Significance
An extensive body of work has revealed that the conformational plasticity of IL-2 plays an important role in targeting the IL-2 receptor signaling axis in both agonist and antagonist mode; however, a more detailed understanding of such processes is needed for actionable value in drug development. Using mouse IL-2 as a model, our current body of work combines methyl-based chemical exchange NMR spectroscopy (CPMG and CEST) with thermodynamic measurements and immunoassays to characterize a global conformational transition to a sparsely populated autoinhibited conformational state, together with its functional significance. Targeting this minor state through mutations and small molecules can be exploited to fine tune the affinity and selectivity for different IL-2 binding partners, toward engineering novel cytokine functions.
Keywords: IL-2, immunomodulation, protein dynamics, NMR, drug design
Abstract
Interleukin-2 (IL-2) is a small α-helical cytokine that regulates immune cell homeostasis through its recruitment to a high-affinity heterotrimeric receptor complex (IL-2Rα/IL-2Rβ/γc). IL-2 has been shown to have therapeutic efficacy for immune diseases by preferentially expanding distinct T cell compartments, and several regulatory T cell (Treg)-biasing anti–IL-2 antibodies have been developed for combination therapies. The conformational plasticity of IL-2 plays an important role in its biological actions by modulating the strength of receptor and drug interactions. Through an NMR analysis of milliseconds-timescale dynamics of free mouse IL-2 (mIL-2), we identify a global transition to a sparse conformation which is regulated by an α-helical capping “switch” at the loop between the A and B helices (AB loop). Binding to either an anti-mouse IL-2 monoclonal antibody (mAb) or a small molecule inhibitor near the loop induces a measurable response at the core of the structure, while locking the switch to a single conformation through a designed point mutation leads to a global quenching of core dynamics accompanied by a pronounced effect in mAb binding. By elucidating key details of the long-range allosteric communication between the receptor binding surfaces and the core of the IL-2 structure, our results offer a direct blueprint for designing precision therapeutics targeting a continuum of conformational states.
Interleukin-2 (IL-2) is a type I α-helical cytokine that functions as a multilineage lymphocyte growth factor (1, 2). IL-2 signals through a high-affinity (10 pM) heterotrimeric receptor complex, consisting of the IL-2Rα (also called CD25), IL-2Rβ (CD122), and γc (CD132) chains, and the intermediate-affinity (1 nM) heterodimeric receptor complex, consisting of the IL-2Rβ and γc chains (Fig. 1A) (1–3). The IL2-Rα subunit is constitutively expressed in high levels on regulatory T (Treg) cells and at lower levels on natural killer (NK) cells and resting effector CD8+ T cells, resulting in differential IL-2 potency between different immune cell compartments (4, 5). From a clinical perspective, both IL-2 agonism and antagonism are of considerable importance, and it has been used for more than three decades toward immunotherapies of malignancies.
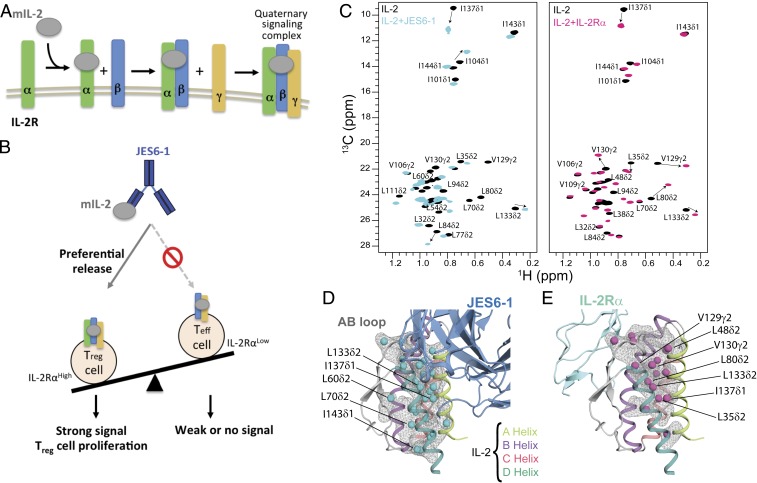
Fig. 1.
A plastic mIL-2 core structure mediates recognition of its binding partners for immune modulation. (A) Schematic of IL-2 cytokine-receptor quaternary complex formation. Assembly of the quaternary complex is thought to occur sequentially, with IL-2 first engaging IL-2Rα with a dissociation constant (Kd) of ∼10−8 M, which increases its affinity for the IL-2Rβ subunit, and finally recruiting the γc subunit to lock down the high-affinity quaternary complex (Kd of ∼10−11 M). (B) Schematic of the proposed mechanism for mIL-2/JES6-1 immunocomplex-mediated selective proliferation of regulatory T (Treg) cells. The JES6-1 Ab (shown as blue scFv) sterically blocks mIL-2 interaction with IL-2Rβ and γc subunits, preventing signaling of IL-2RαLow effector cells (right). However, an exchange mechanism between JES6-1 and the IL-2Rα subunit allows a preferential release of mIL-2 for exclusive signaling on IL-2RαHigh Treg cells, biasing toward an immunosuppressive response (left). (C) Overlay of 1H,13C-HMQC spectra of selectively labeled mIL-2 at the Iδ1-13CH3, L, V proS methyl positions, recorded in the free (black) or as a stoichiometric complex with JES6-1 scFv (cyan) or IL-2Rα receptor (magenta), acquired at 800 MHz, 25 °C. The arrows highlight major chemical shift effects. Selected methyl assignments are noted. (D) Mapping of methyl chemical shift changes on the crystal structure of mIL-2/JES6-1 complex (PDB ID 4YQX) and (E) on the overlaid IL-2Rα subunit from the homologous hIL-2 quaternary complex structure (PDB ID 2B5I). The mIL-2 residues with CSPs > 0.05 ppm are shown with cyan spheres in the schematic representation of JES6-1–bound mIL-2 (blue) and with magenta spheres in IL-2Rα–bound mIL-2 (green).
The known plasticity of IL-2 has been an important parameter in the generation of small molecules that bind to IL-2 and block signaling (6, 7), engineering “superagonist” mutant versions of IL-2 with high affinity for IL-2Rβ (8) and isolation of antibodies that bind to IL-2, resulting in biasing its actions to different immune cell subsets (9–11). Many efforts have partially focused in to improve its therapeutic potential by manipulating its ability to selectively target specific cell types (8, 11–16). In one approach, monoclonal antibodies against IL-2 can alter its properties by binding to a number of distinct conformational epitopes, thereby modifying the interactions of IL-2 with any of the IL-2R subunits resulting in the proliferation of either Treg or effector T (Teff) cells (9, 10). For example, wild-type (WT) mouse IL-2 (mIL-2) can be administered in complex with an anti-mouse IL-2 monoclonal antibody (JES6-1) and used to preferentially induce Treg cell proliferation (12). Subsequent work has validated therapeutic applications of the JES6-1 immunocomplex, which promotes graft tolerance (17, 18) and has shown efficacy in preclinical models of diabetes (19, 20). Thus, the conformational states of IL-2 can be selectively induced to achieve cell-type and receptor selectivity, but more detailed insight is needed into the IL-2 conformational transitions.
Focusing on mouse IL-2 as a model system, our team recently determined the X-ray structure of the mIL-2/JES6-1 complex to elucidate the mechanistic basis for its selective stimulation of Treg over effector cells (10). JES6-1 sterically blocks interactions with the IL-2Rβ and γc subunits, leading to a reduction of signaling effects on IL-2RαLow effector cells. JES6-1 undergoes allosteric exchange with the IL-2Rα subunit where surface-expressed IL-2Rα displaces the JES6-1 antibody and releases the cytokine to signal through the high-affinity heterotrimeric receptor on IL-2RαHigh Treg cells (Fig. 1B). Of particular relevance is the mobility of the AB loop (the loop between α-helices A and B), which has been shown to undergo a large conformational change upon JES6-1 binding. The AB loop is the main binding site for IL-2Rα, and crystallographic studies (10) demonstrated that key residues in the IL-2 AB loop engage the JES6-1 antibody and the IL-2Rα subunit in distinct orientations. Therefore, JES6-1 and IL-2Rα binding are mutually exclusive, leading to bidirectional exchange. Molecular dynamics simulations have suggested a significant conformational rearrangement in the AB loop and BC loop (the loop between α-helices B and C) characterizing the transition from JES6-1–bound to IL-2Rα–bound states (15). Despite the importance of IL-2 in T cell differentiation and homeostasis, a detailed biophysical characterization of its dynamic properties remains elusive.
In the current work, we use methyl-based NMR spectroscopy to establish that, in aqueous solution, free mIL-2 samples an excited state corresponding to an autoinhibitory conformation with distinct functional properties. Using a series of complementary NMR experiments, we characterize a cooperative transition, which involves a concerted motion of an α-helical capping “switch,” linking the AB loop with the hydrophobic core of the mIL-2 structure through a sequential repacking of core side chains. Global quenching of core dynamics, through the introduction of a designed point mutation at the switch region, leads to a three-order-of-magnitude loss of affinity for the JES6-1 antibody and, consistently, a loss of immunomodulatory activity in vitro. Insights derived from our data highlight the potential of targeting specific IL-2 conformational states using mutagenesis or small-molecule binders, to steer the conformational equilibrium toward states with maximum effects on Treg cells for the treatment of autoimmune diseases, and on Teff cells for immunotherapy of cancer.
Results
Long-Range Effects on mIL-2 Core Dynamics upon Recognition of Its Binding Partners.
Due to their hydrophobic character, methyl groups are well-suited NMR probes for monitoring the dynamics of the structural core of proteins (21, 22). The chemical shifts of methyl groups report on side chain rotameric states and local magnetic environment, while their NMR measurement is less influenced by the size of the system under investigation due to favorable relaxation properties (23). To gain insight into conformational changes on mIL-2 induced by its binding partners, we prepared selective 1H/13C MILV (Met, Ile, Leu and Val)-methyl labeled samples on a uniform 12C, perdeuterated background. We obtained NMR assignments of mIL-2 backbone and methyl groups using a combination of transverse relaxation-optimized spectroscopy (TROSY)-based triple-resonance and three-dimensional (3D) out-and-back side chain transfer experiments. Under the conditions of our NMR experiments, mIL-2 is strictly monomeric, as characterized by size exclusion chromatography coupled to multiangle laser light scattering (SEC-MALS) (SI Appendix, Fig. S1). To obtain stereospecific resonance assignments of methyl groups and to reduce spectral overlap observed in the free mIL-2 methyl spectrum, which is typical of α-helical proteins, we also prepared a selectively Iδ1-13C1H3, proS L,V-13C1H3–labeled protein sample (24). This approach allowed us to obtain unambiguous side chain resonance assignments of Met ε1, Ile δ1, Leu δ1/δ2, and Val γ1/γ2 (60 methyl groups in total) (SI Appendix, Fig. S1) (25), distributed throughout the structure, enabling us to comprehensively map the cytokine’s surface and hydrophobic core in the IL-2Rα and JES6-1–bound states (26, 27). Fig. 1C shows 13C-1H band-selective optimized flip angle short transient (SOFAST) heteronuclear multiple quantum coherence (HMQC) of I (LV)proS-methyl labeled mIL-2 unbound (Fig. 1C, black) and as a stoichiometric complex with JES6-1 single-chain Fv (scFv) (Fig. 1C, cyan) or IL-2Rα (Fig. 1C, magenta). The weighted chemical shift perturbations (CSPs) are presented along the mIL-2 sequence in SI Appendix, Fig. S2. In agreement with previous surface plasmon resonance (SPR) measurements (10), under our NMR sample conditions, mIL-2 forms tight complexes with either JES6-1 or IL-2Rα. Exchange between the free and bound states is slow on the chemical shift timescale, as shown by detailed NMR titrations where we observe a unique set of peaks corresponding to the free and bound states with large CSPs (up to 0.6 parts per million [ppm]) between them.
Notably, either receptor or antibody binding leads to measurable conformational changes at the corresponding primary binding sites (for example, L48δ2, L50δ2, and L54δ2 in the AB loop) but also induces long-range effects at remote sites distributed throughout the structure, including L35δ2 and L38δ2 in the A helix, L73δ2, L80δ2, and L86δ2 in the B helix, I101δ1 in the C helix, and V129γ2, V130γ2, L133δ2, and I137δ1 in the D helix. Mapping the most affected methyl groups on the mIL-2 structure (Fig. 1 D and E) highlights a contiguous path connecting the AB loop to the hydrophobic core of the structure. Overall, the methyl CSPs reveal a global response of the mIL-2 structure, indicative of allosteric communication between the primary binding site at the AB loop and core residues of the cytokine (SI Appendix, Fig. S2). Given that the four α-helical bundle core of apo-IL-2 is maintained in the complexed forms with the IL-2Rα receptor or JES6-1 antibody (backbone heavy atom rmsd of 0.8 Å and 2.3 Å, respectively), our results suggest that binding of either JES6-1 or IL-2Rα at the AB loop region induces a remodeling of core side chains. Specifically, the observed methyl chemical shift changes can arise from either changes in the lowest energy rotameric state or through perturbations in ensembles of rotamers sampled by the free and bound forms.
Free mIL-2 Samples a Global Transition to an Excited-State Conformation.
Based on our observation that the 1H-15N transverse relaxation-optimized spectroscopy (TROSY) spectrum of free mIL-2 was of marginal quality (SI Appendix, Fig. S1) with a significant fraction of completely broadened amide resonances (33%), we hypothesized that, in solution, mIL-2 undergoes exchange between different conformational states. The loss of signal for amide resonances corresponding to residues in the AB loop due to 15N line broadening suggests that the loop is switching between multiple backbone conformations with different chemical environments on an intermediate (microseconds to milliseconds [μs–ms]) timescale. This hypothesis is consistent with a recent molecular dynamics (MD) simulation-based model in which free IL-2 samples distinct conformations of the AB loop akin to the crystallographically observed structures in complexes with the JES6-1 antibody and IL-2Rα receptor (10, 15).
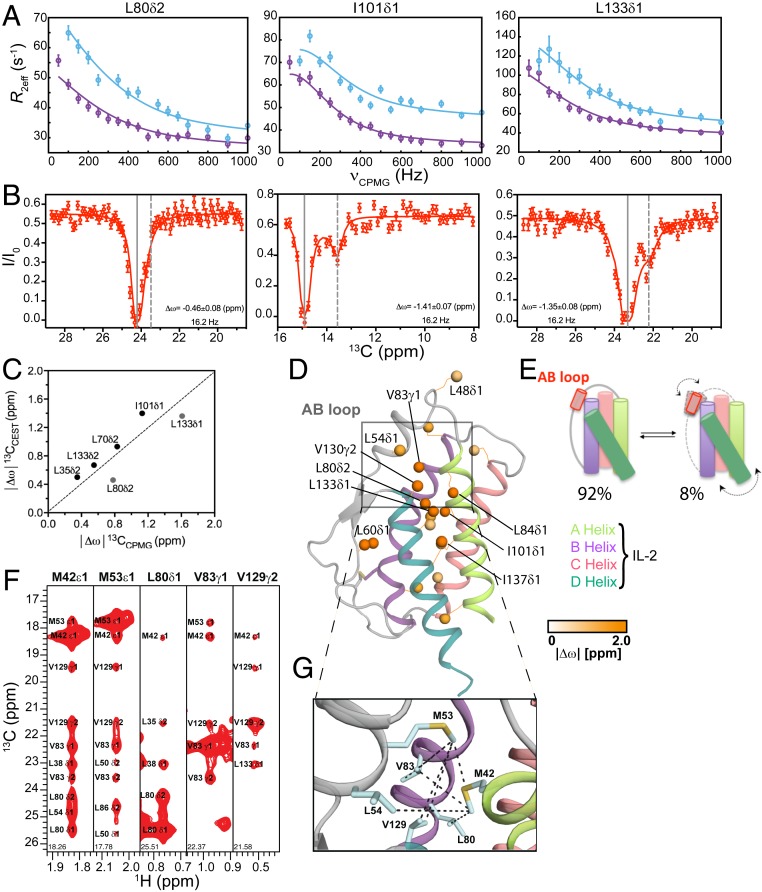
To characterize the conformational landscape sampled by free mIL-2 in solution, we performed a series of 13C chemical exchange saturation transfer (CEST) (28) and 13C single-quantum methyl Carr–Purcell–Meiboom–Gill (CPMG) relaxation dispersion experiments (29). Briefly, in CPMG experiments, the effective transverse relaxation rate (R2,eff) is measured as a function of refocusing pulse frequency (νCPMG), which quenches the effects of conformational exchange, producing so-called dispersion profiles. Such profiles can be fit to extract the rate of exchange between the ground and excited conformational states, kex, the population of the excited state, pE, and the chemical shift differences between the two exchanging states |Δω| (ppm). In CEST, the intensities of protein resonances are measured in the presence of a weak off-resonance radiofrequency (B1) field, typically between 5 and 50 Hz, where a series of two-dimensional (2D) experiments are acquired with a varying offset of the B1 field. When the field offset coincides with the resonance of an exchanging minor state, saturation transfer occurs during a fixed period, leading to an attenuation of the resonance of the major state, which can be readily detected. This enables the indirect observation of the resonance of the otherwise “invisible,” minor state. While, in principle, both experiments can be used to quantify conformational exchange processes in biomolecules (30), CEST is sensitive to motions ranging from ∼50 to 500 s−1 whereas CPMG on the order of 100 to 3,000 s−1. As a result, CEST experiments are typically recorded at a reduced temperature (4 to 10 °C), in order to slow the exchange to a measurable regime.
Due to poor quality of the amide spectra of free mIL-2, we turned to methyl spectra as a readout in both CPMG and CEST experiments. Conformational exchange was observed for the resonances of several methyl groups distributed throughout the mIL-2 structure, as illustrated by representative residues on the B helix (L80δ2), C helix (I101δ1), and D helix (L133δ1) (Fig. 2A). The resonances of 20 methyls exhibiting well-defined CPMG dispersion curves (SI Appendix, Fig. S3A and Table S1) were used in a quantitative analysis. The remaining mIL-2 resonances exhibited low signal-to-noise (S/N) dispersion curves (at 18.8 T magnetic field) or high spectral overlap. Dispersion curves were fitted globally using a two-site exchange model, yielding a kex of 1,000 ± 72 s−1 and an excited state population of 8.0 ± 0.4% (SI Appendix, Fig. S4). Additionally, 13C-CEST profiles for the same residues (L80δ2, I101δ1, and L133δ1) revealed significant dips in the 13C dimension, characteristic of an excited state (Fig. 2B and SI Appendix, Fig. S5). To quantify the extent of structural adaptations upon formation of the excited state for each methyl site participating in the exchange process, we extracted values of the corresponding 13C chemical shift changes, |Δω|, from independent fits of the CPMG relaxation dispersion and CEST datasets. We observed good correlation between the resulting |Δω| values for most methyl probes (Fig. 2C), suggesting that the two datasets are likely reporting on a similar exchange process, with the caveat that the CEST data were recorded at a lower temperature (10 °C relative to the standard temperature of 25 °C used for CPMG data acquisition). Thus, the two observed outliers in the data (L133δ1, L80δ2) could arise from the temperature dependence of the chemical shifts of the major and minor states.
Fig. 2.
Solution dynamics and ground-state structure of free mIL-2 by methyl NMR. (A) Methyl-selective 13C single quantum CPMG relaxation dispersion profiles carried out at two magnetic fields (600 MHz, purple; 800 MHz, blue) are shown for selected residues in the B helix (L80δ2), C helix (I101δ1), and D helix (L133δ1) of WT free mIL-2. Experimental data are shown as small circles in all panels, with errors estimated from the S/N in the raw experimental data. The best-fit lines are shown for a global analysis of 20 methyls with nonflat relaxation dispersion profiles using a two-site conformational exchange model. (B) 13C-CEST profiles carried out at 800 MHz for L80δ2, I101δ1, and L133δ1. Plots of normalized intensity of the ground state resonance relative to a control experiment where the saturating field is highly off-resonance (12 kHz), as a function of the position of the perturbing B1 field (with a measured strength of 16.2 Hz). Uncertainties in the normalized intensity (I/Io) are determined from the root-mean-square deviation in the baseline of the raw CEST profiles where no intensity dips are present (typically, n > 30). Global fits of the CEST data to a two-site model of chemical exchange are shown as solid red lines. The resonances of the major state (gray solid lines) and fitted minor state (gray dashed lines) are indicated, with the resulting chemical shift difference shown in each plot. (C) Linear correlation plot of chemical shifts of the excited state of WT mIL-2 obtained from 13C CPMG (x axis) and 13C CEST (y axis). CPMG and CEST experiments were recorded at 25 °C and 10 °C, respectively. (D) Twenty methyl probes undergoing chemical exchange by CPMG are shown as spheres on the model structure of mIL-2 and colored orange according to the magnitude of fitted |Δω| values. (E) Schematic representation of mIL-2 illustrating the interconversion between a ground state, which is 92% populated, and an invisible, excited state, which is ∼8% populated. The doted arrows represent the coupled conformational transition of the AB loop and the hydrophobic core of mIL-2. (F) 13CM-1HM strips from a 3D CM-CMHM SOFAST NOESY experiment taken at the 13CM coordinates of stereospecifically assigned methyl resonances noted on each panel, showing NOE cross-peaks between the methyl resonances of residues on the AB Loop. (G) Close-up view of the AB loop region from a Rosetta homology-based model of free mIL-2 (using PDB ID 1M47 as a template) showing the network of observed NOEs (black dotted lines), corresponding to the major (ground-state) solution conformation. The pattern of NOEs is consistent with a “closed” conformation of free mIL-2, with where the AB loop is well-packed against the hydrophobic core of the structure.
Methyl groups exhibiting CPMG dispersion curves are plotted on the homology-based model of free mIL-2 in Fig. 2D and color-coded according to the magnitude of the fitted |Δω| values, which report on differences in the local magnetic environment between the major and minor conformations. Large |Δω| values were observed for methyls at the AB loop (L48δ1, L54δ1), in addition to the C terminus of the B helix facing toward the loop (L80δ2, V83γ1, L84δ1) and throughout the hydrophobic core of the structure (I101δ1, V130γ2, L133δ1, and I137δ1). Thus, our CPMG data suggest that free mIL-2 samples a global, cooperative transition to an excited state, which involves a conformational “switch” of the AB loop, coupled to a cooperative change affecting the core methyl groups (Fig. 2E). Notably, a pronounced structural change relative to the free form can be observed in the X-ray structure of JES6-1–bound mIL-2 (10), in which the AB loop (residues Y45 to T55) undergoes an ∼38° rigid-body rotation to adopt a conformation that is optimal for interactions with the antibody surface. Taken together, conformational plasticity of the AB loop is a key component of the solution dynamics of free mIL-2, relevant for the formation of a high-affinity immunomodulatory complex.
To characterize the major conformation sampled by the AB loop in solution, we analyzed methyl nuclear Overhauser effect (NOE) intensities recorded in a 3D CM-CMHM SOFAST nuclear Overhauser effect spectroscopy (NOESY) experiment (Fig. 2F), relative to the corresponding distances observed in 1) a model of free mIL-2 built using the human IL-2 apo-structure as a template (Protein Data bank [PDB] ID code 1M47), 2) the cocrystal structure of mIL-2 in complex with the JES6-1 antibody (Ab) (PDB ID code 4YQX), and 3) a model of the mIL-2 build using the human IL-2/IL-2Rα receptor complex structure as a template (PDB ID code 2B5I). Consistently with the models of mIL-2 in the apo and IL-2Rα receptor-bound states, we find that the network of observed NOEs connecting the methyl groups of M42, M53, L54, L80, V83, and V129 located at the vicinity of the AB loop, (Fig. 2G, black dashed lines) is consistent with a “closed,” well-packed conformation where the side chains of M53 and L54 are making hydrophobic contacts with residues at the hydrophobic core defined by the amphipathic A and B α-helices. This is evident from the observation of several unambiguously assigned short-range (5 Å upper limit) NOEs between the corresponding methyl groups (Fig. 2 D, F, and G). On the contrary, in the antibody-bound mIL-2 structure, the AB loop is found in an “open” conformation, with the methyl groups of M53 and L54 oriented toward the solvent at distances from the core methyls (13 Å), well beyond the NOE detection limit of 10 Å (Fig. 2G and SI Appendix, Table S2). Thus, our NOE data provide strong evidence that the major state of free mIL-2 has a closed AB loop conformation, similar to the X-ray structure of human IL-2 in either the free form or in the complex with IL-2Rα.
A Conserved Hydrogen Bond Network Coupled to the Structural Core Stabilizes the Excited mIL-2 Conformation.
We next sought to characterize the structural features of the excited-state conformation identified by our CPMG and CEST experiments. Excited states play important roles in protein function, including catalysis (31–33) and complex formation via conformational selection or induced fit-type mechanisms (34, 35). However, their de novo structure modeling is challenged by the ambiguity in interpreting methyl chemical shifts. A qualitative comparison of the placement of residues undergoing conformational exchange in free mIL-2 (Fig. 2D) with those perturbed upon formation of the IL-2Rα– and JES6-1–bound states (Fig. 1 D and E) showed a significant overlap, both for residues that are within the AB loop and other binding surfaces (L80δ2 and L86δ2) and also at the core of the structure (L133δ2 and I137δ1), suggesting that the same sites that participate in the formation of the excited state also undergo structural changes upon formation of the mIL-2 tertiary complexes.
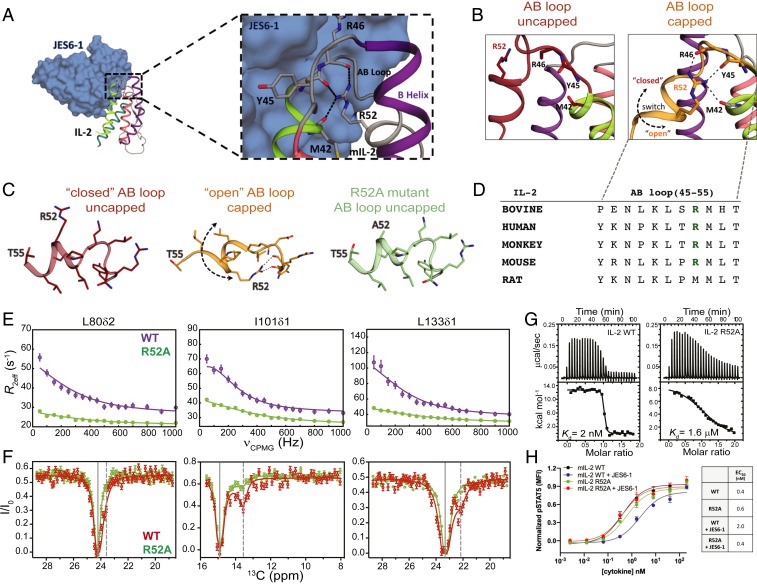
Our NOE-based analysis has shown that the backbone conformation of the AB loop in the major, “closed” state of mIL-2 is inconsistent with the JES6-1–bound structure. Close inspection of the structure reveals that, in the JES6-1 complex, the AB loop instead participates in a network of C-capping interactions with the A helix (Fig. 3 A and D). Specifically, the side chain guanidino group of R52 forms multiple hydrogen bonds with the backbone carbonyl oxygens of M42, Y45 located at the C-terminal end of A helix, and with R46, L48 at the N-terminal part of the AB loop (Fig. 3 B and C). Conversely, in the free structure of the homologous human IL-2 (PDB ID code 1M47, termed “uncapped” state), the R52 side chain is solvent-exposed, and M53 is buried at the hydrophobic interface between the A and B helices. The “closed/uncapped” conformation is optimal for interactions with coreceptors, as exemplified by the IL-2Rα complex where R52 forms a salt bridge with a conserved aspartic acid on the receptor surface (PDB ID code 2B5I). JES6-1 binding therefore induces a transition from a “closed” to “open” AB loop conformation, which exposes the hydrophobic side chains of L50, M53, and L54 toward the surface of the molecule while the side chain of R52 becomes buried. In this “capped” state, the loss of hydrophobic packing contacts is compensated by the formation of multiple C-capping hydrogen bonds with the A helix, and the backbone of the loop itself (Fig. 3 A and C).
Fig. 3.
Effects of a conserved helical capping motif on IL-2 dynamics and function. (A) Crystal structure of mIL-2 (schematic) bound to JES6-1 (blue surface) with a detailed view of the mIL-2/JES6-1 interface on the AB loop and B helix of the cytokine (PDB ID 4YQX). C-capping hydrogen bonds involving the R52 side chain are shown as black lines. (B) The AB loop in mIL-2 adopts either an uncapped “closed” conformation (apo state) or a capped “open” conformation (JES6-1–bound state). The open conformation is stabilized by an intramolecular C-capping interaction between the side chain of R52 and the backbone of M42-Y45-R46. Black dashes indicate hydrogen bonds. The R52 side chain does not contribute to intramolecular stabilizing interactions in the closed conformation. Black dashed arrow shows the direction of loop movement between the closed and open states. (C) Close-up view of the AB loop in the closed and open conformations. The R52A mutation eliminates the C-capping interactions. (D) Sequence conservation pattern in the AB loop region in various IL-2 orthologs. (E) Comparison of 13C-CPMG relaxation dispersion profiles for L80δ2, I101δ1, and L133δ1 of WT mIL-2 (purple) and R52A (green), at 600 MHz and (F) 13C-CEST profiles for WT mIL-2 (red) and R52A (green). Conformational exchange profiles throughput the mIL-2 structure are quenched by the R52A mutation. Experimental errors in CPMG and CEST data are determined as described in Fig. 1. (G) Thermodynamic fingerprints of the interactions between WT and R52A mIL-2 with JES6-1. Isothermal titration calorimetry thermographs and curve fits for titrations. The R52A mutation leads to a three-order-of-magnitude reduction in binding affinity. To minimize enthalpy of solvation effects, all experiments were performed in 20 mM phosphate buffer, pH 7.2, 150 mM NaCl. Data shown are representative of duplicate experiments. (H) STAT5 phosphorylation (pSTAT5) response to WT and R52A mIL-2 in the free state or JES6-1 immunocomplexes treatment in IL-2Rα+ mouse CTLL-2 cells. R52A shows attenuated immunomodulation by the JES6-1 Ab while signaling via IL-2Rα+ is at similar levels to WT. μcal, microcalorie; kcal, kilocalorie; MFI, median fluorescence intensity.
According to our NOE-based analysis, the placement of M53 at the hydrophobic interface between the AB loop and the core of the structure promotes the “closed” loop conformation, which is the major solution state. We sought to determine whether the excited state sampled by free mIL-2 encompasses the C-capping features of the “open” state. If the dynamics observed by NMR correspond to a concerted process, such as an open-to-closed transition of the AB loop, then mutations which destabilize the open state should alter the observed exchange parameters in a uniform manner. To perturb the “open” state conformation, we used the mutant R52A, which eliminates all C-capping interactions (Fig. 3C and SI Appendix, Fig. S6) (36), and compared the resulting CPMG relaxation dispersion and CEST data performed under identical conditions to our established results for the WT. Remarkably, Ala mutation of R52 suppressed the CPMG relaxation dispersion profiles of all methyl probes that were undergoing conformational exchange in the WT form (Fig. 3 E and F and SI Appendix, Fig. S3B). Due to the significantly reduced dispersion range observed for methyl resonances of the R52A mIL-2 mutant, the χ2 surface obtained from a global fit of the CPMG data does not show a well-defined minimum in terms of the estimated population and exchange rate, kex. Consistently, in CEST profiles recorded using the R52A mutant, the second dip was barely observed (I101δ1) or notably absent (L70δ2, I143δ1, L133δ1, L133δ2) (Fig. 3 E and F and SI Appendix, Fig. S5), indicating that the population of the excited state (“open” state) was below the detection limits of the experiment. Both observations are consistent with a significant “dampening” of μs–ms dynamics. The R52A mutation quenched relaxation not only at the side chain methyl but also the backbone amide groups, alleviating resonance broadening and leading to a significant improvement in the quality of the 1H-15N TROSY spectrum (SI Appendix, Fig. S6A). These observations imply the presence of a global conformational exchange process on the µs–ms timescale, involving a switching of the AB loop coupled to a transient twisting or ‘‘breathing’’ of the α-helices. Notably, the conserved C-capping sequence motif at the AB loop is critical for the formation of the excited state.
To determine how destabilization of the excited state affects mIL-2 binding to JES6-1, we used isothermal titration calorimetry (ITC) and measured the binding free energy (ΔG), and its enthalpic (ΔH) and entropic (−TΔS) components both for WT and R52A mIL-2 (Fig. 3 G and H). Consistently with previous SPR measurements (10) and our observation of a tight complex in slow exchange by NMR, WT mIL-2 binds JES6-1 with a high affinity of 2 nM. The interaction is largely entropy-driven, with a positive net enthalpy change of 12.7 kcal⋅mol−1 and a favorable entropic contribution of −24.3 kcal⋅mol−1 at 293 K (Fig. 3H and SI Appendix, Table S3). Notably, R52A binds to JES6-1 with three orders of magnitude lower affinity than WT mIL-2, due to less favorable binding entropy (Fig. 3 G and H). Given that the side chain of R52 forms a salt bridge with E60 of JES6-1 in the X-ray structure of the complex (Fig. 3A), the observed 1,000-fold decrease in binding affinity reflects the combined effects of 1) loss of binding enthalpy and 2) increase in free energy of the “closed” state, which is competent for binding. Thus, our ITC results show that perturbations of the capping hydrogen-bond network coupled to global changes in core dynamics abrogate JES6-1 binding by destabilizing the open conformation, consistently with our NMR measurements showing a shift of the conformational equilibrium toward the closed state.
To determine the functional activity of the R52A mutant, we probed JES6-1 antibody effects on mIL-2–induced signaling in IL-2Rα+ subpopulations of mouse CTLL-2 cells as a surrogate for its effects on IL-2Rαhi immune cell subsets (Fig. 3H). Consistent with our NMR and ITC results, JES6-1 showed a fivefold reduced WT mIL-2–mediated signal transducers and activators of transcription 5 (STAT5) activation. However, STAT5 signaling potency by R52A was independent of JES6-1, as demonstrated by the comparable effective concentration, 50% (EC50) values, suggesting that the weaker affinity to JES6-1 was primarily responsible for the behavior of the R52A mutant (Fig. 3G).
Allosteric Communication in mIL-2 through a Remodeling of Side Chain Rotamers.
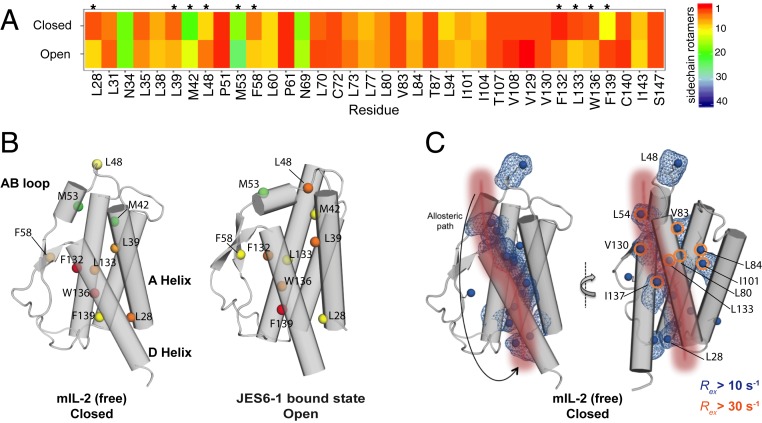
Our methyl-based NMR probes revealed a global response of the mIL-2 structure upon binding to JES6-1, alongside the crystallographically observed conformational changes of the AB loop (Fig. 1 C and D). Consistently, our NMR relaxation measurements suggest a coupling of dynamic motions between the loop and core methyls. We sought to identify a plausible mechanism linking AB loop movement to changes in core side chain packing during transitions between “closed/uncapped” and “open/capped” conformations. To enumerate all possible side chain rotamers that can be adopted by each residue, we performed a global analysis of compatible rotamer pairs using a satisfiability-based approach in Rosetta (Materials and Methods and SI Appendix, Fig. S8) and mapped our results on the free and JES6-1–bound mIL-2 structures (Fig. 4 and SI Appendix, Fig. S9). Using the backbone conformations of the “open” and “closed” states as inputs, our analysis highlights differences in rotamer sets that can be accessed by the free (closed) and JES6-1 bound (open) states. We identified a large set of residues (L28, L39, M42, L48, M53, F58, F132, L133, W136, and F139) (Fig. 4 A and B) spanning the AB loop, A Helix and D Helix, and part of the hydrophobic core. For these residues, the space of rotamers was significantly different between the closed and open states, indicating a plausible remodeling of packing interactions. Specifically, a 10-residue segment (V129 to F139) forming the hydrophobic face of the amphipathic D helix exhibits expansions and contractions in allowed rotamer sets as IL-2 transitions between the two states (SI Appendix, Fig. S8). A concerted motion of the D helix upon binding of IL-2Rα has been highlighted as having a functional role in the formation of the high-affinity heterotrimeric signaling complex (37). These results are consistent with our observed NMR chemical shift mapping of the IL-2Rα and Ab-bound states (Fig. 1 C and D), suggesting that the conformational transition between the free and complexed states can lead to a redistribution of the rotameric states at the hydrophobic core. A plausible allosteric communication network starts at the AB loop on one end of the structure, traverses through the inner core of mIL-2, and ends at the N terminus of the A and D helices adjacent to the binding site of the IL-2Rβ receptor (Fig. 4C). The hydrophobic core residues exhibiting differences in rotamer sets also include sites with significant chemical exchange contributions in our CPMG data, indicating the presence of dynamics at the μs–ms timescale (Fig. 4C, orange circles). Taken together, our results highlight a plausible allosteric communication network in the IL-2 structure mediated via sequential remodeling of side chain packing interactions.
Fig. 4.
Sampling of different ensembles of side chain rotamers in the “open” and “closed” mIL-2 states. (A) Heat map showing the number of allowed side chain rotamers for buried residues in the “closed”- and “open”-state structures. Buried residues were computed using a 10 Å2 solvent accessible surface area threshold. Residues that show a significant difference (>3) in the number of allowed rotamers between closed and open states are highlighted with asterisks (*). (B) Closed/free and open/bound structures showing residues that exhibit a significant difference in side chain rotamer sets, with the total number of allowed rotamers colored as in A. (C) Illustration of a putative allosteric communication network linking the AB loop conformation to the core structure. A sequential path demarcated by residues undergoing side chain remodeling as mIL-2 transitions between the two states is shown with a red patch onto the mIL-2 structure of the closed state, used as a reference. The methyl groups showing significant exchange (Rex) contributions in our CPMG experiments indicating dynamics are highlighted with blue surfaces (Rex > 10 s−1) or orange circles (Rex > 30 s−1).
Skewing the Dynamic Landscape of mIL-2 by Ligand Binding.
The R52A mutation characterized here destabilizes the open mIL-2 conformation by perturbing the C-capping hydrogen-bond network between the AB loop and B helix, leading to quenching of conformational exchange throughout the core of the structure. We hypothesized that a small molecule binding preferentially to the closed AB loop conformation would impact the dynamic landscape of mIL-2 in a similar manner. We used a known compound targeting human IL-2 (hIL-2) (Ro 26-4550), to compete with IL-2Rα binding (7, 38). The cocrystal structure (PDB ID code 1M48) shows that Ro 26-4550 is nestled in a hydrophobic pocket at the interface between the AB loop and B helix, with the hIL-2 side chains of M39, V69, and L72 (mIL-2 residues M53, V83, and L86) packing against the terminal aromatic ring (7). We performed an NMR titration of increasing molar ratios of Ro 26-4550 on isoleucine, leucine, and valine-methyl–labeled WT mIL-2 using 1H-13C HMQC spectra as a readout (Fig. 5A). Under our NMR conditions, we observed the formation of a moderate-affinity (μM range dissociation constant [Kd]) complex in fast exchange with the free mIL-2 form where residues in close proximity to the AB loop (L48δ1, L54δ1, and L60δ1) experience the largest effects (SI Appendix, Fig. S7). However, CSPs extend beyond the AB loop, to sites in the hydrophobic core (L80δ1, V129γ2, V130γ2, L133δ2, and I137δ1) at distances >12 Å from the inhibitor binding site in the cocrystal structure (Fig. 5C). A quantitative NMR line shape analysis of the resonances of V129γ2 and L133δ2 yielded an equilibrium dissociation constant of 39.4 ± 5.5 µM (Fig. 5B), suggesting a two-state cooperative transition impacting all sites in the ligand-bound form. To further examine whether Ro 26-4550 binding affects the µs–ms dynamics of mIL-2, we repeated 13C-CPMG relaxation dispersion experiments, under saturation binding, identical protein concentration and NMR sample conditions, and compared with our established data recorded for the free form (Fig. 5D). Our results indicate that Ro 26-4550 binding strongly dampens relaxation of methyl groups that are undergoing µs–ms conformational exchange throughout the mIL-2 structure. Thus, in agreement with our allosteric network model of mIL-2, stabilization of the closed/uncapped conformation through ligand binding reduces dynamic exchange at the hydrophobic core of mIL-2 as the conformational equilibrium is skewed toward the closed state. The observation of residual exchange at the CPMG timescale suggests that sampling of the open state is still permitted by the presence of the ligand.
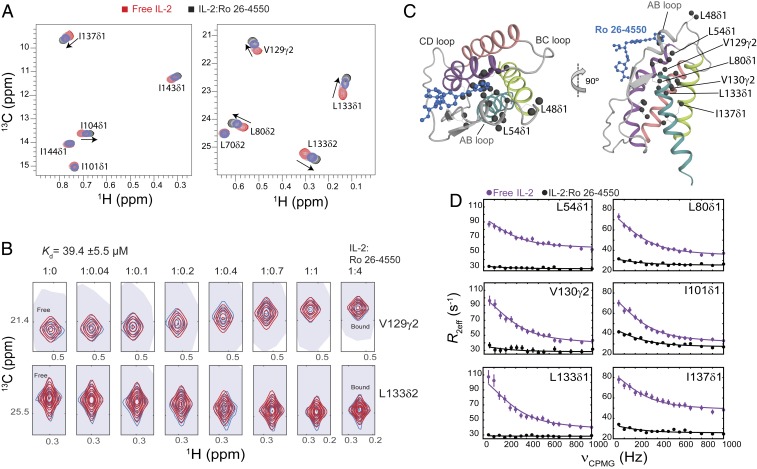
Fig. 5.
Small molecule binding at the AB loop quenches dynamics of mIL-2. (A) The 2D 1H,13C-HMQC spectra of ILV-methyl–labeled mIL-2 in the free state (red), and with increasing concentrations of Ro 26-4550 inhibitor (1:4 molar ratio, shown in black). Data were recorded at 800 MHz, 25 °C. Fast-exchange chemical shift changes are highlighted with arrows for select methyl resonances. (B) NMR line shape analysis for the V129γ2 and L133δ2 methyl resonances using TITAN (Materials and Methods), with indicated equilibrium dissociation constant and errors propagated from the spectral S/N. Recorded NMR spectra are shown in blue, with simulated line shapes in red. Ratios of mIL-2 to inhibitor are indicated in each panel. (C) Two views showing the mapping of residues undergoing significant chemical shift perturbations (CSPs) onto the ribbon representation of mIL-2. Methyl groups with marked CSPs (>0.05 ppm) are shown as black spheres. The inhibitor is shown as a blue ball-and-stick diagram on the overlaid hIL-2 complex structure (PDB ID code 1M48). (D) The 13C single quantum CPMG relaxation dispersion profiles in the absence (purple) and presence of Ro 26-4550 at saturating concentration (black) for selected residues in the AB loop (L54δ1), B Helix (L80δ1), C Helix (I101δ1), and D Helix (V130γ2, L133δ1, I137δ1). CPMG experiments were performed at a 1H field of 600 MHz and 25 °C.
Discussion
The unique conformational plasticity of IL-2 appears to play an important role in targeting the IL-2 receptor signaling axis in both agonist and antagonist mode. However, this plasticity also presents an opportunity to target or otherwise manipulate the conformational landscape of IL-2 for drug discovery, but more detailed insights are needed for actionable value. Evidence for a functional role of such conformational plasticity was provided in previous studies focusing on both human and mouse IL-2. These studies employed small molecule binding (6, 7), antibody engineering (9, 12), or directed mutagenesis (8) to induce minor perturbations in the cytokine’s core α-helical fold, leading to distinct immunomodulatory functions. Specifically, elucidating a link between specific allosteric effects on the IL-2 structure, induced by the JES6-1 and S4B6 antibodies, with the targeting of opposing immune cells types expressing different levels of receptor subunits in vivo has provided a solid foundation for immune modulation through the stabilization of different IL-2 conformational states. Conversely, the high-throughput screening of hIL-2 libraries in yeast led to the identification of a variant, termed “super-2,” comprising a helix that is locked in a conformation which is primed for interactions with the IL-2Rβ subunit, thereby leading to preferential expansion of cells lacking IL-2Rα (8). These results were leveraged to discover a human Treg-based Ab, akin to JES6-1, of significant therapeutic potential (11). Thus, given the sequence and structure conservation of the IL-2 fold across species, the results established for the mouse IL-2 paradigm directly relate to human IL-2, whose use as a multifaceted protein therapeutic (Proleukin) has been hampered by an incomplete understanding of its precise functional properties. Therefore, a detailed characterization of the relevant conformational states and their dynamic interconversion process in solution would provide a road map for drugging IL-2, through a range of approaches.
Our methyl-based NMR measurements from two complementary methodologies, CPMG and CEST, provide strong support that free mIL-2 is highly dynamic at the µs–ms timescale and samples an excited state via a concerted conformational transition of the AB loop and core of the structure. Functionally relevant excited states have been previously described for a range of protein and nucleic acid systems (30) and are typically separated by relatively low energy barriers from the ground state. The inherent plasticity of the AB loop and allosteric cross-talk with the hydrophobic core suggested an avenue for biasing the conformational equilibrium by perturbing key features present in the excited state conformation. Based on our analysis of crystallographically observed snapshots of IL-2, we hypothesized that the excited state might share similar features with the JES6-1 antibody-bound structure, with respect to a stabilizing C-capping motif, and tested this hypothesis through a detailed dynamic, binding, and functional characterization of a designed Ala mutant. In fact, our data demonstrate that the structural basis for such functional complexity may be surprisingly simple: The global dynamic motions of mIL-2 are induced by a conserved C-capping hydrogen-bond network that is connected to the core of the structure and can be switched on and off by a single point mutation (Fig. 3). Taken together, out results show that the IL-2 energy landscape is highly malleable and establish a paradigm for controlling IL-2 cytokine activity through designed changes in protein dynamics.
Analysis of our NMR, ITC, and functional data in the context of the available IL-2 cocrystal structures suggests a general scheme for molecular recognition between IL-2 and its binding partners (Fig. 6). According to this model, free mIL-2 exists in a preequilibrium between closed (major form, 92%) and open conformations (minor form, 8%), with the AB loop and the hydrophobic core engaged in a cooperative motion on the µs–ms timescale. In the closed/uncapped (productive) state, the AB loop is in a conformation that is primed to interact with the IL2-Rα receptor. Formation of a C-capping hydrogen network locks the AB loop in an open (autoinhibited) state, in which the loop adopts a conformation that is optimal for interactions with the JES6-1 antibody (Fig. 6, left). Disruption of the C-capping hydrogen bond network by mutation or small molecule binding shifts the equilibrium toward the closed state and induces global changes in core dynamics which ultimately lead to a reduction in Ab binding by three orders of magnitude (Fig. 6, right). While the “closed” and “open” conformations mediate high-affinity complex formation with the receptor and Ab, respectively, the binding process is likely achieved via the formation of an initial encounter complex where additional induced structural adaptations follow the selection of preexisting conformational states. This model is consistent with the majority of protein–ligand binding examples where both processes contribute to complex formation (34).
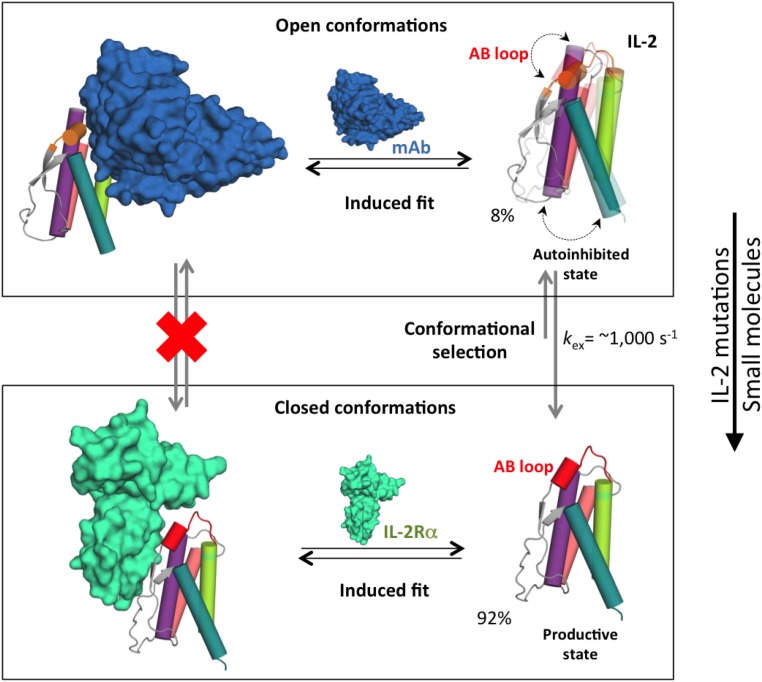
Fig. 6.
Conformational “priming” of free mIL-2 drives high-affinity complex formation. A global conformational transition enables mIL-2 to sample two distinct states that are recognized by IL2-Rα and JES6-1 via additional induced fit steps (black arrows). In the closed conformation, the polar face of the AB loop including R52 is primed to interact with the IL2-Rα receptor. Formation of a C-capping hydrogen bond network by R52 locks the AB loop in an autoinhibitory, open conformation with high affinity for the JES6-1 Ab. As shown in Figs. 3 and 4, point mutations or binding of small molecules can shift the equilibrium to favor the closed conformation, with measurable functional effects (right). Representative exchange parameters and excited-state populations obtained from our NMR data for mIL-2 are indicated. The backbone of the mIL-2 is colored as in Figure 1. Black arrows indicate equilibrium between the free and bound states. Dotted double arrows represent the coupled conformational transition of the AB loop and hydrophobic core of mIL-2. Gray arrows indicate the conformational equilibrium, with the longer arrow pointing to the major state (closed conformation).
A consequence of allosteric cross-talk in the IL-2 fold is that the protein becomes globally sensitive to mutations. Point mutations can induce long-range effects, which may explain the high level of sequence conservation of IL-2 residues removed from the primary receptor recognition sites. Further examples of such long-range modulation are provided by engineered IL-2 agonists. IL-2 superkine (also denoted as “super-2”) functions independently of IL-2Rα and has enhanced binding to IL-2Rβ, despite five of the six mutations being clustered on the BC loop and within the C helix core, removed from the IL-2Rβ primary binding site. Instead, a cluster of hydrophobic substitutions (F80, V85, and V86) induce a subtle change in C helix orientation, leading to an increased affinity for IL-2Rβ. Super-2 exerts a more potent expansion of tumor-specific cytotoxic T cell populations, and a reduced expansion of Treg cells (8). Moreover, Super-2 can be used as a platform for additional mutations that inhibit binding to the γc receptor and, consequently, heterodimerization of IL-2Rβ and γc, thereby acting as a potent antagonist (13). Conversely, to bypass allosteric effects present in the IL-2 fold, a de novo design approach was used to generate a mimetic of IL-2/IL-15 showing an α-helical fold with a well-packed hydrophobic core, denoted Neoleukin-2/15 (Neo-2/15) (16). This synthetic molecule retains the binding surfaces for the IL-2Rβ/γc receptors while also alleviating the requirement of IL-2Rα/IL-15Rα for formation of a productive signaling complex. Neo-2/15 exhibited enhanced antitumor activity in mouse models, was more stable, and lacked detectable immunogenicity (16). These studies highlight the potential for modulating IL-2 function through different design approaches, showing varying levels of responsiveness to regulatory molecules and conditions at the vicinity of the membrane signaling complex.
In summary, our results highlight the role of conformational plasticity at conserved features of the IL-2 fold, acting locally and allosterically to form an “excited” state with discrete functional properties. Our NMR-based characterization offers a window to access otherwise hidden states of the energy landscape that can be then exploited to fine-tune the affinity and selectivity for different IL-2 binding partners toward engineering novel cytokine functions. For example, our established allosteric coupling between the receptor binding sites to the hydrophobic core of the structure presents an opportunity to alter IL-2 function through the introduction of core mutations and induction of allosteric effects without directly affecting the receptor/Ab binding surfaces. Indeed, the allosteric pathway elucidated in detail here using mouse IL-2 as a model system has been recently exploited for human IL-2 to develop Treg-biased antibodies that appear to work by a similar mechanism (11). Given that the IL-2 fold presents a limited surface area with overlapping binding epitopes, our findings have important practical utility from an engineering and synthetic biology perspective.
Materials and Methods
Specific details about mIL-2 NMR sample preparation and stereospecific isotopic labeling, backbone and methyl resonance assignments, relaxation dispersion and 13C CEST experiments, SEC MALS and ITC experiments, side chain rotamer analysis, and STAT5 signaling are outlined in detail in SI Appendix.
Data and Materials Availability.
NMR assignments for the free WT mIL-2, mIL-2 in complex with JES6-1 scFv antibody, mIL-2 in complex with IL-2Rα, and R52A mIL-2 mutant have been deposited in the Biological Magnetic Resonance Data Bank under accession numbers 27969, 27970, 27971, and 27974, respectively.
Supplementary Material
Acknowledgments
This research was supported by the Howard Hughes Medical Institute, by the National Institute of Allergy and Infectious Diseases Grants R01AI143997 and R01AI51321 (to N.G.S. and K.C.G., respectively), by the National Institute of General Medical Sciences Grant R35GM125034 (to N.G.S.), by the National Institute of Diabetes and Digestive and Kidney Diseases Grant UC4DK116264, and by High End Instrumentation (HIE) Grant S10OD018455, which funded the 800 MHz NMR spectrometer at the University of California, Santa Cruz. We would like to acknowledge Hsiau-Wei Lee and Andrew McShan for assistance with recording NMR data, and Sarvind Tripathi and Jeffrey Swan for assistance with SEC-MALS analysis. We thank Enrico Rennella (University of Toronto) for his assistance with the implementation of the 13C-methyl CEST experiment.
Footnotes
The authors declare no competing interest.
Data deposition: Nuclear magnetic resonance assignments for the free wild-type (WT) mouse IL-2 (mIL-2), mIL-2 in complex with JES6-1 single-chain Fv (scFv) antibody, mIL-2 in complex with IL-2 receptor (IL-2Rα), and R52A mIL-2 mutant have been deposited in the Biological Magnetic Resonance Data Bank (BMRB), http://www.bmrb.wisc.edu, under accession numbers 27969, 27970, 27971, and 27974, respectively.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2000419117/-/DCSupplemental.
References
- 1.Boyman O., Sprent J., The role of interleukin-2 during homeostasis and activation of the immune system. Nat. Rev. Immunol. 12, 180–190 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Liao W., Lin J.-X., Leonard W. J., Interleukin-2 at the crossroads of effector responses, tolerance, and immunotherapy. Immunity 38, 13–25 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malek T. R., Castro I., Interleukin-2 receptor signaling: At the interface between tolerance and immunity. Immunity 33, 153–165 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taniguchi T., Minami Y., The IL-2/IL-2 receptor system: A current overview. Cell 73, 5–8 (1993). [DOI] [PubMed] [Google Scholar]
- 5.Malek T. R., Bayer A. L., Tolerance, not immunity, crucially depends on IL-2. Nat. Rev. Immunol. 4, 665–674 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Thanos C. D., DeLano W. L., Wells J. A., Hot-spot mimicry of a cytokine receptor by a small molecule. Proc. Natl. Acad. Sci. U.S.A. 103, 15422–15427 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arkin M. R., et al. , Binding of small molecules to an adaptive protein-protein interface. Proc. Natl. Acad. Sci. U.S.A. 100, 1603–1608 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levin A. M., et al. , Exploiting a natural conformational switch to engineer an interleukin-2 ‘superkine’. Nature 484, 529–533 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Létourneau S., et al. , IL-2/anti-IL-2 antibody complexes show strong biological activity by avoiding interaction with IL-2 receptor alpha subunit CD25. Proc. Natl. Acad. Sci. U.S.A. 107, 2171–2176 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spangler J. B., et al. , Antibodies to interleukin-2 elicit selective T cell subset potentiation through distinct conformational mechanisms. Immunity 42, 815–825 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trotta E., et al. , A human anti-IL-2 antibody that potentiates regulatory T cells by a structure-based mechanism. Nat. Med. 24, 1005–1014 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boyman O., Kovar M., Rubinstein M. P., Surh C. D., Sprent J., Selective stimulation of T cell subsets with antibody-cytokine immune complexes. Science 311, 1924–1927 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Mitra S., et al. , Interleukin-2 activity can be fine tuned with engineered receptor signaling clamps. Immunity 42, 826–838 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arenas-Ramirez N., et al. , Improved cancer immunotherapy by a CD25-mimobody conferring selectivity to human interleukin-2. Sci. Transl. Med. 8, 367ra166 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Spangler J. B., et al. , Engineering a single-agent cytokine/antibody fusion that selectively expands regulatory T cells for autoimmune disease therapy. J. Immunol. 201, 2094–2106 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Silva D. A., et al. , De novo design of potent and selective mimics of IL-2 and IL-15. Nature 565, 186–191 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park Y. H., et al. , Effect of in vitroexpanded CD4(+)CD25(+)Foxp3(+) regulatory T cell therapy combined with lymphodepletion in murine skin allotransplantation. Clin. Immunol. 135, 43–54 (2010). [DOI] [PubMed] [Google Scholar]
- 18.Webster K. E., et al. , In vivo expansion of T reg cells with IL-2-mAb complexes: Induction of resistance to EAE and long-term acceptance of islet allografts without immunosuppression. J. Exp. Med. 206, 751–760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grinberg-Bleyer Y., et al. , IL-2 reverses established type 1 diabetes in NOD mice by a local effect on pancreatic regulatory T cells. J. Exp. Med. 207, 1871–1878 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang Q., et al. , Central role of defective interleukin-2 production in the triggering of islet autoimmune destruction. Immunity 28, 687–697 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruschak A. M., Kay L. E., Methyl groups as probes of supra-molecular structure, dynamics and function. J. Biomol. NMR 46, 75–87 (2010). [DOI] [PubMed] [Google Scholar]
- 22.Sprangers R., Kay L. E., Quantitative dynamics and binding studies of the 20S proteasome by NMR. Nature 445, 618–622 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Ollerenshaw J. E., Tugarinov V., Kay L. E., Methyl TROSY, explanation and experimental verification. Magn. Reson. Chem. 41, 843–852 (2003). [Google Scholar]
- 24.Gans P., et al. , Stereospecific isotopic labeling of methyl groups for NMR spectroscopic studies of high-molecular-weight proteins. Angew. Chem. Int. Ed. Engl. 49, 1958–1962 (2010). [DOI] [PubMed] [Google Scholar]
- 25.De Paula V. S., Sgourakis N. G., Backbone amide and MILV methyl chemical shift assignments of mouse interleukin-2. BioMagResBank. http://www.bmrb.wisc.edu/data_library/summary/index.php?bmrbId=27969. Deposited 7 November 2019.
- 26.De Paula V. S., Sgourakis N. G., ILV(proS) methyl assignment of mIL-2 in complex with JES6-1 scFV antibody. BioMagResBank. http://www.bmrb.wisc.edu/data_library/summary/index.php?bmrbId=27970. Deposited 17 July 2019.
- 27.De Paula V. S., Sgourakis N. G., ILV(proS) methyl assignment of mIL-2 in complex with IL-2Ra (CD25) receptor. BioMagResBank. http://www.bmrb.wisc.edu/data_library/summary/index.php?bmrbId=27971. Deposited 17 July 2019.
- 28.Vallurupalli P., Sekhar A., Yuwen T., Kay L. E., Probing conformational dynamics in biomolecules via chemical exchange saturation transfer: A primer. J. Biomol. NMR 67, 243–271 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Lundström P., Vallurupalli P., Religa T. L., Dahlquist F. W., Kay L. E., A single-quantum methyl 13C-relaxation dispersion experiment with improved sensitivity. J. Biomol. NMR 38, 79–88 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Sekhar A., Kay L. E., NMR paves the way for atomic level descriptions of sparsely populated, transiently formed biomolecular conformers. Proc. Natl. Acad. Sci. U.S.A. 110, 12867–12874 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Henzler-Wildman K. A., et al. , Intrinsic motions along an enzymatic reaction trajectory. Nature 450, 838–844 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Boehr D. D., McElheny D., Dyson H. J., Wright P. E., The dynamic energy landscape of dihydrofolate reductase catalysis. Science 313, 1638–1642 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Whittier S. K., Hengge A. C., Loria J. P., Conformational motions regulate phosphoryl transfer in related protein tyrosine phosphatases. Science 341, 899–903 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hammes G. G., Chang Y. C., Oas T. G., Conformational selection or induced fit: A flux description of reaction mechanism. Proc. Natl. Acad. Sci. U.S.A. 106, 13737–13741 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vogt A. D., Di Cera E., Conformational selection or induced fit? A critical appraisal of the kinetic mechanism. Biochemistry 51, 5894–5902 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Paula V. S., Sgourakis N. G., MILV methyl chemical shift assignments of the R52A mutant of mouse interleukin-2. BioMagResBank. http://www.bmrb.wisc.edu/data_library/summary/index.php?bmrbId=27974. Deposited 17 July 2019.
- 37.Wang X., Rickert M., Garcia K. C., Structure of the quaternary complex of interleukin-2 with its alpha, beta, and gammac receptors. Science 310, 1159–1163 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Tilley J. W., et al. , Identification of a small molecule inhibitor of the IL-2/IL-2Rα receptor interaction which binds to IL-2. J. Am. Chem. Soc. 119, 7589–7590 (1997). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.