Abstract
The tripartite motif (TRIM)-containing proteins have attracted particular attention to their multiple functions in different biological processes. TRIM13, a member of the TRIM family, is a RING domain-containing E3 ubiquitin ligase which plays critical roles in diverse cellular processes including cell death, cancer and antiviral immunity. In this study, a TRIM13 homolog from orange spotted grouper, Epinephelus coioides (EcTRIM13) was cloned and characterized. The full-length of EcTRIM13 cDNA encoded a polypeptide of 399 amino acids which shared 81% identity with TRIM13 homolog from large yellow croaker (Larimichthys crocea). Amino acid alignment analysis showed that EcTRIM13 contained conserved RING finger and B-box domain. Expression patterns analysis indicated that EcTRIM13 was abundant in liver, spleen, kidney, intestine and gill. Moreover, the transcript of EcTRIM13 in grouper spleen was differently regulated after injection with Singapore grouper iridovirus (SGIV) or polyinosin-polycytidylic acid (poly I:C). Under fluorescence microscopy, we observed the tubular structure in wild type EcTRIM13 transfected cells, but the RING domain mutant resulted in the fluorescence distribution was changed and the bright punctate fluorescence was evenly situated throughout the cytoplasm, suggesting that the RING domain was essential for its accurate localization. Overexpression of EcTRIM13 in vitro obviously increased the replication of red spotted grouper nervous necrosis virus (RGNNV), and the enhancing effect of EcTRIM13 on virus replication was affected by the RING domain. Furthermore, the ectopic expression of EcTRIM13 not only negatively regulated the interferon promoter activity induced by interferon regulator factor (IRF) 3, IRF7, and melanoma differentiation-associated protein 5 (MDA5), but also decreased the expression of several interferon related factors. In addition, the overexpression of EcTRIM13 also differently regulated the transcription of pro-inflammatory factors. Together, our results firstly demonstrated that fish TRIM13 exerted negative regulation of antiviral response against nodavirus infection.
Keywords: TRIM13, Grouper, RGNNV, Antiviral, Interferon
Highlights
-
•
EcTRIM13 shared 81% identity with that from large yellow croaker.
-
•
RING domain was essential for its accurate localization of EcTRIM13.
-
•
Overexpression of EcTRIM13 enhanced the replication of RGNNV.
-
•
The ectopic expression of EcTRIM13 negatively regulated the interferon response.
-
•
Overexpression of EcTRIM13 differently regulated the transcription of pro-inflammatory factors.
1. Introduction
The tripartite motif (TRIM)-containing proteins have attracted enough attention to their multiple functions in different biological processes [1], [2]. Especially in antiviral immunity, a number of TRIM proteins have been elucidated to exert crucial roles in response to virus infection recently [3], [4]. Knockdown of TRIM15 decreased retinoic acid-inducible gene-I (RIG-I) induced interferon production and enhanced vesicular stomatitis virus (VSV) replication [5]. Overexpression of TRIM4 potentiated virus-triggered activation of interferon regulator factor (IRF) 3 and IFN-β induction, and finally mildly inhibited VSV replication [6]. Moreover, several TRIM proteins were proposed to restrict human immunodeficiency virus (HIV) or murine leukemia viruses (MLV) replication upon overexpression in vitro [3], [5], [7]. In addition to the antiviral activity to RNA virus, TRIM21 and TRIM5α were also found to exert their antiviral roles against DNA viruses [8].
TRIM13/RFP2, a member of the TRIM family, has been demonstrated to play critical roles in diverse cellular processes including cell death, cancer and antiviral immunity. TRIM13 was firstly demonstrated to act as a RING E3 ubiquitin ligase and play critical roles in ER-associated degradation [9]. Further studies showed that TRIM13 regulated the translocation of caspase-8 to autophagosome and its fusion with lysosome during ER stress [10]. Furthermore, TRIM13 was able to interact with NF-kappa-B essential modulator (NEMO) and modulate its ubiquitination which may regulate IKK complex activity [11]. In addition, TRIM13 not only interacted with both melanoma differentiation-associated protein 5 (MDA5) and RIG-I in vitro, but also functioned as a negative regulator of MDA5-mediated type I IFN production and abolished the resistance against a lethal challenge with encephalomyocarditis virus (EMCV) [12]. Although great progresses have been made in the function of mammalian TRIM13s, no information about fish TRIM13 was available.
Groupers, Epinephelus spp. are important farmed fish species in China and Southeast Asian countries. However, the emergency of virus diseases usually caused heavy economic losses in grouper industry [13], [14], [15]. Red spotted grouper nervous necrosis virus (RGNNV) and Singapore grouper iridovirus (SGIV) infection always caused high mortality in grouper aquaculture [13], [14]. Our previous studies demonstrated several grouper TRIM genes were involved in grouper iridovirus infection. Moreover, grouper TRIM39 and TRIM8 were also confirmed to act as antiviral roles against SGIV and RGNNV [16], [17], [18]. Whether grouper TRIM13 also exerted antiviral roles during grouper virus infection still remained unknown.
In the present study, a TRIM13 homolog from marine fish, orange spotted grouper (Epinephelus coioides), was cloned and characterized. Elucidation of the roles of TRIM13 during fish virus infection will provide new insights into the function of fish TRIMs.
2. Materials and methods
2.1. Fish, cells and viruses
Orange-spotted groupers, E. coioides (50–60 g) were purchased from a marine fish farm, Hainan Province, China. Groupers were kept in a laboratory recirculating seawater system before use. Grouper spleen (GS) cells used in our study were grown in Leibovitz’s L15 medium containing 10% fetal bovine serum (FBS, Gibco) [19]. The stocks of SGIV and RGNNV were prepared in our lab.
2.2. Cloning of EcTRIM13, sequence analysis and plasmid construction
The full length cDNA of EcTRIM13 was amplified using the primers listed in Table 1 . The putative open reading frame (ORF) and deduced amino acid sequence of EcTRIM13 was analyzed using BLAST program in NCBI database. Prediction of the functional domains and motifs was carried out using SMART program. Multiple sequences alignment of TRIM13s was performed using ClustalX1.83 software and edited using GeneDoc program. The phylogenetic tree based on the amino acid sequence was constructed using Mega 4.0 software.
Table 1.
Primers used in this study.
| Name | Sequence (5′–3′) |
|---|---|
| EcTRIM13-ORF-F | ATGGAGCAGCTAGAAGAGGAACT |
| EcTRIM13-ORF-R | CAGCTTACAGCTGCCAATAAAAC |
| EcTRIM13-Flag-F | TAAGGTACCGAATGGAGCAGCTAGAAGAGGAACT |
| EcTRIM13-Flag-R | TACTCTCGAGCAGCTTACAGCTGCCAATAAAAC |
| EcTRIM13-Flag-ΔR-F | TAAGGTACCGAATGCGCAAAGAGAGCCCTCACA |
| C1-EcTRIM13-F | TAACTCGAGCTATGGAGCAGCTAGAAGAGGAACT |
| C1-EcTRIM13-R | AATGGATCCCAGCTTACAGCTGCCAATAAAAC |
| C1-EcTRIM13-ΔR-F | TAACTCGAGCTATGCGCAAAGAGAGCCCTCACA |
| EcTRIM13-RT-F | TCTTGTGCGTAATTTCGATGTGA |
| EcTRIM13-RT-R | GCACGTAGGCTGCAAGGTTGT |
| Actin- RT-F | TACGAGCTGCCTGACGGACA |
| Actin- RT-R | GGCTGTGATCTCCTTCTGCA |
| SGIV MCP- RT-F | GCACGCTTCTCTCACCTTCA |
| SGIV MCP- RT-R | AACGGCAACGGGAGCACTA |
| SGIV ICP-18-RT-F | ATCGGATCTACGTGGTTGG |
| SGIV ICP-18-RT-R | CCGTCGTCGGTGTCTATTC |
| RGNNV RdRp-RT-F | GTGTCCGGAGAGGTTAAGGATG |
| RGNNV RdRp-RT-R | CTTGAATTGATCAACGGTGAACA |
| RGNNV CP-RT-F | CAACTGACAACGATCACACCTTC |
| RGNNV CP-RT-R | CAATCGAACACTCCAGCGACA |
| EcIRF3-RT-F | GACAACAAGAACGACCCTGCTAA |
| EcIRF3-RT-R | GGGAGTCCGCTTGAAGATAGACA |
| EcIRF7-RT-F | CAACACCGGATACAACCAAG |
| EcIRF7-RT-R | GTTCTCAACTGCTACATAGGGC |
| EcTNFα-RT-F | GTGTCCTGCTGTTTGCTTGGTA |
| EcTNFα-RT-R | CAGTGTCCGACTTGATTAGTGCTT |
| EcIL-1β-RT--PF | AACCTCATCATCGCCACACA |
| EcIL-1β-RT-PR | AGTTGCCTCACAACCGAACAC |
| EcIL-8-RT-PF | GCCGTCAGTGAAGGGAGTCTAG |
| EcIL-8-RT-PR | ATCGCAGTGGGAGTTTGCA |
| EcMDA5-RT-PF | ACCTGGCTCTCAGAATTACGAACA |
| EcMDA5-RT-PR | TCTGCTCCTGGTGGTATTCGTTC |
| EcLGP2-RT-F | TGGTGGTACGCTATGGACTGC |
| EcLGP2-RT-R | TTGTAGCTCAGTTATCTTTGTGCGA |
| EcMXI-RT-F | CGAAAGTACCGTGGACGAGAA |
| EcMXI-RT-R | TGTTTGATCTGCTCCTTGACCAT |
| EcISG15-RT-F | CCTATGACATCAAAGCTGACGAGAC |
| EcISG15-RT-R | GTGCTGTTGGCAGTGACGTTGTAGT |
To determine the function of EcTRIM13 in vitro, the wile type of EcTRIM13 and its RING mutant (EcTRIM13-ΔR) were subcloned into pEGFP-N3 and pcDNA3.1-flag vector as described previously. All the primers were listed in Table 1, and the recombinant plasmids (pEGFP-EcTRIM13, pEGFP-EcTRIM13-ΔR, pcDNA-EcTRIM13 and pcDNA-EcTRIM13-ΔR) were all confirmed by DNA sequencing.
2.3. Expression profiles for EcTRIM13 in grouper
To elucidate the tissue distribution pattern of EcTRIM13 in orange-spotted grouper, total RNA was extracted from 11 tissues of 3 healthy groupers, including head kidney, heart, liver, spleen, intestine, muscle, brain, skin, gill, stomach and kidney. The relative expression level of EcTRIM13 in different tissues was detected by quantitative real-time PCR (qRT-PCR) as described in the following. To illustrate the expression changes of EcTRIM13 in response to different stimuli, groupers were injected with PBS, SGIV, poly I:C as described previously [20]. Briefly, poly I:C treated groupers were collected at 0, 3, 6, 12, 24, 48 h post injection, and SGIV treated groupers were collected at 0, 3,12, 24,48, 72 h post injection. At different time points, the spleen of different groups (n > 3) of challenged grouper were collected for RNA extraction and qRT-PCR analysis.
2.4. Cell transfection and reporter gene assay
Cell transfection was performed using Lipofectamine 2000 reagent (Invitrogen) as described previously [17]. Briefly, GS cells were grown in 24-well plates, and then incubated with the mixture of Lipofectamine 2000 and different plasmids for 6 h. Then the fresh medium was added and cells were cultured for further study.
To uncover the effects of EcTRIM13 on the interferon promoter activity, luciferase activity assays were performed as described previously [21]. In brief, GS cells were cultured in 24-well plates, and then cotransfected with 0.1 μg ISRE-Luc/IFN-Luc, and 0.4 μg EcMDA5/EcIRF3/EcIRF7 or 0.4 μg EcTRIM13. The plasmid pRL-TK was transfected to normalize the transfection efficiency. At 48 h post-transfection, cells were harvested and Luciferase activities were examined using a Victor X5 Multilabel plate reader (PerkinElmer).
2.5. Fluorescent microscopy
To explore the subcellular distribution of EcTRIM13 in grouper cells, the plasmids pEGFP-N3, pEGFP-EcTRIM13 or pEGFP-EcTRIM13-ΔR were transfected into GS cells as described above. At 48 h post transfection, cells were fixed using 4% paraformaldehyde. Then cells were stained with 4,6-diamidino-2-phenylindole (DAPI), and observed under fluorescence microscopy.
2.6. qRT-PCR analysis
To clarify whether EcTRIM13 could regulate the viral replication during fish virus, GS cells overexpressing pcDNA-EcTRIM13, pcDNA-EcTRIM13-ΔR and vector (pcDNA3.1-Flag) were incubated with RGNNV for 48 h. Then cells were collected for RNA extraction and qRT-PCR analysis.
Using the SYBR Green realtime PCR Kit (Toyobo), we carried out qRT-PCR in a Roche 480 Real Time Detection System (Roche, German). In brief, each assay was performed in triplicate using the cycling condition as follows: 94 °C for 5 min, followed by 45 cycles of 5 s at 94 °C, 10 s at 60 °C and 15 s at 72 °C. The relative expression levels of virus genes including RGNNV CP (coat protein) and RdRp (RNA-dependent RNA polymerase) gene were analyzed as described previously [17]. The relative expression level of EcTRIM13 in different grouper tissues or host immune related genes in infected cells were also examined using qRT-PCR. All the experiments were carried out in triplicate, and the data shown are mean ± S.D. from one representative experiment. The data were calculated as the folds based on the expression level of targeted genes normalized to β-actin at the indicated time points, and the statistical significances were determined with Student’s t-test. The significance level was defined as p < 0.05 (*).
3. Results
3.1. Sequence characterization of EcTRIM13
After assembly of several EST sequences from grouper spleen transcriptome, we obtained the full length cDNA of EcTRIM13 in this study. The full-length EcTRIM13 cDNA encoded a polypeptide of 399 amino acids which shared 81% identity with TRIM13 homolog from large yellow croaker. Amino acid alignment analysis indicated that EcTRIM13 contained conserved functional domains, including RING finger and B-box domain (Fig. 1 ). Phylogenetic analysis indicated that EcTRIM13 showed the nearest relationship to that of large yellow croaker. All the TRIM13s from different fish species were clustered into one group which was separated from other groups, including amphibian, bird and mammals (Fig. 2 ).
Fig. 1.

Amino acid alignment of TRIM13s from different species. The RING and B-BOX domain were indicated above the sequences. The accession numbers were listed as follows: EcTRIM13, KX258196; Larimichthys crocea, XP_010744461; Danio rerio, NP_001018439; Gallus gallus, XP_015131811; Rattus norvegicus, NP_001012210; Homo sapiens, NP_998755.
Fig. 2.
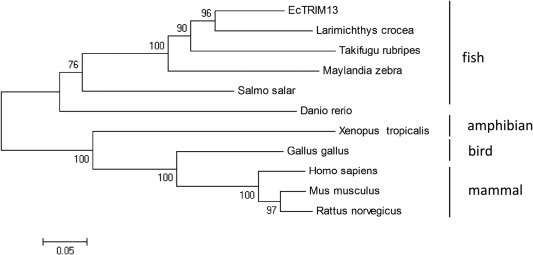
Phylogenetic analysis of EcTRIM13. A neighbor-joining tree was constructed based on the protein sequences of TRIM13-like genes from different species using MEGA 4.0 software. The bootstrap values are indicated at the branch points. Except the accession numbers which were listed in the legends of Fig. 1, the others were listed as follows: Salmo salar, NP_001133371; Maylandia zebra, XP_004551302; Takifugu rubripes, XP_011606878; Xenopus tropicalis, XP_004912024; Mus musculus, NP_075722.
3.2. Tissue distribution and expression profiles of EcTRIM13
To detect the tissue distribution profiles of EcTRIM13 in healthy grouper, the mRNA expression level of EcTRIM13 in different tissues was examined by qRT-PCR. As shown in Fig. 3 A, the predominant expression of EcTRIM13 was found in the liver, spleen, kidney, gill, and intestine.
Fig. 3.

The expression profiles of EcTRIM13 in healthy and challenged grouper. (A) The relative mRNA level of EcTRIM13 in different tissues from healthy groupers. After injection with poly I:C (B), and SGIV (C), the expression levels of EcTRIM13 in grouper spleen were detected using qRT-PCR.
To determine the expression level of EcTRIM13 in response to different stimuli, the transcript of EcTRIM13 in grouper spleen was detected after injection with poly I:C and SGIV using qRT-PCR. Upon challenge with poly I:C, the transcript of EcTRIM13 was decreased at 6 and 12 h, but increased at 24 h post injection (Fig. 3B). Differently, the expression level of EcTRIM13 was only increased at 48 h post-injection (Fig. 3C). Thus, EcTRIM13 was proposed to exert important roles in response to fish virus infection.
3.3. EcTRIM13 encoded a cytoplasmic protein
To determine the subcellular localization of EcTRIM13 in vitro, pEGFP-N3, pEGFP-EcTRIM13 or pEGFP-EcTRIM13-ΔR plasmids was transfected into GS cells and then stained with DAPI at 48 h post transfection. As show in Fig. 4 , the green fluorescence in pEGFP-EcTRIM13 transfected cells showed that EcTRIM13 mostly localized in the cytoplasm of GS cells, and tubular structure were observed in transfected cells. In contrast, the deletion of RING domain displayed the different localization pattern, and only the bright fluorescence punctuates were observed in pEGFP-EcTRIM13-ΔR transfected cells. Differently, in pEGFP-N3 transfected cells, the green fluorescence was distributed throughout the cells. Thus, RING domain was proposed to be essential for the accurate localization of EcTRIM13 in vitro.
Fig. 4.

Subcellular localization of EcTRIM13 in grouper cells. GS cells were transfected with pEGFP-N3, pEGFP-EcTRIM13 or pEGFP-EcTRIM13-ΔR plasmid, and then stained with DAPI. Samples were observed under fluorescence microscopy.
3.4. Ectopic expression of EcTRIM13 in vitro enhanced RGNNV replication
To demonstrate the effects of EcTRIM13 on fish virus replication, we detected the cytopathic effect (CPE) progression and viral gene transcription of RGNNV in infected EcTRIM13 overexpressing cells. As shown in Fig. 5 A, in EcTRIM13 overexpressing cells, the numbers of the vacuoles evoked by RGNNV infection were obviously increased compared to the control cells. In contrast, the vacuoles in EcTRIM13-ΔR overexpressing cells were similar to control cells. Further studies showed that the transcription levels of RGNNV CP and RdRp were significantly increased in EcTRIM13 overexpressing cells compared to control cells. Moreover, the deletion of RING domain reduced the enhancing effect of EcTRIM13 on RGNNV gene transcriptions (Fig. 5B). Thus, the ectopic expression of EcTRIM13 in vitro increased RGNNV replication, and the enhancing effect was dependent on its RING domain.
Fig. 5.
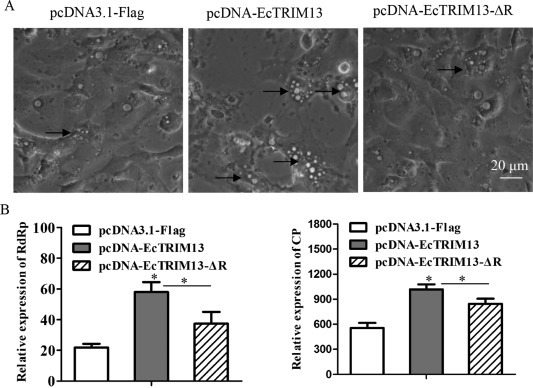
The effects of EcTRIM13 overexpression on virus replication. (A) EcTRIM13 overexpression increased the CPE progression of RGNNV infection. The arrows showed the vacuoles evoked by RGNNV. (B) The effect of EcTRIM13 overexpression on the gene transcription of RGNNV. GS cells overexpressing pcDNA-EcTRIM13, pcDNA-EcTRIM13-ΔR or empty vector were infected with RGNNV for different time intervals. Virus infected cells were collected and the transcription of viral genes, including RGNNV CP and RdRp were evaluated by qRT-PCR analysis. *, p < 0.05.
3.5. Overexpression of EcTRIM13 negatively regulated interferon signaling
To elucidate the effect of EcTRIM13 on interferon signaling pathway, we firstly evaluated the effect of EcTRIM13 overexpression on IFN and ISRE promoter activities using reporter gene assay. As shown in Fig. 6 , overexpression of IRF3, IRF7 and MDA5 all significantly increased the IFN and IFN-stimulated response elements (ISRE) promoter activities in vitro. Cotransfection with EcTRIM13 significantly decreased IRF3, IRF7 and MDA5 induced the activation of IFN and ISRE promoters. While, inhibitory effect on the IFN and ISRE promoter activation was weakened by the RING deletion mutant of EcTRIM13. In addition, we also examined the effects of EcTRIM13 overexpression on the expression of interferon related genes in vitro. Our results showed that the ectopic expression of EcTRIM13 significantly inhibited the expression of IRF3, IRF7, laboratory of genetics and physiology 2 (LGP2), MDA5, MXI and interferon stimulated gene (ISG) 15. In contrast, the inhibitory effects were significantly decreased in the RING mutant co-transfected cells (Fig. 7 ). Thus, we proposed that EcTRIM13 was able to negatively regulate the IFN signaling, and this action was dependent on its RING domain.
Fig. 6.
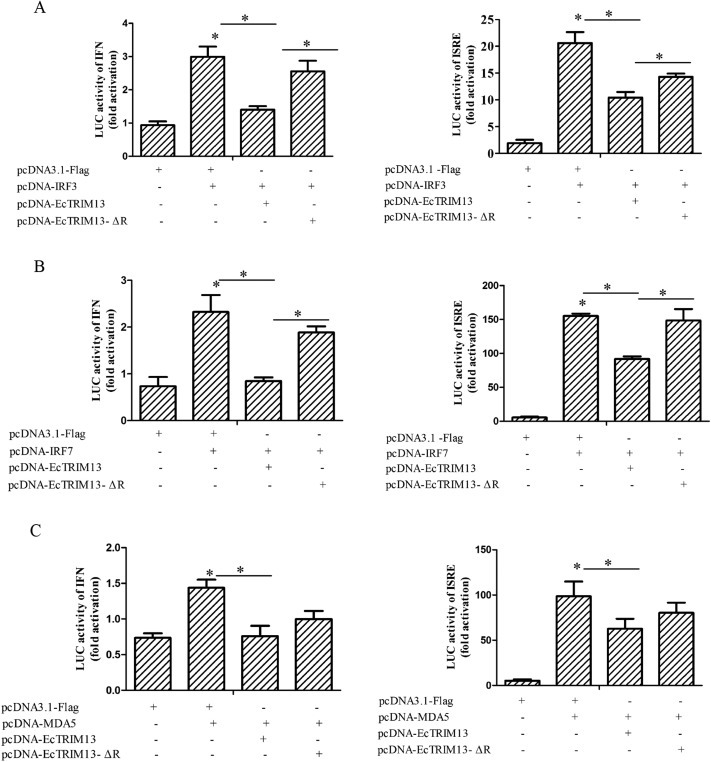
Overexpression of EcTRIM13 negatively regulated interferon immune response. IFN and ISRE promoter activity induced by IRF3 (A), IRF7 (B), and MDA5 (C) were detected using were reporter gene assay. GS cells were co-transfected with IFN-Luc/ISRE-Luc, IRF3/IRF7/MDA5, and EcTRIM13/EcTRIM13-ΔR. The luciferase activity was determined using reporter gene assay.
Fig. 7.
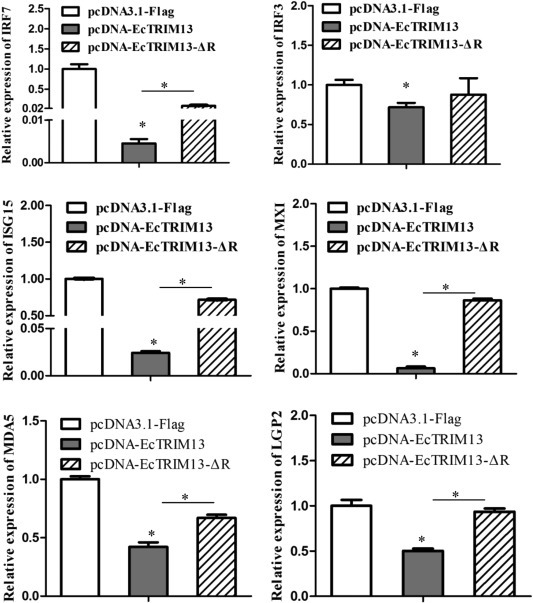
Overexpression of EcTRIM13 regulated the expression levels of interferon related signaling molecules. The relative expression levels of IRF3, IRF7, MDA5, LGP2, MXI and ISG15 in pcDNA-EcTRIM13, pcDNA-EcTRIM13-ΔR or pcDNA3.1-Flag transfected cells were determined by qRT-PCR. The Y-axis represented the fold change of target gene expression in each group relative to that in control group (control vector cells).
3.6. Overexpression of EcTRIM13 differently regulated the transcription of proinflammatory factors
Our previous studies showed that grouper TRIM8 was able to regulate the mRNA expression level of proinflammatory factors [17]. To determine whether EcTRIM13 exerted similar functions in vitro, we detected the changes in the expression level of three cytokines in EcTRIM13 overexpressing cells. As shown in Fig. 8 , the overexpression of EcTRIM13 in GS cells differently regulated the transcripts of these cytokines, including TNFα, IL-6 and IL-1β. In detail, the expression of IL-1β was significantly decreased, but that of IL-8 was significantly increased in EcTRIM13 overexpressing cells compared to the control cells. Differently, the expression of TNFα was moderately reduced compared to the control cells. Consistently, the regulatory effects of EcTRIM13 on the expression of these cytokines were all affected by its RING domain.
Fig. 8.

Overexpression of EcTRIM13 regulated the expression levels of proinflammatory cytokines. The relative expression levels of TNFα, IL-1β and IL-8 in pcDNA-EcTRIM13, pcDNA-EcTRIM13-ΔR or pcDNA3.1-Flag transfected cells were examined by qRT-PCR. The Y-axis represented the fold change of target gene expression in each group relative to that in control group (control vector cells).
4. Discussion
Increased studies have contributed to the notion on the TRIM family proteins due to its important immune regulatory roles of innate immunity [5], [22]. TRIM13, a member of the TRIM family, has been uncovered to play critical roles in different cellular processes including cell death and cancer [10], [11]. However, its roles in response to virus infection and replication still remained largely unknown. Here, we cloned a TRIM13 homolog from fish and characterized its function in iridovirus and nodavirus infection.
Like human TRIMs, fish TRIM proteins also displayed elements of a conserved modular tripartite motif structure consisting of RING domain, one or two zinc binding motifs named B-box and a predicted coiled coil (CC) domain [7]. In our study, EcTRIM13 contained both RING and B-box domain. Sequence analysis indicated that EcTRIM13 shared 81% identity with that from large yellow croaker. Expression patterns analysis indicated that the transcript of EcTRIM13 in grouper spleen was differently regulated after injection with SGIV or poly I:C, suggested that EcTRIM13 might be involved in innate immune response against fish virus infection, like other fish TRIMs [18]. In addition, subcellular localization analysis indicated that EcTRIM13 localized in the cytoplasm of grouper cells and displayed tubular structure. Like EcTRIM39, the deletion of RING significantly affected the accurate intracellular localization of EcTRIM13.
Research efforts so far demonstrated that a substantial number of the TRIM family members have emerged as viral restriction factors or as regulators of pathways in response to virus infection [1], [4]. The ectopic expression of TRIM proteins, including TRIM11, TRIM5, TRIM56, TRIM22, TRIM38 and TRIM68 were able to negatively or positively regulate virus replication in vitro [5], [7], [23], [24], [25], [26], [27], [28], [29]. TRIM56 was found to function as an antiviral host factor that confers resistance to bovine viral diarrhea virus (BVDV), yellow fever virus (YFV), dengue virus serotype 2 (DENV2), and human coronavirus virus (HCoV) OC43 through overlapping and distinct molecular determinants. Moreover, the N-terminal RING domain were always required for the antiviral activity of EcTRIM56 [26], [27]. In contrast, overexpression of TRIM38 or TRIM11 in vitro resulted in enhanced replication of VSV [28], [29]. In our study, the ectopic expression of EcTRIM13 significantly enhanced RGNNV replication, evidenced by the increase of CPE progression and viral gene transcription. Moreover, the deletion of RING domain significantly weakened the enhancing effect of EcTRIM13 on virus replication. The negative regulation of antiviral function was also explored in human TRIM13 after challenge with EMCV [12]. Thus, we speculated that TRIM13 exerted conserved functions in response to RNA virus during the evolution from fish to mammals.
Based on the regulatory effects of TRIM proteins on virus replication, further studies explored the multiple molecular mechanisms underlying the interaction between TRIM proteins and other signaling molecules or their downstream events [6], [27], [29], [30]. Ectopic expression of TRIM11 decreased IFNβ promoter activity induced by the overexpression of RIG-I, mitochondrial antiviral signaling protein (MAVS), or TANK-binding kinase-1 (TBK1). Moreover, TRIM11 overexpression inhibited the phosphorylation of IRF3 and mRNA expression of IFNβ [29]. In addition, TRIM13 was found to interact with both MDA5 and RIG-I in vitro, and negatively regulate MDA5-mediated type I IFN production [12]. In this study, our results indicated that overexpression of EcTRIM13 not only negatively regulated the interferon promoter activity induced by IRF3, IRF7 and MDA5, but also decreased the expression of interferon related genes, including IRF3, IRF7, LGP2, MXI, MDA5, and ISG15. In addition, the ectopic expression of EcTRIM13 differently regulated the expression of IL-6, IL-1β and TNFα. Thus, we proposed that EcTRIM13 not only functioned as a negative regulator of IFN response in the innate antiviral immunity, but also regulated the inflammation response against virus infection. Combined with our previous studies identifying IRF3, MDA5, and ISG15 as antiviral factors against RGNNV infection [20], [31], [32], we speculated that the negative regulation of interferon immune response by EcTRIM13 might contribute directly to the enhancing effect on RGNNV replication.
In summary, a fish TRIM13 homolog was cloned and characterized in this study. EcTRIM13 encoded a cytoplasmic protein, and negatively regulated the interferon immune response which might directly result in the enhancement of RGNNV replication in vitro. Furthermore, overexpression of EcTRIM13 differently regulated the transcription of pro-inflammatory factors. Notably, the deletion of RING domain significantly affected the molecular actions of EcTRIM13. Together, our results demonstrated firstly that fish TRIM13 exerts crucial roles in response to RNA virus infection.
Acknowledgment
This work was supported by grants from the National Natural Science Foundation of China (31172437, 31372566, 31472309), the National Basic Research Program of China (973) (2012CB114404; 2012CB114402), the National High Technology Development Program of China (863) (2014AA093507).
References
- 1.Rajsbaum R., García-Sastre A., Versteeg G.A. TRIMmunity: the roles of the TRIM E3-ubiquitin ligase family in innate antiviral immunity. J. Mol. Biol. 2014;426(6):1265–1284. doi: 10.1016/j.jmb.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hatakeyama S. TRIM proteins and cancer. Nat. Rev. Cancer. 2011;11(11):792–804. doi: 10.1038/nrc3139. [DOI] [PubMed] [Google Scholar]
- 3.Singh R., Patel V., Mureithi M.W., Naranbhai V., Ramsuran D., Tulsi S., Hiramen K., Werner L., Mlisana K., Altfeld M., Luban J., Kasprowicz V., Dheda K., Abdool Karim S.S., Ndung’u T. TRIM5α and TRIM22 are differentially regulated according to HIV-1 infection phase and compartment. J. Virol. 2014;88(8):4291–4303. doi: 10.1128/JVI.03603-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jefferies C., Wynne C., Higgs R. Antiviral TRIMs: friend or foe in autoimmune and autoinflammatory disease? Nat. Rev. Immunol. 2011;11(9):617–625. doi: 10.1038/nri3043. [DOI] [PubMed] [Google Scholar]
- 5.Uchil P.D., Hinz A., Siegel S., Coenen-Stass A., Pertel T., Luban J., Mothes W. TRIM protein-mediated regulation of inflammatory and innate immune signaling and its association with antiretroviral activity. J. Virol. 2013;87(1):257–272. doi: 10.1128/JVI.01804-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yan J., Li Q., Mao A.P., Hu M.M., Shu H.B. TRIM4 modulates type I interferon induction and cellular antiviral response by targeting RIG-I for K63-linked ubiquitination. J. Mol. Cell Biol. 2014;6(2):154–163. doi: 10.1093/jmcb/mju005. [DOI] [PubMed] [Google Scholar]
- 7.Uchil P.D., Quinlan B.D., Chan W.T., Luna J.M., Mothes W. TRIM E3 ligases interfere with early and late stages of the retroviral life cycle. PLoS Pathog. 2008;4(2):e16. doi: 10.1371/journal.ppat.0040016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reszka N., Zhou C., Song B., Sodroski J.G., Knipe D.M. Simian TRIM5alpha proteins reduce replication of herpes simplex virus. Virology. 2010;398(2):243–250. doi: 10.1016/j.virol.2009.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lerner M., Corcoran M., Cepeda D., Nielsen M.L., Zubarev R., Pontén F., Uhlén M., Hober S., Grandér D., Sangfelt O. The RBCC gene RFP2 (Leu5) encodes a novel transmembrane E3 ubiquitin ligase involved in ERAD. Mol. Biol. Cell. 2007;18(5):1670–1682. doi: 10.1091/mbc.E06-03-0248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tomar D., Prajapati P., Sripada L., Singh K., Singh R., Singh A.K., Singh R. TRIM13 regulates caspase-8 ubiquitination, translocation to autophagosomes and activation during ER stress induced cell death. Biochim. Biophys. Acta. 2013;1833(12):3134–3144. doi: 10.1016/j.bbamcr.2013.08.021. [DOI] [PubMed] [Google Scholar]
- 11.Tomar D., Singh R. TRIM13 regulates ubiquitination and turnover of NEMO to suppress TNF induced NF-κB activation. Cell Signal. 2014;26(12):2606–2613. doi: 10.1016/j.cellsig.2014.08.008. [DOI] [PubMed] [Google Scholar]
- 12.Narayan K., Waggoner L., Pham S.T., Hendricks G.L., Waggoner S.N., Conlon J., Wang J.P., Fitzgerald K.A., Kang J. TRIM13 is a negative regulator of MDA5-mediated type I interferon production. J. Virol. 2014;88(18):10748–10757. doi: 10.1128/JVI.02593-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hegde A., Chen C.L., Qin Q.W., Lam T.J., Sin Y.M. Characterization, pathogenicity and neutralization studies of a nervous necrosis virus isolated from grouper, Epinephelus tauvina, in Singapore. Aquaculture. 2002;213:55–72. [Google Scholar]
- 14.Kara H.M., Chaoui L., Derbal F., Zaidi R., de Boisséson C., Baud M., Bigarré L. Betanodavirus-associated mortalities of adult wild groupers Epinephelus marginatus (Lowe) and Epinephelus costae (Steindachner) in Algeria. J. Fish. Dis. 2014;37(3):273–278. doi: 10.1111/jfd.12020. [DOI] [PubMed] [Google Scholar]
- 15.Huang X., Huang Y., Xu L., Wei S., Ouyang Z., Feng J., Qin Q. Identification and characterization of a novel lymphocystis disease virus isolate from cultured grouper in China. J. Fish. Dis. 2015;38(4):379–387. doi: 10.1111/jfd.12244. [DOI] [PubMed] [Google Scholar]
- 16.Huang Y., Huang X., Yan Y., Cai J., Ouyang Z., Cui H., Wang P., Qin Q. Transcriptome analysis of orange-spotted grouper (Epinephelus coioides) spleen in response to Singapore grouper iridovirus. BMC Genomics. 2011;12:556. doi: 10.1186/1471-2164-12-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang Y., Yu Y., Yang Y., Yang M., Zhou L., Huang X., Qin Q. Fish TRIM8 exerts antiviral roles through regulation of the proinflammatory factors and interferon signaling. Fish. Shellfish Immunol. 2016 May 2;54:435–444. doi: 10.1016/j.fsi.2016.04.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang W., Huang Y., Yu Y., Yang Y., Xu M., Chen X., Ni S., Qin Q., Huang X. Fish TRIM39 regulates cell cycle progression and exerts its antiviral function against iridovirus and nodavirus. Fish. Shellfish Immunol. 2016;50:1–10. doi: 10.1016/j.fsi.2016.01.016. [DOI] [PubMed] [Google Scholar]
- 19.Huang X., Huang Y., Sun J., Han X., Qin Q. Characterization of two grouper Epinephelus akaara cell lines: application to studies of Singapore grouper iridovirus (SGIV) propagation and virus-host interaction. Aquaculture. 2009;292:172–179. [Google Scholar]
- 20.Huang X., Huang Y., Cai J., Wei S., Ouyang Z., Qin Q. Molecular cloning, expression and functional analysis of ISG15 in orange-spotted grouper, Epinephelus coioides. Fish. Shellfish Immunol. 2013;34(5):1094–1102. doi: 10.1016/j.fsi.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 21.Huang Y., Ouyang Z., Wang W., Yu Y., Li P., Zhou S., Wei S., Wei J., Huang X., Qin Q. Antiviral role of grouper STING against iridovirus infection. Fish. Shellfish Immunol. 2015;47(1):157–167. doi: 10.1016/j.fsi.2015.09.014. [DOI] [PubMed] [Google Scholar]
- 22.Lascano J., Uchil P.D., Mothes W., Luban J. TRIM5 retroviral restriction activity correlates with the ability to induce innate immune signaling. J. Virol. 2015;90(1):308–316. doi: 10.1128/JVI.02496-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wynne C., Lazzari E., Smith S., McCarthy E.M., Ní Gabhann J., Kallal L.E., Higgs R., Greco A., Cryan S.A., Biron C.A., Jefferies C.A. TRIM68 negatively regulates IFN-β production by degrading TRK fused gene, a novel driver of IFN-β downstream of anti-viral detection systems. PLoS One. 2014;9(7):e101503. doi: 10.1371/journal.pone.0101503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang K., Shi H.X., Liu X.Y., Shan Y.F., Wei B., Chen S., Wang C. TRIM21 is essential to sustain IFN regulatory factor 3 activation during antiviral response. J. Immunol. 2009;182(6):3782–3792. doi: 10.4049/jimmunol.0803126. [DOI] [PubMed] [Google Scholar]
- 25.Versteeg G.A., Rajsbaum R., Sánchez-Aparicio M.T., Maestre A.M., Valdiviezo J., Shi M., Inn K.S., Fernandez-Sesma A., Jung J., García-Sastre A. The E3-ligase TRIM family of proteins regulates signaling pathways triggered by innate immune pattern-recognition receptors. Immunity. 2013;38(2):384–398. doi: 10.1016/j.immuni.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu B., Li N.L., Wang J., Shi P.Y., Wang T., Miller M.A., Li K. Overlapping and distinct molecular determinants dictating the antiviral activities of TRIM56 against flaviviruses and coronavirus. J. Virol. 2014;88(23):13821–13835. doi: 10.1128/JVI.02505-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang J., Liu B., Wang N., Lee Y.M., Liu C., Li K. TRIM56 is a virus- and interferon-inducible E3 ubiquitin ligase that restricts pestivirus infection. J. Virol. 2011;85(8):3733–3745. doi: 10.1128/JVI.02546-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao W., Wang L., Zhang M., Wang P., Yuan C., Qi J., Meng H., Gao C. Tripartite motif-containing protein 38 negatively regulates TLR3/4- and RIG-I-mediated IFN-β production and antiviral response by targeting NAP1. J. Immunol. 2012;188(11):5311–5318. doi: 10.4049/jimmunol.1103506. [DOI] [PubMed] [Google Scholar]
- 29.Lee Y., Song B., Park C., Kwon K.S. TRIM11 negatively regulates IFNβ production and antiviral activity by targeting TBK1. PLoS One. 2013;8(5):e63255. doi: 10.1371/journal.pone.0063255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Versteeg G.A., Benke S., García-Sastre A., Rajsbaum R. InTRIMsic immunity: positive and negative regulation of immune signaling by tripartite motif proteins. Cytokine Growth Factor Rev. 2014 Oct;25(5):563–576. doi: 10.1016/j.cytogfr.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang Y., Huang X., Cai J., OuYang Z., Wei S., Wei J., Qin Q. Identification of orange-spotted grouper (Epinephelus coioides) interferon regulatory factor 3 involved in antiviral immune response against fish RNA virus. Fish. Shellfish Immunol. 2015;42(2):345–352. doi: 10.1016/j.fsi.2014.11.025. [DOI] [PubMed] [Google Scholar]
- 32.Huang Y., Yu Y., Yang Y., Yang M., Zhou L., Huang X., Qin Q. Antiviral function of grouper MDA5 against iridovirus and nodavirus. Fish. Shellfish Immunol. 2016;54:188–196. doi: 10.1016/j.fsi.2016.04.001. [DOI] [PubMed] [Google Scholar]


