Significance
Neutrophils are white blood cells specialized as the first line of host defense in the immune system. One way they protect organisms is through NETosis, in which they expel their DNA to form a web-like trap that ensnares pathogens and promotes clotting. However, NETs also mediate sterile inflammation, causing damage to the body. We used high-resolution live-cell microscopy to characterize the timing of dynamic cellular events leading to NETosis in human and mouse neutrophils and a neutrophil-like cell line. We discovered that NETosis proceeds by a stepwise sequence of cellular events that is conserved across species and requires the activity of the PAD4 enzyme for DNA to be released from the nucleus and cell membrane.
Keywords: neutrophil, innate immunity, microscopy
Abstract
Neutrophil extracellular traps (NETs) are web-like DNA structures decorated with histones and cytotoxic proteins that are released by activated neutrophils to trap and neutralize pathogens during the innate immune response, but also form in and exacerbate sterile inflammation. Peptidylarginine deiminase 4 (PAD4) citrullinates histones and is required for NET formation (NETosis) in mouse neutrophils. While the in vivo impact of NETs is accumulating, the cellular events driving NETosis and the role of PAD4 in these events are unclear. We performed high-resolution time-lapse microscopy of mouse and human neutrophils and differentiated HL-60 neutrophil-like cells (dHL-60) labeled with fluorescent markers of organelles and stimulated with bacterial toxins or Candida albicans to induce NETosis. Upon stimulation, cells exhibited rapid disassembly of the actin cytoskeleton, followed by shedding of plasma membrane microvesicles, disassembly and remodeling of the microtubule and vimentin cytoskeletons, ER vesiculation, chromatin decondensation and nuclear rounding, progressive plasma membrane and nuclear envelope (NE) permeabilization, nuclear lamin meshwork and then NE rupture to release DNA into the cytoplasm, and finally plasma membrane rupture and discharge of extracellular DNA. Inhibition of actin disassembly blocked NET release. Mouse and dHL-60 cells bearing genetic alteration of PAD4 showed that chromatin decondensation, lamin meshwork and NE rupture and extracellular DNA release required the enzymatic and nuclear localization activities of PAD4. Thus, NETosis proceeds by a stepwise sequence of cellular events culminating in the PAD4-mediated expulsion of DNA.
Neutrophils deploy a variety of machineries to fight infection and neutralize pathogens, including phagocytosis and degranulation, as well as the more recently characterized release of neutrophil extracellular traps (NETs) termed NETosis (1). NETs are web-like DNA structures decorated with histones and antimicrobial proteins that are released from stimulated neutrophils. NETs can trap and neutralize or kill pathogens, including bacteria (1), fungi (2), and viruses (3), and propagate inflammatory and immune responses (4). However, NETosis also conveys detrimental effects, including tissue damage during sepsis (5, 6) and thrombosis (7). Furthermore, several autoimmune diseases are associated with high rates of NETosis and/or defects in NET clearance (6), and there is evidence that NETosis promotes cancer (6, 8). Thus, understanding the mechanisms mediating NETosis could facilitate either therapeutic improvement of innate immunity or mitigation of its damaging effects.
The molecular requirements for NETosis have begun to be elucidated. NETosis can be stimulated with a variety of factors, including bacteria or yeast, monosodium urate crystals associated with gout, platelet activating factor, bacterial ionophores or lipopolysaccharides, or can be pharmacologically induced with phorbol ester (9). Regardless of the stimulus, NETosis requires convergence of signaling pathways to mediate the cellular process of chromatin decondensation, which is necessary for NET release (10). Two mechanisms are thought to promote histone release from DNA to mediate decondensation: Neutrophil elastase and other proteases in granules may cleave histones to dissociate them from DNA (11), or PAD4, an enzyme that converts arginine to citrulline, may citrullinate histones, reducing their charge-based interaction with DNA to promote chromatin decondensation (10). The relative importance of proteases and PAD4 for completion of NETosis may be dictated by the cellular stimulus (12) or the species. Indeed, neutrophil elastase is required downstream of the NADPH pathway when NETosis is induced in human neutrophils by phorbol esters or Candida albicans (12), while PAD4 is critical for NETosis in mouse neutrophils stimulated with calcium ionophore or bacteria (13, 14). However, whether different cellular mechanisms are engaged during NETosis in mouse and human neutrophils and whether PAD4 is required for NET release in human neutrophils remains unclear.
Despite advancing knowledge of the molecular requirements for NETosis, less is known about its cellular mechanisms (15). For DNA to be released to the cell exterior during NETosis, it must escape from the nucleus, pass through the cytoplasm containing a network of membranous organelles and cytoskeletal systems, and finally breach the plasma membrane (PM). While it is generally thought that decondensed chromatin is expelled via nuclear envelope (NE) and plasma membrane rupture resulting in neutrophil death (1, 15), some evidence for “vital NETosis” suggests that vesicles containing DNA might be exocytosed to allow neutrophils to survive and retain the capacity for phagocytosis and induction of adaptive immunity after NET release (16, 17). However, little is known about how chromatin breaches organelles and the cytoskeleton to pass through the cytoplasm. There is evidence that actin filaments (18–20) and microtubules (MTs) (19, 21) disassemble during NETosis, yet pharmacological perturbations that either disassemble or stabilize actin can impair NETosis (15, 18, 19), while MT disruption has little effect (19). Thus, an in-depth understanding of the cellular events driving NETosis requires the detailed visualization of various cellular compartments during the entire process.
Here we used time-lapse high-resolution multimode microscopy of living human and mouse blood-derived polymorphonuclear neutrophils (PMNs) as well as differentiated human leukemia (dHL-60) neutrophil-like cells bearing fluorescent markers of organelle and cytoskeletal systems to quantitatively characterize the cellular events mediating NETosis after stimulation with bacterial toxins (ionomycin, lipopolysaccharide [LPS]) or C. albicans. We demonstrate that NETosis proceeds by a well-defined and conserved sequence of cellular events. Characterization of neutrophils derived from PAD4 knockout mice or gene-edited dHL-60 cells demonstrates that PAD4 enzymatic activity and nuclear localization signal are required for efficient DNA decondensation, nuclear lamin meshwork, and NE rupture and extracellular DNA release.
Results
NETosis Starts with Vesiculation Prior to DNA Decondensation, DNA Release into the Cytoplasm, and Extracellular DNA Expulsion in Mouse, Human PMNs, and dHL60 Cells.
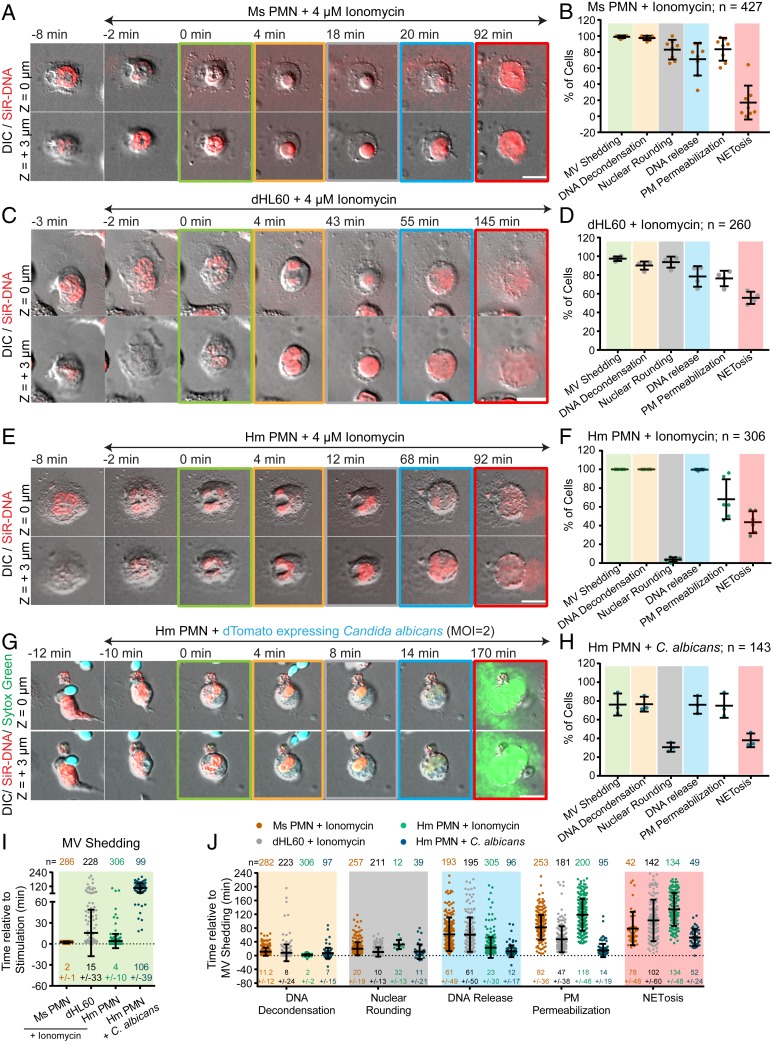
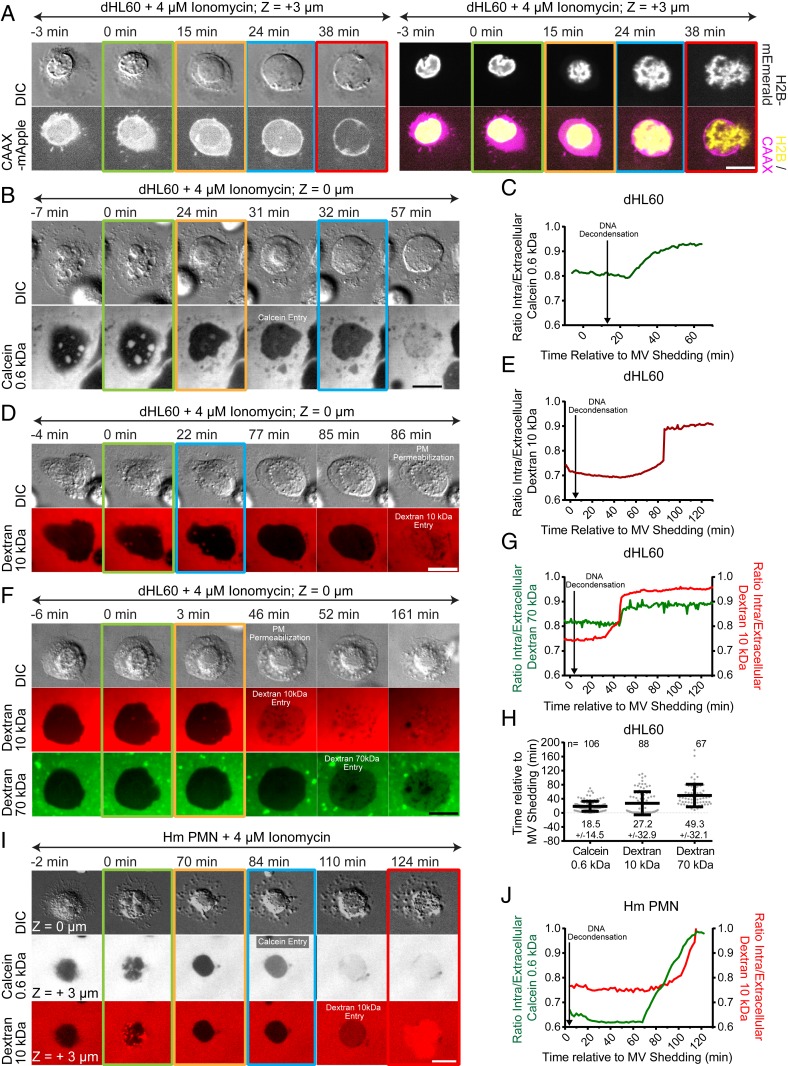
To characterize the cellular events mediating NETosis, we performed high-resolution time-lapse imaging of mouse and human PMNs isolated from fresh blood, as well as dHL-60 cells, upon NETosis stimulation. Cells stained with the DNA-specific vital dye, SiR-DNA, were plated on coverslips for imaging and stimulated with ionomycin. Pairs of differential interference contrast (DIC) and spinning-disk confocal images were acquired at two confocal planes (at the coverslip surface and cell center) at each 1- to 2-min time interval for 4 h. Under these conditions, 17.0%, 44.6%, and 55.6% of ionomycin-stimulated mouse PMNs, human PMNs, and dHL-60 cells observed went on to complete NETosis, respectively, as evidenced by extracellular expulsion of DNA (Fig. 1 B, D, and F). Importantly, unstimulated mouse PMNs did not exhibit DNA expulsion (SI Appendix, Fig. S1A), and the NETosis efficiencies attained during live-cell imaging of dHL-60 cells were similar to those measured in a fixed-timepoint assay (31% in dHL-60; SI Appendix, Fig. S1B) and those previously published for mouse (22) and human (12) PMNs, indicating that our cell isolation and long-term imaging treatments are minimally perturbative to NETosis.
Fig. 1.
NETosis proceeds by vesiculation, DNA decondensation, DNA release from the nucleus into the cytoplasm, and extracellular DNA expulsion in mouse and human blood neutrophils and dHL-60 cells. Mouse (Ms; A, B, I, and J) or human (Hm; E–J) PMNs or dHL-60 cells (C, D, I, and J) stained with far-red SiR-DNA were stimulated with ionomycin (4 μM, A–F, I, and J) or dTomato-expressing C. albicans (C. albicans, MOI of 2, G–J, dTomato shown in blue), and imaged by DIC and confocal microscopy at the coverslip–cell interface (Z = 0 μm) and in the cell center (Z = +3 μm) at 1- to 2-min intervals for 4 h. Time (min) relative to plasma membrane microvesicle (MV) shedding noted. Double-arrow-headed lines indicate presence of ionomycin (A, C, and E) or phagocytosed C. albicans (G); colored boxes around images correspond to color coded cellular events in the graphs. C. albicans experiments (G and H) included Sytox green to highlight extracellular DNA. (A, C, E, and G) Time series of image overlays of DIC (grayscale) and fluorescence of cell, C. albicans (blue in G) and DNA (red in A, C, E, red and green in G) dynamics. (B, D, F, and H) Percent of cells exhibiting MV shedding, DNA decondensation, nuclear rounding, DNA release into the cytoplasm, loss of DIC contrast (PM permeabilization), and extracellular DNA release (NETosis) after stimulation. n = total number of cells, each point = percent of cells in one experiment. (I and J) Timing of MV shedding relative to ionomycin or C. albicans stimulation (I) or of cellular event initiation relative to MV shedding (J). N = total number of cells observed, points = individual cells. In B, D, F, H, I, and J: long bar = mean, short bars = SD. (Scale bars In A, C, E, and G: 10 μm.)
We then examined time-lapse movies to analyze cell dynamics during NETosis. DIC and confocal movies of membrane dynamics in the optical plane at the ventral cell surface of all three cell types revealed that within minutes after ionomycin stimulation, cells rapidly formed many small (∼<2 μm) vesicles that were devoid of fluorescent DNA (Fig. 1 A, C, E, and I and Movie S1). Following vesiculation, cells rounded up and eventually expelled extracellular DNA, marking completion of NETosis. Quantification revealed that although not all cells that vesiculated went on to expel DNA, 100% of cells that completed NETosis had previously formed vesicles. Thus, vesiculation is a hallmark of cellular entry into NETosis (Fig. 1 B, D, F, and I), and shall be used as an easily visualized temporal initiation point for the remainder of this study.
We then analyzed DNA and membrane dynamics in the cells’ central confocal plane. Prior to stimulation, nuclei of all three cell types were lobulated with spatially heterogeneous DNA staining indicative of regions of varied levels of chromatin condensation. Quantification of timing in movies showed that within ∼10 min after ionomycin-induced vesiculation, DNA fluorescence became homogeneous throughout the nucleus (Fig. 1J and Movie S1). Similar fluorescence homogenization was observed during NETosis in dHL-60 cells expressing mEmerald-tagged linker histone H1 (SI Appendix, Fig. S1C and Movie S2), suggesting that these changes correspond to chromatin decondensation. DNA decondensation was followed ∼10 min later by loss of nuclear lobulation and rounding of the nuclear surface in both mouse PMN and dHL-60, while in human PMN, nuclei delobulated but did not fully round up (Fig. 1 E and F). At 20 to 60 min after vesiculation, the sharp DIC contrast band at the rim of the nucleus was lost and DNA expanded rapidly into the cytosol, indicating nuclear rupture (Fig. 1J and Movie S1). After nuclear rounding and before nuclear rupture, there was also a sudden reduction in DIC contrast in the cell periphery (Movie S3), indicating a decrease in cytoplasmic refractive index that likely corresponded to loss of cellular contents. Despite this leakage of cytosol, the peripheral cell membrane appeared intact, and DNA was not released extracellularly until many minutes later, marking the completion of NETosis (Fig. 1 B, D, F, and J). Although not all stimulated cells that exhibited DNA decondensation and leakage of cytosol completed NETosis, 100% of cells that completed NETosis had previously completed these cellular processes. However, while all cells that completed NETosis exhibited nuclear rupture prior to extracellular DNA release, a substantial portion of cells with ruptured nuclei did not go on to release DNA extracellularly (Fig. 1 B, D, and F). Thus, nuclear rupture may be required but not sufficient for NETosis.
To determine whether the sequence of cellular events observed after ionomycin-induced NETosis occurred with other pathogen-derived stimuli, we exposed human PMNs to C. albicans (multiplicity of infection [MOI] of 2) or mouse PMNs to lipopolysaccharide (LPS, 25 μg/mL). 38% of C. albicans-stimulated human PMNs and 34% of LPS-stimulated mouse PMNs completed NETosis. Time-lapse analysis showed that human PMNs exposed to C. albicans phagocytosed actively growing yeast prior to initiating NETosis, and completion of NETosis was very rapid. Quantification indicated that cells that phagocytosed C. albicans or were stimulated with LPS and that completed NETosis exhibited similar membrane and nuclear dynamics as those seen in ionomycin-stimulated cells (Fig. 1 G–J, SI Appendix, Fig. S2 A and B, and Movie S1). Together, these results show that NETosis proceeds by a dynamic, stepwise sequence of cellular events that is conserved, independent of the species or NETosis stimulant.
Microvesicles Containing Cytosolic Components Are Shed from the Plasma Membrane at NETosis Onset.
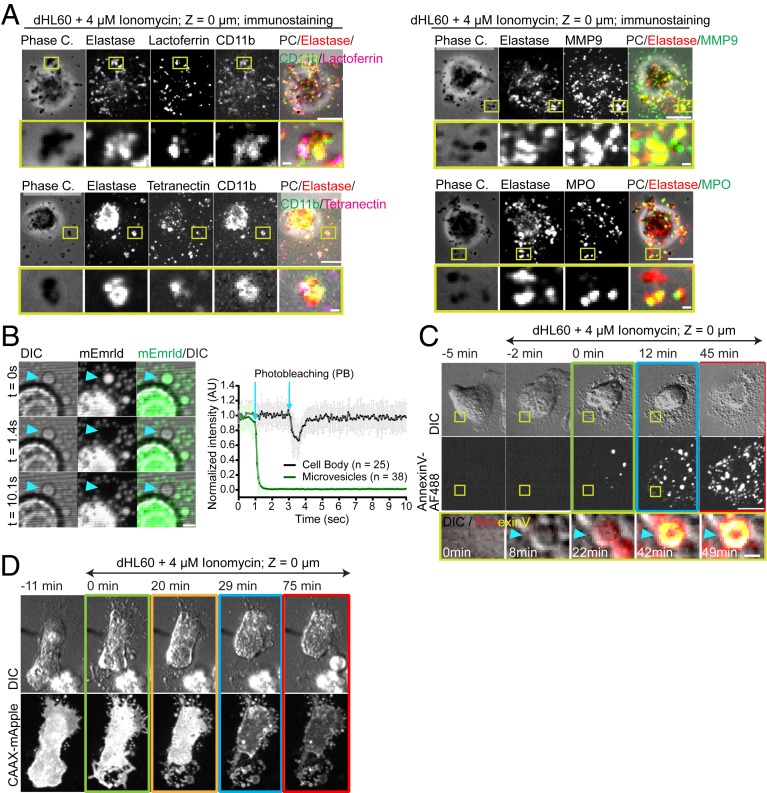
Given the bioactive nature of extracellular vesicles (23), we sought to better characterize the origin and content of the vesicles that appeared in the early stage of NETosis. Immunostaining for granule components in ionomycin-stimulated dHL-60 cells showed that vesicles were positive for markers of primary (neutrophil elastase and myeloperoxidase), secondary (Lactoferrin and CD11b) and tertiary granules (MMP9 and CD11b), as well as secretory vesicles (tetranectin) (Fig. 2A). However, single vesicles often contained markers of multiple granule types, suggesting that vesicles containing cytosolic components, including granules, may be shed from cells. To determine whether vesicles contained cytosol and were physically disconnected from the cell, we expressed soluble mEmerald and performed fluorescence recovery after photobleaching (FRAP). We found that mEmerald localized stably to vesicles, and when their contents were photobleached, the fluorescence did not recover, indicating a lack of continuity between vesicles and the cell body. This was in contrast to bleaching a region of mEmerald in the cell body, which rapidly recovered from the unbleached pool (Fig. 2B and Movie S4). To differentiate between microvesicles and exosomes, we used plasma membrane markers that would be present on microvesicles and absent from exosomes. Imaging dHL-60 cells either expressing the farnesylation signal sequence fused to mApple (CAAX-mApple) or with a fluorescent analog of the phosphatidylserine (PS)-binding protein annexin V added to the media showed that vesicles were positive for both plasma membrane markers (Fig. 2 C and D and Movies S2 and S5). Thus, NETosis is initiated by shedding of microvesicles containing cytosolic contents that form by budding from the plasma membrane.
Fig. 2.
Microvesicles containing cytosolic components are shed from the plasma membrane to initiate NETosis. (A–D) dHL-60 cells stimulated with ionomycin. (A) Phase contrast (grayscale) and immunolocalization of neutrophil elastase (Elastase, red), myeloperoxidase (MPO, green), lactoferrin (magenta), CD11b (green), MMP9 (green), and tetranectin (magenta), fixed 30 min after stimulation. (B, Left) DIC and confocal images of a living dHL-60 cell expressing soluble mEmerald (green) before (Upper row), and after (time noted, Middle and Lower row) photobleaching (PB) of the vesicle (arrowhead). (Scale bar: 2 μm.) (Right) Normalized fluorescence intensity of the bleached region over time in regions in the cell body (black) or in microvesicles (MVs) (green), error bar = SD, arrows = time of photobleaching. (C and D) Cells imaged by DIC and confocal microscopy at the coverslip–cell interface (Z = 0 μm) and in the cell center (Z = +3 μm) at 1- to 2-min intervals for 4 h. Time (min) relative to plasma membrane MV shedding noted. Double-arrow-headed lines indicate presence of ionomycin; boxes around images indicate different cellular events (green: MV shedding; yellow: DNA decondensation; blue: DNA release to the cytosol, red: extracellular DNA release). (C and D) Time series of DIC (Upper) and fluorescence of cells in media containing annexin V-Alexa Fluor 488 (AF488, C) or expressing CAAX-mApple (Lower row, D). C, Bottom row: DIC (grayscale) and annexin V-AF488 (red-hot color scale) overlay. (Scale bar: 10 μm.) (A and C) Yellow boxes (Upper rows, Scale bars: 10 μm.) shown zoomed below (Scale bars: 1 μm.).
ER Vesiculation Is Followed by Nuclear Lamin Meshwork Disassembly, NE Rupture, and Release of DNA into the Cytoplasm, Leading to NETosis.
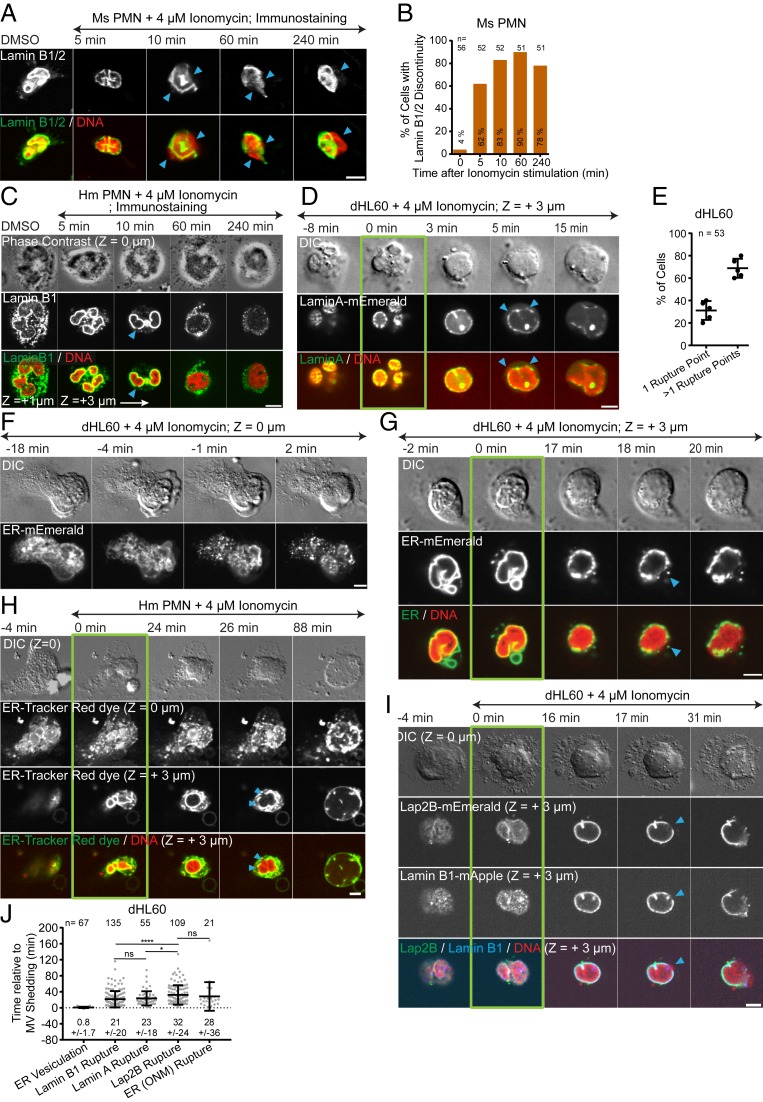
We next focused on how DNA is released from the nucleus into the cytoplasm, which requires passing first through the nuclear lamina and then breaching both membranes of the NE. We first confirmed the expression and localization of lamin isoforms in dHL-60 (24) and human and mouse PMNs (Fig. 3 A and C and SI Appendix, Fig. S3 A–D). Fixation and immunostaining of lamin B1/B2 in mouse PMNs and lamin B1 in human PMNs after ionomycin stimulation of NETosis showed discontinuities in the lamin meshwork detectable at 5 min after stimulation, with extrusion of DNA from the nucleus through the discontinuities and into the cytoplasm visible at 10 min, and disassembly of the lamin meshwork at later timepoints (Fig. 3 A–C). The apparent lag between lamin meshwork disruption and DNA extrusion suggests that the NE may remain intact longer than the lamina.
Fig. 3.
ER vesiculation is followed by nuclear lamina disassembly, NE rupture, and release of DNA into the cytoplasm, leading to NETosis. Mouse (Ms; A and B) or human (Hm; C and H) PMN or dHL60 cells (D–G, I, and J) stimulated with ionomycin and fixed (A and C, time noted) or imaged live (D and F–I) by DIC and confocal microscopy at the coverslip–cell interface (Z = 0 μm) and in the cell center (Z = +3 μm) at 1- to 2-min intervals for 4 h. Time (min) relative to MV shedding noted. Double-arrow-headed lines indicate presence of ionomycin. Arrowheads, lamina or NE rupture points. (D and G–I) Green boxes around images indicate MV shedding. (A and C) Immunofluorescence of lamin B1/B2 in Ms PMN (A, Upper row and Bottom row, green) or of lamin B1 in Hm PMN (C, Middle row and Bottom row, green) and staining of DNA with DAPI (A, 10 min, C) or SiR-DNA (A, DMSO, 5, 60, and 240 min) (Bottom row, red) fixed after ionomycin stimulation at the time noted. (C, Top row) Phase contrast images at Z = 0 μm; Middle and Bottom rows: confocal images at Z = 0 μm and Z = +1 μm, Z = +3 μm. (B) Percent (numbers in bar) of Ms PMN exhibiting lamin meshwork discontinuities, 0 = DMSO control. n = total cells. (D) Time-series of DIC (Top row) and confocal (Lower rows) images of dHL60 cells expressing lamin A-mEmerald (Middle row and Bottom row, green) and stained with SiR-DNA (Bottom row, red). (E) Percent of cells with one or more rupture points. n = total number of cells, each point = percent of cells in one experiment. (F–I) Time series of DIC (Upper rows) and confocal (Lower rows) images of dHL60 cells (F, G, and I) expressing ER-mEmerald (F, Bottom row; G, Middle and Bottom row, green) or coexpressing Lap2β-mEmerald (I, second row and bottom row, green) and lamin B1-mApple (I, third row and bottom row, blue), or live Hm PMN (H) stained with ER-Tracker Red (H, Middle rows and Bottom row, green) and stained with SiR-DNA (G–I Bottom rows red). (J) Timing of initiation of ER vesiculation or Lamin B1 or Lamin A rupture, Lap2β rupture or outer NE rupture (ER [ONM]) relative to MV shedding in dHL-60 cells. n = total number of cell observed, points = individual cells. (E and J) Mean (long bar) and SD (short bar), shown below each plot. ****P < 0.0001; *P < 0.1; ns, nonsignificant. Statistical test: Mann–Whitney U test. (Scale bars in A, C, D, F–I: 5 μm.)
We next sought to determine the sequence of lamin meshwork disruption, NE breakage, and chromatin release into the cytosol by live-cell microscopy. To analyze DNA extrusion relative to lamin meshwork structure, we imaged SiR-DNA-stained dHL-60 cells expressing either lamin A- or B1-mEmerald (Movie S6). Prior to stimulation, lamins formed punctate meshworks at the nuclear periphery (Fig. 3 D and I and SI Appendix, Fig. S3 B and C). After ionomycin-induced microvesicle shedding, DNA decondensation, and nuclear rounding, the lamin meshwork formed holes, often at multiple points (Fig. 3 D, E, and I), followed within 1 to 2 min by DNA extrusion through the holes and disassembly of the lamin meshwork as the DNA rapidly expanded throughout the cytoplasm (Fig. 3 D and I and Movie S6). We analyzed NE dynamics using either a fluorescent endoplasmic reticulum (ER) vital dye (Movie S7) or expressed mEmerald fusions of either the ER-retention signal sequence (ER-5-mEmerald) or the inner nuclear envelope protein, Lap2β (Movie S8). Prior to stimulation, ER markers localized to a reticular network in the cell periphery as well as the NE, and Lap2β-mEmerald localized only to the NE. Upon ionomycin stimulation of dHL-60 cells or LPS stimulation of mouse PMNs and soon after microvesicle shedding, the ER network broke into vesicles (Fig. 3 F–H and J, SI Appendix, Figs. S2C and S4A, and Movie S7) with the NE remaining intact during nuclear rounding and chromatin decondensation (Fig. 3 G and H, SI Appendix, Figs. S2C and S4A, and Movie S8). Subsequent to nuclear rounding, the NE suddenly became discontinuous and rapidly retracted from sites of DNA extrusion into the cytoplasm, signifying NE rupture (Fig. 3 G and H, SI Appendix, Figs. S2C and S4A, and Movie S8). Coexpression of Lap2β-mEmerald and lamin B1-mApple in dHL-60 cells showed that the lamin meshwork became discontinuous slightly before NE rupture (Fig. 3 I and J and Movie S9). We never observed DNA release from the nucleus or cell in NE-derived membrane vesicles (16) or without prior lamin meshwork and NE rupture, and NE or lamin meshwork resealing was never observed. Thus, cells vesiculate their ER networks before decondensing their DNA and rupturing their lamin meshwork, followed by NE rupture to allow chromatin release into the cytoplasm.
The Actin, MT, and Peripheral Vimentin Cytoskeletons Disassemble Prior to NETosis, and F-Actin Disassembly Is Important for NET Release.
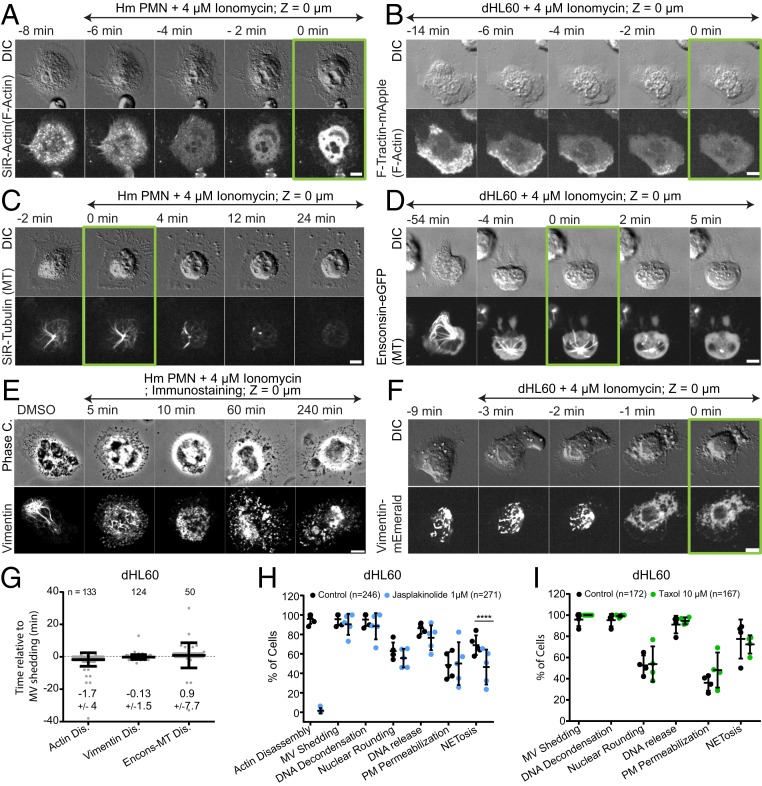
We next sought to examine how chromatin breaches the actin, MT, and vimentin intermediate filament (IF) cytoskeletal networks to allow its extracellular expulsion. To visualize actin, we stained mouse or human PMNs with SiR-actin (Movies S10 and S11) or expressed either F-tractin-mApple (Movie S12) or actin-mEmerald (Movie S13) in dHL-60 cells. MT dynamics were analyzed in mouse and human PMNs stained with SiR-tubulin (Movies S10 and S11) or dHL-60 expressing either the ensconsin MT-binding domain fused to eGFP (Movie S12) or tubulin-mEmerald (Movie S14). Vimentin IFs were visualized in dHL-60 cells expressing vimentin-mEmerald (Movie S12) or by immunofluorescence in human and mouse PMNs. Prior to stimulation in spread neutrophils, actin concentrated in ruffles, uropods and in punctate structures at the cells’ ventral surface, MTs radiated toward the cell periphery from the centrosome, and vimentin IF formed around the nuclear surface and extended into the cytoplasm. Ionomycin (dHL60, mouse, and human PMNs) or LPS (mouse PMNs) stimulation induced rapid redistribution of actin, tubulin, and vimentin from these structures into diffuse cytosolic fluorescence, indicating filament disassembly (Fig. 4 A–G, SI Appendix, Figs. S4 B and C and S2 D and E, and Movies S10–S14). Quantitative analysis of dHL-60 cells showed that actin disassembly occurred just before microvesicle shedding, while MT and vimentin IF disassembly was initiated nearly concurrent with shedding (Fig. 4G). Cytoskeletal disassembly was then followed many minutes later by DNA decondensation, nuclear rounding, and NETosis completion (Movies S13 and S14). Although diffuse vimentin fluorescence after disassembly masked visualization of perinuclear vimentin structures in live cells (Fig. 4F and Movie S12), immunostaining showed that while peripheral vimentin IFs were largely disassembled within 5 min after stimulation, at 10 min some particulate perinuclear vimentin remained (Fig. 4E and SI Appendix, Fig. S4 I and J). Thus, NETosis is initiated by actin disassembly, followed by microvesicle shedding, disassembly of vimentin IFs, and MTs prior to DNA decondensation, nuclear rounding, nuclear lamin meshwork and NE rupture, DNA release into the cytoplasm, and extracellular DNA expulsion.
Fig. 4.
The actin, MT, and vimentin cytoskeletons disassemble prior to NETosis, and F-actin disassembly is important for NET release. Human (Hm) PMN (A, C, and E) or dHL-60 cells (B, D, F, and G–I) stimulated with ionomycin and fixed (time noted, E) or imaged by DIC and confocal microscopy at the coverslip–cell interface (Z = 0 μm) and in the cell center (Z = +3 μm) at 1- to 2-min intervals for 4 h, time relative to microvesicle (MV) shedding noted. Double-arrow-headed lines indicate presence of ionomycin. (A–F) Time-series of DIC (A–D and F) or phase contrast (E) (Top rows) and confocal (Lower rows) images of live (A and C) or fixed (E) Hm PMN or live dHL-60 cells (B, D, and F). (A and B) Actin visualized with far red SiR-actin (A) or expression of F-Tractin-mApple (B). (C and D) MTs visualized with far red SiR-tubulin (C) or expression of Ensconsin-eGFP (D). (E and F) Immunolocalized vimentin (E) or expression of vimentin-mEmerald (F). (A–D and F) Green boxes around images indicate MV shedding. (G) Timing of actin, MT, and vimentin disassembly (Dis) relative to MV shedding. n = total number of cell observed, points represent individual cells, mean (long bar) and SD (short bar) shown below each plot. (H and I) Percent of cells exhibiting actin disassembly, MV shedding, DNA decondensation, nuclear rounding, DNA release into the cytoplasm, loss of DIC contrast (plasma membrane (PM) permeabilization, and extracellular DNA release (NETosis) after stimulation in cells treated with vehicle (control), jasplakinolide (1 μM, H), or taxol (10 μM, I), n = total of cell observed, points = percent of cells in one experiment, long bar = mean, short bars = SD. Statistical test: Fisher’s exact test, ****P < 0.0001. (Scale bars in A–F: 5 μm.)
We then determined the requirement for actin and MT disassembly in NETosis using jasplakinolide or taxol to stabilize actin or MTs, respectively. Pretreatment of dHL-60 cells expressing actin- or tubulin-mEmerald prior to ionomycin stimulation showed that jasplakinolide caused formation of a thick cortical actin network (Movie S13), while taxol (Movie S14) induced peripheral MT bundles, and both structures remained mostly intact throughout NETosis. Quantification revealed that neither treatment affected microvesicle shedding, DNA decondensation, nuclear rounding, DNA release into the cytoplasm, or loss of cytosolic contents. However, while taxol had no effect on extracellular DNA expulsion, jasplakinolide treatment reduced the fraction of cells that completed NETosis (Fig. 4 H and I). Examination of actin dynamics in the subset of jasplakinolide-treated cells that released extracellular DNA showed that DNA escaped through a local disruption in the cortical actin network (Movie S13). Thus, actin disassembly is critical to extracellular DNA release during NETosis.
Plasma Membrane Permeability Is Progressively Increased Prior to Its Rupture and NET Release.
We next sought to determine how DNA breaches the plasma membrane to be released into the extracellular environment. We analyzed plasma membrane dynamics using CAAX-mApple in ionomycin-stimulated dHL-60 cells that were either stained with SiR-DNA or coexpressing histone H2B-mEmerald. This showed that the plasma membrane appeared to remain intact during microvesicle shedding, nuclear rupture, and DNA release into the cytoplasm, after which a large hole was formed concomitant with extracellular DNA expulsion (Fig. 5A and Movie S15). Such plasma membrane and DNA release occurred much later than the decrease in contrast in the cell periphery visualized by DIC microscopy (Movie S3), suggesting that leakage of cytosol may occur by permeabilization of the plasma membrane prior to extracellular DNA release. To test this, we added different sized membrane-impermeant fluorescent markers to the imaging media and monitored their entry into cells stained with SiR-DNA and stimulated with ionomycin (human PMNs or dHL-60) or LPS (mouse PMNs). We found that calcein (0.6 kDa), 10 kDa dextran, and 70 kDa dextran all entered cells much earlier than extracellular DNA release (Fig. 5 B–E and H, SI Appendix, Figs. S4 D and E and S2 F and G, and Movies S16 and S17). Quantification of dHL-60 cells showed that calcein, 10 kDa dextran, and 70 kDa dextran entered cells on average at ∼20, ∼30, and ∼50 min after microvesicle shedding. Addition of either calcein together with 10 kDa dextran or 10 kDa dextran together with 70 kDa dextran showed that calcein entered cells before 10 kDa dextran (human PMNs, Fig. 5 I and J and Movie S17), which preceded 70 kDa dextran entry (dHL-60 cells, Fig. 5 F and G and Movie S17). Thus, during NETosis, the plasma membrane becomes permeable to increasingly larger macromolecules over time, allowing their entry into and leakage from cells prior to plasma membrane rupture and expulsion of DNA.
Fig. 5.
Plasma membrane becomes permeable to progressively larger molecules prior to plasma membrane rupture and NET release. dHL-60 (A–H) or human (Hm) PMN (I and J) stimulated with ionomycin imaged by DIC and confocal microscopy at the coverslip–cell interface (Z = 0 μm) and in the cell center (Z = +3 μm) at 1- to 2-min intervals for 4 h, time relative to MV shedding noted. Double-arrow-headed lines indicate presence of ionomycin; colored boxes around images correspond to color coded cellular events in the graphs in Fig. 1. (A) Time series of DIC (Upper Left) and fluorescence images of a dHL-60 cell coexpressing CAAX-mApple (Lower Left and Right, magenta) and H2B-mEmerald (Upper Right and Bottom Right, yellow). (B, D, F, and I) Time series of DIC (Upper row) and fluorescence (Lower rows) images of dHL-60 (B, D, and F) or Hm PMN (I) cells and calcein (622 Da, B and I, grayscale), Alexa Fluor 647 (D and F, red) or Alexa Fluor 594 (I, red), 10 kDa dextran, or Oregon Green 488 70 kDa (F, green) added to the media. (C, E, G, and J) Normalized intracellular fluorescence of calcein (C and J, green), 10 kDa (E, G, and J, red) or 70 kDa (G, green) dextran over time (in min). Time points of DNA decondensation noted. (H) Onset of calcein, 10 or 70 kDa fluorescent dextran cellular entry relative to MV shedding in dHL-60 cells. n = total number of cell observed, points = individual cells, mean (long bar) and SD (short bar) shown below each plot. (Scale bars in A, B, D, F, and I: 10 μm.)
PAD4 Is Critical to DNA Decondensation, NE Rupture, and Extracellular DNA Expulsion in Mouse PMN.
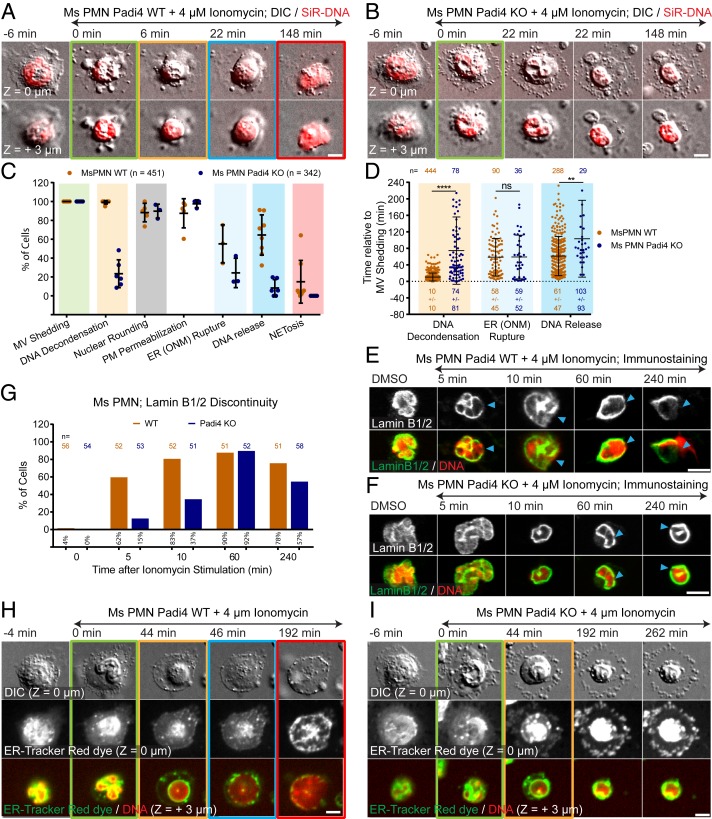
We next sought to determine the role of the arginine deiminase PAD4 in mediating the cellular events of NETosis. PMNs from wild-type (WT) and PAD4 knockout (KO) mice were stained with SiR-DNA, SiR-Actin, SiR-Tubulin, or ER-dye to visualize the DNA, actin, and MT cytoskeletons, or endoplasmic reticulum and NE, respectively, and stimulated with ionomycin. Time-lapse microscopy showed that similar to WT PMNs, PAD4 KO PMNs initiated NETosis by disassembling their actin cytoskeleton before shedding microvesicles (SI Appendix, Fig. S4F, Fig. 6 A and B, and Movie S18), followed by ER vesiculation, MT disassembly, nuclear rounding, and plasma membrane permeabilization (Fig. 6 A–D, H, and I, SI Appendix, Fig. S4 G and H, and Movies S18 and S19). However, in contrast to WT, PAD4 KO cells were deficient in DNA decondensation, NE rupture, DNA release into the cytoplasm, and extracellular NET release (Fig. 6 B–D and I and Movies S18 and S19). The few PAD4 KO cells that completed NETosis had temporal delays in DNA decondensation and extracellular DNA release (Fig. 6D). Immunostaining of vimentin IFs and lamin B1/B2 showed that PAD4 KO cells exhibited delays in the disassembly of peripheral vimentin IFs (SI Appendix, Fig. S4 I and J) and formation of lamin meshwork discontinuities (Fig. 6 E–G). Thus, PAD4 is required for efficient remodeling of the vimentin IF network, DNA decondensation, lamin meshwork and NE rupture, and extracellular DNA release.
Fig. 6.
PAD4 is critical to DNA decondensation, NE rupture, and extracellular DNA expulsion in mouse PMN. PMN from Padi4 WT or Padi4 KO mice (Ms) stimulated with ionomycin, stained with SiR-DNA, and imaged live (A, B, H, and I) by DIC and confocal microscopy at the coverslip–cell interface (Z = 0 μm) and in the cell center (Z = +3 μm) at 2-min intervals for 4 h, time relative to MV shedding noted, or fixed (E and F) after stimulation (time noted). Double-arrow-headed lines above images indicate presence of ionomycin, colored boxes around images correspond to color coded cellular events in the graphs (C). (A, B, H, and I). Time series of image overlays of DIC (Top rows, grayscale) and SiR-DNA (Bottom rows, red). (C) Percent of cells exhibiting MV shedding, DNA decondensation, nuclear rounding, loss of DIC contrast (PM permeabilization) and outer nuclear membrane (ER(ONM)) rupture in cells stained with ER-Tracker Red, DNA release into the cytoplasm, and extracellular DNA release (NETosis) after ionomycin stimulation. n = total cells, point = percent of cells in one experiment. (D) Timing relative to MV shedding of DNA decondensation, ER (ONM) rupture and DNA release. n = total cells observed, point = individual cells, mean (long bar) and SD (short bar) shown below each plot. **** and ** for P value < 0.0001, and <0.01, respectively; ns, nonsignificant, Mann–Whitney U test. (E and F) Immunofluorescence of lamin B1/B2 (Upper row and Bottom row, green) and DAPI (at 10 min) or SiR-DNA (in DMSO, and at 5, 60, and 240 min) staining of DNA (Bottom row, red) in fixed WT (E) or PAD4 KO Ms PMN (F). Arrowheads, rupture points in the lamina. (G) Percent (numbers below bars) of cells with discontinuities in lamin B1/B2 meshwork (time after stimulation noted, 0 = DMSO control). n = total cells. (H and I) Cells stained with ER-Tracker Red (Middle row and Bottom row, green) and SiR-DNA (Bottom row, red). (Scale bars in A, B, E, F, H, and I: 5 μm.)
PAD4 Localizes Predominantly to the Nucleus and Its Enzymatic and Nuclear Localization Activities Mediate Efficient DNA Decondensation, NE Rupture, and Extracellular DNA Expulsion in Human dHL-60.
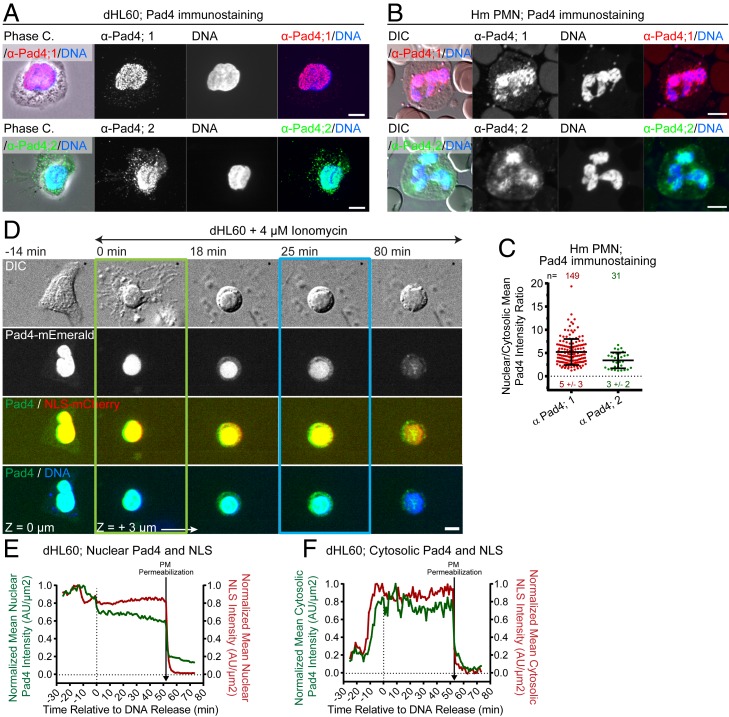
While the importance of PAD4 in mouse PMN NETosis is well documented (13, 14), the role of PAD4 in human neutrophils remains controversial (14, 25). We thus first analyzed PAD4 localization by immunostaining in dHL-60 and human PMNs and its dynamics by expression of PAD4-mEmerald in dHL-60 cells. This showed that PAD4 primarily localized to the nucleus of resting cells as assessed by colocalization with Hoechst- or SiR-stained DNA (Fig. 7 A–C) or with coexpressed mCherry fused to the nuclear localization signal sequence (NLS-mCherry) which served as a soluble nucleoplasmic marker (Fig. 7D and Movie S20). Imaging SiR-DNA-stained dHL-60 cells expressing PAD4-mEmerald after ionomycin stimulation showed that PAD4 remained largely in the nucleus through microvesicle shedding, DNA decondensation, and nuclear rounding. However, prior to nuclear rupture, PAD4 began accumulating in the cytoplasm, followed by rapid nuclear release along with DNA at nuclear rupture (Fig. 7 D and F). Coexpression of PAD4-mEmerald with NLS-mCherry showed that the two initially colocalized in the nucleus and simultaneously accumulated in the cytosol after DNA decondensation but prior to nuclear rupture (Fig. 7 D–F and Movie S20). This indicates that loss of NE integrity, rather than regulated PAD4 export caused its release from the nucleus. In addition, both PAD4-mEmerald and NLS-mCherry were released to the extracellular environment prior to plasma membrane rupture and DNA expulsion (Fig. 7 D and F). Thus, during NETosis, nuclear PAD4 is first released into the cytoplasm by NE permeabilization after DNA decondensation but before nuclear rupture, then released extracellularly by plasma membrane permeabilization, and finally the remaining PAD4 exits the cell along with NET expulsion.
Fig. 7.
PAD4 localizes to the nucleus and enters the cytosol prior to nuclear rupture. dHL-60 (A, D, E, and F) or human (Hm) PMN (B and C) fixed (A and B) or stimulated with ionomycin and imaged live by DIC and confocal microscopy (D) at the coverslip–cell interface (Z = 0 μm) and in the cell center (Z = +3 μm) at 2-min intervals for 4 h, time relative to MV shedding noted. (A and B) Phase-contrast (A, phase C), or DIC (B) and confocal images of DAPI staining of DNA (blue) and immunolocalization of PAD4 by two different antibodies (α-Pad4; 1 [red] and 2 [green]). (C) Mean nuclear to cytosolic ratio of PAD4 intensity from immumostaining. n = total cells observed, points = individual cells, mean (long bar) and SD (short bar) below each plot. (D) Time series of a dHL-60 coexpressing PAD4-mEmerald (Middle and Bottom, green), and NLS-mCherry (third row, red) and SiR-DNA (Bottom row, blue). Double-arrow-headed lines indicate presence of ionomycin, boxes around images indicate cellular events (green: MV shedding; blue: DNA release to the cytosol). (E and F) Normalized mean nuclear (E) and cytosolic (F) intensity over time of PAD4-mEmerald and NLS-mCherry measured from the cell in (D). Loss of DIC contrast (PM permeabilization) noted. (Scale bars in A, B, and D: 5 μm.)
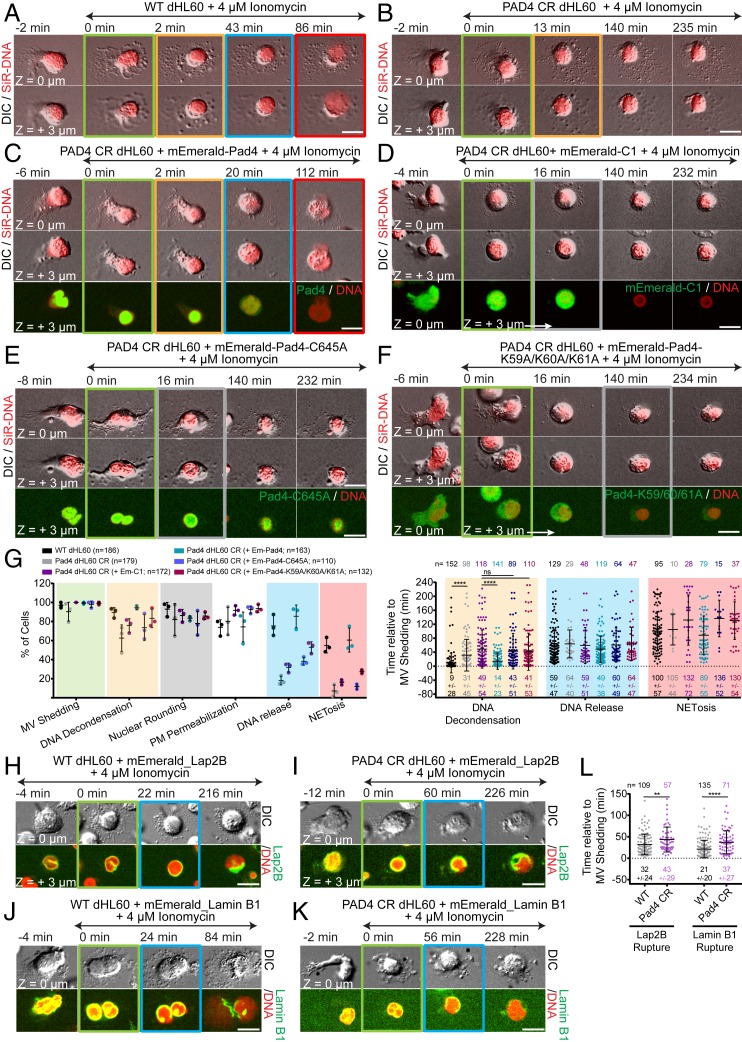
To address the requirement for PAD4 in NETosis in human neutrophils, we generated an HL-60 cell line, PAD4 CR dHL-60, using CRISPR-Cas9 technology. This resulted in near complete loss of PAD4 protein (SI Appendix, Fig. S5 A and B), and had no effect on HL-60 differentiation into neutrophil-like cells, as assessed by reactive oxygen species production after PMA stimulation (20 nM, ref. 26), SI Appendix, Fig. S5 C and D). Stimulation of SiR-DNA-stained WT and PAD4 CR dHL-60 cells with ionomycin showed that PAD4 CR dHL-60 cells exhibited a significant delay in DNA decondensation and largely failed in nuclear rupture and extracellular DNA expulsion (Fig. 8 A, B, and G and Movie S21). Analysis of PAD4 CR dHL-60 cells expressing Lap2β-mEmerald or laminB1-mEmerald showed that for the small fraction of cells that completed NETosis, both NE and lamina rupture was delayed (Fig. 8 H–L). Importantly, reexpressing PAD4-mEmerald in PAD4 CR dHL-60 cells rescued the defects, while expressing mEmerald alone did not (Fig. 8 C, D, and G and Movie S22). Thus, PAD4 is critical for DNA decondensation, nuclear lamina, and NE rupture, and DNA expulsion in both mouse and human neutrophils.
Fig. 8.
PAD4 enzymatic and nuclear localization activities mediate efficient DNA decondensation, NE rupture and extracellular DNA expulsion in human dHL-60. dHL-60 (WT; A, G, H, J, and L) or PAD4 CR dHL-60 (B–G, I, K, and L), stained with SiR-DNA, stimulated with ionomycin, and imaged by DIC and confocal microscopy at the coverslip–cell interface (Z = 0 μm) and in the cell center (Z = +3 μm) at 2-min intervals for 4 h, time in min relative to MV shedding noted on images. Double-arrow-headed lines indicate presence of ionomycin, colored boxes around images correspond to color-coded cellular events in the graphs. (A–F and H–K) Time series of image overlays of DIC (grayscale) and SiR-DNA (red). (C–F) PAD4 CR dHL-60 cells expressing mEmerald-PAD4 (C; Bottom row, green), mEmerald (D; Bottom row, green), mEmerald-PAD4-C645A (E; Bottom row, green) or mEmerald-PAD4-K59A/K60A/K61A (F; Bottom row, green). (G, Left) Percent of cells exhibiting MV shedding, DNA decondensation, nuclear rounding, loss of DIC contrast (PM permeabilization), DNA release to the cytoplasm, and extracellular DNA release (NETosis); (G, Right) Timing relative to MV shedding of DNA decondensation, DNA release to the cytoplasm, and NETosis after ionomycin stimulation. n = total cells observed, point = percent of cells in one experiment (Left), or individual cells (Right). Long bar = mean, short bars = SD shown below each plot (Right). (H–K) WT or PAD4 CR dHL-60 cells expressing Lap2β-mEmerald (H and I; Bottom rows, green) or lamin B1-mEmerald (J and K; Bottom rows, green). (L) Timing relative to MV shedding of the Lap2β or laminB1 rupture. n = total cells observed, points = individual cells, mean (long bar) and SD (short bar) shown below each plot. (G and L) ****P < 0.0001; **P < 0.01; ns, nonsignificant; (G Left and L): Mann–Whitney U test. (Scale bars in A–F and H–K: 10 μm.)
To test if the citrullination or nuclear localization activities were required for the roles of PAD4 in NETosis, we rescued PAD4 CR HL-60 cells with mEmerald-tagged PAD4 mutants specifically defective in these functions. PAD4 is rendered enzymatically dead by a C645A substitution (27), while its nuclear localization is disrupted by the triple K59A/K60A/K61A substitution (28). Expressed PAD4-C645A-mEmerald localized predominantly to the nucleus (Fig. 8E), while PAD4-K59A/K60A/K61A-mEmerald was mostly cytosolic (Fig. 8F). In ionomycin-stimulated PAD4 CR dHL-60 cells, expression of neither PAD4-C645A-mEmerald nor PAD4-K59A/K60A/K61A-mEmerald rescued the defects in DNA decondensation, nuclear rupture, and extracellular DNA release (Fig. 8 E–G and Movie S22) induced by loss of PAD4. This indicates that efficient DNA decondensation, nuclear rupture, and DNA expulsion requires PAD4 enzymatic activity in the nucleus in human neutrophils.
Discussion
In this study we performed the systematic characterization of the timing of dynamic cellular events leading to NETosis in human and mouse primary blood neutrophils, as well as in neutrophil-like dHL-60 cells. We show that NETosis proceeds by a stepwise sequence of cellular events that is conserved between mouse and human primary neutrophils, as well as in dHL-60 cells, thus establishing this cell line as an important model system for in vitro studies of the cellular mechanisms of NETosis. We demonstrate that NETosis begins with the rapid disassembly of the actin cytoskeleton, followed by shedding of plasma membrane microvesicles containing granules and cytosolic components, disassembly and remodeling of the MT and vimentin IF cytoskeletons, ER vesiculation, chromatin decondensation and nuclear rounding, progressive plasma membrane and NE permeabilization, nuclear lamin meshwork and then NE rupture to release chromatin into the cytoplasm, and finally PM rupture and discharge of extracellular chromatin (SI Appendix, Fig. S6). We find that certain cellular events always precede other events, suggesting a requirement for a precise sequence for progression through NETosis. For example, cytoskeletal disassembly, microvesicle shedding and ER vesiculation always occurred before nuclear rupture, while plasma membrane permeabilization could occur before or after nuclear rupture but always preceded extracellular DNA release. We found no evidence of extracellular DNA release in shed microvesicles or without catastrophic rupture of the nucleus or plasma membrane, and we never observed cell viability after NET release, independent of the stimulus or cell system examined. Analysis of mouse PMN and human dHL-60 cells lacking PAD4 demonstrate its required role in efficient DNA decondensation, NE rupture, and extracellular DNA release, and prompt timing of lamin meshwork and vimentin IF disassembly, and mutant add-back experiments show that the nuclear localization and enzymatic activities of PAD4 are critical to these roles.
The conservation of cellular events between mouse and human PMNs and neutrophil-like dHL-60 cells with multiple different stimuli suggests that despite possible divergences in signaling pathways mediating the process (12), NETosis follows a very strict cellular mechanism. Thus, targeting cellular pathways might be a better approach for controlling NETosis progression than targeting the various divergent signaling pathways. Despite the similarities, the three cell types showed some differences; for instance, while mouse PMNs and dHL-60 cells exhibited loss of nuclear lobularity and full nuclear rounding during DNA decondensation, human PMN nuclear lobularity decreased without full rounding up. This could result from differences in lamin expression (SI Appendix, Fig. S3A) promoting differences in nuclear mechanical properties (29).
Our study shows that the plasma membrane undergoes dramatic changes during NETosis, including shedding of microvesicles and regulated permeabilization. Microvesicles could act as important systemic messengers, signaling stress (30) or contributing to disease (31), including the promotion of thrombosis (32). The progressive increase in size of molecules entering the cell suggests a stepwise permeabilization mediated by different membrane pores that could allow escape of specific bioactive molecules like cytokines, citrullinated peptides, or proteins prior to NET release. For example, release of PAD4 from cells prior to NETosis could mediate ADAMTS13 citrullination, preventing clearance of von Willebrand factor from vessel walls, thus enhancing NET attachment and thrombosis (7). The mechanism of permeability is also of interest, with gasdermin D, that forms large pores and is critical for NETosis (20, 33, 34), being a good candidate for mediating the late stages of plasma membrane permeability during NETosis.
Our documentation of rapid disassembly of the actin, MT, and vimentin cytoskeletons and remodeling of nuclear lamins after stimulation of NETosis is reminiscent of the cytoskeletal remodeling at mitosis (35–37) and further supports a role for mitotic kinases in NETosis (21). However, NETosis stimulants also trigger intracellular calcium influx (9, 38, 39), and it is well known that calcium can directly or indirectly promote the disassembly of MTs (40), actin (41), and vimentin filaments (42), suggesting an important role for calcium in progression through NETosis. Our finding that actin disassembly was required for plasma membrane rupture and extracellular NET release indicates that molecules mediating actin disassembly could serve as therapeutic targets for NETosis inhibition.
Our systematic analysis of the role of PAD4 in the cellular events of NETosis utilized genetically modified mouse PMN and dHL-60 human cells, precluding the lack of specificity or reversibility of PAD4 inhibitors (14, 25). We demonstrate that PAD4 is critical for efficient DNA decondensation, lamin meshwork and NE rupture and extracellular DNA release in mouse PMNs and dHL-60 cells. However, we found that PAD4 plays a more prominent role in efficient DNA decondensation in mouse PMN than in dHL-60 cells. This could be attributed to the residual (∼5%) PAD4 in the gene-edited HL-60 cell line, or proteases may be more important for DNA decondensation in human than in mouse cells (11). We also show that both the citrullination activity and nuclear localization signal of PAD4 are required for NETosis, strongly supporting the notion that PAD4 enzymatic activity in the nucleus, potentially through histone citrullination (10), mediates NETosis. Finally, since PAD4 depletion delays the disassembly of lamins and vimentin, both of which are known to maintain nuclear mechanical integrity (43, 44), it is possible that citrullination may regulate intermediate filament disassembly to promote nuclear rupture during NETosis.
Materials and Methods
Cells.
HL-60 cells (ATCC CCL-240) were cultured at 37 °C and 5% CO2 in RPMI 1640 plus l-glutamine with 25 mM Hepes, 1% penicillin and streptomycin (P/S), and 15% heat-inactivated FBS and differentiated to neutrophil-like cells by addition of 1.3% of DMSO. Human and mouse neutrophils were isolated from fresh blood on Percoll gradients. C. albicans were cultured at 30 °C in YPD media with 1% P/S. Cells were resuspended in imaging media (RPMI 1640 lacking phenol red with 25 mM Hepes and 1% antibiotics) before imaging. For more details, see SI Appendix, Materials and Methods.
cDNA Expression Vectors.
cDNAs encoding CaaX-mApple, H1-mEmerald, ER-5-mEmerald, Lamin B1-mApple, Lamin A-mEmerald, F-tractin-mApple, G-actin-mEmerald, Tubulin-mEmerald, Vimentin-mEmerald, H2B-mEmerald, and mEmeral-C1 were the kind gift of the late Mike Davidson, Florida State University, Tallahassee, FL; eGFP-Ensconsin (3X-GFP-EMTB) was a kind gift from Chloe Bulinski, Columbia University, New York, NY; pSpCas9-2A-GFP vector was a kind gift from James Anderson’s laboratory, NHLBI/NIH, Bethesda, MD. Human PADi4 mRNA (pCMV3-N-His-PADi4, HG11072-NH) was inserted into a mEmerald-C1 vector and the enzymatically dead (mEmerald-PAD4-C645A) and the NLS dead (mEmerald-PAD4-K59A/K60A/K61A) mutants were generated by in vitro site-directed mutagenesis. For more details, see SI Appendix, Materials and Methods.
Generation of Gene-Edited HL-60 PAD4 Crispr Cell Line.
The generation of the PAD4 Crispr line was performed as described (45) using the guide RNA Guide1: 3′→5′ TC ACA CGG ATC AAT GTC CCC TGG. For more details, see SI Appendix, Materials and Methods.
Immunofluorescence.
Cells in imaging media were incubated on cleaned coverslips for 5 min at 37 °C and 5% CO2 before the addition of DMSO or ionomycin (4 μM); further incubation and then indirect immunofluorescence were performed. For more details, see SI Appendix, Materials and Methods.
dHL-60 NETosis Endpoint Assay.
NETosis endpoint assay was performed as already described (22). For more details, see SI Appendix, Materials and Methods.
Western Blot.
Western blots were performed as previously described (46). For more details, see SI Appendix, Materials and Methods.
Nitroblue Tetrazolium Assay for Reactive Oxygen Species Production.
The nitroblue tetrazolium (NBT) assay was performed as described (27). For more details, see SI Appendix, Materials and Methods.
RT-PCR for PADi4 mRNA Detection.
RNA from WT and PAD4 CR dHL-60 cells was extracted and RT-PCR was performed using Gapdh as loading control. The following primers were used: PAD4 P1 Fw: 5′-ACT TCT TCA CAA ACC ATA CAC TGG-3′; PAD4 P1 Rev: 5′-CCT CGA GTT ACA TAG CCA AAA TCT-3′; PAD4 P2 Fw: 5′-GTG TTC CAA GAC AGC GTG GT-3′; PAD4 P2 Rev: 5′-GTT TGA TGG GAA ACT CCT TCA G-3′; and GAPDH Fw: 5′-ACC CAG AAG ACT GTG GAT GG-3′; GAPDH Rev: 5′-CCC CTC TTC AAG GGG TCT AC-3′. For more details, see SI Appendix, Materials and Methods.
Microscopy.
Imaging was performed on a Nikon Eclipse Ti or Ti2 equipped with a Yokogawa CSU-X1 or CSU-W1 spinning disk scanhead, a Nikon A1R resonance-scanning confocal microscope, a Zeiss LSM 510 laser scanning confocal microscope, or a Zeiss Axiovert 200M wide-field fluorescence microscope. For live cell imaging, microscopes were equipped with stage-top incubators. For more details, see SI Appendix, Materials and Methods.
Image Analysis.
Image analysis was performed from DIC and fluorescence overlay time-lapse movies. Single cells were followed over time to assess the percentage and timing of events. A detailed description of the quantification procedure can be found in SI Appendix, Materials and Methods.
Statistical Analysis.
Details on statistical tests can be found in SI Appendix, Materials and Methods.
Material and Data Availability.
Raw and processed data (Movies, Excel, and GraphPad files), cell lines, and cDNA expression vectors are available upon request to the corresponding author.
Supplementary Material
Acknowledgments
We thank the Marine Biological Laboratory Whitman Fellows program (D.D.W.); Nikon Instruments for loan of equipment (C.M.W.); the Nikon Fellows program (D.D.W.); Schwanna Thacker, William Shin, Nihal Altan-Bonnet, Robert Fischer, and Ana Pasapera; the National Heart, Lung, and Blood Institute (NHLBI) flow cytometry, light microscopy, and animal facilities; and the NHLBI blood bank. This work was funded by the NHLBI Division of Intramural Research (C.M.W. and H.R.T.); NIH R01 GM106023 and NIH P01 GM096971 (R.D.G., A.E.G., R.Q., M.K., and A.V.); NIH T32 CA08062160038140 (M.K. and A.V.); and NIH R35 HL135765 (D.D.W. and S.L.W.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. G.P.D. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1909546117/-/DCSupplemental.
References
- 1.Brinkmann V., et al. , Neutrophil extracellular traps kill bacteria. Science 303, 1532–1535 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Urban C. F., Reichard U., Brinkmann V., Zychlinsky A., Neutrophil extracellular traps capture and kill Candida albicans yeast and hyphal forms. Cell. Microbiol. 8, 668–676 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Saitoh T., et al. , Neutrophil extracellular traps mediate a host defense response to human immunodeficiency virus-1. Cell Host Microbe 12, 109–116 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Sorvillo N., Cherpokova D., Martinod K., Wagner D. D., Extracellular D. N. A., Extracellular DNA NET-works with dire consequences for health. Circ. Res. 125, 470–488 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clark S. R., et al. , Platelet TLR4 activates neutrophil extracellular traps to ensnare bacteria in septic blood. Nat. Med. 13, 463–469 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Jorch S. K., Kubes P., An emerging role for neutrophil extracellular traps in noninfectious disease. Nat. Med. 23, 279–287 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Fuchs T. A., et al. , Extracellular DNA traps promote thrombosis. Proc. Natl. Acad. Sci. U.S.A. 107, 15880–15885 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Demers M., et al. , Cancers predispose neutrophils to release extracellular DNA traps that contribute to cancer-associated thrombosis. Proc. Natl. Acad. Sci. U.S.A. 109, 13076–13081 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gupta S., Chan D. W., Zaal K. J., Kaplan M. J., A high-throughput real-time imaging technique to quantify NETosis and distinguish mechanisms of cell death in human neutrophils. J. Immunol. 200, 869–879 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y., et al. , Histone hypercitrullination mediates chromatin decondensation and neutrophil extracellular trap formation. J. Cell Biol. 184, 205–213 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Papayannopoulos V., Metzler K. D., Hakkim A., Zychlinsky A., Neutrophil elastase and myeloperoxidase regulate the formation of neutrophil extracellular traps. J. Cell Biol. 191, 677–691 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kenny E. F., et al. , Diverse stimuli engage different neutrophil extracellular trap pathways. eLife 6, e24437 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li P., et al. , PAD4 is essential for antibacterial innate immunity mediated by neutrophil extracellular traps. J. Exp. Med. 207, 1853–1862 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lewis H. D., et al. , Inhibition of PAD4 activity is sufficient to disrupt mouse and human NET formation. Nat. Chem. Biol. 11, 189–191 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuchs T. A., et al. , Novel cell death program leads to neutrophil extracellular traps. J. Cell Biol. 176, 231–241 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pilsczek F. H., et al. , A novel mechanism of rapid nuclear neutrophil extracellular trap formation in response to Staphylococcus aureus. J. Immunol. 185, 7413–7425 (2010). [DOI] [PubMed] [Google Scholar]
- 17.Krishnamoorthy N., et al. ; National Heart, Lung, and Blood Institute Severe Asthma Research Program-3 Investigators , Neutrophil cytoplasts induce TH17 differentiation and skew inflammation toward neutrophilia in severe asthma. Sci. Immunol. 3, eaao4747 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Metzler K. D., Goosmann C., Lubojemska A., Zychlinsky A., Papayannopoulos V., A myeloperoxidase-containing complex regulates neutrophil elastase release and actin dynamics during NETosis. Cell Rep. 8, 883–896 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neubert E., et al. , Chromatin swelling drives neutrophil extracellular trap release. Nat. Commun. 9, 3767 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sollberger G., et al. , Gasdermin D plays a vital role in the generation of neutrophil extracellular traps. Sci. Immunol. 3, eaar6689 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Amulic B., et al. , Cell-cycle proteins control production of neutrophil extracellular traps. Dev. Cell 43, 449–462.e5 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Wong S. L., et al. , Diabetes primes neutrophils to undergo NETosis, which impairs wound healing. Nat. Med. 21, 815–819 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raposo G., Stoorvogel W., Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 200, 373–383 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paulin-Levasseur M., Giese G., Scherbarth A., Traub P., Expression of vimentin and nuclear lamins during the in vitro differentiation of human promyelocytic leukemia cells HL-60. Eur. J. Cell Biol. 50, 453–461 (1989). [PubMed] [Google Scholar]
- 25.Jones J. E., et al. , Synthesis and screening of a haloacetamidine containing library to identify PAD4 selective inhibitors. ACS Chem. Biol. 7, 160–165 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blair O. C., Carbone R., Sartorelli A. C., Differentiation of HL-60 promyelocytic leukemia cells monitored by flow cytometric measurement of nitro blue tetrazolium (NBT) reduction. Cytometry 6, 54–61 (1985). [DOI] [PubMed] [Google Scholar]
- 27.Arita K., et al. , Structural basis for Ca(2+)-induced activation of human PAD4. Nat. Struct. Mol. Biol. 11, 777–783 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Nakashima K., Hagiwara T., Yamada M., Nuclear localization of peptidylarginine deiminase V and histone deimination in granulocytes. J. Biol. Chem. 277, 49562–49568 (2002). [DOI] [PubMed] [Google Scholar]
- 29.Lammerding J., et al. , Lamin A/C deficiency causes defective nuclear mechanics and mechanotransduction. J. Clin. Invest. 113, 370–378 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hugel B., Martínez M. C., Kunzelmann C., Freyssinet J.-M., Membrane microparticles: Two sides of the coin. Physiology (Bethesda) 20, 22–27 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Genschmer K. R., et al. , Activated PMN exosomes: Pathogenic entities causing matrix destruction and disease in the lung. Cell 176, 113–126.e15 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hrachovinová I., et al. , Interaction of P-selectin and PSGL-1 generates microparticles that correct hemostasis in a mouse model of hemophilia A. Nat. Med. 9, 1020–1025 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Chen K. W., et al. , Noncanonical inflammasome signaling elicits gasdermin D–dependent neutrophil extracellular traps. Sci. Immunol. 3, eaar6676 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Ruan J., Xia S., Liu X., Lieberman J., Wu H., Cryo-EM structure of the gasdermin A3 membrane pore. Nature 557, 62–67 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chou Y.-H., Bischoff J. R., Beach D., Goldman R. D., Intermediate filament reorganization during mitosis is mediated by p34cdc2 phosphorylation of vimentin. Cell 62, 1063–1071 (1990). [DOI] [PubMed] [Google Scholar]
- 36.Lancaster O. M., et al. , Mitotic rounding alters cell geometry to ensure efficient bipolar spindle formation. Dev. Cell 25, 270–283 (2013). [DOI] [PubMed] [Google Scholar]
- 37.Heald R., McKeon F., Mutations of phosphorylation sites in lamin A that prevent nuclear lamina disassembly in mitosis. Cell 61, 579–589 (1990). [DOI] [PubMed] [Google Scholar]
- 38.Morgan A. J., Jacob R., Ionomycin enhances Ca2+ influx by stimulating store-regulated cation entry and not by a direct action at the plasma membrane. Biochem. J. 300, 665–672 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alpizar Y. A., et al. , TRPV4 activation triggers protective responses to bacterial lipopolysaccharides in airway epithelial cells. Nat. Commun. 8, 1059 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O’Brien E. T., Salmon E. D., Erickson H. P., How calcium causes microtubule depolymerization. Cell Motil. Cytoskeleton 36, 125–135 (1997). [DOI] [PubMed] [Google Scholar]
- 41.Janmey P. A., Phosphoinositides and calcium as regulators of cellular actin assembly and disassembly. Annu. Rev. Physiol. 56, 169–191 (1994). [DOI] [PubMed] [Google Scholar]
- 42.Yoshida H., Murachi T., Tsukahara I., Degradation of actin and vimentin by calpain II, a Ca2+-dependent cysteine proteinase, in bovine lens. FEBS Lett. 170, 259–262 (1984). [DOI] [PubMed] [Google Scholar]
- 43.Dahl K. N., Kahn S. M., Wilson K. L., Discher D. E., The nuclear envelope lamina network has elasticity and a compressibility limit suggestive of a molecular shock absorber. J. Cell Sci. 117, 4779–4786 (2004). [DOI] [PubMed] [Google Scholar]
- 44.Patteson A. E., et al. , Vimentin protects cells against nuclear rupture and DNA damage during migration. J. Cell Biol. 218, 4079–4092 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ran F. A., et al. , Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pasapera A. M., et al. , Rac1-dependent phosphorylation and focal adhesion recruitment of myosin IIA regulates migration and mechanosensing. Curr. Biol. 25, 175–186 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.