Abstract
The impact of viral co‐infections and recently discovered viruses on the epidemiology of respiratory infections in children is still unclear. To simultaneously detect viruses that are involved in the aetiology of respiratory infections, we used a DNA/RNA microarray assay that identifies 17 different viruses or viral subtypes. Rhinopharyngeal washes were taken from 611 children (aged 1 month to 14 years) who presented in the emergency department with respiratory infections from June 2010 to June 2011 and were treated as outpatients (299, 48.9%) or hospitalized (312, 51.1%). Lower respiratory tract infection was diagnosed more often in hospitalized children (68% versus 36%, p 0.001). Of 397 children in which microarrays detected viral infection (70.1%), a single virus was found in 228 (57.4%) and two or more viruses in 169 (42.5%). The most prevalent viruses among children with positive samples were respiratory syncytial virus (RSV) in 225 (56.6%), parainfluenza virus (PIV) in 118 (29.7%), rhinovirus (RV) in 73 (18.4%), followed by influenza in 56 (14.1%), adenoviruses in 31 (7.8%), bocavirus in 25 (6.3%), human metapneumovirus in 15 (3.7%) and enteroviruses in 12 (3%). Most common viral co‐infections were RSVA–RSVB in 46 children (27.2%), RSV–Influenza in 20 (11.8%), RSV–RV in 18 (10.6%) and PIV–RV in 13 (7.7%). Multiple logistic regression analysis revealed that viral co‐infections were associated with increased probability for hospitalization (OR 1.52, 95% CI 1.01–2.29, p 0.04), and previous pneumococcal vaccination was associated with decreased probability for hospitalization (OR 0.52, 95% CI 0.33–0.81, p 0.004). We conclude that viral co‐infections are involved in a significant proportion of children with an acute respiratory infection and may increase the severity of clinical presentation and the risk for hospitalization.
Keywords: Children, co‐infection, epidemiology, infection, microarrays, respiratory, viral
Introduction
Respiratory infections represent a major public health problem because of their high incidence and ease of spread in the community [1]. Acute respiratory tract infections (ARTIs) are associated with significant morbidity and are the commonest reason for outpatient visits and hospitalizations among young children [2, 3]. Viruses are a leading cause of ARTIs with an epidemiological variability depending on climate, season and regions [4].
Viral aetiology is still unclear in a significant proportion of ARTIs and the role of viral co‐infections is controversial [5, 6]. During the past decade, new respiratory viruses, such as human metapneumovirus and human bocavirus, have been discovered. The impact of these viruses on the global epidemiology of ARTIs has not been fully elucidated [7, 8]. Moreover, human rhinoviruses and enteroviruses, previously identified in childhood upper respiratory tract infections, have been suspected as major aetiological agents of lower respiratory tract infections like bronchiolitis and pneumonia in infants [9, 10].
Viral respiratory diagnosis has traditionally relied on antigen detection and virus isolation. The lack of sensitivity of antigen detection, the delay in the results with virus isolation and the limitations of detecting only cultivable viruses usually made the diagnosis of respiratory viruses incomplete [11].
Detection and identification of viral respiratory agents, however, has improved in the last decade as the result of new molecular techniques. Molecular technology has increased sensitivity and accuracy, and the development of multiplex amplifications makes it possible to detect a broad panel of viruses simultaneously. Indeed, the use of a reverse transcription (RT) ‐PCR DNA/RNA microarray system allows the rapid and accurate detection of conventional and newly discovered viral respiratory pathogens in paediatric patients [12].
The purpose of this study was to investigate the role of viral co‐infections in ARTIs and to examine possible differences in the aetiology, clinical features and epidemiology of such infections between non‐hospitalized and hospitalized children, using a DNA/RNA microarray assay.
Materials and Methods
Patients and samples
A prospective study of respiratory tract infections in children was performed in the First Department of Paediatrics, University of Athens, ‘Aghia Sophia’ Children’s Hospital. The study population included children aged from 1 month to 14 years who presented in the emergency department with symptoms of ARTIs and were treated as outpatients or hospitalized between June 2010 and June 2011. Children were hospitalized on the basis of the clinical severity, as determined by the judgement of the attending physician. Demographic and clinical data, and nasopharyngeal washes were taken from outpatient children in the emergency department. For hospitalized children, samples were taken at the time of admission to exclude hospital‐acquired infections and clinical data were taken during their hospitalization and abstracted from medical records. Children with chronic heart, liver, or kidney disease, cystic fibrosis, or diabetes were reported as children with chronic health problems. Children with at least one dose of conjugated pneumococcal vaccine were regarded as vaccinated.
The ARTIs were classified into three categories: (i) upper respiratory tract infections (URTIs) were diagnosed when rhinitis, pharyngitis, laryngitis or otitis media were present in the absence of lower respiratory tract infection (LRTI) signs; (ii) LRTIs were diagnosed in the presence of signs of lower airway involvement (tachypnoea, dyspnoea, wheezing, rales) or a positive chest X‐ray in the absence of URTIs; and (iii) URTIs plus LRTIs were diagnosed with signs and symptoms from both upper and lower respiratory tract such as laryngotracheobronchitis, rhinitis and wheezing etc.
The study was approved by the hospital’s Ethics Committee and nasopharyngeal samples were collected after informed consent was obtained from the children’s parents and assent was given by the children. The samples were tested using an RT‐PCR DNA/RNA microarray system (CLART® Pneumovir DNA arrays assay; Genomica, Coslada, Madrid, Spain) in the Department of Cytopathology, University Hospital Attikon.
Sample collection
A nasopharyngeal lavage sample was obtained from each child using a standardized procedure. Each respiratory specimen was taken by introducing 5 mL of sterile saline solution into the nasal fossa, maintaining the patient’s head leant backwards and then collect the solution in a sterile container placed below nasal fossae, inclining the patient’s head. Samples were stored and processed according to the manufacturer’s instructions.
Nucleic acid extraction and DNA array assay
Total nucleic acid was extracted from 200 μL of each clinical specimen. The CLART® Pneumovir DNA arrays assay detects and characterizes 17 common human viruses causing respiratory infections in an 8‐h procedure after nucleic acid extraction. These viruses are: influenza virus (INFL) A, B and C; parainfluenza virus (PIV) 1, 2, 3 and 4 (subtypes A and B); respiratory syncytial virus type A (RSVA); respiratory syncytial virus type B (RSVB); rhinovirus (RV); human metapneumovirus (HMPV) (subtypes A and B); enterovirus (EV) (Echovirus); adenovirus (ADV); coronavirus (HCoV) (subtype 229E) and bocavirus (HBoV).
The detection is based on the amplification of specific fragments of the viral genome using RT‐PCR for amplification of a specific 120–330‐base‐pair fragment of the viral genome. During a 5‐h RT‐PCR/PCR amplification, the amplified products were labelled with biotin. Following amplification, hybridization with specific probes immobilized sites of the micro‐array. After incubation with a streptavidin–peroxidase conjugate, the addition of tetramethylbenzidine induced the appearance of an insoluble product that precipitated at the hybridization sites on the micro‐array. The results were processed by a microarray reader using specific software provided by the manufacturers, which allowed the automatic detection and interpretation of the results, giving a full and specific diagnosis of each analysis in a report.
Statistical analysis
Initially, descriptive statistics for the data were calculated and the possible association of the hospitalization with the examined variables was univariance tested using the Chi‐square or the Wilcoxon tests. In the next step multivariate analysis was also performed through multiple logistic regression analysis where, the hospitalization status was considered as the dependent variable and the age (categorical, three categories with baseline being 60+ months), sex (categorical with baseline: boys), a family member with respiratory viral infection (categorical with baseline: no), chronic health problem (categorical with baseline: no), number of wheezing episodes per year (ordered with an increment of two episodes), fever on presentation (categorical with baseline: no), bronchodilator use on presentation (categorical with baseline: no), antibiotic use on presentation (categorical with baseline: no), influenza vaccination (categorical with baseline: no) and pneumococcal vaccination (at least one dose) (categorical with baseline: no) as potential confounders. Finally the viral co‐infection was also introduced into the above model as an additional variable (categorical with baseline: no). Statistical analysis was performed with SAS, (version 9.2).
Results
Demographic and clinical characteristics
Children’s demographic and clinical characteristics according to their hospitalization status are presented in Table 1. A total of 611 children (boys 56%) with ARTIs were included in the study. In all, 312 (51.1%) were hospitalized and 299 (48.9%) were treated as outpatients. The median age was 28 months (IQR 10–59) but hospitalized children were younger (p 0.0001). An URTI was diagnosed in 235 children (38%), LRTI in 320 (53%) and URTI plus LRTI in 54 (9%). Hospitalized children were more often diagnosed with LRTI (p 0.001), had a chronic health problem (p 0.002), had a positive history for previous wheezing episodes (p 0.01), were using bronchodilators on presentation (p 0.0001) and were less vaccinated for pneumococcus (p 0.004).
Table 1.
Distribution of 611 children with respiratory tract infection by demographic and clinical characteristics and by hospitalization status.
| Parameter | Hospitalized (n = 312) n (%) | Non hospitalized (n = 299) n (%) | p‐value derived from Wilcoxon* or Chi‐square test |
|---|---|---|---|
| Age (months) (mean ± SD) | 34.0 ± 36.62 | 49.8 ± 42.84 | 0.0001* |
| Male | 180 (57.7) | 164 (54.9) | 0.48 |
| Family member with ARTI | 154 (49.4) | 127 (42.5) | 0.09 |
| Chronic health problem | 36 (11.5) | 11 (3.7) | 0.0003 |
| Wheezing episodes per year (mean ± SD) | 1.5 ± 2.38 | 1 ± 1.74 | 0.01* |
| Fever on presentation | 224 (71.8) | 187 (62.5) | 0.01 |
| Bronchodilator use on presentation | 78 (25.0) | 17 (5.7) | 0.0001 |
| Antibiotic use on presentation | 72 (23.1) | 63 (21.1) | 0.55 |
| No pneumococcal vaccination | 111 (35.6) | 62 (20.8) | 0.0001 |
| No influenza vaccination | 279 (89.4) | 274 (91.6) | 0.35 |
| Days of symptoms on presentation (mean ± SD) | 3.5 ± 2.3 | 3.1 ± 1.9 | 0.19* |
| Detection of virus | 203/289 (70.2) | 194/277 (70.0) | 0.99 |
| Viral co‐infection | 97/289 (47.7) | 72/277 (37.1) | 0.05 |
ARTI, acute respiratory tract infection; Chronic health problem, chronic heart, liver, or kidney disease, cystic fibrosis or diabetes.
Viral detection
Adequate sample was obtained from 566 out of 611 children (289/312 hospitalized and 277/299 non‐hospitalized). One or more respiratory viruses were detected in 397 of them (70.1%). Among the positive samples, a single virus was identified in 228 (57.4%), two viruses in 111 (28%), and three or more viruses in 58 (14.6%). Detection rates observed among different age groups were for children 1–12 months 73.6% (120/163), for 1–5 years 72.3% (191/265), and for 5–14 years 62.3% (86/138), (p 0.061).
The most prevalent viruses during the study period were RSV in 225 (56.6%) children, PIV in 118 (29.7%) and RV in 73 (18.4%), followed by INFL in 56 (14.1%), ADV in 31 (7.8%), HBoV in 25 (6.3%), HMPV in 15 (3.7%), EV in 12 (3%), and HCoV Type 229 in 2 (0.5%) (Fig. 1).
Figure 1.
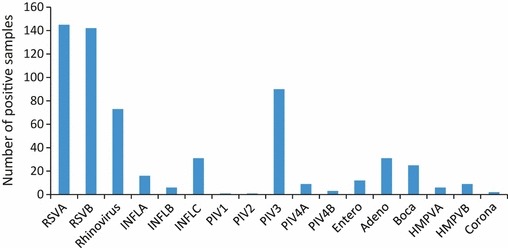
Number of positive samples for 17 different viruses or subtypes as single infections or co‐infections detected by microarrays in 397 children with respiratory tract infections.
Viral co‐infections were detected in 169 children (42.5%). The frequency of co‐infections were for infants <12 months 33.7% (55/163) and for children >12 months 28.3% (114/403) (p 0.119). The most frequent viral co‐infections were RSVA‐RSVB in 46 children (27.2%), RSV–INFL in 20 (11.8%), RSV–RV in 18 (10.6%), PIV–RV in 13 (7.7%), PIV–INFL–RSV in 9 (5.3%), RSV–PIV in 8 (4.7%), RSV–HBoV 5 in (3%), PIV–INFL in 5 (3%), PIV–RSV–ADV in 5 (3%), RSV–HMPV in 4 (2.3%), RSV–ADV in 3 (1.7%), PIV–ADV in 3 (1.7%), and PIV–INFL–ADV in 3 (1.7%) (Fig. 2).
Figure 2.
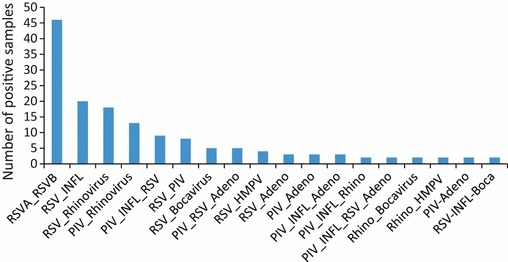
Viral co‐infections that were detected by microarrays in 397 children with respiratory tract infections.
There was no difference in the distribution of viruses between non‐hospitalized and hospitalized children, with the exception of RSV infection, which was detected more frequently in the latter (p 0.011).
Risk factors for hospitalization
Logistic regression analysis showed that the presence of viral co‐infection increased the risk for hospitalization (OR, 1.52; 95% CI, 1.01–2.29; p 0.04). When this analysis was performed only in children who had tested positive for at least one virus (n = 397) the increased risk for hospitalization remained (OR, 1.67; 95% CI, 1.061–2.65; p 0.026). Other risk factors associated with hospitalization were age <12 months (OR, 4.51; 95% CI, 2.60–7.81; p 0.0001), chronic health problem (OR, 3.48; 95% CI, 1.61–7.52; p 0.002), history of wheezing episodes (two more episodes, OR, 1.33; 95% CI, 1.08–1.64; p 0.01), fever on presentation (OR, 2.7; 95% CI, 1.8–4.07; p 0.0001) and bronchodilator use on presentation (OR, 4.50; 95% CI, 2.45–8.24; p 0.0001) (Table 2). In contrast, a negative association of pneumococcal vaccination with risk for hospitalization (OR, 0.52; 95% CI, 0.33–0.81; p 0.004) was found. Among hospitalized children there was no statistically significant association of viral co‐infections with use of bronchodilators and fever on presentation or length of hospitalization.
Table 2.
Multiple logistic analysis derived odds ratios (OR) and 95% confidence intervals (95% CI) for hospitalization by demographic and clinical characteristics.
| Variables | Category or increment | OR | 95% CI | p‐value | |
|---|---|---|---|---|---|
| Age | <12 months | 4.51 | 2.60 | 7.81 | 0.0001 |
| 12–59 | 1.45 | 0.92 | 2.30 | 0.11 | |
| 60+ | Baseline | ||||
| Sex | Female | 1.00 | 0.70 | 1.44 | 0.99 |
| Male | Baseline | ||||
| Family member with ARTI | Yes | 1.03 | 0.72 | 1.49 | 0.86 |
| No | Baseline | ||||
| Chronic health problem | Yes | 3.48 | 1.61 | 7.52 | 0.002 |
| No | Baseline | ||||
| No. of wheezing episodes per year | Two more | 1.33 | 1.08 | 1.64 | 0.01 |
| Fever on presentation | Yes | 2.70 | 1.80 | 4.07 | 0.0001 |
| No | Baseline | ||||
| Bronchodilator use on presentation | Yes | 4.50 | 2.45 | 8.24 | 0.0001 |
| No | Baseline | ||||
| Antibiotic use on presentation | Yes | 0.93 | 0.60 | 1.45 | 0.75 |
| No | Baseline | ||||
| Influenza vaccination | Yes | 1.83 | 0.99 | 3.36 | 0.06 |
| No | Baseline | ||||
| Pneumococcal vaccination | Yes | 0.52 | 0.33 | 0.81 | 0.004 |
| No | Baseline | ||||
| Additionally introduced variable, available for 566 children | |||||
| Viral co‐infection | Yes | 1.52 | 1.01 | 2.29 | 0.04 |
| No | Baseline | ||||
ARTI, acute respiratory tract infection; Chronic health problem, chronic heart, liver, or kidney disease, cystic fibrosis or diabetes; Pneumococcal vaccination, at least one dose.
Discussion
Broadening viral diagnosis in respiratory tract infections may help clinicians to decrease unnecessary prescriptions of antibiotics, implement early antiviral treatments when available, and prevent virus transmission.
In the present study we used a DNA/RNA microarray assay that allows detection and identification simultaneously of 17 types of common and newly discovered human viruses. This method is versatile and greatly expands the spectrum of detectable viruses in a single assay, while simultaneously allowing discrimination among viral subtypes [13].
To the best of our knowledge this is the largest series of children studied prospectively with the aim of detecting single or multiple respiratory viral infections by microarray technology. Few studies have evaluated the use of microarrays for the detection of multiple respiratory viruses in children [11, 12, 14]. The detection rate in these studies, depending on the clinical syndrome and the assay used, was between 58 and 85%, which is similar to our study (70.1%).
One of the drawbacks of virus detection by genetic amplification are false negatives, which are mainly the result of poor quality of the extracted DNA/RNA caused by an insufficient quantity of the initial sample, degradation of the virus genetic material because of inadequate storage of the sample or its loss during extraction. The presence of inhibitors of the enzyme mixture (reverse transcriptase and DNA polymerase) in samples in which virus detection is going to be performed (sputum, respiratory secretions etc.) is another possible explanation. Nevertheless, the assay used in our study eliminated such false negatives with the addition of an internal control that was extracted along with any possible viruses present in the sample and was subsequently amplified in the reaction tubes, confirming the correct efficiency of both the extraction and the PCR amplification [15].
In our study a significant proportion of viral co‐infections were detected among children with positive samples (42.5%) and were associated with increased possibility for hospitalization. The prevalence of co‐infections in other studies ranged between 15% and 44% in children seen in the hospital or emergency department with diverse types of respiratory tract infections and detection methods [16, 17, 18, 19, 20, 21, 22, 23, 24] .
There are conflicting results whether multiple viral infections are associated with a more severe clinical presentation compared with a single infection [25]. Semple et al. [21] reported a ten‐fold increase in the relative risk of admission to a paediatric intensive‐care unit for mechanical ventilation caused by severe bronchiolitis in patients with dual infection with HMPV and RSV. Paranhos‐Baccala et al. [1] showed that the presence of more than one pathogen, and moreover, the association of RSV with rhinoviruses or HMPV, might influence the natural course of bronchiolitis. Other researchers found that children with HMPV/RSV co‐infection were more likely to develop pneumonia, but they did not manifest increased disease severity as defined by the duration of hospitalization and need for oxygen supplementation [26]. In contrast, other investigators failed to demonstrate more serious disease with dual infections [27, 28, 29].
The detection of multiple viruses raises the question if all viruses that are detected are responsible for the clinical symptoms or if they are innocent bystanders. These multiple viruses can be successive infections and molecular tests could detect a persistent genome in the absence of viral activity. The DNA microarray assay employed is only a qualitative technique not able to identify the predominant viral agent that could be considered as the cause of the acute respiratory syndrome. Quantitative PCR methods are probably a better tool to discriminate between the active virus and prolonged presence of previous viruses [6, 17].
Interestingly, pneumococcal vaccination was associated with a decreased probability for hospitalization. Pneumococcal interaction with viral pathogens is well established and can increase the clinical severity of respiratory infections [30, 31].
The present study has limitations because it lasted 1 year and some viruses have different circulation in the community in consecutive years. Moreover, there was no quantification of the viral load or detection of bacterial co‐infection. The specific microarray assay that was used has limited published clinical validation, especially in the paediatric population. A significant number of RSVA–RSVB co‐infections were detected that need further exploration with specific RT‐PCR assays to confirm the specificity of the assay for these viral subtypes. Additionally, the assay may underestimate the presence of some viruses like coronaviruses because it can detect only Coronavirus subtype 229E. In a significant proportion of children that present with symptoms of respiratory infection, no viral or bacterial pathogen can be detected by conventional or molecular methods. This implies that either molecular methods need further improvement for their sensitivity or there are still other viruses to be discovered.
The issue of the association of viral co‐infection with the severity of clinical presentation remains controversial. There is a compelling need to perform multi‐centre studies with common protocols, which will last several years and will measure the viral load for different viruses in co‐infections. Comparison of viral detection in a control group with asymptomatic children may help to understand which viruses can be innocent rhinopharyngeal colonizers.
In conclusion, in our study viral co‐infections in children with respiratory tract infections increased the risk for hospitalization. Simultaneous detection of respiratory pathogens with microarrays may elucidate the interplay between viruses, bacteria and the host and may determine which viral pathogens affect the clinical severity of respiratory infections.
Transparency Declaration
The author declare no conflicts of interest related to this manuscript.
Acknowledgments
This work was presented in part at the Interscience Conference on Antimicrobial Agents and Chemotherapy, held in San Francisco, USA, September 2012.
References
- 1. Paranhos‐Baccala G, Komurian‐Pradel F, Richard N, Vernet G, Lina B, Floret D. Mixed respiratory virus infections. J Clin Virol 2008; 43: 407–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fabbiani M, Terrosi C, Martorelli B et al. Epidemiological and clinical study of viral respiratory tract infections in children from Italy. J Med Virol 2009; 81: 750–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Regamey N, Kaiser L, Roiha HL et al. Viral etiology of acute respiratory infections with cough in infancy: a community‐based birth cohort study. Pediatr Infect Dis J 2008; 27: 100–105. [DOI] [PubMed] [Google Scholar]
- 4. Armstrong GL, Conn LA, Pinner RW. Trends in infectious disease mortality in the United States during the 20th century. JAMA 1999; 281: 61–66. [DOI] [PubMed] [Google Scholar]
- 5. Henrickson KJ, Hoover S, Kehl KS, Hua W. National disease burden of respiratory viruses detected in children by polymerase chain reaction. Pediatr Infect Dis J 2004; 23: S11–S18. [DOI] [PubMed] [Google Scholar]
- 6. Franz A, Adams O, Willems R et al. Correlation of viral load of respiratory pathogens and co‐infections with disease severity in children hospitalized for lower respiratory tract infection. J Clin Virol 2010; 48: 239–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. van den Hoogen BG, Osterhaus DM, Fouchier RA. Clinical impact and diagnosis of human metapneumovirus infection. Pediatr Infect Dis J 2004; 23: S25–S32. [DOI] [PubMed] [Google Scholar]
- 8. Kesebir D, Vazquez M, Weibel C et al. Human bocavirus infection in young children in the United States: molecular epidemiological profile and clinical characteristics of a newly emerging respiratory virus. J Infect Dis 2006; 194: 1276–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jacques J, Bouscambert‐Duchamp M, Moret H et al. Association of respiratory picornaviruses with acute bronchiolitis in French infants. J Clin Virol 2006; 35: 463–466. [DOI] [PubMed] [Google Scholar]
- 10. Papadopoulos NG, Moustaki M, Tsolia M et al. Association of rhinovirus infection with increased disease severity in acute bronchiolitis. Am J Respir Crit Care Med 2002; 165: 1285–1289. [DOI] [PubMed] [Google Scholar]
- 11. Frobert E, Escuret V, Javouhey E et al. Respiratory viruses in children admitted to hospital intensive care units: evaluating the CLART® pneumovir DNA array. J Med Virol 2011; 83: 150–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Renois F, Talmud D, Huguenin A et al. Rapid detection of respiratory tract viral infections and coinfections in patients with influenza‐like illnesses by use of reverse transcription‐pcr DNA microarray systems. J Clin Microbiol 2010; 48: 3836–3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang D, Coscoy L, Zylberberg M et al. Microarray‐based detection and genotyping of viral pathogens. Proc Natl Acad Sci USA 2002; 99: 15687–15692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Falchi A, Turbelin C, Andreoletti L et al. Nationwide surveillance of 18 respiratory viruses in patients with influenza‐like illnesses: a pilot feasibility study in the French sentinel network. J Med Virol 2011; 83: 1451–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dimopoulos G, Lerikou M, Tsiodras S et al. Viral epidemiology of acute exacerbations of chronic obstructive pulmonary disease. Pulm Pharmacol Ther 2012; 25: 12–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Freymuth F, Vabret A, Cuvillon‐Nimal D et al. Comparison of multiplex pcr assays and conventional techniques for the diagnostic of respiratory virus infections in children admitted to hospital with an acute respiratory illness. J Med Virol 2006; 78: 1498–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Aberle JH, Aberle SW, Pracher E, Hutter HP, Kundi M, Popow‐Kraupp T. Single versus dual respiratory virus infections in hospitalized infants: impact on clinical course of disease and interferon‐γ response. Pediatr Infect Dis J 2005; 24: 605–610. [DOI] [PubMed] [Google Scholar]
- 18. Jartti T, Lehtinen P, Vuorinen T et al. Respiratory picornaviruses and respiratory syncytial virus as causative agents of acute expiratory wheezing in children. Emerg Infect Dis 2004; 10: 1095–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Calvo C, Garcia‐Garcia ML, Blanco C et al. Multiple simultaneous viral infections in infants with acute respiratory tract infections in Spain. J Clin Virol 2008; 42: 268–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Canducci F, Debiaggi M, Sampaolo M et al. Two‐year prospective study of single infections and co‐infections by respiratory syncytial virus and viruses identified recently in infants with acute respiratory disease. J Med Virol 2008; 80: 716–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Semple MG, Cowell A, Dove W et al. Dual infection of infants by human metapneumovirus and human respiratory syncytial virus is strongly associated with severe bronchiolitis. J Infect Dis 2005; 191: 382–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stempel HE, Martin ET, Kuypers J, Englund JA, Zerr DM. Multiple viral respiratory pathogens in children with bronchiolitis. Acta Paediatr 2009; 98: 123–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nascimento MS, Souza AV, Ferreira AV, Rodrigues JC, Abramovici S, Silva Filho LV. High rate of viral identification and coinfections in infants with acute bronchiolitis. Clinics (Sao Paulo) 2010; 65: 1133–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Martin ET, Kuypers J, Wald A, Englund JA. Multiple versus single virus respiratory infections: viral load and clinical disease severity in hospitalized children. Influenza Other Resp Viruses 2012; 6: 71–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sly PD, Jones CM. Viral co‐detection in infants hospitalized with respiratory disease: is it important to detect? J Pediatr (Rio J) 2011; 87: 277–280. [DOI] [PubMed] [Google Scholar]
- 26. Caracciolo S, Minini C, Colombrita D et al. Human metapneumovirus infection in young children hospitalized with acute respiratory tract disease: virologic and clinical features. Pediatr Infect Dis J 2008; 27: 406–412. [DOI] [PubMed] [Google Scholar]
- 27. Wilkesmann A, Schildgen O, Eis‐Hubinger AM et al. Human metapneumovirus infections cause similar symptoms and clinical severity as respiratory syncytial virus infections. Eur J Pediatr 2006; 165: 467–475. [DOI] [PubMed] [Google Scholar]
- 28. Garcia‐Garcia ML, Calvo C, Martin F, Perez‐Brena P, Acosta B, Casas I. Human metapneumovirus infections in hospitalised infants in Spain. Arch Dis Child 2006; 91: 290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bezerra PG, Britto MC, Correia JB et al. Viral and atypical bacterial detection in acute respiratory infection in children under five years. PLoS ONE 2011; 6: e18928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Techasaensiri B, Techasaensiri C, Mejias A, McCracken GH Jr, Ramilo O. Viral coinfections in children with invasive pneumococcal disease. Pediatr Infect Dis J 2010; 29: 519–523. [DOI] [PubMed] [Google Scholar]
- 31. Vu HT, Yoshida LM, Suzuki M et al. Association between nasopharyngeal load of streptococcus pneumoniae, viral coinfection, and radiologically confirmed pneumonia in Vietnamese children. Pediatr Infect Dis J 2011; 30: 11–18. [DOI] [PubMed] [Google Scholar]


