Abstract
The SARS-nsp13 protein was identified as an mRNA cap1 methyltransferase. In this study, the nsp13 gene was cloned from the SARS-CoV PUMC02 strain viral RNA by RT-PCR, and inserted into the expression plasmid pET30a(+). The recombinant plasmid pET30a(+)-nsp13 was confirmed by restriction enzymes and sequencing analysis, and transformed into Escherichia coli BL21(DE3). The His-tag-fused protein was expressed by induction of 0.5 mM IPTG and purified by a single Ni2+ affinity chromatography. The protein was validated by western blot and MS analysis. A large quantity of the nsp13 protein obtained with this method may be useful for further study of its structure and function.
Keywords: Severe acute respiratory syndrome (SARS), nsp13 (SARS-CoV methyltransferase), Molecular cloning, Expression, Purification, Mass spectrometric identification
In the spring of 2003, a severe epidemic disease broke out in China and quickly spread to more than 30 countries. It led to over 8000 infected cases and as many as 774 deaths [1]. This disease was designated as severe acute respiratory syndrome or SARS by the World Health Organization (WHO) and the pathogen was identified as a novel coronavirus, named SARS-associated coronavirus (SARS-CoV) [2], [3], [4]. The SARS-CoV is a single-stranded plus-sense RNA virus with a genome of about 30 kb in length that has a 5′ cap structure and a 3′ polyadenylation tail. Upon infection of an appropriate host cell, the 5′-most open-reading frame (ORF) of the viral genome is translated into a large polyprotein that is cleaved by proteases to release several nonstructural proteins (nsps), including nsp2 (3C-like proteinase), nsp9 (RNA-dependent RNA polymerase, RdRp), nsp10 (NTPase/HEL), and so on [5], [6]. Among these nsps, the nsp13 protein has been predicted as an mRNA cap 1 methyltransferase by 3D jury system [7].
Methylated 5′-terminal cap structures have been described in most eukaryotic and many viral mRNAs. The methylated cap structure is essential for efficient initiation of translation and mRNA stability [8]. In all cap structures, including the “minimal” cap 0 (m7G(5′)ppp(5′)N), an N7-methylguanosine (m7G) is attached through a 5′–5′ triphosphate bridge to the penultimate nucleoside. In some molecules, additional 2′-O-ribose methylations are found at the penultimate and the antepenultimate nucleosides, forming the cap 1 (m7G(5′)ppp(5′)Nm) and cap 2 (m7G(5′)ppp(5′)NmpNm) structures, respectively [9], [10]. The enzyme that catalyzes the cap 1 structure is a (nucleoside-2′-O-)-methyltransferase (2′-O-MTase), which usually utilizes S-adenosyl-l-methionine (AdoMet) as the methyl donor. The SARS-nsp13 protein belongs to the ancient family of AdoMet-dependent ribose 2′-O-MTase. The enzymatic role of the protein was confirmed by the presence of the conserved tetrad of residues K–D–K–E essential for mRNA cap 1 formation [7].
Viruses adopt different capping strategies based on their replication cycles and hosts, but most of them have the same cap structure as cellular mRNA. Viruses with cytoplamic replication cycle have to synthesize their 5′-terminal cap structures with their own methyltransferases. The order of methyl transfer reactions is variable, in some viruses the cap 0 structure is necessary for cap 1 methylation [10], and in other viruses the cap 1 is a preferred substrate for the cap 0 MTase [11], [12]. So viral cap-forming enzymes (cap 0 and/or cap 1) are potential targets for antiviral drugs that would interfere with capping of pathogen mRNAs but spare the host capping enzymes. Exploring the structure and function of the SARS-CoV nsp13 protein will be extremely useful for the development of anti-SARS agents.
In this study, we cloned the nsp13 gene by RT-PCR from SARS-CoV PUMC02 strain genomic RNA, and constructed a recombinant expression vector pET30a(+)-nsp13. The His-tag-fused SARS-nsp13 protein was then expressed in Escherichia coli, purified through a single step affinity chromatography, and finally identified by both western blot and mass spectrometry. A large quantity of the nsp13 protein can be obtained with this method.
Materials and methods
Reagents
Restriction enzymes were purchased from New England Biolabs (Berverly, MA). The expand reverse transcriptase and modified bovine trypsin (sequencing grade) were obtained from Roche (Mannheim, Germany). Pfu DNA polymerase and T4 DNA ligase were from Shanghai Sangon Co (Shanghai, China). Trizol reagent was from Invitrogen (Carlsbad, CA). The plasmid extraction kit and DNA agarose gel cleanup kit were obtained from BioDev (Beijing, China). Ni2+ -NTA resin was purchased from Qiagen (Valencia, CA). Nitrocellulose membrane was purchased from Amersham Life Science (Newington, NH), and His-tag monoclonal antibody was from Novagen (Darmstadt, Germany).
Construction of nsp13 gene in the expression vector pET30a(+)
All cloning techniques including PCR, restriction digestion, ligation, and E. coli transformation were performed as previously published [13].
SARS-CoV (PUMC02 strain) RNA was extracted with Trizol reagent according to the manufacturer’s instruction. The reverse transcription was performed by the expand reverse transcriptase with the primer TTGTT AACAA GAATA TCACT TGAAA CCAC, which is proximately downstream of the nsp13 gene. The SARS nsp13 cDNA was subsequently amplified by PCR, using the following primers, nsp13 forward, 5′-CGGGA TCCGC AAGTC AAGCG TGGCA AC-3′, and nsp13 reverse 5′-CCGCT CGAGT TAGTT GTTAA CAAGA ATATC AC-3′, the sequences underlined are the recognition sites of the restriction enzymes BamHI and XhoI. After digestion with BamHI and XhoI, the PCR product was purified with gel cleanup kit and inserted between the BamHI and XhoI sites of the vector pET30a(+). The recombinant vector pET30a(+)-nsp13 was identified by restriction digestion and the SARS-nsp13 insert was verified by sequencing. E. coli DH5α was used for amplification of the recombinant plasmid and E. coli BL21(DE3) was transformed for induced expression of the His-tagged SARS-nsp13 protein.
Expression and purification of SARS-nsp13 protein
A single colony of E. coli BL21(DE3) cells transformed with the recombinant expression plasmid pET30a(+)-nsp13 was inoculated in a tube containing 2 ml of LB medium with 50 μg/ml kanamycin, and cultured overnight at 37 °C in a shaking incubator. The culture was transferred to a 1–L flask containing 200 ml of the same medium. The flask was shaken at 37 °C until the OD600 of the culture was about 0.6–0.8. The expression of SARS-nsp13 protein was induced by the addition of 0.5 mM of isopropyl β-d-thiogalactoside (IPTG). After induction for 5 h at 25 °C, the cells were harvested by centrifugation at 4000g, 4 °C for 30 min. The supernatant was discarded and the cell pellet was washed, frozen, resuspended in PBS (pH 8.0), and then disrupted by sonication. The lysed cells were centrifuged at 12,000g, 4 °C for 30 min. The supernatant was filtered through a 0.45 μm syringe filter. Ni2+ chelating resin (200 μl) was equilibrated with 5 ml of sterile deionized water, 2 ml of 50 mM NiSO4, and 5 ml of PBS. The supernatant was passed through the column, followed by washing the column with 5 ml of PBS and 5 ml of Wash Buffer (20 mM imidazole in PBS), respectively. The protein of interest was then eluted with 1–2 ml of elution buffer (200 mM imidazole in PBS). The eluted fractions were analyzed by 12% SDS–PAGE.
Immunoblot analysis of SARS-nsp13 protein by His-tag monoclonal antibody
The protein sample was resolved on 12% SDS–PAGE gel and transferred onto nitrocellulose membrane using the Bio-Rad trans-blot apparatus. The membrane was incubated for 2 h at room temperature in Buffer I (5% skim milk, 0.02% Tween-20), His-tag monoclonal antibody was then added at a dilution of 1:1000, and the membrane was incubated at 4 °C overnight. Three washes were followed by addition of alkaline phosphatase-conjugated horse anti-mouse IgG at a dilution of 1:1000 in Buffer II (5% skim milk, 150 mM NaCl, 50 mM Tris–Cl, and pH 7.5), the membrane was incubated for 2 h at room temperature. After three washes with PBS, the membrane was immerged into Buffer III (100 mM NaCl, 5 mM MgCl2, and 100 mM Tris–Cl, pH 9.5) containing alkaline phosphatase substrate of 5-bromo-4-chloro-3-indoxyl phosphate (BCIP) and nitroblue tetrazolium (NBT). The blot was allowed to develop and the reaction was stopped by washing the membrane in distilled water.
Mass spectrometric identification of SARS-nsp13 protein
Excised gel band of interest was in-gel digested with modified bovine trypsin [14]. Excess Coomassie brilliant blue stain was removed by washing twice with 1 ml of 100 mM NH4HCO3, 50% acetonitrile for 1 h at room temperature. Then clear gel pieces were vacuum dried completely using a SpeedVac concentrator (Savant). The dried gel pieces were re-hydrated by adding 10 μl of digestion buffer (50 mM NH4HCO3) containing 0.5 μg of trypsin. After allowing 30 min for gel pieces to completely absorb the solution, an additional 200 μl of digestion buffer without trypsin was added to fully immerse the gel pieces. Digestion continued for about 16 h at 37 °C. The digestion buffer, now containing extracted peptides, was carefully removed and transferred into a clean microfuge tube. The gel pieces were re-extracted twice by adding 100 μl of 0.1% TFA in 50% acetonitrile and incubating for 30 min at room temperature. The pooled extracts were vacuum concentrated to reduce the volume to less than 10 μl. A volume of 0.5 μl of matrix solution (α-cyano-4-hydroxycinnamic acid saturated in 50% v/v acetonitrile/0.5% v/v TFA) was mixed with the same volume of the extract, then applied onto stainless steel target. Mass spectra were recorded in positive ion reflection mode of a matrix assisted laser desorption ionization-time of flight (MALDI-ToF) Voyager DE PRO (Applied Biosystems). Peptide masses obtained were searched against NCBInr database without any species limit using the Mascot search engine available online (www.matrixscience.com).
Results and discussion
Construction of the expression vector pET30a(+)-nsp13
The SARS-nsp13 gene was amplified by RT-PCR (Fig. 1 A), the amplified product was about 900 bp, which was accordant to the theoretical length of nsp13 gene. The recombinant plasmid was digested by the restriction enzymes BamHI and XhoI. Agarose gel electrophoresis of the digest revealed a DNA band at about 900 bp (Fig. 1B). The inserted fragment was verified by sequencing, the result was shown in Fig. 2 , which was identical to the published SARS-nsp13 gene sequence in NCBI and in the correct reading frame with the His-tag in the vector.
Fig. 1.
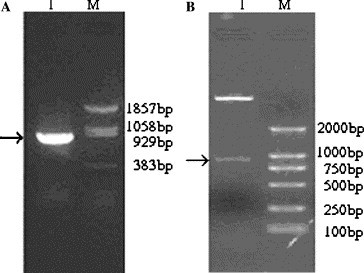
PCR product of SARS-CoV PUMC02 strain-nsp13 gene (A) and the recombinant expression vector pET30a(+)-nsp13 digestion with restriction enzymes BamHI and XhoI (B).
Fig. 2.

Sequencing result of the cloned SARS-CoV PUMC02 strain-nsp13 gene and the translated amino acid sequence. The bold and italic letters indicate the conserved tetrad of residues K–D–K–E.
Expression and purification of SARS-nsp13 protein
Escherichia coli BL21(DE3) was transformed with the recombinant expression vector pET30a(+)-nsp13, and SARS-nsp13 protein was abundantly expressed after induction by IPTG . As shown in Fig. 3 A, some of the target protein existed in the pellet. Lowering culture temperature and reducing the concentration of IPTG did not change the solubility (data is not shown here). The soluble fraction was loaded onto the Ni2+ -NTA affinity column, and the homogeneous protein was successfully purified with elution buffer after washing thoroughly. The purified protein was further analyzed with His-tag monoclonal antibody, and a specific blot band was detected at the corresponding position (Fig. 3B). The predicted molecular mass of the SARS-nsp13 protein was 33.4 kDa, and there were 30 additional amino acids for the His-tag in the N-terminal of the expressed fusion protein. These extra amino acids increased molecular mass of the expressed target protein by about 3.3 kDa. Apparent molecular mass of the purified protein was about 37 kDa, which was expected. The yield from 200 ml of bacterial culture was about 1.3 mg of purified SARS-nsp13 protein after a single Ni2+ affinity chromatographic step. The recovery of SARS-nsp13 protein was about 26% (Table 1 ).
Fig. 3.
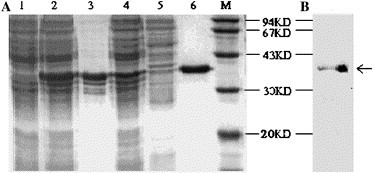
(A) 12% SDS–PAGE analysis of the expression and purification of SARS-nsp13 protein. 1, uninduced; 2, induced; 3, pellet; 4, supernatant; 5, flow-through; 6, purified nsp13 protein by Ni2+-NTA affinity column; and M, Low molecular weight protein marker. (B) Western blot analysis of the SARS-nsp13 protein by His-tag monoclonal antibody.
Table 1.
Purification scheme of SARS-nsp13 protein expressed in E. coli BL21(DE3) with pET30a(+)-nsp13 (200 ml culture)
| Step | Total protein (mg) | nsp13 protein (mg) | Recovery nsp13 (%) |
|---|---|---|---|
| Extraction | 67.8 | 4.9 | 100 |
| Ni2+-affinity column | 1.3 | 1.3 | 26 |
MALDI-ToF MS identification of SARS-nsp13 protein
The MALDI-ToF mass spectrum of tryptic digest of the gel band was shown in Fig. 4 . The achieved peptide masses were searched against NCBInr database without any species limit and with peptide mass tolerance of ±0.1 Da using the Mascot search engine. The first candidate protein, with a score of 94, was SARS nsp16 protein (GI:30133975), a putative ribose 2′-O-methyltransferase (the sequence was identical to that of the detected nsp13 protein here), and was the only one having a score greater than 75. The second candidate protein had a score of 67, which was lower than the significant score of 75. Seven peptide fragments were matched with tryptic peptides of nsp13 protein, and the sequence coverage was 26%. The covered portions in nsp13 protein were underlined in Fig. 5 . The results above indubitably determined the identity of the recombinant His-tagged SARS-nsp13 protein.
Fig. 4.
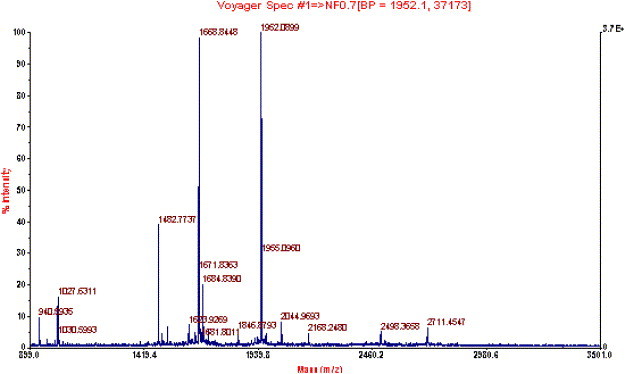
MALDI-ToF mass spectrum of the tryptic digest of His-tagged SARS-nsp13 protein.
Fig. 5.

Matched peptide fragments in SARS-nsp13 protein. The matched portions are shown in bold and underlined.
In conclusion, we have succeeded in the molecular cloning of SARS-nsp13, and have transformed the recombinant plasmid pET30a(+)-nsp13 into E. coli BL21(DE3). The purified His-tagged nsp13 protein has been identified by both western blot and MALDI-ToF mass spectrometry. The expression and purification procedures in this study have provided a simple and efficient method to obtain pure nsp13 protein of SARS-CoV in large quantities. The nsp13 protein obtained can be used for further study of its structure and function.
Acknowledgments
We thank Dr. Anping Ni for providing the SARS-CoV PUMC02 strain virus and Dr. Junming Zhang for critical reading of the manuscript. This work was supported by grant from the National Program for Key Basic Research Project (2001CB510206).
References
- 1.World Health Organization. Summary of probable SARS cases with onset of illness from 1 November 2002 to 31 July 2003. Available at: http://www.who.int/csr/sars/country/table2003-9-23/en/
- 2.Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.E., Humphrey C.D., Shieh W.J., Guarner J., Paddock C.D., Rota P., Fields B., DeRisi J., Yang J.Y., Cox N., Hughes J.M., LeDuc J.W., Bellini W.J., Anderson L.J. SARS Working Group, A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 3.Drosten C., Gunther S., Preiser W., vander Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., Berger A., Burguiere A.M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.C., Muller S., Rickerts V., Sturmer M., Vieth S., Klenk H.D., Osterhaus A.D., Schmitz H., Doerr H.W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 4.Poutanen S.M., Low D.E., Henry B., Finkelstein S., Rose D., Green K., Tellier R., Draker R., Adachi D., Ayers M., Chan A.K., Skowronski D.M., Salit I., Simor A.E., Slutsky A.S., Doyle P.W., Krajden M., Petric M., Brunham R.C., McGeer A.J. National Microbiology Laboratory, Canada; Canadian Severe Acute Respiratory Syndrome Study Team, Identification of severe acute respiratory syndrome in Canada. N. Engl. J. Med. 2003;348:1995–2005. doi: 10.1056/NEJMoa030634. [DOI] [PubMed] [Google Scholar]
- 5.Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H., Tong S., Tamin A., Lowe L., Frace M., DeRisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Gunther S., Osterhaus A.D., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 6.Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S., Khattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M., Freeman D., Girn N., Griffith O.L., Leach S.R., Mayo M., McDonald H., Montgomery S.B., Pandoh P.K., Petrescu A.S., Robertson A.G., Schein J.E., Siddiqui A., Smailus D.E., Stott J.M., Yang G.S., Plummer F., Andonov A., Artsob H., Bastien N., Bernard K., Booth T.F., Bowness D., Czub M., Drebot M., Fernando L., Flick R., Garbutt M., Gray M., Grolla A., Jones S., Feldmann H., Meyers A., Kabani A., Li Y., Normand S., Stroher U., Tipples G.A., Tyler S., Vogrig R., Ward D., Watson B., Brunham R.C., Krajden M., Petric M., Skowronski D.M., Upton C., Roper R.L. The Genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 7.von Grotthuss M., Wyrwicz L.S., Rychlewski L. mRNA cap-1 methyltransferase in the SARS genome. Cell. 2003;113:701–702. doi: 10.1016/S0092-8674(03)00424-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shuman S. Structure, mechanism, and evolution of the mRNA capping apparatus. Prog. Nucleic Acid Res. Mol. Biol. 2001;66:1–40. doi: 10.1016/s0079-6603(00)66025-7. [DOI] [PubMed] [Google Scholar]
- 9.Bisaillon M., Lemay G. Viral and cellular enzymes involved in synthesis of mRNA cap structure. Virology. 1997;236:1–7. doi: 10.1006/viro.1997.8698. [DOI] [PubMed] [Google Scholar]
- 10.Furuichi Y., Shatkin A.J. Viral and cellular mRNA capping: past and prospects. Adv. Virus Res. 2000;55:135–184. doi: 10.1016/S0065-3527(00)55003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Testa D., Banerjee A.K. Two methyltransferase activities in the purified virions of vesicular stomatitis virus. J. Virol. 1977;24:786–793. doi: 10.1128/jvi.24.3.786-793.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hammond D.C., Lesnaw J.A. Functional analysis of hypomethylation variants of the New Jersey serotype of vesicular stomatitis virus. Virology. 1987;160:330–335. doi: 10.1016/0042-6822(87)90003-1. [DOI] [PubMed] [Google Scholar]
- 13.Sambrook J., Russell D.W. third ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2001. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- 14.Shevchenko A., Wilm M., Vorm O., Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]


