Highlights
-
•
Healthy hippocampal spatio-temporal dynamics were mapped with functional MRI.
-
•
Temporal lobe epilepsy alters network dynamics of the hippocampus.
-
•
Hippocampal signal variance may affect network dynamics and alter static connectivity.
Keywords: Epilepsy, Fmri, Dynamics, Functional connectivity
Abbreviations: FC, functional connectivity; Fmri, functional Magnetic resonance imaging; TLE, temporal lobe epilepsy
Abstract
Mesial temporal lobe epilepsy (TLE) is characterized by transient abnormal electrical activity originating in the hippocampus. The objective of this study was to characterize dynamic spatio-temporal fluctuations in hippocampal network connectivity in mTLE using functional connectivity (FC) mapping in 41 unilateral mTLE patients (28 right, 13 left) and 56 healthy control participants using 3T MRI. Dynamic FC was computed across the scan using sliding 60-s windows. This was compared to static FC computed using the whole 10-min functional MRI scan, and to the variance in the hippocampal functional MRI signal. Four states of healthy hippocampal dynamic FC were identified and compared to TLE patients. TLE patients fluctuated between these four states, but the hippocampus ipsilateral to the seizure focus spent more time in a state distinguished by lower prefrontal and parietal FC than the dominant healthy state. Increased time spent in this state was associated with increased impairment in static FC and increased variance in the hippocampal functional MRI signal. Overall, this work provides evidence that increases in variance in signal fluctuations occurring at the seizure focus in the hippocampus in patients with mTLE may contribute to disruptions in healthy FC network dynamics within an fMRI scan that contribute to decreases in static hippocampal FC. These alterations result in decreased hippocampal connectivity to bilateral prefrontal and parietal regions in TLE which may be related to behavior and cognitive impairments in these patients. Therefore, characterization of an individual patient's hippocampal dynamics at different time scales may provide more specific spatio-temporal profiles of network impairment that may be related to hippocampal dysfunction in TLE.
1. Introduction
Mesial temporal lobe epilepsy (TLE) is characterized by transient abnormal electrical activity originating in the hippocampus and possibly other mesial temporal structures (Engel, 2001). This not only occurs during clinical seizures, but is also detected between seizures using scalp and intracranial electroencephalography (EEG) (Cendes et al., 2000; Tao et al., 2005). Quantifying how this activity affects the hippocampus and its relationship with the rest of the brain may elucidate mechanisms of hippocampal related functional and neurocognitive impairment (Helmstaedter and Elger, 2009) and inform treatment. The most direct method to map the hippocampal and related network activity is intracranial EEG, however this is not feasible to perform in healthy humans and lacks spatial resolution and coverage as compared to neuroimaging methods. A non-invasive method to indirectly quantify these effects with relatively high spatial resolution across the brain is Magnetic Resonance Imaging (MRI) functional connectivity (FC) network mapping (Rogers et al., 2007).
There have been many findings of altered functional networks in TLE using FC, including those specifically involving the seizure focus in the hippocampus (Bettus et al., 2010; Maccotta et al., 2013; Haneef et al., 2014; Englot et al., 2016). Functional network differences have been shown to relate to neurocognitive impairment (Holmes et al., 2014; Roger et al., 2019) and surgical treatment outcome (He et al., 2017; Morgan et al., 2017, 2019). The unstated assumption of these types of analyses is that the functional network measured is the average or static activity over the imaging session. However, it is known that healthy hippocampal function requires a complex and unique spatio-temporal framework of activation (Knierim, 2015; Cooper and Ritchey, 2019). Direct measurement of local field potentials of the hippocampus have detected three main types of rhythms involved in this process (Colgin, 2016) including theta (Buzsáki, 2002; Bohbot et al., 2017) (~4–12 Hz), sharp-wave ripple complexes (Bragin et al., 1999) (~80–160 Hz ripples superimposed on 0.01–3 Hz sharp waves), and gamma (~25–100 Hz) band oscillations (Bragin et al., 1995). Currently, no functional MRI (fMRI) methods can measure these signals. But, evidence shows that it is the coupling of these hippocampal oscillations across distributed brain regions that supports hippocampal functions such as the organization of memories in studies of rodents (Hyman et al., 2010; Myroshnychenko et al., 2017). And, while the oscillations occur at significantly higher frequencies, the phase coherence of the coupling of these signals fluctuates on the order of seconds. This suggests that dynamic fMRI network mapping (Lurie et al., 2020) within a MRI session may provide a more temporally specific network characterization, while still preserving its spatial resolution and coverage. In fact, dynamics in healthy hippocampal FC have been described across a scan session (Zhong et al., 2019). In TLE, it has been shown that the variance in FC across an MRI session is increased between the hippocampus and many individual regions including those in sensorimotor, frontal and parietal cortices compared to controls (Laufs et al., 2014), but the simultaneous temporal and spatial characteristics of the hippocampal functional networks have yet to be determined.
The purpose of this work is to examine the effects of TLE on hippocampal dynamics across distributed networks. To do this we took a systematic approach to individually examine several postulates. First, we quantified healthy hippocampal network spatio-temporal dynamics by identifying structured dynamic patterns of FC or transient “states” within the 10-min MRI session in a training set of control participants (Allen et al., 2014; Vidaurre et al., 2017). These were validated by comparing to an independent cohort of healthy controls. Second, we quantified the degree to which hippocampal networks in left and right TLE fluctuated between these healthy states. Third, we determined the relationship between dynamic FC network fluctuations and the static FC averaged over the 10-min MRI session. This would allow interpretation of existing literature of static FC in this context. Finally, the relationship between fMRI hippocampal signal fluctuations and deviation from healthy network states was quantified. This is based on the assumption that transient abnormal activity inherent in the hippocampus in TLE contributes to the changes in hippocampal network fluctuations. Taken together, the aim of these studies is to offer evidence to support the conclusion that TLE results in hippocampal fMRI signal fluctuations that are reflected in alterations in healthy dynamic oscillations of FC networks across the brain. And, that these dynamic alterations are associated with, but not completely characterized by, standard static FC changes across the network.
2. Materials and methods
2.1. Participants
This study included 41 suspected unilateral mesial TLE patients (28 right TLE, 13 left TLE) as determined by long-term video electroencephalography (EEG) of ictal events localizing to anterior/mesial temporal regions, unilateral mesial temporal lobe hypometabolism on positron emission tomography (PET), hippocampal sclerosis on standard MRI, and seizure semiology consistent with TLE (Table 1). Note that six patients did not have hippocampal sclerosis visually detected by clinician on MRI. However, in four of these cases, pathology on the resected specimen confirmed gliosis in the resected tissue. Of the remaining two patients, one of these had resection with an Engel I-d one year outcome. The other has not had resection. Exclusion criteria included structural abnormalities outside the mesial temporal lobe. For comparison, 56 healthy control participants were also enrolled in the study. These control participants were divided into two groups. One group contained a training set of 20 participants from which we computed the healthy dynamic FC states (9 M, age mean ± std = 37.8 ± 13.0 years). These were chosen randomly to be comparable in age to the remaining 36 control participants (23 M, age mean ± std = 41.1 ± 12.5 years) and the TLE patients. Patients were recruited from all patients from the Vanderbilt University Medical Center epilepsy program who met the inclusion criteria and agreed to participate between 2012 and 2019. The protocol was approved by the Vanderbilt University Institutional Review Board. All participants gave informed consent.
Table 1.
Patient characteristics.
| Right TLE (n = 28) | Left TLE (n = 13) | |
|---|---|---|
| M/F | 12/16 | 9/4 |
| Age (years: mean ± std) | 39.6 ± 10.3 | 37.6 ± 15.2 |
| Duration (years: mean ± std) | 20.1 ± 13.6 | 23.3 ± 16.4 |
| MRI HS or hippocampal gliosis on pathology (n,%) | 26, 93% | 13, 100% |
| Lateralizing PET hypometabolism (n,%) | 23, 82% | 10, 77% |
| Localizing video scalp EEG of ictal events (n,%) | 24, 85% | 12, 92% |
M = male; F = female; HS = hippocampal sclerosis including T2 hyperintensity.
2.2. Imaging
All participants underwent a 3T MRI session using a 32-channel head coil. Images acquired included: (1) T1-weighted MRI for inter-subject normalization and regional and tissue segmentation (1 mm3), (2) T1-weighted MRI for spatial normalization acquired in the same slice orientation as the functional images (1 × 1 × 3.5 mm with 0.5 mm gap), (3) T2*-weighted functional MRI (fMRI) Blood Oxygenation Level Dependent MRI at rest with eyes closed for functional connectivity (34 axial slices, echo time = 35 ms, repetition time = 2 s, 3 × 3 × 3.5 mm with a 0.5 mm gap, 10 min). The fMRI was performed twice sequentially on each participant – scan 1 and scan 2. Throughout this work, analyses were performed on scan 1, and then results were tested for within-subject reproducibility using scan 2. Using the MRI scanner integrated pulse oximeter and the respiratory belt, simultaneous physiological monitoring of cardiac and respiratory fluctuations was acquired at a sampling frequency of 500 Hz.
2.3. Static FC quantification
Functional MRI images were preprocessed as described in previous work (Morgan et al., 2019) and briefly described here. Using SPM8 software [http://www.fil.ion.ucl.ac.uk/spm/software/spm8/] and MATLAB 2019a (The MathWorks, Inc, Natick, MA), all functional MRI were preprocessed as follows: slice timing correction, motion correction, physiological noise correction using the Retrospective Correction of Physiological Motion Effects in functional MRI (RETROICOR) protocol (Glover et al., 2000) using the pulse oximeter and respiratory belt time series, spatial normalization to the Montreal Neurological Institute template via the T1-weighted datasets, and spatial smoothing using a 6 × 6 × 6 mm full width, half maximum Gaussian kernel. Then the fMRI time series were temporally band-pass filtered at 0.0067 Hz to 0.1 Hz (Cordes et al., 2001).
A total of 87 regions of interest were identified on the 1 mm3 T1-weighted images using the Multi-Atlas algorithm (Asman and Landman, 2013). This algorithm uses a set of manually labeled atlases (Neuromorphometrics, Inc. Somerville, MA, USA) that followed the brainCOLOR (https://mindboggle.info/braincolor/) segmentation. The accuracy of the algorithm has been found to be comparable to the FreeSurfer algorithm (Fischl, 2012) when manual segmentation is used as a gold standard, but is more robust to global failures than FreeSurfer in older adults and/or those with larger ventricles as in some of the patients in this cohort (Huo et al., 2016). Our regions included those across the frontal, parietal, temporal and occipital lobes, as well as subcortical structures and the right and left hippocampus. The preprocessed functional MRI time series were averaged across each region of interest and scaled by dividing by the mean of the first image. Then a partial Pearson correlation between the right hippocampus time series and the mean time series of each of the other 86 regions was computed using motion and mean white matter time series as confounds. The correlations were then transformed using the Fisher Z transform (Fisher, 1915). The right hippocampal FC was the resulting vector of connectivity values from the right hippocampus to each of the other 86 regions. This was then repeated to determine the left hippocampal FC for each participant for each fMRI scan. The standard static hippocampal FC was computed using the entire 10-min time series.
The static FC of the right and left hippocampus to each of the other 86 regions were compared across three groups using one-way ANOVA. In addition, the FC to the hippocampus was averaged across six left and right hemisphere groups of regions including prefrontal, parietal, occipital, temporal, motor/somatosensory, and subcortical regions (“lobe” analysis). The lobe FC values were also compared between groups using one-way ANOVA. Unless otherwise stated, the three groups were defined as controls (n = 36, excluding the training set), right TLE (n = 28) and left TLE (n = 13).
To enable comparisons of individual patients to static healthy control hippocampal FC, the mean and standard deviation of the hippocampal FC over all controls (both groups, n = 56) was computed. Then for each right and left TLE patient, the mean control FC was subtracted from the participant's FC and divided by the standard deviation. This resulted in a hippocampal static FC vector in units of standard deviation from control mean static FC. The sum of the absolute value of this vector represents the individual participant's hippocampal static FC difference from healthy control. This can be compared to measures of dynamic FC in future sections.
2.4. Dynamic FC quantification
The dynamic FC was then computed to quantify the temporal fluctuations of the hippocampal FC using the sliding window approach (Chang and Glover, 2010) with 60-s windows translated by one volume acquisition time (2 s). This resulted in the hippocampus FC for each window, similar to the whole brain FC dynamics described by Allen et al. (2014).
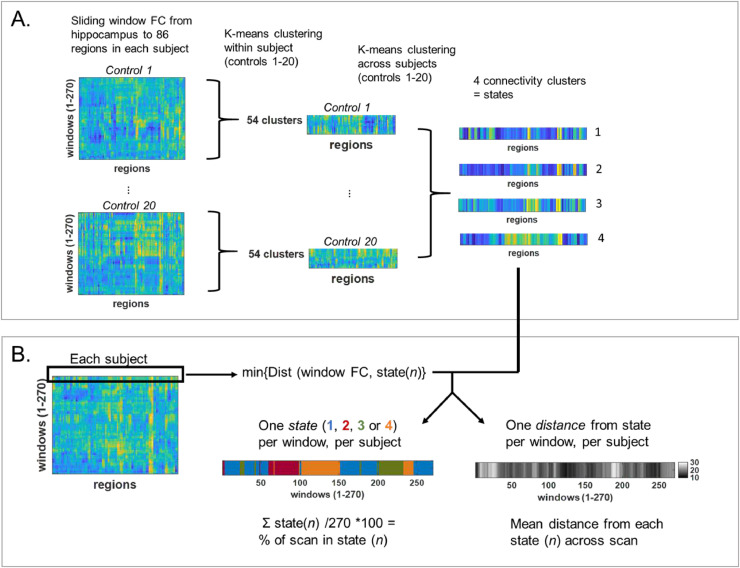
The first objective from these dynamic FC measures was to determine healthy dynamic spatio-temporal brain states in the training control participants. To compute the healthy states, a two-step approach (Allen et al., 2014) was implemented (Fig. 1A). In step 1, a within-subject analysis reduced the 270 windowed hippocampal FCs to 54 (5x reduction) using Matlab k-means clustering using the squared Euclidean norm metric in each participant. In step 2, a between-subject analysis combined the 54 hippocampal clusters from each participant by repeating the k-means clustering to compute four resulting hippocampal FC measures which will be referred to as healthy states (R1, R2, R3, R4). The process was repeated with the left hippocampus (L1, L2, L3, L4).
Fig. 1.
Dynamic FC processing. (A) Two-step k-means clustering approach to identify four hippocampal FC states across 20 healthy control participants using 60-s sliding windows. (B) Determination of brain state and Euclidean norm (distance) from state for each window of hippocampal FC in each participant. FC = functional connectivity.
The second objective was to quantify how well these states represented the windowed dynamic FC in a unique set of healthy controls and patients, as well as how much time within the scan each participant spent in the different states (Fig. 1B). To do this the dynamic windowed hippocampal FC was computed for each participant for each window. From each of these, the Euclidean norm to each of the four healthy states was determined. The participant was considered in the state with the minimum Euclidean norm (distance) at that window. For each participant, the percent of scan time spent in each window and the average distance from each state was quantified. This was repeated with the left hippocampus using the four left hippocampal healthy states. The healthy states were then also compared to the scan 2 data in each participant.
After identification of the four healthy dynamic FC states of the right hippocampus in the training set, it was important to investigate the generalizability of the states to other controls. To do this a two-sample t-test was used to compare the two groups of controls – the 20 training controls and the other 36. Two measures were compared: the mean distance to the state and percent of scan time spent in each state. One test was performed for each of the four states.
To determine if the identified control states accurately represented states in the three participant groups equally, the average distance to each state was compared across groups using one-way ANOVA (i.e. distance to state R1: controls vs. right TLE vs. left TLE). To determine whether the groups had different dynamic right hippocampal FC characteristics than the healthy controls, the percent of total scan time spent in each state was compared across groups using one-way ANOVA (i.e. time spent in state R1: controls vs. left TLE vs. right TLE). This was repeated for the left hippocampus and for scan 2 data.
Next, ipsilateral and contralateral networks were considered by combining both hippocampi. Time spent in each state was compared between controls (right and left hippocampus), ipsilateral networks (right hippocampus in right TLE group and left hippocampus in left TLE group) and contralateral networks (right hippocampus in left TLE group and left hippocampus in right TLE group) using one-way ANOVA for each state (i.e. state R1/L1: controls vs. ipsilateral vs. contralateral).
It was also hypothesized that changes in dynamic FC were associated with alterations detected in standard static FC. To investigate this the Spearman linear correlation between the time spent in each state and each participant's difference from static healthy hippocampal FC was computed across all patients. The analysis was repeated for left hippocampus and scan 2.
2.5. Hippocampal time series
Last, the relationship between the average hippocampal fMRI time series and hippocampal FC state was investigated. Based on the hypothesis that rapid fluctuations in hippocampal activity may drive state transitions, the variance (units are fraction of whole brain mean signal) of this hippocampal time series was chosen as parameter of interest. First, the Spearman correlation between the variance across the whole scan and the percent time spent in each state was computed across the three groups. Next, the average variance of the hippocampal time series within windows during each state was compared across states using one-way ANOVA (i.e. variance in state R1 vs. R2 vs. R3 vs. R4). This was computed for each group separately to determine if the participants within a particular group had higher signal variance within a specific state compared to other states. These analyses were repeated with the left hippocampus and scan 2.
3. Results
3.1. Static hippocampal FC
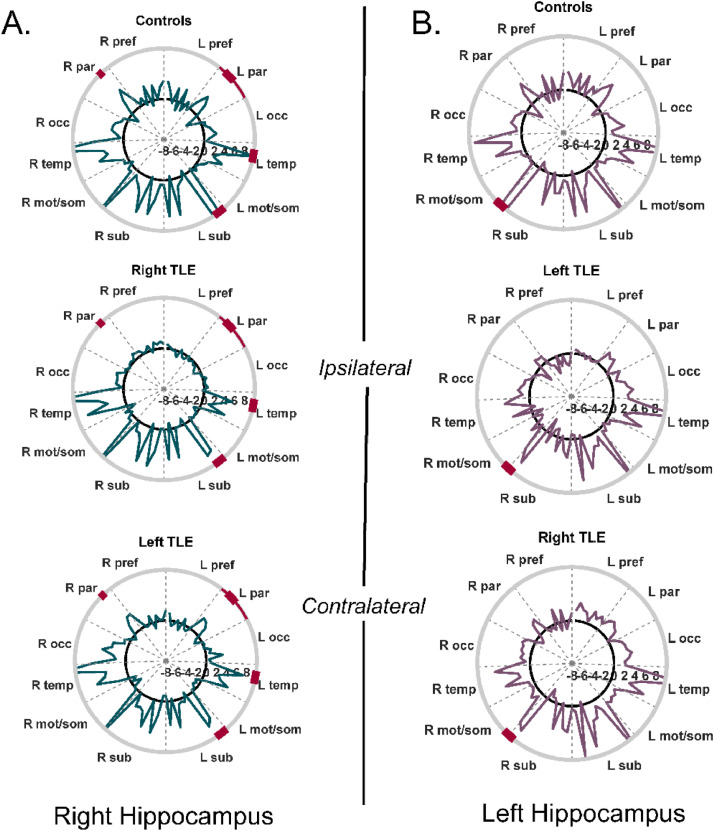
In order to subsequently compare to dynamic FC, the static FC from the right hippocampus to 86 other regions across the brain was computed and averaged across the group of controls, right TLE and left TLE participants using scan 1 (Fig. 2A). This was repeated for the left hippocampus (Fig. 2B). Groups were compared using one-way ANOVA with statistical significance thresholded at p < 0.0006 to account for Bonferroni correction for 86 tests of each hippocampus. The FC from the right hippocampus was less in the right TLE patients than the controls in the left angular gyrus, right and left posterior cingulate, right and left precuneus, left parahippocampal gyrus and the left hippocampus (ANOVA: p < 0.0005, post-hoc t-test: p < 0.001). These individual region differences are indicated in Fig. 2 as thick red marks on the outer circles. The lobe averaged FC was compared between groups using one-way ANOVA with the statistical threshold set to 0.0042 to account for the 12 lobes. The FC from the right hippocampus to the left parietal lobe was decreased in right TLE compared to controls (ANOVA: p < 0.003, post-hoc t-test: p = 0.007). These are indicated by the thinner red marks on the outer circles. There were no differences detected in left hippocampus FC averaged over lobes between subject groups.
Fig. 2.
Static hippocampal FC over 10-min fMRI scan. (A) Average right hippocampal FC across group of controls (top, n = 36), right TLE (middle, n = 28) and left TLE (bottom, n = 13). (B) Average left hippocampal FC across group of controls (top, n = 36), left TLE (middle, n = 13) and right TLE (bottom, n = 28). A total of 87 regions were included. Labels on the circumference indicate lobes. Solid black line indicates 0. Negative values of connectivity are indicated inside the black line. Thick red circumference indicates individual regional differences between groups for each hippocampus. Thin red circumference indicates lobe different between groups for each hippocampus. R = right, L = left, pref = prefrontal, par = parietal, occ = occipital, temp = temporal, mot/som = motor and somatosensory, sub = subcortical. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.2. Dynamic hippocampal FC
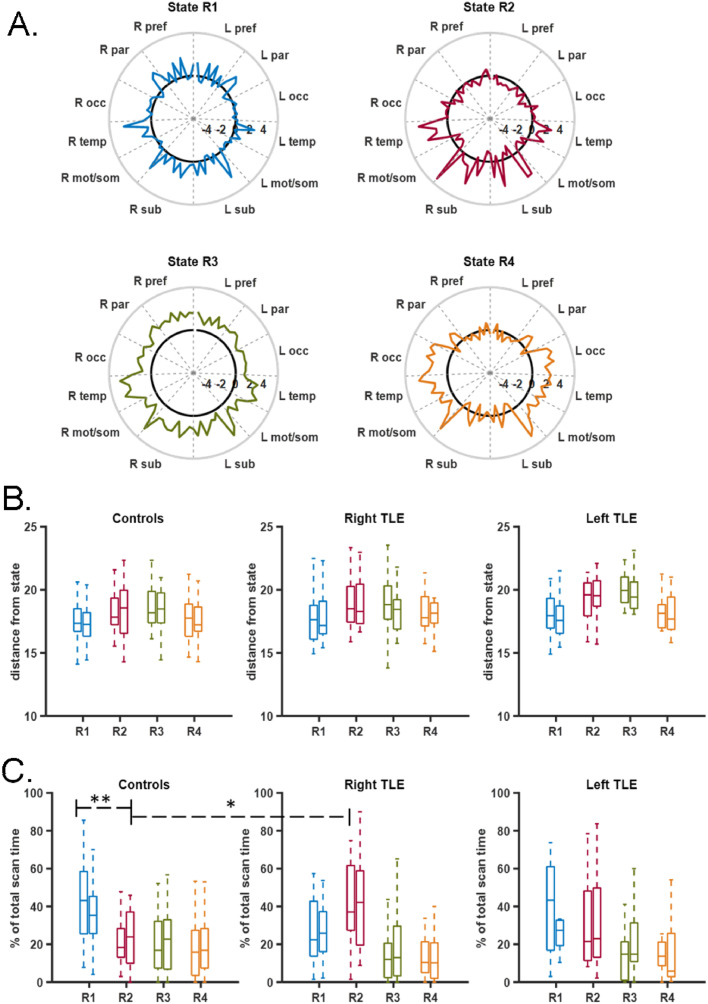
Dynamic FC was computed as the FC from the hippocampus to each of the 86 other regions for each 60-s sliding window. Using the 20 training healthy control participants (scan 1), four different states of network FC from each hippocampus (right hippocampus: R1, R2, R3, R4; left hippocampus: L1, L2, L3, L4) were identified using the k-means clustering of the 60-s windows (Figs. 3A and 4A). Then, in each of the other participants, in each scan, in each 60-s window, one state was identified as having the smallest Euclidean norm (distance) from the hippocampal FC in that window. The distance to the state and the percent of the total scan time spent in each state for each participant for each scan was computed.
Fig. 3.
Dynamic right hippocampal FC states. (A) Four right hippocampal FC states (R1, R2, R3, R4) identified in 20 training control participants. Labels on the circumference indicate groups of regions. Solid black line indicates 0. R = right, L = left, pref = prefrontal, par = parietal, occ = occipital, temp = temporal, mot/som = motor and somatosensory, sub = subcortical. Colors in A correspond to colors in B and C. (B) Euclidean norm (distance) of hippocampal FC from each state for participants in each group. There were no differences between groups for any state. (C) Percent of scan time spent in each state indicated in A for participants in each group. * indicates controls spent less time in state R2 than the right TLE group (ANOVA: p < 0.0001, post-hoc t-test: p < 0.0001, 95% confidence interval Controls-Right TLE [−32.2%, −10.9%]). ** indicates controls spent more time in state R1 than in state R2 (paired t-test R1 vs. R2: p < 0.0001, R1-R2 [12.5%, 29.5%]). Similar results for scan 2. In B and C) Groups are controls (left, n = 36), right TLE (middle, n = 28) and left TLE (right, n = 13). Left and right bar in each indicates scan 1 and scan 2, respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
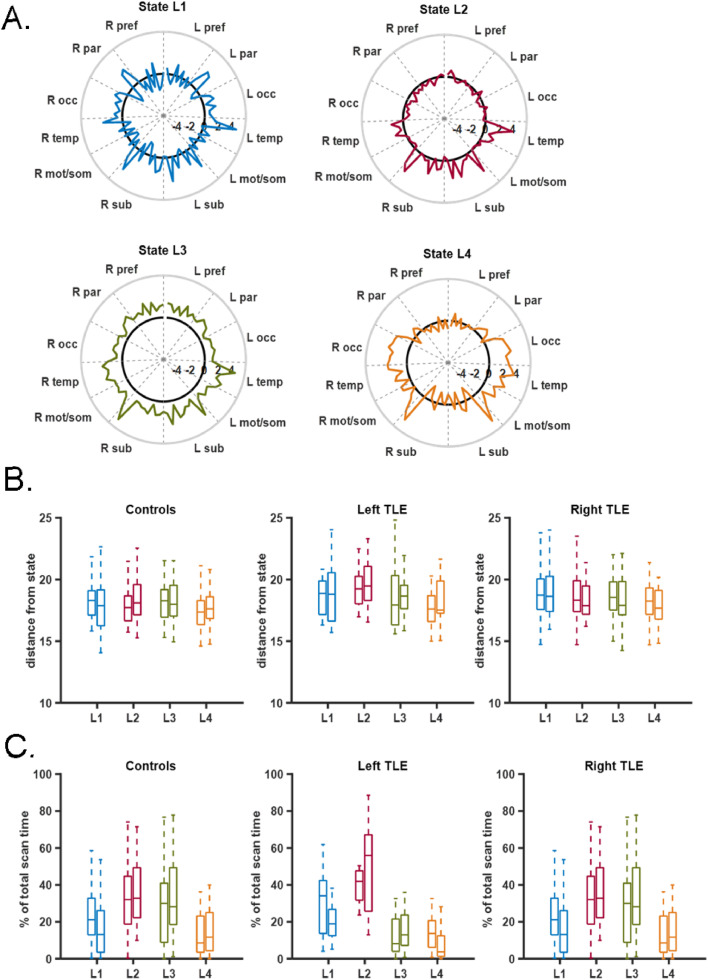
Fig. 4.
Dynamic left hippocampal FC states. (A) Four left hippocampal FC states (L1, L2, L3, L4) identified in 20 training control participants. Labels on the circumference indicate groups of regions. Solid black line indicates 0. R = right, L = left, pref = prefrontal, par = parietal, occ = occipital, temp = temporal, mot/som = motor and somatosensory, sub = subcortical. Colors in A correspond to colors in B and C. (B) Euclidean norm (distance) of hippocampal FC from each state for participants in each group. There was no difference between groups for any state. (C) Percent of scan time spent in each state indicated in A for participants in each group. There was no difference between groups for any state. In (B) and (C) Groups are controls (left, n = 36), left TLE (middle, n = 13) and right TLE (left, n = 13). Left and right bar in each indicates scan 1 and scan 2, respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
When comparing the 20 training controls to the other 36 controls, no difference in distance to state or time spent in state was detected for right or left hippocampus. Similarly, no difference was detected for scan 2. These results support the idea that the healthy hippocampal FC states are generalizable to other healthy participants, and that they can then be compared to patient groups.
Next, the dynamics of the three groups were compared. Using one-way ANOVA for each state, there was no difference detected in distance from state between groups for the right hippocampus (Fig. 3B) and the left hippocampus (Fig. 4B). The results were the same for scan 2. These suggest that the healthy states describe the dynamic states of all three groups equally well. Furthermore, we also compared the distances of the remaining second, third and fourth closest state between groups to determine whether the minimum distance reflects a meaningful separation between states. Results showed no detectable difference between groups for any of the states.
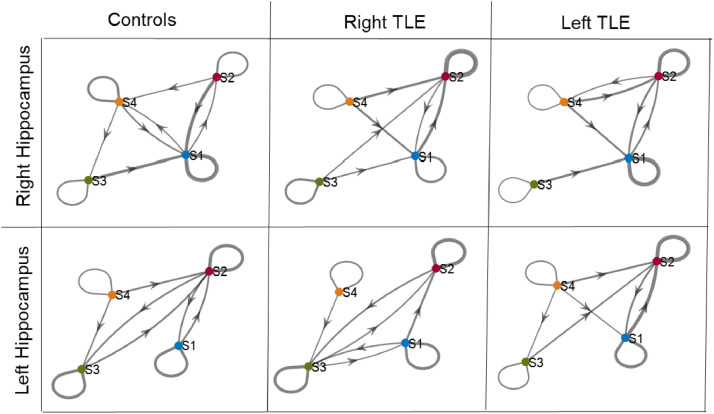
Percent of scan time spent in each state was compared across groups using one-way ANOVA with statistical significance determined by p < 0.0125 to account for Bonferroni correction of the four states. This threshold was used for all four state analyses. For the right hippocampus, the controls spent less time in state R2 than the right TLE group (Fig. 3C) (ANOVA: p < 0.0001, post-hoc t-test: p < 0.0001, Controls-Right TLE [−32.2%, −10.9%]; brackets indicate 95% confidence interval of the difference). In fact, the controls spent more time in R1 than in R2 (paired t-test R1 vs. R2: p < 0.0001, R1-R2 [12.5%, 29.5%]. Using scan 2, the findings were similar. The controls spent less time in state R2 than the right TLE group (ANOVA: p = 0.002, post-hoc t-test: p = 0.001, Controls-Right TLE [−30.2%, −6.4%]). And, the controls spent more time in R1 than in R2 (paired t-test R1 vs. R2: p = 0.0014, R1-R2 [5.0%, 19.3%]). For the left hippocampus there was no differences detected in percent of scan time spent between groups within each state (Fig. 4C). The results were the same for scan 2. This may suggest that we are unable to detect differences in the left hippocampus between the controls and left TLE due to lack of power in the smaller left TLE sample size. Therefore, we then grouped by ipsilateral and contralateral hippocampi to investigate this issue. When considering ipsilateral vs contralateral networks, indeed the ipsilateral networks spent more time in state R2/L2 than the controls networks (Fig. 3C middle, 4C middle) (ANOVA: p < 0.0001, post-hoc t-test: p < 0.0001, Controls-Ipsilateral [−24.0%, −7.4%]). The finding also was true for scan 2 (ANOVA: p = 0.001, post-hoc t-test: p < 0.001, Controls-Ipsilateral [−23.4%, −5.0%]). No differences were found in state R1/L1, R3/L3 or R4/L4. To illustrate the temporal sequence of state transitions across the groups, the average percent of transitions from one state to the next across time was computed for each patient. This is shown in Fig. 5.
Fig. 5.
Transition between states for each group. The four states are represented by the colored circles (S1, S2, S3, S4) and the arrows represent direction of transition between states. Same state transitions are represented by self-loops at the state. Line thickness represents percent of transitions of that state moving to the connected state. A threshold of 15% of transitions are shown. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
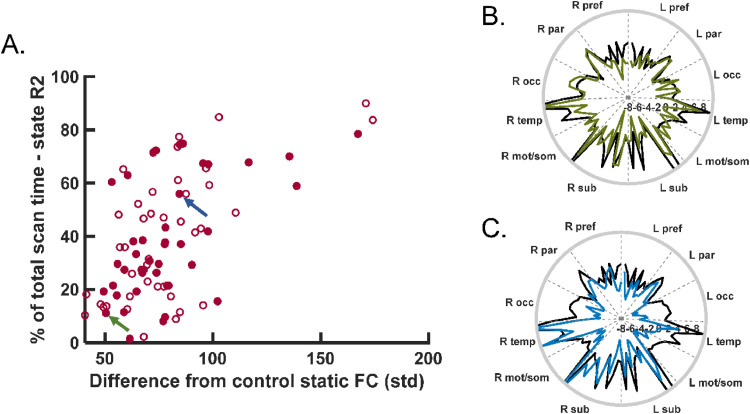
When comparing static right hippocampal FC to dynamic FC in all patients together, increased time spent in state R2 was related to greater distance from the healthy control static FC (Spearman: p < 0.001, ρ = 0.50) (Fig. 6A, filled), and a decreased time spent in state R1 (Spearman: p = 0.001, ρ = −0.48). Fig. 6B shows an example of a left TLE patient in which the right hippocampus static FC is similar to the control static FC, where the time spent in state R2 is 11.1%. Additionally, Fig. 6C illustrates an example of a right TLE patient in which the right hippocampus static FC has a greater difference to control static FC and the time spent in state R2 is 55.9%. No relationships between static and dynamic FC were found in the left hippocampus. Using scan 2, similar relationships were found. In right hippocampus, time spent in state R2 increased with increased distance from control static FC (Spearman: p < 0.001, ρ = 0.51) (Fig. 6, open), but only a trend between decreased time spent in state R1 with increased distance from control static FC (Spearman: p = 0.05, ρ = −0.29). No relation was detected in left hippocampus in scan 2.
Fig. 6.
Static vs. dynamic right hippocampal FC. (A) Difference from control static FC increases as time spent in state R2 increases across all TLE patients. Filled and open points are from scan 1 (Spearman: p < 0.001, ρ = 0.50) and scan 2 (Spearman: p < 0.001, ρ = 0.51), respectively. Green and blue arrow indicate participants shown in parts B and C, respectively. (B) Example of left TLE patient with low static FC distance to control (50.4) and low percent time spent in state R2 (11.1%). (C) Example of right TLE patient with high static FC distance from control (84.5) and high percent time spent in state R2 (55.9%). (B) and (C) black lines represent mean of healthy control static hippocampal FC. Green and blue lines show individual participant. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.3. Hippocampal time series
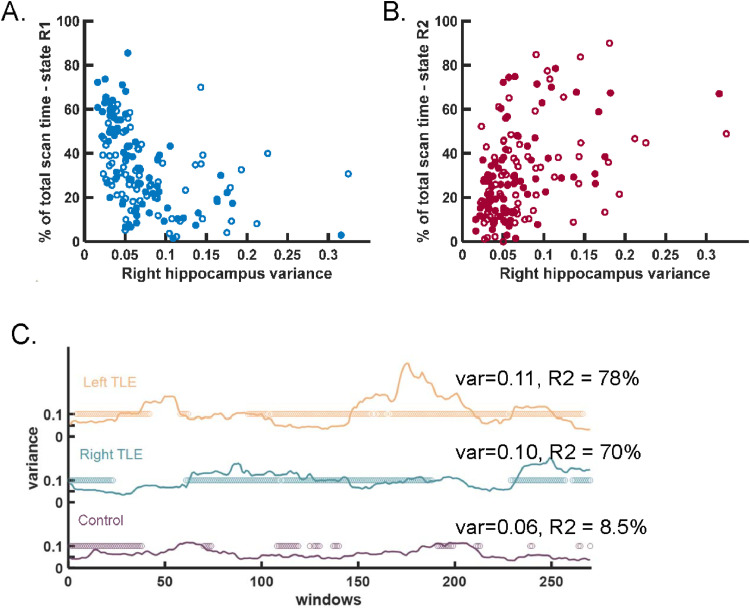
Next, the relationship between the hippocampal fMRI time series variance across the scan and the percent of scan time spent in each state was quantified. For the right hippocampus, the variance of the time series was negatively correlated with time spent in state R1 (Spearman: p < 0.0001, ρ = −0.70) and positively correlated with time spent in state R2 (Spearman: p < 0.0001, ρ = 0.45) (Fig. 7A, B). The same relationships were detected in scan 2 (State R1: Spearman: p < 0.0001, ρ = −0.44; State R2: Spearman: p < 0.001 ρ = 0.41). For the left hippocampus, the time series variance was negatively correlated with time spent in state L1 (Spearman: p < 0.0001, ρ = −0.44), but only weakly associated in scan 2 (Spearman: p = 0.08, ρ = −0.19).
Fig. 7.
Hippocampal time series variance related to percent of time spent in hippocampal FC state. (A) Variance across the 10-min time series was negatively correlated with time spent in state R1 in scan 1 (Spearman: p < 0.0001, ρ = −0.70) (filled). Similar results for scan 2 (open). (B) Variance across the 10-min time series was positively correlated with time spent in state R2 in scan 1 (Spearman: p < 0.0001, ρ = 0.45) (filled). Similar results for scan 2 (open). (C) Example of variance across each window of a healthy control participant (purple, bottom), right TLE patient (middle, cyan) and left TLE patient (top, yellow). The circles indicate the windows in which the participant is in state R2. The variance across the scan (var) and percent of scan spent in state R2 (R2) are indicated for each. No window to window relationship between variance and percent time spent in state R2. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
It is also of interest to investigate the relationship between state and variance within the scan (i.e. window to window changes). To do this, variance across the 60-s windows was computed, and the variance in windows in each state were compared. Based on the results in Fig. 7A and B, the variance of windows in state R1/L1 may be expected to be lower than those in state R2/L2. However, there was no difference in variance of right or left hippocampal signal during windows of one state compared to any other state within each group in scan 1 or scan 2. Fig. 7C shows an example of the variance in each 60-s window of the right hippocampus in a healthy control, a right TLE patient and a left TLE patient. The windows that the participant was determined to be in state R2 are indicated as circles. This illustrates the increased variance and time spent in state R2 in the two patients compared to the control participant, but there is no detectable within-scan (window to window) temporal relationship between variance and state.
4. Discussion
This study characterized static and spatio-temporal dynamic hippocampal FC networks in mesial TLE in relation to healthy network FC. We hypothesized that healthy hippocampal connectivity will have dynamic fluctuations representing the spatiotemporal circuit dynamics that support functions like memory and learning (Hyman et al., 2010; Basu and Siegelbaum, 2015; Knierim, 2015; Bohbot et al., 2017; Myroshnychenko et al., 2017). Thus, the first objective was to characterize healthy dynamics of hippocampal FC. To do this, four FC states representing structured, transient whole brain network connectivity (Allen et al., 2014; Vidaurre et al., 2017; Zhong et al., 2019) were identified from each hippocampus in a set of 20 healthy training participants. To understand generalizability and enable comparisons to other participants, the states were compared to 36 healthy control participants separate from those in whom the states were identified. There was no difference detected in distance from each state between the two sets of controls. Similarly, there was no difference detected in percent of scan time spent in each state. These findings suggest that the healthy control states identified in the training set were representative of general healthy states.
Then, we hypothesized that TLE will contribute additional unique dynamic hippocampal network FC fluctuations to the control dynamics. To test this the healthy FC states were compared to hippocampal FC dynamics in right and left TLE patients. Results showed that the patients exhibited the same hippocampal FC states as the controls, as demonstrated by the lack of difference detected in the distance from the state in any group. On the other hand, the percent scan time spent in each state was different between participant groups, mostly in the right hippocampus networks. In controls, the state R1 was the dominant state. However, in right TLE the time spent in state R2 was greater than in controls, and state R1 was not the dominant state. While the same findings were not detected individually for the left hippocampus, when both hippocampi were considered together the ipsilateral networks (right hippocampus in right TLE and left hippocampus in left TLE) had increased time spent in state R2/L2 than controls.
From Figs. 3A and 4A, it can be seen that state R2/L2 have decreased hippocampal connectivity to bilateral prefrontal and parietal regions compared to state R1/L1. The decreased time spent with hippocampal connection to these regions (Kim, 2019) and networks (Doucet et al., 2013; Holmes et al., 2014), known to be essential in memory encoding and retrieval, may contribute to the memory related cognitive impairments seen in TLE (Helmstaedter and Elger, 2009). Additionally, these state changes may reflect interruptions in the oscillation coupling between hippocampus and prefrontal regions necessary for memory function (Hyman et al., 2010; Myroshnychenko et al., 2017).
After establishing that percent time spent in state R2/L2 was associated with the ipsilateral hippocampal FC network changes in TLE, the relationship between this timing and alteration in static hippocampal FC was identified. Alteration in static hippocampal FC in each TLE patient was quantified as the Euclidean norm from the mean static FC across all 56 healthy control participants in units of standard deviations. This distance increased as percent scan time spent in state R2 increased across all TLE patients in the right hippocampus, revealing that changes in dynamics may contribute to impairments in standard static measures of hippocampal FC in TLE. Note that we showed decreases in static hippocampal FC across the parietal lobe, and specifically in the precuneus in TLE compared to controls. This is consistent with more time spent in the state where FC to prefrontal and parietal regions is minimal, however, our results are not a direct measure of causality between the static and dynamic FC.
Finally, the identified hippocampal FC network alterations in states R1 and R2 were related to hippocampal signal fluctuations. Increased variance of the right hippocampal oxygen level dependent fMRI signal across the scan was associated with increased time spent in state R2, and decreased with time spent in state R1 across all three groups. However, no direct relationships between within-scan (window to window) signal variance and state were detected. These findings imply that greater fluctuations in hippocampal activity across the 10-min scans are associated with dynamic FC changes, but not directly in phase in the 60-s window time scale. This suggests other contributing factors to these state changes. Our increased variance is complementary to Laufs et al. who detected increased hippocampal variance in fMRI signal and dynamic FC to precuneus, supplementary motor and sensorimotor and frontal cortices in TLE compared to controls (Laufs et al., 2014).
There are some limitations of this work that need to be considered. First, the number of patients was not balanced between left and right TLE. This may have contributed to the more significant findings in the right hippocampus. Second, the 60-s sliding window was chosen somewhat arbitrarily. It has been shown that detection is biased towards state durations equal to window length (Shakil et al., 2016), but the exact effect on these data are unknown. For comparison Laufs et al. used 30 s windows (Laufs et al., 2014), while Allen et al. used 44 s windows (Allen et al., 2014). Similarly, the k-means clustering was performed in two steps (Allen et al., 2014) and four final states were identified (Vergara et al., 2017). The within subject cluster number (k1) and the between subject cluster number (k2 = final states) were varied (k1 = 27, 54, 90, 135; k2 = 3, 4, 5, 6) and tested for difference between the two controls sets. It was found that k2 other than 4 showed differences between the control sets. It was also found that varying k1 had little effect on this outcome, but that k1 = 54 and 90 had most repeatable results. This procedure is not validated and other approaches have not been explored. Third, we do not have long term surgical outcomes for all patients on which to confirm the diagnosis of unilateral mesial TLE in all cases. Fourth, the direct effects of the altered hippocampal anatomy in the patients on the dynamic connectivity results shown in this work remain unknown. Finally, no simultaneous scalp EEG was collected during the fMRI acquisition, which might have provided some information on arousal during the scan and the transient spiking at the hippocampus, although most likely only a fraction of the spiking would be detected at the scalp (Tao et al., 2005).
Conversely, however, a second fMRI scan acquired in the same imaging session with the same acquisition parameters and was used to examine within-subject reproducibility. All analyses comparing the states to the participants were repeated in scan 2 with the same results. The correlations between hippocampal fluctuations and state were also reproduced in scan 2.
In conclusion, these investigations have presented evidence that increases in transient pathological signal fluctuations occurring at the seizure focus in the hippocampus in patients with TLE may contribute to disruptions in healthy FC spatio-temporal network dynamics within a fMRI scan. Specifically, TLE patients spend more time in a particular network state characterized by decreased bilateral prefrontal and parietal connectivity compared to the dominant healthy state. Furthermore, these dynamic FC alterations are associated with decreases in static hippocampal FC and are associated with overall increases in variance in the fMRI signal. We propose that characterization of an individual patient's hippocampal dynamics at different time scales may provide more specific spatio-temporal profiles of network impairment that may be related to hippocampal dysfunction in TLE.
Funding
This work was supported by National Institutes of Health R01 NS075270, R01 NS108445, and R01 NS110130 to V.L.M., and R00 NS097618 to D.J.E. and R01 NS112252 to D.J.E and C.C.
CRediT authorship contribution statement
Victoria L. Morgan: Conceptualization, Methodology, Investigation, Formal analysis, Writing - original draft, Writing - review & editing, Funding acquisition. Catie Chang: Investigation, Formal analysis, Writing - review & editing, Funding acquisition. Dario J. Englot: Investigation, Formal analysis, Writing - review & editing, Funding acquisition. Baxter P. Rogers: Methodology, Investigation, Formal analysis, Writing - review & editing.
Declaration of Competing Interests
None.
References
- Allen E.A., Damaraju E., Plis S.M., Erhardt E.B., Eichele T., Calhoun V.D. Tracking whole-brain connectivity dynamics in the resting state. Cereb. Cortex. 2014;24(3):663–676. doi: 10.1093/cercor/bhs352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asman A.J., Landman B.A. Non-local statistical label fusion for multi-atlas segmentation. Med. Image Anal. 2013;17(2):194–208. doi: 10.1016/j.media.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu J., Siegelbaum S.A. The corticohippocampal circuit, synaptic plasticity, and memory. Cold Spring Harb. Perspect. Biol. 2015;7(11) doi: 10.1101/cshperspect.a021733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettus G., Bartolomei F., Confort-Gouny S., Guedj E., Chauvel P., Cozzone P.J. Role of resting state functional connectivity MRI in presurgical investigation of mesial temporal lobe epilepsy. J. Neurol. Neurosurg. Psychiatry. 2010;81(10):1147–1154. doi: 10.1136/jnnp.2009.191460. [DOI] [PubMed] [Google Scholar]
- Bohbot V.D., Copara M.S., Gotman J., Ekstrom A.D. Low-frequency theta oscillations in the human hippocampus during real-world and virtual navigation. Nat Commun. 2017;8:14415. doi: 10.1038/ncomms14415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragin A., Engel J., Jr., Wilson C.L., Fried I., Buzsaki G. High-frequency oscillations in human brain. Hippocampus. 1999;9(2):137–142. doi: 10.1002/(SICI)1098-1063(1999)9:2<137::AID-HIPO5>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Bragin A., Jando G., Nadasdy Z., Hetke J., Wise K., Buzsaki G. Gamma (40-100Hz) oscillation in the hippocampus of the behaving rat. J. Neurosci. 1995;15(1 Pt 1):47–60. doi: 10.1523/JNEUROSCI.15-01-00047.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzsáki G. Theta oscillations in the hippocampus. Neuron. 2002;33(3):325–340. doi: 10.1016/s0896-6273(02)00586-x. [DOI] [PubMed] [Google Scholar]
- Cendes F., Li L.M., Watson C., Andermann F., Dubeau F., Arnold D.L. Is ictal recording mandatory in temporal lobe epilepsy?: Not when the interictal electroencephalogram and hippocampal atrophy coincide. JAMA Neurol. 2000;57(4):497–500. doi: 10.1001/archneur.57.4.497. [DOI] [PubMed] [Google Scholar]
- Chang C., Glover G.H. Time-frequency dynamics of resting-state brain connectivity measured with fMRI. Neuroimage. 2010;50(1):81–98. doi: 10.1016/j.neuroimage.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colgin L.L. Rhythms of the hippocampal network. Nat. Rev. Neurosci. 2016;17(4):239–249. doi: 10.1038/nrn.2016.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper R.A., Ritchey M. Cortico-hippocampal network connections support the multidimensional quality of episodic memory. eLife. 2019;8:e45591. doi: 10.7554/eLife.45591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes D., Haughton V.M., Arfanakis K., Carew J.D., Turski P.A., Moritz C.H. Frequencies contributing to functional connectivity in the cerebral cortex in "resting-state" data. Ajnr. 2001;22(7):1326–1333. [PMC free article] [PubMed] [Google Scholar]
- Doucet G., Osipowicz K., Sharan A., Sperling M.R., Tracy J.I. Hippocampal functional connectivity patterns during spatial working memory differ in right versus left temporal lobe epilepsy. Brain Connect. 2013;3(4):398–406. doi: 10.1089/brain.2013.0158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel J., Jr. Mesial temporal lobe epilepsy: what have we learned? Neurosci. 2001;7(4):340–352. doi: 10.1177/107385840100700410. [DOI] [PubMed] [Google Scholar]
- Englot D.J., Konrad P.E., Morgan V.L. Regional and global connectivity disturbances in focal epilepsy, related neurocognitive sequelae, and potential mechanistic underpinnings. Epilepsia. 2016;57(10):1546–1557. doi: 10.1111/epi.13510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B. FreeSurfer. Neuroimage. 2012;62(2):774–781. doi: 10.1016/j.neuroimage.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R.A. Frequency distribution of the values of the correlation coefficient in samples from an indefinitely large population. Biometrika. 1915;10(4):507–521. [Google Scholar]
- Glover G.H., Li T.Q., Ress D. Image-based method for retrospective correction of physiological motion effects in fMRI: retroicor. Magn. Reson. Med. 2000;44(1):162–167. doi: 10.1002/1522-2594(200007)44:1<162::aid-mrm23>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Haneef Z., Lenartowicz A., Yeh H.J., Levin H.S., Engel J., Stern J.M. Functional connectivity of hippocampal networks in temporal lobe epilepsy. Epilepsia. 2014;55(1):137–145. doi: 10.1111/epi.12476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Doucet G.E., Pustina D., Sperling M.R., Sharan A.D., Tracy J.I. Presurgical thalamic “hubness” predicts surgical outcome in temporal lobe epilepsy. Neurology. 2017;88(24):2285–2293. doi: 10.1212/WNL.0000000000004035. [DOI] [PubMed] [Google Scholar]
- Helmstaedter C., Elger C.E. Chronic temporal lobe epilepsy: a neurodevelopmental or progressively dementing disease? Brain. 2009;132(10):2822–2830. doi: 10.1093/brain/awp182. [DOI] [PubMed] [Google Scholar]
- Holmes M.J., Folley B.S., Sonmezturk H.H., Gore J.C., Kang H., Abou-Khalil B. Resting state functional connectivity of the hippocampus associated with neurocognitive function in left temporal lobe epilepsy. Hum. Brain Mapp. 2014;35(3):735–744. doi: 10.1002/hbm.22210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo Y., Plassard A.J., Carass A., Resnick S.M., Pham D.L., Prince J.L. Consistent cortical reconstruction and multi-atlas brain segmentation. Neuroimage. 2016;138:197–210. doi: 10.1016/j.neuroimage.2016.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman J., Zilli E., Paley A., Hasselmo M. Working memory performance correlates with prefrontal-hippocampal theta interactions but not with prefrontal neuron firing rates. Front. Integr. Neurosci. 2010;4(2) doi: 10.3389/neuro.07.002.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. Neural activity during working memory encoding, maintenance, and retrieval: a network-based model and meta-analysis. Hum. Brain Mapp. 2019;40(17):4912–4933. doi: 10.1002/hbm.24747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knierim J.J. The hippocampus. Curr. Biol. 2015;25(23):R1116–R1R21. doi: 10.1016/j.cub.2015.10.049. [DOI] [PubMed] [Google Scholar]
- Laufs H., Rodionov R., Thornton R., Duncan J.S., Lemieux L., Tagliazucchi E. Altered fMRI connectivity dynamics in temporal lobe epilepsy might explain seizure semiology. Front. Neurol. 2014;5(175) doi: 10.3389/fneur.2014.00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurie D.J., Kessler D., Bassett D.S., Betzel R.F., Breakspear M., Keilholz S. Questions and controversies in the study of time-varying functional connectivity in resting fMRI. Netw. Neurosci. 2020;4(1):30–69. doi: 10.1162/netn_a_00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maccotta L., He B.J., Snyder A.Z., Eisenman L.N., Benzinger T.L., Ances B.M. Impaired and facilitated functional networks in temporal lobe epilepsy. NeuroImage Clin. 2013;2:862–872. doi: 10.1016/j.nicl.2013.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan V.L., Englot D.J., Rogers B.P., Landman B.A., Cakir A., Abou-Khalil B.W. Magnetic resonance imaging connectivity for the prediction of seizure outcome in temporal lobe epilepsy. Epilepsia. 2017;58(7):1251–1260. doi: 10.1111/epi.13762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan V.L., Rogers B.P., Anderson A.W., Landman B.A., Englot D.J. Divergent network properties that predict early surgical failure versus late recurrence in temporal lobe epilepsy. J. Neurosurg. 2019:1–10. doi: 10.3171/2019.1.JNS182875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myroshnychenko M., Seamans J.K., Phillips A.G., Lapish C.C. Temporal dynamics of hippocampal and medial prefrontal cortex interactions during the delay period of a working memory-guided foraging task. Cereb. Cortex. 2017;27(11):5331–5342. doi: 10.1093/cercor/bhx184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roger E., Pichat C., Torlay L., David O., Renard F., Banjac S. Hubs disruption in mesial temporal lobe epilepsy. A resting-state fMRI study on a language-and-memory network. Hum. Brain Mapp. 2019 doi: 10.1002/hbm.24839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers B.P., Morgan V.L., Newton A.T., Gore J.C. Assessing functional connectivity in the human brain by fMRI. Magn. Reson. Imaging. 2007;25(10):1347–1357. doi: 10.1016/j.mri.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakil S., Lee C.-.H., Keilholz S.D. Evaluation of sliding window correlation performance for characterizing dynamic functional connectivity and brain states. Neuroimage. 2016;133:111–128. doi: 10.1016/j.neuroimage.2016.02.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao J.X., Ray A., Hawes-Ebersole S., Ebersole J.S. Intracranial EEG substrates of scalp EEG interictal spikes. Epilepsia. 2005;46(5):669–676. doi: 10.1111/j.1528-1167.2005.11404.x. [DOI] [PubMed] [Google Scholar]
- Vergara V.M., Mayer A.R., Damaraju E., Calhoun V.D. The effect of preprocessing in dynamic functional network connectivity used to classify mild traumatic brain injury. Brain Behav. 2017;7(10):e00809. doi: 10.1002/brb3.809. n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidaurre D., Smith S.M., Woolrich M.W. Brain network dynamics are hierarchically organized in time. Proc. Natl. Acad. Sci. 2017;114(48):12827–12832. doi: 10.1073/pnas.1705120114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Q., Xu H., Qin J., Zeng L.L., Hu D., Shen H. Functional parcellation of the hippocampus from resting-state dynamic functional connectivity. Brain Res. 2019;1715:165–175. doi: 10.1016/j.brainres.2019.03.023. [DOI] [PubMed] [Google Scholar]