Abstract
An ongoing outbreak of severe respiratory pneumonia associated with the 2019 novel coronavirus has recently emerged in China. Here we report the epidemiological, clinical, laboratory and radiological characteristics of 19 suspect cases. We compared the positive ratio of 2019-nCoV nucleic acid amplification test results from different samples including oropharyngeal swab, blood, urine and stool with 3 different fluorescent RT-PCR kits. Nine out of the 19 patients had 2019-nCoV infection detected using oropharyngeal swab samples, and the virus nucleic acid was also detected in eight of these nine patients using stool samples. None of positive results was identified in the blood and urine samples. These three different kits got the same result for each sample and the positive ratio of nucleic acid detection for 2019-nCoV was only 47.4% in the suspect patients. Therefore, it is possible that infected patients have been missed by using nucleic acid detection only. It might be better to make a diagnosis combining the computed tomography scans and nucleic acid detection.
Keywords: 2019 Novel coronavirus pneumonia, Clinical diagnosis, Nucleic acid amplification test
Most human coronavirus infections in the past 20 years were not regarded as highly pathogenic to human beings until the outbreak of severe acute respiratory syndrome and Middle East respiratory syndrome coronaviruses (Zhong et al., 2003, Drosten et al., 2003, Fouchier et al., 2003). Although coronaviruses are broadly distributed in humans and animals, knowledge of non-segmented positive sense RNA viruses is limited (Cui et al., 2019, Woo et al., 2012). At the end of 2019, the China office of the World Health Organization (WHO) reported a cluster of pneumonia cases in Wuhan City, China, and the causative pathogen was identified one week later as a novel coronavirus (2019-nCoV) (Wu et al., 2020, Zhou et al., 2020). China’s National Health Commission provided guidance to laboratories, and WHO has named this disease COVID-19 (C. Wang et al., 2020; D. Wang et al., 2020).
A total of 19 suspected cases were collected at Sichuan Provincial People’s Hospital (ten patients) and Sichuan Mianyang 404 Hospital (nine patients). All study procedures conformed to the Declaration of Helsinki, and the protocol was accepted by the Institutional Review Board and the Ethics Committee of Sichuan Provincial People’s Hospital. Each participant participated in the study voluntarily and provided signed informed consent. We collected four kinds of samples: oropharyngeal swabs, blood, urine, and stool samples from the 19 cases for nucleic acid detection. We reviewed all patients’ medical histories, clinical charts, nursing records, physical findings, and computed tomography (CT) scans, and the hematological, biochemical, radiological, and microbiological investigation results were recorded and analyzed.
Almost all the suspected patients had symptoms of respiratory disease and two had diarrhea. Oropharyngeal swab specimens were obtained and sent for detection of viral respiratory pathogens by nucleic acid amplification testing (NAAT). All of the 19 cases were reported as negative for all other known pathogens tested, including influenza A and B, parainfluenza, respiratory syncytial virus, rhinovirus, adenovirus, and four common coronavirus strains known to cause illness in humans (HKU1, NL63, 229E, and OC43). Stool samples from the two diarrhea cases were tested for common diarrheal pathogens to rule out other causes (To et al., 2019).
Viral nucleic acid was extracted from the specimens following a common workflow and stored at −80 °C. Two highly conserved sequence regions (ORF1b and N) in rotavirus were selected for primers and probes design. Three different 2019-NCoV Fluorescent RT-PCR Kits with different manufacturers but almost the same detection efficiency were used for real-time-PCR assay, including GeneoDx (GZ-TRM2, China), Maccura (Sichuan, China) and Liferiver (W-RR-0479-02, China).
As Table1 shows, the median age was 33 years, and 57.89% were women. According to the results of the oropharyngeal swab NAAT, nine patients were confirmed to be infected with 2019-nCoV, and the other ten cases were negative for 2019-nCoV based on the nucleic acid test results. We found that all nine confirmed patients and five out of the ten negative cases showed bilateral distribution of patchy shadows and patchy ground-glass opacities in CT scans (Figure 1 ). To avoid false negative results, we recollected oropharyngeal swab specimens for these negative cases and reconducted the 2019-nCoV nucleic acid tests for three consecutive days. However, the results all remained negative.
Table 1.
2019-nCoV nucleic acid detection results of the 19 cases in different samples and characteristics index of these cases.
| Age (years) | Sex | CT scan results are abnormal | Presenting symptoms and signs |
Nucleic acid test of 2019-nCoV |
Several laboratory plasma data |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fever | Cough | Fatigue | Diarrhea | Throat swabs | Stool sample | Urine sample | Blood sample | Lymphocyte count (cells/L) (1.0–3.2 × 10⁹) | Hematocrit (0.35–0.45) | Activated partial thromboplastin time (s); (23.3–32.5) | Fibrinogen (g/dL); (1.80–3.50) | C-reactive protein (mg/L); (0.0–5.0) | Urea (mmol/L); (2.8–8.1) | ||||
| Patient 1 | 62 | Female | + | – | + | + | + | + | + | – | – | 1.74 | 0.355 | 27 | 4.04(↑) | 9.56(↑) | 4.19 |
| Patient 2 | 45 | Female | + | + | + | + | – | + | + | – | – | 0.901(↓) | 0.371 | 34.6(↑) | 4.33(↑) | 22.56(↑) | 2.70(↓) |
| Patient 3 | 59 | Female | + | + | + | – | – | + | + | – | – | 1.065(↓) | 0.331(↓) | 34.2(↑) | 4.75(↑) | 24.6(↑) | 2.86(↓) |
| Patient 4 | 33 | Female | + | + | – | – | – | + | + | – | – | 1.52 | 0.308(↓) | 33.2 | 2.49 | 37.13 | 2.7 |
| Patient 5 | 34 | Male | + | + | – | – | – | + | - | – | – | NA | NA | 39.0(↑) | 3.59(↑) | NA | 3.89 |
| Patient 6 | 43 | Male | + | + | + | + | – | + | + | – | – | NA | NA | 29.1 | 4.03(↑) | 9.46(↑) | 3.0(↓) |
| Patient 7 | 26 | Male | + | + | + | + | – | + | + | – | – | 0.900(↓) | 0.137(↓) | 37.7 | 3.69(↑) | 20.24(↑) | 3.0(↓) |
| Patient 8 | 18 | Female | + | + | – | – | – | + | + | – | – | 1.97 | 0.36 | 30.5 | 2.34 | 0.94 | 4.1 |
| Patient 9 | 25 | Male | – | + | + | – | – | + | + | – | – | 0.490(↓) | 0.380(↓) | 33.4 | 3.91 | 22.66(↑) | 3.9 |
| Suspect cases 1 | 31 | Male | + | + | + | + | – | – | – | – | – | 1.117 | 0.359(↓) | 34.1(↑) | 2.5 | 146.64(↑) | 2.78(↓) |
| Suspect cases 2 | 33 | Male | + | + | – | – | + | – | – | – | – | 1.712 | 0.507(↑) | 33.9(↑) | 1.20(↓) | 19.60(↑) | 3.82 |
| Suspect cases 3 | 33 | Male | + | + | + | – | – | – | – | – | – | 1.564 | 0.464 | 34.5(↑) | 0.90(↓) | 105.93(↑) | 5.35 |
| Suspect cases 4 | 39 | Male | + | – | – | + | – | – | – | – | – | 1.444 | 0.489 | 31.3 | 4.48(↑) | 8.27(↑) | 2.49(↓) |
| Suspect cases 5 | 50 | Female | + | – | + | + | – | – | – | – | – | 1.766 | 0.37 | 31.3 | 2.27 | 2.3 | 2.9 |
| Suspect cases 6 | 38 | Female | + | + | + | + | – | – | – | – | – | 1.07 | 0.371 | 32.1 | 3.80(↑) | 38.95(↑) | 3.71 |
| Suspect cases 7 | 31 | Female | + | + | + | - | – | – | – | – | – | 1.588 | 0.304(↓) | 31.3 | 2.96 | 4.78 | 3.14 |
| Suspect cases 8 | 34 | Female | + | – | – | – | – | – | – | – | – | 1.597 | 0.416 | 30.2 | 0.50(↓) | 1.25 | 2.98 |
| Suspect cases 9 | 23 | Female | + | – | + | + | + | – | – | – | – | 1.534 | 0.443 | 29.7 | 3.96(↑) | 2.11 | 2.97 |
| Suspect cases 10 | 8 | Female | – | + | + | – | – | – | – | – | – | 2.846 | 0.378 | NA | NA | 20.52(↑) | NA |
+ = Positive, – = negative, ↑ = above normal range, ↓ = below normal range.
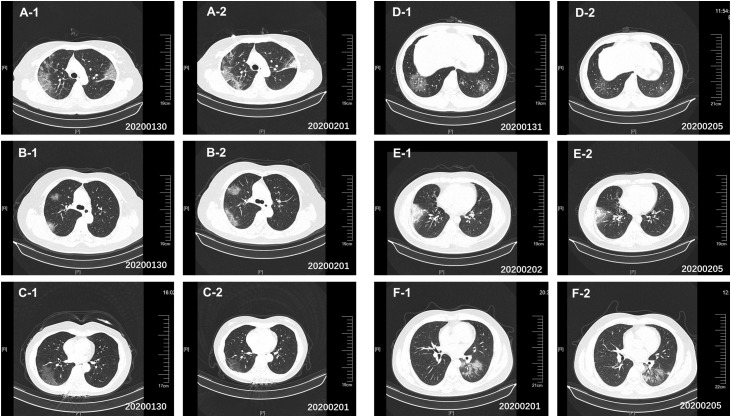
Figure 1.
CT scans of the 2019-nCoV nucleic acid–detected positive patients. Increasing and multifocal ground-glass changes were visible. (A) Patient 1, January 30, 2020 (hospital day 2, illness day 5, A-1); February 1, 2020 (hospital day 4, illness day 7, A-2). (B) Patient 2, January 30, 2020 (hospital day 2, illness day 5, B-1); February 1, 2020 (hospital day 4, illness day 7, B-2). (C) Patient 3, January 30, 2020 (hospital day 1, illness day 6, C-1); February 1, 2020 (hospital day 3, illness day 8, C-2). (D) Suspected case 1, January 31, 2020 (hospital day 1, illness day 4, D-1); February 1, 2020 (hospital day 6, illness day 9, D-2). (E) Suspected case 3, February 1, 2020 (hospital day 1, illness day 6, E-1); February 5, 2020 (hospital day 5, illness day 10, E-2). (F) Suspected case 4, January 31, 2020 (hospital day 1, illness day 2, F-1); February 1, 2020 (hospital day 4, illness day 5, F-2).
Therefore, we extracted RNA from the blood, urine, and stool of all 19 cases to determine whether the 2019-nCoV could be detected by NAAT (Table 1). In the nine confirmed patients, eight stool samples showed positive results for 2019-nCoV; interestingly, the virus could still be detected in stool samples from patients without diarrhea symptoms. However, the other ten cases showed negative results for 2019-nCoV in stool samples, and all of the 19 cases showed negative results for 2019-nCoV in both blood and urine samples. To avoid false results, we used three different kits to test samples and got the same result for each sample.
Although no nucleic acid positives were detected in serum, we cannot say that the virus will not enter the blood; it might be at a low concentration. None of the patients assessed in this study were diagnosed with viremia, but it has previously been reported that viruses have been detected in the sera of patients with viremia who were infected with other coronaviruses (C. Wang et al., 2020; D. Wang et al., 2020). The number of positive oropharyngeal swab samples was very close to the number of positive stool samples, and eight stool samples tested positive in nine patients who were confirmed using the oropharyngeal swab NAAT. This may indicate that feces may be capable of transmitting infection (further study is needed to determine whether the whole virus is found in feces or just pieces of nucleic acid) even if the patient does not have diarrhea.
In this study, we compared the positive ratio of 2019-nCoV NAAT from different samples, and the positive ratio of nucleic acid detection for 2019-nCoV was only 47.4%. Therefore, precise diagnosis of COVID-19 seemed very difficult by relying on nucleic acid detection alone. It might be better to reach a diagnosis by combining CT scans and NAAT results, and this may be very important for the prevention and control of COVID-19.
Statements
This work has no conflict of interest. The protocol was accepted by the Institutional Review Board and the Ethics Committee of Sichuan Academy of Medical Sciences & Sichuan Provincial People’s Hospital. This work was supported by Sichuan Science and Technology Program (No. 2020YFS0014 to ZL Yang). No funding organization played role in the study design or conduct.
Acknowledgments
We thank the patients, the nurses and clinical staff who are providing care for the patients.
Contributor Information
Xingxiang Yang, Email: 350686908@qq.com.
Yi Shi, Email: shiyi1614@126.com.
Zhenglin Yang, Email: zhenglin.yang@hsc.utah.edu.
References
- Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17(3):181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348(20):1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- Fouchier R.A., Kuiken T., Schutten M., van Amerongen G., van Doornum G.J., van den Hoogen B.G. Aetiology: Koch’s postulates fulfilled for SARS virus. Nature. 2003;423(6937):240. doi: 10.1038/423240a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To K.K.W., Yip C.C.Y., Lai C.Y.W., Wong C.K.H., Ho D.T.Y., Pang P.K.P. Saliva as a diagnostic specimen for testing respiratory virus by a point-of-care molecular assay: a diagnostic validity study. Clin Microbiol Infect. 2019;25(3):372–378. doi: 10.1016/j.cmi.2018.06.009. [DOI] [PubMed] [Google Scholar]
- Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. Lancet. 2020;395(10223):470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020 doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Lam C.S., Lau C.C., Tsang A.K., Lau J.H. Discovery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J Virol. 2012;86(7):3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong N.S., Zheng B.J., Li Y.M., Poon, Xie Z.H., Chan K.H. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People’s Republic of China, in February, 2003. Lancet. 2003;362(9393):1353–1358. doi: 10.1016/S0140-6736(03)14630-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]