Significance
Bacteria use the nucleotide cyclic di-3′,5′-adenosine monophosphate (c-di-AMP) for adaptation to changing environments and host–pathogen interactions. Enzymes for nucleotide synthesis and degradation and proteins for binding of the second messenger are key components of signal transduction pathways. It was long unknown how the majority of Actinobacteria, one of the largest bacterial phyla, stop c-di-AMP signals and which proteins bind the molecule to elicit cellular responses. Here, we identify a c-di-AMP phosphodiesterase that bacteria evolved to terminate c-di-AMP signaling and a protein that forms a complex with c-di-AMP in Streptomyces. We also demonstrate that balance of c-di-AMP is critical for developmental transitions from filaments to spores in multicellular bacteria.
Keywords: c-di-AMP, Streptomyces, phosphodiesterase, development, osmostress
Abstract
Antibiotic-producing Streptomyces use the diadenylate cyclase DisA to synthesize the nucleotide second messenger c-di-AMP, but the mechanism for terminating c-di-AMP signaling and the proteins that bind the molecule to effect signal transduction are unknown. Here, we identify the AtaC protein as a c-di-AMP-specific phosphodiesterase that is also conserved in pathogens such as Streptococcus pneumoniae and Mycobacterium tuberculosis. AtaC is monomeric in solution and binds Mn2+ to specifically hydrolyze c-di-AMP to AMP via the intermediate 5′-pApA. As an effector of c-di-AMP signaling, we characterize the RCK_C domain protein CpeA. c-di-AMP promotes interaction between CpeA and the predicted cation/proton antiporter, CpeB, linking c-di-AMP signaling to ion homeostasis in Actinobacteria. Hydrolysis of c-di-AMP is critical for normal growth and differentiation in Streptomyces, connecting ionic stress to development. Thus, we present the discovery of two components of c-di-AMP signaling in bacteria and show that precise control of this second messenger is essential for ion balance and coordinated development in Streptomyces.
Bacteria use mono-, di-, and trinucleotides as second messengers to control fundamental physiological functions in response to signal sensing (1). Among these molecules, cyclic di-3′,5′-adenosine monophosphate (c-di-AMP) is the only nucleotide messenger that must be precisely balanced since both its depletion and overproduction can be toxic (2). Its core function is to control cellular integrity by setting homeostasis of osmolytes that in many bacteria are used for osmoregulation (3, 4). Changes in external osmolarity trigger water fluxes across the membrane, which can lead to cell dehydration or swelling and, finally, collapse or burst when osmobalance mechanisms fail to respond properly (5). As a key component of these mechanisms, c-di-AMP directly targets transport systems for osmoactive and osmoprotective substances such as potassium ions and low-molecular-weight compatible solutes in many bacteria (6–10).
c-di-AMP also plays a central role in host–pathogen interactions and bacterial virulence (11). Secreted c-di-AMP is recognized by hosts’ innate immunity receptors STING, DDX41, ERAdP, and RECON to regulate type I interferon immune response and NF-κB pathways, respectively (12–16). Modulation of intracellular c-di-AMP has been reported to affect virulence of Streptococcus pyogenes (17), Listeria monocytogenes (18), Streptococcus pneumoniae (19), and Mycobacterium tuberculosis. Thus, the molecule is considered an attractive antimicrobial target (20).
c-di-AMP synthesis out of two ATP molecules is catalyzed by the diadenylate cyclase (DAC) activity of the DisA_N domain (Pfam PF02457), which was identified in the structural and biochemical analysis of the DNA integrity scanning protein A (DisA) of Thermotoga maritima (21). DisA is mainly present in sporulating Firmicutes and Actinobacteria (22) and has a conserved domain organization consisting of an N-terminal DAC domain and a C-terminal DNA-binding helix–hairpin–helix domain separated by a linker region (21). c-di-AMP hydrolysis is mediated by the DHH-DHHA1 domain containing the Asp–His–His motif. The multidomain membrane-associated GdpP protein in Bacillus subtilis was the first characterized DHH-DHHA1-type phosphodiesterase (PDE) (23). In addition, HD domains, which contain a catalytic His–Asp motif and were first identified in the PgpH protein in L. monocytogenes, also degrade c-di-AMP (18).
Most Actinobacteria contain DisA for c-di-AMP synthesis; however, the majority of them do not encode DHH-DHHA1 domain–containing or HD-type c-di-AMP PDEs. Hence, we wondered how actinomycetes balance intracellular c-di-AMP levels. Within Actinobacteria, Streptomyces are the most extensively studied mycelial organisms and the richest natural source of antibiotics (24). For growth and reproduction, Streptomyces undergo a complex developmental life cycle, which involves the conversion between three morphologically and physiologically distinct forms of cell existence. During exponential growth, they proliferate by extension and branching of vegetative hyphae. The switch to the stationary phase and onset of the reproductive phase are marked by the erection of aerial hyphae. These filaments elongate and divide into unigenomic prespore compartments that ultimately mature into chains of spores. Completion of the developmental program is easily visible by eye since mature Streptomyces spores accumulate a spore pigment. For example, our model species, the chloramphenicol producer S. venezuelae, is characterized by a green spore pigment such that colonies turn green at the end of the life cycle (25, 26). Importantly, antibiotic production and morphological differentiation are coregulated in Streptomyces. Hence, studying their developmental biology also provides a better understanding of the control of their secondary metabolism.
In this work, we identified and characterized the PDE superfamily protein AtaC as a c-di-AMP–specific hydrolase unrelated to canonical c-di-AMP PDEs. AtaC is broadly distributed in bacteria and the only known c-di-AMP PDE in most actinomycetes. Among others, pathogens such as the causative agent of pneumonia, S. pneumoniae, contain an AtaC homolog that we characterize here to be a functional c-di-AMP hydrolase. Our biochemical and structural analyses show that AtaC is a monomeric Mn2+-dependent PDE with high affinity for c-di-AMP. Moreover, we provide direct biochemical evidence that Streptomyces DisA is an active DAC and that c-di-AMP produced by DisA is crucial for survival under ionic stress conditions. Further, we show that accumulation of c-di-AMP in the S. venezuelae ataC mutant results in profound developmental and growth defects and report the identification of the RCK_C domain (RCK stands for regulator of conductance of K+) containing protein CpeA as a c-di-AMP binding protein in Streptomyces. Overall, in this study we identified and functionally characterized core components of c-di-AMP signaling in Streptomyces and linked c-di-AMP regulation to ion homeostasis to control differentiation in multicellular bacteria.
Results
DisA Synthesizes c-di-AMP in S. venezuelae.
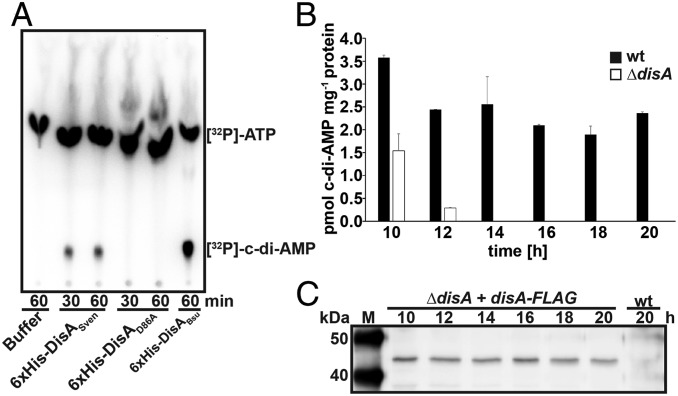
DisA is the sole DAC protein encoded in the S. venezuelae genome and is conserved in all sequenced Streptomyces strains. To demonstrate DisA DAC activity, we purified N-terminally His-tagged DisA and an inactive DisAD86A that carries an alanine instead of aspartate in the active site (21). We included His-tagged B. subtilis DisA (DisABsu) as a positive control for enzymatic activity (21). [32P]-labeled ATP was added as a substrate for in vitro DAC assays, and the reactions were separated by thin-layer chromatography (TLC). DisA synthesized c-di-AMP, whereas the mutated DisAD86A failed, demonstrating that S. venezuelae DisA is a functional DAC, which requires the conserved catalytic aspartate D86 for activity (Fig. 1A).
Fig. 1.
DisA is an active diadenylate cyclase in vitro and in vivo. (A) TLC of DAC assay with purified 6xHis-DisASven and 6xHis-DisAD86A and [32P]-ATP as the substrate. Migration of [32P]-ATP in buffer is shown in lane 1. 6xHis-DisABsu served as a positive control for DAC activity. (B) Intracellular c-di-AMP levels in S. venezuelae wild type (wt) and ΔdisA during late vegetative growth (10 to 12 h), early sporulation (14 to 16 h), and sporulation (18 to 20 h). Data are presented as the mean of biological replicates ± SD (n = 3). (C) Expression profile of DisA-FLAG in a disA mutant complemented with disA-FLAG under the control of the disA promoter grown in liquid sporulation medium (MYM). DisA-FLAG was detected using a monoclonal anti-FLAG antibody. Wild type served as a negative control.
In vivo, DisA is the major source for c-di-AMP during onset of sporulation (14 to 16 h) and the sporulation phase (18 to 20 h) (Fig. 1B) (27). However, we reproducibly detected low c-di-AMP levels in ΔdisA during vegetative growth (10 and 12 h), suggesting that S. venezuelae might contain a non–DAC domain enzyme capable of c-di-AMP production (Fig. 1B). The presence of c-di-AMP throughout the wild-type S. venezuelae life cycle suggested that disA expression is constitutive. To confirm this, we complemented the disA mutant by chromosomal insertion of a C-terminally FLAG-tagged disA under control of its native promoter. Using a monoclonal anti-FLAG antibody, we detected constant DisA-FLAG expression in all developmental stages, which correlated with c-di-AMP production in the wild type under the conditions tested (Fig. 1C and SI Appendix, Fig. S1A).
Altogether, our data show that DisA is a functional DAC in vitro and in vivo and the major enzyme for c-di-AMP production during S. venezuelae sporulation.
The Phosphodiesterase Superfamily Protein AtaC (Vnz_27310) Degrades c-di-AMP.
Streptomycetes do not possess PDEs with a DHH-DHHA1 domain or a PgpH-type HD domain, known to degrade c-di-AMP in other bacteria (18, 23), raising the question of how S. venezuelae removes c-di-AMP from the cytoplasm. To find a potentially novel c-di-AMP PDE, we used interproscan (28) to search for Pfam PF01663, which is associated with putative type I phosphodiesterases/nucleotide pyrophosphatases. Among others, we found two proteins (Vnz_27310 and Vnz_31010) belonging to the phosphodiesterase and metallophosphatase superfamilies, respectively, that we selected for in vitro PDE activity tests.
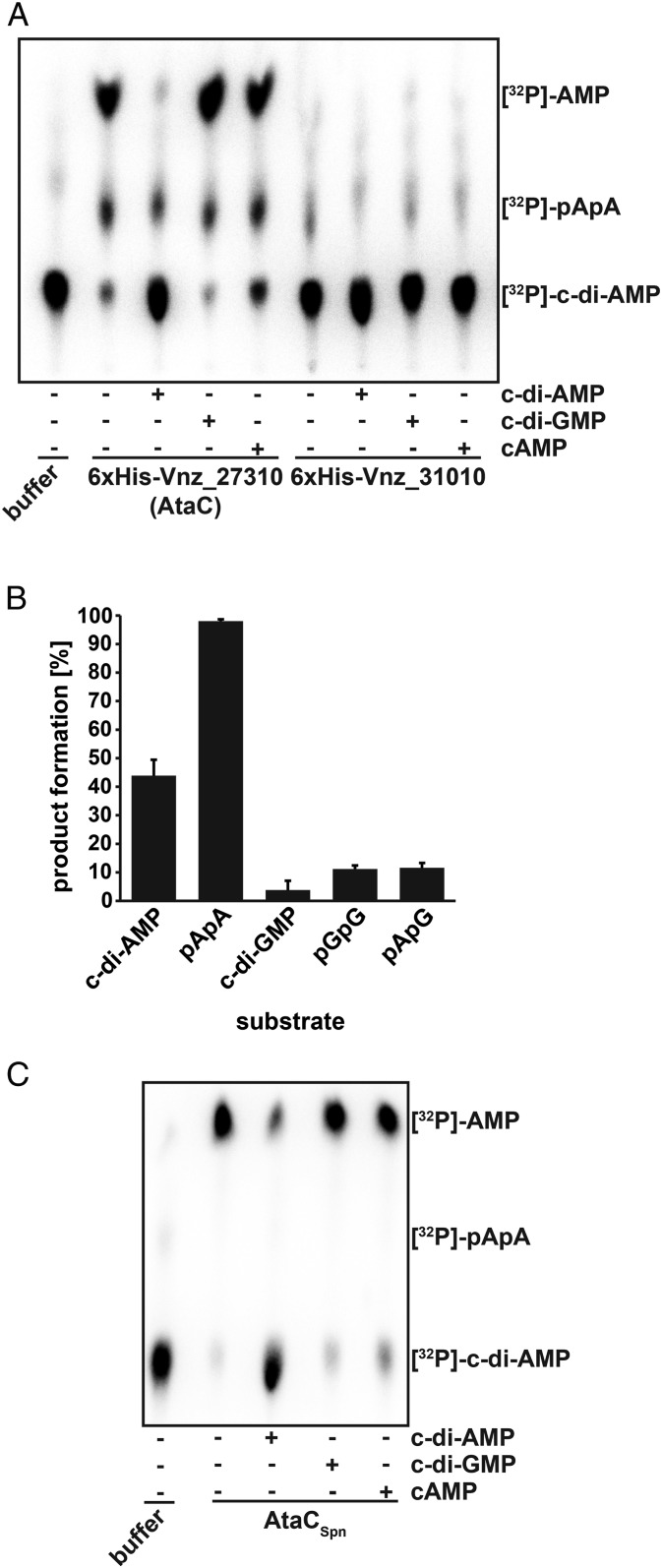
Purified N-terminally His-tagged Vnz_27310 and Vnz_31010 were assayed in vitro using [32P]-labeled c-di-AMP as a substrate. While we could not detect [32P]-c-di-AMP cleavage activity for Vnz_31010, Vnz_27310 clearly degraded c-di-AMP to 5′-pApA and finally to AMP (Fig. 2A), so that we named Vnz_27310 AtaC for actinobacterial PDE targeting c-di-AMP. The addition of unlabeled c-di-AMP but not of c-di-GMP or cAMP competed with [32P]-c-di-AMP and led to reduced cleavage of the radiolabeled substrate, showing specificity for c-di-AMP (Fig. 2A). We analyzed the kinetics of c-di-AMP hydrolysis activity of AtaC using anion exchange chromatography assays and determined a catalytic rate constant (kcat) of 0.2 s−1 (SI Appendix, Fig. S2 A and B), while only a negligible c-di-GMP hydrolysis activity was detected (SI Appendix, Fig. S2C). We also compared AtaC-dependent hydrolysis of the linear dinucleotides 5′-pApG and 5′-pGpG to the hydrolysis of 5′-pApA and observed a high hydrolysis activity for 5′-pApA (kcat = 2.1 s−1), whereas the other substrates tested were only degraded to a small extent (Fig. 2B and SI Appendix, Fig. S2 D–F).
Fig. 2.
AtaC is a c-di-AMP-specific PDE. TLC of the PDE assay of AtaC and Vnz_31010 from S. venezuelae (A) and S. pneumoniae (AtaCSpn) (C) with [32P]-c-di-AMP. Radioactively labeled c-di-AMP in buffer migrates as shown in lane 1. In samples used for competition, unlabeled c-di-AMP, c-di-GMP, or cAMP (indicated by +) were added in excess before starting the reaction with [32P]-c-di-AMP. (B) AtaC activity assay by ion exchange chromatography runs on a 1 mL Resource Q column of the reaction products after 1 h incubation from 100 µL reactions containing 100 nM AtaC + 250 µM c-di-AMP, 5′-pApA, c-di-GMP, 5′-pGpG, or 5′-pApG (n = 3).
Using the PATRIC database (https://www.patricbrc.org), we examined the distribution of the here discovered c-di-AMP PDE (PGF_00172869) and found at least 5,374 prokaryotic species containing homologs to AtaC (Dataset S1), including pathogens such as S. pneumoniae and M. tuberculosis. AtaC from S. pneumoniae (AtaCSpn; sequence ID: CVN04004.1) and from M. tuberculosis (AtaCMtu; sequence ID: CNE38097.1) share 41 and 47%, respectively, identical residues with AtaC from S. venezuelae. In agreement with the high degree of protein identity, enzyme assays data shown in Fig. 2C demonstrate that AtaCSpn is a PDE that hydrolyzes c-di-AMP and AtaCMtu likely has the same function.
In summary, we identified and functionally characterized a c-di-AMP hydrolase in Streptomyces and a c-di-AMP signaling component in pathogens and showed that AtaC is a conserved phosphodiesterase that efficiently and specifically hydrolyzes c-di-AMP to AMP via the intermediate 5′-pApA.
AtaC Is a Monomeric Mn2+-Dependent Phosphodiesterase.
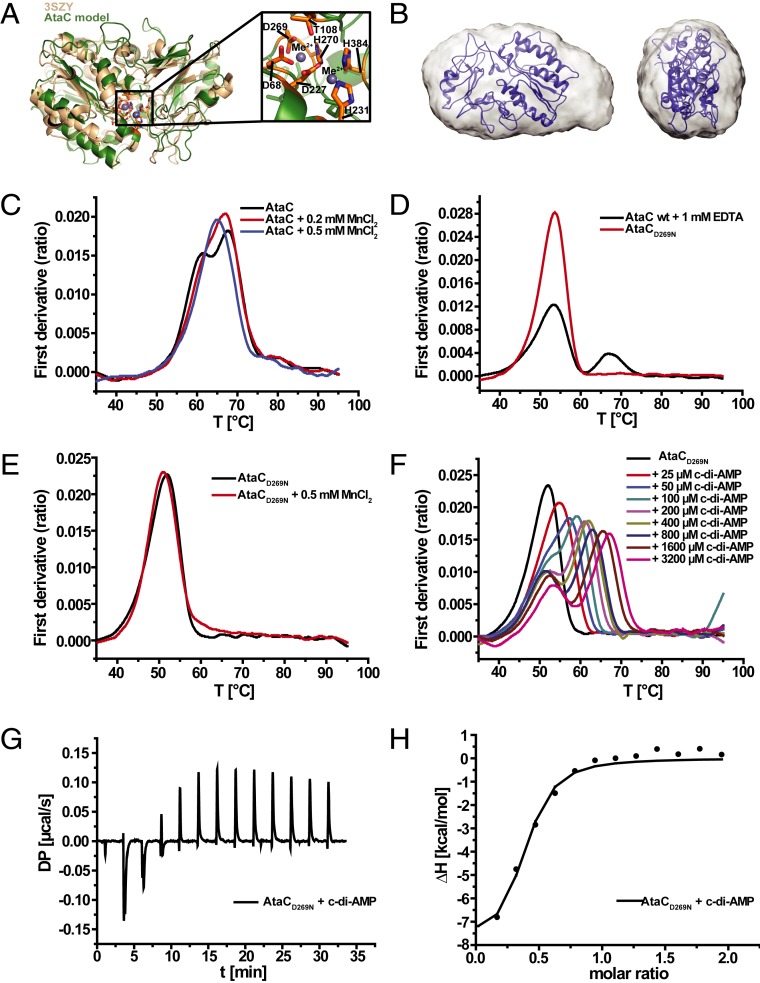
To further characterize the c-di-AMP hydrolysis mechanism of AtaC and to gain some structural insights into this PDE, we used HHpred (29) and found two close structural homologs. The core domain of a phosphonoacetate hydrolase (PhnA) from Sinorhizobium meliloti 1021 (ref. 30 and Protein Data Bank [PDB] code 3SZY) showed the highest similarity and served as a template for the structural model of AtaC including the putative active site. The predicted active site comprises three aspartates (D68, D227, and D269), three histidines (H231, H270, and H384), and one threonine (T108) (Fig. 3A).
Fig. 3.
AtaC is a monomeric Mn2+-dependent PDE. (A) Model of AtaC obtained from HHpred/MODELER (green) superimposed with best-match 3SZY (beige). The close-up shows the predicted active site, annotated with all of the most conserved residues. (B) Modeled structure from A superimposed with the final averaged and filtered ab initio shape (16 ab initio models averaged) from SEC-SAXS with the front view (Left) and side view (Right). (C) nanoDSF thermal shift first-derivative curves of 10 µM apo AtaC (black), 10 µM AtaC + 0.2 mM MnCl2 (red), and 10 µM AtaC + 0.5 mM MnCl2 (blue). (D) nanoDSF thermal shift first-derivative curves of 10 µM AtaC + 1 mM EDTA (black) and 10 µM AtaCD269N (red). (E) nanoDSF thermal shift first-derivative curves of 10 µM AtaCD269N (black) and AtaCD269N + 0.5 mM MnCl2 (red). (F) nanoDSF thermal shift first-derivative curves of 10 µM AtaCD269N + c-di-AMP (25 to 3,200 µM). (G) ITC measurement raw data of 23 µM AtaCD269N mutant titrated with 231 µM c-di-AMP. (H) Binding curve and fit of ITC titration of the AtaCD269N mutant with c-di-AMP (KD = 949 ± 360 nM; n = 3).
Our size exclusion chromatography (SEC) coupled multiangle laser light scattering data show that AtaC is a monomer in solution with a determined molecular weight of 43.7 kDa (SI Appendix, Fig. S3A). The calculated ab initio shape of AtaC from SEC-SAXS (size exclusion coupled small-angle X-ray scattering) data superimposes well with the HHpred model structure (Fig. 3B), and the measured SAXS curve of AtaC is similar to the theoretical scattering curve of our AtaC model (SI Appendix, Fig. S3 B–D), indicating that AtaC and PhnA have a similar shape in solution.
The enzymatic reaction of the PhnA-class hydrolases is known to be catalyzed by two metal ions in the active site (30), so we tested the metal binding for AtaC by thermal unfolding assays using a nano differential scanning fluorimetry (nanoDSF) assay and observed protein stabilization upon addition of manganese ions (Mn2+) (Fig. 3C). Based on the structural similarity to PhnA, we identified potential metal-binding residues in AtaC and generated a variant, AtaCD269N, that we expected to lack Mn2+ coordination but retain nucleotide binding, as shown for DHH-DHHA1-type PDEs (23, 31). nanoDSF data confirmed the stability of AtaCD269N with a melting temperature comparable to the wild-type protein when incubated with ethylenediaminetetraacetic acid (EDTA) (Fig. 3D). Moreover, AtaCD269N behaved identically to the wild-type protein during purification and final SEC. In line with our predictions, AtaCD269N failed to bind Mn2+ (Fig. 3E) and did not hydrolyze c-di-AMP, as shown using ion exchange chromatography-based assays (SI Appendix, Fig. S4A). However, AtaCD269N was still capable of c-di-AMP binding, as confirmed by nanoDSF experiments that showed a shift in the melting curve with increasing ligand concentration (Fig. 3F). Using isothermal titration calorimetry (ITC) analysis, we determined the dissociation constant (KD) of AtaCD269N for c-di-AMP to be 949 ± 360 nM, whereas binding of c-di-GMP could not be detected (Fig. 3 G and H and SI Appendix, Fig. S4B).
Altogether, our combined structural analysis and biochemical data strongly suggest that AtaC uses a similar metal ion-dependent mechanism as its structural homolog PhnA for substrate cleavage.
AtaC Hydrolyzes c-di-AMP In Vivo.
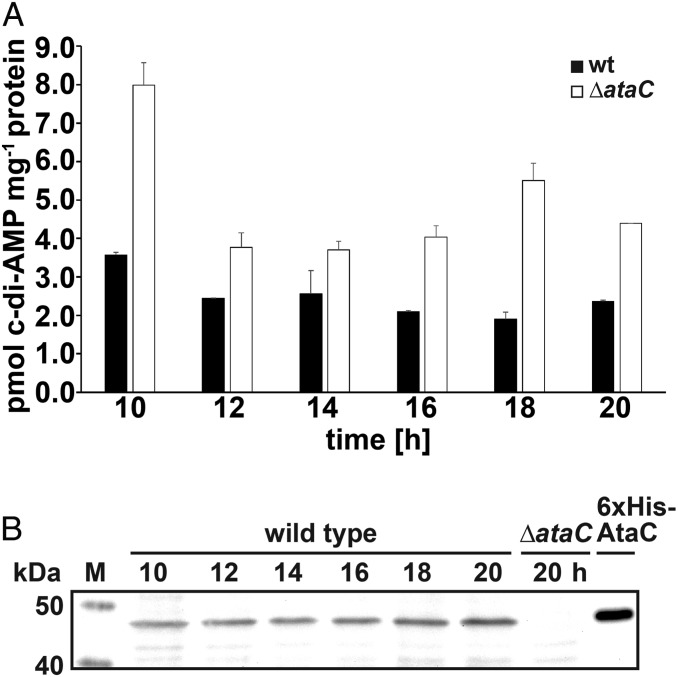
We quantified c-di-AMP in cell extracts isolated from wild-type S. venezuelae and the ataC null mutant using liquid chromatography tandem mass spectrometry (LC-MS/MS). Our data show that c-di-AMP levels are elevated in the ataC mutant during all developmental stages when compared to the wild type, demonstrating that AtaC degrades c-di-AMP in vivo and thus is an important component of c-di-AMP metabolism in S. venezuelae (Fig. 4A). Western blot analysis showed that AtaC levels slightly increase during the life cycle (Fig. 4B and SI Appendix, Fig. S1B).
Fig. 4.
AtaC hydrolyzes c-di-AMP in vivo and is expressed during the life cycle of S. venezuelae. (A) Intracellular c-di-AMP levels in S. venezuelae wild type and ΔataC during late vegetative growth (10 to 12 h), early sporulation (14 to 16 h), and sporulation (18 to 20 h). Data are presented as the mean of biological replicates ± SD (n = 3). (B) Expression profile of AtaC in S. venezuelae wild type grown in a liquid sporulation medium (MYM). AtaC was detected using a polyclonal anti-AtaC antiserum. Protein samples harvested from ΔataC served as negative control. Purified 6xHis-AtaC served as the positive control.
Inactivation of AtaC Delays S. venezuelae Development.
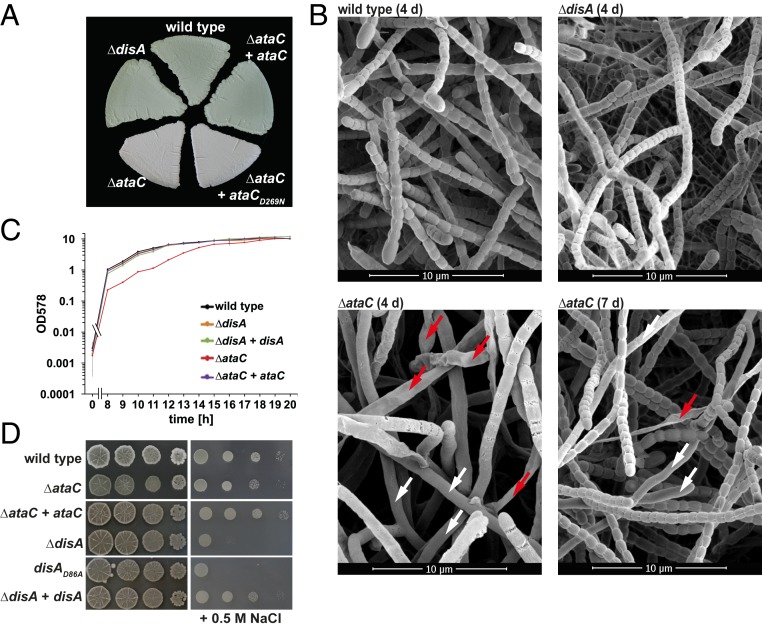
To investigate the physiological functions of disA and ataC and thus of c-di-AMP in S. venezuelae, we first analyzed the developmental phenotypes of mutant strains. Colonies of S. venezuelae ΔdisA became green (Fig. 5A), and scanning electron microscopy (SEM) confirmed that the disA mutant produced spore chains with morphology identical to those of the wild type (Fig. 5B). Thus, neither the DisA protein nor the c-di-AMP produced by DisA is required for differentiation.
Fig. 5.
Mutagenesis of c-di-AMP-metabolizing enzymes impacts development and ionic stress resistance in S. venezuelae. (A) Green morphologies of S. venezuelae wild type and ∆disA indicate the formation of mature spores after 4 d of growth at 30 °C on a solid sporulation medium (MYM agar). S. venezuelae ∆ataC failed to accumulate the spore pigment and remained white after the same incubation time. The wild-type ataC allele complements the phenotype of ∆ataC, while the enzymatically inactive variant ataCD269N does not. (B) Scanning electron micrographs showing that after 4 d of incubation on MYM, S. venezuelae wild type and ∆disA formed spores, but ∆ataC consisted predominantly of nonsporulating aerial hyphae (white arrows) and formed flat, likely lysed hyphae (red arrows). After 7 d of growth, ∆ataC produced wild-type–like spore chains, but occasional nondifferentiated and lysed hyphae were still detectable. (C) Deletion of ataC leads to a growth defect in S. venezuelae. c-di-AMP mutants were grown in a liquid sporulation medium (MYM) at 30 °C, and optical density was measured at 578 nm. ΔataC growth is delayed by 3 h and can be restored by expression of the wild-type allele under the control of its native promoter from the attBΦBT1 site. (D) Osmotic stress resistance of c-di-AMP mutants. Serial dilutions of spores were spotted on NA without additional salt or supplemented with 0.5 M NaCl and grown at 30 °C for ∼2 d. ΔdisA and disAD86A (expressing inactive DisA) are hypersensitive to salt stress.
In contrast, the ataC mutant showed a severe delay in development. After 4 d, the ΔataC strain developed aerial hyphae but did not turn green like the wild type (Fig. 5A), and SEM imaging showed mainly undifferentiated aerial hyphae, in contrast to the fully sporulated hyphae seen in the wild type (Fig. 5B). Moreover, many of the aerial hyphae of the ataC mutant had lysed. After extended incubation (7 d), the aerial hyphae of the ataC mutant had largely sporulated, with sporadic nondifferentiated and lysed filaments still detected (Fig. 5B).
The lysed hyphae seen in the SEMs led us to analyze the growth of the ∆ataC strain in liquid maltose–yeast extract–malt extract (MYM) medium. As shown in Fig. 5C, the ataC mutant grew slower than the wild type in the exponential phase but reached a similar final optical density at 578 nm after 20 h. Notably, deletion of disA had no effect on growth (Fig. 5C). Using a heat resistance assay, we found that neither spores formed by the ataC mutant nor those produced by ∆disA and disAD86A strains were defective in spore viability (SI Appendix, Fig. S5A).
We could fully complement the defects of ∆ataC in development and growth by introduction of the ataC wild-type allele under the control of its native promoter from the pIJ10770 vector (32) that integrates into the chromosomal attBΦBT1 site (Fig. 5 A and C and SI Appendix, Fig. S5B). In contrast, expression of ataCD269N, which cannot cleave c-di-AMP (SI Appendix, Fig. S4A) from the same integrative vector, did not restore the developmental defects caused by ataC deletion (Fig. 5A), showing that the cleavage of c-di-AMP by AtaC is crucial for normal development of Streptomyces.
Altogether, these results demonstrate that elevated levels of c-di-AMP impair growth and development, whereas reduced levels of c-di-AMP do not affect differentiation under standard growth conditions.
The disA Mutant Is More Susceptible to Ionic Stress.
Since regulation of osmotic balance is a major function of c-di-AMP in many bacteria (3), we next investigated the osmotic stress resistance of strains with altered c-di-AMP levels due to mutations in either ataC or disA. We spotted serially diluted spores on nutrient agar (NA) medium plates supplemented with 0.5 M NaCl and a control plate without extra added NaCl. On both plates, growth of the ∆ataC strain was slightly impaired, resulting in smaller colony size compared to the wild type (Fig. 5D). This phenotype reflects the growth defect of the mutant (Fig. 5C) and could be complemented by expression of the wild-type allele from pIJ10770 (Fig. 5D).
In contrast, when grown on NA plates containing 0.5 M NaCl, ΔdisA and disAD86A showed pronounced reduction in growth. Expression of wild-type disA from pIJ10770 fully complemented the growth defect of ΔdisA (Fig. 5D). The identical ΔdisA and disAD86A phenotypes demonstrate that c-di-AMP produced by DisA is crucial for osmotic stress resistance in S. venezuelae (Fig. 5D).
In summary, our data revealed that accumulation of c-di-AMP due to ataC inactivation delays development and slows down Streptomyces growth in the exponential phase. On the other hand, depletion of c-di-AMP due to disA inactivation renders S. venezuelae highly susceptible to ionic stress.
The RCK_C Domain Protein CpeA (Vnz_28055) Binds c-di-AMP.
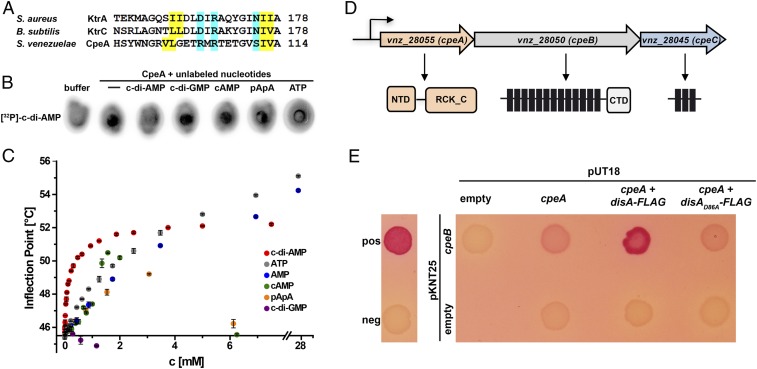
RCK_C domains are established direct targets of c-di-AMP that have the (I/L)(I/L)X2DX1RX5N(I/L)(I/L) signature for ligand binding (Fig. 6A) (33). We found the RCK_C domain protein CpeA (Vnz_28055) with a putative c-di-AMP binding motif (Fig. 6A) in 93 Streptomyces species for which complete genome sequences are available (34). We purified N-terminally His-tagged CpeA and applied differential radial capillary action of ligand assays (DRaCALA) to probe interaction between CpeA and c-di-AMP. DRaCALA allows visualization of protein-bound radiolabeled ligand as a concentrated spot or ring after the application of the protein-ligand mixture onto nitrocellulose (35). With this assay, we confirmed that CpeA binds [32P]-labeled c-di-AMP (Fig. 6B).
Fig. 6.
CpeA (Vnz_28055) binds c-di-AMP and interacts with CpeB (Vnz_28050) in a c-di-AMP-stimulating manner. (A) Alignment of the c-di-AMP binding regions in RCK_C domains was generated using Clustal Omega (49). C-di-AMP binding residues in KtrA (S. aureus; ref. 50), KtrC (B. subtilis; ref. 51), and conserved amino acids in CpeA are highlighted. Amino acids that form the hydrophobic patch are shown in yellow; residues involved in hydrophilic coordination are highlighted in cyan. (B) CpeA binds [32P]-c-di-AMP in DRaCALAs. Binding of the radiolabeled ligand is indicated by dark spots centered on the nitrocellulose. In competition assays, excess (100 µM) of unlabeled c-di-AMP, c-di-GMP, cAMP, 5′-pApA, or ATP was added to the binding reaction containing [32P]-c-di-AMP and 6xHis-CpeA. (C) Inflection points from nanoDSF thermal shift assays of 20 µM CpeA with different concentrations of c-di-AMP, ATP, AMP, cAMP, 5′-pApA, and c-di-GMP (0 to 10 mM) at a heating rate of 1.5 K/min. Shown are mean values of n = 3 independent experiments with SD. (D) cpeA (vnz_28055), cpeB (vnz_28050) and cpeC (vnz_28045) form an operon in S. venezuelae. CpeA has an N-terminal domain (NTD) of unknown function and a C-terminal RCK_C domain. CpeB is a predicted structural homolog of the Na+/H+ antiporter NapA (36). It consists of 13 transmembrane (TM) domains and a cytosolic fraction at the C terminus (CTD). CpeC is a predicted membrane protein with 3 TM domains. (E) Adenylate cyclase–based two-hybrid assays revealing that CpeA and CpeB interact in vivo and that c-di-AMP production by coexpressed DisA-FLAG stimulates protein–protein interaction. Using pKNT25 and pUT18, the T25 and T18 fragments of adenylate cyclase were attached to the C termini of CpeB and CpeA, respectively. disA-FLAG and disAD86A-FLAG were expressed from pUT18-cpeA. The leucin zipper part of the yeast GCN4 protein was used as a positive control. Spotted cotransformants were grown for 24 h at 26 °C.
In competition experiments, we found that unlabeled c-di-AMP interfered with binding of [32P]-c-di-AMP to CpeA, while excess of c-di-GMP, cAMP, 5′-pApA, or ATP still allowed [32P]-c-di-AMP-CpeA complex formation (Fig. 6B). NanoDSF analysis revealed that addition of c-di-AMP in the micromolar range significantly increased the melting point of CpeA. In contrast, high concentrations of other nucleotides such as 5′-pApA, ATP, AMP, or cAMP were needed for CpeA stabilization to the same extent, and c-di-GMP even destabilized the protein (Fig. 6C and SI Appendix, Fig. S6). Taken together, these data indicate that CpeA specifically binds c-di-AMP. To determine the KD, we analyzed c-di-AMP binding through surface plasmon resonance (SPR) assays. Since we faced protein instability problems during CpeA coupling to the chip and using ITC, we applied biotinylated c-di-AMP on chip and CpeA as a ligand in SPR analysis. We determined a KD of 37 µM, probably reflecting an upper limit for the KD as the biotin at the 2′-OH of the ribose moiety likely interferes with binding of c-di-AMP to CpeA (SI Appendix, Fig. S7A). SAXS and static light scattering experiments showed that CpeA is a stable dimer in solution and does not dissociate at concentrations down to 260 nM, as observed in analytical size exclusion chromatography (SI Appendix, Fig. S7 B–E), independent of the presence of c-di-AMP. In summary, we could identify CpeA as a c-di-AMP binding protein in the genus Streptomyces.
cpeA forms a conserved operon with cpeB (vnz_28050). Some Streptomyces species, such as S. venezuelae, contain the small open reading frame cpeC (vnz_28045) in the same operon (Fig. 6D). CpeB is a structural homolog of the sodium/proton antiporter NapA (PDB code 5BZ3_A) from Thermus thermophilus (36), as predicted with 100% probability using HHpred (29). To test whether CpeA and CpeB form a functional interacting unit, we used a bacterial two-hybrid system in which an interaction between bait and target proteins reconstitutes a functional adenylate cyclase (Cya) that allows the Escherichia coli cya mutant to utilize maltose as a carbon source (37). We found that CpeA and CpeB form a complex (Fig. 6E). The interaction between these two proteins was strongly enhanced when enzymatically active DisA-FLAG was coexpressed, while coexpression of the inactive DisAD86A-FLAG variant had no effect (Fig. 6E), demonstrating that c-di-AMP produced by DisA stimulates CpeA-CpeB interaction. These data are consistent with our model that c-di-AMP controls ion transport activity of CpeB by stimulating complex formation with the regulatory unit, CpeA. Thus, we renamed Vnz_28055-28045 to CpeABC for cation proton exchange components A, B, and C. However, c-di-AMP–dependent cation flux via CpeB remains to be proven.
Discussion
In this work, using the chloramphenicol producer S. venezuelae as a model and a combination of bioinformatic, biochemical, structural, and genetic analyses, we identified AtaC as a c-di-AMP–specific PDE. AtaC is widely distributed in bacteria and represents the only c-di-AMP PDE in the majority of Actinobacteria and a c-di-AMP signaling component in pathogens, such as S. pneumoniae (Fig. 2 and Dataset S1).
AtaC is a soluble, single-domain phosphodiesterase superfamily protein that is monomeric in solution (SI Appendix, Fig. S3). In solution, AtaC is structurally similar to the alkaline phosphatase superfamily domain of the C–P bond-cleaving enzyme PhnA from S. meliloti 1021 (Fig. 3A) (30). As described for DHH-DHHA1 domain-containing proteins GdpP and DhhP and the HD domain PDE PgpH, AtaC binds Mn2+ to hydrolyze c-di-AMP, and we showed that residue D269 participates in metal ion coordination and is crucial for hydrolysis activity (Fig. 3 C–E) (18, 23, 38). AtaC has a kcat of 0.2 s−1, which is comparable to the reported kcat of GdpP (0.55 s−1). Hydrolytically inactive AtaCD269N has a dissociation constant of 0.9 µM, which is highly similar to the KD of wild-type PgpH (0.3 to 0.4 µM) (Fig. 3 G and H) (18, 23). Since we determined the AtaC dissociation constant using a protein carrying the D269N mutation lacking Mn2+ coordination, the KD value represents an upper limit as the metal ions bound by the wild-type protein likely contribute to protein stability and c-di-AMP binding. However, while PgpH- and GdgP-type PDEs hydrolyze c-di-AMP exclusively to the linear 5′-pApA, AtaC cleaves c-di-AMP and the intermediate product 5′-pApA to AMP, which has also been shown for some DhhP-type PDEs (Fig. 2 A and B and SI Appendix, Fig. S2 A, B, and D) (18, 23, 38). The substrate specificity of AtaC is strictly dependent on two adenosine bases as it shows only weak hydrolysis activity for 5′-pApG and 5′-pGpG, in contrast to the DhhP-type PDE TmPDE, which does not distinguish between different nucleobases (Fig. 2B and SI Appendix, Fig. S2 E and F) (31).
In Streptomyces, AtaC and the DAC DisA are important regulators of c-di-AMP (Figs. 1B and 4A). On standard growth medium, elevation of intracellular c-di-AMP in ∆ataC interferes with growth and ordered hyphae-to-spores transition, while reduction of the second messenger in ∆disA does not have any noticeable consequences on these cell functions. On the other hand, when incubated at high external NaCl concentrations, ∆disA is severely inhibited in growth, whereas ∆ataC grows similarly to the wild type (Fig. 5), indicating that c-di-AMP stimulates an osmoprotective function. We found that the RCK_C domain protein CpeA binds c-di-AMP and that ligand binding induces interaction between CpeA and CpeB, a structural homolog of the Na+/H+ antiporter NapA from T. thermophilus and a member of the large monovalent cation/proton antiporter (CPA) superfamily (36) (Fig. 6). Na+/H+ antiporters exist in all living cells, where they regulate intracellular pH, sodium levels, and cell volume (39). In some bacteria, Na+/H+ antiporters use the proton motive force to extrude sodium out of the cell and are activated at alkaline pH (40). However, in Staphylococcus aureus, the CPA family transporter CpaA has a cytosolic RCK_C domain that binds c-di-AMP to regulate transport activity (6, 41). Similarly, the regulatory RCK_C domain proteins KtrA and KtrC bind c-di-AMP to control the activity of the corresponding potassium transport units KtrB and KtrD, respectively (33). Thus, in agreement with this general concept and our data, we propose that c-di-AMP sensed by the regulatory RCK_C domain protein CpeA induces CpeA-CpeB complex formation to activate sodium export via CpeB in Streptomyces. At low c-di-AMP, CpeB is presumably inactive, allowing accumulation of toxic Na+ ions in the cell and leading to growth defects of ∆disA on NaCl-containing medium. However, on the other hand, likely constant activity of CpeB at high c-di-AMP in ∆ataC may result in continuous proton influx affecting intracellular pH and thus important cellular functions, causing growth and developmental defects.
In summary, in this study we identified AtaC as a component of c-di-AMP metabolism in bacteria and uncovered CpeA as a potential link between c-di-AMP and ion balance in Streptomyces. In nature, these bacteria primarily inhabit the upper layer of the soil, where they often face fluctuating osmotic conditions through desiccation and rainfall. The c-di-AMP pathway described here is likely crucial for adaptation in such a challenging ecosystem.
Materials and Methods
For a full explanation of the experimental protocols, see Extended Experimental Procedures in SI Appendix.
Bacterial Strains and Plasmids.
All strains, plasmids, and oligonucleotides used in this study are listed in SI Appendix, Table S1. Plasmids and strains were constructed as described in SI Appendix.
Protein Overexpression and Purification.
E. coli BL21 (DE3) pLysS and Rosetta (DE3) were used for protein overexpression. Cultures were grown in the presence of required antibiotics at 37 °C and induced with isopropyl-β-d-thiogalactopyranosid (IPTG) in the logarithmic phase and transferred for growth at 16 °C overnight. Strains overexpressing 6xHis-AtaC, 6xHis-AtaCD269N, 6xHis-Vnz_31010, and 6xHis-AtaCSpn were supplemented with MnCl2 (18). Cultures were harvested and lysed using a French press, and the proteins were purified via nickel–nitrilotriacetic acid (Ni-NTA) chromatography. 6xHis-DisA variants and 6xHis-CpeA were dialyzed twice in DisA cyclase buffer (ref. 42 and SI Appendix), and tested PDEs were dialyzed twice in PDE buffer (SI Appendix) with 5 to 10% glycerol (18) at 4 °C. Dialyzed proteins were stored at −20 °C. For characterization of biophysical properties of 6xHis-AtaC and 6xHis-AtaCD269N, the protein elution was concentrated prior to size exclusion chromatography, flash frozen in liquid nitrogen, and stored at −80 °C.
Biochemical Characterization of DisA and AtaC Variants.
Biochemical assays using radioactive-labeled substrates were conducted as described in ref. 34. For DAC assays, 5 µM 6xHis-tagged DisASven, DisAD86A, or DisABsu were incubated with 83 nM [32P]-ATP (Hartmann Analytic) in DisA cyclase buffer. For PDE assays, 100 nM 6xHis-AtaC or 8 µM 6xHis-Vnz_31010 were mixed with 2 nM [32P]-c-di-AMP (Hartmann Analytic, synthesized using purified 6xHis-DisABsu) in PDE buffer. For competition, 100 µM unlabeled c-di-AMP, c-di-GMP, or cAMP were added on ice prior to starting the PDE reactions with [32P]-c-di-AMP.
Alternatively, enzymatic activity of 6xHis-AtaC and 6xHis-AtaCD269N was detected by separation of nonlabeled reaction products by anion exchange chromatography as described in ref. 31. Reaction solutions contained 50 mM Tris (pH = 7.5), 20 mM NaCl, 100 µM MnCl2, 62.5 to 2,000 µM ligand (c-di-NMP, 5′-pNpN; N = A or G), and 100 nM to 10 µM of 6xHis-AtaC and were incubated at 37 °C for 1 h. The reaction was stopped by separating the reaction products from the protein by ultrafiltration (Centricon, 30 kDa cutoff). The filtrate was diluted to 500 µL with running buffer A (50 mM Tris, pH 9) and loaded on a 1 mL Resource Q anion exchange column (GE Healthcare Life Sciences). A linear gradient to 40% running buffer B (50 mM Tris, 1 M NaCl, pH = 9) over 20 column volumes was used to separate the nucleotides. The product peaks were identified by comparison to nucleotide standards, c-di-NMP, pNpN, N = A or G, obtained from BioLog.
Differential Radial Capillary Action of Ligand Assay.
DRaCALAs were performed using 5 µg of purified 6xHis-CpeA (Vnz_28055) as described in Roelofs et al. (35) with minor modifications. For competition, reactions were incubated with 42 nM [32P]-c-di-AMP for 5 min at room temperature prior to addition of 100 µM of nonlabeled nucleotides. Samples were spotted on nitrocellulose after 10 min reaction at room temperature.
Western Blotting.
For detection of FLAG-tagged DisA, Western blot analysis was performed as described in ref. 34 using 5 µg total protein of S. venezuelae ∆disA expressing the FLAG-tagged disA allele from the ՓBT1 integration site under the control of the native promoter. Anti-FLAG primary antibody (Sigma) and the anti-mouse IgG-HRP (Thermo Fisher Scientific) were used for detection. AtaC was detected in the wild-type strain (10 µg total protein) using polyclonal rabbit anti-AtaC antiserum as the primary antibody (generated by Pineda GmbH using purified 6xHis-AtaC) and donkey anti-rabbit-HRP as the secondary antibody (GE Healthcare). Enhanced chemiluminescent detection reagent (Perkin-Elmer) was used for visualization.
c-di-AMP Extraction and Quantification.
The nucleotide extraction protocol from ref. 2 was adapted to Streptomyces. Wild-type, ΔdisA, and ΔataC strains were grown in MYM. Samples for c-di-AMP extraction and for determination of the protein concentration were taken every 2 h after initial growth for 10 h. c-di-AMP was extracted using acetonitrile/methanol from cells disrupted with the BeadBlaster (Biozym). Samples were analyzed using LC-MS/MS as described in ref. 2.
Bacterial Adenylate Cyclase Two-Hybrid (BACTH) Assays.
The BACTH system was used to assay c-di-AMP–dependent protein–protein interaction between CpeA and CpeB in vivo (37). Plasmids expressing C-terminal fusions of CpeA and CpeB to T18 and T25 fragments of cyaA from Bordetella pertussis, respectively, were transformed into E. coli W3110 lacking cya (43). disA-FLAG or disAD86A-FLAG was introduced into the pUT18-cpeA plasmid directly downstream of the T18 fragment (for details see SI Appendix). Cotransformants were spotted on MacConkey agar supplemented with maltose (1%), ampicillin (100 µg/mL), and kanamycin (50 µg/mL). Red colonies indicate cAMP-dependent fermentation of maltose, which occurs upon direct interactions of the proteins fused to the otherwise separate adenylate cyclase domains.
Small-Angle X-Ray Scattering.
Size-exclusion chromatography coupled small-angle X-ray scattering data (44, 45) for AtaC was collected at beamline P12 operated by the European Molecular Biology Laboratory (EMBL) Hamburg at the PETRA III storage ring of the Deutsches Elektronen Synchroton (DESY), Hamburg, Germany. CHROMIXS of the ATSAS Suite (46) was used for analysis and processing of the chromatogram results. In brief, after choosing an appropriate buffer region and averaging of the respective frames, the protein scattering frames from the elution peak were buffer subtracted and averaged. Buffer-corrected SEC-SAXS data and batch samples (for buffer subtraction, etc.) were analyzed using PRIMUS of the ATSAS suite. The theoretical scattering curve of the AtaC model derived from HHpred/MODELER was obtained using CRYSOL (47). Ab initio models were calculated using DAMMIF and averaged using DAMAVER as described earlier (31).
Label-Free Thermal Shift Assays (nanoDSF).
Label-free thermal shift assay experiments of AtaC were performed with a Tycho NT.6 instrument (NanoTemper Technologies). The samples were heated in a glass capillary at a rate of 30 K/min, and the internal fluorescence at 330 and 350 nm was recorded. Data analysis, data smoothing, and calculation of derivatives was done using the internal evaluation features of the Tycho instrument. Thermal shift assays with CpeA were performed in a Prometheus NT.48 (NanoTemper Technologies) with a 1.5 K/min heating rate (20 °C to 95 °C) in 20 mM Tris HCl (pH = 7.5), 160 mM NaCl, 5% glycerol, and 10 mM MgCl2.
Bioinformatic Characterization of AtaC and Its Abundance in Prokaryotes.
AtaC was identified as a member of the phosphodiesterase family of proteins by annotation of the S. venezuelae genome with interproscan (version 5.27-66.0; ref. 28) and searching for proteins harboring type I phosphodiesterase/nucleotide pyrophosphatase domain (Pfam PF01663).
Scanning Electron Microscopy.
SEM was performed as previously described (48).
Data Availability Statement.
Small-angle scattering data have been deposited in the Small Angle Scattering Biological Data Bank (SASBDB) with accession numbers SASDH25 (AtaC; https://www.sasbdb.org/data/SASDH25) and SASDH35 (CpeA; https://www.sasbdb.org/data/SASDH35). All data discussed in the paper will be made available to readers.
Supplementary Material
Acknowledgments
We are grateful to Mark J. Buttner and Fabian M. Commichau for helpful discussion and critical reading of the manuscript and thank Matt Bush for technical support with scanning electron micrographs. We thank the staff of the EMBL Hamburg beamline P12 at PETRA3 (EMBL/DESY, Hamburg, Germany) for outstanding scientific support. We also acknowledge Anna-Lena Hagemann and Annette Garbe for technical support with LC-MS/MS funded by the Deutsche Forschungsgemeinschaft (DFG) Priority Program SPP 1879 (Grant KA 730/9-1). We acknowledge the use of the Prometheus instrument in the Bioanalytics unit of the Ludwig-Maximilians-Universität München Biocenter and discussions with Franziska Tippel and Beate Kern (NanoTemper Technologies). Research in G.W.’s lab is funded by DFG Grant GRK1721 and the DFG Priority Program SPP 1879 (Grant WI 3717/3-1). Research in N.T.’s lab is funded by the DFG Emmy Noether Program (Grant TS 325/1-1) and the DFG Priority Program SPP 1879 (Grants TS 325/2-1 and TS 325/2-2).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: Small-angle scattering data have been deposited in the Small Angle Scattering Biological Data Bank (SASBDB) with accession numbers SASDH25 (AtaC) and SASDH35 (CpeA).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1917080117/-/DCSupplemental.
References
- 1.Hengge R., et al. , Recent advances and current trends in nucleotide second messenger signaling in bacteria. J. Mol. Biol. 431, 908–927 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Gundlach J., et al. , An essential poison: Synthesis and degradation of cyclic di-AMP in Bacillus subtilis. J. Bacteriol. 197, 3265–3274 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Commichau F. M., Gibhardt J., Halbedel S., Gundlach J., Stülke J., A delicate connection: c-di-AMP affects cell integrity by controlling osmolyte transport. Trends Microbiol. 26, 175–185 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Gundlach J., et al. , Control of potassium homeostasis is an essential function of the second messenger cyclic di-AMP in Bacillus subtilis. Sci. Signal. 10, eaal3011 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Bremer E., Krämer R., Responses of microorganisms to osmotic stress. Annu. Rev. Microbiol. 73, 313–334 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Corrigan R. M., et al. , Systematic identification of conserved bacterial c-di-AMP receptor proteins. Proc. Natl. Acad. Sci. U.S.A. 110, 9084–9089 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schuster C. F., et al. , The second messenger c-di-AMP inhibits the osmolyte uptake system OpuC in Staphylococcus aureus. Sci. Signal. 9, ra81 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gundlach J., et al. , Sustained sensing in potassium homeostasis: Cyclic di-AMP controls potassium uptake by KimA at the levels of expression and activity. J. Biol. Chem. 294, 9605–9614 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pham H. T., et al. , Enhanced uptake of potassium or glycine betaine or export of cyclic-di-AMP restores osmoresistance in a high cyclic-di-AMP Lactococcus lactis mutant. PLoS Genet. 14, e1007574 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quintana I. M., et al. , The KupA and KupB proteins of Lactococcus lactis IL1403 are novel c-di-AMP receptor proteins responsible for potassium uptake. J. Bacteriol. 201, e00028-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Devaux L., Kaminski P. A., Trieu-Cuot P., Firon A., Cyclic di-AMP in host-pathogen interactions. Curr. Opin. Microbiol. 41, 21–28 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Woodward J. J., Iavarone A. T., Portnoy D. A., c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science 328, 1703–1705 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McFarland A. P., et al. , RECON-dependent inflammation in hepatocytes enhances Listeria monocytogenes cell-to-cell spread. mBio 9, e00526-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parvatiyar K., et al. , The helicase DDX41 recognizes the bacterial secondary messengers cyclic di-GMP and cyclic di-AMP to activate a type I interferon immune response. Nat. Immunol. 13, 1155–1161 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barker J. R., et al. , STING-dependent recognition of cyclic di-AMP mediates type I interferon responses during Chlamydia trachomatis infection. mBio 4, e00018-13(2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xia P., et al. , The ER membrane adaptor ERAdP senses the bacterial second messenger c-di-AMP and initiates anti-bacterial immunity. Nat. Immunol. 19, 141–150 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Fahmi T., Faozia S., Port G. C., Cho K. H., The second messenger c-di-AMP regulates diverse cellular pathways involved in stress response, biofilm formation, cell wall homeostasis, SpeB expression, and virulence in Streptococcus pyogenes. Infect. Immun. 87, e00147-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huynh T. N., et al. , An HD-domain phosphodiesterase mediates cooperative hydrolysis of c-di-AMP to affect bacterial growth and virulence. Proc. Natl. Acad. Sci. U.S.A. 112, E747–E756 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bai Y., et al. , Two DHH subfamily 1 proteins in Streptococcus pneumoniae possess cyclic di-AMP phosphodiesterase activity and affect bacterial growth and virulence. J. Bacteriol. 195, 5123–5132 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dey R. J., et al. , Inhibition of innate immune cytosolic surveillance by an M. tuberculosis phosphodiesterase. Nat. Chem. Biol. 13, 210–217 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Witte G., Hartung S., Büttner K., Hopfner K. P., Structural biochemistry of a bacterial checkpoint protein reveals diadenylate cyclase activity regulated by DNA recombination intermediates. Mol. Cell 30, 167–178 (2008). [DOI] [PubMed] [Google Scholar]
- 22.Corrigan R. M., Gründling A., Cyclic di-AMP: Another second messenger enters the fray. Nat. Rev. Microbiol. 11, 513–524 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Rao F., et al. , YybT is a signaling protein that contains a cyclic dinucleotide phosphodiesterase domain and a GGDEF domain with ATPase activity. J. Biol. Chem. 285, 473–482 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chater K. F., Recent advances in understanding Streptomyces. F1000Res. 5, 2795 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bush M. J., Tschowri N., Schlimpert S., Flärdh K., Buttner M. J., c-di-GMP signalling and the regulation of developmental transitions in streptomycetes. Nat. Rev. Microbiol. 13, 749–760 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Tschowri N., Cyclic dinucleotide-controlled regulatory pathways in Streptomyces species. J. Bacteriol. 198, 47–54 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.St-Onge R. J., et al. , Nucleotide second messenger-mediated regulation of a muralytic enzyme in Streptomyces. Mol. Microbiol. 96, 779–795 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Quevillon E., et al. , InterProScan: Protein domains identifier. Nucleic Acids Res. 33, W116–W120 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hildebrand A., Remmert M., Biegert A., Söding J., Fast and accurate automatic structure prediction with HHpred. Proteins 77 (suppl. 9), 128–132 (2009). [DOI] [PubMed] [Google Scholar]
- 30.Agarwal V., Borisova S. A., Metcalf W. W., van der Donk W. A., Nair S. K., Structural and mechanistic insights into C-P bond hydrolysis by phosphonoacetate hydrolase. Chem. Biol. 18, 1230–1240 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Drexler D. J., Muller M., Rojas-Cordova C. A., Bandera A. M., Witte G., Structural and biophysical analysis of the soluble DHH/DHHA1-type phosphodiesterase TM1595 from Thermotoga maritima. Structure 25, 1887–1897.e4 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Schlimpert S., et al. , Two dynamin-like proteins stabilize FtsZ rings during Streptomyces sporulation. Proc. Natl. Acad. Sci. U.S.A. 114, E6176–E6183 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schrecker M., Wunnicke D., Hänelt I., How RCK domains regulate gating of K+ channels. Biol. Chem. 400, 1303–1322 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Al-Bassam M. M., Haist J., Neumann S. A., Lindenberg S., Tschowri N., Expression patterns, genomic conservation and input into developmental regulation of the GGDEF/EAL/HD-GYP domain proteins in Streptomyces. Front. Microbiol. 9, 2524 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roelofs K. G., Wang J., Sintim H. O., Lee V. T., Differential radial capillary action of ligand assay for high-throughput detection of protein-metabolite interactions. Proc. Natl. Acad. Sci. U.S.A. 108, 15528–15533 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee C., et al. , A two-domain elevator mechanism for sodium/proton antiport. Nature 501, 573–577 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karimova G., Pidoux J., Ullmann A., Ladant D., A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. U.S.A. 95, 5752–5756 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ye M., et al. , DhhP, a cyclic di-AMP phosphodiesterase of Borrelia burgdorferi, is essential for cell growth and virulence. Infect. Immun. 82, 1840–1849 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Padan E., Venturi M., Gerchman Y., Dover N., Na(+)/H(+) antiporters. Biochim. Biophys. Acta 1505, 144–157 (2001). [DOI] [PubMed] [Google Scholar]
- 40.Krulwich T. A., Sachs G., Padan E., Molecular aspects of bacterial pH sensing and homeostasis. Nat. Rev. Microbiol. 9, 330–343 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chin K. H., et al. , Structural insights into the distinct binding mode of cyclic di-AMP with SaCpaA_RCK. Biochemistry 54, 4936–4951 (2015). [DOI] [PubMed] [Google Scholar]
- 42.Christen M., Christen B., Folcher M., Schauerte A., Jenal U., Identification and characterization of a cyclic di-GMP-specific phosphodiesterase and its allosteric control by GTP. J. Biol. Chem. 280, 30829–30837 (2005). [DOI] [PubMed] [Google Scholar]
- 43.Herbst S., et al. , Transmembrane redox control and proteolysis of PdeC, a novel type of c-di-GMP phosphodiesterase. EMBO J. 37, e97825 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jeffries C. M., et al. , Preparing monodisperse macromolecular samples for successful biological small-angle X-ray and neutron-scattering experiments. Nat. Protoc. 11, 2122–2153 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blanchet C. E., et al. , Versatile sample environments and automation for biological solution x-ray scattering experiments at the P12 beamline (PETRA III, DESY). J. Appl. Crystallogr. 48, 431–443 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Franke D., et al. , ATSAS 2.8: A comprehensive data analysis suite for small-angle scattering from macromolecular solutions. J. Appl. Crystallogr. 50, 1212–1225 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Svergun D. I., Barberato C., Koch M. H. J., CRYSOL-A program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 28, 768–773 (1995). [Google Scholar]
- 48.Tschowri N., et al. , Tetrameric c-di-GMP mediates effective transcription factor dimerization to control Streptomyces development. Cell 158, 1136–1147 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Madeira F., et al. , The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47, W636–W641 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim H., et al. , Structural studies of potassium transport protein KtrA regulator of conductance of K+ (RCK) C domain in complex with cyclic diadenosine monophosphate (c-di-AMP). J. Biol. Chem. 290, 16393–16402 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rocha R., Teixeira-Duarte C. M., Jorge J. M. P., Morais-Cabral J. H., Characterization of the molecular properties of KtrC, a second RCK domain that regulates a Ktr channel in Bacillus subtilis. J. Struct. Biol. 205, 34–43 (2019). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Small-angle scattering data have been deposited in the Small Angle Scattering Biological Data Bank (SASBDB) with accession numbers SASDH25 (AtaC; https://www.sasbdb.org/data/SASDH25) and SASDH35 (CpeA; https://www.sasbdb.org/data/SASDH35). All data discussed in the paper will be made available to readers.