Significance
Acetylation is an important feature of hemicellulose, altering the physical properties of the plant cell wall and limiting enzyme accessibility. Removal of acetyl groups from β-mannan is a key step toward efficient utilization of this glycan as a carbon source for gut microbiota and in biorefineries. We present detailed insight into mannan deacetylation by two highly substrate-specific acetyl-mannan esterases (AcMEs) from a prevalent gut commensal Firmicute, which cooperatively deacetylate complex galactoglucomannan. The three-dimensional structure of RiCE17 with mannopentaose in the active site has a unique two-domain architecture including a CBM35 and a SGNH superfamily hydrolytic domain. Discovery of β-mannan-specific esterases improves the understanding of an important step in dietary fiber utilization by gut commensal Firmicutes.
Keywords: acetyl esterases, galactoglucomannan, Roseburia intestinalis, gut microbiota, biorefining
Abstract
β-mannans and xylans are important components of the plant cell wall and they are acetylated to be protected from degradation by glycoside hydrolases. β-mannans are widely present in human and animal diets as fiber from leguminous plants and as thickeners and stabilizers in processed foods. There are many fully characterized acetylxylan esterases (AcXEs); however, the enzymes deacetylating mannans are less understood. Here we present two carbohydrate esterases, RiCE2 and RiCE17, from the Firmicute Roseburia intestinalis, which together deacetylate complex galactoglucomannan (GGM). The three-dimensional (3D) structure of RiCE17 with a mannopentaose in the active site shows that the CBM35 domain of RiCE17 forms a confined complex, where the axially oriented C2-hydroxyl of a mannose residue points toward the Ser41 of the catalytic triad. Cavities on the RiCE17 surface may accept galactosylations at the C6 positions of mannose adjacent to the mannose residue being deacetylated (subsite −1 and +1). In-depth characterization of the two enzymes using time-resolved NMR, high-performance liquid chromatography (HPLC), and mass spectrometry demonstrates that they work in a complementary manner. RiCE17 exclusively removes the axially oriented 2-O-acetylations on any mannose residue in an oligosaccharide, including double acetylated mannoses, while the RiCE2 is active on 3-O-, 4-O-, and 6-O-acetylations. Activity of RiCE2 is dependent on RiCE17 removing 2-O-acetylations from double acetylated mannose. Furthermore, transacetylation of oligosaccharides with the 2-O-specific RiCE17 provided insight into how temperature and pH affects acetyl migration on manno-oligosaccharides.
Enzymatic acetylation and deacetylation are involved in numerous processes in nature. N-acetylation of lysine in histones is crucial for the control of gene transcription (1); acetylation or deacetylation of N-acetylglucosamine and N-acetylmuramic acid in microbial peptidoglycan affects its degradation by lysozyme (2). The efficacy of penicillins is also linked to acetylations, e.g., penicillin resistance caused by bacteria expressing cephalosporin esterases (3). Acetylations are important constituents in some of the most abundant polymers on Earth, like chitin, pectin, xylan, and β-mannan (4, 5). Enzymes that process these polymers have adapted to a wide array of structurally diverse targets. Studying the structure-function relationships of acetyl esterases and transferases involved in these processes is needed to shed light on this important and very common covalent modification.
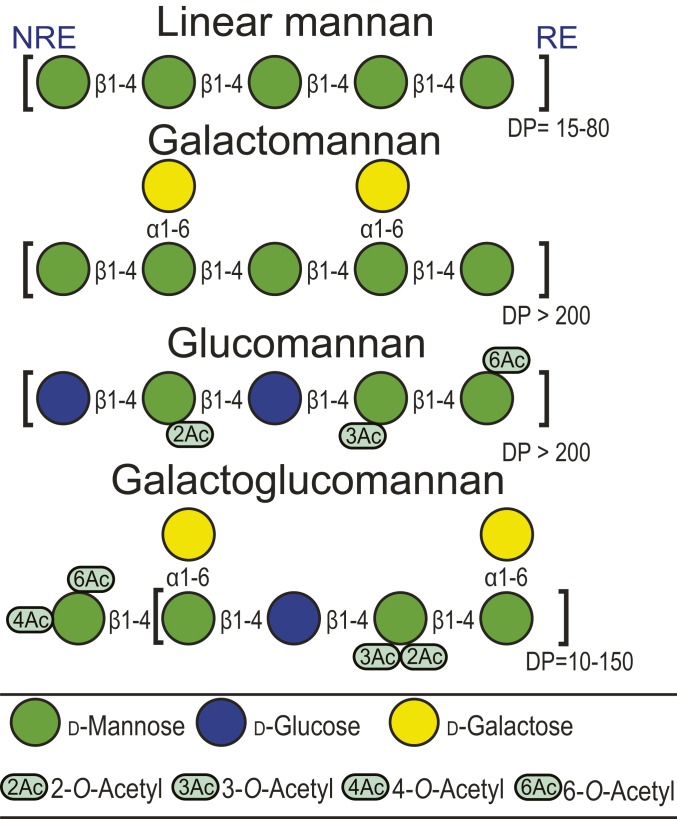
β-mannans are polysaccharides with a backbone consisting of β-1,4-linked d-Manp units (Fig. 1) that serve as energy storage and contribute as structural support in the primary and secondary cell walls of plants. The backbone can be interspersed with β-1,4-linked d-Glcp, as in konjac glucomannan (Amorphophallus konjac), decorated with α-1,6-d-Galp as in carob galactomannan (Ceratonia siliqua), or contain both glucose and galactose such as the galactoglucomannan (GGM) in Aloe vera (AV) and Norway spruce (Picea abies) (6–8). Furthermore, mannans may contain substantial amounts of 2-O-, 3-O-, and 6-O-acetylations on any mannose residue in the chain, as well as 4-O-acetylations (rare) on the nonreducing end of the chain.
Fig. 1.
A cartoon presentation of various types of mannan found in plants. Glucomannans like konjac glucomannan generally contain a low degree of acetylation (DA = 0.07). Galactoglucomannans contain various DAs; the spruce mannan and the A. vera mannan used in this study have a DA of 0.35 and 0.86, respectively. NRE and RE denotes nonreducing and reducing end. Adapted with permission from ref. 22, which is licensed under CC BY 4.0.
Depending on the plant origin, mannans differ in their monosaccharide composition as well as degree and location of acetylations (9). A unique characteristic of the 2-O-acetyl groups of mannans is their relative orientation: 2-O-acetylations are axial, as opposed to other common structural polysaccharides, like xylan, pectin, and chitin, that all have acetylations in the equatorial plane of the sugar ring.
Acetylation of hemicelluloses affects their biological functions and a range of physicochemical properties and is thought to be a defense mechanism developed as a part of an evolutionary arms race against pathogens (5) as well as contributing to aggregation of cellulose microfibrils (10). Acetylations affect the solubility of the polysaccharides by restricting the formation of hydrogen bonds between chains in solution. Effects of mannan acetylations on the viscosity of solutions make them effective thickeners and stabilizers (9, 11) in the food and feed industry where mannans such as guar gum and konjac are commonly used (12, 13). Furthermore, the immunostimulatory properties of AV mannan, a common ingredient in nutraceuticals and cosmetics, have been linked to its high degree of acetylation (8). In the context of biorefining, acetylations reduce fermentability (14) and enzymatic degradability, and thus pose a limitation on the utilization of mannans as a feedstock for fermentation. In this respect, deacetylation of mannan by chemical pretreatment or enzymatic treatment with acetyl esterases is known to improve fermentation yields. On the other hand, the recalcitrance resulting from the presence of acetylations also restricts the number of gut microbes able to consume hemicellulose. This may be beneficial as a means of providing selective growth of bacteria when hemicellulose is considered as an ingredient of food and feed.
Gut microbes are highly adapted to fermenting diet-derived complex carbohydrates and have evolved complex machineries called polysaccharide utilization loci (PUL) to utilize these carbohydrates (15, 16). Bacteroidetes and Firmicutes are the main polysaccharide-degrading commensal bacteria in the human gut (17, 18). β-mannans constitute a small but significant part of human diet that can be found in a wide range of common food products such as coffee (19) and tomatoes (20). This may explain why mannan degradation is identified as one of the core pathways in the human gut microbiome (21). Some species of Firmicutes, such as Roseburia intestinalis, have developed sophisticated enzymatic machineries for complete β-mannan degradation; this PUL quite frequently contains carbohydrate esterases (CEs) (22).
The esterases deacetylating xylan and mannan characterized so far have been classified together as acetylxylan esterases (EC 3.1.1.72). CEs are common in bacterial and fungal genomes and are often found in PUL together with glycoside hydrolases (GH) (22, 23). This highlights their biological role as accessory enzymes facilitating the processing of glycans. CEs are classified into 17 families according to the Carbohydrate-Active enZYmes (CAZy) database (24), in which members of families 1 to 7, 15, and 16 have been reported to be active on hemicelluloses in plant biomass. All CEs, with the exception of family CE4, are serine-histidine hydrolases (5). Esterases from the CE2 family share a two-domain architecture consisting of a catalytic SGNH hydrolase superfamily domain with a conserved catalytic dyad, and an accessory jelly roll domain (25). The two-domain structure results in an open active site able to accommodate a variety of substrates. So far, functionally characterized CE2 esterases have been largely nonspecific; activities have been reported on 2-O-, 3-O-, and 4-O- acetylated xylan, konjac glucomannan (26), and 6-O-acetylated mannan (5). Some CE2 esterases have been considered specific to 6-O-acetylations, which is the position they transacetylate when using vinyl acetate as the acetate donor (27). Enzymatic deacetylation of mannans has previously been described in literature (28, 29), but no esterase exclusively active on mannan has been reported.
Here, we report a detailed functional characterization of RiCE2 and RiCE17—two acetyl mannan esterases (AcMEs) from R. intestinalis. The esterases have complementary substrate specificities and act in tandem to deacetylate glucomannan and galacto(gluco)mannan. Furthermore, we report the structure of an AcME with a mannopentaose cocrystallized in the active site, revealing how the two-domain architecture of RiCE17 facilitates specificity toward the axially oriented 2-O-acetylation.
Results and Discussion
A gene cluster in R. intestinalis encoding mannan-degrading enzymes (22) contains two acetyl esterases, RiCE17 and RiCE2. RiCE2 is a 349 amino acid CAZy family 2 esterase, which shares 29.46% identity with the xylan esterase CjCE2C (also known as CE2C;CJA_2889 and Axe2B) of Cellvibrio japonicus (UniProt accession no B3PC75) (26). Analysis of the RiCE2 sequence with InterProScan (30) identified the C-terminal residues 163 to 375 in RiCE2 as an SGNH hydrolase domain, while residues 1 to 159 constitute a galactose-binding superfamily domain. This two-domain architecture is characteristic for CE2 family esterases (25). BlastP searches (31) of RiCE17 (372 amino acids) returned very low sequence homology to any characterized esterases (20% sequence similarity with acetyl xylan esterase Axe2 of Geobacillus stearothermophilus) (32). Despite the low similarity, the sequence still appeared to be a promising esterase candidate, due to its location in the R. intestinalis mannan PUL, and the N-terminal SGNH hydrolytic domain. Contrary to the CE2 family esterases, where the SGNH domains are present in the C terminus, the RiCE17 hydrolytic domain is present in the N-terminal residues 35 to 209. InterProScan (30) was not able to predict a function for the 163 amino acid C-terminal domain of RiCE17. This previously uncharacterized sequence organization captured our interest for further structural exploration.
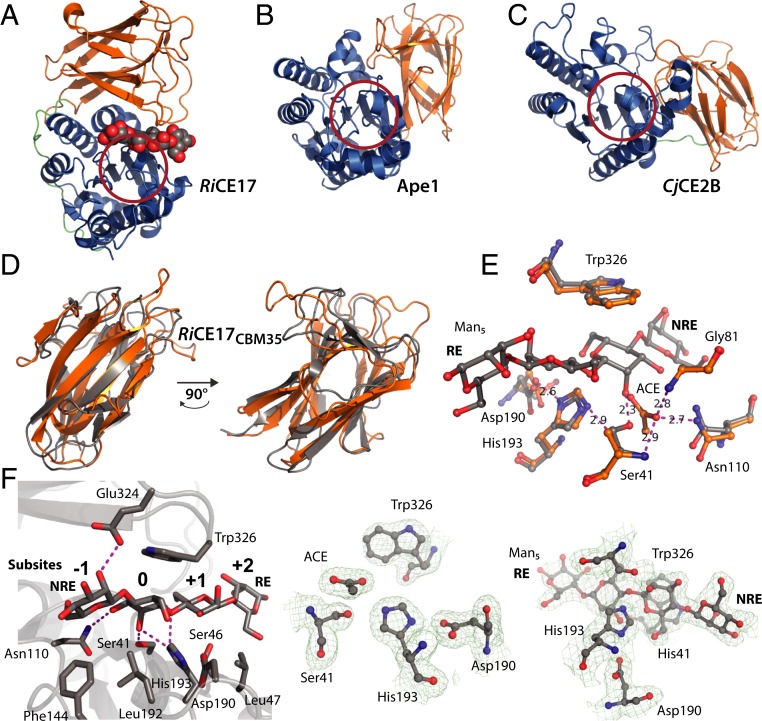
Crystal structures for the RiCE17 with acetate (Protein Data Bank [PDB] ID code 6HFZ, 1.75 Å resolution) and mannopentaose (PDB ID code 6HH9, 2.4 Å resolution) in the enzyme active site reveal several structural features. For the structure with mannopentaose, four mannose units were observed in the electron density maps. The enzyme consists of two domains connected by a linker of about 20 amino acids (Fig. 2A). The N-terminal domain (ca. 210 amino acids) displays an α/β-hydrolase (SGNH-hydrolase) fold while the C-terminal domain (ca. 140 amino acids) comprises a jelly-roll beta-sandwich fold. Structural alignment of the complete RiCE17 structure against the PDB using PDBeFold (33) returned hits with low scores, indicating this is a hitherto unidentified fold. The two structures with the highest scores were Ape1, a peptidoglycan O-acetylesterase from Neisseria meningitidis (Fig. 2B) (PDB ID code 4K40, Q-score 0.17) (34), and the CjCE2B (UniProt accession no. F7VJJ8) carbohydrate esterase from C. japonicus (Fig. 2C) (PDB ID code 2W9X, Q-score 0.12) (26). It is apparent that the second domain of these three enzymes adopts different positions in space when aligning their catalytic SGNH domains (Fig. 2 A–C). These differences are not due to domain flexibility. For the Ape1 enzyme, the second domain is inserted in the middle of the SGNH domain sequence and is kept in place by two short linkers consisting of three to four amino acids. The second domain in the CjCE2B enzyme, located at the N-terminal end of the SGNH domain, also adopts a very different position than that found for the C-terminal domain in RiCE17, connected to the SGNH domain via an eight-residue linker.
Fig. 2.
Comparison with RiCE17 homologs and structural details of RiCE17. (A) Crystal structure of RiCE17, with four of the mannose residues present in mannopentaose (spheres) visible in the electron density map. The catalytic domain (blue) is connected to the CBM35 domain (orange) by a linker region (green). (B and C) Display of the structural homologs Ape1 and CjCE2B that are closest to RiCE17 in the PDB database, respectively. The catalytic domains (blue) in B and C are oriented to superimpose with the RiCE17 catalytic domain in A. The red rings in A–C indicate where the residues of the catalytic triad are located. In D, the RiCE17cbm35 domain (orange) is superimposed on the CtCBM35 domain (gray) and shown at two rotations, demonstrating the highly similar folds. (E) Display of the RiCE17 superposed with acetate (orange carbons, ACE denotes acetate) or mannopentaose (gray carbons) in the active sites. In both structures, Asp190 form a hydrogen bond with His193 (2.6 Å). In the acetate bound structure, the His193-Nε2 is hydrogen bonded with the Ser41-OH O-atom (2.9 Å). The O-atom of the acetate molecule pointing toward the oxyanion hole is hydrogen bonded by Ser41-N (2.9 Å), Asn110-Nδ2 (2.7 Å), and Gly81-N (2.8 Å), indicated by dotted magenta lines to the acetate molecule. In the mannopentaose bound structure, the hydrogen bond between His193-Nε2 and the Ser41-OH O-atom is replaced with a Ser41-OH O-atom hydrogen bond to the C2-OH group of the mannose residue in the esterase catalytic site (2.3 Å). (F) Several charged or polar residues form hydrogen bonds to the mannopentaose ligand (Ser41, Asn110, His193, and Glu324). Ser46 may interact with the ligand through a water bridge. The hydrophobic residues Leu47, Phe144, Leu192, and Trp326 are part of the ligand-protein contact surface. Trp326 plays an important role, stacking on Top of the mannose residue in the active site. The subsites (−1, 0, +1, and +2) are indicated with bold numbers. The electron density maps of the active sites are shown to the right in F. The nonreducing end (denoted NRE) and the reducing end (denoted RE) are labeled in the manno-oligosaccharide ligand to aid interpretation of the figures.
Structural alignment searches using the C-terminal, noncatalytic domain of RiCE17, identified a carbohydrate-binding module 35 (CBM35), similar to that of Clostridium thermocellum (Fig. 2D) (PDBid 2W1W, Q-score 0.39) (35). CBM35 modules have previously been demonstrated to bind decorated mannans (36). The RiCE17CBM35 domain appears to be involved in substrate recognition and binding, forming a lid over the SGNH-domain active site (Fig. 2). Several hydrophobic amino acids, Ile79, Phe86, Ala89, Ile263, Leu330, Leu332, and Ile367, contribute to stabilize the interdomain interaction together with the three pairs of charged or polar amino acid residues Glu119-His338, His116-Glu337, and Gln85-Thr334 (Fig. 2).
In the active site of the SGNH domain, three residues Ser41, His193, and Asp190 form the hydrogen-bonded catalytic triad, and the amide nitrogen atoms of Ser41 and Gly81 and the Nδ2 of Asn110 line the oxyanion hole (Fig. 2E). The high 2-O-acetylation specificity of RiCE17 can be explained by the ligand bound structure. Amino acids from both the SGNH and CBM35 domains form specific interactions with the mannopentaose and align it such that the C2-OH group is only 2.3 Å from the Ser41-OH group (O-O distance), close to the oxyanion hole, which forms a cavity that fits an acetyl group. In the apo-RiCE17 structure, we can clearly see an acetate molecule in the oxyanion hole (Fig. 2E), and this arrangement could represent a product complex. Such binding of acetate in absence of carbohydrate substrate has also been observed for Ape1 (34). The binding of acetate indicates how intermediates in the reaction may be positioned and provides structural insight useful for further mechanistic studies.
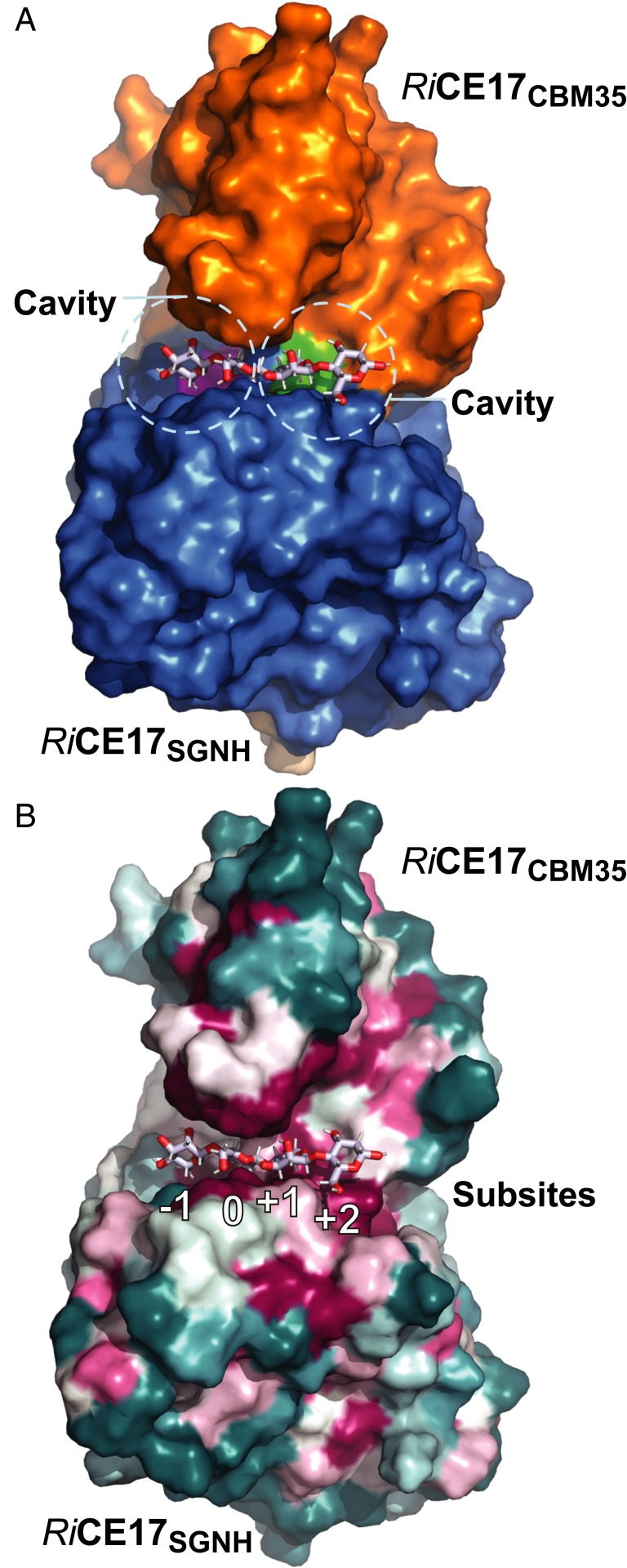
Fig. 2F shows interactions between RiCE17 and the mannopentaose ligand. Two amino acids from the RiCE17CBM35 domain contribute to ligand binding; namely Glu324 and Trp326. The Glu324 forms a hydrogen bond to the O-2 hydroxyl group of the mannose at the nonreducing end next to the active site [subsite −1 according to Hekmat et al. (37)]. Accordingly, if the mannose unit in subsite −1 is occupied by a glucose residue, as could be the case when galactoglucomannans are processed, the Glu324 hydrogen bond to the O-2 hydroxyl group would be lost. This may lead to lower affinities to substrates containing a glucose unit in subsite −1. Aromatic amino acids are often involved in protein-carbohydrate interactions, and in RiCE17 the Trp326 amino acid side chain stacks on top of the mannose unit in the active site. Interestingly, the CBM35 appears to be more than an accessory domain involved in the orientation of glycans in the active site. A truncated version of the enzyme, containing only the catalytic domain, was not active either on natural substrates, or pNP (p-nitrophenol or 4-nitrophenol) acetate (SI Appendix, Fig. S1). An observed lack of activity of the SGNH domain indicates that it does not have an inherent esterase activity alone, but rather is adapted to 2-O-deacetylation by interaction with the CBM35 domain. The activity appears to depend on the two domains forming a confining clamp (Fig. 3A) that provides substrate interactions and aids protein stability. This hypothesis was further supported by the results of thermal shift assays (SI Appendix, Fig. S2), which showed a decrease in stability for the SGNH domain (as compared with the complete RiCE17), and a lack of folding for the CBM35 domain. Interestingly, the two-domain arrangement results in cavities or clefts on each side of the active site indicated by the two green and magenta patches in Fig. 3A. Both of these cavities are arranged so that galactose residues linked to the mannose backbone (corresponding to −1 and +1 subsites) at the O-6 position would point toward these spaces. This feature may be important for recognizing galactose decorated mannans and may also influence the substrate specificity of the enzyme.
Fig. 3.
The 3D structures of substrate and product complexes of RiCE17. (A) The magenta and green patches in the interface between the RiCE17 catalytic domain and the RiCBM35 domain indicate cavities on the enzyme surface that may bind galactose residues decorating galactoglucomannan at the C6 position. In B the conservation score derived by ConSurf is projected on the RiCE17 surface (highly conserved amino acid residues [magenta], semiconserved [white], and variable residues [blue] are indicated). The highly conserved residues are concentrated around the substrate-binding site. Subsites are labeled below the substrate according to the nomenclature suggested by Hekmat et al. (37).
Since mannan degradation is a common process in many microbial ecosystems, we searched for microbes carrying protein sequences related to RiCE17. Searching the UniProtKB database with a Hidden Markov Model (HMM, detailed description in the SI Appendix) of both domains of RiCE17, we identified 482 protein sequences with the same two-domain architecture. Sequence alignment using MView showed that the catalytic triad (Ser41, His193, and Asp190) and the Trp326 of the carbohydrate-binding module (CBM) domain were conserved in the 90% consensus sequence (SI Appendix, Fig. S3). The HMM search allowed us to examine the prevalence of RiCE17 homologs among bacterial taxa and identify the conserved regions, in order to assess if they corresponded to functionally important structural features. A subset of the 121 closest homologous sequences was selected from the HMM results to create a dataset manageable by the ConSurf server. The selection was made by using HMM hits below >10e−120 corresponding to blast e-value ∼1e−90 and resulted in a set of homologs sharing the two-domain architecture of RiCE17. The conservation pattern was projected on the RiCE17 protein surface using the ConSurf server (Fig. 3B) (38). It is apparent that both the SGNH and CBM35 domains contribute with conserved residues around the substrate-binding site. The residues Ser41, Asn110, His193, Leu47, Phe144, and Leu192 of the SGNH domain and Trp326 of the CBM35 domain are all conserved and are interacting with the substrate in the active site.
The mannan PUL in R. intestinalis encodes an extracellular mannanase, RiGH26, which breaks down mannan into oligosaccharides, which are then internalized via an adenosine triphosphate (ATP) binding cassette (ABC) transporter, deacetylated by the esterases, and further metabolized (22). To approximate these conditions, mannans used in activity testing were hydrolyzed with RiGH26 prior to deacetylation. The two esterases were active on a wide range of mannans but not on structurally similar substrates such as acetylated xylan, cellulose monoacetate, or chitin oligosaccharides (SI Appendix, Fig. S4). A selection of mannose-based substrates was used to examine the impact of galactosylations (Norway spruce mannan, Fig. 4A), mannose units with multiple acetylations (A. vera mannan), and different acetylation distributions (konjac mannan, and its chemically acetylated version, See SI Appendix, Fig. S4) on enzyme activity. Neither RiCE2 nor RiCE17 were active on birch xylan (SI Appendix, Fig. S4F) [as well as its hydrolysate produced with a GH10 xylanase from R. intestinalis (23)], cellulose monoacetate, or chitopentaose (SI Appendix, Fig. S4 D and E). Despite the annotation as a xylan esterase, the RiCE2 completely lacked activity on xylan. This may be due to subtle structural features of the RiCE2’s active site that affect the substrate specificity. The activity data presented herein support this notion and demonstrate that both RiCE2 and RiCE17 have evolved to specifically catalyze deacetylation of various mannans. The apparent substrate specificity for the two esterases was the same on mannans, glucomannans, and galactoglucomannans: there was a partial deacetylation seen whenever one of the esterases was used on its own (red and blue traces Fig. 4A and SI Appendix, Fig. S4 A–C). When the two esterases were used together, a complete deacetylation was observed (purple traces Fig. 4A and SI Appendix, Fig. S4 A–C).
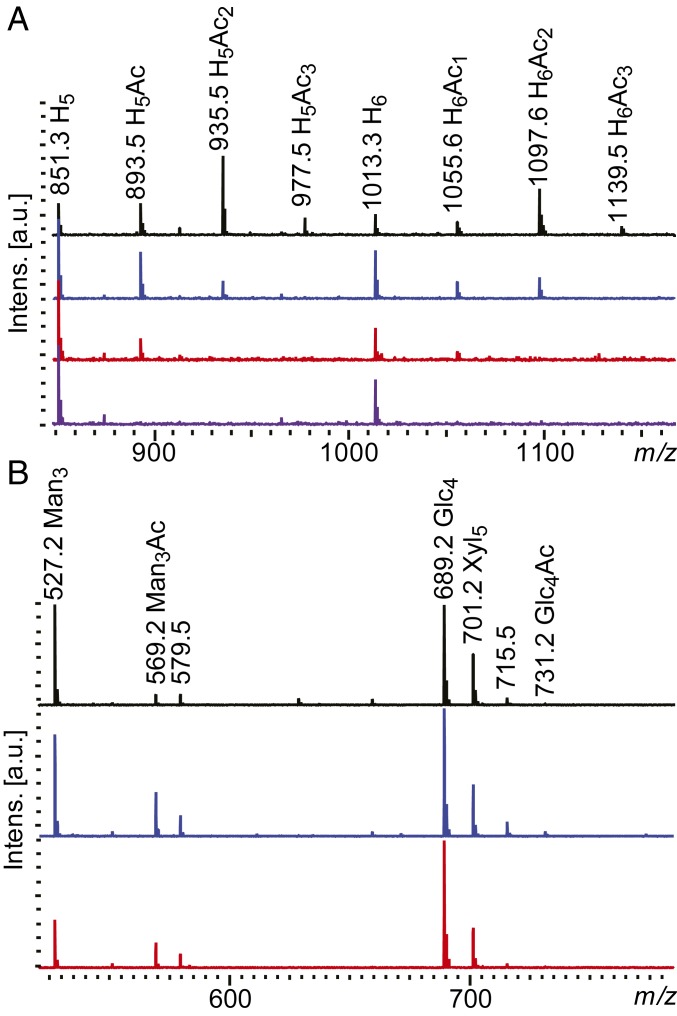
Fig. 4.
MALDI-ToF spectra of deacetylation and transesterification reactions. (A) Ten milligrams per milliliter of spruce GGM hydrolysate (untreated in black) was treated with RiCE2 (in blue), RiCE17 (in red), and the two enzymes in combination (purple). When either of the esterases was used, a decrease in the intensities of peaks signifying acetylated oligosaccharides was observed. Using the two enzymes in combination lead to a complete deacetylation of spruce mannan hydrolysate. (B) Transesterification of a mixture of 1 mg/mL of mannotriose, cellotetraose, and xylopentaose with a mixture of vinyl acetate, propionate, and butyrate. No spontaneous esterification was observed (black trace). Out of all possible substrate combinations, both RiCE2 (in blue) and RiCE17 (in red) produced acetylated mannotriose as the primary product. RiCE2 produced a minor amount of acetylated cellotetraose (m/z 731) as a by-product. H-, hexose; Glc-, glucose; Man-, mannose; Xyl-, xylose; Ac-, acetylation.
Commercially available and in-house produced substrates contain a wide range of acetylations present in a variety of positions on a mannose unit, and different locations in the oligosaccharide chain, making it challenging to precisely determine the specificity of esterases. In transesterification reactions, nonacetylated substrates are treated with esterases in the presence of an appropriate acyl donor, resulting in an amount of highly specific acetylated oligosaccharides consistent with the free energy of the reaction. Transesterification with vinyl acetate has been used as a measure of substrate specificity of acetyl esterases before (39). To further explore the substrate specificity of Roseburia esterases, we used vinyl-acetate, -propionate, and -butyrate as acyl donors. Both enzymes were able to transacetylate and transpropylate (although to a much lesser extent than acetylate) but not transbutylate mannotriose (Fig. 4 and SI Appendix, Fig. S5). Both esterases were able to transacetylate all mannose-based substrates tested, including manno-oligosaccharides DP 2 to 6, 61-α-d-galactosyl-mannotriose, 63, 64-α-d-galactosyl-mannopentaose (SI Appendix, Fig. S6).
As a means of demonstrating specificity, transacetylation experiments were performed on a mixture of mannotriose, cellotetraose, and xylopentaose, with vinyl-acetate, -propionate, and -butyrate as ester donors. Acetylated mannotriose was the only product observed in the matrix-assisted laser desorption ionization-time of flight (MALDI-ToF) spectra (Fig. 4B), clearly indicating the mannan acetylesterase specificity of RiCE17 and RiCE2.
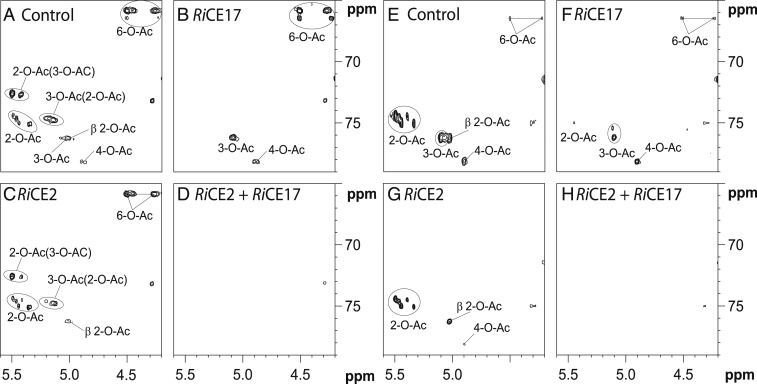
The partial deacetylation observed in Fig. 4 and SI Appendix, Fig. S4 warranted more detailed analysis of the deacetylation preferences of the esterases. NMR analysis (Fig. 5 and SI Appendix, Table S1) revealed that the partial deacetylation seen in RiCE17-treated samples was due to the fact that RiCE17 exclusively removed all 2-O-acetylations from AV and Norway spruce GGM (Fig. 5 B and F, respectively). RiCE2 removed the 3-O- and 4-O-acetylations as well as some of the 6-O-acetylations from single acetylated mannose units in the same substrates (Fig. 5 C and G). Importantly, in the AV samples which contained a high number of double acetylated mannose, RiCE2 was not able to remove the 3-O- and 6-O-acetylations present on the same mannose as a 2-O-acetylation (Fig. 5C). Adding RiCE17 to the solution of AV after the RiCE2 enzymatic treatment and vice versa lead to a complete deacetylation of both substrates (Fig. 5 D and H), indicating that the two esterases have interdependent specificities, and are both required for complete deacetylation of mannans.
Fig. 5.
The two-dimensional (2D)1H-13C heteronuclear single quantum correlation (HSQC) NMR spectra for area of interest of AV mannan RiGH26 hydrolysate (A–D) and Norway spruce mannan RiGH26 hydrolysate (E–H) treated with the esterases. (A) AV hydrolysate without enzyme addition, contained a high degree of acetylation on all possible positions, and a high degree of double acetylation. (B) When treated with RiCE17, the peaks corresponding to 2-O-acetylations disappear, and the shift values for peaks corresponding to 3-O- and 6-O-acetylations change due to the removal of 2-O-acetylations from double acetylated mannoses. (C) Treatment with RiCE2 removed all of the nonreducing end 4-O-acetylations, the majority of 3-O-acetylations, and some of the 6-O-acetylations. (D) Treatment with both esterases at the same time removed all acetylations. (E) Spruce mannan hydrolysate without enzyme addition contained prevalently 2-O-, some 3-O- acetylations, and a lower degree of 4-O- and 6-O-acetylations. (F and G) Both enzymes exhibited similar activity on spruce mannan as on AV. (H). Acetylations in parenthesis signify the adjacent acetylations on double acetylated mannose (2-O-Ac(3-O-Ac) - -2-O-acetylation adjacent to a -3-O-acetylation). Chemical shifts included in SI Appendix, Table S1.
RiCE2 is a broad specificity esterase capable of removing 3-O-, 4-O-, and 6-O-acetylations, although its activity is limited by the presence of 2-O-acetylations as one of multiple acetylations on a single mannose. This dependence of RiCE2 on RiCE17 was also observed in the reaction rates calculated from time-resolved NMR analysis of reactions with AV as the substrate. Reaction rates showed that RiCE2 was much slower when 2-O-acetylations were present (Table 1). The presence of 3-O- and 6-O- acetylations on the same mannose residues containing 2-O-acetylation did not affect the apparent kcat of RiCE17, while the turnover rate of RiCE2 more than doubled when 2-O-acetylations were removed (Table 1). In vivo, the enzyme pair would act in complement, with the specialist RiCE17 removing the 2-O-acetylations that limit the activity of the generalist RiCE2.
Table 1.
Turnover rates and specific activities on oligosaccharides and pNP-acetate
| kcat [s−1] | Specific activity | |
| Time resolved NMR data on A. vera GGM | ||
| RiCE17 | 4.8 | |
| RiCE2 after RiCE17 | 4.7 | |
| RiCE2 | 1.8 | |
| RiCE17 after RiCE2 | 4.5 | |
| Deacetylation of Norway spruce GGM | ||
| RiCE17 | 73.5 | 1.7 |
| RiCE2 | 76.7 | 1.9 |
| RiCE2 + RiCE17 | 41.7 | — |
| Deacetylation of pNP-acetate | ||
| RiCE17 | .93 | |
| RiCE2 | 40.00 |
Turnover rate in time-resolved NMR analysis of deacetylation determined based on the acetate released in the initial 15 min of reaction. Two identical samples of AV mannan hydrolyzed with RiGH26 mannanase were prepared and treated with: 1) 62.5 nM loading of RiCE2 for 16 h, at 298.1 °K, then 10 nM loading of RiCE17; 2) 10 nM loading of RiCE17 for 16 h, then 62.5 nM loading of RiCE2. Turnover rate and specific activity (nanomole acetate/s/µg enzyme) of esterases on Norway spruce GGM RiGH26 hydrolysate were determined based on the amounts of acetate released from samples of 100 mg/mL of substrate in 30 min. Turnover rates of both esterases on pNP-acetate appear lower than on oligosaccharides. Notably, the difference between catalytic activity on AV mannan and Norway spruce GGM cannot be directly compared due to largely different reaction conditions (NMR experiments on AV mannan were run at 298.1 °K/25 °C and pD 5.9 in a 5 mm NMR tube whereas the Norway spruce GGM reactions were run at 30 °C, pH 7.0 and 600 rpm shaking).
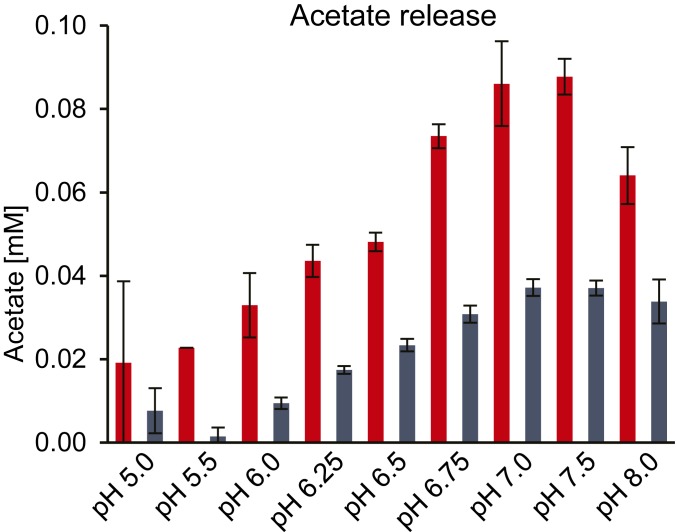
Activity assays using pNP-acetate, which is a commonly used substrate when screening for esterase activities showed that both esterases appear to be most active near neutral pH (Fig. 6). Notably, pNP-acetate was a highly inadequate substrate for RiCE17 with the apparent turnover rate nearly two orders of magnitude lower than on Norway spruce GGM (Table 1). This is not surprising, considering the topology of the RiCE17 active site and the structural difference between the planar phenol ring of pNP and the axially oriented 2-O-acetylation on d-Manp; however, this result highlights the necessity for careful selection of substrates and methods when measuring esterase activity.
Fig. 6.
The pH optima of RiCE17 (in red) and RiCE2 (in blue) as determined by the amounts of acetate released from pNP-acetate at different pH. Error bars indicate SD between triplicates.
To further investigate the different catalytic activity with pNP-acetate observed for RiCE17 and RiCE2, molecular dynamics simulations were employed to map the interaction between this artificial substrate and the enzyme active sites. In SI Appendix, Fig. S7 A–C, the measured atomic fluctuations (B-factors) indicate that when RiCE17 is bound to Man4, the CBM35 domain displays less disorder (SI Appendix, Fig. S7A) compared to the RiCE17-pNP-acetate complex (SI Appendix, Fig. S7B). No force was applied to keep the Man4 molecule in the active site during the 100 ns simulations; however, it was necessary to pull the pNP-acetate molecule toward the catalytic Ser residue to keep it associated with the active site in both the RiCE17 model (SI Appendix, Fig. S7B) and the CjCE2B model (SI Appendix, Fig. S7C). When pulling the pNP-acetate into the active site of RiCE17 it appeared to increase flexibility of the CBM35 domain compared to the crystal structure (SI Appendix, Fig. S7B). On the other hand, pulling of the pNP-acetate into the active site of CjCE2B (SI Appendix, Fig. S7C), no increased disorder of the active site surroundings were observed when comparing to the crystal structure. Assuming that CjCE2B, a homolog to RiCE2, is a suitable model for RiCE2, the difference in activity between the CE17 and CE2 enzymes can be explained by easier access to the active site for pNP-acetate in the CE2 enzyme compared to the CE17 enzyme. The loss of activity toward pNP-acetate when the CBM35 domain of RiCE17 is removed indicates that the shape of the active site is deformed by this modification. This could occur e.g., by disruption of the interactions between the amino acid side chains constituting the catalytic triad, or the removal of the CBM resulting in a flat and nonfunctional substrate-binding surface.
Both esterases showed near identical kcat and specific activities (Table 1) on RiGH26 hydrolyzed Norway spruce GGM substrate. In deacetylation rate measurements at pH 7.0 and 35 °C (approximating intracellular conditions of a gut commensal bacterium), at equal loadings (25 nM final concentration) of each esterase, the apparent total rate of deacetylation was lower than when 50 nM of either esterase was used on its own. The lower resulting rate of deacetylation suggests that the esterases were competing for substrate, an effect observed previously in lignocellulose-degrading enzyme mixtures (40). Reaction rates obtained in time-resolved NMR showed an increase in the RiCE2 activity after removal of 2-O-acetylations. This effect was not observed in the activity on Norway spruce GGM (Table 1) since the substrate used had far fewer double acetylated mannose units.
There is a growing body of evidence that acetylations on hemicellulose are prone to migration on sugar rings and also across glycosidic linkages (41, 42). This phenomenon requires particular attention when characterizing acetylesterases, since the distribution of acetylations will change under certain conditions. Elevated pH and temperature (>90 °C) are known to induce acetyl migration on mannose and xylose (41, 43, 44). In experiments conducted on monosaccharides, it has been shown that the acetyl groups migrate at pH > 6.0. In both d-glucose and d-galactose, acetylations initially present in the 2-O-position appear to move in a “clockwise” (2-O-→3-O-→4-O-) direction at pD >7 (43, 45), while at pD <3.0, d-galactose was shown to deacetylate without the acetyl group migrating (43). Notably, we observed that migration happens also on the polymers and at even less severe conditions. This phenomenon changes the properties of mannans as substrates and as biorefining feedstocks. At pH 5.9 and 20 °C, RiCE17 transacetylated all three Manp units of mannotriose, exclusively in the 2-O- position (SI Appendix, Fig. S8A) without any observed migration occurring. Exposure to just 60 °C for 1 h at pD 5.9 (SI Appendix, Fig. S8B), incubation at pD 7.4 at room temperature (SI Appendix, Fig. S8C), or incubation pD 7.4 at 60 °C (SI Appendix, Fig. S8D) caused a decrease in the signals for 2-O-acetylations and an appearance of signals for 3-O-, 4-O-, and 6-O-acetylations, the latter two only in the nonreducing end mannose. At these conditions, the acetylations on the nonreducing end migrated in the same clockwise 2-O- → 6-O- direction as described before on Galp monosaccharides (43). The glycosidic bond prevents migration from 3-O-→ 6-O- and thus limits the migration from 2-O-/3-O-→6-O- for the reducing end and the intrachain Manp. Migration progressed until an apparent equilibrium of acetyl distribution was reached (SI Appendix, Fig. S8), with a proportion of the acetylations remaining on the 2-O- position. Using the 2-O- deacetylation specificity of RiCE17, we were able to produce a sample of Norway spruce GGM with all 2-O-acetylations removed. We examined the acetylation levels in solutions of Norway spruce GGM exposed to migration-invoking conditions, then treated with RiCE17 to remove newly accumulated 2-O-acetylations. Both pH increase (SI Appendix, Fig. S9) and heating to 60 °C for 1 h (SI Appendix, Fig. S10) induced a redistribution of acetylations resulting in new 2-O-acetylations. Treatment with RiCE17 and repeated migration resulted in a complete removal of acetylations by only removing the 2-O- bound acetyls. This indicates that the acetyl migration does not proceed in a particular direction, but rather is a redistribution process. A detailed description of acetyl migration is provided in the SI Appendix.
Concluding Remarks.
This study provides detailed insight into enzymatic deacetylation of β-mannans by the abundant human gut commensal bacterium R. intestinalis. The deacetylation apparatus of R. intestinalis consists of the highly specialized 2-O-acetylation specific RiCE17, and RiCE2 that removes all remaining types of acetylations. Both enzymes are necessary for complete deacetylation, and their activities are complementary. Both esterases have a two-domain structure with a SGNH superfamily hydrolytic domain and an accessory domain—a galactose-binding superfamily domain in RiCE2 and a CBM35 in RiCE17.
Selective activity of RiCE17 was instrumental in our study of acetyl migration on manno-oligosaccharides. We have demonstrated that acetyl migration proceeds toward the 2-O- position when the acetylations in that position are removed from an oligosaccharide. Our results show explicitly that the choice of substrate and reaction conditions are crucial for studying the specificity of hemicellulose esterases.
The potential for the industrial application of this enzyme pair is quite apparent. With the activity on gluco- and galacto(gluco)mannans, the pair could be applied to various mannan-containing feedstocks to reduce the recalcitrance of mannans, modify the viscosity of mannans in solution, or to rationally design oligosaccharides with distinct esterification patterns via specific deacetylation or transesterification.
Materials and Methods
Enzymes.
Recombinant RiCE17 (ROSINTL182_05471) and RiCE2 (ROSINTL182_05473) were produced in Escherichia coli BL21 Star (DE3) as described previously (22). Truncated versions of RiCE17 and seleno-l-methionine substituted RiCE17 were produced as described in SI Appendix, Materials and Methods.
Substrates.
Galactoglucomannan from Norway spruce (P. abies) and glucuronoxylan from Birch (Betula pubescens) were produced in-house from dried wood chips. The wood was milled into <2 mm particles, and steam exploded at 200 °C (10 min reactor residence time) in 5 to 6 kg batches. The liquid soluble fraction containing the hemicelluloses was extracted by washing the steam-exploded material with MilliQ water in a 50 µm pore WE50P2VWR bag filter (Allied filter Systems). The liquid fraction of hemicellulose was then filtered through a 5 kDa spiral wound polysulphone/polyethersulphone polyester ultrafiltration membrane (GR99PE, Alfa Laval) using a pilot scale filtration system Model L (GEA Filtration, Denmark). The fraction retained by the membrane was concentrated, collected, and freeze dried to become the Norway spruce GGM sample. Birch xylan was produced according to the protocol described in Biely et al. (47). The 61-α-d-galactosyl-mannotriose; 63, 64-α-d-galactosyl-mannopentaose, penta-N-acetylchitopentaose, mannotriose, mannotetraose, mannopentaose, konjac and carob mannans were purchased from Megazyme. A. vera mannan (Acemannan) was purchased from Elicityl. Norway spruce GGM and A. vera mannan were hydrolyzed with RiGH26 in an unbuffered solution. Samples were then filtered through a prewashed 1 mL Amicon Ultracel 3kDa ultrafiltration device (Merck KGaA) to remove the hydrolase and freeze dried.
Activity Analysis.
Activities of the enzymes were tested by adding 1 µM of enzyme to 10 mg/mL solutions of carbohydrate substrates listed above. All substrate solutions were prepared with 20 mM sodium phosphate pH 5.9 to prevent acetyl migration. Reactions were incubated overnight in 20 mM sodium phosphate pH 5.9 at 30 °C with 700 rpm shaking. In some analyses, MilliQ water was used instead of buffer to reduce background signals in MALDI-ToF mass spectrometry (MS) as both enzymes were found to be active without buffers.
Temperature and pH Optima Using 4-Nitrophenyl Acetate (pNP-Acetate).
In order to determine the pH and temperature optima for the enzymes, reactions with 0.5 mM pNP-acetate were prepared using 50 mM sodium phosphate buffer (Sigma-Aldrich) in the pH range 5.5 to 8.0 (Sigma-Aldrich). The pH optimum reactions were incubated at 25 °C. Due to the difference in deacetylation rate of pNP-acetate by the two enzymes, 1 nM final concentration of RiCE2 and 0.1 µM of RiCE17 were used in the pNP-acetate experiments. Standard plots of 4-nitrophenol (p-nitrophenol) were prepared at each pH. To determine the optimum pH, 99 µL of sample mixture containing 1 mM of pNP-acetate in each of the buffers were added to the wells of a 96-well plate. Enzyme solution (1 µL) was then added to the sample mixture and the reaction followed by measuring the absorbance at 405 nm at 1-min intervals. All experiments were performed in triplicate, with two blanks for each condition set.
MALDI-ToF Analysis.
MALDI-ToF analysis of hydrolysis products was conducted on an UltraFlextreme MALDI-ToF instrument (Bruker Daltonics GmbH) equipped with a nitrogen 337 nm laser beam. Samples were prepared by mixing 2 μL of a 9 mg/mL solution of 2,5-dihydroxybenzoic acid (Sigma-Aldrich) in 30% acetonitrile (VWR) to an MTP 384 ground steel target plate (Bruker Daltonics GmbH) and 1 µL of hydrolysate (0.1 to 1 mg/mL). Sample drops were then dried under a stream of warm air.
Crystallography, preparative high-performance liquid chromatography (HPLC), quantification of acetylation, transesterification, and NMR experiments are described in SI Appendix, Materials and Methods.
Data Availability Statement.
All data required to evaluate the paper’s conclusions are present in the paper and/or the SI Appendix.
Supplementary Material
Acknowledgments
Cellulose monoacetate with a degree of acetylation of 0.6 was a kind gift from Qi Zhou (KTH Royal Institute of Technology, Stockholm). A chemically acetylated konjac glucomannan prepared according to Bååth et al. (46) was kindly provided by Francisco Vilaplana. Norwegian Research council grant nos. 244259, 240967, 208674/F50, 226244, and 226247 supported this work.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. W.A.v.d.D. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank archive, http://www.wwpdb.org/ (RiCE17: PDB ID 6HFZ; and RiCE17 in complex with mannopentaose: PDB ID 6HH9).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1915376117/-/DCSupplemental.
References
- 1.Strahl B. D., Allis C. D., The language of covalent histone modifications. Nature 403, 41–45 (2000). [DOI] [PubMed] [Google Scholar]
- 2.Ragland S. A., Criss A. K., From bacterial killing to immune modulation: Recent insights into the functions of lysozyme. PLoS Pathog. 13, e1006512 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sonawane V. C., Enzymatic modifications of cephalosporins by cephalosporin acylase and other enzymes. Crit. Rev. Biotechnol. 26, 95–120 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Gille S., Pauly M., O-acetylation of plant cell wall polysaccharides. Front. Plant Sci. 3, 12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biely P., Microbial carbohydrate esterases deacetylating plant polysaccharides. Biotechnol. Adv. 30, 1575–1588 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Willfor S., et al. , Characterisation of water-soluble galactoglucomannans from Norway spruce wood and thermomechanical pulp. Carbohydr. Polym. 52, 175–187 (2003). [Google Scholar]
- 7.Lundqvist J., et al. , Isolation and characterization of galactoglucomannan from spruce (Picea abies). Carbohydr. Polym. 48, 29–39 (2002). [Google Scholar]
- 8.Simões J., Nunes F. M., Domingues P., Coimbra M. A., Domingues M. R., Mass spectrometry characterization of an aloe vera mannan presenting immunostimulatory activity. Carbohydr. Polym. 90, 229–236 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Singh S., Singh G., Arya S. K., Mannans: An overview of properties and application in food products. Int. J. Biol. Macromol. 119, 79–95 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Cosgrove D. J., Jarvis M. C., Comparative structure and biomechanics of plant primary and secondary cell walls. Front. Plant Sci. 3, 204 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Willfor S., Sundberg K., Tenkanen M., Holmbom B., Spruce-derived mannans–A potential raw material for hydrocolloids and novel advanced natural materials. Carbohydr. Polym. 72, 197–210 (2008). [Google Scholar]
- 12.Du X. Z., Li J., Chen J., Li B., Effect of degree of deacetylation on physicochemical and gelation properties of konjac glucomannan. Food Res. Int. 46, 270–278 (2012). [Google Scholar]
- 13.Mudgil D., Barak S., Khatkar B. S., Guar gum: Processing, properties and food applications-A review. J. Food Sci. Technol. 51, 409–418 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tester R. F., Al-Ghazzewi F. H., Mannans and health, with a special focus on glucomannans. Food Res. Int. 50, 384–391 (2013). [Google Scholar]
- 15.Martens E. C., Chiang H. C., Gordon J. I., Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe 4, 447–457 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reeves A. R., Wang G. R., Salyers A. A., Characterization of four outer membrane proteins that play a role in utilization of starch by Bacteroides thetaiotaomicron. J. Bacteriol. 179, 643–649 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cockburn D. W., Koropatkin N. M., Polysaccharide degradation by the intestinal microbiota and its influence on human health and disease. J. Mol. Biol. 428, 3230–3252 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Flint H. J., Scott K. P., Duncan S. H., Louis P., Forano E., Microbial degradation of complex carbohydrates in the gut. Gut Microbes 3, 289–306 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simões J., Nunes F. M., Domingues Md R. M., Coimbra M. A., Structural features of partially acetylated coffee galactomannans presenting immunostimulatory activity. Carbohydr. Polym. 79, 397–402 (2010). [Google Scholar]
- 20.Lahaye M., Quemener B., Causse M., Seymour G. B., Hemicellulose fine structure is affected differently during ripening of tomato lines with contrasted texture. Int. J. Biol. Macromol. 51, 462–470 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Lloyd-Price J., et al. , Strains, functions and dynamics in the expanded human microbiome project. Nature 550, 61–66 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.La Rosa S. L., et al. , The human gut Firmicute Roseburia intestinalis is a primary degrader of dietary β-mannans. Nat. Commun. 10, 905 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leth M. L., et al. , Differential bacterial capture and transport preferences facilitate co-growth on dietary xylan in the human gut. Nat. Microbiol. 3, 570–580 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Lombard V., et al. , A hierarchical classification of polysaccharide lyases for glycogenomics. Biochem. J. 432, 437–444 (2010). [DOI] [PubMed] [Google Scholar]
- 25.Nakamura A. M., Nascimento A. S., Polikarpov I., Structural diversity of carbohydrate esterases. Biotechnol. Res. Innovation 1, 35–51 (2017). [Google Scholar]
- 26.Montanier C., et al. , The active site of a carbohydrate esterase displays divergent catalytic and noncatalytic binding functions. PLoS Biol. 7, e71 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Topakas E., et al. , Carbohydrate esterases of family 2 are 6-O-deacetylases. FEBS Lett. 584, 543–548 (2010). [DOI] [PubMed] [Google Scholar]
- 28.Tenkanen M., Puls J., Ratto M., Viikari L., Enzymatic deacetylation of galactoglucomannans. Appl. Microbiol. Biotechnol. 39, 159–165 (1993). [Google Scholar]
- 29.Tenkanen M., Thornton J., Viikari L., An acetylglucomannan esterase of Aspergillus oryzae; purification, characterization and role in the hydrolysis of O-acetyl-galactoglucomannan. J. Biotechnol. 42, 197–206 (1995). [DOI] [PubMed] [Google Scholar]
- 30.Finn R. D., et al. , InterPro in 2017-beyond protein family and domain annotations. Nucleic Acids Res. 45, D190–D199 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Altschul S. F., et al. , Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lansky S., et al. , A unique octameric structure of Axe2, an intracellular acetyl-xylooligosaccharide esterase from geobacillus stearothermophilus. Acta Crystallogr. D Biol. Crystallogr. 70, 261–278 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Krissinel E., Henrick K., Multiple alignment of protein structures in three dimensions. Lect. Notes Comput. Sci. 3695, 67–78 (2005). [Google Scholar]
- 34.Williams A. H., et al. , Visualization of a substrate-induced productive conformation of the catalytic triad of the Neisseria meningitidis peptidoglycan O-acetylesterase reveals mechanistic conservation in SGNH esterase family members. Acta Crystallogr. D Biol. Crystallogr. 70, 2631–2639 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Montanier C., et al. , Evidence that family 35 carbohydrate binding modules display conserved specificity but divergent function. Proc. Natl. Acad. Sci. U.S.A. 106, 3065–3070 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boraston A. B., Bolam D. N., Gilbert H. J., Davies G. J., Carbohydrate-binding modules: Fine-tuning polysaccharide recognition. Biochem. J. 382, 769–781 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hekmat O., Tokuyasu K., Withers S. G., Subsite structure of the endo-type chitin deacetylase from a deuteromycete, colletotrichum lindemuthianum: An investigation using steady-state kinetic analysis and MS. Biochem. J. 374, 369–380 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ashkenazy H., et al. , ConSurf 2016: An improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 44, W344–W350 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kremnicky L., Mastihuba V., Cote G. L., Trichoderma reesei acetyl esterase catalyzes transesterification in water. J. Mol. Catal. B Enzym. 30, 229–239 (2004). [Google Scholar]
- 40.Andersen N., et al. , Hydrolysis of cellulose using mono-component enzymes shows synergy during hydrolysis of phosphoric acid swollen cellulose (PASC), but competition on avicel. Enzyme Microb. Technol. 42, 362–370 (2008). [Google Scholar]
- 41.Mastihubová M., Biely P., Lipase-catalysed preparation of acetates of 4-nitrophenyl β-D-xylopyranoside and their use in kinetic studies of acetyl migration. Carbohydr. Res. 339, 1353–1360 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Lassfolk R., et al. , Acetyl group migration across the saccharide units in oligomannoside model compound. J. Am. Chem. Soc. 141, 1646–1654 (2019). [DOI] [PubMed] [Google Scholar]
- 43.Roslund M. U., et al. , Acyl group migration and cleavage in selectively protected β-d-galactopyranosides as studied by NMR spectroscopy and kinetic calculations. J. Am. Chem. Soc. 130, 8769–8772 (2008). [DOI] [PubMed] [Google Scholar]
- 44.Kabel M. A., de Waard P., Schols H. A., Voragen A. G., Location of O-acetyl substituents in xylo-oligosaccharides obtained from hydrothermally treated Eucalyptus wood. Carbohydr. Res. 338, 69–77 (2003). [DOI] [PubMed] [Google Scholar]
- 45.Brecker L., Mahut M., Schwarz A., Nidetzky B., In situ proton NMR study of acetyl and formyl group migration in mono-O-acyl D-glucose. Magn. Reson. Chem. 47, 328–332 (2009). [DOI] [PubMed] [Google Scholar]
- 46.Arnling Bååth J., et al. , Mannanase hydrolysis of spruce galactoglucomannan focusing on the influence of acetylation on enzymatic mannan degradation. Biotechnol. Biofuels 11, 114 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Biely P., et al. , Mode of action of acetylxylan esterases on acetyl glucuronoxylan and acetylated oligosaccharides generated by a GH10 endoxylanase. Biochim. Biophys. Acta 1830, 5075–5086 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data required to evaluate the paper’s conclusions are present in the paper and/or the SI Appendix.