Abstract
Molecular markers have been used as a tool for diagnostic approaches, staging, and evaluation of therapeutic responses in patients with cancer. Cancer molecular markers can also help clinicians to make decision on therapy and prognosis evaluation at the time of diagnosis. In the early diagnosis of breast cancer (BC), estrogen and progesterone receptors (ER/PR) expression levels should be determined through immunohistochemistry (IHC). In molecular genetics, there are some important tissue-based markers that can also be found in blood, such as Mammaglobin (MAM), Cytokeratin 19 (CK19), Mucin (MUC), Proto-oncogene (c-Myc), antigen Ki-67 (Ki67), and Carcinoembryonic antigen (CEA). In this study, the positive level of the marker genes in both blood and tissue of the BC patients were compared using reverse transcription polymerase chain reaction (RT-PCR) method. In addition, the importance of blood vs. tissue-based markers in BC diagnosis also demonstrated and is discussed.
CEA (O), ERβ, CK19 and, c-Myc molecular markers were significantly different between blood of normal and patients while there was no significant difference of these markers in tissue samples.
Blood-based biomarkers can be used for the early diagnosis of BC. Comparing blood versus tissue-based biomarkers indicated that there are correlations between markers in blood and tissue, since blood markers can be substitute to tissue markers in BC patients in the future.
Keywords: Molecular biology, Cancer research, Health sciences, Human genetics, Oncology, Breast cancer, Detection, Biomarkers, Clinical follow up
Molecular Biology; Cancer Research; Health Sciences; Human Genetics; Oncology; Breast cancer; detection; biomarkers; clinical follow up
1. Introduction
Molecular biomarkers expression is responsible for the normal physiological functions in the cells [1]. Deregulation of gene expression in the cells that constitutively promotes cell proliferation, growth, differentiation, and apoptosis can increase the risk of developing cancer [1]. Cancer can be occurred due to various diseases, which are triggered by biomarkers [2]. Biomarkers are made by the cells, in response to different conditions [2]. They can basically be found in the fluids, such as blood or tissues [2]. Tumor markers are ideally used to diagnose cancer in people with no symptoms, and for early diagnosis, as well [3]. Tumor markers can be measured in the blood or tissue of the cancerous cells [3]. Common molecular biomarkers that have been used as tumor markers are classified into two groups including tissue and blood markers, which are listed in this report along with their importance. Some types of cancers can proliferate and spread faster, and are less likely to respond to the treatment [4]. Markers which are able to predict tumor behavior, are particularly important, because they can provide the useful tool for clinical management, assistance in diagnosis, staging, evaluation of therapeutic response, and diagnosis of recurrence and metastases [5]. Physical characteristics of tumors, like size, histological grade, and the number of metastatic lymph nodes have been considered for decades. Tumors are classified based on the origin of the tumor and the most well-established molecular markers found in the breast with prognostic and therapeutic values. Breast tumors are classified by the estrogen and progesterone receptors (ER/PR) expression levels via immunohistochemistry (IHC). According to the breast tissue pathology, different clinical, pathological, and molecular classifications are characterized. The main clinical application of steroid hormone receptors, i.e. ER and PR are in patients with invasive breast cancer (IBC) and using for treatment. There are two main forms of ER, including ERα and ERβ, with opposite effects on proliferative response [5].
A glycoprotein, Carcinoembryonic antigen (CEA) is indicative of tumor size in breast cancer (BC) and nodal involvement [1]. It acts by organizing tissue architecture and regulating different signal transduction, while aberrant expression leads to the development of human malignancies. It has also been expressed in many cancers [6].
Human epidermal growth factor receptor-2 (HER2) is structurally related to epidermal growth factor receptors (EGFRs), which is a trans-membrane tyrosine kinase receptor [1]. In 20–30% of the BCs, this oncogene expression level is amplified. HER2 expression level can be qualified in benign BC patients, due to the HER2 extracellular shedding into the circulation [1]. HER2 measurement has the main clinical use in predicting and response to anti-HER2 therapy [7, 8]. In all patients with primary IBC, HER2 gene amplification or overexpression should be determined for treatment.
Mammaglobin (MAM) is a protein found in mammary tissue and in the serum, as well. MAM has been proposed as a biomarker to diagnose BC, given that patients exhibit an increased amount of the protein in their serum and tumor tissue compared with healthy individuals [9, 10, 11, 12].
Cytokeratin 19 (CK19) which belongs to a family of keratins is normally expressed in more than 95% of breast carcinomas [13, 14, 15].
Antigen Ki-67 (Ki67) is a labile nuclear protein that is linked to the cell cycle [1]. It is not expressed in resting cells; however, it can be expressed in proliferating phases of cell cycle (mid-G1, S, G2, and M). It has been demonstrated that there is a strong correlation between Ki67 and tumor proliferation index [2]. It has been shown that the elevated levels of Ki67 are independently associated with BC patients’ treatment outcome. There is a correlation between the high Ki67 values with poor treatment outcome in patients with BC [16].
Mucin 1 (MUC1) is a trans-membrane protein with two subunits that expresses in normal epithelial cells. It is overexpressed in 90% of BC patients [15]. MUC1 expression can lead to a poor prognosis in many different types of carcinomas. A soluble form of MUC1, which can be seen due to its shedding into the circulation, is cancer antigen 15-3 (CA 15-3). It is a common tumor marker, with a prognostic value like CEA marker. MUC1 is overexpressed in human breast carcinomas, and it is detectable at the elevated levels in the serum of patients [17].
The Myc proto-oncogene (c-Myc) is a helix-loop-helix zipper protein which its activation can lead to transcriptional activation or repression of the other genes [18]. c-Myc expression level can indicate a strong correlation with ER status, metastases, high histological grade, and positive nodal status [19, 20]. It has been considered that c-Myc amplification can represent a clinically predictive factor in metastatic BC [19].
This study aimed at comparing blood- and tissue-based biomarkers to obtain the most validated biomarkers in BC.
2. Materials and methods
2.1. Study design
A total of 64 female BC patients who referred to the Milad hospital through 2011–2013, were enrolled in this non-randomized study (Table 1). Moreover, 50 healthy age-matched female volunteers with no history of BC served as the control group. Patients were analyzed for molecular receptor expression levels on the primary tumor (Table 2). The inclusion criteria were as follows: BC diagnosis prior onset of the treatment process and the written informed consent. Exclusion criterion was the secondary primary malignancy. The informed consent was obtained from all patients for using their blood and tissue samples. The samples were collected using protocols approved by the review board. The study was approved by the ethics committee of the Milad hospital (No. 9003). Authors confirm that all methods were performed in accordance with the relevant guidelines and regulations based on ethics committee.
Table 1.
Clinical-pathological characteristics.
| Characteristic | N | % | |
|---|---|---|---|
| Patients | 64 | 100 | |
| Mean age=49.06 ± 1.25 (range 23-70 years) | |||
| 30 > age | 1 | 2 | |
| 30–40 | 11 | 17 | |
| 40–50 | 25 | 39 | |
| 50< | 27 | 42 | |
| Menopausal status | |||
| Pre | 22 | 34 | |
| Post | 42 | 66 | |
| Histologic subtype | |||
| Invasive Ductal Carcinoma (IDC) | 50 | 78 | |
| Invasive Ductal Carcinoma (NOS Type) | 11 | 17 | |
| Invasive Lobular Carcinoma (ILC) | 3 | 5 | |
| Histologic grade | |||
| І | 6 | 10 | |
| ІІ | 36 | 56 | |
| ІІІ | 18 | 28 | |
| Unknown | 4 | 6 | |
| Stage | |||
| 1 | 6 | 9 | |
| 2 | 45 | 70 | |
| 3 | 9 | 14 | |
| 4 | 3 | 5 | |
| Unknown | 1 | 2 | |
| Size of the tumor[cm] | |||
| 2 ≥ T1 | 11 | 17 | |
| T22-5 | 39 | 61 | |
| T3>5 | 13 | 20 | |
| Unknown | 1 | 2 | |
| The state of lymph node | |||
| Negative | 32 | 50 | |
| Positive | 25 | 39 | |
| Unknown | 7 | 11 | |
| Tissue marker status | |||
| Her2 | Positive (+) | 21 42 |
33 67 |
| Negative (-) | |||
| P53 | Positive (+) | 18 46 |
28 72 |
| Negative (-) | |||
| PR | Positive (+) | 43 21 |
67 33 |
| Negative (-) | |||
| ER | Positive (+) | 46 18 |
72 28 |
| Negative (-) | |||
| Ki67 | Positive (+) | 51 13 |
80 20 |
| Negative (-) | |||
| Triple-positive | 12 | 19 | |
| HER2-enriched | 8 | 12.5 | |
| Basal like | 5 | 8 | |
N: number of subjects; PR: Progesterone receptor; ER: Estrogen receptor; HER2: human epidermal growth factor receptor 2; triple-positive: ER/PR/HER2 positive; HER2-enriched: ER/PR negative&HER2positive; Basal like: triple-negative (TN) or (ER/PR/HER2 negative).
Table 2.
Comparison of biomarker expression in blood and tissue samples.
| Control healthy subjects |
Breast cancer patients |
||||
|---|---|---|---|---|---|
| Blood (n = 50) |
Blood (n = 64) |
Tissue (n = 64) |
|||
| Biomarker | N(+) | N(+) | N(+) | P1 | P2 |
| CEA(O) | 11 | 33 | 25 | 0.001∗ | NS |
| erbB2 | 6 | 30 | 51 | 0.000∗ | 0.000∗ |
| MAM | 18 | 30 | 27 | NS | NS |
| ERβ | 0 | 13 | 16 | 0.001∗ | NS |
| CK19 | 37 | 63 | 61 | 0.000∗ | NS |
| Ki67 | 22 | 10 | 56 | 0.001∗ | 0.000∗ |
| CEA(i) | 40 | 45 | 39 | NS | NS |
| Muc | 44 | 56 | 62 | NS | 0.000٭ |
| c-Myc | 4 | 31 | 26 | 0.000∗ | NS |
P1: Chi-Square test was used between the blood of healthy individual and breast cancer patients; P2: Chi-Square test was used between the blood and the tumor of breast cancer patients; ∗: Significant at 0.05(p < 0.05); NS: Non Significant.
2.2. Immunohistochemistry analysis of the primary tumor
IHC of the tissues obtained prior treatment. ER, PR, HER2, tumor protein p53 (p53), and Ki-67 expression levels were evaluated. ER and PR positivity was defined as ≥ 10% in positive tumor cells by nuclear staining. Fluorescent in situ hybridization was performed to determine HER2 positivity. The stained cells percentage for Ki-67 and p53 were counted and determined by the number of positive cells divided by the total number of counted tumor cells. The elevated expression level of Ki-67 was defined as ≥ 10%. The ER, PR, and HER2 statuses of the primary tumor were obtained from pathology laboratories.
2.3. Sampling of the blood and tissue
5 ml of the blood were collected for isolation of RNA, using the blood collection tubes and stored at 4 °C until further analysis. Blood samples were taken from the patients and healthy subjects (n = 114).
Fresh human breast tumor samples were obtained from the Milad hospital (Experimental and Clinical Research Center). Tissue samples were immediately transferred after surgeries and stored at -80 °C. 61 samples with invasive ductal carcinoma (IDC) obtained from the patients with breast reduction surgery. All samples were reviewed for histological classification according to the nuclear grade and classified as low, intermediate, and high. In addition, the TNM classification of malignant tumors and hormone receptor statuses were also determined.
2.4. Molecular biomarkers detection
Breast cancer-associated gene positive levels in the blood and tissue samples were analyzed using reverse transcription PCR (RT-PCR). RNA samples were isolated from tumor tissue and blood samples analyzed by PCR. The primer sets for the CEA, erb-B2, MAM, ER, CK19, Ki67, MUC, and c-Myc transcripts were provided [21, 22]. PCR was performed with the HotStarTaq Master Mix (Qiagen GmbH), following preparation of the cDNA. Glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH) was used as internal PCR positive control. The thermal profile used for the RT-PCR was as follows: After a 15-min denaturation at 95 °C, 30 cycles of PCR were carried out by denaturation at 94 °C for 30 s, annealing, extension at 60 °C for 30 s, and elongation for 30 s at 72 °C. Subsequently, termination of the reaction was carried out at 72 °C for 5 min followed by storing the samples at 4 °C. Visualization of the PCR fragments was carried out with a BioRad gel documentation system [22].
2.5. RNA isolation and amplification analysis
RNA extraction from samples was performed using RNeasy mini kit (Qiagen, Hilden, Germany) by DNase I digestion in accordance with the manufacturer's guide. The RNA was isolated with RNeasy Tissue Mini Kit (Qiagen). RNA quality was checked by the Nano drop spectrophotometer and amplification was carried out using 50ng of the total RNA.
2.6. RT-PCR (reverse transcription-polymerase chain reaction)
The RNA One-Step RT-PCR System (Bioneer) was used. The procedure was performed in accordance with the manufacturer's guide. For RNA, cDNA synthesis was done using Oligo (dT) primers. The expression of Glyceraldehyde 3-phosphate dehydrogenase as the housekeeping gene (GAPDH) and as an internal control was considered.
2.7. Statistical analysis
Statistical analysis was performed by SPSS 18 (SPSS Inc., Chicago, IL, USA). The statistical differences between the patients and control groups were analyzed. A Chi-squared test was used to evaluate the relationship between biomarkers frequencies, the primary tumor, and clinico-pathological factors. p < 0.05 was considered statistically significant.
3. Results
3.1. Characteristics of patients
As shown in Table 1, a total number of 64 patients were enrolled in the study through 2011–2013. Most patients had IDC, and they were assessed and classified based on the ER-positive (72%), PR-positive (67%), and HER2-negative (67%) tumors.
3.2. Blood and tissue-based biomarkers
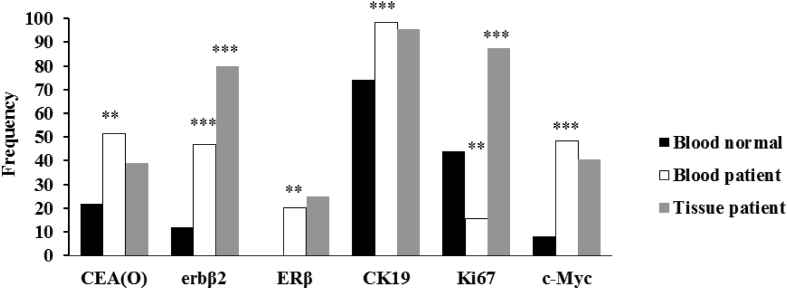
The positive markers for both groups are shown in Table 2. These markers were CEA(O), erb B2, ERβ, CK19, Ki67 and c-Myc in the blood of the control group which was different from the patients with BC and this difference was statistically significant (p < 0.00). There was a significant difference (p < 0.00) between erb B2, Muc, and, Ki67 biomarkers in the blood and tissue samples of the BC patients. CEA (O), ERβ, CK19 and, c-Myc molecular markers were significantly different between blood of normal and patients while there was no significant difference of these markers in tissue samples (Figure 1).
Figure 1.
CEA (O), erbβ2, ERβ, CK19, Ki67 and c-Myc markers frequency in blood and tissue of samples. The markers differ significantly between the blood of normal and patient samples but no difference was shown between the tumor and blood of patients for CEA (O), ERβ, CK19, and c-Myc markers. An asterisk indicates the statistically significant difference between the blood of healthy individual and breast cancer patients as measured by Chi-Square test (∗∗ = p < 0.01 and ∗∗∗ = p < 0.001).
3.3. Biomarkers and clinicopathological features
The expression levels of mRNA biomarkers in the peripheral blood of the BC patients were assessed for pathological features. Comparing the grades І and II with grade III (P2), erb B2 biomarker was significant (p = 0.00; Table 3). The erb B2 and ERβ biomarkers were significant in the stage І compared with the stage II (P3) (P=0.04). CEA (i) was the only significant biomarker in the stage II compared with the stages III and IV (P4) (p = 0.00). Comparing T1 and T2 (P6) tumors, erb B2 and ERβ biomarkers were significant (p = 0.05, and 0.00, respectively), whereas erb B2 biomarker was significant in T1 compared to T3 tumors (P7) (p = 0.00).
Table 3.
Clinicopathological features association with biomarkers expression in the peripheral blood of breast cancer patients.
| peripheral blood biomarkers | P1 | P2 | P3 | P4 | P5 | P6 | P7 | P8 |
|---|---|---|---|---|---|---|---|---|
| CEA(O) | 0.20 | 0.57 | 0.47 | 0.94 | 0.50 | 0.87 | o.68 | 0.70 |
| erb B2 | 0.97 | 0.00٭ | 0.04٭ | 0.53 | 0.17 | 0.05٭ | 0.04٭ | 0.42 |
| MAM | 0.33 | 0.06 | 0.87 | 0.83 | - | 0.55 | 0.21 | 0.12 |
| ERβ | 0.49 | 0.21 | 0.04٭ | 0.44 | 0.29 | 0.02٭ | 0.10 | 0.66 |
| CK19 | 0.34 | 0.12 | 0.08 | 0.60 | 0.14 | 0.64 | - | - |
| Ki67 | 0.82 | 0.58 | 0.75 | 0.09 | 0.14 | 0.81 | 0.43 | 0.15 |
| CEA(i) | 0.11 | 0.11 | 0.25 | 0.00٭ | 0.29 | 0.28 | 0.45 | 0.75 |
| Muc | 0.57 | 0.93 | 0.29 | 0.52 | 0.46 | 0.69 | 0.90 | 0.25 |
| c-Myc | 0.63 | 0.57 | 0.41 | 0.27 | - | 0.41 | 0.21 | 0.98 |
P value: Chi-Square test; P1: Age <50 compared to ≥50; P2: Grade І& II compared to III; P3: Stage І compared to II; P4: Stage II compared to III & IV; P5: Stage I compared to III & IV; P6:T1 compared to T2, 3; P7:T1 compared to T3; P8: Node-negative compared to Node-positive.
∗ Significant (Pvalue≤0.05).
Biomarkers expression levels in the BC tumor tissues and their association with the stage and size of the tumor were assessed with the pathological features (Table 4). The erb B2, MAM, CK19, and CEA (i) biomarkers was significant by comparing the grades І and II with grade III (P2) (p = 0.00, 0.03, 0.00 and 0.00, respectively). No significant biomarker association was observed by comparing stage І with stage II, stage II with stages III and IV, and stage I with stages III and IV (P3, P4, P5). CEA (i) biomarker was also significant in T1 tumor size is comparison with T2 (P6) (p = 0.02). ERβ and CEA (i) biomarkers was significant in T1 tumor size compared with T3 (P7) (p = 0.03 and 0.04, respectively). erb B2 and MAM biomarkers was significant in the node-negative than node-positive patients (P8) (p = 0.05 and 0.03, respectively). In addition, there was an association between mRNA expression of the erb B2 biomarker with the age (younger than 50 years old) (P1).
Table 4.
Clinicopathological features association with biomarkers expression in the tumor tissue in breast cancer patients.
| Tumor tissue biomarkers | P1 | P2 | P3 | P4 | P5 | P6 | P 7 | P8 |
|---|---|---|---|---|---|---|---|---|
| CEA(O) | 0.71 | 0.64 | 0.21 | 0.34 | 0.08 | 0.84 | 0.72 | 0.13 |
| erb B2 | 0.04٭ | 0.00٭ | 0.54 | 0.67 | - | 0.35 | 0.90 | 0.05٭ |
| MAM | 0.49 | 0.03٭ | 0.30 | 0.83 | 0.31 | 0.25 | 0.85 | 0.03٭ |
| ERβ | 0.77 | 0.82 | 0.07 | 0.24 | 0.50 | 0.06 | 0.03٭ | 0.25 |
| CK19 | 0.48 | 0.00 | 0.08 | 0.30 | 0.14 | 0.5 | - | 0.86 |
| Ki67 | 0.57 | 0.61 | 0.34 | 0.63 | 0.46 | 0.41 | 0.10 | 0.75 |
| CEA(i) | 0.06 | 0.00٭ | 0.17 | 0.42 | 0.18 | 0.02٭ | 0.04٭ | 0.06 |
| Muc | 0.12 | 0.50 | 0.71 | 0.52 | 0.46 | 0.20 | 0.90 | 0.10 |
| c-Myc | 0.13 | 0.39 | 0.30 | 0.48 | 0.18 | 0.35 | 0.18 | 0.80 |
P value: Chi-Square test; P1: Age <50 compared to ≥50; P2: Grade І& II compared to III; P3: Stage І compared to II; P4: Stage II compared to III & IV; P5: Stage I compared to III & IV; P6:T1 compared to T2, 3; P7:T1 compared to T3; P8: Node-negative compared to Node-positive.
∗ Significant (Pvalue≤0.05).
3.4. Hormone receptors and biomarkers expression
Biomarkers expression in blood and tumor tissue samples, and its association with hormone receptors, ER, PR, and IHC markers such as HER2, P53 and Ki67 were also assessed. In blood samples (Table 5), there was a significant association between erb B2 biomarker and PR receptor and P53 (p = 0.00). c-Myc was the only significant biomarker in HER2 positive pathologic samples identified by IHC(p=0.00). CEA (O), ERβ, and CEA (i) biomarkers expression levels were significant in Ki67 positive tissue samples assessed by IHC (p = 0.02, 0.00 and 0.03, respectively). In tissue samples (Table 6), CEA (O) biomarker expression level was associated with ER receptor positive samples identified by IHC (p = 0.02). There was a significant association between Ki67 biomarker and PR receptor (p=0.00). CEA (O) and ERβ biomarker expression levels identified through RT-PCR was significant in the P53 positive tissue samples indicated by IHC (p = 0.00 and 0.02, respectively). There was an association between expression levels of Ki67 positive cells diagnosed by IHC in tissue samples and ERβ, CEA (i), and Ki67 biomarkers found via RT-PCR (p = 0.00).
Table 5.
Hormone receptors association with biomarkers expression in the blood patients.
| Biomarker |
ER |
PR |
HER2 |
P53 |
Ki67 |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| N (+)/Total | N (+) | P | N (+) | P | N (+) | P | N (+) | P | N (+) | P | |
| CEA(O) | 33/64 | 24/46 | 0.87 | 22/43 | 0.92 | 12/21 | 0.53 | 12/18 | 0.13 | 30/51 | 0.02٭ |
| erb B2 | 30/64 | 23/46 | 0.42 | 22/43 | 0.00٭ | 11/21 | 0.53 | 14/18 | 0.00٭ | 22/51 | 0.23 |
| MAM | 30/64 | 21/46 | 0.56 | 20/43 | 0.73 | 9/21 | 0.65 | 10/18 | 0.38 | 24/51 | 0.95 |
| ERβ | 13/64 | 8/46 | 0.35 | 8/43 | 0.62 | 7/21 | 0.07 | 4/18 | 0.81 | 7/51 | 0.00٭ |
| CK19 | 63/64 | - | - | - | - | - | - | - | - | - | - |
| Ki67 | 54/64 | 39/46 | 0.88 | 36/43 | 0.83 | 17/21 | 0.59 | 14/18 | 0.36 | 44/51 | 0.40 |
| CEA(i) | 45/64 | 32/46 | 0.83 | 30/43 | 0.89 | 16/21 | 0.47 | 12/18 | 0.69 | 39/51 | 0.03٭ |
| Muc | 56/64 | 41/46 | 0.52 | 40/43 | 0.06 | 17/21 | 0.49 | 17/18 | 0.29 | 46/51 | 0.19 |
| c-Myc | 31/64 | 21/46 | 0.47 | 19/43 | 0.20 | 18/21 | 0.00٭ | 10/18 | 0.47 | 27/51 | 0.15 |
∗ Significant (Pvalue≤0.05).
Table 6.
Hormone receptors association with biomarkers expression in the tumor tissue of breast cancer patients.
| Biomarker |
ER |
PR |
HER2 |
P53 |
Ki67 |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| N (+)/Total | N (+) | P | N (+) | P | N (+) | P | N (+) | P | N (+) | P | |
| CEA(O) | 25/64 | 22/46 | 0.02٭ | 20/43 | 0.08 | 10/21 | 0.32 | 12/18 | 0.00٭ | 20/51 | 0.96 |
| erb B2 | 51/64 | 36/46 | 0.65 | 35/43 | 0.62 | 18/21 | 0.40 | 16/18 | 0.25 | 41/51 | 0.78 |
| MAM | 27/64 | 22/46 | 0.14 | 21/43 | 0.12 | 9/21 | 0.94 | 7/18 | 0.06 | 23/51 | 0.35 |
| ERβ | 16/64 | 11/46 | 0.74 | 11/43 | 0.87 | 8/21 | 0.09 | 8/18 | 0.02٭ | 7/51 | 0.00٭ |
| CK19 | 61/64 | - | - | - | - | - | - | - | - | - | - |
| Ki67 | 56/64 | 42/46 | 0.14 | 41/43 | 0.00٭ | 17/21 | 0.26 | 15/18 | 0.52 | 48/51 | 0.00٭ |
| CEA(i) | 39/64 | 31/46 | 0.09 | 29/43 | 0.12 | 10/21 | 0.12 | 10/18 | 0.58 | 36/51 | 0.00٭ |
| Muc | 2/64 | - | - | - | - | - | - | - | - | - | - |
| c-Myc | 25/64 | 18/46 | 0.98 | 16/43 | 0.66 | 11/21 | 0.12 | 9/18 | 0.26 | 22/51 | 0.18 |
∗ Significant (Pvalue≤0.05).
4. Discussion
Tumor markers appeared to have an improved survival result. In certain treatments, tumor markers can help to select the right treatment option. For example, HER2 expression in tumor tissues of BC can be used to consider Trastuzumab (Herceptin®) as a precision medicine. Tumor markers can also be used to follow-up of the patients, such as monitoring for cancer recurrence. Accordingly, tumor markers should be measured at the diagnosis, before, during, and after treatment.
Several women with BC all over the world are treated with cytotoxic chemotherapy based on standard pathologic and clinical features of the nodal metastatic status which currently is regarded as the main factor for chemotherapy. This can lead to the many cases of overtreatment of not only BC patients, but also other cancer types. Avoiding ineffective treatments, including cytotoxic chemotherapy in modern adjuvant therapy, is the main objective of identification of primary biomarkers. Molecular markers in BC also help therapists to find the right therapy and evaluate prognosis at the diagnosis. There are BC protein markers for clinical application validated by the American Society of Clinical Oncology (ASCO), Food and Drug Administration (FDA), European Medicines Agency (EMA), and National Institute for Health and Clinical Excellence (NICE) [23]. For example, ER and PR are tissue-based markers in IHC analysis, which were approved in 1999 for prognosis and therapy response by the FDA, EMA, and NICE [23]. Her-2 receptor is also a tissue-based marker, which was confirmed by IHC analysis by the FDA, EMA, and NICE in 1998 to find the right treatment [23]. CA 15-3 and CA 27.29 proteins were approved by the FDA and EMA in 1997, using serum immunoassay, which are appropriate candidates for monitoring responses to the therapy [23]. CEA is also recommended by the ASCO, for monitoring patients through the active therapy [24].
Such approved data plays an important role to help therapists and patients to find whether chemotherapy should be prescribed or not, and how it can determine the average risk of survival and recurrence [19]. Gene expression biomarkers, such as DNA biomarkers and RNA biomarkers, and proteins found in the blood or tissues can be used to have a specific molecular signature in predicting prognosis. It can also be used to find the therapy option for each case in personalized cancer therapy [25]. Breast carcinoma subtypes based on histopathological, molecular, and clinical features has been defined. Molecular subtypes are as follows:
-
-
Luminal A, ER+ and/or PgR + HER2, Ki-67 low CK8/18+
-
-
Luminal B, (HER2 negative) ER+ and/or PgR+, Ki-67high
-
-
Luminal B, (HER2 positive) ER+ and/or PgR + Any Ki-6 or HER2+
-
-
HER2-enriched, HER2 + ER_ and PgR_ CK5/6 + GRB7+ ERBB2, GRB7 Poor differentiation
-
-
Basal-like, ER_ and PgR_, HER2_ EGFR + CK5/6 + CK14 + CK17 + HER1+
In addition, the importance of blood versus tissue-based markers in BC diagnosis should be regarded in the guidelines and should also be included in the international and national protocols to be compared and considered. Biopsy is the most common and first method to diagnose cancer, by checking the cancer cells in pathology testing. According to histopathological point of view, 80% of the breast tumors are ductal tumors and 10–15% is invasive lobular carcinoma (ILC) [1]. The most important markers have characterized by IHC-based tumor markers in BC, considering three main hormone receptors, including ER, PR and, HER2 [11]. Retrospective studies have demonstrated that patients with ER– and/or PR-containing tumor cells tend to have a better outcome than those without the receptors [11]. According to the European Group on Tumor Markers (EGTM), ER and PR should be measured in all newly diagnosed primary invasive breast cancers [11]. The measurement of the circulating MUC levels, as determined by the CA15-3 assay, has been approved by the FDA, and has been used to monitor the clinical course of patients with BC through the therapy to diagnose early recurrence. Numerous studies have examined the overexpression of MUC1 as a marker in breast tumors. MUC1-induced gene expression patterns can predict significant decreases in disease-free and overall survival [26]. MUC1 is associated with ER and can antagonize the inhibitory effects of treatment by Tamoxifen as a breast cancer drug [26]. CA 15-3 as a tumor marker to diagnose metastasis is a more efficient marker compared with CEA. However, both serum markers (CA 15-3 and CEA) have suggested for early diagnosis of metastasis [1]. Nowadays, CA125/MUC16 as an ovarian tumor cell marker also used as a biomarker in epithelial ovarian carcinoma [27].
The world health organization (WHO) and EGTM have developed guidelines for biomarkers in BC [23]. The use of tissue biomarker in BC has also considered by the American Association of Cancer Research (AACR) and ASCO guidelines. Serum biomarkers may be used for monitoring along with the history of disease, physical examinations, and diagnostic imaging. Using CA15-3, CA27-29, and CEA for monitoring patients during therapy to find the ongoing systemic therapy has introduced in the ASCO biomarker guidelines [23].
It has been reported that there is a significant association between the high Ki67 levels and tumor response to chemotherapy [16]. Indeed, Ki67 may be used in combination with established prognostic factors such as CEA for determining prognosis, according to EGTM recommendation [16, 28].
Gene expression profiling derived from RNA, was collected and extracted from the frozen tumors in primary cancer tissues or blood in patients with cancer. There are different biomarkers arrays, such as Oncotype DX assay (21-gene assay) or MammoPrint assay (70-gene signature test), based on microarrays to define a panel for genomic array of biomarkers [29]. Both Oncotype DX and MammoPrint quantify the recurrence risk score and they are predictive. Mammoprint is the only FDA-approved BC gene array [30].
A specific gene expression panel can provide a new hope for predicting a pattern for an individual patient prognosis [31]. Gene panels evaluate patients’ samples for alterations in some or all of these assessed genes. Molecular signature by biomarker arrays can be provided for accurate treatment and prognosis.
In this study, blood-based biomarkers were compared with tissue-based biomarkers for the first time, and it was shown that there are different significant biomarkers in each analysis. In blood, significant soluble biomarkers such as CEA (O), CK19, ER and, c-Myc can be found, whereas erb B2 and Ki67 are more important in tissue-based biomarkers. erb B2 expression level was also associated with the higher grades of the tumor as a biomarker in tissue; however, more investigations with a larger sample size are needed.
Based on molecular, immune, and histopathological characterization, among BC subtypes, Ki67 is significantly associated with CEA (O), ERβ, and CEA (i) in the blood, whereas in tissue, it is significantly associated with the ERβ, ki67, and CEA (i). In this regard, P53 subtype is significantly associated with erb B2 in blood, while in tissue it is significantly associated with CEA (O) and ERβ. erb B2 and c-Myc biomarkers are significantly associated in blood for ER, PR, and Her2 subtypes, whereas CEA (O) and Ki67 biomarkers are significantly associated for ER, PR, and Her2 subtypes in tissue. Therefore, biomarkers in tissue and blood can represent histopathology subtypes or hormone receptors with prognosis and predictive values. They can easily be applied and used in cancer therapy and follow-up in cancer patients who need considerable evaluations in the future.
Declarations
Author contribution statement
M. Oloomi: Conceived and designed the experiments; Wrote the paper.
N. Moazzezy: Performed the experiments.
S. Bouzari: Analyzed and interpreted the data.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
The authors wish to thank all patients who participated in the study. Besides, it is necessary to thank Dr. Maria Hashemian in specimen collection from Milad hospital.
References
- 1.Banin Hirata B.K., Oda J.M., Losi Guembarovski R., Ariza C.B., de Oliveira C.E., Watanabe M.A. Molecular markers for breast cancer: prediction on tumor behavior. Dis. Markers. 2014;2014:513158. doi: 10.1155/2014/513158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duffy M.J. Clinical use of tumor biomarkers: an overview. Klin. Biochem. Metab. 2017;25(46):157–161. No. 4. [Google Scholar]
- 3.Loke S.Y., Lee A.S.G. The future of blood-based biomarkers for the early detection of breast cancer. Eur. J. Cancer. 2018 Mar;92:54–68. doi: 10.1016/j.ejca.2017.12.025. [DOI] [PubMed] [Google Scholar]
- 4.Duffy M.J., McDermott E.W., Crown J. Blood-based biomarkers in breast cancer: from proteins to circulating tumor cells to circulating tumor DNA. Tumor Biol. 2018 May;40(5):1–11. doi: 10.1177/1010428318776169. [DOI] [PubMed] [Google Scholar]
- 5.Jameera Begam A., Jubie S., Nanjan M.J. Estrogen receptor agonists/antagonists in breast cancer therapy: a critical review. Bioorg. Chem. 2017 Apr;71:257–274. doi: 10.1016/j.bioorg.2017.02.011. [DOI] [PubMed] [Google Scholar]
- 6.Rizeq B., Zakaria Z., Ouhtit A. Towards understanding the mechanisms of actions of carcinoembryonic antigen-related cell adhesion molecule 6 in cancer progression. Cancer Sci. 2018 Jan;109(1):33–42. doi: 10.1111/cas.13437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roskoski Robert., Jr. Small molecule inhibitors targeting the EGFR/ErbB family of protein-tyrosine kinases in human cancers. Pharmacol. Res. 2019;139:395–411. doi: 10.1016/j.phrs.2018.11.014. [DOI] [PubMed] [Google Scholar]
- 8.Roskoski R., Jr. ErbB/HER protein-tyrosine kinases: structures and small molecule inhibitors. Pharmacol. Res. 2014 Sep;87:42–59. doi: 10.1016/j.phrs.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 9.Al Joudi F.S. Human mammaglobin in breast cancer: a brief review of its clinical utility. Indian J. Med. Res. 2014 May;139(5):675–685. [PMC free article] [PubMed] [Google Scholar]
- 10.Ghersevich S., Ceballos M.P. Mammaglobin A: review and clinical utility. Adv. Clin. Chem. 2014;64:241–268. [PubMed] [Google Scholar]
- 11.Villanueva H., Grimm S., Dhamne S., Rajapakshe K., Visbal A., Davis C.M., Ehli E.A., Hartig S.M., Coarfa C., Edwards D.P. The emerging roles of steroid hormone receptors in ductal carcinoma in situ (DCIS) of the breast. J. Mammary Gland Biol. Neoplasia. 2018 Dec;23(4):237–248. doi: 10.1007/s10911-018-9416-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nguyen H.M., Dao M.Q. Detection of human mammaglobin mRNA in breast cancer cells among Vietnamese women. Breast Cancer (Dove Med Press) 2019 Mar 18;11:143–150. doi: 10.2147/BCTT.S193777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saloustros E., Mavroudis D. CTCs in primary breast cancer (II) Recent Results Cancer Res. 2012;195:187–192. doi: 10.1007/978-3-642-28160-0_17. [DOI] [PubMed] [Google Scholar]
- 14.Tryfonidis K., Kafousi M., Perraki M., Apostolaki S., Agelaki S., Georgoulias V., Stathopoulos E., Mavroudis D. Detection of circulating cytokeratin-19 mRNA-positive cells in the blood and the mitotic index of the primary tumor have independent prognostic value in early breast cancer. Clin. Breast Cancer. 2014 Dec;14(6):442–450. doi: 10.1016/j.clbc.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 15.Park H.S., Han H.J., Lee S., Kim G.M., Park S., Choi Y.A., Lee J.D., Kim G.M., Sohn J., Kim S.I. Detection of circulating tumor cells in breast cancer patients using cytokeratin-19 real-time RT-PCR. Yonsei Med. J. 2017 Jan;58(1):19–26. doi: 10.3349/ymj.2017.58.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Penault-Llorca F., Radosevic-Robin N. Ki67 assessment in breast cancer: an update. Pathology. 2017 Feb;49(2):166–171. doi: 10.1016/j.pathol.2016.11.006. [DOI] [PubMed] [Google Scholar]
- 17.Duffy M.J., Shering S., Sherry F., McDermott E., O'Higgins N. CA 15-3: a prognostic marker in breast cancer. Int. J. Biol. Markers. 2000 Oct-Dec;15(4):330–333. doi: 10.1177/172460080001500410. [DOI] [PubMed] [Google Scholar]
- 18.Eilers M., Eisenman R.N. Myc's broad reach. Genes Dev. 2008 Oct 15;22(20):2755–2766. doi: 10.1101/gad.1712408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Todorović-Raković N., Nešković-Konstantinović Z., Nikolić-Vukosavljević D. C-myc as a predictive marker for chemotherapy in metastatic breast cancer. Clin. Exp. Med. 2012 Dec;12(4):217–223. doi: 10.1007/s10238-011-0169-y. [DOI] [PubMed] [Google Scholar]
- 20.Naab T.J., Gautam A., Ricks-Santi L., Esnakula A.K., Kanaan Y.M., DeWitty R.L., Asgedom G., Makambi K.H., Abawi M., Blancato J.K. MYC amplification in subtypes of breast cancers in African American women. BMC Canc. 2018 Mar 9;18(1):274. doi: 10.1186/s12885-018-4171-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oloomi M., Bouzari S., Mohagheghi M.A., Khodayaran-Tehrani H. Molecular markers in peripheral blood of Iranian women with breast cancer. Cancer Microenviron. 2013 Apr;6(1):109–116. doi: 10.1007/s12307-012-0118-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oloomi M., Moazzezy N., Bouzari S. Modulation of molecular biomarker expression in response to chemotherapy in invasive ductal carcinoma. BioMed Res. Int. 2018 Feb 12;2018:7154708. doi: 10.1155/2018/7154708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zeidan B.A., Townsend P.A., Garbis S.D., Copson E., Cutress R.I. Clinical proteomics and breast cancer. Surgeon. 2015 Oct;13(5):271–278. doi: 10.1016/j.surge.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 24.Lang J.E., Wecsler J.S., Press M.F., Tripathy D. Molecular markers for breast cancer diagnosis, prognosis and targeted therapy. J. Surg. Oncol. 2015 Jan;111(1):81–90. doi: 10.1002/jso.23732. [DOI] [PubMed] [Google Scholar]
- 25.Lam S.W., Jimenez C.R., Boven E. Breast cancer classification by proteomic technologies: current state of knowledge. Cancer Treat Rev. 2014 Feb;40(1):129–138. doi: 10.1016/j.ctrv.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 26.Kufe D.W. Mucins in cancer: function, prognosis and therapy. Nat. Rev. Cancer. 2009 Dec;9(12):874–885. doi: 10.1038/nrc2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Szymańska B., Lukaszewski Z., Hermanowicz-Szamatowicz K., Gorodkiewicz E. A biosensor for determination of the circulating biomarker CA125/MUC16 by Surface Plasmon Resonance Imaging. Talanta. 2020 Jan 1;206:120187. doi: 10.1016/j.talanta.2019.120187. [DOI] [PubMed] [Google Scholar]
- 28.Pagaza-Straffon C., Marchat L.A., Herrera L., Díaz-Chávez J., Avante M.G., Rodríguez Y.P., Arreola M.C., López-Camarillo C. Evaluation of a panel of tumor-associated antigens in breast cancer. Cancer Biomarkers. 2019 Dec 6:1–5. doi: 10.3233/CBM-190708. [DOI] [PubMed] [Google Scholar]
- 29.Falato C., Tobin N.P., Lorent J., Lindström L.S., Bergh J., Foukakis T. Intrinsic subtypes and genomic signatures of primary breast cancer and prognosis after systemic relapse. Mol. Oncol. 2016 Apr;10(4):517–525. doi: 10.1016/j.molonc.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kittaneh M., Montero A.J., Glück S. Molecular profiling for breast cancer: a comprehensive review. Biomark. Cancer. 2013 Oct 29;5:61–70. doi: 10.4137/BIC.S9455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kittaneh M., Montero A.J., Glück S. Molecular profiling for breast cancer: a comprehensive review. Biomark. Cancer. 2013 Oct 29;5:61–67. doi: 10.4137/BIC.S9455. [DOI] [PMC free article] [PubMed] [Google Scholar]