Graphical abstract
Keywords: Organ model, Pharmacological testing in vitro, 3D printing, Excised human tissue, Alternatives to animal testing, Tissue engineering
Abstract
3D organ models have gained increasing attention as novel preclinical test systems and alternatives to animal testing. Over the years, many excellent in vitro tissue models have been developed. In parallel, microfluidic organ-on-a-chip tissue cultures have gained increasing interest for their ability to house several organ models on a single device and interlink these within a human-like environment. In contrast to these advancements, the development of human disease models is still in its infancy. Although major advances have recently been made, efforts still need to be intensified. Human disease models have proven valuable for their ability to closely mimic disease patterns in vitro, permitting the study of pathophysiological features and new treatment options. Although animal studies remain the gold standard for preclinical testing, they have major drawbacks such as high cost and ongoing controversy over their predictive value for several human conditions. Moreover, there is growing political and social pressure to develop alternatives to animal models, clearly promoting the search for valid, cost-efficient and easy-to-handle systems lacking interspecies-related differences.
In this review, we discuss the current state of the art regarding 3D organ as well as the opportunities, limitations and future implications of their use.
1. Basic and preclinical pharmacological research – state of the art
Animal models, particularly rodents, are the gold standard for basic and preclinical research in medical and pharmacological science [1]. From a regulatory perspective, their use is mandatory in the transition from preclinical to clinical studies. As such, they have been beyond discussion for several decades. Recently however, increasing criticism evolved from ethical and scientific concerns. About 80% of potential therapeutics fail in clinical trials despite efficacy and safety in preclinical studies [2]. Potential underlying reasons include poor characterization of the relevant animal models [3], a lack of sufficient experimental quality within in vivo studies [4] and distinct interspecies-related differences to humans such as anatomy, (patho)physiology and immunology. For example, in dermatological research mouse models are predominantly used despite clear interspecies-related differences and a mere ∼ 30% overlap of skin-associated genes between mice and men [5]. Such disparities exist for almost all human organs.
Many diseases that occur in humans do not naturally in other animals, requiring artificial disease induction to model the disease. Even if animals show similar disease phenotypes, it remains unclear if the underlying pathogenesis is comparable or identical to that in humans. For instance, mice normally do not develop atopic dermatitis (AD), one of the most common skin diseases of industrialized nations. Poor homologies between the transcriptomic profile of murine AD models and human AD have been revealed [6]. Nevertheless, most preclinical research is executed in mice because human-based models are lacking. Other prominent examples are viral infections of the human respiratory tract that many animals are not susceptible to, in particular for mice. Differences in antiviral immunity, the layout of anatomical barriers and divergence in receptor repertoires prevent successful infection [7]. In extreme cases, animal models show distorted pathogenic profiles. For instance, the mouse model SARS coronavirus infection demonstrates severe encephalitis absent from the human pathogenesis [8].
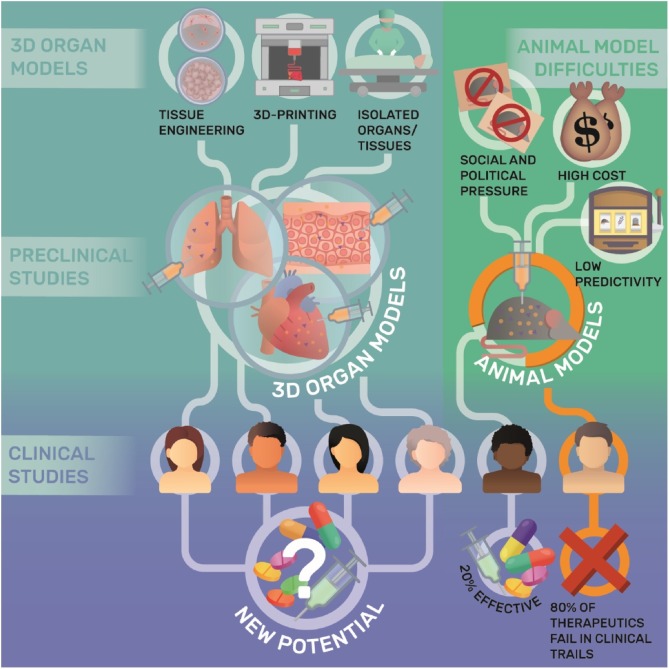
Due to a lack of concordance and reproducibility, the translational value of preclinical animal models has been questioned in several publications (Fig. 1 ) [4,6,9] including Seok and coworkers, who demonstrated the lack of predictive value mouse models hold for inflammatory diseases [10]. Although it must be noted the latter study is controversially discussed [1,11], these publications, at the very least, highlight the difficulty and caution required in correctly deriving information from animal studies.
Fig. 1.
Chances and limitations of 3D organ and animal models in drug development.
Given the high costs of drug development and their high failure rates in the clinical phase, alongside the remaining unmet medical needs, scientists must question the suitability of present approaches to pharmacological research (Fig. 1). Several proposals have been made overcome the limitations of animal testing. One ever-present consideration is the imperative nature of proper study design and model choice in the light of recent findings.
Another equally important strategy is the development of alternative, human-based models for basic and preclinical research. This area has recently gained increasing attention as a result of ethical pressures to develop alternatives to animal models as well as scientific necessity. In this review, we discuss the ‘state of the art’ of organ models, their limitations and regulatory aspects, with a focus on human epithelia and liver. Advancements in organoids and organs-on-a-chip devices are not covered since these topics have been reviewed comprehensively elsewhere [12,13].
2. Implications of biomedical modelling
Models are used in almost every scientific discipline since experiments with real systems cannot be performed for practical or ethical reasons. Hence, a clear understanding how models differ from and simulate real systems is required. Both show stochastic behavior and, consequently, give non-identical results and conclusions. However, the closer a model reflects a real system the more valid it becomes, leading to ask how to design or choose a model to validly confirm or reject a hypothesis proposed about a real system? This raises a further question: do investigators in biomedical research propose hypotheses about real systems/diseases or about the model itself - unconsciously? For example, an immunologist might analyze principles of viral antigen presentation in a well-established mouse model. A virologist, on the other hand, needs data about clinical outcomes to understand the interaction between viruses and the human host, a goal not set out by the immunologist when establishing the mouse model. As a consequence, subsequent results may lead to wrong conclusions regarding the clinical situation, causing uncertainty in their predictive value for translational research.
If research aims to gain knowledge without aiming transferability to human disease, the validity of the model for the real system would be subordinated. However, a primary aim of biomedical research 'indeed is’ the translation from preclinical to clinical work, for which well-characterized models validated against the human situation are necessary. Identifying and characterizing human disease must involve work within both, the real system and their models, in an iterative process. Knowledge of human disease itself is indispensable in validating the theoretical and functional use of a model. This also implies that model validity cannot be achieved by determining absolute indicators, as one extreme, nor by establishing an unrelated model as another. Instead, validation is individual and gradual for every model and requires an open, continuous negotiation between all stakeholders and their goals, where again a ‘back-to-basic’ focus involving meta-research of biomedical modeling must be established. This is especially important since human-based organ models are set to bridge an apparent gap from basic research to preclinical testing. As such, the field of (disease) models will likely become even more heterogeneous and complex (e.g. 2D/3D, cell lines/primary tissue, microfluidic communication, animals/human). To ensure model validity, the principal methods of neighboring disciplines such as computational science [14,15] will become increasingly important and may indeed lead to standardized procedures to calibrate and verify model robustness.
3. Human-based organ models
3.1. Healthy and diseased 3D organ models
3.1.1. Conventional tissue engineering (TE) approaches and 3D bioprinting
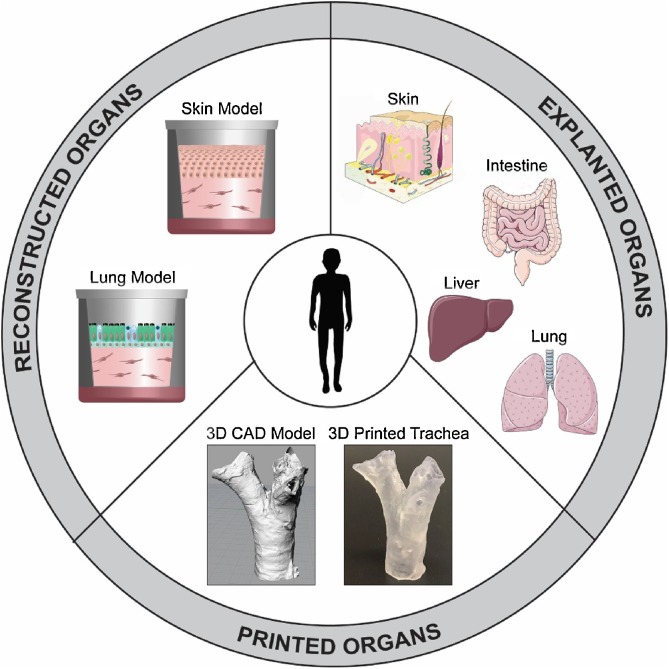
The complex three-dimensional (3D) organization of living tissues enables crosstalk between different cell types, their extracellular matrices, and other organs. Therefore, 3D organ models (Fig. 2 ) that emulate tissue characteristics better than simple 2D monolayer cultures are invaluable to investigations of more complex biological phenomena, as has already been demonstrated for several organs [[16], [17], [18]].
Fig. 2.
Overview of different approaches towards 3D organ models.
Reconstructed models of almost all organs of the human body have been described. They are typically generated by sequential cell seeding into cell culture inserts or porous 3D scaffolds. Of these, bone and cartilage [19], and epithelia such as skin [20,21], or lung [22] are currently the most advanced, while models of more complex tissues such as the brain are at earlier stages of development. Notably, organ models may vary in their complexity and, hence, predictivity. Further, it is important to define the correct culture conditions for reconstructed organs. For instance, cultivation at the air-liquid interface is crucial to the correct formation of epithelia [23]. Others such as cartilage or bone require submersed cultivation and exposure to biomechanical forces [24] such as can be implemented through dynamic conditions within bioreactors. Another essential consideration is the cell source. Many models rely on cell lines due to their tractability, low costs, and availability. Although these models do have merit in some applications including toxicological screenings, the use of primary cells is imperative when aiming for human-based models in preclinical pharmacological research. In this context, stem cells and iPSC will likely play an increasingly important role in the future, especially when aiming for disease models, as they already do for organoids [25]. So far, in vitro modeling of diseases most commonly relies on modulation of disease-associated genes [26], co-cultivation of normal and diseased cells [27], supplementation of disease-associated stimuli [28], or infections with microorganism [29,30]. Overall, however, the development of disease models is still in its infancy.
Organ structures can also be reconstituted by recellularizing scaffolds derived from excised animal organs [31] or non-transplantable human organs [32] or by 3D bioprinting. Detergents are used to remove all cells leaving a dECM composed of collagen fibers and proteinaceous factors facilitating cell growth, differentiation and function after repopulation with human cells [33]. The choice of ECM is pivotal to the functionality of tissue models. For example, Hedström et al. decellularized bronchial airway sections from COPD patients and repopulated them with normal human bronchial epithelial cells [34] resulting in differential gene expression and altered activity of upstream mediators associated with COPD pathophysiology. This highlights the impact of biochemical cues within dECM in healthy and diseased states.
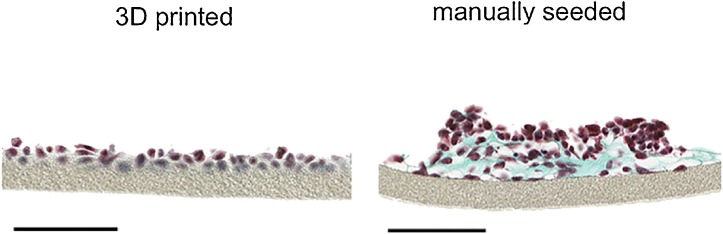
Common problems in recellularizing scaffold include the availability of normal human dECM, discrepancies between the sizes of human cells and animal matrices, and the precise cell positioning during recellularization. 3D bioprinting offers features that overcome some of these bottlenecks, allowing one to print singularized cells in a highly automated and spatio-temporally controlled manner. Bioprinting has been employed for the fabrication of several organ models including human skin [35,36], lung [37,38] and liver [39] and recently also soft functional tissues including muscle [40] or kidney [41]. To take one example, engineered skin tissue requires specific cell-cell and cell-matrix interactions as well as precise positioning of cell layers. 3D bioprinting has the potential to fulfill these requirements by assembling cells and matrix components at the density observed in the native tissue [42]. Various printing technologies have been employed to produce skin models. For example, Ng et al. printed pigmented skin models by adding melanocytes [35]. The models exhibited similar constitutive pigmentation as the skin donors and did not anisotropically contract during maturation, as is often encountered with manually prepared models. Within this vein of thought, an alveolar barrier model composed of a bilayer of immortalized endothelial cells and lung epithelial cells, sandwiched between thin layers of Matrigel™, has been printed layer-by-layer in a highly reproducible manner. Compared to the morphology of the manually arranged bilayers (Fig. 3 ), the printed model features a homogenous cell distribution without cell aggregation [37].
Fig. 3.
Brightfield micrographs of 3D printed and manually seeded paraffin embedded histological cross sections of a 2 layered air-blood barrier model stained with Masson-Goldner trichrome coloration after 3 days in culture. Cytoplasm is stained red, collagen fibers of the ECM Matrigel™ green and cell nuclei dark brown. Note the lack of cell organization in the manually seeded compared to the bioprinted model. Scale bars 100 μm. Reproduced in accordance with the Creative Commons Public License (http://creativecommons.org/licenses/by-nc-nd/4.0/) from [34]. Copyright 2015, Springer Nature. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
Nevertheless, since freshly printed tissues essentially lack any cell-cell and cell-matrix contacts, bioprinted models require tissue maturation similar to conventional TE approaches. The maturation process required to yield a truly biomimetic and functional tissue is time-consuming, and its success is highly dependent on the properties of the applied bioink, as well as biophysical and biochemical cues. Although bioprinting organ models indeed appears promising, in its current state, it must be considered an evolving technology that must overcome several limitations before it can be widely employed. One such limitation is the insufficient resolution with which currently available bioprinters are able to spatially position cells, as compared to the compositions found in native organs. Moreover, focus has been primarily placed on cell lines given their ease of use, comparatively robust nature, and inexpensive price. More advanced models will clearly require the use of primary cells. Another bottleneck is the technical difficulty printing more than one cell type poses, for which many studies have only used a single cell type. Addressing the latter, an increasing number of publications report bioprinting of various cell types recently [37,43,44]. This approach, however, is associated with some problems such as special needs of each cell type with regard to the bioink and the necessity to establish suitable co-culture conditions after the printing itself for tissue maturation. Moreover, although cell positioning can be achieved easily, cell migration and proliferation cannot be controlled which may ultimately impact tissue functionality. Overall, this approach requires further investigations.
3.1.2. Isolated human organs and organ sections
The ex vivo use of excised human organs/tissues obtained from biopsies or surgery is another approach for preclinical research. These organs/tissues may also prove extremely valuable for the validation of TE organ models, since an original human reference seems crucial to the validation of tissue structure, cellular composition and (patho)physiological responses. However, the availability of excised human material is a major bottleneck. On the rare occasion that complete human organs can be obtained for experimental use, they are normally end-stage disease organs. Moreover, experiments with explanted organs require complex equipment (e.g. for organ perfusion) and extensively trained personnel. Nevertheless, the advantages of these models have already been demonstrated, as exemplified with explanted lungs. Important physiologic parameters such as the mechanical strain acting on lung type II cells, as well as surfactant production can be preserved in isolated, ventilated and perfused human lungs. The fact that an Organ Care System Lung device (TransMedics Inc; Andover, MA, USA) is under evaluation as a portable normothermic ex vivo lung perfusion and ventilation system for donor lung preservation highlights the high organ quality achievable in such systems [45]. Additionally, the conjunction of Organ Care Systems with non-transplantable organs/ tissues supplied by institutions such as the International Institute for the Advancement of Medicine (Edison, NJ, USA) may greatly expand the use of high quality organs for biomedical research. However, the accessibility for explanted organs is still restricted and costly. Moreover, the conventionally used systems lack the ability to mimic organ specific environments, limiting periods of investigation to hours.
In contrast to whole human organs, the use of small tissue pieces or organ slices may increase the number of investigable samples. Here, peripheral resected tissue not needed for diagnosis and considered clinically normal may be used. This seems particularly favorable since tissue sections thinner ≤ 200 μm are easier to maintain ex vivo than whole organ explants. Nevertheless, a reproducible, artefact-free preparation of thin sections in parallel plane, without disruptions to tissue integrity is a challenge per se. In particular, for the preparation of highly elastic (e.g. lung) or fragile (e.g. brain) tissues, specialized techniques such as high-pressure water jet cutting need to be employed. Interestingly, such tissues may be integrated more easily into perfusion chambers or microfluidic systems for preclinical studies that may increase their possible cultivation periods [46]. Tissue sections reflect the spatiotemporal behavior of functional units within human organs without the inclusion of systemic body responses such as immune cell recruitment or hormonal effects. Consequently, researchers can primarily investigate local responses. Nevertheless, tissue sections still retain their native immune cells; a major advantage over bioprinted or TE organs. Furthermore, recent data indicate that cryopreservation of tissue slices is possible without major impairment upon cell viability or tissue functionality [47]. This approach also seems interesting for direct testing of individual patient responses to a specific drug, of great relevance to the emerging field of personalized medicine.
4. Current applications of human-based test models in pharmacological research: epithelial and liver models
The application of 3D organ models is well established in toxicological screening and basic research [48,49]. Their use is clearly less well explored in preclinical drug testing, where, currently, spheroidal organoids are more established [50]. Nonetheless, organ models provide greater control over the tissue environment and spatial organization of cells, and may, therefore, prove beneficial. Indeed, the pharmaceutical industry has considerably extended their activities in the development of organ (disease) models, although their use in actual preclinical drug testing is still in its infancy and has not yet yielded a substantial replacement for animal testing.
Notably, although epithelial models of the skin or the upper airways belong to the most advanced organ models, publications describing their use for drug testing are scarce. Most studies reporting novel epithelial models conclude that these are promising tools for preclinical testing in vitro [[51], [52], [53]] without presenting any data to support this. More studies still have been published with regard to drug delivery approaches [54,55], but without including clear pharmacological readouts. Although the importance of these studies is undoubted, the next step must be the assessment of the actual value of organ models for in vitro drug testing including the identification of valid readout parameters, definition of their limitations, and rigorous comparison against the relevant in vivo situation. So far, only a handful of studies have performed drug testing in skin models, and of these, only 1–2 drugs were included [[56], [57], [58], [59], [60]], the experimental settings varied significantly between them, and the choices of readout parameters have not been sufficiently justified.
Initial data are also available on the applicability of bioprinted models in drug testing, most of which have been liver models. For example, a liver model consisting of patient-derived hepatocytes, stellate cells, and endothelial cells [39] was used to study the hepatotoxic effects of the antibiotics levofloxacin and trovafloxacin. Notably, trovafloxacin had been withdrawn from the market previously due to liver toxicity. While trovafloxacin’s hepatotoxic potential did not become evident in standard pre-clinical models, results akin to the clinical situation were seen in the bioprinted liver model. Concordantly, the treatment of an iPSCs-based liver model with rifampicin, a strong inductor of the hepatic cytochrome P450 system, induced the expected upregulation of metabolic enzymes [61]. Such approaches may help to predict drug-drug interactions in the future. However, these initial results should not disguise the fact that bioprinted organ models are far from being routinely used for in vitro drug testing. As a whole, this field’s current major goal is the improvement of bioprinted models aiming for systems that mimic the architecture and composition of the emulated organs as closely as possible.
Isolated whole human organs seem promising for the assessment of novel cell-based medical strategies including the therapeutic application of stem cells that may specifically profit from testing in complex settings. A pair of studies demonstrated the potential of isolated human lungs for as a model to study the effects mesenchymal stem cells have on alveolar fluid clearance, inflammatory responses, and bacterial infections [62,63]. Another study used isolated human lung lobes to investigate pharmacodynamic effects of a novel long-acting ß2-agonist in comparison to salbutamol [64]. Interestingly, the determined onset of drug action was consistent with other preclinical data. However, the duration of the bronchodilatatory effect could not be studied since the viability of the lung lobes declined following 2–3 h study. Another drawback was apparent the lack of dose-response after application of the bronchoconstrictory positive control.
Human organ slices cultivated ex vivo may hold greater potential for preclinical applications due to their easier accessibility. For instance, there is recurrent evidence of their tractability in the study of infectious disease. Pathogens such as Streptococcus pneumoniae are strictly human-specific, and viruses such as influenza A or MERS coronavirus show great genetic adaptation determining their pathogenicity and host tropism. Hence, the use of wild-type pathogens is of utmost importance for the study of infectious diseases and the development of novel therapies. Such investigations must be conducted in tissue from the addressed host (e.g. humans). For example, human lung slices have permitted the study viral tropism and mediator generation of pathogens in humans [65,66]. Moreover, these have also proven valuable for the identification of new therapeutic targets, as recently demonstrated for influenza A- associated pneumococci co-infections [67], and the testing of preventive interventions, as exemplified by certain vaccines [68]. Similarly, human liver slices (hLS) have proven valuable to the study of viral life cycles, the assessment of antiviral drugs efficacy [69], and monitoring of local innate immune responses [70]. An elegant study [71] tested 8 approved drugs with known liver-related effects in hLS by assessing relevant markers of oxidative stress, mitochondrial function, and drug metabolism. The observed characteristics induced by reference drugs may help to evaluate the safety and dosing of new compounds thereby selecting drug candidates with the widest safety margin. Potential antifibrotic drugs for early onset liver fibrosis were also investigated in hLS [72]. Interestingly, contradictory results were obtained between studies in human tissue and rat liver slices, indicating species-specific drug effects.
Overall, the above-mentioned studies on human lung and liver slices indicate the potential of human tissues in the study human diseases ex vivo. However, it has also become clear that use of explanted organs or tissue slices for drug testing is still at an early stage of methodological development and has not yet been explored satisfactorily for wider application.
5. Limitations
Despite the progress in the development of organ models, several challenges remain unsolved. So far, only animal models can enable the investigation of drug actions in a complex, living organism. Almost all organ models lack vascularization, regenerative capacity, immune systems, and inter-tissue crosstalk. These bottlenecks may be addressed by advancements in multi-organ approaches such as microfluidic chips and bioreactors that combine several tissues within a human-like environment [73]. These setups may also help to overcome the lack of adequate biomechanics provided by the mainly static cultivation of organ models. The physical properties of the tissue microenvironment, as well as the external forces that act upon them, alter cell behaviors, tissue organization and cell-generated forces. Hence, mechanical forces are important for tissue development as demonstrated for several organs including the lung [74] and skin [75]. In addition, the development of human disease models per se are still in their infancy. This likely owes, at least in part, to the enormous expense and specific knowledge required to build a meaningful human disease model, which has restricted the development of such models to a limited number of groups [76]. With regard to excised organs/tissues, limited and uncertain availability, as well as complex ethical issues, are common restrictions for their broad use; the elaboration of cryopreservation and biobanking, however, may help to overcome these barriers.
Another point of consideration is the difficulty in comparing organ models made by protocols in which cell types, culture media and ECM differ significantly from one another. Results from the separate systems may produce contradictory or non-reproducible data, as are frequently seen in animal studies. One approach to overcome this shortage may be the development of tissue banks derived from iPSCs which allow repeated use of primary, human-based material (healthy and diseased) providing a genetically identical background for then generated tissues. On the other hand, this approach again may limit the biodiversity reflected in generating organ models from several donor sources, a clear advantage to clinical translation. Another obstacle to widespread employment is the current lack of capacities for large-scale, reproducible 3D organ production. Furthermore, the resolution of bioprinting needs to be improved to accurately mimic the biological architecture; currently, organ models are typically scaled up in size, which causes a bias as the scaffold size is increased while the cell size remains unchanged.
Last but not least, the complex architecture of organ models and the investigation of cell-specific responses require the identification of suitable readout parameters and the implementation of novel techniques. Here, sophisticated microscopic techniques such as (lattice) light sheet and spectral microscopy [77], and further elaboration of multiphoton imaging hold a great potential. Similarly, multi-marker analysis of small sample volumes, tissue microdissection combined with single-cell –Omics, innovative live-cell tissue labelling [78], and robust bioinformatics pipelines may be key technologies for efficient data readouts.
6. Regulatory aspects
In general, European regulatory authorities are positively disposed towards the use of organ (disease) models in preclinical studies, recognizing the necessity of closing the current translational gap. Although pharmaceutical companies are intensifying their efforts in this area, they are still at an early stage, and barely any applications employing organ disease models for pharmacological testing have been submitted Toxicological in vivo data are still mandatory when entering the clinical phase, which is regulated by law, to provide the required safety for the study subjects. For pharmacological testing, case-by-case decisions are possible, and the medical need plays a pivotal role. If a novel drug has the potential for significant medical benefit, it is possible to enter the clinical phase without presenting in vivo data. This also holds true if no (valid) preclinical in vivo model exists, as is the case for several conditions including severe keratinization disorders of human skin, certain tumors, HIV infections and viral lung infections. The development of in vitro organ models for these conditions may facilitate their implementation in the preclinical and regulatory routine since the demand is explicitly high.
7. Concluding remarks & future perspectives
Organ models hold the potential to revolutionize preclinical research, although there is still a long road ahead and the predictivity of most organ models has yet to be proven. Pharmaceutical companies and regulatory authorities have recognized the importance of this technology and are highly engaged with them. Scientists must learn from previous mistakes and combine efforts to improve preclinical-to-clinical translation. In this regard political and financial support is pivotal. Furthermore, joint efforts of expert groups in academia, pharmaceutical industry, and regulatory authorities are now needed to approach and overcome current bottlenecks. Accordingly, governments initiatives are required to promote not only the development of organ (disease) models but also their implementation in preclinical drug testing, otherwise the full potential of organ models may never be realized.
Conflict of interest
None.
Acknowledgments
Financial support by the Berlin-Brandenburg research platform BB3R and the “Bundesinstitut für Risikobewertung” (BfR; 1328-566) is gratefully acknowledged by S.He. This work was also supported by the German Research Foundation (DFG SFB-TR84) to A.C.H. and S.Hi. (B6 and TF1) and A.C.H (Z1a) and the German Federal Ministry of Education and Research (BMBF, RAPID-Network to A.C.H. and S.Hi and FKZ13N13523 to M.W.). Further financial support to J.K. from the “Stiftung zur Förderung der Erforschung von Ersatz- und Ergänzungsmethoden zur Einschränkung von Tierversuchen” (Stiftung SET) and the “Bundesinstitut für Risikobewertung” (1328-568) is gratefully acknowledged. Moreover, we thank Dr. Andrea Marzoll and Dr. Roland Frötschl from the Federal Institute for Drugs and Medical Devices in Germany for their valuable help and insights in decision processes of regulatory authorities. Dr. Guy Yealland’s help in language editing is gratefully acknowledged and we are thankful to Johanna Berg, Thomas Hiller and Anna Löwa for providing images of 3D printed models and figure design.
Glossary
- 3D
Three-dimensional
- COPD
Chronic obstructive pulmonary disease
- dECM
Decellularized extracellular matrix
- ECM
Extracellular matrix
- hLS
Human liver slices
- iPSC
Induced pluripotent stem cell
- TE
Tissue engineering
References
- 1.Osuchowski M.F., Remick D.G., et al. Abandon the mouse research ship? Not just yet! Shock. 2014;41:463–475. doi: 10.1097/SHK.0000000000000153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pound P., Bracken M.B. Is animal research sufficiently evidence based to be a cornerstone of biomedical research? BMJ. 2014;348:g3387. doi: 10.1136/bmj.g3387. [DOI] [PubMed] [Google Scholar]
- 3.Perrin S. Preclinical research: make mouse studies work. Nature. 2014;507:423–425. doi: 10.1038/507423a. [DOI] [PubMed] [Google Scholar]
- 4.Couzin-Frankel J. When mice mislead. Science. 2013;342(922-923):925. doi: 10.1126/science.342.6161.922. [DOI] [PubMed] [Google Scholar]
- 5.Gerber P.A., Buhren B.A., et al. The top skin-associated genes: a comparative analysis of human and mouse skin transcriptomes. Biol. Chem. 2014;395:577–591. doi: 10.1515/hsz-2013-0279. [DOI] [PubMed] [Google Scholar]
- 6.Ewald D.A., Noda S., et al. Major differences between human atopic dermatitis and murine models, as determined by using global transcriptomic profiling. J. Allergy Clin. Immunol. 2017;139:562–571. doi: 10.1016/j.jaci.2016.08.029. [DOI] [PubMed] [Google Scholar]
- 7.Saturni S., Contoli M., et al. Models of respiratory infections: virus-induced asthma exacerbations and beyond. Allergy Asthma Immunol. Res. 2015;7:525–533. doi: 10.4168/aair.2015.7.6.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Netland J., Meyerholz D.K., et al. Severe acute respiratory syndrome coronavirus infection causes neuronal death in the absence of encephalitis in mice transgenic for human ACE2. J. Virol. 2008;82:7264–7275. doi: 10.1128/JVI.00737-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leist M., Hartung T. Inflammatory findings on species extrapolations: humans are definitely no 70-kg mice. Arch. Toxicol. 2013;87:563–567. doi: 10.1007/s00204-013-1038-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seok J., Warren H.S., et al. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc. Natl. Acad. Sci. U. S. A. 2013;110:3507–3512. doi: 10.1073/pnas.1222878110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takao K., Miyakawa T. Genomic responses in mouse models greatly mimic human inflammatory diseases. Proc. Natl. Acad. Sci. U. S. A. 2015;112:1167–1172. doi: 10.1073/pnas.1401965111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fatehullah A., Tan S.H., et al. Organoids as an in vitro model of human development and disease. Nat. Cell Biol. 2016;18:246. doi: 10.1038/ncb3312. [DOI] [PubMed] [Google Scholar]
- 13.Ronaldson-Bouchard K., Vunjak-Novakovic G. Organs-on-a-chip: a fast track for engineered human tissues in drug development. Cell Stem Cell. 2018;22:310–324. doi: 10.1016/j.stem.2018.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsioptsias N., Tako A.A., et al. Model validation and testing in simulation: a literature review. Presented at the 5th Student Conference on Operational Research (SCOR 2016) 2016 [Google Scholar]
- 15.Sargent R.S. Verification and validation of simulation models. Johansson S.J.B., Montoya-Torres J., Hugan J., Yücesan E., editors. Winter Simulation Conference. 2010 [Google Scholar]
- 16.Nichols J.E., Niles J.A., et al. Modeling the lung: design and development of tissue engineered macro- and micro-physiologic lung models for research use. Exp. Biol. Med. (Maywood) 2014;239:1135–1169. doi: 10.1177/1535370214536679. [DOI] [PubMed] [Google Scholar]
- 17.DesRochers T.M., Suter L., et al. Bioengineered 3D human kidney tissue, a platform for the determination of nephrotoxicity. PLoS One. 2013;8 doi: 10.1371/journal.pone.0059219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hopkins A.M., DeSimone E., et al. 3D in vitro modeling of the central nervous system. Prog. Neurobiol. 2015;125:1–25. doi: 10.1016/j.pneurobio.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee H.P., Gu L., et al. Mechanical confinement regulates cartilage matrix formation by chondrocytes. Nat. Mater. 2017;16:1243–1251. doi: 10.1038/nmat4993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blunder S., Ruhl R., et al. Alterations in epidermal eicosanoid metabolism contribute to inflammation and impaired late differentiation in FLG-mutated atopic dermatitis. J. Invest. Dermatol. 2017;137:706–715. doi: 10.1016/j.jid.2016.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huber B., Link A., et al. Integration of mature adipocytes to build-up a functional three-layered full-skin equivalent. Tissue Eng. Part C Methods. 2016;22:756–764. doi: 10.1089/ten.tec.2016.0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marrazzo P., Maccari S., et al. 3D reconstruction of the human airway mucosa in vitro as an experimental model to study NTHi infections. PLoS One. 2016;11 doi: 10.1371/journal.pone.0153985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gordon S., Daneshian M., et al. Non-animal models of epithelial barriers (skin, intestine and lung) in research, industrial applications and regulatory toxicology. Altex. 2015;32:327–378. doi: 10.14573/altex.1510051. [DOI] [PubMed] [Google Scholar]
- 24.Lee P.S., Eckert H., et al. Developing a customized perfusion bioreactor prototype with controlled positional variability in oxygen partial pressure for bone and cartilage tissue engineering. Tissue Eng. Part C Methods. 2017;23:286–297. doi: 10.1089/ten.TEC.2016.0244. [DOI] [PubMed] [Google Scholar]
- 25.Crespo M., Vilar E., et al. Colonic organoids derived from human induced pluripotent stem cells for modeling colorectal cancer and drug testing. Nat. Med. 2017;23:878. doi: 10.1038/nm.4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eckl K.M. Update: advanced methods in three-dimensional organotypic tissue engineering for congenital ichthyosis and other rare keratinization disorders. Br. J. Dermatol. 2014;171:1289–1290. doi: 10.1111/bjd.13450. [DOI] [PubMed] [Google Scholar]
- 27.Berroth A., Kuhnl J., et al. Role of fibroblasts in the pathogenesis of atopic dermatitis. J. Allergy Clin. Immunol. 2013;131:1547–1554. doi: 10.1016/j.jaci.2013.02.029. [DOI] [PubMed] [Google Scholar]
- 28.Danso M.O., van Drongelen V., et al. TNF-alpha and Th2 cytokines induce atopic dermatitis-like features on epidermal differentiation proteins and stratum corneum lipids in human skin equivalents. J. Invest. Dermatol. 2014;134:1941–1950. doi: 10.1038/jid.2014.83. [DOI] [PubMed] [Google Scholar]
- 29.Yonker L.M., Mou H., et al. Development of a primary human co-culture model of inflamed airway mucosa. Sci. Rep. 2017;7:8182. doi: 10.1038/s41598-017-08567-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Essaidi-Laziosi M., Brito F., et al. Propagation of respiratory viruses in human airway epithelia reveals persistent virus-specific signatures. J. Allergy Clin. Immunol. 2018;141:2074–2084. doi: 10.1016/j.jaci.2017.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wagner A., Rohrs V., et al. Use of a three-dimensional humanized liver model for the study of viral gene vectors. J. Biotechnol. 2015;212:134–143. doi: 10.1016/j.jbiotec.2015.08.012. [DOI] [PubMed] [Google Scholar]
- 32.Mazza G., Al-Akkad W., et al. Rapid production of human liver scaffolds for functional tissue engineering by high shear stress oscillation-decellularization. Sci. Rep. 2017;7:5534. doi: 10.1038/s41598-017-05134-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pati F., Jang J., et al. Printing three-dimensional tissue analogues with decellularized extracellular matrix bioink. Nat. Commun. 2014;5:3935. doi: 10.1038/ncomms4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hedstrom U., Hallgren O., et al. Bronchial extracellular matrix from COPD patients induces altered gene expression in repopulated primary human bronchial epithelial cells. Sci. Rep. 2018;8:3502. doi: 10.1038/s41598-018-21727-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ng W.L., Qi J.T.Z., et al. Proof-of-concept: 3D bioprinting of pigmented human skin constructs. Biofabrication. 2018;10:025005. doi: 10.1088/1758-5090/aa9e1e. [DOI] [PubMed] [Google Scholar]
- 36.Kim B.S., Kwon Y.W., et al. 3D cell printing of in vitro stabilized skin model and in vivo pre-vascularized skin patch using tissue-specific extracellular matrix bioink: a step towards advanced skin tissue engineering. Biomaterials. 2018;168:38–53. doi: 10.1016/j.biomaterials.2018.03.040. [DOI] [PubMed] [Google Scholar]
- 37.Horvath L., Umehara Y., et al. Engineering an in vitro air-blood barrier by 3D bioprinting. Sci. Rep. 2015;5:7974. doi: 10.1038/srep07974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berg J., Hiller T., et al. Optimization of cell-laden bioinks for 3D bioprinting of an influenza infection model. Sci. Rep. 2018;8:13877. doi: 10.1038/s41598-018-31880-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nguyen D.G., Funk J., et al. Bioprinted 3D primary liver tissues allow assessment of organ-level response to clinical drug induced toxicity in vitro. PLoS One. 2016;11 doi: 10.1371/journal.pone.0158674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Choi Y.-J., Park S.J., et al. Muscle-derived extracellular matrix on sinusoidal wavy surfaces synergistically promotes myogenic differentiation and maturation. J. Mater. Chem. B. 2018;6:5530–5539. doi: 10.1039/c8tb01475b. [DOI] [PubMed] [Google Scholar]
- 41.Homan K.A., Kolesky D.B., et al. Bioprinting of 3D convoluted renal proximal tubules on perfusable chips. Sci. Rep. 2016;6:34845. doi: 10.1038/srep34845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ng W.L., Wang S., et al. Skin bioprinting: impending reality or fantasy? Trends Biotechnol. 2016;34:689–699. doi: 10.1016/j.tibtech.2016.04.006. [DOI] [PubMed] [Google Scholar]
- 43.Park J.A., Yoon S., et al. Freeform micropatterning of living cells into cell culture medium using direct inkjet printing. Sci. Rep. 2017;7:14610. doi: 10.1038/s41598-017-14726-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Norona L.M., Nguyen D.G., et al. Editor’s highlight: modeling compound-induced fibrogenesis in vitro using three-dimensional bioprinted human liver tissues. Toxicol. Sci. 2016;154:354–367. doi: 10.1093/toxsci/kfw169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Warnecke G., Van Raemdonck D., et al. Normothermic ex-vivo preservation with the portable Organ Care System Lung device for bilateral Lung transplantation (INSPIRE): a randomised, open-label, non-inferiority, phase 3 study. Lancet Respir. Med. 2018;6:357–367. doi: 10.1016/S2213-2600(18)30136-X. [DOI] [PubMed] [Google Scholar]
- 46.Kress S., Baur J., et al. Evaluation of a miniaturized biologically vascularized scaffold in vitro and in vivo. Sci. Rep. 2018;8:4719. doi: 10.1038/s41598-018-22688-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bai Y., Krishnamoorthy N., et al. Cryopreserved human precision-cut lung slices as a bioassay for live tissue banking. A viability study of bronchodilation with bitter-taste receptor agonists. Am. J. Respir. Cell Mol. Biol. 2016;54:656–663. doi: 10.1165/rcmb.2015-0290MA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pridgeon C.S., Schlott C., et al. Innovative organotypic in vitro models for safety assessment: aligning with regulatory requirements and understanding models of the heart, skin, and liver as paradigms. Arch. Toxicol. 2018;92:557–569. doi: 10.1007/s00204-018-2152-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Caddeo S., Boffito M., et al. Tissue engineering approaches in the design of healthy and pathological in vitro tissue models. Front. Bioeng. Biotechnol. 2017;5:40. doi: 10.3389/fbioe.2017.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilkinson D.C., Alva-Ornelas J.A., et al. Development of a three-dimensional bioengineering technology to generate lung tissue for personalized disease modeling. Stem Cells Transl. Med. 2017;6:622–633. doi: 10.5966/sctm.2016-0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Duque-Fernandez A., Gauthier L., et al. A 3D-psoriatic skin model for dermatological testing: the impact of culture conditions. Biochem. Biophys. Rep. 2016;8:268–276. doi: 10.1016/j.bbrep.2016.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Desmet E., Ramadhas A., et al. In vitro psoriasis models with focus on reconstructed skin models as promising tools in psoriasis research. Exp. Biol. Med. (Maywood) 2017;242:1158–1169. doi: 10.1177/1535370217710637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Harrington H., Cato P., et al. Immunocompetent 3D model of human upper airway for disease modeling and in vitro drug evaluation. Mol. Pharm. 2014;11:2082–2091. doi: 10.1021/mp5000295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schweinlin M., Rossi A., et al. Human barrier models for the in vitro assessment of drug delivery. Drug Deliv. Transl. Res. 2017;7:217–227. doi: 10.1007/s13346-016-0316-9. [DOI] [PubMed] [Google Scholar]
- 55.Fytianos K., Chortarea S., et al. Aerosol delivery of functionalized gold nanoparticles target and activate dendritic cells in a 3D lung cellular model. ACS Nano. 2017;11:375–383. doi: 10.1021/acsnano.6b06061. [DOI] [PubMed] [Google Scholar]
- 56.Zoschke C., Ulrich M., et al. The barrier function of organotypic non-melanoma skin cancer models. J. Control. Release. 2016;233:10–18. doi: 10.1016/j.jconrel.2016.04.037. [DOI] [PubMed] [Google Scholar]
- 57.Wufuer M., Lee G., et al. Skin-on-a-chip model simulating inflammation, edema and drug-based treatment. Sci. Rep. 2016;6:37471. doi: 10.1038/srep37471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Faway E., Cambier L., et al. Modeling dermatophytosis in reconstructed human epidermis: a new tool to study infection mechanisms and to test antifungal agents. Med. Mycol. 2017;55:485–494. doi: 10.1093/mmy/myw111. [DOI] [PubMed] [Google Scholar]
- 59.Hönzke S., Gerecke C., et al. Tailored dendritic core-multishell nanocarriers for efficient dermal drug delivery: a systematic top-down approach from synthesis to preclinical testing. J. Control. Release. 2016;242:50–63. doi: 10.1016/j.jconrel.2016.06.030. [DOI] [PubMed] [Google Scholar]
- 60.Smits J.P.H., Niehues H., et al. Immortalized N/TERT keratinocytes as an alternative cell source in 3D human epidermal models. Sci. Rep. 2017;7:11838. doi: 10.1038/s41598-017-12041-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ma X., Qu X., et al. Deterministically patterned biomimetic human iPSC-derived hepatic model via rapid 3D bioprinting. Proc. Natl. Acad. Sci. U. S. A. 2016;113:2206–2211. doi: 10.1073/pnas.1524510113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McAuley D.F., Curley G.F., et al. Clinical grade allogeneic human mesenchymal stem cells restore alveolar fluid clearance in human lungs rejected for transplantation. Am. J. Physiol. Lung Cell Mol. Physiol. 2014;306:L809–815. doi: 10.1152/ajplung.00358.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee J.W., Krasnodembskaya A., et al. Therapeutic effects of human mesenchymal stem cells in ex vivo human lungs injured with live bacteria. Am. J. Respir. Crit. Care Med. 2013;187:751–760. doi: 10.1164/rccm.201206-0990OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gnadt M., Trammer B., et al. Comparison of the bronchodilating effects of inhaled beta(2)-agonists after methacholine challenge in a human lung reperfusion model. Eur. J. Pharm. Biopharm. 2012;81:617–626. doi: 10.1016/j.ejpb.2012.04.013. [DOI] [PubMed] [Google Scholar]
- 65.Chan R.W., Hemida M.G., et al. Tropism and replication of Middle East respiratory syndrome coronavirus from dromedary camels in the human respiratory tract: an in-vitro and ex-vivo study. Lancet Respir. Med. 2014;2:813–822. doi: 10.1016/S2213-2600(14)70158-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hocke A.C., Becher A., et al. Emerging human middle East respiratory syndrome coronavirus causes widespread infection and alveolar damage in human lungs. Am. J. Respir. Crit. Care Med. 2013;188:882–886. doi: 10.1164/rccm.201305-0954LE. [DOI] [PubMed] [Google Scholar]
- 67.Berg J., Zscheppang K., et al. Tyk2 as a target for immune regulation in human viral/bacterial pneumonia. Eur. Respir. J. 2017;50 doi: 10.1183/13993003.01953-2016. [DOI] [PubMed] [Google Scholar]
- 68.Temann A., Golovina T., et al. Evaluation of inflammatory and immune responses in long-term cultured human precision-cut lung slices. Hum. Vaccin. Immunother. 2017;13:351–358. doi: 10.1080/21645515.2017.1264794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lagaye S., Shen H., et al. Efficient replication of primary or culture hepatitis C virus isolates in human liver slices: a relevant ex vivo model of liver infection. Hepatology. 2012;56:861–872. doi: 10.1002/hep.25738. [DOI] [PubMed] [Google Scholar]
- 70.Wu X., Roberto J.B., et al. Precision-cut human liver slice cultures as an immunological platform. J. Immunol. Methods. 2018;455:71–79. doi: 10.1016/j.jim.2018.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vickers A.E., Ulyanov A.V., et al. Liver effects of clinical drugs differentiated in human liver slices. Int. J. Mol. Sci. 2017;18 doi: 10.3390/ijms18030574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Westra I.M., Mutsaers H.A., et al. Human precision-cut liver slices as a model to test antifibrotic drugs in the early onset of liver fibrosis. Toxicol. In Vitro. 2016;35:77–85. doi: 10.1016/j.tiv.2016.05.012. [DOI] [PubMed] [Google Scholar]
- 73.Maschmeyer I., Lorenz A.K., et al. A four-organ-chip for interconnected long-term co-culture of human intestine, liver, skin and kidney equivalents. Lab Chip. 2015;15:2688–2699. doi: 10.1039/c5lc00392j. [DOI] [PubMed] [Google Scholar]
- 74.Nelson C.M., Gleghorn J.P., et al. Microfluidic chest cavities reveal that transmural pressure controls the rate of lung development. Development. 2017;144:4328–4335. doi: 10.1242/dev.154823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rübsam M., Mertz A.F., et al. E-cadherin integrates mechanotransduction and EGFR signaling to control junctional tissue polarization and tight junction positioning. Nat. Commun. 2017;8:1250. doi: 10.1038/s41467-017-01170-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hönzke S., Wallmeyer L., et al. Influence of Th2 cytokines on the cornified envelope, tight junction proteins, and ß-defensins in filaggrin-deficient skin equivalents. J. Invest. Dermatol. 2016;136:631–639. doi: 10.1016/j.jid.2015.11.007. [DOI] [PubMed] [Google Scholar]
- 77.Valm A.M., Cohen S., et al. Applying systems-level spectral imaging and analysis to reveal the organelle interactome. Nature. 2017;546:162–167. doi: 10.1038/nature22369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Johnson B.P., Vitek R.A., et al. Vital ex vivo tissue labeling and pathology-guided micropunching to characterize cellular heterogeneity in the tissue microenvironment. Biotechniques. 2018;64:13–19. doi: 10.2144/000114626. [DOI] [PMC free article] [PubMed] [Google Scholar]