Abstract
The structures of snake venom metalloproteases (SVMPs) are proposed to be useful models to understand the structural and functional relationship of ADAM (a disintegrin and metalloprotease) which are membrane-anchored proteins involved in multiple human diseases. We have purified, sequenced and determined the structures of two new P-III SVMPs – atragin and kaouthiagin-like (K-like) from Naja atra. Atragin exhibits a known C-shaped topology, whereas K-like adopts an I-shaped conformation because of the distinct disulfide pattern in the disintegrin-like (D) domain. K-like exhibits an enzymatic specificity toward pro-TNFα with less inhibition of cell migration, but atragin shows the opposite effect. The specificity of the enzymatic activity is indicated to be dominated mainly by the local structures of SVMP in the metalloprotease (M) domain, whereas the hyper-variable region (HVR) in the cysteine-rich (C) domain is involved in a cell-migration activity. We demonstrate also a pH-dependent enzymatic activity of atragin that we correlate with the structural dynamics of a Zn2+-binding motif and the Met-turn based on the structures determined with a pH-jump method. The structural variations between the C- and I-shapes highlight the disulfide bond patterns in the D domain of the ADAM/adamalysin/reprolysins family proteins.
Keywords: ADAM, P-III SVMP, ADAMalysin, Metzincin, Zinc-binding motif
1. Introduction
P-III snake venom metalloproteases (SVMPs) 1 comprising the metalloprotease (M), disintegrin-like (D), and cysteine-rich (C) domains belong to the ADAM/adamalysin/reprolysin family (Fox and Solange, 2005, Gomis-Ruth, 2003, Paine et al., 1992, White, 2003). The proportion of SVMPs in the elapid venom is much lower than that in the viper venom (∼30%) (Calvete et al., 2007, Li et al., 2004, Sanz et al., 2008), resulting in the limited investigation on SVMPs in the elapid venom in contrast to the extensive study in the viper venom. The viper P-III SVMPs can inhibit platelet aggregation (Fox and Bjarnason, 1995, Wang et al., 2003, Zhou et al., 1996) and involve the digestion of ECM molecules and the cleavage of some plasma proteins that are crucial for hemostasis (Kamiguti et al., 1996, Serrano et al., 2007), whereas the elapid P-III SVMPs show various functions. For example, cobrin from Naja naja has been identified as a human complement component C3-cleaving protease (O’Keefe et al., 1988); mocarhagin from Naja mossambica can cleave the PSGL-1 and GPIb components of a GPIb-IX-V complex (Ward et al., 1996); mikarin from Micropechis ikaheka can activate prothrombin (Gao et al., 2002); atrahagin from Naja atra has a histamine-releasing activity (Wei et al., 2006). The SVMPs of both types belong to metzincins with a consensus zinc binding sequence, HEXXHXXGXXHD, and one conserved methionine-containing turn in the M domain (Gomis-Ruth, 2003). The zinc atom is ligated by three histidines with a distance of about 2–2.5 Å and by one glutamate residue in the consensus zinc binding sequence. The zinc-binding motif is responsible for the substrate cleavage (Ramos and Selistre-de-Araujo, 2006). The regulation of its activity is needed in consideration of its diverse substrate specificity and the lack of the proteolytic activity in the gland.
The crystal structures of three P-III SVMPs from the viper venom – including a P-IV lectin containing SVMP, reclassified as the P-IIId based on a lack of the P-IV mRNA transcript in the venom – indicate a dynamic C-shaped MDC modular architecture (Igarashi et al., 2007, Takeda et al., 2006, Takeda et al., 2007). The widely conserved cysteine residues in P-III SVMPs and mammalian ADAMs have been observed to reveal the MDC domain architecture in the ADAM/adamalysin/reprolysins family (Takeda et al., 2006), even though the disulfide patterns of P-I, P-II and P-III SVMPs are known to vary significantly and might play a role in the post-translational proteolytic process and their venom complexity (Fox and Serrano, 2008). The notable RGD loop in the disintegrin domain of the P-II class becomes modified into a cysteine-contained loop (XXCD) in the P-III disintegrin-like domain (Ramos and Selistre-de-Araujo, 2006). Much evidence indicates that the functional region, which was previously attributed to the D domain, is located at the C domain to interact with the VWA domain (Pinto et al., 2007, Serrano et al., 2006) existing in integrin molecules while the C domain has been indicated to be involved in multiple biological functions and the target selection (Kamiguti et al., 2003, Serrano et al., 2005, Serrano et al., 2006). Further functional and structural characterizations of other P-III SVMPs are thus warranted.
ADAMs (a disintegrin and metalloproteases) are transmembrane proteins from various species of ∼40 kinds in total, of which 19 adam genes exist in humans (Seals and Courtneidge, 2003, White, 2003). Both ADAM and P-III SVMPs belong to the ADAM/adamalysin/reprolysins family. The shedding events by these ADAM molecules can produce cytokines and growth factors. For example, ADAM17 (TACE; tumor necrosis factor-α converting enzyme) can release TNFα as one important immunomodulatory and pro-inflammatory cytokine (Moss et al., 1997), and TGFα (Peschon et al., 1998). ADAM9 is implicated in releasing HB-EGF (heparin-binding EGF) from membrane-anchored HB-EGF that can bind to the α3β1 integrin to inhibit the proliferation of neighboring cells (Raab and Klagsbrun, 1997) and be involved in the EGF receptor transactivated by G-protein-coupled receptors (Prenzel et al., 1999). ADAM10 (Kuzbanian) can shed Delta affecting the Delta/Notch pathway (Lieber et al., 2002, Pan and Rubin, 1997), which is critical in the development of the nervous system. The shedding events by ADAMs are also correlated with multiple human diseases – cancer, asthma, cardiac hypertrophy, SARS (severe acute respiratory syndrome), etc. (Asakura et al., 2002, Haga et al., 2008, Van Eerdewegh et al., 2002, Wu et al., 1997). Although the shedding events of ADAMs are crucial, the full ADAM structure is not yet available. Our current understanding of the shedding mechanism of ADAMs relies on a model based on the available crystal structures of the viper P-III SVMPs (Igarashi et al., 2007, Muniz et al., 2008, Takeda et al., 2006, Takeda et al., 2007, Zhu et al., 2009). Further investigation of the structures and functions of SVMPs from other sources, such as the elapid venom can aid not only the development of therapeutics for envenomation from various species of snakes but also the future toxin-based drug discovery. In this study, we have purified, sequenced and determined the crystal structures of two newly identified P-III SVMPs named atragin and Kaouthiagin-like (K-like) from N. atra. The structural information about their MDC architectures provides an improved understanding of shedding events of ADAMs and various biological activities, including the proteolytic specificity against pro-TNFα and the effect on cell migration from atragin and K-like.
2. Experimental
2.1. Purification of two P-III SVMPs atragin and K-like from N. atra
The crude venom of Taiwan cobra (acquired from the Tainan snake farm in Taiwan) was lyophilized. This lyophilized crude venom was dissolved in a phosphate buffer (50 mM, pH 7.4) containing NaCl (0.5 M) and then centrifuged (9000g) for 15 min. The supernatant was filtered through a 0.22-μm filter and then loaded into a G75 column (GE Healthcare). The HMF (high-M W fraction) from the eluted proteins was dialyzed against the phosphate buffer (50 mM, pH 8.5). Dialyzed proteins were loaded into a Hitrap Heparin HP column (GE Healthcare) and eluted with a phosphate buffer (50 mM, pH 8.5) with a gradient of NaCl from 0 to 2 M. Purified venom proteins were analyzed by SDS–PAGE (10%) and visualized with Coomassie blue staining.
2.2. Primary sequence determination
The purified atragin was sent outside (Mission Bio. Co., Taiwan) to determine the N-terminal sequence, from which we obtained a sequence of ten amino acids (TNTPEQDRYL) from the N-terminal. A search of the sequence similarity with NCBI-BLAST (Basic Local Alignment Search Tool) disclosed two homologous venom proteins: cobrin from N. naja and mocarhagin from N. mossambica. The total RNA was extracted from the snake gland of Taiwan cobra (purchased from the Tainan snake farm) using the TRIzol reagent (GIBCO BRL). The cDNA was prepared with a reverse transcription reaction using reverse transcriptase (Superscript II, Invitrogen). The forward primer (5′-CACCAGAAGAGCTCAGGTTGGCTTGGAA-3′) and the reverse primer (5′-AGAAATAGGATGATCATTTAGTGAGAATCGAAAGT-3′) were synthesized according to the conserved nucleotide sequence in the untranslated region (UTR) of mRNAs between cobrin and mocarhagin. The polymerase chain reaction (PCR) products of two kinds were acquired with Taq DNA polymerase (Invitrogen), and subsequently ligated into PET102 vectors (Invitrogen). The nucleotide sequences and deduced primary sequences of atragin and K-like were determined on sequencing.
2.3. Crystallization
Initial crystallization conditions of purified atragin (10 mg/mL) and K-like (10 mg/mL) were screened with crystallization kits (Hampton and Emerald). We obtained crystals of atragin with PEG6000 (7%) in a sodium citrate buffer (100 mM, pH 5) using a vapor-diffusion hanging-drop method at 20 °C in 3 days, and crystals of K-like with ethandiol (8%) and PEG8000 (10%) in a Hepes buffer (100 mM, pH 6.8) at 20 °C in 2 weeks. The gradients of pH and PEG concentrations were varied to yield the best crystals suitable for X-ray diffraction of satisfactory quality.
2.4. Absorption and fluorescence spectra of atragin crystals
Crystals of atragin grown in the acid buffer (citrate, pH 5.0) were back-soaked and transferred into a neutral solution (Hepes, 100 mM, pH 7.4) for 10–20 s for the experiment of the pH jump. The crystals with and without the pH-jump procedure were dipped in ethandiol (15%, in citrate, pH 5.0, and Hepes, pH 7.4, respectively) for 10 s, and then frozen with liquid nitrogen. With the crystals under a nitrogen stream at 100 K, we conducted the energy-scanned experiment with X-ray near the K-absorption edge of Zn and the fluorescence signals from these two crystals were recorded with a PIPS detector (Canberra) at the beamline NSRRC-13B1 (Hsinchu, Taiwan).
2.5. Data collection and structure determination
Crystals of atragin dipped briefly in ethandiol (15%) were immediately frozen with liquid nitrogen. With these frozen crystals under a nitrogen stream at 100 K, we collected X-ray diffraction data at a wavelength of 1.0 Å on beamlines NSRRC_13B1/C1 (Hsinchu, Taiwan); the anomalous data of the zinc absorption edge were collected at a wavelength of 1.2822 Å on the beamline SPring8_12B2 (Harima, Japan). Crystals of K-like were dipped in glycerol (20%) and other procedures were the same as for atragin. The space groups of both crystals belong to P43212 with different unit-cell dimensions: for atragin, a = 91.65 Å and c = 124.24 Å; for K-like, a = 63.00 Å and c = 273.43 Å. The data were indexed and processed with HKL2000 program (Otwinowski and Minor, 1997). Molecular replacement was performed using the M–D domain (a.a. 185–513) and the C domain of VAP1 (2ERO) as the search model with MOLREP in suite CCP4 (CCP4, 1994). Several cyclic model refinements were performed according to density maps of |2F o − F c| and |F o − F c| coefficients with CNS v1.2 (Brunger et al., 1998) and the Maximum-Likelihood method using default weights with REFMAC5 (Murshudov et al., 1998). The flexible parts of the M and C domains in the structures, which will be discussed later, mainly cause the high root-mean-square deviations from ideal bond lengths and angles at medium–high resolutions. The final R/R free of atragin was 22.40/23.37% at a resolution of 2.5 Å. The structures of the M and C domains of atragin then served as search models using molecular replacement to obtain an initial model of K-like. The D domain structure of K-like was manually constructed, followed by several cyclic refinements. The final R/R free of K-like was 23.36/24.11% at a resolution of 2.3 Å.
During the crystallography refinement of atragin, the Zn2+ atom was placed near the zinc-binding motif based on the |2F o − F c| composite omit density map and Zn-anomalous difference-Fourier map; it exhibited a large temperature B-factor (Table 1 ). To ensure the correct assignment of this Zn2+ instead of a water molecule in the electron density, we performed an additional refinement cycle using REFMAC5 with a water molecule to replace the Zn2+ in both acidic and neutral conditions after the pH jump. The results showed that the replacement of water in acidic and neutral conditions significantly decreased the B-factors from 107.3 to 61.0 Å2 and 51.5 to 11.0 Å2, respectively. The B-factor (11.0 Å2) of the replacing water was much lower than those of the nearby water molecules (averaged B-factor ∼50 Å2) in the neutral condition, indicating that the maximum electron density was properly assigned as Zn2+ rather than a water molecule. In the acidic condition, the B-factor (61.0 Å2) of the replacing water was still slightly larger than those of the neighboring waters, suggesting that the Zn2+ atom exhibits much dynamic motion.
Table 1.
Crystal diffraction and structural statistics.
| Atragin (PDB ID 3K7L) | K-like (PDB ID 3K7N) | |
|---|---|---|
| I. Crystal data | ||
| Wavelength (Å) | 1.00 | 1.00 |
| Temperature (K) | 100 | 100 |
| Resolution range (Å) (outermost shell) | 30–2.5 (2.59–2.5) | 30–2.3 (2.38–2.3) |
| Space group | P43212 | P43212 |
| Unique reflections | 17,673 | 25,453 |
| Completeness (%) | 96.0 (100) | 99.3 (100) |
| I/(σI) | 30.0 (4.0) | 18.2 (3.5) |
| Average redundancy | 19.9 | 7.3 |
| Rsym (%) | 7.1 (36.0) | 7.4 (12.1) |
| Mosaicity (°) | 0.67 | 0.86 |
| Unit-cell parameters (Å) | a = 91.65, c = 124.24 | a = 63.00, c = 273.27 |
| No. of protein molecules per A.U. | 1 | 1 |
| II. Refinement results | ||
| Final refinement | R = 22.40%, Rfree = 23.37% | R = 23.36%, Rfree = 24.11% |
| rmsd bond length (Å) | 0.0326 | 0.0294 |
| rmsd bond angles (°) | 2.75 | 2.56 |
| B-factors: | ||
| Protein | 49.5 | 51.2 |
| Zn2+ | 107.3 | 51.5 |
| Water | 46.4 | 50.2 |
| Total number of residues (built) | 422 (409) | 397 (396) |
| Total number of protein atoms (non-hydrogen) | 3207 | 3067 |
| Total number of N-acetyl glucosamine atoms | 14 | 56 |
| Total number of Ca2+/Zn2+ | 3/1 | 3/1 |
| Total number of water molecules | 75 | 117 |
| Ramachandran plot: | ||
| Most favored (%) | 89.4 | 93.7 |
| Allowed (%) | 10.1 | 6.3 |
| Disallowed (%) | 0.5 (F355, C358) | 0 |
Rsym = ∑h ∑i [|Ii(h) − 〈I(h)〉|/∑h ∑i Ii(h)], where Ii is the ith measurement and 〈I(h)〉 is the weighted mean of all measurements of I(h).
Reflections of 2σI cutoff were applied in generating the statistics. R = ∑h | Fo − Fc|/∑h Fo, where Fo and Fc are the observed and calculated structure factor amplitudes of reflection.
2.6. Dynamic light scattering (DLS) of atragin and K-like
DLS analysis (Zetasizer Nano-S, Malvern Instruments) was performed at 25 °C. DLS data in each set for purified proteins (0.8 mg/mL) of atragin and K-like in 10 mM Tris buffer (1 mL, pH 7.4) were analyzed with DTS software. All protein solutions were filtered through a membrane (porosity 0.22 μm) and centrifuged (10 min at 10,000 rpm) to remove dust before admission to the sampling cell (DTS0012-disposable sizing cuvette) for measurements of DLS. The populations of the hydrodynamic radius were calculated with correlation functions from an average of 20 measurements.
2.7. The metalloprotease activity on pro-TNFα assays
The assays of the peptide cleavage were performed with a fluorogenic peptide substrate, Abz-LAQAVRSSSR-Dpa (SDP-3818-PI, Peptide International). A typical reaction solution (10 mL) containing Tris–HCl (50 mM, pH 7.4), NaCl (25 mM), glycerol (4%), and the peptide substrate (10 μM) was incubated with various venom samples (10 μg/mL) – crude venom, CRISP, aPLA2, atragin, and K-like, respectively, at 37 °C for 2 h. Each venom component was purified from the crude cobra venom (from the Tainan Snake Farm) as described previously (Wang et al., 2004, Yang and Chang, 1988). ADAM17/TACE (10 nM, PF133, Calbiochem) and BSA (10 μg/mL, A6003, Sigma) served as controls. All assays were performed in 96-well plates; the increased fluorescence was monitored with exciton at λ ex 320 nm and emission at λ em 420 nm on a fluorescence plate reader (Wallace Victor 140, Perkin-Elmer). The proteolytic fragments were purified with reverse-phase HPLC (System Gold, Beckman); the cleavage site of the resulting peptide fragment was confirmed with mass spectra. For the protein substrate-cleavage assay, the pro-TNFα fusion protein substrates (1012-PS, R&D) were mixed with each enzyme in a reaction buffer (Tris–HCl, 50 mM, pH 7.4) containing NaCl (100 mM), and incubated at 37 °C for 6 h. The molar ratio of the enzyme to the substrate in the reaction is 1:1. The reactions were terminated on boiling with SDS. The products were sequentially analyzed by SDS–PAGE followed by silver staining.
2.8. The effect of pH on enzymatic activity and fluorescence spectroscopy
The enzymatic activity of atragin was analyzed with an azocaseinolytic assay. The azocasein (from Sigma Co., 2% in 100 mM Hepes at pH 7.5 and 7.0; 100 mM MES at pH 6.5 and 6.0; 100 mM acetic acid at pH 5.5 and 5.0, respectively) was incubated with SVMP atragin (5 μg) at 37 °C for 2 h in the reaction volume (30 μL). The reaction was terminated on adding trichloroacetic acid (30 μL, 0.4 M) for 30 min. Tubes were centrifuged at 500g for 15 min; the absorbance of supernatant solutions was recorded against a reaction blank at λ 440 nm. The fluorescence data of tyrosine in atragin proteins at varied pHs (2 mM in 100 mM Hepes, pH 7.5 and 7.0; 100 mM MES, pH 6.5 and 6.0; 100 mM acetic acid pH 5.5 and 5.0, respectively, total volume 3 mL) were recorded (λ em = 305 nm and λ ex = 275 nm) with a fluorimeter (Varian).
2.9. Transwell cell migration assay
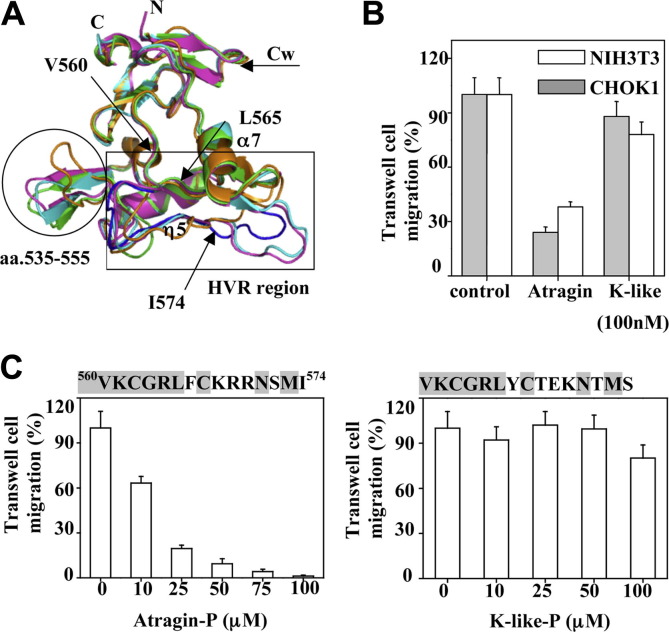
Polycarbonate membranes of the cell culture insert (Becton Dickinson) with a pore size of 8.0 μm were coated overnight at 4 °C in a solution of fibronectin (10 μg/mL). After incubation with BSA (1%, 37 °C), a DMEM medium supplemented with FBS (0.5%), BSA (0.1%), and SVMPs (100 nM) or their peptides atragin-P and K-like-P (at various concentrations) derived from the HVR (peptides synthesized by protection of cysteine residues with acetamidomethylated group) was applied to the lower chamber of a 24-well plate. Cells (mouse fibroblast NIH 3T3 cells or Chinese hamster ovary CHOK1 cells) in a suspension containing 5 × 104 cells in the same medium were added to each upper transwell. After incubation (37 °C, 5 h), cells were fixed with formaldehyde (4%) in PBS and stained in methanol (30%), acetic acid (10%), and crystal violet blue (1.0%) for 10 min. The membrane was washed in the same solution without Coomassie Blue for 10 min. The images of cell migration were captured with an inverted microscope, and the migrated cells on the membrane were counted in three microscopic fields per transwell. Experiments were performed in duplicate.
3. Results and discussion
3.1. Purification and sequencing of two new P-III SVMPs from N. atra
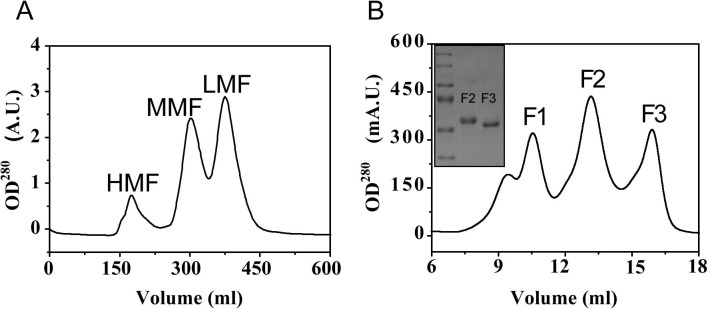
Two SVMPs from N. atra acquired with gel filtration and subsequent heparin affinity chromatography were characterized as atragin (M W 49.7 kDa and theoretical pI 8.6) and K-like (M W 49.3 kDa and theoretical pI 6.7) (Fig. S1). The cDNA-deduced amino acid sequences of atragin and K-like were determined using a molecular-cloning method (Fig. S2) and the protein sequences were validated with mass spectra (data not shown). The sequence search of the two proteins with NCBI-BLAST confirmed that both atragin and K-like belong to P-III SVMP containing the M, D, and C domains with 60% identities (Fig. S3). The signal peptides and prodomains of these two SVMPs are identical in the primary sequences (Fig. S2). The N-terminal sequencing shows that atragin beginning with amino acids TNTPEQ shares a sequence identity of 96% with cobrin (accession: AAF00693) from N. naja, whereas K-like beginning with QDRYLQ shares a sequence identity of 91% with kaouthiagin (accession: P82942) from Naja kaouthia. In total only six cysteine residues, but no seventh cysteine residue, were found in the M domain of both atragin and K-like SVMPs (Fig. S3), confirming the previous observations that elapid SVMPs lack a seventh cysteine in the M domain (Fox and Solange, 2005, Ito et al., 2001). Consistent with the seventh cysteine residue being correlated with the autolytic activity in SVMPs from the viper venom (Fox and Solange, 2005), there was also no detectable autolytic activity in our two SVMP preparations. The sequence alignment of the two toxins shows a notable difference at the D domain region: atragin contains a.a. 442–458 with four cysteine residues whereas this segment is absent in K-like (Fig. S3). Notably, K-like contains one RGD motif (a.a. 538–540 according to the sequence number of atragin) in the C domain that is similar to kaouthiagin (Fig. S3) (Fox and Bjarnason, 1995, Ito et al., 2001), which is potentially involved in integrin-binding (Bridges and Bowditch, 2005, Xiong et al., 2002), but such a motif was not found in atragin. Structurally, the relation between the RGD motif on the C domain of K-like and integrin molecules is worthy of address.
3.2. Overall MDC architectures
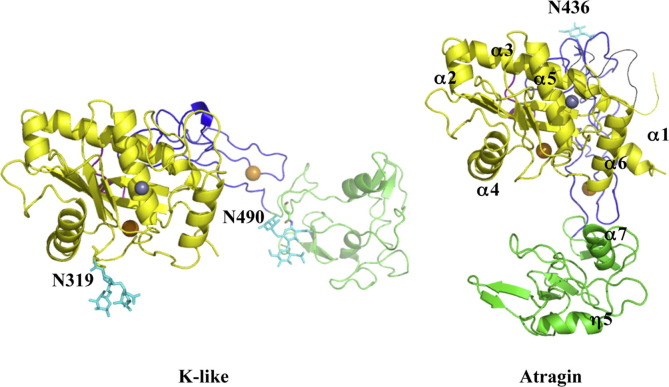
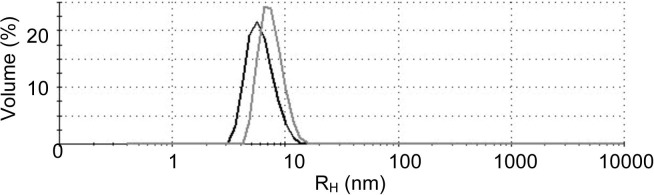
With X-ray crystallography, we determined the structures of two SVMPs – atragin and K-like. The data collection and refinement statistics are listed in Table 1. In a Ramachandran plot, all main-chain dihedral angles of residues are in the most favored or additionally allowed regions except Phe355 and Cys358 of atragin. An inspection of the atragin structure shows that these two residues are near the dynamic loop (a.a. 361–373). The overall architecture of atragin shows a C-shaped structure similar to the first P-III SVMP structure of VAP1 (Takeda et al., 2006), whereas that of K-like exhibits an I-shaped structure (Fig. 1 ). To exclude an influence of crystal packing making such a large conformational variation between atragin and K-like, we measured the hydrodynamic radius of atragin (peak at 6.1 nm) and K-like (peak at 7.5 nm) using dynamic light scattering (DLS) (Fig. S4). This data shows that K-like molecules have an area exposed to solvent larger than those of atragin, consistent with the dimensions of the determined crystal structures of atraign (63 × 52 × 67 Å3) and K-like (47 × 42 × 92 Å3). Notably, whereas the recognition site of the C domain faces toward to the catalytic cleft of the M domain with a linear orientation in the C-shaped structure of atragin, the activity sites of the C and M domains form a non-linear orientation in the I-shaped structure of K-like. These structural features suggest that the target recognized by the C domain differs from the substrate molecules catalyzed by the M domain or the substrate molecules of K-like exhibit a long non-linear conformation. The K-like structure contains two N-glycans located at the M and C domains, respectively (Asn319, Asn490), of which that at Asn319 is near the active site (Fig. 1) and differs from the common glycosylated site of hemorrhagin near the C-terminal of the M domain (Ramos and Selistre-de-Araujo, 2004). In contrast, there is only one N-glycan site (Asn436) on the D domain of the atragin structure. In view of an increasing importance of the post-translational modification of proteins for their biological functions, it will be of interest to investigate how the glycosylations in various domains might affect the biological activities of SVMPs.
Fig. 1.
Crystal structures of two SVMPs – atragin and K-like from Naja atra. The overall architectures of atragin and K-like from Naja atra are presented with metalloprotease domains aligned in the similar orientation. Both SVMP structures contain an M (yellow), a linker domain (pink), a D (blue), and a C (green) domain. N-glycans are colored in cyan. Gray spheres represent zinc ions and orange spheres represent calcium ions located at Ca(I):E208/D292/N398, Ca(II):N413/D417/E420/D423, and Ca(III):D474/E477/D489. A black line represents the missing loop containing the conserved Met-turn in the atragin structure.
3.3. M domain
As shown in Fig. 2 A, the overall M-domain structures of four P-III SVMPs show a great similarity with rmsd 2.3 Å between atragin and K-like. The M-domain structure of atragin exhibits a rmsd (1.85 and 1.92 Å) greater than that of K-like (0.92 and 0.89 Å) in comparison with those of VAP1 and VAP2, respectively. Six α-helices and five stranded β-sheets constitute the M domain of the atragin structure with one cleft comprising the zinc-binding motif H341EXXH345XXGXXH351D to accommodate the substrate binding for proteolysis. In contrast, in the lack of an initial N-terminal α-helix, only five α-helices are found in the M domain of the K-like structure (Fig. 1). The long helix α3 is located at the back, helices α1 and α6 are located at the right, and α2 as well as α4 are located at the left with respect to the center cleft. One zinc atom is readily observable as being ligated by the side chains of His341, His345 located at the α5-helix and His351 at the following turn as being responsible for the catalytic reaction (Gomis-Ruth, 2003). Opposing the active-site cleft, Ca2+ (I) are observable in structures of both atragin and K-like but invisible in the VAP1 structure because of the substitution of Glu208 to Lys (Takeda et al., 2006).
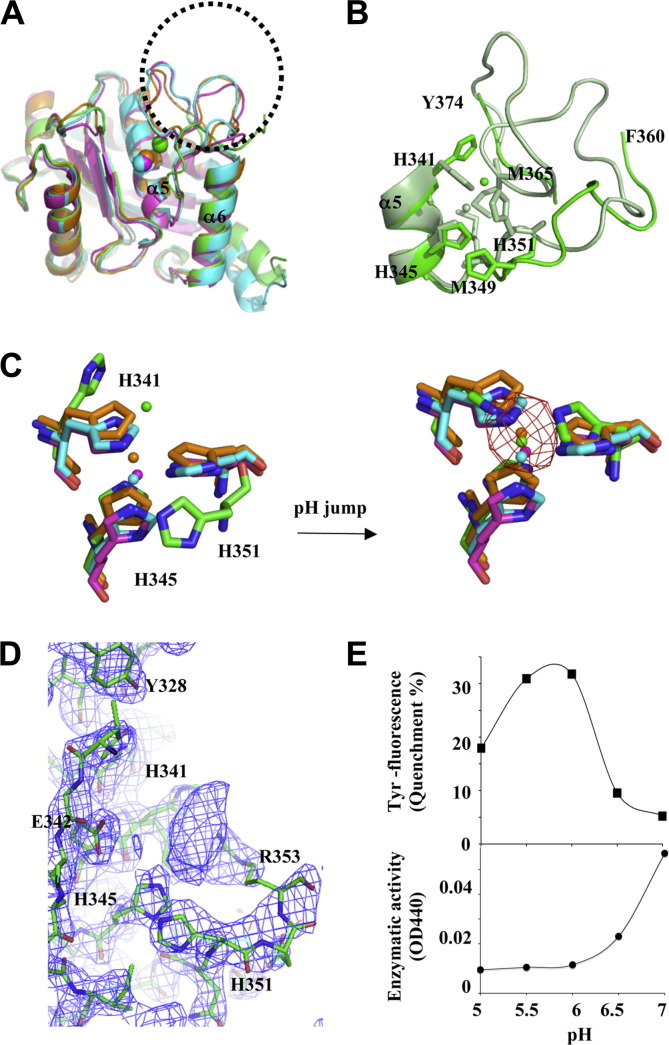
Fig. 2.
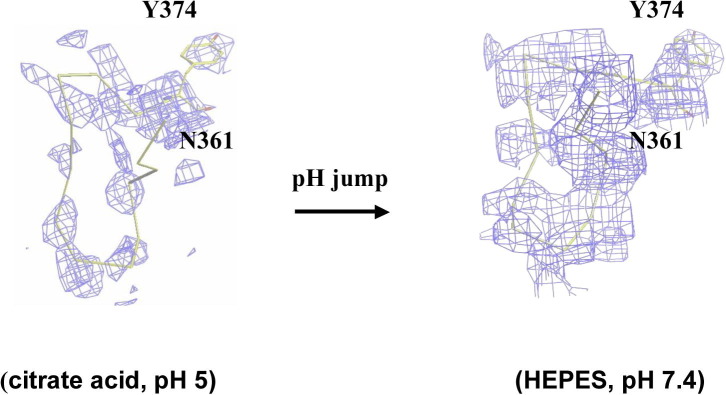
The pH-dependent zinc-binding motif on the metalloprotease domain and the associated proteolytic activity. (A) Superimposition of M domains of four P-III SVMPs: atragin (green), K-like (orange), VAP1 (cyan), and VAP2 (pink). The major variable region is located at the loop containing the Met-turn (enclosed by the dotted circle), which is disordered in the atragin structure. (B) The atragin structure in the acidic crystal (pH 5.0) (green) reveals an unconventional zinc-binding motif resulting in the missing loop from Asn361 to Ala373, whereas in a neutral crystal (pH 7.4) (light green), the binding motif switches to the conventional state and the electron density of the missing loop appears to allow model building. (C) Superimposition of three histidine residues in zinc-binding motifs of four P-III SVMPs shows the unconventional zinc-binding motif of atragin (green) in acidic crystallization conditions (citric acid, pH 5.0) and the conventional motif of atragin in the soaked neutral condition (Hepes, pH 7.4). The zinc anomalous difference-Fourier map (cutoff = 5σ, red mesh) confirms the zinc location in the neutral condition but not in the acidic condition. (D) The composite omit |2Fo − Fc| map (cutoff = 1.5σ, blue mesh) shows the well fit of three histidine residues in an unconventional binding motif and the putative range of the zinc position which is unobservable in the anomalous difference-Fourier map in the acidic pH crystal. The side chain of His341 has two conformations, and the backbone of His351 is repelled compared with a conventional zinc-binding motif. (E) The pH effect on the Tyr fluorescence and enzymatic activity of atragin. At pH 5.0 (acidic crystallization buffer), the enzymatic activity decreased to ∼20%, and the Tyr fluorescence also decreased ∼20% relative to assays at the neutral pH (pH 7.4). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.3.1. The pH-dependent zinc-binding motif
The venom gland lumen appears at an acidic pH, and lacks the proteolytic activity in the presence of citrate (58–125 mM) (Odell et al., 1998) and the tripeptide pyroglutamyl-lysyl-tryptophan enzymatic inhibitor in the gland (Marques-Porto et al., 2008). Our crystals of atragin happened to be grown in a citrate buffer (100 mM, pH 5), a medium similar to the environment of the venom gland lumen. In contrast, most previously reported crystal structures of SVMPs, including K-like (100 mM Hepes, pH 6.8), were solved near at a neutral pH. This circumstance provides an opportunity to understand the structural basis of the mechanism of the self-proteolytic regulation of SVMPs.
As shown in Fig. 2A and B, there is no detectable electron density at the flexible loop (a.a. 361–373) containing the so-called Met-turn (Bode et al., 1993) between helices α5 and α6 of atragin at an acidic pH, relative to those of previous SVMP and the K-like structure at the neutral pH. The lack of a Met-turn is not due to the self-proteolytic activity, as indicated by the SDS–PAGE analysis and mass spectra (data not shown). The best fit of the electron density of atragin at an acidic pH reveals a possible double conformation of the side chain of His341 and an elliptically diffuse electron density near the original ligation position of Zn2+ (Fig. 2D). We speculate that, under an acidic condition (pH 5.0), the protonation of histidines causes a structural change of the main chain of His351 and a double conformation of the side chain of His341 because of the repulsive force between the imidazole side chains of the histidines and Zn2+. This conformational alteration induces Zn2+ to have a large temperature B-factor (Table 1), as shown in the |2F o − F c| composite omit map (Fig. 2D), even though this character is imperceptible in the anomalous difference-Fourier map.
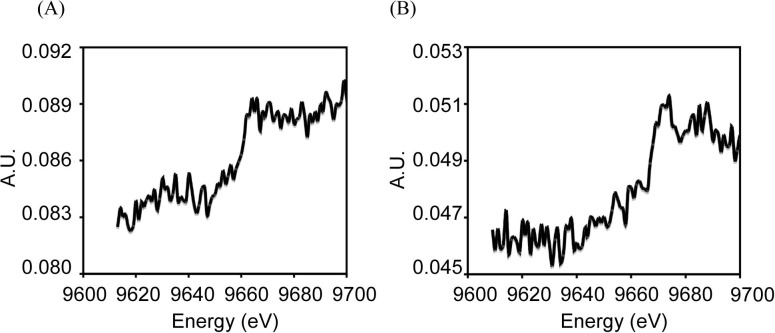
To prove this hypothesis, we transferred the crystals grown at an acidic pH into a neutral solution (Hepes, 100 mM, pH 7.4) for 10–20 s. The data collection and structural refinement statistics of crystals after the pH jump were shown in Table S1. The structures from these soaked crystals show that the three histidines in the zinc-binding motif under the acidic condition converted into the conventional conformation state and that Zn2+ was firmly located from the anomalous difference-Fourier map (Fig. 2C) (data collected at a wavelength 1.2822 Å at the K-absorption edge of zinc). The X-ray absorption and fluorescence-emission spectra of atragin crystals under both acidic and neutral conditions show similar signals and profiles of the zinc atom, indicating that Zn2+ exist in both crystal forms (Fig. S6). These results indicate that the pH switches do not alter the binding of zinc atoms in the structures but that the dynamic Met-tern and histidines (His341 and His351) of the zinc-binding motif with multi-conformations result in the unfixed positions of Zn2+ atoms. Moreover, the electron density of the flexible loop that is absent under acidic conditions becomes observable to allow the model building (Fig. S5), and the conserved Met365 in the Met-turn among metzicins makes the hydrophobic contact with Met349 beneath the pyramidal base of three histidines (Fig. 2B). The hydrophobic contact might be crucial for the enzymatic activity, corresponding to the previous mutagenesis study on protease C from Erwinia chrysanthemi showing that only the substitutions of hydrophobic residues for the methionine on the Met-turn produce the enzymatic activity (Hege and Baumann, 2001).
To examine the structural effect of the zinc-binding motif on the enzymatic activity, we measured the influence of pH on the activity and monitored the conformational modification using Tyr374 at the end of the flexible loop (a.a. 361–373) as a fluorescence probe. The other 16 tyrosines in the atragin show no obvious conformational change in the previous pH-jump experiment. As shown in Fig. 2E, under an acidic pH, the enzymatic activity of atragin decreases greatly, and the fluorescence intensity of Tyr alters significantly. The data indicate that both the zinc-binding motif and the Met-turn undergo dynamic structural alternations in the M domain under a condition of low pH to account for the small enzymatic activity.
3.3.2. Specificity on the proteolysis of pro-TNFα
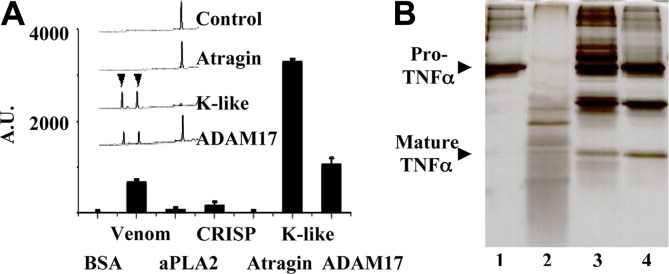
Moura-da-Silva et al. demonstrated that two viper venom SVMPs from Bothrops jararaca and a 54-kDa protein from Echis pyramidum leakeyi can induce cell necrosis on releasing TNFα (Moura-da-Silva et al., 1996). To examine whether the elapid atragin and K-like can also release TNFα, we used the fluorometric peptide Abz-LAQAVRSSSR-Dpa, which was derived from the linker domain of pro-TNFα, for a cleavage test. As shown in Fig. 3 A, K-like digests the fluorometric peptide as ADAM17 exhibits, whereas atragin shows no such effect. To confirm the TACE activity of K-like, we used a pro-TNFα fusion substrate to validate the activity in the SDS–PAGE analysis. The result indicates that the pro-TNFα digestion pattern of K-like is the same as that of ADAM17 (Fig. 3B), supporting the conclusion that the TACE activity of K-like is the same as that of ADAM17.
Fig. 3.
The enzymatic effect of atragin and K-like on Pro-TNFα. (A) Fluorometric measurements of the pro-TNFα-derived peptide (Abz-LAQAVRSSSR-Dpa) cleavage in the presence of various snake venom components. Both crude venom and purified K-like SVMP show a TACE activity as great as ADAM17 but not the other venom proteins. The HPLC profile (inserted panel) of the peptide fragments digested with the metalloprotease indicates that the specificity of K-like on a TNFα-derived peptide is identical to that of ADAM17. (The cleavage site of the peptide was identified with mass spectra.) (B) The SDS–PAGE analysis of cleavage patterns of pro-TNFα fusion substrates with two SVMPs. Lane 1: the pro-TNFα fusion substrate for the control; lane 2: pro-TNFα randomly digested by atragin; lane 3: TNFα released by K-like; lane 4: TNFα released by ADAM17.
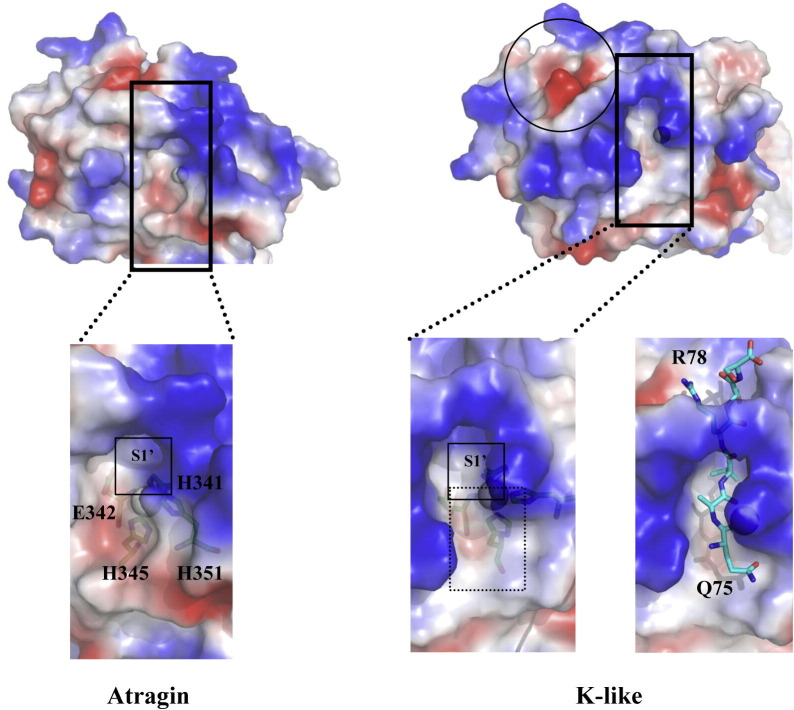
A close inspection of the cleft active sites in the M domain of the two P-III SVMP structures reveals that the S1′ site of K-like is more hydrophobic than that of atragin (Fig. S7). A hydrophobic S1′ site is critical for the pro-TNFα cleavage at Ala76–Val77 by ADAM17 (Maskos et al., 1998) and the drug design for TACE (Lu et al., 2008). We suggest that the hydrophobic S1′ site of K-like could accommodate Val77, and a negatively charged region formed by Asp301, Asp327, and Tyr328 (Fig. S7) of K-like could also accommodate the positively charged side chain of Arg78 of the pro-TNFα linker peptide. With the additional hydrophobic regions (S1 and S2 sites) to accommodate P1 and P2 (Ala76 and Gln75) of the substrate (Fig. S7) relative to atragin, the result of the peptide docking of QAVRSS might explain the specificity of K-like in the proteolytic activity of pro-TNFα (Fig. S7). The data indicates that the TACE activity of the two SVMPs is dominated mainly by the structures and hydrophobicity of the cleft active sites of M domains.
3.4. D domain
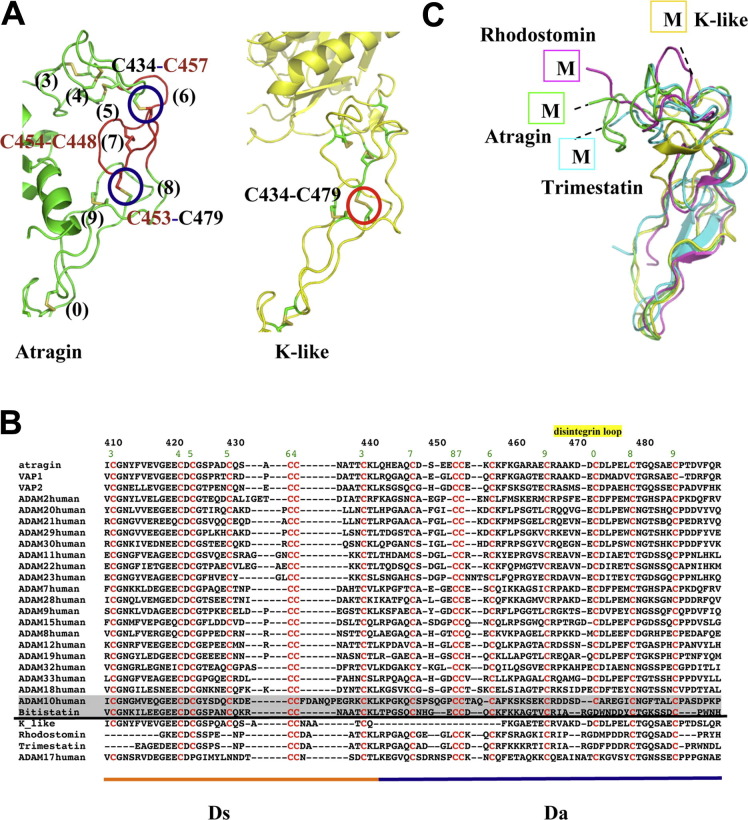
The D domain (a.a. 410–493, according to the sequence number of atragin) following the M domain and the linker S region (a.a. 401–409) plays an important role in the relative orientation of the M and C domains in the architectures of P-III SVMPs. As shown in Fig. 4 A and B, atragin maintains a C-shaped architecture as its structure adopts a similar D-shoulder (Ds) domain (a.a. 410–442) with three disulfide bonds (pairs of 3, 4, and 5) and a D-arm (Da) domain (a.a. 443–493) with another three disulfide bonds (pairs of 7, 8, and 9) constituting the C-shaped arm as in VAP1 and VAP2 (Igarashi et al., 2007, Takeda et al., 2006). Furthermore, one disulfide bond (a pair of 6) connects these two subdomains and another disulfide bond (a pair of 0) connects Da and C-wrist (Cw) comprising one cystine residue in the disintegrin-like loop (XXCD) that cause it to become inaccessible for integrin-binding as in VAP1 and VAP2. In the structure of K-like, the disulfide patterns alter significantly as one long segment of 17 amino acids is absent in comparison of the sequence (a.a. 442–458) of atragin (Fig. 4A). Hence, Cys434 forms a new disulfide bond with Cys462 (corresponding to Cys434 and Cys479, respectively, in the sequence of atragin) in the K-like structure (Fig. 4A). The re-shuffle of the disulfide bond pattern in the D domain affects the C-shaped arm and causes the K-like structure to adopt an I-shaped architecture.
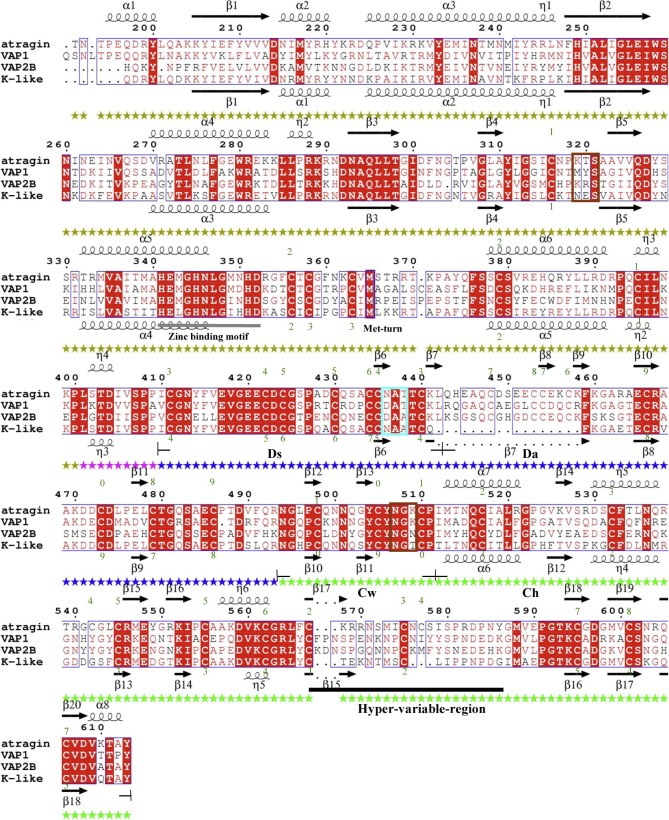
Fig. 4.
Disulfide bond patterns in Ds and Da domains of ADAMalysin family proteins affect the relative orientations of M and C domains. (A) The disulfide pattern in the D domain causes great conformational changes of two SVMPs. In the atragin structure, there are totally eight disulfide bonds in the D domain (numbers in black), of which C434–C457 and C453–C479 are enclosed in blue circles, whereas in the K-like structure, one new disulfide bond C434–C479 is formed and enclosed in red. The absent segment of K-like is colored in red in the atragin structure. (B) Sequence alignment of Ds and Da domains derived by ADAM/adamalysin/reprolysins family proteins and disintegrins. Above the black line, protein sequences with the same cysteine arrangement are aligned, whereas, below the line, protein sequences with significantly different disulfide bond pattern are aligned. The location of disintegrin-loop (XXCD) is labeled in yellow and the green number represents the disulfide pairs in the atragin structure. Bitistatin and human ADAM10 are highlighted by the shadow because their disulfide pairs are also different from conventional ones (e.g., atragin) despite of the same arrangement of csyteins with ADAMs. (C) Four protein structures with different disulfide bond patterns of D domains have distinct orientations of putative metalloprotease domains. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The C-shaped scaffold has been suggested to play an important role in the substrate recognition and shedding in proteins of the ADAM/adamalysin/reprolysins family (Takeda et al., 2006). The sequence alignment of the D domains in human ADAMs reveals that all ADAMs, except for ADAM17, have an arrangement of conserved cysteine residues in the D domain the same as atragin (Fig. 4B). Furthermore, K-like and the disintegrins, such as rhodostomin and trimestatin, contain varied numbers of cysteine residues in the primary sequences of the D domain, revealing an important structural role in the disulfide pattern to account for the relative orientations of the putative M and D domains (Fujii et al., 2003) (Fig. 4B and C).
The disulfide pattern in the D domain also possibly varies from the same protein sequence as the cases of DC domains of VAP2B and processed catrocollastatin C (Fox and Serrano, 2008), or from the same arrangement of cysteine residues in the primary sequences. For example, the MALDI-TOF mass analysis of Bitistatin shows that the disulfide pairs are different from conventional ADAMs (the same disulfide pairs with atragin or VAP1) mainly on the Ds domain, even though they consist of similar conserved cysteine residues in the D domain (Calvete et al., 2000) (Fig. 4B). Moreover, the structure of the DC domain in ADAM10 shows that another disulfide bond pair Cys435–Cys457 was formed, rather than Cys434–Cys457 (disulfide bond pair 6) in atragin, and the varied Cys434 and Cys435 are also located at the Ds domain (Janes et al., 2005). The Ds domain of ADAM10 is, however, mainly disordered, indicating that the Ds domain is dynamic. Furthermore, the primary sequence alignment on D domains of proteins in the ADAM/adamalysin/reprolysins family and disintegrins shows that the significant variation is located at the Ds domain (Fig. 4B). The great difference in the Ds domain increases the opportunity of re-shuffling disulfide pairs in ADAM/adamalysin/reprolysins family proteins, which is verified by the novel finding on the distinct disulfide patterns of K-like and ADAM10.
3.5. C domain
As shown in Fig. 5 A, the superimposed C domains comprising the C-wrist (Cw) (a.a. 494–511) and C-hand (Ch) (a.a. 512–613) of four PIII-SVMPs reveal the major structurally distinct region to be located at the hyper-variable region, HVR (a.a. 567–586), which has been suggested to play an important role in the target selection based on the C-shaped scaffold in the MDC domain (Takeda et al., 2006). Another structurally variable region at the loop (a.a. 535–555) remains to be characterized (Fig. 5A). The C domain of atragin consists of seven disulfide bonds, in contrast to the six conserved disulfide bonds revealed in the VAP1 and VAP2 structures. The additional disulfide bond Cys542–Cys575 in atragin is connected to the HVR, indicating that it might play a crucially functional role. Notably, one important RGD motif (a.a. 538–540 according to the sequence number of atragin) located at the end of the η4 helix of K-like in a position similar to kaouthiagin (Ito et al., 2001) is inaccessible for integrin molecules (Fig. S8), implying that K-like could bind to the integrin through another non-RGD region, such as the HVR (Pinto et al., 2007). We cannot, however, exclude a possibility that such a loop containing an RGD motif can be exposed accessibly for integrin-binding through a large conformational alteration. In fact, the B-factor distributions of atragin and K-like show that most parts of C-domains exhibit flexible features with higher B-factors (Fig. S8). Accordingly, a close inspection of the water distributions and molecular surfaces of both proteins reveals that the dynamic C domain and the regions near the Met-turn loop of the M-domain result in fewer water molecules bound in these areas.
Fig. 5.
The inhibitory effect of atragin, K-like and peptides derived from the HVR in C domains, respectively, on cell migration. (A) Superposition of C domains from four P-III SVMP structures colored as Fig. 2A. The Cw domain structures are similar and rigid among SVMPs, whereas the Ch domains show larger variations, especially in the HVR (in the rectangle). The HVR of atragin is indicated in blue. Another notably variable region is at amino acids 535–555 (in the circle). The peptide residues, V560KCGRLFCKRRNSMI574, are indicated by arrows. (B) The inhibitory effect on cell NIH3T3 and CHOK1 migration with atragin and K-like, respectively. (C) The inhibitory effect on cell NIH3T3 migration with two peptides atragin-P and K-like-P derived from HVRs of atragin and K-like, respectively.
3.5.1. Different cell-migration inhibitory activities of these two SVMPs
Jararhagin, a P-III SVMP from the viper venom, has been shown to inhibit platelet aggregation and to stimulate the migration of epithelial cells (Costa and Santos, 2004). The peptide sequence derived from the HVR of jararhagin can also inhibit platelet aggregation by binding to the VWA domain (Kamiguti et al., 2003, Pinto et al., 2007). How the two SVMPs from the elapid venom might affect such similar activities is therefore of interest. As shown in Fig. 5B, atragin exhibits an inhibitory activity toward cell migration of both NIH3T3 and CHOK1 much stronger than K-like, rather than stimulation. To examine whether the amino acid sequence from the HVR of the two related SVMPs might be responsible for the inhibitory effect, we synthesized two peptides according to their respective HVR regions and tested the effect on cell migration. Despite nine of fifteen amino-acid residues being the same for the two peptides, the one derived from the HVR of atragin shows a clear inhibitory effect (Fig. 5C). A comparison of two peptide sequences and the related 3D structures shows that the consensus sequence V560KCGRL565 is located at an inaccessible region in both structures, whereas the variable sequences (F566CKRRNSMIAtragin/YCTEKNTMSK-like) correspond to the HVR (Fig. 5A). Our result further supports the idea that the HVR in SVMPs is exposed for the target recognition and might play important functional roles toward cell migration.
3.6. Implication of ADAM shedding mechanism
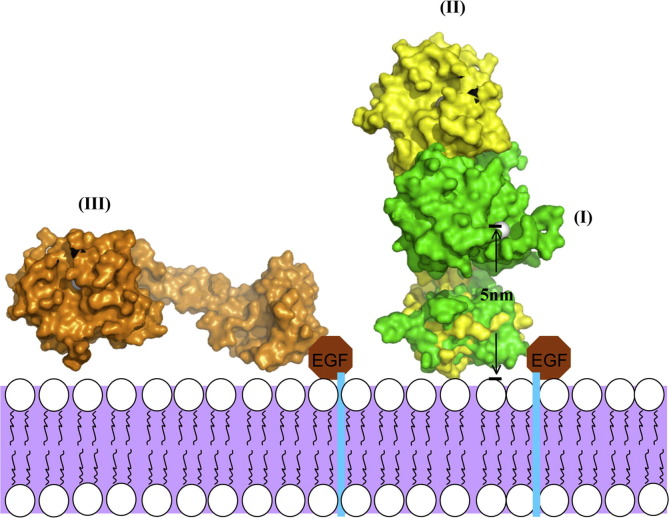
As we report above, the distinct disulfide bond pattern in the D domain alters greatly the relative orientations of the M and C domains in proteins of the ADAM/adamalysin/reprolysins family. The varied relative orientations of the M and C domains might explain some shedding processes of ADAMs that are difficult to be interpreted based on only a C-shaped scaffold. For example, in the event of the pro-TNFα cleavage by ADAM17, an experiment of a deletion on the linker domain of pro-TNFα indicated that the linking domain must contain at least nine amino acids (length ∼3.5 nm maximally) for the cleavage (Tang et al., 1996). The ADAM with a C-shaped scaffold can shed molecules with a height of ∼5 nm (Fig. 6 , I), which is little too great for ADAM 17 to act on the linker domain of pro-TNFα. Additionally, MICA (a MHC-encoded molecule) has been identified to be released by ADAM17 and ADAM10 on cancer cells. The shedding event depends on the stalk length that ranges from 9 to 18 residues, rather than on the properties of amino acids of the MICA stalk (Waldhauer et al., 2008). The length of nine residues is also somewhat too short for the C-shaped scaffold. Several other shedding events of ADAMs, including the growth hormone receptor, L-selectin, ACE, and ICAM-1, also reflect the importance of the stalk lengths, which is the distance from the membrane, rather than the sequences of the stalks (Hinkle et al., 2004, Migaki et al., 1995, Sadhukhan et al., 1999, Tsakadze et al., 2006). These biological phenomena suggest that some ADAMs might adopt another strategy to shed shorter substrates by altering the disulfide pairs on the D domain as in the case of K-like (Fig. 6, III) or by a geometric flexibility through the dynamic Ds domain.
Fig. 6.
Implications for shedding models of ADAMs based on two SVMP structures from Naja atra. The shedding models of ADAMs are placed on the membrane (purple region). The shedding model (I) is based on the C-shaped MDC architecture, whereas the shedding models (II, III) are based on the I-shaped MDC architecture. The EGF-like domains are shown as brown octagons. The distance between the membrane and zinc ions (gray spheres) in the cleft active site is about 5 nm in the model (I). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Another unexplained shedding event is that ADAM10 can recognize EphA3 and shed ephrinA5 in a trans fashion (Janes et al., 2005). According to the geometric constraint of the C-shaped scaffold, such a shedding effect can occur only if the C-shaped ADAM can undergo a significant dynamic alteration of the structure. For example, the OG domain of MMP9/gelatinase B adopts a spring-like action to confer a flexibility between the recognition and proteolysis of a substrate (Rosenblum et al., 2007), or the I-shaped K-like adopts another orientation through the re-shuffle of its disulfide pairs in the D domain as presented in this work (Fig. 6, II).
In summary, we report the crystal structures of two newly identified SVMPs – atragin and K-like – from the elapid venom of N. atra. The MDC domains of K-like adopts an I-shaped scaffold because of its novel patterns of disulfide bonds in the ADAM/adamalysin/reprolysins family, providing direct evidence to support the structural role of the D domain in orientating the M and C domains for its function. Using a pH-jump method, we report also the structural difference of the M domain of atragin under acidic and neutral conditions and correlate its structural variation with the proteolytic activity. The two newly identified P-III SVMPs exhibit distinct activities in terms of the substrate specificity of pro-TNFα and the inhibition of cell migration.
The nucleotide sequences and deduced primary sequences (atragin: FJ177517; K-like: FJ177516) have been deposited in the GeneBank (http://www.ncbi.nlm.nih.gov/Genbank/index.html), and the atomic coordinates and structure factors (code, atragin: 3K7L; K-like: 3K7N) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
Acknowledgments
This work was supported in part by National Science Council (NSC) Grants 94-2313-B-213-001, 95-2313-B-009-001-MY, and National Synchrotron Radiation Center (NSRRC) Grants 963RSB02, 973RSB02 (Chun-Jung Chen) and NSC Grants 96-2311-B-007-007, 96-2113-M-007-023-MY2, and NSRRC Grant 973RSB08 (Wen-guey Wu). We are indebted to Dr. Yuch-Cheng Jean, Dr. Chun-Hsiung Chao, and the supporting staffs at beamlines BL13B1 and BL13C1 at the National Synchrotron Radiation Research Center (NSRRC) for the technical assistance. We are also grateful to Mr. Ping-Chen Shi for all his contribution to this work.
Footnotes
Abbreviations used: SVMP, snake venom metallorprotease; ADAM, a disintegrin and metalloprotease; ECM, extracellular matrix; PSGL-1, P-selectin glycoprotein ligand-1; SARS, severe acute respiratory syndrome; MMP, matrix metalloprotease; TACE: tumor necrosis factor-α converting enzyme; MALDI-TOF; matrix-assisted laser-desorption ionization-time-of-flight; NSRRC, National Synchrotron Radiation Research Center; DLS, dynamic light scattering; ACE, angiotensin-converting enzyme; ICAM-1, intercellular adhesion molecule-1.
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.jsb.2009.11.009.
Contributor Information
Wen-guey Wu, Email: wgwu@nsrrc.org.tw.
Chun-Jung Chen, Email: cjchen@nsrrc.org.tw.
Appendix A. Supplementary data
Supplementary Fig. S1.
The purification profiles of two P-III SVMPs atragin and K-like. (A) The gel filtration chromatography of crude venom from Naja atra through G75 column. Crude venoms are separated into three major populations named high-MW fraction (HMF), medium-MW fraction (MMF), and low-MW fraction (LMF). Both atragin and K-like in this study are in the HMF. (B) The affinity chromatography of HMF through the Hitrap Heparin HP column. HMF is further separated into three major peaks named F1, F2 and F3. All the purification steps are monitored at 280 nm. SDS–PAGE analysis (inset) shows lane 1, molecular mass marker; lane 2, K-like (F2); and lane 3, atragin (F3). All lanes are under reducing conditions.
Supplementary Fig. S2.
Nucleotide sequences and deduced primary sequences of two PIII-SVMPs – atragin and K-like. (A) The nucleotide sequence and deduced primary sequence from cDNA of atragin. (B) The nucleotide sequence and deduced primary sequence from cDNA of K-like. N-glycosylation sites of these two PIII-SVMPs – atragin and K-like are indicated in the gray shadow.
Supplementary Fig. S3.
The sequences alignment of two elapid SVMPs – atragin and K-like from Naja atra with two viper SVMPs – VAP1 and VAP2. Two P-III SVMPs, atragin and K-like, from Naja atra comprise an M domain (yellow star in the bottom), a linker domain (pink star), a D domain (blue star), and a C domain (green star). Two brown squares represent two N-glycosylation sites of K-like in the M domain and in the C domain, respectively, and one cyan square represents only one N-glycosylation site of atragin in the D domain. Disulfide pairs are represented by the numbers in green.
Supplementary Fig. S4.
Dynamic light scattering spectra of atragin and K-like. The hydrodynamic radius of K-like (gray line, peak 7.5 nm) is larger than atragin (black line, peak 6.1 nm).
Supplementary Fig. S5.
The effect of pH-jump on the missing loop (a.a. 361–373) comprising the Met turn of atragin. After the experiment of pH jump, the density of missing loop (a.a. 361-–373, shown in the yellow sticks) appears. The |2Fo − Fc| map was shown in blue and the unclear density of Tyr374 under the acid condition becomes clear in the neutral condition, which indicates the conformation of Tyr374 is more dynamic in the acid condition than that in the neutral condition. The |2Fo − Fc| model map was calculated by atragin structure omitting the missing loop (a.a. 361–373).
Supplementary Fig. S6.
Fluorescence spectra of atragin crystals scanned by the X-ray near the K-absorption edge of Zn. (A) The fluorescence spectrum of atragin crystal at pH 5. (B) The fluorescence spectrum of atragin crystal at pH 7.4 after pH jump.
Supplementary Fig. S7.
The electrostatic surfaces of two SVMP M domains. Gray spheres represent zinc atoms responsible for the catalytic reaction. The close view of the cleft active sites of two SVMPs and S1′ sites are enclosed with black squares. The hydrophobic regions (S1 and S2) in the K-like structure accommodating P1 and P2 residues of substrate are indicated with a dashed line. The negatively charged region enclosed in circles can accommodate Arg78 according to the docking model of K-like and the peptide Q75AVRS derived from the linker domain of pro-TNFα at the right.
Supplementary Fig. S8.
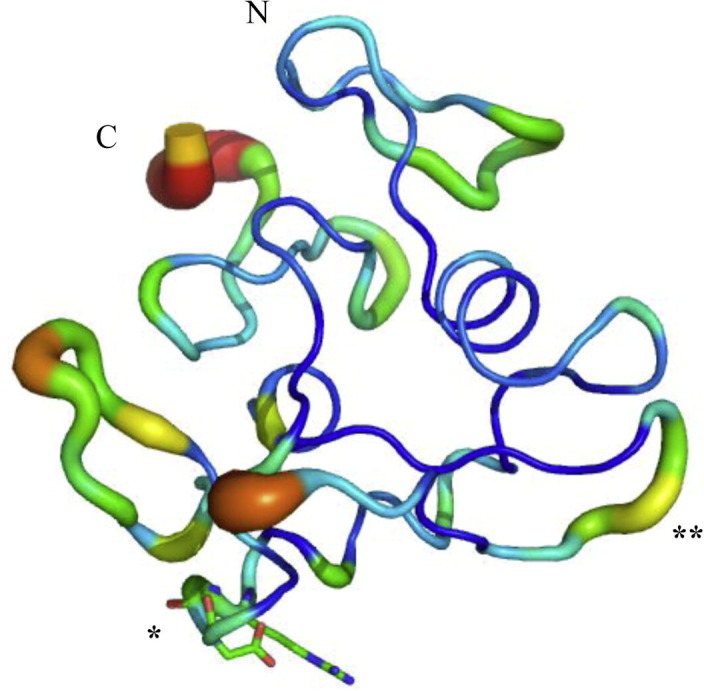
The C domain of K-like displayed with the diameter and the color (blue to red) according to the B-factor values. Structurally, the RGD-motif labeled with ∗ is inaccessible for the binding of integrin-molecules and the HVR labeled with ∗∗ is another possible region for the binding of integrin-molecules.
Crystal diffraction and structural refinement statistics of atragin (after pH jump).
References
- Asakura M., Kitakaze M., Takashima S., Liao Y., Ishikura F., Yoshinaka T., Ohmoto H., Node K., Yoshino K., Ishiguro H., Asanuma H., Sanada S., Matsumura Y., Takeda H., Beppu S., Tada M., Hori M., Higashiyama S. Cardiac hypertrophy is inhibited by antagonism of ADAM12 processing of HB-EGF: metalloproteinase inhibitors as a new therapy. Nat. Med. 2002;8:35–40. doi: 10.1038/nm0102-35. [DOI] [PubMed] [Google Scholar]
- Bode W., Gomis-Ruth F.X., Stockler W. Astacins, serralysins, snake venom and matrix metalloproteinases exhibit identical zinc-binding environments (HEXXHXXGXXH and Met-turn) and topologies and should be grouped into a common family, the ‘metzincins’. FEBS Lett. 1993;331:134–140. doi: 10.1016/0014-5793(93)80312-i. [DOI] [PubMed] [Google Scholar]
- Bridges L.C., Bowditch R.D. ADAM–integrin interactions: potential integrin regulated ectodomain shedding activity. Curr. Pharm. Des. 2005;11:837–847. doi: 10.2174/1381612053381747. [DOI] [PubMed] [Google Scholar]
- Brunger A.T., Adams P.D., Clore G.M., DeLano W.L., Gros P., Grosse-Kunstleve R.W., Jiang J.S., Kuszewski J., Nilges M., Pannu N.S., Read R.J., Rice L.M., Simonson T., Warren G.L. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Calvete J.J., Juarez P., Sanz L. Snake venomics. Strategy and applications. J. Mass Spectrom. 2007;42:1405–1414. doi: 10.1002/jms.1242. [DOI] [PubMed] [Google Scholar]
- Calvete J.J., Jurgens M., Marcinkiewicz C., Romero A., Schrader M., Niewiarowski S. Disulphide-bond pattern and molecular modelling of the dimeric disintegrin EMF-10, a potent and selective integrin alpha5beta1 antagonist from Eristocophis macmahoni venom. Biochem. J. 2000;345(Pt 3):573–581. [PMC free article] [PubMed] [Google Scholar]
- CCP4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Costa E.P., Santos M.F. Jararhagin, a snake venom metalloproteinase-disintegrin, stimulates epithelial cell migration in an in vitro restitution model. Toxicon. 2004;44:861–870. doi: 10.1016/j.toxicon.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Fox J.W., Bjarnason J.B. Atrolysins: metalloproteinases from Crotalus atrox venom. Methods Enzymol. 1995;248:368–387. doi: 10.1016/0076-6879(95)48024-2. [DOI] [PubMed] [Google Scholar]
- Fox J.W., Solange M.T.S. Structural considerations of the snake venom metalloproteinases, key members of the M12 reprolysin family of metalloproteinases. Toxicon. 2005;45:969–985. doi: 10.1016/j.toxicon.2005.02.012. [DOI] [PubMed] [Google Scholar]
- Fox J.W., Serrano S.M. Insights into and speculations about snake venom metalloproteinase (SVMP) synthesis, folding and disulfide bond formation and their contribution to venom complexity. FEBS J. 2008;275:3016–3030. doi: 10.1111/j.1742-4658.2008.06466.x. [DOI] [PubMed] [Google Scholar]
- Fujii Y., Okuda D., Fujimoto Z., Horii K., Morita T., Mizuno H. Crystal structure of trimestatin, a disintegrin containing a cell adhesion recognition motif RGD. J. Mol. Biol. 2003;332:1115–1122. doi: 10.1016/s0022-2836(03)00991-4. [DOI] [PubMed] [Google Scholar]
- Gao R., Manjunatha Kini R., Gopalakrishnakone P. A novel prothrombin activator from the venom of Micropechis ikaheka: isolation and characterization. Arch. Biochem. Biophys. 2002;408:87–92. doi: 10.1016/s0003-9861(02)00447-2. [DOI] [PubMed] [Google Scholar]
- Gomis-Ruth F.X. Structural aspects of the metzincin clan of metalloendopeptidases. Mol. Biotechnol. 2003;24:157–202. doi: 10.1385/MB:24:2:157. [DOI] [PubMed] [Google Scholar]
- Haga S., Yamamoto N., Nakai-Murakami C., Osawa Y., Tokunaga K., Sata T., Yamamoto N., Sasazuki T., Ishizaka Y. Modulation of TNF-alpha-converting enzyme by the spike protein of SARS-CoV and ACE2 induces TNF-alpha production and facilitates viral entry. Proc. Natl. Acad. Sci. USA. 2008;105:7809–7814. doi: 10.1073/pnas.0711241105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hege T., Baumann U. The conserved methionine residue of the metzincins: a site-directed mutagenesis study. J. Mol. Biol. 2001;314:181–186. doi: 10.1006/jmbi.2001.5125. [DOI] [PubMed] [Google Scholar]
- Hinkle C.L., Sunnarborg S.W., Loiselle D., Parker C.E., Stevenson M., Russell W.E., Lee D.C. Selective roles for tumor necrosis factor alpha-converting enzyme/ADAM17 in the shedding of the epidermal growth factor receptor ligand family: the juxtamembrane stalk determines cleavage efficiency. J. Biol. Chem. 2004;279:24179–24188. doi: 10.1074/jbc.M312141200. [DOI] [PubMed] [Google Scholar]
- Igarashi T., Araki S., Mori H., Takeda S. Crystal structures of catrocollastatin/VAP2B reveal a dynamic, modular architecture of ADAM/adamalysin/reprolysin family proteins. FEBS Lett. 2007;581:2416–2422. doi: 10.1016/j.febslet.2007.04.057. [DOI] [PubMed] [Google Scholar]
- Ito M., Hamako J., Sakurai Y., Matsumoto M., Fujimura Y., Suzuki M., Hashimoto K., Titani K., Matsui T. Complete amino acid sequence of Kaouthiagin, a novel cobra venom metalloproteinase with two disintegrin-like sequence. Biochemistry. 2001;40:4503–4511. doi: 10.1021/bi0022700. [DOI] [PubMed] [Google Scholar]
- Janes P.W., Saha N., Barton W.A., Kolev M.V., Wimmer-Kleikamp S.H., Nievergall E., Blobel C.P., Himanen J.P., Lackmann M., Nikolov D.B. Adam meets Eph: an ADAM substrate recognition module acts as a molecular switch for ephrin cleavage in trans. Cell. 2005;123:291–304. doi: 10.1016/j.cell.2005.08.014. [DOI] [PubMed] [Google Scholar]
- Kamiguti A.S., Hay C.R., Zuzel M. Inhibition of collagen-induced platelet aggregation as the result of cleavage of alpha 2 beta 1-integrin by the snake venom metalloproteinase jararhagin. Biochem. J. 1996;320(Pt 2):635–641. doi: 10.1042/bj3200635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiguti A.S., Gallagher P., Marcinkiewicz C., Theakston R.D., Zuzel M., Fox J.W. Identification of sites in the cysteine-rich domain of the class P-III snake venom metalloproteinases responsible for inhibition of platelet function. FEBS Lett. 2003;549:129–134. doi: 10.1016/s0014-5793(03)00799-3. [DOI] [PubMed] [Google Scholar]
- Li S., Wang J., Zhang X., Ren Y., Wang N., Zhao K., Chen X., Zhao C., Li X., Shao J., Yin J., West M.B., Xu N., Liu S. Proteomic characterization of two snake venoms: Naja naja atra and Agkistrodon halys. Biochem. J. 2004;384:119–127. doi: 10.1042/BJ20040354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber T., Kidd S., Young M.W. Kuzbanian-mediated cleavage of Drosophila Notch. Genes Dev. 2002;16:209–221. doi: 10.1101/gad.942302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z., Ott G.R., Anand R., Liu R.Q., Covington M.B., Vaddi K., Qian M., Newton R.C., Christ D.D., Trzaskos J., Duan J.J. Potent, selective, orally bioavailable inhibitors of tumor necrosis factor-alpha converting enzyme (TACE): discovery of indole, benzofuran, imidazopyridine and pyrazolopyridine P1′ substituents. Bioorg. Med. Chem. Lett. 2008;18:1958–1962. doi: 10.1016/j.bmcl.2008.01.120. [DOI] [PubMed] [Google Scholar]
- Marques-Porto R., Lebrun I., Pimenta D.C. Self-proteolysis regulation in the Bothrops jararaca venom: the metallopeptidases and their intrinsic peptidic inhibitor. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2008;147:424–433. doi: 10.1016/j.cbpc.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Maskos K., Fernandez-Catalan C., Huber R., Bourenkov G.P., Bartunik H., Ellestad G.A., Reddy P., Wolfson M.F., Rauch C.T., Castner B.J., Davis R., Clarke H.R., Petersen M., Fitzner J.N., Cerretti D.P., March C.J., Paxton R.J., Black R.A., Bode W. Crystal structure of the catalytic domain of human tumor necrosis factor-alpha-converting enzyme. Proc. Natl. Acad. Sci. USA. 1998;95:3408–3412. doi: 10.1073/pnas.95.7.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migaki G.I., Kahn J., Kishimoto T.K. Mutational analysis of the membrane-proximal cleavage site of L-selectin: relaxed sequence specificity surrounding the cleavage site. J. Exp. Med. 1995;182:549–557. doi: 10.1084/jem.182.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss M.L., Jin S.L., Milla M.E., Bickett D.M., Burhart W., Carter H.L., Chen W.J., Clay W.C., Didsbury J.R., Hassler D. Cloning of a disintegrin metalloproteinase that processes precursor tumour-necrosis factor-α. Nature. 1997;385:733–736. doi: 10.1038/385733a0. [DOI] [PubMed] [Google Scholar]
- Moura-da-Silva A.M., Laing G.D., Paine M.J., Dennison J.M., Politi V., Crampton J.M., Theakston R.D. Processing of pro-tumor necrosis factor-alpha by venom metalloproteinases: a hypothesis explaining local tissue damage following snake bite. Eur. J. Immunol. 1996;26:2000–2005. doi: 10.1002/eji.1830260905. [DOI] [PubMed] [Google Scholar]
- Muniz J.R., Ambrosio A.L., Selistre-de-Araujo H.S., Cominetti M.R., Moura-da-Silva A.M., Oliva G., Garratt R.C., Souza D.H. The three-dimensional structure of bothropasin, the main hemorrhagic factor from Bothrops jararaca venom: insights for a new classification of snake venom metalloprotease subgroups. Toxicon. 2008;52:807–816. doi: 10.1016/j.toxicon.2008.08.021. [DOI] [PubMed] [Google Scholar]
- Murshudov G.N., Vagin A.A., Dodson E.J. Refinement of macromolecular structures by the Maximum-Likelihood method. Acta Cryst. 1998;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- O’Keefe M.C., Caporale L.H., Vogel C.W. A novel cleavage product of human complement component C3 with structural and functional properties of cobra venom factor. J. Biol. Chem. 1988;263:12690–12697. [PubMed] [Google Scholar]
- Odell G.V., Ferry P.C., Vick L.M., Fenton A.W., Decker L.S., Cowell R.L., Ownby C.L., Gutierrez J.M. Citrate inhibition of snake venom proteases. Toxicon. 1998;36:1801–1806. doi: 10.1016/s0041-0101(98)00084-1. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Paine M.J., Desmond H.P., Theakston R.D., Crampton J.M. Purification, cloning, and molecular characterization of a high molecular weight hemorrhagic metalloprotease, jararhagin, from Bothrops jararaca venom. Insights into the disintegrin gene family. J. Biol. Chem. 1992;267:22869–22876. [PubMed] [Google Scholar]
- Pan D., Rubin G.M. Kuzbanian controls proteolytic processing of Notch and mediates lateral inhibition during Drosophila and vertebrate neurogenesis. Cell. 1997;90:271–280. doi: 10.1016/s0092-8674(00)80335-9. [DOI] [PubMed] [Google Scholar]
- Peschon J.J., Slack J.L., Reddy P., Stocking K.L., Sunnarborg S.W., Lee D.C., Russell W.E., Castner B.J., Johnson R.S., Fitzner J.N., Boyce R.W., Nelson N., Kozlosky C.J., Wolfson M.F., Rauch C.T., Cerretti D.P., Paxton R.J., March C.J., Black R.A. An essential role for ectodomain shedding in mammalian development. Science. 1998;282:1281–1284. doi: 10.1126/science.282.5392.1281. [DOI] [PubMed] [Google Scholar]
- Pinto A.F., Terra R.M., Guimaraes J.A., Fox J.W. Mapping von Willebrand factor A domain binding sites on a snake venom metalloproteinase cysteine-rich domain. Arch. Biochem. Biophys. 2007;457:41–46. doi: 10.1016/j.abb.2006.10.010. [DOI] [PubMed] [Google Scholar]
- Prenzel N., Zwick E., Dau H., Leserer M., Abraham R., Wallasch C., Ullrich A. EGF receptor transactivation by G-protein-coupled receptors requires metalloproteinase cleavage of proHB-EGF. Nature. 1999;402:884–888. doi: 10.1038/47260. [DOI] [PubMed] [Google Scholar]
- Raab G., Klagsbrun M. Heparin-binding EGF-like growth factor. Biochim. Biophys. Acta. 1997;1333:F179–F199. doi: 10.1016/s0304-419x(97)00024-3. [DOI] [PubMed] [Google Scholar]
- Ramos O.H., Selistre-de-Araujo H.S. Snake venom metalloproteases – structure and function of catalytic and disintegrin domains. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2006;142:328–346. doi: 10.1016/j.cbpc.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Ramos O.H.P., Selistre-de-Araujo H.S. Comparative analysis of the catalytic domain of hemorrhagic and non-hemorrhagic snake venom metallopeptidases using bioinformatic tools. Toxicon. 2004;44:529–538. doi: 10.1016/j.toxicon.2004.07.005. [DOI] [PubMed] [Google Scholar]
- Rosenblum G., Van den Steen P.E., Cohen S.R., Grossmann J.G., Frenkel J., Sertchook R., Slack N., Strange R.W., Opdenakker G., Sagi I. Insights into the structure and domain flexibility of full-length pro-matrix metalloproteinase-9/gelatinase B. Structure. 2007;15:1227–1236. doi: 10.1016/j.str.2007.07.019. [DOI] [PubMed] [Google Scholar]
- Sadhukhan R., Santhamma K.R., Reddy P., Peschon J.J., Black R.A., Sen I. Unaltered cleavage and secretion of angiotensin-converting enzyme in tumor necrosis factor-alpha-converting enzyme-deficient mice. J. Biol. Chem. 1999;274:10511–10516. doi: 10.1074/jbc.274.15.10511. [DOI] [PubMed] [Google Scholar]
- Sanz L., Ayvazyan N., Calvete J.J. Snake venomics of the Armenian mountain vipers Macrovipera lebetina obtusa and Vipera raddei. J. Proteomics. 2008;71:198–209. doi: 10.1016/j.jprot.2008.05.003. [DOI] [PubMed] [Google Scholar]
- Seals D.F., Courtneidge S.A. The ADAMs family of metalloproteases: multidomain proteins with multiple functions. Genes Dev. 2003;17:7–30. doi: 10.1101/gad.1039703. [DOI] [PubMed] [Google Scholar]
- Serrano S.M., Jia L.G., Wang D., Shannon J.D., Fox J.W. Function of the cysteine-rich domain of the haemorrhagic metalloproteinase atrolysin A: targeting adhesion proteins collagen I and von Willebrand factor. Biochem. J. 2005;391:69–76. doi: 10.1042/BJ20050483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano S.M., Wang D., Shannon J.D., Pinto A.F., Polanowska-Grabowska R.K., Fox J.W. Interaction of the cysteine-rich domain of snake venom metalloproteinases with the A1 domain of von Willebrand factor promotes site-specific proteolysis of von Willebrand factor and inhibition of von Willebrand factor-mediated platelet aggregation. FEBS J. 2007;274:3611–3621. doi: 10.1111/j.1742-4658.2007.05895.x. [DOI] [PubMed] [Google Scholar]
- Serrano S.M., Kim J., Wang D., Dragulev B., Shannon J.D., Mann H.H., Veit G., Wagener R., Koch M., Fox J.W. The cysteine-rich domain of snake venom metalloproteinases is a ligand for von Willebrand factor A domains: role in substrate targeting. J. Biol. Chem. 2006;281:39746–39756. doi: 10.1074/jbc.M604855200. [DOI] [PubMed] [Google Scholar]
- Takeda S., Igarashi T., Mori H. Crystal structure of RVV-X: an example of evolutionary gain of specificity by ADAM proteinases. FEBS Lett. 2007;581:5859–5864. doi: 10.1016/j.febslet.2007.11.062. [DOI] [PubMed] [Google Scholar]
- Takeda S., Igarashi T., Mori H., Araki S. Crystal structures of VAP1 reveal ADAMs’ MDC domain architecture and its unique C-shaped scaffold. EMBO J. 2006;25:2388–2396. doi: 10.1038/sj.emboj.7601131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang P., Huang M.C., Klostergaard J. Length of the linking domain of human pro-tumor necrosis factor determines the cleavage processing. Biochemistry. 1996;35:8226–8233. doi: 10.1021/bi952183l. [DOI] [PubMed] [Google Scholar]
- Tsakadze N.L., Sithu S.D., Sen U., English W.R., Murphy G., D’Souza S.E. Tumor necrosis factor-alpha-converting enzyme (TACE/ADAM-17) mediates the ectodomain cleavage of intercellular adhesion molecule-1 (ICAM-1) J. Biol. Chem. 2006;281:3157–3164. doi: 10.1074/jbc.M510797200. [DOI] [PubMed] [Google Scholar]
- Van Eerdewegh P., Little R.D., Dupuis J., Del Mastro R.G., Falls K., Simon J., Torrey D., Pandit S., McKenny J., Braunschweiger K., Walsh A., Liu Z., Hayward B., Folz C., Manning S.P., Bawa A., Saracino L., Thackston M., Benchekroun Y., Capparell N., Wang M., Adair R., Feng Y., Dubois J., FitzGerald M.G., Huang H., Gibson R., Allen K.M., Pedan A., Danzig M.R., Umland S.P., Egan R.W., Cuss F.M., Rorke S., Clough J.B., Holloway J.W., Holgate S.T., Keith T.P. Association of the ADAM33 gene with asthma and bronchial hyperresponsiveness. Nature. 2002;418:426–430. doi: 10.1038/nature00878. [DOI] [PubMed] [Google Scholar]
- Waldhauer I., Goehlsdorf D., Gieseke F., Weinschenk T., Wittenbrink M., Ludwig A., Stevanovic S., Rammensee H.G., Steinle A. Tumor-associated MICA is shed by ADAM proteases. Cancer Res. 2008;68:6368–6376. doi: 10.1158/0008-5472.CAN-07-6768. [DOI] [PubMed] [Google Scholar]
- Wang S.H., Shen X.C., Yang G.Z., Wu X.F. CDNA cloning and characterization of Agkistin, a new metalloproteinase from Agkistrodon halys. Biochem. Biophys. Res. Commun. 2003;301:298–303. doi: 10.1016/s0006-291x(02)03001-2. [DOI] [PubMed] [Google Scholar]
- Wang Y.L., Goh K.X., Wu W.G., Chen C.J. Purification, crystallization and preliminary X-ray crystallographic analysis of a cysteine-rich secretory protein (CRISP) from Naja atra venom. Acta Crystallogr. D Biol. Crystallogr. 2004;60:1912–1915. doi: 10.1107/S0907444904019766. [DOI] [PubMed] [Google Scholar]
- Ward C.M., Vinogradov D.V., Andrews R.K., Berndt M.C. Characterization of mocarhagin, a cobra venom metalloproteinase from Naja Mocambique Mocambique, and related proteins from other elpidae venoms. Toxicon. 1996;34:1203–1206. doi: 10.1016/0041-0101(96)00115-8. [DOI] [PubMed] [Google Scholar]
- Wei J.F., Mo Y.Z., Qiao L.Y., Wei X.L., Chen H.Q., Xie H., Fu Y.L., Wang W.Y., Xiong Y.L., He S.H. Potent histamine-releasing activity of atrahagin, a novel snake venom metalloproteinase. Int. J. Biochem. Cell Biol. 2006;38:510–520. doi: 10.1016/j.biocel.2005.10.011. [DOI] [PubMed] [Google Scholar]
- White J.M. ADAMs: modulators of cell–cell and cell–matrix interactions. Curr. Opin. Cell Biol. 2003;15:598–606. doi: 10.1016/j.ceb.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Wu E., Croucher P.I., McKie N. Expression of members of the novel membrane linked metalloproteinase family ADAM in cells derived from a range of haematological malignancies. Biochem. Biophys. Res. Commun. 1997;235:437–442. doi: 10.1006/bbrc.1997.6714. [DOI] [PubMed] [Google Scholar]
- Xiong J.P., Stehle T., Zhang R., Joachimiak A., Frech M., Goodman S.L., Arnaout M.A. Crystal structure of the extracellular segment of integrin αvβ3 in complex with an Arg-Gly-Asp ligand. Science. 2002;296:151–155. doi: 10.1126/science.1069040. [DOI] [PubMed] [Google Scholar]
- Yang C.C., Chang L.S. Role of the N-terminal region in phospholipases A2 from Naja naja atra (Taiwan cobra) and Naja nigricollis (spitting cobra) venoms. Toxicon. 1988;26:721–731. doi: 10.1016/0041-0101(88)90279-6. [DOI] [PubMed] [Google Scholar]
- Zhou Q., Dangelmaier C., Smith J.B. The hemorrhagin catrocollastatin inhibits collagen-induced platelet aggregation by binding to collagen via its disintegrin-like domain. Biochem. Biophys. Res. Commun. 1996;219:720–726. doi: 10.1006/bbrc.1996.0301. [DOI] [PubMed] [Google Scholar]
- Zhu Z., Gao Y., Zhu Z., Yu Y., Zhang X., Zang J., Teng M., Niu L. Structural basis of the autolysis of AaHIV suggests a novel target recognizing model for ADAM/reprolysin family proteins. Biochem. Biophys. Res. Commun. 2009;386:159–164. doi: 10.1016/j.bbrc.2009.06.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal diffraction and structural refinement statistics of atragin (after pH jump).