Abstract
Chemokines and their receptors have been reported to drive immune cells into tumours or to be directly involved in the promotion or inhibition of the development of tumours. However, their expression in regional lymph node (LN) tissues in melanoma patients remains unknown. The present study investigated the relationship between the expression of mRNA of chemokines and their receptors and clinicopathology of the regional LN tissues of skin cutaneous melanoma (SKCM) patients available in The Cancer Genome Atlas. The relationship between chemokines and their receptors and the composition of immune cells within the tumour was analysed. In SKCM regional LN tissues, the high expression of 32 types of chemokines and receptors, namely CCL2, 4‐5, 7‐8, 13, 22‐25, CCR1‐9, CXCL9‐13, 16, CXCR3, 5, 6, XCL1‐2 and XCR1 in LN was associated with favourable patient prognosis. Conversely, high expression of CXCL17 was an indicator of poor prognosis. The expression of mRNA for CXCL9‐11, 13, CXCR3, 6, CCL2, 4, 5, 7, 8, 25, CCR1, 2, 5, and XCL1, 2 in regional LN tissues was positively correlated with the fraction of CD8‐positive T cells and M1 macrophages, and was negatively correlated with M0 macrophages. CCR4, 6‐9, CCL13, 22, 23 and XCR1 were positively correlated with the fraction of memory B cells and naive T cells, and negatively correlated with M0 macrophages and resting mast cells, suggesting that chemokines and their receptors may affect the prognosis of patients by guiding immune cells into the tumour microenvironment to eliminate tumour cells.
Keywords: CC, chemokine receptors, chemokines, CX3C, CXC, tumour immunity, XC
1. INTRODUCTION
In many malignancies, including skin cutaneous melanoma (SKCM), enhanced infiltration of the tumour by an immune cell is typically associated with good prognosis.1, 2 Tumour‐infiltrating lymphocytes (TIL) represent the response of the host organism to the tumour. When a tumour develops, the body can react by mobilizing the immune system, and the prognosis of the patient depends on whether the immune cells can generate an effective anti‐tumour response. The destruction of the tumour is dependent on the ability of immune cells to migrate to the site of its location and infiltration of the cancerous tissue. Tumour microenvironment (TME) comprises diverse cells types, including cancer stromal cells, fibroblasts, lymphocytes, granulocytes, macrophages, mast cells (MCs), natural killer cells (NKs), dendritic cells (DCs) and myeloid‐derived suppressor cells (MDSCs).3 In most cases, the presence of B lymphocytes, cytotoxic CD8‐positive T lymphocytes, NKs, ‘M1‐like’ macrophages and high numbers of DCs are indicative of a favourable outcome. CD8‐positive T cells are the main effector of anti‐tumour immunity. They recognize and destruct tumour cells carrying specific antigens, which are the product of the expression of mutated genes.4, 5 Conversely, ‘M2‐like’ macrophages, granulocytes, MCs, MDSCs, immature DCs, regulatory T cells (Tregs) and TH17 lymphocyte high density are associated with poor prognosis.6
Lymph nodes (LNs) are an integral part of the immune system in humans and are essential for the maintenance of effective immune responses. LNs are penetrated by networks of fibres formed by fibroblastic reticular cells (FRCs). These structures provide a basis for the transport of small molecules such as chemokines and soluble antigens with molecular mass less than 70 kD, enabling the mediation of inflammatory response or immune function by chemokines.7 Chemokines, the largest family of cytokines, constitute a class of low molecular weight secreted proteins capable of inducing directional migration of cells. When immune cells and tissue cells, including fibroblasts, endothelial cells and epidermal cells are induced by stimuli such as growth factors, interferons, and viral and bacterial products, different chemokines can be secreted.8, 9 In the TME, both tumour and immune cells express chemokines, which can lead to the spread of tumour cells. On the other hand, chemokines can promote the entry of specific immune cells into tumours, facilitating the anti‐tumour response and improving the prognosis of patients.1, 10 In this regard, the outcome of the disease in SKCM patients has been demonstrated to depend on the infiltration of lymphocytes into the tumour in SKCM patients, a process that is affected by chemokine or cytokine gradients.11, 12 The generation of an effective anti‐tumour immune response depends on the synergy between different immune cells, and their transport and distribution are co‐ordinated by the interaction between chemokines and their receptors. For example, CCL19 and CCL21 chemokines activate naive T cells, B cells, mature DC cells and NK cells via the CCR7 receptor, inducing their migration to secondary lymphoid organs (SLO).13 Immature DCs express CXCR1, CCR1, CCR2 and CCR6 receptors, and inflammatory chemokines acting as their ligands recruit these cells to the site of inflammation.14 B cells express the chemokine receptor CXCR5, and the ligand of CXCR5 promotes the homing of B cells into LNs.14 CD8‐positive T cells express the chemokine receptor CXCR3, which, when bound by the chemokine ligands CXCL9 and 10, drives their migration to the tumour.15 Increased levels of CXCL9‐11 are associated with a higher number of CD8‐positive T cells infiltrating the tumour, decreased metastatic activity and improved survival of cancer patients.16
CIBERSORT, an analytical tool for estimating the relative abundance of different cell types based on RNA transcripts, allows calculating cell infiltration in tissues based on gene expression profile data. In comparison with traditional methods, CIBERSORT has the advantage of simultaneous assessment of multiple types of infiltrating cells. This approach is not affected by the expression of the same surface marker by different cell types. Moreover, samples can be easily processed and stored in a standardized manner, alleviating problems that negatively affect the quality of data collected at different times and locations.17, 18 The results obtained using CIBERSORT to calculate lymphocytic infiltration are consistent with the data generated by flow cytometry, and this methodology has been applied to the study of multiple diseases.17, 18 The basic matrix in CIBERSORT, LM22, permits the relative quantitation of 22 cell types, including T cells, naive and memory B cells, plasma cells and subpopulations of myeloid cells.19
The research on the role of chemokines in SKCM is sporadic. The number of studies on chemokines and chemokine receptors in metastatic regional LN tissue is limited as well. In view of the paucity of relevant information, the present study focused on the relationship between mRNA expression and clinical pathology of chemokines and their receptors in regional LN tissues of 221 patients with The Cancer Genome Atlas (TCGA) SKCM. CIBERSORT was used to analyse the infiltration of SKCM by CD8‐positive and CD4‐positive T cells, DCs, B cells, macrophages, MCs, and NK cells and establish the relationship between chemokines and their receptors in the LN tissues of SKCM patients and the fraction of the immune cells in the tumour.
2. MATERIALS AND METHODS
2.1. Data acquisition
The SKCM gene expression data set available on the TCGA website (https://cancergenome.nih.gov) was downloaded from the University of California, Santa Cruz, Xena website (http://xena.ucsc.edu/). The Xena website contains TCGA ‐SKCM RNA‐seq data that are consistent with the content of the TCGA and have the additional advantage of being easier to download and organize. The data have been standardized for the RSEM (RNA‐Seq by Expectation‐Maximization) conversion. Together, 472 gene expression data sets from 470 patients were retrieved (some gene expressions data sets were taken from the same patient).
The 470 cases comprised 221 cases of LN tissue of SKCM patients, 103 cases of in situ SKMC tissue, 68 cases of distant metastatic tissue, 74 cases of adjacent tissue and three cases of not identified tissue sources. Subsequently, the gene expression data of LN tissue were screened for analysis, and complete gene expression values and clinical information of 221 SKCM patients were obtained. The clinical data included height, weight, age, BIM, pathological grade, TMN staging. See Clinical Study flow chart (Figure 1) and Clinicopathological features (Table 1).
Figure 1.

Study flowchart
Table 1.
Clinicopathological features of melanoma patients
| Variable | n | % |
|---|---|---|
| Sex | ||
| Female | 83 | 37.56 |
| Male | 138 | 62.44 |
| Age (y) | ||
| <55 | 107 | 48.42 |
| ≥55 | 111 | 50.23 |
| Missing | 3 | 1.36 |
| Pathological stage | ||
| Stage 0/I/II | 94 | 42.53 |
| Stage III/IV | 103 | 46.61 |
| Missing | 24 | 10.86 |
| T stage | ||
| T3/T4 | 81 | 36.65 |
| Tis/T0/T1/T2/T3 | 103 | 46.61 |
| TX/missing | 37 | 16.74 |
| N stage | ||
| N0 | 103 | 46.61 |
| N1/N2/N3 | 100 | 45.25 |
| NX/missing | 18 | 8.14 |
| M stage | ||
| M0 | 201 | 90.95 |
| M1 | 8 | 3.62 |
| Missing | 12 | 5.43 |
Abbreviations: M, distant metastasis; N, Lymph node metastasis; T, Local invasion stage.
2.2. Evaluation of immune cell components using CIBERSORT
The CIBERSORT website (http://cibersort.stanford.edu) provides R language computing source code, as well as the basic matrix (LM22). The R language programs include preprocessCore and BiocManager package. The statistical rank was set to 1000 (recommended value is >100) in the R language program, and quantile normalization was disabled. Subsequently, the lymphocyte infiltration ratio of 22 distinct cell types was calculated, with the sum of fractions equal 1. P‐value was determined for the tissue infiltration score in each patient, and P < .05 were considered to be statistically significant.
2.3. LASSO scores for chemokine and its receptor mRNA expression
Survival analysis and univariate Cox regression were used to screen for chemokines and receptors with prognostic value, since strong multicollinearity may be present between chemokines and their receptors. Therefore, the LASSO scoring based on the selected chemokines and receptors was performed first, and subsequently, multi‐factor Cox regression coefficients were calculated to establish a risk‐scoring model.20
2.4. Statistics
To compare differences between groups calculated by survival analysis or Cox regression, continuous variables (including age, chemokine and its receptor mRNA expression value, and the LASSO score) had to be converted into two categorical variables. For the expression of chemokine and its receptor mRNA, and for the LASSO score, the ‘survminer’ package in the R language was used to calculate the best cut‐off value higher than the cut‐off value for the high expression group and lower than the cut‐off value for the low expression group.
Heatmaps were drawn using EXCEL 2016 (Microsoft Corp) and Adobe Illustrator (Adobe Inc). The R software was used to perform the screening of genetic and clinical data screening, as well as statistical calculations. Wilcoxon rank sum test was used to determine differences in clinical pathology and mRNA expression data; P < .01 was considered statistically significant. Correlation between chemokines and their receptors was established by Spearman rank correlation analysis using the ‘cor.test’ function in R software; P < .01 was considered statistically significant. For survival analyses, the Kaplan‐Meier method with log‐rank test was used and the survival curves were plotted by the R software.
3. RESULTS
3.1. Relationship between the expression of chemokines and their receptors mRNA and clinicopathological data
The expression of chemokines and their receptors mRNA in SKCM regional LN tissues did not differ significantly among the T, M, N and AJCC staging (P > .01; Table 2).
Table 2.
Compare of the differences between the expression of RNA of chemokines and their receptors and clinicopathological data
| Gene | T | N | M | Pathological stage | Gene | T | N | M | Pathological stage |
|---|---|---|---|---|---|---|---|---|---|
| CCL1 | 0.25 | 0.88 | 0.79 | 0.72 | CCR7 | 0.9 | 0.29 | 0.96 | 0.6 |
| CCL2 | 0.06 | 0.34 | 0.52 | 0.65 | CCR8 | 0.68 | 0.17 | 0.36 | 0.41 |
| CCL3 | 0.47 | 0.56 | 0.6 | 0.96 | CCR9 | 0.36 | 0.12 | 0.59 | 0.38 |
| CCL4 | 0.12 | 0.23 | 0.38 | 0.51 | CCR10 | 0.33 | 0.05 | 0.28 | 0.05 |
| CCL5 | 0.51 | 0.3 | 0.27 | 0.68 | CXCL1 | 0.25 | 0.06 | 0.62 | 0.11 |
| CCL7 | 0.04 | 0.43 | 0.49 | 0.45 | CXCL10 | 0.08 | 0.35 | 0.96 | 0.71 |
| CCL8 | 0.05 | 0.13 | 0.49 | 0.27 | CXCL11 | 0.16 | 0.43 | 0.86 | 0.67 |
| CCL11 | 0.75 | 0.59 | 0.82 | 0.77 | CXCL12 | 0.67 | 0.43 | 0.27 | 0.4 |
| CCL13 | 0.11 | 0.14 | 0.98 | 0.43 | CXCL13 | 0.45 | 0.98 | 0.31 | 0.77 |
| CCL14 | 0.5 | 0.95 | 0.36 | 0.88 | CXCL14 | 0.39 | 0.56 | 0.96 | 0.86 |
| CCL15 | 0.84 | 0.65 | 0.48 | 0.83 | CXCL16 | 0.15 | 0.13 | 0.82 | 0.36 |
| CCL16 | 0.4 | 0.86 | 0.94 | 0.7 | CXCL17 | 0.76 | 0.43 | 0.85 | 0.52 |
| CCL17 | 0.57 | 0.82 | 0.61 | 0.6 | CXCL2 | 0.05 | 0.33 | 0.14 | 0.48 |
| CCL18 | 0.86 | 0.07 | 0.49 | 0.35 | CXCL3 | 0.48 | 0.74 | 0.16 | 0.91 |
| CCL19 | 0.62 | 0.55 | 0.95 | 0.97 | CXCL5 | 0.52 | 0.6 | 0.26 | 0.71 |
| CCL20 | 0.48 | 0.28 | 0.97 | 0.51 | CXCL6 | 0.48 | 0.8 | 0.79 | 0.93 |
| CCL21 | 0.86 | 0.77 | 0.91 | 0.99 | CXCL8 | 0.83 | 0.75 | 0.37 | 0.56 |
| CCL22 | 0.33 | 0.43 | 0.82 | 0.86 | CXCL9 | 0.17 | 0.58 | 0.76 | 0.86 |
| CCL23 | 0.32 | 0.11 | 0.39 | 0.36 | CXCR1 | 0.69 | 0.8 | 0.86 | 0.69 |
| CCL24 | 0.11 | 0.75 | 0.61 | 0.21 | CXCR2 | 0.69 | 0.66 | 0.91 | 0.82 |
| CCL25 | 0.19 | 0.81 | 0.56 | 0.32 | CXCR3 | 0.33 | 0.32 | 0.33 | 0.56 |
| CCL26 | 0.98 | 0.26 | 0.27 | 0.45 | CXCR4 | 0.32 | 0.43 | 0.29 | 0.47 |
| CCL27 | 0.84 | 0.69 | 0.63 | 0.6 | CXCR5 | 0.97 | 0.46 | 0.89 | 0.83 |
| CCL28 | 0.41 | 0.87 | 0.35 | 0.64 | CXCR6 | 0.16 | 0.27 | 0.41 | 0.46 |
| CCR1 | 0.01 | 0.17 | 0.55 | 0.37 | CXCR7 | 0.23 | 0.25 | 0.47 | 0.49 |
| CCR2 | 0.42 | 0.27 | 0.21 | 0.44 | CX3CL1 | 0.84 | 0.61 | 0.47 | 0.56 |
| CCR3 | 0.72 | 0.23 | 0.88 | 0.34 | CX3CR1 | 0.36 | 0.27 | 0.72 | 0.35 |
| CCR4 | 0.96 | 0.4 | 0.43 | 0.58 | XCL1 | 0.17 | 0.05 | 0.24 | 0.3 |
| CCR5 | 0.14 | 0.35 | 0.43 | 0.55 | XCL2 | 0.22 | 0.04 | 0.35 | 0.23 |
| CCR6 | 0.88 | 0.84 | 0.26 | 0.97 | XCR1 | 0.6 | 0.45 | 0.35 | 0.53 |
Compare the differences between the expression of RNA of chemokines and their receptors and clinicopathological data were analysed by Wilcoxon rank test. P < .01 was considered statistically significant. T: Tis/T0/T1/T2 (n = 103) VS T3/T4 (n = 81), N: N0 (n = 103) VS N1/N2/N3 (n = 100), M: M0 (n = 201) VS M1 (n = 8), Pathological Stage: 0/I/II (n = 94) VS III/IV (n = 103).
Abbreviations: M, distant metastasis; N, lymph node metastasis; T, local invasion stage; VS, versus.
3.2. The effect of expression of chemokines and their receptors mRNA on the survival of SKCM patients
To compare the survival of SKCM patients with high and low expression of chemokines and their receptors mRNA, univariate Cox regression was performed, and survival curves were plotted (Table 3; Figures 2, 3, 4, 5 and 6). The difference in survival between the groups was statistically significant (P < .01). Among the 33 analysed genes, only in the case of CXCL17 the high expression group had worse survival prognosis than the low expression group. For the remaining 32 chemokine/receptor, high expression was associated with a better survival rate of the patients.
Table 3.
Univariate COX risk regression analysis
| Gene | Hazard ratio | 95% CI | P‐value | Gene | Hazard ratio | 95% CI | P‐value |
|---|---|---|---|---|---|---|---|
| CCL1 | 2.59 | 1.26‐5.32 | .01 | CCR6 | 3.55 | 2.11‐5.97 | <.001 |
| CCL11 | 0.64 | 0.44‐0.95 | .027 | CCR7 | 1.81 | 1.22‐2.68 | .003 |
| CCL13 | 2.35 | 1.53‐3.61 | <.001 | CCR8 | 2.18 | 1.49‐3.2 | <.001 |
| CCL14 | 0.66 | 0.45‐0.97 | .036 | CCR9 | 1.92 | 1.28‐2.89 | .002 |
| CCL15 | 0.77 | 0.48‐1.22 | .261 | CX3CL1 | 0.75 | 0.52‐1.1 | .145 |
| CCL16 | 0.81 | 0.54‐1.2 | .286 | CX3CR1 | 1.74 | 1.11‐2.73 | .016 |
| CCL17 | 2.25 | 1.09‐4.63 | .028 | CXCL1 | 0.7 | 0.4‐1.22 | .209 |
| CCL18 | 0.68 | 0.46‐1.02 | .065 | CXCL10 | 2.58 | 1.73‐3.86 | <.001 |
| CCL19 | 1.52 | 1.03‐2.25 | .036 | CXCL11 | 2.2 | 1.46‐3.32 | <.001 |
| CCL2 | 2.19 | 1.46‐3.27 | <.001 | CXCL12 | 2.03 | 1.33‐3.22 | .001 |
| CCL20 | 1.56 | 0.79‐3.09 | .203 | CXCL13 | 3 | 1.92‐4.7 | <.001 |
| CCL21 | 0.76 | 0.52‐1.11 | .156 | CXCL14 | 1.45 | 0.85‐2.47 | .173 |
| CCL22 | 2.13 | 1.38‐3.3 | .001 | CXCL16 | 3.35 | 1.47‐7.65 | .004 |
| CCL23 | 2.77 | 1.76‐4.36 | <.001 | CXCL17 | 0.41 | 0.25‐0.67 | <.001 |
| CCL24 | 1.84 | 1.19‐2.84 | .006 | CXCL2 | 1.85 | 1.23‐2.79 | .003 |
| CCL25 | 2.43 | 1.65‐3.59 | <.001 | CXCL3 | 1.53 | 0.97‐2.43 | .067 |
| CCL26 | 1.73 | 1.07‐2.8 | .025 | CXCL5 | 0.65 | 0.38‐1.13 | .129 |
| CCL28 | 1.36 | 0.91‐2.03 | .128 | CXCL6 | 0.6 | 0.37‐0.96 | .034 |
| CCL3 | 1.54 | 1.03‐2.28 | .034 | CXCL9 | 2.56 | 1.72‐3.79 | <.001 |
| CCL4 | 2.14 | 1.44‐3.16 | <.001 | CXCR1 | 0.67 | 0.45‐1 | .052 |
| CCL5 | 2.44 | 1.54‐3.89 | <.001 | CXCR2 | 0.63 | 0.41‐0.99 | .047 |
| CCL7 | 1.72 | 1.17‐2.53 | .006 | CXCR3 | 2.09 | 1.43‐3.07 | <.001 |
| CCL8 | 3.31 | 1.93‐5.67 | <.001 | CXCR4 | 1.49 | 0.94‐2.34 | .088 |
| CCR1 | 2.26 | 1.54‐3.32 | <.001 | CXCR5 | 1.67 | 1.14‐2.43 | .008 |
| CCR10 | 1.73 | 1.09‐2.76 | .02 | CXCR6 | 3.27 | 2.03‐5.28 | <.001 |
| CCR2 | 2.52 | 1.69‐3.75 | <.001 | CXCR7 | 0.7 | 0.46‐1.08 | .103 |
| CCR3 | 1.88 | 1.28‐2.76 | .001 | XCL1 | 1.9 | 1.29‐2.8 | .001 |
| CCR4 | 1.81 | 1.24‐2.65 | .002 | XCL2 | 2.45 | 1.67‐3.61 | <.001 |
| CCR5 | 2.24 | 1.53‐3.29 | <.001 | XCR1 | 2 | 1.26‐3.18 | .003 |
P < .01 was considered statistically significant.
Abbreviation: 95% CI, 95% confidence interval.
Figure 2.
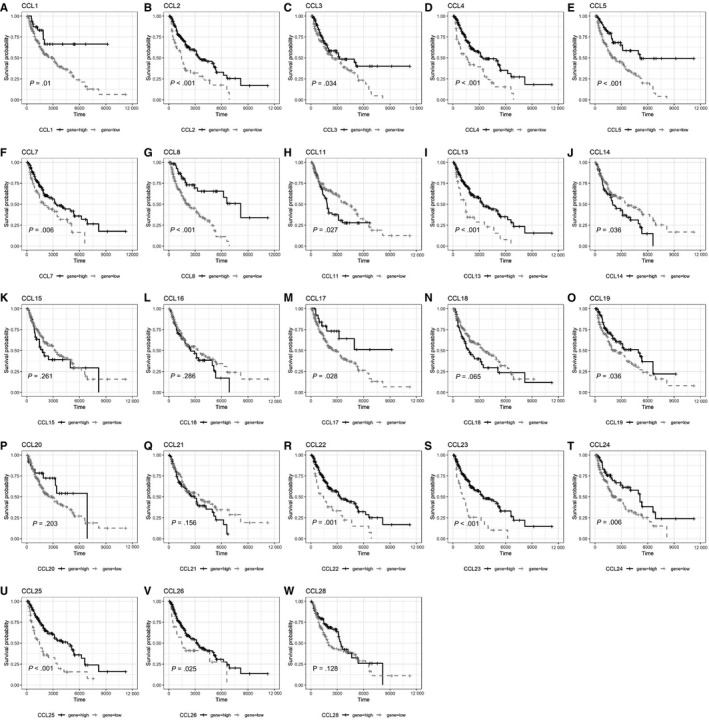
Survival curve of CCL subgroup A, CCL1, B, CCL2, C, CCL3, D, CCL4, E, CCL5, F, CCL7, G, CCL8, H, CCL11, I, CCL13, J, CCL14, K, CCL15, L, CCL16, M, CCL17, N, CCL18, O, CCL19, P, CCL20, Q, CCL21, R, CCL22, S, CCL23, T, CCL24, U, CCL25, V, CCL26, W, CCL28
Figure 3.
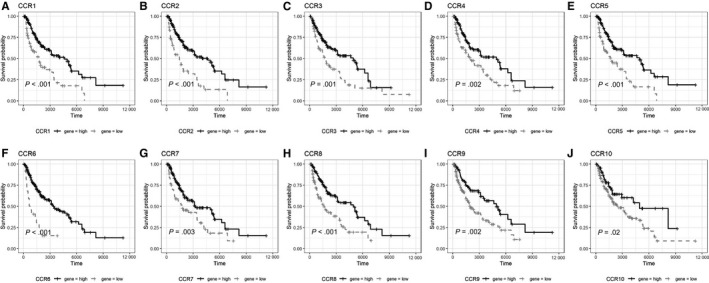
Survival curve of CCR subgroup A, CCR1, B, CCR2, C, CCR3, D, CCR4, E, CCR5, F, CCR6, G, CCR7, H, CCR8, I, CCR9, J, CCR10
Figure 4.
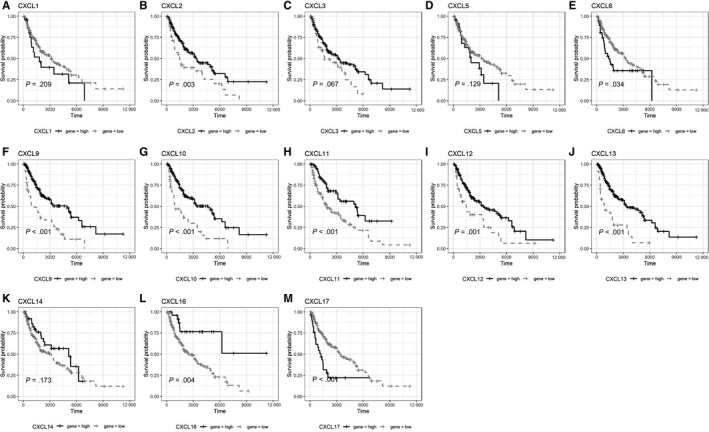
Survival curve of CXCL subgroup A, CXCL1, B, CXCL2, C, CXCL3, D, CXCL5, E, CXCL6, F, CXCL9, G, CXCL10, H, CXCL11, I, CXCL12, J, CXCL13, K, CXCL14, L, CXCL16, M, CXCL17
Figure 5.
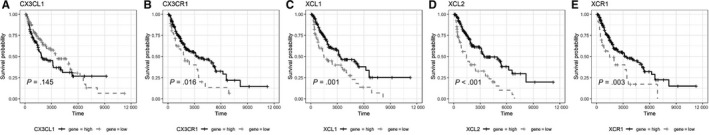
Survival curve of CXCR subgroup A, CXCR1, B, CXCR2, C, CXCR3, D, CXCR4, E, CXCR5, F, CXCR6, G, CXCR7
Figure 6.

Survival curve of CX3CL/CX3CR and XCL/XCR A, CX3CL1, B, CX3CR1, C, XCL1, D, XCL2, E, CCR1
3.3. The relationship between LASSO scores for the expression of mRNA of chemokines and their receptors and the survival of SKCM patients
Univariate screening of the 32 chemokine/receptor (CCL2, 4‐5, 7‐8, CCL22‐25, CCR1‐9, CXCL2‐3, 5, 9‐13, 16, XCL1‐2, and XCR1), for which higher expression was associated with a better prognosis. As there was a significant correlation between these 32 genes (Tables S1‐S3, Figure S1), multicollinearity between chemokines/receptors leads to bias in multivariate COX analysis. Therefore, LASSO score was calculated before multivariate COX analysis. A total of four chemokine/receptor gene pairs were included in the LASSO score: CCL8, CCL2, CXCL10 and CCL16 (Figure 7A,B). The formula obtained was as follows: LASSO score = −(0.097 * CCL8 expression) − (0.038 * CCL2 expression) − (0.023 * CXCL10 expression) − (0.001 * CCL16 expression). The mean value of the Lasso score was −1.36 ± 0.47 (range: −2.46 to 0.43). And those four of the chemokines were negatively correlated with the LASSO score.
Figure 7.
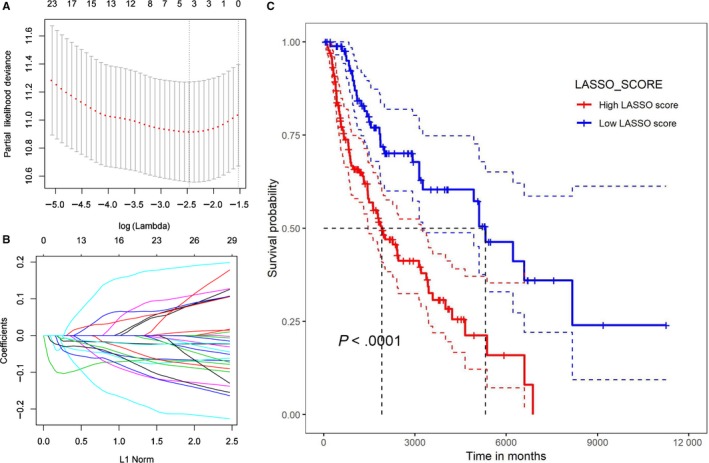
LASSO score A, Results of the LASSO model after 10‐fold cross‐validation. B, Distribution of LASSO coefficients for 22 types of immune cells. C, Survival curves of patients with high and low LASSO scores
Univariate survival analysis was then performed with respect to the LASSO score. The cut‐off value of the LASSO score (−1.080674) was calculated using the R software and was used to divide the patient groups into a high LASSO score group and a low LASSO score group. Survival curves were plotted and indicated that the median survival time in the high LASSO score group was 1460, and 5107 days in the low LASSO score group (P < .001 by the log‐rank test; Figure 7C).
3.4. Relationship between the chemokine/receptor and infiltration of tumour by immune cells
The CIBERSORT algorithm was utilized to calculate the infiltration scores of 22 types of immune cells in LN tissue of SKCM patients. We then calculated the relationship between chemokine/receptor and tumour immune cell fraction (Figure 8; Table S3).
Figure 8.

Relationship between chemokines and their receptors and tumour‐infiltrating immune cells
3.5. Multivariate survival analysis
Univariate COX regression analysis showed that the survival of SKCM patients differed in a statistically significant manner with age T, N, CXCL17 and LASSO scores (P < .01). However, the patients with the M1 stage pathology were too small to evaluate the statistical significance of potential differences. Multivariate analysis showed that T stage, N stage, CXCL17 and LASSO scores had independent prognostic value (P < .01) (Table 4).
Table 4.
Multivariate Cox regression analysis
| Factors | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P value | HR | 95% CI | P value | |
| Sex | 1.19 | 0.79‐1.79 | .406 | |||
| Age | 1.99 | 1.34‐2.96 | .001 | 1.68 | 1.06‐2.67 | .027 |
| T | 0.45 | 0.3‐0.69 | <.001 | 0.53 | 0.33‐0.84 | .007 |
| N | 2.15 | 1.42‐3.26 | <.001 | 1.9 | 1.45‐8.15 | <.001 |
| M | 1.53 | 0.48‐4.85 | .471 | |||
| CXCL17 | 0.41 | 0.25‐0.67 | <.001 | 0.4 | 0.22‐0.73 | .003 |
| LASSO score | 0.39 | 0.25‐0.6 | <.001 | 0.39 | 0.24‐0.65 | <.001 |
4. DISCUSSION
Chemokines, the key mediators of the immune response, are essential for the recruitment of many different types of cells to TME.9 To identify their functions in SCKM, we first was assessed between the survival of the patients and the expression of chemokines and their receptors genes, using the data for SCKM regional LN tissue available in the public TCGA database. The survival prognosis of the patients with high expression of 32 chemokines and receptors (CCL2, 4‐5, 7‐8, CCL22‐25; CCR1‐9; CXCL2‐3, 5, 9‐13, 16; XCL1‐2, XCR1) was found to be better than in the low expression group. Subsequently, a multivariate COX analysis was performed since the strong correlation among these 32 chemokines/receptors might have compromised the validity of the multivariate COX regression analysis due to multiple linearity. Additionally, LASSO regression based on these 32 chemokines was computed, and analysis was performed using a comprehensive LASSO score. This approach has shown that the LASSO score was an independent prognostic factor. Also, this analysis explained the basis for the absence of a significant difference in chemokine/receptor classification in different pathological stages, suggesting that these 32 chemokine/receptor genes have independent survival prognostic significance.
Among the chemokines and receptors analysed, we found CCR4, 6‐9, CCL13, ‐22, ‐23, and XCR1 were positively correlated with the fractions of memory B cells and the naive T cells, and negatively correlated with M0 type macrophages and resting type MCs. A study on the correlation between the mature B cells in the tumour and the prognosis of malignant SKCM documented that that humoral immunity participates in the anti‐tumour defence.21, 22 Immunohistochemistry of human SKCM samples demonstrated that most tumour tissues contain a large number of infiltrating CD20‐positive cells, which are considered to be B lymphocytes, which are dispersed in the matrix surrounding the tumour.22 The percentage of B cell infiltration in and around the tumour was also positively correlated with patient survival.22, 23 Naive T cells express CCR7 and control the migration of immune cells to SLO. This process is mediated by the CCL19 and CCL21 receptors. Thus, CCR7 plays an important role in lymphocyte homing to LNs and spleen.24 It has been demonstrated that in CCR7‐deficient mice T lymphocytes are absent in the lymphoid white pulp of LNs and spleen, and are present only in the red pulp of the spleen.25, 26 Tumour‐associated MCs can release a variety of cytokines, chemokines and growth factors, promoting tumour development by enhancing angiogenesis and remodelling tumour extracellular matrix.27, 28, 29
The current study demonstrated also that CXCL9‐11, CXCR3, 6, CCL2, 4, 5, 7, 8, 25, CCR1, 2, 5, and XCL1, 2 are positively correlated with the fractions of M1 macrophages and CD8‐positive T cells, and negatively correlated with M0 macrophages. CD8‐positive T cells recognize tumour cell antigens and drive anti‐tumour response by secreting effector cytokines, releasing cytotoxic molecules such as granzyme B and perforin, and inducing apoptosis in tumour cells.30 The CXCL9‐11/CXCR3 axis regulates CD8‐positive T cell migration, differentiation and activation.16, 31 CD8‐positive T cells express CXCR3 and are capable of invading into tumours when activated by chemokines. Elevated levels of CXCL9 and CXCL10 are associated with an increased number of tumour‐infiltrating CD8‐positive T cells, decreased cancer metastasis and increased survival of cancer patients.32, 33 Experiments on CXCR3 knockout mice bearing B16 melanoma demonstrated a critical role for CXCR3 in the migration of CD8‐positive cells. These transgenic mice exhibited significant tumour growth and decreased survival.34 Analysis of CD8‐positive T cells early in tumour development has a better prognostic value than the traditional staging. Most patients with stage I and stage II cancer lack T cell infiltration and are prone to disease recurrence within 5 years. T cell infiltration of the tumour is correlated with longer disease‐free survival in stage III cancer patients.35 In addition, CXCR3 and its ligands CXCL9 and CXCL10 are closely related to the TH1 immune response, and CXCR3 mediates the anti‐tumour response by recruiting into tumours NK cells, CD4‐positive Th1 cells and CD8‐positive cytotoxic T lymphocytes tumours. M1 macrophages express and secrete pro‐inflammatory peptides, chemokines and other effector molecules, including IL‐1, IL‐6, TNF, IL‐23 and i‐NOS, contributing to the development of anti‐tumour response.36 The chemokine CCL2 is a major player in this process, and its binding to the CCR2 receptor directly mediates monocyte recruitment to inflammation site and primary tumour tissue.30 In addition, a prolonged survival was observed in metastatic SKCM patients with high expression of CCL4, CCL5, CXCL9, CXCL10 and CXCL1137 and related to patients with higher response to ipilimumab treatment.38 The CCR5‐deficient mice use of CCR5 blockers was associated with a decrease in Treg cells, which have a tumour‐promoting effect.39
CXCL17 is highly expressed in a variety of cancer cells, recruiting MDSCs into tumours and partially promoting tumour growth by enhancing angiogenesis.40 These findings are consistent with the shorter survival of SKCM patients with high CXCL17 documented in the current work.
In conclusion, we have demonstrated that high expression of 32 chemokines and receptors (CCL2, 4‐5, 7‐8, CCL22‐25, CCR1‐9, CXCL2‐3, 5, 9‐13, 16, XCL1‐2 and XCR1) in SKCM regional LN tissue is associated with a good prognosis, which may be related to the attraction of immune cells to the TME and elimination of tumour cells. Conversely, high expression of CXCL17 is indicative of a poor prognosis.
Some limitations of the study should be acknowledged. The validity of the conclusions reached should be confirmed by an investigation involving a larger number of cases. Further analysis is also needed for each subgroup; this was not performed due to a limited number of cases in each subgroup. Other potential factors, such as the size of the tumour and the impact of anti‐cancer drugs on chemokines, need further consideration, as does the interaction between chemokines and their receptors and other signalling pathways. Finally, due to the retrospective nature of this study, the choice of offset cannot be avoided.
Immunotherapy is currently used to treat cancer, but this type of therapy is effective only for specific populations of patients.41, 42, 43, 44 Novel, more powerful treatments are urgently needed. Understanding the role of chemokines in tumour resistance to immunologic defences of the body is essential for the development of new targeted therapeutics in the future.
CONFLICT OF INTEREST
There are no conflicts of interest.
AUTHOR CONTRIBUTIONS
ZX and PF.Q designed the experiments; LQ and MH.Y analysed the data; XT.F, PF.Q and ZX wrote the paper. All other authors participated in revising the paper and finalizing the paper. All authors read and approved the final manuscript.
Supporting information
ACKNOWLEDGEMENTS
This work was supported in part by the National Natural Science Foundation of China (No: 81760344). We analysed raw data that are published in The Cancer Genome Atlas (TCGA) (https://cancergenome.nih.gov).
Xiong T‐F, Pan F‐Q, Liang Q, et al. Prognostic value of the expression of chemokines and their receptors in regional lymph nodes of melanoma patients. J Cell Mol Med. 2020;24:3407–3418. 10.1111/jcmm.15015
Xiong and Pan are contributed to the work equally and should be regarded as co‐first authors.
Contributor Information
Haiyan Mo, Email: mhygx@163.com.
Xiang Zhou, Email: 1360749244@qq.com.
DATA AVAILABILITY STATEMENT
All data sets generated/analysed for this study are included in the manuscript and the Supplementary Files.
REFERENCES
- 1. Jacquelot N, Duong CPM, Belz GT, Zitvogel L. Targeting chemokines and chemokine receptors in melanoma and other cancers. Front Immunol. 2018;9:2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lee N, Zakka LR, Mihm MC Jr, Schatton T. Tumour‐infiltrating lymphocytes in melanoma prognosis and cancer immunotherapy. Pathology. 2016;48:177‐187. [DOI] [PubMed] [Google Scholar]
- 3. Spranger S, Sivan A, Corrales L, Gajewski TF. Tumor and host factors controlling antitumor immunity and efficacy of cancer immunotherapy. Adv Immunol. 2016;130:75‐93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Molon B, Cali B, Viola A. T cells and cancer: how metabolism shapes immunity. Front Immunol. 2016;7:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Joyce JA, Fearon DT. T cell exclusion, immune privilege, and the tumor microenvironment. Science. 2015;348:74‐80. [DOI] [PubMed] [Google Scholar]
- 6. Takeuchi Y, Nishikawa H. Roles of regulatory T cells in cancer immunity. Int Immunol. 2016;28:401‐409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Riedel A, Shorthouse D, Haas L, Hall BA, Shields J. Tumor‐induced stromal reprogramming drives lymph node transformation. Nat Immunol. 2016;17:1118‐1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bachelerie F, Ben‐Baruch A, Burkhardt AM, et al. International Union of Basic and Clinical Pharmacology. [corrected]. LXXXIX. Update on the extended family of chemokine receptors and introducing a new nomenclature for atypical chemokine receptors. Pharmacol Rev. 2014;66:1‐79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lazennec G, Richmond A. Chemokines and chemokine receptors: new insights into cancer‐related inflammation. Trends Mol Med. 2010;16:133‐144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rusakiewicz S, Semeraro M, Sarabi M, et al. Immune infiltrates are prognostic factors in localized gastrointestinal stromal tumors. Cancer Res. 2013;73:3499‐3510. [DOI] [PubMed] [Google Scholar]
- 11. Jacquelot N, Enot DP, Flament C, et al. Chemokine receptor patterns in lymphocytes mirror metastatic spreading in melanoma. J Clin Invest. 2016;126:921‐937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zlotnik A, Yoshie O. The chemokine superfamily revisited. Immunity. 2012;36:705‐716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Esche C, Stellato C, Beck LA. Chemokines: key players in innate and adaptive immunity. J Invest Dermatol. 2005;125:615‐628. [DOI] [PubMed] [Google Scholar]
- 14. Mueller SN, Germain RN. Stromal cell contributions to the homeostasis and functionality of the immune system. Nat Rev Immunol. 2009;9:618‐629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mikucki ME, Skitzki JJ, Frelinger JG, et al. Unlocking tumor vascular barriers with CXCR3: Implications for cancer immunotherapy. Oncoimmunology. 2016;5:e1116675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tokunaga R, Zhang W, Naseem M, et al. CXCL9, CXCL10, CXCL11/CXCR3 axis for immune activation – a target for novel cancer therapy. Cancer Treat Rev. 2018;63:40‐47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen B, Khodadoust MS, Liu CL, Newman AM, Alizadeh AA. Profiling tumor infiltrating immune cells with CIBERSORT. Methods Mol Biol. 2018;1711:243‐259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Petitprez F, Vano YA, Becht E, et al. Transcriptomic analysis of the tumor microenvironment to guide prognosis and immunotherapies. Cancer Immunol Immunother. 2018;67:981‐988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453‐457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Datta S, Le‐Rademacher J, Datta S. Predicting patient survival from microarray data by accelerated failure time modeling using partial least squares and LASSO. Biometrics. 2007;63:259‐271. [DOI] [PubMed] [Google Scholar]
- 21. Chiaruttini G, Mele S, Opzoomer J, et al. B cells and the humoral response in melanoma: the overlooked players of the tumor microenvironment. Oncoimmunology. 2017;6:e1294296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ladanyi A, Kiss J, Mohos A, et al. Prognostic impact of B‐cell density in cutaneous melanoma. Cancer Immunol Immunother. 2011;60:1729‐1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ladanyi A. Prognostic and predictive significance of immune cells infiltrating cutaneous melanoma. Pigment Cell Melanoma Res. 2015;28:490‐500. [DOI] [PubMed] [Google Scholar]
- 24. Kerdiles YM, Beisner DR, Tinoco R, et al. Foxo1 links homing and survival of naive T cells by regulating L‐selectin, CCR7 and interleukin 7 receptor. Nat Immunol. 2009;10:176‐184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Forster R, Davalos‐Misslitz AC, Rot A. CCR7 and its ligands: balancing immunity and tolerance. Nat Rev Immunol. 2008;8:362‐371. [DOI] [PubMed] [Google Scholar]
- 26. Forster R, Schubel A, Breitfeld D, et al. CCR7 coordinates the primary immune response by establishing functional microenvironments in secondary lymphoid organs. Cell. 1999;99:23‐33. [DOI] [PubMed] [Google Scholar]
- 27. Qu Z, Liebler JM, Powers MR, et al. Mast cells are a major source of basic fibroblast growth factor in chronic inflammation and cutaneous hemangioma. Am J Pathol. 1995;147:564‐573. [PMC free article] [PubMed] [Google Scholar]
- 28. Grutzkau A, Kruger‐Krasagakes S, Baumeister H, et al. Synthesis, storage, and release of vascular endothelial growth factor/vascular permeability factor (VEGF/VPF) by human mast cells: implications for the biological significance of VEGF206. Mol Biol Cell. 1998;9:875‐884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ribatti D, Crivellato E. Mast cells, angiogenesis, and tumour growth. Biochim Biophys Acta. 2012;1822:2‐8. [DOI] [PubMed] [Google Scholar]
- 30. Murray T, Fuertes Marraco SA, Baumgaertner P, et al. Very late antigen‐1 marks functional tumor‐resident CD8 T cells and correlates with survival of melanoma patients. Front Immunol. 2016;7:573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liu J, Wang C, Ma X, et al. High expression of CCR5 in melanoma enhances epithelial‐mesenchymal transition and metastasis via TGFbeta1. J Pathol. 2018. [DOI] [PubMed] [Google Scholar]
- 32. Zhang L, Conejo‐Garcia JR, Katsaros D, et al. Intratumoral T cells, recurrence, and survival in epithelial ovarian cancer. N Engl J Med. 2003;348:203‐213. [DOI] [PubMed] [Google Scholar]
- 33. Sato E, Olson SH, Ahn J, et al. Intraepithelial CD8+ tumor‐infiltrating lymphocytes and a high CD8+/regulatory T cell ratio are associated with favorable prognosis in ovarian cancer. Proc Natl Acad Sci USA. 2005;102:18538‐18543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chheda ZS, Sharma RK, Jala VR, Luster AD, Haribabu B. Chemoattractant receptors BLT1 and CXCR3 regulate antitumor immunity by facilitating CD8+ T cell migration into tumors. J Immunol. 2016;197:2016‐2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Galon J, Costes A, Sanchez‐Cabo F, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960‐1964. [DOI] [PubMed] [Google Scholar]
- 36. Mantovani A, Sica A. Macrophages, innate immunity and cancer: balance, tolerance, and diversity. Curr Opin Immunol. 2010;22:231‐237. [DOI] [PubMed] [Google Scholar]
- 37. Messina JL, Fenstermacher DA, Eschrich S, et al. 12‐Chemokine gene signature identifies lymph node‐like structures in melanoma: potential for patient selection for immunotherapy? Sci Rep. 2012;2:765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ji RR, Chasalow SD, Wang L, et al. An immune‐active tumor microenvironment favors clinical response to ipilimumab. Cancer Immunol Immunother. 2012;61:1019‐1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schlecker E, Stojanovic A, Eisen C, et al. Tumor‐infiltrating monocytic myeloid‐derived suppressor cells mediate CCR5‐dependent recruitment of regulatory T cells favoring tumor growth. J Immunol. 2012;189:5602‐5611. [DOI] [PubMed] [Google Scholar]
- 40. Weinstein EJ, Head R, Griggs DW, et al. VCC‐1, a novel chemokine, promotes tumor growth. Biochem Biophys Res Commun. 2006;350:74‐81. [DOI] [PubMed] [Google Scholar]
- 41. Hodi FS, O'Day SJ, McDermott DF, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363:711‐723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Topalian SL, Hodi FS, Brahmer JR, et al. Safety, activity, and immune correlates of anti‐PD‐1 antibody in cancer. N Engl J Med. 2012;366:2443‐2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wolchok JD. PD‐1 Blockers. Cell. 2015;162:937. [DOI] [PubMed] [Google Scholar]
- 44. Atay C, Kwak T, Lavilla‐Alonso S, et al. BRAF targeting sensitizes resistant melanoma to cytotoxic T cells. Clin Cancer Res. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data sets generated/analysed for this study are included in the manuscript and the Supplementary Files.


