Significance
RNA helicases exert mechanical force that changes RNA configurations in many essential cellular pathways, e.g., during mRNA maturation or assembly of ribosomes. DEAH helicases work by translocating along RNA and thereby unwind RNA duplexes or dissociate bound proteins. Because DEAH proteins are poor enzymes without intrinsic selectivity for target RNAs, they require adapter proteins that recruit them to functional sites and enhance their catalytic activity. One essential class of DEAH activators is formed by G-patch proteins, which bind helicases via their eponymous glycine-rich motif. We solved the structure of a G-patch bound to helicase DHX15. Our analysis suggests that G-patches tether mobile sections of DEAH helicases together and activate them by stabilizing a functional conformation with high RNA affinity.
Keywords: G-patch proteins, DEAH/RHA helicase, ribosome biogenesis, splicing
Abstract
RNA helicases of the DEAH/RHA family are involved in many essential cellular processes, such as splicing or ribosome biogenesis, where they remodel large RNA–protein complexes to facilitate transitions to the next intermediate. DEAH helicases couple adenosine triphosphate (ATP) hydrolysis to conformational changes of their catalytic core. This movement results in translocation along RNA, which is held in place by auxiliary C-terminal domains. The activity of DEAH proteins is strongly enhanced by the large and diverse class of G-patch activators. Despite their central roles in RNA metabolism, insight into the molecular basis of G-patch–mediated helicase activation is missing. Here, we have solved the structure of human helicase DHX15/Prp43, which has a dual role in splicing and ribosome assembly, in complex with the G-patch motif of the ribosome biogenesis factor NKRF. The G-patch motif binds in an extended conformation across the helicase surface. It tethers the catalytic core to the flexibly attached C-terminal domains, thereby fixing a conformation that is compatible with RNA binding. Structures in the presence or absence of adenosine diphosphate (ADP) suggest that motions of the catalytic core, which are required for ATP binding, are still permitted. Concomitantly, RNA affinity, helicase, and ATPase activity of DHX15 are increased when G-patch is bound. Mutations that detach one end of the tether but maintain overall binding severely impair this enhancement. Collectively, our data suggest that the G-patch motif acts like a flexible brace between dynamic portions of DHX15 that restricts excessive domain motions but maintains sufficient flexibility for catalysis.
RNA helicases remodel RNA structures in an adenosine triphosphate (ATP)-dependent manner in a multitude of cellular processes, in all steps from synthesis of RNAs to their eventual degradation (1–4). They are key enzymes in the reorganization of RNA–protein complexes (RNPs) because they exert mechanical force that is required for transitions in the conformations of RNPs (5, 6).
DEAH/RHA helicases form an abundant family of enzymes in eukaryotes [Homo sapiens (hs), 15 proteins; Saccharomyces cerevisiae (sc), seven proteins (7)], which play prominent roles in RNA metabolism (2). These enzymes share a conserved core of tandem RecA domains (RecA1 and RecA2), which form a split active site for ATP hydrolysis at their interface (5, 7). This core is flanked by a common set of C-terminal domains (winged helix [WH], Ratchet, and oligonucleotide/oligosaccharide-binding fold [OB-fold]) and varying N termini (7) (Fig. 1A). The auxiliary C-terminal domains cooperate with the RecA domains by completing an RNA binding channel, which has been suggested to allow DEAH/RHA helicases to keep a stable grip on the RNA over multiple ATP hydrolysis cycles (5, 8, 9). They unwind RNA duplexes or strip RNA from proteins using the energy of ATP hydrolysis to move along the RNA in a 3′ to 5′ direction (6, 10). Recent crystal structures of DEAH/RHA helicases in different states have led to a mechanistic model for RNA translocation (8, 11–17): RecA2 carries out an opening and closing motion during transitions between the nucleotide-free and -bound states, respectively. In these two conformational extremes, five (open) vs. four (closed) RNA bases are stacked in the RNA binding channel between a long β-hairpin in RecA2 (β14 to β15 in hsDHX15/scPrp43; SI Appendix, Fig. S1) and a conserved loop in RecA1 (termed “Hook-turn”). This means that during progression into the open state, the β-hairpin and two other RNA-binding patches in RecA2 (termed “Hook-loop” and “motif V”; SI Appendix, Fig. S1) have to shift 1 nucleotide (nt) toward the 5′ end of the RNA. Thus, when the RecA domains close back up, at the start of the next hydrolysis cycle, the RNA is pushed by 1 nt through the RNA channel.
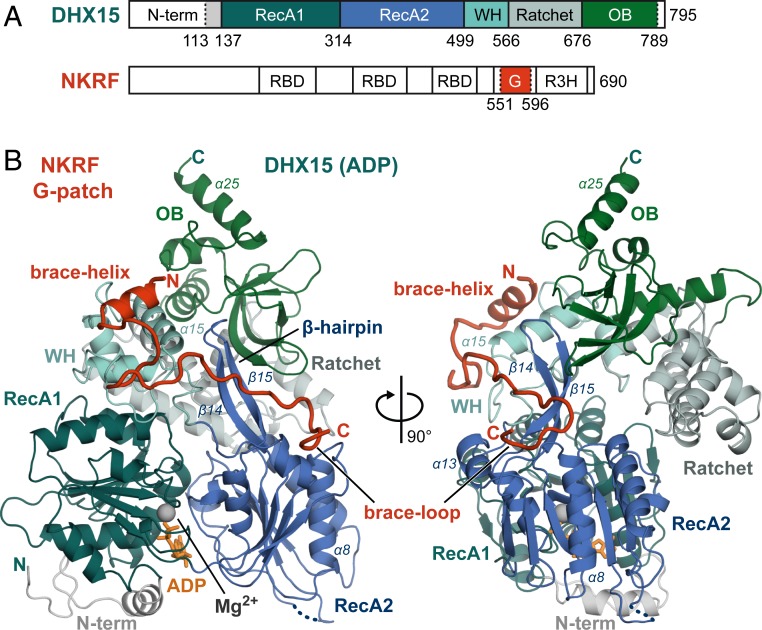
Fig. 1.
The NKRF G-patch tethers two domains of DHX15 together. (A) Domain organization of hsDHX15 and hsNKRF, with domains contained in the expressed constructs highlighted in color. The positions of RecA1, RecA2, WH, Ratchet, and OB domains in DHX15 are shown. In addition to the G-patch motif, NKRF has three RNA binding domains (RBD) and another nucleic acid binding domain termed R3H. Borders of expressed constructs are indicated by dotted lines. (B) Overview of the DHX15–G-patch complex. Domains are colored as in A, and important secondary structure elements described in the text are labeled. The magnesium ion and ADP molecule in the ATPase active site are shown in gray and orange, respectively. An unmodeled loop is indicated by a dotted line.
Members of the DEAH family perform essential tasks in two central RNP maturation pathways: splicing and ribosome biogenesis (2, 3, 18, 19). Both pathways require a large number of accessory factors to assemble in a step-wise manner on a primary transcript [i.e., pre-messenger RNA (pre-mRNA) and pre-ribosomal RNA (pre-rRNA)] to form dynamic catalytic machineries (20, 21). In splicing, a machinery termed spliceosome removes noncoding introns from pre-mRNAs between flanking exons by an excision–ligation mechanism (22). In ribosome assembly, dedicated processomes have to cut four rRNAs to size, extensively modify them, fold them into the correct tertiary structure, and incorporate ∼80 ribosomal proteins (23). During both elaborate processes, helicases facilitate extensive and dynamic remodeling of RNA secondary structures, as well as binding and dissociation of small nuclear or small nucleolar (sno) RNPs (2, 3, 18, 19). Four DEAH enzymes in splicing (hsDHX16/scPrp2, hsDHX38/scPrp16, hsDHX8/scPrp22, and hsDHX15/scPrp43) were assigned to specific steps in the progression of splicing particles from one catalytic intermediate to the next. Because they are all located at the periphery of spliceosomal complexes, they have been suggested to function analogous to a winch: From the exterior, they pull the RNA through the bulk of the spliceosome and via this increased tension disrupt pre-mRNA duplexes within the spliceosomal core (24).
One dual-purpose helicase that is essential in both ribosome biogenesis and splicing is the DEAH family member hsDHX15/scPrp43. In splicing, it is required for the disassembly and recycling of the spliceosome after a completed catalytic cycle, dissociating the spliceosome modules from the intron lariat (25). Furthermore, it is involved in proofreading of correct splice-site selection by facilitating disruption of stalled splicing intermediates (26, 27). In ribosome biogenesis, it has been demonstrated to release snoRNAs from progenitors of the large ribosomal subunit in yeast (28) and to function in early (human) or late (yeast) steps of small subunit maturation (29–32).
Most RNA helicases lack intrinsic specificity for their target RNAs and need to be recruited to specific sites of action by adapter proteins, that often double as direct enhancers of their otherwise poor helicase activity (5, 33, 34). One large and diverse class of DEAH helicase activators is the G-patch protein family named after a ∼45-amino-acid-long glycine-rich motif (Fig. 2 and SI Appendix, Fig. S2A) that directly binds and activates the helicases (35). More than 20 different G-patch proteins have been assigned in humans (five in yeast). In addition, the motif has been detected in a number of viruses, among them human endogenous retrovirus K (SI Appendix, Fig. S2A) (35, 36). Apart from the conserved motif and a general association with RNA pathways, G-patch proteins have little in common. They display great variety in length, domain composition, and cellular localization. Consequently, they might form the molecular basis of the multifunctionality that has been recorded for several RNA helicases by recruiting them to diverse cellular compartments or pathways (37). Nine G-patch interactors have been shown to directly enhance DHX15/Prp43 enzymatic activity, and most could be specifically assigned to function in splicing or ribosome biogenesis (SI Appendix, Fig. S2A) (32, 38–42). One of these adapters is NKRF (Fig. 1A), an RNA-binding protein, which was recently identified to function in human ribosome biogenesis as a tether between pre-rRNA and DHX15/Prp43, as well as XRN2 (32).
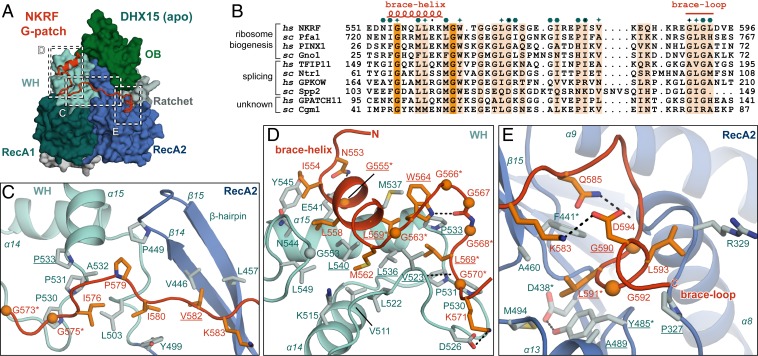
Fig. 2.
Conserved G-patch residues are crucial for forming hydrophobic interactions. (A) Overview of the complex with three different areas of G-patch interaction with DHX15 indicated by dotted frames, which are displayed in detail in C–E. Position D corresponds to G-patch site 1, while position E indicates G-patch site 2, and C marks the intervening linker. (B) Sequence alignment of G-patch proteins from yeast and human that have been assigned as activators of DHX15/Prp43 or DHX16/Prp2. Respective hs and sc homologs are grouped, and the corresponding cellular process is indicated. Brace-helix and brace-loop positions as found in NKRF are labeled on top of the alignment. Dark and light orange highlight residues that are identical and more than 70% similar, respectively. Petrol hexagons mark amino acids involved in DHX15 interactions, while stars mark mutated residues. (C) Interface of the G-patch linker region crossing over to RecA2 via the β-hairpin. (D) Interactions of the N-terminal G-patch brace-helix (red/orange) on the WH domain (cyan/light blue). Names of residues mutated in this study are underlined, and residues mutated in previous studies are marked by asterisks. Hydrogen bonds are indicated by black dotted lines. (E) Binding of the C-terminal G-patch brace-loop to the RecA2 domain.
Despite their great relevance for central RNA metabolism, the molecular basis and mechanism of helicase stimulation by G-patch proteins have remained obscure. Here, we determined the crystal structure of human DHX15 in complex with the G-patch motif of NKRF. The G-patch stretches along the DHX15 surface, where it acts like a flexible brace that tethers two mobile parts of the protein together. Addition of the G-patch peptide to DHX15 increases its RNA affinity, as well as ATPase and helicase rates. Conversely, mutations in the G-patch that maintain its connection to DHX15, but disrupt tethering, abolish this enhancement. In combination, our results suggest that G-patch proteins promote DEAH catalysis by restricting their conformational freedom to a configuration that is compatible with strong RNA binding, but leave enough flexibility for motions required for ATP hydrolysis and RNA translocation. Furthermore, our structural and biochemical analysis rationalizes G-patch selectivity for a subset of DEAH helicases.
Results
The G-Patch Motif of NKRF Is Sufficient for DHX15 Binding.
Full-length (fl) hsNKRF has been shown to bind hsDHX15 directly (32). To verify their direct interaction, we coexpressed both full-length proteins in Escherichia coli and tested for reciprocal copurification from lysates. Maltose binding protein (MBP)-tagged DHX15 copurified with glutathione S-transferase (GST)–NKRF independent of the affinity matrix used (SI Appendix, Fig. S3A). Since both proteins contain multiple domains and unstructured regions (Fig. 1A) that might hamper crystallization, we aimed at obtaining a minimal NKRF–DHX15 complex. hsDHX15 lacking the N-terminal unstructured extension (DHX15ΔN; residues 113–795) has been crystallized in isolation (43), and, given that the equivalent N terminus is not essential in yeast (44), we used the same construct boundaries (Fig. 1A). For NKRF, two different shortened constructs were designed based on sequence conservation around the consensus G-patch motif (residues 551–596) and tested for direct DHX15ΔN binding. DHX15ΔN copurified both with NKRF 541-615 and 541-603 without appreciable loss in affinity, suggesting that the shorter construct comprises all residues contributing to DHX15 binding (SI Appendix, Fig. S3B). Thus, we used the shorter construct (referred to as NKRF G-patch from here on) for large-scale purification of the complex. DHX15ΔN and NKRF G-patch formed a stable and homogenous complex that copurified over three different purification steps and eluted as a single peak from a size-exclusion column (SI Appendix, Fig. S3C).
Structure of the NKRF G-Patch in Complex with DHX15.
We crystallized the minimal complex both in the absence and presence of adenosine diphosphate (ADP) (Fig. 1B) and solved the structure by molecular replacement with hsDHX15 as the search model (43). We refined the apo structure at 2.21 Å (Rwork 21.0%; Rfree 25.1%) and the structure with ADP at 1.85 Å (Rwork 20.4%; Rfree 24.4%) with good statistics (SI Appendix, Table S1). For DHX15ΔN, we observed density for amino acids 113–789 (apo and ADP) with the exception of a few surface loops in RecA2. Furthermore, ADP was well ordered in the active site between the RecA domains (SI Appendix, Fig. S4D). In both structures, extra density for the G-patch was well defined and could be modeled with residues 551–596 (apo) and 553–593 (ADP), respectively (SI Appendix, Fig. S4 A and B), which means that both structures contained the entire conserved G-patch motif (Fig. 2B). To ensure correct assignment of the G-patch sequence, we collected a native anomalous diffraction dataset (single-wavelength anomalous dispersion of sulfur atoms; SI Appendix, Table S1). Using phases from a preliminary model lacking the G-patch, we calculated an anomalous difference density map to locate the sulfur atoms (SI Appendix, Fig. S4C). Clear anomalous density was visible for G-patch M562, which is the only sulfur-containing residue in the motif (Fig. 2 B–E).
The NKRF G-Patch Is a Molecular Brace between Two DHX15 Domains.
As predicted by sequence analysis (35), the G-patch peptide is mostly unstructured, apart from a short N-terminal α-helix. It stretches across the back side of the RNA binding channel of DHX15, effectively tethering WH and RecA2 domain together (Figs. 1B and 2A). The N-terminal G-patch helix (brace-helix) packs perpendicularly on the long helix (α15) of the WH domain (Figs. 1B and 2D). Residues of the subsequent linker region stack into hydrophobic pockets on the WH domain’s surface and bind across the β-hairpin (β14, β15) of the RecA2 domain (Figs. 1B and 2C). The C-terminal brace-loop also stacks into a hydrophobic pocket on top of RecA2, between the central β-sheet and two α-helices (α8 and α13) (Figs. 1B and 2E). Interestingly, the eponymous conserved glycine residues of the G-patch make very few contacts to the helicase themselves (Fig. 2 B–E and SI Appendix, Fig. S2A). Instead, they mostly lend the necessary flexibility to the peptide backbone that allows conserved hydrophobic residues to immerse into hydrophobic surface cavities. One notable exception is the almost-invariable G555, which is located at the closest contact point between the brace-helix and α15 of the WH domain (Fig. 2D) and, thus, enables tight packing of the two helices.
The completely invariable G563 at the end of the brace-helix is, without exception, followed by an aromatic amino acid (W564 in hsNKRF; Fig. 2B and SI Appendix, Fig. S2A). Here, the glycine facilitates a backbone conformation that allows the aromatic side chain to flip under the G-patch main chain and reach back toward the brace-helix (Fig. 2D). Consequently, the aromatic ring ends up sandwiched between a bottom layer formed by a composite hydrophobic patch from WH (P533 and M537) and brace-helix (L559) and a top layer formed by the peptide backbone of the G-patch (peptide bond between T565 and G566). This unusual flipped-back conformation is further stabilized by a hydrogen bond between the W564 aromatic nitrogen and a downstream main-chain carbonyl oxygen (from G567). The hydrogen bond also bends the downstream peptide in a turn around the aromatic side chain and lets the subsequent highly conserved L569 fold back into the same hydrophobic cavity on WH α15 that is already used by the brace-helix. Therefore, the intriguing “under-over-turn” conformation of the G-patch results in a dense packing of several distant hydrophobic side chains into the same helicase pocket.
The brace-loop at the C terminus makes only one contact with a hydrophobic pocket of RecA2, where it embeds the highly conserved L591 (Fig. 2 B and E). Analogous to the N-terminal portion of the motif, the required turn in the peptide is facilitated by a hydrogen bond between the main chain and a hydrophilic side chain (Q585).
The DHX15 contacts of the intervening G-patch stretch are far less extensive, consistent with its poorer evolutionary conservation (Fig. 2B). However, one contact that might be of functional relevance is formed by conserved V582, which binds across the back of the β-hairpin of DHX15 (Fig. 2C). This interaction pushes the backbone of β14 by 2 Å into the RNA-binding channel and, consequently, moves the highly conserved K445 on the inside of the β-hairpin further into the RNA channel by 3.8 Å (SI Appendix, Fig. S5 A and B). When no nucleotide is bound in the active site and the two RecA domains take up the open conformation, K445 is the side chain that is closest to the first base of the five RNA nucleotides in the RNA channel. Therefore, in homologous DEAH/RHA helicases, the analogous residues form hydrogen bonds to the first base (8, 11–13). Although the conformational change is relatively small, we consistently observed it in both structures of DHX15 (apo and ADP-bound). Given that the β-hairpin forms one of the two bookends, between which the four or five RNA bases within the RNA channel are stacked (8, 11), this movement could be significant for RNA binding or translocation of the helicase.
The G-Patch Binding Mode Is Likely Analogous at DHX15/Prp43 in the Spliceosome.
Given that the G-patch peptide is not involved in crystal packing (smallest distance to a neighboring molecule’s side chain >5 Å; SI Appendix, Fig. S4E), the observed configuration is most likely not a crystallization artifact. To infer whether the hsNKRF G-patch conformation is representative for other G-patch partners of DHX15/Prp43 in splicing, we inspected available data from cryo-electron microscopy (cryoEM) and cross-linking mass spectrometry (XL-MS). scPrp43 has undergone XL-MS with scNtr1 (ortholog of hsTFIP11) (Fig. 2B), an established G-patch partner in the spliceosome (45). From the scNtr1 G-patch motif, two lysines on the brace-helix (K60/K67) were cross-linked to the C terminus of scPrp43 (α25 of OB domain and its C-terminal extension). Since the G-patch brace-helix directly neighbors the OB domain of hsDHX15, these cross-links are consistent with our structure. The corresponding residues in hsDHX15 are at a distance of 18–28 Å from the G-patch N terminus (Cα atoms), in agreement with XL-MS distance restraints (<26–30 Å for disuccinimidyl suberate cross-linking) (46). In the assembled yeast intron-lariat spliceosome (ILS), the same cross-link (scNtr1 K60–scPrp43 K737) has been detected (47). Furthermore, in the electron-density map of the scILS, clear extra density is visible both on the WH and on the RecA2 domain (SI Appendix, Fig. S6A) that had not been possible to model due to low local resolution of the map in the ILS periphery (47). Superpositions with the DHX15–G-patch structure show a good fit of the map with the brace-helix and its downstream loop on WH, as well as the brace-loop on RecA2 (SI Appendix, Fig. S6A). Together, these data suggest that TFIP11/Ntr1 G-patch binds DHX15/Prp43 during splicing in a similar conformation as NKRF during ribosome biogenesis.
The Two G-Patch Binding Sites on DHX15 Are of Different Relevance for Complex Formation.
We next mutated the interaction sites between DHX15 and NKRF G-patch to further validate the interface in solution using copurification from E. coli lysates. Single-point mutations addressed either the brace-helix binding site (G-patch site 1) or the brace-loop binding site (G-patch site 2) (Fig. 3). Hydrophobic residues were mostly substituted for glutamates, firstly to ensure disruption of the fuzzy hydrophobic interfaces and, secondly, to improve hydration of the newly water-accessible surfaces upon complex dissociation. Furthermore, several homologous DEAH helicases carried Glu substitutions in the two G-patch sites (e.g., DHX37; SI Appendix, Figs. S2B and S7C). Thus, the mutations could help us to judge whether these homologs could support G-patch binding.
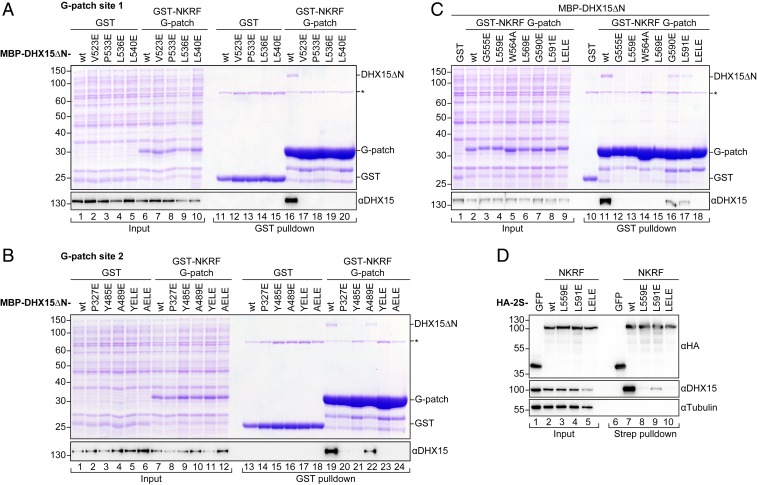
Fig. 3.
Mutations in the two binding sites have different effects on complex formation. (A–C) Coomassie-stained gels of copurification assays of MBP–DHX15ΔN and GST–NKRF G-patch coexpressed in E. coli. Input (0.03%) and eluates (4.4%) were loaded. GST served as a control. DHX15 expression was verified by Western blotting using an antibody against DHX15. The asterisk marks an unspecific band that copurifies with GST from E. coli lysates in all conditions. DHX15 mutations in G-patch sites 1 and 2 are shown in A and B, respectively. YELE corresponds to double mutant Y485E, L536E, while AELE stands for A489E, L540E. Mutations in the G-patch motif are shown in C. (D) Western blots of coprecipitation assays from HEK293T cells overexpressing HA-2S-NKRF or HA-2S-GFP as a control. Samples were treated with RNase A and tested for copurification of endogenous DHX15. Tubulin served as a loading control. For anti-HA blots, 0.75% of input and 5% of eluates were loaded, while 0.25% of input and 15% of eluates were loaded for anti-DHX15 and anti–α-tubulin blots.
All mutations on G-patch site 1, either on DHX15ΔN or NKRF G-patch, fully disrupted complex formation in vitro (Fig. 3 A and C, lanes 12 to 15). Interestingly, removal of the central tryptophan side chain next to the brace-helix (W564A) was also sufficient to prevent G-patch binding, consistent with the important role of this aromatic residue for the overall conformation of the motif (Fig. 2D). Previously, several studies have mutated corresponding residues on G-patch site 1 in yeast and human DHX15/Prp43 or different G-patch proteins (indicated by * in Fig. 2D). These mutations led to loss of helicase interaction and activation, or defects in splicing, ribosome biogenesis, and cell growth, in agreement with a general importance of this site for helicase–G-patch binding (32, 38, 40, 41, 48).
In contrast, mutations on G-patch site 2 were generally less disruptive for complex formation (Fig. 3 B and C, lanes 16 and 17). Only two mutants in the DHX15 RecA2 pocket (P327E and Y485E) showed no binding in copurification assays, while a third mutant (A489E) and both mutations on the G-patch side (G590E and L591E) retained around half the affinity. Interaction was lost when these mutations were combined with a mutation in G-patch site 1 (Fig. 3B, lanes 23 and 24, mutants Y485E, L536E [YELE] and A489E, L540E [AELE], and Fig. 3C, lane 18, mutant L559E, L591E [LELE]). A similar behavior was observed for the binding of NKRF fl in human cells to endogenous DHX15. We overexpressed NKRF with an N-terminal hemagglutinin–TwinStrep-tag (HA-2S) in HEK293T cells and performed pulldowns on StrepTactin resin in the presence of RNase A (Fig. 3D). While DHX15 interaction was completely lost for the brace-helix mutant (G-patch site 1, L559E, lane 8), the brace-loop mutant still showed a decreased interaction (G-patch site 2, L591E, lane 9). Consistently, the double mutant (LELE, lane 10) abolished binding, despite its correct localization to nucleoli (SI Appendix, Fig. S3D) (32). These observations confirmed that the G-patch is the main contributor to NKRF–DHX15 binding in human cells, as suggested before (32). In keeping with a decreased effect on complex formation, several mutants on G-patch site 2 in scPrp43 (marked by * in Fig. 2E) have been found in a previous study to lead to milder growth defects and only conferred lethality in combination with Ntr1 G-patch site 1 mutants (41).
Collectively, these data suggest that the two portions of G-patch motifs have a stronger and a weaker binding site on DEAH helicases, involving brace-helix/WH and brace-loop/RecA2, respectively.
The G-Patch Reinforces a Helicase Conformation Consistent with Stable RNA Binding.
Previously, several structures of Prp43 from the fungus Chaetomium thermophilum in the presence and absence of bound RNA substrate have highlighted the conformational flexibility of isolated DEAH helicases (17). When no RNA is bound, the C-terminal domains (WH, Ratchet, and OB) can turn relative to the RecA domains by 40°, thereby opening up and disrupting the RNA channel, leaving only a shallow RNA binding groove on top of the RecAs (Fig. 4 A and C). This conformational change has been suggested to facilitate RNA loading into the helicase. Analogous observations have been made for DHX36, a DEAH/RHA helicase involved in G-quadruplex unfolding, which shows a similar C-terminal domain movement by 28° in the absence of nucleic acid (12).
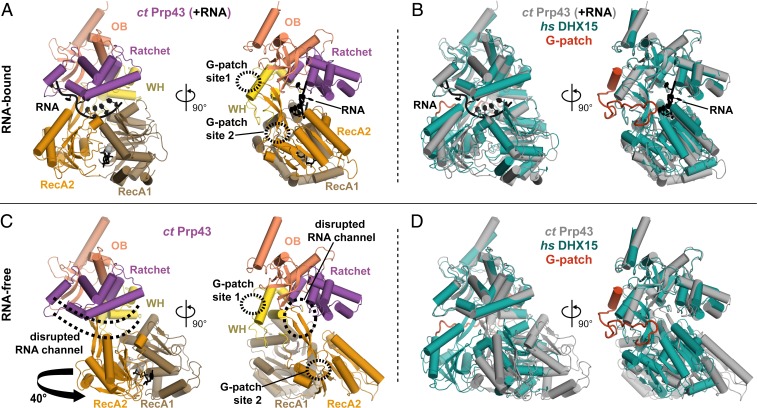
Fig. 4.
G-patch binding stabilizes a DHX15 conformation consistent with RNA binding. (A and C) Different conformations of ctPrp43 recorded in the presence (A) or absence (C) of RNA are shown [PDB ID codes 5lta and 5ltk (17)]. The location of G-patch sites 1 and 2 on Prp43 according to superpositions with the DHX15-G-patch complex are indicated by dashed circles. RNA and ADP present in A are shown in black. The location of the RNA binding channel as displayed in A is indicated in C by dashed lines. Rotation of the RecA domains necessary to transform the RNA-bound structure into the RNA-free structure is indicated by an arrow. (B and D) Superpositions of the DHX15–G-patch complex with the structures shown in A and C, respectively.
Superposition of the DHX15–G-patch complex with the two conformations of ctPrp43 showed good agreement with the RNA bound configuration (rmsd 2.3Å over 672 out of 676 residues). Importantly, the path length between the two G-patch sites was the same in the ctPrp43–RNA structure and the DHX15–G-patch complex. In contrast, in the RNA-free conformation, WH was shifted away from RecA, and the path between the two sites was elongated by 5.1 Å. Given that the Cα distance covered by two amino acids in a peptide is 3.8 Å, this means that the G-patch would need to stretch by more than one additional residue to remain bound to both sites. Therefore, comparison of available structures suggests that G-patches stabilize DEAH helicases in a conformation with an intact RNA channel and, thus, maintain high RNA affinity.
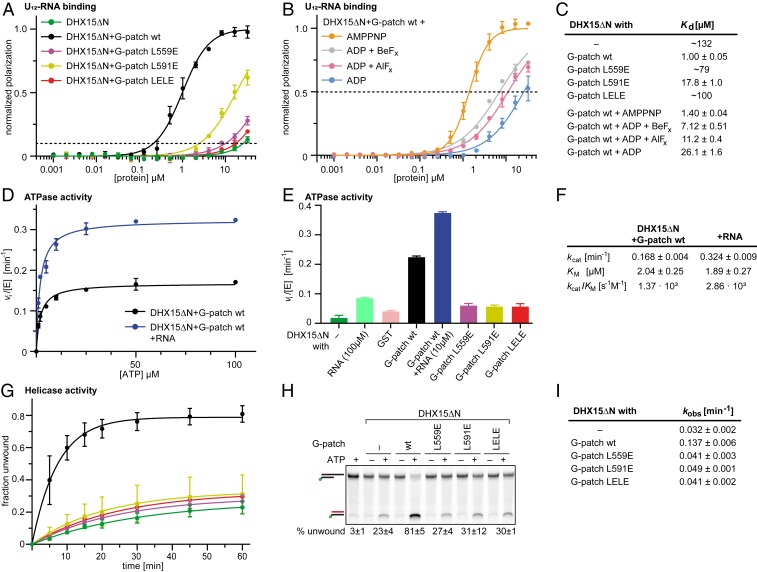
Consistent with this interpretation, 25-fold increased RNA affinity has been observed for scPrp43 in the presence of the G-patch protein scCmg1 or the scNtr1 G-patch motif (37). We therefore tested whether mutations in the NKRF G-patch would influence RNA affinity of DHX15ΔN using fluorescence polarization assays (Fig. 5 A and C). We used the shortened construct in all biochemical assays due to low protein yields that were obtained with DHX15 fl. The wild type (wt) G-patch motif increased DHX15ΔN RNA affinity ∼100-fold. As a control that the DHX15 N terminus does not significantly influence the observed effect, we also measured RNA affinity of DHX15 fl upon G-patch addition and detected a similar improvement of RNA binding (SI Appendix, Fig. S3 E and F). Since the G-patch is too remote from the RNA channel (closest distance to superposed RNA >10 Å), it is unlikely that the increase was due to direct interactions between G-patch and RNA. Mutation of the brace-helix (L559E) that abolished DHX15 binding in pulldowns drastically reduced this increase in RNA affinity to ∼1.5-fold. Similarly, mutation of the brace-loop (L591E) conferred only an ∼7-fold stimulation of RNA binding, despite the fact that this mutant still supported partial DHX15 association in copurification assays, probably via the brace-helix. Therefore, disconnecting domain tethering by G-patch from its association with DHX15 led to an ∼17-fold impairment. Combining the mutations in both G-patch sites (LELE) completely abrogated the enhancing effect of G-patch addition.
Fig. 5.
NKRF G-patch–mediated enhancement of DHX15 RNA affinity and ATPase activity depends on efficient domain tethering. (A) Fluorescence polarization experiments of FAM-U12 RNA with His10–DHX15ΔN in the absence or presence of GST–NKRF G-patch wt or mutants. The dashed line indicates 10% normalized polarization. Average values of triplicate measurements were plotted, and error bars correspond to SDs. (B) Fluorescence polarization experiments with DHX15ΔN and GST-NKRF G-patch wt in the presence of different ATP analogs or ADP (100 µM). The dashed line indicates 50% normalized polarization. Average values of triplicate measurements were plotted, and error bars correspond to SD. (C) RNA dissociation constants (Kd) with SEM determined by fitting curves in A and B by linear regression. For curves that do not reach 50% normalized polarization, the fitted constants were rounded to two digits and listed as approximate values. (D and F) Michaelis–Menten plot of ATPase activity of His10–DHX15ΔN in complex with GST–NKRF G-patch wt with (blue) or without (black) RNA. Measurements were carried out in triplicate, and error bars represent SD. Michaelis–Menten parameters and corresponding SEM were determined by linear regression. (E) Initial ATP hydrolysis rates at 2 mM ATP, normalized for enzyme concentrations for His10–DHX15ΔN with or without GST–NKRF G-patch wt or mutants, or GST as a control. Error bars indicate SD of triplicate measurements. (G and I) RNA duplex unwinding assays with His10–DHX15ΔN in the presence or absence of GST–NKRF G-patch wt or mutants using FAM-labeled dsRNA with a 3′ single-stranded overhang. Time courses were measured in triplicate, and error bars indicate SD. Rate constants and respective SEM were determined by linear regression. (H) Native PAGE of representative samples from the unwinding assay after 60-min reaction time detecting FAM-fluorescence signal. Samples without ATP or without protein were used as controls for RNase contamination. The top bands correspond to duplex RNA substrate, while the lower bands stem from unwound RNA that is quenched with a cDNA oligo.
To ensure that the improved RNA affinity was not simply due to the small conformational change in the β-hairpin of DHX15 that was brought about by the middle section of the G-patch (SI Appendix, Fig. S5 A and B), we mutated both the Lys residue in the RNA binding channel (DHX15 K554A) and the G-patch residue that pushes against the β-hairpin backbone (NKRF G-patch V582G) (SI Appendix, Fig. S5 C–E). Both mutations did not hamper G-patch binding and had only minor effects on RNA affinity (1.2-fold reduction and 1.2-fold increase, respectively), demonstrating that the movement of K554 inside the RNA binding channel cannot account for the strong RNA binding of the DHX15–G-patch complex.
Together, the biochemical and structural data indicate that efficient clamping of WH and RecA2 domain by the NKRF G-patch is required to enhance RNA affinity and that this is achieved by restricting the flexibility of DHX15/Prp43 domains to a conformation that can stably associate with the RNA target.
The G-Patch Can Follow RecA2 Transitions Required for Nucleotide Binding.
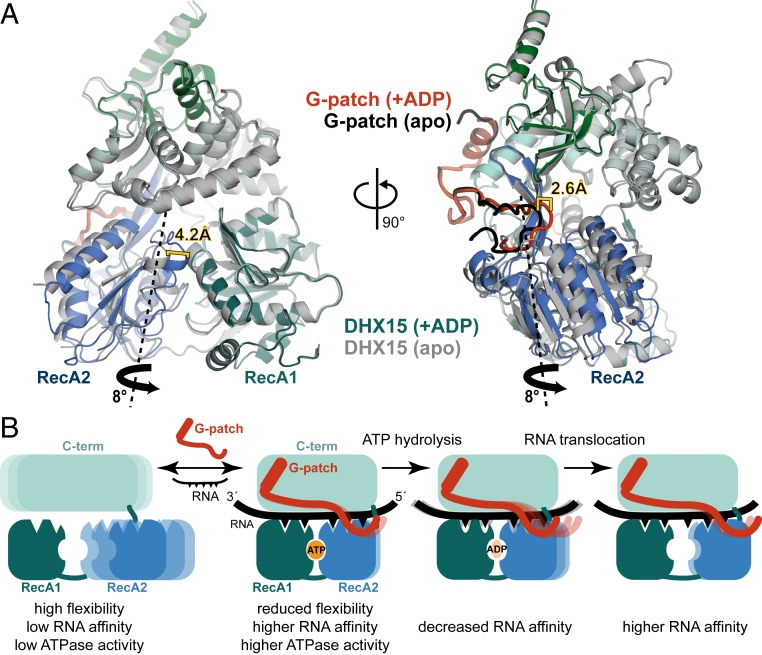
In addition to flexibility of the C-terminal domains, the RecA domains have also been shown to undergo conformational changes during the ATPase cycle (8, 12). To address whether the observed G-patch binding mode would be compatible with the RecA movement that is required for ATP binding and, ultimately, hydrolysis, we compared the DHX15–G-patch apo structure to the complex bound by ADP (Fig. 6A). In the absence of ADP, RecA2 was moved away 4.2 Å from RecA1 via rotation of 8° around a virtual axis through the back of RecA2 and the β-hairpin. The G-patch brace-loop followed this movement of RecA2, meaning that it had to shift by 2.6 Å to the side. This domain twist resulted only in a minor change in the distance between G-patch sites on DHX15 (0.4 Å). Thus, the structures suggest that domain clamping by G-patch is consistent with ATPase activity.
Fig. 6.
G-patch binding retains RecA2 motions in DHX15 induced by ADP binding. (A) Superposition of the DHX15–G-patch complex without (apo; gray/black) or with ADP bound in the active site (blue-green/red). The RecA2 domain rotation required to transform the apo into the ADP-bound structure is indicated by a thick black arrow, while the imaginary rotation axis through the back of RecA2 is shown as a dashed line. Shifts in RecA2 and the G-patch brace-loop are indicated. Domain movements were analyzed by using the Dyndom server (79). (B) Suggested model for G-patch–mediated stimulation of DEAH RNA-helicases. The C-terminal domains including the WH are depicted in cyan, and RecA1 and RecA2 are colored in green and blue, respectively. Mobility is indicated by multiple lighter-shaded schematics.
To test this hypothesis, we analyzed the effect of G-patch addition on DHX15 ATPase activity using a continuous coupled ATPase assay (Fig. 5 D–F) (49). Due to overall low ATPase activities of DHX15 and a limiting amount of protein, we could only determine ATP hydrolysis rates at a saturating ATP concentration to compare G-patch mutants (2 mM, as determined by Michaelis–Menten graph for G-patch wt; Fig. 5 D and F). Assembling DHX15ΔN with GST-tagged G-patch improved ATP hydrolysis rates ∼6-fold compared to the addition of GST alone. As a control, we also measured the effect on DHX15 fl where we observed an ∼4-fold acceleration (SI Appendix, Fig. S3G), in agreement with a previous study (32). Consistently, qualitatively similar rate accelerations have been reported for different G-patch proteins and different DEAH helicases (16, 17, 37, 39, 45, 50, 51). However, when the G-patch was mutated in either brace-helix, brace-loop, or both, the ATPase enhancement was completely abolished (Fig. 5E). This observation leads to two conclusions: Firstly, G-patch clamping as observed in our structures does not interfere with ATPase activity, since relieving the tether to G-patch site 2 does not improve ATP hydrolysis rates but, on the contrary, decreases them. Secondly, restricting flexibility by G-patch binding seems not only conducive to RNA affinity, but also to ATP turnover.
One explanation could come from comparing RecA positions in previous DEAH structures without G-patch. Here, the conformational extremes of apo vs. nucleotide-bound state were much more pronounced, and the two RecA domains relaxed to significantly more open conformations in the apo state (8, 11–13). We judged the distance between RecA1 and RecA2 by measuring the spacing between the Cα of two conserved residues that sandwich the nucleotide in the active site, an Arg in RecA1 (R204 in hsDHX15; SI Appendix, Fig. S4D) and a Phe in RecA2 (F401 in hsDHX15). In a variety of DEAH apo structures, distances were 18.4 to 21.6 Å vs. 13.4 Å in hsDHX15–G-patch (apo), and RecA2 domains were rotated by ∼14° vs. 8° in hsDHX15–G-patch (apo). On the other hand, published nucleotide-bound conformations were similar, showing distances of 10.4 to 12.8 Å vs. 10.9 Å in hsDHX15–G-patch (ADP). One caveat is, of course, that these measurements stem from different helicases, and all were observed within crystal packing, which might distort domain orientations. However, because the compared structures come from different crystal environments, it is unlikely that they are all skewed in the same manner. In addition, the distances are quite consistent, even though they were recorded from different proteins. In balance, this indicates that the observed RecA distances are representative of the true conformational space.
In summary, the available data suggest that DHX15 RecA2 shuffles between less extreme open and closed states when G-patch is bound, which stimulates ATPase activity.
Next, we analyzed the effect of RNA binding on ATPase activity (Fig. 5 D–F and SI Appendix, Fig. S3H). Addition of single stranded U10-RNA improved ATP hydrolysis rates both in the absence (4.7-fold) and presence of G-patch (1.7-fold), similar to what was demonstrated for multiple DEAH helicases (8, 11, 17, 32, 38, 39, 45, 50–54). Maximal rates were obtained when both RNA and G-patch are bound together to DHX15. Interestingly, both RNA and G-patch have a stronger effect (fold change) on ATPase activity when the respective other is absent, i.e., the first activator dampens the effect of the second activator, which could suggest that RNA and G-patch stimulate ATP hydrolysis using a similar mechanism, potentially by shifting the conformational equilibrium of the enzyme toward an “ATPase-competent” state.
Tethering of DHX15 Domains by the G-Patch Stimulates RNA Duplex Unwinding.
In cells, the important reactions that DHX15 has to carry out are RNA translocation and duplex unwinding, which are coupled to ATP hydrolysis. Thus, we also tested the effect of G-patch tethering of DHX15 WH and RecA2 domains on its RNA helicase activity using a gel-based readout (Fig. 5 G–I and SI Appendix, Fig. S3I). DHX15ΔN separated double-stranded RNA (dsRNA) with a single-stranded overhang at the 3′ end four times faster when G-patch was present, in keeping with previous observations (32). Consistent with the negative effect of G-patch mutations on ATPase activity, mutations in either the brace-helix (L559E), the brace-loop (L591E), or both (LELE) completely abrogated the stimulation of RNA helicase activity. This observation suggests that by stabilizing a conformation with high RNA affinity and gearing RecA2 movements toward higher ATP hydrolysis efficiency, the G-patch improves RNA translocation speed. In addition, our data lend further support to the model that the RecA2 domain movements in DEAH/RHA-helicases, which occur during ATP hydrolysis, are the same that lead to shifting of the RNA through the helicase channel.
Finally, we also tested whether nucleotide binding to the active site affected RNA affinity of the DHX15ΔN–G-patch complex (Fig. 5 B and C). Addition of the stable ATP analog adenylyl-imidodiphosphate (AMPPNP) left RNA affinity basically unchanged. However, supplementing the complex with analogs of ATP hydrolysis states [ADP·BeFx for a state similar to the ground state, ADP·AlFx for a transition state (55)] or ADP (as a product after phosphate release) led to a stepwise sevenfold, 11-fold, and >20-fold reduction of RNA affinity, respectively, in keeping with observations for other DEAH helicases (8, 11, 16, 51). This reduction of RNA interaction after ATP hydrolysis could be important to loosen the grip on RNA enough so that RecA2 can translocate toward the 5′ end by 1 nt, before RNA is tightly bound again after ADP dissociation.
Discussion
Mechanistic Model for G-Patch Activation of DEAH RNA Helicases.
Our structural and biochemical analysis of hsDHX15 activation by G-patch protein hsNKRF together with available data from the literature suggests a tentative model for G-patch action on DEAH RNA-helicases (Fig. 6B): The helicases are flexible in isolation, transiently sampling different conformations. Only a limited population of molecules is in the right orientation at any given time to stably bind RNA and hydrolyze ATP. This could be rationalized as an autoinhibition by domain mobility. G-patch protein binding leads to a restriction of this flexibility and stabilization of a configuration with high RNA affinity. Simultaneously, G-patch proteins recruit the respective helicase to its target RNP. Flexible tethering of the helicase domains still provides enough room for RecA domain movements required for ATP binding and RNA translocation. During ATP hydrolysis, the affinity for RNA is reduced by smaller changes in the RecA domains, while opening of the RNA binding channel is prevented by the G-patch partner. This way, RecA2 can move outward and shift upstream by 1 nt, without losing the overall grip on RNA. After release of ADP, RNA affinity increases again, so when RecA2 closes up with RecA1 in the next step upon binding of a new ATP molecule, this movement pushes the RNA through the channel.
A Common Theme for Activators of DEAH and DEAD-Box Helicases?
DEAD-box helicases are closely related to DEAH helicases and also share important roles in RNA metabolism (2, 5, 56). They are more minimal in their domain composition, having only the core RecA domains as a common feature. A subset of DEAD-box helicases is activated by proteins carrying an MIF4G domain (5, 34). Our data suggest several parallels between the activation modality of G-patch and MIF4G proteins: The two RecA domains of DEAD-box proteins are also flexible in isolation, but need to come together in a defined orientation to compose the ATPase active site and the RNA binding surface (5). MIF4G activators help to stabilize this configuration by bridging both RecA domains. Thus, in essence, they act similar to the G-patch motif by restricting flexibility. In a further analogy to G-patches, the MIF4G domains have a weaker and stronger contact site on RecA1 and RecA2, respectively. This allows the two domains to still undergo restricted movements that are necessary for ATP hydrolysis and RNA duplex distortion. Unlike the G-patch motif, the low-affinity site of MIF4G has to detach from RecA1 to allow full domain closure, because MIF4G is a completely folded and, thus, more rigid activator than the extended, largely unstructured G-patch. Thus, in a unified model, both enhancers disabled excessive domain motions of their target DEAH or DEAD-box helicases into unproductive states while maintaining a sufficient degree of flexibility for catalysis.
Are G-Patch Activators Selective for a Subset of DEAH/RHA Helicases?
So far, only two DEAH proteins (DHX15/Prp43 and DHX16/Prp2) have been demonstrated to become directly activated by G-patch proteins (36). Available structural and biochemical evidence suggests that DHX16/Prp2 is bound by its G-patch activator (GPKOW/Spp2) in an analogous configuration as DHX15: First, the G-patch sites in the structure of ctPrp2 are essentially conserved (SI Appendix, Fig. S7A). Second, cryoEM densities of yeast and human Bact spliceosomes (57, 58) revealed clear additional density on DHX16/Prp2 G-patch site 1 that could be explained by a bound G-patch brace-helix (SI Appendix, Fig. S6 B and C). Third, XL-MS of the scBact complex was consistent with both scSpp2 brace-helix and brace-loop binding on WH and RecA2 (57). Consistently, in a parallel study, the Ficner group found an almost identical binding mode for ctSpp2 to ctPrp2 (59).
Based on sequence comparison alone (SI Appendix, Fig. S2B), we would hypothesize that, apart from DHX15 and DHX16, the only other DEAH helicase that can bind G-patch motifs is DHX35, which was found in the hsC spliceosome (60). If at least two helicases can bind the same G-patch motif, how, then, is specificity ensured to recruit the right enzyme to the correct pathway and reaction step? The answer could lie in the divergent N- and C-terminal extensions of DEAH helicases. At least some of them have been shown to interact with other spliceosomal proteins [e.g., a C-terminal helix of Prp22 binds to Prp8 (61)] and could thus provide another layer of specificity to find the right location on an RNP. In addition, other specific surface pockets on the DEAH enzymes could be recognized by different binding partners and help to recruit them to the right target.
For the other two splicing DEAH helicases, DHX38/Prp16 and DHX8/Prp22, evidence from the literature compared with our data could point toward an activation mode analogous to G-patch stimulation, but potentially mediated by other types of activators (i.e., intramolecular by their N-terminal extensions or other binding partners): Both proteins displayed modified binding pockets (SI Appendix, Figs. S2B and S7B), each with one disruptive substitution in site 1 (hsDHX38 E917/scPrp22 K869 corresponding to hsDHX15 P533E; Fig. 3A). However, cryoEM maps of their respective spliceosomal complexes (61–64) showed strong additional density on both binding sites and some even for the linker (SI Appendix, Fig. S6 D–G). The peptides responsible for these extra densities remain to be uncovered. Yet, this evidence leads us to speculate that all splicing DEAH helicases are bound by an internal or external stimulator on the two “G-patch sites,” when they are incorporated into spliceosomal complexes.
Other DEAH/RHA helicases such as DHX37 are reported to be activated by non–G-patch proteins (53, 65) or—such as MLE/DHX9—have no known activator. DHX37 is involved in small ribosomal subunit assembly just like DHX15. Murine (mm) DHX37 shows several deleterious substitutions in the G-patch sites (e.g., equivalent to hsDHX15 L540E; Fig. 3A and SI Appendix, Fig. S7C), indicating that G-patch proteins are unlikely to bind. Consistently, the recorded interaction motif in DHX37-activator UTP14A shares no sequence homology with the G-patch motif (11).
Drosophila (dm) MLE/hsDHX9 has an extended N terminus, which harbors two double-stranded RNA binding domains (dsRBDs) (15). dsRBD2 bridges between WH and RecA2 domain, thus precluding G-patch binding to the same surface (SI Appendix, Fig. S7D). In fact, dsRBD2 could functionally replace activation by G-patches by taking over the same domain-tethering role, acting as an intramolecular enhancer domain. In agreement with this hypothesis, dmMLE can efficiently unwind RNA duplexes in vitro (41, 51), and activity is lost upon deletion of dsRBD2. These data highlight that several DEAH/RHA-family members have evolved divergent mechanisms of activation by gaining different surface patches or additional N-terminal domains, which allows for specific recruitment and activation of these otherwise unspecific enzymes to different pathways at defined steps.
Materials and Methods
Antibodies.
The following antibodies and dilutions were used for Western blotting: horseradish peroxidase (HRP-linked anti-HA antibody [Roche; 1:5,000]), rabbit polyclonal anti-DHX15 antibody (Bethyl Laboratories, catalog no. A300-390A; 1:15,000), mouse monoclonal anti–α-tubulin antibody (Proteintech; catalog no. 660311-Ig; 1:5,000), HRP anti-mouse (Sigma, catalog no. A9044; 1:5,000), and HRP anti-rabbit (Sigma, catalog no. A9169, 1:10,000). For immunofluorescence, polyclonal rabbit anti-ENP1 (described in ref. 66; kind gift from Ulrike Kutay, ETH Zürich, Zürich, Switzerland; dilution 1:10,000) and monoclonal rat anti-HA (Roche, catalog no. 11867423001; 1:200) were used as primary antibodies, Alexa Fluor 488-labeled goat anti-rabbit (Invitrogen, catalog no. A11008; 1:300) and Alexa Fluor 633-labeled goat anti-rat (Invitrogen, catalog no. A21094; 1:300) were used as secondary antibodies.
Expression Plasmids.
Full-length hsDHX15 was generated by amplification of the protein-encoding sequence using gene-specific primers and a plasmid from the human open reading frame library (hORFeome Version 5.1, ID: 13273) as a template. hsDHX15ΔN (residues 113 to 795) was amplified from the full-length sequence by using primers AAAAAACTCGAGCAGTGCATTAATCCGTTCACC and AAAAAAGCTAGCTCAGTACTGTGAATATTCCTTGG.
Full-length hsNKRF was amplified from human complementary DNA (cDNA). The hsNKRF G-patch (541 to 603) was generated by using primers AAAAAACATATGGCAGAGGAGGCTTACAAACAG and AAAAAAGGATCCTCACTTGGCAATTTTATTCACCCTCTC.
For coexpression of hsDHX15ΔN and hsNKRF G-patch proteins in E. coli, hsDHX15ΔN was cloned into XhoI–NheI sites of the plasmid pnEA-NpM. hsNKRF G-patch was cloned into NdeI–BamHI sites of pnYC-NpG. Both expression vectors were derived from the pET-MCN vector series (67) and contain an N-terminal MBP-tag or GST-tag, respectively, followed by a 3C protease site.
To express full-length hsDHX15 and hsDHX15ΔN in insect cells, constructs were cloned by ligation-independent cloning (LIC) using primers CCCTTCCCAATCCAATTCGATGTCCAAGCGGCACCG, CCCTTCCCAATCCAATTCGCAGTGCATTAATCCGTTCACC, and TTATCCACTTCCAATGTTATTATCAGTACTGTGAATATTCCTTGG, respectively. Both constructs were cloned into a modified pFastBac vector of the MacroBac series (68) containing an N-terminal deca-histidine-tag followed by a 3C protease site. LIC cloning and assembly reactions were essentially performed as described in ref. 68 with slight modifications: Annealing was performed by combining 1 μL of 50 mM ethylenediaminetetraacetate (EDTA), 3 μL of LIC-treated vector, and 6 μL of LIC-treated PCR products. The reaction was then annealed in a thermocycler slowly cooling from 50 to 10 °C. Subsequent steps of bacmid generation and isolation were carried out according to the Bac-to-Bac expression system (Invitrogen) using the manufacturer’s protocol. Viruses were produced in Sf9 cells (ThermoFisher) grown in SF-4 Baculo Express medium (BioConcept).
For the expression of hsNKRF constructs in human cells, hsNKRF fl was cloned into the BamHI–XhoI site of a pcDNA5/FRT/TO-vector (Invitrogen), which contained an N-terminal HA-tag followed by a TwinStrep-tag II (69)
Point mutations were introduced into hsDHX15 and hsNKRF constructs by using the QuikChange method (Agilent). All plasmids were verified by sequencing.
Protein Expression and Purification.
For crystallization and copurification assays, proteins were expressed in E. coli. MBP–hsDHX15ΔN and GST–hsNKRF G-patch were coexpressed in BL21star (DE3) cells (ThermoFisher) in ZY autoinduction medium (70) at 20 °C for 10 h. After harvest, pelleted cells were resuspended in lysis buffer (50 mM Hepes, pH 7.5, 200 mM NaCl, 2 mM dithiothreitol [DTT], 10% glycerol, and 2 mM MgCl2) supplemented with cOmplete EDTA-free protease inhibitor mixture (Roche), 1 mg/mL lysozyme (Sigma), and 5 μg/mL DNase I (Roche) and lysed by passing the suspension through a LM10 Microfluidizer. Cell lysate was cleared by centrifugation and subsequent filtration (0.45 μm). The lysate was incubated with preequilibrated glutathione Sepharose beads (Amersham Biosciences) for 1 h at 4 °C. Beads were washed with lysis buffer, and proteins were eluted with lysis buffer containing 25 mM l-glutathione. The eluate was diluted to a low-salt buffer (50 mM Hepes, pH 7.5, 100 mM NaCl, 2 mM DTT, 10% glycerol, and 2 mM MgCl2), and protein tags were cleaved with human rhinovirus 3C protease overnight. The complex was then purified over a heparin column (HiTrap Heparin HP, GE Healthcare), equilibrated in heparin buffer (50 mM Hepes, pH 7.5, 100 mM NaCl, 2 mM DTT, 10% glycerol, and 2 mM MgCl2), and eluted over a linear gradient to 1 M NaCl. Finally, the complex was purified by using a gel-filtration column (Superdex 200, GE Healthcare) and eluted in size-exclusion buffer (10 mM Hepes, pH 7.5, 200 mM NaCl, 2 mM DTT, and 5% glycerol). Fractions containing the complex were concentrated to ∼2 mg/mL, flash frozen in liquid nitrogen, and stored at −80 °C.
For biochemical studies, His10–hsDHX15 constructs were expressed in High Five insect cells (ThermoFisher) in SF-4 Baculo Express medium (BioConcept) for 48 h after virus infection. After harvest, cell pellets were resuspended in lysis buffer (with 2 mM β-mercaptoethanol instead of DTT) supplemented with 20 mM imidazole, cOmplete EDTA-free protease inhibitor mixture, and 5 μg/mL DNase I. Cells were lysed by sonication, and debris was removed by centrifugation. The cleared lysate was filtered (0.45 μm) prior to purification by nickel-affinity chromatography (HiTrap Chelating HP, GE Healthcare). The column was washed with lysis buffer and eluted over a linear gradient to a concentration of 500 mM imidazole. Proteins were further purified over heparin and gel-filtration columns and stored as described above.
For biochemical studies, GST-hsNKRF G-patch wt or mutant constructs were expressed in BL21star (DE3) in lysogeny broth (LB) medium at 20 °C for 10 h after induction with 1 mM isopropyl β-d-1-thiogalactopyranoside. Proteins were purified over glutathione beads and a gel-filtration step as described above.
Crystallization.
Crystals of the hsDHX15(113-795)–NKRF G-patch (541-603) complex were obtained by mixing 0.2 μL of complex (2 mg/mL) with 0.2 μL of reservoir at 18 °C via sitting-drop vapor diffusion after 3 d in a MORPHEUS protein crystallization screen (Molecular Dimensions). Most promising crystals grew in a condition containing 0.02 M sodium formate, 0.02 M ammonium acetate, 0.02 M trisodium citrate, 0.02 M sodium potassium l-tartrate, 0.02 M sodium oxamate, 0.1 M N,N-Bis(2-hydroxyethyl)glycine/Tris, pH 8.5, 12.5% (weight [w]/volume [v]) polyethylene glycol (PEG) 1000, 12.5% (w/v) PEG 3350, and 12.5% (v/v) MPD. Crystals of the protein complex with ADP in the active site were obtained by mixing the protein complex (2.3 mg/mL) supplemented with 25 μM U10-RNA (Mycrosynth) and 1 mM ADP with the reservoir solutions at a 1:1 ratio. Crystals grew in a MORPHEUS condition containing 0.02 M 1,6-hexanediol, 0.02 M 1-butanol, 0.02 M (RS)-1,2-propanediol, 0.02 M 2-propanol, 0.02 M 1,4-butanediol, 0.02 M 1,3-propanediol, 0.1 M 2-(N-morpholino)ethanesulfonic acid/imidazole, pH 6.5, 10% (w/v) PEG 8000, and 20% (v/v) ethylene glycol. Crystals were frozen in liquid nitrogen directly from the drop.
Data Collection and Structure Refinement.
Datasets were collected on an EIGER 16M detector at the PXI beamline and on a PILATUS 2M-F detector at the PXIII beamline of the Swiss Light Source (Paul Scherrer Institute, Villigen, Switzerland). Diffraction images were recorded at a wavelength of 1 Å for the native dataset and 2.066 Å for the anomalous diffraction dataset. Diffraction of crystals extended to 2.21 Å for the complex without ADP and to 1.85 Å for the complex containing ADP. Data were processed with XDS (71), and phasing was achieved by molecular replacement of the DHX15 structure [Protein Data Bank (PDB) ID code 5XDR (43)] using Phaser (72). Iterative cycles of model building in COOT (73) and refinement performed in Phenix (74) against the native dataset were then used to build the G-patch peptide and complete the structure. Both structures were refined by using eight Translation/Libration/Screw groups as automatically defined in Phenix based on B-factor analysis. In both structures, DHX15 residues 113–789 could be modeled apart from the following missing loop residues: (apo) 376–383, 406–413, and 431–432; and (ADP) 408. For NKRF, residues 551–596 (apo) and 553–593.
Polder (75) and omit maps were calculated in Phenix. CCP4 (76) programs CAD and FFT were used to calculate anomalous difference density maps.
Copurification Assays.
For copurification assays of proteins expressed in E. coli wt and mutant constructs of MBP–hsDHX15ΔN (113–795), GST–hsNKRF Gpatch (541–603), and controls, respectively, were coexpressed in BL21Star (DE3) (ThermoFisher) in 100 mL of LB as described above. The pelleted cells were resuspended in 5 mL of PD buffer (50 mM Hepes, pH 7.5, 200 mM NaCl, and 2 mM DTT) supplemented with cOmplete EDTA-free protease inhibitor mixture (Roche), 1 mg/mL lysozyme (Sigma), and 5 μg/mL DNase I (Roche) and were lysed by sonication. The lysate was cleared by centrifugation (8,600 × g, 30 min, 4 °C), and the soluble fraction was passed through a syringe filter (0.45 μm). To 1.5 mL of cleared lysate, 75 μL of glutathione Sepharose beads (50% slurry; Amersham Biosciences) were added and incubated for 20 min at 4 °C on a rotator. The beads were washed three times in PD buffer to remove unbound protein. Bound proteins were eluted by the addition of PD buffer containing 25 mM glutathione. Input and eluate samples were analyzed by sodium dodecyl sulfate (SDS)/polyacrylamide gel electrophoresis (PAGE) and visualized by Coomassie staining and Western blot.
For coprecipitation assays from human cells, 2.7 × 106 HEK 293T cells were seeded in 10-cm dishes and transfected 24 h later by using the calcium phosphate method. To express HA-2S–tagged hsNKRF variants, cells were transfected with 20 μg of plasmid. As a negative control, 5 μg of HA-2S–tagged enhanced green fluorescent protein (eGFP) was transfected together with 15 μg of empty vector. Two days posttransfection, cells were harvested on ice in wash buffer (50 mM Hepes, pH 7.9, 150 mM NaCl, 2 mM DTT, 0.5 mM EDTA, 10% glycerol, 0.2% Triton X-100, and 1.5 mM MgCl2) supplemented with cOmplete EDTA-free protease inhibitor mixture (Roche). Cells were lysed by sonication (10 strokes, 20% duty cycle, and three output control) and subsequently treated with RNase A for 30 min on ice. The cell lysate was clarified by centrifugation at 16,000 × g for 15 min at 4 °C. The cleared lysate was incubated for 30 min at 4 °C with 50 μL of Strep-Tactin Superflow resin (IBA Lifesciences) on a rotator. Beads were washed three times with wash buffer. Bound proteins were eluted in 100 μL of protein sample buffer. For further analysis, proteins were separated by SDS/PAGE and detected by Western blot using the described antibodies.
RNA Binding Assays.
RNA binding affinities of His10–hsDHX15 (ΔN and full-length) in dependence of GST–hsNKRF G-patch variants and in the presence of different adenosine nucleotides were determined via fluorescence polarization using a CLARIOstar microplate reader (BMG Labtech). Experiments were carried out in buffer containing 20 mM Hepes (pH 7.5), 150 mM NaCl, 5% glycerol, and 2 mM MgCl2 in a total reaction volume of 30 μL. Binding to 5′-6-fluorescein amidites (FAM)–labeled U12 RNA (Microsynth) at a concentration of 10 nM was monitored by using protein concentrations ranging from 1 nM to 10 μM. For RNA binding measurements containing adenosine nucleotides, ADP, ADP⋅BeFx (from mixing NaF and BeCl2 solutions in a 3:1 molar ratio), ADP⋅AlFx (from mixing NaF and AlCl3 solutions in a 4:1 molar ratio), and AMPPNP (Sigma) were used at final concentrations of 100 μM. All measurements were performed with an excitation wavelength of 482 nm and were detected five times at 530 nm. All experiments were performed in triplicate. The collected fluorescence polarization values were normalized to 1 and fitted according to ref. 77 by using GraphPad Prism.
ATPase Activity Assays.
For measurements of ATPase activity, all assay components except for ATP were mixed in ATPase buffer (10 mM Hepes–NaOH, pH 7.5, and 200 mM KCl): 1 mM phosphoenolpyruvate, 0.5 mM nicotinamide adenine dinucleotide, 1 mM DTT, 20 mM MgCl2, 12 U of pyruvate kinase, and 18 U of lactate dehydrogenase. All reactions contained 5 μM His10–hsDHX15 (ΔN and full-length), and indicated reactions also contained GST–hsNKRF G-patch variants (5 μM final concentration) or U10–RNA (concentrations varying between 2 and 100 μM as indicated) and were carried out in 96-well plates (BRANDplate, pureGrade). Assays were started by adding the desired amount of ATP in 20 μL of ATPase buffer to 80 μL of reaction mix. Absorption at 340 nm was monitored by using a Synergy 2 plate reader (BioTek Instruments) over 60 min at 25 °C with one measurement per minute. Absorption values were adjusted to 1-cm path length by the instrument and converted to concentrations using the Beer–Lambert law with an extinction coefficient at 340 nm of 6,220 M−1⋅cm−1. Initial velocities were determined by linear regression in the concentration vs. time plot using the Origin software. Rates were corrected by the signal change observed in the absence of ATP. Data were fitted by nonlinear regression using GraphPad Prism. ATP titrations (vi/[E0] vs. [ATP]) were fitted with the Michaelis–Menten equation and kinetic parameters extracted. RNA titrations (vi/[E0] vs. [U10-RNA]) were fitted by using a dose–response relationship, and the half-maximal effective concentration (EC50) value was extracted.
RNA Helicase Assays.
Fluorescently labeled RNA duplex (Microsynth) with a single-stranded 3′ overhang was used as a substrate similar to ref. 45. Strands were annealed by mixing 60 µM unlabeled RNA (5′-GCGCCUACGGAGCUGGUGGCGUAGGCGCU17-3′) with 30 µM 3′-6-FAM–labeled RNA [5′-CACCAGCUCCGUAGGCGC-(FAM)-3′] in 20 mM Hepes (pH 7.5). The two strands were incubated at 80 °C for 5 min and cooled step-wise to 20 °C in a thermocycler. Unwinding reactions contained His10–DHX15ΔN (400 nM), dsRNA (200 nM); a fourfold excess of a competitor DNA (5′-GCGCCTACGGAGCTGGTG-3′), if indicated; GST–NKRF G-patch variants (800 nM) and reaction buffer (50 mM Hepes, pH 7.5, 50 mM KCl, 5% glycerol, 2 mM MgCl2, and 0.4 U/µL RiboLock RNase Inhibitor [Thermo Scientific]) in a total of 50 µL. Reactions were performed at 37 °C and started by adding ATP in reaction buffer to a final concentration of 2 mM. At indicated time points, 4-µL aliquots were removed, and the reaction was quenched with 16 µL of stop buffer (5 mM EDTA, 500 mM KCl, and 2× RNA loading dye [13% Ficoll 400, 2× Tris–borate–EDTA (TBE), 0.05% bromophenol blue, and 0.05% xylene cyanol]). The 2-µL samples were separated on a 10% native TBE–acrylamide gel, and imaged on a Typhoon FLA 9500 (GE Healthcare). Band intensities of duplexed and unwound FAM-RNA oligos were quantified with Image Lab. All experiments were carried out in triplicate. The averages of the fraction of unwound RNA were plotted against time and fitted with a single exponential equation according to ref. 78 using nonlinear regression in GraphPad Prism to extract rate constants.
Immunofluorescence Analysis.
Immunofluorescence analysis was essentially performed as described in ref. 66. HeLaK cells were transiently transfected for 24 h by using X-treme Gene 9 (Roche) according to the manufacturer’s protocol. Cells were fixed in 4% paraformaldehyde (PFA) and permeabilized in 0.1% Triton X-100, 0.02% SDS, and 1× phosphate-buffered saline (PBS) for 5 min. After blocking for 2 h with 10% goat serum and 2% bovine serum albumin (BSA)/PBS, primary antibodies diluted in blocking solution were added to the cells and incubated in the dark for 2 h. Cells were washed three times in 2% BSA/PBS prior to 30-min incubation with secondary antibodies diluted in blocking solution. After three washes with 2% BSA/PBS, cells were washed once with 0.1% Triton X-100, 0.02% SDS, and 1× PBS and fixed with 4% PFA. Residual PFA was removed by washing with PBS before incubation with Hoechst stain (0.5 mg/L in PBS). After a final wash with PBS, the coverslips were mounted in VectaShield (Vector Laboratories) onto microscope slides. Images were taken with a Zeiss 780 upright laser-scanning confocal microscope and a 63× oil objective (ScopeM, ETH Zürich). Excitation at 405, 488, and 633 nm was used for DAPI and Alexa Fluor 488 and 633, respectively.
Supplementary Material
Acknowledgments
We thank beamline scientists at the Swiss Light Source (Paul Scherrer Institute, Switzerland) for assistance with X-ray diffraction measurements; Nenad Ban for access to crystallization equipment; Frédéric Allain for sharing lab infrastructure; Ulrike Kutay for sharing the Enp1 antibody, pcDNA5-HA-2S vectors, and access to equipment; Kevin Sabath for setting up crystallization plates, help with data collection, and comments on the manuscript; Ivo Zemp and Kerstin Dörner for advice on immunofluorescence experiments; and Kerstin Dörner, Maria Hondele, Emil Dedic, and Sébastien Campagne for critical reading of the manuscript. This work was supported by the Swiss National Science Foundation (SNSF) through the National Center for Competence in Research “RNA & Disease” (S.J.) and SNSF Grant 31003A_179498 (to S.J.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: Coordinates for the structures have been deposited in the Protein Data Bank, https://www.wwpdb.org/ (PDB ID codes 6SH7 [apo] and 6SH6 [ADP]).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1913880117/-/DCSupplemental.
References
- 1.Bleichert F., Baserga S. J., The long unwinding road of RNA helicases. Mol. Cell 27, 339–352 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Bourgeois C. F., Mortreux F., Auboeuf D., The multiple functions of RNA helicases as drivers and regulators of gene expression. Nat. Rev. Mol. Cell Biol. 17, 426–438 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Cordin O., Beggs J. D., RNA helicases in splicing. RNA Biol. 10, 83–95 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jarmoskaite I., Russell R., RNA helicase proteins as chaperones and remodelers. Annu. Rev. Biochem. 83, 697–725 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ozgur S., et al. , The conformational plasticity of eukaryotic RNA-dependent ATPases. FEBS J. 282, 850–863 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Pyle A. M., Translocation and unwinding mechanisms of RNA and DNA helicases. Annu. Rev. Biophys. 37, 317–336 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Fairman-Williams M. E., Guenther U.-P., Jankowsky E., SF1 and SF2 helicases: Family matters. Curr. Opin. Struct. Biol. 20, 313–324 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hamann F., Enders M., Ficner R., Structural basis for RNA translocation by DEAH-box ATPases. Nucleic Acids Res. 47, 4349–4362 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walbott H., et al. , Prp43p contains a processive helicase structural architecture with a specific regulatory domain. EMBO J. 29, 2194–2204 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singleton M. R., Dillingham M. S., Wigley D. B., Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 76, 23–50 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Boneberg F. M., et al. , Molecular mechanism of the RNA helicase DHX37 and its activation by UTP14A in ribosome biogenesis. RNA 25, 685–701 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen M. C., et al. , Structural basis of G-quadruplex unfolding by the DEAH/RHA helicase DHX36. Nature 558, 465–469 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen W.-F., et al. , Molecular mechanistic insights into Drosophila DHX36-mediated G-quadruplex unfolding: A structure-based model. Structure 26, 403–415.e4 (2018). [DOI] [PubMed] [Google Scholar]
- 14.He Y., Staley J. P., Andersen G. R., Nielsen K. H., Structure of the DEAH/RHA ATPase Prp43p bound to RNA implicates a pair of hairpins and motif Va in translocation along RNA. RNA 23, 1110–1124 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prabu J. R., et al. , Structure of the RNA helicase MLE reveals the molecular mechanisms for uridine specificity and RNA-ATP coupling. Mol. Cell 60, 487–499 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Robert-Paganin J., et al. , Functional link between DEAH/RHA helicase Prp43 activation and ATP base binding. Nucleic Acids Res. 45, 1539–1552 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tauchert M. J., Fourmann J.-B., Lührmann R., Ficner R., Structural insights into the mechanism of the DEAH-box RNA helicase Prp43. eLife 6, e21510 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin R., Straub A. U., Doebele C., Bohnsack M. T., DExD/H-box RNA helicases in ribosome biogenesis. RNA Biol. 10, 4–18 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodríguez-Galán O., García-Gómez J. J., de la Cruz J., Yeast and human RNA helicases involved in ribosome biogenesis: Current status and perspectives. Biochim. Biophys. Acta 1829, 775–790 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Will C. L., Lührmann R., Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 3, a003707 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klinge S., Woolford J. L. Jr, Ribosome assembly coming into focus. Nat. Rev. Mol. Cell Biol. 20, 116–131 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matera A. G., Wang Z., A day in the life of the spliceosome. Nat. Rev. Mol. Cell Biol. 15, 108–121 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baßler J., Hurt E., Eukaryotic ribosome assembly. Annu. Rev. Biochem. 88, 281–306 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Semlow D. R., Blanco M. R., Walter N. G., Staley J. P., Spliceosomal DEAH-box ATPases remodel pre-mRNA to activate alternative splice sites. Cell 164, 985–998 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arenas J. E., Abelson J. N., Prp43: An RNA helicase-like factor involved in spliceosome disassembly. Proc. Natl. Acad. Sci. U.S.A. 94, 11798–11802 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koodathingal P., Novak T., Piccirilli J. A., Staley J. P., The DEAH box ATPases Prp16 and Prp43 cooperate to proofread 5′ splice site cleavage during pre-mRNA splicing. Mol. Cell 39, 385–395 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mayas R. M., Maita H., Semlow D. R., Staley J. P., Spliceosome discards intermediates via the DEAH box ATPase Prp43p. Proc. Natl. Acad. Sci. U.S.A. 107, 10020–10025 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bohnsack M. T., et al. , Prp43 bound at different sites on the pre-rRNA performs distinct functions in ribosome synthesis. Mol. Cell 36, 583–592 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Combs D. J., Nagel R. J., Ares M. Jr, Stevens S. W., Prp43p is a DEAH-box spliceosome disassembly factor essential for ribosome biogenesis. Mol. Cell. Biol. 26, 523–534 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lebaron S., et al. , The splicing ATPase prp43p is a component of multiple preribosomal particles. Mol. Cell. Biol. 25, 9269–9282 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leeds N. B., Small E. C., Hiley S. L., Hughes T. R., Staley J. P., The splicing factor Prp43p, a DEAH box ATPase, functions in ribosome biogenesis. Mol. Cell. Biol. 26, 513–522 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Memet I., Doebele C., Sloan K. E., Bohnsack M. T., The G-patch protein NF-κB-repressing factor mediates the recruitment of the exonuclease XRN2 and activation of the RNA helicase DHX15 in human ribosome biogenesis. Nucleic Acids Res. 45, 5359–5374 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Silverman E., Edwalds-Gilbert G., Lin R.-J., DExD/H-box proteins and their partners: Helping RNA helicases unwind. Gene 312, 1–16 (2003). [DOI] [PubMed] [Google Scholar]
- 34.Sloan K. E., Bohnsack M. T., Unravelling the mechanisms of RNA helicase regulation. Trends Biochem. Sci. 43, 237–250 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Aravind L., Koonin E. V., G-patch: A new conserved domain in eukaryotic RNA-processing proteins and type D retroviral polyproteins. Trends Biochem. Sci. 24, 342–344 (1999). [DOI] [PubMed] [Google Scholar]
- 36.Robert-Paganin J., Réty S., Leulliot N., Regulation of DEAH/RHA helicases by G-patch proteins. BioMed Res. Int. 2015, 931857 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heininger A. U., et al. , Protein cofactor competition regulates the action of a multifunctional RNA helicase in different pathways. RNA Biol. 13, 320–330 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen Y.-L., et al. , The telomerase inhibitor Gno1p/PINX1 activates the helicase Prp43p during ribosome biogenesis. Nucleic Acids Res. 42, 7330–7345 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lebaron S., et al. , The ATPase and helicase activities of Prp43p are stimulated by the G-patch protein Pfa1p during yeast ribosome biogenesis. EMBO J. 28, 3808–3819 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Niu Z., Jin W., Zhang L., Li X., Tumor suppressor RBM5 directly interacts with the DExD/H-box protein DHX15 and stimulates its helicase activity. FEBS Lett. 586, 977–983 (2012). [DOI] [PubMed] [Google Scholar]
- 41.Tanaka N., Aronova A., Schwer B., Ntr1 activates the Prp43 helicase to trigger release of lariat-intron from the spliceosome. Genes Dev. 21, 2312–2325 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoshimoto R., Kataoka N., Okawa K., Ohno M., Isolation and characterization of post-splicing lariat-intron complexes. Nucleic Acids Res. 37, 891–902 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murakami K., Nakano K., Shimizu T., Ohto U., The crystal structure of human DEAH-box RNA helicase 15 reveals a domain organization of the mammalian DEAH/RHA family. Acta Crystallogr. F Struct. Biol. Commun. 73, 347–355 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martin A., Schneider S., Schwer B., Prp43 is an essential RNA-dependent ATPase required for release of lariat-intron from the spliceosome. J. Biol. Chem. 277, 17743–17750 (2002). [DOI] [PubMed] [Google Scholar]
- 45.Christian H., Hofele R. V., Urlaub H., Ficner R., Insights into the activation of the helicase Prp43 by biochemical studies and structural mass spectrometry. Nucleic Acids Res. 42, 1162–1179 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Merkley E. D., et al. , Distance restraints from crosslinking mass spectrometry: Mining a molecular dynamics simulation database to evaluate lysine-lysine distances. Protein Sci. 23, 747–759 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wan R., Yan C., Bai R., Lei J., Shi Y., Structure of an intron lariat spliceosome from Saccharomyces cerevisiae. Cell 171, 120–132.e12 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Zang S., et al. , GPKOW is essential for pre-mRNA splicing in vitro and suppresses splicing defect caused by dominant-negative DHX16 mutation in vivo. Biosci. Rep. 34, e00163 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Agarwal K. C., Miech R. P., Parks R. E. Jr, Guanylate kinases from human erythrocytes, hog brain, and rat liver. Methods Enzymol. 51, 483–490 (1978). [DOI] [PubMed] [Google Scholar]
- 50.Fourmann J.-B., Tauchert M. J., Ficner R., Fabrizio P., Lührmann R., Regulation of Prp43-mediated disassembly of spliceosomes by its cofactors Ntr1 and Ntr2. Nucleic Acids Res. 45, 4068–4080 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Warkocki Z., et al. , The G-patch protein Spp2 couples the spliceosome-stimulated ATPase activity of the DEAH-box protein Prp2 to catalytic activation of the spliceosome. Genes Dev. 29, 94–107 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tanaka N., Schwer B., Mutations in PRP43 that uncouple RNA-dependent NTPase activity and pre-mRNA splicing function. Biochemistry 45, 6510–6521 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Choudhury P., Hackert P., Memet I., Sloan K. E., Bohnsack M. T., The human RNA helicase DHX37 is required for release of the U3 snoRNP from pre-ribosomal particles. RNA Biol. 16, 54–68 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tanaka N., Schwer B., Characterization of the NTPase, RNA-binding, and RNA helicase activities of the DEAH-box splicing factor Prp22. Biochemistry 44, 9795–9803 (2005). [DOI] [PubMed] [Google Scholar]
- 55.Petsko G. A., Chemistry and biology. Proc. Natl. Acad. Sci. U.S.A. 97, 538–540 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gilman B., Tijerina P., Russell R., Distinct RNA-unwinding mechanisms of DEAD-box and DEAH-box RNA helicase proteins in remodeling structured RNAs and RNPs. Biochem. Soc. Trans. 45, 1313–1321 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rauhut R., et al. , Molecular architecture of the Saccharomyces cerevisiae activated spliceosome. Science 353, 1399–1405 (2016). [DOI] [PubMed] [Google Scholar]
- 58.Haselbach D., et al. , Structure and conformational dynamics of the human spliceosomal Bact complex. Cell 172, 454–464.e11 (2018). [DOI] [PubMed] [Google Scholar]
- 59.Hamann F., et al. , Structural analysis of the intrinsically disordered splicing factor Spp2 and its binding to the DEAH-box ATPase Prp2. Proc. Natl. Acad. Sci. U.S.A. 117, 2948–2956 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jurica M. S., Licklider L. J., Gygi S. R., Grigorieff N., Moore M. J., Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. RNA 8, 426–439 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu S., et al. , Structure of the yeast spliceosomal postcatalytic P complex. Science 358, 1278–1283 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fica S. M., et al. , Structure of a spliceosome remodelled for exon ligation. Nature 542, 377–380 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhan X., Yan C., Zhang X., Lei J., Shi Y., Structure of a human catalytic step I spliceosome. Science 359, 537–545 (2018). [DOI] [PubMed] [Google Scholar]
- 64.Galej W. P., et al. , Cryo-EM structure of the spliceosome immediately after branching. Nature 537, 197–201 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhu J., Liu X., Anjos M., Correll C. C., Johnson A. W., Utp14 recruits and activates the RNA helicase Dhr1 to undock U3 snoRNA from the preribosome. Mol. Cell. Biol. 36, 965–978 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zemp I., et al. , Distinct cytoplasmic maturation steps of 40S ribosomal subunit precursors require hRio2. J. Cell Biol. 185, 1167–1180 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Haffke M., et al. , Characterization and production of protein complexes by co-expression in Escherichia coli. Methods Mol. Biol. 1261, 63–89 (2015). [DOI] [PubMed] [Google Scholar]
- 68.Gradia S. D., et al. , MacroBac: New technologies for robust and efficient large-scale production of recombinant multiprotein complexes. Methods Enzymol. 592, 1–26 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wyler E., et al. , Tandem affinity purification combined with inducible shRNA expression as a tool to study the maturation of macromolecular assemblies. RNA 17, 189–200 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Studier F. W., Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41, 207–234 (2005). [DOI] [PubMed] [Google Scholar]
- 71.Kabsch W., XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McCoy A. J., et al. , Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 74.Adams P. D., et al. , PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liebschner D., et al. , Polder maps: Improving OMIT maps by excluding bulk solvent. Acta Crystallogr. D Struct. Biol. 73, 148–157 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Winn M. D., et al. , Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rossi A. M., Taylor C. W., Analysis of protein-ligand interactions by fluorescence polarization. Nat. Protoc. 6, 365–387 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Özeş A. R., Feoktistova K., Avanzino B. C., Baldwin E. P., Fraser C. S., Real-time fluorescence assays to monitor duplex unwinding and ATPase activities of helicases. Nat. Protoc. 9, 1645–1661 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Girdlestone C., Hayward S., The DynDom3D webserver for the analysis of domain movements in multimeric proteins. J. Comput. Biol. 23, 21–26 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.