Abstract
Severe acute respiratory syndrome (SARS) is an acute respiratory illness, which has broken out in China. It has been known that SARS coronavirus (SARS_CoV) is a novel human coronavirus and is responsible for SARS infection. Belonging to one of the major proteins associated with SARS_CoV, SARS 3C-like protease (SARS_3CLpro) functions as a cysteine protease engaging in the proteolytic cleavage of the viral precursor polyprotein to a series of functional proteins required for coronavirus replication and is considered as an appealing target for designing anti-SARS agents. To facilitate the studies regarding the functions and structures of SARS_3CLpro, in this report the synthetic genes encoding 3CLpro of SARS_CoV were assembled, and the plasmid was constructed using pQE30 as vector and expressed in Escherichia coli M15 cells. The highly yielded (∼15 mg/L) expressed protease was purified by use of NTA-Ni2+ affinity chromatography and FPLC system, and its sequence was determined by LC/MS with the residue coverage of 46.4%.
Keywords: Severe Acute Respiratory syndrome (SARS), SARS coronavirus, 3C like protease, Molecular cloning, Expression, Purification, Mass spectrometric characterization
From the end of the year 2002 to the June of the year 2003, one severe epidemic disease called severe acute respiratory syndrome (SARS) broke out severely in China, and SARS infection has also spread to more than 30 countries. By using biophysical and biochemical techniques such as electron microscopy, virus-discovery microarrays containing conserved nucleotide sequences characteristic of many virus families, randomly primed RT-PCR, and serological tests, it has been determined that SARS coronavirus (SARS_CoV) is responsible for SARS infection [1], [2], [3]. Coronavirus is a positive-stranded RNA virus with halo or corona appearance if viewed under a microscope and involves the largest viral RNA genomes known to date. The studies have suggested that SARS_CoV is a previously unknown coronavirus, which belongs neither to a mutant of any known coronavirus nor a recombinant of known coronaviruses; it is believed to be a novel human coronovirus possibly originated from a non-human host [4], [5].
Proteolytic processing of viral polyproteins is a key step in the replication cycle of many positive-strand RNA viruses and such processing is performed by the encoded proteases [6], [7]. It has been known that the replicase gene for encoding the proteins required for coronavirus replication and transcription encompasses more than 20,000 nucleotides [8], [9] and encodes two overlapping polyproteins, pp1a (replicase 1a, around 450 kDa) and pp1ab (replicase 1ab, around 750 kDa), which feature the sequence motifs of both papain-like cysteine protease and the 3C-like protease (3CLpro) [10], [11]. Recently, the genome sequencings deposited in the GenBank (http://www.ncbi.nlm.nih.gov/) for the SARS_CoV from different SARS patients have laid a potent foundation for the research of SARS pathogenesis and anti-SARS drug design [12], [13], [14]. The fact has been demonstrated that the important proteins associated with the SARS_CoV infection involve the RNA polymerase, the spike (S) glycoprotein, the envelope (E) protein, the membrane (M) protein, the nucleocapsid (N) protein, and the main protease, 3C-like (3CLpro) protease [15], [16]. As the viral main protease, 3CLpro functions as a protease to control the activities of the coronavirus replication complex [17], [18].
It has been concluded from the previous research data that the 3CLpro-mediated processing pathways are conserved in coronaviruses. Coronavirus main proteases employ conserved cysteine and histidine residues in the catalytic site and lack acidic active site residue [6], [19], [20], [21]. The results have also confirmed that for coronavirus main proteases their substrate specificities are also well defined, with the known cleavage sites involving bulky hydrophobic residues (mainly leucine) at the P2 position, glutamine at the P1 position, and small aliphatic residues at the P1′ position [17], [18]. In addition, the recent determination of the crystal structures for human coronavirus (strain 229E) 3CLpro and for an inhibitor complex of porcine coronavirus (transmissible gastroenteritis virus, TGEV) 3CLpro also confirms a remarkable degree of conservation of the substrate binding sites for coronavirus 3CLpro [16]. In fact, the studies have already shown that 3CLpro is a useful target for screening anti-virus agents [18], [22], [23]. Like other 3CLpro, it is hopeful that SARS_3CLpro will surely become an appealing target in discovering new agents for the treatment of SARS [16].
Therefore, based on the aforementioned facts, it seems to be very important to express and purify large amount of the SARS_3CLpro for its structural and functional research purposes. In our previous work [24], [25], we reported a 3D model of SARS_3CLpro and its inhibitor design by virtual screening, as well as the cloning, expression, and purification of the E protein of SARS_CoV. In this article, we would like to present the results describing the molecular cloning, expression and purification of 3CLpro of SARS_CoV, and the preliminary study on its mass spectrometric characterization is also reported.
Materials and methods
Chemicals, enzymes, and the vector pQE30
The restriction and modifying enzymes in this work were purchased from TaKaRa and the vector pQE30, the bacterial strains M15 and DH5α were from Qiagen. Trizol and Superscript II reverse transcriptase were purchased from Gibco. Trypsin (sequencing grade) was purchased from Sigma. The chelating affinity column and lower molecular weight (LMW) marker were purchased from Amersham–Pharmacia Biotech. All other chemicals were from Sigma in analytical grade.
Bacterial strains and culture media
Escherichia coli DH5α was utilized for propagation of plasmids. DH5α was maintained on LB agar plates and grown at 37°C, while M15 was cultured on LB agar plates containing kanamycin (25 mg/L). For agar plates, Bacto agar was added to the media to a final concentration of 1.5% (w/v). Ampicillin was added to the media at a final concentration of 100 mg/L for the selection of transformants. E. coli M15 was chosen as the host for gene expression. The strains were maintained in LB medium including 15% glycerol at −80°C. Ampicillin and kanamycin as antibiotics were added to the media at a final concentration of 100 and 25 mg/L, respectively.
Cloning of SARS_ 3CLpro gene in pQE30
All cloning techniques including PCR, restriction digestion, ligation, E. coli transformation, and plasmid DNA preparation were according to the literature method [26].
SARS_CoV (isolate BJ01) RNA was extracted with Trizol reagent according to manufacturer’s instruction (www.genehub.net/trizol.htm). The reverse transcription was performed with the random priming method by the Superscript II reverse transcriptase. The SARS_3CLpro cDNA was subsequently amplified by PCR, using the following primers: 3CLf (5′-GGGGGATCCACCATGAGTGGTTTTAGGAAAATGGCA-3′) and 3CLr (5′-GGGAAGCTTTTGGAAGGTAACACCAGAGCA-3′). After digestion with BamHI and HindIII, the PCR product was inserted into the BamHI and HindIII sites of the vector pQE30 (Qiagen). The residues in the expression tag are “MRGSHHHHHHGSTM”. The SARS_3CLpro insert was verified by sequencing.
Expression and purification of SARS_3CLpro
Escherichia coli M15 cells transformed with the plasmid pQE30-SARS_3CLpro were grown in 100 ml LB medium containing ampicillin (100 mg/L) and kanamycin (25 mg/L) at 37°C overnight and then inoculated into 1 L LB supplemented with both the antibiotics. The expression of SARS_3CLpro was induced by the addition of 0.5 mM of isopropyl β-d-thiogalactoside (IPTG). After induction for 5 h at 18°C, the cells were harvested by centrifugation at 4000g, 4 °C for 30 min. The pellet was washed, frozen, and then disrupted by sonication against Buffer A (20 mM Tris–HCl, 0.5 M NaCl, and 5 mM imidazole, pH 8.0). The lysed cells were centrifuged at 14,000g at 4 °C for 1 h. Keep the supernatant and discard the pellet. A 1-ml HiTrap Ni2+ chelating column was equilibrated with 10 ml of sterile deionized water, 50 mM NiSO4, and finally 10 ml Buffer A. The supernatant was passed over the column at a flow rate of 5 ml/min, followed by washing it with 20 ml Buffer A and 20 ml Buffer B (80 mM imidazole in Buffer A), respectively. The protease of interest was eluted with 10 ml Buffer C (20 mM Tris–HCl, 0.5 M NaCl, and 0.5 M imidazole, pH 8.0) and then purified further by gel filtration using a HiTrap 16/60 Sephacryl S100 column pre-equilibrated with Buffer D (5 mM dithiothreitol, 150 mM NaCl, and 10 mM Tris–HCl, pH 7.5) through an FPLC system (Pharmacia). The highly purified His-tagged SARS_3CLpro with the yield of 15 mg/L was obtained.
In-gel digest and peptide extraction
The protocol used for the in-gel digest in this study was modified according to the literature method described by Yu et al. [27]. The gel band of interest (SARS_3CLpro) was exercised from the Coomassie-stained SDS–PAGE gel with a steel scalpel and destained in an Eppendorf tube by washing sequentially with 100 μl of 30% CH3CN/100 mM ammonium bicarbonate. The washing step was repeated until the gel bands were clear. And then the gel band was completely dried by a Speed-Vac Vacuum centrifuge apparatus (Savant, Holbrook, NY) and cut into small pieces. The dried pieces were re-swollen by adding about 30 μl of 50 mM ammonium bicarbonate (pH 8.3). The volume added was to the minimum necessary to completely cover the gel pieces and then trypsin was added to the ratio of enzyme to sample in 1:20 (w/w). The gel pieces were incubated at 37 °C for 12–16 h.The tryptic peptides were extracted by adding 30 μl solution containing 60% CH3CN/0.1% TFA and vortexing for 4 min before removing the solution. This extraction step was performed three times with the same solution. The extraction solution was pooled together in a 0.5 ml Eppendorf tube and evaporated to 10–20 μl by Speed-Vac Vacuum centrifuge apparatus.
LC-ion trap-MS and MS/MS
The LC/MS system used for analyzing tryptic peptides was a combination of HP1100 (Agilent, Cheshire, UK) LC system with LCQ-DECA Mass Spectrometry (Thermofinnigan, San Jose, CA). A microbore reverse phase column (C8 50×1.0 mm ID, 7 μm, ABI RP300) was used for LC separation. Solvent A was 0.1% FA in 100% (v/v) water and solvent B was 0.1% FA in 100% (v/v) CH3CN. The gradient started at 5% B, held for 2 min, and went linearly to 80% B in 50 min. The peptide mixture was injected into the column by an autosampler and separated at a flow rate of 200 μl/min. The fractions were detected by PDA (TSP UV6000) and directly introduced on-line into ESI source. The operating condition was optimized with standard solution provided by manufactures and the working parameters of ion source were as follows: capillary temperature, 200 °C; spray voltage, 5 kV; capillary voltage, 15 V; and sheath gas flow rate, 20 arb. To get more mass spectra within an LC peak, two types of scan modes, full scan and MS/MS (with data-dependent), were used for acquiring more data points. The scan mass range was from m/z 400 to m/z 2000 and the collision energy was set at 38%.
Results and discussion
Construction of the expression vector pQE30-SARS_3CLpro
The SARS_3CLpro PCR product verified by sequencing was digested with BamHI and HindIII, and then inserted into the BamHI and HindIII sites of the vector pQE30. E. coli M15 cells transformed with the plasmid pQE30-SARS_3CLpro were used for the expression of His-tagged SARS_3CLpro.
Expression and purification of SARS_3CLpro
Based on the optimization of the expression and purification method of SARS_3CLpro from E. coli, the homogeneous protein was successfully isolated by two chromatographic steps. The SDS–PAGE analysis of the purification of SARS_3CLpro is shown in Fig. 1 and the purification scheme of SARS_3CLpro expressed in E. coli transformed with pQE30-SARS_3CLpro in 1 L culture is listed in Table 1 .
Fig. 1.
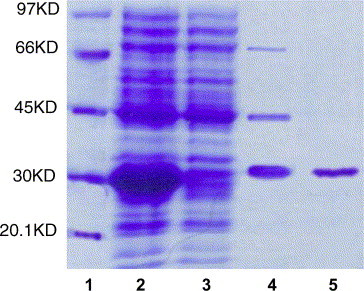
SDS-PAGE analysis of the purification of SARS_3CLpro (1, Marker; 2, supernatant; 3, pellet; 4, purified by NTA-Ni2+ affinity column; and 5, after purified by FPLC system).
Table 1.
Purification Scheme of SARS_3CLpro expressed in E. coli transformed with pQE30-SARS_3CLpro (1 L culture)
| Step | Total protein (mg) | SARS_3CLpro (mg) | Purification factor | 3CLpro yield (%) |
|---|---|---|---|---|
| Extraction | 500 | 50 | 1 | 100 |
| Ni2+-affinity column | 50 | 40 | 8 | 80 |
| Gel filtration | 15 | 15 | 10 | 30 |
From these results it can be seen that the use of pQE30-SARS_3CLpro plasmid and expression in E. coli M15 cell can produce a large amount of soluble SARS_3CLpro. The purification procedure is also easy to be handled.
LC/MS and LC/MS/MS analysis of SARS_3CLpro and tryptic peptides
The result of data search using MS/MS raw data of tryptic peptides from gel band shows that the 3CLpro is the first candidate with a summary score of 456.5, which is much higher than that of the second candidate (score 46.5), and it also shows that eleven peptides (T1–T11) were matched with tryptic peptides of 3CLpro (Fig. 2 ).
Fig. 2.
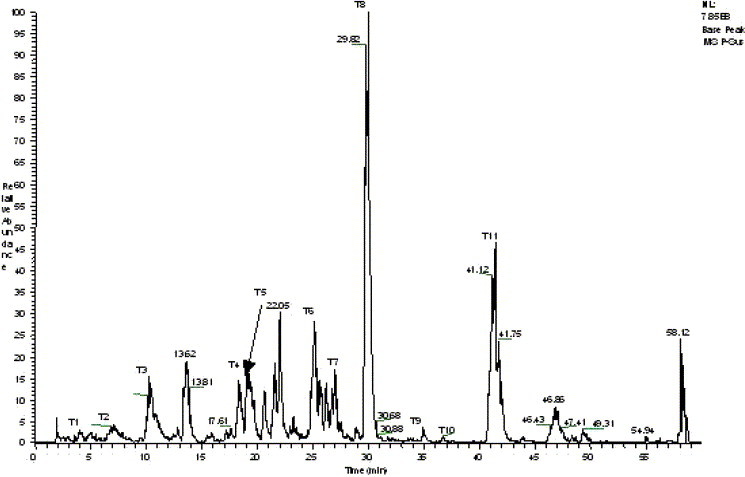
Base peak chromatogram of the peptides from LC/MS analysis of tryptic digests of SARS_3CLpro.
The MS/MS spectrum of doubly charged precursor ion of T3 peptide at m/z 566.2 was displayed as an example in Fig. 3 . Most of the b and y ions were detected (>80%). The total number of amino acids contained within the eleven tryptic peptides was 145 (Scheme 1 ), with the protein coverage at 46.4%.
Fig. 3.
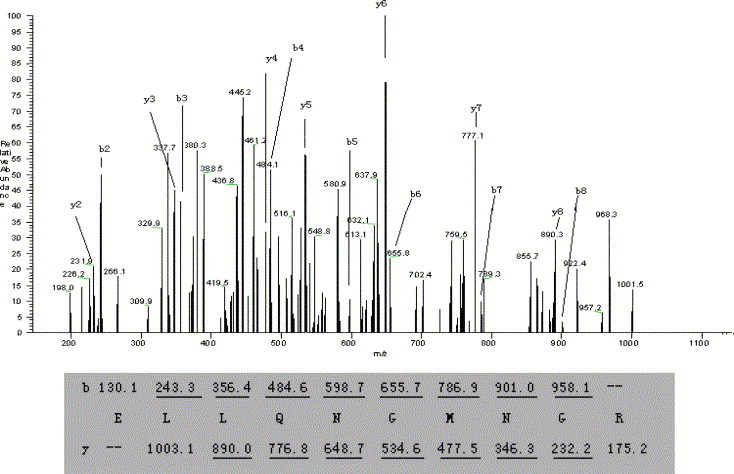
MS/MS spectrum of the doubly charged precursor ion with m/z 566.2(T3) at a retention time of 10.25 min. A sequence is confirmed from the labeled b- and y-ions in the spectrum. Ions observed in the spectrum are underlined and assigned.
Scheme 1.

Sequence of SARS_3CLpro showing the coverage of the protein obtained by mass spectrometry of in-gel tryptic digest. (Fragments of the sequence resolved by LC/MS/MS are shown in boldface and underline, and the expression tag is highlighted in a pane.)
The molecular weight of 3CLpro was also determined by the LC-MS system. The LC condition was based on that above described for peptide separation. An LC peak at a retention time of 18 min was observed (data not shown). Mass spectrum corresponding to this LC peak gave a multiple charge ion (Fig. 4 ). The molecular weight was obtained by deconvolution algorithm in “sequest” program. The measured mass of 3CLpro protease is 35831 (Fig. 5 ) and the difference between measured and theoretical mass (mw.35832) was only 1 dalton. These results completely determine the identity of the expressed SARS_3CLpro in this work.
Fig. 4.
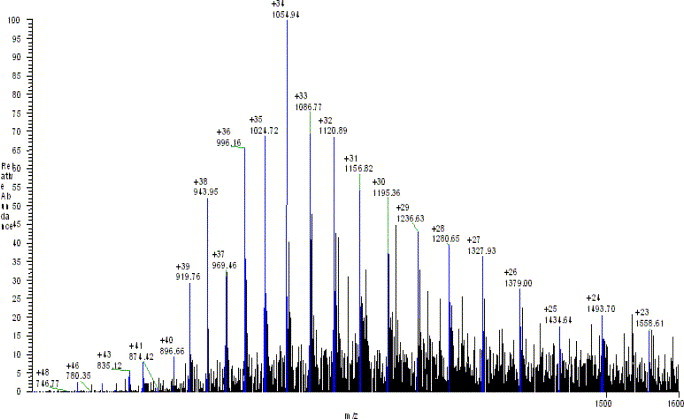
Multiple charge ions of SARS_3CLpro protease.
Fig. 5.
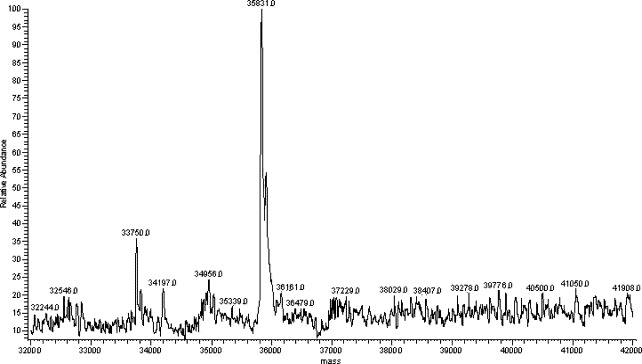
Molecular weight of SARS_3CLpro protease.
In conclusion, in this work we have succeeded in the molecular cloning of pQE30-SARS_3CLpro, and with this plasmid using E. coli as expression system a large amount of purified His-tagged SARS_3CLpro protease has been obtained by NTA-Ni2+ affinity chromatography. The achieved protease may be surely used for screening its crystallized conditions for X-ray crystallographic analysis.
Acknowledgements
The authors are grateful to the Chinese Military Medical Academy for providing the SARS-CoV (BJ01) virus. This work was supported by the 863 Hi-Tech Program (Grants 2001AA235051 and 2001AA235071), Shanghai Basic Research Project from the Shanghai Science and Technology Commission (Grant 02DJ14070), the National Natural Science Foundation of China (Grants 29725203 and 20072042), the State Key Program of Basic Research of China (Grant 2002CB512802), and the special programs of oppugning SARS from the Ministry of Science and Technology, Chinese Academy of Sciences, National Natural Science Foundation of China, and Shanghai Science and Technology Commission.
References
- 1.Kathryn V.H. SARS coronavirus: a new challenge for prevention and therapy. J. Clin. Invest. 2003;111:1605–1609. doi: 10.1172/JCI18819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peiris J.S, Lai S.T, Poon L.L, Guan Y, Yam L.Y, Lim W, Nicholls J, Yee W.K, Yan W.W, Cheung M.T, Cheng V.C, Chan K.H, Tsang D.N, Yung R.W, Ng T.K, Yuen K.Y. SARS study group, Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fouchier R.A, Kuiken T, Schutten M, Van Amerongen G, Van Doornum G.J, Van Den Hoogen B.G, Peiris M, Lim W, Stohr K, Osterhaus A.D. Aetiology: Koch’s postulates fulfilled for SARS virus. Nature. 2003;423:240. doi: 10.1038/423240a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kathryn V.H. SARS-associated coronavirus. N. Engl. J. Med. 2003;348:1948–1951. doi: 10.1056/NEJMp030078. [DOI] [PubMed] [Google Scholar]
- 5.B.S. Kamps, C. Hoffmann, “SARS Reference” second ed. Available from www.SARSReference.com 2003, Flying Publisher
- 6.Ziebuhr J, Heusipp G, Siddell S.G. Biosynthesis, purification, and characterization of the human coronavirus 229E 3C-like proteinase. J. Virol. 1997;71:3992–3997. doi: 10.1128/jvi.71.5.3992-3997.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dougherty W.G, Semler B.L. Expression of virus-encoded proteinases: functional and structural similarities with cellular enzymes. Microbiol. Rev. 1993;57:781–822. doi: 10.1128/mr.57.4.781-822.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eleout J.F, Rasschaert D, Lambert P, Levy L, Vende P, Laude H. Complete sequence (20 kilobases) of the polyprotein-encoding gene 1 of transmissible gastroenteritis virus. Virology. 1995;206:817–822. doi: 10.1006/viro.1995.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thiel V, Herold J, Schelle B, Siddell S.G. Viral replicase gene products suffice for coronavirus discontinuous transcription. J. Virol. 2001;75:6676–6681. doi: 10.1128/JVI.75.14.6676-6681.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herold J, Raabe T, Schelle-Prinz B, Siddell S.G. Nucleotide sequence of the human coronavirus 229E RNA polymerase locus. Virology. 1993;195:680–691. doi: 10.1006/viro.1993.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee H.J, Shieh C.K, Gorbalenya A.E, Koonin E.V, Monica N, Tuler J, Bagdzhadzhyan A, Lai M.M.C. The complete sequence (22 kilobases) of murine coronavirus gene 1 encoding the putative proteases and RNA polymerase. Virology. 1991;180:567–582. doi: 10.1016/0042-6822(91)90071-I. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rota P.A, Oberste M.S, Monroe S.S, Nix W.A, Campagnoli R. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 13.Marra M.A, Jones S.J.M, Astell C.R, Holt R.A, Brooks-Wilson A. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 14.Holmes K.V, Enjuanes L. Perspective: The SARS coronavirus: A postgenomic era. Science. 2003;300:1377–1378. doi: 10.1126/science.1086418. [DOI] [PubMed] [Google Scholar]
- 15.Krokhin O, Li Y, Andonov A, Feldman H, Flick R, Jones S, Stroeher U, Bastien N, Dasuri K.V.N, Cheng K, Simonsen J.N, Perreault H, Wilkins J, Ens W, Plummer F, Standing K.G. Mass spectrometric characterization of proteins from the SARS virus: a preliminary report. Mol. Cell. Proteomics. 2003;2:346–356. doi: 10.1074/mcp.M300048-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anand K, Ziebuhr J, Wadhwani P, Mesters J.R, Hilgenfeld R. Coronavirus main protease (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300:1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- 17.Ziebuhr J, Herold J, Siddell S.G. Characterization of a human coronavirus (strain 229E) 3C-like proteinase activity. J. Virol. 1995;69:4331–4338. doi: 10.1128/jvi.69.7.4331-4338.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anand K, Plam G.J, Mesters J.R, Siddell S.G, Ziebuhr J, Higenfeld R. Structure of coronavirus main protease reveals combination of a chymotrypsin fold with an extra alpha-helical domain. EMBO J. 2002;21:3213–3224. doi: 10.1093/emboj/cdf327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu D.X, Brown T.D. Characterization and mutational analysis of an ORF 1a-encoding proteinase domain responsible for proteolytic processing of the infectious bronchitis virus 1a/1b polyprotein. Virology. 1995;209:420–427. doi: 10.1006/viro.1995.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ziebuhr J, Snijder E.J, Gorbalenya A.E. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 2000;81:853–879. doi: 10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]
- 21.Hegyi A, Ziebuhr J. Conservation of substrate specificities among coronavirus main proteases. J. Gen. Virol. 2002;83:595–599. doi: 10.1099/0022-1317-83-3-595. [DOI] [PubMed] [Google Scholar]
- 22.Kim J.C, Spence R.A, Currier P.F, Lu X, Denison M.R. Coronavirus protein processing and RNA synthesis is inhibited by the cysteine protease inhibitor E64d. Virology. 1995;208:1–8. doi: 10.1006/viro.1995.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Someya Y, Takeda N, Miyamura T. Identification of active-site amino acid residues in the Chiba virus 3C-like protease. J. Virol. 2002;76:5949–5958. doi: 10.1128/JVI.76.12.5949-5958.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiong B, Gui C.S, Xu X.Y, Luo C, Chen J, Luo H.B, Chen L.L, Li G.W, Sun T, Yu C.Y, Yue L.D, Duan W.H, Shen J.K, Qin L, Shi T.L, Li Y.X, Chen K.X, Luo X.M, Shen X, Shen J.H, Jiang H.J. A 3D model of SARS_CoV 3CL protease and its inhibitors design by virtural screening. Acta Pharmacol. Sin. 2003;24:497–504. [PubMed] [Google Scholar]
- 25.Shen X, Xue J.H, Yu C.Y, Luo H.B, Qin L, Yu X.J, Chen J, Chen L.L, Xiong B, Yue L.D, Cai J.H, Shen J.H, Luo X.M, Chen K.X, Shi T.L, Li Y.X, Hu G.X, Jiang H.L. Small envelope protein E of SARS: cloning, expression, purification, CD determination, and bioinformatics analysis. Acta Pharmacol. Sin. 2003;24:505–511. [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch E.F, Maniatis T. second ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. Molecular Cloning: a Laboratory Manual. [Google Scholar]
- 27.Yu L, Zeng R, Shao X, Wang N, Xu Y, Xia Q. Identification of differentially expressed proteins between human hepatoma and normal liver cell lines by two-dimensional electrophoresis and liquid chromatography-ion trap mass spectrometry. Electrophoresis. 2000;21:3058–3068. doi: 10.1002/1522-2683(20000801)21:14<3058::AID-ELPS3058>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]


