Abstract
Defects in the proteasome can result in pathological proteinopathies. However, the pathogenic role of sex‐ and tissue‐specific sensitivity to proteotoxic stress remains elusive. Here, we map the proteasome activity across nine tissues, in male and female mice, and demonstrate strong sexual dimorphism in proteasome activity, where females have significantly higher activity in several tissues. Further, we report drastic differences in proteasome activity among tissues, independently of proteasome concentration, which are exacerbated under stress conditions. Sexual dimorphism in proteasome activity is confirmed in a SOD1 ALS mouse model, in which the spinal cord, a tissue with comparatively low proteasome activity, is severely affected. Our results offer mechanistic insight into tissue‐specific sensitivities to proteostasis stress and into sex differences in the progression of neurodegenerative proteinopathies.
Keywords: amyotrophic lateral sclerosis, gender differences, proteasome, protein aggregates, ubiquitin
Subject Categories: Molecular Biology of Disease; Post-translational Modifications, Proteolysis & Proteomics;
Proteasome activity mapping across nine tissues in female and male mice reveals sex‐ and tissue‐specfic differences in proteostasis capacity and susceptibility to protein aggregation.
Introduction
Defects in the activity of the proteasome are linked to several pathologies including cardiac dysfunction, cancer, and autoimmune diseases 1, 2, 3, 4. Polymorphisms in subunits of the proteasome are associated with type 2 diabetes, myocardial infarction, and coronary disease 5, 6. In addition, the natural decline in proteasome activity with age has also been associated with age‐related conditions, especially neurodegenerative diseases 7, 8. In the latter, differential prevalence and rate of disease progression are observed between women and men, but the reason for such sexual dimorphism remains unknown.
The ubiquitin–proteasome cascade affects nearly all cellular processes and involves a large family of ubiquitin ligases, as well as several different types of ubiquitin chains 9, 10, 11. Among these, linkage of lysine 48 ubiquitin (Ub‐K48) chains targets proteins for degradation by the proteasome. The Ub‐K48‐proteasome cascade confers an essential housekeeping function by degrading proteins in response to specific signaling and protein misfolding. The 26S proteasome is composed of a regulatory particle, which contains various subunits and associated proteins, including ubiquitin receptors, deubiquitnation enzymes and unfoldases, and a catalytic core, which contains three distinct proteolytic activities. The beta‐2, beta‐5, and beta‐1 subunits of the beta rings of the catalytic core encode for the trypsin‐like, chymotrypsin‐like, and caspase‐like activities, respectively 12, 13.
In addition to the standard proteasome, tissue‐specific proteasomes have been identified in the cortical thymic epithelium and the testes. The thymoproteasome and the spermatoproteasome show altered activities 1, 3. Notably, the thymoproteasome only has slightly lower chymotrypsin‐like activity, yet whole body replacement of the standard beta‐5 for the thymus‐specific beta‐5 subunit results in accelerated aging 14. Therefore, small differences in only one of the three proteolytic activities of the proteasome are sufficient to have a large impact on health and longevity. This observation raises the interesting possibility that even small differences in proteasome activity between males and females may contribute to the overall extended longevity in women compared to men or explain differential rate of progression in some diseases between sexes. We therefore initiated a study to map proteasome activity across tissues and gender.
Results and Discussion
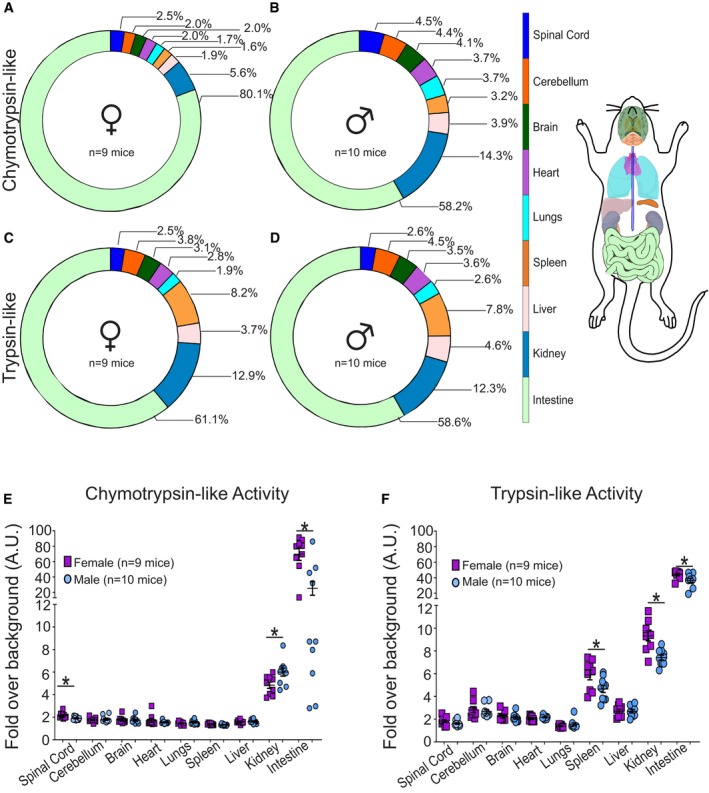
We harvested spinal cord, cerebellum, brain, heart, lung, spleen, liver, kidney, and small intestine from FvBN mice at the age of 3 and 5 months (defined as young). The chymotrypsin‐like and the trypsin‐like activities of the proteasome were measured using activity‐specific peptides linked to 7‐amino‐4‐methylcoumarin (AMC), which allows for the quantification of released AMC upon cleavage and a direct correlation with the specific proteasome activity. The caspase‐like activity could not be determined as we found this activity does not survive freezing and thawing process. First, we analyzed the relative activities of each tissue and found major differences in the distribution of the chymotrypsin‐like activity between females and males. In females, 80.1% of the chymotrypsin‐like activity is associated with the small intestine (Fig 1A), in contrast to 58.2% in males (Fig 1B). To ensure that the much higher activity in the intestine is due to the proteasome and not the secretion of trypsin in this tissue, we inhibited the proteasome with bortezomib and demonstrated that the recorded activity is proteasome‐dependent (Appendix Fig S1A). Other tissues also show sex‐specific differences. For example, in males 14.3% of chymotrypsin‐like activity is associated with the kidney (Fig 1B), compared to only 5.8% in females (Fig 1A). In sharp contrast, we found that the distribution of the trypsin‐like activity is largely identical between males and females (Fig 1C and D). These results are surprising since the catalytic core of the proteasome contains equal numbers of β2 and β5 subunits, and therefore, the distributions of chymotrypsin‐like activity and trypsin‐like activity were expected to be similar. To further analyze these differences, the activities of individual mice were plotted. This analysis confirmed that most tissues have very low chymotrypsin‐like activity, while the kidney and intestine have high activity (Fig 1E). Interestingly, a wide range in chymotrypsin‐like activity was observed in the intestine of males (Fig 1E) while the activity between mice in all other tissues closely clustered. To further, explore this variability in intestine of males, we interrogated the gene expression profile in the intestine of males and females using the GTEX dataset. We found that while the gene expression profile between females largely clustered together, the global gene expression profile in males varies widely (Appendix Fig S1B). Therefore, the variability in chymotrypsin‐like activity we observed in the intestine in males specifically (Fig 1E) is consistent with the sex‐specific heterogeneity of gene expression in this tissue.
Figure 1. Organism‐wide proteasome activity mapping reveals sexual dimorphism in a number of tissues.
-
A–DProportional distribution of chymotrypsin‐like activity of the proteasome across nine tissues from female (n = 9) and male (n = 10) mice. Proportional distribution of trypsin‐like activity of the proteasome across nine tissues from female (n = 9) and male (n = 10) mice. Graphical representation of color code of tissues is analyzed.
-
E, FChymotrypsin‐like or trypsin‐like activity of the proteasome across nine tissues in individual mice. Each data point (n = 9 females and n = 10 males) indicates data from a single mouse performed in triplicate. Each central line and error bar indicate mean ± SEM. *P < 0.05 by unpaired two‐tailed Student's t‐test comparing male and female samples.
Further analysis of the chymotrypsin‐like activity between males and females revealed that, with the exception of the kidney, females show higher activity than males in the spinal cord and intestine (Fig 1E) and also higher trypsin‐like activity in spleen, kidney, and intestine (Fig 1F). Therefore, the mapping of the proteasome activity across multiple tissues and sexes suggests that females show an overall higher proteostasis capacity.
To interrogate the biological impact of these sex‐specific differences in proteasome activity, we analyzed the accumulation of total proteins linked to ubiquitin lysine 48 chains (Ub‐K48). The spinal cord and intestine were selected, as they are the tissues with the lowest and the highest activities, respectively, and females have higher activities than males in both tissues. First, we confirmed that as observed for the intestine, the elevated chymotrypsin‐like activity in the spinal cord in females is due to the proteasome, as treatment with bortezomib abolished this activity (Appendix Fig S1C).
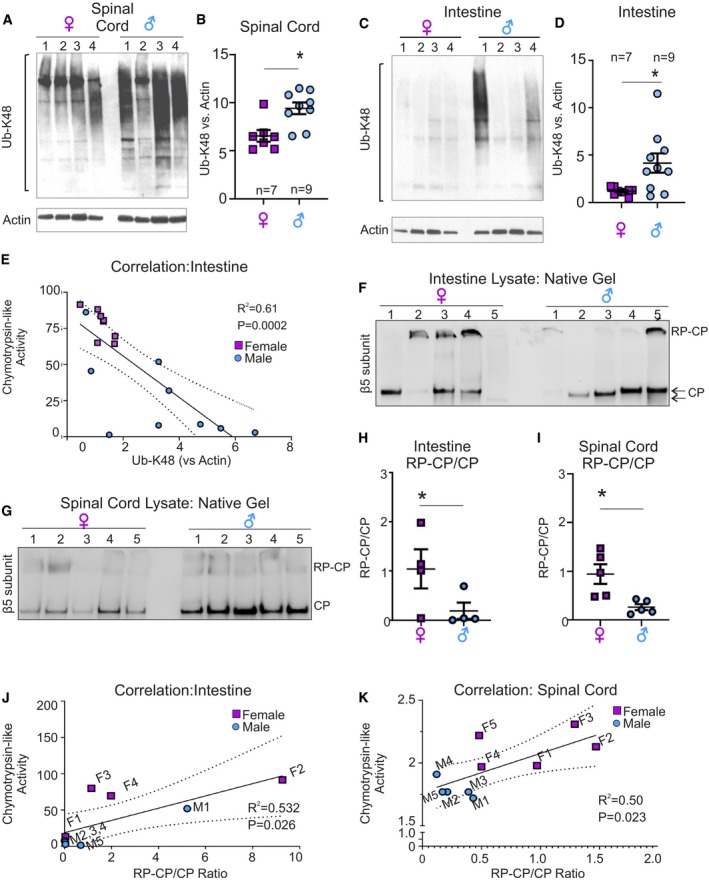
We found that in both spinal cord and intestine, the accumulation of Ub‐K48‐linked proteins is lower in females compared to males (Fig 2A–D), which is consistent with the higher proteasome activity in females (Fig 1E and F). Addition of a de‐ubiquitinated enzymes (DUB) inhibitor did not alter the pattern of ubiquitination (Appendix Fig S2).
Figure 2. Spinal cord and intestine show sexual dimorphism in Ub‐K48‐inked proteins and proteasome assembly.
-
A–DWestern blot analysis for Ub‐K48‐linked proteins in spinal cord (A) and intestine (C) lysates from male and female mice. (B, D) Quantification of panels (A) and (C), respectively. Values were normalized to actin as a loading control. Each central line and error bar indicate mean ± SEM of a biological replicates.
-
ECurve of correlation between chymotrypsin‐like activity and Ub‐K48‐linked proteins in the intestine, where variability between mice is observed.
-
FNative gel followed by Western blot of β5 subunit to distinguish unassembled catalytic core (CP) or assembled with regulatory core (RP) in male (n = 5) and female (n = 5) mice in intestine.
-
GNative gel followed by Western blot of β5 subunit to distinguish unassembled catalytic core (CP) or assembled with regulatory core (RP) in male (n = 5) and female (n = 5) spinal cord.
-
HQuantification of the ratio of assembled (RP‐CP) versus unassembled (CP) proteasome in the intestine of males and females. Each central line and error bar indicate mean ± SEM of a biological replicates (n = 5 per group).
-
IQuantification of the ratio of RP‐CP versus CP proteasome in the spinal cord of males and females. Each central line and error bar indicate mean ± SEM of a biological replicates (n = 5 per group).
-
JCorrelation curve between chymotrypsin‐like activity and RP‐CP/CP ratio in individual female (purple) and male (blue) intestine.
-
KCorrelation curve between chymotrypsin‐like activity and RP‐CP/CP ratio in individual female (purple) and male (blue) spinal cord.
Again, a wide variability was observed in the intestine of males (Fig 2C and D). To determine whether the variability among males in chymotrypsin‐like activity measured using a reporter as readout (Fig 1E) matched the levels of Ub‐K48 proteins determined by Western blot (Fig 2C), we correlated the proteasome activity of individual males to the level of Ub‐K48 in the same mouse. We found a significant correlation between the level of Ub‐K48 and proteasome activity in individual mice (Fig 2E). Collectively, these results indicate tissues and sex‐specific differences in proteasome capacity.
Having established these unexpected differences, we next aimed at determining if they reflect differences in the amount of proteasome subunits. Since the largest differences were observed for the chymotrypsin‐like activity, we have monitored the levels using an antibody against the β5 subunits to quantify the proteasome. However, the amount of β5 subunit in the lungs (representative low‐activity tissue) and intestine (representative high‐activity tissue) in females was similar (Appendix Fig S3A and B) and did not correlate with the measured proteasome activity in these tissues. In males, which have lower activity than females in the intestine (Fig 1E), the levels of β5 subunit tended to be higher (Appendix Fig S3A and B). Therefore, we concluded that the differences in activity are not due to the amount of proteolytic proteasome subunits. Since the activity of the proteasome can be modulated by multiple factors including assembly between the catalytic core (CP) and the regulatory subunits (RP), we performed native gels to gain insight into the differences in proteasome species that may exist between females and males. We found that in both the intestine (Fig 2F) and the spinal cord (Fig 2G), where the proteasome activity is higher in females than males, the ratio of assembled (RP‐CP) to unassembled (CP) catalytic core is significantly higher in females (Fig 2H and I). In addition, a strong correlation between this ratio and the activity of the proteasome was observed (Fig 2J and K).
Further, consistent with the variability in proteasome activity in male intestine (Figs 1E and 2C), analysis of the migration of the catalytic core also revealed variation in molecular weight among males (Fig 2F). Since this analysis was performed on native gels, the differences in electrophoretic mobility are large and likely to represent the presence of alternative binding partners/modifiers, further supporting the variability in proteasome in the male intestine. In addition, side‐by‐side analysis of spinal cord and intestine samples in females revealed that the levels of unassembled proteasome correlate with the low and high activity of the proteasome in these tissues, respectively (Appendix Fig S4).
Collectively, the mapping of the proteasome revealed an unexpected heterogeneity between tissues and sexes. These differences may reflect variable compositions of the proteasome in different tissues. In agreement with this possibility, a heatmap of the expression of 43 proteasome genes in 37 tissues, created using a human dataset from the Human Protein Atlas Project, revealed a wide variation in the expression of proteasome genes among tissues (Appendix Fig S5). These observations are in agreement with the increasing appreciation of the diversity of proteasome species 15, 16. When the combination of various subunits, variants, regulators, and post‐translational modifications is considered, it is estimated that theoretically 5 × 105 subtypes of proteasome may exist 17. While this estimation is difficult to prove experimentally, our results support the wide diversity of proteasome species, not only among different tissues, but also between sexes.
Several studies reported a decline in proteasome activity and proteostasis during aging by comparing samples from young and elderly humans or animals 18, 19, 20, 21, 22. However, studies focusing on proteostasis in middle age are lacking. This represents a significant gap in knowledge since the success of interventions aimed at maintaining proteasome activity and preventing proteinopathies would be significantly increased if administrated before irreversible protein aggregates are formed. Further, whether proteasome activity differences exist between tissues and sexes in the setting of aging has never been tested. We therefore compared the activity of the proteasome in nine tissues of female and male mice aged 10–15 months. We found that in some tissues the chymotrypsin‐like activity of older mice does not decline compared to young mice in both males and females (Fig 3A), while other tissues show a sex‐specific decline (Fig 3B and C). Only two tissues, heart and spinal cord, showed statistically significant age‐related decline in both sexes (Fig 3D). The pattern of decline over aging was different for the trypsin‐like activity (Appendix Fig S6). Collectively, this analysis revealed that the decline in proteasome activity during aging is not uniform across tissues and sexes (Fig 3E and F). Interestingly, the spinal cord is the only tissue showing consistent changes between sexes. In the spinal cord, the chymotrypsin‐like activity decreased with age, while the trypsin‐like activity increased. Further, the variability in proteasome activity among mice also tended to increase with age (Fig 3Ac males, Bb females, Appendix Fig S6 Ba females, Cb females, Cc females and males). These multiple variations raise the possibility that, in addition to sex and tissue differences, aging may also introduce additional heterogeneity in the array of proteasome species.
Figure 3. Proteasome activity shows differential decline with aging across tissues and sexes.

-
A–DComparison of chymotrypsin‐like activity from female (n = 9) and male (n = 10) mice in young (3–5 months old) or older (10–15 months old, female (n = 6) and male (n = 7)) groups. Each point indicates a single mouse. (A) Tissues that did not show significant decline, (B) tissues that show significant decline in females only, (C) tissues that show significant decline in males only, or (D) in both males and females. Each central line and error bar indicate mean ± SEM of a biological replicates (n = 9 young females, n = 6 older females, n = 10 young males, n = 7 older males). P values obtained by unpaired two‐tailed Student's t‐test comparing young and older samples are indicated.
-
E, FSummary Venn diagrams indicating tissues that had significantly different chymotrypsin‐like (E) or trypsin‐like (F) activity, when comparing young and older mice. Arrows indicate the direction of change (up and down indicate increase and decrease, respectively) in female (blue arrows) or male (red arrows) mice.
We then reasoned that, since stress can promote protein misfolding, the impact of the differences in proteasome activity between males and females may be exacerbated under these conditions. To investigate the functional consequences of the sex‐ and tissue‐specific differences in proteasome activity under stress conditions, we again focused on the spinal cord and intestine. To explore this possibility, males and females were exposed to heat shock at 40°C for 20 min (Fig 4A). Four hours later, mice were sacrificed and tissues harvested to assess the levels of Ub‐K48‐linked proteins. We found that after heat shock a drastic elevation was observed in the soluble fraction of male intestine, but only a mild effect was observed in females (Fig 4B and C). Ub‐K48‐linked protein level elevation in the insoluble fractions was even more marked than in the soluble fractions (Fig 4D and E). The drastic accumulation of Ub‐K48‐linked proteins under heat shock is in line with the recent observation that the small intestine is extremely poor in small heat‐shock proteins 23, which are critical in reducing accumulation of protein aggregates 24.
Figure 4. Sex‐specific differences in accumulation of protein aggregates following heat shock.
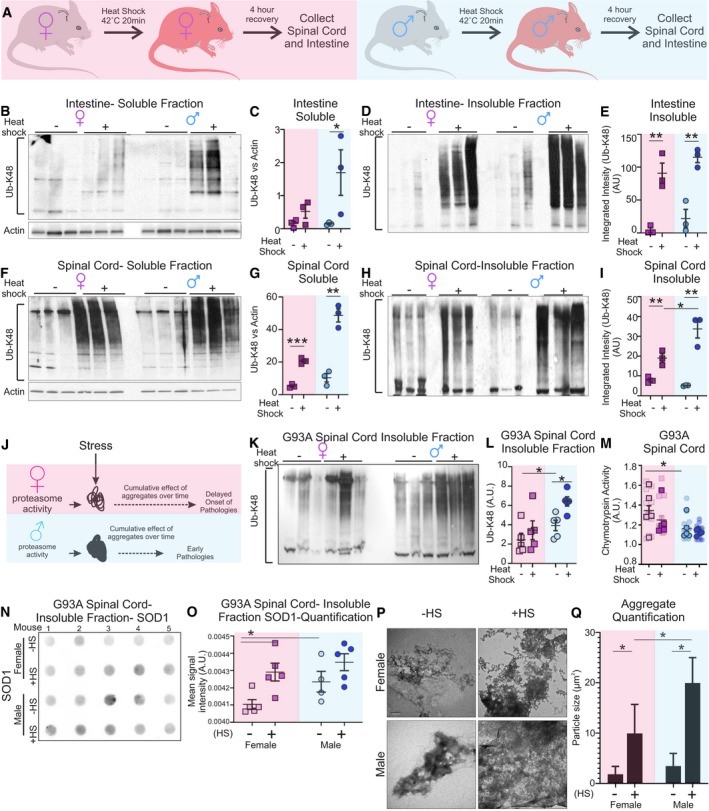
- Experimental design of treatment by heat shock in mice.
- Western blot analysis of Ub‐K48 proteins in the soluble fractions of intestine lysates from mice plus or minus heat shock.
- Quantification of panel (B). Each central line and error bar indicate mean ± SEM of a biological replicates (n = 3 biological replicates per group).
- Western blot analysis of Ub‐K48 proteins in the insoluble fraction of intestine lysates from mice plus or minus heat shock.
- Quantification of (D). Each central line and error bar indicate mean ± SEM of a biological replicates (n = 3 biological replicates per group).
- Western blot analysis of Ub‐K48 proteins in the soluble fractions of spinal cord lysates from mice plus or minus heat shock.
- Quantification of (F). Each central line and error bar indicate mean ± SEM of a biological replicates (n = 3 biological replicates per group).
- Western blot analysis of Ub‐K48 proteins in the insoluble fraction of spinal cord lysates from mice plus or minus heat shock.
- Quantification of (H). Each central line and error bar indicate mean ± SEM of a biological replicates (n = 3 biological replicates per group).
- Model of gender‐specific protein aggregate accumulation following stress.
- Western blot of Ub‐K48 proteins in the insoluble fraction of protein lysates of SOD1‐G93A mouse spinal cord plus or minus heat shock.
- Quantification of panel (K). Each central line and error bar indicate mean ± SEM of a biological replicates (n = 5 biological replicates per group).
- Chymotrypsin‐like proteasome activity in spinal cord lysates from SOD1‐G93A mice. Each central line and error bar indicate mean ± SEM of a biological replicates (n = 5 biological replicates per group). Bordered points indicate biological replicates, and un‐bordered points indicate technical replicates.
- Filter trap assay of protein lysates from the insoluble fraction of SOD1‐G93A spinal cord lysates plus or minus heat shock.
- Quantification of panel (N). Each central line and error bar indicate mean ± SEM of a biological replicates (n = 5 biological replicates per group).
- Representative transmission electron micrographs of protein aggregates in the insoluble fraction of SOD1‐G93A spinal cord lysates plus or minus heat shock. Scale bar equals 600 nm.
- Quantification of (P) (n = 9 technical replicates per group).
In the spinal cord, where the baseline activity of the proteasome is much lower than in the intestine, heat shock led to an increase in the accumulation of Ub‐K48‐linked proteins in both females and males and in both the soluble (Fig 4F and G) and insoluble fractions (Fig 4H and I). However, the increase was more marked in males than females. These results suggest that small sex‐specific differences in proteasome activity in the spinal cord correlate with accumulation of protein aggregates in males under heat‐shock stress (Fig 4J). This observation raises the possibility that under pathological conditions that favor the formation of protein aggregates in the spinal cord, males may show accelerated disease progression and/or earlier onset compared to females.
The SOD1‐G93A transgenic mouse is an established model of familial amyotrophic lateral sclerosis (ALS), a disease, which affects the motor neurons of the spinal cord and is pathologically characterized by protein aggregates. Further, in this model females show a late onset of symptoms and survive longer than males 25. We performed the heat‐shock treatment in male and female SOD1‐G93A mice and harvested their spinal cord for analysis. We found that males show a higher baseline accumulation of Ub‐K48‐linked proteins compared to females (Fig 4K and L). We also analyzed Ub‐K48‐linked proteins in non‐transgenic mice. This analysis confirmed that males show a higher baseline accumulation of K48‐linked proteins than females (Appendix Fig S7A and B). The sex difference was exacerbated in SOD1‐G93A mice (Appendix Fig S7C and D). In SOD1‐G93A mice, heat shock leads to an increase in Ub‐K48‐linked proteins in both females and males, but the increase is more marked in males (Fig 4K and L). Further, we found that heat shock causes a decrease in the activity of the chymotrypsin‐like proteasome activity (Fig 4M) and of the trypsin‐like activity (Appendix Fig S8). As SOD1 aggregates in this ALS mouse model, we next analyzed SOD1 aggregation. Using filter trap assays, we observed an increase in the aggregates in males compared to females at baseline and in both sexes after heat shock (Fig 4N and O). As an independent measure of aggregation, we perform electron microscopy to visualize aggregates. We found that spinal cords of males contain larger and more electron‐dense aggregates compared to females (Fig 4P and Q). Further, in both sexes, heat shock exacerbated the accumulation of large aggregates, but the effect of heat shock was more pronounced in males (Fig 4P and Q). Taken together, these results strongly suggest that females have a greater ability to cope with proteotoxic stress.
Collectively, the results of the current study reveal sex‐ and tissue‐specific differences in proteasome capacity, which is in agreement with the recent recognition of tissue‐specific proteome and heat‐shock protein expression patterns 23, and suggest that the sex‐ and tissue‐specific activities of the proteasome represent another layer of complexity of organismal proteostasis. Since we did not find correlations between proteasome activity and the amount of proteolytic proteasome subunits, our data suggest that the composition of the proteasome differs between tissues and sexes. This interpretation is in line with the notion of proteasome diversity, which arises from multiple potential sources. Notably, tissue‐specific β5 subunits, inducible beta subunits, such as those induced by interferon gamma, different regulatory particles (19S, 11S, or PA200), which are known to modulate the activity of the proteasome and to associate with the catalytic core at either one or both ends, or yet one regulatory particle of each type creating hybrid proteasomes. More specifically, interferon gamma promotes the expression of three immunosubunits, β1i, β2i, and β5i. Incorporation of these subunits in newly formed catalytic core of the proteasome leads to the formation of the immunoproteasome that shows distinct proteolytic activity compared to the standard proteasome 4. In addition, while the caspase‐like activity of the proteasome could not be confidently determined, it remains entirely possible that this activity also differs. Therefore, the caspase‐like activity and the formation of immunoproteasome may both contribute to the proteasome diversity between tissues and sexes reported in the current study.
Further, the expression of inhibitors of the proteasome, assembly factors, and chaperones that modulate the activity or favor the integration of one subunit over another during the assembly of the proteasome, and lastly an array of transiently proteasome‐associated proteins also modulate its activity 15, 16. In addition, stress‐induced components have been identified, such as those induced by treatment with arsenite, suggesting the existence of stress‐proteasomes, although the exact number of stress‐proteasomes remains unclear 26, 27.
Overall, our results support the notion that not only the array of proteasome composition may indeed be extremely large but further differs among tissue types, between sexes and may also be altered with aging. Appreciating the sex‐ and tissue‐specific differences in proteasome is critical to our understanding of its role in aging and proteostasis‐related diseases. Finally, since inhibition of the proteasome has shown efficacy in some cancer types but not in others 28, 29, our results highlight the need to tailor the choice of proteasome inhibitors to specific tissue and cell types in order to develop precision medicine approaches and increase therapeutic success.
Materials and Methods
Mice
All experiments were approved by the Mt. Sinai Institutional Animal Care and Use Committee (IACUC) and performed according to the principles of laboratory animal care outlined in NIH publication No. 86‐23, revised 1985 edition. Wild‐type FVBN mice harvested between 3 and 5 months of age were considered “young”, while mice harvested between 10 and 15 months of age were considered “old”. B6SJL‐TG_SOD1*G93A)1Gur/J and non‐transgenic (NTG) B6SJL mice are available from the Jackson Laboratory and maintained according to IACUC‐approved methods. SOD1‐G93A and NTG control mice were sacrificed in average at 69 days of age, before observable symptoms manifested. All collected tissue was immediately frozen on dry ice and stored for later analysis.
Heat shock
Heat shock was performed as previously described 30. Briefly, four to five wild‐type FVBN mice were kept in a ventilated box maintained at 41–43°C for 20 min using a standard heating lamp. The internal temperature of the box was monitored with and alcohol thermometer suspended in the center of the box. Mice were returned to their normal colony conditions and tissues harvested 4 h after following treatment. Body temperatures were monitored prior and after heat shock using rectal thermometers and showed resting temperatures of 37.24 ± 0.26°C (n = 4) and of 39.53 ± 0.41°C (n = 4) after heat shock.
Tissue collection, soluble, and insoluble fraction preparation
Frozen tissue powders were generated using pestle and mortar on dry ice. Tissues were lysed in NP‐40 lysis buffer (50 mM Tris, 250 mM NaCl, 5 mM EDTA, 0.5% NP‐40, 50 mM NaF, 1 mM DTT plus protease inhibitors) on ice using a 1:4 volume‐to‐volume ratio of tissue powder to lysis buffer. All samples were subjected to probe sonication (fisher scientific FB505) with 2–3 rounds of 1‐s intervals at 20% amplitude on ice. Protein lysates were centrifuged at 20,800 g at 4°C for 20 min. The top layer (soluble fraction) was transferred to a new tube, and the pellet contained the insoluble fraction. Pellet volumes were estimated by pipette, and an equivalent volume of TEN buffer (10 mM Tris–HCl, 1 mM EDTA, 100 mM NaCl plus protease inhibitors) was added to make a 1:1 pellet:TEN buffer suspension. Samples were then sonicated twice with 1‐s intervals at 20% amplitude on ice. 5 μl of this suspension was then added to an equal volume of 5× loading buffer (1 M Tris–HCl, 25% glycerol, 2% SDS, 5% 2‐mercaptoethanol, 0.1% bromophenol blue) and heated to 95°C for 5 min before being separated by electrophoresis.
For DUB inhibition, PR‐619 (Selleck Chemicals) (20 μM) was added to NP‐40 lysis mix during tissue lysis.
Western blotting
Western blot analysis was performed using the Bio‐Rad Criterion Cell Midi blot system. Proteins were collected as described above and quantified using the Bio‐Rad Protein Assay Dye method (cat. no. 500‐0006) before being separated on a 10% or 4–20% gradient TGX™ Precast Midi (Bio‐Rad, cat. no. 5671094) polyacrylamide gel and transferred to a nitrocellulose blotting membrane (GE Healthcare Life Sciences). Insoluble protein fractions were suspended in a 1:1 volume match with TEN buffer (10 mM Tris–HCl, 1 mM EDTA, 100 mM NaCl plus protease inhibitors) and sonicated for 1‐s intervals at 20% amplitude on ice. 5 or 10 μl of the suspension was added to an equal volume of 5× loading buffer (1 M Tris–HCl, 25% glycerol, 2% SDS, 5% 2‐mercaptoethanol, 0.1% bromophenol blue) and heated to 95°C for 5 min before being separated by electrophoresis. Following transfer, membranes were blocked in 5% non‐fat dry milk suspended in Tris‐buffered saline plus 0.1% Tween 20 (TBST) for 45–60 min at room temperature and probed with primary antibodies against Ub‐K48 (EMD‐Millipore, cat. no. 05‐1307), Omi/HtrA2 (BioVision, cat no. 3497‐100), Actin (Santa Cruz Biotechnology, cat. no. sc‐47778), β5 proteasome subunit (Enzo Life Sciences, cat no. BML‐PW8895‐0025), and Tubulin (UofIowa, cat no. 12G10) overnight at 4°C. All primary antibodies were suspended in 2.5% milk TBST. After three washes in TBST, blots were probed with horseradish peroxidase‐conjugated anti‐mouse (Kindle Biosciences, cat. no. 1005) or anti‐rabbit (Kindle Biosciences, cat no. R1006) antibodies and detected using enhanced chemiluminescence (Millipore or Kindle Biosciences). Images of developed blots were captured either using film and then digitized with a flatbed scanner or using the KwikQuant Imager system (Kindle Biosciences, cat. no. D1001). Band quantification was performed in ImageJ using the built‐in densitometry tools.
Native gel
Native gels were performed similarly to as has been described 31. Tissues were snap‐frozen and ground with a pestle and mortar. Tissue powders (~50 μl) were then titurated using a 25G syringe on ice in native tissue lysis buffer (50 mM Tris–HCl (pH 8.0), 5 mM MgCl2, 0.5 mM EDTA, and 1 mM ATP) with three 1 s vortex pulses. Lysates were then clarified by centrifugation at 4°C for 25 min at 20,000 × g. Clarified supernatants were transferred to a new tube and quantified as described for Western blot. 20 μg of protein was loaded per well along with 6× native protein loading buffer (Bio‐Rad cat # 1610738) into a 4–20% PAGE gel (Bio‐Rad cat # 5671094) in native running buffer (90 mM Tris base, 90 mM boric acid, 5 mM MgCl2, 0.5 mM EDTA, 1 mM ATP‐MgCl2) and resolved for 3.5 h at 100 V on ice. The gel was then soaked in transfer buffer (25 mM Tris base, 192 mM glycine) containing 1% SDS for 5 min, then soaked for 10 min in transfer buffer alone. Proteins were then transferred to a nitrocellulose membrane in transfer buffer at 250 mA for 90 min on ice. Probing was performed as described above.
Proteasome fluorescence assay
Proteasome activity was assessed as previously described 32. 10 μg of tissue protein lysates was added to proteasome activity assay buffer (50 mM Tris–HCl, pH 7.5) along with 10 μM of either chymotrypsin (Suc‐LLVY‐AMC) (Calbiochem, cat. no. 539142), trypsin (Ac‐RLR‐AMC) (Boston Biochem, cat. no. s290) or caspase (Ac‐GPLD‐AMC) (Enzo Life Sciences, cat no. BML‐AW9560‐0005) fluorogenic proteasome substrates. Reactions were incubated at 37°C protected from light for 3 h. Following incubation, samples were transferred to black walled 96‐well plates (Greiner Bio‐One, cat no. 655097). Release of free 7‐amino‐4‐methylcoumarin (AMC) was determined using a SpectraMax M5e Microplate Reader (Molecular Devices) with excitation at 380 nm and emission recorded at 460 nm. Reported values are the fold increase over a no‐protein control. For treatment with bortezomib, 10 μg of protein lysate was preincubated with either 2 mM bortezomib or an equivalent volume of DMSO at 4°C for 1 h before adding reaction buffer containing substrate.
Filter trap assay
Insoluble fractions of spinal cord lysates were suspended in TEN buffer and quantified using the Bio‐Rad Protein Assay Dye System (cat. no. 500‐0006). 25 μg of each sample was brought to an equal volume in TEN buffer. 5% Trition X‐100 in Tris‐buffered saline (TBS) was then added to each sample to a final concentration of 0.5% and incubated on ice for 10 min. Samples were then added to the filter trap assembly (Bio‐Rad) and passed through a nitrocellulose membrane with a pore size of 0.2 μm (Optitran). After a short transfer (15 s) or long transfer (1 min), the membrane was removed, washed in TBST, and blocked with 5% milk in TBST for 1‐h shaking at room temperature. The membrane was then probed with a primary antibody against SOD1 (Santa Cruz Biotechnology, cat. no. sc‐17767) overnight. Blots were then visualized as described above for Western blotting.
Electron microscopy
5 μl of insoluble protein fraction suspensions was spotted and air‐dried on a carbon‐coated grid for transmission electron microscopy, then stained with UranyLess (Electron Microscopy Sciences, cat no. 22400), and counterstained with lead citrate. Images were captured using a Hitachi 7500. Images were analyzed in ImageJ. Aggregate sizes were determined by thresholding each image and quantifying the area occupied by protein aggregates in each field.
Statistics
Data on graphs are represented as the mean ± SEM. Student's t‐test was used to ascertain a significant difference in mean when comparing two groups.
Proteasome subunit expression in human tissue
Data were obtained from the Human Protein Atlas project: (https://www.proteinatlas.org/download/rna_tissue.tsv.zip). Reported transcript per million (TPM) values for genes in the KEGG_PROTEASOME gene set (M10680) was recorded for each tissue and organized into a heat map using GraphPad Prism 8. Principal component analysis was conducted using the BioJupies tool (https://doi.org/10.1016/j.cels.2018.10.007) and RNAseq data collected from 20‐ to 29‐year‐old male (n = 14) or female (n = 12) in the Genotype‐Tissue Expression (GTEx) project.
Author contributions
The project was conceived by DG, ECJ, and GM. Experiments were planned by DG, ECJ, and GM. Experiments were performed by ECJ, NS, MG, TCK, SRG, GC, and DZ. Data were analyzed and interpreted by DG, ECJ, and GM. The manuscript was written by DG, ECJ, and GM with comments from all authors.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Review Process File
Acknowledgements
This work was funded by NIH R01NS084486 award to D. Germain and G. Manfredi and by NIH R01NS062055 award to G. Manfredi. We thank Dr. Csaba Konrad (BMRI, Weill Cornell Medicine) for building the heat‐shock equipment.
EMBO Reports (2020) 21: e48978
Contributor Information
Giovanni Manfredi, Email: gim2004@med.cornell.edu.
Doris Germain, Email: doris.germain@mssm.edu.
References
- 1. Tanaka K, Mizushima T, Saeki Y (2012) The proteasome: molecular machinery and pathophysiological roles. Biol Chem 393: 217–234 [DOI] [PubMed] [Google Scholar]
- 2. Rousseau A, Bertolotti A (2018) Regulation of proteasome assembly and activity in health and disease. Nat Rev Mol Cell Biol 19: 697–712 [DOI] [PubMed] [Google Scholar]
- 3. Murata S, Takahama Y, Kasahara M, Tanaka K (2018) The immunoproteasome and thymoproteasome: functions, evolution and human disease. Nat Immunol 19: 923–931 [DOI] [PubMed] [Google Scholar]
- 4. Schmidt M, Finley D (2014) Regulation of proteasome activity in health and disease. Biochim Biophys Acta 1843: 13–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gomes AV (2013) Genetics of proteasome diseases. Scientifica (Cairo) 2013: 637629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cleynen I, Vazeille E, Artieda M, Verspaget HW, Szczypiorska M, Bringer MA, Lakatos PL, Seibold F, Parnell K, Weersma RK et al (2014) Genetic and microbial factors modulating the ubiquitin proteasome system in inflammatory bowel disease. Gut 63: 1265–1274 [DOI] [PubMed] [Google Scholar]
- 7. Gandhi J, Antonelli AC, Afridi A, Vatsia S, Joshi G, Romanov V, Murray IVJ, Khan SA (2019) Protein misfolding and aggregation in neurodegenerative diseases: a review of pathogeneses, novel detection strategies, and potential therapeutics. Rev Neurosci 30: 339–358 [DOI] [PubMed] [Google Scholar]
- 8. Briston T, Hicks AR (2018) Mitochondrial dysfunction and neurodegenerative proteinopathies: mechanisms and prospects for therapeutic intervention. Biochem Soc Trans 46: 829–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ciechanover A (1994) The ubiquitin‐proteasome proteolytic pathway. Cell 79: 13–21 [DOI] [PubMed] [Google Scholar]
- 10. Hershko A, Ciechanover A (1998) The ubiquitin system. Annu Rev Biochem 67: 425–479 [DOI] [PubMed] [Google Scholar]
- 11. Song L, Luo ZQ (2019) Post‐translational regulation of ubiquitin signaling. J Cell Biol 218: 1776–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Collins GA, Goldberg AL (2017) The logic of the 26S proteasome. Cell 169: 792–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Budenholzer L, Cheng CL, Li Y, Hochstrasser M (2017) Proteasome structure and assembly. J Mol Biol 429: 3500–3524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tomaru U, Takahashi S, Ishizu A, Miyatake Y, Gohda A, Suzuki S, Ono A, Ohara J, Baba T, Murata S et al (2012) Decreased proteasomal activity causes age‐related phenotypes and promotes the development of metabolic abnormalities. Am J Pathol 180: 963–972 [DOI] [PubMed] [Google Scholar]
- 15. Dahlmann B (2016) Mammalian proteasome subtypes: their diversity in structure and function. Arch Biochem Biophys 591: 132–140 [DOI] [PubMed] [Google Scholar]
- 16. Morozov AV, Karpov VL (2018) Biological consequences of structural and functional proteasome diversity. Heliyon 4: e00894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang X, Zhao Z, Luo Y, Chen G, Li Z (2011) Gel‐based proteomics analysis of the heterogeneity of 20S proteasomes from four human pancreatic cancer cell lines. Proteomics Clin Appl 5: 484–492 [DOI] [PubMed] [Google Scholar]
- 18. Baraibar MA, Liu L, Ahmed EK, Friguet B (2012) Protein oxidative damage at the crossroads of cellular senescence, aging, and age‐related diseases. Oxid Med Cell Longev 2012: 919832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang X, Yen J, Kaiser P, Huang L (2010) Regulation of the 26S proteasome complex during oxidative stress. Sci Signal 3: ra88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bajorek M, Finley D, Glickman MH (2003) Proteasome disassembly and downregulation is correlated with viability during stationary phase. Curr Biol 13: 1140–1144 [DOI] [PubMed] [Google Scholar]
- 21. Labbadia J, Morimoto RI (2014) Proteostasis and longevity: when does aging really begin? F1000Prime Rep 6: 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Morimoto RI, Cuervo AM (2014) Proteostasis and the aging proteome in health and disease. J Gerontol A Biol Sci Med Sci 69(Suppl 1): S33–S38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sala AJ, Bott LC, Morimoto RI (2017) Shaping proteostasis at the cellular, tissue, and organismal level. J Cell Biol 216: 1231–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Walther DM, Kasturi P, Zheng M, Pinkert S, Vecchi G, Ciryam P, Morimoto RI, Dobson CM, Vendruscolo M, Mann M et al (2015) Widespread proteome remodeling and aggregation in aging C. elegans . Cell 161: 919–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Heiman‐Patterson TD, Deitch JS, Blankenhorn EP, Erwin KL, Perreault MJ, Alexander BK, Byers N, Toman I, Alexander GM (2005) Background and gender effects on survival in the TgN(SOD1‐G93A)1Gur mouse model of ALS. J Neurol Sci 236: 1–7 [DOI] [PubMed] [Google Scholar]
- 26. Stanhill A, Haynes CM, Zhang Y, Min G, Steele MC, Kalinina J, Martinez E, Pickart CM, Kong XP, Ron D (2006) An arsenite‐inducible 19S regulatory particle‐associated protein adapts proteasomes to proteotoxicity. Mol Cell 23: 875–885 [DOI] [PubMed] [Google Scholar]
- 27. Yun C, Stanhill A, Yang Y, Zhang Y, Haynes CM, Xu CF, Neubert TA, Mor A, Philips MR, Ron D (2008) Proteasomal adaptation to environmental stress links resistance to proteotoxicity with longevity in Caenorhabditis elegans . Proc Natl Acad Sci USA 105: 7094–7099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Guerrero‐Garcia TA, Gandolfi S, Laubach JP, Hideshima T, Chauhan D, Mitsiades C, Anderson KC, Richardson PG (2018) The power of proteasome inhibition in multiple myeloma. Expert Rev Proteomics 15: 1033–1052 [DOI] [PubMed] [Google Scholar]
- 29. Bross PF, Kane R, Farrell AT, Abraham S, Benson K, Brower ME, Bradley S, Gobburu JV, Goheer A, Lee SL et al (2004) Approval summary for bortezomib for injection in the treatment of multiple myeloma. Clin Cancer Res 10: 3954–3964 [DOI] [PubMed] [Google Scholar]
- 30. Vallanat B, Anderson SP, Brown‐Borg HM, Ren H, Kersten S, Jonnalagadda S, Srinivasan R, Corton JC (2010) Analysis of the heat shock response in mouse liver reveals transcriptional dependence on the nuclear receptor peroxisome proliferator‐activated receptor alpha (PPARalpha). BMC Genom 11: 16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Elsasser S, Schmidt M, Finley D (2005) Characterization of the proteasome using native gel electrophoresis. Methods Enzymol 398: 353–363 [DOI] [PubMed] [Google Scholar]
- 32. Riar AK, Burstein SR, Palomo GM, Arreguin A, Manfredi G, Germain D (2017) Sex specific activation of the ERalpha axis of the mitochondrial UPR (UPRmt) in the G93A‐SOD1 mouse model of familial ALS. Hum Mol Genet 26: 1318–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Review Process File


