Abstract
Malectin is a carbohydrate-binding lectin protein found in the endoplasmic reticulum (ER). It selectivity binds to Glc2-N-glycan and is involved in a glycoprotein quality control mechanism. Even though malectin may play a role in immunity, its role in innate immunity is not fully known. In the present study, we identified and characterized the malectin gene from Hippocampus abdominalis (HaMLEC). We analyzed sequence features, spatial expression levels, temporal expression profiles upon immune responses, bacterial and carbohydrate binding abilities and anti-viral properties to investigate the potential role of HaMLEC in innate immunity. The molecular weight and isoelectric point (pI) were estimated to be 31.99 kDa and 5.17, respectively. The N-terminal signal peptide, malectin superfamily domain and C-terminal transmembrane region were identified from the amino acid sequence of HaMLEC. The close evolutionary relationship of HaMLEC with other teleosts was identified by phylogenetic analysis. According to quantitative PCR (qPCR) results, HaMLEC expression was observed in all the examined tissues and high expression was observed in the ovary and brain, compared to other tested tissues. Temporal expression of HaMLEC in liver and blood tissues were significant modulated upon exposure to immunogens Edwardasiella tarda, Streptococcus iniae, polyinosinic:polycytidylic and lipopolysaccharide. The presence of carbohydrate binding modules (CBMs) of bacterial glycosyl hydrolases were functionally confirmed by a bacterial binding assay. Anti-viral activity significantly reduced viral hemorrhagic septicemia virus (VHSV) replication in cells overexpressing HaMLEC. The observed results suggested that HaMLEC may have a significant role in innate immunity in Hippocampus abdominalis.
Keywords: Glycoprotein, Hippocampus abdominalis, Immune challenge, Malectin
Highlights
-
•
Malectin was identified and characterized from Big-belly seahorse (HaMELC).
-
•
HaMLEC was ubiquitously expressed in healthy Big-belly seahorse tissues.
-
•
HaMLEC expression was modulated upon immune challenge.
-
•
Recombinant HaMLEC (rHaMLEC) was able to bind to Gram (+) and (−) bacteria.
-
•
Over expression down regulate the viral transcription in vitro.
1. Introduction
Proteins depend on several mechanisms to form biologically active conformations [[1], [2], [3]]. More than half of all proteins are predicted to be glycoprotein in nature. Molecular chaperones and folding factors enclosed by the endoplasmic reticulum (ER) have a vital role in glycoprotein biogenesis [4,5]. N-linked oligosaccharide moieties are glycoprotein ligands that aid in various recognition processes. This is a common modification that contributes to diverse functions that include: stabilizing proteins against proteolysis and denaturation, modulating immune responses, balancing protein turnover and mediating protein-pathogen interactions. Most eukaryotic proteins consist of a clearly defined oligosaccharide core unit, which is comprised of three glucoses, nine mannoses and two N-acetyl-glucosamines (Glc3Man9GlcNAc2) [6].
Calreticulin and calnexin are intracellular lectin proteins, which can bind to glycoproteins to keep them bound to the ER until they are ready for migration [7]. N-glycosylation in the ER is initiated by transfer of Glc3Man9GlcNAc2 (G3M9) to asparagine (Asn) residues within Asn-X-Ser/Thr sequences in newly synthesized peptides from lipid-linked intermediates that are catalyzed by oligosaccharyltransferase (OST) [8]. The transmembrane protein, glucosidase I, rapidly removes the outermost glucose from G3M9. After this glucose is removed, G2M9 binds to malectin. Malectin is an ER-resident lectin protein, first identified in Xenopus laevis, that is involved in glycoprotein quality control. The name “malectin” was designated because the first disaccharide binds to this protein is maltose. Nuclear magnetic resonance (NMR) studies show that the malectin structure is similar to that of carbohydrate-binding modules (CBMs) in bacterial glycosyl hydrolases [9]. Unique selectivity of the ER intermediate Glc2Man9GlcNAc2 (G2M9) N-glycan to human malectin, with an acid dissociation constant (Ka) of 1.97 × 105 M−1, was confirmed by frontal affinity chromatography and in vitro membrane-based binding assays [10]. Recent studies show that malectin may be involved in innate immunity [11,12]. The malectin gene in scallops (Chlamys farreri) is involved in innate immunity [13]. Moreover, malectin stimulates M1/M2 macrophage polarization and inhibition on malectin expression by single nucleotide polymorphisms (SNPs) of MLEC gene leads to cerebral palsy [14]. The polarization of M1/M2 macrophage plays an essential role in immune system by regulating inflammatory responses [15].
Seahorses generally have low mobility, are sparsely distributed and slowly recolonize overexploited areas, have low fertility and require long durations of parental care. These behaviors and traits of seahorses reduce their environmental fitness and make them vulnerable to overfishing or habitat damage [16,17]. They reside in temperate and tropical coastal waters. They are commonly used in traditional Chinese medicine, tonics and as decorative species in aquariums [17]. Several countries, including Australia, New Zealand and China, are involved in commercial seahorse aquaculture [18,19]. However, seahorses are species that are included in the “convention on international trade in endangered species of wild fauna and flora” [18]. Hippocampus abdominalis (H. abdominalis) can differentiate into other seahorses by deepening of the trunk, and formation of tail rings and dorsal fin rays [18]. Various seahorse diseases were identified by observations in aquaculture and laboratory studies [20]. Mortality caused by Philasterides dicentrarchi was identified in aquarium seahorses that was used in public exhibition [21]. Fungal infection in H. abdominalis and mycobacterial infection in Hippocampus reidi have been identified [22]. Lymphocystis caused by iridovirus infection was reported in Hippocampus comes [23]. Understanding the immune system and immune responses will provide insight into diminishing mortality and diseases in seahorse species. Therefore, molecular identification, characterization and functional studies of immune system components may present an efficient way to maintain successful seahorse aquaculture.
In this study, we identified malectin from big-belly seahorse H. abdominalis (HaMLEC) and performed molecular characterization, as well as, spatial and temporal expression analysis. Structural similarity of HaMLEC with CBMs of bacterial glycosyl hydrolases were analyzed by evaluating bacterial binding and agglutination abilities of HaMLEC. Anti-viral activity of HaMLEC was analyzed using a fathead minnow (FHM) cell line overexpressing HaMLEC.
2. Materials and methods
2.1. Identification of seahorse malectin cDNA sequences
A previously constructed transcriptome library of H. abdominalis was used to identify the HaMLEC cDNA sequence. Identification was done by using the Basic Local Alignment Search Tool (BLAST) from the National Center for Biotechnology Information (NCBI) [24]. The Roche GS FLX 454 platform was used to build the transcriptome database of H. abdominalis [25]. HaMLEC cDNA sequence was amplified using HaMLEC-F1 and HaMLEC-R1 primers (Table 1 ), and its sequence was verified by cloning into T-vector PMD20, followed by capillary sequencing (Macrogen, Korea).
Table 1.
Sequences of primers used for HaMLEC amplification and qPCR.
| Primer Name | Purpose | Sequence (5′→3′) |
|---|---|---|
| HaMLEC-F1 | Gene amplification for cloning to T-vector PMD20 | AAGTGTTCCAGACTGAGACCTCCAC |
| HaMLEC-R1 | CTCAATGCGCTTCACACAAGCAGAC | |
| HaMLEC-F2 | Gene amplification for cloning to pMAL-c5X | GAGAGAggatccATGCAGCGGCTCACGG |
| HaMLEC-R2 | GAGAGAaagcttTCACAGCCGACAGAGACAGAACAAA | |
| HaMLEC-F3 | Gene amplification for cloning to pcDNA™3.1(+) | GAGAGAggtaccATGGAGCGGCTCACGGC |
| HaMLEC-R3 | GAGAGActcgagTCACAGCCGACAGAGACAGAACAAAG | |
| HaMLEC -qF | qPCR for HaMLEC expression | TTGGAAGGCAAACTCGGTAAAGCC |
| HaMLEC -qR | ATAGTCGCCTTCCTCACGTATTGGG | |
| 40S ribosomal protein S7-qF | qPCR for 40S ribosomal protein S7 | GCGGGAAGCATGTGGTCTTCATT |
| 40S ribosomal protein S7-qR | ACTCCTGGGTCGCTTCTGCTTATT | |
| EF1α_qF | qPCR internal reference of FHM cells | GGCTGACTGTGCTGTGCTGAT |
| EF1α_qR | GTGAAAGCCAGGAGGGCATGT | |
| GVHSVF | qPCR VHSV G protein | TACAACATCACCCTGCCCAACC |
| GVHSVR | GACCACCCTGTGATCATGTGTCC | |
| MVHSVF | qPCR VHSV M protein | CTGGTTCGCCTATTCCAGAGTGC |
| MVHSVR | GGTCCAGGTAAGTGGCCTTTGC | |
| NVHSVF | qPCR VHSV N protein | TGTCTCAGATCAGTGGGAAGTACGC |
| NVHSVR | GGACCTCAGCGACAAGTTCGG | |
| PVHSVF | qPCR VHSV P protein | CGACAACATACTCTCCATCC |
| PVHSVR | CCAAGTGCTCTCTCATTCC | |
| LVHSVF | qPCR VHSV L protein | CAAGTGCGGACACGATCAATCCC |
| LVHSVR | TGAGGAAAGGGCAACCATTCGC | |
| NVVHSVF | qPCR VHSV NonVirion protein | TCTCCACTTGTCCTTCGC |
| NVVHSVR | TCTCGAAGAAGTCTGTAGCG |
2.2. Molecular profiling of HaMLEC using bioinformatic tools
The open reading frame (ORF) and amino acid sequences of HaMLEC were determined by Unipro UGENE software (version 1.31.1) [26]. Physiochemical properties of HaMLEC were predicted by inputting the amino acid sequence into the ExPAsy ProtParam tool [27]. Online servers, ExPASy PROSITE [28] and Motif Scan (http://myhits.isb-sib.ch/cgi-bin/motif_scan) were used to predict conserved domains and motifs of HaMLEC. Protein sequence identity and similarity of HaMLEC with known malectin protein sequences were determined using the EMBOSS Needle Pairwise Sequence Alignment Tool (https://www.ebi.ac.uk/Tools/psa/emboss_needle/) [29]. Multiple sequence alignments of HaMLEC was performed with malectin sequences from various taxonomies using the ClustalW 2.0 program [30]. Construction of a phylogenetic tree was performed using the sequences of HaMLEC and other malectin proteins with 5000 bootstrap replicates in MEGA (version 7.0.26) software with the neighbor-joining method [31].
2.3. Acclimatization of experimental seahorse and tissue sampling
Seahorses were obtained from the Korean Marine Ornamental Fish Breeding Center (Jeju, South Korea). Seahorses with an average body weight of 8 g were acclimatized for one week in laboratory aquarium tanks (300 L) with average water temperature and salinity values of 18 °C ± 2 °C and 34% ± 0.6%, respectively. To analyze tissue-specific expression of HaMLEC, six healthy seahorses (3 males and 3 females) were dissected to collect fourteen different tissues, including: blood, liver, kidney, spleen, heart, brain, pouch, muscle, intestine, ovary, stomach, gills, skin and testis. Peripheral blood was obtained from experimental seahorses by cutting the tail and immediately pelleted by centrifugation at 4 °C at 3000 × g for 10 min. All of the isolated tissues were immediately snap-frozen in liquid nitrogen and kept at −80 °C for further steps.
2.4. Immune challenge experiment
Acclimatized seahorses were divided into five groups to perform immune challenge experiments. Four groups were separately challenged with live gram-positive (Streptococcus iniae) and gram-negative (Edwardsiella tarda) bacteria and two immune stimulants, lipopolysaccharide (LPS) from Escherichia coli (E. coli) 055:B5 (Sigma, St. Louis, MO, USA) and polyinosinic:polycytidylic (poly I:C) acid (Sigma, USA). The control group was injected with phosphate-buffered saline (PBS). During the challenge experiment, each group containing 35 seahorses was treated separately by intraperitoneal injection with 100 μL of 1 × 105 CFU/μL of S. iniae, 5 × 103 CFU/μL E. tarda, 1.5 μg/μL poly I:C and 1.5 μg/μL LPS prepared in PBS. Five individuals in each group were sacrificed 0 h, 3 h, 6 h, 12 h, 24 h, 48 h and 72 h post-injection and liver and blood tissues were obtained as mentioned in section 2.3.
2.5. Total RNA extraction and cDNA synthesis
Total RNA extraction was carried out using isolated tissues described in section 2.3. Tissues were prepared using equal weights of samples from each of the six seahorses to determine distribution in different tissues. Tissues collected from five treated seahorses at different time points were pooled for challenge experiment analysis. Total RNA extraction and purification was performed from prepared tissues using RNAiso plus total RNA extraction reagent (TaKaRa, Japan) and RNeasy Mini kit (Qiagen, USA), respectively. The concentration of purified RNA was assessed using a Multiskan™ GO Microplate Spectrophotometer at 260 nm (Thermo Scientific, USA). Purified RNA was subject to electrophoresis on a 1% agarose gel to determine the quality of purified RNA samples. Then, first-strand cDNA was synthesized from 2.5 μg of extracted RNA using Prime Script™ II 1st strand cDNA synthesis kit (TaKaRa, Japan). Synthesized cDNA samples were diluted 40-fold and stored at −80 °C.
2.6. Transcriptional analysis of HaMLEC by qRT-PCR
Quantitative real-time polymerase chain reaction (qRT-PCR) was used to analyze HaMLEC mRNA expression of cDNA samples prepared as described in section 2.5. Primers for HaMLEC and 40S ribosomal protein S7 gene-specific primers (Table 1) were designed using the PrimerQuest Tool from the IDT online server (https://sg.idtdna.com/Primerquest/Home/Index) [32]. cDNA was synthesized to test tissue distribution and temporal immune response by qRT-PCR, which was performed using Thermal cycler Dice® Real Time system III (TaKaRa, Japan). Preparation of reaction mixtures and qPCR temperature profile were as reported in our previous studies [33]. qRT-PCR was performed for 40S ribosomal protein S7 from H. abdominalis (accession number KP780177) as the internal control gene using the same conditions. Relative HaMLEC mRNA expression was determined with the 2-ΔΔCT method using the average of results from experiments performed in triplicate [34]. HaMLEC expression levels, following immune challenges, were normalized to determine tissue distribution and relative temporal expression. Tissue distribution and relative temporal expression were calculated with respect to the tissue with lowest expression and expression in the relevant PBS injected control, respectively.
2.7. Construction of eukaryotic and prokaryotic expression vectors
Prokaryotic and eukaryotic expression plasmids were constructed to study the functional role of HaMLEC through in vitro and in vivo experiments, respectively. A recombinant HaMLEC (rHaMLEC) expression vector was constructed to express rHaMLEC fused to a maltose binding protein (MBP) in a prokaryotic expression system using a modified version of our protocol as described in our previous studies [33]. Briefly, the ORF of HaMLEC was amplified by PCR using cDNA from ovarian tissue. During PCR amplification, restriction enzyme sites for BamHI and HindIII were added to the ORF by gene-specific primers HaMLEC-F3 and HaMLEC-R3 (Table 1), respectively. A PCR mixture was prepared to a total volume of 50 μL with nuclease-free water and included: 5 μL of 10 × ExTaq buffer, 4 μL of 2.5 mM dNTP, 10 pmol forward primer, 10 pmol reverse primer, 80 ng cDNA template and 5U ExTaq™ DNA polymerase (TaKaRa, Japan). PCR was performed using a TaKaRa thermal cycler with the following temperature conditions: initial denaturation at 94 °C for 5 min; 30 cycles at 94 °C for 30 s, 58 °C for 30 s and 72 °C for 50 s; and a final extension at 72 °C for 7 min. Agarose gel electrophoresis was carried out to separate the amplified gene product, and the excised band was purified from the gel using an AccuPrep™ Gel Purification Kit (Bioneer, Korea). The gene products and pMAL-c5X vector (New England Biolabs, USA) were separately digested with BamHI and HindIII restriction enzymes (TaKaRa, Japan) at 37 °C for 3 h. Digestion products were purified by gel electrophoresis with a 1% agarose gel. The digested gene product and vector were mixed in a 1:5 ratio in ligation Mighty Mix (TaKaRa, Japan). Ligation was carried out at 16 °C for 30 min, followed by overnight incubation at 4 °C. Finally, the ligated product was transformed into E. coli DH5α competent cells. The recombinant clones were selected with ampicillin and validated by sequencing (Macrogen, Korea). Sequencing confirmed that pMAL-c5X/HaMLEC was successfully transformed into E. coli BL21 (DE3) competent cells for protein expression (New England Biolabs, USA).
For in vivo experiments, the ORF of HaMLEC was cloned into a pcDNA™3.1(+) vector (Invitrogen™, USA) using KpnI and XhoI restriction sites. Cloning of CpcDNA™3.1(+)/HaMLEC was conducted as described for pMAL-c5X/HaMLEC by changing respective gene-specific primers (Table 1) and corresponding enzymes, KpnI and XhoI (TaKaRa, Japan). Subsequent to E. coli DH5α transformation, recombinant plasmids were validated by sequencing (Macrogen, Korea).
2.8. Prokaryotic overexpression and purification of rHaMLEC protein
E. coli BL21(DE3) transformed with pMAL-c5X/HaMLEC plasmids were incubated at 37 °C in Luria-Bertani (LB) medium, supplemented with 0.2 g/L glucose and 100 μg/mL ampicillin, at 200 rpm. The culture was induced using a 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) solution (Promega, USA) when the optical density (OD600) value was 0.6. The culture was further incubated for 8 h at 20 °C, shaking at 200 rpm. The culture was cooled on ice for 30 min after the induction process and centrifuged at 1200 × g for 30 min at 4 °C. The harvested pellet was washed by resuspension in column buffer at pH 7.4 (20 mM Tris-HCl, 200 mM NaCl and 1 mM DTT) and centrifuged at 1200 × g for 20 min at 4 °C. The pellet was kept overnight at −20 °C. The frozen pellet was thawed in cold running water and resuspended in column buffer. The cells were lysed by lysozyme treatment at a final concentration of 1 mg/mL, followed by cold sonication. The lysate was centrifuged at 9000 × g for 20 min at 4 °C, and the supernatant was incubated for 10 min with 1 mL of amylose resin (New England Biolabs, USA) before adding it to the column. After the addition of resin and supernatant mix, the column was washed with column buffer. Finally, the rHaMLEC was eluted using elution buffer (10 mM maltose in column buffer). The concentrations of rHaMLEC in eluted fragments were measured using a Bradford assay with bovine serum albumin (BSA) as the standard [35]. Protein purity was verified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with a 12% acrylamide gel.
2.9. Bacterial agglutination activity of rHaMLEC
Five bacterial strains, including 3 gram-negative (E. coli, E. tarda, Vibrio tapetis) and 2 gram-positive (S. iniae, Streptococcus parauberis) were used to elucidate agglutination ability of rHaMLEC, as described in a previous study [36]. Briefly, the bacterial strains were grown to mid-logarithmic phase, harvested and washed twice with tris-buffered saline (TBS). In a 96-well plate, 25 μL of bacterial suspension (1 × 106 CFU/mL) was incubated with 25 μL of protein (200 μg/mL) at room temperature for 1 h. Bacterial agglutination was observed using a light microscope (40X objective with 10X eyepiece magnification, Leica DMIL LED, Germany).
2.10. Microbial binding activity assay
An enzyme-linked immunosorbent assay (ELISA) was performed to evaluate bacterial binding activity of rHaMLEC. Briefly, bacterial suspensions (E. tarda, Lactococcus garvieae, Vibrio anguillarum, S. iniae, Vibrio harveyi, S. parauberis and E. coli) were prepared to a final concentration of 1 × 107 CFU/mL in coating buffer (30 mM Na2CO3 and 70 mM NaHCO3 at pH 9.0). In a 96-well ELISA plate, 50 μL of prepared bacterial suspension was added and incubated overnight at 4 °C. After washing with 1% TTBS solution (TBS with 0.1% Tween 20), the plate was blocked with 200 μL of BSA solution (1 mg/mL BSA in 0.1% TTBS) for 1 h at 37 °C. The plate was then rinsed with 1% TTBS, and respective wells were treated with 100 μL of 10 ng/μL rHaMLEC or 10 ng/μL rMBP. After 2 h incubation at 37 °C, the plate was washed three times with 1% TTBS and incubated for 2 h with 50 μL of mouse anti-MBP antibody (1:5000, New England Biolabs, USA) at 37 °C. The plate was rinsed again with 1% TTBS, and 50 μL of horseradish peroxidase (1:3000, HRP) conjugated goat anti-mouse secondary antibody (YOUNG IN, Frontier, South Korea) was added. After 2 h incubation at 37 °C, the plate was washed five times with 1% TTBS. Finally, the HRP substrate solution TMB was added for detection according to the manufacturer's protocol (Sigma, USA). OD was measured at 450 nm using Multiskan™ GO Microplate Spectrophotometer (Thermo Scientific).
2.11. Cell culture and transfection
FHM cells were cultured at 25 °C in Leibovitz's L-15 medium supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin and 100 μg/mL streptomycin. In order to perform the antiviral activity assay, constructed pcDNA™3.1(+)/HaMLEC and empty pcDNA™3.1(+) vectors were transiently transfected into FHM cells in separate wells of a 6-well plate. The transfection was performed with XtremeGENE™ 9 DNA transfection reagent (Roche, Germany) using the manufacturer's instructions. Viral hemorrhagic septicemia virus (VHSV) maintenance and propagation was performed as previously described [37].
2.12. Antiviral activity assay
VHSV (FWando05), isolated in Korea from VHSV-infected olive flounder (Paralichthys olivaceus), was used for this study [38]. FHM cells that were transfected with either pcDNA™3.1(+)/HaMLEC or pcDNA™3.1(+) empty vector were incubated at 25 °C for 24 h to allow for HaMLEC overexpression. Three wells of each condition were maintained in the 6-well plate. Each well was infected with 0.01 multiplicity of infection (MOI) of VHSV, and one well from each condition was maintained as an uninfected control. The cells were incubated at 20 °C, collected at 24 h post-infection and frozen at −80 °C until RNA extraction. Samples were prepared as triplicates. RNA extraction, cDNA synthesis and qPCR were performed as mentioned in sections 2.5, 2.6. Expression of internal control gene, elongation factor-1-α (EF-1α; accession number AY643400) and five VHSV genes, nucleoprotein (NP; AGS83377.1), phosphoprotein (PP; AGS83378.1), matrix protein (MP; AGS83379.1), glycoprotein (GP; AGS83380.1), non-virion (NV; AGS83381.1) and RNA-dependent RNA polymerase (L; AGS83380.1) were measured in FHM cells. qPCR detection of HaMLEC, EF-1α, and respective viral proteins were performed using the primer sets: HaMLEC-qF and HaMLEC-qR, EF1α_qF and EF1α_qR, GVHSVF and GVHSVR, MVHSVF and MVHSVR, NVHSVF and NVHSVR, PVHSVF and PVHSVR, NVVHSVF and NVVHSVR, and LVHSVF and LVHSVR (Table 1).
2.13. Statistical analysis
The microbial binding assay and qPCR were performed in triplicate, and represented as mean ± standard deviation (SD). Statistical significance (p < 0.05) was determined using the Student's t-test to compare the control with experimental conditions. The statistical significance of tissue distribution results was determined using one-way analysis of variance (ANOVA) using Tukey's multiple comparisons.
3. Results
3.1. Comprehensive molecular characterization of HaMLEC
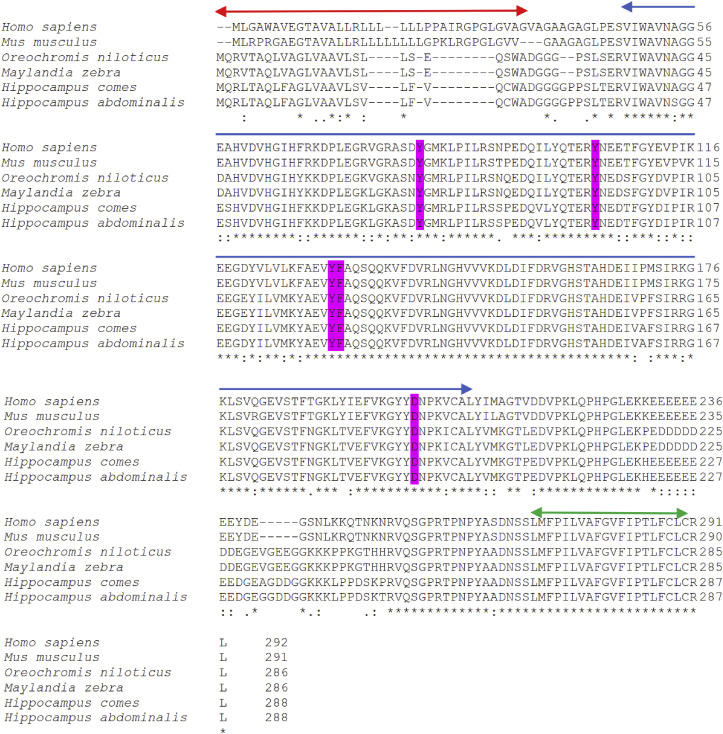
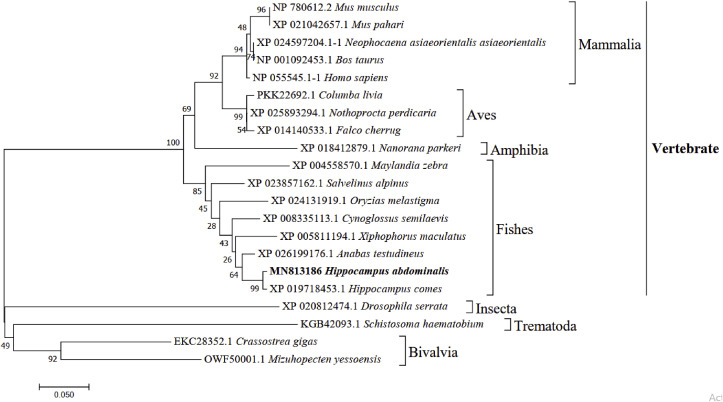
The HaMLEC cDNA sequence contains an ORF of 867 bp, which encodes 288 amino acids (Fig. 1 ). The molecular weight and isoelectric point (pI) of HaMLEC were estimated as 31.99 kDa and 5.17, respectively. The signal peptide was found with a cleavage site between amino acids (aa) 26 and 27. The HaMLEC sequence information were submitted to NCBI-GenBank database with the accession number MN813186. In silico study of the protein sequence revealed that the N-terminal signal peptide, malectin superfamily and C-terminal transmembrane region were positioned between 1 and 26 aa, 39–199 aa and 267–286 aa, respectively (Fig. 2 ). Pairwise alignment of HaMLEC showed high similarity (99.3%) and identity (99.0%) with Hippocampus comes. Additionally, HaMLEC sequence identity analysis with Anabas testudineus, Maylandia zebra, Homo sapiens, Nanorana parkeri and Crassostrea gigas were 92.4%, 85.8%, 71.9%, 61.6% and 41.7%, respectively (Table 2 ). According to phylogenetic analysis, HaMLEC subclustered with fish species and exhibited a close evolutionary relationship with Hippocampus comes (Fig. 3 ).
Fig. 1.
The cDNA and deduced amino acid sequence of HaMLEC. The start codon and stop codon of the ORF sequence are marked with bold letters and are underlined. The amino acid sequence of each codon and the stop codon are marked with the one letter amino acid code and asterisk (*). The N-terminal signal peptide, highly conserved malectin superfamily and C-terminal transmembrane helix regions are marked with green, purple, and red bold letters, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Fig. 2.
Multiple sequence alignment of HaMLEC with other homologues. N-terminal signal peptide (aa 1–26), highly conserved malectin superfamily (aa 39–199) and C-terminal transmembrane helix (aa 267–286) regions are marked with red, blue and green arrows, respectively. The amino acids mediating interaction with carbohydrates are highlighted in pink. The asterisk (*), colon (:) and period (.) below the alignment indicated the positions of fully conserved, strongly similar, and weak similar amino acid residues, respectively. Accession numbers for respective species are as follows: NP_055545.1 (Homo sapiens), NP_780612.2 (Mus musculus), XP_005473061.1 (Oreochromis niloticus), XP_004558571.1 (Maylandia zebra) and XP_019718453.1 (Hippocampus comes). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Table 2.
Pairwise sequence alignment of HaMLEC with its various orthologs.
| Scientific Name | Accession No | Taxon | Amino acid | Identity | Similarity |
|---|---|---|---|---|---|
| Hippocampus comes | XP_019718453.1 | Fish | 288 | 99.0 | 99.3 |
| Anabas testudineus | XP_026199176.1 | Fish | 288 | 92.4 | 95.8 |
| Cynoglossus semilaevis | XP_008335113.1 | Fish | 288 | 88.5 | 94.8 |
| Oryzias melastigma | XP_024131919.1 | Fish | 288 | 87.2 | 94.1 |
| Xiphophorus maculatus | XP_005811194.1 | Fish | 288 | 87.2 | 94.8 |
| Maylandia zebra | XP_004558570.1 | Fish | 288 | 85.8 | 94.4 |
| Salvelinus alpinus | XP_023857162.1 | Fish | 292 | 83.9 | 89.4 |
| Bos taurus | NP_001092453.1 | Mammalia | 293 | 74.4 | 84.3 |
| Homo sapiens | NP_055545.1-1 | Mammalia | 303 | 71.9 | 80.9 |
| Mus musculus | NP_780612.2 | Mammalia | 298 | 71.1 | 82.2 |
| Neophocaena asiaeorientalis asiaeorientalis | XP_024597204.1-1 | Mammalia | 306 | 70.3 | 80.1 |
| Nanorana parkeri | XP_018412879.1 | Amphibia | 333 | 61.6 | 72.4 |
| Nothoprocta perdicaria | XP_025893294.1 | Aves | 288 | 59.4 | 67.4 |
| Columba livia | PKK22692.1 | Aves | 288 | 58.7 | 67.4 |
| Mizuhopecten yessoensis | OWF50001.1 | Bivalvia | 307 | 46.3 | 62.5 |
| Crassostrea gigas | EKC28352.1 | Bivalvia | 290 | 41.7 | 53.8 |
| Drosophila serrata | XP_020812474.1 | Insecta | 362 | 37.3 | 52.8 |
| Schistosoma haematobium | KGB42093.1 | Trematoda | 300 | 25.0 | 35.3 |
Fig. 3.
Phylogenetic tree for different malectin orthologs with HaMLEC amino acid sequence using the neighbor-joining method. The values are shown at each branch node, indicating corresponding bootstrap values.
3.2. Spatial mRNA expression of HaMLEC
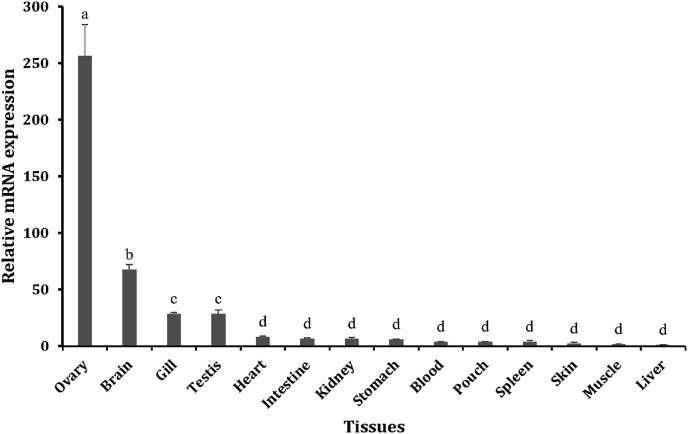
Tissue-specific analysis of transcription may provide information about divergent functionality of HaMLEC throughout the body in normal physiological conditions. HaMLEC expression was detected in all tested tissues. Elevated levels of HaMLEC mRNA expression were detected in the ovary. Levels were also elevated in the brain and gill tissues, and expression was lowest in liver tissues (Fig. 4 ).
Fig. 4.
Tissue-specific HaMLEC expression in vivo. Expression levels were analyzed by qPCR with 40S ribosomal protein S7 as a reference gene. Expression level in liver tissues was considered the basal level for comparing data. The relative mRNA expression of HaMLEC was analyzed according to the Livak method and data are presented as mean ± SD (n = 3). Statistical analysis was performed using triplicate data (2-ΔΔCt values obtained through Livak method analysis) by one-way ANOVA by Tukey's comparison (p < 0.05) and the statistically different expressions are indicated with different lowercase letters.
3.3. Analysis of HaMLEC expression levels after immune challenge
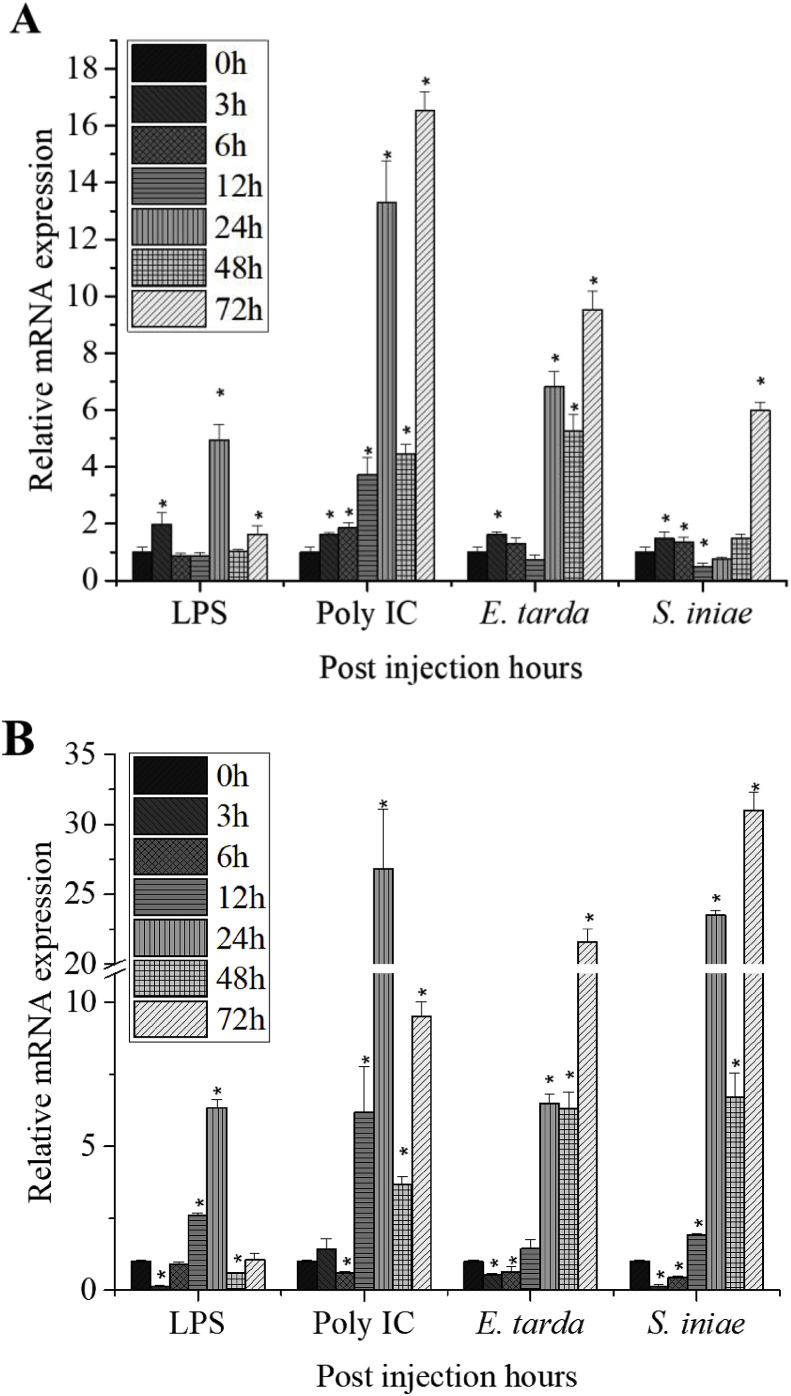
To analyze time-dependent immune responses, HaMLEC mRNA expression levels were determined by qPCR post-immune challenge (p.i). Variation in temporal expression of HaMLEC mRNA in blood tissue is shown in Fig. 5 A qPCR results from the LPS stimulated group showed significant up-regulation at 3 h, 24 h and 72 h p.i. The poly I:C-injected group showed significant up-regulation at all tested time points. The E. tarda-injected group showed significant up-regulation at 3 h, 24 h, 48 h and 72 h p.i. The S. iniae-injected group showed significant down-regulation at 12 h p.i, but significant up-regulation at 3 h, 6 h and 72 h p.i. Furthermore, higher fold changes in expression were observed in all four groups at later time points (24 h, 48 h and 72 h) compared to early time points (3 h, 6 h and 12 h).
Fig. 5.
Temporal expression levels of HaMLEC in (A) seahorse blood and (B) liver tissues after immune challenge with LPS, poly I:C, E. tarda and S. iniae. Mean values ± SD (n = 3) are represented by vertical bars. Significant differences (p < 0.05) between experimental and control (0 h) conditions are noted with asterisks (*).
Temporal expression of HaMLEC mRNA transcripts in liver tissue after immune challenge is shown in Fig. 5B. At early time points (3 h and 6 h), significant down-regulation was observed with all four stimulants. After 24 h, significant up-regulation was observed with all four stimulants and remained up-regulated with poly I:C, E. tarda, and S. iniae throughout later experimental time points (48 h and 72 h). The LPS-stimulated group only showed significant down-regulation at 48 h after immune challenge.
3.4. Overexpression and purification of rHaMLEC
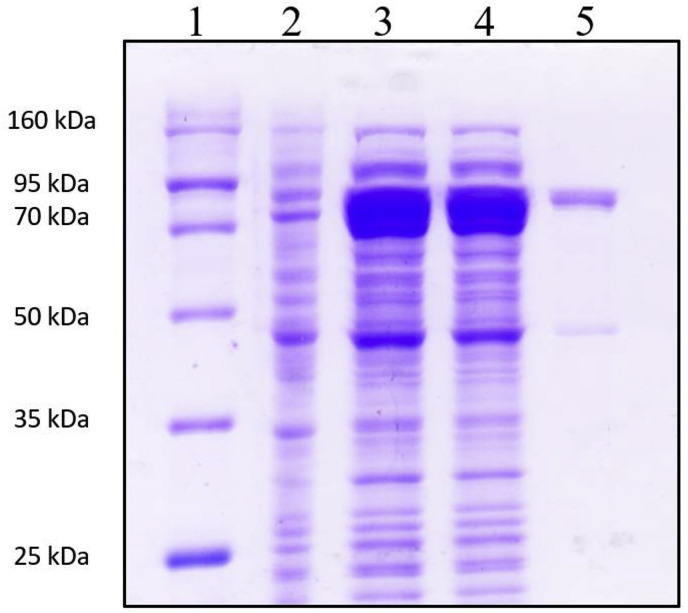
rHaMLEC was overexpressed in E. coli BL21(DE3) cells and purified using a pMAL protein fusion and purification system (New England Biolabs, USA). The following samples were analyzed by SDS-PAGE throughout the purification steps: total protein extract after sonication, supernatant after centrifugation, and purified protein (Fig. 6 ).
Fig. 6.
SDS-PAGE validation of purified rHaMLEC fusion protein. Lane 1 = protein size marker (Enzynomics-Korea); Lane 2 = supernatant from E. coli BL21 (DE3) transfected with rHaMLEC-pMAL-c5X; Lane 3 = total extract from IPTG-induced E. coli BL21 (DE3) cells transfected with rHaMLEC-pMAL-c5X; Lane 4 = supernatant of IPTG-induced E. coli BL21 (DE3) cells transfected with rHaMLEC-pMAL-c5X; Lane 5 = purified rHaMLEC-MBP fusion protein.
3. 5. Functional protein studies
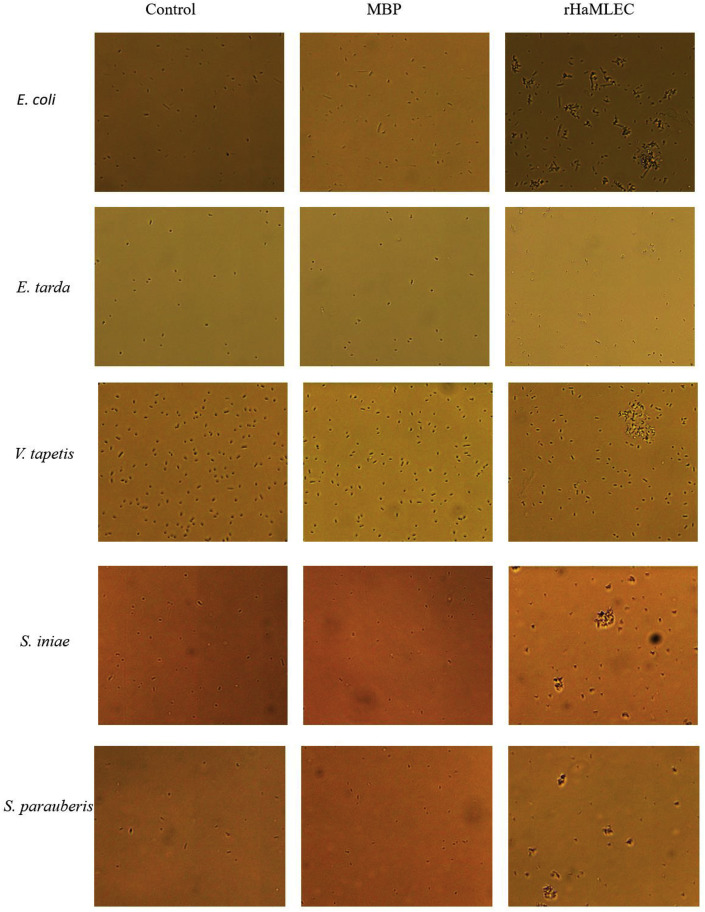
3.5.1. Agglutination of rHaMLEC on microbial pathogens
Bacterial agglutination activity was observed for rHaMLEC with gram-positive (E. coli, E. tarda, V. tapetis) and gram-negative (S. iniae, S. parauberis) bacterial strains. Agglutination was not observed with controls (rMBP and TBS). Images of bacterial agglutination acquired with light microscope are shown in Fig. 7 .
Fig. 7.
Bacterial agglutination by rHaMLEC. Bacterial strains were cultured at mid-logarithmic phase and harvested. Bacterial concentration was adjusted to 1 × 105 CFU/mL in TBS and mixed with 1 μg/mL protein. Agglutination of bacterial strains were observed under a light microscope.
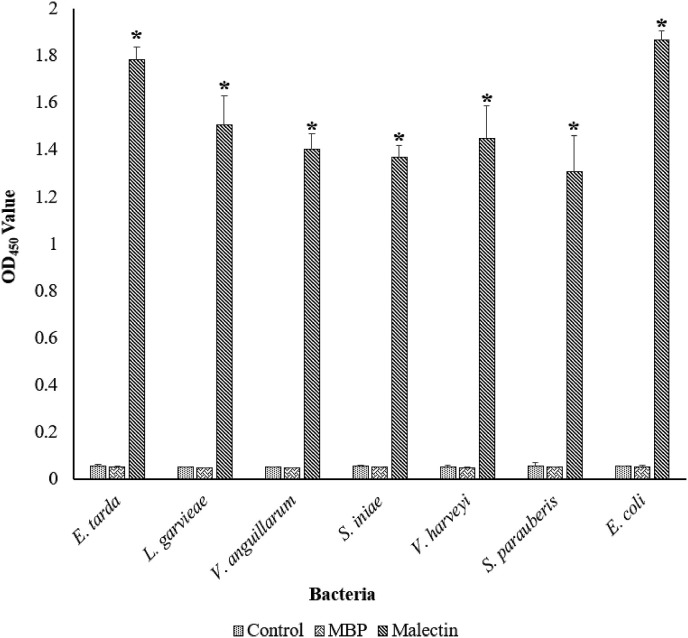
3.5.2. Bacterial binding activity of rHaMLEC
Microbial binding activity of rHaMLEC was investigated by indirect ELISA with different bacteria (E. tarda, L. garvieae, V. anguillarum, S. iniae, Vibrio harveyi, S. parauberis and E. coli). Significant OD450 values against bacteria with rHaMLEC are shown in Fig. 8 . As expected, the OD450 value in controls (rMBP and elution buffer) were negligible compared to rHaMLEC.
Fig. 8.
Assessing bacterial binding activity of rHaMLEC by ELISA. A 96-well ELISA plate was coated with 50 μL of bacterial suspensions at 1 × 107 CFU/mL and treated with 100 μL of 10 ng/μL rHaMLEC and rMBP. Mouse anti-MBP primary antibody and horseradish peroxidase (HRP)-conjugated secondary goat anti-mouse antibody were used. Bacterial binding was detected with HRP substrate solution TMB, and the OD450 was measured using a Multiskan™ GO Microplate Spectrophotometer.
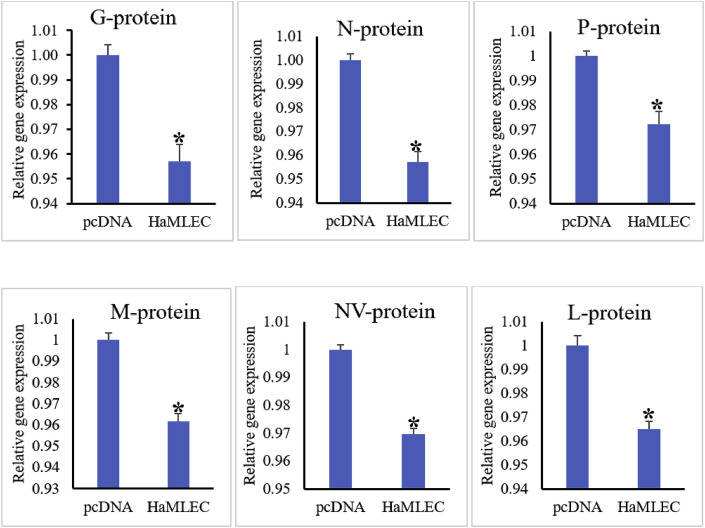
3.5.3. Reduction in virus transcription by HaMLEC
Significant down-regulation of VHSV gene transcript expression was observed at 24 h post-infection in FHM cells overexpressing HaMLEC (Fig. 9 ). Expression of all VHSV genes were down-regulated in HaMLEC/pcDNA™3.1(+)-transfected cells compared to pcDNA™3.1(+)-transfected cells. Overexpression of HaMLEC in cells transfected with HaMLEC/pcDNA™3.1(+) compared to cells transfected with pcDNA™3.1(+) was confirmed by qPCR with HaMLEC-qF and HaMLEC-qR primers.
Fig. 9.
Reduction in virus transcription by HaMLEC at 24 h post-viral inoculation. pcDNA™3.1(+)/HaMLEC and empty vector pcDNA™3.1(+) were transfected into FHM cells and treated with 0.01 MOI of VHSV. Cell samples were collected at 24 h post-viral infection, and transcription of viral proteins was examined by qPCR using FHMEF1α and viral-specific primers and analyzed using the livak method. Mean values ± SD (n = 3) are represented by vertical error bars. Expression of all viral proteins analyzed in this assay were significantly lower (p < 0.05) in pcDNA™3.1(+)/HaMLEC-transfected samples compared to pcDNA™3.1(+)-transfected samples.
4. Discussion
Malectin is a lectin protein that is localized to the ER and plays a role in glycoprotein quality control [9]. Significantly elevated malectin transcription was observed in ER-stress induced HEK293 cells by tunicamycin [10]. Recent studies indicate the potential role of malectin in immunity [[11], [12], [13]]. However, information about malectin in innate immunity is limited and no previous studies on teleost malectin have been reported. In this study, we characterized the malectin gene from H. abdominalis. Tissue-specificity and temporal expression of HaMLEC were analyzed, and functional assays were performed to predict its potential role in innate immunity. Previous studies revealed that malectin localizes to the ER membrane [9]. Accumulation of misfolded proteins in the ER induces the highest expression of malectin. Malectin and ribophorin-I form complex structures for selective retention of misfolded glycoproteins during ER quality control (ERQC) and transports them to the cytoplasm for ER-associated degradation (ERAD). Once malectin becomes highly expressed relative to ribophorin I expression under ER stress, the proportion of free malectin will increase and bind to G2M9 of glycoproteins. To reduce ER stress, the free malectin glycoprotein complex transports to the Golgi [39].
Malectin domains and the sequence identity of HaMLEC compared with other homologues were predicted by bioinformatics methods. High sequence identity was observed with teleosts, followed by other vertebrates, and less than 50% sequence identity with selected invertebrates. HaMLEC showed a close relationship with teleosts by phylogenetic analysis. The highly conserved malectin super family sequence, and the amino acids mediating the carbohydrate interaction, were observed in HaMLEC, as well as, with other vertebrate malectins through multiple sequence alignment (Fig. 2). Bioinformatic studies confirm similarity of vertebrate malectin sequences with the HaMLEC sequence. Moreover, HaMLEC was closely related to malectin of H. comes by phylogenetic analysis and pairwise sequence alignment.
In order to investigate the potential function of HaMLEC in H. abdominalis, tissue-specific expression of HaMLEC was analyzed by qPCR. HaMLEC mRNA transcripts were detected in all tested tissues. HaMLEC expression was highest in the ovary, followed by in the brain, gill and testis. Highest malectin expression was recorded in mature ovarian follicles in Danio rerio [40]. Ubiquitous distribution of malectin was observed in adult Xenopus laevis and has been implicated in embryonic development, preventing late organogenesis by inducing apoptosis or necrosis by inhibiting proliferation of malectin-expressing tissue [41]. The role of malectin in embryogenesis may explain why HaMLEC expression is highest in the ovaries. Gills are always exposed to the external environment and are more susceptible to pathogenic infection. In rainbow trout (Oncorhynchus mykiss), increased immune gene expression was observed during Ichthyophthirius multifiliis infection [42]. Disease outbreak in a turbot farm after Chryseobacterium scophthalmum infection was reported in Scotland by Mudarris and Austin, where they recovered dense bacterial cultures from the gills, brain and other organs [43]. Viral infection was also recorded in the brain of fish [44]. Moreover, recent studies on gilthead seabream and European sea bass proved that nervous necrosis virus (NNV) colonizes and produces infectious particles in male testis [45]. Highest expression of HaMLEC in the above tissues suggests that it might be involved in innate immune function in seahorses.
In order to understand the role of HaMLEC during pathogenic attack, temporal HaMLEC mRNA expression was investigated in two important immune tissues, blood and liver, following immune stimulation. The majority of plasma glycoproteins are synthesized in the liver [46]. Parenchymal hepatocytes occupy 70–85% of the liver, which actively synthesize large amounts of protein, including important innate immune proteins that are released into the blood [47]. Blood from the gut passes through the liver before it reaches the heart [48], therefore, foreign material exposure is greater in the liver. Certain types of white blood cells are a part of the immune system. Moreover, this consists of chemicals and proteins, such as interferon, complement proteins and antibodies. Some of these components can act against foreign substances directly, and the remaining help immune cells [49]. Significant increase in HaMLEC expression following exposure to all four stimulants implied that HaMLEC is involved in the immune system. However, information regarding the mechanism of the role of malectin in immune responses is insufficient. HaMLEC up-regulation in response to poly I:C in blood was significantly higher compared to the other stimulants, suggesting that HaMLEC might play an important role during viral infection. Significant changes in HaMLEC expression in blood and liver tissue following immune stimulation further indicates that HaMLEC might play an important role in innate immunity in H. abdominalis.
Lectin and carbohydrate interactions play an essential role in initiating host defense mechanisms [50]. They play a significant role in cell proliferation, phagocytosis, agglutination, signal transduction, opsonization, apoptosis and metastasis [[51], [52], [53]]. We performed bacterial binding and bacterial agglutination assays to analyze CBM structures in HaMLEC. Significant bacterial binding activity of HaMLEC was observed with a wide range of bacterial species, and negligible bacterial binding activity with MBP was observed compared to the control. Moreover, agglutination activity of HaMLEC with different bacteria validated the function of CBM-like structures in HaMLEC. The results from the agglutination assay indicted that rHaMLEC has the ability to recognize and bind a wide range of gram-positive (E. coli, E. tarda, V. tapetis, V. anguillarum) and gram-negative (S. iniae, S. parauberis) bacterial strains.
Most viruses typically require protein folding machinery of ER in the host to correctly fold their glycoprotein structures [[54], [55], [56], [57]]. Glycosylation has been examined in viral envelope glycoproteins, such as human immunodeficiency virus-1 (HIV-1) [58], Ebola virus glycoprotein (GP) [59], hemagglutinin glycoprotein (HA) of influenza virus [60], coronavirus glycoprotein spike (S) [61] and glycoprotein complex (GPC) of Lassa virus [62]. Glycosylation has also been examined in glycans that contain important secreted proteins, such as non-structural protein-1 (NS1) of flaviviruses [63], secreted GP of Ebola [64] and secreted glycoprotein G (sgG) of herpes simplex virus (HSV) [65]. Malectin selectively binds to glycoproteins in the calnexin chaperone system, without affecting glycoprotein entry and retention. During the ER stress condition, secretion of defective gene products is inhibited by interference of malectin, that might be the aberrant function generated in ER quality control [66]. Broad-spectrum antiviral development studies targeted partial inhibition of ER-folding mechanisms to prevent viral glycoprotein folding [67,68]. A possible role of malectin in glycoprotein quality control was suggested when a complex formed by malectin with ribophorin I was identified [69]. In addition, the association of malectin with newly synthesized misfolded influenza virus HA conformers also has been recorded [66].
To confirm the role of HaMLEC in preventing viral replication, we performed an antiviral activity assay with VHSV in FHM cells. VHSV causes viral haemorrhagic septicemia (VHS) disease in more than 70 species worldwide, including freshwater and marine fish [70]. Since 2001, VHS has affected aquaculture of olive flounder in Korea and caused substantial economic loss [71]. More than 90 marine and freshwater fish species in the world are affected by severe disease and mortality caused by VHSV [72,73]. VHSV is a negative-sense single-stranded RNA virus, which belongs to genus Novirhabdovirus and family Rhabdoviridae. VHSV has an 11 kb negative-sense RNA genome of linear single-stranded, consisting of 6 genes in the order 3′-N-P-M-G-NV-L-5′ encoding five structural proteins: nucleoprotein (N), phosphoprotein (P), matrix protein (M), glycoprotein (G), RNA polymerase (L) and a nonstructural protein (NV) [[74], [75], [76]]. Significant down-regulation of expression was observed for these 6 viral genes in FHM cells overexpressing HaMLEC, further validating that HaMLEC may have a significant role in viral replication in H. abdominalis.
In the current study, the identified HaMLEC was subjected to molecular analysis of its sequence and function. In silico analysis of HaMLEC confirms sequence similarity with known malectin sequences from other species. Analysis of spatial and temporal expression patterns of HaMLEC showed differential HaMLEC transcription in different tissues, and expression changes in liver and blood upon immune challenge. Bacterial binding and agglutination ability of HaMLEC confirmed the presence of CBM-like structures. Antiviral properties of HaMLEC were confirmed with VHSV. Collectively, the above results suggested that HaMLEC may play a significant role in innate immunity of big-belly seahorse.
CRediT authorship contribution statement
Sarithaa Sellaththurai: Conceptualization, Methodology, Investigation, Writing - original draft. K.A.S.N. Shanaka: Validation, Writing - review & editing. D.S. Liyanage: Formal analysis, Writing - review & editing. Hyerim Yang: Data curation. Thanthrige Thiunuwan Priyathilaka: Supervision. Jehee Lee: Resources, Writing - review & editing, Supervision, Project administration, Funding acquisition.
Acknowledgments
This research was a part of the project titled ‘Fish Vaccine Research Center’, funded by the Ministry of Oceans and Fisheries, Korea and supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2019R1A6A1A03033553).
References
- 1.Anfinsen C.B. Principles that govern the folding of protein chains. Science. 1973;181:223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- 2.Hartl F.U. Molecular chaperones in cellular protein folding. Nature. 1996;381:571–580. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- 3.Hartl F.U., Hayer-Hartl M. Converging concepts of protein folding in vitro and in vivo. Nat. Struct. Mol. Biol. 2009:574–581. doi: 10.1038/nsmb.1591. [DOI] [PubMed] [Google Scholar]
- 4.Apweiler R., Hermjakob H., Sharon N. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim. Biophys. Acta Gen. Subj. 1999;1473:4–8. doi: 10.1016/s0304-4165(99)00165-8. [DOI] [PubMed] [Google Scholar]
- 5.Schr M., Kaufman R.J. vol.569. 2005. pp. 29–63. (ER Stress and the Unfolded Protein Response). [DOI] [PubMed] [Google Scholar]
- 6.Helenius A., Aebi M. Roles of N-linked glycans in the endoplasmic reticulum. Annu. Rev. Biochem. 2004;73:1019–1049. doi: 10.1146/annurev.biochem.73.011303.073752. [DOI] [PubMed] [Google Scholar]
- 7.Gahmberg C.G., Tolvanen M. Why mammalian cell surface proteins are glycoproteins. Trends Biochem. Sci. 1996;21:308–311. [PubMed] [Google Scholar]
- 8.Kelleher D.J., Gilmore R. An evolving view of the eukaryotic oligosaccharyltransferase. Glycobiology. 2006;16:47–62. doi: 10.1093/glycob/cwj066. [DOI] [PubMed] [Google Scholar]
- 9.Schallus T., Jaeckh C., Fehér K., Palma A.S., Liu Y., Simpson J.C., Mackeen M., Stier G., Gibson T.J., Feizi T., Pieler T., Muhle-Goll C. Malectin: a novel carbohydrate-binding protein of the endoplasmic reticulum and a candidate player in the early steps of protein N-glycosylation. Mol. Biol. Cell. 2008;19:3404–3414. doi: 10.1091/mbc.E08-04-0354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Y., Hu D., Yabe R., Tateno H., Qin S.-Y., Matsumoto N., Hirabayashi J., Yamamoto K. Role of malectin in Glc2Man9GlcNAc2-dependent quality control of 1-antitrypsin. Mol. Biol. Cell. 2011;22:3559–3570. doi: 10.1091/mbc.E11-03-0201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang M., Wang B., Jiang K., Liu M., Shi X., Wang L. A mitochondrial manganese superoxide dismutase involved in innate immunity is essential for the survival of Chlamys farreri. Fish Shellfish Immunol. 2018;72:282–290. doi: 10.1016/j.fsi.2017.11.010. [DOI] [PubMed] [Google Scholar]
- 12.Mu Y., Li M., Ding F., Ding Y., Ao J., Hu S., Chen X. De novo characterization of the spleen transcriptome of the large yellow croaker (Pseudosciaena crocea) and analysis of the immune relevant genes and pathways involved in the antiviral response. PloS One. 2014;9 doi: 10.1371/journal.pone.0097471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang M.Q., Wang B.J., Liu M., Jiang K.Y., Wang L. 2019. The First Identification of a Malectin Gene (CfMal) in Scallop Chlamys Farreri : Sequence Features and Expression Profiles; pp. 25–33. [Google Scholar]
- 14.Shi W., Zhu Y., Zhou M., Ruan Y., Chen X., Chen X. Malectin gene polymorphisms promote cerebral palsy via M2-like macrophage polarization. Clin. Genet. 2018;93:794–799. doi: 10.1111/cge.13149. [DOI] [PubMed] [Google Scholar]
- 15.Jia J., Shi X., Jing X., Li J., Gao J., Liu M., Lin C.I., Guo X., Hua Q. BCL6 mediates the effects of Gastrodin on promoting M2-like macrophage polarization and protecting against oxidative stress-induced apoptosis and cell death in macrophages. Biochem. Biophys. Res. Commun. 2017;486:458–464. doi: 10.1016/j.bbrc.2017.03.062. [DOI] [PubMed] [Google Scholar]
- 16.Seahorse P., Centre F. Life history and ecology of seahorses : implications for conservation and management. J. Fish. Biol. 2004;65:1–61. [Google Scholar]
- 17.Vincent A.C.J., Giles B.G., Czembor C.A., Foster S.J. Trade in seahorses and other syngnathids in countries outside Asia. Fish. Cent. Res. Reports. 2011;19:2. [Google Scholar]
- 18.Lourie S. a, Foster S.J., Cooper E.W.T., Vincent A.C.J. A guide to the identification of seahorses, north. 2004. http://www.traffic.org/species-reports/traffic_species_fish29.pdf 23.
- 19.Woods C. 2007. Aquaculture of the Big-Bellied Seahorse Hippocampus abdominalis Lesson 1827 (Teleostei: Syngnathidae) pp. 19–21. Thesis. [Google Scholar]
- 20.Koldewey H.J., Martin-Smith K.M. A global review of seahorse aquaculture. Aquaculture. 2010;302:131–152. [Google Scholar]
- 21.Sang P.S., Jee E.H., Dennis K.G., Ji H.K., Casiano H.C.J., Jin W.J., Se C.P. Identification of scuticociliate Philasterides dicentrarchi from indo-pacific seahorses Hippocampus kuda. Afr. J. Microbiol. Res. 2011;5:738–741. [Google Scholar]
- 22.Koldewey H. Syngnathid husbandry in public aquariums. Proj. Seahorse. 2005:32–34. [Google Scholar]
- 23.FAO Fisheries & aquaculture - cultured aquatic species information programme - Hippocampus comes (Cantor, 1849), (n.d.). http://www.fao.org/fishery/culturedspecies/Hippocampus_comes/en (accessed September 6, 2019).
- 24.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 25.Oh M., Umasuthan N., Elvitigala D.A.S., Wan Q., Jo E., Ko J., Noh G.E., Shin S., Rho S., Lee J. First comparative characterization of three distinct ferritin subunits from a teleost: evidence for immune-responsive mRNA expression and iron depriving activity of seahorse (Hippocampus abdominalis) ferritins. Fish Shellfish Immunol. 2016;49:450–460. doi: 10.1016/j.fsi.2015.12.039. [DOI] [PubMed] [Google Scholar]
- 26.Okonechnikov K., Golosova O., Fursov M., Varlamov A., Vaskin Y., Efremov I., German Grehov O.G., Kandrov D., Rasputin K., Syabro M., Tleukenov T., Unipro U.G.E.N.E. A unified bioinformatics toolkit. Bioinformatics. 2012;28:1166–1167. doi: 10.1093/bioinformatics/bts091. [DOI] [PubMed] [Google Scholar]
- 27.Gasteiger E., Hoogland C., Gattiker A., Duvaud S., Wilkins M.R., Appel R.D., Bairoch A. Protein identification and analysis tools on the ExPASy server. Proteomics Protoc. Handb. 2005:571–607. [Google Scholar]
- 28.Sigrist C.J.A., de Castro E., Cerutti L., Cuche B.A., Hulo N., Bridge A., Bougueleret L., Xenarios I. New and continuing developments at PROSITE. Nucleic Acids Res. 2012;41:D344–D347. doi: 10.1093/nar/gks1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rice P., Longden L., Bleasby A. EMBOSS: the European molecular biology open software suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 30.Larkin M.A., Blackshields G., Brown N.P., Chenna R., Mcgettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., Thompson J.D., Gibson T.J., Higgins D.G. Clustal W and clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 31.Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Owczarzy R., Tataurov A.V., Wu Y., Manthey J.A., McQuisten K.A., Almabrazi H.G., Pedersen K.F., Lin Y., Garretson J., McEntaggart N.O., Sailor C.A., Dawson R.B., Peek A.S. IDT SciTools: a suite for analysis and design of nucleic acid oligomers. Nucleic Acids Res. 2008;36:163–169. doi: 10.1093/nar/gkn198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sellaththurai S., Priyathilaka T.T., Lee J. Molecular cloning, characterization, and expression level analysis of a marine teleost homolog of catalase from big belly seahorse (Hippocampus abdominalis) Fish Shellfish Immunol. 2019;89:647–659. doi: 10.1016/j.fsi.2019.03.064. [DOI] [PubMed] [Google Scholar]
- 34.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 35.Bradford Marion M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 36.Thulasitha W.S., Umasuthan N., Whang I., Nam B.H., Lee J. Antimicrobial response of galectin-1 from rock bream Oplegnathus fasciatus: molecular, transcriptional, and biological characterization. Fish Shellfish Immunol. 2016;50:66–78. doi: 10.1016/j.fsi.2016.01.010. [DOI] [PubMed] [Google Scholar]
- 37.Tharuka M.D.N., Priyathilaka T.T., Yang H., Pavithiran A., Lee J. Molecular and transcriptional insights into viperin protein from Big-belly seahorse (Hippocampus abdominalis) Fish Shellfish Immunol. 2019;86:599–607. doi: 10.1016/j.fsi.2018.12.006. [DOI] [PubMed] [Google Scholar]
- 38.Isshiki T., Nishizawa T., Kobayashi T., Nagano T., Migazaki T. An outbreak of VHSV (viral hemorrhagic septicemia virus) infection in farmed Japanese flounder Paralichthys olivaceus in Japan. Dis. Aquat. Org. 2001;47:87–99. doi: 10.3354/dao047087. [DOI] [PubMed] [Google Scholar]
- 39.Yang Q.P., Fu M.F., Gao H., Yamamoto K., Hu D., Qin S.Y. Subcellular distribution of endogenous malectin under rest and stress conditions is regulated by ribophorin i. Glycobiology. 2018;28:374–381. doi: 10.1093/glycob/cwy034. [DOI] [PubMed] [Google Scholar]
- 40.Mlec - malectin precursor - Danio rerio (Zebrafish) - mlec gene & protein. https://www.uniprot.org/uniprot/A9C3P0 n.d., accessed.
- 41.Jäckh C. 2008. Aus Heidelberg, Transcription Factor Networks Directing Pancreas Development in Xenopus laevis; p. 131. [Google Scholar]
- 42.Syahputra K., Kania P.W., Al-Jubury A., Marnis H., Setyawan A.C., Buchmann K. Differential immune gene response in gills, skin, and spleen of rainbow trout Oncorhynchus mykiss infected by Ichthyophthirius multifiliis. PloS One. 2019;14 doi: 10.1371/journal.pone.0218630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mudarris M., Austin B. Systemic disease in turbot Scophthalmus maximus caused by a previously unrecognised Cytophaga like bacterium. Dis. Aquat. Org. 1989;6:161–166. [Google Scholar]
- 44.Project A. 2018. Fish and Shrimp Viruses Session 2 : Fish Viruses. [Google Scholar]
- 45.Valero Y., Arizcun M., Esteban M.Á., Bandín I., Olveira J.G. 2015. Nodavirus Colonizes and Replicates in the Testis of Gilthead Seabream and European Sea Bass Modulating its Immune and Reproductive Functions; pp. 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ross G., Schachter H. Biosynthesis of glycoprotein by liver. The incorporation in vivo of 14C-glucosamine into protein-bound hexosamine and sialic acid of rat liver subcellular fractions. J. Biol. Chem. 1966;241:5408–5418. http://www.jbc.org/ accessed. [PubMed] [Google Scholar]
- 47.Gao B., Jeong W.-I., Tian Z. Liver: an organ with predominant innate immunity. Hepatology. 2007;47:729–736. doi: 10.1002/hep.22034. [DOI] [PubMed] [Google Scholar]
- 48.Svistounov D., N S., C V., Warren A., C A., Fraser R., Le Couteur D.G. Dyslipidemia - from Prev. To Treat. InTech; 2012. Liver sinusoidal endothelial cells and regulation of blood lipoproteins; pp. 263–278. [Google Scholar]
- 49.Immune response: MedlinePlus medical encyclopedia. https://medlineplus.gov/ency/article/000821.htm n.d., accessed.
- 50.Ni Y., Tizard I. Lectin-carbohydrate interaction in the immune system. Vet. Immunol. Immunopathol. 1996;55:205–223. doi: 10.1016/s0165-2427(96)05718-2. [DOI] [PubMed] [Google Scholar]
- 51.Nauta A.J., Castellano G., Xu W., Woltman A.M., Borrias M.C., Daha M.R., van Kooten C., Roos A. Opsonization with C1q and mannose-binding lectin targets apoptotic cells to dendritic cells. J. Immunol. 2004;173:3044–3050. doi: 10.4049/jimmunol.173.5.3044. [DOI] [PubMed] [Google Scholar]
- 52.Dutta S., Sinha B., Bhattacharya B. Characterization of a galactose binding serum lectin from the Indian catfish , clarias batrachus : possible involvement of fish lectins in differential recognition of pathogens. 2005;141:76–84. doi: 10.1016/j.cca.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 53.Tasumi S., Ohira T., Kawazoe I., Suetake H., Suzuki Y., Aida K. Primary structure and characteristics of a lectin from skin mucus of the Japanese eel. Anguilla japonica. 2002;277:27305–27311. doi: 10.1074/jbc.M202648200. [DOI] [PubMed] [Google Scholar]
- 54.Alonzi D.S., Scott K.A., Dwek R.A., Zitzmann N. Iminosugar antivirals : the therapeutic sweet spot. 2017:571–582. doi: 10.1042/BST20160182. 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mehta A., Lu X., Block T.M., Blumberg B.S., Dwek R.A. Hepatitis B virus (HBV) envelope glycoproteins vary drastically in their sensitivity to glycan processing: evidence that alteration of a single N-linked glycosylation site can regulate HBV secretion. Proc. Natl. Acad. Sci. Unit. States Am. 1997;94:1822–1827. doi: 10.1073/pnas.94.5.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hammond C., Braakman I., Helenius A. Role of N-linked oligosaccharide recognition, glucose trimming, and calnexin in glycoprotein folding and quality control. Proc. Natl. Acad. Sci. Unit. States Am. 1994;91:913–917. doi: 10.1073/pnas.91.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Block T.M., Lu X., Platt F.M., Foster G.R., Gerlich W.H., Blumberg B.S., Dwek R.A. Secretion of human hepatitis B virus is inhibited by the imino sugar N-butyldeoxynojirimycin. Proc. Natl. Acad. Sci. Unit. States Am. 1994;91:2235–2239. doi: 10.1073/pnas.91.6.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Struwe W.B., Chertova E., Allen J.D., Seabright G.E., Watanabe Y., Harvey D.J., Medina-Ramirez M., Roser J.D., Smith R., Westcott D., Keele B.F., Bess J.W., Sanders R.W., Lifson J.D., Moore J.P., Crispin M. Site-specific glycosylation of virion-derived HIV-1 env is mimicked by a soluble trimeric immunogen. Cell Rep. 2018;24:1958–1966. doi: 10.1016/j.celrep.2018.07.080. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee J.E., Fusco M.L., Hessell A.J., Oswald W.B., Burton D.R., Saphire E.O. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature. 2008;454:177–182. doi: 10.1038/nature07082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kobayashi Y., Suzuki Y. Evidence for N-glycan shielding of antigenic sites during evolution of human influenza A virus hemagglutinin. J. Virol. 2012;86:3446–3451. doi: 10.1128/JVI.06147-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ritchie G., Harvey D.J., Feldmann F., Stroeher U., Feldmann H., Royle L., Dwek R.A., Rudd P.M. Identification of N-linked carbohydrates from severe acute respiratory syndrome (SARS) spike glycoprotein. Virology. 2010;399:257–269. doi: 10.1016/j.virol.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sommerstein R., Flatz L., Remy M.M., Malinge P., Magistrelli G., Fischer N., Sahin M., Bergthaler A., Igonet S., ter Meulen J., Rigo D., Meda P., Rabah N., Coutard B., Bowden T.A., Lambert P.-H., Siegrist C.-A., Pinschewer D.D. Arenavirus glycan shield promotes neutralizing antibody evasion and protracted infection. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1005276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Flamand M., Megret F., Mathieu M., Lepault J., Rey F.A., Deubel V. Dengue virus type 1 nonstructural glycoprotein NS1 is secreted from mammalian cells as a soluble hexamer in a glycosylation-dependent fashion. J. Virol. 1999;73:6104–6110. doi: 10.1128/jvi.73.7.6104-6110.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mohan G.S., Li W., Ye L., Compans R.W., Yang C. Antigenic subversion: a novel mechanism of host immune evasion by Ebola virus. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1003065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gorander S., Mbwana J., Lyamuya E., Lagergard T., Liljeqvist J.-A. Mature glycoprotein G presents high performance in diagnosing herpes simplex virus type 2 infection in sera of different Tanzanian cohorts. Clin. Vaccine Immunol. 2006;13:633–639. doi: 10.1128/CVI.00051-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Galli C., Bernasconi R., Soldà T., Calanca V., Molinari M. Malectin participates in a backup glycoprotein quality control pathway in the mammalian ER. PloS One. 2011;6 doi: 10.1371/journal.pone.0016304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dwek R.A., Butters T.D., Platt F.M., Zitzmann N. Targeting glycosylation as a therapeutic approach. Nat. Rev. Drug Discov. 2002;1:65–75. doi: 10.1038/nrd708. [DOI] [PubMed] [Google Scholar]
- 68.Chang J., Guo J., Du Y., Block T. Imino sugar glucosidase inhibitors as broadly active anti-filovirus agents. Emerg. Microb. Infect. 2013;2:1–7. doi: 10.1038/emi.2013.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Qin S.Y., Hu D., Matsumoto K., Takeda K., Matsumoto N., Yamaguchi Y., Yamamoto K. Malectin forms a complex with ribophorin i for enhanced association with misfolded glycoproteins. J. Biol. Chem. 2012;287:38080–38089. doi: 10.1074/jbc.M112.394288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pierce L.R., Stepien C.A. Evolution and biogeography of an emerging quasispecies: diversity patterns of the fish Viral Hemorrhagic Septicemia virus (VHSv) Mol. Phylogenet. Evol. 2012;63:327–341. doi: 10.1016/j.ympev.2011.12.024. [DOI] [PubMed] [Google Scholar]
- 71.Kim W.-S., Kim S.-R., Kim D., Kim J.-O., Park M.-A., Kitamura S.-I., Kim H.-Y., Kim D.-H., Han H.-J., Jung S.-J., Oh M.-J. An outbreak of VHSV (viral hemorrhagic septicemia virus) infection in farmed olive flounder Paralichthys olivaceus in Korea. Aquaculture. 2009;296:165–168. [Google Scholar]
- 72.Manual of Diagnostic Tests for Aqatic Animals. fourth ed. 2003. pp. 86–87. [Google Scholar]
- 73.Einer-jensen K., Ahrens P., Forsberg R., Lorenzen N. 2018. Evolution of the Fish Rhabdovirus Viral Haemorrhagic Septicaemia Virus; pp. 1167–1179. [DOI] [PubMed] [Google Scholar]
- 74.Kurath G., Leong J.C. Characterization of infectious hematopoietic necrosis virus mRNA species reveals a nonvirion rhabdovirus protein. J. Virol. 1985;53:462–468. doi: 10.1128/jvi.53.2.462-468.1985. http://www.ncbi.nlm.nih.gov/pubmed/4038520 accessed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schütze H., Mundt E., Mettenleiter T.C. Complete genomic sequence of viral hemorrhagic septicemia virus, a fish rhabdovirus. Virus Gene. 1999;19:59–65. doi: 10.1023/a:1008140707132. http://www.ncbi.nlm.nih.gov/pubmed/10499451 accessed. [DOI] [PubMed] [Google Scholar]
- 76.Schutze H., Enzmann P.-J., Mundt E., Mettenleiter T.C. Identification of the non-virion (NV) protein of fish rhabdoviruses viral haemorrhagic septicaemia virus and infectious haematopoietic necrosis virus. J. Gen. Virol. 1996;77:1259–1263. doi: 10.1099/0022-1317-77-6-1259. [DOI] [PubMed] [Google Scholar]