Significance
Pre-messenger RNA (pre-mRNA) splicing is a crucial step in eukaryotic gene expression. The recognition of the splice sites in pre-mRNA transcripts is initiated by the essential splicing factor U2AF2 that binds to the poly-pyrimidine tract (Py-tract) RNA upstream of exons to assemble the spliceosome. Py-tract sequences are often degenerate, with a wide range of binding affinities and activity. Here, we demonstrate that autoinhibitory intramolecular interactions of a linker region and the RNA binding domains of U2AF2 establish binding selectivity for strong Py-tracts. Disrupting the linker interactions results in dispersed binding to weak Py-tracts and impacts on splicing fidelity. This demonstrates that the binding specificity of RNA binding proteins can involve flanking regions of the canonical RNA binding domains.
Keywords: splicing, protein–RNA interactions, U2 auxiliary factor, structural biology, iCLIP
Abstract
The recognition of cis-regulatory RNA motifs in human transcripts by RNA binding proteins (RBPs) is essential for gene regulation. The molecular features that determine RBP specificity are often poorly understood. Here, we combined NMR structural biology with high-throughput iCLIP approaches to identify a regulatory mechanism for U2AF2 RNA recognition. We found that the intrinsically disordered linker region connecting the two RNA recognition motif (RRM) domains of U2AF2 mediates autoinhibitory intramolecular interactions to reduce nonproductive binding to weak Py-tract RNAs. This proofreading favors binding of U2AF2 at stronger Py-tracts, as required to define 3′ splice sites at early stages of spliceosome assembly. Mutations that impair the linker autoinhibition enhance the affinity for weak Py-tracts result in promiscuous binding of U2AF2 along mRNAs and impact on splicing fidelity. Our findings highlight an important role of intrinsically disordered linkers to modulate RNA interactions of multidomain RBPs.
Pre-mRNA splicing is an essential mechanism in eukaryotic gene expression. Alternative splicing greatly contributes to proteome diversity of higher eukaryotes by differential inclusion of specific exons or usage of distinct splicing boundaries (1–3). A critical early step involves defining the exon/intron boundaries in the pre-mRNA transcripts through the recognition of cis-regulatory RNA motifs by splicing factors. This entails the challenging task of sequentially identifying degenerate key motifs, namely the 5′ splice site, BPS (branch point site), and 3′ splice site, of a given intron, similar to finding needles in a haystack (1). During early spliceosome assembly, in the so-called complex E, the U2AF (U2 auxiliary factor) heterodimer recognizes the Py (polypyrimidine)-tract and the downstream dinucleotide AG in the 3′ splice site by its large (U2AF2, also known as U2AF65) and small (U2AF1) subunit, respectively (4–10). In addition, SF1 (splicing factor 1) binds the BPS (11–13) (Fig. 1A). U2AF2 comprises two canonical RNA recognition motif domains (RRM1 and RRM2) that are connected by a short linker sequence (together referred to as RRM1,2) and mediate RNA binding by a population shift from closed to open states of RRM1,2. An atypical RRM variant in U2AF1, the so-called UHM (U2AF homology motif; Fig. 1A), recognizes a UHM ligand motif (ULM) in the N-terminal region of U2AF2 to form the U2AF heterodimer (14–17).
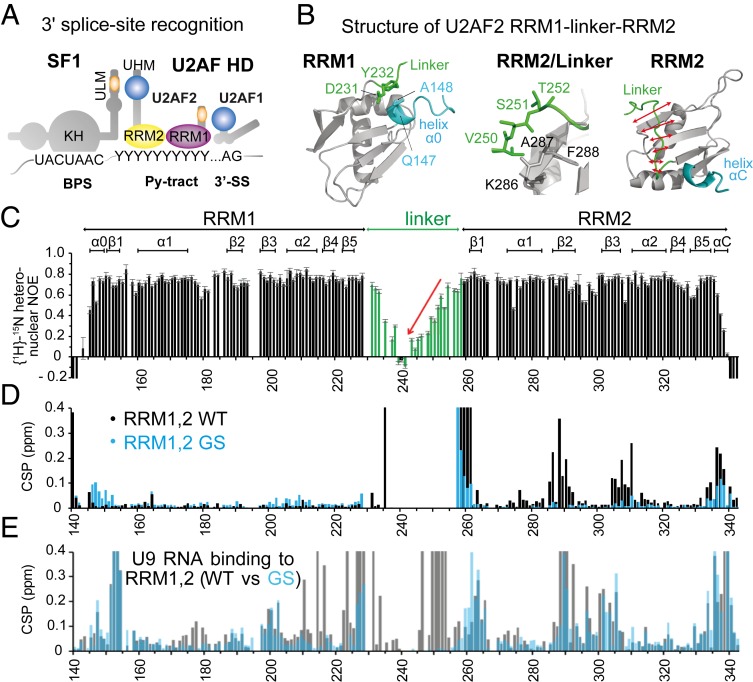
Fig. 1.
Overview of 3′ splice site recognition and structural features of U2AF2 RRM1,2. (A) Overview of cis-regulatory elements (BPS, Py-tract, AG dinucleotide) and splicing factors (SF1, U2AF2, U2AF1) that cooperate in 3′ splice site recognition in human introns. Asterisks denote phosphorylation sites. (B) NMR structures of U2AF2 RRM1 and RRM2 obtained for the redefined U2AF2 RRM1,2 protein (residues 140 to 342). RRM1 (Left) is preceded by a short helical turn (α0), which is in proximity to the N-terminal residues of the RRM1,2 linker, also showing reduced flexibility. RRM2 (Right) shows that the C-terminal region of the linker packs against the β-sheet surface that is also involved in RNA binding. RRM2 has a short C-terminal helical turn (helix αC, blue). (C) NMR {1H}-15N heteronuclear relaxation data for U2AF2 RRM1,2 show that the central linker is highly flexible, but exhibits increased rigidity as it approaches the N-terminal end of the RRM2 fold. (D) Chemical shift differences comparing wildtype (WT; black) and the linker GGS mutation (GS; cyan) of U2AF2 RRM1,2 vs. the individual RRM1 and RRM2 domains. (E) Chemical shift perturbation for WT (black) and GS (cyan) U2AF2 RRM1,2 upon addition of U9 RNA.
The recognition of diverse Py-tract sequences with various binding strengths by U2AF2 documents its important role for 3′ splice-site selection. However, the fact that natural Py-tract sequences can be quite degenerate, with a wide range of binding affinities to U2AF and thus efficiency to promote spliceosome assembly, raises the question how U2AF2 ensures fidelity of splicing. For some splice sites, U2AF2 selectivity arises from its interaction with U2AF1, which was shown to aid U2AF2 in binding to weaker Py-tracts at the 3′ splice site by providing additional interactions with the dinucleotide AG in a cooperative manner (5–7). However, U2AF2 was also reported to bind independently of a downstream AG at intronic and exonic binding sites, which can be functional and contribute to distal regulation of splicing (18, 19) and RNA export (20). In order to investigate the different arrangements of U2AF2 binding, we recently introduced in vitro iCLIP (individual-nucleotide resolution UV cross-linking and immunoprecipitation) as an efficient method to measure the intrinsic binding of U2AF2 RRM1,2 across hundreds of binding sites in in vitro transcripts (21). Interestingly, we found that, at ∼50% of Py-tracts, the binding of recombinant U2AF2 RRM1,2 mirrors U2AF2 in vivo binding. This suggests that a major part of U2AF2’s RNA binding preference is inherent to the two RRM domains and their linker region.
At the molecular level, we previously found that U2AF2 RNA binding specificity relies on a dynamic population shift from closed to open conformations. Structural studies combining NMR spectroscopy, SAXS, computational analysis, and FRET revealed that, in the absence of RNA, the U2AF2 RRM1,2 domains exist mainly in a dynamic arrangement of closed, inactive states (9, 10, 22). This dynamic equilibrium is shifted from inactive closed states to the open, RNA-bound domain arrangement depending on the “strength,” i.e., overall binding affinity, of the RNA ligand. This population shift correlates the Py-tract strength with the efficiency of splicing of a given intron.
Molecular and structural details have been reported for how strong Py-tracts are recognized by the open U2AF2 conformation (8, 9), but the molecular mechanisms that reduce binding to weak, presumably nonfunctional, Py-tracts are poorly understood. An open question also concerns the role of the intrinsically disordered linker region connecting RRM1 and RRM2 of U2AF2. This linker is flexible in solution and not directly involved in RNA recognition (9, 22), but its potential role for modulating U2AF2 binding specificity is unknown.
Here, we combine structural biology with iCLIP experiments to elucidate the molecular mechanisms underlying the RNA binding specificity of U2AF2. An NMR-based solution structure of free U2AF2 RRM1,2 reveals that the intrinsically disordered linker weakly interacts with the RNA binding surface of RRM2 and thus exhibits an autoinhibitory function by competing with RNA binding to RRM2. Mutations that impair linker autoinhibition result in a significantly increased nonspecific binding to dozens of weak Py-tracts in natural pre-mRNA transcripts in vitro and impair splicing fidelity in vivo. Our results thus suggest that the linker autoinhibition serves as an intrinsic proofreading mechanism that enhances the fidelity of Py-tract RNA recognition and suppresses nonproductive U2AF2 binding to sites that are not bona fide splicing substrates. This demonstrates an unexpected molecular mechanism that underlies the RNA binding selectivity of the splicing factor U2AF2 during early steps of spliceosome assembly.
Results
Solution Structure of Free U2AF2 RRM1,2 Reveals a Dynamic Linker–RRM2 Interaction.
Previous studies on the RNA binding of U2AF2 mostly focused on RRM1 and RRM2, but suggested a potential role of the intervening linker region (residues 231 to 258) for regulating the dynamic RNA recognition by U2AF2 (8, 9, 22). Notably, the amino acid sequence in the RRM1–RRM2 linker is evolutionarily conserved in length and in the presence of multiple hydrophobic aliphatic residues, suggesting a potential functional relevance (SI Appendix, Fig. S1). We therefore set up to study its structure and function in more detail.
In order to study the conformation of RRM1–RRM2 and the role of the connecting linker, we first delimited the functionally relevant RNA binding region of U2AF2. We have previously shown that the addition of the U2AF1-binding ULM region (residues 88 to 147) to a short construct of RRM1,2 (residues 148 to 342) noticeably enhances the binding affinity for Py-tract U9 RNA (10). To identify the region that confers this additional affinity, we compared NMR spectra of various U2AF2 RRM1,2 constructs and measured the RNA binding affinity by isothermal titration calorimetry (ITC; SI Appendix, Table S1 and Figs. S2 and S3). This analysis showed that the region comprising residues 140 to 342 (RRM1,2 from here on) captures the complete RNA binding affinity of U2AF2 (SI Appendix).
We next determined the three-dimensional structure of U2AF2 RRM1,2 using solution NMR (Fig. 1B and SI Appendix, Fig. S4 A–C and Table S2) (23, 24). The NMR ensemble of RRM1,2 shows that the individual RRM1 and RRM2 domains are well-defined but do not adopt a specific domain arrangement in solution, as shown previously (22). The RRM domain folds are very similar to those reported previously (SI Appendix, Fig. S4D). However, our structures of the unbound RRM1 and RRM2 domain reveal two notable features. First, two additional short helices form at the N terminus of RRM1, helix α0, and at the C terminus of RRM2, helix αC (Fig. 1B). In particular, the orientation of the N-terminal helix is stabilized by interactions with residues at the N-terminal region of the RRM1–RRM2 linker (D231/Y232). Therefore, the higher RNA binding affinity of the larger RRM1,2 construct suggests a potential role for these elements in enhancing RNA binding. Second, the C-terminal region of the linker (residues 250 to 259) adopts a well-defined conformation in the solution structure and binds to RRM2, supported by 84 distance restraints derived from proton–proton nuclear Overhauser enhancements (NOEs) between the linker and RRM2. In detail, three residues (V250/S251/T252) are packed against to the N-terminal end of strand β2 in RRM2 (K286/A288/F288) in an antiparallel manner, while the rest of this region (residues 253 to 259) interacts with residues in the RRM2 β-sheet.
The conformational flexibility of the two flanking helices and the RRM1–RRM2 linker of RRM1,2 was assessed by 15N-relaxation NMR experiments (Fig. 1C). Consistent with the structural analysis, the N-terminal helix α0 in RRM1 exhibits reduced flexibility at nanosecond times scales, reflected by {1H}-15N heteronuclear NOE values that are only slightly lower compared to the core RRM domains. The RRM1–RRM2 linker is flexible, as indicated by low heteronuclear NOE values. The highest flexibility is observed for the N-terminal region of the linker (residues 235 to 249), beyond the residues (D231/Y232) that stabilize helix α0. Consistently, NMR signals corresponding to these N-terminal linker residues (residues 235 to 245) in NMR spectra of the constructs, RRM1–linker and linker–RRM2, superimpose well with those in the spectrum of RRM1,2, indicative of their intrinsically disordered nature and absence of contacts to RRM1 or RRM2 (SI Appendix, Fig. S3 A and B). This agrees well with sequence-based prediction of disordered residues using IUPred2A (25) (SI Appendix, Fig. S3C), in which the N-terminal region of the linker exhibits the highest IUPred2A scores, while residues in the C-terminal region of the linker are predicted to have propensity to being structured. Indeed, our experimental heteronuclear NOE values display a gradual increase in the C-terminal region of the linker (residues 250 to 258) toward the RRM2 domain, indicative of limited flexibility (Fig. 1C). This is consistent with the interaction of the C-terminal region of the RRM1–RRM2 linker with the RNA binding interface of RRM2, as seen in our structure (Fig. 1B), although in a dynamic manner. The interaction of the C-terminal linker region with RRM2 is further supported by comparing NMR spectra of U2AF2 RRM1,2 with the individual RRM1 and RRM2 domains. Significant chemical shift differences are observed mainly for NMR signals of residues in RRM2 (Fig. 1D), suggesting that the linker residues transiently interact with the core RRM2 fold. Altogether, these observations underline that, in the absence of RNA, the RRM1–RRM2 linker of U2AF2 is largely disordered, but its C-terminal region shows intramolecular interactions with RRM2. Most surprisingly, our structure shows that the linker binds close to and partially overlapping with the RNA binding interface identified by NMR titrations (Fig. 1E and SI Appendix, Fig. S5), i.e., the N-terminal end of strand β2, the C-terminal end of strand β3, and the C-terminal helix, that are partially overlapping with the RNA binding interface.
The Linker Plays an Autoinhibitory Role in Py-Tract RNA Binding.
The interaction with the RNA binding interface of RRM2 suggests a potential role of the RRM1–RRM2 linker in regulating RNA binding. To further explore this, we removed the linker/RRM2 contacts by replacing the core linker region (residues 233 to 257) with Gly-Gly-Ser repeats of the same length (RRM1,2-GS; Fig. 2A). In addition, V254 was mutated to proline, as the amide proton of V254 is involved in numerous interlinker/RRM2 contacts in the free RRM1,2 structure. The superposition of NMR spectra of RRM1,2-GS and RRM1,2-WT shows significant, nontrivial chemical shift differences for the residues in RRM2 that interact with the linker (Fig. 1D and SI Appendix, Fig. S5B). These chemical shift differences are similar to those comparing RRM1,2-WT and RRM2 alone, i.e., in the absence of the linker (Fig. 1D, black). On the contrary, NMR signals of RRM2 superimpose very well with the corresponding region in RRM1,2-GS (Fig. 1C, cyan), consistent with the absence of linker interactions in both cases. Collectively, these data show that RRM1,2-GS comprises two functional RRM domains but lacks the contacts of the RRM1,2 linker to RRM2.
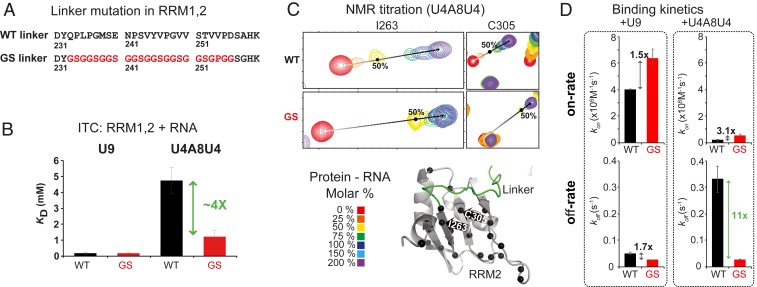
Fig. 2.
RNA binding of U2AF2 RRM1,2-WT and RRM1,2-GS to weak and strong Py-tract RNA. (A) Residues in the RRM1,2 linker are replaced by Gly-Gly-Ser, and V254P to remove transient linker/RRM2 interactions. (B) Dissociation constants KD for the binding of RRM1,2-WT and RRM1,2-GS to strong (U9) and weak (U4A8U4) Py-tract RNAs determined by ITC. (C) NMR titrations showing spectral changes for residues in RRM2 that are located in proximity to the linker upon binding the weak Py-tract RNA. (D) Kinetic rates for the interaction of RRM1,2-WT and -GS with RNAs determined by switchSENSE.
Next, we used ITC to assess the potential role of the linker in modulating the binding of U2AF2 to strong and weak Py-tract RNAs (U9 and U4A8U4, respectively; Fig. 2B). When titrating U9 RNA, no significant difference in binding affinity is observed in the absence of the linker/RRM2 interaction (RRM1,2-GS; KD = 170 nM) when compared to RRM1,2-WT (KD = 185 nM). Strikingly, when RRM1,2-GS is titrated with the weak Py-tract RNA (U4A8U4), we observe a substantial increase (>fourfold) in the binding affinity compared to the RRM1,2-WT (Fig. 2B). We therefore reasoned that the competition of linker and RNA for RRM2 favors binding of strong over weak Py-tracts.
To structurally map the differences of RRM1,2-WT and -GS in RNA binding, we analyzed the NMR chemical shift changes of amide signals in titration experiments with strong and weak Py-tract RNAs. Overall, in all four combinations, similar chemical shift perturbations are observed that map around the expected RNA binding interfaces, including the RNP sequence motifs in the β-sheets of RRM1/2 and the N-/C-terminal helical extensions of RRM1 and RRM2 (Fig. 1E and SI Appendix, Fig. S5C). Interestingly, upon RNA binding, additional chemical shift perturbations are observed in RRM1,2-GS (i.e., for the amides of F288 and I310) compared to RRM1,2-WT (SI Appendix, Fig. S6A), where these residues are shielded by the linker interaction. These data further corroborate the autoinhibitory role of the linker, and show that the dynamic linker/RRM2 interaction reduces the RNA binding affinity of RRM1,2. The linker thereby proofreads against the binding of weak RNA ligands by directly competing for the RNA binding interface on the RRM2. Of note, the autoinhibitory role of the linker is also recapitulated in the context of the minimal U2AF heterodimer (SI Appendix, Fig. S6B), indicating that the presence of the small subunit U2AF1 does not affect or modulate the RRM2/linker interaction.
Finally, to assess the binding kinetics of the RNA interactions, we determined experimental on- and off-rates (kon, koff) for the binding of U2AF2 RRM1,2-WT and RRM1,2-GS using SwitchSENSE (Methods). Consistent with NMR titrations (SI Appendix, Supplementary Text and Figs. S6 and S7 and Fig. 2C), we observed the largest differences between RRM1,2-WT and RRM1,2-GS in the off-rates upon binding to the weak Py-tract RNA (Fig. 2D and SI Appendix, Fig. S8), where presence of the linker interaction increases the off-rate 11-fold. There is also an effect on the on-rate, which is 3-fold smaller in the wildtype protein, as the linker interaction can effectively compete with binding of a weak Py-tract RNA and thus reduce the on-rate of complex formation. The off-rate–driven binding inhibition for weak Py-tract RNAs can be rationalized by realizing that the linker competition occurs within an existing macroscopic complex of the protein and RNA. As the linker interacts only with a part of the RNA-binding interface of RRM2, the RNA can interact with the flanking, accessible region on the surface of RRM2. The linker interaction inhibits some, but not all, contacts of a weak Py-tract RNA ligand with RRM2, thus resulting in a short-lived complex (faster off-rate). This effect is much reduced for strong Py-tracts, which can effectively outcompete the transient linker interaction with the RRM surface. The overall contribution of on- and off-rates affords a higher apparent binding affinity for RRM1,2-GS, i.e., in the absence of the linker interaction, which is consistent with the significantly increased binding affinity determined by ITC for RRM1,2-GS binding to weak Py-tracts (Fig. 2B).
To further dissect the molecular features that establish the autoinhibitory role of the linker, we analyzed whether the linker/RRM2 interaction directly alters the RNA binding of RRM2 in the absence of RRM1. To this end, we tested U2AF2 fragments comprising the RRM2 domain with and without the linker, namely linker–RRM2 (residues 231 to 342) and RRM2 (residues 258 to 342), respectively (Fig. 3A). The presence of the linker reduces the binding affinity of RRM2 for the weak Py-tract even more than seen for the tandem RRM1,2 construct (10- vs. 4-fold; Fig. 3B). Interestingly, the presence of the linker also reduces the binding affinity for the strong Py-tract fourfold. This supports that the dynamic linker/RRM2 interaction limits the steric accessibility of RRM2 (Fig. 3B). However, considering that binding to strong Py-tracts is not affected in the tandem RRM1,2 construct, the dynamic interaction of the linker with RRM2 must be significantly weaker than the RNA binding affinity of RRM2 for strong Py-tracts, but comparable to the interaction with weak Py-tracts. Thus, in the presence of RRM1, binding cooperativity of the tandem RRM1,2 domains reduces the autoinhibitory role of the linker and thereby leads to a binding preference for strong Py-tracts, as seen in Fig. 2B.
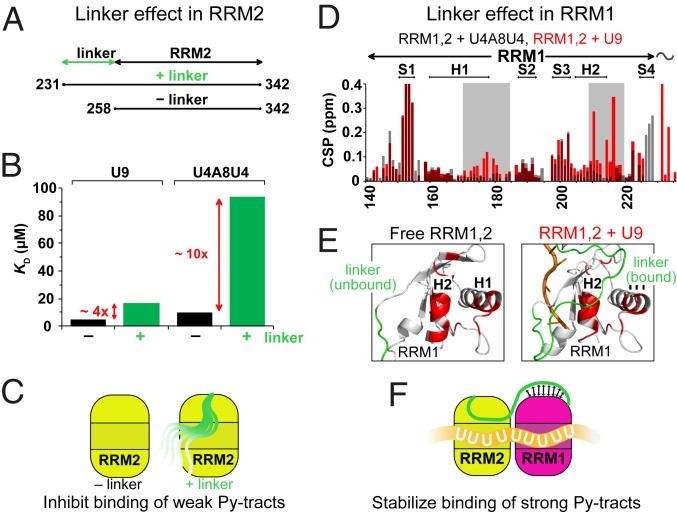
Fig. 3.
Molecular features that enable proofreading of U2AF2 RRM1,2 against weak Py-tracts. (A) Constructs used (RRM2 and linker–RRM2) to study the role of the linker. (B) Dissociation constants KD for the binding of RRM2 and linker–RRM2 to strong (U9) and weak (U4A8U4) Py-tract RNAs determined by ITC. (C) Role of the C-terminal region of the RRM1,2 linker: the C-terminal region of the linker competes with binding of weak Py-tracts. (D) NMR chemical shift perturbations (CSPs) in the RRM1 region upon binding of RRM1,2-WT to strong (U9) and weak (U4A8U4) Py-tract RNAs (cf. SI Appendix, Fig. S5). Significantly stronger CSPs are observed for residues (gray box) that contact the RNA upon binding to strong Py-tract RNA. (E) The N-terminal region of the RRM1,2 linker is flexible in the unbound protein, but weakly stabilizes RNA interactions when bound to a strong Py-tract RNA. The distinct CSPs upon binding to the strong Py-tract RNA shown in D are highlighted in red on a cartoon presentation of RRM1. (F) Role of the N-terminal region of the RRM1,2 linker: the N-terminal region of the linker may stabilize binding of strong (U-rich) Py-tract RNAs.
Careful comparison of the NMR titrations (RRM1,2-WT/GS to strong/weak Py-tracts) indicates an additional contribution of the linker and RRM1 in discriminating strong vs. weak Py-tracts (Fig. 3C and SI Appendix, Fig. S5C). Unique chemical shift changes seen for the titration of RRM1,2-WT with the strong Py-tract RNA map to the surface of RRM1 (Fig. 3D). Interestingly, this region corresponds to contacts between the N-terminal region of the linker and RRM1 that are seen in the crystal structure of RNA-bound U2AF2 (8), but absent in the free U2AF2 RRM1,2 structure (Fig. 3E). This suggests that the linker-to-RRM1 interaction is only induced when the two domains are juxtaposed on a long, i.e., strong, Py-tract and implicates its potential role in reinforcing the recognition of strong Py-tracts by pulling the two domains together.
Taken together, distinct molecular features of the RRM1–RRM2 linker modulate the RNA binding of U2AF2. Autoinhibition of the C-terminal region of the linker with RRM2 preferentially reduces the binding of U2AF2 to weaker, nonspecific Py-tracts. When binding to strong, high-affinity Py-tract RNAs, the N-terminal region of the linker appears to slightly stabilize the RNA interactions. Both effects work together in establishing an efficient proofreading against binding to weak RNA ligands by U2AF2 in vitro.
SAXS Analysis Indicates a Compaction of U2AF2 RRM1,2 upon RNA Binding.
We then tested how the linker affects the U2AF2 domain arrangement in solution using SAXS (small angle X-ray scattering). As expected, in the absence of RNA, a larger maximum particle distance, Dmax, is observed for RRM1,2-GS, indicating that the tandem domains can sample more extended conformations in the absence of linker interaction (SI Appendix, Fig. S9). Upon binding to strong Py-tracts, the overall shape of RRM1,2-WT and RRM1,2-GS protein–RNA complexes becomes significantly more compact, as indicated by changes in the pairwise distance distributions (SI Appendix, Fig. S9A) and the reduced maximum Dmax and radii of gyration Rg values (SI Appendix, Fig. S9B).
The RNA-induced compaction is stronger for U9-bound RRM1,2-WT compared to RRM1,2-GS. This likely reflects that the GS-linker is completely disordered and mainly extended because, in contrast to RRM1,2-WT, linker interactions with RRM1 and RRM2 are not present. Notably, upon binding weak Py-tracts, even more extended conformations are observed for both RRM1,2-WT and RRM1,2-GS proteins. As the stoichiometry of these complexes is 1:1 based on the SAXS data, this likely reflects that the two short U4 binding sites in the weak U4A8U4 Py-tract RNA may individually bind to the two domains, while not requiring that RRM1,2 adopts a fully closed state. In addition, considering the intrinsically weaker RNA binding affinity of RRM1, it is conceivable that RRM1 only partially contributes to RNA binding in the complex, in which the unbound fraction of RRM1 is detached from RRM2. Intriguingly, the overall dimensions and compactness of the U2AF2 RRM1,2 domains and their complexes with strong and weak Py-tract RNAs approximately correlate with their binding affinities, such that a more compact conformation is associated with a higher binding affinity (lower KD; Fig. 2B). This suggests that a compact arrangement of RNA binding domains and cognate cis-regulatory RNA binding motifs is important for a high RNA binding affinity. The overall compact state of the 3′ splice site recognition complex may thus be an important feature of the role of U2AF2 in spliceosome assembly.
Loss of Linker Autoinhibition Results in Dispersed U2AF2 Binding in Human Transcripts.
In order to examine how the linker/RRM2 interaction in U2AF2 affects binding to natural RNA sequences, we performed in vitro iCLIP experiments (21). This method allows us to measure the binding of recombinant proteins across hundreds of sites in in vitro transcripts of human genes to reveal how RBPs interpret the pre-mRNA sequence. We quantified the binding of RRM1,2-WT and RRM1,2-GS (1 μM) to an equimolar mixture of nine in vitro transcripts, which resemble endogenous pre-mRNAs (0.75 nM each) (26). We measured a total of 424 binding sites (Methods). As exemplified in the MAT2A transcript (Fig. 4A), the in vitro binding maps for RRM1,2-WT resemble in vivo iCLIP experiments with endogenous U2AF2 (21, 27). However, for RRM1,2-GS, in which the linker/RRM2 interaction is abolished, the in vitro iCLIP landscape markedly changes, such that many binding sites are strongly reinforced or newly appear in introns and exons (Fig. 4A). This results in a noticeably more dispersed binding of U2AF2, as exemplified by the fact that significantly more binding sites share the majority of all cross-link events from RRM1,2-GS compared to RRM1,2-WT or U2AF2 in vivo (Fig. 4B).
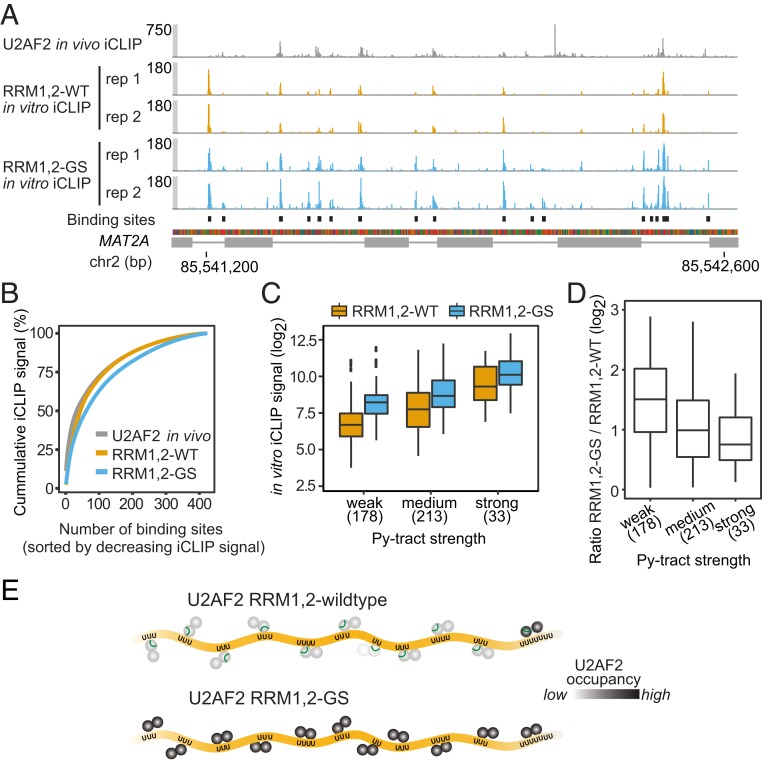
Fig. 4.
In vitro iCLIP analysis confirms the role of the linker region in proofreading against weak Py-tracts. (A) U2AF2 RRM1,2-GS shows increased binding throughout introns. Regulated binding sites show strong differences between in vivo and in vitro U2AF2 binding. Genome browser view of U2AF2 in vivo iCLIP (gray) as well as U2AF2 RRM1,2 (orange) and RRM1,2-GS (blue) in vitro iCLIP on MAT2A. In vitro iCLIP was performed with 1 μM RRM1,2(-GS) and an equimolar pool of nine in vitro-transcribed RNAs. (B) RRM1,2-GS disperses across binding sites. Cumulative fraction of iCLIP signal across binding sites of decreasing strength in the respective dataset. (C) Linker mutation increases RRM1,2 binding. Box plot to compare RRM1,2 and RRM1,2-GS binding (iCLIP signal) on binding sites with weak, medium, and strong Py-tracts. Number of binding sites in each category is given below. (D) Linker mutation shows strongest impact on weak Py-tracts. Box plots of log2-transformed ratios of normalized RRM1,2-GS over RRM1,2 in vitro iCLIP read counts. Binding site categories as in C. (E) The autoinhibitory linker interaction in U2AF2 RRM1,2-wildtype ensure specific binding to bona fide Py-tract sequences in pre-mRNA. Lack of this selectivity filter in the U2AF2 RRM1,2-GS mutant leads to nonspecific, more promiscous binding.
In order to test whether the linker interaction particularly impacts at weak Py-tracts, we stratified the binding sites by their associated Py-tract strength, which directly correlates with U2AF2 affinity (21) (SI Appendix, Fig. S10). As expected, the in vitro iCLIP data show most RRM1,2-WT binding on strong Py-tracts, which gradually decreases at medium and weak Py-tracts (Fig. 4C). We find that, compared to RRM1,2-WT, the binding of RRM1,2-GS is elevated in all three categories. Notably, the removal of the linker interaction shows the strongest effect on weak Py-tracts, which, on average, increase by 2.7-fold, while strong Py-tracts are only mildly affected (1.6-fold; Fig. 4D). Together, these observations support that the presence of the linker interaction suppresses binding to weak Py-tracts and highlights its autoinhibitory role in the context of natural RNA sequences (Fig. 4E).
Linker Autoinhibition Promotes Splicing Fidelity In Vivo.
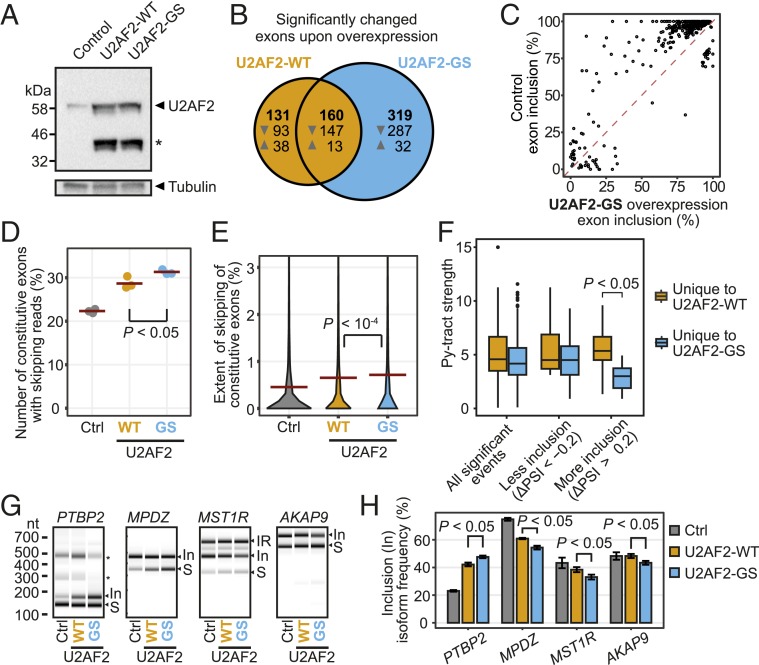
In order to investigate the functional role of the linker, we overexpressed full-length U2AF2-WT and the mutant version U2AF2-GS lacking the linker interaction in human HeLa cells (Fig. 5A). We monitored splicing changes by sequencing rRNA-depleted total RNA (28). Overexpression (OE) of U2AF2-WT triggers significant changes in 291 cassette exons (false discovery rate [FDR] < 0.05; Methods and Fig. 5B). In 240 out of 291 cases (82%), the inclusion of the exon is reduced, indicating that excessive U2AF2 partially interferes with splicing. In line with the more dispersed RRM1,2-GS binding in vitro, the splicing changes are even more pronounced when overexpressing the mutant version U2AF2-GS, which leads to a significant reduction in 434 exons (out of 479 regulated exons, 91%; Fig. 5 B and C). Interestingly, U2AF2-GS OE not only affects alternative exons, but also triggers an increased skipping of constitutive exons (Fig. 5 D and E). We hypothesize that the widespread reduction in exon inclusion represents squelching effects, whereby the promiscuous binding of U2AF2 quenches the recognition of bona fide splice sites. By contrast, the few exons that strongly increase in inclusion specifically upon U2AF2-GS OE (11 exons with ΔPSI > 20%, FDR < 0.05) are associated with weaker Py-tracts than exons that also react to U2AF2-WT OE (Fig. 5F and SI Appendix, Fig. S11). This suggests that stronger U2AF2-GS binding to these Py-tracts promotes recognition of the corresponding alternative exons.
Fig. 5.
Overexpression of U2AF2-WT and U2AF2-GS impairs constitutively spliced exons. (A) Western blot documents overexpression of U2AF2-WT and U2AF2-GS in human HeLa cells. Tubulin served as loading control. An asterisk marks a U2AF2 cleavage product. (B) U2AF2-WT OE leads to down-regulation of alternatively spliced exons, which is further augmented by U2AF2-GS. Venn diagram comparing significantly regulated exons (FDR < 0.05) upon OE of U2AF2-WT (orange) or U2AF2-GS (blue). Up- and down-regulated exons (indicated by arrowheads) are given in each section. (C) Most down-regulated exons show more than 80% inclusion in untreated cells. Scatterplot of inclusion levels (in percent spliced-in; PSI) in untreated HeLa cells (control) and upon U2AF2-GS OE. (D) More constitutive exons are skipped upon U2AF2-GS OE. Dot plot showing the percentage of constitutive exons (n = 37,052) with at least one junction-spanning read that supports skipping upon overexpression of U2AF2-WT (orange) or U2AF2-GS (blue) versus empty vector (gray, Ctrl). P value < 0.05 for all pairwise comparisons by Student’s t test. Only the P value for the selected comparison is shown. (E) Constitutive exons show an increased extend of skipping upon U2AF2-GS OE. Violin plot shows the distribution of percent skipping for constitutive exons as in E. Dashed lines indicate the mean value. P value < 0.001 for all pairwise comparisons by Student’s t test. Only the P value for the selected comparison is shown. (F) Strongly down-regulated exons upon U2AF2-GS OE have weaker Py-tracts. Box plot displays the Py-tract strengths for significantly regulated events (FDR < 0.05) with |ΔPSI| > 20% that are uniquely regulated upon overexpression of U2AF2-WT (orange) or U2AF2-GS (blue). P value from Student’s t test with Bonferroni correction. Only the P value for the selected comparison is shown. Boxes represent quartiles, center lines denote 50th percentile, and whiskers extend to most extreme values within 1.5× the interquartile range. Extended plot with exon numbers in each category is shown in SI Appendix, Fig. S11. (G) Reporter minigenes confirm effects of U2AF2-GS OE on splicing. Semiquantitative RT-PCR analyses of four alternatively spliced exons (PTBP2 exon 10, MPDZ exon 18, MST1R exon 11, AKAP9 exon 19) upon overexpression of U2AF2-WT (orange) or U2AF2-GS (blue) versus empty vector (gray; Ctrl). Gel views of capillary electrophoresis of the PCR products for exon inclusion (In) and skipping (“S”) marked on the right. Intron retention products (IR; MST1R) and unassigned isoforms (asterisks; PTBP2) are marked. (H) Quantification of alternative splicing changes in G. Bar diagrams depict the mean inclusion level in each sample. Error bars represent SD of mean; n = 3. n.s., P values from Student’s t test with Benjamini–Hochberg correction. Only P values for the selected comparisons are shown.
Reporter assays confirmed that U2AF2-GS OE can influence exon inclusion in both directions. For instance, PTBP2 exon 10 inclusion is significantly enhanced, whereas inclusion of MPDZ exon 18, MST1R exon 11, and AKAP9 exon 19 is reduced (Fig. 5 G and H). Importantly, the effects are significantly stronger for U2AF2-GS OE compared to U2AF2-WT OE for all tested minigenes (P value < 0.05, Student’s t test). More generally, the observed effects support that U2AF2-GS represents a gain of function rather than a loss of function, since the same effects are triggered to a lesser extent by U2AF2-WT. Altogether, these results corroborate that removal of the autoinhibitory RRM2–linker interaction impairs exon inclusion in vivo.
Discussion
The recognition motifs of RBPs are short in general and often quite degenerate (29), suggesting that a given RBP may bind to a large number of motifs throughout the transcriptome. It is thus difficult to rationalize how RBPs can bind to specific transcript locations and regulate distinct functions. Specific RNA recognition is particularly important in the context of splicing when splice sites need to be found in the large sequence space of pre-mRNAs. Among the RBPs binding the 3′ splice site, the essential splicing factor U2AF2 exhibits highest affinity and thereby forms the nucleation point for spliceosome assembly (1, 30). It is therefore critical for the cell to precisely define where and when U2AF2 binds to pre-mRNA.
Given that Py-tracts are highly abundant in the transcriptome, proofreading mechanisms have evolved to ensure U2AF2 selectivity. For instance, hnRNP A1 (31) and DEK (32) have been shown to clear U2AF2 binding at Py-tracts that are not followed by an AG dinucleotide. However, it remains unclear how U2AF2 itself discriminates between weak and strong Py-tract RNAs. Here, we discover an unexpected autoinhibitory mechanism that significantly enhances U2AF2’s RNA binding specificity for strong Py-tracts. Our solution structure of free U2AF2 RRM1,2 identifies so far unknown regulatory roles of three structural features, namely short N- and C-terminal helices flanking the core RNA binding region of U2AF2 and the intrinsically disordered RRM1–RRM2 linker, that modulate the RNA recognition. The two short helices flanking RRM1,2 are already preformed in the free protein and modulate RNA binding by U2AF2 in distinct ways. First, the additional N-terminal helix contributes as an additional RNA binding interface to the rest of RRM1,2 domains (consistent with contacts seen in a crystal structure with U-rich RNA) (8). Second, the linker/RRM1 interaction may stabilize RNA binding of strong Py-tracts by mediating few additional contacts and stabilizing a compact arrangement of RRM1,2, which is only induced upon binding strong Py-tracts.
Third, and most importantly, the dynamic interaction of the C-terminal region of the RRM1,2 linker to the RNA binding surface of RRM2 results in autoinhibition of RNA binding. This is a key feature to explain the binding preference of U2AF2 to strong Py-tract RNAs by disfavoring interactions with weaker Py-tracts. In the case of weak, low-affinity Py-tracts that span over a longer sequence and in which consecutive pyrimidine stretches are separated by noncognate nucleotides, the binding selectivity will be mediated by RRM2. We have previously shown that RRM2 has a much higher intrinsic RNA binding activity compared to RRM1 (9), and thus cooperative contributions by RRM1 to RNA binding are initially less relevant. Therefore, the dynamic linker-to-RRM2 interaction effectively weakens the initial interaction of RRM2 with short, less pyrimidine-rich RNA sequences. The binding selectivity of the U2AF2 RRMs for Py-tract RNAs is thus established by an intrinsic pyrimidine binding preference of RRM2 that is tuned by the autoinhibitory linker interaction.
Our findings on the role of the U2AF2 RRM1,2 linker and previous data (9, 22) suggest that U2AF2 needs to adopt a compact arrangement of its tandem RRM domains when bound to RNA to achieve high-affinity binding and promote efficient downstream function. Thus, the compactness of the U2AF2 RNP (ribonucleoprotein complex) appears to control the efficiency of spliceosome assembly and thereby the splicing capacity of the downstream exon. One possible explanation is that the RNP arrangement ensures close spatial proximity of the complex E components at the 3′ splice site.
Our results demonstrate that linker autoinhibition is a key factor in the binding selectivity of U2AF2. Importantly, removing the linker interactions results in dispersed binding to weaker Py-tracts within introns and impacts on splicing fidelity. A tight regulation against such spurious U2AF2 binding is particularly important, since even deep intronic U2AF2 binding events can activate cryptic 3′ splice sites (27) or impact on bona fide splicing (19). Even though weak Py-tracts are generally disfavored, they still occur at alternative exons, where they allow for flexible modulation of alternative splicing. This can be achieved, e.g., by stabilization of U2AF2 binding at weak Py-tracts by additional cofactors (10, 21).
The RRM is the most abundant and extensively studied RNA binding fold, comprised of a four-stranded β-sheet with two helices packed against one side. The predominant and canonical RNA binding interface involves the β-sheet surface, which can recognize diverse RNA sequences by varying key amino acids within the limit of RNP sequence conservation (33). However, among the ∼250 reported experimental RRM/RNA complex structures, numerous RRMs exhibit additional structural elements (β-strands, loop extensions) that can provide further contacts and contribute to RNA binding specificity. For example, the interaction of a C-terminal extension with the canonical RRM fold, or the formation of a fifth β-strand pairing with the canonical β2 strand, are prominent extensions of the basic RRM fold (33). Interestingly, the linker-to-RRM2 interaction in U2AF2 mainly involves hydrophobic residues, reminiscent of the C-terminal extension (SI Appendix, Fig. S12). However, unlike in PTBP1 and hnRNP L RRM domains, U2AF2 facilitates this interaction through the C-terminal region of the RRM1,2 linker that precedes RRM2. The structural resemblance of these auxiliary components is intriguing and shows how extensions and linker regions flanking the basic RRM fold can modulate RNA interactions and binding specificity. Thus, our findings highlight an important role of linkers flanking RNA binding domains to modulate RNA interactions in multidomain RNA binding proteins and potentially open the door for understanding the role of auxiliary structural features embedded in many other RRM domains.
It is noteworthy that the central region of the U2AF2 linker is intrinsically disordered and highly flexible in solution. This feature is presumably required to enable the tandem RRMs to sample a large conformational space to support the dynamic population shift from an ensemble of closed states to the open domain arrangement when bound to strong RNAs (9).
In this study, we demonstrate how a combination of structural biology with large-scale mapping of RNA interactions by in vitro and in vivo iCLIP is a powerful approach to dissect molecular mechanisms and identify crucial structural features of RNA binding proteins. We expect that this unique combination of detailed structural insights and genome-wide studies will prove important to study the posttranscriptional regulation of gene expression and offer opportunities to specifically interfere with the underlying protein–RNA interactions in the future.
Methods
Multiple Sequence Alignment.
Protein sequences were aligned using MUSCLE (Multiple Sequence Comparison by Log-Expectation; EMBL-EBI) (34), using the Jalview graphical interface (35) to highlight the key conserved hydrophobic and aromatic residues in the linker region as well as the generally conserved residues of RRM1 and RRM2 in the vicinity of the linker. The aligned proteins are Homo sapiens, NP_009210; Danio rerio, NP_991252; Xenopus laevis, AAH44032; Bos taurus, NP_001068804; Tetraodon nigroviridis, CAF97922.1; Macaca mulatta, NP_476891; Mus musculus, EDL31287; Anopheles gambiae, XP_311994; Aedes aegypti, XP_001662443; Ciona intestinalis, XP_002130386; Drosophila melanogaster U2AF50, NP_476891; D. melanogaster LS2, AAF46969; Apis mellifera, XP_026299544; Caenorhabditis elegans, NP_001022967; Nicotiana plumbaginifolia, XP_016512340; Arabidopsis thaliana, AAB80661; Oryza sativa, ABR26075; Triticum aestivum, AAY84881; and Schizosaccharomyces pombe, NP_595396.
Protein Expression.
The genes encoding the previous and present U2AF2 RRM1,2 variants (residues 148 to 342 and 140 to 342, respectively) as WT or GS mutant (residues 233 to 257 replaced with Gly-Gly-Ser repeats of the same length and V254 mutated to proline) were cloned into the pETM11 vector (obtained from EMBL) encoding a His tag followed by TEV cleavage site. Recombinant proteins were expressed in Escherichia coli BL21(DE3) cells in standard media or minimal M9 media supplemented with 1 g/L 15NH4Cl and 2 g/L 13C-glucose. Protein expression and purification was done as described previously (9). After growth of bacterial cells up to an OD600 of 0.7 to 0.8, protein expression was induced by 1.0 mM IPTG following overnight expression at 18 °C. Cells were resuspended in 30 mM Tris/HCl, pH 7.5, 500 mM NaCl, 10 mM imidazole supplemented with protease inhibitors and lysed by sonication. The cleared lysate was loaded on Ni‐NTA resin and washed with an additional 25 mM imidazole followed by elution with additional 500 mM imidazole. After cleavage of the tag by His‐tagged TEV protease at 4 °C overnight, samples were reloaded on Ni‐NTA resin to remove the tag, TEV protease, and uncleaved protein. All protein samples were further purified by size-exclusion chromatography on a HiLoad 16/60 Superdex 75 column (GE Healthcare) equilibrated with NMR buffer (20 mM sodium phosphate, pH 6.5, 100 mM NaCl, 2 mM DTT).
NMR Spectroscopy.
NMR experiments were recorded at 298 K on 900-, 800-, and 600-MHz Bruker Avance NMR spectrometers equipped with cryogenic triple resonance gradient probes. NMR spectra were processed by TOPSPIN3.5 (Bruker) or NMRPipe (36), then analyzed using Sparky (T. D. Goddard and D. G. Kneller, SPARKY 3, University of California, San Francisco). Samples were measured at 0.5 to 1 mM concentration in NMR buffer with 10% D2O added as lock signal. Backbone resonance assignments of U2AF2 RRM1,2 alone and in complex with RNAs were obtained from a uniformly 15N,13C-labeled protein in the absence and presence of saturating concentrations of RNA. Standard triple resonance experiments HNCA, HNCACB, and CBCA(CO)NH (37) were recorded at 600 MHz. The 15N relaxation experiments (38) were recorded on a 600-MHz spectrometer at 25 °C. The 15N T1 and T1ρ relaxation times were obtained from pseudo-3D HSQC-based experiments recorded in an interleaved fashion with 12 different relaxation delays (21.6, 86.4, 162, 248.4, 345.6, 432, 518.4, 669.6, 885.6, 1,144.8, 1,404, and 1,782 ms) for T1 and 8 different relaxation delays (5, 7, 10, 15, 20, 25, 30, and 40 ms) for T1ρ. Two delays in each experiment were recorded in duplicates for error estimation. Relaxation rates were extracted by fitting the data to an exponential function using the relaxation module in NMRViewJ (39).
Structure Calculations.
Automatic NOE assignments and structure calculations were initially performed using CYANA3 (40). NOEs initially assigned by CYANA were manually inspected with the corresponding hydrogen bond pattern, backbone dynamics, and the dihedral restraints derived and based on the consensus of SSP (41) and 13C secondary chemical shifts using TALOS+ (42). Final structures were refined using NOE distance and dihedral angle restraints in explicit water (43) using ARIA1.2 (44) and CNS (45). Structural quality was evaluated using ProcheckNMR (46) and PSVS (47). Ribbon representations and the electrostatic surface potential were prepared with PYMOL (DeLano Scientific). Ensemble structural r.m.s. deviations were calculated using MolMol (48). Structural statistics are reported in SI Appendix, Table S2.
Isothermal Titration Calorimetry.
All of the ITC measurements were performed on MicroCal PEAQ-ITC (Malvern Panalytical) using nonisotopically labeled U2AF proteins and U9 and U4A8U4 RNA oligonucleotides, as described earlier, in 20 mM sodium phosphate, pH 6.5, 50 mM NaCl, at 25 °C. U2AF proteins in the cell (concentration ranges, 10 to 30 μM) were titrated with RNAs (concentration of 150 to 400 μM) for the final molar ratios of 1:1.2 to 1:3, adjusted depending on the affinity of the interaction. The SD values were estimated from the repetitions of each experiment as indicated in SI Appendix, Table S1.
Small Angle X-Ray Scattering.
Small angle X-ray scattering was measured on a Rigaku BIOSAXS1000 instrument equipped with a Cu-Kα rotating anode and a Pilatus 100K detector. Transmission was measured with a photodiode beamstop. For q calibration, a silver behenate sample was used. Samples were measured at 25 °C in four 15-min frames checked for beam damage and averaged. Circular averaging normalization to transmission and solvent subtraction was made with SAXSLab V 3.02. To exclude concentration-dependent effects, three concentrations of 4, 6, and 8 mg/mL were measured and compared. Rg values and P(r) functions were calculated with the ATSAS package V 2.8.0 (49). RNA-bound samples were consistently prepared by adding 1.8 times molar excess of RNA ligands to 225 μM protein. Given the overall binding affinities, the scattering curves thus also reflect a ca. 20% contribution from unbound excess RNA. The scattering from the unbound RNA has some minor effect on the shape of the P(r) distribution. However, it does not affect the overall (maximum) dimension of the complex, which is used to assess and compare the compactness of the complexes.
Kinetic Experiments.
Kinetic binding experiments of U2AF2 to RNA probes were carried out using a DRX switchSENSE platform on an MPC2-48–2-Y1-S sensor chip (Dynamic Biosensors). TE40 buffer (10 mM NaPi, pH 7.4, 40 mM NaCl, 50 μM EDTA, 50 μM EGTA, 0.05% Tween 20) served as running buffer.
The RNA samples (U9, 5′-UUUUUUUUU-ATC AGC GTT CGA TGC TTC CGA CTA ATC AGC CAT ATC AGC TTA CGA CTA-3′; U4A8U4, 5′-UUUUAAAAAAAAUUUU-ATC AGC GTT CGA TGC TTC CGA CTA ATC AGC CAT ATC AGC TTA CGA CTA-3′) used in these experiments were synthesized with a generic 48-mer switchSENSE immobilization DNA sequence at the 5′-end (italic), complementary to the surface-grafted DNA on the switchSENSE chip. All oligonucleotides used for switchSENSE experiments were synthesized by Ella Biotech. The immobilization of the RNA probes was carried out using the standard functionalization routing of the DRX instrument.
The association and dissociation kinetics of U2AF2 variants to the RNA probes were measured under a constant flow rate of 2,000 µL/min. During the dissociation, the flow channel was rinsed with running buffer. Binding traces were recorded at the indicated U2AF2 concentration using the dynamic measurement mode at a sampling rate of 1 data point per second. Binding traces were recorded at the indicated U2AF65 concentration using the dynamic measurement mode at a sampling rate of 1 data point per second. For evaluation, the kinetic parameters were extracted using a monoexponential fit model using switchANALYSIS software (Dynamic Biosensors).
Differences in the absolute apparent binding affinity values (KD) are likely attributable to the different experimental setups and conditions in switchSENSE and ITC experiments.
In Vitro iCLIP.
Experiments were performed as previously described (21). Briefly, 1 µM of recombinant RRM1,2-WT (residues 140 to 342), which is an extension of the previously used version (residues 148 to 342), or RRM1,2-GS (residues 233 to 257 replaced with Gly-Gly-Ser repeats of the same length) was mixed with a pool of nine different in vitro transcripts (CD55, C4PBP, MAT2A, MYC, MYL6, NF1, PAPD4, PCBP2, and PTBP2; 0.75 nM each) (21) in binding buffer. The in vitro iCLIP experiments for both proteins were performed in quadruplicates. NUP133 in vitro transcript bound with RRM12-WT was spiked into each in vitro mix to normalize the samples. The in vitro mixtures were irradiated with 50 mJ/cm2 UV at a wavelength of 254 nm and immunoprecipitated with a monoclonal anti-U2AF2 antibody (Sigma; cat. no. U4758). All in vitro mixtures were further processed with the established in vitro iCLIP protocol (21). Multiplexed iCLIP libraries were sequenced on an Illumina MiSeq sequencing system (76-nt single-end reads).
U2AF2 Overexpression and RNA Sequencing.
Full-length U2AF2-WT and U2AF2-GS (residues 233 to 257 replaced with Gly-Gly-Ser repeats of the same length and V254 mutated to proline) were cloned into pcDNA5 vectors to create overexpression constructs. Two micrograms of the constructs were transfected into HeLa cells at 90% confluency using Lipofectamine 2000 (Thermo Fisher Scientific) according to the manufacturer’s instructions. An empty pcDNA5 vector was transfected as a negative control. The cells were harvested 24 h posttransfection, and the overexpression was confirmed by using Western blot with a monoclonal anti-U2AF2 antibody (Sigma; cat. no. U4758). Total RNA was extracted from the harvested cells using an RNeasy Plus Mini Kit (Qiagen) according to the manufacturer’s instructions. The RNA-seq libraries were prepared from the total RNA with Illumina’s TruSeq Stranded Total RNA LT Sample Prep Kit following the standard protocol with Ribo-Zero rRNA depletion (part no. 15031048 Rev. E). Libraries were prepared with a starting amount of 1,000 ng and amplified in 10 PCR cycles. Libraries were profiled in a HS DNA chip on a 2100 Bioanalyzer (Agilent Technologies) and quantified using the Qubit dsDNA HS AssayKit on a Qubit 2.0 Fluorometer (Life Technologies). All libraries were pooled together in equimolar ratio and sequenced on one NextSeq500 High Output FC, SR for 1× 160 cycles plus seven cycles for the index read.
In Vitro iCLIP Data Processing.
Basic sequencing quality checks were applied to all reads using FastQC (version 0.11.5; https://www.bioinformatics.babraham.ac.uk/projects/fastqc). Afterward, reads were filtered based on sequencing qualities (Phred score) of the barcode region. Only reads with at most one position with a sequencing quality <20 in the experimental barcode (positions 4 to 7) and without any position with a sequencing quality <17 in the random barcode (positions 1 to 3 and 8 to 9) were kept for further analysis. Remaining reads were demultiplexed based on the experimental barcode on positions 4 to 7 using Flexbar (version 3.0.0) (50) without allowing mismatches.
All following steps of the analysis were performed on all individual samples after demultiplexing. Remaining adapter sequences were trimmed from the right end of the reads using Flexbar (version 3.0.0), allowing up to one mismatch in 10 bp, requiring a minimal overlap of 1 bp of read and adapter. After trimming off the adapter, the barcode is trimmed off from the beginning of the reads (first 9 bp) and added to the header of the read, such that the information is kept available for downstream analysis. Reads shorter than 15 bp were removed from further analysis. Trimmed and filtered reads were mapped to the human genome (assembly version hg38/GRCh38) and its annotation based on GENCODE release 25 (51) using STAR (version 2.5.4b) (52). When running STAR, up to two mismatches were allowed, soft-clipping was prohibited at the 5′ ends of reads, and only uniquely mapping reads were kept for further analysis. Following mapping, all samples were reduced to 100,000 randomly selected uniquely mapped reads each (downsampling) to facilitate direct comparisons. Afterwards, duplicate reads were marked using the dedup function of bamUtil (version 1.0.13), which defines duplicates as reads whose 5′ ends map to the same position in the genome (https://github.com/statgen/bamUtil). Subsequently, marked duplicates with identical random barcodes were removed since they are considered technical duplicates, while biological duplicates showing unequal random barcodes were kept. Resulting bam files were sorted and indexed using SAMtools (version 1.5) (53). Based on the bam files, bedgraph files were created using bamToBed of the BEDTools suite (version 2.25.0) (54), considering only the position upstream of the 5′ mapping position of the read, since this nucleotide is considered as the cross-linked nucleotide. bedgraph files were then transformed to bigWig file format using bedGraphToBigWig of the UCSC tool suite (55).
In order to account for sample-specific effects such as sequencing depth and processing effects, read counts per nucleotide were normalized by the number of reads mapping to the spike-in (NUP133 in vitro transcript cross-linked to RRM1,2-WT), which was added to the mixture of in vitro transcripts after separate UV cross-linking, plus a constant to adjust scales. Identification of binding sites was performed on normalized in vitro iCLIP data (merged data for RRM1,2-WT and RRM1,2-GS) by iteratively identifying 9-nt windows with the highest cumulative signal and sufficient enrichment over a region-wise uniform background distribution (exceeding a uniform background signal in all four replicates of at least one U2AF variant) as previously described (21). This procedure yielded a total of 424 binding sites across the 9 in vitro transcripts that were bound by RRM1,2-WT and/or RRM1,2-GS. In order to account for different transcript abundances in vivo, the in vivo iCLIP signal on binding sites identified in vitro was represented as summed binding site signal over intron signal scaled by intron width (Fig. 5B).
The Py-tract strength at each binding site was determined as follows. The extended binding site region (9-bp binding site ± 5-bp flanking sequence) was screened for the window with the highest scoring Py-tract via sliding windows of increasing width (widths 5 to 19 bp). The Py-tract strength of each window was calculated as the χ2 test statistic with 1 degree of freedom, comparing the observed number of pyrimidines with the expected number based on the assumption of uniform nucleotide distribution. Py-tract scores ranged from 0 to a maximum achievable score of 9.5. Py-tract scores were classified as “weak,” “medium,” or “strong” when falling within [0,3], (3, 6.5] and (6.5, 9.5], respectively. Correlation of Py-tract strength and U2AF2 affinity (SI Appendix, Fig. S11) was evaluated on the subset of binding sites that overlapped (by at least 7 bp) with the previously determined binding sites (21) (303 out of 424 binding sites). In vivo iCLIP data were taken from a previous study (21, 27) by merging the replicates (sample lujh32).
U2AF2 Overexpression Data Processing.
RNA-seq libraries were sequenced on an Illumina NextSeq500 (160-nt single-end reads), yielding 44.4 to 49.6 million reads per sample. In order to remove potential adapter fragments at the 3′ ends of reads while maintaining equal read lengths for downstream analyses, all reads were trimmed at the 3′ end to 100 bp. Sequencing quality was checked using FastQC (version 0.11.5; https://www.bioinformatics.babraham.ac.uk/projects/fastqc). All reads were mapped to the human genome (assembly version hg38/GRCh38) and its annotation (GENCODE release 25) (51) with the splice-aware alignment software STAR (version 2.5.4b) (52) with up to 4% allowed mismatches and an overhang of at most 99 base pairs at each splice junction. Reads were sorted using SAMtools (version 1.5) (53). Uniquely mapped reads were summarized per gene using featureCounts from the subread package (version 1.5.1) (56). Differential splicing analysis was performed using rMATS (version 4.0.2) (57). Significantly changing cassette exons upon U2AF2-WT and U2AF2-GS overexpression were identified with false discovery rate (FDR) <0.05. Py-tract strength at 3′ splice sites of cassette exons was determined as described earlier, screening a 39-nt region upstream of the AG dinucleotide.
Analysis of Skipped Constitutive Exons.
Constitutive exons were defined as all exons not overlapping with introns in any other transcript based on GENCODE release 25 (gencode.v25.annotation.gtf), using only protein-coding genes and excluding level 3 annotation and overlapping genes. First and last exons of each transcript were removed. This procedure yielded an initial set of 113,142 constitutive exons. Junction-spanning reads supporting exon inclusion or skipping were extracted from the STAR output (bam file) such that each N operation in the CIGAR string of a genomic alignment was interpreted as a potential junction. Inclusion of a constitutive exon was calculated based on the number of reads using the 5′ splice site and 3′ splice site (“inclusion reads”), taking the average of both numbers. Skipping was derived based on all junction-spanning reads connecting any annotated splice sites and completely overlapping the respective exon. Percent skipping was calculated as skipping reads over inclusion plus skipping reads times 100 for each sample and then averaged over replicates. For the analysis in Fig. 5 D and E, constitutive exons were required to show inclusion reads in all nine RNA-seq samples (three replicates of control, U2AF2-WT, and U2AF2-GS OE), and, to restrict to well-expressed genes, to have at least 43 inclusion reads in control samples (average over replicates), corresponding to the median in control samples over all constitutive exons. This yielded 37,052 constitutive exons.
Minigene Reporter Assays.
Minigene reporters were constructed to test the effect of U2AF2-WT and U2AF2-GS overexpression on four alternative exons: PTBP2 exon 10 (genomic locus chr1:96,804,170–96,806,896; vector backbone PCDNA5; internal plasmid ID pR020; sequence alteration g.96804751_96804752_insT), MST1R exon 11 (chr3:49,895,665–49,896,404; pCDNA3; pS003; g.49896117C>T, g.49896107T>G), AKAP9 exon 19 (chr7:92,040,557–92,043,053; pCDNA5; pAB009; no alterations), and MPDZ exon 18 (chr9:13,183,336–13,189,061; pCDNA5; pAB010; g.13187763_13187764_tgaaaaggcagatacacgttt, g.13187313G>T, g.13185150G>A, g.13185060T>C, g.13184482T>C). All coordinates are based on human genome version GRCh38/hg38. Transfection of a minigene together with a vector expressing U2AF2-WT, U2AF2-GS, or empty vector was carried out for 24 h using Lipofectamine 2000 (Thermo Fisher Scientific) according to the manufacturer’s recommendations. MCF-7 or HeLa cells were used for the experiments with the MST1R minigene or the three other minigenes (PTBP2, AKAP9, MPDZ), respectively. For splicing measurements, total RNA was extracted using an RNeasy Plus Mini Kit (Qiagen) and reverse-transcribed into cDNA via oligo(dT)18 primers. Splicing products were amplified with minigene-specific forward primers (MST1R, oS66, TGCCAACCTAGTTCCACTGA; PTBP2, oL172, AGCTGGTGGCAATACAGTCC; AKAP9, oA65, GAGAGACAGCGAGAAGACCAGG; MPDZ, oA61, GGGACTGTGAGAATAGGAGTTGC) and a common reverse primer (oL39, GCAACTAGAAGGCACAGTCG). Visualization was performed with a 2200 TapeStation system (Agilent Technologies) to obtain the molar ratio of each splicing product (% integrated area).
Data Availability.
Atomic coordinates and NMR data for the unbound U2AF2 RRM1,2 structure have been deposited in the Protein Data Bank (https://www.wwpdb.org/) with accession code 6TR0 and in the BMRB with accession code 34466. All in vitro iCLIP and RNA-seq data generated in this study have been submitted to the Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under the SuperSeries accession number GSE126694. The in vivo iCLIP data are available under the accession number GSE99688.
Supplementary Material
Acknowledgments
We thank Juan Valcárcel, Sophie Bonnal, and members of the Sattler and König groups for inspiring discussions; Ivona Bagdiul (University of Geneva) for cloning of initial constructs; and Sam Asami and Gerd Gemmecker (TUM) for NMR support. We gratefully acknowledge access to measurement time at the Bavarian NMR Center, the SAXS facility of the SFB1035 at the TUM Chemistry Department, and the IMB Genomics Core Facility and the use of its Illumina NextSeq500 (INST 247/870-1 FUGG) and Illumina MiSeq. This work was supported by the Deutsche Forschungsgemeinschaft through Grants SPP1935 (Sa 823/10-1 to M.S. and KO 4566/2-1 to J.K.) and SFB1035 and GRK1721 (to M.S.) and by the Helmholtz Association Initiative and Networking Fund under project number ZT-I-0003 (to H.-S.K. and M.S.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. B.S.T. is a guest editor invited by the Editorial Board.
Data deposition: Atomic coordinates and NMR data for the unbound U2AF2 RRM1,2 structure have been deposited in the Protein Data Bank (https://www.wwpdb.org/; accession code 6TR0) and Biological Magnetic Resonance Bank (http://www.bmrb.wisc.edu; accession code 34466). All in vitro iCLIP and RNA-seq data generated in this study have been submitted to the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/) under the SuperSeries accession number GSE126694. The in vivo iCLIP data are available via GEO under the accession number GSE99688.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1913483117/-/DCSupplemental.
References
- 1.Wahl M. C., Will C. L., Lührmann R., The spliceosome: Design principles of a dynamic RNP machine. Cell 136, 701–718 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Nilsen T. W., Graveley B. R., Expansion of the eukaryotic proteome by alternative splicing. Nature 463, 457–463 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fu X. D., Ares M. Jr, Context-dependent control of alternative splicing by RNA-binding proteins. Nat. Rev. Genet. 15, 689–701 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Valcárcel J., Gaur R. K., Singh R., Green M. R., Interaction of U2AF65 RS region with pre-mRNA branch point and promotion of base pairing with U2 snRNA [corrected]. Science 273, 1706–1709 (1996). [DOI] [PubMed] [Google Scholar]
- 5.Merendino L., Guth S., Bilbao D., Martínez C., Valcárcel J., Inhibition of msl-2 splicing by Sex-lethal reveals interaction between U2AF35 and the 3′ splice site AG. Nature 402, 838–841 (1999). [DOI] [PubMed] [Google Scholar]
- 6.Wu S., Romfo C. M., Nilsen T. W., Green M. R., Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature 402, 832–835 (1999). [DOI] [PubMed] [Google Scholar]
- 7.Zorio D. A., Blumenthal T., Both subunits of U2AF recognize the 3′ splice site in Caenorhabditis elegans. Nature 402, 835–838 (1999). [DOI] [PubMed] [Google Scholar]
- 8.Agrawal A. A., et al. , An extended U2AF(65)-RNA-binding domain recognizes the 3′ splice site signal. Nat. Commun. 7, 10950 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mackereth C. D., et al. , Multi-domain conformational selection underlies pre-mRNA splicing regulation by U2AF. Nature 475, 408–411 (2011). [DOI] [PubMed] [Google Scholar]
- 10.Voith von Voithenberg L., et al. , Recognition of the 3′ splice site RNA by the U2AF heterodimer involves a dynamic population shift. Proc. Natl. Acad. Sci. U.S.A. 113, E7169–E7175 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berglund J. A., Chua K., Abovich N., Reed R., Rosbash M., The splicing factor BBP interacts specifically with the pre-mRNA branchpoint sequence UACUAAC. Cell 89, 781–787 (1997). [DOI] [PubMed] [Google Scholar]
- 12.Berglund J. A., Fleming M. L., Rosbash M., The KH domain of the branchpoint sequence binding protein determines specificity for the pre-mRNA branchpoint sequence. RNA 4, 998–1006 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Z., et al. , Structural basis for recognition of the intron branch site RNA by splicing factor 1. Science 294, 1098–1102 (2001). [DOI] [PubMed] [Google Scholar]
- 14.Corsini L., et al. , U2AF-homology motif interactions are required for alternative splicing regulation by SPF45. Nat. Struct. Mol. Biol. 14, 620–629 (2007). Correction in: Nat. Struct. Mol. Biol.14, 785 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Kielkopf C. L., Rodionova N. A., Green M. R., Burley S. K., A novel peptide recognition mode revealed by the X-ray structure of a core U2AF35/U2AF65 heterodimer. Cell 106, 595–605 (2001). [DOI] [PubMed] [Google Scholar]
- 16.Loerch S., Kielkopf C. L., Unmasking the U2AF homology motif family: A bona fide protein-protein interaction motif in disguise. RNA 22, 1795–1807 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Selenko P., et al. , Structural basis for the molecular recognition between human splicing factors U2AF65 and SF1/mBBP. Mol. Cell 11, 965–976 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Lim K. H., Ferraris L., Filloux M. E., Raphael B. J., Fairbrother W. G., Using positional distribution to identify splicing elements and predict pre-mRNA processing defects in human genes. Proc. Natl. Acad. Sci. U.S.A. 108, 11093–11098 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shao C., et al. , Mechanisms for U2AF to define 3′ splice sites and regulate alternative splicing in the human genome. Nat. Struct. Mol. Biol. 21, 997–1005 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gama-Carvalho M., Barbosa-Morais N. L., Brodsky A. S., Silver P. A., Carmo-Fonseca M., Genome-wide identification of functionally distinct subsets of cellular mRNAs associated with two nucleocytoplasmic-shuttling mammalian splicing factors. Genome Biol. 7, R113 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sutandy F. X. R., et al. , In vitro iCLIP-based modeling uncovers how the splicing factor U2AF2 relies on regulation by cofactors. Genome Res. 28, 699–713 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang J. R., et al. , Transient electrostatic interactions dominate the conformational equilibrium sampled by multidomain splicing factor U2AF65: A combined NMR and SAXS study. J. Am. Chem. Soc. 136, 7068–7076 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Kang H.-S., Sattler M., Solution structure of U2AF2 RRM1,2. Protein Data Bank in Europe. https://www.ebi.ac.uk/pdbe/entry/pdb/4YH8. Deposited 17 December 2019.
- 24.Kang H.-S., Sattler M., Solution structure of U2AF2 RRM1,2. Biological Magnetic Resonance Data Bank. http://www.bmrb.wisc.edu/data_library/summary/index.php?bmrbId=34466. Deposited 17 December 2019.
- 25.Mészáros B., Erdos G., Dosztányi Z., IUPred2A: Context-dependent prediction of protein disorder as a function of redox state and protein binding. Nucleic Acids Res. 46, W329–W337 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kang H. S., et al. , RNA-seq data / GSE126694. Gene Expression Omnibus (GEO). https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE126694. Deposited 18 February 2019.
- 27.Zarnack K., et al. , Direct competition between hnRNP C and U2AF65 protects the transcriptome from the exonization of Alu elements. Cell 152, 453–466 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kang H. S., et al. , iCLIP data / GSE126694. Gene Expression Omnibus (GEO). https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE126694. Deposited 18 February 2019.
- 29.Jankowsky E., Harris M. E., Specificity and nonspecificity in RNA-protein interactions. Nat. Rev. Mol. Cell Biol. 16, 533–544 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berglund J. A., Abovich N., Rosbash M., A cooperative interaction between U2AF65 and mBBP/SF1 facilitates branchpoint region recognition. Genes Dev. 12, 858–867 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tavanez J. P., Madl T., Kooshapur H., Sattler M., Valcárcel J., hnRNP A1 proofreads 3′ splice site recognition by U2AF. Mol. Cell 45, 314–329 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Soares L. M., Zanier K., Mackereth C., Sattler M., Valcárcel J., Intron removal requires proofreading of U2AF/3′ splice site recognition by DEK. Science 312, 1961–1965 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Blatter M., et al. , The signature of the five-stranded vRRM fold defined by functional, structural and computational analysis of the hnRNP L protein. J. Mol. Biol. 427, 3001–3022 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Madeira F., et al. , The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47, W636–W641 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Waterhouse A. M., Procter J. B., Martin D. M., Clamp M., Barton G. J., Jalview Version 2–A multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Delaglio F., et al. , NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 (1995). [DOI] [PubMed] [Google Scholar]
- 37.Sattler M., Schleucher J., Griesinger C., Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog. Nucl. Magn. Reson. Spectrosc. 34, 93–158 (1999). [Google Scholar]
- 38.Farrow N. A., Zhang O., Forman-Kay J. D., Kay L. E., Comparison of the backbone dynamics of a folded and an unfolded SH3 domain existing in equilibrium in aqueous buffer. Biochemistry 34, 868–878 (1995). [DOI] [PubMed] [Google Scholar]
- 39.Johnson B. A., Blevins R. A., NMR View: A computer program for the visualization and analysis of NMR data. J. Biomol. NMR 4, 603–614 (1994). [DOI] [PubMed] [Google Scholar]
- 40.Güntert P., Automated structure determination from NMR spectra. Eur. Biophys. J. 38, 129–143 (2009). [DOI] [PubMed] [Google Scholar]
- 41.Marsh J. A., Singh V. K., Jia Z., Forman-Kay J. D., Sensitivity of secondary structure propensities to sequence differences between alpha- and gamma-synuclein: Implications for fibrillation. Protein Sci. 15, 2795–2804 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shen Y., Delaglio F., Cornilescu G., Bax A., TALOS+: A hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR 44, 213–223 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Linge J. P., Williams M. A., Spronk C. A., Bonvin A. M., Nilges M., Refinement of protein structures in explicit solvent. Proteins 50, 496–506 (2003). [DOI] [PubMed] [Google Scholar]
- 44.Linge J. P., O’Donoghue S. I., Nilges M., Automated assignment of ambiguous nuclear overhauser effects with ARIA. Methods Enzymol. 339, 71–90 (2001). [DOI] [PubMed] [Google Scholar]
- 45.Brünger A. T., et al. , Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 (1998). [DOI] [PubMed] [Google Scholar]
- 46.Laskowski R. A., Rullmannn J. A., MacArthur M. W., Kaptein R., Thornton J. M., AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8, 477–486 (1996). [DOI] [PubMed] [Google Scholar]
- 47.Bhattacharya A., Tejero R., Montelione G. T., Evaluating protein structures determined by structural genomics consortia. Proteins 66, 778–795 (2007). [DOI] [PubMed] [Google Scholar]
- 48.Koradi R., Billeter M., Wuthrich K., MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graph. 14, 51–55, 29–32 (1996). [DOI] [PubMed] [Google Scholar]
- 49.Petoukhov M. V., et al. , New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Cryst. 45, 342–350 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dodt M., Roehr J. T., Ahmed R., Dieterich C., FLEXBAR-flexible barcode and adapter processing for next-generation sequencing platforms. Biology (Basel) 1, 895–905 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Harrow J., et al. , GENCODE: The reference human genome annotation for The ENCODE project. Genome Res. 22, 1760–1774 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dobin A., et al. , STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li H. et al.; 1000 Genome Project Data Processing Subgroup , The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Quinlan A. R., Hall I. M., BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kent W. J., Zweig A. S., Barber G., Hinrichs A. S., Karolchik D., BigWig and BigBed: Enabling browsing of large distributed datasets. Bioinformatics 26, 2204–2207 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liao Y., Smyth G. K., Shi W., featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 57.Shen S., et al. , rMATS: Robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl. Acad. Sci. U.S.A. 111, E5593–E5601 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and NMR data for the unbound U2AF2 RRM1,2 structure have been deposited in the Protein Data Bank (https://www.wwpdb.org/) with accession code 6TR0 and in the BMRB with accession code 34466. All in vitro iCLIP and RNA-seq data generated in this study have been submitted to the Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under the SuperSeries accession number GSE126694. The in vivo iCLIP data are available under the accession number GSE99688.