Abstract
Recent developments in molecular diagnostic tools have led to the easy and rapid detection of a large number of rhinovirus (HRV) strains. However, the lack of clinical and epidemiological data hampers the interpretation of these diagnostic findings. From October 2009 to January 2011, we conducted a prospective study in hospitalized children from whom samples were taken for the detection of respiratory viruses. Clinical, epidemiological and microbiological data from 644 patients with 904 disease episodes were collected. When HRV tested positive, strains were further characterized by sequencing the VP4/VP2 region of the HRV genome. HRV was the single respiratory virus detected in 254 disease episodes (28%). Overall, 99 different serotypes were detected (47% HRV‐A, 12% HRV‐B, 39% HRV‐C). Patients with HRV had more underlying pulmonary illness compared with patients with no virus (p 0.01), or patients with another respiratory virus besides HRV (p 0.007). Furthermore, cough, shortness of breath and a need for oxygen were significantly more present in patients with HRV infection. Particularly, patients with HRV‐B required extra oxygen. No respiratory symptom, except for oxygen need, was predictive of the presence of HRV. In 22% of HRV‐positive disease episodes, HRV infection was hospital acquired. Phylogenetic analysis revealed several clusters of HRV; in more than 25% of these clusters epidemiological information was suggestive of transmission within specific wards. In conclusion, the detection of HRV may help in explaining respiratory illness, particular in patients with pulmonary co‐morbidities. Identifying HRV provides opportunities for timely implementation of infection control measures to prevent intra‐hospital transmission.
Keywords: Infection control, nosocomial transmission, respiratory infection, rhinovirus, sequence analysis
Introduction
In recent years, human rhinoviruses (HRVs) have been increasingly recognized as a potential cause of acute otitis media, bronchiolitis, asthma and pneumonia in children 1, 2, 3. The development of sensitive and rapid molecular techniques markedly improved the detection rate of HRV and revealed the high genetic diversity. Over 150 serotypes of HRV have been described, classified into three main species: HRV‐A, HRV‐B and HRV‐C 4. HRV‐C is the most recently discovered species and is thought to contribute more to recurrent wheezing and exacerbations of asthma compared with HRV‐A and HRV‐B 1, 5. Also, recently published data suggest that HRV‐A and HRV‐C cause more severe illness than HRV‐B, with the greatest virulence during the winter 6. Currently, molecular diagnostics are increasingly integrated into routine practice, allowing detection and quantification of HRV, and also raising questions about their value in direct patient care and infection control. We performed a hospital‐based prospective study to determine the clinical, epidemiological and viral characteristics associated with HRV infection in children.
Materials and Methods
Patients and sample collection
In October 2009 a prospective study of respiratory infections in hospitalized children was initiated at the University Medical Center Groningen (UMCG), the Netherlands. The UMCG is a tertiary referral hospital with more than 1300 beds in the northern region of the country. Demographic, clinical and microbiological data were systematically collected from all children under 18 years of age, from whom respiratory samples were taken for the detection of 15 respiratory viruses (influenza A/B, respiratory syncytial virus A/B, coronavirus 229E/NL63/OC43, para‐influenzavirus type 1–4, metapneumovirus, adenovirus, bocavirus and rhinovirus). Samples positive for HRV were further characterized by sequence analysis of the VP4/VP2 region. Information on the presence of bacterial respiratory pathogens was included when bacteriological culture was performed on the same day, 1 day before or 1 day after the virological sample was taken.
Clinical data were collected using a standardized case record form, with items regarding the presence of an underlying chronic illness (pulmonary, cardiovascular, gastrointestinal or neurological) and/or immune suppression (transplantation, malignancy or immune suppressive therapy), clinical symptoms (fever, cough, shortness of breath, otitis media, wheezing, vomiting, diarrhoea, need for oxygen or mechanical ventilation), treatment (antibiotics, antivirals or inhalation therapy), clinical diagnosis (upper respiratory tract infection (pharyngitis, coryza or otitis media) or lower respiratory tract infection (pneumonia, bronchiolitis, exacerbation of asthma or croup)) and outcome. Underlying pulmonary illness included asthma, congenital pulmonary illness or anatomic malformations, cystic fibrosis and bronchopulmonary disease. Cardiovascular disease was divided into inborn or acquired heart disease. Patients with partial resection of the bowel or failure to thrive, and those who were waiting for a liver transplantation, were categorized as having gastrointestinal disease as an underlying illness. Neurological disease was not further specified.
Epidemiological data were gathered to determine whether the respiratory infection was community or hospital acquired, including measures that were taken in the hospital to prevent further transmission of respiratory viruses. Hospital‐acquired HRV infection was defined as a first day of illness 2 or more days after admission to the hospital. Infection control measures consisted of a combination of droplet and contact precautions (gown, gloves and mask for healthcare workers during patient care, and patient in a single room) and were installed when HRV was detected.
The study was approved by the local Medical Ethical Committee of the UMCG. Informed consent was obtained from a parent or guardian.
Real‐time PCR and sequencing
The majority of samples (91%) arrived at the laboratory within 1 day after collection. Samples were divided into aliquots and stored at 4°C if PCR testing was performed on the same or the next day. Longer storage was carried out at −80°C. In general, PCR testing was performed on a daily basis, providing results within 48 h after arrival of the sample at the laboratory.
All respiratory samples, nasopharyngeal swabs, aspirates or sputum, were tested by a laboratory‐developed (LDT) real‐time PCR, as has been described elsewhere 7. For rhinovirus detection, a real‐time LDT‐PCR was introduced using the SuperScript® III Platinum® One‐Step qRT‐PCR Kit, (Life technologies, Carlsbad, CA, USA). All reactions were performed with Phocine Distemper Virus as an internal control in a total volume of 25 μL containing 12.5 μL of 2× reactionmix, 0.5 μL SuperScript® III RT/Platinum® Taq mix, 0.5 μL of 1:10 Rox reference dye, 300 nm of each forward primer, 600 nm reverse primer and 100 nm of each probe, and 5 μL of genomic RNA template. Primers and probes used are listed in Table 1. The addition of this large set of primers and probes is performed in a specific reaction to avoid bias and favouring against other respiratory viruses. Our rhinovirus PCR has been optimized during recent years based on the available genetic information, in particular regarding species C. By re‐optimizing the assay with new primers and probes, together with the use of Invitrogen SuperScript enzymes, we detected retrospectively more HRV‐positive samples at a lower cycle threshold (Ct) value. To ensure that these HRVs were not enteroviruses, all samples were sequenced. We only detected HRV, eventually 99 serotypes, as well as enterovirus 68, which is genetically identical to HRV.
Table 1.
Primers and probes for HRV detection used in this study
| Primers/probes | Sequence (5′→ 3′) | Positiona | Tm |
|---|---|---|---|
| Rhino‐fwdB‐mod‐TM | GGTGTGAAGACTCGCATGTGC | 408–427 | 60.1 |
| Rhino‐fwdA‐mod‐TM | GGTGTGAAGAGCCCCGTGTG | 408–426 | 62.4 |
| Rhino‐fwd‐C‐TM | GGTGTGAAGAGCCNANTGYGCTC | 408–429 | 58.9 |
| Rhino‐fwd‐D‐TM | GGTGYGAAGANCCNANTGTGC | 408–427 | 58.9 |
| Rhino‐fwd‐E‐TM | GGTGTGAAGACYTGCATGTGC | 408–427 | 57.9 |
| Rhino‐fwd‐F‐TM | GGTGTGAAGAGYCNCGTGTGCT | 408–428 | 58.1 |
| Rhino‐rev3 | CCAAAGTAGTYGGTYCCRTCCC | 523–544 | 58.4 |
| Rhino‐Probe‐TM | TCCTCCGGCCCCTGAATGCG | 438–457 | 70.2 |
| Rhino‐Probe3 | TCCTCCGGCCCCTGAATGTGG | 438–458 | 69.1 |
Tm, melting temperature.
Primer positions are given according to the orientation of the primer; numbers are given according to an HRV‐A16 reference strain (GenBank no. L24917).
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
The Ct value (the number of amplification cycles needed for a PCR to become positive) was used as relative estimate for the amount of HRV present in the samples.
Characterization of HRV was carried out by amplification and sequencing of a 549 nucleotide fragment spanning the hypervariable part of the 5′NTR, the entire VP4 gene and the 5′ terminus of the VP2 gene as described before 8, 9. For phylogenetic analysis a fragment of 395–401 bp was aligned with Clustal W 2.0 and phylogenetic trees were constructed by the neighbour‐joining method using MEGA 4.0 with the maximum likelihood model and complete deletion for missing data. The HRV sequences derived from this work have been submitted to GenBank (accession numbers JQ042307‐JQ042680).
Data analysis
For data analysis, the first respiratory sample of each episode of infection was included. An episode of infection, or disease episode, was defined as a time period starting with a first day of illness and ending with discharge from the hospital or resolution of clinical symptoms, during which a respiratory sample was taken for viral diagnostics. Statistical analyses were performed using SPSS software version 20.0, (IBM, Armonk, NY, USA). For normally distributed continuous variables, parametric tests were used (Student t‐test). The distribution of categorical variables in comparison groups was analysed using the chi‐squared test. The association of HRV with chronic underlying illness and symptoms was determined using binary and multinomial logistic regression (with no infection, HRV mono‐infection, HRV mixed infection and other respiratory infection as the dependent variable). A two‐sided p‐value <0.05 was considered statistically significant.
Results
Patients' characteristics
From October 2009 till January 2011, 644 unique patients were included, with 904 disease episodes; 366 (57%) were male and 278 (43%) female. Characteristics of the patients with no virus detected (n = 242, with 341 disease episodes), those with only HRV (n = 162, 254 disease episodes) and those with another respiratory virus than HRV (n = 157, 195 disease episodes) are summarized in Table 2. In 83 patients (111 disease episodes), mixed infection of HRV with one or more other respiratory viruses was found, most frequently adenovirus (37 episodes), RSV‐A/B (35 episodes), bocavirus (20 episodes) and influenza A virus (11 episodes). These patients did not differ significantly from the ones with HRV mono‐infection (Table 2).
Table 2.
Characteristics of patients with no respiratory virus detected (PCR negative), with HRV mono‐ or mixed infection and with a respiratory virus other than HRV
| PCR negative, N = 242 (%) | HRV mono‐infection, N = 162 (%) | HRV mixed infection, N = 83 (%) | Other respiratory virus, N = 157 (%) | |
|---|---|---|---|---|
| Sex | ||||
| M | 135 (55.8) | 92 (56.8) | 51 (61.4) | 88 (56.1) |
| F | 107 (44.2) | 70 (43.2) | 32 (38.6) | 69 (43.9) |
| Age (months) | ||||
| Mean | 31.6 | 32.7 | 24.2 | 36.6 |
| Median | 1.0a | 11.0 | 13.0 | 13.0 |
| Underlying illness | 142 (58.7) | 108 (66.7)b | 45 (54.2) | 69 (43.9) |
| Pulmonary | 52 (21.5) | 54 (33.3)c | 19 (22.9) | 31 (19.7) |
| Cardiovascular | 50 (20.7) | 37 (22.8) | 12 (14.4) | 15 (9.6)d |
| Gastro‐intestinal | 33 (13.6) | 14 (8.6) | 8 (9.6) | 8 (5.1) |
| Neurology | 21 (8.7) | 15 (9.3) | 7 (8.4) | 13 (8.2) |
| Immune suppressione | 36 (14.9) | 36 (22.2) | 11 (13.3) | 14 (8.9) |
p <0.0001.
HRV mono‐infection vs. other virus: p <0.001.
HRV mono‐infection vs. PCR negative p 0.01, vs. other virus p 0.007.
Other virus vs. PCR negative p 0.003, other virus vs. HRV mono‐infection p 0.001.
Malignancy, transplantation, use of immune‐suppressive therapy.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Patients with HRV infection had more underlying pulmonary illness, mainly caused by asthma (39%) or congenital pulmonary illness/anatomical malformations (44%), compared with PCR‐negative patients (p 0.01) or patients with another respiratory virus than HRV (p 0.007).
Although the majority of patients were under the age of 5 years (80%), the prevalence of underlying illness was significantly higher in patients older than 5 years: in patients with HRV mono‐infection, 86% had co‐morbidities, compared with 64% in those under 5 (p 0.03); for PCR negatives, this was 83% vs. 56% (p 0.001); and in patients with another respiratory virus, 70% had a chronic underlying disease, compared with 38% in those under 5 (p 0.002). These differences in prevalence of chronic underlying disease were mainly caused by a significantly higher prevalence of chronic respiratory disease in the older age category (data not shown).
Clinical symptoms
To determine the relationship between HRV and clinical symptoms, PCR‐negative disease episodes were compared with those with an HRV mono‐infection. Because patients with HRV had significantly more chronic respiratory disease, we stratified disease episodes for this underlying condition. HRV‐positive patients who also had a chronic respiratory disease, experienced significantly more cough, shortness of breath and a need for oxygen compared with PCR negatives. However, in HRV‐positive patients without an underlying pulmonary illness, only cough and fever were significantly more present (Table 3).
Table 3.
Clinical symptoms in disease episodes of patients with HRV mono‐infection compared with PCR‐negative patients
| Symptoms | Pulmonary underlying illness | No pulmonary underlying illness | ||
|---|---|---|---|---|
| PCR negative, N = 83 episodes (%) | HRV mono‐infection, N = 98 episodes (%) | PCR negative, N = 238 episodes (%) | HRV mono‐infection, N = 139 episodes (%) | |
| Fever | 29 (34.9) | 48 (49.0) | 111 (46.6) | 82 (59.0)a |
| Cough | 13 (15.7) | 38 (38.8)b | 28 (11.8) | 35 (25.2)c |
| Dyspnoea | 18 (21.7) | 54 (55.1)d | 53 (22.3) | 34 (24.5) |
| Diarrhoea | 4 (4.8) | 12 (12.2) | 27 (11.3) | 24 (17.3) |
| Vomiting | 6 (7.2) | 12 (12.2) | 27 (11.3) | 19 (13.7) |
| Oxygen need | 31 (37.3) | 62 (63.3)b | 57 (23.9) | 29 (20.9) |
| Mechanical ventilation | 34 (41.0) | 30 (30.6) | 91 (38.2) | 40 (28.8) |
p 0.02 (HRV positives vs. PCR negatives in patients with no underlying pulmonary illness).
p 0.001 (HRV positives vs. PCR negatives in patients with underlying pulmonary illness).
p 0.001(HRV positives vs. PCR negatives in patients with no underlying pulmonary illness).
p <0.001 (HRV positives vs. PCR negatives in patients with underlying pulmonary illness).
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
In multivariate analysis, the presence of a chronic respiratory disease remained positively associated with HRV mono‐infection (OR, 2.06; CI, 1.43–2.97). This association was unique for HRV mono‐infections and was not found for HRV mixed infections or other respiratory viral infections. Besides, in patients with a chronic respiratory illness a need for oxygen was exclusively related to HRV detection (odds ratio 3 for HRV mono‐infection, and 3.3 for mixed infection). Cough and shortness of breath were equally strongly associated with the detection of other respiratory viruses as with HRV (Table 4).
Table 4.
Association of clinical symptoms with the detection of HRV and other respiratory viruses in patients with chronic respiratory disease
| Symptoms | HRV mono‐infection (OR, 95% CI) | HRV co‐detection (OR, 95% CI) | Other respiratory virus (OR, 95% CI) |
|---|---|---|---|
| Fever | 1.37 (0.69–2.71) | 2.62 (1.02–6.70) | 4.19 (1.68–10.42) |
| Cough | 2.52 (1.17–5.4) | 2.7 (1.03–7.16) | 3.77 (1.53–9.28) |
| Dyspnoea | 4.12 (2.10–8.09) | 6.74 (2.68–16.94) | 6.13 (2.60–14.44) |
| Diarrhea | 2.04 (0.55–7.57) | 1.43 (0.28–7.25) | 1.36 (0.31–5.94) |
| Vomiting | 1.33 (0.43–4.14) | 1.60 (0.39–6.58) | 2.50 (0.72–8.72) |
| Need for oxygen | 3.02 (1.63–5.58) | 3.29 (1.38–7.84) | 1.39 (0.66–2.92) |
| Mechanical ventilation | 0.63 (0.33–1.19) | 0.19 (0.06–0.61) | 0.46 (0.20–1.05) |
OR, odds ratio; CI, 95% confidence interval.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Patients with an HRV infection were more often diagnosed with an exacerbation of asthma compared with PCR negatives and patients with another respiratory virus, although the difference with the latter group was not statistically significant (Table 5). In contrast, pneumonia and bronchiolitis were more often associated with the detection of respiratory viruses other than HRV.
Table 5.
Clinical diagnosis in patients with HRV compared with patients who were PCR negative or who had another respiratory virus (% of episodes)
| Clinical diagnosis | PCR negative (%) | HRV infection (%) | Other respiratory virus (%) |
|---|---|---|---|
| Upper respiratory tract infection | 13.5 | 52.8a | 49.7a |
| Otitis media | 0.9 | 1.2 | 3.6 |
| Exacerbation asthma | 0.6 | 5.1b | 1.0 |
| Bronchiolitis | 0.9 | 2.0 | 15.4c |
| Pneumonia | 10.9 | 12.2 | 18.5d |
p <0.001 compared with PCR negative.
p 0.008 compared with PCR negative, p 0.12 compared with other respiratory virus.
p <0.001 compared with PCR negative and HRV positive.
p 0.02 compared with PCR negative, p 0.04 compared with HRV positive.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
The median length of stay in the hospital was significantly shorter for patients who had any respiratory virus compared with patients with no virus (7 days vs. 25 days, p <0.001). Also, patients with HRV had to stay longer in the hospital compared with patients with another respiratory virus (median length of stay 11 days vs. 6 days, p <0.001).
The median Ct value of HRV in HRV mono‐infection was 24, significantly lower than the median Ct value of HRV in HRV mixed infections (28, p <0.001). However, we found no relationship between the presence of symptoms and the relative amount of virus in the samples.
Bacteriology and antibiotic use
Antibiotic use did not differ between the three patient groups (in 52% of PCR negatives, 50% of those with HRV and 53% with another respiratory virus, antibiotics were given). Also, the duration of antibiotic therapy was comparable (median duration of 7 days). In more than 60% of the disease episodes, no bacteriological culture was recorded around the time the virological sample was taken. When bacteriological cultures were taken, significantly more H. influenzae was found in patients with HRV (17.4%, p 0.04) and in patients with another respiratory virus (19.4%, p 0.02) compared with the patients who had no respiratory virus detected (3.6%). S. pneumoniae was more often found in patients with another respiratory virus (19.4%) than in patients with HRV (2.2%, p 0.02). The use of antibiotics was strongly associated with the clinical diagnosis of pneumonia and the submission of samples for bacteriological culture: antibiotics were prescribed in 94% of the patients with pneumonia and in 75% of patients who had samples collected for bacteriological culture (both p <0.001). However, the outcome of the bacteriological culture (positive or negative) did not influence the use of antibiotics (p 0.5), nor did the presence of a chronic underlying illness (p 0.6).
HRV characterization
HRV was found throughout the study period, with the least frequent detection during February, July and August 2010 and peaks in spring (2010) and autumn (2009 and 2010) (Fig. 1). However, during the winter period (December and January) HRV was detected in 40–60% of the samples positive for respiratory viruses.
Figure 1.

Monthly distribution of the number of respiratory specimens taken in our patient population, the number of HRV detections (either as mono‐infection or mixed infection) and the number of detected respiratory viruses other than HRV (non‐HRV positive).
Sequence analysis of HRV was possible in 303 out of 365 HRV‐positive disease episodes; 221 episodes with HRV mono‐infection and 82 with HRV mixed infection. The reason for unsuccessful sequence analysis was the low amount of HRV present in the samples.
Overall, 99 different serotypes were detected, most often species HRV‐A (47%) and HRV‐C (39%) (Fig. 2). In 12% of the episodes, HRVs were detected belonging to species HRV‐B. We did not observe a trend in circulating serotypes. Almost 50 patients were identified with serial infections with different serotypes or with different species. Three patients had detection of serotypes belonging to HRV‐A, HRV‐B and HRV‐C consecutively.
Figure 2.
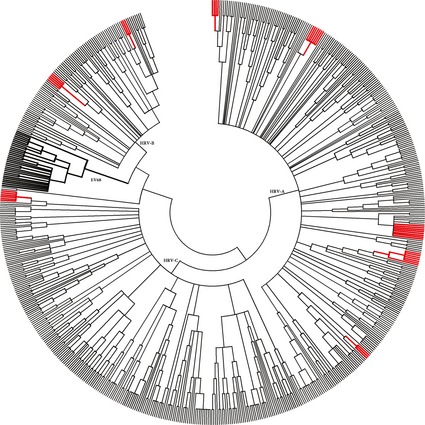
Phylogenetic tree of detected HRV serotypes in this study. In red are the clusters of infection based on epidemiological and virological information. In bold are the clusters of EV68, as described before 8.
In patients with HRV‐B, more underlying pulmonary illness was present (54%) compared with those with HRV‐A and HRV‐C (32% and 39%), although the difference was only statistically significant for the comparison of HRV‐B with HRV‐A (p 0.045). No significant differences were seen in clinical symptoms related to the species of HRV, except for the need for oxygen, which was more present in patients with HRV‐B (p 0.012). Patients with HRV‐C had a shorter duration of stay in the hospital than patients with HRV‐A (median 6 days vs. 10 days for HRV‐A, p 0.009) and HRV‐B (median 16 days, p 0.008).
Nosocomial infection
Based on epidemiological data (first day of illness in relation to admission to the hospital), hospital‐acquired infection was present in 81 out of 365 HRV‐positive episodes (22%), with a first day of illness ranging from 3 to 242 days after admission. In 69 of 81 episodes (85.2%) HRV was detected as a mono‐infection, compared with 179 out of 273 (66%) community‐acquired HRV infections (p <0.001). Even when the definition of hospital‐acquired infection was adjusted to a first day of illness 5 days or more after admission, still 20% of the HRV‐positive disease episodes were nosocomial related. No differences were observed in the age distribution in community and hospital‐acquired HRV infections. Although HRV‐B was the least frequently detected HRV species, equal proportions of HRV‐A and HRV‐B infection were hospital acquired (33.3% resp. 38.5%), while HRV‐C was in only 18.0% of the HRV‐positive episodes acquired during hospitalization. Due to the small number of HRV‐B infections, these differences did not reach statistical significance.
Using phylogenetic analysis, 41 clusters of three or more isolates with identical sequences could be identified during this study period. Nine of the 41 clusters were formed by three or more consecutively taken isolates from one patient per cluster. Two clusters of EV68 were identified, corresponding with an upsurge of EV68‐related respiratory tract infections detected in the autumn of 2010, as described previously 8. We collected more detailed information about the wards patients were admitted to, first day of illness, date of admission and sample date in the remaining 30 clusters, to see whether the clustering of patients based on the virological analysis was supported by epidemiological data. In eight clusters epidemiological information was suggestive of transmission of HRV on the same hospital ward (Fig. 2).
Discussion
In this large hospital‐based prospective cohort study into viral respiratory infections in children, HRV was the most commonly detected respiratory virus, in the majority as HRV mono‐infections. Also, high nosocomial infection rates were observed. HRV infection was associated with substantial respiratory illness, especially among children with pre‐existing pulmonary disease.
Although the association of HRV with asthma has been well established in recent years, the occurrence of HRV infections in our study was associated with chronic respiratory illness in general 5, 10, 11. Whether these children are more vulnerable to the acquisition of HRV, or whether they have more symptomatic illness related to HRV, remains to be elucidated. In patients with a chronic respiratory condition, only the need for oxygen was predictive for HRV; cough and shortness of breath were not only associated with the detection of HRV, but also with the detection of other respiratory viruses. Thus, respiratory symptoms alone are not sufficient to predict the presence of an HRV infection.
Several studies tried to relate viral load to the presence or absence of symptoms. Although a positive relationship between a higher viral load and more (serious) respiratory symptoms has been reported, others were not able to reproduce these findings 11, 12, 13, 14, 15. Adequate quantification of HRV in respiratory samples is currently limited by several factors, as addressed in recent reports 16, 17. One major limitation is the lack of a reference standard for the quantification of all HRV types. Secondly, HRV viral load is affected by the sample type and the method of collection and detection 18. Thus, although in our study no relationship between the relative amount of virus (expressed as Ct value) and the presence of clinical symptoms could be demonstrated, the interpretation of the Ct value in relation to severity of illness remains to be determined and is probably, with the current limitations in quantification of HRV, hardly feasible.
HRV was detected year round, with the least frequent detection in February, July and August 2010. Although peaks in detection were seen in autumn and spring, as is known from the literature, HRV was still detected in around half of the samples positive for respiratory viruses in the remaining months, including the winter period 6, 17, 19. However, as most studies suggest variation of HRV detection by season and year, our study period may well have been too short to fully understand the seasonality of HRV in our hospital.
Antibiotic use and duration of antibiotic therapy did not differ between the patients with and without a respiratory viral infection, suggesting that the decision to prescribe antibiotics was not influenced by the outcome of viral diagnostics, as has been observed elsewhere 20, 21. This might be explained by concerns over getting a bacterial co‐infection. Indeed, antibiotic use was associated with submission of samples for bacteriological culture, assuming a suspicion of bacterial infection in those patients. However, although bacteria known to cause (secondary) respiratory infections in infants were more frequently cultured in patients with a respiratory viral infection compared with patients with no viral infection (H. influenzae, S. pneumoniae), the outcome of the bacteriological cultures did not influence the use of antibiotics. Because the collection of samples for bacteriological culture was not standardized, but dependent on the judgement of the clinician, and carried out in only a minority of patients, these results might have been biased and more studies are needed to confirm our findings.
The high prevalence of co‐morbidities and the associated vulnerability of our patient population could have lowered the threshold for prescribing antibiotics. However, we did not find a positive association between antibiotic therapy and the presence of a chronic underlying illness. We did find a strong correlation between antibiotic therapy and pneumonia as the clinical diagnosis. This may suggest that clinical signs and symptoms were more important in deciding on antibiotic therapy than patient characteristics and microbiological results.
Sequence analysis revealed a high diversity of different HRV serotypes, as reported before 22, 23, 24. In our population, species HRV‐A and HRV‐C were detected in equal amounts, and dominated over HRV‐B. We observed no species‐specific pattern of clinical illness, except that HRV‐B was associated with a higher need for oxygen. Besides, patients with HRV‐A and HRV‐B had to stay in the hospital significantly longer than patients with HRV‐C. Until now, studies that addressed the relationship between species and severity of illness reported HRV‐B as being the least virulent compared with HRV‐A and HRV‐C 6, 17. However, recently, Miller et al. also showed that patients with HRV‐B were more likely to require supplemental oxygen and had a longer duration of stay in the hospital 19. Also, patients with HRV‐B tended to have higher disease severity scores. The reason for the different observations regarding HRV‐B‐associated severity of illness is not clear. Our study and the study of Miller et al. included hospitalized children, whereas in a recent study by Lee et al., who showed a clear distinction in severity of illness between HRV‐A/C and HRV‐B, the majority of patients were not admitted to the hospital 6. Also, we observed a tendency for more chronic respiratory illness in patients with an HRV‐B infection, possibly explaining the higher need for oxygen. These findings may suggest that HRV‐B is associated with either asymptomatic carriage or mild respiratory disease, and that host factors such as pre‐existing pulmonary disease contribute to more serious, symptomatic respiratory illness caused by HRV‐B.
One of the most remarkable findings of our study is the frequency of hospital‐acquired HRV infection: in more than 20% of the HRV‐positive disease episodes, patients had a first day of illness more than 2 days after admission to the hospital. Phylogenetic analysis revealed several clusters of HRV with identical sequences; in more than 25% of these clusters epidemiological information was suggestive of transmission of the virus on specific wards. Although HRV‐B is the least frequently found species in our study, almost 40% of HRV‐B infections are acquired in the hospital. This suggests that the hospital environment is for some reason more favourable for transmission of HRV‐B. Also, if HRV‐B is indeed associated with asymptomatic carriage or mild respiratory disease, this would facilitate transmission within the hospital, only causing clear respiratory symptoms in patients with pre‐existing pulmonary illness.
Outbreaks of rhinovirus infections have been described in long‐term care facilities, but very little is known about the frequency of hospital‐acquired HRV infection 25, 26. Transmission mainly occurs via droplets or contact and although measures to prevent transmission were taken the moment HRV was detected in respiratory samples, infectiousness is probably highest in the early stages of disease, before the outcome of diagnostic tests is available and thus before infection control measures were taken. These findings advocate the implementation of infection control measures driven by the presence of respiratory symptoms instead of the outcome of a laboratory test. However, further investigations are needed to determine the contribution of healthcare workers, other patients and visitors in nosocomial transmission of HRV.
The present study has some limitations. Being a tertiary referral hospital could have biased our findings towards more serious illness, as is reflected by the characteristics of the included patients: more than 50% of the children had a chronic underlying illness, and even in the ones that had no respiratory virus detected, signs and symptoms of serious illness were present (e.g. mechanical ventilation). Furthermore, information on bacterial co‐infection was based on cultures taken just around the time the viral sample was taken. Information on the presence of bacterial pathogens outside this timeframe was not included. Also, we had no information about the presence of so‐called atypical bacterial pathogens (e.g Mycoplasma or Chlamydophila). However, given the high prevalence of HRV infection in our study population and the expected low prevalence of atypical pathogens, we assume that this limitation does not influence the main conclusions of our study 20, 21.
In conclusion, HRVs are capable of causing serious respiratory disease in children hospitalized in a tertiary referral hospital, particularly in patients with pulmonary co‐morbidities. Nosocomial infection occurs frequently. Most notably, HRV species B may have features that facilitate transmission within the hospital. Although antiviral therapy is not yet available for patients infected with HRV, the detection and identification of these viruses could help in explaining respiratory illness. Also, identifying HRV provides opportunities for implementing timely and accurate infection control measures to prevent further transmission. Our study illustrates the value of sequence analysis not only in gaining insight into the genetic diversity of rhinoviruses, but also as a tool in defining transmission routes within the hospital and in the detection of sources of nosocomial infection.
Transparency Declaration
The authors declare no conflicts of interest.
Acknowledgements
We greatly acknowledge the contribution to this study of Monique van Linschoten, who collected all clinical data. We thank Renze Borger and Reina van der Heide for their work in optimizing the HRV PCR and performing phylogenetic analysis. Part of this work is presented at the annual congresses of the European Society of Clinical Virology and the European Society for Pediatric Infectious Diseases.
Clin Microbiol Infect 2013; 19: E435–E442
References
- 1. Arden KE, Mackay IM. Newly identified human rhinoviruses: molecular methods heat up the cold viruses. Rev Med Virol 2010; 20: 156–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Peltola V, Waris M, Osterback R, Susi P, Hyypia T, Ruuskanen O. Clinical effects of rhinovirus infections. J Clin Virol 2008; 43: 411–414. [DOI] [PubMed] [Google Scholar]
- 3. Ruuskanen O, Lahti E, Jennings LC, Murdoch DR. Viral pneumonia. Lancet 2011; 377: 1264–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Palmenberg AC, Spiro D, Kuzmickas R et al Sequencing and analyses of all known human rhinovirus genomes reveal structure and evolution. Science 2009; 324: 55–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gern JE. The ABCs of rhinoviruses, wheezing, and asthma. J Virol 2010; 84: 7418–7426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lee WM, Lemanske RF Jr, Evans MD et al Human rhinovirus species and season of infection determine illness severity. Am J Respir Crit Care Med 2012; 186: 886–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bosis S, Esposito S, Niesters HG et al Role of respiratory pathogens in infants hospitalized for a first episode of wheezing and their impact on recurrences. Clin Microbiol Infect 2008; 14: 677–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rahamat‐Langendoen J, Riezebos‐Brilman A, Borger R et al Upsurge of human enterovirus 68 infections in patients with severe respiratory tract infections. J Clin Virol 2011; 52: 103–106. [DOI] [PubMed] [Google Scholar]
- 9. Savolainen C, Blomqvist S, Mulders MN, Hovi T. Genetic clustering of all 102 human rhinovirus prototype strains: serotype 87 is close to human enterovirus 70. J Gen Virol 2002; 83(Pt 2): 333–340. [DOI] [PubMed] [Google Scholar]
- 10. Friedlander SL, Busse WW. The role of rhinovirus in asthma exacerbations. J Allergy Clin Immunol 2005; 116: 267–273. [DOI] [PubMed] [Google Scholar]
- 11. Message SD, Laza‐Stanca V, Mallia P et al Rhinovirus‐induced lower respiratory illness is increased in asthma and related to virus load and Th1/2 cytokine and IL‐10 production. Proc Natl Acad Sci USA 2008; 105: 13562–13567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Franz A, Adams O, Willems R et al Correlation of viral load of respiratory pathogens and co‐infections with disease severity in children hospitalized for lower respiratory tract infection. J Clin Virol 2010; 48: 239–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jansen RR, Schinkel J, Dek I et al Quantitation of respiratory viruses in relation to clinical course in children with acute respiratory tract infections. Pediatr Infect Dis J 2010; 29: 82–84. [DOI] [PubMed] [Google Scholar]
- 14. Utokaparch S, Marchant D, Gosselink JV et al The relationship between respiratory viral loads and diagnosis in children presenting to a pediatric hospital emergency department. Pediatr Infect Dis J 2011; 30: e18–e23. [DOI] [PubMed] [Google Scholar]
- 15. van Elden LJ, Sachs AP, van Loon AM et al Enhanced severity of virus associated lower respiratory tract disease in asthma patients may not be associated with delayed viral clearance and increased viral load in the upper respiratory tract. J Clin Virol 2008; 41: 116–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schibler M, Yerly S, Vieille G et al Critical analysis of rhinovirus RNA load quantification by real‐time reverse transcription‐PCR. J Clin Microbiol 2012; 50: 2868–2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jacobs SE, Lamson DM, St George K, Walsh TJ. Human rhinoviruses. Clin Microbiol Rev 2013; 26: 135–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Meerhoff TJ, Houben ML, Coenjaerts FE et al Detection of multiple respiratory pathogens during primary respiratory infection: nasal swab versus nasopharyngeal aspirate using real‐time polymerase chain reaction. Eur J Clin Microbiol Infect Dis 2010; 29: 365–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Miller EK, Williams JV, Gebretsadik T et al Host and viral factors associated with severity of human rhinovirus‐associated infant respiratory tract illness. J Allergy Clin Immunol 2011; 127: 883–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wishaupt JO, Russcher A, Smeets LC, Versteegh FG, Hartwig NG. Clinical impact of RT‐PCR for pediatric acute respiratory infections: a controlled clinical trial. Pediatrics 2011; 128: e1113–e1120. [DOI] [PubMed] [Google Scholar]
- 21. Huijskens EG, Biesmans RC, Buiting AG, Obihara CC, Rossen JW. Diagnostic value of respiratory virus detection in symptomatic children using real‐time PCR. Virol J 2012; 9: 276–422X‐9‐276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Huang T, Wang W, Bessaud M et al Evidence of recombination and genetic diversity in human rhinoviruses in children with acute respiratory infection. PLoS ONE 2009; 4: e6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Savolainen C, Mulders MN, Hovi T. Phylogenetic analysis of rhinovirus isolates collected during successive epidemic seasons. Virus Res 2002; 85: 41–46. [DOI] [PubMed] [Google Scholar]
- 24. Wisdom A, Kutkowska AE, McWilliam Leitch EC et al Genetics, recombination and clinical features of human rhinovirus species C (HRV‐C) infections; interactions of HRV‐C with other respiratory viruses. PLoS ONE 2009; 4: e8518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wenzel RP, Deal EC, Hendley JO. Hospital‐acquired viral respiratory illness on a pediatric ward. Pediatrics 1977; 60: 367–371. [PubMed] [Google Scholar]
- 26. Longtin J, Marchand‐Austin A, Winter AL et al Rhinovirus outbreaks in long‐term care facilities, Ontario, Canada. Emerg Infect Dis 2010; 16: 1463–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]


