Abstract
Background
Viral aetiologies are the most common cause of central nervous system (CNS) infections. Approximately one-half of CNS infections remain of undetermined origin. High-throughput sequencing (HTS) brought new perspectives to CNS infection investigations, allowing investigation of viral aetiologies with an unbiased approach. HTS use is still limited to specific clinical situations.
Objectives
The aim of this review was to evaluate the contribution and pitfalls of HTS for the aetiologic identification of viral encephalitis, meningoencephalitis, and meningitis in CNS patient samples.
Sources
PubMed was searched from 1 January 2008 to 2 August 2018 to retrieve available studies on the topic. Additional publications were included from a review of full-text sources.
Content
Among 366 studies retrieved, 29 used HTS as a diagnostic technique. HTS was performed in cerebrospinal fluid and brain biopsy samples of 307 patients, including immunocompromised, immunocompetent paediatric, and adult cases. HTS was performed retrospectively in 18 studies and prospectively in 11. HTS led to the identification of a potential causal virus in 41 patients, with 11 viruses known and ten not expected to cause CNS infections. Various HTS protocols were used.
Implications
The additional value of HTS is difficult to quantify because of various biases. Nevertheless, HTS led to the identification of a viral cause in 13% of encephalitis, meningoencephalitis, and meningitis cases in which various assays failed to identify the cause. HTS should be considered early in clinical management as a complement to routine assays. Standardized strategies and systematic studies are needed for the integration of HTS in clinical management.
Keywords: Diagnosis, Encephalitis, High-throughput sequencing, Meningitis, Meningoencephalitis, Virus
Introduction
Meningitis, encephalitis, and meningoencephalitis are caused by various pathogens, but viral aetiologies are the most common cause [1], [2], [3], [4]. Among these, enterovirus (EV), herpes simplex type 1 and 2 (HSV-1 and HSV-2), and varicella zoster virus (VZV) are the most frequent viruses associated with encephalitis, meningoencephalitis, and meningitis in paediatric and adult populations [1], [2], [4], [5], [6], [7]. The prevalence of other viruses varies according to the geographical location and immune status of the patient.
During the last two decades, the implementation of molecular assays as a complement to serological assays, immunohistochemistry, and culture have improved the diagnosis of viral central nervous system (CNS) infections. Nevertheless, these assays have limitations because of their targeted approach. Apart from technical limitations, the diagnosis of viral CNS infections is subject to several issues, such as the type of sample (cerebrospinal fluid (CSF) or brain biopsy), the timeline of sample collection, and the different pathogenic mechanisms of viruses. Despite technical progress, approximately one-half of encephalitis, meningoencephalitis, and meningitis cases remain of unknown origin [1], [2], [5], [7].
High-throughput sequencing (HTS) has brought new perspectives to CNS infection investigations. Although HTS has been recently integrated in encephalitis management guidelines [8], its use is still limited to specific clinical situations or research. The contribution of HTS warrants a better appraisal for further implementation in CNS infection management. This narrative review aims to evaluate the contribution and pitfalls of HTS for the aetiologic identification of viral encephalitis, meningoencephalitis, and encephalitis in paediatric and adult patients.
Methods
A comprehensive PubMed search was conducted from 1 January 2008 to 2 August 2018 to identify human studies using the following MeSH and keywords research algorithm: ‘((central nervous system infection OR cerebrospinal fluid OR central nervous system) AND sequencing) AND virus’. Additional publications were identified from a review of full-text sources. The title and abstract of each citation were screened by two reviewers and assessed for eligibility by detailed analysis. Inclusion criteria were studies including patients with encephalitis, meningoencephalitis, or meningitis of unknown origin and reporting the use of HTS for the aetiologic identification of a viral origin in CNS samples. Exclusion criteria were reviews, animal studies, other CNS diseases, and studies addressing only technical aspects.
Results
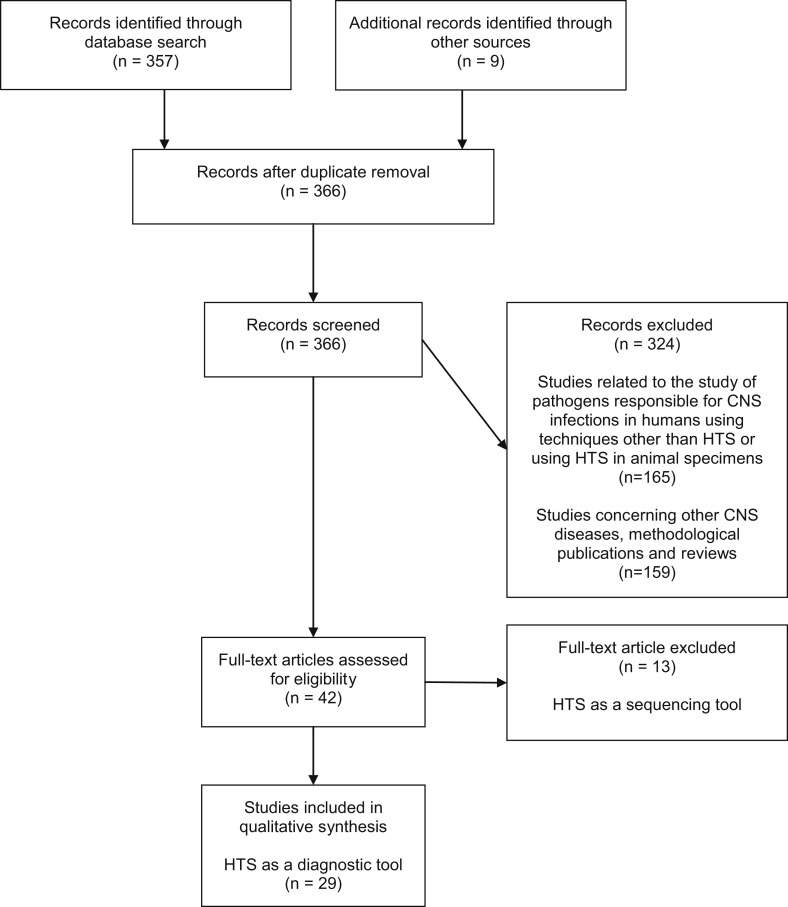
A total of 366 references were retrieved (Fig. 1 ). Screening led to the exclusion of 324 references. Among the 42 studies retained, 13 were excluded in which HTS was performed as a sequencing technique [9], [10], [11], [12], [13], [14], [15], [16], [17], [18], [19], [20], [21]. In these studies, HTS was performed as a sequencing technique after virus identification with (RT)-PCR assays (HSV-1, echovirus 18, Ebola virus, Toscana virus, EV D68 and 71, bornavirus, dengue virus serotype 3), focusing on their phylogenic classification and the comprehension of their epidemiology [9], [10], [11], [12], [13], [14], [15], [16], [17], [18], [19], [20], [21]. Twenty-nine studies were selected for qualitative analysis (19 case reports, ten case series) (Table 1 ).
Fig. 1.
Flowchart of study selection. HTS, high-throughput sequencing; CNS, central nervous system.
Table 1.
Characteristics of studies included in the narrative review
| Reference | Timing of HTS use | Characteristics of patients with CNS samples screened with HTS |
HTS |
Confirmatory assays | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Geographical origin | Number of patients | Immunocompromised patients | Paediatric or adult patients | Age | Diagnosis | Clinical sample analysed | Probable viral cause of CNS disease (number of patients) | Platform | Nucleic acid extraction methods | Report of control use | |||
| Naccache SN et al. Clin Infect Dis. 2015 [33] | Prospective | Europe | 1 | Yes | Adult | 42 yo | Encephalitis | CSF and brain tissue | Novel HAstV-VA1 (1) | HiSeq 2500 | RNA/DNA | Positive control | RT-PCR, in situ hybridization |
| Frémond M-L et al. J Pediatric Infect Dis Soc. 2015 [23] | Prospective | Europe | 1 | Yes | Paediatric | 14 yo | Encephalitis | Brain tissue | Novel HAstV-VA1 (1) | Ion Proton | RNA | No | RT-PCR |
| Salzberg SL et al. Neurol Neuroimmunol Neuroinflamm. 2016 [39] | Prospective | North America | 2 | Yes (1),No (1) | Adult | 44 yo, 67 yo | Encephalitis | Brain tissue | EBV (1) | MiSeq | DNA | No | in situ hybridization |
| Wilson MR et al. Ann Neurol. 2017 [30] | Prospective | Oceania | 1 | Yes | Adult | 34 yo | Chronic meningoencephalitis | CSF and brain tissue | Cache Valley virus (1) | HiSeq 4000 | RNA | Negative control | RT-PCR, viral culture, immunohistochemistry |
| Wilson MR et al. Am J Transplant. 2017 [40] | Prospective | North America | 1 | Yes | Paediatric | 14 yo | Meningoencephalitis | CSF | WNV (1) | HiSeq 2500 | RNA | Negative control | Serology |
| Morfopoulou S et al. Acta Neuropathol. 2017 [41] | Prospective | Europe | 1 | Yes | Paediatric | 1.5 yo | Chronic encephalitis | Brain tissue | Mumps virus (vaccine strain) (1) | NextSeq 500 | RNA | No | RT-PCR, immunohistochemistry, serology |
| Chiu CY et al. Emerg Infect Dis. 2017[42] | Prospective | North America | 1 | Yes | Adult | 68 yo | Encephalitis | CSF | St Louis encephalitis virus (1) | Not specified | RNA/DNA | No | RT-PCR, serology, viral culture |
| Palacios G. et al. N Engl J Med. 2008 [26] | Retrospective | Oceania | 2 | Yes | Adult | 63 yo and 64 yo | Encephalitis | CSF and brain tissue | LCMV-related virus (2) | GSL FLX | RNA | No | RT-PCR, serology |
| Quan PL et al. Emerg Infect Dis. 2010 [47] | Retrospective | North America | 1 | Yes | Paediatric | 15 yo | Encephalitis | Brain tissue | Human astrovirus undetermined (1) | GSL FLX | RNA | No | RT-PCR, immunohistochemistry immunofluorescence |
| Brown JR et al. Clin Infect Dis. 2015 [31] | Retrospective | Europe | 1 | Yes | Paediatric | 1.5 yo | Encephalitis | Brain tissue | Novel HAstV-VA1 (1) | MiSeq | RNA | Positive control | RT-PCR, immunohistochemistry |
| Morfopoulou S et al. N Engl J Med. 2016 [25] | Retrospective | Europe | 1 | Yes | Paediatric | 11 mo | Encephalitis | Brain tissue | Human coronavirus OC43 (1) | HiSeq 2500 | RNA | No | RT-PCR, immunohistochemistry |
| Sato M et al. J Clin Virol. 2016 [24] | Retrospective | Asia | 1 | Yes | Paediatric | 4 yo | Encephalitis | CSF | Novel HAstV-MLB1 (1) | MiSeq | RNA/DNA | No | RT-PCR |
| Lum SH et al. Transpl Infect Dis. 2016 [43] | Retrospective | Europe | 1 | Yes | Paediatric | 4 mo | Encephalitis | Brain tissue | Novel HAstV-VA1 (1) | HiSeq 2500 | RNA | No | RT-PCR |
| Lipowski D et al. J Infect Dis. 2017 [29] | Retrospective | Europe | 3 | Yes | Adult | Median 48 yo (range 27–54 yo) | Encephalitis | CSF and brain tissue | TBEV (3) | HiSeq 1500 | RNA | No | RT-PCR |
| Cordey S et al. Emerg Infect Dis. 2016 [22] | Retrospective | Europe | 1 | No | Adult | 21 yo | Meningitis | CSF | Novel HAstV-MLB2 (1) | HiSeq 2500 | RNA/DNA | No | RT-PCR |
| Benjamin LA et al Emerg Infect Dis. 2011 [46] | Retrospective | Asia | 12 | No | Paediatric | 2 yo, 3 yo Other paediatric patients <16 yo |
Encephalitis | CSF | Parvovirus 4 (2) | GS FLX Titanium | RNA/DNA | No | PCR, serology |
| Perlejewski K et al. J Virol Methods 2015 [35] | Retrospective | Europe | 1 | No | Adult | 60 yo | Encephalitis | CSF | HSV1 (1) | MiSeq | RNA | Negative and positive controls | PCR |
| Guan H et al. J Neurovirol. 2016 [36] | Retrospective | Asia | 4 | No | Adult | Median 43.5 yo (range 31–64 yo) | Meningoencephalitis | CSF | HSV-1 (2), HSV-2 (1), VZV (1) | BGISEQ-100 | DNA | No | PCR |
| Kawada J et al. Sci Rep. 2016 [48] | Retrospective | Asia | 18 | No | Paediatric | Median 3 yo (range 3 mo to 15 yo) | Encephalitis | CSF | Mumps virus (1), coxsackie A9 virus (2) | MiSeq | RNA/DNA | Positive control | RT-PCR |
| Arden KE et al. J Med Virol. 2017 [49] | Retrospective | Oceania | 1 | No | Adult | 51 yo | Encephalitis | CSF | Toscana virus (1) | HiSeq 2500 | RNA | No | None |
| Tan le V et al. MBio. 2013 [28] | Retrospective | Asia | 125 | NA | Paediatric and adult | NA | Encephalitis | CSF | CyCV-VN (2) | FLX genome sequencer | RNA/DNA | No | PCR |
| Phan TG et al. Virology. 2015 [27] | Retrospective | Asia | 62 | NA | Paediatric and adult | Paediatric patients: 2 mo to 12 yo Adult patients: 15 to 72 yo |
Encephalitis | CSF | Gemycirculavirus (3), CyCV-VN (1) | MiSeq | DNA | No | PCR |
| Chan BK et al. PLoS One. 2014 [34] | Retrospective | North America | 7 | NA | NA | NA | Encephalitis | Brain tissue | HSV-1 (3), MeV (2) | HiSeq 2000 | RNA | Negative and positive controls | PCR, RT-PCR |
| Phan TG et al. Arch Virol. 2016 [37] | Retrospective | North America | 53 | NA | Paediatric | 6 yo Other patients: NA |
Encephalitis | CSF | HuCSFDV1 (1) | MiSeq | RNA/DNA | No | PCR |
| Greninger AL et al. Genome Med. 2015 [32] | Prospective | North America | 1 | No | Paediatric | 15 yo | Encephalitis | CSF and brain tissue | None | MiSeq | RNA | Negative control | – |
| Wilson MR et al. Ann Neurol. 2015 [45] | Prospective | North America | 1 | Yes | Adult | 74 yo | Encephalitis | CSF | None | HiSeq 2500 | RNA | Negative control | – |
| Wilson MR et al. N Engl J Med. 2014 [44] | Prospective | North America | 1 | Yes | Paediatric | 14 yo | Meningoencephalitis | CSF | None | MiSeq | DNA | Negative control | – |
| Christopeit M et al. Ann Hematol. 2016 [38] | Prospective | Europe | 1 | Yes | Adult | 65 yo | Encephalitis | CSF | None | Not specified | RNA | Negative and positive controls | – |
| Mongkolrattanothai K et al. J Pediatric Infect Dis Soc. 2017 [50] | Retrospective | North America | 1 | No | Paediatric | 11 yo | Encephalitis | CSF | None | HiSeq | RNA/DNA | Negative and positive controls | – |
HTS, high-throughput sequencing; CNS, central nervous system; CSF, cerebrospinal fluid; y, years old; m, months old; NA, not available. HAstV, human astrovirus; EBV, Epstein-Barr virus; WNV, West Nile virus; LCMV, lymphocytic choriomeningitis virus; TBEV, tick-borne encephalitis virus; CyCV-VN, cyclovirus Viet-Nam; HSV, herpes simplex virus; VZV, varicella zoster virus; MeV, measles virus; HuCSFDV1, CSF-associated densovirus 1.
Characteristics of patients with CNS samples screened with HTS
Fourteen and 13 studies concerned paediatric and adult cases, respectively, and two studies concerned both populations (Table 1). HTS was performed in 307 cases (52 adults; 123 paediatric (<18 years); 132 cases with no information on age). Twenty-five studies reported patient age (median age, 14 years; range, 3 months to 68 years). Diagnostic criteria for encephalitis, meningoencephalitis, and meningitis were inconsistently reported. Immune status was reported for 60 patients and comprised 20 immunocompromised (nine paediatric, 11 adults) and 40 immunocompetent patients (22 paediatric, eight adults). Studies came from a wide range of regions: Europe (ten), North America (ten), Asia (six), and Oceania (three) (Table 1).
Clinical samples analysed with HTS
When brain specimens were available, pathological examination provided proof of diagnosis of encephalitis. CSF analysis results were reported in 25 patients of 20 studies, with the white blood cell count ranging from 1 to 494 cell/mm3 in encephalitis and meningoencephalitis cases and 915 cell/mm3 in the only meningitis case [22]; three publications reported normal CSF analysis without any description [23], [24], [25]. HTS was performed on individual CSF and brain specimens in 129 and 21 patients, respectively. CSF samples of 162 patients were pooled for HTS analysis [26], [27], [28]. HTS was performed on CSF and brain specimens in five patients [26], [29], [30], [31], [32]. Positive results on both samples were obtained in a Cache Valley virus chronic meningoencephalitis case [30] and positive results on brain biopsies only were reported in two human astrovirus (HAstV)-VA1 and tick-borne encephalitis (TBEV) cases [29], [33]. In one patient, HTS analysis did not identify a viral cause, but Balamuthia mandrillaris was identified in both CSF and brain biopsy [32]. Despite limitations because of publication bias, the overall diagnostic yield for a viral aetiology according to sample type was estimated to be higher for brain specimens (16/21 (76.2%) positive samples) than for CSF samples (26/291 (8.9%) positive samples).
HTS use in microbiological investigations
Detailed microbiological investigations performed prior to HTS varied according to local practice and were not reported in seven studies [24], [34], [35], [36], [37], [38], [39]. In most studies, viruses identified with HTS were not part of the microbiological work-up, except in three cases where HTS identified a virus for which diagnostic assays were negative during routine investigation. These included a West Nile virus (WNV) identified in the CSF sample of a renal transplant recipient with meningoencephalitis and a negative serological assay [40]. HSV-1 was identified in an encephalitis case [35]. A vaccine strain of mumps virus was identified in a brain specimen of an immunosuppressed child with chronic encephalitis in whom (RT)-PCR for mumps on a CSF sample was negative as the assay did not target vaccine strains [41].
HTS was performed retrospectively in 18 studies and prospectively as part of the initial work-up in 11 case reports with an impact on the clinical management of three immunocompromised patients: a child with encephalitis associated with HAstV-VA1 [23]; an adult with encephalitis associated with HAstV-VA1 [33]; and an adult with chronic meningoencephalitis associated with Cache Valley virus [30]. Turnaround times were reported in ten studies [23], [31], [33], [36], [39], [40], [42], [43], [44], [45]. Among publications in which HTS was used prospectively, turnaround times ranged from 48 hours to 7 days [23], [33], [39], [40], [42], [44], [45].
Virus identification with HTS
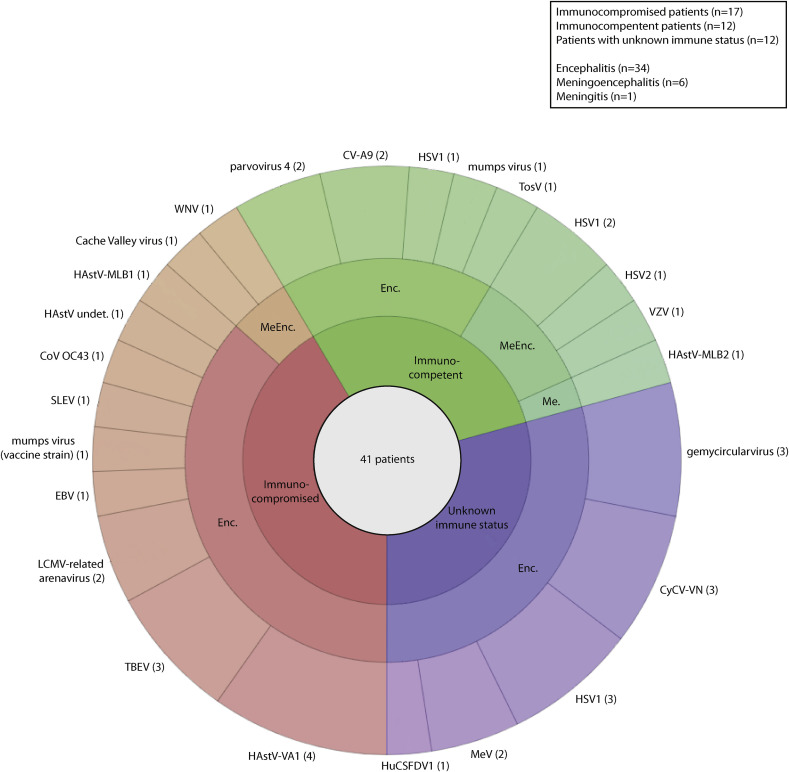
HTS performed on pooled or individual samples and/or subsequent confirmatory assays allowed the identification of a potential causal virus in 41 of 307 patients (13.4%), comprising 15 paediatric cases (eight immunocompromised cases), 17 adult cases (nine immunocompromised cases) and nine cases for whom age was not specified; median age was 21 years (range 3 months to 68 years). Fig. 2 shows the distribution of viruses identified according to patient immune status and clinical manifestations. HTS allowed the identification of viruses previously unknown or unexpected as a cause of CNS infection (n = 10) and thus not screened during diagnostic investigations (Table 1). These included parvovirus 4 (two) [46], human coronavirus OC43 (one) [25], and novel HAstV-MLB2 (one) [22] identified in immunocompetent patients. A mumps virus vaccine strain (one) [41], HAstV (undetermined specie; one) [47], HAstV-VA1 (four) [23], [31], [33], [43], and HAstV-MLB1 (one) [24] were identified in immunocompromised patients. A gemycircularvirus was also identified, but its causal role in encephalitis is under debate [27]. Three novel viral species or strains were identified in CSF samples of patients with encephalitis (Table 1): human CSF-associated densovirus 1 (HuCSFDV1) [37]; cyclovirus Viet-Nam (CyCV-VN) [28]; and lymphocytic choriomeningitis virus (LCMV)-related arenavirus [26]. HTS analysis also identified viruses known to be responsible for CNS infections (n = 11) and not screened or detected by routine assays (HSV-1, HSV-2, VZV, Epstein–Barr virus (EBV), TBEV, WNV, Cache Valley, Saint Louis encephalitis, Toscana, mumps, measles, and coxsackie A9 virus) [29], [30], [34], [35], [39], [40], [42], [48], [49] (Table 1).
Fig. 2.
Distribution of viruses identified with high-throughput sequencing and/or subsequent confirmatory assays in the cerebrospinal fluid and brain samples of 41 patients with a potential viral cause of central nervous system infection, according to immune status and clinical manifestations. The number of patients in whom each virus has been identified is mentioned in brackets. Enc., encephalitis; MeEnc., meningoencephalitis; Me., meningitis; CyCV-VN, cyclovirus Viet-Nam; HSV, herpes simplex virus; MeV, measles virus; HuCSFDV1, human CSF-associated densovirus 1; CV-A9, coxsackie virus A9; TosV, Toscana virus; VZV, varicella zoster virus; HAstV, human astrovirus; TBEV, tick-borne encephalitis virus; LCMV, lymphocytic choriomeningitis virus; EBV, Epstein–Barr virus; SLEV, Saint Louis encephalitis virus; CoV, coronavirus; WNV, West Nile virus; undet., undetermined.
HTS protocols and controls
Most studies performed nucleic acid extraction protocols dedicated to RNA, or RNA and DNA. Thirteen RNA and seven DNA viral species were identified (Table 1). One study reported the identification of HSV-1 in a CSF sample after RNA extraction protocol [35]. Six studies where HTS analysis was not restricted to the detection of viruses resulted in the identification of bacterial (Brucella melitensis and Leptospira santarosai) [44], [50], mycobacterial (Mycobacterium tuberculosis) [39], and parasitic (Balamuthia mandrillaris) [32], [45] or fungal (Candida tropicalis and Fusarium solani and oxysporum) [38] pathogens.
The use of controls was not systematically reported. Nine studies reported various negative control samples, such as brain specimens without encephalitis [34], CSF samples from patients without infection [30], [32], [35], [38], [40], [45], serum samples, and water or elution buffer [30], [44], [45], [50]. Viral sequences of negative controls were not consistently described. Positive controls, such as CSF or serum samples positive for DNA or RNA viruses, were rarely reported or used [33], [35], [38], [48], [50].
To address the specificity of HTS results, other techniques were performed to confirm HTS results in all studies, except one [49]. (RT)-PCR assays were performed in samples from 37 patients and were positive in at least one sample in 36 patients. HTS results were also confirmed with serological assays [26], [40], [41], [46], immunohistochemistry, and in situ hybridization [25], [30], [31], [33], [39], [41], [47]. Viral culture confirmed the presence of a replicative Saint Louis encephalitis virus in a CSF sample [42], but was unsuccessful concerning a Cache Valley virus [30].
Discussion
Contribution of HTS in identifying viral causes of encephalitis, meningoencephalitis and meningitis
HTS offers the possibility of investigating viral aetiologies of CNS infections by an unbiased approach when work-up according to guidelines fails to identify a causal pathogen. Based on the studies retrieved, its diagnostic yield for a viral aetiology is difficult to estimate, particularly because of publication bias (high number of case reports), methodological heterogeneity, and a lack of systematic prospective studies. When focusing specifically on case series, the diagnostic yield for the identification of a viral cause was approximately 10%, but this result should be interpreted with caution in the light of the evolution of the technique from 2008 to 2017. Among the studies reviewed, the HTS contribution is evident not only for the identification of a potential causal virus in CNS infections of unknown origin, but also in the detection of novel or divergent viruses [26], [28], [37]. Similar to other techniques, the type of sample used for analysis is of particular importance. Despite diverse HTS protocols and publication bias, HTS seemed to have a higher diagnostic yield in brain specimens than in CSF samples. The diagnostic yield was particularly low in two studies where HTS was performed on CSF supernatant [27], [28]. HTS also shows its clinical value in situations where the viral pathogenic mechanisms and specific clinical situations impairs the results of conventional assays [40]. Among immunocompetent patients, HTS led to the identification of viruses not previously associated with CNS infections (parvovirus 4, CyCV-VN, gemycircularvirus, and novel HAstV-MLB2) [22], [27], [28], [46].
HTS clinical impact was mainly demonstrated among immunocompromised patients, with most studies dedicated to this population. It was performed prospectively in 11 cases and led to a change in clinical management in three [23], [30], [33]. The rapid decision to perform HTS, short HTS turnaround times, and the efficient interpretation of results were determinant for the management of these latter patients. Among immunocompromised patients, HTS contributed to the detection of viruses for which no assay was performed during conventional work-up: viruses known to cause CNS infections (TBEV, WNV, Cache Valley virus, Saint-Louis encephalitis virus, EBV) [29], [30], [39], [40], [42], viruses not known to be responsible for CNS infections (novel HAstV-VA1, HAstV-MLB1, human coronavirus OC43, mumps virus vaccine strain) [23], [24], [25], [31], [33], [41], [43], [47], and novel viruses (LCMV-related arenavirus) [26]. Focusing on novel HAstV, HTS brought new insights in our understanding of their association with CNS infections [22], [23], [24], [31], [33], [43]. Furthermore, all (RT)-PCR assays performed retrospectively confirmed HTS results, thus highlighting the specificity of HTS.
Issues of HTS in identifying viral causes of encephalitis, meningoencephalitis and meningitis
In the absence of standardization, the methodological heterogeneity of studies is striking, not only concerning pre-analytic steps, but also HTS per se, with the use of diverse HTS platforms, single or paired protocols, as well as diverse bioinformatic pipelines and databases. The use of positive controls as quality controls was only reported in seven studies [31], [33], [34], [35], [38], [48], [50]. Addressing the issue of contamination, only a few studies reported the use of negative controls [30], [32], [34], [35], [38], [40], [44], [45], [50]. Viral sequences assigned to viruses not considered as the cause of CNS infection were not consistently performed: 12 studies provided a description for one CNS sample or more [25], [27], [29], [30], [32], [33], [34], [35], [38], [40], [44], [50]. For most of these viral sequences, no interpretation of results was explicitly provided. Among reads of viruses known to cause infections in humans, Anelloviridae [51] and Herpesviridae were the most described in four and seven samples, respectively; human pegivirus reads were identified in one sample. The genome of the torque teno virus, a member of the Anelloviridae family, and human pegivirus have been identified in CNS samples, but without any association with a CNS disease so far [52], [53], [54], [55], [56], [57]. Other viral sequences were mostly assigned to viruses infecting plants or non-vertebrates and were considered to be reagent contaminants. The minimal description of these HTS ‘background’ results impairs the comprehension of the composition of the CNS virome. Furthermore, the integration of HTS results in the clinical context is of particular importance and the absence of standardization of any reporting methods precludes an objective interpretation of these results.
Finally, HTS-negative results could be interpreted in the context of several clinical and technical aspects that could impact on the sensitivity of the method. First, from a clinical point of view, differences in diagnostic yield from a biopsy compared with CSF samples could be explained by several factors: patient selection (cases of encephalitis); the type of sample (e.g. multiple pooled post-mortem brain samples); and the timeline of sampling in the context of encephalitis (biopsy positivity could possibly be less affected by time than CSF). From a technical point of view, it should be considered that this narrative review includes studies from 2008 to 2018 and thus takes into account the tremendous evolution of the HTS technique over this last decade. Several technical issues need to be considered for the interpretation of negative results: pre-analytic steps (e.g. the use of fresh, frozen, or paraffin-embedded samples for analysis, extraction protocols, fragmentation methods, library preparation, paired-end versus single-end protocols); sequencing depth; sequencing platforms (Table 1); and the analysis of HTS raw data (e.g. mapping software, viral databases, and pipeline precision).
Perspectives for HTS implementation in the management of encephalitis, meningoencephalitis and meningitis
This review highlights that the use of HTS in investigations concerning a viral cause of encephalitis, meningoencephalitis, and meningitis could extend not only to immunocompromised, but also to immunocompetent patients. Considering the selection and publication bias of the literature reviewed here, the negative predictive value for the aetiologic identification of viral encephalitis, meningoencephalitis, and meningitis is difficult to quantify and further studies are needed. HTS needs to be integrated in clinical management as a second-line technique or in parallel to first-line investigations when a standard work-up according to guidelines [8], [58] and additional investigations considering local epidemiology and specific clinical situations fail to identify a causal agent. Brain biopsy should also be considered. Furthermore, HTS is of particular interest for the screening of a large panel of viruses, particularly to avoid a restricted screening of low-volume clinical samples, such as in paediatric patients. HTS brought new perspectives to the investigations of infectious diseases. Notably, its unbiased approach is of particular interest in samples that would not usually be tested in specific syndromes. Its use may not only be restricted to CNS samples, but also extended to other clinical samples. This is illustrated by the positive results of (RT)-PCR assays performed on blood or plasma samples collected at the time of neurological manifestations, which allowed the identification of the same virus detected by HTS in CNS samples (parvovirus 4, LCMV-related arenavirus, novel HAstV-VA1, novel HAstV-MLB2) [22], [26], [31], [46]. This could be of particular interest when a cerebral biopsy cannot be performed and a disseminated infection occurs or is suspected, particularly in immunocompromised patients.
An early decision to perform HTS, short HTS turnaround times, and an efficient interpretation of results are major issues for allowing HTS to contribute to clinical management. For prospective HTS use in clinical routine, this timeframe should be as short as possible for clinical decision-making. Among publications in which HTS was used prospectively, reported turnaround times ranged from 48 hours to 7 days [23], [33], [39], [40], [42], [44], [45].
HTS use is still restricted to a limited number of diagnostic laboratories considering the cost of analysis and informatics infrastructures needed (e.g. costs of sequencing platforms, computing resources, data storage). Despite the expanding use of HTS in clinical microbiology, the surprisingly low number of studies retrieved for this review might be explained for several reasons, including financial limitations when considering the costs of the analysis, the need for shorter turnaround times, and the limitations cited above.
Addressing the question of the proof of causality, particularly in the context of pathogen discovery, Lipkin proposed several criteria for pathogen causality with grading certainty according to confirmation with serological assays or culture for instance [59]. Most studies confirmed HTS results with (RT)-PCR assays and a few with cell culture, serological assays, and immunohistochemistry. Thus, HTS should be implemented in clinical routine in association with other diagnostic tests. In most studies, the approach to establish causality was not explicitly described, but was reported as the temporal association of clinical manifestations and the identification of viral sequences of a specific virus using HTS on CNS samples at the time of manifestations. This process was only described in few studies. Similar to other molecular tests such as (RT)-PCR, the detection of viral sequences or genome in a clinical sample should be interpreted with caution in the clinical context. In a near future, the process for the establishment of causality in HTS analysis should be more transparent and should comprise multidisciplinary sessions involving infectious disease specialists and bioinformatics experts, not only for results concerning viruses unexpected to cause CNS infections. Finally, for HTS implementation in clinical routine, the question of standardization has to be addressed concerning HTS protocols, data analysis algorithms, reference databases and quality controls, and further prospective studies are needed [60].
Conclusion
This review shows that HTS contributed to the identification of potential viral aetiologies of encephalitis, meningoencephalitis, and meningitis of unknown origin in approximately 13% of cases and is of particular interest in immunocompromised patients. This unbiased or semi-unbiased approach led to the identification of novel viruses, viruses known or not expected to cause CNS infections. Standardized strategies are needed for the further implementation of HTS in clinical management. In centres where available, the decision to perform HTS should be considered early in the management of encephalitis, meningoencephalitis, and meningitis in as a second-line technique or in parallel to recommended investigations.
Transparency declaration
The authors have no conflicts of interest to declare. No funding was received for this study.
Acknowledgments
The authors would like to acknowledge Rosemary Sudan (Geneva University Hospitals, Switzerland) for editorial assistance.
Editor: F. Allerberger
References
- 1.Glaser C.A., Honarmand S., Anderson L.J., Schnurr D.P., Forghani B., Cossen C.K. Beyond viruses: clinical profiles and etiologies associated with encephalitis. Clin Infect Dis. 2006;43:1565–1577. doi: 10.1086/509330. [DOI] [PubMed] [Google Scholar]
- 2.Mailles A., Stahl J.P., Steering C., Investigators G Infectious encephalitis in France in 2007: a national prospective study. Clin Infect Dis. 2009;49:1838–1847. doi: 10.1086/648419. [DOI] [PubMed] [Google Scholar]
- 3.Granerod J., Ambrose H.E., Davies N.W., Clewley J.P., Walsh A.L., Morgan D. Causes of encephalitis and differences in their clinical presentations in England: a multicentre, population-based prospective study. Lancet Infect Dis. 2010;10:835–844. doi: 10.1016/S1473-3099(10)70222-X. [DOI] [PubMed] [Google Scholar]
- 4.Bartt R. Acute bacterial and viral meningitis. Continuum (Minneap Minn) 2012;18:1255–1270. doi: 10.1212/01.CON.0000423846.40147.4f. [DOI] [PubMed] [Google Scholar]
- 5.de Ory F., Avellon A., Echevarria J.E., Sanchez-Seco M.P., Trallero G., Cabrerizo M. Viral infections of the central nervous system in Spain: a prospective study. J Med Virol. 2013;85:554–562. doi: 10.1002/jmv.23470. [DOI] [PubMed] [Google Scholar]
- 6.Logan S.A., MacMahon E. Viral meningitis. BMJ. 2008;336:36–40. doi: 10.1136/bmj.39409.673657.AE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kupila L., Vuorinen T., Vainionpaa R., Hukkanen V., Marttila R.J., Kotilainen P. Etiology of aseptic meningitis and encephalitis in an adult population. Neurology. 2006;66:75–80. doi: 10.1212/01.wnl.0000191407.81333.00. [DOI] [PubMed] [Google Scholar]
- 8.Stahl J.P., Azouvi P., Bruneel F., De Broucker T., Duval X., Fantin B. Guidelines on the management of infectious encephalitis in adults. Med Mal Infect. 2017;47:179–194. doi: 10.1016/j.medmal.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 9.Bondre V.P., Sankararaman V., Andhare V., Tupekar M., Sapkal G.N. Genetic characterization of human herpesvirus type 1: full-length genome sequence of strain obtained from an encephalitis case from India. Indian J Med Res. 2016;144:750–760. doi: 10.4103/ijmr.IJMR_747_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krumbholz A., Egerer R., Braun H., Schmidtke M., Rimek D., Kroh C. Analysis of an echovirus 18 outbreak in Thuringia, Germany: insights into the molecular epidemiology and evolution of several enterovirus species B members. Med Microbiol Immunol. 2016;205:471–483. doi: 10.1007/s00430-016-0464-z. [DOI] [PubMed] [Google Scholar]
- 11.Jacobs M., Rodger A., Bell D.J., Bhagani S., Cropley I., Filipe A. Late Ebola virus relapse causing meningoencephalitis: a case report. Lancet. 2016;388:498–503. doi: 10.1016/S0140-6736(16)30386-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Magurano F., Baggieri M., Gattuso G., Fortuna C., Remoli M.E., Vaccari G. Toscana virus genome stability: data from a meningoencephalitis case in Mantua, Italy. Vector Borne Zoonotic Dis. 2014;14:866–869. doi: 10.1089/vbz.2014.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boland T.A., McGuone D., Jindal J., Rocha M., Cumming M., Rupprecht C.E. Phylogenetic and epidemiologic evidence of multiyear incubation in human rabies. Ann Neurol. 2014;75:155–160. doi: 10.1002/ana.24016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reinke S.N., Resch L., Maingat F., Branton W., Jackson A.C., Holt R. Metagenomic and metabolomic characterization of rabies encephalitis: new insights into the treatment of an ancient disease. J Infect Dis. 2013;207:1451–1456. doi: 10.1093/infdis/jis479. [DOI] [PubMed] [Google Scholar]
- 15.Nougairede A., Bichaud L., Thiberville S.D., Ninove L., Zandotti C., de Lamballerie X. Isolation of Toscana virus from the cerebrospinal fluid of a man with meningitis in Marseille, France, 2010. Vector Borne Zoonotic Dis. 2013;13:685–688. doi: 10.1089/vbz.2013.1316. [DOI] [PubMed] [Google Scholar]
- 16.Cordey S., Bel M., Petty T.J., Docquier M., Sacco L., Turin L. Toscana virus meningitis case in Switzerland: an example of the ezVIR bioinformatics pipeline utility for the identification of emerging viruses. Clin Microbiol Infect. 2015;21 doi: 10.1016/j.cmi.2014.11.010. 387 e1-4. [DOI] [PubMed] [Google Scholar]
- 17.Greninger A.L., Naccache S.N., Messacar K., Clayton A., Yu G., Somasekar S. A novel outbreak enterovirus D68 strain associated with acute flaccid myelitis cases in the USA (2012-14): a retrospective cohort study. Lancet Infect Dis. 2015;15:671–682. doi: 10.1016/S1473-3099(15)70093-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marlinge M., Crespy L., Zandotti C., Piorkowski G., Kaphan E., Charrel R.N. Afebrile meningoencephalitis with transient central facial paralysis due to Toscana virus infection, southeastern France, 2014 [corrected] Euro Surveill. 2014;19:20974. doi: 10.2807/1560-7917.es2014.19.48.20974. [DOI] [PubMed] [Google Scholar]
- 19.Duong V., Mey C., Eloit M., Zhu H., Danet L., Huang Z. Molecular epidemiology of human enterovirus 71 at the origin of an epidemic of fatal hand, foot and mouth disease cases in Cambodia. Emerg Microbes Infect. 2016;5:e104. doi: 10.1038/emi.2016.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoffmann B., Tappe D., Hoper D., Herden C., Boldt A., Mawrin C. A variegated Squirrel Bornavirus associated with fatal human encephalitis. N Engl J Med. 2015;373:154–162. doi: 10.1056/NEJMoa1415627. [DOI] [PubMed] [Google Scholar]
- 21.Dhenni R., Karyanti M.R., Putri N.D., Yohan B., Yudhaputri F.A., Ma'roef C.N. Isolation and complete genome analysis of neurotropic dengue virus serotype 3 from the cerebrospinal fluid of an encephalitis patient. PLoS Negl Trop Dis. 2018;12 doi: 10.1371/journal.pntd.0006198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cordey S., Vu D.L., Schibler M., L'Huillier A.G., Brito F., Docquier M. Astrovirus MLB2, a new gastroenteric virus associated with meningitis and disseminated infection. Emerg Infect Dis. 2016;22:846–853. doi: 10.3201/eid2205.151807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frémond M.L., Perot P., Muth E., Cros G., Dumarest M., Mahlaoui N. Next-generation sequencing for diagnosis and tailored therapy: a case report of astrovirus-associated progressive encephalitis. J Pediatr Infect Dis Soc. 2015;4:e53–e57. doi: 10.1093/jpids/piv040. [DOI] [PubMed] [Google Scholar]
- 24.Sato M., Kuroda M., Kasai M., Matsui H., Fukuyama T., Katano H. Acute encephalopathy in an immunocompromised boy with astrovirus-MLB1 infection detected by next generation sequencing. J Clin Virol. 2016;78:66–70. doi: 10.1016/j.jcv.2016.03.010. [DOI] [PubMed] [Google Scholar]
- 25.Morfopoulou S., Brown J.R., Davies E.G., Anderson G., Virasami A., Qasim W. Human coronavirus OC43 associated with fatal encephalitis. N Engl J Med. 2016;375:497–498. doi: 10.1056/NEJMc1509458. [DOI] [PubMed] [Google Scholar]
- 26.Palacios G., Druce J., Du L., Tran T., Birch C., Briese T. A new arenavirus in a cluster of fatal transplant-associated diseases. N Engl J Med. 2008;358:991–998. doi: 10.1056/NEJMoa073785. [DOI] [PubMed] [Google Scholar]
- 27.Phan T.G., Mori D., Deng X., Rajindrajith S., Ranawaka U., Fan Ng T.F. Small circular single stranded DNA viral genomes in unexplained cases of human encephalitis, diarrhea, and in untreated sewage. Virology. 2015;482:98–104. doi: 10.1016/j.virol.2015.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tan le V., van Doorn H.R., Nghia H.D., Chau T.T., Tu le TP., de Vries M. Identification of a new cyclovirus in cerebrospinal fluid of patients with acute central nervous system infections. MBio. 2013;4 doi: 10.1128/mBio.00231-13. e00231-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lipowski D., Popiel M., Perlejewski K., Nakamura S., Bukowska-Osko I., Rzadkiewicz E. A cluster of fatal tick-borne encephalitis virus infection in organ transplant setting. J Infect Dis. 2017;215:896–901. doi: 10.1093/infdis/jix040. [DOI] [PubMed] [Google Scholar]
- 30.Wilson M.R., Suan D., Duggins A., Schubert R.D., Khan L.M., Sample H.A. A novel cause of chronic viral meningoencephalitis: Cache Valley virus. Ann Neurol. 2017;82:105–114. doi: 10.1002/ana.24982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brown J.R., Morfopoulou S., Hubb J., Emmett W.A., Ip W., Shah D. Astrovirus VA1/HMO-C: an increasingly recognized neurotropic pathogen in immunocompromised patients. Clin Infect Dis. 2015;60:881–888. doi: 10.1093/cid/ciu940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Greninger A.L., Messacar K., Dunnebacke T., Naccache S.N., Federman S., Bouquet J. Clinical metagenomic identification of Balamuthia mandrillaris encephalitis and assembly of the draft genome: the continuing case for reference genome sequencing. Genome Med. 2015;7:113. doi: 10.1186/s13073-015-0235-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Naccache S.N., Peggs K.S., Mattes F.M., Phadke R., Garson J.A., Grant P. Diagnosis of neuroinvasive astrovirus infection in an immunocompromised adult with encephalitis by unbiased next-generation sequencing. Clin Infect Dis. 2015;60:919–923. doi: 10.1093/cid/ciu912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chan B.K., Wilson T., Fischer K.F., Kriesel J.D. Deep sequencing to identify the causes of viral encephalitis. PLoS One. 2014;9 doi: 10.1371/journal.pone.0093993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perlejewski K., Popiel M., Laskus T., Nakamura S., Motooka D., Stokowy T. Next-generation sequencing (NGS) in the identification of encephalitis-causing viruses: unexpected detection of human herpesvirus 1 while searching for RNA pathogens. J Virol Methods. 2015;226:1–6. doi: 10.1016/j.jviromet.2015.09.010. [DOI] [PubMed] [Google Scholar]
- 36.Guan H., Shen A., Lv X., Yang X., Ren H., Zhao Y. Detection of virus in CSF from the cases with meningoencephalitis by next-generation sequencing. J Neurovirol. 2016;22:240–245. doi: 10.1007/s13365-015-0390-7. [DOI] [PubMed] [Google Scholar]
- 37.Phan T.G., Messacar K., Dominguez S.R., da Costa A.C., Deng X., Delwart E. A new densovirus in cerebrospinal fluid from a case of anti-NMDA-receptor encephalitis. Arch Virol. 2016;161:3231–3235. doi: 10.1007/s00705-016-3002-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Christopeit M., Grundhoff A., Rohde H., Belmar-Campos C., Grzyska U., Fiehler J. Suspected encephalitis with Candida tropicalis and Fusarium detected by unbiased RNA sequencing. Ann Hematol. 2016;95:1919–1921. doi: 10.1007/s00277-016-2770-3. [DOI] [PubMed] [Google Scholar]
- 39.Salzberg S.L., Breitwieser F.P., Kumar A., Hao H., Burger P., Rodriguez F.J. Next-generation sequencing in neuropathologic diagnosis of infections of the nervous system. Neurol Neuroimmunol Neuroinflamm. 2016;3:e251. doi: 10.1212/NXI.0000000000000251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilson M.R., Zimmermann L.L., Crawford E.D., Sample H.A., Soni P.R., Baker A.N. Acute West Nile virus meningoencephalitis diagnosed via metagenomic deep sequencing of cerebrospinal fluid in a renal transplant patient. Am J Transplant. 2017;17:803–808. doi: 10.1111/ajt.14058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morfopoulou S., Mee E.T., Connaughton S.M., Brown J.R., Gilmour K., Chong W.K. Deep sequencing reveals persistence of cell-associated mumps vaccine virus in chronic encephalitis. Acta Neuropathol. 2017;133:139–147. doi: 10.1007/s00401-016-1629-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chiu C.Y., Coffey L.L., Murkey J., Symmes K., Sample H.A., Wilson M.R. Diagnosis of fatal human case of St. Louis encephalitis virus infection by metagenomic sequencing, California, 2016. Emerg Infect Dis. 2017;23:1964–1968. doi: 10.3201/eid2310.161986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lum S.H., Turner A., Guiver M., Bonney D., Martland T., Davies E. An emerging opportunistic infection: fatal astrovirus (VA1/HMO-C) encephalitis in a pediatric stem cell transplant recipient. Transpl Infect Dis. 2016;18:960–964. doi: 10.1111/tid.12607. [DOI] [PubMed] [Google Scholar]
- 44.Wilson M.R., Naccache S.N., Samayoa E., Biagtan M., Bashir H., Yu G. Actionable diagnosis of neuroleptospirosis by next-generation sequencing. N Engl J Med. 2014;370:2408–2417. doi: 10.1056/NEJMoa1401268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilson M.R., Shanbhag N.M., Reid M.J., Singhal N.S., Gelfand J.M., Sample H.A. Diagnosing Balamuthia mandrillaris encephalitis with metagenomic deep sequencing. Ann Neurol. 2015;78:722–730. doi: 10.1002/ana.24499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Benjamin L.A., Lewthwaite P., Vasanthapuram R., Zhao G., Sharp C., Simmonds P. Human parvovirus 4 as potential cause of encephalitis in children, India. Emerg Infect Dis. 2011;17:1484–1487. doi: 10.3201/eid1708.110165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Quan P.L., Wagner T.A., Briese T., Torgerson T.R., Hornig M., Tashmukhamedova A. Astrovirus encephalitis in boy with X-linked agammaglobulinemia. Emerg Infect Dis. 2010;16:918–925. doi: 10.3201/eid1606.091536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kawada J., Okuno Y., Torii Y., Okada R., Hayano S., Ando S. Identification of viruses in cases of pediatric acute encephalitis and encephalopathy using next-generation sequencing. Sci Rep. 2016;6:33452. doi: 10.1038/srep33452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arden K.E., Heney C., Shaban B., Nimmo G.R., Nissen M.D., Sloots T.P. Detection of Toscana virus from an adult traveler returning to Australia with encephalitis. J Med Virol. 2017;89:1861–1864. doi: 10.1002/jmv.24839. [DOI] [PubMed] [Google Scholar]
- 50.Mongkolrattanothai K., Naccache S.N., Bender J.M., Samayoa E., Pham E., Yu G. Neurobrucellosis: unexpected answer from metagenomic next-generation sequencing. J Pediatr Infect Dis Soc. 2017;6:393–398. doi: 10.1093/jpids/piw066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Spandole S., Cimponeriu D., Berca L.M., Mihaescu G. Human anelloviruses: an update of molecular, epidemiological and clinical aspects. Arch Virol. 2015;160:893–908. doi: 10.1007/s00705-015-2363-9. [DOI] [PubMed] [Google Scholar]
- 52.Maggi F., Bendinelli M. Human anelloviruses and the central nervous system. Rev Med Virol. 2010;20:392–407. doi: 10.1002/rmv.668. [DOI] [PubMed] [Google Scholar]
- 53.Maggi F., Fornai C., Vatteroni M.L., Siciliano G., Menichetti F., Tascini C. Low prevalence of TT virus in the cerebrospinal fluid of viremic patients with central nervous system disorders. J Med Virol. 2001;65:418–422. doi: 10.1002/jmv.2051. [DOI] [PubMed] [Google Scholar]
- 54.Pollicino T., Raffa G., Squadrito G., Costantino L., Cacciola I., Brancatelli S. TT virus has a ubiquitous diffusion in human body tissues: analyses of paired serum and tissue samples. J Viral Hepat. 2003;10:95–102. doi: 10.1046/j.1365-2893.2003.00417.x. [DOI] [PubMed] [Google Scholar]
- 55.Kriesel J.D., Hobbs M.R., Jones B.B., Milash B., Nagra R.M., Fischer K.F. Deep sequencing for the detection of virus-like sequences in the brains of patients with multiple sclerosis: detection of GBV-C in human brain. PLoS One. 2012;7 doi: 10.1371/journal.pone.0031886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu Z., Zhang Y., Wei F., Xu M., Mou D., Zhang T. Detection of GB virus C genomic sequence in the cerebrospinal fluid of a HIV-infected patient in China: a case report and literature review. Epidemiol Infect. 2016;144:106–112. doi: 10.1017/S0950268815001326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hardie D., Smuts H. Human pegivirus-1 in the CSF of patients with HIV-associated neurocognitive disorder (HAND) may be derived from blood in highly viraemic patients. J Clin Virol. 2017;91:58–61. doi: 10.1016/j.jcv.2017.04.007. [DOI] [PubMed] [Google Scholar]
- 58.Tunkel A.R., Glaser C.A., Bloch K.C., Sejvar J.J., Marra C.M., Roos K.L. The management of encephalitis: clinical practice guidelines by the Infectious Diseases Society of America. Clin Infect Dis. 2008;47:303–327. doi: 10.1086/589747. [DOI] [PubMed] [Google Scholar]
- 59.Lipkin W.I. The changing face of pathogen discovery and surveillance. Nat Rev Microbiol. 2013;11:133–141. doi: 10.1038/nrmicro2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schlaberg R., Chiu C.Y., Miller S., Procop G.W., Weinstock G., Professional Practice C. Validation of metagenomic next-generation sequencing tests for universal pathogen detection. Arch Pathol Lab Med. 2017;141:776–786. doi: 10.5858/arpa.2016-0539-RA. [DOI] [PubMed] [Google Scholar]