Significance
This work offers a unique portrayal of yeast mitochondria through the characterization of its absolute proteome. The study of biophysical changes in the mitochondrial network associated with proteome profiling, throughout yeast growth and the transition from fermentative to respiratory metabolism, lays out the crucial role this organelle has in balancing the overall metabolic status of the cell. Using proteomic mass spectrometry, state of the art fluorescence microscopy, and lipidomics analysis, these data provide a highly quantitative description of key mitochondrial processes across three states of metabolism. In particular, the work highlights the significant contribution of functional and structural remodeling occurring during the diauxic shift of this subcellular organelle.
Keywords: mitochondria, absolute proteomics, Saccharomyces cerevisiae, diauxic shift
Abstract
Saccharomyces cerevisiae constitutes a popular eukaryal model for research on mitochondrial physiology. Being Crabtree-positive, this yeast has evolved the ability to ferment glucose to ethanol and respire ethanol once glucose is consumed. Its transition phase from fermentative to respiratory metabolism, known as the diauxic shift, is reflected by dramatic rearrangements of mitochondrial function and structure. To date, the metabolic adaptations that occur during the diauxic shift have not been fully characterized at the organelle level. In this study, the absolute proteome of mitochondria was quantified alongside precise parametrization of biophysical properties associated with the mitochondrial network using state-of-the-art optical-imaging techniques. This allowed the determination of absolute protein abundances at a subcellular level. By tracking the transformation of mitochondrial mass and volume, alongside changes in the absolute mitochondrial proteome allocation, we could quantify how mitochondria balance their dual role as a biosynthetic hub as well as a center for cellular respiration. Furthermore, our findings suggest that in the transition from a fermentative to a respiratory metabolism, the diauxic shift represents the stage where major structural and functional reorganizations in mitochondrial metabolism occur. This metabolic transition, initiated at the mitochondria level, is then extended to the rest of the yeast cell.
In the eukaryotic model organism Saccharomyces cerevisiae, glucose fermentation is a high-flux process that has proven to be catalytically more efficient than respiration, in terms of adenosine triphosphate (ATP) production per protein mass (1). The presence of glucose induces a catabolite repression response (CRR) that prevents utilization of alternative carbon sources by down-regulating the transcription of essential metabolic genes like the ones required for mitochondrial biogenesis, oxidative phosphorylation, and the tricarboxylic acid (TCA) cycle (2). When extracellular glucose is exhausted, yeast cells undergo a highly regulated transition from fermentative to respiratory metabolism; the turning point between these two distinct metabolic states are known as the diauxic shift (3) (Fig. 1A). The switch toward respiratory metabolism begins gradually, when extracellular glucose is still available in the medium (Fig. 1 B and C). This is followed by major metabolic reorganization and adaptation to growth on ethanol, as determined by the concomitant activation of both the TCA cycle and the gluconeogenic pathway. At this stage, a marked change in the cellular protein pool is also accompanied by a decrease in growth rate (1, 4). According to Zampar et al. (5), at the cellular level, progression toward the diauxic shift is characterized by three main steps: 1) a decline in glycolytic flux and a down-regulation of phosphofructokinase and pyruvate kinase ahead of glucose exhaustion; 2) a reversion of carbon flux through glycolysis after glucose depletion and an up-regulation of the glyoxylate cycle; and, finally, 3) an inhibition of the pentose phosphate pathway (PPP) and a reconfiguration of alternative strategies for nicotinamide adenine dinucleotide phosphate (NADPH) regeneration (5).
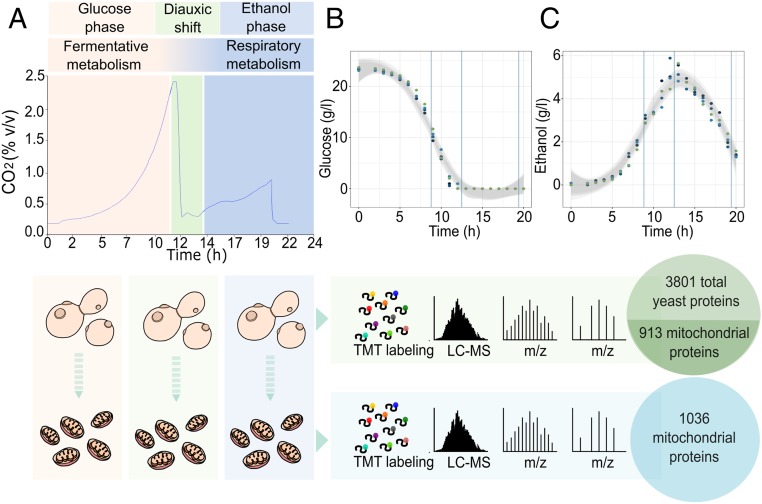
Fig. 1.
Overview of the experimental design. (A) The experimental workflow showing the CO2 profile of yeast grown in controlled batch cultivation. Cells and mitochondria samples were then digested with trypsin, and the resulting peptide samples were labeled with isobaric TMT mass tags and analyzed using an Orbitrap Fusion Tribrid mass spectrometer. (B and C) Extracellular glucose and ethanol concentrations were measured at intervals of 2 h throughout the experiment. Shaded areas represent the 95% confidence intervals for each curve, with vertical lines corresponding to the time chosen for the yeast cell harvesting and mitochondrial isolation (number of biological replicates: 4). LC-MS: liquid chromatography–mass spectrometry; m/z: mass-to-charge ratio.
As semiautonomous organelles, mitochondria are considered a center of many essential metabolic processes, and, as such, they are tightly interconnected to other cellular functions and pathways that take place in separate cellular compartments (6). In particular, they are dynamic and interconnected organelles, which undergo continuous events of fusion and fission to form a transitional tubular structure known as the mitochondrial network (7). By being the site of cellular respiration, mitochondria are highly influenced by changes in cellular growth conditions. The shift from fermentative to respiratory metabolism, therefore, triggers a substantial remodeling of mitochondrial metabolism, as well as the shape, structure, and volume of the mitochondrial network (6). To date, there have been several studies focusing on the metabolic transition from exponential to stationary phase in S. cerevisiae, with some of the most recent studies describing the transcriptional regulation that occurs and relative quantification of the yeast cell proteome during the diauxic shift (5, 8–11). Fewer studies, however, have performed comparative proteomic analysis of yeast using mitochondria isolated from cells grown on different carbon sources, these principally being fermentable and respiratory nonfermentable (12–15). To accurately understand the physiological adaptation during these growth phases and highlight the central role played by the mitochondrial network, we performed an absolute quantitative analysis of the mitochondrial proteome isolated throughout diauxic yeast growth. This study applies an approach toward studying the role of mitochondria in yeast cells, providing a realistic representation of the contribution of this organelle in the progression from a fermentative to a respiratory metabolism, illustrating the Janus-faced function of mitochondria in these distinct metabolic states.
Results
Absolute Proteomic Characterization of Yeast Mitochondria.
To evaluate the progressive adaptation of the mitochondrial proteome, yeast cultures were grown in a controlled culture environment using batch bioreactors. Samples were collected in biological triplicates at 9, 13, and 20 h after inoculation (Fig. 1A), with cells at these time points undergoing fermentative metabolism, the diauxic shift, and respiratory metabolism, respectively. Mitochondria isolation was performed via differential centrifugation followed by additional sample concentration, according to a preexisting protocol, as described in ref. 16. The samples were then tested via Western blot analysis to confirm the purity of the isolated mitochondria (SI Appendix, Fig. S1A).
For estimation of the absolute protein abundance, we applied a method developed by Schwanhäusser et al. (17) using tandem mass tag (TMT) 10-plex for isobaric mass tag labeling to enable simultaneous quantification of our replicates and the reference sample. This procedure is based on the assumption that the sum of all peptide intensities per protein, divided by the theoretical number of tryptic peptides is proportional to protein concentration (17). Absolute quantification was achieved using standards from UPS2, a set of distinct reference proteins of known concentration. The absolute proteomic data obtained gave a high level of replicate reproducibility (SI Appendix, Fig. S1 C–F), with proteome profiling resulting in the identification of 3,801 proteins across cell samples (∼65% of the expressed proteome) with a total of 3,700 proteins detected for the isolated mitochondrial samples. Despite the high number of proteins detected in the mitochondrial samples, a low level of contaminating, cytosolic proteins were identified, indicating the high purity of our mitochondrial fractions. Among the 3,700 proteins identified, in total, 1,036 were previously reported as mitochondrial (Dataset S1). These identifications were through both high-throughput studies and manual curation according to the Saccharomyces Genome Database (SGD) (18), with 824 proteins being identified as mitochondrial through manual curation alone. Additionally, two recently published studies, assigning a mitochondrial proteome of 901 and 986 proteins, respectively, were used as references for defining mitochondrial proteins (12, 15). Our approach, which enabled 50 previously undescribed proteins to be identified subsequently shows a 5% increase relative to previous studies.
Alongside our proteomic data acquisition, we aimed to characterize comprehensively several other cell parameters across the different metabolic states. Therefore, along with measuring cell dry weight (CDW) (SI Appendix, Fig. S1B), we collected data on cell number and cell volume for each time point, using an electrical current exclusion cell-counter and analyzer (CASY). Here, cell volume during the glucose phase and the diauxic shift were similar (41.4 ± 0.98 fL and 39.3 ± 1.99 fL, respectively). When the cells shifted to respiratory metabolism in the ethanol phase, the cell volume reduced to about half the volume measured at fermentative growth (21.7 ± 4.7 fL) (SI Appendix, Fig. S2 A–C) paired with a slower growth rate, as observed in postdiauxic yeast cells.
Determination of Mitochondrial Volume and Mass and Their Variations during the Diauxic Shift.
To reveal the structural changes occurring at the mitochondrial level, we performed in vivo measurements of mitochondrial volume before, during, and after the diauxic shift. For this, we engineered the S. cerevisiae yeast strain CEN.PK113-7D, integrating the pMitoLoc plasmid (19) into its genome. CEN.PK113-7D MitoLoc constitutively expresses two fluorescent proteins, GFP and mCherry, bearing specific mitochondrial targeting sequences derived from the N-terminal localization sequence of the ATPase subunit 9 (preSU9) of Neurospora crassa and the cytochrome C oxidase 4 (preCOX4), respectively. The import of preCOX4-mCherry is strictly dependent on the presence of an active membrane potential, and import is, therefore, proportional to the membrane potential, causing the preCOX4-mCherry to relocate to the cytoplasm upon weakening of the membrane potential (19). The preSU9 is a well-established strong mitochondrial presequence that is less sensitive to the membrane potential than other presequences and can drive import even at low membrane potential (19–22). The import strength of the preSU9 presequence has been demonstrated in experiments using addition of increasing concentrations of the uncoupler cyanide m-chlorophenyl hydrazone (CCCP) in order to gradually decrease the membrane potential (19, 22). In these experiments, the coupling of proteins to preSU9 enables import even at low membrane potentials where other presequences, such as preCOX4, are insufficient to drive translocation. Based on these principles, through application of MitoLoc, we were able to visualize mitochondrial morphology, determine the mitochondrial volume, and, at the same time, qualitatively ensure the mitochondrial functional integrity was intact. The term mitochondrial morphology implies the ability of this organelle to adopt a variety of shapes, from small spheres and short or elongated tubules to reticular (net-like) networks, based on cell type and their metabolism (23). This intrinsic plasticity of mitochondria is regulated mainly by fission and fusion events, as well as branching/debranching and extension/retraction of outer and inner mitochondrial membranes, all operated by molecular effectors (23).
It has been demonstrated that changes in mitochondrial shape has a crucial role associated with mitochondrial respiration. The morphological visualization of yeast cells expressing the MitoLoc genes were performed using a Nikon A1 confocal microscope (n = 50 cells per each condition). Z-stacks were acquired and analyzed using NIS-Elements Confocal software enabling automatic deconvolution, three-dimensional projections, and, finally, volume measurement of the whole mitochondrial network (Fig. 2B).
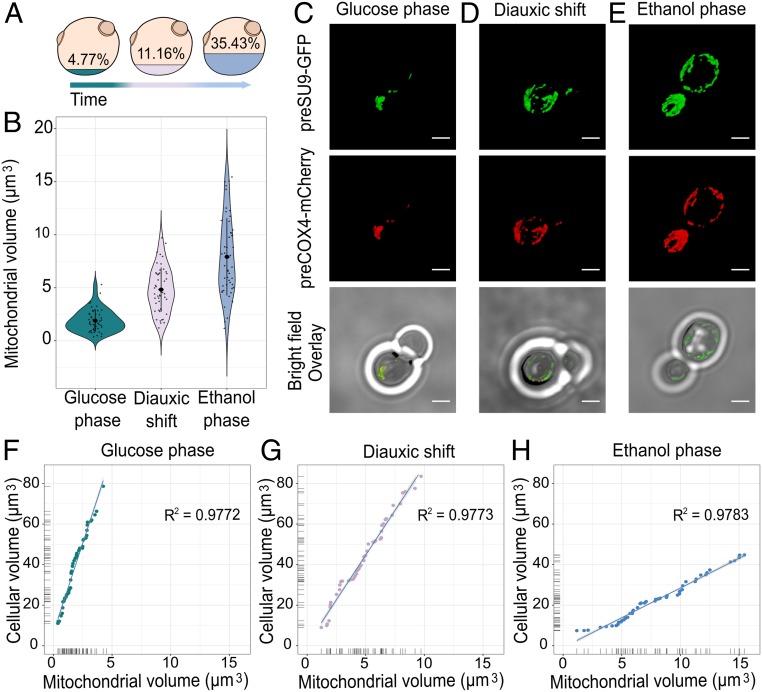
Fig. 2.
Tracking mitochondrial volume during cell growth. (A) Representation of the cellular volume (cubic microns) occupied by the mitochondrial network during the different stages of growth. (B) Mitochondrial volume variation during glucose phase, diauxic shift, and ethanol phase (n = 50 cells per condition). (C–E) Mitochondrial network morphology of yeast cells as determined by confocal microscopy. Cells are expressing the mitochondrial membrane potential (MMP)-dependent and MMP-independent preCOX4-mCherry and preSU9-GFP (“MitoLoc”), which are imported into the mitochondria in the different growth stages. (Scale bar, 2 µm.) (F–H) Correlation of cell volume-to-mitochondrial volume during the three growth stages (n = 50 cells per condition, with samples including budding and nonbudding cells).
Analysis of the mitochondrial volume revealed that as cells begin to respire, the mitochondrial network dramatically expands, occupying 4.77% of the whole cellular volume during fermentation, increasing to 11.16% during the diauxic shift, and then to 35.43% when they have fully transitioned to respiratory metabolism in the ethanol phase of growth (Fig. 2 A and B). This would suggest that significant changes in the size of mitochondria that arise begin to occur during the diauxic shift, at the time of glucose depletion and before the cells start assimilating ethanol at the start of the respiratory phase. This contrasts with what we observed for cell volume, which decreases only after the cells complete the diauxic shift. The volume of mitochondria, therefore, appears to be strictly bounded to the presence and depletion of carbon-source availability for the cell. In fact, it has recently been shown that following the exhaustion of the carbon source, it is possible to recognize, in a heterologous population, quiescent and senescent cells based solely on the structure of their mitochondrial network. These findings subsequently tighten the association between the role and functionality of mitochondria with processes of cellular aging, whereby the latter cells are known to increase their composition of senescent cells showing defects in mitochondrial morphology (24).
Additionally, we found a strong correlation between the cell volume and mitochondrial volume under all of the three growth phases studied, with increases in cell size being reflected by a concomitant increase in their mitochondrial network (Fig. 2 F–H). These findings are in line with what was already published by Rafelski et al. (25) and are consistent with the observations that mitochondria play a central role in regulating cell size (26).
Based on previous studies of the biophysical properties of mitochondria, it is possible to approximate that the yeast cell and the mitochondrial network have comparable densities (27, 28) (SI Appendix, Fig. S2D). Given this assumption and the data collected on cell mass, cell volume, and mitochondrial volume, we were able to calculate the mass of the mitochondrial network for each condition. The data obtained on the mitochondrial mass at the three stages of yeast growth were then used to calculate the absolute protein abundance at the mitochondrial level.
Identification of Significant Changes in Protein Abundance during the Glucose Phase, Diauxic Shift, and Ethanol Phase of Yeast Growth.
To assess overall changes in the mitochondrial proteome as cells transition from fermentative to respiratory metabolism, we used biological triplicate proteomic data for significance testing of the protein abundance changes in a pairwise manner. Student’s t test was used to calculate the significance of the changes in protein abundance along with their log2 fold changes (log2FC) between the three different conditions (Dataset S3). To classify the changes in abundance, we set a cutoff of P < 0.05 and an absolute log2FC > 1. This resulted in a total of 428 proteins being significantly regulated when comparing the diauxic shift and the glucose phase (413 up-regulated and 15 down-regulated), 309 being significantly regulated when comparing the ethanol and the glucose phase (273 up-regulated and 36 down-regulated), while only 16 proteins were found to be significantly regulated when comparing the ethanol phase and the diauxic shift (2 up-regulated and 14 down-regulated) (Dataset S4) (Fig. 3 A–C). Although only 16 proteins were significantly regulated between the ethanol phase and the diauxic shift, given the defined log2FC cutoff, 82 proteins were found to be significantly down-regulated (P < 0.05) (Dataset S4).
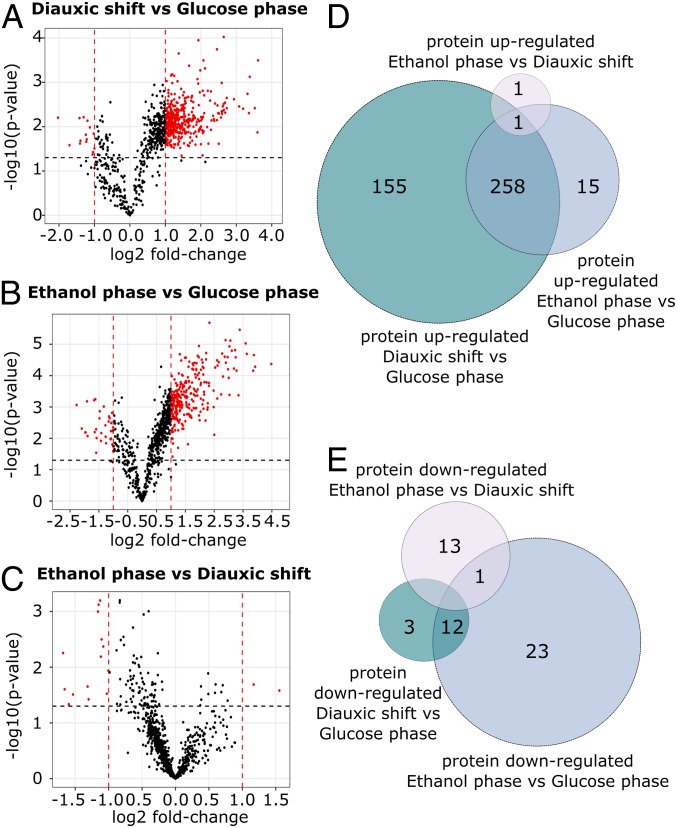
Fig. 3.
Differentially expressed proteins in the three metabolic phases of yeast growth. (A–C) Volcano plots showing differentially expressed proteins identified through the comparison of the three metabolic phases of yeast growth. Log-transformed P values (t test) are plotted against log-transformed fold change. The horizontal dashed line marks a P value of 0.05. Vertical dashed lines indicate a log2 fold change of ±1. (D and E) Venn diagrams depicting the number of proteins specifically up-regulated and down-regulated identified through comparison of the differentially expressed protein in the three phases of yeast growth. The specific identifications are reported in Dataset S4.
To identify some general trends with regard to the time point of regulation, we compared those proteins with significant changes in abundance across the different phases (Fig. 3 D and E). Of the 482 unique proteins significantly regulated during any of the stages, 272 proteins were shared and significantly regulated, both when comparing the diauxic shift and the glucose phase and when comparing the ethanol and glucose phase (259 up-regulated and 13 down-regulated) (Fig. 3 D and E) (Dataset S4). To identify trends among processes being regulated, we next ran a gene ontology (GO)-term enrichment analysis on biological processes (Benjamini–Hochberg corrected P value: <0.05) using YeastMine (29). The results of this enrichment analysis showed that among the 272 significantly regulated proteins, the TCA cycle, the respiratory chain, and ATP synthase, as well as mitochondrial transporters, were the most represented processes (Dataset S5). These proteins are up-regulated in the diauxic shift and follow a further, although small, up-regulation throughout the ethanol phase, indicating that these proteins are part of an early response to glucose exhaustion that lasts throughout the entire diauxic shift. These results are in good agreement with findings previously reported by Murphy et al. (11), who investigated the temporal changes of the S. cerevisiae proteome during the diauxic shift. In this study, the TCA cycle and oxidative phosphorylation were found to be part of an early response to glucose exhaustion, with an up-regulation of protein abundances of these processes showing to occur throughout the diauxic shift.
From pairwise comparison of the three metabolic phases, we additionally identified significantly regulated proteins that were uniquely up- and down-regulated in each phase, respectively, enabling a sharper overview of what processes may be uniquely associated with the different metabolic stages of yeast growth (Dataset S4). Of the proteins with a significant change in abundance in the diauxic shift, versus the glucose phase, 155 were up-regulated, while only 3 were down-regulated (these being the Altered inheritance of mitochondria protein Aim6, the Phospholipid-transporting ATPase Dnf1 and the cell wall protein Ecm33). Among the 155 specifically up-regulated, several proteins of notable interest include the Dynamin-like GTPase Mgm1, Mitochondrial distribution and morphology protein Mdm31, Most of the ribosomal protein (e.g., Mrp1, Mrp13, Mrpl1, etc.), Mitochondrial translation factor Atp22, Required for respiratory growth protein 1 Rrg1, Cytochrome b translational activator protein Cbs1, Cytochrome oxidase assembly factors Coa1 and Coa4, and the Mitochondrial respiratory-chain complexes assembly protein Yta12.
When considering the group of significantly regulated proteins in the ethanol phase versus the glucose phase, we detected 15 proteins that were uniquely up-regulated versus 23 that were down-regulated proteins specifically for this group. Among the proteins specifically up-regulated, we found the mitochondrial Acyl carrier protein Acp1, the Cytochrome c oxidase assembly protein Cox16, the Dynamin-related protein Dnm1, and MIOREX complex component 3 Mrx3. The larger group of down-regulated proteins include the Iron-sulfur cluster assembly protein 2 Isu2, ATP-dependent molecular chaperone Hsc82, Phosphoglycerate mutase 1 Gpm1, and Membrane-anchored lipid-binding protein Lam1.
Finally, the group of ethanol phase versus diauxic shift includes only a few unique significantly regulated proteins, with only one up-regulated protein and 13 down-regulated proteins. The single up-regulated protein consists of the probable secreted beta-glucosidase Uth1 involved in the regulation of mitochondrial biogenesis (30). While the group of down-regulated proteins includes the Ubiquinone biosynthesis protein Coq9, Ubiquinone biosynthesis O-methyltransferase Coq3, Cytochrome c iso-2 Cyc7, and the Stationary phase gene 1 and 4 proteins Spg1 and Spg4.
Through this analysis, it was possible to point out the high number of proteins specifically up-regulated in the diauxic shift (155 against the 15 proteins up-regulated in the ethanol phase versus glucose phase). Among the processes that are specifically up-regulated during the diauxic shift, we identified proteins involved in mitochondria morphology (i.e., MGM1), as well as in energy metabolism. This confirms the crucial role of this metabolic stage in the regulation of a variety of processes associated with the transition from a fermentative to respiratory metabolism.
Finally, we looked at a group of proteins that are significantly up-regulated in the diauxic shift compared with the glucose phase but have a log2FC < 1 when comparing the ethanol phase and the glucose phase. GO-term enrichment analysis on biological processes (Benjamini–Hochberg corrected P value: <0.05) showed that this group of proteins was significantly enriched in GO terms including mitochondrial gene expression, mitochondrial translation, mitochondrial organization, protein targeting to mitochondria, and respiratory-chain complex assembly, indicating that a reprogramming of the mitochondrial network and biogenesis-related functions are essential at an early stage of the adaptation of mitochondria to a respiratory metabolism (Dataset S5). These processes were not identified as significantly enriched among the regulated proteins found by Murphy et al. (11). We speculate that the analysis of the cellular proteome alone, compared with our analysis of the mitochondrial proteome specifically, performed by Murphy et al. and other studies (8–11) does not sufficiently portray changes occurring at the mitochondrial level, which are often involving less abundant, membrane-bounded mitochondrial proteins groups. On top of providing an accurate representation of mitochondrial processes during the diauxic shift, our study, therefore, additionally highlights the benefit of focusing on the mitochondrial proteome specifically and alongside studying the cell proteome (Dataset S2), when analyzing metabolic processes that directly involve this organelle.
Stoichiometric Evaluation of ATP Synthase Subunits and Remodeling of the Cristae Structure during the Diauxic Shift.
ATP synthase is a highly conserved complex responsible for the formation of ATP via a rotary mechanism. Through the application of a stoichiometric model, the F1Fo-ATP synthase has been identified as the key flux-controlling enzyme for the respiratory metabolism in S. cerevisiae (1, 31). Therefore, to validate the quality of our absolute proteomic data, we investigated the subunit stoichiometry of the multiprotein ATP synthase complex during the three metabolic phases (Fig. 4 A and B and SI Appendix, Fig. S3 A and B).
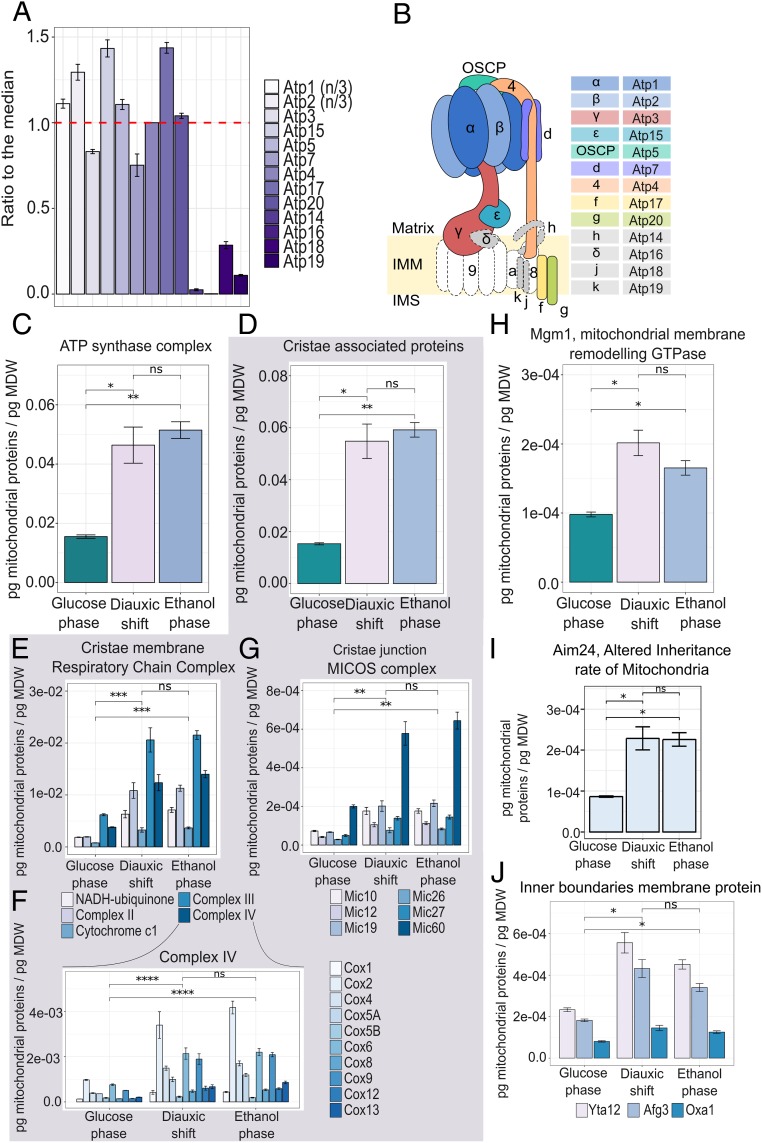
Fig. 4.
Increasing proteome allocation toward respiratory metabolism is coupled to increased cristae formation. (A) Stoichiometry of the subunits of ATP synthase, in the glucose phase, expressed as the ratio to the median, taking the theoretical stoichiometry of ATP1 and ATP2 (three subunits per complex) into account. Data represent the means ± SD of three biological replicates. (B) Overview of ATP synthase subunit composition in yeast. Subunits in color were detected in this study; subunits in gray were detected with low coverage and quantification accuracy, possibly due to their presence in the inner mitochondrial membrane. Subunits in white were not detected. (C) Absolute abundance of the ATP synthase complex. Data represent the means ± SD of three biological replicates. Statistical analyses were performed using paired t test. (D–I) Absolute abundance of the cristae-associated proteins (D), with detailed analyses on the absolute abundance of the Respiratory Chain Complexes (E); Protein of the Complex IV of the Respiratory Chain (F); MICOS complex (G); the protein Mgm1, Dynamin-like GTPase (H); the protein Aim24, Altered inheritance rate of Mitochondria (I); and inner boundaries membrane proteins Yta12, Alg3, and Oxa1 (J). Data represent the means ± SD of three biological replicates. Statistical analyses were performed using paired t test. Statistically significant differences are indicated as follows: ns (not significant), P > 0.05; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
To achieve this, we calculated each subunit’s ratio to its median abundance using its copy number per cell abundance detected. For comparison, we also included proteins whose quantification could not be considered reliable, given the low number of peptides identified (Fig. 4 A and B and SI Appendix, Fig. S3 A and B). Overall, these results showed that despite the changing conditions, cells maintained the ratio of subunits in the stoichiometry required for complex formation, with over 75% of the total complex-forming proteins being within −1.7 and 1.0 log2FC of the median in the glucose phase, −2.5 and 1.0 log2FC in the diauxic shift, and −1.8 and 1.0 log2FC in the ethanol phase (SI Appendix, Fig. S3 F and G).
Next, we analyzed protein allocation across the three growth phases, focusing on the ATP synthase complex and “cristae associated proteins,” the latter including the respiratory-chain complex located in the cristae membranes and the MICOS complex situated in the cristae junctions (Fig. 4 C, D, F, G, and I).
From the analysis of the ATP synthase, we found that the onset of mitochondrial respiratory metabolism in the ethanol phase is marked by an ∼threefold increase in abundance of this complex (after summing all subunits) compared with growth on glucose. This increase in mitochondrial respiration, as reflected by ATP synthase becoming more abundant, was also coupled to a concomitant ∼threefold increase in the abundance of cristae-associated proteins, linking the transformation of mitochondrial functions to the remodeling of their morphology (Fig. 4 D, F, G, and I). Furthermore, we observed a drastic increase in protein abundance predominantly during the diauxic shift, with a 2.9-fold increase in the ATP synthase complex during the transition from glucose phase to diauxic shift versus a 0.4-fold increase from diauxic shift to ethanol phase. From the analyses of the respiratory-chain complex and the MICOS complex, we also observed the same trend, with proportional increases occurring mainly as cells transition from the glucose phase to the diauxic shift. As well, in the overall analyses of the cristae associated protein, we found a 3.5-fold increase from glucose phase to diauxic shift and a 0.3-fold increase from diauxic shift to ethanol phase (Fig. 4 D, F, G, and I).
The analyses of proteins located in the inner membrane boundaries (e.g., Yta12, Afg3, and Oxa1) (Fig. 4J) show the highest levels of these proteins during the diauxic shift, followed by constant or a slightly lower allocation in the subsequent respiratory phase. These proteins are required in the assembly of mitochondrial enzymes complexes (Yta12 and Afg3) and the insertion of mitochondrial and some nuclear-encoded proteins in the inner mitochondrial membrane (Oxa1) (32, 33). This suggests changes in this part of the mitochondria that principally occur during the diauxic shift may perform a regulatory role in initiating structural changes necessary for respiratory metabolism.
It has been previously established that a tight link exists between the structure and function of mitochondria (23–26). Indeed, within the inner mitochondrial membranes, the cristae structure has been shown to be crucial for the function of the respiratory chain, due to its ability to 1) modulate organization of respiratory-chain components into supercomplexes (SCs) and 2) affect the structural stability of these SCs (34). Among the proteins involved in the cristae structure, OPA1 (Mgm1 S. cerevisiae homolog), located in the inner membrane boundaries, also known as the master cristae shape regulator, plays a critical role in the stability and assembly of the respiratory-chain SCs (35, 36). By looking at the distribution of this protein in the three different metabolic phases considered in this study, we notice a significant increase in its abundance, mainly in the diauxic shift (with a two-fold increase from the glucose phase to the diauxic shift versus 1.7 fold between the glucose phase and ethanol phase) (Fig. 4E). As well as OPA1, the MICOS complex also plays a critical role in connecting structural changes to this organelle’s function. In particular, this complex is important for securing the architecture of mitochondrial membranes, by holding the cristae junction and stabilizing the cristae curvature (37). The stability of MICOS, in turn, is also aided by the inner membrane protein Aim24 (Fig. 4H), which, if deleted, results in impaired respiration, destabilization of the MICOS complex, as well as an alteration in cardiolipin (CL) acylation pattern (38). From the analysis of Aim24’s distribution during the three metabolic phases of the yeast growth, we observe again as for the other aforementioned proteins, an increase in this protein’s allocation during the diauxic shift that remains constant during the ethanol phase (Fig. 4H). Altogether, these data support the hypothesis that the diauxic shift is the phase in which most of the structural and functional rearrangements in the mitochondria occur, in preparation for maximum respiratory function during growth on ethanol.
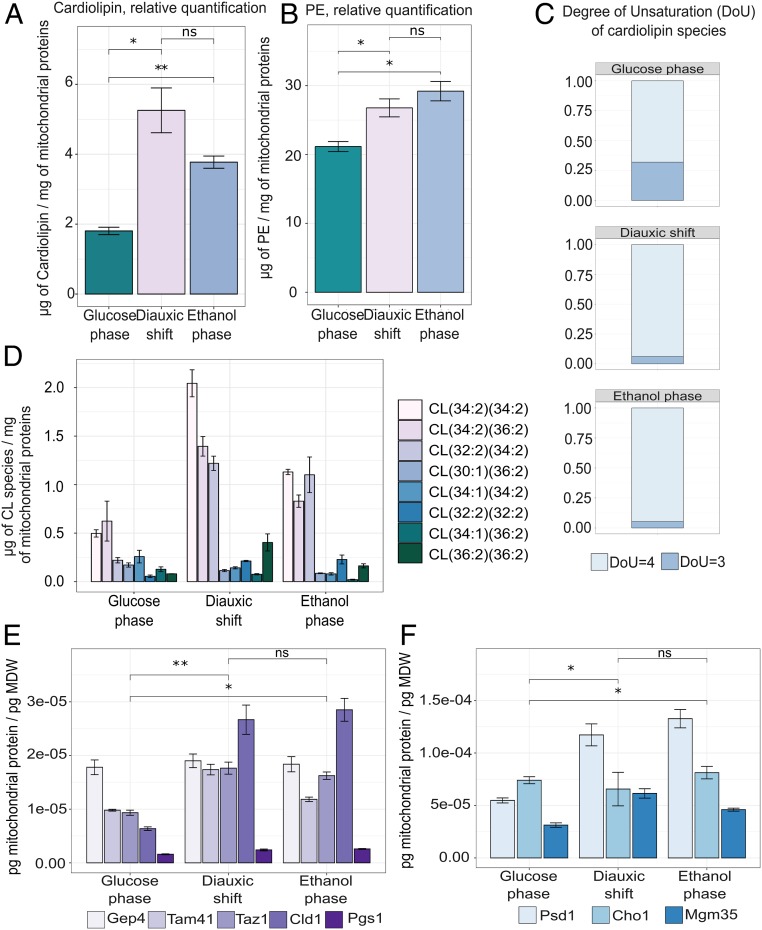
To reconcile further what we found for cristae at the protein level, with its function in respiration, we next performed lipidomics on the crude mitochondrial extract to see if the lipid composition of the cristae was also changing in line with mitochondrial metabolism. Our lipidomics data revealed that the mitochondrion-specific glycerophospholipid CL increased in abundance during the diauxic shift (from 1.81 ± 0.18 μg/mg to 5.25 ± 1.11 μg/mg), followed by a decrease (to 3.77 ± 0.30 μg/mg) as cells reached the ethanol phase (Fig. 5A), probably due to the majority of mitochondrial remodeling occurring already during the diauxic shift. Due to its conical shape, CL is known to be abundant in areas of high membrane curvature; moreover, this lipid has been implicated in the stabilization of individual complexes of the respiratory chain (39). We found that as cells shift toward respiration, the acyl-chain composition of the CL species also moves toward a higher degree of unsaturation (Fig. 5 C and D). It has previously been speculated that the increase in CL unsaturation enables an increase in membrane fluidity, in turn allowing cristae to increase in curvature and accommodate more ATP synthase for respiration (39). This observation toward CL reconfiguration was further supported by the proteomic data, which showed an up-regulation of the Mitochondrial CL-specific phospholipase Cld1; the Mitochondrial phosphatidylglycerolphosphatase (PGP phosphatase) Gep4; the Phosphatidylglycerolphosphate synthase Pgs1, a rate-limiting step in the synthesis of CL; the Mitochondrial phosphatidate cytidylyltransferase (CDP-DAG synthase) Tam41, also required for CL biosynthesis; and the Lysophosphatidylcholine acyltransferase Taz1, involved in the CL acyl-chain remodeling (Fig. 5E). These proteins, which are strictly involved in the generation of the mitochondrial distinctive glycerophospholipid CL, have a peak allocation during the diauxic shift, reflecting the higher concentration of CL during this metabolic phase shown in Fig. 5A.
Fig. 5.
The changing abundance and unsaturation of CL as cells progress to respiratory postdiauxic growth. (A) Abundance of CL scaled to picogram of mitochondrial dry weight (MDW), calculated as the mean ± SD of three biological replicates, based on liquid chromatography–mass spectrometry (LC-MS) analysis of isolated mitochondria. Statistical analyses were performed using paired t test. (B) Degree of unsaturation of the CL moieties, calculated as (2C + 2 + N − X − H)/2, where C is the number of carbon atoms, N is the number of nitrogen atoms, X is the number of halogen atoms (F, Cl, Br, I), and H is the number of hydrogen atoms. (C) Distribution of CL species with different acyl-chain composition, obtained from LC-MS analysis of lipids from mitochondria isolated in the different stage of growth. Data were calculated as the means of three biological replicates. Individual CL species are represented by different bar fill colors. (D) Absolute abundance of the identified enzymes involved in CL biosynthesis and CL modifying the acyl-chain composition of CL, Gep4, Tam41, Taz1, Cld1, and Pgs1 in the three stages of growth. Data represent the means ± SD of three biological replicates. Statistical analyses were performed using paired t test. (E) Abundance of PE scaled to picogram of MDW, calculated as the mean ± SD of three biological replicates, based on LC-MS analysis of isolated mitochondria. Statistical analyses were performed using paired t test. (F) Absolute abundance of the identified enzymes involved in PE metabolism, Psd1, Cho1, and Mgm35, in the three stages of growth. Data represent the means ± SD of three biological replicates. Statistical analyses were performed using paired t test. All absolute abundances are scaled to picogram of MDW. Statistical analyses were performed using paired t test. Statistically significant differences are indicated as follows: ns (not significant), P > 0.05; *P < 0.05; **P < 0.01.
Furthermore, the increase in unsaturation documented in Fig. 3 B and C is in line with published evidence that acyl-chain unsaturation occurs alongside an increase in respiratory activity (40). A recent study hypothesizes that the increased expression of oxidative phosphorylation complexes creates a stress in the lipid bilayer and is responsible for the induction of the CL remodeling by Cld1 and Taz1. The increased content in tetra-unsaturated CL species would, therefore, stabilize the lipid–protein interactions necessary for the correct functionality of protein complexes residing in the mitochondrial membrane (39).
Alongside CL, the other non–bilayer-forming phospholipid, phosphatidylethanolamine (PE), has been demonstrated to play a role in the maintenance of mitochondrial structure and function (41). Indeed, a synergistic activity between CL and PE has been shown to exist, with both being required for optimal mitochondrial import activity and for the respiratory-chain complex to function (42). However, in contrast to what is observed for CL (Fig. 5A), PE appears to accumulate mostly during the ethanol phase, most likely to compensate for reduced levels of CL (Fig. 5B). These data are also reflected by the increased allocation of Psd1, the leading mitochondrial enzyme involved in the synthesis of PE (Fig. 5F). Altogether, integration of the absolute mitochondrial proteome with lipidomic profiling, performed on isolated mitochondria, supports the position of CL and PE as regulators of mitochondrial respiratory function. Additionally, these data highlight modifications in the membrane composition occurring during the diauxic shift, in particular, the acyl-chain remodeling of CL, a feature that is connected to mitochondria-induced biogenesis and proliferation, which are taking place during this metabolic phase.
A study published by Casanovas et al. (10) that employed a jointed proteomic and lipidomic approach to analyze proliferating S. cerevisiae cells, showed, in alignment with our findings, an increase of CL levels during the diauxic shift, together with an up-regulation of Taz1 and an activation of CL-remodeling. However, the increase in PE that we observed during the diauxic and postdiauxic phase was not reported by Casanovas et al., who conversely report an increase of PE, alongside other phospholipids during fermentation. This discrepancy is possibly due to additional measurement of phospholipid molecular species present in the cell, outside the mitochondria, that may confound interpretation for how much is derived solely from this organelle. For example, the presence of a second PE-synthesizing enzyme (Psd2) located in the endoplasmic reticulum (ER), would be able to supply the cells with additional required phospholipids (43). This approach may, therefore, preclude the accurate quantification of mitochondrial PE, while a targeted analysis of isolated mitochondria enables a more probable perspective of lipid dynamics in this organelle specifically.
Mitochondrial Absolute Proteome Allocation and Variation of the Organelle’s Role Amid Diauxic Growth.
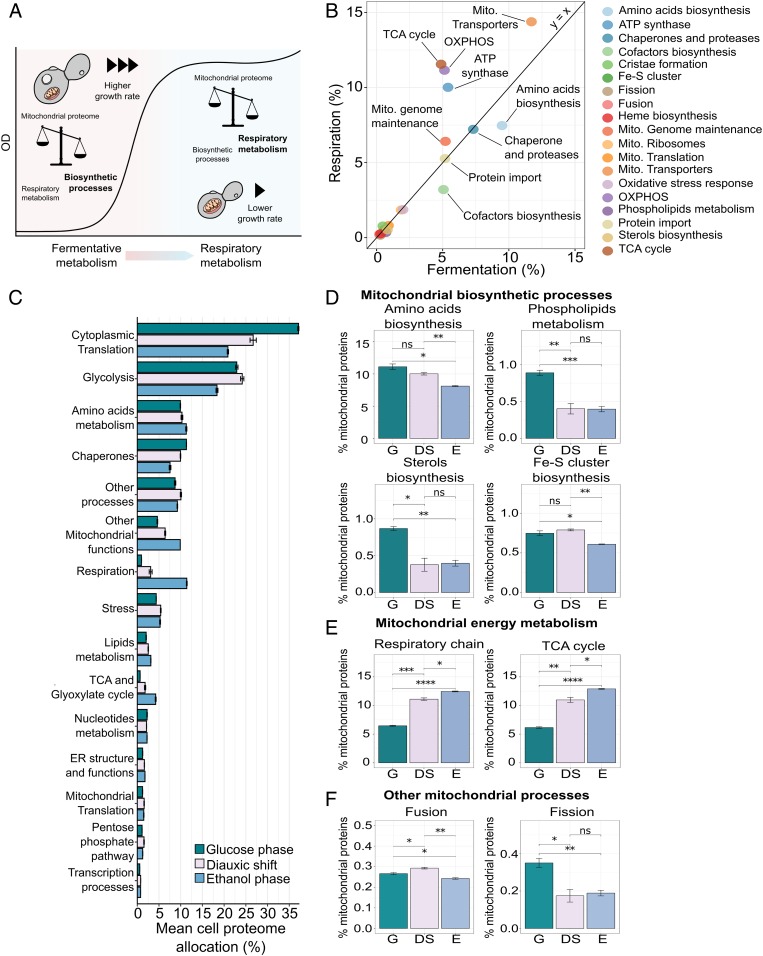
To understand how the mitochondrial proteome is reallocated for respiration in the postdiauxic phase, we split the proteome into 19 groups based on each protein’s molecular function. We then analyzed the relative abundance of these groups in the respiratory versus fermentative growth phase. Overall, the mitochondrial proteome showed a general shift in proteome allocation from biosynthetic processes, at the high growth rate, to energy-related processes, such as oxidative phosphorylation, at a lower growth rate (Fig. 6 A and B). Similarly, to analyze the allocation of the whole-cell proteome, (Fig. 6C), we divided the proteome into 14 groups of similar function based on GO (44, 45) annotation and Kyoto Encyclopedia of Genes and Genomes pathways (KEGG) (46) (Dataset S6). These groups covered ∼90% of the proteome mass-wise, and the remaining proteins were assigned to an additional group (other processes). By examining the allocation between the groups in the different growth stages, we found that during exponential growth, the proteome is allocated mostly toward translation-related proteins (∼37%) and glycolytic proteins (∼20%). As the cells transitioned to respiratory metabolism, however, we saw a decrease in allocation toward translation by ∼20%, in line with previous work that showed the proportion of translation-related proteins in the proteome to scale with growth rate (47).
Fig. 6.
The transition to respiratory metabolism is associated with a shift in mitochondrial proteome allocation from biosynthetic to energy-related processes. (A) Overview of the reallocation of the mitochondrial proteome during the transition to postdiauxic growth. (B) Comparison of the overall mitochondrial proteome allocation in respiratory and fermentative growth. Protein allocation was calculated as the mean percentage allocation of three biological replicates, with proteins grouped based on function. (C) Allocation of the whole cellular proteome in the three stages of growth, calculated as the mean percentage allocation. Data are means ± SD of three biological replicates. The specific identifications are reported in Dataset S6. (D–F) Allocation of selected mitochondrial processes. Data are means ± SD of three biological replicates. Statistical analyses were performed using paired t test. Statistically significant differences are indicated as follows: ns (not significant), P > 0.05; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. Fe-S, iron-sulfur; G: glucose phase; DS: diauxic shift; E: ethanol phase.
Along with this decrease, there is a noticeable shift in allocation (from 6 to 26%) toward respiratory functions in the cell, the TCA cycle, and other mitochondrial processes (Fig. 6C). However, in contrast to what has been observed for the mitochondrial proteome, in the analysis of the whole-cell proteome, the allocation to mitochondrial functions and energy metabolism is more pronounced during the ethanol phase and to a lesser extent in the diauxic shift. In fact, by just looking at the whole-cell proteome, the diauxic shift seems to represent an intermediate state between the glucose and ethanol phase with respect to the allocation of proteins associated with mitochondrial energy metabolism. For instance, considering the whole-cell proteome, we measured an increase of 48% in allocation toward respiratory-chain proteins between the diauxic shift and ethanol phase.
However, when focusing solely on the mitochondrial proteome, the difference between the diauxic shift and the ethanol phase for the same class of proteins is only 10.7% (Fig. 6 C and E). This observation is further clarified when comparing side-by-side the percentage allocation of the respiratory-chain proteins in the mitochondrial proteome and the cellular proteome (SI Appendix, Fig. S4A). In the mitochondrial proteome, we observe a higher allocation during the glucose phase, followed by a rapid increase (of 43.7%) in proteome allocation for respiratory proteins in the diauxic shift and a smoother transition (with a 10.4% increase) to the ethanol phase. In contrast, by looking at the cellular proteome, we see a milder increment (of 18.6%) between glucose phase and diauxic shift, followed by a drastic rise (of 72.9%) in the protein complexes’ allocation from the diauxic shift to the ethanol phase (SI Appendix, Fig. S4A). These results clearly highlight that subcellular level proteomics is required to reveal organelle-specific dynamics, which may be less contrasting in terms of protein abundance at the cellular level. To show organelle-specific changes in proteome allocation and remove potential biases in protein abundance from analyzing the entire cellular proteome at once, we further investigated proteomics data generated from the analysis of isolated mitochondria. Once again, we looked at the total allocation of 19 reported functional groups, as well as each group respectively, in order to highlight the precise protein distribution during the glucose phase, diauxic shift, and ethanol phase (Fig. 6 D–F, SI Appendix, Fig. S5A, and Dataset S7).
The allocation of some specific functional groups of mitochondrial proteins, including those involved in mitochondrial-specific amino acid, sterol, and phospholipid biosynthetic processes, was shown to be significantly enriched during the fermentative growth stage, compared with the ethanol phase (Fig. 6D and SI Appendix, Fig. S5A). Regarding the phospholipid metabolism, in particular, we see an opposite trend between the biosynthetic pathways connected to the synthesis of CL and PE, involved in the regulation of respiratory mitochondrial functions (Fig. 5 A, B, E, and F), in comparison with the remaining proteins that are involved in the metabolism of bilayer-forming phospholipids. This is highlighted in more detail in SI Appendix, Fig. S5I by looking at the allocation of the proteins Cds1, Cpt1, Opi3, Pgc1, and Pis1, which are respectively involved in the synthesis of CDP-diacylglycerol from phosphatidic acid (PA), the biosynthesis of phosphatidylcholine, the regulation of phosphatidylglycerol (PG) accumulation, and the synthesis of phosphatidylinositol (PI).
The differential distribution in allocation based on these proteins’ specific processes suggests three things: during fermentation, mitochondria are predominantly supporting cell proliferation by supplying important building blocks and metabolic intermediaries; secondly, proteins related to mitochondrial energy metabolism are most active after fermentation, with a steep increase during the diauxic shift that continues through to the ethanol phase. Finally, these observations confirm the dual activity of this organelle as a central hub for supplying biosynthetic precursors as well as for energy production (Fig. 6 D and E).
Aligned with this hypothesis, proteins involved in the structural constitution of mitochondrial ribosomes, and, to a lesser extent, the regulation of translation, are more abundant during the diauxic shift (Fig. 6B and SI Appendix, Fig. S5 E and G). This pattern can be justified by the fact that the core subunits of the respiratory-chain complexes are synthesized locally by mitochondrial ribosomes (48). Otherwise, we observed that some functional protein groups (Heme biosynthesis and proteins involved in the mitochondrial genome maintenance; SI Appendix, Fig. S5 C and H), maintain a more consistent level of expression in the glucose phase and thorough the diauxic shift to reach the ethanol phase, with a slight decrease in the allocation of the oxidative stress response-related protein during the ethanol phase (SI Appendix, Fig. S5F). Similarly, we find that despite the tight connection between the energetic state of mitochondria and its import machinery, changes in allocation toward proteins involved in mitochondrial protein import, including chaperones and proteases, are independent of these two metabolic phases (Fig. 6B and SI Appendix, Fig. S5K).
Until now, the regulatory dynamics of the mitochondria’s import system have remained unknown, with the complexes involved considered as being constitutionally expressed and maintaining a constant import capacity. Recently, however, it has been shown that the functionality of the whole TOM complex is regulated by cytosolic kinases (49), and not by changes in the protein allocation of the TOM import system, which we also observe here for the latter (SI Appendix, Fig. S5K). Furthermore, the analysis of the other major import machinery proteins SAM, TIM23, and TIM23 shows a similar trend (SI Appendix, Fig. S5K). This kinase involvement subsequently indicates a direct link between fermentative metabolism and the inactivation of Tom70 through phosphorylation (50). In fact, an increase in glucose concentration is known to trigger cAMP accumulation and cAMP-dependent protein kinase (PKA) activation. The kinase phosphorylates a binding pocket of Tom70 blocking its interaction with the chaperone protein carrying the precursor protein and consequently arresting the import (50). In addition to the analysis of the complexes constituting the mitochondrial import machinery, we have investigated the allocation of the chaperones and proteases, both having a crucial role in assisting the process of protein import into the mitochondria (SI Appendix, Fig. S5J). The overall distribution of these proteins throughout the three metabolic phases considered shows an increase during the diauxic shift and a quasiidentical allocation during the glucose phase and the ethanol phase. A more in-depth look at some of the key proteases and chaperones identified in this study shows again a higher, although not significant, allocation during the diauxic shift and a similar distribution during fermentative and respiratory growth (SI Appendix, Fig. S5J).
Finally, with respect to the dynamics between fusion and fission events, our results suggest fission to predominantly impact network structural remodeling during these three states in metabolism. We find a more substantial fraction of the mitochondrial proteome to be directed toward fission events during the glucose phase, relative to those related to fusion, in line with cells having a more fragmented mitochondrial network under this condition (51). Indeed, this change in proteome allocation toward fission-related proteins was shown to be significantly (P < 0.01) higher than the increase in fusion as cells began to respire following glucose exhaustion, confirming that the instigation of fission events is a predominant driver in the structural adaptation of mitochondria to a respiratory metabolism (Fig. 6F).
Discussion
In this study, we provide a thorough analysis of mitochondrial functions during yeast’s diauxic growth, encompassing the mitochondrial metabolic role coupled with their distinctive dynamic morphological adaptations. By using absolute quantitative subcellular proteomics alongside state-of-the-art optical imaging and lipidomics, we could track the transition of mitochondria from their role as a central biosynthetic hub during fast growth to a primary energy generator, focused on respiratory energy metabolism at lower growth rates. In fact, during fermentative growth, we observed an increase in proteins associated with the production of amino acids, sterols, and phospholipids, which are synthesized by the mitochondria and constitute essential sources of intermediaries for other critical cellular functions, such as translation and membrane biogenesis and proliferation. We found the shift toward respiratory metabolism to be contrastingly characterized by a reduction in biosynthetic functions in favor of processes linked to energy generation.
Through our approach, we combined the absolute proteomic data, obtained at different growth stages, together with biophysical properties of this organelle (volume, structure, and mass), which was crucial for parametrization and interpretation of the proteomic data. In fact, by analyzing the copy number of mitochondrial proteins per cell alone, without taking into account the changes in mitochondrial size and mass, we could only observe an increase in the mitochondrial proteome during the diauxic shift and especially in the ethanol phase, as a direct consequence of the expansion of the mitochondrial network (SI Appendix, Fig. S6A). Additionally, the structural rearrangements undertaken by the organelle could be clarified at the functional system-level. For example, we found a role of fission proteins that act as a control mechanism toward mitochondrial expansion during fermentative metabolism, which complements the regulation of this organelle’s expansion via posttranslational modifications, as speculated previously (51). As well as a decrease in fission events, changes in CL composition to higher unsaturation levels and an increase in ATP synthase appear to be cornerstone features of mitochondrial adaptation to metabolic constraints on the cell, underlining the inherently dynamic nature of this organelle (52).
The data presented in this study also suggest that as cellular metabolism switches from fermentation to respiration, the major structural and functional adaptations occurring at the mitochondrial level appear to be completed during the diauxic shift, with relatively fewer changes occurring in proteome allocation as cells shift from the diauxic shift to respiration. We, therefore, propose that the diauxic shift should not be defined as only a simple intermediate phase between the main metabolic stage of fermentation and respiration. Instead, it should be considered as a separate phase responsible for hosting significant reorganization events related to mitochondrial function, in particular, with respect to its role in early-stage adaptation of mitochondria to a respiratory metabolism. In contrast, at the cellular level, there shows to be a more gradual transition of the protein pool from fermentation to respiration with the diauxic shift constituting an intermediary metabolic state. This would indicate that the reorganization of yeast metabolism principally occurs during the diauxic shift and is initiated at the mitochondrial level.
In summary, this study highlights two major trends in the metabolic reorganization of mitochondria occurring during the yeast growth and the transition from fermentation to respiration. The first trend consists of the drastic up-regulation of energy-related processes occurring during the diauxic shift, which are continuously up-regulated to reach a further, although minor, increase during the ethanol phase. The second trend encompasses mitochondrial biosynthetic processes related to, for example, amino acid and phospholipids, as well as cofactor biogenesis. These biosynthetic functions are most prevalent during the glucose phase to sustain exponential cell growth.
In addition, we observed that processes like mitochondrial genome maintenance and oxidative stress response are up-regulated during the diauxic shift, followed by a decrease in the ethanol phase. Similarly, when looking at the overall protein allocation for functions associated with the protein imports (including chaperones and proteases), we also observe a significantly higher protein distribution during the diauxic shift. However, by looking closely at the individual proteins and complexes, the protein allocation during the three metabolic phases of relevant import complexes (i.e., TOM, TIM23, and SAM) appear evenly distributed, and the increase observed in the diauxic shift for individual proteases and chaperones are not significant.
In conclusion, by offering a highly quantitative insight into proteome allocation of mitochondria, this study provides an essential advancement in the systems analysis of eukaryotic subcellular dynamics. This can be used to complement experiments of genome-scale metabolic modeling that incorporate enzymatic constraints at the subcellular level (53). Furthermore, they constitute an important resource to highlight the mitochondrial role in bioenergetic and biosynthetic metabolism, as well as their involvement in the aging process and neurodegenerative diseases.
Materials and Methods
The details of the experimental model, media and growth conditions, high-performance liquid chromatography analysis of exometabolites, cell confocal microscopy, isolation of mitochondria, mitochondrial lipidomics, and the proteomics analysis are provided in SI Appendix, Supplementary Materials and Methods.
Data Availability.
The mass spectrometry proteomics datasets have been deposited in the ProteomeXchange Consortium via the Proteomics Identification Database partner repository (54) with the dataset identifiers PXD012802 and PXD012803. All data pertaining to this study, as well as any code used for analysis, are available from the corresponding author upon request.
Supplementary Material
Acknowledgments
We acknowledge The Proteomics Core Facility at the University of Gothenburg, Sweden. The Proteomics Core Facility is grateful to the Inga-Britt and Arne Lundbergs Forskningsstiftlese for the donation of the Orbitrap Fusion Tribrid mass spectrometry instrument. We acknowledge the Chalmers Mass Spectrometry Infrastructure at Chalmers University of Technology, Sweden, in particular, Maria Vizcaino, for support with the mitochondrial lipidomic analysis. This investigation was financially supported by the Knut and Alice Wallenberg Foundation and the Novo Nordisk Foundation.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. N.P. is a guest editor invited by the Editorial Board.
Data deposition: All mass spectrometry proteomics data reported in this paper have been deposited in the ProteomeXchange Consortium via the Proteomics Identification Database partner repository (dataset identifiers PXD012802 and PXD012803).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1918216117/-/DCSupplemental.
References
- 1.Nilsson A., Nielsen J., Metabolic trade-offs in yeast are caused by F1F0-ATP synthase. Sci. Rep. 6, 22264 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dickinson J. R., Schweizer M., Metabolism and Molecular Physiology of Saccharomyces cerevisiae (CRC Press, 2020). [Google Scholar]
- 3.Brauer M. J., Saldanha A. J., Dolinski K., Botstein D., Homeostatic adjustment and metabolic remodeling in glucose-limited yeast cultures. Mol. Biol. Cell 16, 2503–2517 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Radonjic M., et al. , Genome-wide analyses reveal RNA polymerase II located upstream of genes poised for rapid response upon S. cerevisiae stationary phase exit. Mol. Cell 18, 171–183 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Zampar G. G., et al. , Temporal system-level organization of the switch from glycolytic to gluconeogenic operation in yeast. Mol. Syst. Biol. 9, 651 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malina C., Larsson C., Nielsen J., Yeast mitochondria: An overview of mitochondrial biology and the potential of mitochondrial systems biology. FEMS Yeast Res. 18, foy040 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Westermann B., Bioenergetic role of mitochondrial fusion and fission. Biochim. Biophys. Acta 1817, 1833–1838 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Costenoble R., et al. , Comprehensive quantitative analysis of central carbon and amino-acid metabolism in Saccharomyces cerevisiae under multiple conditions by targeted proteomics. Mol. Syst. Biol. 7, 464 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galdieri L., Mehrotra S., Yu S., Vancura A., Transcriptional regulation in yeast during diauxic shift and stationary phase. OMICS 14, 629–638 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casanovas A., et al. , Quantitative analysis of proteome and lipidome dynamics reveals functional regulation of global lipid metabolism. Chem. Biol. 22, 412–425 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Murphy J. P., Stepanova E., Everley R. A., Paulo J. A., Gygi S. P., Comprehensive temporal protein dynamics during the diauxic shift in Saccharomyces cerevisiae. Mol. Cell. Proteomics 14, 2454–2465 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morgenstern M., et al. , Definition of a high-confidence mitochondrial proteome at quantitative scale. Cell Rep. 19, 2836–2852 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohlmeier S., Kastaniotis A. J., Hiltunen J. K., Bergmann U., The yeast mitochondrial proteome, a study of fermentative and respiratory growth. J. Biol. Chem. 279, 3956–3979 (2004). [DOI] [PubMed] [Google Scholar]
- 14.Renvoisé M., et al. , Quantitative variations of the mitochondrial proteome and phosphoproteome during fermentative and respiratory growth in Saccharomyces cerevisiae. J. Proteomics 106, 140–150 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Vögtle F.-N., et al. , Landscape of submitochondrial protein distribution. Nat. Commun. 8, 290 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meisinger C., Pfanner N., Truscott K. N., Isolation of yeast mitochondria. Methods Mol. Biol. 313, 33–39 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Schwanhäusser B., et al. , Global quantification of mammalian gene expression control. Nature 473, 337–342 (2011). [DOI] [PubMed] [Google Scholar]
- 18.Cherry J. M., et al. , Saccharomyces Genome Database: The genomics resource of budding yeast. Nucleic Acids Res. 40, D700–D705 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vowinckel J., Hartl J., Butler R., Ralser M., MitoLoc: A method for the simultaneous quantification of mitochondrial network morphology and membrane potential in single cells. Mitochondrion 24, 77–86 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmidt B., Wachter E., Sebald W., Neupert W., Processing peptidase of Neurospora mitochondria. Two-step cleavage of imported ATPase subunit 9. Eur. J. Biochem. 144, 581–588 (1984). [DOI] [PubMed] [Google Scholar]
- 21.Westermann B., Neupert W., Mitochondria-targeted green fluorescent proteins: Convenient tools for the study of organelle biogenesis in Saccharomyces cerevisiae. Yeast 16, 1421–1427 (2000). [DOI] [PubMed] [Google Scholar]
- 22.Martin J., Mahlke K., Pfanner N., Role of an energized inner membrane in mitochondrial protein import. Delta psi drives the movement of presequences. J. Biol. Chem. 266, 18051–18057 (1991). [PubMed] [Google Scholar]
- 23.Galloway C. A., Lee H., Yoon Y., Mitochondrial morphology-emerging role in bioenergetics. Free Radic. Biol. Med. 53, 2218–2228 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laporte D., Gouleme L., Jimenez L., Khemiri I., Sagot I., Mitochondria reorganization upon proliferation arrest predicts individual yeast cell fate. eLife 7, e35685 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rafelski S. M., et al. , Mitochondrial network size scaling in budding yeast. Science 338, 822–824 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miettinen T. P., Björklund M., Mitochondrial function and cell size: An allometric relationship. Trends Cell Biol. 27, 393–402 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Bryan A. K., Goranov A., Amon A., Manalis S. R., Measurement of mass, density, and volume during the cell cycle of yeast. Proc. Natl. Acad. Sci. U.S.A. 107, 999–1004 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pollak J. K., Munn E. A., The isolation by isopycnic density-gradient centrifugation of two mitochondrial populations from livers of embryonic and fed and starved adult rats. Biochem. J. 117, 913–919 (1970). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Balakrishnan R., et al. , YeastMine–An integrated data warehouse for Saccharomyces cerevisiae data as a multipurpose tool-kit. Database (Oxford) 2012, bar062 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Camougrand N. M., Mouassite M., Velours G. M., Guérin M. G., The “SUN” family: UTH1, an ageing gene, is also involved in the regulation of mitochondria biogenesis in Saccharomyces cerevisiae. Arch. Biochem. Biophys. 375, 154–160 (2000). [DOI] [PubMed] [Google Scholar]
- 31.Yoshida M., Muneyuki E., Hisabori T., ATP synthase–A marvellous rotary engine of the cell. Nat. Rev. Mol. Cell Biol. 2, 669–677 (2001). [DOI] [PubMed] [Google Scholar]
- 32.Lee S., Lee H., Yoo S., Kim H., Molecular insights into the m-AAA protease-mediated dislocation of transmembrane helices in the mitochondrial inner membrane. J. Biol. Chem. 292, 20058–20066 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stuart R., Insertion of proteins into the inner membrane of mitochondria: The role of the Oxa1 complex. Biochim. Biophys. Acta 1592, 79–87 (2002). [DOI] [PubMed] [Google Scholar]
- 34.Cogliati S., et al. , Mitochondrial cristae shape determines respiratory chain supercomplexes assembly and respiratory efficiency. Cell 155, 160–171 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Faelber K., et al. , Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1. Nature 571, 429–433 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arnarez C., Marrink S. J., Periole X., Molecular mechanism of cardiolipin-mediated assembly of respiratory chain supercomplexes. Chem. Sci. 7, 4435–4443 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rampelt H., Zerbes R. M., van der Laan M., Pfanner N., Role of the mitochondrial contact site and cristae organizing system in membrane architecture and dynamics. Biochim. Biophys. Acta Mol. Cell Res. 1864, 737–746 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Harner M. E., et al. , Aim24 and MICOS modulate respiratory function, tafazzin-related cardiolipin modification and mitochondrial architecture. eLife 3, e01684 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu Y., et al. , Assembly of the complexes of oxidative phosphorylation triggers the remodeling of cardiolipin. Proc. Natl. Acad. Sci. U.S.A. 116, 11235–11240 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Budin I., et al. , Viscous control of cellular respiration by membrane lipid composition. Science 362, 1186–1189 (2018). [DOI] [PubMed] [Google Scholar]
- 41.Basu Ball W., Neff J. K., Gohil V. M., The role of nonbilayer phospholipids in mitochondrial structure and function. FEBS Lett. 592, 1273–1290 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Joshi A. S., Thompson M. N., Fei N., Hüttemann M., Greenberg M. L., Cardiolipin and mitochondrial phosphatidylethanolamine have overlapping functions in mitochondrial fusion in Saccharomyces cerevisiae. J. Biol. Chem. 287, 17589–17597 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schuiki I., Schnabl M., Czabany T., Hrastnik C., Daum G., Phosphatidylethanolamine synthesized by four different pathways is supplied to the plasma membrane of the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1801, 480–486 (2010). [DOI] [PubMed] [Google Scholar]
- 44.Ashburner M., et al. , Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.The Gene Ontology Consortium , The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 47, D330–D338 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kanehisa M., Goto S., KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Metzl-Raz E., et al. , Principles of cellular resource allocation revealed by condition-dependent proteome profiling. eLife 6, e28034 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Derbikova K. S., Levitsky S. A., Chicherin I. V., Vinogradova E. N., Kamenski P. A., Activation of yeast mitochondrial translation: Who is in charge? Biochemistry (Mosc.) 83, 87–97 (2018). [DOI] [PubMed] [Google Scholar]
- 49.Harbauer A. B., Zahedi R. P., Sickmann A., Pfanner N., Meisinger C., The protein import machinery of mitochondria-a regulatory hub in metabolism, stress, and disease. Cell Metab. 19, 357–372 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Li J., Qian X., Hu J., Sha B., Molecular chaperone Hsp70/Hsp90 prepares the mitochondrial outer membrane translocon receptor Tom71 for preprotein loading. J. Biol. Chem. 284, 23852–23859 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Otera H., Ishihara N., Mihara K., New insights into the function and regulation of mitochondrial fission. Biochim. Biophys. Acta 1833, 1256–1268 (2013). [DOI] [PubMed] [Google Scholar]
- 52.Mishra P., Chan D. C., Metabolic regulation of mitochondrial dynamics. J. Cell Biol. 212, 379–387 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sánchez B. J., et al. , Improving the phenotype predictions of a yeast genome-scale metabolic model by incorporating enzymatic constraints. Mol. Syst. Biol. 13, 935 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Perez-Riverol Y., et al. , The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 47, D442–D450 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics datasets have been deposited in the ProteomeXchange Consortium via the Proteomics Identification Database partner repository (54) with the dataset identifiers PXD012802 and PXD012803. All data pertaining to this study, as well as any code used for analysis, are available from the corresponding author upon request.