplastic anemia.
Conclusions: This is the first study assessing the efficacy and tolerability of linezolid in children with infectious complications of oncohematological diseases. Linezolid was highly effective when used as a component of empirical therapy of the febrile neutropenia. Haematological toxicity of linezolid was low, even when baseline suppression of hemopoiesis was present, allowing full compliance with treatment schedule.
P1257: Chryseobacterium indologenes bacteraemia in a diabetic child. Case report
C. Iaria, G. Stassi, G.B. Costa, G. Crisafulli, A. Cascio (Messina, I)
Background: Chryseobacterium indologenes is a non-fermentative Gram-negative bacillus that is a rare pathogen in humans. Its occurrence in diabetic children has never been reported. In this report, we describe a case with C. indologenes bacteraemia possibly associated with the use of a peripheral venous catheter.
Case presentation: A 2-year-old boy with type I diabetes mellitus admitted for coma due to cerebral oedema and treated successfully with insulin, dexamethasone, mannitol and KCl, on the tenth day presented with fever of 40°C, agitation, restlessness, lack of appetite, somnolence, fatigue. Pulse rate 90 per min, respiratory rate 20 per min. Laboratory studies revealed a white blood cell count of 4900/mm3 with 67% neutrophils and 27% lymphocytes. Cultures of blood yielded C. indologenes. Treatment with ceftriaxone was started before the culture results were achieved, and was continued after susceptibility test results C. indologenes were obtained. The patient became afebrile after 48 hours, and his general conditions improved within 36 hours. The infection did not recur.
Conclusions: This is the third case of bacteraemia outside of Asia due to C. indologenes and the first occurred in a diabetic child not otherwise immunocompromised. Our case indicates that C. indologenes infection can occur in diabetic children without ventilator or central venous catheter and might be treated with a single agent after in vitro susceptibility tests have been performed.
P1258: Antibiotic control in a paediatric teaching hospital
N. Goldman, A. Vergison (Brussels, B)
Objectives: To evaluate the impact of antibiotics control policies in a 170-beds paediatric teaching hospital between 2001 & 2003. The aims were to (1) decrease broad spectrum antibiotics use, (2) promote rapid switch to oral treatment, (3) reduce spectrum of the antibiotics used for the treatment of community acquired pneumonia (CAP).
Methods: Written guidelines for the management of CAP, set up by all concerned specialists, were implemented in the hospital in 2002. Since march 2002, seminars on antibiotic resistance and rationalized antibiotic prescription took place twice yearly. Objectives were regularly discussed with the infectious disease specialist in the different wards. Antibiotic consumption was analysed through pharmacy billing data. Data were expressed in DDD/100 patient-days (PD) and retrieved for each department (excepted for oncology, nephrology and neonatology).
Results: Amoxicillin-clavulanic acid (AC) use decreased from 89 to 48 DDD/100 PD during the studied period. The reduction was marked in the general paediatrics and pneumology department (30 to 9 DDD/100 PD). Both Oral and IV use of AC decreased (41 to 24 DDD/100 PD and 48 to 24 DDD/100 PD for oral and IV use respectively). Oral cefuroxime-axetil increased from 3 to 47 DDD/100 PD and IV cefuroxime from 11 to 26 DDD/100 PD. Penicillin consumption raised from 5 to 9 DDD/100 PD and altogether, narrow spectrum antibiotics (ampicillin, penicillin oxacillin and amoxicillin) increased from 40 to 47 DDD/100 PD. Oral antibiotics (AC, cefuroxime-axetil, amoxicillin and ciprofloxacin) increased from 51 to 82 DDD/ 100PD, mainly explained by the rise of cefuroxime (share from 6 to 58% of total oral drugs). The IV drugs decreased from 156 to 140 DDD/100 PD and total antibiotics consumption was slightly increased (207 DDD/100 PD in 2001 versus 222 DDD/100 PD in 2003. Broad spectrum antibiotics use decreased (133 to 85 DDD/ 100 PD), mainly due to decrease in AC and ceftriaxone which went down from 17.4 to 9.3 DDD/100 PD.
Conclusion: 1. Antibiotics use analysis showed adherence to new CAP guidelines with change in ceftriaxone, AC, cefuroxime and penicillin prescription pattern. 2. Oral drugs increased and IV antibiotics decreased. 3. Broad spectrum drugs decreased and narrow spectrum ones increased. These data provide interesting information feedback for all hospital physicians and should encourage the implementation of more concerted guidelines.
P1259: Determinants of under- and overuse of antibiotics in primary care patients with acute otitis media
A.E. Akkerman, M.M. Kuyvenhoven, J.C. van der Wouden, T.J.M. Verheij (Utrecht, Rotterdam, NL)
Objectives: In the Netherlands, antibiotic prescribing rates for acute otitis media (AOM) are very low compared to other countries. Some publications suggested a relation between decline in antibiotic use and a rise in complications of respiratory tract infections. Therefore it is important to study not only overprescription but also underprescription of antibiotics, and which clinical factors are associated with both phenomena. Thus, a study was undertaken to assess the appropriateness of antibiotic treatment in AOM in a low prescribing country like the Netherlands and to assess clinical patient characteristics that cause both under- and overprescribing of antibiotics in cases of AOM.
Methods: During four weeks in the winter of 2002/2003, 146 Dutch GPs in the middle region of the Netherlands included all patients with acute ear complaints. They registered patient demographics, clinical presentation (signs and symptoms), severity of illness, whether they thought patients expected antibiotic treatment, diagnosis and management. Using key issues of current guidelines on AOM as benchmark, we assessed the appropriateness of antibiotic prescribing in cases of AOM. In addition, we assessed the association between clinical patient characteristics and under- or overprescribing of antibiotics in cases of AOM.
Results: In seven out of ten patients with acute ear complaints antibiotic treatment was according to the guidelines. In 17% of all consultations antibiotics were indicated but not prescribed (underprescribing) and in 12% antibiotics were not indicated but prescribed (overprescribing). The lower the GP assessed the severity of illness, if there was no worsening since the previous contact, if the duration of symptoms prior to the consultation was short, or if the patient had only one inflammation sign, more underprescribing occurred. If the GP assessed a high severity of illness, if the patient was coughing, or if the GP thought the patient expected an antibiotic, then the GP prescribed more often antibiotics without indication (overprescribing).
Conclusion: Both incorrect interpretation of signs and symptoms and perceived expectation of patients were correlated with incorrect antibiotic management in acute otitis media.
P1260: Clarithromycin, a good alternative to tetracyclines, in the treatment of Mediterranean spotted fever in children. A randomised trial
E. Anton, T. Munoz, B. Font, M.D. Ferrer, I. Sanfeliu, F. Segura (Sabadell, E)
Background: In vitro and in vivo studies demonstrated the high efficacy of doxycycline in the treatment of MSF. However, it is convenient to have another alternatives available for patients who are allergic to tetracyclines, pregnant, or for children under 8 years. Based on good results in vitro we considered of interest to evaluate the clinical efficacy of clarithromycin in the treatment of MSF.
Methods: Randomized therapeutic study. Group A received clarithromycin 500 mg/12 h for 5 days in adults and 15 mg/kg/12 h for 5 days in children. Group B received the treatment of choice which was, doxycycline (200 mg/12 h for 1 day) in adults and doxycycline (5 mg/kg/12 h for 1 day) or josamycin (50 mg/kg/12 h for 5 days) in children. Inclusion criteria: patients who had the compatible clinical with MSF and the presence of the primary lesion (tache noire) or a positive serology against R. conorii. Exclusion criteria: patients who had received effective antibiotic treatment or whose symptoms appeared 8 or more days before enrolment in the study.
Results: There were randomized 48 patients. There were excluded 14 patients, 10 from group A and 4 from group B. 34 patients were evaluated. 20 males. Mean age: 38.4 years. 12 were under 14 years. 33 patients presented fever. The exanthema was presented in 30 cases. The tache noire was observed in 29 cases. The diagnosis was confirmed by serology in 31 cases. 14 patients received clarithromycin (group A) and 20 doxycycline or josamycin (group B). The interval between the onset of the symptoms and the start of treatment was 3.6 ± 2.2 days in group A and 3.6 ± 1.9 days in group B (p = NS). The time taken for disappearance of the fever after treatment was 2.57 ± 1.7 days in group A vs 2.19 ± 1.4 days in group B (p = NS). The symptoms had disappeared at 4.35 ± 2.5 days in group A vs 4.52 ± 3.4 days in group B (p = NS). There were no adverse reactions to the treatment or relapses in either group.
Conclusion: Clarithromycin could be considered as an alternative in the treatment of MSF.
Mechanisms of antibiotic resistance in Gram-negative bacillie in Gram-negative bacilli
P1261: Fitness-cost and silencing of antibiotic resistance-coding mobile genetic elements in Escherichia coli
V.I. Enne, A.A. Delsol, C.M. Hayes, J.M. Roe, P.M. Bennett (Bristol, UK)
Objectives: Many questions remain unanswered about the fate of antibiotic resistance (AbR) in drug-free environments. We examined the fitness cost and expression of five AbR-coding mobile genetic elements in the absence of drug selection.
Methods: Plasmids RP1, pUB307 and N3, the transposon Tn1 and the ICE element R391 were separately introduced into 345-2 RifR, a rifampicin-resistant derivative of a recent porcine isolate. The insertion site of Tn1 was identified using PCR and DNA sequencing. The fitness cost of each AbR element was assessed in vitro by pairwise growth competition and in vivo by monitoring the number of CFU/g of faeces regularly for 21 days following inoculation of six seven-week old organic piglets with 1010 CFU. The bacteria were recovered by plating onto MacConkey agar containing rifampicin and retention of the AbR elements monitored by replicating onto agar containing the appropriate antibiotics. Isolates that did not express the expected resistance profile were characterised by PCR and DNA sequencing.
Results: Tn1 inserted into a cryptic DNA sequence adjacent to the rhsD gene. The insertion site was 2.1 kb away from an independent insertion made in a previous study, where we found that Tn1 enhanced host fitness. The Tn1 insertion described here had no effect on fitness in vitro and in vivo. The plasmid RP1 had a 3.3 ± 0.9% fitness cost per generation in vitro but was recovered at the same rate from the pig gut as the plasmid-free strain (p = 0.09). Similarly the plasmid pUB307, which is RP1 without Tn1, had an in vitro fitness cost of 1.9 ± 0.8% but no effect on fitness in vivo (p = 0.21). The plasmid N3 had a large fitness cost of 9.1 ± 1.8% in vitro and was also recovered at lower rates in vivo (p = 0.04). The ICE element R391 had a neutral effect on fitness in vitro and in vivo. Most AbR element-carrying strains recovered from the animals retained resistance to the relevant antibiotics, except three RP1-carrying isolates, which lost resistance to ampicillin, tetracycline and kanamycin. These isolates retained the plasmid including intact wild-type resistance gene and promoter sequences, indicating the genes have been silenced.
Conclusions: The fitness impact exerted upon E. coli 345–2 RifR by carriage of AbR elements is variable depending on the element, and can also be absent. The advantage sometimes conferred by Tn1 appears dependent on the insertion site. Silencing of resistance genes encoded on RP1 can occur in vivo.
P1262: Development of a protein based test for multiple antibiotic-resistant Salmonella typhimurium
N.G. Coldham, L.P. Randall, L.J.V. Piddock, M.J Woodward (Surrey, Birmingham, UK)
Objectives: The multiple antibiotic resistance (Mar)phenotype is characterised by resistance to different classes of antibiotics (e.g. fluoroquinolones, beta-lactams and tetracycline), disinfectants and certain dyes. The development of this phenotype is considered to be an important step in the transition to high level antibiotic resistance of clinical significance. Active efflux by proteins of the AcrA/B-TolC system has been closely associated with the Mar phenotype and development of high level resistance to fluoroquinolones. There is no single accepted test in widespread use for the Mar phenotype. The objective of the present study was to identify target proteins for the development of a simple test for the Mar phenotype.
Methods: Previous proteomic studies (Coldham and Woodward, 2004) have demonstrated the presence of proteins associated with the Mar phenotype in the cell envelope fraction derived from S. typhimurium (strain SL1344). Cell envelope proteomes were prepared from S. typhimurium following treatment with the fluoroquinolones ciprofloxacin and enrofloxacin (0.032 and 0.008 μg/ml) and analysed by 2-dimensional gel electrophoresis and 2-dimensional HPLC-mass spectrometry.
Results: The number of proteins detected in 2D gels derived from control cultures was significantly (p < 0.05) reduced following treatment with ciprofloxacin (31.2 ng/ml) from 296 ± 77 to 153 ± 36 (mean + 1 SD) respectively. However, increased expression (p < 0.05) of 17 cell envelope proteins was detected. The greatest increase in expression, by up to 10 fold (p < 0.0001), was observed in the AtpA, AtpB, AtpC and AtpH protein sub units of the F1F0-ATP synthase proton pump complex. Increased expression of other proteins including the outer membrane channel protein TolC, organic solvent tolerance protein Imp, the outer membrane protein OmpA and the porin OmpB was also observed. Analysis of the cell envelope proteome by 2D-HPLC-MSn provided a more complete assessment and identification scores suggested increased expression of many other proteins including AcrA and AcrB.
Conclusion: The F1F0-ATP synthase proton pump complex provides the motive force for efflux activity and may modulate the membrane potential for porin selective molecular influx and presents a possible target protein complex for Mar. The expression of other outer membrane proteins, such as phospholipase A, was increased after treatment with fluoroquinolones and may provide alternative targets.
P1263: Molecular epidemiology and mechanisms of resistance to several antimicrobial agents in sporadic Salmonella spp. strains causing acute gastroenteritis in Cuba
R. Cabrera, J. Ruiz, M. Ramírez, L. Bravo, A. Fernández, A. Aladueña, A. Echeíta, J. Gascón, P. Alonso, J. Vila (Barcelona, E; Havana, CUB; Madrid, E)
Objective: Determine the antimicrobial susceptibility and the molecular mechanisms of resistance of Salmonella spp. strains causing acute gastroenteritis in Cuba and determine the potential dissemination of a resistant clone.
Methods: A total of 34 Salmonella strains isolated from feces of patients with acute gastroenteritis isolated from different regions of Cuba in 2002 were received and processed in the laboratory of Clinical Microbiology of Clínic Hospital in Barcelona. The antimicrobial susceptibility to 9 antibiotics: ampicillin, amoxicillin/clavulanic acid, nalidixic acid, tetracycline, trimethoprim/sulphametoxazole, chloramphenicol, gentamicin, ciprofloxacin and spectinomycin, was determined using the agar dilution method. The molecular mechanisms of resistance to several antimicrobial agents were detected by PCR and the chloramphenicol acetyl transferase activity by a colorimetric assay. Analysis of molecular epidemiology was performed by pulsed field gel electrophoresis using the low frequency restriction enzyme XbaI.
Results: Twenty-two strains presented resistance, 64% was multirresistant. The serotype Typhimurium phagetype 104 was the most frequent and presented similar genetic profiles of PFGE. High levels of resistance to tetracycline (53%), spectinomycin (50%), ampicillin (44%) and chloramphenicol (41%) were found. Resistance to tetracycline was associated with the tet G and tet A genes. Resistance to ampicillin was due to the presence of β-lactamases, mainly the CARB type. The floR gene was the main mechanism of resistance to chloramphenicol. Among the susceptible strains, six belong to the serotype Agona were epidemiologically related.
Conclusions: The presence of two main clones was detected in Cuba, with the widespread dissemination of a multiresistant clone of Salmonella enterica serotype Typhimurium DT 104 and an antimicrobial susceptible clone of Salmonella enterica serotype Agona in two separate regions in the country.
P1264: Molecular characterisation of antibiotic resistance in clinical isolates of Pseudomonas putida
T. Horii, Y. Shiba, H. Muramatsu, A. Takeshita, M. Maekawa (Hamamatsu, Nagoya, JP)
Objectives: Infections caused by Pseudomonas putida are mostly reported in compromised patients such as newborns and cancer patients. Recently, emergence of resistance to carbapenems and other antibiotics have been reported. In the present study, we examined susceptibilities and characterized resistance to betalactams and fluoroquinolones in clinical isolates of P. putida.
Methods: Six clinical isolates of P. putida from the different patients and Pseudomonas aeruginosa PAO-1 were used. These isolates showed different pulsed-field gel electrophoresis genotypes. MICs were determined by an agar dilution method as described by the National Committee for Clinical Laboratory Standards. Detection of the metallo-beta-lactamase genes, blaIMP-1, blaVIM-1 and blaVIM-2, was carried out by polymerase chain reaction amplification. Outer membrane protein profiles were characterized by SDS-PAGE. Nucleotide sequences of the quinolone resistance-detemining regions (QRDRs) of the gyrA, gyrB and parC genes were determined in 6 isolates of P. putida and P. aeruginosa PAO-1.
Results: Five P. putida isolates showed resistance to betalactams, including ampicillin (>128 mg/L), ceftazidime (>128 mg/L) and carbapenems (8→128 mg/L) such as imipenem and meropenem. Four isolates showed high resistance to fluoroquinolones (>128 mg/L), including norfloxacin, levofloxacin, sparfloxacin, gatifloxacin and pazufloxacin. The MIC range for sitafloxacin was between <0.25 and 8 mg/L. Four isolates resistant to both beta-lactams and fluoroquinolones also showed minocycline resistance (>16 mg/L). One isolate was susceptible to both beta-lactams and fluoroquinolones. All of carbapenem-resistant isolates had the blaIMP-1 gene. In one isolate highly resistant to all carbapenems (>128 mg/L) except sitafloxacin (8 mg/L) SDS-PAGE showed reduced production of a protein band, which was identified as OprD by amino acid sequencing. Some specific mutations conferring fluoroquinolone resistance were found in the QRDRs of GyrA (Thr83Ile), GyrB (Glu469Asp) and ParC (Ser87Leu) in P. putida isolates.
Conclusion: Five of 6 isolates of P. putida were resistant to carbapenems because of acquisition of the metallo-bata-lactamase gene. Our results suggested that reduced production of OprD was associated with increse in MICs of carbapenems. Four isolates showing resistance to fluoroquinolones except sitafloxacin had a combination of 3 amino acid alterations in the QRDRs of GyrA, GyrB and ParC.
P1265: Porin expression among clinical isolates of Escherichia coli showing AmpC-hyperproduction phenotype
J.W. Huizing, M.C. Conejo, G. Amblar, A. Pascual, L. Martínez-Martínez, E.J. Perea (Seville, Santander, E)
Objective: To evaluate outer membrane protein (OMP) profiles of 80 clinical isolates of E. coli showing an AmpC hyperproduction phenotype and to relate them with susceptibility to antimicrobial agents.
Methods: Eighty clinical isolates of E. coli collected in the laboratory of Microbiology, University Hospital Virgen Macarena of Seville, in the period 1999–2001, were studied. They represented sixty different clones, as determined by REP-PCR. AmpC hyperproduction phenotype was defined as co-resistance to ampicillin, cephalotin, cefoxitin, and amoxicillin-clavulanic acid, in the absence (NCCLS guidelines) of extended spectrum beta-lactamase expression. To analyze OMP profile bacterial cells were grown in nutrient broth (NB; low osmolarity medium) and in Mueller Hinton broth (MHB; high osmolarity medium). Cell envelopes were obtained from lysed cells by sonication. OMPs were isolated as sodium-lauryl sarcosynate insoluble material. The proteins were resolved by SDS-PAGE through 10% acrylamide-6 M urea gels. E. coli K12 and derivatives lacking either or both OmpF and OmpC were used as controls. The OMP pattern was compared with the resistance phenotype to betalactams, fluoroquinolones, aminoglycosides, tetracycline and cotrimoxazole, as determined by broth microdilution (NCCLS guidelines).
Results: An OMP profile similar to that of E. coli K12 (expression or both OmpF and OmpC in NB and downregulation of OmpF in MHB) was found in 33 (41%) strains Absence of OmpF and expression of OmpC in both NB and MHB was observed in 23 (29%) strains. Expression of both OmpF and OmpC in both NB and MHB was observed in 11 (14%) strains. Loss of both porins was only found in one strain. A variety of profiles were noted for the remaining 12 (15%) strains. All strains were susceptible to cefepime, carbapenems and amikacin, but for other agents susceptible and resistant strains were included in every profile group, with a wide range of MICs in every group.
Conclusions: 1. OMP profile among clinical isolates of E. coli showing an AmpC hyperproduction phenotype is diverse, with only 41% of strains showing a profile similar to that of E. coli K-12. 2. There was a great variation in MIC values of the different antimicrobial agents for strains with the same porin profile.
P1266: Carbapenem-resistant, CTX-M-producing Klebsiella pneumoniae in the UK
M.E. Ward, N. Woodford, M. Warner, R. Pike, M.-F.I. Palepou, M.E. Kaufmann, J. Turton, I. Eltringham, D.M. Livermore (London, UK)
Objectives: From June-August 2004, ARMRL received 4 carbapenem-resistant isolates of K. pneumoniae from separate patients on the Liver ICU of a London tertiary care hospital for investigation. Two isolates were from blood cultures, one from bronchial washings and one from a wound swab. All patients had clinical infections and were treated with broad-spectrum antibiotics, including colistin. One patient with bacteraemia died shortly after beginning treatment, but there was no other attributable mortality. We sought to define the resistance mechanisms of the isolates.
Methods: Isolates were compared by PFGE of XbaI-digested genomic DNA and analysed with BioNumerics software. MICs were determined and interpreted according to BSAC guidelines. Isolates were screened for beta-lactamase genes, including those for known carbapenemases by PCR. Imipenem hydrolysis was investigated by spectrophotometry. Iso-electric focusing (IEF) was performed to visualise beta-lactamase. Outer membrane profiles were examined by SDS-PAGE analysis.
Results: The four isolates were 95% similar by PFGE and represented a single strain. Their antibiogram was complex. Resistance was evident to ertapenem (MIC > 16 mg/L), but less marked to meropenem (MIC 4–16 mg/L) and imipenem (1–8 mg/L). MICs were unaffected by EDTA, and no carbapenemase activity or genes were detected by spectrophotometry or PCR, respectively. Resistance to all cephalosporins tested, including cefepime (MIC > 64 mg/L), was not reduced significantly by clavulanate; isolates were also highly resistant to cefoxitin (MIC > 64 mg/L). Isolates were resistant to gentamicin, tobramycin and ciprofloxacin. All were susceptible to colistin, and 3 showed borderline susceptibility to amikacin (MIC 4 mg/L). Two beta-lactamases were apparent by IEF, and were consistent with the group 1 blaCTX-M and blaOXA-1-like alleles detected by PCR; blaSHV was also detected. Neither IEF nor PCR yielded evidence of AmpC production. SDS-PAGE showed the loss of a major outer membrane protein (OMP).
Conclusions: Impermeability, associated with OMP loss, combined with the production of group 1 CTX-M and OXA-1-like enzymes appears to underlie the complex pattern of beta-lactam resistance in this K. pneumoniae strain. However, the lack of potentiation of cephalosporins by clavulanate is unusual for CTX-M producing klebsiellae, as is the carbapenem resistance. Limited treatment options for these more resistant variants poses a significant clinical problem.
P1267: Characterisation of a multiresistant mutator strain of Pseudomonas aeruginosa from an ICU patient
B. Henrichfreise, I. Wiegand, B. Wiedemann (Bonn, D)
Objectives: Hypermutation is a common feature of P. aeruginosa from cystic fibrosis patients with chronic infections. Little is known about mutator strains from patients with acute infections among which hypermutation seems to be rare. The aim of our study was (I) to characterize the molecular alteration responsible for the mutator phenotype and (II) to investigate mutation-mediated resistance in a multiresistant mutator strain of P. aeruginosa from an ICU patient.
Methods: A multiresistant P. aeruginosa strain was isolated from a wound swab from a 63-year old patient in an IC unit of a German hospital in 2004. The strain showed resistance to Levofloxacin and Piperacillin and intermediate resistance to Imipenem, Ceftazidime, and Gentamicin. Hypermutation was detected by disk diffusion tests. Mutation frequency was determined on selective rifampicin agar. The mismatch repair system genes mutS, mutL and uvrD were amplified and sequenced. Effluxpump overexpression was detected by testing levofloxacin MICs both with and without the effluxpump inhibitor MC-270,110. MexR coding for the repressor of the mexABoprM operon and the QRDR of gyrA and parC and were amplified and sequenced.
Results: The mutation rate of the strain was 2.26 × 10−6. Sequencing of the mismatch repair system genes revealed a 1 bp deletion (A1250) in mutL resulting in a frameshift. The levofloxacin MIC was reduced from 128 mg/L to 2 mg/L in presence of MC-270,110 indicating effluxpump overexpression and alterations in type II topoisomerases. In mexR a N insertion after L52 was detected. Two changes in gyrA (T83I, D87G) and one change in parC (S80L) were found.
Conclusions: This is the first genetic characterisation of a multiresistant mutator strain of P. aeruginosa from an ICU patient with an acute infection. A new mutation in mutL of P. aeruginosa responsible for the mutator phenotype was found. A mutator phenotype can confer a selective advantage in stressful environments. So, one could consider coselection of the mutator and the mutation-mediated resistance phenotype in this strain. Further work will be necessary to understand the underlying mechanisms of this evolutionary process in a strain of a patient with an acute infection.
P1268: A novel class 1 integron containing an aadA aminoglycoside resistance gene cassette resulting from in vivo homologous recombination
M.E. Cano, I. de Benito, J.M. García-Lobo, J. Agüero (Santander, E)
Objective: To describe a new variant of the aadA gene encoding resistance to aminoglycosides, as the result of a sequence reassortment between the aadA2 and aadA1a gene cassettes.
Methods: A total of 78 clinical isolates of Yersinia enterocolitica collected from different patients in our area between 1994 and 2003, were analysed for the presence of class 1 integrons. Identification and susceptibility testing were performed with the Microscan Walkaway System (Dade Behring). Susceptibility to chloramphenicol, nalidixic acid, tetracycline, kanamycin, streptomycin and spectinomycin was determined by disk-diffusion. Strains were screened by PCR using primers specific for IntI1 gene, and gene cassettes were identified by sequencing the PCR products of the integron variable regions obtained with primers 5′CS and 3′CS.
Results: Sixty out of the 78 Y. enterocolitica isolates (77%) showed the presence of integrons, and they could all be grouped into two clusters of 1000 bp (45 isolates) and 1900 bp (15 isolates) according to the two patterns obtained from the amplification of the variable region. The sequencing of the 1000 bp products revealed the presence of a new aadA gene cassette the sequence of which was a mosaic built up from pieces from both aadA2 and aadA1a. All the isolates were resistant to streptomycin, spectinomycin and chloramphenicol, and susceptible to gentamicin, tobramycin, amikacin and kanamycin.
Conclusions: The new aadA gene described here enlarges the reported examples of integron containing gene cassettes. Furthermore, nucleotide sequence analysis revealed that new gene variants can be produced in nature by exchange mechanisms probably mediated by homologous recombination.
P1269: Sulfamethoxazole resistance genes in uropathogenic E. coli
J. Shchepetova, K. Truusalu, E. Sepp, M. Mikelsaar (Tartu, EST)
Increasing antimicrobial resistance among E. coli (Ec) causing urinary tract infection (UTI) is a serious worldwide problem. The treatment with co-trimoxazole (ST), a combination of sulfamethoxazol (Sulfa) and trimetoprim, often does not prevent recurrent UTI (rUTI). The sensitivity to antibiotics of consecutive rUTI isolates is usually detected by disk-diffusion or E-test, but it does not always reflect the presence of resistance genes. Sulfa resistance in enterobacteria arises from acquisition of two additional genes, sulI and sulII, encoding the dihydropteroate synthetase. These genes are located on different types of mobile elements: sulI is associated with type I integrons and sulII with non-conjugative plasmids. However, there are few data on the prevalence of Sulfa resistant genes and integron 1 in community-acquired Ec strains isolated from children with rUTI. One of the putative reasons for rUTI may lie in non-accordance of pheno- and genotypical results of antibiotic susceptibility determination, resulting in non-efficient antimicrobial treatment. The aim of this study was to determine the prevalence of Sulfa resistance genes (Int1, SulI and SulII) among Ec causing rUTI in children.
Material and methods: Altogether 29 consecutive Ec isolates from 10 rUTI patients (Tartu University Children Hospital) with changes in antibiotic resistance to ST were applied. Ec resistance to Sulfa was detected by E-test (Bio Mereux). DNA was extracted using QIAamp DNA Mini Kit (QIAGEN). Int1, sulI and sulII genes were detected by PCR (Polymerase Chain Reaction).
Results: 20 out of 29 (65.5%) Ec isolates were resistant to Sulfa (MIC3512 mg/L). Int 1 was present in 15 out 29 (51.7%) Ec isolates. SulI and/or sulII genes were detected in all resistant Ec isolates. Three different combinations of genes were found: 9 isolates contained sulI and sulII; 3 isolates only sulI and 8 isolates only sulII gene. Among 9 phenotypically sensitive Ec strains 3 isolates (MIC 8 mg/L) carried sulI and sulII genes while 6 strains (MIC 12–32 mg/L) did not contain any of the three genes.
Conclusions: This study demonstrates that the resistance genes of Sulfa (sulI and sulII) are widely distributed in Ec strains from children with rUTI. Half of the rUTI causing strains carry integron 1 providing them with capability to obtain multiresistance. The detection of Sulfa gene in Ec might be of importance in prevention of rUTI.
P1270: Plasmid transfer from Klebsiella pneumoniae to Escherichia coli
S. Schjørring, C. Struve, K.A. Krogfelt (Copenhagen, DK)
Objectives: The increased number of gastrointestinal infections and the rapid increase in antibiotic bacterial resistance, makes investigation of antibiotic resistance development during intestinal colonization a very important issue. The objectives are to investigateaKlebsiellapneumoniaestrain'sabilitytoconjugatewith Escherichia coli, in vitro in vivo, and to characterise the transconjugants by plasmid profiling and restriction fragment analysis.
Methods: The donor K. pneumoniae ATCC 700721 strain was a multi-resistant mucoid strain, which contains three large plasmids (100–150 kbp.) and two small ones (<7 kbp.). The recipient was an E. coli MG1655 StrepR, RifR. The in vitro conjugation was performed on solid and liquid media, whereas the in vivo conjugations were performed during colonisation of the large intestine of streptomycin/ampicillin treated mice. Transconjugants were isolated from both in vitro and in vivo experiments. A combination of plasmid purification (which purify large plasmids in a way that allows following restriction cutting), plasmid profiles, and restriction fragment analysis, was used to characterise the transconjugants obtained from these experiments.
Results: In vitro conjugation between K. pneumoniae and E. coli showed transfer of either only 1 or all 5 plasmids depending on the conditions. When K. pneumoniae was used to colonise the large intestine, it was shown that K. pneumoniae transferred one of its plasmids containing antibiotic resistance genes to an indigenous E. coli strain. When mice were co-infected with K. pneumoniae and the laboratory E. coli strain, it was seen that K. pneumoniae again transferred its plasmid with antibiotic resistance to E. coli. Both the laboratory strain and the indigenous E. coli with the newly acquired plasmids were tested for their conjugative ability and it was seen that they both had obtained conjugative plasmids from K. pneumoniae. Furthermore, the plasmids were characterised by restriction fragment analysis confirming, that all the transconjugants plasmids were identical with the plasmids from the original donor; the K. pneumoniae ATCC 700721.
Conclusion: K. pneumoniae ATCC 700721 contains conjugative plasmids, and conjugation is occurring in a high frequency both in vivo and in vitro. The type of plasmid, which is conjugated depends on the environmental conditions.
P1271: Accuracy of the Microscan Walkaway system for identifying P. aeruginosa strains carrying class 1 integrons with MICs of cefepime higher than those of ceftazidime
E. Ugalde, A.B. Campo-Esquisabel, M.E. Cano, J. Calvo-Montes, J. Agüero, L. Martínez-Martínez (Santander, E)
Objectives: MICs of Cefepime (FEP) against P. aeruginosa are usually similar to those of Ceftazidime (CAZ), but isolates expressing efflux pumps and containing PSE-1 or OXA enzymes may present MICs of FEP higher than those of CAZ. In this study we have evaluated the Microscan Walkaway system (W/A) for detecting P. aeruginosa isolates with any of the following phenotypes: 1. CAZ susceptible (S) and FEP intermediate (I); 2. CAZ-S and FEP resistant (R); 3. CAZ-I and FEP-R. The presence of Class 1 integrons in these isolates was also investigated.
Methods: From May to Sept. 2004, all P. aeruginosa isolates (one per patient) I or R to FEP and S to CAZ, and the first 30 strains I or R to both CAZ and FEP were evaluated. MICs of FEP and CAZ were determined by reference microdilution (NCCLS guidelines). Then clinical categories obtained with W/A for organisms with any of the previously indicated phenotypes were compared with those obtained by the reference method. Class 1 integrons were detected by PCR using primers specific for the type 1 integrase gene and primers specific for conserved regions 5′ and 3′ of class 1 integrons. Gene cassettes were identified by sequencing the amplicons obtained from two different isolates for each integron type found.
Results: The following phenotypes of CAZ/FEP and number of isolates were identified by the W/A system as: S/I:9; S/R:4; I/ I:15; R/I:4, and R/R:11. Reference microdilution confirmed that 23 isolates were more susceptible to CAZ than to FEP. The W/A correctly identified 13 out of these isolates (53.5% sensitivity). Reference microdilution confirmed higher susceptibility to CAZ than to FEP in all 13 isolates detected by W/A (100% specificity). For the 23 isolates, the W/A did not cause any Major (false resistance) error for CAZ or FEP. The phenotypes of CAZ/FEP for the 10 isolates not recognized by W/A were I/I (n = 6), R/I (n = 3) or R/R (n = 1). A class 1 integron was identified in 18 out of the 23 (78.3%)isolates: 13 with a 1600 bp integron, coding for an OXA protein which shares the highest aminoacid identity with group II OXA enzymes (77%), 3 with a 1200 bp integron, coding for PSE-1, and 2 in which PCR failed to amplify any integron conserved region.
Conclusions: The W/A system is very specific, but poorly sensitive, for recognition of P. aeruginosa more susceptible to CAZ than to FEP. In our centre, this phenotype is often associated to the presence of integrons coding for an OXA enzyme or PSE-1.
P1272: Non-beta-lactamase mediated beta-lactam resistance in Stenotrophomonas maltophilia
V. Gould, D. Miller, R.A. Howe, M. Avison (Bristol, UK)
Objectives: Stenotrophomonas maltophilia is an important, emerging, Gram-negative, nosocomial pathogen that can cause bacteraemias, respiratory tract colonisation and other organ-specific infections. Isolates have the potential to mutate to multidrug resistance (MDR), which has been linked to overexpression of the efflux pump SmeDEF. Beta-lactam resistance is thought to be due to beta-lactamase overexpression since SmeDEF does not pump out beta-lactams.
Methods: Betalactam resistant mutants of S. maltophilia isolates K279a and N531 were selected using either ceftazidime or meropenem. MICs of various classes of antimicrobial agents were determined by E-test using Muller-Hinton agar. Betalactamase induction was attempted using cefoxitin challenge (10 mg/L, 2 h) and specific betalactamase assays were performed using nitrocefin with (L2) or without (L1+L2) EDTA (500 mM). RNA extraction and RT-PCR for smeE and smeF used kits from Quiagen.
Results: From parent isolate K279a one high level meropenem-resistant (MIC > 512) and one high level ceftazidime resistant mutant (MIC = 16) were selected. From parent isolate N531 one high level meropenem resistant mutant was selected (MIC > 512). Betalactamase assays in the absence of inducer revealed that none of these mutants over-express beta-lacta-mase. Indeed, unlike the parent strains, betalactamase expression in the mutants was not inducible under the conditions tested. MICs of other classes of antimicrobials revealed increased MICs of fluoroquinolones against the betalactam resistant mutants. RT-PCR confirmed that smeDEF are not over-expressed in any of the mutants.
Conclusions: These data suggest that betalactam resistance is part of an MDR phenotype in some S. maltophilia mutants. The mechanism involved appears to alter drug accumulation rates, since betalactam inducer was not able to reach a concentration high enough to stimulate betalactamase expression. The mechanism is most likely to be effluxpump mediated, suggesting the existence of a previously uncharacterised efflux pump in S. maltophilia.
P1273: First complete sequence of a penicillin-binding protein of the genus Aeromonas
A. Schiefer, K. Westphal, I. Wiegand, B. Wiedemann (Bonn, D)
Objectives: Members of the genus Aeromonas are human pathogens causing several infections especially to immunocompromised patients and patients with severe underlying diseases. Antibiotic resistance is an increasing problem for effective therapeutic strategies and it is important to understand the mechansims of resistance. Betalactam antibiotics are known to bind to and inhibit PBPs which leads to a disturbance of the peptidoglycan metabolism. Until now there is only little information about the PBPs of Aeromonas veronii. Eight PBPs with different molecular masses were identified. Tn5 insertion mutants of Aeromonas that over-express betalactamases have a disruption within one of two different genes. The partial sequences of both gene products have been assigned functions as DD-carboxypeptidases. We identified the complete sequence of one of the inactivated genes encoding a DD-carboxypepti-dase with homology to PBP4 from different species, in order to create a basis for further studies of the cell wall metabolism and betalactamase induction.
Methods: Purified genomic DNA from one Tn5 insertion mutant was digested with EcoRI or SalI. The DNA-fragments were ligated into an equal linearized vector, pSU18 and transformed into E. coli DH5a. The selected colonies were investigated for recombinant plasmids with the correct insert. Sequences were determined using primer walking strategy.
Results: The sequenced fragment codes for an open reading frame of 481 amino acids with a calculated MW of 52 kDa. This MW is in good agreement with the MW of PBP4 from E. coli, Vibrio and Shewanella where the MW ranges between 52-56 kDa. The deduced protein shows homology to PBP4 from different species, a bifunctional enzyme with both DD-carboxypeptidase and D-D-endopeptidase function. All characteristic active-site fingerprints of PBPs are located in the sequence.
Conclusions: This is the first complete sequence of a PBP of the genus Aeromonas. The disruption of this pbp-gene leads to overexpression of beta-lactamases in Aeromonas veronii. This result will be the starting point for further studies of the role of PBPs in peptidoglycan metabolism and beta-lactamase induction of Aeromonas veronii.
P1274: Metronidazole resistance in nim-positive and nim-negative B. fragilis strains after several passages on metronidazole containing Columbia agar plates
R. Schaumann, S. Petzold, A.C. Rodloff (Leipzig, D)
Objectives: Recent data show an emergence of B. fragilis strains resistant against fluoroquinolones and in part against other antimicrobial agents. The aim of the present study was to investigate inducible metronidazole (MTR) resistant in B. fragilis strains.
Methods: Of 18 B. fragilis strains (including 4 nim-positive reference strains and one ATCC strain) MIC values for MTR were determined by E-test and analysed for nim-genes (A-D) by PCR. After that bacterial suspension were incubated on supplemented columbia agar plates containing MTR at twice the MIC-value of the specific strain tested and incubated under anaerobic conditions for 48 h. After incubation growing bacteria were harvested and cultivated and thereafter incubated at 4 times MIC. This procedure was repeated with increasing antibiotic concentrations. The resulting MIC-values were confirmed by E-test.
Results: The MIC-values for MTR of the four nim-positive reference strains ranged from 3 to 8 mg/L. Three clinical isolates of B. fragilis strains showed MIC-values >256 mg/L. In all 3 strains the nim-gene was detected by PCR. The B. fragilis ATCC 25285 strain was nim-negative with a MIC-value of 0.19 mg/L. The other 10 clinical isolates of B. fragilis were also nim-negative. MIC-values ranged from 0.25 to 0.75 mg/L. The nim-positive reference strains showed after few passages MIC-values >256 mg/L for MTR. After several passages on MTR containing agar, all other B. fragilis strains including the ATCC 25285 strain exhibited MIC values of 8 to 256 mg/L.
Conclusion: MTR resistance can be selected not only in nim-positive B. fragilis strains but also in nim-negative strains. This suggests that mechanisms other than nim are involved in MTR resistance. These findings underscore the importance of susceptibility testing of anaerobes even in routine laboratories.
P1275: Dissemination of nitroimidazole resistance determinants among Gram-negative anaerobic bacteria in Greece
A. Katsandri, J. Papaparaskevas, A. Pantazatou, L.S. Tzouvelekis, G.L. Petrikkos, N.J. Legakis, A. Avlamis on behalf of The Hellenic Study Group on Gram-Negative Anaerobic Bacteria
Objectives: The surveillance of the incidence of nitroimidazole resistance (nim) determinants among Gram-negative anaerobic clinical isolates in Greece.
Materials and methods: A total of 185 Gram-negative anaerobic bacteria collected from eight hospitals in Athens, Greece, were tested for metronidazole resistance using the E-test method on brucella blood agar plates. Incubation in a ChelLab Anaerobic Chamber was performed for 48 hours. Interpretation of the results was according to NCCLS guidelines. For quality control the strains B. fragilis ATCC25285 and B. thetaiotaomicron ATCC29741 were used. Strains having an MIC > 1 mg/L were screened for nim genes by PCR using the nim3/nim5 set of primers. PCR products were stained with ethidium bromide and documented under UV illumination. Sequencing was performed in an ABI 3100 genetic analyzer using the BigDye terminator kit. The Blast search programme of the National Center for Biotechnology Information was used to search the Gene Bank for significant alignments.
Results: A total of 16 isolates having an MIC > 1 mg/L were detected (four Bacteroides fragilis group, eight Prevotella spp., one Bacteroides spp. non-fragilis and three miscellaneous). Three, three, two, one and seven strains had MICs of 2, 4, 8, 16 and 32 mg/L or higher, respectively. Eight strains were positive by PCR for the 458 bp amplicon, of which three had a metronidazole MIC of 4 to 16 mg/L (lower than the current NCCLS breakpoint). DNA sequencing revealed that one, two, two and three strains harboured nimA, nimC, nimD and nimE genes, respectively. No strain was detected harbouring the nimB gene. No particular relationship was detected regarding species or MIC distribution and nim gene class.
Conclusions: Nitroimidazole resistance determinants were detected among both metronidazole resistant and susceptible Gram-negative anaerobic bacteria in Greece. This study confirms the widespread occurrence of nim genes and demonstrates that ‘silent’ nim genes can be detected by PCR in metronidazole susceptible isolates. Members of The Hellenic Study Group on Gram-Negative Anaerobic Bacteria are: A. Avlamis, C. Koutsia-Karouzou, C. Kontou-Kastelanou, A. Pangalis, E. Papafrangas, E. Trika-Grafakos, H. Malamou-Ladas and A. Vogiatzi.
P1276: Morphological changes in Clostridium difficile during exposure to metronidazole
T. Peláez, R. Alonso, L. Alcalá, J. Martínez-Alarcón, E. Cercenado, M. Rodriguez-Créixems, E. Bouza (Madrid, E)
Background: We report a high prevalence (6%) of metronidazole (MTZ) resistance in Clostridium difficile in our institution. Resistance to MTZ seems to be heterogeneous and unstable. We observed morphological alterations in the strains growing in the presence of metronidazole.
Objective: The aim of this study was to characterize these morphological changes and to study the stability of the altered phenotypes after MTZ removal.
Methods: We selected a total of 18 isolates which were resistant to metronidazole (MICs from 16 to 64 mg/L). C. difficile ATCC 9689 was also included as a control. Brain Heart Infusion broth tubes (B.H.I.) were prepared with 4 and 8 mg/L MTZ. Brucella plates without antibiotic were also used. Bacterial morphology was evaluated by microscopy (Gram-stained) and also by plate colonies. A volume of 0.1 mL, from a 0.5 McFarland culture, was used to inoculate the B.H.I. tubes. Broths were incubated at 37°C in an anaerobic chamber for 10 days. Every two days, the cultures were screened by microscopy and by serial passages performed on Brucella plates without antibiotic. Plates were further incubated under the same conditions and observed for 10 days.
Results: The ATCC 9689 C. difficile strain was not able to grow in the presence of any of the MTZ concentrations. Gram stains of MTZ-resistant C. difficile strains, grown in the presence of both assayed MTZ concentrations, revealed Gram-positive coccoid forms that co-existed, at first with typical Gram-positive bacillary forms, which became progressively predominant during the 10 day-period. Subcultures of the antibiotic B.H.I. tubes on Brucella plates rendered colonies with an atypical white appearance and round shape. Colonies reverted progressively to typical C. difficile phenotypes and after 7–10 days’ incubation in free-MTZ Brucella plates, they showed their characteristic pleomorphic and yellow–green, ground glass appearance. Gram staining of the reverted phenotypes showed the typical spore-forming, Gram-positive bacilli.
Conclusions: The presence of MTZ induces morphological changes in C. difficile that can easily be reverted in antibiotic-free medium. A similar phenomenon has been described in another genus (Helicobacter pylori), and is related to a decrease in intracellular ATP levels, although further studies are needed to explain this phenomenon in C. difficile.
P1277: A reporter gene system for the identification and characterisation of multiple antibiotic resistance (mar) in E. coli associated with altered expression of the AcrAB-TolC drug efflux pump
N. Matthiessen, P. Heisig (Hamburg, D)
Objectives: The increasing prevalence of bacterial resistance to antibiotics is a worldwide problem for the therapy of infectious diseases. Among the three basic mechanism leading to antibiotic resistance, i.e. alteration of the target, inactivation of the drug, and reduced accumulation of the drug at the target site, the latter has become the most important resistance-mechanism due to the following reasons: (i) it is the most abundant mechanism detectable in both Gram-negative and Gram-positive bacteria, (ii) it often mediates multi drug resistance (mdr) to a broad range of – unrelated – drug classes, (iii) it can favor the acquisition of additional mechanisms of resistance. Most frequently mdr is achieved by one of several mutations resulting in the deregulation of mdr efflux pump expression. The major multidrug efflux pump in E.coli AcrAB-TolC is a constitutively expressed tripartite complex consisting of a RND-type transporter AcrB, a membrane fusion protein AcrA and an outer membrane channel TolC. The expression of acrAB is regulated by a local repressor AcrR and known global regulator systems MarRAB, SoxRS and Rob. Since any mutation inactivating AcrR or a global regulator, like MarR may result in acrAB overexpression, detection of a resistance mutation requires extensive sequencing and additional susceptibility tests using different antibiotics.
Methods: Thus, a reporter gene system has been developed that senses alterations in the expression of the AcrAB-TolC efflux pump caused by induction and/or mutation/deletion of the local and/or global regulators. Briefly, the luciferase gene luc of the firefly Photinus pyralis as a reporter gene was fused to the promotor pacrAB by a modified PCR technique (SOEing) and inserted into the plasmid pBR322.
Results: Firefly luciferase as a reporter is advantageous due to the high sensitivity with no background activity, wide range of applicability, ease of use and cost efficiency. Alterations in the expression of acrAB either due to the presence of inductors or mutations in marR could be detected as increased luciferase activities by this reporter gene system.
Conclusions: Thus, this newly developed reporter gene system can be used to identify acrR mutants and to quantify alterations of the expression of the acrAB operon. Moreover, it is a useful tool to study under different environmental conditions the expression and to screen for inductors or inhibitors of the AcrAB-TolC efflux pump.
P1278: Expression of mRNA for efflux pump proteins in Pseudomonas aeruginosa strains from cystic fibrosis patients in relation to antibiotic resistance
S Islam, H. Oh, S. Jalal, O. Ciofu, N. Hoiby, B. Wretlind (Stockholm, S; Copenhagen, DK)
Objectives: Multidrug efflux system plays a prominent role in resistance to several antibiotics in Pseudomonas aeruginosa. To date, 7 different MDR efflux systems have been characterized in P. aeruginosa: MexAB-OprM, MexCD-OprJ, MexEF-OprN, MexXY-OprM, MexJK-OprM, MexVW-OprM and MexHI-OpmD. The aim of the present investigation was to discern to which extent overexpression of efflux pumps contributes to antimicrobial resistance, with particular interest on quinolone and aminoglycoside resistance in P. aeruginosa strains from CF-patients.
Methods: Twenty Pseudomonas aeruginosa isolates collected from six cystic fibrosis (CF) patients, aged 27 to 33, in 1994 (9 isolates) and 1997 (11 isolates) at the CF Center, Copenhagen, Denmark, were studied. MICs to norfloxacin, ciprofloxacin, amikacin, tobramycin, tetracycline, ceftazidime, piperacillin/ tazobactam, meropenem and imipenem were determined using E-test. The relative expression of mRNA for four efflux pump proteins was determined by real-time PCR and correlated with susceptibilities to two fluoroquinolones and seven other antimicrobial agents.
Results: A strain was considered to hyperproduce mRNA for pump proteins if cDNA level was >5x strain PAO1. MexY mRNA overproduction (18/20 strains) did not seem to achieve clinically relevant levels of resistance to quinolones but was correlated with decreased susceptibility to aminoglycosides. MexB overproduction (3/20 strains, <23× PAO1) did not mediate any significant quinolone resistance, but seemed to decrease susceptibilities to other antimicrobials. High-level overexpression of MexD (4/20) affected quinolone susceptibility. No clear correlation between the R82L alteration in 16/20 strains in NfxB (regulates MexCD-OprJ) and MexD mRNA hyperproduction was seen. MexF mRNA expressed at high levels (6/20) was correlated with resistance to quinolones and imipenem.
Conclusion: Elevated production of mRNA for the four efflux pumps tested correlated not only to decreased susceptibility to quinolones, but also to other antimicrobials including penems, aminoglycosides and beta-lactams.
Public health, surveillance and geographic information systems
P1279: International Circumpolar Surveillance of invasive bacterial diseases, 2000–2004
T. Cottle, M. Bruce, D. Parks, S.L. Deeks, T. Tam, K. Brinklov, Jensen, A. Nystedt, A.J. Parkinson (Anchorage, AK, USA; Ottawa, ON, CAN; Nuuk, GRL; Lulea, S)
Background: The International Circumpolar Surveillance (ICS) system conducts population-based surveillance of invasive bacterial diseases in Greenland (GN), Northern Canada (N Can), Northern Sweden (N Swe) and in the U.S. Arctic (Alaska [AK]).
Methods: Isolates from patients with invasive diseases caused by Haemophilus influenzae (Hi), Neisseria meningitidis (Nm), Group A Streptococcus (GAS), and Group B Streptococcus (GBS) were forwarded to reference laboratories in Alaska (2000–2004), Canada (2000–2004), Greenland (2001–2003), and Sweden (2003) for confirmation and serotyping. Clinical and demographic information were collected using standardized surveillance forms. Data reported for 2004 is preliminary.
Results: The total numbers of reported cases were 110 Hi, 43 Nm, 153 GAS, and 130 GBS. Crude annualized rates of invasive disease per 100,000 population varied by country and organism (Table 1). AK Native and N Can Aboriginal people had consistently higher rates of disease (except GBS in N Can) than non-Aboriginals. Of the 105 Hi cases that were serotyped, 20 (19%) were Hib [AK 15 cases (rate 0.48), N Can 5 cases (rate 0.80)] and age ranged from <1 to 69 years; most Hib disease occurred in persons <2 years of age (AK = 53%, N Can = 80%). Thirty-three (31%) Hi cases were serotype a (Hia) [AK 10 cases (rate 0.32), N Can 23 cases (rate 3.68)]. No Hia cases were reported in AK during 2000, 2001 and 2004 to date; in 2002 and 2003, rates in AK were 0.62 and 0.92, respectively. In N Can, Hia cases were reported during each year from 2000–2004; rates were 2.99, 7.03, 3.13, 2.34 and 2.82 respectively. Case fatality ratios were higher in AK than N Can and GN for invasive disease caused by both Hi (AK = 18%, N Can = 7.4%) and Nm (AK = 12.2%, N Can = 0%, GN = 0%).
Table 1.
Rates* of Invasive Disease, ICS Data, 2000–2004
| Country | Hi | Nm | GAS | GBS |
|---|---|---|---|---|
| AK | 2.04 | 1.00 | 4.02 | 3.43 |
| GN | 0 | 4.72 | 0.59 | 1.77 |
| N Can | 7.37 | 0.48 | 4.33 | 2.56 |
| N Swe | 0.39 | 0.39 | 0.39 | 1.97 |
annualized crude rate per 100,000
Conclusion: Aboriginal peoples of AK and N Can have high rates of invasive bacterial disease caused by Hi, Nm, GAS and GBS. Overall rates of Nm disease are higher in GN than AK, N Can and N Swe. Cases of invasive Hib disease continue to occur in children <2 years of age. Rates of Hia appear to be elevated in N Can; this trend merits further surveillance.
P1280: Enter-net Italia: integrated medical and veterinary surveillance of salmonellosis in Italy
A.M. Dionisi, P. Galetta, E. Filetici, I. Benedetti, S. Arena, L. Busani, C. Graziani, A. Ricci, V. Cibin, L. Barco, M.C. Dalla Pozza, I. Luzzi (Rome, Padua, I)
Objectives: The laboratory surveillance of Salmonella in Italy has a high level of integration among involved institutions. It is represented by the Enter-net system, coordinated by the Istituto Superiore di Sanità, which concerns notifications from human cases, food items and environmental sampling, and the Entervet network, coordinated by the Istituto Zooprofilattico Sperimentale delle Venezie, which collects information on isolations from animals and food of animal origin.
Methods: The networks use harmonised microbiological methods and share their databases. Data on Salmonella serotypes, phagetypes and antimicrobial resistance are collected with epidemiological informations. Each year, the network collects and analyzes data on about 6000 human isolates and 2000 isolates each from animals, foods and environment. More than 70% of the human isolates is represented by S. typhimurium (ST) and S. enteritidis. Subdivision within phagetypes is becoming necessary to elucidate outbreaks during investigations because, since the 1990s, some phagetype of epidemiological importance have predominated: S. enteritidis phagetype 4 and S. typhimurium phagetype 104 are the more common phagetypes among human and animal isolates. Molecular typing may be an important subtyping tool for isolates belonging to the same phagetype and Pulsed-Field Gel Electrophoresis analysis is considered the gold standard method.
Results: An accurate analysis of the Enter-net and Enter-vet databases allowed us to detect an increase during 2002 and 2003 in the prevalence of S. typhimurium non phagetypable (NT) and of an atypical monophasic strain, defined as 4,5,12: i,-, both in human cases and in veterinary samples. ST NT accounted for the 25% of the ST isolated in humans during this period and for the 20% of the ST isolated from sample of swine origin. The resistance to Ampicillin, Streptomycin, Sulphonamides, and Tetracycline is the typical profile found in a high percentage of human and swine strains such as PFGE shows the same profile of restriction for the most part of human and swine ST NT isolates.
Conclusions: The Italian surveillance integrated network for Salmonella represents an important database for the study of Salmonella infection epidemiology. Epidemiological data together with serotyping, phagetyping and molecular typing of Salmonella isolates from human and animal sources provide further information for a better estimate of risk factor for human infections.
P1281: Knowledge and attitude of Iranian healthcare worker about Crimean Congo haemorraghic fever
M. Mardani, M. Rahnavardi, M. Rajaeinejad, K. Holakouie Naieni, F. Pourmalek (Tehran, IR)
Background: Crimean-Congo Haemorrhagic Fever (CCHF) is a potentially fatal viral infection caused by a tick-born virus, nosocomial outbreaks with high mortality among hospital staff been documented. Sporadic cases occur through In 1999, 3 health care workers (HCWs) in Iran were infected with CCHF virus following contact with one suspected case of CCHF that one of them died. Systan-Baluchestan (south-east of Iran) and Isfahan (centre of Iran) provinces are now endemic area of CCHF infection in Iran. HCWs of these two provinces are frequently in contact with CCHF cases.
Materials & Methods: 191 pre-tested and self-administered questionnaires were filled by HCWs who work in admitting wards of hospitals of Systan-Baluchestan and Isfahan provinces of Iran. Collected data was analysed to determine the level of knowledge and attitude of these staff and to distinguish the predicting factors of knowledge and attitude.
Results: 82% of these HCWs had good knowledge on CCHF, while 83% showed acceptable attitude towards the disease. Knowledge is directly associated with attitude (p < 0.03). Those in higher job rank had better knowledge (p < 0.001) and higher attitude (p < 0.01). The most common used source of data on CCHF was ‘Poster and pamphlet’ (32.2%) among these HCWs. Those who used ‘Poster and pamphlets’ had higher knowledge (p < 0.05). No significant difference was seen among different sex, job or provinces groups in using ‘Poster and pamphlets’. The most common type of contact to CCHF patients was ‘Intact skin to blood’ contact in 37.3% of enrolled HCWs, while 10% had ‘Percutaneous’ contact with CCHF cases. Working in Systan-Baluchestan province was accompanied with higher risk of contact with CCHF cases (p < 0.05).
Conclusion: We conclude that enrolled HCWs in Systan-Baluchestan and Isfahan provinces of Iran had acceptable knowledge and attitude. Improvement of knowledge via ‘Poster and pamphlet’ could be an effective modality in these setting. Health authority should pay more attention on Systan-Baluchestan province to provide it with sufficient and effective universal protection to lower nosocomial transmission risk of CCHF in HCWs of this area.
P1282: Seroprevalence of rubella among puerperae in an area of North-Eastern Italy
A. Beltrame, L. Driul, G. Rorato, L. Scudeller, C. Negri, D. Marchesoni, P. Viale (Udine, I)
Objectives: The proportion of susceptible individuals represent one of the main elements of the decisional workup for a vaccination campaign. In particular, eradication of congenital rubella is achievable only when protective levels of antibodies are maintained among women of reproductive age. The aim of our study was to assess adherence to and results of rubella serological screening among pregnant women in an area of Italy, Country where vaccination is not compulsory.
Methods: Puerperae delivering at the Teaching Hospital of Udine were prospectively enrolled and interviewed by means of a standardised CRF recording demographic and obstetric data, number of living children, history of childhood viral exanthema, history of vaccination, prenatal screening for rubella and appropriate rE-testing (i.e. monthly rE-testing for IgG negative women and no additional testing for IgG positive women). Descriptive statistics were obtained; comparisons were made by t-test, Mann-Whitney or ≑ 2 tests as appropriate.
Results: From February to July 2004, 568 puerperae were enrolled. Mean age was 33 years (range 18 to 47 years). Country of origin was Italy in 494 (87%), Eastern Europe in 35 (6%), Africa in 16 (3%), America in 12 (2%), Asia in 6 (1%), Western Europe in 5 (1%). First pregnancies were 301 (37%). A history of rubella infection was reported in 314 (55.4%) and of rubella vaccination in 130 (22.9%). Prenatal screening tests for rubella was available for 414 women (73.2%), with no difference between Italians and other nationalities (73% vs 76%, p = NS). Serologic evidence of susceptibility to rubella infection was found in 44 (9.7%). The proportion of susceptible women did not vary across different ages (18–39 yrs 9.8%, >39 yrs 8.8%, P = NS) and nationality groups (Italians 9.7%, other 9.6%, P = NS). Correct rE-testing in pregnancy occurred in 365 cases (64.5%). Among 44 susceptible women, 24 (54.5%) had been pregnant at least once before (8 had two child or more). No acute infection during pregnancy was diagnosed.
Conclusion: In our study, prenatal screening rates for rubella in puerperae was unsatisfactory. Moreover a proportion of nearly 10% of susceptible pregnant women results in a high risk of rubella infection in pregnancy. No correlation was found between age, nationality and rubella susceptibility. Besides more than half of rubella susceptible puerperae had one child or more. These results also suggest a need for improved postpartum vaccine implementation.
P1283: Spatial distribution and registry based case-control analysis of Campylobacter infections in Denmark, 1991–2001
S. Ethelberg, J. Simonsen, P. Gerner-Smidt, K.E.P. Olsen, K. Mølbak (Copenhagen, DK)
Objective: To examine the potential of environmental sources contributing to Campylobacter infections in Denmark, using geographical analyses.
Methods: We analysed available information on the place of living of all registered laboratory confirmed domestically acquired cases of campylobacteriosis in Denmark over a period of 11 years. The study was performed as a register-based case control study; 15 controls for each case were selected from the national population register, individually matched on age, gender and county of residence. A total of 22,066 cases were compared to 318,958 controls and variables relating to geography and the addresses of living were analysed by logistic regression.
Results: Three factors were independently associated with an increased risk of infection: 1) Type of housing. Relative to the one-family house, there was a decrease in odds of housing typical of cities and an increase in odds of housing typical of rural areas. 2) Living in areas with a low population density. This was assessed by counting the people living in a 1 km2 square surrounding each case and control subject. Children were largely responsible for the increased odds ratio associated with areas with a low population density. 3) The municipality of residence. After adjustment for type of housing and population density the odds of living in different municipalities (274 in Denmark) varied (p < 0.0001) and there was no apparent order in low/high risk municipalities when they were visualized on a map.
Conclusions: This is the largest study so far made of the geography and type of housing of Campylobacter cases. We found that children living in non-urban areas are at increased risk of Campylobacter infections. Under the assumption that risk foods (i.e. fresh chicken in particular) are equally distributed across the country, the results indicate that exposures via animals and the environment are the sources of a substantial proportion of sporadic infections of Campylobacter among children in the countryside. Furthermore, contaminated drinking water is a likely explanation of the finding of varying risks between municipalities, since people in different municipalities have different water suppliers in Denmark.
P1284: Use of spatial analysis on a survey of H. influenzae and S. pneumoniae isolated from children in an urban area of southeastern Brazil
A.M. Monteiro, C.M. Mendes, J.L. Sampaio, C. Oplustil, P. Garbes, P. Lima, L. Pedneault, G. Camara, C.R. Kiffer (Sao Jose dos Campos, Sao Paulo, Rio de Janeiro, BR; Brussels, B)
Objectives: This study examines the spatial distribution and patterns of H. influenzae and S. pneumoniae and their resistance patterns isolated from children of an urban area in Brazil.
Methods: Spatial analyses were used on a population of children under 7 years old with symptomatic upper respiratory tract infections (URTI) submitted to routine cultures of respiratory specimens, as part of a surveillance study on the prevalence of resistance among H. influenzae and S. pneumoniae in the city of São Paulo, Brazil. Patients were only included in the study if they had a positive culture result for the above pathogens. S. pneumoniae isolates were tested against penicillin and other antimicrobials with Minimum Inhibitory Concentrations (MICs) determined by E-test methodology. Interpretative criteria used were those described by NCCLS document M100-S14. H. influenzae isolates were tested for beta-lactamase production by a chromogenic cephalosporin method. Cases and their respective resistance patterns were geocoded in a digital map and the spatial analysis technique applied was the Kernel function method in order to explore possible cluster formations.
Results: There were 111 S. pneumoniae and 146 H. influenzae isolates from this population. Only 50 S. pneumoniae cases (16 with a penicillin MIC in the intermediate or resistant range), and 59 cases of H. influenzae (8 beta-lactamase producers) were geocoded, due to apparent random geocoding technique losses. Preliminary spatial analysis method (Kernel function) based on geographical information systems (GIS) showed a possible non-random pattern located in the eastern region of the city.
Conclusion: Kernel function is an initial exploratory technique for interpolating and smoothing point events and is mainly used for identifying possible cluster formations. Further analyses are required for precisely determining the existence of S. pneumoniae and H. influenzae clusters and their related risk factors. However, it seems that there is a specific transmission pattern of bacterial pathogens within a population under elevated risk for resistance. GIS and spatial methods can be applied to better understand epidemiological patterns and to discriminate target areas for public health interventions.
P1285: Human tuberculosis in the Ile-de-Cayenne, French Guiana. Risk maps for better prevention and effective control strategies
V. Guernier, X. Deparis, J-M. Fotsing, J-F. Guégan (Montpellier, Cayenne, Orléans, F)
Objectives: In French Guiana, tuberculosis (TB) incidence appear to be great, but many problems prevent from effective TB control in this region. One way of improving TB control is to increase case-finding rates and therefore the proportion of cases treated by the identification of population at risk for tuberculosis. The aim of the study presented here is to determine vulnerability index of populations living in the Ile-de-Cayenne for tuberculosis, in order to produce risk map for tuberculosis.
Methods: A geographic information system (G.I.S.) is a tool that allows to organize and analyze data that can be referenced spatially, i.e. data that can be tied to a physical location. Many types of data have a spatial aspect, including epidemiological studies. A digital map from IGN and a Spot-5 satellite image were included in the G.I.S. as a cartographic base. In addition, data that should be used to assess the degree of insalubrity of the urban area was included, as well as demographic data that can influence transmission rate. These different layers were cross-linked to assess the vulnerability index of populations. The cartography of tuberculosis cases was combined to the vulnerability map in order to adjust the model of risk map for tuberculosis.
Results: The distribution of the 387 TB cases reported from 1 January 1996 to 31 December 2003 have been monitored by GPS using home locations of the patients. A vulnerability index of populations have been derived from a qualitative analysis of both urban landscape and socioeconomic data. Using a G.I.S. and geostatistical tools, we cross-linked variables as habitat typology (derived from air photo interpretation), population density, or type of urban sewerage systems, which are indeed indicative of existing socio-spatial inequalities. A map of tuberculosis risk among populations was established, coupling the cartography of tuberculosis cases with the cartography of vulnerability.
Conclusion: This framework will allow to reveal some transmission patterns of tuberculosis in the Ile-de-Cayenne, providing support for the development of health policies and programmes in French Guiana. It will also offer the unique opportunity to dispose of an impressive environment-health database which will serve to regional health authorities for health planification and forecasting.
P1286: Prevention of infectious diseases in the Aral Sea region
Re. Khaydarov, Ra. Khaydarov (Tashkent, UZB)
According to announcements of the Ministry of Public health of Republic of Uzbekistan - a quality of portable water is the main reason of infectious diseases in the Aral Sea Region, where fresh water resources are limited and unevenly distributed, and drinking water often contains extraordinary large numbers of pathogenic bacteria. The water disinfecting method presented in this work is based on the destructive impact of low concentrations of metal ions on bacteria in water. During the disinfection process, alloyed electrodes are placed into the water body and a current applied to the electrode causes the release of metal ions. The metal ions bind to the bacterial cell wall, causing its disruption and lyses. The efficacy of using different metal ions (Ag+, Cu2+, Au) combinations (within the limits of current drinking water regulations) for killing typhoid–paratyphoid, Legionella pneumophila, Salmonella, V. Cholerae etc. has been examined. The cultivation, culture enrichment and the testing bacteria were performed following the Standard Methods for the Examination of Water and Wastewater (American Public Health Association, 1995) for the evaluation of disinfection. Tests which were carried out by various independent labs and universities during the period of 1999 through 2003 have shown the dependence of bacteria killing time against metal ion concentration, different initial bacteria concentrations (from 103 to 1012 CFU/L), and the influence of different ion (Cl−, SO, S2−, Fe2+, Fe3+) concentrations on the disinfection process. The best disinfection is obtained by using an alloy of silver/copper/gold composition with concentrations of metals in the ratio 70–90%/10–30%/0.1–0.2%, respectively. In the Aral Sea region (Uzbekistan) water disinfecting devices that were based on the developed method were installed on several hundreds manual water pumps, that allowed to decrease community-acquired infectious diseases in this region.
Endocarditis
P1287: Epidemiology of infective endocarditis in Hungary
A. Heczey, G. Prinz, é. Bán (Budapest, HUN)
Objectives: In Hungary, statistical data of infective endocarditis (IE) is unknown; therefore, a prospective study was conducted. The goal of this investigation was to determine the most common pathogens, predisposing factors, affected valves, the mean time from the first symptoms to the established diagnosis, the incidence of vascular phenomena, the percentage when surgery is needed, and the total in-hospital mortality.
Methods: Between October 1, 2002 and September 30, 2004. 80 patients were identified and examined by one member of our Department. Patients were either hospitalized in our Institution or were encountered through infectious disease consultations. The diagnosis of infective endocarditis was made by using the modified Duke Criteria. All cases were definite IE.
Results: The number of patients: 80 (52 men, 28 women). Mean time to established diagnosis from the first symptoms: 6.28 weeks (SD: 6.8). Average age: 58.1 The distribution of isolated pathogens was: Enterococcus spp. 19 cases (23.75%); Staphylococcus aureus 15 cases (18.75%); viridans streptococci 8 cases (10%); Group B Streptococcus 5 (6.25%); Streptococcus bovis 4 cases (5%); Coagulase negative Staphylococcus 3 cases (3.75%); Streptococcus pneumoniae 2 cases (2.5%); Lactobacillus 2 cases (2.5%); Haemophilus spp. 2 cases (2.5%); plus 4 other pathogens one case each. 15 cases were haemoculture negative. The affected valves were: only mitral: 28 cases (35%), only aorta 26 cases (32.5%), only tricuspidal: 3 cases (3.75%), aortic and mitral: 18 cases (22.5%), aortic, mitral and tricuspidal: 1 case (1.25%). The predisposing factors of IE were: previous valve disease: 25 cases (31.25%), prosthetic valve: 23 (28.75%), pacemaker: 9 cases (11.25%). IE in intravenous drug users or HIV infected patients was not present in this period. Vascular phenomena were observed in 35 patients (43.75%). Cerebral embolism: 17 (21.25%), spleen infarction: 5(6.25%), pulmonary embolism 4(5%), lower extremitiy embolism 2 (2.5%). Janeway phenomen: 6(7.5%). Surgical intervention was performed in 45 (56.25%) cases. The overall mortality of IE was 22.5%, among the medically treated patients 25.71%, among the surgically and medically treated patients: 20%.
Conclusion: Enterococcus spp. became the leading pathogen of IE in adults in Hungary. Other data is not significantly different from previously published epidemiological studies.
P1288: Epidemiologic and clinical aspects of infective endocarditis in Turkey
H. Leblebicioglu, H. Yilmaz, Y. Tasova, E. Alp, R. Saba, R. Caylan, M. Bakir, A. Akbulut, B. Arda, S. Esen on behalf of the Endocarditis Study Group
Objective: The aim of our study was to establish the etiology, risk factors of infective endocarditis (IE) and determine the prognostic factors for adverse outcome during hospital admission in a Turkish population.
Material and Methods: Between January 2002 and January 2004, the clinical and laboratory features of 112 consecutive adult patients (>18 years) with a diagnosis of IE who referred to infectious diseases clinics/departments of 17 teaching hospitals in Turkey were evaluated. Cases of IE were defined according to modified Duke Criteria. Mortality was defined as death occurring within 30 days or during hospital stay period. Univariate and multivariate analysis of factors predicting a fatal outcome were performed.
Results: 112 consecutive patients presented with 101 definite and 11 probable IE episodes defined according to the modified Duke Criteria. The mean age was 45.2 ± 19.9.50% of the patients were male. 90 (60.4%) of 112 patients have risk factors for infective endocarditis and 48 (42.9%) of them have ≥ 2 risk factors. 49.1% of patients have cardiac risk factors. Blood cultures were positive in 94 (83.9%) cases. Staphylococci were the most common agents (50.0%), followed by streptococci (28.7%) and enterococi (16.0%). Native cardiac valves were present in 93 (83%) of the episodes of suspected IE. Valvuler involvement was present in 103 (92%) patients, the mitral valve, alone or in combination with other valves, was affected in 70 (62.5%). Echocardiography was able to detect vegetations in 105 patients (93.8%). The mortality was 28.6%. Three factors were independently associated with mortality; haemodialysis OR: 14.5 (95% CI: 1.5–138.2), mobile vegetation OR: 4.7 (95% CI: 1.5–15.4) and mental alteration OR: 4.1 (95% CI: 1.1–15.6).
Conclusion: Mortality is still high in IE. Our data indicate that patients with altered mental status or mobile vegetation or being on haemodialysis had poorer prognosis.
P1289: Blood culture negative endocarditis. Analysis of 59 cases
J.D. Ruiz-Mesa, J.M. Reguera, A. Alarcón, F. Miralles, A. Plata, A. Muñoz, A. Villalobos, J. Pachón, J.D. Colmenero (Malaga, Seville, E)
Objectives: To know the clinical features and possible etiologic agents of the blood culture negative endocarditis (BCNE).
Methods: We have carried out a descriptive cross sectional study in a serie of patients with Infectious Endocarditis (IE) diagnosed from January 1985 to December 2001 in two tertiary hospitals in the south of Spain excluding those who were intravenous drug user. All the patients presented a clinically definitive IE or pathologically proven according to the Duke criteria.
Results: Fifty nine (16.4%) of 359 cases of IE included were categorized as BCNE affecting in 45 (76.3%) and 14 patients (23.7%) to native and prosthetic valves respectively. The mean age was 44.5 ± 14.8 with a range from 11 to 73. Forty one (69.5%) patients were male and 18 female. The most affected valve was aortic in 33 cases (55.9%), mitral in 17 (28.8%) and both valves in 9 (15.3%). A previous valvular disease was found in 39 cases (66.1%). An infectious focus was suspected in 16 (27.1%) patients. The symptoms were fever (96%), chills (78%), sweating (65%), malaise (69%), new murmur (27.1%) and splenomegaly (26%). In a total of 23 patients (38.9%) we detected vascular phenomena and immunological phenomena were present in 9 patients (15.5%). Thirty five patients (59.32%) developed heart failure. We performed at least two blood cultures in all of patients. Antibiotics were used previously in 29 (49.1%) patients. Fifteen cases had positive serologic test for C. burnetti and 4 for Brucella. All of patients had a transthoracic echocardiogram (TTE) and 16 (27.1%) had a transesophageal echocardiogram (TEE). Valvular regurgitation were seen in 52 (88.1%)and vegetations in 37 cases (62.7%). Surgery was undertaken in 31 cases (52.5%) during the hospital admission and in 5 cases later.
Congestive heart failure was the main reason for surgery (52.5%). Surgery allowed us to confirm the etiologic agents in 11 cases (2 Aspergillus, 1 Mucor). The global mortality ratio was 20.3%.
Conclusions: 1) Antibiotics taken before a IE diagnosed is the main factor for the negativity of the blood culture. 2) Serologic tests for Brucella and C. burnetti might be considered in BCNE mainly in endemic areas. 3) The histologic and microbiologic examination of the valves after the surgery is so much important to identify the etiologic agent. 4) Molecular techniques may be an interesting alternative diagnostic test for IE caused by bacteria that usually give a negative blood culture.
P1290: Culture negative endocarditis: the value of 16S rDNA PCR and sequencing
A.A. van Zwet, S. Riemens, L. Laterveer, M. Kooistra (Groningen, Meppel, NL)
Objectives: An essential element in the diagnosis of bacterial endocarditis is the finding of positive bloodcultures. In patients with endocarditis successive negative bloodcultures can be misleading and finally at best, by exclusion, the inadequate diagnosis of a so-called ‘culture negative endocarditis’ is made. In these special cases 16S rDNA PCR and sequencing of a blood sample can be of great value as is demonstrated in the following case history.
Patient and Methods: A 64 years old man presented himself at our hospital with complaints of fatigue, fever and weight loss since a couple of weeks. Eight years ago he had received an aortic valve bioprosthesis. Under the working diagnosis of a bacterial endocarditis several blood- and a bone marrow samples were taken for culture. Incubation time was extended, however, all cultures remained negative. Since trans-esophageal-echoscopy of the valves could not confirm the diagnosis of an endocarditis, uncertainty concerning the diagnosis was growing. Finally, a 3 ml EDTA blood sample was taken for 16S rDNA PCR and sequencing. DNA was extracted from the sample by a bead-beating/silica/guanidinium thiocyanate procedure. Amplification was performed with universal bacterial primers. A positive signal was found and subsequent sequencing revealed a Bartonella species. A positive Bartonella serology (IgG titer 500) confirmed our 16S rDNA PCR finding and the diagnosis of a Bartonella endocarditis was made. Patient was treated with a combination of antibiotics.
Conclusion: Direct detection of bacterial DNA in an EDTA blood sample using broad-range PCR and subsequent DNA sequencing is an important diagnostic tool, particularly helpful in cases where bacteraemia is suspected but bloodcultures nevertheless remain negative.
P1291: Evaluation of a real-time universal 16S rRNA gene PCR and sequencing method for diagnosis of infective endocarditis directly from heart valve tissue
M. Marín, P. Muñoz, M. Rodríguez-Creixems, L. Alcalá, C. Sánchez, E. Bouza on behalf of the Gregorio Marañón Hospital Endocarditis Study Group
Objective: Traditionally, infective endocarditis (IE) has been microbiologically diagnosed by culture. In recent years, the diagnostic importance of PCR has been proven. Our objective is to evaluate the usefulness of a real-time universal PCR method followed by sequencing in explanted heart valves for IE diagnosis, in the routine of a clinical microbiology laboratory, compared with conventional microbiological culture of blood (BC) or heart valve tissue (HVT).
Methods: One hundred and three HVT from 100 patients were studied. Twenty-two HVT belonged to 20 patients with definite EI. Eighty patients were free of IE and their 80 HVT were included as negative controls. Universal 16S rRNA gene PCR was made with primers PSL and P13P by real-time PCR in a Light-Cycler instrument using Sybr-green and 35 PCR cycles. Positive samples were subsequently sequenced for identification. The results obtained were compared with those of microbiological cultures of HVT and BC. Analytical sensitivity was assessed by extracting DNA from tenfold dilutions of a suspension of Streptococcus oralis spiked in a culture-negative HVT.
Results: All but one of the HVT from patients free of IE gave negative PCR results. All culture-positive samples were also positive by PCR and in all cases microorganisms identified from HVT by this molecular method matched with those isolated by culture methods. The causative organisms of IE were diagnosed only by PCR in two patients whose microbiological cultures were negative. All culture-positive samples amplified before 27 PCR cycles. One PCR positive sample that amplified at cycle 29, belonged to a patient without EI and gave a mixed sequence, being considered contaminated. The median time of analysis to obtain a PCR result was 2.5 hours and to a bacterial identification by sequencing was 2 days. The analytical sensitivity of this assay was 100 cfu/mg. Sensitivity, specificity, positive predictive and negative predictive values of this real-time PCR method were respectively: 100%, 98.76%, 95.6% and 100%.
Conclusions: This method of universal real-time PCR followed by direct sequencing applied to resected heart valves has proved to be more sensitive, specific and rapid than conventional culture methods. This real-time assay makes it possible to predict IE by the number of cycles of PCR at which samples of HVT amplify. Supported in part by \\‘Red Española de Investigación en Patología Infecciosa’ (REIPI - C03/14).
P1292: Clinical spectrum of endocarditis in patients with cancer: a case-control evaluation of culture-positive (CPE) and culture-negative (CNE) disease, 1994–2004
S.W. Yusuf, S. Ali, A. Tong, J. Swafford, J.B. Durand, D.P. Kontoyiannis, K.V. Rolston, I.I. Raad, A. Safdar (Houston, USA)
Background: Spectrum of endocarditis in patients with cancer is not certain. We sought to evaluate characteristics of culture-positive (CPE) and culture-negative (CNE) in patients with cancer.
Methods: A retrospective evaluation of all transthoracic (TTE) and transesophageal echocardiograms (TEE) during these 10 years was undertaken after obtaining IRB approval. A positive result included demonstration of vegetation along the heart valve.
Results: Forty-five (7%) of 654 patients had a positive result; 26 (58%) had positive concurrent blood culture, whereas 19 patients (42%) blood cultures remained sterile. Among CPE, Staphylococcus species (9 Staphyococcus aureus, and 6 coagulase-negative staphylococcus) were common, followed by 4 Enterococcus spp. The median age was 55 ± 18.6 years (range, 14 to 84) in 30 (67%) men and 15 (33%) women. Hematologic malignancies were observed in 23 (51%); among solid-organ cancers gastrointestinal malignancies were common (N = 8; 18%) followed by lung cancer (N = 4; 9%). All 45 patients had TTE, while TEE was done in 22 (49%) of 45 patients. All 16 (100%) patients with negative TTE had a diagnosis of endocarditis made on TEE, whereas one patient with positive TTE had a negative TEE (p < 0.0001). Eighteen (78%) of 23 patients with CVC had CPE compared with 8 (36%) of 22 without CVC had a positive blood culture (OR = 6.30; 95% CI: 1.69–23.53; p < 0.0062). Similarly, 24 of 45 patients with endocarditis had received antineoplastic therapy within 60 days of infection diagnosis (OR = 3.24; 95% CI: 0.94–11.12; P = 0.0619). Presence of severe neutropenia (<500 cells/uL) was evenly present in CPE (n = 6 of 26) and patients with CNE (n = 4 of 19). Interestingly, embolization to brain more common in patients with CNE (n = 7 of 19; 37%), compared to 3 (12%) of 26 with CPE (P = 0.07). Septic emboli to lung occurred in patients with CPE (4 of 26, 15%; p < 0.04); all 4 patients had endocarditis due to Staphylococcus aureus. Seven of 8 patients who were candidates for valve replacement surgery underwent operation; of these 6 had CPE (3 had valvular Abscesses). Endocarditis was attributed to death in 6 (31.6%) of 19 CNE patients, whereas only 1 (3.9%) death was attributed to endocarditis in CPE group (n = 26; p < 0.03).
Conclusions: CNE was more severe disease, which may be related to delayed diagnosis and possibly suboptimum antimicrobial therapy. Presence of CVC has appeared as an important predictor of CPE in these high-risk cancer patients.
P1293: Outcomes of treatment of infective endocarditis with home intravenous antibiotic therapy
M.A. Goenaga, C. Garde, M. Millet, J.A. Carrera, E. Arzellus, A. Cuende (San Sebastian, E)
Objective: Evaluate outcomes of patients (pts) with EI treated with home intravenous antibiotic therapy (HIVAT).
Methods: IE cases referred for HIVAT were identified though our HIVAT registry. Information was extracted from hospital records with follow up of all cases.
Results: 23 cases of IE, in 22 pts, were treated with HIVAT. 86% male, 14% female. Age range 26–89 yrs, mean 59 yrs. Underlying cardiac pathologies were present in 14 cases. All pts were evaluated by an internal or infectious disease physician in hospital. All patients were medically stable, 2pts had required surgical intervention. 7 (30%) cases were on prosthetic valve (5), defibrillator wire (1) or pacemaker wire (1). Causative microorganisms were S. viridans (10), MSSA (3), S. epidermidis (3), E. faecalis (2), M. morganii (1), G. haemolysens (1), P. aeruginosa (1) blood culture negative (2). All cases, less one, the treatment was initiated in hospital. Antimicrobial agents used were ceftriaxone (7), vancomicin (5), penicillin G (3), ampicillin (3), cloxacillin (3), ceftazidime (1), teicoplanin (1). Aminoglycosides were associated in 10 cases. The mean hospital length of stay was 14.5 d (range 0–36 d) and at home 28 d (range 2–125 d). The mean 65% (range 14–100%) of treatment was realized at home. 22 cases (95.6%) ended treatment at home with good clinical response and 1 case (blood culture negative, valve prosthetic) return to hospital due fever. 4 cases (17%) had iv access problems. One patient develop a benign intracraneal hypertension. There were no serious IE or HIVAT complications. In the follow up (median 46 months, range 5–89 m) 21 pts are cured, one pts died not related IE.
Conclusions: In our group, after a carefully inpatient selection, pts with IE can be treated with safe at home. Prosthetic valve disease or other microorganisms than S. viridans had not worse outcomes.
P1294: Non-toxigenic Corynebacterium diphtheriae as a cause of bacterial endocarditis in children with congenital heart defects
M.G. Dove, R.P. Loxton, E.J. Olivier, B. Prinsloo (Pretoria, ZA)
Introduction: Non-toxigenic strains of C. diphtheriae have been increasingly recognised as a cause of invasive disease. Bacteraemia and endocarditis caused by these strains have been reported with increasing frequency. Other invasive diseases such as septic arthritis, splenic abscesses and mycotic cerebral aneurisms have also been described. To date at least 67 cases of non-toxigenic C. diphtheriae causing endocarditis have been reported worldwide. Most of these cases were in either native or prosthetic heart valves and in IV drug abusers.
Objective: We report on 4 cases of infective endocarditis caused by non-toxigenic C. diphtheriae in patients with underlying congenital heart defects, occurring at the Pretoria Academic Hospital over the past 7 years.
Method: Endocarditis was confirmed by the presence of diphtheroid organisms in the blood cultures of all the patients. A final diagnosis was made on microscopy (Gram and Albert's stains), culture on blood agar and Hoyle's medium, in-house biochemical tests substantiated by API CORYNE (Biomerieux) An elek test for toxin production was performed on all isolates.
Results: The ages of the 4 patients were between 3 and 16 years. Positive blood cultures (Bactec 9240 System) were obtained from all patients on multiple occasions. Characteristic ‘Chinese letter’ arrangements of the bacilli were seen on both Gram and Alber's stains, as were metachromatic granules. In-house biochemical tests validated by API CORYNE confirmed all organisms to be C. diphtheriae var gravis. Elek tests in all cases indicated no toxin production. Sensitivities to a number of antibiotics (ampicillin, penicillin, erythromycin, gentamicin, piperacillin and cefuroxime) were determined by the Kirby-Bauer disc diffusion method. With the exception of penicillin and Ampicillin resistance in one patient, all antibiotics tested were sensitive. Patients were treated with penicillin and gentamicin parenterally and all survived without complications.
Conclusion: Non-toxigenic C. diphtheriae is an infectious pathogen, and detection of coryneform bacteria in the blood can no longer be dismissed as contamination and must be investigated. Failure to recognise this pathogen can delay final diagnosis and initiation of appropriate chemotherapy. Species identification is important as mortality differs with the different biotypes. The importance of this organism as emergent pathogen should not be underestimated.
P1295: Aerococcus urinae – a rarely detected pathogen of infective endocarditis
M. Slany, P. Pavlik, J. Cerny, T. Freiberger (Brno, CZ)
Objectives: Aerococcus urinae is a rarely reported pathogen, possibly due to difficulties in the identification of the organism. A. urinae is a Gram-positive coccus that grows in pairs and clusters as alpha-hemolytic colonies on blood agar. Because of these characteristics A. urinae is often misidentified as a streptococcus, enterococcus, or staphylococcus. In addition, there were also reported several cases of blood culture negative infections due to A. urinae. Most infections are mild, but serious ones such as endocarditis and septicemia can occur. To our best knowledge, the total number of thirteen cases of infective endocarditis including eight fatal cases have been described in the world literature so far.
Methods: Silica based DNA isolation from aortic valve tissue sample and broad range 16S rRNA PCR followed by sequencing analysis was used.
Results: Here we report a case of blood culture negative and culture negative aortic valve endocarditis caused by A. urinae in 69-year-old male. Patient was successfully treated with surgical aortic valve replacement and ceftriaxon (4 g daily) applied for 12 weeks.
Discussion: Most patients infected with A. urinae are elderly males with predisposing conditions who present initially with urinary tract infections (UTI). However, our patient developed infective endocarditis in the absence of UTI symptoms.
Conclusion: Broad range 16S rRNA PCR is shown to be a power tool in detection of bacterial pathogens in culture negative cases of infective endocarditis. This work was supported by grant of the MZ CR No. 209775.
P1296: Trends in antimicrobial susceptibilities of viridans group streptococci isolated in patients with infective endocarditis from 1990 to 2003
F. Marco, Y. Armero, C. García de la Mària, E. Amat, M. Almela, A. Moreno, X. Claramonte, N. Pérez, C.A. Mestres, J.M. Gatell, M.T. Jiménez de Anta, J.M. Miró (Barcelona, E)
Objective: To know the current incidence of resistance to penicillin and other antibiotics in viridans group streptococci (VGS) isolated in patients with endocarditis in our institution from 1990 to 2003.
Methods: A total of 107 consecutive VGS strains isolated from patients with infective endocarditis (IE) were included in the study. Antimicrobial agents tested were: penicillin (PN), ampicillin (AM), ceftriaxone (CR), imipenem (IM), vancomycin (VA), teicoplanin (TP), erythromycin (EE), clindamycin (CD), telithromycin (TL), quinupristin/dalfopristin (Q/D), linezolid (LZ), ciprofloxacin (CP), moxifloxacin (MX), levofloxacin (LV) and gatifloxacin (GA). MICs were determined following the NCCLS recommendations. Tolerance to glucopeptides and high-level resistance to gentamicin (GN) and streptomycin (ST) was also studied. In addition, the combination of PN, LZ or LV plus GN was investigated in selected isolates (Penicillin MIC >1 mcg/ mL) by time-killing curves and the checkerboard method.
Results: The species identified were as follows: S. bovis (26%), S. oralis (25%), S. mitis (21%), S. sanguis (7%), anginosus group (8%) and 13% miscellaneous (S. mutans, S. salivarius, S. gordonii and S. parasanguis). MICs 50/90 (mcg/mL) were: PN (0.06/0.5), AM (0.12/1), CR (0.06/0.5), IM (0.015/0.12), VA (0.5/1), TP (0.06/0.12), ER (0.12/256), CD (0.06/256), TL (0.007/2), Q/D (0.5/2), LZ (1/1), CP (1/2), MX (0.12/0.25), LV (0.5/1), GA (0.25/0.25). Overall, 18.7% of the isolates had reduced PN susceptibility (MIC range: 0.25–4 mcg/ml). Among these, S. mitis, S. oralis and S. sanguis isolates had the highest prevalence of PN resistance, whereas all S. bovis and S. mutans isolates were susceptible. Only 3 isolates (2 S. mitis and 1 S. oralis) showed a MIC of 4 mcg/ml, but it was notable that they were identified during the last 3 years. All isolates were susceptible to VA, TP and LZ. Tolerance to VA and TP was found in 83 isolates (77.6%). High-level resistance to GN and ST was detected in 1 (0.9%) and 11 (10.3%) isolates, respectively. Among the three combinations tested in PN resistant isolates, only LZ + GN showed synergistic activity.
Conclusion: Decreased susceptibility to betalactam antibiotics is a common feature in VGS isolated in patients with infective endocarditis in our institution.
Enteric infection
P1297: Standard inoculum for a broth microdilution method for antimicrobial sensitivity testing of Campylobacter jejuni
V. Gindonis, M. Hakkinen, H. Nummi, A.-L. Myllyniemi (Helsinki, FIN)
Objectives: Antimicrobial resistance in campylobacters has become a subject of great concern as resistant strains are more and more commonly found in samples of both human and animal origin. C. jejuni ATCC 33560-strain has been confirmed to be a suitable quality control (QC) strain for campylobacters. However, so far only tentative ranges of minimal inhibitory concentrations (MICs) for this strain are available. In this study a broth microdilution method (VetMICTM) was validated for C. jejuni. The manufacturer's method guideline for VetMICTM Camp-plates (SVA, Sweden) was modified to standardize the size of the inoculum.
Methods: A nephelometer and colony counts were used to measure inoculum densities. Inoculum sizes were determined for bovine intestinal isolates and repeatedly for the QC strain using both pre-incubation and direct inoculation. The QC strain was tested in 28 independent VetMICTM-procedures using pre-incubation in broth and 25 times by direct inoculation.
Results: No correlation was detected between colony counts and McFarland levels by nephelometer. It was concluded that standardizing the inoculum size could not be based on this, for other bacteria commonly used method. However, it was noticed that growing campylobacters in Brucella broth for 24 hours yielded consistently approximately 10 to the 8th CFU/ml. The mean cell density for bovine isolates was 8.2 log10 CFU/ml with the standard deviation (SD) of 0.3 when campylobacters were pre-incubated in broth (n = 140). By direct inoculation the mean cell density was 8.9 log10 CFU/ml (SD 0.2, n = 156). Cell densities for the QC strain were 8.3 log10 CFU/ml (SD 0.2, n = 24) and 8.9 log10 CFU/ml (SD 0.2, n = 27) respectively. Pre-incubation in broth yielded more accurately the target inoculum size of 10 to the 8th CFU/ml. All VetMICTM-results of the QC strain for erythromycin, nalixidic acid, oxytetracyclin and gentamicin by both methods were within tentative QC ranges given by NCCLS. Slight differences could be seen in distributions of results between methods. Based on the results we set QC limits of 2–8 mg/l for ampicillin and 0.25–0.5 mg/l for enrofloxacin to be used in our laboratory for the modified method.
Conclusion: The results suggest that the modified broth microdilution method presented here is reliable and reproducible for testing antimicrobial resistance in C. jejuni.
P1298: Diagnosis of Salmonella gastro-intestinal infections by LPS-based ELISA, a follow-up study
M. Strid, K. Mølbak, T. Dalby, K.A. Krogfelt (Copenhagen, DK)
Objectives: Diagnosis of human gastrointestinal salmonellosis is traditionally done by fecal culturing and agglutination assays. An automatized ELISA is an obvious candidate for a more reliable, fast and easy method of diagnosis. An indirect ELISA employing LPS antigens was developed and evaluated.
Methods: Two automatized indirect ELISAs employing commercially available Salmonella enteritidis (SE) LPS, respectively Salmonella typhimurium (ST) LPS were developed. Both IgA, IgM and IgG antibodies were detected. Sera from 153 Danish patients diagnosed with infection by Salmonella enteritidis and from 150 Danish patients diagnosed with infection by Salmonella typhimurium were obtained and analysed. Blood-samples were collected at approximately one, three, six and twelve months after onset of salmonellosis. Cut-off values were defined as the mean plus two standard-deviations when analysing sera from 164 healthy, Danish blood-donors.
Results: The developed ELISAs proved reliable, and sensitivities of 96% for both analyses were reached when sera collected less than 35 days after onset of salmonellosis were analysed. The found detection rates at approx. one, three, six and twelve months after onset were: 95%, 85%, 62% and 40% for the SE-patients and 89%, 55%, 40% and 16% for the ST-patients. Crossreactions between the two types of anti-LPS antibodies were found, as well as few crossreaction to antibodies against other gastrointestinal bacteria.
Conclusion: The LPS-based ELISA was found to detect recent cases of gastrointestinal salmonellosis with a sensitivity of 96%. This makes it a likely candidate for routine diagnosis. The method was incapable of discriminating between SE- and ST-patients; but a mix-ELISA incorporating both antigens could however be a very promising means of a combined high-sensitivity, fast serologic diagnosis of these two, the commonest, Salmonella infections.
P1299: Evaluation of the LOUIS test for the presumptive reporting of Salmonella from selective agars employing hydrogen sulphide as a discriminatory character and as a confirmatory test from chromogenic agars
G. Wilson (Stirling, UK)
Objectives: The LOUIS test is a rapid biochemical based screening protocol, which can be used to facilitate the rapid identification of Salmonella. The test has previously been evaluated for use with Lactose non-fermenting colony types. The purpose of this study was to evaluate its use with media that employ hydrogen-sulphide as a discriminatory character and also to evaluate its use in colony confirmation from Salmonella chromogenic media.
Methods: Over a 6 month period 500 out patient stool samples were inoculated onto 4 different agar types: SSI enteric (Statens Serum Institute, Denmark), Salmonella-Shigella, SMID2 (both bioMerieux, France) and ABC (LabM, England). A selenite F broth (Oxoid, England) was inoculated and after incubation cultured to the 4 agars. This gave rise to 161 H2S positive and 93 chromogenic positive isolates for testing. Isolates were inoculated to 1 ml of sterile saline to give a suspension equivalent to at least McFarland No. 4. Suspensions were dispensed (0.2 ml) in to 3 test tubes, a nutrient agar slope (Oxoid, England) and a purity plate. Individual reagent tablets (Rosco, Denmark) were then added to the tubes and incubated at 37°C/3 h. A new algorithm (Table 1) was developed for use with H2S positive colonies and the original algorithm (Table 2) was used for isolates from chromogenic media. The API rapid ID32E gallery (bioMerieux, France) was used for full biochemical confirmation. Polyvalent and group specific sera (Murex, England) were used for serological identification.
Results: The application of the new algorithm to isolates from SSI and SS agars correctly identified 96 isolates as Salmonella and discarded 61 isolates, leaving 4 unknown isolates for full identification. None were significant. When applied to isolates from chromogenic media the original algorithm correctly discarded 9 isolates as non-significant. Of the 83 Salmonella profiles produced 2 were false positives (1 ONPG negative Hafnia alvei and 1 atypical E. coli). One unknown isolate required full identification (Serratia spp). Overall the LOUIS test had a sensitivity and specificity for Salmonella identification of 99.9 (CI of 97.9–100) and 97.4 (CI of 91-99.3).
Table 1.
LOUIS test algorithm for H.S positive colonies
| LDC | ONPG | URE | IND | Possible indentification | STEP 1 | STEP 2 | |
|---|---|---|---|---|---|---|---|
| + | − | − | − | Salmonella | Confirm by Serology (O and H antigens) | Positive | Negative serology. |
| Full biochemical ID | Discard | ||||||
| − | − | − | − | Possible LDC negative Salmonella | API rapid ID32E (4h) | Confirm with serology | Negative serology. Discard |
| − | − | + | + | Proteus spp or Morganella morganii | Discard | ||
| − | − | + | + | ||||
| − | + | − | − | Citrobacter spp | Discard |
* Discard any other combination or reactions
Table 2.
Original LOUIS test algorithm
| LDC | CNPG | URE | IND | Possible identification | STEP 1 | STEP 2 | |
|---|---|---|---|---|---|---|---|
| + | + | − | − | + E. coli | Discard | ||
| + | − | − | + | ||||
| − | − | + | + | Proteus spp or | Discard | ||
| − | − | + | − | Morganella morganii | |||
| + | − | − | − | Salmonella | Confirm by serology (O and H antigens) | Positive API rapid ID32E (4h) | Negative serology: Discard |
| − | − | − | − | Shigella spp (possible LDC negative Salmonella) | API rapid ID32E (4h) | Confirm by serology | Negative serology: Discard |
| − | − | − | + | Shigella spp | API rapid ID32E (4h) | Confirm by serology | Negative serology: Discard |
| − | + | − | − | Shigella sonnei or Sh. dsyenteriae 1 | Confirm by serology | Positive API rapid ID32E (4h) | Negative serology: Discard |
± Discard any other combination of reactions
Conclusions: Early presumptive reporting of Salmonella can be achieved with a positive LDC profile with confirmatory serology 3 h after isolation on chromogenic media and also on media that utilise hydrogen sulphide as a discriminatory character.
P1300: Comparison of three Clostridium difficile toxin assays, C. difficile GDH-antigen EIA, C. difficile GDH-PCR, bacterial culture and cytotoxicity assay for the diagnosis of C. difficile-associated diarrhoea
K. Sachs, G. Ackermann, A.C. Rodloff (Leipzig, D)
Objectives: Clostridium difficile- associated diarrhoea (CDAD) remains the leading cause of nosocomial-acquired diarrhoea. Prolonged hospital stay and diagnostic and therapeutic procedures due to CDAD cause additional costs. The present study had the aim to assess the value of different assays to detect C. difficile infections among patients with nosokomial diarrhoea.
Methods: 377 stool samples from patients from a tertiary care hospital were investigated for C. difficile toxins using three Toxin A/B assays (RIDASCREEN, r-biopharm; Premier C. difficile Toxin A&B, Meridian; Immunocard Toxins A&B, Meridian), a C. difficile GDH antigen EIA (C. DIFF CHEK, Techlab), C. difficile GDH PCR, stool culture and cytotoxicity assay. Toxin assays and GDH-antigen EIA were performed according to the instructions of the manufacturer. Stool culture was done using cycloserin-cefoxitin-fructose agar.
Results: The toxin A/B positive results received from the three toxin assays were as follows: RIDASCREEN, 55 (14.6%); Premier Toxin A&B, 87 (23%); Immunocard Toxin A&B, 106 (28%). GDH-antigen was detected in 121 (32%) stool samples using the C. DIFF CHEK and in 100 (26.5%) specimens by PCR. C. difficile grew from 66 stool samples (17.5%). Specific cytopathic effect in cell culture due to toxin B of C. difficile was detected in 137 (36.3%) stool preparations.
Conclusion: Higher sensitivity and faster protocols make new C. difficile tests more attractive for routine diagnostics. The performance of the Immunocard Toxin A&B assay and the two GDH-antigen tests were excellent. Nevertheless, C. difficile culture is still necessary for further investigations as for toxigenicity, antimicrobial susceptibility and typing.
P1301: Evaluation of three commercial tests for the rapid diagnosis of Clostridium difficile-mediated antibiotic-associated diarrhoea: a routine laboratory perspective
Y. Miendje, O. Vandenberg, F. Crockaert, M. Gerard, A. Dediste, P. Retore, G. Mascart, G. Zissis (Brussels, B)
Objectives: Clostridium difficile is the most important infectious cause of nosocomial diarrhoea in industrialized countries. Early diagnosis is associated with better prognosis, therefore rapid laboratory diagnosis is highly desirable. Therefore, we have compared under routine conditions three commercial Clostridium difficile toxin immunoassays (EIA) (Premier Toxins A&B – Meridian (PTAB), Immunocard Toxins A&B – Meridian (ICTAB) and C. difficile Toxin A test – Oxoid (TAO)) allowing a rapid diagnosis of C. difficile associated diarrhoea (CDAD) directly on the stool specimen.
Methods: From January 04 to November 04, all stool samples submitted for routine investigation of C. difficile infection to our hospital laboratory serving four University Hospitals in Brussels, Belgium were evaluated with the 3 EIAs. For comparison purpose, the combination of the culture carried out on cycloserine-cefoxitin-fructose agar and the demonstration of the toxigenicity in cell lines Hep2 performed directly from the stool or from the C. difficile isolate ('second look') was considered as the gold standard.
Results: Of the 234 stool specimens from 188 patients included in the study, 25 were found positive (by the toxigenicity assays) for C. difficile, giving an overall recovery rate of 10.7%. The sensitivity and specificity of the tested methods were respectively, 88% and 97.6% for the PTAB test, 92% and 100% for the ICTAB test and 60.0% and 99.5% for the TAO test. The positive and negative predictive values found were respectively, 81.5% and 98.6% for the PTAB, 100.0% and 99.1% for the ICTAB and 93.8% and 95.4% for the TAO.
Conclusion: These data suggest that the PTAB, ICTAB and TAO assays are acceptable tests for the rapid diagnosis of CDAD due to high negative predictive value but are not equivalent to the cytotoxin assay. Because, the ICTAB is the most sensitive and specific EIA evaluated, we recommend this test for a rapid screening of patient with nosocomial diarrhoea.
P1302: Multicentre evaluation of a new rapid Tox A+B test (Meridian ICTAB) for the diagnosis of Clostridium difficile-associated diarrhoea
J. Van Broeck, J. Verhaegen, D. Van Kerkhoven, B. Van Meensel, M. Delmée (Brussels, Leuven, B)
Objective: We evaluated the performances of a new rapid enzyme immunoassay (Tox A+B) for the detection of Clostridium difficile Toxins A and B (Immunocard Toxin A&B – ICTAB, Meridian Cincinnati, Ohio USA) on stool specimens received in two university hospitals. Tox A+B was compared to a toxin A latex immunoassay (Tox A) (C. difficile toxin A test, Oxoid Basingstoke England) and to the standard procedure in use in the laboratories which comprises faecal cytotoxin detection and faecal toxigenic culture.
Methods: Stools were from inpatients older than 2 years and suffering from nosocomial diarrhoea following antimicrobial- or chemotherapy in two large university hospitals (St Luc-UCL hospital in Brussels and Gasthuisberg-KULeuven hospital in Leuven). Tox A+B and Tox A were performed according to the respective manufacturer's instructions. Faecal tissue culture assay was performed using HeLa cells. Toxigenic culture consisted of culture of faeces on CCFA medium followed, if the culture was positive, by toxin detection on colonies using the Tox A and Tox A+B assays. Toxigenic culture was considered the gold standard.
Results: A total of 533 stools from 398 patients were included. Culture was positive in 62 cases (11.6%) and 50/62 (80.6%) strains were shown to be toxigenic. The following table summarizes the main results. The assays’ sensitivity, specificity, PPV and NPV were respectively: Cytotoxin assay : 56%, 100%, 100%, 95.6% Tox A: 66%, 98.6%, 82.5%, 96.5% Tox A+B: 88%, 99.1%, 91.7%, 98.8% Among the 50 stools with a positive toxigenic culture, 24 were positive for the three toxin assays, 9 were positive for Tox A and Tox A+B, 4 for Tox A+B and faecal cytotoxin, 8 for Tox A+B only and 7 for none of the three tests. Among the 4 specimens with positive Tox A+B and negative culture, one was from a patient who had a positive stool culture ten days before.
| Toxigenic culture |
Faecal cytotoxin |
Tox A |
Tox A + B |
|||
|---|---|---|---|---|---|---|
| + | − | + | − | + | − | |
| Pos N = 50 | 28 | 22 | 33 | 17 | 44 | 6 |
| Neg N = 483 | 0 | 433 | 7 | 476 | 4 | 479 |
Conclusion: Tox A+B was the most sensitive and highly specific assay for the detection of C. difficile toxins in faecal specimens. Moreover, compared to other detection methods, Tox A+B is particularly fast, easy to perform and has the added benefit of detecting both toxins.
P1303: A new approach to laboratory diagnostic of infectious gastroenteritis
B. Olesen, D.S. Hansen, H. Tybring, A. Hansen, K.E.P. Olsen, B.G. Bruun (Hillerød, Copenhagen, DK)
Objectives: In order to optimize use of laboratory facilities and ensure flexibility in relation to current epidemiology, a new approach to laboratory diagnosis of infectious gastroenteritis was applied: choice of examinations for the various pathogens was defined by the demographic, clinical and epidemiological information submitted on the request form.
Methods: From April 1-August 31, 2004, hospitals and general practitioners submitted a request form with the following information together with the stool sample(s): 1) acute diarrhoea or persistent diarrhoea (duration > 2 weeks); 2) bloody stools; 3) recent history of foreign travel; 4) > 2 persons with similar symptoms in the surroundings; and 5) nosocomial infection. Based on this information, microbiological analyses were performed according to predefined algorithms. Examination for Salmonella, Campylobacter, Yersinia, Shigella, Clostridium difficile and Vibrio spp. were done by culturing. Verotoxin-producing E. coli (VTEC), enteropathogenic E. coli (EPEC), enterotoxigenic E. coli (ETEC) and enteroinvasive E. coli (EIEC) were identified by PCR of virulence genes and serotyping. Rota- and adenovirus were detected by antigen tests, norovirus by PCR and parasites by microscopy.
Results: In the study period our department examined 2,463 stool samples from 1,242 patients. Preliminary results indicate that 501 patients had acute diarrhoea, 13% of these were infected with Campylobacter, 3% with Salmonella, and 1% each with VTEC, ETEC and rotavirus; 573 patients had persistent diarrhoea, 4% of these were infected with Campylobacter, 1% each with Salmonella and Giardia lamblia; 260 patients had a history of recent foreign travel, 8% of these were infected with Campylobacter, 5% with Salmonella, 4% with Giardia and 3% with ETEC; 77 patients had bloody stools, 16% of these were infected with Campylobacter, 7% with VTEC and 1% with Salmonella. An outbreak of VTEC O157:H7 occurred among visitors to a local petting farm. Sheep and goats on the farm were found to be colonized with O157:H7 with the same distinct PFGE pattern as isolates from the patients.
Conclusions: The new approach to microbiological examination of stools according to predefined criteria has a number of advantages: better patient management; collection of microbiological data on defined patient groups, hopefully optimizing use of laboratory facilities; and flexibility regarding adaptation to current epidemiological knowledge.
P1304: Genetic profiling, clonal diversity and antibiotic susceptibility in Escherichia coli O157:H7 isolated in Cataluña (Spain)
A. Benavente, N. Larrosa, E. Roselló, J. Blanco, A. Echeita, R. Bartolomé (Barcelona, Lugo, Madrid, E)
Objectives: To detect the major virulence genes, the clonal diversity, phage types and antibiotic susceptibility evolution of 30 Escherichia coli O157:H7 strains isolated from patients with gastroenteritis in Catalunya for the last 13 years (1990–2003).
Methods: The main virulence genes from E. coli O157:H7 were characterized by classic PCR with specific primers for the phage-encoded cytotoxins stx1 and stx2 genes (shiga toxins 1 and 2), eae-gamma1 (specific O157:H7 intimin) and the plasmidic ehxA gene (enterohemolysin). We also studied the chromosomal encoded eae (intimin) gene and the plasmid encoded pO157 gene (pCDV419 plasmid). The serotypes were confirmed by multiplex PCR, detecting the somatic antigen O157 (rfbEO157 gene) and the flagellar antigen H7 (fliCH7 gene). The epidemiologic subtyping was performed by pulsed field electrophoresis (PFGE) method. Cleavege of the agarose-embedded total genomic DNA was achieved with XbaI. Phage typing (16 strains) was carried out in the Enterobacterial Department of the Carlos III Health Institute. The sensitivity of the 30 strains was assayed with 23 antibiotics using the Kirby-Bauer method.
Results: PCR showed that 21 (70%) isolates harboured both stx1 and stx2 genes (stx1+/stx2+) whereas 8 (26.6%) only carried stx2 gene (stx1-/stx2+). Only 1 (3.3%) strain carried the stx1 gene at all (stx1+/stx2-). The specific O157:H7 eae-gamma1 gene, the pCDV419 plasmid, the enterohemolysin, as well as the somatic and flagellar antigens, were detected in all the strains (100%). The PFGE studies showed 29 diferent macrorrestriction patterns (1 strain non-typeable), while phage typing showed 6 (3 strains non-typeable) profiles: phage types 2, 8, 14, 31, 54 and 87. Nevertheless, 9 strains (56.2%) could be grouped in only 3 phage-types. The 60% (18/30) of the strains were susceptible to all the antibiotics assayed. Moreover, 8 (26.6%) were resistant to cotrimoxazol, 8 (26.6%) to doxicycline, 4 (13.3%) to ampicillin, 3 (10%) to nalidixic acid, 2 (6%) to cloramphenicol and 1 (3.3%) to kanamycin. The 26.6% (8 strains) were multirresistant.
Conclusions: In our geographical area, like in other countries the greatest part of the E. coli O157:H7 strains, encodes the shiga toxin 2 and more rarely the shiga toxin 1. Phage typing is a less discriminative than PFGE as a typing method because strains with the same phage type, showed different macrorrestriction PFGE profiles. All the strains remain quite sensitive.
P1305: Eight new E. coli O groups that include verocytotoxin-producing E. coli
F. Scheutz, T. Cheasty, D. Woodward, H.R. Smith (Copenhagen, DK; London, UK; Winnipeg, CAN)
Objectives: Serotype and investigate virulence factors and occurrence of the two temporary E. coli O group strains OX3 and OX7 and six presumptive new O groups in order to establish them as test strains for O groups O174-O181.
Methods: Serotyping using agglutination tests and immunochemistry, and virulence typing of isolates from our strain collections combined with a review of the literature on new types.
Results: O174 and O175 were originally isolated from cases of human diarrhoea. The O174 strain is negative for known virulence genes, the O175 strain is positive for astA and daaC. Six new strains produced Verocytotoxin and were positive for vtx1, vtx2 or both genes. Additional virulence genes associated with diarrhoeal disease in humans were found in four of the strains. O176 and O177 strains were isolated from calves, O178 and O181 strains from meat, the O179 strain from human bloody diarrhoea, and the O180 strain from swine.
Conclusion: The agglutination tests, the epidemiological and clinical data were more than sufficient for the establishment of O groups O174 through O181 as part of the existing serotyping scheme for E. coli and will serve to characterise strains of E. coli belonging to both diarrhoeagenic E. coli (DEC), extraintestinal pathogenic E. coli (ExPEC) and nonpathogenic E. coli.
P1306: Ongoing cluster of Salmonella typhimurium PT U291 infections in Austria, starting September 2003
D. Schmid, P. Much, A.M. Pichler, C. Kornschober, C. Berghold, F. Allerberger (Vienna, Graz, A)
Objectives: S. typhimurium phage type U291 has not been documented in Austria before Sept. 2003. Since then, this phage type has spread to all but one of the nine provinces in Austria. We describe this cluster.
Methods: From Sept. 2003 till Nov. 2004 187 laboratory-confirmed cases of infection with PT U291 were identified by the National Reference Laboratory. Investigations revealed another 45 epidemiologically related clinical cases, and included consultation of the other European Salmonella Reference laboratories via Enternet.
Results: The cluster consists of three outbreaks and a further 100 laboratory confirmed cases without common links. The three outbreaks (one hotel-associated continuous source outbreak, one family point source outbreak, and one wedding celebration-associated point source) comprised 30, 7, and 86 clinical cases, respectively. The hotel-associated outbreak comprised 24 laboratory-confirmed cases and 6 epidemiologically related clinical cases, and occurred between 05.10 and 11.10.2003. The cases were Austrian and German tourists, and employees of a hotel in a western Austrian province. Case series data indicated that illness was primarily associated with tiramisu, but no epidemiological or microbiological evidence for this hypothesis was provided. The family outbreak affected seven persons between 21.10 and 22.10.2003 in the same province. Cases were linked by common exposure to a self-made tiramisu on 20.10.2003. Food examinations revealed no salmonella. The third outbreak having affected two eastern neighbouring provinces in Austria was associated with a wedding celebration (Turkish ethnicity) on 4 Sept. 2004. A total of 86 of approx. 230 wedding guests met the case definition criteria. Non-human isolates of the causative agent were found in samples of abraded walnuts and pistachios, and also in the sample of the refiner obtained from that bakery having provided the wedding cake, which was the most likely source of infection. No Salmonella was identified in the food samples taken from the restaurant, where the weeding celebration has taken place.
Conclusions: In absence of S. typhimurium PT U291 in most of Europe, the increasing occurrence of this previously rare phage type in Austria points to locally produced food as the culprit. This cluster also underlines the importance of the zoonoses directive, requiring adequate epidemiological and microbiological studies of food-borne outbreaks, for disease prevention.
P1307: Prevalence of intestinal parasites and enteropathogens micro-organisms in outpatients of a central are of Madrid, Spain
M.A. Orellana, M. Aramendi, G. Galera, M.T. Sanchez, L. Garcia, C. Manzanares, M.A. Amerigo (Madrid, E)
Background: The intestinal parasitic and enteric infections are a public health problem that must be controlled in each area. The inmigration and international travels have increased the number of these process.
Objectives: To know the prevalence of intestinal parasites and enteropathogens microorganisms infections in outpatients of Center Area from Madrid (Spain).
Methods: −3.875 coprocultures and 4.391 parasite studies were examined between February 2.003 and September 2.004. COPROCULTURE: The stools were inoculated in XLD, Campylosel and CYN-agar(Biomerieux), and Selenito F (Pronadisa). Adenovirus and rotavirus were detected by immunocromatographie COMBI-STRIP(FASTIA). PARASITE STUDY: The stools were concentrated using formalin-ethyl-acetate(Biosepar, Germany); criptosporidium was detected by CRYPTO-STRIP method (FASTIA) and enterobius vermicularis using perineal samples(Graham test).
Results: The prevalence of positive coprocultures was:9.47%. The frequency was: Salmonella 191(52.0%), Campylobacter sp 131(35.7%), Rotavirus 26(7.1%), Adenovirus 13(3.5%), Shigella sp.2(0.5%), Yersinia sp.2(0.5%) and Aeromona sp.2(0.5%). Among Samonellas the most frequent was S. enteritidis (111), followed by S. group D(51), Salmonella sp.(10), S. paratyphi B(9), S. group C(5), S. paratyphi A(3) and S. choleraesuis (2). The overall prevalence of intestinal parasitisms was: 17.10%. The frequency was: Blastocystis hominis 377 (56.9%), Entamoeba Coli 109 (16.4%), Giardia intestinalis 106 (16.0%), Criptosporidium parvum 16 (2.4%), Iodamoeba butschlii 11 (1.6%), Ascaris lumbricoides 10 (1.5%), enterobius vermicularis 8(1.2%), Estrongiloides estercolaris 6 (0.9%), Endolimax nana 6 (0.9%), Himenolepsis nana 4 (0.6%), Tenia sp 4 (0.6%), E.histolytica/dispar 2 (0.3%), Trichuri trichura 2 (0.3%)and Anquilostoma duodenale 1 (0.1%). Two or more parasites/sample were found in 5.9%.
Conclusion: Salmonella was the enteropathogen more frequent isolated, followed by Campylobacter. The high prevalence of intestinal parasites in our area is a public health problem and controls are necessary to improve the prevention and contain their spread.
P1308: Serotypes and antibiotic resistance of enteric Salmonella isolated in outpatients in a central area of Madrid, Spain
M.A. Orellana, G. Galera, M.T. Sanchez, M. Aramendi, L. Garcia, C. Manzanares, M.A. Amerigo (Madrid, E)
Objective: To study the resistance patterns and serotypes of enteric Salmonella isolated in outpatients in Center Area of Madrid (Spain).
Methods: 305 enteric Salmonellas strains were isolated from coproculture between January 2002 to September 2004. Identification and susceptibility were performed by Walkaway MicroScan System(DADE-Behring)according to NCCLS recommendation. The antimicrobial agents tested were: Ampicillin/ Sulbactam(A/S), Gentamicin(Gm), Cotrimoxazol(T/S), Piperacillin(Pi), Meropenem(Me), Amoxicillin/A.clavuláinico(AMC), Ciprofloxacin(Cp), and Ampicillin(Am). The seropyping was tested using antisera for flagelar and somatic antigens(Difco and Biorad).
Results: The serotypes of Salmonellas isolated were: S. enteritidis 162 (53.1%), S.group D 78 (25.6%), S. paratyphi B 28(9.2%), Salmonella sp.21 (6.9%), S. choleraesuis 7 (2.3%), S. group C 6 (1.9%) and S. paratyphi A 3 (0.9%). The resistance to antimicrobials tested was: Am 20.9%, T/S 3.9%, Gm 2.3%, A/S 20.9%, Pi 20.3%, Ti 20.9%, AMC 4.9%, Cp 0.6% and Me 0%. The 75.4% of the strains studied were susceptible to all the antibiotics tested. The most frequent antibiotic resistance patterns were: A/S, Ti, Pi, Am: 18.8%; A/S, Ti, Pi, AMC, Am:2.9%; A/S, T/S, Ti, Pi, Am:1.6% and 8.2% showed a different resistance pattern. Among the different serotypes the susceptibility to all antibiotic tested was: S. enteritidis 82.7%, S. group D 75.6%, S. paratyphi B 25%, Salmonella sp. 76.2%, S. choleraesuis 100%, S. group C 100% and S. paratyphi A 33.3%.
Conclusions: S. enteritidis was the serotype isolated more frequent. The 75% of Salmonellas studied were susceptible to all antibiotics tested. Ampicillin, Ampicillin/Sulbactam, Piperacillin and Ticarcillin were the antimicrobials less sensitive. The antibiotic resistance pattern more frequent was: A/S, Ti, Pi, Am. The serotype with more resistance to antibiotic tested was S. paratyphi B.
P1309: Distribution of Salmonella enterica serovar Enteritidis phage types in the Slovak Republic during 1995–2004
L. Majtanova, V. Majtán (Bratislava, SVK)
Objectives: In the last few decades, Salmonella enterica serovar Enteritidis (S. Enteritidis) has emerged as a major cause of food-borne illness worldwide. Nearly 90% of human salmonelloses in the SR are caused by this serovar. The object of this study was to describe the distribution of S. Enteritidis phage types which occurred from January 1995 to October 2004 in the SR.
Methods: Two thousand five hundred and sixty six strains were phage typed during 1995–2004. The strains were isolated from both human and food sources. The phage types were identified according to Ward et al. (1987) in the National Reference Center for phage typing of Salmonellae (head: Dr. L. Majtáinová).
Results: 2481 strains (96.7%) were typeable and belonged to 23 different phage types (PT). PT8 (62.8%) predominated in the SR. Majority from outbreaks as well as from sporadic cases were caused by strains of this PT. The second most frequently isolated phage type was PT4 (9.2%). Further phage types according to their occurrence were PT6 (4.0%), PT2 (3.9%), PT1 (3.8%), PT13a (2.1%), PT15 (1.2%), PT9a (0.7%), PT21 (0.6%). Other phage types have been occurred with low frequencies in all sources investigated. In addition, strains of RDNC (react but not conform to designated phage types) and untypeable (UT) were isolated at frequencies of 5.5% and 3.3%, respectively.
Conclusion: The distribution of phage types among sporadic cases of Enteritidis isolates was similar to that among outbreak-associated cases. This may suggest the common reservoirs for both cases of salmonelloses. From the public health view it as necessary to control on an international basis the spread of infection caused by this serovar as well as the spread of the individual phage types. This surveillance can be helpful, together with molecular typing techniques, to clarify the epidemiology of S. Enteritidis.
Acknowledgements: This work was supported by Science and Technology Assistance Agency under the contract No. APVT-21–052602.
P1310: Non-typhoid Salmonella bacteraemia in a Danish county: a population-based prognostic follow-up study
K.O. Gradel, H.C. Schønheyder, L. Pedersen, H.T. Sørensen, R.W. Thomsen, M. Nørgaard, H. Nielsen (Aalborg, Aarhus, DK)
Objectives: We performed a population-based prognostic follow-up study of Non-typhoid Salmonella (NTS) bacteraemia patients in North Jutland County, Denmark. Data on age, co-morbidity, clinical presentation, medicine use, and microbiological factors were evaluated as prognostic determinants.
Methods: In North Jutland County, Denmark, all bacteraemias are recorded in a research database, from which we extracted all 111 patients with NTS bacteraemia during a 10-year period, 1994–2003. The Hospital Discharge Registry (all discharge diagnoses), the Prescription Registry (redemption of prescription drugs), the Central Population Registry (data on domicile, migrations, and deaths) and medical hospital records were used as data sources, all linked by the unique Danish personal identification number. Medical records could not be retrieved from one patient and one was a non-Danish resident. The primary outcomes were mortality within 30 and 180 days of the first NTS positive blood sample.
Results: The incidence rate (mean 0.22/100,000 person years) increased steadily from age 40–49 years (0.17) until age beyond 90 years (1.46). Twelve (11%) and 24 (22%) patients died within 30 and 180 days, respectively. The 15 patients in whom a secondary focus was diagnosed did not have significantly higher 30-d (OR = 0.6 (95% confidence intervals 0.02–4.7)) or 180-d (OR = 1.4 (0.3–5.3)) mortality. The 30-d mortality rates (MRs) were 2/51 (3.9%), 5/41 (12.2%), and 5/17 (29.4%) in patients having co-morbidity scores (Charlson index) of 0, 1–2, and ≥3, respectively (p = 0.014), a distinctive trend maintained after 180 d (0 p: 3/ 51 (5.9%); 1–2 p: 12/41 (29.3%); ≥3 p: 9/17 (52.9%) (p = 0.0001)). Patients with major gastro-intestinal (GI) disorders did not have higher mortality (30-d OR = 0.7 (0.03–6.3); 180-d OR = 1.2 (0.2–5.5)). The 180-d MRs and density of bacteraemia measured by numbers of positive blood culture bottles were positively correlated. No other microbiological factors (e.g., serotype, antibiotic resistance) were associated with mortality.
Conclusions: The presence of co-morbid diseases was a major determinant of mortality, whereas age per se, microbiological factors and GI related conditions seemed less important.
P1311: Food poisoning outbreak at a bank cafeteria: the investigation report
N. Hanna, A. Sarafian, M. Mokbel, Z. Daoud, S. Adib (Beirut, LBN)
An investigative group was called on 26/5/04 by the Ministry of Public Health to investigate a food poisoning outbreak which had occurred at the central office of one of the biggest banks in Lebanon. The first symptoms of gastro-enteritis compatible with food poisoning appeared among bank employees in the evening of 17/4/04. On that day, many employees had eaten at the bank cafeteria. All of those with signs and symptoms shared the fact that they had eaten the main dish of ‘poulet aux nouilles’. In at least one case, the ‘poulet aux nouilles’ was the only food item consumed at the cafeteria. Of the 32 contacted persons, 26 people eventually reported signs and symptoms compatible with food poisoning; The attack rate for this outbreak was thus very high at 81%, suggesting a very large infective dose ingested in the incriminated dish. Most commonly reported signs are shown in Table 1. In one case, severe septicemia occurred. These findings are compatible with acute salmonellosis. The diagnosis of salmonellosis was confirmed with stool and blood cultures within 48–72 hours after hospital admission of first cases. Investigation of the source: All 18 kitchen workers were subjected to a sampling of rectal and nasal mucosa to be cultured at the laboratory of the Saint-Georges Hospital (HSG) in Beirut. Results confirm the absence of Salmonella carriage in all. At the time of the outbreak, water was provided from a tanker company. Water samples were repeatedly done on May 18 and 21 yielding high fecal coliform counts but no Salmonella. Different food samples were collected from different batches. A portion of left-overs of the ‘poulet aux nouilles’ dish which was sent out on 18/5/04 revealed the presence of Salmonella on 7/6/04, thus confirming the contamination of the dish. As per the chicken analysis, two specimens heavily grew Salmonella from the surface and internal tissues of raw chicken. Genetic studies have been done on the different isolated strains from chicken, leftovers, and patient, showing clear genetic similarity (Salmonella enteritidis) and suggesting therefore the same origin: chicken.
Frequency of signs and hospitalization following the outbreak (N = 32)
| SIGNS/SYMPTOMS | n | % |
|---|---|---|
| Diarrhea | 23 | 88.5 |
| Fever | 22 | 84.6 |
| Abdominal pain | 21 | 80.8 |
| Vomiting | 16 | 61.0 |
| Loss of consciousness | 3 | 11.5 |
Conclusion: Based on our results, the source of contamination was raw chicken. It is important to protect the public from contaminated raw chicken, and to investigate sources of contamination at the chicken farm level.
P1312: An outbreak of gastroenteritis due to Aeromonas hydrophila
M. Carev, D. Tandara, P. Rizvan, Z. Barisic, K. Sisko Kraljevic, E. Borzic (Split, HR)
Objectives: Our goal was to report an outbreak of Aeromonas hydrophila gastrointestinal infection due to consumption of contaminated water, in the rural area of Dalmatia County, Croatia. This report warns about diagnostic difficulties and importance of finding Aeromonas in both water and human stool samples.
Methods: Our epidemiologist personally interviewed 30 symptomatic and 36 asymptomatic villagers. Four human stools (13.3%) from the diseased were analysed on: Salmonella spp., Shigella spp., Yersinia spp., Vibrio spp., Campylobacter spp., Rota and Adenoviruses, intestinal protozoa and helmint eggs. Six water samples were examined bacteriologically, virologically and parasitologically.
Results: All diseased were drinking water from local drinking water distribution system, with pipelines very distant from the water source. Main symptoms were abdominal pain, watery diarrhoea and tiredness. Most of the patients recovered completely after 48 hrs and were only treated symptomatically. 28 persons (78%) from control group didn't drink that water at all. All stool samples were negative on viruses, parasites and bacterial pathogens. The result of water analyses was negative for viruses and intestinal parasites. Bacteriological analyses of first water sample showed that the number of aerobic mesophilic and psychrophilic bacteria per ml was higher than standard. A. hydrophila was isolated from Cefsulodin Irgasan Novobiocin (CIN) agar (>1800 CFU/100 ml). Further analysis of the last two stool samples included additional testing of pink colonies from CIN agar. They were transferred on Kligler Iron Agar and incubated at 37°C and 25°C. Oxydase and catalase tests were positive and Api 20E and BBL crystals confirmed isolates as A. hydrophila. Stool and water isolates showed same susceptibility patterns for antibiotics (resistance to amoxicillin and cefazolin). This was the first confirmed isolation of these bacteria as cause of an outbreak in our County. After cleaning, chlorination and connection to new water source, water samples were in accordance to the standards. There were no new cases.
Conclusion: Identical biochemical and antimicrobial susceptibility patterns of human and water strains, with positive clinical findings and epidemiological data showed us that this small outbreak was due to A. hydrophila. This conclusion was supported by negative tests for other pathogens. Real prevalence of this pathogen is still unknown and probably underestimated.
P1313: Antimicrobial resistance and molecular typing of Salmonella isolates from food
S. Valdezate, M. Arroyo, R. González, R. Ramiro, S. Herrera-León, M.A. Usera, J.A. López Portolés, A. Echeita (Madrid, E)
Objectives: To study the clonal relationship and the antimicrobial resistance showed by strains of different serotypes of Salmonella spp. isolated from food for human consumption during a one-year period.
Methods: Antimicrobial susceptibility to 23 antibiotics was determined by disk diffusion in 380 Salmonella spp. isolates submitted to LNRSSE from all Spanish regions with the following distribution by serotypes: Enteritidis (n = 176), Typhimurium (n = 60) and other serotypes (n = 144). Molecular epidemiology of Enteritidis and Typhimurium serotypes was investigated using PFGE and computerized numerical analysis of the data. Phage typing was also performed.
Results: Analysis was carried out with strains without epidemiological link. Analyzed Enteritidis strains (n = 96) were mainly from eggs and derived (27%) and from poultry and derived (24%), detecting 16 phage types (PT) with predominance of PT1 (39%). They were identified 9 pulsetypes, with a similarity genetic range of 81–96%, emerging a frequent clone (74%). Analyzed Typhimurium strains (n = 53) were from sausage and cold meat (21%), pig and derived (15%), and from poultry and derived (15%), detecting 12 DTs, with predominance of DT104 (28%) and U302 (19%). They were identified 13 pulsetypes, with a similarity genetic range of 64–86%, emerging a frequent clone (34%). Antimicrobial resistance rates (NCCLS, M100-S13) for the strains of Enteritidis, Typhimurium, and other 49 different serotypes (n = 115) were, respectively: ampicillin (8, 62 and 14%), spectinomycin (99, 87 and 100%), streptomycin (1, 53 and 59%), gentamicin (1, 4 and 0%), tobramycin (1, 4 and 0%), amikacin (1, 0 and 0%), netilmicin (1, 4 and 0%), nalidixic acid (41, 22 and 14%), tetracycline (16, 72 and 31%), sulphonamide (7, 62 and 21%), trimethoprim-sulphamethoxazole (7, 19 and 14%) and cloramphenicol (0, 51 and 9%). All the strains were susceptible to the other tested betalactams and fluorquinolones.
Conclusion: High clonality of food strains of Enteritidis serotype, showing higher discrimination index (ID) the phage typing (0.81) than PFGE (0.44) with a high nalidixic acid resistance (42%). High efficiency of PFGE in the serotype Typhimurium, with similar ID in both techniques (0.86), noteworthy resistances higher than 50% for ampicillin, streptomycin, tetracycline, sulphonamide and cloramphenicol. Strains of other serotypes showed minor resistance rates.
P1314: A survey on drug resistance in Shigella organisms in 3 hospitals in Tehran over a six month period
L. Chamani, M.A. Noyan Ashraf, S. Asadi (Tehran, IR)
Objectives: Shigellosis is a common disease in Asian countries esp in warm months. Knowledge of the pattern of resistance in a given population which changes by time is useful. We designed this study to evaluate drug resistance in Shigella spp in 3 hospitals of Tehran in order to find the suitable empirical antibiotic therapy in shigellosis.
Methods: It was a cross sectional prospective study in three different hospitals of Tehran from March to April 2003. Fresh stool samples of patients with dysentery were cultured on MacConkey agar and Hectoen enteric agar and incubated overnight at 37 degrees. Lactone negative colonies were transferred to triple sugar iron agar and lysine-iron agar slants and reincubated. Those with characteristic reaction (alkaline salt, acid butt, and no gass) were tested biochemically and then serologically identified with shigella grouping and typing antisera. Drug Resistance was evaluated in cultured Shigella organisms by disk diffusion susceptibility testing.
Results: Shigella spp. Were cultured in 96 of 1476 stool cultures (6.5%). The antibiotic susceptibility testing results were as bellow:- Shigella spp. Were completely(100%) sensitive to Ciprofloxacin, Ofloxacin & Norfloxacin and 96% sensitive to Nalidixic Acid. 1. They were 100% sensitive to Ceftriaxone and Cefotaxime but 85% to Ceftizoxime and 80% to Cefazolin. 2. There was 100% resistance to Ampicilline and 85% resistance to Cotrimoxazole.- They were 54% sensitive,12% resistant and 34% intermediatly resistant to Gentamicin.
Conclusion: It seems that new Fluoroquinolones for adults and Nalidixic Acid for chidren are suitable empirical antibiotic therapy of shigellosis in Tehran. Ceftriaxone and Cefotaxime are suitable for severe cases and hospitalized patients and ceftizoxime & cefazoline may be useful alternatively. Ampicilline and Cotrimoxazole should not be used for treatment any more. Supported by the BIB, Avesina Research Institute, Tehran, Iran.
P1315: Severe pseudomembranous colitis mimicking an acute abdomen in elderly patients
L. Legout, L. Bernard, D. Gasselin, M. Assal, P. Rohner, P. Hoffmeyer (Geneve, CH; Paris, F)
Background: Pseudomembranous colitis is a life threatening complication of broad spectrum antibiotic therapy caused by Clostridium difficile. The frequency of pseudomembranous colitis with potential fatal outcome is underestimated especially in elderly patients.
Patients and methods: We report 5 cases of pseudomembranous colitis in elderly patients who had an adynamic ileus mimicking an acute abdomen. C. difficile was identified by toxins and culture. Plain films of the abdomen and CT were performed for all patients.
Results: 5 females (median age: 83 years- range: 76–95) were admitted in hospital for pneumonia (n = 1), osteomyelitis (n = 2), diarrhoea (n = 1), elective orthopaedic surgery (n = 1) and received a betalactamin (n = 4), or clindamycin (n = 1). Their co-morbidity were lupus (n = 2), diabetes mellitus (n = 1), cancer (n = 1), renal chronic failure (n = 1). They had fever (n = 5), bad general condition (n = 5), cramping abdominal pains (n = 5) following by an acute abdominal (n = 5). The diarrhoea was only present at the beginning of medical history for 2 patients. The biological exams found:white-cell count: 30.7 G/L (15.4–49.7); neutrophils polynuclear count: 25.8 G/L (10.141.2), platelets count: 481 G/L (313–622); C-reactive protein: 235 mg/l (180–300); creatinine:129 μmol/l (82–270); urea: 8.5 mmol/l (4–17.8). The liver tests and chest x-ray were all normal. Plain films of the abdomen showed a megacolon (n = 2). Abdomen CT suspected a volvulus of caecum (n = 1), diverti-culitis (n = 1), found wall thickening of the transverse colon (n = 4), ascites (n = 1) but no perforation. 2 patients had have an abdominal laparoscopic exploration. After 4 days of treatment with metronidazole, the outcome was fatal for 2 patients due to heart failure.
Discussion and conclusions: The diagnosis of pseudomembranous colitis must be evocated in elderly patients especially if an history of antibiotic therapy even a short course (for example perioperative prophylaxis) is found and cramping abdominal pain is associated with a high white cell count (> 25 G/L). If surgical treatment is required, the overall mortality increase.
P1316: Characterisation of Campylobacter jejuni/coli strains isolated in Serbia and Montenegro
B. Miljkovic-Selimovic, L. Ng, D. Woodward, L. Price, T. Babic, M. Ratkovic-Jankovic, L. Ristic, B. Kocic (Nis, CS; Winnipeg, CAN)
Objective: Campylobacter jejuni (C. jejuni), and Campylobacter coli (C. coli), represent the main cause of bacterial diarrhoea in developed countries and one of the most frequent causes of enterocolitis in developing countries. After C. jejuni infection, severe chronic sequelae may occur, such as reactive arthritis, post-infective neuropathy, Guillain-Barree syndrome (GBS) and Miller-Fisher Syndrome (MFS) In addition, some serotypes are more often associated with GBS and MFS. Serotyping based on heat stable antigens, (HS, Penner serotyping) is methods for the investigation of the clonality of the C. jejuni/coli strains isolated in patients with diarrhoea and in patients with post-infective neuropathy. In addition, there is lack of evidence about data related to serotype distribution for some geographical areas and also for GBS associated strains.
Methods: In this study, we have characterized a strain of thermophilic Campylobacter isolated in a patient with GBS, 37 strains of thermophilic Campylobacters isolated in patients with diarrhoea in Nis, and 6 strains from the collection of the Institute for Immunobiology and Virology “Torlak”, Belgrad. Strains presumptively identified as campylobacters were differentiated to the species level by a combination of biotyping tests and by the use of a PCR-based RFLP test. HS serotyping was performed using a passive hemagglutination test using erythrocytes sensitized with heat extracted antigens and antisera.
Results: The GBS associated strain was identified as Campylobacter jejuni, biotype II, O19. Among the isolates from Nis, C. jejuni dominated over C. coli and biotype I dominated over other biotypes. In these C. jejuni strains, diversity of HS serotypes were detected (O:1, O:2, O:2,66, O:3, O:3,50, O:4 complex, O:6,57, O:8, O:8,17, O:9,21,58, O:10, O:11…). as well as among C. coli strains (O:4,28,32, O:14,34, O:24, O:34, O:49, O:64,66). In the 6 strains collected at Torlak, all isolates were identified as C. coli. All of these isolates were identified as biotype I and three different HS serotypes were found (O:14,34, O:34, O:49).
Conclusion: The investigation of HS serotypes in isolated C. jejuni and C. coli strains, confirmed their clonal diversity indicating epidemiologcaly unrelated origin of the strains. Detected HS serotype (O:19) of the GBS associated C. jejuni, is the most often described one in this post-infective complication.
P1317: Tigecycline compared with imipenem/cilastatin in the treatment of complicated intra-abdominal infections
E.J. Ellis-Grosse, E. Loh on behalf of the Tigecycline 301 Study Group
Objective: Due to their diverse bacteriology and emergence of bacterial resistance, treatment of complicated intra-abdominal infections (cIAI) represents a clinical challenge. The efficacy of tigecycline (TGC) monotherapy, a novel, expanded broad-spectrum glycylcycline, was compared with imipenem/cilastatin (IMI/CIS) in adult hospitalized patients with cIAI.
Methods: In this double-blind, phase 3, multinational trial, patients were stratified by disease severity (APACHE II score ≤15 vs >15 but <31), and randomly assigned to IV TGC (100 mg loading, then 50 mg q12h) or IV IMI/CIS adjusted for body weight (500/500 mg q6h for ≥70 kg) for 5–14 days. Clinical response at test-of-cure (TOC, 12–44 days after therapy) for microbiological evaluable (ME) and microbiological modified intent-to-treat (m-mITT) populations were co-primary efficacy endpoints in which cure/failure responses were determined.
Results: Of 825 mITT patients who received more than 1 dose of study drug, 621 (75%) comprised the m-mITT cohort (309 TGC, 312 IMI/CIS) and 502 (61%) were ME (247 TGC, 255 IMI/CIS). Treatment groups were balanced with respect to demographic/ baseline medical characteristics. Patients were predominately males (65%) with a mean age of 44 years. The primary diagnoses for the mITT group were complicated appendicitis (52%), perforation of intestine (10%), gastric/duodenal perforation (10%), cholecystitis (8%), and intraabdominal abscess (8%). The median duration of therapy was 7 days. For the ME group, clinical cure rates at TOC were 80.6% (199/247) for TGC vs 82.4% (210/255) for IMI/CIS (95% CI = −8.4, 5.1; p < 0.001). Corresponding clinical cure rates for the m-mITT cohort were 73.5% (227/309) for TGC vs 78.2% (244/312) for IMI/CIS (95% CI = −11.0, 2.5; p < 0.001). The most commonly reported adverse events for TGC and IMI/CIS were nausea (31.0% and 24.8%) and vomiting (25.7% and 19.4%).
Conclusions: TGC is an expanded-broad-spectrum IV glycylcycline with activity against gram-positive, gram-negative and anaerobic pathogens, including strains resistant to commonly used antibiotics. TGC met statistical criteria for non-inferiority to the comparator IMI/CIS and appears to be safe and effecacious in the treatment of hospitalized patients with cIAI.
P1318: Detection of viral, bacterial and parasitological RNA or DNA of nine intestinal pathogens in faecal samples archived as part of the Infectious Intestinal Disease Study: assessment of the stability of target nucleic acid
C.F.L. Amar, C. East, J. Gray, E. Maclure, J. McLauchlin (London, UK)
Objective: The purpose of this study was to apply PCR based procedures to assess the stability of pathogen specific nucleic acid sequences present in frozen and archived faecal samples of the English Infectious Intestinal disease (IID).
Methods: Faecal samples were collected from cases and controls as part of the IID study and were stored as frozen suspensions for eight to 12 years. Samples were selected from the archive where either Cryptosporidium, Giardia, Salmonella, Campylobacter, enteroaggregative Escherichia coli (EAggEC), enterotoxigenic Clostridium perfringens, rotaviruses, noroviruses or sapoviruses had been previously detected. A generic nucleic acid extraction method to recover RNA or DNA was used. Complementary DNA was generated from RNA by reverse transcription with random priming. Block based and real time PCR assays were used to amplify and detect gene fragments from each of these pathogens.
Results: The percentage of target detected was as follows: Giardia duodenalis 68%, Cryptosporidium 96%, Campylobacter 98%, Salmonella 98%, enterotoxigenic C. perfringens 34%, EAggEC 93.3%, rotavirus 95%, norovirus 73% and sapovirus 85%.
Conclusion: This study has shown that nucleic acid could be extracted and specific sequences amplified and detected from archived faecal samples. The IID archive therefore represents a valuable resource for further studies, especially the investigation of the samples where no pathogens had previously been detected.
P1319: Norovirus associated gastroenteritis at a tertiary hospital in Greece
P. Karabogia-Karafillidis, M. Orfanidou, A. Strouza, E. Vagiakou-Voudris, H. Malamou-Lada (Athens Papagos, GR)
Objectives: To report the first laboratory diagnosed cases of Norovirus associated gastroenteritis at a tertiary hospital in Athens, Greece.
Methods: During two months period (1/9/04–1/11/04) 45 stool specimens obtained from 44 patients and one healthcare worker with acute gastroenteritis were investigated for the presence of enteropathogens (Salmonella, Shigella, Campylobacter, Yersinia, E. coli O 157-H7, C. difficile) by conventional methods. Norovirus was detected by an EIA, antigen assay (IDEIA TM Norovirus, Dako Cytomation) which differentiates Norovirus genotypes G1 and G2. Moreover EIA was applied in stool specimens of 10 healthy controls.
Results: Noroviruses were detected in 15/45 (33.3%) patients with acute gastroenteritis. None of the healthy controls was found to be positive for Noroviruses. G1 was defined in two (2/ 15, 13%) outpatients. Both G1+G2 were detected in three (3/15, 20%) hospitalized patients while G2 was defined in 10 (10/15, 66.7%). Two were outpatients and three were hospitalized in various wards. Moreover, the five remaining cases (three inpatients, one visitor and one health worker) were observed in the Nephrology ward in one week period. Salmonella enteritidis and C. difficile were isolated in four patients.
Conclusions: These are the first laboratory diagnosed cases of Norovirus associated acute gastroenteritis in Greece. Most of the cases were hospital-acquired infections caused mainly by G2 Norovirus. An outbreak was observed at the Nephrology department. No false positive results were found in the healthy controls. ELISA is a rapid and useful test for early diagnosis of Norovirus gastroenteritis and prevention of Norovirus spread in the hospital environment.
P1320: Intracellular expression of B subunit of heat-labile enterotoxin of ETEC in Saccharomyces cerevisiae
M. Ahangarzadeh Rezaee, A. Rezaee, A. Salmanian, M. Moazeni, S. Najar Peerayeh, M. Arzanlou (Tehran, IR)
Objectives: Heat-labile enterotoxin (LT) of ETEC, consists of A and B subunits. Heat-labile enterotoxin B subunit (LTB) has been used in many scientific applications. LTB is a subunit vaccine candidate against diarrhoea caused by enterotoxigenic E. coli. It has been already expressed in several bacterial and plant systems. In this study, we expressed LTB in yeast Saccharomyces cerevisiae.
Methods: In order to construction of yeast expressing vector for LTB protein, the eltB gene encoding LTB was amplified from a human origin enterotoxigenic E. coli DNA by PCR. The expression plasmid pLTB83 was constructed by inserting eltB gene into pYES2 shuttle vector immediately downstream of the GAL1 promoter. The new construct was introduced into S. cerevisiae cells. The cells were induced in the presence of 2% galactose.
Results: Immunoblot analysis showed the presence of LTB in yeast lysate and indicated that the yeast-derived LTB protein was antigenically indistinguishable from bacterial LTB protein. The monomeric LTB protein molecules (11.6 KDa) were the dominant molecular species detected in yeast total soluble protein.
Conclusions: Since, whole recombinant yeast introduced as a new vaccine formulation, expression of LTB in S. cerevisiae may offer an effective and inexpensive strategy to protect people especially in developing countries against ETEC and cholera.
P1321: Expression of B subunit of cholera toxin in Saccharomyces cerevisiae
M. Arzanlou, A. Rezaee, N. Shahrokhi, A. Zavaran Hosseini, M. Ahangarzadeh Rezaee (Tehran, IR)
Objectives: Cholera toxin, secreted by Vibrio cholera, consists of A and B subunits. Cholera toxin B subunit (CTB) has been used in many scientific applications. CTB is a subunit vaccine candidate against cholera. It has been already expressed in several bacterial and plant systems.
Methods: In this study, in order to construction of yeast expressing vector for CTB protein, the ctxB gene (309 bp) encoding CTB was amplified from Vibrio cholera genomic DNA by PCR. The expression plasmid pCTB83 was constructed by inserting ctxB gene into pYES2 shuttle vector immediately down stream of the GAL1 promoter. The new construct was introduced into S. cerevisiae cells. The cells were induced in the presence of 2% galactose.
Results: Immunoblat analysis showed the presence of CTB in yeast lysate and indicated that the yeast-derived CTB protein was antigenically indistinguishable from bacterial CTB protein. The monomeric CTB protein molecules (11.6 KDa) were the dominant molecular species detected in yeast total soluble protein.
Conclusions: Since, whole recombinant yeast introduced as new vaccine formulation, expression of CTB in S cerevisiae may offer an effective and inexpensive strategy to protect people against cholera and enterotoxigenic E. coli in high risk areas.
Respiratory tract infection
P1322: Relationship between bacterial colonisation, exacerbations and current smoking in 575 patients with chronic obstructive pulmonary disease
L.H. Mygind, J. Vestbo, J.J. Christensen, C. Pedersen, N. Frimodt-Møller, S.S. Pedersen (Odense, DK; Manchester, UK; Copenhagen, DK)
Objectives: Patients with COPD are often colonised with bacteria in their lower respiratory tract, but the clinical relevance is debatable. One small study has shown, that colonisation in stable state was related to exacerbation frequency. Previous studies have found a correlation between current smoking and the degree of colonization, which could not be reproduced in a more recent study. We want to investigate the degree of colonisation and compare the findings with exacerbations and smoking habits.
Methods: 575 COPD patients with a previous hospitalization due to an exacerbation were included in the study. Inclusion criteria were: age (49 years, FEV1 (60% predicted, FEV1/FVC < 70% and (4 weeks in a clinically stable state. Lung function was measured and spontaneous sputum collected. A severe exacerbation was defined as one requiring hospitalisation.
Results: Age 50–88 (median 71): FEV1 0.18–3.19 l (median 0.90 l, 39% predicted) and FVC 0.57–4.43 l (median 2.00 l). 266 (46%) patients could produce sputum samples: 231 samples were considered representative of lower airways. 77(33%) patients were colonised with potentially pathogenic bacteria (PPB) and 154 patients had growth of respiratory non-pathogenic bacteria (non-PB). The frequencies of PPB were: Haemophilus influenzae 26, Moraxella catarrhalis 18, Streptococcus pneumoniae 18, Enterobacteriaceae 21, Pseudomonas aeruginosa 7. In 13 patients more than one bacterium were found. The patients with PPB had 1.7 mean exacerbations/year compared to 1.5 in patients with non-PB. The patients were stratified according to severity of COPD: 18% were moderate (FEV1 (50%), 56% were severe (FEV1 (30% to (50%) and 26% were very severe (FEV1 (30%). In the very severe COPD group patients with PPB and non-PB had 2.2 and 1.9 and in the severe COPD group patients had 1.5 and 1.4 mean exacerbations/year, respectively. There was not a higher frequency of colonisation in current smokers as compared to ex-smokers neither in the whole group nor when stratified for severity.
Conclusion: 46% of the 575 stable COPD patients spontaneously produced sputum, and of these 33% had pathogenic bacteria. There was no correlation between colonisation and current smoking. The patients with colonisation had an increased number of severe exacerbations, even when stratified for severity of COPD, suggesting that the presence of bacteria is clinically relevant. The next step will be to perform intervention studies aimed at reducing colonisation.
P1323: Predictors of pneumonia in patients with lower respiratory tract infection in primary care
A. Holm, J. Nexoe, L.P. Nielsen, N. Obel, S.S. Pedersen, C. Pedersen (Odense, DK)
Objectives: The majority of out-patients with lower respiratory tract infection (LRTI) is treated with antibiotics, although the benefit of this is questionable. The objective of our study was to identify predictors of pneumonia that could contribute to the identification of patients in need of antibiotic treatment. This may lead to a reduction in antibiotic prescribing in the group of patients with non-pneumonic LRTI.
Methods: We prospectively studied 364 consecutive outpatients with LRTI. Patient characteristics were registered, a chest x-ray was taken, and blood samples were drawn for measuring C-reactive protein (CRP) and leukocytes. Pneumonia was defined by a transient infiltrate on chest X-ray.
Results: Pneumonia was found in 48 patients (13%). The pneumonic patients were older with a median age of 61 years compared to 48 years in the non-pneumonic patients. There were no differences in the occurrence of cough, dyspnoea, chest pain or auscultative abnormalities, but sputum production was less common in pneumonic compared to non-pneumonic patients (69% vs. 83%; p = 0.03). The median CRP was higher in pneumonic patients (73 mg/l vs. 11 mg/l; p (0.01), as was the median leukocyte count (10 × 109/l vs. 8 × 109/l; p (0.01). There was no significant difference in the median temperature between groups (37.5°C vs. 37.3°C). The median respiratory frequency was higher (21/min vs. 18/min; p (0.01), and the oxygen saturation lower (0.95 vs. 0.98; p (0.01) in the pneumonic patients.
Conclusions: Pneumonic patients differed from non-pneumonic patients in being older and presenting less often with a productive cough. The respiratory frequency was higher, and the oxygen saturation lower. A promising predictor was found in the measure of CRP found to be higher in pneumonic patients. A point-of-care-test for CRP is available for use in primary care. This may guide the general practitioner in identifying patients with LRTI in need of antibiotic treatment.
P1324: Community outbreaks of legionellosis in Catalonia, 1990–2003
J. Alvarez, A. Dominguez, M. Sabrià, L. Ruiz, N. Torner, J. Caylà, I. Barrabeig, M.R. Sala, P. Godoy, N. Camps, S. Minguell for the Working Group on Communicable Diseases of the RCESP
Objectives: 1. To analyze the characteristics of the community-acquired outbreaks notified in Catalonia during the period 1990–2003 2. To compare the characteristics of the outbreaks of the period 1990–1996, with those of the period 1997–2003 when the urinary antigen test was widely used.
Materials and methods: Epidemiological records of all community outbreaks of legionellosis reported to the Department of Health of the Generalitat of Catalonia (6.3 million inhabitants), Spain, excluding those of nosocomial origin, during the period 1990–2003 were studied. An epidemic outbreak was considered as the appearance of two or more epidemiologically-related cases of legionellosis. Descriptive variables were recorded for each outbreak and a comparative analysis made of outbreaks occurring between 1990 and 1996 (period I) and 1997 and 2003 (period II). χ2 test was used to compare qualitative variables.
Results: Ninety outbreaks were studied. The origin was determined solely by epidemiological data in 47 (52.2%), and by epidemiological data and molecular biology in 6 (6.7%). The origin could not be determined in the remaining outbreaks. Cooling towers were involved in 34.4% of the outbreaks. The annual evolution of the number of outbreaks has been progressively greater from 1990 with a clear increase from 1999 onwards. 71.1% of the outbreaks occurred in the period June–October. The total number of people affected in all 90 outbreaks was 558 and the median number affected per outbreak was 3 (range 2–113). The average age of those affected was 57.3 years (SD 10.8). Hospitalization was required in 463 (82.9%) cases and 27 people died (global case-fatality rate 4.8%). The outbreaks during period I were smaller, the average age of patients was higher and the outbreak was detected later. The case-fatality rate was significantly higher in period I than in period II (12.2% versus 4.1%). In period I, the diagnostic methods used were BCYE and serology, whereas 97.1% of the outbreaks in period II were diagnosed by the urinary antigen test.
Conclusion: In Catalonia, the number and size of LD outbreaks has risen clearly from the 1990s to the present. However, the beginning of the outbreaks was detected sooner and the case fatality rate has fallen significantly. The use of more sensitive diagnostic tests for Legionella and the rapid establishment of appropriate treatment may explain these results.
P1325: High prevalence of rhinovirus in lower respiratory tract specimens
C. Minosse, M. Selleri, S. Zaniratti, G. Cappiello, F.N. Lauria, R. Longo, P. Roselli, S. Antonelli, E. Schifano, M. Visca, M. Cava, A. Spanò, M.R. Capobianchi, V. Puro (Rome, Vasto, I)
Objectives: To assess the prevalence of respiratory viruses in the lower respiratory tract specimens of hospitalized patients and their potential role on respiratory disease outcomes.
Methods: Sputum and endotracheal aspirate (EA)samples consecutively sent to the microbiology laboratories of two general hospitals in Italy were collected during April–October 2004. Sputum was considered valid if it contained more than 25 polymorphonuclear leucocytes per field. In addition to routine microbiological tests (i.e. direct sputum examination and culture), clinical specimens were analysed by PCR or RT-PCR for the presence of eleven different viruses (Influenza A and B, Metapneumovirus, Adenoviruses, Parainfluenza 1, 2 and 3, RSV, Rhinovirus (RV), Coronaviruses OC43 e 229E), as well as Clamydia and Mycoplasma pneumoniae and Legionella pneumophilia. A nested PCR was used for the detection of RV. Positive and negative controls were included in each PCR and RT-PCR assay. Patients’ clinical data are correlated with microbiological and virological results.
Results: On a total of 60 specimens collected, 17 of 36 (47%) sputum and 12 of 24 (50%) EA were positive for RV. In an additional case, isolated Parainfluenza 1 was detected; no other viruses were identified. RV-positive patients had pneumonia (7 cases), exacerbation of chronic obstructive pulmonary disease (COPD, 9 cases) or other respiratory events; mean age was 62 years (median 69); 30% had mixed bacterial infections. No statistically significant differences were found among RV positive and negative patients with regard to respiratory events and disease outcomes.
Conclusions: Lower respiratory tract infections are a leading cause of morbidity whose etiology remains undetermined in more than 50% of cases. Our study shows an unexpected high prevalence of RV RNA in lower respiratory tract specimens obtained by adult patients with pneumonia, exacerbation of COPD or other respiratory events. Such a high rate of RV infection has not been reported before PCR application. The study is consistent with recent evidences on paediatric population, showing that, when sensitive detection methods are used, the prevalence of RV is much higher than previously thought and suggest that RV can infect the lower respiratory tract. Further and larger studies are needed to assess the role of RV as a leading cause of low respiratory tract infections. Funded by Ministero della Salute Ricerca Finalizzata e Ricerca Corrente IRCCS
P1326: Evaluation of 2 amplification methods and 4 serological tests for the detection of Chlamydia pneumoniae in patients with community-acquired pneumonia
K. Loens, D. Ursi, L. Daniëls, H. Goossens, M. Ieven (Edegem, B)
Objectives: Studies comparing serology and nucleic acid amplification methods for diagnosis of C. pneumoniae (CP) in CAP are rare. The aim of this study was to evaluate PCR, nucleic acid sequence-based amplification (NASBA), microimmunofluorescence (MIF) and 3 different EIA assays for the detection of CP in patients with CAP.
Methods: 195 paired and 18 acute phase sera from 213 patients with community acquired pneumonia (CAP) were available. MIF (IgM and IgG) (Focus Technologies) and 3 different EIA's were evaluated: Medac IgM and IgG EIA (Medac); AniLabsystems IgM and IgG EIA (Biomedical Diagnostics); and Euroimmun IgM, and IgG EIA (Biognost). PCR and NASBA were applied to respiratory specimens from all 213 patients collected on admission to the hospital. An expanded gold standard was used to calculate the sensitivities and specificities of all tests: (1) positive by PCR and NASBA or (2) positive by 1 amplification test and at least one serological test (either IgM or a seroconversion or significant rise of IgG antibodies), or (3) a seroconversion or significant rise of IgG antibodies in at least two different EIA's.
Results: 21 patients met the criteria of the expanded gold standard. The sensitivities of the IgM assays ranged from 29% to 57% for IgM in the acute phase serum and from 33% to 71% for IgM in the convalescent serum sample. IgG seroconversion or a significant rise in titre ranged from 43% to 81.0%. Low sensitivities were obtained for PCR and NASBA: 24% and 29%, respectively. The specificities ranged from 83% to 99.5% for IgM in the acute phase serum, from 81% to 99% for IgM in the convalescent phase serum, and from 90% to 97% for IgG. The specificities of PCR and NASBA were 100% and 99.5%, respectively. The CP IgM assay with the best combined values for sensitivity and specificity was the Medac EIA. The best CP IgG assay was the Anilabsystems EIA. CP PCR and NASBA positive patients had usually higher rises in IgG titre than CP PCR and NASBA negative patients. The MIF test had the lowest sensitivity and specificity scores in all serologic tests.
Conclusion: Substantial differences between the performances of the assays were found. Amplification tests seem to be less sensitive. However, since important discordances between the different serological tests were observed, both the sensitivities and specificities of these tests have to be questioned.
P1327: Invasive procedures and aetiological diagnosis in severe community-acquired pneumonia: a case study of 46 patients
S. Cordani, A. Manna, M. Vignali, A. Carpenzano, E. Battolla, PA. Canessa (Sarzana, I)
Objectives: The aim of this study was to improve the etiological diagnostic accuracy in patients with severe community-acquired pneumonia (sCAP) using an invasive procedure (IP).
Methods: Over a 2-year period, 46 patients showing clinical and radiological characteristics of severe CAP and failing to respond to antimicrobial therapy were studied. 33 males and 13 females with a mean age of 60 years and ranging between the ages of 21 and 84 years were admitted either to the Pneumology Department or Intensive Care Unit (ICU). Severe pneumonia was defined by clinical criteria determined according to FINE score class IV or V. In conjunction to initial routine serological and microbiological diagnostic tests on blood, urine and sputum (if available), Bronchoalveolar Lavage (BAL) or Transthoracic Fine Needle Agoaspiration (TTFNA) was performed to identify the responsible infecting pathogen. Granulocytes (50% and a bacterial culture Cutoff (10.000 CFU/ml defined infectious pneumonia.
Results: Of the 46 patients enrolled, 10 presented a noninfectious lung disease that mimicked CAP. The final differential diagnoses were: 3 lung carcinoma, 1 ARDS, 2 BOOP, 1 T cell lung lymphoma, 1 drug parenchymal damage, 1 Wegener's Granulomatosis, and 1 unknown diagnosis but granulocytes (50% in BAL ruled out infectious pneumonia. Of the remaining 36 patients, 1 had a malignancy overlapping infectious pneumonia. By means of invasive procedure, an etiological diagnosis was achieved in 13 cases. M. tuberculosis was isolated in 4 of these cases. Therapeutic treatment was modified in 9 cases.
Conclusions: Invasive procedure significantly improved the diagnostic accuracy in patients with sCAP and established the differential diagnosis in noninfectious and unusual infectious pneumonia that mimicked sCAP. Though not routinely used, IP had a substantial impact on the etiological diagnosis and decisively influenced change in therapeutic strategy in a selected number of cases. The implementation of IP proved to be a safe and effective means of reducing therapeutic failure in sCAP. Identification of unsuspected or resistant pathogens would otherwise have been unattainable and patient survival reduced.
P1328: The unexpected findings of pertussis by 16S rDNA sequencing in bronchoalveolar lavage fluid in an immunocompromised patient in an endemic setting
S. Dudman, T.0. Jonassen, M. Steinbakk (L0renskog, N)
Objectives: The immunization of infants has reduced the prevalence of pertussis in Norway, however in 1997 an epidemic of whooping cough started to spread in older children and adolescents throughout the country. Pertussis is a notifiable disease and in 2002 a total of 3182 cases were reported. The diagnosis of this disease can be made by the isolation of B. pertussis or detection by PCR from respiratory secretions, as well as using serological methods (ELISA) at our laboratory. This paper reports the results from the different methods used, and describes a case where pertussis was found unexpectedly.
Methods: All cases of pertussis were included from January 1st to October 31st 2004, and the positive rates were calculated according to the different laboratory methods. For the case of pertussis detected in bronchoalveolar lavage fluid, medical records were obtained.
Results: A total number of 458 patients were diagnosed with pertussis during the observation period. In some patients, the organism could be found in two different specimens. 391 (15%) out of 2614 samples were found positive by serological methods. 74 (8%) out of 954 samples were found to be PCR positive. In 6 (5%) of the 110 cultured samples, the B. pertussis could be isolated from nasopharyngeal secretions. In one case the bacteria was isolated from a blood culture. In addition, one case was detected in bronchoalveolar lavage fluid, this patient suffered from chronic lymphatic leukaemia. He was admitted to hospital because of a fever, dyspnoea and a dry cough, and was diagnosed with interstitial pneumonia. 16S rDNA sequencing performed directly on the culture negative bronchoalveolar lavage fluid showed B. pertussis. The patient was successfully treated with Chlarithromycin.
Conclusions: In view of the epidemic levels of whooping cough, it is important to routinely screen for this disease even in patients not showing typical symptoms of pertussis but are suffering from coughing.
P1329: Mycoplasma pneumoniae pneumonia in hospitalised patients
A. Anastasovska, I. Kondova Topuzovska, K. Marangozova, M. Arapova, I. Vidinic (Skopje, MK)
Objective: To determinate incidence of the acute Mycoplasma pneumoniae infection in hospitalized patients with low respiratory tract infection.
Methods: A total of 93 children and 72 adult hospitalized patients with CAP within one year period (2002/2003) were included in our study. The mean age was 4.6 years (range:1–17) for children and 45 years (range:18–74) for adults. From children there were 57 (61.29%) male and 36 (38.70%) female and from adults there were 47 (65%) male and 25 (35%) female. Diagnosis of Mycoplasma pneumoniae infection was established by serological confirmation by detection of IgM, IgG or total antibody against Mycoplasma pneumoniae Ag with Pneumoslide M (Vircell) IF test and/or Mycoplasma pneumoniae IgM, IgG (Vircell) ELISA test.
Results: Acute Mycoplasma pneumoniae infection as a cause of CAP from 93 hospitalized children was detected in 11 (11.83%) patients and from 72 adult hospitalized patient was detected in 6 (8.33%) patients. There was no significant discrepancy, from urban or rural distribution of these patients. From children in 1 of 11 patients with Mycoplasma pneumoniae infection, coinfection with Coxiella burnetti was detected and in 2 with Influenza B. In the group of adults there was no detected coinfection with some other pathogen of low respiratory tract infections.
Conclusions: Acute Mycoplasma pneumoniae infection takes significant place among low respiratory infections, in adults with 8.33% of the patients hospitalized with CAP and in the group of hospitalized children with CAP in 11.83%. Further analyze in few years period will give us dates to determine prevalence and epidemiological model of acute Mycoplasma pneumoniae low respiratory infection in this region
P1330: Prevalence of nosocomial pneumonia in severe trauma patients (according to autopsy data)
L. Stratchounski, A. Jdanuk, I. Gudkov, E. Ryabkova (Smolensk, RUS)
Objectives: Nosocomial pneumonia (NP) is one of most serious infectious complications in severe trauma patients, especially in those who receive mechanical ventilation. Data on the prevalence and etiology of NP in severe trauma patients are scarce as diagnostics of NP is difficult and in some cases NP remains undiagnosed. Thus, the objective of our study was to assess prevalence and etiology of NP in died patients with severe trauma.
Methods: Retrospective analysis of medical records and autopsy protocols of died patient from 2 traumatology and 1 neurosurgery units of Clinical Hospital of Emergency Medicine.
Results: Among 9771 trauma patients hospitalized in 2001–03 the overall mortality rate was 4.7%. Autopsy was performed in 411/458 (89.7%) patients. Morphological signs of NP was found in 138 (33.6%) patients. NP was most common in patients with head trauma (79%), chest trauma (26.8%) and proximal femoral fractures (10.9%). Mean age was 51.9 ± 17.6 (21–89) years. Impaired conciseness presented in 97.8% patients; 81.9% patients were admitted to ICU, 78.3% patients were mechanically ventilated. Bilateral pneumonia was observed in 84.7% patients, pleurisy – in 19.6%, right-sided pneumonia – in 9.5% and left sided pneumonia – in 5.8%. In spite chest X-ray was performed in all patients, it failed to reveal 34.1% cases of NP intravitally. Microbiological investigation both intravitally and postmortem was performed in 21.7% cases. Among pathogens isolated Acinetobacter spp. (20%), Pseudomonas aeruginosa (20%), Klebsiella pneumoniae (14%) and Proteus mirabilis (8%) were the most common.
Conclusion: NP is a frequent infectious complication in severe trauma patients that difficult to diagnose intravitally. The most common pathogens of NP in this category of patients are gram-negative bacteria that should be taken into consideration for choice of empirical antimicrobial therapy.
P1331: X-ray changes of Chlamydia pneumoniae pneumonia
I. Kondova Topuzovska, G. Kondov, Z. Trajkovski, D. Dimitriev, A. Anastasovska (Skopje, MK)
A prospective study was performed that comprised 407 hospitalized adult patients with community acquired pneumonia (CAP). Chlamydia pneumoniae (CP) as a cause of CAP was detected in 13.27% (n = 54) and it was a single cause in 10.32% (n = 42). The diagnosis was made serologically with EIA and MIF assays. Mean age of the patients with CP pneumonia (n = 42) was × = 47, 57 years (s = 18.26). There were 66.7% (n = 28) males. Clinical, laboratory and radiographic findings were analysed and compared with a control group of patients with bacterial etiology of the CAP (n = 80). The analysis of radiographic changes prior to admission in the hospital of those patients with Chlamydia pneumoniae pneumonia revealed that only half of them had interstitial infiltration (χ2 ≥ 100, p ≤ 0.001) and the remaining had alveolar or mixed infiltration. Beside of domination of diffuse extension of changes in 71.4%, in 16.7% of the patients with CP pneumonia there was lobar and in 11.9% segmental extension of the infiltration in the lungs (χ2 = 10.54, p = 0.0144). Pleural effusion was found in 7.1% (χ2 = 1.177, p = 0.758) of the patients with CP pneumonia and hilar adenopathy in 23.8% (χ2 = 1.959, p = 0.16). Partial regression of the changes was found in 4.8% and contra lateral progression in 2.4% (χ2 = 2.85, p = 0.24). Infiltrations were more frequently in lower lobes either left or right (χ2 = 3.5857, p = 0.3098). In order to increase the percentage of patients with CP pneumonia who would be initially treated with adequate antimicrobial therapy, multifactor regression and discrimination factors analyses were done based on the results obtained. This was followed by postulating diagnostic algorithm for making more exact diagnosis – Chlamydia pneumoniae pneumonia.
P1332: Surveillance of probably bacterial pneumonia in children less than 5 years old in two geographical areas in Argentina
R. Ruvinsky, A. Gentile, F. Gentile, C. Gil, J. Kupervaser, M. Quiriconi, J. Bakir, J. Pace, S. Garcia, J. Andrews on behalf of the Argentine Pneumoniae Working Group
Background: Bacterial pneumonia represents an important cause of morbimortality in children (5 ys old in Latin American countries.
Objectives: (a) Determine the incidence of PBP cases in Concordia and Paraná Departments (CPD) and Pilar Department (PD), based in WHO standardized radiological criteria (b) evaluate pediatricians and radiologist concordance.
Methods: Population: CPD 44,892 and PD 27,209 children (5 ys old. Inclusion criteria was all children (5 ys with clinicalradiological signs of PBP, from 1-11-2002 to 30-04-2004 (18 months), belonging to these Departments (CPD n = 1000), PD n = 525). Chest-Rx was digitalized and sent to the reference radiologist, who confirmed the presence of consolidation. Serotyping and CIM's of S. pneumoniae isolated from blood or pleural fluid culture, was made in the National Reference Laboratory. Concordance on consolidation between pediatrician and radiologist independent diagnosis was analysed.
Results: Total of pneumonia cases with chest-Rx consolidation: CPD 509/1000 (47.8%) PD 294/525 (56.0%); Incidence/100,000 children (5 ys per year was: Concordia 1.189.0; Paraná 709.7 and Pilar 962.9. Hospitalized children: CPD 69.5%, PD 63.6%. Low and medium social-economical level: CPD 90.4%, PD 84.4%; Age (2 ys: CPD 65.8% and PD 68.7%; undernutrition: 6.2% in both centres; previous antibiotic therapy (3 months): CPD 27.1%, PD 10.9%. Bacterial isolation: CPD 5.9% (S.pn 5.6%) and PD 9.2% (Spn 4.6%). the most frequent serotypes: 14, 5, 6B, 7F and 23F. Letality: CPD 0.8% and PD 0.5%. Consolidation concordance: CPD 791/1000 (79.1%) and PD 307/446 (68.8%)
Conclusions: (a) High incidence of pneumonia with consolidation in (5 years (b) Low rate of bacteriological documentation as expected in pneumonia; (c) Consolidation concordance between pediatricians and radiologist was moderate
P1333: Colonisation of lower airways with potential pathogenic bacteria in healthy patients and patients with chronic lung diseases
U. Møller Weinreich, J. Korsgaard (Aalborg, DK)
Introduction: Patients with chronic lung disease often suffer from repeated lower respiratory tract infections. For patients with chronic obstructive lung diseases alone 25000 patients are admitted to hospital in Denmark each year.
Material: From January 1st 2003 until June 30th 2004 a total of 151 adult patients underwent bronchoscopy with bronchial lavage and bacterial culture. Patients with ongoing infection with recent (within 24 hours) antibiotic treatment (15) and/or significant leucocytosis (>11 × 109/l) (19) or patients with a final diagnosis of carcinoma (20) were excluded so that 97 adult patients with a final diagnosis of normal (27), recent lower respiratory tract infection (22), chronic obstructive lung diseases and/or lung fibrosis (29) and patients with a final diagnosis of bronchiectasies (19) were included in this retro-spective analysis.
Methods: Patients were investigated with fibre optic bronchoscopy and investigated with bronchial lavage with 10 ml of isotonic NaCl and the aspirate send for semiquantitative bacterial culture at the department of microbiology. Culture was performed with 10 and 1 μl of lavage fluid on separate halfs of chocolate agar plates and results expressed as colony forming units (cfu) per ml lavage fluid, and identification as potential pathogenic bacteria (ppb) or normal respiratory tract flora.
Results: 43 (44%) of 97 patients were grown with ppb with 4 patients grown with 2 ppb's and 1 patient grown with 3 ppb's adding up to 49 separate isolates of ppb's among the 43 patients. The most frequent isolate was H. Influenzae (20) followed by S. Pneumoniae (15), other gram negative bacteria (8) and 6 isolates of S. Aureus. The frequency varied from 15% with ppb among normal (4/27), to 34% (10/29) in patients with stable disease with COPD and/or fibrosis, to 61 % in patients with recent (weeks to months) lower respiratory tract infection, while the highest colonization rate was found among patients with bronchiectasies (15/19) where 79% were colonized with ppb in the lower airways. The mean cfu/ml of ppb was 2000 in normal patients, 39,000 in patients with recent infection, 35,000 in patients with COPD and 87,000 in patients with bronchiectasi (p < 0.001)
Conclusion: Healthy persons are rarely (15%) colonized with ppb while patients with chronic lung diseases in stable fase (34–79%) and patients with recent lower respiratory tract infection (61%) frequently are colonized with ppb in the lower airways.
P1334: Bacterial causes of community-acquired pneumonia and antibiotic resistance on adult patients hospitalised during 2002–2003
M. Zoumberi, C. Papaefstathiou, A. Karaitianou, A. Koutoumanis, S. Botsari, G. Kouppari (Athens, GR)
Objective: To study the bacterial causes of community acquired pneumonia in patients who were hospitalized during 2002–2003. The antibiotic resistance in adult patients is also to be studied.
Methods: Of the 268 patients (193 male and 75 female) who were hospitalized with lower respiratory tract infection, 235 sputum cultures, 33 bronchotracheal secretions cultures and 125 urine cultures were sent to be examined for detection of S. pneumoniae and Legionella antigens. All specimens were cultured on appropriate media for aerobic and anaerobic microorganisms and smears were performed by Gram stain. The identification of the isolated bacteria was performed by standard methods and the API-system (Bio Merieux). The susceptibility testing was carried out by disk diffusion method and E-test (AB Biodisk Solna Sweden). The detection of Legionella and S. pneumonia in urine samples was performed by immunochromatographic assay. (Now, binax)
Results: One hundred and seventeen patients had community-acquired pneumonia, 121 suffered from some other infection of the lower respiratory system. Of the patients with community-acquired pneumonia were isolated in order of appearance: S. pneumoniae (27.3%), H. influenzae (20.5%), P. aeruginosa (15.4%), K. pneumoniae (9.4%), L. pneumophila (7.7%), H. parainfluenzae (7.7%), S. maltophilia (4.3%), S. aureus (2.6%), and others (5.1%). Of S. pneumoniae isolates 30% were intermediate susceptible to Penicillin, 0% were resistant to cefotaxime, 0% to vancomycin, 0% to ofloxacin and 5% to clindamycin. Of H. influenzae isolates 2.4% were intermediate susceptible to ciprofloxacin, (4.7%) were resistant to ampicillin, 0% to cefotaxime, (19%) to co-trimoxazole, and 0% to imipenem. Of P. aeruginosa isolates 32% were resistant to ceftazidime, 42% to ciptofloxacin, 40% to gentamycin and 42% to imipenem.
Conclusion: The most common isolated bacteria in patients with community-acquired pneumoniae were: S. pneumoniae, H. influenzae, P. aeruginosa, H. pneumoniae and L. pneumophila. Among strains of S. pneumoniae 30% were intermediate susceptible to Penicillin and all of them were susceptible to Vancomycin.
P1335: The prevalence of Mycoplasma pneumoniae in hospitalised children with lower respiratory infections
I. Varzakakos, G. Tapaki, H. Papavasileiou, E. Damaskopoulou, K. Avgerinakou, G. Giorzatou, A. Voyatzi (Athens, GR)
Objectives: The aim of this study was 1. To determine the prevalence of Mycoplasma pneumoniae IgG and IgM antibodies in relation to age in hospitalized children with confirmed community – acquired pneumonia (CAP) or extrapulmonary symptoms. 2. To provide epidemiological data for the seasonal distribution and the annual incidence of Mycoplasma pneumoniae infections.
Materials and methods: A total of 1074 patients from 2 to 14 years old with symptoms and signs compatible with CAP were enrolled during a 4-year period (2001–2004). Thorax radiography and paired sera were obtained from each patient and the course of illness was monitored uniformly. Specific IgG and IgM antibodies were measured by enzyme linked immunosorbent assay (Remel, USA) and Immunocard based IgM EIA (Meridian France). The children were divided in three age groups: Group A: 2–6 years old Group B: 7–12 years and Group C: more than 12 years. The serum CRP concentration was also determined by nephelometry.
Results: IgG and IgM antibodies were detected in a total of 337(31.8%) children out of 1074 patients. IgM antibodies were found only in 191 patients (17.8%). No significant difference between sexes was found. Significantly higher frequency of M. pneumoniae infections was recorded during the two last winter months and the springtime. Seroprevalence to M. pneumoniae was found to increase in the group B, 7–12 years old, (101 patients, 52.8%). The incidence per year ranged between 10.7% and 29.2%. The majority of the hospitalized children dominated high temperatures, cough, headache and chest pain, while 27 patients developed extrapulmonary manifestations. 13 cases with rash, 8 cases with pleural effusion, 3 with gastrointestinal manifestations, 2 with otitis media and 1case with erythema nodosum.
Conclusion: The results show the high incidence of M. pneumoniae in the etiology of lower respiratory infections in Greek hospitalized children and especially in children aged more than 7 or more years old while the extrapulmonary manifestations are not rare. The importance of timely diagnosis and treatment is obvious.
P1336: Demography and Fine score of community-acquired pneumonia in adults admitted to hospital in Spain
E. Pérez-Trallero, F. Baquero, J. Torres, C. García-Rey, J.M. Nogueira on behalf of the NACER Group
Objectives: Despite of its considerable frequency there is little epidemiological information about the demography of adults with community-acquired pneumonia (CAP) admitted to hospitals and their Fine scores in large and recent Spanish series.
Methods: Retrospective review of the hospital charts of patients admitted to 10 geographically scattered hospitals with the diagnosis of CAP over a 1-year period in Spain. Subjects included were those with diagnosed with CAP at discharge from 1 Nov 01 to 31 Oct 02.
Results: Overall, data were available from 3233 patients. The 10 hospitals have 8523 beds (5.4% of the nation capacity). The occupation rate of adult CAP with hospital admission was 37.9/ 100 beds/year. Males represented a 64% of the sample with a mean, median and modal age of 66.6, 71 and 78 years. Up to 44% of the sample were 80-year-old or older, 27% had an age between 60 and 79, 16% between 40 and 59, and 13% between 18 and 39. Comorbidities were very common since up to 83% reported at least one of them. Of these, their frequency was as follows: previous hospital admittance in the last 5 years 22%; COPD 13%; diabetes mellitus 10%; smoker 10%; previous episode of pneumonia 8%; malignancy 7%; immunosuppression, chronic heart failure and cerebrovascular disease 5% each; hepatopathy 4%; alcoholism and nephropathy 3% each; others 2%. Antibiotics were already being taken before admission in 26.2% (penicillins 8.6%; macrolides 6.2%; quinolones 5.6%; cephalosporins 3.9%). The data of proper influenza and/or pneumococcal vaccination was of no use in the medical management of these patients. The antecedent of vaccination was acknowledged in 0.6% and 0.8% of the subjects studied. Only 0.2% of the subjects were participating in a clinical trial of CAP. Mean, median and modal length of hospital stay for the CAP episode was 11.3, 9 and 8 days, respectively. Five per cent of the subject entered the ICU with a corresponding mean, median and modal stay of 9.3, 5 and 1 day, respectively. Fine variables were properly recorded in only 1722 subjects (53.3%), whose distribution is broken down in the Table.
| LOS |
||||||
|---|---|---|---|---|---|---|
| Score | N (%) | % hypoxemia | % hypotension | Mean | Median | Mode |
| Total | 1722 | 49.7 | 8.3 | 11.1 | 9 | 8 |
| I | 201 (11.7) | 24.9 | 5.0 | 8.2 | 7 | 4 |
| II | 255 (14.8) | 26.7 | 9.0 | 9.1 | 8 | 6 |
| III | 357 (20.7) | 44.5 | 5.0 | 10.9 | 9 | 8 |
| IV | 623 (36.2) | 58.4 | 8.2 | 12.5 | 10 | 8 |
| V | 286 (16.5) | 74.8 | 8.2 | 12.1 | 10 | 10 |
Conclusions: The occupation rate of hospital admitted adult CAP was 37.9/100 beds/year. There was a male predominance and a clear trend age disbalance as 71% of the patients were at least 60 years old. 26.2% of the subjects were already taking antibiotics when admitted. 47.2% of the subjects were admitted to hospital with very low Fine scores (I–III), but hypoxemia occurred in 34.1% of them.
P1337: Hospital differences in the diagnostic microbiological workup in patients admitted to hospital with community-acquired pneumonia in Spain
E. Pérez-Trallero, J.L. Pérez, C. García-Rey, R. Landínez, J. Garau on behalf of the NACER Group
Objectives: The processing of microbiological samples is known to yield different results depending on many external factors some of them logistic in nature. We sought to assess the differences in microbiological workup in patients with community-acquired pneumonia (CAP) admitted to 10 Spanish hospitals.
Methods: Retrospective review of the microbiological workup, its results and diagnosis at discharge of patients admitted to the hospital with the diagnosis of CAP over a 1-year period (1 Nov 01 to 31 Oct 02) in 10 geographically scattered hospitals in Spain. Data were available from 3233 patients.
Results: Blood cultures were drawn in 57.2% of the patients, and of these, 78.7% were taken before antimicrobial treatment. Sputa were collected from 41.7% of the cases, and of these, only 43.2% were taken before therapy. Overall etiology was unknown in 77.6% of the subjects. Among the 22.4% etiologically diagnosed, S. pneumoniae was the main cause (58.8%), followed by Legionella spp. (9.4%), H. influenzae (6.4%), M. pneumoniae (5.3%), S. aureus (4.1%), P. aeruginosa (3.6%), and others (12.5%). Microbiological performance (%) was very different among hospitals. See Table.
| Centre | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| Etiology known | 7.1 | 39.0 | 31.6 | 20.0 | 16.5 | 13.8 | 23.5 | 23.5 | 13.9 | 12.3 |
| Blood culture (properly taken) | 66.3 (80.9) | 62.2 (91.9) | 82.6 (98.4) | 56.6 (79.7) | 29.4 (54.7) | 66.7 (92.8) | 59.9 (93.7) | 422 (66.2) | 45.5 (50.4) | 606 (78.0) |
| Sputa obtained (properly taken) | 28.1 (19.1) | 63.3 (58.1) | 19.1 (68.2) | 45.5 (45.8) | 47.5 (45.0) | 42.0 (46.2) | 59.4 (47.8) | 38.4 (18.6) | 26.4 (31.6) | 46.8 (52.1) |
Conclusions: (1) Microbiological confirmation was obtained in only 22.4% of the patients and diagnostic performance varied considerably among centres (Mean: 20.1%; Min: 7.1%, Max: 39.0%). (2) Large differences were observed among hospitals in the number and the quality of the diagnostic microbiological procedures done. (3) Globally, only 57.2% of the adult patients with CAP admitted to hospital had blood cultures done (range: 29.4–82.6%). Besides centres with a lower proportion of blood cultures ((55%) seemed also to have done them more frequently at the wrong time, and in fact, blood was drawn before starting antibiotic therapy in 78.7% of the episodes. Therefore, a proper blood sample for microbiological processing occurred in 45.0% (4) Success in obtaining sputa from patients was also diverse, but even in those providing sputum for microbiological exam a considerable proportion was done while on antibiotic treatment. Mean proportion of sputa obtained was 41.7%, and properly obtained in 43.2%.
P1338: Clinical presentation of community-acquired pneumonia in adults admitted to hospital in Spain
J.L. Pérez, J. Ruiz, J.E. Martín-Herrero, R. Dal-Ré, J. Garau on behalf of the NACER Group
Objectives: To assess the frequency of classical signs and symptoms of acute pneumonia in a large series of adult patients with community-acquired pneumonia (CAP) that required hospital admission.
Methods: Retrospective review of the hospital charts of patients admitted to the hospital with the diagnosis of CAP over a 1-year period in 10 geographically scattered hospitals in Spain. All patients admitted from 1 November 2001 to 31 October 2002 were included. Data were available from 3233 patients.
Results: Fever defined as an axillary temperature >38°C was present in only 38.7% of subjects, and its presence decreased in patients with high risk Fine score (groups IV–V) compared with low risk Fine score (groups I–III) (35.0% vs. 45.4%; p (0.0001). Leukocytosis was present in 68.9% and the sputa were purulent in 53.8%. An abrupt onset was recorded in 34.7%. Hypoxemia occurred in 45.2%, hypotension in 9.3% and mechanic ventilation was needed in 3.1% of the subjects. Based on the above results, the estimated probability of the classic association of fever, leukocytosis, purulent sputum and abrupt onset, typical of classic pneumococcal pneumonia would only have taken place in 5% of the adults patients with CAP admitted to the hospital. As for chest X-ray films, 85% of the cases were unilateral, and 77.6% were monolobar. A predominant alveolar pattern occurred in 80.3%, an interstitial in 7.2%, pleural effusion in 11.3% and only 1.1% presented cavitation.
Conclusions: (1) In real practice classic clinical presentation of bacterial pneumonia does not fit well with that presented in patients with CAP admitted to hospital. Isolated signs and symptoms have not a good sensibility as predictors of admission to hospital in CAP. (2) Only 5% of subject can be expected to present a classic pneumococcal presentation. (3) An alveolar chest X-ray pattern was predominant in our experience. Pleural effusion was not uncommon at all since it occurred in one tenth of subjects. (4) The proportion of patients with a body temperature >38°C decreases as Fine store increases.
P1339: Development of a prediction rule for hospitalisation or death in elderly primary care patients with a lower respiratory tract infection
J. Bont, E. Hak, A.W. Hoes, M.J.M. Bonten, T.J.M. Verheij on behalf of ESPRIT
Objectives: Acute lower respiratory tract infections (LRTIs) in elderly are of great concern to general practitioners since their course is often more complicated. Classification into low- or high risk may improve medical care, hence reducing unnecessary treatment and target monitoring and therapy more efficiently. Our objective was to develop a prediction rule for 30-day hospitalization or death in elderly primary care patients with a LRTI.
Methods: To develop a prediction rule we retrospectively analysed easily obtainable medical data from 3166 episodes of physician-attended LRTI including pneumonia, acute bronchitis and exacerbations of COPD in patients ≥65 years of age. Characteristics were identified that were predictive for 30-day hospitalisation or mortality. We subsequently developed a prediction rule with the use of logistic regression.
Results: Hospitalization or death in 30 days, occurred in 274 (8.7%) of all episodes (2.4% all cause mortality). Increasing age, male gender, hospitalization in 12 month prior to diagnosis, heart failure, diabetes, use of oral glucocorticoids, use of antibiotics in the prior month and a diagnosis of pneumonia or an exacerbation of COPD were predictors for hospitalization or death. Patients with ≥3 points had a 97 % chance of an uncomplicated course. Patients with ≥8 points had a 30% chance of hospitalization or death.
Conclusions: This prediction rule discerns elderly patients with high or low risk for hospitalization or death. Classification into risk groups can help the general practitioner to adjust his preventive and therapeutic decisions to the expected prognosis. This might lead to fewer complications and lower costs.
P1340: Identification of a major immunogenic polypeptide of Mycoplasma pneumoniae
Z. Greenberg, O. Dvir, J. Schlesinger, M. Lee (Ashdod, IL)
Objectives: Mycoplasma pneumoniae (MP) is an etiological agent responsible for 10–30% of community-acquired pneumonia cases. Pneumonia due to MP is labeled under atypical pneumonia infections. MP may be detected in all parts of the respiratory system, and its effects are well recognized and documented. In respect to diagnosis and treatment, the most prominent structural feature of MP is the lack of a cell wall. It has been shown that surface-exposed polypeptides elicit immunogenic response, in particular those that are involved in the attachment organelle of MP. This attachment organelle is composed of a complex of polypeptides, in which P1 Cytadhesin Protein has a major role. Due to its high immunogenicity P1 is a paradigm for utilizing a definitive antigen in sero-diagnostical systems. On the way to produce either a recombinant- or a peptide-based antigen of a similar nature, a modified extraction procedure of membranous proteins has been employed.
Methods: MP cells were exposed to the non-ionic detergent Triton X-114. The same detergent was applied then on the separated membranes, extracting insoluble proteins in a temperature-dependent way. The antigenic activity of the revealed extract was tested in ELISA with characterized sera, utilizing detection systems specific to IgG, IgM and IgA.
Results: The insoluble phase of the Triton X-114 extract was comprised of a single 65 kDa polypeptide, as shown in SDS-PAGE. The soluble phase of the extraction, as well as the currently used antigen in SeroMP test, has shown multiple bands. Comparison in ELISA between the three preparations revealed that the antigenic activity of the insoluble extract is comparable to that of the current assay. This result was repeated for IgG, IgM and IgA, meaning that the majority of the antigenic activity in the insoluble fraction may be attributed to the 65 kDa polypeptide.
Conclusions: The current observation is supported by previous works that have indicated the same size of polypeptide as a detergent extractable and being associated with the attachment organelle. We show here the antigenic potential of the 65 kDa extracted in our modified procedure, which reveals a comparable activity as the current assay. This finding identifies the 65 kDa polypeptide as an appropriate candidate to be used in diagnostic tests.
P1341: Characterisation of Legionella pneumophila isolates from geographically different Bulgarian regions
I. Tomova, S. Panaiotov, V. Levterova, I. Ivanov, T. Kantardjiev (Sofia, BG)
Nowadays legionellae are known as common water bacteria with ability to cause outbreaks and epidemics of both: Legionnaires’ disease and Pontiac fever. In the time of intensive traveling of people the laboratories must be capable to type environmental and human isolates and to give clear answers about the source of infection. In Bulgaria there were no data concerning the type-scope of the L. pneumophila strains circulating in the potable waters. We present here the results from a pilot study for the typing L. pneumophila (Lp) strains isolated from water samples from three geographically different Bulgarian regions. After isolation of the strains they were subjected to characterization by the use of serogroup specific techniques, monoclonal antibody and Amplified Fragment Length Polymorfism (AFLP) typing methods. Water samples collected from buildings located in one west and two east Bulgarian regions revealed presence of variety L. pneumophila isolates. A total of twenty six Lp strains were isolated. The predominant serogroup was Lp serogroup 1, but there were also 7 isolates belonging to Lp serogroup 6, Lp serogroup 8 and Lp serogroup 15. Co-contamination of water samples with different Lp serogroups was observed. The most frequent Lp serogroup 1 monoclonal subtypes were Knoxville and Allentown. The obtained AFLP profiles show good discriminatory power. Our results from this pilot study point that Lp serogroup 1 is not so rare finding in our potable waters as it was considered in the past based on a restricted study in one town Lp serogroup 15 was isolated for the first time in our country during this study. The necessity for use of a complex of methods for clear epidemiological typing was demonstrated. The results show that more extensive studies are needed to give clear picture of the environmental distribution of Lp types in Bulgarian waters and to compare Lp types from environmental sources with those from clinical cases of Legionnaires’ disease.
P1342: Clinical and economical efficacy of microbiology investigations in hospitalised patients with pneumonia
T. Chernenkaja, M. Bogdanov, A. Podolcev (Moscow, RUS)
Objectives: To evaluate clinical and economical efficacy of microbiological investigations (MI) in hospitalised patients with pneumonia.
Methods: The records of hospitalized pneumonia patients were screened for MI and results of antibiotic treatment (AT). 222 records with MI were selected for years 2000–2002. There were evaluated presence of clinically significant pathogens, the time between MI and beginning of AT, accordance between pathogens sensitivity (SN) to antibiotics and real AT, results of AT. The MI were interpreted as utilized if SN was available before of within 3 days after the beginning of AT according SN. The MI were interpreted as non-utilised if pathogens were not identified, or SN was available 3 days after beginning of AT, or there were discrepancy between SN and real AT. Clinical effectiveness of AT was determined in the case of utilised and non-utilised MI. Also it was evaluated the number of MI needed to determine SN; the formula ‘Number needed to screen’ (NNS) was used to calculate the number of MI with SN needed to prevent one case of ineffective AT. The cost of total number of MI was compared with the buying costs of second-line antibiotics such as 3rd, 4th generation cephalosporins, vancomycin and carbapenems for course of treatment. The other costs associated with ineffective first AT course were not calculated.
Results: In the case of pneumonia etiologically significant pathogens were identified in 44% of probes, it means that 2.27 MI were done to get one SN. In the case of utilised and non-utilised MI the effectiveness of AT was 95% and 47% respectively, based on these figures NNS calculation demonstrate that 2.09 MI with SN were needed to prevent one case of ineffective AT. So the total number of MI needed for prevention of one case of ineffective AT was 4.74 (2.2 multiply 2.09). The costs of 4.74 MI were significantly less than the buying costs of any second-line antibiotics.
Conclusion: MI significantly increase the effectiveness of AT of pneumonia. In the hospital were investigation was performed the direct buying costs of second-line antibiotics were higher than cost of MI. To increase the effectiveness of MI it is necessary to reduce number of MI without pathogens, MI with wrong SN and MI which are not used by physicians for selection of AT. In the settings with other cost structure to evaluate economical effectiveness of MI it may require to calculate the full costs associated with ineffective AT.
P1343: The outcome of non-hospital pneumonia depending on pre-hospital tactics during patient's first application for medical aid
A. Vertkin, A. Naumov (Moscow, RUS)
Aim: Elaboration, introduction and evaluation of the effectiveness of clinical recommendations for treating patients with non-hospital pneumonia on the pre-hospital stage.
Methods: The research was carried out in 1081 patients with community acquired pneumonia (CAP). The patients were divided into 3 groups: I − 431, patients diagnosed with CAP, in a number of cases, was not confirmed while in hospital; II − 391, the tactics of treating CAP on the pre-hospital stage did not correspond to the clinical recommendations; III − 259 patients with confirmed CAP, the diagnosis of which and the pre-hospital treatment corresponded to the clinical recommendations. The patients of all the groups were comparable.
Results: On the pre-hospital stage the clinical picture was evaluated only in 4% of the patients, on average. The underestimation of the clinical picture caused hyper diagnostics of CAP on the pre-hospital stage, whereas in hospital, the absence of characteristic complaints and physical signs of CAP allowed to evaluate the situation adequately and to change the diagnosis. On the pre-hospital stage the patients of group II did not have the clinical criteria of CAP (70.3%)in the accompanying documents, their concomitant diseases were not revealed (81.4%), the anamnesis and data about the previous treatment were not collected, the adequate treatment of CAP was not carried out (antibacterial drugs were not administered in any of the cases). The risk of unfavorable termination was known to be not evaluated in 47% of the cases, and the hospitalization of more than one third of the patients was non-profile. Besides, the index of the pre-day lethality came to 12.1% and the general lethality came to 4.6%. in this group. In group III all the patients had the symptoms of CAP which allowed to evaluate the degree of the disease severity correctly, to administer the therapy of antibiotics – amoxicillin (Phlemoxin Solutab) in the case of non-severe pneumonia and ceftriaxon in case of severe pneumonia and to carry out correct sorting of the patients. The pre-day lethality came to 3.1% and the general lethality came to 2.3%. in this group. The time of group III patients’ stay in hospital came to 17 ± 3.4 days, and that of group II patients came to 21 ± 4.7 days (p (0.05). Moreover, the necessity of antibacterial therapy changing in group III, when in hospital, occurred only in 13.1% of the observation, whereas in group II it happened in 34.3% of the cases.
P1344: Comparison the bacteriology and beta-lactamase production of the tonsils and adenoids surface and core flora
I. Ozcan, I. Mumcuoglu, N. Balaban, I. Taylan, I. Baran, A. Ulusoy, H. Dere (Ankara, TR)
Objectives: Adenoidectomy and tonsillectomy, indicated for children with recurrent or persistent symptoms of infection or hypertrophy, are among the most frequent operations performed in children. This study was carried out for investigating the microbial surface and core flora of the tonsils and adenoids.
Methods: Core and surface cultures were taken from the tonsils and adenoids of the 40 patients at the time of the surgery for tonsillectomy and adenoidectomy. Patients’ ages ranged from 3 to 13 years (mean age 6.6 years). Specimens were inoculated onto 5% sheep blood, chocolate, EMB agar and Haemophilus medium (BioMerieux/France). For anaerobic bacteria, core samples were inoculated onto anaerobic blood agar and incubated in Genbaganaer pockets (Biomerieux/France). The anaerobic plates examined at 48 and 96 hours. Aerobic organisms were identified by conventional methods and anaerobic organism were identified by VITEK ANI (Biomerieux/France) systems. Chocolate agar and Haemophilus medium (HAEM) were also compared for Haemophilus influenza identification.
Results: One hundred and thirty three aerobic bacterial isolates were recovered from surface of the tonsils, 111 from core of the tonsils, 128 from surface of the adenoids and 74 from core of the adenoids. Eight anaerobic bacteria were identified from core of the tonsils and 14 from core of the adenoids. The most frequently isolated microorganisms were alpha-haemolytic streptococci, Neisseria spp, and H. influenza. Beta-lactamase producing bacteria (BLPB) were found in all patients. The most frequently BLPB were Staphylococcus aureus (100% of the isolates), H. influenza (58% of the isolates), and Neisseria spp (45.2% of the isolates). Potential pathogenic microorganisms (beta-haemolytic streptococci, S. aureus, H. influenza and S. pneumoniae) were isolated in 33 patients.
Conclusion: This study demonstrates a polymicrobial aerobic–anaerobic flora in both adenoids and tonsils. There was a close relationship between the bacteriology of the tonsil and adenoid core and surface flora. We could not find any statistical difference between the bacteriology and BLPB of the tonsil and adenoid core and surface flora. HAEM and chocolate agar were also found similar for H. influenza but for H. parainfluenza a statistical difference was found.
P1345: Microbial colonization of laryngectomy stomas
M. Villar Vidal, E. Eraso, L. Madariaga, C. Marcos, A. Acha, J. Aguirre, G. Quindós (Bizkaia, E)
Objective: To investigate the microbial colonization of the stoma of laryngectomy patients.
Patients and methods: Nineteen consecutive patients who had previously undergone laryngectomy were recruited from the Odontology Clinic of our University. Swabs were taken from the laryngectomy stoma site. Microbiological culture and isolation were performed following standard procedures in Columbia agar and Candida ID2 chromogenic agar plates (bioMérieux, France). Identification of isolates was done by catalase production detection, differential growth in Mannitol–Salt agar, Slidex Staph Plus latex reagent and ID 32 STAPH (bioMérieux). Slidex MRSA detection test (bioMérieux) was used for the evaluation of methicillin resistance.
Results: Despite no clinical sign of infection, 13 patients were carriers of potentially pathogenic microorganisms (68.4%). Staphylococcus aureus was detected in the stoma of 10 patients (52.6%). Methicillin-resistant S. aureus (MRSA) were not isolated. Staphylococcus epidermidis was isolated from three patients (15.8%). Staphylococcus xylosus was isolated from the stoma of a patient (5.3%) also colonized by S. aureus. Candida albicans and yeast species were not isolated from these clinical specimens.
Conclusion: We have found a high incidence of colonization with Staphylococci in laryngectomy stomas with no clinical signs of infection. These microorganisms could be the potentially source for superficial or deep infections.
P1346: The epidemiology of peritonsillar abscess disease in Northern Ireland
B. Hanna, R. McMullan, G. Gallagher, S. Hedderwick (Belfast, UK)
Objectives: 1. To describe the epidemiology of peritonsillar abscess disease in Northern Ireland. 2. To investigate the impact of the nature of microbiological sampling on culture results. 3. To describe the impact of culture and sensitivity results on individual patient treatment and for guiding empirical antibiotic therapy.
Methods: Retrospective review of cases of peritonsillar abscess identified by diagnostic coding in three centres in Northern Ireland between August 2001 and July 2002.
Results: 128 patients with confirmed peritonsillar abscess were treated as inpatients accounting for 1 in 10,000 per year of the population in the hospitals’ catchment area. The mean age was 26.4 years (range 9–78). Sixty-nine (54%) patients were male; the mean length of hospital stay was 3 days. Needle aspirates, swabs of pus, throat swabs and blood were submitted for microbial culture. Culture yield was greatest from needle aspirates, and was similar even with prior antibiotic exposure, although the relative frequency of pathogens was different in the group who had received prior antibiotics. Beta-haemolytic streptococci were the most common isolates, however a variety of pathogens were implicated. Throat swabs and blood cultures were typically unhelpful. The results of culture and sensitivity did not affect individual patient treatment, but reviewing the sensitivities demonstrated frequent resistance of isolates of Group A beta-haemolytic streptococci to macrolide antibiotics. Heterophil antibody testing was routine and revealed that Epstein–Barr Virus infectious mononucleosis had a prevalence of 1.8% in this population.
Conclusion: We support the view that aspirates of pus from peritonsillar abscesses should be periodically cultured to guide empirical antibiotic management since performing cultures in every case may be unnecessary. Patients who have taken antibiotics prior to aspiration may be included in such surveillance.
P1347: Single-dose azithromycin microspheres versus three-day azithromycin for the treatment of group A beta-haemolytic streptococcal (GABHS) pharyngitis/tonsillitis in adults and adolescents
S. Kegel, C. McCoig, D. Jorgensen (Erkner, D; Madrid, E; New London, USA)
Objective: Antibiotic therapy for GABHS pharyngitis is recommended for earlier symptom resolution, prevention of complications, and reduced spread of disease. Azithromycin (500 mg QD for 3 days) is an effective treatment for documented GABHS pharyngitis in adults. A novel microsphere formulation of azithromycin now makes it possible to administer a full course of therapy as a single dose while maintaining tolerability and optimizing compliance. The objective of this study was to test the hypothesis that a single 2.0 g dose of azithromycin microspheres is bacteriologically noninferior to 3 days of azithromycin (500 mg QD for 3 days) when used to treat adults and adolescents with GABHS pharyngitis/tonsillitis.
Methods: This was a Phase III multicentre, randomized, double-blind, double-dummy trial conducted in North America, Europe, and India. The primary endpoint was bacteriologic response at Test of Cure (TOC; Day 24–28) in the Bacteriologic Per Protocol (BPP) population. The secondary endpoints were clinical response at TOC and safety.
Results: Five hundred ninety-eight subjects were enrolled in the study; of the 594 treated subjects 420 (70.7%) were included in the BPP population. Bacteriologic eradication was achieved in 86.3% (177/205) and 81.4% (175/215) subjects in the azithromycin microspheres and 3-day azithromycin groups, respectively at TOC (95% CI (2.1, 12.0). At Long-Term Follow Up (LFTU; Day 38–45), bacteriologic recurrence was observed in 5.5% (9/163) subjects in the azithromycin-microspheres group, compared with 7.7% (12/156) subjects in the 3-day azithromycin group. Clinical cure was observed in 99.0% (203/205) of subjects in the azithromycin microspheres group and 96.7% (208/215) in the 3-day azithromycin group. Both treatments were well tolerated and most adverse events were mild to moderate in intensity. The most frequent adverse event (AE) was diarrhoea/loose stools, which occurred in 11% of both treatment groups. No subjects in either group discontinued treatment due to treatment-related AEs.
Conclusion: A single 2.0 g dose of azithromycin microspheres is as effective and well tolerated as 3 days of azithromycin (500 mg QD) for treating GABHS pharyngitis in adults and adolescents.
P1348: Predicting prognosis and effect of antibiotic treatment in rhinosinusitis
A. De Sutter, M.B. Lemiengre, G. van Maele, M. van Driel, M. De Meyere, T. Christiaens, J. De Maeseneer (Ghent, B)
Background: In dealing with patients with suspected rhinosinusitis, family physicians have to rely mainly on history, physical examination and plain radiographs. Yet, evidence of the value of this information for the management is sparse. The aim of this study was to examine whether in patients with suspected rhinosinusitis illness duration and/or the effect of antibiotic treatment can be predicted on basis of clinical symptoms/signs or radiology.
Methods: Participants were 300 patients with suspected rhinosinusitis participating in an RCT comparing amoxicillin with placebo. By means of Cox regression we assessed the association between the presence at baseline of rhinosinusitis symptoms/ signs or an abnormal radiograph and the subsequent illness course. By testing for interactions we investigated whether the presence at baseline of any of these symptoms/signs could predict a beneficial effect of antibiotic treatment.
Results: ‘Poor general condition’ {Hazard ratio 0.77(0.60–0.99)} and ‘reduced productivity’ {(HR 0.68(0.53–0.88)} at baseline were independently associated with a prolonged course. None of the classical sinusitis-like symptoms, nor abnormalities on radiography had any prognostic value. Prognosis also remained unchanged whether or not the patient was treated with antibiotics, regardless of his baseline symptoms.
Conclusions: In a representative group of patients with suspected acute rhinosinusitis in FP, neither the presence of sinusitis symptoms/signs nor an abnormal radiograph provided information with regard to the prognosis or effect of amoxicillin treatment. Patients who felt poorly at baseline, or didn't feel able to work, needed more time to recover, but this could not be influenced by amoxicillin.
P1349: Effects of moxifloxacin and clarithromycin on the chlamydial gene expression in treatment-refractory persistent Chlamydia pneumoniae infection
J. Rupp, V. Wobbe, M. Maass (Lübeck, D)
Objectives: Chlamydia pneumoniae causes chronic infections that have been related to asthma bronchiale and atherosclerosis. These chronic infections cannot be eradicated by short-term antimicrobial treatment due to a non-replicating persistent state of C. pneumoniae that survives and continues to synthesize mRNA in the presence of antibiotics. It is unknown if antibiotics modify the gene expression profile of these persistent chlamydiae in a clinically relevant manner. Therefore, we analysed the differential expression of selected genes in the presence of moxifloxacin and clarithromycin, which both are active in acute chlamydial infection, using a model of chlamydial persistence in peripheral blood monocytes (PBMC).
Methods: Persistent infection with C. pneumoniae was initiated in PBMC in established methodology. Infected cells were continuously exposed to moxifloxacin (3.1 μg/ml) or clarithromycin (2 μg/ml) or kept in antibiotic-free medium. Differential gene expression of selected C. pneumoniae target genes (type III secretion: LcrD, YscL, YscN; inclusion membrane: IncA, IncC; amino acid metabolism; TyrB, GlyA) was quantitatively analysed between 1 and 192 h after infection by RT-PCR using the LightCycler system.
Results: In antibiotic-free medium gene expression levels varied individually over time for each gene analysed. However, all target genes showed a 5- to 9-fold upregulation of mRNA peaking at 120 h under exposure to moxifloxacin. This was less prominent under clarithromycin.
Conclusion: Our initial expectation to further reduce transcription levels in persistent C. pneumoniae by adding antibiotics proved wrong. The surprisingly clear upregulation of gene expression under moxifloxacin rather indicated a general regulatory effect of the drug on the pathogen. This is the first demonstration that chlamydiae in the persistent state are not completely inert and show a reaction to antibiotics. It is under investigation whether protein synthesis is upregulated in parallel. The potential clinical relevance of this finding regarding host cell response and course of the infection remains to be analysed in in vivo models for which this study provides the basis.
P1350: The application of a validated risk score safely reduces the rate of hospital admission in patients with community-acquired pneumonia presenting to the emergency department
L. Richeldi, F. Marchetti, M. De Guglielmo, D. Giovanardi, L. Fabbri (Modena, Verona, I)
Objectives: Patients receiving a diagnosis of community acquired pneumonia (CAP) in the hospital Emergency Department (ED) need to be carefully evaluated to be treated as in- or out-patients. The Pneumonia Score Index (PSI) is a validated score predicting the short term risk of death in patients with CAP and could therefore assist in deciding whether a single patient with CAP needs to be hospitalized or not.
Methods: We implemented a computer-based critical decision pathway for the management of patients with CAP, based on the PSI score and a dedicated software (GesPOrEx©). Briefly, all adults aged >18 years with provisional diagnosis of CAP were eligible for inclusion in the study. All patients received written information about their diagnosis of pneumonia and their treatment plan. CAP was defined as the presence of new pulmonary infiltrate on chest Rx and symptoms consistent with pneumonia, including cough, dyspnea, change in sputum, pleuritic chest pain. The PSI score was then calculated for all patients meeting eligibility criteria. Patients with scores of 90 points or lower were recommended for outpatients treatment, whereas those with higher scores were recommended for hospital admission. The PSI score was used only as a guide to the admission decision and did not superseded clinical judgment. The follow-up consisted of two visits, within 10 days and about 1 month after discharge from the hospital.
Results: The protocol was applied to 117 consecutive patients with CAP presenting at our ED. Compared to the previous year, we detected a significant 37% reduction (p < 0.001, 95% CI 26–49%) in the rate of admissions to the hospital of patients with CAP. Moreover, the length of stay in the hospital showed a trend toward reduction (from 9 ± 2 days before protocol implementation, to 7 ± 5 days). In the 3 months follow up, we did not detect any re-hospitalization in patients treated as out-patients and the rate of cure was similar before and after the protocol implementation.
Conclusion: We estimated that the application of this critical pathway generated a potential saving of about 110.00 Euros in one year. Interestingly, after the study end the trend towards a reduction in admission rate for CAP patients was maintained, thus suggesting that the use of the PSI score entered clinical practice with persistent beneficial effects on clinically safe and cost-effective management of CAP patients.
P1351: Adult community-acquired pneumonia. Evaluation of the antibiotherapy proposed in an emergency department and analysis of the impact of training
L. Piglione, V. Blanc, L. Lerousseau, D. Rafidiniaina, C. Rotomondo (Antibes, F)
Objectives: We evaluated the quality of adult community-acquired pneumonia (CAP) probabilist antibiotherapy within an Emergency Department and the influence of a targeted training on this prescription.
Methods: Study took place in a medium sized general hospital. All the files related to patients admitted for the diagnosis of CAP and hospitalized were reviewed in a prospective way by a local committee of independent Experts during the first half-year 2003 (T1) and the first half-year 2004 (T2), and compared. Between the two periods, the prescribing physicians have been trained (educational sessions based on the guidelines for pneumonia care of the French Society of Infectious Diseases (SPILF)). Three major quality indicators were used: initial molecule selection, dosage and time to first dose. Cost of antibiotherapy was calculated.
Results: 53 files were selected during T1 and 42 during T2. The 2 groups were well matched (age, sex, Fine's score). Inadequate initial molecule selection decreased from 43% to 29% (NS), inadequate dosage decreased from 4% to 0% (NS). Time to first dose was <4 hours in the 2 groups. Between T1 and T2, the prescription of amoxicillin increased by 15%, at the expense of fluoroquinolones (−13%) and of the 3rd-generation cephalosporins (−9%). Macrolides prescription remained stable. Antibiotherapy cost decreased by a 2.5 fold ratio (p < 0.05)
Conclusions: Our work demonstrates that, even if the dosage errors are rare and the antibiotic therapy is always early, there was a mediocre adhesion to the recommendations as regards to the choice of the molecule, before the intervention. Even though results obtained with our targeted training are modest, they are encouraging as they increased the use of guidelines recommended antibiotherapy, and leaded to significant cost gain. The development of different educational strategies is essential for the application of good medical practices for a better patient's management from the moment they are admitted in the Emergency Department.
P1352: Moraxella catarrhalis replacement in the nasopharynx of asymptomatic children
A. Sulikowska, P. Grzesiowski, W. Hryniewicz (Warsaw, PL)
Objective: The aim of the study was to analyse, using phenotypic and molecular methods, the dynamics of nasopharyngeal carriage of Moraxella catarrhalis in asymptomatic children sampled at two time points with a 4-month interval.
Methods: 77 children (43 from an orphanage and 34 from a day care centre – DCC) were examined twice, first in Winter and again in the following Spring. Clinical data and information about sex, age, socio-economic status, respiratory tract infections and antibiotic treatment within 3-months prior the sampling, were collected by questionnaire. Nasopharyngeal swabs were processed and M. catarrhalis was identified by standard procedures. The β-lactamase production was determined using the nitrocefin test. Pulsed-field gel electrophoresis (PFGE) of BcuI-digested M. catarrhalis DNA was performed to determine the relatedness among isolates.
Results: Altogether 58 M. catarrhalis isolates were identified; 35 were derived from children from the orphanage and 23 from the DCC. Thirty children (17 from the orphanage and 13 from the DCC) were colonised in Winter and 28 (18 from the orphanage and 10 from the DCC) in Spring. Fourteen children (9 from the orphanage and 5 from the DCC) were colonised by M. catarrhalis both in Winter and Spring. Out of them only one child was colonised by the same strain during the first and second sampling. In most cases, M. catarrhalis isolated at 2 different points of the year from the same child were unrelated. All isolates of M. catarrhalis were b-lactamase producers.
Conclusions: Colonisation of nasopharynx by M. catarrhalis represents a dynamic process – bacteria are acquired, eliminated and re-acquired over a period of 3–4 months. Long-term colonization (over 4 months) involved only 1 of the 77 investigated children.
P1353: Characterisation of invasive and non-invasive Haemophilus influenzae-isolates in a highly vaccinated population
B Gröndahl (Mainz, D)
Objectives: Characterization of invasive and non-invasive Haemophilus influenzae (Hi)-strains has not been done since the implementation of general vaccination against Hi type b in Germany.
Methods: Invasive isolates were acquired through a population based, nationwide surveillance using 2 detection systems. Colonizing, non-invasive isolates were isolated from nasopharyngeal aspirates of hospitalized children with acute respiratory diseases.
Results: From 09/2001 to 08/2004 a total of 175 invasive isolates were collected, of which 160 (91.4%) could be evaluated. Most strains were non-capsulated (n = 99; 56.6%). The most frequent capsular type was type b (n = 39; 22.3%), followed by types f (n = 19; 10.9%), e (n = 3; 1.7%) and a (n = 1; 0.6%). Types c and d were not detected. There was no increase of ‘non-b'-isolates over the observation period. More capsulated Hi (13%) were detected by PCR than by agglutination (n = 62 vs. n = 54). The most frequent biotype was type I (n = 65; 37.1%), followed by type II (n = 51; 29.1%) and type III (n = 26; 14.9%). Only a small proportion of isolates produced beta-lactamase (n = 6; 3.4%). In the same period 102 non-invasive, colonizing Hi were isolated. All strains but one (capsular type e) were non-capsulated. No Hi type b could be detected. The most frequent biotype was type II (n = 41; 42.7%), followed by type I (n = 23; 24.0%) and III (n = 20; 20.8%). Only four isolates produced beta-lactamase (3.9%).
Conclusions: Haemophilus influenzae type b as colonizing or as invasive pathogen has almost disappeared in Germany. Replacement was not observed. Invasive or colonizing Hi-isolates rarely produced beta-lactamase (3.4 – 3.9%).
P1354: Chlamydia-like micro-organism, Simkania negevensis, as a possible emerging pathogen in Finland?
M. Paldanius, J. Hietala, M. Leinonen, M. Uhari, P. Saikku (Oulu, FIN)
Objectives: Simkania negevensis, environmental Chlamydia-like intracellular bacterium, has been shown, in a few reports, to be associated to respiratory infections in infants and in adults (1). There is no data available on the prevalence of S. negevensis antibodies or infections from Finland. We developed microimmunofluorescence (MIF) method for the measurement of S. negevensis antibodies and tested paired sera obtained from Finnish children with infectious episode using this method.
Methods: Serum samples from 262 children were screened for IgG, IgA and IgM antibodies by in-house MIF test utilizing urografin purified formalinized bacteria of S. negevensis, ATCC strain (VR-1471) as antigens. The incubation time was overnight. If the patient had three sera, the middle serum sample was selected and when only paired sera were available, the last serum was selected for the screening. All S. negevensis positive screenings (a titer of ≥8) were tested with paired sera (or three sera). The presence of IgM antibodies (after removing IgG antibodies with Gullsorb reagent) or four-fold titer rise in IgG or IgA between paired sera were considered diagnostic for acute infection.
Results: The prevalence of S. negevensis antibodies was 19 % for IgG, 0 % for IgA and 5.7 % for IgM. Acute S. negevensis infection was diagnosed in altogether 23 (8.8%) of 262 children: in six cases by IgG seroconversions and in 18 cases by the presence of IgM antibodies.
Conclusion: S. negevensis antibodies were demonstrated for the first time in Finnish children. Serological diagnosis of acute S. negevensis infection was obtained in 9% of children. Further studies using e.g. direct demonstration of S. negevensis by culture and PCR in clinical samples are needed to elucidate the pathogenetic significance and clinical picture associated to this bacterium. MIF method developed in this study seems to be suitable for the measurement of S. negevensis antibodies and there seems to be no cross-reactivity with Chlamydia pneumoniae antibodies when using this test. 1. Friedman MG, Dvoskin B, Kahane S. Infections with the Chlamydia-like microorganism Simkania negevensis, a possible emerging pathogen. Review. Microbes Infect 5:1013–21, 2003
P1355: Macrolide treatment failure in pulmonary Nocardia nova infection
Y. Keynan, H. Sprecher, G. Weber (Haifa, IL)
Objectives: To describe failure of Clarithromycin in the treatment of pulmonary Nocardia Nova infection, despite in vitro susceptibility to macrolides.
Patient and Methods: A 61-year-old male with diabetes mellitus, non Hodgkin's lymphoma after chemotherapy, chronic adrenal replacement therapy (due to lymphomatous infiltration of the adrenals), presented with persistent cough in the previous 3 month. Weakly acid fast bacilii were detected in sputum and treatment with ethambutol, rifampin and clarithromycin was given for presumed MOTT infection. Although the patient reports strict compliance with drug therapy for three weeks, neither symptomatic nor radiographic improvement was noted. Nocardia spp. was isolated from sputum culture. DNA was extracted from bacterial colonies. A 439-bp fragment encompassing the hsp-65 gene was PCR-amplified and subsequently digested with BstEII and HaeIII endonucleases. Bacterial DNA was also PCR-amplified using a primer pair specific for conserved regions of 16S rRNA. The nucleotide sequence of the resulting amplicons was established and analysed using the BLAST software.
Results: PCR-RFLP analysis revealed a restriction pattern compatible with that of N. Nova. This identification was ultimately confirmed as N. Nova, (99% homology) based on BLAST analysis of the bacteria 16S rRNA sequence. The isolate was sensitive to erythromycin. The patient received trimethoprim/sulfamethoxazole with gradual clinical and radiologic improvement. After three month of follow-up he remains well.
Conclusions: Nocardia Nova may account for 20% of Nocardia isolates identified as N. asteroides. N. Nova strains were previously noted for their susceptibility to ampicillin, cephalosporins and erythromycin. A macrolide was suggested as an alternative to sulfonamide in nonresponders or those with allergy to sulfa. The apparent disparity between in vitro susceptibility to macrolides and clinical failure warrants cautious use of these antibiotics in N. nova infections. Sequence based identification allows for accurate separation of N. nova from other related species.
Pneumococcal pneumonia
P1356: Changing antimicrobial susceptibility patterns among S. pneumoniae and H. influenzae from Brazil: report from the SENTRY Antimicrobial Surveillance Programme 1998–2003
H. Sader, A. Gales, R. Jones (North Liberty, USA; Sao Paulo, BR;)
Background: Although antimicrobial resistance (R) rates among S. pneumoniae (SPN) and H. influenzae (HI) have increased significantly in most countries in the last years, most studies from Brazil report relatively low R rates among these pathogens. We analysed the susceptibility (S) patterns of SPN and HI from Brazil (6 years) for significant trends.
Methods: 729 SPN and 566 HI collected from 1998–2003, mainly from respiratory tract and bloodstream infections, were susceptibility (S) tested by NCCLS broth microdilution methods against >30 drugs and the results analysed by year.
Results: Results are summarized below:
| % S by year (SPN/HI) |
||||||
|---|---|---|---|---|---|---|
| Antimicrobial | 1998 | 1999 | 2000 | 2001 | 2002 | 2003 |
| Penicillin (PEN) | 84 (3)a/10b | 83 (6)a/13b | 75 (5)a/7b | 82 (8)a/16b | 78 (8)a/16b | 72 (10)a/ |
| Erythromycin (ERY)/Clarithromycin | 85/90 | 88/100 | 91/92 | 90/91 | 94/90 | 91/94 |
| Clindamycin (CLI) | 94/- | 95/- | 96/- | 95/- | 100/- | 98/- |
| Gatifioxacin/Levofloxacin | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 |
| Chloramphenicol (CHL) | 99/96 | 98/99 | 99/97 | 100/95 | 99/93 | 100/96 |
| Tetracycline (TET) | 72/96 | 75/84 | 82/95 | 78/95 | 86/98 | 85/96 |
| Cotrimoxazole | 50/53 | 52/53 | 51/55 | 40/48 | 39/54 | 39/57 |
% RCMI (> 2 mg/L) to PEN.
β-lactamase producing strains.
Conclusions: R to PEN has increased markedly among SPN over 6 years (from 3 to 10%). R to T/S also escalated from 50 to 61%, but S to ERY, CLI and TET significantly decreased among SPN strains. R to the antimicrobials tested remained very stable among HI with only some year-to-year variations. R to the newer fluoroquinolones was not detected and CHL showed an excellent spectrum (>95% S) against both pathogens.
P1357: Contemporary activity of gemifloxacin and other fluoroquinolones tested against Streptococcus pneumoniae isolated during 1999–2004 in Europe and America
D. Biedenbach, J. Ross, H. Sader, T. Fritsche, A. Fuhrmeister, R. Jones (North Liberty, USA)
Objectives: To determine the potency of gemifloxacin (GEMI) tested against S. pneumoniae (SPN) for the years 1999–2004. The evaluation of GEMI compared to other currently marketed fluoroquinolones (FQ) will also be made including mechanisms of resistance (R).
Methods: During a six year period (1999–2004), a total of 5892 SPN isolates were collected from medical centres on three continents and tested for antimicrobial susceptibility (S) using reference NCCLS broth microdilution methods and interpretive criteria (M100-S15, 2005). The antimicrobial agents tested included five FQs: GEMI, ciprofloxacin (CIPRO), levofloxacin (LEVO), gatifloxacin (GATI) and moxifloxacin (MOXI). Analysis of the quinolone resistance determining region (QRDR) was performed for 35 FQ- R strains (LEVO MIC at >4 mg/L).
Results: The activity of GEMI against SPN over a six year period is shown in the table: During the six years, the rank order of potency (MIC90, mg/L) for the FQs was GEMI (0.03) > MOXI (0.12–0.25) > GATI (0.5) > LEVO (1–2) > CIPRO (2). All FQ median and modal potencies remained stable over the monitored interval. SPN isolates with CIPRO MIC values ≥4 mg/L ranged from 1.6 to 2.1% with no detectable trend towards increasing R across all regions. The most common QRDR mutations among strains with CIPRO and LEVO MIC values ≥4 mg/L were: gyrA (S83F or T), parC (S79F or T and D83N) and parE (I460V). Against these isolates, GEMI maintained the most potent activity at MIC50/90 values of 0.5/1 mg/L and a MIC range of only 0.25–2 mg/L.
| GEMI-MIC (mg/L) |
% by category |
||||
|---|---|---|---|---|---|
| Year (no. tested) | 50% | 90% | S | I | R |
| 1999 (554) | ≤0.03 | ≤0.03 | 99.6 | 0.4 | 0.0 |
| 2000 (508) | ≤0.03 | ≤0.03 | 99.6 | 0.2 | 0.2 |
| 2001 (1.435) | 0.016 | 0.03 | 99.4 | 0.3 | 0.3 |
| 2002 (1.502) | 0.016 | 0.03 | 99.5 | 0.3 | 0.2 |
| 2003 (890) | 0.016 | 0.03 | 100.0 | 0.0 | 0.0 |
| 2004 (1.003) | 0.016 | 0.03 | 99.2 | 0.7 | 0.1 |
| All years (5.892) | 0.016 | 0.03 | 99.5 | 0.3 | 0.2 |
Conclusions: Among the FQs tested, GEMI was clearly the most potent agent and remains a valuable candidate for treating multidrug R SPN, an increasingly observed respiratory-tract pathogen. During the study years, FQ-R did not increase significantly and S rates to GEMI remained at >99.2% and was unrelated to increasing R among beta-lactam and macrolide antimicrobials.
P1358: Resistance patterns of S. pneumoniae and H. influenzae isolated from children with upper respiratory tract infection in Southern Brazil − 2003 and 2004
C.M. Mendes, A.C. Arruda, C. Oplustil, J.L. Sampaio, P. Garbes, P. Lima, L. Pedneault, A. Gonçalves, C. Dias, A. Barth, C.R. Kiffer (Sao Paulo, Rio de Janeiro, BR; Brussels, B; Porto Alegre, BR)
Objective: Establish the susceptibility profile of S. pneumoniae and the beta-lactamase production of H. influenzae isolated from children with upper respiratory tract infection (URTI) from 3 cities in Brazil.
Methods: Samples (one per patient) were selected from patients under 7 years old during 2003 and 2004. Only patients with clinical diagnosis of URTI and a positive culture result for S. pneumoniae or H. influenzae were included in the study. Clinical data related to age, gender, diagnosis and samples were described. S. pneumoniae isolates were tested against penicillin, amoxicillin, amoxicillin/clavulanic acid, cefuroxime, cefaclor, and azithromycin. Minimum Inhibitory Concentrations (MICs) were determined by E-test methodology (testing conditions 35C; 5%CO2; 20–24 hours). Interpretative criteria used were those described by NCCLS documents M100-S14 for all antimicrobials, except for azithromycin (AB Biodisk). H. influenzae isolates were tested for beta-lactamase production by a chromogenic cephalosporin method.
Results: There were 137 S. pneumoniae and 170 H. influenzae isolates from children under 7 years old. Demographic data are presented in the table. Of H. influenzae, 10.6% were betalactamase producers. Among S. pneumoniae, 66.4% were susceptible (S), 23.4% intermediate (I) and 10.2% resistant (R) to penicillin (MIC90 = 1.5 μg/mL). As for azithromycin, 90.5% were S, 1.5% I, and 8% R (MIC90 = 4 μg/mL). Only one isolate (0.7%) was resistant to amoxacillin (MIC = 3.0 μg/mL).
Demographic data – survey on children with clinical diagnosis of URTI and a positive culture result for S. pneumoniae or H. influenzae
| S. pneumoniae (137) n(%) | H. influenzae (170) n(%) | ||
|---|---|---|---|
| Age Group: | |||
| < 1 year-old | 14 (10.2%) | 16 (9.4%) | |
| 1 to 2 years-old | 49 (35.8%) | 55 (32.4%) | |
| 3 to 4 years-old | 45 (32.8%) | 58 (34.1%) | |
| 5 to 7 years-old | 29 (21.1%) | 41 (24.1%) | |
| Gender: | |||
| male | 59 (43.1%) | 91 (53.5%) | |
| female | 78 (56.9%) | 79 (46.5%) | |
| Clinic: | |||
| outpatient | 110 (80.3%) | 151 (88.8%) | |
| inpatient | 6 (4.4%) | 8 (4.7%) | |
| not available | 21 (15.3%) | 11 (6.5%) | |
Conclusions: A significant rate of beta-lactamase production in H. influenzae was detected. The prevalence of penicillin intermediate/resistant S. pneumoniae, was high and more common compared with previous years. Empiric therapy with penicillins alone or in low dose should be avoided in this population. Amoxicillin would still be appropriate to treat pneumococcal infections (despite penicillin resistance) and so would be co-amoxiclav.
P1359: Evidence for the emergence of non-vaccine types causing invasive pneumococcal disease in Spain
E. Cercenado, C. Arenas, A. Fenoll, E. Bouza (Madrid, Majadahonda, E)
Objectives: S. pneumoniae (Sp) is an important cause of morbidity and mortality in children and in adults. Vaccination reduces carriage and prevents the disease. In a nationwide point prevalence study, 147 institutions from all areas of Spain collected all Sp isolated during one week (February 16–22th, 2004). The isolates were sent to a central laboratory for susceptibility testing and serotyping. In this study we analyze the distribution of serotypes and the resistance to antimicrobials of Sp isolated from children and adults.
Methods: A total of 360 Sp isolates were identified. Susceptibility testing was performed by the broth microdilution method in cation-adjusted Mueller-Hinton broth with 5% lysed horse blood following NCCLS guidelines. Serotyping was performed by standard methods.
Results: The isolates belonged to 251 adults (70%) and 109 children. Origins were: respiratory tract (48%), blood (24%), ear (12%), conjunctiva (7%), CSF (4%), other sterile fluids (3%) and miscellaneous (2%). Penicillin resistance (I+R) was 42%, and erythromycin resistance was 36%. A total of 34 isolates were non-typeable and were excluded for further analysis. The most frequent serotypes (St) were 3, 19F, and 19A. St 14, 19F, and 23F were the most frequent among the penicillin-resistant strains, and St 3 among susceptible strains. The most frequent St of the 96 invasive isolates (blood and CSF) were 14 and 19F, in children and in adults, respectively. Among the 96 invasive isolates, 84% corresponded to St included in the 23-valent (23-V) vaccine, and 46% to St included in the 7-valent (7-V) vaccine. The most frequent non-vaccine St were 6A, 16, 31, 34, 35B, and 35F. A total of 106 (45%)of Sp isolated from adults belonged to St frequently isolated from children (6, 14, 18, 19, and 23). Considering Sp isolated from all origins, the estimated coverage of the 7-V vaccine was 40% in children and 38% in adults; and the estimated coverage of the 23-V vaccine was 77% in adults. Considering only invasive isolates, the estimated coverage of the 7-V vaccine was 61% in children and 41% in adults; and the estimated coverage of the 23-V vaccine was 79% in adults.
Conclusions: These results confirm the high rates of resistance of Sp to penicillin and erythromycin, the spread to adults of St frequently isolated from children, the evidence for the emergence of non-vaccine types causing invasive pneumococcal disease, and the moderate coverage of the 7-valent pneumococcal vaccine.
P1360: Incidence, predisposing conditions and outcome of invasive pneumococcal infections in Finland
P. Klemets, O. Lyytikäinen, P. Ruutu, J. Ollgren, P. Nuorti (Helsinki, FIN)
Objectives: To determine the incidence of invasive pneumococcal infections (IPI) in patient groups recommended to receive pneumococcal polysaccaride vaccine (PPV23) as well as outcome of the illness.
Methods: All laboratories performing blood and cerebrospinal fluid (CSF) cultures submitted data on isolation of Streptococcus pneumoniae from blood or CSF during 1995–2002. Information on vital status, comorbidities and denominator data on persons at risk were obtained from the Population Information System, National Hospital Discharge Registry, Cancer Registry, National Social Insurance Institution, and National Infectious Disease Registry. The patient's national identity code was used for linking databases. Only the first episode of IPI was included in the analysis.
Results: A total of 4357 episodes of IPI were identified (incidence 10.6 cases per 100,000 population). Highest incidence of IPI was seen in patients with haematological malignancy (547 per 100,000 population), organ or bone marrow transplantation (164), males aged ≥55 with chronic obstructive pulmonary disease (143) and persons with HIV (130). The overall case-fatality proportions (CFPs) at 7, 28 and 90 days after the positive culture were 9%, 12% and 16%, respectively. At 28 days the CFPs ranged from 1% to 25% in different age groups. In patients aged ≥50 they were significantly higher in men than in women (20% vs 15%; p < 0.01). Patients with non-hematological malignancy and alcoholism had the highest CFPs at 28 days, 31% and 27%, respectively. In patients with hematological malignancy the 28-day CFP was 16% and 6% among healthy adults aged 18–64 years. In adults aged 18–64 years, 931 (42%) of 2216 cases had an underlying condition for which PPV23 is recommended.
Conclusion: The incidence and outcome of IPI varied considerably in different patient groups. However, the patient groups with highest rates of IPI were not always the same as those at highest risk of death. The overall mortality associated with IPI was nearly twice as high 90 days after infection compared with the first week, suggesting the contribution of pre-existing illness or long-term sequelae. Among non-elderly adults, the high prevalence of underlying conditions for which PPV23 is recommended emphasizes the importance of vaccinating groups at highest risk of disease and death.
P1361: Usefulness of prognostic score systems – Pneumonia Severity Index (PSI), CURB-65, and Ewig scores – in community-acquired bacteraemic pneumococcal pneumonia
C. Spindler, Å. örtqvist (Stockholm, S)
Objectives: Predictive score systems for CAP have all been developed for the heterogeneous group of ‘all’ CAP, but none have been evaluated for pneumonia caused by a single pathogen. Streptococcus pneumoniae is the most common cause of CAP and the most common cause of fatal pneumonia. We have prospectively studied the accuracy of three score systems, PSI, CURB-65, and Ewig's score, for predicting need of ICU-treatment, and death due to community-acquired bacteraemic pneumococcal pneumonia (BPP).
Methods: All adult patients (n = 114) with invasive pneumococcal disease (IPD) and x-ray verified pneumonia at Karolinska and Danderyd Hospitals, Stockholm, Sweden, 1999–2000, were included. 88 patients were treated in the dpt of Infectious Diseases (DID) and 26 patients in other dpts (ODs). Severity scores were calculated according to the original publications and the independent prognostic importance of different variables was analysed by multiple regression analyses.
Results: PSI > III, CURB-65 > 1, and presence of 1 major or >1 minor risk factor in Ewig's score all had a high sensitivity, but somewhat less good specificity, for predicting fatal outcome (Table). The area under the Receiver Operating Characteristics curves for predicting death were between 0.83 and 0.85 for all three tests. The death rate was 12% (14/114), but patients treated in DID had a significantly lower rates than those treated in ODs, 5% (4/88) vs. 35% (10/26) (p < 0.0001). Also within the same severity score strata mortality was lower among patients treated in DID than in ODs. The fatality for a PSI-score of IV-V was 9/20 in OD vs. 4/33 in DID (p = 0.01). No other factors explaining this difference were found in the multiple regression analyses.
| Mortality |
IUC-treatment |
|||||
|---|---|---|---|---|---|---|
| PSI IV-V | CURB-65 (2-5) | Ewig (1 major or >1 minor) | PSI IV-V | CURB-65 (2-5) | Ewig (1 major or >1 minor) | |
| Sensitivity | 100% | 92% | 85% | 95% | 85% | 90% |
| Specificity | 60% | 67% | 84% | 64% | 70% | 90% |
| PPV | 25% | 27% | 41% | 36% | 38% | 67% |
| NPV | 100% | 99% | 98% | 98% | 96% | 98% |
Conclusions: All score-systems were useful for predicting need of ICU-care and risk for death due to BPP. PSI was the most sensitive, but CURB-65 score more easy to use. That the death rate was 3–4 times higher for patients treated in OD, compared to those treated in DID, despite the same severity score, illustrates that comparisons of mortality between different patient materials must be interpreted with caution, even if the analysis is based on calculation of severity of disease.
P1362: Prevalence of international clones among invasive isolates from Portugal
I. Serrano, J. Melo-Cristino, M. Ramirez (Lisbon, P)
Objective: To evaluate the prevalence of international clones recognized by the Pneumococcal Molecular Epidemiology Network (PMEN) among invasive isolates from Portugal.
Methods: A collection of 465 invasive isolates from Portugal (1999–2002) described recently (Serrano, I., et al. 2004. Invasive Streptococcus pneumoniae from Portugal: implications for vaccination and antimicrobial therapy. Clin Microbiol Infect 10:652–6.) was characterized using a combination of macrorestriction profiling, using SmaI and pulsed field gel electrophoresis (PFGE), and multi-locus sequence typing. Serotypes 14, 1, 3, 4, 8, 9V, 23F, 7F, 19A and 12B were the 10 most prevalent overall by decreasing rank order. A total of 12% of the strains were recovered from children <2 years old.
Results: By combining the PFGE data with the sequence types (ST) of 104 isolates we were able to identify the genetic lineages of the majority of the strains. We found 66 STs, including 20 novel STs, corresponding to 47 different lineages by e-BURST analysis. Out of the 26 clones currently recognized by the PMEN we found representatives of 5 among our collection: Spain23F-1, Spain6B-2, Spain9V-3, England14-9, Poland6B-20, Greece6B-22 and Colombia23F-26. The clones Greece6B-22 and Spain6B-2 grouped into a single PFGE cluster including 57% (n = 8) of the isolates expressing serotype 6B. An additional 3 isolates belonged to the same cluster as the clone Poland6B-20, such that 79% of the isolates expressing serotype 6B belonged to either one of the clones. In serotype 9V all the isolates (n = 21) belonged to the same cluster as clone Spain9V-3. This same clone accounted for the majority (n = 52) of the serotype 14 isolates. Since the cluster including the England14-9 clone encompassed 9 strains, a total of 98% of the strains expressing serotype 14 belonged to PMEN clones. Among serotype 19A strains, clone Spain23F-1 accounted for 41% (n = 7) of the isolates. The clones Colombia23F-26 (n = 17) and Spain23F-1 (n = 3) accounted for 91% of all 23F isolates. The PMEN clones accounted for 87% of penicillin non-susceptible (PNS) strains, including all resistant isolates, as well as the majority (61%) of the strains resistant to erythromycin.
Conclusion: International clones recognized by the PMEN accounted for 26% of all isolates in our collection including 87% of PNS and 61% of erythromycin resistant strains. Strains belonging to clone Spain9V-3, expressing either serotype 9V or 14, constituted 16% of all isolates.
P1363: Pneumococcal bacteraemia in children in northern Taiwan: clinical, serotyping and genotyping analysis
C.L Lin, Y.C Huang, C.-H Chiu, T.Y Lin (Kweishan, Taoyuan, TW)
Objectives: To assess the association among the clinical presentations of children with Streptococcus pneumoniae bacteraemia and the serotypes and genotypes of the isolates.
Methods: Children less than 18 years of age with S. pneumoniae bacteraemia from Chang Gung Children Hospital were identified retrospectively from January, 2001, to December, 2003. Demographic data, clinical features were reviewed. Clinical isolates were studied by standard microbiologic methods, Quellung test and pulsed-field gel electrophoresis.
Results: A total of 77 patients with 78 disease episodes and 79 isolates were included. The mean age was 2.5 yrs (1mo-9 yr) and 46 (59%) children were male. Clinical spectrums included occult bacteraemia (22/78, 28.2%), uncomplicated pneumonia (16/78, 20.5%), complicated pneumonia (15/78, 19.2%), meningitis (13/78, 16.7%), septic shock (5/78, 6.4%), septic arthritis (3/78, 3.8%), mastoiditis (1/78, 1.2%), and nosocomial bacteraemia (3/78, 3.8%). 82% of the isolates were penicillin nonsusceptible (MIC > 0.1 μg/mL). Nine children (12%) died, and 6 of them had underlying diseases. A total of 7 serotypes were found and three predominant serotypes, including 23F (30%), 14 (28%), and 6B (27%), were found. Serotype 14 accounted for 14 (45%) of 31 isolates from patients with pneumonia. Serotype 23F isolates (71%) presented more resistant to penicillin than serotype 14 (18%). 31 PFGE types were identified. There were only two genotypes with 10 or more isolates (15 and 10). Fourteen of 15 isolates of genotype 3 were serotype 23F. All 10 isolates of genotype 1 were serotype 14 and 7 of them caused pneumonia.
Conclusions: Pneumonia and occult bacteraemia were the two most common presentations. Three were three major serotypes. Relative clusters in two genotypes were identified with one clone associated with serotype 14 and pneumonia.
P1364: Levofloxacin is effective in the treatment of RTIs caused by penicillin-susceptible and penicillin-resistant Streptococcus pneumoniae (SELECT Study)
J. Yamakita, M.D. O'Hare (Tokyo, JP; Bournemouth, UK)
Objectives: To retrospectively assess the efficacy of levofloxacin for respiratory tract infections (RTIs) caused by penicillin-susceptible Streptococcus pneumoniae (PSSP) or penicillin-nonsusceptible S. pneumoniae (PNSSP).
Methods: 18 investigators in Europe (Belgium 2; France 6; Germany 1; Italy 4; Spain 5) collected data on 100 cases of levofloxacin-treated RTIs caused by S. pneumoniae (49 PNSSP [18 PRSP and 31 PISP], 51 PSSP). Each patient was treated with levofloxacin (i.v./oral), at a dosage of 500 mg once or twice daily for a planned minimum 7 day duration. S. pneumoniae was to have been isolated within 2 days prior to the start of levofloxacin treatment. Clinical outcome was analysed according to penicillin susceptibility. Other factors examined included medical history, primary clinical diagnosis, other antimicrobials prescribed, and number of days spent in hospital.
Results: There were no major differences in the subject demography or medical history between patients with PSSPs and PNSSPs. 90/100 subjects (90%) were clinical successes; two subjects (2%) were clinical failures (one in each group) and eight subjects (8%) died due to initial infection (4 in PSSP subjects and 4 in PNSSP subjects). Clinical success was equally common among patients with PSSPs (46/51 subjects, 90.2%) and PNSSPs (44/49 subjects, 89.8%). For all cases in total, there was no difference between clinical success rate of subjects with PSSP infection and those with PNSSP infection.
Conclusions: Levofloxacin is highly effective in the treatment of RTIs caused by S. pneumoniae, regardless of penicillin susceptibility, with 90.2% success in PSSPs and 89.8% success in PNSSPs. This is very similar to success rates in clinical trials and thus there is no evidence of a decline in efficacy over the seven years since the launch of levofloxacin in Europe. The usefulness of levofloxacin therapy for RTI due to PNSSP is clearly demonstrated.
P1365: Bacteriological outcomes with pharmacokinetically enhanced amoxicillin/ clavulanate (2000/125 mg) in patients with community-acquired respiratory infection caused by Streptococcus pneumoniae, including drug-resistant strains
S. Miller, M. Twynholm, E. Berkowitz, S. Gormley, A. White, L.A. Miller, C. Jakielaszek (Harlow, Greenford, UK; Collegeville, USA)
Objectives: To assess the bacteriological efficacy of pharmacokinetically enhanced amoxicillin/clavulanate 2000/125 mg (PKE AMX/CA) against Streptococcus pneumoniae infections, particularly strains resistant to penicillin+ ≥1 other antibacterial class (drug-resistant, DRSP), which are now of global clinical concern.
Methods: Bacteriological data were obtained from patients in the bacteriology per-protocol populations (≥1 pathogen isolated at screening and adherence to study protocol) of 13 clinical studies of PKE AMX/CA (community-acquired pneumonia [CAP], 6; acute bacterial sinusitis [ABS], 3; acute exacerbations of chronic bronchitis [AECB], 4). Data from 840 patients with S. pneumoniae isolated at screening were analysed to assess overall bacteriological efficacy against S. pneumoniae. MICs of AMX/CA (2:1 ratio based on AMX content), penicillin and 16 other antibacterials were determined, and isolate susceptibility assessed based on NCCLS breakpoints. Isolates resistant to penicillin (PRSP, MIC ≥2 mg/L) and ≥1 other antibacterial class were considered DRSP. Success, as defined in the individual trials, was the eradication or presumed eradication, based on clinical evidence, of the initial pathogen. Failure was the persistence/recurrence of the initial pathogen in a repeat culture sample or, in the absence of a repeat sample, clinical evidence of persistence/recurrence. Data from the follow-up visit of each study were used. Exact methodology was used to determine 95% confidence intervals (CIs).
Results: PKE AMX/CA achieved bacteriological success in 94.2% (791/840; 95% CI 92.4, 95.7) of patients with S. pneumoniae isolates in the pooled analysis. Against DRSP strains, success was 97.6% (81/83; 95% CI 91.6, 99.7) and against non-DRSP was 93.8% (710/757; 95% CI 91.8, 95.4). PKE AMX/CA was successful against 46/46 isolates with an AMX/CA MIC of 2/1 mg/L, 14/14 with an AMX/CA MIC of 4/2 mg/L and 9/11 with AMX/CA MICs of 8/4–16/8 mg/L. Bacteriological success against DRSP was 97.7% (43/44; 95% CI 88.0. 99.9) in CAP, 100% (33/33; 95% CI 89.4, 100) in ABS and 83.3% (5/6; 95% CI 35.9, 99.6) in AECB.
Conclusion: PKE AMX/CA was highly bacteriologically successful against S. pneumoniae, including DRSP, across all three respiratory indications studied, and against strains with elevated AMX/CA MICs. PKE AMX/CA should, therefore, be effective in treating S. pneumoniae respiratory infections, even when PRSP or DRSP are suspected.
P1366: Study of antibiotic resistant commensal and invasive Pneumococci isolated in Romania between 1996–2003
M. Pana, M. Ghita, I. Nistor, R. Papagheorghe, N. Popescu, G. Bancescu, M. Andrei, V. Ungureanu (Bucharest, RO)
Objective: To study the antibiotic resistance in Romanian Pneumococci isolated from carriers and ill people between 1996 and 2003.
Methods: 1109 strains of Streptococcus pneumoniae coming from blood, CSF, tracheal aspirate, pleural fluid, middle ear fluid and 376 strains coming from nasopharynx from HIV infected children were isolated between 1996 and 2003 at the National reference Center for Streptococcus. The strains were serotyped with in house co-agglutinant reagents prepared with pneumococcal antigens from Serum Staten Institute Copenhagen). The isolates were tested for susceptibility (MICs) to the following antibiotics: Penicillin(Pc), erythromycin(Em), cephalothin(Kf), cefuroxim(cxm), cefotaxim(Ctx), amoxicillin(Amx), trimethoprim/sulfamethoxazole(Sxt), ofloxacin(Ofx), vancomicin(va) by standard dilution MIC testing with Steers replicator.
Results: Breakpoints were used as proposed by NCCLS 2004. Invasive strains of Pneumococci showed lower levels of antibiotic resistance (40%Pc, 22%Kf, 7.3%Cxm, 5%Ctx, 6%Amx, 25%Em, 2.5%Ofx, 69%Sxt) against strains from carriers which revealed high levels of resistance (65%Pc, 27%Kf, 10.8%Cxm, 6%Ctx, 8.7%Amx, 59%Em, 4%Ofx, 74%Sxt). No resitant strain to Va was found. The pneumococcal strains isolated from carriers belonged only to few serotypes: 23, 14, 19, and 6 closely correlated with the antibioresistance. The most frequently serotypes encountered in invasive strains were: 8, 7, 1, 19, 14, 23, 6.
Conclusion: During the study period the most efficient antibiotics were: Cxm, Ctx, Amx and Ofx. There is an urgent need in Romania for the surveillance, prevention and control of antibiotic resistant Pneumococci and to enhance the use of an efficient pneumococcal vaccine.
Pneumonia in non-HIV immunosuppressed patients
P1367: Pneumocystis jiroveci-pneumonia in immununodeficient patients without AIDS
T. Blum, A. Roth, H. Mauch, H. Lode (Berlin, D)
Introduction: Patients with pneumonia resistant to treatment prove to be a common problem in chest Hospitals. Pneumocystis jiroveci (PJ) should always be thought of as an opportunistic pathogen in case of potential, especially T-cell-related immunodeficiency – even if AIDS-disease is not apparent. We report on cases of PJP without associated AIDS-disease in a chest hospital.
Objectives: The aim of the study was to investigate frequency, diagnostic procedures, course and outcome of PJP without associated AIDS-disease in our hospital.
Methods: In a retrospective study, we evaluated hospitalized patients presenting with pneumonia during January 1st 2003 and August 31st 2004 in our hospital. The identification of the cases with PJP was based upon discharge diagnosis as well as our microbiological database (confirmed PJ in broncho-alveolar lavage by immunofluorescence-test (IFT) and/or PCR).
Results: The diagnosis of PJP without associated AIDS-disease could be made in 7 out of 506 patients (1.4%) hospitalized because of proven pneumonia. All 7 patients were treated with immunosuppressive medication prior to admission (indications: pulmonary fibrosis, COPD, and cerebral edema respectively). CD4-cell-counts were substantially decreased (CD4 cells <200/ μl) in 3 out of 6 cases. To establish the diagnosis of PJP a PJ-PCR (using broncho-alveolar lavage as material) was necessary in 5 cases – in all of these cases the PJ-IFT proved falsely-negative. Severe hypoxemia could successfully bridged by non-invasive Ventilation in 2 patients, l patient had to be ventilated invasively. The mortality-rate was 28.6 % (2 out 7 patients). See table 1 for details.
Table 1.
| Initials | Age | Sex | Comorbidity | Immunosuppressive medication prior to admission | CD4 cell count (per microliter) | PJ-IFT | PJ-PCR | Outcome |
|---|---|---|---|---|---|---|---|---|
| 1 E.B. | 68 | female | Idiopathic pulmonary fibrosis | prednisolone 30 mg daily azathioprine 75 mg daily | 264 | − | + | deceased |
| 2 R.B. | 80 | female | COPD, cerebral edema | dexamethasone 12 mg daily | 46 | − | + | alive |
| 3 K.E. | 66 | male | Idiopathic pulmonary fibrosis | prednisolone 20 mg daily azathioprine 75 mg daily | 490 | + | + | alive |
| 4 R.H. | 55 | male | Idiopathic pulmonary fibrosis | prednisolone 30 mg daily, cyclophosphamide 150 mg daily | 147 | + | + | alive |
| 5 J.H. | 57 | male | COPD | none | 240 | − | + | alive |
| 6 D.K. | 70 | male | Idiopathic pulmonary fibrosis | prednisolone 20 mg daily azathioprine 150 mg daily | 16 | − | + | alive |
| 7 D.T. | 56 | male | NSCLC | dexamethasone 12 mg daily | not done | − | + | deceased |
Conclusion: PJP is an important differential diagnosis and at the same time a severe pulmonary complication in immunodeficient patients. We were able to confirm the high mortality rates of 30–60 % in PJP-patients without associated AIDS-disease published in literature. As expected, PJ-PCR was superior to PJ-IFT and should therefore be performed on a routine basis for diagnosing PJP. Non-invasive Ventilation demonstrated to be a worthwhile therapeutic Option for bridging severe hypoxemia in patients with PJP.
P1368: Molecular epidemiology of Pneumocystis jirovecii carriage in idiopathic interstitial pneumonia
C. de la Horra, F.J. Medrano, M. Montes-Cano, N. Respaldiza, I. Mateos, V. Friaza, S. Vidal, J. Martin-Juan, E. Rodriguez-Becerra, J.M. Varela, E. Calderon (Seville, E)
Introduction: The Idiopathic Interstitial Pneumonia (IIP) is one of the most severe pulmonary diseases. Initial chronic inflammation leading to parenchymal fibrosis is the common characteristic of this process; and infectious agents are the possible cause for the initial inflammation. It has been proved that Pneumocystis jirovecii (Pj) mediate an inflammatory response, increasing the permeability of alveolo-capillary membrane and, causes diffuse alveolar damage. Therefore, pulmonary carriage of Pj could play a role in the pathogenesis of the disease. However, the molecular epidemiology of Pj in this kind of patients is still unknown.
Objective: To study the prevalence and molecular epidemiology of Pj carriers in IIP patients.
Patients and Methods: The study included 95 consecutive patients with a final diagnosis of IIP. DNA was isolated from 80 bronchoalveolar lavage and 15 oropharyngeal wash samples. A nested PCR protocol was used to analyse the presence of Pj. The polymorphisms at mt LSU rRNA region were studied by direct sequencing. A touchdown-PCR protocol followed by RFLP with AccI and HaeIII was assayed to determinate mutations at DHPS gene.
Results: The prevalence of Pj carriage was 31.5% among IIP patients. Only 17 patients were successfully sequenced at mt LSU rRNA gene. The genotype 85C/248C was founded in 15 (88.2%) and the genotype 85T/248C in two cases (11.8%). The analysis of DHPS mutations shows a low rate of resistant strains 1/16 (6%) among IIP patients.
Conclusions: There is a high prevalence of Pj carriers among IIP. The genotype 1 (85C/248C) is amply distributed in the lung of IIP patients. We found a low rate of mutations in DHPS gene, related to sulpha-resistance drugs. New studies will be necessary to elucidate the role of Pj colonisation in this pathology. This study was partially supported by the Spanish Ministry of Science and Technology (ref.: SAF2003–06061) and Thematic Network for Joint Research (G03/90)
P1369: P. carinii in sputum of asymptomatic HIV seronegative oncologic patients
A. Georgouli, E. Chinou, P. Apostolakopoulos, M. Alexiou, E. Skouteli, P. Golemati (Athens, GR)
Objectives: To investigate the presence of ‘asymptomatic carriers’ of P. carinii among oncologic patients without HIV.
Methods: Induced sputum specimens from 438 HIV seronegative asymptomatic oncologic patients (196 M, 242 F, of mean age 59 years), were studied for the detection of P. carinii cysts and trophozoites using a direct immunofluorescent procedure. Hematologic malignancy in 286 (65.3%) and solid tumors in 152 (34.7%). Four patients were the underlying diseases, while 83.8% of them were undergoing chemotherapy during the last six months.
Results: P. carinii cysts and trophozoites were detected in 82 out of 438 of our patients (18.7%). There was no significant difference between the patient with hematologic malignancy (19.2%, 55/286) and those with solid tumors (17.8%, 27/152). All 82 of patients with presence of P.carinii in sputum had undergone chemotherapy during the last 6 months and 73 of them (89%) were also receiving long-term corticosteriod therapy (mean duration: 3 months).
Conclusion: The prevalence of P. carinii colonization is quite high among oncologic patients. This may imply a higher risk of pneumonia in these patients due to reactivation of the organism.
P1370: Prevalence and genotypes characterisation of Pneumocystis jirovecii colonisation among cystic fibrosis patients in Spain
I. Mateos, J. Dapena, C. de la Horra, M.A. Montes-Cano, V. Friaza, N. Respaldiza, F.J. Medrano, E. Calderon, J.M. Varela (Seville, E)
Introduction: Pneumocystis jirovecii carriers may exist among cystic fibrosis (CF) patients due to their underlying pulmonary diseases, but their role in the natural history of the disease is poorly understood. The prevalence and molecular epidemiology patients still remains unknown. Moreover, the knowledge of P. jiroveci genotypes distribution may help for understanding the epidemiology of this pathogen.
Objective: To describe the prevalence and genotypes distribution of P. jirovecii in a population of CF Spanish patients.
Methods: A cross-sectional study was performed in 100 CF patients, including 12 lung transplant recipients. P. jirovecii was identify by nested PCR the mt LSU rRNA gene in respiratory samples (sputum or oropharyngeal washes). The genotype was analysed by direct sequencing at positions 85 and 248.
Results: The overall prevalence of P. jirovecii colonization among CF patients was 24%. The prevalence in lung transplant recipients was higher than in non transplanted CF patients. The polymorphism 85C/248C (42.8%) and 85T/248C (28.5%) were predominant, and also genotype 85A/248C (21.4%) was identified; in one case we founded a mix of genotypes. The colonisation is higher in CF patients under 18 years old. Besides, 35.7% patients receiving prophylaxis with cotrimoxazol and 17.2% with azithromycin were colonized for P. jirovecii but none of them developed Pneumocystis pneumonia (PcP) for a year follow-up. The analysis shows concordance in the colonization status between brother groups.
Conclusions: They are a high prevalence of P. jirovecii carriers among CF patients in Spain. These data suggest that chemoprophylaxis with Cotrimoxazol and Azithromycin may prevent PcP but not avoid colonization. The concordance observed among brothers support the idea of a common source of infection or person-to-person transmission. This study was partially supported by the Spanish Ministry of Health (FIS 03/1743) and by the Consejeria de Salud de la Junta de Andalucia (Research Project 32/02)
P1371: Distribution of Pneumocystis jirovecii genotypes in patients with different pulmonary diseases
M. Montes-Cano, J.M. Varela, C. de la Horra, N. Respaldiza, I. Mateos, V. Friaza, S. Vidal, J. Martin-Juan, E. Rodriguez-Becerra, F.J. Medrano, E. Calderon (Seville, E)
Introduction: In recent years, asymptomatic carriers of Pneumocystis jirovecii has been identified among immunocompetent patients with different pulmonary diseases as Chronic Obstructive Pulmonary Disease (COPD) or Idiopathic Interstitial Pneumonia (IIP) subjects. The knowledge of genotypes distribution may help for understanding the role of the microorganism in the natural history of these pathologies. However, the genotypes distribution of P. jirovecii status in these diseases remains unknown.
Objectives: To describe P. jiroveci genotypes distribution in COPD vs IIP patients based on mt LSU rRNA polymorphisms and DHPS mutations
Patients and Methods: The study analysed the respiratory samples from 36 COPD patients and 17 IIP subjects colonised by P. jiroveci. The genotypes at mt LSU rRNA gene were analysed by nested-PCR using and direct sequencing at 85 and 248 nucleotide positions. DHPS mutations were assayed by RFLP: Touchdown-PCR at and digestion with AccI and HaeIII.
Results: Based on the analysis of mt LSU rRNA and DHPS gene we found:
| COPD |
IIP |
|||
|---|---|---|---|---|
| Genotypes mt LSU rRNA | N = 38 | % | N = 17 | % |
| 85C/248C | 15 | 41.6 | 15 | 82.2* |
| 85A/248C | 4 | 11.1 | 0 | 0 |
| 85T/248C | 15 | 41.6 | 2 | 11.8 |
| Mix | 2 | 5.5 | 0 | 0 |
(p = 0.004, OR = 10.5, CI 95 % 1.9—103)
| COPD |
IIP |
|||
|---|---|---|---|---|
| Genotypes DHPS | N = 15 | % | N = 16 | % |
| wt/wt | 10 | 66.6 | 15 | 94 |
| 55/wt | 1 | 6.6 | 0 | 0 |
| wt/57 | 2 | 13.3 | 1 | 6 |
| 55/57 | 1 | 6.6 | 0 | 0 |
| Mix | 1 | 6.6 | 0 | 0 |
Conclusions: The genotypes distribution at mt LSU rRNA is significantly different among the groups studied. In COPD patients exists a high rate of sulfa-resistence respect to the IIP patients, probably related to previous exposition to sulfa-drugs. New studies are required to elucidate the role of the genotypes in the evolution of patients. This study was partially supported by the Spanish Ministry of Science and Technology (ref.: SAF2003–06061) and Thematic Network for Joint Research (G03/90)
P1372: Pneumonia in cancer patients
M. Aguilar-Guisado, E. Cordero, J. Cisneros, I. Espigado, M. Noguer, R. Parody, J. Pachón (Seville, E)
Introduction: Pneumonia in cancer patients is frequent and mortality is high.
Objectives: To analyse the epidemiology, aetiology, clinical features and outcome of pneumonia in cancer patients, including these with neutropenia.
Patients and method: Prospective study of all patients with cancer admitted in Oncology and Haematology Departments that had a pneumonia. Study period: from November 2002 to August 2004.
Results: Eighty-five cases in 73 patients were included, 61% males. The median age was 53 years (range from 16–84 years). Pneumonia incidence was 33 episodes for each 1000 admissions. Most of episodes (71.8%) were in patients with lymphoma (30.6%) and acute leukaemia (21%). 23.5% were blood stem-cell transplant recipients. 66% of episodes were community acquired. 60% had neutropenia at the diagnosis of pneumonia, and 14% were bacteriemic. An aetiologic diagnosis was established in 46% of episodes. Bacterial aetiology was predominant (n = 21, 55%), followed by fungal (n = 17, 45%). The most frequent bacteria were Pseudomonas aeruginosa (n = 8, 21%), Streptococcus pneumoniae (n = 4, 10%), Legionella pneumophila (n = 2, 5%) and Fusobacterium nucleatum (n = 2, 5%). The aetiology of invasive mycosis were aspergillosis (n = 13, 34%, definitive n = 3, probable n = 1, and possible n = 9), and Pneumocystis jiroveci (n = 4, 10%). Diagnostic yield of blood cultures was 14%. TC had diagnostic usefulness in 37%. The global mortality at 30th day was 32%, and in patients with neutropenia was 33%. The presence of neutropenia (p = 0.02; RR 1.9; CI95% 1.05–3.3), bacteraemia (p < 0.001; RR 4.6; IC95%: 2.6–7.9) and multilobar extension (p = 0.01; RR 3.1; CI95%: 1.5–6.5) were associated with severe clinical features in the first 48 hours. The presence of initial respiratory failure was the only independent factor of adverse outcome selected by multivariate analysis (p = 0.001; RR 5.4; CI95%: 1.4–20.6).
Conclusions: The incidence of pneumonia in cancer patients is high. Aetiology is eestablished in one half of episodes. Bacteria are the most frequent aetiology, and P. aeruginosa the more frequent specie. Aspergillus spp make up one third of the episodes. Mortality of pneumonia in these patients is high and associated with initial respiratory failure.
P1373: Nocardiosis in a teaching hospital in the Central Anatolia region: treatment and outcome
O. Yildiz, E. Alp, B. Tokgoz, B. Tucer, B. Aygen, B. Sumerkan, A. Couble, P. Boiron, M. Doganay (Kayseri, TR; Lyon, F)
Objectives: To evaluate the main characteristics of the patients with nocardiosis retrospectively.
Methods: A retrospective study was carried out from January 2001 to February 2004 in a University Hospital. We analysed predisposing factors, clinical features, radiological findings, antimicrobial susceptibility patterns, treatments, duration of treatments and outcomes of the patients with nocardiosis. Treatment was started empirically with ceftriaxone (4 g/d) plus amikacin (1 g/d) and modified according to the antimicrobial susceptibility pattern and was continued for 6–12 months.
Results: Nocardia species were isolated from consecutive nine patients. In six of the nine patients (67%), predisposing factors were identified. The most common predisposing factor included receiving immunosuppressive therapy (67%), cadaveric kidney transplant (44%), diabetes mellitus (22%) and systemic lupus erythematosus (11%). Clinical syndromes of nocardial infection seen were pulmonary infection in three patients, cerebral infection in five patients and disseminated infection in one patient. All previously healthy patients developed cerebral nocardiosis due to Nocardia farcinica. The initial antibiotics used were not appropriate in four of the nine patients (44%). However, only two of these four patients experienced relapse and one with relapse died of disease. Overall mortality in our patients was 33%; in two cases death was due to the Nocardia infection.
Conclusions: Despite its relative rarity, Nocardia is an important cause of infection, especially for patients with a predisposing factor. The use of imipenem or meropenem in combination with amikacin is suggested for initial therapy of serious Nocardia infections. The duration of therapy should be protracted for 12 months for preventing relapse.
P1374: Stenotrophomonas maltophilia: changing spectrum of bacterial pneumonia in cancer patients with low-suspicion of S. maltophilia infection
G. Aisenberg, J. Tarrand, K.V. Rolston, D. Kontoyiannis, I. Raad, A. Safdar (Houston, USA)
Background: S. maltophilia, a nonfermentative gram-negative bacteria is often associated with serious ventilator-associated pneumonia. We sought to determine characteristics of S. maltophilia lung infections in cancer patients who had low-suspicion of S. maltophilia infection.
Methods: All S. maltophilia from respiratory samples during 1998–2004 were evaluated retrospectively. Patients with established risk of S. maltophilia infection such as critical care unit (CCU) stay, mechanical ventilation (MV), neutropenia (ANC <500 cells/μL), HIV infection and underlying structural lung disease were excluded.
Results: In 40 patients, median age was 54 ± 16 years, 27 (68%) were male, 10 patients (25%) had severe lymphocytopenia (<500 cells/μL), APACHE II score was 10 ± 4, and 9 (33%) of 27 with hematologic malignancies had acute leukaemia. Fourteen (82%) of 17 BMT recipients had received allogeneic grafts and were being treated for chronic graft-versus-host disease (cGVHD; systemic corticosteroids >60 mg daily prednisolone or equivalent dose). Infection occurred 184 ± 325 days (range, 35–1458 days) after BMT. 15 patients (38%) had refractory or advanced cancer, 7 (18%) had diabetes mellitus and 4 (10%) had COPD. Twenty-three patients (58%) had nosocomial infection. None had either prior S. maltophilia orointestinal, genitourinary tract colonization or concomitant S. maltophilia bacteraemia. Cough was common (n = 24; 60%), dyspnea and fever occurred in 22 (55%) and 16 (40%) patients, respectively. Thirty-three patients (83%) presented with pneumonia while receiving broad-spectrum systemic antibiotics for 7 ± 31 days. Most S. maltophilia (n = 37; 93%) were susceptible to trimethoprim-sulfamethoxazole and ticarcillin-clavulanic acid (n = 26; 65%). Fifteen patients (38%) did not need hospitalization. In 25 hospitalized patients, 6 (15%) required CCU admission and 5 MV for 3 ± 5 days. Eight (20%) deaths were attributed to S. maltophilia pneumonia; presence of young age (37 vs 59 years; p < 0.004), prolonged hospitalization (42 vs 14 days; p < 0.01), and high APACHE II score (13 vs 10; P = 0.05) were associated with increased mortality. By univariate analysis, presence of acute leukaemia, APACHE II score >16, MV, CCU stay and cGVHD were poor predictors of outcome, by stepwise logistic regression analysis, only the later 2 emerged as significant prognosticators of death.
Conclusions: S. maltophilia pneumonia was a serious lung infection in these non-neutropenic, non-CCU patients with cancer.
P1375: Usual interstitial pneumonia associated with cytomegalovirus infection after percutaneous transluminal coronary angioplasty
M. Falagas, M. Rizos, S. Tsiodras, A. Betsou, P. Foukas, A. Michalopoulos (Athens, GR)
Background: Ventilator associated pneumonia due to CMV is a largely unexpected but probably underestimated diagnosis.
Methods: We describe a patient with usual interstitial pneumonia associated with cytomegalovirus infection after percutaneous transluminal coronary angioplasty.
Results: A 71-year-old woman, with history of diabetes mellitus, was admitted to the ICU of our hospital due to acute myocardial infarction. Percutaneous transluminal coronary angioplasty was performed. Two days following ICU discharge, the patient became febrile 38.5°C with non-productive cough and progressive dyspnoea. Three days later, she was readmitted to the ICU due to severe dyspnoea and type I respiratory failure. The patient was intubated and admission-CXR revealed diffuse infiltrates in both lungs and pleural effusions. The patient was treated with intravenous piperacillin-tazobactam, ofloxacin and teicoplanin. She remained febrile 39°C with no improvement. A chest CT-scan demonstrated confluent opacities in the right upper and middle lobes and in the left lower lobe as well as airbronchograms, an extensive right pleural effusion and a pathological swelling of pretracheal lymph nodes. Serologic tests for CMV revealed positive IgG antibodies, with no IgM antibodies present. An open lung biopsy revealed distortion of lung parenchyma, moderate inflammation and patchy fibrosis with a subpleural accentuation. The fibrotic areas consisted of dense collagen with focal ‘honeycomb’ pattern alternating with areas of relatively normal alveolar parenchyma. There were also focal alveolar macrophage accumulation, smooth muscle proliferation and focal subpleural fatty metaplasia. The overall pattern was consistent with usual interstitial pneumonia. Some epithelial type II pneumocytes showed atypia with abundant cytoplasma and large pleomorphic nuclei harbouring intranuclear inclusions consistent with CMV infection. Despite treatment with ganciclovir the patient died two weeks later from severe ARDS and multiple organ failure.
Conclusion: Clinicians should be aware of CMV associated severe bilateral pneumonia after cardiac procedures even in non-transplanted patients. Correct diagnosis depends on clinical awareness in the appropriate setting along with proof of viral infection.
Detection of methicillin resistance
P1376: Rising MRSA occurrence in Central Norway – importance of swab sites
T. Jacobsen, A.W. Børseth, L. Marstein, O. Scheel (Trondheim, N)
Objectives: The MRSA epidemic in Norway is steadily growing with a more than 10-fold increase in reported cases since 1997. In the last years, a significant increase in MRSA-cases in nursing homes has been reported. In August 2003 a more sensitive method for MRSA detection has been introduced in the reference laboratory for Central Norway at St Olav Hospital. In this same period, we have observed a growing incidence of MRSA in the throat, both alone and combined with findings at other sites of the body. The aim of the present study was to investigate the importance of including throat swabs in MRSA screening.
Materials and Methods: We have examined the occurrence of all new patients positive for MRSA in any site A (in the period January 1st 1997-October 22nd 2004, and B) in the period from August 1st 2003 to October 22nd 2004, the same period the new MRSA detection broth has been in used. Only the first positive specimen-set from new patients, both carriage and infections, were included. In period A the specimens were grown on a selective mannitol agar containing 6% NaCl (2% NaCl after October 2002) and Oxacillin 4 mg/L, whereas in period B the specimens were grown in a selective broth containing phenol red mannitol with aztreonam 75 mg/L and ceftizoxime 5 mg/L. Isolates of MRSA were verified by PCR detection of nuc and mecA genes by PCR in both periods.
Results: The total number of new MRSA positive patients has increased from 1 (1997) to 71 (until Oct. 22nd 2004), mainly in nursing home residents. Among 88 new MRSA positive individuals in period B, 48 persons had throat swabs taken. 23 (47.9%) grew MRSA in throat combined with other sites. Five (10.4%) grew MRSA in throat only. Among 50 persons examined with perineum swabs, only 12 (24.0 %) were positive combined with other MRSA positive locations, but none were MRSA positive in perineum only.
Discussion: In the last eight years, we have experienced a 70-fold increase in new MRSA positive patients in Central Norway. Many outbreaks have not been thoroughly cleared because of wrong examination sites. Based on the finding of MRSA positive swabs from 5 persons from throat and none from perineum as the only location for MRSA, we conclude that throat swabs must be a part of routine MRSA screening.
P1377: Evaluation of oxoid mannitol salt agar with cefoxitin for the detection of methicillin-resistant Staphylococcus aureus from surveillance specimens
B.M. Willey, P. Akhavan, N. Kreiswirth, K. Wong, O. Imas, A. McGeer, T. Mazzulli, S. Poutanen, J. Strauss, G. Small, I. Edwards, N. Nelson, M. Skulnick (Toronto, CAN)
Objectives: Challenged by an increasing demand for MRSA surveillance and declining resources, clinical laboratories require sensitive and specific screening media to ensure a rapid turn-around-time (TAT) to notification and a low false-positivity rate if costs and MRSA are to be controlled. This study compares routine Mannitol Salt agar with 4 μg/mL oxacillin (MSO) to Oxoid's Mannitol Salt (MSF) and Analine Blue Salt (ABF) agars, both containing 10 μg cefoxitin per mL.
Methods: 1343 surveillance swabs from 750 patients (427 nasal, 409 rectal, 305 nasal/axilla/groin/perineum, 161 wound, 41 other) were prospectively screened for MRSA on MSO, MSF and ABF agars. Each media was incubated in ambient air at 35°C, and read independently at 24 and 48 h. Yellow colonies from MSO and MSF, and blue colonies from ABF were tested for capsular antigens 5 and 8 (Pastorex Staph Plus, BioRad), tube coagulase and PBP2a (Denka Seiken) to identify MRSA. Positive PBP2a reactions were confirmed using the NCCLS 6 μg/mL oxacillin screen agar. Discrepant strains were reconfirmed to be MRSA and typed by SmaI PFGE.
Results: Overall 78 MRSA were recovered within 48 h: 73 from MSF, 43 from MSO and 32 from ABF, generating relative sensitivities of 93.6%, 55.1% and 41%, respectively. The MSF grew 30 (41%) of its 73 MRSA within 24 h, compared to 11 (34.3%) on ABF and 8 (18.6%) on MSO. 54 (74%) of strains growing on MSF could be provisionally identified as MRSA directly from the plate allowing for same day notification of infection control: 17 within 24 h and 37 within 48 h. Notably, when discrepancies between media were identified (>48 h), 23 additional MRSA could be recovered from mixed blue colonies on ABF and 3 mannitol non-fermenting MRSA were identified from the MSF. Also, 25 MRSA (representing 4 distinct clones prevalent in Toronto) were recovered on MSO after broth enrichment from the original swabs that initially failed to yield MRSA on routine MSO. From the 1265 MRSA-negative swabs, breakthrough coagulase negative Staphylococci required workup at 24 h and 48 h on 45 and 278 AB, 38 and 219 MSF and 8 and 216 MSO plates, respectively, resulting in overall specificities of 84.9% for MSO, 83.1% for MSF and 74.9% for ABF.
Conclusion: MSF was significantly more sensitive than MSO and ABF (p < 0.0001) without loss of specificity or increase in cost. In addition, the TAT was markedly improved on MSF as 74% of MRSA were able to be identified directly from the plate at 24 h.
P1378: Evaluation of BBL CHROMagar MRSA for detection of methicillin-resistant Staphylococcus aureus from surveillance swabs
B.M. Willey, A. McGeer, T. Mazzulli, S. Poutanen, J. Strauss, A. Tyler, K. Wong, O. Imas, P. Akhavan, N. Kreiswirth, G. Small, I. Edwards, N. Nelson, M. Skulnick (Toronto, CAN)
Objectives: Highly sensitive screening media and a rapid turnaround-time (TAT) to notification is imperative in the control of MRSA. Good specificity is also paramount as false positives significantly increase labour and material costs. This study compares selective CHROM agar MRSA with 6 μg/mL cefoxitin (CFOX) to routine mannitol salt agar with 4 g/mL oxacillin (MSOX) in a population with <5% prevalence.
Methods: MRSA screens from 750 patients (427 nasal, 409 rectal, 305 nasal/axilla/groin/perineum, 161 wound, 41 other) were planted to CFOX and MSOX on receipt. The plating order was alternated every 200 swabs. CFOX were incubated in the dark at 35°C. Media were read independently at 24 and 48 h. Mauve colonies from CFOX and yellow from MSOX were tested for capsular antigens 5 and 8 (Pastorex Staph Plus, BioRad), tube coagulase and PBP2a (Denka Seiken) to identify MRSA. Positive PBP2a reactions were confirmed using the NCCLS 6 μg/mL oxacillin screen agar. Identities of discrepant strains were confirmed and typed by SmaI PFGE.
Results: From 1343 swabs, 78 (5.8%) grew MRSA on at least one medium (71 CFOX; 43 MSOX; 36 both) generating a relative sensitivity of 91% for CFOX and 55% for MSOX. The identities of 35 MRSA that failed to grow on MSOX were confirmed and 25 of these (from 4 distinct PFGE clones) were grown on MSOX from the original swabs after broth enrichment. There was no difference in MRSA growth rate between media: CFOX grew 17% MRSA at 24 h and 83% at 48 h; MSOX grew 18.6% at 24 h and 81.4% at 48 h. From the 1265 MRSA-negative swabs, breakthrough coagulase negative Staphylococci required workup at 24 h and 48 h on 1 and 28 from CFOX and 8 and 216 from MSOX, resulting in 24 h/48 h specificities of 99.9%/97.8% for CFOX and 99.4%/85.4% for MSOX, respectively. MRSA was identified directly from CFOX in 66.2% cases, allowing for same day notification to infection control of 10 MRSA at 24 h and 39 at 48 h, while 19 were notified at 72 h, and 3 at 96 h. By comparison, all 43 MRSA from MSOX were cultured to blood agar prior to identification, and of the 5 new MRSA cases that required rapid notification, 3 were notified at 72 h, and 1 each at 4 and 5d (38 cases had been previously identified).
Conclusion: While the CFOX plates were more costly, they were significantly more sensitive (p < 0.0001) and specific (p < 0.0001) than MSOX resulting in less material and labour costs. The TAT to notification appears reduced due the ease of working directly from the CFOX plate.
P1379: Comparison of MRSA ID medium and enrichment broth culture for detection of methicillin resistant Staphylococcus aureus carriers by muco-cutaneous surveillance cultures
C. Nonhoff, M.J. Struelens, A. Brenner, N. Legros, C. Thiroux, O. Denis (Brussels, B)
Objectives: To evaluate the performance of a new chromogenic agar medium, MRSA ID (BioMérieux), for detection of methicillin resistant Staphylococcus aureus (MRSA) from surveillance cultures of muco-cutaneous swab specimens in patients admitted to a 860-bed teaching hospital.
Methods: Hospitalized patients (n = 331) were screened for MRSA carriage by sampling swabs from nares (n = 363), throat (n = 47), perineum (n = 46) and skin (n = 35). Swabs were inoculated into nutrient broth (SB) supplemented with 7.5% NaCl, Columbia sheep blood agar with cefoxitin disk (30 μg) (SBA) and MRSA ID agar at 35°C. SB broths were sub-cultured after 24 h onto SBA with cefoxitin disk (30 μg) and MRSA ID agar. S. aureus isolates were identified by coagulase test. Susceptibility to oxacillin was determined by cefoxitin disk method according to NCCLS. Identification and oxacillin resistance were confirmed by PCR for 16S rRNA, nuc and mecA genes.
Results: MRSA strains were isolated in 55 (11%) specimens, either from primary MRSA ID (n = 53), primary SBA (n = 37), secondary MRSA ID (n = 53), or secondary SBA (n = 48), respectively. Sensitivities/specificities of the different media were: 96%/99% for primary MRSA ID, 63%/85% for primary SAB, 96%/99% for secondary MRSA ID and 86%/85% for secondary SAB. The combination of MRSA ID agar with salt enrichment broth did not increase the screening sensitivity in contrast with SBA. The median times to MRSA identification were 3 days for both agars (range 2–4 days) and 3 days after enrichment broth (range 3–4 days). Sixty-four per cent MRSA recovered from MRSA ID were detected after 24 h incubation.
Conclusions: MRSA ID was found to be a sensitive and specific medium for the screening of MRSA carriage in hospitalized patients. Prolonged incubation of MRSA ID for 48 h increased the yield of this medium by 36%. According to the results obtained with the MRSA ID screen plate, it does not require the additional phase of enrichment broth. The direct consequences are a gain of time-to-result of 24 h as well as an important simplification of methodology for the culture surveillance of MRSA.
P1380: Evaluation of 10, 30 and 60 mcg cefoxitin neo-sensitabs for prediction of methicillin resistance in S. aureus
R. Skov, A.R. Larsen, N. Frimodt-Møller (Copenhagen, DK)
Objective: Cefoxitin paperdisks have been shown to accurately predict methicillin resistance in S. aureus. In this paper we evaluate 10, 30 and 60 mcg Cefoxitin Neo-Sensitabs tablets for predicting methicillin resistance using a challenge set of S. aureus.
Methods: A total of 196 recently isolated S. aureus all tested for presence of the mecA gene (EVIGENE, SSI, Denmark) were investigated. The mecA positive isolates (N = 62) were a challenge set consisting of the most heterogeneous resistant MRSA isolates found in a recent investigation (Skov et al, J Antimicrob Chemother). The isolates represented 39 different PFGE types (at least one band difference) and included isolates with oxacillin MIC <2 mg/L. The mecA negative isolates were consecutive blood culture isolates (N = 99) as well as the mecA negative isolates with the smallest cefoxitin zones in a recent evaluation (N= 35). S. aureus ATCC 29213 and 25923 were included as control strains in each run. All isolates were tested with confluent growth and 18–22 h incubation at 35–37°C on Mueller Hinton Agar (BBLII, Becton Dickinson, US) using 10, 30 and 60 mcg Neo-Sensitabs tablets (Rosco, Taastrup, Denmark). Inhibition zone diameters were measured to the nearest millimetre at the inner zone edge–disregarding a faint haze, if present.
Results: Using a breakpoint of R < 19 mm (cefox 10); R < 24 mm (cefox 30) and R < 29 mm (cefox 60) two, three and two mecA positive isolates tested false susceptible, respectively. Five, four and six mecA negative isolates tested false resistant. Zone edges for the 10 mcg tablet were more distinct and easier to read. For S. aureus ATCC 29213 the following zone diameters were found 21–23 mm, 27–29 mm and 29–33 mm, respectively and for S. aureus ATCC 25923 22–23 mm; 27–29 mm, and 32–33 mm, respectively.
Conclusions: All three cefoxitin Neo-Sensitabs performed with high accuracy using this challenge set and as good as the results obtained in a recent evaluation of a 10 mcg paperdisk. The cefoxitin 10 mcg Neo-Sensitabs performed slightly better than the 30 or 60 mcg and zone edges were easier to read why we will suggest the use of 10 mcg tablet.
P1381: Evaluation of lipovitellin-salt-mannitol-agar with an oxacillin and cefoxitin disc as primary screening medium for methicillin-resistant Staphylococcus aureus
M. Boudewijns, K. Lagrou, J. Verhaegen (Leuven, B)
Introduction: Numerous primary media have been advocated for the enhanced detection of methicillin-resistant Staphylococcus aureus (MRSA). Recently, a lipovitellin-salt-mannitol-agar (LSM) with an oxacillin disk has been described. On this medium, detection of MRSA isolates is enhanced due to production of a lecithinase, which results in enlarged colonies surrounded by a zone of opacification. Also, the use of a cefoxitin disk method has recently been described. It has been reported as being superior to the oxacillin disk method.
Aim: To evaluate the performance of LSM with a 1 μg oxacillin disk and a 30 μg cefoxitin disk (LSMoc) as selective and differential primary medium for detection of MRSA from screening specimens.
Materials and methods: Screening specimens (swabs of nose and perineum) were obtained from a patient population at the intensive care unit. The LSMoc medium was compared with an in house method using colistin-nalidixic acid agar (CNA) with 5% horse blood by performing parallel inoculation. S. aureus isolates were presumptively identified by colonial morphology and confirmed by coagulase tube test and Vitek 2 ID-GPC testcard. Methicillin resistance on the LSMoc medium was presumptively detected by reading the zones of inhibition around both disks and confirmed by disk diffusion and PBP2’ latex agglutination assay.
Results: 149 (19.7%) of 754 screening specimens were positive for MRSA by one or both methods. Sensitivity of the LSMoc medium (90%) was higher than sensitivity of the in house method (76%) (p < 0.01). However, in contrast to the CNA medium, the LSMoc medium required an additional 24–48 hours of incubation before it could be examined for the presence of MRSA. The LSMoc medium misidentified 22 isolates as MRSA (specificity 96.4%). These were mostly coagulase-negative Staphylococci, mainly belonging to the species S. capitis, which gave opacification. There was no difference in the number of MRSA isolates detected on the LSMoc medium by use of the oxacillin or cefoxitin disk.
Conclusion: Use of the LSMoc medium increases the yield of MRSA, due to added enrichment for growth and ease of visual recognition of MRSA isolates. However, the medium still requires confirmation of presumptive MRSA isolates and requires prolonged incubation.
P1382: Two evaluation methods for screening specimens to detect methicillin-resistant Staphylococcus aureus
Y. Martín, A. Rezusta, J. Castillo, M.J. Revillo (Zaragoza, E)
Objectives: Rapid assessment of clinical specimens for the presence of MRSA is an important part to control measures for infections taken to reduce the spread of MRSA and thus, to decrease hospitalization costs. The purpose of this study was to evaluate the sensitivity, specificity and the optimal incubation time from patient specimens by using ORSAB (Oxacillin Resistance Screening Agar Base) as a primary culture medium compared with those obtained by using a previous enrichment broth medium, Tryptone Soy Broth added with 75 μg/ml of aztreonam (TSB Azt).
Methods: Every MRSA screening swab was inoculated initially onto ORSAB and incubated aerobically for 48 h at 35°C; the plates were examined after 24 and 48 h. At the same, each specimen was introduced in TSB Azt. After 24 h of incubation, they were subcultured onto ORSAB, following the same process described previously. All mannitol-fermenting colonies were confirmed as S. aureus by using a latex agglutination system (Pastorex Staph Plus Bio Rad). Resistance to methicillin was determined by the disk diffusion method according to the NCCLS.
Results: Out of 404 studied samples, 90.35% were nose swabs. MRSA was detected in 15% of these samples. The sensitivity and specificity obtained when ORSAB alone was used were 96.6 and 48.3% respectively. When we previously used TSB Azt, the results were 94.9 and 49.5%. While 93.0% of MRSA was detected after 24 h of incubation in the first case, it was 98.2% after using an enrichment broth medium.
Conclusions: ORSAB gave a positive result in 93.0% of the samples after 24 h of incubation. The three MRSA isolated only from direct culture onto ORSAB belonged to samples which had a low amount of microorganisms. Low specificity was especially due to Staphylococcus haemolyticus, very common in nose samples. With such a high number of nose samples, our study showed that the use of TSB Azt gave a minimum increase of sensitivity.
P1383: Multiresistant bacteria screening: clinical evaluation of MRSA ID, a new chromogenic medium for the screening of methicillin-resistant Staphylococcus aureus
M.E. Reverdy, S. Orenga, J.M. Roche, S. Delorme, J. Etienne (Lyon, La Balme les Grottes, Craponne, F)
Objective: Nasal carriage of Methicillin-Resistant S. aureus (MRSA) is an important cause of nosocomial infection. Screening of patients during admission is recommended to reduce the incidence of MRSA. The aim of the study was to evaluate the new MRSA ID media (bioMérieux) for rapid isolation and identification of MRSA.
Methods: MRSA ID was compared to ORSAB medium (Oxoid). CNA sheep blood agar (COL) was used as the reference method. A total of 278 nasal swabs were inoculated directly on the media which were incubated 24 and 48 h at 35°C. Green colonies on MRSA ID, blue on ORSAB and typical S. aureus colonies on COL were regarded as presumptive MRSA isolates. Confirmation was performed using a coagulase test and PCR for mecA gene.
Results: A total of 45 swabs were positive for MRSA. After 24 h incubation, 42, 38 and 42 MRSA strains were isolated on MRSA ID, ORSAB and COL respectively. MRSA isolates produced colonies of intense color on chromogenic media. The sensitivity of detection of MRSA ID (93.3%) was higher than ORSAB (84.4%). The number of false positives (FP) on ORSAB was higher (25 coagulase-negative Staphylococc --CNS, and 2 methicillin-sensitive S. aureus) compared to MRSA ID (3 CNS). If only taking into account colony color, the predictive positive value (PPV) was 93.3% for MRSA ID, 58.5% for ORSAB. If the coagulase test result was included, the PPV value of ORSAB became 95%. After 48 h, MRSA ID enabled the recovery of 1 MRSA strain more (43 strains) whereas 5 strains more were isolated on ORSAB (43 strains). However the PPV values of both media decreased because MRSA ID produced many pale green colonies of CNS, and ORSAB in the same way. Depending on the interpretative reading of the laboratory, FP on MRSA ID were easily differentiated from MRSA isolates.
Conclusion: MRSA ID enables rapid and definitive identification of MRSA isolates. Sensitivity of MRSA ID after 24 h was higher than that of ORSAB. Reading of MRSA ID after 48 hrs is not recommended due to the possibility of FP results. The use of MRSA ID does not require any supplementary testing after 24 h, whereas additional tests are necessary for ORSAB media.
P1384: Is disk diffusion method reliable for detection of methicillin-resistant Staphylococcus aureus?
M. Rahbar (Tehran, IR)
Objective: The aim of this study was to evaluate using of home made oxacillin for detection of MRSA
Methods: 96 clinical isolates of S. aureus strains in Milad hospital were chosen for detection of MRSA Disk diffusion method was performed as recommended by National Committee for Clinical Laboratory Standards (NCCLS M2-A8) Collectively three kinds of xoacillin disks from different sources were tested. Two kinds of disks were home made (we called with sources of A and B) and the third was from other country (source c). E-test was selected as reference method. all test performed at the same condition.
Results: By E-test method of 96 strains of S. aureus 51 strains were MRSA and 45 strains were susceptible to methicillin. Oxacillin disks from company A showed 94 MRSA, and two intermediate strains. There was not any susceptible strain by using this product. By using other home made oxacillin disk(B) 60 strains were MRSA, 21 strains intermediate and 22 strains susceptible to methicillin. By using oxacillin disks from other country(C) of 96 tested S. aureus isolates 52 strains were MRSA, 8.strain intermediate and 36 strains susceptible. Unfortunately, home made disks especially from A company showed very low sensitivity and specificity.
Conclusions: This study reveals that in our country using of disk diffusion method is not reliable for detection of MRSA. This may be due to limitation in sources of oxacillin disks and very poor in quality of these products. It is strongly recommended that using of other methods such as E-test and oxacillin screen agar methods are highly preferable.
P1385: Evaluation of the new ATB STAPH and rapid ATB STAPH devices for oxacillin resistance detection in Staphylococci
V. Vernet-Garnier, J. Madoux, C. De Champs, L. Mullier, G. Zambardi (Reims, LA-Balme-les-Grottes, F)
Objectives: Reliable, simple, rapid and low cost phenotypic methods are still needed for the oxacillin resistance detection in Staphylococci. Due to an heterogeneous expression, this detection is still challenging as demonstrated by the various protocols that have been proposed: NaCl complementation (2–5%), decreased temperature (30°C) or increased length of incubation (48 h), increased inoculum size, specific agar screen test, breakpoints redefinition according to the species and use of alternative markers (cefoxitin, moxalactam). The aim of this study is to evaluate how perform the oxacillin test included in both ATB STAPH (ref. 14329) and rapid ATB STAPH (ref. 14479) devices (bioMérieux) versus the mecA gene detection (gold standard) and the oxacillin (5 mcg) and cefoxitin (30 mcg) disk diffusion tests (OXA DD, FOX DD).
Methods: Two-hundred S. aureus strains were tested with the various methods (100 mecA+, 100 mecA−). DD tests were performed according to the French expert guidelines (CA-SFM 2004). For FOX DD, when an Intermediate (non conclusive) category was obtained, the result was considered S or R according to, respectively, the absence or presence of the mecA gene. An additional set of 100 coagulase negative Staphylococci (CNS) was tested with ATB STAPH only (50 mecA+, 50 mecA−). Oxacillin MIC and population analysis were also performed for the mecA+ strains not detected by the ATB devices.
Results: Sensitivity and specificity results for oxacillin resistance detection of each method, versus mecA, are shown in the Table.
|
S.aureus (n=200) |
All species (n = 300) |
|||||
|---|---|---|---|---|---|---|
| ATB STAPH (24 hrs) | rapid ATB STAPH (4.5 hrs) | OXA DD (24 hrs) | OXA DD (48 hrs) | FOX DD* (24 hrs) | ATB STAPH (24 hrs) | |
| Sensitivity (%) | 95 | 93 | 58 | 85 | 96 | 96 |
| Specificity (%) | 93 | 99 | 100 | 100 | 100 | 95 |
For the FOX DD test, non conclusive results were obtained for 6% of the strains.
Conclusions: This study highlights that ATB STAPH and rapid ATB STAPH devices perform as well as the FOX DD test and are superior to the OXA DD test for the detection of oxacillin resistance in S. aureus. ATB STAPH performances are still good when CNS are tested. The resistance is missed only for strains expressing heterogeneous resistance, as demonstrated by the low oxacillin MICs (≤1 mg/L) and the population analysis profiles. All methods show very good specificity, except ATB STAPH for which some S. aureus strains are overcalled resistant. Based on the 2004 CA-SFM guideline, the FOX DD test is sometimes non conclusive and this requires additional testing
P1386: Detection of methicillin resistance in Staphylococci employing the Uro-Quick system
S. Roveta, S. Cagnacci, F. Cavallini, D.L. Rocca, A. Marchese, E.A. Debbia (Genoa, I)
Objectives: Uro-Quick system has been employed to detect methicillin-resistance (met-R) in S. aureus and coagulase-negative Staphylococci (CoNS). In order to achieve full agreement between the antibiotic susceptibility results obtained by the reference method (NCCLS) and the Uro-Quick system, the optimal experimental conditions (inoculum size, time of incubation, antibiotic and NaCl concentration) were determined for S. aureus and CoNS respectively.
Methods: 72 met-R Staphylococci including different species (42 S. aureus and 30 CoNS, in which 12 S. epidermidis) heterogeneous in their expression of resistance to β-lactam agents were screened with the Uro-Quick System. S. aureus ATCC 29213 (mecA negative) and S. aureus ATCC 43300 (mecA positive) were used for quality control. Oxacillin (in appropriate concentration following the NCCLS breakpoints) was added in a vial containing 2 ml of suspension of strain to tested (a drug-free vial was used as control). After an opportune time of incubation the instrument printed the results: no growth and a growth curve like the control are representative of a susceptible and resistant strain respectively.
Results: The best results were obtained using Mueller-Hinton broth and a concentration of 106 cells/ml for S. aureus and 5×106 for CoNS. All the 42 S. aureus tested grown within 5 hours of incubation and the met-R phenotype was correctly detected within 6 hours in 100% of strains (66.7% and 93.3% in 4 and 5 hours respectively). The 97% of CoNS strains grown within 10 hours of incubation, an insufficient growth within this period of time was observed for 1 strain of S. haemolyticus only. Met-R was correctly detected within 10 hours in 100% of strain grown at this time (55%, 64% and 95% in 6, 7 and 8 hours respectively).
Conclusion: On the basis of the present findings, the Uro-Quick system appears to be useful for the rapid detection of met-R Staphylococci. Excellent results were obtained on S. aureus, concerning CoNS our result suggest that there are differences in the growth rate among the various members of this group and in the incubation time necessary for the met-R detection (more isolates of the respective species must be analysed to reach generalized conclusion).
P1387: Comparison of different techniques to detect methicillin resistance in Staphylococci and evaluation of Vitek2 P523AST cards
ö. Akan, S. Uysal (Ankara, TR)
Laboratory diagnosis of methicillin resistance has been a problem and every system should be evaluated before introduction to routine laboratory practice. In this study the results of comparison of disk diffusion (oxacillin 1 mcg disks) (DD), oxacillin agar screen (OAT) tests, miniapi staph 5 AST system (biomerieux) (MA), and Vitek 2 AST p-523 cards (biomerieux) (VT) for detection of methicillin resistance in clinical Staphylococci is presented. MIC values using E-test (AB biodisk) were used as reference. NCCLS procedures and manufacturers’ recommendations were used. There was no difference to detect methicillin resistance in DD, OAT, MA and E-tests among 151 S. aureus except false susceptibility in OAT in 2 strains (1.3%) and DD in one strain (0.7%). Randomly selected 50 methicillin resistant 43 methicillin sensitive strains were studied using VT and 13 (14%) strains showed false resistance. Among them 12 showed discordance in oxacillin screening wells and MIC detection wells and could not pass advanced expert system and only 1 (1.1%) was real false positive. When same validation is applied to 141 coagulase negative Staphylococi (CNS) (116 S. epidermidis, 25 CNS) false negativity was more common in 16 (11.3%) OAT (not recommended by NCCLS) and one strain (0.7%) in DD. 5 strains (3.5%) were falsely resistant by MA. For VT validation among randomly selected 81CNS strains (58 S. epidermidis, 23 CNS) only one strain (0.7%) was falsely resistant. 10 (7.1%) strains showed discordance in oxacillin screening wells and MIC detection wells. When we ommitted oxacillin screening well (because of its high antibiotic content to detect resistant CNS) false positivity in 3 strains still remained. Antibiotic susceptibility tests were more compatible in S. aureus isolates. Results obtained from VT shows that this system can be adopted to our routine laboratory practice, but it should be used with complementary tests both for S. aureus and CNS in case of discordant interpretative results between two wells for oxacillin resistance.
P1388: Comparison of oxacillin resistance screen agar base with real-time PCR and conventional culture for rapid detection of methicillin-resistant Staphylococcus aureus
A. Anders, G. Geis, S. Gatermann (Bochum, D)
Objectives: Rapid and accurate identification of methicillin-resistant S. aureus (MRSA) is crucial for therapy and management of infected and colonized patients. The use of conventional culture techniques requires prolonged incubation and time-consuming isolation procedures followed by identification and susceptibility testing. An accurate diagnosis may take up to five days. The most early point of time for reporting MRSA by conventional methods is after 48 hours. The aim of this study is to evaluate the new Oxacillin resistance screen agar base (ORSAB) and a 24-hour PCR-method for the identification of MRSA in clinical specimen and compare it to the conventional culture.
Methods: Swabs from various sites were suspended in 1 ml of 0.9 % NaCl and equal aliquots were used for the three methods. The methods were a) ORSAB (Oxoid) b) real-time PCR detecting the mecA gene and a S. aureus-specific marker after an incubation in a selective broth and c) the conventional culture consisting of a blood agar plate and tryptic soy broth containing 10% NaCl. The ORSAB, blood agar plates and tryptic soy broth were inspected after 20–24 and 44–48 hours. PCR was performed after 20–24 hours.
Results: We examined 1024 swabs for the presence of MRSA. With the PCR, there were 80 positive MRSA results that could be reported to the clinician after 24 hours. After 48 hours 38 MRSA could be reported from the ORSAB whereas conventional culture with a sheep blood agar plate yielded 67 MRSA. The final evaluation after 5 days resulted in 197 MRSA from blood agar combined with enrichment broth, 156 MRSA from ORSAB and 80 MRSA from PCR. No MRSA could be detected in 789 samples.
Conclusions: Real-time PCR for detection of MRSA is a useful method for providing MRSA results within a very short period of time. The demand for a very sensitive method can only be met with a longer incubation time and the use of an enrichment broth which accounted in our study for additional 25% positive results.
P1389: Co-colonisation of methicillin-resistant coagulase-negative Staphylococci and methicillin-susceptible Staphylococcus aureus strains: risk for misidentification in molecular detection assays
K. Becker, I. Pagnier, B. Schuhen, F. Wenzelburger, A.W. Friedrich, G. Peters, C. von Eiff (Munster, D)
Objectives: Detection of carriers of methicillin-resistant Staphylococcus aureus (MRSA) is crucial for infection control purposes. Numerous molecular methods, often simultaneously in multiplex format, to identify the mecA gene coding for methicillin resistance (MR) and genes specific for the species S. aureus have been introduced. However, application of molecular methods for detection of MRSA directly from specimens may be influenced by co-colonization with MR-coagulase-negative Staphylococci (CoNS) carrying the mecA gene. Studies investigating the frequency of the co-colonization with methicillin-susceptible S. aureus (MSSA) and MR-CoNS on the skin and mucous membranes with the potential risk of false-positive results in molecular MRSA detection tests are missing.
Methods: A total of 249 nasal swabs were collected directly at admission to the cardiac surgery department at the University Hospital of Muenster as well as from known MRSA carriers (n = 14). Only one swab per patient was included. Staphylococcal isolates were identified by standard microbiological procedures and confirmed by molecular methods. Methicillin resistance was determined by mecA amplification.
Results: Growth of staphylococcal isolates were found in 232 (93.2%) of the nasal swabs comprising 66 S. aureus isolates consisting of 45 MSSA and 21 MRSA isolates and 311 isolates of CoNS (S. epidermidis, n = 216; S. haemolyticus, n = 31, other CoNS, n = 64). Overall, 130 (41.8%) CoNS isolates were tested methicillin resistant (MR-S. epidermidis, n = 98; MR-S. haemolyticus, n = 19, other MR-CoNS, n = 13). Co-colonization of the anterior nares with MSSA and MR-CoNS (MR-S. epidermidis, n = 6; MR-S. haemolyticus, n = 1; other MR-CoNS, n = 1) were found in 8 (3.2%) of the nasal swabs tested.
Conclusion: Nasal co-colonization with MSSA and MR-CoNS was found in a low percentage. However, in a low MRSA setting, false-positive MRSA test results may lead to a couple of efforts in eliminating the patient's putative MRSA colonization and in additional infection control measures and, consequently, in a substantial increase in costs and personnel workload.
P1390: Specific detection of methicillin-resistant Staphylococcus aureus directly from clinical specimen by real-time polymerase chain reaction
P. Goffinet, A. Louahabi, N. Hougardy (Arlon, B)
Objectives: Although Staphylococcus aureus (S. aureus) is relatively easy to cultivate, culture methods require 24 h incubation and conventional identification methods may yield false-positive or false-negative results. Standard susceptibility and penicillin-binding protein latex agglutination tests are time-consuming because they require colonies to be isolated from 24 h culture or more. The correct identification of S. aureus and the detection of the mecA gene based on molecular methods is considered as a gold standard in the determination of methicillin-resistant S. aureus (MRSA). Many PCR for simultaneous detection of S. aureus specific gene target and mecA gene were developed, but cannot be applied for the direct detection of MRSA from nonsterile specimens such as nasal samples without an isolation step. A more comprehensive real time PCR assay that targets both an S. aureus-specific gene and the mecA gene within a single PCR tube reaction using only one couple of primer and two adjacent fluorescent probes was established and evaluated.
Methods: The validation of the assay was performed using 12 MRSA strains and 31 other strains of Staphylococci (not MRSA). A positive result was obtained for all MRSA strains and not with the other strains. 437 screening swabs from anterior nares and throat were obtained from patients hospitalized in the intensive care unit. S. aureus strains were identified according to their characteristic growth morphologies, Gram stain characteristics, reaction to catalase, coagulase production, colour of colonies on SAID culture media (Biomerieux) and LightCycler staphylococcus kit (Roche Diagnostics). Resistance to oxacillin was measured with an AST-P536 card on the VITEK-2 instrument (Biomerieux) and/or LightCycler MRSA detection kit (Roche Diagnostics). In order to gain time, we included an automated DNA extraction protocol on a LC MagNA Pure instrument and we realised the real-time PCR on a LightCycler instrument.
Results: 24 MRSA could distinctly be detected by this PCR assay, Vitek-2 and/or LightCycler Staphylococcus-MRSA detection kits. From these, 14 were detected both by PCR and Vitek-2, 4 only by Vitek-2 and 6 only by the new PCR assay. The per cent of agreement is 97.7%.
Conclusion: This real-time PCR assay represents a rapid and powerful method which can be used for the detection of MRSA directly from specimens containing a mixture of staphylococci. The turn-around-time for the whole process is only 4 hours.
P1391: Detection of MRSA directly from blood cultures using real-time PCR
P. Della-Latta, P. Pancholi, F. Wu (New York City, USA)
Objectives: Methicillin-resistant Staphylococcus aureus (MRSA) septicemia is a life-threatening infection challenging both clinicians and laboratorians. Early detection of MRSA sepsis is of utmost importance to decrease patient morbidity and mortality, reduce empiric use of vancomycin and permit effective decisions for patient management. Real-time PCR using the IDI-MRSA (Infectio Diagnostic) commercial assay and various home-brew (HB) PCR assays have been used to detect MRSA directly from both colonized and infected patients. This study 1) evaluated a modification of the IDI-MRSA in comparison to a HB PCR assay for direct detection of MRSA from newly positive blood culture bottles 2) compared the performance of a modification of the IDI assay to conventional culture techniques 3) contrasted time to results of molecular and non-molecular procedures.
Methods: Aliquots of blood culture bottles (BACTEC 9240, Becton-Dickinson) obtained from 368 patients at Columbia University Medical Center that signaled positive and that Gram stained as gram-positive cocci (GPC) in clusters, were tested with the IDI-MRSA assay and a HB PCR using the SmartCycler (Cepheid). The IDI system detects the staphylococcal cassette mecA – orfX amplicon; whereas the HB assay detects the nuc and mecA genes. The introduced modifications of the IDI system, which included one NaOH wash and two dilution steps with Tris-buffer, were necessary to decrease false-positive results due to high bacterial counts. The molecular methods were compared to culture using the MicroScan Walkaway SI (Dade Behring, IL) for identification and the oxacillin screen plate.
Results: MRSA was detected from 37 of 368 (10%) of blood culture sets tested. The sensitivity, specificity, positive predictive and negative predictive values for IDI-MRSA compared to culture were 100%, 99.4%, 94.9% and 100%, respectively. Discordance of IDI and culture was observed in 0.5% of patients and was resolved using HB PCR. The IDI assay, as opposed to HB, accurately distinguished MRSA from the 0.4% blood cultures that were mixed with S. aureus and other bacteria, i.e. coagulase-negative Staphylococci (CoNS). MRSA detection by IDI was obtained within 2–24 hr compared to 48–72 hr by culture.
Conclusions: The IDI real-time assay using the SmartCycler can be successfully adapted for accurate, same-day detection of MRSA directly from positive blood culture bottles, regardless of the co-existence of CoNS.
P1392: Molecular diagnosis of Staphylococcus aureus isolates
N.S. Mariana, V. Neela, S. Zamberi, A.R. Raha, R. Rozita, S. Radu (Selangor, MYS)
Objective: Frequent outbreaks of Staphylococcus aureus infections worldwide emphasizes urgent need for rapid and reliable detection methods for S. aureus, especially in hospital settings and community. The ultimate aim of the study is to identify potential diagnostic marker and to develop DNA based assays that can be used in any microbiological laboratories for the rapid and reliable detection of S. aureus from pure culture and also directly from clinical samples.
Methods: To select a genetic target for diagnostic purpose, S. aureus isolates obtained different hospitals in Malaysia were fingerprinted using rep (repetitive element sequence) primers. From rep fingerprint, a marker band present in all isolates was cloned and sequenced. A primer pair to amplify a 197 bp fragment and an oligonucleotide probe was designed from the most conserved region. Primers and probe were tested against 89 local S. aureus isolates and 1 ATCC to validate the ubiquity. The specificity of molecular assays were verified by using genomic DNA from a battery of gram-positive and gram negative bacterial species. The ability of assays to detect S. aureus directly from clinical samples was also tested. PCR and membrane assays were compared for ubiquity, specificity, and rapidity.
Results: The rep fragment sequenced with universal M13 primers determined the size of marker as 489 bp, showed high similarity (>95%) to GAP (glyceraldehyde-3-phosphate Dehydrogenase) Operon in S. aureus genome. The molecular assays developed with innovated primers and probe was very successful, as none of other non-S. aureus species showed positive signal confirming the ubiquity and specificity. PCR assay was more efficient in detection of S. aureus direct from clinical samples, membrane assay required purified DNA. In terms of rapidity, both assays produced results in 2–3 hrs.
Conclusion: The development of simple and rapid molecular assays with innovated primers and probe, specific and ubiquitous for S. aureus that can even detect S. aureus directly from clinical samples could be applied in any microbiological laboratory for fast diagnosis of S. aureus infection. The novel achievements made in this study will allow the faster and correct identification of S. aureus, establishes the effective antibiotic therapy and thereby controls and prevent future outbreaks.
P1393: Development of a membrane based assay for the identification of Staphylococcus aureus at species level and detection of multiple drug resistant isolates
N.S. Mariana, V. Neela, A.R. Raha (Selangor, MYS)
Objectives: The incidence of Staphylococcus aureus infection has increased in recent years, due to the spread of multidrug-resistant isolates. Conventional identification based on biochemical characteristics and determining the antibiotic resistance based on antibiotic susceptibility pattern can identify S. aureus, but the rapidities and efficiencies of these methods need to be improved. Rapid and direct identification of this bacterium from clinical samples (urine) would be useful for the early diagnosis of S. aureus infection in the clinical microbiology laboratory. This study incorporates the development of a single tube PCR system (multiplex) and membrane based assay for the identification of S. aureus and detection of multiple antibiotic resistant isolates directly from clinical samples.
Methods: Seventy isolates of S. aureus from different hospitals in Malaysia were studied. The single tube PCR system for the identification of S. aureus and detection of methicillin, gentamycin, erythromycin and vancomycin resistant genes was carried out using a set of published primers. The S. aureus was isolated directly from urine samples by boiling method. The sensitivity of the system was determined by serially diluting the S. aureus culture, an aliquot of each dilution was subjected to DNA isolation and the purified DNA was used as template for PCR. The specificity of the system was verified using a panel of gram positive and gram negative isolates. A membrane based assay was developed based on DNA–probe hybridization technique.
Results: The single tube PCR system yielded products of 108 bp for S. aureus (identity fragment), 139 bp for erythromycin, 174 for gentamycin and 533 bp for methicillin resistant genes, when sequenced showed 100% homology. None of the isolates amplified vancomycin resistant gene. The assay was very specific for S. aureus as none of the other gram negative and positive isolates amplified the identity fragment. The sensitivity level achieved with urine samples was 1 CFU with 25 cycles of amplification. Chemiluminescence detection of hybrid obtained from membrane based assay with a minimum concentration of 160 ng probe confirmed the identity of S. aureus and detected the multiple drug resistant isolate.
Conclusions: This study demonstrates the practical value, sensitivity, rapidity, specificity and accuracy of the molecular based method for the diagnosis and thereby treatment of S. aureus infections with the right choice of drugs.
P1394: Efficient genotyping of methicillin-resistant Staphylococcus aureus using a combination of binary markers and single nucleotide polymorphisms
F. Huygens, A.J. Stephens, G.R. Nimmo, J.M. Schooneveldt, G.W. Coombs, E.P. Price, P.M. Giffard (Brisbane, AUS)
Objectives: Staphylococcus aureus continues to be a significant human pathogen. Several lineages of methicillin resistant S. aureus (MRSA) are common agents of nosocomial infections, and recent years have seen the appearance and rapid spread of MRSA clones capable of causing community acquired infections. The objective of this study was to develop an efficient S. aureus genotyping method that is carried out using real-time PCR technology. The method was designed to provide an epidemiological fingerprint and also allow inference of clinically relevant aspects of the phenotype, through identification of the lineage of the genome backbone using SNPs in combination with the testing for the presence of genes that exhibit binary variability.
Methods: The basis of our approach to designing genotyping methods is the computerised analysis of comparative sequence databases in order to identify minimal sets of genetic polymorphisms that provide the desired degree of resolving power. Such analyses are carried out using the computer programme ‘Minimum SNPs’ which assembles sets of single nucleotide polymorphisms (SNPs) empirically on the basis of maximisation of the Simpsons Index of Diversity (D). SNPs were interrogated using allele specific PCR in the real time format (kinetic PCR).
Results: Analysis of the S. aureus multilocus sequence typing (MLST) database yielded seven single nucleotide polymorphisms (SNPs) that provide a D of 0.95 with respect to the MLST database i.e. if two sequence types are chosen at random from the database, there is a probability of 0.95 that they will differ at one or more of the SNPs. Recalculation of published S. aureus MLST-based studies has shown that a similar level of discrimination is obtained with actual collections of isolates. The SNP profiles are highly concordant with clonal complexes. A kinetic PCR method for interrogating the SNPs was shown to be robust, although a modified SNP set gave better results with crude genome preparations. The SNPs gave nine profiles with a 107 Australian MRSA isolates. The informative power of a number of binary genes in combination with the SNPs was tested, and it was found that testing for presence of the pvl, sdrE and cna genes, together with three mecA-associated plasmids increased the number of types to 21.
Conclusion: A rationally designed S. aureus genotyping approach suitable for routine use on real-time PCR or medium density chip-based platforms has been demonstrated.
P1395: Development of a green fluorescent protein based reporter gene system in Staphylococcus epidermidis
G.C. Franke, S. Dobinsky, M.A. Horstkotte, J.K.-M. Knobloch, D. Mack, H. Rohde (Hamburg, D; Swansea, UK)
Objectives: Based on its biofilm forming capacity Staphylococcus epidermidis is the most frequent cause of foreign-body related infections. Several specific factors mediating primary attachment and accumulation have been characterized. In order to analyze the concerted action of these factors methods for in situ expression analysis are demanded. The aim of the present study was to establish a Green Fluorescent Protein (GFP) based reporter gene system in Staphylococcus epidermidis.
Results: Therefore we cloned gfpmut3.1 (Clontech) into pASI under the control of a xylose-inducible promotor and introduced the resulting construct into S. epidermidis 1457. However, under xylose induction no significant increase of fluorescence intensity compared to the un-induced control was detected. This finding may be attributed to inefficient translation initiation at the natural Shine-Delgarno (SD) sequence. Indeed, by coupling different SD-sequences from well-characterized S. epidermidis genes in front of the gfp start codon the crucial impact of this element for gfp translation initiation was demonstrated: whereas a strong fluorescence signal was obtained in presence of the hld SD sequence, almost no signal was detected with the sarA SD sequence. The influence of different SD sequences on transcription and translation of gfpmut3.1 was also analysed in real-time transcription and semi-quantitative western blotting experiments. In all constructs investigated an almost identical gfp transcription level was found regardless of the SD sequence present. In contrast, western blot analysis using an anti-GFP antibody revealed huge differences in GFP amounts that corresponded to the respective quantitative fluorescence signal. The calculated half-life in S. epidermidis 1457 is 6.9 h. Importantly, expression of gfpmut3.1 did not interfere with the biofilm-positive phenotype of this strain.
Conclusions: In conclusion, using an appropriate SD sequence for optimal translation initiation gfpmut3.1 offers an attractive system for monitoring gene expression in S. epidermidis.
Infections and diagnosis in non-HIV immunosuppressed patients
P1396: Isolation of Bordetella pertussis in blood culture from a patient with multiple myeloma
M. Trøseid, T. Jonassen, M. Steinbakk (Lørenskog, N)
Bordetella pertussis is a fastidious aerobic, Gram-negative coccobacillus that causes the classical disease of whooping cough. The clinical manifestations of pertussis are due to the toxins that are produced by the organism locally and not by bacterial invasion. Isolation from blood cultures is extremely rare and to our knowledge this is the third reported case of Bordetella pertussis bacteraemia (1,2). All cases have been reported in immunocompromised patients. The patient was a 63 year old man with a 2 year history of multiple myeloma, initially treated with autologous bone marrow transplantation and currently treated with chemotherapy due to disease progression. One weak after treatment with vincristine, adriamycin and dexamethasone (VAD) he developed a productive cough and fever and was admitted to medical department. On admission C-reactive protein was 131 mg/l, WBC count and chest x-ray were normal and blood cultures were initially negative. He was treated for 5 days with intravenous cefotaxime for suspected bronchopneumonia and discharged in good condition. The patient was readmitted 8 days later with dry cough, hoarseness and fever. C-reactive protein was 270 mg/l and WBC count was 12.7*10 9/l. Chest X-ray was normal and blood cultures were negative. Again bronchopneumonia was suspected, and he was given intravenous penicillin for 7 days. Eventually, blood culture from the first admission revealed growth of an atypical Gram-negative rod from an aerobic bottle. The bacterium was identified as Bordetella pertussis by sequencing of the 16S rRNA gene. The patient was discarged in good condition and treated for 10 days with oral clarithromycin. Norway has experienced an outbreak of whooping cough since 1997 with more than 3000 reported cases annually. Systemic infections may occur in the immunocompromised host. Growth in blood culture may be slow, and prolonged incubation is recommended.
-
1
Janda WM, Santos E, Stevens J, Celig D, Terrile L, and Schreckenberger PC. Unexpected Isolation of Bordetella pertussis from a Blood Culture. Journal Clin Microbiol 1994; 32:2851–3.
-
2
Centers for Disease Control and Prevention. Fatal case of Unsuscpected Pertussis Diagnosed from a Blood Culture-Minnesota. MMWR 2004;53:131–2.
P1397: Current spectrum of bacterial infections in patients with haematological malignancies
D. Yadegarynia, K.V. Rolston (Tehran, IR; Houston, USA)
Background: Patients with hematologic malignancies (HM) develop bacterial infections often. Unfortunately most institutions include only bloodstream infections (BSI) when describing the spectrum of infection in such patients, despite the fact that BSI account for only 25–35% of infections. Also most data bases do not include polymicrobial infections either. These omissions give an incomplete and erroneous impression regarding the spectrum of bacterial infection.
Objective: To determine the overall spectrum of bacterial infection (not just – BSI or monomicrobial infections) in patients with underlying HM, with or without neutropenia.
Methods: A retrospective review of microbiological and clinical records of patients with HM over a 1 year period (October 2002 to October 2003).
Results: 311 patients with bacterial infections were identified. Median age was 65 y (range 20–80 y). Acute leukemias were most common − 41%, followed by lymphomas − 32%, chronic leukaemia − 18%, and myelomas − 5%. Only 44% were neutropenic, and 45% were on antibacterial prophylaxis. Sites of infection included respiratory tract − 38%, BSI (including catheter-related) − 38%, urinary tract − 10%, skin and skin structure − 8%, and gastrointestinal tract − 5%. 221 episodes (71%) were monomicrobial and 90 (29%) were polymicrobial. Gram-positive bacteria accounted for 67% of monomicrobial BSI but only 45% of infections overall (i.e. when polymicrobial infections were also included in the spectrum). Gram-negative bacteria accounted for 33% of monomicrobial BSI, but only 26% of infections overall. More than 85% of polymicrobial infections had a gram-negative component, with P. aeruginosa being the single most common species isolated. The most common gram-positives were coagulase-negative staphylococci, S. aureus, viridans group streptococci, and Enterococcus spp. The most common gram-negatives were E. coli, P. aeruginosa, S. maltophilia, and Klebsiella spp.
Conclusions: Patients with HM develop bacterial infections even when not neutropenic, and despite prophylaxis. Gram-positives are not as predominant overall as they are in BSI. Polymicrobial infections and S. maltophilia infections are more frequent now than in the past.
P1398: Adequacy of trimethoprim-sulfamethoxazole for prevention of urinary tract infection in renal transplant recipients
C.S. Moon, S.H. Kim, H.K. Ki, K.S. Ko, W.S. Oh, K.R. Peck, N.Y. Lee, J.-H. Song on behalf of the Asian Pacific Research Foundation for Infectious Disease
Objective: To evaluate the adequacy of trimethoprim-sulfamethoxazole (TMP-SMX) for prevention of urinary tract infection(UTI) in patients underwent renal transplantation in the setting where TMP-SMX resistance is common.
Methods: All patients underwent renal transplantation at Samsung Medical Center from January 1998 and August 2002 were included with the completion of 2 year-follow-up. TMP-SMX was prophylactically administered during 12 months after renal transplantation. Their medical records and microbiologic data were reviewed.
Results: A total of 336 patients were enrolled. (male to female ratio 191:145, mean age 39 ± 10 years). 146 episodes of UTI were observed in 104 patients (31%) within 2 years after renal transplantation. 52 episodes (36%) developed during post-transplantation 30 days, and 87 episodes (60%) during post-transplantation 6 months. UTI occurred predominantly in female recipients (124 episodes, p = 0.00001). There was no difference in the incidence of UTI with regard to the kind of immunosuppressants (p = 0.69) or graft rejections (p = 0.85). Among the isolated strains, E. coli (51%) was the most common, followed by Enterococcus spp. (12%), Pseudomonas spp. (7%), Enterobacter spp. (6%), Coagulase-negative staphylococci (6%), Klebsiella spp. (5%), and Streptococcus spp. (1%). Among 75 E. coli isolates, rates of resistance to TMP-SMX, ciprofloxacin, ampicillin/sulbactam, and ceftriaxone were 63%, 35%, 34%, and 1%. There was no difference in mortality rate related with the occurrence of UTI (p = 0.75).
Conclusion: Despite of high prevalence of TMP-SMX-resistant E. coli, the incidence of UTI in renal recipients receiving TMP-SMX are similar to that in the setting where TMP-SMX resistance is uncommon. Nevertheless, because most episodes of UTI occur within 1 or 6 months of transplantation, further study is warranted to evaluate if additional preventive strategies during early period is needed.
P1399: Prospective evaluation of procalcitonin in adults with febrile episodes after allogeneic hematopoietic stem cell transplantation
M. Ortega, M. Rovira, X. Filella, J.A. Martínez, M. Almela, J. Puig, E. Carreras, J. Mensa (Barcelona, E)
Background: We have previously demonstrated that procalcitonin (PCT) values higher than 3 ng/mL were associated with fungal invasive infections (IFI) in patients with persistent fever after five days of empirical antibiotherapy during neutropenic phase after Hematopoietic Stem Cell Transplantation (HSCT). Objective: To analyse if PCT is an independent and early diagnostic marker of IFI in high risk adult patients undergoing allogeneic HSCT.
Patients and methods: PCT levels were determined in 129 febrile episodes developed by 65 consecutive patients receiving an allogeneic HSCT. There were 72 episodes during neutropenic period and 57 within the second and third phase of immune recovery after HSCT. All febrile episodes were classified according to the final diagnosis in: fever of unknown origin, microbiological or clinical documented infection and noninfectious febrile episodes. The microbiological or clinical documented infection were classified as possible, probable or proven IFI according to the EORTC/MSG criteria. The values of PCT were not included in the classification criteria.
Results: The final diagnosis of febrile episodes were: fever of unknown origin in 58 cases, microbiological or clinical documented infection in 54 and non-infectious fever in 17. Among the 54 patients with a documented infection, 29 did not fulfil any IFI criteria, and 8, 15 and 2 had a possible, probable and proven IFI, respectively. Eleven out of 25 IFI cases occurred during the neutropenic phase (neutrophil absolute count, NAC, < 0.5 × 109/L) and 14 cases during the non-neutropenic phase of HSCT. Mean (ng/mL) (±SD) values of PCT on the first day of fever among neutropenic patients were: 1.0 (±0.3) in patients with infections other than IFI (n = 19); 0.9 (±1.3) in possible IFI (n = 5); 1.2 (±2.1) in probable IFI (n = 5) and 0.63 in proven IFI (n = 1). Mean values of PCT on the first day of fever among non-neutropenic patients were: 1.5 (±0.9) in infections other than IFI (n = 10); 6.1 (±3.4) in possible IFI (n = 3); 9.7 (±6.1)* in probable IFI (n = 10) and 6.3 in proven IFI (n = 1), (*p = 0.013, Kruskal-Wallis; OR: 1.1, IC 95%: 0.92–2.1).
Conclusions: During the non-neutropenic phases of allogeneic HSCT, a high PCT value on first day of fever is associated significatively with IFI. PCT monitoring could be an early and useful help to the differential diagnosis of febrile episodes in patients with high risk of IFI.
P1400: Agrobacterium radiobacter peritonitis in a patient on continuous ambulatory peritoneal dialysis
H. Konstantakopoulou-Papadaki, S. Botsari, M. Sonician, D. Vlasopoulos, G. Kouppari (Athens, GR)
Objective: The presentation of Agrobacterium radiobacter peritonitis in an adult under continuous ambulatory peritoneal dialysis (CAPD).
Methods: Peritoneal fluid specimens were examined for cells count and were cultured in Bactec Plus culture vials (Bactec plus, Becton Dickinson). After centrifugation the specimen was also cultured on appropriate media for aerobic and anaerobic microorganisms and smears were performed by Gram stain. The identification of bacterium was carried out by standard methods and API-32E, API 32GN (bioMerieux). The susceptibility testing was performed by disk diffusion method.
Case report: A male patient aged 87 years, at final stage of renal failure under CAPD for 15 months and deficient renal function, was admitted to hospital because of the appearance of a cloudy peritoneal dialysate effluent and nausea. The patient had a history of three episodes of peritonitis caused by Gram negative bacteria (Klebsiella pneumoniae and Pseudomonas aeruginosa, Flavomonas oryzihabitans, Acinetobacter johnsonii). The patient was successfully treated with IP tobramycin and recovered completely. On day of admission the peritoneal analysis showed 3100 WBC/ìl (96% neutrophills). The patient was treated empirically with IP tobramycin. Forty eight hours after the incubation of peritoneal fluid gram negative bacterium was isolated. Tests for oxidase and catalase were positive. The hydrolysis test of esculine was positive. The bacterium was identified as Agrobacterium radiobacter. The bacterium was susceptible to cefuroxime, ceftriaxone, ciprofloxacin, imipenem, tetracycline and gentamicin and resistant to ampicillin, trimethoprime-sulfamethoxazole, cephalothin and tobramycin. The patient was treated with cefuroxime 750mgx3 IP for three weeks and he recovered completely.
Conclusion: Agrobacterium radiobacter is a microorganism found in the environment and more particularly in several kinds of soils and is considered a plant pathogen. In rare cases of immunocompromised patients and especially for patients with transcutaneous catheters or implanted biomedical protheses, Agrobacterium radiobacter has been regarded as the cause of bacteramia and peritonitis.
P1401: Erysipelothrix rhusiopathiae. Septic arthritis of the knee
M. Lyons, A. Kirk, G. Channon, S. Thomas, B. Reddy (High Wycombe, UK)
Introduction: E. rhisiopathiae is a rare pathogen found worldwide. It is a commensal of a wide variety of vertebrate and invertebrate species but its major reservoir is domestic swine. The greatest commercial impact of infection is due to swine erysipelas. Serious infection in humans in extremely rare and associated with cutaneous infection in those who have had occupational exposure. Clinical manifestations include brain abscesses, osteomyelitis and chronic arthritis. Systemic infection can often be complicated by infective endocarditis, in which the mortality rate appears to be higher than for other organisms. We report a rare case of E. rhusiopathiae joint infection.
Case Report: A 74 year old male patient on long term treatment for rheumatoid arthritis presented to clinic with an isolated knee sepsis, associated with a raised CRP and WCC. The knee was aspirated and E. rhusiopathiae isolated from culture broth after failing to grow on initial culture media. There were no other systemic manifestations. No direct occupational exposure was reported. A repeat knee aspirate failed to grow E. rhusiopathiae, but a third aspirate taken at arthroscopy once more isolated the organism. It was assumed that a soft tissue infection with E. rhusiopathiae had led to a bacteraemia which seeded into the rheumatoid knee joint. He underwent prolonged treatment, with a six week course of Clindamycin. Unfortunately, a low grade chronic infection persisted leading to complete disorganisation of the joint. A joint replacement is being considered.
Discussion: This report is of interest because of the exceptionally unusual nature of the pathogen and because of the lack of systemic symptoms or history of any social or occupational exposure to explain the presence of the organism. Diagnosis of E. rhusiopathiae was problematical but is essential to ensure effective treatment as most strains are resistant to vancomycin, which is often used empirically to treat bacteraemia due to gram positive organisms.
P1402: Invasive nocardiosis: emergence of new species and usefulness of susceptibility testing
M. Hallin, F. Kidd, O. Denis, S. Rottiers, C. Thiroux, M.J. Struelens, F. Jacobs (Brussels, B)
Objective: The aim of the study was to review clinical and microbiological features of the nocardial infections diagnosed in our 858-bed academic hospital over the past 3 years.
Patients and Methods: All patients with at least one sample culture positive for Nocardia sp between May 2000 and August 2004 were included. Underlying conditions, immunosuppressive treatments, site of infection, clinical, microbiological and radiological features, treatment and outcome were compiled. The identification was based on the morphology by Gram and modified Kinyoun stains, aspect of the colonies on blood agar, phenotypical tests and amplification-sequencing of a species-specific region of the 16S rDNA. Susceptibility tests were performed by E-test on Mueller-Hinton agar and interpreted according to NCCLS criteria.
Results: Nocardia sp was recovered from 8 patients (5 males and 3 females, mean age 52 y.). All patients had predisposing factors: 4 were solid organ transplant recipients and 4 were treated with glucocorticosteroids. Infection involved the lung in 3 patients, CNS in 2, muscles and soft tissues in 1, and was disseminated in 2. The species identification of the isolates were N. farcinica (3), N. nova (3) and newly described N. cyriacigeorgici (2). Phenotypic and genotypic identification methods were concordant in all cases. All isolates were susceptible to trimethoprim/sulfamethoxazole (TMP/SMX) and amikacin and all but one N. farcinica strain (MIC imipenem/meropenem >32 μg/ml) were susceptible to carbapenems. Patients were treated with high doses of oral or i.v. TMP/SMX (3), i.v. meropenem (3), and the combination of TMP/SMX and meropenem (1). All patient rapidly improved except the patient infected with the N. farcinica strain resistant to carbapenem, until his initial meropenem treatment was replaced by high doses of i.v. TMP/SMX.
Conclusions: Nocardia can cause various clinical syndromes in immunocompromised patients. Early microbiological diagnosis of nocardial infections enabled rapid appropriate management and favorable outcome of all patients. Furthermore, the initial clinical failure associated with carbapenem resistance of a N. farcinica strain underlines the need for rapid and reliable identification of Nocardia to species level and susceptibility testing.
P1403: Microbiologic and clinical features of N. asteroides infections
M. Makarona, H. Moraitou, S. Karabela, G. Dimopoulos, E. Fragouli, S. Kanavaki (Athens, GR)
Objectives: Lower respiratory system represents the most frequent site of nocardial infection. However, as the immunocompromised population is increasing, the disseminated nocardial disease is not rare. Worldwide, Nocardia asteroides accounts for the majority of infections. The aim of our study was to present our experience, regarding the N. asteroides isolations, in our Hospital.
Methods: During 2000–2004, in the Microbiology Laboratory of ‘Sotiria’ Chest Diseases Hospital of Athens, 8 strains of N. asteroides were isolated, from 6 male and 2 female patients. Four of the patients had a localized pulmonary infection, while 4 disseminated. The disseminated infection concerned 3 cases of pulmonary empyema and 1 of cutaneous abscess of the left arm. All patients were immunocompromised: 3 of them suffering from malignancies, 2 from diabetes mellitus, one from collagenic disease, one from pulmonary tuberculosis and one from ITP. The microbiologic testing included a combination of methods:
-
•
Colonial and microscopic morphology:using the acid-fast Kinyoun-modified stain
-
•
Simple biochemical and hydrolysis testing: casein hydrolysis, gelatin liquefaction and decomposition of tyrosine, xanthine and hypoxanthine
-
•
Antibiotic susceptibility profiles: susceptibility to cefamandole and tobramycin.
Results: A high degree of correspondence between biochemical characteristics and antibiotic susceptibility patterns was noted. All strains were identified as N. asteroides, by both biochemical and susceptibility pattern criteria. Patients were treated either with cotrimoxazole, high doses of imipenem or minocyclin. Three patients died, two suffering from lung cancer and one of ITP.
P1404: Fournier's gangrene: a life-threatening clinic disorders in uncontrolled type II diabetes mellitus patients
O. Oncul, C. Erenoglu, H. Uluutku, M.L. Akin, S. Cavuslu, T. Celenk (Istanbul, TR)
Objectives: Fournier's gangrene is a serious skin and soft tissue infectious-necrotising process in the peri-neogenital area affecting adults, usually in their sixties or seventies. Isolated flora from cultures of the necrotic lesion is commonly multi-microbial. Although an idiopathic condition is considered in the past, today a genitourinary, anorectal or dermal triggering factor can be identified in most patients. There are a series of systemic host debilitating disorders such as diabetes mellitus, chronic alcohol abuse, and malignant neoplasia that are associated with this condition and may be considered risk factor to suffer this disease. In this study, 11 patients with FG, eight of whom had uncontrolled DM were investigated for risk factors, clinical signs, laboratory findings and prognosis.
Methods: In this study, 11 FG patients who were hospitalized in Gulhane Military Medical Academy Haydarpasa Training Hospital Department of General Surgery Service were presented between 1998 to 2004. Their age, clinical presentation, predisposing factors, microbiology testing, management and prognosis were studied. Broad -spectrum parenteral antibiotics, early and aggressive surgical debridement of the necrotic areas and hiperbaric oxygene therapy were applied to all cases.
Results: The mean age was 39.2 (21–79) of 11 FG cases. The average time from begin the symptoms to apply to hospital was 7.2 (3–17) days. The most prominent associated disease was diabetes, affecting 72.7 per cent of the patients, other potential contributing factors included trauma and surgical operations in three cases. The fasting blood glucose levels, HbA1C levels of the diabetic patients were 182–522 mg/dl and 8.1–10.5, respectively. Bacterial culture results revealed a single organism in 28.6%, and more than one organism in 71.4% of the cases. The most frequently isolated bacteriae were Escherichia col, Streptococcus pyogenes and Klebsiella pneumoniae from quantitative tissue cultures. The overall mortality rate was 45 percent. However, the mortality rate among diabetics was 62.5 per cent (P = 0.001).
Conclusion: Fournier's gangrene is a very serious life-threatening disorder in especially diabetic patients despite of early aggressive debridement with the use of appropriate antimicrobial therapy.
P1405: Relationship between uropathogenicity of Escherichia coli and host compromise status
E. Moreno, A. Andreu, T. Pérez, M. Sabaté, G. Prats (Barcelona, E)
Objectives: To evaluate the role that compromise status play in the uropathogenicity of E. coli strains, we set virulence factors and ECOR phylogenetic group against patients with and without compromise status.
Methods: 50 E. coli strains from patients with pyelonephritis and negative blood culture and 50 from urinary bacteraemia were analysed for ECOR phylogenetic groups: A, B1, B2 and D; virulence factors (VF) genes: papA, papGI, papGII, papGIII, fimH, afa/draBC, sfa/focDE, hlyA, cnf1, iutA, fyuA, kpsMII, ibeA, traT, malX by PCR. O antigens associated to UTI (O1, O2, O4, O6, O7, O18 and O83) were determined using agglutination microtechnique. Compromise status, obtained from medical records, included immnunocompromise and/or local predisposing factors to urinary tract infection.
Results: 58% patients with pyelonephritis and 68% with urosepsis were compromised. Pyelonephritic strains from healthy patients belonged to: group B2 76% and group D 24%, and from compromised patients: groups A and B1 10%, group B2 59% and group D 21%. Urosepsis strains from normal patients belonged to: group A 6% and group B2 94%, and from compromised patients: group A 21%, group B1 6%, group B2 56% and group D 18%. E. coli strains from noncompromised patients were associated to higher virulence score: pyelonephritis 7.52 virulent traits and urosepsis 8.50 than strains from compromised host: pyelonephritis 6.59 virulent traits and urosepsis 7.41. Despite this, only papGII was significantly more prevalent in pyelonephritic strains from healthy patients vs compromised (p < 0.05); whereas malX, papA, kpsMII and fyuA were somewhat more prevalent in normal people with both pyelonephritis and urosepsis that in people with compromise, while the remain traits were similary distributed. Strains from group B2 showed a high virulence score with a mean of 8.34 traits, followed by group D (6.06), group B1 (4.80) and A (4.64) (P = 0.004, group B2 vs others).
Conclusions: E. coli strains from normal hosts were predominantly from the ECOR phylogenetic group B2, harbouring a great number of virulent traits and consequently showed a high virulence score; whereas strains from phylogenetic group A and B1, posses few virulent traits and only infect patients with compromise status. However the substantially prevalence of group B2 strains, with a lot of virulent traits, among compromised host, could explain the lack of associations between this condition and reduced virulence.
P1406: Microbiological aspects of peritonitis in patients undergoing continuous ambulatory peritoneal dialysis
S. Maraki, A. Georgiladakis, Z. Gitti, E. Nioti, M. Epanomeritakis, Y. Tselentis (Heraklion, GR)
Objective: Peritonitis is the most important complication of continuous ambulatory peritoneal dialysis (CAPD). We reviewed the incidence and the etiology of CAPD-associated peritonitis.
Methods: A total of 468 peritoneal fluid samples from 76 adult patients on CAPD were examined from January 2001 to October 2004. Effluent samples were cultured on appropriate solid and liquid media after 10 min centrifugation. In addition 10 ml of fluids were inoculated in aerobic and anaerobic VITAL culture vials (BioMerieux, Marcy-l’ Etoile, France). Enumeration of WBC was done using a standard counting chamber. The identification of microorganisms was performed using standard methods and the API systems (BioMerieux).
Results: A total of 102 out of the 468 dialysates were positive (21.8%). Two of the positive samples were polymicrobial. Gram-positive organisms accounted for 68.3% of the infections of which coagulase negative staphylococci (CNS) were the commonest (35.6%). Gram-negative bacteria were found in 21.1% of the positive samples, anaerobic bacteria in 1.9%, and fungi in 8.7%.
Conclusion: CAPD-associated peritonitis was most commonly caused by coagulase negative staphylococci. Prompt identification of the causative agents is essential for the appropriate management of microbial peritonitis in patients on CAPD.
P1407: Neisseria sicca peritonitis in patient on continuous ambulatory peritoneal dialysis: a case of the ‘nonpathogenic’ Neisseriae infection
C. Papaefstathiou, M. Zoumberi, D. Arvanitis, D. Vlasopoulos, G. Kouppari (Athens, GR)
Objectives: To report an unusual case of peritonitis caused by N. sicca in a continuous ambulatory peritoneal dialysis (CAPD) patient.
Methods: Peritoneal effluent specimens were examined for WBC count and detection of microorganisms by Gram stained direct smears. They were cultured on appropriate media after centrifugation and were also inoculated in Bactec culture vials (Becton Dickinson). The identification of the microorganism was performed by standard methods, biochemical characteristics, colonies morphology and API NH system (bioMerieux). Susceptibility testing was carried out by disc diffusion method according to the NCCLS performance standard.
Case report: A female 57-year-old CAPD patient end stage renal failure, was admitted to the hospital because of peritoneal effluent turbidity. She started CAPD eleven months ago. She was suffering from systemic lupus erythematosus for 17 years and was on corticosteroids for 15 years. That was the 1st episode of peritonitis for her. On admission WBCs count of peritoneal fluid was 2.600/ μl (95% neutrophils) and was decreased to 1.000/μ l and 200/μl at second and third day respectively. On admission was administered cefazolin 1.5 gr I.P. and afterward 300 mg × 6 I.P. for the next 3 days. Gram (-) diplococci (intracellular or not) were seen on direct smears by gram stain. From two peritoneal effluent cultures Gram(-) diplococcus, oxidase positive and strict aerobic was isolated. The colonies were white, opaque, wrinkled, adhesive, peaked. The nitrate reduction was negative. With API NH (biotype:7101, %id:99,8 :1,00 -very good identification) the strain was identified as Neisseria spp group, which included the species N. sicca, N. mucosa I. subflava. The strain was differentiated from I. subflava on the base of colony morphology and from N. mucosa of nitrate reduction. The strain was beta-lactamase negative and resistant to erythromycin, trimethoprime/sulphamethoxazole, clindamycin and susceptible to penicillin, cephalosporines, aminoglycosides, ampicillin, ciprofloxacin and tetracycline. The patient was treated with cefazolin 300 mg × 4 I.P per day for the 18 next days and recovered completely.
Conclusions: ‘Nonpathogenic’ Neisseria spp can cause severe disease in immunocompromised patients. This is the first case report of CAPD peritonitis with Neisseria sicca in Greece and the third world wide.
P1408: Antibiotic resistance of enterococci in faecal samples from cancer patients
E. Chinou, P. Apostolakopoulos, E. Skouteli, A. Georgouli, P. Golemati (Athens, GR)
Objectives: The aim of this study was to determine the prevalence of antibiotic resistance of isolated enterococci in fecal samples from oncologic patients.
Methods: A total of 38 strains of enterococci were isolated in fecal cultures from 234 oncologic patients undergoing peripheral blood stem cell transplantation (PBSCT) or bone marrow transplantation (BMT) during one year (2003). The fecal samples were inoculated into bile esculin agar plates with and without 6 mgr/lt vancomycin and into an enrichment bile-esculin broth supplemented with 4 mgr/lt vancomycin. The identification of the isolated bacteria was performed by standard methods and the Api-strep system. The susceptibility testing was carried out by disk diffusion method according to the NCCLS guideline and by E-test for vancomycin and teicoplanin. All patients were febrile, with diarrhoea and were treated with antimicrobial agents before stool cultures.
Results: From 38 strains of isolated enterococci, 28 strains were identified as E. faecalis, 8 strains as E.faecium and two strains as Enterococcus sp. A total of 19 strains were found resistant to ampicillin (50%) 15 to high level gentamycin (39. 5%), 27 to ciprofloxacin (71%), 21 to tetracycline (55%) and 5 to quinopristin / dalfopristin (13%). All the isolated strains were sensitive to vancomycin, teicoplanin and linezolid. 7 of the 8 isolated strains of E.faecium were observed multidrug resistant and sensitive only to glycopeptides, linezolid and quinopristin / dalfopristin.
Conclusions: The present study indicates that the prevalence of V.R.E. in fecal samples from oncologic patients appears to be very low. Ciprofloxacin, a frequently used antimicrobial agent in cancer patients, was the less active of the antibiotics tested. In general, the good activity of the glycopeptides and linezolid tested shows promise for the combination therapy required in enterococcal infection in immunocompromised patients.
P1409: Microbiological findings of early infections in allogeneic or autologous stem cell transplantation
O. Yildiz, I. Sari, F. Altuntas, E. Alp, B. Aygen, O. Coban Yuksel, B. Sumerkan (Kayseri, TR)
Objectives: Infections are a major cause of morbidity and mortality in patients undergoing high-dose therapy and subsequent autologous or allogeneic stem cell transplantation (SCT), despite antimicrobial prophylaxis, use of growth factors and newer antimicrobial drugs.
Methods: We compared the incidence of early infectious complications between autologous SCT and allogeneic SCT recipients in a single centre over a 6-year period (between January 1997 and June 2004) in 164 consecutive adult patients. Infections occurring within the 30 days after transplant were defined as early infections. Forty-two patients were allografted and 157 autografted. Antimicrobial prophylaxis was mainly quinolones, fluconazole, acyclovir and trimethoprim/sulfamethoxazole.
Results: Within the first 30 days, 120 of 164 patients (73.2%) developed febrile neutropenia episodes. Infections were documented in 78 patients (47.6%). Patients undergoing allogeneic SCT tended to have more documented infections compared to recipients of autologous SCT (78.6% vs 36.9%, respectively; p < 0.001). The most frequent infection was bacteremia (41%), followed by urinary infection (19.2%), pneumonia (16.7%) and catheter-related infection (15.4%). Pathogens isolated in 66.7% of the febrile neutropenia episodes were mostly gram-positive organisms (50.6%), followed by gram-negative rods (46.9%) and Candida spp. (2.4%). Predominant pathogens were coagulase-negative staphylococcus (26.5%), E. coli (24.1%) and S. aureus (16.9%). Infections were responsible for 2.4% of deaths after transplantation within the 30 days in all patients. Early mortality associated with infection was 4.8% after allogeneic SCT and 1.6% after autologous SCT (p < 0.005).
Conclusions: Febrile episodes are the most frequent complication of both autologous and allogeneic SCT and gram-positive isolates remain the main pathogen in these patients Methicillin resistance is increasing and glycopeptides remain the only choice for treating such infections. The high incidence of febrile episodes and bacteraemia may be due to the lack of efficacy of antimicrobial prophylaxis. Although the infection rate is high, measures taken to prevent and treat infections result in very low rates of mortality from infection in SCT patients. Studies reporting local microbiological findings are necessary because they support an antibiotic choice for prophylaxis or therapy more accurately than reports from other areas.
AMPL/SU: ampicillin/sulbactam, CTAX cefotaxine, CTAZ: ceftazidime, CEPI: cefepime, AMK: amikacin, CIPX ciprofloxacin, PIPC/TZ: piperacillin/tazobactam, CPER/SU: cefoperazone/sulbactam, IMIP:imipenem.
AMPI/SU: ampicillin/sulbactam, TMP/SMX: trimethoprim/sulphamethoxazole, METC: methicillin, GEN: gentamicin, VAN: vancomycin, CLI: clindamycin, CTIN: cefalothin
P1410: Microbiologically documented infections following peripheral blood stem cell transplantation
F. Altuntas, O. Yildiz, B. Eser, E. Alp, I. Sari, M. Cetin, A. Unal (Kayseri, TR)
Objectives: To assess the isolation rate of bacterial and fungal causative agents in early and late infections in patients who underwent peripheral blood stem cell transplantation (PBSCT).
Methods: Conditioning and the pre-engraftment period were defined as the early period; the post-engraftment period until one year was defined as the late period. This study was performed to evaluate early and late infections in 114 patients who underwent PBSCT (84 autologous, 30 allogeneic) in a single institution in 1997 until 2003. All the patients received antibiotic prophylaxis (ciprofloxacin, acyclovir, fluconazole and TMP/ SMX orally) and hematopoietic growth factors during neutropenia. Febrile patients received i.v. imipenem or cefepime plus amikacin or ceftazidime plus amikacin.
Results: A total of 117 episodes with microbiologically documented infections were seen 90 of 114 patients and 79% of the patients experienced at least one febrile episode with microbiologically documented infections during their post-transplant course. Of these episodes, 69 (59%) were in the early period and 48 (41%) were in the late period. In the early period, 38.8% of causative organisms were gram positive, 51.5% were gram negative and 7.7% were fungi. The most common pathogens were Coagulase-negative Staphylococcus (CoNS) and E. coli in the early period. In the late period, 44.6% of causative organisms were gram positive, 44.6% were gram negative and 6.8% were fungi. CoNS and E. coli were also the most commonly isolated agents in this period. A total of 19 microbiologically documented catheter infections were seen, of 11 were in the early period and of 8 were in the late period. The most common pathogen was CoNS in catheter related infections. Resistance to methicillin was detected 47.4% of S. aureus and 86.5% of CoNS isolates.
Conclusions: The isolation rate was in accordance with previous reports; similar percentages of gram positive and gram negative isolates were found in patients with underwent PBSCT. A remarkably low rate of viridans group streptococci and fungi were observed. The spectrum of pathogens detected in these cases serves as the basis for recommendations on the choice of empiric antimicrobial treatment regimens. Therefore, studies reporting local microbiological findings as well as their susceptibility profiles are necessary. We suggest that local microbiologic surveillance should be known before empiric antimicrobial therapy is started in each institution.
P1411: Detection of human polyomaviruses (BKV and JCV) infection by PCR assay in patients after haematopoietic progenitor cell transplantation
K. Rybka, E. Gorczynska, D. Turkiewicz, J. Toporski, K. Kalwak, A. Dyla, A. Chybicka (Wroclaw, PL)
Objectives: Two human polyomaviruses BK (BKV) and JC (JCV) are common in population. Following the primary infection both viruses establish latency in renal tissue and in B lymphocytes, 60–90% adults are asymptomatic polyomavirus carriers. Polyomavirus–related disease is largely associated with immunological impairment. JCV is the causative agent of the progressive multifocal leukoencephalopathy in AIDS patients as well as in other immune compromised hosts. In patients undergoing renal or bone marrow transplantation the reactivation of both polyomaviruses may result in hemorrhagic cystitis (HC). Late-onset viral HC mainly related to BKV reactivation is a significant cause of post-transplant morbidity. The aim of the study was to evaluate the frequency and clinical implication of human polyomaviruses infections in children who underwent hematopoietic progenitor cell transplantation (HPCT).
Methods: Polyomavirus DNA was detected in plasma and urine with PCR-based assays. Viral DNA was extracted from plasma or urine samples by digestion with proteinase K and purification on Qiagen columns according to manufacturer's protocol. The primers pair amplified a 176-bp sequence from BKV genome and a 173-bp sequence from JCV genome. Digestion of the PCR products with the BamH1 prior to electrophoresis was used to discriminate between BKV and JCV sequences.
Results: A regular screening for polyomavirus infections was performed in 103 children who underwent HPCT. BKV was detected in urine of 39 children (37.9%) and in serum of 8 patients (7.8%). JCV was detected in urine of only 4 patients (3.9%) and in plasma of 1 patient. The most frequent clinical manifestation probably related to HPV infection was hemorrhagic cystitis, however almost 50% of HPV positive patients was asymptomatic.
Conclusion: In conclusions we demonstrated that rapid identification of viral agents may allow to initiate effective therapy and is imperative to polyomavirus infections following hematopoietic stem cell transplantation.
P1412: Monitoring of herpesvirus DNA load in children after allogeneic hematopoietic stem cell transplantation − EBV and CMV may be sufficient
P. Hubacek, O. Cinek, S. Voslarova, K. Rakova, M. Zajac, P. Keslova, P. Sedlacek, J. Stary (Prague, CZ)
Objectives: Herpesvirus reactivation can cause a life threatening disease in hematopoietic stem cell transplant (HSCT) recipients. Our work aimed at assessing the prevalence and clinical correlates to reactivation of seven herpesviruses in a single paediatric cohort.
Methods: Between January 2001 and September 2004, 88 children and adolescents (aged median 8.7 yrs, range 0.2–20.5 yrs) underwent allogeneic HSCT. We tested 2383 blood samples drawn weekly for first three months after HSCT and less frequently thereafter. Viral loads were tested using real-time quantitative PCR, and normalised to 100000 human genomic equivalents. Results of EBV, CMV and HHV6 were available within one day, while HSV, VZV and HHV7 were tested retrospectively in 1405 representative samples. The provisional positivity threshold was set to 1000 normalised copies.
Results: The positivity threshold was exceeded by EBV load in 51 (2.1%) samples from 17 (19%) children, by CMV load in 71 (3.0%) samples from 17 (19%) children, by HHV6 load in 19 (0.8%) samples from 10 (11%) children. Based on the viral load results and clinical evaluation, therapy was instituted in 3/17 EBV-positive patients (anti-CD20 antibody), 17/17 CMV positive and 1/10 HHV6 positive patient (ganciclovir, foscavir, cidofovir). Of these, only one patient died of CMV pneumonia. Among retrospectively tested samples, the threshold was exceeded by HHV7 only (11 samples from 5 children, no clinical correlate). HSV and VZV were detectable in minute quantities only.
Conclusions: PCR monitoring of viral load has proven useful for guiding preemptive therapy for EBV or CMV, as shown by only one death among 88 transplanted children. On the other hand, despite relatively high prevalence of HHV6 or HHV7, their prospective monitoring is unlikely to improve the prognosis of the patients, as the viruses seem to be relatively harmless. The absence of significant load of HSV and VZV may by associated with preventive administration of acyclovir and may therefore be specific for centres using this prophylaxis. The work is supported by grant of Ministry of Health of Czech Republic No. 7459
P1413: Quantitative PCR for EBV DNA in paediatric renal transplant patients
K. Harvey-Wood, A. Balfour, J. Beattie, C. Williams (Glasgow, UK)
Introduction: Epstein-Barr virus (EBV)-induced post-transplant lymphoproliferative disorder (PTLD) has been reported in between 1%-10% of all paediatric renal transplant recipients. PTLD is a heterogeneous group of conditions characterised by EBV-driven proliferation of B-lymphocytes in the face of impaired T-cell immune surveillance. We have performed longitudinal surveillance EBV DNA levels in renal transplant patients to attempt to correlate virological findings with changes in blood levels of EBV DNA.
Aim: To determine the usefulness of longitudinal measurement of EBV levels in paediatric patients following renal transplantation.
Methods: EBV in whole blood was quantified using real time PCR and clinical data on the patients collected.
Results: We have found no correlation between the absolute level of EBV DNA and the development of PTLD. All patients who developed PTLD have positive PCR results but a number of patients without PTLD also had raised levels of EBV DNA.
Conclusion: Longitudinal Quantitative measurement of EBV DNA allows the diagnosis of PTLD to be considered but in our population we have been unable as yet to set either and absolute cut off or rate of change of count which could be said to be diagnostic of PTLD.
P1414: Taxonomic structure and susceptibility of Candida spp. in surgical ICU of cancer hospital
I. Petukhova, Z.V. Volkova, E. Malysheva, N. Dmitrieva (Moscow, RUS)
Objective: To study pattern and isolation rate of Candida spp. in surgical ICU.
Methods: Candida spp. isolated from cancer pts in ICU after extensive and combined operations for esophageal, gastric, colorectal cancer etc., were analysed. ‘Chromagar’ and semi-automatic analyzer ‘ATB-Expression’ were used for identification.
Results: 626 bacterial and fungal strains from 817 pathologic materials were isolated, including 121 Candida spp. (19.3%). C.albicans was isolated in 58 of 121 cases (47.9%). 2 strains C.nonalbicans were isolated from blood. Wounds were infected with mixed microflora including Candida spp. in patients who have previously received 2 or 3 lines of antimicrobial therapy and all pts had C.non-albicans from wound discharge. 28 C. albicans and 16 C. non-albicans were isolated from sputum and 26 C. albicans and 27 C. non-albicans were isolated from bronchoscopic materials in pts with clinical and radiographic evidence of pneumonia. Prevalence of C.albicans in sputum may be explained as this material was taken from less severely ill patients, than bronchoscopy, which was undertaken in critically ill patients. Urine was colonized with 4 C. albicans and 13 C. non-albicans. All pts had urinary cateters and didn't have clinical evidences of urinary infection. C. non-albicans consisted of C. glabrata − 30 (24.8%), C. parapsilosis − 13 (10.8%), C. krusei − 6.0%), C.inconspicua/norvegensis − 5 (4.1%), C. tropicalis − 3 (2.5%), ñ. kefyr − 2 (1.7%), ñ. globosa, C. sake, C. lusitaniae, C. dubliniensis − 1 strain each (0.8% each). Susceptibility testing performed in 3 more often isolated strains were the follows: All tested strains of C. albicans were susceptible to amphotericin B, flucitozine, miconazole. Intermediate susceptibility was seen to ketokonazole, econazole and nystatin (each − 2%) and resistance was revealed to nystatin (2%). In C. glabrata 22% of strains had intermediate susceptibility to ketokonazole, no resistant strains were found. C. parapsilosis was resistant and had intermediate susceptibility to miconazole in 50%, to econazole − 63% and was resistant to nystatin − 10% and amphotericin B − 10%.
Conclusion: ICU patients often suffer from severe infectious complications and are treated with broad spectrum antibiotics, resulting in fungal colonization and superinfections. The cause Candida non-albicans colonization and infections may be extensive and inappropriate prophylactic use of fluconazole.
P1415: Candidemia in Brazilian cancer patients
A.C. Pasqualotto, D.D. Rosa, L.C. Severo (Porto Alegre, BR)
Objectives: To review all cases of candidemia that affected cancer patients in our medical centre over a 9-year period to assess demographic features, etiology, therapy, and outcome of the infection.
Methods: Retrospective cohort study developed in Santa Casa Complexo Hospitalar, Brazil, during 1995 and 2003. Medical charts were reviewed to record clinical and demographic characteristics presented in the period of 30 days before collection of the first blood sample positive for Candida.
Results: During the period of study, 74 patients (38.7%) with nosocomial candidemia had cancer. Solid tumors occurred in 77.0% and most of them were localized (63.8%). Species other than C. albicans caused 59.1% of episodes of candidemia in cancer patients. In comparison with other patients, candidemia in cancer patients was more frequently associated with neutropenia, mucositis, and port-a-cath. Patients without cancer had higher exposure to invasive procedures, large spectrum antibiotics and surgery. Previous steroids use, chemotherapy, and cefepime use were more common in patients with haematological neoplasia, in comparison with solid tumors. However, major surgeries were more common in patients with solid cancers (47.4% and 0.0%), mainly in gastrointestinal tract. Overall mortality was 50.3%.
Conclusions: Patients with candidemia may have different predisposing factors to acquire the infection when stratified according to baseline diseases. More studies are needed to emphasize specific risk factors for candidemia in patients with solid tumors. Following a worldwide trend, species other than Candida albicans were the main etiology of candidemia in this study. Therefore, continuous epidemiologic monitoring is necessary to follow further changes in the patterns of candidal infections.
Mechanisms of resistance to quinolones
P1416: Plasmid mediated quinolone resistance gene among ESBL-producing E. coli and K. pneumoniae in Spain
E. Valverde Romero, T. Parras Padilla, A. Herrero Hernández, J. Fernández Gorostarzu, J.A. García-Rodríguez, J.L. Muñoz Bellido (Salamanca, E)
Introduction: Fluoroquinolone resistance has been usually associated to two main mechanisms, both in Gram positive and in Gram negative bacteria: mutations in gyrase and/or topoisomerase IV (mainly in their subunits A) and efflux mechanisms. Recently, a third, plasmid-mediated mechanism has been described. The gene responsible for this resistance, qnr, has been found in clinical isolates of Klebsiella pneumoniae and Escherichia coli. A previous study has shown that qnr was present in 8% of quinolone-resistant clinical isolates of E. coli from Shanghai, China. A recent study in the USA has shown the presence of qnr in 11.1% of fluoroquinolone-resistant K. pneumoniae, in two cases associated to a SHV-7 ESBL. We have determined the presence of this gene in 125 ESBL producing E. coli and K. pneumoniae isolated in SPAIN during 2004.
Methods: We determined by PCR and sequencing the presence of the qnr gene in plasmid DNA in 125 ESBL-producing E. coli and K. pneumoniae clinical isolates. ESBLs had been previously characterized by isoelectric focusing, PCR and sequencing.
Results: No E. coli isolates harboured qnr gene. qnr was found in one K. oxytoca isolate producing SHV-12 ESBL. The qnr sequence was identical to the sequence previosuly described by other authors. Nevertheless, the isolate was intermediate to nalidixic acid (16 mg/l), and susceptible to norfloxacin and ciprofloxacin (<0.1 mg/l). No mutations were found in gyrA and parC genes.
Comments: qnr gene is still infrequent among ESBL-producing microorganisms, but can be occasionally found. No qnr harbouring, ESBL-producing enterobacteria had been previously described in Spain. In this case, qnr is not enough, in absence of topoisomerase mutations, to produce fluoroquinolone resistance, though mutant selection might be more frequent than in strains which do not harbour qnr.
P1417: Effect of the plasmid encoded quinolone resistance determinant in P. stuartii on the selection of strains with clinical resistance to ciprofloxacin
I. Wiegand, I. Luhmer-Becker, B. Wiedemann (Bonn, D)
Objectives: The influence of the plasmid encoded quinolone resistance determinant qnr in E. coli and K. pneumoniae strains with different single target gene mutations, efflux levels and porin patterns has already been studied. None of the strains was clinically resistant to ciprofloxacin. P. stuartii strains show a slightly higher ciprofloxacin MIC in their natural sensitive population than the studied E. coli and K. pneumoniae strains. As we had detected qnr in three different clinical P. stuartii isolates we wanted to determine the influence of qnr in this species on the ability to select ciprofloxacin resistant mutants.
Methods: Qnr was transferred to P. stuartii ATCC29914 RifR using the filter mating technique with the qnr-positive strain P. vulgaris Pv123 as donor. A qnr-positive transconjugand (TCPs123) was selected and used for further studies. MIC values were determined by E-test according to the manufacturers instructions. Mutants were selected using inocula of more than 1 × 1010 cells plated on LB agarplates containing ciprofloxacin concentrations of 2×–32× the MIC.
Results: Transconjugands were obtained with a conjugation frequency of 1.2 × 10−5. MIC values are presented in table 1. The mutant prevention concentration was determined to be 16x the MIC for both strains. Some mutants that were picked showed normal colony sizes but also minor subpopulations of small colony variants (SCVs). These were shown to be stable on antibiotic free medium for at least three passages. MIC values of four or more representatives of each colony type per strain were uniform within a small range, the median value is given in table 1.
Table 1.
MIC value of parent strains and ciprofloxacin selected mutants
| Strain | P. stuartii ATCC 29914 RifR | Mutants of P. stuartii ATCC 29914 RifR normal colony size | Mutants of P. stuartii ATCC 29914 RifR SCV | TCPs 123 | Mutants of TCPs 123 normal colony size | Mutants of TCPs 123 SCV |
|---|---|---|---|---|---|---|
| Ciprofloxacin MIC [mg/L] | 0.03 | 0.25 | 3 | 0.5 | 6 | >32 |
Conclusions: For P. stuartii the combination of qnr with a single mutational event can lead to clinical resistance. Thus it is likely that with a single quinolone treatment such single step mutants can be selected. Species other than E. coli and Klebsiella spp. may be an important reservoir for the qnr resistance determinant. Their role in the clinical setting remains to be determined.
P1418: Evidence of fluoroquinolone-resistant mechanism associated with efflux pump and outer membrane protein(OMP) in Neisseria gonorrhoeae
J. Yoo, B. Kim, C. Yoo, W. Seong (Seoul, KOR)
Objectives: Ciprofloxacin resistant N.gonorrhoeae in Korea have been dramatically increased 1% in 1999 to 87% in 2003. Although it was known to be mainly caused by mutation within gyrA and parC, the fact which another mechanisms involved in the resistance to fluoroquinolones was well known in other organisms. This study was designed to investigate the evidence of another mechanisms related to resistance to ciprofloxacin in N.gonorrhoeae.
Methods: Mutants derived from 2 parent strains which isolated in Korea were obtained after several in vitro selection on GC agar supplemented with increasing concentrations of ciprofloxacin. Amino acid substitutions within the quinolone resistance-determining region (QRDR) of GyrA, ParC were determined. SDS-PAGE and 2D gel electrophresis of outer membrane proteins (OMP) were performed.
Results: A significant reduced sensitivity to ciprofloxacin was observed in the selected mutants. MICs of ciprofloxacin for 76mu10 and 92mu13 strains were increased seven-, eleven-fold higher than those of wild type strains respectively. Mutants derived from 76/WT showed the resistance to PEN, TE and CRO either. However, It was observed that mutants from 92/ WT had reduced sensitivity to AZI, TE and increased sensitivity to SPT. On addition of CCCP, a proton motive force uncoupler, MIC of ciprofloxacin for 92mu13 strain was most greatly reduced by 8- fold. The sequence analysis of the gyrA has demonstrated a single mutation at 91(S91Y) or 95(D95N) except 1 strain but no mutations in parC for all mutant. 76mu10 strain only showed both point mutation at 91,95 in gyrA. SDS-PAGE analysis of the OMPs of 76mu10 strain revealed remarkable alteration in the expression of proteins which located in the range from 29 to 50 kDa. And also in 2D analysis, it showed over thirty spots which is changed in amount five-fold. Interestingly, OMPs of 92mu13 strain showed little changes comparing to wild type.
Table 1.
Susceptibilities of gonococcal strains to various antimicrobial agents and amino acid changes in GyrA and MtrR promotor
| Strain | MIC (μg/ml) |
Mutations in |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Pen | AZI | TE | SPT | CRO | OFL | CIP | CIP+cccp70 | gyrA | mtrR promoter | |
| 76/WT | 4 | 0.25 | 2 | 16 | 0.03 | 0.5 | 0.12 | 0.03 | 91Tyr | A del |
| 76mu6 | 16 | 0.25 | 8 | 16 | 0.12 | 8 | 4 | 1 | 91Tyr | A del |
| 76mu10 | 16 | 0.25 | 8 | 8 | 0.25 | 8 | 16 | 4 | 91Tyr, 95Asn | A del |
| 92/WT | 0.25 | 0.12 | 0.12 | 16 | 0.008 | 0.06 | 0.004 | ND | – | – |
| 92mu6 | 0.5 | 1 | 2 | 8 | 0.015 | 4 | 2 | 0.03 | 95Asn | – |
| 92mu13 | 0.5 | 1 | 2 | 4 | 0.008 | 4 | 8 | 0.03 | 95Asn | – |
Abbreviation; PEN: Penicillin, AZI: Azithromycin, TE: Tetracycline, SPT: Spectinomycin, CRO: Ceftriaxone, OFL: Ofloxacin, CIP: Ciprofloxacin, CCCP: Carbonyl cyanide tri-chlorophenyl hydrazone
Conclusion: We induced the two kinds of mutants in which probably had different fluoroquinolone resistant mechanisms. Although they had one mutation in gyrA, it was strongly suggested that the main cause of resistance to ciprofloxacin was reduction of OMPs or increase of efflux pump by OMPs analysis and genetic analysis.
P1419: Streptococcus agalactiae highly resistant to quinolones
M. Rebollo, E. Miró, A. Ribera, M. Alvarez, F. Navarro, B. Mirelis, P. Coll (Barcelona, E)
Objectives: To characterise the mechanism of resistance to fluoroquinolones in two out of 162 strains of Streptococcus agalactiae isolated between January 2003 and November 2004.
Methods: Susceptibility studies were performed using a microdilution method (Sensititre R) following NCCLS guidelines, and E-test following the manufacturer's procedures. A clonal relationship between the two strains was ruled out by pulsed-field gel electrophoresis. Mutational alterations in the QRDR of gyrA and parC were investigated by PCR and sequencing using the dideoxy method with the Thermo SequenaseTM CyTM5 Dye Terminator Sequencing kit and the Automatic Laser Fluorescent DNA Sequencer.
Results: The first ciprofloxacin-resistant S. agalactiae strain (P0162) was recovered in October 2003; the second (P0425) was isolated in September 2004. Both strains were isolated from urological patients previously treated several times with ciprofloxacin. The two ciprofloxacin-resistant S. agalactiae isolates were also resistant to tetracycline and susceptible to penicillin, vancomycin, erythromycin, clindamycin, quinupristin-dalfopristin, chloramphenicol and rifampicin. MICs values of quinolones for P0162 and P0425 strains were: ciprofloxacin >32 mg/L, norfloxacin >256 mg/L, levofloxacin 8 mg/L and >32 mg/L, sparfloxacin 4 mg/L and >32 mg/L, moxifloxacin 0.5 mg/L and 2 mg/L, and clinafloxacin 0.25 mg/L and 0.75 mg/L, respectively. Both quinolone-resistant S. agalactiae isolates showed the same mutation in parC (Ser79Phe), as well as an additional mutation in gyrA. The P0162 strain showed Glu85Ala whereas P0425 strain presented Glu85Lys.
Conclusion: This is the first report of S. agalactiae quinolone resistant strains in Spain, whose mechanism of resistance was mutations in parC and gyrA. Both strains were isolated from patients treated with ciprofloxacin, and no clonal relationship was observed among them.
P1420: Novel gyrB and parE mutations detected in the quinolone-resistance-determining-region of clinical quinolone-resistant Pseudomonas aeruginosa
S. Ferreira, T.R. Walsh, S. Mendo (Aveiro, P; Bristol, UK)
Objectives: The aim of the present study was to characterize mutations occurring in quinolone-resistance-determining-region (QRDR) region of the gyrA, gyrB, parC and parE genes of the 35 clinical ciprofloxacin and pefloxacin resistant Pseudomonas aeruginosa isolated from patients in a Hospital from central Portugal.
Methods: Nucleotide sequences of the PCR-amplified gyrA, gyrB, parC and parE fragments were determined using an automated DNA sequencer. The Biological Sequence Alignment Editor, BioEdit version 7.0.0 was used for DNA and amino acid sequence alignments. Sequences obtained were compared to others deposited in the EMBL Genebank.
Results: DNA sequences were analysed for mutations leading to amino acid changes associated with fluoroquinolone resistance. Nine strains harboured a mutation (Thr → Ile) in the codon 83 of gyrA. Some silent mutations were also found. Four of the P. aeruginosa strains harboured a gyrB mutation that occurred in codon 464 leading to an amino acid substitution (Ser → Tyr). Two strains showed a mutation in codon 465 (Gly → Arg), also leading to an amino acid substitution. However compared to the sequence of the gyrB fragment obtained from P. aeruginosa PAO1, some silent mutations were found. In parC, four strains exhibited amino acid substitution (Ser→Leu) in codon 87. A new mutation occurred in codon 35 leading to an amino acid substitution (Asp → Glu) in two of the strains. Silent mutations were also found in the parC QRDR region. In the nucleotide sequence of the par E gene a few amino acid replacements were found. Those changes occurred in codons 431 (Leu → Val), 483 (Glu → Gln), 487 (Ala → Pro), 530 (Ala → Pro), 538 (Gly → Val) and 544 (Gln → His). Two of these replacements are novel (codon 483, Glu→Gln and codon 487, Ala → Pro) and both occurred in the same strain. This replacement occurred inside the highly conserved motif EGDSA. Furthermore, silent mutations were also found.
Conclusion: GyrA mutants containing a Thr-83-Ile substitution showed higher levels of resistance to fluoroquinolones than mutants containing a different single point mutation. Novel mutations were detected in gyrB and parE genes. Also, the results obtained for parC and par E genes revealed that mutations in type II topoisomerase subunit B are not as rare as they used to be.
P1421: Single mutations in parC gene among levofloxacin susceptible clinical isolates of Streptococcus pneumoniae
E. Mateo, R. Alonso, R. Cisterna (Vitoria, Bilbao, E)
Objectives: Levofloxacin (LVX) is recommended for the treatment of community-acquired pneumonia because increasing multidrug resistance in Streptococcus pneumoniae. The incidence of levofloxacin resistance in clinical isolates of S. pneumoniae is relatively low. LVX resistance requires at least 2 mutations in the quinolone resistance determining region (QRDR) of topoisomerase IV and DNA gyrase. The purpose of this study was to determine the prevalence of single QRDR mutations in parC gene of S. pneumoniae.
Methods: Sixty-nine levofloxacin-susceptible pneumococci (MICs 1–2 mg/L) were isolated at the Microbiology Service of Hospital de Basuto, Bilbao (Spain) during 2004. Eight isolates showed a LVX MIC of 2 mg/L and 61 isolates showed a LVX MIC of 1 mg/L. We used a PCR-RFLP assay to screen 27 randomly chosen pneumococcal isolates with levofloxacin MIC of 1 mg/L and 8 isolates with a MIC of 2 mg/L for mutations known to confer resistance (parC: S79, D83; gyrA: S81, E85). The QRDR region of parC of isolates with suspected mutations was amplified by PCR and its DNA sequence determined.
Results: Of the 27 strains with LVX MICs of 1 mg/L, no strains had a S79 or D83 ParC change. Among 8 strains with LVX MICs of 2 mg/L, 5 strains (62.5%) had a S79 or D83 ParC change. Four isolates showed single parC mutation (S79F, D83G, D83V). Only one isolate showed double parC mutation (S79F + K137N). No mutations in gyrA gene were detected in any strains.
Conclusions: Fluoroquinolone resistance among S. pneumoniae remains low, however, an increase in isolates containing a parC mutation has been observed. The selection of parC mutations could be related to the use of levofloxacin to treat S. pneumoniae infections.
P1422: Mutations in parC and gyrA genes among levofloxacin resistant clinical isolates of Streptococcus pneumoniae
R. Alonso, E. Mateo, F. Calvo, R. Cisterna (Vitoria, Galdácano, Bilbao, E)
Objectives: The incidence of levofloxacin resistance in clinical isolates of S. pneumoniae is relatively low. LVX resistance requires at least 2 mutations in the quinolone resistance determining region (QRDR) of topoisomerase IV and DNA gyrase. The purpose of this study was to characterize among recent clinical isolates of S. pneumoniae the mutations that conferred resistance to levofloxacin.
Methods: Twenty-four levofloxacin-resistant pneumococci (MICs > 4 mg/L) isolated from respiratory samples during 2004 were analysed. Minimal inhibitory concentrations (CMIs) to levofloxacin, gatifloxacin, moxifloxacin, and gemifloxacin were determined by agar dilution method using Mueller–Hinton agar with 5% horse blood. The QRDR regions of parC and gyrA were amplified by PCR and their DNA sequence determined.
Results: Levofloxacin resistance was associated with combinations of at least two amino acid substitutions, most commonly involving ParC Ser79Phe (23/25), and GyrA Ser81Tyr (20/25). Of the 24 strains, three strains showed single parC mutation (S79F) and a double gyrA mutation (S81Y + E85G). One strain showed a K137N substitution in ParC and no mutations were found in the QRDR of gyrA. This strain may be harbour mutations in gyrB gene. None of the 24 strains was susceptible to gatifloxacin; twelve and twenty isolates were susceptible to moxifloxacin (CMI < 4 mg/L) and gemifloxacin (CMI < 1 mg/ L), respectively.
Conclusions: The results suggest that resistance to levofloxacin require at least two substitutions of amino acids within the QRDR region of gyrase and topoisomerase IV, and that there is considerable cross-resistance among fluoroquinolones associated with these changes.
P1423: Mechanisms of quinolone resistance in P. aeruginosa strains with efflux pump overexpression
B. Henrichfreise, I. Wiegand, B. Wiedemann (Bonn, D)
Objectives: The aim of our study was to determine whether additional mechanisms of quinolone resistance (target modification via mutation or protection) can be found in P. aeruginosa (PA) strains with effluxpump overexpression (EPO).
Methods: 4 levofloxacin-resistant strains of PA were collected in a German hospital in 2004. EPO was verified by testing Levofloxacin (LEV) MICs using broth microdilution both with and without the effluxpump inhibitor (EPI) MC-270,110. The QRDR of gyrA and parC, furthermore mexR and the intergenic region between mexR and mexA (ir) containing the shared operator-promotor region of mexA-mexB-oprM and mexR were amplified and sequenced. Also, a screening for qnr was conducted. Clonal identity was investigated by multiple-locus variable number of tandem repeat analysis (MLVA).
Results: The phenotypic and genotypic characterisation of all strains is shown in Table 1. In 3 strains (1–3) whose LEV MIC was reduced by EPI to the wildtype (wt) level of 0.125 mg/L no aminoacid changes in GyrA and ParC were found. In strain 4 two changes in GyrA and one change in ParC were detected. All strains showed mutations in mexR but no mutations in ir. The exchange of V126 to E in strains 2 and 3 was previously described in LEV-susceptible isolates. Qnr was not detected. MLVA indicated no clonal relationship among all strains.
Table 1.
| Strain | 1 | 2 | 3 | 4 |
|---|---|---|---|---|
| Lev MIC | 16 | 8 | 8 | 128 |
| Lev MIC + EPI | 0.125 | 0.125 | 0.125 | 2 |
| gyrA/GyrA | wt | wt | Wt | T83to1 D87toG |
| parC/ParC | wt | wt | silent mutation codon 80 | S80 to L |
| maxR/MaxR | c insertion after g75 | V126toE | V126toE | N insertion after L52 |
Conclusions: For strain 1 and 4 nalB mutations were observed. The EPO phenotype in strains 2 and 3 could be caused by nalC or by overexpression of other EPs. Contrary to usual findings in our strains no correlation between EPO and target mutation was found.
P1424: Analysis of sequential isolates of Pseudomonas aeruginosa from cystic fibrosis patients, repeatedly treated with ciprofloxacin by mutant prevention concentration, minimal inhibitory concentration and pulsed field gel electrophoresis
J. Blondeau, L. Blondeau, C. Williams (Saskatoon, CAN; Glasgow, UK)
Objective: MPC measures the propensity of an antimicrobial (AM) compound to select for AM resistance based on drug concentrations required to block growth of first-step resistant mutants. Cystic fibrosis patients (CFP) are frequently infected with Pseudomonas aeruginosa (PA) and require repeat courses of AM therapy. As AM therapy may precipitate AM resistance, we tested sequential PA isolates from CFP, repeatedly treated with Cpx, by MIC and MPC.
Methods: Sequential isolates were collected over a 3-yr period. PA isolates were tested to Cpx and levofloxacin (Lfx) by microbroth dilution in accordance with NCCLS guidelines. For MPC testing, 10 billion organisms were applied to agar plates containing drug and incubated 24–48 hr. The lowest concentration preventing growth was the MPC. PA strains were compared by pulsed field gel electrophoresis (PFGE) using Spe I.
Results: Patient 1 (P1) (10 isolates) received 6 courses of Cpx (500–750 mg bid) and patient 2 (P2) 5 courses over the period collected (3 yrs). MICs for Cpx and Lfx for P1 and P2 ranged 0.031–1 μg/ml and 0.25-2 μg/ml respectively; MPCs ranged 1–4 μg/ml (79% ≤ 2 μg/ml) and 2–16 μg/ml (64% ≥ 8 μg/ml) respectively. PFGE profile for P1 isolates were identical while P2 had 3 different strains. Cpx therapy did result in increased MPC values. Lfx MPC values were higher than Cpx in every instance.
Conclusion: This represents first report of MPC testing on sequential PA isolates where AM history was available. MPCs remained constant to Cpx over the duration of organism isolations. MPCs to Lfx were high and beyond achievable and sustainable drug concentrations. As P1, P2 have never received Lfx, yet MPC values were so high, suggests Lfx will more readily select for quinolone resistant PA. MPC can be used to monitor changes in susceptibility and as such guide appropriate selection of AM therapy.
P1425: Detection of a quinolone resistance-mediating gyrA mutation in a fluoroquinolone susceptible Salmonella live vaccine strain
A. Preisler, P. Heisig (Hamburg, D)
Objectives: The production of effective vaccines instead of developing new antibiotics is a promising approach to circumvent rapid development of bacterial resistance. Beside an attenuated Salmonella typhi live vaccine strain used in human, S. typhimurium strain TAD Salmonella vacT is being used as live vaccine strain in food-producing chicken. Attenuation of virulence has been achieved by random mutagenesis of the wildtype S. typhimurium DT009 strain M415 followed by selection for reduced virulence. Epidemiological markers of vaccine strain include reduced susceptibilities to rifampicin (rifR) and nalidixic acid (nalR) and the loss of tensid tolerance, presumably associated with altered membrane permeability (marker rtt). Curiously, the vaccine strain retains a high susceptibility to fluoroquinolones and macrolides. The present study aimed at understanding the molecular basis for this phenotype.
Methods: TAD Salmonella vacT was characterized by determination of antibiotic susceptibility pattern, generation time, DNA-supercoiling degree, and DNA-sequence of the entire gyrA genes as well as the quinolone resistance-determining regions (QRDRs) of the remaining fluoroquinolone target genes gyrB, parC, and parE -encoding the subunits B of gyrase or A and B of topoisomerase IV, respectively.
Results: Compared to its parent, TAD Salmonella vacT showed a reduced growth rate, but no significant changes in the DNA supercoiling. DNA sequence analysis of the complete gyrA gene revealed three novel mutations affecting codons 59 (trp–arg), 75 (gly–ala), and 867 (ser–ile) and one known point mutation at position 87 (asp–gly). Although the mutation at codon 87 is known to mediate quinolone resistance, the vaccine strain is highly susceptible to fluoroquinolones like sparfloxacin, and ciprofloxacin and only moderately resistant to nalidixic acid.
Conclusions: Two hypotheses might -alone or in combination-explain this phenotype (1) At least one of the novel gyrA mutations compensates for the fluoroquinolone resistance mutation (asp-87-gly) and (2) an increase in drug accumulation, resulting from the inactivation of a multi-drug efflux pump (presumably associated with the rtt marker) allows for a higher accumulation of fluoroquinolones -and macrolides- in the cell. Our results point to a combination of both mechanisms and therefore provide a molecular basis to understand the reported stability of the live vaccine in vitro and in the field.
P1426: Identification of plasmid-mediated quinolone resistance in enterobacterial isolates in Turkey
P. Nordmann, H. Nazik, B. Ongen, H. Mammeri, L. Poirel (Le Kremlin Bicetre, F; Istanbul, TR)
Objectives: The plasmid-mediated quinolone-resistance determinant QnrA has been identified recently from enterobacterial isolates in USA, China, Thailand, Korea, The Netherlands and France. These strains were resistant to nalidixic-acid and most of them produced expanded-spectrum beta-lactamases. Our objective was to evaluate the prevalence of the qnr gene in nalidixic-acid resistant enterobacterial isolates recovered at the University Hospital of Istanbul, Turkey, since it is located just between mainland Europe and Asia.
Methods: PCR with primers specific for the qnrA-like gene was used for screening. Primers specific for the different ESBL genes were used to identify the corresponding beta-lactamase genes. Mating-out assays were attempted to demonstrate the transferability of the Qnr determinant. Combinations of primers were used to identify the qnr-surrounding sequences. Eighty-eight nalidixic-acid resistant enterobacterial strains isolated in 2004 were studied. Among them, 51 were ESBL producers.
Results: The qnrA gene was identified in two ESBL-positive isolates (1% of the ESBL (+) strains), an Enterobacter cloacae and a Citrobacter freundii isolate. The E. cloacae isolate expressed an SHV-5-like ESBL that was encoded on a plasmid that co-transferred the nalidixic-acid resistance in addition to resistance to kanamycin, tobramycin, chloramphenicol, trimethoprim, and sulfonamides. The C. freundii isolate possessed also a conjugative plasmid that harboured the qnrA gene associated with the blaVEB-1 gene and that conferred also resistance to streptomycin, tobramycin, chloramphenicol, rifampin, and sulfonamides. In both cases, the qnrA gene was located downstream of the Orf513 recombinase gene, as previously identified in other qnrA-positive isolates.
Conclusion: This study emphasizes that the qnrA gene conferring resistance to nalidixic acid is present in distantly-located European countries. Interestingly, the qnrA gene was found in association with blaVEB-1 gene, that corresponds to the first identification of this latter ESBL in Turkey.
P1427: Norfloxacin as an alternative marker of decreased susceptibility to fluorquinolones in Salmonella enterica
L. López, A. Gisaeus, A. Pascual (Seville, E; Malmö, S)
Objective: Treatment failure with fluorquinolones has been a problem in cases when the salmonella strains show resistance to Nalidixic acid (NAL). The Ciprofloxacin (CIP) MIC value increase if the salmonella strain is NAL resistant so several authors have proposed a change of the Ciprofloxacin susceptibility breakpoint from ≤1 to ≤ 0.125 mg/L for Salmonella enterica. The VITEK system is extensively used as automated susceptibility system but NAL testing is not included and the lowest CIP dilution in the VITEK cards is 0.25 mg/L. The aim of this study is to trace Nalidixic acid resistance in Salmonella enterica isolates with the VITEK system, using the Norfloxacin MIC value as a marker of nalidixic acid resistance.
Material and methods: Forty-two clinical Salmonella enterica isolates were collected during July–August 2004. The susceptibility tests were achieved by: 1) using AST-N020 cards and the VITEK system (bioMérieux) according to manufacturer, 2) testing NAL, CIP, Norfloxacin (NOR) and Ofloxacin (OFX) with the microdilution broth method according to NCCLS, and 3) testing discs with NAL 30 microg (Oxoid), CIP 5 microg (Oxoid) and NOR 10 microg (Oxoid) with the diffusion agar method according to NCCLS.
Results: Thirty-seven (88%) Salmonella enterica isolates belonged to serogroup D, 4 (10%) to serogroup B and 1 (2%) to serogroup C. With the Vitek system five of the strains tested had a NOR MIC <0.5 mg/L and 37 showed an increased MIC value for NOR (2 mg/L), but although within the NCCLS limits for susceptibility. The five strains with Norfloxacin VITEK MIC<0.5 mg/L were NAL susceptible (average 25, 8 mm and ≤0.03 mg/L) and all the isolates with Norfloxacin VITEK MIC 2 mg/L were NAL resistant (average 6 mm and ≥256 mg/L). For Ciprofloxacin the lowest VITEK MIC value ≤0.25 mg/L is represented for both the Nalidixic acid resistance and susceptible strains. The 37 NAL-R strains showed higher NCCLS MIC values, ranging from 0.125–0.5 mg/L for Ciprofloxacin, 0.54 mg/L for Norfloxacin and 0.25–1 mg/L for Ofloxacin, than the NAL-S strains (MIC ≤0.06 mg/L for Ciprofloxacin, Norfloxacin and Ofloxacin).
Conclusion: The results of this study show that Norfloxacin MIC >0.5 mg/L should be considered as a marker of Nalidixic acid resistant Salmonella enterica strains when using VITEK 2 system for susceptibility testing.
P1428: Factors affecting resistance to quinolones mediated by plasmids containing the qnrA gene
J.M. Rodriguez-Martinez, C. Velasco, A. Pascual, I. Garcia, L. Martinez-Martinez (Seville, Santander, E)
Objectives: The effect of copy number and transcriptional level of qnrA on plasmid-mediated quinolone resistance, and the effect of quinolones in inducing qnrA expression were analysed in four clinical strains of Klebsiella pneumoniae (UAB1, N5, 1960 and 1132 strains) and in Escherichia coli transconjugants derived from UAB1, N5 and 1960. No transconjugants containing qnrA from strain 1132 are available.
Methods: Southern-RFLP analysis was also performed to investigate basic structural details of plasmids containing qnrA. Copy number of qnrA was determined by dot blot hybridization. Transcriptional studies were carried out using total RNA isolated from transconjugants grown in the absence or in the presence of ciprofloxacin or moxifloxacin at 0.2, 0.4, 0.8 or 1 × MIC.
Results: qnrA is located in plasmids with similar mobility and identical Southern- RFLP (in all four plasmids, the qnrA probe hybridized with one EcoRI fragment of 4.1 Kb). MICs of quinolones against transconjugants from the same donor were identical, but MICs against transconjugants from different parental strains varied within a 4-fold range. qnrA copy numbers in strains UAB1 and 1132 were 8 times higher than in strains 1160, and 2.7 times higher than in strain N5. No differences in qnrA copy number in the E. coli transconjugants were observed. The amount of qnrA transcripts was at least 5-fold greater in the transconjugant from UAB1 than in the transconjugants from strain N5 or 1960 without quinolone induction. Differences in transcription of qnrA in different transconjugants were also noted at both basal levels and after induction by quinolones. A dose-response study of the inducing effect of ciprofloxacin on qnrA transcription was performed with transconjugants from strains UAB1, N5 and 1960, with a maximum effect at 0.4 XMIC. After induction with ciprofloxacin at 0.4 × MIC, transcription of qnrA increased 4.3, 5.1 and 2.6 times over baseline in transconjugants from strains UAB1, N5, and 1960, respectively. Induction of qnrA by ciprofloxacin was 1.04, 2.2 and 2.03 times higher than that caused by moxifloxacin against transconjugants from UAB1, N5 and 1960, respectively.
Conclusion: Regulation of qnrA expression may be similar in all four strains. There are differences in qnrA copy number in strains containing this gene. Transcriptional analysis revealed that both ciprofloxacin and moxifloxacin are able to induce qnrA gene, with ciprofloxacin being a better inducer than moxifloxacin.
P1429: The use of norfloxacin 10 mcg as a screening disc to predict fluoroquinolone resistance in Streptococcus pneumoniae
C. Borén, R.W. Smyth, W. Hryniewicz, G. Kahlmeter, E. Sadowy (Växjö, S; Warsaw, PL)
Objectives: The use of sensitive screen discs for the detection of antibiotic class resistance is becoming increasingly important. A nalidixic acid disc (30 mcg) has been successfully used for the detection of fluoroquinolone (FQ) resistance in Enterobacteriaceae, Haemophilus spp and Neisseria spp. Cefpodoxime has been used for the detection of ESBL and most recently cefoxitin for the detection of MRSA. We compared the sensitivity of norfloxacin (10 mcg) and ciprofloxacin (1 and 5 mcg) discs for the detection of fluoroquinolone resistance in a collection of FQ sensitive and resistant Streptococcus pneumoniae.
Methods: A collection of S. pneumoniae strains with (from Poland) and without (from Sweden) fluoroquinolone resistance mechanisms were tested with antibiotic discs, norfloxacin 10 mcg and ciprofloxacin 1 and 5 mcg, from Oxoid UK. Mueller–Hinton Agar (Oxoid, UK) with 5% defibrinated sheep blood and IsoSensitest Agar (Oxoid, UK) with 5% defibrinated horse blood and NAD were used. Mueller–Hinton agar was investigated with confluent and semi-confluent inoculum while ISA was tested with a semi-confluent inoculum. Ciprofloxacin MIC-values (E-test, AB Biodisk, Sweden) were determined on ISA. All plates were incubated at 35–37 C in 5% carbon dioxide for 18–24 h before measuring the inhibition zones. Strains exhibiting ciprofloxacin MIC-values of 0.125–2.0 mg/L were considered part of the wild type MIC distribution as defined by the EUCAST epidemiological cut-off value (www.eucast.org).
Results: Strains with MIC ≥ 32 mg/L were reliably detected with all methods and discs. Most difficult to reliably separate from the wild type distribution were strains with MIC 4.0 mg/L. Irrespective of agar and inoculum the norfloxacin 10 mcg disc more easily distinguished strains with MIC 4 mg/L from strains of ≤2 mg/L. Ciprofloxacin 1 mcg was slightly inferior to the norfloxacin disc but much more reliable than ciprofloxacin 5 mcg.
Conclusion: Using methods based on NCCLS and BSAC/ SRGA methodologies, norfloxacin 10 mcg and ciprofloxacin 1 mcg were preferable to ciprofloxacin 5 mcg in the detection of fluoroquinolone resistance in S. pneumoniae. We suggest that fluoroquinolone resistance surveillance is performed with one of these discs and that ciprofloxacin MIC-determination is performed on strains identified by the screen test.
Antibacterial susceptibility studies – II
P1430: Comparative in vitro activity of several antimicrobial agents against different Nocardia species
Y. Glupczynski, C. Berhin, M. Janssens, G. Wauters (Yvoir, Brussels, B)
Background: Treatment of Nocardia spp. continues to be challenging and there have been only few surveys on the susceptibility of the different Nocardia species to antibiotics. We therefore studied the in vitro activity of 11 antimicrobials against 81 clinical isolates of Nocardia spp.
Methods: Most isolates were isolated from clinical specimens in different Belgian centres between 1990 and 2004. Identification of all isolates was confirmed to the species level by conventional phenotypical tests and by 16S rDNA sequencing. Species identified were: N. farcinica (37), N. nova (16), N. cyriacigeorgica (12), N. brasiliensis (6), N. abscessus (5), N. paucivorans (2), N. carnea, N. niigatensis and N. asiatica (1 each). The known origins of the isolates were: respiratory samples (34 isolates), wound/soft tissue infections (13 isolates), blood (7 isolates), brain abscesses (4 isolates), CSF (1 isolate). Antimicrobial susceptibility testing was performed by E-test on Mueller Hinton agar. MIC values were read after 48 and 72 h incubation. 18 additional reference strains (DSM and NCTC collections) belonging to 12 different Nocardia spp. were also included in the testing.
Results: The MIC values (mg/L) and range of activities of selected antibiotics tested against Nocardia spp. are summarized in the Table.N. farcinica and N. brasiliensis showed the highest MIC values to the different classes of antibiotics tested while N. nova and N. abscessus appeared as the most susceptible species on the average. Overall, we found a rather close correlation between phenotypic susceptibility/resistance profiles and species identification.
| Antimicrobials | MIC range (mg/L) | MIC50 | MIC90 |
|---|---|---|---|
| Amoxicillin | 0.016->256 | 12 | 32 |
| Amoxi/Clav. | 0.016->256 | 2 | 8 |
| Ceftriaxone | 0.032->256 | 8 | >256 |
| Imipenem | 0.002->32 | 0.5 | 2 |
| Ciprofloxacin | 0.032->32 | 4 | >32 |
| Clarithromycin | <0.016->256 | 8 | 96 |
| Tobramycin | <0.016-24 | 4 | 12 |
| Amikacin | 0.016-2 | 0.38 | 1 |
| Minocycline | 0.016-4 | 1.5 | 3 |
| Linezolid | 0.047-3 | 1 | 2 |
| Cotrimoxazole | 0.002-0.75 | 0.064 | 0.38 |
Conclusion: On the whole, cotrimoxazole, amikacin, imepenem, minocycline and linezolid displayed the highest intrinsic activity. The excellent activity of linezolid in particular shows promises for the treatment of infections due to Nocardia sp. owing to its good oral availability.
P1431: Antibiotic susceptibility profiles of glucopeptide intermediate Staphylococcus species isolated from patients with bacteraemia
D. Mylona-Petropoulou, G. Ganderis, E. Yfantis, M. Damala, H. Malamou-Lada (Athens, GR)
Objectives : To investigate the frequency of glycopeptide intermediate Staphylococcus species (GISS) and their susceptibility to other antibiotics.
Methods: BetweenJanuary2001 toNovember2004theantibiotic susceptibility pattern of coagulase − negative Staphylococci (CNS) with reduced susceptibility to glycopeptides, isolated from patients with bacteraemia, hospitalized in the General Hospital of Athens ‘G. Gennimatas', was analysed. The isolates were recognized by the VITEK II automated system (bio Merieux, France), by the brain heart infusion agar screening plate with 6 mg/L vancomycin and confirmed by the E test method (AB Biodisk, Sweden) according to the manufacturer's recommendations.
Results: A total of 4270 strains of Staphylococci were isolated from consecutive blood cultures during this period: Three hundred two (7,07%) S. aureus and 1614 (37,8%) CNS. Seventy three (4,5%) strains (64 S. epidermidis, 7 S. haemolyticus and 2 S. hominis) demonstrated reduced susceptibility to glycopeptides. Four strains were intermediate sensitive (MIC 8–12 mg/L) and one strain was resistant (MIC: 32 mg/L) to vancomycin. Fourty seven strains were intermediate sensitive (MIC: 16 mg/L) and 26 strains were resistant (MIC: 32–256 mg/L) to teicoplanin. It is worthwhile to notice that all the strains with reduced susceptibility to vancomycin were resistant to teicoplanin. Six out of 73 strains (8,2%) were sensitive to oxacillin. The resistance rate (%) of 73 GISS strains to other antibiotics was as follows: gentamicin (87,7), tobramycin (91,8), rifampin (32,8) trimethoprime / sulfamethoxazole (38,3), ofloxacin (84,9), while all GISS strains demonstrated sensitivity to quinupristin / dalfopristin and linezolid.
Conclusion: Glycopeptide resistance in Staphylococci is becoming more common in our hospital. Laboratories should use proper diagnostic techniques to detect such strains, because these bacteria may play an important role in therapeutic failure of serious Staphylococcal infections.
P1432: Methicillin-resistant coagulase-negative staphylococci with increased resistance to glycopeptides
A. Bisiklis, F. Tsapara, S. Alexiou-Daniel (Thessaloniki, GR)
Objectives: The purpose of our study was to inquire if there were Vancomycin resistant subpopulations among coagulase-negative staphylococci, which indicate intermediate or homogeneous resistance to Teicoplanin.
Methods: We have studied four Staphylococci epidermidis and a Staphylococcus haemolyticus. These strains were isolated during second semester in 2003, from two blood cultures, two venous catheters cultures and a sample culture from operative injury. These cultures were performed in common nutritive substrates. The indentification and determination of antibiotic sensitivity was performed by VITEK 2 (BioMérieux). The basic features of strains were the homogeneous resistance to Oxacillin, the homogeneous or intermediate resistance to Teicoplanin and their sensitivity to Vancomycin. Oxacillin resistance and Vancomycin sensitivity were confirmed by the classical method of E-test in Müller Hinton agar, with inoculation turbidity 0.5 of MacFarland scale and for 24 hours incubation in 37°C. The determination of subpopulations with increased Glycopeptide-resistance was performed by macromethod of E-test in Brain Heart Infusion agar, with inoculation turbidity 2.0 of MacFarland scale and for 48 hours incubation in 35°C.
Results: Three Staphylococci epidermidis indicated Vancomycin MIC 2 μg/ml and one 4 μg/ml with the classical method of E-test. With the macromethod of E-test, the first three strains indicated MIC 8 μg/ml, 8 μg/ml, 12 μg/ml and the fourth strain 12 μg/ml. Staphylococcus haemolyticus indicated Vancomycin MIC 3 μg/ml and 32 μg/ml with the classical method and macromethod, respectively. Teicoplanin MIC ranged from 24 μg/ml to over 256 μg/ml for Staphylococcus haemolyticus.
Conclusions: a) We have observed that, all strains, which were Vancomycin-sensitive consistently with Vitek and classical method of E-test, revealed subpopulations with increased Vancomycin-resistance.b)The appearance of high Teicoplanin MIC for all strains and high Vancomycin MIC for S. haemolyticus consinstently with macromethod of E-test was remarkable. c) We suggest that, when Vancomycin is used as a treatment choice we should seriously consider Oxacillin and Teicoplanin resistance as determined by routine methods. The presence of subpopulation with heterogeneity resistance is possible to cause treatment failure.
P1433: In vitro evaluation of effective antibiotic lock therapy for treatment of catheter-related infections caused by Staphylococcus species
J.Y. Lee, K.S. Ko, W.S. Oh, N.Y. Lee, J.-H. Song, K.R. Peck (Seoul, KOR)
Objective: Antibiotic lock therapy (ALT) is recently recommended for conserving catheters in treatment of central venous catheter (CVC)-related infections. We performed this study to investigate the adequate antibiotics, the concentration of antibiotics, and treatment duration in ALT.
Methods : We evaluated the bacterial killing activity of vancomycin, teicoplanin, ciprofloxacin, rifampin, cefazolin, gentamicin, nafcillin, and erythromycin against biofilms of S. aureus (SA204 and SA195) and S. epidermidis (ATCC35983 and ATCC35984). The effectiveness of the antibiotic locks was assayed after exposure to antibiotics (1, 5, and 10 mg/ml) for 1, 3, 5, 7, 10, or 14 days by using in vitro model of biofilms on polyurethane (PU) film. Biofilm bacteria were quantified by the determination of viable counts.
Results: Significant biofilm killings were not achieved with cefazolin, nafcillin, gentamicin, and erythromycin at any of the time intervals examined.
| Musc on of CFU/sheet (mg/ml) |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lock period (day) | Control |
VAN |
TEC |
CIP |
HIF |
||||||||
| 1 | 5 | 10 | 1 | 5 | 10 | 1 | 5 | 10 | 1 | 5 | 10 | ||
| 0 | 6.6 × 104 | ||||||||||||
| 1 | 1.5 × 104 | 2.2 × 104 | 1.1 × 104 | ND | 9.1 × 101 | 1.1 × 106 | 2.7 × 101 | 4.1 × 104 | 4.8 × 101 | ND | 1.7 × 101 | 0 | 0 |
| 3 | 2.9 × 104 | 1.0 × 104 | 9.7 × 104 | ND | 5.6 × 101 | 4.9 × 104 | 3.1 × 101 | 1.4 × 101 | 0ND | 1.3 × 106 | 0 | 0 | |
| 5 | 3.6 × 104 | 1.8 × 104 | 0 | ND | 1.2 × 101 | 1.6 × 101 | 8.0 × 101 | 8.2 × 101 | 0 | ND | 0 | 0 | 0 |
| 7 | 1.4 × 1028 | 5.7 × 101 | 0 | ND | 3.0 × 101 | 0 | 0 | 1.4 × 101 | 0 | ND | 0 | 0 | |
| 10 | 1.1 × 1028 | 6.2 × 101 | 0 | ND | 0 | 0 | 0 | 0 | 0 | ND | 0 | 0 | 0 |
| 14 | 2.4 × 1028 | 3.0 × 101 | 0 | 0 | 0 | 0 | 0 | ND | 0 | 0 | 0 | ||
Conclusion: These data suggest that ALTs for less than 7 days with vancomycin, teicoplanin, ciprofloxacin, and rifampin (5 mg/ml of each) can be effectively used in treating catheter-related infections caused by S. epidermidis and S. aureus. It warrants prospective clinical trials for the evaluation of clinical efficacy.
P1434: Synergism between ultrasound and ceftazidime in the phagocytosis of Pseudomonas aeruginosa
N. Kashef, Q. Behzadian Nejad, M. Sattari, M. Mokhtari (Tehran, IR)
Objectives: Pseudomonas aeruginosa is an opportunistic pathogen with innate resistance to many antibiotics, predominantly infecting patients with defects in antibacterial defenses. Ultrasound is currently used in medical practice for diagnostic and therapeutic purposes. A recent application of ultrasound is in drug delivery. There are many reports in the literature suggesting that ultrasound activates, potentializes, or makes more effective some pharmacological agents. In this study, we investigated the effect of ultrasound and sMICs of ceftazidime on phagocytosis of Pseudomonas aeruginosa (ATCC 27853) separately and in combination.
Methods: The susceptibility of bacteria to murine macrophage killing was examined following exposure to ceftazidime and ultrasound (at a frequency of 1 MHz and power output of 0.25 W). Bacteria were added to macrophage-containing polypropylene tubes. After incubation, centrifugation, and washing of macrophages to remove adherent but unphagocytized organisms, cells were lysed by distilled water. Each sample was plated and after incubation, the number of viable colonies was counted.
Results: In vitro, pretreatment of bacteria with sMICs of antibiotic and ultrasound separately resulted in an enhancement of macrophage phagocytosis and killing of the organisms (p < 0.0001). But there was a notable enhancement effect manifested by increased nonopsonic killing following pretreatment of bacteria with those two independent variables in combination (p < 0.0001).
Conclusion: These results showed that simultaneous application of ultrasound and ceftazidime has some efficacy in inactivating Pseudomonas aeruginosa, and improves phagocyte activation. The physical mechanism of inactivation by ultrasound alone appears to be transient cavitation, but the mechanism by which ultrasound enhanced antibiotic action may be due to perturbation of the cell membrane or to stress responses by the bacteria. The mechanism of synergistic effect of ultrasound and ceftazidime is unknown and many more attempts should be made.
P1435: Occurrence of resistance to wide spectrum of antimicrobial agents in clinical isolates of the genus Acinetobacter
E. Sodomova, D. Michalkova-Papajova, M. Vrabelova-Poczova, D. Rovna, M. Kettner (Bratislava, SVK)
Objectives: The aim of the study was to determine the occurrence of resistance to 33 antimicrobial agents in Acinetobacter clinical isolates.
Methods: Clinical isolates included in the study originated from Hospital Ruzinov, Bratislava (Slovakia). The majority of 37 isolates tested were obtained from Burn Department (35.1 %) and from Surgical Department (29.7 %) of this hospital and were isolated from burn infections (29.7 %) and wounds (27.0 %). The isolates were identified by NEFERMtest24 (Pliva-Lachema, Czech Republic) and selected as resistant to clinically used beta-lactam antibiotics. Resistance to antimicrobial agents was determined by standard disk diffusion method according to the NCCLS recommendations. The following antimicrobials were tested: mezlocillin, ticarcillin, piperacillin, carbenicillin, ampicillin-sulbactam, piperacillin-tazobactam, ticarcillin-clavulanic acid, ceftazidime, cefepime, cefoperazone, cefotaxime, ceftriaxone, ceftizoxime, moxalactam, imipenem, meropenem, aztreonam, gentamicin, amikacin, tobramycin, netilmicin, tetracycline, doxycycline, minocycline, ciprofloxacin, levofloxacin, lomefloxacin, norfloxacin, ofloxacin, gatifloxacin, chloramphenicol, trimethoprim-sulfamethoxazole, sulfonamides.
Results: In the set of 37 clinical isolates 100.0 % were resistant to mezlocillin, 86.5 % to ticarcillin, 100.0 % to piperacillin, 100.0 % to carbenicillin, 10.8 % to ampicillin-sulbactam, 43.2 % to piperacillin-tazobactam, 32.4 % to ticarcillin-clavulanic acid, 56.8 % to ceftazidime, 21.6 % to cefepime, 100.0 % cefoperazone, 75.7 % to cefotaxime, 75.7 % to ceftriaxone, 86.5 % to ceftizoxime, 75.7 % to moxalactam, 0.0 % to imipenem, 0.0 % to meropenem, 59.5 % to aztreonam, 100.0 % to gentamicin, 62.1 % to amikacin, 10.8 % to tobramycin, 5.4 % to netilmicin, 100.0 % to tetracycline, 54.1 % to doxycycline, 45.9 % to minocycline, 100.0 % to ciprofloxacin, 78.4 % to levofloxacin, 100.0 % to lomefloxacin, 100.0 % to norfloxacin, 43.2 % to ofloxacin, 83.8 % to gatifloxacin, 100.0 % to chloramphenicol, 59.5 % to trimethoprim-sulfamethoxazole and 86.5 % to sulfonamides.
Conclusions: More than 75 % of the clinical isolates were resistant to 20 or more from 33 antimicrobial agents tested. Carbapenems (imipenem and meropenem) were the only antimicrobial agents effective to all clinical isolates.
P1436: Time-kill studies of linezolid, piperacillin-tazobactam, imipenem, amikacin, trimethoprim-sulfametoxazole and moxifloxacin alone or in combination against Nocardia spp.
M.F. Tripodi, R. Fortunato, G. Ruggiero, S. Cuccurullo, R. Utili (Naples, I)
Objectives: Nocardia may cause severe infections in immunocompromised hosts. Treatment has been largely based on sulfonamide agents. The aim of the study was to evaluate the in-vitro bactericidal activity of several antibiotic combinations against Nocardia spp.
Methods: Seven Nocardia strains (N. asteroides, 3; N. brasiliensis, 2; N. farcinica 1, N. spp. 1) were isolated from pulmonary lesions in heart-transplant patients. In vitro activity of antibiotic combinations was evaluated by time-kill method on Mueller Hinton broth II (MHBII) using the following antibiotics alone or in combination with each other at the following peak-serum concentrations (mg/l): LZD, 20; piperacillin-tazobactam (TZP), 200; imipenem (IMP), 43; trimethoprim-sulphametoxazole (SXT), 3.4/17; amikacin (AN), 38; moxifloxacin (MFX), 4.3. Time-kill studies were performed in MHBII using an inoculum of 1 × 106 cfu/ml, incubated at 37°C for 72 h. Viability counts were performed at 0,24, and 72 h, by plating 10 fold dilutions onto blood agar plates (BioMèrieux, France). The bactericidal activity was defined as a decrease major or equal to 3 log10 cfu/ml in the viable count at 24, 72 h compared with the initial inoculum. Synergism or antagonism were defined as a decrease or increase major or equal to 2 log10 cfu/ml in the viable count with the combination at 24, 72 h compared with the most active agent alone.
Results: Only AN, IMP, MFX, as single drug, showed a bactericidal activity in 7, 4 and 2 strains respectively, all other agents were bacteriostatic. For antibiotic combinations, high rate of bactericidal activity was observed with: IMP + AN and IMP + MFX in all seven strains; TZP + MFX, TZP + AN and MFX + AN in 6 strains; AN + SXT in 5. Among the bactericidal combinations, synergism was observed for TZP with MFX or SXT in 2 and 1 strain respectively. For the other combinations, synergism was not evaluable due to high bactericidal activity of AN, IMP and MFX. Linezolid behaved always as bacteriostatic agent and was bactericidal only in one case with AN and MFX, respectively, but was antagonistic with AN and IMP in 7 and 2 strains, respectively. All the other antibiotic combinations were bacteriostatic.
Conclusion: To date there is no standardized treatment for nocardosis. Based on these data the combinations of IMP and AN or MFX appears suitable as initial treatment of invasive nocardiosis. LZD, MFX and SXT may represent a choice for sequential oral therapy as single drugs.
P1437: Intracellular activity of ampicillin, azithromycin, telithromycin, ciprofloxacin and moxifloxacin against non-typeable Haemophilus influenzae
C. Kratzer, W. Graninger, A. Buxbaum, A. Georgopoulos (Vienna, A)
Objectives: Non-typeable Haemophilus influenzae (NTHi), a well-known, major respiratory tract pathogen, has recently been shown to be able to invade and persist inside human epithelial cells. In this study we analysed the intracellular in vitro activity of ampicillin, azithromycin, telithromycin, ciprofloxacin and moxifloxacin against NTHi.
Methods: Confluent normal human bronchial epithelial (NHBE) cells were loaded with 100 bacteria per epithelial cell for 2 h. Extracellular H. influenzae were killed by gentamicin, and the intracellular bacteria were incubated with fresh invasion medium, supplemented with selected antimicrobial agents at concentrations of 1 and 10 mg/l for further 4 and 8 h. As final step the invasion medium was removed and the number of intracellular viable bacteria was determined after lysis of the epithelial cells.
Results: Moxifloxacin showed the highest bactericidal efficacy against intracellular NTHi, followed by ciprofloxacin, azithromycin and telithromycin. During the first 8 h, ampicillin was not able to influence the intracellular survival of H. influenzae in comparison to a control without antibiotic.
Conclusions: When compared with the other tested antibiotics, moxifloxacin was most effective in killing intracellular NTHi. Therefore, moxifloxacin, which combines high extracellular and intracellular activity, can be seen as an important tool for the treatment of RTIs.
P1438: Antibiotic susceptibility of Fusobacterium necrophorum strains causing serious infections
R. Hannula, L. Bevanger (Trondheim, N)
Objectives: F. necrophorum causes abscesses, mainly in the oropharynx, and Lemierre's syndrome (postanginal sepsis), a rare life-threatening disease in young, previously healthy adults. Long-term antibiotic treatment is often needed. We studied the susceptibility pattern for F. necrophorum in 13 available clinical isolates in our hospital from January 1991 until October 2004.
Methods: Four blood-stream isolates and nine other clinical isolates, mainly from abscesses, were available for testing. The bacteria were cultured on fastidious anaerobic agar (FAA) and were identified by their colony morphology, microscopic morphology, chartreuse fluorescence, lipase positivity and biochemically by the rapid ID 32 A (bioMérieux). Bacteria suspended in Brain Heart broth of 1 McFarland turbidity were swabbed on PDM agar with 5% defibrinated horse blood. E-test strips (BIODISK) were applied on the dry agar surface. The plates were incubated anaerobically for 48 hours before reading the results. The strains were tested for penicillinase activity with nitrocephin.
Results: Three strains were resistant for penicillin but did not produce penicillinase. MIC values for ciprofloxacin were generally higher than for other antibiotics. The colony size on these agar plates was increased compared to the colonies on the other plates, suggesting that the growth was stimulated by ciprofloxacin. E-test results for imipenem showed a high MIC due to hazy colonies in the inhibition zone.
| Antibiotics | MIC50 (mg/l) | MIC90 (mg/l) | MIC range (mg/l) |
|---|---|---|---|
| Penicillin G | 0.016 | >32 | 0.004–256 |
| Metronidazole | 0.125 | 0.5 | 0.016–2 |
| Clindamycin | 0.032 | 0.064 | 0.016–0.125 |
| Imipenem | 1.5 | >32 | 0.004–32 |
| Ciprofloxacin | 2 | 6 | 0.75–32 |
| Cefotaxime | 0.016 | 0.064 | 0.016–256 |
| Doxycyclin | 0.064 | 0.19 | 0.016–0.75 |
Conclusions: E-test results indicate resistance to penicillin and imipenem in some F. necrophorum strains. Clindamycin and metronidazole seem to be good therapy alternatives. MIC values for ciprofloxacin were high and ciprofloxacin is not a choice for therapy. Doxycyclin and cefotaxime might be used as second line therapy. MIC testing should always be performed in serious anaerobic infections.
P1439: In vitro activities of various antibiotics, alone and in combination with colistin methanesulfonate against Pseudomonas aeruginosa strains isolated from cystic fibrosis patients
C. Bozkurt, A. Gerceker (Istanbul, TR)
The in vitro activities of various antibiotics, either alone or in combination with colistin methanesulfonate were assessed using Pseudomonas aeruginosa strains isolated from cystic fibrosis patients. According to minimum inhibitory concentration (MIC) values, 100%, 98%, 96% and 84% of the isolates were found susceptible to amikacin, colistin methanesulfonate, meropenem and ceftazidime, respectively. The minimum bactericidal concentrations were generally equal to or twice greater than those of the MICs. The in vitro activities of antibiotics in combination were determined by microbroth chequerboard technique and results were interpreted by fractional inhibitory concentration (FIC) index. With a FIC index of <0.5 as borderline, synergistic interactions were more frequent in combinations where amikacin was involved than those with colistin methanesulfonate. No antagonism was observed.
P1440: A comparative in vitro analysis of amoxicillin-clavulanic acid and seven comparative agents in 15,521 paediatric respiratory isolates
S. Bouchillon, J. Johnson, D. Hoban, D. Butler, S. Min, L. Miller, D. Payne (Schaumburg, Collegeville, USA)
Background: Several large scale adult surveillance studies have demonstrated the ongoing effectiveness of amoxicillin-clavulanic acid (A/C) even after 20 years of clinical use. This is a retrospective collection of data comprising over 15,000 paediatric respiratory isolates collected from several surveillance studies conducted in 54 countries over a period of 5 years. The in vitro activity of A/C and 7 comparators are reported for S. pneumoniae (SPN), H. influenzae (HFLU), M. catarrhalis (MCAT) and S. pyogenes (SPY). Not all antimicrobials were tested in all studies, or against all isolates.
Methods: Regional reference laboratories used broth microdilution technique according to NCCLS guidelines.
Results: A/C at the Augmentin ES-600 PK/PD breakpoint of 4 mcg/mL for SPN, was the most active agent tested with 96.5% susceptible including 83.5% and 88.6% susceptible for PRSP and ERSP, respectively.
| Drug/MIC90/%Sus | H.influenzae | S. pneumoniae | PRSP | ERSP |
|---|---|---|---|---|
| Amox/Clav | 2/99.2 | 2/92.3a 96.5b | 8/67.1a/83.5b | 8/81.5a/88.6b |
| Amp | >16/74.6 | 4/0.0 | 8/0.0 | 8/ns |
| Pen | >16/na | 2/60.6 | 4/0.0 | 4/18.6 |
| Cefuroxime | 2/96.7 | 8/65.2 | 8/0.7 | 8/26.0 |
| Azithro | 2/99.1 | >32/71.4 | >32/23.0 | >32/0.3 |
| Erythro | 8/na | >32/71.2 | >32/21.5 | >32/0.0 |
| TMP/SMX | >4/76.5 | 8/50.2 | >8/13.8 | 8/24.5 |
NCCLS breakpoint of 2/1 mcg/mL applied for conventional Augmentin.
PK breakpoint of 4/2 mcg/mL applied for Augmentin ES-600.
Conclusions: A/C activity was comparable to or better than comparator agents against all the key respiratory pathogens including resistant strains involved in paediatric respiratory infections.
P1441: Activity of telithromycin and oral comparators against 20,886 Canadian respiratory tract pathogens isolated from 1997–2003
D.J. Hoban, L.P. Palatnick, B. Weshnoweski, A. Wierzbowski, K. Nichol, G.G. Zhanel (Winnipeg, CAN)
Objectives: Community-acquired respiratory tract infections (RTIs) caused by Streptococcus pneumoniae (SPN), Haemophilus influenzae (HI), Moraxella catarrhalis (MC), and Streptococcus pyogenes (SPY) are commonly treated with empiric antibiotic therapy. Surveillance has documented increasing prevalence of betalactam-, erythromycin-, trimethoprim-sulfamethoxazole-, and fluoroquinolone-resistant RTI pathogens. In vitro, telithromycin, a ketolide antibacterial, has been shown to be active against RTI pathogens. This study reviews the activity of telithromycin against RTI pathogens collected in Canada over the past 6 years.
Methods: As part of an ongoing national RTI surveillance study, the MICs for telithromycin and comparative oral agents to over 20.000 clinically significant RTI pathogens collected across Canada from 1997–2002 were determined according to NCCLS guidelines (2003).
Results: MIC90 for penicillin, levofloxacin, azithromycin, clarithromycin, and telithromycin to SPN (8378), HI (7489), MC (2497) and SPY (2522) were, respectively: SPN: 1, 1, 1, 0.5, and 0.008 ig/ml; HI: 8, 0.12, 2,16, and 4 ig/ml; MC: 8, 0.12, 0.25, 0.5, and 0.12 ig/ml; and SPY: 0.03, 1, 0.5, 0.12, and 0.008 ig/ml.
Conclusion: In vitro, telithromycin was active against RTI pathogens isolated in Canada since 1997, including multidrug resistant isolates.
P1442: In vitro susceptibility of methicillin-resistant Staphylococcus aureus to daptomycin, vancomycin, and other antibiotics
Z. Samra, O. Ofir, H. Shmuely (Petach -Tiqua, IL)
Background: Methicillin-resistant Staphylococcus aureus (MRSA) is becoming increasingly prevalent as both a nosocomial and a community-acquired pathogen. Vancomycin is the traditional drug of choice, but decreasing susceptibility to vancomycin and other glycopeptides has been recently reported. Daptomycin, a lipopeptide antibiotic now in phase III clinical trials, is rapidly bactericidal in vitro against a range of gram-positive organisms, including MRSA.
Aim: To test the in vitro activity of daptomycin and other antibiotic agents on clinical isolates of MRSA.
Material and methods: Susceptibility of 200 MRSA isolates recovered from 200 hospitalized patients were test:thirty-two isolates from blood, 168 from wound and other body fluids. Activity of vancomycin, daptomycin, fusidic acid, trimethoprim sulfamethoxazole, tetracycline, rifampicin, chloramphenicol, gentamicin, clindamycin, erythromycin and ofloxacin was tested by disk diffusion method on Mueller-Hinton agar according to NCCLS criteria. For daptomycin a breakpoint of > /=18 mm was used to define susceptibility. S. aureus ATCC 25923 was used for quality control.
Results: All isolates were sensitive to vancomycin and daptomycin. Sensitivity to other antibiotics was: fusidic acid −97.9%, trimethoprim sulfamethoxazole −96.9%, tetracycline −91.7%, rifampicin −86.6%, chloramphenicol −46.9%, gentamicin −37.6%, clindamycin −22.2%, erythromycin −20.6% and ofloxacin −73.1%.
Conclusion: These results indicate that the activity of daptomycin against clinical MRSA isolates is equal to vancomycin. Daptomycin is given once daily and may be considered as an alternative treatment for patients with vancomycin resistance strains or those who can not tolerate vancomycin. Among the other antibiotics fusidic acid and trimethoprim sulfamethoxazole showed the best activity.
P1443: In vitro activities of gatifloxacin, moxifloxacin and linezolid versus bloodstream isolates of methicillin-sensitive and methicillin-resistant Staphylococcus aureus at a Canadian tertiary-care centre
T. Karnauchow, M. Binns, W. Wobeser, K. Suh, G. Evans (Kingston, CAN)
Objectives: Staphylococcus aureus bacteraemia is a significant burden to patients and institutions in terms of morbidity and financial cost. Treatment of S. aureus bacteremias generally requires 2-4 weeks of IV antibiotics. Newer oral agents with anti-staphylococcal activity may offer alternative approaches to treatment and enhance patient quality of life by shortening the duration of IV therapy. As a first step toward investigating this possibility, we evaluated the in vitro activities of gatifloxacin, moxifloxacin and linezolid versus bloodstream isolates of methicillin-sensitive (MSSA) and methicillin-resistant S. aureus (MRSA) recovered from patients at Kingston General Hospital (KGH).
Methods: MICs for gatifloxacin, linezolid, moxifloxacin, ciprofloxacin, rifampin, cloxacillin, cefazolin, and vancomycin were determined against 200 randomly selected non-duplicate MSSA (05/01–05/03) and 50 consecutive non-duplicate MRSA (05/00–06/03) blood isolates. Agar dilution and MIC interpretations were performed in accordance with NCCLS guidelines (M7-A4; M100-S14).
Results:
| MSSA MIC (n = 200) |
MRSA MIC (n = 50) |
|||||
|---|---|---|---|---|---|---|
| Range (mg/mL) | MIC50 | MIC90 | Range (mg/mL) | MIC50 | MIC90 | |
| Cefazolin | 0.25–1.0 | ≤0.5 | ≤1 | 8.0–>256 | ≤256 | >256 |
| Ciprofloxacin | 0.25–>64 | ≤0.5 | ≤0.5 | 32–>64 | >64 | >64 |
| Cloxacilin | 0.125–16 | ≤0.25 | ≤0.25 | 8.0–>128 | >128 | >128 |
| Gatifloxacin | 0.0625–4.0 | ≤0.0625 | ≤0.125 | 4 | ≤4 | ≤4 |
| Linezolid | 2 | ≤2 | ≤2 | 2 | ≤2 | ≤2 |
| Moxifloxacin | 0.025–2.0 | ≤0.025 | ≤0.0625 | 4 | ≤4 | ≤4 |
| Rifampin | 0.0075–0.0175 | ≤0.0075 | ≤0.0375 | 0.0075–>128 | ≤32 | ≤32 |
| Vancomycin | 0.5–1.0 | ≤0.5 | ≤1 | 1 | ≤1 | ≤1 |
Conclusions: Bloodstream isolates of MSSA from KGH demonstrate in vitro susceptibility to gatifloxacin, linezolid, and moxifloxacin*. MRSA isolates exhibit intermediate susceptibility to gatifloxacin and moxifloxacin* but are fully susceptible to linezolid. Linezolid may be an effective oral agent for treatment of MSSA and MRSA bacteraemia, but use of the newer fluoroquinolones may be limited to treatment of MSSA bacteraemia. Further studies are needed to clarify the role that these agents may play in the treatment of S. aureus bacteraemia.*(low MICs for S. aureus; no moxifloxacin interpretive criteria exist).
P1444: Daptomycin tested against 915 bloodstream isolates of viridans group streptococci (eight species) and Streptococcus bovis
J. Streit, J. Steenbergen, G. Thorne, J. Alder, R. Jones (North Liberty, Lexington, USA)
Background: To evaluate the activity of daptomycin tested against numerous species of viridans group streptococci (VgS) and Streptococcus bovis which are pathogens associated with wound infections, sepsis, cellulitis, endocarditis, abscesses and dental caries. The incidence of pencillin-resistant (R) or MLSB-R strains among VgS often varies by species, and large collections of strains may be required to assess an antimicrobial true clinical susceptibility (S).
Methods: The activity of daptomycin was compared to seven other antimicrobial classes using reference broth microdilution (NCCLS, M7-A6) and disk diffusion (M2-A8) methods tested against 915 streptococci (815 VgS strains, 66 to 107 per species; 100 S. bovis). Mueller-Hinton broth was supplemented with 2–5% LHB and up to 50 mg/L Ca ++.
Results: Among VgS and S. bovis, 99.9% of isolates were daptomycin-S (breakpoint at ≤ 1 mg/L; MIC90, 0.06 − 1 mg/L). In contrast, penicillin (65.5–98.1% S), macrolides (48.6 – 88.7%) and tetracycline (35.0 – 93.9%) activity varied widely between species. Erythromycin was least active, in contrast linezolid (99.1 – 100.0% S), vancomycin (100.0%) and quinupristin/dalfopristin (99.0 – 100.0%) were equally active as daptomycin, but less potent. Intermethod categorical agreement between daptomycin and linezolid (comparison agent) disk and microdilution tests was very high, each showing near complete S (99.9%).
Conclusions: Daptomycin was demonstrated to be an active agent that has clinically usable potency against these nine streptococcal species. The highest recorded daptomycin MIC was only 2 mg/L (1 strain; 0.1%). These results show that daptomycin would be an excellent candidate for further clinical trials targeting serious systemic infections caused by VgS/S. bovis.
P1445: Study of species affiliation and susceptibility to antibiotics of micro-organisms, isolated from urine of patients with benign prostatic hyperplasia
M. Sredkova, V. Grigorova, S. Stratev, K. Dragoev, O. Mihaylov (Pleven, BG)
Objectives: To determine the species affiliation and susceptibility to antibiotics of microorganisms, isolated from urine of patients with benign prostatic hyperplasia (BHP), treated in the University Hospital, Pleven, Bulgaria.
Methods: Total 186 samples of morning urine, cultured with calibrated loops according to routine semi-quantitative method from 109 patients with BHP were tested during a 1-year period (2003). Ninety-five strains were isolated totally (30 before and 65 after operative intervention) from 44 patients. The isolated microorganisms were identified by conventional routine methods and the automated miniAPI System (bioMerieux). E. coli strains were screened for ESBLs producing by the double disc synergy method. Susceptibility to antimicrobials was tested by a disk diffusion method according to NCCLS recommendations.
Results: The most frequently isolated microorganisms were: E. coli (38.9%), P. aeruginosa (21.1%) and E. faecalis (14.7). All strains E. coli and P. aeruginosa were susceptible to carbapemens, and enterococci − to vancomycin. The isolated strains of enteroccoci post-operatively had a high level of resistance to gentamicin, ciprofloxacin and tetracycline (66.7, 66.7 and 77.8%, respectively). The distribution of ESBLs products among E. coli strains, isolated after operative intervention was 86.2%. E. coli strains were highly resistant to ampicillin, amoxicillin/clavulanat (93.1 and 93.1%, respectively); to cephalosporins, II and III generation (86.2 and 86.2% respectively); to gentamicin and ciprofloxacin (82.7 and 82.7%, respectively). P. aeruginosa strains were highly resistant to azlocillin and piperacillin (73.3 and 73.3%, respectively), gentamicin and ciprofloxacin (86.7 and 93.3%, respectively).
Conclusions: E. coli are the most frequent agents of urinary tract infections (UTI) in patients having BPH (38.9%), followed by P. aeruginosa (21.1%) and E. faecalis (14.7%). All strains E. coli and P. aeruginosa were susceptible to carbapemens, andenterococci -- to vancomycin, which determines the pointed antimicrobial agents as strategic in the acceptable choice of medicines for UTI in patients having benign prostatic hyperplasia.
P1446: Clinical Pseudomonas spp.: potential factors of pathogenicity and resistance to antimicrobials
A. Hostacka, I. Ciznar, L. Slobodnikova, D. Kotulova (Bratislava, SVK)
Objectives: Resistance to 17 antimicrobials, surface hydrophobicity, motility, biofilm, production of N-acylhomoserine lactone signal molecules (C4-HSL, 3-oxo-C12-HSL) and response to oxidative stress were analysed in 50 clinical Pseudomonas spp. strains (94% P. aeruginosa). The data represent a part of the project focused to analysis of association between resistance to antimicrobials and pathogenicity in bacteria.
Methods: MICs were estimated by modifications of the standard colorimetric method. Potential factors of pathogenicity were evaluated in vitro using test for hydrophobicity (adherence to xylene), motility (0.35% agar), production of biofilm (microtiter plate assay), HSLs (biosensors: C. violaceum 026 and A. tumefaciens NTL4) and response to oxidative stress evoked by hydrogen peroxide.
Results: The strains demonstrated the greatest level of resistance in addition to natural resistance to cefotaxime (90%). Isolates in the range of 42–56% were resistant to aminoglycosides and ciprofloxacin, of 26–38% to cephalosporins. On the other hand, 98% of the strains remained susceptible to meropenem, 92% to piperacillin/tazobactam and 84% to piperacillin. The majority of the strains (74%) manifested hydrophilic character. Higher zones of motility showed 12 isolates (in average 54.8 mm) as compared to the other ones (31.1 mm). One third of the strains (32%) produced higher amount of biofilm quantified by measuring of absorbance of solubilized crystal violet (0.2–0.46) than the rest of isolates (0–0.19). All, but two strains produced 3-oxo-C12-HSL and in 46% of samples were detected C4-HSL. Only four isolates with higher biofilm production showed both types of HSLs. Majority of the strains (68%) manifested higher resistance to hydrogen peroxide as compared to the rest of strains. The group of strains resistant to aminoglycosides and ciprofloxacin revealed significantly higher number of hydrophobic strains as compared to sensitive ones. Such association was not found among the rest of the tested parameters.
Conclusion: The data obtained in this study indicated that the resistance to antimicrobials in Pseudomonas spp. isolates was not always associated with the changes in the production of the pathogenicity factors.
P1447: Study of synergetic action of dioxidine and isoniazid on the structure of tubercle bacillus in vitro
L.F. Stebaeva, L.Y. Krylova, R.G. Glushkov, A.O. Turaev (Moscow, RUS)
Objectives: While isoniazid (I) actively effects upon mycobacteria and dioxidine (D) effects upon gram-negative and gram-positive bacterium, we have studied its complicated effect on tubercle bacillus in vitro.
Methods: Effect of the compounds were studied by morphometric and electromicroscopic methods. For our experiment we used the following component: 100 D + 50 I and 100 D and 250 I. To be compared, we used I in dose of 7, 8, 15, 6 or 31.2 mcg/ml and in a dose of 15.6–31.2 mcg/ml.
Results: D and I result in significant changes in a structure of mycobacteria. All changes clearly depend on a dose. However, as a rule, we can find perservered bacteria even if compounds are highly dose in culture. In our experiment, where D and I are in conjunction, cells of mycobacteria are dramatically changed. Upon minimal concentration of compounds, bacteria are heavily vacuolisated, their basic mass is destroyed, the cells can not be stained. If the dose is increased up to 31.2 mcg/ml bacteria are determined by contour shadows. If D and I are used in ratio 1:2.5 we can not find living bacteria in the culture.
Conclusion: Our study shows that the conjunction of dioxidine and izoniazid is effective on mycobacteria. Changes in mycobacteria are more crucial compared to the changes in case when only izoniazid is used. Addition of dioxidine allows us to decrease a dose of izoniazid, with the same effect to diminish the possibility to obtain resistant strains of bacteria.
P1448: Tazobactam, more than a beta-lactamase inhibitor against the Bacteroides fragilis group
K.E. Aldridge (New Orleans, USA)
Objectives: Tazobactam(Tz), a penicillanic acid sulphone derivative, is a potent beta-lactamase inhibitor of beta-lactamase enzymes produced by a variety of aerobic and facultatively anaerobic bacteria when combined with piperacillin(Pip). This synergistic action is also demonstrated by Pip-Tz against anaerobes but little data are available detailing the activity of Tz alone. This study establishes the inherent in vitro activity of Tz against B. fragilis group pathogens.
Methods: A total of 449 clinical isolates of the B. fragilis group were tested against Tz, Pip, Pip-Tz, cefoxitin(Fox), and cefotaxime(Tax) using an NCCLS-recommended broth microdilution method. All isolates were beta-lactamase positive by the nitrocephin assay. MIC values were determined at 48 hr as the lowest concentration of each antimicrobial that inhibited the visible growth of each isolate.
Results: Tz alone showed remarkable inherent activity against the test isolates overall. Using inhibitory concentrations of = / < 4.8, and 16 mg/L for comparison Tz inhibited 86%, 97%, and 98% of B. fragilis isolates, compared to 25%, 50%, and 100% of B. distasonis; 42%,74%, and 100% of B. thetaiotaomicron; 23%, 56%, and 86% of B. ovatus, and 50%, 83%, and 100% of B. vulgatus at the same concentrations. At 16 mg/L Tz was more active than Pip(44% to 82%), Fox(36% to 75%) and Tax (36% to 75%) for the same species using their established NCCLS breakpoints. Pip-Tz inhibited >99% of the test isolates at = / < 32 mg/L.
Conclusions: Tz alone exhibits potent inherent anti-B. fragilis group activity and was more active than Pip, Fox and Tax. These data are particularly significant therapeutically in polymicrobial infections since for the combination Pip-Tz the Pip is excreted more rapidly than Tz and Tz becomes the predominant component in serum after 8 to 10 hr which could enhance or extend the anti-Bacteroides coverage in the presence of low concentrations of Pip. Pharmacologic studies have shown that, although dose dependent Tz concentrations reach a % time above MIC of >50% against the majority of B. fragilis group isolates with susceptible MIC values.
P1449: Bactercidal activity of daptomycin, tigecycline, teicoplanin and vancomycin against community-associated and multidrug-resistant methicillin-resistant Staphylococcus aureus
B. Tsuji, M. Rybak, C. Cheung, M. Amjad (Detroit, USA)
Background: The epidemiology of S. aureus is rapidly changing. MRSA has traditionally been confined to institutional settings and the occurrence of multi-drug resistance is a cause for concern. However, even more concerning is the increasing prevalence of MRSA in the community. Therapeutic options for these infections are limited. DAP is a cyclic lipopeptide recently released in the USA for clinical use in the treatment of serious Gram-positive infections. TIG (GAR-936) is a parenteral investigational glycylcyline derivative of minocycline with a wide spectrum of activity including MRSA. We examined the inhibitory and bactericidal activity of DAP, TIG, TEI, and VAN against CA-MRSA and MDR-MRSA.
Methods: One-hundred clinical stains of CA-MRSA and Fifty-Five MDR-MRSA (Resistant to at least four antimicrobials including oxacillin) defined by CDC definition and SCCmec type were evaluated. ATCC 29213 was used as a control. Molecular methods were used to determine SCCmec type and the presence of PVL and mecA gene. Minimum inhibitory concentrations and minimum bactericidal concentrations were performed per NCCLS. Bactericidal activity was evaluated using time kill experiments using two randomly selected strains from each group.
Results: MIC50 (Range) of 100 CA-MRSA (SCCmec IV) for DAP: 0.25(0.12–1.0), TIG: 0.12(0.06–1.0), TEI: 0.5(0.25–4.0) and VAN: 2.0(0.25–2). For CA-MRSA the MBC ranged for DAP:0.12–1.0, TIG:0.12–16.0, TEI:0.25–4.0, VAN:0.25–4.0. MIC50 (Range) of 55 MDR-MRSA (Sccmec II) for DAP: 0.50(0.25–2.0), TIG: 0.25(0.06–1.0), TEI: 1.0(0.25–4.0) and VAN: 2.0(0.5–4.0). For MDR-MRSA the MBC ranged for DAP:0.25–2.0, TIG:0.25–16, TEI:0.5–4.0, VAN:1.0–4.0. Overall TIG and DAP exhibited low MIC against all 155 strains. Time kill experiments demonstrated that DAP = VAN > TIG = TEI (p < 0.05) against CA-MRSA. FOR MDR-MRSA DAP > VAN > TEI > TIG. DAP achieved 99.9% kill as early as 4 h against all strains.
| CA-MRSA (n = 100) |
MDR-MRSA(n = 55) |
|||||
|---|---|---|---|---|---|---|
| MIC50 | MBC Range | Δ Log10 CFU/ml | MIC50 | MBC range | Δ Log10 CFU/ml | |
| DAP | 0.25 | 0.12–1 0 | −3.88 | 0.50 | 0.25–2 | −4.12 |
| TIG | 0.12 | 0.12–16.0 | −2.37 | 0.25 | 0.12–16.0 | −2.59 |
| TEI | 0.5 | 0.25–4.0 | −1.86 | 1.0 | 0.5–4.0 | −3.82 |
| VAN | 2.0 | 0.25–4.0 | −3.78 | 2.0 | 1.0–4.0 | −3.97 |
Conclusions: Given the limited therapeutic options for serious CA-MRSA and MDR-MRSA infections, DAP and TIG may offer additional options as they demonstrated good activity against clinical strains of CA-MRSA and MDR-MRSA. Against all isolates, 99.9% kill was achieved as early as 4 h for DAP against all isolates and TIG achieved > 2 log10 CFU/ml kill. Further investigations are warranted.
P1450: Bactericidal activity of ciprofloxacin in the presence of chloramphenicol against two Escherichia coli mutants
S. Roveta, B. Repetto, S. Cagnacci, A. Marchese, E.A. Debbia (Genoa, I)
Objectives: Quinolones are potent bactericidal agents. While their mechanism of killing is poorly understood, one interesting feature is represented by the fact that, under anaerobic conditions, the growth of bacteria is inhibited while their viability is not affected. A similar effect is observed when these drugs are associated to protein synthesis inhibitors. In this study was evaluated the quinolone susceptibility in presence of chloramphenicol on two mutant strains previously described (Int. J. Antimicrob. Agents 2001, 17:S161–2) susceptible to nalidixic acid under anaerobic environment.
Methods: The mutations were co-transferred with Tn10 inserted at 28.5 min of the E. coli map (P1-mediated transduction performed by standard method) into wilde-type strain SA224. Time-kill experiments, under both aerobic and anaerobic condition, were carried out with two different quinolones (ciprofloxacin or nalidixic acid 5×MIC) alone or in combination with chloramphenicol (50 mg/l) following the procedures suggested by the NCCLS and reported elsewhere (Antimicrob. Agents Chemother. 2002, 46:4022).
Results: The transductants revealed the same phenotypes as the original mutants: susceptibility to nalidixic acid under anaerobic conditions (assessed by time kill) and elongated cells formation during the aerobic growth, generation time about 65 min in comparison to 25 min of the control. Time kill experiment under aerobic environment revealed that the two transductants were also susceptible to ciprofloxacin but not nalidixic acid in the presence of chloramphenicol.
Fig. 1.
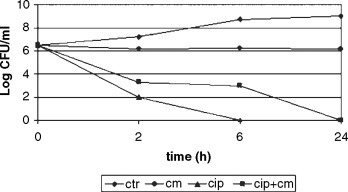
Bactericidal effect of ciprofloxacin in presence of chloramphenicol (mean of 10 different experiments)
Fig. 2.
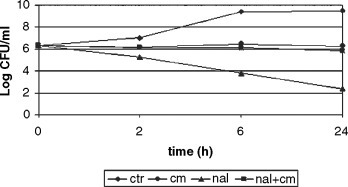
Bacteriostatic effect afnalidixic acid in presence of cho-loromphenicol (mean of 10 different experiments)
Conclusion: These results suggest a possible role of bacterial topoisomerase (topA) in the anaerobic susceptibility to nalidixic acid of the mutants. Further studies are under way in order better characterize these strains.
Antibiotic-resistant community-acquired pathogens – I
P1451: Worldwide antimicrobial susceptibility patterns of inducible Enterobacteriaceae causing intraabdominal infections: results from SMART 2003
F. Baquero, V. Satishchandran, C. Harvey, H. Teppler, M. DiNubile (Madrid, E; West Point, USA)
Objectives: SMART (Study for Monitoring Antimicrobial Resistance Trends) is an ongoing global antimicrobial surveillance programme focused on clinical isolates from intraabdominal infections (IAI). The aim of this interim analysis was to assess antimicrobial susceptibility patterns among inducible Enterobacteriaceae from 4 different regions of the world during 2003.
Methods: 71 centres in the North America (NA), Latin American (LA), Europe (EU) & Asia-Pacific (A/P) tested the in vitro activity of 12 antimicrobial drugs commonly used to treat IAI against consecutive unique Enterobacteriaceae spp. likely to harbour inducible beta-lactamases isolated from IAI according to NCCLS guidelines. All Enterobacter, Serratia, Citrobacter & Providencia spp., & Morganella morganii were presumed to have inducible beta-lactamases. Isolates recovered within 48 hours of hospitalization were considered community-acquired (CA).
Results: 830 isolates of inducible Enterobacteriaceae from 798 patients were tested, of which 282 (34%) were CA. The most common species were Enterobacter spp. (52%), Citrobacter spp. (24%) & M. morganii (14%). The % susceptible are shown by region:
| NA (N = 163) | LA (N=113) | EU (N = 354) | A/P (N = 200) | |
|---|---|---|---|---|
| Ertapenem | 94 | 91 | 98 | 98 |
| Imipenem | 98 | 98 | 98 | 99 |
| Meropenem | 99 | 98 | 99 | 100 |
| Cefoxitin | 23 | 30 | 29 | 27 |
| Ceftriaxone | 75 | 71 | 79 | 60 |
| Ceftazidime | 71 | 69 | 75 | 59 |
| Cefepime | 92 | 88 | 97 | 89 |
| Piperacillin-Tazobactam | 82 | 78 | 85 | 80 |
| Tobramycin | 93 | 79 | 93 | 77 |
| Amikacin | 99 | 86 | 99 | 91 |
| Levofloxacin | 90 | 80 | 90 | 87 |
| Ciprofloxacin | 87 | 72 | 88 | 81 |
Conclusion: Among inducible Enterobacteriaceae causing IAI, resistance rates for most antibiotics tested were modestly higher in A/P & LA than NA & EU. Carbapenems, including Group I agents like ertapenem, were the most reliably active drugs in vitro. Because these spp. often harbour a common chromosomal Type 1 cephalo-sporinase that can be reversibly induced by betalactam antibiotics, they may appear misleadingly susceptible to some cephalosporins.
P1452: Antimicrobial susceptibility among Enterobacteriaceae causing intraabdominal infections in Europe: results from SMART 2003
F. Baquero, J. Chow, T. Snyder, V. Satishchandran, C. Harvey, M. DiNubile (Madrid, E; West Point, USA)
Objectives: SMART (Study for Monitoring Antimicrobial Resistance Trends) is an ongoing global antimicrobial surveillance programme unique in its focus on isolates from intraabdominal infections (IAI). The objective of this subanalysis was to assess antimicrobial susceptibility patterns among Enterobacteriaceae recovered at participating European sites during 2003.
Table for P1452
| Ertapenem | Imipenem | Meropenem | Cefoxitin | Cefriaxone | Ceftazidime | Cefepime | Tobramycin | Amikacin | Piperacillin tazobactam | Levofloxacin | Ciprofloxacin | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Enterobacteriaceae | 99 | >99 | >99 | 82 | 91 | 92 | 96 | 93 | 99 | 91 | 86 | 84 |
| E. coli | >99 | 100 | >99 | 94 | 95 | 96 | 96 | 92 | 99 | 94 | 81 | 80 |
| Klebsiella | 99 | 99 | 99 | 92 | 89 | 94 | 94 | 91 | 98 | 85 | 93 | 89 |
Methods: 32 centres in 7 European countries each tested the in vitro activity of 12 antimicrobial drugs commonly used to treat IAI against consecutive non-duplicate Enterobacteriaceae isolated from intraabdominal specimens using microdilution techniques according to NCCLS guidelines and breakpoints. Production of extended-spectrum beta-lactamases (ESBL) was confirmed in isolates with a MIC of ceftriaxone, ceftazidime, or cefepime ≥2 μg/mL by comparing cefepime MICs with and without clavulanate. Pathogens recovered within 48 hours of hospitalization were considered community-acquired (CA).
Results: A total of 1948 Enterobacteriaceae were recovered from 1713 patients. E. coli (1106 isolates; 57%) & Klebsiella spp. (341 isolates; 18%) were the most common isolates. The per cent susceptible according to labeled MIC breakpoints (without regard to ESBL production) are reported below. A total of 68 E. coli (6%) & 45 Klebsiella spp. (13%) produced an ESBL. Of the CA isolates, 16/ 511 E. coli (3%) & 7/129 Klebsiella spp. (5%) were ESBL-producers.
Conclusions: Among these Enterobacteriaceae causing IAI at selected sites across Europe in 2003, resistance rates to cefoxitin, levofloxacin, and ciprofloxacin exceeded 10%. ESBL-producing pathogens were recovered from some CA IAI. Carbapenems and amikacin were the most reliably active drugs in vitro in this study.
P1453: 5-year trends in resistance among community-acquired lower respiratory tract isolates of S. pneumoniae from the UK and Ireland
R. Reynolds, D. Felmingham on behalf of the BSAC Working Party on Respiratory Resistance Surveillance
Objective: To identify trends in antimicrobial resistance in S. pneumoniae, correcting for possible confounding factors.
Methods: 3584 lower respiratory tract isolates of Streptococcus pneumoniae were collected from a total of 27 laboratories in the UK and Ireland over the five winters from 1999–2000 to 2003–2004. Isolates were excluded if they were duplicates within 2 weeks, or from samples collected > 48 hours after hospitalisation, or from patients with cystic fibrosis. MICs were determined centrally using the BSAC agar dilution method and interpreted by BSAC criteria. Logistic regression models were fitted separately for penicillin-non-susceptibility and tetracycline-, erythromycin- and ciprofloxacin-resistance (PEN-NS, TET-R, ERY-R & CIP-R).
Results: Table 1 shows unadjusted resistance rates. The final models for PEN-NS, TET-R and ERY-R included centre, age group, and year as a linear function. The final model for CIP-R included centre, age group, and year as a categorical variable. Sex, specimen type (sputum/other) and care setting (GP/nursing home/hospital) were not required in any of the models. Isolates with missing data (n = 25) were excluded. For PEN-NS, TET-R and ERY-R in Ireland, there was a significant linear trend of falling resistance. The odds ratios (OR) in Table 2 show the factor by which the odds of resistance change each year. The estimated reduction in odds was 23% per year for PEN-NS, 29% per year for TET-R and 24% per year for ERY-R. There was no significant trend in the UK. For CIP-R, there was no consistent trend over time in Ireland or the UK.
Table 1.
| Resistance | Year | N | PEN-NS % >0.06 mg/L | TET-R % >1 mg/L | ERY-R % >0.5 mg/L | CTP-R % >2 mg/L |
|---|---|---|---|---|---|---|
| Ireland | 1999–2000 | 66 | 40.9 | 24.2 | 27.3 | 3.0 |
| 2000–2001 | 59 | 42.4 | 25.4 | 23.7 | 3.4 | |
| 2001–2002 | 68 | 36.8 | 16.2 | 25.0 | 16.2 | |
| 2002–2003 | 89 | 24.7 | 10.1 | 12.4 | 15.7 | |
| 2003–2004 | 76 | 14.5 | 7.9 | 9.2 | 13.2 | |
| UK | 1999–2000 | 595 | 7.4 | 7.2 | 11.8 | 5.7 |
| 2000–2001 | 608 | 7.1 | 6.3 | 10.0 | 5.3 | |
| 2001–2002 | 631 | 4.6 | 4.8 | 10.8 | 7.9 | |
| 2002–2003 | 683 | 6.4 | 5.7 | 9.7 | 2.2 | |
| 2003–2004 | 709 | 5.9 | 6.2 | 10.6 | 11.7 |
Table 2.
| Trend/year | Ireland |
UK |
||||
|---|---|---|---|---|---|---|
| OR | 95% CI | P | OR | 95% CI | P | |
| PEN-NS | 0.77 | 0.63, 0.94 | 0.009 | 0.95 | 0.86, 1.06 | 0.39 |
| TET-R | 0.71 | 0.55, 0.91 | 0.008 | 0.94 | 0 84, 1.05 | 0.27 |
| EEY-R | 0.76 | 0.60, 0.96 | 0.024 | 0.98 | 0.90,1.07 | 0.68 |
Conclusion: Trends in resistance in S. pneumoniae differed between Ireland and the UK, and between classes of antimicrobials. Resistance to penicillin, tetracycline and erythromycin decreased in Ireland but not to a similar extent in the UK, and there was no trend in ciprofloxacin resistance. Reasons for these differences should be sought.
P1454: Influence of age on resistance in community-acquired lower respiratory tract isolates of S. pneumoniae from the UK and Ireland
R. Reynolds, D. Felmingham on behalf of the BSAC Working Party on Respiratory Resistance Surveillance
Objective: To examine the relationship between patient age and antimicrobial resistance in S. pneumoniae.
Methods: 3584 lower respiratory tract isolates of Streptococcus pneumoniae were collected from a total of 27 laboratories in the UK and Ireland over the five winters from 1999–2000 to 2003–2004. Isolates were excluded if they were duplicates within 2 weeks, or from samples collected >48 hours after hospitalisation, or from patients with cystic fibrosis. MICs were determined centrally using the BSAC agar dilution method and interpreted by BSAC criteria. Logistic regression models for penicillin-non-susceptibility and tetracycline-, erythromycin- and ciprofloxacin-resistance (PEN-NS, TET-R, ERY-R & CIP-R) were fitted, correcting for age, year and collecting laboratory.
Results: Age was unknown for 4 patients. The median age was 62. PEN-NS, TET-R and ERY-R were modelled with age group as a categorical variable using the 40–49 group as baseline; resistance was least in this group and greater in both younger and older patients, as shown by the odds ratios (ORs) in Table 1. CIP-R was modelled with a linear function of age; ORs corresponding to the median age within each age group are shown. Moxifloxacin-resistance (MXF-R, MIC > 1 mg/L) was too rare for logistic modelling, but was also strongly associated with age. Of the 29 MXF-R isolates (all with CIP MICs >8 mg/L), 25 (86%) were from patients aged ≥ 60 (Fisher's exact p < 0.001), and none was from a patient aged <50. Observed resistance rates for all isolates and for the baseline age group are shown in Table 2.
Table 1.
| Odds Ratios | ||||||
|---|---|---|---|---|---|---|
| Age Group, years | Median Age in Group | N | PEN-NS > 0.06 mg/L | TET-R > 1 mg/L | ERY-R > 0.5 mg/L | CIP-R > 2 mg/L |
| 0–4 | 0 | 233 | 1.66 | 1.54 | 1.38 | 0.61 |
| 5–19 | 11 | 130 | 1.57 | 1.40 | 1.43 | 0.69 |
| 20–39 | 33 | 402 | 1.20 | 1.60 | 1.80 | 0.88 |
| 40–49 | 45 | 352 | 1 | 1 | 1 | 1 |
| 50–59 | 55 | 527 | 2.07 | 2.68 | 2.31 | 1.11 |
| 60–69 | 65 | 759 | 1.52 | 1.62 | 1.84 | 1.24 |
| 70–79 | 74 | 746 | 2.09 | 2.28 | 2.04 | 1.37 |
| >80 | 84 | 431 | 2.72 | 1.82 | 2.30 | 1.53 |
Table 2.
| Observed Resistance | ||||||
|---|---|---|---|---|---|---|
| Age Group | N | PEN-NS % | TET-R % | ERY-R % | CIP-R % | MXF-R% |
| All | 3580 | 8.7 | 7.0 | 11.4 | 7.1 | 0.8 |
| 40–49 | 352 | 5.1 | 3.7 | 6.3 | 6.3 | 0 |
Conclusion: Patient age is a significant predictor of resistance in S. pneumoniae. Isolates from both young and elderly patients had increased risk of resistance to PEN, TET and ERY, but fluoroquinolone resistance increased linearly with age. Resistance to MXF was rare in the UK and Ireland, and was seen only in isolates from patients aged over 50.
P1455: Enhanced bacteraemia surveillance of EARSS pathogens in Ireland 2004
A. Oza, S. Murchan, R. Cunney (Dublin, IRL)
Objectives: Aim of this presentation is to provide an update on a study investigating the factors that affect bacteraemia caused by key pathogens isolated in Irish hospitals. The study is primarily focused on those pathogens exhibiting antibiotic resistance and the results are used to supply timely feedback to contributing laboratories.
Method: A sample of hospital laboratories participating in the European Antimicrobial Resistance Surveillance System (EARSS) provided additional information on bloodstream infections arising from EARSS pathogens on a quarterly basis from the start of 2004. The additional information included patient demographic and hospital administrative data, risk factors, primary source and secondary foci. This information was linked to the resistance data for each isolate and hospital activity denominator data were used to calculate rates.
Results: Data for quarters 1 and 2 resulted in 433 matched records from 7 representative hospital laboratories. Analyses of this dataset AND data received for quarters 3 and 4 of 2004 will be presented. Preliminary results indicated that most infections occurred in the younger and older (≥65 years) age groups. Malignancies were identified in a third of the patients. Recent surgery, ICU-stay and haemodialysis were particularly associated with MRSA bacteraemia. Respiratory tract infection was the most common source for S. pneumoniae bacteraemia, urinary tract for E. coli, and intra-abdominal/gastrointestinal tract for both E. coli and enterococci (including VRE). Different predominant sources of S. aureus infection were associated with different levels of methicillin-resistance: skin/soft tissue (low proportion of MRSA), CVC (medium) and respiratory tract (high).
Conclusions: The findings so far support the fact that hospital function can influence bacteraemia rates as a result of treating patients with different risk factor profiles. The analyses provided through this surveillance programme could be used to make informed and targeted infection control decisions.
P1456: Relationship between Cotrimoxazole use and resistance in Streptococcus pneumoniae, Haemophilus influenzae and Moraxella catarrhalis
P. Karpanoja, S. Huikko, T. Voipio, P. Paakkari, M. Bergman, P. Huovinen, H. Sarkkinen for the Finnish Study Group for Antimicrobial Resistance FiRe
Objectives: The combinations of sulfonamides and trimethoprim (Cotrimoxazoles) are mostly (81 %) prescribed for respiratory tract infections in Finnish health care centres (Rautakorpi et. al, 2001, Scand J Infect Dis 33:920-926). The aim of this study was to analyse the relationship between Cotrimoxazole resistance and Cotrimoxazole consumption among the three major respiratory tract pathogens Streptococcus pneumoniae, Haemophilus influenzae and Moraxella catarrhalis.
Methods: The consumption rates of Cotrimoxazole (sulfamethoxazole/trimethoprim and sulfadiatzine/trimethoprim) between 1997 and 2002 were obtained from the Finnish National Agency for Medicines. The sale figures for pharmacies, which correspond to the use of the antibiotic in community health care, were used in this analysis. The consumption is expressed as defined daily doses per 1000 inhabitants per year (DDD). The resistance data for the three pathogens were collected annually between 1998 and 2003 from 27 clinical microbiology laboratories (FiRe -network). The resistance rates are expressed as percentages. Figures based on at least 30 tested isolates per laboratory were included. A linear mixed model for repeated measures was used. A random effects model with time and consumption as fixed and intercept as random effect was fitted.
Results: The total use of Cotrimoxazole compounds has declined in Finland during the study period from 0,98 to 0,57 DDD. The annual resistance rates in the whole country were between 14.1-21.4 % in S. pneumoniae, 9.7-18.7 % in H. influenzae and 3.2-14.5 % in M. catarrhalis. A positive association (p = 0.04) was found between the level of resistance and the consumption of Cotrimoxazole in S. pneumoniae. The change in resistance was not significant (p = 0796). In H. influenzae the resistance decreased significantly (p = 0.001). However, this could not be explained by consumption (p = 0.616). In M. cartarrhalis the change in resistance was not significant (p = 0.358), and the level of resistance could not be explained by drug use (p = 0.532).
Conclusions: The resistance of Cotrimoxazole correlates to consumption only in S. pneumoniae. However, consumption is not a strong exogenous variable.
P1457: Analysis of trends in antimicrobial resistance in Staphylococcus aureus and Escherichia coli in Ireland, 1999–2004
S. Murchan and the Irish EARSS Steering Group on behalf of the Irish EARSS participants
Objectives: To analyse antimicrobial resistance (AMR) trends from surveillance data collected in Ireland, as part of the European Antimicrobial Resistance Surveillance System (EARSS), for Staphylococcus aureus and Escherichia coli, two pathogens commonly associated with bacteraemia.
Methods: Routine susceptibility data on invasive isolates of S. aureus and E. coli were collected from participating laboratories (EARSS coverage as of mid-2004 was ∼95% of the Irish population). Data were collated, analysed and fed back to laboratories and public health professionals. The trends over time, either quarterly or annual as appropriate, in the proportions of isolates resistant to key antibiotics were analysed using statistical methods {Linear Regression with Analysis of Variance (ANOVA) and Chi-Squared Test for Trend (Chi2)}.
Results: Between Jan'99 and Jun'04, data were submitted on 4786 S. aureus isolates, of which 1983 (41.4%) were resistant to methicillin. The annual proportions over this period ranged from 38.9% in 1999 to 42.1% in 2003 and 42.8% up to Jun'04. ANOVA showed that there was a linear relationship with resistance increasing by ∼1% per year (P = 0.01). Chi2 suggested that the increase was of borderline significance (P = 0.06). Amongst MRSA isolates, there was a highly significant decrease in the proportion of resistance to gentamicin from 58.4% in 1999 to 13.1% in 2003 (p < 0.001). Between Jan'01 and Jun'04, data were submitted on 2432 E. coli isolates, of which 8.4% were resistant to ciprofloxacin. The quarterly proportions over this period ranged from 2.9% in Quarter 2 (Q2) 2001 to 13.4% in Q2 2004. ANOVA showed that there was a linear relationship with resistance increasing by ∼1% per quarter (P = 0.014). Chi2 suggested that the increase was highly significant (p < 0.001). No significant trends were observed in resistance to 3rd-generation cephalosporins (2.5% of isolates) or gentamicin (3.7%).
Conclusions: Methicillin resistance in S. aureus remains an important public health problem in Ireland and has increased over the past five years, although the increase is of doubtful significance. Ciprofloxacin resistance in E. coli has increased significantly since surveillance began in 2001. Further studies are warranted to understand the reasons for this. As part of any surveillance system, continued and appropriate analysis of the data for trends is essential. The data presented here highlight the changing profile of AMR in Ireland over time.
P1458: Antimicrobial susceptibility in respiratory bacterial pathogens among children in Greenland: little antibiotic resistance in spite of high antibiotic use
A. Koch, M. Tvede, L. Høgh Andersen, T. Hjuler, M. Melbye (Copenhagen, DK)
Objectives: In Alaskan natives the prevalence of penicillin-resistant S. pneumoniae isolates is high. In Greenland respiratory tract infections in children are frequent, and antibiotic use is high. As local data on antimicrobial resistance hardly exist in Greenland, the objective of his study was to obtain such information.
Methods: A population-based cohort of children aged 0–4 years was followed in Sisimiut, Greenland, for a two-year period (1996–1998). Throat swabs were taken when symptoms of respiratory tract infection presented and every half-year without symptoms. In case of acute purulent ear discharge a swab was taken from the middle ear. Swabs were placed in Stuart's transport medium and sent by airmail to the University Hospital, Copenhagen, for culturing. Antibiotic susceptibility was determined by the disk diffusion method with Danish Blood Agar and Rosco New Sensitabs. Antimicrobial susceptibility was defined as susceptible, intermediate, or resistant. Only the first isolate from each child was used for analysis.
Results: Overall, 1624 swabs from 376 children (81% of cohort) were cultured and 2560 isolates identified. All S. pneumoniae (n = 153) Isolates were susceptible to Penicillin, Ampicillin, Erythromycin and other drugs tested. Only 1 of all 270 S. pneumoniae isolates was inter-mediately susceptible to Penicillin. Of 237 H. influenzae isolates 92% were susceptible to Ampicillin, and 94% susceptible to Erythromycin. All Ampicillin resistant strains were beta-lactamase producing. Of 228 beta-haemolytic streptococcal isolates 98% were susceptible to Erythromycin. Of 35 Moraxella spp. isolates 49% were susceptible to Ampicillin and 51% resistant, while 100% were susceptible to Erythromycin. There was no Meticillin resistant S. aureus (MRSA). The distribution was unchanged if only throat swabs were analysed.
Conclusions: Antibiotic resistance is low in Greenland and similar to that in Scandinavia. With respect to Penicillin, resistance is less in Greenland than in Alaskan natives. Surveillance of antimicrobial susceptibility in Greenland is warranted. In 2000 Greenland joined the International Circumpolar Surveillance of infectious diseases.
P1459: Patterns of antibiotic resistance in pathogens causing respiratory tract infections: results of the 2002–2003 PROTEKT Italy survey
A. Marchese, F. Ardito, S. Stefani, G. Fadda, G. Nicoletti, G.C. Schito (Genoa, Rome, Catania, I)
Objective: PROTEKT Italy, an extension of PROTEKT International Survey, is an ongoing 3-year surveillance study established to assess the susceptibility of community-acquired respiratory pathogens circulating in this Country.
Methods: 52 (year 1: 2002) and 46 (year 2: 2003) Clinical Microbiology Laboratories provided 1056 and 1086 S. pyogenes, 848 and 798 S. pneumoniae, 463 and 518 methicillin-susceptible S.aureus (MSSA), 317 and 349 H.influenzae, 207 and 196 M. catarrhalis respectively. In vitro susceptibility to 20 (year 1) and 19 (year 2) antibiotics was determined according to NCCLS-approved microdilution methods and breakpoints (2004).
Results: High rates of macrolide resistance (30.0%, year 1) and (34.0%, year 2) were observed in S.pyogenes. Telithromycin (92.7%-92.8% susceptibility) was active on the great majority of these macrolide-refractory strains. Total pneumococcal penicillin-resistance was stable during the study period ranging from 23.7% to 23.4%, while lack of susceptibility to macrolides increased from 40.0% to 43.4%. The most active drugs were: telithromycin (99.7–99.1%), levofloxacin (97.3–97.1%), rifampin (96.7–98.9%), amoxicillin (93.8–96.6%) and ceftriaxone (88.9–93.7%). Percentages of ampicillin-resistance in H. influenzae were 26.5% (year 1) and 20.6% (year 2). For the first time in 2003 two ampicillin-resistant beta-lactamase negative (0.6%) isolates were recovered. High rates of resistance were found for clarithromycin (36.9–25.7%) and co-trimoxazole (13.6–14.1%). The remaining drugs tested (ciprofloxacin, levofloxacin, moxifloxacin, azithromycin, telithromycin, amoxicillin-clavulanate, chloramphenicol, tetracycline, 2nd and 3rd generation cephalosporins) were highly active: 100–94.6% of susceptible strains. InM. catarrhalis the susceptibility rates ranged from 22.7–18.9% (ampicillin) and 77.3–81.1% (co-trimoxazole) to 100% (amoxicillin-clavulanate, ceftriaxone, cefixime, telithromycin, azithromycin, ciprofloxacin, levofloxacin and moxifloxacin). Against MSSA the most active drugs (100%-92% susceptibility) were teicoplanin, rifampin, co-trimoxazole, ceftriaxone, cefaclor, telithromycin and levofloxacin.
Conclusions: Antibiotic resistance among community-acquired respiratory pathogens circulating in Italy is on the rise in comparison to the values described in previous National Surveys (1997–2000). Globally, telithromycin, levofloxacin, amoxicillin-clavulanate and ceftriaxone are the most active drugs.
P1460: Incidence and antibiotic susceptibility of pathogens causing severe community-acquired and nosocomial infections in Italy
G.C. Schito, E.A. Debbia, G. Fadda, F. Ardito, E. Magliano, G. Nicoletti, V.M. Nicolosi, A. Pantosti, G. Rezza, A. Cassone, E. Garaci (Genoa, Rome, Milan, Catania, I)
Objectives: To evaluate the incidence and antibiotic susceptibility of pathogens isolated from severe infections in a nationwide survey in Italy.
Methods: During 2003, 44 Clinical Microbiology Laboratories isolated bacterial pathogens from hospitalized patients with severe infections Strains were sent to three centres in Genoa, Rome and Catania for confirmation of identity. All pathogens were then forwarded to a Laboratory in Switzerland for quality-controlled, centralized susceptibility testing. MICs were determined employing NCCLS-approved methodologies and breakpoints. Gram-positive and Gram-negative bacteria were tested against 12 and 10 antimicrobial molecules respectively.
Results: Septicaemia, all-cause pneumonia and CVC-associated infections accounted for >50% of all infections, and S. aureus, P. aeruginosa and E. coli together represented nearly 50% of the 4702 isolates (2516 Gram-negative and 2186 Gram-positive). This preliminary report describes the major findings of 759 and 612 tests carried out on the two group of strains. Extended spectrum beta-lactamase-producers in E.coli increased, in comparison with previous national studies, from 6.3 to 16.7%, in K. pneumoniae from 14 to 23.7%, in Enterobacter from 32.2 to 53%, remaining stable to 30% in P.mirabilis. Cefotaxime resistance ranged from 13.2 (S. marcescens) to 14,3% (M. morganii), and reached 84.5% in P. stuartii. In P.aeruginosa the level of resistance to 3rd generation cephalosporins (3rdGC) decreased from 14 to 4% and the same was true for imipenem (23.3 to 11.9%). Lack of susceptibility to ciprofloxacin increased from 32.8 to 38.5%. For A. baumannii and B.cepacia levels of resistance were respectively 13.3 and 73.8% for 3rdGC, 13.3 and 7.2% for piperacillintazobactam, 22.5 and 71.4% for imipenem, and 29 and 92.9% for ciprofloxacin. Among Gram-positive pathogens the major drug-resistance phenotypes included macrolides (31.8%) in S. pyogenes, penicillin (20.0%) and macrolides (35.0) in S.pneumoniae, methicillin in S. aureus (37.1) and in S. epidermidis (86.7%). In E. faecalis ampicillin displayed very low (0.4%) and vancomycin and linezolid no resistance traits.
Conclusions: These preliminary findings indicate a worrying negative evolution towards acquisition of drug resistance in bacterial pathogens causing severe infections in Italy. Knowledge of current in vitro potency of the available antibiotic classes may help clinicians select an effective initial empiric therapy.
P1461: Patterns of susceptibility of Streptococcus pneumoniae and Haemophilus influenzae at a university hospital
Z. Daoud, N. Hanna, A. Cocosaki, N. Hakime (Beirut, LBN)
Streptococcus pneumoniae and Haemophilus influenzae are important pathogens in many community and hospital acquired respiratory infections. Until early 1990s nearly all Streptococci were uniformly susceptible to penicillin, and then the first reports of Penicillin Resistant Pneumococcus (PRSP) emerged. On the other hand, Life-threatening invasive infections caused by Haemophilus influnezae are more commonly caused by encapsulated type b strains, however, localized infections are often treated empirically, a knowledge of antibiotic resistance determined on the basis of systematic surveillance studies is essential.
Objective: The objective of this study was to determine the MICs of the invasive strains of S. pneumoniae and Haemophilus influenzae isolated at Saint George Hospital for the period of three years extending between June 2001 and June 2004.
Results and comments: Streptococcus pneumoniae: In total 116 strains were tested against erythromycin, 18 % of them were non susceptible to erythromycin. More than 22 % of the PNSP showed susceptibility to erythromycin. All tested strains were highly susceptible to the 3rd generation cephalosporins as well as to quinolones. Haemophilus influenzae: 60 % of the total isolates (n = 129) were resistant to ampicillin. 62.5% of the total isolates (n = 135) were b-lactamase negative. 6 isolates (2.5%) were b-lactamase negative, however, resistant to Ampicillin (BLNAR) and only 1 isolate (0.16%) was b-lactamase positive but resistant to Amox/clav (BLPACR). The prevalence of invasive S. pneumoniae was seasonal with clear peaks during winter. The percentage of penicillin non susceptible S. pneumoniae among these strains displayed no seasonality. The prevalence of invasive and penicillin non susceptible S. pneumoniae was highest in children 4 years and younger. This emphasizes the importance of prudent antimicrobial use of antibiotics and vaccination in this age group. The proportion of erythromycin resistance among Penicillin non susceptible S. pneumoniae was not very high in comparison to other studies done mainly in Europe (EARSS). PNSP are still in general highly susceptible to Levofloxacin, Cefotaxime, and Imipenem. The problem of Ampicillin resistance among our isolates of H. influenzae is important (60%) and is complicated by the b-lactamase-negative strains which were resistant to ampicillin (2.5%) by some other mechanism perhaps elaboration of altered penicillin-binding proteins.
S. pneumoniae: Minimum Inhibitory Concentrations
| Antibiotic | % S | Mean MIC-S | %R | Mean MIC-R |
|---|---|---|---|---|
| Cefuroxime | 95.2 | 0.33 | 4.8 | 3 |
| Cefotaxime | 95.2 | 0.3 | 4.8 | 3 |
| Amox-clav | 100 | 0.1 | 0 | – |
| TMX | 47.6 | 0.22 | 58.4 | 14.2 |
| Imipenem | 100 | 0.08 | 0 | – |
| Erythromycin | 82 | 0.06 | 18 | 0.38 |
| Peni G | 46 | 0.02 | 54 | 0.51 |
| Levofloxacine | 100 | 0.04 | 0 | 5.2 |
H. influenzae: Minimum Inhibitory Concentrations
| Antibiotic | %S | Mean MIC-S |
|---|---|---|
| Cefuroxime | 94.6 | 0.78 |
| Ceftriaxone | 100 | 0.42 |
| Amox-clav | 94.6 | 0.8 |
| Chloramphenicol | 97.2 | 0.55 |
| TMX | 78 | 0.13 |
| Ampicilline | 60 | 0.29 |
| Levofloxacine | 100 | 0.46 |
P1462: Exotic pets as possible reservoirs of ciprofloxacin-resistant Salmonella typhimurium infecting children
I. Casin, F. Weill, J. Breuil, R. Lailler, J. Croizé, J. Darchis, E. Collatz (Paris, Villeneuve-Saint Georges, Maisons-Alfort, Grenoble, Compiégne, F)
Objectives: Considering that high-level resistance to ciprofloxacin (≥2 mg/L; CipR) in human isolates of salmonellae is still rare in Europe, we analysed the molecular characteristics of 12 CipR strains isolated from seven children and one adult in order to explore a possible epidemiological relationship with two CipR strains isolated from a parrot and a snake. Pet animals of these species have been reported as reservoirs of salmonellae sporadically transmitted to humans.
Methods: The strains were characterized by serotyping, phage typing, antimicrobial susceptibility testing, with or without an efflux pump inhibitor (EPI), and pulsed-field gel electrophoresis (PFGE). Polymerase chain reaction and DNA sequencing were used to identify integron-borne gene cassettes and mutations in the GyrA, GyrB, ParC and ParE genes.
Results: All 14 S. enterica strains were of serotype Typhimurium and of a phage type 12 variant. All were resistant to multiple antibiotics (AmpStr/SpcTetCmpSul) including Cip (≥16 mg/L) and, with one exception, Gen and Tmp. CipR was associated in each case with mutations in GyrA (Ser83Phe, Asp87Asn), GyrB (Ser464Phe) and ParC (Ser80Arg) and moderately decreased quinolone efflux. The 13 Gen-, Tmp-resistant strains carried oxa-30 and had a ca. 2 kb integron with aadA2 and dfrXII cassettes, while the Gen-, Tmp-susceptible strain contained oxa-30 and aadA2 cassettes, also in a ca. 2 kb integron. Using XbaI, the PFGE profiles of all strains were indistinguishable, while the use of BlnI revealed 3 discrete subgroups (with 1- or 2-band differences): a) 3 isolates, from 2 children and the snake which was identified as the source of infection; b) 10 isolates, from 4 children, 1 adult and the parrot, without documented contact; c) the Gen-, Tmp-susceptible isolate from a child who had been in contact with a pet snake.
Conclusion: While the food chain is the most common route of animal-to-human transmission of salmonellae which may be resistant to multiple antibiotics including fluoroquinolones, the present study raises the possibility that exotic pet animals may become a reservoir of such strains which may be transmitted especially to young children.
P1463: Changes in antimicrobial associated resistance, susceptibility patterns and ‘resistance load’ in 35,956 consecutive UTI Escherichia coli 1993–2003
A. Wimmerstedt, G. Kahlmeter (Växjö, S)
Objectives: Antimicrobial resistance is most often expressed as individual resistance rates for a species against a defined drug. The objective of this study was to present alternative models to describe resistance development.
Materials and Methods: During 1993–2003, 35 956 Escherichia coli from urinary tract infections were systematically categorised for susceptibility to a fluoroquinolone, trimethoprim, ampicillin, cefadroxil, mecillinam and nitrofurantoin using the methodology and interpretive criteria of the Swedish Reference Group of Antibiotics. Three models were analysed. ‘Associated resistance’ was presented as the resistance rate of each drug in the presence and absence of resistance to each of the other drugs as previously described (JAC, 2003; 52, 128). ‘Susceptibility pattern changes’ were described as a change in rank order of patterns and in the number of different patterns over time. ‘Resistance load’ was expressed by transforming the susceptibility pattern of each strain into a numerical value: the susceptibility pattern SSSSSR corresponded to 2 (0 + 0 + 0 + 0 + 0 + 2) and the pattern RSSIRS to 5 (2 0 + 0 + 1 + 2 + 0). The minimum and maximum ‘resistance load’ were 0 and 12, respectively.
Results: Associated resistance was high all through the observation period. As an example, trimethoprim resistance in E.coli resistant and sensitive to ampicillin was 28% versus 3% in 1993 and 43% versus 4% in 2003. The five most common susceptibility patterns were the same over the 10 years whereas the number of different patterns increased from 28 in 1993 to 49 in 2003. The proportion of E. coli without any sign of decreased susceptibility to the 6 drugs decreased from 79.0% in 1993 to 76.6% in 2003. The mean resistance load in E.coli with decreased susceptibility to one or more of the drugs increased from 3.0 in 1993 to 3.4 in 2003. The proportion of isolates with resistance load of 6 or more increased from 0.8% in 1993 to 2.2% in 2003.
Conclusion: Antimicrobial resistance is traditionally expressed as rates over time. More information is obtained by presenting changes in total ‘resistance load’ and associated resistance over time. The increasing number of susceptibility patterns is yet another sign of resistance development and it emphasises the difficulties faced in empirical antimicrobial therapy.
P1464: Increase of resistance to nalidixic acid among four clinically important Enterobacteriaceae pathogens in three Central European countries
M. Kresken, D Hafner on behalf of the Working Group for Antimicrobial Resistance of the Paul-Ehrlich-Society for Chemotherapy
Objectives: Since 1984 when the first fluoroquinolone (FQ) was introduced in Europe the consumption of the FQ has markedly increased. To evaluate the influence of the increasing consumption on the prevalence of resistance to nalidixic acid (NAL) and FQ in clinical isolates of four clinically important members of the family Enterobacteriaceae we reviewed the susceptibility data from two collaborative studies conducted in 1984 (prior to the introduction of the first FQ) and in 2001 by the Working Group for Antimicrobial Resistance of the Paul-Ehrlich-Society for Chemotherapy.
Methods: Isolates of Enterobacter cloacae (ECL), Escherichia coli (ECO), Klebsiella pneumoniae (KPN), and Proteus mirabilis (PMI) collected in 1984 and 2001 from 23 and 26 laboratories, respectively, located in Austria, Germany, and Switzerland, were included. MICs were determined using the broth microdilution method according to the standard of the German DIN.
Results: In 1984 and 2001, a total of 1,640 (198 ECL, 834 ECO, 263 KPN, 345 PMI) and 1,348 (234 ECL, 619 ECO, 268 KPN, 227 PMI) isolates, respectively, were tested. Resistance to NAL markedly increased in all species between 1984 and 2001, i. e. from 2.0% to 17.5% in ECL, from 1.6% to 19.2% in ECO, from 12.2% to 21.6% in KPN, and from 2.6% to 15.9% in PMI. Of the NAL-resistant ECO isolates collected in the 2001 survey (n = 116), 26.1% were susceptible and 71.4% resistant to ciprofloxacin (CPFX). Susceptibility rates of CPFX for ECL, KPN, and PMI were 53.4%, 43.9%, and 30.6%, respectively. In the 2001 survey, NAL-resistant isolates of all species exhibited lower susceptibility rates to other therapeutic classes than NAL-susceptible isolates. Notably in ECO, susceptibilities to cefotaxime, ceftazidime, piperacillin-tazobactam, gentamicin, and tobramycin were significantly lower (p < 0.05) among NAL-resistant isolates (89.9%, 93.3%, 85.7%, 68.9%, 71.4%) than among NAL-susceptible isolates (99.2%, 99.0%, 94.2%, 91.2%, 95.0%). In contrast, cefepime, carbapenems, and amikacin retained in vitro activity against NAL-resistant ECO isolates. Similar trends were observed for the other three species.
Conclusion: Resistance to NAL (and FQ) has markedly increased among Enterobacteriaceae pathogens recovered from patients in hospitals located in Germany, Austria, and Switzerland. We strongly recommend a judicious use of FQ in order to combat the spread of FQ resistance in Enterobacteriaceae.
P1465: E. coli and S. aureus – antimicrobial resistance surveillance and the effect of excluding duplicate isolates in a consecutive database 1990–2003
M. Sundqvist, G. Kahlmeter (Växjö, S)
Objectives: The NCCLS and ESGARS (ESCMID Study Group on Antimicrobial Resistance Surveillance) have recommended exclusion of duplicate isolates in studies on antibiotic resistance. The procedure is not yet standardized. The present study compared outcome of cut-off levels at 30 and 365 days on 14 years of surveillance data for E.coli and S.aureus.
Materials and methods: All consecutive isolates of E. coli (n = 62380) and S. aureus (n = 24743) from routine susceptibility testing at our laboratory (1990–2003) were included. Duplicates were identified and excluded based on a unique personal identification number, species and cut-off time (30 and 365 days) from first isolate. Susceptibility testing was performed as recommended by the Swedish Reference Group of Antibiotics and did not change over the years except for fluoroquinolones (norfloxacin 1990–1993, ciprofloxacin 1994–2000, nalidixic acid 2001–2003).
Results: The effects on resistance rates of excluding duplicate isolates (DI) were small despite the fact that almost one third of the isolates were excluded through the 365 days exclusion algorithm. Except for fluoroquinolones, resistance rates in E. coli decreased when DI were excluded on the basis of 30 days but increased when DI were excluded on the basis of 365 days. Resistance in S. aureus tended to decrease when duplicates were excluded independent of cut-off time.
Conclusion: E. coli and S. aureus are two of our most important pathogens and as such common in resistance surveillance. Although the effects on resistance rates of exclusion of duplicate isolates were minor and not statistically significant in the present study we suggest that the exclusion cut-offs should match the timeline, i.e. if rates are presented as yearly figures, the exclusion cut-off should be 365 days, and so on. We furthermore believe that the effect of excluding duplicates should be presented in conjunction with presentation of surveillance data. Our data suggests that E. coli re-infection (infection with the same species more than 30 days after the first incident) is mainly caused by new and less resistant strains whereas patients who have acquired resistant strains of S. aureus continue to be colonized with the same strain.
P1466: Haemophilus influenzae antibiotics resistance in Greek patients (2001–2004)
S. Kanavaki, M. Makarona, H. Moraitou, K. Kastrisiou, S. Arbilia, A. Malgarinou, S. Triantafyllou, S. Karabela (Athens, GR)
Objectives: H. influenzae represents one of the commonest pathogens, in community-acquired respiratory infections. Recently, there is a great concern worldwide regarding the increased prevalence of beta-lactamase producing strains, the increasing incidence of resistance to ampicillin and macrolides, the emergence of ampicillin resistant non-beta-lactamase producing isolates (BLNAR), as well as the isolation of betalactamase producing strains resistant to amoxicillin+clavulanic acid (BLPACR). The aim of our study was to investigate the susceptibility profiles of H. influenzae isolated in ‘Sotiria’ District General Hospital, in the period 2001–2004.
Methods: All strains were isolated from sputum samples, in the Microbiology Laboratory of ‘Sotiria’ Chest Diseases Hospital of Athens. In total, 421 strains of H. influenzae were isolated during the whole study period: 119 in 2001–2002, 194 in 2002–2003 and 118 in 2003–2004. All isolates were tested for beta-lactamase production as well as for susceptibility to ampicillin, amoxicillin, amoxicillin + clavulanic acid (A/C), erythromycin, trimethoprime/sulfamethoxazole, ciprofloxacin and moxifloxacin, by the disk diffusion method, according to standard procedures.
Results: An increasing incidence of beta-lactamase producing strains was noted, rising from 10.9% in 2001–2002 to 19.4% in 2003–2004. In addition, there was an increase in ampicillin resistance rates, from 10.9% in the period 2001–2002, to 23.1% for the period 2003–2004, when 5/108 (4.6%) BLNAR strains were isolated. As a result to the presence of BLNAR isolates, the resistance rates to A/C was raised from 0% in the beginning, to 4.6% to the end of the period of study. Erythromycin resistance increased from 49.5% to 58.0%, while trimethoprime/sulfamethoxazole resistance decreased from 26.9% to 13.4%. No isolate was found resistant to quinolones.
Conclusions: From the beginning of 2001 to the end of 2004, a significant increase of H. influenzae resistance to ampicillin, amoxicillin, A/C and macrolides was noted, attributable to betalactamase production, as well as the emergence of BLNAR.
P1467: Resistance to antimicrobial agents in human Salmonella isolated in Bulgaria, 1999–2004
G. Asseva, P. Petrov, I. Ivanov, T. Kantardjiev (Sofia, BG)
Objective: To analyse the distribution of resistant Salmonella and resistance mechanisms among most frequently encountered serovars in Bulgaria: S. enteritidis, S. typhimurium, S. corvallis.
Methods: Culture, biochemical tests and serotyping for identification of strains. Screening for resistance to 14 antimicrobial agents: Cefotaxime, Cefoxitin, Carbenicillin, Ceftazidime, Cefuroxime, Cephalothin, Ampicillin, Amoxicillin/Clavulanic acid, Gentamicin, Tetracycline, Chloramphenicol, Ciprofloxacin, Nalidixic acid, Trimethoprim/Sulfamethoxazole with the standard Bauer-Kirby disk-diffusion method. Double disk synergy method (Jarlier et al.) was applied to determine ESBLs production. Transfer of bla-CTX-M and bla-TEM genes has been studied with experimental conjugation. For PCR detection of bla-CTX-M and bla-TEM genes specific primers have been used.
Results: For a study period 200 Salmonella strains out of 2123 were resistant to one and more antimicrobial agents;23 resistant strains were isolated from 163 confirmed cases during outbreaks and 177 of resistant strains were causing sporadic cases of human illness or carrier state. 104 ESBLs producers have been detected:5 S. enteritidis, 1 S. typhimurium, 1 S. isangi and 97 S. Corvallis with types of extended-spectrum beta-lactamases CTX-M3, TEM and SHV. All ESBLs producing strains were multiresistant to 9, 10 and 11 antimicrobial agents. Bla-CTX-M3 and bla-TEM genes were successfully transferred into a recipient E. coli C1A strain simultaneously with genes coding for resistance to aminoglycosides and sulfonamides (for bla-CTX-M3) and genes coding for resistance to aminoglycosides and chloramphenicol (for bla-TEM.PCR amplification revealed bla-CTX-M3 gene in S. Enteritidis and bla-TEM in S. corvallis. Before 1999 all S. Enteritidis were susceptible to all antimicrobial agents tested. In this study salmonellae have revealed resistance to at least one antimicrobial agent, most frequently to Nalidixic acid and Trimethoprim/sulfamethoxazole. Resistance to Nalidixic acid combined with retained susceptibility to ciprofloxacin in S. enteritidis is suggestive for mutations in the chromosomal gyrA. Resistance to ampicillin in S. Enteritidis could be explained with widely distributed plasmids in European countries including Bulgaria. Selection of multiresistant bla-TEM producing S. Corvallis is probably unique for our country.
Conclusions: Diversity of resistance genes are widely distributed among the leading causative agents of human salmonelloses in Bulgaria.
P1468: Gram-positive bacteria from diabetic foot ulcers and resistance to currently used antibiotics in a Greek general hospital
E. Platsouka, E. Perivolioti, M. Dimoutsos, H. Dimoudia, M. Lekka, Z. Doriza, O. Paniara (Athens, GR)
Objective: Patients with diabetic foot ulcers are usually treated with various antibiotics for a long period of time. In order to monitor the development of bacterial resistance because of the long treatment, Gram-positive isolates from diabetic foot ulcers were collected and tested to currently used antibiotics, such as glycopeptides, linezolid and quinopristin-dalfopristin.
Methods: One hundred eighty nine diabetic patients with foot ulcers were included. Aerobic and anaerobic cultures from the depth of the wound were obtained after surgical debridement. The isolates were identified by commercial ID panels. Susceptibility testing was performed by broth microdilution method (MIC panels) and E-test according to NCCLS guidelines.
Results: Of the two hundred forty one isolated bacteria, 81 were Staphylococcus aureus, 59 Enterococcus faecalis, 53 Staphylococcus epidermidis, 24 Streptococcus viridans, 6 Enterococcus faecium, 2 Streptococcus agalactiae and 16 Peptostreptococcus spp. Twenty-two (27.2%) of the S. aureus and 19 (35.8%) of the S. epidermidis isolates were methicillin resistant. Neither vancomycin-resistant enterococci nor glycopeptide-intermediate staphylococci were found. Three (3.7%) of the S. aureus isolates were found resistant to quinopristin-dalfopristin. Except for E.faecalis isolates (naturally resistant), we found two E.faecium strains resistant to quinopristin- dalfopristin. No resistance to linezolid was detected.
Conclusion: The most frequently isolated Gram (+) bacteria were S. aureus(33.6%) and E. faecalis (24.5%). The proportion of staphylococci resistant to methicillin is moderate. No resistance to glycopeptides and linezolid was detected. The resistance of S. aureus and E. faecium to quinopristin-dalfopristin is low. A close monitoring of isolates from diabetic foot ulcers is necessary as local conditions in these lesions might promote selection of resistant strains
P1469: Microbiological efficacy of antibiotics in patients with and without antibiotic pre-treatment of community-acquired pneumonia
T. Chernenkaja, M. Bogdanov (Moscow, RUS)
Objectives: To evaluate microbiological efficacy of antibiotics in patients with and without antibiotic pre-treatment of community acquired pneumonia (CAP).
Methods: In patients hospitalized with CAP in 2000–2003 it was evaluated the sensitivity of pathogens to erythromycin (ERY), amoxicillin (AMO), azythromycin (AZY) and 3rd generation cephalosporins (CEPH, cefotaxime or ceftriaxon) in sputum or blood or bronchial aspirate. Only the probes received within 3 first days of hospitalization were analysed; pathogens from patients with destructive pneumonia were excluded; atypical pathogens were not investigated. The identification and sensitivity tests were performed according NCCLS for respective period by disc-diffusion method and at ATB Expression (BioMerieux). The patients’ records were screened to evaluate the possible antibiotic treatment of current CAP before hospitalization. The records which do not contain clear statement for antibiotic pre-treatment or absence of such pre-treatment were excluded from analysis. The pathogens from patients with and without antibiotic pre-treatment were compared. The null hypothesis about absence of difference in antibiotics sensitivity in treated and non-treated patients was tested by Hi2.
Results: The number of etiologically insignificant pathogens increased from 29% in non-treated group (NTG) to 42% in antibiotic treated group (TG). Among etiologically significant pathogens the frequency of Enterobacteriaceae spp. grows from 12% in NTG to 23% in TG, changes in other pathogens were not significant on available sample. Total number of strains sensitive to AMO and AZY were significantly higher in NTG (69.5% and 75.9% respectively) than in TG (54.2% and 66.7% respectively). The drop in sensitivity to ERY in TG from 26.3% to 22.9% in TG and to CEPH from 89.5% to 87% in NTG and TG respectively were not significant. The investigated set of pathogens as well as their sensitivity could be shifted against other settings because the hospital admission policy facilitates the entrance of auto plant employees.
Sensitivity of pathogens in community acquired pneumonia in patients with and without antibiotic pretreatment
| Patients without antbiotic pretreatment |
Patients after antibotic pretreatment |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| % of sensitive strains |
% of sensitive strains |
|||||||||
| Pathogens | Abs. | AMO± | ERY | AZY± | 3rd CEPH | Abs. | AMO | ERY | AZY | 3rd CEPH |
| S. -pneumoniae | 13 | 92.3 | 100.0 | 100,0 | 92.3 | 3 | 66.7 | 100.0 | 100.0 | 66.7 |
| S. pyogenes | 4 | 100.0 | 100.0 | 100,0 | 100.0 | 4 | 75.0 | 100.0 | 100.0 | 75.0 |
| H. influenzae | 26 | 88.5 | 0.0 | 100.0 | 100,0 | 10 | 80.0 | 0.0 | 100.0 | 100.0 |
| Other Haemophilus sp. | 21 | 90.5 | 00 | 1000 | 100.0 | 11 | 90.9 | 0.0 | 100.0 | 100.0 |
| E. coli | 5 | 60.0 | 0.0 | 0,0 | 100.0 | 1 | 100.0 | 0.0 | 0.0 | 100.0 |
| Kl. pneumoniae | 4 | 0.0 | 0.0 | 0,0 | 100.0 | 5 | 0.0 | 0.0 | 0.0 | 100.0 |
| Other Enterobacteriacae | 2 | 50.0 | 0.0 | 0,0 | 100.0 | 5 | 20.0 | 0.0 | 0.0 | 80.0 |
| P. aeruginosa | 3 | 0.0 | 0.0 | 0,0 | 0.0 | 3 | 0.0 | 0.0 | 0.0 | 0.0 |
| Other non-fermenting bacteria | 1 | 0.0 | 0.0 | 0,0 | 0.0 | 0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Enterococcus sp | 1 | 100.0 | 0.0 | 0,0 | 0.0 | 0 | 0.0 | 0.0 | 0.0 | 0.0 |
| S. aureus | 11 | 9.1 | 45.5 | 456 | 72.7 | 4 | 0.0 | 75.0 | 75.0 | 100.0 |
| Staph. coaq (−) | 4 | 50.0 | 75.0 | 75.0 | 75.0 | 2 | 50.0 | 50.0 | 50.0 | 100.0 |
| Total | 95 | 69.5 | 26.3 | 75.8 | 89.5 | 48 | 54.2 | 22.9 | 66.7 | 87.5 |
* ± Total number of sensitive strains for amoxicillin and azythromycin significantly higher in patients without antibiotic pretreatment, p < 0.05
Conclusion: The AMO and AZY could be acceptable empirical choice antibiotics in CAP in NTG of hospitalized patents while in TG preferential choice is CEPH. ERY and other macrolides without anti-Haemophilus activity do not poses sufficient microbiological efficacy even in non-treated patients.
P1470: Microbiological efficacy of antibiotics in patients with different severity scores of community-acquired pneumonia
T. Chernenkaja, M. Bogdanov (Moscow, RUS)
Objectives: To evaluate microbiological efficacy of antibiotics in patients with different severity of community acquired pneumonia (CAP).
Methods: In patients hospitalized with CAP in 2000–2003 it was evaluated the sensitivity of pathogens to erythromycin (ERY), amoxicillin (AMO), azythromycin (AZY) and 3rd generation cephalosporins (CEPH, cefotaxime or ceftriaxon) in sputum or blood or bronchial aspirate. Only the probes received within 3 first days of hospitalization were analysed; pathogens from patients with destructive pneumonia were excluded; atypical pathogens were not investigated. The identification and sensitivity tests were performed according NCCLS for respective period by disc-diffusion method and at ATB Expression (BioMerieux). Modified Fine scores were used to evaluate the severity of pneumonia. The pathogens from patients with severity scores less than 70 (1st and 2nd classes) and more than 70 scores (3rd and 4th classes) were compared. The null hypothesis about absence of difference in antibiotics sensitivity in patients with different severity of CAP was tested by Hi2.
Results: In lower severity group (LSG) the predominant pathogens were Haemophilus spp. (56%), in higher severity group (HSG) they drop to 34%. The frequency of Streptococcus spp., non-fermenting gram-negative bacteria and Staphylococcus spp. was increased from 10%, 5% and 10% in LSG to 17%, 10% and 18% respectively in HSG. Nevertheless on the available sample there were no significant differences in total number of sensitive strains between severity groups; in LSG sensitivity was 64.8% to AMO, 17.6% to ERY, 73.6% to AZY and 87.9% to CEPH, in HSG it was 55,8% to AMO, 26,3% to ERY, 60.0% to AZY, 83.2%to CEPH. The investigated set of pathogens as well as their sensitivity could be shifted against other settings because the hospital admission policy facilitates the entrance of auto plant employees.
Sensitivity of pathogens in community acquired pneumonia of different severity
| Less than 70 scores |
70 and more scores |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Abs. | % of sensitive strains |
Abs. | % of sensitive strains |
|||||||
| Pathogens | AMO | ERY | AZY | 3rd CEPH | AMO | ERY | AZY | 3rd CEPH | ||
| S.pneumoniae | 6 | 66.7 | 100.0 | 100.0 | 66.7 | 11 | 100.0 | 100.0 | 100.0 | 100.0 |
| S. pyogenes | 3 | 66.7 | 100.0 | 100.0 | 66.7 | 5 | 100.0 | 100.0 | 100.0 | 100.0 |
| H. influenzae | 25 | 88.0 | 0.0 | 100.0 | 100.0 | 18 | 88.9 | 0.0 | 100.0 | 100.0 |
| Other Haemophilus sp. | 26 | 88.5 | 0.0 | 100.0 | 100.0 | 14 | 92.9 | 0.0 | 100.0 | 100.0 |
| E. coli | 5 | 80.0 | 0.0 | 0.0 | 100.0 | 4 | 50.0 | 0.0 | 0.0 | 100.0 |
| Kl. pneumoniae | 5 | 0.0 | 0.0 | 0.0 | 100.0 | 7 | 0.0 | 0.0 | 0.0 | 100.0 |
| Other Enterobacteriacae | 5 | 20.0 | 0.0 | 0.0 | 80.0 | 7 | 28.6 | 0.0 | 0.0 | 85.7 |
| P. aeruginosa | 5 | 0.0 | 0.0 | 0.0 | 0.0 | 5 | 0.0 | 0.0 | 0.0 | 0.0 |
| Other non-fermenting bacteria | 0 | 0.0 | 0.0 | 0.0 | 0.0 | 5 | 0.0 | 0.0 | 0.0 | 0.0 |
| Moraxella sp. | 0 | 0.0 | 0.0 | 0.0 | 0.0 | 1 | 0.0 | 100.0 | 100.0 | 100.0 |
| Enterococcus sp | 2 | 100.0 | 0.0 | 0.0 | 0.0 | 1 | 100.0 | 0.0 | 0.0 | 0.0 |
| S. aureus | 6 | 0.0 | 83.3 | 83.3 | 100.0 | 13 | 7.7 | 38.5 | 38.5 | 76.9 |
| Staph. coag(−) | 3 | 33.3 | 66.7 | 66.7 | 100.0 | 4 | 50.0 | 75.0 | 75.0 | 75.0 |
| Total | 91 | 64.8 | 17.6 | 73.6 | 87.9 | 95 | 55.8 | 26.3 | 60.0 | 83.2 |
Conclusion: Fine severity scores could not predict microbiological efficacy of antibiotics in hospitalized patients with CAP. The total number of pathogens sensitive to AMO and macrolides without anti-Haemophilus activity are not sufficient to legitimize these antibiotics as a fist line treatment of hospitalized patients with CAP; sensitivity to AZY is marginally acceptable; CEPH provide the best microbiological efficacy.
P1471: Resistance patterns of selected respiratory tract pathogens in Poland
A. Skoczynska, M. Kadlubowski, A. Klarowicz, W. Hryniewicz (Warsaw, PL)
Objective: To obtain the antimicrobial susceptibility data on major pathogens responsible for community acquired respiratory tract infections (RTI) in Poland.
Methods: In 2002–03 from 35 medical centres located in different parts of Poland 834 isolates were collected: 275 S. pneumoniae, 267 H. influenzae, 80 M. catarrhalis and 212 S. pyogenes. The first three species were isolated from patients with community acquired lower RTIs. S. pyogenes was isolated from patients with pharyngitis/tonsilitis. Isolates were identified to the species level by standard procedures and MICs were determined by the broth microdilution method according to the NCCLS guidelines.
Results: Among H. influenzae isolates 5.2% were resistant to ampicillin, via production of beta-lactamases, except 1 BLNAR isolate. All H. influenzae isolates were susceptible in vitro to amoxicillin/clavulanic acid, 3rd generation cephalosporins, azithromycin, fluoroquinolones and chloramphenicol, whereas 92.1%, 93.3%, 98.5% and 54.3% were susceptible to cefaclor, cefprozil, tetracyclines and trimethoprim/sulfamethoxazole, respectively. Although 83.6% of S. pneumoniae isolates were susceptible to penicillin G and this percentage remained similar during the two years tested, in the second year the proportion of resistant isolates with MIC ≥ 2 mg/l increased from 50 to 81%. The susceptibility of S. pneumoniae was as follow: amoxicillin (99.3%), cefaclor (82.5%), 3rd generation cephalosporins (94.2%), cefprozil (87.3%), macrolides (88.4%), ciprofloxacin (94.5%), levofloxacin (99.6%), clindamycin (91.3%), tetracyclines (82.2%), trimethoprim/sulfamethoxazole (46.2%) and rifampin (99.3%). Among M. catarrhalis isolates 91.3% were found to produce betalactamases. A significant number of S. pyogenes isolates was nonsusceptible to tetracycline (25.9%) and erythromycin (10.8%).
Conclusions: The most prevalent bacterial etiologic agents responsible for community acquired lower RTIs in Poland are S. pneumoniae and H. influenzae. Although the percentage of pneumococci nonsusceptible to penicillin G is stable, a higher proportion of isolates fully resistant to penicillin G may eliminate this drug from RTIs therapy caused by this pathogen. A very high percentage of S. pneumoniae and H. influenzae isolates was non-susceptible to trimethoprim/sulfamethoxazole.
P1472: Trends in antimicrobial susceptibility and serotype distribution of nontyphoidal Salmonella from chickens in four European countries during 1998–2004
A. de Jong, S. Friederichs (Leverkusen, D)
Objectives: In many countries, nontyphoidal Salmonella is a leading cause of food-borne illness in humans. Salmonellosis is usually self-limiting but antimicrobial treatment is recommended for severe illness, with fluoroquinolones and third-generation cephalosporins as drugs of choice. As concern was raised recently regarding poultry as a major source of fluoroquinolone-resistant Salmonella spp., a surveillance comprising two large EU regions was conducted.
Methods: In one programme, caecal samples were randomly taken at a Belgian slaughterhouse from chickens raised in Belgium, France or the Netherlands. Caecal contents of 5 birds were pooled to provide a single sample per flock. In another, carcass meat samples were collected from chickens processed in several processing plants across Germany. In total, 2856 Salmonella strains were isolated. Standardized susceptibility testing to ciprofloxacin (CP) and nalidixic acid (NA) as well as non-quinolones including ampicillin (AM), cefotaxime (CT), chloramphenicol (CA), gentamicin (GM), streptomycin (S), tetracycline (TE) and trimethoprim/sulfadiazine (TS) was performed by agar dilution. Resistance was assessed, if applicable, using NCCLS criteria.
Results: In all, 52 different serotypes were identified. The serotype prevalences differed strikingly between the two geographical areas as well as in time. In both programmes, resistance to CP was absent, except for one isolate (MIC 4 μg/ml). Decreased susceptibility was apparent but did not deteriorate, as indicated by stable CP MIC90 values around 0.25 μg/ml, and 32 and 9 % resistance to NA. Among the serotypes with decreased susceptibility S. hadar, S. virchow, S. blockley and S. paratyphi B were relatively the most frequent. In contrast, resistance to AM, S, TE and TS amounted to 46, 27, 33 and 18 % in the Belgian collection and to 16, 11, 16 and 12 % in Germany. CA resistance was 8 and 4%, respectively; GM resistance did not exceed 1 % in both programmes. Of 1832 isolates thus far tested for CT susceptibility, none have been resistant (MIC 50/90 of 0.12 to 0.25 μg/ml).
Conclusions: Resistance among Salmonella spp. from chickens varied for non-quinolones from 0 % for CT to considerable higher rates for some older drugs, while resistance to CP, particularly important for treating invasive salmonellosis in humans, approached zero. Decreased CP susceptibility varied markedly with the different serotypes, which prevalences differed notably in site and in time.
P1473: Integron class 1-determined antibiotic resistance in Enterobacteriaceae from blood stream infections in a Danish county
N. Norskov-Lauritsen, D. Sandvang, H.C. Schonheyder (Aarhus, Copenhagen, Aalborg, DK)
Objectives: To determine antibiotic resistance gene cassettes associated with class 1 integrons (Int1) in Enterobacteriaceae isolated from blood.
Methods: Consecutive blood isolates 1996–1998 of Enterobacteriaceae (N = 1362) from the county of Northern Jutland were investigated. Isolates expressing resistance to sulphonamides (N = 351) were examined for presence of Int1 by PCR amplification of a conserved fragment. Gene cassettes inserted in class 1 integrons were PCR amplified with primers located up- and downstream of the recombination site in Int1. Amplicons were allocated to types according to restriction fragment length polymorphism (RFLP) after digestion with MspI or HinfI. The gene cassettes were identified by sequencing of a representative from each type.
Results: Resistance to sulphonamides were expressed in 295 of 940 isolates of Escherichia coli (30%). One hundred and twenty-five were positive for presence of Int1, and they could be divided into 13 RFLP types. Resistance to sulphonamides were expressed in 56 of 422 isolates of non-E. coli (13%). Twenty-three were positive for presence of Int1, and they could be divided into 13 RFLP types, six unique and seven which were also found in E. coli.A total of 19 distinct Int1 types with one to three gene cassettes were found in the 148 isolates. Four types accounted for 72% of the isolates, and two of these were associated only with E. coli. The predominant Int1-determined antibiotic resistance was directed towards streptomycin/spectinomycin (seven distinct genes in 127 isolates) and trimethoprim (eight distinct genes in 70 isolates). Six isolates carried a sat1 gene conferring resistance to the veterinary antibiotic streptotricin. The number of class 1 integrons carrying genes conferring resistance to β-lactam antibiotics (four) or gentamicin (three) was conspicuously low.
Conclusions: Presence of class 1 integrons and acquired resistance to sulphonamides were 2–3 times more prevalent in E. coli than in other representatives of Enterobacteriaceae. Int1-determined antibiotic resistance in this collection of isolates was predominantly directed towards streptomycin/spectinomycin and trimethoprim, i. e. antibiotics which has been in clinical use for a long time.
P1474: Molecular basis of the high resistance rates unrelated to antimicrobial exposure detected in faecal Escherichia coli from the human population of a remote rural Bolivian community
L. Pallecchi, C. Lucchetti, A. Bartoloni, F. Bartalesi, H. Gamboa Barahona, A. Mantella, S. Cresti, M. Strohmeyer, F. Paradisi, G.M. Rossolini (Siena, Florence, I; Camiri, BOL)
Background: In a recent study, unexpectedly high resistance rates unrelated to heavy antimicrobials consumption were detected in the commensal E. coli of the population of a very remote rural community in Bolivia. The purpose of this study was to analyse the molecular basis of the drug-resistance in isolates collected from that setting.
Methods: 113 drug-resistant E. coli isolates, collected from 72 healthy individuals, were subjected to clonal analysis by means of RAPD. Isolates presenting the most common multi-drug resistance (MDR) pattern (ampicillin, tetracycline, chloramphenicol, and trimethoprim-sulfamethoxazole: A/T/C/S pattern) (n = 35) were investigated for the nature and transferability of resistance determinants, and for plasmid profiles.
Results: RAPD results identified 19 clonal groups, with one largely prevalent (34 isolates). Intra-familial clonal clustering was often observed, and in one case the same MDR strain was detected in the fecal samples of all the members of a family (8 subjects). Heterogeneous resistance phenotypes were observed within most clonal groups, underlying a relevant role of transferable resistance determinants. Clonal and plasmid variability was observed in the 35 MDR isolates showing the A/T/C/S resistance pattern:i) 20 belonged to the most prevalent clonal group, showed colicin production and harboured the tetA gene. Transferability of resistance determinants could not be detected in these strains;ii) 7 belonged to a different clonal group and harboured a conjugative plasmid carrying the tetA gene linked to beta-lactam, chloramphenicol and trimethoprim-sulfamethoxazole resistance determinants and to genes involved in colicin production;iii) 8 belonged to further 5 different clonal groups, did not show colicin production and harboured tetB genes, often located on conjugative plasmids with different restriction profiles and different carriage of resistance determinants.
Conclusions: The clonal and plasmid diversity suggests that the presence of MDR isolates in this particular setting is due to a mechanism more complex than simple dissemination of a single clone or a single resistance plasmid occasionally introduced into the community.
Fungal diagnostics
P1475: Sequence-based identification of zygomycetes species of medical interest
P. Schwarz, S. Bretagne, A.S. Delannoy, F. Dromer, O. Lortholary, E. Dannaoui (Paris, F)
Objectives: Zygomycosis is a life-threatening disease with increasing incidence in immunocompromised patients. Several fungi are responsible for these infections but identification to the species level remains difficult by conventional mycological techniques. The aim of this study was to investigate the usefulness of rDNA sequencing for species identification of zygomycetes.
Methods: 34 isolates, mostly of clinical origin, belonging to the order Mucorales were tested. These included the 7 species (belonging to 5 genera) most commonly responsible for zygomycosis in humans. Mycelium was grown in liquid RPMI 1640 medium and complete genomic DNA was extracted with CTAB/Chloroform. PCR-amplification of the ITS1-5.8S-ITS2 rDNA region was performed with the primer set V9D (5′-3′, TTAAGTCCCTGCCCTTTGTA) and LS266 (3′-5′, GAGTCGAGTTGTTTGGGAATGC). After amplification, both strands sequencing of the fungal DNA was performed and sequences were aligned and compared with ClustalW.
Results: Sequences of ca. 600–800 bp were obtained and analysis showed that all species studied (except Absidia corymbifera) were homogeneous with ≥ 99% similarity between strains within a given species. In contrast, sequence variability between genera (similarities of ≤68%) and between species (similarities of ≤ 82%) allowed a precise identification to the species level. In particular it was possible to differentiate Rhizopus oryzae from R. microsporus and to discriminate between the different Mucor spp. Only the sequences for the 5 A. corymbifera were heterogeneous with 60 to 100% similarities between strains. Interestingly, none of the studied species owned the universal fungal primer sequence ITS 2 / ITS 3.
Conclusions: The results showed that sequencing of the ITS1-5.8S-ITS2 region is a reliable molecular tool for identification of agents of zygomycosis to the species level. For strains identified as A. corymbifera, a higher number of isolates have to be studied. The molecular data obtained here are promising for development of new diagnosis methods of zygomycosis in patients.
P1476: Molecular identification of zygomycetes species in tissues during experimental zygomycosis
P. Schwarz, S. Bretagne, A.S. Delannoy, F. Dromer, O. Lortholary, E. Dannaoui (Paris, F)
Objectives: Culture of infected tissues during zygomycosis is often negative. In these cases the only available diagnostic tool is histopathology demonstrating large, non-septated hyphae characteristic of zygomycetes. Nevertheless, identification at the genus or species level is not possible by conventional histopathology. The aim of this study was to evaluate the usefulness of rDNA sequencing for species identification in tissues of mice infected with various zygomycetes.
Methods: Mice were infected intravenously with nine different species of the order Mucorales (Rhizopus oryzae, R. microsporus var rhizopodiformis, Absidia corymbifera, Mucor indicus, M. circinelloides, M. racemosus, Rhizomucor pusillus, Cunninghamella bertholletiae, and Syncephalastrum racemosum). Different inocula were tested ranging from 104 to 107 sporangiospores/mouse. Mice challenged with C. bertholletiae and S. racemosum received 100 mg*kg−1*d−1 of hydrocortisone subcutaneously before infection. Mice were sacrificed 3–4 days post infection. Brain and kidneys were aseptically removed, homogenized and complete genomic DNA was extracted with CTAB/Chloroform. Fungal DNA was PCR-amplified with the universal fungal primer set V9D (5′-3′, TTAAGTCCCTGCCCTTTGTA) and LS266 (3′-5′, GAGTCGAGTTGTTTGGGAATGC). After amplification, both strands sequencing of the fungal DNA was performed and sequences were aligned and compared with ClustalW.
Results: Active infection demonstrated by the presence of hyphae in brain and kidneys was obtained for all tested species except for M. racemosus and fungal DNA was amplified directly from tissues with all the eight former species. Furthermore for six species, sequencing of the ITS1-5.8S-ITS2 region allowed identification of the infecting strain to the species level while technical problems impaired analysis of the sequences of C. bertholletiae and S. racemosum.
Conclusions: We set-up animal models of zygomycosis for the most common species responsible for infections in humans and demonstrated that extraction, amplification and sequencing of fungal DNA is possible directly from infected tissues. These animal models are valuable to evaluate new diagnostic tools for zygomycosis. Further studies with paraffin-embedded tissue samples are warranted.
P1477: Molecular identification of black grain mycetoma agents
M. Desnos, S. Bretagne, A.S. Delannoy, F. Dromer, O. Lortholary, E. Dannaoui (Paris, F)
Objective: Black-grain mycetoma are subcutaneous chronic infections mostly seen in tropical countries. Several dematiaceous fungi can be causative agents of this disease. Identification of these fungi with standard mycological procedures is difficult because of poor or delayed sporulation. Furthermore it is time-consuming and required expertise restricted to some reference laboratories. Thus new diagnostic tools are needed. The aim of this study was to assess the accuracy of molecular identification of the different fungi responsible for black-grain mycetoma.
Methods: A total of 48 strains, mostly of clinical origin, have been used including 11 Madurella mycetomatis, 4 Madurella grisea, 13 Leptosphaeria senegalensis, 4 Leptosphaeria tompkinsii, 5 Pyren-ochaeta romeroi, 4 Curvularia lunata, and 7 Exophiala jeanselmei. Strains were cultured in liquid RPMI medium and DNA was extracted from the mycelium by DNEasy plant kit (Qiagen). ITS1-5.8S-ITS2 region DNA was then PCR amplified by using the universal fungal primers LS266 and V9D. Both strands sequencing was performed, sequences were aligned with ClustalW and both intra- and inter-species sequence similarities were assessed. Aligned sequences were also used to generate phylogenetic trees.
Results: M. mycetomatis appeared to be a homogeneous species, with 91 to 100% homologies between strains (except for one strain that differed significantly and raises the issue of its identification). Similarly, few intra-species variations were found for C. lunata and E. jeanselmei, with 93 to 100% homologies between strains. L. senegalensis and L. tompkinsii showed intra-species similarities of >99% but similarity between the two species was < 90%. In contrast, P. romeroi and M. grisea appeared heterogeneous with intra-species similarities of 40 to 100% and 53 to 100%, respectively. Inter-genera and inter-species variations were important (except between P. romeroi and M. grisea) with sequence homologies of < 80% between genera.
Conclusions: P. romeroi and M. grisea are heterogeneous species whereas within the other species a high degree of sequence homologies are observed. These results show that sequencing of ITS region is a suitable tool for identification of black grains mycetoma agents usually difficult to identify by standard phenotypic methods.
P1478: Rapid diagnosis of PCP and resistance to co-trimoxazole using real-time PCR
K.E. Templeton, J. Gooskens, E.C.J. Claas, E.J. Kuijper (Leiden, NL)
Objectives: To compare conventional microbiological staining methods with conventional PCR and real-time PCR methods for diagnosis Pneumocystis jiroveci in patients suspected for pneumocystis pneumonia (PCP) and to assess positive samples for the presence of co-trimoxazole(Co-T) resistant mutations.
Methods: Eighty-four sequential bronchoalveolar lavage samples from patients analysed by methanamine silver and Giemsa staining methods were stored at −70°C and subsequently analysed by PCRs. The BALs were collected over a 20-month period. The real-time PCR was designed to the dihydropteroate synthase (DHPS) to be specific for P. jiroveci and enable sequencing of the PCR product to determine resistance mutations at nucleotide positions 165 and 171. A conventional PCR to the mitochondrial large sub-unit rRNA described by Wakefield et al (Lancet. 1990;336:451-3) was also performed on all 84 BALs.
Results: The staining methods showed that 16/84 (19%) patients had PCP. The real-time PCR and the conventional PCR both detected P. jiroveci in 19/84 (22.6%). All the samples positive by the staining method were positive and the 3 additional positives were detected by both PCR methodologies. Using the staining methods as the ‘Gold Standard’, the sensitivity, specificity, positive predictive value and negative predictive value for the PCR methods were 100%, 96%, 84% and 100% respectively. The mean cycle threshold (Ct) value for the 16 stain positives was 31.5 and for the 3 stain negative/PCR positives was 41.5. Analysis of the clinical records and microbiological results of the 3 discrepant samples showed that no other microbiological agent was found and PCP was the most likely clinical diagnosis. The 19 positives were all sequenced and no resistance mutations were found. The time to perform the different methods was 2.5 hours, 3 hours and 5 hours for the staining methods, real-time PCR and conventional PCR. The sequencing could be performed directly after the real-time PCR and results were available in one working day.
Conclusions: Real-time PCR for P.jiroveci can provide rapid, sensitive and specific diagnosis for PCP in BAL samples. Additionally the Ct value may be useful in determining the amount of infection. Using this method targeting the DHPS gene rapid results can also be obtained for resistance to Co-T.
P1479: Identification of Trichosporon mucoides isolates using ID-32C tests (BioMérieux) and direct DNA sequencing of internally transcribed spacer (ITS) 1 and ITS2 of rDNA
U. Nawrot, P-A Fonteyne, F. Fauche, N. Nolard (Wroclaw, PL; Brussels, B)
Objectives: The identification system provided by BioMérieux distinguish three out of the 35 species published so far for to the genus Trichosporon. The aim of this study was to investigate the genetical homogeneity of the strains identified according to ID-32C as Trichosporon mucoides (Guého, 1992).
Strains: The study was performed on 74 strains from the BCCM-IHEM collection from clinical (56) environmental (10) or (8) unknown origin. The strains had been classified according to obsolete identification schemes as T. beigelii (38 strains), T. mucoides (22), T. cutaneum (8), T. ovoides (2) and T. sp (4). As quality control typical strains T. mucoides IHEM13922, T. ovoides IHEM 19546, T. moniliiforme IHEM 19572, T. jirovecii IHEM 19577, and T. asahii IHEM 19578 were used.
Methods: The ID-32C tests were prepared according to manufacturer description. The results were read after 48 h, 72 h and 6 days of incubation at 30 degree C by use ATB automatic reader (Bio Merieux, France). The complete DNA sequence of the ITS locus in the rRNA genes was established trough direct sequencing of PCR products. A ‘molecular’ identification was proposed on the basis of comparison with sequences available in public databases.
Results: After two days incubation, the ID-32C tests identified 68 strains as T. mucoides (all test positive except laevulinate assimilation) and 6 strains as C. humicolus (all tests positive). Interestingly, after 6 days incubation time a shift from T. mucoides to C. humicolus was observed for 11 strains. The analysis of ITS sequence indicated 10 different sequevars for the 74 strains. Four sequevars representing 68 strains were considered compatible with T. mucoides, with one of them also identical to the ITS sequence published for T. dermatis (Sugita, 2002). The remaining 6 sequevars were considered as compatible with T. asahii, T. jirovecii, T. laibachii, T. moniliforme, or T. debeurmanianum.
Conclusion: ID32C and ITS were compatible for 62 strains. Six strains identified as T. mucoides had incompatible ITS sequences. Six strains with T. mucoides ITS sequences had incompatible ID32C results and were identified as C. humicolus. Considering ITS sequencing as the golden standard ID32C had a sensitivity of 91% and specificity of 91%.
P1480: Molecular characterisation of beta-tubulin gene in dermatophyte pathogen Trichophyton rubrum
S. Rezaie, K. Zomorodian, M. Rezaian (Tehran, IR)
Trichophyton rubrum (T. rubrum) is an cosmopolitan arthropophilic dermatophyte which causes skin and nail infection. The most of antifungal drugs available for clinical use are targeted four molecules including: beta-glucan, sterol-alpha-demethylase, ergosterol and DNA/RNA synthesis. The limited selection of effective antifungal agents combined with the emergence of drug resistance, has resulted in a critical need for development new drugs directed against novel molecular targets. In the present study we tried to Characterize the Gene encoded Beta-tubulin protein in this fungus which can be used as a potential molecular targets of novel drugs. Clinical isolated of T. rubrum were cultured on liquid medium and the Nucleic Acids (DNA & RNA) were isolated from obtained mycelial mass by standard methods. The sequences of known Beta-tubulin genes in other fungi were aligned and pairs of 21 nt primers were designed from highly conserved regions. Using mentioned primers, we amplified predicted molecules by using genomic DNA as well as cDNA of T.rubrum as PCR templates. By the time, 1200 nucleotides have been sequenced from this new gene which encodes a polypeptide with 400 amino acids. Nucleotide sequence comparison in gene data banks (NCBI, NIH) for both the partial DNA and its deduced amino acid sequence revealed significant homology with members of the eukaryotic Beta-tubulin genes. The amino acid sequence of the encoded protein was about 82% identical to the sequence of Beta-tubulin proteins from other fungi. Considering the role of Tubulin protein in formation of mitotic spindle structure in the cells, the Beta-tubulin gene may be applied in antifungal research fields. Moreover it can be used to determining the molecular action of some drugs including mitotic spindle structure in fungi, leading to metaphase arrest. The probable mechanisms of Beta-tubulin expression inhibition as well as definition of its possible role in the physiological function of T. rubrum are still under investigation.
P1481: A rapid PCR-RFLP assay for identification of 11 medically important Candida species, Trichosporon asahii, Cryptococcus neoformans, Saccharomyces cerevisiae, Hansonula anomala and Geotrichum candidum
H. Mirhendi, K. Makimura (Tehran, IR; Tokyo, JP)
Opportunistic yeast infections have various clinical features and are caused by several Candida species and by several other yeasts belong to other genera. The emergence of these mycoses is increasing, for example deep-seated candidiasis has increased dramatically in recent years. Rapid identification of yeast isolates to the species level in the clinical laboratory is necessary to more rapid and effective antifungal therapy and to facilitate hospital infection control measures. Conventional phenotype-based methods for identifying yeasts species are often difficult and time consuming. Molecular biological techniques provide useful alternative methods. In this study, ITS1-5.8S-ITS2 region of the fungal rRNA genes were amplified in 31 yeast standard strain and 137 clinical Candida isolates with the universal primers ITS1 and ITS4. Digestion the PCR product by the restriction enzymes HpaII and AcsI allowed us to clearly identify C. albicans, C. glabrata, C. parapsilosis, AC. tropicalis, AC. krusei, C. dubliniensis, C. guilliermondii, C. lusitania, C. rogosa, C. famata, C. kefyr, Trichosporon asahii, Cryptococcus neoformans, Saccharomyces cerevisiae, Hansonula anomala and Geotrichum candidum. All clinical isolates were checked by cultivation on Chrome-agar Candida. This panel of PCR-restriction enzyme could be rapid, simple and useful in diagnostic and epidemiological studies of Candida and candidiasis and the infections caused by other yeasts, without any requirement of expensive equipments.
P1482: A simple PCR-RFLP method for identification and differentiation of eleven Malassezia species
M. Mahzounieh, H. Mirhendi, K. Makimura, K. Zommorodian, M. Keivanlu (Shahrekord, Tehran, IR; Tokyo, JP)
Malassezia species are part of resident skin flora of human and other warm-blooded vertebrates. These yeasts are associated with various superficial diseases including pityriasis versicolor, seborrheic dermatitis and folliculitis, as well as nosocomial bloodstream infection in paediatric care units. Although various DNA-based molecular methods have been described, a simple, reliable and cost effective method is still needed for differentiation of Malassezia species. In this study, a PCR-RFLP method using one primer pair to amplification of a 580 bp fragment related to 26S rDNA and using 2 restriction enzymes, CfoI and BstF51 was developed to clearly identify and differentiate 11 Malassezia species including M. furfur, M. pachydermatis, M. globosa, M. obtuse, M. restricta, M. sympodialis, M. dermatis, M. slooffiae, M. nana, M. japonica and M. yamatoensis. Malassezia type and standard strains were examined to verify the method. Thirteen clinical isolates were also identified in this study. The results of PCR-RFLP analysis of clinical isolates were completely comparable with those from DNA sequencing techniques. This method enables the cost effective, rapid and reliable identification of Malassezia species and therefore it could be suitable for laboratory applications.
P1483: Mitochondrial DNA variability of human pathogenic Trichoderma isolates
Z. Antal, J. Varga, L. Kredics, A. Szekeres, L. Manczinger, C. Vàgvölgyi, E. Nagy (Szeged, HUN)
Objectives: Trichoderma species are filamentous fungi commonly found in soils. Although Trichoderma species have been known for a long time as nonpathogenic microorganisms, in the last two decades some Trichoderma strains were identified as causative agents of opportunistic fungal infections with increasing frequency. Among them, Trichoderma longibrachiatum isolates are reported predominantly to cause health problems in humans ranging from localized infections to fatal disseminated diseases. The aim of this study was to investigate and compare the mitochondrial DNA variability of T. longibrachiatum strains isolated from clinical and soil samples.
Methods: Mitochondrial DNA (mtDNA) RFLPs of 8 saprophytic and 9 clinical T. longibrachiatum isolates were examined. For mtDNA characterization total DNA was isolated from mycelia and digested with BsuRI and Hin6I restriction enzymes. The mtDNA profiles were converted to a similarity matrix, and genetic distances were calculated to create a dendrogram.
Results: Based on Hin6I RFLP profiles, the saprophytic T. longibrachiatum isolates belonged to 3 different haplotypes, while clinical isolates exhibited 7 different haplotypes. Using BsuRI, the saprophytic and clinical isolates belonged into 3 and 5 different haplotypes, respectively. Interestingly, some saprophytic isolates collected in Hungary exhibited the same mtDNA patterns as isolates from Egypt and UK. The sizes of the mtDNAs varied between 35.1 and 39.5 kBp. This size variability is possibly caused by loss of introns or intergenic sequences. Although on the dendrogram constructed based on mtDNA RFLP profiles 5 clinical isolates form a well-defined group, other saprophytic and clinical isolates do not form separate clusters.
Conclusion: The high intraspecific variability of mtDNAs could be partly due to the higher evolutionary rates of mtDNAs compared to nuclear DNA. Accordingly, the discriminatory power of the mtDNA RFLP method was found to be higher than that of some previously used techniques including sequence analysis of the ITS region, isoenzyme analysis or carbon source utilization tests. Besides, the method is more reliable than the frequently used RAPD technique. Nevertheless the saprophytic and clinical isolates did not form separate clusters on the dendrogram, suggesting that every environmental isolate could have the capacity to cause infection. This work was supported financially by grant F037663 of the Hungarian Scientific Research Found.
P1484: A novel rapid vaginal yeast infection test
Z. Greenberg, M. Illouz, R. Margalit, J. Kopelowitz, M. Shaposhnikov, O. Babai, B. Chertof, M. Lee (Ashdod, IL)
Objectives: Vulvovaginal candidiasis (VVC) is thought to be one of the most common causes of vaginal infections. Currently, the diagnosis of VVC relies on microscopy, which has been shown to have a limited sensitivity and may be subject to significant operator-error. Yeast cultures, the gold standard for diagnosing VVC, are expensive, underutilized, and may take a week to give positive results. A rapid test for vaginal yeast would greatly aid in the diagnosis of VVC, but no such test currently exists. The need for a rapid test is further emphasized in view of availability of over-the-counter anti-fungal medications. The use of a home-test will avoid misuse of medications and may reduce health-care costs.
Methods: Savyon Diagnostics has recently finalized the development of a diagnostic kit for rapid detection of vaginal Candida infections. The test is intended to be used as a laboratory, point of care and over-the-counter test, meaning that vaginal secretion samples may be obtained through either patient or physician sampling. The kit is composed of a sampling swab and a unique detection device, which houses a test strip, functioning as a lateral flow immuno-chromatographic-based test. The novel device enables extraction of the Candida antigens from the sampling swab, in-device dilution, transfer of the sample liquid into the detection strip via an innovative mechanism, and providing clear results in short time. All in two steps.
Results: The test provides clear, reliable and unchangeable results within 5 minutes. It detects Candida albicans, which is the most abundant species, but also others like C. glabrata, the secondary in abundance. Sensitivity and specificity have been shown to be comparable to the results of yeast culture taken as a reference method. The results are also in accordance with clinical symptoms. Precision tests further validate the test quality. The test kit has been proven to be user friendly, as shown by the similarity between the results obtained by physician sampling vs. patient self-sampling. Surveys have shown high contentment of potential users in regard to the device operation as well as its innovative shape.
Conclusions: A novel kit including a unique device has been developed to fill the widely desired need for a rapid and reliable diagnosis of vaginal yeast infections. The kit may be used either at home or in the clinic, may avoid misuse of antifungal drugs, and save health-care costs.
P1485: Rapid detection of yeast infections in patients with febrile neutropenia by PCR-RFLP
F. Ruzicka, V. Hola, R. Horvath, I. Kocmanova, J. Sterba, R. Tejkalova, M. Votava, V. Woznicova (Brno, CZ)
Yeasts are frequent pathogens in patients with febrile neutropenia. Very serious complication is Candida sepsis for the patients, with the mortality rate up to 60%. Early detection and identification of fungal pathogens is essential for initiation of effective antifungal therapy. Conventional methods of detection and identification of the infectious agent, such as cultivation, microscopy and biochemical identification, are time consuming and may take several days or even weeks. For that reason the use of fast methods, which are able to detect fungal infection within hours, is essential for successful treatment of infection. The detection of fungal DNA or fungal antigens are the perspective methods. Three hundred and three blood samples from 57 patients with febrile neutropenia were examined by a PCR-based assay. The primers for detection of ITS2 sequence and adjacent regions were prepared. The identification of captured agents was performed by Amplification Product Length Polymorphism and by Restriction Fragment Length Polymorphism methods. A total number of positive samples was 15. (C. albicans (8), C. krusei (1), C. glabrata (2), R. rubra (2) and 2 unidentifiable fungi. The same agent was detected repeatedly in 3 patients. In 2 patiens the agent isolated from second blood culture was different from that isolated from the first one. In 4 cases out of 10 the results corresponded well with results of detection of mannan antigen or antimannan antibodies in serum of the patients. In no cases the blood sample was positive by cultivation. The possible cause of negative cultivation was antifungal therapy in most of these patients which could supress the growth of yeasts in blood cultures. Our results suggest that the combination of PCR detection of fungal DNA with the proof of mannan antigen and antimannan antibodies can help in the diagnostics of fungal blood stream infections, especially in patients under antimycotic therapy. This work was supported by the grant agency IGA MZ NR/7980-3.
P1486: Value of antigen detection in the diagnosis of invasive aspergillosis
A. Grapsa, S. Kartali, M. Kantzanou, D. Margaritis, S. Karavassiliadou, M. Panopoulou, E. Archondidou, G. Bourikas (Alexandroupolis, GR)
Introduction: Invasive aspergillosis (IA) has been a significant cause of life-threatening opportunistic infections in immunosuppressed hosts. Early diagnosis is important to achieve the best outcome for these patients; however, definite proof often is difficult to obtain due to counterindicated invasive procedures. Serum galactomannan detection is considered to be a useful test for early diagnosis and follow–up of invasive aspergillosis.
Methods: This study evaluated the specificity and sensitivity of the detection of galactomannan(GM) for the diagnostic of IA in 98 adult patients hospitalized in our Hematology unit. Patients were considered to have confirmed or probable invasive aspergillosis, based on clinical and radiological data. Serial screening of Aspergillus GM circulating antigen was evaluated using a double sandwich ELISA assay (Platelia Aspergillus, BioRad, France) on 760 sera. Test positivity was defined in accordance with the manufacturer's recommendations.
Results: Among the patients studied, 10 (10.2%) presented with confirmed IA (n = 8 patients) or probable IA (n = 2 patients). Seven patients (7.1%) having a positive result (OD index >1.5) in two consecutive Platelia Aspergillus tests were considered galactomannan − positive cases. No antigens were detected in the sera from one patient with confirmed IA. Latex agglutination assay of galactomannan was positive in both patients with probable IA. In patients without IA, 9 of 88 had positive antigenemia. Sensitivity (70%), specificity (89%), were comparable to those of larger series. Circulating antigens were not detected in the control group, composed of healthy adults. The gradual rise in the antigen titer in consecutive samples is a very strong indication that an infection is present and should be taken into account when interpreting the results.
Conclusion: The detection of the circulating Aspergillus galactomannan antigen by a sandwich enzyme-linked immunosorbent assay (ELISA) is one of the most promising method to diagnose IA in at-risk patients. The presence of antigen has a good diagnostic value mainly when there is an increase in the titer on two consecutive sera samples. A repeated negative result is a strong argument against the diagnosis of IA.
P1487: Inhibition controls are not always necessary for PCR on human samples
T. Barkham, V. Rajagopal (Singapore, SGP)
Objective: To assess the need for inhibition controls in diagnostic PCR on human serum and on blood cultures in BactAlert FAN media.
Methods: Dengue RT-PCR was performed daily for 6 months on RNA extracted from serum (Qiagen viral RNA minikit). The PCR kit (Artus GmbH Germany) included an internal control for every sample and was performed on a lightcycler (Roche). We performed an in-house multiplex gel based PCR assay with primers aimed at C.albicans, C. glabrata, C.tropicalis and C. parapsilosis. The PCR was applied, without DNA extraction, to the supernatant from BactAlert FAN bottles that had been left to stand to allow the charcoal particles to settle. These bottles had flagged positive and been shown to contain yeasts by Gram's stain. Neat and doubling dilutions in saline to 1:8 were tested.
Results: The inhibition control was positive for all sera tested with the Artus Dengue RT-PCR kit, meaning that the PCR process was successful without significant inhibition in any of the samples. Inhibition was detected using neat broth from BactAlert FAN bottles but was not detected at a 1:2 dilution.
Conclusion: Inhibition of PCR performed on sera is not common and it is unnecessary to control for inhibition. PCR on the supernatant from BactAlert FAN bottles is subject to inhibition. However, PCR was successful on all supernatants diluted 1:2. This assay was used on samples known to contain yeasts, demonstrated by Gram's stain, so an absent signal would suggest inhibition or a species not recognised by the primers. While a negative result might delay the identification to species level and refinement of antifungal chemotherapy, it would not delay the diagnosis of candidaemia. This data supports the view that inhibition controls are unnecessary for PCR applications validated to perform adequately. Furthermore, inhibition is a considerable problem with bacterial cultures but it is not standard practice to detect and report this inhibition, so it would be inconsistent to make inhibition controls obligatory for nucleic acid tests.
P1488: Comparison of histoplasma antigen EIA performed at MiraVista Diagnostics and Statens Serum Institute shows good correlation
M.C. Arendrup, L.J. Wheat (Copenhagen, DK; Indianapolis, USA)
Objective: Histoplasmosis is a systemic fungal infection caused by inhaling the spores of the dimorphic fungus Histoplasma capsulatum. The Histoplasma antigen EIA is useful for the diagnosis of histoplasmosis but is not widely available outside the United States. The Histoplasma antigen EIA was first described in 1986 and has been performed for clinical diagnostic purposes since 1988; it is currently available at MiraVista Diagnostics (MVD). Recently, a pilot kit was produced at MVD and sent to Statens Serum Institute (SSI) for a comparative study that would assess the feasibility of offering the Histoplasma antigen EIA to the European medical community at SSI.
Methods: A test panel of 18 samples included two negative controls, one high positive control, one low positive control, five healthy donor urine samples, five urine samples from known histoplasmosis cases, and four samples of a dilution series of galactomannan antigen purified from Histoplasma capsulatum cultures was evaluated at SSI and MVD on two separate days. Briefly, the EIA is a sandwich style system that uses rabbit polyclonal antibody to Histoplasma antigen as capture; plates are then blocked and allowed to dry. Test samples are added, allowed to incubate and washed off. Next, the same polyclonal antibody used as capture but containing a biotin label is added followed by streptavidin-horse radish peroxidase. After incubation, the conjugates are washed off, and (3,3′,5,5′-tetramethyl benzidine) TMB is added as substrate. The reaction is stopped using 2N H2SO4, and the endpoint optical density (OD) is read at 450 nm-630 nm.
Results: Intralaboratory correlation was good with a single sample of purified galactomannan antigen testing positive on day one and negative on day two in both SSI and MVD labs. Interlaboratory correlation was excellent, comparing results from both labs for day two, each of the 18 samples and controls resulted in an identical result interpretation according to these guidelines where results are expressed as EIA units (EU): <1.0 EU is negative (n = 8); >1.0 − <2.0 EU is weak positive (n = 1); >2.0 − <10.0 EU is moderate positive (n = 4); and >10.0 EU is high positive (n = 5). All results remained within these categories at both laboratories.
Conclusion: The excellent correlation of results supports the feasibility of developing a Histoplasma antigen EIA test kit that can be used for reference testing at SSI.
P1490: Blind subculture increases only marginally detection of fungaemia in high-risk patients
M. S0gaard, U. Hjort, T. H0jbjerg, H.C. Sch0nheyder (Aalborg, DK)
Objectives: The ability to detect fungaemia with bacteriological blood culture (BC) media and an automated system may be questioned. We investigated retrospectively whether blind subculture on the 3rd day increased detection of fungaemia with the BacT/Alert system or accelerated the diagnosis in high risk patients.
Methods: This historical cohort study was conducted at a university hospital and its affiliated hospitals. During a period of app. 5 years (Nov. 1998 to Dec. 2003) BCs were subcultured on the 3rd day of incubation if patients based on clinical and microbiological assessment were believed to be at high risk for fungaemia. For adult patients each BC set comprised one standard aerobic, one FAN aerobic and one standard anaerobic bottle (Bac T/Alert system, bioMerieux); aerobic bottles were vented at reception, and the incubation period was 6.7 days. All data were recorded in a departmental information system.
Results: 79.165 BCs were drawn during the study period. 359 (0.45%; 95% confidence interval (CI): 0.41-0.50%) yielded growth of yeast species (C. albicans 62.8%, C. glabrata 14.9%, C. parapsilosis 9.0%, C. tropicalis 3.3%, C. krusei 3.3%, other yeast species 6.8%). A total of 2154 BCs were selected for subculture originating from 170 patients, 89 of whom were admitted to ICUs. 102 BCs (4.7%; 95% CI: 3.9–5.7%) from 51 patients turned out yeast positive. However, blind subculture was instrumental in detecting yeasts in only 26 BCs (25.5%). Eight BCs (7.8%) were negative on blind subculture, but one or more bottles were detected positive by the BacT/Alert system during continued incubation. In 68 BCs (66.7%) growth of yeasts was detected during the first three days of incubation. Contrary to our expectations, we found time to detection to be shorter in cases not selected for subculture. In this group 84.9% (95% CI: 79.9–89.1%) of the yeast positive BCs were detected within 3 days as compared to 66.7% (95% CI: 56.6–75.7%) for the subculture group as stated above.
Conclusion: Patients believed to be at high risk and thus selected for the subculture group proved to have an approximately 10 times increased rate of yeast positive BCs. However, blind subculture on the 3rd day disclosed only a quarter of yeast positive BCs in this group. Due to the very limited increase in detection we do not recommend blind subculture as a precaution in patients at high risk for fungaemia.
ELISA-based diagnostics
P1491: Clinical-laboratory correlation study on the diagnosis of Echinococcus granulosus in Albania E. Cani (Tirana, AL)
Objective: To evaluate the role of different serological tests in the diagnosis of Echinococcosis in correlation with the data obtained from clinical and imaging examination.
Methods: A total of 358 people, 139 males and 219 females, underwent a serological testing for the diagnosis of Echinococcosis in the Laboratory of Parasitology of IPH, Tirana, from September 1997 to May 2001. Among them 325 were adults and 33 children of ages 0–14 years old. Everyone had been examined by a clinician and was checked by ultrasound imaging for the presence of one or more cysts in one or several organs. Their symptoms consisted of: light or severe pain in the upper right abdominal side or other body parts, weakness, weight loss etc. We filled out a questionnaire and collected a blood sample from each person. The sera were analysed depending on the availability of the diagnostic kit-s in the lab. The ELISA IgG and IHA tests were performed in our laboratory respectively in 331 and 94 sera. While 194 sera were sent for a second testing by Western-Blott in the Institute of Parasitology, Bern, CH. 8 patients had been tested again after surgery to follow-up the titer of antibody. 9 people had been tested for a second and third time to follow-up the presence of antibody.
Results: Out of 358 people examined, 164 resulted infected with the parasite Echinococcus granulosus. They were 94 women, 55 men and 8 children. Most of patients infected belonged to the group ages of 35–44 and 45–54 years old. The three serological tests used: ELISA IgG, IHA and Western-Blott gave a 100% correlation. The specifity of these tests reach to 94% and the sensitivity to 87%. The comparison of surgery data with those of serology revealed a 100% correlation. However, the ultrasound results failed in two cases.
Conclusions: Echinococcosis is a parasitosis that presents a big problem for Public Health in Albania. Based in our data the incidence of infection is very consistent every year. The parasite can affect adults and children as well. Women are infected approximately two times more than men due to longer exposure to domestic animals and the parasite. There is not a clear difference between urban and rural areas in terms of where the prevalence of the disease is higher. The reason is probably a persistant migration of the population from one area to another. The laboratory diagnosis of Echinococcosis based on one or more serological tests has a great importance for the treatment of the patients.
P1492: Evaluation of ImmunoCard STAT! immunoassay in the detection of Giardia lamblia and Cryptosporidium parvum specific antigens
I. Wybo, D. Piérard, M. Reynders, J. Breynaert, L. Covens, D. Cnudde, S. Lauwers (Brussels, B)
Objectives: The ImmunoCard Stat ! Cryptosporidium/Giardia assay (Meridian Inc.) is a solid-phase immunochromatographic assay for the rapid detection and differentiation of Giardia lamblia and Cryptosporidium parvum antigens in fecal specimens. The objective of this study was to compare performance of this assay with routine microscopic examination in the in- and outpatient population of our 675 bed University hospital.
Methods: 653 consecutive fecal specimens summitted to our laboratory between January 2002 and October 2004 were analysed. Stool samples were concentrated according to the method of Ritchie and microscopic examination was performed for the detection of cysts and trophozoites of Giardia (Lugol's Iodine stain) and for the oocysts of Cryptosporidium (Modified acid-fast stain). The ImmunoCard Stat ! Cryptosporidium/Giardia assay (Meridian Inc.) was performed on unconcentrated stool samples according to recommendations of the manufacturer. Assay results were read after 10 minutes. As prescribed visible test lines of any intensity were interpreted as positive.
Results: Giardia lamblia. The prevalence of Giardia-positive stools by microscopy (considered as ‘gold standard’) was 4.7% (31/653). Giardia antigen was detected in all these samples except one. Ten additional samples were antigen positive. Sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) of the Giardia immunoassay were 96.8% (30/31; 95% confidence interval (CI) 81.5–99.8), 98.4 % (612/622; 95% CI 97–99.2), 75% (30/40; 95% CI 58.5–86.8) and 99.8 (612/613; 95% CI 98.9–100) respectively. Cryptosporidium parvum. Oocysts of Cryptosporidium were detected in 2.1% (14/ 653) fecal samples. In all samples except one the Cryptosporidium antigentest was positive as well. Another 19 samples were antigen positive. Sensitivity, specificity, PPV and NPV of the Cryptosporidium immunoassay were 92.9% (13/14; 95% CI 64.2–99.6), 97 % (620/639; 95% CI 95.3–98.1), 40.6% (13/32; 95% CI 24.2–59.2) and 99.8%(620/621; 95% CI 99.0–100) respectively.
Conclusions: As compared to microscopy as gold standard for both Giardia and Cryptosporidium detection, sensitivity, specificity and NPV of the antigentest were very favourable. The rapid assay was easy to perform and less labour-intensive. Low PPV could be explained by the low prevalence of both pathogens in our population or by overrating of the false positives because of the use of a non-optimal gold standard.
P1493: Yersinia outer membrane protein- and cell wall-specific antibodies in sera of healthy adult Hungarians
Å. Sonnevend, é. Czirók, T. Pál (Al Ain, UAE; Budapest, HUN)
Objectives: The aim of the study was to investigate the presence of anti-Yop IgG and IgA antibodies in healthy blood donors in Hungary, i.e. in a country with a reportedly low incidence of yersiniosis (1.0–1.4/100.000/year) and to compare the data to the prevalence of cell wall-specific agglutinins.
Methods: Sera of 112 healthy Hungarian blood donors were collected between December 1999 and January 2000. The specimens were tested by Yop-specific IgG and IgA ELISAs (Mikrogen, Germany). Samples were also subjected to tube agglutination using Y. enterocolitica O3, O9 and Y. pseudotubercolusis I-II-III-IV-V antigens. Specimens exhibiting agglutination with yersinia antigens were also tested with brucella and salmonella OB and OD antigens.
Results: Of the 112 sera tested 46 (41%) exhibited a positive anti-Yop reaction in the IgG, and 17 (15.1 %) in the IgA class, while the boundary figures were 7 (6.2%) and 6 (5.3%), respectively. All the IgA positive and boundary samples were also positive for IgG. Samples positive in the IgA class gave a significantly stronger anti-Yop IgG reaction (184.4 ± 70.3 U/ml) compared to those positive in the IgG class, only (62.13 ± 53.6 U/ml) (p < 0.001). There was very little correlation between the ELISA and agglutination data. Only one specimen showing a clear positive agglutination reaction with the Y. enterocolitica O3 antigen exhibited also a positive IgG, a boundary IgA, and an equivocal agglutination titre with Y. pseudotuberculosis IV antigen. Further 3 samples showed equivocal titres with various Y. pseudotuberculosis antigens with varying ELISA results in the IgG and IgA classes. No agglutination with Brucella or Salmonella antigens were observed in any of the samples tested.
Conclusions: The anti-Yop ELISA data showed very little, if any, correlation with the results of cell wall antigen-specific agglutination. The prevalence of anti-Yop IgG and IgA antibodies in Hungary was similar to those found in some Western European countries with a considerably higher incidence of yersiniosis. Further, more extensive seroepidemiological studies should clarify whether the reported incidence of yersinia infections in Hungary reflects the real rate of encounter with the pathogens.
P1494: Application of ELISA and confirmative Western Blot assay for the detection IgG and IgA antibodies against plasmid-coded secretory proteins of pathogenic Yersinia strains in serodiagnosis of paediatric yersiniosis
A. Kostoula, C. Bobogianni, G. Vrioni, I. Sionti, V. Papatolis, S. Levidiotou (Ioannina, GR)
Objectives: Y. enterocolitica and Y. pseudotuberculosis infections are manifested as enterocolitis, mesenteric lymphadenitis, reactive arthritis and erythema nodosum. Important factors in the pathogenicity of the yersinias are associated with the presence of virulence plasmid coded for release proteins (Yersinia outer membrane proteins, YOPs). The detection of antibodies to these proteins is a highly specific and very sensitive method for the serological diagnosis of all forms of yersiniosis. The aim of this study was the use of an enzyme immunoassay (ELISA) in combination with a western blotting assay for the detection of antibodies against YOPs of pathogenic Yersinia strains in serodiagnosis of yersiniosis.
Methods: A total number of 171 sera collected from equal number of children, aged from 5–12 years, referred as probable cases of yersiniosis were tested. These children were hospitalised in the Paediatric Clinic of University Hospital of Ioannina (NW Greece) during a 4-year period (2001–2004). In a first step, all sera were examined using the Yersinia enterocolitica ELISA IgG/IgA test (Genzyme Virotech GmbH), which detects specific antibodies against antigens of the 70 Kb virulence plasmid of pathogenic Yersinia. In a second step, positive ELISA-results were confirmed by a western blotting assay (Yersinia ViraBlot IgG, IgA test), which detects antibodies against YOPs 51, 44, 41, 37, 35, 33 and 23 kD.
Results: 65 out of 171 children were found positive with ELISA for detection IgG/IgA antibodies [53 IgG(+) and IgA(+), 8 IgG(+), 4 IgA(+)]. Main clinical manifestations of these children were fever and abdominal pain. Mesenteric lymphadenitis confirmed by ultrasonography accounted for one third of cases and erythema nodosum occurred in 5 cases. Yersinia ViraBlot IgG and IgA tests confirmed the results for 65 ELISA-positive sera: at least two clear bands from 51, 44, 41, 37, 35, 33 or 23 kD for IgG and isolated clear band 35 kD or at least two clear bands 51, 44, 41, 37, 35, 33 or 23 kD for IgA antibodies. All children received antibiotics and all had an excellent outcome. As a conclusion, serological examination for yersiniosis should be proceeded as two-test-approach: in a first step the specific ELISA is recommended and in the second step positive ELISA-results should be confirmed by the more specific western blot assay. For the final clinical diagnosis, all results from these tests must be correlated with clinical history and epidemiological data.
P1495: The presence of IgM, IgA and IgG antibodies against chlamydial lipopolysaccharide in circulating blood immunocomplexes
Z. Medkova, P. Hejnar (Brno, Olomouc, CZ)
Objectives: The aim of the study was to evaluate 1. the presence of different isotypes of antibodies (IgA, IgG, IgM) against chlamydial lipopolysaccharide (LPS) in blood circulating immunocomplexes; 2. the influence of freezing and defreezing on the blood levels of the described isotypes of antibodies (Abs).
Methods: 100 human sera showing the presence of specific anti-chlamydial LPS IgA and the absence of the IgG in the initial testing (the same specificity; 21 sera were anti-LPS IgM positive) were collected. The sera were stored at −20 degrees of Celsius temperature, defrosted at room temperature and divided into two halves. In one aliquot the Abs were unbound from circulating blood immunocomplexes (CIC) by polyethylenglycol precipitation (PEGP) and the both aliquoths were then tested for the presence of anti-chlamydial LPS IgA, IgG and IgM respectively. The diagnostic sets Chlamydia-rELISA IgA, IgG, IgM (medac, Wedel) were used.
Results: With a mere defreezing IgG developed in 23 and IgM in 24 sera, whereas IgA developed only in 72 samples. After completion of PEGP IgG were demonstrated in 21 sera, IgM in 18 and IgA only in eight. On the whole, after a mere defreezing and PEGP IgG Abs developed in 34 blood samples (29,4% correspondence between methods), IgM in 30 (40% correspondence) and IgA in 72 (only 11,1% correspondence).
Conclusions: With the use of the presented method the specific IgG bound in CIC were demonstrated in about one third of the samples (and IgM bound in CIC in one tenth of sera). A mere freezing and defreezing caused destruction of free IgA and IgM in about 30% sera. PEGP resulted in destruction of free IgA and IgM in about 90% and 40% tested samples, respectively. Thus, the interpretation of the chlamydial serology is more complicated then we could expect, and next research in that area is needed.
P1496: Evaluation of the performance of the new VIDAS C. difficile Toxin A/B test
A. Carricajo, A.C. Vautrin, N. Fonsale, M.C. Bastide, A. Foussadier, G. Aubert (Saint Etienne, Marcy l'Etoile, F)
Objective: Evaluate the performance of the VIDAS C. difficile Toxin A/B (VIDAS CDAB) test (bioMérieux, Marcy l'étoile, France) for the detection of Clostridium difficile A and B toxins directly in stool specimens.
Methods: 62 fresh stool specimens sent to the laboratory for suspected of diarrhoea due to C. difficile were tested in parallel using the culture test on CCFA media (bioMérieux) and the VIDAS CDAB test (bioMérieux). The C. difficile strain isolated from a positive C. difficile culture was further rE-tested with VIDAS CDAB (determination of the toxigenic status of the strain). In case of discrepant results (negative culture/positive VIDAS CDAB), a cytotoxicity test (ECP) of the stool specimen was performed using cell culture. The clinical status of the samples was determined by the concordant results of at least two of the tests performed: VIDAS CDAB, positive culture of a toxinogenic strain and ECP.
Results: Among the 62 stool specimens, 29 were found to be negative and 33 positive. Among the 29 negative stool specimens: 18 were negative with both the culture and the VIDAS CDAB tests, 9 were positive in culture with a non-toxinogenic strain of C. difficile (negative result with the VIDAS CDAB test), and 2 gave false positive results with the VIDAS CDAB test (positive in culture with a non-toxinogenic strain of C. Difficile (negative result with the VIDAS CDAB test)). Among the 33 positive stool specimens: 25 were positive both with the VIDAS CDAB tests (among which 6 were negative in culture and positive in ECP), 5 gave equivocal results with the VIDAS CDAB test, and 3 gave negative results with the VIDAS CDAB test.
Conclusion: The VIDAS CDAB test enables rapid and reliable detection of the toxinogenic C. difficile strains with a specificity of 93.1% (27/29) and a sensitivity of 89.3% (25 /28). Although conducted on a reduced number of fresh specimens, this survey is representative of the performance obtained on a significant number of frozen specimens (evaluation performed during the development of the VIDAS CDAB reagent).
P1497: Evaluation and comparison of indirect immunofluorescent antibody test and direct agglutination test for the diagnosis of visceral leishmaniasis
D. Dorostcar Moghaddam, H. Hejazi, M. Ghasemi (Isfahan, IR)
Objectives: Human visceral leishmaniasis (HVL) is endemic in several foci in IRAN, such as Ardebil and Fars provinces (in North western and south part of IRAN) and in some region as sporadic. Visceral leishmaniasis in Iran is Mediterranean type and the causative agent is leishmania infantum and its main reservoir is dog.
Methods: In this study direct agglutination test (DAT) was compared with indirect fluorescent antibody test (IFAT) for the diagnosis of visceral leishmaniasis in patients suspected of kala-azar. A total of 70 serum samples collected from suspected kala-azar patients mainly in the kala-azar endemic areas. The leishmania infantum antigens (MHO/TN/80/IPTi) for these studies were prepared in Department of parasitology, school of medicine, Isfahan university of medical sciences. The principal phases of the procedure from making DAT antigen were mass production of promastigotes of leishmania in the RPMI1640 + fetal bovine serum, Trypsiniznation of parasites, staining with coomassie blue and fixing with formaldehyde. The human serum samples were tested by DAT, as well as, by IFAT, with the L.infantum antigen prepared in our laboratory.
Results: The sero positive rate (SPR) with DAT in titers of > 1:3200 was 91.4% and with IFAT in titers of > 1:80 was 94.3%. Geometric means of reciprocal titers (GMRT) were 6309 for DAT and 692 for IFAT. Therefore, as the titers of > 1:3200 are usually considered positive in DAT. The titers of > 1:80 were regarded as positive in IFAT. The coincidence of the two tests were 92%.
Conclusions: These results showed that a simple local laboratory with one or two trained technicians is quite sufficient for DAT, serodiagnosis and serological survey of kala-azar in an endemic area. According to the results of these studies, it seems that in Kala azar endemic areas, the clinical symptoms of Visceral leishmaniasis, particularly among the children with DAT antibody titers equal or >1:3200 is a good indication for specific treatment of Kala-azar.
P1498: Evaluation of three serodiagnostic methods: radioimmunoassay, indirect haemagglutination and immunoelectrodiffusion in human hydatidosis and the principal subclass specific immunoglobulin
D. Dorostcar Moghaddam, A. Andalib, M. Amrollahei (Isfahan, IR)
Objective: Several techniques have been developed for the serodiagnosis of hydatid disease. As emphasized by the scientists the percentage of positive results depends partly on technique utilized and the localization of the cyst. Approximately 10% of sera from patients with hydatidosis give false negative reactions. This relatively high figure necessitates the use and comparison of several diagnostic techniques for hydatidosis. In the present study we have attemped to investigate further a solid phase radioimmunoassay for the diagnosis and compare with indirect heamagglutination (IHA) and immunoelectrodiffusion (IED).
Materials and Methods: Sera from 29 patients who had clinically confirmed hydatid disease were tested by radioimmunoassay (RIA), IHA and IED. In 21 patients the localization was hepatic, in 6 pulmonary, and in 2 both hepatic and pulmonary. In addition, 15 sera from patients with clinically confirmed hydatidosis, but which were negative by IHA and IED, were also tested By RIA. The 40 control sera were obtained from apparently healthy blood donors.
Results: In the present study by RIA, IHA and IED approximately 80% of the patients with clinically suspected hydatid disease were confirmed serologicaly. The sensitivity of the three methods was similar. The Principal subclass of specific antihydatid immunoglobulin was IgG and high levels of specific anti-IgE found in two out of the five patients studied.
Conclusion: It is concluded that for a satisfactory serodiagnosis of hydatid disease the RIA and IED should both be used and that fuether work should be done on the purification of hydatid antigens to improve the sensitivity of the Radioimmunoassay without loss of specificity.
P1499: Development of an enzyme-linked immunosorbent assay specifically detecting anti-polysaccharide intercellular-adhesin antibodies
H. Rohde, J. Klettke, N. Siemssen, L. Frommelt, M.A. Horstkotte, C. Burdelski, J.K.-M. Knobloch, D. Mack (Hamburg, D; Swansea, UK)
Objectives: Staphylococci, especially Staphylococcus epidermidis and Staphylococcus aureus, are important pathogens in the context of implanted medical device infections. The ability to establish multilayered biofilms on the surface of foreign bodies significantly contributes to the pathogenicity of these species. The polysaccharide intercellular adhesin (PIA), synthesized by icaADBC encoded proteins, mediates biofilm accumulation in S. epidermidis and S. aureus. Therefore, PIA appears to be a suitable antigen for novel diagnostic tools. In order to measure the humoral immune response appropriate systems for the specific detection of anti-PIA-antibodies are demanded.
Results: We here describe the development of an enzyme-linked immuno sorbent assay (ELISA) specifically detecting anti-PIA antibodies in human sera. 96-well Maxisorp plates (Nunc) were coated with purified PIA. Using these PIA-coated plates, an IgG titer of 1:8000 was detected in the serum of a rabbit immunized with purified PIA. In contrast, in the pre immune serum 16fold lower titers were detected. Absorption of the anti-PIA antiserum against PIA-positive S. epidermidis 1457 lead to an 84% reduction of the serum reactivity, whereas absorption against PIA-negative 1457-M10 had no effect, demonstrating the specificity of the test. This was confirmed by competition ELISA experiments where purified PIA but not pneumococcal or meningococcal capsule polysaccharides specifically inhibited binding of the antibodies to immobilized PIA up to 68%. Using this ELISA, in serum samples from 10 patients with endoprosthesis-associated infections due to clonally independent icaADBC- and PIA-positive S. epidermidis strains anti-PIA-antibody titers ranging from 1:20000 – 1:36000 were detected. In contrast, in a collection of serum samples from patients with endoprosthesis-related infections due to icaADBC-negative S. epidermidis and from healthy blood donors IgG titers were only 1:2000–1:6000.
Conclusions: Due to the widespread presence of icaADBC in pathogenic staphylococci PIA appears as a suitable antigen for novel diagnostic tools in foreign-body infections. Using specific ELISA anti-PIA-IgG titers in patients with icaADBC-positive S. epidermidis infections were significantly higher than in a control collective. Measurement of anti-PIA IgG appears as a suitable tool for improving the diagnosis of staphylococcal foreign-body infections.
Clostridium difficile
P1500: Characterisation of Clostridium difficile associated disease recently observed in a Canadian tertiary care hospital
A. Labbee, D.R. MacCannell, L. Poirier, J. Pepin, T.J. Louie, M. Laverdiere (Montreal, Sherbrooke, Calgary, CAN)
Many hospitals in Montreal have recently experienced an important increase in the incidence of Clostridium difficile associated disease (CDAD). At Maisonneuve-Rosemont hospital, it has increased from 10 per 1000 admissions in 2002 to 30 per 1000 admissions in 2004.
Objectives: To characterize and compare the Clostridium difficile (CD) isolates involved during two distinct periods.
Methods: CD isolates were collected from consecutive cytotoxin positive stools of patients (one isolate per patient) who presented diarrhoea during two distinct periods: Nov 2000-Mar 2001 (pre-epidemic period) and Oct 2003-Jan 2004 (epidemic period). Minimal inhibitory concentrations (MICs) against metronidazole were determined using an agar dilution method. Strains from each period were ribotyped and probed for binary toxin (CDT), and the ermB MLSB resistance determinant. Clinical charts were reviewed.
Results: Among the 206 isolates, 56 were recovered in 2000–1 and 150 in 2003–4. MICs against metronidazole ranged from 0.25 to 4 mg/L and tended to be higher in 2003–4 (median 2 mg/L) than in 2000–1 (median 1 mg/L). Two principal epidemic ribopatterns were identified, and were annotated as strains ‘A’ (tcdA+/tcdB+/ermB+/CDT-) and ‘B’ (tcdA+/tcdB+/ermB-/CDT+), respectively. In 2000–1. 46 (82%) of the isolates were clonally related (ribotype ‘A’), and 10 (18%) of mixed patterns. In 2003–4, by contrast, a binary toxin-positive straintype (ribotype ‘B’) dominated, accounting for 114 (76%) strains, with only a small subset of ribotype ‘A’ (n = 26; 17%) and mixed types (n = 10; 7%) remaining. Median MICs against metronidazole tended to vary according to the ribotype identified: 2 mg/L for ribotype ‘B’, 1 mg/L for ribotype ‘A’ and 0.5 mg/L for other ribotypes. Chart review of 170 patients hospitalized at the time of their CDAD episode revealed that 167 (98%) had received antibiotics in-hospital within the preceding 2 months. At least one relapse of CDAD occurred in 58 (34%) patients. Overall mortality at 30 days after the first CDAD episode was 22%, 13% and 13% with ribotype ‘B’, ribotype ‘A’ and other ribotypes, respectively.
Conclusion: The CDAD in our hospital are both predominantly clonal in nature, and the epidemic strain associated with each period was genetically distinguishable, with the 2003-4 epidemic strain causing an increased incidence of CDAD. Overall mortality at 30 days was higher for patients infected with ‘B’ strain, but statistically not significant.
P1501: Epidemiological examination of Clostridium difficile isolates from different parts of Hungary
G. Terhes, E. Urban, J.S. Brazier, J. Soki, E. Nagy (Szeged, HUN; Cardiff, UK)
Objectives: The aims of this study were to continue and extend our previous investigations, in which the main ribotypes were determined at a local university hospital. In the present survey, we determined the prevalence of different ribotypes of C. difficile strains, and detected the distributions of these types in three Hungarian regions.
Methods: 105 C. difficile strains were isolated in 5 Hungarian laboratories from diarrhoeal faeces of both inpatients and outpatients. The presence of toxin genes such as those of toxin A (tcdA), toxin B (tcdB) and actin-specific ADP-ribosyl-transferase (cdtA and cdtB) were detected by PCR in the Szeged laboratory. The ribotypes of these strains were determined by PCR ribotyping method at the ARL in Cardiff.
Results: A total of 31 ribotypes were detected among the 105 tested C. difficile strains: 5 ribotypes were distinct from all previously described types, suggesting that these are new types. The most common types in Hungary were ribotype 014 (24.8%) and ribotype 002 (13.3%), while in the UK, the most predominant type was ribotype 001. The distributions of the examined ribotypes differed in the different Hungarian regions: ribotype 012 was frequent (20.7%) in South Hungary, at the same time in the Budapest region, this ribotype was rare, while in West Hungary, we did not detect this type during the examined period. In West Hungary and the Budapest region, the most frequent type was ribotype 014 (28.9% and 29% respectively).
Conclusion: Numerous different methods have been used internationally to study C. difficile strains of different origins. Comparisons are therefore difficult. The most frequently applied and most useful typing method is the PCR amplification of rRNA intergenic spacer regions, which is a discriminative, reproducible and rapid technique for determination of the different types of C. difficile. Our present survey and previous studies have revealed that the presence and distribution of C. difficile ribotypes vary from country to country, and also depend on the site of isolation and the period in which the tested strains were isolated. We have now shown that there can additionally be regional differences within a given country. This work was supported by a Hungarian Eötvös Scholarship and grant TO32385 from the Hungarian National Research Foundation (OTKA).
P1502: Tn916-Tn1545-like elements in Clostridium difficile clinical isolates
P. Spigaglia, V. Carucci, F. Barbanti, P. Mastrantonio (Rome, I)
Objectives: In C. difficile, tetracycline resistance is predominantly due to a tet(M) gene. This gene has been shown to be carried by Tn5397 in the clinical strain C. difficile 630, whereas by a Tn916-like element in the environmental strain C. difficile 42373. These two elements are related, but very different in their integration/excision module and can not be co-present in the same cell. The aim of the study was to examine Clostridium difficile clinical isolates for the presence of the Tn916 and Tn1545 like elements.
Methods: Detection of tet(M), erythromycin resistant gene erm(B) and the integrase gene int (markers for the Tn916) was performed by PCR, as well as the analysis of the genetic arrangement of the elements. The amplified fragments were used as probes for hybridisation assays on genomic DNA of C. difficile isolates, after digestion with HindIII. The nucleotide sequence of tet(M) genes was also analysed. Antibiotic susceptibility of the strains was assessed by the E-test method.
Results: Eighteen C. difficile isolates were positive for tet(M) and int and the signals obtained using these genes as probes overlapped. Ten isolates showed one hybridising band of either 6.3, 9.0 or 14 kb. Seven isolates showed two hybridising bands of 6.3 and 9.0 kb and one strain two banda of 6.0 and 9.0 kb, indicating the presence of two copies of a Tn916-Tn1545 like elements. Heterogeneity in these elements was observed. The E-test results indicated that isolates with two copies of tetracycline resistance elements were resistant (88%) or inducibly resistant to tetracycline (12%), whereas isolates with one copy were resistant (36%), inducibly resistant (18%) or susceptible (46%) to this antibiotic. Three isolates were also erm(B)-positive and resistant to erythromycin. The bands obtained using tet(M) and erm(B) probes overlapped at 9.0 kb, indicating the presence of a Tn1545-like element. The association of these genes was also confirmed by PCR. Seven different alleles were identified sequencing the tet(M) gene of prototype strains among those examined.
Conclusion: Heterogenic elements of the Tn1545-Tn916 family are harboured by C. difficile clinical isolates showing different phenotypes for tetracycline. These elements carry different tet(M) alleles and can be found in one or two copies in C. difficile chromosome. Tn1545-like elements, have been detected for the first time in this pathogenic bacterium.
P1503: Clostridium difficile among patients in department of surgery
G. Martirosian, A. Szczesny, J. Silva Jr. (Katowice, PL; Sacramento, USA)
Nosocomial outbreaks of C. difficile-associated diarrhoea were described in different hospital units. Patients undergoing surgery are under special control because of necessity of antibiotic treatment. The aim of this study was to determine the frequency of C. difficile infection among patients hospitalized in Department of Surgery and to compare isolated strains by phenotypic and genotypic characteristics. During period of 11 months 318 stool samples taken from patients with gastrointestinal problems, hospitalized in Department of Surgery of Medical Center UC Davis and 62 environmental samples were cultured for isolation of C. difficile strains. Thirty (9.4%) C. difficile strains were isolated from stool samples cultured on selective CCCA plates (bioMerieux, France) with sodium taurocholate and 6 (9.6%) strains from environmental samples using Rodac plates with TCCA. Toxigenicity of isolated strains was determined by using of Tox A/B ELISA test (TechLab, USA) and PCR (primenrs YT 28/YT29 and YT18/YT19 correspondingly for toxin A and B genes). All isolated strains were compared by AP-PCR and PCR-ribotyping. Twenty two toxigenic (A+/B+), 5 nontoxigenic (A−/B−) and 3 toxin A-positive/toxin B-negative C. difficile strains were detected among patients'isolates and 4 toxigenic and 2 toxin A-positive/toxin B-negative strains - among environmental samples. Twenty five out of 30 C. difficile-positive patients had performed surgery before C. difficile testing. Majority of C. difficile-positive patients were treated previously by cephalosporins and penicillins with inhibitors of beta-lactamases. Among patients, infected by toxigenic strains significantly higher leukocytosis and longer duration of fever were observed. Seven strains isolated from patients’ fecal samples and one strain isolated from environment demonstrated high level resistance to erythromycin and clindamycin (MIC > 256 μg/ml) in E-test. The results obtained by AP-PCR and PCR-ribotyping revealed genetic heterogeneity among the strains isolated from patients’ fecal samples. However, similarity was observed among environmental strains and strains isolated from patients’ fecal samples. The resistance of isolated C. difficile strains to clindamycin and erythromycin (E-test, AB Biodisk) indicated possibility of transmission in the hospital strains with macrolide-lincosamide-streptogramin B (MLS-B) resistance type.
P1504: Introduction of TcdA-negative, TcdB-positive Clostridium difficile in a general hospital in Argentina
R.J. van den Berg, M.C. Legaria, A. de Breij, E.R. van der Vorm, J.S. Brazier, E.J. Kuijper (Leiden, NL; Buenos Aires, RA; Amsterdam, NL; Cardiff, UK)
Objectives: Clostridium difficile is the major causative agent of nosocomial antibiotic associated diarrhoea and pseudomembranous colitis. The main virulence factors are two toxins produced by this pathogen: TcdA and TcdB. Clinically important TcdA-negative, TcdB-positive (A-B+) C. difficile strains from different countries have occasionally been reported, but epidemics are very rare.
Methods: In a prospective study to determine the incidence of CDAD in a 200-bed general hospital in Argentina, all faecal samples of symptomatic patients with a positive immunoassay (Premier Cytoclone A/B or Triage Micro C. difficile panel) and/ or a positive cytotoxicity assay were cultured for the presence of C. difficile. The isolated strains were investigated for the presence of tcdA, tcdB, and erm(B) genes. All isolates were further typed using PCR-ribotyping and amplified fragment length polymorphism (AFLP). Additionally, a toxinotyping method was optimised to recognize 24 toxinotypes and was also applied on the clinical isolates.
Results: The incidence of CDAD per 1,000 admissions varied from 5.9 (2000), to 10.9 (2001), 6.9 (2002) and 4.7 (2003), respectively. Most patients were diagnosed at the Departments of Internal Medicine (57%), Pulmonology (13/%), and Intensive Care (10%). Of all C. difficile isolates, the percentage A-B+ isolates increased from 12.5% in 2000, 58.1% in 2001, 87.9% in 2002, to 96% in 2003. Comparison of 37 patients with CDAD due to A+B+ strains with 80 patients with CDAD due to A-B+ strains, revealed no significant differences with respect to age and gender, department, underlying disease, severity of CDAD, or relapse rate of the patients. All 80 A-B+ C. difficile isolates belonged to toxinotype VIII and PCR-ribotype 017, and 89% were resistant to clindamycin due the presence of the erm(B) gene. The A-B+ isolates could be divided in clusters using AFLP and belonged to different clusters then the A+B+ strains.
Conclusion: In a period of 4 years, A-B+ strains completely replaced A+B+ strains. This shift was not accompanied with a change in incidence, affected patient group, clinical presentation or relapse rate. All strains belonged to one PCR-ribotype and toxinotype, but could be distinguished in smaller clusters using AFLP. Clinicians should be aware of the widespread distribution of the clindamycin resistant A-B+ strains belonging to toxinotype VIII and PCR-ribotype 017.
P1505: C. difficile pseudo-outbreak in a general hospital
V. Stempliuk, V. Borrasca, M. Araujo, C. Ciola, A. Cataldi, S. Costa, M. Souza Dias (Sao Paulo, BR)
C. difficile -associated diarrhoea (CDAD) is the most common cause of nosocomial diarrhoea. While healthy adults have a 3% colonization rate, in-hospital colonization can increase to 30%. Few Brazilian hospitals have the capacity to detect C. difficile toxins and its epidemiology in Brazil is scarcely known. Hospital Sirio-Libanes is a private 250-bed community hospital with predominantly surgical and oncologic patients. In March 2002 the number of cases CDAD cases increased from a mean of 2/ month to 7,6/ month. An outbreak was suspected and control measures were initiated (contact precautions, chlorine based environmental desinfection and staff orientation).
Objective: Describe the investigation of an outbreak of C. difficile associated diarrhoea.
Methods: Detection of toxin A and B (C. difficile Detection test - toxin A and B - Meridian). Toxin − positive stool samples were cultivated in anaerobic environment in non-selective blood agar. Clostridium difficile was diagnosed by 16S rRNA PCR. Genotyping was done by AP-PCR with arbitrary primer T-7.
Results: From 3/2002 until 12/2003 138 patients were diagnosed with CDAD, 96 (70%) became symptomatic during their hospital stay and 42 (30%) were admitted with diarrhoea − 34 from the community and 8 from other hospitals. At least 10/138 referred they had not used antimicrobial agents in the previous 2 months. The incidence of new ‘hospital acquired’ cases was 0.41% in 2002 and 0.42% in 2003. Clostridium spp. was isolated in 20 samples and C. difficile confirmed in 16 samples. Genotyping by AP-PCR revealed 13 different band patterns.
Conclusion: Although epidemiological data indicated an outbreak and possible cross infection, genotyping excluded this possibility. Control measures may be more efficiently directed at restriction of antimicrobial use then measures to contain cross infection.
P1506: Laboratory diagnosis of antibiotic associated diarrhoea in hospitalised patients (Clostridium difficile or Clostridium perfringens) − preliminary study in Poland
H. Pituch, P. Obuch-Woszczatynski, D. Wultanska, F. Meisel-Mikolajczyk, M. Luczak (Warsaw, PL)
Although Clostridium difficile is the most commonly identified pathogen in antibiotic associated diarrhoea (AAD), the majority of the current cases remain undiagnosed. C. perfringens (CPEnt+) was first implicated as a cause of AAD in 1984. The main virulent factor is a 35 kDa protein-enterotoxin (CPEnt). CPEnt is encoded by the cpe gene, can be detected in faecal samples by the Vero cell line and EIA commercial test. In the present study 52 faecal specimens from patients hospitalised in University Hospital in Warsaw and gastroenterology units of different Polish hospitals, submitted for C. difficile testing were studied for C. perfringens infection. C. difficile toxins were detected with commercial immunoassay test TOX A/B (TechLab, USA). For detection enterotoxin C. perfringens Enterotoxin Test (TechLab, USA) was used. All specimens were inoculated onto CCCA medium for detection of C. difficile and TSN medium for detection C. perfringens. To detect cpe gene PCR was performed. We used the E-test to asses for susceptibility to metronidazole. C. difficile toxins (TcdA and/orTcdB) were detected in 39 faecal samples. However, enterotoxin (CPEnt) of C. perfringens was detected in 16 faecal samples. In 16 gave positive tests results for both C. difficile and C. perfringens toxins. From the same samples was cultured 16 C. difficile and 15 C. perfringens strains. In the present study 75% of the specimens were positive for C. difficile TcdA/TcdB toxins, 31% were positive for CPEnt C. perfringens and 28% gave positive test results for both C. difficile and C. perfringens toxins. We were found relatively a small number of enterotoxigenic C. perfringens (CPEnt) strains. All strains C. difficile and C. perfringens were susceptible for metronidazole. The occurrence of C. perfringens (CPEnt) as etiologic agents of nosocomial diarrhoea is not known in Poland. In our laboratory, we have found a suprisingly significant number of C. perfringens enterotoxin (CPEnt) positive stools. We conclude that C. perfringens is a potential cause of AAD in Poland.
Chlamydia trachomatis
P1507: Detection of Chlamydia trachomatis from self-collected vaginal swabs in commercial sex-workers
M.V. Boost, P. Leung, A. Grey, C. Lo (Hong Kong, HK)
Objectives: To determine the level of chlamydial infection in female commercial sex-workers (CSW) in Hong Kong, and the acceptability of self-collected vaginal swabs.
Method: CSW were contacted by an outreach worker who informed them of the risks of untreated chlamydial infection and offered screening by use of a self-collected vaginal swab. Those willing to participate were instructed how to collect the specimen, and provided with a dacron swab, a sterile empty tube, and an envelope in which to place the tube. Subjects were asked to complete a short questionnaire on sexual practices and medical check-ups for sexually transmitted disease (STD). Specimens were returned to the laboratory within 36 h. C. trachomatis was detected using a previously validated in-house PCR method. The tip of the swab was cut off using sterile scissors and placed into a 2-ml microcentrifuge tube to which 600≥of PBS (pH 7.4) was added. The tube was vortexed for 30 s, and then the swab pressed against the wall of the tube to squeeze out the fluid before discarding it using sterile forceps. The fluid was centrifuged at 13,000 rpm for 20 min, the supernatant discarded and the pellet resuspended PCR buffer containing proteinase K and Tween-20. It was incubated at 55°C for 1 h and then heated at 80°C to inactivate the proteinase K. Ten microlitres of the treated sample were used for PCR using primers for C. trachomatis. Thirty cycles of 30 s at 94°C followed by 1 min at 60 °C and 1 min at 72°C were used for amplification, with final annealing of 7 min at 72°C and 7 min at 15°C. The amplified DNA was run on a 1% agarose gel and visualized with ethidium bromide. Positive and negative controls were used on each occasion.
Results: Of 180 CSW invited to participate, specimens were collected from 163, none of whom were aware of a current infection with C. trachomatis. Nine specimens were positive, representing a 5% infection rate. Seventy per cent of the CSW reported usually using condoms with clients.
Conclusion: The level of infection in CSW in Hong Kong was relatively low compared to other countries. This may reflect regular condom use. Self-collected vaginal swabs and anonymous screening were well-accepted by the CSW as many reported fear of stigmatization and possible legal repercussions if they attended STD clinics.
P1508: The effect of freezing, washing and high-speed centrifugation on the outcome of BD ProbeTec Chlamydia analysis on urine samples
G. Lisby, H. Westh (Hvidovre, DK)
Objectives: When urine samples for Chlamydia trachomatis (CT) are not analysed immediately, it has been standard procedure to freeze the samples at −20°C until analysis. On known CT-positive and CT-negative patient samples, we valuated the impact of freezing, washing or not washing the unfrozen samples as well as the impact of centrifugating frozen samples at 10.000 xg prior to analysis.
Methods: All urine samples were analysed (no freezing) with the BD ProbeTec system. The remaining sample was frozen at −20 C for inclusion in this study. We carried out three ‘arms’ of investigation: ‘Arm 1′: Frozen positive samples and frozen negative samples were thawed and reanalysed after washing. ‘Arm 2’: Refrigerated positive and negative samples were analysed without washing. ‘Arm 3′: frozen positive and frozen negative samples were thawed and analysed after centrifugation at 10.000 xg and washing. We tested the effect of centrifugation at 10.000 xg to determine whether 10.000 xg is more efficient than the traditional 3,000 xg to sediment the CT organisms. Due to cellular lysis caused by the freeze/thaw cyclus, the intracellular CT will be found in the extracellular phase, and might not sediment completely at 3,000 xg.
Results: In ‘arm 1′, 181 positive samples were frozen, thawed and reanalysed after washing. 162 (89.5%) remained positive, 13 (7.2%) became negative and six (3.3%) showed inhibition. Of the 182 negative samples, 166 (91.2%) produced a negative result, while 15 (8.2%) showed inhibition and one (0.6%) was borderline positive. In ‘arm 2′, of the 90 positive samples analysed without a washing step, 85 (94.5%) remained positive and five (5.5%) became negative. In 91 negative samples, 70 (76.9%) produced a negative result, while 21 (23.1%) showed inhibition. In ‘arm 3′, data are pending.
Conclusion: Compared to standard testing of urine samples by the BD ProbeTec system, (unfrozen, washed/centrifugated (3,000 xg) samples), freezing at °20C followed by washing caused loss of signal in 7.2% of positive samples as well as inhibition in 3.3% of positive samples and 8.2% of negative samples. Also compared to standard testing, omitting the washing step caused 5.5% of the known positive samples to become negative and of the known negative samples, 23.1% showed inhibition.
Overall conclusion: 1. Freezing samples reduces sensitivity and causes low-grade inhibition. 2. Omitting the washing step in unfrozen samples causes extensive inhibition.
P1509: Comparison of inhibition rates for vaginal and cervical swabs and urine for the diagnosis of Chlamydia trachomatis by Aptima Combo 2, ProbeTec ET and Amplicor
M. Chernesky, D. Jang, M. Smieja, S. Chong, K. Luinstra, C. MacRitchie, W. Cai, B. Hayhoe (Hamilton, Toronto, CAN)
Background: Inhibitors of nucleic acid amplification (NAA) tests in clinical specimens and analytical test sensitivity may play a role in the ability of diagnostic assays for Chlamydia trachomatis to detect infected patients. Some manufacturers provide amplification controls to monitor NAA inhibition. The objectives were (a) to determine the analytical sensitivity of APTIMA COMBO 2® (AC2), ProbeTecTM ET (PT) and AMPLICOR® (AMP) assays and to produce sensitive internal controls for spiking clinical specimens; (b) to test first void urines (FVU), cervical swabs (CS), and vaginal swabs (VS) for inhibitors and C. trachomatis nucleic acids from each patient.
Methods: C. trachomatis L2 434 isolate was propagated in McCoy cells. Aliquots were titrated in each assay for swabs and FVU, and dilutions where 16/16 replicates were positive were used for spiking clinical samples. A total of 301 women signed a consent for collection of three CS, three VS and an FVU (30 ml). Each sample was tested spiked and unspiked in the AC2 (Gen-Probe), PT (Becton Dickinson) and AMP (Roche Diagnostics) assays for C. trachomatis.
Results: The analytical sensitivities determined by one of 16 replicates being positive, were as follows: PT 10–5 to 10–6, AMP 10–5 to 10–6, AC2 10–8. Percent inhibition rates in FVU, VS and CS were 27.6, 2.5, 2.5 for PT, 12.6, 4.5, 3.8 for AMP and 0.4, 1.2, 0.4 for AC2. Overall, there were 70 infected women. The percentage of patients positive for C. trachomatis by specimen type were as follows: by VS, CS and FVU there were 98.6, 91.4, 85.7 by AC2; 67.1, 65.7, 67.1 by PT and 64.3, 60.0, 57.1 by AMP. The differences between the AC2 results and the other two assays were statistically significant (p < 0.001).
Conclusions: Because of different analytical sensitivities and inhibition rates, more positive patients were identified by the AC2 test. Most patients had more than one specimen type positive.
P1510: Diagnostics of urinogenital chlamidiosis common forms in young people
Y. Lobzin, A. Poznyak (Saint-Petersburg, RUS)
Objectives: To study clinical-laboratory manifestations of chlamidiosis common forms 1002 patients at the age of 18–32 were observed and treated for acute (urethritis, cervicitis) n = 193, localized (prostatitis, vesiculoprostatitis, adnexitis, salpingitis), n = 340 forms of infection, Reiter disease n = 274, acute and chronic sinusitis, associated with Chlamydia trachomatis, n = 95, meningitis and encephalitis, n = 100.
Methods: The complex of laboratory methods was used: PCR, culture method, DIF, indirect immunofluorescence reaction, EIA, complement fixation reaction, electronic microscopy. Chlamydia trachomatis and other agents were revealed in clinical material from primary (brush cytology of the urethra and the cervical part of the uterine neck, prostatic secretion, ejaculation, urinary sediment) and secondary (brush cytology of the throat, conjunctiva and rectum, sputum, synovial membrane biopsy material, synovial fluid, maxillary sinuses contents, liquor) foci. To reveal Chlamydia and other sexually transmitted infections venous blood test (including white blood cells concentrate) was made. To specify the localization and manifestation of chlamidiosis primary and secondary foci ultrasonic scanning of the small pelvis organs, radioisotope scintigraphy and single-photon emission CT were carried out.
Results: Urinogenital chlamidiosis can manifest as common infections (24.6% of cases) with hematogenic dissemination and involvement of different organs and systems. Development of these forms increases with the time interval the urinogenital system was involved. In such a form of infection Chlamydia trachomatis is associated with other sexually transmitted infections in 76.4% of cases. Clinical-laboratory criteria of common chlamidiosis diagnostics are based on a finding of chlamydia in blood, presence of extraurogenital foci of infection in different organs and tissues (joints, CNS, eyes, ENT-organs, respiratory organs, distal part of the intestines and others), general infectious intoxication syndrome, immune impairment and intestinal dysbacteriosis.
Conclusion: Usage of clinical material from the urinogenital tract cannot solve all the problems of diagnostics of different urinogenital chlamidiosis forms. Important clinical criterion of common urinogenital chlamidiosis is the presence of chlamydia in blood. For detailed localization of secondary foci of chlamidiosis it is advisable to use radioisotope scintigraphy.
P1511: Vaginal swabs detect more chlamydial infections than do urine specimens
J. Schachter, M. Chernesky (San Francisco, USA; Hamilton, CAN)
Objectives: Recently the FDA cleared use of vaginal swabs (VS) in screening for Chlamydia trachomatis (CT) and Neisseria gonorrhoeae in Gen-Probe Incorporated's APTIMA Combo 2 Assay (AC2) for both organisms. First catch urine (FCU) has been considered the noninvasive specimen of choice for screening. We reanalysed data from a clinical trial assessing the performance of AC2 to determine the best specimen for screening for chlamydial infections.
Methods: FCUs and two cervical swabs (CS) were collected from 1464 women attending STD, family planning and Ob/Gyn clinics. The FCU and one CS were tested by AC2 and FCU and the other CS by the BDProbeTecTM ET System CT Assay (BD Diagnostic Systems). Patient collected VS (PVS) specimens were also tested by AC2. All positive results were confirmed using an alternate target amplification test specific for CT, the APTIMA® CT Assay.
Results: The AC2 assay had 174 positive FCU and 171 (98.3%) were confirmed. The ProbeTec assay had 155 positive FCUs and 144 (92.9%) were confirmed. There were 195 positive CS by AC2 assay and 194 (99.5%) were confirmed. The ProbeTec assay had 159 positive CS specimens and 157 (98.7%) were confirmed. There were 208 positive PVS by AC2 and 204 (98%) were confirmed.
Conclusions: AC2 was more sensitive than the ProbeTec assay. It identified about 20% more infected women than the ProbeTec assay with CS (194 vs 157 positive specimens, 24% more) and FCUs (171 vs 144, 19% more). Testing PVS with AC2 identified at least as many infected women as did testing of CS (204 vs 194) and yielded more positives than did testing of FCUs (204 vs 171, 19% more). VS is a non-invasive specimen which allows for self-collection. VS can be collected without performing a pelvic exam, and testing VS will detect more infections. For these reasons, it is clear that vaginal swabs should be considered the specimens of choice for screening for C. trachomatis in women.
P1512: International external quality assessment for C. trachomatis using EIA and molecular methods
A. Rossouw, V. Chalker, P. Patel, H. Vaughan, H. Seyedzadeh, V.L.A. James (London, UK)
C. trachomatis is a sexually transmitted pathogen that causes genital tract infections in both men and women. Clinical symptoms may be absent or silent, delaying treatment and leading to serious consequences such as infertility. In 1991, UK NEQAS for Microbiology introduced an international EQA scheme for the detection of C. trachomatis by EIA and since the use of molecular methods increased in clinical laboratories more challenging specimens were included in the distributions.
Objective: To summarise results of an international EQA scheme for C. trachomatis and compare the performance of EIA and molecular methods used in the detection of C. trachomatis in specimens with varying EB/mL consistent with those found in urine samples.
Methods: A total of 9 positive and 7 negative 0.5 mL specimens were produced in varying concentration of C. trachomatis (L2 strain in buffer solution with 0.05% Bronidox) and distributed to participants in 8 panels. Participants were instructed to examine 0.1 mL of each specimen for the presence of C. trachomatis and report results within 21 days.
Results: Overall detection of C. trachomatis in the positive specimens by all methods varied from 32.3 to 83.6%. Detection by molecular methods (66.4–98.6%) was significantly higher than EIA methods (2.3 – 69.2%). Risk difference analysis showed that all methods were significantly more likely to detect C. trachomatis when present at higher concentrations. Analysis of specific molecular assays indicated Abbott LCx® detected 72.7–100%, BDProbeTeET detected 31.4–100% and the Roche* methods detected 84.5–100%. Participants using the BDProbeTecET were less likely to give positive results at dilutions of 1:1200 and greater, whereas Abbott LCx™ was less likely to give positive results at dilution of 1:2500 and higher and the Roche methods were more likely to report a positive result on less concentrated specimens. False positive reporting was rare with the negative specimens (19/1985; 0.3–1.5%) and more common with EIA users.
Conclusion: Detection was excellent at higher concentrations of C. trachomatis irrespective of the method used. However, at lower concentrations molecular methods are superior to EIA based assays for the detection of C. trachomatis (L2 strain) in simulated EQA specimens. * Includes all participants stating Roche, Roche AMPLICOR® CT/NG or Roche COBAS AMPLICOR® CT/NG.
P1513: Detection of Chlamydia trachomatis using an automated PCR-based system
A. Babic-Erceg, G. Vojnovic, S. Ljubin Sternak (Zagreb, HR)
Objectives: Chlamydia trachomatis urogenital infection is the most common bacterial sexually transmitted disease in both, men and women. It causes cervicitis, salpingitis, urethritis, pelvic inflamatory disease, infertility, ectopic pregnancy and chronic pelvic pain. Laboratory testing for C. trachomatis continues to be a challenge to clinical laboratories. Usual methods, enzyme immunoassay, direct fluorescent assay, even isolation in cell culture, lack sensitivity when compared with nucleic acid amplification methods.
Methods: A PCR Cobas Amplicor CT/NG method; Roche Diagnostics, semi automated test was used. The Cobas Amplicor is a two-segment thermal cycler which automates amplification, denaturation, hybridisation, incubation, washing, colorimetric endpoint and results reporting. Specimen preparation was the only step conducted manually. Cervical and urethral swabs were transported in 2-SP transport medium, and first-voided urine specimens in the sterile containers.
Results: In a three month period, total of 1270 specimens from outpatients were examined. There were 1166 cervical swabs, 87 urethral swabs (25 female, 62 male), and 17 urine samples (8 female, 9 male). The prevalence of C. trachomatis infection was 2.1% (25 cervical specimens, 1 male urethral swab and 1 female urine sample). In a hundred cervical swabs and 17 urin samples, no inhibition was observed.
Conclusion: Our results in a short period of time show low prevalence of C. trachomatis infection. Lack of inhibition is due to an adequate transport medium used and pooling of specimens.
P1514: Results of early performance studies of the COBAS TaqManTM CT (TaqMan) test for detecting Chlamydia trachomatis in male and female urine specimens on the Roche COBAS TaqManTM 48 analyzer
D.V. Ferrero, D.E. Schultz, J. Cuerpo, S. Stewart, R. Dalal, S.A. Willis (Stockton, USA)
Objective: We evaluated Roche's next generation polymerase chain reaction (PCR) test, the COBAS TaqManTM CT test (Roche Molecular Diagnostics) for use in clinical and public health laboratories. This test is not yet commercially available. The COBAS TaqManTM 48 analyzer incorporates automated realtime amplification and detection of DNA or RNA for up to four simultaneous detections. Our study compares TaqMan performance in detecting Chlamydia trachomatis (CT) from first catch urine (FCU) specimens to the COBAS AMPLICORTM CT/NG (AMPLICOR) (Roche Molecular Diagnostics) and the BD ProbeTecTM ET (ProbeTec) (Becton Dickinson Company) nucleic acid amplification systems.
Methods: FCU specimens were collected from patients attending STD and family planning clinics. Specimens were collected, transported and tested according to the manufacturer's recommendations. Specimens were tested by the ProbeTec, AMPLICOR, and TaqMan tests for CT. TaqMan was compared to ‘infected patient status’, which defined a patient as infected whenever any two comparator results were positive.
Results: FCU specimens were obtained from 406 patients. The overall TaqMan sensitivity for FCU when compared to infected patient status was 97.2% (70/72). The overall TaqMan specificity for FCU when compared to infected patient status was 99.0% (298/301). There were 31 inhibitory results in the ProbeTec test, 4 in the AMPLICOR test and 3 in the TaqMan test.
Conclusion: The TaqMan test produced excellent overall results when compared to the AMPLICOR and the ProbeTec tests in sensitivity and specificity in detecting CT from FCU. The TaqMan test had fewer inhibited test results than either the ProbeTec or the AMPLICOR tests. The TaqMan test demonstrated a faster turnaround time than COBAS AMPLICOR CT/NG Test. These excellent results indicate the TaqMan test to be robust nucleic acid amplification next generation technology.
P1515: The role of Chlamydia trachomatis urinary tract infection in children
I. Choroszy-Krol, R. Bednorz, M. Frej-Madrzak, D. Teryks-Wolyniec, B. Kowalska-Krochmal (Wroclaw, PL)
Objectives: Chlamydia trachomatis (C. trachomatis) is an important patogen (agent?) responsible for variety of genitourinary diseases in children and in adults. Since C. trachomatis infection requires specific treatment and prevention it should be taken into account in practice. The aim of our study was to determine the prevalence of chlamydial urethritis in children with different urinary tract diseases.
Methods: In 195 children and adolescents aged 14 to 18 years (115 girls and 79 boys) with urinary tract infection (UTI) urethral swabs were taken. It those with positive result for C. trachomatis blood and urine were collected. Bacteriological evaluation was done using MicroTrak Chlamydia trachomatis Direct Specimen Test (Trinity Biotech plc, Ireland). Urine samples were tested for chlamydia DNA using the polymerase chain reaction (DNA-Gdansk II s.c. Polska).
Results: C. trachomatis infection was detected in 99 of 195 (50.7%) patients (64 of 116 (55.1%) girls and 35 of 79 (44.3%) boys. C. trachomatis DNA in urine was detected in 16 of 23 (69.5%) patients (8 of 14 (57.1%) girls and 7 of 9 (77.7%) boys.
Conclusions: High prevalence (50.7%) of C. trachomatis in urethral swabs indicates that chlamydia has a significant role in UTI in children.
P1516: Comparison of Chlamydia and Mycoplasma incidence between different female populations in Greece
M. Simou, K. Avdeliodi, S. Tzortzatou, T. Lazaraki, H. Kada (Athens, GR)
Objectives: To determine the incidence of Chlamydia trachomatis and Mycoplasma infection in female population of reproductive age after the significant increase of foreign women (immigrants) in Greece during the last years.
Methods: 3152 cervical and vaginal smears of women in reproductive age who attended the Outpatient Dpt. of our Hospital from January 2003 till November 2004, were tested for the presence of Chlamydia trachomatis and Mycoplasmas. From the total of 3152 women, 2773 were Greek and 379 were immigrants. The detection of C. trachomatis was performed by ligase chain reaction method (Lcx Abbott) and Probe Tec assay (Becton Dickinson). Mycoplasma identification and sensitivity test was performed using the Mycofast Screening Evolution 2 kit (International Microbio, France).
Results: From the total of 3152 women a percentage of 4,15% were positive for C. trachomatis. The prevalence of C. trachomatis among Greek (3.4%) and immigrants (9.2%) was statistically significant (p < 0.001). From the total of 3152 women a percentage of 27.2% were positive for mycoplasmas. Greek women were positive in a percentage of 25.3% and immigrants were positive in a percentage of 42.2% (p < 0.001).
Conclusions: The incidence of chlamydia and mycoplasma infection has increased during the last years compared to our previous studies where the prevalence of Chlamydia trachomatis was 3.8% and the prevalence of Mycoplasma was 24.2%. The increase of chlamydia and mycoplasma infection in general female population is due to the increase of immigrants who attended the Outpatient Dpt. during the last years. Care should be taken to discover and treat all chlamydia positive and mycoplasma positive women with appropriate antimicrobials in order to diminish the danger of infection and related problems in conception and fertility.
P1517: Antibiotic susceptibility of Mycoplasma hominis and Ureaplasma urealyticum strains isolated from symptomatic and asymptomatic women
M. Svabic-Vlahovic, I. Dakic, I. Cirkovic (Belgrade, CS)
The examinations was carried out on the 400 symptomatic and asymptomatic women, aged 15–59, middle value 29.3. 274 of them had in the history pregnancy, four of them were pregnant in the moment of examination, and 112 passed examination because of infertility treatment. Among the examined patients 102 (50.25%) had signs of cervicitis or inflamations of vaginal mucose or disorders in secretion. 84 of examined women had in anamnesis treatment with antibiotics, and in 36 cases we established that board spectrum antibiotics was used. Isolation and identification of U. Urealyticum (UU) and M. hominis (MH) were carried out on home made media, and for 40 samples we used two media - home made media and BioMerieux (ist 2) media. Presence of C. trachomatis in cervical swobs was examined by direct IF (BioMerieux). Other bacteria and microorganisms (Candida) are isolated by standard methodology.
Results: UU was isolated from 146 patients (36.5%) and MH from 38 patients (9.5%). From the groupe of women with signs of inflamation, UU was isolated in 37%, MH in 23%. In this group 2% patients had in same time both microorganisms (MH + UU). C trachomatis was present in 16 patients, but eight of them were with local inflamation. 64 patients had more than 8 PMN in direct smear and 11 vaginosis were confirmed (Nugent score), 10 from 11 vaginoses were present among group with local disorders. 66 patients had Gram negative bacteria, 22 Enterococcus spp. and 12 S. agalactiae. MICs was carried out for all isolated strains in microtitration plates. Among 146 strains of UU 11 were resistant to Doxicyclin, 3 to Ofloxacin, 9 to Ciprofloxacin (but 28 strains were with MIC value of 2). There were no strains resistant to Tetracycline. 38 MH strains were tested: 3 were resistant to Doxyciclin, 1 to Tetrracycline, 8 to Ciprofloxacin, 3 to Ofloxacin.
P1518: Aetiology of urethritis among Yemeni patients
A.Y. AL-Haifi, I. Alshami, A. Aljaufy, T. Salam, S.A.H. Awad (Sanaa, YE; Manchester, UK)
Objectives: There are no previous or recent trends in study the aetiology and epidemiological characteristics of specific microorganisms causing urethritis in Yemen; therefore we designed this study to determine the prevalence and susceptibility rate of different causes of urethritis infection among Yemeni patients.
Methods: During 11 months we investigated a total of 420 patients with urethritis (55 males and 365 females) attending the Yemeni hospitals with a clinical presentation of urethritis. The study subjects include 93.8% married, 93% monogamy, 87.9% with genital discharge, 37.9% with lower abdominal pain, 46.9 with urinary retention and dysurea or urinary tract infection, 18.8% with joint pain, 8.8% travelling abroad and 6.4% with partner complain urethral discharge. Etiology was defined with special methods of diagnosis including direct examination, cytological test, culture, Immunoserological and immunochromatographic tests.
Results: N. gonorrhoea, C. trachomatis, and T. vaginalis represent only 15.2% of the etiological agents of urethritis in yemen. The other 84.2% of urethritis could be attributed to other infectious agents or other causes of inflammation. There were 22 cases of N. gonorrhoea (5.2%), 30 cases of C. trachomatis (7.1%) and 12 cases of T. vaginalis urethritis (2.9%). There was mixed infection (0.95%) for N. gonorrhoea and C. trachomatis, (0.7%) for N. gonorrhoea and T. vaginalis and (0.95%) for C. trachomatis and T. vaginalis.
Conclusion: Urethritis and other STDs are difficult to assess in Yemen due to lack of recording system in hospitals and clinics and lack of facilities for conducting and confirming the diagnosis. Knowledge about STDs and their methods of transmission and protection system is limited. Education about the methods of transmission and protection are necessary for elimination of STDs and urethritis.
P1519: Optimisation and evaluation of the detection of Chlamydia trachomatis from sperm and urine specimens by combining an automated extraction protocol and a commercial real-time PCR assay
F. Grattard, S. Thamm, H. Tuzet, A. Ros, E. Gaillard, B. Pozzetto (Saint Etienne, F; Wiesbaden, D; Rungis, F)
Objectives: To optimise and evaluate the use of an automated extraction on m1000 apparatus (Abbott) before performing realtime PCR using the RealArt C. trachomatis TM PCR kit (Abbott) for a rapid and sensitive detection of Chlamydia trachomatis (CT) in sperm and urine specimens. In France, the detection of CT in sperm is mandatory before assisted reproductive techniques are performed, justifying the need to have validated methods to test such specimens known to contain PCR inhibitors.
Methods: An automated protocol was defined on the m1000 apparatus and applied to the extraction of urine and sperms previously tested positive by the Cobas Amplicor test (Roche Diagnostics) and kept frozen at −20°C. The quality of the extraction on sperm samples was evaluated by the quantification of the beta-globin gene by an in-house real-time PCR. The sensitivity of CT detection was evaluated on sperm samples spiked with dilutions of a positive cloned PCR product included in the PCR kit. In the final protocol, the internal control of the PCR kit was incorporated in the sample tube on the m1000 apparatus in order to control the whole extraction protocol. These extraction and PCR conditions were then evaluated in a prospective study involving 300 clinical specimens of sperm (n = 285) or urine (n = 15).
Results: In the preliminary experiments, the specimens detected positive by Cobas Amplicor and kept frozen were all found positive by combining the automated extraction protocol and real-time PCR. The extraction protocol was found suitable either on crude sperms or on sperm pellets, with a reproducible quality of extraction on the m1000 apparatus, as evaluated after quantification of the human beta-globin gene. The sensitivity of detection in sperms was at least of 1 copy/microlitre elution volume of positive control. In the prospective study, 5 positive specimens were detected, one in urine (6.6%) and 4 in sperm (1.4%). The inhibition rate in sperm was 1.4% (4/285); a pretreatment of samples with proteinase K and SDS removed these inhibitors. The whole duration of the process, including automated extraction and real-time PCR testing, was 4 hours for at least 30 samples.
Conclusion: The m1000 apparatus was found suitable and convenient for the rapid and automated extraction of DNA from sperm and urine specimens before detection of CT by real-time PCR.
P1520: Simultaneous isolation of Chlamydia trachomatis, Neisseria I, Mycoplasma genitalium and Ureaplasma urealyticum from urine, using chlamCAP, an automated sample preparation system based on magnetic particle separation
J. Lysén, H.K. Høidal Berthelsen, M. Anglès d'Auriac, M. Espelund, U.H. Refseth (Oslo, N)
Objectives: Chlamydia trachomatis and Neisseria gonorrhoeae infections are a worldwide public health problem with a combined estimated incidence of over 75 million cases. For C. trachomatis alone approximately 4 mill new cases is registered every year. Sexually transmitted diseases are often caused by more than one microorganism. For instance, co-infections of C. trachomatis with N. gonohorroeae may be as high as 20% in certain countries. Of nonchlamydial, nongonococcal urethrisis, Mycoplasma genitalium and Ureaplasma urealyticum are considered to be the microorganisms most commonly involved. M. genitalium has been directly implicated in numerous genitourinary tract pathologies, and U. urealyticum has been associated with acute prostatatis and preterm delivery in connection with pregnancy.
Methods: Here, we present a new system, chlamCAP, for simultaneous sample preparation of DNA for all four bacteria mentioned above from urine samples followed by four separate PCR amplifications using specific primers. With the new method, the bacteria is initially adsorbed to uniquely coated paramagnetic particles and magnetically separated from the sample. A rapid lysis at room temperature releases DNA, which is adsorbed onto the same magnetic particles. After washing, the robot transfers purified DNA to microwells for PCR analysis.
Results: AC. trachomatis positive urine sample was spiked with N. gonorrhoeae, M. genitalium, U. urealyticum in all combinations of four different concentrations, with the highest concentration at approximately 10.000 CFU/ml. All samples were positive for C. trachomatis indicating that the presence of the other bacteria does not negatively interfere with the capture of this bacterium.
In most samples, all four species could be detected. Moreover, increased concentration of N. gonorrhoeae had a positive effect on the detection of the other species, which can be described as interspecies hitchhiking.
Conclusion: In summary, the results obtained demonstrate that the chlamCAP system can be used to simultaneously detect all these STD species. This new system represents a solution for analysis of the bacteria most frequent involved in STD using urine samples, and could be an important contribution to reduce their prevalence.
P1521: A new flexible automated sample preparation method based on paramagnetic particles for the isolation of Chlamydia trachomatis from urine samples, for downstream analysis using three different naat systems
M. Anglès d'Auriac, M. Espelund, T. Engen, S. Jeansson, G. Størvold, S. Hjelmevoll, S.A. Nordbø, I.J. Haugen, U.H. Refseth (Oslo, Tromsø, Trondheim, N)
Objectives: Chlamydia trachomatis is the leading cause of sexually transmitted disease worldwide. It is important to improve diagnostic methods using non-invasive sample collection to favor increased testing. With the current available methods, swab still prevails over urine sampling mainly due to higher NAAT inhibition and the necessity of centrifugation of urine samples.
Method: A method avoiding centrifugation, chlamCAP (Genpoint, Norway), initially developed for C. trachomatis has been automated using a customized Tecan Miniprep 75 robotic system for DNA preparation from urine samples. A multicentre study is presented where urine samples were analysed for C. trachomatis using the automated chlamCAP system combined with three different NAAT 1) BDProbeTecTM ET (Becton Dickinson), 2) validated in-house Taqman PCR and 3) Cobas Amplicor (Roche). Evaluation was performed using the results produced in parallel by the on-site reference DNA preparation methods and a third independent method was used to resolve discrepant results. With this new method, bacterial cells are initially adsorbed to uniquely coated paramagnetic particles and magnetically separated from the urine, removing NAAT inhibiting substances. After lysis at RT and washing, the robot transfers purified DNA, resuspended in the appropriate buffer, to the final NAAT recipient.
Results: With the BDProbeTecTM ET SDA as downstream analysis system, specificity obtained for both sample preparations was 99.9%, whereas a sensitivity of 98.8% and 97.6% was obtained for BDProbeTecTM ET and chlamCAP, respectively. No inhibition was observed using the magnetic particles. For the in-house developed Taqman analysis sensitivity was 95% and 98.3% for on-site reference sample preparation and chlamCAP, respectively. For the Cobas Amplicor analysis, sensitivity was 93.1 and 98.3% and specificity at 99.7% and 100%, using Cobas Amplicor and chlamCAP procedure for DNA isolation, respectively.
Conclusion: The clear improvements brought by this new automated DNA preparation method will help promote the choice of urine as sample material, alleviating the need of a physician for sample collection. Finally, its compatibility towards diverse NAAT and potential for DNA preparation of other STD organisms such as Neisseria gonorrhoeae, Mycoplasma genitalium and Ureaplasma urealyticum, enables the development of a universal automated DNA preparation platform suitable for most STD laboratories.
Hepatitis A, B and E viruses
P1522: Efficacy of hepatitis B immunoprophylaxis in children with malignant diseases at the time of chemotherapy
O. Geludcova, N. Predeina, M. Rusanova, I. Borodina (Moscow, Chelyabinsk, RUS)
Objectives: To estimate efficacy of the hepatitis B immunoprophylaxis in children with malignant diseases.
Materials and methods: 250 children with different malignancy at the age from 0 to 16 years (median 6 years) which received the chemotherapy were included in the study. 124 patients received an active immunization by the recombinant vaccines «Engerix B» or «HB-Vax II» by scheme: 0–1-2–6 months, 10 mkg − 21 patients, 20 mkg − 92 patients. 13 patients received a combined immunoprophylaxis - specific antibody «Hepatekt» 20 MU/kg by scheme: 0–1-2–3-4–5 months along with «HB-Vax II», 0–1-2–6 months, 10 mkg. Vaccination was conducted in the children without serologic marker of the hepatitis B at the first 3–7 day after the diagnosis of malignancy. 113 children with malignancy not received the specific immunoprophylaxis, and formed a control group.
Results: After the 6 months, the level of the protective antibodies (anti-HBs) was exceeded 10 mMU/ml (the titre median 15.3 mMU/ml) in 66% of children, which received 20 mkg vaccines, and in 25% of children, which received 10 mkg vaccines (g < 0.05). The titre of protective antibodies in 13 patients, which received a combined immunoprophylaxis, was revealed earlier (g < 0.05). The titre median of anti-HBs after 3, 6 and 12 months were accordingly 193.5 mMU/ml, 126.2 mMU/ ml and 47.4 mMU/ml. The contamination of the hepatitis B in group with specific immunoprophylaxis was 13.7%, in control group − 41%, (g < 0.05).
Conclusion: Vaccination in children with malignancy at the time of chemotherapy in the redoubled dose or combined immunoprophylaxis were effective.
P1523: Quantification of serum HBV-DNA with Cobas Taqman for the diagnosis, monitoring and prognosis of chronic hepatitis B virus infection
A.S. Hadziyannis, H. Panopoulou, A. Laras, E.S. Hadziyannis, G. Colucci, S.J. Hadziyannis (Athens, GR; Rotkreuz, CH)
Objectives: Currently commercially available techniques for the detection and quantification of serum HBV-DNA present a number of problems in their application for the diagnosis, monitoring and treatment of chronic HBV infection. The objective of our study was to evaluate in this context the efficacy of the new High Pure System Viral Nucleic Acid/COBAS TaqMan HBV Test (Roche Molecular Systems).
Methods: 400 sera from individuals with current or past HBV infection, chronic HDV infection and subjects unexposed to the hepatitis B virus were tested with the COBAS TaqMan HBV Test. The results were compared to the findings of the COBAS Amplicore HBV Monitor Test (Roche) and our in house Real Time PCR.
Results: The TaqMan test correlated best with our PCR (r = 0.98, p < 0.001). Its specificity was very high, with the TaqMan Test being negative in the samples of persons unexposed to the virus. In samples from patients with low level viraemia, its sensitivity was much higher, picking up 69% of PCR negative and 94% of monitor negative samples. Among patients diagnosed as long-term inactive HBsAg carriers, 95% of the samples were positive with levels ranging from 32 to 106 copies/ml, whereas 16% and 20% of the samples were negative with PCR and monitor respectively. Low HBV viraemia levels were also picked up in patients with markers of past infection, who were negative by the compared PCR methods. All patients with HDV infection had detectable HBV-DNA with a maximum of 15,000 copies/ml. Data from serial samples of patients undergoing antiviral treatment showed that many individuals were wrongly classified as virologic responders on the basis of non detectability with the compared methods, when in fact HBV-DNA was positive or exhibited a breakthrough increase under therapy.
Conclusions: The COBAS TaqMan HBV Test is a sensitive method for detection and quantification of HBV-DNA in serum and can be used for a better monitoring of treatment results. When sequential samples are tested, this method can predict earlier a subsequent breakthrough of the response to antiviral therapy.
P1524: Hepatitis B virus genotype distribution in Greek population and economic immigrants and association with HBeAg status
M. Theodorou, I. Petinelli, A. Mavrommati, A. Tsellou, M. Menexidou, A. Toliopoulos, A. Papanastasiou, A. Chounda, L. Boniatsi, E. Sagana (Athens, GR)
Introduction: HBV strains have been classified into seven genotypes designated A–G based on an intergenotypic nucleotide divergence exceeding 8% of the complete genome. HBV genotypes seem to have a characteristic geographic distribution. More over the majority of Greek patients with CHB are HBeAg (()/HBeAb(+). During the last decade Greece has hosted a great number of economic immigrants.
Aim: Our intention is to assess the prevalence of HBVgenotypes and associate them with HBeAg status.
Patients/Methods: 83 patients (63.8% Greeks, 15.6% Albanians and 20.5% Eastern Europeans–Asians) with clinical profile of chronic HBV infection, HBV DNA(+). HBV Genotype was identified by nested PCR of HBV pol BC and reversed hybridisation (INNO-LIPAINNOGENETICS). HBeAb and HBeAg were determined by MEIA (AXSYM ABBOTT). HBV DNA was quantified by nucleic acid amplification, hybridization to type specific probes and detected by Elisa (Amplicor Monitor ROCHE).
Results: 1) Genotype distribution is as follows; in Greek patients: D 94.3%, A 3.8%, A/D 1.9%. In Albanian patients: D 92.3%, A/D 7,7% and in Eastern European-Asian patients: D 88.2%, A 5.9%, C 5.9%. 2) Genotype D associates with 92.2% HBeAg (() and Genotype A associates with 33,3% HBeAg ((). 3) Genotype D detection among different ethnic groups is: in Greek 94.3%, in Albanian 92.3%, in Eastern European–Asian patients 88.2%.
Conclusions: Our results suggest that: 1) the distribution of the HBV genotype does not differ among Greeks and economic immigrants. 2) Genotype D predominates in Greeks and host population (92.7%) while genotypes A, A/D, C appear to insignificant percentages. 3) Genotype D is strongly associated (92.2%) with HBeAg negative/HBeAb positive CHB infection while genotype A does not seem to be related with HBeAg negative/HBeAb positive CHB infection.
P1525: Evaluation of two commercial HBsAg assays for low-end sensitivity to hepatitis B surface antigen
J. van Helden, O. Lentzen (Mönchengladbach, D)
Objectives: The development of immunoassays with the highest possible sensitivity for detecting those HBV infections most commonly encountered in the routine clinical setting is a major challenge for manufacturers. Currently, the diagnostic sensitivity of most commercial HBsAg tests is below 0.5 ng/ml. However, false negative results are still obtained. This may be due to the presence of HBsAg below the detection limit of the assay, which could occur in early acute disease or in chronic and convalescent phases of the disease. Mutant HBV strains with altered antigens have been shown to affect the reactivity of the antigens with some commercial HBsAg assays. The incidence of these viral variants appears to be limited to chronically infected individuals especially those under medically or naturally induced immune pressure.
Methods: The present study was designed to evaluate the low-end sensitivity of two commercial HBsAg immunoassays, the Bayer ADVIA Centaur® HBsAg assay and Abbott AxSYM® HBsAg assay. The following materials were utilized to evaluate HBsAg assay sensitivity: PEI standards for HBsAg ad/ay subtypes, one BBI HBsAg (ad/ay) sensitivity panel, 3 low-titer and mixed-titer panels, BBI, BCP and NABI HBsAg seroconversion panels, and the Teragenix Hepatitis B Precore Mutant panel.
Results: The Bayer ADVIA Centaur HBsAg assay has a better low-end sensitivity and detects the presence of HBsAg at an earlier stage than the Abbott AxSYM HBsAg assay, thus reducing the window period. The study results also indicate that both assays detect all HBV precore mutants. Results are summarised in Tables 1 and 2.
Table 1.
HBsAg Sensitivity
| HBsAg Material + dilution used | Bayer ADVIA Centaur HBsAg s/co [Cut-off = 1] | Abbott AxSYM HBsAg s/co [Cut-off = 2] | ||
|---|---|---|---|---|
| PEI Standard HBsAg ay 1.10,000 0.01 U/mL | 1.12 | POS | 1.86 | NEG |
| BBI PHA 106-09 1:2 | 2.11 | POS | 1.78 | NEG |
| HBsAg −0.06 IU/mL | 2.34 | 1.87 | ||
| BBI PHA 106-11 1:4 | 1.54 | POS | 1.84 | NEG |
| HBsAg −0.05 lU/mL | 1.66 | 1.91 | ||
| BBI PHA 807-06 1:5 | 2.19 | POS | 1.84 | NEG |
| HBsAg ad- 0.03 IU/mL | 2.25 | 1.97 | ||
| BBI PHA 807- 19 undiluted | 8.63 | POS | 1.97 | NEG |
| HbsAg ay- 0.05 IU/mL | 7.85 | 1.85 | ||
| BBI PHA 807- 20 undiluted | 1.32 | POS | 1.75 | NEG |
| HBsAg ay- 0.02 IU/mL | 1.45 | 1.68 | ||
Table 2.
Comparison of HBV sero conversion panels
| Panel ID | HBsAg Positive Result From Initial Draw Date |
ADVIA centaur AxSYM Assay vs. Assay |
||
|---|---|---|---|---|
| ADVIA Centaur HBsAg Assay (Days) | AxSYM HBsAg Assay (Days) | Difference in Bleeds* | Difference in Days | |
| BCP 62433 | 19 | 22 | +1 | +3 |
| BCP 62675 | 19 | 19 | 0 | 0 |
| BCP 63568 | 14 | 10 | −1 | −4 |
| BCP 62825 | 26 | 29 | +1 | +3 |
| BCP 64121 | 27 | 27 | 0 | 0 |
| BCP 63133 | 36 | 36 | 0 | 0 |
| BCP 61291 | 112 | 98 | −3 | −14 |
| PHM 933 | 7 | 7 | 0 | 0 |
| PHM 931 | 19 | 19 | 0 | 0 |
| PHM 932 | 50 | 61 | +1 | +11 |
| PHM 930 | 0 | 3 | +1 | +3 |
Conclusions: The improved low-end sensitivity of the ADVIA Centaur HBsAg provides a clinically important advantage for routine HBsAg testing.
P1526: Hepatitis B vaccination in chronic renal failure patients with a 4-week schedule
M. Tato, J.L. Teruel, M. Fernandez-Lucas, J. Chacon, M.L. Mateos (Madrid, E)
Objectives: Standard vaccination schedule against hepatitis B consists of three doses of hepatitis B vaccine (20 mcg each), at days 0, 30, 180 (sometimes a fourth booster dose is necessary at day 360). In chronic renal failure patients, the schedule is the same but doses are doubled (40 mcg). Recently, a shorter schedule has been reported to confer the same seroprotection rate as the standard schedule in the general population. This schedule consists of three doses of vaccine, at days 0, 14, 28. The aim of this study was to evaluate the efficacy of a shorter schedule for hepatitis B vaccination and to compare it with the standard schedule in chronic renal failure patients.
Patients and methods: 56 patients on haemodialysis were vaccinated against HBV with the classic schedule: 40 mcg of Engerix (GlaxoSmithKline, S.A.) i.m. in the deltoid region. Eighteen chronic renal failure patients (creatinine clearance 12–27 ml/min/1.73 m2) and 33 patients on haemodialysis received three doses of 40 mcg of Engerix with the short schedule. In non-responders, a fourth dose was administered six months after the third. Antibodies to HBsAg (anti-HBs) were measured with an immunoenzyme assay (Assxym, Abbott Diagnostics). More than 10 UI/l of anti-HBs two months after the first dose of vaccine was considered as protective.
Results: Forty-three of the 56 patients (77%) vaccinated with the classic schedule seroconverted (>10 UI/l of anti-HBs). Eleven of the 18 patients (61%) with chronic renal failure and 10 (30%) of the 30 hemodialysis patients vaccinated with the short schedule seroconverted. Among hemodialysis patients, the response rate was lower in patients vaccinated with the short schedule (30% and 77%, p < 0.001). In 16 non-responders, 9 seroconverted after the fourth dose of vaccine.
Conclusion: In chronic renal failure patients, the short schedule of vaccination against hepatitis B is less efficient than the standard 6-months schedule. A 4-week schedule may be indicated only in patients who have no time to receive a vaccination with a classic schedule. Our data indicate that a booster dose of HBV vaccine will be needed in most of chronic renal failure patients.
P1527: A study of HBV vaccination among anti-HBs negative hospital health care workers
B.-M. Shin, H.M. Yoo, S.M. Yoo, S.K. Park (Sanggye, KOR)
Objective: Needle stick injury is not a rare accident among hospital health care workers. Hepatitis B virus (HBV) infection is one of the main infectious agent through needle stick injury. Therefore, HBV vaccination is strongly recommended for HBV exposure risk group. Through the surveillance we studied how many HCWs could get anti-HBs after vaccination and analysed the effects of HBV vaccination in Korean HCWs.
Methods: We surveyed 571 hospital HCWs (doctors, nurses, aid-nurses and technicians) about previous HBV vaccination history and tested HBsAg, anti-HBs with titer and HBcAb, IgG using enzyme immunoassay. After that, we recommended HBV vaccinations according to 3 cycle schedules (0.1 and 2 months) in 118 cases of anti-HBs negative HCWs.
Results: Of 571 persons, 118 showed anti-HBs negative results. Among them, 58 persons had completely finished 3 cycle HBV vaccination programme. The ratio of male to female was 11:47(19%:81%). The positive rate of anti-HBs were 65.5%(38), 81.0%(47) and 93.1%(54) after 1st, 2nd and 3rd cycle respectively. However, the anti-HBs titers were not associated with vaccination cycles. The 4 cases of anti-HBs negative persons had no previous vaccination histories and HBcAb were also negative. The HBcAb negative group showed remarkably higher anti-HBs titers compared with HBcAb positive group (622 IU/mL vs. 52 IU/mL) though their anti-HBs formation rates were lower than HBcAb positive group (92% vs. 100%)
Conclusions: The lower rate of programme performance (49.2%) needed further regular education about risk of HBV infection among HCWs. The result of HBV vaccination was excellent after 3 cycles and could reduce the risk of HBV infection among HCWs. According to anti-HBs titers, no further vaccination was needed if anti-HBs was acquired during the any vaccination cycles. The different results about anti-HBs formation and the titers of anti-HBs between HBcAb positive and negative group suggested pre-existing under-detectable anti-HBsAb and different memory B cell functions.
P1528: A study of HBV viral marker with HBV vaccination histories in healthcare workers in case of needle stick injury
B.-M. Shin, H.M. Yoo, S.K. Park, A.S. Lee (Sanggye, KOR)
Objective: Needle stick injury associated with HBV is not a rare accident among hospital healthcare workers (HCWs). The prevalence of HBV infection was reported as 5–8% in Korea. Therefore basic data about prevalence of HBV viral markers (HBsAg, HbsAb and HBcAb) with vaccination histories in health care workers are very useful for the guideline of hospital infection control and HCW benefit.
Methods: We tried to survey 725 HCWs including doctors, nurses, aid-nurses and technicians and 571 HCWs (123 male and 448 female) participated in the surveillance and answered about previous HBV vaccination histories. HBsAg, HBsAb (Elecsys 2010, Roche) and HBcAb, IgG (Centaur, Bayer) were analysed with enzyme immunoassay.
Results: The positive rate of HBsAg and HBsAb was 2.4% and 76.8% respectively. HBsAg(−)/HBsAb(−) cases were 20.7%. The range of HBsAb positive rates were 64.3%-79.6% according to the occupational divisions. The division of nurse was the most (80%) and group of doctors (64%) were the least prevalent HBsAb positive groups respectively. Of all cases, 24.2% did not have HBV vaccination histories and 68.8% had experienced more than 1 time of HBV vaccination. The HBsAb titers between 100 and <1000 IU/mL were most popular in HBsAb positive cases. Among 571 cases, 74.1% showed negative HBcAb, IgG and 76.1% of them had HBsAb. The cases showing positive HBcAb also represented 78.1% positive in HBsAb tests.
Conclusion: The prevalence of HBsAg and HBsAb in HCWa in a Korean hospital was similar to general population on Korea. We need more education or announcement for the group of doctors about HBV vaccination. About 75% of hospital HCWs had not been exposed to HBV mainly because of HBV vaccinations. However, no difference was found between vaccination group and non-vaccination group in HBsAb positivity, suggesting Korea is still high prevalent area about HBV infection. Therefore, we need to practice HBV vaccination programme for about 20% of HCWs in case of HBV exposures, such as a needle stick injury.
P1529: Short-term use of lamivudine for patients with severe acute hepatitis B, a series of cases
A. Radulescu, D. Tatulescu, M. Turdean, V. Zanc (Cluj-Napoca, RO)
Objectives: There are limited data on the use of lamivudine for patients with acute hepatitis B. We present our experience with the use of lamivudine in seven patients with severe acute hepatitis B. Short-term use of lamivudine was considered based on almost complete viral suppression within 2–3 weeks and on practical issues (prompt recovery and drug availability).
Methods: Noncomparative prospective study on the use of lamivudine in severe hepatitis B. Diagnosis of fulminant hepatitis B was established based on accepted criteria: clinical signs, portal encephalopathy, high aminotransferases levels, prothrombin levels of less than 50%. Severe hepatitis B was diagnosed upon prolonged and worsening evolution. Serologic tests were performed for hepatitis B surface antigen, Ig M antibodies to hepatitis B core antigen also for hepatitis A and C. Lamivudine at a dose of 100 mg per day was introduced since hepatitis was severe and progressive under supportive treatment. Patients were monitored till hospital discharge and the routine follow-up was ensured.
Results: Fulminant hepatitis B developed in three healthy individuals (27–50 years) and in two cases (50, 62 years), having liver injury due to isoniazid-rifampin and alcohol abuse, respectively. One severe cholestatic hepatitis B occurred in a 62-year-old patient at three months after surgery for cholelitiasis. After initiation of lamivudine astonishing clinical recovery occurred in six cases, the aminotransferases and billirubin levels decreased quickly and patients were discharged well in three weeks without lamivudine treatment excepting the two cases, with previous liver injury who continued the treatment for 3–6 months. After drug cessation, no clinical signs and biochemical tests suggested a rebound. AgHBs seroconversion was documented in three patients at 3 and 6 months, all six patients being well with normal biochemical findings. In a healthy 33-year-old woman, who developed aggressive fulminant hepatitis due to strenuous physical effort and use of anti-inflammatory drugs before onset lamivudine was postponed. No benefit was observed and she died before liver transplantation after seven days of treatment with lamivudine.
Conclusion: Short-term use of lamivudine in severe hepatitis B induced a prompt recovery and sustained serological response. Our data encourage the use of this safe drug that seems to be effective in short regimens covering the period of hepatic failure.
P1530: Detection and characterisation of a hepatitis B surface antigen mutant in a heart transplant patient
A. Boel, A. Vankeerberghen, H. De Beenhouwer (Aalst, B)
Objective: Analysis of mutations in the surface gene (S-gene) of a Hepatitis B Virus (HBV)-mutant in a Heart Transplant (HTx) patient and comparison of the performance of different commercial serological assays for HBV surface antigen (HBsAg) detection for this case.
Methods: HBsAg testing was done with Vidas HbsAg and HbsAg-Ultra (bioMérieux), Advia Centaur HBsAg (Roche), Axsym HbsAg (V2) (Abbott), Abbott Prism HBsAg (Abbott) and Access AgHBs (Analis). HBV surface antibodies (HBsAb), HBV e antigen and antibody (HBeAg and HBeAb) were tested with Vidas (bioMérieux), HBV core antibodies (HBcAb) with Access (Analis). DNA was extracted from serum and amplification was done by real time PCR with primers directed to the precore/core gene of HBV. For analysis of the S-gene sequence, cycle sequencing was done with primers targeting the small S-gene.
Results: Before HTx, the patient was HBsAg and HBV DNA negative but HBcAb and HbsAb positive. Three years after HTx, elevated liver tests were found repeatedly but serological investigation for HAV, HCV, CMV, HSV and EBV was negative. Real time PCR detected high load of HBV-DNA. None of the above mentioned serological assays was able to detect HBsAg in the serum of the HTx patient. However, HBcAb, HBsAb, HBeAg as well as HBeAb were positive. The HBV small surface antigen sequences were analysed by cycle sequencing and compared with NCBI Genbank sequences. This revealed the presence of multiple missense mutations: 15 mutations (compared to AY236162) were found in the major hydrophilic loop (aa 98–169), the most antigenic part of the small surface antigen.
Conclusion: The virus escaped detection by all commercial serological HBsAg assays tested. The majority of the antibodies used in these assays are directed against the small surface antigen, and binding is strongly influenced by the presence of missense mutations at crucial loci present in the viral DNA. Future serological assays for detection of HBsAg should take mutations in the major hydrophilic loop into account in an attempt to detect as much HBV mutants as possible.
P1531: Complete genome sequence and phylogenetic relatedness of hepatitis B virus isolates from Iran
S. Amini-Bavil-Olyaee, R. Sarrami-Forooshani, M. Azizi, A. Adeli, F. Sabahi, M. Abachi, F. Mahboudi (Tehran, IR)
Objectives: To date, no study has been carried out by the hepatitis B virus (HBV) complete genome sequence from Iranian HBV infected patients. The objective of this study was to investigate phylogenetic analysis, genome organization and genotype characterization of HBV strains which obtained from Iranian chronic infected subjects and compared with HBV genotypes reported from the Middle East countries.
Methods: HBsAg-positive sera were collected from the five Iranian chronic HBV carriers. The complete HBV genome was amplified using two novel primers that have introduced Hind III and EcoR I restriction enzyme cleavage sites. The HBV full-length amplicon was cloned into pCR®2.1 plasmid and then sequenced. The Iranian HBV genome sequences were compared with 23 human HBV genome sequences. Alignment was achieved using CLUSTALX software, genetic distance was estimated using the Kimura two-parameter algorithm and then phylogenetic trees were constructed by the neighbor-joining method. Recombination was investigated using SimPlot, BootScanning programme and a web-based genotyping tool for viral sequences.
Results: Results showed that the five strains were closely related to each other, with 97–100% nucleotide similarity. Phylogenetic analysis based on the complete genome sequence revealed that all stains were classified into genotype D. The S gene encoded Arg122, Pro127 and Lys160 corresponding to subtype ayw2. All strains had nucleotide lengths of 3182 bp except the IR-P4 strain with a unique 3185 bp and with a Phe89 insertion in the × gene. The intragenotypic divergence of the complete genome sequence of Iranian strains was 1.8% and the intergenotypic to the genotype D was 3.8% and to other genotypes was 7.9–15.4%. The Middle East countries HBV gene sequences analyses findings showed that HBV genotype D subtype ayw2 is the most dominant. Results showed that Saudi Arabia isolates has the most similarity to Iranian isolates and then Turkish, Egyptian and Yemeni isolates ranked next, respectively.
Conclusion: This is the first report of the full-length nucleotide sequences and genome organization investigation of the HBV isolates from Iran. This study revealed that the HBV genotype D, subtype ayw2 is present in the Iranian infected patients. A unique amino acid insertion in the × gene of the one Iranian strain was detected with an unprecedented length of 3185 bp. The update Middle East HBV geographical genotypic distribution map was depicted.
P1532: Autochthonous sporadic acute hepatitis E cases in Madrid, Spain
A. Molina, J.L. Patier, V. Moreira, J. Chacon, F. Baquero, M.L. Mateos (Madrid, E)
Introduction: Hepatitis E virus (HEV) infection is an important cause of epidemic and acute sporadic hepatitis in developing countries where HEV is considered endemic. In Western Europe hepatitis E virus (HEV) infection is rare and only imported cases have been reported. Recently, autochthonous cases in some industrialized countries have emerged in patients who had never travelled to endemic areas. The aim of this study was to describe nine autochthonous sporadic cases of HEV infection in Madrid (Spain).
Methods: From June 1999 to September 2004, nine patients with acute hepatitis and IgG anti-HEV antibodies in serum specimens were detected at our hospital. Viral markers for HAV, HBV and HCV were negative with an EIA assay (Assxym, Abbott Diagnostics). IgG and IgM anti-HEV antibodies were studied by EIA (Bioelisa HEV IgG and Bioelisa HEV IgM, Biokit; Barcelona, Spain). Detection of RNA-HVC and DNA-HVB was performed with TaqManR (Roche Diagnostics). All patients had increased aminotransferase levels and symptoms of acute hepatitis. Patients were questioned about epidemiological risk factors related to HEV infection such as travelling to endemic areas or contact with animals.
Results: Anti-HEV IgG and IgM antibodies were found in all patients. Seven patients were males and two females, mean age 61.4 years (46–80) with mean ALT values of 1418 (84–5727). No risk factors for HEV infection were found except in two patients: one who worked in a slaughter-house and another who was an occasional horse rider. Patients denied to have travelled to endemic areas.
Conclusions: In endemic areas, HEV infections are most frequently found in young adults. Our patients were older than those reported by other authors. Given that some animals may be reservoirs of HEV and animal-to-human transmission occurs, we think that two of our patients had a risk factor for HEV infection. Although HEV infection is very rare in non-endemic areas, it must be considered in the diagnosis of acute hepatitis with negative viral markers for HAV, HBV and HCV, even if patients have never been in endemic areas. HEV infection must be considered as emergent in our country because of the increasing immigration and geographical situation.
P1533: Seroepidemiology of hepatitis A and hepatitis B virus in Luxembourg
J. Mossong, L. Putz, S. Patiny, F. Schneider (Luxembourg, LUX)
Objectives: A prospective seroepidemiological survey was carried out in Luxembourg in 2000–2001 to determine the antibody status of the Luxembourg population against hepatitis A virus (HAV) and hepatitis B virus (HBV). One of the objectives of this survey was to assess the impact of the hepatitis B vaccination programme, which started in May 1996 and included a catch-up campaign for all adolescents aged 12–15.
Methods: Venous blood from 2679 individuals were screened for the presence of antibodies to hepatitis A virus antigen and antibodies to hepatitis B surface antigen (HBsAg) using an enzyme immunoassay. Samples positive for HBsAg antibody were tested for antibody to hepatitis B core antigen (anti-HBc) using a chemiluminiscent microparticle immunoassay to distinguish between individuals with past exposure to vaccine or natural infection.
Results: The estimated age-standardized anti-HAV seroprevalence was 42% in the population above four years of age. Seroprevalence was age-dependent and highest in adult immigrants from Portugal and former Yugoslavia. The age-standardized prevalence of anti-HBsAg and anti-HBc was estimated at 19.7% and 3.16%, respectively. Anti-HBsAg seroprevalence exceeding 50% was found in the cohorts targeted by the routine hepatitis B vaccination programme, which started in 1996.
Conclusions: Our study illustrates that most young people in Luxembourg are susceptible to HAV infection. The hepatitis B vaccination programme is having a substantial impact on population immunity in children and teenagers.
P1534: Hepatitis A outbreak in a school observed after an explosion in the sewer system
G. Sengoz, M. Bakar, T. Colakoglu, E. Celiikkol, S. Kiliçarslan, S. Uz, E. Tuncer (Istanbul, TR)
Objectives: Hepatitis A is a classical example of infections transmitted by fecal-oral route and its prevalence is closely related with the socioeconomical status. In societies showing moderate endemicity, primary transmission route is personal contact, on the other hand water originated outbreaks may be seen.
Methods and Results: In this study, a hepatitis A outbreak observed in a local school following an explosion in the sewer system in a street in Istanbul Sariyer on September 27th 2004, is investigated. 32 children attending this school and living at homes around had hepatitis A, clinically, the first case seen 25 days after the explosion. Age distribution of the children is 1–18 and the mean age is 8.4. The first case was detected on October 22nd and 21 cases were reported in the first 6 days and 32 cases were seen in 17 days. Water samples were taken and fecal contamination was searched at the school in order to find out the origin. The canteen and the toilets were also searched. All the students were checked up to find out new cases. Teachers and students were educated about the disease and hygiene by doctors of Health Group Presidency, continuity of education was provided by a doctor in charge throughout the outbreak and other schools around were checked and educated by cooperation with Directory of National Education. Origin research was also performed at homes of all cases and the families were informed about the disease and quarantine precautions. ISKI was quickly informed about the subject and controls by taking water examples and documentations were also done by ISKI.
Conclusion: The dimensions of an outbreak of an infectious disease transmitted by water fecal-orally in a metropolis of 11 million population may be great. In our country report of hepatitis A is obligatory. Because of this, the origin of the disease is searched soon after reported cases and the isolation of the cases is provided. Hepatitis A vaccine is not included in the National Vaccination Programme of our country. If there is not a serious risk to have the disease in childhood, there is no need for passive immunization. All the contacts were evaluated in this respect, also. The outbreak was under control at the end of the first week by the precautions and education at the school with a population of 1400.
P1535: Prevalence of anti-HAV antibodies in different age groups in Spain
S. Muñoz Criado, E. Rodríguez Tarazona, A. Rodríguez Achaerandio, J.L. Muñoz Bellido, J.A. García-Rodríguez, M.C. Sáenz González (Salamanca, E)
Introduction: The hepatitis A virus (HAV) usually spreads by the fecal-oral route, and is frequently associated with poor hygiene habits and conditions. The prevalence of HAV decreases when hygiene conditions improve. IgG antibodies against HAV last for years (frequently lifetime) in patients who have suffered hepatitis A, and seroprevalence can so be used to know the frequency of contact with the HAV in a group. We have studied the seroprevalence of IgG antibodies against HAV in sera from non-selected, healthy individuals, obtained in 1997 and 2004, in order to know the seroprevalence in different age groups and the evolution of seroprevalence.
Methods: 250 sera samples obtained in 1997 and 250 sera obatined in 2004 from healthy patients were studied. Both sera groups were organized in 5 age group including 50 sera each, as follows: group 1: patients 18-25 years old, group 2: 26-35 years old, group 3: 36–15, group 4: 46-55 and group 5: 56-65. Sera were studied by EIA (HAV Liaison, Saluggia, Italy.
Results: Appear in Table 1. In 1997, more than 50% of patients between 26 and 35 years old, and almost all patients older than 36 years had anti-HAV antibodies, while only 28% of patients between 18 and 25 years were positive. In sera obtained in 2004, the seroprevalence has changed significantly. The seroprevalences in groups 1, 2 and 3 are much lower (8% vs. 28%, 22% vs. 56%, and 58% vs. 98%), while a seroprevalence of 100% remain in groups 4 and 5.
| Seroprevalence rate (%) |
||
|---|---|---|
| Age groups | 1997 | 2004 |
| 1 | 28 | 8 |
| 2 | 56 | 22 |
| 3 | 98 | 58 |
| 4 | 96 | 100 |
| 5 | 100 | 100 |
Comments: Seroprevalence of anti-HAV antibodies has been very high in adult individuals in Spain, but is decreasing very fast. At this moment, seroprevalence remains around 100% only in patients older than 45 years, and is around 50% in patients between 36 and 45 years, but is becoming very infrequent in young adults. This situation shall be had in account in order to stablish vaccination protocols against HAV.
Herpes virus
P1536: Antibodies to varicella zoster virus: no role in prognosis of Herpes zoster
W. Opstelten, G.A. van Essen, A.J.M. van Wijck, K.G.M. Moons, C.J. Kalkman, A.M. van Loon, T.J.M. Verheij on behalf of ESPRIT
Objectives: To measure antibodies (IgA, IgM, and IgG) to varicella-zoster (VZ) virus in patients with clinical herpes zoster (HZ) and to relate these findings to clinical symptoms of acute HZ and to the occurrence of postherpetic neuralgia (PHN).
Methods: Prospective study of elderly (>50 years) patients presenting to their general practitioner with acute HZ (rash <1 week). Personal and demographic data were registered from patients and pain was assessed using visual analogue scale and quality of life questionnaires. Blood samples were collected at baseline and one week later and analysed for IgA, IgG, and IgM. Pain was assessed at the initial visit and subsequently after 1, 2, 3, 4, 8, 12, and 26 weeks.
Results: 270 patients were included. In 26 (10%) of them HZ could not be serologically confirmed. A significant association was found between titers of IgA and IgG at the initial visit and age and severity and duration of rash. There was no significant association between reported pain and titers of antibodies. Serological data did not contribute to the prediction of occurrence of PHN.
Conclusions: The clinical diagnosis of HZ made by the general practitioner can be serologically confirmed in most cases. The titers of antibodies are correlated with the severity and duration of the rash. They are, however, not associated with the intensity of pain, neither are they predictive for the duration of pain.
P1537: Herpes zoster in hospitalised Turkish children
N. Dalgic, E. Ciftci, Z. örgerin, S. öncel, N. Belet, E. Ince, ü. Dogru (Ankara, TR)
Objectives: To investigate clinical features and outcome of herpes zoster in hospitalized children.
Methods: All records of the children hospitalized for herpes zoster between 1998 and 2004 in the Department of Paediatric Infectious Diseases at Ankara University Medical School, were retrospectively evaluated. Data about age, dermatome involvement, underlying disease, history of previous varicella, the interval between varicella and herpes zoster, herpes zoster-associated pain, treatment and outcomes of the patient were recorded.
Results: During the period of 6 years, 19 children with herpes zoster were identified. Ten out of 19 (52%) were male. The mean age at presentation was 119.3 ± 55.5 months (ranges from 9 months to 15 years). Established underlying diseases for childhood herpes zoster were found in 17 (89%) of 19 cases. The most common underlying diseases were lymphoma (6 patients) and acute lymphoblastic leukaemia (5 patients). No history of varicella was recalled in 9 (47%) of 19 cases. The interval between varicella and herpes zoster ranged from 4 to 144 months. Involved dermatomes were thoracic (10 patients), servical (7 patients) and sacral (2 patients). Two dermatomal involvement was detected in only two children. Two children experienced mild pain during the eruption, without requiring specific therapy. In 3 (15%) of 19 cases, herpes zoster-associated complications including pneumonia, hepatitis and keratocon-junctivitis were noted. None of the patients was immunized against varicella. The mean period between the onset of rash and the initiation of acyclovir treatment was 2.4 ± 1.1 days (range 1–5 days). Eighteen of 19 cases received intravenous acyclovir treatment. Five of these patients had received sequential parenteral/oral acyclovir treatment, whereas only one patient received only oral treatment. Acyclovir treatment was discontinued because of renal toxicity in one child. The mean treatment period was 7.7 ± 1.0 days (range 7–10 days) in 18 patients. No cases with post herpetic neuralgia were recorded.
Conclusion: Acyclovir treatment started on time, is very important for the prevention of progress of the disease. Acyclovir treatment is so effective that acyclovir prophylaxis seems unnecessary to prevent or modify clinical varicella in immunocompromised children. Varicella immunization is important for prevention of varicella and herpes zoster.
P1538: Effects of acyclovir on nitric oxide and cell death in herpesvirus type 1 and 2 infected HEp-2 cells
H. Baskin, Z. Yazici, Y. Baskin, N. Olgun, A. Ozkul, I.H. Bahar (Izmir, Samsun, Ankara, TR)
Objectives: Herpesviruses are among the most ‘successful’ human pathogens. Inhibition of apoptosis is a common strategy of viral pathogenesis that favors virus replication and may contribute to oncogenesis. The herpes simplex viruses 1 and 2 (HSV1, HSV2) code for a variety of proteins that cooperate in blocking apoptosis triggered by virus infection. Nitric oxide (NO), a potentially toxic signal molecule, has been implicated in a wide range of diverse pathophysiological process. The crosstalk between cell destructive and protective signaling pathways, their activation or inhibition under the modulatory influence of NO will determine the possible role of NO in apoptotic cell death and/or survival. Acylovir's effect mechanism targets at the viral DNA polymerase, acts as a chain terminator. Its first phosphorylation step is catalyzed by the virus encoded thymidine kinase. Acyclovir's interactions with cell death and NO seem to be very important because of ‘post treatmental cognitive functions’ that may be encountered due to treatment processes for encephalitis, meningitis cases.
Methods: HEp-2 cells were infected by HSV1 (KOS strain) (and UV-inactivated Mock of the same strain) and HSV2 (G strain) (and UV-inactivated Mock of the same strain). Infected cells were treated by non apoptotic doses of acyclovir (48.8; 24.4; 12.2; 6.1; 3; 1.5 μg/mL), respectively. Following 24 hours of infection cells were detected by Hoechst 33342 and propidium iodide for apoptosis/necrosis, and culture supernatants by Griess reagent for NO.
Results: NO responses were significantly higher in HSV 1 infected cells in 48.4; 24.4; 12.2, but after 6.1, in 3 and in 1.5 NO responses were higher in HSV2 infected cells. Also in HSV1 infected cells with 48.4; 24.4; 12.2 doses of acyclovir, apoptotic responses were higher than HSV2. But necrosis responses were significantly higher in HSV2 infected cells (48.8; 24.4; 12.2; 6.1; 3 μg /mL).
Conclusion: In this study, it is shown that there are significant differences between HSV 1 and 2 in apoptosis/necrosis, and NO responses. It is assumed that involvement of acyclovir in doses higher than 6 μg/mL may trigger the necrotic pathway in HSV2 infected cells, the same compound seems to cause statistically significant higher NO response in HSV1 infected cells in contrast to those infected with HSV2. These results deserve further studies in order to explain the effects of acyclovir in treatment processes especially in ‘cognitive functions’.
P1539: Clinical evaluation and significance of herpes viruses DNA amplification in the central nervous system of neurological patients (2000–2004)
J. Reina, M.D. Macia, O. Gutierrez, E. Ruiz de Gopegui (Palma de Mallorca, E)
Objectives: To retrospective study the clinical manifestations and evaluate the Herpes viruses DNA amplification significance in the CNS of patients with neurological symptoms.
Methods: From January 2000 to September 2004, we studied the CSF of all patients with different clinical symptoms of neurological disease. A commercial nested PCR (Real, Durvitz, Valencia), which amplifies simultaneously the HSV, VZV, CMV, EBV and HHV-6, was used for the detection of Herpes viruses genome. All clinical samples were also inoculated in the MRC-5 cell line for the isolation of entero viruses. The clinical characteristics of patients with a PCR positive were reviewed.
Results: During the study period, 745 CSFs were analysed. Of these, 38 (5.1%) were considered positive, corresponding to different patients. Positive percentages varied over the study period from 1.2% in 2003 to 7.8% in 2004. The 38 CSFs were positive for: 25 HSV (65.7%), 10 CMV (26.3%), 2 VZV (11.1%) and 1 HHV-6 (5.5%). Of the 38 patients, 22 (57.8%) were adults and 16 (42.2%) were children (<15 years); 26 (68.4%) were male and 12 (31.6%) female (p < 0.05). The definitive clinical diagnosis of the HSV-positive patients were: 10 (40%) encephalitis, 6 (24%) lymphocytic meningitis, 3 (12%) intracraneal hemorrhage, 2 (8%) tuberculosis meningitis, and 4 (16%) other neurological syndromes; in this group 4 (16%) patients were HIV-positive. Of the 10 CMV-positive patients, 3 (30%) presented with a congenital CMV infection, 3 (30%) were HIV-positive, 2 (20%) had a lymphocytic meningitis, and 2 (20%) other neurological syndromes. The 2 (100%) VZV-positive patients were children with a clinical post-varicella encephalitis. The only HHV-6 positive patient was a child of 4 years with exanthem subitum with neurological manifestations (convulsions).
Conclusions: The correlation between Herpes viruses genome amplification in the CSF and the clinical study of the patients analysed (clinical significance) was 64% in the HSV, 80% in CMV and 100% in VZV and HHV-6, with an overall correlation of 71%. Of the CMV and HSV-positive patients, 30% and 16% respectively were HIV-positive (p < 0.05). The PCR is a highly sensitive technique although in some cases the results obtained are difficult to relate with clinical symptoms, especially in tuberculosis meningitis and cerebral hemorrhage (false positive).
P1540: The crystal structure of HCMV UL44 and analysis of its interaction with UL54: new targets for drug discovery
A. Loregian, B. Appleton, J.M. Hogle, D.M. Coen, G. Palu (Padua, I; Boston, USA)
The human cytomegalovirus DNA polymerase is composed of a catalytic subunit, UL54, and an accessory protein, UL44, which has been proposed to act as a processing factor. We have solved the crystal structure of UL44 to 1.85 A resolution. Remarkably, the structure revealed a dimer of UL44 shaped like a C-clamp that could partially surround double-stranded DNA. Each monomer resembles herpes simplex virus UL42, with the dimer interface near the base of a region of UL44 identified in the crystal structure as the ‘connector loop’. Analytical ultra-centrifugation and gel filtration measurements demonstrated that UL44 also forms a dimer in solution, and substitution of large hydrophobic residues along the homodimer interface with alanine disrupted dimerization and decreased DNA binding. In addition, to define individual residues in UL44 and UL54 that are crucial for interacting with each other, we have engineered several mutations both in the C-terminal region of UL54 and in the connector loop of UL44. Substitution of alanine for Ile135 in UL44 and for Leu1227 or Phe1231 in UL54 greatly impaired both the UL54/UL44 interaction in pull-down assays and lonGChain DNA synthesis, identifying these residues as crucial for subunit interaction. Using isothermal titration calorimetry, we quantitatively measured the binding of peptides corresponding to the extreme C-terminus of UL54 to UL44. We found that a peptide corresponding to last 22 residues of UL54 bound specifically UL44 in a 1:1 complex with a dissociation constant of approximately 0.7 iM, and that substitution at UL44 residue 135 completely abolished binding to the peptide. Thus, the results identify specific side chains that appear to be crucial for UL54/ UL44 interaction. This information and the structure of UL44 may aid in the rational design of new anti-HCMV compounds which act by disrupting the UL44/UL44 or the UL54/UL44 interaction.
P1541: Development of a real-time NASBA for herpes simples virus 1+2
B. Deiman, F. Jacobs, S. Vermeer, C. Schrover, D. van Strijp (Boxtel, NL)
Objectives: The aim was to develop and evaluate a Herpes simplex virus assay based on NASBA amplification and including real-time detection with molecular beacons. In this study, the performance of this new test is evaluated.
Methods: HSV DNA is isolated using a semi automated magnetic extraction method and the NucliSens® miniMag. An internal control is added to the sample prior to nucleic acid extraction. Primers are directed against the POL-gene region of the HSV genome and both HSV 1 and 2 and the internal control are amplified with the same primer set. The assay includes three molecular beacon probes: two genotype-specific molecular beacon probes for the detection of HSV 1 and HSV 2 specifically, and one additional beacon for the detection of the internal control. Amplification reactions were performed in a NucliSens® EasyQ Analyser allowing real-time detection.
Results: HSV 1, HSV 2 and IC are detected in one and the same reaction using a three-label approach. In coinfections, both HSV 1 and HSV 2 are detectable. Using serial dilutions of in vitro DNA, HSV detection is shown down to ≤10 copies in amplification for both HSV 1 and HSV 2. Good results are obtained with the QCMD panel: HSV was detectable down to 1–6 × 102 Geq/ml (lowest concentration) and the genotypes of the HSV samples were identified correctly. In addition, HSV is detectable in clinical samples (CSF, swabs). VZV and EBV are not detectable with this assay. With the real-time assay, HSV in CSF samples is detectable within approximately 3.0 hours.
Conclusions: The data showed that the real-time and internally controlled HSV assay is a rapid, sensitive and specific qualitative assay for the detection of HSV 1 and HSV 2 in one and the same reaction. The use of standardized reagents offers considerable benefits in a routine setting for the clinical management of patients with HSV infections. This assay will bring same-day results, enabling early and accurate clinical intervention leading to better patient care and considerable overall health care cost savings.
P1542: Development of type specific based glycoprotein G assay to identify of HSV-2 infection
M. Jamalidoust, F. Fotouhi Chahooki, H. Soleimanjahi, M. Roostaee (Tehran, IR)
Objective: Herpes Simplex Virus type-2 (HSV-2) is the main cause of genital herpes infection. Its prevalence is increasing worldwide and varies widely with generally higher rate in developing than developed countries and urban than rural areas. For studying prevalence and seroepidemology survey, HSV gG-2 was prepared in prokaryotic system to recognize asymptomic and unknown symptomic individuals.
Methods: HSV-2 Iranian isolate was propagated in HeLa cell line. The viral genome was extracted by phenol-chloroform, and used as template in nested polymerase chain reactions (n-PCR) to amplify gG-2 gene. The amplified gene was cloned into a cloning vector (pTZ57R/T) after sequencing. In the next step competent E-coli DH5a was transformed with constructed plasmid. The resulted white colonies were selected in the presence of ampicilin; X-gal and IPTG. The recombinant plasmid (pTZ57R-gG2) was extracted and purified in the large scale after restriction enzyme analysis. The 1.1 Kb fragment was extracted and purified from LMP gel after treated with BamHI and HindIII enzymes. The isolated gene was subcloned into appropriate sites of pET32 (a) plasmid as an expression vector. The protein production was confirmed by SDS-PAGE and western blotting after induction by IPTG using a polyclonal antibody.
Result: The HSV-2 gG gene was confirmed by sequencing. The results of western blotting analysis revealed that glycoprotein G of Herpes Simplex Virus type 2 was expressed efficiently in the prokaryotic system.
Conclusion: The pure gG-2 can be used as a candidate antigen instead of crude whole HSV-9+2 in the serological tests such as ELISA.
P1543: Markers of inflammatory process in genital virus infection
A.V. Shurshalina, L.A. Marchenko, G.T. Sukhikh (Moscow, RUS)
Objectives: Severe genital herpes may be the reason of infertility, recurrent pregnancy loss and IVF failures.
Methods: 103 women (mean age 30.8 ± 0.82) with severe genital herpes (mean recurrence per year 7.5 ± 0.6) were included in study. We tested cervical samples for cytokines (IL-6, IL-10, TNF-alfa, IFN-gamma) and serum samples for CD69+, CD25+ and HLA-DR+ during recurrence, remission and after 8-month Valaciclovir treatment in the dose of 500 mg per day. Control group was ten healthy women without genital herpes and HSV antibodies G and M in serum.
Results: During herpes recurrence, the local levels of TNF-alfa increased twice and 8 times as much as compared to remission and the controls, IFN-gamma − 1.5 and 2.3 times, CD69+ − 1.4 and 3.3 times, CD25+ − 1.4 and 2.5 times, HLA-DR+ − 1.6.and 2.5 times (p < 0.05). IL-10 decreased 4 and 3.3 times as less. After Valaciclovir treatment, the cervical levels of TNF-alfa decreased by 60%, CD69+ − 26%, HLA-DR+ − 23.6% (p < 0.05).
Conclusion: very high levels of inflammatory markers in cervix and serum during severe genital herpes confirm an important role of HSV in genital inflammation, cervical and endometrial pathology, and allow to consider genital herpes as an inflammatory process.
P1544: Herpes simplex virus – causative agent, or innocent bystander?
K. Soltysova, E. Bronska, D. Picha, V. Maresova (Prague, CZ)
Objectives: Herpes simplex virus (HSV) is a well-known causative agent of sporadic necrotising encephalitis. Encephalitis occurs during HSV virus primoinfections and reactivations. HSV neuroinfections may also have milder forms that often remain unnoticed if the patient's cerebrospinal fluid (CSF) is not directly tested for the HSV DNA presence.
Methods: Medical records of 1800 patients hospitalized with neuroinfections from 1998 to 2003 were retrospectively searched for examination results indicating the presence of HSV DNA in CSF. 89 patients were examined. The presence of HSV was tested using the nested PCR method at the National Reference Laboratory in Prague. Out of these patients, 6 cases of immunocompetent adults were identified, which themselves contained another etiological agent. The following data was collected: severity of the symptoms, the hospitalization length, permanent neurological sequels, and the results of neuroimaging methods including EEG. The clinical course of dual infections was then compared to all the imunocompetent adults hospitalized with neuroinfection in 2001, 185 in total.
Results: 14 cases of HSV DNA were identified, including the 6 dual infections mentioned above. Among the 6 cases, 4 corresponded to the tick-born encephalitis agent (serologically confirmed), 1 case to neuroborreliosis, and 1 case to purulent meningitis. The 6 patients were 27–80 years old, with an average age of 58. All showed a substantial neurological involvement during the entrance examinations. The average hospitalization period for the dual infections was 14 days longer than in the 185 reference patients. Focal neurological sequels, typical for HSV encephalitis, were confirmed only in 1 case, the other 5 showed normal CT results. All 6 patients were treated with Acyclovir at least for 10 days. Despite the dual infection and severe entrance neurological involvement, the treatment showed positive results.
Conclusion: The number of patients suffering from dual infections is underestimated. Reactivation of herpes viruses is one cause of the dual infections. Since dual infections have additive effects, it is desirable to identify these cases and to cure them causatively.
P1545: Increasing evidence of primary cytomegalovirus infection in otherwise healthy adults
R. Manfredi, L. Calza, F. Chiodo Bologna, I
Introduction: The increased availability of serology-biomolecular assays for acute Cytomegalovirosis (C), allows us to include these assays in the workout of fever of unknown origin (FUO) of immunocompetent adults. A retrospective survey of patients (p) diagnosed with primary C after referring to our centre because of a FUO associated with a broad spectrum of constitutional signs-symptoms was performed.
Patients and Methods: Since the year 2001,103 p aged 14–42 years (64 females), were assessed for FUO.C serology was carried out in the 1st-step workout, together with haematological–biochemistry studies, and assays for collagen vascular and autoimmune disorders, thyroid function, and microbiology studies for mononucleosis, toxoplasmosis, HIV, community-acquired respiratory, urinary, and stool pathogens, and tuberculosis. All p underwent C serology for IgM-IgG search, a minority of p had viral DNA-antigen search.
Results: Of 103 p,14 (13.6%) had a positive IgM search for C, confirmed in 4 cases by biomolecular-antigen assays. A crossreaction with EBV serology was found in 8 p, but IgM antibody titres against EBV were lower and rapidly declined during a 6–12 month follow-up. An altered leukocyte count and differential was always present, and lymphocyte subsets showed a transient imbalance of CD4/CD8 cell ratio, a decrease of rate and absolute number of CD4 cells (250–580/μL), and an expansion of CD8 subset. Underlying signs-symptoms included fever in all p associated with a mononucleosis-like syndrome in 11 p, usually characterized by a moderate lymphadenopathy and pharyngodinia. In 10 cases a moderate (2–4-fold) elevation of transaminases was present and hepatosplenomegaly was confirmed by ultrasonography. In 3 p only fever, fatigue and anorexia were the predominant signs and symptoms. A normalization of hepatic enzymes and leukocyte differential preceded the disappearance of lymphadenopathy and hepatosplenomegaly, while positive C IgM search lasted until 36 months in 1 p (mean time to disappearance:10–26 months).
Discussion: Primary C is a self-limiting disease, whose apparent increase in frequency is probably attributable to the easier access to specific laboratory testing. Although no treatment is indicated in otherwise immunocompetent p, clinicians should maintain an elevated suspicion for primary C when a FUO is of concern, since C may last significantly and p with missed diagnosis are at risk of a vicious circle between an apparently unexplained disease, and the need of further examinations.
P1546: Risk factors and seroprevalence of serum antibody to human herpesvirus 8 among HIV seropositive and seronegative adults from Croatia
O. Dakovic Rode, J. Begovac, S. Zidovec Lepej (Zagreb, HR)
Objectives: Human herpesvirus 8 (HHV-8) IgG antibodies precede and strongly predict the development of Kaposi's sarcoma (KS), but HHV-8 requires additional factors to exert its effect in KS development. The aim of this study was to estimate possible predictors of HHV-8 infection.
Methods: A cross-sectional study was conducted at the University Hospital for Infectious Diseases in Zagreb from May 1999 till August 2001. A total of 166 serum samples from adult HIV-infected patients and 219 from blood donors were analysed for IgG antibodies to HHV-8 by enzyme-linked immunosorbent assay (ELISA) using whole virus lysate as antigen (ABI, Columbia, USA). We analysed HHV-8 infection according to gender, age, geographic origin (Mediterranean part of Croatia compared to other parts of the country), HIV-infection and herpes simplex 2 (HSV-2) infection determined by a gG2-specific ELISA (DiaSorin, Saluggia Italy). Bivariate and multivariate logistic regression was used to evaluate association between HHV-8 and investigated parameters.
Results: The prevalence of HHV-8 was 13.3% among HIV-infected patients and 4.1% in blood donors. There were no differences among HHV-8 prevalence according to gender and geographic origin. HHV-8 prevalence among HIV-infected patients was significantly higher in older age (OR 4.0; 95%CI 1.39–14.29). Independent predictors of HHV-8 infection included HIV-infection (OR 2.72; 95%CI 1.11–6.65), HSV-2 infection (OR 2.53; 95%CI 1.09–5,89) and male gender (OR 5.0; 95%CI 1.12–20.00)
Conclusion: HHV-8 infection was independently associated with HIV, HSV-2 and male gender. Our results suggest that sexual transmission of HHV-8 is the most important mode of infection with HHV-8 in adults from Croatia.
P1547: Enzyme immunoassay for the detection of anti-Kaposi's sarcoma-associated herpes virus IgG antibodies based on new mosaic protein
N. Kornienko, M. Gladysheva, T. Ulanova, V. Puzyrev, A. Burkov, L. Loginova, A. Obriadina (Nizhniy Novgorod, RUS)
Background: Kaposi's sarcoma-associated herpes virus (or human herpes virus type 8 HHV-8) is a recently discovered gamma-herpesvirus associated with 4 clinical and epidemiological variants of Kaposi sarcoma (classic, endemic, iatrogenic, and acquired immunodeficiency virus-associated), primary effusion lymphoma and multicentric Castleman's disease.
Objective: The aim of this study was to learn the antigenic properties of new recombinant mosaic protein and to develop and evaluate a screening enzyme immunoassay (EIA) for the detection of anti HHV-8 IgG activity in serum specimens.
Materials and methods: Mosaic of 2 antigenic domains from the proteins encoded by open reading frames 65 (140–170 aa) and K8.1 (32–62 aa) of HHV-8 was produced as GST fusion protein to develop an assay for the detection of anti HHV-8 antibodies. Assay conditions were optimized to reduce the possibility of false positive and false negative results. To validate the specificity and sensitivity of new EIA sera from HIV-infected individuals (n = 163), children (0–15 years) (n = 170), patients with sexual transmitted diseases (STD) (n = 136), and from European normal blood donors (n = 1349) were tested. Serum samples from KS patients (n = 30) were initially tested as positive by immunofluorescence assay with LANA protein. All specimens were additionally tested for IgG anti-HHV-8 activity by commercially available EIA (Vecto HHV-8-IgG-strip, Russia).
Results: 27 out of 30 HHV-8-positive samples were positive on the novel EIA. Assay sensitivity was calculated at 90%. Coincidence with commercially available EIA was 98.21%. The percentages of positive reactivity in all investigated groups were as follows: 119 for health blood donors, 1.76 for children, 2.21 for STD-patients and 3.38 for HIV-infected. Specificity of the assay was around 98.2%-98.8% and there were no significant differences between health donors/children and groups at highest risk of acquiring HHV8 infection.
Conclusion: The artificial mosaic protein used in this study demonstrated significant potential as diagnostic reagent. The new EIA is highly specific diagnostic assay for the detection of anti-HHV-8 activity in serum specimens and may be useful tool for studies of HHV-8 epidemiology. KSHV seroprevalence in the Russian European population is low both among health donors and HIV-infected people.
P1548: Human herpes virus 8 infection and pulmonary hypertension in lung transplantation candidates
E. Nicastri, C. Vizza, S. Cicalini, F. Carletti, R. Badagliacca, R. Poscia, F. Fedele, N. Petrosillo (Rome, I)
Objectives: Human herpes virus-8 (HHV-8) has been detected in biological samples of patients affected by Kaposi Sarkoma, Castleman disease and primary-effusion B-Cell lymphomas. HHV-8 has been recently described in the pathogenesis of idiopathic pulmonary arterial hypertension (IPAH). We conducted a seroprevalence study aimed at detecting antibodies to HHV-8 among a population of lung transplantation candidates; we assessed the HHV-8 antibodies seroprevalence among pulmonary hypertension (PH) patients with and without IPAH, and we compared them with non-PH patients.
Methods: From January 2001 to February 2004, 75 patients referred to the Department of Cardiovascular and Respiratory Sciences of the University of Rome La Sapienza for clinical and serological evaluation for lung transplantation were consecutively enrolled in the study. The diagnosis of PH was based on international criteria. We performed assays for antibodies directed to lytic antigens of HHV-8 in plasma samples using an indirect immune fluorescent assay based on BCBL-1 cell line. Samples reactive at 1:40 dilution in the anti-lytic test were considered as positive.
Results: Thirty-three patients out of 75 transplant candidates had significant PH (mean pulmonary arterial pressure >25 mmHg. Sixteen of them had IPAH, the ramining had secondary-PH. HHV-8 antibodies were detected in 9 out of 75 patients (11.5%). Three patients with HIV infection were HHV-8 Ab negative. A difference in the HHV-8 seroprevalence was found between the PH patients (3.0%) and the patients without PH (19.0%). Particularly, among the 33 patients with PH, one patient out of 16 with IPAH (6.3%) had serological HHV-8 antibodies, whereas no patient with secondary-PH had HHV-8 antibodies. Among the 42 patients with no clinical or diagnostic evidence of PH, five patients out of 29 with cystic fibrosis (17.2%), and three patients out of 13 with interstitial lung disease (23.1%) had HHV-8 antibodies, all of them affected by idiopathic pulmonary fibrosis. No difference in the HHV-8 seroprevalence rate was reported neither in patients with cystic fibrosis nor in patients with interstitial lung disease
Conclusions: A 11.5% HHV-8 prevalence rate similar to that found in the Italian general population was reported among candidates to lung transplantation. No relationship between HHV-8 infection and PH, both idiopathic and secondary, was shown.
P1549: Molecular epidemiology of Kaposi sarcoma herpesvirus in Cuban and German patients with Kaposi's sarcoma (KS) and asymptomatic sexual contacts of Cuban KS patients
V. Kouri, A. Marini, R. Doroudi, M. Rodriguez, V. Capo, S. Resik, O. Blanco, B. Mantecon, U. Hengge (Havana City, CUB; Dusseldorf, D)
Objectives: Since the discovery of Kaposi sarcoma herpesvirus (KSHV) in 1994, many questions arose about the origin, transmission and pathogenesis of KSHV. The ORF K1 gene is used to define five major subtypes A-E, with 15–30% of amino acid variability. There is not previous reports of ORF K1 Real Time assay.
Methods: We analysed the K1 gene phylogenetically in 17 Cuban and 19 German KS tissues (most HIV+) by nested PCR and sequencing. In addition, we studied for the first time-24 PBMC specimens from Cuban asymptomatic sexual contacts of KS patients for the presence of circulating KSHV DNA by Real-Time PCR and also we sequenced those whose results. Moreover, we compared KSHV viral load between PBMC and KS tissues.
Results: Ten of 24 (41.7%) PBMC samples showed KSHV infection by the K1 nested PCR, reinforcing that KSHV is sexually transmitted. Phylogenetic characterization revealed the majority of German samples belonging to subtypes C (n = 13) and A (n = 6), as has been previously described for European KSHV strains. Cuban samples showed a wide range of subtypes, including A (n = 17.4 were A5), C (n = 7) and B (n = 2), and one newly described subtype E. Subtypes A5 and B have previously been reported as African genotypes and E subtype has been recently found in Amerindians from Brazil and Ecuador. Furthermore, Real-time PCR detected 5 additional patients with very low KSHV viral load in PBMC that were not detectable with nested PCR. The KSHV viral load in KS tissues was much more higher than in PBMC of asymptomatic infected patients, there was also correlation among KSHV antibodies and the two DNA amplification methods.
Conclusions: The subtype variability reflects the mixed ethnic background of Cubans. This new DNA amplification assay (Real Time PCR) represents a reliable technique to identify individuals at risk for developing KS lesions as well as following KSHV viral load during therapy.
New drugs: in vitro studies
P1550: In vitro activity of plectasin, an antimicrobial peptide from the fungus Pseudoplectania nigrella
P. Mygind, K. Schnorr, M. Hansen, C. Sonksen, D. Raventos, B. Christensen, J. Vind, D. Yaver, H.-H. Kristensen (Bagsvaerd, DK; Davis, USA)
Objectives: Plectasin (NZ2000) is a newly discovered defensin-type antimicrobial peptide of 40 amino acids isolated from the saprophytic ascomycete fungus Pseudoplectania nigrella. Plectasin is active against several Gram-positive bacteria including drug resistant strains but shows very poor activity against Gram-negative bacteria. In addition, Plectasin has shown activity in murine pneumonia and peritonitis models against clinical isolates of Streptococcus pneumoniae when the peptide is dosed intraveneously.
Methods: Antimicrobial activity and spectrum were assessed by determining the minimum inhibitory and bactericidal concentrations in cationic-adjusted Mueller-Hinton broth using the NCCLS guidelines. Bactericidal activity was characterized by time kill experiments at several concentrations.
Results: Purified Plectasin showed potent activity against a range of Gram-positive bacteria with MICs as low as 0.25 μg/ml against clinical isolates of S. pneumonia including penicillin, tetracycline, chloramphenicol and trimethoprim resistant isolates. Plectasin is cidal as demonstrated by MBCs that are equivalent to the MICs and has rapid kill kinetics in time kill experiments. After 24 hours incubation in 90% human serum, Plectasin is stable as measured by both ELISA and antimicrobial activity.
Conclusions: Plectasin is a novel antimicrobial peptide that displays potent antimicrobial activity against Gram-positive bacteria including drug resistant organisms. The peptide is rapidly cidal against all Gram-positive bacteria but inactive towards Gram-negative bacteria. Results reported here on serum stability as well as in vivo animal models suggest that Plectasin may have potential as a systemic therapeutic agent against Gram-positive bacteria.
P1551: In vitro evaluation of the peptide deformylase inhibitor LBM415 in combination with aztreonam to determine the synergistic potential against Gram-positive and Gram-negative pathogens
D. Biedenbach, R. Jones, T. Fritsche (North Liberty, USA)
Objective: To evaluate the interactions between LBM415 (LBM) and aztreonam (AZT) in vitro. LBM is a peptide deformylase inhibitor (PDI), a class of compounds with a novel mechanism of action against Gram-positive (GP) organisms and some respiratory tract Gram-negative (GN) fastidious pathogens that is currently in clinical development.
Methods: A checkerboard broth microdilution test method was used to test 108 clinical isolates: Enterobacteriaceae (ENT; 81), nonfermentative GN (NFGN; 15), N. meningitis (NM; 2), Staphylococci (included MRSA and MR-CoNS; 6) and Enterococcus spp. (4). Tests were performed using Mueller-Hinton broth (supplemented with lysed horse blood for NM). The following definitions were used in characterizing combination MIC results: antagonism (ANT), ≥4-fold increase of both agents; synergy (SYN), ≥4-fold decrease of both agents; partial SYN (PSYN), ≥4-fold decrease of 1 agent/2-fold decrease of the other; additive (ADD), 2-fold decrease of both agents; indifference (IND), no decrease or only a 2-fold decrease or increase for 1 agent.
Results: No ANT was observed for any isolate or tested combination. All 10 GP pathogens showed IND to the combination of LBM and AZT. The ENT showed a significant enhanced activity with 69–100% of each species giving additive or greater interaction results (exception, S. marcescens). Among Klebsiella spp. (KSP) isolates, 53% displayed SYN, E. coli (EC) and Enterobacter spp. (EBS) showed 56% PSYN or SYN and 80% of P. mirabilis (PM) showed PSYN. Among the NFGN, Acinetobacter (ASP) isolates showed 60% SYN and 40% PSYN, whereas only 27% of P. aeruginosa were PSYN or ADD. S. maltophilia (40%) and B. cepacia (20%) isolates displayed ADD effects; the rest were IND to the combination. One of the NM showed an ADD effect.
Conclusions: The combination of LBM and AZT provided predominant SYN or PSYN interactions against many GN organisms (ASP, KSP, EC, EBS and PM). Several strains of other GN species showed at least an ADD interaction effect. These preliminary studies demonstrate that PDIs may display enhanced activity when tested in combination with a GN active agent and a complete lack of ANT in either GP or GN pathogens, potentially expanding the spectrum of activity of both agents. The unique mechanism of action of PDIs warrants further investigation into their potential role in combination therapy.
P1552: Antimicrobial activity of LBM415 (NVP PDF-713) against a recent (2003) international collection of Gram-positive and respiratory tract pathogens
A. Watters, R. Jones, M. Stilwell, T. Fritsche (North Liberty, USA)
Objective: To evaluate the spectrum and potency of LBM415 (LBM; formerly NVP PDF-713) when tested against an international collection of common bacterial pathogens comprising the 2003 LBM surveillance programme. LBM is the first member of the peptide deformylase inhibitor (PDI) class being considered for clinical trials for treatment of community-acquired respiratory tract infections and serious infections caused by antimicrobial-resistant Gram-positive cocci.
Methods: Non-duplicate, clinically-significant bacterial isolates (10,903 strains) were collected from more than 70 medical centres participating in the global surveillance programme, and originated from North America, Europe, South America and Asia geographic areas. All isolates were susceptibility (S) tested using NCCLS broth microdilution methods and interpretive criteria, where applicable. LBM was compared with representative agents used in the empiric or directed therapy of the targeted indications or species.
Results: The antimicrobial activity of LBM is shown in the table: Antibiogram characteristics of the collection include: 28% oxacillin-resistant (OR) SA, 76% OR CoNS, 34% penicillin-non-susceptible SPN, 22% vancomycin-R ENT, and 21% ampicillin-R HI. LBM was highly active against Staphylococci, Streptococci, Enterococci and MCAT, resulting in ≥99% inhibition at ≤4 mg/L, as well as 93% inhibition of HI strains at the same concentration. No differences were noted in the activity of LBM between S and R subsets of SA, CoNS, SPN, ENT and HI for antimicrobials such as oxacillin, penicillin, ampicillin, macrolides, vancomycin and fluoroquinolones. While regional differences were apparent with some comparator agents, activity of LBM did not vary between geographic samples.
| Organisms (no. tested) | MIC (mg/L) |
Cum. % of strains inhibited at MIC (mg/L): |
||||
|---|---|---|---|---|---|---|
| 50% | 90% | ≤1 | 2 | 4 | 8 | |
| S. aureus (SA; 3,955) | 0.5 | 1 | 99 | 99 | 100 | – |
| Coag-neg staphylococci (CoNS; 1,485) | 0.5 | 1 | 95 | 99 | 100 | – |
| S. pneumoniae (SPN, 736) | 1 | 2 | 76 | 93 | 100 | – |
| Viridans group streptococci (134) | 0.25 | 1 | 99 | 100 | – | – |
| Beta-haemolytic streptococci (356) | 0.5 | 1 | 96 | 100 | – | – |
| Enterococci (ENT, 2,062) | 2 | 4 | 48 | 87 | 99 | 100 |
| E.faecalis (1,432) | 2 | 4 | 37 | 83 | 99 | 100 |
| E. faecium (553) | 1 | 2 | 70 | 96 | 100 | – |
| H. influenzae (HI 1,855) | 2 | 4 | 49 | 78 | 93 | 99 |
| M. catarrhalis (MCAT, 108) | 0.25 | 0.5 | 99 | 100 | – | – |
Conclusions: LBM displays a broad-spectrum of activity against the common Gram-positive pathogens, including R subsets, with no regional differences in potency. Some fastidious Gram-negative species (HI, MCAT) were also inhibited by LBM. As a class of antimicrobials, the PDI compounds exhibit a spectrum of activity and unique mode of action that promises to be a significant advance in chemotherapy.
P1553: A baseline assessment of telavancin's activity against a collection of Gram-positive isolates, including resistant phenotypes
D.C. Draghi, R. Blosser, M. Jones, D.F. Sahm (Herndon, USA; Hilversum, NL)
Objectives: As new antimicrobial agents are developed, it is important to examine their in vitro activity against a collection of organisms for which they are targeted. Telavancin (TLV) is a rapidly bactericidal lipoglycopeptide with multiple mechanisms of action that is in Phase 3 development for the treatment of serious infections caused by gram-positive pathogens. The current study examines the in vitro activity of TLV against 101 staphylococci, Enterococci, and Streptococci, including clinically-relevant resistant phenotypes.
Methods: 50 Staphylococcus aureus (SA), 11 coagulase-negative Staphylococci (CNS), 10 Enterococcus faecalis (EF), 10 Enterococcus faecium (EM), 10 Streptococcus pneumoniae (SP), and 10 other Streptococcus spp. (STR) were tested against TLV and comparator agents by broth microdilution (BMD; NCCLS, M7-A6, 2003) and E-test. BMD and E-test MICs were analysed on an isolate-per-isolate basis, and MICs within plus or minus one doubling dilution were considered correlative.
Results: Against the SA tested, TLV had an MIC range of 0.12–4 mg/L. MICs were similar against both MSSA (0.12–1 mg/L) and MRSA (0.12–1 mg/L). TLV MICs to vancomycin (VAN)-nonsusceptible (NS; intermediate and resistant) SA (n = 4) were 2–4 mg/L and against daptomycin-NS SA (n = 3) were 0.5–2 mg/L. TLV showed MICs against CNS of 0.03–0.5 mg/L with consistent activity against oxacillin-susceptible (S) (0.03–0.5 mg/L) and -resistant (R) (0.12–0.25 mg/L) strains. Against VAN-S EF and EM, TLV showed MICs of 0.25–0.5 mg/L and 0.06–0.5 mg/L, respectively and elevated MICs of 0.25-8 mg/L and 4–8 mg/L against VAN-R strains. TLV showed low MICs against all Streptococci (≤0.06 mg/L), including penicillin-R and multidrug-R SP (MICs = 0.015 mg/L). TLV MICs generated using E-test were similar to those generated using BMD. Correlations between E-test and BMD were 84% for SA, 100% for CNS, 100% for EF, 90% for EM, 100% for SP, and 100% for STR.
Conclusions: TLV showed potent in vitro activity against the gram-positive organisms tested, including isolates with clinically-relevant resistant phenotypes.
P1554: Antimicrobial activity of daptomycin tested against Gram-positive strains collected from European medical centres in 2004
H. Sader, T. Fritsche, R. Jones (North Liberty, USA)
Background: Daptomycin (DAP) is a recently FDA approved lipopeptide with activity against relevant Gram-positive pathogens. We evaluated the in vitro activity of DAP tested against recent clinical isolates collected in Europe in 2004.
Methods: A total of 2103 consecutive strains were collected in 23 medical centres located in 12 European countries. The main pathogens evaluated were: S. aureus [SA; 1172 isolates, 27% oxacillin (OXA)-resistant (R)]; coagulase-negative Staphylococci (CoNS; 404, 79% OXA-R), E. faecalis (EF; 195; 2% vancomycin [VAN]-R), E. faecium (EFM; 90, 14% VAN-R), beta-haemolytic Streptococcus spp. (BHS; 150), and viridans group Streptococcus spp. (VGS; 73). The strains were tested by NCCLS broth microdilution methods in Mueller-Hinton broth supplemented to 50 mg/L calcium. Numerous comparators were also tested.
Results: DAP activity is summarized in the table: All isolates, except 2 VAN-S EFM, were inhibited at DAP MIC of ≤4 mg/L. DAP and linezolid were the most active agents against VAN-R Enterococci. All CoNS and 99.7% of SA isolates evaluated were inhibited at DAP MIC ≤1 mg/L. DAP was highly active against BHS (MIC50, ≤0.06 mg/L) and 97% of VGS strains were inhibited at DAP MIC ≤1 mg/L (susceptible).
| DAP MIC (mg/L) |
%S | |||
|---|---|---|---|---|
| Organism (no. tested) | 50% | 90% | Range | |
| SA OXA-3 (860) | 0.25 | 0.5 | ≤0.06–2 | 99.9 |
| OXA-R (312) | 0.5 | 0.5 | 0.12–2 | 99.0 |
| CoNS OXA-S (85) | 0.5 | 0.5 | 0.12–1 | 100.0 |
| OXA-R (319) | 0.5 | 0.5 | ≤0.06–1 | 100.0 |
| EF(195) | 1 | 1 | ≤0.06–4 | 100.0 |
| EFM VAN-G (77) | 4 | 4 | 1—8 | 97.4 |
| VAN-R (13) | 1 | 4 | 1—4 | 100.0 |
| Enterococcus spp. (19) | 1 | 4 | 0.5–4 | 100.0 |
| BHS (150) | ≤0.06 | 0.25 | ≤0.06–0.5 | 100.0 |
| VGS (73) | 0.25 | 1 | ≤0.06–2 | − |
Conclusions: R to other compounds did not influence the high DAP activity against Staphylococci, Enterococci or Streptococci. DAP showed high potency and broad spectrum against recent clinical isolates of facultative Gram-positive cocci isolated in European medical centres, including multi-drug resistant subsets.
P1555: Comparative in vitro activity of XRP 2868, a new, oral streptogramin
D. Felmingham, M.J. Robbins, J. Shackcloth, C. Dencer, L. Williams, C. Couturier, A. Bryskier (London, UK; Romainville, F)
Objective: XRP 2868 is an investigational, oral streptogramin comprised of a 70:30 mixture of RPR 132552A (a pristinamycin IIB derivative) and RPR 202868 (a pristinamycin 1A derivative). In this study, the in vitro activity of XRP 2868 was determined in comparison with Synercid, pristinamycin and erythromycin A, against bacterial pathogens causing respiratory tract and skin/ soft tissue infections.
Methods: Minimum inhibitory concentrations (MIC) of XRP 2868 and comparators were determined using appropriate NCCLS methods.
Results: Results from the study are summarised in Table 1. XRP 2868 was highly active against isolates of Staphylococcus aureus, Streptococcus pneumoniae, and Streptococcus pyogenes, including isolates resistant to erythromycin A by a variety of genetic mechanisms. XRP 2868 was also very active against isolates of Haemophilus influenzae with improved activity when compared with Synercid and pristinamycin.
Table 1.
| Species | N | MIC50/MIC90 (mg/L) |
|||
|---|---|---|---|---|---|
| XRP 2868 | Synercid | Pristinamycin | Erythromycin A | ||
| S. aureus (MS) | 41 | 0.12/0.25 | 0.5/0.5 | 0.25/0.25 | 0.25/>64 |
| S. aureus (MR) | 53 | 0.25/0.5 | 0.5/1 | 0.5/1 | >64/>64 |
| S. pneumoniae | 106 | 0.25/0.5 | 1/4 | 0.5/1 | >32/>32 |
| S. progenies | 60 | 0.06/0.06 | 0.25/0.25 | 0.12/0.12 | >64/>64 |
| Streptococcus spp | 63 | 0.12/0.5 | 1/4 | 0.25/0.5 | 0.06/0.12 |
| E. faecalis | 36 | 1/4 | 8/16 | 2/8 | >64/>64 |
| E. faecium | 38 | 0.25/0.5 | 0.5/2 | 0.25/0.5 | >64/>64 |
| L. monocytogenes | 23 | 0.5/0.5 | 1/1 | 0.5/0.5 | 0.12/0.12 |
| A. haemolyticum | 9 | ≤0.015-0.03* | 0.12-0.25* | ≤0.03-0.06* | ≤0.015-0.03* |
| H. influenzae | 52 | 0.5/1 | 4/8 | 2/4 | 4/8 |
| H. parainfluenzae | 20 | 2/8 | 32/32 | 8/16 | 2/8 |
| M. catarrhalis | 30 | 0.06/0.12 | 1/1 | 0.25/0.5 | 0.12/0.12 |
| B. pertussis | 10 | 0.03-0.06 | 0.12-0.25 | all 0.06 | 0.06-0.12 |
| N meningitidis | 20 | 0.03/0.06 | 0.25/0.5 | 0.06/0.12 | 0.12/0.25 |
≤10 isolates, MIC range only.
Conclusions: XRP 2868 is a suitable candidate for further investigation as a potential oral, anti-infective therapeutic agent.
P1556: Antimicrobial profile of the new biocide akacid
A. Buxbaum, C. Kratzer, W. Graninger, A. Georgopoulos (Vienna, A)
Objectives: Akacid is a new member of the polymeric biguanide family of disinfectants. It was especially developed to enhance the antimicrobial activity of this class with significantly less toxicity. This paper describes the wide range of biocidal activity and low potential for induction of resistance of akacid.
Methods: A total of 370 recent clinical isolates from patients with documented infections in hospitals located in Austria was studied. The organisms tested by reference methods included: Staphylococcus aureus (98), S. epidermidis (9), Enterococcus faecalis (37), Klebsiella spp. (45), Escherichia coli (65), Salmonella spp. (6), Shigella spp. (2), Yersinia enterocolitica (1), Acinetobacter spp. (4), Helicobacter pylori (7), Proteus spp. (7), Pseudomonas aeruginosa (59), Stenotrophomonas maltophilia (4), Mycobacterium spp. (6), Legionella pneumophila (3), Candida spp. (10), Aspergillus spp. (7). Additionally, spores of Bacillus subtilis ATCC 6633 and B. anthracis CH10 (anthrax spores reg.no. G112/WET/ACT 36/47) were tested. Also, in-vitro selection of resistance to akacid was carried out on 30 strains.
Results: Akacid showed good activity against both gram-positive and gram-negative bacteria, regardless of their antibiotic resistance-profile. Generally, MICs of tested isolates were the same for antibiotic-sensitive and multiresistant strains. Potent activity was also observed against L. pneumophila, Mycobacterium spp. and against spores of B. subtilis and B. anthracis. Good antifungal efficacy could be recorded against Candida spp. and Aspergillus spp. Bactericidal action was observed at 1 × MIC, that is that MIC values equaled MBC values. The in-vitro selection of resistance test showed no increase in MIC values in any isolate after 30 passages.
Conclusions: This study clearly demonstrates the excellent antimicrobial properties of akacid. Akacid was highly active against bacteria (including Mycobacterium spp. and L. pneumophila), spores and fungi, without being affected by the resistance profile of the strains. These properties, together with a very low potential to select for resistance, make akacid a promising tool for the battle against rising resistance and therapeutic failures.
P1557: Selective and effective antimicrobial properties of newly synthesised transition metal ions chelates with N, N-Dimethylbiguanide (DMGB)
R. Olar, M. Badea, M. Balotescu, R. Cernat, M. Bucur, V. Lazar (Bucharest, RO)
Biguanidine derivatives and their transition metal compounds are known to exhibit interesting biological properties, such as glucose lowering agents, analgesic, antimalarial and antimetabolite by competition for folic acid. In this study a series of newly synthesised complexes of type M(DMBG)2×2·nH2O ((1) M:Ni, X:CH3COO, n = 0; (2) M:Cu, X:CH3COO, n = 2; (3) M:Ni, X:ClO4, n = 0; (4) M:Cu, X:ClO4, n = 0) and previously characterised for their thermal stability and the coordination geometry by a range of techniques, including elemental and thermal analysis, IR, EPR and electronic reflectance spectra were screened in vitro for their ability to inhibit the microbial growth. The modifications evidenced in IR spectra were correlated with the presence of DMBG as chelate through N1 and N4. The EPR and electronic spectra are typical for a square-planar surrounding of Ni (II) and respectively an distorted octahedral one for Cu (II). The in vitro antimicrobial testings were performed by broth microdilution method, in order to establish the minimal inhibitory concentration (MIC), against Gram-positive (Bacillus subtilis, Listeria monocytogenes, Staphylococcus aureus), Gram-negative (Psedomonas aeruginosa, Escherichia coli, Klebsiella pneumoniae, Salmonella enteritidis), as well as Candida sp., using both reference and clinical, multidrug resistant strains. The antimicrobial effects were comparatively evaluated for the final compounds and for the transition metals salts. Our results showed that the transition metals salts exhibited MIC > 256 mcg/ml, for all tested strains. The tested compounds exhibited a specific antimicrobial activity, both concerning the microbial spectrum and the MIC value. The MICs values ranged between 1024 and 16 mcg/ml. The compounds no. (1) and (4) were highly active against Listeria monocytogenes (MIC < 2 mcg/ml). The compound no. (4) showed intermediate activity against Psudomonas and Bacillus strains (MIC 128 mcg/ml) and compound no. (2) against Salmonella (MIC 128 mcg/ml). The other tested strains (S. aureus, Escherichia coli and Candida) were resistant to all of the metal complexes exhibiting MICs > 256 mcg/ml. Our studies demonstrated that among the multiple biological activities of biguanidine complex combinations, they could also exhibit some effective antimicrobial properties against selected bacterial strains.
P1558: In vitro activity of human intestinal anaerobic bacteria against Nisin
E. Wahlund, E. Sillerström, C.E. Nord (Stockholm, S)
Objectives: Nisin is a new bacteriocin used as a food preservative. Resistance to Nisin is not associated with increased resistance to antibiotics. Nisin will be used in animal foods and it is therefore important to investigate if Nisin has an impact on the normal human intestinal microflora. The aim of the present investigation was to study the antimicrobial activity of Nisin against anaerobic bacteria isolated from the normal human intestinal microflora.
Methods: Anaerobic strains were isolated from the human intestinal normal microflora in healthy subjects at the Karolinska University Hospital, Stockholm, Sweden. All strains were identified according to techniques such as morphological tests, biochemical tests and gas-liquid chromatography. The antibacterial susceptibility tests were performed by the agar dilution method according to NCCLS. The minimum inhibitory concentration (MIC) was defined as the lowest concentration of the drug that inhibited growth. The appearance of a single colony or of a barely visible haze was disregarded. Six control strains were used for monitoring the antibacterial susceptibility test: Bacteroides fragilis ATCC 25285, Bacteroides thetaiotaomicron ATCC 29741, Clostridium perfringens ATCC 13124, Eubacterium lentum ATCC 43055, Escherichia coli ATCC 25922, and Enterococcus faecalis ATCC 29212. The Nisin agent was obtained from ImmuCell Corporation, Portland, Maine, USA.
Results: All Clostridium perfringens strains (11) were susceptible to Nisin. The minimum inhibitory concentrations were 0.25–0.5 mg/l. Eleven Eggerthella lentum strains showed high susceptibility to Nisin with minimum inhibitory concentrations of 0.125 mg/l. The Nisin agent was active against Lactobacilli (10) with minimum inhibitory concentrations between 0.125–16 mg/ l. All Peptostreptococci (10) except one strain were highly susceptible to Nisin (0.064–1.0 mg/l). The Bacteroides fragilis strains (12) had high minimum inhibitory concentration values for Nisin (>256 mg/l). Most Fusobacteria (9) showed high minimum inhibitory concentration values for Nisin (>256 mg/l). Three strains had lower minimum inhibitory concentration values (0.5–16 mg/l).
Conclusions: The present study showed that Nisin had good activities against anaerobic Gram-positive bacteria but was less active against anaerobic Gram-negative bacteria isolated from the human normal intestinal microflora.
P1559: In vitro activity of tigecycline and comparative agents against Enterococci in Sweden
L.E. Nilsson, G. Kronvall, M. Walder, M. Sörberg (Linköping, Stockholm, Malmö, Solna, S)
Background: Tigecycline is a new glycylcycline, which has been shown to have potent activity against Enterococci. The activity of tigecycline and comparative against clinical isolates from intensive care units (ICUs) and other hospital wards in three university hospitals in Sweden were determined.
Methods: A total of 609 initial clinical isolates of Enterococci were collected during November 2003-October 2004 at ICUs and other hospital wards at university hospitals in Linköping, Stockholm and Malmö. The isolates were identified to Enterococcus faecalis (N = 395) and Enterococcus faecium (N = 214), respectively. MICs were determined with E-test on Mueller Hinton broth and the breakpoints of the Swedish Reference Group for Antibiotics (www.srga.org) were use for all antibiotics except for tigecycline (tentative breakpoints ≤2 μg/ml)
Results: A total of 4% of the E. faecium isolates were vancomycin resistant (VRE). The van B gene was detected in these isolates. High level gentamicin resistance (HLGR) was seen among a total of 18% and 13% of the E. faecalis and the E.faecium isolates, respectively.
| Susceptibility ICU/non-ICU (%) | ||
|---|---|---|
| Antibiotic | E. faecalis | E. faecium |
| Tigecycline | 100/100 | 100/100 |
| Ampicillin | 99/99 | 17/8 |
| Linezolid | 98/99 | 98/99 |
| Piperacillin/Tazobactam | 99/100 | 7/7 |
| Vancomycin | 100/100 | 93/98 |
Conclusion: E. faecalis showed a high susceptibility against all the tested antibiotics both in the ICUs and the other hospital wards. Tigecycline, linezolid and vancomycin were the only antibiotics that showed high susceptibility against most E. faecium.
P1560: In vitro activity of tigecycline and comparative agents against Staphylococci in Sweden
L.E. Nilsson, G. Kronvall, M. Walder, M. Sörberg (Linköping, Stockholm, Malmö, Solna, S)
Background: Tigecycline is a new glycylcycline, which has been shown to have potent activity against Staphylococci. The activity of tigecycline and comparative against clinical isolates from intensive care units (ICUs) and other hospital wards in three university hospitals in Sweden were determined.
Methods: A total of 877 initial clinical isolates of Staphylococci were collected during November 2003-October 2004 at ICUs and other hospital wards at university hospitals in Linköping, Stockholm and Malmö. The isolates were identified to Staphylococcus aureus (N = 443) and Coagulase Negative Staphylococci (CoNS) (N = 433), respectively. MICs were determined with E-test on Mueller-Hinton agar and the breakpoints of the Swedish Reference Group for Antibiotics (www.srga.org) were use for all antibiotics except for tigecycline (tentative breakpoints S ≤2 μg/ml).
Results: Only a total of 2% of the S. aureus isolates were oxacillin resistant. The mec A gene was detected in these isolates.
| Susceptibility ICU/non-ICU (%) | ||
|---|---|---|
| Antibiotic | S. aureus | Coagulase Negative Staphylococci |
| Tigecycline | 100/100 | 100/100 |
| Oxacillin | 97/98 | 34/48 |
| Clindamycin | 99/96 | 54/70 |
| Fusidic acid | 91/93 | 55/71 |
| Linezolid | 100/100 | 100/100 |
| Rifampicin | 99/99 | 86/97 |
| Vancomycin | 100/100 | 100/100 |
Conclusion: The incidence of MRSA was low. Tigecycline, linezolid and vancomycin showed a high susceptibility both against oxacillin resistant and susceptible S. aureus and CoNS. The other antibiotics show lower susceptibility against CoNS especially at the ICUs.
P1561: In vitro activity of tigecycline and comparative agents against selected species of Enterobacteriacae in Sweden
L.E. Nilsson, M. Walder, G. Kronvall, M. Sörberg (Linköping, Malmö, Stockholm, Solna, S)
Background: Tigecycline is a new glycylcycline, which has been shown to have potent activity against most species of Enterobacteriacae.
Objective: The activity of tigecycline and comparative agents against clinical isolates from intensive care units (ICUs) and other hospital wards in three university hospitals in Sweden were determined.
Methods: A total of 1662 initial clinical isolates of Enterobacteriaecae were collected during November 2003-October 2004 at ICUs and other hospital wards at university hospitals in Linköping, Stockholm and Malmö. MICs were determined with E-test on Mueller-Hinton agar at each site. The breakpoints of the Swedish Reference Group for Antibiotics (www.srga.org) were use for all antibiotics except for tigecycline (tentative breakpoint S ≤2 μg/ml). Detection of ESBLs was made by E-test using cefepime, cefotaxime and ceftazidime ± clavulanic acid.
Results: The ESBL phenotype among Escherichia coli, Klebsiella pneumoniae, Klebsiella oxytoca and Enterobacter spp was detected in 1.8, 1.2, 3.9 and 0.2% of the isolates, respectively.
| Susceptible isolates ICU/non ICU (%) |
||||
|---|---|---|---|---|
| Antibiotic | E. coli | K. pneumoniae | K. oxytoca | Enterobacter spp |
| Tigecycline | 100/100 | 93/98 | 100/100 | 91/96 |
| Cefepime | 98/99 | 98/100 | 100/96 | 84/92 |
| Cefotaxime | 98/99 | 98/100 | 100/96 | 67/81 |
| Ceftazidime | 98/99 | 98/100 | 100/100 | 75/89 |
| Cefuroxime | 92/94 | 87/94 | 100/92 | 50/53 |
| Ciprofloxacin | 93/90 | 83/88 | 100/96 | 88/92 |
| Gentamicin | 98/98 | 98/100 | 100/100 | 99/100 |
| Imipenem | 99/100 | 100/99 | 100/100 | 96/98 |
| Meropenem | 100/99 | 100/100 | 100/100 | 89/96 |
| Pip/Tazo | 98/99 | 97/100 | 100/94 | 78/93 |
Conclusion: Tigecycline showed a high susceptibility against all the selected species both in the ICUs and the other hospital wards. The susceptibility of Enterobacter spp against ciprofloxacin, the cephalosporins, piperacillin/tazobactam and meropenem was lower especially at the ICUs.
P1562: Comparative in vitro activity of AVE6971A and AVE2221A, novel inhibitors of DNA topisomerase IV, against Staphylococcus aureus
M.J. Robbins, A. Bryskier, D. Felmingham (London, UK; Romainville, F)
Objective: AVE6971A and AVE2221A are both novel inhibitors of DNA-topoisomerase IV, an important antibacterial target in Gram-positive bacteria. In this study, the in vitro activity of AVE6971A and AVE2221A was determined against populations of methicillin-susceptible and -resistant, recent clinical isolates of Staphylococcus aureus.
Methods: Minimum inhibitory concentrations (MIC) of AVE6971A, AVE2221A, Synercid, pristinamycin, teicoplanin, linezolid and moxifloxacin were determined using the NCCLS agar dilution method.
Results: Results are summarised in Table 1. AVE6971A and AVE2221A exhibited potent antibacterial activity against both methicillin-susceptible and -resistant isolates of Staphylococcus aureus. This level of activity was generally similar to Synercid and pristinamycin and 2–4 fold greater than that of teicoplanin and linezolid. AVE6971A and AVE2221A were 4–8 times less active than moxifloxacin against fluoroquinolone-susceptible isolates of Staphylococcus aureus but, importantly, retained activity against fluoroquinolone-resistant strains of this species.
Table 1.
| Antimicrobial | methicillin susceptible Staphylococcus aureus (n = 41), MIC (mg/L) |
methicillin resistant Staphylococcus aureus (n = 53), MIC (mg/L) |
||||
|---|---|---|---|---|---|---|
| 50% | 90% | range | 50% | 90% | range | |
| AVE 6971A | 0.5 | 0.5 | 0.06–1 | 0.5 | 1 | 0.12–2 |
| AVE 2221A | 0.25 | 0.25 | 0.06–0.25 | 0.25 | 0.25 | 0.06–0.5 |
| Erythromycin A | 0.25 | >64 | 0.25–>64 | >64 | >64 | 0.25–>64 |
| Synercid | 0.5 | 0.5 | 0.25–1 | 0.5 | 1 | 0.12–1 |
| Pristinamycin | 0.25 | 0.25 | 0.12–0.5 | 0.5 | 1 | 0.12–1 |
| Teicoplanin | 1 | 1 | 0.25–2 | 1 | 2 | 0.5–8 |
| Linezolid | 2 | 2 | 0.5–2 | 2 | 2 | 1–32 |
| Moxifloxacin | 0.06 | 0.12 | 0.03–8 | 2 | 8 | 0.06—16 |
Conclusion: AVE6971A and AVE2221A are interesting molecules worthy of further investigation as candidates for development as anti-staphylococcal compounds particularly in view of the current problem of diminishing therapeutic options for the treatment of infection caused by methicillin and multi-drug resistant strains of Staphylococcus aureus.
P1563: In vitro activity of garenoxacin in comparison with other fluoroquinolones against isolates of E. coli, P. aeruginosa, and S. aureus, including genetically defined fluoroquinolone-resistant mutants
A. Heisig, T. Claußen, P. Heisig (Hamburg, D)
Objectives: Enhanced research activites aimed at overcoming the increasing incidence of fluoroquinolone (FQ) resistant bacterial pathogens and at extending the spectrum of activity to Gram-positive pathogens have resulted in the development of the new 6-desfluoro-quinolone garenoxacin (GRN, former BMS-284756).
Methods: To evaluate its in-vitro activity in comparison to 6-fluoroquinolones levofloxacin (LVX), gatifloxacin (GAT), and ciprofloxacin (CIP), MICs of garenoxacin (GRN) were determined for susceptible isolates of E. coli, P. aeruginosa, and S. aureus and a series of isogenic mutants carrying different combinations of known resistance mutations that affect either target, DNA gyrase and topoisomerase IV, as well as multiple drug resistance (mdr) efflux pumps.
Results: For susceptible isolates and most low-level resistant mutants of E. coli, the MICs of GRN and CIP were between 0.008/ 0.015 and 1/2 μg/ml, respectively, and thus, slightly lower than those of GAT and LVX (0.03–4). The activity of GRN against high-level resistant mutants of E. coli was comparable to CIP but higher to LVX, and GAT, while GRN (MIC 0.015–4 μg/ml) was superior to all tested FQs (MICs 0.03→256 μg/ml) against susceptible and resistant S. aureus. The susceptibility of wild-type P. aeruginosa for GRN and GAT was comparable (MIC 0.5–1 μg/ml), but higher for CIP and LVX (MICs 0.25 and 0.5 μg/ml). Overexpression of mdr efflux pumps MexAB, MexCD, and MexEF had a similar impact on all FQs and GRN increasing the MIC by 2–3 serial dilution steps.
Conclusions: Garenoxacin is a promising new quinolone derivative that combines a high activity against both, Gram-positive and Gram-negative, pathogens including clinically relevant mutants of S. aureus with multiple resistance mutations.
P1564: In vitro activity of garenoxacin, a novel Des-F(6)-quinolone, and other orally administered antimicrobials tested against 50,217 Enterobacteriaceae collected wordwide by the SENTRY Antimicrobial Surveillance Programme (1999–2003)
H. Sader, T. Fritsche, M. Stilwell, R. Jones (North Liberty, USA)
Objective: To compare the activity of garenoxacin (GRN; formerly BMS284756) with selected antimicrobials against a worldwide collection of Enterobacteriaceae (ENT). GRN, unlike recently marketed fluoroquinolones (FQ), lacks fluorine at the C-6 position.
Methods: The isolates were consecutively collected from >70 medical centres from bloodstream, respiratory, urinary and skin and soft tissue infections and tested by reference broth microdilution methods according to NCCLS guidelines. A GRN susceptible (S) breakpoint of <2 mg/L was applied for comparison purposes only.
Results: The results of the major organism groups tested follows: GRN showed excellent activity against this large collection of ENT (MIC50, 0.06 mg/L) and 87% of isolates were inhibited at ≤2 mg/L. The in vitro activity of GRN was similar to that of ciprofloxacin (CIP) against most ENT species, the exceptions were Serratia and indole-positive Proteus where CIP was slightly more active. In general GRN and CIP showed higher potency and a broader spectrum than orally administered betalactams or TMP/SMX. GRN was also highly active against E. coli O157:H7 and Yersinia enterocolitica (MIC90, 0.25 mg/L for both).
| Organism (no. tested) | GRN (MIC 50/% S) | Ciprofloxacin | Levofloxacin | %S Amox/Clav | Cefuroxime | TMP/SMXa |
|---|---|---|---|---|---|---|
| E. coli (22,698) | ≤0.03/87 | 87 | 88 | 81 | 76 | 76 |
| Klebsiella (10,513) | 0.12/90 | 91 | 92 | 80 | 71 | 86 |
| Enterobacter spp. (5,759) | 0.12/86 | 88 | 90 | 4 | 21 | 89 |
| Salmonella spp. (2,985) | 0 06/99 | 99 | 99 | 89 | 62 | 92 |
| P. mirabilis (2391) | 0.5/78 | 83 | 89 | 91 | 91 | 80 |
| Serratia spp. (2385) | 1/75 | 89 | 93 | 3 | 1 | 91 |
| Citrobacter spp. (1,206) | 0.12/87 | 90 | 92 | 40 | 57 | 90 |
| Indole-pos. Proteae (1,109) | 0.5/74 | 80 | 82 | 23 | 16 | 80 |
| Shigella spp (787) | ≤0.03/>99.9 | 100 | 100 | 65 | 96 | 32 |
| Total (50,217) | 0.06/87 | 89 | 90 | 67 | 63 | 81 |
TMP/SMX = Trimethoprim/sulfamethoxazole
Conclusions: GRN in vitro activity was similar to that of the most commonly used FQs and superior to other listed orally administered antimicrobials when tested against over 50,000 global ENT isolates.
P1565: Potency of garenoxacin, a new des-F(6)-quinolone, tested against community-acquired respiratory tract infection pathogens worldwide (29,837 Strains): report from the SENTRY Antimicrobial Surveillance Programme (1999–2003)
R. Jones, T. Fritsche, M. Stilwell, H. Sader (North Liberty, USA)
Objective: To evaluate the spectrum and potency of garenoxacin (GRN; formerly BMS284756) tested against typical community-acquired respiratory tract infection (CARTI) pathogens isolated from clinic patients emphasizing S. pneumoniae (SPN), H. influenzae (HI) and M. catarrhalis (MCAT). A comprehensive collection of 29,837 isolates was assessed from Europe, Asia and the Americas, each susceptibility (S) tested by reference methods in centralized laboratories.
Methods: All testing were performed by NCCLS M7-A6 (2003) methods and results were interpreted by M100-S15 (2005). The breakpoint for GRN was defined as S at ≤1 mg/L for comparison with fluoroquinolones (FQs): gatifloxacin (GATI) S at ≤1 mg/L; levofloxacin (LEVO) S at ≤2 mg/L (≤1 mg/L used for comparisons here); and moxifloxacin (MOXI) S at < mg/L. The QRDR regions of gyrA, parC and parE were sequenced in those strains that had a GRN MIC ≥0.25 mg/L and/or were R to FQs.
Results: The SPN strains (11,878) were characterized as follows: penicillin non-S at 35.1% (R at 19.4%; this rate was higher than community-acquired, hospitalized pneumonia cases); erythromycin-R at 29.7%; and ciprofloxacin (CIPRO)-R (>4 mg/L; Chen et al., 1999) at 3.9% = possible QRDR mutations. HI strains (12,569) overall had ampicillin (AMP)-R at 22.8% and beta-lactamase-negative AMP-R (BLNAR) at 1.0% with 98/132 of these strains from Asia (Japan). MCAT (5,390) strains were dominantly beta-lactamase-positive (PEN-R; 95.3%). The following table lists key GRN and FQ MIC results: FQR SPN strains (GRN MIC, 1→4 mg/L) had multiple QRDR mutations (gyrA at S83F or T, parC at S79F or T or D83N and parE at I460V). One FQR HI had QRDR mutations (gyrA at S84L and parC at E88K). The overall rank order of activity versus SPN using MIC90 (mg/L) results was: GRN (0.06) > MOXI (0.25) > GATI (0.5) > LEVO(1) > CIPRO (2).
| MIC(mg/L) |
% inhibited at ≤ 1 mg/L |
|||||
|---|---|---|---|---|---|---|
| Organism (no. tested) | 50% | 90% | GRN | GATI | LEVO | MOXI |
| SPN | ||||||
| PEN-S (7,703) | 0.06 | 0.06 | 99.9 | 99.4 | 97.4 | 99.4 |
| PEN-I (1,865) | 0.06 | 0.06 | 99.8 | 98.9 | 96.9 | 99.3 |
| PEN-R (2,310) | 0.06 | 0.06 | 99.9 | 93.2 | 96.4 | 96.4 |
| CIPRO-R (460) | 0.06 | 1 | 97.8 | 76.1 | 37.4 | 80.7 |
| HI | ||||||
| Amp-S (9,707) | ≤0.03 | ≤0.03 | >99.9 | >99.9 | >99.9 | >99.9 |
| Amp-R (2,862) | ≤0.03 | ≤0.03 | 100.0 | 100.0 | 100.0 | 100.0 |
| BLNAR (132) | ≤0.03 | ≤0.03 | 100.0 | 100.0 | 100.0 | 100.0 |
| MCAT | ||||||
| PEN-S (254) | ≤0.03 | ≤0.03 | 100.0 | 100.0 | 100 0 | 100.0 |
| PEN-R (5,136) | ≤0.03 | ≤0.03 | 100.0 | 100.0 | 100.0 | 100.0 |
Conclusions: GRN was the most active quinolone tested against a global collection of 29,837 typical CARTI pathogens and was at lease 4-fold more active than MOXI against SPN isolates. GRN should be a welcome addition to our antimicrobial formularies for ambulatory care treatment of FQR and multidrug-R species associated with CARTI.
P1566: Garenoxacin activity against isolates from patients hospitalised with community-acquired pneumonia (Streptococcus pneumoniae, Haemophilus influenzae and Moraxella catarrhalis; 3,087 strains): report from the SENTRY Antimicrobial Surveillance Programme (1999)
R. Jones, T. Fritsche, H. Sader, M. Stilwell (North Liberty, USA)
Objective: To evaluate the potency of garenoxacin (GRN), a new des-F(6)-quinolone, susceptibility (S) when tested against organisms isolated from patients hospitalized with community-acquired respiratory tract infections (CARTI) i.e. S. pneumoniae (SPN), H. influenzae (HI) and M. catarrhalis (MCAT). Organisms from 1999-2003 were available for study, each isolated in Europe, the Americas or Asia; and their origin was lower respiratory tract cultures. Although, MCAT was monitored, it was not isolated in sufficient numbers for analysis.
Methods: Consecutive, non-duplicate cultures of SPN (1,444) and HI (1,643) were tested by reference NCCLS broth microdilution methods with concurrent QC, and interpretation guided by NCCLS M100-S15 (2005). Comparison agents numbered >20 and included four fluoroquinolones (FQs): ciprofloxacin (CIPRO), gatifloxacin (GATI), levofloxacin (LEVO) and moxifloxacin (MOXI). The QRDR regions of gyrA, parC and parE were sequenced in those strains with elevated GRN MICs (≥0.25 mg/L) or resistant (R) to other FQs.
Results: The SPN collection was characterized as follows: penicillin-S at 70.1% (R rate, 16.6%); macrolide-R at 26.1%; clindamycin-R at 11.9%; and amoxicillin/clavulanate- and ceftriaxone (CTRI)-R at 1.4 and 1.1%, respectively. Rank order of FQ activity was (MIC90, mg/L): GRN (0.06) > MOXI (0.12) > GATI (0.5) > LEVO (1, 0.8% R) > CIPRO (2). The HI collection was defined as follows: ampicillin (AMP)-R at 20.3%; beta-lactamase-negative AMP-R at 1.5% (nearly 80% of strains from Japan; rate was <0.5% for all other regions); GRN MIC50/ 90 at ≤0.008/0.016 mg/L; and one FQ-R strain had multiple QRDR mutations. See table for co-R analysis among SPN: Co-R trends were observed among beta-lactams, macrolides, tetracyclines, TMP/SMX, but not for FQs. An increase in LEVO-R among penicillin-R SPN was observed. Sixty-one % of all SPN GRN MIC values were 0.06 mg/L, a dominant MIC mode.
| Penicillin category |
|||
|---|---|---|---|
| Parameter (1,444 strains) | S | Intermediate | R |
| GRN | |||
| MIC50 (mg/L) | 0.06 | 0.06 | 0.06 |
| MIC90 (mg/L) | 0.06 | 0.06 | 0.06 |
| % ≤1 mg/L | 100.0 | 100.0 | 99.6 |
| CIPRO MIC > 4mg/L(%) | 3.9 | 4.7 | 3.3 |
| LEVO-R (%) | 0.6 | 0.7 | 1.6 |
| CTRI-R (%) | 0.1 | 2.1 | 6.2 |
Conclusions: GRN exhibited superior activity (lower MIC results) against SPN and HI isolates associated with pneumonias in CARTI patients; 2- to 32-fold more potent than other FQs. SPN and HI isolates with GRN MIC values of more than 0.25 mg/L were very rare (11/3,087; 0.4%) and secondary to numerous QRDR mutations in gyrA, parC and E. GRN appears to be an excellent candidate to treat multi-drug R CARTI pathogens in hospitalized patient populations.
P1567: Potency and spectrum of garenoxacin tested against an international collection of skin and soft tissue infection pathogens: report from the SENTRY Antimicrobial Surveillance Programme (1999–2003)
T. Fritsche, H. Sader, R. Jones (North Liberty, USA)
Objective: To evaluate the spectrum and potency of garenoxacin (GRN), a novel des-F(6)-quinolone, against a large international collection (9,227) of Gram-positive and -negative bacterial pathogens that cause skin and soft tissue infections (SSTI).
Methods: Consecutive, non-duplicate bacterial isolates were collected from 1999 to 2003 from patients with documented community-acquired or nosocomial SSTI in >70 medical centres participating in the SENTRY Programme in North America (36.1%), Europe (24.1%), Latin America (16.1%) and the Asia-Pacific region (23.7%). All isolates were tested using NCCLS broth microdilution methods against GRN, the currently marketed fluoroquinolones (FQ) ciprofloxacin (CIPRO), levofloxacin (LEVO), gatifloxacin (GATI) and representative comparator agents used for the empiric therapy of SSTI.
Results: The Table lists the potency and cumulative inhibition rates for GRN against the top 10 (by frequency) SSTI pathogens: Published data demonstrate that the agents causing SSTI are comprised of a distinct set of Gram-positive cocci and Gram-negative aerobic and facultative bacilli. GRN was the most potent agent tested against SA, and was at least 2-fold more potent than GATI (MIC50, 0.06 mg/L) and 8-fold more potent than either CIPRO or LEVO (MIC50, 0.25 mg/L). Furthermore, GRN was 4-to 8-fold more potent than the FQ against BHS and viridans group Streptococci (VGS), and 2- to 4-fold more potent against ESP. GRN was comparable with CIPRO, LEVO and GATI against EC KSP and ASP, but less active than these agents against PSA.
| MIC (mg/L) |
% inhibited at MIC (mg/L) |
|||
|---|---|---|---|---|
| Organism (# tested) | 50 | ≤1 | 2 | 4 |
| S. aureus (SA; 3,790) | ≤0.03 | 87 | 93 | 97 |
| P. aeruginosa (PSA; 1, 080) | 2 | 41 | 61 | 70 |
| E. coli (EC; 864) | 0.03 | 83 | 84 | 84 |
| Enterococcus spp, (ESP;650) | 0.25 | 63 | 71 | 86 |
| Klebsiella spp. (KSP; 465) | 0.12 | 85 | 88 | 90 |
| Enterobacter spp. (ENT; 463) | 0.12 | 86 | 88 | 90 |
| β-haemolytic streptococci (BHS; 420) | 0.06 | 100 | – | – |
| Coagulase neg. staphylccocci (CoNS; 365) | 0.12 | 73 | 86 | 95 |
| P. mirabilis (PM: 238) | 0.5 | 81 | 82 | 84 |
| Acinetobacter spp. (ASP; 210) | 2 | 48 | 50 | 60 |
Conclusions: GRN was the most potent FQ when tested against SA, BHS, VGS and ESP, and was similar in activity to these agents against other species including EC, KSP and ASP. The in vitro data suggest that GRN maybe superior to the FQ for the treatment of SSTI infections caused by Staphylococci and Streptococci, warranting further clinical studies.
P1568: Antimicrobial susceptibility of 5,859 non-Enterobacteriaceae Gram-negative organisms other than Pseudomonas aeruginosa tested against the novel Des-F(6)-quinolone, garenoxacin (SENTRY Antimicrobial Surveillance Programme, 1999–2003)
H. Sader, T. Fritsche, M. Stilwell, R. Jones (North Liberty, USA)
Objective: To compare the antimicrobial activity of garenoxacin (GRN) and selected antimicrobial agents against 5,859 non-enteric Gram-negative organisms other than P. aeruginosa collected as part of the SENTRY Antimicrobial Surveillance Programme (1999–2003).
Methods: The isolates were consecutively collected at >70 medical centres on six continents from bloodstream, respiratory, urinary and skin and soft tissue infections and tested by NCCLS broth microdilution methods. A GRN susceptible (S) breakpoint of ≤2 mg/L was applied for comparison purposes only.
Results: The results of major organism groups tested: Overall, GRN (MIC50, 1 mg/L; 59% S) was more active than ciprofloxacin (MIC50, 2 mg/L; 49% S), ceftazidime (MIC50, 8 mg/L; 56% S), piperacillin/tazobactam (MIC50, 32 mg/L; 45% S) and amikacin. GRN was the most active compound tested against Chryseobacterium spp. GRN was also highly active against Aeromonas spp. (MIC90, 2 mg/L; 95% S), N. meningitides (MIC90, ≤0.03 mg/L; 100% S) and P. multocida (MIC90, ≤0.03 mg/L; 98% S). Additionally, 83% of S. maltophilia strains were inhibited at 4 mg/L of GRN.
| Cum % inhibited at GRN MIC (mg/L) |
|||||
|---|---|---|---|---|---|
| Organism (no. tested) | MIC 50 | <0.5 | 1 | 2 | 4 |
| Alcaligenes/Achromobacter spp. (170) | >4 | 5 | 7 | 9 | 19 |
| Acinetobacter spp. (3,260) | 2 | 46 | 49 | 51 | 60 |
| Aeromonas spp. (387) | 0.12 | 79 | 88 | 95 | 97 |
| Burkholderia spp. (180) | 4 | 6 | 18 | 38 | 56 |
| Chryseobacterium spp. (59) | 0.25 | 81 | 90 | 93 | 95 |
| N. meningitidis (130) | ≤0.03 | 100 | 100 | 100 | 100 |
| P. multocida (59) | ≤0.03 | 97 | 97 | 98 | 100 |
| S. maltophilia (1,449) | 2 | 22 | 44 | 66 | 83 |
| Total (5,859) | 1 | 43 | 51 | 59 | 70 |
Conclusions: GRN exhibited reasonable in vitro activity against many of the rarely isolated Gram-negative species and maybe an alternative therapy for infections caused by these ‘difficult to treat’ organisms.
P1569: Activity of ceftobiprole against recent clinical isolates of Enterobacteriaceae from respiratory infections from hospitalised patients in Europe and USA
M. Heep, D.F. Sahm, D.C. Draghi, M.E. Jones (Basel, CH; Herndon, USA)
Objectives: Ceftobiprole (BPR), formerly BAL9141, is the first of a new class of anti-MRSA cephalosporins and has broad-spectrum activity towards most clinically relevant bacterial pathogens. This study aimed at evaluating the in vitro activity of BPR and other beta-lactams against Enterobacteriaceae isolates from respiratory infections, including extended spectrum betalactamase (ESBL) producing strains.
Methods: A total of 372 organisms were studied, comprising non-repeat isolates derived from quality respiratory specimens from ICU and inpatients from Europe and USA during 2001–4. The species distribution was: 20 Citrobacter freundii, 7 Citrobacter koseri (diversus), 17 Enterobacter aerogenes, 60 Enterobacter cloacae, 60 Escherichia coli, 25 Klebsiella oxytoca, 60 Klebsiella pneumoniae, 60 Proteus mirabilis, and 62 Serratia marcescens. MICs of BPR, cefepime, cefepime/clavulanate cefotaxime cefotaxime/clavulanate, cefoxatin, ceftazidime ceftazidime/clavulanate, and ciprofloxacin were determined by broth microdilution according to NCCLS guidelines. BPR breakpoints used were 4 mg/L susceptible, 8 mg/L intermediate and 16 mg/L resistant.
Results: Overall BPR displayed excellent activity against Enterobacteriaceae. MIC distributions show 92.2% of isolates were inhibited at a concentration 4 mg/L (cefepime 96.2%, cefotaxime, 84.7%). Overall BPR demonstrated limited activity against putative ESBL producers (38.1% inhibited at 4 mg/L) but good activity, superior to cefotaxime or ceftazidime, against putative high–level AmpC producers (66.7% inhibited at 4 mg/ L). MIC90 values to drugs tested are shown.
| MIC (μg/ml) |
||||
|---|---|---|---|---|
| Agent | Range | MIC50 | MIC90 | (%S) |
| Ceftobiprole | ≤0.015 ->32 | 0.06 | 4 | (92.2) |
| Cefepime | ≤0.015 ->128 | 0.003 | 1 | (97.8) |
| Cefepime/clavulanate | ≤0.25 - 128 | ≤ 0.25 | ≤ 0.25 | – |
| Cefotaxime | ≤ 0.03 ->128 | 0.06 | 32 | (85.5) |
| Cefotaxime/clavulanate | ≤ 0.25 ->128 | ≤ 0.25 | 16 | – |
| Cefoxitin | ≤ 0.5 -> 32 | 4 | > 32 | (59.9) |
| Ceftazidime | ≤ 0.06 ->128 | 0.12 | 32 | (87.4) |
| Ceftazidime/clavulanate | ≤ 0.25 -> 128 | ≤ 0.25 | 16 | – |
| Ciprofloxacin | ≤ 012 ->2 | ≤ 0.12 | 2 | (89.5) |
Conclusion: In addition to its anti-MRSA activity, the spectrum of activity of BPR resembles that of 3rd generation cephalosporins and cefepime, demonstrating activity against Enterobacteriaceae associated with hospital acquired respiratory infections.
P1570: Ceftobiprole activity against epidemic strains of methicillin-resistant Staphylococcus aureus from hospitalised patients in Belgium
O. Denis, A. Deplano, C. Nonhoff, R. De Ryck, S. Rottiers, M.J. Struelens (Brussels, B)
Background: Ceftobiprole (formerly BAL9141, Basilea Pharmaceutica Ltd., Switzerland) is a new broad-spectrum cephalosporin in clinical development with potent bactericidal activity on Gram-positive cocci including beta-lactam resistant Staphylococci. We compared its antimicrobial activity with that of 18 antimicrobial agents against methicillin-resistant Staphylococcus aureus (MRSA) strains collected during a national survey conducted in 2003 in Belgian hospitals.
Methods: In 2003, 521 MRSA strains were collected from 112 hospitals and confirmed by PCR for 16S rRNA, nuc and mecA genes. Strains were genotyped by pulsed field gel electrophoresis (PFGE) after SmaI macrorestriction, SCCmec type, and MLST. MICs of 19 antimicrobials were determined by agar dilution methods according to NCCLS. Vancomycin susceptibility was further evaluated on vancomycin screen agar.
Results: The proportion of MRSA strains resistant to aminoglycosides ranged from 0.1% for amikacin to 44% for tobramycin; 63% were resistant to erythromycin and 40% to clindamycin. More than 90% of strains were susceptible to tetracycline, minocycline, fusidic acid, rifampin and cotrimoxazole. Ciprofloxacin resistance rate was 98%, high-level mupirocin resistance (MIC > 524 μg/ml) 3.4%, respectively. No strains resistant to glycopeptides, linezolid, or ceftobiprole were detected. The MIC50, MIC90 and range of MIC for ceftobiprole were: 0.5, 2, 0.06–4 μg/ml. By molecular typing, 90% of MRSA strains belonged to 9 epidemic genotypes: PFGE type B2 ST45-SCCmec IV (n = 251), A20 ST8-SCCmec IV (n = 97), G10 ST5-SCCmec II (n = 27), A21 ST8-SCCmec IV (n = 26), C1 ST5-SCCmec II (n = 13), C3 ST5-SCCmec IV (n = 12) A1 ST247-SCCmec I (n = 12), D8 ST228-SCCmec I (n = 11) and L1 ST22-SCCmec IV (n = 10).
Conclusion: The new parenteral cephalospirin, ceftobiprole, showed an excellent activity against all MRSA strains from hospitalized patients in Belgium. These strains belonged to 9 genotypes of five pandemic MRSA lineages associated with nosocomial infections.
P1571: In vitro activity of AVE1330A, a novel β-lactamase inhibitor, in combination with aztreonam or ceftazidime against ceftazidime-resistant isolates of species of the Enterobacteriaceae
J. Shackcloth, L. Williams, J. Northwood, A. Bryskier, D. Felmingham (London, UK; Romainville, F)
Objective: To determine the ability of AVE1330A, an investigational, non β-lactam, β-lactamase inhibitor, to protect ceftazidime and aztreonam against hydrolysis by various β-lactamases.
Methods: Minimum inhibitory concentrations (MIC) of ceftazidime and aztreonam, alone and in combination with AVE1330A at a fixed concentration of 4 mg/L, and, for ceftazidime only, in fixed ratio combinations of 4:1 and 4:2 (antimicrobial : inhibitor), were determined using the NCCLS agar dilution method for isolates of ceftazidime-resistant species of the Enterobacteriaceae (n = 45), Klebsiella spp expressing SHV-ESBL (n = 24) and Escherichia coli transformants expressing TEM and OXA B-lactamases (n = 4).
Results: Results are summarised in Table 1. At a fixed concentration of 4 mg/L, AVE1330A was a potent inhibitor of unclassified β-lactamases responsible for ceftazidime and aztreonam resistance in species of the Enterobacteriaceae including; Escherichia coli (n = 12), Citrobacter freundii (n = 6), Klebsiella pneumoniae (n = 12), Enterobacter cloacae (n = 13) and Morganella morganii (n = 2) with the great majority of isolates inhibited by ≤1 mg/L of either antimicrobial in combination with the inhibitor. AVE1330A inhibited the activity of TEM 6, 9, 10, and OXA 5 in ceftazidime-resistant Escherichia coli transformants expressing these enzymes, reducing MIC to ≤1 mg/L. AVE1330A restored the activity of ceftazidime and aztreonam (MIC ≤2 mg/L and ≤1 mg/L respectively) against isolates of Klebsiella spp expressing SHV-ESBL. Fixed ratio combinations of ceftazidime and AVE1330A at 4 : 1 and 4 : 2, respectively demonstrated similar inhibitory activity against the β-lactamase producing isolates. However, MIC were generally 2–8 fold higher when compared to those achieved with ceftazidime plus a 4 mg/L fixed concentration of AVE1330A.
Table 1.
| Aztreonam MIC range (mg/L) |
Ceftazidime MIC range (mg/L) |
||||||
|---|---|---|---|---|---|---|---|
| Species | n | Alone | +4mg/L AVE 1330A | Alone | + 4mg/L AVE1330A | 4:1 ratio with AVE1330A | 4:2 ratio with AVE1330A |
| E. coli (ceftazidimeR) | 12 | 4->64 | ≤0.06-0.12 | 16->128 | ≤0.03-1 | 0.25-16 | 0.25-8 |
| Citrobacter freundii (ceftazidimeR) | 6 | 4-64 | 0.12-0.25 | 32->128 | 0.25-1 | 0.5-4 | 0.5-2 |
| Klebsiella pneumoniae (ceftazidimeR) | 12 | 2->64 | ≤0.06-0.05 | 12B- > 128 | ≤0.03-1 | 1-4 | 1-4 |
| Enterobacter cloacae (ceftazidime R) | 13 | 0.5->64 | ≤0.06-1 | 16- > 128 | 0.25-8 | 2-8 | 0.5-8 |
| Morganella morganii (ceftazidimeR) | 2 | 2/8 | ≤0.06/0.25 | 16/64 | 0.12/0.25 | 0.5/2 | 0.25/0.5 |
| E. coli (TEM 9 transformant) | 1 | 16 | 0.12 | >128 | 0.5 | NT | NT |
| E. coli (TEM 9 transformant) | 1 | 64 | ≤0.06 | >128 | 0.5 | NT | NT |
| E. coli (TEM 9 transformant) | 1 | 64 | <0.06 | >128 | 0.5 | NT | NT |
| E. coli (TEM 10 transformant) | 1 | 32 | 0.12 | >128 | 0.5 | NT | NT |
| E. coli (OXA 5 transformant) | 1 | 16 | 0.12 | 64 | 0.5 | NT | NT |
| Klebsiella spp (SHV-ESBL producers) | 24 | 4->64 | ≤0.06-1 | 8->64 | 0.015-2 | 0.5-8 | 0.25-8 |
Conclusion: AVE1330A is a potent inhibitor of a wide range of β-lactamases, including SHV-ESBL and restores potent activity to ceftazidime and aztreonam against ceftazidime-resistant isolates of species of the Enterobacteriaceae.
P1572: High affinity for PBP 2′ enables RO490–8463 (CS-023), a unique guanidine-pyrrolidine carbapenem, to achieve good in vitro activity against MRSA, which is confirmed in murine infection models
E. Namba, H. Inoue, T. Koga, T. Fukuoka, I. Simpson (Tokyo, JP; Ely, UK)
Objectives: RO49O-8463 (RO) is a carbapenem with a unique guanidine-pyrrolidine side chain which confers exceptional broad-spectrum activity against diverse hospital pathogens, including MRSA. This study was undertaken to determine (1) the affinity (IC50, mg/L) of RO for the PBPs of MRSA and (2) efficacy (% survival) of RO in immunocompetent and neutropenic mice infected with MRSA.
Methods: MICs were determined to NCCLS guidelines. Affinities of RO, imipenem (IMI) and meropenem (MPM) for the PBPs of MSSA (ATCC 6538P) and MRSA 123 were determined using a competitive assay with 14C-labelled penicillin G. In vivo efficacy of RO was assessed in two systemic infection models, using immunocompetent (Model 1) or neutropenic (Model 2) mice, with dosing regimens designed to simulate human plasma pharmacokinetics. In Model 1, 4-week-old male immunocompetent SPF ddY mice were challenged intraperitoneally with 1.5 × 109 cfu/mouse of MRSA 12394. In Model 2, 5-week-old male neutropenic SPF ddY mice were challenged intraperitoneally with 2.3 × 103 cfu/mouse of MRSA 12394 (MIC of RO, 8 mg/L). In both models, RO was subcutaneously administered at 30 minute intervals in fractionized, decreasing doses to simulate human plasma concentrations after a single intravenous infusion at doses of 1500, 1000, 750, 500 and 250 mg (10 mice/dose). Mortality was monitored for 5 days after infection.
Results: For MRSA 123, RO exhibited substantially lower MIC (8 mg/L) and PBP affinities (5.3 mg/L) than IMI (32 mg/L and 170 mg/L) and MPM (32 mg/L and 130 mg/L). In Model 1, all untreated immunocompetent mice died within 3 days. RO achieved survival rates of 50% at 250 mg, 80% at 500 and 750 mg and 100% at >1000 mg. In Model 2, 90% of untreated neutropenic mice died within 3 days. RO achieved survival rates of 70% at 250 mg, 90% at 500 and 750 mg and 100% at >1000 mg.
Conclusions: RO represents a new class of guanidine-pyrrolidine carbapenems with good activity against MRSA, which may be attributable to high affinity for PBP 2′. In immunocompetent and neutropenic murine models infected with MRSA, RO achieved >80% survival at doses simulating the pharmacokinetics of a single 500 mg infusion in humans.
P1573: Activity and pharmacodynamics of RO490–8463 (CS-023), a carbapenem with activity against MRSA
M.E. Jones, D.P. Nicolau, J.L. Kuti, C. Nightingale, D.C. Draghi, R.K. Flamm, D.F. Sahm (Hilversum, NL; Hartford, Herndon, USA)
Objectives: R0490–8463 is a unique carbapenem, containing guanidine and pyrrolidine rings, and demonstrates broad-spectrum activity against diverse hospital pathogens including MRSA. This study was designed to establish activity against a representative selection of nosocomial pathogens from EU and US hospitals, comprising S. aureus (SA), coagulase-negative Staphylococci (CNS), P. aeruginosa (PA), S. pneumoniae (SP), E. coli (EC) and K. pneumoniae (KP). For SA, Monte Carlo (MC) simulations were used to determine target attainments (TA) of different dosage regimens for RO at specific MICs.
Methods: RO and comparators were tested against 363 SA (58 MSSA/305 MRSA), 43 CNS, 275 PA, 159 SP, 121 EC and 132 KP, using NCCLS guidelines (2004). Isolates were non-repeat clinical isolates collected through 2004. Using a 5000 subject MC simulation, TA for achieving free drug concentrations above the MIC (>40% dosing interval) was assessed for SA using pharmacokinetic data from Phase I studies.
Results: For MSSA/MRSA respectively, RO MIC90s (μg/ml) were 0.25/8 (US) and 0.12/16 (EU) and for CNS, 2 (US) and 4 (EU). For IMP-S/IMP-R PA respectively, RO MIC90s (μg/ml) were 0.5/ 8 (US) and 2/16 (EU). For PEN-S/PEN-R SP MIC90s (mg/ml) were ≤0.008/0.5 for both regions. RO showed good activity against ESBL and AmpC producing EC and KP (0.12–0.25, US; 0.03, EU). For SA, TA for RO regimens of 750 mg q12 h/750 mg q8 h were 81.4%/96.3% respectively for US and 59.3%/80.9% respectively for EU isolates. For SA, TA for RO regimens of 1500 mg q12 h/1500 mg q8 h were 96.0%/99.8% respectively for US and 80.9%/98.7% respectively for EU isolates.
Conclusions: RO demonstrated broad spectrum in vitro activity against resistant phenotypes commonly encountered within the hospital setting, including MRSA. To achieve bactericidal exposures for MRSA, MC simulations support the use of 750 mg q8 h or 1500 mg q12 h in the US and 1500 mg q8 h in EU, as a 1 hour infusion.
P1574: Delayed resistance selection for doripenem when passaging P. aeruginosa isolates with an aminoglycoside
D. Biedenbach, H. Huynh, R. Jones (North Liberty, USA)
Background: doripenem (DOR) (MIC50, 0.5 mg/L), a parenteral carbapenem, has activity equal or superior to meropenem (MIC50, 0.5) and imipenem (MIC50, 1) against PSA. This study determined the resistance selection of DOR ± gentamicin (GENT) during subinhibitory passaging using 6 P. aeruginosa (PSA) strains.
Methods: PSA bloodstream strains (2003) with DOR MICs at or near the proposed susceptible breakpoint (<4 mg/L). Broth microdilution methods were used to establish baseline MICs for DOR (2-8) and GENT (4→256). After overnight incubation, the organism suspension, one well below the MIC was transferred into fresh media and allowed to grow (0.5 McFarland) then inoculated into panels containing DOR ± GENT (MIC/4; not to exceed 4 mg/L). Isolates were passaged over 7 days.
Results: The table shows baseline and 7 day passaging results: After 7 days, strain MIC increases were: DOR alone at >4X (3), 2X (1), NC (2); + GENT at 4X (1), 2X (2), NC (3). No strain was resistant to DOR + GENT (MIC, >16 mg/L) and enhanced combination activity was noted for nearly all strains during the first 3 passages.
| DOR/+GENT MIC (mg/L) by passage |
||||||||
|---|---|---|---|---|---|---|---|---|
| Strain* | Baseline | Day 1 | 2 | 3 | 4 | 5 | 6 | 7 |
| 30-3232A | 2 | 2/2 | 2/1 | 2/2 | 4/4 | 2/4 | 4/4 | 2/8 |
| 107-3347A | 4 | 4/2 | 4/2 | 16/4 | >16/4 | 16/4 | >16/4 | >16/8 |
| 45-11372A | 4 | 4/2 | 16/4 | 16/4 | >16/8 | >16/4 | >16/8 | >16/8 |
| 75-3075A | 4 | 8/2 | 8/2 | 8/2 | 8/4 | 8/4 | 8/4 | 16/4 |
| 24-3338A | 8 | 8/2 | 8/2 | 8/4 | 16/8 | 16/8 | 16/8 | 16/8 |
| 38-12060A | 8 | 8/8 | 8/8 | 16/8 | 16/8 | 8/8 | 8/8 | 8/8 |
Conclusions: The combination of a co-drug (GENT) with DOR against PSA provided enhanced activity during early passaging and minimized the number of DOR-R strains. DOR and aminoglycosidecombinations maybeaneffectivetreatmentregimenfor infections caused by PSA with elevated carbapenem MIC values.
P1575: Heterologous expression of lysK encoding a lysin with anti-MRSA activity from the genome of the anti-staphylococcal phage K
A. Coffey, W. Meaney, G. Fitzgerald, P. Ross, S. O'Flaherty (Cork, Fermoy, IRL)
Objectives: The emergence of antibiotic resistant Staphylococci has prompted the need for alternative antibacterial controls other than antibiotics. Thus the objective of this study was to develop and evaluate a non-antibiotic antimicrobial with activity against Staphylococcus aureus.
Method, background and approach: The bacteriophage K was previously shown to exhibit a very broad host range against Staphylococcus and lysed a range of methicillin-resistant S. aureus (MRSA) strains from Irish hospitals, hetro-vancomycin and vancomycin resistant S. aureus (hVRSA and VRSA) and also teicoplanin resistant strains (Coffey et al., 2004 Use of polyvalent anti-staphylococcal bacteriophages for the biocontrol of methicillin-resistant Staphylococcus aureus and other Staphylococci. Clinical Microbiology and Infection 10 (Suppl.3): 135–136. 14th ECCMID). The phage was subjected to genome sequencing (O'Flaherty et al. 2004, Genome of staphylococcal phage K: a new lineage of Myoviridae infecting Gram-positive bacteria with a low G-C content. Journal of Bacteriology, 186). Genome analysis revealed a linear DNA genome of 127,395 base pairs, which encodes 118 putative ORFs. The gene encoding the lysin protein was identified and standard molecular biology techniques were utilized to clone the lysin protein.
Results: Bioinformatic analysis of the bacteriophage lysin (lysK) gene indicated that it was interrupted by an intron. This observation was confirmed by RT-PCR, and the resulting cDNA was found to encode 495 amino acid protein. The lysK gene was heterologously over-expressed in both Escherichia coli (using the T7 promoter) and Lactococcus lactis (using a nisin-inducible promoter) with a view to generating cell factories for lysin production. Results from zymograms with dead staphylococcal cells indicated that active recombinant LysK product was generated in both cases exhibiting cell wall degrading activity against coagulase negative Staphylococci, MRSA strains, a VRSA and S. aureus associated with bovine infections.
Conclusions: The bacteriophage lysin cloned in this study exhibits activity against drug resistant S. aureus. Thus it is a promising candidate as an antimicrobial for eliminating problematic S. aureus strains.
P1576: Circumventing resistance to beta-lactam antibiotics in Staphylococcus aureus
C. Fuda, S. Mobashery (Notre Dame, USA)
Objective: Emergence of methicillin-resistant Staphylococcus aureus (MRSA) has created challenges in treatment of nosocominal infections. S. aureus normally produces four PBPs, which are susceptible to modification by β-lactam antibiotics, an event that leads to bacterial death. The gene product of mecA from MRSA is a penicillin-binding protein (PBP) designated PBP2a. PBP2a is refractory to the action of all available β-lactam antibiotics. Furthermore, PBP2a is capable of taking over the functions of the other PBPs of S. aureus in the face of the challenge by β-lactam antibiotics. The basis for resistance of PBP2a to inhibition by β-lactam antibiotics has been studied by us. These methodologies have been applied to investigations of three cephalosporins from ABRI (Antibiotic Research Institute, Vienna), Sandoz, which were specifically developed for inhibition of PBP2a in MRSA.
Methods: The mecA gene was cloned and expressed in E. coli, and PBP2a was purified to homogeneity. Also, synthetic cell wall fragments have been produced for these studies.
Results: The kinetic parameters for interactions of typical β-lactam antibiotics and PBP2a were evaluated. PBP2a manifests resistance to inhibition by available β-lactam antibiotics by attenuation of the microscopic rate constant for acylation (k2) and by elevation of the dissociation constants (Kd). The two factors working in concert result in drug resistance. The ABRI cephalosporins, developed for treatment of MRSA, exhibit lower Kd values and k2/Ks ratios were approximately one to two orders of magnitude higher than those of other β-lactams. We have documented that binding of portions of the cell wall outside of the active site of PBP2a facilitates the opening of the active site (from a closed conformation), such that PBP2a becomes more predisposed to modification by the ABRI antibiotics, manifesting in increases in k2/Ks of over 1000-fold compared to typical β-lactam antibiotics.
Conclusions: The recent emergence of variants of MRSA resistant to oxazolidinone and glycopeptide antibiotics has created challenges in treatment of certain strains of S. aureus, a disconcerting situation clinically. The studies from our laboratory provide mechanistic details for the inhibition of PBP2a. The results with the ABRI cephalosporins demonstrate how the problem presented by this unique PBP can be circumvented, presenting opportunities in antibiotic design.
P1577: Telavancin possesses low potential for resistant mutant selection in serial passage studies of Staphylococcus aureus and Enterococci
K.M. Krause, B.M. Benton, D.L. Higgins, K. Kaniga, M. Renelli, P.K. Humphrey (South San Francisco, USA)
Objectives: Serial passage selection studies are useful for predicting the development of endogenous antibiotic resistance in the clinical setting. Telavancin (TLV) is a rapidly bactericidal lipoglycopeptide with multiple mechanisms of action against Gram-positive bacteria. Previous serial passage mutant selection experiments performed using solid media failed to generate stable TLV-resistant isolates from Staphylococcus aureus or Enterococci. In the present study we report results of multi-step selection studies performed in liquid media.
Methods: Selection for spontaneous resistance was performed by serial passage inoculation of liquid media containing a range of TLV concentrations with 106 CFU/mL of each of 7 strains of MRSA (ATCC 33591, MCJ-25, MCJ-10, KPB-08, S-1, SFVA-09, MED-121); MSSA ATCC 29213, VSE E. faecalis ATCC 29212, VanA-type VRE E. faecalis MGH-01 and E. faecium CDC-01, and E. faecium KPB-01 for 20 consecutive passages. Isolates with increased TLV MIC were characterized for stability of resistance, susceptibility to other agents, and growth rate.
Results: Mutants with TLV MICs elevated 2–4X that of parental levels were selected from each strain. However, TLV failed to select mutants with MICs >4× that of parental strains following 20 passages. No major changes were observed in the growth properties of isolates with reduced susceptibility to telavancin. Two mutants with TLV MIC 4× that of their parental strains (32 μg/mL) were selected from the VRE strains. These mutants exhibited reduced susceptibility to daptomycin at or near the same fold MIC increase observed for telavancin, however, no significant cross-resistance to other antimicrobial agents was observed. Mutant phenotypes were stable upon extended subculture (approximately 300 generations) in drug-free medium.
Conclusions: High-level resistance to TLV was not selected from glycopeptide-sensitive organisms, including MRSA, MSSA, or VSE, following continuous exposure for 20 consecutive passages. Mutants with 4X-reduced susceptibility to TLV were selected from 2 out of the 3 VRE strains. These data suggest that TLV possesses a low potential for resistant mutant selection.
New drugs: in vivo studies
P1578: Population pharmacokinetic analysis of dalbavancin
M. Buckwalter, M. Stogniew, J.A. Dowell (King of Prussia, USA)
Objectives: Dalbavancin is a novel, second generation lipoglycopeptide antibiotic in late stage clinical development for complicated skin and skin structure infections (cSSSI). Its distinct pharmacokinetic characteristics support once weekly dosing. A population pharmacokinetic analysis was preformed to estimate patient pharmacokinetic parameters and to determine the significance of possible covariates.
Methods: A population pharmacokinetic analysis was performed that included 1668 dalbavancin concentrations from 532 patients across 3 completed phase 2 and 3 clinical trials. The majority of patients (n = 502) had SSSI with suspected or confirmed Gram-positive bacterial pathogens. The remaining patients (n = 30) had catheter-related bloodstream infections. Most patients received a weekly dalbavancin administration: 1000 mg on Day 1 and a second dose of 500 mg on Day 8. Mixed-effects models were evaluated using the First Order (FO) and FO Conditional Estimation with Interaction (FOCEI) maximum likelihood estimation method in NONMEM. Possible covariates examined on the model included patient demography and concomitant medication use.
Results: The pharmacokinetic model determined for dalbavancin was a two-compartment model with interpatient variability (IPV) described on all the parameters. The typical value and IPV (CV%) of clearance (CL) was 0.0571 L/h (18.0%) and was influenced by body surface area (BSA) and creatinine clearance (CLCR). Patients in the studies had CLCR of greater than 50 mL/min. Volume of distribution of the central compartment (V1) was 4.15 L (24.5%) and was influenced by BSA. The intercompartmental clearance and the peripheral volume of distribution were 0.476 L/h and 11.4 L, respectively. The estimated values for the predominant half-life and volume of distribution at steady state derived from this model were 8.5 days and 15.7 L, respectively. Although BSA and CLCR were identified as sources of variability on CL, and BSA as a source of variability on V1, the addition of these covariates described less than 25% of the IPV.
Conclusion: Dalbavancin pharmacokinetic parameters in patients were predictable, demonstrating low IPV. BSA and CLCR were determined to be statistically important covariates on the model, but had no clinically significant effect on drug concentrations. No dosage adjustment is required for dalbavancin based on the covariates examined in the model, including BSA, patient weight, or CLCR (>50 mL/min).
P1579: A randomised, double-blind, placebo-controlled, parallel-design trial of multiple doses of tolevamer in healthy male volunteers
D. Davidson, A. Porzio, P. Nealon, J. Peppe (Waltham, USA)
Background: Tolevamer potassium-sodium (tolevamer) is a novel, high molecular weight, non-antibiotic polymer developed to treat Clostridium difficile associated diarrhoea (CDAD). The current form has been modified from an earlier compound, tolevamer sodium, by the addition of potassium to prevent the anionic polymer from exacerbating hypokalemia by binding and excreting intestinal potassium in the stool. Tolevamer sodium was shown to resolve the symptoms of CDAD with similar efficacy as vancomycin in a large Phase 2 study. Tolevamer is currently in Phase 3 development.
Objective: To determine the safety and tolerability of liquid tolevamer given at four different dose levels (6.0 g, 9.0 g, 12.0 g, and 15.0 g) to healthy male volunteers.
Methods: A randomized, double-blind, placebo-controlled, parallel-design, multiple dose study consisting of a screening period, a 9 day treatment period and a 1 week follow-up period was conducted. For each dose group, 10 subjects were randomized: 8 to active drug, and 2 to matching placebo. Half the subjects in each dose group received a loading dose (single dose equivalent to a total daily dose) on Day 1, while the other half received their loading dose on Day 9. All subjects received tolevamer t.i.d. on Days 2 through 8. Subjects received a controlled diet averaging 6600 mg/day potassium.
Results: Tolevamer was well tolerated with all forty subjects completing the study. All adverse events were of mild intensity, transient and resolved without sequelae. No dose relationship was apparent. Those considered related to study treatment were gastrointestinal disorders. Flatulence was the most common event. No clinically relevant changes were found in safety laboratory values, vital signs, or physical examination. Analysis of 24-hour urine collections demonstrated that potassium balance was achieved with tolevamer with the exception of a small reduction in mean potassium excretion from baseline to endpoint in the 15.0 g/day dose group of approximately 10 mmol potassium/day. No dose response was evident in urinary potassium excretion, and serum potassium was not adversely affected in any dose group.
Conclusion: Oral treatment with a daily dose of up to 15.0 g tolevamer divided t.i.d. and as a loading dose was safe and well tolerated by healthy male volunteers.
P1580: Tolerability and efficacy of N-chlorotaurine in infections of different body sites: results of phase II clinical trials
M. Nagl, A. Neher, B. Teuchner, R. Höpfl, W. Gottardi (Innsbruck, A)
Objectives: N-chlorotaurine (NCT) is a long-lived oxidant produced by human leukocytes during inflammation. Its crystalline sodium salt can be synthetized chemically, and it has broad spectrum antimicrobial activity against bacteria, fungi, and viruses. Because of mild activity and therefore low toxicity NCT is thought to be useful as an antiseptic applicable to different body sites. We performed three clinical phase II studies to evaluate efficacy and tolerability in otitis externa, epidemic keratoconjunctivitis, and chronic leg ulcers with a purulent coating.
Methods: Aqueous 1% NCT (55 mM) solution was applied topically for 5–7 days in all three indications. All studies were designed with a randomized test (NCT) and control group (standard medication). The otitis externa and epidemic kerato-conjunctivitis study were double-blind. Evaluation was done clinically using a scoring system for subjective and objective symptoms and microbiologically.
Results: NCT was well tolerated in all indications, little burning in the eye for a few minutes in few patients like sweat and significantly less burning (p < 0.05) than by chloramine T in purulent crural ulcers were the only side effects. Granulation and re-epithelialization in ulcers appeared earlier in the NCT group (p < 0.05). NCT was equally effective in removal of the purulent coating than the stronger antiseptic chloramine T. The clinical symptoms of external otitis decreased 2 days earlier in the test group than in the control group treated with a combination of polymyxin B, neomycin, and hydrocortisone (p < 0.01). Regarding epidemic conjunctivitis, severe courses caused mainly by adenovirus type 8 showed a reduction of symptoms after 3–5 days, which was earlier than in the control group mock treated with gentamicin (p < 0.05).
Conclusion: The mild antiseptic NCT is excellently tolerated and effective in infections of different body sites. As an endogenous amino acid derivative toxic and allergic side effects have never been observed, and it can be applied without additives. Because of these advantages NCT can be estimated as a very promising antimicrobial agent in human medicine.
P1581: Ertapenem as initial antimicrobial monotherapy for patients with typical community-acquired pneumonia
C. Petrogiannopoulos, G. Svoukas, K. Papamichael, A. Deliousis, K. Kanakis, P. Papasava, N. Kostakos, A. Zaharof (Athens, GR)
Objectives: Ertapenem, a Group 1 carbapenem, is a once-a-day parenteral a-lactam antibiotic which is used as initial antimicrobial monotherapy for treatment of community-acquired pneumonia (CAP). The aim of our study was to evaluate the clinical response and the safety profile of ertapenem.
Methods: We studied 64 patients (34 female, 30 male) of 61. mean age (range 18–9) with CAP that was proved clinically, radiographically and after blood test examinations. All blood cultures were negative, while sputum cultures were positive for Streptococcus pneumoniae in 6 patients and Klebsiella pneumoniae in 2 patients. The patients recieved 1 gr ertapenem intravenous (iv) daily, as initial antimicrobial therapy, for 5 days at least. Everyday, haematological and biochemical tests were made in order to evaluate the clinical response and the safety profile of ertapenem. Clinically improving patients were switched to oral antibiotic therapy after 5 days.
Results: After 5 days therapy with ertapenem 61 patients (95,3%) had clinical and haematological improvment, concerning the reduction of fever, WBC from 14000 ± 1700-7800 ± 1200 and CRP value from 9.1 ± 1.1-2.2 ± 0.8. The most common drug-related adverse experiences reported during parenteral therapy in patients treated with ertapenem were diarrhoea (6.1%), infused vein complication (4.7%), nausea (3.1%) and headache (1.6%), while liver function tests were normal before and after treatment.
Conclusion: These data provided evidence for the efficacy of ertapenem as initial antimicrobial monotherapy for patients with CAP. Ertapenem, given 1 gr once a day by iv infusion was generally well tolerated and had overall a very good safety and tolerability profile.
P1582: Treatment of chronic MRSA infections using a novel aqueous extract of allicin (AB1000)
R.R. Cutler, P.D. Josling, N.J. Bennett (London, Rye, UK)
Allicin is recognised as the main bioactive agent from Allium sativum or garlic. This compound is highly active but generally unstable. Using a cold aqueous extraction method, we have obtained a novel extract of allicin (AB1000) that we have reported is stable and highly active in vitro against methicillin resistant Staphylococcus aureus (MRSA). Due to national publicity of AB1000, patients with long standing unresolved MRSA infections requested this agent for treatment. MRSA is commonly related to delayed closure for many chronic and acute wounds. This is associated with high levels of bacteria in tissues but they can also through toxin secretion. These toxins can cause local necrosis and disrupt the delicate balance of critical mediators such as cytokines and proteases necessary for healing progression. We present initial findings from three patients who have completed a course of treatment. These courses consisted of capsules (450 mg, 3 per day); spraying liquid AB1000 (1000 μg ml−1) onto the affected areas once per day and applying AB1000 Cream (500 μgml−1) to the infected area once daily. Patients were screened, nasal and wound for MRSA prior and during treatment. All patients were nose and wound swab MRSA positive prior to treatment. All were over 60 years of age and had either major surgery or long term skin infections leading to the formation of ulcers infected by MRSA. Two of the MRSA infections were community acquired and one hospital acquired. The strains isolated from each patient were tested in vitro against AB1000 and all were susceptible. Patients reported an improvement in their condition after 2 and 6 weeks treatment and the infections resolved in 3–4 months. Although the timescales required for treatment may be longer than those normally required using antibiotics, the initial relief from weeping ulcers and pain was much quicker. It should be noted these the patients had been receiving unsuccessful treatment with antibiotics for months or years prior to treatment with AB1000. A possible reason for the initial relief from symptoms could relate to the reported activity of garlic extracts to neutralise bacterial exoenzymes in vitro. This could account for the findings that patients got relief from their symptoms before the MRSA were fully removed from the lesion site.
P1583: In vitro and in vivo antibacterial activity of DA-7218, a new oxazolidinone, against Staphylococci and Enterococci
S. Choi, T. Lee, E. Kang, T. Son, W. Im, J. Rhee, W. Kim, D. Yong, J.H. Yum, K. Lee (Yongin-Si, Seoul, KOR)
Objectives: With the approval of linezolid as the first therapeutically acceptable oxazolidinone, efforts have been made to identify new oxazolidinones with improved antibacterial properties. This study was performed to establish the in vitro and in vivo antibacterial activity of DA-7218 against Staphylococci and Enterococci compared to linezolid.
Methods: MICs were determined against S. aureus, CNS, E. faecalis and E. faecium using NCCLS agar dilution method. In vivo protection tests of DA-70218 were carried out in a murine systemic infection against S. aureus, CNS, E. faecalis and E. faecium. Drugs were administrated intravenous (i.v.) or orally (p.o.).
Results: MIC90 values of DA-7157, an active metabolite of DA-7218, were 0.5 μg/ml for both methicillin-susceptible and -resistant Staphylococci and 0.25–0.5 μg/ml for both vancomycin-susceptible and -resistant Enterococci, and they were 4- to 8-fold lower than those of linezolid. ED50s of DA-7218 were 2–4 fold and 4–8 fold lower than those of linezolid upon p.o. and i.v. administration in staphylococcal and enterococcal systemic infections, respectively.
Conclusions: DA-7157 showed 4–8 fold better in vitro antibacterial activity than linezolid against Staphylococci and Enterococci, and DA-7218 had excellent in vivo efficacies against staphylococcal and enterococcal infections.
P1584: Inhibition of ampicillin-induced resistance by targeted recombinant beta-lactamase monitored by PFGE and antibiograms of Escherichia coli
S. Mentula, T.A. Virtanen, J. Harmoinen, M. Carson, E. Könönen (Helsinki, Espoo, FIN)
Objectives: Targeted recombinant beta-lactamase (TRBL) has been designed to degrade beta-lactam antibiotic residues in the gastrointestinal tract and thereby inhibit the emergence of resistance. In the present study the changes in resistance rates and persistence of different intestinal Escherichia coli strains were followed in beagles during ampicillin treatment with or without TRBL.
Methods: Eighteen laboratory beagles were randomised to receive ampicillin (AMP), ampicillin + TRBL (AMP/TRBL) or placebo (PLA). Susceptibility of 961 intestinal E. coli isolates, collected before during and after the treatment, was tested against 9 antimicrobial agents by the NCCLS disk diffusion method. Selected 402 isolates were typed using pulsed-field gel electrophoresis (PFGE) after XbaI restriction of total DNA.
Results: Vast genetic heterogeneity, including 25 different PFGE types displaying 19 different resistance patterns, was detected. The beagles harboured on average 5 PFGE types during the study and shared several common clones. Although the number of resistant isolates increased and similar resistance patterns were detected in all treatment groups, no single ampicillin-resistant PFGE type was widely spread between groups. Ampicillin resistance was found in 6 PFGE types and in 9 different combinations of co-resistance. The proportion of resistant isolates (98%, 36% and 28%) and number of dogs (6, 5 and 3) carrying ampicillin-resistant strains during the treatment was higher in AMP group than in AMP/TRBL and PLA groups, respectively. New resistant PFGE types appeared and extensively replaced the previous types in the AMP group but there were 2 cases where a previously susceptible strain acquired resistance during the treatment. In the AMP/TRBL group the slight increase in resistance was due to enrichment of resistant strains in dogs that already carried resistance. In the PLA group new resistant PFGE types appeared but their proportions remained moderate.
Conclusions: TRBL seems to inhibit the selection pressure by preserving the genetic diversity of intestinal E. coli populations. Increase in resistance is due to selection of heterogeneous resistant strains rather than emergence of resistance among previously susceptible strains.
Pharmacokinetics/pharmacodynamics of anti-staphyloccal drugs
P1585: Kinetics of linezolid release from polymethylmethacrylate beads
P. Anguita Alonso, M. Rouse, K.E. Piper, D.J. Jacofsky, D.R. Osmon, A.D. Hanssen, J.M. Steckelberg, R. Patel (Rochester, USA)
Objective: To study the release of linezolid from polymethylmethacrylate (PMMA) beads loaded with three different concentrations of antimicrobial.
Methods: PMMA beads were prepared loading Surgical Simplex Pe radiopaque bone cement with 2.5%, 7.5% and 15% (w/w) linezolid. Each bead was placed in a continuous flow chamber with 1 ml Krebs Ringer buffer, flowing at 1 ml/h at 37°C for 48 h. The antimicrobial concentrations were determined in triplicate by disk diffusion antimicrobial bioassays (lower detection limit was 5 mcg/ml). The detection time, Cmax, Tmax, AUC0-infinity and per cent antimicrobial recovered from the beads were calculated.
Results: Results are listed in the table as mean ± standard deviation of three values. No antimicrobial greater than the limit of detection of the assay was present for 2.5% linezolid loaded beads. Elution parameters of linezolid were similar for beads containing 7.5 and 15% of antimicrobial.
| Antimicrobial concentration | Time initially detectable (h) | Time no longer detectable (h) | Peak concentration mcg/ml | Peak time (h) | AUC (mcgh/ml) | Percent initial amount detected |
|---|---|---|---|---|---|---|
| Linezolid 2.5% | – | – | <5 | – | – | – |
| Linezolid 7.5% | 1 ± 0 | 18 ± 9 | 64 ± 39 | 1±1 | 315 ± 144 | 17 ± 6 |
| Linezolid 15% | 1 ± 0 | 19 ± 4 | 65 ± 31 | 1±0 | 330 ± 64 | 18 ± 5 |
| Vancomycin 7.5% | 3±0 | 23 ± 1 | 19 ± 4 | 4±0 | 180 ± 8 | 11 ± 0 |
From Clin Orthop Rel Res 2002;403:49
Conclusions: Linezolid peak concentration and AUC were greater than calculated for vancomycin with similar detectable number of hours. In vivo studies with linezolid loaded PMMA are warranted.
P1586: Predicted AUC/MIC using a simple vancomycin nomogram exceeds proposed pharmacodynamic targets for Staphylococcus aureus infections
S. Davis, M. Rybak, B. Domagala, P. McKinnon (Detroit, Grand Rapids, St. Louis, USA)
We previously reported on a validated vancomycin (V) dosing nomogram (NM) with minimal trough level monitoring that achieves clinical outcomes equivalent to traditional pharmacokinetic dosing. A proposed target for V in the treatment of S. aureus (SA) is an AUC/MIC of ≥400. [Moise-Broder et al. Clin Pharmacokinet 2004]
Objective: To determine if a simple nomogram for dosing V achieves an AUC above proposed pharmacodynamic (PD) targets for SA infections.
Methods: 516 patients who received V per NM from 1996–2003 were evaluated. Demographics, antibiotic therapy, microbiology, and clinical outcomes were collected. Predicted area under the curve (AUC24), Cmax, and Cmin were extrapolated from patient-specific data, and clinical outcomes were assessed. Recent SENTRY data representing 35,488 SA isolates from the US demonstrates an MIC90 of 1. [Fritsche T et al. Int J Antimicrob Agents 2004] Therefore, for MIC values reported as <2 mcg/mL, a value of 1 was assumed in AUC/MIC calculations.
Results: The study population (n = 516) was 63% male, average age 49 ±17 years. Average Clcr was 82±28mL/min, and average duration of therapy was 9.2 ± 7.5 days. 57% of patients received 1 g every 12 h, 19% 1 g every 8 h, 12% 1 g every 24 h, 6% 0.5 g every 12 h, and 6% other doses. Average predicted AUC24 was 532 ± 213, and Cmin was 15.1 ± 8.1. Actual Cmin in patients with troughs was similar, 15.7 ± 10.4. AUC was noted to be slightly higher in patients with lower Clcr. For those with Clcr of 31–60 ml/min, AUC was (mean, 95%CI) 642 (592–692); 61–90 ml/min was 565 (540–588), and >90 ml/min was 421 (403–438). In the subset of patients with documented SA infections (n = 201), 76% were methicillin-resistant, and all isolates had a vancomycin MIC <2. Bacteremia accounted for 46% of infections, while 30% were skin/soft tissue, 15% lung, and 9% other infections. Figure 1 presents clinical and microbiologic outcomes of SA infection by groupings of AUC/MIC.
Conclusions: This previously developed V nomogram exceeds adequate Cmin and AUC/MIC targets for the treatment of SA infections and produces reliable clinical success rates. Use of the NM can obviate the need for routine serum concentration monitoring of V in all patients. In patients with higher Clcr, AUC was slightly lower but still exceeded proposed PD targets.
P1587: In vitro and in vivo release of daptomycin or vancomycin from polymethacrylate beads
M. Rouse, E. Hall, K.E. Piper, M. Jacobson, A.D. Hanssen, D.J. Jacofsky, R. Patel (Rochester, USA)
Objective: Polymethylmethacrylate (PMMA) is used as a local antimicrobial delivery in the treatment and prevention of bone/ joint infection. We developed an in vitro assay to determine release profiles of antimicrobial agents from PMMA. In this study we compared the in vitro release kinetics of daptomycin or vancomycin in a continuous flow chamber with in vivo release in a rat model.
Methods: Daptomycin or vancomycin (7.5% w/w) was loaded into the powder component of PMMA; the liquid monomer added and mixed. Antimicrobial loaded PMMA was formed into 3 mm beads and allowed to polymerize for 24 h. The effect of PMMA components and polymerization on the activity of daptomycin or vancomycin was determined by homogenizing loaded beads in 2 ml of Kreb's Ringer buffer and assaying the buffer for antimicrobial activity. In vitro antimicrobial release kinetics were determined by placing a bead in a chamber containing 1 ml of Kreb's Ringer buffer, with 1 ml/h Kreb's Ringer buffer flowing through the chamber. Buffer eluted from the system was collected hourly and assayed for antimicrobic using a bioassay. Each bead/antimicrobial was tested in triplicate. In vivo antimicrobial release kinetics were performed by surgically implanting single beads into bone defects in the proximal left tibia of rats. Three rats with a daptomycin loaded bead and three with a vancomycin loaded bead were sacrificed at 2, 4, 6, 8, 10, 14, 18 and 21 d after placement. Bone tissue within 5 mm of the bead was recovered, homogenized in 2 ml Kreb's Ringer buffer, incubated 24 h, and assayed for antimicrobic. The per cent released listed is total antimicrobial detected in buffer or tissue/total amount of antimicrobial loaded into each bead.
Results: 100% of daptomycin and 89% of vancomycin loaded into PMMA beads was detected immediately after polymerization. Pharmacokinetic results are shown the mean concentrations in the table.
| Elultion mode | Cmax | Tmax | Percent released | AUCn-2 | |
|---|---|---|---|---|---|
| Daptomycin | invitro | 62 mcg/ml | 6h | 18% | 272 h × mcg/ml |
| invivo | 178 mcg/g | 4d | 19% | 621 d × mcg/g | |
| Vancomycin | invitro | 19 mcg/ml | 4h | 11% | 180 h × mcg/ml |
| invivo | 49 mcg/g | 4d | 7% | 206 d × mcg/g |
Conclusion: The peak daptomycin or vancomycin concentration observed in vivo was approximately three-fold the peak observed in vitro. Daptomycin loaded PMMA produced higher antimicrobial concentrations than did vancomycin loaded PMMA in vitro and in vivo.
P1588: Comparative pharmacodynamics of daptomycin and vancomycin with Staphylococcus aureus: multiple-dose simulations using an in vitro dynamic model
S. Zinner, M. Smirnova, I. Alferova, I. Lubenko, S. Vostrov, Y. Portnoy, A. Firsov (Cambridge, USA; Moscow, RUS)
Objective: To compare the pharmacodynamics of daptomycin and vancomycin, killing kinetics of S. aureus were studied in five-day treatment courses over a wide range of ratios of the 24-h area under the curve (AUC) to MIC.
Methods: A clinical isolate S. aureus 866 with MIC of daptomycin of 0.35 mg/L and MIC of vancomycin of 0.7 mg/L was exposed to once-daily daptomycin and twice-daily vancomycin for five consecutive days. Mono-exponential concentration-time profiles were simulated with half-lives of 9 h (daptomycin) and 6 h (vancomycin) over a 16-fold range of the AUC/MIC ratio: from 16–256 h. The antimicrobial effect was expressed by its intensity (IE–the area between the control growth curve in the absence of antibiotics and the time-kill/regrowth curve observed in the presence of antibiotic). The cumulative effect of each treatment was determined from time zero to the time the effect no longer could be detected, i.e. the time after the last antibiotic dose at which the number of antibiotic-exposed bacteria reached at least 9 LogCFU/ml. This value was the cutoff level used to determine IE.
Results: With both daptomycin and vancomycin, bacterial regrowth followed initial killing over the entire range of the simulated AUC/MIC ratio. Anti-staphylococcal effects of the antibiotics depended on the AUC/MICs: the higher the AUC/ MIC, the lower numbers of surviving organisms and the later the regrowth. The IE-log AUC/MIC plots were linear and virtually superimposed for daptomycin and vancomycin. Based on the IE-AUC/MIC relationship of the IE, the anti-staphylococcal effect of the proposed therapeutic dose of daptomycin (4 mg/kg; AUC/MIC 1100 h for S. aureus 866) is 20% greater than that of two 1-g doses of vancomycin given at a 12-h interval (AUC/MIC 570 h).
Conclusion: These findings predict greater anti-staphylococcal efficacy of clinically achievable AUC/MICs of daptomycin relative to vancomycin.
P1589: Relative abilities of daptomycin and vancomycin to prevent the enrichment of resistant Staphylococcus aureus mutants in an in vitro dynamic model
A. Firsov, I. Alferova, M. Smirnova, I. Lubenko, S. Vostrov, E. Mirchink, Y. Portnoy, S. Zinner (Moscow, RUS; Cambridge, USA) Objectives: The mutant selection window (MSW) hypothesis has been successfully tested in our recent study with S. aureus 866 (MIC of DAP 0.35 mg/L and MIC of VAN 0.70 mg/L) exposed to daptomycin (DAP) and vancomycin (VAN) over a wide range of the 24-h area under the curve (AUC) to MIC. To explore if the AUC/MIC relationships of resistance are bacterial strain-independent, a less susceptible S. aureus 10 (MIC of DAP 1.1 mg/L and MIC of VAN 2.2 mg/L) was tested using the same model.
Methods: Five-day treatments were mimicked with once-daily DAP (half-life 9 h) and twice-daily VAN (half-life 6 h). S. aureus 10 was exposed to a series of pharmacokinetic profiles of DAP and VAN that provide peak concentrations equal to the MIC, between the MIC and the mutant prevention concentration (MPC), i.e., within the MSW, and above the MPC (AUC/MIC 16–256 h). MPCs of DAP and VAN were 5.5 and 10.9 mg/L, respectively. Changes in the susceptibility of S. aureus 10 to DAP or VAN were examined by repeated MIC determinations at each 24 h and in the end of treatment. To determine the resistance frequency (f), the surviving organisms were counted by plating the specimens on agar containing 2 × and 4 × MIC of DAP or VAN.
Results: Both in terms of susceptibility testing and the population analysis, selection of the resistant mutants occurred at DAP and VAN concentrations that fell into the MSW (AUC/ MIC from 32 to 128 h) but not at concentrations out of MSW (AUC/MIC 16 and 256 h). Bell-shape AUC/MIC relationships of resistance of S. aureus 10 were similar to those observed with DAP- and VAN-exposed S. aureus 866 as well as with S. aureus exposed to fluoroquinolones. Based on these findings, an AUC/ MIC ratio that may protect from the selection of resistant mutants was predicted at 256 h for both DAP and VAN. For S. aureus 10, this protective value is less than AUC/MIC provided by usual 4 mg/kg dose of DAP (350 h) but not by two 1-g doses of VAN (180 h).
Conclusions: Clinically achievable concentrations DAP may be more effective than those of VAN in prevention of the selection of resistant S. aureus. Also, these findings support the MSW hypothesis.
P1590: The influence of Staphylococcus aureus accessory gene regulator function on the development of glycopeptide heteroresistance in an in vitro pharmacodynamic model
B. Tsuji, M. Rybak (Detroit, USA)
Objective: The development of glycopeptide heteroresistance in patients with S. aureus infections has been associated with prolonged exposure to sub-therapeutic levels of vancomycin. S. aureus which display accessory gene regulator (agr) dysfunction have been associated with persistent bacteraemia and may be predisposed to develop glycopetide resistance: sub-inhibitory concentrations of vancomycin have selected for heteroresistance in vitro in agr-null group II S. aureus. We studied the effect of administering varying exposures of vancomycin and the development intermediate-level glycopeptide resistance in agr positive and negative group II S. aureus in an in vitro pharmacodynamic model (IVPM).
Methods: One agr+ group II (RN6607) and the respective agr–group II, null derivate (RN9120) strain of S. aureus were obtained from the Network on Antimicrobial Resistance in Staphylococcus aureus (NARSA). MICs were determined by E-test & microdilution according to NCCLS. An IVPM was used to simulate regimens of vancomycin of 1 g, 750 mg, 500 mg, 250 mg and 125 mg q12 h (Targeted Peak/Trough concentrations 40/10.0, 30/7.5, 20/5.0, 10/2.5 and 5/1.2 μg/ml). Simulations were performed in triplicate and bacterial quantification occurred over 72 h. The development of vancomycin heteroresistance was evaluated at 24, 48 and 72 h.
Results: Pre-exposure vancomycin MIC were 1.0 against both agr+ group II and agr− group II strains. Against agr+ II S. aureus, exposure to all simulated of vancomycin regimens did not result in the development of resistance over 72 h. Post-exposure MICs for all regimens were ≤2.0 μg/ml. Against agr− II, over 72 h, exposure to vancomycin 1 g q12 h (24 h Area Under Concentration Curve (AUCfree24/MIC:510 μg/ml/hr) and 750 mg q12 h (AUCfree24/MIC: 382) resulted in post-exposure MICs of 1–3 μg/ml. Increased post-exposure MICs up to 3 μg/ml were noted secondary to exposure to 500 mg (AUCfree24/MIC: 264) and 250 mg (AUCfree24/MIC: 123). Exposure to 125 mg (AUCfree24/MIC:67) resulted in the development of vancomycin heteroresistance, as post-exposure MICs increased to 6 μg/ml.

Conclusions: Sub-optimal exposures of vancomycin resulted in the development of glycopeptide heteroresistance in agr-null group II S. aureus. These findings suggest that prolonged, low-level vancomycin concentrations or decreased vancomycin penetration in sequestered infections may provide conditions that select for glycopeptide heteroresistance.
P1591: Killing kinetics of Staphylococcus aureus exposed to linezolid, alone and in combination with doxycycline, in an in vitro dynamic model
M. Smirnova, I. Alferova, I. Lubenko, S. Vostrov, Y. Portnoy, E. Isakova, S. Zinner, A. Firsov (Moscow, RUS; Cambridge, USA)
Objective: To predict the possible advantages of linezolid/ doxycycline combinations, their pharmacodynamics were studied with S. aureus in an in vitro dynamic model that mimics human pharmacokinetics of the antibiotics.
Methods: A clinical isolate of S. aureus at a starting inoculum of 8 Log CFU/ml was exposed to twice-daily linezolid (half-life 6 h), alone and in combination with once-daily doxycycline (half-life 15 h) for five consecutive days. The MIC of linezolid was 1.56 mg/L and MIC of doxycycline was 6.25 mg/L. With linezolid, a clinically attainable ratio of the 24-h area under the curve (AUC) to MIC (200 h) was simulated. To achieve the same AUC/MIC ratio with linezolid/doxycycline, a 60-h AUC/MIC of linezolid was combined with a 140-h AUC/MIC of doxycycline. In addition, similar five-day treatments were performed with linezolid at AUC/MIC of 60 h alone. To provide simultaneous mono-exponential elimination of linezolid and doxycycline with different half-lives, a previously described dynamic model was modified according to Blaser and Zinner.
Results: Regardless of the simulated AUC/MIC, linezolid alone produced a relatively slow initial reduction of the starting inoculum (8 Log CFU/ml), with minimal numbers of surviving organisms on the second day and regrowth on the fifth day of treatment. AUC/MIC-dependent killing was observed beginning from the third day when numbers of surviving organisms were significantly lower after linezolid exposure at AUC/MIC of 200 than 60 h. The respective intensities of the anti-staphylococcal effect (IEs) were 180 and 110 (Log CFU/ml) × h. The linezolid/doxycycline combination exhibited a more rapid initial killing followed by much lower numbers of surviving organisms with later regrowth. The respective IE was 1.7 times greater than mono-therapy with linezolid at the same AUC/MIC (200 h) and 2.7 times greater than at linezolid's AUC/MIC of 60 h.
Conclusions: These data predict the ability of doxycycline to enhance anti-staphylococcal effects of linezolid.
Infections in neutropenic patients
P1592: Empirical antibiotics against Gram-positive infections for febrile neutropenia. Systematic review and meta-analysis of randomised controlled trials
M. Paul, S. Borok, A. Fraser, L. Vidal, L. Leibovici (Petah Tikva, IL)
Background: The use of glycopeptides for febrile neutropenia has increased, following the prevalence of gram-positive (GP) infections among these patients. Withholding empirical glycopeptide treatment is ecologically wise stipulating that this does not result in increased mortality. We performed a systematic review and meta-analysis of trials assessing the addition of anti-GP treatment in febrile neutropenia.
Methods: Included were randomised trials comparing one antibiotic regimen with or without placebo against the same regimen with additional antibiotics against GP infections. We searched Medline, Embase, the Cochrane Library, references and conference proceedings. No date, language, age or publication limits were imposed. Two reviewers extracted data independently. Relative risks and 95% confidence intervals were estimated using the fixed effect model. Values <1 favor the addition of anti-GP antibiotics.
Results: Thirteen trials and 2392 participants were included. Antibiotics against gram-positive infections included glycopeptides in 9 studies., cephalotin in 2, flucloxacillin and trimethoprim-sulphamethoxazole one each. Eleven studies randomised patients at onset of febrile neutropenia, and 2 studies randomised patients with persistent fever at 72–96 hours. No significant reduction in all-cause mortality was seen with the addition of antibiotics against gram-positive infections (figure), RR 0.86 [0.58–1.26]. Overall failure was equivalent in both study arms, RR 1.00 [0.79–1.27] while failure including modifications occurred more frequently in the control arm due to empirical addition of glycopeptides, RR 0.76 [0.68–0.85]. Amphotericin was added more frequently to the intervention arm in non-blinded trials, RR 1.51 [0.80–2.83] but not in double-blind studies, RR 0.99 [0.75–1.33]. The subgroup of patients with documented GP infections was assessed and no difference in mortality or failure was detected. Meta-regression demonstrated no association between the rate of GP bacteraemia and the Clinical Outcome by primary infection (ITT)* relative risk for mortality or failure. Empirical glycopeptides were associated with a significant reduction in bacterial superinfections, RR 0.38 [0.24–0.59], more specifically gram-positive superinfections, RR 0.21 [0.11–0.37], and increased nephrotoxicity, 1.43 [1.06–1.94].
Conclusions: Addition of glycopeptides to the antibiotic treatment of febrile neutropenic patients, in general, can be safely deferred for the documentation of resistant GP infections.
P1593: Linezolid in febrile neutropenic oncology patients
G. Martinelli, C. Hartman, L. Leonard, K.J. Tack (Milan, I; Ann Arbor, USA)
Objectives: Gram-positive bacteria are common pathogens in neutropenic oncology patients. Linezolid (LZD) is effective for the treatment of infections caused by Gram-positive pathogens including methicillin resistant Staph spp., glycopeptide resistant Enterococcus spp., and multi-drug resistant Strep pneumoniae. LZD was compared to vancomycin (VAN) in a randomized, double-blind, multi-centre trial in febrile neutropenic oncology patients.
Methods: Eligible oncology patients with febrile neutropenia (absolute neutrophil count <500/mm3) and a proven or suspected Gram-positive infection received LZD (600 mg) or VAN (1 g) IV q12 h for 10–28 days. Test of cure (TOC) assessment was conducted 7 days posttherapy. Clinical efficacy was analysed using sponsor defined clinical outcome.
Results: 605 patients were treated (304 LZD, 301 VAN); groups were similar with respect to demographics, comorbidities, severity/type of illness, and degree of neutropenia. Hematologic/lymphatic cancers were the most frequently observed (95.4% LZD, 93.7% VAN). Average duration of treatment was similar between groups (LZD 11.4 days; VAN 11.5 days). Mean time until defervescence (oral temperature ≤ 37.5°C) in LZD patients was 6.4 days vs 6.7 days in VAN patients (p = 0.54). Overall mortality rate at 16 days posttherapy was 5.6% for LZD patients vs 7.6% for VAN (p = 0.31). Overall clinical success rates at FU were comparable between LZD and VAN in the intent-to-treat (ITT) population (57.0% vs 52.9%, p = 0.32). Clinical success rates within the subgroups of primary infections were also similar between LZD and VAN (Table 1). Microbiologic outcomes were similar in the two groups. Clinical laboratory results for hematology and biochemistry tests, such as change from baseline and clinically abnormal values, were similar between groups. Drug-related adverse events (DRAEs) occurred significantly less frequently for LZD patients than for VAN patients (52/303 [17.2%] vs 72/300 [24.0%], p = 0.04). Nausea, vomiting, rash, erythema, and diarrhoea were the most frequently reported DRAEs for both treatment groups. No DRAE was reported significantly more often in the LZD group, however VAN patients reported a significantly higher percentage of drug-related kidney failure (LZD 0/303 vs VAN 4/300, p = 0.04).
Clinical Outcome by primary infection (ITT)*
| Linezolid [% (n/N)] | Vancomycin [% (n/N)] | P-Value | |
|---|---|---|---|
| BAC | 48.3 (42/87) | 41.0 (34/83) | 0.34 |
| CRI | 63.3 (19/30) | 59.3 (16/27) | 0.75 |
| FUO | 59.3 (51/86) | 56.8 (50/88) | 0.74 |
| PNE | 60.0 (15/25) | 50.0 (10/20) | 0.50 |
| SSI | 72.7 (16/22) | 61.1 (11/18) | 0.44 |
| UTI | 50.0 (1/2) | 66.7 (2/3) | 0.71 |
| Other | 56.3 (18/32) | 62.2 (23/37) | 0.62 |
Does not include patients designated as missing or indeterminate at TOC BAC (bacteremia of unknown source), CRI (vascular catheter related infections), FUO (fever of uncertain origin), PNE (pneumonia), SSI (skin/skin structure infection), UTI (urinary tract infection)
Conclusion: LZD is as effective as VAN and is a safe therapeutic option for treating febrile neutropenic oncology patients with proven or suspected Gram-positive infections of varying underlying etiologies.
P1594: UK Survey of antibiotic treatment of febrile neutropenia
H. Ziglam, K. Gelly, W. Olver (Dundee, UK)
Objectives: Is to perform a nationwide survey to define the different practice in managing febrile neutropenia in haematology units.
Methods: The design was a single round audit questionnaire was sent out to the selected haematologist of 220 haematology units in the UK. Questions were asked regarding both antimicrobial chemotherapy of choice and antimicrobial prophylaxis in managing febrile neutropenia.
Results: Responses were received from 167 (76%) of the 220 haematology units providing care to patients with febrile neutropenia. Febrile neutropenia is empirically treated with piperacillintazobactam or carbapenem monotherapy by 10% of haematologists. Overall, dual therapy (i.e., an aminoglycoside plus a piperacillim-tazobactam) is prescribed by 72% of haematologists. When response to initial empirical therapy does not occur after 3–4 days, 32% of haematologists may add glycopetide (vancomycin or teicoplanin) or 29.1% may change to carbapenem and glycopeptide. Forty-eight per cent of haematologists will routinely add conventional amphotericin to the ongoing antibiotics in persistent neutropenia, compared to 39% using liposomal amphotericin. Granulocyte colony-stimulating factor is frequently used as an adjuvant in managing febrile neutropenia (47% of haematologists). For antifungal prophylaxis, fluconazole is prescribed by 57% of haematologist followed by itraconazole by 33%. Forty-seven of haematologists use antifungal prophylaxis in all neutropenic patients compared to 17% whom used antifungals only in patients with neutrophil less than 0.1 × 109/L.
Conclusions: Rationalization of using antimicrobial chemotherapy in the management of febrile neutropenia, by adopting nationally agreed practice guidelines, could significantly reduce costs and improve patient's care.
P1595: Ciprofloxacin versus an aminoglycoside as combination therapy to a beta lactam for the treatment of febrile neutropenia: a meta-analysis of randomised controlled trials
I. Bliziotis, A. Michalopoulos, S. Kasiakou, G. Samonis, C. Christodoulou, S. Chrysanthopoulou, M. Falagas (Athens, Iraklion, GR)
Background: Ciprofloxacin has been used extensively for prevention and treatment of infections in neutropenic patients in both the outpatient and inpatient setting.
Objectives: To compare the effectiveness and toxicity of ciprofloxacin versus an aminoglycoside (both in combination with a beta lactam) for the treatment of febrile neutropenia in the inpatient setting.
Design: Meta-analysis of randomized controlled trials (RCTs).
Data sources: Data for this meta-analysis were identified from PubMed (1/1950 to 10/2004), Current Contents, Cochrane central register of controlled trials, references from relevant articles, and abstracts presented in international conferences.
Study selection: RCTs comparing ciprofloxacin with an aminoglycoside as combination therapy to a beta lactam for the treatment of febrile neutropenia and reporting effectiveness, mortality, and/or toxicity data were included in the analysis. Studies that examined quinolones as monotherapy, the combination of a quinolone with an aminoglycoside, use of quinolones in the outpatient setting from the start of treatment, or quinolones other than ciprofloxacin were excluded.
Data extraction: Data for 3 primary and 2 secondary outcomes were extracted by two investigators.
Data synthesis: A total of 8 RCTs were included in the analysis. Better outcomes (some with statistical significance) were observed with ciprofloxacin/beta-lactam compared to aminoglycoside/beta-lactam combination; clinical cure without modification of the initial regimen (OR 1.32, 95% CI 1.00–1.74), clinical cure in the subset of patients with documented infections (OR 1.56, 95% CI 1.05–2.31), all cause mortality (OR 0.85, 95% CI 0.54–1.35), withdrawal of the study drugs due to toxicity (OR 0.87, 95% CI 0.57–1.32), and nephrotoxicity (OR 0.30, 95% CI 0,16–0.59). The ciprofloxacin-containing combination was also associated with better clinical cure in the subset of RCTs with non-low-risk patients (OR 1.38, 95% CI 1.01–1.88).
Limitations: The study results may not be extrapolated to patients who received prior a quinolone for prevention of infection nor to patients who receive per os or intravenous treatment for febrile neutropenia in the outpatient setting.
Conclusions: The combination of ciprofloxacin with a betalactam antibiotic should be considered as an important therapeutic option in hospitalized febrile neutropenic patients who have not been receiving a quinolone for prevention of infections.
P1596: Blood cultures statistic study in febrile neutropenic cancer patients
E. Chinou, A. Apostolakopoulos, A. Georgouli, E. Skouteli, P. Golemati (Athens, GR)
Objectives: The aim of this study is to evaluate the frequency and susceptibility patterns of bacterial pathogens isolated from blood cultures in febrile neutropenic cancer patients hospitalized in an anticancer hospital over a two years period.
Methods: A total number of 586 blood cultures were examined, using the automated Bact/Alert Microbial Detection System (aerobic FAN and anaerobic FN). After classic subculture methods, all isolates were characterized at species level by the Api 20 system. Minimal inhibitory concentrations were performed using the semi automated method Microscan according to NCCLS recommendations. All the investigated patients were neutropenic (WBC <1 × 10 to the power of 9 per liter) with fever (>38°C) and were under antimicrobial and antifungal therapy at the time of blood culture.
Results: Over the two years period (2002–2003) 586 blood cultures were performed on the basis of the physicians request from 81 patients hospitalized in the two oncology and one hematologist ward of our hospital. The mean length of neutropenia was 13 days. The pathogens were isolated from 64 blood cultures in 21 patients (25%). Among the 22 bacterial strains isolated from bloodstream infections, 72.7% were Gram negative strains (n = 16), 18,1% Gram positive cocci (n = 4), one strain L. monocytogenes and one strain C. kruzei. In the group of Gram-negative strains E. coli was dominant with an increase antibiotic resistance (7/9 resistant to Amoxicillin / Clavulanic acid and 5/9 resistant to Ciprofloxacin). All the Gram-negative rods were highly susceptible to Imipenem (15/16), Meropenem (16/16) and Piperacillin/tazobactam (16/16). In the group of Gram-positive bacteria glycopeptide resistant strains were no found. All the Staphylococci strains were susceptible to Methicillin.
Conclusions: We observed rising trends in the number of Gram-negative isolates and in antibiotic resistance of all gram-negative rods. The empiric therapy with wide range antibiotics in febrile neutropenic cancer patients seems to be the optimal option, but the monitoring of antibiotic susceptibility of bacterial strains is still required.
P1597: Epidemiological, clinical and prognostic features of febrile neutropenia in cancer patients
M. Aguilar-Guisado, E. Cordero, J. Cisneros, I. Espigado, R. Parody, J. Pachón (Seville, E)
Objective: To describe epidemiology, aetiology, clinical and prognostic features of febrile neutropenia (FN) in patients with solid or haematological neoplasms.
Method: Prospective observational study of all FN episodes, among patients admitted in Oncology and Haematology Departments during 21 months (November 2002-August 2004) with either haematological or solid neoplasm of any localization.
Results: We studied 292 episodes of FN in 227 patients. 50.3% were males. Median age was 51 years (16–84). 79% had a haematological neoplasm, 21% a solid neoplasm, and 21% were blood stem-cell transplantation recipients. Most frequent malignat disease was acute leukaemia (36%) followed by lymphoma (24%). 22% had a chronic disease different from the neoplasm. A diagnosis of infection was established in 59% of episodes (N = 173), and in 48.5% of them an aetiological diagnosis was established. More frequent infections were pneumonia (30%), catheter infection (22%), primary bacteraemia (20%) and urinary tract infections (9%). 73 patients (42%) had bacteraemia. Bacteria (82%) were the most common infectious aetiology, followed by mixed infections by fungi and bacteria (14%) and fungal infections alone (4%). Between bacteria, the most frequent were: Staphylococcus coagulase-negative (30%), Escherichia coli (20%), Pseudomonas aeruginosa (13%), Staphylococcus aureus (9%), Enterococcus faecium (3%) and Proteus mirabilis (3%). Between fungi the most frequent were Aspergillus spp (13%) and Pneumocystis jiroveci (3%). 21% of FN presented wtih severe clinical features, among them the most frequent were arterial hypotension (16%) and acute renal failure (8%). FN episodes with a diagnosis of infection had a higher frequence of severe clinical features in comparison with these with unknown aetiology (p = 0.003; RR 2.18; IC 95%: 1.3–5.3). Mortality at 30th day of episode was 17%. Pneumonia (p = 0.04;RR 2.2; IC 95%: 1.1–4.8), aetiological diagnosis of infection (p < 0.01;RR 2.3; IC 95%: 1.2–4.5), and septic shock (p < 0.001; RR 10; IC 95%: 3.5–28.9) were adverse outcome factors identified by multivariate analysis.
Conclusions: Infection is the cause of at least 59% of FN episodes in patients with cancer. The aetiology of infection is established in one half of episodes, with a predominance of bacteria. Pneumonia and catheter infection are the most frequent infection foci. Pneumonia, aetiological diagnosis of infection and septic shock are adverse outcome factors in patients with FN and cancer.
P1598: Bloodstream infections in adult patients with malignant blood disorders and neutropenia: microbial spectrum and antimicrobial susceptibility pattern
S. Levidiotou, P. Bouranta, H. Gesouli, G. Vrioni, C. Pappa, V. Alymara, K.L. Bourantas (Ioannina, GR)
Objectives: Bloodstream infections are a major cause of morbility and mortality among patients with haematological disorder. The aim of the present study was the bacterial spectrum and antimicrobial susceptibility pattern of organisms causing bloodstream infections among patients hospitalized in a hematology centre.
Methods: A total of 90 episodes of bacteraemia and fungaemia in 63 patients were identified. All patients were treated in the haematology ward during a 5-year period (2000–2004) and had an underlying haematological disorder (lymphoma, leukaemia and myeloma). There were 42 male and 21 female patients, aged 17–80 years old.
Results: A total of 101 microbial strains were isolated in 90 episodes. Fifty-one (50.5%) isolates were Gram-negative bacteria, forty-seven (46.5%) Gram-positive bacteria (46.5%) and 3% yeasts. Polymicrobial bacteraemia was observed in 11 from 90 (12.2%) episodes. The dominated pathogens were coagulase-negative Staphylococci (CNS) 24%, Pseudomonas aeruginosa 21%, E. coli 16%, Enterococcus spp. 8% and S. aureus 7%. The mortality rate was 30.15% (19 patients) being higher in patients with bacteraemia caused by P. aeruginosa, CNS and Klebsiella spp. Oxacillin resistance was detected in 43% of S. aureus isolates and in 63% of CNS. Vancomycin resistance was detected only in one strain E. faecium (12.5%). E. coli strains produced ESBL in 2/16 (12.5%). Among P. aeruginosa isolates, one strain was resistant to all tested agents, 14.3% were susceptible only to colistin and 9.5% only to carbapenems and colistin.
Conclusion: The identification of the microbiology profile of bloodstream infections and antimicrobial susceptibility pattern of isolated strains may help in managing these infections in haematology patients, and reviewing infection control and antibiotic policies.
P1599: Incidence of bacteraemia in patients with haematological and other malignancies after placement of central venous catheter
G. Pedersen, M. Nørgaard, H.C. Schønheyder, H. Larsson, M. Kiiveri, E. Stolberg, P.W. Andersen, H.T. Sørensen (Aalborg, Aarhus, DK)
Objective: Patients with cancer, and in particular patients with haematological malignancies are susceptible to infections including bacteraemia. Central venous access devices are used for intravenous chemotherapy in these patients, but data on the risk of bacteraemia are few. We therefore estimated the incidence of first time bacteraemia and type of microorganisms after placement of the first central venous catheter in patients with haematological (haem) and other types of cancer ('other').
Methods: We conducted the study in North Jutland County from 2000 to 2002 based on data from three databases: the Danish Cancer Registry (cancer patients of age >14 years), the Hospital Discharge Registry (central venous catheter and co-morbidity) and the Bacteraemia Research Database (bacteraemia). Three different types of central venous access devices were used: short-term catheters (S) and two long-term catheters: Hickmann (H) and Port-a-cath (P).
Results: A total of 362 patients were identified, 107 in the haem group and 252 in the ‘other’ group. The predominant malignancies were breast cancer (n = 121), non Hodgkin lymphoma (n = 32), ovarian cancer (n = 29), and acute myeloid leukaemia (n = 29). Compared with patients in the haem group, we found a predominance of elderly females in the ‘other’ group. The distribution of first central venous catheters was: S = 70 (haem: 46), H = 53 (all were haem patients) and P = 239 (haem: 8). A total of 83 patients presented with bacteraemia (haem: 55); the distribution in relation to catheters was: S = 33 patients, H = 26 patients and P = 24 patients. The incidence rate of bacteraemia according to catheter type was S = 8.3/person-year, H = 2.0/ person-year, and P = 0.2/person-year. Incidence rate ratio (IRR) for bacteraemia in patients in the haem group compared with the ‘other’ group was 13.4 (95% CI: 7.9–22.7) adjusted for age, sex and comorbidity. The most frequent microorganisms were Staphylococcus aureus (n = 20), polymicrobial (n = 17), Escherichia coli (n = 13) and coagulase-negative staphylococci (n = 9). Since all H. catheters were in the haem group and nearly all P. catheters in the ‘other’ group, we could not rule out the significance of catheter type.
Conclusion: Patients with haematological malignancies have a 13-fold increased incidence of bacteraemia compared with other cancer patients. The significance of catheter type warrants further study.
P1600: Aetiology and clinical outcome of blood-stream infections in patients with haematological malignancies in two Danish university hospitals 2000–2003
J.K. Møller, B. Gahrn-Hansen, B.G. Bruun (Skejby, Odense, Hillerød, DK)
Objectives: To compare etiologies of blood-stream infections according to haematological diagnosis and clinical outcomes of dominating microbiological species/groups at two Danish haematological departments.
Methods: A total of 995 blood-stream infections detected in 622 patients between 2000 and 2003 in the two departments were included. Blood-stream infections caused by coagulase-negative staphylococci, coryneforms and propionibacteria were excluded. Bactec® (OUH) or BacT/ALERT® (AUH) blood culture systems were used for blood culturing. Subcultivation and identification was done using standard microbiological laboratory methods. Haematological diagnoses and clinical outcomes were recorded retrospectively by electronic data analysis.
Results: Distributions of haematological diagnoses at the two departments were approximately the same, except for myelomatosis that comprised 19% in AUH and 11% in OUH. We detected a total of 652 and 412 isolates in 609 and 386 blood-stream infections in 360 and 262 patients at AUH and OUH, respectively. A similar proportion of episodes in both hospitals (6%) were polymicrobial. The following differences between microbiological etiology were found: S. pneumoniae, non-haemolytic streptococci, and Gram-negative enterics occurred more frequently at AUH, while enterococci and Gram-negative non-enterics occurred more frequently at OUH. Bacteremia with S. pneumoniae was found in < 1% of AML patients and in about 25% of myelomatosis patients, while E. coli + K. pneumoniae bacteraemia occurred in about 60% of AML patients and 20% of myelomatosis patients. The mortality rate was highest in P. aeruginosa and fungal infections.
Conclusion: It is important to perform local surveillance of blood culture etiology and clinical outcomes of blood stream infections, as these vary between haematological departments. Differences in mortality for specific microorganisms necessitate continuing considerations regarding optimal antimicrobial and adjuvant therapy.
P1601: Fatal acute liver failure due to varicella zoster virus infection in a child with acute lymphoblastic leukaemia
A.M. Spanaki, E. Markaki, N. Anagnostatou, P. Smirnaki, E. Vasilaki, N. Katzilakis, E. Barbounakis, G. Briassoulis, M. Kalamnti (Heraklion, GR)
Varicella Zoster Virus (VZV) is frequently associated with mild hepatitis and rarely with acute liver failure. Immunologic impairment is a significant predisposing factor.
Objectives: We report a 4 years-old girl with Acute Lymphoblatic Leukemia (ALL) and acute fatal liver necrosis after VZV infection.
Methods/Results: She was the second child of the family, with a negative history of VZV disease or vaccination. ALL had diagnosed six months ago, the child was treated according to the ALL - BFM 2000 protocol and was then in complete remission. On the time of admission, she was three weeks on dexamethasone (10 mgr/m2/day) (re-induction). The child was first admitted in the Hematology/Ongology Department with severe abdominal pain, fever (38°C) and elevated transaminases. Next day, cutaneous lesions appeared on the chest and face, suggesting VZV infection, and antiviral therapy with acyclovir was immediately initiated. Two days later the patient was transferred to the PICU with severe clinical and laboratory deterioration, high fever, tachypnea with positive chest x-ray (right low lobe consolidation with pleural effusion), abdominal distention, liver enlargement, gross haematuria and GI bleeding, vesicular rash on the chest, face and scalp and generalized petechiae and echymoses. The patient had pancytopenia, marked elevated hepatic enzymes and bilirubin, hypoalbuminemia and severe disseminated intravascular coagulopathy. She was continuously transfused with FFP, RBC, PLT and cryoprecipitate. The child was intubated in hours, because of progressive respiratory and neurological deterioration. Renal failure was developed. Assistance was asked from a liver transplant team and the little girl was urgently transferred to a special department for emergency liver transplantation. Unfortunately, two days after transportation, the child developed deep coma and severe hypotension and died. VZV antibody levels were negative, but VZV DNA was detected from liver biopsy and blood specimen, with PCR.
Conclusions: Immunocompromised children are potentially threatened of infections, such as highly contagious chickenpox. The mortality of VZV- induced hepatic failure is very high. Only few patients survived with liver transplantation.
P1602: Usefulness of procalcitonin measurement in the diagnosis of febrile neutropenia
S. Blanco, J. Sancho, C. Prat, B. Xicoy, J.A. Domínguez, C. Ferrà, M. Giménez, J. Grau, E. Martró, M. Batlle, M.D. Sánchez, L. Matas, J. Ribera, V. Ausina (Barcelona, E)
The diagnosis of infection in patients with neutropenia secondary to chemotherapy is difficult because clinical signs are unspecific, being fever the only manifestation of sepsis in most of the cases.
Objective: 1. To study the usefulness of procalcitonin (PCT) measurement for the diagnosis of systemic bacterial infection in neutropenic patients with fever. 2. To establish baseline values of PCT in patients with haematological malignancies at the time of admission as well as at the onset of neutropenia.
Patients and Methods: 43 patients with diagnosis of haematological malignancies who underwent induction chemotherapy or were on conditioning regimens for stem cell transplantation were included. Sera samples on the day of admission, at the beginning of neutropenia, the first day of fever and daily for 6 days were collected. Immunoluminometric assay was used for PCT measurement. Episodes of fever were classified according to the clinical and microbiological diagnosis in three groups: (1) documented bacteraemia; (2) bacterial infection without bacteraemia; and (3) fever of unknown origin, with negative cultures.
Results: PCT mean levels at the time of admission and at the onset of neutropenia were 0.337 and 0.33 ng/ml, respectively. Group 1: fifteen patients presented documented bacteraemia. The isolated microorganisms were Escherichia coli (4 patients), Klebsiella pneumoniae (1), coagulase-negative Staphylococcus (5), Streptococcus mitis (1), Staphylococcus aureus (1), Streptococcus salivarius (1) and Lactobacillus sp (1). PCT mean levels at 24–48 hours from the beginning of fever were 3.681 ng/ml in gram-negative and 1.73 ng/ml in gram-positive infections. Group 2: nine patients were included (urinary tract infection in 4, respiratory tract infection in 4 and catheter-related in one) with mean PCT level of 1.51 ng/ml. Group 3: twenty patients presented fever of unknown origin, related with mucositis in eleven of them. PCT mean level was 0.51 ng/ml. Significative differences were only observed between patients with gram-negative bacteraemia and patients with fever of unknown origin (p < 0.05)
Conclusions: PCT levels remain at normal values in patients with haematological malignancies at the time of admission and at the beginning of neutropenia. Determination of PCT is useful in the diagnosis of neutropenic fever from infectious origin especially when gram-negative bacteraemia. Supported in part by Grants FIS P1/020098 and predoctoral grant from SEIMC.
P1603: Association between the serum chemokine RANTES, platelets and infection in patients with haematological malignancy
M. Ellis, B. al-Ramadi, U. Hedstrom, C. Frampton, H. Alizadeh, J. Kristensen (Al Ain, UAE)
Objectives: To describe changes in the leucocyte trafficking chemokine RANTES in haematological pts undergoing chemotherapy, it's relation to platelets and outcome from infection.
Methods: A prospective observational study of 40 pts with haematological cancer receiving chemotherapy. Venous blood samples were collected at 6am daily throughout treatment. Serum RANTES was determined by ELIZA (R&D systems).
Results: At start of chemotherapy RANTES mean (SD) concentrations were 6561 ± 3290 and fell progressively to a nadir of 1436 ± 2038 pg/ml. Corresponding platelet concentrations fell from 104 (IQR 43–205) to 5.5 (IQR 4–8) × 109/L. The correlation between platelets and RANTES was highly significant (r = 0.82 (IQR 0.72–0.89)), confirming platelets as the major source of RANTES. During episodes of SIRS/sepsis or severe sepsis/shock mean RANTES levels were lower at 3394 and 2939 respectively (p < 0.01). Bacteremic pts also had lower RANTES levels (3022 vs 5111 pg/ml, p < 0.01) and platelet concentrations (18.1 vs 36.3, p = 0.07). In pts who survived the level at recovery was 6433 pg/ml, not significantly different from pre-chemotherapy values. 63 of 67 infection episodes resolved despite persistent low RANTES levels. However 4 of the 40 pts died (aspergillosis, candidosis, pneumonia or infectious colitis) − their RANTES concentrations were extremely low (1629) and irrecoverable. RANTES levels began to recover 4.5 days before platelet levels (p < 0.001) and ‘bursts’ of RANTES were observed during thrombocytopenia, suggesting an additional extra-platelet reservoir for RANTES. Platelet transfusions temporarily elevated the RANTES concentrations. When examined simultaneously within a nested mixed model regression analysis, platelets were the only independent variable among steroids, hematopoietic colony stimulating factor, recombinant human interleukin-11, sepsis status and neutropenia associated with RANTES concentration. The following figure illustrates the relation between RANTES and platelets in 1 pt.
Conclusions: A significant hypo-RANTES immunologic environment is created by chemotherapy, driven by accompanying thrombocytopenia, but does not affect the ability of most patients with infection to recover and questions the role of RANTES in such situations. However patients who die from sepsis uniformly have profoundly low RANTES concentrations − whether RANTES is a surrogate marker or important for adequate host response remains a moot point.
Procalcitonin, other markers of infection
P1604: Evaluation of procalcitonin as a diagnostic marker of bacteraemia
S. Levidiotou, E. Papantoniou, A. Charitou, V. Charitou, H. Sakkas, D. Papamichael (Ioannina, GR)
Objectives: Procalcitonin (PCT) was reported to be a reliable marker for severe bacterial infections. The aim of the present study was to clarify the diagnostic value of PCT in patients with bacteraemia.
Methods: We prospectively determined the PCT level in 41 adult patients with bacteraemia hospitalized for a period of 4 months (September to December 2004). In 25 of those patients the bacteraemia was caused by Gram (+) bacteria: coagulase-negative Staphylococci (CNS) 12, Staphylococcous aureus 2, Enterococcus spp. 4, Pneumoniococcus 3, Clostridium perfingens 1, Corynebacterium acnes 3. In the remaining 16 patients the bacteraemina was caused by Gram (-) bacteria: Klebsiella spp. 6, E. coli 5, Pseudomonas aeruginosa 2, Acinetobacter baumannii 3. Serum PCT was estimated with an assay based on immunochemiluminescence (BRAHMS Diagnostica, Berlin, Germany).
Results: Among patients with bacteraemia caused by Gram (-) bacteria the PCT levels (ng/ml) were 2–10 ng/ml (n = 6) and >10 ng/ml (n = 10). In patients with bacteraemia caused by CNS the PCT levels were <0.5 ng/ml (n = 6) and 0.5–2 ng/ml (n = 6). By S. aureus the PCT levels were >10 ng/ml. By Enterococcus spp., Pneumoniococcus, Clostridium perfingens and Corynebacterium acnes the PCT levels were >2 ng/ml.
Conclusion: Bacteraemia associated with CNS may fail to elevate serum procalcitonin levels while bacteraemia associated with Gram (-) bacteria elevate significantly. More studies are needed to confirm our findings.
P1605: Procalcitonin measurement in post-surgical neonates: a guide to antibiotic management
E. Broadis, J. Tong, C. Davis, C. Lucas, C. Williams (Glasgow, UK)
Measurement of plasma C-reactive protein (CRP) has become an established means of monitoring response to antimicrobial therapy. However raised levels are indicative of inflammatory processes in general and do not specifically signal infection, this could result in a tendency to over-prescribe antibioticsIn adult patients Procalcitonin (PCT) measurement has specifically differentiated infection from other inflammatory processes but to date paediatric data, especially in neonates, is limited.
Aim: to assess the potential for PCT as a guide to infection in post-surgical neonates.
Methods: A prospective study was undertaken in post-surgical, intensive care babies between September 2003 and March 2004. Plasma CRP and PCT measurements were performed in parallel on a daily basis. Blood cultures were performed as clinically indicated.
Results: 60 patients (24–42 weeks gestation) underwent 138 operations/procedures. 726 PCT/CRP assays were performed and 198 blood cultures were taken.
-
•
Normal PCT levels consistently accompanied normal CRP levels
-
•
PCT was not elevated in the absence of other inflammatory markers.
-
•
Of 65 positive blood cultures, PCT was normal in only 3 instances in each of which the CRP remained <30 mg/l, platelet and white cell counts were normal and potential contaminants were cultured.
-
•
70% of PCT levels measured on day 6 post surgery had normalised, in comparison to 30% of CRP levels
Conclusions:
-
•
Culture confirmed bacterial sepsis is reliably accompanied by a raised PCT level in neonatal patients.
-
•
Earlier cessation of antimicrobial therapy based on sequential PCT measurements may be possible.
P1606: Plasma procalcitonin levels and urinary tract infections among elderly patients
M. Belechri, D. Dimou, N. Ioakimides, N. Marougas, C. Kotsovassilis, P. Tsiodra (Athens, GR)
Objectives: To evaluate whether measurements of PCT levels, can help distinguish acute fever due to UTI from fever of other non-microbial inflammatory processes, among elderly patients treated in an internal medicine department.
Methods: We prospectively examined 77 patients with acute fever admitted in our department. Variables recorded included patient demographics and principal diagnosis. PCT, C-reactive protein (CRP), Erythrocyte Sedimentation Rate (ESR) and white blood cell count (WBC) were measured. Blood samples were collected 24–48 hours after the presence of fever. Patients were distinguished according to aetiological diagnosis, based on clinical assessment and laboratory results, to those with fever due to urinary track infection (UTI) (Group A) and those with fever of other inflammatory processes (Group B). Statistical tests applied were Student's t-test and Chi-Square.
Results: Of 77 study patients, 37 (48%) had fever due UTI and 40 (52%) due to other inflammatory processes. 32 (42%) were males, 45 (58%) females and mean age of patients studied was 73 years (±SD 7.6), The mean plasma concentrations of PCT in patients with UTI were 1.16 ng/ml (±SD 1.76) vs 0.41 ng/ml (±SD 0.5) in patients of Group B (p = 0.01). Patients in Group A, had as expected, higher values of ESR, 58 mm/h (±SD 33.4) vs 90 mm/h (±SD 28.9) (p < 0.0001) and CRP compared with patients in Group B, although the last did not reach statistical significance (p = 0.07). The mean values of WBC did not differ between the two groups, while percentage of neutrophil differ at a significant level (p = 0.002).
Conclusions: Among hospitalized elderly patients with fever, levels of PCT seem to be higher among those with UTI compared to patients with fever of other inflammatory processes. PCT can probably be used as an indicator of bacterial infection among older patients with acute fever treated in a department of internal medicine.
P1607: Procalcitonin trend identifies critically ill patients at high risk of mortality
J. Jensen, M. Tvede, L. Heslet, T. Hartvig Jensen, K. Espersen, P. Steffensen (Copenhagen, DK)
Objectives: Serum procalcitonin (PCT) has been shown to be an accurate predictor of sepsis, and elevation of PCT blood levels occurs early in the course of a bacterial infection. We investigated the PCT trend to evaluate this as a predictor of mortality in the intensive care unit (ICU).
Methods: We enrolled all patients admitted to a multidisciplinary Intensive Care Unit in 2002. Daily PCT measurements, were carried out during the study period. Antimicrobial therapy was, among other factors, guided by the PCT course. The clinical end point was all cause mortality during the stay in the intensive care unit and in a 90-day follow up period of time.
Results: PCT measurements evaluated in 472 critically ill patients. Overall mortality of critically ill patients in the one-year study was 19.1%. Odds Ratio for mortality in the ICU for patients with increasing compared to decreasing PCT trend after reaching a level of 1.0 ng/ml was after one day: 2.1 (95% CI 1.2–3.7), after two days: 3.4 (95% CI 1.7–7.0) and after three days: 6.0 (95% CI 2.6–14.1).
Conclusion: PCT trend increase for one to three days is associated with high and increasing risk of mortality in the ICU. PCT trend may be a useful tool in fast stratification of critically ill patients in high and low risk patients.
P1608: Procalcitonin PCT and other routine measured factors of bacterial infections
I. Morañska, J. Niscigorska, A. Boron-Kaczmarska (Szczecin, PL)
Objective: Procalcitonin PCT is one of the recently accepted markers of the systemic inflammatory response to severe bacterial infections, especially to sepsis. The aims of this study were: 1. to analyse level of PCT in patients with confirmed sepsis and other infections with systemic clinical manifestations, 2. to assess possible correlations between PCT and other parameters used in routine diagnosis of infections.
Methodology: We have analysed 28 patients with sepsis and systemic inflammatory response syndrome (SIRS), 20 men and 8 women, aged 17–80 years, mean 47,8. In all patients we have measured concentration of PCT (ng/ml), C-reactive protein CRP (mg/l), fibrinogen (mg/dl) and counts of leukocytes (K/μl) and platelets (K/μl). All measurements were made immediately after admission to hospital, before antibiotic treatment. In the statistical analysis W Shapiro-Wilk test, Pearson's and rang Spearman's correlation coefficients were used. P values below 0,05 determined statistical significance.
Results: 1) The studied population contained 13 patients with confirmed sepsis (3-Staph. aureus, 5-Str. pneumoniae, 1- Neisseria meningitidis, 2-Salmonella typi C, 1- Staph. hominis, 1-Leptospira icterohaemorrhagiae), 2 patients with bacterial meningitis, 2 patients with malaria, 11 patients with signs of SIRS caused by bacterial infections without positive blood cultures. 2) We observed high concentrations of PCT (>10 ng/ ml) in 11 patients with sepsis, 2 patients with malaria, 1 patient with bacterial meningitis and 5 patients with SIRS in course of localized bacterial infection. 3) CRP concentration ranges from 13,8 to 654,32 mg/l, fibrinogen from 115,9 to 1094 mg/dl, WBC from 2,39 to 39,2 K/μl, PLT from 11,8 to 304 K/μl. 4) We did not find any correlations between concentration of PCT and other inflammatory parameters like CRP, WBC, PLT and fibrinogen.
Conclusions: 1) Our study confirmed previous observations that PCT is a very characteristic, early and sensitive parameter of severe systemic bacterial or parasitic infections. 2) PCT is an independent factor of severe systemic infections and did not correlate with other routine measured parameters like CRP, WBC, PLT and fibrinogen. 3) In our opinion measurement of PCT is very important in an early differential diagnosis of sepsis and should be made in clinical daily practice.
P1609: Comparison of cerebrospinal fluid procalcitonin levels with serum C-reactive protein in the diagnosis of meningitis
Z. Ozkurt, S. Erol, M. Ertek, K. Ozden, M.A. Tasyaran (Erzurum, TR)
Objective: Procalcitonin (PCT) is a new marker connected to bacterial infection. The aim of the study was to evaluate clinical usefulness of serum procalcitonin measurements in the differential diagnosis of purulent versus tuberculosis and viral meningitis in adults.
Methods: A prospective study including adult patients who were suspected of having meningitis and who were admitted to an Department of Infectious Diseases. The diagnosis of meningitis was based on clinical findings, Gram staining, culture, and chemical analysis of CSF. The value of cerebrospinal fluid (CSF) PCT for differentiating between acute purulent, tuberculous and viral meningitis was assessed and compared to serum C-reactive protein (CRP) values. Thirty patients with acute purulent meningitis, 30 with tuberculous menengitis, 29 with viral meningitis, and 10 patients with a noninflammatory central nervous system diseases as the controls were included in this study. All CSF samples were taken at patient admission and PCT was detected by immunoluminometric assay (PCT LUMI-TEST, Brahms Diagnostica, Germany).
Results: Mean CSF PCT concentrations were 0.18 ng/mL in purulent menengitis, 0.10 ng/mL in tuberculous menengitis and 0.10 ng/mL in viral meningitis, and 0.03 ng/mL in control. There was no any statistical difference in CSF PCT values between acute purulent, tuberculous and viral meningitis groups (P > 0.05). Mean serum CRP values were 19.1 mg/dL in purulent meningitis, 5.9 mg/dL in tuberculous meningitis, 2.1 mg/dL in viral meningitis and 0.3 mg/dL in the control group (p = 0.000).
Conclusion: Serum CRP values were found useful, but not CSF PCT values, for differential diagnosis of meningitis. Additionally, CRP is a practical and an inexpensive diagnostic test.
P1610: Value of body temperature, white blood cell count and C-reactive protein for the diagnosis of intensive care unit-acquired infections: a prospective study
E. Akinci, A. Erbay, A. çolpan, H. Bodur, M.A. çevik, S.S. Eren (Ankara, TR)
Objectives: Markers of infection like fever, high C-reactive protein (CRP) and leukocytosis may be associated with noninfectious causes like trauma, surgical operations and inflammations. Due to the patients in intensive care units are subjected to multiple inflammatory stimuli, the diagnosis of intensive care unit-acquired infections is difficult. The aim of this prospective study was detection of the value of CRP, white blood cell count (WBC) and body temperature for the diagnosis of intensive care unit-acquired infections.
Patients and Methods: The patients staying for more than 72 hours in intensive care unit and had no infection during admission were included in the study. They were visited daily and data were recorded in to the individual forms. CRP was measured within 72 hours of admission and thereafter every three days. The patients were classified as ‘infected’ if infections were identified and as ‘noninfected’ if no infection was identified during their stay in the intensive care unit. The highest body temperature in the day (Tmax), daily WBC count and CRP values were compared between the two groups statistically. STATA 7.0 (College station TX) programme was used for statistical analysis.
Results: Sixty-seven patients were included in the study. Of these patients, 31 were within the infected group and 36 were within the noninfected group. In univariate analysis, CRP, Tmax and WBC were significantly high for the infected patients (p < 0.05). In multivariate analysis, only CRP was associated with infection significantly and independently (OR:1.037, %95CI:1.019–1.054, p < 0.001). The presence of CRP > 50 mg/L yielded sensitivity as % 75, specificity % 96.3, positive predictive value % 96.7 and negative predictive value % 72.2. After development of infection, the increase in CRP and Tmax was statistically significant (p < 0.05). For 27 (87%) infected patients, increase in CRP level was greater than 25%.
Conclusion: This study concludes that CRP is the most useful parameter for the diagnosis of intensive care unit-acquired infections. According to this result, routine measurement of CRP level in intensive care units and follow up the changes in the serum concentration of CRP are recommended.
P1611: Erythrocyte sedimentation rate and C-reactive protein are useful preoperative predictors of the absence of prosthetic hip and knee infection
A. Trampuz, K.E. Piper, D.R. Hanssen, D.R. Osmon, J.M. Steckelberg, R. Patel (Rochester, USA)
Objective: Leukocyte count, erythrocyte sedimentation rate (ESR) and C-reactive protein (CRP) are often obtained on patients with prosthetic joints prior to arthroplasty surgery. We evaluated the value of these parameters for the diagnosis of prosthetic joint infection (PJI).
Methods: We prospectively analysed preoperative leukocyte count, ESR and CRP level in patients prior to revision or resection knee or hip arthroplasty at Mayo Clinic, Rochester, MN between January 1998 and November 2004. PJI was defined as growth of the same microorganism from at least two specimens, synovial fluid or periprosthetic purulence, acute inflammation on histopathologic evaluation, or the presence of a sinus tract. Sensitivity, specificity, positive predictive values (PPV) and negative predictive values (NPV) were calculated using 2 × 2 contingency tables.
Results: 231 preoperative blood specimens from 227 patients prior to total knee (n = 192) or hip (n = 37) replacement were analysed. The median age was 70 years (range, 41–101 years); 55% were male. The primary diagnosis was osteoarthritis (n = 209), inflammatory joint disease (n = 13), trauma (n = 8) and osteonecrosis (n = 1). Causative organisms were coagulase-negative staphylococci (n = 26), Staphylococcus aureus (n = 8), enterococci (n = 3), Propionibacterium acnes (n = 3), corynebacteria (n = 3), viridans group streptococci (n = 2), mycobacteria (n = 2) and gram-negative bacilli (n = 2). Three infections were polymicrobial and 12 were culture-negative. 39 patients (61%) had received antimicrobial treatment in the month prior to surgery. Table 1 shows the median (range) leukocyte count, ESR and CRP in those with aseptic failure versus PJI. Sensitivity, specificity, PPV and NPV for PJI (with 95% confidence interval) are summarized in Table 2.
Conclusion: Leukocytes did not reliably predict PJI. The combination of normal ESR (30 mm/h or less) and CRP (1.0 mg/dl or less) predicted the absence of PJI in 92% of patients in our study.
| Parameter | Aseptic failure (n = 167) | PJI(n = 64) |
|---|---|---|
| Leukocytes, × 109/1 | 6.7 (3.1–14.7) | 7.8 (4-14.7) |
| ESR, mm/h | 10 (1-75) | 43 (3-123) |
| CRP level, mg/dl | 0.3 (0-2.7) | 3.1 (0.3–20.2) |
| Cutoff Values | Sensitivity | Specificity | PPV | NPV |
|---|---|---|---|---|
| Leukocytes >10 × 109/1 | 17 (8-27) | 93 (89-97) | 48 (27-68) | 75 (69-81) |
| ESR > 30 mm/h | 66 (53-78) | 90 (85-95) | 72 (60-84) | 87 (81-92) |
| CRP > 1.0 mg/dl | 77 (65-88) | 84 (77-90) | 66 (54-77) | 90 (85-95) |
| ESR > 30 mm/h or CRP > 1.0 mg/dl | 84 (74-94) | 80 (73-87) | 64 (53-75) | 92 (87-97) |
P1612: Serum neopterin levels in patients with active pulmonary tuberculosis, in patients with brucellosis and healthy individuals
S. Cesur, S. Aksaray, H. Irmak, G. Tarhan, T. Aslan, I. Ceyhan, E. Guvener (Ankara, TR)
Objectives: Neopterin is a sensitive indicator for activation of cell-mediated immune reactions and thus, determination of neopterin concentrations in various body fluids is of diagnostic marker in a variety of infectious diseases in which T lymphocytes and macrophages are involved. Cell mediated immune reactions play important roles in intracellular bacteria such as Mycobacterium tuberculosis and Brucella species. The aim of this study was to compare the serum neopterin levels in patients with active pulmonary tuberculosis, in patients with brucellosis and healthy subjects.
Methods: Thirty-nine patients with active pulmonary tuberculosis (10 females, 29 males, mean age: 40.6) 16 patients with brucellosis (7 females, 9 males, mean age: 35.8) and 39 healthy subjects (16 females, 23 males, mean age: 41.2) were included in this study. The patients who had another infection, malignity, autoimmune disease, surgical intervention and recently blood transfusion were not included the study. Serum samples were collected from the patients and control groups and stored at)20°C until analysed. Neopterin concentrations were measured by ELISA according to the protocol of manufacturer by IBL (Hamburg, Germany).
Results: Mean neopterin levels were 18.56 ± 14.23 nmol/L in patients with active pulmonary tuberculosis, 33.82 ± 22.27 nmol/L in patients with brucellosis and 9.87 ± 2.90 nmol/L in healthy subjects, respectively. Serum neopterin levels were found to be significantly higher in patients with active tuberculosis and in patients with brucellosis compared with healthy controls (p < 0.001). Besides, there were differences between patients with active pulmonary tuberculosis and patients with brucellosis (p < 0.001).
Conclusion: This study showed that neopterin levels increased both in patients with active pulmonary tuberculosis and in patients with brucellosis. Neopterin can be used in follow up the patients who had chronic infectious diseases such as brucellosis and tuberculosis.
Diagnosis of viral infections: new methods
P1613: Automated extraction of RNA and DNA from different specimen types feeding the detection of several markers in the field of molecular diagnostics
S. Thamm, K. Beuselinck, T. Bourlet, S. Kares, R. Karhu, H. Mairhofer, H. Nitschko, B. Pozzetto, J. van Eldere, G. Michel (Wiesbaden, D; Leuven, B; St. Etienne, F; Tampere, FIN; Munich, D)
Objective: A recently introduced automated generic RNA and DNA extraction system, m1000, enables a throughput of 48 samples in less than 2 hours. Several studies have shown that m1000 is a suitable automated extraction system for many different sample types feeding molecular diagnostics assays.
Methods: Unlinked surplus specimens from the clinical routine were used. Cross contamination studies were designed with high titer samples interspersed between negative samples giving a checkerboard pattern on the m1000 subsystem. Diagnostic assays used during these studies were either commercially available or homebrew assays validated for the laboratory routine.
Results: No cross-contamination was observed in using HIV RNA of 5–7 log copies/ml, HCV RNA of 5–7 log IU/ml, and polyomavirus DNA of 8 log copies/ml. A total of 437 serum or plasma samples were processed for RNA extraction using LCx HIV or HCV RNA Quantitative assays with 97.6–99.0% agreement to Cobas Amplicor Monitor HIV/HCV assays. Coextraction of HIV/HCV RNA from 28 serum or plasma samples revealed no impairment of LCx assay results when compared to monoextraction. Reaching a 100% agreement between manual and automated extraction on 53 initially positive urine samples on Cobas Amplicor Ct showed the feasibility of extracting bacterial DNA of C. trachomatis with m1000. Genomic DNA extraction from 31 whole blood samples for homebrew HLA-B27 typing revealed a 100% agreement to the manual extraction. Automated extraction of viral DNA of HBV from 25 serum or plasma samples revealed a correlation coefficient of r = 0.96 compared to the manual extraction. 2–7 replicates of HBV quantitation standards were tested with homebrew real-time PCR after automated extraction with excellent linearity and a 100% detection rate of all concentration levels down to 100 IU(ml. Viral DNA has been successfully extracted from spinal fluid, urine, cell culture supernatant, BAL, nasopharyngeal washing, breast milk, biopsy, leukocytes, and bone marrow samples for CMV, HSV, or polyomaviruses JC and BK testing with homebrew real-time PCR (n = 48).
Conclusion: Taken together, these studies have shown that m1000 provides a versatile solution for automated extraction of viral RNA or DNA, and bacterial or genomic DNA from 12 different specimen types feeding 8 different molecular diagnostics assays in the field of hepatitis B/C, HIV, sexually transmitted diseases, transplantation, and HLA typing.
P1614: Evaluation of automated extraction of RNA and DNA for high throughput ‘in-house’ quantitative real-time PCR assays
K. Beuselinck, J. Van Eldere (Leuven, B)
Objective: Nucleic acid (NA) extraction has become the most critical and labour intensive step in NA based diagnostics. The performance of NA based diagnostics is primarily dependent on the NA extraction yield, purity and the amount of sample that can be extracted. Recently, Abbott introduced the m1000, a fully automated generic RNA and DNA extraction system that uses magnetic micro-particle processing allowing extraction of up to 48 samples in 2 hours.
Methods: In this study, the performance of the m1000 as a front-end extraction system for high throughput ‘in-house’ quantitative real-time PCR assays (HCV, HBV, CMV, EBV) was analysed and compared to manual extraction for plasma and serum samples (HCV and HBV) and EDTA-blood samples (CMV and EBV).
Results: Linearity of extraction was tested on dilution series of HCV and HBV reference material. The correlation coefficient for standard curves based on repeated extraction runs of these dilutions was 0.970 + 0.060 for HCV and 0.971 + 0.025 for HBV, indicating a linear extraction between 100 − 1.0 × 105 HCV IU/ml and 100 − 1.0 × 106 HBV IU/ml. Intra- and inter-run variability was not significantly different and below 0.2 log10 IU/ml for 2.98 − 5.28 log10 HCV IU/ml and 2.70 − 5.20 log10 HBV IU/ ml. Correlation between automated and manual extraction was very good. For HCV, the correlation coefficient was 0.91 and the mean difference in viral load was 0.128 log10 IU/ml (SD: 0.44 log10 IU/ml). For HBV the correlation coefficient was 0.98 and the mean difference in viral load 0.612 log10 HBV IU/ml (SD: 0.52 log10 IU/ml). Cross-contamination was tested with strongly positive polyomavirus samples (8.10 log10 copies/ml) and polyomavirus-negative samples. No cross-contamination was observed. The turn-around time for 48 samples was 2.5 h and hands-on time was 0.5 h compared to 4.4 h for manual extraction. The list-price reagent cost was around 11.6 euro per sample compared to 8.0 euro for manual extraction (HCV and HBV).
Conclusion: Automated extraction as offered by this system offers high reliability of extraction at a total cost that is inferior to manual extraction.
P1616: Performance of a new amplification based assay for quantification of human immunodeficiency virus-1 (HIV-1) RNA
C. Wagner, C. Lin, S. Bhade, R. Torres, S. Nicol, J. Surtihadi, G. Gorrin (Berkeley, USA)
Objectives: A new quantitative HIV-1 RNA kinetic amplification assay is under development. Analytical performance characteristics of this assay were evaluated, including: sensitivity, specificity, linearity, and the ability to detect group M and O subtypes. Data from these evaluations are presented.
Methods: Viral RNA was extracted from panels and donor samples using an automated silica-bead based method, after which reverse transcription, amplification, and detection steps were performed. The assay includes an internal control (IC) and chemical contamination control using recombinant cod UNG (rcUNG). Linear range, precision, and sensitivity were evaluated by testing serial dilution panels prepared from HIV-18E5/LAV Clade B virus (quantified by VERSANT HIV-1 RNA 3.0 (bDNA)) spiked into pooled normal human plasma. Panels ranged in concentration from 10 to 107 copies/mL. Assay specificity was evaluated by testing plasma from 360 HIV-1 antibody-negative donors. The ability of the assay to detect all HIV-1 group M (AH) and O subtypes was evaluated with both transcripts quantified by phosphate analysis and OD260, and panels prepared from electron microscopy quantified HIV-1 group M (A to H) and O cultured isolates (Boston Biomedica, Inc.). Multiple subtype A, B, C, and O isolates were tested in the evaluation.
Results: The linear range of the assay was determined to be from 24 to 13,000,000 copies/mL, with 100% specificity. HIV-1 transcripts and virus panels representing all Groups M and O subtypes were detected at “d 50 copies/mL. Total %CV of quantitation was “T 25% above 1000 copies/mL, and “T 50% between 50 and 1000 copies/mL. The IC worked across the entire range of the assay.
Conclusion: The assay was demonstrated to detect RNA of all group M (A-H) and O subtypes down to 50 copies/mL. Sensitivity (24 copies/mL) and specificity (100%) of the new automated HIV-1 viral load assay under development were both excellent. The assay internal control and chemical contamination control methods were effective.
P1617: Evaluation of a new Dot Blot assay for confirmation of human immunodeficiency virus type 1 (HIV-1) and 2 (HIV-2) infections using recombinant p24, gp41, gp120 and gp36 antigens
M. Ravanshad, F. Sabahi, F. Mahboudi, M. Roostaee, A. Kazemnejad (Tehran, IR)
A Dot Blot assay using four recombinant proteins corresponding to human immunodeficiency virus type 1 (HIV-1) and type 2 (HIV-2) gene products was developed to confirm the presence of antibodies to HIV-1 and 2 in sera reactive in screening ELISAs. Serum samples for testing were obtained from healthy seronegative blood donors and from different categories of HIV-infected individuals (asymptomatic HIV-infected, and AIDS). A positive reaction was defined as reactivity against gag (p24) and at least one other env (either gp41 or gp120) HIV gene products; negative result was defined as no reaction with any antigen; and indeterminate result was defined as reactivity with gag (p24) or with env (gp41 or gp120) alone. None of the 180 serum samples from healthy seronegative blood donors gave a positive result, and only 4 of these samples (2.2%) gave indeterminate results. The recombinant HIV Dot blotting assay identified seropositive individuals with a high degree of accuracy; none of the 125 HIV-seropositive subjects had a negative test result. Reactivity with these antigens, demonstrated 100% sensitivity and specificity in distinguishing seronegative from seropositive sera. All seronegative and seropositive samples were tested with the commercially available Western blot. The developed in-house HIV Dot blot assay accurately identified more seropositive and seronegative samples and had fewer indeterminate results than did commercial Western blot (as interpreted by CDC criteria).
P1618: Hepatitis B virus DNA testing by nested-PCR, PCR-ELISA and hybrid capture II from a blood and blood products
J.-H. Lim, H.-S. Jung, S.-E. Choi, S.-N. Kim, C.-M. Hong, S.-N. Park, H.-K. Min, S.-H. Hong (Seoul, KOR)
The purpose of this study is to establish Nested-PCR for the detection of hepatitis B virus (HBV) in blood and blood products. The primer pair set was designed to amplify 513 bp in S-region of HBV genome in the first PCR and 233 bp of first PCR amplicon with Rubisco (internal control) in the second PCR. To assess the specificity of PCR results, all the samples were tested cross-reactivity or interference in the assay. In case of HBV spiked blood products such as immunogloubulin and coagulation factors, this method could detect HBV DNA up to 62.5 IU/ml. Nested-PCR was compared with PCR-ELISA and hybrid capture II (HC-II), the PCR-ELISA showed a sensitivity of 96% (HC-II; 77%) and a specificity of 98% (HC-II; 100%) (p < 0.005). The results of the study show that Nested-PCR and PCR-ELISA could be used equally in the management for HBV detection in blood and blood products.
P1619: An in vitro assay for flavivirus NS2B/NS3 serine protease
M. Bessaud, C.N. Peyrefitte, B. Pastorino, M. Grandadam, H.J. Tolou (Marseille, F)
Objectives: The genus Flavivirus comprises more than 70 viruses. Many of them cause severe human diseases. The genome of flavivirus, a single strand RNA molecule with positive polarity, is translated into a large polyprotein that is processed into structural and non-structural proteins by both viral and cellular proteases. The virus-encoded protease is a binary complex constituted by the NS3 protein and its co-factor, NS2B. As the viral protease plays a critical role in the virus replication cycle, it represents one of the main targets for antiviral therapy against members of the Flavivirus genus. The aim of this study was to develop an in vitro assay using the protease of several flaviviruses belonging to different groups in order to characterize their enzymatic properties, such as temperature, pH and salt-sensitivity and substrate specificity.
Methods: Sequences encoding the viral proteinase were located on the genome of 8 flaviviruses. NS2B/NS3 proteinase were expressed as hexahistidine-tagged recombinant proteins and then purified by immobilized-metal affinity chromatography. Their enzymatic properties were characterized in vitro using BAPNA, a chromogenic substrate for trypsin-like proteases.
Results: The protease moiety of the 8 flaviviruses were successfully produced and purified. 5 of them exhibited activity towards BAPNA. Effect of temperature, ionic strength and pH on enzymatic activity were determined. Our results suggest that the hydrophilic domain of NS2B is necessary for proteolytic activity.
Conclusion: The system we developed will allow us to establish a screening test so as to identify or to design inhibitors active as antiviral drugs against one or more pathogenic flaviviruses. The lack of activity of 3 of the 8 proteases we assayed could indicate slight differences in flaviviral proteases’ selectivity and activity.
P1620: Diagnosis of dengue infection by enzyme-linked immunosorbent assay and reverse transcription-nested polymerase chain reaction from dried urine spots on filter paper
J. Pupaibool, N. Jaimchariyatam, S. Krajiw, K. Arunyingmongkol, P. Suandork, O. Prommalikit, R. Sittichai, A. Nisalak, C. Pancharoen, U. Thisyakorn, W. Kulwichit (Bangkok, TH)
Objectives: Polymerase chain reaction (PCR) and enzyme-linked immunosorbent assay (ELISA) from serum are used for diagnosis of dengue infection. However, in certain areas of the world, these are not readily available. Storage of specimens on filter paper to be tested later at a centre is an attractive way to confirm a clinical diagnosis or for an epidemiologic study. We performed a pilot study to see if urine spots on filter paper can be used for dengue diagnosis.
Methods: Adults and children admitted to King Chulalongkorn Memorial Hospital suspected of dengue infection were enrolled. Final diagnosis of dengue infection was based on the standard serum ELISA assay. Patients with negative serology served as controls. Urine specimens were collected twice, 5 days to 3 weeks apart, spotted on filter paper and stored at 4 degrees for a minimum of 10 days before testing. RT-nested PCR and ELISA were performed on extracts from the spots. Our ELISA criteria for urine were single IgM > 10 units for primary dengue infection or single IgG > 10 units or 2-fold increase in IgG titer with the second titer >10 units for secondary dengue infection.
Results: 139 patients were enrolled. 64 and 71 specimens were analysed by RT-PCR and ELISA, respectively. The results are summarised in the Table:
| Patients |
||||||
|---|---|---|---|---|---|---|
| Method | Adult | Children | Sensitivity | Specificity | PPV | NPV |
| RT-PCR | 36 | 28 | 21/51 cases 41.18% | 11/13 cases 84.62% | 21/23 cases 91.30% | 11/41 cases 26.83% |
| ELISA | 25 | 46 | 49/60 cases 81.67% | 10/11 cases 90.91% | 49/50 cases 98.00% | 10/21 cases 47.62% |
If 3 primary dengue cases were excluded, the sensitivity of ELISA for secondary cases were 84.21%.
Conclusions: This is the first study using dried urine samples on filter paper for dengue diagnosis. In this study, fresh urine was spotted onto filter paper in the laboratory, and cross contamination was possible. Realistically, this process would be done on a near-patient basis with vastly lower likelihood of such contamination and thus better results of RT-PCR. Collection of urine is less invasive than that of blood and is especially suitable for children. This method would be highly applicable in epidemiologic studies, such as during dengue outbreaks. Due to a low number of primary cases in an endemic area such as Thailand, a study elsewhere is needed to assess the utility of urine spots in such cases.
P1621: A rapid sequencing based prototype assay for detection of high risk HPV strains in cervical samples
N. Marlowe, R. Bruce, M. Owens, T. White, M. Zoccoli (Alameda, USA)
Background: Human papillomavirus (HPV) is one of the most common causes of sexually transmitted diseases resulting in an estimated 288,000 deaths yearly from cervical cancer worldwide. Of more than 30 types found in the anogenital tract, at least 13 are considered high risk (HR), as they are significantly associated with progression to invasive cervical cancer. In an effort to accommodate high throughput screening for HR HPV types, we recently developed a prototype HPV assay using a Single Base Dye Primer Profiling (SBDPP) approach. In the present study, we evaluate prototype assay performance on 74 patient samples.
Objective: To evaluate SBDPP assay for the detection of HR HPV strains in cervical samples.
Methods: Genomic DNA purified from 74 retrospective cervical samples was used for this feasibility study. Fifty-four out of 74 were previously typed successfully with the Digene Hybrid Capture® 2 (HC2) assay. The remaining 20 samples yielded inconclusive results with the HC2 assay. Using consensus PCR primer sequences conserved among HPV types, a fragment of the L1 region was amplified in the presence of an intercalating dye. HPV-positive samples were identified by amplification Ct values and dissociation curve melting profiles, sequenced with a rapid SBDPP protocol and analysed on the ABI PRISM® 3100 Genetic Analyzer. HR HPV types were identified by their unique single base distribution profiles.
Results: The presence of HR HPV types was determined in 28/ 74 clinical samples using the SBDPP assay. HPV types were confirmed by NCBI BLAST (v2.2.9) analysis using four-color sequencing data. Twenty-four of the 54 HC2-typed samples were typed as HR HPV, while 30 were HPV-negative/low risk HPV. Four of the twenty previously untyped samples were genotyped as HR HPV. Type determinations for 24 HR HPV samples were concordant between the SBDPP and HC2. Of the 28 cervical samples identified as HR types, 5 samples were determined to be HPV mixed infections.
Conclusion: This feasibility study demonstrates the potential of the SBDPP assay to type patients harbouring HR HPV strains. Genotyping results obtained with the prototype assay are highly comparable to the four-color DNA sequencing data. These findings confirm that a rapid SBDPP method can be used for high-throughput screening of cervical samples for the presence of HR HPV types.
P1622: DNA microarray format for detection of human papillomavirus: discrepancies between techniques applied to detect HPV DNA
M. Dominguez-Gil, R. Ortiz de Lajarazu Leonardo, J. Eiros, A. Curiel, M. Moreno, S. Jimenez, L. Sobrino, I. Gracia, A. Tenorio, C. Labayru, M. Ortega, B. Hernandez, A. Rodriguez Torres (Valladolid, E)
Introduction: The large variety of techniques available for detection and subtyping of the multitude of HPV types illustrates the fact that no single technique provides the ultimate and complete solution to this effect.
Objective: Explain the diagnostic correlation between the techniques applied to detect HPV DNA and typing in cervical samples from women suspected to have ginecological lession selected to HPV.
Methods: A total of 40 genital smears with HPV evidences were selected by a solution hybridisation assay (Hybrid Capture, Digene, Gaithersburg, USA). The applied hybridisation technique classifies the virus on those associated to a low or moderated-high risk of cell transformation. We analysed the agreement between these results and those obtained by PCR and subsequent typing by means of digestion with restriction enzymes (HPVfast, Genomica S.A.U., Madrid, Spain). We used a Papillomavirus Clinical Arrays (Genomica S.A.U., Madrid, Spain).
Results: The results obtained by means of hybridisation were distributed as follows: 47.5% of the smears revealed HPV of moderated-high risk of cell transformation, 12.5% was associated to a HPV of low risk of transformation and the remaining 40% were positive on both groups of HPV. PCR revealed the presence of HPV DNA in 40% of analysed smears (N = 40). The 40% of moderated-high risk HPV was not detected by means of PCR. The 15% was not detected by means of DNA Microarray Format (N = 20). The 100% (N = 4) among low-risk HPV and the 75% (N = 16) of those positive for both HPV groups were not detected by means of PCR. The 100% of low-risk HPV and those positive for both HPV groups was detected by means of DNA Microarray Format.
Conclusions: The detection of HPV DNA by hybridisation was associated in a greater percentage to positive results than it actually applied to the PCR Technique used. However hybridisation and DNA Microarray Format were in more agreement. The PCR identification technique was not able to detect a signification number of HPV positive samples, mainly on the HPV H-m risk and less for the others. Then there is a better correlation between DNA Microarray Format and DNA hybridisation for detection and subtyping of human papillomavirus and more discrepancies among PCR and hybridation
P1623: Evaluation of human papillomavirus PCR detection in urine and cervical swabs of patients attending a colposcopy unit: a viable screening alternative?
A. Daponte, S. Pournaras, I. Mademtzis, I. Kristo, A.N. Maniatis, I. Messinis (Larissa, GR)
Objectives: To evaluate the performance of Human Papillomavirus (HPV) PCR detection in urine compared with cervical swabs, among patients attending a colposcopy unit. The appropriate detection of HPV in urine would serve as a viable alternative for screening and prevention of cervical cancer.
Methods: Paired urine and cervical specimens were collected from 60 consecutive patients referred to a colposcopy clinic. Urinalysis and urine microscopy was performed for evaluation of potential PCR inhibitors and specimen epithelial cell content. In-house PCR and a commercial PCR for the detection of HPV 16 and 18, as well as a commercial multiplex PCR for HPV 6, 16, 18, 31 and 33 were performed in all samples. To test the sensitivity of HPV detection in urine, urine samples were spiked with cell lines infected with HPV.
Results: The in-house PCR exhibited the highest detection rate. Cervical samples were tested positive for the oncogenic HPV types 16 or 18 in 21 of the 47 (44.7%) cases of women that proved to have pre-malignant or malignant lesion by histology. 14 of the 47 (29.8%) urine samples, with histologically proven pre-malignant or malignant lesions were positive for HPV16 or 18. In all cases the respective cervical samples were positive for the same HPV type. The HPV detection concordance was 14/21 (66.7%). The HPV detection concordance was better in histological diagnoses of cancer and high-grade lesions compared to low-grade lesions. The sensitivity of HPV detection in urine was similar to that in distilled water.
Conclusions: This study is one of the few that examined the presence of HPV in urine samples from patients referred to a colposcopy unit. High HPV type-specific concordance between cervical and urine samples and good sensitivity of urine detection are reported. Only specimens with two or more epithelial cells in 400 × magnification are worth processing with the more expensive HPV detection method. This report supports the possibility that self-testing for HPV, using urine, will one day give to low-income groups and specific populations like young adolescents a good screening alternative.
Diagnosis of viral infections: respiratory and herpes viruses
P1624: Significance of rapid detection methods versus serological tests in human respiratory viruses isolation
R. Ortiz de Lejarazu Leonardo, L. Sobrino, A. Tenorio, I. Gracia, M. Domínguez-Gil (Valladolid, E)
Objectives: Descriptive, retrospective and observational study to research the incidence of respiratory viruses in our environment and to evaluate the most useful samples and detection methods.
Methods: The period of the study was 2000 January to 2004 May. We received 2763 diverse samples registering patient data (sex, age, source); 1694 serum for complement fixation test (CFT) and 1069 diverse samples for rapid detection methods (RDM): shell vial culture, direct immunofluorescence (DIF), enzyme-linked fluorescent assay (ELFA) and immunochromatographic test (ICT).
Results: We obtained 386 positive results, 227 in men and 159 in women. Classification by ages: 0–1 year old, 197 positive cases; 1–2 years old, 48; 2–14 years old, 48; 14–30 years old, 5; 30–65 years old, 16; more than 65 years old, 56. Isolated viruses: influenza A virus (Flu A), 96; influenza B virus (Flu B), 24; respiratory syncytial virus (RSV), 226; parainfluenza virus (PIV), 25; adenovirus (ADV), 14. Monthly evolution of samples received and positive cases shows that patterns of serum and the other samples are different. The 1st graphic represents the monthly sending of serum that does not follow a seasonal curve on the contrary than the other samples represented in the 2nd graphic; it occurs the same when we look at the positive results distribution. Samples: Serum was positive in 73 cases; nasopharyngeal wash in 276; pharyngeal smear in 33; Bronchoalveolar wash and tracheal aspirate, 4. Detection methods: CFT was positive in 73 cases; Shell vial culture was positive in 276, but in 132 was the unique method that achieve an isolation; ELFA in 134; DIF in 86; ICT was positive in 11 cases, but was only used since 2004 January. Rapid methods combined accurate 313 positive cases.
Conclusions: Children are the most affected by human respiratory viruses, according with the bibliography. RSV and Flu A were the predominant viruses detected. The maximal monthly positively results for respiratory viruses reached in cold season but PIV do not followed a seasonal curve. Rapid detection methods are more sensitive than CFT. Based on these results, we recommend to take nasopharyngeal washes to combine rapid detection methods and accurate the best rate of virus isolation.
P1625: Evaluation of a real-time NASBA assay for the detection of influenza A and B viruses in clinical respiratory samples
J.N. Telles, G. Billaud, A. Lefeuvre, M. Perret, D. Thevenot, F. Komurian-Pradel, G. Vernet, A. Troesch, D. Floret, B. Lina, G. Paranhos-Baccalà (Marcy l'Etoile, Lyon, F)
Objectives: bioMérieux is developing a real-time NASBA assay to detect Influenza A and B RNA in different kind of respiratory clinical samples. The Nuclisens EasyQ Influenza A+B assay, in combination with Nuclisens MiniMag sample preparation system, was evaluated on 89 nasal/throat swabs from hospitalized children during 2003/2004 influenza season at the Paediatric Emergency Department of Edouard Herriot hospital (HEH-Lyon, France). The assay is designed to detect in a single tube, a wide range of both Influenza A and B viruses by targeting conserved regions of the genome. The aim of this study was to evaluate the real-time NASBA sensitivity against classical cell culture method.
Methods: 89 Nasal/throat swabs in transport medium from children 0 to 16 years age were used for the evaluation of Influenza viruses presence. RNA extraction was performed using the bioMérieux Nuclisens MiniMag instrument and reagents. Extracted RNA were submitted to NASBA amplification and detection using the Nuclisens EasyQ Influenza A+B assay. The obtained data were analysed by Nuclisens Director 2.0 software. The results were then compared to the cell culture results already performed on the samples.
Results: To be confident in the sensitivity of real-time NASBA assay, we showed that no interference was observed with Influenza A and B strains spiked in different negative swabs. Among 89 swabs tested in this study, real-time NASBA detected 10 (11.2%) samples for Influenza A and 2 (2.2%) samples for Influenza B. Comparatively, by cell culture method only 5 (5.6%) samples were identified as Influenza A and none as Influenza B. Interestingly, 1 Influenza A positive sample identified by cell culture was found negative in real-time NASBA.
Conclusion: In this study, we showed that Nuclisens EasyQ Influenza A+B assay detected 50% more Influenza A virus than cell culture method. Moreover, real-time NASBA detected 2 Influenza B positive samples which were not detected by cell culture. In conclusion, our Nuclisens EasyQ Influenza A+B assay demonstrated clinical relevance in samples from hospitalized children suffering from respiratory diseases.
P1626: Evaluation of a protein microarray assay for detecting antibodies to SARS-coronavirus
B.M. Willey, N. Kreiswirth, H. Zhu, G. Jona, X. Zhu, M. Snyder, A. McGeer, Y. Li, M. Cameron, W. Gold, A. Tyler, T. Mazzulli (Toronto, CAN; New Haven, USA; Winnipeg, CAN)
Objectives: Several diagnostic tests for the detection of SARS antibodies from serum have been previously evaluated, including ELISA and IIFT assays. This study evaluated a novel protein chip assay, the Coronavirus Protein Microarray (CoV-PM), that has been developed for the detection of antibodies to multiple coronaviruses, including SARS-CoV, comparing it to the currently accepted commercially available gold standard assay, the EUROIMMUN SARS-CoV IgG Indirect Immunofluorescence test (IIFT).
Methods: The CoV-PM comprised of all the SARS-CoV proteins and protein fragments, along with proteins from 5 additional coronaviruses that infect human, cats, mice and cows. The arrays were used to probe a selection of 399 previously validated, challenge samples of human sera for antibodies to SARS-CoV that included samples from the same patients from different dates. These sera included 40 SARS-CoV IgG-negatives (neg) from acute SARS cases (ACUT), 164 SARS-CoV IgG positive (pos) from convalescent cases (CONV), 112 SARS-CoV-neg from patients with other respiratory illness (RESP), and 84 from healthcare workers (HCW; 17 SARS-CoV IgG pos; 66 neg). Each grid on the CoV-PM was probed with 1 μL serum/patient, reacted with fluorescently labeled antibodies and signals were detected using a standard microarray scanner. Scans were analysed by two readers using a programme developed to predict the nature of each serum.
Results: Of the 181 SARS-CoV-pos sera, 168 were read as positive by CoV-PM by both readers (93.3% sensitivity), while 8 CONV sera were read as CoV-PM pos by one reader but not by the other, whereas 16 and 7 were called neg and inconclusive by both readers. Interestingly, in the majority of cases, SARS-CoV pos sera that tested discrepant by CoV-PM were CoV-PM pos in other samples from the same patients from later dates. 64 HCW, 40 ACU and 100 RESP from 218 SARS-CoV-neg sera were CoV-PM-neg (specificity 99.1%), with SARS-CoV antibodies found in 1 HCW and 1 RESP while 2 others were inconclusive. Some of the SARS-CoV pos sera also reacted with 229E proteins, suggesting concurrent or prior infection with this coronavirus; some sera within this selection tested positive for 229E/OC43 by Western blot assay.
Conclusions: The CoV-PM appears to be a sensitive and specific method for the detection of SARS-CoV antibodies from serum. The added value of its ability to detect antibodies to other coronaviruses requires further evaluation.
P1627: Comparison of Roche MagNA Pure Total Nucleic Acid Isolation Kit and LC SARS-CoV Quantification assay against Qiagen RNeasy Kit and artus RealArt HPA-Coronavirus LC RT PCR assay for detecting SARS-CoV RNA in autopsy tissue
B.M. Willey, P. Akhavan, N. Kreiswirth, G. Farcas, K. Kain, S. Poutanen, T. Mazzulli (Toronto, CAN)
Objectives: The occurrence of new cases of SARS-associated Coronavirus (SARS-CoV) infection in China after the outbreak was declared over emphasized the continued need for rapid and reliable molecular tests for the detection of SARS-CoV. This study compared the ability of the Roche MagNA Pure Total Nucleic Acid Isolation Kit and LC SARS-CoV Quantification Assay (MP-RSA) to the Qiagen RNeasy Kit and artus RealArt HPA-Coronavirus LC RT PCR Assay (QIA-ACA) for the detection of SARS-CoV in autopsy tissues.
Methods: SARS-CoV RT-PCR amplification using the Roche LightCycler 1.2 (LC) instrument was performed on 296 homogenized tissue samples (79 SARS-CoV positives - all from known SARS cases that met the CDC/WHO case definition, and 217 SARS-CoV-negatives; 162 lung, 22 spleen, 20 liver, 21 kidney, 19 large bowel,19 small bowel, 15 lymph node, 18 skeletal muscle). Using aliquots from the same tissue homogenates, RNA extractions and SARS-CoV detections were performed using QIA-ACA and MP-RSA methods. MP extracts that gave MP-RSA-negative results from QIA-ACA-positive samples were rE-tested by ACA to determine if the MP or the RSA had failed: ACA-positives would indicate a false-negative RSA result, whereas ACA-negative would indicate a MP extraction failure or degraded sample. If the ACA was negative, a new QIA extract was made from which both PCR assays were redone. If the ACA and the RSA were negative, the RNA was presumed degraded and the sample excluded from the study.
Results: The QIA-ACA detected SARS-CoV in 79 of 296 samples, whereas the MP-RSA detected SARS-CoV in only 32 of these samples. The MP-RSA did not detect any SARS-CoV in the 190 QIA-ACA SARS-negative samples. ACA detected SARS-CoV in 27 of 47 discrepant MP-RSA-negative/QIA-ACA-positive samples. The 20 remaining samples that were MP-ACA-negative were re-extracted by QIA from the initial homogenate and rE-tested. 15 of these 20 new extracts were ACA positive and 5 that were ACA-negative were excluded from the study as the samples were presumed to be degraded. On rE-testing these with the RSA, only 4 of the 15 were found to be SARS-CoV-positive.
Conclusion: In comparison to the QIA-ACA, the MP-RSA was substantially less sensitive, detecting SARS-CoV in only 46% of the tissue samples from confirmed SARS cases. The specificity of the combined assays was 100%. The lack of RSA sensitivity may be due to evolutionary mutations. Future sequencing of the target region (pol gene) may be necessary.
P1628: Analysis of respiratory viruses and new viruses with combined cell culture/PCR testing
C. Larcher, H.P. Huemer (Innsbruck, A)
The prevalence of respiratory viruses in hospitalised patients is rather unclear. Due to the rather large effort of reverse transcriptase (RT)-PCR methods, the poor sample quality of many specimens and the sensitivity of most rapid antigen detection tests, many viruses are not diagnosed. With nucleic acid tetection methods there is also question, whether there are acute replicating viruses or residual genetic material detected. In this presentation we describe our results from a study using a combined cell culture/PCR method suitable for detection of productive infection of different respiratory viruses. By cultivation of a total of 266 nasopharyngeal secretions and bronchioalveolar lavages (BAL) from children hospitalized in the local paediatrics department and from adult transplant patients on HEp-2 human larynx epithelial and MA-104 monkey kidney cells we were able to detect also slow growing viruses like human metapneumovirus (HMPV). The method was also able to detect Parainfluenza Virus or Rhinoviruses and to a certain extent Adenoviruses. Additionally Influenzaviruses, Respiratory-Syncytial Virus (RSV) and Adenoviruses were detected by commercial antigen detection assays. The epidemiology of respiratory viruses in Western Austria for the winter season 2003/04 reveals a Parainfluenza 1 outbreak as confirmed by sequencing right before the main Flu season, a relatively weak RSV season and many of HMPV isolations with some double infections as indicated below. Additionally the samples were also tested for ‘exotic’ agents like Mimiviridae. Mimiviruses are a new species which has been described most recently in water amoeba and their nearest relatives appear to be aquatic Phycodnaviruses. As replication of Mimivirus similar to the Leginella spp., which also replicate in amoeba, has been suggested to occur also in alveolar cells of humans we decided to test our patient samples using a ‘suicide-PCR’ protocol. Disappointingly enough we did neither detect this type of virus in our respiratory samples, nor in samples from water tanks containing amoeba species.
P1629: Rapid detection of parainfluenza virus in respiratory samples: a 3-year experience
P. Bruynseels, W.A. Verstrepen, M. Boers, A. Mertens (Antwerp, B)
Introduction: Parainfluenzavirus (PI) is a paramyxovirus causing acute respiratory illness, mostly in children. Four human PI have been described, PIV-I and PIV-II tend to produce epidemics in autumn and early winter, PIV-III has peak activity in spring and summer. PIV-IV usually causes mild upper respiratory disease.
Objectives: To evaluate the indirect immunofluorescence assay (IIF) with pooled PI- I, II & III monoclonal antibodies (MAB) (Chemicon International) performed directly on respiratory specimens as compared to conventional shell vial culture (SVC) on VERO cells.
Methods: 2272 nasopharyngeal aspirates of children presenting with respiratory symptoms between January 2002 and June 2004 were processed. PI detection was done with both the IIF performed directly on the specimen and IFF after SVC on VERO cells. Simultaneous detection of other common respiratory viruses was done using the appropriate techniques.
Results: PI was detected in 116/2272 (5.1%)samples. 86 PI were detected by IFF performed directly on the sample (sensitivity 74.1%). In 5 IFF positive samples, no virus could be detected by the conventional SVC (specificity 99.8%). Positive (94.5%) and negative (98.6%) predictive values were excellent.
Conclusion: IIF performed directly on respiratory specimens may prove to be a valuable technique for rapid PI detection in children presenting with respiratory illness, given the high specificity and predictive values of the test. Negative IFF results should be confirmed by conventional detection methods.
P1630: Development of a real-time NASBA respiratory syncytial virus assay
B. Deiman, F. Jacobs, C. Schrover, S. Vermeer (Boxtel, NL)
Objective: The aim was to develop and evaluate a Respiratory Syncytial virus assay based on NASBA amplification and including real-time detection with molecular beacons. In this study the performance of this new test is evaluated.
Methods: RSV RNA is isolated using a semi automated magnetic extraction method and the NucliSens® miniMag. An internal control is added to the sample prior to nucleic acid extraction. Primers are directed against the F-gene region of the RSV genome and both RSV A and B and the internal control are amplified with the same primer set. One generic molecular beacon probe is designed to detect both RSV A and B. An additional beacon is designed for the detection of the internal control. Amplification reactions were performed in a Nucli-Sens® EasyQ Analyser allowing real-time detection.
Results: Using serial dilutions of in vitro RNA, the 95% hit rate of the real-time RSV assay was found to be 62 copies in isolation for both RSV A and RSV B. Using tissue culture samples, the assay sensitivity was approximately 0.01 TCID50 in isolation for RSV A and 0.1 TCID50 in isolation for RSV B. No cross reactivity was observed with PIV2, PIV3, hMPV A1, hMPV A2, hMPV B1, hMPV B2, or measles. With the real-time assay, RSV in nose swab samples is detectable within approximately 3,0 hours.
Conclusions: The data showed that the real-time RSV assay is a rapid and sensitive qualitative assay for the detection of RSV. The use of standardized reagents offers considerable benefits in a routine setting for the clinical management of patients with RSV infections.
P1631: Intrathecal synthesis of specific antibodies in facial nerve palsy
J. Bednarova, P. Stourac, H. Stroblova, A. Sevcikova (Brno, CZ)
Objectives: According to literature at least part of idiopathic facial nerve palsies is caused by herpetic viruses, for example varicella zoster virus in Ramsay-Hunt syndrome. The aim of our study was to detect intrathecal synthesis of specific IgG antibodies against varicella zoster virus in patients with idiopathic facial nerve palsy and to evaluate the importance of this examination for establishing the etiology of this disease and subsequent treatment.
Methods: We investigated a cohort of 30 patients: 15 patients were diagnosed as idiopathic facial nerve palsy and 15 patients were diagnosed as neuroborreliosis (NB) with facial nerve palsy as a part of their clinical symptoms. Serum and CSF samples were analysed at each patient. We used the diagnostic kit of Human Company, Germany (Varicella-Zoster Virus Human ELISA IgG Antibody Test) and the diagnostic kit of Test-Line Company, Clinical Diagnostics, Czech Republic (EIA Borrelia garinii IgG) for the detection of specific IgG antibodies. The intrathecal synthesis was evaluated as specific antibody index − AI according to Reibeŕs method.
Results: Intrathecal synthesis of IgG antibodies against varicella zoster virus was detected in 20% patients with idiopathic facial nerve palsy. All these patients had negative intrathecal antiborrelia antibody synthesis. In the NB subgroup all patients had negative intrathecal antibody synthesis against varicella zoster virus. The statistical significance of specific antibody intrathecal synthesis was confirmed by Spearman rank coefficient and Wilcoxon test.
Conclusion: Our statistically supported data emphasize the importance of intrathecal synthesis of specific antibodies as a diagnostic method in patients with idiopathic facial nerve palsy. It reveals a subgroup of patients with viral ethiology who can thus profit from specific antiviral treatment.
P1632: Second quality control for the detection of herpes simplex virus in cerebrospinal fluid in Belgium
D. Ursi, K. Loens, H. Goossens, M. Ieven (Edegem, B)
Objectives: A second quality control was organized in Belgium in 2004 to test the ability of laboratories to detect Herpes simplex virus (HSV) in cerebrospinal fluid (CSF). This panel also included a positive sample artificially inhibited by the addition of fraxiparine to address the issue of possible false negative PCR results.
Methods: The panels were prepared in a pool of CSF specimens. Each panel consisted of 10 coded specimens: 4 negative specimens, 1 specimen containing cytomegalovirus (CMV), 3 specimens containing HSV type 1 at a concentration of 4.102, 7.5, 102 and 3.75. 103 TCID50/ml on Vero cells and 2 specimens containing HSV type 2 at a concentration of 4.102 and 4.103 TCID50/ml. One specimen (containing HSV type1 at a concentration of 7.5. 102 TCID50/ml) was artificially inhibited by the addition of fraxiparine at a concentration of 356 U/ml. Panels were sent to 13 laboratories together with a questionnaire concerning the methods employed.
Results: All laboratories sent their results and all but one laboratory answered the questionnaire. All used an ‘in house’ PCR. All but one laboratory used real-time PCR and all but two laboratories did monitor inhibition. Overall, 97% of the specimens were correctly reported All positive specimens were correctly identified. False positive results were reported in 6.6% of the negative specimens by 3/13 participants. Six laboratories had an assay that distinguishes between HSV type 1 and 2. The different types were correctly distinguished by all. The specimen containing CMV was never reported positive. Remarkably, the different assays seem to be influenced at varying degrees by inhibitory factors. Only 7/13 (54%) participants reported the specimen containing fraxiparine as positive despite the inhibition. Six of these labs used the same QiaAmp DNA kit (Qiagen) for extraction and one lab did not specify the extraction method. Five laboratories reported the fraxiparine containing specimen as inconclusive because inhibited and one participant who did not monitor inhibition reported this specimen as false negative. Only 1/6 labs used the QiaAmp DNA kit (Qiagen) for extraction.
Conclusions: The results of this second quality control for the detection of HSV in CSF specimens were excellent. The most striking conclusion of this quality control was the difference in PCR results obtained with a positive specimen artificially inhibited, being probably a reflection of the different DNA extraction methods used.
P1633: Rapid diagnosis herpes simplex genital infection using real-time PCR after automated extraction nucleic acid
M.C. Nogales, M. Ramirez, C. Serrano, I. Pueyo, E. Martin-Mazuelos (Cadiz, E)
Objectives: Herpes simplex virus genital infections have been increased in the last years. A sensitive and specific method for detecting herpes simplex virus type 1 (HSV-1) and herpes simplex virus type 2 (HSV-2) is important for diagnosing genital infections and to avoid HSV-1 and HSV-2 transmission. The aim of these study is to show our short experience on the diagnosis of HSV-1 and HSV-2 genital infection using real-time PCR after automated extraction nucleic acid.
Methods: A total of 86 specimens (44 genital ulceration/vesicle, 21 endocervical, 14 urethral and 7 rectal) were collected from 61 patients (33 women and 28 men). All of them ask for the begining of symptoms and none have risk factor. All the patients were followed at STD clinic, Seville, Spain. The sample were collected following the manufacters rules. Magnapure was used to extraction nucleid acid. These products were used for determinating genital infection by using Light-cycler real-time PCR. (Roche Diagnostic System LC HSV1/2 detection). Positive and negative controls were used in all cases.
Results: Of the total of the samples, 37.2% (32/86) were positive to HSV. Genital lesions and vesicles were positive in 61% (27/ 44), 19% (4/21) endocervival swabs, 7.1% (1/14) urethral swabs. Of the all positive results in 23 cases were detected HSV-1 (71.8%), in 6 cases (18.8%) HSV-2, and in 3 cases (9.4%). All the patients with urethral and endocervical swabs positive have genital lesion positive too. All the HSV positive were in patients with primary infection.
Conclusion: 1. Magnapure is an easy, effective, and non-time consuming method for automated extraction nucleid acid of the direct sample. 2. Real-time PCR using LC allows the detection of HSV1 y 2 on genital lesion and it is possible to distinguish between two types clearly.3. In our study the most profitable sample for the diagnosis of herpes simplex genital infection was genital ulcerations and vesicles.
P1634: Human herpesvirus-6, Epstein-Barr virus and cytomegalovirus viral load in cases of sudden death in children. An approach with real-time PCR assay
A. Fernández-Rodríguez, R. álvarez-Lafuente, M.P. Suárez-Mier, B. Aguilera, S. Ballesteros, G. Vallejo, J. Gómez (Madrid, E)
Objectives: The aim of this work was to determine the viral load of three ubiquitous viruses, Human Herpesvirus-6 (HHV-6), Epstein-Barr virus (EBV), and Cytomegalovirus (CMV), with a quantitative real-time polymerase chain reaction (PCR) assay, in 22 paraffin-embedded tissues from 9 cases of sudden children death (SCD) in which viruses had been previously detected by other techniques (serology, viral culture or nested PCR).
Methods: The QIAamp DNA Mini Kit (Qiagen) was used for the extraction of DNA from 22 paraffin-embedded tissues from 9 cases of SCD, according to the manufacturer's protocol. DNA concentration was determined by reading the optical density at 260 nm. Two blank reactions were extracted together with each set of ten samples in order to evaluate a possible cross-reaction. For the detection of HHV-6, EBV and CMV genomes, quantitative real-time PCR was carried out with specific sets of primers and TaqMan probes; a beta-actin PCR was carried out as an internal control to assure the quality of the DNA of each sample and its suitability for PCR. Each sample was analysed in duplicate for each one of the viruses and the internal control.
Results: The viral loads (in copies/μg of DNA) were as follows: Patient 1 (1.7 years) had HHV-6 in liver (19.3) and in spleen (25) and CMV in spleen (24.8), and was negative for the three herpesviruses in lung. Patient 2 (2.6 years) had CMV in liver (30.6), and was negative for the three herpesviruses in larynx and lung. Patient 3 (2 years) had EBV in amygdale (28.5), and CMV in amygdale (49.3) and kidney (34.3). Patients 4 (3 months) and 5 (a newborn) were negative for the three herpesviruses in lung. Patient 6 (3 months) had HHV-6 in spleen (2.1), and was negative for the three viruses in lung. Patient 7 (2.2 years) had HHV-6 in amygdale (34.8) and EBV in lung (36.8). Patient 8 (1.2 years) had CMV in spleen (78.1), and was negative for the three viruses in lung. Patient 9 (3 months) had HHV-6 in lymphoid tissue (45.4) and EBV in heart (24.9).
Conclusion: We have shown that quantitative real-time PCR could be an important tool in the diagnosis of viral infections in SCD cases, since its sensitivity allowed us to find the presence of pathogens that other assays did not detect because of their low yields. We emphasise the need to establish a viral load threshold in order to make an accurate interpretation of the presence of herpesviruses in tissues.
P1635: The contribution of IgG avidity in the diagnosis of CMV infection
P. Koudounis, M. Pagkalou, G. Triantafillou, P. Rozi (Serres, GR)
Objectives: The evaluation of measurement of Cytomegalovirus immunoglobulin G avidity for distinguishing recent CMV infection from the past.
Methods: Blood samples were taken from 262 patients attending the Outpatient's Department or who were hospitalized in the General Hospital of Serres with symptoms and signs of recent CMV infection in 18 months (7/2002 to 12/2003). The presence of specific CMV antibodies was determined using the microparticle enzyme immunoassay (ABBOTT-AXSYM system) and confirmed with the enzyme linked fluorescent assay (Vidas). IgG CMV avidity test also made in Vidas system (bioMerieux, France).
Results: A total of 262 patients (126 men-70 women-66 children) suggesting recent CMV infection, were examined. Thirty four (first group) had both IgM and IgG positive tests, while 6 (second group) had only IgM positive antibodies. The use of the IgG avidity test on the patients of the first group excluded recent primary infection (less than 3 months) in 27 out of 34 patients (79%). In the remaining 7, recent infection was confirmed by the second method of detection IgM antibodies. In all the above patients that were examined, a second blood sample was taken after 3 weeks. The last results of the 6 patients (second group) for IgM antibodies were negative.
Conclusion: The CMV IgG avidity is a useful diagnostic tool in laboratory confirmation of acute infection.
P1636: Screening for cytomegalovirus infection in lung-transplant patients: viral load or virus isolation?
K. Van Vaerenbergh, W. Laffut, M. Develter, G. Verleden, J. Van Eldere (Leuven, B)
Objective: Cytomegalovirus is a major cause of morbidity and mortality in transplant patients. Efforts have focused on identifying patients at risk for disease (especially D+R-) before the onset of symptoms, in order to start pre-emptive antiviral treatment. In this study, we evaluated the use of quantitative real-time Taqman PCR on whole blood versus isolation of CMV from urine and respiratory tract specimens.
Methods: 71 lung-transplant recipients were prospectively studied. At 400 time points PCR and rapid detection of CMV by shell vial assay (SVA, urine and throat) were performed on linked samples.
Results: Concordance of the three test results was obtained in 76% (negative in 69%, positive in 7%). In 12.5% of the samples studied, PCR was positive with negative isolations. In 5.5% a negative PCR was obtained, with at least one positive isolation. Positivity of only one isolation site combined with a positive PCR was observed in a minority of cases (5% urine, 1% throat). Patients with consecutive low copy numbers in PCR ((500 c/ml) had intermittently positive isolations, with the best concordance between urine isolation and viral load. Rarely, isolations remained completely negative. For patients with continuously positive viral loads, there was a good correlation of PCR and isolations, although occasionally an isolation remained negative. In only 2 patients (both R+) three consecutive positive urine cultures were observed, with a negative viral load. For both patients there was no evidence of CMV disease. Other discordances between PCR (negative) and isolation (positive) were preceded or followed by positive PCR.
Conclusions: In this patient population of lung-transplant recipients, viral load and virus isolation in urine were highly concordant.
P1637: Two-year analysis of prospective CMV-PCR surveillance data in lung transplant recipients
J. Harwood, A. Krause, K. Gould, J. Dark, P. Corris (Newcastle upon Tyne, UK)
Description: CMV is a major cause of mortality and morbidity in transplant (tx) recipients. We commence weekly Quantitative CMV PCR (QPCR) surveillance on our CMV positive (+) recipients four weeks post-tx for three months. The CMV mismatch (MM) recipients commence weekly QPCR for three months, two weeks after the cessation of prophylactic ganciclovir (GCV), which is given for the first three months post-tx in this group. CMV (recipients receive no prophylaxis. During the surveillance period, pre-emptive therapy (Rx) with oral GCV is given if QPCR is (1 × 104 copies/ml and the patient is asymptomatic. IV GCV is given in symptomatic patients above this cutoff.
Objectives: 1. To estblish whether there is a correlation between clinical symptoms associated with CMV disease and the viral load of CMV. 2. To review the present viral load cut-off above which pre-emptive Rx is considered in asymptomatic patients to prevent development of disease.
Methods: We conducted a prospective analysis of patients’ notes who underwent lung tx between Novemeber 2001 and November 2003. Thirty eight patients (>18 years) and one patient (<18 years) who survived six months were included in the study.
Results: Of all the patients, six remained QPCR negative throughout the study (3 CMV MM, 3 CMV+). Twelve patients had low level QPCR levels, 103-104 copies/ml, of which one MM patient developed symptoms and required pre-emtpive Rx. Eleven patients had QPCR levels of 104-105 copies/ml, of which 4 (1 MM, 3 CMV+) received IV GCV for symptomatic disease. Nine patients had QPCR levels (105 copies/ml (4MM, 5 CMV+) of which 3 received treatment for symptomatic disease.
Conclusions: We did not find any association between viral load level and development of CMV disease. Only 9 patients developed CMV disease requiring treatment, whereas 23 patients had raised QPCR levels with no disease. This data suggests that our current cut off of (104 copies/ml is appropriate for the initiation of pre-emptive therapy in asymptomatic patients and is effective in preventing disease.
P1638: Evaluation of a new IgM test for detection of cytomegalovirus IgM antibody
P. Chen, S. Binder, C. Hixson, R. Kaul, K. Johnson, S. Garcia, F. Torres (Hercules, USA)
Objectives: Cytomegalovirus (CMV) is a ubiquitous pathogen in humans. CMV infection can be asymptomatic in immunocompetent adults, or life threatening in newborns and immunocompromised individuals. CMV IgM is a sensitive and specific indicator of on-going and recent infection, but may not be specific for primary infection due to the persistence of CMV IgM in some adults. IgG avidity testing has been useful in identifying and excluding non-acute infection when the IgM tests were discrepant (R. Kaul. et al. J. Clin. Microbiol. in press). We developed a CMV IgM assay specifically to detect IgM antibody to CMV using Bio-Rad's fully automated, random access BioPlex 2200 immunoassay analyzer (BioPlex) which measures multianalytes in a single test. This study examines the performance of the BioPlex CMV IgM assay in conjunction with a CMV IgM assay and an avidity test.
Methods: A total of 186 patient samples were tested on the BioPlex and characterized by bioMerieux VIDAS CMV IgM and IgG assays (Marcy-l'Etoile, France) as follows: 2 were CMV IgM positive and IgG negative, 38 were both CMV IgM and IgG positive, 86 were CMV IgG positive and IgM negative, and 60 were both CMV IgM and IgG negative. The CMV IgG positive samples were also tested by the Dade Behring Enzygnost Anti CMV IgG avidity kit (Marburg, Germany).
Results: Compared to the VIDAS CMV IgM assay, the BioPlex CMV IgM assay results yielded a sensitivity of 90.0% (36/40) and specificity of 95.9% (140/146). Six false positive samples on the BioPlex exhibited a low IgG avidity indicating primary infection, and 4 false negative samples had a high IgG avidity suggesting non-acute infection. The sensitivity and specificity of the BioPlex CMV IgM assay increased to 97.7% and 100%, respectively, when the ten discrepant samples were resolved by the avidity test results. In the samples with low IgG avidities, the sensitivity of CMV IgM assay was 96.2% (25/26) for the BioPlex versus 73.1% (19/26) for the VIDAS. One of the low IgG avidity samples was IgM negative in both the VIDAS and BioPlex CMV IgM assay. In the samples with high IgG avidities, the sensitivities of the VIDAS and BioPlex IgM assays were 19.4% (19/98) and 15.3% (15/98), respectively.
Conclusion: The BioPlex 2200 CMV IgM assay described here is sensitive and specific for detecting CMV IgM antibody, and exhibited an ability to detect CMV IgM in specimens containing low avidity IgG antibodies.
Biofilms: pathogenesis and antibiotic susceptibility
P1639: The effect of atmosphere and inoculum size on Staphylococcus epidermidis biofilm density
J.S. Cargill, M. Upton (Manchester, UK)
Objectives: Staphylococcus epidermidis is a potential cause of nosocomial infection, and its pathogenicity is related to its ability to form a biofilm. The aim of this study was to assess the variation in biofilm density of Staphylococcus epidermidis strains (including the reference strain RP62A) at different concentrations and in different atmospheres.
Methods: Staphylococcus epidermidis strains were cultured overnight in typtone soya broth with an additional 0.25% w/v glucose (TSB). Sterile flat-bottomed polystyrene 96-well cell culture plates were prepared with 100 μl of fresh TSB in 10 columns. 100 μl of overnight culture was added to an empty column, and serial dilutions performed in the TSB containing wells, with 100 μl discarded from the final dilution. One column was left blank as a reference. The plates were then incubated at 36°C for 20 hours before the wells were emptied by pipette, washed three times in 200 μl phosphate-buffered saline and air dried. They were then stained with 0.4% crystal violet solution for ten minutes before rinsing under gently running water. Biofilm density was measured as the optical density at 450 nm (OD > 0.12, weak positive; OD > 0.24, strong positive). 200 μl of 1:100 dilutions of overnight culture were added to the columns of 2 96-well plates. One set was incubated in normal atmosphere and the other in a 5% carbon dioxide enhanced atmosphere. Biofilm density was assessed as above. Each experiment was repeated three times.
Results: For most strains the biofilm density of the serial dilutions remained approximately constant at dilutions above 1:4, although some showed a gradual reduction in density above this level. One strain showed a pronounced drop in biofilm density between 1:8 and 1:32 dilution. Eight of fifteen strains showed biofilm positive results above 1:4 dilution in all three repeats. Nine of fifteen strains showed increased biofilm densities in a normal atmosphere while four strains showed increased density in when grown in the presence of elevated carbon dioxide. Two strains did not show statistically significant (Mann–Whitney test) overall differences.
Conclusion: This study shows that the biofilm density of some Staphylococcus epidermidis strains are affected by the dilution, and hence inoculum size, and the atmosphere. The results vary in a strain dependant manner.
P1640: Desferrioxamine increases Staphylococcus epidermidis biofilm density
J.S. Cargill, M. Upton (Manchester, UK)
Objectives: Staphylococcus epidermidis is a potential cause of nosocomial infection, and its ability to form a biofilm is important in pathogenicity. This study was designed to evaluate the change in biofilm density of clinical isolates and the reference strain RP62A in the presence of an iron source and the iron chelator desferrioxamine.
Methods: Staphylococcus epidermidis strains were cultured overnight in typtone soya broth with an additional 0.25% w/v glucose (TSB). Overnight cultures were diluted 1:50 in fresh TSB, and 100 il used to seed the columns of sterile flat-bottomed polystyrene 96-well cell culture plates. Serial dilutions of Sigma iron supplement were prepared from a maximum of 4 ml/l and 100 il added to the wells. This gave 1:100 dilutions of staphylococci with reducing concentrations of iron supplement from 2 ml/l. A reference control was provided by a 1:100 dilution without iron supplement. After 20 hours incubation the plates were washed in triplicate in phosphate buffered saline and stained with crystal violet. Biofilm density was assessed by recording the optical density at 450 nm (OD > 0.12, weak positive; OD > 0.24, strong positive). Some of the strong positive strains were tested in the presence of desferrioxamine with 2 ml/l iron supplement in the same manner as above, together with set of replicates without iron supplement.
Results: 3/15 strains were biofilm negative at all iron concentrations. 2/15 strains gave weak positive to negative results at all iron concentrations. 4/15 strains were weak to strong positive without iron, and showed maximum biofilm density at 1.0 ml/l of iron supplement. The remaining 6/15 were strong positive and showed maximum density at 2 ml/l. Three of these strains were tested with desferrioxamine and showed increased density in the presence of the iron chelator.
Conclusions: Iron supplementation increases the biofilm density of biofilm positive Staphylococcus epidermidis strains. Some strains show increased biofilm density in the presence of desferrioxamine. Even though desferrioxamine has previously been shown to inhibit the growth of S. epidermidis, it may be acting as a staphylococcal siderophore.
P1641: Low levels of vancomycin increase the density of Staphylococcus epidermidis biofilms
J.S. Cargill, M. Upton (Manchester, UK)
Objectives: Staphylococcus epidermidis is a potential cause of nosocomial infection, and its pathogenicity is related to its ability to form a biofilm. The antibiotic vancomycin is often used to treat Gram-positive infections, although resistance can be found in some staphylococcal strains. The aim of this study was to assess the change in biofilm density of a variety of clinical isolates and the reference strain RP62A in the presence of vancomycin.
Methods: Biofilm forming Staphylococcus epidermidis strains were cultured overnight in typtone soya broth with an additional 0.25% w/v glucose (TSB). Overnight cultures were diluted 1:50 in fresh TSB, and 100 μl used to seed five columns of sterile flat-bottomed polystyrene 96-well cell culture plates (two strains per plate). Various concentrations of sterile filtered vancomycin in TSB were added to four columns per strain, with TSB alone added to the remaining column. Four different repeats of the experiment were performed, giving results for 1:100 dilutions of eight strains with 0 (control), 40, 20, 10, 5 μg/ ml vancomycin (1 set), 0, 8, 4, 2, 1 μg/ml (2 sets) and 0, 5, 4, 3, 2 μg/ml (1 set). Each set comprised of eight replicates of each condition. The plates were then incubated at 36°C for 20 hours before the wells were emptied by pipette, washed three times in 200 μl phosphate-buffered saline and air dried. They were then stained with 0.4% crystal violet solution for ten minutes before rinsing under gently running water. Biofilm density was measured as the optical density at 450 nm (OD > 0.12 taken as positive). Repeats of the same condition were combined by expressing the results as a multiple of the control condition. Statistical significance was assessed using the Mann–Whitney test (p < 0.5 taken as significant).
Results: Five of the eight strains showed statistically significant increases in biofilm density in the presence of low vancomycin concentrations, resulting in biofilm-positive results in 95% of wells (see table).
| Strain | Vancomycin* (μg/ml) | Relative biofilm density** | Mann-Whitney p-value |
|---|---|---|---|
| RP62A | 3.0 | 0.99 (0.34) | 0.370 |
| s2 | 4.0 | 1.46 (0.11) | <0.001 |
| s4 | 0.5 | 0.82 (0.17) | <0.001 |
| s5 | 2.0 | 1.96 (1.69) | 0.020 |
| s41 | 3.0 | 1.12 (003) | <0.001 |
| s79 | 1.0 | 0.83 (0.12) | <0.001 |
| s159 | 3.0 | 1.14 (0.16) | 0.020 |
| s163 | 3.0 | 1.26 (0.06) | <0.001 |
Vancomycin concentration at which 95% of test wells gave biofilm-positive results.
Mean (SD) of the biofilm density expressed as a multiple of the result at a vancomycin concentration of 0.
Conclusion: This study has shown that the biofilm density of some strains of S. epidermidis increased in the presence of low concentrations of vancomycin. It is unknown whether this reflects increased biofilm-forming ability of less sensitive clones within a population, an effect on the cell wall of the organisms, or modulated gene expression.
P1642: Regulation of biofilm formation by SigmaB is a common mechanism in Staphylococcus epidermidis and is not mediated by the SigmaB dependent sarA transcript
S. Jäger, D. Mack, M.A. Horstkotte, H. Rohde, J.K. Knobloch (Hamburg, D)
Objectives: The alternative sigma factor SigmaB was identified as a major regulator of biofilm formation in Staphylococcus epidermidis 1457, whereas for S. aureus only a minor impact of SigmaB on biofilm formation could be observed. To exclude an individual behavior of S. epidermidis 1457 we further investigated in this study the influence of SigmaB on biofilm formation and sarA transcription in the methicillin susceptible clinical isolate S. epidermidis 8400 as well as in the methicillin resistant clinical strain 1057.
Methods: Mutants with various deletions within the sigB operon were characterized phenotypically by a quantitative biofilm assay and by a PIA specific immunofluorescence assay. Transcription analysis of the genes icaA, icaR, sarA, and asp23 was performed using quantitative real-time PCR.
Results: All mutants with dysfunctional SigmaB activity displayed a decreased biofilm formation, whereas for mutants with inactivation of the antisigmafactor RsbW an increased biofilm formation was observed. Transcriptional analysis revealed that icaA transcription was down-regulated in SigmaB negative mutants while icaR transcription was up-regulated. However, the observed differences of icaR transcription in these mutants did not always reach a significant level, indicating that additional SigmaB dependent regulators are involved in regulation of biofilm expression. Except for the mutant with inactivation of RsbW in S. epidermidis 8400, which displayed a significant increase of icaA transcription congruent with a strong increase of biofilm formation mutants with inactivation of the antisigmafactor displayed only minor transcriptional differences. Analysis of sarA transcription revealed no or only minor differences compared to the respective wild types.
Conclusions: The similar influence on phenotypes as well as a comparable transcriptional regulation of icaR and icaA by the alternative sigma factor SigmaB in three independent genetic backgrounds of clinical S. epidermidis isolates suggests that regulation of biofilm formation by SigmaB is a general feature in S. epidermidis. Therefore, the SigmaB dependent regulatory pathway could be a promising target for new strategies to prevent foreign body-associated infections. The lack of significant differences of sarA transcription indicates that the SigmaB dependent sarA transcript is not involved in the observed phenotypic changes.
P1643: Foreign body infection and ‘wild type’ Staphylococcus epidermidis − Potential of biofilm formation and antimicrobial resistance
B. Parschalk, A. Grisold, F. Yassin, H. Adametz, W. Graninger, E. Presterl (Vienna, Graz, A)
Objectives: Foreign body infections (FBI) due to Staphylococcus epidermidis have become major clinical problems associated with considerable morbidity and costs. Biofilm formation and resistance to multiple antibiotics are the major obstacles for a successful treatment. The aim of the study was to test Staphylococcus epidermidis isolates from patients with verified foreign body infections i) for their ability to form biofims and ii) for their susceptibility to 12 standard antibiotics, and compare these to ‘wild type’ Staphylococcus epidermidis isolated from the skin of healthy volunteers.
Methods: Sixty patients with verified FBI were evaluated. Hundred and twenty-nine blood culture isolates from 60 patients with FBI, and 52-skin isolates from healthy, not hospital-associated, volunteers were analysed. Identification of Staphylococcus epidermidis was done using routine methods. To exclude doubles FBI isolates were genotyped by PFGE. The susceptibility testing was done using disk diffusion method according to the NCCLS. Antimicrobial agents tested were penicillin, oxacillin, erythromycin, clindamycin, gentamicin, amikacin, vancomycin, fosfomycin, fusidic acid, rifampicin, ciprofloxacin, trimethoprim and linezolid. Biofilm formation was tested using a microtiter plate biofilm model.
Results: Among the 129 FBI isolates 86 strains with distinct PFGE profiles were identified. Biofilm formation was detected in 86.4% of FBI strains and in 76.9% of the ‘wild type’ isolates (not significant). More than 30% of all FBI strains were resistant to penicillin, oxacillin, erythromycin, clindamycin, ciprofloxacin and trimethoprim. The 52 ‘wild type’ Staphylococcus epidermidis were generally more susceptible than the clinical isolates (p < 0.05): However, 46.2 % of these controls were resistant to penicillin and to erythromycin, 17.3% were resistant to clindamycin.
Conclusions: Biofilm formation is present to the same extent in both, FBI-associated and ‘wild type’ Staphylococcus epidermidis strains demonstrating the potential of skin flora to cause FBIs. Although it is well known that nosocomial Staphylococcus epidermidis are resistant to multiple antibiotics, the ‘wild type’ isolates show alarming resistance to erythromycin and clindamycin possibly reflecting the abundant use of macrolides for minor infections in the population.
P1644: barAB a new regulatory gene locus mediating NaCl induction of Staphylococcus epidermidis biofilm formation
J.K. Knobloch, K. Bartscht, S. Jäger, H. Rohde, M.A. Horstkotte, D. Mack (Hamburg, D)
Objectives: Biofilm expression in Staphylococcus epidermidis is a major virulence factor and attachment to polymeric surfaces leads to reduced susceptibility against antimicrobial substances. The alternative sigma factor SigmaB is an important regulator of biofilm formation in S. epidermidis. However, biofilm formation is under influence of additional yet uncharacterized regulators, which were examined in this study.
Methods: The Tn917 insertion of the biofilm-negative mutant M12 generated in the biofilm-positive genetic background of S. epidermidis 1457 was identified by arbitrary PCR. The transcription of the icaADBC locus, essential for biofilm formation as well as of the inactivated genes was investigated. The function of the inactivated genes was characterized by complementation studies under different environmental conditions.
Results: In M12 Tn917 was inserted at the distal end of an orf homologous to the regulatory gene purR in Bacillus subtilis. The gene upstream of purR is conserved between S. epidermidis, B. subtilis, and S. aureus furthermore two additional orfs (orf3 and orf4) homologous to B. subtilis, and S. aureus are located downstream of the Tn917 insertion site. Within the purR genes of S. epidermidis, B. subtilis, and S. aureus a highly conserved motif homologous to the consensus sequence for SigmaB dependent promoters was identified upstream of Tn917. The SigmaB dependent transcription of the orf3 and orf4 could be demonstrated in mutants with inactivation of sigB. In M12 transcription of the biofilm associated genes icaADBC was abolished, whereas the transcription of their negative regulatory gene icaR was unaffected. Complementation with the single genes showed no effect in M12, whereas complementation of M12 with orf3 and orf4 together was able to restore biofilm formation under high osmotic conditions. Interestingly, under standard conditions used for biofilm formation as well as under biofilm induction with subinhibitory concentrations of ethanol no biofilm formation was observed in the complemented mutant.
Conclusions: We identified two new regulators of S. epidermidis biofilm formation, which we designate now as biofilm accumulation regulators A and B (barAB). BarA and B act as positive regulators of icaADBC transcription independent of the regulatory gene icaR. BarAB is a SigmaB dependent gene locus, which is at least partially responsible for the NaCl induction of biofilm formation in S. epidermidis.
P1645: Role of Fur and transferrin binding protein in Staphylococcus epidermidis during in vitro and in vivo biofilm formation
C. Massonet, J. Van Eldere (Leuven, B)
Objectives: To determine whether GADPH (transferrin binding protein-TBP) and its regulator (Fur) play a role in the biofilm formation by Staphylococcus epidermidis.
Methods: A biofilm forming strain of S. epidermidis, isolated from a patient with proven catheter-related infection was used. RNA and DNA isolations were performed as described by S. Vandecasteele et al. (Biochem. Biophys. Res. Commun. 2002. 291:528–534). For In vitro studies bacteria were grown overnight in BHI or RPMI, without iron (fRPMI) and re-incubated in fRPMI or fRPMI with 1 μm FeCl3 or fRPMI with apotransferrin (iron free; 20 μg/ml) or fRPMI with holotransferrin (iron-saturated; 20 μg/ml). For in vivo studies a subcutaneous rat model was used. Samples were quantified through real-time PCR described by S. Vandecasteele et al. (Biochem. Biophys. Res. Commun. 2002. 291:528–534).
Results: During the growth of the bacteria, the expression of fur and tbp for planktonic bacteria was higher in an iron depleted medium and lower in an iron rich medium. In vitro studies in an iron depleted medium show a statistically significant (2-way ANOVA, Bonferonni, N = 12) up regulation of fur expression in sessile bacteria in comparison with planktonic bacteria. In an iron rich environment there is no variation in the expression of fur between sessile and planktonic bacteria. In fRPMI with holotransferrin or apotransferrin the expression of fur for sessile bacteria in comparison with that of planktonic bacteria is significantly up regulated in fRPMI with holotransferrin and down regulated in medium with apotransferrin. The results obtained for tbp are similar. In vivo, the expression of fur is down regulated in the beginning, followed by an increase of expression after one week. The expression of tbp is very low and stays constant over a period of two weeks. (Statistically significant: 1-way ANOVA, Bonferroni, N = 16).
Conclusion: According to in vitro results, the lack of iron has a greater impact on sessile bacteria than on planktonic bacteria. Holotransferrin and apotransferrin also affect the expression of fur and tbp in sessile bacteria, but further investigation is required. In vivo results show that fur plays a role in the late stages of the biofilm.
P1646: Effect of iron on iron-regulated genes in biofilm-associated Staphylococcus epidermidis
C. Massonet, R. Merckx, J. Van Eldere (Leuven, B)
Objectives: To determine whether iron and iron-related genes (sirR, coding for the regulator; sitA, coding for ATP binding protein; sitB, coding for membrane protein; sitC, coding for lipoprotein) play a role in biofilm formation of Staphylococcus epidermidis.
Methods: A biofilm-forming strain of S. epidermidis, isolated from a patient with proven catheter-related infection was used. RNA and DNA isolations were performed as described by S. Vandecasteele et al. (Biochem. Biophys. Res. Commun. 2002. 291:528–534). For In vitro studies iron was removed from RPMI medium and experiments were performed with 1 μm FeCl3 added or without iron. For in vivo studies a subcutaneous rat model was used. Samples were quantified through real-time PCR described by S. Vandecasteele et al. (Biochem. Biophys. Res. Commun. 2002. 291:528–534).
Results: In vitro results show that in an iron-limited environment, the planktonic form of S. epidermidis produces siderophores and grows slower than in an iron-rich environment. In vitro, during an 18 hours observation period, the expression of sirR stays constant and is independent of iron concentration. The expression of sitA, sitB and sitC on the other hand is higher in medium with low iron content than in the medium with added iron. In vitro studies also indicate that sirR has a statistically (2-way ANOVA, Bonferroni, N = 12) significant higher expression in sessile bacteria compared to planktonic bacteria. The expression of sitC is significantly (2-way ANOVA, Bonferroni, N = 9) higher for sessile bacteria than for planktonic bacteria in an iron-limited medium. The expression of sitC in an iron-rich medium is identical for both forms. In vivo (1-way ANOVA, Bonferroni, N = 16), the expression of sirR is high and remains stable over a time period of two weeks. SitC, sitA and sitB are also highly expressed during this two weeks time period.
Conclusion: The constant high in vivo expression of sitABC and the regulator sirR could indicate a role of these genes in the biofilm growth mode. In vitro results show an iron-regulated expression of sitABC. The expression of sirR (in vitro) is not iron-regulated but a specific regulation mechanism is not yet known.
P1647: Role of virulence regulator genes in Staphylococcus epidermidis biofilm formation
V. Pintens, S. Vandecasteele, R. Merckx, J. Van Eldere (Leuven, B)
Objective: To monitor gene expression of Staphylococcus epidermidis regulator loci during early in vitro and in vivo biofilm formation.
Methods: AgrA, RNAIII, sarA, rsbU, rsbV and sigB expression in vitro was examined at 0, 10, 30, 60, 120 and 180 min after inoculation in 0.9% NaCl in planktonic (n = 68) and sessile (n = 70) bacterial cultures. Gene expression during in vivo biofilm formation was evaluated over two weeks. Catheter fragments inoculated with S. epidermidis were implanted subcutaneous in rats as described by S. Vandecasteele et al. (Biochem Biophys Res Commun. 2002; 291: 528–534). Catheters (n = 295) were explanted 0, 15, 30, 60, 90, 120, 240, 360, 720, 1440, 2880, 5760, 10080 and 20160 min after implantation. Expression was determined by TaqmanTM real-time PCR as described by S. Vandecasteele (Biochem Biophys Res Commun. 2002; 291: 528–534).
Results: In vitro expression of rsbU, the first gene of the sigB operon, is significant higher in sessile than in planktonic bacteria. Relative expression levels of RNAIII and sarA are higher in planktonic than in sessile bacteria. There is no significant difference between in vitro expression of agrA, sigB and rsbV in sessile versus planktonic bacteria. In vivo, expression of all genes reaches a peak level between 1 and 2 h after implantation (0.5–1.5 log10 times the expression level at implantation) and subsequently decreases towards its lowest level at 6 h. For agrA, expression remains stable during the whole observation period; expression of RNAIII and sarA decreases and remains thereafter at a very low level. SigB expression remains at a high level for the whole observation period, whereas expression of rsbV and rsbU increases. RsbU is expressed to a significantly higher degree than the other sigB operon genes.
Conclusion: In contrast to S. aureus, the sigB operon and particularly rsbU seem to be important factors in S. epidermidis biofilm formation.
P1648: Staphylococcal biofilm as a target for linezolid, vancomycin, oxacillin and/or lysostaphin
E. Walencka, B. Sadowska, S. Rozalska, W. Hryniewicz, B. Rozalska (Lodz, Warsaw, PL)
Objectives: Medical device-associated infections, frequently caused by staphylococci, are related to biofilm formation. Chemotherapy of such infections is seldom fully effective, due to high biofilm resistance. A number of alternative approaches to antimicrobial treatment have been proposed. The aim was to study the activity of antibiotics in monotherapy or in combination with lysostaphin, towards S. aureus biofilm.
Methods: S. aureus: ATCC29213 (MSSA), A3 (clinical MRSA), 1474/01 (clinical hVISA), were used as planktonic cultures or 24-hour-old biofilm built in the wells of microplate, in the chambers of a LabTekII chamber slide or on the polyethylene catheter. MICs of linezolid, vancomycin, oxacillin and lysostaphin for planktonic bacteria were determined according to the standards of NCCLS. BIC/BEC (Biofilm Inhibitory/Eradicating Concentration) were estimated by the MTT assay measuring active metabolism of bacteria that survived a single or cyclical dose of antimicrobials (antibiotic alone or in combination with subMIC lysostaphin given earlier or together with antibiotic). The integrity of biofilm treated with antimicrobials was also examined: in catheter study: visually by TTC reduction assay, in chambers of a LabTekII chamber slide: by staining with FITC and confocal fluorescence microscopy.
Results: We have demonstrated the effectiveness of lysostaphin in the treatment (3–24 hours) of biofilm built not only on the flat surface (wells of microplate, chambers of a LabTekII chamber slide) but also on catheter extra- and intraluminal surfaces. MIC/BIC of lysostaphin were 0.25, 8/16 mg/L for MSSA and MRSA, 0.06, 4/8 mg/L for hVISA, respectively. The synergistic effect of subMIC lysostaphin + oxacillin was observed for MSSA and MRSA biofilms (BICoxa/Lin dropped after 24 h from >128 to <4 mg/L) but such effect was not demonstrated for hVISA strain (BICOxa was still >128 mg/L). SubMIC lysostaphin + -linezolid or + vancomycin, given as 3 cycles therapy, were effective in disruption of MSSA, MRSA, hVISA or MSSA and MRSA biofilms, respectively. Thus, lysostaphin in combination with antibiotics, was effective in preventing the biofilm rebuilding (BEC) in experiments lasting 5 days.
Conclusions: Our study indicates that staphylococcal biofilm eradication could be achieved by treatment with lysostaphin alone or in combination with selected antibiotics. However, complete biofilm removal needs the cyclical therapy Supported by Grant 3PO4C08124 (KBN)
P1649: Zinc concentration and porin profile of P. aeruginosa forming biofilms on siliconized latex urinary catheters
M.C. Conejo, I. García, L. Picabea, á. Pascual (Seville, E)
Objective: Zinc compounds are used in the siliconized latex urinary catheters (SLUCs) manufacture. Studies in vitro have shown that Zn elutes from SLUCs and determines decreased susceptibility to carbapenems in planktonic Pseudomonas aeruginosa. This effect is associated with OprD loss. P. aeruginosa is able to adhere efficaciously to the surface of SLUC forming biofilms. The objective of this study was to evaluate the relevance of this phenomenon in P. aeruginosa forming biofilms on SLUCs.
Methods: Biofilms of P. aeruginosa PAO1 were grown on 1 cm SLUCs segments in Mueller–Hinton broth for 24 hours. Bacterial biofilms were detached by sonication and Zn accumulation in biofilms was determined by inductively coupled plasma atomic emission spectrometry (ICP-AES). SLUC segments without biofilms were used as control. To study the outer membrane proteins (OMPs) of bacterial biofilms, cells were disrupted by sonication (40 kHz, two minutes in a sonication water bath) and OMPs were isolated as sodium-lauryl sarcosynate insoluble material and the proteins were resolved by SDS-PAGE. OMP pattern was compared to that expressed by planktonic bacteria grown in Mueller–Hinton broth supplemented or not with Zinc Chloride (40 mg of Zn/L).
Results: P. aeruginosa PAO1 SLUC biofilms did not accumulate Zn. Zn values reached by bacterial biofilms (1.67 ± 0.32 μg/g of catheter) were similar to that those obtained with the control (1.88 ± 0.27 μg/g of catheter). P. aeruginosa PAO1 forming biofilms lost OprD2 and expressed OprD3, showing the same OMP profile as planktonic bacteria in the presence of Zn.
Conclusion: P. aeruginosa PAO1 SLUC biofilms did not expressed OprD, the porin responsible for carbapenem entry into P. aeruginosa. Zn eluted from SLUCs did not accumulate by PAO1 biofilms.
P1650: Diversity in adhesion, initial biofilm formation and motility abilities among clonal lineages of nosocomial strains of Pseudomonas aeruginosa
A.P. Fonseca, P. Correia, A.F. Fonseca, R. Tenreiro, J.C. Sousa (Porto, Lisbon, P)
Objectives: The objective of this study was to test for potential constraints of genotypes on several virulence parameters and to examine the extent of phenotypic diversity in the context of clonal lineages (CLs) among nosocomial strains of P. aeruginosa.
Methods: A combination of two genomic typing systems, the minisatellite-primed PCR (MSP-PCR) and the enterobacterial repetitive consensus sequence PCR (ERIC-PCR), was used to discriminate the 96 strains collected at a Portuguese Central Hospital and isolated from 5 different sources. The data was analysed using BioNumerics software. Bacterial adhesion to polystyrene (PS2h) and initial biofilm formation (PS6h) were studied by growing bacteria in LB in a modified microtiter-plate assay. Strains were screened for their capacity to adhere to hydrophobic biomaterials such as silicone and hexadecane (CSH) using a biphasic separation method. The opportunistic P.aeruginosa were also screened for their capacity to swim (flagella) and twitch (pili). All tests were run in triplicate.
Results: Our combined genotypic and phenotypic analysis revealed extensive diversity in all the parameters within CLs of P. aeruginosa strains and no significant differences among CLs. However, small clusters of strains with relatively high PS2h and PS6h abilities can be found; one specific group containing 2 strains of CL1 (similarity 70%) are worth mentioning; a group of 2 strains from CL7 (similarity 60%) had low PS2h and PS6h abilities. Positive correlations (Spearman's) were observed between PS2h and PS6h (0.748, p < 0.01), between PS2h and CSH (0.233, p < 0.05), between PS6h and CSH (0.228, p < 0.05) and between PS6h and twitching (0.220, p < 0.01). The strain with highest adhesion value was one of the most efficient initial biofilm producer and other strain had the same behavior but had the lowest value. The strain with highest CSH was also one of the most efficient in adhesion to silicone and the strain with lowest adhesion was the same for the two parameters.
Conclusion: Our results suggest extensive diversity, namely by wide variations in virulence parameters among strains within a CL and no significant differences between CLs in all the virulence parameters. These results also suggest the importance of hydrophobicity in adhesion ability to abiotic surfaces, the importance of twitching in biofilm formation and the correlation between PS2h and PS6h ability.
Acknowledgments: This study was supported by Fundação Calouste Gulbenkian.
P1651: Comparative study of antibiotic susceptibility levels of some clinical strains of P. aeruginosa as planktonic and adherent growing in biofilms cells
V. Lazar, M. Balotescu, M. Bucur, O. Banu, R. Cernat, D. Bulai (Bucharest, RO)
Background: Besides the well known genetic antibiotic resistance of Pseudomonas aeruginosa strains, the growing biofilms on medical implants are responsible for persistent infections, being much less susceptible to antimicrobial agents than are their planktonic counterparts. This recalcitrance could be due to self-aggregation and bacterial adherence to different substrata, generation of a protecting exopolysaccharidic mathrix limiting the antibiotic diffusion in the biofilm, underexpression of porins, overexpression of multidrug resistance pumps, the presence of persisters that survive in the presence of an antibiotic that inhibits their growth. For highlighting some of the above-mentioned factors, we have investigated the antibiotic resistance levels of P. aeruginosa planktonic cells comparatively with cells growing in biofilms developed on central venous catheter pieces imersed in nutrient broth (original experimental model) or included in agar mimicking the biofilm mathrix.
Methods: The study was performed on some P. aeruginosa strains isolated from central venous catheter related infections in patients admitted for cardiovascular surgery. The strains were firstly submitted to antimicrobial testings performed by standard disk diffusion method and 25 strains susceptible to aminoglycosides, beta-lactams and quinolones were further tested by two experiemental models for antibiotic susceptibility of bacterial cells included in biofilms formed in liquid and solid media. The strains were also studied for slime production and for cell hydrophobicity.
Results: Our results showed that the P. aeruginosa cells growing in biofilms are significantly less susceptible to antibiotics, in a dose-response (increased MICs and MBCs) and time-dependent manner. This aspect was particularly evidenced and statistically confirmed for aminoglycosides, which poorly penetrate the biofilm, due to their policationic structure. The majority of the tested strains produced high amounts of slime, evidenced after safranin staining of the biofilm developed on glass tube walls, expressed both at 37°C and 4°C and well correlated with the presence of bacterial capsule. All the tested strains exhibited high hydrophobicity levels evidenced by decreased absorbance values of the aqueous phase of bacterial cultures in the presence of paraxilen and also by the ability to self-aggregate, raising the risk of colonizing the inert materials and of catheter related persistent infections.
P1652: Biofilm formation of nosocomial Acinetobacter baumannii strains
F. Can, O. Kurt Azap, M. Demirbilek, F. Timurkaynak, G. Karabay, H. Arslan (Ankara, TR)
Acinetobacter baumannii an important nonocomial pathogen, usually found on various surfaces of the hospital environment. Biofilm formation is a virulence factor for A. baumannii. In this study, 17 blood isolates of A. baumannii were studied for the ability to form biofilms on the surface of polystyrene and ultrastructural cell wall properties of biofilm-formatted strains were evaluated.
Methods: A total of 17 A. baumannii strains isolated from blood cultures of different patients in intensive care units between 2003 and 2004 were selected. The ability of Acinetobacter strains to form biofilm was determined on polystyrene microtiter plates using Brain Heart Infusion supplemented with 0.25% Glucose as a growth medium. Additionally, transmission electron microscopy of strains was performed for the presence of fimbria, cell wall thickness and, an amorphous material cover the cell described for some other bacteria.
Results: Biofilm formation was detected in 9 (52.9%) of A. baumannii strains. Ultra structural analysis showed that fimbriae were found all of the isolates while the thickest layers of amorphous material was observed around the biofilm-positive strains.
Conclusion: Biofilm formation of A. baumannii strains from patients with bacteriemia was found high. The results obtained in electron microscopy studies suggested that amorphous material covers the bacteria probably plays an important role of the maintenance of the bacterium in the hospital environment and protect it against the defense mechanisms of the host.
P1653: Strains of Enterobacter cloacae isolated in hospital environment: ability to adherence to Hep-2 cell line and to biofilm forming
K.W. Pancer, S. Fila, A. Trzcinska, A.E. Laudy, T. Wernik, E. Mikulak, A. Gliniewicz, H. Stypulkowska-Misiurewicz (Warsaw, PL)
Bacteria Enterobacter cloacae were observed as etiological agents of nosocomial infections in Poland, especially hospital-acquired pneumonia (9–11%) and urinary tract infections (3–7%). Moreover the majority of E. cloacae strains isolated from patients with HAP, hospitalized at ICUs showed resistance to ceftazidime. The aim of our study was the examination of virulence properties like: adherence to human cell line and forming biofilm of 15 strains of E. cloacae isolated from patients (infection or colonisation) and of 11 strains isolated from the body surface of German cockroaches collected in hospitals. The examination of ability to adherence to human cell line (Hep-2) was done according Knutton et al. method. Three patterns of adhesion were observed: A. aggregative; B. localized and C. diffuse adhesion. Moreover determination of activity of 3 selected chemical disinfectant to bacteria growing as a planctonic form (by evaluation MICs) and growing as a biofilm on catheter (effectiveness of working solution) was done. Patterns of adhesion to Hep-2 cell line varied among tested strains. The aggregative adhesion was observed among 2 strains of E. cloacae colonising patients and one isolated from infections. No one of 11 E. cloacae strains isolated from the surface of cockroaches showed this pattern of adhesion. According to statistical analysis the significant number of E. cloacae strains isolated from cockroaches presented lack of adhesion to Hep-2 cell line or diffuse adhesion in comparison to strains collected from patients. Totally 65% E. cloacae strains showed mannose-sensitive mechanism of adherence to Hep-2 cell line (71% of strains from the colonised patients, 62.5% of strains that caused infection and 63.6% of strains from the surface of cockroaches). 27% of the strains showed stronger adhesion in presence of mannose. Only one E. cloacae strain did not form the biofilm on catheter. The majority of tested strains growing 5 days on catheter were resistant to working solution of potassium persulfate, glucoprotamin and sodium dichloroisocyjanurate. However, E. cloacae strains that caused infection were the only resistant to all 3 disinfectants working solution, even if the MIC values were similar to the MICs of these strains from patient colonisation or from cockroaches. The study on virulence markers presented by E. cloacae hospital strains should be continued.
P1654: The development of crystalline Proteus mirabilis biofilms on Foley catheters
S.D. Morgan, D.J. Stickler (Cardiff, UK)
Objective: The management of many patients undergoing long-term bladder catheterisation is complicated by the encrustation and blockage of their catheters. The problems result from infection by urease producing bacteria. The organisms colonize the catheters, the urease induces alkaline conditions under which calcium and magnesium phosphates precipitate and the resulting crystalline biofilm blocks the flow of urine. The aim of this study was to investigate the early stages of biofilm formation on a range of catheter types.
Methods: A laboratory model of the catheterised bladder was supplied with urine and infected with Proteus mirabilis NSM6, a clinical isolate from an encrusted catheter. Models were fitted with five different types of catheters. After incubation for various periods up until the times they blocked, catheters were removed from the models for examination. Scanning electron microscopy and X-ray microanalysis were used to follow the colonization of catheters by bacteria and crystalline material.
Results: All-silicone, hydrogel-coated latex and silicone-coated latex catheters all acquired a layer of crystalline material within 4 h. These deposits consisted predominantly of calcium phosphate. Subsequently, they become colonized by large numbers of bacilli and the rapidly developing crystalline biofilm blocked the catheters within 12.5 h-42.5 h. In the case of a nitrofurazone-impregnated all-silicone catheter, although the initial process was slower, with the crystalline layers being first observed at 12 h, catheter blockage occurred at 29 h. Triclosan-impregnated silicone catheters however, showed little sign of encrustation or biofilm formation and were still draining urine freely at 7 days when the experiment was terminated.
Conclusion: The catheters currently available for long-term bladder management were rapidly colonized by crystalline P. mirabilis biofilm. Triclosan-impregnated catheters however were able to resist encrustation and biofilm formation in vitro for up to 7 days.
P1655: The role of capsules in Klebsiella pneumoniae biofilm formation
C. Struve, S. Molin, K.A. Krogfelt (Copenhagen, DK)
Objectives: K. pneumoniae is an important opportunistic pathogen and frequent cause of hospital infections. K. pneumoniae cells are characteristically surrounded by a pronounced polysaccharide capsule, which has been proven to be an important virulence factor for the bacteria. The ability of bacteria to form biofilms is recognized to play a role in the pathogenicity of many bacterial species. In biofilms bacteria are organised in microcolonies embedded in a matrix of exo-cellular polymeric substances (EPS). The EPS is considered to stabilize the biofilm structure and protect the bacteria against host defence mechanisms and the actions of antibiotics. In this study we investigate the role of the polysaccharide capsule on K. pneumoniae biofilm formation.
Methods: The gene-cluster encoding the synthesis of the K16 capsule was cloned and sequenced from the clinical K. pneumoniae isolate 3091, and an isogenic non-capsulated mutant was constructed. The wildtype and mutant were tagged chromosomally with fluorescent markers and grown in continuous flow-cell systems to characterize their biofilm formation using confocal laser scanning microscopy.
Results: The wildtype stain was found to form thick irregularly shaped biofilms. The biofilms mainly consisted of loose clusters of bacteria, up to 40 μm thick. Although the biofilm covered most of the substratum, open areas were also present within and between the bacterial clusters. The non-capsulated mutant was also able to form biofilms; however the architecture differed from that of the wildtype. The non-capsulated mutant spread as a confluent thin layer of cells covering the substratum. The vast majority of the biofilm was less than 5 μm thick, and only occasionally thicker bacterial clusters resembling the wildtype biofilm phenotype were observed.
Conclusions: K. pneumoniae cells are able to form thick biofilms when grown in continuous flow systems. The polysaccharide capsule was not found to be essential for biofilm formation per se but its absence significantly affected biofilm development and architecture. Future studies may reveal the influence of capsules on biofilm resistance against host defence systems and actions of antibiotics.
P1656: The biofilm formation by Klebsiella strains isolated from various infections on chemically different catheters
B.M. Maczyñska, D.S. Smutnicka, A.P.M. Przondo-Mordarska, M.B. Bartoszewicz, U.K. Kasprzykowska (Wroclaw, PL)
Objectives: Klebsiella bacilli are frequently cases of nosocomial infections associates with usage urinary (CAUTI) and intravenous catheters (CR-BSI) due to its ability to biofilm formation on biomaterials. The aim of our study was the correlation between pathogenic properties in clinical Klebsiella strains (the capsule, the types of adhesins), the type of using biomaterials and degree of in vitro formation the biofilm structure on the catheters.
Material and Methods: The 69 clinical Klebsiella strains producing several distinct types of fimbria were tested (30 isolated from UTI, 23 from blood and 16 from other clinical samples). Influence of capsule on ability of the biofilm formation was estimated using uncapsulated mutants and their capsulated variants. In the investigations the urinary catheters-polyvinyl chloride Nelaton, latex Foley, all-silicone Foley and venous catheter-polyurethan Cavafix were tested. In vitro biofilm formation was evaluated by Richard method. In this assay, soluble colourless TTC is reduced to insoluble red formazan by electron transfer associated with active oxidative bacterial metabolism and is precipitated intracellulary. This assay were validated by electron microscopy.
Results: All investigated strains showed the ability of formation of the biofilm on the catheters after 24 hours incubation. For most of strains the value of fomazan reduction were evaluated like as intermediate or strong. The highest level of biofilm formation showed the strains to venous catheters from polyurethan and urinary catheters from polyvinyl chloride. The lowest adhesion rate was observed in case of all-silicone Foley catheters. The capsulated strains made the biofilm weaker then the uncapsulated strains. The presence of the characteristic biofilm structure on catheters was confirmed in electron microscopy.
Conclusions: The level of adhesion in clinical Klebsiella bacilli to biomaterials was associated with the chemical type of catheters. The capsule absence leads in case of investigated strains to the stronger biofilm formation on the surface of biomaterials. The various Kebsiella strains showed in electron microskopy various structures of biofilms The correlation between the type of expressed fimbria and degree of biofilm formation was not observed in this investigation.
Acknowledgments: This study was supported by the Polish Committee of Scientific Studies (KBN Grant No 4 P05A 064 24).
P1657: Biofilm formation in uropathogenic Escherichia coli clinical isolates
S.M. Soto, J.A. Martínez, A. Smithson, J.P. Horcajada, J. Mensa, M.T. Jiménez de Anta, J. Vila (Barcelona, E)
Objectives: Biofilm is composed of surface-attached cells and excreted exopolymeric substances. Bacterial biofilm infections are particularly problematic because sessile bacteria are more resistant to antibiotics and host immune responses, leading to therapeutic difficulties. The objective of this study was to analyse the capacity of biofilm formation of E. coli clinical isolates causing different urinary tract infections (cystitis-CT, pyelonephritis-PNA, and prostatitis-PT).
Methods: A total of 151 uropathogenic E. coli strains (44 from cystitis, 75 from pyelonephritis, and 32 from prostatitis) were analysed. Capacity of ‘in vitro’ biofilm formation was determined by bacterial growth in minimal glucose medium, stained with 1% violet crystal, and washed with PBS. The cells were dissolved with DMSO and the plate was automatically read in a spectrophotometer. The HC-91255 strain was used as a control strain and the experiments were made in duplicate.
Results: Biofilm formation was noted in 19 (43%), 30 (40%), and 20 (63%) strains from patients with CT, PNA, and PT, respectively. The ability to form biofilm was significantly more frequent in PT than in PNA strains (p = 0.03), and showed a trend to be significantly more frequent than in CT strains (p = 0.09). No differences between CT and PNA strains regarding biofilm production were noted (p = 0.7). After pooling CT and PNA strains together, biofilm formation was the only characteristic which was significantly associated with strains causing PT in both univariate and multivariate analysis (OR = 2.38, 95% CI = 1.06–5.35, p = 0.03).
Conclusions: Biofilm formation is significantly more prevalent among strains of E. coli involved in acute prostatitis than in those causing other urinary tract infections. Biofilm formation can be relevant in promoting persistent prostate infection; and given the high frequency of biofilm production among strains causing acute prostatitis, the treatment of these patients should be conducive to the eradication of microbial biofilms.
P1658: Environmental maltose and glucose concentrations regulate biofilm formation in Enterococcus faecalis
J. Huebner, A. Kropec, S. Koch, R. Creti (Freiburg, D; Boston, USA; Rome, I)
Objectives: Biofilm formation in E. faecalis seems to play an important role in a number of enterococcal infections, but little is know regarding the underlying mechanisms. We wanted to identify environmental signals that regulate biofilm formation in enterococcus.
Methods: We have recently identified a genetic locus involved in biofilm formation of E. faecalis. Biofilm formation was measured using a polystyrene plate assay and media supplemented with oligosaccharides. The expression of genes was measured by real-time PCR and the locus was sequenced. Homology was assessed using BLAST and related programmes.
Results: The genetic locus identified by us is also involved in maltose uptake and metabolism. The expression of genes in this locus is regulated by maltose and glucose in the growth medium indicating that nutritional oligosaccharides may promote or inhibit biofilm formation. While the wild-type strain was able to produce biofilm in medium containing glucose or maltose, two mutants (one transposon mutant and one deletion mutant) showed opposite effects. A transposon mutant showed a reduced biofilm formation when grown in medium containing 1% glucose while a deletion mutant produced more biofilm when grown in glucose, but was unable to form biofilm when maltose was added to the growth medium. The sugar-binding transcriptional regulator bopD seems to bind to an operator upstream of the bop locus and a consensus binding sequence shown to bind to the maltose repressor MalR is present in the non-coding region upstream of bopA.
Conclusions: The biofilm-positive phenotype of the wildtype strain seems to facilitate colonization of enterococci in the gut and the presence of oligosaccharides in food may regulate biofilm formation and therefore colonization of enterococci in the gastrointestinal system.
P1659: Biofilm formation and morphology of group A streptococci isolates of patients with sepsis or necrotising fasciitis
E. Presterl, M. Eder, S. Reichmann, R. Gattringer, A.M. Hirschl (Vienna, A; Golm, D)
Objectives: Group A streptococci may cause severe sepsis associated with multiorgan failure and high mortality. Additionally, they are the primary cause of necrotizing fasciitis. The objective of this study was to evaluate the ability to form biofilms and the morphology of these biofilms produced by GAS isolated from blood of patients with sepsis and from the tissue of patients with necrotizing fasciitis.
Methods: Twenty-six GAS isolates from blood cultures of patients with sepsis and bacteraemia, and 26 isolates from tissue of patients with necrotizing cellulitis and fasciitis were collected. They were isolated and identified using routine methods, and frozen at −73°C in BHI broth with 3% glycerol. To test their ability to form biofilms, the isolates were grown in microtiter plates in tryptone soya broth for 24 or 48 hours. To examine the morphology of the biofilm formation the isolates were either examined unfixed or fixed with 2% glutaraldehyd using electrone microscope scanning (Philips XL30 ESEM). For quantification they were fixed with 2% glutaraldehyd and dyed with 1% crystal violet to measure the mean optical density (OD using a routine microtiter-plate-reader at 550 nm wavelength. To calculate differences in the frequency in the groups the Fisher's exact test and to calculated differences in the OD the Mann–Whitney U-Test were used.
Results: Seventeen out of 26 isolates from patients with sepsis and 14 out of 26 isolates fom patients with necrotizing fasciitis formed biofilms (difference not significant). Scanning electron studies confirmed biofilm formation however it looked somewhat scarce in isolates from necrotizing fasciitis. For the biofilm forming isolates, the OD of isolates from sepsis were significantly higher than the OD of isolates from necrotizing fasciitis (0.996 versus 0.578, p < 0.001).
Conclusions: GAS isolates from both, bloodstream and tissue, had ability to form biofilms. Biofilms of blood stream isolates were significantly denser than biofilms of the tissue isolates. This surplus opacity of blood stream isolates was confirmed by the scanning electron microscopy studies which revealed a more scanty looking morphology of the tissue GAS isolates. The lesser density of the biofilms of GAS isolated from the tissue might be important for the pathogenesis of necrotizing fascitiis probably facilitating to the rapid spread of the bacteria within the tissue.
P1660: Inhibition of Candida growth and biofilm formation on polyurethanes by fluconazole adsorption
G. Donelli, I. Francolini, V. Ruggeri, E. Guaglianone, A. Piozzi (Rome, I)
Objectives: Recent attempts to prevent device-related infections included several strategies among which catheter coating with antibiotics resulted to be one of the most promising. However, so far only sporadic studies were designed to prevent fungal colonization of devices presumably because of the only recently described ability of Candida species to form biofilms. In this study we report in vitro experiments on the efficacy of coating of newly synthesized polyurethanes with the antifungal drug fluconazole in preventing polymer colonization and biofilm formation by Candida albicans.
Methods: Polymers used in this study are three synthesized urethane polymers having different functional groups in the side-chain: hydroxyl groups, primary amino groups and tertiary amino groups. Fluconazole was adsorbed on round shaped disks made of the above described polyurethanes. The kinetics of fluconazole release from polymers, either containing or not albumin as pore forming agent, was studied by keeping fluconazole-loaded polymeric disks in water for increasing times up to 8 days. The antifungal activity of polymers was studied by the Kirby–Bauer test and scanning electron microscopy.
Results: Among the tested polymers, the most hydrophilic ones were able to adsorb higher drug amounts by establishing ‘hydrogen bond’ and ‘van der Waals’ interactions. The kinetics of fluconazole release from polymers was influenced by the degree of polymer swelling in water and resulted significantly improved by the albumin incorporation in polyurethanes which increased polymer porosity. In our best experimental in vitro model consisting of an hydrophilic polymeric disk (average weight 250 mg) impregnated with 62.5 mg albumin and 62.5 mg fluconazole, the Candida albicans growth was inhibited, as evidenced by the Kirby–Bauer test, and biofilm formation on polymeric surface was not observed up to 8 days, as evidenced by scanning electron microscopy.
Conclusion: Overall, data obtained from our newly synthesized functionalized polyurethanes, treated with albumin and loaded with fluconazole, seem to be very promising in the perspective to develop medical devices refractory to Candida colonization.
P1661: In vitro activity of caspofungin against Candida biofilms
C. Cocuaud, M.-H. Rodier, G. Daniault, C. Imbert (Poitiers, F)
Candidiasis can be associated with the formation of biofilms on bioprosthetic surfaces and the intrinsic resistance of Candida albicans biofilms to the most commonly used antifungal agents has been demonstrated.
Objectives: In this study, we report on the antifungal activity of two concentrations of caspofungin on C. albicans and C. parapsilosis biofilms with different ages of maturation.
Methods: Fifteen strains of C. albicans (ten strains were susceptible to fluconazole in vitro and five strains were resistant to this antifungal agent) and six strains of C. parapsilosis (all were susceptible to fluconazole in vitro) were studied. The antifungal activity of caspofungin was assessed by looking for a significant inhibition of the metabolic activity of yeasts included in biofilms, after the antifungal treatment. Biofilms were obtained in vitro, on silicone catheters.
Results: Caspofungin used at MIC did not modify the metabolic activity of yeasts, whatever the Candida species and the maturation age of biofilms. The use of a therapeutic concentration of caspofungin (2 mg/L) induced a significant decrease in the metabolism of the all tested strains of C. albicans and C. parapsilosis, independently of the maturation age of biofilms. This high antifungal potency of caspofungin on C. albicans biofilms was observed independently of the susceptibility of yeasts to fluconazole.
Conclusion: This study demonstrated that caspofungin used at MIC was not efficient to reduce Candida biofilms but it suggested that caspofungin used at 2 mg/L could represent a good Candidate in the prevention of candidiasis associated with silicone medical devices. Our results also suggested that fluconazole resistance of yeasts did not modify caspofungin activity.
P1662: Effect of short-time desiccation on biofilm-associated bacteria
I. Turetgen, E.I. Sungur, A. Cotuk (Istanbul, TR)
Objectives: Microorganisms tend to form biofilms consisting of cells embedded in a highly hydrated, extracellular polymeric matrix. Biofilms may be responsible for a wide variety of nosocomial infections. Sources of biofilm-related infections can include the surfaces of catheters, medical implants, dental unit water lines or other types of devices, as well as cooling towers, shower heads. The biofilm protects its inhabitants from antimicrobial agents, pH alterations, and confers protection against desiccation. It is possible to encounter desiccation in natural or man-made environments through evaporation, flowing-off or system shutdown. However biofilm-associated bacteria can survive for a while in the absence of water within systems. In this study the survival of heterotrophic bacteria, sulphate reducing bacteria (SRB) and amoeba were evaluated against short time desiccation.
Methods: Biofilms were allowed to grow for 30 and 60 days on stainless steel coupons. For the desiccation experiment, coupons were taken out from the reactor and left air-dried. Desiccated cells were rehydrated in 10 ml of sterile phosphate buffer after 6, 24, 48, 72, 96, 168 hours and biofilm were removed from coupons. For enumeration of heterotrophic bacterial count, samples are plated on R2A agar for 10 days at 27°C. Postgate medium B was used for SRB cultures. For amoeba isolation biofilm homogenates were spread on non-nutrient agar plate overlaid with Eschericha coli.
Results: After 72 h of desiccation of 30 d old biofilm, SRB growth was seen while no heterotrophic bacterial growth was observed. Whereas on 60 d old biofilm, heterotrophic bacterial growth was observed after 96 h, however SRB growth was seen after 168 h. No significant differences were found between zero time and 24 h counts regarding heterotrophic bacteria.
Discussion: Heterotrophic bacteria are located at the biofilm-water interface; whereas anaerobic bacteria niched in the deeper layers of the biofilm. The ability of some pathogenic bacteria to survive within the cysts of amoebas is suggested as a possible mechanism by which the organism evades disinfection and spreads to colonize new environments. Bacterial resistance to desiccation has been reported frequently, but the maximal tolerance of microorganisms in the air-dried state is still unknown. The study showed that diurnal absence of water could not affect biofilm-associated microorganisms significantly.
P1663: Effects of food preservatives on oral streptococci biofilm generated in a biofilm reactor
A. Al-Ahmad, T.M. Auschill, K. Kobberstad, G. Braun, M. Sendic, E. Hellwig, N.B. Arweiler (Freiburg, D)
Objectives: The aim of this laboratory study was to evaluate the influence of the preservatives sodium benzoate, potassium sorbate and sodium nitrite on mixed species biofilms generated by oral streptococci.
Methods: A modified biofilm reactor (Bio Surfaces Technologies, Corp., Bozeman, USA) was used to build up a biofilm on bovine enamel slabs (BES). A mixture of six oral streptococci (S. mutans, S. mitis, S. oralis, S. sanguis, S. salivarius and S. sobrinus) was investigated to generate biofilms over a period of four days. Eight slabs each (with the adhering biofilms) were treated with one of the following solutions for one minute: a) Saline (0.9%, negative control), b) 0.2% chlorhexidine digluconate (CHX, positive control), c) 0.1% sodium benzoate (SB), d) 0.1% potassium sorbate (PS) and e) 0.06 sodium nitrite (SN). After treatment four slabs each were washed in saline and a vital-dead staining was conducted on the adhering biofilms. The slabs were then analysed in situ for biofilm thickness and vitality using confocal laser scanning microscopy (CLSM). From the other four slabs biofilms were dispersed in saline and the colony forming units (CFUs) on Columbia blood agar as well as total bacterial counts using DAPI-staining were determined.
Results: The biofilm thickness of the different tested BES was between 12.3 and 35.8 μm in average. The vitality of the negative control was determined to be 86.14 ± 4.6% and 37.27 ± 8.11% for positive control. For sodium benzoate and potassium sorbate a vitality of 74.39 ± 8.08% and 71.11 ± 6.93% was measured, respectively. The treatment with sodium nitrite led to a vitality of 66.73 ± 6.36%. The mean log10 CFUs of untreated biofilm was determined to be 7.83 ± 0.14 and 7.28 ± 0.33 for the biofilm treated with CHX. After treatment with SB and PS the mean log10 CFUs was 7.20 ± 0.20 and 6.98 ± 0.24, respectively. For biofilm treated with SN a mean log10 CFUs was determined to be 7.12 ± 0.40. The plating efficiency was determined as followed: 56.16 ± 8.78 for the negative control, 24.97 ± 13.30 for the positive control, 22.89 ±11.71 for SB treated biofilm, 22.82 ± 13.19 for PS treated biofilm and 19.06 ± 10.81 for SN treated biofilm.
Conclusions: Our results indicate that legal concentrations of the tested preservatives may have antibacterial effects on oral streptococci. An anti-cariogenic influence of food preservatives can be suggested and should be confirmed in in situ studies using intraoral biofilms.
P1664: Dynamics of biofilm formation as a function of temperature and nutrition
V. Hola, F. Ruzicka, M. Votava (Brno, CZ)
Chronic infections caused by biofilm-forming staphylococci represent serious medical problem. Their higher prevalence now-adays is associated with more frequent use of artificial implants and medical devices. Surface of these implants facilitates adhesion of bacteria, which than form biofilm. The knowledge of biofilm formation dynamics is fundamental for the understanding of processes running in the biofilm layer. The aim of this study was to determine with mathematical models the differences in biofilm formation in different conditions and to determine the minimum time and conditions necessary for the development of the homogenous and matured biofilm layer suitable for the antibiotic testing. The Staphylococcus epidermidis biofilm-positive strains were used in this study. As the positive control, strain CCM 7221 was used. The biofilm was grown on tissue culture microtiter plates. The strains were cultivated in different concentrations of glucose and at different temperatures. Each strain was cultivated simultaneously in 4 wells. In the first 48 hrs the data were collected every hour, than up to 96 hrs every 6 hrs. After cultivation the wells of microtiter plates were stained with crystal violet and the biofilm formation was assessed spectrophotometrically. Data were processed with the Multiple Analysis of Covariance (MANCOVA) and mathematical models were evaluated with Regression Analysis. For the image analysis and visualisation of the biofilm in different stages of formation, specific dyes (alcian blue, acridin orange, Rylux etc.) were also used. All tested strains showed better growth of the biofilm at the temperature of 37°C in the nutrient-richer environment. The model can be simply described as follows: the first signs of bacterial adhesion were visible after 2–4 hrs of cultivation, the first homogenous but a very thin layer was visible after 5 hrs, after 7 hrs the biofilm layer was cca 3-times thicker. After 10 hrs the biofilm layer seemed to be mature–the changes in the thickness were not so evident after this time. After the cultivation longer than 34–42 hrs the parts of the biofilm layer started to detach, so the biofilm became unhomogenous. In the regression modelling, other nutrient and temperature conditions showed also different influence on the formation of biofilm. For the antibiotic testing, the biofilm cultivation for 12 hrs should be sufficient. The study was supported by the grants IGA MZ 7980-3 and FRVS 0448-2004.
P1665: Electron microscopic, bacteriological and clinical studies of biliary stent blockage
A. Yosry, H. Yehia, M. Diab, H. El-Khayat, A. Negm (Cairo, Giza, EGY)
Objective: Biliary stenting is a common treatment for the palliation of malignant obstructive jaundice. However, stent occlusion due to sludge adhesion remains an important clinical problem. For more understanding of the mechanism of such occlusion, the present work dealt with studying the bacteriological characteristics of bile samples.
Methods: Taken from 40 patients with extra hepatic cholestasis before biliary decompression(group1). Also, bile samples and sludge material from13 patients presenting with recurrent jaundice due to late stent blockage (group2), were processed for microbiological and transmission electron microscopic study (TEM). Bacteriological analysis included isolation and identification of various bacterial species as well as detection of B-glucouronidase and several esterase activities (19enzymes)of the isolated species.
Results: Bacteriological analysis disclosed a wider prevalence of monomicrobial Gram negative isolated (80.7%) than Gram positive isolates (19.3%) among patients of group1. The TEM examination of sludge material (group2) revealed the presence of clusters of Gram negative bacteria embedded in an amorphous matrix. The visualized microcolonies of bacterial growth were confined to the same morophotype for each case. Bacteriological analysis confirmed the TEM results and showed that the most frequent isolated species were Escherichia coli(E.coli) (41.6%), Samonella arizona (S. arizona) (25%) Klebsiella species (16.6%) and Enterobocter(8.3%) Although S. arizona isolated from blockage stents were the most abundant producers (14 out of 19) and they elaborated 4 out of 5 such enzymes, yet they recorded the longest patency duration among the blocked tudied stents. By TEM it was noticed that the biofilm matrix changed according to the type of bacteria detected. The biofilm matrix in S. arizona cases was shown to be fenestrated. It looked thick and more etectron dense when compared to the matrix of E. coli cases. The latter showed a clealr thin matrix, more electron lucent and detected fibres were mostly thin.
Conclusion: The present work confirms the pivotal role of Gram negative bacteria in stent blockage therefore antibiotics against Gram negative bacteria are mandatory 24 hours before endoscopic retrogade cholangiopancreatography (ERCP). Also, the current study highlights the relation between the type of microorganism and the morphology of the formed biofilm matrix which may be an important underlying factor in stent blockage and its duration of patency.
P1666: Lethal biofilm formation in a biliary stent
H. Krogh Johansen, N. Høiby (Copenhagen, DK)
Objective: Medical implants are widely used in a variety of clinical treatments, but they frequently become colonised by opportunistic bacteria, which form biofilm on the device surface. We describe this medical problem in a 57-yr old alcoholic man who had a biliary stent inserted because of intra- and extrahepatic bile stasis, probably caused by a cystic process in the pancreatic head that blocked the flow.
Methods: A 7 cm long teflon coated polyurethane biliary stent was inserted endoscopically in the common bile duct. Antibiotic prophylaxis or treatment was not used and the patient was discharged in healthy condition after 5 days.
Results: Two weeks after insertion of the biliary stent the patient was readmitted with sepsis, severe abdominal pain and increasing liver enzymes. Ultrasound scan revealed intrahepatic bile stasis, a large inflamed abscess in the pancreatic head, a fistula between the abscess and the common bile duct and blockage of the lumen of the stent. The stent was replaced by two 10 cm long stents. The patient was treated with ampicillin, metronidazole and gentamicin for only 3 days. Ampicillin susceptible E. coli were cultured from the blood. After 5 days the patient was discharged in healthy condition. Three weeks after the second stents were inserted the patient was admitted emergently with septic shock and treated with ceftriaxone, netilmicin and metronidazole. After 12 hours of intensive treatment the patient died. E. coli and K. oxytoca were cultured from the blood, pancreas, and the stents. Pulsed-field-gelelectrophoresis was performed and the E. coli cultured during the two septic episodes had the same pattern. This indicates that the bacteria escaped the brief antibiotic treatment probably because of biofilm formation.
Conclusions: This case underlines the need for antibiotic prophylaxis when bile stents are inserted into infected areas and continuous antibiotic therapy when patients with a medical implant experience a septic episode.


