Abstract
Benzimidazoles (BZ) have been the anthelmintic of choice for controlling Nematodirus battus infections since their release in the 1950s. Despite heavy reliance on this single anthelmintic drug class, resistance was not identified in this nematode until 2010 (Mitchell et al., 2011). The study aimed to explore the prevalence of BZ-resistance mutations in N. battus from UK sheep flocks using deep amplicon sequencing and pyrosequencing platforms. Based on evidence from other gastrointestinal nematodes, resistance in N. battus is likely to be conferred by single nucleotide polymorphisms (SNP) within the β-tubulin isotype 1 locus at codons 167, 198 and 200. Pyrosequencing and deep amplicon sequencing assays were designed to identify the F167Y (TTC to TAC), E198A (GAA to GCA) and F200Y (TTC to TAC) SNPs. Nematodirus battus populations from 253 independent farms were analysed by pyrosequencing; 174 farm populations were included in deep amplicon sequencing and 170 were analysed using both technologies. F200Y was the most prevalent SNP identified throughout the UK, in 12–27% of the populations tested depending on assay, at a low overall individual frequency of 2.2 ± 0.6% (mean ± SEM, based on pyrosequencing results). Four out of the five populations with high frequencies (>20%) of the F200Y mutation were located in NW England. The F167Y SNP was identified, for the first time in this species, in four of the populations tested at a low frequency (1.2% ± 0.01), indicating the early emergence of the mutation. E198A or E198L were not identified in any of the isolates. Results obtained were comparable between both techniques for F200Y (Lins’ CCC, rc = 0.96) with discrepancies being limited to populations with low frequencies. The recent emergence of resistance in this species will provide a unique opportunity to study the early stages of anthelmintic resistance within a natural setting and track its progress in the future.
Keywords: Anthelmintic resistance, Benzimidazole, β-tubulin, Nematodirus battus, Amplicon sequencing, Pyrosequencing
Graphical abstract
Highlights
-
•
Early emergence of benzimidazole (BZ) resistance in Nematodirus battus.
-
•
F200Y mutation most prevalent SNP identified.
-
•
First identification of F167Y mutation associated with BZ resistance in Nematodirus.
-
•
Good agreement between deep amplicon sequencing and pyrosequencing platforms.
1. Introduction
Nematodirosis, caused by the species Nematodirus battus, is a major cause of severe diarrhoea and mortality in young lambs (Thomas and Stevens, 1956), predominantely in the spring. Benzimidazole (BZ) compounds are favoured for N. battus control because of their high efficacy, lack of reports of resistance and the high safety index (Abbott et al., 2012). BZ-resistance has been described in Nematodirus spathiger and Nematodirus filicollis (Middleberg and McKenna, 1983; Martin et al., 1985; Obendorf et al., 1991; Oliver et al., 2016), but had not been detected in N. battus. One hypothesis is based on the distinct hatching behaviour of the parasite, which creates a large refugia, e.g. eggs survive at pasture for prolonged periods of time and hatch in a synchronised manner. However, recently, BZ-resistance was described and characterised in a UK N. battus population (Mitchell et al., 2011; Morrison et al., 2014).
Given the prevalence of BZ-resistance in other gastro-intestinal nematode (GIN) species co-infecting sheep in the UK, the use of BZ compounds to control N. battus early in the grazing season has also been advocated because it protects the other drug classes from additional selection pressure for resistance development at a time of year where refugia would be considered low. The development of effective and sustainable control strategies for N. battus in the UK currently relies on previous history/experience and Nematodirus forecasting tools (www.scops.org.uk/forecasts/nematodirus-forecast/) to predict the risk of exposure and the use of faecal egg count reduction tests to assess anthelmintic efficacy. The use of molecular tools to test anthelmintic efficacy provides an opportunity for rapid, accurate results to be generated from a single sample without the need to treat animals and collect post-treatment samples and, can be conducted at a time when the risk of acute disease is low or absent.
Single nucleotide polymorphism (SNP) mutations at codons 167, 198 and 200 of the β-tubulin isotype-1 gene have been associated with BZ-resistance in several nematode species of veterinary importance (Kwa et al., 1994; Silvestre and Cabaret, 2002; Silvestre and Humbert, 2002; Ghisi et al., 2007). Evaluation of the primary BZ-resistant N. battus isolate identified that the F200Y mutation was involved in conferring resistance in this species (Morrison et al., 2014).
Pyrosequencing is a well-established technique for the detection of SNP mutations associated with anthelmintic resistance (von Samson-Himmelstjerna et al., 2009; Morrison et al., 2014; Ramunke et al., 2016), and has been widely utilised in genotype prevalence studies worldwide (Redman et al., 2015; Chaudhry et al., 2015a, Chaudhry et al., 2015b; Chaudhry et al., 2016; Ramunke et al., 2016). This technique provides detailed information from the analysis of individual parasites. Deep amplicon sequencing is a highly versatile tool which provides a wealth of data, given the development of different analysis pipelines, this can be used for a wide range of research and diagnostic purposes. Recent applications of this technology for veterinary nematodes include nemabiome analysis; the study of nematode species from pooled faecal samples (Avramenko et al., 2015) and the detection of BZ-resistance SNPs in multiple ovine trichostrongylid nematode species including Teladorsagia circumcincta, Haemonchus contortus and Haemonchus placei (Avramenko et al., 2019; Sargison et al., 2019). Application of this technique for detection and quantification of BZ-resistance SNPs in N. battus could provide a high-throughput alternative to pyrosequencing, allowing for analysis of up to 384 populations, run in a single pool.
The aim of the present study was to explore the prevalence of β-tubulin isotype-1 mutations associated with BZ-resistance in N. battus from UK sheep flocks to create a baseline measurement from which to monitor future development of anthelmintic resistance. In addition, two methodologies; pyrosequencing and deep amplicon sequencing, were compared, to test their suitability for the detection and quantification of BZ-resistance mutations in N. battus.
2. Materials and methods
2.1. Sample collection
N. battus populations were collected between 2011 and 2016. Samples were collected in a non-stratified independent approach but attempts were made to balance for perceived spatial bias. A number of samples were submitted by Animal and Plant Health Agency (APHA) and Scotland's Rural College (SRUC) surveillance centres from across the UK. Additional samples were collected opportunistically in 2015 in a non-random manner, including farms from the local region surrounding the initial case of BZ-resistance in this species. Sampling in 2016 was targeted to regions which were under-represented in the biobank of N. battus isolates collected, but which appeared to have significant sheep densities (sheep density data from the Office for National Statistics in 2009 was mapped using QGIS; data source Geo-wiki). Farms in the target regions were contacted via local advisors, veterinarians and the Animal and Horticulture Development Board (AHDB).
During farm visits, fresh lamb faecal samples were collected from the ground, at least 10 per farm or field. Faecal samples were placed in individual plastic bags with excess air removed to suspend development during transport, once in the laboratory, samples were stored at 4 °C prior to processing. Samples which were submitted by SRUC and APHA surveillance centres were packaged in air-tight containers for postage. Sample collection packs were posted to farmers who volunteered to submit samples for the present study. Farmers were instructed to collect 10 fresh lamb faecal samples from the ground, sealing each in an individual zip lock bag with excess air removed, samples were then packaged following royal mail guidelines for biological samples and posted back to the laboratory where samples were stored at 4 °C prior to processing.
2.2. Sample preparation
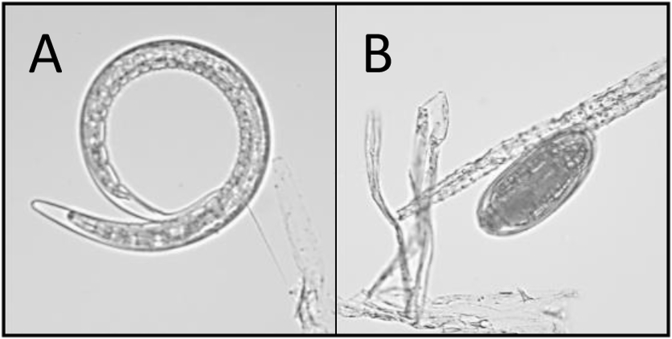
To confirm the presence of eggs and estimate infection intensity, faecal egg counts (Jackson and Christie, 1972) were carried-out on all individual samples. Faeces were pooled per population for processing; a procedure based on differential sieving and flotation was used to extract eggs from faeces. Briefly, faecal samples from each farm were homogenised in tap water and thoroughly washed over stacked sieves; 212 μm, 125 μm and 53 μm to remove large particles. The filtrate containing the N. battus eggs were separated from the fine faecal debris using centrifugal salt flotation (saturated sodium chloride solution; specific gravity 1.2). The eggs were washed with copious quantities of tap water to remove remaining NaCl and placed into non-air-tight containers with tap water. These egg cultures were stored at ambient room temperature (18–22 °C), protected from direct sunlight to allow for larval development. Cultures were monitored microscopically for development and hatching. Larvated eggs (Fig. 1A) and third stage larvae (L3) (Fig. 1B) were concentrated and fixed in ethanol (final concentration >70% EtOH) prior to DNA extraction.
Fig. 1.
Picture of a larvated Nematodirus battus egg (A) and third-stage larvae (B).
2.3. DNA extraction
To generate individual parasite genotypes by pyrosequencing individual egg/larvae DNA lysates were prepared from 257 farm populations. The ethanol-fixed larvated eggs/infective larvae (L3) were first re-hydrated in 1x phosphate-buffered solution (PBS) for 30 min. Thirty individual eggs/larvae from each farm population were picked in 1 μl PBS, using a new pipette tip per egg/larvae, into individual wells of a 96 well plate (Axygen, USA), containing 15 μl lysis buffer (50 mM KCl, 2.5 mM MgCl2, 10 mM Tris (pH 8.3) 0.45% Nonidet P-40, 0.45% Tween 20, 0.01% Gelatine; Kwa et al., 1995). To facilitate hatching the populations containing eggs were subjected to eight freeze/thaw cycles (30 s freeze in liquid nitrogen followed by 1 min incubation at 100 °C) to weaken the egg shells. A further 15 μl of lysis buffer containing 0.2 mg/ml proteinase K was added to each well of the plate for both egg and larvae samples. Plates were placed at 56 °C for 12 h, and then incubated at 95 °C for 10 min to deactivate the proteinase K. Three lysate negative controls were included per plate, containing lysis buffer and enzyme. Crude lysates were used directly as template in PCR reactions.
To generate deep amplicon sequencing data from bulk farm samples, DNA was extracted from pools of 500–1000 larvated eggs and larvae (L3) per farm. Ethanol was removed from the parasite material, eggs/larvae were pelleted by centrifugation (16,000×g for 4 min) and the supernatant removed. The eggs/larvae were then washed three times in lysis buffer to remove any remaining ethanol, and then re-suspended in a final volume of 150 μl. Following the final wash step, samples containing eggs were subjected to eight cycles of freeze/thaw (30 s liquid nitrogen/1 min at 100 °C) to crack the outer shell of the egg for enzyme digestion. Proteinase K (Promega, USA) was then added to each sample (final concentration of 0.8 mg/ml) and incubated at 56 °C 12 h and 95 °C for 10 min. Unlike the individual larvae DNA samples, these crude lysates were put through a silica column (DNA extraction kit, Zymo, USA) following the manufacturers protocol and eluted in 100 μl 1xTE buffer.
2.4. Individual parasite pyrosequencing
The N. battus-specific codon 198/200 SNP assay used was previously described by Morrison et al. (2014). A 197bp fragment, spanning exons 4 and 5 and intervening intron of the β-tubulin isotype-1 locus was PCR amplified to analyse the 198/200 SNP and a second 104bp fragment spanning the same locus was amplified for the codon 167 assay. DNA lysates of individual eggs/larvae from 257 populations were amplified and analysed with the codon 198/200 assay and 18 populations with the codon 167 assay. PCR reactions for P198/200 and P167 were conducted using NovaTaq Hot start master mix (Merck, USA) in 50 μl volumes containing 0.185 μM reverse primer, 0.2 μM biotinylated forward primer, 4.5 mM MgCl2, 25 μl 2 x buffer and 4 μl of template DNA. PCR reactions were incubated at 95 °C for 10 min followed by 45 cycles at 94 °C for 30s, anneal for 30 s at 58 °C for the P198/200 assay or 54 °C for P167, and 72 °C for 30s with a final extension phase at 72 °C for 10 min. The amplified DNA fragment was analysed using the N. battus-specific codon 198/P200 pyrosequencing assay (Pyromark ID, Qiagen, Germany). The same pyrosequencing protocol was used for the P167 genotyping. Pyrosequencing primers for analysis of the P167 SNP were designed using the PSQ assay design v1.0 (Biotage) (Fig. 3A, Supplementary Table S1) with the following dispensation order of nucleotides (GTCATAGCT). To monitor for contamination during DNA preparation and genotyping a total of five negative controls were included per 96-well plate; three lysate and two PCR negative controls. Amplification and pyrosequencing were repeated if any evidence of contamination was observed in these wells and the populations were discarded from the dataset if negative controls were not clear. In addition, populations were only retained for further analysis if at least 24 eggs/larvae (from a total of 30; 80% threshold) produced a strong enough pyrosequencing signal to produce a genotype.
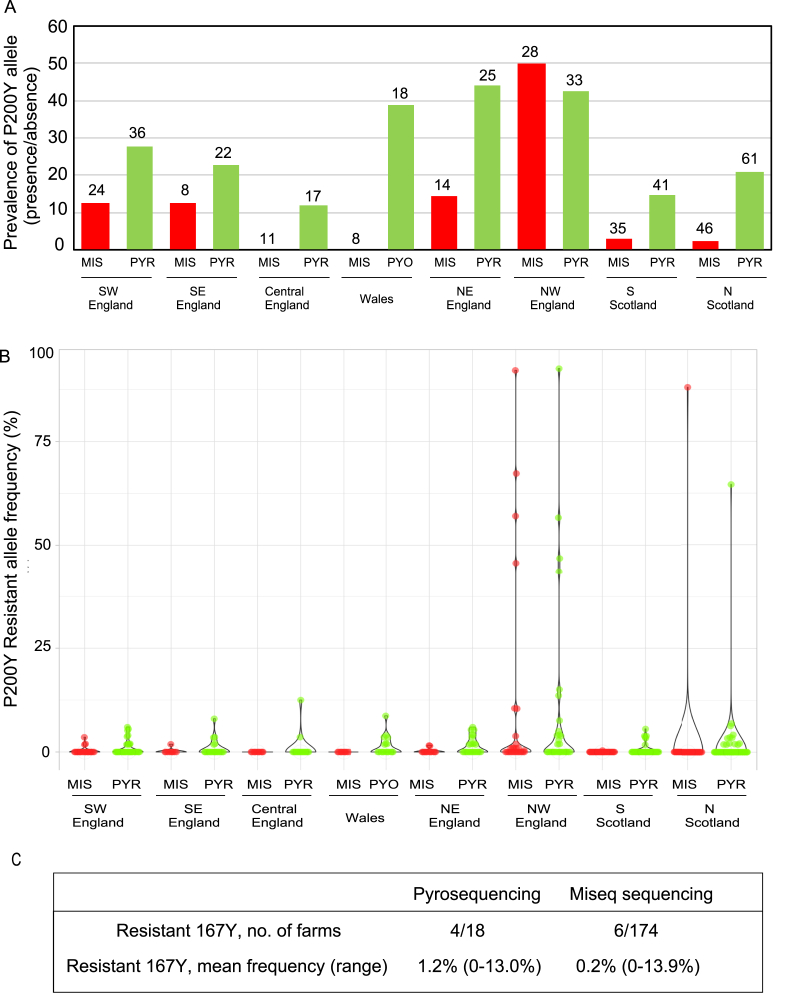
Fig. 3.
Frequency of resistant mutations at codons 167 and 200 in Nematodirus battus populations on UK farms obtained from pyrosequencing of individual larvated eggs and larvae (n = 253 populations) (PYR) and deep amplicon sequencing bulk egg/larval DNA preparations (n = 174 populations) (MIS). Note not all farms were examined by both techniques
Panel A. Number of farm populations in which resistant alleles were identified at codon 200 (y-axis) by deep amplicon sequencing (MIS) and pyrosequencing (PYR), separated by geographic region (x-axis), with the total numbers of farms analysed by each technology indicated in parentheses above the bars.
Panel B. Frequency of resistant mutation at codon 200 displayed as a violin plot with the 95% confidence interval of the median frequency per geographic region indicated.
Panel C. Frequencies of resistant mutation at codon 167 summarised with the total number of farms analysed by each assay (pyrosequencing n = 18/deep amplicon sequencing n = 174).
2.5. Next-generation amplicon sequencing using the Illumina MiSeq platform
Deep amplicon sequencing of the β-tubulin isotype-1 gene was carried-out on DNA extracted from pools of 500–1000 eggs/larvae with two separate amplicons. A total of 174 N. battus populations had sufficient parasite material for inclusion. To allow for a direct comparison with the pyrosequencing data the first was the same amplicon used for the P198/P200 pyrosequencing assay (Fig. 2A, Supplementary Table S1). These primers produced an amplicon of 198bp and contained the codons P198/P200. Primers were designed to generate a second, larger amplicon (321bp) containing all three codons, P167, P198 and P200 (Fig. 2A, Supplementary Table S1). The PCR products produced with these primers included adaptor sequences that enabled the attachment of unique barcodes to individual samples in a two-step PCR approach and thus allowed samples to be pooled together to create amplicon libraries for sequencing.
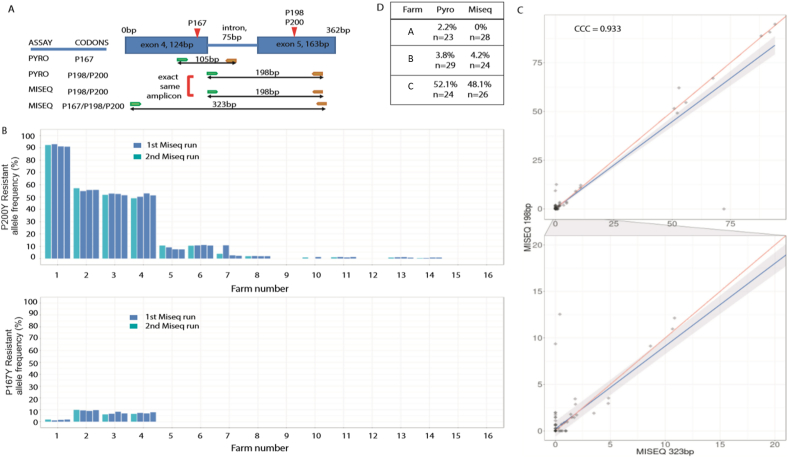
Fig. 2.
Reproducibility of deep amplicon sequencing to determine the frequency of resistance in Nematodirus battus from bulk larvae DNA preparations.
Panel A. Schematic representation of location of primers used and size of amplicons produced for the deep amplicon sequencing assays and how they relate to the amplicons generated for the pyrosequencing of individual N. battus larvae.
Panel B. Frequency of benzimidazole resistance associated mutations at codons 167 and 200 of the isotype 1 β-tubulin gene generated by deep amplicon sequencing (323bp amplicon) from pools of 500–1000 N. battus larvae per sample. Amplification and sequencing was independently conducted four times for each of the sixteen farms to determine reproducibility of assay.
Panel C. Scatter-plot comparing the frequencies of the benzimidazole resistance associated mutation at codon 200 generated by deep amplicon sequencing of the two different amplicons (198bp and 323bp) spanning codon 200. Frequencies generated from sequencing the larger 323bp amplicon on the x-axis and frequencies generated from sequencing the smaller 198bp amplicon on the y-axis. The upper plot is over the range of frequencies of 0–100% and the lower plot over the range of frequencies 0–20% to better visualize the low frequency samples. Lin's Agreement analysis shown (0.933).
Panel D. Frequency of resistant mutation at codon 200 generated from deep amplicon sequencing (amplicon, 323bp) and pyrosequencing populations of individual eggs/larvae (n = 23–29) from three different farms clearly showing consistency between assays when analysing the exact same template.
To test the reproducibility of deep amplicon sequencing, technical replicates were analysed from sixteen farm populations; amplification and sequencing was conducted a total of four times on each of these samples as described below and the allele frequencies compared.
Deep amplicon sequencing was also conducted on DNA extracted from 30 individual eggs/larvae from three farm populations for direct comparison with pyrosequencing results. Sample preparation and DNA extraction was conducted as described above for individual parasite analysis using pyrosequencing.
The initial PCR reactions were performed in 25 μl reaction volumes using Kappa HiFi Hotstart PCR kit (5 μl 5X buffer, 10 mM dNTP mix, 10 μM Nb forward primer + adapter sequence, 10 μM Nb reverse primer + adapter sequence, 0.5U Kappa HiFi hotstart polymerase, 4–10 μl of template DNA or molecular grade water for PCR negative controls) with the following cycling conditions; 95 °C for 2 min followed by 30 cycles of 98 °C for 20s, 62 °C for 15s and 72 °C for 15s, final extension phase at 72 °C for 2 min. To remove unused primers, dNTPs and contaminants, the amplicons were purified with AMPure XP beads. Illumina barcodes were added to the first round adapter PCR product using a second-round amplification. PCR reactions were performed in 25 μl reaction volume using Kappa HiFi Hotstart PCR kit (5 μl 5X buffer, 10 mM dNTP mix, 10 μM forward primer (S502 – S522), 10 μM reverse primer (N701 – N729), 0.5U Kappa HiFi hotstart polymerase, 3 μl of first round PCR product and molecular grade water) with the following cycling conditions; 98 °C for 45s followed by seven cycles of 98 °C for 20s, 62 °C for 20s and 72 °C for 2 min. Unique combinations of barcoded primers were added to each PCR reaction to allow for sample identification after sequencing, using primer set Nextera XT index kit V2 set (Illumina, USA) (Supplementary Table S2). Second round amplicons were bead purified to remove unwanted contaminants. Individual samples were quantified and pooled to create a normalised library (50 ng of each sample). The library was quantified by qPCR using the Illumina library Quantification Kit and Universal qPCR Mix (Kappa Biosystems) that uses absolute quantification against supplied DNA standards. The library was then diluted to 4 nM for denaturation, combined with 20% PhiX and loaded onto the Miseq sequencer following standard procedures.
2.6. Analysis pipeline for Illumina sequence data
The analysis pipeline was adapted from a previously published method (Avramenko et al., 2019). Initially, the software package, Mothur v1.41.0 (Schloss et al., 2009) was used to assign specific sequences to their respective nematode species of origin. In brief, forward and reverse sequences were merged to produce contigs, these contigs were then filtered and sequences were removed on the basis of contig size (<200bp and >450bp) and the presence of ambiguities in the overlapping region. The sequences were then aligned to a database of nematode isotype-1 β-tubulin sequences (Supplementary Data S1; for details see Avramenko et al., 2019) and removed if they did not align to at least 10% of any other sequences with at least 60% similarity. The remaining sequences were assumed to be solely β-tubulin isotype-1 sequences and were taxonomically classified with the k-nearest-neighbour method (k = 3) to generate a list of sequences belonging to each species from each sample. Returning to the original raw data, paired-end reads from each sample were merged with the default settings in PEAR 0.9.6 (Zhang et al., 2014) to create contiguous FASTQ sequences. The species list generated by Mothur was then used to divide the FASTQ sequences from each sample generated by PEAR into individual species-specific FASTQ files.
To identify non-synonymous mutations in codons 167, 198 and 200 the species-specific FASTQ files were aligned with BWA 0.7.12 to their species reference sequences (consensus isotype-1 β-tubulin sequences with known susceptible haplotypes at codon 167, 198 and 200; Supplementary Data S2) using default settings (Li et al., 2009). The resulting aligned SAM files were converted to BAM files (SAMtools 0.1.19, Li et al., 2009) and the missing headers and sample information was retrieved with Picard Tools 1.139 (Broad Institute: https://boardinstitute.github.io/picard). This generated a reference sequence directory that could be used for variant calling. Variants from each species were called with VarDict 1.5 (settings: amplicon-aware variant calling, 0.1% minimum frequency and 40 maximum allowed mismatches). Variants with greater than 200 reads for a respective species per sample were translated and annotated using the SnpEff 4.3.1 bioinformatic tool (Cingolani et al., 2012). SnpSift 4.3.1 was also used to sort variants and only variants with moderate to high effects (i.e. non-synonymous and frame-shift mutations) were kept. For the purposes of this paper, only Nematodirus battus variants resulting in changes at codons 167, 198, and 200 were analysed further.
2.7. Statistical analysis
Allele frequencies of technical replicates prepared from sixteen farm populations analysed by deep amplicon sequencing were compared using a Kruskal Wallis test in SPSS v24 (IBM) and the resistant allele frequencies obtained from pyrosequencing and deep sequencing were compared by Lin's Concordance Correlation Coefficient, calculated in SPSS with the syntax provided by https://gjyp.nl/marta/Lin.sps. rc (CCC) values > 0.8 are classified as a near perfect agreement (rc = 1, would represent perfect agreement). Populations were divided into geographical regions for analysis (map boundaries detailed in Supplementary Fig. S1). Binomial logistic regression analysis was carried out to calculate the increase in risk of identifying F200Y resistant alleles between regions using pyrosequencing results (R version 3.2.5). Hardy-Weinberg analysis was performed to determine whether the P200 locus was under active selection at the point of the study. Observed and expected homo/hetero-zygote frequencies were compared using a chi-squared analysis, performed in Microsoft Excel.
3. Results
3.1. Reproducibility of the deep amplicon sequencing assay
To evaluate the technical reproducibility of the amplicon sequencing assay to estimate the frequency of resistant mutations, sixteen bulk farm samples were sequenced on two separate occasions several months apart using freshly-generated amplicons from the same archived DNA preparation on each occasion. The frequencies of both the 167Y and 200Y mutations were found to be very similar between separate sequencing runs (Fig. 2B). In addition, the same amplicon was barcoded with three different sets of indices on the same run and treated as separate samples throughout the bioinformatic pipeline to investigate the effect barcoding may have on the outcome of the assay. Frequencies of the 167Y mutation (H = 0.006, p < 0.02) and the 200Y mutation (H = 0.124, p < 0.05) were found to have a low variance (Fig. 2B) and showed that the deep sequencing was technically robust producing consistent results both within a sequencing run (same amplicon is used) and between sequencing runs when independent amplicons from the same DNA preparation was used.
3.2. Comparison of two separate deep amplicon sequencing assays and negating primer-induced bias
Different primer pairs were used to amplify two distinct amplicons; the first 323bp in length was designed to encompass all three resistant mutations (codons 167, 198 and 200) and second was 198bp in length which was exactly the same amplicon used for the pyrosequencing assay for codons 198 and 200 (Fig. 2A). The use of the same amplicon for both pyrosequencing and deep sequencing enabled a direct comparison between the technologies to be made at least for codon 198 and 200 by eliminating any potential differences in primer-pair amplification efficiency or haplotypes targeted. The allele frequencies of 200Y derived from the two deep sequencing assays proved very similar (H = 0.293, p < 0.05) with only 2.5% of samples producing more than a 5% difference in resistant allele frequency between the two sequencing assays (Lins’ CCC, rc = 0.933; Fig. 2C). This shows that the primers used in the pyrosequencing and amplicon sequencing assays do not contribute significantly to any potential differences observed in the frequencies of the resistance mutations.
3.3. Comparison of individual parasite genotyping by pyrosequencing and deep amplicon sequencing
Initially a small sub-set of three farm populations were selected to examine the potential of the two technologies to genotype populations of individual eggs/larvae and estimate frequency of the 200Y resistant mutation on each of the farms. DNA was prepared from between 23 and 29 individual eggs/larvae and genotyped at codon 200 by pyrosequencing and by deep amplicon sequencing. The amplicon sequencing successfully amplified 98.4% of the individual parasites previously genotyped via pyrosequencing, also confirming that they were N. battus. Comparison of the frequency of the 200Y resistant mutation within each of these farm populations indicated good agreement between pyrosequencing and deep amplicon sequencing of individual parasites when the same template was used for each assay (Fig. 2D).
3.4. Prevalence and frequency of resistant mutations in N. battus populations in the UK
Pyrosequencing was performed on 257 Nematodirus battus populations using individual larvated egg/larvae (n = 30). Four populations failed to reach the 80% threshold, therefore results represent the remaining 253 populations. Within the 257 populations, 174 were analysed by deep amplicon sequencing using DNA made from pools of 500–1000 larvated eggs and larvae, including the four populations removed from pyrosequencing analyses. A total of 170 populations were analysed using both platforms. The following results include all populations analysed by each platform respectively. The resistant allele at codon 200 was identified on 68 of 253 (26.8%) farms tested throughout the UK by pyrosequencing and 22 of 174 (12.6%) farms by deep amplicon sequencing (Fig. 3A). The number of farms where the resistant mutation was identified was highest for the NW of England where it was found on 14 of the 33 farms (42.4%) for pyrosequencing and 14 of the 28 farms (50.0%) for deep amplicon sequencing. The overall individual allele frequency was low; (mean ± SEM) 2.2 ± 0.6% by pyrosequencing and 2.3 ± 0.9% using deep sequencing but geographical separation of the data revealed that the F200Y resistant allele frequency was 9.01% ± 3.6% on the basis of pyrosequencing and 10.5% ± 4.5% on the basis of deep amplicon sequencing in the NW of England (Fig. 3B). The wide distribution of the F200Y resistant allele was analysed by logistic regression analysis revealing that it was almost three times more likely in the NW of England compared to other regions of the UK (estimate 2.7; 95% CI. 2.1–3.5, p < 0.001; Fig. 3B). The resistant allele frequency was also found at high frequency on one particular farm in the North of Scotland; 64.6% for pyrosequencing and 88.0% for deep amplicon sequencing (Fig. 3B).
Deep amplicon sequencing identified the F167Y mutation in six farm isolates of the 174 tested. The overall frequency of F167Y was low; 0.2 ± 0.07%, range 0–13.9%. A subset of 18 N. battus farm populations were analysed at codon 167 by pyrosequencing. Of the 18 populations tested, the F167Y mutation was identified in 4 populations ranging from 3 to 13% (Fig. 3C). No BZ-resistance associated mutations were identified at codon 198 (E198A or E198L) in any of the populations tested by either pyrosequencing or deep amplicon sequencing.
3.5. Hardy-Weinberg analysis
The F167Y mutation was found to be mutually exclusive with F200Y, i.e. double heterozygous individuals were identified but no double homozygous resistant individuals were observed. Heterozygote: homozygote ratios were obtained from pyrosequencing for codon 200. The genotype ratios were compared with the Hardy-Weinberg equilibrium to evaluate whether the loci were under active selection at the time of sampling. Analysis concluded that codon 200 was departed from the Hardy-Weinberg equilibrium, consistent with the locus being under selection (χ2 = 833, p < 0.001).
3.6. Overall comparison of next-generation amplicon sequencing and pyrosequencing results
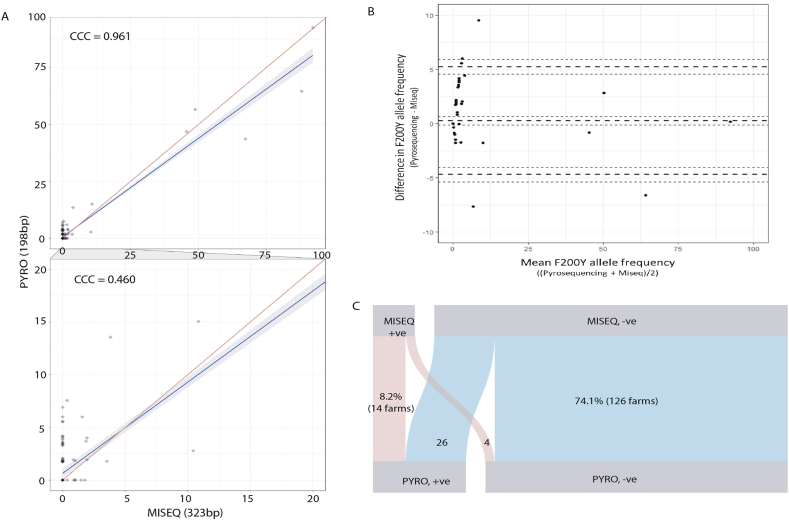
N. battus populations from 170 farms were successfully analysed by both deep amplicon sequencing and pyrosequencing at the F200Y locus. Results were largely comparable between the two technologies. Pyrosequencing identified the resistant F200Y mutation in 40 of 170 analysed farm populations with a mean resistant allele frequency of 2.6 ± 0.8% (±SEM) (range 0–92.6%). The F200Y mutation was identified in 22 of the 170 farm populations by deep sequencing; mean resistant allele frequency 2.3 ± 0.9% (range 0–92.1%). Comparison of the pyrosequencing and deep amplicon sequencing F200Y allele frequencies by Lins' concordance analysis found significant agreement between the results (Lins' Concordance Correlation Coefficient, rc = 0.961, Fig. 4A). Repetition of this analysis with just data below a threshold frequency of 20% revealed much weaker agreement between the results of single egg/larvae pyrosequencing and bulk parasite deep amplicon sequencing (Lins’ CCC, rc = 0.460, Fig. 4A) highlighting variation between the two assays at lower frequencies. Another standard approach to compare the paired results is to use a Bland-Altman plot that plots the mean F200Y allele frequency (x-axis) against the difference in F200Y allele frequency between the two technologies (y-axis): data points at the lower frequencies both above and below the origin on the y-axis highlight the discrepancies (Fig. 4B). On the basis of presence or absence with a 1% threshold a parallel-set plot revealed that the two assays agreed for 140 out of a total of 170 farms (82.4% of farms). Suspected resistant alleles; where one technology identified the F200Y mutation and the other did not, occurred in 17.7% of farm isolates, suspect resistant alleles were more commonly identified by pyrosequencing than deep sequencing; in 15.3% (26/170 farms) and 2.4% (4/170 farms) of populations respectively (Fig. 4C). The resistant allele frequencies detected in suspect resistant populations were relatively low; 1–1.8% resistant alleles in deep amplicon sequencing and 1.7–7.6% in pyrosequencing. Although the F200Y SNP was identified in a greater number of regions by pyrosequencing compared to deep amplicon sequencing, both technologies identified resistant alleles in the regions which were highlighted as confirmed or suspected focal regions of resistance; North West England and North Scotland respectively.
Fig. 4.
Comparison of the two different assays, pyrosequencing populations of individual egg/larvae and deep amplicon sequencing of bulk egg/larval DNA preparations to estimate frequency of resistant mutations in the N. battus β-tubulin isotype-1 gene.
Panel A. Scatter-plot of frequencies of resistant mutation at codon 200 generated from deep amplicon sequencing and pyrosequencing. Frequencies generated from sequencing on the x-axis and frequencies generated from pyrosequencing on the y-axis. Lin's Agreement analysis shows (0.961) that overall there is very little difference between the results generated from the two different assays except at the lower frequencies (<20%) when Lins' Agreement estimate drops to 0.460.
Panel B. Bland-Altman plot comparing F200Y allele frequency results obtained from pyrosequencing vs. deep amplicon sequencing. The mean F200Y allele frequency (x-axis) was plotted against the difference in F200Y allele frequency between the two technologies (y-axis) to explore the variation observed. The dashed lines represent the mean difference and standard deviations with the respective 95% confidence intervals of each indicated by dotted lines.
Panel C. Parallel-sets plot displaying the relative proportions of samples that are either positive i.e. the resistant mutation (200Y) is present on a farm or negative i.e. the resistant mutation is absent on that farm, at a threshold of 1%. The cross-over regions represent the relative proportion of samples that disagree between the assays and were categorised as suspected resistant farms (where one technology identified the F200Y mutation and the other did not). Suspect resistant alleles were more commonly identified by pyrosequencing than deep sequencing.
4. Discussion
The results of the current study have demonstrated that benzimidazole resistance mutations are emerging but are at an early stage of dissemination in UK populations of N. battus. Although N. battus has been previously believed to be refractory from the development of anthelmintic resistance, we have identified two different BZ-resistance associated SNPs in UK populations of this parasite species - at codons 167 and 200 of the β-tubulin isotype 1 gene - by both pyrosequencing and next-generation amplicon sequencing. F200Y was found to be the predominant mutation, as is most often the case in other ovine GIN species (Barrere et al., 2013; Chaudhry et al., 2014; Ramunke et al., 2016; Avramenko et al., 2019). Analysis of the resistance and suscepible allele ratios is consistent with this SNP being under selection in N. battus populations although further analysis on population structure and/or neutral control loci is needed to confirm this. In this study F200Y was identified in populations from throughout the UK, in 12–27% of the populations tested, depending on the methodology used, albeit at a very low individual resistant allele frequency overall (∼2%) compared with the allele frequencies observed in other strongyle nematode species (Ramunke et al., 2016). The F167Y SNP was identified for the first time in this species but, the prevalence was found to be extremely low, detected at an allele frequency greater than 1% in populations originating from only four independent farms. These results suggest that this mutation has very recently emerged in this species.
BZ-resistance in H. contortus and T. circumcincta is at an advanced stage in the UK and previous work has provided evidence that F200Y has likely arisen multiple independent times (Redman et al., 2015). In contrast, the overall low frequency of resistance alleles together with the strong regional distribution of those few farms with high F200Y mutation frequencies suggests that resistance is at an early stage of development in the UK and focused in the NW of England. Dissemination of resistance within this region and beyond is possibly mediated by animal movements and trade. This is analogous to the situation described for the relatively rare E198A mutation in Southern India for which the molecular evidence suggests early spread from a single source (Chaudhry et al., 2015a, Chaudhry et al., 2015b). Understanding the emergence of anthelmintic resistance in parasitic nematodes of veterinary importance has generally been hampered by resistance being at a late stage by the time it has been detected and studied. The detection of BZ-resistance at such an early stage of the emergence in N. battus in the UK provides a unique opportunity to study its development and dissemination, using both the sample set presented here and resampling populations in the future.
The local spread of resistant alleles between farms in focal regions may be linked with animal trade, wildlife movement or weather events such as flooding to move parasites from one farm to another. Wildlife are frequently observed grazing in livestock fields and have been shown to carry GIN typically associated with sheep and cattle (Pato et al., 2013; Chintoan-Uta et al., 2014). N. battus can establish viable infections in wild deer and rabbits (Dunn, 1965; Boag, 1972), making wildlife reservoirs a possible route of local transmission. A previous study demonstrated the passage of a BZ-resistant isolate of Haemonchus contortus in roe deer which produced viable eggs capable of infecting cattle and sheep (Chintoan-Uta et al., 2014), and so at least in principal, wild deer have the potential to contribute to the spread of BZ-resistant N. battus between farms. A review of historic complaints to the Agricultural Development and Advisory Service regarding damage to agricultural land or crops by wild deer indicated that red deer were most commonly associated with pasture and forage crop damage whilst other species such as fallow and roe deer were linked to loss of cereals, arable crops and orchard damage (Putman and Moore, 1998). A recent study of the geographic distribution of red deer in the UK illustrated that the North West of England, where high F200Y resistant allele frequencies were identified in the current study, have been inhabited by wild red deer consistently since the study began in 2007 (British Deer Society, 2016). Further research would be required to assess the distribution of N. battus within the wild deer population.
Another interesting question is why the emergence of anthelmintic resistance has been much slower in N. battus compared to many other GIN species, including other Nematodirus species (Oliver et al., 2016). The delay in resistance emergence may be related to distinct differences in life history traits between the species. For example, N. battus is typically restricted to a narrow spring infection, the large population active at one time following synchronised hatching may provide a large refugia, thus slowing the expansion of resistance. Alternatively, if there were a greater fitness cost of resistance alleles, or genetically linked polymorphisms, in N. battus compared to other species this could be responsible for the delay.
Within this study we also compared data on BZ-resistance SNP frequencies based on pyrosequence genotyping and deep amplicon sequencing using the Illumina Miseq platform. There is currently no ‘gold standard’ method for detecting and measuring SNP mutations. However, in the current study we demonstrated that both pyrosequencing and next–generation amplicon sequencing were valuable methods for analysing the presence and frequency of SNPs in N. battus. Results were similar between the technologies with a high level of agreement overall. However, there was some discrepancy between the platforms at low resistance allele frequencies. The technologies agreed on the presence or absence (using a threshold of 1%) of the F200Y SNP in 82.4% of the populations tested. However, 17.7% of the populations were reported to contain the resistance allele by one assay and not the other. This discrepancy exclusively occurred at low allele frequencies. Resistance alleles were reported in 26 farm populations by pyrosequencing but not by deep amplicon sequencing (F200Y, frequency range 1.7–7.6%). The reverse was true for four populations (F200Y, frequency range 1.0–1.8%). The reasons for the variation observed between platforms, with the pyrosequencing suggesting that more populations have low frequency resistance alleles than the MiSeq, is unclear at this point. Pyrosequencing was performed on 30 individual parasites per population whilst deep amplicon sequencing used pools of 500–1000 and so it is unlikely that the lack of detection by the amplicon sequencing was due to resistant individuals in the population not being sampled. Also, we ruled out the possibility that the amplicon sequencing assay might be missing specific haplotypes that were detected by pyrosequencing assay by using identical primers to generate the 198bp amplicons for both amplicon and pyrosequencing assays (Fig. 2A). Finally, although extensive measures were taken to minimize contamination risk at each stage of the process, and negative controls included in each sample set, it is not possible to completely rule out rare template contamination events. The consequences of rare contamination events will differ depending on the methodologies and platforms used. For example, in the case of pyrosequencing, where 30 individual larvae were genotyped per population, even if there were as few as 1 in 300 samples being positive due to template contamination, that would result in as many as 1 in 10 populations being falsely called as positive for the presence of low frequency resistance mutations. In contrast, if 1 in 300 samples were positive due to template contamination, the deep amplicon sequencing assay would only falsely suggest 1 in 300 populations were positive for the resistance mutation since only a single PCR is performed per population. However, in the case of amplicon sequencing, such a false positive could lead to a large number of sequence Illumina reads that carry the mutation and so falsely suggest the presence of the resistance mutation at a high frequency in the population. Consequently, the same template contamination events would have different effects on the false positive rate between the two different methodologies. It would lead to a larger number of false positive populations for the resistance mutation being suggested by individual egg/larvae pyrosequencing but these mutations appear to be present at low frequency in the affected populations. In contrast, the deep amplicon sequencing would suggest fewer false positive populations but could falsely suggest the resistance mutations was present at a high frequency in the smaller number of affected populations. These issues illustrate that more work needs to be undertaken to develop protocols and controls to account for and mitigate rare template contaminations events when applying these assays to large scale surveillance particularly when resistance is at an early stage of emergence.
Although there was variation in results between the two methods for low frequency alleles, our results show that both methods are valuable diagnostic tools. Previous research in other nematode species suggests clinical drug inefficacy is believed to require resistant allele frequencies to be above 10–30% (von Samson-Himmelstjerna et al., 2009; Cudekova et al., 2010). Hence, either method would be sufficient as a tool to diagnose possible anthelmintic resistance following suspected drug failure. For other research applications, each method has its own advantages and disadvantages depending on the nature of the question being asked. For example, the ability to screen a large number of pooled samples by next-generation amplicon sequencing from a single MiSeq run by using unique barcoded primer pairs makes this method better suited to large-scale projects or central diagnostic/research laboratories. However, for smaller scale studies, or if individual genotypes are required to provide information on hetero- and homozygous genotype ratios, the pyrosequencing platform provides a much more cost effective method.
5. Conclusions
Analysis of a large number of UK N. battus populations identified two SNPs previously associated with BZ-resistance in other GIN species; at codons 167 and 200 of the β-tubulin isotype 1 gene. F200Y was identified in approximately a quarter of the populations tested, at very low frequency on most farms on which it was detected. Four out of the five farms with a high frequency were located in the NW of England, possibly indicating an early emergence of resistance from that region of the country. The F167Y was also identified but only at a very low frequency suggesting an even earlier emergence of this mutation. The results discussed here provide a benchmark of resistance in this species, providing a unique opportunity to follow the progress of BZ-resistance in the future.
Next-generation amplicon sequencing and pyrosequencing assays were successfully developed for the detection and quantification of SNPs associated with BZ-resistance in N. battus. Results from the two methods were comparable for the F200Y mutation overall, believed to be key in conferring BZ-resistance in the species. Although, there were some discrepancies for low frequency alleles, our results suggest that both methods can be utilised to monitor the development and dissemination of resistance SNPs in N. battus and help confirm BZ-resistance in cases of suspected clinical drug failure.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This research was funded by the Animal and Horticulture Development Board PhD studentship fund and The Scottish Government's Rural and Environment Science and Analytical Services Division (RESAS) Strategic Research Programme 2016–2021. Additional funding was awarded from Moredun Research Institute Diagnostic Pillar to cover travel costs to visit the University of Calgary and reagent costs for next-generation amplicon sequencing analysis were kindly covered by Dr John Gilleard, University of Calgary. The authors would like to thank EPIC is the Centre of Expertise on Animal Disease Outbreaks. Thanks also to Iain Richards, Judith Lees, Donald McClean, Ian Gill, Eric Morgan, Nor Azlina Abdul Aziz and the other farmers, vets, Suitably Qualified Persons and advisors who assisted in sample collection throughout the project.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijpddr.2020.03.001.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Abbott K.A., Taylor M., Stubbings L.A. 4th ed. 2012. Sustainable Worm Control Strategies for Sheep. [Google Scholar]
- Avramenko R.W., Redman E.M., Lewis R., Yazwinski T.A., Wasmuth J.D., Gilleard J.S. Exploring the gastrointestinal "nemabiome": deep amplicon sequencing to quantify the species composition of parasitic nematode communities. PLoS One. 2015;10 doi: 10.1371/journal.pone.0143559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avramenko R.W., Redman E.M., Melville L., Bartley Y., Wit J., Queiroz C., Bartley D.J., Gilleard J.S. Deep amplicon sequencing as a powerful new tool to screen for sequence polymorphisms associated with anthelmintic resistance in parasitic nematode populations. Int. J. Parasitol. 2019;49:13–26. doi: 10.1016/j.ijpara.2018.10.005. [DOI] [PubMed] [Google Scholar]
- Barrere V., Falzon L.C., Shakya K.P., Menzies P.I., Peregrine A.S., Prichard R.K. Assessment of benzimidazole resistance in Haemonchus contortus in sheep flocks in Ontario, Canada: comparison of detection methods for drug resistance. Vet. Parasitol. 2013;198:159–165. doi: 10.1016/j.vetpar.2013.07.040. [DOI] [PubMed] [Google Scholar]
- Boag B. Helminth parasites of the wild rabbit Oryctolagus cuniculus (L.) in north east England. J. Helminthol. 1972;46:73–78. doi: 10.1017/s0022149x00022136. [DOI] [PubMed] [Google Scholar]
- British Deer Society . 2016. Deer Distrobution Study 2016.https://www.bds.org.uk/index.php/research/deer-distribution-survey [online] available at. [accessed 12th December 2018] [Google Scholar]
- Chaudhry U., Miller M., Yazwinski T., Kaplan R., Gilleard J. The presence of benzimidazole resistance mutations in Haemonchus placei from US cattle. Vet. Parasitol. 2014;204:411–415. doi: 10.1016/j.vetpar.2014.05.025. [DOI] [PubMed] [Google Scholar]
- Chaudhry U., Redman E.M., Abbas M., Muthusamy R., Ashraf K., Gilleard J.S. Genetic evidence for hybridisation between Haemonchus contortus and Haemonchus placei in natural field populations and its implications for interspecies transmission of anthelmintic resistance. Int. J. Parasitol. 2015;45:149–159. doi: 10.1016/j.ijpara.2014.09.002. [DOI] [PubMed] [Google Scholar]
- Chaudhry U., Redman E.M., Raman M., Gilleard J.S. Genetic evidence for the spread of a benzimidazole resistance mutation across southern India from a single origin in the parasitic nematode Haemonchus contortus. Int. J. Parasitol. 2015;45:721–728. doi: 10.1016/j.ijpara.2015.04.007. [DOI] [PubMed] [Google Scholar]
- Chaudhry U., Redman E.M., Ashraf K., Shabbir M.Z., Rashid M.I., Ashraf S., Gilleard J.S. Microsatellite marker analysis of Haemonchus contortus populations from Pakistan suggests that frequent benzimidazole drug treatment does not result in a reduction of overall genetic diversity. Parasites Vectors. 2016;9:349. doi: 10.1186/s13071-016-1624-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P., Platts A., Wang L.L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., Ruden D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. 2012;6:80–92. doi: 10.4161/fly.19695. (Austin) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintoan-Uta C., Morgan E.R., Skuce P.J., Coles G.C. Wild deer as potential vectors of anthelmintic-resistant abomasal nematodes between cattle and sheep farms. Proc. Biol. Sci./R. Soc. 2014;281:20132985. doi: 10.1098/rspb.2013.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cudekova P., Varady M., Dolinska M., Konigova A. Phenotypic and genotypic characterisation of benzimidazole susceptible and resistant isolates of Haemonchus contortus. Vet. Parasitol. 2010;172:155–159. doi: 10.1016/j.vetpar.2010.04.022. [DOI] [PubMed] [Google Scholar]
- Dunn A.M. The gastro-intestinal helminths of wild ruminants in Britain. I. Roe deer, Capreolus capreolus capreolus. Parasitology. 1965;55:739–745. [PubMed] [Google Scholar]
- Ghisi M., Kaminsky R., Maser P. Phenotyping and genotyping of Haemonchus contortus isolates reveals a new putative candidate mutation for benzimidazole resistance in nematodes. Vet. Parasitol. 2007;144:313–320. doi: 10.1016/j.vetpar.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Jackson F., Christie M. Quantitative recovery of floatable helminth eggs from 1 g. of ruminant faeces for counting followed by hatching for identification. Trans. R. Soc. Trop. Med. Hyg. 1972;66:546. [PubMed] [Google Scholar]
- Kwa M.S., Veenstra J.G., Roos M.H. Benzimidazole resistance in Haemonchus contortus is correlated with a conserved mutation at amino acid 200 in beta-tubulin isotype 1. Mol. Biochem. Parasitol. 1994;63(2):299–303. doi: 10.1016/0166-6851(94)90066-3. [DOI] [PubMed] [Google Scholar]
- Kwa M.S., Veenstra J.G., Van Dijk M., Roos M.H. Beta-tubulin genes from the parasitic nematode Haemonchus contortus modulate drug resistance in Caenorhabditis elegans. J. Mol. Biol. 1995;246:500–510. doi: 10.1006/jmbi.1994.0102. [DOI] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin P.J., Anderson N., Jarrett R.G. Resistance to benzimidazole anthelmintics in field strains of Ostertagia and Nematodirus in sheep. Aust. Vet. J. 1985;62:38–43. doi: 10.1111/j.1751-0813.1985.tb14230.x. [DOI] [PubMed] [Google Scholar]
- Middleberg A., McKenna P.B. Oxfendazole resistance in Nematodirus spathiger. N. Z. Vet. J. 1983;31:65–66. doi: 10.1080/00480169.1983.34971. [DOI] [PubMed] [Google Scholar]
- Mitchell S., Mearns R., Richards I., Donnan A.A., Bartley D.J. Benzimidazole resistance in Nematodirus battus. Vet. Rec. 2011;168:623–624. doi: 10.1136/vr.d3584. [DOI] [PubMed] [Google Scholar]
- Morrison A.A., Mitchell S., Mearns R., Richards I., Matthews J.B., Bartley D.J. Phenotypic and genotypic analysis of benzimidazole resistance in the ovine parasite Nematodirus battus. Vet. Res. 2014;45:116. doi: 10.1186/s13567-014-0116-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obendorf D.L., Nicholls J., Koen T., Lacy E. Benzimidazole resistant Nematodirus species in tasmania. Aust. Vet. J. 1991;68:72–73. doi: 10.1111/j.1751-0813.1991.tb03142.x. [DOI] [PubMed] [Google Scholar]
- Oliver A., Pomroy W.E., Leathwick D.M. Benzimidazole resistance in Nematodirus spathiger and Nematodirus filicollis in New Zealand. N. Z. Vet. J. 2016;64:201–206. doi: 10.1080/00480169.2016.1149117. [DOI] [PubMed] [Google Scholar]
- Pato F.J., Vazquez L., Diez-Banos N., Lopez C., Sanchez-Andrade R., Fernandez G., Diez-Banos P., Panadero R., Diaz P., Morrondo P. Gastrointestinal nematode infections in roe deer (Capreolus capreolus) from the north west of the Iberian Peninsula: assessment of some risk factors. Vet. Parasitol. 2013;196:136–142. doi: 10.1016/j.vetpar.2013.01.027. [DOI] [PubMed] [Google Scholar]
- Putman R.J., Moore N.P. Impact of deer in lowland Britain on agriculture, forestry and conservation habitats. Mamm Rev. 1998;28:141–163. [Google Scholar]
- Ramunke S., Melville L., Rinaldi L., Hertzberg H., de Waal T., von Samson-Himmelstjerna G., Cringoli G., Mavrot F., Skuce P., Krucken J., Demeler J. Benzimidazole resistance survey for Haemonchus, Teladorsagia and Trichostrongylus in three European countries using pyrosequencing including the development of new assays for Trichostrongylus. Int. J. Parasitol.: Drugs Drug Resist. 2016;6:230–240. doi: 10.1016/j.ijpddr.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redman E., Whitelaw F., Tait A., Burgess C., Bartley Y., Skuce P.J., Jackson F., Gilleard J.S. The emergence of resistance to the benzimidazole anthlemintics in parasitic nematodes of livestock is characterised by multiple independent hard and soft selective sweeps. PLoS Neglected Trop. Dis. 2015;9 doi: 10.1371/journal.pntd.0003494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargison N.D., MacLeay M., Morrison A.A., Bartley D.J., Evans M., Chaudhry U. Development of amplicon sequencing for the analysis of benzimidazole resistance allele frequencies in field populations of gastrointestinal nematodes and Ali et al 2019 Emergence and the spread of the F200Y benzimidazole resistance mutation in Haemonchus contortus and Haemonchus placei from buffalo and cattle. Int. J. Parasitol.: Drugs Drug Resist. 2019;10:92–100. doi: 10.1016/j.ijpddr.2019.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss P.D., Westcott S.L., Ryabin T., Hall J.R., Hartmann M., Hollister E.B., Lesniewski R.A., Oakley B.B., Parks D.H., Robinson C.J., Sahl J.W., Stres B., Thallinger G.G., Van Horn D.J., Weber C.F. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvestre A., Cabaret J. Mutation in position 167 of isotype 1 beta-tubulin gene of Trichostrongylid nematodes: role in benzimidazole resistance? Mol. Biochem. Parasitol. 2002;120:297–300. doi: 10.1016/s0166-6851(01)00455-8. [DOI] [PubMed] [Google Scholar]
- Silvestre A., Humbert J.F. Diversity of benzimidazole-resistance alleles in populations of small ruminant parasites. Int. J. Parasitol. 2002;32:921–928. doi: 10.1016/s0020-7519(02)00032-2. [DOI] [PubMed] [Google Scholar]
- Thomas R.J., Stevens A.J. Some Observations on Nematodirus disease in Northumberland and Durham. Vet. Rec. 1956;68:471–475. [Google Scholar]
- von Samson-Himmelstjerna G., Walsh T.K., Donnan A.A., Carriere S., Jackson F., Skuce P.J., Rohn K., Wolstenholme A.J. Molecular detection of benzimidazole resistance in Haemonchus contortus using real-time PCR and pyrosequencing. Parasitology. 2009:1–10. doi: 10.1017/S003118200800543X. [DOI] [PubMed] [Google Scholar]
- Zhang J., Kobert K., Flouri T., Stamatakis A. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics. 2014;30(5):614–620. doi: 10.1093/bioinformatics/btt593. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.