Abstract
Dynamical features of cell signaling are the essence of living organisms. To understand animal development, it is fundamental to investigate signaling dynamics in vivo. Robust reporters are required to visualize spatial and temporal dynamics of enzyme activities and protein-protein interactions involved in signaling pathways. In this review, we summarize recent development in the design of new classes of fluorescent reporters for imaging dynamic activities of proteases, kinases, and protein-protein interactions. These reporters operate on new physical and/or chemical principles; achieve large dynamic range, high brightness, and fast kinetics; and reveal spatiotemporal dynamics of signaling that is correlated with developmental events such as embryonic morphogenesis in live animals including Drosophila and zebrafish. Therefore, many of these reporters are great tools for biological discovery and mechanistic understanding of animal development and disease progression.
Introduction
DNA sequencing and mass spectrometry-based technologies have identified a large number of genes and proteins and much higher number of protein-protein interactions (PPI) in cells and model organisms. Together with RNA interference and recent CRISPR, these technologies have moved our understanding of signal transduction from linear pathways to integrated networks. However, dynamical features of signaling, which are the essence of living organisms, are largely unknown especially in animals because of technical challenges [1–7]. For example, many fundamental questions remain unanswered, including: how do the proteins and their interactions drive the dynamics of flow of information through signaling networks? How does signaling dynamics regulate animal development? In cell and developmental biology, it remains open questions about how dynamics of an individual pathway can elicit distinct cellular outcomes in the same cell type, and how multiple pathways can crosstalk and form signaling networks to regulate cellular processes. The development of fluorescent reporters, including reporters of PPI and activity reporters of enzymes in the signaling pathways such as proteases and kinases, enables us to visualize and analyze information flow and dynamics of cell signaling in living cells and animals with unprecedented spatiotemporal resolution and can help address these key issues.
Fluorescence imaging of cell signaling in living animals is challenging because of tissue autofluorescence, cell heterogeneity and rapid shape and position changes. Therefore, an ideal fluorescent reporter for in vivo imaging should have large dynamic range (i.e. large fluorescence change) and high brightness, in addition to be genetically encoded requiring no exogenous cofactors. Furthermore, to detect rapid dynamics of cell signaling, the reporters should have fast kinetics (i.e. rapid response time). Recently, several groups including ours have applied new physical and/or chemical principles and designed new classes of fluorescent reporters. Here, we will introduce several of them, including activity reporters of proteases and kinases, as well as PPI reporters.
Genetically encoded and multicolor fluorogenic reporters of protease activity
Proteases play fundamental roles in almost every major biological process. Human genome contains ~600 proteases and homologs, similar to the number of protein kinases (~500) [8–10]. Changes to proteolytic systems lead to many diseases including cancer, neurodegenerative and cardiovascular diseases [11–14]. To image protease activity in living animals, recently several new types of multicolor fluorogenic protease reporters have been developed, including the near-infrared fluorogenic protease reporter iCasper[15], the GFP-based green fluorogenic protease reporters ZipGFP[16] and FlipGFP[17], and the mCherry-based red fluorogenic protease reporter FlipCherry[17].
iCasper.
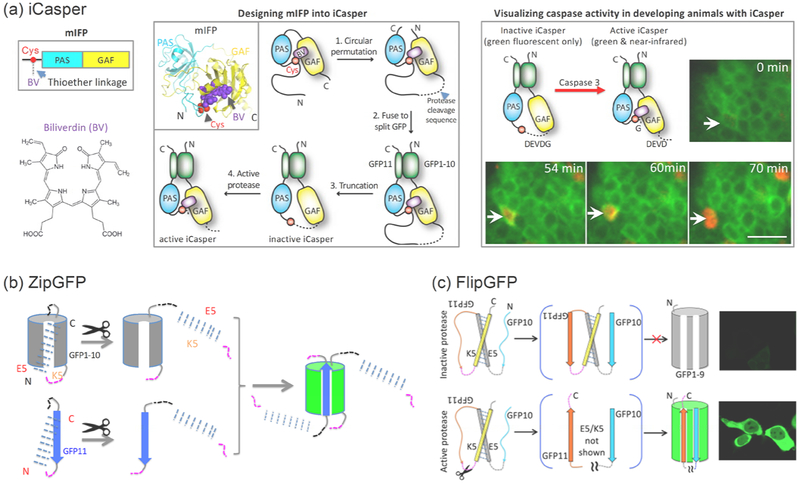
iCasper is developed by redesigning a naturally monomeric near-infrared fluorescent protein (mIFP) [18] so that the chromophore incorporation is regulated by protease activity (Figure 1a) [15]. iCasper achieves large dynamic range (dark-to-bright) and fast kinetics (~10 seconds). By incorporating the specific cleavage sequence of executioner caspases, iCasper has been used for imaging caspase activity and apoptosis in cells and animals. In vivo imaging using iCasper shows that it reveals spatiotemporal coordination between apoptosis and embryonic morphogenesis, as well as dynamics of apoptosis during brain tumorigenesis in Drosophila[15]. Because of modular design, iCasper can be used to engineer reporters of other proteases by incorporating protease-specific cleavage sequence.
Figure 1. Multicolor fluorogenic protease reporters.
(a) Near-infrared fluorogenic protease reporter iCasper is designed from a naturally monomeric near-infrared fluorescent protein mIFP. mIFP is composed of a PAS domain followed by a GAF domain. It is engineered from a truncated bacterial phytochrome (BphP). Top left, primary structure of mIFP. Bottom left, chemical structure of the chromophore biliverdin. Middle panel, schematic diagram showing how mIFP is designed into iCasper. It has been well known that for a BphP, the chromophore biliverdin (BV) non-covalently binds to GAF, and then forms a thioether bond with a catalytic cysteine at the N-terminal region. The key design principle of iCasper is that the length of the cleavage sequence, which connects the catalytic cysteine to the C-terminus of GAF, is shorter than the distance between the cysteine and the GAF so that the cysteine is displaced from the binding cavity of the chromophore. Upon protease activation and cleavage, the cysteine is released and returns to the BV binding site, forming the thioether bond with BV, which restores the near-infrared fluorescence. iCasper incorporates BV as the chromophore and becomes fluorescent within ten seconds. Right panel, iCasper containing caspase cleavage sequence visualizes caspase activity and apoptosis during embryonic development of Drosophila. (b) A bipartite GFP-based protease reporter ZipGFP, which is based on caging or “zipping” binding cavity of GFP1–10 and GFP11 with heterodimeric coiled coils E5 and K5. The protease cleavage sequence is shown in magenta. Reprinted with permission from reference16. Copyright 2019 Elsevier Inc. (c) A tripartite GFP-based fluorogenic protease reporter FlipGFP by flipping a beta strand of GFP. A tripartite mCherry-based red fluorogenic protease reporter FlipCherry has also been developed using the same principle, see text for details. GFP1–9 is the fragment of GFP containing the 9 beta-strands (1st to 9th) and the central alpha-helix. The central alpha-helix contains three amino acids that form the chromophore. GFP10 and GFP11 are the 10th and 11th beta-strand of GFP, respectively. GFP11 contains the highly conserved Glu222 that is critical for chromophore maturation. Reprinted with permission from reference17. Copyright 2019 American Chemical Society.
Successful development of iCasper based on the unique protein-chromophore interactions suggests that bacterial phytochrome (BphP)-derived near-infrared fluorescent proteins (IFPs) may provide a new and promising scaffold for designing many fluorogenic reporters, including calcium and membrane potential sensors. Indeed, recently Campbell’s group has designed mIFP into a near-infrared calcium reporter NIR-GECO[19]. Furthermore, thousands of BphPs available in the protein sequence database increase our choice of promising scaffolds. Such genetically encoded fluorogenic reporters will be ideal in visualizing spatiotemporal dynamics of cell signaling in vivo.
ZipGFP.
A GFP-based fluorogenic protease reporter, dubbed ZipGFP, has been developed by caging or “zipping” the two parts of the self-assembling split GFP so that its self-assembly is regulated by protease activity (Figure 1b) [16]. ZipGFP-based TEV protease reporter increases fluorescence 10-fold after activation by the protease. A ZipGFP-based executioner caspase reporter visualizes apoptosis in live zebrafish embryos with spatiotemporal resolution. Thus, ZipGFP-based caspase reporter will be useful for monitoring apoptosis during animal development and for designing reporters of proteases beyond the executioner caspases.
FlipGFP and FlipCherry.
To further increase the dynamic range, FlipGFP is rationally designed by flipping a beta strand of the GFP (Figure 1c) [17]. Upon protease activation and cleavage, the beta strand is restored, leading to reconstitution of the GFP and fluorescence. FlipGFP-based TEV protease reporter achieves 100-fold fluorescence increase upon protease activation. FlipGFP has quantum yield 0.66, which is 2.6-fold higher than that of ZipGFP. A FlipGFP-based executioner caspase reporter visualizes apoptosis in live zebrafish embryos with spatiotemporal resolution. FlipGFP also visualizes apoptotic cells in the midgut of Drosophila. Therefore, FlipGFP-based caspase reporter will be useful for monitoring apoptosis during animal development and for designing reporters of proteases beyond caspases. The design strategy of FlipGFP has been further applied to a red fluorescent protein mCherry for engineering a red fluorogenic protease reporter FlipCherry.
FPX, VC3AI, and CA-GFP.
In addition to the above protease reporters, there are several other protease reporters, including but not limited to: FPX that is based on dimerization-dependent fluorescent protein exchange[20], VC3AI that is based on protein splicing of the circularly permuted Venus[21], CA-GFP that utilizes an oligomeric peptide[22]. These new types of reporters achieve higher dynamic range than FRET-based protease reporters.
How to apply the above reporters to new proteases?
Because of the modular design, the above protease reporters including iCasper, ZipGFP and FlipGFP, can be used to design activity reporters of many other proteases. However, several points may be considered. First, length of the consensus cleavage sequence for the specific protease need to be within the working range of each reporters. For iCasper, it should be within (including) 11 amino-acid (aa). Most protease cleavage sequence is shorter than 11aa. But if the cleavage sequence is 12aa or longer, the dynamic range of iCasper will be smaller (i.e. background fluorescence before protease cleavage will appear) [15]. On the other hand, if the cleavage sequence is shorter than 11aa, it is suggested to add flexible linker (glycine, serine) to flank both ends of the cleavage sequence. For FlipGFP, the working range of the cleavage sequence has not been comprehensively tested, but the ideal length is 7aa. Second, certain proteases may bind specific sequence or domain of substrate for its recruitment. In this case, one may need to fuse the docking sequence or domain to the reporter, in addition to the cleavage sequence. Third, if the dynamic range is the top priority while designing a new protease activity reporter, one may choose iCasper since it achieves the largest dynamic range. Fourth, if the reporter brightness after protease cleavage is the priority, one may choose FlipGFP, which achieves the highest brightness with quantum yield 66% after protease activation. Lastly, when simultaneous imaging of two proteases’ activities is desired, one can either use both iCasper and FlipGFP, or simply use ZipGFP. For the latter case, one could design cleavage sequence into each “zipped” part of ZipGFP. In this way, ZipGFP’s 10-fold fluorescence increase will not be obtained until both proteases are activated.
Together, the new class of protease reporters will be important tools for visualizing spatiotemporal dynamics of protease activities in living cells and animals. Furthermore, these multicolor protease reporters enable simultaneous visualization of dynamic activities of multiple proteases in the signaling network.
A new class of genetically encoded fluorescent reporters of kinase activity
Protein kinases play critical roles in signal transduction, and protein phosphorylation is an essential type of post-translational modification[23–25]. Furthermore, temporal dynamics of kinase signaling can elicit distinct cellular responses[26,27]. Therefore, mechanistic understanding of animal development and tissue biology will benefit from visualizing dynamics of kinase activity in physiological contexts[1,28,29]. An ideal kinase reporter for in vivo imaging should have large dynamic range, high brightness and fast kinetics in order to detect when and in which tissues and cells an endogenous kinase is activated during animal development. Recently, several groups have developed new types of kinase activity reporters. Here we will focus on a GFP phase separation-based kinase reporter called SPARK (Separation of Phases-based Activity Reporters of Kinases) [30], and briefly mention other types of reporters. SPARK has been demonstrated in Drosophila and zebrafish for imaging endogenous kinases’ activities, and is so far the most sensitive activity reporters of kinases in terms of dynamic range, brightness and signal pattern with second-to-minute kinetics.
SPARK.
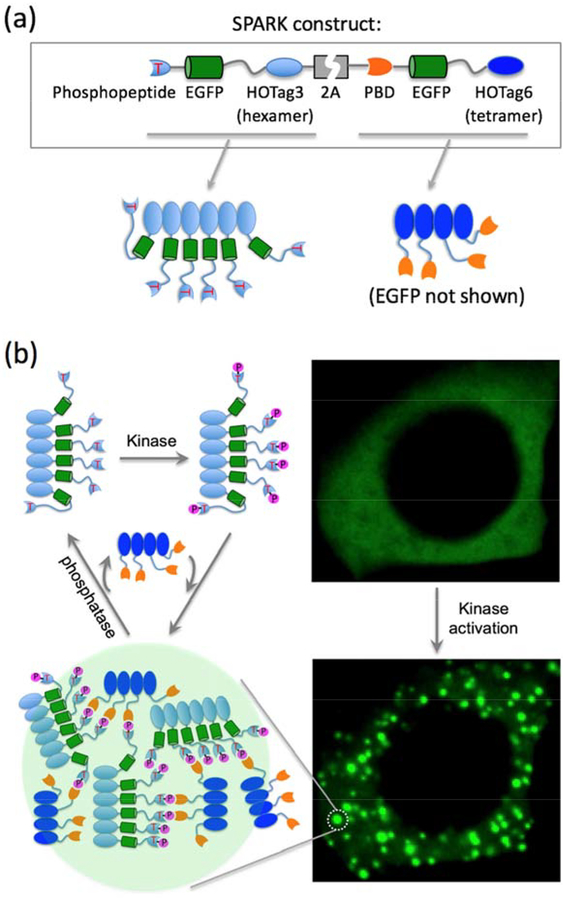
The guidance of designing SPARK is that fluorescence change of a kinase reporter is determined by kinase activity-dependent changes to the brightness of a digital fluorescence image, which is dependent on fluorophore’s intrinsic brightness including quantum yield and extinction coefficient, and is also proportional to the number of “imaged” fluorophores per pixel. Förster resonance energy transfer (FRET)-based kinase reporters rely on changes of intrinsic brightness of donor and/or acceptor fluorophores via FRET[31–33]. A different approach is to design reporters that change the number of “imaged” fluorophores per pixel. Based on this approach, we have designed a new class of kinase reporters called SPARK that, upon kinase activation, phase separate and form highly concentrated droplets[30]. SPARK is designed based on multivalent PPI-driven protein phase separation (Figure 2a): 1) to introduce multivalency, we utilized de novo designed homo-oligomeric coiled coils (named as HOTag); 2) to sense kinase activity, we introduce kinase activity-dependent PPI – a consensus kinase substrate sequence (a phosphopeptide) and a phosphopeptide-binding domain; 3) to obtain fluorescence signal, we incorporated EGFP (enhanced GFP) into the reporter.
Figure 2. GFP phase separation-based activity reporter of kinases.
(a) Top, DNA construct of SPARK. Bottom, cartoon showing two parts of the SPARK. HOTag (homo-oligomeric tag) is de novo designed coiled coils (~30 amino acids). (b) Schematic showing multivalent interaction-driven phase separation of SPARK upon kinase phosphorylation. Right panel, fluorescence images of a HEK293 cell expressing PKA-SPARK upon protein kinase A (PKA) activation. Adapted with permission from reference30. Copyright 2019 Elsevier Inc.
Upon activation of the endogenous kinase, the kinase substrate peptide is phosphorylated and then interacts with phosphopeptide-binding domain, the resulted multivalent (via HOTag) PPI leads to highly concentrated (10X) EGFP droplets (Figure 2b). Therefore, when kinase is inactive, SPARK is evenly distributed in the cell and is as bright as EGFP. When the endogenous kinase is activated, SPARK is phosphorylated, forming fluorescent droplets that are 10-fold brighter. The intensive brightness and punctal signal pattern enable SPARK to robustly detect kinase activity in vivo. SPARK has been used to visualize dynamic kinase signaling in Drosophila and zebrafish. SPARK has ~20-second response time and is reversible, with no perturbation of kinase signaling and no apparent toxicity in transgenic animals[30]. The modular design of SPARK allows straightforward design of reporters of other kinases by swapping the kinase substrate sequence and the phosphopeptide-binding domain.
Other kinase reporters.
Recently developed kinase reporters include but not limited to: an excitation ratiometric kinase sensor ExRai[34], fluorescence anisotropy-based reporters FLARE[35], GFP translocation-based reporter KTR[36]. These reporters achieve better signal than the conventional FRET-based kinase reporters. However, performance of these reporters for imaging dynamic kinase signaling in animals such as Drosophila and zebrafish will need to be seen in future biological applications.
How to apply SPARK to new kinases?
The modular scaffold of SPARK can be used to design activity reporters of most if not all 518 kinases. For example, my group has now designed SPARK-based activity reporters of ~ 20 kinases including serine/threonine and tyrosine kinases (unpublished). While in principle SPARK can be applied to most kinases, there are several points that should be considered. First, the consensus phosphopeptide should be specific for the kinase. This ensures specificity of the SPARK signal. Second, certain kinases, such as ERK and LIMK, bind docking sequence or domain of their substrate for its recruitment. In this case, the docking sequence or domain should be appropriately included into the SPARK design, in addition to the phosphopeptide sequence. Third, the phosphopeptide binding domain needs to be appropriately chosen for the specific phosphopeptide. Lastly, while SPARK is mainly expressed in the cytoplasm, one can add nuclear localization signal to SPARK in order to detect certain kinases’ activities in the nucleus (unpublished observation).
Protein-protein interaction reporters
Cells must be able to sense and respond to their environment for fate decisions. Signal transduction from extracellular cues to intracellular effector proteins confers cells in an organism with the ability to appropriately decide when they should proliferate, grow, divide, differentiate, migrate, or perform other cellular functions[29,37,38]. The coordinated signal relay depends on PPI between signaling molecules including extracellular hormones, cell surface receptors, intracellular scaffold and adaptor proteins, enzymes, transcription factors, and other effector proteins[38–40]. Therefore, PPIs play fundamental roles in signal transduction.
Here we will focus on several new types of PPI reporters, which have fast kinetics so that they can detect dynamical features of PPIs. These reporters also have one or several additional features that enable robust imaging of PPIs, including: 1) they have no background fluorescence; 2) they are reversible, so that they can detect protein-protein dissociation, which contributes to the transient nature of many PPIs; 3) they can detect weak PPIs, which have been seen in a large portion of PPIs and play critical roles in dynamic cell signaling.
Tripartite GFP and Trio with no background fluorescence.
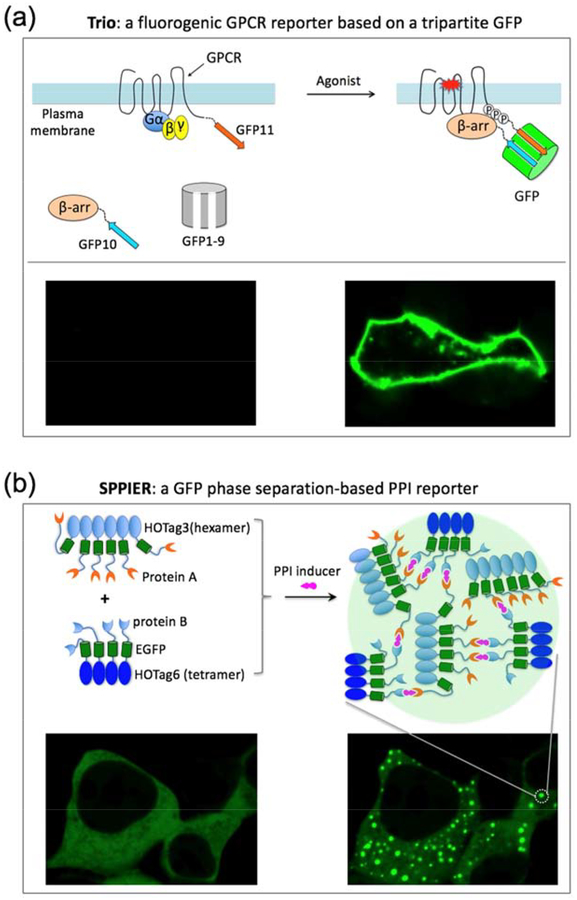
A tripartite split-GFP system was developed to detect PPIs without background fluorescence[41]. The three parts do not self-assemble until two of them are in close proximity. Recently, this system has been used in developing a dark-to-bright fluorogenic GPCR/β-arrestin interaction assay named Trio (Figure 3a) [42]. Trio has been applied to identify several key residues at the c-terminal region of the Gαq-coupled protease-activated receptor-1 (PAR1) that are essential for recruiting β-arrestin upon activation[42].
Figure 3. Fluorogenic and phase separation-based protein-protein interaction reporters.
(a) Tripartite GFP-based GPCR and beta-arrestin interaction assay Trio. GFP1–9 is the fragment of GFP containing the 9 beta-strands (1st to 9th) and the central alpha-helix. The central alpha-helix contains three amino acids that form the chromophore. GFP10 and GFP11 are the 10th and 11th beta-strand of GFP, respectively. GFP11 contains the highly conserved Glu222 that is critical for chromophore maturation. The three parts self-assemble only when two of them are in close proximity. Upon GPCR/beta-arrestin interaction, GFP10 and GFP11 are brought together in close proximity. Then GFP1–9 spontaneously assembles with GFP10/GFP11, reconstituting GFP, leading to chromophore maturation and fluorescence. (b) GFP phase separation-based PPI reporter SPPIER. HOTag (homo-oligomeric tag) is de novo designed coiled coils (~30 amino acids). SPPIER visualizes small molecule-induced dynamic PPIs in living cells including immunomodulatory drug-induced interaction between cereblon and Ikaros, CC885-induced interaction between cereblon and GSPT1, PROTAC-induced interaction between E3 ubiquitin ligases and target proteins, as well as nutlin-induced dissociation between MDM2 and p53. Adapted with permission from reference47. Copyright 2019 American Chemical Society.
Trio has been expanded to image other GPCR/β-arrestin interactions in live cells, including protease-activated receptor-2 (PAR2), β2 adrenergic receptor (β2AR), neurokinin-1 receptor (NK1R), and μ-type opioid receptor (MOR) [42]. The GPCR superfamily is the largest receptor family in the animal kingdom, with more than 800 GPCRs encoded in the human genome[43]. Thus, this new assay Trio, which achieves spatiotemporal resolution, will be an important tool for the GPCR field.
Split UnaG-based reversible PPI reporters.
While Trio is able to detect PPI without background fluorescence, it is irreversible, presumably due to the tight structure of GFP. To solve this problem, recently my group has designed a reversible green fluorogenic PCA (protein complementation assay, also known as BiFC) [44] based on the crystal structure of UnaG, a recently discovered green fluorescent protein that has a scaffold different from GFP[45]. In living mammalian cells, the non-fluorescent fragments complement and rapidly become fluorescent, and lose fluorescence when PPI is inhibited. This reversible UnaG-based PPI reporter (uPPI) utilizes bilirubin (BR) as the chromophore and requires no exogenous cofactor. With further improvement of its brightness, uPPI will find many applications in visualizing spatiotemporal dynamics of PPI.
GFP phase separation-based reporters that detect small molecule-induced weak PPIs.
Small molecules that modulate PPI are important tools for biological investigation and therapeutic intervention[46]. To visualize small molecule-induced PPIs in living cells, a fluorophore phase separation-based PPI assay named SPPIER has been developed[47]. Upon small molecule-induced PPI, SPPIER rapidly forms highly fluorescent GFP droplets (Figure 3b). SPPIER has been demonstrated in detecting immunomodulatory drugs (IMiDs)-induced PPI between cereblon and the transcription factor Ikaros. The cereblon-based degradation of Ikaros accounts for the clinical efficacy of IMiDs against multiple myeloma[48–50]. SPPIER also detects IMiDs analog (e.g. CC-885)-induced PPI between cereblon and GSPT1. SPPIER can be modified to image small molecule-induced protein-protein dissociation, such as nutlin-induced dissociation between MDM2 and p53. SPPIER is similar to a previous assay named Fluoppi[51]. While Fluoppi uses tetrameric fluorescent proteins, SPPIER is not limited to oligomeric fluorescent proteins.
Conclusion and outlook
Visualizing dynamic cell signaling in vivo is fundamental in understanding animal development and disease because dynamical features of signaling are the essence of living organisms. Cell signaling is transduced via PPIs from external cues to the appropriate intracellular effector proteins including proteases and kinases. Genetically encoded fluorescent reporters are ideal in imaging dynamic signaling in living cells, which have been demonstrated by many FRET-based reporters. For robust imaging of dynamic signaling in living animals, fluorescent reporters should achieve large dynamic range, high brightness and fast kinetics. In the past decade, many new types of fluorescent reporters have been developed based on new physical and chemical principles. Unlike most FRET-based reporters, many of these new reporters robustly visualize dynamic cell signaling during animal development. Because of large number and types of effector proteins and even larger number of PPIs, more new types of fluorescent reporters should be developed to expand the biology toolkit. These new reporters should be developed to address the specific biological needs because of various types of effector proteins (e.g. small GTPases, kinases), as well as various types of PPIs (e.g. strong, weak and transient PPIs). Tool developers and biologists should work closely for demonstration, improvement, and application of the new reporters.
ACKNOWLEDGMENTS
This work was supported by National Institute of General Medical Sciences (NIGMS) R35 GM131766, R01 GM127664, R01 GM115399 (to X.S.). We apologize for no or brief mention of several newly developed fluorescent reporters due to page limitations.
Footnotes
Conflict of interest statement
Xiaokun Shu is on two patents describing a naturally monomeric infrared fluorescent protein (mIFP), and the infrared fluorogenic protease reporter (iCasper); and on a patent application describing the phase separation-based PPI reporters.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Doupe DP, Perrimon N: Visualizing and Manipulating Temporal Signaling Dynamics with Fluorescence-Based Tools. Sci Signal 2014, 7:re1–re1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pollard TD: No Question about Exciting Questions in Cell Biology. PLoS Biology 2013, 11:e1001734–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nurse P, Hayles J: The Cell in an Era of Systems Biology. Cell 2011, 144:850–854. [DOI] [PubMed] [Google Scholar]
- 4.Feller SM: The dawn of a new era in cell signalling research. Cell Commun Signal 2010, 8:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pawson T, Warner N: Oncogenic re-wiring of cellular signaling pathways. Wiki 2007, 26:1268–1275. [DOI] [PubMed] [Google Scholar]
- 6.Taniguchi CM, Emanuelli B, Kahn CR: Critical nodes in signalling pathways: insights into insulin action. Nat Rev Mol Cell Biol 2006, 7:85–96. [DOI] [PubMed] [Google Scholar]
- 7.Pires-daSilva A, Sommer RJ: The evolution of signalling pathways in animal development. Nat Rev Genet 2003, 4:39–49. [DOI] [PubMed] [Google Scholar]
- 8.Turk B, sbreve an Turk Du, Turk V: EMBO Member’s ReviewProtease signalling: the cutting edge. EMBO J 2012, 31:1630–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lopez-Otin C, Bond JS: Proteases: Multifunctional Enzymes in Life and Disease. J Biol Chem 2008, 283:30433–30437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Puente XS, Sanchez LM, Gutiérrez-Fernández A, Velasco G, Lopez-Otin C: A genomic view of the complexity of mammalian proteolytic systems. Biochem Soc Trans 2005, 33:331–334. [DOI] [PubMed] [Google Scholar]
- 11.Eisenberg D, Jucker M: The Amyloid State of Proteins in Human Diseases. Cell 2012, 148:1188–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mason SD, Joyce JA: Proteolytic networks in cancer. Trends Cell Biol 2011, 21:228–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kessenbrock K, Plaks V, Werb Z: Matrix Metalloproteinases: Regulators of the Tumor Microenvironment. Cell 2010, 141:52–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vassar R, Kovacs DM, Yan R, Wong PC: The Beta-Secretase Enzyme BACE in Health and Alzheimer’s Disease: Regulation, Cell Biology, Function, and Therapeutic Potential. Journal of Neuroscience 2009, 29:12787–12794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.••.To T-L, Piggott BJ, Makhijani K, Yu D, Jan Y-N, Shu X: Rationally designed fluorogenic protease reporter visualizes spatiotemporal dynamics of apoptosis in vivo. Proceedings of the National Academy of Sciences 2015, 112:3338–3343. [DOI] [PMC free article] [PubMed] [Google Scholar]; The paper describes a dark-to-bright near-infrared fluorogenic protease reporter iCasper. By incorporating specific cleavage sequence of executioner caspases, iCasper reveals spatiotemporal dynamics of caspase activity and apoptosis signaling during embryonic morphogenesis in Drosophila.
- 16.To T-L, Schepis A, Ruiz-González R, Zhang Q, Yu D, Dong Z, Coughlin SR, Shu X: Rational Design of a GFP-Based Fluorogenic Caspase Reporter for Imaging Apoptosis In Vivo. Cell Chemical Biology 2016, 23:875–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.•.Zhang Q, Schepis A, Huang H, Yang J, Ma W, Torra J, Zhang S-Q, Yang L, Wu H, Nonell S, et al. : Designing a Green Fluorogenic Protease Reporter by Flipping a Beta Strand of GFP for Imaging Apoptosis in Animals. J Am Chem Soc 2019, 141:4526–4530. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study describes a green fluorogenic protease reporter FlipGFP. It achieves 100-fold fluorescence increase upon protease activation. FlipGFP has been demonstrated in imaging spatiotemporal dynamics of caspase activities in zebrafish and Drosophila.
- 18.Yu D, Baird MA, Allen JR, Howe ES, Klassen MP, Reade A, Makhijani K, Song Y, Liu S, Murthy Z, et al. : a naturally monomeric infrared fluorescent protein for protein labeling. Nat Methods 2015, 12:763–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.•.Qian Y, Piatkevich KD, Larney BM, Abdelfattah AS, Mehta S, Murdock MH, Gottschalk S, Molina RS, Zhang W, Chen Y, et al. : A genetically encoded near-infrared fluorescent calcium ion indicator. Nat Methods 2019, doi: 10.1038/s41592-018-0294-6. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors engineered a naturally monomeric near-infrared fluorescent protein mIFP into a near-infrared calcium reporter. This reporter has been demonstrated in imaging calcium signaling in brain tissue.
- 20.•.Ding Y, Li J, Enterina JR, Shen Y, Zhang I, Tewson PH, Mo GCH, Zhang J, Quinn AM, Hughes TE, et al. : Ratiometric biosensors based on dimerization-dependent fluorescent protein exchange. Nat Methods 2015, 12:195–198. [DOI] [PMC free article] [PubMed] [Google Scholar]; The paper describes a new class of fluorescent reporters based on exchange of heterodimeric partners of green and red fluorescent proteins. This technology is applied to design many reporters including a caspase reporter, which acheives 8-fold red-to-green ratio change in living cells.
- 21.Zhang J, Wang X, Cui W, Wang W, Zhang H, Liu L, Zhang Z, Li Z, Ying G, Zhang N, et al. : Visualization of caspase-3-like activity in cells using a genetically encoded fluorescent biosensor activated by protein cleavage. Nature Communications 2013, 4:2157. [DOI] [PubMed] [Google Scholar]
- 22.Nicholls SB, Chu J, Abbruzzese G, Tremblay KD, Hardy JA: Mechanism of a Genetically Encoded Dark-to-Bright Reporter for Caspase Activity. J Biol Chem 2011, 286:24977–24986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hunter T: Tyrosine phosphorylation: thirty years and counting. Curr Opin Cell Biol 2009, 21:140–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pawson T, Scott JD: Protein phosphorylation in signaling--50 years and counting. Trends Biochem Sci 2005, 30:286–290. [DOI] [PubMed] [Google Scholar]
- 25.Manning G: The Protein Kinase Complement of the Human Genome. Science 2002, 298:1912–1934. [DOI] [PubMed] [Google Scholar]
- 26.Toettcher JE, Weiner OD, Lim WA: Using Optogenetics to Interrogate the Dynamic Control of Signal Transmission by the Ras/Erk Module. Cell 2013, 155:1422–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marshall CJ: Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell 1995, 80:179–185. [DOI] [PubMed] [Google Scholar]
- 28.Sopko R, Perrimon N: Receptor Tyrosine Kinases in Drosophila Development. Cold Spring Harb Perspect Biol 2013, 5:a009050–a009050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hynes NE, Ingham PW, Lim WA, Marshall CJ, Massagué J, Pawson T: Signalling change: signal transduction through the decades. Nat Rev Mol Cell Biol 2013, 14:393–398. [DOI] [PubMed] [Google Scholar]
- 30.••.Zhang Q, Huang H, Zhang L, Wu R, Chung C-I, Zhang S-Q, Torra J, Schepis A, Coughlin SR, Kornberg TB, et al. : Visualizing Dynamics of Cell Signaling In Vivo with a Phase Separation-Based Kinase Reporter. Mol Cell 2018, 69:334–345.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]; The paper describes GFP phase separation-based kinase reporters SPARK. When kinase is inactive, SPARK is as bright as EGFP with homogenous distribution in the cell. When kinase is activated, SPARK rapidly (within 20-second) forms intensely bright puncta with 10-fold fluorescence increase at single pixel level. SPARK is reversible and has been demonstrated in visualizing oscillatory and dynamic kinase signaling in living cells and animals including Drosophila and zebrafish.
- 31.Kiyokawa E, Aoki K, Nakamura T, Matsuda M: FRET sensors summary. Annu Rev Pharmacol Toxicol 2011, 51:337–358. [DOI] [PubMed] [Google Scholar]
- 32.Piston DW, Kremers G-J: Fluorescent protein FRET: the good, the bad and the ugly. Trends Biochem Sci 2007, 32:407–414. [DOI] [PubMed] [Google Scholar]
- 33.Fairclough RH, Cantor CR: The use of singlet-singlet energy transfer to study macromolecular assemblies. Meth Enzymol 1978, 48:347–379. [DOI] [PubMed] [Google Scholar]
- 34.•.Mehta S, Zhang Y, Roth RH, Zhang J-F, Mo A, Tenner B, Huganir RL, Zhang J: Single-fluorophore biosensors for sensitive and multiplexed detection of signalling activities. Nat Cell Biol 2018, doi: 10.1038/s41556-018-0200-6. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study describes excitation ratiometric kinase reporters using cpGFP flanked by kinase-specific phosphopeptide and phosphopeptide-binding domain. These reporters achieve higher dynamic range than FRET-based reporters and have been demonstrated in imaging multiple signaling events in single cells.
- 35.Ross BL, Tenner B, Markwardt ML, Zviman A, Shi G, Kerr JP, Snell NE, McFarland JJ, Mauban JR, Ward CW, et al. : Single-color, ratiometric biosensors for detecting signaling activities in live cells. eLife 2018, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.••.Regot S, Hughey JJ, Bajar BT, Carrasco S, Covert MW: High-Sensitivity Measurements of Multiple Kinase Activitiesin Live Single Cells. Cell 2014, 157:1724–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]; The paper describes a GFP translocation-based kinase reporter KTR. Upon kinase phosphorylation, KTR shuttles between the cytoplasm and nucleus. KTR achieves higher dynamic range than FRET reporters and reveals spatiotemporal dynamics of kinase activities in individual cells.
- 37.Welch CM, Elliott H, Danuser G, Hahn KM: PERSPECTIVES. Nat Rev Mol Cell Biol 2011, 12:749–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scott JD, Pawson T: Cell Signaling in Space and Time: Where Proteins Come Together and When They’re Apart. Science 2009, 326:1220–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pawson T, Nash P: Protein-protein interactions define specificity in signal transduction. Gene Dev 2000, 14:1027–1047. [PubMed] [Google Scholar]
- 40.Alberts B: The cell as a collection of protein machines: preparing the next generation of molecular biologists. Cell 1998, 92:291–294. [DOI] [PubMed] [Google Scholar]
- 41.Cabantous S, Nguyen HB, Pédelacq J-D, Koraïchi F, Chaudhary A, Ganguly K, Lockard MA, Favre G, Terwilliger TC, Waldo GS: A New Protein-Protein Interaction Sensor Based on Tripartite Split-GFP Association. Sci. Rep 2013, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Q, Zheng Y-W, Coughlin SR, Shu X: A rapid fluorogenic GPCR-β-arrestin interaction assay. Protein Science 2018, 52:6366–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lefkowitz RJ: A Brief History of G-Protein Coupled Receptors (Nobel Lecture). Angew. Chem. Int. Ed 2013, 52:6366–6378. [DOI] [PubMed] [Google Scholar]
- 44.To T-L, Zhang Q, Shu X: Structure-guided design of a reversible fluorogenic reporter of protein-protein interactions. Protein Science 2016, 25:748–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kumagai A, Ando R, Miyatake H, Greimel P, Kobayashi T, Hirabayashi Y, Shimogori T, Miyawaki A: A Bilirubin-Inducible Fluorescent Protein from Eel Muscle. Cell 2013, 153:1602–1611. [DOI] [PubMed] [Google Scholar]
- 46.Burslem GM, Crews CM: Small-Molecule Modulation of Protein Homeostasis. Chem Rev 2017, 117:11269–11301. [DOI] [PubMed] [Google Scholar]
- 47.•.Chung C-I, Zhang Q, Shu X: Dynamic Imaging of Small Molecule Induced Protein–Protein Interactions in Living Cells with a Fluorophore Phase Transition Based Approach. Anal Chem 2018, doi: 10.1021/acs.analchem.8b03476. [DOI] [PMC free article] [PubMed] [Google Scholar]; The paper describes a GFP phase transition-based protein-protein interaction reporter named SPPIER using de novo designed homo-oligomeric coiled coils. SPPIER visualizes small molecule-induced weak and dynamic PPIs in living cells.
- 48.Krönke J, Udeshi ND, Narla A, Grauman P, Hurst SN, McConkey M, Svinkina T, Heckl D, Comer E, Li X, et al. : Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science 2014, 343:301–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu G, Middleton RE, Sun H, Naniong M, Ott CJ, Mitsiades CS, Wong K-K, Bradner JE, Kaelin WG: The myeloma drug lenalidomide promotes the cereblon-dependent destruction of Ikaros proteins. Science 2014, 343:305–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ito T, Ando H, Suzuki T, Ogura T, Hotta K, Imamura Y, Yamaguchi Y, Handa H: Identification of a primary target of thalidomide teratogenicity. Science 2010, 327:1345–1350. [DOI] [PubMed] [Google Scholar]
- 51.•.Watanabe T, Seki T, Fukano T, Sakaue-Sawano A, Karasawa S, Kubota M, Kurokawa H, Inoue K, Akatsuka J, Miyawaki A: Genetic visualization of protein interactions harnessing liquid phase transitions. Sci. Rep 2017, doi: 10.1038/srep46380. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors describe a GFP phase transition-based protein-protein interaction reporter using tetrameric fluorescent proteins.