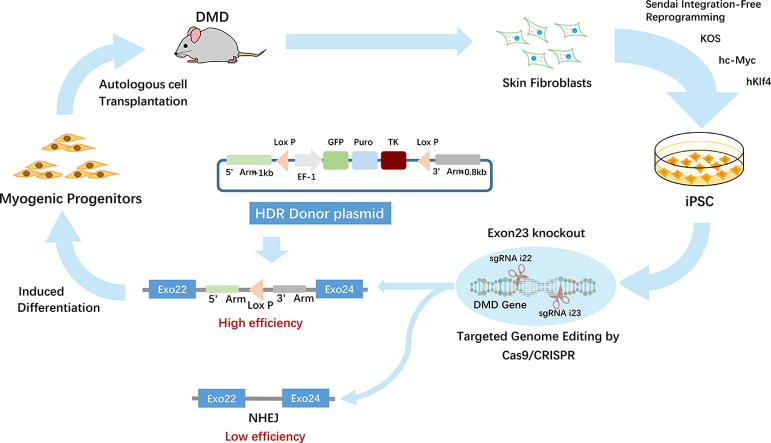
Graphical abstract
Keywords: Homology directed repair (HDR), Induced pluripotent stem cells, interspaced short palindromic repeats/CRISPR-associated 9 (CRISPR/Cas9), Duchenne muscular dystrophy (DMD), myogenic progenitor cells (MPC)
Abstract
Duchenne muscular dystrophy (DMD) is a progressive myopathic disease caused by mutations in the gene encoding dystrophin protein that eventually leads to the exhaustion of myogenic progenitor cells (MPC). Autologous induced pluripotent stem cells (iPSCs) provide an endless source of MPC, which can potentially replenish the progenitor cell pool, repair muscle damage, and prevent DMD progression. Deletion of mutant exon 23 (ΔEx23) with clustered regularly interspaced short palindromic repeats/CRISPR-associated 9 (CRISPR/Cas9) gene-editing technology can correct dystrophin gene expression in iPSCs. However, successful exon23 deletion and clonal isolation are very inefficient (~3%), and manual selection of each iPSC clone and genotyping to identify ΔEx23 is labor-intensive. To overcome these obstacles, we added a homology-directed repair (HDR) donor vector, which carries floxed fluorescent protein and antibiotic selection genes, thus allowing us to identify ΔEx23 iPSC with donor selective gene integration. Our results indicate that the HDR-mediated targeted integration enables ΔEx23 iPSC identification; the HDR donor vector increased the recognition efficiency of clonal isolation (>90% as confirmed by Sanger sequencing). After removal of the inserted genes by Cre-mediated recombination followed by doxycycline (Dox)-induced MyoD induction, ΔEx23 iPSC differentiated into MPC with restored dystrophin expression in vitro. Importantly, transplanted ΔEx23 iPSC-MPC express dystrophin in the muscles of a mouse model of DMD (Mdx mice). In conclusion, the use of HDR donor vector increased the efficiency of ΔEx23 gene correction by CRISPR/Cas9, and facilitate the identification of successfully edited iPSC clones for cell therapy of DMD.
1. Introduction
Duchenne muscular dystrophy (DMD) is a severe, progressive genetic disorder affecting about one in 3500 newborn boys, who on average lose the ability to ambulate between 7 and 13 years of age and become wheelchair dependent at approximately 15 years of age [1]. DMD is caused by mutations in the DMD gene leading to loss of dystrophin protein and damage to muscle fibers during contraction. This, in turn, leads to muscle inflammation, which inhibits muscle regeneration and, ultimately, damaged muscles are replaced by fibrotic and adipose tissue [2]. DMD patients often die in their 20 s due to heart failure or respiratory failure due to diaphragm dysfunction [3].
Because DMD is an X-linked recessive hereditary disease, it cannot be cured. Anti-inflammatory drugs and antioxidants are the two main approaches to reducing muscle damage in DMD patients [4]. Recently, exon skipping therapy targeting pre-mRNA transcripts using modified antisense oligonucleotides (such as eteplirsen and drisapersen) has been used to treat DMD patients, and both have been shown to slightly increase the expression of dystrophin protein [5], [6], [7].
Since muscle damage caused by DMD is caused by a lack of functional dystrophin, overexpression of dystrophin by gene therapy is a promising approach to restoring dystrophin expression. Adeno-associated virus (AAV) is safe for use in humans and exhibits muscle tropism, but it cannot carry a gene the size of DMD (2.4 Mb) [8]. To overcome this obstacle, scientists have developed the micro-dystrophy protein (μDMD) gene with partial DMD function to adapt to the AAV vector [9]; even so, as an episomal virus, rAAV-mediated μDMD expression is gradually lost due to muscle turnover [10]. Thus, treatment must be repeated to maintain the expression of μDMD in the muscle. Since rAAV-treated patients will produce neutralizing antibodies after the first treatment, the utility of rAAV gene therapy is likely to be limited in DMD patients [11].
CRISPR/Cas9-mediated gene editing is a novel technology that can be used to induce double-stranded DNA breaks at specific locations in the genome to correct mutations at the genomic DNA (gDNA) level, thus avoiding the need for repeated gene therapy to maintain RNA and protein expression [12]. Recent studies have shown that the deletion of mutant exons by rAAV-mediated SaCas9 and two gRNA vectors can restore the dystrophin reading frame in infected muscle cells in mouse models [13], [14], [15]. The main challenge with the rAAV approach is that rAAV is unable to infect muscle progenitor cells (MPC), such as myoblasts or satellite cells, so the regenerating skeletal muscle cells still carry the mutated dystrophin gene after injury. Therefore, patients will gradually experience loss of muscle function even after successful rAAV-mediated Cas9/gRNA gene therapy [16].
DMD can also be considered a stem cell disease since MPCs are depleted by repeated muscle loss-regeneration-muscle loss cycles [17], [18]. Therefore, cell transplantation with genetically corrected autologous stem cells is a promising strategy for treating DMD. The rationale for this approach is based on the following facts: (1) reprogramming of iPSCs from adult cell sources, such as skin fibroblasts, is feasible [19]; (2) autologous MPC can be induced from iPSC by overexpression of muscle-specific transcription factors, such as MyoD [20], [21], and iPSC are characterized by self-renewal and thus represent an infinite source of stem cells [22]; (3) Cas9-mediated gene editing is highly effective in proliferative stem cells compared to non-divided muscle cells [23].
To delete the mutant exon23 of DMD gene in Mdx mice-derived iPSCs for cell therapy, we previously used a cas9-based non-homologous end joining (NHEJ) approach to test the efficiency of deletion of mutation exon23. Although all iPSC clones expressed Cas 9 and two gRNAs after lentiviral vector transduction followed by antibiotic selection, the deletion efficiency was only about 3% (3 corrected clones from a total of 94 picked clones infected with Cas9 and two gRNAs). Therefore, the NHEJ method is inefficient. CRISPR/Cas9-induced double-stranded DNA breaks can be repaired by either NHEJ or homologous direct repair (HDR); the latter occurs only in dividing cells (e.g., stem/progenitor cells), but not post-mitotic cells (e.g., cardiomyocytes and skeletal muscle cells) [24]. We hypothesized that an HDR-based reporter gene knock-in (KI) strategy could help us enrich exon23-deleted iPSC by replacing mutant exon23 with selective genes (e.g., fluorescent protein and antibiotic resistance gene). Our results indicate that engineered HDR donor vectors coupled with the CRISPR/Cas9 system help identify the correct iPSC clones by genetic enrichment, thereby increasing the efficiency of DMD gene correction.
2. Materials and methods
2.1. Collection of adult Dmdmdx mouse skin fibroblasts
Adult mouse tail skin from Dmdmdx mice (The Jackson Laboratory) were minced into small pieces and subjected in Dulbecco’s minimal essential medium/Ham’s F12 (DMEM/F12) containing 0.1% collagenase IV (Worthington Biochemical Corporation, LS004189) and 1 U/ml of dispase (STEMCELL Technologies, 7923) for 2 h at 37 °C in a 5% CO2 incubator. The explants were plated on gelatin-coated dishes and cultured for 10 d in fibroblast culture medium (Dulbecco modified Eagle medium [DMEM] with 10% fetal calf serum [FBS], 100 IU/mL penicillin G, 100 μg/ml streptomycin, 0.25 μg/mL amphotericin B, 2 mM L-glutamine, 0.10 mM nonessential amino acids and 0.05 mM β-mercaptoethanol) at 37 °C and 5% CO2.
2.2. Generation of iPS cells from Dmdmdx mouse skin fibroblasts
Fifty thousand skin fibroblasts were plated per well on a 6-well plate two days before infection, and medium containing 5:5:3 mix of lentiviral vectors expressing KOS, hc-Myc, and hKlf4 was applied to cells. The medium was replaced with fresh fibroblast medium 24 h after transfection, and cells were cultured for one week with medium exchange every other day. The medium was then replaced with mouse ES cell culture medium (Dulbecco modified Eagle medium [DMEM] with 10% fetal calf serum [FBS], 100 IU/mL penicillin G, 100 μg/ml streptomycin, 0.25 μg/mL amphotericin B, 2 mM L-glutamine, 0.10 mM nonessential amino acids and 0.05 mM β-mercaptoethanol, 1x mouse recombinant Leukemia Inhibitory Factor (LIF) (ESGRO® Supplement, Millipore), 1x MEK/GSK-3 inhibitor supplement (MEK/GSK-3 Inhibitor Supplement, Millipore). The putative iPS cell colonies were identified and chosen using morphological selection criteria. Mouse iPS cell colonies were dissociated with TVP solution (PBS with 1% chicken serum, 0.025% trypsin, and 1.27 mM EDTA) and passage onto a 0.1% gelatin-coated dish containing mouse ES cell culture medium, which was replaced every other day.
2.3. Teratoma formation
To assess the pluripotency and tumorigenic potential of iPS cells generated from Dmdmdx skin fibroblasts, we injected 5 × 105/30 μl iPS cells into hind limb muscles of 8-week-old immunocompromised NOD SCID gamma mice (the Jackson Laboratory). After two weeks, teratomas were dissected and fixed in 4% paraformaldehyde. Samples were OCT-embedded and cut into 5 µm sections using a cryostat and processed with Immunofluorescence staining.
2.4. Construction of DMD-exon 23-deficient stable cell line using CRISPR/Cas9 and homologous-directed recombination (HDR) targeting vector system
Plasmid lenti-CRISPRv2-blast (Addgene plasmid no.83480) was gift from Mohan Babu, and lentiGuide-Hygro-iRFP670 (Addgene plasmid no. 99377) were gifts from Kristen Brennand [25]. Two pairs of gRNA oligos targeting intron-flanked dystrophin exon 23 were designed using the online program (http://crispor.tefor.net/crispor.py). The designed pairs are as follows:
i22sense:
5′-CACCGTTAAGCTTAGGTAAAATCAA-3′;
i22anti-sense:
5′-AAACTTGATTTTACCTAAGCTTAAC-3′;
i23sense:
5′-CACCGAGTAATGTGTCATACCTTCT-3′;
i23anti-sense:
5′-AAACAGAAGGTATGACACATTACTC-3′.
Plasmid lenti-CRISPRv2-blast and lenti-Guide-Hygro-iRFP670 were digested by BsmB1/Esp3I, and the annealed oligos were then cloned into the vectors (annealed oligos i22 for plenti-CRISPRv2-blast and i23 for plenti-Guide-Hygro-iRFP670), using the Quick Ligation Kit (NewEngland BioLabs). To make the lentivirus, plentiCRISPR V2-Blast-gRNAi22 and plentiGuide-Hygro-iRFP670-gRNAi23 were separately co-transfected with packaging plasmid psPAX2 and envelope plasmid pMD2.G into 293FT cells (Thermo Scientific). Packaged lentiviruses were collected from the supernatant. Mouse iPSCs were then transfected with lenti-CRISPR V2-blast-gRNAi22, lenti-Hygro-iRFP670-gRNAi23, and control (empty vector: lenti-CRISPR V2-blast and lenti-Hygrp-iRFP670). Transfected miPSCs were selected in mES medium containing 2.5 µg/mL blasticidin (Sigma) and100 µg/mL hygromycin B (Invitrogen) for 72 h after nucleofection and maintained until large resistant colonies became visible.
A ~1 kb 5′ homology arm and a ~0.8 kb 3′ homology arm were synthesized (BIOMATIK) and cloned into a homologous-directed recombination targeting vector (System Biosciences, HR210-PA-1) at 5́ and 3́ multiple cloning sites separately.
1.5 μg of homologous-directed recombination targeting vector was transfected into 1x 105 Cas9/gRNA-infected iPSCs by electroporation at 1200 V 40 ms (one pulses). The HDR integrated iPSCs were selected in mES medium containing 1 μg/mL puromycin (MP Biomedicals) for 48 h. The LoxP flanked selection cassette was removed with Cre recombinase mRNA (MACS, 130-101-113) using Lipofectamine™ MessengerMAX™ Transfection Reagent (ThermoFisher). Finally, the iPSC were treated with 5 µg/mL ganciclovir (ThermoFisher) for 48 h to kill cells carrying the cassette, which express the hsv-tk gene.
2.5. Differentiation of mouse iPS cells to myogenic progenitor cells (MPC) via enhanced MyoD variant
Plasmid lenti-TRE-VP64-mouse-MyoD-T2A-dsRedExpress2 (Addgene, 60625, a gift from Charles Gersbach [26] was co-transfected with packaging plasmid psPAX2 and envelope plasmid pMD2.G into 293FT cells. Packaged lentiviruses were collected from the supernatant. For transfection, the cell medium was replaced with viral supernatant supplemented with 4 μg/mL Polybrene. The viral supernatant was changed 72 h later. Then cells were selected in 1 μg/mL puromycin to obtain a pure population of transfected cells. Cells were expanded in standard growth medium supplemented with puromycin. Selected cells were grown to confluence, and MyoD transgene expression was induced by supplementing the medium with 3 μg/mL doxycycline (Fisher BioReagents) for at least ten days. Cells were given fresh media supplemented with doxycycline every two days.
2.6. Cell-based genotyping
Genomic DNA was extracted from miPSCsCas9-Ctrl, miPSCsCas9-gRNA-HDR and miPSCsCas9-gRNA-HDR-CRE using QIAamp DNA Blood Mini Kit (QIAGEN), following the manufacturer’s protocol. Extracted DNA was amplified using the primers listed in Table 1 (Exon23, β-actin, copGFP HR210) and PCR was performed on a T100TM Thermal Cycler (BIO-RAD) with PrimeSTAR Max Premix (Takara) using the following protocol: 98 °C for 1 min, 35 cycles of 98 °C for 10 s, 60 °C for 15 s, 72 °C for 30 s, and a final extension at 72 °C for 1 min. The detection of the resulting PCR products was performed by conventional 2% agarose gel electrophoresis in 1 × TAE buffer.
Table 1.
Primer sequence.
| PCR primers | |
| Exon23-Forward | 5′-CCAAGAAAGCACCTTCAGAAATATG-3′ |
| Exon23-Reverse | 5′-TTTGGCAGCTTTCCACCA-3′ |
| β-actin-Forward | 5′-CCGTAAAGACCTCTATGCCAAC-3′ |
| β-actin-Reverse | 5′-AGGAGCCAGAGCAGTAATCT-3′ |
| copGFP HR210-Forward | 5′-TCTACCACTTCGGCACCTA-3′ |
| copGFP HR210-Reverse | 5′-CGGATGATCTTGTCGGTGAA-3′ |
| MYF5-Forward | 5′-GGTAGCAGGCTGTGAGTTG-3′ |
| MYF5-Reverse | 5′-GATTGCTTGTCCAGCATTGTG-3′ |
| Nanog-Forward | 5′-AACCAAAGGATGAAGTGCAAGCGG-3′ |
| Nanog-Reverse | 5′-TCCAAGTTGGGTTGGTCCAAGTCT-3′ |
| GAPDH-Forward | 5′-TGACAAGCTTCCCATTCTCG-3′ |
| GAPDH-Reverse | 5′-CCCTTCATTGACCTCAACTACAT-3′ |
| DMD3′HDR-Forward | 5′-CGATCCCGTGCCACCTT-3′ |
| DMD3′HDR-Reverse | 5′-GGGAAGGAAATATGGCAGAAATTAAACA-3′ |
| DMD5′HDR-Forward | 5′-ACATGTCTTATCAGTCAAGAGATCA-3′ |
| DMD5′HDR-Reverse | 5′-CGAGAAGCGTTCAGAGGAAA-3′ |
| DMD-PostCRE-Forward | 5′-ACATGTCTTATCAGTCAAGAGATCA-3′ |
| DMD-PostCRE-Reverse | 5′-AGAAGTCAATGTAGGGAAGGAAATA-3′ |
2.7. Sanger DNA sequencing
PCR products corresponding to miPSCsCas9-Ctrl, miPSCsCas9-gRNA-HDR and miPSCsCas9-gRNA-HDR-CRE were purified using the QIAEX II Gel Extraction Kit (QIAGEN, 20051), according to the manufacturer’s recommendations, and were ligated to the pCR™ 2.1-TOPO® TA vector using TOPO® TA Cloning® Kits (Invitrogen). Stellar chemically competent cells (Takara) were transformed with the products of ligation after overnight incubation at 37 ◦C in an agar plate with 100 µg/mL carbenicillin (Millipore) and 64 µl 25 mg/ml X-GaL (MilliporeSigMa). The white clones were selected randomly and grown in 5 ml of terrific broth medium (Fisher BioReagents) supplemented with 100 µg/ml carbenicillin. Plasmids were extracted using the QIAprep Spin Miniprep Kit (QIAGEN) following the protocol provided by the manufacturer. The selected inserts were subjected to Sanger sequencing service (GENEWIZ).
2.8. qRT-PCR
For qRT-PCR, total RNA was extracted from cells using RNAzol (Molecular Research Center, INC), and approximately 1 µg of total RNA was used for cDNA synthesis using the RevertAid RT Reverse Transcription Kit (Thermo scientific) according to the manufacturer's instructions. Real-time qPCR was performed on a CFX96TM Real-Time System (BIO-RAD) with PowerUP SYBR Green Master Mix (Thermo Scientific, A25742) and the primers indicated in Table 1 (MYF5, Nanog, GAPDH) using the following protocol: 50 °C for 2 min, 95 °C for 2 min, 50 cycles of 95 °C for 15 s, 60 °C for 1 min. All samples were normalized to glyceraldehyde 3-phosphate dehydrogenase GAPDH and conducted in triplicate.
2.9. MPC transplantation
Male immunodeficient Prkdcscid Dmdmdx mice (Jackson Laboratory) were anesthetized with ketamine/xylazine (HENRY SCHEIN ANIMAL HEALTH). A 30 μl solution containing 5 × 105 MPCs from iPSCCas9-Ctrl, iPSCCas9-gRNA-HDR or iPSCCas9-gRNA-HDR-CRE in DMEM was injected into the tibialis anterior (TA) muscle of each 8-week-old mouse. Four weeks post-cell transplantation, TA muscles were harvested and fixed in 4% paraformaldehyde. Samples were OCT-embedded and cut into 5 µm sections using a cryostat and processed with immunofluorescence staining. All animal handling and surgical procedures were performed by a protocol approved by the Augusta University Institutional Animal Care and Use Committee (IACUC). Mice were fed a standard diet and water ad libitum.
2.10. Immunofluorescence assays and confocal imaging
For cell staining, cells were plated on Corning BioCoat Poly-D-Lysine/Laminin Culture Slides (CORNING) and fixed with 4% paraformaldehyde. The slides were blocked with 5% goat serum for one hour, followed by mouse IgG-blocking solution from the M.O.M. kit (Vector Laboratories) according to the manufacturer’s instruction. The cells were then incubated with mouse anti-stage-specific embryonic antigen-1 (SSEA1) antibody (1:100; Cell Signaling), rabbit anti-homolog A (Lin28) antibody (1:400; Cell Signaling), rabbit anti-SRY-box 2 (SOX2) antibody (1:500; Abcam), rabbit anti-MyoD antibody (1:50; Santa Cruz), rabbit anti-Myogenic factor 6 (MYF6) antibody (1:200; AVIVA SYSTEMS BIOLOGY), or rabbit anti-dystrophin antibody (1:300; Thermo) at 4 °C overnight. Primary antibodies were resolved via secondary staining with Alexa488-conjugated goat-anti-mouse antibody (1:400, Invitrogen) or Alexa555-conjugated goat-anti-rabbit antibody (1:400, Invitrogen). Slides were mounted using HardSet Antifade Mounting Medium with DAPI (Vector Laboratories).
For tissue staining to assess the pluripotency of iPS cells generated from Dmdmdx mouse skin fibroblast cells, two weeks post cell therapy, mouse teratomas were harvested, embedded in OCT compound, snap frozen, cut into 5-μm sections, fixed with 4% paraformaldehyde, blocked with 5% goat serum for 1 h and immunostained with rabbit anti-α-fetoprotein (AFP) antibody (1:50; Thermo), rabbit anti-α-smooth muscle actin antibody (1:50; Cell Signaling) or rabbit anti-tyrosine hydroxylase (TH) antibody (1:50; SANTA CRUZ, sc-14007) at 4 °C overnight. Primary antibodies were resolved via secondary staining with the Alexa555-conjugated goat-anti-rabbit antibody (1:400, Invitrogen). Nuclei were counterstained with DAPI (Vector Laboratories).
To detect dystrophin expression in Dmdmdx mouse models, we remove TA muscles from immunodeficient Dmdmdx mouse 4 weeks after the injection of dox-induced MPCs from iPSCCas9-Ctrl, iPSCCas9-gRNA-HDR or iPSCCas9-gRNA-HDR-CRE. Tissues were embedded in OCT compound, snap frozen, cut into 5-μm sections, fixed with 4% paraformaldehyde, blocked with 5% goat serum for 1 h and immunostained with rabbit anti-dystrophin (1:300; Thermo) antibody. Primary antibodies were resolved via secondary staining with the Alexa555-conjugated goat-anti-rabbit antibody (1:400, Invitrogen). Nuclei were counterstained with DAPI (Vector Laboratories). All imaging was performed on a Zeiss 780 upright confocal microscope (Carl Zeiss).
2.11. Statistical analyses
Data are expressed as mean ± standard deviation (SD). Comparisons between groups were made by one-way analysis of variance, P < 0.05 was considered statistically significant.
3. Results
3.1. Generation and phenotypic characterization of Dmdmdx mouse skin fibroblast cells-derived induced pluripotent stem cells (iPSC)
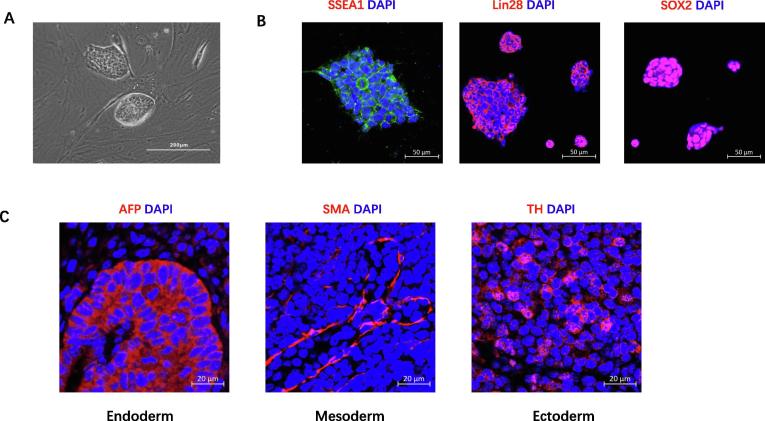
To generate iPSC cells, we used integration-free reprogramming vectors expressing KOS, hc-Myc, and hKlf4 to reprogram skin fibroblast from Dmdmdx mice. Colonies with mouse ES dome-shaped morphology were selected and expanded. The colonies exhibited large nuclear-cytoplasmic ratios, defined borders, and prominent nucleoli (Fig. 1A). The iPSC colonies stained positively for SSEA1, Lin28 and SOX2, pluripotency markers for mouse embryonic stem cells, by immunofluorescent microscopy (Fig. 1B). To investigate whether skin fibroblast cell-derived iPS cells can differentiate into cell types representing the three germ layers, we injected iPS cells into the muscle of NOD/SCID mice. After two weeks, the tumor mass (teratoma) was harvested and processed for histology. Within the teratomas, immunostaining demonstrated that the iPSC had differentiated into cells representative of all three germ layers in vivo, including liver cells of endoderm (AFP), smooth muscle cells of mesoderm (SMA), and adrenergic neuron cells of ectoderm (TH) (Fig. 1C).
Fig. 1.
Characterization of Dmdmdx skin fibroblast-derived iPS cells. (A) Representative image of ES-like colonies (scale bar = 200 µm); (B) Immunostaining of iPS cells for the pluripotency markers SSEA1 (green), Lin28 (red), SOX2 (red). Nuclei were stained with DAPI (blue) (scale bar = 50 µm); (C) Immunofluorescent staining for AFP (endoderm), SMA (mesoderm), and tyrosine hydrolase (TH) (ectoderm) in teratoma 2 weeks after iPSC injection (scale bar = 20 µm). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2. DMD exon23 deletion mediated by CRISPR/Cas9 and homologous-directed recombination (HDR) targeting vector system
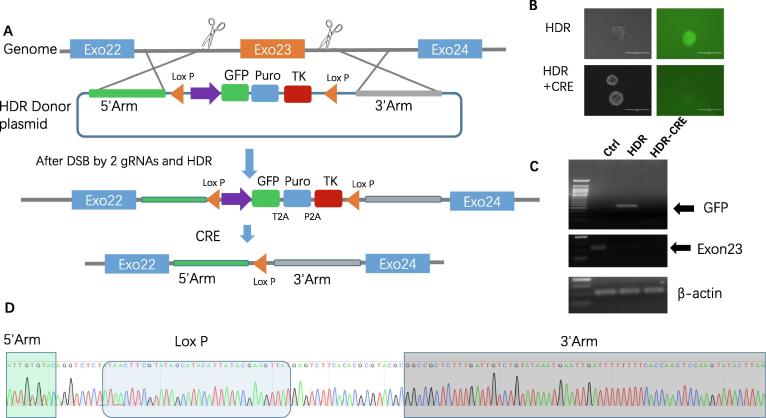
We designed two gRNAs that flank the mutant exon23 after Cas9-mediated double-stranded breaks (DSB) (Fig. 2A). Next, we incorporated the homology arms at the 5́ and 3́ end of the cutting site into a floxed EF-1 promoter-driven triple marker selection cassette (LoxP + Insulator Sequence + EF-1 + GFP-T2A-Puro-P2A-HSV-TK-PolyA + Insulator Sequence + LoxP) and transfected the vector into iPSC for HDR-based targeting to exon23 (Fig. 2A) through electroporation. The EF-1 promoter-driven GFP and Puro selection markers can facilitate the selection of HDR-modified iPSC (Fig. 2B), followed by the use of Cre recombinase mRNA to remove the floxed selection cassette (Fig. 2A). Finally, ganciclovir treatment was used to kill those cells without the cassette excision via HSV-tk negative selection. Genomic DNAs extracted from Dmdmdx mouse iPSC Cas9-Ctrl, iPSC Cas9-gRNA-HDR, and iPSC Cas9-gRNA-HDR-CRE were subjected to PCR genotyping. Fig. 2B demonstrated that iPSC Cas9-gRNA-HDR express GFP while the GFP expression loses post-CRE treatment. Consistently, PCR genotyping shows iPSC Cas9-gRNA-HDR have GFP fragment insertion while GFP fragment disappears after CRE treatment, indicating successful homologous-directed recombination of the cassette as described above(Fig. 2C); iPSCCas9-gRNA-HDR and iPSCCas9-gRNA-HDR-CRE have exon 23 deletions, indicating appropriate genomic targeting.
Fig. 2.
Deletion of DMD exon23 using the combination of CRISPR/Cas9 and HDR. (A) Schematic diagram of CRISPR/Cas9 and HDR mediated exon23 deletions. Step1: The Cas9 nuclease targets intron 22 and intron 23 by two gRNAs. Double-stranded breaks (DSBs) by Cas9 results in the excision of the mutant exon23. Step 2: Through HDR, a portion of the open reading frame (ORF) is replaced with the selection cassette containing GFP and puromycin resistance markers. step 3: The dual-marker cassette, flanked by loxP sites, can be excised from the genome by Cre recombinase. (B) Sorted mouse iPSCsCas9-gRNA-HDR and iPSCsCas9-gRNA-HDR-CRE were imaged by fluorescent microscopy using a GFP filter (scale bar = 200 µm). (C) PCR genotyping analysis of GFP and exon23. The arrows indicate the PCR products of GFP and exon23; β-actin serves as a reference. (D) Sequence results from a single clone confirm the deletion of mutant exon23 after the precise integration of the EF-1 driven reporter into the genome. Only the Lox P site remained after Cre recombinase treatment.
Furthermore, to demonstrate unequivocally that the selection cassette was removed without leaving footprint mutations in the genome, we performed Sanger sequencing. Fig. 2D shows that exon23 of dystrophin was deleted and only one Lox P insert remained after Cre recombinase mRNA treatment. By Sanger sequencing, 11 out of a total of 12 clones prior to Cre recombinase mRNA treatment had cassette integration, while 9 out of 12 clones exhibited deletion of the cassette following Cre recombinase treatment, indicating that the use of the HDR donor vector improves the efficiency of isolating correctly engineered clones.
3.3. Myogenic progenitor cell (MPC) differentiation of Dmdmdx mouse skin fibroblast-derived iPS cells
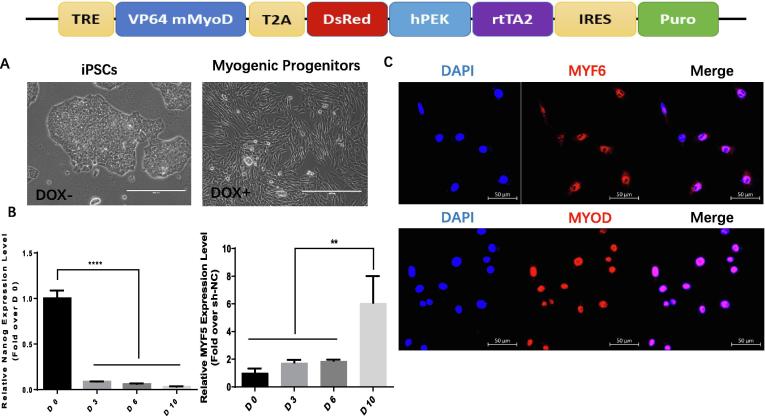
The tetracycline-inducible MyoD expression Tet-ON system was used to induce myogenic differentiation of miPSCs. Fig. 3A shows the morphology of the iPSCs transfected lenti-TRE-VP64-mouse-MyoD-T2A-dsRedExpress2 with or without doxycycline (Dox) treatment to induce MyoD expression. qRT-PCR showed that the mRNA level of Nanog (a pluripotent marker) rapidly decreased, while the expression of MYF5, a skeletal muscle marker, gradually increased after Dox induction (Fig. 3B). Moreover, immunofluorescent staining verified the expression of MYF6 and MyoD in the Dox-induced MPCs (Fig. 3C).
Fig. 3.
Differentiation of mouse iPSCs into the myogenic lineage. (Upper panel) the scheme of LV-TRE-VP64-mMyoD-T2A-dsRed-hPEK-rtTA2-IRES-Puro construct, in which the expression of VP64 mouse MyoD fusion protein is driven by a tetracycline-responsive promoter element (TRE), and reverse transactivator (rtTA2) is driven by human PEK promoter; (A) Representative image of mouse iPSCs (scale bar = 200 µm) and Dox-induced MPCs from mouse iPSCs (scale bar = 400 µm). (B) qRT-PCR depicting the time course of Nanog and MYF5 expression (mRNA) in Dox-treated iPSCsCas9-gRNA-HDR-CRE (****P < 0.0001, D0 vs D3, D6 and D10, n = 3 for Nanog) (**P < 0.01 D10 vs D0, D3 and D6, n = 3 for MYF5). (C) Immunofluorescent analysis of MYF6 and MYOD in Dox-induced MPCs from mouse iPSCs (scale bar = 50 µm).
3.4. Restoring dystrophin expression in MPC in vitro and in vivo
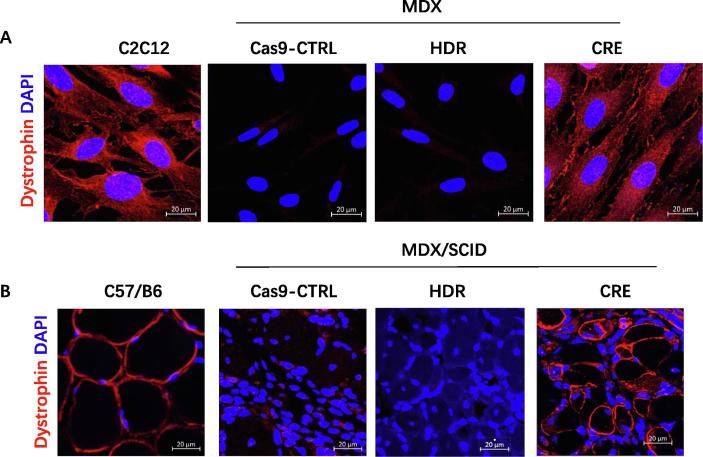
To examine whether MPCs derived from iPSCCas9-gRNA-HDR-CRE could restore the expression of dystrophin protein in cell culture, we performed immunofluorescent staining. As shown in Fig. 4A, dystrophin expression was detected in MPC derived from iPSCCas9-gRNA-HDR-CRE, but not in iPSCCas9-Ctrl or iPSCCas9-gRNA-HDR; C2C12, a wild-type mouse MPC cell line was used a positive control.
Fig. 4.
Restoration of dystrophin expression in vitro and in vivo. (A) Immunofluorescent analysis of dystrophin expression in C2C12 cell line and Dox-induced MPCs from miPSC Cas9-Ctrl, miPSCCas9-gRNA-HDR, and miPSCCas9-gRNA-HDR-CRE (scale bar = 20 µm). (B) Immunofluorescent analysis of dystrophin expression in TA muscles from C57/B6 and MDX/SCID mice 4 weeks after injection with Dox-induced MPCs from miPSCCas9-Ctrl, miPSCCas9-gRNA-HDR and miPSCCas9-gRNA-HDR-CRE (scale bar = 20 µm).
To determine whether cell transplantation with MPCs from iPSCCas9-gRNA-HDR-CRE could restore the expression of dystrophin protein in the muscle of Mdx mice, we intramuscularly injected equivalent numbers of MPCs from iPSCCas9-Ctrl, iPSCCas9-gRNA-HDR or iPSCCas9-gRNA-HDR-CRE into the TA muscle of 8-week-old MDX/SCID mice. Four weeks after cell transplantation, we observed restored dystrophin expression in the muscles injected with MPCs from iPSCCas9-gRNA-HDR-CRE, but not in the control groups; TA muscle from normal wild type C57/B6 mice was used as a positive control for dystrophin staining (Fig. 4B). The regenerated muscle fibers from donor MPC are variable in fiber size and relatively small as compared with the size of muscle cells from wide type mice (C57/B6). These centronucleated regenerating fibers are expressing dystrophin and infiltrated by inflammatory cells. Many dystrophin-expressing muscle fibers have multiple nuclear, suggesting cell fusion, which can be fusion with donor cells or fusion with recipient cells. Since the dystrophin expressing fibers only appeared locally at the injected site; therefore, the percentage of revertant fibers in the Mdx mice is still quite low (~5%).
4. Discussion
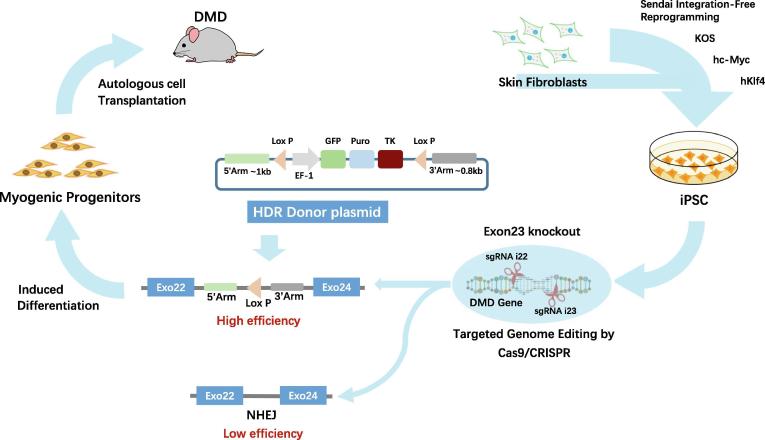
DMD is a severe X-linked myopathy caused by dystrophin deficiency, which ultimately leads to exhaustion of muscle stem cells and impaired muscle contractility. CRISPR/Cas9 gene-editing provides the potential to correct DMD gene defects and restore dystrophin expression in iPSC-derived MPC for autologous cell transplantation without eliciting immune activation. In this study, we demonstrated that the use of HDR donor vector and CRISPR/Cas9 technology to replace mutant exon23 in iPSC, coupled with MPC differentiation and MPC transplantation, can lead to the restoration of dystrophin in Mdx muscles. This strategy provides a viable approach for autologous stem/progenitor cell transplantation to treat DMD patients by restoring dystrophin expression in affected muscles (Fig. 5).
Fig. 5.
Proposed schematic of development and transplantation of autologous iPSC-derived myogenic progenitor cells with CRISPR/Cas9-mediated precise gene correction for treating muscular dystrophy in mice.
Immature C2C12 cells usually do not express dystrophin. However, many evidence supports that C2C12 expresses dystrophin, for examples, Wein et al. [27] performed the assay in C2C12 mouse myoblasts which express dystrophin, and Li et al. [28] reported that myotubes that had strong dystrophin expression in C2C12 cells plated at high density. These results suggest that sufficient cell-cell contact favors cell fusion and, therefore, myotube formation.
In our cell culture experiments, we cultured cells at high density and with 10% FBS, other than 20% FBS, as shown in Fig. 4A, there is enough cell-cell contact, and our staining clearly shows Dystrophin expression on the cell membrane of contacted cells.
Induced MyoD overexpression is a reproducible approach to induce myogenic differentiation of pluripotent stem cells; the differentiation will not be stopped at the progenitor stage. Uchimura et al. [29] used a tetracycline-inducible MyoD overexpression model of myogenic differentiation in human iPSCs (hiPSCs). In addition, Amilon et al. [30] use LV-TRE-WT human MyoD-T2A-dsRedExpress2 system to generate functional myocytes and observed the formation of myotubes that stained for MyHC. In our experiments, we use a similar system to induce MyoD to overexpress in iPSCs to activate muscle differentiation programs in stem cells. Therefore, the MPC derived from MyoD-programmed iPSC can include myogenic progenitors at early, intermediate, and later stages.
Here we report that a lentiviral vector-based Cas9 system using double-cut sgRNAs and HDR based donor vector leads to high-level HDR-mediated precision replacement with the integration of EF-1 promoter-driven GFP, puromycin resistant gene, and HSV-TK cassette in iPSCs. We previously showed that the dual cuts by dual-gRNAs have low efficiency in the deletion of mutant exon23 by non-homologous end joining (NHEJ) repair. In this study, we found that HDR-mediated gene targeting by insertion of the reporter at the targeted gene locus can efficiently enrich iPSC clones with the deletion of mutant exon23.
In our previous study, we tried to use two gRNAs to delete mutant Exon23 from iPSCMdx. However, the efficiency of NHEJ repair in mouse iPSC was extremely low (approximately 3%). To circumvent the tedious process of clonal selection and genotyping, we utilized HDR-mediated CRISPR/Ca9 gene-editing strategy, in which GFP, puromycin resistant gene, and herpes simplex virus thymidine kinase (HSV-TK, driven by EF-1 promoter) is engineered into the vector. With the HDR design, the 1 kb 5′ homology arm (HA) is identical to sequences surrounding DSB created by i22 gRNA, and the 0.8 kb 3′ HA is identical to sequences surrounding DSB created by i23 gRNA. In this study, we chose an EF-1 promoter donor HDR reporter system rather than a promoterless donor HDR reporter system because the DMD gene only expresses in muscle progenitor cells and mature muscle cells, not in undifferentiated pluripotent stem cells; thus, a promoterless HDR reporter system would not permit selection of iPSC in which exon23 was correctly replaced. Insertion of the EF-1 promoter into the target locus drives transgene expression and allows us to select correct clones by FACS or with antibiotic administration. Most importantly, the HDR donor is flanked by two LoxP sites, which make removing the HDR donor feasible by Cre/LoxP recombination. By taking advantage of the triple selection strategy, we achieved efficient exon23 deletion in our cell system. It is important to point out that in designing the HDR donor, it is necessary to mutate the PAM sequence or gRNA seed sequence in 5′HA and 3′HA to prevent inserted HDR deletion by the activated CRISPR/Cas9 system.
Since iPSCs have been widely used for heart and skeletal muscle regeneration, the addition of this HDR donor plasmid step potentially can ease the burden of correctly enriching iPSC clones with proper gene editing, thereby improving the effectiveness of corrective stem cell therapy for genetic disorders.
In conclusion, employing an HDR donor carrying a triple selection cassette can markedly enhance the selection of correctly edited iPSC clones, and the knock-in cassette can be easily removed by Cre recombination. Therefore, an HDR-mediated replacement, insertion, and removal strategy can enhance the efficiency of CRISPR/Cas9 genomic editing of iPSC.
In this study, we demonstrated technology for obtaining iPSC through reprogramming, using HDR-based CRISPR-Cas9 to restore the function of the dystrophin gene, and using inducible myogenic transcription factor to induce muscle differentiation in iPSC. Although muscle progenitor cell transplantation is considered as a potential strategy to replenish stem cell pool in DMD patients, and MPC transplantation have been tested in clinical trials to treat patients with DMD, however, most of these trial failed due to rapid cell death, poor cell engraftment in diseased host muscles, and immune rejection [31], [32], [33], [34]. To improve the survival and engraftment of donor cells in Mdx mice, Muir et al. [35] reported that there is a 70% reduction in donor cells at day 5 and a 94% reduction by day 28 after transplantation into the muscle of Mdx mice, and they tested a prosurvival cocktail which includes heat shock followed by treatment of insulin-like growth factor-1, a caspase inhibitor, a Bcl-XL peptide, a KATP channel opener, basic fibroblast growth factor, Matrigel, and cyclosporine A, and found about a three-fold increase in donor cells in early engraftment in transplanted muscles. In our study, although we used IPSC-derived MPSC, we observed a 5% dystrophin-expressing donor cells in the recipient’s Mdx muscle at four weeks after cell transplantation, which is consistent to Muir LA’s finding. Since the purpose of this study is to use HDR to improve the efficiency of Cas9-mediated gene editing in iPSCs, and we will test the prosurvival cocktail to improve donor cell survival in Mdx muscles in our future study.
CRediT authorship contribution statement
Yue Jin: Investigation, Formal analysis, Data curation. Yan Shen: Investigation. Xuan Su: Investigation. Neal L. Weintraub: Writing - review & editing. Yaoliang Tang: Conceptualization, Methodology, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Tang and Weintraub were partially supported by NIH-AR070029, NIH-HL086555, NIH-HL134354.
References
- 1.Prevalence of Duchenne/Becker muscular dystrophy among males aged 5-24 years – four states, 2007. MMWR Morb Mortal Wkly Rep. 2009;58(40):1119–22. [PubMed]
- 2.Serrano A.L., Munoz-Canoves P. Fibrosis development in early-onset muscular dystrophies: mechanisms and translational implications. Semin Cell Dev Biol. 2017;64:181–190. doi: 10.1016/j.semcdb.2016.09.013. [DOI] [PubMed] [Google Scholar]
- 3.Mosqueira M., Zeiger U., Forderer M., Brinkmeier H., Fink R.H. Cardiac and respiratory dysfunction in Duchenne muscular dystrophy and the role of second messengers. Med Res Rev. 2013;33(5):1174–1213. doi: 10.1002/med.21279. [DOI] [PubMed] [Google Scholar]
- 4.Miyatake S., Shimizu-Motohashi Y., Takeda S., Aoki Y. Anti-inflammatory drugs for Duchenne muscular dystrophy: focus on skeletal muscle-releasing factors. Drug Des Devel Ther. 2016;10:2745–2758. doi: 10.2147/DDDT.S110163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aslesh T., Maruyama R., Yokota T. Skipping multiple exons to treat DMD-promises and challenges. Biomedicines. 2018;6(1) doi: 10.3390/biomedicines6010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Voit T., Topaloglu H., Straub V., Muntoni F., Deconinck N., Campion G. Safety and efficacy of drisapersen for the treatment of Duchenne muscular dystrophy (DEMAND II): an exploratory, randomised, placebo-controlled phase 2 study. Lancet Neurol. 2014;13(10):987–996. doi: 10.1016/S1474-4422(14)70195-4. [DOI] [PubMed] [Google Scholar]
- 7.Kole R., Krieg A.M. Exon skipping therapy for Duchenne muscular dystrophy. Adv Drug Deliv Rev. 2015;87:104–107. doi: 10.1016/j.addr.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 8.Gruntman A.M., Bish L.T., Mueller C., Sweeney H.L., Flotte T.R., Gao G. Gene transfer in skeletal and cardiac muscle using recombinant adeno-associated virus. Curr Protoc Microbiol. 2013 doi: 10.1002/9780471729259.mc14d03s28. Chapter 14:Unit 14D.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duan D., Systemic A.A.V. Micro-dystrophin gene therapy for duchenne muscular dystrophy. Mol Ther. 2018;26(10):2337–2356. doi: 10.1016/j.ymthe.2018.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dupont J.B., Tournaire B., Georger C., Marolleau B., Jeanson-Leh L., Ledevin M. Short-lived recombinant adeno-associated virus transgene expression in dystrophic muscle is associated with oxidative damage to transgene mRNA. Mol Ther Methods Clin Dev. 2015;2:15010. doi: 10.1038/mtm.2015.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zygmunt D.A., Crowe K.E., Flanigan K.M., Martin P.T. Comparison of serum rAAV serotype-specific antibodies in patients with duchenne muscular dystrophy, becker muscular dystrophy, inclusion body myositis, or GNE myopathy. Hum Gene Ther. 2017;28(9):737–746. doi: 10.1089/hum.2016.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang F., Wen Y., Guo X. CRISPR/Cas9 for genome editing: progress, implications and challenges. Hum Mol Genet. 2014;23(R1):R40–R46. doi: 10.1093/hmg/ddu125. [DOI] [PubMed] [Google Scholar]
- 13.El Refaey M., Xu L., Gao Y., Canan B.D., Adesanya T.M.A., Warner S.C. In vivo genome editing restores dystrophin expression and cardiac function in dystrophic mice. Circ Res. 2017;121(8):923–929. doi: 10.1161/CIRCRESAHA.117.310996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nelson C.E., Hakim C.H., Ousterout D.G., Thakore P.I., Moreb E.A., Castellanos Rivera R.M. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science. 2016;351(6271):403–407. doi: 10.1126/science.aad5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bengtsson N.E., Hall J.K., Odom G.L., Phelps M.P., Andrus C.R., Hawkins R.D. Muscle-specific CRISPR/Cas9 dystrophin gene editing ameliorates pathophysiology in a mouse model for Duchenne muscular dystrophy. Nat Commun. 2017;8:14454. doi: 10.1038/ncomms14454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Verhaart I.E.C., Aartsma-Rus A. Therapeutic developments for Duchenne muscular dystrophy. Nat Rev Neurol. 2019;15(7):373–386. doi: 10.1038/s41582-019-0203-3. [DOI] [PubMed] [Google Scholar]
- 17.Dumont N.A., Rudnicki M.A. Targeting muscle stem cell intrinsic defects to treat Duchenne muscular dystrophy. NPJ Regen Med. 2016;1. doi: 10.1038/npjregenmed.2016.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu A., Poddar M., Tang Y., Proto J.D., Sohn J., Mu X. Rapid depletion of muscle progenitor cells in dystrophic mdx/utrophin-/- mice. Hum Mol Genet. 2014;23(18):4786–4800. doi: 10.1093/hmg/ddu194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okita K., Nakagawa M., Hyenjong H., Ichisaka T., Yamanaka S. Generation of mouse induced pluripotent stem cells without viral vectors. Science. 2008;322(5903):949–953. doi: 10.1126/science.1164270. [DOI] [PubMed] [Google Scholar]
- 20.Uchimura T., Otomo J., Sato M., Sakurai H. A human iPS cell myogenic differentiation system permitting high-throughput drug screening. Stem Cell Res. 2017;25:98–106. doi: 10.1016/j.scr.2017.10.023. [DOI] [PubMed] [Google Scholar]
- 21.Shoji E., Woltjen K., Sakurai H. Directed myogenic differentiation of human induced pluripotent stem cells. Methods Mol Biol (Clifton, NJ) 2016;1353:89–99. doi: 10.1007/7651_2015_257. [DOI] [PubMed] [Google Scholar]
- 22.Yoshida Y., Yamanaka S. Induced pluripotent stem cells 10 years later: for cardiac applications. Circ Res. 2017;120(12):1958–1968. doi: 10.1161/CIRCRESAHA.117.311080. [DOI] [PubMed] [Google Scholar]
- 23.Conboy I., Murthy N., Etienne J., Robinson Z. Making gene editing a therapeutic reality. F1000Res. 2018;7 doi: 10.12688/f1000research.16106.1. F1000 Faculty Rev-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang B., Li K., Wang A., Reiser M., Saunders T., Lockey R.F. Highly efficient CRISPR/HDR-mediated knock-in for mouse embryonic stem cells and zygotes. Biotechniques. 2015;59(4) doi: 10.2144/000114339. pp. 201–2, 4, 6–8. [DOI] [PubMed] [Google Scholar]
- 25.Ho S.M., Hartley B.J., Flaherty E., Rajarajan P., Abdelaal R., Obiorah I. Evaluating synthetic activation and repression of neuropsychiatric-related genes in hiPSC-derived NPCs, neurons, and astrocytes. Stem Cell Rep. 2017;9(2):615–628. doi: 10.1016/j.stemcr.2017.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kabadi A.M., Thakore P.I., Vockley C.M., Ousterout D.G., Gibson T.M., Guilak F. Enhanced MyoD-induced transdifferentiation to a myogenic lineage by fusion to a potent transactivation domain. ACS Synth Biol. 2015;4(6):689–699. doi: 10.1021/sb500322u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wein N., Vulin A., Falzarano M.S., Szigyarto C.A.-K., Maiti B., Findlay A. Translation from a DMD exon 5 IRES results in a functional dystrophin isoform that attenuates dystrophinopathy in humans and mice. Nat Med. 2014;20(9):992–1000. doi: 10.1038/nm.3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li B., Lin M., Tang Y., Wang B., Wang J.H. A novel functional assessment of the differentiation of micropatterned muscle cells. J Biomech. 2008;41(16):3349–3353. doi: 10.1016/j.jbiomech.2008.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uchimura T., Otomo J., Sato M., Sakurai H. A human iPS cell myogenic differentiation system permitting high-throughput drug screening. Stem Cell Res. 2017;25:98–106. doi: 10.1016/j.scr.2017.10.023. [DOI] [PubMed] [Google Scholar]
- 30.Amilon K.R., Cortes-Araya Y., Moore B., Lee S., Lillico S., Breton A. Generation of functional myocytes from equine induced pluripotent stem cells. Cell Reprogram. 2018;20(5):275–281. doi: 10.1089/cell.2018.0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chowdhury N., In A.A. Utero stem cell transplantation: potential therapeutic application for muscle diseases. Stem Cells Int. 2017;2017 doi: 10.1155/2017/3027520. 3027520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kallestad K.M., McLoon L.K. Defining the heterogeneity of skeletal muscle-derived side and main population cells isolated immediately ex vivo. J Cell Physiol. 2010;222(3):676–684. doi: 10.1002/jcp.21989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Awaya T., Kato T., Mizuno Y., Chang H., Niwa A., Umeda K. Selective development of myogenic mesenchymal cells from human embryonic and induced pluripotent stem cells. PLoS ONE. 2012;7(12):e51638-e. doi: 10.1371/journal.pone.0051638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Z., Chamberlain J.S., Tapscott S.J., Storb R. Gene therapy in large animal models of muscular dystrophy. ILAR J. 2009;50(2):187–198. doi: 10.1093/ilar.50.2.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muir L.A., Murry C.E., Chamberlain J.S. Prosurvival factors improve functional engraftment of myogenically converted dermal cells into dystrophic skeletal muscle. Stem Cells Dev. 2016;25(20):1559–1569. doi: 10.1089/scd.2016.0136. [DOI] [PMC free article] [PubMed] [Google Scholar]