To the Editor:
The ongoing outbreak of the novel coronavirus (2019-nCoV) pneumonia in Wuhan, China and other regions remains a major public health concern. We thank Dhungana for comments to our study, Zhao et al. (2020), recently published in the International Journal of Infectious Diseases. The estimates on the basic reproduction number, R 0, were carried out early in the outbreak as of January 22, 2020, when the surveillance data and the knowledge of the key epidemiological features of 2019-nCoV were limited.
The assumptions of exponential growth as well as other similar growth patterns are commonly accepted and adopted to capture the growth trends during the early phase of an outbreak (Nishiura and Chowell, 2014, Chowell et al., 2004, Wearing et al., 2005). The exponential growth rate (γ), or the intrinsic growth rate, is estimated from the early epidemic curve and used to calculate the R 0. We repeat the analysis Zhao et al. (2020), γ is estimated at 0.18 (95%CI: 0.14–0.22), 0.15 (95%CI: 0.12–0.18) and 0.11 (95%CI: 0.09–0.13) per day associated with 2-, 4- and 8-fold increase in the reporting rate, respectively. By using the serial interval (SI) estimate (mean ± SD at 7.5 ± 3.4 days) from Li et al. (2020), we found the R 0 at 3.33 (95%CI: 2.17–4.04), 2.69 (95%CI: 2.28–3.17) and 2.13 (95%CI: 1.88–2.42) associated with 2-, 4- and 8-fold increase in the reporting respectively. Our estimates were in line with the WHO estimates in both the early version (2-fold case) and the published version. The key message as we highlighted in the paper is the changes in the reporting rate. This is recently reconfirmed by Tuite and Fishman (Tuite and Fisman, 2020). We thank the editor and Dhungana to give us this opportunity to reclarify our key message that the reporting rate was not constant during the early outbreak and could affect the estimation of R 0. There is indeed a large amount of later confirmed cases that were not counted in the early official daily situation reports Li et al. (2020); Imai et al., 2020; Riou and Althaus, 2020; Zhao et al., 2020b). In other words, if the same reporting standard in the second half of January was applied to the first half of January, the number of cases would be much higher. Other teams either used a retrospective dataset which was not publicly available on January 23, 2020 or used overseas reported cases which were not (to a much less extent) affected by the changes in reporting rate.
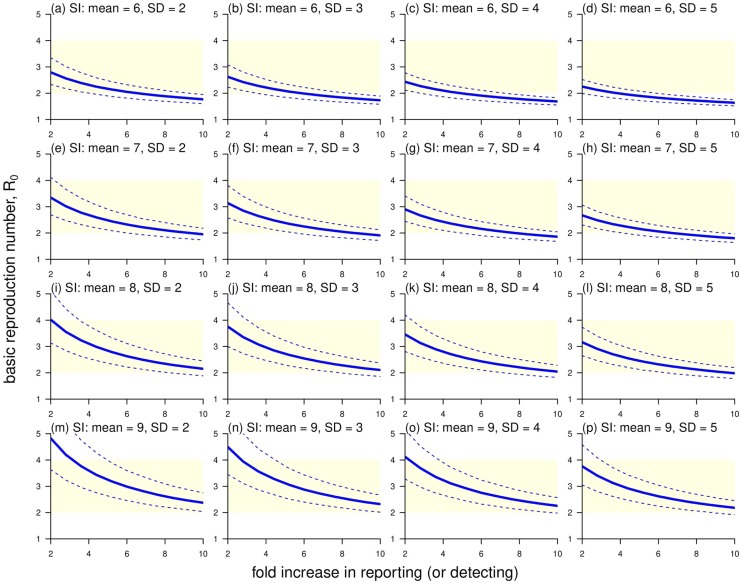
By using the same analysis and dataset as in Zhao et al. (2020), an additional sensitivity analysis on the R 0 estimates and varying SI and reporting rate was conducted and is shown in Figure 1 . We report that R 0 estimates increase while the mean SI increases or the SD of SI decreases. By selecting a mean between 7 and 8 days and SD between 3 and 4 days for SI of 2019-nCoV, the R 0 estimates are largely consistent within a range from 2 to 4 in many existing reports (Li et al., 2020, Imai et al., 2020, Riou and Althaus, 2020, Zhao et al., 2020b, Wu et al., 2020), see panels (f), (g), (j) and (k) Fig. 1. We conclude that our previous estimation and main conclusions in hold based on the reasonable selection of the SI estimates of 2019-nCoV. Not only is our early version (2-fold case) in line with the WHO estimates, but also we pointed out the issue in the reporting rate changes in the official reported cases.
Figure 1.
The estimates of the basic reproduction number, R0, with varying reporting rates, mean and SD of serial interval (SI). The mean of SI, from top to bottom vertically, varies at 6, 7, 8 and 9 days. The SD of SI, from left to right horizontally, varies at 2, 3, 4 and 5 days. The light-yellow area highlights the R0 ranging from 2 to 4 referring to the estimates in (Li et al., 2020, Imai et al., 2020, Riou and Althaus, 2020, Zhao et al., 2020b, Wu et al., 2020). The blue bold curve is the mean estimate, and the blue dashed curves are the 95% confidence interval (95%CI).
recently published in the International Journal o
Ethics approval and consent to participate
The ethical approval or individual consent was not applicable.
Availability of data and materials
All data and materials used in this work were publicly available.
Consent for publication
Not applicable.
Funding
WW was supported by National Natural Science Foundation of China (Grant Number 61672013) and H uaian Key Laboratory for Infectious Diseases Control and Prevention (Grant Number HAP201704), Huaian, Jiangsu, China.
Disclaimer
The funding agencies had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; or decision to submit the manuscript for publication.
Conflict of interests
The authors declared no competing interests.
Authors' contributions
All authors conceived the study, carried out the analysis, discussed the results, drafted the first manuscript, critically read and revised the manuscript, and gave final approval for publication.
Acknowledgement
The authors thank Cindy Y Tian from the Chinese University of Hong Kong for helping in processing the reference files.
Contributor Information
Shi Zhao, Email: zhaoshi.cmsa@gmail.com.
Qianying Lin, Email: qianying.lin@connect.polyu.hk.
Jinjun Ran, Email: jimran@connect.hku.hk.
Salihu S. Musa, Email: salihu-sabiu.musa@connect.polyu.hk.
Guangpu Yang, Email: kennethgpy@link.cuhk.edu.hk.
Weiming Wang, Email: weimingwang2003@163.com.
Yijun Lou, Email: yijun.lou@polyu.edu.hk.
Daozhou Gao, Email: dzgao@shnu.edu.cn.
Lin Yang, Email: l.yang@polyu.edu.hk.
Daihai He, Email: daihai.he@polyu.edu.hk.
Maggie H Wang, Email: maggiew@cuhk.edu.hk.
References
- Chowell G., Hengartner N.W., Castillo-Chavez C., Fenimore P.W., Hyman J.M. The basic reproductive number of Ebola and the effects of public health measures: the cases of Congo and Uganda. J. Theor Biol. 2004;229(1):119–126. doi: 10.1016/j.jtbi.2004.03.006. [DOI] [PubMed] [Google Scholar]
- Imai N., Dorigatti I., Cori A., Riley S., Ferguson N.M. Preprint published by the Imperial College London; 2020. Estimating the potential total number of novel Coronavirus (2019-nCoV) cases in Wuhan City, China.https://www.imperial.ac.uk/mrc-global-infectious-disease-analysis/news--wuhan-coronavirus/ [Google Scholar]
- Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus–Infected Pneumonia. N Engl J Med. 2020 doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiura H., Chowell G. Early transmission dynamics of Ebola virus disease (EVD), West Africa, March to August 2014. Eurosurveillance. 2014;19(36):20894. doi: 10.2807/1560-7917.es2014.19.36.20894. [DOI] [PubMed] [Google Scholar]
- Riou J., Althaus C.L. Pattern of early human-to-human transmission of Wuhan 2019 novel coronavirus (2019-nCoV), December 2019 to January 2020. Eurosurveillance. 2020;25(4) doi: 10.2807/1560-7917.ES.2020.25.4.2000058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuite A.R., Fisman D.N. Reporting, Epidemic Growth, and Reproduction Numbers for the 2019 Novel Coronavirus (2019-nCoV) Epidemic. Ann Internal Med. 2020 doi: 10.7326/M20-0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wearing H.J., Rohani P., Keeling M.J. Appropriate models for the management of infectious diseases. PLoS Med. 2005;2(7):e174. doi: 10.1371/journal.pmed.0020174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J.T., Leung K., Leung G.M. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study. The Lancet. 2020;395(10225):689–697. doi: 10.1016/S0140-6736(20)30260-9. [DOI] [Google Scholar]
- Zhao S., Lin Q., Ran J., Musa S.S., Yang G., Wang W. Preliminary estimation of the basic reproduction number of novel coronavirus (2019-nCoV) in China, from 2019 to 2020: A data-driven analysis in the early phase of the outbreak. Int J Infect Dis. 2020;92:214–217. doi: 10.1016/j.ijid.2020.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S., Musa S.S., Lin Q., Ran J., Yang G., Wang W. Estimating the Unreported Number of Novel Coronavirus (2019-nCoV) Cases in China in the First Half of January 2020: A Data-Driven Modelling Analysis of the Early Outbreak. J Clin Med. 2020;9(2) doi: 10.3390/jcm9020388. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data and materials used in this work were publicly available.