Abstract
Genetically encoded fluorescent protein-based kinase biosensors are a central tool for illumination of the kinome. The adaptability and versatility of biosensors have allowed for spatiotemporal observation of real-time kinase activity in living cells and organisms. In this review, we highlight various types of kinase biosensors, along with their burgeoning applications to complex biological systems. Specifically, we focus on kinase activity reporters used in neuronal systems and whole animal settings. Genetically encoded kinase biosensors are key for elucidation of the spatiotemporal regulation of protein kinases, with broader applications beyond the petri dish.
1.1. Introduction
Since the kinome was first annotated in 2002 [1], our understanding of human kinases has expanded. We now know over 500 kinases encoded in the human genome phosphorylate up to one third of the proteome and mediate nearly all aspects of cellular function [2]. Proteomic strategies have provided detailed annotation of kinases and their molecular targets [3,4], and phosphorylation specific antibodies allow detection of phosphorylation events in cells and tissues. However, these approaches only allow for a “snapshot” of kinase activity, which could miss intricate spatial and temporal regulation involved in signal propagation. Kinase signaling is highly dynamic, requiring tools that can adequately interrogate complex living biological systems [3,4]. Therefore, other in situ approaches are required for probing kinome activity in living cells and organisms within real time.
One such approach are genetically encoded fluorescent protein-based biosensors, which allow for the real-time readout of kinase and phosphatase activity in a living cell or organism. Biosensors are most commonly based on an adaptive molecular “switch” which allows for one biosensor design to be used for many kinases [5,6]. Using molecular switches specific for a kinase of interest enables a single-cell view of kinase activity in complicated systems. Furthermore, biosensors lend themselves to live-cell applications which can be used to study transient and dynamic signaling events. Improvements in biosensor sensitivity leads to observations of subtle biological events which are otherwise overlooked. In this review, we briefly highlight the most recent advances in biosensor designs using fluorescent proteins, focusing on select applications for monitoring the kinome in neurobiological settings and whole animals.
1.2. Genetically encoded fluorescent protein-based biosensors
1.2.1. Design of FRET-based kinase activity reporters
Many biosensors use Förster Resonsance Energy Transfer (FRET) as a readout of kinase activation or activity, by modulating FRET that occurs between two fluorescent proteins (FRET pair) where the FRET donor has emission wavelengths overlapping the FRET acceptor excitation wavelengths. When the FRET pair are within close proximity (<10 nm), the donor nonradiatively transfers energy to the acceptor moiety through their dipole-dipole interactions, resulting in FRET [5]. Most biosensors for kinases are activity reporters (KARs), where a kinase-specific substrate domain and a phosphoamino acid binding domain (PAABD) are flanked by a FRET pair (Figure 1A). Phosphorylation of the kinase-specific substrate induces PAABD binding, resulting in a conformational change of the biosensor and change in FRET efficiency. By changing the PAABD, fluorescent proteins, and peptide linkers between sensing components, improvements can be made to increase the dynamic range and sensitivity of KARs [6–8]. FRET-based biosensors, such as those for protein kinase A (PKA) [6,8–11], protein kinase C (PKC) [12], AMP activated protein kinase (AMPK) [13–15], mechanistic target of rapamycin complex I (mTORC1) [16], protein kinase B/Akt [8,17–19], epidermal growth factor receptor (EGFR) [20–22], Src [22,23], focal adhesion kinase (FAK) [24,25], and more [26] have been invaluable tools for illuminating the kinome.
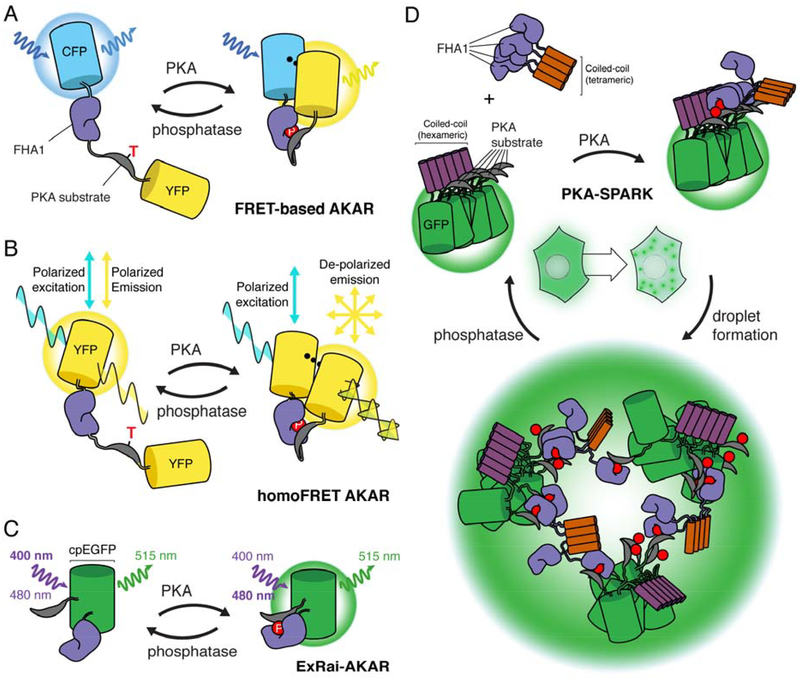
Figure 1: Design of genetically encoded kinase activity reporters for PKA (AKAR).
A, the design of FRET-based AKAR. The FRET pair cyan fluorescent protein (CFP) and yellow fluorescent protein (YFP) flank the phosphoamino acid binding domain, forkhead associated domain 1 (FHA1), and a PKA-specific substrate, with the threonine subject to phosphorylation highlighted. Upon PKA phosphorylation of the biosensor, a phosphatase-reversible conformational change is induced which alters the FRET efficiency. B, design of homoFRET AKAR. Two identical fluorescent proteins, YFP, flank the sensing component of AKAR, with polarized excitation and emission. Upon PKA phosphorylation and biosensor conformational change, YFP emission becomes de-polarized, which is used as the readout of PKA activity. C, design of excitation-ratiometric AKAR (ExRai AKAR), where circularly permutated EGFP (cpEGFP) is between the PKA substrate and FHA1 domains. PKA phosphorylation or phosphatase activity induce conformational changes in ExRai AKAR, resulting in switching of the excitation wavelength of cpEGFP from ~400 nm when the biosensor is unphosphorylated to ~480 nm when the biosensor is phosphorylated. D, design of separation of phases-based activity reporter of kinase (SPARK) for PKA (PKA-SPARK). The PKA-specific substrate is tethered to GFP and a hexameric coiled-coil domain, whereas the FHA1 domain is attached to a tetrameric coiled-coil. Upon PKA phosphorylation, FHA1 binds to the phosphorylated PKA substrate, resulting in phase-separated liquid droplets, which can be reversed by phosphatase activity.
While imaging kinase activity within single cells in vitro can be accomplished with a variety of different imaging modalities [26], imaging endogenous kinase activities within physiologically relevant conditions, such as in tissues and whole animals, is more challenging due to light scattering and phototoxicity, among other issues [27,28]. To overcome these limitations, two-photon fluorescence lifetime imaging microscopy (2p-FLIM), has become a standard technique for visualization of kinase activity [29,30]. As the donor and acceptor molecules move closer or further away from each other leading to changes in FRET, the lifetime of the donor fluorescence decreases or increases, respectively, which is used as a readout for kinase activity. Incorporation of two-photon modality into KARs allows for imaging in tissue slices or whole animal [31]. Optimization of biosensors for 2p-FLIM, through the incorporation of donors with enhanced brightness, photostability, and mono-exponential fluorescence decay [31] enables sensing of kinase activities in subcellular compartments in a native signaling state within tissues and living organisms. Overall, FRET-based approaches are the workhorse of KAR design, and adaptations to their design allow for applications beyond the petri dish.
1.2.2. Multiplexed imaging of the kinome
Kinases do not operate in a vacuum, instead they work in complex networks coordinating upstream and downstream effectors, and crosstalk from other pathways, requiring multiplexing of biosensors for measurement of more than one kinase activity in the single cell. FRET-based sensors with cyan and yellow or green and red fluorescent protein FRET pairs occupy most of the spectrum of visible light, preventing the imaging of other kinases or signaling molecules. Therefore, to observe more than one or two kinase activities within a single cell, other techniques must be deployed.
Multiplexed imaging of kinase activity is possible through modifications to traditional FRET-based KARs. For instance, the use of homotransfer FRET (homoFRET) allows for multiplexed imaging of the kinome. In homoFRET, the donor and acceptor fluorophores are spectrally identical (Figure 1B), and fluorescence polarization microscopy is used to measure changes in FRET efficiency [32]. Another approach is incorporation of near-infrared (NIR) fluorescent protein FRET pairs into KARs [33,34]. In NIR-KARs, two NIR fluorescent proteins, typically with excitation and emission wavelengths between 600–700 nm, are incorporated into a FRET-based KAR. This design allows for red-shifted NIR-KARs to be used alongside cyan-yellow KARs or other opto-genetic tools for perturbation of cellular activities [33]. Thus, FRET-based KARs can be used for multiplexed imaging of kinase activity.
1.2.3. Design of single-fluorophore-based kinase activity reporters
Single-fluorophore biosensors utilize only a single fluorescent protein, and inherently enable multiplexed imaging. In a single-fluorophore kinase biosensor, a circularly permutated fluorescent protein (cpFP) [35] is flanked by a kinase substrate peptide and PAABD. Phosphorylation by the kinase of interest results in a conformational change altering fluorescence intensity of the fluorophore. We recently used this approach to develop single-fluorophore excitation-ratiometric (ExRai) KARs for Akt, PKC, and PKA [36] based on cpEGFP [35,37]. ExRai KARs were found to have two excitation peaks at ~400 nm and ~480 nm, whereby phosphorylation of the kinase substrate domain induces a conformational change within the KAR which shifts the excitation wavelength (Figure 1C). We found ExRai KARs allow for imaging of kinase activity with greater sensitivity and dynamic range compared to FRET-based KARs [36]. Additionally, we developed intensity-based single-fluorophore KARs for PKA and PKC, where conformational changes in the biosensor induced by kinase or phosphatase activity decrease or increase fluorescence intensity of the fluorescent protein. For optimal multiplexing, we took advantage of dimerization-dependent fluorescent proteins [38] to design red single-color KARs for extracellular regulated kinase (ERK) and PKA. Using a combination of these biosensors, imaging of up to six intracellular species within a single cell is possible [36], illuminating the highly complex signaling which occurs within a single cell in response to extracellular perturbations.
1.2.4. Kinase activity as reported by phase separation
Aside from changes in fluorescence intensities or FRET signals, KARs have been made which report kinase activity via phase separation (Figure 1D). Phase separation-based KARs employ multivalent homo-oligomeric coiled coil peptides fused to either fluorescent protein-tagged kinase-specific substrates or PAABDs. Upon phosphorylation by the kinase of interest, the PAABD binds to the phosphorylated substrate. This results in reversible oligomerization of the coiled coils and phase separation as visualized by formation of fluorescent puncta throughout the cytoplasm of the cell [39]. Phase separation-based KARs for PKA and ERK, termed separation of phases-based activity reporter of kinase (SPARK), enable both in cell and in vivo imaging of real-time kinase activity [39] with the possibility to combine with other KARs for multiplexed imaging of kinase activities within a single cell or organism.
1.2.5. Kinase activation reporter design
Aside from probing kinase activity, the kinase itself can be monitored for activation, accomplished through the use of kinase activation reporters. Kinase activation reporters similarly use FRET, but instead of a kinase-specific substrate, the FRET pair flanks the kinase itself. Conformational changes in the kinase result in changes in FRET efficiency, corresponding to kinase activation or inactivation, and have been developed for Akt [40], AMPK [41], aurora kinase A [42], and calcium-calmodulin kinase II [43]. These probes enable direct measurement of kinase activation, instead of a readout of kinase activity through the use of KARs.
1.3. Visualizing the kinome in neuronal systems
1.3.1. Using FRET-based KARs to visualize neuronal PKA activity
PKA is a critical integrator of calcium signaling within neuronal circuits. Upstream of PKA are several G-protein-coupled receptors (GPCRs) and ion channels which respond to neuromodulators and synaptic activity within the brain [44,45]. The influx of calcium and regulation of adenylyl cyclase activity produces cyclic AMP (cAMP) production, which modulates PKA activity [46,47]. PKA goes on to regulate several events within the nervous system, including synaptic transmission, transcription, long-term potentiation, and learning [48]. PKA dysregulation has also been implicated in a variety of neurological disorders and appears to be critical for neurological function [49]. Therefore, several efforts have been made to monitor PKA signaling in the brain.
GPCRs are coupled to different subtypes of trimeric G proteins, such as Gs, Gi and Gq. Among these, Gs- and Gi-coupled GPCRs are known to critically regulate PKA activity within the brain, but it was unknown if Gq-coupled muscarinic receptors could activate PKA. Using a FLIM-based A Kinase Activity Reporter (FLIM-AKAR), Chen et al measured PKA activity in single neurons stimulated by Gαq-coupled muscarinic receptor agonists [50]. Endogenous Gαq-coupled muscarinic receptors were sufficient to activate PKA, and through the use of a PKA inhibitory peptide and pharmacological perturbation of other GPCRs, PKA activity was determined to be required and specific for Gαq-coupled muscarinic receptor-mediated synaptic depression. Further mechanistic explorations using FLIM-AKAR in hippocampal slices found that Gαq-coupled muscarinic receptor signaling activates PKA through either calcium or PKC, in a mechanism specific for Gαq signaling. Furthermore, pharmacologic perturbation of other endogenous Gαq-coupled receptors could activate PKA. Altogether, through the use of FLIM-AKAR and pharmacological perturbation of hippocampal slices using various Gαq agonists and antagonists, the authors revealed a novel role for endogenous Gαq-coupled receptors in the activation of PKA for synaptic depression, revising the known model of PKA signaling within the brain [50].
Using a variant of FLIM-AKAR targeted to microtubules, Ma et al were able to measure PKA activity within locomotive mice [51]. Targeted FLIM-AKAR was expressed in vivo in cortical neurons of mice and imaged using a device implanted in the barrel cortex. Comparing between measurements of resting PKA activity in awake mice and in brain slices, PKA activity was found to be enhanced in vivo. Light anesthesia using isofluorane or the β-adrenergic receptor antagonist propranolol induced reversible depression of PKA activity across all neurons imaged, indicating that basal PKA activity is a hallmark of wakefulness in cortical neurons. In mice running on a treadmill, PKA activity was increased in the motor, visual, and barrel cortexes to varying degrees, directly demonstrating PKA activity is increased during locomotion. The imaging approaches used allowed for single-neuron resolution, and the authors found that each neuron has different amplitudes for PKA activity and kinetics, demonstrating single-cell specificity during locomotion in awake mice [51]. Altogether, these reports establish the use of AKARs to study PKA activity in the brain and expand visualization of the kinome from a petri dish to awake animals.
1.3.2. Imaging PKC dynamics using PKC activation reporters
PKC was initially discovered in the brain and is enriched in both neuronal and glial cells [52]. PKC is activated by calcium and lipids to orchestrate both pre- and post-synaptic signaling through regulation of the cytoskeleton, ion channels, transporters, and GPCRs, which implicates PKC in several neurodegenerative diseases [53]. Upon interaction with calcium, PKC is recruited to the plasma membrane, encountering lipids, and begins to exert its downstream effects [52]. There are three families of PKC, conventional, novel, and atypical [54], which have isozyme-specific localizations throughout the brain [52], and allow for diversity in PKC signaling [55]. Recent work using isozyme-specific FLIM-FRET sensors for PKC has begun to unravel PKC signaling in synaptic plasticity [56].
To monitor isozyme-specific PKC activity, Colgan et al developed two PKC biosensors, to measure kinase activation and kinase activity. To measure kinase activity which occurs upon translocation to the plasma membrane, various PKC isozymes were tagged with a FRET donor. Upon translocation, tagged PKC encounters the plasma membrane-localized FRET acceptor, resulting in changes in FRET efficiency. To monitor kinase activation, the pseudosubstrate domain from a specific isoform of PKC tagged with a FRET acceptor is recruited to the plasma membrane, interacting with the activated membrane-bound PKC isozyme tagged with a FRET donor resulting in a change in FRET [56]. Using this biosensor design in mouse hippocampal neurons, compartmentalized and transient PKC activation was observed in dendritic spines during structural long-term potentiation. Activation of structural plasticity in hippocampal slices was assessed in neurons expressing isozyme-specific PKC reporters, with each classical isozyme of PKC activated within dendritic spines to differing degrees. Using neurons expressing PKC KARs from isozyme-specific knockout mice, the necessity of each PKC isozyme for structural plasticity was assessed. Only the classical PKC isozyme PKCα was required for structural plasticity. PKCα knockout mice exhibited learning deficits, indicating PKCα is necessary for learning. Pharmacological antagonists of N-methyl-D-aspartate (NDMA) receptors and tropomyosin receptor B (TrkB) applied to neurons expressing PKC biosensors reduced PKC activation, whereas glutamate receptor antagonists did not, indicating PKC activity during structural plasticity is mediated by NDMA receptor and TrkB activity [56]. This example highlights the integration of kinase activation reporters to understand kinase regulation and correlate to behavioral studies in mice.
1.4. Perspectives on visualizing the kinome
In the nearly 20 years KARs have been in use, advancement in sensing capabilities from expansion of the probable kinome [26] to use in cell and tissue culture to animals have significantly enhanced our understanding of kinases and their function. We now have the capabilities to visualize subcellular kinase activity in living systems in real time and in whole animals.
Current and future work developing KARs with high dynamic range, improved signal to noise, and low background will allow for KARs to be more useful for measuring kinase activity in whole animals. Advancements in imaging modality will also allow for an enhanced view of the kinome. For example, we have worked on combining super-resolution techniques with KAR design to image the kinome at super-resolution, which will facilitate an expanded spatiotemporal view of kinome activity [57]. Using bioluminescence in KAR design enables in vivo imaging [58–60]. NIR KARs also have in vivo applicability as red-shifted light is able to penetrate further into tissues than visible light. Translocation-based KARs, where the movement of a fluorescent protein from the cytoplasm to the nucleus and reverse is used as a readout of kinase activity, have been deployed in zebrafish [61] and could be combined with 2p technologies for deeper imaging of the kinome on a single cell scale in whole animals.
As we learn more about the kinome, we can expand the use of kinase biosensors to understudied and orphaned kinases both in cells and in vivo. The generalizable design of AKAR proved to be a gateway for kinase biosensors, with biosensors for over 45 kinases available [26]. Implementing the KAR design requires knowledge of kinase-specific phosphorylation motifs. Various experimental and computational approaches have been developed to identify these motifs [62–70], resulting in the identification of putative phosphorylation motifs for over 80 protein kinases [65]. In addition, engineering a functional kinase-dependent molecular switch requires careful selection of a PAABD to match the phosphorylated substrate [65], and new affinity reagents may be engineered on demand [71,72]. In some cases, conformational changes in native substrates can also be taken advantage of to develop KARs [16, 22]. In this respect, continuing advances in our overall understanding of the kinome will speed the development of new KARs, adding spatial and temporal dimensions to kinase networks.
Funding
This work was supported by the National Institutes of Health (NIH/NCI T32 CA009523 to D.L.S. and R01 MH111516, R35 CA197622, R01 DK073368, and R01 GM111665 to J.Z.); the University of California President’s Postdoctoral Fellowship (to D.L.S.); and the Air Force Office of Scientific Research (FA9500-18-1-0051 to J.Z.). The authors declare no competing interests.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading (*)
- 1.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S: The protein kinase complement of the human genome. Science 2002, 298:1912–1934. [DOI] [PubMed] [Google Scholar]
- 2.Ferguson FM, Gray NS: Kinase inhibitors: the road ahead. Nat Rev Drug Discov 2018, 17:353–377. [DOI] [PubMed] [Google Scholar]
- 3.Lottspeich F: Proteomics—An unexpected journey into the complexity of protein structures and functions. EuPA Open Proteomics 2018, 21:1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schubert OT, Röst HL, Collins Ben C, Rosenberger G, Aebersold R: Quantitative proteomics: challenges and opportunities in basic and applied research. Nat Protoc 2017, 12:1289–1294. [DOI] [PubMed] [Google Scholar]
- 5.Day RN, Davidson MW: Fluorescent proteins for FRET microscopy: Monitoring protein interactions in living cells. Bioessays 2012, 34:341–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang J, Hupfeld CJ, Taylor SS, Olefsky JM, Tsien RY: Insulin disrupts β-adrenergic signalling to protein kinase A in adipocytes. Nature 2005, 437:569–573. [DOI] [PubMed] [Google Scholar]
- 7.Lam AJ, St-Pierre F, Gong Y, Marshall JD, Cranfill PJ, Baird MA, McKeown MR, Wiedenmann J, Davidson MW, Schnitzer MJ, et al. : Improving FRET dynamic range with bright green and red fluorescent proteins. Nat Methods 2012, 9:1005–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Komatsu N, Aoki K, Yamada M, Yukinaga H, Fujita Y, Kamioka Y, Matsuda M: Development of an optimized backbone of FRET biosensors for kinases and GTPases. Molecular Biology of the Cell 2011, 22:4647–4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allen MD, Zhang J: Subcellular dynamics of protein kinase A activity visualized by FRET-based reporters. Biochemical and Biophysical Research Communications 2006, 348:716–721. [DOI] [PubMed] [Google Scholar]
- 10.Zhang J, Ma Y, Taylor SS, Tsien RY: Genetically encoded reporters of protein kinase A activity reveal impact of substrate tethering. Proc Natl Acad Sci USA 2001, 98:14997–15002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Depry C, Allen MD, Zhang J: Visualization of PKA activity in plasma membrane microdomains. Mol. BioSyst 2011, 7:52–58. [DOI] [PubMed] [Google Scholar]
- 12.Violin JD, Zhang J, Tsien RY, Newton AC: A genetically encoded fluorescent reporter reveals oscillatory phosphorylation by protein kinase C. J Cell Biol 2003, 161:899–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsou P, Zheng B, Hsu C-H, Sasaki AT, Cantley LC: A Fluorescent Reporter of AMPK Activity and Cellular Energy Stress. Cell Metabolism 2011, 13:476–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sample V, Ramamurthy S, Gorshkov K, Ronnett GV, Zhang J: Polarized activities of AMPK and BRSK in primary hippocampal neurons. Mol. Biol. Cell 2015, 26:1935–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *15.Konagaya Y, Terai K, Hirao Y, Takakura K, Imajo M, Kamioka Y, Sasaoka N, Kakizuka A, Sumiyama K, Asano T, et al. : A Highly Sensitive FRET Biosensor for AMPK Exhibits Heterogeneous AMPK Responses among Cells and Organs. Cell Reports 2017, 21:2628–2638. [DOI] [PubMed] [Google Scholar]; In this work, the authors measured AMPK activity in whole tissues in mice, demonstrating spatial differences in pharmacological AMPK stimulation.
- 16.Zhou X, Clister TL, Lowry PR, Seldin MM, Wong GW, Zhang J: Dynamic Visualization of mTORC1 Activity in Living Cells. Cell Reports 2015, 10:1767–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kunkel MT, Ni Q, Tsien RY, Zhang J, Newton AC: Spatio-temporal Dynamics of Protein Kinase B/Akt Signaling Revealed by a Genetically Encoded Fluorescent Reporter. J. Biol. Chem 2005, 280:5581–5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao X, Zhang J: Spatiotemporal analysis of differential Akt regulation in plasma membrane microdomains. Mol. Biol.Cell 2008, 19:4366–4373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miura H, Matsuda M, Aoki K: Development of a FRET biosensor with high specificity for Akt. Cell Struct. Funct 2014, 39:9–20. [DOI] [PubMed] [Google Scholar]
- 20.Itoh RE, Kurokawa K, Fujioka A, Sharma A, Mayer BJ, Matsuda M: A FRET-based probe for epidermal growth factor receptor bound non-covalently to a pair of synthetic amphipathic helixes. Experimental Cell Research 2005, 307:142–152. [DOI] [PubMed] [Google Scholar]
- 21.Offterdinger M, Georget V, Girod A, Bastiaens PIH: Imaging phosphorylation dynamics of the epidermal growth factor receptor. J. Biol. Chem 2004, 279:36972–36981. [DOI] [PubMed] [Google Scholar]
- 22.Ting AY, Kain KH, Klemke RL, Tsien RY: Genetically encoded fluorescent reporters of protein tyrosine kinase activities in living cells. Proc Natl Acad Sci USA 2001, 98:15003–15008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, Botvinick EL, Zhao Y, Berns MW, Usami S, Tsien RY, Chien S: Visualizing the mechanical activation of Src. Nature 2005, 434:1040–1045. [DOI] [PubMed] [Google Scholar]
- 24.Cai X, Lietha D, Ceccarelli DF, Karginov AV, Rajfur Z, Jacobson K, Hahn KM, Eck MJ, Schaller MD: Spatial and Temporal Regulation of Focal Adhesion Kinase Activity in Living Cells. Molecular and Cellular Biology 2007, 28:201–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seong J, Ouyang M, Kim T, Sun J, Wen P-C, Lu S, Zhuo Y, Llewellyn NM, Schlaepfer DD, Guan J-L, et al. : Detection of focal adhesion kinase activation at membrane microdomains by fluorescence resonance energy transfer. Nature Communications 2011, 2:406–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **26.Greenwald EC, Mehta S, Zhang J: Genetically Encoded Fluorescent Biosensors Illuminate the Spatiotemporal Regulation of Signaling Networks. Chem. Rev 2018, 118:11707–11794. [DOI] [PMC free article] [PubMed] [Google Scholar]; Comprehensive review of genetically encoded biosensor design and application, including kinase and small molecule biosensors. In compliment with this review is a database of biosensors, biosensordb.ucsd.edu
- 27.Benninger RKP, Piston DW: Two-Photon Excitation Microscopy for the Study of Living Cells and Tissues. Curr Protoc Cell Biol 2013, 59:4.11.1–4.11.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rubart M: Two-Photon Microscopy of Cells and Tissue. Circulation Research 2004, 95:1154–1166. [DOI] [PubMed] [Google Scholar]
- 29.Chen Y, Saulnier JL, Yellen G, Sabatini BL: A PKA activity sensor for quantitative analysis of endogenous GPCR signaling via 2-photon FRET-FLIM imaging. Front. Pharmacol 2014, 5:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tao W, Rubart M, Ryan J, Xiao X, Qiao C, Hato T, Davidson MW, Dunn KW, Day RN: A practical method for monitoring FRET-based biosensors in living animals using two-photon microscopy. American Journal of Physiology-Cell Physiology 2015, 309:C724–C735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yasuda R: Imaging intracellular signaling using two-photon fluorescent lifetime imaging microscopy. Cold Spring Harb Protoc 2012, 2012:1121–1128. [DOI] [PubMed] [Google Scholar]
- 32.Snell N, Rao V, Seckinger K, Liang J, Leser J, Mancini A, Rizzo M: Homotransfer FRET Reporters for Live Cell Imaging. Biosensors 2018, 8:89–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **33.Oliinyk OS, Shemetov AA, Pletnev S, Shcherbakova DM, Verkhusha VV: Smallest near-infrared fluorescent protein evolved from cyanobacteriochrome as versatile tag for spectral multiplexing. Nature Communications 2019, 10:279. [DOI] [PMC free article] [PubMed] [Google Scholar]; Here, the authors developed a small, 17 kDa NIR fluorescent protein which is utilized in a NIR FRET-based KAR for PKA and Jnk. Additionally, the authors combine their NIR FRET-based KARs with optogenetic perturbations of kinase signaling.
- 34.Chernov KG, Redchuk TA, Omelina ES, Verkhusha VV: Near-Infrared Fluorescent Proteins, Biosensors, and Optogenetic Tools Engineered from Phytochromes. Chem. Rev 2017, 117:6423–6446. [DOI] [PubMed] [Google Scholar]
- 35.Tian L, Hires SA, Mao T, Huber D, Chiappe ME, Chalasani SH, Petreanu L, Akerboom J, McKinney SA, Schreiter ER, et al. : Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat Methods 2009, 6:875–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **36.Mehta S, Zhang Y, Roth RH, Zhang J-F, Mo A, Tenner B, Huganir RL, Zhang J: Single-fluorophore biosensors for sensitive and multiplexed detection of signalling activities. Nat Cell Biol 2018, 20:1215–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]; Development and application of single-fluorophore KARs, enabling the multiplexing of kinases and small molecules within a single cell.
- 37.Akerboom J, Chen TW, Wardill TJ, Tian L, Marvin JS, Mutlu S, Calderon NC, Esposti F, Borghuis BG, Sun XR, et al. : Optimization of a GCaMP Calcium Indicator for Neural Activity Imaging. Journal of Neuroscience 2012, 32:13819–13840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alford SC, Abdelfattah AS, Ding Y, Campbell RE: A Fluorogenic Red Fluorescent Protein Heterodimer. Chemistry & Biology 2012, 19:353–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **39.Zhang Q, Huang H, Zhang L, Wu R, Chung C-I, Zhang S-Q, Torra J, Schepis A, Coughlin SR, Kornberg TB, et al. : Visualizing Dynamics of Cell Signaling In Vivo with a Phase Separation-Based Kinase Reporter. Molecular Cell 2018, 69:334–346.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]; Development and application of phase-transition KARs for PKA and Erk activity. The authors used the developed KARs to monitor PKA and Erk in flies.
- 40.Ananthanarayanan B, Fosbrink M, Rahdar M, Zhang J: Live-cell molecular analysis of Akt activation reveals roles for activation loop phosphorylation. J. Biol. Chem 2007, 282:36634–36641. [DOI] [PubMed] [Google Scholar]
- *41.Pelosse M, Cottet-Rousselle C, Bidan CM, Dupont A, Gupta K, Berger I, Schlattner U: Synthetic energy sensor AMPfret deciphers adenylate-dependent AMPK activation mechanism. Nature Communications 2019, 10:1038. [DOI] [PMC free article] [PubMed] [Google Scholar]; In this work, the authors developed a genetically encoded AMPK activation reporter used to unveil how AMPK responds to changes in cellular nucleotide concentrations.
- 42.Bertolin G, Sizaire F, Herbomel G, Reboutier D, Prigent C, Tramier M: A FRET biosensor reveals spatiotemporal activation and functions of aurora kinase A in living cells. Nature Communications 2016, 7:4111–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee S-JR, Escobedo-Lozoya Y, Szatmari EM, Yasuda R: Activation of CaMKII in single dendritic spines during long-term potentiation. Nature 2009, 458:299–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nagai T, Yoshimoto J, Kannon T, Kuroda K, Kaibuchi K: Phosphorylation Signals in Striatal Medium Spiny Neurons. Trends in Pharmacological Sciences 2016, 37:858–871. [DOI] [PubMed] [Google Scholar]
- 45.Park P, Kang H, Sanderson TM, Bortolotto ZA, Georgiou J, Zhuo M, Kaang B-K, Collingridge GL: The Role of Calcium-Permeable AMPARs in Long-Term Potentiation at Principal Neurons in the Rodent Hippocampus. Front. Synaptic Neurosci 2018, 10:92–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taylor SS, Zhang P, Steichen JM, Keshwani MM, Kornev AP: PKA: Lessons learned after twenty years. Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics 2013, 1834:1271–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taylor SS, Kim C, Vigil D, Haste NM, Yang J, Wu J, Anand GS: Dynamics of signaling by PKA. Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics 2005, 1754:25–37. [DOI] [PubMed] [Google Scholar]
- 48.Wang H, Peng R-Y: Basic roles of key molecules connected with NMDAR signaling pathway on regulating learning and memory and synaptic plasticity. Military Medical Research 2016, doi: 10.1186/s40779-016-0095-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dagda RK, Banerjee Das T: Role of protein kinase A in regulating mitochondrial function and neuronal development: implications to neurodegenerative diseases. Reviews in the Neurosciences 2015, 26:105–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **50.Chen Y, Granger AJ, Tran T, Saulnier JL, Kirkwood A, Sabatini BL: Endogenous Gαq-Coupled Neuromodulator Receptors Activate Protein Kinase A. Neuron 2017, 96:1070–1083.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors used a FLIM-based PKA KAR to uncover previously unknown PKA activation by Gαq-coupled signaling in single neurons, revising the known model of PKA regulation by GPCRs.
- 51.Ma L, Jongbloets BC, Xiong W-H, Melander JB, Qin M, Lameyer TJ, Harrison MF, Zemelman BV, Mao T, Zhong H: A Highly Sensitive A-Kinase Activity Reporter for Imaging Neuromodulatory Events in Awake Mice. Neuron 2018, 99:665–679.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Callender JA, Newton AC: Conventional protein kinase C in the brain: 40 years later. Neuronal Signaling 2017, 1:NS201600005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Newton AC: Protein kinase C: perfectly balanced. Critical Reviews in Biochemistry and Molecular Biology 2016, 0:208–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Steinberg SF: Structural Basis of Protein Kinase C Isoform Function. Physiological Reviews 2008, 88:1341–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Antal CE, Newton AC: Tuning the signalling output of protein kinase C. Biochm. Soc. Trans 2014, 42:1477–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **56.Colgan LA, Hu M, Misler JA, Parra-Bueno P, Moran CM, Leitges M, Yasuda R: PKCα integrates spatiotemporally distinct Ca2+ and autocrine BDNF signaling to facilitate synaptic plasticity. Nat Neurosci 2018, 21:1027–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]; In this work, the authors used both PKC activation reporters and KARs to determine the isozyme-specificity of PKC within hippocampal neurons.
- 57.Mo GCH, Ross B, Hertel F, Manna P, Yang X, Greenwald E, Booth C, Plummer AM, Tenner B, Chen Z, et al. : Genetically encoded biosensors for visualizing live-cell biochemical activity at super-resolution. Nat Methods 2017, 14:427–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu C, Peter M, Bouquier N, Ollendorff V, Villamil I, Liu J, Fagni L, Perroy J: REV, A BRET-Based Sensor of ERK Activity. Front. Endocrinol 2013, 4:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Goyet E, Bouquier N, Ollendorff V, Perroy J: Fast and high resolution single-cell BRET imaging. Sci Rep 2016, 6:28231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Komatsu N, Terai K, Imanishi A, Kamioka Y, Sumiyama K, Jin T, Okada Y, Nagai T, Matsuda M: A platform of BRET-FRET hybrid biosensors for optogenetics, chemical screening, and in vivo imaging. Sci Rep 2018, 8:8984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *61.Mayr V, Sturtzel C, Stadler M, Grissenberger S, Distel M: Fast Dynamic in vivo Monitoring of Erk Activity at Single Cell Resolution in DREKA Zebrafish. Front. Cell Dev. Biol 2018, 6:3446–10. [DOI] [PMC free article] [PubMed] [Google Scholar]; Here, the authors developed translocation-based reporters of Erk activity for use within whole zebrafish. Using this Erk reporter, the authors measured Erk activity during muscle cell wounding and pharmacological stimulation of Erk signaling.
- 62.Barber KW, Miller CJ, Jun JW, Lou HJ, Turk BE, Rinehart J: Kinase Substrate Profiling Using a Proteome-wide Serine-Oriented Human Peptide Library. Biochemistry 2018, 57:4717–4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hutti JE, Jarrell ET, Chang JD, Abbott DW, Storz P, Toker A, Cantley LC, Turk BE: A rapid method for determining protein kinase phosphorylation specificity. Nat Methods 2004, 1:27–29. [DOI] [PubMed] [Google Scholar]
- 64.Meyer NO, O’Donoghue AJ, Schulze-Gahmen U, Ravalin M, Moss SM, Winter MB, Knudsen GM, Craik CS: Multiplex Substrate Profiling by Mass Spectrometry for Kinases as a Method for Revealing Quantitative Substrate Motifs. Anal. Chem 2017, 89:4550–4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Obenauer JC: Scansite 2.0: proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Research 2003, 31:3635–3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schwartz D, Gygi SP: An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat Biotechnol 2005, 23:1391–1398. [DOI] [PubMed] [Google Scholar]
- 67.Chou MF, Prisic S, Lubner JM, Church GM, Husson RN, Schwartz D: Using Bacteria to Determine Protein Kinase Specificity and Predict Target Substrates. PLoS ONE 2012, 7:e52747–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Corwin T, Woodsmith J, Apelt F, Fontaine J-F, Meierhofer D, Helmuth J, Grossmann A, Andrade-Navarro MA, Ballif BA, Stelzl U: Defining Human Tyrosine Kinase Phosphorylation Networks Using Yeast as an In Vivo Model Substrate. Cell Systems 2017, 5:128–139.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hu J, Rho HS, Newman RH, Zhang J, Zhu H, Qian J: PhosphoNetworks: a database for human phosphorylation networks. Bioinformatics 2013, 30:141–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Newman RH, Zhang J, Zhu H: Toward a systems-level view of dynamic phosphorylation networks. Front. In Gen 2014, 5:263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cetin M, Evenson WE, Gross GG, Jalali-Yazdi F, Krieger D, Arnold D, Takahashi TT, Roberts RW: RasIns: Genetically Encoded Intrabodies of Activated Ras Proteins. Journal of Molecular Biology 2017, 429:562–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stoeber M, Jullié D, Lobingier BT, Laeremans T, Steyaert J, Schiller PW, Manglik A, Zastrow von M: A Genetically Encoded Biosensor Reveals Location Bias of Opioid Drug Action. Neuron 2018, 98:963–976.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]