Significance
Cancer chemotherapy often causes side effects that require modulations in dosing, which then reduce anticancer efficacy. Here, we show that targeted inhibition of gut bacterial enzymes alleviates key stages of gut epithelial damage caused by the cancer drug irinotecan, blunts stark gut microbial compositional shifts caused by irinotecan, and enhances irinotecan’s antitumor effectiveness by reducing its gastrointestinal toxicity.
Keywords: microbiome, chemotherapy, cancer, gastrointestinal toxicity
Abstract
Irinotecan treats a range of solid tumors, but its effectiveness is severely limited by gastrointestinal (GI) tract toxicity caused by gut bacterial β-glucuronidase (GUS) enzymes. Targeted bacterial GUS inhibitors have been shown to partially alleviate irinotecan-induced GI tract damage and resultant diarrhea in mice. Here, we unravel the mechanistic basis for GI protection by gut microbial GUS inhibitors using in vivo models. We use in vitro, in fimo, and in vivo models to determine whether GUS inhibition alters the anticancer efficacy of irinotecan. We demonstrate that a single dose of irinotecan increases GI bacterial GUS activity in 1 d and reduces intestinal epithelial cell proliferation in 5 d, both blocked by a single dose of a GUS inhibitor. In a tumor xenograft model, GUS inhibition prevents intestinal toxicity and maintains the antitumor efficacy of irinotecan. Remarkably, GUS inhibitor also effectively blocks the striking irinotecan-induced bloom of Enterobacteriaceae in immune-deficient mice. In a genetically engineered mouse model of cancer, GUS inhibition alleviates gut damage, improves survival, and does not alter gut microbial composition; however, by allowing dose intensification, it dramatically improves irinotecan’s effectiveness, reducing tumors to a fraction of that achieved by irinotecan alone, while simultaneously promoting epithelial regeneration. These results indicate that targeted gut microbial enzyme inhibitors can improve cancer chemotherapeutic outcomes by protecting the gut epithelium from microbial dysbiosis and proliferative crypt damage.
Irinotecan (CPT-11) is essential for treating colorectal and pancreatic cancers and is frequently administered as a mixture with 5-fluorouracil and/or oxaliplatin. Ongoing clinical trials seek to extend irinotecan to other forms of cancer. However, irinotecan causes both myelosuppression and gastrointestinal (GI) side effects, including mucositis and late-onset diarrhea, which are often dose limiting: 88% of irinotecan patients develop diarrhea, with 30% experiencing acute grade 3 to 4 diarrhea (1–3). This toxicity is frequently refractory to antimotility drugs. Thus, treating irinotecan-induced diarrhea is a significant clinical need (4, 5).
The irinotecan prodrug is activated in vivo to SN38, a potent topoisomerase I inhibitor (6) that retards the growth of rapidly proliferating cells in tumors and the intestinal epithelium, which renews every 5 d. SN38 is detoxified through the addition of glucuronic acid (GlcA) to form SN38-G, a compound marked for elimination via the GI tract. Gut commensal bacterial β-glucuronidase (GUS) proteins remove GlcA from SN38-G. Reactivated SN38 inflicts epithelial damage, resulting in bleeding diarrhea and acute weight loss in animal models (7). We reduced irinotecan-induced gut toxicity using inhibitors that selectively, nonlethally, and potently block the actions of gut bacterial GUS enzymes (8, 9). The utility of GUS inhibition also extends to drugs beyond irinotecan (10–12).
Here, we probe three critical topics related to the in vivo use of GUS inhibitors for irinotecan-induced gut toxicity. First, we examined how quickly irinotecan toxicity appears in the murine GI tract and establish that a single dose of GUS inhibitor can alleviate early gut epithelial damage. Second, we assessed the degree to which GUS inhibition enhances irinotecan’s antitumor effectiveness using two mouse models of cancer. Third, we determined how irinotecan and GUS inhibitor impact murine gut microbiota composition. Together, the data presented provide key mechanistic insights into the abilities of gut bacterial enzyme inhibitors to alleviate drug-induced GI damage and to improve the effectiveness of this widely used cancer chemotherapeutic.
Results
GUS Loop Architecture Dictates SN38-G Processing Efficiency and Inhibition.
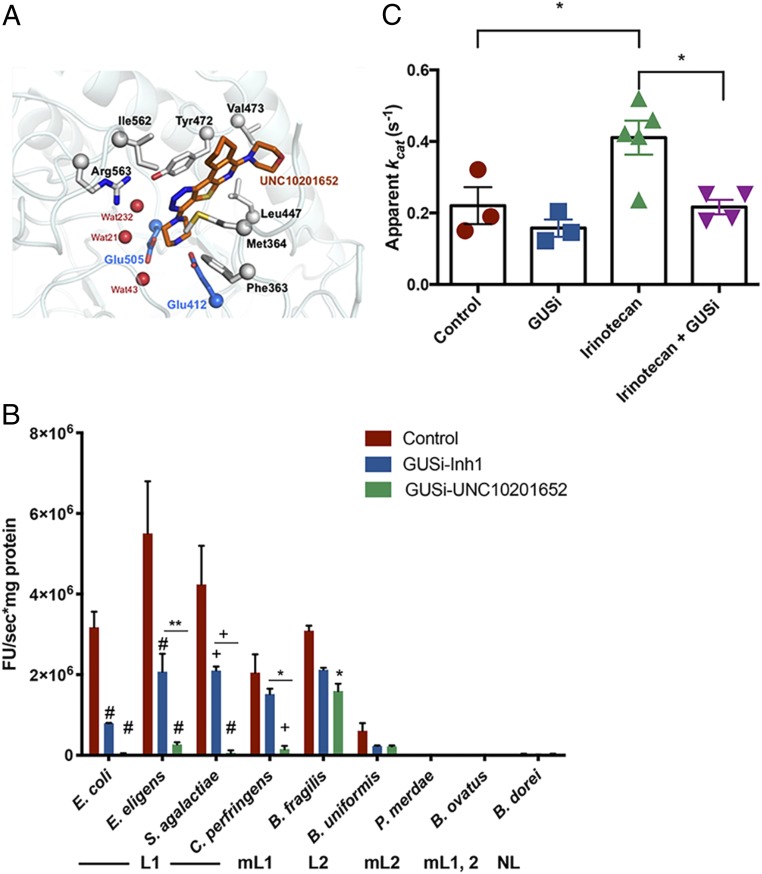
The hundreds of unique gut microbial GUS enzymes mapped to date have been categorized into six functionally distinct groups based on active site architecture (13–20). GUSs are encoded by Bacteroidetes, Firmicutes, and Proteobacteria; among these phyla, Proteobacteria are unique in encoding only Loop 1 (L1) GUS orthologs, which are most efficient with smaller glucuronidated substrates (14). We examined the SN38-G cleavage abilities of a representative in vitro panel of human gut GUS enzymes from these groups (e.g., L1, Mini-Loop 1 [mL1], Loop 2 [L2], etc.) and examined inhibition by two chemotypes: Inhibitor 1 (SI Appendix, Fig. S1A), used in previous studies (8–11, 21, 22), and UNC10201652 (SI Appendix, Fig. S1B), a distinct mechanism-based GUS inhibitor that uniquely forms a covalent inhibitor–glucuronide conjugate at the enzyme’s active site (17). While UNC10201652 has been examined in vitro (17), the in vivo function is studied here. We found that L1 GUS enzymes process SN38-G most efficiently and that the mL1 and L2 enzymes exhibit modest activity, while the remainder of the enzymes showed no activity (Fig. 1A). In terms of inhibition, UNC10201652 was found to be superior to inhibitor 1 (Inh1) in inhibiting GUS-mediated reactivation of SN38-G in vitro, exhibiting stronger potency and broader efficacy against L1 enzymes (Fig. 1A). The crystal structure of UNC10201652 in complex with the L1 GUS from Clostridium perfringens (CpGUS) reveals that this more potent inhibitor makes numerous favorable interactions at the L1 enzyme active site, likely explaining its potency (Fig. 1B and SI Appendix, Fig. S1 C and D). Thus, L1 GUS enzymes most efficiently reactivate SN-38G. UNC10201652 precisely targets gut bacterial L1 GUS orthologs, potently blocking in vitro SN38-G reactivation by these enzymes.
Fig. 1.
Gut microbial GUS structure and function in vitro and in fimo. (A) Active site interactions in the UNC10201652-bound CpGUS. UNC10201652 (orange), water (red), and catalytic residues (blue; Glu505 and Glu412) are shown. (B) In vitro processing of SN38-G by gut microbial GUS enzymes and variable enzymatic inhibition by GUS inhibitors. Error bars are ± SEM. mL1,2, Mini-Loop 1,2; mL2, Mini-Loop 2; NL, no loop. *P < 0.05 by two-way ANOVA with Tukey’s multiple comparisons test; **P < 0.01 by two-way ANOVA with Tukey’s multiple comparisons test; +P < 0.0005 by two-way ANOVA with Tukey’s multiple comparisons test; #P < 0.0001 by two-way ANOVA with Tukey’s multiple comparisons test. (C) Elevated in fimo GUS activity in animals pretreated with irinotecan 24 h prior. Cotreatment with GUSi–UNC10201652 blunts the increase. Error bars are ± SEM. *P < 0.05 by one-way ANOVA with Sidak’s correction for multiple comparisons. E. eligens, Eubacterium eligens; S. agalactiae, Streptococcus agalactiae; B. fragilis, Bacteriodes fragilis; B. uniformis, Bacteriodes uniformis; P. merdae, Parabacteroides merdae; B. ovatus, Bacteriodes ovatus; B. dorei, Bacteriodes dorei.
SN38-G Processing and Inhibition In Fimo.
We recently proposed “in fimo” to describe the experimental examination of stool samples derived from the formal Latin “fimus” for “excrement” (23). In fimo processing of SN38-G occurs in luminal contents of the cecum and proximal and distal colons of irinotecan-treated FVB mice (SI Appendix, Fig. S2 A–C). UNC10201652 was more efficacious than Inh1 at blocking GUS activity in fimo and was thus chosen for further experiments (SI Appendix, Fig. S2 D–F). FVB mice treated with irinotecan, UNC10201652 (hereafter termed “GUSi” [β-glucuronidase inhibitor]), or both irinotecan and GUSi were examined for SN38-G reactivation in fimo after 24 h. A single intraperitoneal (i.p.) injection of irinotecan (50 mg/kg) significantly increased SN38-G processing activity in the cecum (Fig. 1C). While 1 mg/kg GUSi delivered orally (per oral [p.o.]) did not alone alter in fimo SN38-G reactivation, single doses of GUSi and irinotecan together significantly reduced in fimo SN38-G reactivation to levels seen in naïve mice (Fig. 1C). Thus, one irinotecan dose increases in fimo SN38-G reactivation within 24 h, an effect that is blunted by a single dose of GUSi.
GUS Inhibition Protects against Gut Epithelial Damage.
It has previously been established that inhibiting bacterial GUSs does not alter the pharmacokinetics of irinotecan or its key metabolites in mouse serum (9). To determine whether an L1 GUS enzyme (e.g., Escherichia coli GUS) can drive irinotecan-induced gut damage, we monoassociated germ-free mice with either wild-type (WT) E. coli or the isogenic gus gene deletion strain (ΔGUS) (13). One irinotecan dose (50 mg/kg i.p.) increased in fimo GUS activity in WT E. coli-colonized mice but not in ΔGUS E. coli-colonized animals (SI Appendix, Fig. S3 A–C). With repeated irinotecan dosing, WT E. coli-colonized mice had significantly higher levels of fecal lipocalin-2, indicative of gut damage and inflammation, than ΔGUS E. coli-colonized mice (SI Appendix, Fig. S3 D and E). Furthermore, WT E. coli monoassociated mice exhibited marked reduction in intestinal epithelial cell proliferation compared with mice colonized with ΔGUS E. coli (SI Appendix, Fig. S3F). Thus, an L1 gut microbial GUS enzyme is sufficient to drive irinotecan-induced GI tract damage and reduced intestinal epithelial proliferation.
We next validated the ability of orally dosed GUSi to reach the GI tract by treating mice with a single oral dose of 1 mg/kg GUSi and collecting short-term (60, 90, 180 min) (SI Appendix, Fig. S4 A–C) or longer-term (8, 24 h) (SI Appendix, Fig. S4 D and E) cecal and colon contents. GUSi was detected in all intestinal compartments: in the small intestine within 60 min and in the cecum and colon within 180 min. Between 8 and 24 h, cecal levels remained constant, while colonic levels increased by fivefold. Thus, orally dosed GUSi reaches the mouse GI tract.
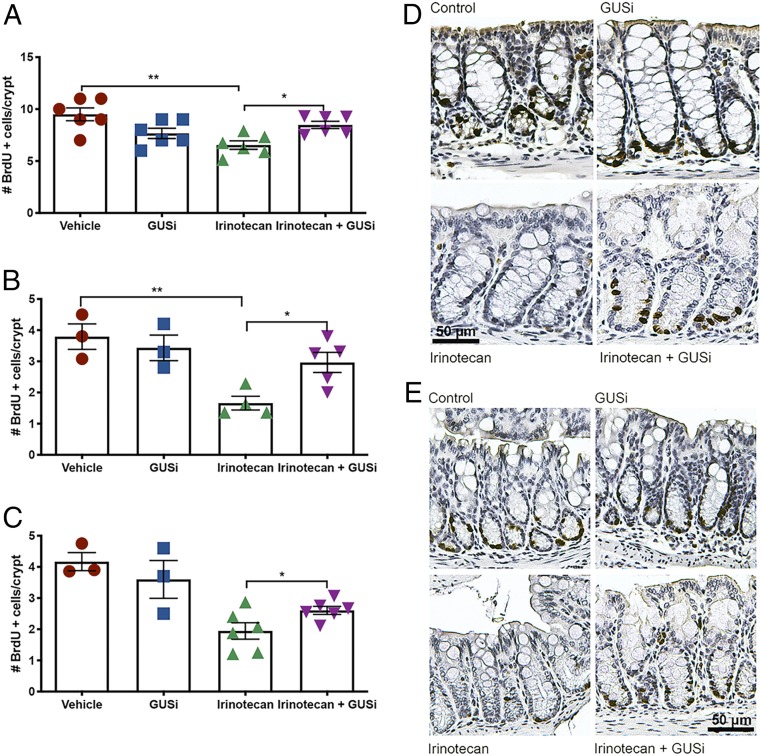
We undertook time course studies in two cohorts containing four groups of age-, weight-, and litter-matched female FVB mice (to match the transgenic breast cancer model used below) treated with a single dose of vehicle, GUSi (1 mg/kg p.o.), irinotecan (50 mg/kg i.p.), or irinotecan plus GUSi. One cohort was euthanized after 24 h, and the second was euthanized after 120 h; both were injected with 5-bromo-2'-deoxyuridine (BrdU) 30 min prior to euthanasia to label proliferating cells. Proliferation was assessed using immunohistochemical detection of BrdU; 24 h after treatment, no differences were observed between treatment groups (SI Appendix, Fig. S5). In contrast, the 120-h data revealed that a single dose of irinotecan significantly decreased levels of proliferative (BrdU+) cells per crypt in the ileum (Fig. 2A), proximal colon (Fig. 2 B and D), and distal colon (Fig. 2 C and E), effects that were blocked by a single concomitant dose of GUSi (Fig. 2). Thus, GUSi prevents irinotecan’s gut damage 5 d after injection. These data help to establish a timeline for gut damage caused by irinotecan—it robustly appears after 5 d, in concordance with the diarrhea phenotype in mice. Together with in fimo data, these in vivo results demonstrate that GUSi blocks two irinotecan-induced effects in the mouse GI tract—increase in gut bacterial GUS activity within 1 d and reduction in GI epithelial cell proliferation within 5 d.
Fig. 2.
GUSi reduces the acute toxicity exerted by irinotecan treatment. Numbers of BrdU+ cells in 10 consecutive crypts were blindly quantified in the (A) ileum, (B) proximal colon, and (C) distal colon. Error bars are ± SEM. *P < 0.05 by one-way ANOVA with Sidak’s multiple comparisons test; **P < 0.01 by one-way ANOVA with Sidak’s multiple comparisons test. Immunohistochemistry to detect BrdU+ cells (brown) in (D) proximal (quantified in B) and (E) distal (quantified in C) halves of colons of mice treated as indicated; nuclei are counterstained with hematoxylin (blue). (Scale bar, 50 μm.)
GUS Inhibition Alleviates Irinotecan-Induced Gut Toxicity in Tumor Xenograft Mice.
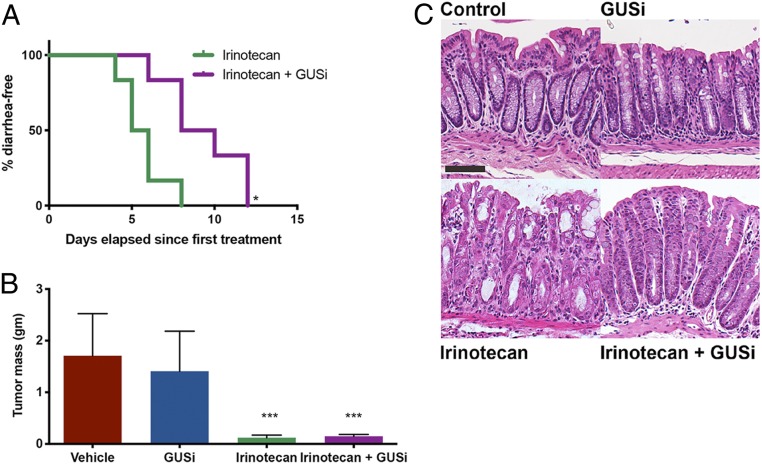
GUSi’s effects on irinotecan’s GI damage and antineoplastic efficacy were tested in vivo using a preclinical model of inflammatory breast cancer. Irinotecan formulations have been examined in mouse models of breast cancer (24), and human clinical trials for breast cancer have used SN38 combination therapies (25). Furthermore, breast cancer models were chosen to avoid confounding effects of microbial dysbiosis caused by the inflammatory tumor microenvironment of colorectal cancer (26, 27). SUM149 cells, representative of triple-negative breast cancer (ER−, PR−, Her2−, BRCA1mut, basal like), readily form tumors when subcutaneous injected in immune-deficient athymic nude mice (28). After palpable tumors were detected, mice were randomized into four groups: control, GUSi (1 mg/kg total delivered via twice daily p.o. administrations), daily injections (all days including weekends) of 50 mg/kg irinotecan i.p., and irinotecan + GUSi. Half of the mice receiving irinotecan alone developed diarrhea in 5 d, and all mice developed diarrhea by day 8; however, significantly fewer animals treated with both irinotecan and GUSi developed diarrhea (Fig. 3A). Ten days following the first treatment, the entire irinotecan-treated cohort required euthanasia due to weight loss; in contrast, mice in the irinotecan + GUSi group were protected against irinotecan-induced weight loss and trended toward living longer with irinotecan (SI Appendix, Figs. S6A and S7). Thus, GUSi alleviates irinotecan-induced diarrhea and weight loss in this xenograft model. Both irinotecan and irinotecan + GUSi cohorts bore significantly reduced tumor volumes and terminal tumor masses compared with vehicle and GUSi cohorts (Fig. 3B and SI Appendix, Fig. S6B). Histological examination of fixed colon tissues revealed greater inflammation and crypt damage in the irinotecan treatment group compared with irinotecan + GUSi or controls (Fig. 3C). Therefore, GUSi affords strong protection against irinotecan-induced gut epithelial cell damage in immune-deficient mice.
Fig. 3.
GUSi maintains irinotecan efficacy in the tumor xenograft model. (A) Kaplan–Meier analysis of animals remaining diarrhea free with GUSi cotreatment with irinotecan. *P < 0.05 by log-rank (Mantel–Cox) test. (B) GUSi cotreatment does not impede the antitumor efficacy of irinotecan. ***P < 0.001 by one-way ANOVA (Sidak multiple comparison test). (C) Early epithelial erosions, loss of goblet cells, dysplastic crypts of Lieberkühn, and increased inflammatory infiltrates in colons of irinotecan-treated mice; GUSi cotreatment preserves intestinal architecture. (Scale bar, 37.5 μm.)
GUS Inhibition Dramatically Improves Irinotecan Efficacy in Tumor-Bearing Genetically Engineered Mouse Model Animals.
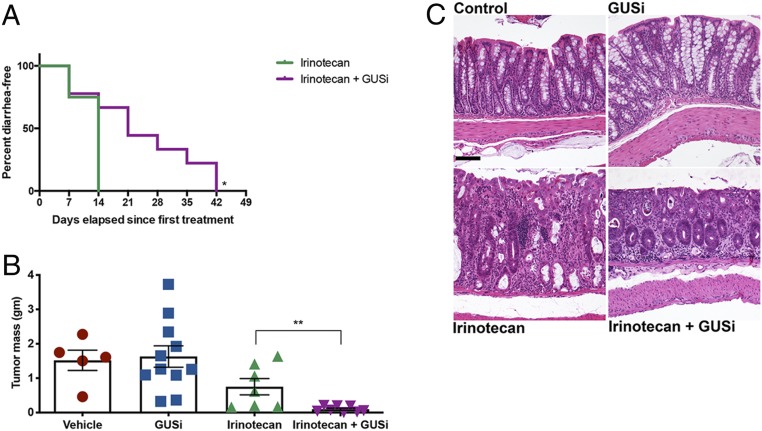
We used the C3Tag genetically engineered mouse model (GEMM) of breast cancer to examine irinotecan dose escalation and the effects of GUSi under those conditions in mice with an intact immune system (29). After palpable tumors were detected (∼100 mm3), mice were randomized into one of four groups: vehicle, GUSi (1 mg/kg total delivered twice daily via p.o. administration), irinotecan (50 mg/kg i.p.), or irinotecan + GUSi. Irinotecan was provided in a dose-intensification strategy, starting with two doses per week for 1 wk, then three doses per week for 2 wk, and finally, five doses per week in all subsequent weeks. All irinotecan-treated animals developed diarrhea within 14 d, while animals receiving irinotecan and GUSi experienced significantly reduced diarrhea, with some animals remaining diarrhea free for a remarkable 42 d (Fig. 4A). Thus, GUSi effectively alleviates irinotecan-induced diarrhea even with irinotecan dose intensification.
Fig. 4.
GUSi improves irinotecan efficacy in C3TAg breast cancer GEMM. (A) Kaplan–Meier analysis of remaining diarrhea free with GUSi cotreatment with irinotecan. P < 0.05 by log-rank (Mantel–Cox) test. (B) Irinotecan reduces tumor masses in C3Tag animals, while IRI + GUSi significantly diminishes tumor masses compared with irinotecan alone. **P < 0.01 by one-way ANOVA with Sidak’s multiple comparisons test. (C) Representative sections of colon revealing no significant microscopic differences between untreated control (Upper Left) and GUSi-treated (Upper Right) mice. Colons of IRI-treated mice (Lower Left) display moderate inflammation and epithelial damage, with the lamina propria expanded by inflammatory cells (arrows), variably sized and irregularly shaped crypts lined by attenuated epithelial cells with lumens containing sloughed epithelial cells and inflammatory cells (filled circles), and a fragmented and irregular overlying mucosal epithelium (filled square). While the colons of IRI + GUSi-treated mice (Lower Right) display inflammation in the lamina propria (arrow) and a few remaining damaged crypts at the superficial surface (filled circle), there is abundant crypt regeneration as the deeper mucosa contains a line of regenerative crypts lined by plump, elongated cells with cytoplasmic basophilia (asterisks) and the overlying epithelium displaying outstretched epithelial cells in the process of repair (open square). (Scale bar, 100 μM.)
Vehicle- and GUSi-treated animals gained weight due to tumor growth (SI Appendix, Fig. S8A), while mice receiving irinotecan consistently lost weight until euthanasia was required for all 28 d after therapy initiation (SI Appendix, Fig. S8F). Vehicle- and GUSi-treated control animals did not develop diarrhea and were followed until tumor size necessitated euthanasia (SI Appendix, Fig. S8F). Irinotecan + GUSi animals lost more weight than the irinotecan treatment group in the first 21 d of the study, but then, they rebounded, recovered their lost weight, and allowed the study to continue to day 49 (SI Appendix, Fig. S8A). Kaplan–Meier analysis reveals that the irinotecan + GUSi treatment group had a significantly better overall survival rate compared with each of the other treatment groups: 43% longer than irinotecan alone and 29% longer than vehicle and GUSi (SI Appendix, Fig. S8B). Thus, GUSi significantly protects against weight loss and increases survival in the C3TAg breast cancer GEMM treated with dose-intensified irinotecan.
The number of tumors observed in the animals in each treatment group did not differ (SI Appendix, Fig. S8D). Primary tumors in the vehicle and GUSi groups averaged 1.5 g in mass and accounted for ≥5% of the animals’ body weight (Fig. 4B and SI Appendix, Fig. S8E). Mice receiving irinotecan alone showed trends toward smaller primary tumors (∼1 g) accounting for only 3% body weight, but these values were not statistically different from the vehicle and GUSi groups (Fig. 4B and SI Appendix, Fig. S8E). In contrast, mice in the irinotecan + GUSi group exhibited dramatically reduced primary tumor masses, uniformly less than 0.1 g or <1% of total body weight, a 15-fold reduction (Fig. 4B and SI Appendix, Fig. S8E). Longitudinal tumor volume measures establish that irinotecan plus GUSi improves tumor regression, resulting in prolonged survival compared with the other groups (SI Appendix, Fig. S8F).
Microscopic examination of colon tissues revealed treatment with GUSi along with irinotecan promoted abundant epithelial regeneration of epithelial damage. Irinotecan treatment alone resulted in moderate inflammatory infiltrates expanding the colonic lamina propria, resulting in crypt destruction, goblet cell loss, and disruption of the overlying epithelium (Fig. 4C). While irinotecan + GUSi treatment animals displayed similar proprial inflammation and evidence of prior crypt destruction, there was pronounced epithelial regeneration characterized by abundant, well-organized regenerative crypts, which was not a feature of the irinotecan-only group at the same time point (Fig. 4C). Indeed, despite the dose-intensification regimen used, mice receiving irinotecan + GUSi exhibited less epithelial damage and improved body weight than mice receiving irinotecan alone. Together, these factors allowed irinotecan + GUSi mice to receive significantly more doses of irinotecan compared with those receiving irinotecan alone (SI Appendix, Fig. S8C). Thus, GUSi enhanced irinotecan-mediated tumor regression, improved animal survival, and reduced GI damage in this immunocompetent cancer GEMM.
GUS Inhibitor Protects against Gut Enterobacteriaceae Expansion in Athymic Mice.
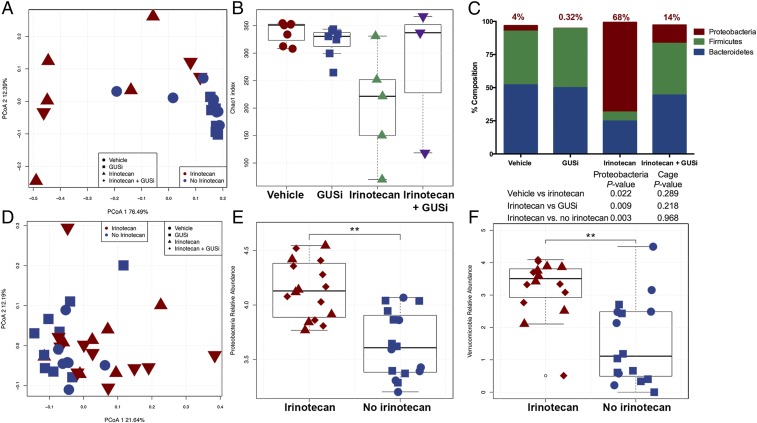
The 16S ribosomal ribonucleic acid (rRNA) sequencing analysis of the colon contents of the immune-deficient athymic mice bearing Sum149 triple-negative breast cancer xenograft tumors was performed. Principal coordinate analysis (PCoA) using UniFrac (30) showed that irinotecan with or without GUSi (irinotecan [IRI] and IRI + GUSi) significantly changed the microbial composition of the athymic mice gut microbiota (PCoA1 P = 0.001) (SI Appendix, Fig. S9A), which could not be attributed to cohousing effects (cage P value = 0.98). Pairwise comparisons between the four treatments also showed that irinotecan-treated mice had a significantly different gut microbiota composition than vehicle- and GUSi-treated mice (SI Appendix, Fig. S10). A significant decrease in alpha diversity (P value = 0.004) as assessed by Chao1 index in animals treated with IRI was observed (Fig. 5A), a drop that is not caused by cohousing (cage P value = 0.97). Mice treated with both IRI + GUSi, however, maintained species alpha diversity to levels similar to that of vehicle controls (Fig. 5A). Thus, GUSi improves gut microbial species alpha diversity when used in conjunction with irinotecan.
Fig. 5.
Irinotecan significantly changes gut microbiota in athymic (A–C) and C3TAg GEMM (D–F) mice. Four different treatments (irinotecan, irinotecan + GUSi, vehicle, and GUSi) divided into two groups (irinotecan: irinotecan alone and irinotecan + GUSi; no irinotecan: vehicle and GUSi) are shown. (A) PCoA analysis of athymic mice gut microbiota generated from UniFrac metric. Irinotecan group gut microbial composition (beta diversity) is significantly different from the no irinotecan group: PCoA1 P value = 0.001, cage P value = 0.98. (B) Alpha diversity analysis showing Chao1 index for the four treatments. Irinotecan and no irinotecan groups are significantly different from each other (P value = 0.004, cage P value = 0.97). Pairwise comparisons showed significant differences between irinotecan and GUSi treatments (P value = 0.01, cage P value = 0.6) and irinotecan and vehicle treatments (P value = 0.005, cage P value = 0.09). (C) Composition summary showing the major phyla detected in the athymic mice microbiota. Significant increase in Proteobacteria abundance is detected in irinotecan-treated mice independent of cage. (D) PCoA analysis of C3TAg GEMM mice gut microbiota generated from UniFrac metric. Irinotecan-treated mice (with and without GUSi) show different microbial composition (beta diversity) than mice that did not receive irinotecan: PCoA1 P value = 0.007, cage P value = 0.68. Proteobacteria (E) and Verrucomicrobia (F) are significantly increased in response to irinotecan treatment (with and without GUSi): Proteobacteria P value = 0.003, cage P value = 0.98; Verrucomicrobia P value = 0.004, cage P value = 0.98, **P < 0.01.
Furthermore, 16s rRNA sequencing analysis revealed that irinotecan causes a striking expansion of gut microbial Proteobacteria in xenografted athymic mice. At the phylum level, the luminal contents of vehicle-treated mice contained 52% Bacteroidetes, 41% Firmicutes, and 4% Proteobacteria (Fig. 5B). By contrast, irinotecan-treated mice exhibited a dramatic increase in the levels of Proteobacteria, up to 68%, and decreases in both Firmicutes (down to 6%) and Bacteroidetes (25%) (Fig. 5B). In mice treated with both irinotecan and GUSi, however, the expansion of Proteobacteria was markedly reduced, down to only 14%, while maintaining near-vehicle treatment levels of Firmicutes (39%) and Bacteroidetes (44%) (Fig. 5B). Finally, GUSi alone was able to reduce the levels of Proteobacteria in the mouse GI tract more than 10-fold, down to 0.32% compared with 4% observed in vehicle-treated mice (Fig. 5B). Thus, in immune-deficient mice, GUSi increases gut microbial diversity, markedly blunts the expansion of Proteobacteria observed in irinotecan-treated animals, and reduces the level of Proteobacteria in the GI tract in mice not treated with irinotecan.
Family-level taxonomic changes reveal that the Proteobacterial expansions observed with irinotecan arose from the Enterobacteriaceae, specifically Salmonella, Escherichia, Shigella, Klebsiella, and Yersinia. These opportunistic pathogens can flourish in inflamed sites of epithelial damage that leak oxygen into the intestinal lumen. Furthermore, the Enterobacteriaceae are the only intestinal taxa that encode a GUS operon containing the gus gene as well as glucuronide transporters (31), which may give these relatively trace Enterobacteriaceae taxa the ability to outcompete the more abundant Firmicutes and Bacteroidetes by increasing GlcA utilization. Importantly, the Enterobacteriaceae only encode L1 GUS enzymes, those that process SN38-G most efficiently and are also most potently inhibited by GUSi.
GUS Inhibition Does Not Alter Gut Microbial Composition in the Immune-Competent GEMM.
Finally, we examined the effects of irinotecan and GUSi on the composition of gut microbiota in the C3Tag GEMM with an intact immune system. We found that irinotecan was the sole driver of changes in gut microbial composition. While the vehicle and GUSi treatment groups seemed similar by PCoA1 analysis, both the irinotecan and irinotecan + GUSi groups were similar to each other and significantly distinct (PCoA1 P value = 0.007) from mice not receiving irinotecan (SI Appendix, Fig. S9B). In addition, unlike the xenograft model, irinotecan does not induce significant changes in the diversity of the gut microbiota in these GEMM animals (SI Appendix, Fig. S11). However, at the phylum level, we found that irinotecan, either alone or in combination with GUSi, led to increases in Proteobacteria (Fig. 5C) as well as Verrucomicrobia (Fig. 5D) compared with groups receiving no irinotecan. The changes observed in the GEMM were not as dramatic as those observed with the athymic mice (Fig. 5B). The increase in levels of Verrucomicrobia, which do not encode L1 GUS enzymes, is associated with a significant increase in Akkermansia muciniphila, a microbe that is typically linked with a healthy gut and the processing of host mucins (32, 33). These first data on the effects of gut microbial GUS inhibitor on the GI microbiota composition reveal differential effects depending on the immune function of the mouse model used.
Discussion
We established that irinotecan toxicity could be alleviated with nonlethal compounds that inhibit gut microbial GUS enzymes without affecting the mammalian GUS ortholog, the product of an essential gene, or affecting the plasma concentration of irinotecan or its metabolites (8, 9, 22). Beyond irinotecan, the intestinal toxicity of NSAIDs, which are also glucuronidated and reactivated by GUS enzymes in the gut, has also been alleviated using targeted microbial GUS inhibitors administered to male and female mice (10, 11, 34). Inh1 has been shown in rats to reduce nonsteroidal antiinflammatory drug (NSAID)-induced intestinal anastomotic leakage (35). Kong et al. (36) have replicated our findings with Inhibitor 1 and have shown that the tricyclic antidepressant amoxapine is a bacterial GUS inhibitor. Although partially blocking GUS activity in fimo, amoxapine reduces the viability of cultured bacteria (37). The novel pyrazolo[4,3-c]quinolones have been reported to inhibit intestinal GUS activity and protect the GI epithelium, further confirming that molecular targeting of a bacterial enzyme can prevent the side effects of irinotecan in vivo (38).
We assembled an atlas of gut microbial GUS enzymes in the human gut microbiome, showing that the 279 unique enzymes can be sorted into six structural categories based on active site features (14). Similar structural categories were also observed in the recently reported “mouse GUSome” (20). Purified representative enzymes revealed that structurally distinct GUS enzymes process chemically distinct substrates (14). Here, we show that L1 GUS proteins are most efficient a reactivating SN38-G and are more effectively inhibited by GUSi (UNC10201652) than Inhibitor 1. Furthermore, we demonstrate that GUSi preferentially inhibits L1 enzymes over other GUS orthologs present in the human gut microbiome. As such, GUSi acts as a targeted inhibitor that targets the GUS enzymes most responsible for SN-38 reactivation in the GI tract.
Intestinal contents of mice treated with irinotecan exhibit higher levels of SN38-G hydrolysis activity than control or GUSi-treated mice, suggesting either an expansion of GUS-expressing bacteria or up-regulated expression of GUS in individual members of the microbiota in response to irinotecan treatment. Intestinal contents of mice treated with irinotecan + GUSi showed significantly reduced levels of SN38-G processing activity. Short-term dosing studies revealed that irinotecan increases in fimo GUS activity within 24 h, an effect blocked by concurrent GUSi treatment. Intestinal proliferation decreases in vivo within 5 d after irinotecan injection, and this effect is blunted by GUSi. In both cases, single doses of irinotecan and GUSi were sufficient to see these effects. In a breast cancer xenograft model with immune-deficient mice, GUSi protected mice from diarrhea and gut damage and promoted recovery from irinotecan-induced weight loss. In the C3TAg breast cancer GEMM with an intact immune system, GUSi exerted significant protection against diarrhea, improved recovery from weight loss, and increased overall survival compared with animals receiving irinotecan alone.
Most dramatically, by reducing intestinal damage as well as preserving body weight, GUSi enabled mice to tolerate significantly more doses of irinotecan, which translated into remarkable tumor regression in the irinotecan plus GUSi group. Our results significantly extend previous research in alleviating irinotecan-induced gut toxicity and improving antitumor efficacy. While GUSi effectively reduces diarrhea and gut damage, as do Inh1 and other chemotypes previously examined (8, 36, 38), the GEMM data presented here demonstrate that GUSi is capable of significantly enhancing therapeutic efficacy of irinotecan. While we have chosen only female mice to assess efficacy in breast cancer models, we have previously reported that GUS inhibitors reduce intestinal ulcers resulting from NSAIDs in both male and female mice (11). In work reported in 2010, Lam et al. (39) reported that the botanical mixture PHY906 alleviated irinotecan-induced gut toxicity by reducing inflammatory infiltrates and plasma proinflammatory cytokines and promoting intestinal progenitor cell proliferation. PHY906 may also inhibit GUS activity given that it contains extracts of the Chinese herb Scutellaria (40) enriched with flavonoids that are known GUS inhibitors (41).
Previous culture- and PCR-based studies had shown that irinotecan induces shifts in gut microbial composition (42, 43), including increases in Proteobacteria. Here, we extend these investigations by using 16S rRNA sequencing to demonstrate that irinotecan causes dramatic expansions in gut Proteobacteria in athymic mice and less dramatic increases in Proteobacteria and Verrucomicrobia, including A. muciniphila, in immune-competent mice. The expansions in athymic mice are likely more dramatic than in immune-competent animals given the importance of the immune system’s response to the intestinal microbiota. The observation that the remarkable increase in Proteobacteria in immune-deficient (athymic) mice can be blunted by GUSi suggests translational implications for patients with local intestinal immune imbalances (e.g., inflammatory disorders) or with systemic immune deficiencies (e.g., HIV infection). Indeed, we were intrigued to see that Proteobacteria levels in athymic mice can be reduced 10-fold (4 to >0.4%) via GUS inhibition even in mice not receiving irinotecan. Proteobacteria, specifically the Enterobacteriaceae, are unique among gut microbial taxa in encoding an operon of genes encoding GUS enzymes that are up-regulated in response to the presence of glucuronidated compounds, allowing these bacterial species to use GlcA for growth (31). By inhibiting Enterobacteriaceae GUS enzymes and blocking access to GlcA, GUSi alone seems capable of blunting the growth of Proteobacteria in the mouse GI tract.
There are several limitations to the current study. For example, we do not examine mouse models of colon or intestinal cancer. We do not extend our in fimo investigations using human samples, either from healthy donors or from patients receiving irinotecan. Finally, we do not delve into the potential implications that limiting the levels of Enterobacteriaceae in the GI tract might have for other intestinal disorders. We will address these limitations in future studies, in some cases using recently described activity-based probe-enabled proteomics (44).
In summary, targeted inhibition of gut microbial GUS enzymes protects the GI tract from epithelial cell toxicity in preclinical models. GUS inhibition could dramatically improve the treatment of human cancer using irinotecan as well as the wide range of glucuronidated chemotherapeutics that cause gut toxicity.
Materials and Methods
Protein Purification, Crystallization, and Structure Determination.
The CpGUS enzyme was expressed and purified as previously described (8). The CpGUS was preincubated with 1 mM UNC10201652 (see below) for 1 h prior to cocrystallization setup. Cocrystals of CpGUS-Inh9 were flash frozen in liquid nitrogen in preparation for X-ray data collection. Diffraction data were collected on the 23-ID beamline at General Medical Sciences and National Cancer Institute Collaborative Access Team (Advanced Photon Source, Argonne National Laboratory). The data collection and refinement statistics are detailed in SI Appendix, Table S1.
Inhibitors.
As previously reported (17), 4-(8-(piperazin-1-yl)-1,2,3,4-tetrahydro-[1,2,3]triazino[4′,5′:4,5]thieno[2,3-c]isoquinolin-5- yl)morpholine, also known as UNC10201652, was synthesized in house at the University of North Carolina at Chapel Hill Center for Integrative Chemical Biology and Drug Discovery. Inhibitor 1 was previously described (8).
β-Glucuronidase Activity Assays.
We used a fluorometric assay to measure processing of SN38-G, which emits strong fluorescence at 420 nm when excited at 230 nm; fluorescence is lost on hydrolysis of SN38-G to SN38. In vitro assays contained 5 μL of purified enzyme, 5 μL of 10× buffer (250 mM (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) [Hepes], 250 mM NaCl, variable pH), 5 μL of SN38-G (final concentration of 15 μM), and 35 μL of water. For in vitro inhibition assays, conditions were the same except that 5 μL of inhibitor (100 μM final) and 30 μL of water were added. Buffer, substrate, and inhibitor were preincubated for 10 min at 37 °C, and the reaction was initiated by the addition of enzyme. For bacterial in cell assays, bacteria were grown overnight in 10 mL of lysogeny broth (LB) in ambient air with shaking. The next morning, 20 μL of each was subcultured into 2 mL of fresh LB for 1 h, after which 1 mM 4-nitrophenyl-β-d-glucuronidase was added for 1 h to induce expression of GUS; 10 μM inhibitor or equal volume of dimethylsulfoxide (DMSO) was added to the cells at the same time, and cultures were grown to an optical density of ∼0.6, after which bacteria were pelleted and washed and cells were lysed. Then, 10 μL of resultant lysate was used to initiate the hydrolysis reaction of 150 μM SN38-G in a reaction buffer composed of 20 mM Hepes and 50 mM NaCl at pH 7.4. For in cell inhibition assays, 10 μM either Inh1 or UNC10201652 was used, with equal volume of DMSO as control. For in fimo assays (23), frozen fecal samples were rehydrated, and bacterial cells were lysed, sonicated, and then clarified; 5 μL of fecal slurry supernatant was used to initiate the hydrolysis reaction of 150 μM SN38-G resuspended in the same buffer.
The initial velocities of the resultant progress curves of the reaction were calculated in MATLAB by linear regression. Initial velocities were then normalized to the protein concentration (in vitro assay), culture optical density (in cell assay), or total fecal protein content (in fimo assay) calculated using a standard Bradford assay. Fluorescence units were converted to concentration by standard curve analysis to generate the final units presented. All statistical analysis was performed on Prism (Graphpad). Statistical tests are indicated in figure legends, and details on the analyses are reported in SI Appendix, Table S6.
Quantification of GUSi and Irinotecan Metabolites.
Sample, standard preparation, and ultra performance liquid chromatography-mass spectrometry methods are found in SI Appendix.
Animal Study Designs.
All animal studies were approved by the University of North Carolina Institutional Animal Care and Use Committee according to the Guide for the Care and Use of Laboratory Animals by the NIH (45). Given that breast cancer primarily afflicts females, female mice were chosen for all experiments. All animals (except for germ-free mice used in monoassociation studies) were maintained in specific-pathogen free conditions in sterile microventilator cages containing corn bedding. All animals were given free access to chow and water, both of which were sterilized for the athymic mice that were housed in sterilized cages. Details on each animal model used and experimental outcomes measured are found in SI Appendix.
The 16S rRNA Amplicon Sequencing.
Isolation of total DNA from stool samples was carried out using the MoBio Powerfecal kit per the manufacturer’s directions. Total bacterial DNA was amplified using primers targeting the V3 to V4 region of the 16S rRNA gene and overhang adapter sequences appended to the primer pair for compatibility with Illumina index and sequencing adapters (46). Each sample was next amplified using a limited cycle PCR program, adding Illumina sequencing adapters to the amplicon target. The final libraries were purified, quantified, and normalized prior to pooling and loading on the MiSeq instrument (Illumina). Automated cluster generation and paired-end sequencing with dual reads were performed. Details on sequence analysis are found in SI Appendix.
Data Availability Statement.
The coordinates and structure factors of C. perfringens in complex with UNC10201652 can be found in the Protein Data Bank (ID code 6CXS). The 16S rRNA sequences have been deposited in the NCBI Sequence Read Archive (submission ID SUB4783842) and the BioProject database (ID code PRJNA505302).
Supplementary Material
Acknowledgments
We thank the staff of the Mouse Phase 1 Unit, the Animal Studies and Histopathology cores at the University of North Carolina at Chapel Hill (UNC), and the National Gnotobiotic Rodent Resource Center for their expert assistance. We also thank Drs. Virginia Miller and Kimberly Walker at the UNC for use of their equipment. R.Z.G. is supported by University of Florida Health Cancer Center funds. This work was supported by the University Cancer Research Fund, the Crohn’s and Colitis Foundation (R.B.S.), United States Department of Agriculture Grant 055336 (S.J.B.), and National Institutes of Health Grants T32-DK007737 (A.P.B.), P30 DK034987 (M.A.A.-P. and R.B.S.), P40 OD010995 (R.B.S.), CA207416 (S.J.B.), CA098468 (M.R.R.), and CA207416 (M.R.R.).
Footnotes
Competing interest statement: M.R.R. is the Scientific Founder of Symberix, Inc., which is developing drugs targeting the human microbiome. B.D.W., who is currently an employee of Symberix, Inc., conducted the work presented here as a graduate student at the University of North Carolina at Chapel Hill.
This article is a PNAS Direct Submission.
Data deposition: The coordinates and structure factors of Clostridium perfringens in complex with UNC10201652 can be found in the Protein Data Bank (ID code 6CXS). The 16S rRNA sequences have been deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (submission ID SUB4783842) and the BioProject database (ID code PRJNA505302).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1918095117/-/DCSupplemental.
References
- 1.Conroy T., et al. ; Groupe Tumeurs Digestives of Unicancer ; PRODIGE Intergroup , FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N. Engl. J. Med. 364, 1817–1825 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Pommier Y., Topoisomerase I inhibitors: Camptothecins and beyond. Nat. Rev. Cancer 6, 789–802 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Peddi P. F., et al. , Multi-institutional experience with FOLFIRINOX in pancreatic adenocarcinoma. JOP 13, 497–501 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Stein A., Voigt W., Jordan K., Chemotherapy-induced diarrhea: Pathophysiology, frequency and guideline-based management. Ther. Adv. Med. Oncol. 2, 51–63 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krishnamurthi S. S., Macaron C., Management of acute chemotherapy-related diarrhea. UpToDate (2017). https://www.uptodate.com/contents/management-of-acute-chemotherapy-related-diarrhea. Accessed 5 February 2018.
- 6.Smith N. F., Figg W. D., Sparreboom A., Pharmacogenetics of irinotecan metabolism and transport: An update. Toxicol. In Vitro 20, 163–175 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Takasuna K., et al. , Involvement of beta-glucuronidase in intestinal microflora in the intestinal toxicity of the antitumor camptothecin derivative irinotecan hydrochloride (CPT-11) in rats. Cancer Res. 56, 3752–3757 (1996). [PubMed] [Google Scholar]
- 8.Wallace B. D., et al. , Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science 330, 831–835 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wallace B. D., et al. , Structure and inhibition of microbiome β-glucuronidases essential to the alleviation of cancer drug toxicity. Chem. Biol. 22, 1238–1249 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.LoGuidice A., Wallace B. D., Bendel L., Redinbo M. R., Boelsterli U. A., Pharmacologic targeting of bacterial β-glucuronidase alleviates nonsteroidal anti-inflammatory drug-induced enteropathy in mice. J. Pharmacol. Exp. Ther. 341, 447–454 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saitta K. S., et al. , Bacterial β-glucuronidase inhibition protects mice against enteropathy induced by indomethacin, ketoprofen or diclofenac: Mode of action and pharmacokinetics. Xenobiotica 44, 28–35 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taylor M. R., et al. , Vancomycin relieves mycophenolate mofetil-induced gastrointestinal toxicity by eliminating gut bacterial β-glucuronidase activity. Sci. Adv. 5, eaax2358 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Biernat K. A., et al. , Structure, function, and inhibition of drug reactivating human gut microbial β-glucuronidases. Sci. Rep. 9, 825 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pollet R. M., et al. , An atlas of β-glucuronidases in the human intestinal microbiome. Structure 25, 967–977.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pellock S. J., Walton W. G., Redinbo M. R., Selecting a single stereocenter: The molecular nuances that differentiate β-hexuronidases in the human gut microbiome. Biochemistry 58, 1311–1317 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Little M. S., et al. , Active site flexibility revealed in crystal structures of Parabacteroides merdae β-glucuronidase from the human gut microbiome. Protein Sci. 27, 2010–2022 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pellock S. J., et al. , Gut microbial β-glucuronidase inhibition via catalytic cycle interception. ACS Cent. Sci. 4, 868–879 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pellock S. J., et al. , Three structurally and functionally distinct β-glucuronidases from the human gut microbe Bacteroides uniformis. J. Biol. Chem. 293, 18559–18573 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pellock S. J., et al. , Discovery and characterization of FMN-binding β-glucuronidases in the human gut microbiome. J. Mol. Biol. 431, 970–980 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Creekmore B. C., et al. , Mouse gut microbiome-encoded β-glucuronidases identified using metagenome analysis guided by protein structure. mSystems 4, e00452-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahmad S., et al. , A high throughput assay for discovery of bacterial β-glucuronidase inhibitors. Curr. Chem. Genomics 5, 13–20 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roberts A. B., Wallace B. D., Venkatesh M. K., Mani S., Redinbo M. R., Molecular insights into microbial β-glucuronidase inhibition to abrogate CPT-11 toxicity. Mol. Pharmacol. 84, 208–217 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhatt A. P., Grillo L., Redinbo M. R., In fimo: A term proposed for excrement examined experimentally. Gastroenterology 156, 1232 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stewart C. F., et al. , Gefitinib enhances the antitumor activity and oral bioavailability of irinotecan in mice. Cancer Res. 64, 7491–7499 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Bardia A., et al. , Sacituzumab govitecan-hziy in refractory metastatic triple-negative breast cancer. N. Engl. J. Med. 380, 741–751 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Sears C. L., Garrett W. S., Microbes, microbiota, and colon cancer. Cell Host Microbe 15, 317–328 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bhatt A. P., Redinbo M. R., Bultman S. J., The role of the microbiome in cancer development and therapy. CA Cancer J. Clin. 67, 326–344 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duncan J. S., et al. , Dynamic reprogramming of the kinome in response to targeted MEK inhibition in triple-negative breast cancer. Cell 149, 307–321 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Green J. E., et al. , The C3(1)/SV40 T-antigen transgenic mouse model of mammary cancer: Ductal epithelial cell targeting with multistage progression to carcinoma. Oncogene 19, 1020–1027 (2000). [DOI] [PubMed] [Google Scholar]
- 30.Lozupone C., Hamady M., Knight R., UniFrac–An online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinf. 7, 371 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Little M. S., Pellock S. J., Walton W. G., Tripathy A., Redinbo M. R., Structural basis for the regulation of β-glucuronidase expression by human gut Enterobacteriaceae. Proc. Natl. Acad. Sci. U.S.A. 115, E152–E161 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Derrien M., Vaughan E. E., Plugge C. M., de Vos W. M., Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int. J. Syst. Evol. Microbiol. 54, 1469–1476 (2004). [DOI] [PubMed] [Google Scholar]
- 33.Cirstea M., Radisavljevic N., Finlay B. B., Good bug, bad bug: Breaking through microbial stereotypes. Cell Host Microbe 23, 10–13 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Boelsterli U. A., Redinbo M. R., Saitta K. S., Multiple NSAID-induced hits injure the small intestine: Underlying mechanisms and novel strategies. Toxicol. Sci. 131, 654–667 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yauw S. T. K., et al. , Microbial glucuronidase inhibition reduces severity of diclofenac-induced anastomotic leak in rats. Surg. Infect. (Larchmt.) 19, 417–423. (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kong R., et al. , Old drug new use–Amoxapine and its metabolites as potent bacterial β-glucuronidase inhibitors for alleviating cancer drug toxicity. Clin. Cancer Res. 20, 3521–3530 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang W., Wei B., Yan R., Amoxapine demonstrates incomplete inhibition of β-glucuronidase activity from human gut microbiota. SLAS Discov. 23, 76–83 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Cheng K. W., et al. , Specific inhibition of bacterial β-glucuronidase by pyrazolo[4,3-c]quinoline derivatives via a pH-dependent manner to suppress chemotherapy-induced intestinal toxicity. J. Med. Chem. 60, 9222–9238 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Lam W., et al. , The four-herb Chinese medicine PHY906 reduces chemotherapy-induced gastrointestinal toxicity. Sci. Transl. Med. 2, 45ra59 (2010). [DOI] [PubMed] [Google Scholar]
- 40.Liu S. H., Cheng Y. C., Old formula, new Rx: The journey of PHY906 as cancer adjuvant therapy. J. Ethnopharmacol. 140, 614–623 (2012). [DOI] [PubMed] [Google Scholar]
- 41.Weng Z. M., et al. , Structure-activity relationships of flavonoids as natural inhibitors against E. coli β-glucuronidase. Food Chem. Toxicol. 109, 975–983 (2017). [DOI] [PubMed] [Google Scholar]
- 42.Lin X. B., et al. , Irinotecan (CPT-11) chemotherapy alters intestinal microbiota in tumour bearing rats. PLoS One 7, e39764 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stringer A. M., et al. , Irinotecan-induced mucositis manifesting as diarrhoea corresponds with an amended intestinal flora and mucin profile. Int. J. Exp. Pathol. 90, 489–499 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jariwala P. B., et al. , Discovering the microbial enzymes driving drug toxicity with activity-based protein profiling. ACS Chem. Biol. 15, 217–225 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.National Research Council , Guide for the Care and Use of Laboratory Animals (National Academies Press, Washington, DC, ed. 8, 2011). [Google Scholar]
- 46.Bartram A. K., Lynch M. D., Stearns J. C., Moreno-Hagelsieb G., Neufeld J. D., Generation of multimillion-sequence 16S rRNA gene libraries from complex microbial communities by assembling paired-end illumina reads. Appl. Environ. Microbiol. 77, 3846–3852 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The coordinates and structure factors of C. perfringens in complex with UNC10201652 can be found in the Protein Data Bank (ID code 6CXS). The 16S rRNA sequences have been deposited in the NCBI Sequence Read Archive (submission ID SUB4783842) and the BioProject database (ID code PRJNA505302).