Significance
The ability of an animal to sense what to eat, such as carbohydrate-rich food, as well as to reject food that may be disadvantageous or even deadly is essential for survival and adaptation to metabolic demands compounded by continuous changes in the environment. Here we investigate evolutionarily conserved pathways central for either choosing or rejecting nutrients based on neuronal system interactions between internal signals and the external milieu. These pathways are likely to be necessary for the ability to perform quick adaptation to the immediate conditions and in general provide clues to the structure of the fly’s internal network governing nutrient sensing.
Keywords: feeding behavior, metabolism, hypothalamus, dopamine
Abstract
The intake of macronutrients is crucial for the fitness of any animal and is mainly regulated by peripheral signals to the brain. How the brain receives and translates these peripheral signals or how these interactions lead to changes in feeding behavior is not well-understood. We discovered that 2 crustacean cardioactive peptide (CCAP)-expressing neurons in Drosophila adults regulate feeding behavior and metabolism. Notably, loss of CCAP, or knocking down the CCAP receptor (CCAP-R) in 2 dorsal median neurons, inhibits the release of neuropeptide F (NPF), which regulates feeding behavior. Furthermore, under starvation conditions, flies normally have an increased sensitivity to sugar; however, loss of CCAP, or CCAP-R in 2 dorsal median NPF neurons, inhibited sugar sensitivity in satiated and starved flies. Separate from its regulation of NPF signaling, the CCAP peptide also regulates triglyceride levels. Additionally, genetic and optogenetic studies demonstrate that CCAP signaling is necessary and sufficient to stimulate a reflexive feeding behavior, the proboscis extension reflex (PER), elicited when external food cues are interpreted as palatable. Dopaminergic signaling was also sufficient to induce a PER. On the other hand, although necessary, NPF neurons were not able to induce a PER. These data illustrate that the CCAP peptide is a central regulator of feeding behavior and metabolism in adult flies, and that NPF neurons have an important regulatory role within this system.
A balanced intake of macronutrients is essential for the fitness of any organism. Deficient intake of macronutrients can cause several problems, such as reduced energy, growth defects, hypoglycemic seizures, and liver or heart damage (1–3). On the other hand, high-sugar intake possibly leads to decreased insulin sensitivity, high-protein consumption leads to kidney disease, and a high-fat diet can cause chronic inflammation, which itself can lead to type 2 diabetes (4–8). While many known regulators of food intake have been implicated in affecting macronutrient consumption, such as neuropeptide Y (NPY), proopiomelanocortin, and galanin, this topic has been difficult to study and various research groups have used different conditions for testing macronutrient selection (9–11). For example, depending on the experimental setup, galanin has been shown to either increase total caloric intake in rats, independent of which macronutrient was presented, or to give a preference for a high-fat diet, with no change in caloric intake (12–14). Consequently, how physiological conditions affect macronutrient intake and interact with the neuronal pathways that determine macronutrient consumption are not well-understood.
In Drosophila, palatable food cues are detected by gustatory receptor neurons (GRNs) expressing gustatory receptor 5a (Gr5a) and gustatory receptor 64f (Gr64f), while toxic compounds, such as bitter substances and high-salt concentrations, are detected by Gr66-expressing GRNs (15–20). Recently, it was shown that during nutrient starvation, neuropeptide F (NPF) regulates a dopaminergic to Gr5a pathway to increase sugar sensitivity in Drosophila adults, while short neuropeptide F (sNPF) signals to Gr66-expressing GRNs to reduce the inhibition to bitter taste as starvation progresses (15). Although it is understood that NPF receptor-expressing neurons are negatively regulated by insulin-like peptides in larvae (21), which neuropeptides or hormones control NPF signaling in adults is not clear.
Until recently, it was believed that crustacean cardioactive peptide (CCAP) neurons were critical for proper larval and pupal ecdysis in Drosophila, as well as maintaining the rhythm of adult ecdysis (eclosion) (22–25). In brief, eclosion hormone (EH) regulates CCAP release within the larval central nervous system and, along with bursicon, a copeptide released from CCAP neurons, it was thought that CCAP initiated ecdysis and shut off preecdysis (23). Recently, the involvement of CCAP in the ecdysis process has come under question, as CCAP-null mutants produce normal adults, with no apparent defects in ecdysis or postecdysis (22). Interestingly, within 24 h of eclosion, most CCAP-expressing neurons undergo programmed cell death and only a pair of neurons in the dorsal median region of the protocerebrum remain in the adult (26). No information has been published on a possible function for these 2 remaining adult CCAP neurons.
While the CCAP neuropeptide has been extensively studied in the regulation of ecdysis (22–25), its function in Drosophila adults has not been explored. Moreover, although there are indications in other species that CCAP may play a role in the regulation of food uptake, direct evidence is lacking (27, 28). We employed extensive genetic, optogenetic, and behavioral techniques to investigate the role of CCAP in adult flies. Using genetic manipulation, we determined that a small subpopulation of CCAP-R–expressing NPF neurons regulate feeding behavior in adults. Our data further suggest that the CCAP neuropeptide also regulates metabolism in Drosophila adults.
Results
CCAP Receptor Expressed in NPF Neurons.
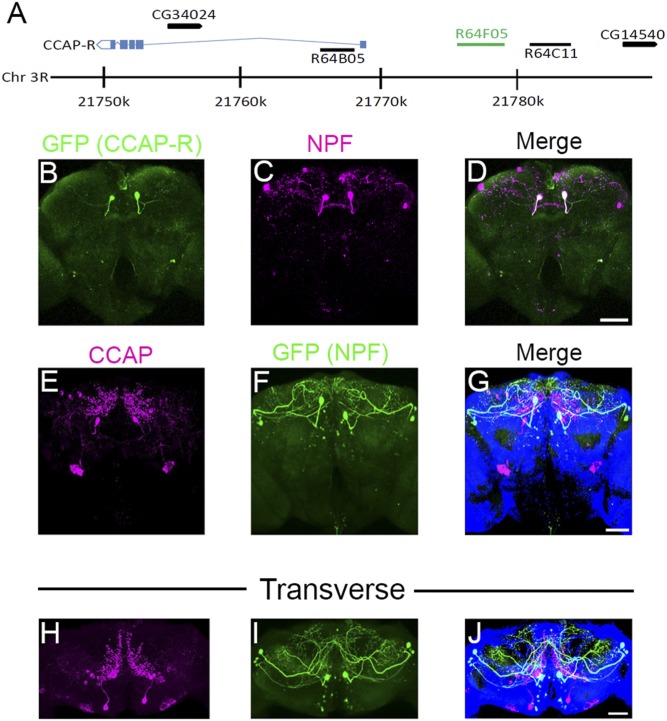
Starvation increases sugar taste sensitivity in Drosophila, partly through the activation of NPF neurons (15). Currently, it is not understood which neuropeptides regulate NPF neurons under starvation conditions. To look for genes that may regulate NPF signaling, the FlyLight database (29) was searched for available GAL4 lines having an expression pattern similar to NPF-expressing neurons. The GAL4 line R64F05 appeared to express in a pattern reminiscent of 2 dorsal median NPF-expressing neurons, designated P1 neurons (30). R64F05-GAL4 was produced by cloning a 3,563-bp fragment from a noncoding region upstream of CCAP-R to drive GAL4 expression (Fig. 1A, green line). To examine the expression pattern, R64F05-GAL4 flies were crossed with UAS-GFP and the brains of 5- to 9-d-old F1 generation adult males were costained for green fluorescent protein (GFP) (Fig. 1B, green) and NPF expression (Fig. 1C, magenta). This revealed that R64F05-GAL4 was exclusively expressed in the dorsal median P1 NPF-expressing neurons (Fig. 1D, overlapping expression seen as white). Two other GAL4 drivers (R64B05 and R64C11), produced by cloning DNA from around the CCAP-R gene, with expression patterns that appeared to overlap dorsal median P1 or dorsal lateral L1-I NPF neurons (Fig. 1A), were also tested. Neither driver showed expression that corresponded to any NPF neurons (SI Appendix, Fig. S1 A and B) (30).
Fig. 1.
Branches of CCAP neurons superimpose NPF neuron cell bodies. (A) Structure of the Drosophila CCAP-R genomic region; blue boxes indicate CCAP-R exons and blue lines indicate introns. Black arrows with corresponding CG numbers above indicate other genes in the region. Sequences cloned to drive GAL4 expression are indicated as black lines with the corresponding FlyLight number above. The genomic sequence used for R64F05-GAL4 is indicated as a green line. (B–D) Dissected R64F05-GAL4;UAS-GFP male adult brains (5 to 9 d posteclosion) were costained with (B) anti-GFP (green) and (C) anti-NPF (magenta). (D) White in the merged picture indicates overlapping GFP (R64F05) and NPF expression. (Scale bar, 50 µm.) Pictures show a representative Z stack, which includes 40 (2-µm) slices. (E–G) Z projection of (E) CCAP (magenta) and (F) GFP (green; NPF-GAL4;UAS-GFP) neurons in dissected male adult brains (10 to 12 d posteclosion). (G) Merged picture showing CCAP (magenta) and GFP (green; NPF) expression. (Scale bar, 50 µm.) The picture is representative of a Z stack, which includes 40 (2-μm) slices. (H–J) Transverse Z projection of (H) CCAP (magenta) and (I) GFP (green; NPF-GAL4;UAS-GFP) neurons in dissected male adult brains (10 to 12 d posteclosion). (J) Merged picture showing CCAP (magenta) and GFP (green; NPF) expression. (Scale bar, 50 µm.)
To determine if CCAP-expressing neurons could be innervating NPF neurons, the brains of 5- to 7-d-old adult males, expressing GFP in an NPF neural pattern (NPF-GAL4>UAS-GFP), were costained for CCAP (Fig. 1E, magenta) and GFP (Fig. 1F, green) expression (31). Two CCAP neurons (Fig. 1 E and G) were evident that sent projections toward the 2 dorsal median P1 NPF neurons (Fig. 1 F and G and Movie S1). By examining the CCAP neural staining in a transverse orientation, it became evident that the CCAP neuronal projections extended throughout the superior medial protocerebrum (Fig. 1H and Movie S2). Furthermore, the projections seemed to track close to the dorsal median P1 NPF neuronal cell bodies (Fig. 1 I and J). Other CCAP expression observed in the brain was most likely background staining, as it was still visible in equally aged CCAPexc7 null males (SI Appendix, Fig. S1C, arrows, and Movie S3).
CCAP Signaling Regulates NPF Neural Activity.
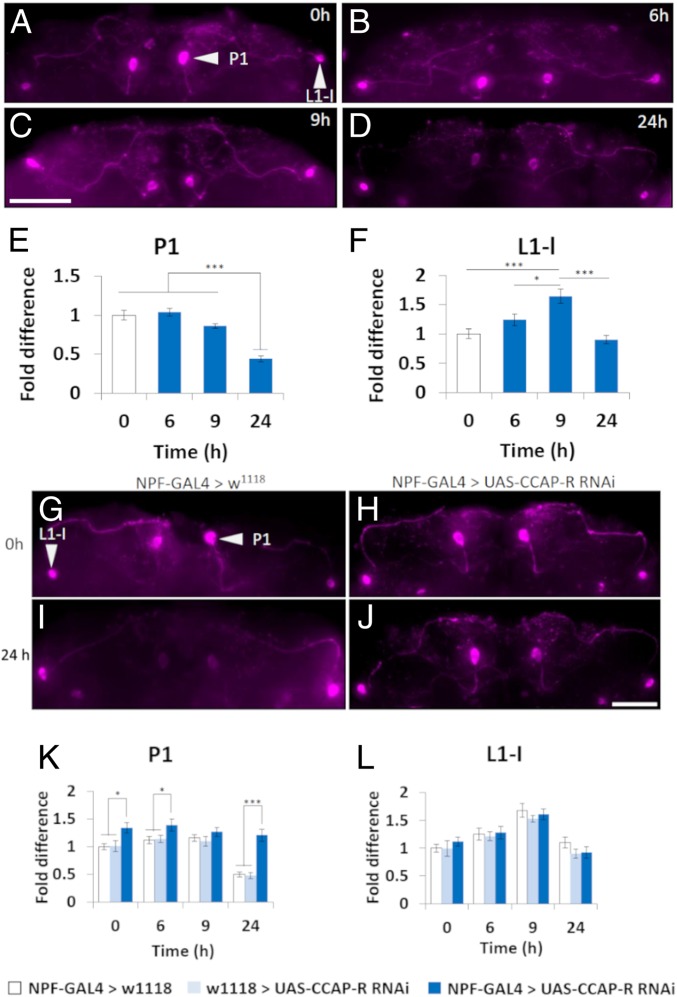
Given that R64F05-GAL4 was only detected in P1 NPF neurons, we wanted to understand if dorsal median (P1) and dorsal lateral (L1-I) NPF neurons reacted differentially under starvation conditions (29). To do this, brains from wild-type male flies, which had undergone either acute (3, 6, and 9 h) or chronic (24 h) starvation, were stained for NPF expression. After 24-h starvation, NPF expression was significantly lower in the dorsal median P1 neurons (Fig. 2 D and E), when compared with flies fed ad libitum (Fig. 2 A and E) (30). In contrast, NPF expression in the dorsal lateral L1-I neurons was significantly higher in flies starved for 9 h (Fig. 2 C and F) but returned to fed levels by 24-h starvation (Fig. 2 D and F) (30). Studying the CCAP neurons revealed a significant reduction in CCAP peptide expression at the 9-h starvation time point (SI Appendix, Figs. S2 and S3).
Fig. 2.
CCAP regulates CCAP-R–expressing NPF neurons. (A–D) Close-up of NPF neurons in whole Drosophila male brains, 5 to 9 d posteclosion, visualizing NPF expression after various times of starvation. (Scale bar, 50 µm.) (E and F) Relative NPF immunofluorescence expression levels in fed flies and after various times of starvation. (E) P1 (dorsal median) NPF neurons. (F) L1-I (dorsal lateral) NPF neurons. NPF neurons of 10 to 12 brains for each condition were investigated (*P < 0.05, ***P < 0.005; initially, a Kolmogorov–Smirnov test of normality was performed before a one-way ANOVA with Tukey’s post hoc test for multiple comparisons between all samples). (G–J) Close-up of NPF neurons in whole Drosophila male brains, control (NPF-GAL4>w1118) and experimental (NPF-GAL4>UAS-CCAP-RRNAi), 5 to 9 d posteclosion, visualizing NPF expression after various times of starvation. (Scale bar, 50 µm.) (K and L) Relative NPF immunofluorescence expression levels in fed flies and after various times of starvation. (K) P1 (dorsal median) NPF neurons. (L) L1-I (dorsal lateral) NPF neurons. NPF neurons of 10 to 12 brains for each condition were investigated (*P < 0.05, ***P < 0.005; initially, a Kolmogorov–Smirnov test of normality was performed before a one-way ANOVA with Tukey’s post hoc test for multiple comparisons between samples within the same time point). Error bars represent SEM.
Next, we used NPF-GAL4 (30, 32) to lower CCAP-R expression, via UAS-CCAP-RRNAi, and repeated the NPF staining under starvation conditions. When CCAP-R expression was inhibited in NPF neurons there was significantly more NPF expressed in the 2 dorsal median P1 neurons under ad libitum feeding, as well as at the 3-, 6-, and 24-h starvation time points (Fig. 2 G–K). No significant effect was observed in the dorsal lateral L1-I NPF neurons (Fig. 2 G–J and L). We repeated the experiment using adult male CCAP nulls (CCAPexc7) (22). Similar to knocking down CCAP-R in the P1 NPF neurons, loss of CCAP expression in the brains of adult males significantly increased NPF expressed in the 2 dorsal median P1 NPF neurons at the 9- and 24-h starvation time points (SI Appendix, Fig. S4 A–E), while having no effect on the dorsal lateral L1-I NPF neurons (SI Appendix, Fig. S4 A–D and F). To understand if CCAP was sufficient to activate NPF neurons, CCAP-GAL4 was crossed to UAS-TrpA1 (CCAP-GAL4>UAS-TrpA1) (24). TrpA1 is a heat-activated thermosensitive cation channel (33). Expressing TrpA1 in CCAP neurons, and maintaining flies fed ad libitum at 29 °C, was sufficient to significantly reduce NPF expression in the 2 P1 dorsal median neurons (SI Appendix, Fig. S5). Interestingly, activating CCAP neurons in flies fed ad libitum significantly increased NPF expression in the dorsal lateral L1-I neurons (SI Appendix, Fig. S5).
CCAP Neurons Regulate Feeding Behavior.
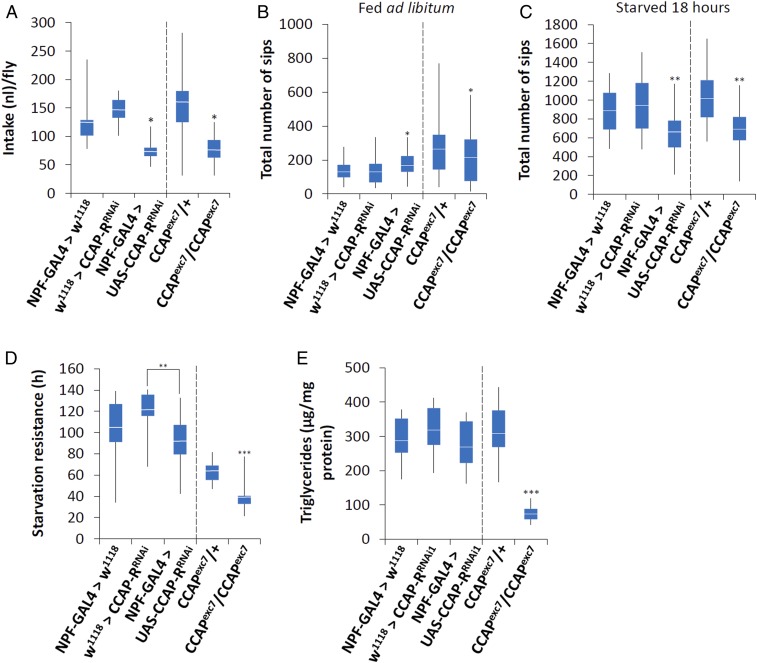
To determine if CCAP had a role in the regulation of feeding behavior, we employed the capillary feeding (CAFE) assay to measure total food intake (34). Over a 24-h period, males where CCAP-R expression was knocked down in NPF neurons, as well as CCAPexc7 null males, ate significantly less than control flies (Fig. 3A). A second CCAP-R RNAi line was also tested to make sure there were no off-target effects due to RNAi (UAS-CCAP-RRNAi2) expression. This line also ate significantly less than controls when crossed to NPF-GAL4 (SI Appendix, Fig. S6 A and B).
Fig. 3.
CCAP regulates feeding behavior. (A) A CAFE assay was used to assess total food intake over a 24-h period in 5- to 9-d-old adult males. Five males were used for each replicate and the assay was repeated at least 10 times for each genotype. The food source was 150 mM sucrose. Initially, a Kolmogorov–Smirnov test of normality was performed. Since some of the samples failed the test, a nonparametric Kruskal–Wallis ANOVA was performed with Dunn’s post hoc test for multiple comparisons (*P < 0.05). (B and C) The flyPAD was used to measure the total number of sips over 1 h using males that were either (B) fed ad libitum or (C) previously starved for 18 h. n is between 32 and 64 and a Kolmogorov–Smirnov test of normality was performed. Since some of the samples failed the test, a nonparametric Kruskal–Wallis ANOVA was performed with Dunn’s post hoc test for multiple comparisons, comparing either NPF-GAL4>w1118, w1118>UAS-CCAP-RRNAi, and NPF-GAL4>UAS-CCAP-RRNAi or CCAPexc7/+ and CCAPexc7 nulls (*P < 0.05, **P < 0.01). (D) A starvation resistance test was performed using the DAMS. In short, 5- to 7-d-old male flies were maintained in a DAMS tube with 1% agarose so they had water but no food. Their activity was monitored and these data were used to calculate starvation resistance. n is between 32 and 64. Initially, a Kolmogorov–Smirnov test of normality was performed, followed by a one-way ANOVA performed with Tukey’s post hoc test for multiple comparisons, comparing either NPF-GAL4>w1118, w1118>UAS-CCAP-RRNAi, and NPF-GAL4>UAS-CCAP-RRNAi or CCAPexc7/+ and CCAPexc7 nulls (**P < 0.01, ***P < 0.005). (E) Triglyceride levels were determined in 5- to 7-d-old male flies that were fed ad libitum. n = 30 males per treatment; the assay was repeated at least 10 times for each genotype. Initially, a Kolmogorov–Smirnov test of normality was performed, followed by a one-way ANOVA with Tukey’s post hoc test for multiple comparisons, comparing either NPF-GAL4>w1118, w1118>UAS-CCAP-RRNAi, and NPF-GAL4>UAS-CCAP-RRNAi or CCAPexc7/+ and CCAPexc7 nulls (***P < 0.005). Error bars represent SEM. An asterisk over one sample with no line connecting to another sample indicates it is significantly different from the other samples.
To understand how satiation state influences feeding behavior, we employed the fly proboscis and activity detector (flyPAD) (35). This apparatus allowed us to detect changes on a minute-by-minute scale in feeding response under 2 different satiety states, fed ad libitum or starved for 18 h. Itskov et al. (35) demonstrated that flies feeding on solid food are not able to pump in the solution but extend the proboscis to take individual sips. A group of sips constitutes a feeding burst (FB), and a group of feeding bursts makes up an activity bout (AB) (35). The flyPAD is able to detect different components of feeding, including individual sips, the number of sips per burst, FB and AB durations, as well as the intervals (FBI and ABI) between various FBs and ABs (35). When satiated, CCAPexc7 null males took significantly fewer sips (Fig. 3B and Table 1), had significantly fewer FBs, significantly longer FBIs (Table 1), and significantly fewer ABs (Table 2) than controls. On the other hand, knocking down CCAP-R in NPF neurons significantly increased the number of FBs and the total number of sips (TSs) in fed flies (Fig. 3B, Table 1, and SI Appendix, Fig. S6C), while the number of ABs was significantly decreased (Table 2). When starved for 18 h, both CCAPexc7 nulls and males where CCAP-R was knocked down in NPF neurons had fewer FBs and took significantly fewer TSs than controls (Fig. 3C, Table 1, and SI Appendix, Fig. S6D).
Table 1.
flyPAD feeding burst data in male flies
| Genotype | Fed ad libitum | Starved 18 h | ||||||||
| FB | FBD | FBI | SPB | TS | FB | FBD | FBI | SPB | TS | |
| CCAPexc7>w1118 | 17.0 ± 2.0 | 0.9 ± 0.1 | 108 ± 25 | 5.0 ± 0.2 | 264 ± 35 | 113 ± 6 | 0.8 ± 0.0 | 1.7 ± 0.1 | 4.6 ± 0.2 | 1008 ± 49 |
| CCAPexc7/CCAPexc7 | 10.9 ± 1.7* | 1.4 ± 0.1*** | 162 ± 31* | 4.1 ± 0.4* | 202 ± 34* | 62.2 ± 3.9*** | 0.9 ± 0.1 | 3.4 ± 0.3*** | 4.0 ± 0.2* | 684 ± 37*** |
| NPF-GAL4>w1118 | 12.5 ± 1.4 | 1.5 ± 0.1 | 64.8 ± 1.1 | 5.8 ± 0.4 | 136 ± 11 | 87.7 ± 7.2 | 1.2 ± 0.1 | 1.4 ± 0.1 | 5.4 ± 0.2 | 881 ± 53 |
| w1118>CCAP-RRNAi | 13.5 ± 2.1 | 1.6 ± 0.2 | 64.5 ± 0.2 | 7 ± 0.6 | 132 ± 19 | 98.7 ± 6.7 | 1.4 ± 0.04 | 1.4 ± 0.1 | 5.1 ± 0.2 | 965 ± 58 |
| NPF-GAL4>CCAP-RRNAi | 19.5 ± 1.5* | 1.1 ± 0.11* | 64.5 ± 8.1 | 4.3 ± 0.3* | 177 ± 12* | 67.5 ± 4.8* | 1.2 ± 0.1 | 1.6 ± 0.1 | 4.8 ± 0.2 | 654 ± 46* |
For all assays, n is between 32 and 64 and a nonparametric Kruskal–Wallis ANOVA was performed with Dunn’s post hoc test for multiple comparisons. *P < 0.05, ***P < 0.005 compared with controls. FBD, feeding burst duration; SPB, sips per feeding burst.
Table 2.
flyPAD activity bout data in male flies starved for 18 h
| Genotype | Fed ad libitum | Starved 18 h | ||||
| AB | ABD | ABI | AB | ABD | ABI | |
| CCAPexc7>w1118 | 20.9 ± 1.3 | 3.1 ± 0.2 | 96.2 ± 21.5 | 62.8 ± 3.2 | 1.8 ± 0.1 | 151 ± 14.4 |
| CCAPexc7/CCAPexc7 | 10.9 1.2*** | 4.2 ± 0.2*** | 187 ± 21.5*** | 58.7 ± 2.4 | 1.2 ± 0.0*** | 242 ± 22.9*** |
| NPF-GAL4>w1118 | 11.4 ± 1.2 | 3.6 ± 0.3 | 56.1 ± 9.9 | 45.8 ± 3.1 | 2.4 ± 0.1 | 218 ± 24 |
| w1118>CCAP-RRNAi | 10.6 ± 1.0 | 3.5 ± 0.3 | 77.9 ± 10.5 | 50.7 ± 3.6 | 2.4 ± 0.2 | 235 ± 58 |
| NPF-GAL4>CCAP-RRNAi | 3.5 ± 0.9*** | 2.6 ± 0.1* | 76.8 ± 9.2 | 33.7 ± 2.6* | 2.5 ± 0.3 | 277 ± 43 |
For all assays, n is between 32 and 64 and a nonparametric Kruskal–Wallis ANOVA was performed with Dunn’s post hoc test for multiple comparisons. *P < 0.05, ***P < 0.005 compared with controls. ABD, activity bout duration.
To determine if the dorsal median P1 NPF neurons were sufficient to control food intake, UAS-CCAP-RRNAi was expressed in these neurons using R64F05-GAL4. No difference in feeding behavior was observed (SI Appendix, Fig. S6E). Next, we expressed the inward-rectifying channel Kir2.1 using either NPF-GAL4 or R64F05-GAL4, which should hyperpolarize the neurons. We then used the flyPAD to look at the feeding behavior of males starved for 18 h. We observed that while inhibiting all NPF neurons (NPF-GAL4) significantly decreased the number of sips (SI Appendix, Fig. S6F), inhibiting only the dorsal median NPF neurons (R64F05-GAL4) did not significantly change the feeding patterns (SI Appendix, Fig. S6G).
Since there was a stronger feeding phenotype when flies were starved, we decided to determine if loss of CCAP, or CCAP-R in NPF neurons, influenced metabolism. To do this, we first carried out a starvation assay. From this it was obvious that loss of CCAP-R in NPF neurons had no significant effect on starvation resistance. Yet, compared with control flies, CCAPexc7 null males were significantly more susceptible to starvation (Fig. 3D). Since both CCAPexc7 nulls and males where CCAP-R has been specifically knocked down in NPF neurons eat significantly less (Fig. 3A), the difference in starvation resistance must be due to another deficiency specific to CCAPexc7 nulls. To test this possibility, we measured triglyceride levels in adult males fed ad libitum. Interestingly, CCAPexc7 nulls had a significant decrease in triglyceride levels compared with all other flies tested (Fig. 3E).
CCAP Neurons Are Necessary and Sufficient to Induce PER Behavior.
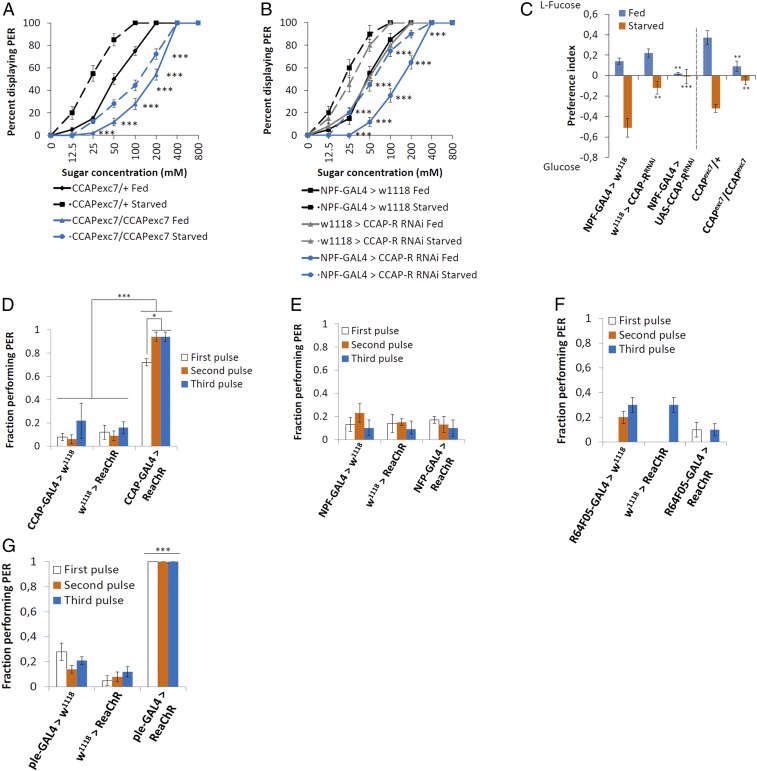
The proboscis extension reflex (PER) assay was used to test whether CCAP neurons, or its receptor (CCAP-R) in NPF neurons, were necessary for a proper behavioral response to a sugar cue. This was performed by observing if the proboscis extended in response to increasing concentrations of a sucrose solution, known as the proboscis extension reflex. In flies fed ad libitum, loss of CCAP significantly impaired the PER across a range of sucrose concentrations (Fig. 4A, solid blue line). Starvation significantly enhanced the PER to sucrose in control males, measured as a leftward shift of the PER vs. sucrose dose–response curve (Fig. 4A, solid vs. dashed black lines). Yet, in starved CCAPexc7 null males the increase in sucrose sensitivity was much less pronounced, although there was a shift toward higher sensitivity levels (Fig. 4A, dashed blue line). Knocking down CCAP-R in NPF neurons also impaired the PER across a range of sucrose concentrations before or after 18-h starvation (Fig. 4B, blue lines).
Fig. 4.
CCAP regulates PER behavior. (A and B) Average (±SEM) fraction of adult males 5 to 9 d old fed ad libitum (solid lines) and 24-h wet-starved flies (dashed lines) exhibiting a PER to the indicated concentrations of sucrose (n = 8 to 10, 10 to 20 flies per group). Asterisks denote statistically significant differences between starved flies and flies fed ad libitum (statistical methods can be found in SI Appendix, Materials and Methods; ***P < 0.005). (C) The flyPAD was used for a 2-choice food test between 100 mM l-fucose and 100 mM glucose in flies that were either fed ad libitum prior to testing (blue lines) or after 18-h starvation (orange lines). n is between 32 and 64 and a Kolmogorov–Smirnov test of normality was performed. Since some of the samples failed the test, a nonparametric Kruskal–Wallis ANOVA was performed with Dunn’s post hoc test for multiple comparisons (**P < 0.01, ***P < 0.005). (D–G) Optogenetic control of PER where the channelrhodopsin ReaChR is expressed in (D) CCAP neurons, (E) all NPF neurons using NPF-GAL4, (F) dorsal median NPF neurons using R64F05-GAL4, or (G) dopaminergic neurons (ple-GAL4). Fractions indicate the number of responders out of the number of flies tested. The photostimulation was performed at 620 nm, with 3 pulses at 1 Hz (100-ms pulse width) (n = 5 or 6 flies, 10 to 20 groups of flies for each genotype tested). Two different tests were performed: 1) Similar pulses were compared between various genetic backgrounds and 2) different pulses were compared within the same genetic background. In all instances, a separate Kolmogorov–Smirnov test of normality was performed for D–G, followed by a one-way ANOVA with Tukey’s post hoc test for multiple comparisons (*P < 0.05, ***P < 0.005). Error bars represent SEM.
We performed a 2-choice test between nutritive versus nonnutritive sugars to further understand if loss of CCAP impaired sugar sensitivity. Using the flyPAD, flies were given the choice between equal molar amounts of l-fucose, sweet but nonnutritive, or glucose, less sweet but nutritive. Satiated control flies preferred to eat the sweeter nonnutritive l-fucose, but switched to the less sweet, more nutritive glucose when they had been starved for 18 h (Fig. 4C). On the other hand, knocking down CCAP-R in NPF neurons and CCAPexc7 null males had no preference for either l-fucose or glucose in either a satiated or starved state (Fig. 4C).
To establish if CCAP signaling was sufficient to induce a PER, red-shifted channelrhodopsin (ReaChR) (36) was expressed specifically in CCAP neurons (CCAP-GAL4>UAS-ReaChR) and optogenetic analysis was performed to analyze the percentage of flies demonstrating a PER. The flies were given 10 pulses of light (620 nm) for 500 ms at 0.2 Hz; this process was repeated 3 times with a 15-s gap between pulse trains. Adult males expressing ReaChR in CCAP neurons yielded a robust PER on the first pulse train, which significantly increased on the second pulse train (Fig. 4D and Movie S4). We also noticed that optogenetic activation induced hyperactivity in the flies (seen as increased leg movements in the video).
Next, we expressed ReaChR in NPF neurons (NPF-GAL4>UAS-ReaChR) to test if they were also sufficient to induce a PER. No PER in response to optogenetic activation was observed (Fig. 4E and Movie S5). NPF-GAL4 is expressed in neurons besides the 2 dorsal median CCAP-R–expressing neurons, some of which could be inhibiting PER; therefore, we expressed ReaChR using the R64F05-GAL4 driver, which only expresses in the 2 dorsal median NPF neurons, and performed the assay again. As with the NPF-GAL4 driver, no PER in response to optogenetic activation was observed (Fig. 4F). Previously, it was shown that Gr5a gustatory neurons are sufficient to induce a PER in response to optogenetic activation (36). Since dopaminergic neurons are supposed to signal between NPF and Gr5a neurons (15), we tested if dopaminergic neurons would respond to optogenetic activation (ple-GAL4>UAS-ReaChR). Pale (ple) is the Drosophila homolog of tyrosine hydroxylase. Similar to CCAP neurons, these flies performed PER in response to optogenetic activation (Fig. 4G and Movie S6).
Discussion
Here we demonstrate that 2 residual CCAP neurons not only signal for food intake but also have a role in regulating metabolism, where they are important for maintaining triglyceride levels. CCAP signaling activates an NPF pathway for proper sensing of sugars for food intake. Of note, flies lacking CCAP, or CCAP-R in 2 dorsal median P1 NPF neurons, are not able to distinguish nutritive from nonnutritive sugar. On the other hand, CCAP signaling to these P1 NPF neurons is not sufficient for the NPF feeding phenotypes, as knocking down CCAP-R specifically in these neurons, or inhibiting these 2 neurons using the inward-rectifying channel Kir2.1, was not able to recapitulate the phenotypes. This hints at other CCAP- and NPF-regulated neurons being involved in the control of feeding behavior. One possibility is the peripheral L1-I NPF neurons, as they did react to starvation by increasing NPF protein levels and exhibited increased NPF protein levels when CCAP neurons were activated. When flies sense palatable food, a reflexive behavior known as the proboscis extension reflex is initiated. We show that CCAP is both necessary and sufficient to induce this reflex. Moreover, dopaminergic neurons are also sufficient to induce this response, but not NPF neurons (Fig. 4 E–G). Thus, we have identified CCAP as a possible key node in regulating feeding behavior.
Previously it was shown that under acute starvation one of the Drosophila NPY homologs, NPF, initiates a response that activates dopaminergic signaling, leading to the sensitization of gustatory neurons (Gr5a) toward sugar taste (15). We show that Drosophila CCAP regulates the activity of the 2 dorsal median P1 NPF neurons and that this is sufficient to control food intake. First, CCAP neurons project toward the 2 dorsal median P1 NPF neurons (Fig. 1 and Movie S1). Second, knocking down CCAP expression, or CCAP-R expression in the dorsal median P1 NPF neurons, increases NPF expression under ad libitum conditions (CCAP-R knockdown), as well as in response to starvation (both CCAP and CCAP-R) (Fig. 2 and SI Appendix, Fig. S3). It is interesting that loss of CCAP-R already significantly influences NPF expression under conditions where flies are fed ad libitum. Possibly, NPF is being released at low levels even under fed conditions. It is known that NPF also regulates sleep and the reward system (37, 38). CCAP may signal upstream of NPF to regulate sleep and reward as well. This possibility should be tested in the future. Furthermore, activating CCAP neurons using thermogenetics was sufficient to reduce NPF expression in the dorsal median P1 NPF neurons, indicating the neurons were more active (SI Appendix, Fig. S4). On the other hand, activating CCAP neurons increased NPF expression in the dorsal lateral L1-I neurons, indicating that activation of CCAP neurons inhibited these NPF-expressing neurons. This increase of NPF in the peripheral L1-I neurons was also observed when flies were starved. From these data, we conclude that activating CCAP neurons in turn activates 2 dorsal median NPF neurons, leading to sugar sensitization (Fig. 4H) (15). Our inability to recapitulate the CCAPexc7 feeding phenotypes by knocking down CCAP-R expression specifically in the dorsal median P1 NPF neurons, or inhibiting these 2 neurons using the inward-rectifying channel Kir2.1, may indicate that the peripheral L1-I NPF neurons also play an important role in regulating food intake. More work is needed to understand CCAP’s possible regulation of the peripheral neurons, as loss of CCAP had no influence on NPF protein levels in these neurons.
We found that CCAP neurons were not only necessary, but also sufficient, to induce the PER. Previously, by the use of optogenetics, it was determined that Gr5a neurons were sufficient to induce the PER (36). On the other hand, although NPF neurons and dopaminergic neurons were determined to be necessary for a proper PER when flies were presented with varying concentrations of sugar, it was not established whether they were sufficient (15). Using optogenetics, we determined that while dopaminergic neurons (ple-GAL4) were able to induce a PER, NPF neurons (NPF-GAL4 or R64F05-GAL4) were not sufficient (Fig. 4 E and F).
Interestingly, adult flies lacking the CCAP peptide had significantly lower triglyceride levels. This was not observed when CCAP-R was specifically knocked down in NPF neurons (Fig. 3 D and E). In order to maintain homeostasis, the brain must process extrinsic and intrinsic information. In Drosophila, different peptides have been shown to regulate these signals, such as diuretic hormone 44 (Dh44) (39, 40), corazonin (Crz) (41), allatostatin A (AstA) (42, 43), and SIFamide (SIFa) (44). Furthermore, similar to mammals, insulin-like peptides and a glucagon-like hormone (AKH) are also involved in regulating feeding behavior (45, 46). Interestingly, Dh44 was shown to be necessary for the fly to sense postprandial nutritional information (40), and in our study we showed that flies lacking either CCAP or CCAP-R in NPF neurons were unable to determine between nutritional and nonnutritional sugars (Fig. 4C). That said, it must be mentioned this experiment only lasted 1 h and longer times may be necessary to truly understand if CCAP is involved in regulating postprandial nutritional signals. Another possibility is that CCAP neurons regulate Crz signaling. Crz is a Drosophila peptide related to mammalian gonadotropin-releasing hormone (47–49). Activation of Crz-expressing neurons was reported to reduce triglyceride levels, while loss of Crz regulation of insulin-producing cells leads to increased triglyceride storage, suggesting that Crz signals to decrease energy reserves (41, 48). It is possible that CCAP signals to inhibit Crz in order to control the decrease in energy reserves under starvation conditions. Furthermore, loss or activation of SIFa neurons in adult flies produced feeding phenotypes very similar to when CCAP signaling is inhibited or activated, meaning there could be an interaction between SIFa and CCAP as well (44).
In summary, our experiments identify 2 CCAP peptidergic neurons as being required to induce feeding behavior via the NPF pathway in adult Drosophila. We suggest that CCAP-expressing neurons regulate feeding behavior and are necessary for the proper sensing of sugars, while also regulating triglyceride levels. Continued studies of these 2 CCAP neurons, their neuronal network, as well as how they regulate feeding behavior and metabolism may help in our understanding of satiety control and how peripheral physiological signals are translated into behavioral changes by the brain.
Materials and Methods
Fly Stocks and Maintenance.
All flies, unless otherwise stated, were maintained on enriched Jazz mix standard fly food (Fisher Scientific) supplemented with yeast extract (VWR). Flies were maintained at 25 °C in an incubator at 60% humidity on a 12-h:12-h light:dark cycle. Flies crossed to GAL4 drivers and controls were raised at 18 °C until the adults emerged. Once collected, adults were raised at either 18 or 29 °C, depending on the experiment, for the appropriate times. All flies were crossed into the same w1118 background. In all assays, the GAL4 drivers and UAS transgenic flies were crossed to w1118 flies and their F1 progeny were used as controls. All fly lines are described in SI Appendix, Materials and Methods.
Immunohistochemistry.
Immunohistochemistry of adult male brains was performed in a similar manner as ref. 50. Detailed information and sources of antibodies and dilutions are described in SI Appendix, Materials and Methods.
RNA Purification, cDNA Synthesis, and qRT-PCR.
RNA purification, complementary DNA (cDNA) synthesis, and qRT-PCR were performed as in ref. 50. Detailed information and primer sequences are described in SI Appendix, Materials and Methods.
Capillary Feeding.
This method was modified from Ja et al. (34). Detailed information is described in SI Appendix, Materials and Methods.
flyPAD Feeding Assays.
Feeding experiments were performed in a similar fashion as Itskov et al. (35). For feeding behavior experiments, 5- to 9-d-old male flies were either fed ad libitum on normal laboratory food or wet-starved for 18 h. Flies were individually transferred to the flyPAD behavioral arena by mouth aspiration and left to feed on 4 μL 150 mM sucrose (Sigma-Aldrich) in 0.9% agarose (Sigma-Aldrich) for 1 h. Throughout the duration of the experiments the flies were maintained at 55 to 60% humidity and 21 to 22 °C, unless otherwise stated.
Preference Testing between 2 Different Sugar Sources.
The feeding experiments for preference testing were performed using the flyPAD (35). The experimental conditions through the duration of the experiments were 55 to 60% humidity and 21 to 22 °C, unless otherwise stated. For the preference feeding behavior experiments, 5- to 7-d-old male flies were either fed on normal laboratory food or wet-starved for 18 h prior to the experiment. The male progeny from the experimental flies, as well as the control flies, were placed at 29 °C once they were collected. Each independent run was performed so that the experimental flies were always tested together with the control groups. Flies were transferred to the flyPAD behavioral arena and allowed to choose freely between 100 mM glucose and 100 mM l-fucose (Sigma-Aldrich) solutions that were dissolved in 1% agarose (Sigma-Aldrich). The flies were left to eat ad libitum for 1 h in the flyPAD, having been provided with 4 μL both 100 mM glucose and 100 mM l-fucose.
Starvation Assay.
The starvation resistance assay was performed in a similar fashion as Hergarden et al. (43).
Starvation resistance was measured by placing 32 (5- to 7-d-old) male flies in individual tubes containing 1% agarose in the Drosophila Activity Monitoring System (DAMS) (TriKinetics). The DAMS assay was performed at 25 °C in an incubator, on a 12-h:12-h light:dark cycle.
Triacylglycerol Determination.
Flies (25 males) were homogenized with 100 μL PBST buffer (1× phosphate-buffered saline with 10% Tween 20), incubated at 70 °C for 5 min, and then centrifuged at maximum speed for 10 min. The supernatant was transferred to a new 1.5-mL microcentrifuge tube and used as samples. Glycerol was used to generate a standard curve with concentrations of 1.0, 0.8, 0.6, 0.4, and 0.2 mg/mL equivalent triolein concentration. Free glycerol reagent (100 μL) was added to 10 μL blank (PBST), standards, or samples and initial absorbance at 540 nm was measured after incubation at 37 °C for 15 min. The concentration of free glycerol in the samples was calculated from the standard curve generated by this initial absorbance value. Then, 20 μL triglyceride reagent was added to each standard and sample and incubated at 37 °C for 15 min. The final absorbance was taken at 540 nm to calculate the triglyceride concentration from the generated standard curve. The protein concentration of each sample was measured by the Bio-Rad Protein Assay Kit. The concentrations of free glycerides and triglycerides in samples (mg/mg of protein) were calculated from 5 replicates (50).
Proboscis Extension Assay.
For the PER assays (51), sucrose solutions were presented to each fly once in order of increasing molarity. Any flies responding to an initial water stimulus were discarded. For a more detailed protocol, including statistical methods, see SI Appendix, Materials and Methods. Optogenetic experiments to activate the PER were performed similar to Inagaki et al. (36). Flies were fed ad libitum on normal laboratory fly food or wet-starved for 18 h before being mounted 5 flies per glass slide using clear tape. Flies were then placed above a high-power LED (light-emitting diode) array (Cree) and PERs were monitored using an ACA640-750UM video camera (Basler). Bouts of PER were counted manually by repeatedly watching film. A bout was defined as beginning when flies start extending their proboscis and ending when they retract the proboscis. Incomplete proboscis extensions were not counted. For CCAP-GAL4, 10 pulses (1 pulse train) of 500-ms photostimulation at 0.2 Hz were delivered. Three trials were performed with a 15-s gap between pulse trains, and flies showing more than 1 PER during this activation period were counted as responders.
Statistical Analysis.
SEM from all replicates of each experiment was calculated. All analysis was performed with GraphPad Prism 4, and used ANOVA with appropriate post hoc analysis for multiple comparisons. Normal distribution was performed using the Kolmogorov–Smirnov test of normality. Samples not passing the Kolmogorov–Smirnov test of normality were analyzed using the Mann–Whitney U test calculator. The type of analysis performed for each assay is specified in the appropriate figure legend.
Data Availability.
We confirm that the data supporting the findings of this study are available within the article and/or SI Appendix.
Supplementary Material
Acknowledgments
We thank Professor John Ewer for his gracious gift of CCAP mutant flies, Professor Fernando Jiménez Díaz-Benjumea for his kind gift of CCAP antibody, and Dr. Pavel Itskov for helping to set up the flyPAD system in our lab. This work was supported by the Swedish Research Council (Vetenskapsrådet), Brain Foundation (Hjärnfonden), Åhlén-Stiftelsen, and Carl Tryggers Stiftelsen.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1914037117/-/DCSupplemental.
References
- 1.Zoungas S. et al.; ADVANCE Collaborative Group , Severe hypoglycemia and risks of vascular events and death. N. Engl. J. Med. 363, 1410–1418 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Crook M. A., Hypoglycemia, hypotriglyceridemia and starvation associated with cardiogenic shock. Nutrition 30, 1093–1094 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Bellou E., et al. , Effect of acute negative and positive energy balance on basal very-low density lipoprotein triglyceride metabolism in women. PLoS One 8, e60251 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Raalte D. H., Verchere C. B., Improving glycaemic control in type 2 diabetes: Stimulate insulin secretion or provide beta-cell rest? Diabetes Obes. Metab. 19, 1205–1213 (2017). [DOI] [PubMed] [Google Scholar]
- 5.da Rosa Lima T., et al. , Effect of administration of high-protein diet in rats submitted to resistance training. Eur. J. Nutr. 57, 1083–1096 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Devassy J. G., et al. , Mixed compared with single-source proteins in high-protein diets affect kidney structure and function differentially in obese fa/fa Zucker rats. Appl. Physiol. Nutr. Metab. 42, 135–141 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Herieka M., Erridge C., High-fat meal induced postprandial inflammation. Mol. Nutr. Food Res. 58, 136–146 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Stanhope K. L., Sugar consumption, metabolic disease and obesity: The state of the controversy. Crit. Rev. Clin. Lab. Sci. 53, 52–67 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keeley R. J., Zelinski E. L., Fehr L., McDonald R. J., The effect of exercise on carbohydrate preference in female rats. Brain Res. Bull. 101, 45–50 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Buff C. E., Shay N. F., Beverly J. L., Noradrenergic activity in the paraventricular nucleus and macronutrient choice during early dark onset by zinc deficient rats. Nutr. Neurosci. 9, 189–193 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Smith B. K., Berthoud H. R., York D. A., Bray G. A., Differential effects of baseline macronutrient preferences on macronutrient selection after galanin, NPY, and an overnight fast. Peptides 18, 207–211 (1997). [DOI] [PubMed] [Google Scholar]
- 12.Tanaka M., Kido Y., Serotonergic regulation of galanin-induced selective macronutrient intake in self-selecting rats. J. Med. Invest. 55, 196–203 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Kyrkouli S. E., Strubbe J. H., Scheurink A. J., Galanin in the PVN increases nutrient intake and changes peripheral hormone levels in the rat. Physiol. Behav. 89, 103–109 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Leibowitz S. F., Regulation and effects of hypothalamic galanin: Relation to dietary fat, alcohol ingestion, circulating lipids and energy homeostasis. Neuropeptides 39, 327–332 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Inagaki H. K., Panse K. M., Anderson D. J., Independent, reciprocal neuromodulatory control of sweet and bitter taste sensitivity during starvation in Drosophila. Neuron 84, 806–820 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park J. H., Kwon J. Y., Heterogeneous expression of Drosophila gustatory receptors in enteroendocrine cells. PLoS One 6, e29022 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thorne N., Bray S., Amrein H., Function and expression of the Drosophila Gr genes in the perception of sweet, bitter and pheromone compounds. Chem. Senses 30 (suppl. 1), i270–i272 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Hiroi M., Meunier N., Marion-Poll F., Tanimura T., Two antagonistic gustatory receptor neurons responding to sweet-salty and bitter taste in Drosophila. J. Neurobiol. 61, 333–342 (2004). [DOI] [PubMed] [Google Scholar]
- 19.Thorne N., Chromey C., Bray S., Amrein H., Taste perception and coding in Drosophila. Curr. Biol. 14, 1065–1079 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Ishimoto H., Tanimura T., Molecular neurophysiology of taste in Drosophila. Cell. Mol. Life Sci. 61, 10–18 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lingo P. R., Zhao Z., Shen P., Co-regulation of cold-resistant food acquisition by insulin- and neuropeptide Y-like systems in Drosophila melanogaster. Neuroscience 148, 371–374 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lahr E. C., Dean D., Ewer J., Genetic analysis of ecdysis behavior in Drosophila reveals partially overlapping functions of two unrelated neuropeptides. J. Neurosci. 32, 6819–6829 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clark A. C., del Campo M. L., Ewer J., Neuroendocrine control of larval ecdysis behavior in Drosophila: Complex regulation by partially redundant neuropeptides. J. Neurosci. 24, 4283–4292 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park J. H., Schroeder A. J., Helfrich-Förster C., Jackson F. R., Ewer J., Targeted ablation of CCAP neuropeptide-containing neurons of Drosophila causes specific defects in execution and circadian timing of ecdysis behavior. Development 130, 2645–2656 (2003). [DOI] [PubMed] [Google Scholar]
- 25.Baker J. D., McNabb S. L., Truman J. W., The hormonal coordination of behavior and physiology at adult ecdysis in Drosophila melanogaster. J. Exp. Biol. 202, 3037–3048 (1999). [DOI] [PubMed] [Google Scholar]
- 26.Lee G. G., Kikuno K., Nair S., Park J. H., Mechanisms of postecdysis-associated programmed cell death of peptidergic neurons in Drosophila melanogaster. J. Comp. Neurol. 521, 3972–3991 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Sakai T., Satake H., Takeda M., Nutrient-induced alpha-amylase and protease activity is regulated by crustacean cardioactive peptide (CCAP) in the cockroach midgut. Peptides 27, 2157–2164 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Mikani A., Watari Y., Takeda M., Brain-midgut cross-talk and autocrine metabolastat via the sNPF/CCAP negative feed-back loop in the American cockroach, Periplaneta americana. Cell Tissue Res. 362, 481–496 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Tokusumi T., et al. , Screening and analysis of Janelia FlyLight Project enhancer-Gal4 strains identifies multiple gene enhancers active during hematopoiesis in normal and wasp-challenged Drosophila larvae. G3 (Bethesda) 7, 437–448 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee G., Bahn J. H., Park J. H., Sex- and clock-controlled expression of the neuropeptide F gene in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 103, 12580–12585 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moris-Sanz M., Estacio-Gómez A., Alvarez-Rivero J., Díaz-Benjumea F. J., Specification of neuronal subtypes by different levels of Hunchback. Development 141, 4366–4374 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Wu Q., et al. , Developmental control of foraging and social behavior by the Drosophila neuropeptide Y-like system. Neuron 39, 147–161 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Sokabe T., Tsujiuchi S., Kadowaki T., Tominaga M., Drosophila painless is a Ca2+-requiring channel activated by noxious heat. J. Neurosci. 28, 9929–9938 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ja W. W., et al. , Prandiology of Drosophila and the CAFE assay. Proc. Natl. Acad. Sci. U.S.A. 104, 8253–8256 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Itskov P. M., et al. , Automated monitoring and quantitative analysis of feeding behaviour in Drosophila. Nat. Commun. 5, 4560 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Inagaki H. K., et al. , Optogenetic control of Drosophila using a red-shifted channelrhodopsin reveals experience-dependent influences on courtship. Nat. Methods 11, 325–332 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chung B. Y., et al. , Drosophila neuropeptide F signaling independently regulates feeding and sleep-wake behavior. Cell Rep. 19, 2441–2450 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pu Y., Zhang Y., Zhang Y., Shen P., Two Drosophila neuropeptide Y-like neurons define a reward module for transforming appetitive odor representations to motivation. Sci. Rep. 8, 11658 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cannell E., et al. , The corticotropin-releasing factor-like diuretic hormone 44 (DH44) and kinin neuropeptides modulate desiccation and starvation tolerance in Drosophila melanogaster. Peptides 80, 96–107 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang Z., et al. , A post-ingestive amino acid sensor promotes food consumption in Drosophila. Cell Res. 28, 1013–1025 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kapan N., Lushchak O. V., Luo J., Nässel D. R., Identified peptidergic neurons in the Drosophila brain regulate insulin-producing cells, stress responses and metabolism by coexpressed short neuropeptide F and corazonin. Cell. Mol. Life Sci. 69, 4051–4066 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hentze J. L., Carlsson M. A., Kondo S., Nässel D. R., Rewitz K. F., The neuropeptide allatostatin A regulates metabolism and feeding decisions in Drosophila. Sci. Rep. 5, 11680 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hergarden A. C., Tayler T. D., Anderson D. J., Allatostatin-A neurons inhibit feeding behavior in adult Drosophila. Proc. Natl. Acad. Sci. U.S.A. 109, 3967–3972 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martelli C., et al. , SIFamide translates hunger signals into appetitive and feeding behavior in Drosophila. Cell Rep. 20, 464–478 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Broughton S. J., et al. , DILP-producing median neurosecretory cells in the Drosophila brain mediate the response of lifespan to nutrition. Aging Cell 9, 336–346 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee G., Park J. H., Hemolymph sugar homeostasis and starvation-induced hyperactivity affected by genetic manipulations of the adipokinetic hormone-encoding gene in Drosophila melanogaster. Genetics 167, 311–323 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Veenstra J. A., Isolation and structure of the Drosophila corazonin gene. Biochem. Biophys. Res. Commun. 204, 292–296 (1994). [DOI] [PubMed] [Google Scholar]
- 48.Zhao Y., Bretz C. A., Hawksworth S. A., Hirsh J., Johnson E. C., Corazonin neurons function in sexually dimorphic circuitry that shape behavioral responses to stress in Drosophila. PLoS One 5, e9141 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kubrak O. I., Lushchak O. V., Zandawala M., Nässel D. R., Systemic corazonin signalling modulates stress responses and metabolism in Drosophila. Open Biol. 6, 160152 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Williams M. J., et al. , The Drosophila ETV5 homologue Ets96B: Molecular link between obesity and bipolar disorder. PLoS Genet. 12, e1006104 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Inagaki H. K., et al. , Visualizing neuromodulation in vivo: TANGO-mapping of dopamine signaling reveals appetite control of sugar sensing. Cell 148, 583–595 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We confirm that the data supporting the findings of this study are available within the article and/or SI Appendix.