Abstract
The positioning of nuclei within the cell is a dynamic process that depends on the cell’s fate and developmental stage and that is adjusted for optimal cell function. This is especially true in skeletal muscle cells, which contain hundreds of myonuclei distributed evenly along the periphery of the muscle cell. Mispositioned myonuclei are often associated with muscle dysfunction and disease. Different mechanisms governing myonuclear positioning are now emerging, with several of the new genes implicated in nuclear movement linked to human muscle disease. Here we discuss the recent advances in myonuclear positioning and its implications for muscle size and function from the view of Drosophila. Additionally, we highlight similarities and differences to mammalian systems and provide connections to human muscle disease.
The Nucleus in Skeletal Muscle
In most textbooks, the nucleus is depicted at the center of the cell. However, nuclei adopt different localizations, depending on the cell’s fate, developmental stage, or specific function (e.g., neurons, epithelia, and skeletal muscle cells). Importantly, in all cell types and cell states, nuclear positioning is an active process and is continuously adjusted for optimal cell function. Changes in nuclear positioning are often associated with cellular dysfunction and disease [1,2]. The position of nuclei in skeletal muscle cells (myofibers) is of particular interest, since these cells are multinucleated, with up to hundreds of nuclei (myonuclei), evenly distributed along the cell surface. Mislocalized myonuclei have been associated with a variety of muscle diseases [3] that are characterized by reduced muscle size, muscle weakness, and decreased muscle function [4]. Many genetic mutations lead to the development of diseases displaying these characteristics, including those in nuclear proteins, which are associated with centronuclear myopathy (CNM) and Emery-Dreifuss muscular dystrophy (EDMD) [4–6]. While mispositioned myonuclei are typical for many muscle diseases, it is still unclear whether these diseases share a common mechanism or are the result of distinct mechanisms that lead to myonuclear mispositioning. A better understanding of the mechanisms underlying the positioning of myonuclei is required to understand and treat these severe disorders (Box 1).
Box 1. Nuclear Mispositioning and Muscle Disease.
Nuclear mispositioning is a hallmark of several muscle diseases, including EDMD and CNM [5,6]. Nevertheless, it was unclear whether mispositioning is a cause or consequence of muscle disease. The analysis of ens mutations in Drosophila suggested a causal link between aberrant nuclear position and muscle dysfunction [15]. Following this study, other genes, such as Dynein and Bsg25D, were found to have mispositioned myonuclei and larval movement defects [19,24–26,31]. Recently, mutations associated with EDMD and CNM in Drosophiia showed similar phenotypes [17]. However, it remains an outstanding question why mispositioned myonuclei in these mutants lead to deficits in muscle function. Several hypotheses merit investigation.
One hypothesis relates specific nuclear localization to specific function within the muscle cell. It has been long recognized that myonuclei in mammals are clustered at the NMJ and express unique RNAs [16,58]. These observations were further developed in Drosophila, where changes in nuclear and nucleolar size, DNA content, and transcriptional activity were associated with different regions of the muscle cell (NMJ, MTJ, others) [44]. Together, these data suggest that myonuclei in these regions are distinct and produce unique products required for optimal muscle function. Hence, myonuclear mispositioning would lead to alterations in region-specific transcriptional programs. This hypothesis could be tested using single nuclear sequencing approaches to define unique transcripts for myonuclei in each region.
A second related hypothesis considers myonuclear position within its cytosolic context, also known as its myonuclear domain [44]. Each myonucleus has MT arrays that maintain the position of individual myonuclei [19,44,52]. These arrays may also function to traffic RNAs and proteins within the domain. One can speculate that RNAs and proteins may not be properly trafficked within the myonuclear domain in the positioning mutants, thus reducing muscle homeostasis. An extreme case can be seen with Bsg25D: misexpression of Bsg25D leads to missing myonuclear MT arrays, mispositioned myonuclei, and decreased muscle size [19]. Future studies in which fluorescently tagged RNA transcripts or proteins are followed by time-lapse imaging in Bsg25D and other positioning mutants will provide further insight to the mechanisms required for nuclear positioning and function.
Translating these findings into treatments for patients suffering from skeletal muscle diseases remains a critical challenge. Clearly, identifying additional mutated gene(s) associated with muscle diseases, such as EDMD and CNM, will enlarge our understanding of disease mechanisms and contributions of myonuclear positioning to disease. Likewise, assessing the extent of global (muscle) and local (myonuclear domain) transcriptional changes in patient samples with aberrant myonuclear positioning will provide possible therapeutic targets and approaches. Lastly, a better understanding of the dynamics of myonuclear positioning in the context of satellite cell-mediated repair may offer new insights to disease treatment.
From the first observation of a nucleus in 1700 [7], our knowledge of nuclear composition, organization, and positioning has continuously evolved. In addition to containing the genomic DNA, nuclei across species and different cell types share many structural similarities. The nucleus is delimited by the nuclear envelope (NE), composed of two lipid bilayer membranes that separate the nucleoplasm from the cytoplasm (Figure 1). The inner nuclear membrane (INM) and outer nuclear membrane (ONM) have distinct compositions and functions and only associate where there is a nuclear pore. Nuclear pore complexes allow for communication between the cytoplasm and nucleoplasm. The nuclear lamina is intimately associated with the INM and is composed of a thin meshwork of intermediate filaments, mostly A- and B-type lamins, that associate with heterochromatin and provide structural support to the nucleus [8]. Linker of the nucleoskeleton and cytoskeleton (LINC) complexes support the structure of the nucleus as well as its interaction with cytoskeletal components [8,9]. LINC complexes are located throughout the NE and consist primarily of transmembrane Sad1 and UNC-84 (SUN) and Klarsicht, Anc-1, and Syne Homology (KASH) proteins. SUN proteins are INM components that associate with the nuclear lamina and other components through their N terminal domains. The C terminal domain of SUN proteins projects into perinuclear space (PNS) and associates with KASH proteins. KASH proteins are present at the ONM and interact with SUN proteins at the PNS through their C terminal domains. The KASH N terminal domain links to the cytoplasmic cytoskeletal networks, including the microtubules (MTs) and actin filaments. By connecting nuclear and cytoplasmic components, the LINC complexes are essential for maintaining the integrity of the nucleus, as well as its position within the cell (Figure 1) [8,9].
Figure 1. Schematic Representation of the Components Present at the Nuclear Envelope of a Drosophila Larval Myonucleus.
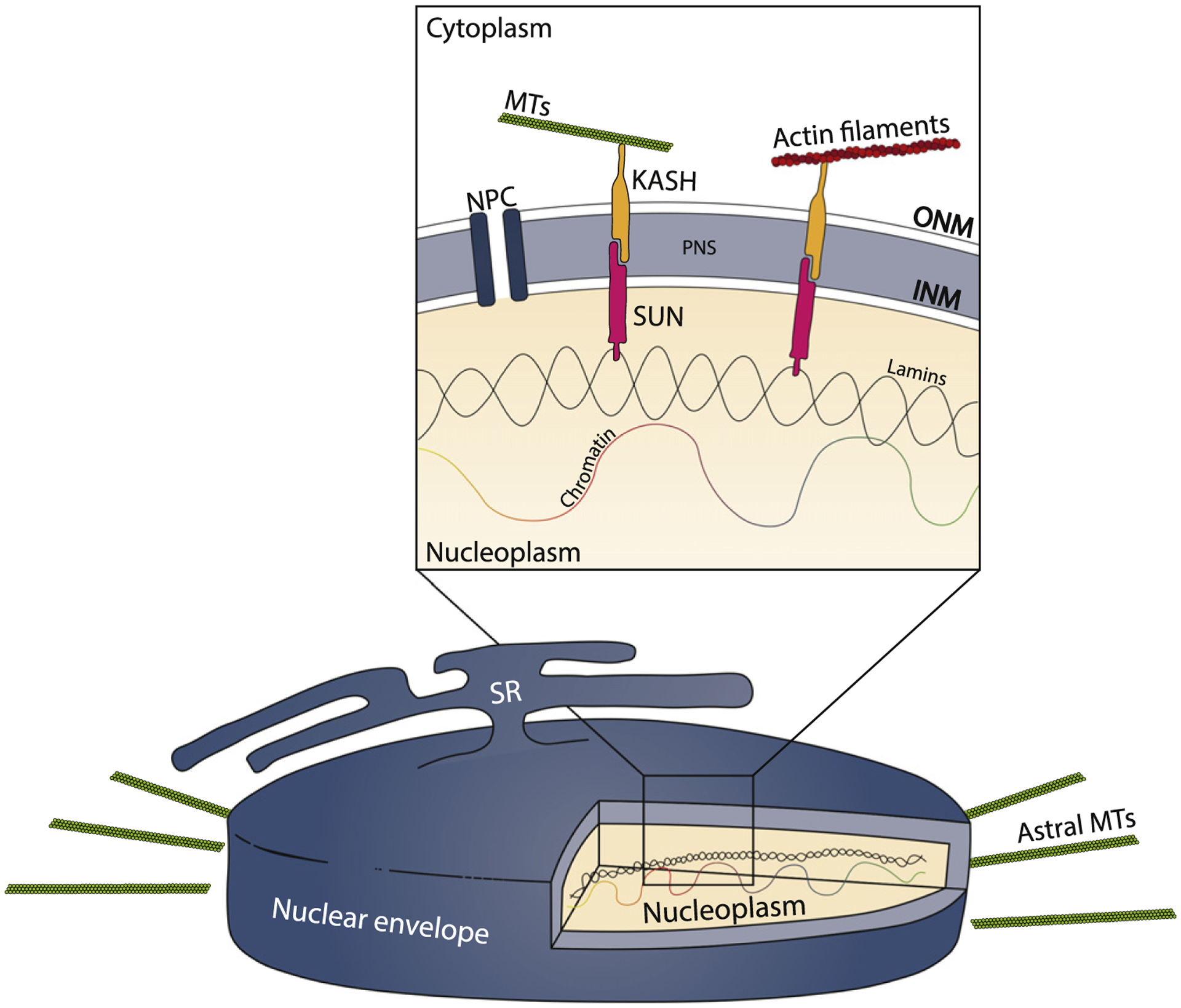
Astral MTs represent the microtubules nucleating from the nuclear envelope in larval muscle. The box highlights a zoomed region of the nuclear envelope. Representative KASH and SUN proteins are shown in orange and magenta, respectively. Abbreviations: KASH, Klarsicht, Anc-1, and Syne Homology; INM, inner nuclear membrane; MTs, microtubules; NPC, nuclear pore complex; ONM, outer nuclear membrane; PNS, perinuclear space; SR, sarcoplasmic reticulum; SUN, Sad1 and UNC-84.
Multinucleated skeletal muscle cells develop through a series of steps that are conserved across invertebrate and vertebrate species. First, muscle progenitors (myoblasts) are specified in the embryonic mesoderm. This is followed by a series of cell-cell fusion events. Each fusion event adds a nucleus and cytoplasmic mass, leading to the formation of syncytial myotubes (Figure 2A; for additional information on muscle cell fusion see [10,11]). Myoblast fusion in the Drosophila embryo requires two types of myoblasts: founder cells (FCs) and fusion-competent myoblasts (FCMs). The FC contains all the information that determines an individual muscle cell’s identity. Each FC fuses with a specific number of FCMs (2–25) and establishes a unique muscle with a specific size, shape, tendon attachment, and innervation [12,13]. Each embryonic muscle is composed of a single muscle cell with a relatively small number of nuclei, which is ideal for investigating molecular events underlying myogenesis in great detail. In mammals, muscles are more complex, consisting of bundles of muscle cells. In addition, mammalian myoblasts appear to be equivalent and fuse to form syncytia that contain up to 100s of nuclei [14]. Nevertheless, myonuclei in both systems undergo a similar series of stereotyped movements during myogenesis and assume positions that maximize their internuclear distances at the surface of mature myofibers.
Figure 2. Schematic Representation of Myonuclear Positioning during Embryonic and Larval Drosophila melanogaster Development.
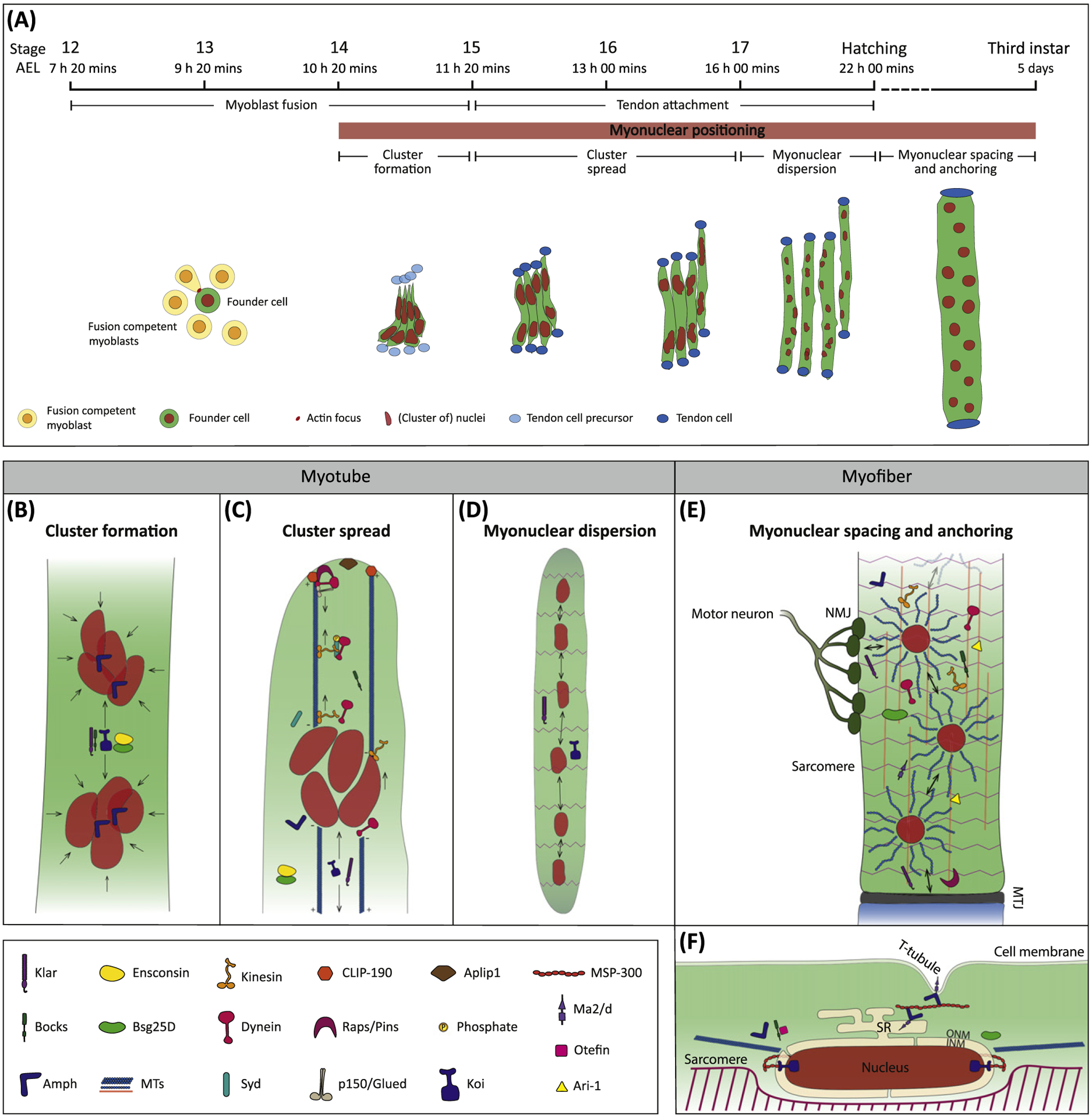
(A) Drosophila myogenesis consists of multiple steps, including myoblast fusion, tendon attachment, and myonuclear positioning. The latter is further divided into cluster formation (B), cluster spread (C), myonuclear dispersion (D), and myonuclear spacing and anchoring (E and F). Development and myonuclear positioning from stages 12 to the end of stage 17 are shown using the embryonic lateral transverse (LT) muscles as examples. From hatching to third instar larva, the muscle represented is a larval ventral longitudinal (VL) muscle. Stage and times correspond to those observed at 25°C. (B) Close-up of the two clusters of myonuclei present in the embryonic LT muscle. Arrows indicate the forces required to maintain the integrity of the two clusters and those to keep them separated. (C) Close-up of a single cluster within an embryonic LT muscle as it moves towards the end of the myotube. Arrows indicate the pulling and pushing forces clustered myonuclei experience as they move. (D) View of a single embryonic LT muscle showing the presence of sarcomeres, which, together with the neuromuscular junction (NMJ) (not shown), indicate that the muscle can experience coordinated contraction. In this panel, the double arrows represent the movements of myonuclei to be equidistant from each other. (E) View of a larval VL myofiber and its interaction with two cell types: motor neuron (dark green), forming the NMJ, and tendon (bottom of the muscle, blue), forming the myotendinous junction (MTJ). The two populations of microtubules (MTs) can be observed: longitudinal MTs run along the length of the myofiber (orange lines) and astral MTs surround the nucleus and extend in multiple directions (blue). The double arrows indicate the distance between myonuclei and the edges of the myofiber. (F) Longitudinal cross-section of a larval VL nucleus for a more detailed view of the localization of proteins involved in myonuclear spacing and anchoring. Note: protein localization in panels B-F represents the function in specific processes or mechanisms, rather than the precise described localization of each protein in the cell. Abbreviations: AEL, after egg laying; INM, inner nuclear membrane; ONM, outer nuclear membrane; SR, sarcoplasmic reticulum.
Over the past several years, researchers have defined different mechanisms that ensure the proper localization of myonuclei through their interactions with the cytoskeleton and the LINC complexes. In this review, we highlight the latest findings on the molecular mechanisms of myonuclear positioning, the relationship between nuclear scaling and positioning in muscle cells, and the functional consequences of myonuclear mispositioning. We focus specifically on the Drosophila melanogaster somatic musculature and highlight similarities and differences to the mammalian systems.
Myonuclear Positioning
D. melanogaster presents an excellent in vivo system to study muscle development, particularly the mechanisms of myonuclear positioning and the interactions of muscle with other cell types, such as the tendons and motor neurons. The fly embryo consists of 12 segments, each hemisegment containing 30 skeletal muscles. The plethora of genetic tools permits the visualization of nuclei from individual muscle cells, both in fixed and live samples at different developmental stages. Therefore, the movements of myonuclei in four dimensions can be readily assessed throughout muscle development.
Based on work with the Drosophila lateral transverse (LT) and ventral longitudinal (VL) muscles, the steps required for proper nuclear positioning were initially described by Metzger et al. [15]. Here, we review this process and subdivide it into four significant steps; (i) cluster formation, (ii) cluster spread, (iii) myonuclear dispersion, and (iv) myonuclear spacing and anchoring (Figure 2). Steps i through iii are assessed during embryonic development, whereas step iv is usually analyzed in the mature myofibers of the fly larva. In mammalian muscle, similar movements have been observed [14,16]. Nuclear positioning in both systems is controlled molecularly by a variety of proteins from different groups, mainly MTs, MT motor and non-motor proteins [also known as MT-associated proteins (MAPs)], actin and actin-associated proteins, and LINC complex proteins. Most proteins play a role in distinct myonuclear movement steps throughout myogenesis, which has been demonstrated in various cellular and animal models, as well as in several human diseases. For a list of proteins and their functions in myonuclear positioning that are included in this review, we refer the reader to Table 1. Below we describe the most recent findings for each step of myonuclear positioning.
Table 1.
Summary of Proteins Involved in Myonuclear Positioning and the Consequences of Their Removal for Muscle Function
| Mammalian | Drosophila | Role in Drosophila | Muscle function consequences (human diseasea; Drosophila) | Refs |
|---|---|---|---|---|
| Nesprin-4 | Klarsicht/Klar | Cooperates with Bocks; cluster formation (separation of nuclei into distinct clusters); myonuclear dispersion (sarcomere assembly); myonuclear spacing and anchoring (regulates myonuclear distances to the NMJ and near the MTJ) | EDMD; affects larval muscle function | [17,18,31,57] |
| Emerin | Bocksbeutel/ Bocks | Cooperates with Klar; cluster formation (separation of nuclei into distinct clusters); cluster spread (viathe cortical pulling pathway); myonuclear spacing and anchoring (in combination with Ote, it regulates Klar expression at the NE; regulates astral MTs) | EDMD; affects larval muscle function | [17,18] |
| SUN2 | Klaroid/Koi | Cluster formation (separation of nuclei into distinct clusters); clusterspread; myonuclear dispersion (sarcomere assembly; regulates Klar expression at the NE) | EDMD disease modifier | [18,31,32,61] |
| Amphiphysin | Amphiphysin/ Amph | Cluster formation (maintains nuclei within clusters); myonuclear spacing and anchoring (necessary for the proper localization of Ma2/d; regulates astral MTs) | CNM; affects larval muscle function | [17,49] |
| MAP7 | Ensconsin/Ens | Positively regulated by Bsg25D; cluster formation; cluster spread; myonuclearspacing and anchoring | Affects larval muscle function | [15,19] |
| Ninein | Blastoderm-specific gene 25D/Bsg25D | Positively regulates Ens; cluster formation; cluster spread; myonuclear spacing and anchoring (regulate astral MTs and MT stability) | Affects larval muscle stiffness and function | [19] |
| Kinesin/Kif5b | Kinesin/Khc | Cluster spread (force exertion of nuclei leading edge and viathe cortical pulling pathway); myonuclear spacing and anchoring (regulates myonuclear distance near the NMJ) | Not reported | [24,25,57] |
| Dynein | Dynein/Dhc | Cluster spread (force exertion of the nuclear lagging edge and via the cortical pulling pathway); myonuclearspacing and anchoring (regulates myonuclear distance near the NMJ) | Affects larval muscle function | [24,25,57] |
| CLIP1/2 | CLIP-190 | Cluster spread (via the cortical pulling pathway; connects the MTs to the cell cortex) | Affects larval muscle function | [24–26] |
| Dynactin 1, p150 subunit | Glued/p150 | Cluster spread (via the cortical pulling pathway; forms acomplexwith Dynein) | Affects larval muscle function | [24,25] |
| G protein-signaling modulator 2 | Partner of inscuteable/pins/raps | Cluster spread (via the cortical pulling pathway; anchors Dynein at the muscle ends); myonuclear spacing and anchoring (regulates the myonuclear position near the MTJ) | Not reported | [26,57] |
| JIP3 | Sunday driver/Syd | Cluster spread (via the cortical pulling pathway; mediates the formation of a Kinesin-Dynein complex) | Affects larval muscle function | [26] |
| JIP1 | Aplip1 | Cluster spread (regulates nuclear translocation via changes in nuclear shape and the cortical pulling pathway; regulates the MT networks) | Not reported | [27] |
| SYNE-1/SYNE-2 | Muscle-specific protein 300 kDa/ MSP-300 | Myonuclear spacing and anchoring (provides elasticity to the perinuclear shield; maintenance of nuclear shape; attachment of the MT asters to the NE; necessary for the proper localization of Ma2/d) | Autosomal-dominant EDMD, EDMD-like phenotypes, arthrogryposis (SYNE-1), autosomal recessive cerebellar ataxia type 1 (SYNE-1); affects larval muscle function, control of glutamate receptor density at the NMJ | [31,49,62] |
| Spectraplakin | Short stop/ Shot | Myonuclear spacing and anchoring (provides rigidity to the perinuclear shield) | Not reported | [48] |
| EB family member 1 | Eb1 | Myonuclear spacing and anchoring (provides rigidity to the perinuclear shield) | Not reported | [48] |
| Emerin | Otefin/Ote | Myonuclear spacing and anchoring (in combination with bocks, it regulates klar expression at the NE) | X-EDMD | [18,63] |
| Muscle-specific α2/δ/Ma2/d | Myonuclear spacing and anchoring (necessary for the sarcoplasmic reticulum-nucleus association) | Affects larval muscle function | [49] | |
| ARIH1 | Ariadne 1/ Ari-1 | Myonuclear spacing and anchoring (proper localization of MSP-300; mono-ubiquitinates Koi) | SMC dysfunction (e.g., thoracic aortic aneurysms) | [50,51] |
Where known.
Cluster Formation
Cluster formation in Drosophila has been investigated primarily in the four finger-shaped LT muscles. The LT muscles establish dorsal-ventral orientation; LT muscles 1,2, and 3 contain between six and eight myonuclei, while LT 4 has between four and six nuclei. The first myonuclear movements in the LTs occur while the myotubes are still undergoing fusion events [stage 14; 10 h 20 min after egg laying (AEL); Figure 2A,B]. As each fusion event occurs, the added nucleus moves to the cell center, becoming part of a cluster. Two closely apposed nuclear clusters can be identified in the ventral center of the LTs at this stage. However, it was not entirely clear how myonuclei associate with a cluster.
Recently, a number of proteins present at the NE have been implicated in the initial myonuclear cluster formation. These include Bocks and Klar, the Drosophila homologs of Emerin and the LINC protein Nesprin-4, respectively, and Koi, the Drosophila SUN2 protein [17,18]. In addition, Amphiphysin, a BAR domain protein linked to vesicle trafficking, is implicated in nucleus-nucleus interactions to maintain cluster cohesion. Although the mechanism of Amphiphysin remains unknown, the authors suggest that it could be MT independent [17]. Mutations in Emerin and Nesprin-4 are linked to EDMD, and Amphiphysin is implicated in CNM, indicating that the crucial roles of these proteins in myonuclear positioning are highly conserved.
One cytoskeletal element involved in cluster formation in Drosophila is Ensconsin (Ens)/MAP7. Ens loss results in the formation of a single cluster instead of two clusters [15]. Recently, Bsg25D, the Drosophila homolog of Ninein, was described to physically interact with and positively regulate Ens activity during myonuclear positioning. Loss of Bsg25D in developing myotubes affects nuclear positioning in embryos that have been sensitized by the partial loss of Ens. However, muscle-specific Bsg25D overexpression leads to a phenotype similar to that of Ens loss. While in mature myofibers Bsg25D and Ens regulate perinuclear MT organization (see below), the mechanisms by which they act during the formation of the initial nuclear clusters remain unclear [19]. To this end, work in mammalian cell culture provides insights. In mammalian myotubes generated in 2D tissue culture, myonuclei are found in the cell center, forming a single cluster. Upon fusion, the newly added myonucleus rapidly moves towards the center in an MT-dependent way. Several key players, including a dynein/dynactin complex, Cdc42, Par6, and Par3, have been shown to be involved in this process in vitro [14,20]. It remains to be tested whether similar mechanisms are at play in early Drosophila myotubes.
An interesting distinction between fly and mammalian muscle with regards to clustering is that, in the latter, myonuclei cluster initially in the cell center during regeneration upon injury, with subsequent movement to the periphery at a later time-point [21]. Muscle repair in mammals is promoted by adult muscle stem cells, called satellite cells, that become activated, proliferate, and subsequently fuse to the regenerating myofiber [22]. Recently, a satellite-like cell population has also been identified in the adult Drosophila muscles. These cells, upon muscle injury, are capable of proliferating and fusing to existing myofibers. However, instead of appearing in the cell center, these new myonuclei were only detected at the muscle cell surface [23]. It would be interesting to investigate the myonuclear movements in Drosophila muscle undergoing repair to better understand this process and compare it with observations in mammalian muscles.
Cluster Spread
In Drosophila embryonic LT muscles, the second step of nuclear positioning involves the movement of the two adjacent nuclear clusters towards the ends of the muscle cell (stages 15–16; 11 h 20 min to 13 h 00 min AEL; Figure 2A,C). This step is perhaps the most well-studied in the Drosophila myonuclear positioning field. Quantification of the cluster distance from the muscle ends relative to muscle length at stage 16 has proven to be effective at assessing even small differences in cluster spread [17,19,24–28]. The activities of the MT motor proteins, Kinesin-1 and cytoplasmic Dynein, as well as their associated proteins, Ens/MAP7, Bsg25D, CLIP-190, Glued, Pins/Raps, Syd, and Aplip1, are necessary for the movement for the nuclear clusters [15,19,24–27]. Both Kinesin-1 and Dynein exert forces directly on the nuclei and from the cell cortex via the MTs, with the latter being termed as the cortical pulling mechanism [24,25]. The localization of both motor proteins was shown to be essential for their correct activity during this step. The proposed mechanism has Syd, a homolog of the mammalian JIP3, mediating the formation of a complex between Kinesin-1 and Dynein and promoting the Kinesin-1- and JNK signaling-dependent transportation of Dynein along the MTs to the cell cortex. The MTs are stably connected to the cell cortex through CLIP-190. Once Dynein is anchored at the muscle end via Raps/Pins, it becomes active to pull myonuclei. The authors further hypothesize that, while Kinesin-1 and Dynein have roles in nuclear dynamics, Syd may also organize a population of both motors near the nucleus before transportation to the cell cortex [26]. The cortical pulling pathway is the most complete mechanism described to date in Drosophila. However, new components are still being discovered, suggesting that other proteins may be involved in this step via this pathway.
Ens is an important regulator of myonuclear positioning through its genetic and physical interaction with the motor protein Kinesin-1 [15]. Given the strong clustering phenotype it produces, Ens is also a potential candidate for the cortical pulling pathway [15]. However, despite being required for Kinesin-1 recruitment to MTs in other cellular contexts, the molecular mechanism of Ens’ action during myonuclear movements in Drosophila is unclear [15,29,30]. Ens also interacts genetically and physically with Bsg25D to regulate cluster spread [19]. However, there is a lack of evidence, such as genetic interactions with motor proteins, to implicate Bsg25D’s direct participation in the cortical pulling pathway. These data suggest that alternative pathways involving these proteins remain to be uncovered.
Several NE components are involved in cluster spread. The genetic interaction of bocks with both Khc and Dhc affects the spreading of myonuclear clusters, suggesting that Bocks regulates myonuclear positioning through the cortical pulling pathway [17]. Moreover, recent work indicates that bocks genetically interacts with klar to properly move the myonuclear clusters [18]. This particular result could be a consequence of defective cluster formation and/or indicate that bocks has roles in more than one mechanism regulating myonuclear positioning. Other proteins associated with the LINC complexes also play a role in cluster spread. In particular, embryos that have homozygous mutations or knockdown constructs for klar or koi fail to establish the appropriate distance between the nuclear clusters [31,32].
As the clusters migrate towards the muscle ends, dynamic movements of individual myonuclei have also been noted within each cluster. These movements include nuclear rotations and shape changes and involve Kinesin-1 and Dynein acting directly on individual myonuclei in a polarized manner: while Kinesin-1 exerts forces on the leading edge, Dynein acts on the nuclear lagging edge [25]. More recently, Aplip1, the Drosophila JIP1, was also shown to be involved in regulating nuclear cluster translocation via changes in nuclear shape [27]. The forces that determine nuclear shape and orientation are independent of the cortical pathway and are required to keep nuclei from changing directions.
An additional aspect of nuclear movement during cluster spread was revealed by Rosen et al. [19]. Detailed time-lapse analyses showed that, while the nuclear clusters were overall moving towards the cell poles (positive movement), this trajectory was frequently disrupted by shortterm movements in the opposite direction (negative movement). At the end of cluster spread, the final position of nuclear clusters resulted from the ratio of positive and negative movements. Strikingly, in different genotypes (e.g., Bsg25D overexpression and Ens loss) the quantification of this movement ratio at stage 15 was highly predictive of the final separation of nuclear clusters at stage 16 [19]. Together with the nuclear rotations and shape changes described above, these suggest that both the manner in which the clusters move and the interactions between myonuclei within clusters are essential for proper cluster spread. A better understanding of the mechanisms that regulate myonuclear dynamics and their impact on nuclear activity is needed.
As many of the proteins involved in the spreading of the myonuclear clusters in Drosophila are associated with the MT network, several studies have examined the MT network integrity and MT dynamics. Analysis of the MT-plus end binding protein EB1 has been used to probe how MT dynamics are affected in response to mutations in genes essential for cluster spread. Current data suggest that the MT network is not perturbed by the absence of most genes discussed in this section, with the exceptions of CLIP-190 and Aplip1 [19,24,26,27]. While CLIP-190 is necessary to facilitate the MT-cortex interactions that allows Dynein to move nuclei towards the muscle ends, the mechanism by which Aplip1 regulates the MT networks remains unknown.
In the Drosophila embryo, myonuclear cluster spread and the establishment of the myotendinous junctions (MTJs) occur simultaneously [33] (Figure 2A). The MTJ, a specialized junction that connects muscles to tendons, is essential for structural support and force transmission. While the mature MTJ is necessary for proper muscle contraction without detachment and allows the muscle cells to recover their shape after contraction [34–37], it is still unknown if and how the interactions between muscles and tendons influence myonuclear cluster movements. In mammalian muscle in vitro, there is no evidence of the two distinct clusters spreading as described in the Drosophila embryo, as myonuclei align in a single file and then spread. While these in vitro data are compelling, 2D muscle cell cultures lack extracellular cues provided by tendons and other cells types, which could be the reason for this observation. Nevertheless, the spreading of myonuclei in mammalian and Drosophila myotubes has several players in common. In mammals, MTs form a bipolar network around myonuclei that is required for proper myonuclear distribution. Similar to the fly, these networks seem to be minimally affected by depletion of Kinesins. Further, Kinesins and Dynein motors, Ens/MAP7, and MTs were shown to be necessary for nuclear 3D rotation and translocation, resulting in the proper distribution of nuclei [15,38–40]. Due to the complexity of skeletal muscles and the intimate interactions with a variety of different cell types, in vivo systems are essential to understanding myogenic processes. Our understanding of the roles that these interactions play in the regulation of myonuclear positioning is still limited, in particular concerning cluster spread, which could be highly dependent on structural cues and stability provided by stable cell-cell contacts.
Myonuclear Dispersion
During stage 17, the last stage of Drosophila embryonic development (16 h 00 min AEL; Figure 2A,D), the myonuclei dissociate from their cluster and disperse along the entire cell. During this stage, the myotubes mature into myofibers through the development of distinct structural and functional features; (i) the sarcomeres, highly conserved contractile units, that are linked to form longitudinal myofibrils and allow for muscle contraction, and (ii) the neuromuscular junction (NMJ), through which the motor neurons signal for muscle contraction. While these and other structural changes occur (e.g., maturation of the MTJs), the Drosophila embryo starts to experience coordinated muscle contractions at this stage. It is tempting to hypothesize that any of the structural changes mentioned above, as well as muscle activity, could promote/cause the dispersion of nuclei along the myofiber. However, we have observed that myonuclear dispersion occurs even in the absence of muscle contractions (unpublished observations), suggesting that a different mechanism is driving this aspect of nuclear positioning.
An alternative mechanism that could promote myonuclear dispersion is the assembly of the sarcomeres and the formation of myofibrils. Before sarcomere assembly, Zasp, a protein present at the sarcomere Z-line, colocalizes with F-actin in puncta at the vicinity of the nucleus in a LINC-dependent manner [32]. This accumulation of Zasp, and the subsequent sarcomere and myofibril formation, only occurs after nuclear positions are determined. Additionally, embryos with defects in nuclear positioning or reduced expression of the LINC complex components, Klar and Koi, show poorly assembled sarcomeres and unstable, incomplete, and/or torn myofibril networks [31,32]. These data indicate that proper nuclear positioning and the presence of LINC complexes are required for stable sarcomere assembly. While this does not exclude the possibility that sarcomerogenesis contributes to nuclear dispersion, it is unlikely that it drives these nuclear movements.
A final attractive hypothesis is that nuclear dispersion is linked to the reorganization of the cytoskeleton. The cytoskeleton is a structural complex present in all cells and it is composed of different classes of filamentous proteins: MTs, actin, and intermediate filaments. An interesting feature of skeletal muscle development is the drastic reorganization of the cytoskeleton, particularly of the MT networks. In myoblasts, MTs nucleate at a single centrosome, which is composed of two centrioles plus several proteins and constitutes the microtubule organizing center (MTOC). During most embryonic nuclear movements, the MT network appears to consist mostly of longitudinal MTs that run along the length of the myofiber. In addition, live imaging of EB1 comets to visualize growing MTs near myonuclei suggests that there is an increase in MT nucleating at the nuclear membranes throughout development (unpublished data). In differentiated myofibers of the Drosophila larva, at least two networks of MTs can be observed: longitudinal MTs that run along the length of the myofiber, as well as MTs nucleating from the nuclear membranes (astral MTs). Astral MTs result from the redistribution of the MTOCs [41]. It is not entirely clear when these MT rearrangements occur or whether they can drive the myonuclear dispersion observed in Drosophila embryonic muscles. It is further unclear if MAPs play a role in this process. Although Ens is essential for cluster spread in Drosophila LT muscles, a certain degree of myonuclear dispersion can be observed in Ens mutant embryos [15]. Similarly, despite the severe defects in cluster spreading, individual myonuclei and some dispersion can be observed in embryos lacking Dynein [24]. The partial recovery of nuclear positioning under these conditions suggests that cluster spread and nuclear dispersion have distinct regulation and that additional regulators of myonuclear dispersion remain to be discovered.
Similar to fly muscles, mammalian muscle cells also relocate their MTOC to the NE. However, these rearrangements occur in postmitotic myoblasts, prior to fusion [14,41]. After fusion, the myonuclei cluster in the cell, then align along the long axis of the cell, before spreading in an MT-dependent way that involves Dynein and Kinesin-1 [16]. Results from mammalian muscle cells and a computational model suggest that MAP7/Ens and Kinesin cooperate to slide antiparallel MTs nucleating from neighboring nuclei [15,42]. Another mechanism proposed to explain myonuclear dispersion in cultured mammalian myotubes involves Nesprin 1 or 2, as part of the LINC complex, recruiting motor proteins to the NE. The authors identified the nucleus as a Kinesin-1 cargo, with Nesprins 1 and 2 working as cargo adaptors. According to their model, nuclear dispersion is achieved by using the antiparallel MT network created by the multiple myonuclei [38,39]. Hence, it appears that MTs and MAPs are involved in nuclear dispersion in Drosophila and mammalian muscle fibers. However, further experiments exploring the rearrangements and dynamics of the cytoskeleton are required to better understand myonuclear dispersion.
Myonuclear Spacing and Anchoring
At the end of Drosophila embryogenesis, the muscles become fully functional and contract in a coordinated manner, allowing the embryo to hatch into a first instar larva. Subsequent larval development (5 days at 25°C) is characterized by dramatic muscle growth and continuous muscle activity manifested as locomotion. At this stage, the VL muscles have been the most studied group of myofibers for this process due to their location, size, shape, and nuclear arrangement [43–45]. VL muscles have a rectangular, flat shape with disc-shaped nuclei peripherally positioned on one side of the cell [44] (Figure 2A,E,F). Due to their simple morphology, they allow for the precise analysis of many muscle cell features, including protein localization, cytoskeletal networks organization, and myonuclear positioning. While the size of individual muscle fibers increases up to 50-fold during larval growth, the number of myonuclei remain the same [46]. To support this rapid growth, the myonuclei increase their DNA content through endoreplication, in a way that linearly scales with the muscle size [44,46]. Although little is known about myonuclear movements after the embryo hatches, the positions that myonuclei adopt along the length of the myotubes at the end of embryonic development are maintained in larval muscles. To maintain proper spacing, the myonuclei have to continuously adjust their positions along the growing cell. To withstand the forces produced during muscle contraction, each nucleus also needs to be anchored at its position.
Myonuclei are protected from intrinsic and extrinsic forces by a flexible perinuclear shield. This shield, which consists of nuclear, cytoskeletal, and LINC complex components, is involved in nuclear spacing as well as anchoring. Some components provide elasticity, such as the Nesprin 1 homolog MSP-300, while others provide rigidity, such as the MT network which is stabilized by Shot, the Drosophila spectraplakin, and EB1 [47,48]. In the absence of this shield, myonuclei show changes in morphology and have reduced levels of nuclear proteins, such as lamins and HP1, and positioning defects [48]. In addition to protecting the integrity of the nucleus itself, components of the perinuclear shield also connect to other muscle cell structures, like the T-tubules and the sarcoplasmic reticulum [49].
Further support for the roles of the LINC components in myonuclear spacing and anchoring has been generated by examining mutations in parkin-like E3 ubiquitin ligase Ariadne-1 (Ari-1). The authors showed that Ari-1 mono-ubiquitinates Koi, a central member of the LINC complex. Mutations in ari-1 resulted in Drosophila larval muscles with myonuclei clusters and morphology defects. Additionally, the authors identified rare variants of the human homolog of ari-1 (ARIH1), which is associated with conditions resulting from smooth muscle cell (SMC) dysfunction, such as thoracic aortic aneurysms. Excitingly, the Drosophila defects could be rescued with human ARIH1, while the rare variants found in patients could not. Due to the presence of abnormal nuclear morphology in SMCs from patients, the authors concluded that ARIH1 is crucial for myonuclear anchoring to the cytoskeleton [50,51]. Collectively, these data reinforce the importance of appropriate myonuclear positioning and anchoring through these complexes, and the use of simpler models, like Drosophila, to study mechanisms of human muscle dysfunction.
The spacing of myonuclei in Drosophila VL muscles is far from random. A recent in silico study proposes that myonuclei maximize their distances with each other by pushing on neighboring myonuclei and the cell membrane via the astral MTs at the NE (Figure 2E) [52]. These data are supported by various in vivo observations. Loss of bocks and Amph resulted in changes in the polarization of the perinuclear MTs and, in some cases, the absence of the MT asters. These MT changes were associated with defects in myonuclear spacing [17]. Similar to the embryo, Bsg25D and Ens also interact to position myonuclei in the larvae. Muscle-specific Bsg25D overexpression also shows severe myonuclear positioning defects in larval VL muscles, a phenotype that can be rescued by Ens expression. Similarly, increased levels of Bsg25D fully disrupts astral MTs in all myonuclei and result in the formation of ectopic MTOCs where Bsg25D and Ens colocalize. An interesting characteristic of polymerized tubulin is its ability to undergo post-translational modifications, which are associated with MT stability. Tubulin acetylation in particular is associated with increased binding of motors to MTs [53]. Bsg25D overexpression causes a decrease in acetylated tubulin, a marker of MT stability found in the longitudinal MTs, which might indicate a loss of MT stability [19]. The stiffness of the muscles was also decreased in Bsg25D overexpression larvae, which the authors attribute to changes in MT organization [19]. These studies clearly highlight the importance of the MT cytoskeleton, in particular the astral MTs, in establishing and maintaining nuclear spacing in the mature myofiber.
Mammalian myofibers in vivo are rod-shaped and display peripheral nuclear localization around the perimeter of the cell. An in vitro mammalian model has recently been developed in which mammalian myotubes mature into a myofiber in culture, complete with myofibrils. Coupled with a theoretical modeling, this in vitro model has revealed that centrally located myonuclei move to the cell periphery in the maturing myofiber. The authors found that this movement was due to centripetal forces generated by myofibril contraction and crosslinking around the nuclei. This process is dependent on the Cdc42-regulated organization of desmin. The nuclear dispersion also relies on the stiffness of nuclei and the activity proteins like Amphiphysin-2 and the Arp2/3 complex proteins, which regulate the deformation and squeezing of nuclei to the muscle periphery [54–56]. Additionally, peripheral nuclear positioning depends on the local accumulation of fibronectin deposited by myofibroblasts [56]. Since several muscle disorders show centrally located myonuclei, understanding how making myonuclei peripheral and possibly keeping them anchored in place is key to discover the mechanisms behind these diseases and generate therapies that help prevent or revert this process.
An important feature of myonuclear spacing and anchoring is that myonuclei in Drosophila and mammalian mature myofibers diversify and adopt specific characteristics dependent on their position within the cell, specifically their proximity to MTJs and NMJ [44,52,57]. In Drosophila larval myofibers, internuclear distances near the NMJ depend on Kinesin-1 and Dynein, and the distance between myonuclei and the NMJ is regulated by Klar [57]. Nuclei that are closer to the NMJ are overall larger and have higher DNA copy number than other nuclei. The opposite is true for the nuclei near MTJ; they are smaller in size with fewer copies of DNA [44]. Additionally, there are more nuclei near the MTJ and their positions within muscle cells appear to be regulated by Klar and Pins, as well as in response to mechanical stimuli [52,57]. Similar to what is observed in Drosophila myofibers, mammalian muscle in vivo also displays two very clear populations of myonuclei that aggregate near NMJs and MTJs [16,58]. These data argue for the similarities between animal models and the important relationships that the muscle establishes with other cell types. However, a better understanding of these relationships using mammalian in vivo models or in vitro coculture models will better clarify the importance of these interactions and the impact that they have on myonuclear spacing and anchoring within the myofiber.
Concluding Remarks
Myonuclear positioning is a complex sequence of steps involving many molecular players active at multiple processes. Here, we subdivide the Drosophila myonuclear positioning into four steps and describe the most recent mechanisms including those players. Overall, the major molecular components are nuclear components, LINC complexes, and cytoskeletal and motor proteins. These processes and the genes that regulate them appear to be highly conserved from fly to mammal. Most importantly, in all models, mispositioned nuclei are associated with a decrease in muscle function (Table 1). Nevertheless, several aspects of myonuclear positioning remain to be addressed, including the impact of muscle cell interactions with other cells types, the links between myonuclei as they move, and the movement of myonuclei during growth and regeneration (see Outstanding Questions). The use of multiple models (Drosophila, mammalian in vitro and in vivo) and approaches (genetic, biochemical, mathematical modeling) will surely uncover the answers.
Highlights.
Myonuclear movement and positioning require several steps with different mechanisms.
Crucial players involve the LINC complexes, microtubule cytoskeleton, and associated proteins.
Defects in myonuclear positioning in Drosophila and mammals share many molecular components.
Mutations in genes linked to centronuclear myopathy and Emery- Dreifuss muscular dystrophy cause myonuclear positioning phenotypes in Drosophila.
Myonuclear mispositioning has been shown to be a hallmark of numerous muscular disorders. For example, centronuclear myopathies are a diverse group of disorders associated with mutations in several different genes [59]. Similarly, EDMD represents a genetically heterogeneous disease. For both of these examples, novel mutations are still being discovered [59,60]. These new mutations open new ways to explore muscle biology as well as offer targets for precise and individualized therapies. From studies using Drosophila to study myonuclear positioning, at least a dozen genetic mutations have been identified to affect muscle function, some of which involve genes directly linked to disease [15,17,24,26,28,31,44,49]. These studies confirm the contribution of proper myonuclear positioning for muscle function and the validity of this model for exploring the mechanisms behind myonuclear position and muscle loss of function (Box 1). Nevertheless, it is the collective data from studies in both the fly and mammals that will clarify the importance of myonuclear positioning.
Outstanding Questions.
Do muscle diseases share a common mechanism or result from different mechanisms?
Are the mechanisms that drive nuclear clustering and positioning during development and regeneration the same?
How do muscle cell interactions with other cell types (e.g., tendons and motor neurons) affect myonuclear positioning?
How does position dictate nuclear identity?
How are myonuclear positions maintained during myofiber growth and contraction?
What are the mechanisms that drive even myonuclear dispersion with the muscle cell?
How do nuclear clusters maintain their cohesion during cluster spread?
When do the rearrangements of the microtubule network occur during fly myogenesis?
Acknowledgments
We thank the members of the Baylies Laboratory, particularly Stefanie Windner and Mridula Balakrishnan, for helpful discussions and for critical reading of the manuscript and for contributing to the figure (Windner, Figure 2, panel A, embryonic muscle schematics). This work was supported by the Portuguese Science and Technology Foundation, Portugal (SFRH/BD/52041/2012) to M.A., the National Institutes of Health (NIH) (GM078318, AR108981) to M.K.B., and National Cancer Institute (P30 CA 008748) core grant to the Memorial Sloan Kettering Cancer Center. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Calero-Cuenca FJ et al. (2018) Dealing with the nucleus during cell migration. Curr. Opin. Cell Biol 50, 35–41 [DOI] [PubMed] [Google Scholar]
- 2.Gundersen GG and Worman HJ (2013) Nuclear positioning. Cell 152, 1376–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dubowitz V and Sewry CA (2007) Definition of pathological changes seen in muscle biopsies In Muscle Biopsy, Elsevier [Google Scholar]
- 4.Folker ES and Baylies MK (2013) Nuclear positioning in muscle development and disease. Front. Physiol 4, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spiro AJ et al. (1966) Myotubular myopathy: persistence of fetal muscle in an adolescent boy. Arch. Neurol 14, 1–14 [DOI] [PubMed] [Google Scholar]
- 6.Sewry CA et al. (2001) Skeletal muscle pathology in autosoma dominant Emery-Dreifuss muscular dystrophy with lamin A/C mutations. Neuropathol. Appl. Neurobiol 27, 281–290 [DOI] [PubMed] [Google Scholar]
- 7.Osorio DS and Gomes ER (2013) The contemporary nucleus: a trip down memory lane. Biol. Cell 105, 430–441 [DOI] [PubMed] [Google Scholar]
- 8.Lee YL and Burke B (2018) LINC complexes and nuclear positioning. Semin. Cell Dev. Biol 82, 67–76 [DOI] [PubMed] [Google Scholar]
- 9.Starr DA (2011) KASH and SUN proteins. Curr. Biol 21, R414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng S et al. (2017) Acting on identity: myoblast fusion and the formation of the syncytial muscle fiber. Semin. Cell Dev. Biol 72,45–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abmayr SM and Pavlath GK (2012) Myoblast fusion: lessons from flies and mice. Development 139, 641–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dobi KC et al. (2015) Specification of the somatic musculature in Drosophila. Wiley Interdiscip. Rev. Dev. Biol 4, 357–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim JH et al. (2015) Mechanisms of myoblast fusion during muscle development. Curr. Opin. Genet. Dev 32, 162–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cadot B et al. (2015) Moving and positioning the nucleus in skeletal muscle-one step at a time. Nucleus 6, 373–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Metzger T et al. (2012) MAP and kinesin-dependent nuclear positioning is required for skeletal muscle function. Nature 484, 120–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roman W and Gomes ER (2018) Nuclear positioning in skeletal muscle. Semin. Cell Dev. Biol 82, 51–56 [DOI] [PubMed] [Google Scholar]
- 17.Collins MA et al. (2017) Emery-Dreifuss muscular dystrophy-linked genes and centronuclear myopathy-linked genes regulate myonuclear movement by distinct mechanisms. Mol. Biol. Cell 2303–2317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mandigo TR et al. (2019) Drosophiia emerins control LINC complex localization and transcription to regulate myonuclear position. J. Cell Sci 132, jcs.235580 [DOI] [PubMed] [Google Scholar]
- 19.Rosen JN et al. (2019) The Drosophila Ninein homologue Bsg25D cooperates with Ensconsin in myonuclear positioning. J. Cell Biol 218, 524–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cadot B et al. (2012) Nuclear movement during myotube formation is microtubule and dynein dependent and is regulated by Cdc42, Par6 and Par3. EMBO Rep. 13, 741–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clark WE (1946) An experimental study of the regeneration of mammalian striped muscle. J. Anat 80, 24–36 [PubMed] [Google Scholar]
- 22.Chargé SBP and Rudnicki MA (2004) Cellular and molecular regulation of muscle regeneration. Physiol. Rev 84, 209–238 [DOI] [PubMed] [Google Scholar]
- 23.Chaturvedi D et al. (2017) Identification and functional characterization of muscle satellite cells in Drosophila. Elife 6, 1–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Folker ES et al. (2012) Muscle length and myonuclear position are independently regulated by distinct Dynein pathways. Development 139, 3827–3837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Folker ES et al. (2014) Translocating myonuclei have distinct leading and lagging edges that require Kinesin and Dynein. Development 141, 355–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schulman VK et al. (2014) Syd/JIP3 and JNK signaling are required for myonuclear positioning and muscle function. PLoS Genet 10, e1004880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Auld AL et al. (2018) Aplip1, the Drosophila homolog of JIP1, regulates myonuclear positioning and muscle stability. J. Cell Sci 131, jcs.205807 [DOI] [PubMed] [Google Scholar]
- 28.Camuglia JM et al. (2018) An RNAi based screen in Drosophiia larvae identifies fascin as a regulator of myoblast fusion and myotendinous junction structure. Skelet Muscle 8, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Métivier M et al. (2019) Dual control of Kinesin-1 recruitment to microtubules by Ensconsin in Drosophila neuroblasts and oocytes. Development 146, 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sung HH et al. (2008) Drosophila Ensconsin promotes productive recruitment of Kinesin-1 to microtubules. Dev. Cell 15, 866–876 [DOI] [PubMed] [Google Scholar]
- 31.Elhanany-Tamir H et al. (2012) Organelle positioning in muscles requires cooperation between two KASH proteins and microtubules. J. Cell Biol 198, 833–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Auld AL and Folker ES (2016) Nucleus-dependent sarcomere assembly is mediated by the LINC complex. Mol. Biol. Cell 27, 2351–2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schweitzer R et al. (2010) Connecting muscles to tendons: tendons and musculoskeletal development in flies and vertebrates. Development 137, 3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gillies AR and Lieber RL (2012) Structure and function of the skeletal muscle extracellular matrix. Muscle Nerve 44, 318–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bentzinger CF et al. (2013) Cellular dynamics in the muscle satellite cell niche. EMBO Rep. 14, 1062–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomopoulos S et al. (2013) Structural Interfaces and Attachments in Biology, Springer [Google Scholar]
- 37.Valdivia M et al. (2017) Mechanical control of myotendinous junction formation and tendon differentiation during development. Front. Cell Dev. Biol 5, 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilson MH and Holzbaur ELF (2012) Opposing microtubule motors drive robust nuclear dynamics in developing muscle cells. J. Cell Sci 125, 4158–4169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson MH and Holzbaur ELF (2015) Nesprins anchor kinesin-1 motors to the nucleus to drive nuclear distribution in muscle cells. Development 142, 218–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gache V et al. (2017) Microtubule motors involved in nuclear movement during skeletal muscle differentiation. Mol. Biol. Cell 28, 865–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bugnard E et al. (2005) Reorganization of microtubule nucleation during muscle differentiation. Cell Motil. Cytoskeleton 60, 1–13 [DOI] [PubMed] [Google Scholar]
- 42.Gimpel P et al. (2017) Nesprin-1α-dependent microtubule nucleation from the nuclear envelope via Akap450 is necessary for nuclear positioning in muscle cells. Curr. Biol 27, 2999–3009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Piccirillo R et al. (2014) Mechanisms of muscle growth and atrophy in mammals and Drosophila. Dev. Dyn 243, 201–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Windner SE et al. (2019) Nuclear scaling is coordinated among individual nuclei in multinucleated muscle fibers. Dev. Cell 49, 48–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kohsaka H et al. (2012) Development of larval motor circuits in Drosophila. Develop. Growth Differ 54, 408–419 [DOI] [PubMed] [Google Scholar]
- 46.Demontis F and Perrimon N (2009) Integration of insulin receptor/Foxo signaling and dMyc activity during muscle growth regulates body size in Drosophila. Development 136, 983–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang S and Volk T (2015) Composite biopolymer scaffolds shape muscle nucleus: insights and perspectives from Drosophila. Bioarchitecture 5, 35–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang S et al. (2015) Nesprin provides elastic properties to muscle nuclei by cooperating with spectraplakin and EB1. J. Cell Biol 209, 529–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reuveny A et al. (2018) Ma2/d promotes myonuclear positioning and association with the sarcoplasmic reticulum. Development 145, dev159558 [DOI] [PubMed] [Google Scholar]
- 50.Tan KL et al. (2018) Ari-1 regulates myonuclear organization together with Parkin and is associated with aortic aneurysms. Dev. Cell 45, 226–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Balakrishnan M and Baylies MK (2018) Myonuclear positioning and aneurysms are LINC’d by ariande. Dev. Cell 45, 149–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Manhart A et al. (2018) Mechanical positioning of multiple nuclei in muscle cells. PLoS Comput Biol 14, 1–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.De Forges H et al. (2012) Interplay between microtubule dynamics and intracellular organization. Int. J. Biochem. Cell Biol 44, 266–274 [DOI] [PubMed] [Google Scholar]
- 54.Falcone S et al. (2014) N-WASP is required for Amphiphysin-2/ BIN1-dependent nuclear positioning and triad organization in skeletal muscle and is involved in the pathophysiology of centronuclear myopathy. EMBO Mol. Med 6, 1455–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roman W et al. (2017) Myofibril contraction and crosslinking drive nuclear movement to the periphery of skeletal muscle. Nat. Cell Biol 19,1189–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roman W et al. (2018) Local arrangement of fibronectin by myofibroblasts governs peripheral nuclear positioning in muscle cells. Dev. Cell 46, 102–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Perillo M and Folker ES (2018) Specialized positioning of myonuclei near cell-cell junctions. Front. Physiol 9, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bruusgaard JC et al. (2003) Number and spatial distribution of nuclei in the muscle fibres of normal mice studied in vivo. J. Physiol 551, 467–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Casar-Borota O et al. (2015) A novel dynamin-2 gene mutation associated with a late-onset centronuclear myopathy with necklace fibres. Neuromuscul. Disord 25, 345–348 [DOI] [PubMed] [Google Scholar]
- 60.Chen Z et al. (2017) A novel SYNE1 gene mutation in a Chinese family of Emery-Dreifuss muscular dystrophy-like. BMC Med. Genet 18, 4–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meinke P et al. (2014) Muscular dystrophy-associated SUN1 and SUN2 variants disrupt nuclear-cytoskeletal connections and myonuclear organization. PLoS Genet. 10, e1004605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Janin A and Gache V (2018) Nesprins and lamins in health and diseases of cardiac and skeletal muscles. Front. Physiol 9,1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Koch AJ and Holaska JM (2014) Emerin in health and disease. Semin. Cell Dev. Biol 29, 95–106 [DOI] [PMC free article] [PubMed] [Google Scholar]


