Significance
Systemic lupus erythematosus (SLE) is an autoimmune disease characterized by pathogenic autoantibodies. CD4+T cells are involved, but the identity of the pathogenic B helper T cells is unclear. Notably, the antiinflammatory cytokine interleukin 10 (IL-10) might promote autoantibody production in SLE. Here, we identified a T cell population that expressed CCR6 and promoted antibody production in a partially IL-10–dependent manner. These CCR6+T cells were distinct from previously identified B helper T cells and expanded in mice developing lupus-like disease and in SLE patients, where they spontaneously induced autoantibodies. They were abundant in lymph nodes of SLE patients, but were excluded from B cell follicles. These findings suggest that extrafollicular, IL-10–producing CCR6+T cells play a pathogenic role in SLE.
Keywords: lupus, interleukin-10, B cell help
Abstract
Interleukin 10 (IL-10) is an antiinflammatory cytokine, but also promotes B cell responses and plays a pathogenic role in systemic lupus erythematosus (SLE). CD4+CCR6+IL-7R+T cells from human tonsils produced IL-10 following stimulation by naïve B cells, which promoted B cell immunoglobulin G (IgG) production. These tonsillar CCR6+B helper T cells were phenotypically distinct from follicular helper T (TFH) cells and lacked BCL6 expression. In peripheral blood, a CCR6+T cell population with similar characteristics was identified, which lacked Th17- and TFH-associated gene signatures and differentiation-associated surface markers. CD4+CCR6+T cells expressing IL-10, but not IL-17, were also detectable in the spleens of cytokine reporter mice. They provided help for IgG production in vivo, and expanded systemically in pristane-induced lupus-like disease. In SLE patients, CD4+CCR6+IL-7R+T cells were associated with the presence of pathogenic anti-dsDNA (double-stranded DNA) antibodies, and provided spontaneous help for autoantibody production ex vivo. Strikingly, IL-10–producing CCR6+T cells were highly abundant in lymph nodes of SLE patients, and colocalized with B cells at the margins of follicles. In conclusion, we identified a previously uncharacterized population of extrafollicular B helper T cells, which produced IL-10 and could play a prominent pathogenic role in SLE.
Interleukin-10 (IL-10) is a tolerogenic cytokine, since it inhibits myeloid cells from producing proinflammatory cytokines and up-regulating MHC (major histocompatibility complex) and costimulatory molecules, which mediate T cell activation (1, 2). For this reason it is considered to be a characteristic cytokine of regulatory T cells, in particular of type 1 regulatory (Tr1) T cells (3, 4). Notably, Tr1 cells inhibit B cell responses, but this inhibitory effect is independent of IL-10 (5, 6). IL-10 production is not unique to regulatory T cells but is also secreted by helper T cells, where it exerts, however, also mainly regulatory functions (7–11). Despite its tolerogenic effects on myeloid cells and T cells, IL-10 is a well-known B cell growth and differentiation factor. Thus, it can promote survival, proliferation, isotype switching, and differentiation of human B cells to plasma cells (1, 12). Indeed, IL-10 is also produced by follicular helper T (TFH) cells (13), an effector T cell subset that is specialized to provide B cell help in B cell follicles for antibody production (14, 15). TFH cells are characterized by the expression of the transcription factor BCL6, the chemokine receptor CXCR5, the coreceptors ICOS and PD1, and the cytokine IL-21 (13, 14, 16–20). Notably, these canonical CXCR5+ICOS+TFH effector cells are absent from peripheral blood, but some circulating memory T cells express CXCR5 and possess superior B helper capacities. They are thus considered TFH-like cells, and can be subdivided according to chemokine receptor expression into subsets of CXCR3+TFH1, CCR6+TFH17, and CXCR3−CCR6−TFH2 (20, 21). Notably, also Th17 cells have been reported to provide B cell help (22, 23), and splenic “natural” Th17 cells from naïve mice induce intestinal IgA production upon homing to Peyer’s patches (24).
SLE is a systemic autoimmune disorder characterized by a breakdown of B cell tolerance and the production of pathogenic autoantibodies (25). In particular, many SLE patients harbor anti-dsDNA (double-stranded DNA) antibodies (26), which form immune complexes, induce interferon (IFN)-α production from plasmacytoid dendritic cells (DCs), and can deposit in the kidney and cause nephritis (27). Importantly, IL-10 has a pathogenic role in SLE in humans and mice (28–30), presumably because it promotes autoreactive B cell responses (28, 31). The identity of pathogenic cells that produce IL-10 and induce B cell autoantibody production in SLE is a field of intense research (32). TFH cells are obvious candidates, given their potent B helper capacities (33), but also other B helper T cell populations were proposed to be involved in lupus (34–36).
We previously showed that CCR6+ memory T cells in human blood were autoreactive and could selectively produce IL-10 upon suboptimal T cell receptor (TCR) stimulation, and speculated that they might have context-dependent regulatory/helper functions (37). Here, we analyzed the role of IL-10–producing CCR6+T cells in B cell responses and lupus. We found that CCR6+T cells in tonsils provided B cell help via IL-10 and accumulated in lupus-like disease in mice and in SLE patients, where they promoted the production of pathogenic autoantibodies.
Results
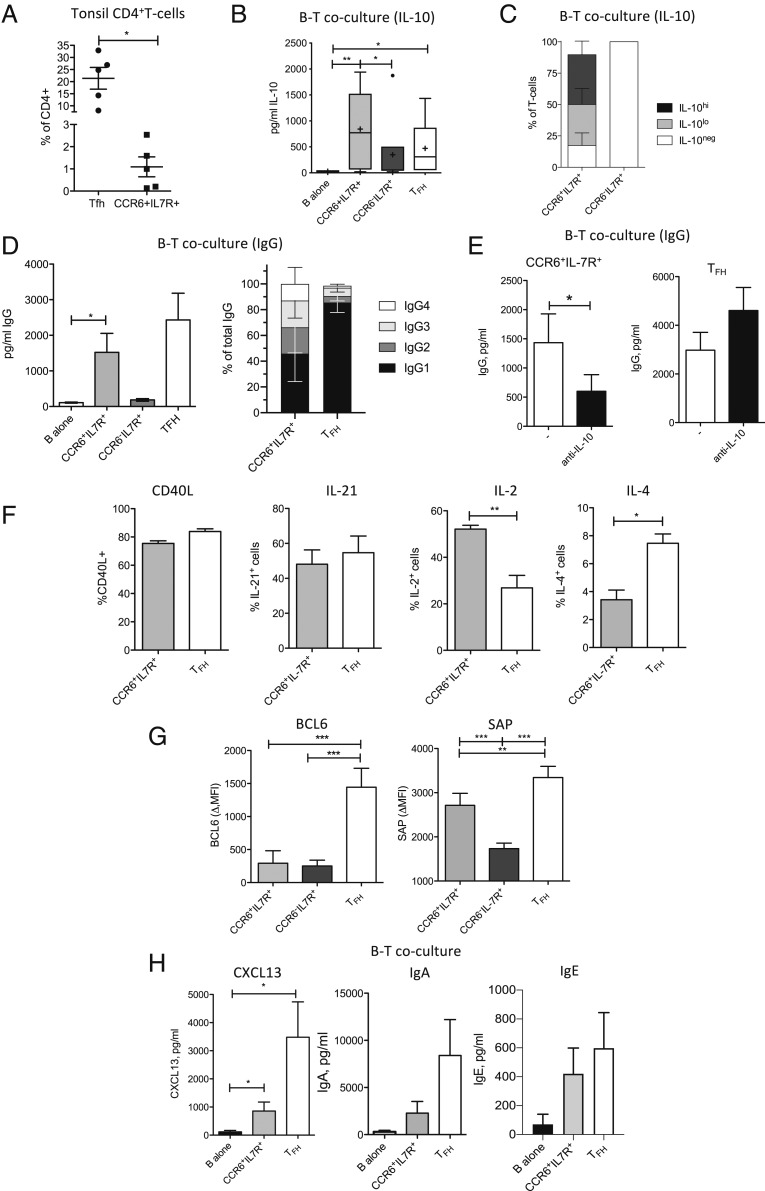
CCR6+IL-7R+T Cells from Human Tonsils Produce IL-10 upon Stimulation with Naïve B Cells.
CCR6+IL-7R+CD25−/loT cells (abbreviated CCR6+IL-7R+) could be identified at low frequencies in human tonsils (Fig. 1A and SI Appendix, Fig. S1A). They were phenotypically distinct from CXCR5+ICOS+TFH cells, since the latter were IL-7R−/lo (38) and lacked CCR6 expression (SI Appendix, Fig. S1B). Moreover, CXCR5- and ICOS-coexpressing cells were hardly detectable among CCR6+IL-7R+T cells (SI Appendix, Fig. S1C). The few tonsillar T cells that coexpressed CCR6, CXCR5, and ICOS were enriched for FOXP3+ cells (SI Appendix, Fig. S1D), indicating that they contained follicular regulatory T cells, and were excluded from further analysis.
Fig. 1.
Human tonsillar CCR6+IL-7R+T cells are distinct from TFH cells and promote B cell responses via IL-10. (A) Frequencies of CXCR5+ICOS+TFH cells and CCR6+IL-7RhiCD25lo/− T cells in human tonsils (n = 5). (B) IL-10 production in cultures of naïve B cells and SEB in the absence or presence of tonsillar CCR6−CXCR5+ICOS+ TFH, CD4+CCR6−IL-7R+, and CCR6+IL-7R+T cell subsets was assessed after 8 d by ELISA (n > 5). (C) Immunofluorescence analysis of IL-10 expression in cocultures of circulating CCR6−IL-7R+T cells or CCR6+IL-7R+T cells and naïve B cells (n = 3). (D) IgG production in tonsillar T–B coculture supernatants was measured by ELISA (n > 5). (D, Left) Concentrations of total IgG (day 8). (D, Right) Relative contributions of IgG1 to 4 to total IgG (day 10). (E) IgG concentrations in cocultures of naïve B cells and tonsillar CD4+CCR6−IL-7R+T cells (n = 10) or TFH cells (n = 5) in the absence or presence of IL-10 neutralization (“anti–IL-10”). (F) CD40L up-regulation (n = 3) and production of IL-21 (n = 6), IL-2 (n = 4), and IL-4 (n = 6) by purified tonsillar CD4+CCR6−IL-7R+T cells or TFH cells following brief PMA and ionomycin stimulation. (G) BCL6 and SAP expression was analyzed in the indicated tonsillar T cell subsets by intracellular staining ex vivo (n = 5). Mean fluorescence intensity is shown; the isotype control was subtracted (ΔMFI). (H) CXCL13 (Left) and IgA (Middle) concentrations in day 6 and IgE concentrations in day 10 culture supernatants (Right) of naïve B cells alone or together with autologous tonsillar CCR6+IL-7R+ or TFH cells and SEB (n = 5). *P < 0.05, **P < 0.005, and ***P < 0.0005 are statistically significant. Error bars show SEM. n indicates the number of analyzed patients; a maximum of two patients were analyzed in the same experiments.
As previously reported for CCR6+T cells from peripheral blood (37), also tonsillar CCR6+IL-7R+T cells could produce IL-10 after suboptimal stimulation with anti-CD3 antibodies, whereas tonsillar CCR6−IL-7R+ control T cells required CD28 costimulation (SI Appendix, Fig. S1E). We analyzed here if CCR6+IL-7R+T cells could produce IL-10 upon TCR activation by naïve B cells, which express only low levels of MHC class II and costimulatory molecules (39) and provide thus also only low levels of T cell stimulation. IL-10 was indeed detected in coculture supernatants of superantigen-loaded naïve B cells and tonsillar CCR6+IL-7R+T cells or TFH cells, but was low or undetectable in control cocultures with CCR6−IL-7R+T cells (Fig. 1B). Similar results were obtained with CCR6+IL-7R+T cells from peripheral blood (SI Appendix, Fig. S1F). Notably, IL-10 protein in B–T cocultures could be detected by immunofluorescence in a major fraction of CCR6+IL-7R+T cells, but not in CCR6−IL-7R+ control T cells (Fig. 1C and SI Appendix, Fig. S1G).
In summary, CCR6+IL-7R+T cells were present in human tonsils, were distinct from TFH cells, and produced IL-10 upon stimulation by naïve B cells.
IL-10 Produced by Tonsillar CCR6+IL-7R+T Cells Promotes B Cell Responses.
Tonsillar CCR6+IL-7R+T cells induced naïve B cells to produce immunoglobulin G (IgG) (Fig. 1D), indicating that they possessed B helper functions. TFH cells induced IgG production as expected very efficiently, whereas CCR6−IL-7R+ control T cells failed to do so (Fig. 1D). Interestingly, while TFH cells consistently induced mainly IgG1, the effect of CCR6+IL-7R+T cells on IgG isotypes was more variable, and IgG1 contributed on average <50% of total IgG (Fig. 1D). Importantly, IgG induction by CCR6+IL-7R+T cells was significantly reduced when IL-10 was neutralized (Fig. 1E). Conversely, IgG induction by TFH cells increased when IL-10 was neutralized (Fig. 1E), consistent with a regulatory role of IL-10 produced by TFH cells (11). CCR6+IL-7R+T cells and TFH cells expressed similar levels of IL-10R (SI Appendix, Fig. S1H), suggesting that different T cell sensitivities for IL-10 were unlikely to play a role. Both tonsillar CCR6+IL-7R+T cells and TFH cells efficiently up-regulated CD40L and produced high amounts of IL-21 (Fig. 1F), consistent with their B helper functions. CCR6+IL-7R+T cells and TFH cells produced, however, different levels of IL-2 and IL-4 (Fig. 1F), which also stimulate B cells. Importantly, only TFH cells expressed high levels of BCL6, but both subsets expressed elevated levels of the intracellular adaptor SAP (Fig. 1G), which promotes stable interactions of T and B cells (40). CCR6+IL-7R+T cells produced also lower amounts of the B cell-attracting chemokine CXCL13 than TFH cells, and were inefficient in inducing IgA (Fig. 1H). However, both subsets induced some IgE (Fig. 1H).
In conclusion, although tonsillar CCR6+IL-7R+T cells and TFH cells both possess B helper functions, they have nevertheless different phenotypes and characteristics. In particular, only IL-10 produced by CCR6+IL-7R+T cells promoted B cell IgG production.
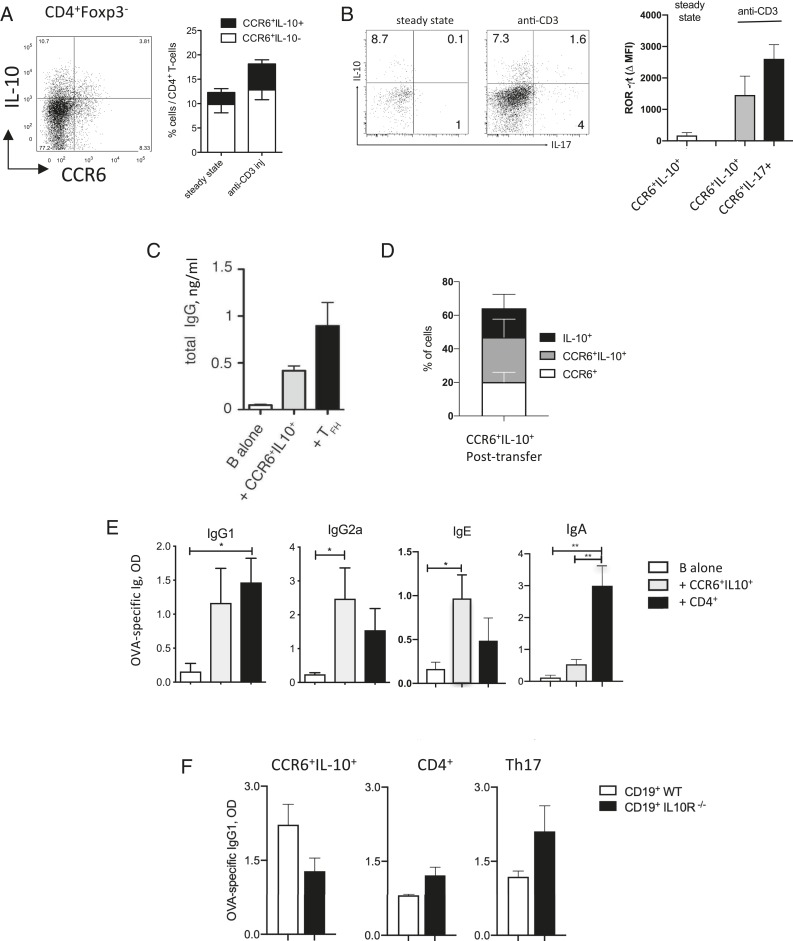
Mouse CCR6+IL-10+T Cells Provide B Cell Help In Vivo.
We next analyzed if cells with the same characteristics were also present in mice. IL-10–secreting CCR6+T cells could indeed be detected in the spleen of IL-10eGFPFoxp3RFP double-reporter mice (Fig. 2A). Analysis of naïve IL-10eGFPxFoxp3RFPxIL-17AKata triple-reporter mice revealed that IL-10eGFP+CCR6+T cells did not produce IL-17 in the steady state (Fig. 2B), indicating that they were distinct from the previously described natural Th17 cells (24, 41). Consistently, RORγt expression in IL-10eGFP+CCR6+ T cells was low in the steady state. However, RORγt increased rapidly upon anti-CD3 injections, a treatment that also induced IL-17 production (Fig. 2B).
Fig. 2.
Murine splenic IL-10+CCR6+T cells provide B cell help in vivo. (A) IL-10 and CCR6 expression by CD4+T cells in the spleen of IL-10/FOXP3 double-reporter FIRxTiger mice. (A, Left) Representative dot plot (steady state). Numbers in quadrants indicate percentages of cells. (A, Right) Mean percentages in the steady state or following anti-CD3 injections (n = 4). (B) IL-17A and IL-10 expression by CCR6+T cells in the spleen of IL-10, FOXP3, and IL-17 triple-reporter mice in the steady state or following anti-CD3 injections. (B, Left) Dot plots show representative IL-10eGFP and IL-17Kata expression. (B, Right) Histograms show mean intracellular ROR-γt expression by CD4+CCR6+IL-10+ or CCR6+IL-10−IL-17+T cells in the spleen (n = 3). (C) In vitro B cell help for IgG production by splenic TFH cells and CCR6+IL-10+T cells isolated from anti-CD3–injected FIRxTiger mice (n = 3). (D) Percentages of CCR6 and/or IL-10eGFP expression in CCR6+IL-10+T cells 12 d after transfer into Rag−/− mice (n = 6). (E) Rag−/− mice were reconstituted with purified CD19+B cells and with either CCR6+IL-10+ or CD4+ control T cells isolated from steady-state spleens of OTIIxFIRxTiger mice. OVA-specific IgG1, IgG2, IgE, and IgA were measured in sera of mice 12 d post vaccination (n = 3). (F) Rag−/− mice were reconstituted with B cells from wild-type or IL-10RDN mice and with either splenic CCR6+IL-10+T cells, total CD4+T cells, or in vitro induced Th17 cells, all isolated from OTIIxFIRxTiger mice. Ova-specific IgG1 production was measured in sera of mice 12 d post vaccination (n = 3). *P < 0.05 and **P < 0.005 are statistically significant. Error bars show SEM. n indicates the number of mice analyzed in independent experiments.
We then investigated whether IL-10–producing CCR6+T cells in mice could provide B cell help. Purified CD4+CCR6+IL-10eGFP+T cells as well as CXCR5+PD1+TFH cells induced by anti-CD3 injections in spleens of IL-10eGFPxFoxp3RFP double-reporter mice up-regulated CD40L expression (SI Appendix, Fig. S2A) and induced B cells to secrete IgG in vitro (Fig. 2C). In order to understand if IL-10–producing CCR6+T cells also provided B cell help in vivo, we reconstituted T cell- and B cell-deficient mice with B cells together with purified splenic CD4+T cells from OTIIxIL-10eGFPxFoxp3RFP double-reporter TCR transgenic mice and immunized with ovalbumin (OVA). Notably, the majority of CCR6+IL-10eGFP+T cells maintained IL-10 reporter and/or CCR6 expression following transfer to Rag−/− mice (Fig. 2D). Importantly, CCR6+IL-10eGFP+T cells isolated from untreated mice induced OVA-specific IgG1, IgG2a, and IgE upon cotransfer with B cells (Fig. 2E) but induced only low levels of IgA, similar to CCR6+IL-7R+T cells from human tonsils (Fig. 1H). Moreover, in vivo help by CCR6+IL-10eGFP+T cells was partially IL-10–dependent, since they induced higher amounts of IgG1 and IgG2A from wild-type B cells than from B cells that lacked a functional IL-10R (Fig. 2F and SI Appendix, Fig. S2B). In contrast, CD4+ control T cells and in vitro differentiated Th17 cells induced IgG in IL-10R–deficient B cells as efficiently as in wild-type B cells (Fig. 2E and SI Appendix, Fig. S2B). Interestingly, however, impaired IL-10R signaling in B cells resulted in all cases in lower serum levels of IgA (SI Appendix, Fig. S2B).
We conclude that IL-10–producing CCR6+T cells are present in the spleens of naïve mice. They are distinct from natural Th17 cells and promote B cell IgG production in a partially IL-10–dependent manner in vivo, and thus have similar features as human tonsillar CCR6+IL-7R+T cells.
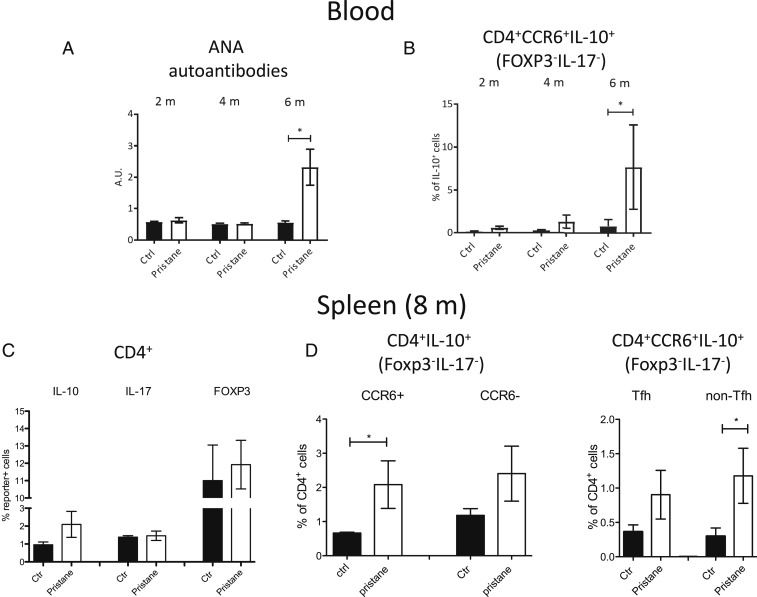
CCR6+IL-10+T Cells Accumulate Systemically in Mice with Lupus-Like Disease.
Since B helper T cells and IL-10 contribute to aberrant B cell responses in lupus, we wondered if IL-10–producing CCR6+T cells were involved. We therefore analyzed IL-17xIL-10xFoxp3 triple-reporter mice that developed lupus-like disease following injection of pristane (42). Anti-nuclear autoantibodies accumulated in the serum after 6 mo after pristane injection (Fig. 3A). Importantly, we also observed a progressive appearance of CCR6+T cells that expressed IL-10, but not IL-17 or FOXP3, in the circulation of mice with lupus-like disease, and these CCR6+IL-10+T cells were significantly increased after 6 mo (Fig. 3B). After 8 mo, splenic CD4+T cells from mice with lupus-like disease had largely unaltered expression of CCR6, IL-17, and Foxp3, whereas IL-10eGFP+ cells were moderately increased (Fig. 3C). Notably, IL-10 was significantly up-regulated among splenic CCR6+T cells that lacked Foxp3 and IL-17 reporter expression (Fig. 3D), and there was virtually no coexpression of IL-17 and IL-10 (SI Appendix, Fig. S2C). Interestingly, the increase of IL-10 production among CCR6+T cells was most prominent among cells with a non-TFH phenotype (Fig. 3D and SI Appendix, Fig. S2D). IL-10 expression increased also in CCR6−T cells and CXCR5+PD1+TFH cells, but these increases were less marked and did not reach statistical significance (Fig. 3D).
Fig. 3.
Murine IL-10+CCR6+T cells accumulate systemically in lupus-like disease. IL-10xIL-17xFOXP3 triple-reporter mice were injected with pristane (n = 5) or left untreated (n = 5) and analyzed in parallel. (A) Anti-nuclear autoantibody (ANA) concentrations were measured in the serum of mice after 2, 4, and 6 mo by ELISA. (B) Kinetics of CD4+CCR6+IL-10+FOXP3−IL-17−T cell accumulation in the peripheral blood of the same mice. (C) Mean percentages of IL-10, IL-17, and FOXP3 reporter expression in splenic CD4+T cells after 8 mo in pristane-injected and noninjected control mice. (D) Frequencies of IL-10eGFP+ cells among CD4+FOXP3−IL-17−T cells according to CCR6 expression (Left) and in the CCR6+ fraction according to TFH phenotype (CXCR5+PD1+; Right) in pristane-injected and noninjected control mice. *P < 0.05 is statistically significant. Error bars show SEM.
We conclude that CCR6+IL-10+ helper T cells expand systemically upon development of autoantibodies in a model of lupus-like disease.
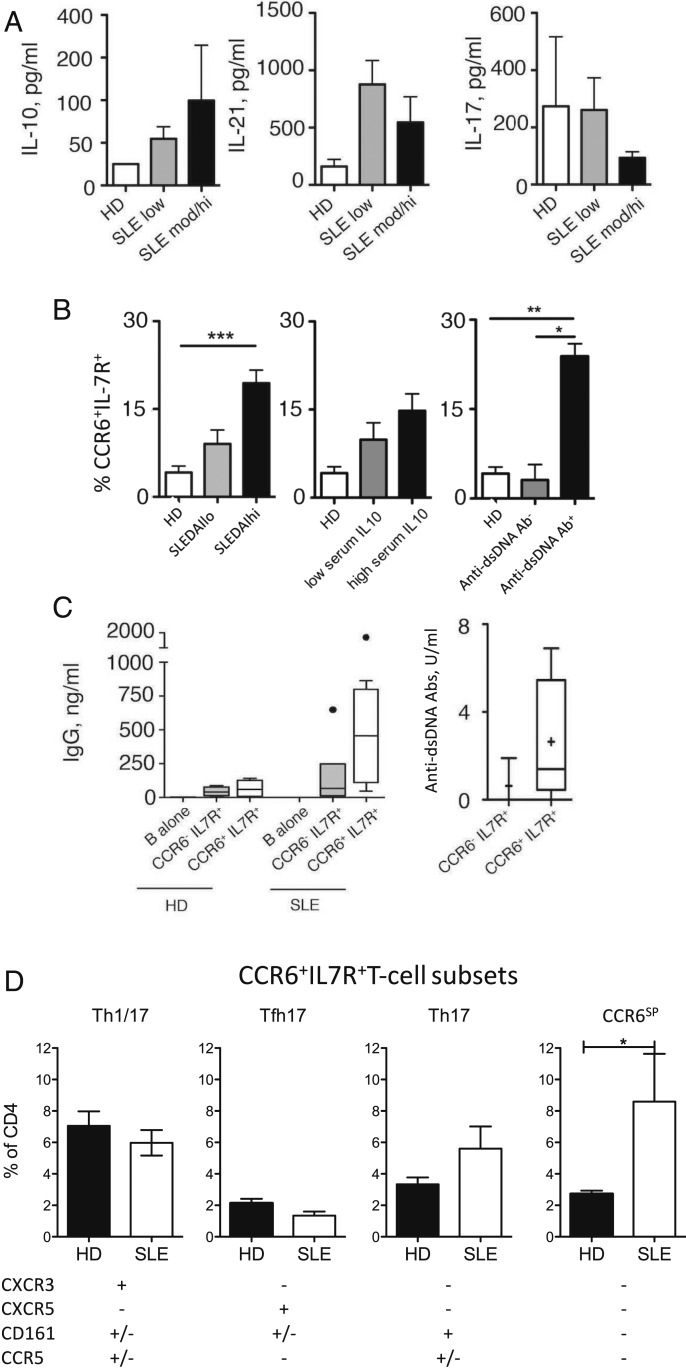
CCR6+IL-7R+T Cells Are Selectively Increased in SLE Patients and Spontaneously Induce Pathogenic Autoantibodies.
We next analyzed whether CCR6+IL-7R+T cells were involved in human SLE. Consistent with previous reports, we detected increased serum levels of IL-10 in a cohort of SLE patients (SI Appendix, Table S1), in particular in those with a higher disease score (Fig. 4A). Conversely, serum IL-17 was not associated with SLE, while IL-21 serum levels were on average highest in SLE patients with a low disease score (Fig. 4A). Importantly, CCR6+IL-7R+T cells were increased in the circulation of SLE patients, and the increase was again more prominent in patients with a higher disease score or with higher IL-10 serum levels (Fig. 4B). Notably, coculture supernatants of B cells and CCR6+IL-7R+T cells from SLE patients contained as expected IL-10 (SI Appendix, Fig. S3A). Strikingly, the strongest association of CCR6+IL-7R+T cells was found with the presence of anti-dsDNA autoantibodies (Fig. 4B). Most important, CCR6+IL-7R+T cells of SLE patients induced spontaneously, that is, in the absence of exogenous TCR agonists, IgG production from autologous B cells (Fig. 4C). Conversely, CCR6−IL-7R+ control T cells from SLE patients and CCR6+IL-7R+T cells from healthy individuals were inefficient. Moreover, we could detect anti-dsDNA antibodies among those spontaneously IgG induced by CCR6+IL-7R+T cells, whereas these pathogenic autoantibodies were lower or undetectable in control supernatants with CCR6−IL-7R+ control T cells from the same patients (Fig. 4C).
Fig. 4.
CCR6+IL-7R+T cells provide help for autoantibody production and are associated with anti-dsDNA autoantibodies in SLE patients. (A) Serum levels of IL-10, IL-17, and IL-21 in healthy donors (HDs) (n = 6) and SLE patients (n = 25) according to disease scores (SLEDAIhi: >5). (B) Frequencies of CCR6+T cells among CD4+IL-7R+T cells in peripheral blood of healthy donors (HDs; n = 16) or SLE patients (n = 20) that were stratified according to the SLEDAI disease score (Left), IL-10 serum levels (Middle), or the presence of anti-dsDNA antibodies (Right). (C) B cells from HDs (n = 4) and SLE patients (n = 8) were cultured with CCR6+IL-7R+ or CCR6−IL-7R+T cells in the absence of exogenous antigens for 14 d and secretion of total IgG (Left) or of anti-DNA autoantibodies in SLE patients (Right) was measured by ELISA and DELFIA, respectively. (D) Frequencies of CCR6+IL-7R+T cell subsets (CXCR3+Th1/17, CXCR5+TFH17, CD161+TH17, and CXCR3−CCR5−CXCR5−CD161−CCR6SPT cells; gating strategy is shown in SI Appendix, Fig. S3B) in peripheral blood of HDs (n = 14) and SLE patients (n = 9). *P < 0.05, **P < 0.005, and ***P < 0.0005 are statistically significant. Error bars show SEM.
Although total CCR6−IL-7R+T cells increased in the circulation of SLE patients, we could not detect any significant increase of previously described subsets of CCR6+IL-7R+T cells, namely of CXCR3+CCR5+/−“Th1/17” cells (43), CXCR5+“TFH17” cells (20), and CD161+CXCR3−Th17 cells (23) (Fig. 4D). Conversely, a previously undescribed population of CCR6+IL-7R+T cells, which lacked all these subset-defining markers (CCR6 “single-positive” cells; CCR6SP; see the gating strategy in SI Appendix, Fig. S3B), was strongly and significantly expanded in SLE patients (Fig. 4D). These CCR6SPT cells possessed only low levels of IL-17–producing capacities (SI Appendix, Fig. S3C), but secreted high levels of IL-10 in cocultures with naïve B cells and induced IgG in a partially IL-10–dependent manner (SI Appendix, Fig. S3 D and E). The expansion of CCR6SPT cells in SLE patients was not only selective among CCR6+IL-7R+T cells but we were also unable to confirm previously reported increases of CCR6−CXCR5+TFH2 (44) and of PD1+CXCR5−CXCR3+Th1 cells (35) (SI Appendix, Fig. S3F).
In conclusion, a previously uncharacterized population of CCR6+IL-7R+T cells accumulates in the blood of SLE patients harboring anti-dsDNA autoantibodies, and may thus contribute to the aberrant production of IL-10 and autoantibodies.
CCR6SPIL-7R+T Cells Lack Signature Genes of TFH and Th17 Cells.
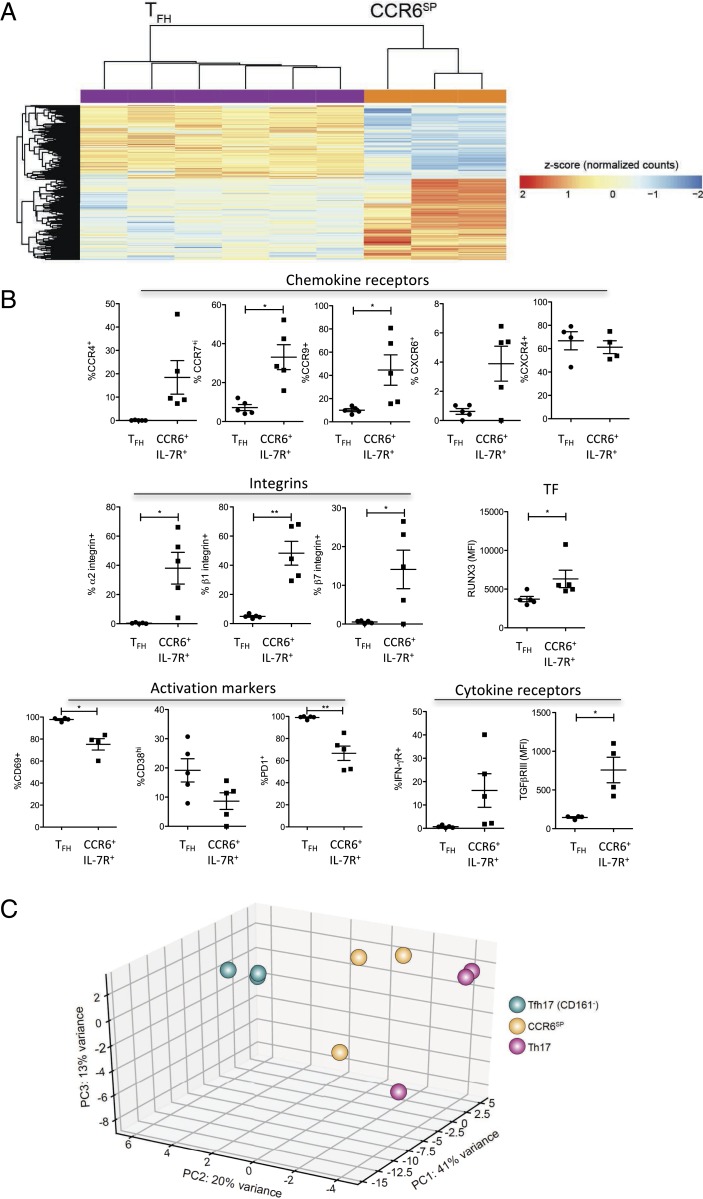
To investigate the biology of CCR6SPIL-7R+T cells, we performed transcriptome analysis by RNA sequencing. Circulating CCR6SPIL-7R+T cells were compared with CD161+CXCR5−Th17 cells and with CD161−CXCR5+TFH17-like cells (20) from the same donors (SI Appendix, Fig. S3B), and with canonical CXCR5+ICOS+TFH cells from tonsils (45) (SI Appendix, Fig. S1A), in order to survey possible groupings through the application of unsupervised approach by principal-component analysis (PCA). The visualization of the cells in the space of the first three principal components showed that the circulating CCR6SPIL-7R+T cells, Th17 cells, and, to a lesser degree, TFH17-like cells were clearly separated from tonsillar TFH cells (SI Appendix, Fig. S4A). We therefore compared CCR6SPIL-7R+T cells with tonsillar TFH cells to identify candidate genes reflecting the distinctive profiles, and then validated genes of interest in tonsils. A large number of differentially expressed genes were identified in CCR6SPIL-7R+T cells as compared with CXCR5+ICOS+TFH (Fig. 5A). A canonical TFH signature, including CXCR5, ICOS, BCL6, IL-21, and PD1, was as expected up-regulated in TFH cells. Conversely, several chemokine receptors, integrins, cytokine receptors, and transcription factors were significantly up-regulated in CCR6SPIL-7R+T cells (Dataset S1). We validated a panel of genes of interest at the protein level by flow cytometry. CCR4, CCR7, CCR9, α2-, β1-, and β7-integrins, IFN-γR1, TGF-βR3, and RUNX3 proteins were indeed expressed at higher levels in tonsillar CCR6+IL-7R+T cells, while the activation-associated surface receptors CD38, CD69, and PD1 were higher on tonsillar TFH cells (Fig. 5B). As control, we analyzed expression of CXCR4, which was not differentially expressed according to RNA sequencing, and found that also surface CXCR4 protein expression levels were similar.
Fig. 5.
CCR6SPT cells lack canonical TFH and Th17 gene signatures. Th17 cells, CCR6SPT cells, and CD161−TFH17-like cells from the peripheral blood of three donors were isolated and RNA was extracted and sequenced. Differential expression of genes was analyzed in CCR6SPT cells as compared with Th17 cells, TFH17-like cells, or tonsillar TFH cells. (A) Heatmap of differentially expressed genes in circulating CCR6SPT cells (n = 3) compared with tonsillar TFH (n = 6). A detailed list of genes is reported in Dataset S1. (B) Validation of selected candidate genes identified in A in tonsillar CCR6+IL-7R+T cells and TFH cells at the protein level by flow cytometry (n = 5). Reported are the percentages of cells expressing five chemokine receptors, three integrins, two cytokine receptors, three activation markers, and one transcription factor (TF; expressed in MFI). (C) Principal-component analysis of Th17 cells, CCR6SPT cells, and CD161−TFH17-like cells. *P < 0.05 and **P < 0.005 are statistically significant.
PCA of the three circulating CCR6+IL-7R+T cell subsets revealed in addition that CCR6SPIL-7R+T cells were localized in between TFH17-like and Th17 cells (Fig. 5C). Four hundred and sixty two genes were differentially expressed in CCR6SPIL-7R+T cells compared with TFH17-like cells (SI Appendix, Fig. S4B and Dataset S2). Notably, among these genes, RAR-γ, HOPX, SOCS2, SOX8, and TGF-βR3 were also up-regulated in CCR6SPIL-7R+T cells as compared with tonsillar TFH cells (Dataset S1). When CCR6SPIL-7R+T cells were compared with Th17 cells, 292 differentially expressed genes were identified (SI Appendix, Fig. S4C). A canonical signature of human Th17 cells, comprising RORC, IL-1R1, and IL-23R, as well as IL-4i1 (46) and musculin (47), was up-regulated in Th17 cells (Dataset S3). Interestingly, CCR6SPIL-7R+T cells expressed instead increased levels of the stemness-associated transcription factor LEF1 and the WNT agonist WNT10A (Dataset S3).
Overall, these results indicate that CCR6SPIL-7R+T cells lack the effector gene signature of TFH and Th17 cells but have instead up-regulated genes involved in migration and stemness.
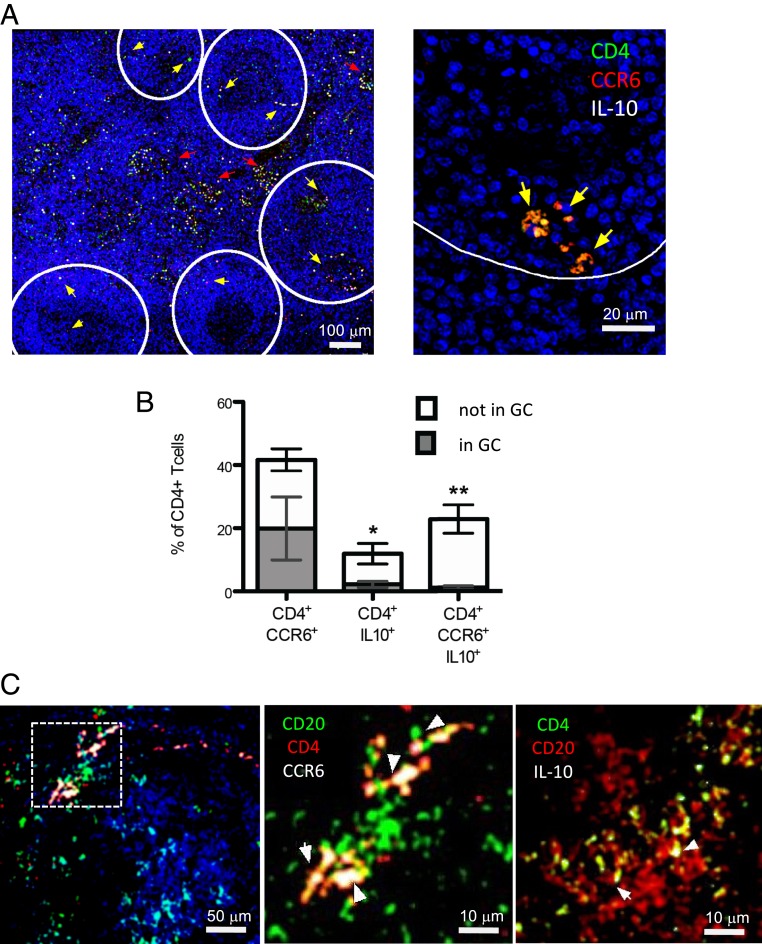
CCR6+IL-10+T Cells Are Abundant in Lymph Nodes of SLE Patients and Interact with B Cells at the Margins of Follicles.
Since B–T interactions occur in secondary lymphoid organs, we finally assessed if CCR6+IL-10+T cells were present in lymph nodes of SLE patients and if they were interacting with B cells. CD4+T cells that coexpressed CCR6 and IL-10 could be clearly detected in all lymph node sections from eight different SLE patients (Fig. 6A). Strikingly, on average, roughly 25% of CD4+T cells coexpressed CCR6 and IL-10 in situ (Fig. 6B). Notably, the majority of IL-10–producing CD4+T cells expressed CCR6, and a major fraction of CCR6+T cells produced IL-10 (Fig. 5B). Conversely, in control lymph nodes, IL-10 was hardly detectable in CCR6+CD4+T cells (SI Appendix, Fig. S5). Importantly, CD4+CCR6+IL-10+T cells in lymph nodes of SLE patients were excluded from germinal centers (Fig. 6 A and B). Nevertheless, we could detect several CD4+T cells that expressed CCR6 or IL-10 in contact with CD20+B cells at the margins of follicles (Fig. 6C).
Fig. 6.
IL-10–producing CCR6+T cells are abundant in lymph nodes of SLE patients and interact with B cells outside of germinal centers. (A, Left) Confocal images of a representative lymph node from lupus patients with scattered CD4+CCR6+IL-10+T cells (indicated by yellow arrows). Follicles are indicated by a white line. (A, Right) Magnification shows triple-positive cells at the boundary of a follicle. (B) Mean percentages of CD4+T cells that express CCR6 and IL-10 alone or in combination inside (gray) and outside (white) of germinal centers (GCs) (n = 8). *P < 0.05 and **P < 0.005 indicate significant enrichment outside of germinal centers. Error bars show SEM. (C) Representative images showing CD20+B cells in close proximity to CCR6+CD4+ (Left and Middle) or IL-10+CD4+T cells (Right).
These findings illustrate that CCR6+IL-10+T cells are highly abundant in lymph nodes of SLE patients and some could interact with B cells, but these interactions occurred rather exclusively outside of germinal centers.
Discussion
IL-10 has a dual role in immune responses, since it inhibits T cell activation but stimulates B cells (32). It is also produced by helper T cells and has a pathogenic role in SLE (28–30), presumably because it stimulates autoreactive B cells (32). Here, we characterized a population of extrafollicular B helper T cells that produced IL-10 and induced autoantibodies, and are thus likely to play a prominent pathogenic role in SLE.
We previously reported that human CCR6+ memory T cells had a peculiar regulation of IL-10, since they produce only little IL-10 upon standard stimulation but could nevertheless produce suppressive IL-10 in response to suboptimal TCR stimulation provided by immature DCs (37). However, optimal stimulation or vaccination antigens like tetanus toxoid, which induces protective antibody responses, induced CD40L and IL-2 in addition. We therefore speculated that CCR6+ memory T cells could have a context-dependent regulatory/helper function and provide help in recall responses (37). Here, we demonstrated that CCR6+T cells with similar characteristics are present in human lymphoid organs and are able to produce IL-10 upon interaction with naïve B cells, which also provide only suboptimal TCR stimulation (39). Importantly, IL-10 produced by CCR6+IL-7R+T cells was functionally relevant, since it promoted IgG production in humans in vitro and also in mice in vivo. Notably, CCR6+IL-10+T cells maintained high IL-10 production in recipient mice, and the majority also expressed CCR6. The finding that only a fraction of cells coexpressed CCR6 and IL-10 post transfer could be explained by the fact that T cell activation induces IL-10 reporter expression (48) but may transiently down-regulate CCR6 (49). Importantly, we provided evidence that IL-10–producing CCR6+T cells represent a distinct T cell population, because they were not only distinct from IL-10–producing regulatory T cell subsets (3, 4), which lack IL-7R expression and suppress B cell responses (5, 6), but also from other B helper T cells like TFH cells and Th17 cells (19, 22). Indeed, they lacked canonical Th17 and TFH gene signatures, but expressed instead genes promoting migration and stemness, consistent with the view that they represent poorly differentiated, central memory-like cells. We showed that in particular TFH cells had several different features including BCL6 expression, cytokine production, and, importantly, their localization. Thus, CCR6+IL-10+T cells in lymph nodes of SLE patients were in contact with B cells at the margins of follicles, suggesting that they mediate extrafollicular B cell help. Extrafollicular B helper T cells have been described previously in human tonsils and also in murine lupus models (34, 36, 50), and our results are consistent with the view that they are relevant in human SLE. Indeed, the fact that immune-suppressive therapy of SLE patients often results in a selective drop of anti-dsDNA autoantibody titers (26) is consistent with a role for short-lived plasma blasts that could be generated in extrafollicular B cell responses.
An important feature of CCR6+T cells is their increased autoreactivity (37, 43), which suggests that they are particularly relevant in autoimmune diseases. Consistently, they increased systemically in lupus-like disease in mice and in SLE patients harboring anti-dsDNA antibodies, where they provided spontaneous help for autoantibody production. Thus, they are likely to give an important contribution to the aberrant production of IL-10 and anti-dsDNA autoantibodies in SLE patients, suggesting that they play a prominent pathogenic role. It is tempting to speculate that the physiological role of these extrafollicular B helper T cells is complementary to TFH cells, that is, to induce a rapid B cell response of lower quality that provides some initial protection, before the slower germinal response generates high-affinity antibodies that persist long-term.
Previous to this report, other B helper T cell populations were proposed to contribute to the pathogenesis of SLE (33, 35), which is a highly heterogeneous disease that can affect different organs (51). In particular, circulating CXCR5+TFH-like subsets in a subset of patients, and, more recently, PD1+Th1 cells, were reported to be systemically increased in SLE patients. However, while a higher fraction of TFH-like cells displayed an activated PD1+CCR7lo phenotype (52) also in our SLE cohort, the frequencies of TFH-like subsets and of PD1+CXCR3+CXCR5−Th1 cells among peripheral blood CD4+T cells were actually reduced. A caveat of PD1+CXCR3+T cells is further that they expressed Eomes and CCR5, and thus overlap with IL-10– and IFN-γ–producing Tr1-like cells (53, 54), which we showed lack CD40L and helper capabilities but suppress B cell responses (5). In any case, PD1+CXCR3+T cells are distinct from the here-identified B helper T cells, since the latter expressed CCR6, but not CXCR3.
In conclusion, we identified here a previously uncharacterized population of extrafollicular B helper T cells, a finding that broadens our understanding of adaptive humoral immune responses. In addition, this potentially pathogenic T cell population might be exploited as prognostic markers of SLE progression and/or activity, and targeting their B helper functions may be a future therapeutic strategy.
Materials and Methods
Human Samples and Patients.
Buffy-coated blood of healthy donors and tonsil specimens that were surgically removed from pediatric patients were obtained from the Fondazione Istituto di Ricovero e Cura a Carattere Scientifico Ospedale Maggiore Policlinico Milano and the Deutsches Rotes Kreuz. The joint ethical committee of the Institute Gaetano Pini and Ospedale Maggiore Policlinico in Milan, and the committee of the Charité University in Berlin, approved the use of blood and tonsil specimens for research purposes (Permission EA1/107/10), and informed consent was obtained from the subjects involved in the study. Information concerning the identity of the involved subjects was not transmitted to the involved research laboratories. Peripheral blood from patients with SLE was obtained from Institute Gaetano Pini. All patients fulfilled the American College of Rheumatology criteria for the classification of SLE, and disease activity was assessed by Systemic Lupus Erythematosus Disease Activity Index 2000 (SLEDAI-2K) (55). Disease activity was ranked according to SLEDAI-2K score: no activity (SLEDAI score 0), low/mild activity (SLEDAI score 1 to 5), moderate activity (SLEDAI score 6 to 10), and high activity (SLEDAI score >11) (SI Appendix, Table S1).
Mice.
C57BL/6Rag1−/−, C57BL/6 CD45.1+, and OTII mice were purchased from The Jackson Laboratory. Foxp3 reporter mice [FIR mice (56)] and IL-10eGFP mice [Tiger mice (48)] were crossed and generated at Yale University. FIRxTiger mice were crossed with OTII mice at Yale University to generate OTIIxFIRxTiger mice. TigerKata IL-10xIL-17A reporter mice and IL-10RDN mice were generated and crossed at Yale University. Age- and sex-matched littermates between 8 and 12 wk of age were used. Animal procedures were approved by the Institutional Animal Care and Use Committee of Yale University.
Flow Cytometry.
Human lymphocytes were stained with antibodies purchased from BD (anti-CD4 [RPA-T4], -CD27 [M-T271], -CD25 [M-A251], -BCL6 [K112-91], -CCR6 [11A9], -CD40L [TRAP1], –IL-10 [JES-19F1], –IL-17 [N49-653], –IL-4 [MP4-25D2], -CD161 [DX12], -CCR5 [3A9], and –β7-integrin [FIB504]), BioLegend (anti-CD210/IL-10R1 [3F9], –β1-integrin [TS2/16], –α2-integrin [P1E6-C5], -CCR4 [L291H4], –IL-2 [MQ1-17H12], -CD69 [FN50], -PD1 [EH12.2H7], and –CCR9 [L053E8]), eBioscience (anti-CD19 [HIB19], –IL-7R [RDR5], -CD25 [BC96], -ICOS [ISA-3], -FOXP3 [PCH101], –IL-21 [3A3-N2], -SAP [XLP-1D12], and -CCR7 [3D12]), and R&D Systems (anti-CXCR5 [51505], -CXCR4 [44717], -CXCR6 [56811], -RUNX3 [527327], and –TGF-βR3 [242]).
Murine lymphocytes were stained with anti-CD4 (GK1.5), anti-CCR6 (29-2I17), anti-TCR (H57-597), anti-CD278 (15F9), anti-CD127 (A7R34), anti-CD154 (MR1), anti-CD19 (6D5), and anti-PD1 (RMP1-30), all purchased from BioLegend. Anti-RORC (B2D) was obtained from eBioscience. Data were analyzed using FlowJo software (TreeStar).
Cell Isolation.
Peripheral blood mononuclear cells (PBMCs) and tonsillar mononuclear cells were isolated by Ficoll-Hypaque gradient (Sigma-Aldrich). CD4+T cell populations were purified by cell sorting on a FACSAria (BD) based on expression of IL-7R, CD25, and CCR6 to a purity of >95%. Subsets of IL-7R+CCR6+CD25− cells were isolated as CXCR3−CD161+/CCR5+Th17, CXCR5+CXCR3−CD161+/−TFH17, and CXCR3−CCR5−CD161−CXCR5−CCR6SP cells. Tonsillar TFH cells were purified by cell sorting based on the expression of CD4, CXCR5, ICOS, and CCR6. Naïve tonsillar B cells were sorted as CD19+CD27− cells.
RNA Isolation and RNA Sequencing.
RNA was isolated using the mirVana Isolation Kit (Ambion). Residual contaminating genomic DNA was removed from the total RNA fraction using Turbo DNA-free (Thermo Fisher). The RNA yields were quantified using the QuantiFluor RNA System (Promega) and RNA quality was assessed by 2100 Bioanalyzer (Agilent). Library construction was performed with Illumina TruSeq RNA Sample Preparation Kit v2 using from 30 to 100 ng of total RNA as starting material. Final libraries were sequenced on an Illumina HiSeq with paired-end 2 × 150-bp read lengths.
RNA-Sequencing Data Analysis.
Quality control of raw reads was performed with FastQC v0.11.2. Adapter removal and trimming were performed using Trimmomatic. Reads were aligned to the reference genome (GRCh38) using STAR 2.5.2b. The output computed (raw read counts) was then used as input for DESeq2 analysis. Subsequent analyses were performed using R (3.4.2). Raw counts were normalized using DESeq2’s function “rlog” and normalized counts were used to perform PCA. To estimate differential expression, the matrix of gene counts was analyzed by DESeq2. Up-regulated/down-regulated genes were selected setting a false discovery rate lower than 5%. RNA-sequencing data reported in this paper have been deposited at https://www.ebi.ac.uk/ena/data/view/PRJEB15005 (tonsillar TFH cells) and https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-8862/ (Th17 cells, TFH-like subsets, and CCR6SPT cells from peripheral blood).
T Cell Cytokine Production.
Intracellular cytokines were detected after stimulating cells for 6 h with 0.1 μM PMA (phorbol myristate acetate) and 1 μg/mL ionomycin (Sigma-Aldrich). Brefeldin A (10 μg/mL; Sigma) was added for the last 2 h of culture. Cells were fixed with 1 to 2% paraformaldehyde and permeabilized with 0.5% saponin, and nonspecific binding was blocked with 10% FCS (fetal calf serum) or 0.5% BSA (bovine serum albumin). Cells were stained with labeled antibodies for cytokines, washed, and analyzed by flow cytometry.
Polyclonal In Vivo Activation of Murine T Cells.
Mice were injected with anti-CD3 (15 μg; 145-2C11) isotype antibody or PBS (phosphate-buffered saline) i.p. (intraperitoneally) two times every other day (53). Splenocytes were isolated 3 h after the last injection according to a standard protocol.
B Helper Assays.
Human or mouse naïve B cells (3 × 104) were cocultured with 104 sorted autologous CD4+T cell subsets in the presence of 1 µg/mL endotoxin-reduced staphylococcal enterotoxin B (Sigma) in RPMI, 5% low-IgG serum. Neutralizing antibodies to IL-10 (Miltenyi) and IL-10R (R&D Systems) were used at a concentration of 10 μg/mL. CXCL13 (BioSupply), IL-10, IgG, IgA (BD), IgG subclasses (Invitrogen), and IgE (Arigo Biolaboratories) were measured by ELISA (enzyme-linked immunosorbent assay) in culture supernatants. In order to detect IgE and IgG1 to 4, supernatants were analyzed after 10 d. No IL-10 was detectable in the presence of blocking anti–IL-10 antibodies, indicating that IL-10 was neutralized.
For in vivo B helper assays, Rag−/− mice were reconstituted i.v. with 2 × 106 wild-type or IL-10R−/−CD19+ B cells and 5 × 105 purified OTII T cell populations. CCR6+IL-10+ T cells or CD4+ control T cells were purified from spleens of naïve OTIIxFIRxTiger mice. Th17 cells were induced in vitro with anti-CD3 and anti-CD28 antibodies in the presence of IL-2 and neutralizing anti–IL-4 and anti–IFN-γ antibodies and the addition of IL-6 (10 ng/mL), IL-23 (20 ng/mL), and TGF-β. Twenty-four hours post reconstitution, mice were immunized i.p. with 50 µg OVA per mouse in alum. Serum OVA-specific IgG or IgG1, IgG2, IgE, or IgA (BD) was analyzed 12 d post immunization by ELISA.
Spontaneous human B helper assay and autoantibody production.
CCR6+IL-7R+T cells (5 to 9 × 104) purified from total PBMCs of SLE patients or healthy donors were cocultured with equal numbers of autologous CD20+B cells from healthy subjects or SLE patients in RPMI supplemented with 10% low-IgG serum and in the absence of exogenous TCR agonists. At day 6, cocultures were supplemented with a half-volume of fresh medium and supernatants were harvested at day 14 and tested for total IgG production by ELISA. For the measurement of autoantibody production, supernatants were diluted 1:2 in QUANTA Lite dsDNA SC ELISA Kit (Inova Diagnostics) dilution buffer and incubated in precoated wells of the same kit. The DELFIA System was used for the detection step of autoantibodies. Wells were washed briefly with PBS, 1% BSA, 0.05% Tween 20 and incubated with eurobium-labeled anti-human IgG (PerkinElmer) for 30 min with slow shaking in the dark. Wells were then washed and incubated with DELFIA enhancement solution (PerkinElmer) for 5 min with slow shaking in the dark and fluorescence was acquired with an Infinite 200 microplate reader (Tecan).
Immunofluorescence and imaging.
Paraffin-embedded lymph node (LN) sections of SLE patients classified as “reactive” without other pathology (n = 8) and control slides from healthy donors were obtained from the Academic Medical Center Pathology Department, Amsterdam. Sections were dewaxed by descending alcohol scale and washed in 1× PBS (GIBCO). After permeabilization in 0.02% Triton X-100 (Sigma) in 1× PBS and 1 h blocking in 10% FCS plus 2% goat serum (Thermo Fisher Scientific) in PBS, sections were stained with primary antibodies diluted 1:100, as follows: rabbit anti-human CD4 (1.5 μg/mL; Abcam; ab133616), rat anti-human IL-10 (1 mg/mL; eBioscience; 16-7108-85), and mouse anti-human CCR6 (1 mg/mL; R&D Systems; MAB195) at 4 °C, overnight in a humid chamber. The following day, slides were washed and labeled with Alexa Fluor (AF)-conjugated secondary antibodies diluted 1:1,000, as follows: goat anti-mouse AF 488, goat anti-rat AF 647, and goat anti-rabbit AF 568 (all from Molecular Probes, Thermo Fisher Scientific) for 1 h at room temperature in a humid chamber. After washing in PBS, cells were counterstained with DAPI (Molecular Probes, Thermo Fisher Scientific) and mounted using ProLong Diamond and ProLong Glass mounting reagents (both from Thermo Fisher Scientific). Image acquisition was performed employing an SP5 laser-scanning confocal microscope (Leica Microsystems) equipped with eight laser lines and four PMT detectors, using a 20× air (numerical aperture [NA] 0.95) or a 63× oil (NA 1.40) objective. Sequential multiparametric acquisition was performed on single specific fields of view (FOVs) or on all sections via an automatized stage and tiling software module. Z sectioning was performed in Z-Galvo System-optimized modality with a 130-nm Z-step indexing for better three-dimensional evaluation. Several FOVs were acquired for each sample (ranging from n = 7 for smaller specimens to n = 15 for larger LN samples). Alternatively, a Nikon Ti equipped with a CREST Optics X-Light-V2 spinning disk and VCS superresolution modules (all from CREST Optics) was used to acquire highly resolved images on specific areas of interest. A 16-LED line (PE-40000; CoolLed) or a 6-LED line (SpectraAura; Lumencore) excitation source was used coupled with an Andor DU-888 EM-CCD camera and with an Andor Zyla 5.5 camera for confocal and superresolution acquisition, respectively; a superresolution TIRF 100×SR oil objective (NA 1.49; Nikon Instruments) was employed on the Nikon platform. Image processing, analysis, and measurements were all performed using NIS-Elements v5 software (Lim) using the specialized “general analysis” module to process, binarize, segment, and count all images in the batch. Imaging data were exported as Excel data files and analyzed via GraphPad Prism software. For IL-10 quantification, a mean fluorescence intensity (MFI) of <30 was set according to the MFI of an isotype control as IL-10−. IL-10hi cells were arbitrarily defined as cells with an MFI >80.
For positioning of T cells in germinal centers, an independent analysis has been performed only on those FOVs in which B cell follicles were clearly evident within LN parenchymal areas, employing the DAPI channel. Thus, a segmentation approach was used to specifically recognize follicular areas within the DAPI image channel. Following morphological segmentation, binary criteria and Boolean operations were applied to measure the numbers of CD4+, CCR6+, and IL-10+ cells together with double- (CD4+CCR6+; CD4+IL-10+) and triple-positive (CD4+CCR6+IL-10+) cells within designated follicular areas. At least n = 8 FOVs in each of which n = 3 follicles could be detected were analyzed for all LN specimens.
Statistics.
Statistical significance was calculated using two-tailed Student’s t test or one-way ANOVA. *P < 0.05, **P < 0.005, and ***P < 0.0005 were regarded as statistically significant. Error bars in all figures show SEM.
Supplementary Material
Acknowledgments
We thank K. Stölzel at Charité University Medicine Berlin for tonsillar samples. This work was supported by the Fondazione Cariplo, National Institute of Molecular Genetics “Romeo ed Enrica Invernizzi,” as well as Italian Lupus Foundation, a Long-Term European Molecular Biology Organization (EMBO) Fellowship (to N.G.), and a Short-Term EMBO Fellowship (to F.F.). R.A.F. is supported by the Howard Hughes Medical Institute.
Footnotes
Competing interest statement: J.G. is a consultant for Biotest for anti–IL-10 therapy in SLE patients.
This article is a PNAS Direct Submission.
Data deposition: RNA-sequencing data reported in this paper have been deposited in the ArrayExpress database, https://www.ebi.ac.uk/arrayexpress/ (accession no. E-MTAB-8862) and the European Nucleotide Archive, https://www.ebi.ac.uk/ena/ (accession no. PRJEB15005).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1917834117/-/DCSupplemental.
References
- 1.Moore K. W., de Waal Malefyt R., Coffman R. L., O’Garra A., Interleukin-10 and the interleukin-10 receptor. Annu. Rev. Immunol. 19, 683–765 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Sabat R., et al. , Biology of interleukin-10. Cytokine Growth Factor Rev. 21, 331–344 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Groux H., et al. , A CD4+ T-cell subset inhibits antigen-specific T-cell responses and prevents colitis. Nature 389, 737–742 (1997). [DOI] [PubMed] [Google Scholar]
- 4.Häringer B., Lozza L., Steckel B., Geginat J., Identification and characterization of IL-10/IFN-gamma-producing effector-like T cells with regulatory function in human blood. J. Exp. Med. 206, 1009–1017 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Facciotti F., et al. , IL-10-producing forkhead box protein 3-negative regulatory T cells inhibit B-cell responses and are involved in systemic lupus erythematosus. J. Allergy Clin. Immunol. 137, 318–321.e5 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Okamura T., et al. , TGF-β3-expressing CD4+CD25(−)LAG3+ regulatory T cells control humoral immune responses. Nat. Commun. 6, 6329 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jankovic D., et al. , Conventional T-bet(+)Foxp3(−) Th1 cells are the major source of host-protective regulatory IL-10 during intracellular protozoan infection. J. Exp. Med. 204, 273–283 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anderson C. F., Oukka M., Kuchroo V. J., Sacks D., CD4(+)CD25(−)Foxp3(−) Th1 cells are the source of IL-10-mediated immune suppression in chronic cutaneous leishmaniasis. J. Exp. Med. 204, 285–297 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McGeachy M. J., et al. , TGF-beta and IL-6 drive the production of IL-17 and IL-10 by T cells and restrain T(H)-17 cell-mediated pathology. Nat. Immunol. 8, 1390–1397 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Esplugues E., et al. , Control of TH17 cells occurs in the small intestine. Nature 475, 514–518 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cañete P. F., et al. , Regulatory roles of IL-10-producing human follicular T cells. J. Exp. Med. 216, 1843–1856 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rousset F., et al. , Interleukin 10 is a potent growth and differentiation factor for activated human B lymphocytes. Proc. Natl. Acad. Sci. U.S.A. 89, 1890–1893 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim C. H., et al. , Subspecialization of CXCR5+ T cells: B helper activity is focused in a germinal center-localized subset of CXCR5+ T cells. J. Exp. Med. 193, 1373–1381 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chtanova T., et al. , T follicular helper cells express a distinctive transcriptional profile, reflecting their role as non-Th1/Th2 effector cells that provide help for B cells. J. Immunol. 173, 68–78 (2004). [DOI] [PubMed] [Google Scholar]
- 15.Ma C. S., et al. , Early commitment of naïve human CD4(+) T cells to the T follicular helper (T(FH)) cell lineage is induced by IL-12. Immunol. Cell Biol. 87, 590–600 (2009). [DOI] [PubMed] [Google Scholar]
- 16.Schaerli P., et al. , CXC chemokine receptor 5 expression defines follicular homing T cells with B cell helper function. J. Exp. Med. 192, 1553–1562 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Breitfeld D., et al. , Follicular B helper T cells express CXC chemokine receptor 5, localize to B cell follicles, and support immunoglobulin production. J. Exp. Med. 192, 1545–1552 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rasheed A. U., Rahn H. P., Sallusto F., Lipp M., Müller G., Follicular B helper T cell activity is confined to CXCR5(hi)ICOS(hi) CD4 T cells and is independent of CD57 expression. Eur. J. Immunol. 36, 1892–1903 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Crotty S., Follicular helper CD4 T cells (TFH). Annu. Rev. Immunol. 29, 621–663 (2011). [DOI] [PubMed] [Google Scholar]
- 20.Morita R., et al. , Human blood CXCR5(+)CD4(+) T cells are counterparts of T follicular cells and contain specific subsets that differentially support antibody secretion. Immunity 34, 108–121 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Locci M. et al.; International AIDS Vaccine Initiative Protocol C Principal Investigators , Human circulating PD-1+CXCR3-CXCR5+ memory Tfh cells are highly functional and correlate with broadly neutralizing HIV antibody responses. Immunity 39, 758–769 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mitsdoerffer M., et al. , Proinflammatory T helper type 17 cells are effective B-cell helpers. Proc. Natl. Acad. Sci. U.S.A. 107, 14292–14297 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Annunziato F., et al. , Phenotypic and functional features of human Th17 cells. J. Exp. Med. 204, 1849–1861 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hirota K., et al. , Plasticity of Th17 cells in Peyer’s patches is responsible for the induction of T cell-dependent IgA responses. Nat. Immunol. 14, 372–379 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gualtierotti R., Biggioggero M., Penatti A. E., Meroni P. L., Updating on the pathogenesis of systemic lupus erythematosus. Autoimmun. Rev. 10, 3–7 (2010). [DOI] [PubMed] [Google Scholar]
- 26.Pisetsky D. S., Anti-DNA antibodies—Quintessential biomarkers of SLE. Nat. Rev. Rheumatol. 12, 102–110 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Rönnblom L., Potential role of IFNα in adult lupus. Arthritis Res. Ther. 12, S3 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peng H., et al. , Role of interleukin-10 and interleukin-10 receptor in systemic lupus erythematosus. Clin. Rheumatol. 32, 1255–1266 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Llorente L., et al. , Clinical and biologic effects of anti-interleukin-10 monoclonal antibody administration in systemic lupus erythematosus. Arthritis Rheum. 43, 1790–1800 (2000). [DOI] [PubMed] [Google Scholar]
- 30.Llorente L., et al. , Role of interleukin 10 in the B lymphocyte hyperactivity and autoantibody production of human systemic lupus erythematosus. J. Exp. Med. 181, 839–844 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geginat J., et al. , IL-10 producing regulatory and helper T-cells in systemic lupus erythematosus. Semin. Immunol. 44, 101330 (2019). [DOI] [PubMed] [Google Scholar]
- 32.Geginat J., et al. , The light and the dark sides of interleukin-10 in immune-mediated diseases and cancer. Cytokine Growth Factor Rev. 30, 87–93 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Blanco P., Ueno H., Schmitt N., T follicular helper (Tfh) cells in lupus: Activation and involvement in SLE pathogenesis. Eur. J. Immunol. (2015). [DOI] [PubMed] [Google Scholar]
- 34.Bentebibel S. E., Schmitt N., Banchereau J., Ueno H., Human tonsil B-cell lymphoma 6 (BCL6)-expressing CD4+ T-cell subset specialized for B-cell help outside germinal centers. Proc. Natl. Acad. Sci. U.S.A. 108, E488–E497 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Caielli S., et al. , A CD4+ T cell population expanded in lupus blood provides B cell help through interleukin-10 and succinate. Nat. Med. 25, 75–81 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Odegard J. M., et al. , ICOS-dependent extrafollicular helper T cells elicit IgG production via IL-21 in systemic autoimmunity. J. Exp. Med. 205, 2873–2886 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rivino L., et al. , CCR6 is expressed on an IL-10-producing, autoreactive memory T cell population with context-dependent regulatory function. J. Exp. Med. 207, 565–577 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lim H. W., Kim C. H., Loss of IL-7 receptor alpha on CD4+ T cells defines terminally differentiated B cell-helping effector T cells in a B cell-rich lymphoid tissue. J. Immunol. 179, 7448–7456 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Liu Y. J., et al. , Memory B cells from human tonsils colonize mucosal epithelium and directly present antigen to T cells by rapid up-regulation of B7-1 and B7-2. Immunity 2, 239–248 (1995). [DOI] [PubMed] [Google Scholar]
- 40.Schwartzberg P. L., Mueller K. L., Qi H., Cannons J. L., SLAM receptors and SAP influence lymphocyte interactions, development and function. Nat. Rev. Immunol. 9, 39–46 (2009). [DOI] [PubMed] [Google Scholar]
- 41.Marks B. R., et al. , Thymic self-reactivity selects natural interleukin 17-producing T cells that can regulate peripheral inflammation. Nat. Immunol. 10, 1125–1132 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leiss H., et al. , Pristane-induced lupus as a model of human lupus arthritis: Evolvement of autoantibodies, internal organ and joint inflammation. Lupus 22, 778–792 (2013). [DOI] [PubMed] [Google Scholar]
- 43.Paroni M., et al. , Recognition of viral and self-antigens by TH1 and TH1/TH17 central memory cells in patients with multiple sclerosis reveals distinct roles in immune surveillance and relapses. J. Allergy Clin. Immunol. 140, 797–808 (2017). [DOI] [PubMed] [Google Scholar]
- 44.Le Coz C., et al. , Circulating TFH subset distribution is strongly affected in lupus patients with an active disease. PLoS One 8, e75319 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ripamonti A., et al. , Repression of miR-31 by BCL6 stabilizes the helper function of human follicular helper T cells. Proc. Natl. Acad. Sci. U.S.A. 114, 12797–12802 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Santarlasci V., et al. , IL-4-induced gene 1 maintains high Tob1 expression that contributes to TCR unresponsiveness in human T helper 17 cells. Eur. J. Immunol. 44, 654–661 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Santarlasci V., et al. , Musculin inhibits human T-helper 17 cell response to interleukin 2 by controlling STAT5B activity. Eur. J. Immunol. 47, 1427–1442 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Kamanaka M., et al. , Expression of interleukin-10 in intestinal lymphocytes detected by an interleukin-10 reporter knockin tiger mouse. Immunity 25, 941–952 (2006). [DOI] [PubMed] [Google Scholar]
- 49.Steinfelder S., et al. , Epigenetic modification of the human CCR6 gene is associated with stable CCR6 expression in T cells. Blood 117, 2839–2846 (2011). [DOI] [PubMed] [Google Scholar]
- 50.Lee S. K., et al. , B cell priming for extrafollicular antibody responses requires Bcl-6 expression by T cells. J. Exp. Med. 208, 1377–1388 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seredkina N., Van Der Vlag J., Berden J., Mortensen E., Rekvig O. P., Lupus nephritis: Enigmas, conflicting models and an emerging concept. Mol. Med. 19, 161–169 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Choi J. Y., et al. , Circulating follicular helper-like T cells in systemic lupus erythematosus: Association with disease activity. Arthritis Rheumatol. 67, 988–999 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alfen J. S., et al. , Intestinal IFN-γ–producing type 1 regulatory T cells coexpress CCR5 and programmed cell death protein 1 and downregulate IL-10 in the inflamed guts of patients with inflammatory bowel disease. J. Allergy Clin. Immunol. 142, 1537–1547.e8 (2018). [DOI] [PubMed] [Google Scholar]
- 54.Gruarin P., et al. , Eomesodermin controls a unique differentiation program in human IL-10 and IFN-γ coproducing regulatory T cells. Eur. J. Immunol. 49, 96–111 (2019). [DOI] [PubMed] [Google Scholar]
- 55.Gladman D. D., Ibañez D., Urowitz M. B., Systemic Lupus Erythematosus Disease Activity Index 2000. J. Rheumatol. 29, 288–291 (2002). [PubMed] [Google Scholar]
- 56.Wan Y. Y., Flavell R. A., Identifying Foxp3-expressing suppressor T cells with a bicistronic reporter. Proc. Natl. Acad. Sci. U.S.A. 102, 5126–5131 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.