Significance
Lens transparency critically requires the abundant accumulation of crystallin proteins, and deregulation of this process causes congenital cataracts in humans. Rbm24 is an RNA-binding protein with highly conserved expression in differentiating lens fiber cells among all vertebrates. We use a zebrafish model to demonstrate that loss of Rbm24 function specifically impedes lens fiber cell differentiation, resulting in cataract formation and blindness. Molecular analyses reveal that Rbm24 interacts with cytoplasmic polyadenylation complex and binds to a large number of lens-expressed messenger RNAs to maintain their stability and protect their poly(A) tail length, thereby crucially contributing to their efficient translation into functional proteins. This work identifies an important mechanism by which Rbm24 posttranscriptionally controls lens gene expression to establish transparency and refraction power.
Keywords: Rbm24, lens, cataract, cytoplasmic polyadenylation, zebrafish
Abstract
Lens transparency is established by abundant accumulation of crystallin proteins and loss of organelles in the fiber cells. It requires an efficient translation of lens messenger RNAs (mRNAs) to overcome the progressively reduced transcriptional activity that results from denucleation. Inappropriate regulation of this process impairs lens differentiation and causes cataract formation. However, the regulatory mechanism promoting protein synthesis from lens-expressed mRNAs remains unclear. Here we show that in zebrafish, the RNA-binding protein Rbm24 is critically required for the accumulation of crystallin proteins and terminal differentiation of lens fiber cells. In the developing lens, Rbm24 binds to a wide spectrum of lens-specific mRNAs through the RNA recognition motif and interacts with cytoplasmic polyadenylation element-binding protein (Cpeb1b) and cytoplasmic poly(A)-binding protein (Pabpc1l) through the C-terminal region. Loss of Rbm24 reduces the stability of a subset of lens mRNAs encoding heat shock proteins and shortens the poly(A) tail length of crystallin mRNAs encoding lens structural components, thereby preventing their translation into functional proteins. This severely impairs lens transparency and results in blindness. Consistent with its highly conserved expression in differentiating lens fiber cells, the findings suggest that vertebrate Rbm24 represents a key regulator of cytoplasmic polyadenylation and plays an essential role in the posttranscriptional control of lens development.
Lens morphogenesis is temporally and spatially coordinated by a multitude of signaling pathways (1–4). The terminal differentiation of lens fiber cells is characterized by the accumulation of extremely abundant crystallin proteins, as well as the disappearance of cell nuclei and other organelles (3, 5). Crystallins belong to a large family of proteins that constitute the highly abundant soluble proteins in lens fiber cells (6–8) and can be grouped into three main classes: α-, β-, and γ-crystallins. α-crystallins are small heat shock proteins that have molecular chaperone activity to ensure the correct folding of other proteins, while β- and γ-crystallins are structural components (7). These proteins play an essential role in lens transparency and refraction, with structure and function particularly conserved among vertebrates (9, 10). Mutations or dysfunctions of crystallin proteins impede lens transparency and lead to different types of cataracts (11), which are the leading cause of human blindness (12). Whereas the transcriptional regulation of lens-specific expression of crystallin genes is relatively well understood, posttranscriptional mechanisms underlying the accumulation of crystallin proteins are largely unclear, especially when transcriptional contribution slows down during denucleation of lens fiber cells.
RNA-binding proteins (RBPs) are implicated in posttranscriptional regulation of gene expression in a variety of biological processes (13–15). Through tissue-specific expression and interaction with specific cofactors, RBP-constituted messenger RNA (mRNA) interactome controls RNA metabolism at multiple levels (16, 17). Several RBPs are linked to congenital cataracts and other lens abnormalities in humans or animal models (18–20). The highly conserved Rbm24 protein contains a single RNA recognition motif (RRM) and exhibits tissue-specific expression patterns in all vertebrate species. It plays an important role in skeletal and cardiac muscle development (21–24) by regulating mRNA stability (25, 26), pre-mRNA splicing (24, 27–30), and mRNA translation (31). Moreover, its expression in head sensory organs, particularly in the lens, is also conserved in vertebrates (23, 32). Recent studies have associated the rbm24 gene with microphthalmia and anophthalmia (33, 34), highlighting its implication in vertebrate eye development. Nevertheless, the molecular and biochemical mechanisms by which it orchestrates lens development remain elusive.
Here we report the functional implication of Rbm24 in lens differentiation using zebrafish embryos. We present the unexpected finding that it is critically required for maintaining the stability and poly(A) tail length of lens-expressed mRNAs. The expression of the rbm24 gene is restricted in differentiating lens fiber cells, and its loss of function severely compromises the accumulation of crystallin proteins in the developing lens. Moreover, Rbm24 binds to a wide spectrum of lens-specific mRNAs and interacts with key components of the cytoplasmic polyadenylation complex. Thus, we uncover an important mechanism by which Rbm24 posttranscriptionally controls lens gene expression through cytoplasmic polyadenylation.
Results
Zebrafish rbm24a Is Expressed in Differentiating Lens Fiber Cells.
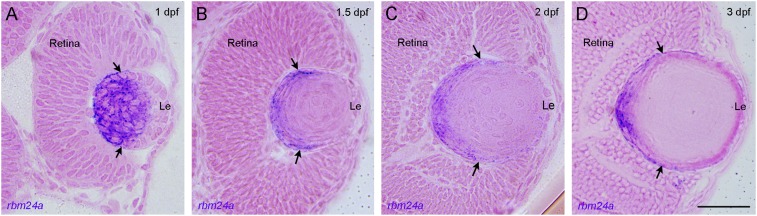
There are two rbm24 paralogs in zebrafish, rbm24a and rbm24b. The expression patterns of rbm24a in the somites, heart, and lens are identical to those of rbm24 in other vertebrates (22, 23), whereas rbm24b mainly shows heart and somitic expression (35). In situ hybridization analysis on ocular sections indicated that rbm24a expression was restricted to differentiating lens fiber cells. At 1 dpf (day postfertilization), rbm24a transcripts were detected in the posterior and central regions of the lens mass, where differentiation of primary fiber cells and initial formation of secondary fiber cells take place (36, 37). They were not present in the anterior lens mass, including the prospective lens epithelium (Fig. 1A). At 1.5 and 2 dpf, rbm24a transcripts were localized in the posterior and lateral regions and remained absent in the lens epithelium (Fig. 1 B and C). At 3 dpf, the transcripts became further restricted to differentiating fiber cells in the posterior border and the lateral zone (Fig. 1D). This highly localized expression pattern suggests that Rbm24a may be involved in the differentiation of lens fiber cells.
Fig. 1.
Expression of zebrafish rbm24a transcripts during lens development. (A) At 1 dpf, the expression of rbm24a transcripts (purple) is localized in differentiating fiber cells at the posterior and central regions of the lens mass. (B–D) From 1.5 to 3 dpf, the expression of rbm24a becomes progressively restricted to the posterior lens border and lateral zone. The transcripts were not present in the anterior lens epithelium (Le) at all stages of lens differentiation. Arrows indicate the anterior region of rbm24a expression, corresponding to the equator. The sections were counterstained with eosin. (Scale bar, 50 µm.)
Zebrafish rbm24a Mutants Display Severe Cataract Phenotype and Defective Lens Differentiation.
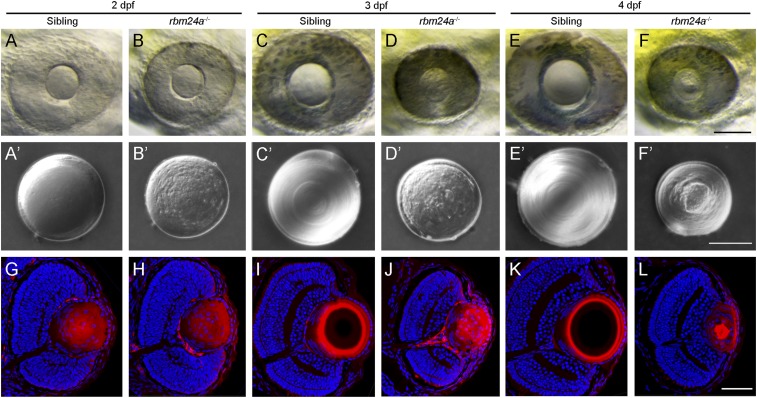
We used TALENs (transcription activator-like effector nucleases) to create mutation in the rbm24a locus and obtained a deletion of eight base pairs in the first exon, which caused a frameshift in the coding region of the RRM and a premature termination of translation (SI Appendix, Fig. S1 A–C). RT-PCR analysis further confirmed the mutation at mRNA level (SI Appendix, Fig. S1D). Homozygous mutants showed complete penetrance of severe heart defects, with heart edema becoming evident at 1.5 dpf. As development proceeds, they also developed microcephaly, which was associated with bilateral microphthalmia (SI Appendix, Fig. S2). Examination of the ocular phenotypes revealed defective lens development. In wild-type (WT) siblings, both the retina and the lens progressively increased in size from 2 to 4 dpf (Fig. 2 A, C, and E), and the lens developed a sleek surface, reflecting the establishment of transparency (Fig. 2 A′, C′, and E′). In time-matched rbm24a mutants, however, the retina and the lens were strongly reduced in size (Fig. 2 B, D, and F), and the lens was particularly coarse and highly irregular, implying a lack of transparency (Fig. 2 B′, D′, and F′). Thus, the mutants showed a severe cataract phenotype. Heterozygous mutants developed normally, while homozygous embryos could not survive beyond 7 dpf, primarily due to heart defects.
Fig. 2.
Cataract phenotype and lens differentiation defects in rbm24a mutants. (A–F) Live images compare eye phenotypes between WT siblings and time-matched rbm24a mutants from 2 to 4 dpf. (Scale bar, 100 µm.) (A′–F′) Differential interference contrast images illustrate the cataract phenotype in surgically dissected lenses. In WT siblings, the lens progressively increases in size, with fiber cells forming regular and refractive concentric layers. In rbm24a mutants, the lens shows no growth, with an irregular surface and a granular aspect. (Scale bar, 50 µm.) (G–L) Staining of lens sections with phalloidin and Hoechst 33342 at the level of the optic nerve compares fiber cell organization and denucleation between WT siblings and rbm24a mutants. (Scale bar, 50 µm.)
Analysis of ocular sections showed no obvious differences between WT siblings and rbm24a mutants at 2 dpf (SI Appendix, Fig. S3 A and B). However, the reduction of lens size and the retention of fiber cell nuclei were particularly evident in rbm24a mutants at 3 and 4 dpf (SI Appendix, Fig. S3 C–F). Terminal deoxynucleotidyl transferase dUTP nick end labeling did not detect obvious increased cell death in the lens of rbm24a mutants when compared with time-matched WT siblings (SI Appendix, Fig. S4 A–C), but few 5-bromodeoxyuridine–positive cells were present in the lens epithelium, indicating reduced cell proliferation (SI Appendix, Fig. S4 D–F). Consistently, there were few lens epithelial cells undergoing cell cycle progression in rbm24a mutants, as revealed by anti-phospho-histone H3 antibody (SI Appendix, Fig. S4 G–I). We then monitored lens differentiation defects by staining ocular sections with phalloidin and Hoechst. At 2 dpf, F-actin and fiber cell nuclei were present similarly in WT siblings and rbm24a mutants (Fig. 2 G and H). At 3 dpf, F-actin was organized as concentric rings at the periphery of the lens, and the central lens nucleus showed weak phalloidin staining and a complete absence of fiber cell nuclei in WT siblings (Fig. 2I). By contrast, disorganized F-actin and degenerating fiber cell nuclei were present in rbm24a mutants (Fig. 2J). At 4 dpf, the lens further differentiated in WT siblings, with complete disappearance of F-actin in the central lens nucleus (Fig. 2K). However, phalloidin-stained large aggregates and Hoechst-positive fragmented nuclei were still present in rbm24a mutants (Fig. 2L). Thus, these defects lead to the development of a cataract phenotype.
Immunofluorescence staining using specific markers was performed to further examine lens differentiation defects. Pax6 transcription factor is localized in the anterior epithelium during lens differentiation (38). When compared with WT siblings from 2 to 4 dpf, this expression pattern was not affected in rbm24a mutants (SI Appendix, Fig. S5 A–F), indicating that Rbm24a is not required for the specification and maintenance of lens epithelium. This is consistent with its absence of expression in these cells. The AQP0 protein marks elongating lens fiber cells (39); its staining pattern clearly revealed a defective elongation of posterior lens fiber cells in rbm24a mutants when compared with time-matched WT siblings (SI Appendix, Fig. S5 G–L). These analyses indicate that mutation of Rbm24a severely impairs the differentiation of lens fiber cells.
Loss of Rbm24a Causes Cataract Formation Independently of Fiber Cell Denucleation.
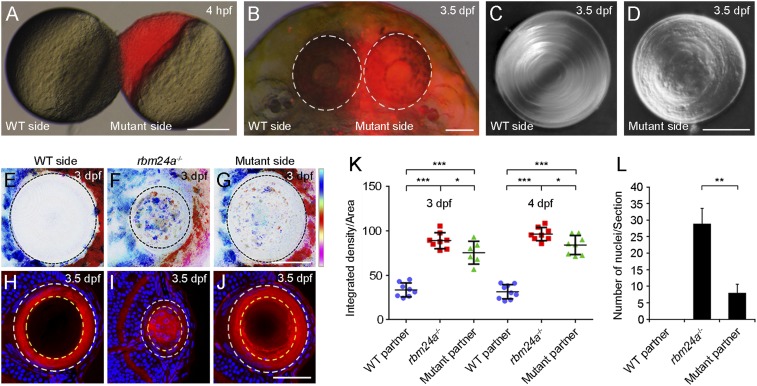
The heart valve defects in rbm24a mutants block blood circulation (Movie S1). Moreover, rbm24a is not expressed in the retina. Thus, the microphthalmia defects could be caused by the absence of blood supply. We examined this possibility by specifically expressing an rbm24a-GFP transgene in the heart of rbm24a mutants under the control of a cmlc2 promoter region (SI Appendix, Fig. S6 A and B). As might be expected, the transgene fully rescued heart and microphthalmia defects (SI Appendix, Fig. S6 C–H), but it did not prevent cataract formation (SI Appendix, Fig. S6 I–K). Furthermore, heterotypic parabiosis, by joining a WT embryo with an rbm24a mutant at 4 hpf (hours postfertilization), restored blood circulation and prevented microphthalmia in the mutant partner (Fig. 3 A and B and Movie S2). However, the mutant lens remained disorganized, lacking a regular and smooth surface (Fig. 3 C and D). The opacity of the mutant lens was confirmed using the confocal reflection mode. In WT embryos at 3.5 dpf, the lens showed no or weak reflected signal (Fig. 3 E and K). By contrast, in rbm24a mutants and rbm24a mutants parabiosed with WT embryos, the lens displayed stronger reflection (Fig. 3 F, G, and K). We also detected the presence of large reflected granules in the lens of rbm24a mutants, which is likely due to defective fiber cell denucleation. Indeed, confocal microscopic analysis following staining with phalloidin and Hoechst indicated that nuclear degradation was severely impaired in rbm24a mutants, whereas this was only weakly delayed in rbm24a mutant partners parabiosed with WT embryos (Fig. 3 H–J and L). These observations strongly suggest that both microphthalmia and altered fiber cell denucleation in rbm24a mutants are an indirect consequence of the absence of blood supply. Thus, loss of Rbm24a should directly impair other processes that promote lens transparency.
Fig. 3.
Rescue of lens fiber cell denucleation but not lens transparency in rbm24a mutants by heterotypic parabiosis. (A) Parabiosis by surgically joining an unlabeled WT embryo and a rhodamine-lysine dextran-labeled rbm24a mutant (red). (Scale bar, 200 µm.) (B) Parabiotic WT and rbm24a mutant partners at 3.5 dpf, with eyes outlined by dashed circles. (Scale bar, 100 µm.) (C and D) Differential interference contrast images show the lens phenotypes in parabiotic WT and mutant partners. (Scale bar, 50 µm.) (E–G) Lens transparency determined by confocal reflection mode, with vertical color bar indicating colors of 10 focal planes at a depth of 30 µm. (Scale bar, 50 µm.) (H–J) Staining of lens sections with phalloidin and Hoechst 33342 shows lens fiber cell denucleation in WT, rbm24a mutants, and rbm24a mutants parabiosed with WT embryos. (Scale bar, 50 µm.) (K) Scatterplot comparing lens transparency by analyzing the integrated density of the scanned lens region in the maximum projection image obtained under the confocal reflection mode. Horizontal lines and error bars represent the mean value ± SD from six to nine independent lenses (*P < 0.05; ***P < 0.001, Student’s t test). (L) Statistical analysis of lens fiber cell denucleation by examining the number of fiber cell nuclei present within 2/3 radius of a lens section at the level of the optic nerve (outlined by dashed yellow circles in H–J). This area shows complete denucleation in WT embryos at 3.5 dpf. The results were obtained from three independent lens sections (**P < 0.01, Student’s t test).
The Overall Abundance of Lens-Specific Transcripts Is Differentially Affected in rbm24a Mutants.
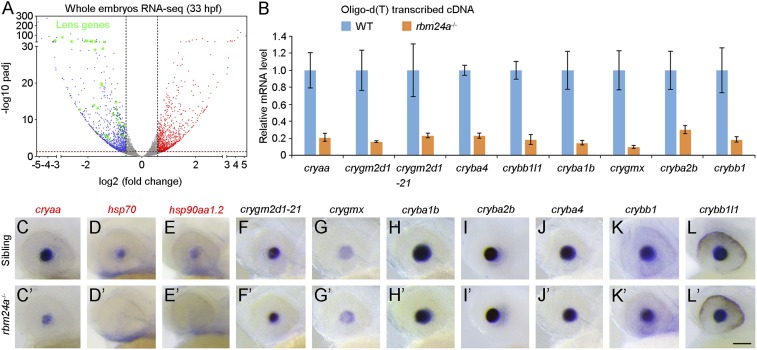
We then performed RNA sequencing (RNA-seq) to analyze Rbm24a-regulated events. Indirect changes were minimized by preparing polyadenylated mRNA samples from whole embryos or head regions at 33 hpf, when morphological and histological alterations of the eye were not apparent. Compared with previously reported muscle-specific alternative splicing events in mice (24), the RNA-seq data revealed that 60.9% (42/69) of them displayed similar alternative splicing patterns in zebrafish (Dataset S1). However, identification of splice junctions did not reveal meaningful differences in alternative splicing events for lens-specific pre-mRNAs between WT embryos and rbm24a mutants (Dataset S2). Rather, differential gene expression analysis indicated a strongly reduced expression level of a large number of lens-related genes in rbm24a mutants (Fig. 4A and SI Appendix, Fig. S7). This was further validated by qRT-PCR analysis with complementary DNAs reverse transcribed using oligo(dT) primer (Fig. 4B).
Fig. 4.
Reduced expression level of polyadenylated crystallin mRNAs in rbm24a mutants. (A) RNA-seq analysis of whole embryos at 33 hpf. Volcano plot shows the down-regulation of lens-related genes coding for heat shock proteins and other crystallins in rbm24a mutants (represented by green dots). Red dots show up-regulated genes, with padj (P value adjusted) less than 0.05 and fold change more than 1.5; blue dots represent down-regulated genes, with padj less than 0.05 and fold change less than 0.67. (B) The qRT-PCR analysis compares crystallin mRNA levels in WT siblings and rbm24a mutants, following reverse transcription using oligo(dT) primer. The results were obtained from three independent experiments, using different batches of embryos. (C–L′) In situ hybridization analysis of crystallin gene expression patterns at 33 hpf. Representative images show strongly reduced expression of cryaa, hsp70, and hsp90aa1.2 in rbm24a mutants. The overall transcript abundance for other crystallin genes does not change. Due to the extremely high level of nucleotide sequence identity, the crygm2d1-21 probe recognizes transcripts of all 21 types of γ-crystallins. (Scale bar, 100 µm.)
Based on the RNA-seq and qRT-PCR data, we selected 10 genes and performed in situ hybridization to compare their expression patterns between WT siblings and rbm24a mutants at 33 hpf. Unexpectedly, only a few genes that encode heat shock proteins, including cryaa, hsp70, and hsp90aa1.2, displayed strongly reduced expression in rbm24a mutants (Fig. 4 C–E and C′–E′). By contrast, a large number of crystallin genes, including crygm2d1-21, crygmx, cryba1b, cryba2b, cryba4, crybb1, and crybb1l1, showed similar expression between WT siblings and rbm24a mutants (Fig. 4 F–L and F′–L′), suggesting that the overall abundance of their transcripts was not affected in rbm24a mutants. Because RNA-seq and qRT-PCR analyses used polyadenylated mRNAs, the results from in situ hybridization raises the possibility that loss of Rbm24a may essentially affect the polyadenylation but not the overall abundance of those crystallin mRNAs coding for structural components.
Rbm24a Binds to a Wide Spectrum of Lens-Expressed mRNAs.
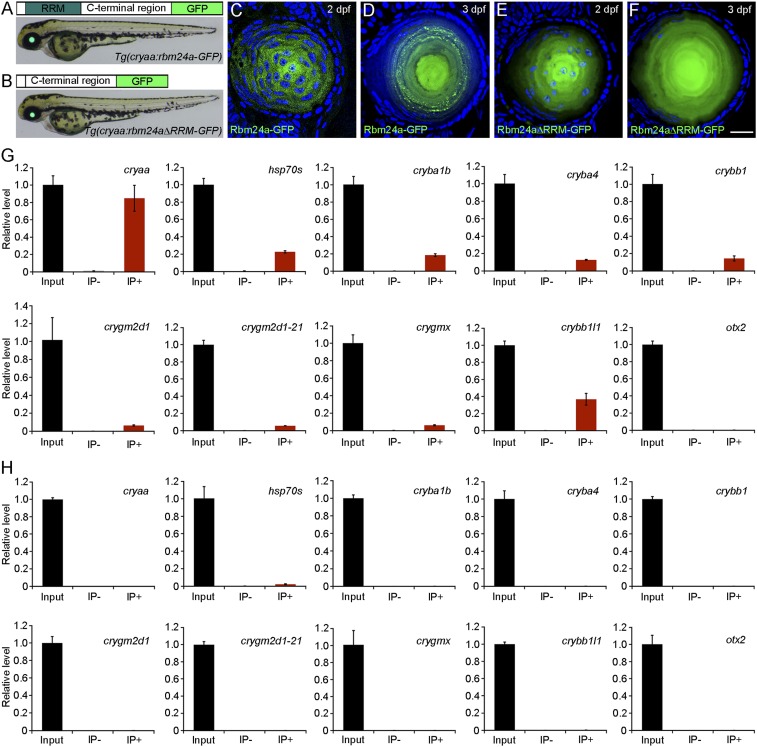
To provide direct evidence of the implication of Rbm24a in regulating crystallin gene expression, we analyzed its interaction with lens-expressed mRNAs by RNA immunoprecipitation and qPCR (RIP-qPCR). This was performed using transgenic lines specifically expressing Rbm24a-GFP or Rbm24aΔRRM-GFP fusion proteins in the lens, under the control of a cryaa promoter region (Fig. 5 A and B). Confocal microscopic analysis indicated that both fusion proteins were localized in the cytoplasm. Interestingly, Rbm24a-GFP showed granular staining at 2 dpf (Fig. 5C) and displayed a punctate pattern in elongating lens fiber cells at 3 dpf (Fig. 5D). This is likely a result of its association with RNAs. In sharp contrast, Rbm24aΔRRM-GFP was present diffusely and uniformly in lens fiber cells at different stages (Fig. 5 E and F), probably due to the lack of binding with RNAs. Indeed, we found that Rbm24a-GFP bound to multiple lens-related transcripts with different efficiency, whose overall abundance was either decreased (cryaa and hsp70) or unchanged (cryba1b, cryba4, crybb1, crygm2d1, crygm2d1-21, crygmx, crybb1l1) in rbm24a mutants (Fig. 5G). The RRM was required for interaction with these transcripts because no PCR products could be detected using transgenic embryos expressing Rbm24aΔRRM-GFP (Fig. 5H). Furthermore, neither Rbm24a-GFP nor Rbm24aΔRRM-GFP interacted with otx2 transcripts, which were not expressed in the lens (Fig. 5 G and H). These results show that Rbm24a specifically interacts with a large number of lens-expressed mRNAs.
Fig. 5.
Rbm24a protein is localized in the cytoplasm of lens fiber cells and specifically binds to lens-expressed mRNAs. (A and B) Transgenic embryos express Rbm24a-GFP or Rbm24aΔRRM-GFP fusion proteins in the lens under the control of a cryaa promoter region. (C–F) Lens sections show differential patterns of cytoplasmic localization of the two fusion proteins at 2 and 3 dpf. (Scale bar, 20 µm.) (G) RIP-qPCR analysis shows the interaction between Rbm24a and lens-expressed mRNAs. For each RIP reaction, 10% of the supernatant was used as input, and the value obtained after PCR amplification was normalized as 1. The remaining supernatant was processed for RNA purification in the presence of anti-GFP antibody (IP+) or anti-myc antibody (IP−). Nonspecific binding of mRNAs to antibody-conjugated beads was controlled by amplification of otx2. The results were obtained from three independent experiments, using different batches of embryos. (H) RIP-qPCR analysis shows the absence of interaction between Rbm24aΔRRM and different mRNAs.
Loss of Rbm24a Reduces the Stability and Poly(A) Tail Length of crystallin mRNAs.
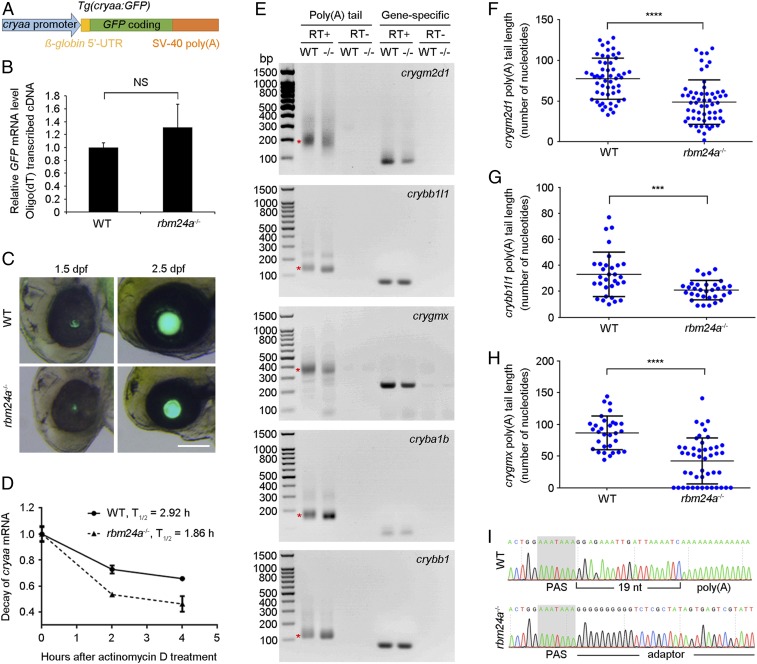
It is unclear whether the reduced overall transcript abundance of lens mRNAs in rbm24a mutants could be caused at the transcriptional level. We thus generated a transgenic line by placing a GFP coding region along with a β-globin leader sequence and SV-40 poly(A) signal under the control of a cryaa promoter region (Fig. 6A). We reasoned that this transgene should faithfully recapitulate the transcriptional activation of the endogenous gene, but the stability and polyadenylation of the resulting transcripts should not be affected in the absence of Rbm24a. Indeed, qPCR analysis following reverse transcription using oligo(dT) primer showed a similar level of GFP expression in WT siblings and rbm24a mutants (Fig. 6B). There were also comparable kinetics and intensity of lens-specific fluorescence in live embryos (Fig. 6C). These indicate that Rbm24a is not involved in the transcriptional control of cryaa, at least not through the promoter region used here. To determine whether loss of Rbm24a affects the stability of cryaa transcripts, head regions dissected from WT siblings and rbm24a mutants were treated with actinomycin D to inhibit de novo RNA synthesis, and the levels of cryaa transcripts at different time intervals were analyzed by qRT-PCR. We found a more rapid decline of cryaa transcripts in rbm24a mutants, with a half-life of about 1.86 h compared to 2.92 h in WT siblings (Fig. 6D). Thus, Rbm24a functions to maintain the stability of a subset of lens mRNAs.
Fig. 6.
Reduced stability and poly(A) tail length of crystallin mRNAs in rbm24a mutants. (A) Schematic representation of the GFP reporter under the control of cryaa promoter. (B) The qRT-PCR analysis compares the expression of GFP transcripts in WT embryos and rbm24a mutants. Bars represent the mean value ± SD from three independent experiments (NS, not significant; Student’s t test). (C) Live images show lens-specific fluorescence in WT siblings and rbm24a mutants. (Scale bar, 100 µm.) (D) Rapid decay of endogenous cryaa mRNA in rbm24a mutants. The initial mRNA levels in WT siblings and rbm24a mutants were normalized as 1 in triplicate experiments. (E) Analysis of poly(A) tail length in crystallin mRNAs. Smear bands from poly(A) PCR products in WT embryos are indicated by asterisks. The corresponding bands from rbm24a mutants (−/−) show a shift in gel mobility. RT+, reverse transcription; RT−, no reverse transcription. (F–H) Scatterplots show differences in poly(A) tail length in three crystallin mRNAs between WT embryos and rbm24a mutants. Horizontal lines and error bars represent the mean value ± SD from all sequenced clones (***P < 0.001; ****P < 0.0001, Student’s t test). (I) Sequencing chromatograms show the complete absence of polyadenylation of crygmx transcripts in rbm24a mutants.
For most other crystallin genes, only their polyadenylated mRNAs showed a decreased expression level in rbm24a mutants. To further validate this unexpected and potentially novel function of Rbm24a, we used poly(A) tail-length assay to examine the polyadenylation state of crygm2d1, crybb1l1, crygmx, cryba1b, and crybb1 mRNAs. Following poly(G/I) tailing and reverse transcription, the poly(A) PCR-amplified products from rbm24a mutants showed a weak shift with respect to those from WT siblings (Fig. 6E). Furthermore, sequence analysis confirmed an overall reduction of poly(A) tails (Fig. 6 F–H). As an example, the average poly(A) tail length for crygmx transcripts in WT embryos was 86 nucleotides, whereas this was reduced to 42 nucleotides in rbm24a mutants (Fig. 6H and Dataset S3). More strikingly, a large number of crygmx transcripts showed a complete absence of poly(A) tail, such that the 3′-untranslated region terminated at the polyadenylation signal (PAS), whereas in WT embryos the poly(A) tail followed the general rule by starting at about 20 nucleotides downstream of this signal (Fig. 6I). These results strongly suggest that Rbm24a is critically required for the elongation or maintenance of poly(A) tail length in a large number of crystallin mRNAs.
Rbm24a Is Required for the Accumulation of Crystallin Proteins.
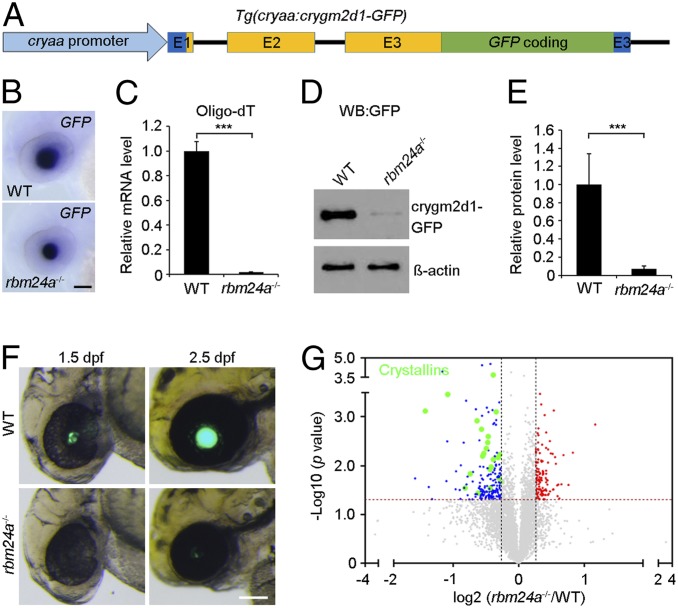
To determine the functional consequence of this reduced polyadenylation on the accumulation of crystallin proteins, we first generated a transgenic line by inserting a GFP coding sequence into the third exon of the crygm2d1 gene, before its stop codon (Fig. 7A). Thus, the expression of the crygm2d1-GFP fusion protein under the control of the cryaa promoter should closely reproduce the actual condition with respect to crygm2d1 mRNA expression and translation. Indeed, in situ hybridization analysis using the GFP probe revealed a similar overall abundance of the fusion transcripts between WT siblings and rbm24a mutants at 2 dpf (Fig. 7B). However, qPCR analysis following reverse transcription using oligo(dT) primer showed that the amounts of polyadenylated crygm2d1-GFP mRNAs were markedly reduced in rbm24a mutants (Fig. 7C), suggesting an absence or severe shortening of the poly(A) tail. Consistently, the expression level of the fusion protein was severely reduced in rbm24a mutants, as determined by Western blot analysis using GFP antibody (Fig. 7 D and E). The decreased translational efficiency of crygm2d1-GFP fusion mRNA was also evident in live embryos. Compared with WT siblings, time-matched rbm24a mutants showed very weak or no lens-specific fluorescence (Fig. 7F). Furthermore, analysis by quantitative proteomics confirmed a global decrease of endogenous crystallin proteins in rbm24a mutants (Fig. 7G, SI Appendix, Fig. S8, and Datasets S4 and S5). Thus, these observations demonstrate a critical requirement for Rbm24a in the accumulation of crystallin proteins.
Fig. 7.
Reduced translation efficiency of crystallin mRNAs in rbm24a mutants. (A) Schematic representation of the crygm2d1-GFP transgene. The three exons (E1, E2, and E3) of the crygm2d1 gene are shown. Dark blue represents 5′- and 3′-untranslated regions that will be present in the fusion transcripts. (B) In situ hybridization analysis using the GFP probe detects a similar expression pattern of the fusion transcripts in WT embryos and rbm24a mutants at 2 dpf. (Scale bar, 100 µm.) (C) qPCR analysis following reverse transcription using oligo(dT) primer shows a residual level of polyadenylated crygm2d1-GFP mRNAs in rbm24a mutants at 2 dpf. The data were obtained from three independent samples of different batches of embryos, and the expression level in WT embryos is set as 1 after normalization to glyceraldehyde-3-phosphate dehydrogenase (***P < 0.001; Student’s t test). (D) Western blot analysis using anti-GFP antibody shows a trace amount of the crygm2d1-GFP protein in rbm24a mutants at 2 dpf. β-Actin is an input control. (E) Quantification of protein bands after normalization to β-actin using three independent Western blots (***P < 0.001; Student’s t test). (F) Live images show marked differences in the lens-specific expression of crygm2d1-GFP between WT embryos and rbm24a mutants at 1.5 and 2.5 dpf. (Scale bar, 100 µm.) (G) Analysis by quantitative proteomics shows a globally decreased accumulation of endogenous crystallin proteins in rbm24a mutants at 2 dpf.
Since restoration of blood circulation in rbm24a mutants led to nearly normal degradation of fiber cell nuclei, the decreased synthesis of crystallin proteins should represent a direct and autonomous effect of Rbm24a mutation on cataract formation. This was supported by cell transplantation experiments (40, 41), in which fluorescently labeled cells were implanted in the presumptive lens placode region of stage-matched recipient embryos (SI Appendix, Fig. S9 A and B). In WT recipients receiving rbm24a mutant donor cells, the lens displayed a cataract-like phenotype at 3.5 dpf, and this caused blindness in the chimeric larvae, as they were unable to prey on the paramecia (SI Appendix, Fig. S9 C–F and Movie S3). We further used the crygm2d1-GFP reporter to monitor fiber cell differentiation. Compared to WT cells, transplanted rbm24a mutant cells integrated into the WT lens failed to express the fusion protein at 1.5 dpf (SI Appendix, Fig. S9 G and H), but they were expected to undergo normal denucleation because of the presence of blood supply. By contrast, WT cells were able to express the fusion protein in rbm24a mutant recipients, regardless of the absence or presence of blood circulation (SI Appendix, Fig. S9 I and J). These results clearly show that Rbm24a regulates the accumulation of crystallins in a cell-autonomous manner.
Rbm24a Interacts with the Cytoplasmic Polyadenylation Complex.
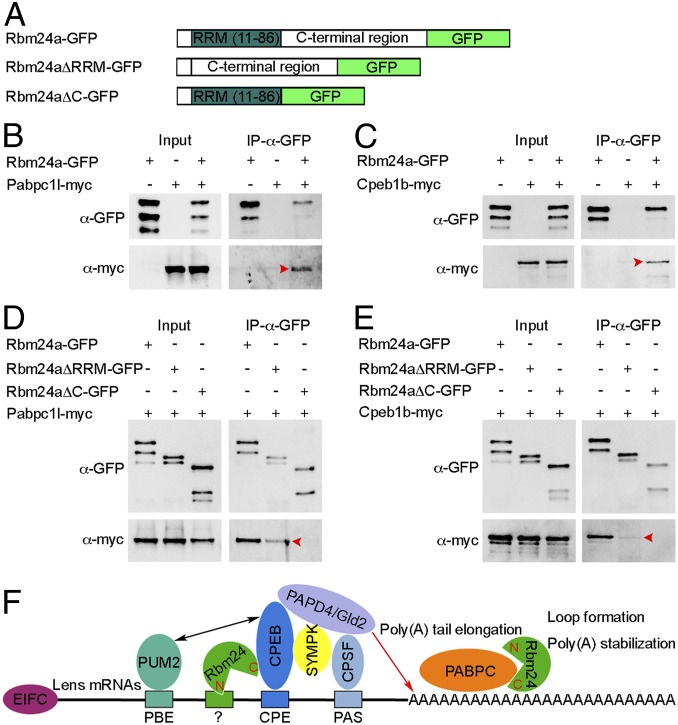
The critical implication of Rbm24a in maintaining poly(A) tail length to promote translational efficiency raises the question of whether and how it interacts with the cytoplasmic polyadenylation complex. Among different components that participate in the formation and function of this complex, cytoplasmic polyadenylation element-binding protein (CPEB) promotes poly(A) tail elongation when it is phosphorylated, and cytoplasmic poly(A)-binding protein (PABPC) stimulates translation and protects poly(A) tail from degradation (42, 43). By coimmunoprecipitation using full-length and truncated Rbm24a proteins (Fig. 8A), we found that GFP-tagged Rbm24a specifically bound to myc-tagged zebrafish Pabpc1l and Cpeb1b (Fig. 8 B and C). In addition, the C-terminal region but not the RRM was directly involved in these interactions (Fig. 8 D and E). Interestingly, the C-terminal region of Rbm24a showed a stronger affinity with Pabpc1l than with Cpeb1b, suggesting that Rbm24a may interact with these proteins via different subdomains and that the binding of the Rbm24a C-terminal region with Cpeb1b may also need the assistance of the RRM. These analyses indicate that Rbm24a may participate in the translational activation of lens-expressed mRNAs by regulating the elongation and the stability of the poly(A) tail through interaction with Cpeb1b and Pabpc1l, respectively (Fig. 8F). Altogether, we have revealed that vertebrate Rbm24 is an important component of the lens mRNA interactome and demonstrated that it represents a regulator of cytoplasmic polyadenylation.
Fig. 8.
Rbm24a physically interacts with Pabpc1l and Cpeb1b through the C-terminal region. (A) Schematic representation of GFP-tagged full-length and truncated Rbm24a proteins. (B and C) The full-length Rbm24a interacts with Pabpc1l and Cpeb1b (arrowheads). (D and E) The C-terminal region of Rbm24a coimmunoprecipitates with Pabpc1l and Cpeb1b (arrowheads), but the interaction with Cpeb1b is weak. (F) Possible function of vertebrate Rbm24 in poly(A) tail elongation and stabilization. In the lens, Rbm24 directly associates with CPEB and promotes the activity of PAPD4/Gld2 poly(A) polymerase in poly(A) tail elongation; it also interacts with PABPC to facilitate loop formation and stabilize poly(A) tail. The RNA motif that recruits Rbm24 remains to be determined. EIFC, eukaryotic translation initiation factor complex; PBE, PUM2-binding element; CPE, cytoplasmic polyadenylation element; PAS, polyadenylation signal; N, Rbm24 N-terminal region; C, Rbm24 C-terminal region.
Discussion
The establishment of lens transparency is coordinated by the accumulation of functional crystallin proteins and degradation of organelles. The transcriptional control of lens differentiation is relatively well documented, but the posttranscriptional regulation remains largely unknown. Particularly, when the transcriptional contribution declines during denucleation of differentiating lens fiber cells, it becomes important to set up a mechanism that promotes the efficient translation of lens mRNAs. However, the cytoplasmic control of protein expression mediated by RBPs is generally less understood mechanistically (42, 43). This work reveals a critical role of vertebrate Rbm24 in regulating cytoplasmic polyadenylation during lens differentiation. We show that loss of Rbm24 impairs the stability and polyadenylation of lens-expressed mRNAs encoding regulatory and structural components in lens fiber cells, thus preventing the accumulation and correct folding of crystallins, which eventually leads to cataract formation.
Due to its multiple expression sites, constitutive knockout of the rbm24a gene in zebrafish produces pleiotropic morphological alterations, including impaired blood circulation that delays eye development and lens denucleation. Our results establish that the reduced eye size and impaired fiber cell nuclear degradation are indirect consequences due to the absence of blood supply. Restoration of blood circulation in rbm24a mutants by transgenic correction of heart defects or by fusion of the circulatory systems through heterotypic parabiosis completely rescues eye size and leads to essentially normal denucleation in lens fiber cells, but this does not prevent the occurrence of the cataract phenotype. A direct consequence of rbm24a mutation in producing lens opacity is unequivocally proved by transplantation of rbm24a mutant cells into the developing lens of WT recipients, which causes cataract formation and leads to blindness. These observations strongly suggest that Rbm24a functions in an autonomous manner to regulate lens differentiation, independently of organelle degradation. Indeed, it is known that lens fiber cell denucleation is controlled by regulated transcription and activity of acidic DNase (44–46). Thus, it will be of interest to investigate how blood supply and DNase activity cooperate in this process.
The lens refractive power is ensured by an extremely high concentration of crystallins (8). Our RNA-seq data indicate that the expression level, but not the alternative splicing of lens-specific genes, is affected in rbm24a mutants. Furthermore, Rbm24a binds to a large number of lens-specific mRNAs and regulates their translation efficiency via distinct mechanisms, but with the same outcome for the accumulation of crystallin proteins. First, Rbm24a is required for maintaining the stability of a subset of lens mRNAs, such as cryaa, hsp70, and hsp90aa1.2. Thus, their rapid decay in rbm24a mutants affects the accumulation of the corresponding proteins that have molecular chaperone activity to ensure the correct folding and solubility of other lens proteins. Second, loss of Rbm24a causes the shortening of poly(A) tail length in those crystallin mRNAs encoding structural components, without affecting the overall RNA abundance. This markedly impairs their translation and clearly represents a mechanism by which Rbm24a regulates mRNA fate during lens fiber cell differentiation. It is well established that one of the important functions of poly(A) tails is the regulation of the translational state through interaction with the cytoplasmic polyadenylation complex and the translation initiation complex (43, 47). During lens differentiation, the rapid degradation of fiber cell nuclei may preclude a continuous transcription of crystallin genes. As a compensatory mechanism, the transcribed mRNAs should be stabilized and/or processed for efficient translation into proteins. Rbm24a may function as a major regulator mediating these critical processes. In the absence of Rbm24a function, both the reduced stability and shortened poly(A) tail length in lens mRNAs prevent the accumulation of crystallin proteins and, consequently, decrease lens refractive power. Thus, our finding emphasizes a predominant role of posttranscriptional control during lens differentiation and supports previous observations showing the involvement of translation initiation machinery in lens development (48).
It has become increasingly evident that cytoplasmic polyadenylation regulates poly(A) tail elongation of nuclear exported mRNAs and plays an important role in translation activation during oogenesis and early embryonic development (49). This process is mediated by cytoplasmic poly(A) polymerases through interaction with specific RBPs, such as CPEB, which recognizes the U-rich cytoplasmic polyadenylation element present in target mRNAs (42, 43, 46). In addition, PABPC interacts with other components of the cytoplasmic polyadenylation complex and promotes binding of the complex to newly produced poly(A) tails. Thus, it can activate translation and prevent mRNAs from deadenylation (50). Based on the requirement of its RRM in binding to a large number of crystallin mRNAs and its C-terminal domain in interaction with CPEB and PABPC, we could postulate that Rbm24a may function as a component in the cytoplasmic polyadenylation machinery to promote efficient translation of lens-specific mRNAs. Although the precise mechanism awaits further investigation, it is possible that the interaction between Rbm24a and CPEB may activate the cytoplasmic polyadenylation complex for the elongation of poly(A) tails; this is consistent with an overall shortening of poly(A) tails in crystallin mRNAs following loss of Rbm24a. In addition, Rbm24a may interact with PABPC to protect lens mRNAs from deadenylation; this is supported by the complete disappearance of poly(A) tail in crygmx transcripts when Rbm24a function is absent. Therefore, we unveil a critical role of vertebrate Rbm24 in cytoplasmic polyadenylation. This unprecedented function of Rbm24 may be not limited to lens differentiation; it may also operate in other tissues or cells where Rbm24 is localized in the cytoplasm, such as the inner ear, nasal epithelium, and myoblasts (23, 32).
Our recent work shows that the mouse Rbm24 protein is first localized in the cytoplasm of lens fiber cells at the posterior region of the lens vesicle and then in the equatorial epithelial cells, which will give rise to the secondary lens fibers (32). As these cells differentiate, they terminally leave the cell cycle, start to elongate, and express numerous genes that help to establish lens transparency (51, 52). Zebrafish rbm24a mRNA is also restricted to differentiating and elongating lens fiber cells, and the Rbm24a protein is localized in the cytoplasm as a punctate pattern, reminiscent of binding with RNAs. Therefore, the conserved expression patterns and the cataract phenotype in rbm24a mutants are fully consistent with a critical role of vertebrate Rbm24 in controlling important events related to lens fiber differentiation. Taken together, evidence now available clearly shows that Rbm24 represents a major posttranscriptional regulator of embryonic cell lineage differentiation. Its cell-type-specific expression in different tissues is fully consistent with a critical role in orchestrating the initiation and maintenance of cell differentiation. Moreover, Rbm24 can act as a multifunctional RBP that governs tissue-specific and also stage-specific regulation of gene expression. These functions are likely accomplished by its dynamic subcellular localization and interaction with specific partners. Thus, it will be important in the future to determine the regulated changes in Rbm24 cellular compartmentalization and its tissue- and stage-specific functions.
In summary, we show that vertebrate Rbm24 plays a key role in controlling the translation efficiency of tissue-specific mRNAs. This may represent a general function of Rbm24 in regulating various differentiation processes. Because of the implication of RBPs in eye development and disease (53) and the similarities in structure and regulation between zebrafish and human lenses (54), understanding the posttranscriptional mechanisms that help to produce high levels of transparent proteins in the lens could open novel perspectives for the prevention and treatment of cataracts. Furthermore, given the highly conserved expression of vertebrate Rbm24 in the lens, it will be particularly interesting to identify human mutations affecting the cis-regulatory elements that disrupt its lens-specific expression and lead to congenital cataracts.
Materials and Methods
Zebrafish and rbm24a Mutants.
Adults of the AB strain were maintained at 28.5 °C in standard housing systems (Haisheng). TALEN repeat variable diresidues targeting the first exon of the rbm24a gene were engineered through the Golden Gate assembly method (55). The constructs were assembled in the modified pCS2+KKR and pCS2+ELD vectors (56). Screening of F0 founder and F1 offspring was performed by sequencing PCR products amplified from tail fin genomic DNA (57). Identified rbm24a heterozygous fish were used for different experiments. Additional materials and methods are available in SI Appendix, SI Materials and Methods.
Data Availability.
The sequencing data reported in this paper have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) database (58, 59), https://www.ncbi.nlm.nih.gov/geo (accession nos. GSE136006 and GSE136003).
Supplementary Material
Acknowledgments
We thank H. Y. Yu, X. M. Zhao, C. B. Liu, and S. Wang from the State Key Laboratory of Microbial Technology for assistance with confocal imaging and histological sectioning. This work was supported by the National Natural Science Foundation of China (Grants 31671509 and 31871451); the National Key R&D Program of China (Grant 2018YFA0801000); the Programs of Shandong University Young Scholars and Qilu Young Scholars; Shandong Provincial Natural Science Foundation (Grant ZR2017BC068); and the Collaboration Programs from Shandong Provincial Key Laboratory of Animal Cell and Developmental Biology (Grant SDKLACD2019014).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The sequencing data reported in this paper have been deposited in National Center for Biotechnology Information Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession nos. GSE136006 and GSE136003).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1917922117/-/DCSupplemental.
References
- 1.Donner A.-L., Lachke S.-A., Maas R.-L., Lens induction in vertebrates: Variations on a conserved theme of signaling events. Semin. Cell Dev. Biol. 17, 676–685 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Graw J., Eye development. Curr. Top. Dev. Biol. 90, 343–386 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Cvekl A., Ashery-Padan R., The cellular and molecular mechanisms of vertebrate lens development. Development 141, 4432–4447 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cvekl A., Zhang X., Signaling and gene regulatory networks in mammalian lens development. Trends Genet. 33, 677–702 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wride M.-A., Lens fibre cell differentiation and organelle loss: Many paths lead to clarity. Philos. Trans. R. Soc. Lond. B Biol. Sci. 366, 1219–1233 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bloemendal H., et al. , Ageing and vision: Structure, stability and function of lens crystallins. Prog. Biophys. Mol. Biol. 86, 407–485 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Wistow G., The human crystallin gene families. Hum. Genomics 6, 26 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bassnett S., Costello M.-J., The cause and consequence of fiber cell compaction in the vertebrate lens. Exp. Eye Res. 156, 50–57 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takemoto L., Sorensen C.-M., Protein-protein interactions and lens transparency. Exp. Eye Res. 87, 496–501 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graw J., Genetics of crystallins: Cataract and beyond. Exp. Eye Res. 88, 173–189 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Graw J., Congenital hereditary cataracts. Int. J. Dev. Biol. 48, 1031–1044 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Liu Y.-C., Wilkins M., Kim T., Malyugin B., Mehta J.-S., Cataracts. Lancet 390, 600–612 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Glisovic T., Bachorik J.-L., Yong J., Dreyfuss G., RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 582, 1977–1986 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ye J., Blelloch R., Regulation of pluripotency by RNA binding proteins. Cell Stem Cell 15, 271–280 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Q., Hu G., Post-transcriptional regulation of the pluripotent state. Curr. Opin. Genet. Dev. 46, 15–23 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marchese D., de Groot N.-S., Lorenzo Gotor N., Livi C.-M., Tartaglia G.-G., Advances in the characterization of RNA-binding proteins. Wiley Interdiscip. Rev. RNA 7, 793–810 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brinegar A.-E., Cooper T.-A., Roles for RNA-binding proteins in development and disease. Brain Res. 1647, 1–8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lachke S.-A., et al. , Mutations in the RNA granule component TDRD7 cause cataract and glaucoma. Science 331, 1571–1576 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dash S., Dang C.-A., Beebe D.-C., Lachke S.-A., Deficiency of the RNA binding protein caprin2 causes lens defects and features of Peters anomaly. Dev. Dyn. 244, 1313–1327 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Siddam A.-D., et al. , The RNA-binding protein Celf1 post-transcriptionally regulates p27Kip1 and Dnase2b to control fiber cell nuclear degradation in lens development. PLoS Genet. 14, e1007278 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H.-Y., Bourdelas A., Carron C., Shi D.-L., The RNA-binding protein Seb4/RBM24 is a direct target of MyoD and is required for myogenesis during Xenopus early development. Mech. Dev. 127, 281–291 (2010). [DOI] [PubMed] [Google Scholar]
- 22.Poon K.-L., et al. , RNA-binding protein RBM24 is required for sarcomere assembly and heart contractility. Cardiovasc. Res. 94, 418–427 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Grifone R., et al. , The RNA-binding protein Rbm24 is transiently expressed in myoblasts and is required for myogenic differentiation during vertebrate development. Mech. Dev. 134, 1–15 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Yang J., et al. , RBM24 is a major regulator of muscle-specific alternative splicing. Dev. Cell 31, 87–99 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Jin D., Hidaka K., Shirai M., Morisaki T., RNA-binding motif protein 24 regulates myogenin expression and promotes myogenic differentiation. Genes Cells 15, 1158–1167 (2010). [DOI] [PubMed] [Google Scholar]
- 26.Xu E., et al. , RNA-binding protein RBM24 regulates p63 expression via mRNA stability. Mol. Cancer Res. 12, 359–369 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ito J., et al. , RBM20 and RBM24 cooperatively promote the expression of short enh splice variants. FEBS Lett. 590, 2262–2274 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Zhang T., et al. , Rbm24 regulates alternative splicing switch in embryonic stem cell cardiac lineage differentiation. Stem Cells 34, 1776–1789 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Cardinali B., et al. , MicroRNA-222 regulates muscle alternative splicing through Rbm24 during differentiation of skeletal muscle cells. Cell Death Dis. 7, e2086 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu J., Kong X., Zhang M., Yang X., Xu X., RNA binding protein 24 deletion disrupts global alternative splicing and causes dilated cardiomyopathy. Protein Cell 10, 405–416 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang M., et al. , Rbm24, a target of p53, is necessary for proper expression of p53 and heart development. Cell Death Differ. 25, 1118–1130 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grifone R., Saquet A., Xu Z., Shi D.-L., Expression patterns of Rbm24 in lens, nasal epithelium, and inner ear during mouse embryonic development. Dev. Dyn. 247, 1160–1169 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Brastrom L.-K., Scott C.-A., Dawson D.-V., Slusarski D.-C., A high-throughput assay for congenital and age-related eye diseases in zebrafish. Biomedicines 7, E28 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dash S., et al. , The master transcription factor SOX2, mutated in anophthalmia/microphthalmia, is post-transcriptionally regulated by the conserved RNA-binding protein RBM24 in vertebrate eye development. Hum. Mol. Genet., 10.1093/hmg/ddz278 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maragh S., et al. , Rbm24a and Rbm24b are required for normal somitogenesis. PLoS One 9, e105460 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dahm R., Schonthaler H.-B., Soehn A.-S., van Marle J., Vrensen G.-F., Development and adult morphology of the eye lens in the zebrafish. Exp. Eye Res. 85, 74–89 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Greiling T.-M., Aose M., Clark J.-I., Cell fate and differentiation of the developing ocular lens. Invest. Ophthalmol. Vis. Sci. 51, 1540–1546 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Imai F., Yoshizawa A., Fujimori-Tonou N., Kawakami K., Masai I., The ubiquitin proteasome system is required for cell proliferation of the lens epithelium and for differentiation of lens fiber cells in zebrafish. Development 137, 3257–3268 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Mochizuki T., Kojima Y., Nishiwaki Y., Harakuni T., Masai I., Endocytic trafficking factor VPS45 is essential for spatial regulation of lens fiber differentiation in zebrafish. Development 145, dev170282 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng X.-N., et al. , Leucine repeat adaptor protein 1 interacts with Dishevelled to regulate gastrulation cell movements in zebrafish. Nat. Commun. 8, 1353 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shao M., Cheng X.-N., Liu Y.-Y., Li J.-T., Shi D.-L., Transplantation of zebrafish cells by conventional pneumatic microinjector. Zebrafish 15, 73–76 (2018). [DOI] [PubMed] [Google Scholar]
- 42.Eckmann C.-R., Rammelt C., Wahle E., Control of poly(A) tail length. Wiley Interdiscip. Rev. RNA 2, 348–361 (2011). [DOI] [PubMed] [Google Scholar]
- 43.Charlesworth A., Meijer H.-A., de Moor C.-H., Specificity factors in cytoplasmic polyadenylation. Wiley Interdiscip. Rev. RNA 4, 437–461 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nishimoto S., et al. , Nuclear cataract caused by a lack of DNA degradation in the mouse eye lens. Nature 424, 1071–1074 (2003). [DOI] [PubMed] [Google Scholar]
- 45.Nakahara M., et al. , Degradation of nuclear DNA by DNase II-like acid DNase in cortical fiber cells of mouse eye lens. FEBS J. 274, 3055–3064 (2007). [DOI] [PubMed] [Google Scholar]
- 46.Cui X., et al. , HSF4 regulates DLAD expression and promotes lens de-nucleation. Biochim. Biophys. Acta 1832, 1167–1172 (2013). [DOI] [PubMed] [Google Scholar]
- 47.Nicholson A.-L., Pasquinelli A.-E., Tales of detailed poly(A) tails. Trends Cell Biol. 29, 191–200 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Choudhuri A., Maitra U., Evans T., Translation initiation factor eIF3h targets specific transcripts to polysomes during embryogenesis. Proc. Natl. Acad. Sci. U.S.A. 110, 9818–9823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reyes J. M., Ross P. J., Cytoplasmic polyadenylation in mammalian oocyte maturation. Wiley Interdiscip. Rev. RNA 7, 71–89 (2016). [DOI] [PubMed] [Google Scholar]
- 50.Goss D.-J., Kleiman F.-E., Poly(A) binding proteins: Are they all created equal? Wiley Interdiscip. Rev. RNA 4, 167–179 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Audette D.-S., Scheiblin D.-A., Duncan M.-K., The molecular mechanisms underlying lens fiber elongation. Exp. Eye Res. 156, 41–49 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mochizuki T., Luo Y.-J., Tsai H.-F., Hagiwara A., Masai I., Cell division and cadherin-mediated adhesion regulate lens epithelial cell movement in zebrafish. Development 144, 708–719 (2017). [DOI] [PubMed] [Google Scholar]
- 53.Dash S., Siddam A.-D., Barnum C.-E., Janga S.-C., Lachke S.-A., RNA-binding proteins in eye development and disease: Implication of conserved RNA granule components. Wiley Interdiscip. Rev. RNA 7, 527–557 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Greiling T.-M., Clark J.-I., New insights into the mechanism of lens development using zebra fish. Int. Rev. Cell Mol. Biol. 296, 1–61 (2012). [DOI] [PubMed] [Google Scholar]
- 55.Cermak T., et al. , Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 39, e82 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lei Y., et al. , Efficient targeted gene disruption in Xenopus embryos using engineered transcription activator-like effector nucleases (TALENs). Proc. Natl. Acad. Sci. U.S.A. 109, 17484–17489 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xing Y.-Y., et al. , Mutational analysis of dishevelled genes in zebrafish reveals distinct functions in embryonic patterning and gastrulation cell movements. PLoS Genet. 14, e1007551 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shao M., Shi D.-L., Rbm24 functions as a critical translational regulator to pretect crystallin proteins accumulation for lens transparency. RNA-seq data of 33 hpf rbm24a−/− and wild-type zebrafish embryo. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?&acc=GSE136006. Deposited 19 August 2019.
- 59.Shao M., Shi D.-L., Rbm24 functions as a critical translational regulator to pretect crystallin proteins accumulation for lens transparency. RNA-seq data of 33 hpf rbm24a−/− and wild-type zebrafish embryo, head explant. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?&acc=GSE136003. Deposited 19 August 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data reported in this paper have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) database (58, 59), https://www.ncbi.nlm.nih.gov/geo (accession nos. GSE136006 and GSE136003).