Highlights
-
•
Development and in-house validation of a multiplex RT-qPCR method.
-
•
Identification and discrimination of Zika and chikungunya viruses.
-
•
Specific and sensitive method applicable to real-life samples.
-
•
High coverage of arbovirus strains/isolates.
-
•
Testing of the method via a new bioinformatics tool on whole genome sequencing data.
Keywords: RT-qPCR, Zika virus, Chikungunya virus, Multiplex, Identification, Discrimination
Abstract
Objective
The re-emergence and spread of tropical viruses to new areas has raised a wave of concern worldwide. In order to treat patients at an early stage and prevent the diffusion of an outbreak, early diagnosis, and therefore fast and adequate detection, is needed. To this end, a multiplex reverse transcription real-time polymerase chain reaction TaqMan method was designed to detect Zika (ZIKV) and chikungunya (CHIKV) viruses simultaneously.
Methods
Two methods targeting different genome segments were selected from the literature for each virus. These were adapted for high genome coverage and combined in a four-plex assay that was thoroughly validated in-house. The SCREENED tool was used to evaluate the sequence coverage of the method.
Results
The full validation approach showed that the new four-plex method allows the specific and sensitive identification and discrimination of ZIKV and CHIKV in routine samples. The combination of two targets per virus allowing almost 100% coverage of about 500 genomes is shown for the first time.
Conclusions
PCR is a reliable user-friendly technique that can be applied in remote areas. Such multiplex methods enable early and efficient diagnosis, leading to rapid treatment and effective confinement in outbreak cases. They may also serve as an aid for surveillance, and the full validation permits easy method-transfer allowing worldwide harmonization.
Introduction
Arboviruses are a group of viruses transmitted by arthropod vectors such as Aedes spp. mosquitos. Over the last decades, several epidemic arboviral diseases have been reported in areas different from their endemic region (tropics/subtropics; Kraemer et al., 2015, Liu-Helmersson et al., 2014) (e.g. Anukumar et al., 2014, Aubry et al., 2015, Paules and Fauci, 2017, Sadarangani and Hsu, 2016, WHO, 2018a, WHO, 2018c, WHO, 2018e, WHO, 2018f). The alphavirus chikungunya virus (CHIKV) and the flavivirus Zika virus (ZIKV) are two such arboviruses which have gained attention due to several outbreaks in recent years (Aamir et al., 2017, Campos et al., 2015, Gregianini et al., 2017, Kabir et al., 2017, Leparc-Goffart et al., 2014, WHO, 2016a). Infections caused by both viruses, although basically asymptomatic, may have severe effects in humans (Burt et al., 2017, Capeding et al., 2013, Hamer and Chen, 2014, Javelle et al., 2015, Joubert et al., 1985, Rampal and Meena, 2007, Reiter, 2010, Schilte et al., 2013, Thiberville et al., 2013, Wielanek et al., 2007), including neurological disorders (Brasil et al., 2016, Broutet et al., 2016, Calvet et al., 2016, Cao-Lormeau et al., 2016, Cauchemez et al., 2016, de Araújo et al., 2016, de Oliveira et al., 2017, Mlakar et al., 2016, Oehler et al., 2014, Parra et al., 2016, Rasmussen et al., 2016, Rodrigues, 2016, WHO, 2016c).
Urbanization, globalization, and global warming (Gubler, 2011, Messina et al., 2015, Whitehorn and Farrar, 2010) have enhanced the expansion of the Aedes aegypti and Aedesalbopictus vectors from their geographic origin to other regions, including Europe (Caminade et al., 2012; Charrel et al., 2014; Deblauwe et al., 2015; Ducheyne et al., 2018; ECDC; Grard et al., 2014; Kraemer et al., 2015; Medlock et al., 2012; Renault et al., 2007; Renault et al., 2007; Rocklöv et al., 2016; Schaffner and Mathis, 2014; Wilder-Smith et al., 2017; Wong et al., 2013). Consequently, three million people are living in Aedes-infested regions (Wilder-Smith et al., 2017), greatly enhancing the risk for ZIKV and CHIKV infections in areas where the local population is immunologically naïve. Additionally, travel-related cases of ZIKV and CHIKV infection have been reported in Europe (Beltrame et al., 2007, Duijster et al., 2016, Maria et al., 2016, Paty et al., 2014, Rezza et al., 2007, WHO, 2018d, WHO, 2018g, Zammarchi et al., 2015), Japan (Shinohara et al., 2016), Australia (Pyke et al., 2014), and Israel (Meltzer et al., 2016). The 2016 ZIKV epidemic in Brazil, spreading rapidly to other South American countries and North America (Armstrong, 2016, Chen and Hamer, 2016, Faria et al., 2016; WHO), also supports this reality. Moreover, autochthonous transmission of CHIKV (Cunha et al., 2017, Delisle et al., 2015, Grandadam et al., 2011, Kabir et al., 2017, WHO, 2016b) and ZIKV (Moi et al., 2017, Musso and Lanteri, 2017, WHO, 2018b, Zanluca et al., 2015) has been observed, as well as sexual transmission (D’Ortenzio et al., 2016, Davidson, 2016, Deckard, 2016, Foy et al., 2011, McCarthy, 2016, Musso et al., 2015b), mother-to-child/perinatal transmission (Besnard et al., 2014, Calvet et al., 2016, Gérardin et al., 2014, Oliveira Melo et al., 2016, Ramful et al., 2007), and transmission via blood/platelet transfusion (Brouard et al., 2008, Magnus et al., 2018, Motta et al., 2016, Musso et al., 2014). These different ways of transmission/infection put the global population at risk, especially as there is currently no vaccine against ZIKV or CHIKV. It is thus of great importance to develop differential diagnostic tools for the early detection of such infections in order to allow a rapid intervention.
In earlier years, methods developed for the detection of ZIKV and CHIKV were mainly based on serological analyses such as plaque reduction neutralization tests (Lanciotti et al., 2007, Lindsey et al., 1976, Shan et al., 2017) and immunoassays such as ELISA tests (EUROIMMUN; Grivard et al., 2007, Huzly et al., 2016, Johnson et al., 2016, Litzba et al., 2008, Martin et al., 2000, Steinhagen et al., 2016, Tsai et al., 2018). Although these techniques are still useful as they allow detection at a later stage, their clear interpretation is rendered difficult by the fact that cross-reactivity with other arboviruses occurs (Duffy et al., 2009, Fagbami, 1979, Kam et al., 2015, Lanciotti et al., 2008).
In the meantime, molecular based methods have gained interest. They are fast, reliable, specific, allow simultaneous identification and quantification, and are efficient in early diagnosis, as viral RNA can be detected in serum in the first week after the onset of clinical illness, as well as in other sample types, e.g., saliva (Barzon et al., 2016, Musso et al., 2015a), urine (Barzon et al., 2016, Gourinat et al., 2015, Shinohara et al., 2016), and semen (Atkinson et al., 2016, Mansuy et al., 2016). Several such assays have already been developed for ZIKV/CHIKV (Balm et al., 2012, Calvert et al., 2017, Faye et al., 2013, Panning et al., 2008, Parida et al., 2007, Patel et al., 2016, Pongsiri et al., 2012, Wang et al., 2016a, Wang et al., 2016b). However, many have been developed for a single region- and/or outbreak-specific strain and thereby lack broad range coverage. Furthermore, they have been developed and published by single laboratories, each using their own experimental conditions, and thus they do not allow a single-plate combination, and specificity studies have often been limited to few other (arbo)viruses. These features make these methods ineffective for a global view of arbovirus diagnosis, and existing methods should thus be adapted or newly designed to allow more straightforward and universal detection.
However, RNA viruses have a high mutation rate (Drake and Holland, 1999, Jenkins et al., 2002), as illustrated by changes in ZIKV (Corman et al., 2016, Faye et al., 2014) and CHIKV (Weaver and Forrester, 2015) sequences throughout the years/epidemics. These mutations can lead to false-negative results when they occur in the annealing sites of the primers/probes. Next generation sequencing (NGS) allows a high volume of sequence data to be obtained, which can be analysed for mutations (Aw et al., 2014). This could be used to help in the verification of existing molecular diagnostic methods and the necessary redesign of primer/probes, and thus help improve on current methods by filling in the gaps.
This article reports the development and in-house validation of a new multiplex RT-qPCR TaqMan method designed for the simultaneous detection and discrimination of ZIKV and CHIKV. To circumvent the possibility of obtaining false-negative results due to affected primer/probe annealing sites (Drexler et al., 2007, Kwok et al., 1990), two pre-existing methods – each targeting a different segment in the genomic sequence – were combined for each virus: nsp4 gene (Panning et al., 2008) and E gene (Lanciotti et al., 2007) for CHIKV; M/A genes and E gene (Lanciotti et al., 2008) for ZIKV. The different method acceptance parameters (specificity, sensitivity, applicability, and practicability) needed to declare a method fit for purpose were evaluated extensively. Additionally, the in silico coverage of a broad range of complete sequences was verified for the first time using whole genome sequencing (WGS) data and a recently developed bioinformatics tool (Vanneste et al., 2018).
Materials and methods
Selection of primer/probe sets
Methods detecting ZIKV/CHIKV were selected from the literature based on three criteria: TaqMan technology, amplicon size (50–150 bp), and targeted sequence. The oligonucleotides were subjected to alignment analysis on a limited set of CHIKV/ZIKV sequences retrieved from NCBI, using Clustal Omega (EMBL-EBI). Primer/probe sequences were adapted if necessary to allow higher sequence coverage. Subsequently, primer/probe complementarities, melting temperature (Tm), and self-annealing were evaluated for each method using Multi Primer Analyzer software with standard settings (Thermo Fisher Scientific).
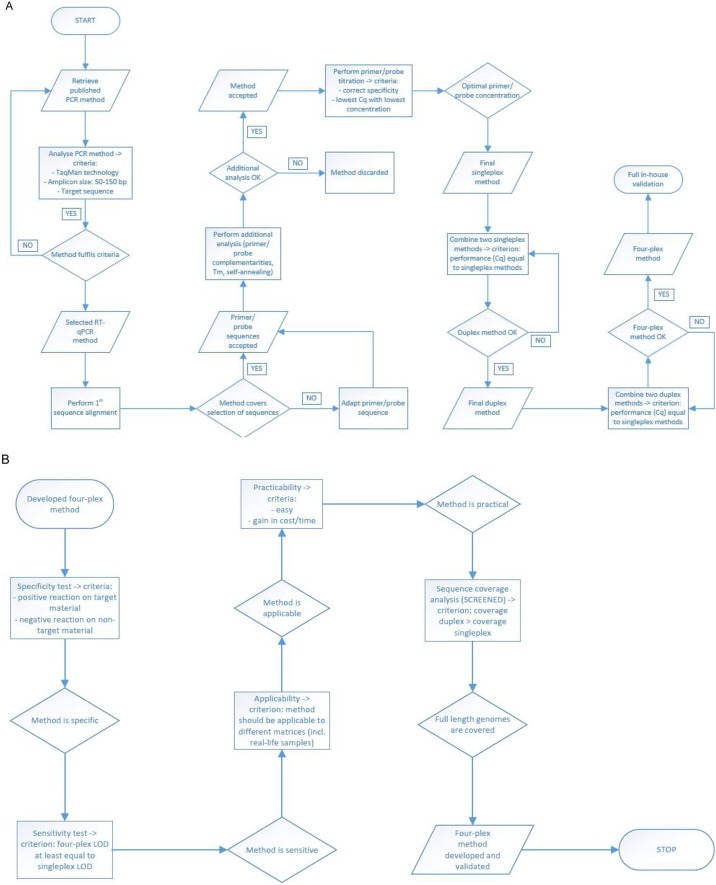
Methods passing this selection stage were tested in a singleplex assay in a primer/probe concentration titration experiment (five primer/probe concentration combinations) using positive reference materials specific for the virus under analysis (Table 1 ). Methods that performed well (i.e., positive reaction on the specific reference materials and no reaction on the negative control, lowest quantification cycle (Cq) value with lowest primer/probe concentration) were combined into a two-plex assay per virus (acceptance criterion: similar performance as the singleplex) and then into the final four-plex assay. The procedure followed is explained in Figure 1 .
Table 1.
Materials used and specificity results using the four-plex RT-qPCR method.
| Species | Type | Strain | Origin | Expected signal |
Obtained signal |
||
|---|---|---|---|---|---|---|---|
| CHIKV | ZIKV | CHIKV | ZIKV | ||||
| Viruses | |||||||
| Chikungunya | RNA | S27 Petersfield straina | AmpliRun® Chikungunya Virus RNA Control; Labconsult (Vircell) | + | − | + | − |
| La Reunion – 2006 | CIBU | + | − | + | − | ||
| ROS | Sciensano (Viral Diseases) | + | − | + | − | ||
| Zika | RNA | MR766 strain (African)a | AmpliRun® Zika Virus RNA Control; Labconsult (Vircell) | − | + | − | + |
| PRVABC59 strain (Asian) | AmpliRun® Zika Virus (Asian Lineage) RNA Control; Labconsult (Vircell) | − | + | − | + | ||
| 7630 Cote d’Ivoire – 1980 | CIBU | − | + | − | + | ||
| Strain H/PF/2013 (clinical isolate) – French Polynesia 2013 | Sciensano (Viral Diseases) | − | + | − | + | ||
| Dengue serotype 1 | RNA | Hawaii strain | AmpliRun® Dengue 1 Virus RNA Control; Labconsult (Vircell) | − | − | − | − |
| DENV1 – Hawaii (sngt vero cells) | CIBU | − | − | − | − | ||
| Dengue serotype 2 | RNA | New Guinea C strain | AmpliRun® Dengue 2 Virus RNA Control; Labconsult (Vircell) | − | − | − | − |
| DENV2 – New Guinea (sngt vero cells) | CIBU | − | − | − | − | ||
| Dengue serotype 3 | RNA | H87 strain | AmpliRun® Dengue 3 Virus RNA Control; Labconsult (Vircell) | − | − | − | − |
| DENV3 – Pat H Birmanie (sngt vero cells) | CIBU | − | − | − | − | ||
| Dengue serotype 4 | RNA | H241 strain | AmpliRun® Dengue 4 Virus RNA Control; Labconsult (Vircell) | − | − | − | − |
| DENV4 – 63632 Birmanie (sngt AP61) | CIBU | − | − | − | − | ||
| Tick-borne encephalitis | RNA | Neudorfl strain | AmpliRun® Tick Borne Encephalitis Virus RNA Control; Labconsult (Vircell) | − | − | − | − |
| TBEV – Langat | CIBU | − | − | − | − | ||
| TBEV – Hypr | CIBU | − | − | − | − | ||
| TBEV – Neudorfl | Sciensano (Viral Diseases) | − | − | − | − | ||
| TBEV – Absettarov | Sciensano (Viral Diseases) | − | − | − | − | ||
| West Nile | RNA | New York-99 strain | AmpliRun® West Nile Virus RNA Control; Labconsult (Vircell) | − | − | − | − |
| WNV – Souche 143 – lineage 1 | CIBU | − | − | − | − | ||
| WNV – Camargue 2002 – lineage 1 | CIBU | − | − | − | − | ||
| WNV – New York 99 – lineage 1 | Sciensano (Viral Diseases) | − | − | − | − | ||
| WNV – lineage 2 | Sciensano | − | − | − | − | ||
| Yellow fever | RNA | 17D strain | AmpliRun® Yellow Fever Virus RNA Control; Labconsult (Vircell) | − | − | − | − |
| Asibi strain | CIBU | − | − | − | − | ||
| FNV | CIBU | − | − | − | − | ||
| 17D strain | CIBU | − | − | − | − | ||
| Japanese encephalitis | RNA | JEV – genotype 1 | CIBU | − | − | − | − |
| JEV – Nakayama (genotype III) | Sciensano (Viral Diseases) | − | − | − | − | ||
| St Louis encephalitis | RNA | / | AmpliRun® St Louis Encephalitis Virus RNA Control; Labconsult (Vircell) | − | − | − | − |
| Western equine encephalitis | RNA | H160/99 strain | AmpliRun® Western Equine Encephalitis RNA Control; Labconsult (Vircell) | − | − | − | − |
| Eastern equine encephalitis | RNA | H178/99 strain | AmpliRun® Eastern Equine Encephalitis RNA Control; Labconsult (Vircell) | − | − | − | − |
| Parainfluenza 1 | RNA | C-35 strain | AmpliRun® Parainfluenza 1 RNA Control; Labconsult (Vircell) | − | − | − | − |
| Influenza A H1 | RNA | A/Brisbane/59/2007 | AmpliRun® Influenza A H1 RNA Control; Labconsult (Vircell) | − | − | − | − |
| Influenza B | RNA | B/Brisbane/60/2008 | AmpliRun® Influenza B RNA Control; Labconsult (Vircell) | − | − | − | − |
| Rhino | RNA | 1059 strain | AmpliRun® Rhinovirus RNA Control; Labconsult (Vircell) | − | − | − | − |
| Corona | RNA | 229E STRAIN | AmpliRun® Coronavirus RNA Control; Labconsult (Vircell) | − | − | − | − |
| Bacteria | |||||||
| Streptococcus salivarius | DNA | Subsp. salivarius Andrewes and Horder | LGC (ATCC® 9759D5™) | − | − | − | − |
| Actinomyces naeslundii | DNA | Thompson and Lovestedt | LGC (ATCC® 12104D5™) | − | − | − | − |
| Lactobacillus jensenii | DNA | Gasser et al. | LGC (ATCC® 25258D™) | − | − | − | − |
| Lactobacillus gasseri | DNA | Lauer and Kandler | LGC (ATCC® 33323D5™) | − | − | − | − |
| Staphylococcus epidermidis | DNA | (Winslow and Winslow) Evans | LGC (ATCC® 35984D5™) | − | − | − | − |
| Staphylococcus aureus | DNA | Subsp. aureus Rosenbach | LGC (ATCC® 700699D5™) | − | − | − | − |
| Streptococcus agalactiae | DNA | Lehmann and Neumann | LGC (ATCC® BAA611D5™) | − | − | − | − |
| Listeria monocytogenes | DNA | 53 XXIII strain | AmpliRun® Listeria Monocytogenes DNA Control; Labconsult (Vircell) | − | − | − | − |
| Salmonella enteritidis | DNA | CDC K-1891 strain (subspecies enterica) | AmpliRun® Salmonella Enteritidis DNA Control; Labconsult (Vircell) | − | − | − | − |
| Salmonella typhi | DNA | / | AmpliRun® Salmonella Typhi DNA Control; Labconsult (Vircell) | − | − | − | − |
| Escherichia coli (VTEC) | DNA | / | AmpliRun® Escherichia Coli (VTEC) DNA Control; Labconsult (Vircell) | − | − | − | − |
| Yeast/fungi | |||||||
| Candida albicans | DNA | (Robin) Berkhout | (ATCC® 14053D™) | − | − | − | − |
| Aspergillus versicolor | DNA | BCCM/IHEM 1994 | Sciensano (IHEM/BCCM collection, Mycology and Aerobiology) | − | − | − | − |
| Aspergillus fumigatus | DNA | MCV-C#10 strain | AmpliRun® Aspergillus Fumigatus DNA Control; Labconsult (Vircell) | − | − | − | − |
| Parasites | |||||||
| Giardia intestinalis | DNA | WB clone C6 | AmpliRun® Giardia Intestinalis DNA Control; Labconsult (Vircell) | − | − | − | − |
| DNA | (Lambl) Alexeieff | LGC (ATCC® 50803D™) | − | − | − | − | |
| Cryptosporidium parvum | DNA | Strain Iowa | LGC (ATCC® PRA67™) | − | − | − | − |
| Entamoeba histolytica | DNA | Schaudinn | LGC (ATCC® 30459D™) | − | − | − | − |
| Human | |||||||
| Human | DNA | / | Thermo Fisher Scientific | − | − | − | − |
CHIKV, chikungunya virus; ZIKV, Zika virus.
Strain used for the determination of the sensitivity of the method (limit of detection, LOD); +: signal at Cq ≤38, −: signal at Cq >38.
Figure 1.
Overview of the workflow used to (A) select the RT-qPCR methods and develop the four-plex method and (B) validate the four-plex RT-qPCR method.
Materials and extraction
Reference RNA and DNA materials were purchased or obtained from sample collections (Table 1).
The QCMD 2016 Zika Virus EQA Pilot Study consisted of 10 samples: two ZIKV-negative sample (one containing CHIKV), seven ZIKV-positive samples (four African, three French Polynesian), and one mixed sample (composed of dengue virus serotype 2 (DENV-2), West Nile virus (WNV), and yellow fever virus (YFV)). Serum, saliva, and urine samples were collected during the 2016 outbreak in New Caledonia by Institut Pasteur de Nouvelle-Calédonie (IPNC) and in 2017 in French Guiana by Institut Pasteur de la Guyane (IPG) from patients presenting symptoms possibly due to a ZIKV/CHIKV infection.
RNA from proficiency test and serum/saliva/urine samples was extracted using the QIAamp Viral RNA Mini Kit (Qiagen) and MagNA Pure LC Total Nucleic Acid Isolation Kit (Roche), respectively.
One-step RT-qPCR
All reactions were performed on a CFX96 Touch™ instrument (Bio-Rad) using the SuperScript III Platinum One-Step Quantitative RT-PCR System (Invitrogen) according to the manufacturer’s instructions. A final reaction volume of 25 μl containing 5 μl template was used. The following cycling conditions were applied: a cDNA synthesis cycle 15 min/50 °C, a hold step 2 min/95 °C, and subsequently 45 cycles of denaturation 15 s/95 °C and annealing/elongation 30 s/60 °C. Nuclease-free water was used as the no-template control (NTC).
Detailed information concerning the oligonucleotides used (Eurogentec) is given in Table 2 .
Table 2.
Primer/probe sets and final concentrations used in the four-plex RT-qPCR method, as well as reference amplicons for each method as used in the SCREENED analysis.
| Primer/probe name | Sequence (5′ → 3′) | Target sequence | Amplicon size (bp) | Location on reference genomea | Final concentration (nM) | Ref. |
|---|---|---|---|---|---|---|
| Target: CHIKV | ||||||
| CHIKV-a-F CHIKV-a-R CHIKV-a-P |
TGATCCCGACTCAACCATCCT GGCAAACGCAGTGGTACTTCCT FAM-TCCGACATCATCCTCCTTGCTGGC-BHQ-1 |
Nsp1 | 83 | 241–323 | 600 600 300 |
Panning et al. (2008) |
| Reference amplicon: TGATCCCGACTCAACCATCCTGGATATTGGTAGTGCGCCAGCAAGGAGGATGATGTCGGACAGGAAGTACCACTGCGTTTGCC | ||||||
| CHIKV-b-F CHIKV-b-R CHIKV-b-P |
TCACTCCCTGTTGGACTTGATAGA TTGACGAACAGAGTTAGGAACATACC HEX-AGGTACGCGCTTCAAGTTCGGCG-BHQ-1 |
E1 | 126 | 6856–6981 | 800 800 400 |
Lanciotti et al. (2007) |
| Reference amplicon: TCACTCCCTGTTGGACTTGATAGAGGCTGCTTTCGGAGAGATTTCCAGCTGTCATCTACCGACAGGTACGCGCTTCAAGTTCGGCGCCATGATGAAATCTGGTATGTTCCTAACTCTGTTCGTCAA | ||||||
| Target: ZIKV | ||||||
| ZIKV-a-F ZIKV-a-R ZIKV-a-P |
TTGGTCATGATACTGCTGATTGC CCYTCCACAAAGTCCCTATTGC TEX-CGGCATACAGYATCAGGTGCATWG GAG-BHQ-2 |
M/A | 77 | 941–1017 | 600 600 300 |
Adapted from Lanciotti et al. (2008) |
| Reference amplicon: TTGGTCATGATACTGCTGATTGCCCCGGCATACAGTATCAGGTGCATTGGAGTCAGCAATAGAGACTTCGTGGAGGG | ||||||
| ZIKV-b-F ZIKV-b-R ZIKV-b-P |
YCGYTGCCCAACACAAG CCACYAAYGTTCTTTTGCAGACAT Cy5-AGCCTACCTTGACAAGCARTCAGACA CTCAA-BHQ-2 |
E | 77 | 1192–1268 | 1000 1000 500 |
Adapted from Lanciotti et al. (2008) |
| Reference amplicon: TCGTTGCCCAACACAAGGTGAAGCCTACCTTGACAAGCAATCAGACACTCAATATGTCTGCAAAAGAACATTAGTGG | ||||||
Generation of an RNA standard
The regions targeted by the two ZIKV and CHIKV methods were amplified from the respective reference materials (Table 1) and cloned into the pBluescript II SK+ plasmid (GeneCust). Additionally, a genetically modified (GM) plant sequence (EURL, 2010) was cloned into the plasmid. The presence of the DNA inserts was confirmed by Sanger sequencing.
The plasmid was linearized by digestion with SpeI. The target sequences were amplified using the TranscriptAid T7 High Yield Transcription Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. The in vitro transcribed RNA was treated with DNAse to digest the plasmid and was then purified using the GeneJET RNA Purification Kit (Thermo Fisher Scientific). The RNA was quantified on a Qubit 3.0 Fluorometer (Thermo Fisher Scientific).
Method acceptance parameters
The specificity of the four-plex assay was tested in two replicates on a list of materials, as shown in Table 1. The RT-qPCR conditions were as described above. For materials where the copy number was known, a dilution of 200 cp/μl was used; for other materials a 1:10 and 1:100 dilution was used. Based on the literature, the Cq cut-off was set at 38. The sensitivity of the assay was determined for each method of the multiplex on a specific material (Table 1) by preparing a dilution series (1000, 100, 50, 10, 5, 1, 0.1 cp) in nuclease-free water and measuring each dilution two-fold in two independent runs under repeatability conditions (ISO24276, 2006). Using samples from a proficiency test (QCMD 2016 Zika Virus EQA Pilot Study) and routine samples, all tested in duplicate, the parameters applicability and practicability were evaluated.
Use of the bioinformatics tool
Coverage of the ZIKV and CHIKV genome sequences by the four chosen methods (individually and in duplex) was analysed using the SCREENED web tool (Vanneste et al., 2018) with default advanced options. A config file containing the oligonucleotide and reference amplicon sequences (Table 2) was used for the analysis. As the reference amplicon sequence input in SCREENED is a codon DNA strand, the CHIKV-a probe, originally designed to anneal to the anti-codon DNA strand, was introduced as its reverse complement. Full-length nucleotide sequences for ZIKV were retrieved from the NCBI Virus Resource database (5/11/2017) allowing for any host, region/country, genome region, and isolation source; and for CHIKV from the NCBI Nucleotide database using ‘“Chikungunya virus”[porgn:__txid37124] AND “complete genome”’ as keywords (5/11/2017). The accession numbers retrieved for both viruses are available as Supplementary material (Tables S1 and S2).
Results
Development of a new four-plex RT-qPCR method
For CHIKV, all four RT-qPCR methods that complied with the set criteria showed good sequence coverage and no oligonucleotide adaptations were necessary. The subsequent analysis showed a good performance for all four methods, as no primer/probe complementarities or self-annealing properties were detected and Tm values complied with standard PCR rules. In the titration experiment, two CHIKV methods performed well and were combined to form a duplex.
For ZIKV, a similar procedure was followed. Four of the five retrieved methods were adapted after the alignment analysis to cover for genetic variation. All five, together with an in-house designed method, showed a good performance in the subsequent analysis. Based on the titration experiment, three methods were discarded (late Cq values for all primer/probe concentrations). The three remaining methods (all adapted from the literature) were combined into three duplex assays, of which one showed the best results.
The two duplex assays, complying with the set criterion, were combined into a four-plex assay, which was subjected to the validation process evaluating the necessary acceptance parameters (Figure 1); the optimal primer/probe concentrations that resulted from the titration experiment were used (Table 2).
Due to the limitation to four fluorophores, no internal control was added to the reaction. However, an external positive (and negative) control should always be included. The constructed RNA standard gave a specific positive reaction with the multiplex assay for all four methods and could thus be used effectively as a positive control. The cloned soybean 356043 GM plant sequence (EURL, 2010) allows testing for contamination; i.e., when a presumed negative control reacts positively with the ZIKV/CHIKV multiplex, running a PCR with the GM-specific primers/probes and its specific cycling programme will allow to determine whether the positivity comes from a contamination with the plasmid.
In-house validation of the new multiplex RT-qPCR method
To ensure a new PCR method is fit for purpose, a number of method acceptance parameters should be tested, such as the specificity and sensitivity, at the very least (cf MIQE guidelines (Bustin et al., 2009)). As no specific guidelines are available for RT-qPCR methods for virus detection, the in-house validation was based on a combination of the methods reported by Saunders et al. (Saunders et al., 2013) and Broeders et al. (Broeders et al., 2014) and the basic specificity principles of ISO 22118 (ISO22118, 2011) and ISO 16140 (AFNOR, 2003).
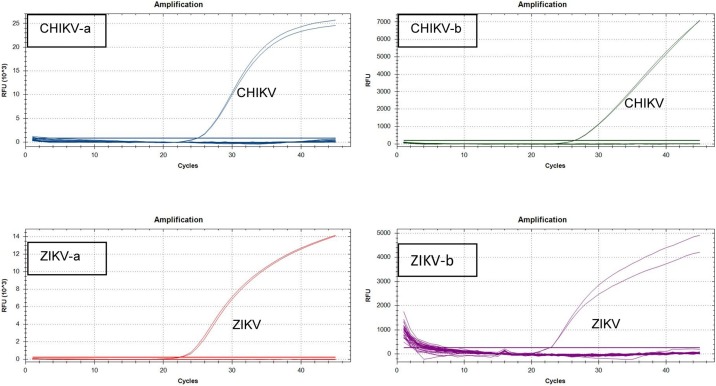
Specificity
For all four methods, all strains of the respective positive reference materials showed a clear specific amplification (Table 1; Figure 2 ). None of the non-target-containing materials gave a specific amplification (Table 1). This list included RNA from arboviruses (different strains) other than ZIKV and CHIKV, RNA from respiratory viruses showing similar symptoms upon infection, human DNA, DNA from bacteria commonly/possibly present in saliva (Kang et al., 2006), DNA from bacteria and yeast putatively found in the urinary tract (Hilt et al., 2014), and DNA from a few other microorganisms (ISO22118, 2011, AFNOR, 2003). Additionally, no unspecific hits were returned in the in silico test (data not shown) performed for each method, first against its counterpart (CHIKV methods versus ZIKV database and vice versa) and subsequently as MegaBlast against the entire NCBI database without any specification for species.
Figure 2.
Amplification curves obtained in the specificity test for each of the four methods constituting the new multiplex RT-qPCR method. For readability of the graphics, a selection of tested materials was made: dengue serotype 1-4, CHIKV, West Nile virus, Yellow Fever virus, St Louis encephalitis virus, Eastern Equine encephalitis virus, Western Equine encephalitis virus, Influenza A H1, Influenza B (all reference materials from Labconsult (Vircell)), ZIKV, Japanese encephalitis virus, Tick borne encephalitis virus (all materials received form CIBU), Streptococcus salivarius, Lactococcus jensenii (both ATCC materials obtained from LGC) and NTC.
Sensitivity
The sensitivity of the multiplex method was assessed via the determination of the limit of detection (LOD) of each of the methods when used as a four-plex. The LOD of each run was set at the lowest copy number at which both replicates were still positive (Cq cut-off = 38), and the LOD of the method was established as the highest LOD over the two runs. According to this, the LOD was set at 5 cp and 50 cp for the CHIKV-a and CHIKV-b methods, respectively, and at 100 cp for both ZIKV methods (Table 3 ).
Table 3.
Results of the sensitivity test for the four-plex RT-qPCR method for the detection of ZIKV and CHIKV.
| 1000 cp | 100 cp | 50 cp | 10 cp | 5 cp | 1 cp | 0.1 cp | NTC | |
|---|---|---|---|---|---|---|---|---|
| CHIKV-a | 28.2–28.2 | 31.9–31.5 | 33.3–32.9 | 35.2–35.9 | 35.7–36.1 | 40.3–ND | ND–ND | ND |
| 28.4–28.5 | 32.2–32.2 | 33.2–33.1 | 35.8–35.7 | 36.8–36.0 | 38.6–ND | ND–ND | ND | |
| CHIKV-b | 30.1–30.0 | 35.0–34.0 | 35.8–36.1 | ND–ND | ND–ND | ND–ND | ND–ND | ND |
| 28.3–28.4 | 31.9–31.5 | 33.0–33.8 | 36.3–ND | ND–ND | ND–ND | ND–ND | ND | |
| ZIKV-a | 31.9–32.6 | 35.1–35.8 | 37.6–36.6 | ND–ND | ND–ND | ND–ND | ND–ND | ND |
| 32.1–32.1 | 34.8–34.5 | 35.7–ND | ND–ND | ND–ND | ND–ND | ND–ND | ND | |
| ZIKV-b | 32.0–33.2 | 36.3–36.9 | 38.4–38.0 | 39.1–39.9 | ND–ND | ND–ND | ND–ND | ND |
| 33.3–33.2 | 37.3–37.4 | 38.2–38.2 | 40.1–ND | ND–ND | ND–ND | ND–ND | ND |
CHIKV, chikungunya virus; ZIKV, Zika virus. Cut-off used = 38. ND: not determined (Cq >45). Results in italic indicate the limit of detection (LOD) of the single run.
Applicability and practicability
To show the applicability of this multiplex RT-qPCR method, it was tested on different matrices: the reference materials used for the development, the plasmid positive control, the samples from the QCMD proficiency test, and routine serum/saliva/urine samples collected from patients suspected to be infected with ZIKV/CHIKV (Table 4 ). A correct outcome was obtained for all materials, i.e., exponential-shaped amplification curves and Cq <38 for CHIKV/ZIKV-containing matrices, and an unspecific curve and Cq >38 considered as negative for non-ZIKV/CHIKV materials.
Table 4.
Results of the ZIKV/CHIKV four-plex RT-qPCR method on real-life samples.
| Sample origin (suspected infection) | Serum |
Urine |
Saliva |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ZIKV-a | ZIKV-b | CHIKV-a | CHIKV-b | ZIKV-a | ZIKV-b | CHIKV-a | CHIKV-b | ZIKV-a | ZIKV-b | CHIKV-a | CHIKV-b | |
| IPNC (ZIKV) | + (35.7) | ND | − (42.2) | − (38.9) | + (31.4) | + (35.1) | ND | ND | + (28.4) | + (29.8) | + (41.2) | ND |
| IPNC (ZIKV) | + (35.9) | ND | ND | ND | + (25.8) | + (28.9) | ND | ND | + (29.8) | + (31.1) | ND | ND |
| IPG (ZIKV) | ND | ND | ND | ND | + (32.0) | + (32.4) | ND | ND | + (36.7) | + (37.6) | ND | ND |
| IPG (CHIKV) | ND | ND | + (18.6) | + (25.9) | / | / | / | / | / | / | / | / |
| IPG (CHIKV) | ND | ND | + (18) | + (27.2) | / | / | / | / | / | / | / | / |
CHIKV, chikungunya virus; ZIKV, Zika virus. Cut-off used = 38. ND: not determined (Cq > 45). /: test not performed. Represented Cq values are the mean of two values.
Concerning practicability, the fact that four methods were combined into one already renders it very time and cost efficient. Additionally, due to the use of the one-step RT-qPCR kit, the method is easy to apply in a routine laboratory, as it can be set up as a simple PCR reaction not needing any additional instrument/infrastructure or training of staff.
Sequence coverage of the multiplex method
For CHIKV, 521 full-length genomes were retrieved and the predictive SCREENED analysis showed that 99.8% and 97.1% were covered by CHIKV-a and CHIKV-b, respectively. CHIKV-a was expected to detect all genomes except KX168429.1 (Asia) due to a two-nucleotide mismatch at the very 3′ end of the reverse primer. CHIKV-b showed slightly smaller in silico inclusivity, but it was predicted to detect genome KX168429.1, while the 15 genomes not detected by CHIKV-b were recognized by CHIKV-a. When the two methods were combined, 100% of the retrieved genomes were thus predicted to be detected (Table 5 ).
Table 5.
Evaluation of CHIKV and ZIKV RT-qPCR methods using the SCREENED web tool. The first column shows the method name. The second column shows the total number of genomes analysed per method. The third, fourth, and fifth columns show the numbers of genomes detected, numbers of genomes not detected, and numbers of genomes where the amplicon could not be retrieved, respectively. The sixth column shows the percentage for the in silico inclusivity per method and per duplex.
| Method name | Number of genomes analysed | Number of genomes detected | Number of genomes not detected | Number of genomes no amplicon found | % in silico inclusivity |
|---|---|---|---|---|---|
| CHIKV-a | 521 | 520 | 1 | 0 | 99.8 |
| CHIKV-b | 521 | 506 | 15 | 0 | 97.1 |
| CHIKV duplex | 521 | 521 | 0 | 0 | 100.0 |
| ZIKV-a | 478 | 474 | 4 | 0 | 99.2 |
| ZIKV-b | 478 | 461 | 17 | 0 | 96.4 |
| ZIKV duplex | 478 | 476 | 2 | 0 | 99.6 |
CHIKV, chikungunya virus; ZIKV, Zika virus.
A total of 478 complete ZIKV genome sequences were selected and both ZIKV-a and ZIKV-b methods performed well, with an in silico inclusivity of 99.16% and 96.44%, respectively. In duplex, only two genomes were predicted to be undetected. For genome KF383115 (Africa), this was due to one nucleotide mismatch within the 3′ end of the reverse primer in both methods. If one mismatch is allowed in this region, or if the length of the 3’end region sensitive to nucleotide mismatches is set at 4 (instead of the default value of 5), this genome would be detected by both methods. Genome KF383118 (Africa) was predicted to be undetected by both ZIKV-a (because of one nucleotide mismatch in the 3′ end of the forward primer-template duplex) and ZIKV-b (because of a nucleotide mismatch of the last nucleotide in the 3′ end of the reverse primer-template duplex). If two mismatches are allowed in this critical region, this genome would be detected by both methods.
Discussion
Rapid, reliable, and early diagnosis is of great importance to enable the correct treatment to be applied for infected individuals and, perhaps even more importantly, to contain the spread of the infection.
Although several RT-qPCR methods for arbovirus detection have been developed, they have some drawbacks for application in a multi-target screening environment: they have often been designed based on a single region/outbreak-specific strain, using particular run conditions, and validation has most often been tested in a restricted way. Parameters such as specificity, sensitivity, practicability, and applicability, often lacking or poorly addressed in the literature, are needed to declare a newly developed method fit for purpose. These parameters should be systematically included in the process of method development; for example, see Broeders et al. and Saunders et al. (Broeders et al., 2014, Saunders et al., 2013). Such method validation, according to well-defined criteria, increases the efficiency of diagnosis as it enables easy method transfer between laboratories, allowing worldwide harmonization.
To exploit the PCR technology in a more efficient way and to meet the current needs in arbovirus detection – i.e., effectiveness, reliability, transferability, and universal application – a new four-plex RT-qPCR assay for the simultaneous detection and discrimination of ZIKV and CHIKV was developed and thoroughly validated in-house. The assay is specific: it only detects the different strains of the targeted arboviruses and none of the non-target materials, including organisms that may be present simultaneously with the infective arbovirus. A sensitivity of 5–100 cp was obtained, which is comparable to the individual original methods. The method can be efficiently applied on proficiency test and routine serum/saliva/urine samples. This is of great importance in view of eventually using non-invasive samples to circumvent the current necessity of venous blood collection. It has indeed already been demonstrated that arboviruses are present in samples other than serum, e.g., saliva (Musso et al., 2015a, Musso et al., 2016, Tauro et al., 2017), urine (El Wahed et al., 2017, Gourinat et al., 2015, Hirayama et al., 2012), and semen (Barzon et al., 2016). It is worth establishing this further on a larger set of samples. The positive detection in these samples also supports the fact that the four-plex method is applicable on strains from different regions/outbreaks.
The plasmid, which was developed in parallel and can be used as a positive control, is easily multipliable; this therefore eliminates the need to buy expensive control material or to purify it in high concentration from viruses for which specific confinement/biosafety conditions need to be taken into account.
The use of two independent target sequences for each virus enhances the reliability of the assay, as false-negative results for the first method due to the occurrence of mutations can be covered by the second method. Indeed, using the SCREENED tool it was shown, for example, that genome KX168429.1, which is undetected by CHIKV-a, is covered by its counterpart CHIKV-b; this proves the added value of designing RT-qPCR methods on two distinct targets for the same virus.
Bio-informatics analysis, as presented here for the first time, using the SCREENED tool, further demonstrates the possible detection of a wide number of strains emerging in different regions and from different outbreaks. This analysis, important for the worldwide application of a detection method, can never be performed so extensively in a laboratory. However, the tool only gives an indication of the performance of an (RT-q)PCR method and laboratory testing is still required. For example, it cannot be excluded that the method will work in practice on genome KF383115, which is undetected by the two ZIKV methods due to a mismatch located in the fifth last nucleotide of the 3′ end of the primer.
The analysis of a large amount of WGS data (obtained via NGS) using the SCREENED tool could further help to verify the performance of the existing molecular diagnostic methods. The observed genetic variability and the detection of mutations could lead to the modification of primers/probes, thereby providing highly universal methods that are applicable on the different occurring strains/isolates and thus helping to cover the gaps (i.e., poor inclusivity) in the current methods.
Overall, the results show the validity of this multiplex RT-qPCR method. The combination of the four chosen methods allows the detection of the specific viruses in different sample types without cross-reactivity and this can easily be implemented in routine settings as a first step in the differential diagnosis of arbovirus infections.
(RT-)qPCR is an easy, user-friendly technique, making it an ideal tool to be implemented at point-of-care (POC) facilities. The development of countertop PCR instruments (e.g., Genesystem Genechecker; Roche Diagnostics Cobas Liat PCR System; Mediphos GenePOC revogene platform; Cepheid GeneXpert) that are affordable, small, easy to operate, run on site, have fast turnaround times, and even include the extraction step, which is often a critical point when working in the field, will prompt early diagnosis, leading to quicker decision-making. The further development of other arbovirus-specific multiplex methods, using the same RT-qPCR one-step kit and cycling conditions as the CHIKV/ZIKV one described here, will aid in building up a multi-target one-plate panel usable in screening analysis, allowing the detection of different targets in an efficient manner and a shorter time. If, in addition, a full in-house validation is performed, the transfer of these methods could enable worldwide harmonization.
This progress, together with extended prevention and information campaigns (e.g., use of social media), will lead to the better and faster control and management of arbovirus infections – and by extension other diseases – which in turn could improve patient health, reduce the risk of transmission, and potentially lower mortality and morbidity, as well as allow worldwide surveillance.
Author contributions
The authors declare that all persons in the list of authors actively contributed to either the design of the study, the acquisition/analysis of data, the drafting of the article, or the revision of the manuscript. All authors also declare approving the final submitted version of the article.
Human and animal rights
The study presented, including the publication of data related to arbovirus infection, was approved by the Comité Consultatif d’Ethique de Nouvelle-Calédonie (February 2015) and by the Comité de Protection des Personnes Ile de France I (March 2015-13851 and January 2018-14793). The human biological samples used in this study were obtained from patients after receiving written informed consent.
Conflict of interest
The authors declare that they have no competing interests.
Acknowledgements
This work was supported by the project ORIENT-EXPRESS financed by the WIV-ISP(presently named Sciensano; RP-PJ project number 0000754) and partially by the Arbo-VIRTUESS project financed by the Actions Concertées Interpasteuriennes(project number ACIP 2014-053). These funding sources had no involvement in the study design, data collection/analysis/interpretation, or writing of the article. The authors are grateful to the service Mycology & Aerology (IHEM/BCCM collection – Sciensano) and B. Lambrecht (Sciensano) for providing materials, and S. Lamoral (Sciensano) for technical support. The authors would also like to thank A. Perilhou and N. Jolly (Centre for Translational Science, Institut Pasteur Paris) for their expertise on ethical issues.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.ijid.2019.12.028.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- Aamir U.B., Badar N., Salman M., Ahmed M., Alam M.M. Outbreaks of chikungunya in Pakistan. Lancet Infect Dis. 2017;17(5):483. doi: 10.1016/S1473-3099(17)30191-3. [DOI] [PubMed] [Google Scholar]
- AFNOR . Organisation internationale de Normalisation; Genève: 2003. Microbiologie des aliments–Protocole pour la validation des méthodes alternatives (ISO 16140-2003) c2003. [Google Scholar]
- Anukumar B., Sapkal G.N., Tandale B.V., Balasubramanian R., Gangale D. West Nile encephalitis outbreak in Kerala, India, 2011. J Clin Virol. 2014;61(1):152–155. doi: 10.1016/j.jcv.2014.06.003. [DOI] [PubMed] [Google Scholar]
- Armstrong P. Travel-associated Zika virus disease cases among US residents—United States, January 2015–February 2016. MMWR Morb Mortal Wkly Rep. 2016;65 doi: 10.15585/mmwr.mm6511e1. [DOI] [PubMed] [Google Scholar]
- Atkinson B., Hearn P., Afrough B., Lumley S., Carter D., Aarons E.J. Detection of Zika Virus in Semen. Emerg Infect Dis. 2016;22(5):940. doi: 10.3201/eid2205.160107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubry M., Teissier A., Roche C., Richard V., Yan A.S., Zisou K. Chikungunya outbreak, French Polynesia, 2014. Emerg Infect Dis. 2015;21(4):724–726. doi: 10.3201/eid2104.141741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aw P.P.K., de Sessions P.F., Wilm A., Hoang L.T., Nagarajan N., Sessions O.M. Dengue. Springer; 2014. Next-generation whole genome sequencing of dengue virus; pp. 175–195. [DOI] [PubMed] [Google Scholar]
- Balm M.N., Lee C.K., Lee H.K., Chiu L., Koay E.S., Tang J.W. A diagnostic polymerase chain reaction assay for Zika virus. J Med Virol. 2012;84(9):1501–1505. doi: 10.1002/jmv.23241. [DOI] [PubMed] [Google Scholar]
- Barzon L., Pacenti M., Berto A., Sinigaglia A., Franchin E., Lavezzo E. Isolation of infectious Zika virus from saliva and prolonged viral RNA shedding in a traveller returning from the Dominican Republic to Italy, January 2016. Euro Surveill. 2016;21(10):30159. doi: 10.2807/1560-7917.ES.2016.21.10.30159. [DOI] [PubMed] [Google Scholar]
- Beltrame A., Angheben A., Bisoffi Z., Monteiro G., Marocco S., Calleri G. Imported chikungunya infection, Italy. Emerg Infect Dis. 2007;13(8):1264–1266. doi: 10.3201/eid1308.070161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnard M., Lastere S., Teissier A., Cao-Lormeau V., Musso D. Evidence of perinatal transmission of Zika virus, French Polynesia, December 2013 and February 2014. Euro Surveill. 2014;19(13):20751. [PubMed] [Google Scholar]
- Brasil P., Pereira J.P., Jr., Moreira M.E., Ribeiro Nogueira R.M., Damasceno L., Wakimoto M. Zika Virus infection in pregnant women in Rio de Janeiro. N Engl J Med. 2016;375(24):2321–2334. doi: 10.1056/NEJMoa1602412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broeders S., Huber I., Grohmann L., Berben G., Taverniers I., Mazzara M. Guidelines for validation of qualitative real-time PCR methods. Trends Food Sci Technol. 2014;37(2):115–126. [Google Scholar]
- Brouard C., Bernillon P., Quatresous I., Pillonel J., Assal A., De Valk H. Estimated risk of Chikungunya viremic blood donation during an epidemic on Reunion Island in the Indian Ocean, 2005 to 2007. Transfusion. 2008;48(7):1333–1341. doi: 10.1111/j.1537-2995.2008.01646.x. [DOI] [PubMed] [Google Scholar]
- Broutet N., Krauer F., Riesen M., Khalakdina A., Almiron M., Aldighieri S. Zika virus as a cause of neurologic disorders. N Engl J Med. 2016;374(16):1506–1509. doi: 10.1056/NEJMp1602708. [DOI] [PubMed] [Google Scholar]
- Burt F.J., Chen W., Miner J.J., Lenschow D.J., Merits A., Schnettler E. Chikungunya virus: an update on the biology and pathogenesis of this emerging pathogen. Lancet Infect Dis. 2017;17(4):e107–e117. doi: 10.1016/S1473-3099(16)30385-1. [DOI] [PubMed] [Google Scholar]
- Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55(4):611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Calvert A.E., Biggerstaff B.J., Tanner N.A., Lauterbach M., Lanciotti R.S. Rapid colorimetric detection of Zika virus from serum and urine specimens by reverse transcription loop-mediated isothermal amplification (RT-LAMP) PLoS One. 2017;12(9) doi: 10.1371/journal.pone.0185340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvet G., Aguiar R.S., Melo A.S.O., Sampaio S.A., de Filippis I., Fabri A. Detection and sequencing of Zika virus from amniotic fluid of fetuses with microcephaly in Brazil: a case study. Lancet Infect Dis. 2016;16(6):653–660. doi: 10.1016/S1473-3099(16)00095-5. [DOI] [PubMed] [Google Scholar]
- Caminade C., Medlock J.M., Ducheyne E., McIntyre K.M., Leach S., Baylis M. Suitability of European climate for the Asian tiger mosquito Aedes albopictus: recent trends and future scenarios. J R Soc Interface. 2012;9(75):2708–2717. doi: 10.1098/rsif.2012.0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos G.S., Bandeira A.C., Sardi S.I. Zika Virus outbreak, Bahia, Brazil. Emerg Infect Dis. 2015;21(10):1885–1886. doi: 10.3201/eid2110.150847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao-Lormeau V.-M., Blake A., Mons S., Lastère S., Roche C., Vanhomwegen J. Guillain-Barré Syndrome outbreak associated with Zika virus infection in French Polynesia: a case-control study. Lancet. 2016;387(10027):1531–1539. doi: 10.1016/S0140-6736(16)00562-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capeding M.R., Chua M.N., Hadinegoro S.R., Hussain I.I., Nallusamy R., Pitisuttithum P. Dengue and other common causes of acute febrile illness in Asia: an active surveillance study in children. PLoS Negl Trop Dis. 2013;7(7):e2331. doi: 10.1371/journal.pntd.0002331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cauchemez S., Besnard M., Bompard P., Dub T., Guillemette-Artur P., Eyrolle-Guignot D. Association between Zika virus and microcephaly in French Polynesia, 2013–15: a retrospective study. Lancet. 2016;387(10033):2125–2132. doi: 10.1016/S0140-6736(16)00651-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrel R.N., Leparc-Goffart I., Gallian P., de Lamballerie X. Globalization of Chikungunya: 10 years to invade the world. Clin Microbiol Infect. 2014;20(7):662–663. doi: 10.1111/1469-0691.12694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.H., Hamer D.H. Zika Virus: rapid spread in the Western Hemisphere. Ann Intern Med. 2016;164(9):613–615. doi: 10.7326/M16-0150. [DOI] [PubMed] [Google Scholar]
- Corman V.M., Rasche A., Baronti C., Aldabbagh S., Cadar D., Reusken C.B. Assay optimization for molecular detection of Zika virus. Bull World Health Organ. 2016;94(12):880–892. doi: 10.2471/BLT.16.175950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha M.S., Cruz N.V.G., Schnellrath L.C., Medaglia M.L.G., Casotto M.E., Albano R.M. Autochthonous transmission of East/Central/South African genotype chikungunya virus, Brazil. Emerg Infect Dis. 2017;23(10):1737–1739. doi: 10.3201/eid2310.161855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Ortenzio E., Matheron S., de Lamballerie X., Hubert B., Piorkowski G., Maquart M. Evidence of sexual transmission of Zika virus. N Engl J Med. 2016;374(22):2195–2198. doi: 10.1056/NEJMc1604449. [DOI] [PubMed] [Google Scholar]
- Davidson A. Suspected female-to-male sexual transmission of Zika virus—New York City, 2016. MMWR Morb Mortal Wkly Rep. 2016;65 doi: 10.15585/mmwr.mm6528e2. [DOI] [PubMed] [Google Scholar]
- de Araújo T.V.B., Rodrigues L.C., de Alencar Ximenes R.A., de Barros Miranda-Filho D., Montarroyos U.R., de Melo A.P.L. Association between Zika virus infection and microcephaly in Brazil, January to May, 2016: preliminary report of a case-control study. Lancet Infect Dis. 2016;16(12):1356–1363. doi: 10.1016/S1473-3099(16)30318-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira W.K., de Franca G.V.A., Carmo E.H., Duncan B.B., de Souza Kuchenbecker R., Schmidt M.I. Infection-related microcephaly after the 2015 and 2016 Zika virus outbreaks in Brazil: a surveillance-based analysis. Lancet. 2017;390(10097):861–870. doi: 10.1016/S0140-6736(17)31368-5. [DOI] [PubMed] [Google Scholar]
- Deblauwe I., Demeulemeester J., De Witte J., Hendy A., Sohier C., Madder M. Increased detection of Aedes albopictus in Belgium: no overwintering yet, but an intervention strategy is still lacking. Parasitol Res. 2015;114(9):3469–3477. doi: 10.1007/s00436-015-4575-z. [DOI] [PubMed] [Google Scholar]
- Deckard D.T. Male-to-male sexual transmission of Zika virus—Texas, January 2016. MMWR Morb Mortal Wkly Rep. 2016;65 doi: 10.15585/mmwr.mm6514a3. [DOI] [PubMed] [Google Scholar]
- Delisle E., Rousseau C., Broche B., Leparc-Goffart I., L’Ambert G., Cochet A. Chikungunya outbreak in Montpellier, France, September to October 2014. Euro Surveill. 2015;20(17) doi: 10.2807/1560-7917.es2015.20.17.21108. [DOI] [PubMed] [Google Scholar]
- Drake J.W., Holland J.J. Mutation rates among RNA viruses. Proc Natl Acad Sci. 1999;96(24):13910–13913. doi: 10.1073/pnas.96.24.13910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler J.F., de Souza Luna L.K., Pedroso C., Pedral-Sampaio D.B., Queiroz A.T., Brites C. Rates of and reasons for failure of commercial human immunodeficiency virus type 1 viral load assays in Brazil. J Clin Microbiol. 2007;45(6):2061–2063. doi: 10.1128/JCM.00136-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducheyne E., Tran Minh N.N., Haddad N., Bryssinckx W., Buliva E., Simard F. Current and future distribution of Aedes aegypti and Aedes albopictus (Diptera: Culicidae) in WHO Eastern Mediterranean Region. Int J Health Geogr. 2018;17(1):4. doi: 10.1186/s12942-018-0125-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy M.R., Chen T.H., Hancock W.T., Powers A.M., Kool J.L., Lanciotti R.S. Zika virus outbreak on Yap Island, Federated States of Micronesia. N Engl J Med. 2009;360(24):2536–2543. doi: 10.1056/NEJMoa0805715. [DOI] [PubMed] [Google Scholar]
- Duijster J.W., Goorhuis A., van Genderen P.J., Visser L.G., Koopmans M.P., Reimerink J.H. Zika virus infection in 18 travellers returning from Surinam and the Dominican Republic, The Netherlands, November 2015–March 2016. Infection. 2016;44(6):797–802. doi: 10.1007/s15010-016-0906-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ECDC. European Centre for Disease Prevention and Control - Mosquito maps; Available from: https://ecdc.europa.eu/en/disease-vectors/surveillance-and-disease-data/mosquito-maps.
- El Wahed A.A., Sanabani S.S., Faye O., Pessôa R., Patriota J.V., Giorgi R.R. Rapid molecular detection of Zika virus in acute-phase urine samples using the recombinase polymerase amplification assay. PLoS Curr. 2017;9 doi: 10.1371/currents.outbreaks.a7f1db2c7d66c3fc0ea0a774305d319e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EMBL-EBI. European Bioinformatics Institute, https://www.ebi.ac.uk/Tools/msa/clustalo/.
- EURL . 2010. Event-specific method for the quantification of soybean event DP-356043-5 using real-time PCR – validated method (CRLVL04/07VP). European Union Reference Laboratory for GM Food and Feed. [Google Scholar]
- EUROIMMUN. https://www.euroimmun.com/products/indications/infektions-serologie/zika-viruses.html.
- Fagbami A.H. Zika virus infections in Nigeria: virological and seroepidemiological investigations in Oyo State. J Hyg (Lond) 1979;83(2):213–219. doi: 10.1017/s0022172400025997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria N.R., Azevedo R., Kraemer M.U.G., Souza R., Cunha M.S., Hill S.C. Zika virus in the Americas: early epidemiological and genetic findings. Science. 2016;352(6283):345–349. doi: 10.1126/science.aaf5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faye O., Faye O., Diallo D., Diallo M., Weidmann M., Sall A.A. Quantitative real-time PCR detection of Zika virus and evaluation with field-caught mosquitoes. Virol J. 2013;10(1):311. doi: 10.1186/1743-422X-10-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faye O., Freire C.C., Iamarino A., Faye O., de Oliveira J.V.C., Diallo M. Molecular evolution of Zika virus during its emergence in the 20th century. PLoS Negl Trop Dis. 2014;8(1):e2636. doi: 10.1371/journal.pntd.0002636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foy B.D., Kobylinski K.C., Foy J.L.C., Blitvich B.J., da Rosa A.T., Haddow A.D. Probable non–vector-borne transmission of Zika virus, Colorado, USA. Emerg Infect Dis. 2011;17(5):880. doi: 10.3201/eid1705.101939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gérardin P., Sampériz S., Ramful D., Boumahni B., Bintner M., Alessandri J.-L. Neurocognitive outcome of children exposed to perinatal mother-to-child Chikungunya virus infection: the CHIMERE cohort study on Reunion Island. PLoS Negl Trop Dis. 2014;8(7):e2996. doi: 10.1371/journal.pntd.0002996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourinat A.C., O’Connor O., Calvez E., Goarant C., Dupont-Rouzeyrol M. Detection of Zika virus in urine. Emerg Infect Dis. 2015;21(1):84–86. doi: 10.3201/eid2101.140894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandadam M., Caro V., Plumet S., Thiberge J.M., Souares Y., Failloux A.B. Chikungunya virus, southeastern France. Emerg Infect Dis. 2011;17(5):910–913. doi: 10.3201/eid1705.101873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grard G., Caron M., Mombo I.M., Nkoghe D., Ondo S.M., Jiolle D. Zika virus in Gabon (Central Africa)–2007: a new threat from Aedes albopictus? PLoS Negl Trop Dis. 2014;8(2):e2681. doi: 10.1371/journal.pntd.0002681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregianini T.S., Ranieri T., Favreto C., Nunes Z.M.A., Tumioto Giannini G.L., Sanberg N.D. Emerging arboviruses in Rio Grande do Sul, Brazil: Chikungunya and Zika outbreaks, 2014‐2016. Rev Med Virol. 2017;27(6):e1943. doi: 10.1002/rmv.1943. [DOI] [PubMed] [Google Scholar]
- Grivard P., Le Roux K., Laurent P., Fianu A., Perrau J., Gigan J. Molecular and serological diagnosis of Chikungunya virus infection. Pathol Biol (Paris) 2007;55(10):490–494. doi: 10.1016/j.patbio.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Gubler D.J. Dengue, urbanization and globalization: the unholy trinity of the 21st century. Trop Med Health. 2011;39(4 SUPPLEMENT):S3–S11. doi: 10.2149/tmh.2011-S05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamer D.H., Chen L.H. Chikungunya: establishing a new home in the Western hemisphere. Ann Intern Med. 2014;161(11):827–828. doi: 10.7326/M14-1958. [DOI] [PubMed] [Google Scholar]
- Hilt E.E., McKinley K., Pearce M.M., Rosenfeld A.B., Zilliox M.J., Mueller E.R. Urine is not sterile: use of enhanced urine culture techniques to detect resident bacterial flora in the adult female bladder. J Clin Microbiol. 2014;52(3):871–876. doi: 10.1128/JCM.02876-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama T., Mizuno Y., Takeshita N., Kotaki A., Tajima S., Omatsu T. Detection of dengue virus genome in urine by real-time reverse transcriptase PCR: a laboratory diagnostic method useful after disappearance of the genome in serum. J Clin Microbiol. 2012;50(6):2047–2052. doi: 10.1128/JCM.06557-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huzly D., Hanselmann I., Schmidt-Chanasit J., Panning M. High specificity of a novel Zika virus ELISA in European patients after exposure to different flaviviruses. Euro Surveill. 2016;21(16) doi: 10.2807/1560-7917.ES.2016.21.16.30203. [DOI] [PubMed] [Google Scholar]
- ISO22118 . International Organization for Standardization; Genève, Switzerland: 2011. Microbiology of food and animal feeding stuffs — Polymerase chain reaction (PCR) for the detection and quantification of food-borne pathogens — Performance characteristics. [Google Scholar]
- ISO24276 . International Organization for Standardization; Genève, Switzerland: 2006. Foodstuffs - Methods of analysis for detection of genetically modified organisms and derived products - General requirements and definitions. [Google Scholar]
- Javelle E., Ribera A., Degasne I., Gauzere B.A., Marimoutou C., Simon F. Specific management of post-chikungunya rheumatic disorders: a retrospective study of 159 cases in Reunion Island from 2006-2012. PLoS Negl Trop Dis. 2015;9(3) doi: 10.1371/journal.pntd.0003603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins G.M., Rambaut A., Pybus O.G., Holmes E.C. Rates of molecular evolution in RNA viruses: a quantitative phylogenetic analysis. J Mol Evol. 2002;54(2):156–165. doi: 10.1007/s00239-001-0064-3. [DOI] [PubMed] [Google Scholar]
- Johnson B.W., Russell B.J., Goodman C.H. Laboratory diagnosis of chikungunya virus infections and commercial sources for diagnostic assays. J Infect Dis. 2016;214(suppl. 5) doi: 10.1093/infdis/jiw274. S471–S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joubert J., Prozesky O., Lourens J., Theron J., Swanevelder C., Meenehan G. Prevalence of hepatitis virus and some arbovirus infections in Kavango, northern SWA/Namibia. South Afr Med J. 1985;67(13):500–502. [PubMed] [Google Scholar]
- Kabir I., Dhimal M., Muller R., Banik S., Haque U. The 2017 Dhaka chikungunya outbreak. Lancet Infect Dis. 2017;17(11):1118. doi: 10.1016/S1473-3099(17)30564-9. [DOI] [PubMed] [Google Scholar]
- Kam Y.W., Pok K.Y., Eng K.E., Tan L.K., Kaur S., Lee W.W. Sero-prevalence and cross-reactivity of chikungunya virus specific anti-E2EP3 antibodies in arbovirus-infected patients. PLoS Negl Trop Dis. 2015;9(1):e3445. doi: 10.1371/journal.pntd.0003445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J.G., Kim S.H., Ahn T.Y. Bacterial diversity in the human saliva from different ages. J Microbiol. 2006;44(5):572–576. [PubMed] [Google Scholar]
- Kraemer M.U., Sinka M.E., Duda K.A., Mylne A.Q., Shearer F.M., Barker C.M. The global distribution of the arbovirus vectors Aedes aegypti and Ae. albopictus. Elife. 2015;4 doi: 10.7554/eLife.08347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok S., Kellogg D.E., McKinney N., Spasic D., Goda L., Levenson C. Effects of primer-template mismatches on the polymerase chain reaction: human immunodeficiency virus type 1 model studies. Nucleic Acids Res. 1990;18(4):999–1005. doi: 10.1093/nar/18.4.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanciotti R.S., Kosoy O.L., Laven J.J., Panella A.J., Velez J.O., Lambert A.J. Chikungunya virus in US travelers returning from India, 2006. Emerg Infect Dis. 2007;13(5):764–767. doi: 10.3201/eid1305.070015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanciotti R.S., Kosoy O.L., Laven J.J., Velez J.O., Lambert A.J., Johnson A.J. Genetic and serologic properties of Zika virus associated with an epidemic, Yap State, Micronesia, 2007. Emerg Infect Dis. 2008;14(8):1232–1239. doi: 10.3201/eid1408.080287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leparc-Goffart I., Nougairede A., Cassadou S., Prat C., De Lamballerie X. Chikungunya in the Americas. Lancet. 2014;383(9916):514. doi: 10.1016/S0140-6736(14)60185-9. [DOI] [PubMed] [Google Scholar]
- Lindsey H.S., Calisher C.H., Mathews J.H. Serum dilution neutralization test for California group virus identification and serology. J Clin Microbiol. 1976;4(6):503–510. doi: 10.1128/jcm.4.6.503-510.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litzba N., Schuffenecker I., Zeller H., Drosten C., Emmerich P., Charrel R. Evaluation of the first commercial chikungunya virus indirect immunofluorescence test. J Virol Methods. 2008;149(1):175–179. doi: 10.1016/j.jviromet.2008.01.004. [DOI] [PubMed] [Google Scholar]
- Liu-Helmersson J., Stenlund H., Wilder-Smith A., Rocklov J. Vectorial capacity of Aedes aegypti: effects of temperature and implications for global dengue epidemic potential. PLoS One. 2014;9(3) doi: 10.1371/journal.pone.0089783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnus M.M., Esposito D.L.A., Costa V.A.D., Melo P.S., Costa-Lima C., Fonseca B. Risk of Zika virus transmission by blood donations in Brazil. Hematol Transfus Cell Ther. 2018;40(3):250–254. doi: 10.1016/j.htct.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansuy J.M., Dutertre M., Mengelle C., Fourcade C., Marchou B., Delobel P. Zika virus: high infectious viral load in semen, a new sexually transmitted pathogen? Lancet Infect Dis. 2016;16(4):405. doi: 10.1016/S1473-3099(16)00138-9. [DOI] [PubMed] [Google Scholar]
- Maria A.T., Maquart M., Makinson A., Flusin O., Segondy M., Leparc-Goffart I. Zika virus infections in three travellers returning from South America and the Caribbean respectively, to Montpellier, France, December 2015 to January 2016. Euro Surveill. 2016;21(6) doi: 10.2807/1560-7917.ES.2016.21.6.30131. [DOI] [PubMed] [Google Scholar]
- Martin D.A., Muth D.A., Brown T., Johnson A.J., Karabatsos N., Roehrig J.T. Standardization of immunoglobulin M capture enzyme-linked immunosorbent assays for routine diagnosis of arboviral infections. J Clin Microbiol. 2000;38(5):1823–1826. doi: 10.1128/jcm.38.5.1823-1826.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy M. 2016. Zika virus was transmitted by sexual contact in Texas, health officials report; p. 352. [DOI] [PubMed] [Google Scholar]
- Medlock J.M., Hansford K.M., Schaffner F., Versteirt V., Hendrickx G., Zeller H. A review of the invasive mosquitoes in Europe: ecology, public health risks, and control options. Vector Borne Zoonotic Dis. 2012;12(6):435–447. doi: 10.1089/vbz.2011.0814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meltzer E., Lustig Y., Leshem E., Levy R., Gottesman G., Weissmann R. Zika Virus Disease in traveler returning from Vietnam to Israel. Emerg Infect Dis. 2016;22(8):1521–1522. doi: 10.3201/eid2208.160480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messina J.P., Brady O.J., Pigott D.M., Golding N., Kraemer M.U., Scott T.W. The many projected futures of dengue. Nat Rev Microbiol. 2015;13(4):230–239. doi: 10.1038/nrmicro3430. [DOI] [PubMed] [Google Scholar]
- Mlakar J., Korva M., Tul N., Popovic M., Poljsak-Prijatelj M., Mraz J. Zika virus associated with microcephaly. N Engl J Med. 2016;374(10):951–958. doi: 10.1056/NEJMoa1600651. [DOI] [PubMed] [Google Scholar]
- Moi M.L., Nguyen T.T.T., Nguyen C.T., Vu T.B.H., Tun M.M.N., Pham T.D. Zika virus infection and microcephaly in Vietnam. Lancet Infect Dis. 2017;17(8):805–806. doi: 10.1016/S1473-3099(17)30412-7. [DOI] [PubMed] [Google Scholar]
- Motta I.J., Spencer B.R., Cordeiro da Silva S.G., Arruda M.B., Dobbin J.A., Gonzaga Y.B. Evidence for transmission of zika virus by platelet transfusion. N Engl J Med. 2016;375(11):1101–1103. doi: 10.1056/NEJMc1607262. [DOI] [PubMed] [Google Scholar]
- Musso D., Lanteri M.C. Zika virus in Singapore: unanswered questions. Lancet Infect Dis. 2017;17(8):782–783. doi: 10.1016/S1473-3099(17)30251-7. [DOI] [PubMed] [Google Scholar]
- Musso D., Nhan T., Robin E., Roche C., Bierlaire D., Zisou K. Potential for Zika virus transmission through blood transfusion demonstrated during an outbreak in French Polynesia, November 2013 to February 2014. Eurosurveillance. 2014;19(14):20761. doi: 10.2807/1560-7917.es2014.19.14.20761. [DOI] [PubMed] [Google Scholar]
- Musso D., Roche C., Nhan T.X., Robin E., Teissier A., Cao-Lormeau V.M. Detection of Zika virus in saliva. J Clin Virol. 2015;68:53–55. doi: 10.1016/j.jcv.2015.04.021. [DOI] [PubMed] [Google Scholar]
- Musso D., Roche C., Robin E., Nhan T., Teissier A., Cao-Lormeau V.M. Potential sexual transmission of Zika virus. Emerg Infect Dis. 2015;21(2):359–361. doi: 10.3201/eid2102.141363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musso D., Teissier A., Rouault E., Teururai S., de Pina J.-J., Nhan T.-X. Detection of chikungunya virus in saliva and urine. Virol J. 2016;13(1):102. doi: 10.1186/s12985-016-0556-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCBI. Virus Resource database, https://www.ncbi.nlm.nih.gov/genomes/VirusVariation/Database/nph-select.cgi?taxid=64320.
- Oehler E., Watrin L., Larre P., Leparc-Goffart I., Lastere S., Valour F. Zika virus infection complicated by Guillain-Barre syndrome–case report, French Polynesia, December 2013. Eurosurveillance. 2014;19(9):20720. doi: 10.2807/1560-7917.es2014.19.9.20720. [DOI] [PubMed] [Google Scholar]
- Oliveira Melo A.S., Malinger G., Ximenes R., Szejnfeld P.O., Alves Sampaio S., Bispo de Filippis A.M. Zika virus intrauterine infection causes fetal brain abnormality and microcephaly: tip of the iceberg? Ultrasound Obstet Gynecol. 2016;47(1):6–7. doi: 10.1002/uog.15831. [DOI] [PubMed] [Google Scholar]
- Panning M., Grywna K., van Esbroeck M., Emmerich P., Drosten C. Chikungunya fever in travelers returning to Europe from the Indian Ocean region, 2006. Emerg Infect Dis. 2008;14(3):416–422. doi: 10.3201/eid1403.070906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parida M.M., Santhosh S.R., Dash P.K., Tripathi N.K., Lakshmi V., Mamidi N. Rapid and real-time detection of Chikungunya virus by reverse transcription loop-mediated isothermal amplification assay. J Clin Microbiol. 2007;45(2):351–357. doi: 10.1128/JCM.01734-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra B., Lizarazo J., Jiménez-Arango J.A., Zea-Vera A.F., González-Manrique G., Vargas J. Guillain–Barré syndrome associated with Zika virus infection in Colombia. N Engl J Med. 2016;375(16):1513–1523. doi: 10.1056/NEJMoa1605564. [DOI] [PubMed] [Google Scholar]
- Patel P., Abd El Wahed A., Faye O., Pruger P., Kaiser M., Thaloengsok S. A field-deployable reverse transcription recombinase polymerase amplification assay for rapid detection of the chikungunya virus. PLoS Negl Trop Dis. 2016;10(9) doi: 10.1371/journal.pntd.0004953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paty M.C., Six C., Charlet F., Heuze G., Cochet A., Wiegandt A. Large number of imported chikungunya cases in mainland France, 2014: a challenge for surveillance and response. Euro Surveill. 2014;19(28):20856. doi: 10.2807/1560-7917.es2014.19.28.20856. [DOI] [PubMed] [Google Scholar]
- Paules C.I., Fauci A.S. Yellow fever—once again on the radar screen in the Americas. N Engl J Med. 2017;376(15):1397–1399. doi: 10.1056/NEJMp1702172. [DOI] [PubMed] [Google Scholar]
- Pongsiri P., Praianantathavorn K., Theamboonlers A., Payungporn S., Poovorawan Y. Multiplex real–time RT–PCR for detecting chikungunya virus and dengue virus. Asian Pac J Trop Med. 2012;5(5):342–346. doi: 10.1016/S1995-7645(12)60055-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyke A.T., Daly M.T., Cameron J.N., Moore P.R., Taylor C.T., Hewitson G.R. Imported Zika virus infection from the Cook Islands into Australia, 2014. PLoS Curr. 2014;6 doi: 10.1371/currents.outbreaks.4635a54dbffba2156fb2fd76dc49f65e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramful D., Carbonnier M., Pasquet M., Bouhmani B., Ghazouani J., Noormahomed T. Mother-to-child transmission of Chikungunya virus infection. Pediatr Infect Dis J. 2007;26(9):811–815. doi: 10.1097/INF.0b013e3180616d4f. [DOI] [PubMed] [Google Scholar]
- Rampal S.M., Meena H. Neurological complications in Chikungunya fever. J Assoc Phys India. 2007;55:765–769. [PubMed] [Google Scholar]
- Rasmussen S.A., Jamieson D.J., Honein M.A., Petersen L.R. Zika virus and birth defects—reviewing the evidence for causality. N Engl J Med. 2016;374(20):1981–1987. doi: 10.1056/NEJMsr1604338. [DOI] [PubMed] [Google Scholar]
- Reiter P. Yellow fever and dengue: a threat to Europe? Euro Surveill. 2010;15(10):19509. [PubMed] [Google Scholar]
- Renault P., Solet J.-L., Sissoko D., Balleydier E., Larrieu S., Filleul L. A major epidemic of chikungunya virus infection on Reunion Island, France, 2005–2006. Am J Trop Med Hyg. 2007;77(4):727–731. [PubMed] [Google Scholar]
- Rezza G., Nicoletti L., Angelini R., Romi R., Finarelli A.C., Panning M. Infection with chikungunya virus in Italy: an outbreak in a temperate region. Lancet. 2007;370(9602):1840–1846. doi: 10.1016/S0140-6736(07)61779-6. [DOI] [PubMed] [Google Scholar]
- Rocklöv J., Quam M.B., Sudre B., German M., Kraemer M.U., Brady O. Assessing seasonal risks for the introduction and mosquito-borne spread of Zika virus in Europe. EBioMedicine. 2016;9:250–256. doi: 10.1016/j.ebiom.2016.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues L.C. Microcephaly and Zika virus infection. Lancet. 2016;387(10033):2070–2072. doi: 10.1016/S0140-6736(16)00742-X. [DOI] [PubMed] [Google Scholar]
- Sadarangani S.P., Hsu L.Y. The 2016 outbreak of Zika in Singapore. Ann Acad Med Singapore. 2016;45(9):381–382. [PubMed] [Google Scholar]
- Saunders N., Zambon M., Sharp I., Siddiqui R., Bermingham A., Ellis J. Guidance on the development and validation of diagnostic tests that depend on nucleic acid amplification and detection. J Clin Virol. 2013;56(3):344–354. doi: 10.1016/j.jcv.2012.11.013. [DOI] [PubMed] [Google Scholar]
- Schaffner F., Mathis A. Dengue and dengue vectors in the WHO European region: past, present, and scenarios for the future. Lancet Infect Dis. 2014;14(12):1271–1280. doi: 10.1016/S1473-3099(14)70834-5. [DOI] [PubMed] [Google Scholar]
- Schilte C., Staikowsky F., Couderc T., Madec Y., Carpentier F., Kassab S. Chikungunya virus-associated long-term arthralgia: a 36-month prospective longitudinal study. PLoS Negl Trop Dis. 2013;7(3):e2137. doi: 10.1371/journal.pntd.0002137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan C., Xie X., Ren P., Loeffelholz M.J., Yang Y., Furuya A. A rapid zika diagnostic assay to measure neutralizing antibodies in patients. EBioMedicine. 2017;17:157–162. doi: 10.1016/j.ebiom.2017.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara K., Kutsuna S., Takasaki T., Moi M.L., Ikeda M., Kotaki A. Zika fever imported from Thailand to Japan, and diagnosed by PCR in the urine. J Travel Med. 2016;23(1):tav011. doi: 10.1093/jtm/tav011. [DOI] [PubMed] [Google Scholar]
- Steinhagen K., Probst C., Radzimski C., Schmidt-Chanasit J., Emmerich P., van Esbroeck M. Serodiagnosis of Zika virus (ZIKV) infections by a novel NS1-based ELISA devoid of cross-reactivity with dengue virus antibodies: a multicohort study of assay performance, 2015 to 2016. Euro Surveill. 2016;21(50) doi: 10.2807/1560-7917.ES.2016.21.50.30426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauro L.B., Bandeira A.C., Ribeiro G., Reis M.G., Pizarro C.P.M., Araujo K. Potential use of saliva samples to diagnose Zika virus infection. J Med Virol. 2017;89(1):1–2. doi: 10.1002/jmv.24696. [DOI] [PubMed] [Google Scholar]
- Thermo Fisher Scientific. https://www.thermofisher.com/be/en/home/brands/thermo-scientific/molecular-biology/molecular-biology-learning-center/molecular-biology-resource-library/thermo-scientific-web-tools/multiple-primer-analyzer.html.
- Thiberville S.D., Moyen N., Dupuis-Maguiraga L., Nougairede A., Gould E.A., Roques P. Chikungunya fever: epidemiology, clinical syndrome, pathogenesis and therapy. Antiviral Res. 2013;99(3):345–370. doi: 10.1016/j.antiviral.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai W.-Y., Youn H.H., Tyson J., Brites C., Tsai J.-J., Pedroso C. Use of urea wash ELISA to distinguish zika and dengue virus infections. Emerg Infect Dis. 2018;24(7):1355. doi: 10.3201/eid2407.171170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste K., Garlant L., Broeders S., Van Gucht S., Roosens N.H. Application of whole genome data for in silico evaluation of primers and probes routinely employed for the detection of viral species by RT-qPCR using dengue virus as a case study. BMC Bioinformatics. 2018;19(1):312. doi: 10.1186/s12859-018-2313-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.M., Ali U.H., Sekaran S.D., Thayan R. Chikungunya virus. Springer; 2016. Detection and quantification of chikungunya virus by real-time RT-PCR assay; pp. 105–117. [DOI] [PubMed] [Google Scholar]
- Wang X., Yin F., Bi Y., Cheng G., Li J., Hou L. Rapid and sensitive detection of Zika virus by reverse transcription loop-mediated isothermal amplification. J Virol Methods. 2016;238:86–93. doi: 10.1016/j.jviromet.2016.10.010. [DOI] [PubMed] [Google Scholar]
- Weaver S.C., Forrester N.L. Chikungunya: evolutionary history and recent epidemic spread. Antiviral Res. 2015;120:32–39. doi: 10.1016/j.antiviral.2015.04.016. [DOI] [PubMed] [Google Scholar]
- Whitehorn J., Farrar J. Dengue. Br Med Bull. 2010;95:161–173. doi: 10.1093/bmb/ldq019. [DOI] [PubMed] [Google Scholar]
- WHO. ZIKV epidemic in Brazil with a fast spreading to other South American countries and North America.
- WHO . 2016. Countries and territories reporting mosquito-borne Zika virus transmission. [Google Scholar]
- WHO . 2016. Disease outbreak news 15 June 2016: Chikungunya – United States of America. [Google Scholar]
- WHO . 2016. Situation report: Zika virus microcephaly and Guillain-Barré syndrome. [Google Scholar]
- WHO . 2018. Disease outbreak news 1 May 2018: Dengue fever – Réunion, France. [Google Scholar]
- WHO . 2018. Disease outbreak news 12 April 2018: Zika virus infection – Viet Nam. [Google Scholar]
- WHO . 2018. Disease outbreak news 15 October 2018: Chikungunya - Sudan. [Google Scholar]
- WHO . 2018. Disease outbreak news 25 August 2018: Chikungunya - France. [Google Scholar]
- WHO . 2018. Disease outbreak news 27 February 2018: Chikungunya – Mombasa, Kenia. [Google Scholar]
- WHO . 2018. Disease outbreak news 27 February 2018: Yellow fever – Brazil. [Google Scholar]
- WHO . 2018. Disease outbreak news 29 September 2018: Chikungunya - Italy. [Google Scholar]
- Wielanek A.C., Monredon J.D., Amrani M.E., Roger J.C., Serveaux J.P. Guillain-Barre syndrome complicating a Chikungunya virus infection. Neurology. 2007;69(22):2105–2107. doi: 10.1212/01.wnl.0000277267.07220.88. [DOI] [PubMed] [Google Scholar]
- Wilder-Smith A., Gubler D.J., Weaver S.C., Monath T.P., Heymann D.L., Scott T.W. Epidemic arboviral diseases: priorities for research and public health. Lancet Infect Dis. 2017;17(3):e101–e106. doi: 10.1016/S1473-3099(16)30518-7. [DOI] [PubMed] [Google Scholar]
- Wong P.S., Li M.Z., Chong C.S., Ng L.C., Tan C.H. Aedes (Stegomyia) albopictus (Skuse): a potential vector of Zika virus in Singapore. PLoS Negl Trop Dis. 2013;7(8):e2348. doi: 10.1371/journal.pntd.0002348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zammarchi L., Tappe D., Fortuna C., Remoli M.E., Gunther S., Venturi G. Zika virus infection in a traveller returning to Europe from Brazil, March 2015. Euro Surveill. 2015;20(23):21153. doi: 10.2807/1560-7917.es2015.20.23.21153. [DOI] [PubMed] [Google Scholar]
- Zanluca C., Melo V.C., Mosimann A.L., Santos G.I., Santos C.N., Luz K. First report of autochthonous transmission of Zika virus in Brazil. Mem Inst Oswaldo Cruz. 2015;110(4):569–572. doi: 10.1590/0074-02760150192. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.