Abstract
The purpose of this investigation was to study the viral aetiology of influenza-like illness (ILI) and acute respiratory tract infection (ARTI) among patients requiring intensive care unit admission.
A cross-sectional retrospective study was carried out in Sicily over a 4-year period. A total of 233 respiratory samples of patients with ILI/ARTI admitted to intensive care units were molecularly analyzed for the detection of a comprehensive panel of aetiologic agents of viral respiratory infections.
About 45% of patients was positive for at least one pathogen. Single aetiology occurred in 75.2% of infected patients, while polymicrobial infection was found in 24.8% of positive subjects. Influenza was the most common aetiologic agent (55.7%), especially among adults. Most of patients with multiple aetiology (76.9%) were adults and elderly. Mortality rates among patients with negative or positive aetiology did not significantly differ (52.4% and 47.6%, respectively).
Highly transmissible respiratory pathogens are frequently detected among patients with ILI/ARTI admitted in intensive care units, showing the occurrence of concurrent infections by different viruses. The knowledge of the circulation of several types of microorganisms is of crucial importance in terms of appropriateness of therapies, but also for the implication in prevention strategies and hospital epidemiology.
Keywords: Influenza-like illness, Viral infection, Co-infection, Intensive care units, Sicily, Italy
1. Introduction
According to the World Health Organization (WHO), influenza virus and acute respiratory tract infections (ARTIs) cause considerable morbidity and mortality worldwide, representing close to 20% of all deaths under 5 years of age, particularly in developing countries [1].
Moreover, the elderly or the immunocompromised subjects with influenza-like illness (ILI) or ARTI are at higher risk for exacerbations in pre-existing chronic conditions or progression to respiratory complications, which can be responsible of hospitalization and admission to an intensive care unit (ICU).
It has been demonstrated that several pathogens are implicated in ILI/ARTI, either of viral or bacterial origin, although specific aetiology often goes undiagnosed [2], [3]. Patients with acute respiratory illness admitted to the ICU, are usually monitored following a standard approach consisting of routine culture and testing of the bacterial agents commonly responsible of community-acquired pneumonia. Conversely, viral agents are generally underestimated in their clinical relevance [4] and laboratory detection is often used as second-line diagnostics or limited to patients negative for other bacterial pathogens, consequently leading to a delayed administration of potentially beneficial antiviral drugs [5].
Additionally, it is well-known that patients showing acute febrile respiratory illnesses may facilitate the spread of infectious agents, especially during the early phase of the disease, often sustained by aerosol-generating procedures able to favor patient-to-patient dissemination in ICU [6] or to healthcare workers (HCWs), visitors, and ultimately the community. This evidence can be further enhanced by well documented low vaccination coverage rates among HCWs versus influenza or other preventable infectious respiratory diseases [7], [8], as well as by an inappropriate habit of working while symptomatic for ILI [9].
Although the impact of influenza infection in intensive care wards have been previously described, particularly during the 2009H1N1 influenza pandemic [10], only a limited number of studies have focused on the complex aetiology of ILI/ARTIs in critically ill patients admitted to an ICU.
In this context, a better understanding of a wide spectrum of viral respiratory pathogens causing acute infections among hospitalized patients in these settings is essential for improving preventive and therapeutic strategies and prioritizing diagnostic efforts.
The purpose of this a 4-year cross-sectional study was to evaluate the burden of ILIs and/or ARTIs among hospitalized patients admitted to intensive care units in Sicily and to determine the specific aetiology of respiratory infections.
2. Materials and methods
2.1. Study design and settings
A cross-sectional retrospective study was carried out, from July 2009 to December 2012, on 253 different biological samples (oropharyngeal swabs or bronco-alveolar lavages for intubated patients) consecutively collected from patients admitted to intensive care unit with clinical symptoms of ILI/ARTI. ILI and ARTI were defined according to the European Centre for Disease Prevention and Control (ECDC) (http://ecdc.europa.eu/en/healthtopics/influenza/surveillance/Pages/influenza_case_definitions.aspx; accessed: 17/11/2015). Twenty-two different hospitals located in Sicily, an Italian region with a population of over 5 million people (http://demo.istat.it/pop2013/index.html) participated to the study.
On the basis of the current national healthcare system of Italy, patients were stratified in three different age-groups: ≤14 years, 15–64 years, and ≥65 years (pediatric, adult, and elderly patients, respectively).
A cross-linkage with the Hospital Discharge Records (HDRs) available for the Sicilian Region (Regional Department of Health, Epidemiological Observatory) was performed in order to confirm all information or to obtain additional epidemiological data concerning International Classification of Disease (ICD9-CM) codes, length of hospital stay, and deaths.
Consequently, the study population consisted of patients admitted to ICUs with ILI/ARTI clinical symptoms for whom HDRs included ICD9-CM codes suggestive of ILI/ARTI as primary or secondary diagnosis. The final coding set was selected according to Marsden-Haug and colleagues [11] with the inclusion of ICD9-CM codes for ARTI (i.e. 481, 482.x).
2.2. Detection of respiratory pathogens
Each respiratory specimen was transported at 4 °C and stored at −80 °C upon arrival before processing at the laboratory of the Clinical Epidemiology Unit of the University Hospital “P. Giaccone” in Palermo (Sicily, Italy).
Samples were analysed for the detection of specific viral genomes belonging to human Respiratory Syncytial virus type A (hRSV-A) and B (hRSV-B), Parainfluenza virus 1–4 (hPIV-1/4), Metapneumovirus A (hMPV-A) and B (hMPV-B), Coronavirus OC43 (hCoV-OC43), HKU1 (hCoV-HKU1), NL63 (hCoV-NL63) and 229E (hCoV-229E), Enteroviruses (hEV), Rhinoviruses (hRV), Parechovirus (hPeV), Influenza viruses type A (FluA), B (FluB) and C (FluC), Bocavirus 1–4 (hBoV-1/4), Adenoviruses (hAdV), WU and KI Polyomaviruses (WUPyV and KIPyV). Nucleic acids were extracted from 140 μL of each sample using the QIAamp Viral RNA Mini Kit or the QIAamp DNA Mini Kit (QIAGEN, Milan, Italy) according to manufacturer's protocol.
The detection of respiratory pathogens was performed with singleplex real-time PCR (rPCR) assays using the TaqMan technology and run on a 7000 Real-Time PCR System platform (Applied Biosystems, Life Technologies); primers and probes sets are described in Supplementary Table 1.
For the detection of RNA viruses, we performed RT-rPCR as a single step using the Superscript III® Platinum® one-step Quantitative RT-PCR System (Invitrogen, Life Technologies), while for DNA viruses, the Platinum® Quantitative PCR Supermix-UDG with ROX (Invitrogen, Life Technologies) was used according to manufacturer's instruction.
Each run included positive and negative controls for each target. For FluA and FluB RT-rPCR assays wild type viruses were used as positive controls, whereas for each other target a plasmid standard was used, after cloning into TOPO® TA vector (Life Technologies) a synthetic construct (GeneArt® Gene Synthesis, Life Technologies) or a PCR product.
2.3. Data analysis
Descriptive statistics were calculated to explore both the burden of ILI/ARTI among ICU patients and the distribution of single and multiple aetiology by gender, age, length of hospital stay, and adverse outcomes.
Categorical variables were described as frequencies and percentages and were compared with the chi-square test or Fisher's exact test where appropriate. Continuous variables were presented as the median and interquartile range and compared using the Student t test once normality was demonstrated; otherwise, the nonparametric Mann–Whitney U test was performed. Cochran–Armitage test for trend was used to compare prevalences across different subgroups. All tests were two-tailed and a p-value of 0.05 was considered statistically significant. All analyses were performed with STATA version 12.1 for Macintosh (Apple) [12].
2.4. Ethics statement
The present work was reviewed and approved by the institutional review board of the University Hospital “P. Giaccone” of Palermo, health data were stored according to the Italian laws on privacy, and the research was conducted following the Helsinki declaration statements. A written informed consent was waived due to the non-interventional design.
3. Results
A schematic diagram of inclusion/exclusion criteria used for the selection of study/reference populations is depicted in Fig. 1 .
Fig. 1.
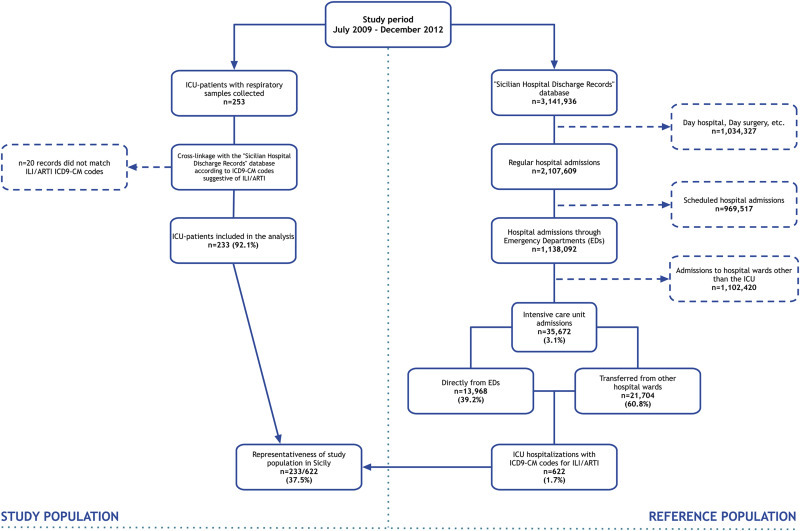
Schematic diagram of study/reference population inclusion and exclusion.
According to the HDRs database of Sicily, a total of 3,141,936 admissions were recorded during the study period, of which 67.1% (n = 2,104,609/3,141,936) were regular hospital admissions and about one-half (54.0%; n = 1,138,092/2,107,609) were through Emergency Departments (EDs).
On the whole, ICU hospitalizations in Sicily accounted for 3.1% (n = 35,672/1,138,092) of total ED-related admissions and about 1.7% (n = 622/35,672) included ICD9-CM codes suggestive of ILI/ARTI in their health records.
During the same period, a total of 253 respiratory samples were collected from ICU patients with ILI/ARTI. Of these, 233 patients (92.1%) were retrospectively found to match the inclusion criteria and were included in the final study group. Therefore, we estimate to have included in the analysis about 40% (n = 233/622) of all Sicilian hospitalized critically ill patients showing an ILI/ARTI presentation.
Table 1 reports the demographic and clinical characteristics of ICU hospitalized patients suffering from ILI/ARTI, according to the status of infection.
Table 1.
Demographic and clinical characteristics of 233 ICU hospitalized patients in Sicily presenting ILI/ARTI, according to the status of infection. Period: July 2009–December 2012.
| Characteristics | Total (% by column) | Negative | Positive (single + co-infection) | Single infection | Co-infection | p-value |
|---|---|---|---|---|---|---|
| ICU hospitalized subjects [n(%)] | 233 (100.0) | 128 (54.9) | 105 (45.1) | 79 (75.2) | 26 (24.8) | |
| Sex [n(%)] | ||||||
| Male | 129 (55.4) | 75 (58.1) | 54 (41.9)b | 41 (75.9) | 13 (24.1) | 0.27b |
| Female | 104 (44.6) | 53 (51.0) | 51 (49.0)b | 38 (74.5) | 13 (25.5) | |
| Age [median (IQR), years] | 52.9 (36.6) | 58.3 (32.5)c | 46.0 (47.6)c | 44.6 (53.0)d | 49.4 (39.4)d | <0.01c, 0.59d |
| Age group (years) [n(%)] | ||||||
| ≤14 | 41 (17.6) | 15 (36.6) | 26 (63.4)e | 20 (76.9) | 6 (23.1) | <0.01e |
| 15–64 | 117 (50.2) | 64 (54.7) | 53 (45.3)e | 41 (77.4) | 12 (22.6) | |
| ≥65 | 75 (32.2) | 49 (65.3) | 26 (34.7)e | 18 (69.2) | 8 (30.8) | |
| Length of hospital stay [median (IQR), days] | 18.5 (25) | 21 (25)f | 18 (28)f | 18 (29)g | 15 (15)g | 0.79f, 0.96g |
| Adverse outcomes [n(%)] | ||||||
| Pneumonia | 162 (69.5)a | 87 (53.7) | 75 (46.3) | 56 (74.7) | 19 (25.3) | |
| Respiratory failure | 105 (45.1)a | 59 (56.2) | 46 (43.8) | 35 (76.1) | 11 (23.9) | |
| ARDS | 59 (25.3)a | 28 (47.5) | 31 (52.5) | 24 (77.4) | 7 (22.6) | |
| Sepsis/Septic shock | 12 (5.2)a | 8 (66.7) | 4 (33.3) | 4 (100.0) | 0 (0) | |
| Death | 84 (36.1)a | 44 (52.4)h | 40 (47.6)h | 32 (80.0) | 8 (20.0) | 0.56h |
Percentages are not mutually exclusive; IQR: Interquartile range; ARDS: Acute Respiratory Distress Syndrome.
p value associated with the comparison of positivity between male and female.
p value associated with the comparison of median age between negative and positive patients.
p value associated with the comparison of median age between groups of patients with single or multiple infection.
p value associated with the test for trend of prevalences of positivity (microorganism detection) vs. age groups.
p value associated with the comparison of length of hospital stay between negative and positive patients.
p value associated with the comparison of length of hospital stay between groups of patients with single or multiple infection.
p value associated with the comparison of median age at death between negative and positive patients.
Overall, there were 129/233 (55.4%) males and 104/233 (44.6%) females (ratio M/F = 1.24). Among all patients, 105/233 (45.1%) were positive for at least one of the selected pathogens, without difference by sex (p = 0.27), most of whom (n = 79/105; 75.2%) had a single infection.
Age ranged between 1 day and 88 years (median 52.9 years, IQR = 36.6 years). Patients with laboratory-confirmed viral aetiology were significantly younger than negative subjects (46.0 years vs. 58.3 years; p < 0.01), but with no significant age-difference between groups with single and multiple infections (44.6 years vs. 49.4 years; p = 0.59, respectively).
Although non-pediatric subjects covered more than 80% of all ILI/ARTI cases in Sicily during the study period, higher positive rates of infection were observed among younger individuals (63.4%), and the proportion of positive subjects significantly decreased with age (p < 0.01).
Length of hospital stay differed neither between negative and positive patients (p = 0.79) nor between subjects showing single or co-infections (p = 0.96).
Pneumonia was the prevalent adverse outcome (n = 162/233; 69.5%), followed by respiratory failure (n = 105/233; 45.1%), and acute respiratory distress syndrome (ARDS) (n = 59/233; 25.3%). Sepsis and/or septic shock was documented in clinical records of 12/233 (5.2%) ICU-patients.
Eighty-four patients with ILI/ARTI presentation deceased and no difference was observed in relation to the aetiology (p = 0.56; 44 vs. 40 deaths among negative and positive subjects, respectively).
In general, ILI/ARTI cases were reported throughout the study period, with seasonal influenza confirmed infections mainly detected during the period October–March, while other different viral aetiologies were also found from August to September and from April to May (for more details see Supplementary Fig. 1). Supplementary Fig. 2 depicts the proportions of laboratory-confirmed samples, according the aforementioned three different age-groups and microorganisms.
Study population with laboratory-confirmed positivity was stratified according to a single or a multiple detection (Table 2 ). About half of all subjects with mono-infections (n = 41/79; 51.9%) were adults. In general, influenza viruses were the most common aetiologic agents (n = 44/79; 55.7%), and influenza A(H1N1)pdm09 prevailed, especially among adults (n = 32/44; 72.7%).
Table 2.
Distribution of single (a) and multiple aetiology (b) among ICU hospitalized patients in Sicily presenting ILI/ARTI, according to age-group. Period: July 2009 to December 2012.
| ICU hospitalized subjects |
|||||
|---|---|---|---|---|---|
| Total (% by column) | ≤14 yrs (% by row) | 15–64 yrs (% by row) | ≥65 yrs (% by row) | ||
| (a) | Microorganism: single aetiology [n(%)] | ||||
| N° of infected subjects | 79 (100.0) | 20 (25.3) | 41 (51.9) | 18 (22.8) | |
| Influenza viruses | 44 (55.7) | 9 (20.5) | 32 (72.7) | 3 (6.8) | |
| A(H1N1)pdm09 | 41 (51.8) | 9 (22.0) | 29 (70.7) | 3 (7.3) | |
| Type A - untypable | 2 (2.5) | 0 (0) | 2 (100.0) | 0 (0) | |
| Type B | 1 (1.3) | 0 (0) | 1 (100.0) | 0 (0) | |
| Human parainfluenza viruses (hPIV) | 11 (13.9) | 3 (27.3) | 3 (27.3) | 5 (45.5) | |
| hPIV-1 | 4 (5.1) | 2 (50.0) | 0 (0) | 2 (50.0) | |
| hPIV-2 | 3 (3.8) | 1 (33.3) | 1 (33.3) | 1 (33.3) | |
| hPIV-3 | 3 (3.8) | 0 (0) | 1 (33.3) | 2 (66.7) | |
| hPIV-4 | 1 (1.3) | 0 (0) | 1 (100.0) | 0 (0) | |
| Human enterovirus (hEV) | 9 (11.4) | 2 (22.2) | 4 (44.4) | 3 (33.3) | |
| Human respiratory syncytial viruses (hRSV) | 6 (7.6) | 3 (66.7) | 0 (0) | 3 (33.3) | |
| hRSV-A | 4 (5.1) | 1 (25.0) | 0 (0) | 3 (75.0) | |
| hRSV-B | 2 (2.5) | 2 (100.0) | 0 (0) | 0 (0) | |
| Human coronaviruses (hCoV) | 4 (5.1) | 1 (25.0) | 2 (50.0) | 1 (25.0) | |
| hCoV-NL63 | 2 (2.5) | 1 (50.0) | 1 (50.0) | 0 (0) | |
| hCoV-HKU1 | 1 (1.3) | 0 (0) | 1 (100.0) | 0 (0) | |
| hCoV-OC43 | 1 (1.3) | 0 (0) | 0 (0) | 1 (100.0) | |
| Human rhinovirus (hRV) | 2 (2.5) | 1 (50.0) | 0 (0) | 1 (50.0) | |
| Human bocavirus type 1 (hBoV-1) | 2 (2.5) | 1 (50.0) | 0 (0) | 1 (50.0) | |
| Human KI polyomavirus (KIPyV) | 1 (1.3) | 0 (0) | 0 (0) | 1 (100.0) | |
| (b) | Microorganism: multiple aetiology [n(%)] | ||||
| N° of co-infected subjects | 26 (100.0) | 6 (23.1) | 12 (46.2) | 8 (30.8) | |
| Influenza A(H1N1)pdm09 + | |||||
| Influenza type C | 1 (3.8) | 0 (0) | 1 (100.0) | 0 (0) | |
| hPIV-2 | 1 (3.8) | 0 (0) | 1 (100.0) | 0 (0) | |
| hPIV-3 | 1 (3.8) | 0 (0) | 1 (100.0) | 0 (0) | |
| hEV | 2 (7.7) | 0 (0) | 2 (100.0) | 0 (0) | |
| hCoV-OC43 | 2 (7.7) | 0 (0) | 1 (50.0) | 1 (50.0) | |
| hCoV-NL63 | 1 (3.8) | 0 (0) | 1 (100.0) | 0 (0) | |
| hMPV-A | 1 (3.8) | 0 (0) | 0 (0) | 1 (100.0) | |
| hMPV-B | 2 (7.7) | 0 (0) | 2 (100.0) | 0 (0) | |
| hRV | 1 (3.8) | 1 (100.0) | 0 (0) | 0 (0) | |
| Influenza type A (untypable) + hMPV-B + hEV | 1 (3.8) | 1 (100.0) | 0 (0) | 0 (0) | |
| Human metapneumoviruses (hMPV) | |||||
| hMPV-A + hMPV-B | 5 (19.2) | 2 (40.0) | 1 (20.0) | 2 (40.0) | |
| hRSV-A + hCoV-OC43 | 1 (3.8) | 1 (100.0) | 0 (0) | 0 (0) | |
| hRSV-B + hCoV-229E | 1 (3.8) | 0 (0) | 0 (0) | 1 (100.0) | |
| hRV + hEV | 3 (11.5) | 1 (33.3) | 1 (33.3) | 1 (33.3) | |
| hPIV-1 + hRV | 1 (3.8) | 0 (0) | 0 (0) | 1 (100.0) | |
| hPIV-3 + hCoV-OC43 | 1 (3.8) | 0 (0) | 0 (0) | 1 (100.0) | |
| hPIV-2 + hMPV-A + hMPV-B + hCoV-NL63 | 1 (3.8) | 0 (0) | 1 (100.0) | 0 (0) | |
Human parainfluenza viruses and enteroviruses had similar detection frequencies (13.3% and 11.4%, respectively), six out of 79 patients (7.6%) were infected with human respiratory syncytial viruses, while in four patients (5.1%) human coronaviruses (n = 2, hCoV-NL63; n = 1, hCoV-HKU1; and n = 1, hCoV-OC43) were found. Finally, 2/79 subjects (2.5%) were positive for human rhinovirus, as well as for human bocavirus type 1; only one patient (1.3%) was positive for human KI polyomavirus.
AdV, hPeV and WUPyV were not detected at all. Most of ICU-patients with laboratory-confirmed polymicrobial aetiology (n = 20/26; 76.9%) were adults and elderly (Table 2b).
4. Discussion
During the 4-year period of this study, more than 3 millions hospitalizations were recorded in Sicily, and the admission rate to intensive care from EDs documented in our area (3.1%) is comparable to that reported in other developed countries [13], [14]. However, many differences still remain because of a high variability of the threshold for ICU-admission, in respect of intensive care services provided in different healthcare systems, clinical management strategies of critically ill patients, resources, and ultimately availability of ICU beds [15].
To date, respiratory infections still represent a major concern and a leading cause of hospital admission to intensive care [13], [15], although the burden and aetiology of influenza-like illnesses and acute respiratory infections has not yet been well-defined [6].
The key finding of this present study was that viral infections affected a large proportion of ICU patients with ILI/ARTI, and the detection rate of 45.1% in our study population was significantly higher than previously reported in other European countries [16].
According to other authors [17], influenza virus was the most frequently detected microorganism, and coincident detection of different viral pathogens constituted about 25% of total infections, fitting well with the results of similar reports [16].
In this regard, a lower detection rate found in other different settings of non-ICU hospitalized patients [18], and the potential synergism between viruses and bacteria, which can lead to more severe disease and excess mortality [19], underline the importance of a complete microbiological investigation, particularly in a such group of severely ill patients at higher risk of death.
Other respiratory pathogens were found in our series, such as human parainfluenza viruses and coronaviruses (13.3% and 5.1%, respectively), which have also been described to cause significant symptoms, morbidity and potential need of intensive care treatment [4]. Surprisingly, microorganisms such as hRV, hRSV, hMPV, commonly reported in several studies and also responsible for admission to ICU and mortality in the elderly [6], [17], were found at very low prevalences and often co-detected with other respiratory agents.
In such a scenario, regardless the frequency of detection of different infectious agents, the knowledge of the circulation of several types of microorganisms may be essential from the standpoint of infection prevention and hospital epidemiology, even more crucially, in intensive care wards. In fact, different situations may be responsible for healthcare facility outbreaks of highly transmissible respiratory pathogens. On one hand, the lack of appropriate isolation strategies of high vulnerable patients with acute febrile respiratory illness prior to diagnosis of potentially infectious agent [6], [20], and on the other hand, the very low compliance with vaccination and coverage rates among HCWs [8], [21], [22], [23], further worsened by improper behaviours such as “presenteeism” (working while ill rather than take sick leave), which is prevalent among HCWs [9], [24], despite recent recommendation from the Centers for Disease Control and Prevention on prevention strategies for ILI in healthcare setting [25].
We are aware of some limitations of this study. First, this was a descriptive retrospective study and no comparison groups at different risk were considered in order to evaluate the potential role of single/multiple aetiology as a cause of ICU hospitalization and severity of disease.
Second, there was a lack of data regarding bacterial aetiology, which did not allow to detail the co-presence of bacterial/viral infections and, consequently, caused an underestimated infection rate for ICU-patients.
Third, in our setting, oropharyngeal samples were mostly collected, which more likely represent the microbiota of the upper airways, being unable to distinguish colonization/infection of the upper respiratory tract from infections of the lower airways. Despite it has been hypothesized an agreement between lower and upper respiratory specimens for the detection of respiratory pathogens [26], it is very difficult to formulate assumptions, in some cases, on pathogenicity of detected microorganisms in the upper respiratory tract [27]. This anatomic site is a reservoir of a diverse community of commensal germs which, in a dynamic process and because of individual risk factors or imbalances in the ecosystem, may change their role to pathogens causing respiratory or invasive disease, also in immunocompetent subjects. Furthermore, immunosuppressed subjects or the elderly [19], who may suffer of pre-existing respiratory tract involvement, pulmonary and cardiac failure, together with low vaccination coverage rates [28], can lead to hospitalization and intensive care treatment.
Nevertheless, despite these limitations, the present study included both a good representativeness of our geographic area, although with a moderate sample size, and the use of comprehensive panel of microorganisms.
Additionally, our study differed from previous published reports in that our population study also included ILI subjects rather than only patients with pneumonia or other severe respiratory symptoms [29], [30] thus depicting the heterogeneity of respiratory agents circulating in intensive care settings.
Our findings confirm that polymicrobial aetiologies are common among patients with ILI/ARTI admitted to ICU, playing a very important role in the overall outcome of vulnerable patients and representing an unrecognized health exposure that puts all ICU-admitted patients at risk.
Early recognition of ILI/ARTI aetiology in ICUs is important in order to implement early infection-control strategies and to assess of drug therapy appropriateness. The approach to critically ill patients with respiratory symptoms needs to evolve to include a public health perspective, also promoting positive habits among HCWs, in addition to the focus on individual diagnosis and treatment.
Conflicts of interest statement
All authors report no conflicts of interest relevant to this article.
Acknowledgements
We would like to thank Dr. Tobias Allander (Karolinske University Hospital, Stockholm, Sweden) for providing us with hBoV-1 plasmid, Dr. Anje Maria Soderlund-Venermo and Dr. Kalle Kantola (University of Helsinki, Finland) for hBoV-2 and hBoV-3 plasmids, and Dr. Krzysztof Pyrc (Jagiellonian University, Krakow, Poland) for hCoV-NL63 plasmid.
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.micinf.2015.11.008.
Contributor Information
Fabio Tramuto, Email: fabio.tramuto@unipa.it.
Carmelo Massimo Maida, Email: carmelo.maida@unipa.it.
Giuseppe Napoli, Email: g.napoli15@virgilio.it.
Caterina Mammina, Email: caterina.mammina@unipa.it.
Alessandra Casuccio, Email: alessandra.casuccio@unipa.it.
Cinzia Cala', Email: cinzia.cala@unipa.it.
Emanuele Amodio, Email: amoema79@libero.it.
Francesco Vitale, Email: francesco.vitale@unipa.it.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
References
- 1.Black R.E., Cousens S., Johnson H.L., Lawn J.E., Rudan I., Bassani D.G. Global, regional, and national causes of child mortality in 2008: a systematic analysis. Lancet. 2010;375:1969–1987. doi: 10.1016/S0140-6736(10)60549-1. [DOI] [PubMed] [Google Scholar]
- 2.Obasi C.N., Barrett B., Brown R., Vrtis R., Barlow S., Muller D. Detection of viral and bacterial pathogens in acute respiratory infections. J Infect. 2014;68:125–130. doi: 10.1016/j.jinf.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amodio E., Tramuto F., Costantino C., Restivo V., Maida C., Calamusa G. Diagnosis of influenza: only a problem of coding? Med Princ Pract. 2014;23:568–573. doi: 10.1159/000364780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miggins M., Hasan A., Hohmann S., Southwick F., Casella G., Schain D. The potential influence of common viral infections diagnosed during hospitalization among critically ill patients in the United States. PLoS One. 2011;6:e18890. doi: 10.1371/journal.pone.0018890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu U.I., Wang J.T., Ho Y.C., Pan S.C., Chen Y.C., Chang S.C. Factors associated with development of complications among adults with influenza: a 3-year prospective analysis. J Formos Med Assoc. 2012;111:364–369. doi: 10.1016/j.jfma.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 6.Sandrock C., Stollenwerk N. Acute febrile respiratory illness in the ICU: reducing disease transmission. Chest. 2008;133:1221–1231. doi: 10.1378/chest.07-0778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amodio E., Tramuto F., Maringhini G., Asciutto R., Firenze A., Vitale F. Are medical residents a “core group” for future improvement of influenza vaccination coverage in health-care workers? A study among medical residents at the University Hospital of Palermo (Sicily) Vaccine. 2011;29:8113–8117. doi: 10.1016/j.vaccine.2011.08.033. [DOI] [PubMed] [Google Scholar]
- 8.Amodio E., Restivo V., Firenze A., Mammina C., Tramuto F., Vitale F. Can influenza vaccination coverage among healthcare workers influence the risk of nosocomial influenza-like illness in hospitalized patients? J Hosp Infect. 2014;86:182–187. doi: 10.1016/j.jhin.2014.01.005. [DOI] [PubMed] [Google Scholar]
- 9.Esbenshade J.C., Edwards K.M., Esbenshade A.J., Rodriguez V.E., Talbot H.K., Joseph M.F. Respiratory virus shedding in a cohort of on-duty healthcare workers undergoing prospective surveillance. Infect Control Hosp Epidemiol. 2013;34:373–378. doi: 10.1086/669857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bramley A.M., Dasgupta S., Skarbinski J., Kamimoto L., Fry A.M., Finelli L. Intensive care unit patients with 2009 pandemic influenza A (H1N1pdm09) virus infection - United States, 2009. Influenza Other Respir Viruses. 2012;6:e134–e142. doi: 10.1111/j.1750-2659.2012.00385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marsden-Haug N., Foster V.B., Gould P.L., Elbert E., Wang H., Pavlin J.A. Code-based syndromic surveillance for influenza-like illness by international classification of diseases, ninth revision. Emerg Infect Dis. 2007;13:207–216. doi: 10.3201/eid1302.060557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.StataCorp: stata statistical software, release 12. StataCorp LP College Station; TX: 2011. [Google Scholar]
- 13.Erkuran M.K., Duran A., Ocak T., Citisli V., Kaya H. The impact of the duration of admission to the emergency room on the mortality of intensive care patients. Niger J Clin Pract. 2014;17:320–323. doi: 10.4103/1119-3077.130233. [DOI] [PubMed] [Google Scholar]
- 14.Flabouris A., Jeyadoss J., Field J., Soulsby T. Association between emergency department length of stay and outcome of patients admitted either to a ward, intensive care or high dependency unit. Emerg Med Australas. 2013;25:46–54. doi: 10.1111/1742-6723.12021. [DOI] [PubMed] [Google Scholar]
- 15.Wunsch H., Angus D.C., Harrison D.A., Linde-Zwirble W.T., Rowan K.M. Comparison of medical admissions to intensive care units in the United States and United Kingdom. Am J Respir Crit Care Med. 2011;183:1666–1673. doi: 10.1164/rccm.201012-1961OC. [DOI] [PubMed] [Google Scholar]
- 16.Goka E.A., Vallely P.J., Mutton K.J., Klapper P.E. Single and multiple respiratory virus infections and severity of respiratory disease: a systematic review. Paediatr Respir Rev. 2014;15:363–370. doi: 10.1016/j.prrv.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goka E.A., Vallely P.J., Mutton K.J., Klapper P.E. Single, dual and multiple respiratory virus infections and risk of hospitalization and mortality. Epidemiol Infect. 2015;143:37–47. doi: 10.1017/S0950268814000302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Roux A., Ewig S., García E., Marcos M.A., Mensa J., Lode H. Mixed community-acquired pneumonia in hospitalised patients. Eur Respir J. 2006;27:795–800. doi: 10.1183/09031936.06.00058605. [DOI] [PubMed] [Google Scholar]
- 19.Bosch A.A., Biesbroek G., Trzcinski K., Sanders E.A., Bogaert D. Viral and bacterial interactions in the upper respiratory tract. PLoS Pathog. 2013;9:e1003057. doi: 10.1371/journal.ppat.1003057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jaeger J.L., Patel M., Dharan N., Hancock K., Meites E., Mattson C. Transmission of 2009 pandemic influenza A (H1N1) virus among healthcare personnel-Southern California. Infect Control Hosp Epidemiol. 2009;2011(32):1149–1157. doi: 10.1086/662709. [DOI] [PubMed] [Google Scholar]
- 21.Maltezou H.C., Maragos A., Katerelos P., Paisi A., Karageorgou K., Papadimitriou T. Influenza vaccination acceptance among health-care workers in Greece: a nationwide survey. Vaccine. 2008;26:1408–1410. doi: 10.1016/j.vaccine.2008.01.049. [DOI] [PubMed] [Google Scholar]
- 22.Castilla J., Martínez-Baz I., Godoy P., Toledo D., Astray J., García S. Trends in influenza vaccine coverage among primary healthcare workers in Spain, 2008–2011. Prev Med. 2013;57:206–211. doi: 10.1016/j.ypmed.2013.05.021. [DOI] [PubMed] [Google Scholar]
- 23.Lindley M.C., Yonek J., Ahmed F., Perz J.F., Williams Torres G. Measurement of influenza vaccination coverage 23 healthcare personnel in US hospitals. Infect Control Hosp Epidemiol. 2009;30:1150–1157. doi: 10.1086/648086. [DOI] [PubMed] [Google Scholar]
- 24.Widera E., Chang A., Chen H.L. Presenteeism: a public health hazard. J Gen Intern Med. 2010;25:1244–1247. doi: 10.1007/s11606-010-1422-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.CDC, Centers for disease control and prevention: prevention strategies for seasonal influenza in healthcare Settings. Available on line at: http://www.cdc.gov/flu/professionals/infectioncontrol/healthcaresettings.htm. Accessed March 5, 2015.
- 26.Hare K.M., Grimwood K., Leach A.J., Smith-Vaughan H., Torzillo P.J., Morris P.S. Respiratory bacterial pathogens in the nasopharynx and lower airways of Australian indigenous children with bronchiectasis. J Pediatr. 2010;157:1001–1005. doi: 10.1016/j.jpeds.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 27.Simons E., Schroth M.K., Gern J.E. Analysis of tracheal secretions for rhinovirus during natural colds. Pediatr Allergy Immunol. 2005;16:276–278. doi: 10.1111/j.1399-3038.2005.00246.x. [DOI] [PubMed] [Google Scholar]
- 28.Catania J., Que L.G., Govert J.A., Hollingsworth J.W., Wolfe C.R. High intensive care unit admission rate for 2013–2014 influenza is associated with a low rate of vaccination. Am J Respir Crit Care Med. 2014;189:485–487. doi: 10.1164/rccm.201401-0066LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Legoff J., Guérot E., Ndjoyi-Mbiguino A., Matta M., Si-Mohamed A., Gutmann L. High prevalence of respiratory viral infections in patients hospitalized in an intensive care unit for acute respiratory infections as detected by nucleic acid-based assays. J Clin Microbiol. 2005;43:455–457. doi: 10.1128/JCM.43.1.455-457.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Daubin C., Parienti J.J., Vincent S., Vabret A., du Cheyron D., Ramakers M. Epidemiology and clinical outcome of virus-positive respiratory samples in ventilated patients: a prospective cohort study. Crit Care. 2006;10:R142. doi: 10.1186/cc5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


