Significance
The development of K-Ras G12C covalent inhibitors suggests that this strategy could lead to therapeutic agents directly targeting Ras. Unfortunately, the overwhelming majority of Ras mutants and all Ral proteins do not have an accessible cysteine residue. Screening of a covalent fragment library containing aryl sulfonyl fluorides led to the discovery of one compound class that formed a covalent bond with noncatalytic residue Tyr-82. Remarkably, a high-resolution crystal structure at 1.2-Å resolution revealed that the compound created a deep hydrophobic pocket that was never previously observed in Ras or Ral GTPases. This binding pocket has a SiteMap DrugScore that is identical to druggable kinase ATP-binding pockets, suggesting that it could be used to develop therapeutics targeting oncogenic Ras lacking cysteine.
Keywords: Ras, covalent inhibitors, tyrosine, Ral
Abstract
Ral (Ras-like) GTPases are directly activated by oncogenic Ras GTPases. Mutant K-Ras (G12C) has enabled the development of covalent K-Ras inhibitors currently in clinical trials. However, Ral, and the overwhelming majority of mutant oncogenic K-Ras, are devoid of a druggable pocket and lack an accessible cysteine for the development of a covalent inhibitor. Here, we report that covalent bond formation by an aryl sulfonyl fluoride electrophile at a tyrosine residue (Tyr-82) inhibits guanine exchange factor Rgl2-mediated nucleotide exchange of Ral GTPase. A high-resolution 1.18-Å X-ray cocrystal structure shows that the compound binds to a well-defined binding site in RalA as a result of a switch II loop conformational change. The structure, along with additional high-resolution crystal structures of several analogs in complex with RalA, confirm the importance of key hydrogen bond anchors between compound sulfone oxygen atoms and Ral backbone nitrogen atoms. Our discovery of a pocket with features found on known druggable sites and covalent modification of a bystander tyrosine residue present in Ral and Ras GTPases provide a strategy that could lead to therapeutic agent targeting oncogenic Ras mutants that are devoid of a cysteine nucleophile.
Ral (Ras-like) GTPases were discovered while searching for RAS-related genes (1). Two Ral GTPases have been identified, RalA and RalB. Like Ras, Ral GTPases cycle between an active GTP-bound and an inactive GDP-bound complex (2). GTP-bound Ral binds to a range of effector proteins triggering signaling through pathways that control multiple cellular processes. Ral effector proteins include RalBP1 (Ral-binding protein 1)/RIP (Ral-interacting protein), Sec5, and exo84 (3, 4). Cycling between GDP- and GTP-bound Ral is facilitated by guanine exchange factors (GEFs) and guanine activating proteins (GAPs) through a standard mechanism that is shared by members of the Ras superfamily (2, 5). This process depends on the flexibility of two regions known as switch I (residues 41–51 in Ral GTPases) and switch II (residues 69–81 in Ral GTPases) (6). GAPs catalyze the hydrolysis of GTP to GDP, while GEFs promote GDP to GTP exchange by favoring conformational states of Ral that favor GTP binding (5). Four of these Ral GEFs, namely RalGDS, Rgl1, Rgl2, and Rgl3, possess a Ras exchange motif (REM), a CDC25 homology domain, and a Ras association domain (RA). All four RalGEFs interact with active GTP-bound Ral through their RA domain, thereby directly activating Ral GTPases and making Ral-RalGEF a major Ras signaling pathway along with PI3K and RAF.
There exists substantial evidence supporting a role for Ral GTPases in cancer that is both dependent and independent of Ras (7–16). Like Ras, there is intense interest in small-molecule Ral antagonists for the development of cancer therapeutics (17). However, the development of small-molecule Ral antagonists has been very challenging. Ral shares identical three-dimensional (3D) structure with Ras (18), and both apo and complex structures of Ral and Ras GTPases are devoid of druggable pockets. The development of small molecules that bind reversibly to Ral or Ras has been achieved for solvent-exposed and shallow pockets, but none of these compounds engage Ral or Ras GTPases at therapeutic doses (19–22). This challenge may have been recently overcome by the development of covalent Ras inhibitors (23, 24). Covalent inhibitors do not require deep hydrophobic pockets to engage a target as long as the reactive group of these compounds can rapidly form a covalent bond with an amino acid side chain. The presence of a cysteine residue in a rare K-Ras mutant (G12C), which occurs in 11–16% of lung adenocarcinomas and 1–4% of pancreatic and colorectal adenocarcinomas, presented a unique opportunity to develop small-molecule K-Ras covalent inhibitors (25–27). Despite their low affinity, these compounds have shown remarkable in vivo efficacy. Three compounds, Amgen AMG 510, Mirati MRTX849, and Janssen ARS-3248, are currently in clinical trials.
In the vast majority of Ras- or Ral-driven tumors, Ral and Ras GTPases do not have a cysteine residue that is amenable to covalent bond formation. However, covalent inhibitors, chemical tools, and approved drugs have been developed to form a bond with other residues, such as tyrosine, serine, lysine, histidine, and methionine (28–41). There are more than 50 covalent FDA-approved drugs, and most react at amino acids other than cysteine, such as penicillin, which uses a β-lactam reactive group to form a covalent bond with a serine residue of the penicillin-binding protein target (42), and the oncology drug bortezomib, which uses a boronic acid warhead to react at a threonine residue on the proteasome (43). Historically most Food and Drug Administration (FDA)-approved covalent inhibitors fall into the category of mechanism-based inhibitors, which correspond to compounds that form a covalent bond with an enzyme active-site catalytic residue (44), or targeted covalent inhibitors, which form a covalent bond with bystander or noncatalytic residues (29). Most of the recently approved covalent inhibitors, such as ibrutinib or afatinib, along with investigational compounds like the K-Ras inhibitors AMG 510, MRTX849, and ARS-3248, are targeted covalent inhibitors that form a covalent bond at bystander cysteine residues. However, several reactive groups have been developed for covalent bond formation at residues other than cysteine (30, 32, 35). Examples include S(VI)-containing groups sulfonyl fluorides, which react at tyrosine, lysine, or serine residues (30, 36, 44–47). Aryl sulfonyl fluorides provide useful tools to 1) identify amino acids that are amenable to covalent bond formation; 2) uncover new pockets that can be used in drug development; 3) provide starting points to develop derivatives with higher affinity and more suitable reactive groups. There are several tyrosine, lysine, and serine residues on Ral GTPases located at the interface between Ral GTPases and their GEFs or effector proteins. Among them is noncatalytic bystander residue Tyr-82, which, in K-Ras is equivalent to Tyr-71, is located near pockets that are the binding site of fragment and small molecules on Ral and Ras.
Here, we screen a library of aryl sulfonyl fluoride fragments to explore the suitability of Tyr-82 for the development of Ral covalent inhibitors. We report the discovery of a small-molecule series that form a covalent bond with Tyr-82 and inhibit Rlg2-mediated Ral nucleotide exchange. A 1.18-Å structure revealed that the compound binds to a binding site that was created as a result of a conformational change of the Ral switch II loop. Calculations reveal that the pocket has a druggability score that is comparable to known druggable binding sites. Preparation of several derivatives along with additional high-resolution structures afforded a limited structure–activity relationship study further confirming the existence of the pocket and the importance of key hydrogen bonding interactions.
Results
Small Molecules Inhibit Ral Nucleotide Exchange by Rgl2.
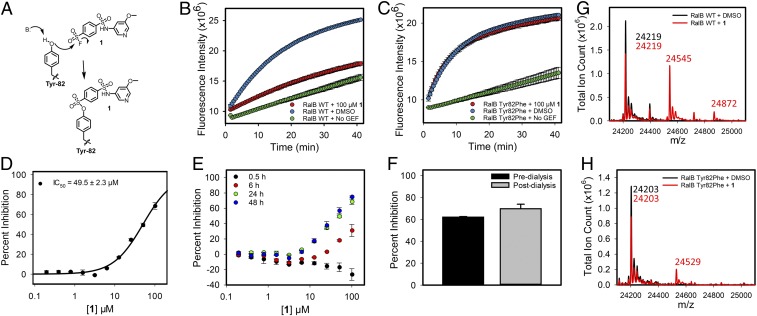
Analysis of the 3D nuclear magnetic resonance structures of RalB in complex with Ral effector protein RalBP1 (PDB ID code: 2KWI) shows the presence of a shallow but well-defined binding site occupied by RalBP1 Trp-430 (SI Appendix, Fig. S1A). In addition, the structure of Ral in complex with a Ral guanine exchange factor Rgl2 (PDB ID code: 5CM8) shows that this binding site is located at the Ral•Rgl2 protein-protein interface (SI Appendix, Fig. S1B). Although the binding site is well-defined, it is not sufficiently deep and hydrophobic to accommodate a small molecule that can engage Ral at therapeutic doses. As a result, we explored the possibility of developing a covalent inhibitor of Rgl2-mediated Ral activation. Although there are no accessible cysteine residues on Ral, there exists a tyrosine (Tyr-82) near the Trp-430 binding site that could provide an opportunity for covalent bond formation with an electrophile. To explore this possibility, we resorted to fragment-based screening using a library of 89 sulfonyl fluoride compounds. A fluorescence-based guanine nucleotide exchange assay was used to measure inhibition of Rgl2-mediated exchange of Ral-bound GDP with fluorescently labeled BODIPY-FL-GDP. The increase in fluorescence intensity of the BODIPY-FL group is measured at 30-s intervals. The exchange is initiated by the addition of Rgl2 and BODIPY-FL-GDP after the compound has been preincubated with Ral. Compound 1 (Fig. 1A) was identified to inhibit the Rgl2-mediated nucleotide exchange of RalB (Fig. 1B). To confirm that inhibition of RalB exchange by Rgl2 was due to covalent bond formation at Tyr-82, the nucleotide exchange study was repeated using RalB Tyr82Phe mutant. Circular dichroism (CD) spectrum of the RalB Tyr82Phe mutant and WT RalB were similar, indicating no significant change in secondary structure as a result of covalent bond formation (SI Appendix, Fig. S2 A and B). The RalB Tyr82Phe mutant showed robust nucleotide exchange by Rgl2 (Fig. 1C). The compound completely lost its ability to inhibit Rgl2-mediated nucleotide exchange of RalB Tyr82Phe mutant (Fig. 1C). A concentration-dependent exchange study was carried out to obtain the concentration of compound required for 50% inhibition of Rgl2-mediated exchange (Fig. 1D). This was done by incubating RalB with varying concentrations of 1 for 24 h at 4 °C, prior to the initiation of nucleotide exchange by the addition of Rgl2 and BODIPY-FL-GDP. The rate constant is calculated by fitting a three-parameter exponential function for each measurement. To determine percent inhibition, the fitting was also done for control dimethyl sulfoxide (DMSO) samples and samples without Rgl2. A plot of percent inhibition versus compound concentration resulted in an IC50 of 49.5 ± 2.3 µM following 24-h incubation at 4 °C (Fig. 1D).
Fig. 1.
Compound 1 inhibits RalB•Rgl2 interaction through covalent reaction at Tyr-82. (A) Proposed reaction mechanism of compound 1 with RalB Tyr-82. (B) Inhibition of Rgl2-mediated guanine nucleotide exchange of RalB by 100 µM 1 after 24-h incubation at 4 °C. (C) Inhibition of Rgl2-mediated guanine nucleotide exchange of RalB Tyr82Phe mutant by 100 µM 1 after 24-h incubation at 4 °C. (D) Concentration-dependent percent inhibition of Rgl2-mediated guanine nucleotide exchange of RalB after 24-h incubation with 1 at 4 °C (mean ± SD, n = 2). (E) Concentration-dependent percent inhibition of Rgl2-mediated guanine nucleotide exchange of RalB after 0.5, 6, 24, and 48 h of incubation with 1 at 4 °C (mean ± SD, n = 2). (F) Percent inhibition of Rgl2-mediated guanine nucleotide exchange of RalB by 100 µM 1 after 24-h incubation at 4 °C followed by 24-h dialysis against assay buffer at 4 °C (mean ± SD, n = 2). (G) Whole-protein ESI mass spectrometry analysis of 4 µM RalB incubated with 50 µM 1 or equivalent volume of DMSO for 12 h at 4 °C. (H) Whole-protein ESI mass spectrometry analysis of 4 µM RalB Tyr82Phe mutant incubated with 50 µM 1 or equivalent volume of DMSO for 12 h at 4 °C.
To further establish that the compound is a covalent inhibitor, a time-dependent study for Rgl2-mediated exchange of RalB nucleotide was carried out (Fig. 1E). At 30 min, there was no inhibition of exchange detected at the range of concentrations that were considered for the compound. At 6 h, the compound inhibited exchange with an IC50 of 153 ± 30 µM. At 24 h, even greater inhibition of RalB nucleotide exchange of Rgl2 was observed. No further increase in the extent of inhibition of the compound occurred from 24 to 48 h. The time-dependent inhibition of exchange confirms that 1 is a covalent inhibitor of RalB activation by Rgl2. Aryl sulfonyl fluorides are considered stable, but it is known that they are prone to hydrolysis over time. The rate of hydrolysis will depend on the physico-chemical properties of substituents on the aromatic ring, as well as the position of these substituents relative to the sulfonyl fluoride moiety. We expect that the hydrolysis of the sulfonyl fluoride warhead will affect the measured IC50 values over time. The effect on IC50 is expected to become larger with time as less compound is available for reaction with Ral GTPase due to hydrolysis. This may explain the lack of further decrease in the IC50 from 24 to 48 h.
Sulfonyl fluorides are considered irreversible inhibitors. The tyrosine oxygen is expected to form a covalent bond with the sulfur atom of the compound displacing the fluorine atom in a substitution reaction (Fig. 1A). Protein dialysis was used to establish that the inhibition of RalB by 1 is irreversible. RalB was incubated with compound for 24 h at 4 °C, followed by 24-h dialysis at 4 °C to remove the presence of excess compound. As shown in Fig. 1F, nucleotide exchange of RalB by Rgl2 was completely inhibited despite the absence of excess compound in solution, confirming that the compound is an irreversible covalent inhibitor of RalB.
Intact protein mass spectrometry was used to further establish that the compound forms a covalent bond with RalB at Tyr-82. Following incubation of RalB with 50 µM compound 1 for 12 h at 4 °C (Fig. 1G), a peak at m/z 24219 was observed corresponding to the RalB protein. Another peak at m/z 24545 (∆ = 326) corresponds to the covalent complex of RalB and 1, which has a molecular weight of 346 g/mol. The formed adduct reflects the fact that a fluorine atom from the compound (∆ = 19) and a hydrogen atom from the protein (∆ = 1) have been eliminated, resulting in a difference of 326. Another small peak at m/z 24872 (RalB + 653) is observed, which corresponds to a complex of RalB with two compound 1 molecules, indicating a secondary reaction site. The same experiment was repeated except that RalB Tyr82Phe was incubated with 1 for 12 h at 4 °C (Fig. 1H). The spectrum shows a peak at m/z 24203 corresponding to RalB Tyr82Phe. Due to the loss of reaction at Tyr-82, the major peak observed for WT RalB (Fig. 1G) is no longer present, and only a minor peak is observed at m/z 24529. These results further confirm that 1 forms a covalent adduct with RalB at Tyr-82. The formation of additional minor peaks may be due to the fact that Ral GTPases may have an additional lower affinity binding site for the fragment-like compounds with an electrophile positioned such that it can form a covalent bond with a sulfonyl fluoride. The fact that the second peak is much smaller than the first peak suggests that the binding mode is not very suitable for covalent bond formation, possibly because the reactive group is not well positioned for reaction, or due to the lower reactivity of the nucleophile. It is also possible that binding of our compound and covalent bond formation at Tyr-82 may change the conformation of RalB and create a new low affinity binding site that can accommodate the fragment compound and result in covalent bond formation. Regardless, the design of derivatives that take advantage of additional binding pockets near the major binding site of the compound will likely eliminate reaction at a secondary site.
High-Resolution Crystal Structures of Covalent Complex Reveals Well-Defined Pocket.
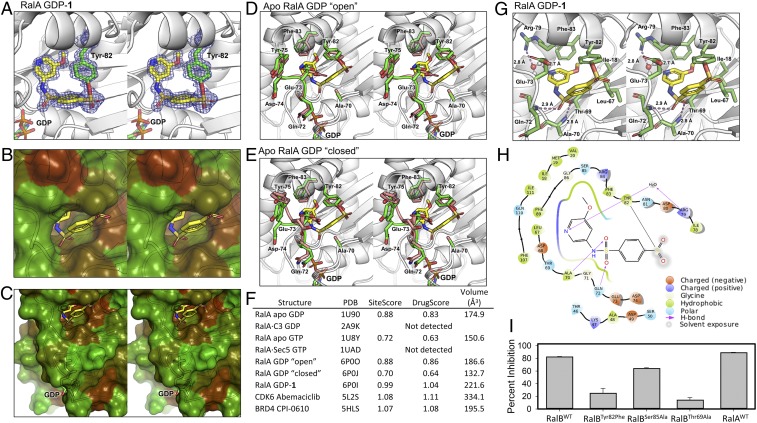
X-ray crystallography was used to determine the binding mode of 1 with Ral GTPase. Attempts to crystallize GDP-bound RalB did not yield quality crystals, while GDP-bound RalA readily crystallized to yield the first X-ray structure of a human RalA GTPase. Since there is high sequence identity between RalA and RalB, we soaked GDP-bound RalA crystals with 1, which led to a high resolution 1.18-Å structure of the covalent complex (SI Appendix, Table S1 and Fig. 2A). In addition to revealing the binding mode of 1, the clear electron density confirmed the presence of a covalent bond between the sulfone sulfur atom of 1 and the hydroxyl oxygen of Tyr-82, further establishing the existence of the covalent Ral–1 complex at Tyr-82. Remarkably, the compound created a well-defined and deep binding site within RalA (Fig. 2 B and C). This binding site is not present in any crystal structure of apo Ras or Ral GTPases or in complexes of these proteins with fragments and compounds.
Fig. 2.
Crystal structure of human RalA in covalent complex with 1. (A) Stereo image of the composite omit electron density map (blue mesh) of compound 1 (sticks with yellow carbon, red oxygen, blue nitrogen, and orange sulfur) covalently bound to RalA Tyr-82 (sticks with green carbon) (PDB ID code: 6P0I). (B) Stereo image of the binding pocket of 1 shown in solvent-accessible surface representation, colored by hydrophobicity (darker brown indicates more hydrophobicity). (C) Same as in B, zoomed out to show location of the pocket with respect to GDP. (D) Stereo image of RalA-1 binding pocket (carbons and loop in green) superimposed with apo RalA (carbons and loop in pink) where the switch II loop of apo RalA is in a conformation that has the pocket more open (PDB ID codes: 6P0I and 6P0O). (E) Stereo image of RalA-1 binding pocket (carbons and loop in green) superimposed with apo RalA (carbons and loop in pink) where the switch II loop of apo RalA is in a conformation that has the pocket closed (PDB ID codes: 6P0I and 6P0J). (F) Druggability scores and volume of representative RalA structures at the binding pocket of 1. Pocket scores and volumes were calculated by Sitemap (49). In comparison, the scores of a FDA-approved kinase inhibitor Abemaciclib and CPI-0610, a compound targeting the bromodomain BRD4 in clinical trials, are shown. (G) Stereo image of RalA–1 complex, highlighting the binding interactions between 1 (sticks with yellow carbons, blue nitrogens, red oxygens, orange sulfurs) and the RalA pocket (stick with green carbons). Hydrogen bonds are displayed in dashes and labeled with distance. (H) Two-dimensional ligand interaction map of covalently bound compound 1 in the pocket at the switch II loop of RalA generated using Maestro. (I) Percent inhibition of Rgl2-mediated guanine nucleotide exchange of RalB, RalB Tyr82Phe mutant, RalB Ser85Ala mutant, Thr69Ala mutant, and RalA by 100 µM 1 after 24-h incubation at 4 °C (mean ± SD, n = 2).
In addition to the RalA–1 complex, we solved two high-resolution crystal structures of human apo RalA (Fig. 2 D and E). These structures highlight the flexibility of the switch II loop, where the segment Ala-70 to Tyr-75 is present in more than one conformation. Data were collected from three different crystals resolutions of 1.55 Å, 1.54 Å (PDB ID code: 6P0O), and 1.31 Å (PDB ID code: 6P0J) to confirm this observation. In the three crystals, the electron density around the Ala-70 to Tyr-75 loop presents as at least two distinct conformations (“open” and “closed”) and are fitted for individual datasets. The “open” conformation of the loop (PDB ID code: 6P0O) is similar to the RalA–1 complex, except for Glu-73, which is flipped into the binding pocket in the apo structure (Fig. 2D). The “closed” conformation of the loop (PDB ID code: 6P0J) is different from the one observed in the RalA–1 complex (Fig. 2E).
The Schrödinger SiteMap program (48) was used to determine the druggability of the pocket occupied by 1 (Fig. 2F). In the apo RalA structures, the volume of the pocket ranges from 150 Å3 (PDB ID code: 1U8Y) to 187 Å3 (PDB ID code: 6P0O). In the RalA–1 complex, the pocket has a volume of 221 Å3. The SiteMap program also provides measures to assess ligand binding and druggability of a pocket known as SiteScore and DrugScore, respectively (49). These scores are calculated using the hydrophobicity and accessibility of a detected binding site. Unlike DrugScore, SiteScore limits the impact of hydrophilicity in charged and highly polar sites. A DrugScore of 1 or above suggests that a pocket possesses physico-chemical properties that are similar to pockets found on druggable sites. The ATP-binding pocket of kinases, which is the active site of many FDA-approved drugs (50, 51), has a DrugScore greater than 1 (49). For example, the ATP-binding pocket of CDK6 bound to the FDA-approved drug abemaciclib (PDB ID code: 5L2S) is 1.1. Another druggable pocket is the acetylated lysine recognition site on bromodomains (52). One example is the druggable pocket of the bromodomain BRD4 occupied by CPI-0610 (PDB ID code: 5HLS), a compound currently in clinical trials, has a DrugScore of 1.08. Similarly, the pocket on RalA that is occupied by compound 1 has a DrugScore of 1.04, suggesting that this pocket has physico-chemical characteristics of a druggable site. It is important to note that these scores do not take into consideration the potential energetic costs associated with the opening of the pocket that result from compound binding.
The RalA–1 structure shows that the compound is anchored by two hydrogen bond interactions between each of its sulfonamide oxygen atoms and backbone amide nitrogen atoms of Ala-70 and Gln-72 (Fig. 2G). These two residues are located on the flexible switch II loop region. In the open conformation structure of apo RalA (Fig. 2D), the backbone nitrogen atoms of Ala-70 and Gln-72 are well positioned to donate to the hydrogen bonds, indicating that the pocket is partially primed for compound 1. Interestingly, Glu-73 is found to have experienced substantial conformational change. In the apo structure, the side chain of the residue is located in the pocket occupied by our covalent inhibitors. In the complex, the side chain is flipped out of the pocket, and it is completely solvent exposed. Analysis of the structure of apo RalA reveals that Glu-73 does not make any favorable hydrogen bonding interactions with other residues. The conformational change experienced by Glu-73 is therefore likely to be thermodynamically favorable considering that increased exposure of the carboxylate moiety to solvent will result in improved solvation free energy. Interestingly, the methoxy group of 1 is located in a region that is occupied by the side chain of Phe-83. In the RalA–1 complex, the Phe-83 side chain rotates from its native orientation that is seen in the apo structure to accommodate the methoxy group of 1. The nitrogen atom of the pyridine ring of 1 forms a water-mediated hydrogen bond with the guanidinium ion of Arg-79. Finally, the compound engages several hydrophobic residues, including Ile-18, Val-20, Ala-48, Leu-67, and Phe-83, through van der Waals interactions (Fig. 2H).
Considering that the structure of 1 was solved with RalA, an exchange study was carried out to confirm that 1 also inhibits RalA, which is highly similar in overall structure to RalB and possesses more than 80% sequence similarity. As shown in Fig. 2I, compound 1 inhibited RalA and RalB to the same extent. To further probe the contribution of individual amino acids within the binding site of 1, the effect of the compound on the rate of nucleotide exchange was tested against two RalB mutants, Ser85Ala and Thr69Ala. Thr-69 comes in direct contact with the compound, while Ser-85 is located in the vicinity but does not come in contact with the compound. As expected, the Ser85Ala mutation did not affect the inhibition of RalB by 1. Thr69Ala mutation, however, which is within the binding pocket, substantially impaired the ability of the compound to inhibit RalB exchange (Fig. 2I).
Compound 1 Selectively Inhibits Ral over Ras.
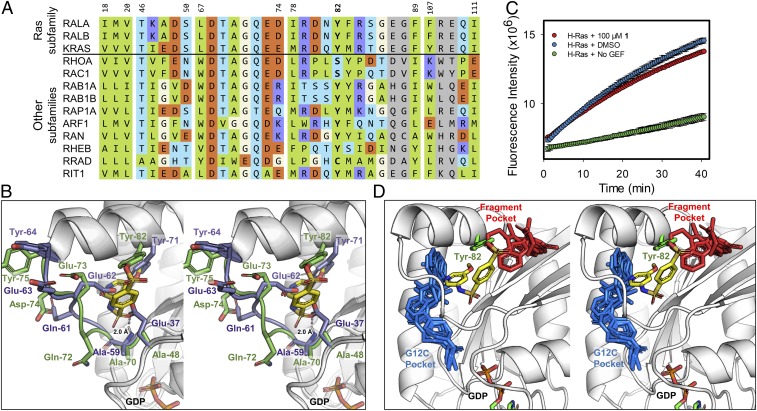
Ral and Ras GTPases have similar tertiary structures. Multiple sequence alignment of RalA and RalB to K-Ras as well as representative members of other GTPases in the Ras superfamily reveal similarities in the amino acid composition of the binding site of compound 1 (Fig. 3A). Superimposition of our RalA–1 complex with the structure of K-Ras shows that K-Ras, like Ral GTPases, possesses a tyrosine residue (Tyr-71) at the same position occupied by Ral Tyr-82 (Fig. 3B). In fact, the sequence alignment in Fig. 3A shows that this tyrosine is present in nearly all of the Ras superfamily GTPases shown in the figure, except for RhoA and Rac1 in the Rho subfamily. The presence of a tyrosine at position 82 suggests that compound 1 should form a covalent bond with K-Ras Tyr-71 and inhibit GEF nucleotide exchange of the GTPase. However, the structures also reveal some differences, such as the presence of a glutamic acid on K-Ras (Glu-37) instead of an alanine residue on Ral at the same position (Ala-48) (Fig. 3B). We tested whether compound 1 inhibited Son of Sevenless (SOS)-mediated nucleotide exchange of K-Ras using a similar fluorescence-based guanine nucleotide exchange assay that we developed for Ral. Compound 1 did not inhibit the SOS-mediated guanine-nucleotide exchange (Fig. 3C). Further examination of K-Ras crystal structures reveals the presence of a hydrogen bond between Glu-37 and the backbone nitrogen of Ala-59. As K-Ras Ala-59 is the equivalent of Ral Ala-70, and the interaction between the sulfone oxygen of 1 and the backbone of Ral Ala-70 is critical, the Glu-37 hydrogen bond may further reduce the ability of 1 to bind to this pocket on K-Ras. These results suggest that Glu-37, which protrudes into the binding site of 1, may block access to the pocket on K-Ras by compound 1 (Fig. 3B). Sequence alignment of representative Ras superfamily members indicates that glutamic acid or phenylalanine are prevalent at this position, while several other members have a glycine residue (Fig. 3A).
Fig. 3.
Compound 1 selectively inhibits Ral over Ras. (A) Multiple sequence alignment of representative members of the Ras superfamily to RalA. Residues are colored according to the scheme in Fig. 2H. Residue numbers are in reference to their respective position on RalA. Multiple sequence alignment was performed using Clustal Omega (v1.2.4) (61). (B) Stereo image of RalA–1 complex (green loops and carbons) superimposed with K-Ras (PDB ID code: 4EPV; purple loops and carbons). RalA residues are labeled in green and K-Ras residues are labeled in purple. (C) Inhibition of SOS-mediated guanine nucleotide exchange of K-Ras by 100 µM 1 after 24-h incubation at 4 °C (mean ± SD, n = 2). (D) Small molecule compounds bound to K-Ras are superimposed onto the binding mode of RalA-1. RalA is shown in white cartoon with the covalently bound RalA-Tyr82 and 1 compound shown as green and yellow sticks, respectively. Compounds that target the G12C mutation of K-Ras (blue sticks; PDB ID codes: 4M22, 5V6S, 5V9U, 5YXZ, 6N2J) and noncovalent fragments of K-Ras (red sticks; PDB ID codes: 4DSO, 4EPT, 4EPV, 4EPW, 4PZY) target adjacent binding pockets on K-Ras.
The binding pocket of compound 1 was compared to the binding sites of small molecules and fragments that were previously cocrystallized with K-Ras (Fig. 3D). Compounds shown in blue-capped sticks bind at the site of the G12C mutation on K-Ras. Compounds shown in red-capped sticks bind to a pocket that accommodates a Trp-430 of RalBP1 on Ral GTPases. The pocket created by our compounds is distinct from these two binding sites. It is located in the middle of the two most common binding sites of existing compounds that have been shown to bind to K-Ras (Fig. 3D). The pocket in our high-resolution structures is also different in its physico-chemical characteristics. It is deep and hydrophobic, while the other two pockets are solvent exposed.
Compound 1 Derivatives and Crystal Structures Confirm Covalent Complex and Binding Site.
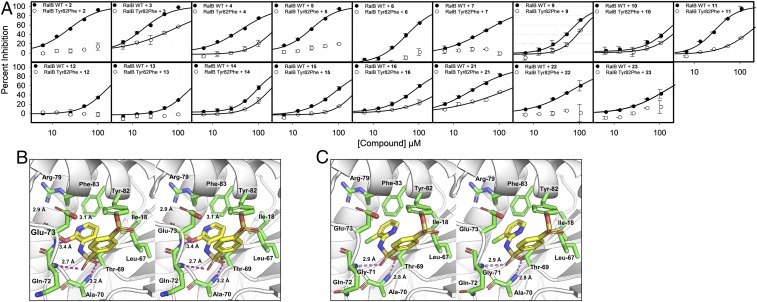
We designed and prepared several derivatives of 1 (SI Appendix, Table S2). The compounds were tested against WT RalB, RalB Tyr82Phe mutant, WT RalA, H-Ras, and K-Ras at a concentration of 50 µM with 24-h incubation at 4 °C (SI Appendix, Fig. S3). Generally, the compounds inhibited WT RalB and RalA with similar potency, while showing weaker inhibition of RalB Tyr82Phe mutant, H-Ras, and K-Ras exchange. Several compounds had substantially lower potency, such as 8, 17–19, and 20. As expected, 17, which was synthesized to confirm that adduct formation was essential to inhibition, did not inhibit any of the proteins. For compounds 18 and 19, the removal of the sulfonamide group lead to loss of inhibition (SI Appendix, Fig. S3). This sulfonamide moiety is crucial as it makes hydrogen bonds with the backbone nitrogen atoms of Ala-70 and Gln-72 on RalB. The fluorosulfate derivative of 1, namely 20, also did not inhibit Ral exchange. This is attributed to the shift in the position of the sulfur due to presence of an additional oxygen atom as well as the low intrinsic reactivity of the fluorosulfate reactive group (30). All other compounds showed significant inhibition of WT RalA and RalB and were tested in a concentration-dependent manner to determine their IC50 at 24 h (Fig. 4A and SI Appendix, Table S2).
Fig. 4.
Derivatives of 1. (A) Concentration-dependent percent inhibition of Rgl2-mediated guanine nucleotide exchange of RalB and RalB Tyr82Phe mutant after 24 h incubation at 4 °C with active derivatives of 1 (mean ± SD, n = 2). (B) Stereo image of RalA–2 complex, highlighting the binding interactions between 2 (sticks with yellow carbons, blue nitrogens, red oxygens, orange sulfurs) and the RalA pocket (stick with green carbons). Hydrogen bonds are displayed in dashes and labeled with distance (PDB ID code: 6P0L). (C) Stereo image of RalA–4 complex, highlighting the binding interactions between 4 (sticks with yellow carbons, blue nitrogens, red oxygens, orange sulfurs) and the RalA pocket (stick with green carbons). Hydrogen bonds are displayed in dashes and labeled with distance (PDB ID code: 6P0M).
Compound 2 was a better inhibitor of WT RalB exchange than 1. Compound 2 showed no inhibition of the RalB Tyr82Phe mutant. A crystal structure of 2 in complex with RalA was determined at 1.30-Å resolution (PDB ID code: 6P0L; SI Appendix, Table S1); the electron density clearly identified the binding mode of the compound (SI Appendix, Fig. S4A). The binding mode of 2 reveals that the compound preserves the interaction from the sulfone to the backbone amides of Ala-70 and Gln-72 (Fig. 4B). As the methoxy group is moved from the meta position to the ortho position, the clash with Phe-83 does not occur. In addition, a hydrogen bond to the backbone amide of Glu-73 is established with the methoxy group at the ortho position. This additional hydrogen bond may explain the twofold improvement in IC50 at 24 h for compound 2 (SI Appendix, Table S2). Compound 3, which features a hydroxyl moiety at the ortho position, was a strong inhibitor of wild-type RalB, with an IC50 of 17.0 ± 7.7 µM at 24 h. However, this improvement in inhibition was accompanied by inhibition of the Tyr82Phe mutant, as well as K-Ras, perhaps indicating off-target reactions.
Compound 4 has a similar IC50 to 1 of 41.7 ± 8.5 µM. An X-ray crystal structure of 4 in complex with RalA was determined at 1.50 Å (SI Appendix, Table S1 and Fig. 4C). The compound was clearly present in the electron density (SI Appendix, Fig. S4B). Compound 4 lacks the methoxy group present in 1. Compound 5 and 6 possess halogen atoms at the meta position, replacing the methoxy group of 1. The chlorine derivative 5 showed a twofold improvement in IC50 compared to 1 (IC50 = 24.4 ± 4.9 µM), while the fluorine derivative 6 had similar IC50 to 1 (51.5 ± 5.2 µM). Crystal structures of RalA-5 and RalA-6 were determined at 1.49 Å and 1.63 Å, respectively (SI Appendix, Table S1). The compounds were clearly present in the electron density (SI Appendix, Fig. S4 C and D). Structures of the bound compounds reveal that the hydrogen bonds with the sulfonamide are maintained and the clash with Phe-83 is alleviated (SI Appendix, Fig. S4 E and F). Both the chlorine of 5 or the fluorine of 6 are located within 3.6–5.0 Å of residues Ile-18, Leu-67, Thr-69, and Phe-83. The chlorine being better suited for van der Waals interaction likely enhances the potency of 5. An additional nitrogen in 7 negates the twofold improvement gained in compound 5. Modification of 5 to include the m-methoxy group of the parent 1, yielded 21, which had no significant improvement in inhibition with an IC50 of 24.0 ± 1.0 µM, while inhibition of RalB Tyr82Phe increased at higher concentrations of compound. Switching the chlorine of 5 to a methoxy group in 22 resulted in weakening of the IC50 to 64.0 ± 4.1 µM. Further modification of 22, with a chlorine at the ortho position in 23, reduced its inhibition even further, leading to an IC50 of 127 ± 11.7 µM.
Substitutions at the para position has detrimental effects to the IC50, probably due to clashes with Phe-83. This is seen in compounds 8-10, 12, and 13. In the case of 12, the change to a nitrogen atom may make the moiety unsuitable in the hydrophobic pocket. The modification of the methoxy group at the meta position in compounds 14–16 resulted in weaker inhibition compared to 1. The larger and more hydrophobic moieties likely result in increased clashes in the pocket.
Discussion
Ral (Ras-like) GTPases are directly activated by Ras GTPases through RalGEFs. Like Ras, binding sites on Ral are shallow and do not have the combination of size, hydrophobic, and hydrophilic characteristics that are required to engage a drug at therapeutic doses. One strategy to overcome the problem of a lack of druggable pockets is to develop covalent inhibitors. Our analysis of Ral and Ras crystal structures in complex with their effector or GEFs revealed the presence of a tyrosine residue on Ral (Tyr-82) and Ras (Tyr-71) at the protein-protein interface. In addition, this tyrosine is located near a binding pocket that accommodates a tryptophan from RalBP1 and small-molecule fragments on K-Ras (53). We hypothesized that small molecules that form a covalent bond at Tyr-82 could disrupt GEF-mediated activation of Ral GTPases. To test our hypothesis, we screened a library of aryl sulfonyl fluoride library to identify potential candidates that would form a covalent bond with Tyr-82. Aryl sulfonyl fluorides have been extensively used to probe tyrosine and lysine residues in proteins (54). Although these compounds are not considered suitable warheads for drugs, they have served as valuable probes to uncover reactive groups and binding sites. Our goal in this work was to determine if Tyr-82 was available for adduct formation and whether the binding sites near the tyrosine were accessible to small molecules for drug development targeting Ral and Ras. The discovery of 1 and derivatives confirmed that Tyr-82 was amenable to adduct formation.
We did not expect that these compounds would uncover a well-defined pocket on Ral GTPases with physico-chemical characteristics that are similar to pockets found on druggable targets. This binding site is not present in any crystal structure of apo Ras or Ral GTPases or in complexes of these proteins with fragments and compounds. Our expectation was that these compounds would occupy the binding pocket of the tryptophan of RalPB1, where several fragments were identified to bind on the same pocket on K-Ras. The apo structure of RalA confirms that the switch II region of Ral is highly flexible, which is also the case among most members of the Ras and Rho GTPase families. Without compound 1, it is unlikely that the pocket would have been identified. The chemical structure of the compound is also likely another reason for the discovery of the pocket. Our structure-activity study based on synthesis of several derivatives and several high-resolution X-ray structures reveals the importance of two key hydrogen-bonding interactions by the sulfonamide oxygen atoms with backbone nitrogen atoms of Ral switch II residues Ala-70 and Gln-72.
Numerous screening campaigns have likely been carried out in the past to identify small-molecule inhibitors of Ral and Ras nucleotide exchange. The most likely reason that these efforts have not identified the binding site reported in this work is that they either screened noncovalent libraries, or in cases where covalent libraries were screened, none used electrophiles that are capable of forming covalent bonds with a tyrosine. It may be possible for a noncovalent compound to bind to the binding site, but such a compound would have to 1) bind at much higher affinity than the fragments we used in this work; and 2) possess suitable binding mode to create the critical hydrogen bonding interactions that were observed between inhibitors reported in this work. To achieve high-affinity, noncovalent inhibitors must bind deeper into the pocket and extend into neighboring G23 binding pocket (equivalent to the G12 binding site on K-Ras). It is possible that none of the screens carried out in the past were diverse enough to contain such compounds. Our work uncovered the pocket because we used covalent fragments. Furthermore, the reactive group that we used here was specifically chosen to react with tyrosine, in an attempt to identify small-molecule Ral or Ras inhibitors that form a covalent bond at a tyrosine, specifically Tyr-82 (Tyr-71 on K-Ras), which is located near the compound binding site that we identified. The formation of a covalent bond at Tyr82 created an anchor that trapped the covalent complex and compensated for the low affinity of the fragments.
The discovery that Tyr-82 is accessible for covalent modification and the presence of a druggable pocket near the residue could have profound implications for the development of therapeutic agents targeting the Ras signaling pathway. The binding mode of 1 and its derivatives provides a strategy to develop Ral GTPase antagonists that can lead to therapeutic agents targeting the Ras signaling pathway. First, the reactive group must exhibit greater stability in buffer as well as plasma and microsomes to be suitable for animal studies. One strategy to stabilize the sulfonyl fluoride reactive moiety, which is known to be prone to hydrolysis, is to introduce substituents on the aromatic ring ortho, meta, or para to the reactive group, which could reduce the electrophilic character of the sulfone, making it less prone to hydrolysis and nonspecific reactions. Another strategy is to replace the sulfonyl fluoride with a more stable moiety such as a fluorosulfate group, which is considered to be much more stable. Second, the binding affinity of the compound can be improved. This can be accomplished through a standard medicinal chemistry approach by adding substituents on the compound to enhance its binding affinity to RalB or by modifying its core structure. Finally, it is important that a covalent inhibitor possess a favorable binding constant (KI) and larger inactivation rate constant (kinact). Generally, the second-order rate constant kinact/KI is considered to be the most important parameter to guide compound optimization. A covalent inhibitor with a cellular IC50 under 1 µM and a 4-h occupancy time-point could be expected to have a kinact/KI of ∼100 M−1⋅s−1. Our fragment-based covalent inhibitors have an IC50 of ∼20 µM at a 24-h time point, which will require a 120-fold improvement in kinact/KI to achieve this level of potency (44). Physiologically relevant values above 1,000 M−1⋅s−1, with sufficiently optimized kinact values, can be good candidates for in vivo experiments (55).
Methods
Protein Expression and Purification.
Details for protein expression and purification of RGL2 (50-514), His-RalA (1-178), His-RalB (12-185), His-H-Ras, His-K-Ras, and HIS-SOS-cat (564-1049) are provided in SI Appendix.
Exchange Assay.
Ten microliters of RalB (2 µM), RalA (2 µM), and Ras (0.8 µM) in an exchange assay buffer (100 mM NaCl, 10 mM MgCl2, 20 mM Tris pH 8, 0.01% IGEPAL CA-630 Sigma, catalog no. I8896) were added to a 384-well, black, round bottom, low volume, polystyrene plate (Corning, catalog no. 4515) and incubated with 2 µL of varying concentrations of compounds in the exchange assay buffer supplemented with 20% vol/vol DMSO for 24 h (unless otherwise specified) at 4 °C. After the incubation, 5 µL of RGL2 (1 µM), SOS (0.4 µM), or buffer was added. Finally, 3 µL of Bodipy-FL-GDP (1.67 µM for RalA/B, 1.0 µM for Ras) was added and the fluorescence was read immediately on an Envision Multilabel Plate Reader (PerkinElmer) using a filter set with excitation and emission wavelengths of 485 and 535 nm, respectively, for 40 min at 30-s intervals. The fluorescence increase over time was fitted to an exponential function:
The rate constant of the exchange was calculated by fitting experimental values for fluorescence intensity and corresponding time. The Initial Fluorescence was estimated from the initial reading of the fluorescence intensity from the experimental control sample without guanine exchange factor (GEF). The extent of binding is the difference between the maximal fluorescence intensity of the DMSO control sample versus the initial fluorescence recorded for the No GEF control sample. Percent inhibition was calculated by comparing the rate constant of the compound inhibited sample versus the maximal DMSO control and the minimal control without GEF. Based on the plot of the percent inhibition versus compound concentration, a four-parameter logistic curve was fit to determine the IC50 values at 24-h incubation time.
Percent inhibition and compound concentration are experimental values. Maximum inhibition is set at 100% as no plateau were achieved. Minimum inhibition value was data-dependent and were mostly found to be near 0%. Due to the fact that the compounds were covalent inhibitors and not classical reversible inhibitors, hillslope value was not constrained.
Protein Mass Spectrometry.
Compounds were incubated with 5 µM RGL2 or Tyr82Phe mutant in buffer (100 mM NaCl, 20 mM Tris pH 8.0, 10 mM MgCl2, 2% DMSO) for 24 h (unless otherwise specified) at 4 °C. After the incubation, the samples were centrifuged at 20,000 × g for 10 min to remove precipitants prior to being injected into a Zorbax 300-SB C3 column (Agilent) on an Agilent 1200 liquid chromatography system (Agilent), using a gradient of buffer A (H2O with 0.1% formic acid) and buffer B (acetonitrile with 0.1% formic acid), and the masses were detected on an Agilent 6520 Accurate Mass Q-TOF.
Compound Stability Assay.
Two hundred micromolar compound 1 was incubated in a buffer containing 20 mM Tris pH 8.0, 100 mM NaCl, 10 mM MgCl2 at 4 °C for varying amounts time. After the incubation, the samples were centrifuged at 20,000 × g for 10 min to remove precipitants prior to being injected into an Agilent EclipsePlus C18 RRHD column (Agilent) on an Agilent 1200 liquid chromatography system (Agilent), using a linear gradient from 100% buffer A (H2O with 0.1% formic acid) to 70% buffer B (acetonitrile with 0.1% formic acid), and the masses were detected on an Agilent 6520 Accurate Mass Q-TOF.
Crystallization and Structure Determination.
RalA.GDP crystals were grown using the hanging-drop vapor-diffusion method with a drop containing 20–25 mg/mL RalA•GDP and reservoir solution (0.2 M calcium acetate pH 5.5 and 18–22% polyethylene glycol 3350) at 20 °C. The crystals appeared after 2 d. RalA–inhibitor complexes were obtained by soaking the crystals overnight in reservoir solution supplemented with 2- to 5-mM compounds. Crystals were harvested and cryo-protected in reservoir solutions supplemented with 20% glycerol or a mix of 10% glycerol and 10% ethylene glycol prior to being flash-cooled in liquid nitrogen. Diffraction data were collected at 100 K at the Beamline station 4.2.2 at the Advanced Light Source (ALS; Berkeley National Laboratory, CA) and were indexed, integrated, and scaled using XDS (56). The structure was solved by molecular replacement using PHASER and the simian RalA model (PDB ID code: 1U8Z). The Autobuild function was used to generate a first model that was improved by iterative cycles of manual building in Coot (57) and refinement using PHENIX (58). MolProbity software (59) was used to assess the geometric quality of the models, and PyMOL (version 2.3.1) was used to generate molecular images. Data collection and refinement statistics are indicated in SI Appendix, Table S1.
Single crystals were used to obtain a complete dataset for each RalA-compound complex. In the case of apo RalA, data from three different crystals were collected and analyzed individually (crystal 1 at resolution 1.55 Å, crystal 2 at 1.54 Å [PBD ID code: 6P0O] and crystal 3 at 1.31 Å [PDB ID code: 6P0J]). For simplicity, two models (PDB ID code: 6P0O “open” and PDB ID code: 6P0J “closed” conformation) representing two distinct conformations were deposited.
Computational Analysis of Binding Sites.
Binding sites were identified and scored as previously described (60). Details of the computational analysis are provided in SI Appendix.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant R01CA197928 (to S.O.M.). We acknowledge use of the Macromolecular Crystallography Facility at the Molecular and Cellular Biochemistry Department, Indiana University Bloomington. We also thank Jay Nix for his assistance during X-ray data collection at beamline 4.2.2, Advanced Light Source (ALS) and Dr. John Strelow for helpful discussions.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, https://www.wwpdb.org/ (PDB ID codes 6P0I, 6P0J, 6P0K, 6P0L, 6P0M, 6P0N, and 6P0O).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1913654117/-/DCSupplemental.
References
- 1.Chardin P., Tavitian A., The ral gene: A new ras related gene isolated by the use of a synthetic probe. EMBO J. 5, 2203–2208 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cherfils J., Zeghouf M., Chronicles of the GTPase switch. Nat. Chem. Biol. 7, 493–495 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Cantor S. B., Urano T., Feig L. A., Identification and characterization of Ral-binding protein 1, a potential downstream target of Ral GTPases. Mol. Cell. Biol. 15, 4578–4584 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Issaq S. H., Lim K. H., Counter C. M., Sec5 and Exo84 foster oncogenic ras-mediated tumorigenesis. Mol. Cancer Res. 8, 223–231 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cherfils J., Zeghouf M., Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol. Rev. 93, 269–309 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Vetter I. R., Wittinghofer A., The guanine nucleotide-binding switch in three dimensions. Science 294, 1299–1304 (2001). [DOI] [PubMed] [Google Scholar]
- 7.Saito R., et al. , Downregulation of Ral GTPase-activating protein promotes tumor invasion and metastasis of bladder cancer. Oncogene 32, 894–902 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Guin S., et al. , Contributions of KRAS and RAL in non-small-cell lung cancer growth and progression. J. Thorac. Oncol. 8, 1492–1501 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Male H., et al. , Inhibition of RalA signaling pathway in treatment of non-small cell lung cancer. Lung Cancer 77, 252–259 (2012). [DOI] [PubMed] [Google Scholar]
- 10.Rybko V. A., et al. , Different metastasis promotive potency of small G-proteins RalA and RalB in in vivo hamster tumor model. Cancer Cell Int. 11, 22 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spiczka K. S., Yeaman C., Ral-regulated interaction between Sec5 and paxillin targets Exocyst to focal complexes during cell migration. J. Cell Sci. 121, 2880–2891 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith S. C., et al. , The metastasis-associated gene CD24 is regulated by Ral GTPase and is a mediator of cell proliferation and survival in human cancer. Cancer Res. 66, 1917–1922 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Singhal S. S., Awasthi Y. C., Awasthi S., Regression of melanoma in a murine model by RLIP76 depletion. Cancer Res. 66, 2354–2360 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Lim K.-H., et al. , Divergent roles for RalA and RalB in malignant growth of human pancreatic carcinoma cells. Curr. Biol. 16, 2385–2394 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Lim K.-H., et al. , Activation of RalA is critical for Ras-induced tumorigenesis of human cells. Cancer Cell 7, 533–545 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Gildea J. J., Harding M. A., Seraj M. J., Gulding K. M., Theodorescu D., The role of Ral A in epidermal growth factor receptor-regulated cell motility. Cancer Res. 62, 982–985 (2002). [PubMed] [Google Scholar]
- 17.Yan C., Theodorescu D., RAL GTPases: Biology and potential as therapeutic targets in cancer. Pharmacol. Rev. 70, 1–11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nicely N. I., Kosak J., de Serrano V., Mattos C., Crystal structures of Ral-GppNHp and Ral-GDP reveal two binding sites that are also present in Ras and Rap. Structure 12, 2025–2036 (2004). [DOI] [PubMed] [Google Scholar]
- 19.Shima F., et al. , In silico discovery of small-molecule Ras inhibitors that display antitumor activity by blocking the Ras-effector interaction. Proc. Natl. Acad. Sci. U.S.A. 110, 8182–8187 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maurer T., et al. , Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity. Proc. Natl. Acad. Sci. U.S.A. 109, 5299–5304 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patgiri A., Yadav K. K., Arora P. S., Bar-Sagi D., An orthosteric inhibitor of the Ras-Sos interaction. Nat. Chem. Biol. 7, 585–587 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan C., et al. , Discovery and characterization of small molecules that target the GTPase Ral. Nature 515, 443–447 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Janes M. R., et al. , Targeting KRAS mutant cancers with a covalent G12C-specific inhibitor. Cell 172, 578–589.e17 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Ostrem J. M., Peters U., Sos M. L., Wells J. A., Shokat K. M., K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature 503, 548–551 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bailey M. H., et al. , Comprehensive characterization of cancer driver genes and mutations. Cell 173, 371–385.e18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lito P., Solomon M., Li L. S., Hansen R., Rosen N., Allele-specific inhibitors inactivate mutant KRAS G12C by a trapping mechanism. Science 351, 604–608 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ostrem J. M., Shokat K. M., Direct small-molecule inhibitors of KRAS: From structural insights to mechanism-based design. Nat. Rev. Drug Discov. 15, 771–785 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Vasudevan A., et al. , Covalent binders in drug discovery. Prog. Med. Chem. 58, 1–62 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Ray S., Murkin A. S., New electrophiles and strategies for mechanism-based and targeted covalent inhibitor design. Biochemistry 58, 5234–5244 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Martín-Gago P., Olsen C. A., Arylfluorosulfate-based electrophiles for covalent protein labeling: A new addition to the Arsenal. Angew. Chem. Int. Ed. Engl. 58, 957–966 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ghosh A. K., Samanta I., Mondal A., Liu W. R., Covalent inhibition in drug discovery. ChemMedChem 14, 889–906 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gehringer M., Laufer S. A., Emerging and Re-emerging warheads for targeted covalent inhibitors: Applications in medicinal chemistry and chemical biology. J. Med. Chem. 62, 5673–5724 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Zhao Q., et al. , Broad-spectrum kinase profiling in live cells with lysine-targeted sulfonyl fluoride probes. J. Am. Chem. Soc. 139, 680–685 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.So W. H., et al. , Site-selective covalent reactions on proteinogenic amino acids. Curr. Opin. Biotechnol. 48, 220–227 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Pettinger J., Jones K., Cheeseman M. D., Lysine-targeting covalent inhibitors. Angew. Chem. Int. Ed. Engl. 56, 15200–15209 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Mortenson D. E., et al. , “Inverse drug discovery” strategy to identify proteins that are targeted by latent electrophiles as exemplified by aryl fluorosulfates. J. Am. Chem. Soc. 140, 200–210 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Long M. J. C., Aye Y., Privileged electrophile sensors: A resource for covalent drug development. Cell Chem. Biol. 24, 787–800 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lagoutte R., Patouret R., Winssinger N., Covalent inhibitors: An opportunity for rational target selectivity. Curr. Opin. Chem. Biol. 39, 54–63 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Jackson P. A., Widen J. C., Harki D. A., Brummond K. M., Covalent modifiers: A chemical perspective on the reactivity of α,β-Unsaturated carbonyls with thiols via Hetero-Michael addition reactions. J. Med. Chem. 60, 839–885 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De Cesco S., Kurian J., Dufresne C., Mittermaier A. K., Moitessier N., Covalent inhibitors design and discovery. Eur. J. Med. Chem. 138, 96–114 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Awoonor-Williams E., Walsh A. G., Rowley C. N., Modeling covalent-modifier drugs. Biochim. Biophys. Acta (BBA) Proteins Proteomics 1865, 1664–1675 (2017). [DOI] [PubMed] [Google Scholar]
- 42.Nicola G., Tomberg J., Pratt R. F., Nicholas R. A., Davies C., Crystal structures of covalent complexes of β-lactam antibiotics with Escherichia coli penicillin-binding protein 5: Toward an understanding of antibiotic specificity. Biochemistry 49, 8094–8104 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Groll M., Berkers C. R., Ploegh H. L., Ovaa H., Crystal structure of the boronic acid-based proteasome inhibitor bortezomib in complex with the yeast 20S proteasome. Structure 14, 451–456 (2006). [DOI] [PubMed] [Google Scholar]
- 44.Powers J. C., Asgian J. L., Ekici Ö. D., James K. E., Irreversible inhibitors of serine, cysteine, and threonine proteases. Chem. Rev. 102, 4639–4750 (2002). [DOI] [PubMed] [Google Scholar]
- 45.Fadeyi O. O., et al. , Covalent enzyme inhibition through fluorosulfate modification of a noncatalytic serine residue. ACS Chem. Biol. 12, 2015–2020 (2017). [DOI] [PubMed] [Google Scholar]
- 46.Hett E. C., et al. , Rational targeting of active-site tyrosine residues using sulfonyl fluoride probes. ACS Chem. Biol. 10, 1094–1098 (2015). [DOI] [PubMed] [Google Scholar]
- 47.Zheng Q., et al. , SuFEx-enabled, agnostic discovery of covalent inhibitors of human neutrophil elastase. Proc. Natl. Acad. Sci. U.S.A. 116, 18808–18814 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Halgren T., New method for fast and accurate binding-site identification and analysis. Chem. Biol. Drug Des. 69, 146–148 (2007). [DOI] [PubMed] [Google Scholar]
- 49.Halgren T. A., Identifying and characterizing binding sites and assessing druggability. J. Chem. Inf. Model. 49, 377–389 (2009). [DOI] [PubMed] [Google Scholar]
- 50.Ferguson F. M., Gray N. S., Kinase inhibitors: The road ahead. Nat. Rev. Drug Discov. 17, 353–377 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Zhang J., Yang P. L., Gray N. S., Targeting cancer with small molecule kinase inhibitors. Nat. Rev. Cancer 9, 28–39 (2009). [DOI] [PubMed] [Google Scholar]
- 52.Ferri E., Petosa C., McKenna C. E., Bromodomains: Structure, function and pharmacology of inhibition. Biochem. Pharmacol. 106, 1–18 (2016). [DOI] [PubMed] [Google Scholar]
- 53.Sun Q., et al. , Discovery of small molecules that bind to K-Ras and inhibit Sos-mediated activation. Angew. Chem. Int. Ed. Engl. 51, 6140–6143 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Narayanan A., Jones L. H., Sulfonyl fluorides as privileged warheads in chemical biology. Chem. Sci. (Camb.) 6, 2650–2659 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Strelow J. M., A perspective on the kinetics of covalent and irreversible inhibition. SLAS Discov. 22, 3–20 (2017). [DOI] [PubMed] [Google Scholar]
- 56.Kabsch W., XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Adams P. D., et al. , PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen V. B., et al. , MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu D., Jalal S. I., Sledge G. W., Meroueh S. O., Small-molecule binding sites to explore protein-protein interactions in the cancer proteome. Mol. Biosyst. 12, 3067–3087 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sievers F., et al. , Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.