Significance
Inactivation of potassium channels controls its mean open time and provides exquisite control over biological processes. In the highly conserved C-type inactivation process, opening of the activation gate causes subsequent inactivation. We test whether the open state of the channel simply has a poor ability to bind the K+ ion. Previously, activated and inactivated states were stabilized using truncations or a significant pH drop. Here, we use the H25R/E118A constitutively open mutant of KcsA and also observe a large drop in potassium binding affinity. This provides strong evidence that channel opening causes an allosteric loss of ion affinity, and that the central feature of this universal channel inactivation process is loss of ion affinity at the selectivity filter.
Keywords: solid-state NMR, ion channels, transmembrane allostery, KcsA
Abstract
Transmembrane allosteric coupling is a feature of many critical biological signaling events. Here we test whether transmembrane allosteric coupling controls the potassium binding affinity of the prototypical potassium channel KcsA in the context of C-type inactivation. Activation of KcsA is initiated by proton binding to the pH gate upon an intracellular drop in pH. Numerous studies have suggested that this proton binding also prompts a conformational switch, leading to a loss of affinity for potassium ions at the selectivity filter and therefore to channel inactivation. We tested this mechanism for inactivation using a KcsA mutant (H25R/E118A) that exhibits an open pH gate across a broad range of pH values. We present solid-state NMR measurements of this open mutant at neutral pH to probe the affinity for potassium at the selectivity filter. The potassium binding affinity in the selectivity filter of this mutant, 81 mM, is about four orders of magnitude weaker than that of wild-type KcsA at neutral pH and is comparable to the value for wild-type KcsA at low pH (pH ≈ 3.5). This result strongly supports our assertion that the open pH gate allosterically affects the potassium binding affinity of the selectivity filter. In this mutant, the protonation state of a glutamate residue (E120) in the pH sensor is sensitive to potassium binding, suggesting that this mutant also has flexibility in the activation gate and is subject to transmembrane allostery.
The extracellular selectivity filter of potassium channels selectively allows potassium ions to cross biological membranes when activated at the intracellular gate by an external stimulus such as voltage or nucleotide binding. Generally, however, the channel spontaneously stops conducting even if potassium ions and activation stimuli are still present (1, 2). This process typically occurs on a millisecond-to-second timescale after activation and is termed “C-type inactivation.” Inactivation is notably modulated by permeant ions and pore blockers (3). Understanding the structural and mechanistic basis of C-type inactivation is expected to create insights into regulation of biomedically important channels such as the HERG channel that regulates the human heart and is significant for drug development (4–6).
KcsA is a prototypical bacterial potassium channel which is pH-activated. The pH “gate” is located at the intracellular face of the channel, formed by a group of key ionizable residues including H25, E118, and E120 (7–9). Like its more complex eukaryotic homologs, KcsA undergoes inactivation on the second timescale after activation by an intercellular pH drop (8, 9). In previous studies, our group and others hypothesized that the site of inactivation is the selectivity filter, where potassium ions are bound and conducted through the membrane (1, 10–12). This mechanism is termed activation-coupled inactivation and involves an allosteric coupling between the selectivity filter and activation gate (e.g., the pH gate in KcsA) that leads to a decrease in potassium affinity at the selectivity filter after activation. A direct measurement of the potassium ion affinity at the selectivity filter by solid-state NMR (SSNMR) was carried out on KcsA embedded in a hydrated lipid environment (13). The potassium ion affinity reduced upon acidification from Kapp = 4 ± 1 μM at pH 7.5 to Kapp = 14 ± 1 mM at pH 3.5. Consequently, following activation of the channel at low pH, a reduction in potassium affinity at the selectivity filter is expected to explain ion loss and conformational change at the selectivity filter, which, in turn, defeats efficient transmission. This is consistent with the electrophysiology studies showing that increasing extracellular potassium ion concentration could decrease the rate of inactivation (10, 11).
To test this model for allosteric coupling between the pH gate and the selectivity filter, we characterize a KcsA mutant that is constitutively open at neutral pH. The pH gate residues H25, E118, and E120 have been suggested to form complex intersubunit and intrasubunit interactions at the cytoplasmic ends of the TM1 and TM2 transmembrane helices (14, 15). A triple mutant H25R/E120A/E118A was shown to have the activation gate open across a broad range of pH values from 4 to 9 (16). Separately, a solution NMR study also showed that H25 is key to the function of the pH gate (17). Subsequently, it was shown that only two mutations (to make the double mutant H25R/E118A) are sufficient to render the channel open at neutral pH (18). Here we use the constitutively open H25R/E118A mutant to test whether the open channel indeed loses its ability to keep potassium ion loaded in the selectivity filter, and to control for nonspecific effects of the low pH used in other studies.
Results
The pH Gate Mutant Adopts Similar Conformations as Compared with Wild Type, for Both Conducting and Collapsed Forms of the Selectivity Filter.
NMR chemical shifts are precise indicators for protein structural and conformational changes. We conducted SSNMR experiments to investigate the structure of the H25R/E118A mutant of KcsA in a hydrated lipid bilayer environment (DOPE:DOPS = 9:1) with a 1:1 (wt/wt) protein to lipid ratio, as in our prior studies of wild-type KcsA (10, 11, 13, 19). The protein is well folded and displays high-quality NMR spectra at both the low (3.5) and high (7.5) pH values, and most chemical shift markers are similar to the corresponding markers for wild type (SI Appendix, Figs. S1 and S2). This confirms that the H25R/E118A mutant is well folded in lipid bilayers. Moreover, like the wild type, the H25R/E118A mutant displays two conformations at the selectivity filter. NMR chemical shifts report on the conformational change that occurs during K+ binding for the wild type, distinguishing the conductive and collapsed conformations seen in various crystal structures, as has been extensively discussed in prior literature. Both states can be observed at low pH and neutral pH, depending on potassium ion concentration. This indicates that the H25R/E118A mutant is well behaved and has the same conformational behavior at the selectivity filter as the wild-type channel.
Opening the pH Gate by Mutation Significantly Lowers Potassium Affinity at the Selectivity Filter at Neutral pH.
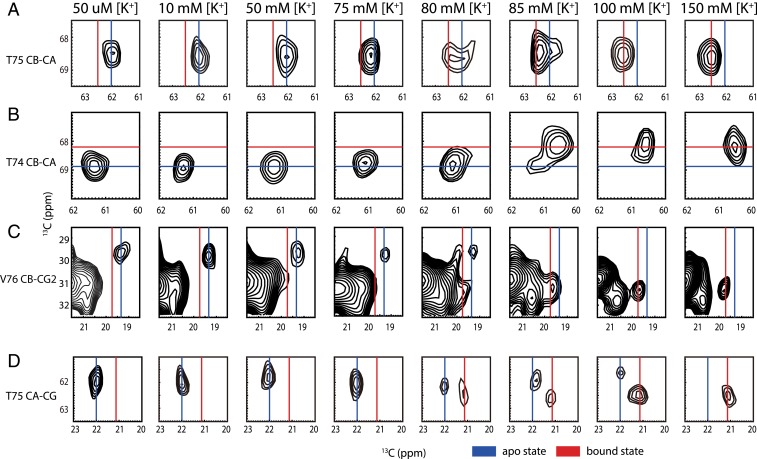
To test for the consequences of an open pH activation gate on function and binding in the selectivity filter, we measured the potassium affinity of the open pH gate channel at neutral pH in H25R/E118A. We used nuclei in residues previously shown to have characteristic chemical shifts in the potassium binding sites, including T75 (CA, CB, CG), T74 (CA, CB, CG), and V76(CB, CG2), to measure the populations of the apo and K+ bound states (SI Appendix, Fig. S3). The two-dimensional (2D) 13C-13C dipolar assisted rotational resonance (DARR) spectra of chemical shift marker residues near the selectivity filter are shown in Fig. 1 for various ambient potassium ion concentrations (at 10, 50, 75, 80, 85, 100, and 150 mM [K+]). The ionic strength was kept constant by compensating with sodium ions, since ionic strength might affect the potassium affinity and function and sodium ions are known to have little interference on potassium channel (20, 21). As is the case for wild-type KcsA, the K+ bound and apo state conformers do not exchange rapidly in this mutant, as evidenced by their resolved peaks when the system is partially titrated (13).
Fig. 1.
[K+] dependence of apo and bound state cross-peaks (marker peaks) near the selectivity filter in 2D 13C-13C correlation spectra of H25R/E118A KcsA at pH 7.5. (A) T75 CB-CA, (B) T74 CB-CA, (C) V76 CB-CG2, and (D) T75 CA-CG show that the marker peaks shift from apo state in low [K+] to bound state in high [K+]. The contour levels of the spectrum were set at 5 times noise level. Marker peaks could be integrated to calculate the population ratio of apo and bound states.
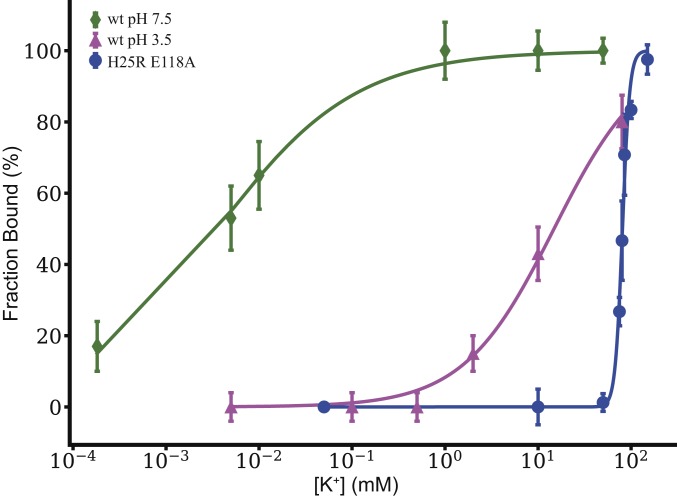
The ratio of potassium-bound KcsA to apo indicated by the NMR spectra is a steep function of ambient [K+] near 80 mM. This titration dependence was fit to a Hill model to extract Kapp (13). When the Hill coefficient was freely varied, Kapp for K+ binding to H25R/E118A KcsA was best fit to 81 ± 1 mM, which is the same order of magnitude as the value for wild-type KcsA at pH 3.5 (14 ± 1 mM) (Fig. 2) (13). These values are in stark contrast with the much tighter affinity for wild-type KcsA at pH 7.5, 4 ± 1 μM. (Details of the simulation of the titration curve are given in SI Appendix, Table S1.) For the H25R/E118A mutant, the transition from the bound to the apo state is steep as compared to that of the wild-type KcsA at pH 3.5, and the best-fit Hill coefficient was >4, in contrast to that for the wild type, which was ∼1. This can be explained as an increase in the cooperativity of the potassium binding process at the selectivity filter in this mutant as compared with the wild type. KcsA has multiple sites for proton binding and for potassium binding, so noncooperative binding is not necessarily expected. Differences in cooperativity with respect to the wild type may be related to the possibility that this mutant’s pH gate is open to a different degree than the wild type, or that it eliminates states with intermediate numbers of ions that occur for the wild type.
Fig. 2.
Potassium affinity of the selectivity filter of KcsA based on NMR is presented as a titration plot, presenting the ratio of bound K+ to bound-plus-apo as a function of ambient [K+]. The H25R/E118A mutant (blue) is compared with previously reported data for wild-type pH 7.5 (green) and wild-type pH 3.5 (magenta) (13). Kapp of the open pH gate mutant was calculated to be 81 ± 1 mM by fitting the data to a Hill binding model, compared with 4 ± 1 μM for wild type at pH 7.5 and 14 ± 1 mM for wild type at pH 3.5 (13). These data show that the H25R/E118A mutant with a constitutively open pH gate exhibits a K+ affinity that is somewhat looser still as compared to the open wild-type KcsA at pH 3.5.
The affinity for this mutant at low pH was probed to test for a nonspecific effect of pH. At 80 mM ambient [K+], the fraction of H25R/E118A channels with K+ bound is ∼40% at both low pH and neutral pH, suggesting that the Kapp of the mutant is relatively pH-insensitive (SI Appendix, Fig. S4) and that nonspecific effects due to pH changes are not large.
In summary, these two very distinct perturbations (pH vs. mutation) appear to affect the thermodynamics at the selectivity filter very similarly, in that they both act to open the channel. This offers strong support for the hypothesis of allosteric activation coupled inactivation.
The pH Gate Is Coupled to Ion Binding in the Constitutively Open Mutant.
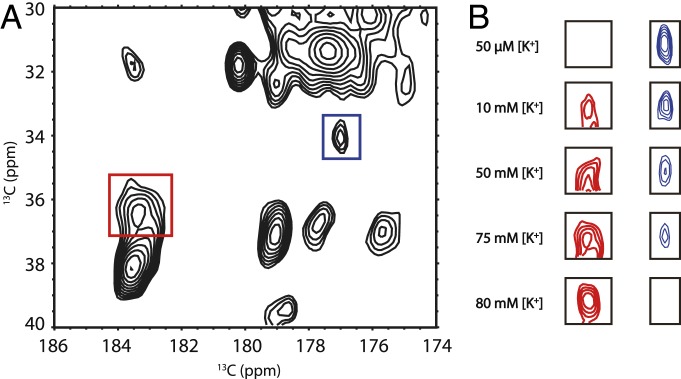
Allosteric coupling between the selectivity filter and the pH gate has been demonstrated in previous work (1, 13, 18, 22–25). Not only does protonation and pH gate opening cause ion loss at the selectivity filter, but changes in ion occupation at the selectivity filter can reciprocally affect the protonation state of the pH gate residues E118 and E120 as well. For wild-type KcsA, the peaks for E118 and E120 in 13C-13C homonuclear SSNMR spectra are congested (11, 13). Thus, after we removed the E118 peak from the spectrum by mutagenesis, we assign the peak at (34.11, 177.30) and (36.15, 183.10) (parts per million [ppm]) to CG−CD peak of residue E120 in the H25R/E118A mutant at high and low [K+], respectively, and we monitored the peak intensity changes as ambient [K+] was increased, as shown in Fig. 3. Changes in the E120 CG−CD peak intensity and position indicate that this residue is protonated at low [K+], and deprotonated at high [K+], while the pH is held constant at a value of 7.5. At 50 μM [K+], E120 CG−CD appears to be essentially fully protonated, while, at 80 mM [K+], the E120 CG−CD appears to be essentially fully deprotonated. Curiously, the titration curve for E120 is not identical to that of the selectivity filter markers, suggesting the existence of K+ ion sites of different affinity. These results show that, in the H25R/E118A KcsA mutant, the pH gate and selectivity gate are allosterically coupled, as in the wild type (11).
Fig. 3.
The effect of [K+] on the E120 residue in the pH gate of the H25R/E118A KcsA mutant at pH 7.5. (A) The 2D 13C-13C correlation spectrum of open pH gate KcsA at 75 mM [K+]. The peaks in the red and blue rectangles are the E120 CG−CD peaks in deprotonated and protonated state, respectively. (B) The protonated and deprotonated state change of the E120 peak across various [K+]. Protonated and deprotonated E120 CG−CD peaks at 50 μM and 10, 50, 75, and 80 mM are shown. At pH 7.5, E120 is protonated at low [K+]; while, at 80 mM [K+] and above, E120 is completely deprotonated. In samples with [K+] larger than 80 mM, the E120 CG−CD peaks are all in the deprotonated state. This protonation state change provides clear evidence for pKa change of the glutamic acid residue.
Electrophysiology of the H25R/E118A/T74S Mutant.
It was previously shown that, although the H25R/E118A mutant is open, it does not conduct under standard electrophysiology conditions (pH 7.0, 100 mM [K+]), because it, like the wild type, inactivates when open. Various mutations can rescue inactivation. For example, E71A prevents inactivation state by stabilizing the conductive conformation of the selectivity filter and is often used as a “background” mutation to study the open activated channel. In recent work (26), we showed that, by manipulating various residues involved in allosteric interactions, we could reduce allosteric coupling and thereby prevent inactivation, providing an alternative “background” mutation for stabilization and studies of the open activated state. (SI Appendix, Fig. S5). These mutations involve a handful of essential participants in allosteric coupling between the activation and inactivation gate that have been characterized using NMR and molecular dynamics (26, 27). T74 is one of the key allosteric participants. The mutation T74S causes a strong reduction in allosteric coupling in KcsA at pH 5.0 and pH 3.5 as shown by NMR, and also reduces the extent of inactivation dramatically, as shown by electrophysiology. Here we show that the T74S/H25R/E118A mutant recovers activity in electrophysiology experiments (SI Appendix, Fig. S5), and therefore it is open and also has partly reduced inactivation. These results further support the hypothesis that the H25R/E118A mutant has a pH-independent open activation gate.
The electrophysiology results additionally support our hypothesis that T74 is crucial for allosteric coupling between the pH gate and the selectivity filter. However, the activity is not completely recovered, and T74S/H25R/E118A triple mutant exhibits 30% open probability from pH 4 to 7, whereas, in E71A, the open probability is close to 100%. Apparently, when the channel is constitutively open (in the H25R/E118A), the T74S mutation reduces but does not fully attenuate allosteric coupling. This is in contrast to the results for E71A and T74S at low pH (i.e., with the wild-type activation gate H25/E118); when low pH rather than the mutation is used to open the channel, the activity was nearly 100%, and no inactivation was evident. The fact that T74S essentially abolishes allosteric coupling when paired with the wild-type pH activation gate, whereas the constitutively open T74S/H25R/E118A triple mutant only partially attenuates the coupling and inactivation, suggests subtle differences in channel energetics comparing the fully protonated (open) wild type and the H25R/E118A mutant. One possible explanation is that, lacking some of the restraints of the hydrogen bonding network present in the wild type, the H25R/E118A mutant may open to a greater degree and thereby encourage inactivation—although this interpretation is purely speculative in the absence of further evidence and investigation.
Discussion
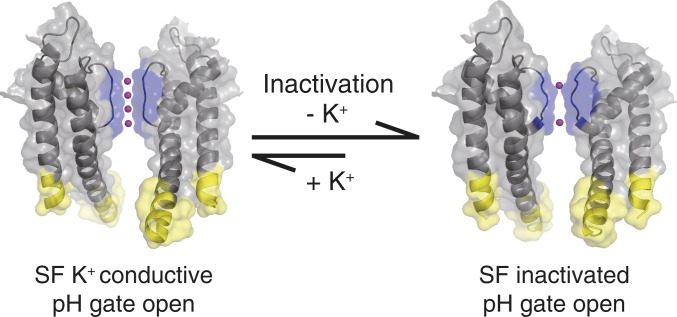
We documented transmembrane allosteric coupling in the inactivation process of a potassium channel using KcsA mutant H25R/E118A with an open activation gate. Specifically, we tested the hypothesis that C-type inactivation involves collapse of the selectivity filter and loss of K+ ions as an allosteric response of the opening of the pH gate (13, 28). By comparing potassium-dependent SSNMR data from the open KcsA mutant to that of the closed wild type, we tested whether the open channel loses its grip on the potassium ion (Scheme 1).
Scheme 1.
The potassium channel KcsA changes are depicted undergoing a conversion from a conductive state (with selectivity filter loaded with K+ ions) to an inactivated K+-depleted state. We hypothesize that such a change provides the basis for spontaneous inactivation and is caused by the allosteric coupling between the pH gate and the selectivity filter. In this work, we characterize the H25R/E118A KcsA mutant where the pH gate is open, to study the K+ binding and inactivation. In our previous work, channel opening was achieved by lowering the pH, raising the possibility of nonspecific effects of pH (13). The conductive state and the inactivated state shown in the scheme are based on Protein Data Bank entries 5VK6 and 5VKE, respectively (44). The selectivity filter pH gate and potassium ions are rendered in blue yellow and magenta, respectively.
The activated state of the channel has been difficult to stabilize (28). Efforts to trap the activated state and the early intermediates of the C-type inactivation process have involved lowering the pH and significant mutations (10, 11, 16, 18, 23, 29, 30). Although lowering the pH seems to be an intuitively correct method to trap the activated and the inactivated state, we had concerns about nonspecific effects of low pH on both protein and essential lipids (31–34). Here, instead of changing the pH, we conducted measurements on a mutant of KcsA with constitutively open pH gate at neutral pH conditions, and effectively eliminated this concern. We observed a reduction in potassium affinity at the selectivity filter for H25R/E118A KcsA by four orders of magnitude when the channel is opened neutral pH. This confirms that opening the pH gate significantly affects the affinity of the potassium ion. Evidently, the pH gate does not directly affect the bound and apo structures of the selectivity filter, as indicated by the NMR spectra of the extracellular markers in and near the selectivity filter. Since the pH gate and the selectivity filter are almost 30 Å away from each other, such a dramatic effect is understood to be an allosteric coupling.
Our results are in contrast with experimental results from isothermal titration calorimetry (ITC) studies on the similar mutant H25R/E118A/E120A with truncated C terminus in a detergent environment. Under these conditions, the affinity for K+ at pH 8 is 0.13 mM, and close to the value for wild-type KcsA measured in the same study (35). We attribute the differences with respect to our work to the detergent and truncation in the ITC experiment, which could affect the stability and thermodynamics of the system, as previously indicated in studies of KcsA and other systems (1, 36). In electrophysiology experiments, KcsA with a truncated C terminus showed altered inactivation (28, 37). By contrast, the SSNMR measurements reported here involve full-length KcsA in detergent-free hydrated bilayers and are therefore more comparable to physiological conditions.
Conclusions
The inactivation process of a potassium channel was explored by contrasting potassium binding in the open vs. closed state using SSNMR. The potassium affinity of the open pH mutant is of the same order of magnitude as the affinity of wild-type KcsA channel under acidic conditions. Thus, we consider that both are good models for the open state and that the open state has a loose affinity for potassium ion. This result further confirms the hypothesis that, in C-type inactivation, the open pH gate leads to ion loss in the selectivity filter. The pKa of E120 in the intracellular pH gate was also shown to be influenced by potassium ion binding at the selectivity filter, providing additional evidence for existing an allosteric coupling network in KcsA.
Materials and Methods
Expression and Purification of 13C,15N-Labeled Open pH Gate KcsA.
The open pH gate H25R/E118A KcsA was recombinantly expressed using a plasmid previously prepared by the Nimigean laboratory (18). The protein expression and purification protocol were based on our former work, with minor modifications (38). Four microliters of the open pH gate KcsA plasmids was added into 50 μL of JM83 competent cells in a culture test tube. The mixture was incubated on ice for 30 min, then heat-shocked for 70 s in a 42 °C water bath and incubated on ice for another 3 min. Then 1 mL of super optimal broth with catabolite repression medium (Invitrogen) was added, and the cells were incubated at 37 °C for 1 h. The transformed cells were plated on Luria broth (LB) agar plates containing 100 μg/mL ampicillin and incubated overnight at 37 °C. Single colonies of the transformed cells were picked and transferred to 4 × 5 mL of LB with 100 μg/mL ampicillin. The preculture was incubated at 37 °C, 250 rpm, when the OD600 reached 1.0, the precultures were transferred into 4 × 1 L LB with 100 μg/mL ampicillin using the same incubation conditions. Once the OD600 reached 0.9, the cells were harvested via centrifugation at 4 °C, 8000 rpm for 15 min and resuspended in 1 L M9 medium with 3.0 g 13C-labeled D-glucose, 0.5 g 15NH4Cl and 100 μg/mL ampicillin. The cells were incubated at 37 °C, 250 rpm for 1 h to recover. The protein expression was induced with anhydrotetracycline (Sigma), and incubated at 20 °C, 330 rpm overnight. The induced cells were harvested via centrifugation at 4 °C at 8,000 rpm for 15 min, resuspended in 100 mM KCl, 50 mM Tris, 2 mM decyl maltoside (DM), pH 7.5 equilibrium buffer and lysed by French Press. The cell membranes were extracted with 30 mM DM at 4 °C overnight. The unlysed cells and membranes were pelleted via centrifugation at 4 °C at 15,000 rpm for 1 h. The protein was purified by nickel affinity column and eluted with 200 mM Imidazole. Imidazole concentration was reduced by buffer exchange with the equilibrium buffer, and the protein solution was concentrated for reconstitution.
Ten milligrams of 9:1 wt/wt DOPE and DOPS (Avanti) were mixed in a 10-mL test tube; the chloroform was evaporated by nitrogen gas flow, and the lipids were resuspended into 100 mM KCl, 50 mM Tris, 2 mM DM, 0.01 mM sodium azide, pH 7.5 buffer at concentration of 10 mg/mL. The 13C,15N-labeled open pH mutant KcsA was reconstituted into 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE)/1,2-dioleoyl-sn-glycero-3-phospho-l-serine (DOPS) liposome at 1:1 protein to liposome weight ratio. The mixture was dialyzed against a solution of 50 mM Tris, x mM KCl, (100 − x) mM NaCl (to compensate ionic strength, except for the 150 mM KCl sample) at pH 7.5 overnight at room temperature, changing three times. The protein/liposome pellets were harvested, then frozen and thawed at −80 °C and room temperature three times to remove bulk water. The final sample was reduced to a volume of ∼30 μL by again carrying out three freeze−thaw cycles (−80 °C to room temperature), and the sample was then packed into a 3.2-mm Bruker rotor, which contained ∼10 mg of mutant KcsA.
SSNMR and Data Analysis.
Magic-angle spinning (MAS) SSNMR spectra were measured on Bruker 750-MHz (17.6 T) and 900-MHz (21.1 T) Avance spectrometers at the New York Structural Biology Center (NYSBC). The MAS rate was 14 kHz, and the variable temperature set point was 264 K. Typical radiofrequency field strengths were 93 kHz to 109 kHz for 1H and 50 kHz to 62.5 kHz for 13C. The 13C chemical shifts were referenced externally to the downfield adamantane CH2 resonance chemical shift at 40.48 ppm (39).
The 2D 13C-13C DARR experiments were measured to obtain homonuclear 13C correlation spectra (40, 41). The DARR mixing time was 15 ms. SPINAL64 decoupling on the 1H channel was 85 kHz to 90 kHz during acquisition (42).
We calculated the population of the bound and apo states as previously (13), using the bound and apo state of marker peaks for T75 CB−CA, T74 CB−CA, T75 CB-CG, and V76 CB-CG, quantifying the integrals using a sum over box method in Sparky (43). Peaks were identified based on our former assignments (11, 43). The bound population was computed as θ = [Ibound/(Ibound + Iapo)] × 100%. Normalized bound ratios of the protein for different ambient K+ concentrations were calculated by averaging the bound ratios of the marker peaks. Data were plotted and fit to a Hill equation in OriginPro: θ = xn/(xn + ), in which n is the Hill coefficient. The average bound population increased from 0% at 10 mM [K+] to 97% at 150 mM [K+]. In the range of 50 mM to 100 mM [K+], the major conformational transition takes place.
Electrophysiology.
T74S/H25R/E118A KcsA was prepared freshly using former protocols and purified with a gravity nickel column and desalting column (GE HiPrep 26/10) on fast protein liquid chromatography (Bio-Rad NGC). Five milligrams of DOPE:1,2-dioleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) 3:1 (wt/wt) liposome were prepared as follows: Lipids in chloroform were dried under nitrogen gas, and pentane was added to remove organic solvents; dry lipids were resuspended by sonication in 1 mL of Hepes swelling buffer while slowly adding 3-[(3-Cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS) detergent until clear. In total, 30 mg of CHAPS was added. Then 2.5 μg of T74S/H25R/E118A KcsA was added to the lipid/detergent solution, and the mixture was incubated at room temperature for 30 min. A biobead column was used to remove the detergent, and the proteoliposomes were eluted at 8 mL and frozen in liquid nitrogen.
Electrophysiology experiments were carried out with a partition diameter of 100 μm. The lower chamber bath was at a pH of 4.0 (succinic acid buffer), and the upper chamber was at a pH of 7.0 (Hepes buffer, 100 mM K+). Data were collected in Clampex and processed in Clampfit.
Data Availability.
Tabulated chemical shifts are provided in SI Appendix.
Supplementary Material
Acknowledgments
We thank Dr. Crina Nimigean of Cornell Weill University for assistance with the electrophysiology experiments. The NMR data were collected at the NYSBC with support from the Center on Macromolecular Dynamics by NMR Spectroscopy, a Biomedical Technology Research Resource supported by the NIH through Grant P41 GM118302. The NYSBC is also enabled by a grant from the Empire State Division of Science Technology and Innovation, and by Office of Research Infrastructure Programs/NIH Facility Improvement Grant CO6RR015495. This work was supported by NIH Grant R01 GM088724 (to A.E.M.).
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1908828117/-/DCSupplemental.
References
- 1.Cuello L. G., et al. , Structural basis for the coupling between activation and inactivation gates in K+ channels. Nature 466, 272–275 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McCoy J. G., Nimigean C. M., Structural correlates of selectivity and inactivation in potassium channels. Biochim. Biophys. Acta 1818, 272–285 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zachariae U., et al. , The molecular mechanism of toxin-induced conformational changes in a potassium channel: Relation to C-type inactivation. Structure 16, 747–754 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Alseikhan B. A., DeMaria C. D., Colecraft H. M., Yue D. T., Engineered calmodulins reveal the unexpected eminence of Ca2+ channel inactivation in controlling heart excitation. Proc. Natl. Acad. Sci. U.S.A. 99, 17185–17190 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Faber E. S., Sah P., Ca2+-activated K+ (BK) channel inactivation contributes to spike broadening during repetitive firing in the rat lateral amygdala. J. Physiol. 552, 483–497 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pau V., Zhou Y., Ramu Y., Xu Y., Lu Z., Crystal structure of an inactivated mutant mammalian voltage-gated K+ channel. Nat. Struct. Mol. Biol. 24, 857–865 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cuello L. G., et al. , Proton-dependent gating in the Streptomyces K+ channel. Biophys. J. 74, A254 (1998). [Google Scholar]
- 8.Heginbotham L., LeMasurier M., Kolmakova-Partensky L., Miller C., Single Streptomyces lividans K+ channels: Functional asymmetries and sidedness of proton activation. J. Gen. Physiol. 114, 551–560 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cortes D. M., Cuello L. G., Perozo E., Molecular architecture of full-length KcsA: Role of cytoplasmic domains in ion permeation and activation gating. J. Gen. Physiol. 117, 165–180 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhate M. P., McDermott A. E., Protonation state of E71 in KcsA and its role for channel collapse and inactivation. Proc. Natl. Acad. Sci. U.S.A. 109, 15265–15270 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wylie B. J., Bhate M. P., McDermott A. E., Transmembrane allosteric coupling of the gates in a potassium channel. Proc. Natl. Acad. Sci. U.S.A. 111, 185–190 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Linder T., de Groot B. L., Stary-Weinzinger A., Probing the energy landscape of activation gating of the bacterial potassium channel KcsA. PLoS Comput. Biol. 9, e1003058 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu Y., Bhate M. P., McDermott A. E., Transmembrane allosteric energetics characterization for strong coupling between proton and potassium ion binding in the KcsA channel. Proc. Natl. Acad. Sci. U.S.A. 114, 8788–8793 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miloshevsky G. V., Jordan P. C., Open-state conformation of the KcsA K+ channel: Monte Carlo normal mode following simulations. Structure 15, 1654–1662 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Cuello L. G., et al. , Design and characterization of a constitutively open KcsA. FEBS Lett. 584, 1133–1138 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thompson A. N., Posson D. J., Parsa P. V., Nimigean C. M., Molecular mechanism of pH sensing in KcsA potassium channels. Proc. Natl. Acad. Sci. U.S.A. 105, 6900–6905 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takeuchi K., Takahashi H., Kawano S., Shimada I., Identification and characterization of the slowly exchanging pH-dependent conformational rearrangement in KcsA. J. Biol. Chem. 282, 15179–15186 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Posson D. J., Thompson A. N., McCoy J. G., Nimigean C. M., Molecular interactions involved in proton-dependent gating in KcsA potassium channels. J. Gen. Physiol. 142, 613–624 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhate M. P., Wylie B. J., Tian L., McDermott A. E., Conformational dynamics in the selectivity filter of KcsA in response to potassium ion concentration. J. Mol. Biol. 401, 155–166 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anderson C. S., MacKinnon R., Smith C., Miller C., Charybdotoxin block of single Ca2+-activated K+ channels. Effects of channel gating, voltage, and ionic strength. J. Gen. Physiol. 91, 317–333 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Islas L. D., Sigworth F. J., Electrostatics and the gating pore of Shaker potassium channels. J. Gen. Physiol. 117, 69–89 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cuello L. G., Cortes D. M., Jogini V., Sompornpisut A., Perozo E., A molecular mechanism for proton-dependent gating in KcsA. FEBS Lett. 584, 1126–1132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Imai S., Osawa M., Takeuchi K., Shimada I., Structural basis underlying the dual gate properties of KcsA. Proc. Natl. Acad. Sci. U.S.A. 107, 6216–6221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thompson A. N., et al. , Molecular interactions involved in KCSA pH gating. Biophys. J. 100, 273 (2011). [Google Scholar]
- 25.Kim D. M., et al. , Conformational heterogeneity in closed and open states of the KcsA potassium channel in lipid bicelles. J. Gen. Physiol. 148, 119–132 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu Y., Zhang D., Rogawski R., Nimigean C. M., McDermott A. E., Identifying coupled clusters of allostery participants through chemical shift perturbations. Proc. Natl. Acad. Sci. U.S.A. 116, 2078–2085 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J., Ostmeyer J., Cuello L. G., Perozo E., Roux B., Rapid constriction of the selectivity filter underlies C-type inactivation in the KcsA potassium channel. J. Gen. Physiol. 150, 1408–1420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cuello L. G., Jogini V., Cortes D. M., Perozo E., Structural mechanism of C-type inactivation in K+ channels. Nature 466, 203–208 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cordero-Morales J. F., Cuello L. G., Perozo E., Voltage-dependent gating at the KcsA selectivity filter. Nat. Struct. Mol. Biol. 13, 319–322 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Chakrapani S., Cordero-Morales J. F., Perozo E., A quantitative description of KcsA gating I: Macroscopic currents. J. Gen. Physiol. 130, 465–478 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dart C., Lipid microdomains and the regulation of ion channel function. J. Physiol. 588, 3169–3178 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakao H., et al. , pH-dependent promotion of phospholipid flip-flop by the KcsA potassium channel. Biochim. Biophys. Acta 1848, 145–150 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Schmidt D., Jiang Q. X., MacKinnon R., Phospholipids and the origin of cationic gating charges in voltage sensors. Nature 444, 775–779 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Valiyaveetil F. I., Zhou Y., MacKinnon R., Lipids in the structure, folding, and function of the KcsA K+ channel. Biochemistry 41, 10771–10777 (2002). [DOI] [PubMed] [Google Scholar]
- 35.Liu S., et al. , Ion-binding properties of a K+ channel selectivity filter in different conformations. Proc. Natl. Acad. Sci. U.S.A. 112, 15096–15100 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Otzen D. E., Protein unfolding in detergents: Effect of micelle structure, ionic strength, pH, and temperature. Biophys. J. 83, 2219–2230 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uysal S., et al. , Crystal structure of full-length KcsA in its closed conformation. Proc. Natl. Acad. Sci. U.S.A. 106, 6644–6649 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bhate M. P., et al. , Preparation of uniformly isotope labeled KcsA for solid state NMR: Expression, purification, reconstitution into liposomes and functional assay. Protein Expr. Purif. 91, 119–124 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morcombe C. R., Zilm K. W., Chemical shift referencing in MAS solid state NMR. J. Magn. Reson. 162, 479–486 (2003). [DOI] [PubMed] [Google Scholar]
- 40.Takegoshi K., Nakamura S., Terao T., C-13-H-1 dipolar-driven C-13-C-13 recoupling without C-13 rf irradiation in nuclear magnetic resonance of rotating solids. J. Chem. Phys. 118, 2325–2341 (2003). [Google Scholar]
- 41.Takegoshi K., Nakamura S., Terao T., C-13-H-1 dipolar-assisted rotational resonance in magic-angle spinning NMR. Chem. Phys. Lett. 344, 631–637 (2001). [Google Scholar]
- 42.Fung B. M., Khitrin A. K., Ermolaev K., An improved broadband decoupling sequence for liquid crystals and solids. J. Magn. Reson. 142, 97–101 (2000). [DOI] [PubMed] [Google Scholar]
- 43.Goddard T. D., Kneller D. G., Sparky 3. https://www.cgl.ucsf.edu/home/sparky/. Accessed 13 March 2020.
- 44.Cuello L. G., Cortes D. M., Perozo E., The gating cycle of a K+ channel at atomic resolution. eLife 6, e28032 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Tabulated chemical shifts are provided in SI Appendix.