Abstract
We evaluated the usefulness of a serum Aspergillus PCR assay for the diagnosis and prognosis of invasive aspergillosis in a study involving 941 patients for a total of 5146 serum samples. Fifty-one patients had proven/probable aspergillosis. We compared galactomannan (GM), PCR and mycologic analysis of pulmonary samples in both neutropenic and nonneutropenic patients. PCR performed in serum yielded 66.7% sensitivity, 98.7% specificity, 75.6% positive predictive value and 98.0% negative predictive value, while the GM index yielded 78.4% sensitivity, 87.5% specificity, 27% positive predictive value and 98.6% negative predictive value. The inclusion of PCR in the European Organization for Research and Treatment of Cancer (EORTC) and the Mycosis Study Group (MSG) mycologic criteria permitted the reclassification of nine other cases from possible to probable aspergillosis and increased the sensitivity to 71.7%. Combining the GM index with serum PCR increased the detection rate of invasive aspergillosis with 88.2% sensitivity. PCR was systematically negative in 16 patients with noninvasive forms of aspergillosis (namely aspergilloma and chronic aspergillosis). Remaining PCR positive after a period of 14 to 20 days of treatment was related to poor outcome at 30 and 90 days. Our results also indicate that, unlike the determination of the GM index, the initial fungus load as determined by PCR was highly predictive of 90-day mortality, with the rate of the latter being 15.8% for patients with <150 copies/mL vs. 73.2% for patients at or above that cutoff (p <0.0001). Therefore, PCR appears to be a powerful and interesting tool for the identification of patients with invasive aspergillosis who might benefit from more intense care.
Keywords: Aspergillus fumigatus, fungal infection, galactomannan, haematology, solid organ transplant
Introduction
Invasive aspergillosis (IA) is a major threat for immunocompromised hosts. Early diagnosis and initiation of appropriate antifungal therapy are essential to improve the prognosis of the disease [1], [2]. However, the diagnosis of IA remains difficult. Currently it is frequently based on a set of host, clinical and mycologic criteria, such as those defined jointly by the European Organization for Research and Treatment of Cancer (EORTC) and the Mycosis Study Group (MSG) [3]. In the EORTC/MSG criteria, galactomannan (GM) or β-d-glucan assays are the only noninvasive mycologic diagnostic methods currently available. Although these assays are of interest and widely used, they lack sensitivity, especially in nonneutropenic patients [4]. They also may be influenced by the use of certain medical devices [5] or antibiotics [6], [7], leading to a large number of false-positive results. GM detection has also been reported in invasive fungal diseases not due to Aspergillus [8], [9], [10]. Moreover, β-d-glucan is present in many fungus species and is therefore not specific to aspergillosis. Other tools have been developed to improve diagnosis, including in particular real-time PCR in blood samples to detect circulating Aspergillus DNA. Several studies have shown the potential interest of this PCR approach, but because of a lack of standardization, it has not yet been included in the EORTC/MSG mycologic criteria. Recently however, a panel of experts has argued for its inclusion in those criteria [11].
For the present study, we aimed at evaluating the performance of an in-house A. fumigatus real-time PCR assay using 1 mL volume of serum for the diagnosis of IA in at-risk patients, both neutropenic and nonneutropenic, and comparing the PCR results with those of the GM assay to determine their 30- and 90-day prognostic contributions.
Materials and Methods
Design
A retrospective single-centre analysis was performed between February 2012 and October 2014 in Hôpital de La Pitié-Salpêtrière, Paris, France.
Patients
All patients who are at risk of IA are routinely subjected to monitoring with a serologic assay for the detection of GM antigen and serologic PCR for the detection of A. fumigatus DNA. For the present study, we focused on patients with proven/probable IA according to EORTC/MSG criteria [3], extended to include the additional criteria of alcoholic liver cirrhosis, a long stay in the intensive care unit and severe acute respiratory distress syndrome as host factors, as already reported [12], [13]. GM either in serum or in bronchoalveolar lavage (BAL) was used as mycologic criterion for probable case. GM determination and PCR results were also available for some patients with noninvasive aspergillosis.
PCR
We used a real-time PCR that targets a previously described 67 bp segment of a 28S ribosomal RNA coding DNA [14], [15], for which the primer sequences were 5′-CTCGCAATGTATCACCTCTCGG3′ and 5′TCCTCGGTCCAGGCAGG-3′ and the probe was 5′-(6FAM)TGTCTTATAGCCGAGGGTGCAATGGG(TAMRA)-3′. DNA extraction was performed on 1 mL of serum with the MagNA Pure Compact large volume kit on a MagNA Pure device (Roche). Elution volume was 50 μL. Amplification was performed on the 7500 Fast Real-Time PCR System (Applied Biosystems). Quantification was achieved using five serial tenfold dilutions of the plasmid PGEMT Easy-Afu28S containing the target. The final PCR result was expressed in numbers of copies per millilitre of serum. We used an internal control in our assay for all wells (TaqMan exogenous Internal Positive Control) as well as an extraction control (albumin gene) for each sample. All PCRs were performed in duplicate. A single positive well was considered to be a positive result.
GM determination
The GM index was determined by enzyme immuno-assay (Bio-Rad) according to the manufacturer's recommendations. A result was considered positive after two determinations, performed on two different assays but on the same sample, showing both an index above 0.5 for serum and 1 for BAL.
Statistical analysis
Tests were performed using GraphPad Prism 5 software and the online site BiostaTGV (http://marne.u707.jussieu.fr/biostatgv/).
Results
Patients
Over the study period, GM assay and A. fumigatus PCR were performed in 970 patients (Fig. 1 ). Clinical data were available for 941 patients (5146 serum samples). A diagnosis of proven or probable IA was made, respectively, for six and 45 patients according to the extended EORTC/MSG criteria. Moreover, a noninvasive form of aspergillosis was diagnosed in 16 patients. Although the study was not focused on an exhaustive identification of all possible cases, we did register diagnoses of IA based on host factors and clinical evaluation for nine other patients, classifying them as possible IA according to the EORTC/MSG criteria. These patients had positive serum PCR.
Fig. 1.
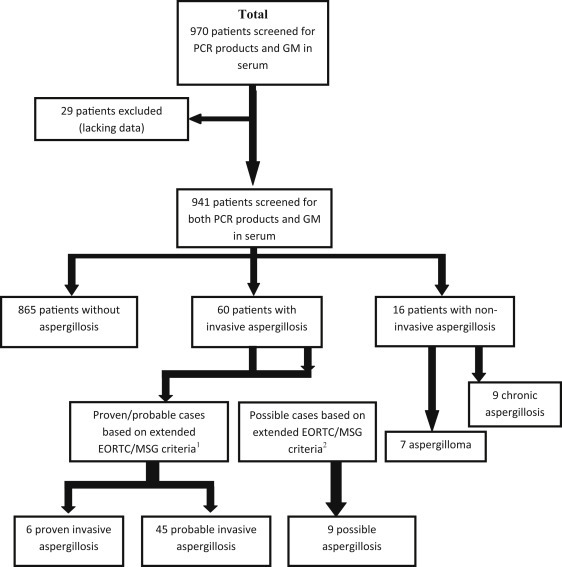
Flow chart showing number of patients and samples. 1Extended EORTC/MSG criteria included host factors as published in 2008 plus several other host factors now recognized as leading to risk of developing IA, namely alcoholic liver cirrhosis, severe acute respiratory syndrome, long stay in intensive care unit and solid organ cancer. 2Study design did not include exhaustive collection of possible cases; we present only possible cases with positive PCR results that were considered as IA by clinicians and treated as such. EORTC/MSG, European Organization for Research and Treatment of Cancer/Mycosis Study Group; IA, invasive aspergillosis.
Characteristics and outcomes of patients with aspergillosis
Among the 51 patients with proven or probable aspergillosis, 19 were female and 32 male. Their median age was 56 years (range 20–82 years). Twenty-two patients (43%) were neutropenic (absolute neutrophil count <500/μL) at the time of diagnosis. Underlying conditions were haematopoietic stem cell transplant (n = 17, 33.3%), haematologic malignancies (n = 13, 25.5%), heart transplantation (n = 9, 17.7%), liver transplantation (n = 3, 5.9%), kidney transplantation (n = 1, 2%) and liver/kidney transplantation (n = 1, 2%). Others risk factors were present in seven patients as follows: severe acute respiratory distress syndrome (n = 2, 3.8%), oncologic diseases (n = 2, 3.8%), alcoholic liver cirrhosis (n = 1, 2%), cardiogenic shock (n = 1, 2%), multiorgan failure and long stay in intensive care unit (n = 1, 2%) (Table 1 ). Overall 3-month mortality was 49%. Neutropenic patients had a rate of mortality of 54.5% (12/22) vs. 44.8% (13/29) for nonneutropenic patients (p 0.49, Fisher's exact test).
Table 1.
Characteristics of patients with proven/probable invasive aspergillosis according to EORTC/MSG criteria and Aspergillus PCR level
| Characteristic | EORTC/MSG criteria |
According to Aspergillus PCR level |
p | ||
|---|---|---|---|---|---|
| Without PCR | With PCR | <150 copies | ≥150 copies | ||
| No. of patients | 51 | 60 | 41 | 19 | |
| Haematologic malignancy | 13 (25.5%) | 18 (30%) | 10 (24.4%) | 5 (26.3%) | 0.87 |
| Haematopoietic stem cell transplant | 17 (33.3%) | 18 (30%) | 10 (24.4%) | 6 (31.6%) | 0.78 |
| Antifungal therapya | 0.6 | ||||
| Azole based | 36 (70.5%) | 42 (70%) | 31 (75.6%) | 11 (57.9%) | |
| Non–azole based | 13 (25.5%) | 16 (26.7%) | 10 (24.4%) | 6 (31.6%) | |
| SOT recipient | 14 (27.4%) | 16 (26.7%) | 9 (22%) | 5 (26.3%) | 0.96 |
| Other | 7 (13.7%) | 8 (13.3%) | 5 (12.2%) | 3 (15.8%) | 0.7 |
| Interval between time of sample and start of targeted antifungal therapy (mean days) | 0.65 | 0.67 | 0.8 | 0.37 | 0.7 |
| 3-month mortality | 25 (49%) | 27 (45%) | 11 (26.8%) | 16 (84.2%) | <0.0001 |
EORTC/MSG, European Organization for Research and Treatment of Cancer/Mycosis Study Group; SOT, solid organ transplant.
Two patients in the >150 copies/mL group died before start of antifungal therapy.
Performance of GM index, mycologic examination of respiratory sample and PCR for diagnosis of proven/probable aspergillosis
A diagnosis of proven/probable IA (according to the extended EORTC/MSG criteria; n = 51) was made for 40 patients by a positive GM index (Fig. 2 ). PCR was positive in 34 patients (including 11 with a positive GM in serum, four with a positive mycologic analysis of respiratory samples, 18 with both positive GM in serum and positive mycologic analysis of respiratory samples and one with an isolated positive GM in BAL), including five with a negative serum GM index. In one patient with a positive PCR result, the diagnosis of probable aspergillosis was based only on a positive GM index in a BAL sample. Sensitivities of the serum GM index, mycologic examination and serum PCR were 78.4% (95% confidence interval (CI) 67.1–89.7), 62.7% (95% CI 49.4–76) and 66.7% (95% CI 53.8–79.6) respectively. Of note, sensitivity of GM and PCR in case of proven aspergillosis was similar (83.3%; 5/6 patients). Among the patients for whom the diagnosis of any clinical form of aspergillosis was excluded by clinicians, there were 108 with positive GM indices, 11 with positive PCR results, and two with positive GM indices and positive PCR results. Specificities of GM and PCR were 87.5% (95% CI 85.3–89.7) and 98.7% (95% CI 97.9–99.5) respectively. PCR yielded 75.6% positive predictive value (PPV) (95% CI 63.1–88.1) and 98.0% negative predictive value (NPV) (95% CI 97.1–98.9), while the GM index yielded 27% PPV (95% CI 19.8–34.2) and 98.6% NPV (95% CI 97.8–99.4). No differences were observed between PCR and GM as concerns the precocity of diagnosis (data not shown).
Fig. 2.
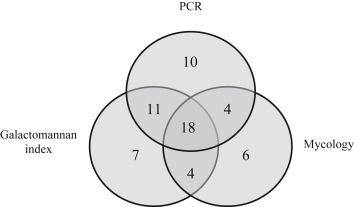
Venn diagram showing data for patients with invasive aspergillosis (n = 60). Diagram shows data for patients for whom PCR products and GM in sera as well as mycologic analysis of respiratory samples were available. Among ten patients with only positive PCR, one was positive for GM in bronchoalveolar lavage samples. GM, galactomannan.
PCR results for noninvasive forms of aspergillosis
Among the 16 noninvasive aspergillosis patients, ten were immunocompetent, three had metastatic malignancy, two had solid organ transplants and one had alcoholic liver cirrhosis. Clinical forms were simple aspergilloma (n = 7), colonization (n = 4), chronic cavitary aspergillosis (n = 4) and chronic bronchitis (n = 1). In addition to clinical condition, host history and radiologic features, patient diagnoses were further supported by mycologic examination (n = 11), presence of anti-Aspergillus antibodies (n = 1) or both (n = 4). None of these patients had positive serum GM or PCR.
Addition of PCR to the mycologic criteria
Of the nine patients treated for IA but classified as possible cases due to a lack of EORTC/MSG mycologic criteria, seven were male and two were female. Six patients were neutropenic (five haematologic malignancies and one haematopoietic stem cell transplant) and three were not (one liver transplantation, one kidney transplantation and one severe acute respiratory distress syndrome). All nine of these patients had positive PCR results.
Thus, considering PCR as a mycologic criterion would have enabled the reclassification of nine patients from possible to probable IA, increasing the number of patients with proven/probable IA to 60 (Table 1, Table 2 ). In this scenario, GM determination and PCR would have been positive in 40 and 43 of the 60 patients respectively, conferring sensitivities of 66.7% (95% CI 54.8–78.6) and 71.7% (95% CI 60.3–83.1) and specificities of 87.7% (95% CI 85.5–89.9) and 98.8% (95% CI 98.1–99.5) for GM and PCR respectively. PCR would yield 79.6% PPV (95% CI 68.9–90.3) and 98.0% NPV (95% CI 97.1–98.9), while the GM index would yield 27% PPV (95% CI 19.8–34.2) and 97.5% NPV (95% CI 96.4–98.6).
Table 2.
Performance of PCR to detect Aspergillus fumigatus in serum, determination of galactomannan index in serum and mycologic examination of respiratory samples for the diagnosis of invasive aspergillosis in 60 patients treated for proven/probable invasive aspergillosis according to extended EORTC/MSG criteria with the addition of PCR in the mycologic criteria
| Method | Neutrophil status | Group | No. of samples with result |
Sensitivity | Specificity | Positive predictive value | Negative predictive value | pb | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Positive | Negative | |||||||||
| Galactomannan | All patients | IAa | 40 | 20 | 66.7 | 87.7 | 27 | 97.5 | ||
| Non-IA | 108 | 773 | ||||||||
| Neutropenic | IA | 19 | 9 | 67.8 | 83.9 | 22 | 97.5 | 0.93 | Compared to nonneutropenic | |
| Non-IA | 67 | 350 | <0.005 | Compared to nonneutropenic | ||||||
| Nonneutropenic | IA | 21 | 11 | 65.6 | 91.2 | 33.9 | 97.5 | |||
| Non-IA | 41 | 423 | ||||||||
| Mycologic examination | All patients | IA | 32 | 14 | 69.6 | NA | NA | NA | ||
| Neutropenic | 8 | 9 | 47 | |||||||
| Nonneutropenic | 24 | 5 | 82.7 | |||||||
| PCR | All patients | IA | 43 | 17 | 71.7 | 98.8 | 79.6 | 98 | ||
| Non-IA | 11 | 870 | ||||||||
| Neutropenic | IA | 23 | 5 | 82.1 | 98.1 | 74.2 | 98.8 | 0.09 | Compared to nonneutropenic | |
| Non-IA | 8 | 409 | 0.09 | Compared to nonneutropenic | ||||||
| Nonneutropenic | IA | 20 | 12 | 62.5 | 99.4 | 87 | 97.5 | |||
| Non-IA | 3 | 461 | ||||||||
EORTC/MSG, European Organization for Research and Treatment of Cancer/Mycosis Study Group; IA, invasive aspergillosis; NA, not available.
Criteria used for classification of IA were those defined jointly by the EORTC/MSG consensus and published in 2008 with additionally inclusion of PCR as a mycologic criteria and minor modifications for host factors (e.g. inclusion of alcoholic liver cirrhosis).
As calculated by chi-square test between neutropenic and nonneutropenic patients.
Neutropenic versus nonneutropenic
Considering these 60 patients, PCR sensitivity tended to be better in neutropenic patients (82.1%) than in nonneutropenic patients (62.5%). The difference did not reach statistical significance (p 0.09), but the power of the test (P = 39%) was insufficient to draw conclusions. PCR specificity did not differ between the two groups. As for the GM index, sensitivity did not differ between the two groups, but specificity was significantly lower in neutropenic patients (83.9%) compared to nonneutropenic patients (91.2%) (p <0.005).
Effect of antifungal therapy
Among our 60 patients with IA, 49 had at least one sample taken before initiation of targeted antifungal therapy. All patients had a sample taken after treatment initiation except for five patients who died early. At the time of diagnosis, 24 patients received antifungal therapy: 12 received empirical therapy with caspofungin and 12 received more targeted anti-Aspergillus therapy with voriconazole or amphotericin B. Among the patients who received antifungal drugs, PCR and GM yielded similar sensitivity of 70.8% (17/24), and there was also no difference according to the antifungal drug provided. Among the 36 patients without antifungal therapy, sensitivity was 72.2% (26/36) for PCR and 63.9% for GM (23/36). The difference is not statistically significant (p 0.6). When comparing patients who did and did not receive antifungal therapy, there was no statistically significant difference of sensitivity for PCR or GM.
Variation of fungus load after treatment initiation and prediction of outcome
Real-time PCR allows the quantification of fungus load, with results expressed as number of Aspergillus gene copies per millilitre of serum. GM results are presented as an index which can be interpreted quantitatively. Considering this, we assessed variations (decreases or increases) in PCR and the GM index to determine their ability to predict treatment efficacy and 30- and 90-day mortality.
Concerning the response to targeted antifungal therapy, we observed no early variations in PCR results because fungus loads did not decrease quickly after the initiation of treatment, even when outcomes were favourable. Moreover, an increase or decrease in the number of copies in the first week was not related to day 30 or day 90 outcome. However, patients who become PCR negative (and one who had cleared more than 99% of the initial load) between day 14 and day 20 after treatment initiation were all alive at day 30 (n = 9) while those who remained PCR positive during this period had poor outcomes, with 80% (4/5) mortality at day 30 (Table 3 ). This result was highly significant (p <0.005, Fisher's exact test). PCR results between days 14 and 20 were also predictive of day 90 outcome (Table 3).
Table 3.
Results of Aspergillus PCR after 14 days of antifungal therapy related to day 30 and day 90 outcome
| Aspergillus PCR result in serum sampled between days 14 and 20 | Outcome at day 30 |
Outcome at day 90 |
||||
|---|---|---|---|---|---|---|
| Alive | Dead | p | Alive | Dead | p | |
| Positive | 1 | 4 | <0.005 | 0 | 5 | <0.05 |
| Negativea | 9 | 0 | 7 | 2 | ||
All patients had positive PCR result at time of diagnosis and had at least one serum sample taken between 14 and 20 days after initiation of targeted antifungal treatment and tested for PCR; p value was determined by Fisher's exact test.
One patient who cleared more than 99% of the initial load was considered negative.
Initial fungus load and prediction of outcome
We also assessed initial fungus loads as predictors of outcome. Receiver operating characteristic curves for the evaluation of fungus load as a marker of 90-day mortality in IA indicated that the 150 copies/mL cutoff offered the most efficient value (Fig. 3 a). Patients with PCR results strictly below 150 copies/mL had a higher probability of survival 90 days after diagnosis (n = 30/41, 73.2% survival) compared to those with PCR results at or above this cutoff (n = 3/19, 15.8% survival, median survival 20 days) (Fig. 3b). This result was highly statistically significant (p <0.0001, log rank test). The test had a hazard ratio of 0.14 (95% CI of ratio 0.05 to 0.34). Similar results were obtained when all patients were considered, i.e. including the 17 patients with negative PCR in the <150 copies/mL group. There were no differences between these two groups in terms of age, sex ratio, underlying diseases or antifungal therapy (Table 1). There were also no differences concerning the interval between sampling and the start of targeted antifungal therapy. In comparison, among patients with initial positive GM, patients with GMs below 2 (the most efficient value) appear to have more favourable outcomes than others, although the difference was not statistically significant (p 0.19, log rank test) (Fig. 3c).
Fig. 3.
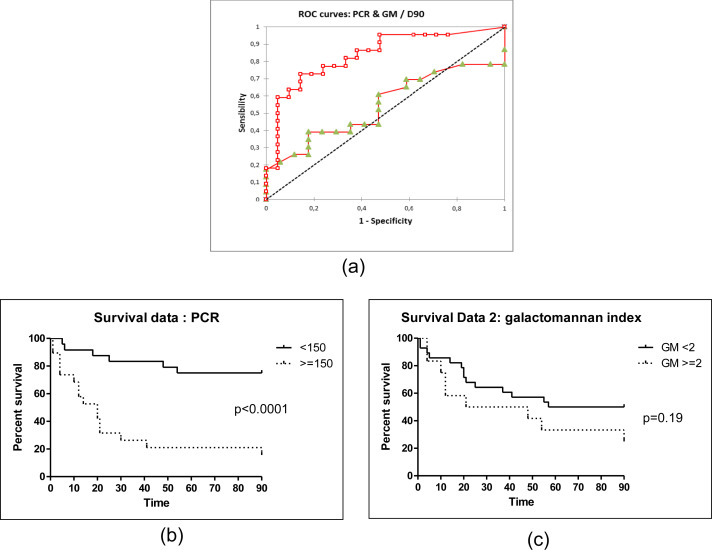
Serum Aspergillus PCR is highly predictive of 90-day mortality in IA. (a) ROC curve for evaluation of PCR (square) or GM (triangle) as marker of 90-day mortality in IA. Cutoff of 150 copies/mL offers most efficient value and area under curve of 0.837. For GM index cutoff of 2 (most efficient value) is related to small area under curve of 0.546. (b) Patients with initial fungus loads <150 copies/mL (n = 41, 73.2% survival) have more favourable outcomes than other patients (n = 19, 15.8% survival); p <0.0001 by log rank (Mantel-Cox) test, hazard ratio 0.14 (95% CI of ratio 0.05 to 0.34). (c) Patients with initial GMs below 2 (n = 28, 50% survival) appear to have more favourable outcomes than others (n = 12; 25% survival), but difference is not statistically significant (p 0.19 by log rank (Mantel-Cox) test; hazard ratio 0.5, 95% CI of ratio 0.20 to 1.29). CI, confidence interval; GM, galactomannan; IA, invasive aspergillosis; ROC, receiver operating characteristic.
Discussion
PCR was originally excluded from the 2008 EORTC/MSG definitions of invasive fungal diseases, but expert consensus now considers that this method is mature enough for its inclusion [11]. Meta-analyses have indeed demonstrated that PCR offers sensitivity ranging from 77 to 88% and specificity from 75 to 94.1%, rates similar to those attained with GM assays [16], [17], [18]. Moreover, PCR permits the use of quality control and interlaboratory checks, and it provides a more robust quantification assessment than does the determination of the GM index.
The results of the present study add to this evidence, with PCR sensitivity reaching 71.7% and specificity 98.7%. In our study, PCR appeared to offer an interesting PPV, i.e. 79.6% for all patients, in contrast to the GM index, where a high-false positive rate, particularly in neutropenic patients, led to a poor overall PPV of 27%. PCR and GM both offered interesting negative predictive values above 97%. PCR was more sensitive in neutropenic patients (82.1%) than it was in nonneutropenic patients (62.5%), but the difference was not significant (p 0.09). Increasing the number of patients could lead to a significant result. Nevertheless, our study shows that PCR is also useful in nonneutropenic patients and nonhaematologic populations such as solid organ transplant recipients. As previously reported, our result indicate that performing both GM determination and PCR on the same sample increases the sensitivity [18].
In 2010 Koo et al. [19] reported a relation between GM and outcome, and in 2012 Bergeron et al. [20] reported that patients with poor outcomes 45 days after the initiation of antifungal treatment had high baseline serum GM. We, however, found only a nonstatistically significant trend for the GM index in the determination of outcomes. Nevertheless, Bergeron et al. also found no association between outcome and Aspergillus DNA detection, while our work supports that the initial level of Aspergillus DNA is highly predictive of the 90-day mortality rate. As concerns our results more specifically, we found that a PCR threshold of 150 copies/mL could discriminate patients with low (below the threshold) or high (above) probabilities of 90-day mortality. PCR thus has a much greater discriminative capacity than does the determination of the GM index. This result could be useful for identifying patients who may benefit from more intensive care and designing further clinical studies. We also showed that PCR quantification is useful in follow-up to predict outcomes. After only 1 week of therapy, an increase or decrease in the number of copies is not relevant for the assessment of treatment efficacy or outcome. However, PCR results after 2 weeks of treatment did appear to have relevance. In our study, patients who had a negative PCR (to which we add one patient who had cleared more than 99% of the initial load) between days 14 and 20 were all alive at day 30, and seven of them were still alive at day 90 vs. none of the five patients who remained PCR positive.
In a work initiated by the European Aspergillus PCR Initiative, White et al. [21] clearly demonstrated that PCR performed on plasma gives better results than PCR performed on serum. In consideration of this evidence, we are now changing our methods. The PCR that we used in the study was specific to the fumigatus species, and thus it would not have amplified the genomes of other species such as flavus, niger or nidulans. This may explain at least in part the relative lack of sensitivity for PCR in our study, as non-fumigatus species may account for more than 25% of disease [22]. We did, however, retrieve the causative species in 32 of our cases; strikingly, all were fumigatus. Going forward, the utility of PCR in IA and the determination of cutoffs will have to be evaluated in other laboratory specimens, such as cerebrospinal fluid and BAL.
Conclusion
Aspergillus DNA detection in serum by PCR is a interesting tool for the diagnosis of IA in both neutropenic and nonneutropenic patients and should be used concomitantly with the GM index in at-risk populations. Moreover, PCR allows quantification of fungus load; our results indicate that a threshold of 150 copies/mL is very powerful to discriminate patients with low (below the threshold) or high (above) probabilities of 90-day mortality. Therefore, initial fungus load and variations in fungus load during treatment as determined by PCR are robust predictive markers of mortality that may be used to identify patients who might benefit from closer and more attentive care.
Transparency Declaration
All authors report no conflicts of interest relevant to this article.
Editor: E. Roilides
Footnotes
Presented in part in French at the Congress of the French Society for Medical Mycology (Société Française de Mycologie Médicale, SFMM), 21 May 2015, where it was awarded the Oral Communication Prize; and as an oral communication at the Trends in Medical Mycology International Congress, October 2015, Lisbon, Portugal
References
- 1.von Eiff M., Roos N., Schulten R., Hesse M., Zuhlsdorf M., van de Loo J. Pulmonary aspergillosis: early diagnosis improves survival. Respiration. 1995;62:341–347. doi: 10.1159/000196477. [DOI] [PubMed] [Google Scholar]
- 2.Walsh T.J., Anaissie E.J., Denning D.W., Herbrecht R., Kontoyiannis D.P., Marr K.A. Treatment of aspergillosis: clinical practice guidelines of the Infectious Diseases Society of America. Clin Infect Dis. 2008;46:327–360. doi: 10.1086/525258. [DOI] [PubMed] [Google Scholar]
- 3.De Pauw B., Walsh T.J., Donnelly J.P., Stevens D.A., Edwards J.E., Calandra T. Revised definitions of invasive fungal disease from the European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group and the National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) consensus group. Clin Infect Dis. 2008;46:1813–1821. doi: 10.1086/588660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meersseman W., Lagrou K., Maertens J., Wilmer A., Hermans G., Vanderschueren S. Galactomannan in bronchoalveolar lavage fluid: a tool for diagnosing aspergillosis in intensive care unit patients. Am J Respir Crit Care Med. 2008;177:27–34. doi: 10.1164/rccm.200704-606OC. [DOI] [PubMed] [Google Scholar]
- 5.Marty F.M., Koo S. Role of (1→3)-beta-d-glucan in the diagnosis of invasive aspergillosis. Med Mycol. 2009;47(Suppl. 1):S233–S240. doi: 10.1080/13693780802308454. [DOI] [PubMed] [Google Scholar]
- 6.Mennink-Kersten M.A., Warris A., Verweij P.E. 1,3-Beta-d-glucan in patients receiving intravenous amoxicillin–clavulanic acid. N Engl J Med. 2006;354:2834–2835. doi: 10.1056/NEJMc053340. [DOI] [PubMed] [Google Scholar]
- 7.Sulahian A., Porcher R., Bergeron A., Touratier S., Raffoux E., Menotti J. Use and limits of (1–3)-beta-d-glucan assay (Fungitell), compared to galactomannan determination (platelia Aspergillus), for diagnosis of invasive aspergillosis. J Clin Microbiol. 2014;52:2328–2333. doi: 10.1128/JCM.03567-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fekkar A., Brun S., D’Ussel M., Uzunov M., Cracco C., Dhedin N. Serum cross-reactivity with Aspergillus galactomannan and cryptococcal antigen during fatal disseminated Trichosporon dermatis infection. Clin Infect Dis. 2009;49:1457–1458. doi: 10.1086/644499. [DOI] [PubMed] [Google Scholar]
- 9.Wheat L.J., Hackett E., Durkin M., Connolly P., Petraitiene R., Walsh T.J. Histoplasmosis-associated cross-reactivity in the biorad platelia Aspergillus enzyme immunoassay. Clin Vaccine Immunol. 2007;14:638–640. doi: 10.1128/CVI.00479-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dalle F., Charles P.E., Blanc K., Caillot D., Chavanet P., Dromer F. Cryptococcus neoformans galactoxylomannan contains an epitope(s) that is cross-reactive with Aspergillus galactomannan. J Clin Microbiol. 2005;43:2929–2931. doi: 10.1128/JCM.43.6.2929-2931.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.White P.L., Wingard J.R., Bretagne S., Loffler J., Patterson T.F., Slavin M.A. Aspergillus polymerase chain reaction: systematic review of evidence for clinical use in comparison with antigen testing. Clin Infect Dis. 2015;61:1293–1303. doi: 10.1093/cid/civ507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prodanovic H., Cracco C., Massard J., Barrault C., Thabut D., Duguet A. Invasive pulmonary aspergillosis in patients with decompensated cirrhosis: case series. BMC Gastroenterol. 2007;7:2. doi: 10.1186/1471-230X-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Azoulay E., Lemiale V., Mokart D., Pene F., Kouatchet A., Perez P. Acute respiratory distress syndrome in patients with malignancies. Intensive Care Med. 2014;40:1106–1114. doi: 10.1007/s00134-014-3354-0. [DOI] [PubMed] [Google Scholar]
- 14.Suarez F., Lortholary O., Buland S., Rubio M.T., Ghez D., Mahe V. Detection of circulating Aspergillus fumigatus DNA by real-time PCR assay of large serum volumes improves early diagnosis of invasive aspergillosis in high-risk adult patients under hematologic surveillance. J Clin Microbiol. 2008;46:3772–3777. doi: 10.1128/JCM.01086-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Challier S., Boyer S., Abachin E., Berche P. Development of a serum-based TaqMan real-time PCR assay for diagnosis of invasive aspergillosis. J Clin Microbiol. 2004;42:844–846. doi: 10.1128/JCM.42.2.844-846.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mengoli C., Cruciani M., Barnes R.A., Loeffler J., Donnelly J.P. Use of PCR for diagnosis of invasive aspergillosis: systematic review and meta-analysis. Lancet Infect Dis. 2009;9:89–96. doi: 10.1016/S1473-3099(09)70019-2. [DOI] [PubMed] [Google Scholar]
- 17.Arvanitis M., Ziakas P.D., Zacharioudakis I.M., Zervou F.N., Caliendo A.M., Mylonakis E. PCR in diagnosis of invasive aspergillosis: a meta-analysis of diagnostic performance. J Clin Microbiol. 2014;52:3731–3742. doi: 10.1128/JCM.01365-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arvanitis M., Anagnostou T., Mylonakis E. Galactomannan and polymerase chain reaction–based screening for invasive aspergillosis among high-risk hematology patients: a diagnostic meta-analysis. Clin Infect Dis. 2015;61:1263–1272. doi: 10.1093/cid/civ555. [DOI] [PubMed] [Google Scholar]
- 19.Koo S., Bryar J.M., Baden L.R., Marty F.M. Prognostic features of galactomannan antigenemia in galactomannan-positive invasive aspergillosis. J Clin Microbiol. 2010;48:1255–1260. doi: 10.1128/JCM.02281-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bergeron A., Porcher R., Menotti J., Poirot J.L., Chagnon K., Vekhoff A. Prospective evaluation of clinical and biological markers to predict the outcome of invasive pulmonary aspergillosis in hematological patients. J Clin Microbiol. 2012;50:823–830. doi: 10.1128/JCM.00750-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.White P.L., Barnes R.A., Springer J., Klingspor L., Cuenca-Estrella M., Morton C.O. Clinical performance of Aspergillus PCR when testing serum and plasma—a study by the European Aspergillus PCR Initiative. J Clin Microbiol. 2015;53:2832–2837. doi: 10.1128/JCM.00905-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steinbach W.J., Marr K.A., Anaissie E.J., Azie N., Quan S.P., Meier-Kriesche H.U. Clinical epidemiology of 960 patients with invasive aspergillosis from the PATH Alliance registry. J Infect. 2012;65:453–464. doi: 10.1016/j.jinf.2012.08.003. [DOI] [PubMed] [Google Scholar]


