Significance
Plant reproduction relies on proper pollen tube growth to reach the female gametes. This process is highly regulated by a family of secreted signaling peptides that are recognized by cell-wall monitoring proteins to enable plant fertilization. Here, we report that these signaling peptides are sensed both by specific cell-wall and membrane-anchored proteins, which together enable cell-wall remodeling and, consequently, pollen tube growth.
Keywords: Arabidopsis, cell-wall integrity, pollen tube, structure biology, peptide signaling
Abstract
Plant reproduction relies on the highly regulated growth of the pollen tube for sperm delivery. This process is controlled by secreted RALF signaling peptides, which have previously been shown to be perceived by Catharanthus roseus RLK1-like (CrRLK1Ls) membrane receptor-kinases/LORELEI-like GLYCOLPHOSPHATIDYLINOSITOL (GPI)-ANCHORED PROTEINS (LLG) complexes, or by leucine-rich repeat (LRR) extensin proteins (LRXs). Here, we demonstrate that RALF peptides fold into bioactive, disulfide bond-stabilized proteins that bind the LRR domain of LRX proteins with low nanomolar affinity. Crystal structures of LRX2–RALF4 and LRX8–RALF4 complexes at 3.2- and 3.9-Å resolution, respectively, reveal a dimeric arrangement of LRX proteins, with each monomer binding one folded RALF peptide. Structure-based mutations targeting the LRX–RALF4 complex interface, or the RALF4 fold, reduce RALF4 binding to LRX8 in vitro and RALF4 function in growing pollen tubes. Mutants targeting the disulfide-bond stabilized LRX dimer interface fail to rescue lrx infertility phenotypes. Quantitative biochemical assays reveal that RALF4 binds LLGs and LRX cell-wall modules with drastically different binding affinities, and with distinct and mutually exclusive binding modes. Our biochemical, structural, and genetic analyses reveal a complex signaling network by which RALF ligands instruct different signaling proteins using distinct targeting mechanisms.
In flowering plants, sexual reproduction depends on the directional growth of pollen tubes to deliver the sperm cells to the female gametes. The polarized, rapid growth of the pollen tube depends on the highly dynamic remodeling of their cell wall. The redundant signaling peptides RALF4 and RALF19 ensure pollen tube integrity and growth by interacting with two distinct protein families, the malectin-domain CrRLK1L membrane receptor kinases (1–4), and the cell-wall monitoring leucine-rich repeat (LRR) extensin proteins (LRXs) (5, 6). Together with ANXUR1/2 and BUDDHA’S PAPER SEAL1/2 (BUPS1/2), the pollen-expressed LORELEI-like GLYCOLPHOSPHATIDYLINOSITOL (GPI)-ANCHORED PROTEINS2 (LLG2) and LLG3 are also required for monitoring pollen tube integrity and are part of the same signaling complex (2, 7). Recent structural data of the ternary RALF23–LLG2–FERONIA (FER) (8) complex suggests a shared mechanism for recognition of RALF peptides by LLG proteins, where the N-terminal region of the peptide is responsible for inducing an allosteric change in LLG to nucleate assembly with the corresponding CrRLK1L receptor (9). LRXs and RALF4/19 functions seem to be directly connected with the regulation of extracellular matrix composition to allow for proper cell-wall homeostasis during pollen tube growth. Deregulation of both RALF4/19 and LRX proteins translates into the unstructured deposition of pectin and changes in wall mechanical properties, leading to defective pollen tubes (5, 6).
While CrRLK1L/LLGs and LRXs are required for proper pollen tube growth, their relative contribution to specific RALF peptide sensing remains to be investigated. Here, we mechanistically dissect how pollen-expressed RALF peptides differentially interact with secreted LRX and membrane-anchored LLG proteins, respectively. Our work reveals that RALFs are folded signaling proteins precisely sensed by LRX proteins, whose bioactivity is determined by their oligomeric state to control pollen tube cell-wall integrity.
Results
There are 37 RALF peptides in Arabidopsis (10), with RALF4/19 being involved in the regulation of pollen tube integrity and growth (3, 5). Mature RALF4/19 peptides are 51 amino acids long and contain four invariant cysteines. We hypothesized that they may be folded proteins, stabilized by intramolecular disulfide bridges. Hence, for biochemical and structural analyses, we expressed RALF4/19 as thioredoxin A fusion proteins by secreted expression in insect cells. We observed secretion of RALF4/19 into the growth cell medium only when coexpressed with members of the LRX family, but neither as a stand-alone protein nor in the presence of the CrRLK1L ANXUR1 (ANX1) (11, 12) (SI Appendix, Fig. S1). Efficient secretion of RALF4/19 required the N-terminal half of LRX8/11 (SI Appendix, Fig. S1). RALF4/19 formed stable complexes with LRX833–400 (amino acids 33 to 400) and LRX1145–415 in size-exclusion chromatography (SEC) experiments (SI Appendix, Fig. S1). The presence of both proteins in the respective complexes was verified by mass spectrometry (SI Appendix, Fig. S2). Next, we biochemically mapped the RALF4 binding site to the predicted LRR core of LRX8 and LRX11 (SI Appendix, Fig. S1).
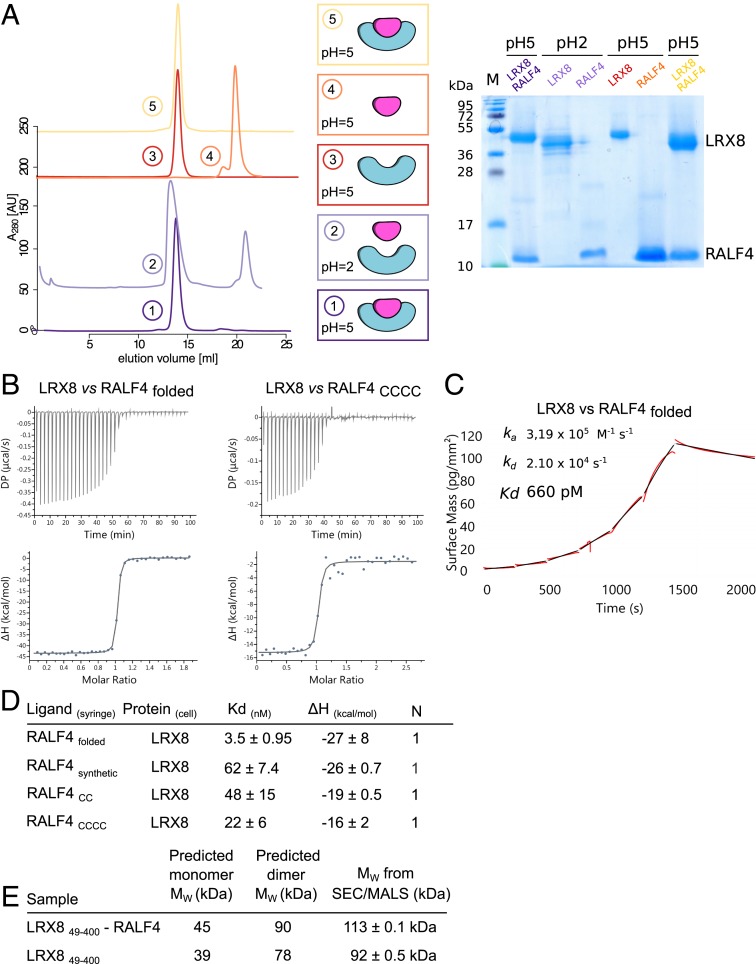
To quantify the interaction of RALF4 and LRX8, we first dissociated the complex at low pH (Materials and Methods) (Fig. 1A). The LRX8–RALF4 complex could be fully reconstituted when shifting the pH to 5.0 (13, 14) (Fig. 1A). To characterize the interaction of LRX8 with RALF4, we titrated the RALF4 protein isolated from insect cells (RALF4folded hereafter) into a solution containing isolated LRX849–400 using isothermal titration calorimetry (ITC). We found that RALF4 tightly binds the LRR core of LRX8 with ∼3.5-nM affinity and a binding stoichiometry (N) of ∼1 (Fig. 1 B and D and SI Appendix, Fig. S3). Next, we used a label-free surface biosensor based on grating-couple interferometry (GCI) (Materials and Methods) (15) to study the binding kinetics between LRX8 and the folded RALF4 peptide (Fig. 1C and SI Appendix, Fig. S1). In GCI experiments, LRX8 binds RALF4folded with a dissociation constant (Kd) of ∼0.5 nM, in good agreement with the ITC experiments. This tight interaction of the peptide to the cell-wall monitoring protein is characterized by a fast association (ka ∼ 4·105 M−1 s−1) and a very slow dissociation rate (kd ∼ 2·104 s−1) (Fig. 1C and SI Appendix, Fig. S1). To assess whether the RALF4 tertiary structure is a major binding determinant, we measured the binding affinity of mutant RALF4 peptides, in which we replaced two (Cys86 and Cys105) or all four cysteines by alanine in a synthetic peptide (RALFCC, RALFCCCC) (Fig. 1 B and D and SI Appendix, Fig. S1). RALF4CC and RALFCCCC bind LRX8 with ∼10-fold reduced affinity, suggesting that LRX8 preferentially senses the folded RALF4 signaling protein but still binds the linearized peptide with nanomolar affinity (Fig. 1 B and D and SI Appendix, Figs. S1 and S3). A synthetic, wild-type (WT) RALF4 peptide (RALF4synthetic) bound LRX8 with lower affinity, Kd of ∼60 nM (Fig. 1D and SI Appendix, Fig. S1), in agreement with previous findings (5). We then compared the folding state of insect cell-expressed RALF4folded with the different synthetic peptides using near-UV circular dichroism spectra (CD). In these experiments, we observed a spectrum characteristic for a disulfide-bond stabilized protein for RALF4folded but not for the RALFCCCC (SI Appendix, Fig. S4). The spectrum of RALF4synthetic indicates a partially folded state (SI Appendix, Fig. S4).
Fig. 1.
Dimeric LRX proteins bind RALFfolded peptides with high affinity. (A) Folded RALF4 peptide can be obtained by complex dissociation. LRX8–RALF4 complex purification, dissociation, and reconstitution. SEC graphs of the different steps are plotted on the Left. From bottom to top: LRX8–RALF4 purified complex at pH 5.0 (dark purple); dissociated LRX8–RALF4 complex at pH 2.0 (light purple). Orange and red runs represent RALF4 and LRX8 peaks, respectively, dialyzed back to pH 5.0. The SEC for the reconstituted complex, using the previously separated proteins, is shown in yellow. Schematics of the different steps, and SDS gel of the corresponding peaks, are shown alongside. (B) ITC of LRX8 vs. RALF4folded and RALF4CCCC. Raw thermograms of ITC experiments are plotted. (C) Binding kinetics of LRX8 vs. RALF4folded obtained from GCI experiments. The sensogram with recorded data are shown in red with the respective fits in black. The summary table contains the corresponding association rate constant (ka), dissociation rate constant (kd), and the dissociation constant Kd. (D) ITC table summaries of LRX8 vs. RALF4 peptides. Kd, (dissociation constant) indicates the binding affinity between the two molecules considered (in nanomoles). The N indicates the reaction stoichiometry (n = 1 for a 1:1 interaction). The values indicated in the table are the mean ± SD of two or three independent experiments. (E) Table of SEC–MALS analysis of apo–LRX8 and LRX8–RALF4 complex. The predicted and measured values are reported in the table for comparison. Values indicated in the table are the mean ± SD of two or three independent measurements.
Next, we assessed the stoichiometry of LRX–RALF complexes. We found LRX8 and LRX11 to be homodimers in the presence or absence of RALF4folded as judged by analytical SEC and multiangle light scattering (MALS) experiments (Fig. 1E and SI Appendix, Fig. S5). The MALS data are consistent with a 2 + 2 complex, with each glycosylated LRX protomer binding one RALF4 molecule.
The fact that LRX proteins appear to specifically sense RALFfolded peptides prompted us to investigate LRX–RALF complex structures. Diffracting crystals were obtained for LRX2–RALF4 and the pollen-specific LRX8–RALF4 complex, determined at 3.2-Å and 3.9-Å resolution, respectively (SI Appendix, Table S1). LRX2–RALF4 crystals contain four RALF4-bound dimers in the asymmetric unit, which closely align with each other, and with the LRX8–RALF4 complex (root mean square deviation [r.m.s.d.] is ∼0.8 Å comparing 357 pairs of corresponding Cα atoms) (Fig. 2A and SI Appendix, Fig. S6). We thus used the higher resolution LRX2–RALF4 structure to further analyze the signaling complex. A structural homology search with the program DALI (16) revealed that the LRX2 protein is closely related to the extracellular domain of known LRR receptor kinases, such as the immune receptor FLS2 (DALI Z-score of 33.5, r.m.s.d. of ∼1.7 comparing 299 corresponding Cα atoms). LRX2 comprises 11 LRRs sandwiched by canonical N- and C-terminal capping domains and a cysteine-rich protrusion that represents the N-terminal part of the extensin domain (Fig. 2 A and D). The LRX2 and LRX8 complex structures confirm that LRX proteins form constitutive dimers, covalently linked by a conserved disulfide bond (Fig. 2A and SI Appendix, Figs. S6 and S7). One RALF4 peptide is bound to each LRX protomer in the dimer, consistent with our MALS and ITC experiments in solution (Figs. 1 B, D, and E and 2A). The LRR core provides a binding platform for RALF4folded (Fig. 2B and SI Appendix, Fig. S6). Despite the moderate resolution of our complex structures, we located difference electron density accounting for the entire RALF4 folded peptide, again supporting its tight interaction with LRX proteins (SI Appendix, Fig. S8). The mature RALF4 protein consists of a short N-terminal alpha-helix, followed by a long loop region that is shaped by two disulfide bonds and a final one-turn 310-helix at the C terminus. The rather unstructured RALF4 peptide adopts a defined conformation when binding to the LRX LRR domain by making extensive contacts with the binding pocket (Fig. 2B and SI Appendix, Figs. S6 and S7). The RALF binding pocket is highly conserved among the known LRX family members, suggesting that other LRX proteins may bind different RALF peptides in a similar conformation (Fig. 2C and SI Appendix, Fig. S7). This particular binding configuration exposes the least conserved regions of the peptide to the solvent (SI Appendix, Fig. S9). In our structures, we also captured the cysteine-rich tail that links the C-terminal LRR capping domain of LRX2 and LRX8 to the cell-wall anchored extensin domain. The cysteine-rich linker adopts similar conformations in the LRX2 and LRX8 structures (SI Appendix, Fig. S6). The cysteine-rich tail folds back onto the LRR domain and connects via a ladder of disulfide bridges with LRRs 7 to 11. Several conserved polar and hydrophobic residues further stabilize this LRR–linker interface (Fig. 2D and SI Appendix, Fig. S7). This suggests that the RALF4 binding domain of LRX proteins is tightly connected to its extensin domain.
Fig. 2.
The LRR core of LRX proteins provides a conserved homodimer interface, a RALF binding pocket, and a stabilized connection to the extensin domain. (A) Front and 90° x axis rotated view of the covalently linked LRX2 homodimer in complex with RALF4 (ribbon diagram). The LRR domain is depicted in blue, the cysteine-rich tail in orange, and the RALF4 peptide is highlighted in pink. The disulfide bridge covalently linking the two LRX protomers is highlighted in yellow. (B) Surface view of LRX2 (color code as in A) along with a cartoon representation of the RALF4 peptide (pink), highlighting the LRX2’s RALF4 binding pocket. N-glycosylations are depicted in yellow. (C) Surface representation of LRX2 colored according to the LRX family amino acid conservation. The black rectangle underlines the conservation of the LRX’s RALF binding pocket. (D) Cartoon representation of the C-terminal part of the LRR core and the Cys tail of LRX2 (color code as in A). Each connecting disulfide bond is formed between a Cys from the LRR domain (depicted in blue) and the Cys-rich tail (highlighted in orange). Interface residues making polar contacts are shown as sticks and hydrogen bonds are depicted as dotted lines (in magenta).
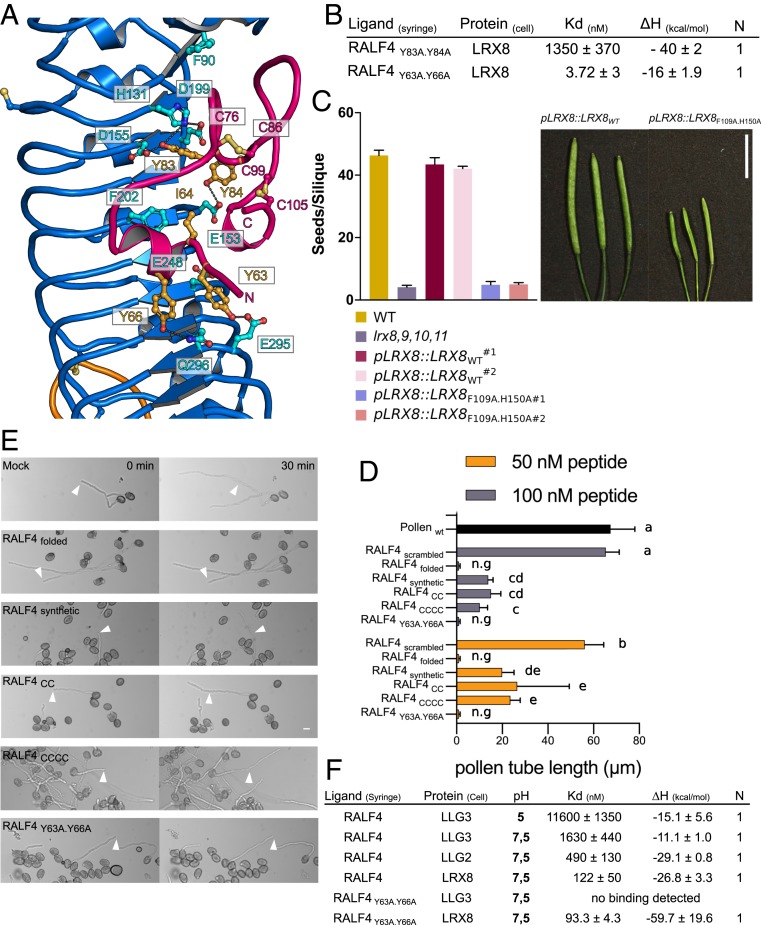
We next dissected the RALF4–LRX8 binding mode. Few interactions between the N-terminal alpha-helix (residues 64 to 69) of RALF4folded, which is a major determinant for LLG binding (9) with the LRR core of LRXs are mediated by RALF4 residues Tyr63, Ile64, and Tyr66 and LRX8 residues Gln314 (Gln296LRX2), Phe221(Phe202LRX2), Glu266 (Glu248LRX2), and Glu313 (Glu295LRX2) (Fig. 3A and SI Appendix, Fig. S10). A RALF4 Tyr63Ala/Tyr66Ala double mutant still bound LRX8 with wild-type affinity, suggesting that the N-terminal helix in RALF peptides is not a major determinant for high-affinity binding to LRX proteins (Fig. 3B, see below). The loop region in RALF4folded (residues 70 to 101) is structurally stabilized and oriented by the Cys76–Cys86 and Cys99–Cys105 disulfide bonds, required for high-affinity binding to LRX8 (Figs. 1 B and D and 3A and SI Appendix, Fig. S3). We identified the conserved RALF4 Tyr83 and Tyr84 to be part of this loop region and directly contacting the LRR core of LRX8 and LRX2 (Fig. 3A). Titration of a RALF4Y83A.Y84A mutant peptide to LRX8 in ITC assays resulted in a much weaker binding affinity (Kd ∼ 1.4 µM) when compared to wild-type RALF4 (Fig. 3B and SI Appendix, Fig. S11). Thus, the loop region forming part of the RALF4–LRX complex interface in our structures is critical for RALF4 binding in vitro.
Fig. 3.
LRX8 bioactivity depends on the interaction with folded RALFs. (A) Close-up view of the LRX2–RALF4 binding pocket. A large surface of the RALF4 peptide (in pink) directly interacts with the LRR core (in blue) of LRX2. Polar contacts of RALF4 with LRX2 are shown as dotted lines (in gray). The two disulfide bridges responsible for the folding of the peptide, C76–C86 and C99–C105, are depicted as sticks in light orange. (B) ITC table summaries of LRX8 vs. RALF4Y83A.Y84A and RALF4Y63A.Y66A mutants. Table summaries show the dissociation constant (Kd), binding stoichiometries (N), and thermodynamic parameters. The values indicated in the table are the mean ± SD of two or three independent experiments. (C) Complementation assay of lrx8,9,10,11 with point mutated LRX8F109A.H150A driven by its endogenous promoter. (C, Left) Seeds per silique of WT, lrx8,9,10,11, pLRX8::LRX8WT lines 1 and 2, and pLRX8::LRX8F109A.H150A in the lrx8,9,10,11 quadruple mutant background, for two independent lines. Data are means ± SEM of 10 siliques. For LRX8 mutant transgenic lines, 16 independent lines were analyzed. (C, Right) Silique images of the corresponding lines. (Scale bar, 10 mm.) (D) Growth rate of wild-type pollen tubes after the addition of RALF4scrambled, RALF4folded, RALF4synthetic, RALF4CC, RALF4CCCC, and RALF4Y63A.Y66A peptides at different concentrations after 30 min. Data were analyzed by one-way ANOVA followed by Tukey’s test as post hoc, considering P ≤ 0.05 as significantly different; data shown are mean ± SEM of three biological replicates, n = 28 each. Same letters represent samples that are not different between each other; n.g, no growth after peptide addition. (E) Effect of RALF4 peptides on wild-type pollen tubes growing in semisolid medium at two time points after adding them to a concentration of 50 nM. Arrowheads indicate the position of the tip of selected pollen tubes at time point 0 min. (Scale bar, 10 µm.) (F) Binding matrix of RALF4folded and RALF4Y63A.Y66A peptides vs. LRX8 and LLG2/3 membrane proteins in acidic and alkaline conditions. ITC table summary with dissociation constants (Kd), binding stoichiometries (N), and thermodynamic parameters (ΔH).
To confirm the binding mode of RALF4 to LRX8, we mutated the LRX surface in contact with the RALF4 loop region. Coexpression of LRX8 Phe109Ala/His150Ala (Phe90LRX2/His131LRX2) and wild-type RALF4 led to a reduced expression of the LRX8 protein in insect cells and secretion of the RALF4 peptide was not detected. This suggests that the mutations in the RALF–LRX loop interface inhibit the formation of a stable LRX8–RALF4 complex. Similar results were obtained in planta, where LRX8 and RALF4 mutant proteins targeting the complex interface failed to interact in coimmunoprecipitation experiments in tobacco (SI Appendix, Fig. S11). To determine whether RALF4 binding to LRX is critical to regulate pollen tube growth and integrity, we complemented a lrx quadruple mutant (lrx8,9,10,11) with the full-length LRX8F109A.H150A mutant expressed under the control of the native LRX8 promoter (SI Appendix, Fig. S12). The mutant failed to rescue the pollen tube growth defect and fertility phenotype (Fig. 3C) despite the proper localization of the C-terminal LRX8 F109A.H150A–mCitrine fusion protein to the cell wall (SI Appendix, Fig. S13).
Altogether, our experiments show that the disulfide bond-stabilized loop in RALF4folded is a major determinant for LRX binding and that binding of the peptide to LRX is required to control pollen tube growth. Based on these findings, we compared the bioactivity of synthetic, folded, and cysteine-mutated RALF4 signaling peptides in pollen tube growth assays. Addition of RALF4 peptides represses pollen tube growth, suggesting that high levels of RALF4 can also deregulate cell-wall integrity and/or sensing, leading to growth arrest (5, 17). The inhibitory effect of RALF4 peptides on pollen tube elongation was quantified using two peptide concentrations: 50 and 100 nM (Fig. 3D). Addition of a sequence-permutated version of RALF4 (RALF4scrambled) had no effect on pollen tube growth, whereas RALF4folded completely inhibited growth at both concentrations. The synthetic and cysteine-mutated RALF4 peptides partially suppressed pollen tube growth (Fig. 3 D and E), in good agreement with our structural and quantitative biochemical data (Figs. 1 B–D and 2A).
It has previously been reported that the CrRLK1Ls FER and THESEUS1 (8, 18) are receptors for RALF peptides with binding affinities in the nanomolar (nM) to the micromolar (µM) range (3, 19–21). Together with FER, the GPI-anchored protein LORELEI, expressed in the female gametophyte, is also required for fertilization (22, 23) and it copulls down with the receptor kinase in vitro (24). In addition, FER has been reported to coimmunoprecipitate with LLG1 in the presence of RALF peptides (24). These data have been supported by the recent crystal structure of the ternary FER–LLG2–RALF23 complex, where the N-terminal region of RALF23 (YISY motif) (25) is recognized by LLG proteins, inducing a conformational change promoting the interaction with the FER receptor-like kinase (9). We thus quantified the direct binding of RALF4folded to the ectodomains of the pollen-expressed LLG2/3 (2, 7), which have also been reported to be part of CrRLK1L–LLG complexes (2). We quantified the interactions in acidic and alkaline conditions since oscillatory influx of H+ is thought to be important for tip growth of the pollen tube (26, 27), and RALFs have been reported to alkalinize the apoplast space (28). We found that LLG3 binds RALF4folded with Kds of ∼11 µM at pH 5.0, ∼4,000 times weaker than to LRX8 at the same pH (Fig. 3F). We next quantified the interaction of RALF4folded with LRX8 and LLG2/3 at pH 7.5. We determined a Kd for RALF4–LRX8 to be ∼0.1 µM (Fig. 3F). RALF4folded bound LLG2 and LLG3 with dissociation constants of ∼0.5 µM and ∼1.5 µM, respectively (Fig. 3F and SI Appendix, Fig. S17). Thus, pH alterations modulate the interaction of RALF4folded with LRX8 and LLGs, with LRX proteins consistently recognizing RALF4folded with higher affinity.
Next, we compared the binding modes for RALF4folded interacting with LRX8 or LLGs. We found that mutations in the RALF4 YISY motif (Tyr63Ala/Tyr66Ala mutant), did not interfere with RALF4folded binding to LRX8, but disrupted the interaction with LLGs, consistent with previous structural and biochemical work (Fig. 3 B and F and SI Appendix, Fig. S18) (9). The mutant RALF4folded Y63A.Y66A protein completely inhibited pollen growth to a similar extent as wild-type RALF4folded, suggesting that RALF4-mediated pollen tube growth arrest may be exclusively regulated by LRX proteins (Fig. 3 D and E). Taken together, our findings suggest that LLGs and LRX proteins sense RALF peptides with mechanistically distinct peptide binding modes. Despite the similar phenotypes induced by mutations in ANX1, BUPS2, LLG2, LLG3, RALF4/19, and pollen-expressed LRX proteins (2, 3, 5–7, 11), we could not observe any simultaneous interactions between RALF4folded, LRX proteins, and LLGs or CrRLK1Ls (SI Appendix, Figs. S14–S16). Thus, RALF4folded appears to interact in a mutually exclusive manner with either LRXs or LLGs.
We next characterized the LRX homodimer, which we find stabilized by a central disulfide bridge in our two complex structures (Figs. 2A and 4A and SI Appendix, Fig. S6). The disulfide bond contributes to a dimer interface formed by highly conserved hydrophobic and polar amino acids (642-Å2 buried surface area) (Fig. 4 A and B and SI Appendix, Figs. S6 and S7 and Table S2). We probed the LRX8 dimer interface by mutational analysis: LRX8 still behaves as a dimer in solution when the disulfide bond was disrupted by mutating Cys157 (Cys138LRX2) to Ala, suggesting that the intermolecular disulfide bond is not essential for the structural integrity of the LRX dimer. Simultaneous mutation of Tyr87 (Phe68LRX2) to Ala and Ala133 (Ala114LRX2) to Phe resulted in a monomer–dimer equilibrium, which we could quantitatively shift to a LRX8 monomer by additionally introducing the Cys157Ala mutation (Fig. 4 C and D and SI Appendix, Fig. S19). Both dimeric and monomeric LRX8 forms retain the capacity to bind RALF4, with the monomer binding the folded peptide with wild-type affinity (∼2.5 nM) (Fig. 4E and SI Appendix, Fig. S19). Crystallographic analysis of the LRX8monomer–RALF4folded complex yielded a 2.3-Å resolution structure of the complex with excellent stereochemistry (SI Appendix, Table S1). In the LRX8monomer complex, RALF4folded binds to the LRX binding surface in a conformation similar to the wild-type LRX8dimer (r.m.s.d. is ∼0.65 Å comparing 327 residues of corresponding Cα atoms) (SI Appendix, Fig. S20). Thus, our biochemical and structural data reveal that mutations affecting the dimer interface of LRX8 specifically alter the oligomeric state of the cell-wall protein, while leaving its interaction with RALF peptides intact.
Fig. 4.
LRX8 functional mechanism requires homodimerization to control pollen tube growth. (A) Close-up view of the dimer interface in LRX2. Details of the interactions between the two protomers (depicted in blue and gray) are highlighted. The Cys (C138) mediating the disulfide bond formation is depicted in yellow. The ionic and hydrogen bonds are shown as dotted lines (in magenta). (B) Side surface representation of LRX2 colored according to LRX family sequence conservation. The black rectangle depicts the LRX dimer interface. (C) The dimer interface of LRX proteins is stabilized by a central disulfide bridge. SEC runs of wild-type LRX8–RALF4, LRX8C157A–RALF4, LRX8Y87A.A133F–RALF4, and LRX8C157A.Y87A.A133F–RALF4. Peaks 1 and 2 correspond to the dimeric and monomeric forms of LRX8, respectively. (D) SEC–MALS summary table for the LRX8C157A–RALF4 and LRX8C157A.Y87A.A133F–RALF4 complexes in solution at pH 5.0, along with the predicted molecular weights. The values indicated in the table are the mean ± SD of two independent measurements. (E) ITC of LRX8C157A.Y87A.A133F vs. RALF4folded. The table summarizes the biophysical values obtained. Kd, (dissociation constant), binding stoichiometries (N), and thermodynamic parameters (ΔH). (F) Shift of the monomer–dimer equilibrium under reducing conditions in solution. LRX8Y87A.A133F–RALF4 run in the absence (orange) and presence (purple) of TCEP. (G) Complementation assays of lrx8,9,10,11 with LRX8 oligomeric mutants. (G, Left) Seeds per silique of WT, lrx8,9,10,11, pLRX8::LRX8WT lines 1 and 2, pLRX8::LRX8Y87A.A133F and pLRX8::LRX8C157A.Y87A.A133F in the lrx8,9,10,11 quadruple mutant background. Data are means ± SEM of 10 siliques. For LRX8 mutant transgenic lines, 16 independent lines were analyzed. (Scale bar, 10 µm.) (G, Right) Siliques images of the corresponding lines. (H) Working model and open questions about RALF-mediated autocrine signaling during pollen tube growth. Physical interactions are depicted by double arrows and genetic relationships by single arrows. Straight lines indicate what is known, and dotted lines indicate speculative relationships. Affinity ranges are marked next to the interaction arrows. Legends of the different components are shown on Top.
To assess the role of the dimer interface and its central cysteine in a changing redox environment, we incubated purified LRX8 and LRX8–RALF4 complex with increasing concentrations of the reducing agent dithiothreitol (DTT) under denaturing conditions. In both cases, the dimer could be converted into monomers (SI Appendix, Fig. S21). In addition, we could also quantitatively shift the oligomeric equilibrium of LRX8Y87A.A133F toward the monomeric species in the presence of a reducing agent in SEC experiments (Fig. 4F). Consistent with these observations, we could detect both oligomeric species of a myc-tagged LRX81–400, when transiently expressed in tobacco leaves, in the absence of DTT (SI Appendix, Fig. S21).
Next, we asked whether the oligomeric state of LRX proteins has functional relevance in planta. We transformed the quadruple lrx8,9,10,11 mutant with monomeric LRX8 (LRX8C157A.Y87A.A133F) and the monomer–dimer variant (LRX8Y87A.A133F) under the control of the native LRX8 promoter (SI Appendix, Fig. S12). Both LRX8 variants were secreted into the cell wall and we could detect expression of both mutants’ proteins in pollen (SI Appendix, Figs. S12 and S13). Like the previously tested LRX8F109A.H150A mutant (Fig. 3C), these oligomeric variants failed to complement the sterility phenotype of the lrx8,9,10,11 quadruple mutant (Fig. 4G), suggesting that both the RALF binding capacity and the dimeric state of LRX8 are required to facilitate the rapid growth of the pollen tube to the ovule.
Discussion
Our work reveals that RALF peptides can exist as small folded signaling proteins, placing them together with other cysteine-rich signaling peptides in plants, such as the LURE peptides controlling pollen tube guidance or the EPF peptides controlling stomatal patterning (29, 30). In the case of RALF peptides, the isolated protein may already be folded, as suggested by a recent NMR study (31), and is fully structured when bound with high affinity to LRX proteins (Fig. 2). It is of note that RALF4, when bound to LRX, adopts a defined conformation exposing a highly basic surface patch, which we speculate could mediate targeted interactions with other proteins or cell-wall components (SI Appendix, Fig. S8). Binding of RALFs to the LRR region could thus affect the overall conformation of the LRX extensin domain, which our structures reveal to be tightly linked to the LRR core. The identity of these interacting factors will be a key question to address in future studies.
Polarized tip growth of pollen tubes has been reported to be driven by waves of acidic pH and reactive oxygen species (ROS) production to allow for the proper cell-wall dynamics (26, 32–35). This correlates with our biochemical findings, where both parameters modulate RALF sensing by LRXs (Figs. 1 and 3) and the oligomeric state of LRX proteins (Fig. 4H). A putative redox-controlled oligomerization of LRX proteins would add an additional layer to the modulation of RALF sensing and signaling. In this respect, it is noteworthy that cell-wall thioredoxins have been identified as putative LRX interactors (36).
Our and previous structural and biochemical data support two distinct targeting mechanism for RALF peptides toward the LRX and CrRKL1L–LLGs signaling modules (9). We demonstrate that these binding events are mutually exclusive in the context of pollen tube growth and, consequently, we did not observe formation of tertiary complexes containing RALFs, LRXs, LLGs, or CrRLK1Ls proteins. This suggests that RALF peptides trigger two parallel, converging but mechanistically distinct signaling pathways. On the one hand, RALF4/19 peptides can signal via the CrRLK1Ls–LLGs to control processes inside the pollen tube through the cytoplasmic receptor-like kinase MARIS (5, 37); on the other hand, RALF4/19 may signal to control processes outside the cell through their interaction with LRXs in the cell wall. This is consistent with the finding that, unlike mutants with reduced RALF4/19 activity (5), mutants lacking multiple pollen-expressed LRXs cannot be suppressed by dominant active MARIS (SI Appendix, Fig. S22) (38). That indicates that also based on genetic evidence, RALF4/19 peptides function in different but converging pathways to fine tune pollen tube integrity.
Our binding data between RALFs, LRXs, and LLGs, together with previous studies (20), suggest that this autocrine signaling cascade could be additionally regulated by pH fluctuations in the cell wall to modulate pollen tube growth (13, 21, 26, 27). Our in vitro data clearly suggest that LRX proteins represent high-affinity sensors for RALFs at acidic pH, while LLGs show a binding preference under neutral/alkaline conditions (Fig. 4H). Our structural, quantitative biochemical, and physiological experiments suggest that LRX proteins preferentially bind folded rather than linear RALF peptides. The N-terminal α-helix conserved among all RALF family members does not play a major role in LRX recognition but has previously been shown to be critical and sufficient for CrRLK1Ls–LLGs complex formation (9). An attractive hypothesis would thus be that cell-wall pH and redox changes alter the conformations and oligomeric states of both RALF ligands and their sensors, allowing different signaling complexes to form (Fig. 4H). RALF peptides may exist in a fully folded oxidized state preferentially bound by LRX proteins. However, in response to a changing cell-wall environment, they may acquire a reduced linearized state preferentially bound by LLG-containing complexes (Fig. 4H). This point should be considered in future studies aimed at dissecting the contributions of LRXs and CrRLK1Ls to the diverse RALF-mediated signaling processes in plants. The RALF signaling system emerges as a unique signaling cascade, in which one family of ligands is recognized by two structurally distinct receptor families, that are differentially modulated but together shape a core developmental process in plants.
Materials and Methods
Protein Expression and Purification.
Codon-optimized synthetic genes for expression in Spodoptera frugiperda (Invitrogen GeneArt), coding for Arabidopsis thaliana LRX2 (residues 1 to 385; At1g62440), LRX8 (residues 33 to 400, 49 to 400, and 49 to 373; At3g19020), LRX11 (residues 45 to 415, 64 to 415, and 64 to 388; At4g33970); LLG2 (residues 24 to 135; At2g20700), LLG3 (residues 24 to 137; At4g28280), BUPS2 (residues 40 to 439; At2g21480) domains were cloned into a modified pFastBac (Geneva Biotech) vector, providing a TEV (tobacco etch virus protease) cleavable C-terminal StrepII-9xHis tag. LRX8 (49 to 400) fused to a noncleavable Avi-tag was also cloned into a modified pFastBac vector harboring the Drosophila BiP secretion signal peptide (39–41). Spodoptera frugiperda codon-optimized RALF4 (residues 58 to 110; At1g28270) and RALF19 (residues 58 to 110; At2g33775) mature peptide sequences were N-terminally fused to TRX A (Thioredoxin A) in a pFastBac vector driven by a 30 K signal peptide (42). For protein expression, Trichoplusia ni Tnao38 cells (43) were coinfected with a combination of LRX and RALF virus with a multiplicity of infection (MOI) of 3 and incubated for 1 d at 28 °C and 2 d at 22 °C at 110 rpm. The secreted complexes were purified from the supernatant by sequential Ni2+ (HisTrap excel; GE Healthcare; equilibrated in 25 mM KPi pH 7.8, 500 mM NaCl) and StrepII (Strep-Tactin Superflow high capacity [IBA Lifesciences] equilibrated in 25 mM Tris pH 8.0, 250 mM NaCl, 1 mM EDTA) affinity chromatography. All proteins were incubated with TEV protease to remove the tags. Proteins were further purified by SEC on a Superdex 200 increase 10/300 GL column (GE Healthcare), equilibrated in 20 mM sodium citrate pH 5.0, 150 mM NaCl. For crystallization and biochemical experiments, proteins were concentrated using Amicon Ultra concentrators (Millipore, molecular weight cut-off 3,000 and 30,000). Proteins were analyzed for purity and structural integrity by sodium dodecyl sulfate– polyacrilamide gel electrophoresis and mass spectrometry. The BirA enzyme was cloned into a modified pETM11 vector providing an N-terminal 6xHis tag and a TEV site. The enzyme was purified from Escherichia coli by Ni2+ affinity chromatography.
Mass Spectrometry.
For LC-MS/MS analysis of LRX8–RALF4, 5 µL of complex (1 mg/mL) was diluted in 20 µL of the following buffer: 50 mM ammonium bicarbonate, 10 mM TCEP (triscarboxyethylphosphine) and 40 mM chloroacetamide. Samples were incubated 45 min in the dark at room temperature (RT), and then digested with 0.1 µg of sequencing-grade trypsin or chymotrypsin (Promega). Samples were incubated at 37 °C for 4 h, and digestion reaction was stopped with 2 µL of 10% formic acid. They were then diluted 10× with loading buffer (2% acetonitrile, 0.05% trifluoroacetic acid) and injected on a Fusion Tribrid orbitrap mass spectrometer (Thermo Fisher Scientific) interfaced to a Dionex RSLC 3000 nano-HPLC. Peptides were separated on a 65-min gradient from 4 to 76% acetonitrile in 0.1% formic acid at 0.25 µL/min on an in-home packed C18 column (75-μm internal diameter × 40 cm, 1.8 μm, Reprosil Pur, Dr. Maisch). Full MS survey scans were performed at 120,000 resolution. In data-dependent acquisition controlled by Xcalibur 4.1 software (Thermo Fisher), a top speed precursor selection strategy was applied to maximize acquisition of peptide tandem MS spectra with a maximum cycle time of 1.5 s. higher-energy collisional dissociation (HCD) (normalized collision energy: 32%) or electron-transfer/higher-energy collisional dissociation (supplemental HCD activation energy: 25%) fragmentation mode were used with a precursor isolation window of 1.6 m/z. MS/MS spectra were acquired at a 15,000 resolution, and peptides selected for MS/MS were excluded from further fragmentation during 60 s. For data analysis, tandem mass spectra were searched using PEAKS software (version 8.5, Bioinformatics Solutions Inc.) against a custom database containing common contaminants and the sequences of the proteins of interest. Mass tolerances used were 10 ppm for the precursors and 0.02 Da for collision-induced dissociation fragments. Activities of proteases considered were semitrypsin or semichymotrypsin (one specific cut) with two missed cleavages. Carbamidomethylation of cysteine was specified as a fixed modification. N-terminal acetylation of protein and oxidation of methionine were specified as variable modifications. PEAKS results were imported into the software Scaffold 4.8.5 (Proteome Software Inc.) for validation of MS/MS-based peptide (minimum 90% probability) and protein (min 95% probability) identifications, dataset alignment, as well as parsimony analysis to discriminate homologous hits.
For LC-MS/MS analysis of LRX11–RALF4, 10 µL of complex (10 mg/mL) was diluted in 20 µL of the following buffer: 8 M urea, 50 mM TEAB (triethylammonium bicarbonate), 5 mM TCEP (triscarboxyethylphosphine), and 20 mM chloroacetamide. After 1 h incubation at RT, the sample was then diluted adding 90 µL of 50 mM TEAB, split into two aliquots of 55 μL and digested with 0.2 µg of trypsin or chymotrypsin. Samples were incubated at 37 °C for 2.5 h, and digestion reaction was stopped with 1 µL of 10% formic acid and 200 µL of loading buffer. They were injected on a Fusion Tribrid orbitrap mass spectrometer interfaced to a Dionex RSLC 3000 nano-HPLC, as above. Full MS survey scans were performed at 120,000 resolution. In data-dependent acquisition controlled by Xcalibur 4.1 software, a top speed precursor selection strategy was applied to maximize acquisition of peptide tandem MS spectra with a maximum cycle time of 0.6 s. HCD (normalized collision energy: 32%) fragmentation mode was used with a precursor isolation window of 1.6 m/z. MS/MS spectra were acquired in the linear trap and peptides selected for MS/MS were excluded from further fragmentation during 60 s. For data analysis, tandem mass spectra were searched using Mascot (Matrix Science, version 2.6.2) against a custom database containing common contaminants and the sequences of the proteins of interest. Mass tolerances used were 10 ppm for the precursors and 0.5 Da for collision-induced dissociation fragments. Activities of proteases considered were semitrypsin (one specific cut) or chymotrypsin, with two or three missed cleavages, respectively. Carbamidomethylation of cysteine was specified as a fixed modification, and oxidation of methionine as variable modification. Mascot results were imported into the software Scaffold for validation of MS/MS-based peptide (minimum 95% probability) and protein (minimum 95% probability) identifications, dataset alignment, as well as parsimony analysis to discriminate homologous hits.
MALS.
MALS was used to assess the monodispersity and molecular weight of the apo LRXs and in complex with RALF4. Samples containing 100 µg of protein were injected into a Superdex 200 increase 300/10 GL column (GE Healthcare) using an HPLC system (Ultimate 3000, Thermo Fisher Scientific) at a flow rate of 0.5 mL min−1, coupled in-line to a multiangle light scattering device (miniDAWN TREOS, Wyatt). Static light scattering was recorded from three different scattering angles. The scatter data were analyzed by ASTRA software (version 6.1, Wyatt). Experiments were done in duplicate with SD indicated in the figures.
Protein Complex Dissociation.
LRX–RALF complexes were obtained in vivo by coexpression in insect cells. The apo proteins were obtained by complex dissociation in vitro using a low pH buffer. After purification of the LRX–RALF complex (see above), the protein complex was incubated for 1 h at 4 °C in the following buffer: 20 mM citric acid pH 2, 150 mM NaCl. Next, the dissociated proteins were separated by gel filtration, using a Superdex 200 increase 10/300 GL column (GE Healthcare) equilibrated in the same buffer. Each protein was then collected separately and dialyzed against 20 mM citric acid pH 5, 150 mM NaCl. To test whether proteins were still folded and functional, proteins were again run in a Superdex 200 increase 10/300 GL column (GE Healthcare) in 20 mM citric acid pH 5, 150 mM NaCl. We also tested, for each batch of dissociated protein, that the complex would be fully reconstituted when mixing the proteins in equimolar proportions. Additionally, we tested the in vivo activity in pollen tube growth assays of the dissociated peptide (folded) versus a synthetic linear version of RALF4 of the same sequence.
ITC.
Experiments were performed at 25 °C using a MicroCal PEAQ-ITC (Malvern Instruments) with a 200-µL standard cell and a 40-μL titration syringe. LRX8 protein and RALF peptides were gel filtrated into either pH 5 ITC buffer (20 mM sodium citrate pH 5.0, 150 mM NaCl) or pH 7.5 ITC buffer (20 mM Hepes pH 7.5, 150 mM NaCl). For the RALF vs. LRX interactions, a typical experiment consisted of injecting 1 μL of a 100-μM solution of the RALF peptides into 10 μM LRX8 (49 to 400) solution in the cell at 150-s intervals. The experiments using RALF, CrRLK1Ls, and LLGs were typically performed with 10 µM protein in the cell and 100 µM ligand in the syringe, and an injection pattern of 2 μL. ITC data were corrected for the heat of dilution by subtracting the mixing enthalpies for titrant solution injections into protein-free ITC buffer. Experiments were done at least in duplicates and data were analyzed using the MicroCal PEAQ-ITC Analysis Software provided by the manufacturer. All ITC runs used for data analysis had an N ranging between 0.8 and 1.3. The N values were fitted to 1 in the analysis.
Protein Biotinylation.
A total of 25 μM of LRX8 (49 to 400) were biotinylated with the biotin ligase BirA (2 μM) (40) for 1 h at 30 °C, in a volume of 200 μL. The buffer used in the reaction was: 25 mM Tris pH 8, 150 mM NaCl, 5 mM MgCl2, 2 mM 2-mercaptoethanol, 0.15 mM biotin, 2 mM ATP. Biotinylated proteins were purified by SEC in 20 mM Hepes pH 7.5, 150 mM NaCl.
GCI.
GCI experiments were performed with the Creoptix WAVE system using 4 PCP WAVE chips (thin quasiplanar polycarboxylate surface [Creoptix]). Chips were first conditioned with borate buffer (100 mM sodium borate pH 9.0, 1 M NaCl [Xantec]). Then, the chips were activated with 1:1 mix of 400 mM N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride and 100 mM N-hydroxysuccinimide (Xantec). Streptavidin (1.5 mg/mL [Sigma]) in 10 mM sodium acetate pH5 (Sigma) was immobilized on the chip surface until saturation was reached, followed by passivation of the surface with bovine serum albumin (BSA) (5 mg/mL [Sigma]) in 10 mM sodium acetate pH 5, and a final quenching step with 1 M ethanolamine pH8 (Xantec). Then, biotinylated LRX8 (ligand) (200 to 300 μg/mL) was captured on the streptavidin surface until the desired density was reached. Two different kinetic analyses were performed to measure the binding of RALF4folded (analyte) to LRX8: a regeneration-free kinetic and a normal kinetic. Both experiments were performed at 25 °C with a 1:2 dilution series from a maximum concentration of 10 nM RALF4folded, in 20 mM citrate pH 5, 250 mM NaCl, 0.01% Tween-20, 0.1% BSA. Blank injections were used for double referencing and a dimethyl sulfoxide (DMSO) calibration curve for bulk correction. Analysis and correction of the obtained data were performed using the Creoptix WAVE control software (correction applied: X and Y offset; DMSO calibration; double referencing). A one-to-one binding model was used for both experiments.
Crystallization and Data Collection.
Crystals of the LRX229–385–RALF4 and LRX833–400–RALF4 complexes developed at room temperature in hanging drops composed of 1.0 μL of protein solution (20 mg/mL) and 1.0 μL of the following crystallization buffers, respectively: 20% (wt/vol) polyethylene glycol (PEG) 3350, 0.1 M Bis-Tris pH 5 and 0.2 M sodium acetate and 17.5% (wt/vol) PEG 8000, 0.1 M Bis-Tris pH 7 and 0.2 M sodium citrate. Drops were suspended above 0.6 mL of crystallization buffer. For data collection, LRX2–RALF4 crystals were transferred into crystallization buffer supplemented with 15% (vol/vol) ethylene glycol and snap frozen in liquid nitrogen. In the case of the LRX8–RALF4 complex, crystals were snap frozen in the presence of 15% glycerol as cryoprotectant.
Crystals of the LRX8C157A.Y87A.A133F mutant in complex with RALF4, grew in 0.3 M ammonium sulfate and 20% (wt/vol) PEG 4000. Crystals of the monomer were cryoprotected using 15% (vol/vol) ethylene glycol and snap frozen in liquid nitrogen. The 3.2-Å, 3.9-Å, and 2.3-Å native datasets, respectively, were collected at beam-line PXIII of the Swiss Light Source. Data processing and scaling were done in XDS (version June 2017) (44).
Circular Dichroism.
Near-UV spectra (250 to 400 nm) were acquired using a Chirascan V100 spectrophotometer (Applied Photophysics) equipped with a 150 W Xenon arc lamp, on a 6.5 µM (1 mg/mL) sample in a quartz cuvette of 1-cm light path (Hellma). Buffer and samples were measured 10 times each. Signal from the buffer was subtracted from the samples and values were then normalized to the absorbance at 278 nm (highest in all cases).
Structure Determination and Refinement.
The structure of the LRX2–RALF4 complex (PDB ID: 6QXP) was determined by molecular replacement using the program PHASER, implemented in the Phenix MRage pipeline (45). Search models were selected using the program HHPRED (46) and tested iteratively in the MRage pipeline. A solution using a fragment of the LRR ectodomain of the plant immune receptor FLS2 (PDB ID: 4MNA) yielded a solution with eight molecules in the asymmetric unit. The search model, which showed 28% sequence identity with LRX2 target was further improved using the program CHAINSAW in CCP4 (47, 48). The final solution was used for noncrystallographic symmetry averaging and density modification in the program PHENIX.RESOLVE (49). The resulting map was readily interpretable and the LRX2–RALF4 complex structure was built and completed by iterative round of manual model building in COOT (50) and restrained TLS in phenix.refine. The structure of LRX8–RALF4 (PDB ID: 6QWN) was solved by molecular replacement in PHASER (51) and using the LRX2–RALF4 as search model. The structure of LRX8C157A.Y87A.A133F–RALF4 (PBD ID: 6TME) was determined by molecular replacement in PHASER, using the LRX2–RALF4 complex as search model. The LRX8–RALF4 and LRX8C157A.A133F.Y87A–RALF4 structures were also built and completed by iterative round of manual modeling in COOT and restrained refinement in phenix.refine. Inspection of the final models with phenix.molprobity (52) reveal good stereochemistry (SI Appendix, Table S1). Diagrams were prepared with PYMOL (https://pymol.org/2/) or CHIMERA (53). Structure-based sequence alignments include a secondary structure assignment calculated with the program DSSP (54). The two-dimensional (2D) ligand-interaction diagram was generated with LIGPLOT (55).
Transient Protein Expression in Tobacco Leaves.
Nicotinia benthamiana plants were grown for 3 wk prior to agroinfiltration. Appropriate Agrobacterium tumefaciens cultures were grown in yeast extract broth media at 28 °C until reaching an optical density of 0.6. Cultures were centrifuged for 10 min at 4,000 rpm. Cells were resuspended in fresh infiltration media (50 mM MES, 2 mM NaH2PO4, 0.5% [m/v] saccharose, 100 µM acetosyringone, pH 5.6) and left on the shaker for 2 h in the dark. For coinfiltration of two Agrobacterium strains, infiltration media with the appropriate Agrobacterium strains harboring the desired constructs were mixed in a 1:1 vol/vol ratio. Infiltration was performed with a syringe. Plants were grown for 2 more days prior to material collection. Leaves were cut and flash frozen in liquid nitrogen. For the plasmolysis experiments, an acetosyringone concentration of 500 µM was used, and plants were imaged 6 d after infiltration. Plants were then infiltrated with a 30% glycerol solution to induce plasmolysis and imaged right after. For LRX8-mCitrine WT and mutants, the excitation and emission wave lengths used were 514 nM and 525 to 550, respectively. For the plasma membrane marker Lti6-mCherry the excitation wavelength was 561 nm and the emission was between 595 and 645 nm. Leaves were imaged with a SP8 Leica confocal microscope. Images were adjusted using ImageJ.
In Vitro Redox Sensitivity Experiments.
To determine the importance of disulfide bridge formation in the LRX homodimers, samples were incubated at 95 °C for 5 min in Laemmli buffers containing DTT 0, 1, 3, 5, or 20 mM, respectively. They were subsequently run on denaturing SDS gels for further size identification. In solution, reducing assays were performed by incubating the LRX8Y87A.A133F protein with and without 5 mM TCEP for 1 h at RT. Samples were then run by size-exclusion chromatography on a Superdex 200 increase 10/300 GL column (GE Healthcare), equilibrated in 20 mM sodium citrate pH 5.0, 150 mM NaCl, in the presence or absence of TCEP.
Protein Extraction, Coimmunoprecipitation, and Western Blot Analysis.
Transiently expressed LRX8 and RALF4 proteins in tobacco were extracted as follow: 300 mg of plant material were crushed in liquid nitrogen and resuspended in 600 µL of extraction buffer (150 mM Tris pH 7.5; 150 mM NaCl; 10% [vol/vol] glycerol; 10 mM EDTA, 0.5% IGEPAL, cOmplete protease inhibitor [Roche]). Samples were centrifuged at 15,000 rpm for 20 min, and the supernatant was then centrifugated twice more to discard residual pellet debris. A total of 50 µL of input sample was kept at this stage, the rest was incubated for 1 h at 4 °C with anti-myc or anti-HA magnetic beads (µMACS, Milteny Biotech). Immunoprecipitation was performed according to manufacturer’s specifications, using the extraction buffer for column washes. Standard Laemmli buffer (56) was added to the input and immunoprecipitated samples prior to gel loading. Finally, Western blot analysis was performed with horseradish peroxidase-coupled anti-myc and anti-HA antibodies (MACS, Milteny Biotech) at 1:2,000 dilution. Detection was performed using WesternBright Sirius (Advansta). Images were taken with a LAS500 BlotImager. For Western blot detection of expressed proteins in insect cells (supernatant and pellet fractions) we used anti-His HRP conjugated from Roche in a 1:2,000 dilution.
Pollen Germination In Vitro Assays.
Open flowers were incubated at 22 °C for 30 min in moist incubation boxes. Then, pollen was bound to xilane-coated slides containing germination medium (0.01% boric acid [wt/vol], 5 mM CaCl 2, 5 mM KCl, 1 mM MgSO4, 10% sucrose, pH 7.5). Pollen grains were preincubated in moist incubation boxes for 30 to 45 min at 30 °C and then transferred to 22 °C from 30 min to 2 h as indicated. After 2 h of in vitro pollen germination, peptides were added to pollen germination medium to a concentration of 50 nM and 100 nM. The effect on pollen tube growth was imaged using a Leica DM 6000 microscope and analyzed using the ImageJ 1.40g software (http://rsb.info.nih.gov/ij). RALF4synthetic, RALF4scrambled, RALF4C86A.C105A, and RALF4C76A.C86A.C99A.C105A synthetic peptides were obtained from PHTD Peptides Industrial Co, Ltd. and Genscript (see sequences below). The rest of the peptides were expressed and produced in insect cells complexed with LRX8 and dissociated as indicated above.
Sequences of the Synthetic Peptides.
Sequence of RALF4synthetic: RRYIGYDALKKNNVPCSRRGRSYYDCKKRRRNNPYRRGCSAITHCYRYAR
Sequence of RALF4scrambled: PTYNSCRRKCKRDRGYAARRYKRYRYVNADIKRNHSGYPCRICSRLYGRN
Sequence of RALF4C86A.C105A (RALF4CC): RRYIGYDALKKNNVPCSRRGRSYYDAKKRRRNNPYRRGCSAITHAYRYAR
Sequence of RALF4C76A.C86A.C99A.C105A (RALF4CCCC): RRYIGYDALKKNNVPASRRGRSYYDAKKRRRNNPYRRGASAITHAYRYAR
Sequence of RALF4Y83A.Y84A: RRYIGYDALKKNNVPCSRRGRSAADCKKRRRNNPYRRGCSAITHCYRYAR.
Generation of Transgenic Lines.
All transgenic lines were generated using the floral dip method (57) with the A. tumefaciens strain GV3101. The construct pLAT52:MRIR240C-YFP, described previously (37), was used for A. thaliana transformation. The construct expressions in pollen tubes were checked and imaged using a Leica DM 6000 microscope. Amplification of LRX8 full-length from transgenic lines was performed using the following primers: Fw-5′: ATG ACC CGA AGA ACA ATG GAG, Rev-5′: ATG GTG TCT CAG GCT TTG GAC TAG. To detect the expression of LRX8-myc mutants, Western blot analysis was performed with horseradish peroxidase-coupled anti-myc (MACS, Milteny Biotech) at 1:2,000 dilution. Seed per silique phenotypes were checked in T1 plants.
Data and Materials Availability.
All data are available in the main text or in SI Appendix, Supplementary Materials.
Supplementary Material
Acknowledgments
We thank V. Olieric for providing beam time and the staff at beam line PXIII of the Swiss Light Source, Villigen, for technical assistance during data collection. We also thank Fabio Spiga and Rony Nehme from Creoptix (Switzerland) for technical assistance when performing and analyzing the GCI experiments. We thank Cyril Lipp (University of Zurich) for help with plant care. Funding: Supported by the University of Lausanne, the University of Zurich, European Research Council grant agreement no. 716358 (J.S.), Swiss National Science Foundation grants 31003A_173101 (J.S.) and CR3213_156724 (U.G.), the Programme Fondation Philanthropique Famille Sandoz (J.S.), and European Molecular Biology Organization long-term fellowship 1004-2017 (S.M.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, http://www.wwpdb.org/ (PDB ID codes 6QXP, 6QWN, and 6TME).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2000100117/-/DCSupplemental.
References
- 1.Boisson-Dernier A., Kessler S. A., Grossniklaus U., The walls have ears: The role of plant CrRLK1Ls in sensing and transducing extracellular signals. J. Exp. Bot. 62, 1581–1591 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Ge Z., et al. , LLG2/3 are co-receptors in BUPS/ANX-RALF signaling to regulate Arabidopsis pollen tube integrity. Curr. Biol. 29, 3256–3265.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ge Z., et al. , Arabidopsis pollen tube integrity and sperm release are regulated by RALF-mediated signaling. Science 358, 1596–1600 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moussu S., Augustin S., Roman A.-O., Broyart C., Santiago J., Crystal structures of two tandem malectin-like receptor kinases involved in plant reproduction. Acta Crystallogr. D Struct. Biol. 74, 671–680 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mecchia M. A., et al. , RALF4/19 peptides interact with LRX proteins to control pollen tube growth in Arabidopsis. Science 358, 1600–1603 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Fabrice T. N., et al. , LRX proteins play a crucial role in pollen grain and pollen tube cell wall development. Plant Physiol. 176, 1981–1992 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng H., et al. , LORELEI-LIKE GPI-ANCHORED PROTEINS 2/3 regulate pollen tube growth as chaperones and coreceptors for ANXUR/BUPS receptor kinases in Arabidopsis. Mol. Plant 12, 1612–1623 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Escobar-Restrepo J.-M., et al. , The FERONIA receptor-like kinase mediates male-female interactions during pollen tube reception. Science 317, 656–660 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Xiao Y., et al. , Mechanisms of RALF peptide perception by a heterotypic receptor complex. Nature 572, 270–274 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Campbell L., Turner S. R., A comprehensive analysis of RALF proteins in green plants suggests there are two distinct functional groups. Front. Plant Sci. 8, 37 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boisson-Dernier A., et al. , Disruption of the pollen-expressed FERONIA homologs ANXUR1 and ANXUR2 triggers pollen tube discharge. Development 136, 3279–3288 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miyazaki S., et al. , ANXUR1 and 2, sister genes to FERONIA/SIRENE, are male factors for coordinated fertilization. Curr. Biol. 19, 1327–1331 (2009). [DOI] [PubMed] [Google Scholar]
- 13.Barbez E., Dünser K., Gaidora A., Lendl T., Busch W., Auxin steers root cell expansion via apoplastic pH regulation in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 114, E4884–E4893 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gjetting K. S. K., Ytting C. K., Schulz A., Fuglsang A. T., Live imaging of intra- and extracellular pH in plants using pHusion, a novel genetically encoded biosensor. J. Exp. Bot. 63, 3207–3218 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kozma P., Hamori A., Cottier K., Kurunczi S., Horvath R., Grating coupled interferometry for optical sensing. Appl. Phys. B 97, 5–8 (2009). [Google Scholar]
- 16.Holm L., Sander C., Dali: A network tool for protein structure comparison. Trends Biochem. Sci. 20, 478–480 (1995). [DOI] [PubMed] [Google Scholar]
- 17.Morato do Canto A., et al. , Biological activity of nine recombinant AtRALF peptides: Implications for their perception and function in Arabidopsis. Plant Physiol. Biochem. 75, 45–54 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Hématy K., et al. , A receptor-like kinase mediates the response of Arabidopsis cells to the inhibition of cellulose synthesis. Curr. Biol. 17, 922–931 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Haruta M., Sabat G., Stecker K., Minkoff B. B., Sussman M. R., A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science 343, 408–411 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stegmann M., et al. , The receptor kinase FER is a RALF-regulated scaffold controlling plant immune signaling. Science 355, 287–289 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Gonneau M., et al. , Receptor Kinase THESEUS1 is a rapid alkalinization factor 34 receptor in Arabidopsis. Curr. Biol. 28, 2452–2458.e4 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Liu X., et al. , The role of LORELEI in pollen tube reception at the interface of the synergid cell and pollen tube requires the modified eight-cysteine motif and the receptor-like kinase FERONIA. Plant Cell 28, 1035–1052 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Capron A., et al. , Maternal control of male-gamete delivery in Arabidopsis involves a putative GPI-anchored protein encoded by the LORELEI gene. Plant Cell 20, 3038–3049 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li C., et al. , Glycosylphosphatidylinositol-anchored proteins as chaperones and co-receptors for FERONIA receptor kinase signaling in Arabidopsis. eLife 4, e06587 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pearce G., Yamaguchi Y., Munske G., Ryan C. A., Structure-activity studies of RALF, rapid alkalinization factor, reveal an essential–YISY–motif. Peptides 31, 1973–1977 (2010). [DOI] [PubMed] [Google Scholar]
- 26.Feijó J. A., Sainhas J., Hackett G. R., Kunkel J. G., Hepler P. K., Growing pollen tubes possess a constitutive alkaline band in the clear zone and a growth-dependent acidic tip. J. Cell Biol. 144, 483–496 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Messerli M. A., Danuser G., Robinson K. R., Pulsatile influxes of H+, K+ and Ca2+ lag growth pulses of Lilium longiflorum pollen tubes. J. Cell Sci. 112, 1497–1509 (1999). [DOI] [PubMed] [Google Scholar]
- 28.Pearce G., Moura D. S., Stratmann J., Ryan C. A. Jr, RALF, a 5-kDa ubiquitous polypeptide in plants, arrests root growth and development. Proc. Natl. Acad. Sci. U.S.A. 98, 12843–12847 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin G., et al. , A receptor-like protein acts as a specificity switch for the regulation of stomatal development. Genes Dev. 31, 927–938 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang X., et al. , Structural basis for receptor recognition of pollen tube attraction peptides. Nat. Commun. 8, 1331 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frederick R. O., et al. , Function and solution structure of the Arabidopsis thaliana RALF8 peptide. Protein Sci. 28, 1115–1126 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Monshausen G. B., Bibikova T. N., Messerli M. A., Shi C., Gilroy S., Oscillations in extracellular pH and reactive oxygen species modulate tip growth of Arabidopsis root hairs. Proc. Natl. Acad. Sci. U.S.A. 104, 20996–21001 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boisson-Dernier A., et al. , ANXUR receptor-like kinases coordinate cell wall integrity with growth at the pollen tube tip via NADPH oxidases. PLoS Biol. 11, e1001719 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaya H., et al. , Ca2+-activated reactive oxygen species production by Arabidopsis RbohH and RbohJ is essential for proper pollen tube tip growth. Plant Cell 26, 1069–1080 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Potocký M., Jones M. A., Bezvoda R., Smirnoff N., Zárský V., Reactive oxygen species produced by NADPH oxidase are involved in pollen tube growth. New Phytol. 174, 742–751 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Zhao C., et al. , Leucine-rich repeat extensin proteins regulate plant salt tolerance in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 115, 13123–13128 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boisson-Dernier A., Franck C. M., Lituiev D. S., Grossniklaus U., Receptor-like cytoplasmic kinase MARIS functions downstream of CrRLK1L-dependent signaling during tip growth. Proc. Natl. Acad. Sci. U.S.A. 112, 12211–12216 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Franck C. M., et al. , The protein phosphatases ATUNIS1 and ATUNIS2 regulate cell wall integrity in tip-growing cells. Plant Cell 30, 1906–1923 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cull M. G., Schatz P. J., Biotinylation of proteins in vivo and in vitro using small peptide tags. Meth. Enzymol. 326, 430–440 (2000). [DOI] [PubMed] [Google Scholar]
- 40.Fairhead M., Howarth M., Site-specific biotinylation of purified proteins using BirA. Methods Mol. Biol. 1266, 171–184 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smakowska-Luzan E., et al. , An extracellular network of Arabidopsis leucine-rich repeat receptor kinases. Nature 553, 342–346 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Futatsumori-Sugai M., Tsumoto K., Signal peptide design for improving recombinant protein secretion in the baculovirus expression vector system. Biochem. Biophys. Res. Comm. 391, 931–935 (2010). [DOI] [PubMed] [Google Scholar]
- 43.Hashimoto Y., Zhang S., Zhang S., Chen Y.-R., Blissard G. W., Correction: BTI-Tnao38, a new cell line derived from Trichoplusia ni, is permissive for AcMNPV infection and produces high levels of recombinant proteins. BMC Biotechnol. 12, 12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kabsch W., Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. D Biol. Crystallogr. 66, 133–144 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bunkóczi G., et al. , Phaser MRage: Automated molecular replacement. Acta Crystallogr. D Biol. Crystallogr. 69, 2276–2286 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Söding J., Biegert A., Lupas A. N., The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33, W244–W248 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stein N., CHAINSAW: A program for mutating pdb files used as templates in molecular replacement. J. Appl. Cryst. 41, 641–643 (2008). [Google Scholar]
- 48.Schwarzenbacher R., Godzik A., Grzechnik S. K., Jaroszewski L., The importance of alignment accuracy for molecular replacement. Acta Crystallogr. D Biol. Crystallogr. 60, 1229–1236 (2004). [DOI] [PubMed] [Google Scholar]
- 49.Adams P. D., et al. , PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Cryst. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 51.McCoy A. J., et al. , Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen V. B., et al. , MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pettersen E. F., et al. , UCSF Chimera: A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 54.Touw W. G., et al. , A series of PDB-related databanks for everyday needs. Nucleic Acids Res. 43, D364–D368 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wallace A. C., Laskowski R. A., Thornton J. M., LIGPLOT: A program to generate schematic diagrams of protein-ligand interactions. Protein Eng. Des. Sel. 8, 127–134 (1995). [DOI] [PubMed] [Google Scholar]
- 56.Laemmli U. K., Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 (1970). [DOI] [PubMed] [Google Scholar]
- 57.Clough S. J., Bent A. F., Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.