Significance
The striatum of the basal ganglia is divided into dorsal and ventral regions. The dorsal striatum regulates movement and cognition, whereas the ventral striatum modulates reward and emotion. The importance of striatal divisions also is reflected in neurological diseases as the dorsal and ventral striata are differentially targeted by Huntington’s disease, Parkinson’s disease, and drug addiction disorders. Despite the significance of these neurological differences, the neural mechanisms underlying the division of the striatum into the dorsal and ventral districts remain elusive. Here, we have identified two key genes Nolz-1 and Dlx1/2, and the interactions between Nolz-1 and Dlx1/2 genes are critical for the developmental processes by which the striatum is divided into dorsal and ventral districts.
Keywords: basal ganglia, dorsal striatum, ventral striatum, nucleus accumbens, olfactory tubercle
Abstract
The striatal complex of basal ganglia comprises two functionally distinct districts. The dorsal district controls motor and cognitive functions. The ventral district regulates the limbic function of motivation, reward, and emotion. The dorsoventral parcellation of the striatum also is of clinical importance as differential striatal pathophysiologies occur in Huntington’s disease, Parkinson’s disease, and drug addiction disorders. Despite these striking neurobiologic contrasts, it is largely unknown how the dorsal and ventral divisions of the striatum are set up. Here, we demonstrate that interactions between the two key transcription factors Nolz-1 and Dlx1/2 control the migratory paths of striatal neurons to the dorsal or ventral striatum. Moreover, these same transcription factors control the cell identity of striatal projection neurons in both the dorsal and the ventral striata including the D1-direct and D2-indirect pathways. We show that Nolz-1, through the I12b enhancer, represses Dlx1/2, allowing normal migration of striatal neurons to dorsal and ventral locations. We demonstrate that deletion, up-regulation, and down-regulation of Nolz-1 and Dlx1/2 can produce a striatal phenotype characterized by a withered dorsal striatum and an enlarged ventral striatum and that we can rescue this phenotype by manipulating the interactions between Nolz-1 and Dlx1/2 transcription factors. Our study indicates that the two-tier system of striatal complex is built by coupling of cell-type identity and migration and suggests that the fundamental basis for divisions of the striatum known to be differentially vulnerable at maturity is already encoded by the time embryonic striatal neurons begin their migrations into developing striata.
The striatum of the basal ganglia regulates a broad range of neurological functions, including movement, reward learning, affect, and cognition, and the dysfunction of the striatum is involved in neurological and psychiatric disorders (1–4). The broad range of striatal function stems from the fact that parallel pathways from the motor, sensory, associative, and limbic cortices run through different regions of the striatum (1, 5–7). The striatum is a two-tier system that comprises cytoarchitecturally similar but functionally distinct dorsal and ventral striata. The dorsal striatum consists of the caudate nucleus and putamen (CP) that controls motor and cognitive function (1, 5). The ventral striatum consists of the nucleus accumbens (NAc) and olfactory tubercle (OT) that regulates the limbic function of motivation, affect, and reward (8–11). Dorsal and ventral striata are differentially involved in neurological diseases. Dorsal striatal circuits are pathological loci of movement disorders including Parkinson's disease and Huntington's disease (12, 13), whereas ventral striatal circuits are the targets of addictive disorders (14). Despite the extensive knowledge of the structure and function of the striatal complex in adulthood, little is yet known about how the dorsal and ventral striata are differentially specified during development.
Because CP, NAc, and OT neurons express similar profiles of transcription factors and neurotransmission-related molecules and exhibit cellular morphology similar to that of medium-sized spiny neurons (10, 15), they may share developmental origins in neurogenesis. This is supported by homotopic transplantation studies, which show that donor-derived cells grafted from the lateral ganglionic eminence ([LGE], striatal anlage) are distributed throughout CP, NAc, and OT neurons of host brains (16). The LGE is divided into dorsal and ventral LGEs (17). The dorsal and ventral LGEs give rise to interneurons in the olfactory bulb and projection neurons in the CP, respectively. It is yet unclear whether the progenitors of CP, NAc, and OT neurons are localized in specific domains within the LGE, and/or they are derived from temporal progression of progenitors that differentiate at different time windows, through combinatorial expression of transcription factor codes delineated in progenitor domains of the LGE (18).
In an attempt to decipher mechanisms underlying the developmental construction of the dorsal and ventral striata, we performed genome-wide comparisons of gene expression patterns of dorsal and ventral parts of the LGE. We identified a number of genes that were differentially expressed in the dorsal and ventral developing striata. We focused on Nolz-1/Znf503/Zfp503 that was expressed at high levels in developing dorsal striata. Here, we report that Nolz-1 plays a pivotal role in the regulation of cell-type specification and neuronal migration in the dorsal and ventral striata during development. Nolz-1 null mutation not only resulted in aberrant differentiation of striatal neurons of the dorsal and ventral striata, but also induced abnormal enlargement of the ventral striatum at the expense of the dorsal striatum. The distorted striatal complex in the Nolz-1 mutant brain was primarily caused by an abnormal Dlx1/2-dependent cell migration, which drove aberrant migration of striatal cells from the dorsal toward the ventral striatum. Therefore, repression of Dlx1/2 signaling in the postmitotic striatal neurons by Nolz-1 is required for normal migration to their dorsal and ventral locations and proper specification of the cell types in the dorsal and ventral striata, which allows the parcellation of the striatal complex into the dorsal and ventral striata.
Results
Identification of Genes Differentially Enriched in the Dorsal or Ventral Striatum during Development.
To search for genes that are differentially expressed in developing dorsal and ventral striata, we dissected dorsal and ventral parts of the LGE, striatal anlage in the E13.5 mouse forebrain (SI Appendix, Fig. S1A) and then performed genome-wide microarray analysis. We compared gene expression profiles between the dorsal and the ventral parts of the LGE. Ingenuity pathway analysis revealed differentially expressed genes in several categories, including sonic hedgehog signaling, fibroblast growth factor (FGF) signaling, transforming growth factor (TGF)-β signaling, human embryonic stem cell pluripotency, transcriptional regulatory network in embryonic stem cells, axonal guidance signaling, and others, and these genes included growth factors and transcription factors (SI Appendix, Table S1). qRT-PCR assays confirmed dorsal-enriched genes Wnt7b and Nolz-1/Znf503, and ventral-enriched genes Acvr2a, fgf14, fgf15, Otx2, Gli1, Zic1, Nrp2, Plxnc1, Nts, and Calb1 (SI Appendix, Fig. S1 A and B).
Among genes enriched in the dorsal striatum of the E13.5 mouse brain, Nolz-1 was of particular interest (SI Appendix, Fig. S1B). We have previously reported that Nolz-1 is a developmentally regulated striatum-enriched gene in the rat brain (19). Nolz-1 is highly expressed in the lateral ganglionic eminence of the striatal anlage. Nolz-1 is not expressed in proliferating progenitors but is expressed in early differentiating striatal neurons. Nolz-1 expression is high in the embryonic striatum, but it is down-regulated in the postnatal striatum (19, 20). We generated floxed Nolz-1fl/fl mice for studying Nolz-1 function (SI Appendix, Fig. S2). Nolz-1fl/fl mice were intercrossed with Protamine-Cre mice to generate germline Nolz-1−/− knockout (KO) mice. Nolz-1−/− KO mice died immediately after birth. Immunostaining of Nolz-1 confirmed the absence of the Nolz-1 protein in the E18.5 Nolz-1 KO striatum (SI Appendix, Fig. S2).
Nolz-1 Mutation Induces Hypoplasia of the Dorsal Striatum but Hyperplasia of the Ventral Striatum.
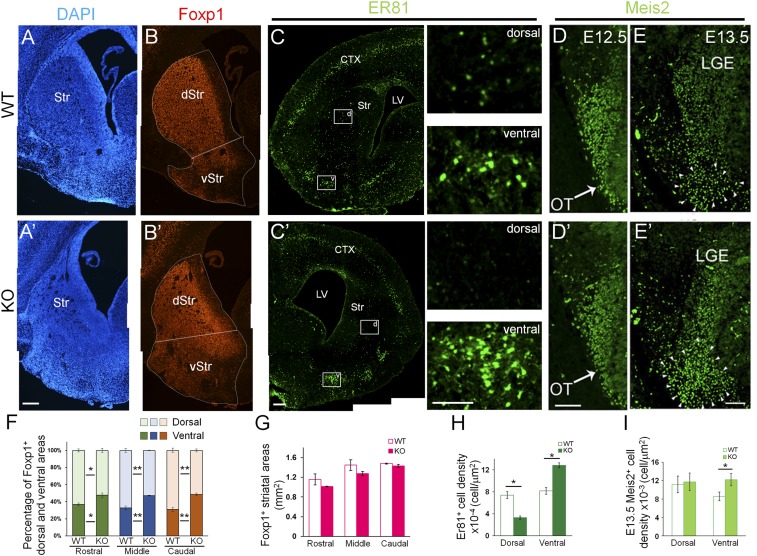
Nolz-1 null mutation induced a dramatically structural alteration in the striatal complex in E18.5 Nolz-1 KO brains. DAPI staining showed that the mutant striatal complex consisted of a smaller dorsal striatum but a larger ventral striatum when compared to wild-type (WT) brains (Fig. 1 A and A′). Because of the structural malformation, it is difficult to use known landmarks to demarcate the dorsal and ventral striata in Nolz-1 KO brains. Here, we define the boundary between the dorsal and the ventral striata by drawing a line between the septoeminential sulcus and the piriform cortex (Fig. 1 B and B'). As Foxp1 was expressed in both dorsal and ventral striata and Foxp1 expression appeared unaltered in the KO striatum, we defined Foxp1+ areas above and below the boundary, respectively, as the dorsal and ventral striata (Fig. 1 B and B'). The Foxp1+ dorsal striatum was decreased in Nolz-1 KO brains. By contrast, the Foxp1+ ventral striatum was markedly increased in KO brains from rostral (R) to caudal (C) levels (Fig. 1 B, B', and F). Note that the total areas of the striatum were not changed in E18.5 Nolz-1 KO brains (Fig. 1G).
Fig. 1.
Abnormal structural alteration in the developing striatum of Nolz-1 KO brains. (A and A') DAPI staining shows the phenotype of the shrunk dorsal striatum and expanded OT of the ventral striatum in E18.5 Nolz-1 KO brains. (B and B') Foxp1+ dorsal and ventral striata, respectively, are decreased and increased in E18.5 Nolz-1 KO brains. (C and C') Larger Er81+ cell clusters are present in the OT of the E18.5 Nolz-1 KO brain compared to WT. Scattered Er81+ cells are nearly absent in the dorsal KO striatum. The boxed regions in D and D′ are shown at high magnification. (D and D') At E12.5, the Meis2 expression pattern in the the LGE is similar between WT and Nolz-1 KO brains. (E and E') By E13.5, the Meis2+ region is increased in the ventral part of the KO LGE. (F) Quantification shows decreases and increases, respectively, in Foxp1+ dorsal and ventral striata of E18.5 Nolz-1 KO brains. (G) The total area of the striatum is not changed in the Nolz-1 KO striatum from R to C levels. (H) Er81+ cells are decreased and increased, respectively, in the dorsal and ventral KO striata. (I) Meis2+ cells are increased in the ventral part of the KO LGE. *P < 0.05; **P < 0.01; n = 3/group. (Scale bars, 200 μm [A–C′], 100 μm [C and C′] for high magnification, 100 μm [D and E′].) Photomicrographs in A–C′ are stitched images.
To investigate the cellular changes in the Nolz-1 KO striatum, we examined Er81 expression (2). In the WT E18.5 striatum, Er81 was expressed in prominent cell clusters in the OT of the ventral striatum, whereas scattered Er81+ cells were present in the dorsal striatum (Fig. 1C). Er81+ cells were markedly increased in the ventral KO striatum (Fig. 1 C' and H). In contrast, Er81+ cells were decreased in the dorsal KO striatum (Fig. 1 C' and H). The formation of an abnormal striatum was detected as early as E13.5 by Meis2 immunostaining. At E12.5, the Meis2 expression pattern in the LGE was similar between WT and Nolz-1 KO brains (Fig. 1 D and D'). By E13.5, the Meis2+ cell population was significantly expanded in the ventral part of the KO LGE (Fig. 1 E, E', and I). Such ventral expansion of the striatal anlage was also evident in Isl1 immunostaining of the E13.5 KO LGE (see Fig. 4F below).
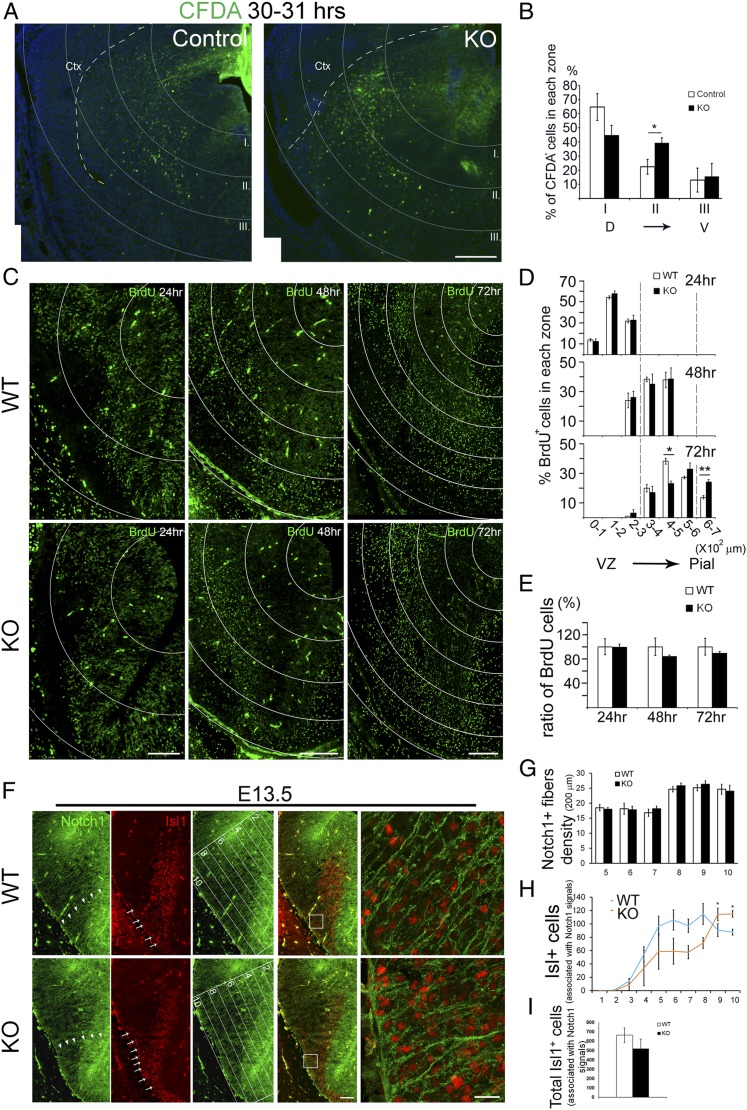
Fig. 4.
In vivo tracing of aberrantly migrating cells in the developing striatum of Nolz-1 KO brains. (A and B) CFDA is locally injected into the dorsal part of the E14.5 striatum, and the brains are harvested 31 h later. The striatum is overlaid with dorsal ventricular zone centered-concentric rings 200 μm apart for cell counting in each zone (zones I–III). Comparing to control brains, more CFDA+ cells migrate ventrolaterally into zone II (400–600 μm) in Nolz-1 KO) brains. (C) The migratory pattern of BrdUE12.5 cells in the striatum at 24, 48, and 72 h after BrdU injection. The distribution of BrdUE12.5 cells in the striatum are traced in concentric rings 100 μm apart. (D) No significant difference of BrdUE12.5 cells was found at 24 and 48 h between WT and Nolz-1 KO brains. By 72 h, BrdUE12.5 cells are decreased in the 400–500-μm region but increased in the 600–700-μm region in KO brains compared to WT brains. (E) The total number of BrdUE12.5 cells at each time point was not different between the WT and the KO brains. (F) Double immunostaining of Notch1 and Isl1 shows that Notch1+ radial glial fibers extend across the LGE as far as into the pial surface of the ventrolateral subpallium (arrowheads) in the WT E13.5 brain. Many Isl1+ cells are associated with radial glial fibers along the trajectory routes into the ventrolateral subpallium where the OT anlage is located (arrows, Top). In the Nolz-1 KO brain, Isl1+ cells are increased in the OT-like region (arrows, Bottom). Confocal images of the boxed regions are shown at high magnification on the far Right. The LGE is divided into 10 equidistant bins from the dorsomedial LGE to the ventrolateral OT anlage as shown in the third panels. (G) The density of Notch1+ radial fibers is not changed in bins no. 5–no. 10 of KO brains. (H) Isl1+ cells associated with Notch1+ radial fibers are increased in bins no. 9 and no. 10 where the mutant OT-like anlage is located compared to that in WT. (I) The total number of Isl1+ cells associated Notch1+ radial fibers is not changed in KO brains. *P < 0.05; **P < 0.01; n = 3/group. (Scale bars, 200 μm [A], 50 μm [C] for 24 h, 100 μm [C] for 48 h, 72 h, 100 μm [F] for low magnification, and 20 μm [F] for high magnification.)
Nolz-1 Promotes Differentiation and Axonal Outgrowth/Navigation of Striatonigral Neurons.
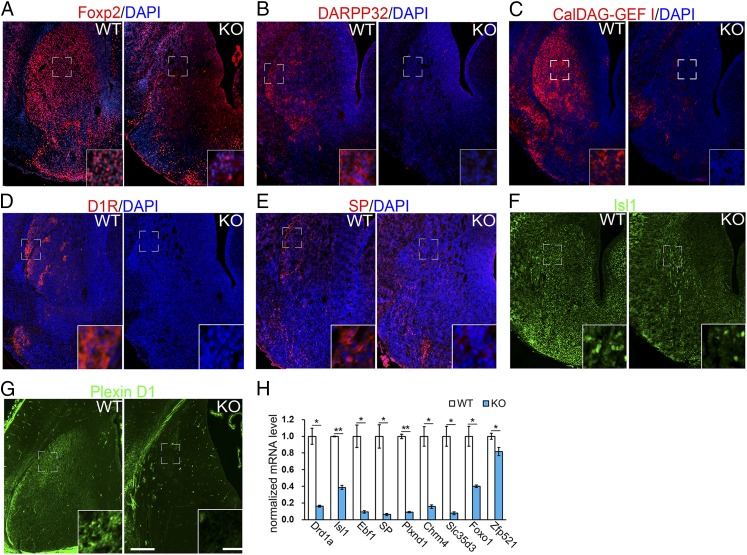
We further examined whether differentiation of striatal neurons was affected in Nolz-1 KO brains by immunostaining with striatal differentiation markers, including Foxp2, DARPP-32, and CalDAG-GEFI (21–23). Foxp2 was significantly reduced in the Nolz-1 KO striatum at E18.5 (Fig. 2A). DARPP-32 and CalDAG-GEFI markers, respectively, of the developing striosomal and matrix compartments, were also markedly decreased in the KO striatum (Fig. 2 B and C).
Fig. 2.
Striatonigral-enriched genes are decreased in the Nolz-1 KO striatum. (A) Foxp2 is decreased in the Nolz-1 KO striatum compared to the WT striatum of E18.5 brains. (B and C) DARPP-32-positive striosomes (B) and CalDAG-GEF I-positive matrix (C) are decreased in the Nolz-1 KO striatum. (D–F) D1R, (D), SP, (E), Isl1 (F), and Plxn D1 (G), markers of striatonigral neurons are decreased in the Nolz-1 KO striatum. The bracketed regions are shown at high magnification in the Insets. (H) The qRT-PCR assay shows significant reductions of striatonigral-enriched genes in the E18.5 Nolz-1 KO striatum. *P < 0.05; **P < 0.01; n = 3/group. (Scale bars, 200 μm [A–G], and 30 μm [A–G] for the Insets.)
Striatal projection neurons consist of striatonigral and striatopallidal neurons (15). Immunostaining showed significant reductions in the dopamine D1 receptor (D1R), substance P (SP), Plexin D1 (PlxnD1), and Isl1 markers of striatonigral neurons (24–27) in the E18.5 Nolz-1 KO striatum (Fig. 2 D–G). Consistently, the qRT-PCR analysis indicated that a repertoire of mRNAs encoding striatonigral-enriched genes, including D1R/Drd1a, Isl1, Ebf1, SP, PlxnD1, Chrm4, Slc35d3, FoxO1, and Zpf521 were markedly decreased in the Nolz-1 KO striatum (Fig. 2H) (24–28).
Anterograde tracing further showed a reduction of DiI-labeled striatonigral axons along the projection pathway from the diencephalon and telencephalic boundaries through the cerebral peduncles to the substantia nigra in the midbrain of Nolz-1 KO brains (SI Appendix, Fig. S3A). We, then, conditionally deleted Nolz-1 in striatonigral cell lineages by intercrossing Nolz-1fl/fl;CAG-CAT-EGFP mice with Isl1-Cre mice (26, 27). The defective striatonigral pathway was evident in Isl1-Cre;Nolz-1fl/fl;CAG-CAT-EGFP conditional KO brains in which marked reduction of striatonigral GFP+ axons was found along the routes of striatonigral axonal projections (SI Appendix, Fig. S3B).
Nolz-1 Suppresses Striatopallidal-Enriched Genes.
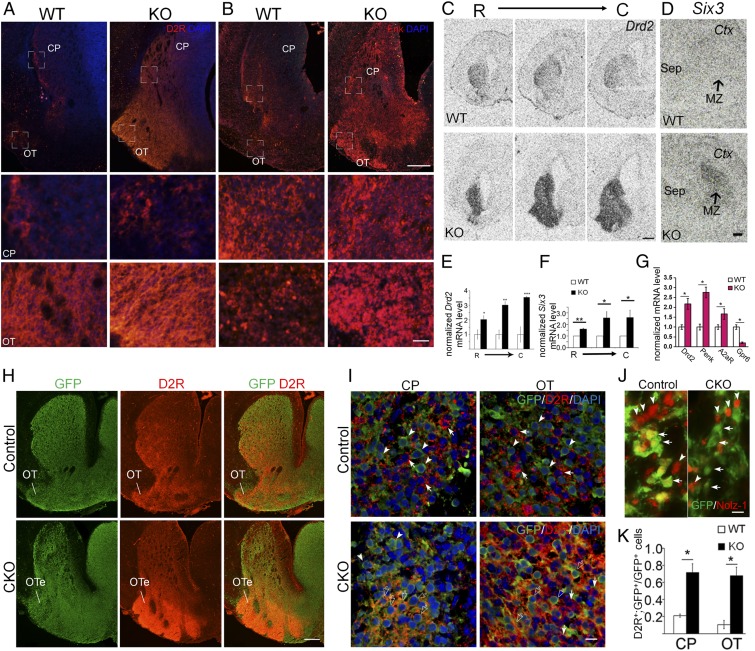
In dramatic contrast to the reduction of normal markers of direct pathway striatonigral-enriched genes, including D1R and SP (Fig. 2), immunostaining showed significant increases in D2R and Met-Enkephalin (Enk), markers of striatopallidal-enriched genes, in the E18.5 Nolz-1 KO striatum (Fig. 3 A and B). In situ hybridization demonstrated robust increases in Drd2 mRNA in the Nolz-1 KO striatum throughout rostrocaudal levels (Fig. 3 C and E). Six3, another gene involved in neurogenesis of D2R-expressing striatal neurons (29, 30), was also markedly increased in the KO striatum (Fig. 3 D and F). The qRT-PCR assays further confirmed significant increases in mRNAs encoding striatopallidal-enriched genes (24), including Drd2 (D2R), preproenkephalin (Penk), and adenosine receptor A2a (A2aR) in the E18.5 Nolz-1 KO striatum, although Gpr6 was decreased in the KO striatum (Fig. 3G).
Fig. 3.
Striatopallidal-enriched genes are derepressed in striatonigral cells of the Nolz-1 KO striatum. (A and B) Dopamine D2 receptor (D2R) and enkephalin (Enk), markers of striatopallidal neurons, are expressed in both the CP and the OT of WT E18.5 brains. D2R and Enk are markedly increased in both CP and OT of Nolz-1 KO brains. The bracketed regions in the CP and OT are shown at high magnification in the second and third rows of the panels. (C–F) In situ hybridization shows robust derepression of Drd2 mRNA (C and E) and Six3 mRNA (D and F) in both dorsal and ventral striata of the Nolz-1 KO brain from R to C levels. (G) qRT-PCR assay confirms up-regulation of striatopallidal-enriched genes in the E18.5 Nolz-1 KO striatum except that Gpr6 is reduced. (H) Deletion of Nolz-1 in Isl1 cell lineages results in hyperplasia of the ventral striatum and hypoplasia of the dorsal striatum in Isl1-Cre;Nolz-1fl/fl;CAG-CAT-EGFP conditional KO (CKO) brains compared to control E18.5 Isl1-Cre;Nolz-1fl/+;CAG-CAT-EGFP brains. (I and J) D2R (arrows) and GFP (arrowheads) are not colocalized in many striatal cells of the CP and OT in control brains. Colocalization of GFP and D2R (empty arrows) is increased in striatal cells of the CP and OT-like regions in CKO brains. (K) Validation of the successful deletion of Nolz-1 in GFP-expressing Isl1 cell lineages in the striatum by the absence of Nolz-1 immunoreactivity (red, arrowheads) in GFP-positive cells (green, arrows) of CKO brains. OTe: enlarged OT. *P < 0.05; **P < 0.01; ***P < 0.001; n = 3/group. (Scale bars, 200 μm [A and B], 50 μm [A and B)] for the Bottom, 500 μm [C], 200 μm [D and H], and 10 μm [I and J].) The low-magnification images in A and B are stitched images.
Conditional Deletion of Nolz-1 in Striatonigral Cell Lineages Up-Regulates Striatopallidal-Enriched Genes.
We further asked whether Nolz-1 null mutation caused derepression of striatopallidal-enriched genes in striatonigral cells by specific deletion of Nolz-1 in striatonigral cell lineages using Isl1-Cre mice (26, 27). Double immunostaining confirmed the successful deletion of the Nolz-1 protein in GFP-expressing Isl1 cell lineages in Isl1-Cre;Nolz-1fl/fl;CAG-CAT-EGFP E18.5 mutant brains (Fig. 3J). D2R and GFP were not colocalized in many striatal cells of the CP and OT in control Isl1-Cre;Nolz-1fl/+;CAG-CAT-EGFP brains (Fig. 3 H and I). By contrast, colocalization of GFP and D2R was significantly increased in striatal cells of CP- and OT-like regions in Isl1-Cre;Nolz-1fl/fl;CAG-CAT-EGFP mutant brains (Fig. 3 I and K). These findings suggest that selective deletion of Nolz-1 in Isl1+ striatonigral cell lineages derepresses striatopallidal genes in striatonigral cells.
Transgenic Overexpression of Nolz-1 Increases and Decreases, Respectively, Striatonigral- and Striatopallidal-Enriched Genes.
In addition to the loss-of-function study, we have also investigated the effects of Nolz-1 overexpression. We intercrossed Dlx5/6-Cre mice with the Cre-dependent Nolz-1 transgenic mice (Nolz-1Tg/+) to conditionally overexpress Nolz-1 in postmitotic striatal neurons. The Cre-dependent Nolz-1 transgenic construct contains a floxed STOP cassette. Upon the Cre-mediated DNA recombination, the full-length Nolz-1 cDNA with a bicistronic ires-driven EGFP reporter gene is transcribed (31). Transgenic overexpression of Nolz-1 resulted in increases in striatonigral gene expression, including Isl1, D1R, SP, and Foxp2 in striatal neurons of Dlx5/6-Cre;Nolz-1Tg/+ newborn mice (SI Appendix, Fig. S4 A–J′), which is complementary to the phenotypes of Nolz-1 KO brains (Fig. 2 A and D–H).
Conversely, transgenic overexpression of Nolz-1 resulted in decreases in striatopallidal gene expression, including D2R and enkephalin in striatal neurons of Dlx5/6-Cre;Nolz-1Tg/+ newborn brains (SI Appendix, Fig. S4 K–N′), which is opposite to the phenotypes of Nolz-1 KO brains (Fig. 3 A and B).
Taken together, the results of Nolz-1 loss-of-function and overexpression studies, Nolz-1 increases and decreases, respectively, striatonigral and striatopallidal genes in the developing striatum.
Conditional Deletion of Nolz-1 in Striatonigral Cell Lineages Is Sufficient to Induce Hypoplasia of the Dorsal Striatum and Hyperplasia of the Ventral Striatum.
The striatal complex in Nolz-1 null mutant brains consisted of a smaller dorsal striatum but a larger ventral striatum when compared to WT brains (Fig. 1 A and A′). Similar structural changes were observed in Isl1-Cre;Nolz-1fl/fl;CAG-CAT-EGFP conditional KO brains (Fig. 3H). Intriguingly, there were streams of GFP+ cells that appeared to extend from the dorsal toward the enlarged ventral mutant striatum (Fig. 3H). These findings indicate that selective deletion of Nolz-1 in differentiating striatonigral cells is sufficient to induce hypoplasia and hyperplasia of the dorsal and ventral striata, respectively.
Cell Proliferation Is Not Altered in the Germinal Zone of the Nolz-1 Mutant Striatal Anlage.
We, then, investigated the mechanisms underlying Nolz-1 mutation-induced alteration of the striatal complex. Structural changes in the Nolz-1 KO striatal complex might result from altered progenitor populations in the striatal anlage. The expression pattern of Ascl1/Mash1 mRNA, a proneural gene (32), was not altered in E13.5 and E16.5 Nolz-1 KO striata (SI Appendix, Fig. S5). We further examined the proliferation of progenitor cells in the LGE by pulse labeling of proliferating cells with bromodeoxyuridine (BrdU) for 1 h. BrdU pulse labeling marked proliferating cells in S phase. Ki67 labeled proliferating cells in the cell cycle progression. The proliferation index (BrdU+;Ki67+ cells/total Ki67+ cells) was not changed in the E12.5 LGE and E15.5 striata of Nolz-1 KO brains (SI Appendix, Fig. S6A). We also assayed the cell cycle exit index 24 h after BrdU injection. The cell cycle exit index (BrdU+;Ki67− cells/total BrdU+ cells) was not changed in the E12.5 LGE and E15.5 striata of Nolz-1 KO brains (SI Appendix, Fig. S6A). Neither did immunostaining of phosphohistone 3 (PH3), a mitotic marker, exhibit changes in the number of PH3+ cells in the ventricular zone (VZ) or subventricular zone (SVZ) of the E12.5 LGE or the E15.5 striatum of Nolz-1 KO brains (SI Appendix, Fig. S6B).
Previous studies have suggested that OT neurons arise at early embryonic stages from the ventral part of the LGE, the septoeminential sulcus, and the rostromedial telencephalic wall (33, 34) and that the ventral parts of the VZ in the LGE and septum give rise to NAc neurons (35). We found no changes in the proliferation index (BrdU+;Ki67+ cells/total Ki67+ cells) within any of these progenitor regions in E12.5 and E13.5 Nolz-1 KO brains (SI Appendix, Fig. S6C).
Cell Apoptosis Is Not Changed in the Nolz-1 Mutant Striatum.
We, then, investigated whether abnormal cell death occurred in the Nolz-1−/− KO striatum by terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assays. No significant changes in TUNEL+ apoptotic cells were found in E12.5, E14.5, and E16.5 KO striata (SI Appendix, Fig. S7 A and B), suggesting that Nolz-1 is not required for striatal cell survival.
Abnormal Redistribution of Striatal Cells from the Dorsal to the Ventral Mutant Striatum.
As cell proliferation and apoptosis were not changed in the Nolz-1 KO striatum, we next tested whether the concurrent hypoplasia of the dorsal striatum and hyperplasia of the ventral striatum resulted from redistribution of striatal cells from the dorsal to the ventral KO striatum. We pulse labeled early-born and late-born striatal cells with BrdU at, respectively, E12.5 and E15.5, and then analyzed the distribution of BrdU-labeled cells at E18.5. Quantitative analysis indicated that the number of BrdUE12.5 cells was decreased by 60% in dorsal KO striatum but was increased by 80% in the ventral KO striatum (SI Appendix, Fig. S8 A and C). Similar results were found with late-born BrdUE15.5 cells in the KO striatum (SI Appendix, Fig. S8 B and D). The total numbers of BrdUE12.5 cells or BrdUE15.5 cells were not markedly changed in the entire KO striatum (SI Appendix, Fig. S8 C and D). These results suggest an abnormal redistribution of striatal cells from dorsal to ventral in the KO striatum.
Aberrant Migration of Striatal Cells from Dorsal to Ventral Mutant Striatum.
The redistribution of BrdU cells implicated abnormal cell migration with an increased number of cells migrating from the dorsal to ventral KO striatum. We examined this hypothesis in three sets of experiments. First, we focally labeled cells in the dorsal part of the germinal zone of the E14.5 LGE with the 5-(and 6)-carboxyfluorescein diacetate (CFDA) cell tracker and then traced the migration of CFDA-labeled cells 31 h later in vivo. Owing to technical limitations, E14.5 was the earliest time point for which we succeeded in this set of experiments. A successful CFDA injection site was defined as an ∼400-μm-radius circle centering at the intensive CFDA-labeled VZ of the dorsal LGE (Fig. 4A). CFDA+ cells were found to migrate from the dorsomedial toward the ventrolateral parts of the WT and Nolz-1 KO LGE. For quantification, we divided the LGE into three equidistant concentric zones (200 μm apart) with the epicenter at the dorsomedial germinal zone where the CFDA tracer was deposited. Zone I was 0–200 μm away from the germinal zone. Zone II was 200–400 μm away from the germinal zone. Zone III was 400–600 μm away from the germinal zone. We counted the percentage of CFDA+ cells within each zone by dividing CFDA+ cells within each zone by the total number of CFDA+ cells (Fig. 4 A and B). The percentage of CFDA+ cells tended to decrease within zone I of Nolz-1 KO LGE. By contrast, the percentage of CFDA+ cells within zone II significantly increased by 76% in Nolz-1 KO relative to control WT LGE (Fig. 4 A and B). These findings indicate that, in the KO LGE, the distribution of CFDA+ cells as a whole shifted ventrolaterally relative to that in WT LGE, suggesting that Nolz-1 null mutation promoted cell migration from the dorsal toward the ventral striatum.
Second, we performed a time-course study to trace the migratory routes of cells by pulse labeling striatal cells with BrdU at E12.5 and then chased the cells at 24, 48, and 72 h following BrdU injection. We plotted the distribution of BrdUE12.5 cells and calculated the percentage of BrdUE12.5 cells in each 100-μm equidistant concentric ring. The results showed no changes in BrdUE12.5 cells at 24 and 48 h in Nolz-1 KO brains. By 72 h, 38% of cells migrated into the 400–500-μm region, approximately the mantle zone (MZ) of the dorsal striatum in WT brains. Comparing to WT brains, the number of BrdUE12.5 cells was decreased by 40% in the 400–500-μm region (Fig. 4 C and D), but the number of BrdUE12.5 cells was increased by 75% in the 600–700-μm region where the presumptive mutant OT was located (Fig. 4 C and D). As the total numbers of BrdUE12.5 cells were not changed in KO brains at 24, 48, and 72 h compared to WT brains (Fig. 4E), these results suggest that, by 72 h, some BrdUE12.5 mutant cells do not stop migration, but instead, continue migrating ventrally toward OT-like regions.
Third, by immunostaining of Notch1, a marker of radial glia (36), we found that many Notch1+ radial glial fibers extended from the VZ of the LGE through the MZ into the ventrolateral subpallium where the presumptive OT anlage was located in WT E13.5 brains (Fig. 4F). A similar cytoarchitecture of Notch1+ radial glia was found in the Nolz-1 KO subpallium, and the density of Notch1+ radial fibers was not changed in the ventral LGE of KOs (Fig. 4 F and G). Because Isl1+ cell lineages contributed to the major part of the ventral striatum (Fig. 3H), we then asked whether Isl1 cell lineages might migrate along radial glial fibers into the ventral striatum. Consistent with this hypothesis, in the E13.5 WT brain, many Isl1+ cells were closely associated with Notch1+ fibers along the trajectory routes of radial glia (Fig. 4 F and H), suggesting that Isl1+ cells generated in the LGE proper may migrate along with radial glial processes into the OT anlage. For quantification, we divided the subpallium into 10 equidistant bins with the top bin no. 1 covering the VZ of the dorsomedial LGE, and the bottom bin no. 10 covering the ventrolateral subpallium that contained the OT anlage and then counted the Isl1+ cells associated with Notch1+ radial glial processes in each bin. When compared to E13.5 WT brains, in mutants, the number of Isl1+ cells associated with radial fibers tended to be lower from bins no. 1 to no. 8 but was significantly increased by, respectively, 26% and 31% in bins no. 9 and no. 10 where the mutant OT anlage was located (Fig. 4H). However, there were no significant changes in the total number of Isl1+ cells associated with Notch1+ fibers that were found in the KO LGE (Fig. 4I). These results indicate a redistribution of Isl1+ cells from the dorsal LGE toward the ventral LGE in the Nolz-1 KO brains.
We further attempted to clarify the identity of Isl1+ cells in the mutant OT by double immunostaining for Isl1 and calbindin, a marker of OT neurons at early stages (33). The number of calbindin+ cells was not changed in E13.5 Nolz-1 KO brains (SI Appendix, Fig. S9 A and B). Nor was the percentage of Isl1+;calbindin+ cells/calbindin+ cells altered in KO brains (SI Appendix, Fig. S9 A and C). The fact that Isl1+ cells were increased in mutant OT-like regions (Fig. 4H), but Isl1+;calbindin+ cells were not changed in mutant OT-like regions, suggested that many Isl1+ cells that immigrated into the mutant OT-like regions did not differentiate into canonical calbindin+ OT neurons.
Taken together, the results of these three sets of experiments, including the increases in CFDA+ migrating cells, pulse-labeled BrdUE12.5 cells, and Isl1+ cells associated with radial glia in the ventral KO striatum, consistently support the hypothesis that Nolz-1 null mutation induces aberrant cell migration from the dorsal toward the ventral striatum.
Nolz-1 Suppresses Dlx Genes in the Developing Striatum.
The formation of the abnormal ventral striatum was detected as early as E13.5 as evidenced by the expansion of the Meis2+ domain (37) in the ventral striatal anlage of Nolz-1 KO brains (Fig. 1 E and E'). To identify molecular mechanisms underlying the abnormal development of the mutant striatal complex, we screened a repertoire of striatum-enriched genes in Nolz-1 KO brains. We found that Dlx homeobox genes were dramatically derepressed in the KO striatum. In situ hybridization showed that, in contrast to restricted expression of Dlx1 and Dlx2 to VZ and SVZ of the WT striatum (38, 39), Dlx1 and Dlx2 mRNAs were significantly up-regulated in differentiated MZ but not in VZ/SVZ germinal zones of the E16.5 Nolz-1 KO striatum (Fig. 5 A and B). Dlx5 was increased in the MZ of the R KO striatum (Fig. 5C). Consistently, we found that, in the WT E13.5 striatum, the majority of Dlx1 and Dlx2 mRNAs were not colocalized with the Nolz-1 protein (Fig. 5 D and E). Together, these findings implicate that Nolz-1 suppresses Dlx1, Dlx2, and Dlx5 expression in the developing striatum.
Fig. 5.
Nolz-1 directly transcriptional represses Dlx1 and Dlx2 in the developing striatum. (A–C) In situ hybridization shows that Dlx1 (A), Dlx2 (B), and Dlx5 (C) mRNAs are markedly increased in the MZ (arrows) of the E16.5 Nolz-1 KO striatum compared to the WT striatum. (D) Double Dlx1 in situ hybridization and Nolz-1 immunostaining show Dlx1 mRNA (red puncta) and Nolz-1 protein (green nuclei, arrowheads) are not colocalized in the E13.5 WT striatum. (E) Nor are Dlx2 mRNA (red puncta) colocalized with the Nolz-1 protein (green nuclei, arrowheads) in the E13.5 WT striatum. (F) Overexpression of Nolz-1 by in utero electroporation decreases Dlx1 mRNA (arrowheads) and Dlx2 mRNA (arrows) in the VZ and SVZ of the E15.5 LGE. (G) The ChIP-seq binding motif analysis derives a putative Nolz-1 binding motif as AATTA. (H) Three AATTA motifs are located in the I12b region. (I) The ChIP-qPCR assay detects a strong immunoprecipitated signal in the I12b region with a Nolz-1 antibody at the E15.5 striatum compared to the controls of the Nolz-1 antibody preabsorbed with Nolz-1 recombinant protein (RP) and IgG (Nolz-1 Ab vs. RP: 104 ± 11 vs. 8 ± 3; Nolz-1 Ab vs. IgG: 104 ± 10 vs. 1). (J) The reporter gene assay shows that transfection of the pcBIG-Nolz-1-ires-EGFP plasmid into E15.5 cultured striatal cells decreases pGL3-I12b-c-fos-Luc reporter gene activity. Mutations of the three motifs of AATTA to GGCCG (MT3) but not the motif of AGGAT to ATTTT (MT1) abolish the repression of reporter gene activity by Nolz-1. Ctx, cerebral cortex; LGE; MGE, medial ganglionic eminence; Sep, septum. MZ, SVZ, VZ., *P < 0.05; **P < 0.01; ***P < 0.001; n = 3/group. (Scale bars, 400 μm [A–C], 200 μm [D and E] for low magnification, 10 μm [D and E] for high magnification, 5 μm [F].)
Overexpression of Nolz-1 Suppresses Dlx1/2 Expression in the Germinal Zone of the Developing Striatum.
As Dlx1 and Dlx2 mRNAs were up-regulated in the MZ of the Nolz-1 KO striatum (Fig. 5 A and B), we tested whether overexpression of Nolz-1 could inhibit Dlx1 and Dlx2 expressions in the developing striatum of WT brains. Indeed, overexpression of Nolz-1 by in utero electroporation resulted in 61% and 41% decreases in Dlx1 and Dlx2 mRNAs, respectively, in the VZ and SVZ of E15.5 WT LGE (Fig. 5F). This finding supports Nolz-1 as a suppressor of Dlx gene expression.
Nolz-1 Represses Dlx1/2 Genes through Inhibition of the I12b Intergenic Enhancer Activity.
We next asked whether Nolz-1 directly transcriptionally repressed Dlx1/2 genes. It has been shown that transgenic reporter mice carrying the I12b intergenic enhancer of Dlx1/2 exhibit the reporter gene pattern similar to the endogenous Dlx1/2 pattern in telencephalon (40), which suggests that endogenous transcriptional regulators may act through the I12b intergenic enhancer to regulate Dlx1/2 expression. Chromatin immunoprecipitation (ChIP)-qPCR detected a robust immunoprecipitated signal in the I12b region with a Nolz-1 antibody from the E15.5 striatum, and the specificity of the ChIP signals was demonstrated by the reduction of the ChIP signal in the group of antigen preabsorbed Nolz-1 antibodies (Fig. 5I). These results suggest that Nolz-1 is capable of binding to the I12b intergenic enhancer in vivo. ChIP-seq binding motif analysis derived a putative Nolz-1 binding motif as “AATTA” in Dlx1 and Dlx2 genes (Fig. 5G). We found three AATTA motifs in the I12b region (Fig. 3H). We then tested whether Nolz-1 could interact with Dlx1/2 through the I12b enhancer using a reporter gene assay. Transfection of the pcBIG-Nolz-1-ires-EGFP plasmid into the E15.5 striatal cell culture decreased 45% of pGL3-I12b-c-fos-Luc reporter gene activity compared to that of the mock control pcBIG-ires-EGFP (Fig. 5J). Moreover, mutations of the three motifs of AATTA to “GGCCG” (MT3) in the I12b region significantly reduced the repression of reporter gene activity by Nolz-1 (Fig. 5J). We noted that a conserved zebrafish Nlz binding site “AGGAT” (41) was present in the antisense strand of the I12b region, but mutation of the AGGAT motif to “ATTTT” (MT1) in our experiments did not affect reporter gene activity (Fig. 5J). Collectively, our findings suggest that Nolz-1 directly suppresses Dlx1/2 through inhibition of the I12b enhancer activity.
Knocking down Dlx1/2 Gene Expression Rescues the Abnormal Pattern of Cell Migration in the Nolz-1 KO Striatum.
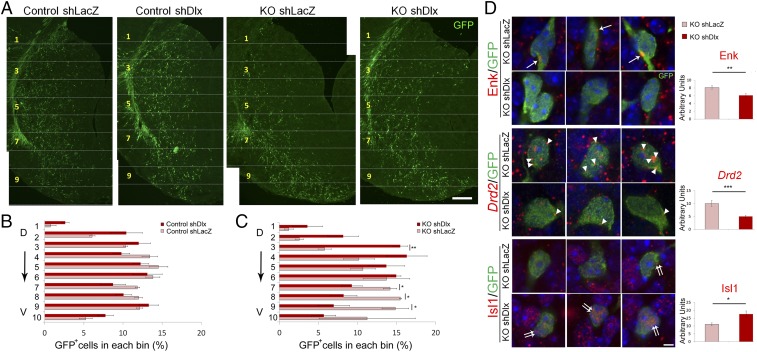
Dlx1 and Dlx2 genes are not only essential for tangential migration of cortical GABAergic interneurons (42, 43), but also important for striatal cell migration (44). Because Dlx1 and Dlx2 were markedly up-regulated in the differentiated MZ of the Nolz-1 KO striatum, we postulated that abnormal increases in Dlx1 and Dlx2 expressions might promote aberrant cell migration. To test this hypothesis, we knocked down Dlx1 and Dlx2 by electroporating short hairpin RNA interference (shRNAi) constructs into the Nolz-1 KO E13.5 striatum to see whether this could restore normal cell migration. We tested by qRT-PCR the knockdown effects and specificity of Dlx1 and Dlx2 shRNAis and confirmed them (SI Appendix, Fig. S10). GFP reporter gene plasmids were coelectroporated into the E13.5 LGE of Nolz-1 KO brains, and brains were harvested at E18.5. The striatum was again divided into 10 equidistant bins along the dorsoventral axis. Electroporation of Dlx1/2 shRNAi plasmids markedly decreased the percentage of GFP+ cells in the ventral bins of the KO striatum (bin no. 7 by 34%, bin no. 8 by 47%, and bin no. 9 by 53%) compared to results for control plasmids (Fig. 6 A and B). By contrast, in the dorsal KO striatum, knockdown of Dlx1/2 increased GFP+ cells by 167% in bin no. 3 (Fig. 6 A and C). These results suggest a Dlx1/2-dependent mechanism underlying aberrant cell migration in the Nolz-1 KO striatum.
Fig. 6.
Knocking down Dlx1/2 genes alleviates abnormal patterns of cell migration and differentiation in the Nolz-1 KO striatum. (A) shDlx1/2 plasmids are coelectroporated with GFP reporter plasmids into the E13.5 LGE by in utero electroporation. The migratory pattern of shDlx1/2-GFP+ cells is analyzed at E18.5. In control brains, the distribution pattern of shDlx1/2-GFP+ cells is similar to that of mock control shLacZ-GFP+ cells. In Nolz-1 KO brains, the number of shDlx1/2-GFP+ cells is decreased in the ventral striatum compared to that of shLacZ-GFP+ cells. (B and C) The striatum is divided into 10 bins along the dorsoventral axis. The percentage of shDlx1/2-GFP+ cells in each bin does not differ from that of shLacZ-GFP+ cells in control brains (B). In Nolz-1 KO brains (C), the percentage of shDlx1/2-GFP+ cells is decreased in the ventral striatum (bins nos. 7–9) but is concurrently increased in the dorsal striatum (bin no. 3) compared to that of shLacZ-GFP+ cells. (D) Electroporation of shDlx1/2 plasmids into the E13.5 Nolz-1 KO LGE decreases Enk (arrows) and Drd2 mRNAs (arrowheads) but increases Isl1 mRNA expression (double arrows) in the E18.5 KO striatum compared to the mock control of shLacZ. D, dorsal; V, ventral. *P < 0.05; ***P < 0.001; n = 3/group. (Scale bars, 100 μm [A] and 5 μm [D].)
Overexpression of Dlx2 in the WT Striatum Phenocopies Aberrant Cell Migration in the Nolz-1 KO Striatum.
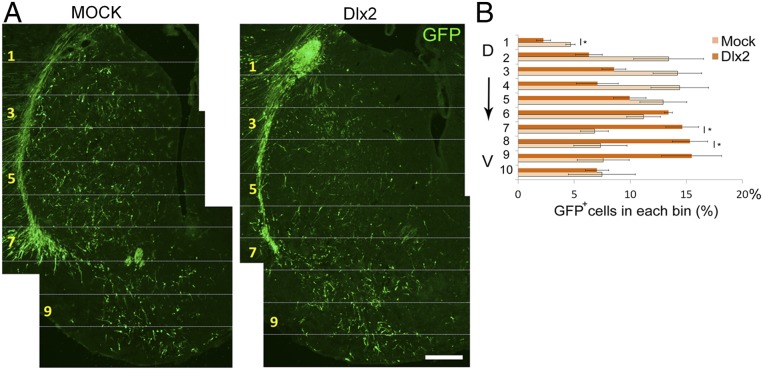
If deranged Dlx1/2 signaling plays a key role in directing aberrant mutant cell migration, then overexpression of Dlx1/2 in the WT striatum might mimic the mutant migratory phenotype. We, therefore, coelectroporated a Dlx2 cDNA construct with a GFP reporter gene into the E13.5 WT LGE. Unlike the generally even distribution of GFP+ cells along the dorsoventral axis of the E18.5 striatum in mock controls, the distribution of GFP+ cells was modified from the dorsal to the ventral striatum with Dlx2 overexpression. The percentage of GFP+ cells decreased by 52% in bin no. 1 of the dorsal striatum (Fig. 7 A and B) but increased, respectively, by 115% in bin no. 7 and 109% in bin no. 8 of the ventral striatum (Fig. 7 A and B). This dorsal-to-ventral shifted pattern of cell migration was reminiscent of the aberrant pattern of cell migration found in the Nolz-1 KO striatum (Figs. 2 A and B and 6A). These results indicate that overexpression of Dlx2 is sufficient to bias cell migration from the dorsal striatum toward the ventral striatum and further suggest that deranged Dlx1/2 signaling is likely to play a causal role in priming aberrant cell migration in the Nolz-1 KO striatum.
Fig. 7.
Overexpression of the Dlx2 gene in the WT striatum phenocopies aberrant cell migration in the Nolz-1 KO striatum. (A) Dlx2 expression plasmids are coelectroporated with the GFP reporter gene into the E13.5 LGE by in utero electroporation. The migratory pattern of GFP+ cells is analyzed at E18.5. In mock controls, GFP+ cells are distributed in both the dorsal and the ventral striatum. In Dlx2 overexpressed brains, more GFP+ cells migrate toward the ventral striatum compared to mock control brains. (B) The striatum is divided into 10 bins along the dorsoventral axis. Quantification shows decreased GFP+ cells in the dorsal striatum (bin no. 1) but increased GFP+ cells in the ventral striatum (bins no. 7 and no. 8) in Dlx2 overexpressed brains. D, dorsal; V, ventral. *P < 0.05 and n =3/group. (Scale bar, 100 μm.)
Knocking down Dlx1/2 Gene Expression Alleviates Abnormal Differentiation of Striatal Neurons in the Nolz-1 Null KO Striatum.
Given that knocking down Dlx1/2 alleviated abnormal mutant cell migration, we asked whether it could also reverse abnormal increases and decreases, respectively, in striatopallidal and striatonigral gene expressions in the Nolz-1 null KO striatum (Fig. 6D). Indeed, electroporation of Dlx1/2 shRNAi plasmids with a GFP reporter gene into the E13.5 KO LGE resulted in a 50% decrease in Drd2 signals in Drd2+;GFP+ cells and a 25% decrease in Enk signals in Enk+;GFP+ cells in the E18.5 Nolz-1 null KO striatum relative to control plasmids (Fig. 6D). By contrast, Isl1 signals in Isl1+;GFP+ cells were increased by 59% in the null KO striatum (Fig. 6D). These results indicate that knocking down Dlx1/2 in mutant cells can partially reverse the aberrant differentiation of striatal neurons in the Nolz-1 KO striatum.
Overexpression of Dlx2 in the WT Striatum Recapitulates Aberrant Differentiation of Nolz-1 KO Striatal Neurons.
The above rescue experiment by Dlx1/2 knockdown suggests that Nolz-1 deletion-induced abnormal up-regulation of Dlx1/2 leads to decreases and increases, respectively, in striatonigral and striatopallidal gene expressions. We then asked whether misexpression of Dlx1/2 in the WT striatum could recapitulate the aberrant differentiation of the Nolz-1 knockout phenotypes. We performed in utero electroporation to overexpress Dlx2 in developing striatal neurons at E13.5. We found that Dlx2 overexpression decreased striatonigral genes of Isl1, D1R, SP, and Foxp2 (SI Appendix, Fig. S11A) but increased striatopallidal genes of D2R and enkephalin (SI Appendix, Fig. S11B). These results again support the hypothesis that Nolz-1-Dlx1/2 signaling regulates the differentiation of striatonigral and striatopallidal neurons.
Altered Gene Expression Patterns in the Ventral Striatum of Nolz-1 KO Brains.
Finally, to characterize the cell types in the mutant ventral striatum, we examined ventral striatum-enriched genes compared to dorsal striatum-enriched genes that were identified by our microarray analysis and from the Allen Brain Atlas. Ventral striatum-enriched genes Acvr2a and Npy1r were decreased in the ventral striatum of E18.5 Nolz-1 KO brains (SI Appendix, Fig. S12A). Dorsal striatum-enriched genes, Ebf1 and Astn2, remained at low levels in the ventral KO striatum, although Ebf1 and Astn2 were decreased in the dorsal KO striatum (SI Appendix, Fig. S12A). Expression of Sox1, a gene required for cell migration in the ventral striatum (45), was reduced in the ventral KO striatum (SI Appendix, Fig. S12B). Expression of Brn4, which was expressed at high levels in the OT and the junction between dorsal and ventral striata in WT brains (46), was decreased in the KO striatum (SI Appendix, Fig. S12C). Neuropilin 2 (Nrp2) is expressed in the OT but not in the CP (46). In the WT basal forebrain, a continuous Nrp2+ band extended from the septum across the OT to the piriform cortex. By contrast, an indented band that composed weak Nrp2+ regions was found in mutant OT-like regions (SI Appendix, Fig. S12D), suggesting that cells in the expanded mutant OT regions may not differentiate normally or that OT expansion may arise from cells aberrantly immigrating from other regions. Taken together with the results of biased cell migration toward the ventral striatum in Nolz-1 KO brains, many of the immigrant cells appear not to differentiate properly into the cell types characteristic of the ventral striatum.
Discussion
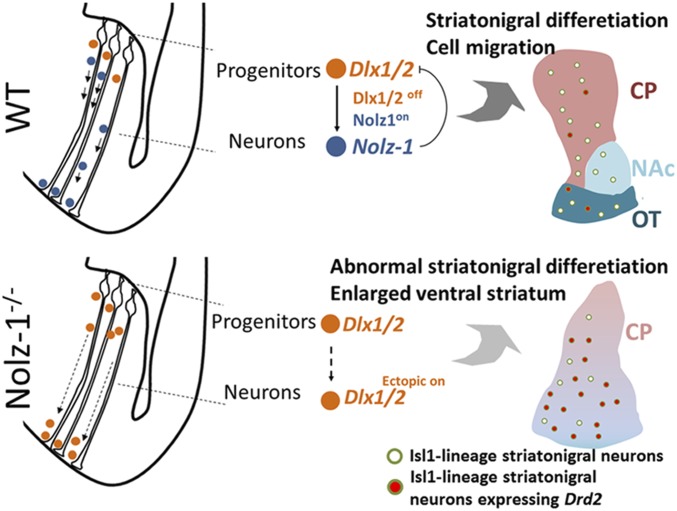
Our study not only describes Nolz-1-Dlx1/2 signaling-mediated regulation of cell types and differentiation, but also informs the fundamental question of how dorsal and ventral districts of the striatum are formed. Our study has uncovered a mechanism by which Nolz-1, by suppressing Dlx1/2 in postmitotic neurons, controls differential migration of striatal neurons into the dorsal and ventral locations and thereby set up the dorsal and ventral divisions of the striatum during development (Fig. 8). This paper provides mechanistic accounts of the developmental process underlying parcellation of the striatal complex into the dorsal and ventral districts. Our study also suggests that the regulatory process of cellular migration is coupled to the differentiation programs to set up the cell type and regional specificity during development.
Fig. 8.
Working hypothesis. Schematics showing migration and differentiation of striatal cells in striatal anlage at early (Left) and late (Right) stages of development. (Top) In the WT brain, Dlx1/2 is expressed in striatal progenitors. Repression of Dlx1/2 by Nolz-1 (Dlx1/2off;Nolz-1on) in postmitotic neurons is required for normal migration and differentiation of striatal neurons. (Bottom) In the Nolz-1−/− KO brain, persistent expression of Dlx1/2 (Dlx1/2ectopic on) in postmitotic neurons promotes aberrant cell migration from the dorsal toward the ventral striatum. These postmitotic neurons are of Isl1+ striatonigral cell lineages, but they ectopically express the striatopallidal gene Drd2 in the absence of Nolz-1.
Developmental Cell Lineages of the CP, NAc, and OT.
It is yet unclear whether striatal neurons in the CP, NAc, and OT are derived from the same or distinct cell lineages. In terms of the time windows of neurogenesis, previous birthdating studies have shown that neurons in the CP are born E13-P3, whereas NAc and OT neurons become postmitotic, respectively, at E15-P3 and E12-P0 in the rat brain (47, 48). In the mouse brain, OT and CP neurons are born, respectively, at E11-E15 and ∼E11.5–E18.5 (34, 49). Therefore, the time windows of neurogenesis in the dorsal and ventral striata are overlapping except that NAc neurons are born later than CP and OT neurons. Much of the information on the spatiotemporal origins of the CP, NAc, and OT is based on analyses of mutant mice. Regarding temporal segregation of striatal progenitor cells, it has been proposed that Ascl1/Mash1-expressing early-born LGE cells produce some OT neurons and striosomes/patch neurons of the CP, whereas Ascl1/Mash1 and Dlx1/2-expressing late-born cells generate other OT neurons, matrix neurons of the CP, and NAc neurons (50, 51). This reasoning is based on the findings that genetic mutations of Ascl1/Mash1 and Gsh2/Gsx2 result in loss of Enk- and D2-expressing NAc and ventrolateral telencephalon, including the OT in mutant brains (52, 53). In terms of spatial segregation of striatal progenitor cells, Gsh2 mutant studies implicate that the dorsolateral LGE gives rise to OT neurons at early developmental stages because the ventral striatum, particularly, Isl1+ and Sox1+ OT, is reduced in Gsh2 and Gsh1/2 mutants in which the dorsolateral LGE cells are misspecified to pallial cells (54, 55). We found Notch1+ radial glial fibers extending from the LGE across the subpallium as far as into the OT anlage as early as E12.5, suggesting that striatal cells generated in the LGE may migrate along radial glial fibers en route through the LGE into the OT at early stages of development. This result is consistent with a recent study showing a ventral migratory route from the ventral LGE to the OT primordium (34).
Aberrant Dlx1/2-Dependent Cell Migration in Nolz-1 Mutant Brains.
It has been suggested that Gsh1/2 and Ascl1/Mash1 regulate the generation of early-born striatal cells that contribute to the OT, striosomes/patch compartments, and NAc (50–55). In line with this hypothesis, Nolz-1 mutation resulted in the loss of the striosomal marker DARPP-32 and redistribution of BrdU12.5 striosomal cells from the dorsal toward the ventral striatum. Therefore, a hypothetical scenario is that, at the early stages of development, Nolz-1-dependent cell migratory mechanisms regulate early-born striatal cells that are differentially sorted into the CP and the OT. In the absence of Nolz-1, striatal cells destined for striosomes in the CP are aberrantly directed toward the OT.
What are Nolz-1-dependent mechanisms of cell migration? Based on the finding that knocking down aberrantly up-regulated Dlx1/2 expression restored a normal pattern of cell migration in the Nolz-1 KO striatum, we propose that Nolz-1-mediated repression of Dlx1/2 in postmitotic striatal neurons plays an essential role in controlling differential cell migration to the dorsal and ventral striata. Rubenstein’s group has previously demonstrated the importance of Dlx1 and Dlx2 in striatal cell migration. Double KOs of Dlx1 and Dlx2 result in impaired migration of late-born striatal cells from the SVZ into the MZ, although early-born striatal cells are not affected (44). Dlx1 and Dlx2 are expressed by cells at the transition from proliferation to terminal differentiation in the VZ and the SVZ, and they are down-regulated in the MZ of striatal anlage (39). Our present study has extended Rubenstein group's findings by identifying Nolz-1 as a key gene in restricting Dlx1/2 expression to the germinal zone. Because Nolz-1 deletion-induced up-regulated expression of Dlx1/2 in the differentiated MZ causes aberrant migration and differentiation of striatal neurons, timely and precise down-regulation of Dlx1/2 in the postmitotic MZ is important for proper striatal cell migration and differentiation. Rubenstein's group has further demonstrated that Dlx1/2 promotes tangential cell migration of cortical interneurons by inhibiting axon and dendrite growth through repressing the p21-activated serine/threonine kinase PAK3 (56). It would be of interest to see whether similar mechanisms underlie Dlx1/2-mediated migration of striatal cells.
Cell-Type Identity in the Ventral Striatum of Nolz-1 KO Brains.
Given the increased migration toward the ventral striatum in Nolz-1 KO brains, it raises a possibility that dorsal striatal cells may take on the identity of NAc cells when they are ectopically located in the ventral KO striatum. We attempted to test this hypothesis with NAc markers. However, NAc markers, such as dopamine D3 receptors, were expressed at very low levels in embryonic NAc (57), which prevented us from testing this possibility. For the developmental origin of the OT, it has been proposed that the OT is a striatal–pallidal structure in which the medium-sized cell population is related to the striatum, whereas the pallidal-like cell population is invaded from the ventral pallidum (10, 58). The reduction in the expression of a set of ventral striatum-enriched genes, including Acvr2a, Npy1r, Sox1, and Brn4, and the altered pattern of Nrp2 expression in the ventral KO striatum (SI Appendix, Fig. S12 A–D) do not support the possibility that dorsal striatum-derived cells differentiate into ventral striatal cell types in mutant OT-like regions. Nor do the dorsal-derived mutant cells retain the ability to differentiate into proper dorsal striatal cell types in the ventral mutant striatum as the dorsal striatum-enriched genes Ebf1 and Astn2 remained at low levels in the ventral KO striatum (SI Appendix, Fig. S12A). These results suggest that Nolz-1-dependent mechanisms are essential for the proper specification of cell-type identity in the striatal complex.
Conditional Deletion of Nolz-1 in Isl1 Cell Lineages Recapitulates Major Striatal Phenotypes of Nolz-1 KO Brains.
We demonstrate here that cell-type specific deletion of Nolz-1 in Isl1+ striatonigral cell lineages using Isl1-Cre mice not only leads to abnormal cell migration, but also causes derepression of the striatopallidal genes D2R and Enk in striatonigral neurons, which normally express little or none of these typical striatopallidal (indirect pathway) genes. Concomitantly, there were decreases in the expression of typical direct pathway striatonigral-enriched genes. It is not clear whether the mutant cells are respecified into striatopallidal neurons. Morphological changes in the Nolz-1 KO striatum rendered us unable to determine whether the axons of GFP+ striatonigral mutant neurons were rerouted to innervate the globus pallidus. Interestingly, our previous Isl1 KO mouse study has shown that deletion of Isl1 resulted in derepression of striatopallidal genes and loss of striatonigral gene expression in striatonigal neurons (27), which is similar to that in Nolz-1 KO mice. Our present study indicates that Nolz-1 is upstream of Isl1 as Isl1 is reduced in the Nolz-1 null KO striatum. Since Isl1 KO and Nolz-1 KO mice share similar phenotypes of striatonigral differentiation, Nolz-1 is likely to promote differentiation of striatonigral neurons through Isl1.
Conclusion
Recent studies using the single cell-RNA sequencing technique suggest that generic transcriptional programs in mitotic progenitors of ganglionic eminences and, subsequently, diverse transcriptional programs in early postmitotic neurons direct diverse trajectories of cortical interneurons during development (59, 60). The mixed model of spatial and temporal regulation of cell lineages suggests that predetermined genetic programs as well as environmental interactions shortly after the interneurons become postmitotic are crucial for the generation of diversity of cell types. Indeed, a recent study has shown that both genetic factors and migratory routes are important for the production of different cortical interneurons with distinct axonal projection patterns (61). The genetic codes of Nolz-1 and Dlx1/2 permit differential migration to dorsal and ventral locations, and environmental signals in migratory routes and destinations may promote diversification of cell types in the CP, NAc, and the OT to construct specific circuits that underlie distinct striatal functions.
Concerning the functional aspect of the dorsal and ventral striata, the striatonigral direct pathway, and the striatopallidal indirect pathway in the dorsal striatum preferentially control movement, whereas their counterparts in the ventral striatum preferentially regulate reward-seeking aversive responses and emotion. These differences are by no means complete as research shows, but it is significant that many of the connections of these two different subdivisions are also different. Our study reveals a fundamental transcriptional cascade by which the dorsal and ventral striata are determined at the early stages of development. An important question is how the circuit wiring is differentially laid out to support the functional diversity of the dorsal and ventral striata. Concerning neurological diseases, the dorsal striatum appears to be a main pathological target in neurodegenerative diseases, e.g., the nigrostriatal pathway innervating the dorsal striatum is the major etiological site in Parkinson’s disease, and neuronal degeneration primarily occurs in the dorsal striatum of Huntington’s disease. In both of these disorders, the ventral striatum is relatively spared until the late stages of the disorders. It would be of interest to explore whether different developmental trajectories of dorsal and ventral striatal neurons as highlighted here render them vulnerable or resilient to these or other disorders. The powerful genome-editing technology of CRISPR/Cas9 may then be amenable to the treatment of the disorders.
Materials and Methods
Animals.
The mice were maintained in the Animal Center of National Yang-Ming University (NYMU, Taiwan). The protocol for animal use was approved by the Institutional Animal Care and Use Committee in NYMU and conformed to the NIH Guide for the Care and Use of Laboratory Animals (62). Details of the generation of Nolz-1 KO mice are described in the SI Appendix.
Immunohistochemistry.
Immunohistochemistry of embryonic and newborn brain tissues was performed as previously described (27). Details of immunohisochemisty are available in the SI Appendix.
In Situ Hybridization.
The sections of WT and littermate mutant brains were mounted on the same slides to ensure they were processed under the same conditions. In situ hybridization was performed with 35S-UTP-labeled or digoxigenin-labeled probes as previously described (27, 63). The information of the riboprobes for in situ hybridization is available in the SI Appendix.
Data Availability.
All data are included in the paper and SI Appendix.
Supplementary Material
Acknowledgments
We thank Drs. D. Anderson, P. Arlotta, A.C. Chang, M. Colbert, J. Cordell, E.B. Crenshaw III, D. Duboule, K. Kobayashi, O. Marin, and J.-Y. Yu for providing the reagents and transgenic mice, A.M. Graybiel for reading and editing the paper, and the Transgenic Mouse Model Core Facility of the National Core Facility for Biopharmaceuticals (NCFB), Ministry of Science and Technology (MOST) for help in the generation of Nolz-1 mutant mice. This work was supported by National Science Council Grants NSC95-3112-B-010-014, NSC96-3112-B-010-007, NSC97-3112-B-010-005, NSC99-2311-B-010-005-MY3, NSC101-2321-B-010-021, NSC102-2321-B-010-018, Ministry of Science and Technology Grants MOST103-2321-B-010-009, MOST104-2311-B-010-010-MY3, MOST-107-2321-B-010-002, MOST107-2320-B-010-041-MY3, MOST-108-2321-B-010-002, and MOST-108-2321-B-007-003-MY2, (F.-C.L.), Postdoctoral Fellowship Grants MOST104-2811-B-010-026 (K.-M.L.), MOST106-2811-B-010-031, MOST107-2811-B-010-518, and MOST108-2811-B-010-525 (S.-Y.C.), and a Brain Research Center Grant from the Higher Education Sprout Project, Ministry of Education in Taiwan (F.-C.L.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1921007117/-/DCSupplemental.
References
- 1.Alexander G. E., DeLong M. R., Strick P. L., Parallel organization of functionally segregated circuits linking basal ganglia and cortex. Annu. Rev. Neurosci. 9, 357–381 (1986). [DOI] [PubMed] [Google Scholar]
- 2.Wichmann T., DeLong M. R., Functional and pathophysiological models of the basal ganglia. Curr. Opin. Neurobiol. 6, 751–758 (1996). [DOI] [PubMed] [Google Scholar]
- 3.Crittenden J. R., Graybiel A. M., Basal Ganglia disorders associated with imbalances in the striatal striosome and matrix compartments. Front. Neuroanat. 5, 59 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bostan A. C., Strick P. L., The basal ganglia and the cerebellum: Nodes in an integrated network. Nat. Rev. Neurosci. 19, 338–350 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Middleton F. A., Strick P. L., Basal ganglia and cerebellar loops: Motor and cognitive circuits. Brain Res. Brain Res. Rev. 31, 236–250 (2000). [DOI] [PubMed] [Google Scholar]
- 6.Haber S. N., Corticostriatal circuitry. Dialogues Clin. Neurosci. 18, 7–21 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hunnicutt B. J., et al. , A comprehensive excitatory input map of the striatum reveals novel functional organization. eLife 5, e19103 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Russo S. J., et al. , The addicted synapse: Mechanisms of synaptic and structural plasticity in nucleus accumbens. Trends Neurosci. 33, 267–276 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grueter B. A., Rothwell P. E., Malenka R. C., Integrating synaptic plasticity and striatal circuit function in addiction. Curr. Opin. Neurobiol. 22, 545–551 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heimer L., Zahm D. S., Alheid G. F., “Basal Ganglia” in The Rat Nervous System, Paxinos G., Ed. (Academic Press, Inc., San Diego, 1995), vol. 2, pp. 579–628. [Google Scholar]
- 11.Nestler E. J., et al. , Neurobiology of depression. Neuron 34, 13–25 (2002). [DOI] [PubMed] [Google Scholar]
- 12.Kalia L. V., Lang A. E., Parkinson’s disease. Lancet 386, 896–912 (2015). [DOI] [PubMed] [Google Scholar]
- 13.McColgan P., Tabrizi S. J., Huntington’s disease: A clinical review. Eur. J. Neurol. 25, 24–34 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Volkow N. D., Morales M., The brain on drugs: From reward to addiction. Cell 162, 712–725 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Gerfen C. R., Wilson C. J., “The basal ganglia” in Handbook of Chemical Neuroanatomy. Vol.12: Integrated Systems of the CNS. Part III, Swanson L. W., Björklund A., Hökfelt T., Eds. (Elsevier Science B.V., 1996), pp. 371–468. [Google Scholar]
- 16.Wichterle H., Turnbull D. H., Nery S., Fishell G., Alvarez-Buylla A., In utero fate mapping reveals distinct migratory pathways and fates of neurons born in the mammalian basal forebrain. Development 128, 3759–3771 (2001). [DOI] [PubMed] [Google Scholar]
- 17.Stenman J., Toresson H., Campbell K., Identification of two distinct progenitor populations in the lateral ganglionic eminence: Implications for striatal and olfactory bulb neurogenesis. J. Neurosci. 23, 167–174 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flames N., et al. , Delineation of multiple subpallial progenitor domains by the combinatorial expression of transcriptional codes. J. Neurosci. 27, 9682–9695 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang C. W., et al. , Identification of a developmentally regulated striatum-enriched zinc-finger gene, Nolz-1, in the mammalian brain. Proc. Natl. Acad. Sci. U.S.A. 101, 2613–2618 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ko H. A., Chen S. Y., Chen H. Y., Hao H. J., Liu F. C., Cell type-selective expression of the zinc finger-containing gene Nolz-1/Zfp503 in the developing mouse striatum. Neurosci. Lett. 548, 44–49 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Takahashi K., Liu F. C., Hirokawa K., Takahashi H., Expression of Foxp2, a gene involved in speech and language, in the developing and adult striatum. J. Neurosci. Res. 73, 61–72 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Ouimet C. C., Miller P. E., Hemmings H. C. Jr, Walaas S. I., Greengard P., DARPP-32, a dopamine- and adenosine 3′:5′-monophosphate-regulated phosphoprotein enriched in dopamine-innervated brain regions. III. Immunocytochemical localization. J. Neurosci. 4, 111–124 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawasaki H., et al. , A Rap guanine nucleotide exchange factor enriched highly in the basal ganglia. Proc. Natl. Acad. Sci. U.S.A. 95, 13278–13283 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lobo M. K., Karsten S. L., Gray M., Geschwind D. H., Yang X. W., FACS-array profiling of striatal projection neuron subtypes in juvenile and adult mouse brains. Nat. Neurosci. 9, 443–452 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Lobo M. K., Yeh C., Yang X. W., Pivotal role of early B-cell factor 1 in development of striatonigral medium spiny neurons in the matrix compartment. J. Neurosci. Res. 86, 2134–2146 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Ehrman L. A., et al. , The LIM homeobox gene Isl1 is required for the correct development of the striatonigral pathway in the mouse. Proc. Natl. Acad. Sci. U.S.A. 110, E4026–E4035 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu K. M., Evans S. M., Hirano S., Liu F. C., Dual role for Islet-1 in promoting striatonigral and repressing striatopallidal genetic programs to specify striatonigral cell identity. Proc. Natl. Acad. Sci. U.S.A. 111, E168–E177 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waclaw R. R., et al. , Foxo1 is a downstream effector of Isl1 in direct pathway striatal projection neuron development within the embryonic mouse telencephalon. Mol. Cell. Neurosci. 80, 44–51 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lavado A., Oliver G., Six3 is required for ependymal cell maturation. Development 138, 5291–5300 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu Z., et al. , SP8 and SP9 coordinately promote D2-type medium spiny neuron production by activating Six3 expression. Development 145, dev165456 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang S. L., et al. , Ectopic expression of Nolz-1 in neural progenitors promotes cell cycle exit/premature neuronal differentiation accompanying with abnormal apoptosis in the developing mouse telencephalon. PLoS One 8, e74975 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma Q., Sommer L., Cserjesi P., Anderson D. J., Mash1 and neurogenin1 expression patterns define complementary domains of neuroepithelium in the developing CNS and are correlated with regions expressing notch ligands. J. Neurosci. 17, 3644–3652 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.García-Moreno F., López-Mascaraque L., de Carlos J. A., Early telencephalic migration topographically converging in the olfactory cortex. Cereb. Cortex 18, 1239–1252 (2008). [DOI] [PubMed] [Google Scholar]
- 34.Martin-Lopez E., Xu C., Liberia T., Meller S. J., Greer C. A., Embryonic and postnatal development of mouse olfactory tubercle. Mol. Cell. Neurosci. 98, 82–96 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heimer L., et al. , The accumbens: Beyond the core-shell dichotomy. J. Neuropsychiatry Clin. Neurosci. 9, 354–381 (1997). [DOI] [PubMed] [Google Scholar]
- 36.Gaiano N., Nye J. S., Fishell G., Radial glial identity is promoted by Notch1 signaling in the murine forebrain. Neuron 26, 395–404 (2000). [DOI] [PubMed] [Google Scholar]
- 37.Toresson H., Parmar M., Campbell K., Expression of Meis and Pbx genes and their protein products in the developing telencephalon: Implications for regional differentiation. Mech. Dev. 94, 183–187 (2000). [DOI] [PubMed] [Google Scholar]
- 38.Price M., Lemaistre M., Pischetola M., Di Lauro R., Duboule D., A mouse gene related to Distal-less shows a restricted expression in the developing forebrain. Nature 351, 748–751 (1991). [DOI] [PubMed] [Google Scholar]
- 39.Porteus M. H., et al. , DLX-2, MASH-1, and MAP-2 expression and bromodeoxyuridine incorporation define molecularly distinct cell populations in the embryonic mouse forebrain. J. Neurosci. 14, 6370–6383 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ghanem N., et al. , Regulatory roles of conserved intergenic domains in vertebrate Dlx bigene clusters. Genome Res. 13, 533–543 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brown J. D., et al. , Expression profiling during ocular development identifies 2 Nlz genes with a critical role in optic fissure closure. Proc. Natl. Acad. Sci. U.S.A. 106, 1462–1467 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anderson S. A., Eisenstat D. D., Shi L., Rubenstein J. L., Interneuron migration from basal forebrain to neocortex: Dependence on Dlx genes. Science 278, 474–476 (1997). [DOI] [PubMed] [Google Scholar]
- 43.Marín O., Rubenstein J. L., Cell migration in the forebrain. Annu. Rev. Neurosci. 26, 441–483 (2003). [DOI] [PubMed] [Google Scholar]
- 44.Anderson S. A., et al. , Mutations of the homeobox genes Dlx-1 and Dlx-2 disrupt the striatal subventricular zone and differentiation of late born striatal neurons. Neuron 19, 27–37 (1997). [DOI] [PubMed] [Google Scholar]
- 45.Ekonomou A., et al. , Neuronal migration and ventral subtype identity in the telencephalon depend on SOX1. PLoS Biol. 3, e186 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Long J. E., et al. , Dlx1&2 and Mash1 transcription factors control striatal patterning and differentiation through parallel and overlapping pathways. J. Comp. Neurol. 512, 556–572 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bayer S. A., Neurogenesis in the rat neostriatum. Int. J. Dev. Neurosci. 2, 163–175 (1984). [DOI] [PubMed] [Google Scholar]
- 48.Bayer S. A., Altman J., Directions in neurogenetic gradients and patterns of anatomical connections in the telencephalon. Prog. Neurobiol. 29, 57–106 (1987). [DOI] [PubMed] [Google Scholar]
- 49.Liao W. L., et al. , Modular patterning of structure and function of the striatum by retinoid receptor signaling. Proc. Natl. Acad. Sci. U.S.A. 105, 6765–6770 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yun K., Garel S., Fischman S., Rubenstein J. L., Patterning of the lateral ganglionic eminence by the Gsh1 and Gsh2 homeobox genes regulates striatal and olfactory bulb histogenesis and the growth of axons through the basal ganglia. J. Comp. Neurol. 461, 151–165 (2003). [DOI] [PubMed] [Google Scholar]
- 51.Kelly S. M., et al. , Radial glial lineage progression and differential intermediate progenitor amplification underlie striatal compartments and circuit organization. Neuron 99, 345–361.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Casarosa S., Fode C., Guillemot F., Mash1 regulates neurogenesis in the ventral telencephalon. Development 126, 525–534 (1999). [DOI] [PubMed] [Google Scholar]
- 53.Corbin J. G., Gaiano N., Machold R. P., Langston A., Fishell G., The Gsh2 homeodomain gene controls multiple aspects of telencephalic development. Development 127, 5007–5020 (2000). [DOI] [PubMed] [Google Scholar]
- 54.Toresson H., Potter S. S., Campbell K., Genetic control of dorsal-ventral identity in the telencephalon: Opposing roles for Pax6 and Gsh2. Development 127, 4361–4371 (2000). [DOI] [PubMed] [Google Scholar]
- 55.Toresson H., Campbell K., A role for Gsh1 in the developing striatum and olfactory bulb of Gsh2 mutant mice. Development 128, 4769–4780 (2001). [DOI] [PubMed] [Google Scholar]
- 56.Cobos I., Borello U., Rubenstein J. L., Dlx transcription factors promote migration through repression of axon and dendrite growth. Neuron 54, 873–888 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sokoloff P., Giros B., Martres M. P., Bouthenet M. L., Schwartz J. C., Molecular cloning and characterization of a novel dopamine receptor (D3) as a target for neuroleptics. Nature 347, 146–151 (1990). [DOI] [PubMed] [Google Scholar]
- 58.Heimer L., A new anatomical framework for neuropsychiatric disorders and drug abuse. Am. J. Psychiatry 160, 1726–1739 (2003). [DOI] [PubMed] [Google Scholar]
- 59.Mi D., et al. , Early emergence of cortical interneuron diversity in the mouse embryo. Science 360, 81–85 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mayer C., et al. , Developmental diversification of cortical inhibitory interneurons. Nature 555, 457–462 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lim L., et al. , Optimization of interneuron function by direct coupling of cell migration and axonal targeting. Nat. Neurosci. 21, 920–931 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.National Research Council , Guide for the Care and Use of Laboratory Animals (National Academies Press, Washington, DC, ed. 8, 2011). [Google Scholar]
- 63.Liao W. L., Tsai H. C., Wu C. Y., Liu F. C., Differential expression of RARbeta isoforms in the mouse striatum during development: A gradient of RARbeta2 expression along the rostrocaudal axis. Dev. Dyn. 233, 584–594 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are included in the paper and SI Appendix.