Abstract
Our prior studies showed that dysfunctional plasmacytoid dendritic cells (pDCs) contribute to multiple myeloma (MM) pathogenesis. Specifically, pDC interactions with tumor and T/NK effector cells in the bone marrow (BM) milieu induce immune suppression and MM cell proliferation. Delineation of the mechanism(s) mediating pDC–MM–T–NK cell interactions will identify novel therapeutic targets to both enhance cytotoxicity and anti-MM immunity. Here, we utilized gene expression profiling (GEP) to show that pDC–MM interactions trigger upregulation of immunosuppressive tryptophan catabolic kynurenine (Kyn) pathway. In particular, we show that Kyn pathway enzyme kynurenine-3-monooxygenase (KMO) is upregulated during pDC–MM interactions. Using our coculture models of patient autologous pDC–T–NK–MM cells, we show that pharmacological blockade of KMO activates pDCs and triggers both MM-specific cytotoxic T-cell lymphocytes (CTL) and NK cells cytolytic activity against tumor cells. Furthermore, we show that simultaneous inhibition of Kyn pathway and immune checkpoint PD-L1 enhances antitumor immunity and cytotoxicity in MM. Our preclinical data therefore provide the basis for novel immune-based therapeutic approaches targeting Kyn metabolic pathway enzyme KMO, alone or in combination with anti-PD-L1 Ab, to restore anti-MM immune responses in MM.
Introduction
Multiple myeloma (MM) accounts for 10% of all hematologic malignancies and affects 30,200 new individuals annually in the United States, highlighting the need for identification of factors in the host-MM bone marrow (BM) microenvironment that facilitate MM progression and drug resistance [1, 2]. Our studies demonstrated a key role of plasmacytoid dendritic cells (pDCs) in the pathophysiology of MM [3, 4]. For example, analysis of MM patient BM biopsies with multiparametric flow cytometry and immunoflourescence assays showed increased infiltration of immunologically dysfuntional pDCs in MM BM [3]. Furthermore, interactions of pDCs with tumor cells and immune effector T and NK cells in the BM milieu confer immune suppression, protect tumor cells from therapy-induced cytotoxicity, and induce MM cell proliferation [3, 4]. Identification of tumor-promoting and immunosuppressive mechanism(s) triggered by pDC–MM interactions will help design interventional therapeutic strategies to restore anti-MM immunity and enhance MM cytotoxicity.
Prior studies have linked dysregulated kynurenine (Kyn) metabolic pathway to tumorigenesis and immunosuppression [5–8]. Kyn is a major component of tryptophan catabolism pathway. Specifically, indoleamine 2,3-dioxygenses (IDO1/IDO2) and tryptophan 2,3-dioxygenase (TDO) catalyze conversion of tryptophan into Kyn, which in turn, is further metabolized into 3-hydroxyanthranilic acid (3-HK), 3-HAA, and quinolinic acid via enzymatic activity of kynurenine-3-monooxygenase (KMO). Overexpression of KMO or IDO1 triggers depletion of tryptophan and hyperaccumulation of Kyn metabolites, which promote differentiation of naive T cells to immunosuppressive T regulatory cells [9–12]. Moreover, overexpression of Kyn pathway enzymes negatively mediates the bidirectional signaling axis between T cells and antigen-presenting cells, as well as stimulates tumor growth [10]. While the above findings show that Kyn pathway enzymes mediate tumor cell escape from innate and adaptive immune activity, the role of these enymes in MM, especially in the presence of accessory cells in the MM BM milieu, remains undefined.
In this study, we analyzed genetic changes in MM cells after coculture with pDCs using DNA microarray-based gene expression profiling (GEP) studies. pDC–MM interactions induce transcription of enzymes in the immunosuppressive Kyn metabolic pathway. To assess the functional significance of these findings, we used our pDC/MM, pDC/T, or pDC/NK cells coculture models, and show that targeting Kyn pathway enzyme KMO generates MM-specific cytotoxic T-cell lymphocyte (CTL) activity and NK cell cytotoxicity against MM cells. Moreover, the combination of KMO inhibitor with anti-PD-L1 Ab enhances antitumor immunity and cytotoxicity in MM.
Materials and methods
Cell culture and reagents
MM cells were cultured in 10% FBS plus RPMI-1640 medium supplemented with antibiotics. MM–pDCs were cocultured either in DCP-MM medium (Mattek Corp., Ashland, MA) or complete RPMI-1640 medium supplemented with IL-3 (Peprotech Inc., Rocky Hill, NJ, USA). CD3-PE/FITC/APC; CD4-FITC/PE or APC-Cy7; CD8-APC/FITC, CD56-PE; CD123-PE/PE-Cy5/FITC; and CD138-FITC/PE/APC were obtained from BD Biosciences (San Jose, CA). BDCA-2-FITC and CD11c-APC were obtained from Miltenyi Biotec (Auburn, CA); CD303-; CD304-; CD107a; and PD-L1-BV421 were purchased from Biolegend. All immunomagnetic separation kits were purchased from Miltenyi Biotec. The CellTrace Violet and CellTracker Green flow assay kits were obtained from Life Technologies (USA). Functional-grade PD-L1 blocking antibody (antihuman PD-L1, clone MIH1) was obtained from eBiosciences [4]. Ro 61–8048 [13] and INCB 024360 were purchased from Selleck Chemicals. WST-1 Cell Proliferation Reagent was purchased from Clontech Laboratories, Inc. (USA).
Purification of MM patient BM pDCs, T cells, NK cells, and CD138+ tumor cells
All studies using MM patient samples were performed following IRB-approved protocols at Dana-Farber Cancer Institute/Brigham and Women’s Hospital, Boston, USA. Informed consent was obtained from all patients in accordance with Helsinki protocol, and patient samples were de-identified prior to their use in experiments. pDCs were purified from both BM and peripheral blood mononuclear cells (PBMCs) using CD304 (BDCA-4/Neuropilin-1) microbeads kit (Miltenyi Biotec). The purity of pDCs (CD3-, CD14-, CD19-, CD20-, CD56-, CD11c-, and MHC-II/CD123/BDCA-2+) was confirmed, as previously described [3, 4]. The flow cytometry raw data were analyzed using FACS Diva (BD Biosciences) and FlowJo (Tree Star Inc., USA). MM patient cells were purified (>95% purity) by positive selection using CD138 Microbeads kit. CD8+ T cells and NK cells were purified using negative selection immunomagnetic separation techniques, as previously described [4].
DNA microarray analysis
MM cells were cocultured with pDCs (pDC:MM; 1:5 ratio) for 72 h; MM cells were then separated from the cocultures using CD138 microbeads, and then subjected to gene expression analysis using HGU133 plus 2.0 plus Affymetrix oligonucleotide microarrays. The raw data CEL files were obtained using Affymetrix Microarrays Suite 5.0 software. GeneChip 5.0 (Affymetrix, Santa Clara, CA) was utilized to scan, quantify, and analyze the scanned image. Gene expression patterns for MM cells cultured with or without pDCs were compared, and heat maps were generated (>1.5-fold change in transcript levels was considered significant, CI > 95%). The raw microarray data are provided at the website Gene Expression Omnibus. Accession Number “GSE17407”.
Cell viability assays, flow cytometry analysis, and transient transfections
Cell viability was assessed by MTT or WST assay, as previously described [4]. MM cells were cocultured with pDCs for 24 h; cells were then stained with KMO-FITC and CD138 Abs, followed by multicolor flow analysis to quantify KMOhi MM cell populations. MM.1S cells were transfected with KMO-siRNA using TransIT-X2 (Mirus Bio) transfection system.
Cytotoxic T lymphocyte (CTL) and NK cell activity assays
MM patient BM CD8+ T or NK cells were cocultured with autologous pDCs at 1:10 (pDC:T/NK) ratio, in the presence or absence of KMO inhibitor Ro 61–8048 (ref.13) (100 nM) or IDO1 inhibitor [14] (100 nM) for 3 days; pre-stained MM cells were added for 24 h (E/T ratio 10:1, T:MM), followed by quantification of viable MM cells by FACS. NK-cell mediated cytotoxicity was assessed using flow cytometry-based CFSE-stained MM cell lysis assays and CD107a degranulation assays, as previously described [15].
Statistical analysis
Student’s t test was utilized to derive statistical significance. The minimal level of significance was p < 0.05 (Graph Pad PRISM version 6, La Jolla, California, USA).
Results and discussion
pDC–MM interactions trigger transcription of kynurenine (Kyn) pathway enzyme kynurenine-3-monooxygenase (KMO) in MM cells
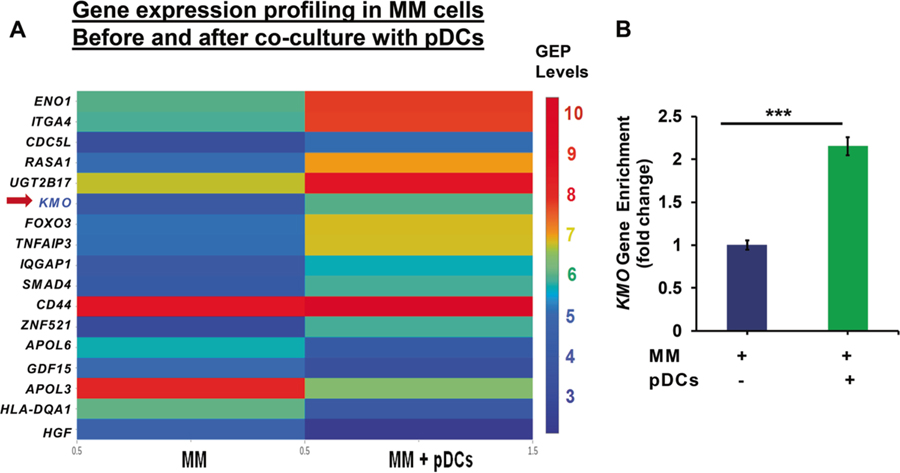
MM.1S cells were cocultured with MM patient pDCs (pDC: MM; 1:5 ratio) for 72 h; MM.1S cells were then separated from the cocultures using CD138 microbeads, and subjected to gene expression analysis using HGU133 plus 2.0 plus Affymetrix oligonucleotide microarrays. Expression patterns for MM cells cultured with or without pDCs were compared, and a heat map was generated (>1.5-fold change in transcript levels was considered significant, CI > 95%). The results demonstrate that pDCs induce alterations in transcription of several genes in MM cells, including those in immunosuppressive metabolic pathways (Fig. 1a). For example, pDCs upregulate KMO, a key molecule in the immunosuppressive kynurenine–tryptophan catabolic pathway, in MM cells (Fig. 1a, b; 2.153-fold increased expression versus MM alone; n = 3; CI > 95%). Although our data show an increase in other metabolic pathway enzymes (e.g., ENO1, CD44, or ITGA4/CD49d), we focused on examining the role of KMO since (1) a maximal induction of KMO was noted during pDC–MM interactions versus other molecules; (2) the role of KMO is undefined in the context of MM–pDC interactions; (3) the assessment of functional significance of KMO is feasible due to availability of a specific pharmacological inhibitor; and (4) other reports provide evidence for a role of KMO in inhibiting T/NK cells antitumor activity and promoting tumor progression [9–12]. Moreover, gene expression profile analysis of KMO from a publicly available GEP database showed that the patients with low KMO expression have increased survival rate compared with patients with high KMO expression (Supplementary Fig. S1). Importantly, KMO levels are the highest in patients with relapsed/refractory MM compared with normal healthy donor plasma cells, patients with newly diagnosed MM, or individuals with monoclonal gammopathy of underdetermined significance (Supplementary Fig. S2). Chromosome 1 abnormalities play an important role in the pathogenesis of MM; and importantly, KMO is located on chromosome 1: band: 1q43: 241.53–241.6 Mb. We therefore analyzed the MMRF CoMMpass data focusing on chromosome 1. In the absence of 1q21 gain, KMO expression (RNA: FPKM:-fragments per kilobase of transcript per million mapped reads) is upregulated from baseline to disease progression state. Interestingly, KMO expression (RNA: FPKM) is 1.4 times higher in patients with trisomy of chr1 versus patients with normal 1q21 (Supplementary Fig. S3).
Fig. 1.
Gene expression profiling of MM cells in the presence or absence of pDCs. a DNA microarray analysis: MM cells were cocultured with pDCs for 48 h; separated using anti-CD138 antibody by flow sorting, and harvested. Poly RNA was subjected to microarray analysis using HG-U133 plus 2.0 plus Affymetrix chip [3]. Data processing: the CEL files were obtained using Affymetrix Microarrays Suite 5.0 software. GeneChip 5.0 (Affymetrix, Santa Clara, CA) was utilized to scan, quantify, and analyze the scanned image. GeneChip software automatically calculated intensity values for each probe cell, and marked a presence or absence call for each mRNA. Algorithms in the software used probe cell intensities to calculate an average intensity for each set of probe pairs representing a gene, which correlates with the amount of mRNA. Gene expression patterns for MM cells cultured in the presence versus absence of pDCs were compared, and a heat map was generated (>1.5-fold change in transcript is considered significant, CI > 95%). The expression profile of KMO transcript is shown. b Quantification of KMO expression: Normalized KMO gene enrichment in pDC–MM cell coculture versus MM cells alone is presented [2.15-fold upregulation; n = 3; CI > 95%]
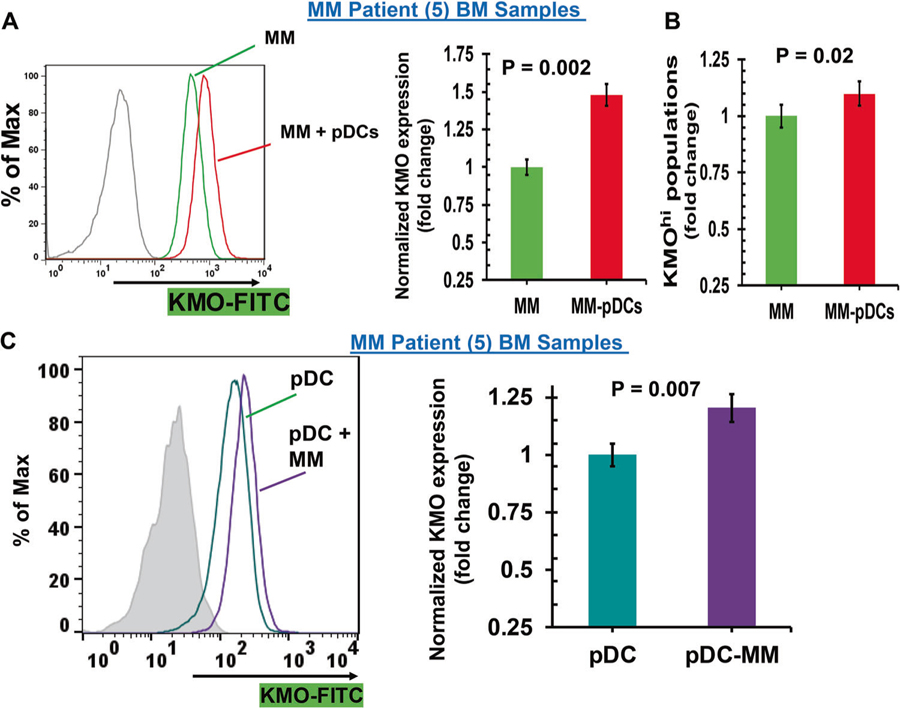
To evaluate the biologic significance and potential clinical relevance of our microarray data, we examined the alterations in KMO expression levels using multicolor flow cytometry in MM patient autologous tumor-pDCs coculture systems. Results show that both tumor cells and patient pDCs constitutively express KMO; and importantly, that pDC–MM cell coculture further increases KMO expression in MM cells (2–3-fold versus MM; = 0.006) (Fig. 2a, scatter plot and bar graph). Moreover, cocultures of pDCs and MM cells led to a significant increase in KMO+ MM cell populations (~1.2-fold; p < 0.05) (Fig. 2b). As for MM cells, an increase in KMO levels was observed in pDCs after coculture with MM cells (MFI: 1.2-fold; p = 0.007) (Fig. 2c, scatter plot and bar graph). Examination of IDO1, a molecule upstream of KMO in Kyn pathway, showed similar results (data not shown). pDCs from normal healthy donors also increase KMO expression in MM cells, albeit to a much lesser extent than MM patient pDCs (Supplementary Fig. S4). Furthermore, coculture of MM cells with patient-BM CD138-negative cells also increases KMO expression in MM cells (Supplementary Fig. S5). In addition, forced overexpression of KMO in MM cells further increases pDC-triggered MM cell growth (Supplementary Fig. S6). Together these findings suggest activation of the Kyn pathway during pDC–MM interactions.
Fig. 2.
Modulation of KMO expression during pDC–MM cell interactions (a) pDCs were cocultured with autologous patient MM cells for 24 h, followed by multicolor flow analyses to assess the pDC-induced change in KMO expression on MM cells. CD138+ MM cells were examined using flow cytometry, and median fluorescence intensity (MFI) of KMO expression was determined using anti-KMO Ab conjugated to FITC, both in the presence or absence of pDCs. Left panel: representative histograms shows KMO expression in MM cells cultured in the presence (red) and absence (green) of pDCs. [Black histogram: isotype control Ab]. Right panel: normalized KMO expression in MM cells cultured in the presence and absence of pDCs. Data was quantified from the histogram analyses shown in the left panel (mean ± SD; p < 0.05; data obtained from analysis of five MM patient BM samples). b pDCs were cocultured with autologous patient MM cells for 24 h, followed by multicolor flow analysis to determine the pDC-induced change in the KMOhi MM cell population [mean ± SD; p < 0.05; data obtained from five MM patient BM samples]. c MM patient BM-pDCs were cocultured with autologous MM cells for 24 h, followed by multicolor flow analyses to determine change in KMO expression on pDCs. pDCs (CD304/CD123/CD303+) were examined by flow analysis, and median fluorescence intensity (MFI) was determined for KMO expression on pDCs, using anti-KMO-FITC Ab, both in the presence or absence of MM cells. Left panel: representative histograms shows KMO expression in MM–BM–pDCs cultured in the presence (violet) and absence (blue–green) of MM cells. [Shaded black histogram: isotype control Ab]. Right panel: normalized KMO expression in MM-patient pDCs cultured in the presence or absence of patient MM cells. Data was quantified from the histogram analyses shown in the left panel (mean ± SD; p < 0.05; data obtained from five MM patient BM samples)
Inhibition of KMO activates MM pDCs and increases T cell proliferation
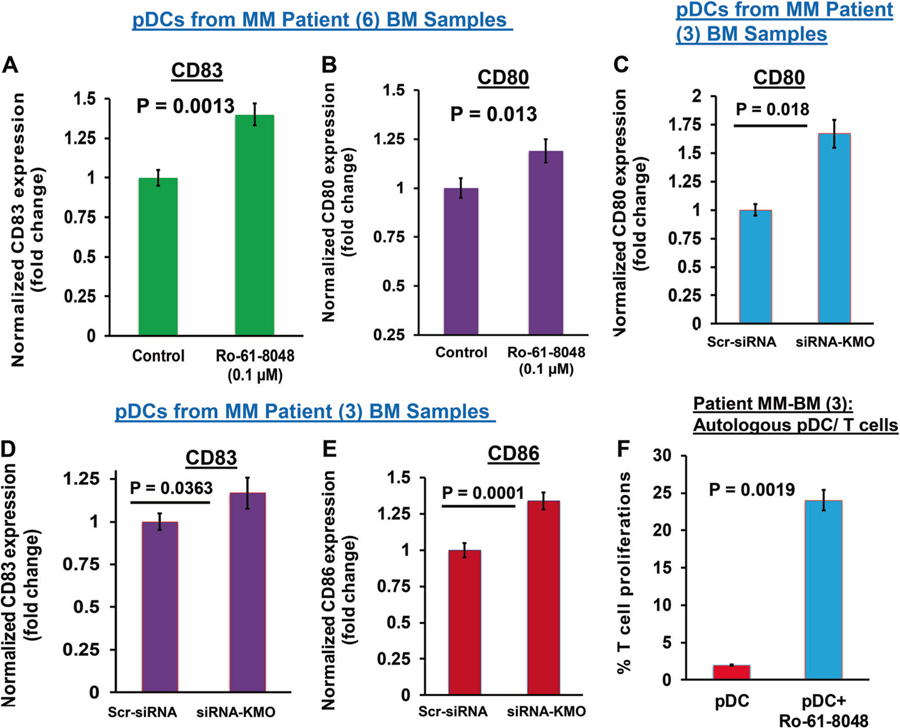
We have previously shown that pDCs from MM patients have significantly impaired ability to induce T cell proliferation versus normal pDCs [3, 4, 15–17]. In our studies, elevated expression of immune checkpoint PD-L1 and toll-like-receptor (TLR-9/TLR-7) negatively regulate pDCs immune function; conversely, both TLR9/TLR7 agonists and anti-PD-L1 Abs activate pDCs and restore the ability of pDCs to induce T cell proliferation [4, 15, 17]. Our current finding that pDC–MM cell interactions upregulate KMO in pDCs, coupled with other studies reporting immunosuppressive role of KMO, implicate involvement of Kyn pathway in dysregulated pDCs immune response in MM. We therefore next assessed whether KMO inhibition affects maturation and immune function of pDCs in MM. For these studies, we treated pDCs from MM patients with nontoxic concentrations of a specific inhibitor of KMO Ro 61–8048 (0.1–0.2 µM) [13], and examined changes in activation/maturation markers on pDCs. Ro-61–8048 triggers upregulation of CD83 and CD80 on pDCs (Fig. 3a, b, respectively).
Fig. 3.
KMO blockade activates pDCs and enhances pDC-triggered T cell proliferation (a, b) MM patient pDCs were treated with Ro-61–8048 (100 nM) for 24 h, followed by multicolor staining and flow cytometric analysis of pDC activation/maturation markers CD83 and CD80. Fold change in Ro-61–8048-treated versus untreated is presented (mean ± SD; p < 0.05; Data obtained from six MM patient samples). c–e MM patient pDCs were transfected with KMO-siRNA using TransIT-X2 transfection reagent as per manufacturer’s protocol (Mirus Bio, USA). The KMO-siRNA-transfected pDCs were cultured in complete DCP medium for 48 h, and then stained with fluorophore-conjugated Abs against CD80, CD83, and CD86, followed by flow cytometry analysis. Bar graph: quantification of MFI for each marker, and data is shown as fold change between scr-siRNA- versus KMO-siRNA-transfected pDCs. Analysis was performed using three MM patient samples (mean ± SD; CD80: p = 0.018; CD83: p = 0.0363; CD86: p = 0.0001). Student’s t test was utilized to derive statistical significance. The minimal level of significance was p < 0.05 (GraphPad PRISM version 6, La Jolla, California, USA). f pDCs from MM patients were cocultured with autologous T cells at 1:10 (pDC:T) ratio in the presence or absence of Ro 61–8048 (100 nM) for 3 days, and viable CD8+ T cells were quantified using CellTrace Violet-Cell Proliferation Kit by FACS (mean ± SD; P < 0.05, three MM patient BM samples)
To confirm our findings obtained from biochemical inhibition, we performed genetic studies using RNA interference strategy: MM patient pDCs or MM cells were transfected with KMO-siRNA, followed by either flow cytometric analysis of pDC maturation markers or coculture experiments for measuring pDC-induced MM cell growth. Genetic knockdown (KD) of KMO in MM patient pDCs significantly increases CD80, CD83, and CD86 expressions in pDCs, suggesting that (similar to biochemical inhibition) KMO-KD triggers activation of pDCs (Fig. 3c–e). As a control, transfection with siRNA-KMO significantly reduced KMO levels (Supplementary Fig. S7A). In cocultures with pDCs, KMO-siRNA-transfected MM cells showed significantly reduced growth versus scr- siRNA or non-transfected MM cells (Supplementary Fig. 7B). Importantly, KMO blockade restores the ability of MM patient pDCs to induce proliferation of autologous T cells (Fig. 3f). Together, these findings show that blockade of KMO induce pDCs activation and increases pDCs-mediated autologous T cell response.
KMO blockade triggers pDC-induced T and NK cell-mediated anti-MM activity
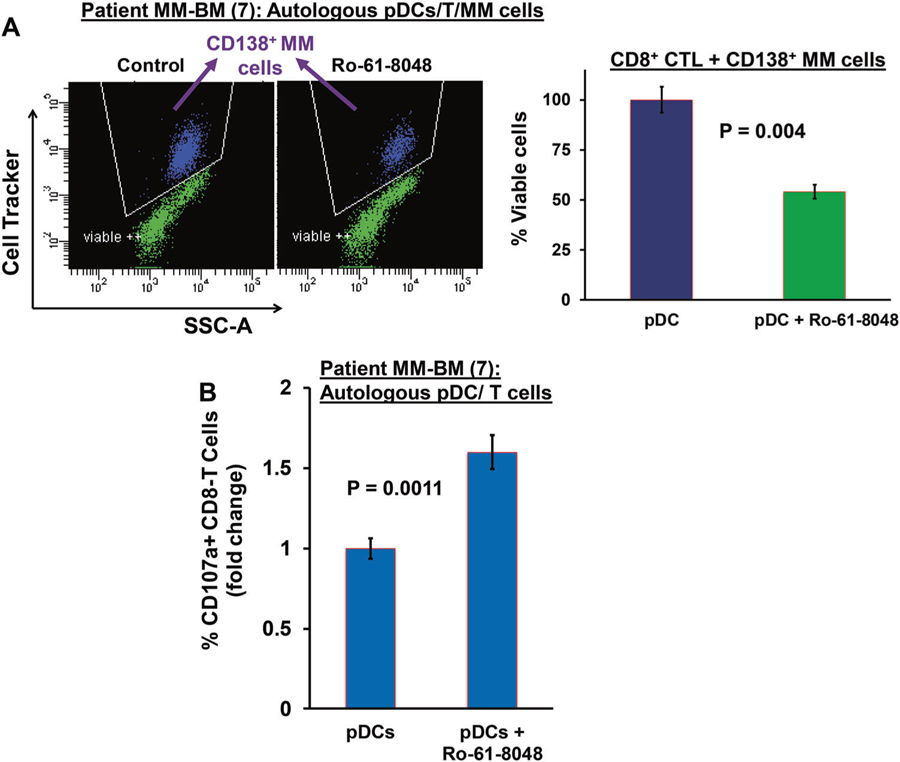
Recent studies showed promising therapeutic potential of KMO blockade in cancers and many immunological diseases [18–22]. To determine whether pharmacological inhibition of KMO alters pDC-mediated T and NK cell anti-MM immune responses in the MM-BM milieu, we next utilized our coculture models of host-MM tumor cells, pDCs, T cells, and NK cells. These coculture models of patient pDCs, T cells, or NK cells with autologous MM cells have been useful in validating immune checkpoint PD-L1 as therapeutic target, as well as supporting ongoing clinical trials of anti-PD-L1 Abs in MM [3, 4, 16, 23, 24]. Using these models, we next examined whether KMO blockade can trigger the generation of MM-specific pDC-induced cytotoxic T lymphocytes ex vivo. Freshly isolated CD8+ T cells from MM patient BM were cultured with autologous pDCs in the presence of Ro-61–8048 or DMSO control for 3 days; cells were then washed to remove drug and resuspended in complete medium. Autologous MM cells were then added, and viable MM cells quantified after 24 h coculture. As shown in Fig. 4a, Ro-61–8048 triggers robust MM-specific CD8+ CTL activity (~2.0-fold versus control; p = 0.009), evidenced by decreased viable MM cell populations. In concert with these findings, Ro-61–8048 increases CD107a+ degranulated CD8 + CTLs (Fig. 4b, CD107a + CD8 + T:1.6 fold Ro-61–8048-treated versus untreated; p = 0.0011). Among seven patient samples analyzed, five patients had MM resistant to bortezomib, dexamethasone, and lenalidomide therapies; and two patients had newly diagnosed untreated MM.
Fig. 4.
KMO inhibition triggers pDC-induced MM-specific CD8 + CTLs. a MM patient BM CD8+ T cells were cocultured with autologous pDCs (n = 7) at 1:10 (pDC: T) ratio in the presence or absence of Ro 61–8048 (100 nM) for 3 days. After washing to remove Ro 61–8048, cells were cultured with autologous MM cells prestained with CellTracker Violet (T/MM; 10:1 ratio) 24 h, followed by 7-AAD staining and quantification of CTLs-mediated MM cell lysis by FACS. Left panel: representative FACS scatter plot showing the decrease in number of viable CellTracker-positive MM cells. Right panel: bar graph shows quantification of CD8+ CTLs-mediated MM cell lysis, reflected in CD138 + MM cell viability, using data obtained in left panel (data obtained from seven MM patient BM samples; mean ± SD; p < 0.05). b MM patient pDCs and autologous T cells (1:10 pDC/T ratio) were treated with DMSO control or Ro 61–8048 (100 nM), followed by degranulation assay to assess surface CD107a+ T cells by multiparameter flow cytometry. The plot shows percentage of surface CD107a+ T cells, indicating degranulated CTLs (data obtained from seven MM patient BM samples; mean ± SD; p < 0.05)
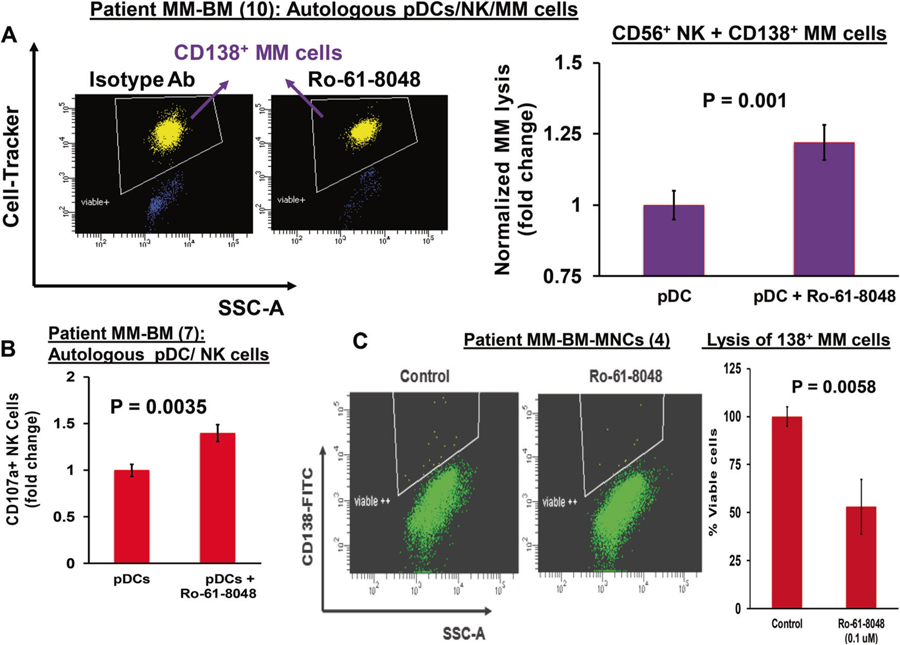
Besides pDCs, NK cells in MM also show reduced immune function [25, 26]. We therefore next examined whether KMO inhibition alters anti-MM activity of NK cells using our coculture model of autologous pDCs–NK cells–MM cells. Freshly purified NK cells from MM patient BM were cocultured with autologous pDCs in the presence of Ro-61–8048 or DMSO control for 3 days; cells were then washed to remove drug and resuspended in complete medium. Autologous MM cells were then added, and percent MM cell lysis was quantified at 24 h. Ro-61–8048 significantly increased NK cell cytolytic activity against MM cells (Fig. 5a, scatter plot and bar graph). Moreover, Ro-61–8048 increases CD107a+ degranulated NK cells (Fig. 5b). Our data therefore show that elevated KMO in pDCs contributes to immunosuppressive pDCs–NK cell interactions; and conversely, that inhibition of KMO restores anti-MM activity of NK cells.
Fig. 5.
KMO inhibition triggers pDC-induced NK cell-mediated lysis of MM cells (a) MM patient BM NK cells were cocultured with autologous pDCs at 1:10 (pDC:NK) ratio in the presence or absence of Ro 61–8048 (100 nM) for 3 days. After washing to remove Ro 61–8048, cells were cultured with autologous MM cells prestained with CellTrace violet (10:1 NK:MM ratio) for 24 h, followed by 7-AAD staining and quantification of MM cell lysis by FACS [pDC:NK ratio; 1:10; effector/target; 10:1]. Left panel: representative FACS scatter plot showing a decrease in number of viable CellTrace Violet-positive MM cells. Right panel: bar graph shows quantification of NK-mediated MM cell lysis using data obtained in left panel. The fold change was obtained after normalization with control, and normalized MM cell lysis in the Ro 61–8048-treated versus untreated is presented (data obtained from ten MM patient BM samples; mean ± SD; p < 0.05). b MM patient pDCs and autologous NK cells (1:10 pDC/NK ratio) were treated with DMSO control or Ro 61–8048 (100 nM), followed by degranulation assay to assess surface CD107a expression in CD3−/CD56+ NK cells by multiparameter flow cytometry. The plot shows percentage of surface CD107a+ NK cells, indicating degranulated cytotoxic NK cells (data obtained from seven MM patient BM samples; mean ± SD; p < 0.05). c MM patient total BM-MNCs were treated with Ro 61–8048 (100 nM) for 2 days, and multicolor flow analysis was utilized to assess MM cell lysis. CD138+ MM cells were selected based on their staining of CD138-FITC Ab and quantified. Left panel: representative FACS scatter plot showing a decrease in number of viable FITC-positive MM cells. Right panel: bar graph shows quantification of CD138+ MM cells in left panel. The fold change was obtained after normalization with control data, and the bar graph is presented as percentage of viable cells in the presence and absence of Ro-61–8048. (Data obtained from four MM patient BM samples; mean ± SD; p < 0.05)
We next further examined whether KMO inhibitor Ro-61–8048 retains its ability to generate anti-MM activity even in the presence of accessory cells in the BM milieu. Specifically, we evaluated its activity using total BM-MNCs from seven MM patients. As shown in Fig. 5c, KMO triggers a significant decrease in viable MM cells in total BM-MNCs (50% ± 6% decrease in viable CD138+ cells; p < 0.0035). Importantly, these findings show that inhibition of Kyn pathway enzyme KMO induces anti-MM activity even in the MM–BM microenvironment. This anti-MM effect of Ro-61–8048 in BM-MNCs likely reflects both abrogation of Kyn-pathway-mediated immune suppressive signaling triggered by pDCs–MM cell interactions, as well as simultaneous activation of pDC-induced CTL-and NK cell-mediated cytotoxicity against MM cells. Together, our results show that blockade of KMO activates pDCs and restores both pDC-induced T and NK cell-mediated cytolytic activity against MM cells.
Combination of anti-PD-L1 Ab and Ro-61–8048 enhances T cell-and NK cell-mediated MM-specific cytotoxic activity
Our previous studies showed pDC–MM cell interactions upregulate immune-inhibitory molecules including programmed cell death ligand-1 (PD-L1) on pDCs and MM cells, which further blunt the PD-1-expressing T- and NK-cell responses [3, 4, 16, 27, 28]. These data, together with our present findings indicate that pDC–MM interactions induce two distinct immunosuppressive signaling pathways: one via induction of immune checkpoints, and the second via the kyn metabolic pathway. We therefore hypothesized that simultaneous blockade of both immune checkpoint PD-L1 and Kyn pathway enzyme KMO may be more potent in restoring anti-MM immunity than either alone. We specifically examined the anti-MM immune response to combined blockade of KMO and PD-L1. Freshly isolated MM patient BM CD8+ T cells were cocultured with autologous pDCs at 1:10 (pDC: T) ratio in the presence of anti-PD-L1 Ab (5 µg/ml), Ro-61–8048 (100 nM), or PD-L1 Ab plus Ro-61–8048 for 3 days; and then evaluated in CTL assays against autologous MM cells (E:T ratio 10:1, T:MM), as in Fig. 4. The combination of Ro-61–8048 and anti-PD-L1 Ab triggers more robust autologous MM-specific CD8 + CTL activity than anti-PD-L1 Ab alone (1.5–2.0-fold increase in combination regimen versus anti-PD-L1 Ab alone) (Fig. 6a). Moreover, this combination regimen induces cytotoxicity against allogeneic HLA-A2 + U266 cells (data not shown). Finally, the combination of Ro-61–8048 plus anti-PD-L1 Ab triggered significantly increased NK-cell mediated MM cell lysis than either Ro-61–8048 (p = 0.045) or anti-PD-L1 Ab (p = 0.001) alone (Fig. 6b). Consistent with our data, studies in other cancers have also shown promising antitumor activity of anti-PD-L1 Ab and an inhibitor of tryptophan-Kyn pathway enzyme IDO1 [29].
Fig. 6.
Combination treatment with anti-PD-L1 Ab and Ro-61–8048 enhances T cell-and NK cell-mediated MM-specific cytotoxic activity (a) MM patient BM CD8+ T cells (n = 7) were cocultured with autologous pDCs (pDC:T; 1:10 ratio) in the presence of anti-PD-L1 Ab (5 µg/ml), Ro-61–8048 (100 nM), or Ro-61–8048 plus anti-PD-L1 Ab for 3 days. After washing to remove drugs, cells were cultured with autologous MM cells prestained with CellTracker Violet (T/MM; 10:1 ratio) for 24 h, followed by 7-AAD staining and quantification of CTLs-mediated MM cell lysis by FACS. Left panel: representative FACS scatter plot shows a decrease in number of viable CellTracker Violet-positive MM cells. Right panel: bar graph shows quantification of CD8+ CTLs-mediated MM cell lysis, reflected in % viable MM cells, using data obtained in left panel. Percentage of viable MM cells for each treatment versus control (isotype Ab) is presented (data obtained from seven MM patient BM samples; mean ± SD; p < 0.05). b NK cells from MM patient BM (n = 10) were cocultured with autologous pDCs (1:10 pDC: NK ratio) in the presence anti-PD-L1 Ab (5 µg/ml), Ro-61–8048 (100 nM), or Ro-61–8048 plus anti-PD-L1 Ab for 3 days. After washing to remove drugs, cells were cultured with autologous MM cells pre-stained with CellTracker Violet (10:1 NK: MM ratio) for 24 h, followed by 7-AAD staining and quantification of MM cell lysis by FACS. Left panel: representative FACS scatter plot shows the decrease in number of viable CellTracker Violet-positive MM cells. Right panel: bar graph shows quantification of NK-mediated MM cell lysis using data obtained in left panel. The fold change was obtained after normalization with control, and normalized MM cell lysis in each treated sample versus control (isotype Ab) is presented (data obtained from ten0 MM patient BM samples; mean ± SD; p < 0.05)
The mechanism(s) whereby KMO expression is increased during pDC–MM interaction remains to be elucidated in ongoing studies. An earlier study [30] indicated that an increase in KMO substrate Kyn or cytokines such as IL-6 or TNF-α may increase KMO levels. Our prior study showed that pDC–MM interactions induce both IL-6 and TNF-α levels [3]. Moreover, both IL-6 and TNF are elevated in the MM BM milieu and promote MM cell growth and survival [31, 32]. It is therefore likely that these cytokines upregulate KMO in pDCs and MM cells, which in turn enhances conversion of Kyn into immunosuppressive metabolites 3-HAA and 3-HK in the MM BM milieu.
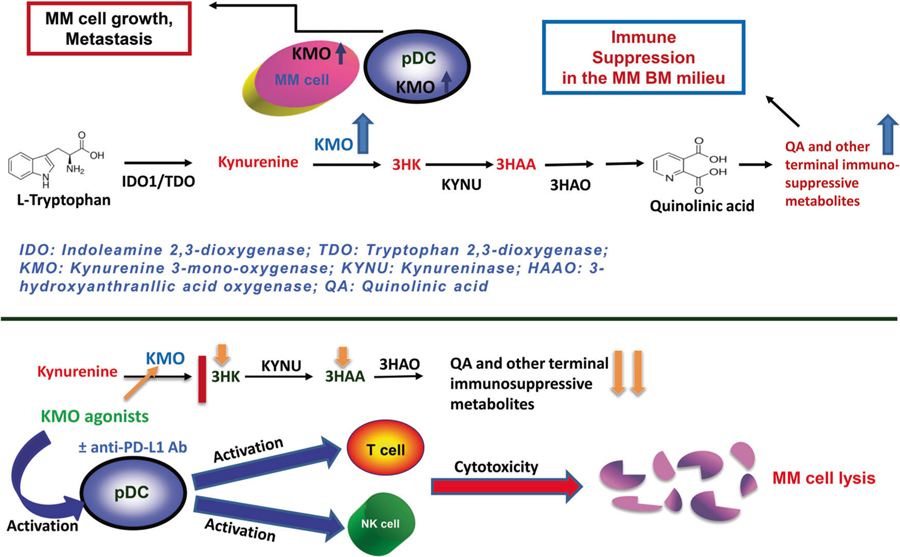
Collectively, we show that (1) pDC–MM interactions induce transcription of Kyn pathway enzyme KMO in MM cells; (2) both pDCs and MM cells express KMO, and pDC–MM interactions further increase KMO levels in both cells; (3) blockade of KMO using a biochemical inhibitor Ro-61–8048 activates MM patient pDCs and enhances their ability to trigger autologous T cell proliferation; (4) Ro-61–8048 triggers MM-specific CD8+ CTL activity, as well as NK-cell-mediated cytolytic activity against autologous MM cells; and (5) the combination of Ro-61–8048 and anti-PD-L1 Ab triggers more potent MM-specific CD8+ CTL and NK-cell cytolytic activity against autologous tumor cells than either agent alone. Overall, our preclinical data provides the basis for novel immune-based therapeutic approaches (Fig. 7) targeting Kyn pathway enzyme KMO, either alone or in combination with anti-PD-L1 Ab, to enhance MM cytotoxicity and restore anti-MM immunity
Fig. 7.
Schematic representation depicting the role of KMO in MM (a) pDCs trigger growth, survival, and drug resistance in MM cells. Tryptophan pathway: tryptophan is degraded into kynurenine (KYN) (and downstream metabolites 3 hydroxykynurenine [3HK], 3-hydroxyanthranilic acid [3-HAA], and quinolinic acid [QA]). Tryptophan catabolic Kyn pathway is upregulated in MM, as evidenced by high KMO levels in MM cells and pDCs. Importantly, pDCs–MM interactions further upregulate KMO in MM cells, resulting in accumulation of terminal immunosuppressive metabolites mediating immunosuppression of MM-T and NK cells in the MM BM milieu. b Therapeutic potential of KMO as an immuno-metabolic target: treatment of MM-pDCs by KMO antagonists, either alone, or in combination with anti-PD-L1 Abs, generates anti-MM-specific CD8 + CTL activity, as well as enhances NK-cell-mediated anti-MM cell cytotoxicity
Supplementary Material
Acknowledgements
The grant supports for this investigation were provided by “Dr Miriam and Sheldon Adelson Medical Research Foundation” and by the National Institutes of Health Specialized Programs of Research Excellence (SPORE) grant P50100707, R01CA207237, and RO1 CA050947. KCA is an American Cancer Society Clinical Research Professor.
Footnotes
Supplementary information The online version of this article (https://doi.org/10.1038/s41375–019-0558-x) contains supplementary material, which is available to authorized users.
Conflict of interest KCA is on Advisory board of Celgene, Millenium-Takeda, Gilead, Janssen, and Bristol Myers Squibb, and is a Scientific Founder of Oncopep and C4 Therapeutics. DC is a consultant to Stemline Therapeutic, Inc., and equity owner in C4 Therapeutics. The remaining authors declare no conflict of interest.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Anderson KC. Multiple myeloma. Hematol Oncol Clin North Am. 2014;28:xi–xii. [DOI] [PubMed] [Google Scholar]
- 2.An G, Acharya C, Feng X, Wen K, Zhong M, Zhang L, et al. Osteoclasts promote immune suppressive microenvironment in multiple myeloma: therapeutic implication. Blood. 2016;128: 1590–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chauhan D, Singh AV, Brahmandam M, Carrasco R, Bandi M, Hideshima T, et al. Functional interaction of plasmacytoid dendritic cells with multiple myeloma cells: a therapeutic target. Cancer Cell. 2009;16:309–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ray A, Das DS, Song Y, Richardson P, Munshi NC, Chauhan D, et al. Targeting PD1-PDL1 immune checkpoint in plasmacytoid dendritic cell interactions with T cells, natural killer cells and multiple myeloma cells. Leukemia. 2015;29:1441–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jacobs KR, Castellano-González G, Gilles J, Guillemin GJ, Lovejoy DB. Major developments in the design of inhibitors along the kynurenine pathway. Curr Med Chem. 2017;24:2471–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith JR, Jamie JF, Guillemin GJ. Kynurenine-3-mono-oxygenase: a review of structure, mechanism, and inhibitors. Drug Discov Today. 2016;21:315–24. [DOI] [PubMed] [Google Scholar]
- 7.Platten M, von Knebel Doeberitz N, Oezen I, Wick W, Ochs K. Cancer immunotherapy by targeting IDO1/TDO and their downstream effectors. Front Immunol. 2015;5(Article673):1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheong JE, Sun S. Targeting the IDO1/TDO2–KYN–AhR pathway for cancer immunotherapy challenges and opportunities. Trends Pharmacol Sci. 2018;39:307–25. [DOI] [PubMed] [Google Scholar]
- 9.Amaral M, Levy C, Heyes DJ, Lafite P, Outeiro TF, Giorgini F, et al. Structural basis of kynurenine 3-monooxygenase inhibition. Nature. 2013;496:382–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jin H, Zhang Y, You H, Tao X, Wang C, Jin G, et al. Prognostic significance of kynurenine 3-monooxygenase and effects on proliferation, migration, and invasion of human hepatocellular carcinoma. Sci Rep. 2015;5:10466 10.1038/srep10466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Le Maux Chansac B, Missé D, Richon C, Vergnon I, Kubin M, Soria JC, et al. Potentiation of NK cell-mediated cytotoxicity in human lung adenocarcinoma: role of NKG2D-dependent pathway. Int Immunol. 2008;20:801–10. [DOI] [PubMed] [Google Scholar]
- 12.Murakami Y, Hoshi M, Imamura Y, Arioka Y, Yamamoto Y, Saito K. Remarkable role of indoleamine 2,3-dioxygenase and tryptophan. Mediatiors Inflamm. 2013;2013:391984 10.1155/2013/391984. 9 pages [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao J, Yao L, Xia T, Liao X, Zhu D, Xiang Y. Biochemistry and structural studies of kynurenine 3-monooxygenase reveal allosteric inhibition by Ro 61–8048. FASEB J. 2018;32:2036–45. [DOI] [PubMed] [Google Scholar]
- 14.Amobi A, Qian F, Lugade AA, Odunsi K. Tryptophan catabolism and cancer immunotherapy targeting IDO mediated immune suppression In: Kalinski P, editors. Tumor immune microenvironment in cancer progression and cancer therapy. Advances in experimental medicine and biology, vol 1036 Springer: Cham; 2017. p.129–144. [DOI] [PubMed] [Google Scholar]
- 15.Ray A, Das DS, Song Y, Hideshima T, Tai YT, Chauhan D, Anderson KC. Combination of a novel HDAC6 inhibitor ACY-241 and anti-PD-L1 antibody enhances anti-tumor immunity and cytotoxicity in multiple myeloma. Leukemia . 2018;32:843–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ray A, Das DS, Song Y, Macri V, Richardson P, Brooks CL, et al. A novel agent SL-401 induces anti-myeloma activity by targeting plasmacytoid dendritic cells, osteoclastogenesis and cancer stem-like cells. Leukemia 2017;31:2652–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ray A, Tian Z, Das DS, Coffman RL, Richardson P, Chauhan D, et al. A novel TLR-9 agonist C792 inhibits plasmacytoid dendritic cell-induced myeloma cell growth and enhance cytotoxicity of bortezomib. Leukemia. 2014;28:1716–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giorgini F, Huang SY, Sathyasaikumar KV, Notarangelo FM, Thomas MA, Tararina M, et al. Targeted deletion of kynurenine 3-monooxygenase (KMO) in mice: a new tool for studying kynurenine pathway metabolism in periphery and brain. J Biol Chem. 2013;288:36554–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwarcz R, Bruno JP, Muchowski PJ, Wu HQ. Kynurenines in the mammalian brain: when physiology meets pathology. Nat Rev Neurosci. 2012;13:465–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Costantino G. New promises for manipulation of kynurenine pathway in cancer and neurological diseases. Expert Opin Ther Targets. 2009;13:247–58. [DOI] [PubMed] [Google Scholar]
- 21.Zwilling D, Huang SY, Sathyasaikumar KV, Notarangelo FM, Guidetti P, Wu HQ, et al. Kynurenine 3-monooxygenase inhibition in blood ameliorates neurodegeneration. Cell. 2011;145:863–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Campesan S, Green EW, Breda C, Sathyasaikumar KV, Muchowski PJ, Schwarcz R, et al. The kynurenine pathway modulates neurodegeneration in a Drosophila model of Huntington’s disease. Curr Biol. 2011;21:961–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar SK. Checking in: T cells against multiple myeloma. Blood. 2017;130:1175–6. [DOI] [PubMed] [Google Scholar]
- 24.Badros A, Hyjek E, Ma N, Lesokhin A, Dogan A, Rapoport AP, et al. Pembrolizumab, pomalidomide and low dose dexamethasone for relapsed/refractory multiple myeloma. Blood. 2017;1410:1189–97. [DOI] [PubMed] [Google Scholar]
- 25.Jenkins MR, Rudd-Schmidt JA, Lopez JA, Ramsbottom KM, Mannering SI, Andrews DM, et al. Failed CTL/NK cell killing and cytokine hypersecretion are directly linked through prolonged synapse time. J Exp Med. 2015;212:307–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pittari G, Vago L, Festuccia M, Bonini C, Mudawi D, Giaccone L, et al. Restoring natural killer cell immunity against multiple myeloma in the era of new rugs. Front Immunol. 2017;8:1444 10.3389/fimmu.2017.01444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Görgün G, Samur MK, Cowens KB, Paula S, Bianchi G, Anderson JE, et al. Lenalidomide enhances immune checkpoint blockade-induced immune response in multiple myeloma. Clin Cancer Res. 2015;1521:4607–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Woods DM, Sodre AL, Villagra A, Sarnaik A, Sotomayor EM, Weber J. HDAC inhibition upregulates PD-1 ligands in melanoma and augments immunotherapy with PD-1 blockade. Cancer Immunol Res. 2015;3:1375–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prendergast GC, Malachowski WJ, Mondal A, Scherle P, Muller AJ. Indoleamine 2,3-dioxygenase and its therapeutic inhibition in cancer. Int Rev Cell Mol Biol 2018;336:175–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Connor TJ, Starr N, O’Sullivan JB, Harkin A. Induction of indolamine 2,3-dioxygenase and kynurenine 3-monooxygenase in rat brain following a systemic inflammatory challenge: a role for IFN-gamma? Neurosci Lett. 2008;441:29–34. [DOI] [PubMed] [Google Scholar]
- 31.Chauhan D, Catley L, Li G, Podar K, Hideshima T, Velankar M, et al. A novel orally active proteasome inhibitor induces apoptosis in multiple myeloma cells with mechanisms distinct from Bortezomib. Cancer Cell. 2005;8:407–19. [DOI] [PubMed] [Google Scholar]
- 32.Hideshima T, Chauhan D, Richardson P, Mitsiades C, Mitsiades N, Hayashi T et al. NF-kappa B as a therapeutic target in multiple myeloma. J Biol Chem. 2002;277:16639–47. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.