Abstract
Serum levels of IgG, IgM and IgA against severe acute respiratory distress syndrome (SARS)‐associated coronavirus (SARS‐CoV) were detected serially with the use of immunofluorescent antibody assays in 30 patients with SARS. Seroconversion for IgG (mean 10 days) occurred simultaneously, or 1 day earlier, than that for IgM and IgA (mean 11 days for both). IgG could be detected as early as 4 days after the onset of illness. The earliest time at which these three antibodies reached peak levels was similar (mean 15 days). A high IgG level (1:800) could persist for > 3 months. The kinetics of neutralisation antibodies obtained with 100× the tissue culture infective dose (TCID50) of the SARS‐CoV TW1 strain in five patients with SARS nearly paralleled those for IgG. There were no significant differences in the kinetics of the IgG, IgM and IgA responses between patients with or without underlying medical disease, steroid or intravenous immunoglobulin therapy, or mechanical ventilation.
Keywords: Coronavirus, IgA, IgG, IgM, neutralisation antibody, SARS
Introduction
Severe acute respiratory distress syndrome (SARS) is an emerging infection that has affected more than 8000 patients in many countries [1]. This highly contagious infection has a propensity to spread to healthcare workers and household members, and may also cause outbreaks in the community [2, 3, 4, 5, 6, 7]. As of 5 July 2003, when Taiwan was declared free of SARS by the World Health Organization, 346 laboratory‐confirmed SARS cases had been reported, and 37 (11%) of these patients had died [1].
The first SARS patient in Taiwan was identified in the National Taiwan University Hospital (NTUH) on 25 February 2003, and 76 patients with SARS were eventually identified in this hospital during the outbreak [2, 7, 8, 9]. Among these patients, 18 had microbiological evidence of infection with SARS‐associated coronavirus (SARS‐CoV), including positive RT‐PCR and real‐time RT‐PCR assays from respiratory or serum samples. In all patients, an indirect enzyme‐linked immunosorbent assay (ELISA) revealed IgG antibody against SARS‐CoV in serum samples collected 28–35 days after the onset of fever.
The aim of this study was to evaluate the chronological evolution of IgM, IgA, IgG and neutralisation (NT) antibodies following SARS‐CoV infection of 30 patients who were treated at NTUH during the epidemic.
Patients and methods
Patients
Of the 76 SARS patients for whom serial serum samples were preserved, 30 were included in this study. Sera from these 30 patients (6–12 samples from each patient) were collected from < 7 days to 2–3 months after the onset of illness (defined as first appearance of fever with body temperature ≥ 38.3°C). The patients were aged 25–80 years (mean 43 years). Four patients had underlying disease, namely diabetes mellitus (n = 2), hypertension (n = 1) and chronic hepatitis B virus carriage (n = 1), while the other patients were previously healthy. Sputum or throat swab specimens from 12 of these patients were positive for SARS‐CoV RNA.
Immunofluorescent antibody assays
Specific antibodies (IgG, IgM and IgA) to SARS‐CoV were determined with two different immunofluorescent antibody (IFA) assays: an in‐house assay using whole‐cell lysate of infected Vero E6 cells as an antigen, or a commercial kit (Anti‐SARS‐CoV‐IIFT; Euroimmun, Lübeck, Germany) [6, 10]. For the in‐house IFA assay, spot slides were prepared by applying 10 µL of Vero E6 cell suspension, either infected or non‐infected with the SARS‐CoV TW1 strain (GenBank accession no. AY291451). Slides were dried and fixed in acetone. The conjugates used were goat anti‐human IgG, IgM and IgA conjugated to fluorescein isothiocyanate (Organon Teknika‐Cappel, Turnhout, Belgium). The starting dilutions of serum specimens were 1:25 for the in‐house IFA and 1:10 for the Euroimmun kit. Before determination of IgM and IgA antibodies with IFA, IgG antibodies were removed from patient sera by immunosorption with anti‐human IgG, using either a Eurosorb kit (Euroimmun) with the commercial IFA assay, or a Gullsorb kit (Meridian Bioscience, Cincinnati, OH, USA) with the in‐house assay. The cut‐off values for a positive result were 1:25 for the in‐house IFA and 1:10 for the commercial IFA kit [2, 10].
ELISA
IgG antibody against SARS‐CoV was also measured with an indirect ELISA, with recombinant nucleocapsid as the coated antigen (SARS‐96 (TMB); General Biologicals, Hsin‐Chu, Taiwan) [10, 11]. The cut‐off value for a positive IgG result by ELISA was 0.26 [10, 11].
Control sera
Controls comprised 200 paired sera from patients with community‐acquired pneumonia seen at NTUH from October 2001 to December 2002, 70 sera from hospitalised patients with acute respiratory distress syndrome treated in 2002 at the hospital, and ten sera from ten pregnant women obtained during routine pre‐labour check‐ups in 2002. The control sera were tested for the presence of IgG, IgM and IgA by the three methods described above.
NT antibody assay
Briefly, sera from five patients were incubated at 56°C for 30 min, and then diluted two‐fold in cell culture medium (modified Eagle medium). Aliquots (50 µL) of diluted sera (from four‐fold to 516‐fold) were added to 50 µL of cell culture medium containing 100× the tissue culture infective dose (TCID50) of the SARS‐CoV TW1 strain on a 96‐well microtitre plate and incubated at 37°C for 2 h in CO2 5% v/v. Finally, 100 µL of Vero E6 cells (2.5 × 105/mL) was added to each well of the plate. The plates were incubated at 37°C for 3–5 days in CO2 5% v/v and examined daily for a cytopathic effect. On day 5, the highest dilution of serum that completely inhibited 100× TCID50 of SARS‐CoV was recorded as the NT titre. NT assays were performed in triplicate with negative control sera from healthy volunteers.
Results
All control sera were negative for IgG by ELISA, and for IgG, IgM and IgA by IFA. The time required for seroconversion, as determined by the two IFA assays for IgG, IgM and IgA, and by the two IFA assays and ELISA for IgG, was nearly identical among these patients (Fig. 1). Tests for IgG, IgM or IgA were negative until at least 3 days after the onset of illness in all 30 patients (Fig. 1a). Assays for all three specific antibodies were positive for at least 19 days after the onset of illness in these patients. Seroconversion of IgG (mean 10 days) occurred at the same time, or 1 day earlier, than for IgM and IgA (mean 11 days for both). The time required for the first peak level of these three antibodies was similar (mean 15 days) (Fig. 1b). The highest levels of IgG, IgM and IgA were 1:6400, 1:640, and 1:1280, respectively. All patients were positive for IgG for > 28 days (1:400–1:1600), and one patient had a high level of IgG (1:800) at 100 days after the onset of illness. The levels of IgM and IgA started to decline 3–4 weeks after the onset of illness, and remained at low levels (1:40–1:80) at 12 weeks after onset.
Figure 1.
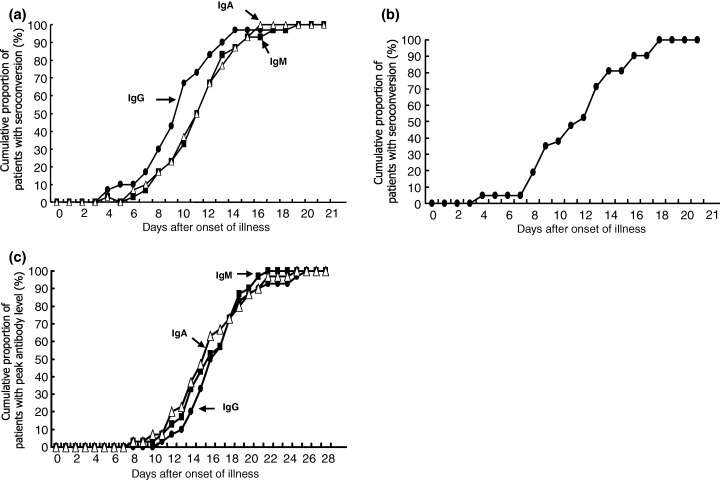
Chronological evolution of IgG, IgM, and IgA antibodies to SARS‐associated coronavirus (SARS‐CoV) in 30 patients with SARS. (a) Cumulative proportion against time required for seroconversion to IgG, IgM and IgA according to immunofluorescent antibody (IFA) assays. (b) Cumulative proportion against time required for seroconversion to IgG according to ELISA. (c) Cumulative proportion against time required for peak levels of antibodies according to IFA assays.
In addition to treatment with ribavirin (used for 29 of the 30 patients), 28 patients received intravenous methylprednisolone (1–11 days, mean 6 days, after the onset of illness, and 2–4 days before any IgG response), 21 received intravenous immunoglobulin (2–12 days, mean 6 days, after the onset of illness), and nine were given mechanical ventilation (4–12 days, mean 8 days, after the onset of illness) following respiratory failure. There were no significant differences in the kinetics of the IgG, IgM and IgA response between patients with or without underlying medical disease, steroid or intravenous immunoglobulin therapy, or mechanical ventilation. However, two patients had early IgG seroconversion (1:50 and 1:100, respectively) on day 4 of illness before starting corticosteroid therapy (day 5), and one patient had IgG antibody (1:50) on day 5 of illness when corticosteroid therapy was started.
NT antibody appeared on days 10–12 (mean 1:32), increased thereafter, and peaked (1:128–1:256) on days 18–24. In four patients, the NT antibody titre remained at 1:32 or 1:64 at 2 months after onset, and was 1:64 on day 100 of the illness. Figs 2a and 2b illustrate the characteristic changes in IgG, IgM, IgA and NT antibodies against SARS‐CoV in two previously healthy patients during the acute and convalescent stages of the disease.
Figure 2.
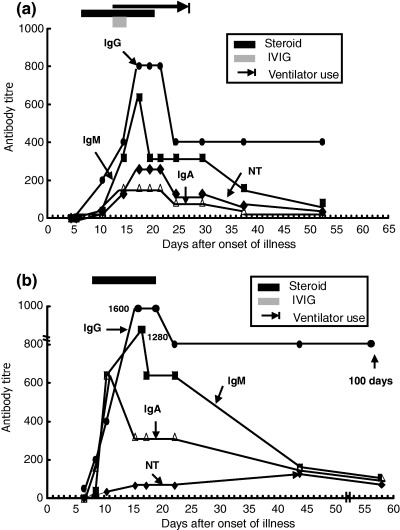
Dynamic changes of IgG, IgM, IgA and neutralisation (NT) antibodies against SARS‐CoV in two previously healthy patients with SARS. (a) Antibody changes in a SARS patient who received corticosteroid, intravenous immunoglobulin (IVIG) and mechanical ventilation. (b) Antibody changes in a SARS patient who received corticosteroid therapy only.
Discussion
The results of this study demonstrate five important points. First, although previous reports have indicated that specific IgG is not detected in SARS patients until days 7–9 of the disease [2, 11, 12, 13, 14, 15, 16], the present study showed clearly that IgG seroconversion can start as early as 4 days after the onset of illness. Second, in contrast to the profiles of antibody responses against acute virus infections, and a previous finding on humoral immunity (IgG and IgM) to SARS [9], a simultaneous or earlier IgG response against SARS‐CoV TW1, compared with IgM or IgA, was observed. This indicates that detection of IgM or IgA would not provide earlier evidence for SARS‐CoV infection than detection of IgG. Third, the study showed the presence of low levels of IgM and IgA at 100 days after the onset of illness, although a previous study showed the disappearance of IgM after 12 weeks [14]. Fourth, the presence of underlying disease (diabetes mellitus, hypertension, chronic hepatitis B virus carriage) and the use of immunosuppressive or immunomodulatory agents did not influence the dynamics of the antibody response (i.e., the times required to achieve seroconversion and peak antibody levels) [2]. Finally, the presence of high levels of specific IgG and NT antibodies to SARS‐CoV in the late convalescent stage (2 to at least 3 months after the onset of illness) suggests that passive immunisation with convalescent plasma or concentrated SARS‐CoV IgG antibody from recovered SARS patients might be an option for the treatment of SARS.
The reason for a simultaneous or slightly earlier IgG response is unclear. The time at which these isotypes were detected might depend more on the test employed than the actual timing of the IgM‐to‐IgG switch. Theoretically, this should not happen if these patients had a true primary response to SARS, and should be accelerated if it was a secondary response. One possibility is that some patients were infected with SARS‐CoV before the documented febrile episodes during the epidemic. This is difficult to exclude, but is extremely unlikely. A previous study of emergency room workers revealed a similar antibody subclass response in two patients with mild symptoms (fever with body temperature < 38.3°C) and one asymptomatic worker [10]. Alternatively, the observation could reflect the low sensitivity of IFA assays for the detection of IgM or IgA. It is known that detection of IgM or IgA without the separation of IgG yields higher rates of false‐positive and false‐negative results. In the present study, IgG was absorbed before detection of IgM and IgA in two different IFA assays. Accordingly, the sensitivity of the IgM and IgA assays after IgG absorption, rather than the biology of the host response, might contribute to this phenomenon. Therefore, the serological response of SARS‐CoV‐infected patients might need to be examined with the use of more sensitive methods or different antigens.
Theoretically, steroids are more likely to interfere with an established immune response than its initiation, and the present study did not observe a delay in the primary immune response with steroid therapy. Previous observations have suggested that the upsurge of IgG antibody to SARS‐CoV correlates with the clinical worsening of pneumonia [2, 7, 14]. However, in the present study, patients whose pulmonary condition improved after corticosteroid therapy had similar IgG profiles to patients who later developed respiratory failure necessitating ventilator support. An over‐exuberant host response, as well as other immunopathological processes, might contribute to a worsening of disease and progressive lung damage.
Acknowledgments
We are indebted to many members of the front‐line medical and nursing staff and laboratory personnel of the National Taiwan University Hospital for their management of these patients. We thank D.‐S. Chen for his critical review and constructive comments on this manuscript.
References
- 1. World Health Organization. Severe Acute Respiratory Syndrome (SARS): summary table of SARS cases by country, 1 November 2002. http://www.who.int/csr/sars/country/table2003_09_23/en/ (accessed 26 September 2003).
- 2. Hsueh PR, Hsiao CH, Yeh SH et al. Microbiologic characteristics, serologic response, and clinical manifestations in severe acute respiratory syndrome, Taiwan. Emerg Infect Dis 2003; 9: 1163–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ksiazek TG, Erdman D, Goldsmith CS et al. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med 2003; 348: 1953–1966.DOI: 10.1056/NEJMoa030781 [DOI] [PubMed] [Google Scholar]
- 4. Lee N, Hui D, Wu A et al. A major outbreak of severe acute respiratory syndrome in Hong Kong. N Engl J Med 2003; 348: 1986–1994.DOI: 10.1056/NEJMoa030685 [DOI] [PubMed] [Google Scholar]
- 5. Poutanen SM, Low DE, Henry B et al. Identification of severe acute respiratory syndrome in Canada. N Engl J Med 2003; 348: 1995–2005.DOI: 10.1056/NEJMoa030634 [DOI] [PubMed] [Google Scholar]
- 6. Tsang KW, Ho PL, Ooi GC et al. A cluster of cases of severe acute respiratory syndrome in Hong Kong. N Engl J Med 2003; 348: 1977–1985.DOI: 10.1056/NEJMoa030666 [DOI] [PubMed] [Google Scholar]
- 7. Wang JT, Sheng WH, Fang CT et al. Clinical manifestations, laboratory findings, and treatment outcomes of SARS patients. Emerg Infect Dis 2004; 10: 818–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hsueh PR, Chen PJ, Hsiao CH et al. Patient data, early SARS epidemic, Taiwan. Emerg Infect Dis 2004; 10: 489–493. [DOI] [PubMed] [Google Scholar]
- 9. Hsueh PR, Yang PC. Severe acute respiratory syndrome (SARS)—an emerging infection of the 21st century. J Formos Med Assoc 2003; 102: 825–839. [PubMed] [Google Scholar]
- 10. Chang WT, Kao CL, Chung MY et al. SARS exposure and emergency department workers. Emerg Infect Dis 2004; 10: 1117–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huang LR, Chiu CM, Yeh SH et al. Evaluation of antibody responses against SARS coronavirus nucleocapsid or spike proteins by immunoblotting or ELISA. J Med Virol 2004; 73: 338–346.DOI: 10.1002/jmv.20096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Drosten C, Gunther S, Preiser W et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med 2003; 348: 1967–1976.DOI: 10.1056/NEJMoa030747 [DOI] [PubMed] [Google Scholar]
- 13. Li G, Chen X, Xu A. Profile of specific antibodies to the SARS‐associated coronavirus. N Engl J Med 2003; 349: 508–509.DOI: 10.1056/NEJM200307313490520 [DOI] [PubMed] [Google Scholar]
- 14. Peiris JS, Chu CM, Cheng VC et al. Clinical progression and viral load in a community outbreak of coronavirus‐associated SARS pneumonia: a prospective study. Lancet 2003; 361: 1767–1772.DOI: 10.1016/S0140-6736(03)13412-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Peiris JS, Lai ST, Poon LL et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 2003; 361: 1319–1325.DOI: 10.1016/S0140-6736(03)13077-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wu HS, Chiu SC, Tseng TC et al. Serologic and molecular biologic methods for SARS‐associated coronavirus infection, Taiwan. Emerg Infect Dis 2004; 9: 304–310. [DOI] [PMC free article] [PubMed] [Google Scholar]


