Abstract
Every year, more than 10 million pilgrims arrive in the Kingdom of Saudi Arabia for the Hajj or Umrah. Crowding conditions lead to high rates of respiratory infections among the pilgrims, representing a significant cause of morbidity and a major cause of hospitalization. Pre- and post-Hajj nasal specimens were prospectively obtained from a paired cohort (692 pilgrims) and from nonpaired cohorts (514 arriving and 470 departing pilgrims) from 13 countries. The countries of residence included Africa (44.2%), Asia (40.2%), the United States (8.4%) and Europe (7.2%). Nasal specimens were tested for 34 respiratory pathogens using RT-PCR. A total of 80 512 PCRs were performed. The prevalence of viruses and bacteria increased, from 7.4% and 15.4% before the Hajj to 45.4% and 31.0% after the Hajj, respectively, due to the acquisition of rhinovirus, coronaviruses (229E, HKU1, OC43), influenza A H1N1, Streptococcus pneumoniae, Haemophilus influenzae and Staphylococcus aureus. We did not identify Middle East respiratory coronavirus carriage. At arrival, the prevalence of several viruses was clearly dependent on the pilgrim's country of origin. After Hajj participation, these viruses were isolated among pilgrims from all countries, with few exceptions. No significant differences were observed between paired and nonpaired cohort results. Our results strongly suggest that, given the particularly crowded conditions during the rituals, an international mass gathering such as the Hajj may contribute to the globalization of respiratory pathogens after the cross-contamination of pilgrims harbouring pathogens that easily spread among participants. Influenza and pneumococcal vaccination, face mask use and hand hygiene should be considered in the context of the Hajj.
Keywords: Bacteria, globalization, Hajj, mass gathering, nasal carriage, viruses
Introduction
Every year, more than 10 million pilgrims from over 180 countries arrive at Makkah for the Hajj or Umrah. The number of pilgrims undertaking the Hajj has increased by a factor 5 from 1920 to 2012 [1]. One of the major public health concerns in relation to mass gatherings is the importation or exportation of infectious diseases, spread between attendees and the local population and further spread to home countries through the returning participants [2]. The crowding conditions within a confined area during the Hajj leads to high rates of upper and lower respiratory infections among the pilgrims, which represent a significant cause of morbidity and a major cause of hospitalization [3]. Recent studies conducted among French pilgrims showed rapid acquisition of respiratory viruses and bacteria carriage, particularly rhinovirus and Streptococcus pneumoniae [4], [5], [6], [7]. In the present report, we investigate the nasal carriage of a large panel of 34 respiratory pathogens among pilgrims arriving in the Kingdom of Saudi Arabia (KSA) from 13 countries on four continents and among pilgrims departing from KSA after participating in the Hajj. The aim of the study was to identify changes in colonization before and after the pilgrimage and to investigate circulation and potential for globalization of these pathogens using two distinct cohort survey designs based on previous works [4], [5], [6], [7], [8], [9].
Methods
Participants and study design
Pilgrims participating in the 2013 Hajj were recruited upon arrival at the Jeddah airport (October 2 to October 7) and after performing the Hajj, at the Mina encampment before leaving the KSA (October 16 to October 24). The selection of the country of residence was based on representative countries from each continent and the availability of nationalities during the sampling dates. Potential adult participants were asked to participate in the study on a voluntary basis. Three groups of pilgrims were investigated. One group was included in a paired cohort survey in which the same pilgrims were evaluated before and after participating in the Hajj. The two other groups were included in a nonpaired cohort survey: one group consisted of pilgrims recruited before the Hajj, and the other consisted of different pilgrims of the same nationalities recruited after the Hajj.
Upon inclusion, the participants were interviewed by investigators using a standardized questionnaire that collected information on demographics, vaccination status and medical history. A post-Hajj questionnaire that collected clinical data and information on compliance with face mask use was completed by the same team of investigators. The protocol was approved by the Saudi Ministry of Health ethical review committee. The study was performed in accordance with the good clinical practices recommended by the Declaration of Helsinki and its amendments. All participants provided oral informed consent.
Respiratory specimens
Nasal swabs were collected at arrival and at Mina from each participant (anterior nares) using commercial rigid cotton-tipped swab applicators (Remel, USA), placed in universal transport medium (Remel) at the time of collection and stored in a freezer at −80°C within 48 hours of collection. The specimens were transported on dry ice to France for analysis. Each swab was processed manually. One milliliter of medium was pipetted automatically (Beckman NX automate) to prepare 96-well microplates for total (DNA + RNA) nucleic acid extraction and long-term storage. To avoid unnecessary testing, a dual aliquot was prepared from 100 μL of nasal swab origin. The total nucleic acids were then purified using the Macherey-Nagel Nucleospin-96 kit. The extract was then split for testing for the presence of viruses and bacteria.
Detection of respiratory viruses
The real-time PCR detection of viruses was performed (a) using the FTD Respiratory Pathogens 21-plus kit (Fast Track Diagnostics, Luxembourg) for influenza A (FLUA)/H3N2, 2009 FLUA/H1N1 (A/H1N1), influenza B (FLUB), human rhinovirus (HRV), human coronavirus (HCoV) NL63, 229E, OC43, HKU1, human parainfluenza virus (HPIV) 1 through 4, human metapneumovirus A/B (HMPV), human bocavirus, human respiratory syncytial virus A/B (HRSV), human adenovirus (HAdV), human enterovirus from all groups (HEV) and parechovirus (HPeV), and (b) using single-plex assays for Middle East respiratory coronavirus [10], [11], human cytomegalovirus [12] and influenza C virus [13], with internal controls as described [14].
Detection of respiratory bacteria and Pneumocystis jirovecii
All real-time PCRs were performed using a 7900HT-thermocycler (Applied Biosystems, USA) and QuantiTect-Probe PCR Kit (Qiagen, France) according to the manufacturer's recommendations. Each sample was tested for Streptococcus pneumoniae, Haemophilus influenzae, Klebsiella pneumoniae, Staphylococcus aureus, Neisseria meningitidis, Coxiella burnetii, Bordetella pertussis, Mycoplasma pneumoniae, Legionella pneumophila, Streptococcus pyogenes, Salmonella spp., Pneumocystis jirovecii and Chlamydia pneumoniae with the primers and probes listed in Supplementary Table S1. A negative control (sterile water) and positive control (DNA from the bacterial strain) were included in each run.
Statistical analysis
Differences in the proportions were tested by Pearson's chi-square, McNemar's chi-square or Fisher's exact tests when appropriate. All statistical tests were two-sided. Percentages and odds ratio (OR) with 95% confidence interval (CI) estimations and comparisons were carried out using the R 2.8.1 environment (The R Foundation for Statistical Computing, Austria; http://www.r-project.org). A p value of ≤0.05 was considered significant.
Results
Characteristics of the study participants
A total of 1206 individuals were recruited at arrival to participate in the study, and all responded to the pretravel questionnaire (Supplementary Table S2). Of these, 692 were included in the paired cohort survey, and 514 individuals were included in the nonpaired cohort survey (arrival group). A total of 470 additional individuals were included in the nonpaired cohort survey after the Hajj (departure group). Overall, the M/F sex ratio was 1.9, with a mean age of 50 years (age range, 18–88 years). The countries of residence included Africa (44.2%), Asia (40.2%), the United States (8.4%) and Europe (7.2%). Documentation about chronic conditions was missing in most cases. Among the individuals for whom information was available, 21.9% declared having been vaccinated against influenza in 2013 and 1.2% against invasive pneumococcal diseases during the 5 past years. Of 1162 pilgrims investigated after the Hajj, 61.9% reported influenza-like illness, and 45.5% took antibiotics.
Overall detection of respiratory pathogens
A total of 80 512 PCRs were performed (Table 1 ). Overall, the prevalence of viruses increased from 7.4% of the pre-Hajj specimens to 45.4% of the post-Hajj specimens testing positive for at least one virus. This increase was statistically significant for HRV and HCoV 229E in both cohorts and for FLUA H1N1, HCoV HKU1 and OC43 in the nonpaired cohort survey. The overall acquisition rate calculated from the paired cohort was 49.1% (34.1% and 14.6%, respectively, for HRV and HCoV 229E). FLUA, FLUB, HCoV NL63, HPIV 2, 3 and 4, HEV, HMPV, HRSV, HAdV and HPeV were also acquired during the stay in KSA by a low proportion of the participants. None of the participants tested positive for influenza C virus, Middle East respiratory coronavirus or cytomegalovirus at any point during the study period.
Table 1.
Overall prevalence of respiratory viruses, bacteria and Pneumocystis jirovecii among participants
| Respiratory pathogen | Paired cohort |
Nonpaired cohort |
|||||
|---|---|---|---|---|---|---|---|
| Arrival, n (%) of positive specimens | Departure, n (%) of positive specimens | p | Acquisition rate, n (%) positive specimens at departure among individuals negative at arrival | Arrival, n (%) of positive specimens | Departure, n (%) of positive specimens | p | |
| Viruses | |||||||
| Influenza virus A/H3N2 | 4 (0.6%) | 14 (2.0%) | 0.71 | 13 (1.9%) | 6 (1.2%) | 8 (1.7%) | 0.48 |
| Influenza virus 2009 A(H1N1) | 1 (0.1%) | 11 (1.6%) | 0.32 | 10 (1.4%) | 1 (0.2%) | 14 (3.0%) | <0.001 |
| Influenza virus B | 0 | 0 | — | 0 | 0 | 1 (0.2%) | 0.48 |
| Influenza virus C | 0 | 0 | — | 0 | 0 | 0 | — |
| Human parainfluenza viruses 1 | 0 | 0 | — | 0 | 2 (0.4%) | 0 | 0.50 |
| Human parainfluenza viruses 2 | 0 | 2 (0.3%) | — | 2 (0.3%) | 1 (0.2%) | 2 (0.4%) | 0.61 |
| Human parainfluenza viruses 3 | 2 (0.3%) | 1 (0.1%) | — | 1 (0.1%) | 0 | 1 (0.2%) | 0.61 |
| Human parainfluenza viruses 4 | 0 | 1 (0.1%) | — | 1 (0.1%) | 1 (0.2%) | 0 | 1 |
| MERS coronavirus | 0 | 0 | — | 0 | 0 | 0 | — |
| Human coronavirus 229E | 6 (0.9%) | 101 (14.6%) | <0.001 | 101 (14.6%) | 5 (1.0%) | 48 (10.2%) | <0.001 |
| Human coronavirus NL63 | 2 (0.3%) | 14 (2.0%) | 0.71 | 13 (1.9%) | 0 | 1 (0.2%) | 0.48 |
| Human coronavirus HKU1 | 3 (0.4%) | 9 (1.3%) | 0.16 | 9 (1.3%) | 1 (0.2%) | 7 (1.5%) | 0.03 |
| Human coronavirus OC43 | 1 (0.1%) | 11 (1.6%) | 0.34 | 11 (1.6%) | 2 (0.4%) | 9 (1.9%) | 0.02 |
| Human cytomegalovirus | 0 | 0 | — | 0 | 0 | 0 | — |
| Human enterovirus | 4 (0.6%) | 3 (0.4%) | — | 3 (0.4%) | 4 (0.8%) | 5 (1.1%) | 0.74 |
| Human metapneumovirus | 0 | 1 (0.1%) | — | 1 (0.1%) | 1 (0.2%) | 3 (0.6%) | 0.35 |
| Human respiratory syncytial virus | 4 (0.6%) | 5 (0.7%) | — | 5 (0.7%) | 3 (0.6%) | 1 (0.2%) | 0.63 |
| Human rhinovirus | 15 (2.2%) | 238 (34.4%) | <0.001 | 236 (34.1%) | 16 (3.1%) | 145 (30.9%) | <0.001 |
| Human adenovirus | 0 | 4 (0.6%) | — | 4 (0.6%) | 0 | 2 (0.4%) | 0.23 |
| Human bocavirus | 1 (0.1%) | 0 | — | 0 | 1 (0.2%) | 0 | 1 |
| Parechovirus | 0 | 1 (0.1%) | — | 1 (0.1%) | 0 | 0 | — |
| At least one virus | 42 (6.1%) | 343 (49.6%) | <0.001 | 340 (49.1%) | 44 (8.6%) | 212 (45.1%) | <0.001 |
| Bacteria | |||||||
| Bordetella pertussis | 0 | 0 | — | 0 | 0 | 0 | — |
| Mycoplasma pneumoniae | 0 | 0 | — | 0 | 0 | 0 | — |
| Neisseria meningitidis | 0 | 2 (0.3%) | — | 2 (0.3%) | 0 | 3 (0.6%) | 0.11 |
| Streptococcus pneumoniae | 39 (5.6%) | 88 (12.7%) | <0.001 | 83 (12.0%) | 24 (4.7%) | 64 (13.6%) | <0.001 |
| Streptococcus pyogenes | 0 | 0 | — | 0 | 0 | 0 | — |
| Klebsiella pneumoniae | 20 (2.9%) | 27 (3.9%) | 0.09 | 27 (3.9%) | 24 (4.7%) | 26 (5.5%) | 0.54 |
| Legionella pneumophila | 0 | 0 | — | 0 | 0 | 0 | — |
| Salmonella spp. | 0 | 0 | — | 0 | 0 | 0 | — |
| Staphylococcus aureus | 39 (5.6%) | 55 (7.9%) | <0.001 | 52 (7.5%) | 32 (6.2%) | 46 (9.8%) | 0.04 |
| Coxiella burnetii | 0 | 1 (0.1%) | — | 1 (0.1%) | 0 | 0 | — |
| Chlamydophila pneumoniae | 0 | 0 | — | 0 | 0 | 0 | — |
| Haemophilus influenzae | 16 (2.3%) | 81 (11.7%) | <0.001 | 79 (11.4%) | 12 (2.3%) | 67 (14.3%) | <0.001 |
| At least one bacteria | 101 (14.6%) | 201 (29.0%) | <0.001 | 196 (28.3%) | 83 (16.1%) | 158 (33.6%) | <0.001 |
| Pneumocystis jirovecii | 0 | 0 | — | 0 | 0 | 0 | — |
| At least one pathogen | 136 (19.7%) | 430 (62.1%) | <0.001 | 425 (61.4%) | 121 (23.5%) | 306 (65.1%) | <0.001 |
Overall, the prevalence of bacteria increased from 15.4% of the pre-Hajj specimens to 31.0% of the post-Hajj specimens testing positive for at least one bacterial pathogen. The increase was statistically significant for S. pneumoniae, H. influenzae and S. aureus in both cohorts. The overall acquisition rate calculated from the paired cohort was 28.3% for bacteria (12.0%, 11.4% and 7.5%, respectively, for S. pneumoniae, H. influenzae and S. aureus). K. pneumoniae, N. meningitidis and C. burnetii were also acquired during the stay in KSA by a low proportion of the participants (0.1–3.9%). None of the participants tested positive for B. pertussis, M. pneumoniae, S. pyogenes, L. pneumophila, Salmonella spp., C. pneumoniae or P. jirovecii at any point during the study period.
Multiple carriage was common, accounting for 40% of the positive post-Hajj specimens (two pathogens, 29.3%; three pathogens, 9.0%; four pathogens, 1.6%; five pathogens, 0.1%). The most frequent associations were HRV/H. influenzae, followed by HRV/S. pneumoniae, S. pneumoniae/H. influenzae, HRV/HCov 229E and HRV/S. aureus (Supplementary Fig. S1).
Detection of respiratory pathogens by country of residence
At arrival, the prevalence of several viruses was clearly dependent on the pilgrim's country of origin (Fig. 1, Fig. 2 ). H1N1 was found only among pilgrims arriving from Indonesia. HCoVs were isolated among pilgrims arriving from Africa and Southeast Asia only, with the exception of HCoV HKU1, which was also isolated among pilgrims from Albania. In contrast, HRV and FLUA/H3N2, to a lesser extent, and several bacteria including S. pneumoniae, H. influenzae, K. pneumoniae and S. aureus were found in pilgrims arriving from a wide range of regions.
Fig. 1.
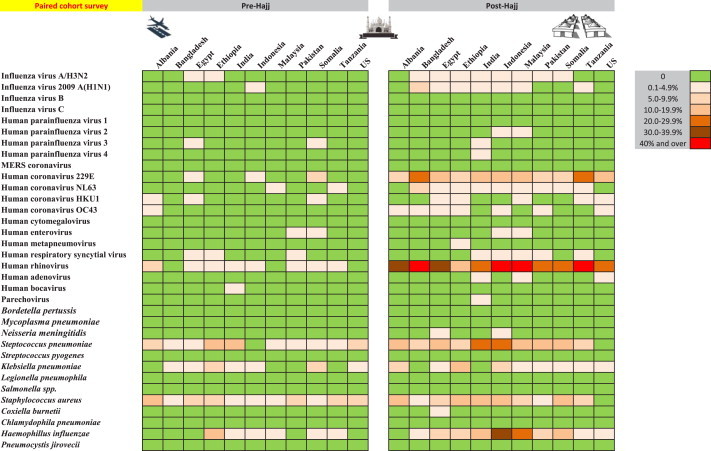
Nasal carriage prevalence of 34 respiratory pathogens among a paired cohort of 692 pilgrims from 11 countries before and after participating in the 2013 Hajj.
Fig. 2.
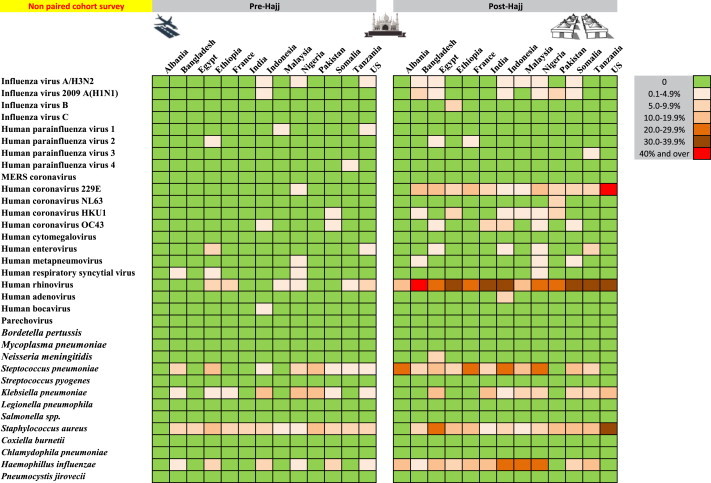
Nasal carriage prevalence of 34 respiratory pathogens between two distinct groups of pilgrims from 13 countries sampled before (n = 514) and after (n = 470) participating in the 2013 Hajj (nonpaired cohort survey).
After participation in the Hajj, these viruses and bacteria were isolated among pilgrims from all countries, with few exceptions. The acquisition rate of HRV calculated from the paired cohort was significantly higher among the pilgrims from Indonesia (OR 3.6, CI 1.32–7.93, p 0.01) and Bangladesh (OR 2.67, CI 1.01–7.32, p 0.05) compared to the pilgrims from the United States. The acquisition rate of H. influenzae was also higher among the pilgrims from Indonesia (OR 23.4, CI 4.31–438, p 0.003).
Other factors
The effects of age, sex, chronic conditions, vaccination status, face mask use, occurrence of influenza-like illness symptoms and antibiotic use on the acquisition rate of the pathogens most frequently isolated was investigated in the paired cohort. We found that age was associated with the acquisition of HCoV NL63 (OR 1.2 by year of age, CI 1.04–1.33, p 0.02) and S. pneumoniae (OR 1.03 by year of age, CI 1.002–1.06, p 0.04) and that diabetes was associated with the acquisition of HCoV NL63 (OR 29.2, CI 1–837, p 0.04) and H. influenzae (OR 3.6, CI 1.16–11.2, p 0.025).
From the nonpaired cohort, we found a significant negative association between K. pneumoniae carriage and the occurrence of influenza-like illness symptoms in the departing pilgrims (OR 0.11, CI 0.01–0.63, p 0.03).
Discussion
We found that the mass gathering of the Hajj pilgrimage is associated with the nasal acquisition of respiratory pathogens in more than 60% of the pilgrims. In this multinational survey, we confirm our previous observations made among French pilgrims that HRV, non-Middle East respiratory coronaviruses, FLUA viruses and S. pneumoniae account for the majority of pathogens acquired by pilgrims [4], [5], [6], [7]. Additionally, we demonstrate that H. influenzae and S. aureus are also among the most frequent pathogens for which nasal carriage is increased after participation in the Hajj. These pathogens may be introduced into the pilgrims' home countries upon their return, thus contributing to their potential international spread. We did not identify Middle East respiratory coronavirus carriage, confirming previous research [6], [9], [15], [16].
We provide for the first time a global picture of the circulation of respiratory pathogens at the Hajj. We observed three distinct epidemiological patterns. The first pattern was characterized by the introduction into KSA of pathogens isolated at low rates among pilgrims arriving specifically from one or two countries (Fig. 3 and Supplementary Fig. S2). Some of these pathogens then spread to pilgrims from a large number of countries, with a slight increase in overall prevalence. This was particularly obvious for A/H1N1, which was found in two Indonesians only upon arrival but spread to 25 pilgrims from Africa, Central Asia and Southeast Asia after the Hajj. A survey conducted in a separate and larger paired cohort of French pilgrims at the same period also showed the acquisition of A/H1N1 after the Hajj, whereas the pilgrims sampled before leaving France were negative [7]. This indicates that the acquisition resulted not only from human-to-human transmission through close contact within each group of pilgrims from the same country but was also acquired from other pilgrims, given the extremely high crowding density to which individuals from many parts of the world are exposed when performing Hajj rituals. Contamination originating from an environmental source may also have played a role.
Fig. 3.
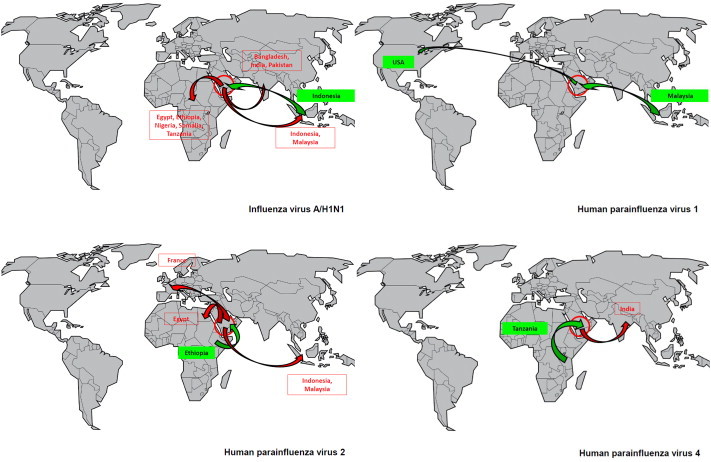
Nasal carriage of A/H1N1 virus and human parainfluenza virus 1, 2 and 4 among pilgrims arriving in the KSA (n = 1206) and departing from the KSA from (n = 1162) after participating in the 2013 Hajj and according to country of residence.
Other viruses were specifically isolated among rare pilgrims from one or two countries, but their spread was more limited or nil (HPIVs, HMPV, human bocavirus). The second epidemiologic pattern was that of cosmopolitan pathogens (Fig. 4 and Supplementary Figs. S3 and S4), including HRV, S. pneumoniae, H. influenzae, K. pneumoniae and S. aureus. These pathogens were isolated in 2% to 6% of the pilgrims arriving from almost all countries, with further amplification among the entire population of pilgrims after the Hajj in an epidemic mode, with acquisition rates of 11% to 34%. Some viruses such as HRV appeared to be spread among a very high proportion of pilgrims, as exemplified by the Bengali pilgrims, who were all negative at arrival but presented a prevalence of 50% after the Hajj. FLUA/H3N2, HCoVs, HEV and HRSV showed comparable patterns of transmission, but their geographical spread and/or prevalence were lower. Finally, a third pattern was observed, whereby pathogens were sporadically isolated among departing pilgrims but were never isolated among arriving ones (Fig. 5 and Supplementary Fig. S2). This group included FLUB, HAdV, HPeV N. meningitidis and C. burnetii. This pattern may be explained by an overall low prevalence of pathogens among adults worldwide (FLUB) [17] or by a preferential circulation of pathogens in KSA (C. burnetii) [18].
Fig. 4.
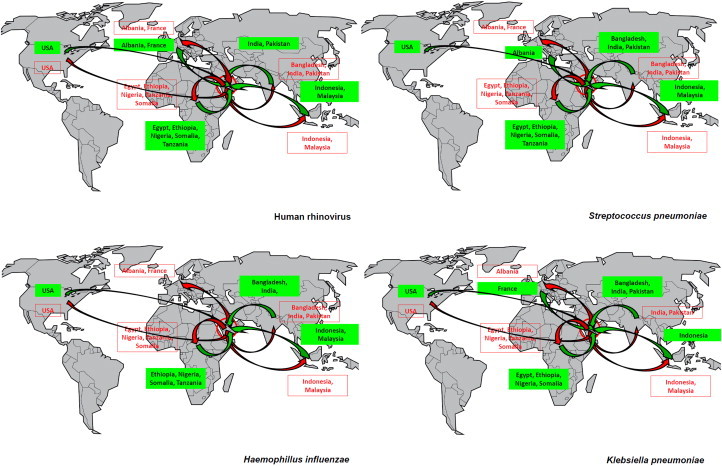
Nasal carriage of human rhinovirus, Streptococcus pneumoniae, Haemophilus influenzae and Klebsiella pneumoniae among pilgrims arriving in the KSA (n = 1206) and pilgrims departing from the KSA (n = 1162) after participating in the 2013 Hajj and according to country of residence.
Fig. 5.
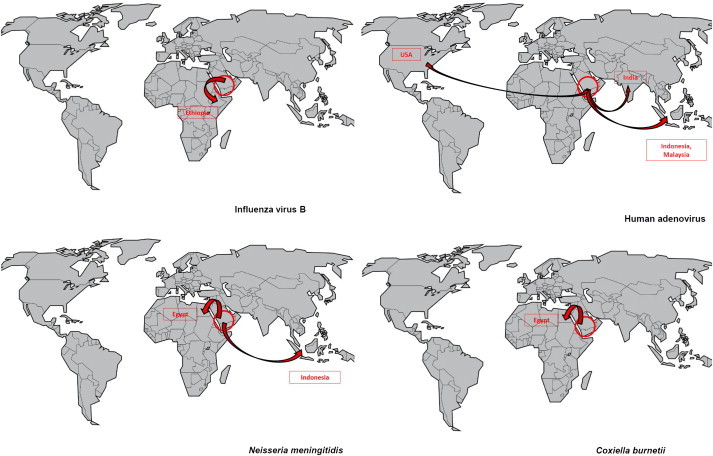
Nasal carriage of influenza B virus, human adenovirus, Neisseria meningitidis and Coxiella burnetii among pilgrims arriving in the KSA (n = 1206) and pilgrims departing from the KSA (n = 1162) after participating in the 2013 Hajj and according to country of residence.
The purpose of this work was not to demonstrate whether the pathogens detected in the respiratory specimens were responsible for the observed symptoms. Nasal carriage was observed in asymptomatic pilgrims in certain cases. Furthermore, swabs were taken from the anterior nose and may thus be more representative of colonization than acute infection. Nevertheless, we postulate that “Hajj cough” may result from infection of the respiratory tract by various respiratory viruses, notably including HRV and HCoV229E, which are known to cause mild or serious lower respiratory tract infections [19], [20].
The results obtained from the paired and nonpaired cohort led to similar conclusions. The paired cohort survey design, however, allows the calculation of the acquisition rates of pathogens, whereas the nonpaired cohort survey only allows for comparing the prevalence of pathogens at arrival and departure. We found that the nonpaired cohort survey results slightly underestimated the increase in the prevalence of viruses after participation in the Hajj compared to the paired cohort survey results. This discrepancy was not observed for bacterial carriage.
Among the pathogens most frequently acquired by pilgrims, three account for vaccine-preventable diseases, including influenza, S. pneumoniae and H. influenzae infections. Thus, the availability of the seasonal influenza vaccine to all target individuals attending the Hajj should be highlighted [21], [22]. Additionally, vaccination with a conjugate pneumococcal vaccine should be considered in individuals with medical risk factors for invasive pneumococcal disease [23]. Studies are needed to evaluate the potential effect of H. influenzae vaccine within the context of the Hajj. Nonpharmaceutical intervention including face mask use and hand hygiene should be also considered [24].
Our study has some limitations. The included population represented only a small percentage of the total number of pilgrims attending the 2013 Hajj; hence, the results cannot be extrapolated to the entire pilgrim population. The number of pilgrims from Europe and the United States was limited, and no pilgrims from the Middle East were included. In addition, data on chronic health conditions, preventive measures and symptoms during the Hajj were difficult to obtain and were available only for a limited number of subjects. Nevertheless, our study investigated a much larger and geographically more diverse cohort than previous studies [4], [5], [6], [7]. Overall, these results strongly suggest that, given the particularly crowded conditions during the rituals, an international mass gathering such as the Hajj may contribute to the globalization of common respiratory pathogens after the cross-contamination of pilgrims harbouring pathogens that easily spread among participants [25].
Transparency declaration
All authors report no conflicts of interest relevant to this article.
Editor: E. Bottieau
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.cmi.2015.02.008.
Appendix A. Supplementary data
The following are the Supplementary data related to this article:
References
- 1.Memish Z.A., Zumla A., Alhakeem R.F., Assiri A., Turkestani A., Al Harby K.D. Hajj: infectious disease surveillance and control. Lancet. 2014;383:2073–2082. doi: 10.1016/S0140-6736(14)60381-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abubakar I., Gautret P., Brunette G.W., Blumberg L., Johnson D., Poumerol G. Global perspectives for prevention of infectious diseases associated with mass gatherings. Lancet Infect Dis. 2012;12:66–74. doi: 10.1016/S1473-3099(11)70246-8. [DOI] [PubMed] [Google Scholar]
- 3.Al-Tawfiq J.A., Zumla A., Memish Z.A. Respiratory tract infections during the annual Hajj: potential risks and mitigation strategies. Curr Opin Pulm Med. 2013;19:192–197. doi: 10.1097/MCP.0b013e32835f1ae8. [DOI] [PubMed] [Google Scholar]
- 4.Benkouiten S., Charrel R., Belhouchat K., Drali T., Salez N., Nougairede A. Circulation of respiratory viruses among pilgrims during the 2012 Hajj pilgrimage. Clin Infect Dis. 2013;57:992–1000. doi: 10.1093/cid/cit446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benkouiten S., Gautret P., Belhouchat K., Drali T., Salez N., Memish Z.A. Acquisition of Streptococcus pneumoniae carriage in pilgrims during the 2012 Hajj. Clin Infect Dis. 2014;58:e106–e109. doi: 10.1093/cid/cit749. [DOI] [PubMed] [Google Scholar]
- 6.Gautret P., Charrel R., Benkouiten S., Belhouchat K., Nougairede A., Drali T. Lack of MERS coronavirus but prevalence of influenza virus in French pilgrims after 2013 Hajj. Emerg Infect Dis. 2014;20:728–730. doi: 10.3201/eid2004.131708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benkouiten S., Charrel R., Belhouchat K., Drali T., Nougairede A., Salez N. Respiratory viruses and bacteria among pilgrims during the 2013 Hajj. Emerg Infect Dis. 2014;11:1821–1827. doi: 10.3201/eid2011.140600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Memish Z.A., Assiri A.M., Hussain R., Alomar I., Stephens G. Detection of respiratory viruses among pilgrims in Saudi Arabia during the time of a declared influenza A(H1N1) pandemic. J Travel Med. 2012;19:15–21. doi: 10.1111/j.1708-8305.2011.00575.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Memish Z.A., Assiri A., Almasri M., Alhakeem R.F., Turkestani A., Al Rabeeah A.A. Prevalence of MERS-CoV nasal carriage and compliance with the saudi health recommendations among pilgrims attending the 2013 Hajj. J Infect Dis. 2014;25:186–190. doi: 10.1093/infdis/jiu150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corman V.M., Müller M.A., Costabel U., Timm J., Binger T., Meyer B. Assays for laboratory confirmation of novel human coronavirus (hCoV-EMC) infections. Euro Surveill. 2012;17:20334. doi: 10.2807/ese.17.49.20334-en. [DOI] [PubMed] [Google Scholar]
- 11.Corman V.M., Eckerle I., Bleicker T., Zaki A., Landt O., Eschbach-Bludau M. Detection of a novel human coronavirus by real-time reverse-transcription polymerase chain reaction. Euro Surveill. 2012;17:20285. doi: 10.2807/ese.17.39.20285-en. [DOI] [PubMed] [Google Scholar]
- 12.Griscelli F., Barrois M., Chauvin S., Lastere S., Bellet D., Bourhis J.H. Quantification of human cytomegalovirus DNA in bone marrow transplant recipients by real-time PCR. J Clin Microbiol. 2001;39:4362–4369. doi: 10.1128/JCM.39.12.4362-4369.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faux C.E. Influenza type C. In: Carter I.W.J., Schuller M., James G.S., Sloots T.P., Halliday C.L., editors. PCR for clinical microbiology. An Australian and international perspective. Springer Verlag; New York: 2010. pp. 311–312. [Google Scholar]
- 14.Ninove L., Nougairede A., Gazin C., Thirion L., Delogu I., Zandotti C. RNA and DNA bacteriophages as molecular diagnosis controls in clinical virology: a comprehensive study of more than 45,000 routine PCR tests. PLoS One. 2011;6:e16142. doi: 10.1371/journal.pone.0016142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gautret P., Charrel R., Belhouchat K., Drali T., Benkouiten S., Nougairede A. Lack of nasal carriage of novel corona virus (HCoV-EMC) in French Hajj pilgrims returning from the Hajj 2012, despite a high rate of respiratory symptoms. Clin Microbiol Infect. 2013;19:315–317. doi: 10.1111/1469-0691.12174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Al-Tawfiq J.A., Zumla A., Memish Z.A. Travel implications of emerging coronaviruses: SARS and MERS-CoV. Travel Med Infect Dis. 2014;12:422–428. doi: 10.1016/j.tmaid.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paul Glezen W., Schmier J.K., Kuehn C.M., Ryan K.J., Oxford J. The burden of influenza B: a structured literature review. Am J Public Health. 2013;103:e43–e51. doi: 10.2105/AJPH.2012.301137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Almogren A., Shakoor Z., Hasanato R., Adam H.M. Q fever: a neglected zoonosis in Saudi Arabia. Ann Saudi Med. 2013;33:464–468. doi: 10.5144/0256-4947.2013.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacobs S.E., Lamson D.M., St George K., Walsh T.J. Human rhinoviruses. Clin Microbiol Rev. 2013;26:135–162. doi: 10.1128/CMR.00077-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gerna G., Campanini G., Rovida F., Percivalle E., Sarasini A., Marchi A. Genetic variability of human coronavirus OC43-, 229E-, and NL63-like strains and their association with lower respiratory tract infections of hospitalized infants and immunocompromised patients. J Med Virol. 2006;78:938–949. doi: 10.1002/jmv.20645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haworth E., Barasheed O., Memish Z.A., Rashid H., Booy R. Prevention of influenza at Hajj: applications for mass gatherings. J R Soc Med. 2013;106:215–223. doi: 10.1258/jrsm.2012.120170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Memish Z.A., Al-Rabeeah A.A. Health conditions of travellers to Saudi Arabia for the pilgrimage to Mecca (Hajj and Umra) for 1434 (2013) J Epidemiol Glob Health. 2013;3:59–61. doi: 10.1016/j.jegh.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rashid H., Abdul Muttalif A.R., Mohamed Dahlan Z.B., Djauzi S., Iqbal Z., Karim H.M. The potential for pneumococcal vaccination in Hajj pilgrims: expert opinion. Travel Med Infect Dis. 2013;11:288–294. doi: 10.1016/j.tmaid.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 24.Benkouiten S., Brouqui P., Gautret P. Non-pharmaceutical interventions for the prevention of respiratory tract infections during Hajj pilgrimage. Travel Med Infect Dis. 2014;12:429–442. doi: 10.1016/j.tmaid.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Charrel R.N. Hajj, Umrah, and other mass gatherings: which pathogens do you expect? Beware of the tree that hides the forest! Travel Med Infect Dis. 2014;12:418–419. doi: 10.1016/j.tmaid.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


