Abstract
Structural variant (SV) differences between human genomes can cause germline and mosaic disease as well as inter-individual variation. De-regulation of accurate DNA repair and genomic surveillance mechanisms results in a large number of SVs in cancer. Analysis of the DNA sequences at SV breakpoints can help identify pathways of mutagenesis and regions of the genome that are more susceptible to rearrangement. Large-scale SV analyses have been enabled by high throughput genome-level sequencing on humans in the past decade. These studies have shed light on mechanisms and prevalence of complex genomic rearrangements. Recent advancements in both sequencing and other mapping technologies as well as calling algorithms for detection of genomic rearrangements have helped propel SV detection into population-scale studies, however some regions of the genome are still inaccessible for the majority of methods. Here, we discuss the genomic organization of simple and complex SVs, the molecular mechanisms of their formation, and various ways to detect them. We also introduce methods for characterizing SVs and their consequences on human genomes.
Keywords: Structural variant, high throughput sequencing, DNA repair, transposon, cancer, bioinformatic approaches
INTRODUCTION
The Human Genome Reference (HGR) sequence has come a long way in the almost two decades since its development (Lander et al. 2001), however the ability for this reference to convey differences common in the human population is still somewhat lacking. The current haploid nucleotide-level resolution HGR is approximately 3 billion nucleotides, and still contains unfinished gaps and repeat regions (Genome Reference Consortium Human Build 38; GRCh38/hg38). Aligning regions between any human genome and the HGR yields alterations in the individual genome. If differences between individuals span more than 50 contiguous nucleotides or base pairs (bp) the change is considered a structural variant (SV) (Audano et al. 2019; Chaisson et al. 2019; Korbel et al. 2007; Sudmant et al. 2015). The mechanisms of SV formation are diverse, but these events are largely due to errors in DNA replication (polymerase slippage events), mobilization of transposable elements, or mis-repair of DNA double strand or single-ended double strand breaks (DSBs) (Hastings et al. 2009b; Scully et al. 2019; Weckselblatt and Rudd 2015). Even though shorter rearrangements such as indels (insertions and deletions shorter than 50 bp) and single nucleotide variants (1bp mismatches; SNVs) are more prevalent in human genomes (1000 Genomes Project Consortium et al. 2010), SVs can have a larger impact on human genomes and phenotypes by the alteration of many nucleotides in a single event (Conrad et al. 2010; Iafrate et al. 2004; Lupski et al. 1992). SVs are more likely to affect coding regions, or even more than one gene (Stankiewicz and Lupski 2010).
With advancements in technology, cytogenetic techniques are used less often for ascertainment of SVs in favor of high throughput sequencing (HTS). HTS is used to call a variety of SV types, but importantly is also able to identify smaller genomic alterations and copy number neutral events (1000 Genomes Project Consortium et al. 2010; 1000 Genomes Project Consortium et al. 2015; Shendure and Ji 2008; Tattini et al. 2015). Understanding the different types of SV and their inherent complexities is critical for calling them in large-scale datasets, and knowing the limitations of a given methodology for calling different types of variants (Table 1). Even though HTS techniques can identify most SV types, they often rely on computational tools to detect SVs with respect to the HGR. This can result in false positive or false negative errors because either the reads produced are shorter than the size of the SV (short-read techniques) or the reads have a higher error rate (long-read techniques). While sequencing techniques identify the bulk of SVs present in a genome, they often do not directly call breakpoint junctions at nucleotide-level precision, especially without de novo assembly (Cameron et al. 2017; Chaisson et al. 2015a; Wala et al. 2018). Identifying precise SV breakpoints allows the inference of mechanisms involved in rearrangements (Conrad et al. 2010). In addition to the identification of SVs, further work is required for the characterization of the cellular and organismal phenotypes resulting from a genomic rearrangement as well as the precise mechanisms underlying a given SV.
Table 1.
Summary of techniques for SV detection
| TECHNIQUE | Detectable SV types | Resolution | Key features |
|---|---|---|---|
| Karyotyping | Large CNV, TRA, aneuploidies | >3 Mbp | Staining |
| FISH | CNV, TRA, CGR | 10 kbp | Fluorescent probes |
| aCGH | CNV, CGR | 5–10 kbp | hybridization |
| SNP Array | CNV, LOH, CGR | 100 bp | SNP probes |
| Illumina short-read Sequencing | Simple SVs, CGR | bp | Paired-end reads |
| WES | Simple SVs | >50 bp (generally more than 3 exons) | Protein coding region |
| 10X genomics | Simple SVs | bp | Gel-bead in Emulsion |
| PacBio SMRT Sequencing | Simple SVs, CGR | bp | CCS and CLR |
| Oxford Nanopore Sequencing | Simple SVs, CGR | bp | Ultra-long reads |
| Bionano Optical mapping | Simple SVs, CGR | >500 bp | Long contigs |
| Strand-seq | INV, SCE | bp | Template strand |
| Transcriptome Sequencing | TRA; Fusion genes | bp | RNA |
TYPES OF STRUCTURAL VARIANTS
Simple Chromosomal Rearrangements
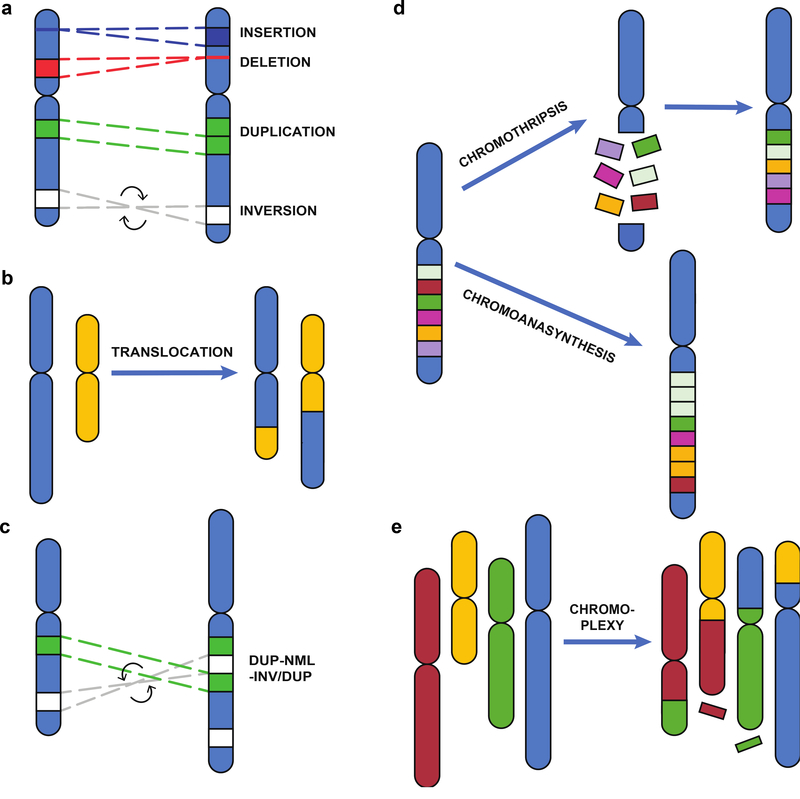
SVs are classified as either simple or complex events. Simple SVs are comprised of rearrangements with two breakpoints and often one resultant junction. Simple SVs include Deletion (DEL), Insertion (INS), Duplication (DUP), Inversion (INV; two junctions) (Fig. 1a), and Translocation (TRA; two junctions) events (Fig. 1b). A combination of two or more of these events (either of the same or different types) results in greater than two breakpoints and a complex genomic rearrangement. The mechanisms underlying both simple and complex SVs are numerous (Hastings et al. 2009b; Scully et al. 2019). These mechanisms include simple ligation reactions such as Non-Homologous End-Joining (NHEJ) (Shrivastav et al. 2008) or Microhomology Mediated End Joining (or Alt-NHEJ) (Nussenzweig and Nussenzweig 2007). Additionally, erroneous homologous recombination repair can result in Non-Allelic Homologous Recombination (NAHR) between ectopic repeats during meiosis (Liu et al. 2011b) or Single Strand Annealing (SSA), and replication-based mechanisms such as Microhomology Mediated Break Induced Replication (MMBIR) (Hastings et al. 2009a). Template switching and replication slippage (Lee et al. 2007; Sheen et al. 2007) can also lead to genomic rearrangements. Finally, transposon mobility can give rise to insertions in human genomes, and occasionally transposition is accompanied by further genomic alterations at the target site (Beck et al. 2010; Gardner et al. 2017; Gilbert et al. 2002; Kazazian et al. 1988).
Fig. 1. Classification of Structural Variants.
The diagram portrays a sample genome containing the indicated structural variant (right) compared to the reference genome (left). Simple rearrangements can a. occur on a single chromosome (insertion, deletion, duplication, and inversion) or b. can involve two chromosomes (translocation). c. Complex SVs contain multiple rearrangements that arise from a single event. Duplication-Normal-Inversion/Duplication (DUP-NML-INV/DUP) events encompass two duplicated (green and gray) regions of a chromosome. One gray fragment is inverted on the sample genome. Chromoanagenesis involves numerous rearrangements occurring in a single event. d. Chromothripsis is caused by a single shattering event followed by fragment reassembly, with loss of few fragments (maroon). Chromoanasynthesis results in multiple copy number changes and higher order genomic amplifications. e. Chromoplexy causes intra-chromosomal translocations involving multiple chromosomes, accompanied with fragment loss at few junctions (maroon and green).
Diploid human genomes have a standard copy number of two for each locus, and a loss (DEL: 0/1 copy) or gain (DUP: 3 copies, TRP: 4 copies, or higher amplification) of a genomic region is identified as a Copy Number Variant (CNV) (Conrad et al. 2010). CNVs and other SV types are determined with respect to the copy number and orientation of the HGR. Duplicated segments of the genome can be present next to the original content (tandem), or further away (interspersed). In addition to CNVs, other simple rearrangements are copy number neutral, including inversions and some translocations. When a segment of a chromosome is in the opposite orientation in comparison to the reference genome it represents an inversion (INV) in the individual. Translocations (TRA) occur when there is an exchange of genetic content between two chromosomes or distal regions within the same chromosome. Translocations can alter the genomic content (unbalanced) or result in a copy number neutral rearrangement (balanced). Robertsonian translocations are a sub-type that occurs when two acrocentric chromosomes fuse at the centromere, resulting in one fewer chromosome but maintenance of the net genomic content (Therman et al. 1989). Integration of genomic sequence into a region that was lacking this DNA in the reference is known as an insertion. Mobile elements, also known as transposons, insert pseudo-randomly across the genome (Flasch et al. 2019), leading to disruption of genes (McClintock 1950) and comprise approximately one fourth of the SVs differentially present in human genomes (Gardner et al. 2017). Mobile Element Insertion (MEI) events can cause disease (Kazazian et al. 1988), and are generally caused by a subset of these elements that are still active in human genomes (Beck et al. 2010; Brouha et al. 2003). MEIs result in both deletions (absence of an insertion or empty site) and insertions of mobile elements with respect to the HGR.
Complex Genomic Rearrangements
Although a majority of rearrangements are simple, complex genomic rearrangements (CGRs) involve several stretches of DNA from one or more chromosomes, and therefore contain two or more genomic breakpoints (Quinlan and Hall 2012; Zhang et al. 2009a). CGRs can be formed as a result of a single mutational event. They are composed of a combination of simple SVs, and are often confounding for calling algorithms because there is not enough information available in the data to differentiate complex rearrangements from series of simple rearrangements (Quinlan and Hall 2012). Moreover, without knowledge of inheritance patterns of a given SV, it is hard to distinguish simple rearrangements occurring in close proximity from a complex rearrangement (Zhang et al. 2009a).
Complex SVs can contain multiple changes in copy number state. Some of the genomic patterns of CGRs are duplication-normal-duplication (DUP-NML-DUP) (Brand et al. 2015; Gu et al. 2015), Triplication (TRP) (Liu et al. 2012), DUP–NML–INV/DUP (Fig. 1c) (Zhang et al. 2009b), and DUP-TRP/INV-DUP (Carvalho et al. 2011). These can occur through a variety of mechanisms such as FoSTeS or MMBIR (Zhang et al. 2009b) and are often accompanied by additional complexity at the junctions of the rearrangements, such as small insertions and deletions, polymerase slippage events and SNVs (Carvalho and Lupski 2016; Carvalho et al. 2013; Conrad et al. 2010). Insertional TRAs result in a copy number alteration distal from the original location of the sequence, and are often accompanied by small complexities (Gu et al. 2016; Kang et al. 2010; Neill et al. 2011).
Chromoanagenesis encompasses massive chromosomal rearrangements, including chromothripsis, chromoplexy, and chromoanasynthesis, that occur due to a single catastrophic event (Holland and Cleveland 2012; Pellestor 2019). Chromothripsis involves the shattering of a portion of the genome into multiple pieces followed by the joining of those pieces into a novel order with either no loss of genomic content (balanced chromothripsis) or loss of genomic content (unbalanced chromothripsis) (Kloosterman et al. 2011) (Fig. 1d). Chromothripsis primarily happens de novo and is more commonly observed in osteosarcoma, neuroblastoma and a few other types of cancer (Stephens et al. 2011). Chromothriptic events can involve hundreds of breakpoints confined to just one chromosome where the junctions are mated, signifying that they occur by NHEJ (Kloosterman et al. 2012). Chromoplexy is similar to chromothripsis in the sense of breakage and rearrangements of genomic segments, but involves more chromosomes and fewer breakpoints (Fig. 1e), and can occur multiple times during cancer evolution (Shen 2013). Chromoanasynthesis involves multiple copy number alterations within a single chromosome arm that are likely mediated by replication-based mechanisms (Fig. 1d) (Liu et al. 2011a).
STRUCTURAL VARIANT IDENTIFICATION
Identification of SVs has significantly evolved over the past decade, from the use of light microscopy to detect large (> 3 Mbp) rearrangements from metaphase spreads to the modern era of computational calling of SVs using genomic sequencing techniques (all ranges of SV size are detectable). Even though advancements in technologies have helped to achieve cost-effective and reliable whole genome SV analysis, it is difficult to capture all types of SVs with any one technology, especially at a reasonable cost. Moreover, the repetitive content of mammalian and plant genomes make it challenging to identify SVs. Some studies use a combination of more than one technique in predicting and validating variants. At present, chromosomal rearrangements are commonly detected by chromosomal banding, hybridization, or sequencing techniques, with the latter increasing in popularity and superseding other methodologies in recent years.
Chromosomal banding techniques
Karyotyping uses staining to form a unique banding pattern across chromosomes (Caspersson et al. 1968; Trask 2002). The intensity of dye incorporation depends on the DNA content of the chromosomal locus; giemsa dye incorporates readily in heterochromatic, A/T rich genomic regions, and less so in euchromatic, G/C rich regions. Banded, condensed chromosomes from multiple metaphase cells are then imaged using a light microscope to identify insertions, deletions, and translocations, as well as aneuploidies (O’Connor 2008). The location of an SV is noted based on the affected bands and their relative locations within the condensed chromosomes. Due to its low-resolution, karyotyping is better suited at recognizing large (>3 Mbp) chromosomal alterations, and is especially powerful at identifying copy number neutral translocation events.
Hybridization and mapping techniques
Fluorescence in situ Hybridization (FISH) uses fluorescent probes that hybridize to complementary chromosomal DNA. After hybridization, metaphase-stage cells are imaged with a microscope to quantify the number of fluorescent signals present and to identify potential mis-localization of a signal (Hu et al. 2014). Standard FISH techniques can detect rearrangements with a resolution of 100 kbp and longer, and have high accuracy and low false positive rates when compared to other techniques (Cui et al. 2016). FISH techniques can detect rearrangements detected by banding techniques but with much higher resolution. Additionally, these techniques are especially useful in identifying translocations (Pinkel et al. 1988) and sub-telomeric rearrangements (Kallioniemi et al. 1992; Linardopoulou et al. 2005). FISH has also been used to characterize chromothripsis events; multi-color FISH and multi-color banding FISH (Mackinnon and Campbell 2013) have resolutions as low as 1 kbp. A new technique, termed Cas9-mediated FISH (Deng et al. 2015) is used in conjunction with fiber-FISH (Ersfeld 2004), and is capable of marking highly repetitive regions in the genome.
Comparative Genomic Hybridization (CGH) uses hybridization of genomic DNA from two individuals (patient vs. reference or tumor vs normal) to oligonucleotides or bacterial artificial chromosomes to identify the differences (CNVs) between them. The two genomes are fragmented and labeled with different fluorescent dyes and their corresponding signal ratio is measured and normalized. Based on the ratio (log2) of the fluorescence gains and losses of the genomic content between samples can be inferred (Kallioniemi et al. 1992). Array CGH (aCGH) now commonly uses an array of oligonucleotides to carry out large-scale CGH at higher resolution (Conrad et al. 2010; Iafrate et al. 2004). With the help of computational algorithms, aCGH can be used to detect multiple CNVs in a single experiment, even examining the whole genome at an ~1 kbp resolution in a single experiment. aCGH cannot identify copy neutral SVs or loss of heterozygosity (LOH) events without additional SNP data (Wiszniewska et al. 2014).
Single Nucleotide Polymorphism arrays (SNP arrays) work in an analogous manner to aCGH, but use SNP-containing, allele-specific oligos in the hybridization array (Cooper et al. 2008). SNP arrays can also measure allele frequency, which allows the detection of certain copy neutral events like uniparental disomy or loss of heterozygosity that occurs due to consanguinity or during CNV in cancer. SNP arrays are predominantly used for genotyping, however they can be used to identify CNVs (Wang et al. 2007) and LOH (Carvalho et al. 2015) that accompany some complex SVs. SNP arrays lack the ability to identify inversions and translocations.
High-throughput optical mapping (Bionano genomics) uses fluorescent labeling of nicking restriction enzyme sites along a long stretch of DNA (300 kbp – 3 Mbp) to create optical images (maps), which are then processed to extract the read information (Teague et al. 2010). The reads are de novo assembled locally and are then compared to a reference genome to identify regions containing genomic rearrangements (Chan et al. 2018). Bionano genomics have developed a platform for high throughput optical mapping. Bionano optical mapping can identify DELs (>500 bp), INS (>500 bp), DUP (>30 kbp), INV (30 kbp) and TRA (>50 kbp), however it does not resolve breakpoints at a nucleotide level. Bionano Access software contains informatic tools for identifying and visualizing simple SVs, yet has not been extended to complex SV events.
Hybridization techniques have difficulty quantifying higher-order copy number gains (3 or 4 or 5 copies) and do not identify SV breakpoints at a base-pair level. Hybridization techniques have been used to conduct cost-effective CNV calling on hundreds of individuals, and are still effective tools for assessing CNVs. With advancements in sequencing technologies, large-scale discovery of SVs using whole genome sequencing (WGS) or HTS has become more feasible and cost effective. Thus, clinical and diagnostic settings are adapting to the use of HTS methods for SV/CNV calling.
Sequencing techniques
Advancements in HTS technologies have made it possible to identify various types of SVs across an individual genome in a single experiment. Genomic sequencing methodologies are commonly divided into either short-read or long-read techniques. Identifying SVs using HTS can involve either a comparison of sequence reads to a reference, or increasingly, a de novo assembly of a genome followed by comparison to a reference. For the former, the HTS analysis involves two-steps; the reads are first aligned or mapped to a reference genome and then based on evidence indicating the differences between the reference and the sample genome, SVs are inferred. Sequencing techniques can identify SVs with basepair-level resolution.
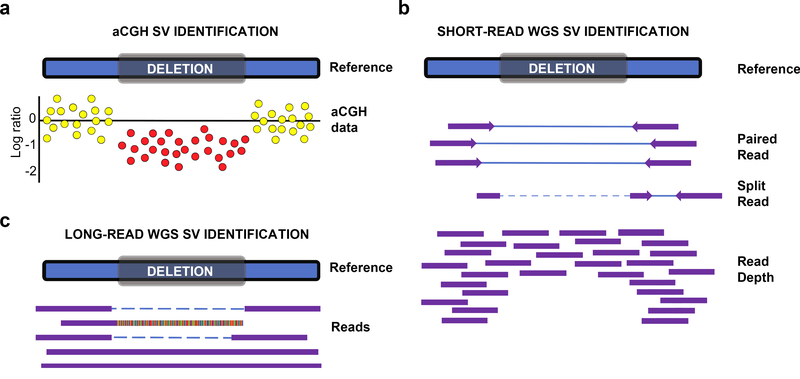
Short-read HTS generally begins with DNA is broken down into smaller fragments and size selected (library insert size). Both ends of the fragments are sequenced (paired-end) (Korbel et al. 2007). The paired-end reads are then computationally mapped against a reference. The change in insert size, along with the sequences and orientation of the reads and the depth of coverage of a locus, helps the algorithms to infer the type of SV that has occurred in the given locus. Illumina paired-end WGS with reads of 75–150 bp is the most commonly used approach for SV detection via short-read HTS. Short-read techniques primarily focus on four types of evidence for identifying genomic rearrangements; read pair based (SV present between the sequenced paired-end reads), split read based (SV present on the sequenced portion of the paired read), read depth based (number of reads mapping to a given locus in comparison to the broader genomic coverage) (Fig. 2b) and assembly based (contigs are produced from read and compared to the reference) (Guan and Sung 2016; Kosugi et al. 2019). SV calling tools use either one or a combination of the four methods mentioned above to detect SVs. Short-read techniques have the lowest error rate per basepair, however read lengths in the ~100 bp range make calling of some SVs challenging.
Fig. 2. Heterozygous deletion detection.
A depiction of deletion detection by aCGH, short-read and long-read whole genome sequencing (WGS) technologies. a. In aCGH, colored dots represent the signal ratio between the reference and sample for hybridization to a probe at that locus. Normal signals are represented with yellow dots and loss in signal is represented with red. On a log scale, copy neutral regions are indicated as 0, heterozygous deletions are −1 and homozygous deletions are −2. b. Using short-read WGS data, deletion signatures are identified using paired-end reads (read spans the deletion), split-reads (read containing the deletion; breakpoint precision), and read-depth (number of reads around the deletion locus; better suited at differentiating zygosity) information. c. In long-read WGS data, reads can span the entire deletion or can extend into the deletion and therefore represent split reads events.
Alignment tools
BWA MEM (Li 2013), Bowtie 2 (Langmead and Salzberg 2012), LAST (Kielbasa et al. 2011), Bowtie (Langmead et al. 2009)
Variant Calling tools
Pindel (Ye et al. 2018), SVABA (Wala et al. 2018), novoBreak (Chong and Chen 2018), GROM (Smith et al. 2017), MELT (Gardner et al. 2017), Manta (Chen et al. 2016), Genome STRiP (Handsaker et al. 2015), Lumpy (Layer et al. 2014), Breakdancer (Fan et al. 2014), Delly (Rausch et al. 2012), ForestSV (Michaelson and Sebat 2012), Control-FREEC (Boeva et al. 2012), CREST (Wang et al. 2011), CNVnator (Abyzov et al. 2011)
Long-read HTS is currently used largely to overcome SV detection in challenging loci, such as highly repetitive genomic regions or lengthy segmental duplications (Sedlazeck et al. 2018a). Short reads often lack the ability to span such regions. Long-read sequencing produces reads long enough to span the entire size of the majority of genomic SVs, and even enables the assembly of highly repetitive regions (Fig. 2c). Single-molecule real-time (SMRT) sequencing by Pacific Biosciences (PacBio) and Nanopore sequencing by Oxford Nanopore Technologies (ONT) are the two most commonly used long-read sequencing techniques. PacBio sequences the read with the help of fluorescence (Zero Mode Waveguides), whereas ONT uses fluctuation in current when the DNA passes through their specially designed nanopore. Long-read HTS data have error rates of ~5–15% per base. These errors are pseudorandom, however PacBio has a predilection for 1bp indels, and ONT has issues with correctly representing homopolymeric tracts of DNA. ONT and PacBio are both working towards reducing their technological error-rates, through pore modulation or through deriving consensus sequences from multiple passages around a zero-mode waveguide (Wenger et al. 2019). SV calling tools use split-reads, soft-clipped reads, clustering, and de novo assembly.
Alignment tools
NGMLR (Sedlazeck et al. 2018b), minimap2 (Li 2018), Canu (assembly) (Koren et al. 2017), BWA-MEM (Li 2013), LAST (Kielbasa et al. 2011)
Variant calling tools
Sniffles (Sedlazeck et al. 2018b), pbsv (https://github.com/PacificBiosciences/pbsv), Picky (Gong et al. 2018), SMRT-SV (Chaisson et al. 2015a)
Linked reads (10X genomics) is useful both for haplotyping genomes and for situations with limited DNA to perform genome-wide sequencing (McTaggart et al. 2018). In particular, long-read sequencing requires a substantial input DNA. With 10X sequencing, a fraction of the fragmented DNA (300 genomic equivalents, or 1 nanogram of high molecular weight DNA) gets embedded into gel beads containing unique barcodes (Gel-bead in Emulsion; GEM). This significantly reduces the chance that GEMs contain DNA from same locus. Sequencing the barcoded short DNA sequences results in a linked-read that provides enough physical coverage with lower sequence coverage and can create long, haplotype specific contigs (Zheng et al. 2016). 10X genomics can identify DEL, DUP, INV and TRA events. Long Ranger is an in-house software suite that aligns reads and calls SVs using 10X genomics data. Long Ranger uses Lariat (https://github.com/10XGenomics/lariat) for aligning linked reads.
Strand-seq, or single-cell DNA template strand sequencing, separates the two DNA strands (Watson strand and Crick strand) on each chromosome and then conducts Illumina paired-end sequencing (Sanders et al. 2017). In a single cell, DNA replication is carried out in the presence of bromodeoxyuridine (BrdU) forming two daughter cells with BrdU incorporated in one of strands. UV photolysis causes nicking at BrdU incorporated sites. During PCR amplification for library formation after UV exposure, only the strand lacking incorporated BrdU (template strand) is amplified. Resulting libraries preserve the directionality of the template strand. Several single-cell libraries, are pooled and sequenced using Illumina paired-end sequencing (Falconer et al. 2012). Strand-seq can identify sister chromatid exchange events and misoriented contigs (Falconer and Lansdorp 2013), and has also been useful in the identification of inversions (Sanders et al. 2016). Bioinformatic Analysis of Inherited Templates (BAIT) software (Hills et al. 2013) is used to bin reads based on which strand in the reference they map to. The R Packages invertR and breakpointR (Porubsky et al. 2019) use Strand-seq data to identify inversions and other genomic perturbations based on the change in strand state.
Targeted sequencing methodologies
Whole Exome Sequencing (WES) is similar to whole-genome techniques, but focuses only on protein coding (~1.5%) regions of genomic DNA. After DNA extraction, exonic sequences are enriched prior to sequencing (Ku et al. 2012). WES is better at identifying SNVs and large CNVs present within the coding regions of the genome. Transcriptome Sequencing, also known as RNA-seq, queries reverse transcribed RNA or RNA directly, and can be used for the identification of SVs in coding regions. RNA-seq for SV analysis primarily focuses on identifying fusion genes and mutations within the transcript (Wang et al. 2009). Both WES and RNA-seq are widely used in cancer research (tumor-normal comparison) and for the identification of germline, non-somatic SVs, in particular fusion gene formation (Heyer et al. 2019; Schroder et al. 2019). Other targeting techniques include panel approaches, locus specific capture techniques (Wang et al. 2015), or PCR-free CRISPR/Cas9 targeted sequencing approaches (Gabrieli et al. 2017; Hoijer et al. 2018; Tsai et al. 2017) can be used for short or long-read sequencing specific to a location or loci in the genome. These approaches have been invaluable for performing long-read HTS for clinically relevant cases (Mantere et al. 2019).
WES tools
CN-Learn (Pounraja et al. 2019), XHMM (Fromer et al. 2012), CANOES (Backenroth et al. 2014), ADTEx (Amarasinghe et al. 2014), CONTRA (Li et al. 2012), ExomeCNV (Sathirapongsasuti et al. 2011), and VarScan2 (Koboldt et al. 2012)
Transcriptome sequencing tools
SQUID (Ma et al. 2018), Arriba (Uhrig et al. 2018), and CNVkit-RNA (Talevich and Shain 2018)
De novo assembly
The reads generated using HTS are shorter than a chromosome. In order to call variants between genomes with many of the previously mentioned approaches, reads from HTS aligned to the HGR, and discrepancies between the genomes are called as variants. While reference-based techniques are primarily used to call SVs from HTS data, they are not without shortcomings (Nagarajan and Pop 2013). The current reference Genome (GRCh38) still contains gaps, unfinished centromeric and telomeric regions, and does not reflect population-level diversity. Additionally, more than half of the human genome is made up of repeats, which can cause mismapping of reads in highly repetitive regions (e.g. segmental duplications). The generation of reads that are longer than segmental duplications and that can stretch for hundreds of thousands of base pairs into centromeric regions can help overcome this limitation (Jain et al. 2018; Wenger et al. 2019). In order to build a genome without a reference sequence, the HTS data then need to be computationally assembled; this process is known as de novo assembly (Paszkiewicz and Studholme 2010; Sedlazeck et al. 2018a). De novo assembly uses overlapping sequences (Overlap-Layout-Consensus) or graph-based approaches (De Bruijn or string graph) to construct contigs (contiguous reads stitched together) from HTS based on the similarity between reads (Nagarajan and Pop 2013). The benefit of such approaches includes the resultant contigs from either unmapped or relatively low mappability reads that would not be utilized in traditional reference-based approaches (Chaisson et al. 2015b). Read length and coverage are key factors influencing the performance of de novo assembly (Nagarajan and Pop 2013). Long-read sequencing technologies are more effective for de novo assembly approaches, however they also are more expensive. De novo assembly using long-read HTS at higher depth of coverage may lead to a reference genome with few existing gaps, however additional technologies are currently needed for finishing the assembly (Bionano or 10x Genomics), and there are some chromosomal loci that may remain intractable to current methods, particularly in centromeric regions (Miga et al. 2019). Population specific variants are generally missed when a single reference is used globally (Popejoy and Fullerton 2016; Rosenfeld et al. 2012). De novo assembly can reveal novel sequences in a population that are absent from current reference genomes, and these can include genic regions (Eisfeldt et al. 2019; Seo et al. 2016; Sherman et al. 2019; Shi et al. 2016). The ability to identify so many novel regions, particularly in non-European individuals, emphasizes the need for a reference genome that considers the diversity in human species or population specific reference genome.
Alignment tools
Flye (Kolmogorov et al. 2019), Peregrine (Chin and Khalak 2019), Wtdbg2 (Ruan and Li 2019), SGVar (Tian et al. 2018), Fermikit (Li 2015), MHap (Berlin et al. 2015), MaSuRCA (Zimin et al. 2013), SOAPdenovo2 (Luo et al. 2012), Cortex (Iqbal et al. 2012)
Ensemble approaches for SV identification
Performing a variety of HTS techniques on the same genome has shown that PacBio SMRT sequencing achieves a three-fold increase in the number of SV calls when compared with Illumina paired-end sequencing. PacBio SMRT sequencing also had a higher concordance with ONT sequencing (92% outside tandem repeats and 83% inside tandem repeats). In order to improve the sensitivity and accuracy of SV identification with short-read HTS, a multi-caller approach is often used (Becker et al. 2018; English et al. 2015; Jeffares et al. 2017; Zarate et al. 2018). In a recent study detailing SV calling in three trios, the utility of multiple HTS and other genomic approaches were compared (Chaisson et al. 2019). They found that long-read data are better suited for the identification of long, intact transposon insertions, as well as SVs in segmental duplications and repetitive loci in the genome. For simple CNVs longer than 50 kbp Bionano appeared to be the best strategy. Strand-seq technology has higher sensitivity and accuracy at identifying inversions longer than 50 kbp, whereas short-read and long-read HTS are better at identifying shorter inversions.
VALIDATION AND CHARACTERIZATION OF STRUCTURAL VARIANTS
Although SV detection has significantly advanced over the last two decades, the ability to accurately define precise breakpoint junctions for SVs has lagged behind. Discerning exact SV breakpoint junctions can pave the way for inferences about the underlying mechanisms producing the rearrangement, and understanding the erroneous repair mechanisms involved in formation of the variant (Carvalho and Lupski 2016; Conrad et al. 2010). To validate a given SV, the first step involves the verification of its presence, followed by identification of the precise locations of the its breakpoint junctions. The presence of an SV can be visualized using tools such as the Integrative Genomics Viewer (IGV), which allows examination of coverage and read support (paired-reads and split reads) to infer the presence of an SV from a single or multiple HTS datasets (Robinson et al. 2011). Other genomic information (repeats, segmental duplications, gaps, genic regions) present in a given locus can also to viewed to deduce potential effects of the SV.
Traditional PCR, karyotyping, or hybridization techniques can be used to confirm the presence of SVs identified by HTS (see identification section for more on these methods). Digital PCR measures the copy number state of a locus and accurately quantifies CNVs (Hindson et al. 2011). Partitioning of the sample into tens of thousands of droplets increases detection sensitivity for rare events. Sanger sequencing is still considered a gold standard for identifying precise breakpoint junctions of the SVs, however even PCR and Sanger sequencing can be error prone, and can be difficult in some repetitive regions of the genome. Designing unique primers to some genomic loci is difficult, and nested PCR or amplification in repetitive loci can result in false positive products (Ji et al. 1994).
Performing PCR and Sanger sequencing on every SV identified in a sample is labor intensive and expensive. Using orthogonal sequencing or genomic analysis methods can reduce the number of false positive SVs in a sample (Chaisson et al. 2019). Assembly-based variant calling tools such as SVABA (Wala et al. 2018), GRIDSS (Cameron et al. 2017), and SMRT-SV (Chaisson et al. 2015a), are more precise at locating breakpoint junctions, so the future for junction identification without extensive PCR analysis is bright.
In addition to the identification of SVs, the field is moving towards understanding the impact of SVs on gene expression and on what drives SV formation in germline events and oncogenesis. To this end, a few groups have started to probe the impact of SVs on gene expression (Chiang et al. 2017) and on GWAS (Goubert et al. 2019; Payer et al. 2017). The difficulty of performing these studies lies in both inadequate SV detection, and in the number of individual genomes for whom reliable SV calling has been performed. SVs additionally can affect TAD domains, leading to phenotypes in the individuals carrying them (Lupianez et al. 2015).
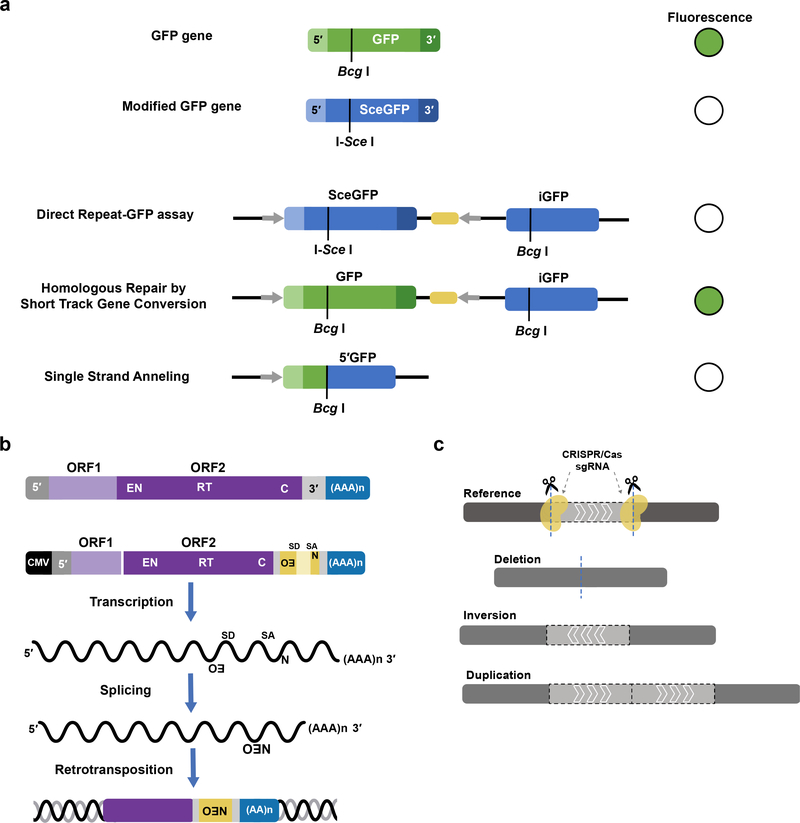
Aside from their effects on the genomes and humans in which they reside, SVs can arise via diverse mechanisms. In cultured mammalian cells, assays to detect and mechanistically interrogate SV formation have only recently been developed. The use of drug resistance cassettes was superseded in 1999 with the use of recombination-reconstituted eGFP markers (Pierce et al. 1999) (Fig. 3a), and subsequently numerous assays utilizing similar cassettes have examined the role of DNA repair in generating genomic instability (Scully et al. 2019; Willis et al. 2014). Recently, the generation of breaks followed by sequencing at the site-specific lesion has allowed thorough examination of class switching and translocation junctions (Chiarle et al. 2011; Frock et al. 2015). Furthermore, the mechanisms and prevalence of transposon mobility has been investigated extensively using both selectable marker and eGFP-based assays for retrotransposition (Fig. 3b) (Moran et al. 1996; Ostertag et al. 2000). Finally, cell culture and animal models of deletion and duplication events can lend credence to their impact on human genomes. Isogenic cell culture models with and without a given SV can be used to interrogate changes to gene expression. Mouse models of deletions and duplications can be used to establish genotype-phenotype associations with SV (Fig. 3c) (Kraft et al. 2015).
Fig. 3. Characterization of structural variants.
a. A fluorescence and puromycin-dependent DNA double strand break (DSB) assay (Pierce et al. 1999). Direct Repeat-GFP (green fluorescent protein) containing cells contain an iSceGFP (modified GFP with an iSceI cut site instead of a BcgI site) and iGFP (5′ and 3′ truncated GFP) separated by puromycin-N-acetyltransferase gene (yellow). Expression of I-SceI in SceGFP induces a DSB. Homologous repair by short track gene conversion restores GFP fluorescence, whereas repair by single strand annealing results in a truncated GFP (no fluorescence). b. LINE-1 retrotransposition assay (Moran et al. 1996). A model of a full-length LINE-1 is illustrated. To test for activity, the element is cloned into a vector with an antisense retrotransposition indicator mneoI cassette (yellow; neomycin phosphotransferase gene with a separate promoter) inserted into the 3′ UTR, and a CMV promoter (black) driving transcription of the LINE-1 a. SD: splice donor site, SA: splice acceptor site. In cell culture, when the LINE-1 element on the vector is transcribed from the 5′ end, the intron is spliced out of the neo cassette, and upon retrotransposition and insertion into the genome the cells express the neo gene, and are therefore G418 resistant. c. In vivo modelling of SV in mouse embryonic stem cells and resultant generation of mice using CRISPR/Cas. Cas9 proteins and synthetic guide RNA target the locus of interest and induce a DNA DSB. Deletion, inversion, and duplication SVs are induced upon resolution of the DSB (Kraft et al. 2015).
DISCUSSION
Advancements in HTS technologies over the past two decades have greatly improved the quantity and breadth of chromosomal rearrangements discovered. These improvements also decreased the cost and DNA input requirements for HTS methods. Long-read sequencing has several significant advantages over short-read methods, but the cost per base and the requirement of significantly larger quantities of DNA is restrictive for many research projects. In many clinical settings, having access to the amount of DNA needed might not be feasible (for instance, biopsy samples of tumors), and many of the samples are likely to consist of fragmented DNAs, leading to inadequate long-read sequencing data. As long-read sequencing moves to the fore and is performed on many more samples, examining the individual variation present in these currently “dark” regions of human genomes will be fascinating, some of the loci examined are likely to be of clinical interest. With short-read sequencing data, there are thousands and thousands of whole genome data sets available, and much still needs to be done to utilize this information more thoroughly. A comprehensive comparison of short- and long-read HTS as well as other genomic approaches allows the identification of regions that are called competently with Illumina sequencing and currently available SV calling tools, and can pinpoint loci to further refine short-read SV callers. Of particular importance is the use of ensemble approaches in conjunction with tools such as FUSOR SV (Becker et al. 2018), SURVIVOR (Jeffares et al. 2017), and BEDtools (Quinlan 2014) to help merge information from multiple callers and improve the sensitivity of SV calls on large-scale datasets. With multiple WGS technologies available, development of algorithms that utilize data from orthogonal techniques in parallel to detect SVs could be highly valuable. Synergizing SNPs with SVs to infer haplotypes, inheritance of an SV, and LOH associated with a rearrangement would be helpful for mechanistic inferences. Furthermore, refinement of split-read de novo assembly tools (Pounraja et al. 2019; Wala et al. 2018) for breakpoint analysis will also aid in understanding the underlying mechanisms behind SV formation. Finally, a few studies have started to query whether linear reference genomes without variation information hinder the progress of variant calling (Eisfeldt et al. 2019; Sherman et al. 2019). Graph genomes may be able to overcome the failure of the latest reference genome to capture the diversity of the human population (Rakocevic et al. 2019).
With the increase in accuracy and length of sequencing techniques we are on the cusp of obtaining sequences spanning human genomic gaps, centromeres, and other large repeats, and completing high quality genomes for multiple individuals. Once we obtain finished genomic maps of diverse genomes, one of the next frontiers will likely be studying the implications of variation on the genomes and phenotypes of those that harbor them; some studies have already been focusing on the phenotypic effects of more common SVs. The next two decades of genomics research are likely to pave the way for a better understanding of both the mechanisms driving human structural mutations and also their consequences.
ACKNOWLEDGEMENTS
We thank members of the Beck lab for reading and editing the review, in particular Alex V. Nesta. This work was supported in part by National Institute of General Medical Sciences grants R00GM120453 and R35GM133600 and startup funds from the University of Connecticut Health and the Jackson Laboratory to Christine R. Beck.
ABBREVATIONS
- GRCh38
Genome Reference Consortium Human Build 38
- HGR
Human Genome Reference
- HTS
High Throughput Sequencing
- WGS
Whole Genome Sequencing
- SNV
Single Nucleotide Variant (1 bp)
- InDel
Insertion and Deletion (2–49 bp)
- SV
Structural Variant (> 50 bp)
- DSB
Double Strand Break
- CNV
Copy Number Variant
- bp
Base Pair
- DEL
Deletion
- INS
Insertion
- MEI
Mobile Element Insertion
- DUP
Duplication
- TRP
Triplication
- INV
Inversion
- TRA
Translocation
- CGR
Complex Genomic Rearrangement
- LOH
Loss of Heterozygosity
- SNP
Single Nucleotide Polymorphism
- NHEJ
Non-Homologous End Joining
- NAHR
Non-Allelic Homologous Recombination
- FoSTeS
Fork Stalling and Template Switching
- MMBIR
Microhomology Mediated Break Induced Replication
- SSA
Single Strand Annealing
- FISH
Fluorescence in Situ Hybridization
- aCGH
Array Comparative Genomic Hybridization
- ONT
Oxford Nanopore Technologies
- SMRT
Single Molecule Real Time
- BrdU
Bromodeoxyuridine
- GEM
Gel-bead in Emulsion
- IGV
Integrative Genomics Viewer
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of a an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
REFERENCES
- 1000 Genomes Project Consortium et al. (2010) A map of human genome variation from population-scale sequencing Nature 467:1061–1073 doi: 10.1038/nature09534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1000 Genomes Project Consortium et al. (2015) A global reference for human genetic variation Nature 526:68–74 doi: 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abyzov A, Urban AE, Snyder M, Gerstein M (2011) CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing Genome Res 21:974–984 doi: 10.1101/gr.114876.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amarasinghe KC, Li J, Hunter SM, Ryland GL, Cowin PA, Campbell IG, Halgamuge SK (2014) Inferring copy number and genotype in tumour exome data BMC Genomics 15:732 doi: 10.1186/1471-2164-15-732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audano PA et al. (2019) Characterizing the Major Structural Variant Alleles of the Human Genome Cell 176:663–675 e619 doi: 10.1016/j.cell.2018.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backenroth D et al. (2014) CANOES: detecting rare copy number variants from whole exome sequencing data Nucleic Acids Res 42:e97 doi: 10.1093/nar/gku345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck CR et al. (2010) LINE-1 retrotransposition activity in human genomes Cell 141:1159–1170 doi: 10.1016/j.cell.2010.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker T et al. (2018) FusorSV: an algorithm for optimally combining data from multiple structural variation detection methods Genome Biol 19:38 doi: 10.1186/s13059-018-1404-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlin K, Koren S, Chin CS, Drake JP, Landolin JM, Phillippy AM (2015) Assembling large genomes with single-molecule sequencing and locality-sensitive hashing Nat Biotechnol 33:623–630 doi: 10.1038/nbt.3238 [DOI] [PubMed] [Google Scholar]
- Boeva V et al. (2012) Control-FREEC: a tool for assessing copy number and allelic content using next-generation sequencing data Bioinformatics 28:423–425 doi: 10.1093/bioinformatics/btr670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand H et al. (2015) Paired-Duplication Signatures Mark Cryptic Inversions and Other Complex Structural Variation Am J Hum Genet 97:170–176 doi: 10.1016/j.ajhg.2015.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouha B, Schustak J, Badge RM, Lutz-Prigge S, Farley AH, Moran JV, Kazazian HH Jr. (2003) Hot L1s account for the bulk of retrotransposition in the human population Proc Natl Acad Sci U S A 100:5280–5285 doi: 10.1073/pnas.0831042100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron DL et al. (2017) GRIDSS: sensitive and specific genomic rearrangement detection using positional de Bruijn graph assembly Genome Res 27:2050–2060 doi: 10.1101/gr.222109.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho CM, Lupski JR (2016) Mechanisms underlying structural variant formation in genomic disorders Nat Rev Genet 17:224–238 doi: 10.1038/nrg.2015.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho CM et al. (2013) Replicative mechanisms for CNV formation are error prone Nat Genet 45:1319–1326 doi: 10.1038/ng.2768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho CM et al. (2015) Absence of heterozygosity due to template switching during replicative rearrangements Am J Hum Genet 96:555–564 doi: 10.1016/j.ajhg.2015.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho CM et al. (2011) Inverted genomic segments and complex triplication rearrangements are mediated by inverted repeats in the human genome Nat Genet 43:1074–1081 doi: 10.1038/ng.944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspersson T et al. (1968) Chemical differentiation along metaphase chromosomes Exp Cell Res 49:219–222 doi: 10.1016/0014-4827(68)90538-7 [DOI] [PubMed] [Google Scholar]
- Chaisson MJ et al. (2015a) Resolving the complexity of the human genome using single-molecule sequencing Nature 517:608–611 doi: 10.1038/nature13907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaisson MJ, Wilson RK, Eichler EE (2015b) Genetic variation and the de novo assembly of human genomes Nat Rev Genet 16:627–640 doi: 10.1038/nrg3933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaisson MJP et al. (2019) Multi-platform discovery of haplotype-resolved structural variation in human genomes Nat Commun 10:1784 doi: 10.1038/s41467-018-08148-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S et al. (2018) Structural Variation Detection and Analysis Using Bionano Optical Mapping Methods Mol Biol 1833:193–203 doi: 10.1007/978-1-4939-8666-8_16 [DOI] [PubMed] [Google Scholar]
- Chen X et al. (2016) Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications Bioinformatics 32:1220–1222 doi: 10.1093/bioinformatics/btv710 [DOI] [PubMed] [Google Scholar]
- Chiang C et al. (2017) The impact of structural variation on human gene expression Nat Genet 49:692–699 doi: 10.1038/ng.3834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiarle R et al. (2011) Genome-wide translocation sequencing reveals mechanisms of chromosome breaks and rearrangements in B cells Cell 147:107–119 doi: 10.1016/j.cell.2011.07.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin C-S, Khalak A (2019) Human Genome Assembly in 100 Minutes bioRxiv:705616 doi: 10.1101/705616 [DOI] [Google Scholar]
- Chong Z, Chen K (2018) Structural Variant Breakpoint Detection with novoBreak Methods Mol Biol 1833:129–141 doi: 10.1007/978-1-4939-8666-8_10 [DOI] [PubMed] [Google Scholar]
- Conrad DF et al. (2010) Origins and functional impact of copy number variation in the human genome Nature 464:704–712 doi: 10.1038/nature08516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper GM, Zerr T, Kidd JM, Eichler EE, Nickerson DA (2008) Systematic assessment of copy number variant detection via genome-wide SNP genotyping Nat Genet 40:1199–1203 doi: 10.1038/ng.236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui C, Shu W, Li P (2016) Fluorescence In situ Hybridization: Cell-Based Genetic Diagnostic and Research Applications Front Cell Dev Biol 4:89 doi: 10.3389/fcell.2016.00089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Shi X, Tjian R, Lionnet T, Singer RH (2015) CASFISH: CRISPR/Cas9-mediated in situ labeling of genomic loci in fixed cells Proc Natl Acad Sci U S A 112:11870–11875 doi: 10.1073/pnas.1515692112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisfeldt J, Martensson G, Ameur A, Nilsson D, Lindstrand A (2019) Discovery of Novel Sequences in 1,000 Swedish Genomes Mol Biol Evol doi: 10.1093/molbev/msz176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- English AC et al. (2015) Assessing structural variation in a personal genome-towards a human reference diploid genome BMC Genomics 16:286 doi: 10.1186/s12864-015-1479-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ersfeld K (2004) Fiber-FISH: fluorescence in situ hybridization on stretched DNA Methods Mol Biol 270:395–402 doi: 10.1385/1-59259-793-9:395 [DOI] [PubMed] [Google Scholar]
- Falconer E et al. (2012) DNA template strand sequencing of single-cells maps genomic rearrangements at high resolution Nat Methods 9:1107–1112 doi: 10.1038/nmeth.2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconer E, Lansdorp PM (2013) Strand-seq: a unifying tool for studies of chromosome segregation Semin Cell Dev Biol 24:643–652 doi: 10.1016/j.semcdb.2013.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X, Abbott TE, Larson D, Chen K (2014) BreakDancer: Identification of Genomic Structural Variation from Paired-End Read Mapping Curr Protoc Bioinformatics 45:15 16 11–11 doi: 10.1002/0471250953.bi1506s45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flasch DA et al. (2019) Genome-wide de novo L1 Retrotransposition Connects Endonuclease Activity with Replication Cell 177:837–851 e828 doi: 10.1016/j.cell.2019.02.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frock RL, Hu J, Meyers RM, Ho YJ, Kii E, Alt FW (2015) Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases Nat Biotechnol 33:179–186 doi: 10.1038/nbt.3101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromer M et al. (2012) Discovery and statistical genotyping of copy-number variation from whole-exome sequencing depth Am J Hum Genet 91:597–607 doi: 10.1016/j.ajhg.2012.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabrieli T, Sharim H, Michaeli Y, Ebenstein Y (2017) Cas9-Assisted Targeting of CHromosome segments (CATCH) for targeted nanopore sequencing and optical genome mapping bioRxiv:110163 doi: 10.1101/110163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner EJ et al. (2017) The Mobile Element Locator Tool (MELT): population-scale mobile element discovery and biology Genome Res 27:1916–1929 doi: 10.1101/gr.218032.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert N, Lutz-Prigge S, Moran JV (2002) Genomic deletions created upon LINE-1 retrotransposition Cell 110:315–325 doi: 10.1016/s0092-8674(02)00828-0 [DOI] [PubMed] [Google Scholar]
- Gong L et al. (2018) Picky comprehensively detects high-resolution structural variants in nanopore long reads Nat Methods 15:455–460 doi: 10.1038/s41592-018-0002-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goubert C, Zevallos NA, Feschotte C (2019) Contribution of unfixed transposable element insertions to human regulatory variation bioRxiv:792937 doi: 10.1101/792937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu S et al. (2016) Mechanisms for Complex Chromosomal Insertions PLoS Genet 12:e1006446 doi: 10.1371/journal.pgen.1006446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu S et al. (2015) Alu-mediated diverse and complex pathogenic copy-number variants within human chromosome 17 at p13.3 Hum Mol Genet 24:4061–4077 doi: 10.1093/hmg/ddv146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan P, Sung WK (2016) Structural variation detection using next-generation sequencing data: A comparative technical review Methods 102:36–49 doi: 10.1016/j.ymeth.2016.01.020 [DOI] [PubMed] [Google Scholar]
- Handsaker RE, Van Doren V, Berman JR, Genovese G, Kashin S, Boettger LM, McCarroll SA (2015) Large multiallelic copy number variations in humans Nat Genet 47:296–303 doi: 10.1038/ng.3200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Ira G, Lupski JR (2009a) A microhomology-mediated break-induced replication model for the origin of human copy number variation PLoS Genet 5:e1000327 doi: 10.1371/journal.pgen.1000327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Lupski JR, Rosenberg SM, Ira G (2009b) Mechanisms of change in gene copy number Nat Rev Genet 10:551–564 doi: 10.1038/nrg2593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer EE et al. (2019) Diagnosis of fusion genes using targeted RNA sequencing Nat Commun 10:1388 doi: 10.1038/s41467-019-09374-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hills M, O’Neill K, Falconer E, Brinkman R, Lansdorp PM (2013) BAIT: Organizing genomes and mapping rearrangements in single cells Genome Med 5:82 doi: 10.1186/gm486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindson BJ et al. (2011) High-throughput droplet digital PCR system for absolute quantitation of DNA copy number Anal Chem 83:8604–8610 doi: 10.1021/ac202028g [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoijer I et al. (2018) Detailed analysis of HTT repeat elements in human blood using targeted amplification-free long-read sequencing Hum Mutat 39:1262–1272 doi: 10.1002/humu.23580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland AJ, Cleveland DW (2012) Chromoanagenesis and cancer: mechanisms and consequences of localized, complex chromosomal rearrangements Nat Med 18:1630–1638 doi: 10.1038/nm.2988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L et al. (2014) Fluorescence in situ hybridization (FISH): an increasingly demanded tool for biomarker research and personalized medicine Biomark Res 2:3 doi: 10.1186/2050-7771-2-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iafrate AJ et al. (2004) Detection of large-scale variation in the human genome Nat Genet 36:949–951 doi: 10.1038/ng1416 [DOI] [PubMed] [Google Scholar]
- Iqbal Z, Caccamo M, Turner I, Flicek P, McVean G (2012) De novo assembly and genotyping of variants using colored de Bruijn graphs Nat Genet 44:226–232 doi: 10.1038/ng.1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M et al. (2018) Linear assembly of a human centromere on the Y chromosome Nat Biotechnol 36:321–323 doi: 10.1038/nbt.4109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffares DC et al. (2017) Transient structural variations have strong effects on quantitative traits and reproductive isolation in fission yeast Nat Commun 8:14061 doi: 10.1038/ncomms14061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji W, Zhang XY, Warshamana GS, Qu GZ, Ehrlich M (1994) Effect of internal direct and inverted Alu repeat sequences on PCR PCR Methods Appl 4:109–116 [DOI] [PubMed] [Google Scholar]
- Kallioniemi A, Kallioniemi OP, Sudar D, Rutovitz D, Gray JW, Waldman F, Pinkel D (1992) Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors Science 258:818–821 doi: 10.1126/science.1359641 [DOI] [PubMed] [Google Scholar]
- Kang SH et al. (2010) Insertional translocation detected using FISH confirmation of array-comparative genomic hybridization (aCGH) results Am J Med Genet A 152A:1111–1126 doi: 10.1002/ajmg.a.33278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian HH Jr., Wong C, Youssoufian H, Scott AF, Phillips DG, Antonarakis SE (1988) Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man Nature 332:164–166 doi: 10.1038/332164a0 [DOI] [PubMed] [Google Scholar]
- Kielbasa SM, Wan R, Sato K, Horton P, Frith MC (2011) Adaptive seeds tame genomic sequence comparison Genome Res 21:487–493 doi: 10.1101/gr.113985.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman WP et al. (2011) Chromothripsis as a mechanism driving complex de novo structural rearrangements in the germline Hum Mol Genet 20:1916–1924 doi: 10.1093/hmg/ddr073 [DOI] [PubMed] [Google Scholar]
- Kloosterman WP et al. (2012) Constitutional chromothripsis rearrangements involve clustered double-stranded DNA breaks and nonhomologous repair mechanisms Cell Rep 1:648–655 doi: 10.1016/j.celrep.2012.05.009 [DOI] [PubMed] [Google Scholar]
- Koboldt DC et al. (2012) VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing Genome Res 22:568–576 doi: 10.1101/gr.129684.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolmogorov M, Yuan J, Lin Y, Pevzner PA (2019) Assembly of long, error-prone reads using repeat graphs Nat Biotechnol 37:540–546 doi: 10.1038/s41587-019-0072-8 [DOI] [PubMed] [Google Scholar]
- Korbel JO et al. (2007) Paired-end mapping reveals extensive structural variation in the human genome Science 318:420–426 doi: 10.1126/science.1149504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM (2017) Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation Genome Res 27:722–736 doi: 10.1101/gr.215087.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S, Momozawa Y, Liu X, Terao C, Kubo M, Kamatani Y (2019) Comprehensive evaluation of structural variation detection algorithms for whole genome sequencing Genome Biol 20:117 doi: 10.1186/s13059-019-1720-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft K et al. (2015) Deletions, Inversions, Duplications: Engineering of Structural Variants using CRISPR/Cas in Mice Cell Rep 10:833–839 doi: 10.1016/j.celrep.2015.01.016 [DOI] [PubMed] [Google Scholar]
- Ku CS et al. (2012) Exome versus transcriptome sequencing in identifying coding region variants Expert Rev Mol Diagn 12:241–251 doi: 10.1586/erm.12.10 [DOI] [PubMed] [Google Scholar]
- Lander ES et al. (2001) Initial sequencing and analysis of the human genome Nature 409:860–921 doi: 10.1038/35057062 [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2 Nat Methods 9:357–359 doi: 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome Genome Biol 10:R25 doi: 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layer RM, Chiang C, Quinlan AR, Hall IM (2014) LUMPY: a probabilistic framework for structural variant discovery Genome Biol 15:R84 doi: 10.1186/gb-2014-15-6-r84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JA, Carvalho CM, Lupski JR (2007) A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders Cell 131:1235–1247 doi: 10.1016/j.cell.2007.11.037 [DOI] [PubMed] [Google Scholar]
- Li H (2013) Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv e-prints [Google Scholar]
- Li H (2015) FermiKit: assembly-based variant calling for Illumina resequencing data Bioinformatics 31:3694–3696 doi: 10.1093/bioinformatics/btv440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H (2018) Minimap2: pairwise alignment for nucleotide sequences Bioinformatics 34:3094–3100 doi: 10.1093/bioinformatics/bty191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J et al. (2012) CONTRA: copy number analysis for targeted resequencing Bioinformatics 28:1307–1313 doi: 10.1093/bioinformatics/bts146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linardopoulou EV, Williams EM, Fan Y, Friedman C, Young JM, Trask BJ (2005) Human subtelomeres are hot spots of interchromosomal recombination and segmental duplication Nature 437:94–100 doi: 10.1038/nature04029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Carvalho CM, Hastings PJ, Lupski JR (2012) Mechanisms for recurrent and complex human genomic rearrangements Curr Opin Genet Dev 22:211–220 doi: 10.1016/j.gde.2012.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P et al. (2011a) Chromosome catastrophes involve replication mechanisms generating complex genomic rearrangements Cell 146:889–903 doi: 10.1016/j.cell.2011.07.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Lacaria M, Zhang F, Withers M, Hastings PJ, Lupski JR (2011b) Frequency of nonallelic homologous recombination is correlated with length of homology: evidence that ectopic synapsis precedes ectopic crossing-over Am J Hum Genet 89:580–588 doi: 10.1016/j.ajhg.2011.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R et al. (2012) SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler Gigascience 1:18 doi: 10.1186/2047-217X-1-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupianez DG et al. (2015) Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions Cell 161:1012–1025 doi: 10.1016/j.cell.2015.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski JR et al. (1992) Gene dosage is a mechanism for Charcot-Marie-Tooth disease type 1A Nat Genet 1:29–33 doi: 10.1038/ng0492-29 [DOI] [PubMed] [Google Scholar]
- Ma C, Shao M, Kingsford C (2018) SQUID: transcriptomic structural variation detection from RNA-seq Genome Biol 19:52 doi: 10.1186/s13059-018-1421-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackinnon RN, Campbell LJ (2013) Chromothripsis under the microscope: a cytogenetic perspective of two cases of AML with catastrophic chromosome rearrangement Cancer Genet 206:238–251 doi: 10.1016/j.cancergen.2013.05.021 [DOI] [PubMed] [Google Scholar]
- Mantere T, Kersten S, Hoischen A (2019) Long-Read Sequencing Emerging in Medical Genetics Front Genet 10:426 doi: 10.3389/fgene.2019.00426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B (1950) The origin and behavior of mutable loci in maize Proc Natl Acad Sci U S A 36:344–355 doi: 10.1073/pnas.36.6.344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McTaggart AR et al. (2018) Chromium sequencing: the doors open for genomics of obligate plant pathogens Biotechniques 65:253–257 doi: 10.2144/btn-2018-0019 [DOI] [PubMed] [Google Scholar]
- Michaelson JJ, Sebat J (2012) forestSV: structural variant discovery through statistical learning Nat Methods 9:819–821 doi: 10.1038/nmeth.2085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miga KH et al. (2019) Telomere-to-telomere assembly of a complete human X chromosome bioRxiv:735928 doi: 10.1101/735928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH Jr. (1996) High frequency retrotransposition in cultured mammalian cells Cell 87:917–927 doi: 10.1016/s0092-8674(00)81998-4 [DOI] [PubMed] [Google Scholar]
- Nagarajan N, Pop M (2013) Sequence assembly demystified Nat Rev Genet 14:157–167 doi: 10.1038/nrg3367 [DOI] [PubMed] [Google Scholar]
- Neill NJ et al. (2011) Recurrence, submicroscopic complexity, and potential clinical relevance of copy gains detected by array CGH that are shown to be unbalanced insertions by FISH Genome Res 21:535–544 doi: 10.1101/gr.114579.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussenzweig A, Nussenzweig MC (2007) A backup DNA repair pathway moves to the forefront Cell 131:223–225 doi: 10.1016/j.cell.2007.10.005 [DOI] [PubMed] [Google Scholar]
- O’Connor C (2008) Karyotyping for Chromosomal Abnormalities. Nature Education 1(1):27 [Google Scholar]
- Ostertag EM, Prak ET, DeBerardinis RJ, Moran JV, Kazazian HH Jr. (2000) Determination of L1 retrotransposition kinetics in cultured cells Nucleic Acids Res 28:1418–1423 doi: 10.1093/nar/28.6.1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paszkiewicz K, Studholme DJ (2010) De novo assembly of short sequence reads Brief Bioinform 11:457–472 doi: 10.1093/bib/bbq020 [DOI] [PubMed] [Google Scholar]
- Payer LM et al. (2017) Structural variants caused by Alu insertions are associated with risks for many human diseases Proc Natl Acad Sci U S A 114:E3984–E3992 doi: 10.1073/pnas.1704117114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellestor F (2019) Chromoanagenesis: cataclysms behind complex chromosomal rearrangements Mol Cytogenet 12:6 doi: 10.1186/s13039-019-0415-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce AJ, Johnson RD, Thompson LH, Jasin M (1999) XRCC3 promotes homology-directed repair of DNA damage in mammalian cells Genes Dev 13:2633–2638 doi: 10.1101/gad.13.20.2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkel D, Landegent J, Collins C, Fuscoe J, Segraves R, Lucas J, Gray J (1988) Fluorescence in situ hybridization with human chromosome-specific libraries: detection of trisomy 21 and translocations of chromosome 4 Proc Natl Acad Sci U S A 85:9138–9142 doi: 10.1073/pnas.85.23.9138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popejoy AB, Fullerton SM (2016) Genomics is failing on diversity Nature 538:161–164 doi: 10.1038/538161a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porubsky D, Sanders AD, Taudt A, Colome-Tatche M, Lansdorp PM, Guryev V (2019) breakpointR: an R/Bioconductor package to localize strand state changes in Strand-seq data Bioinformatics doi: 10.1093/bioinformatics/btz681 [DOI] [PubMed] [Google Scholar]
- Pounraja VK, Jayakar G, Jensen M, Kelkar N, Girirajan S (2019) A machine-learning approach for accurate detection of copy number variants from exome sequencing Genome Res 29:1134–1143 doi: 10.1101/gr.245928.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR (2014) BEDTools: The Swiss-Army Tool for Genome Feature Analysis Curr Protoc Bioinformatics 47:11 12 11–34 doi: 10.1002/0471250953.bi1112s47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM (2012) Characterizing complex structural variation in germline and somatic genomes Trends Genet 28:43–53 doi: 10.1016/j.tig.2011.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakocevic G et al. (2019) Fast and accurate genomic analyses using genome graphs Nat Genet 51:354–362 doi: 10.1038/s41588-018-0316-4 [DOI] [PubMed] [Google Scholar]
- Rausch T, Zichner T, Schlattl A, Stutz AM, Benes V, Korbel JO (2012) DELLY: structural variant discovery by integrated paired-end and split-read analysis Bioinformatics 28:i333–i339 doi: 10.1093/bioinformatics/bts378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP (2011) Integrative genomics viewer Nat Biotechnol 29:24–26 doi: 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld JA, Mason CE, Smith TM (2012) Limitations of the human reference genome for personalized genomics PLoS One 7:e40294 doi: 10.1371/journal.pone.0040294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan J, Li H (2019) Fast and accurate long-read assembly with wtdbg2 bioRxiv:530972 doi: 10.1101/530972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders AD, Falconer E, Hills M, Spierings DCJ, Lansdorp PM (2017) Single-cell template strand sequencing by Strand-seq enables the characterization of individual homologs Nat Protoc 12:1151–1176 doi: 10.1038/nprot.2017.029 [DOI] [PubMed] [Google Scholar]
- Sanders AD, Hills M, Porubsky D, Guryev V, Falconer E, Lansdorp PM (2016) Characterizing polymorphic inversions in human genomes by single-cell sequencing Genome Res 26:1575–1587 doi: 10.1101/gr.201160.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathirapongsasuti JF et al. (2011) Exome sequencing-based copy-number variation and loss of heterozygosity detection: ExomeCNV Bioinformatics 27:2648–2654 doi: 10.1093/bioinformatics/btr462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder J, Kumar A, Wong SQ (2019) Overview of Fusion Detection Strategies Using Next-Generation Sequencing Methods Mol Biol 1908:125–138 doi: 10.1007/978-1-4939-9004-7_9 [DOI] [PubMed] [Google Scholar]
- Scully R, Panday A, Elango R, Willis NA (2019) DNA double-strand break repair-pathway choice in somatic mammalian cells Nat Rev Mol Cell Biol doi: 10.1038/s41580-019-0152-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedlazeck FJ, Lee H, Darby CA, Schatz MC (2018a) Piercing the dark matter: bioinformatics of long-range sequencing and mapping Nat Rev Genet 19:329–346 doi: 10.1038/s41576-018-0003-4 [DOI] [PubMed] [Google Scholar]
- Sedlazeck FJ, Rescheneder P, Smolka M, Fang H, Nattestad M, von Haeseler A, Schatz MC (2018b) Accurate detection of complex structural variations using single-molecule sequencing Nat Methods 15:461–468 doi: 10.1038/s41592-018-0001-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo JS et al. (2016) De novo assembly and phasing of a Korean human genome Nature 538:243–247 doi: 10.1038/nature20098 [DOI] [PubMed] [Google Scholar]
- Sheen CR et al. (2007) Double complex mutations involving F8 and FUNDC2 caused by distinct break-induced replication Hum Mutat 28:1198–1206 doi: 10.1002/humu.20591 [DOI] [PubMed] [Google Scholar]
- Shen MM (2013) Chromoplexy: a new category of complex rearrangements in the cancer genome Cancer Cell 23:567–569 doi: 10.1016/j.ccr.2013.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shendure J, Ji H (2008) Next-generation DNA sequencing Nat Biotechnol 26:1135–1145 doi: 10.1038/nbt1486 [DOI] [PubMed] [Google Scholar]
- Sherman RM et al. (2019) Assembly of a pan-genome from deep sequencing of 910 humans of African descent Nat Genet 51:30–35 doi: 10.1038/s41588-018-0273-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L et al. (2016) Long-read sequencing and de novo assembly of a Chinese genome Nat Commun 7:12065 doi: 10.1038/ncomms12065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrivastav M, De Haro LP, Nickoloff JA (2008) Regulation of DNA double-strand break repair pathway choice Cell Res 18:134–147 doi: 10.1038/cr.2007.111 [DOI] [PubMed] [Google Scholar]
- Smith SD, Kawash JK, Grigoriev A (2017) Lightning-fast genome variant detection with GROM Gigascience 6:1–7 doi: 10.1093/gigascience/gix091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankiewicz P, Lupski JR (2010) Structural variation in the human genome and its role in disease Annu Rev Med 61:437–455 doi: 10.1146/annurev-med-100708-204735 [DOI] [PubMed] [Google Scholar]
- Stephens PJ et al. (2011) Massive genomic rearrangement acquired in a single catastrophic event during cancer development Cell 144:27–40 doi: 10.1016/j.cell.2010.11.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudmant PH et al. (2015) An integrated map of structural variation in 2,504 human genomes Nature 526:75–81 doi: 10.1038/nature15394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talevich E, Shain AH (2018) CNVkit-RNA: Copy number inference from RNA-Sequencing data bioRxiv:408534 doi: 10.1101/408534 [DOI] [Google Scholar]
- Tattini L, D’Aurizio R, Magi A (2015) Detection of Genomic Structural Variants from Next-Generation Sequencing Data Front Bioeng Biotechnol 3:92 doi: 10.3389/fbioe.2015.00092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teague B et al. (2010) High-resolution human genome structure by single-molecule analysis Proc Natl Acad Sci U S A 107:10848–10853 doi: 10.1073/pnas.0914638107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therman E, Susman B, Denniston C (1989) The nonrandom participation of human acrocentric chromosomes in Robertsonian translocations Ann Hum Genet 53:49–65 doi: 10.1111/j.1469-1809.1989.tb01121.x [DOI] [PubMed] [Google Scholar]
- Tian S, Yan H, Klee EW, Kalmbach M, Slager SL (2018) Comparative analysis of de novo assemblers for variation discovery in personal genomes Brief Bioinform 19:893–904 doi: 10.1093/bib/bbx037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trask BJ (2002) Human cytogenetics: 46 chromosomes, 46 years and counting Nat Rev Genet 3:769–778 doi: 10.1038/nrg905 [DOI] [PubMed] [Google Scholar]
- Tsai Y-C et al. (2017) Amplification-free, CRISPR-Cas9 Targeted Enrichment and SMRT Sequencing of Repeat-Expansion Disease Causative Genomic Regions bioRxiv:203919 doi: 10.1101/203919 [DOI] [Google Scholar]
- Uhrig S, Fröhlich M, Hutter B, Brors B (2018) PO-400 Arriba – fast and accurate gene fusion detection from RNA-seq data ESMO Open 3:A179–A179 doi: 10.1136/esmoopen-2018-EACR25.426 [DOI] [Google Scholar]
- Wala JA et al. (2018) SvABA: genome-wide detection of structural variants and indels by local assembly Genome Res 28:581–591 doi: 10.1101/gr.221028.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J et al. (2011) CREST maps somatic structural variation in cancer genomes with base-pair resolution Nat Methods 8:652–654 doi: 10.1038/nmeth.1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K et al. (2007) PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data Genome Res 17:1665–1674 doi: 10.1101/gr.6861907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M et al. (2015) PacBio-LITS: a large-insert targeted sequencing method for characterization of human disease-associated chromosomal structural variations BMC Genomics 16:214 doi: 10.1186/s12864-015-1370-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M (2009) RNA-Seq: a revolutionary tool for transcriptomics Nat Rev Genet 10:57–63 doi: 10.1038/nrg2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weckselblatt B, Rudd MK (2015) Human Structural Variation: Mechanisms of Chromosome Rearrangements Trends Genet 31:587–599 doi: 10.1016/j.tig.2015.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenger AM et al. (2019) Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome Nat Biotechnol 37:1155–1162 doi: 10.1038/s41587-019-0217-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis NA, Chandramouly G, Huang B, Kwok A, Follonier C, Deng C, Scully R (2014) BRCA1 controls homologous recombination at Tus/Ter-stalled mammalian replication forks Nature 510:556–559 doi: 10.1038/nature13295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiszniewska J et al. (2014) Combined array CGH plus SNP genome analyses in a single assay for optimized clinical testing Eur J Hum Genet 22:79–87 doi: 10.1038/ejhg.2013.77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye K, Guo L, Yang X, Lamijer EW, Raine K, Ning Z (2018) Split-Read Indel and Structural Variant Calling Using PINDEL Methods Mol Biol 1833:95–105 doi: 10.1007/978-1-4939-8666-8_7 [DOI] [PubMed] [Google Scholar]
- Zarate S et al. (2018) Parliament2: Fast Structural Variant Calling Using Optimized Combinations of Callers bioRxiv:424267 doi: 10.1101/424267 [DOI] [Google Scholar]
- Zhang F, Carvalho CM, Lupski JR (2009a) Complex human chromosomal and genomic rearrangements Trends Genet 25:298–307 doi: 10.1016/j.tig.2009.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Khajavi M, Connolly AM, Towne CF, Batish SD, Lupski JR (2009b) The DNA replication FoSTeS/MMBIR mechanism can generate genomic, genic and exonic complex rearrangements in humans Nat Genet 41:849–853 doi: 10.1038/ng.399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng GX et al. (2016) Haplotyping germline and cancer genomes with high-throughput linked-read sequencing Nat Biotechnol 34:303–311 doi: 10.1038/nbt.3432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimin AV, Marcais G, Puiu D, Roberts M, Salzberg SL, Yorke JA (2013) The MaSuRCA genome assembler Bioinformatics 29:2669–2677 doi: 10.1093/bioinformatics/btt476 [DOI] [PMC free article] [PubMed] [Google Scholar]