Significance
Despite sharing the first four reactions, coutilization of the Wood–Ljungdahl pathway (WLP) with the glycine synthase-reductase pathway (GSRP) and reductive glycine pathway (RGP) to fix C1 compounds has remained unknown. In this study, using Clostridium drakei, we elucidated the role of the GSRP and RGP in the presence of the WLP, via a genome-scale metabolic model, RNA-seq, 13C isotope-based metabolite-tracing experiments, biochemical assays, and heterologous expression. Overall, the data suggested the pathways are functional under autotrophic conditions. Along with the WLP, GSRP and RGP convert CO2 to glycine and then to acetyl-phosphate and serine, which then obtain ATP by producing acetate and operate with limited reducing power. This is a unique coutilization of the pathways under autotrophic conditions in acetogens.
Keywords: CO2 fixation, acetogen, Wood–Ljungdahl pathway, systems biology, glycine synthase-reductase pathway
Abstract
Among CO2-fixing metabolic pathways in nature, the linear Wood–Ljungdahl pathway (WLP) in phylogenetically diverse acetate-forming acetogens comprises the most energetically efficient pathway, requires the least number of reactions, and converts CO2 to formate and then into acetyl-CoA. Despite two genes encoding glycine synthase being well-conserved in WLP gene clusters, the functional role of glycine synthase under autotrophic growth conditions has remained uncertain. Here, using the reconstructed genome-scale metabolic model iSL771 based on the completed genome sequence, transcriptomics, 13C isotope-based metabolite-tracing experiments, biochemical assays, and heterologous expression of the pathway in another acetogen, we discovered that the WLP and the glycine synthase pathway are functionally interconnected to fix CO2, subsequently converting CO2 into acetyl-CoA, acetyl-phosphate, and serine. Moreover, the functional cooperation of the pathways enhances CO2 consumption and cellular growth rates via bypassing reducing power required reactions for cellular metabolism during autotrophic growth of acetogens.
The linear Wood–Ljungdahl pathway (WLP) in anaerobic acetogens is considered the most energetically efficient pathway to convert CO2 to formate and then into acetyl-CoA. With this advantage, acetogens are considered to be the most promising industrial platform to produce biofuels and chemical commodities through synthesis gas fermentation (1–4). Although gene composition and arrangement of the WLP vary among acetogens, the WLP-coding genes are well-conserved, along with two genes encoding a partial glycine synthase: the glycine cleavage system H protein (gcvH) and dihydrolipoyl dehydrogenase (lpdA) genes (5–8). The glycine synthase pathway was initially proposed for the utilization of CO2 under autotrophic growth conditions (2). While the two genes are well-conserved in the gene cluster, other genes in the glycine synthase pathway are missing in many acetogen genomes, which raises questions regarding a potential functional role of these enzymes under autotrophic growth conditions. Following synthesis, glycine can be reduced to acetyl-phosphate (acetyl-P), which is likely to be converted into acetate by acetate kinase (ackA), thereby producing one ATP, termed the glycine synthase-reductase pathway (GSRP). Alternatively, serine hydroxymethyltransferase (SHMT) converts the produced glycine to serine, which then becomes transformed to pyruvate and biomass (9–11). Recently, an artificial metabolic pathway constructed with glycine synthase and SHMT, termed the reductive glycine pathway (RGP), has shown the capability of fixing CO2 using alternative electron donors (12, 13). Despite sharing common reactions and the presence of genes encoding a partial glycine synthase, the functional role of the pathway in the presence of intact WLP has remained uncertain.
In this study, we elucidated the role of the GSRP and RGP in pure-cultured Clostridium drakei SL1T during autotrophic growth. Initially, assembly of the C. drakei genome revealed the coexistence of the GSRP, RGP, and WLP, which were then utilized for reconstruction of a genome-scale metabolic model (GEM) to predict metabolic flux through the core carbon pathways. Subsequently, transcriptome analysis revealed the transcriptional activation of genes encoding the pathways; this was then validated by using 13C isotope-based metabolite-tracing experiments, biochemical assays, and genetic engineering. Based on the integration of the data, we concluded that C. drakei coutilizes the GSRP, RGP, and WLP to fix CO2, and then converts it into acetyl-CoA or acetate during autotrophic growth.
Results
Genes and Metabolic Pathways for Carbon Fixation in the C. drakei Genome.
The C. drakei genome was assembled completely, resulting in a 5.7-Mbp complete genome with 29.7% GC content (SI Appendix, Figs. S1 and S2 and Datasets S1 and S2) (14). Following the assembly, we identified 5,144 genes, which were composed of 5,024 coding sequences, 30 ribosomal RNAs, and 90 transfer RNAs (Dataset S3; see SI Appendix, Text S2 for details). The C. drakei genome encodes three formate dehydrogenase (FDH) genes, one WLP gene cluster, one ATP synthase gene cluster, and one CO dehydrogenase (CODH) gene cluster (Fig. 1A and Dataset S3) (15, 16). The third FDH (B9W14_20090) was located downstream of hydrogenase- and NAD(P)H-coding genes, indicating a similar genomic composition to that found in Clostridium autoethanogenum, which was reported to reduce formate using charged ferredoxin (Fd) and NAD(P)H (17). A single WLP gene cluster consisting of 15 genes associated with the carbonyl and the methyl branches was identified, which is identical to the gene composition in the Clostridium species (Fig. 1B; see SI Appendix, Text S3 for details) (6, 7). Energy conservation systems play essential roles in the autotrophic growth of acetogens by orchestrating the required reduction power for the WLP reactions and generating ATP (18, 19). In the C. drakei genome, hydrogenase complex, Fd–NAD+ oxidoreductase complex (Rnf), ATP synthase complex, electron transfer flavoprotein (ETF), and electron-bifurcating transhydrogenase (Nfn)-coding gene clusters were identified, which are similar to those in C. autoethanogenum and Clostridium ljungdahlii (Fig. 1B and SI Appendix, Fig. S3; see SI Appendix, Text S4 for details).
Fig. 1.
Comparative autotrophic metabolic pathway analysis of C. drakei. (A) Comparison of proteins associated with the Wood–Ljungdahl pathway and energy conservation system between C. drakei and 14 acetogenic bacteria. The proteins in C. drakei are set as a reference and analyzed using Basic Local Alignment Search Tool (BLAST). The similarity score is colored as indicated on the color key, and absent of the target protein is colored in dark gray. (B) The Wood–Ljungdahl pathway gene cluster and glycine synthase and glycine synthase-reductase gene cluster arrangement across acetogens. Based on the phylogenetic distance the gene clusters in acetogens were compared, and significant similarity between strains is indicated by a gray line (E-value threshold of 10E-5). Genes are indicated by an arrow and colored according to function. Borders between genes or clusters are shown with break lines. A, Acetobacterium; Ac, Acetohalobium; C, Clostridium; E, Eubacterium; M, Moorella; P, Peptoclostridium; T, Treponema; Th, Thermacetogenium; Thr, Thermoanaerobacter.
In the C. drakei genome, the glycine synthase cluster consists of glycine cleavage system T protein (gcvT) and two glycine dehydrogenase subunits (gcvPA and gcvPB), along with gcvH and lpdA, which are located in the WLP gene cluster (Fig. 1B and Dataset S3). The first four reactions of the glycine synthase pathway are similar to the methyl branch of the WLP, converting CO2 to methylene-THF (tetrahydrofolate), which then forms glycine from methylene-THF and CO2 along with NADH and ammonia (20–22). Following the synthase, the GSRP and the RGP convert the glycine into acetyl-P or serine catalyzed by glycine reductase or SHMT, respectively, which is the essential difference between the pathways (Fig. 2A). For GSRP, the coding gene clusters for glycine reductase are composed of six genes: thioredoxin reductase (trxA and trxB), which donates electrons from NADPH; selenoprotein glycine reductase A (grdA), which integrates essential subunits and then reduces glycine from the trxAB subunits; glycine reductase B (grdB), which binds to the α-carbon of glycine; and glycine reductase C (grdC and grdD), which accepts acetyl-esters and then phosphorylates these into acetyl-P (Fig. 1B and Dataset S3). Conservation of glycine synthase and the GSRP-associated gene clusters in the other acetogens was examined, revealing only five acetogens containing intact glycine synthase and the GSRP clusters (Fig. 1B). In addition, for the RGP, glyA encoding the SHMT was located distant from glycine synthase and the GSRP gene clusters (Dataset S3).
Fig. 2.
Construction of the genome-scale metabolic network model of C. drakei (iSL771). (A) C. drakei flux distribution obtained from a Markov chain Monte Carlo. The top boxes represent the flux value from the heterotrophic condition. The bottom boxes represent the flux value from the autotrophic condition. (B) Flux distributions of CO2-fixing pathway-associated reactions in wild-type and knockout strains. (C) Acetyl-CoA production ratio in wild-type and knockout strains.
Based on these analyses, C. drakei is predicted to convert CO2 into formate via FDH and then into methylene-THF using the first four reactions of the methyl branch of the WLP during autotrophic growth. Subsequently, methylene-THF diverges to form glycine or acetyl-CoA by using glycine synthase or the remaining WLP enzymes, respectively. Next, the formed glycine is either converted into serine or acetyl-P via the RGP or GSRP, respectively (Fig. 2B). Although the pathways are not conserved across most acetogens, the functionality of the genes substantiates that the pathways could be utilized during autotrophic growth, which needs to be validated.
Reconstruction of a Genome-Scale Metabolic Network of C. drakei.
To predict the functionality of the metabolic pathways, we reconstructed a GEM (iSL771) that mathematically calculates cellular metabolism in a given condition (SI Appendix, Fig. S4A; see SI Appendix, Text S5 for details) (23–26). The model iSL771 represents an acetogen genome-scale metabolic model with a genome size exceeding 5.0 Mbp, which is composed of 771 genes, 922 reactions, and 854 metabolites. Following reconstruction, the iSL771 model was then experimentally validated for the functional capability for growth and production rates in a heterotrophic condition using fructose and an autotrophic condition using H2/CO2. The experimental and simulated growth rates were 0.184 and 0.178 h−1 for the fructose condition and 0.044 and 0.046 h−1 for the H2/CO2 condition, respectively (SI Appendix, Fig. S4B). The respective experimental and simulated acetate production rates were 4.524 and 4.490 mmol⋅g dry weight (gDW)−1⋅h−1 and 3.278 and 3.291 mmol⋅gDW−1⋅h−1 for the fructose and the H2/CO2 conditions, respectively (SI Appendix, Fig. S4B). The comparisons between the experimental and in silico data indicate that the model predicted fairly accurate growth and production rates under both conditions.
Then, the genome-scale metabolic flux states of C. drakei under the growth conditions were predicted with the GEM iSL771 and Markov chain Monte Carlo sampling with 100,000 sampling points (Dataset S4) (27–29). According to the obtained flux values, the gluconeogenesis flux superseded the glycolytic flux under the H2/CO2 condition, observing an initial increased flux value of reactions from acetyl-CoA to pyruvate, and then to phosphoenolpyruvate and 3-phosphoglyceric acid (Fig. 2A and Dataset S4). In addition, the predicted WLP flux was active in the presence of both fructose and H2/CO2, with higher WLP fluxes in the H2/CO2 condition by at least twofold. Prior to the GSRP and RGP pathways, a flux distribution of glycine synthase in the H2/CO2 condition was 2.09, which diverged from the prior reactions, that is, the first four reactions associated with the WLP (Fig. 2A and Dataset S4). For the GSRP, in the H2/CO2 growth condition, glycine was converted to acetyl-P with a flux distribution of 1.98, higher than the value in the fructose condition (0.55). For the RGP, glycine was converted to serine using SHMT encoded by glyA with a flux distribution of 0.10, and then to pyruvate catalyzed by serine dehydratase encoded by sda with a flux distribution of 0.04 (Fig. 2A and Dataset S4). The fluxes of the RGP and GSRP reactions potentially produce pyruvate via CO2 fixation without using the Fd-utilizing por reaction, which reduces a usage of reduction power. Overall, the in silico analysis demonstrated that C. drakei coutilizes the WLP, GSRP, and RGP to fix CO2 into acetyl-CoA, acetyl-P, or serine, respectively (Fig. 2A).
After determining the flux distribution, each reaction associated with the CO2-fixing pathways was investigated. Among the seven WLP reactions, constraining the fluxes of the first five reactions critically affected the growth of the model, indicating the reactions are potentially essential to strain growth under the autotrophic condition (SI Appendix, Fig. S4C). Interestingly, constraining the carbon monoxide dehydrogenase/acetyl-CoA synthase (CODH/ACS) reaction, which plays an essential role in synthesizing acetyl-CoA, did not inhibit the growth of the model, suggesting an alternative pathway that could replace the role of the CODH/ACS reaction (SI Appendix, Fig. S4C). To determine an alternative reaction, a flux distribution of glycine reductase (GLYR) that synthesizes acetyl-P and then potentially acetyl-CoA was examined, with the result, similar to the CODH/ACS flux reaction, of constraining GLYR flux that did not affect the growth of the model (SI Appendix, Fig. S4C). Despite being growth-ineffective reactions, CODH/ACS and GLYR showed inversely correlated flux values, indicating the two reactions balance fluxes to maintain biomass by rerouting the pathway in the absence of one reaction (SI Appendix, Fig. S4D).
To predict the feasibility of the glycine synthase pathways during CO2 fixation in the absence of the WLP, we modified the model by removing the CODH/ACS (ΔCODH/ACS), GLYR (ΔGLYR), or glycine synthase (ΔGLYS) reaction and compared it with the wild-type iSL771 model (WT), with the result that removing two reactions simultaneously was predicted to have no in silico growth. Compared with the WT, fluxes of the first four WLP reactions in the ΔGLYR model were relatively similar, and fluxes of the three latter reactions of the WLP were enhanced (Fig. 2B). Similarly, the ΔGLYS model predicted an increase of the three latter reactions, but the first four reactions were decreased compared with the WT (Dataset S4). In the ΔGLYS model, the three WLP latter reaction fluxes were increased due to the absence of an alternative route and maximizing WLP fluxes. In the ΔCODH/ACS model, compared with the WT, the first four WLP reaction fluxes were increased, but the latter reaction fluxes were negligible (Fig. 2B). In addition, the ΔCODH/ACS model maximized the first four fluxes and constrained the latter reactions to avoid a dead-end route, resulting in increased GLYS and GLYR reaction fluxes to reroute the metabolic pathway. Furthermore, the acetyl-CoA production route varied across the constructed models. In WT, acetyl-CoA production was attributed to CODH/ACS and pta, which in the latter case converts acetyl-P synthesized via GLYR to acetyl-CoA (Fig. 2C). Based on the simulation result, the ΔCODH/ACS model and ΔGLYR model were predicted to produce acetyl-CoA by rerouting the fluxes to pta and CODH/ACS reactions (Fig. 2C). In addition, GLYR and the RGP are predicted to be functional under the autotrophic condition to synthesize acetyl-P or pyruvate, which raises an additional question of whether the pathways are functional in vivo.
Transcriptional Response of C. drakei in the Autotrophic Condition.
To validate the in silico prediction, we measured the transcriptomic changes induced by the heterotrophic and autotrophic conditions using RNA-seq (sequencing) (SI Appendix, Figs. S5 and S6 and Dataset S5; see SI Appendix, Text S6 for details). Following RNA-seq experiments, differentially expressed gene (DEG) analysis identified 693 up-regulated and 651 down-regulated genes (SI Appendix, Fig. S7 and Datasets S6 and S7) (30). Gene expression of FDH- and WLP-encoding genes was significantly up-regulated in the H2/CO2 condition (Fig. 3A). Of the three FDHs, the first (B9W14_06825) and third (B9W14_20090) were transcriptionally up-regulated with fold changes of 3.02 and 4.04 (adjusted P value for DESeq2 package results; DESeq P < 7.10 × 10−36), respectively. A minimum fold change of 5.10 (DESeq P < 1.26 × 10−92) of hytE (B9W14_20070) located upstream of the third FDH (B9W14_20090) suggested that the FDH utilizes nearby hydrogenase genes (B9W14_20060–B9W14_20085) to reduce CO2 into formate (Fig. 3A, SI Appendix, Fig. S8, and Datasets S8 and S9). All of the genes associated with the carbonyl branch of the WLP were up-regulated with a minimum fold change of 1.86 (DESeq P < 2.81 × 10−12) for cooC (B9W14_22305). For the methyl branch of the WLP, all of the genes were significantly up-regulated with a minimum fold change of 2.14 (DESeq P < 5.04 × 10−13) for fhs (B9W14_22300). Consistent with the expression levels of the WLP-coding genes, a transcriptional abundance of the glycine synthase-coding gene (B9W14_22245) that was located in the WLP gene cluster was increased by 2.52 (DESeq P < 5.02 × 10−19).
Fig. 3.
Experimental validation of the glycine synthase-reductase pathway. (A) Transcription changes of the Wood–Ljungdahl pathway, glycine synthase-reductase pathway, and reductive glycine pathway under heterotrophic growth and autotrophic growth in C. drakei. The stack is composed of three boxes; the top and middle boxes represent normalized RNA reads from heterotrophic and autotrophic conditions, respectively. The bottom box indicates fold change, as autotrophic expression over heterotrophic expression. Asterisks represent an insignificant change of the corresponding gene. (B) Overview of glycine and serine production by using fructose or CO2 as a carbon source via the Wood–Ljungdahl pathway, glycine synthase-reductase pathway, reductive glycine pathway, and glycolytic pathway. (C) 13C-label metabolite-tracing experiment to validate the glycine synthase-reductase pathway. “H” represents samples obtained from [U-13C]fructose, and “A” represents samples obtained from 13CO2. Dark blue color bars indicate spectra of unlabeled carbon; light brown indicates spectra of [13C]fructose and [13C]CO2 spectra. (D) Relative labeled and unlabeled fractions of glycine and serine in samples obtained from the autotrophic compared with the heterotrophic condition. Error bars shows SD (n = 3). (E) Thioredoxin reductase activity measured using lysate extracted from C. drakei that had been cultured in heterotrophic (H) and autotrophic (A) conditions, from biological triplicates. The error bars represent SD (n = 6). Calculation of the unit is described in SI Appendix, Materials and Methods. Asterisks represent a significant change of the corresponding genes (**P < 0.01).
Notably, we observed that transcription levels of the GSRP-associated genes were significantly up-regulated in the H2/CO2 condition (Dataset S6). For glycine synthase, all of the associated genes were up-regulated with a minimum fold change of 1.87 (DESeq P < 3.25 × 10−13) for gcvT, along with the gene expression of gcvH and lpdA located in the WLP cluster, with a minimum fold change of 2.52 and 2.55 (DESeq P < 1.41 × 10−15), respectively (Fig. 3A and Datasets S8 and S10). Moreover, gene expression of the second and third GLYR clusters (B9W14_09375–09410 and B9W14_09465–09495) was significantly up-regulated with a minimum fold change of 7.48 (DESeq P < 1.88 × 10−41) and 5.08 (DESeq P < 3.24 × 10−13) for grdC (B9W14_09490) and grdD (B9W14_09410), respectively. In the first GLYR cluster (B9W14_00350–00380), five genes were transcriptionally up-regulated, with the other three genes remaining unchanged. Taken together, the DEG analysis results showed that C. drakei transcriptionally activates the WLP cluster along with the GSRP-associated genes, indicating that fixed CO2 formed into acetyl-CoA by using the WLP whereas the synthesized glycine, via the GSRP, is likely reduced to acetyl-P under the autotrophic condition.
13C-Labeling Experiment Confirmed Activation of the Glycine Synthesis Metabolic Flux under the Autotrophic Growth Condition.
Based on the in silico analysis and the transcriptional analysis, C. drakei was strongly expected to coutilize the WLP and the GSRP to fix CO2. To confirm the functional incorporation of the two pathways, we examined the path of 13C quantitatively using [U-13C]fructose or 13CO2 as a carbon source (Fig. 3B). [U-13C]Fructose-supplemented C. drakei showed a 100% fraction of unlabeled carbon for glycine (Fig. 3C). The basal medium for C. drakei contained 2 g/L yeast extract, which is required for the bacterium to proliferate. It is possible that the presence of the yeast extract supplemented unlabeled amino acids to C. drakei, resulting in unlabeled glycine in the heterotrophic condition.
In contrast, 47.3% of detected serine was labeled, demonstrating that C. drakei actively biosynthesizes serine in the presence of fructose (Fig. 3C). In C. drakei, serine can be synthesized from glycine synthase or the glycolytic pathway by converting 3-phosphoglycerate to 3-phosphohydroxypyruvate and then finally to serine. With a number of charged carbons, serine synthesis likely derived from the glycolytic pathway that utilized [U-13C]fructose rather than from glycine, which was unlabeled (Fig. 3C). Conversely, labeled glycine was observed in the presence of 13CO2, demonstrating that C. drakei generates glycine using glycine synthase (Fig. 3C). Subsequently, the direction of glycine was checked by investigating the 13C labeling of serine, which demonstrated that 7.8% of serine was labeled, much lower than in the heterotrophic condition. In addition, a fold change of glycine over serine in the strain under the autotrophic condition was 4.48, higher than the change detected under the heterotrophic condition with 1.25-fold change (Fig. 3D). Decreases in the labeled serine fraction and glycine-to-serine ratio indicate that serine synthesis may be limited in the autotrophic condition. Taken together, consistent with the genome-scale metabolic network prediction, the 13C-labeling experiment results demonstrated that the glycine synthase reaction fixes CO2 and synthesizes glycine.
Glycine Reductase Pathway Converts the Synthesized Glycine to Acetyl-P.
The 13C-labeling experiments validated the role of glycine synthase; however, the downstream direction of glycine during autotrophic growth remained unclear. For the GSRP, thioredoxin reductase activity plays an important role in reducing glycine. We hypothesized that if the activity of thioredoxin reductase in autotrophically grown cells increased compared with that in the heterotrophic condition, more thioredoxin would be available to potentially reduce glycine into acetyl-P, as demonstrated in the RNA-seq results. To test this hypothesis, thioredoxin reductase activity in cells grown under both conditions was measured using a colorimetric assay, resulting in average activity units of 3.76 × 10−3 and 2.82 × 10−2 for the fructose and the H2/CO2 conditions, respectively. This result demonstrates a significant difference between the two conditions by a minimum change of fourfold (P = 1.3 × 10−3) (Fig. 3E). Taken together, the results validated that C. drakei activates not only the WLP but also the GSRP during the autotrophic growth condition, and then utilizes thioredoxin for reduction of glycine into acetyl-P, rather than serine.
Glycine Reductase Pathway Enhances the CO2 Consumption Rate under the Autotrophic Growth Condition.
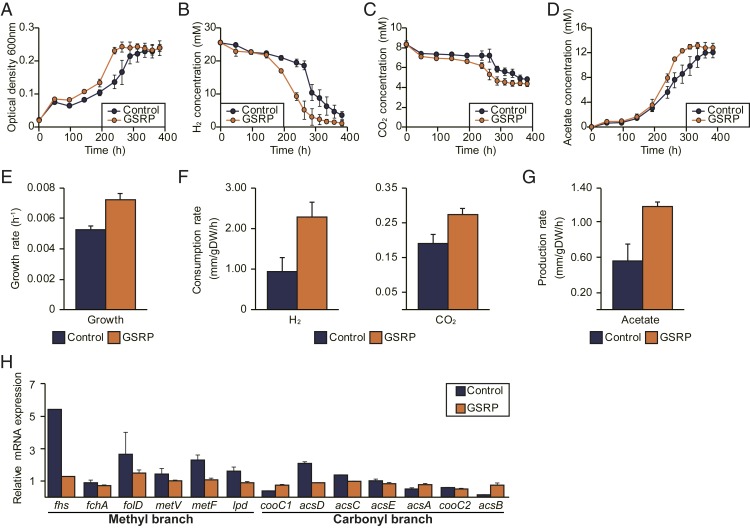
Despite the validations, a phenotypical effect of coexistence of the pathways remains unclear. To confirm the functional role of a pathway, a reconstruction of C. drakei to modify the pathways is required to confirm the prior result, but, unfortunately, developing a genetic modification tool for the strain was infeasible. As an alternative, a genetically modifiable acetogen, the Eubacterium limosum ATCC 8486 strain with the absence of GSRP-coding genes in its genome, was examined for the heterogeneous introduction of the pathway (8, 31). To confirm the functional role, the GSRP-encoding gene cluster from the C. drakei genome was cloned into a plasmid, which was then introduced into E. limosum (the GSRP strain), and the same plasmid backbone without the gene cluster was introduced into E. limosum as a control strain (SI Appendix, Fig. S9A). Using the control and the GSRP strain, growth, consumption, and production profiles were measured to understand the effect of the pathway in the presence of the WLP. According to the growth profile, the maximum cell densities of the control and the GSRP strains were 0.241 and 0.243, respectively, indicating the introduction of the GSRP does not enhance maximum biomass produced under the condition (Fig. 4A). Similarly, a difference of total consumed H2 and CO2 between the strains was insignificant, and likewise for amounts of acetate produced by the strains (Fig. 4 B–D).
Fig. 4.
Phenotypical effect of the glycine synthase-reductase pathway. To compare phenotypical changes, genes encoding the glycine synthase-reductase pathway from C. drakei were introduced into E. limosum, which then was cultured under the autotrophic growth condition. (A–D) Cell density (A), H2 consumption (B), CO2 consumption (C), and acetate production (D) of E. limosum with glycine synthase-reductase pathway-coding genes (GSRP) and without the genes (Control) were measured during autotrophic growth. (E–G) Growth rate (E), H2 and CO2 consumption rate (F), and acetate production rate (G) by the control and GSRP strains were compared. (H) Gene expression of the methyl and carbonyl branches of the Wood–Ljungdahl pathway in both strains was compared. In all panels, dark blue bars indicate results obtained from the control strain and orange bars indicate results obtained from the GSRP strain. All error bars in the panels represent SD (n = 3).
Despite the similar biomass production, the growth rate of the GSRP strain was faster than that of the control strain, with growth rates of 0.00719 and 0.00523 h−1, respectively (Fig. 4E). Similar to the growth rates, the consumption rates of H2 and CO2 by the GSRP strain were 2.291 and 0.276 mmol⋅gDW−1⋅h−1, respectively, which were much higher than the consumption rates of 0.943 and 0.191 mmol⋅gDW−1⋅h−1 by the control strain, respectively (Fig. 4F). Consistent with the results, the acetate production rate by the GSRP strain was higher than the rate by the control strain, with 1.191 and 0.559 mmol⋅gDW−1⋅h−1, respectively (Fig. 4G). To validate the result, gene expression of the GSRP- and the WLP-coding genes in the strains was measured using qRT-PCR. As expected, gene expression of the GSRP was observed only in the GSRP strain and not in the control strain, indicating genes encoding the GSRP were transcriptionally active under the autotrophic growth condition (SI Appendix, Fig. S9B). In addition, the WLP gene expression in the control strain was transcriptionally more active than in the GSRP strain (Fig. 4F), likely due to the absence of genes encoding the GSRP leading to more energy available for WLP gene expression (SI Appendix, Fig. S9C).
Taken together, although the GSRP did not affect the autotrophic growth capacities in the acetogen, with an additional CO2-fixing route the introduction of the pathway enhanced CO2 and H2 consumption rates that led to an increase of the production rate of acetate, which provided energy available for the cell that eventually altered the growth rate of the acetogen.
Discussion
Among phylogenetically and physiologically diverse acetogens, several contain the GSRP and the RGP coupled with the WLP (6, 22, 32). Despite sharing the first four reactions, coutilization of the WLP with the GSRP and RGP in acetogens to fix C1 compounds has remained unknown. In this study, to ensure the functional role of the GSRP and RGP in the autotrophic growth condition, the genome-scale metabolic network model iSL771 was reconstructed and predicted metabolic flux values of the reactions from methylene-THF to glycine to acetyl-P, along with all WLP reactions. Furthermore, transcriptomic analysis, 13C-label metabolite-tracing experiments, biochemical activity measurements, and heterologous expression of the GSRP-coding genes further revealed that the GSRP in the autotrophic condition was fully functional in the presence of the WLP.
In terms of energy conservation, similar to the WLP, the GSRP gains ATP by converting acetyl-P into acetate, thereby recouping the ATP invested at the beginning of the pathway. For creating a chemical gradient that leads to ATP synthesis, the GSRP is predicted to be less efficient than the WLP due to bypassing methylene-THF, which translocates ions across the membrane, resulting in a lower chemical gradient generated (33). However, the GSRP helps to maintain the organism even at low reduction potential, such as fixing CO2 in low-ATP environments, during the acetogenesis reaction in autotrophic conditions (2). The WLP and the GSRP utilize similar electron carriers, but additional reduced Fd for the WLP and NADPH is used for the GSRP. Comparing the reduction power difference, NADPH has a higher redox potential than reduced Fd, indicating under the limited reduced Fd available condition the strain is likely to operate the GSRP to fix CO2. Another advantage of utilizing the identified pathways is the synthesis of pyruvate using the RGP. For producing pyruvate, the conventional WLP catalyzes por using reduced Fd, but the RGP circumvents the usage of reduction and converts serine to pyruvate. Overall, we hypothesized that C. drakei balances the reduction potential, which is critical during autotrophic growth, by coutilizing the WLP, GSRP, and RGP, which were calculated by in silico and RNA-seq analysis.
Further, to balance reduction power, Nfn-, ETF-, Rnf-, and ATP synthase-coding gene clusters were identified in the genome, which are responsible for energy conservation in C. drakei. Among these, the Nfn complex, which utilizes reduced Fd to reduce NADP+, showed transcriptional abundance during autotrophic growth (Dataset S9). This result is consistent with a previous report that mutation of the Nfn complex in C. autoethanogenum retards growth in autotrophic conditions (34). The NADPH generated from the complex is likely to reduce thioredoxin to cooperate with glycine reductase in C. drakei. In addition, among all ETFs, only lactate dehydrogenase (LDH)-bound ETF, which reduces oxidized Fd from NADH, was transcriptionally activated (Dataset S9). It is unclear why the gene expression increased despite the absence of lactate. One speculation is that acetogen is capable of bifurcating various metabolites using the LDH complex, suggesting that in the presence of energy-deficient conditions the cluster is transcriptionally activated to receive electrons from other metabolites (35). Accordingly, the energy conservation-associated proteins mentioned are vital in 0 net ATP-gaining environments to balance redox couples to generate energy.
Notably, among the seven hydrogenase clusters, two were significantly up-regulated. Prior to the transcriptional analysis, it had been speculated that in the presence of high-level hydrogen the majority of the hydrogenases would be activated to utilize available hydrogen. However, the other five hydrogenases exhibited low gene expression levels in both conditions, indicating that they may not oxidize hydrogen in the autotrophic condition. Of the three FDH clusters, only two were up-regulated, whereas membrane-bound hydrogen-producing formate hydrogen lyase remained unchanged. Similarly, all of the other hydrogenase-coding genes that were located within the formate hydrogen lyase-coding gene cluster were inactive in the autotrophic condition, indicating that the hydrogenases specifically cooperate with other enzymes nearby. Moreover, the oxidoreductase-coding gene was absent in the first FDH cluster, which raised the question of how the protein accepts electrons. However, the transcriptionally up-regulated seleno-dependent FDH, which is bound by NADPH-reducing hydrogenase, was expected to exhibit higher catalytic activity than FDH without selenocysteine (36). Based on these results, C. drakei contains the complex energy conservation system to functionally utilize pathways to fix CO2 by balancing various electron carriers and the introduction of the GSRP of C. drakei enhanced C1 compound consumption and growth rates in other acetogen.
Although the 13C-label metabolite-tracing experimental data validate the synthesis of glycine from CO2 using the pathways, lowering the unlabeled glycine proportion would improve the quality of the data and strongly support the proposed conclusion, which requires a yeast extract-independent C. drakei strain. Overall, we confirmed the coutilization of the pathways during autotrophic conditions, which represents genetic modules to guide strain engineering for advancing the CO2-fixing capability of the strain.
Materials and Methods
Bacterial Strains and Growth Conditions.
C. drakei SL1T was obtained from the Leibniz Institute DSMZ (German Collection of Microorganisms and Cell Cultures). Culture was performed anaerobically at 30 °C in 100 mL DSM 135 medium (pH 7.0). Medium composition and culture conditions are described in SI Appendix. A purged H2/CO2 (80:20) at a pressure of 200 kPa in 50 mL of headspace provided autotrophic growth condition and 5 g/L fructose provided heterotrophic growth conditions. All of the experiments were done with biological duplicates.
DNA Isolation.
At exponential growth, the cell pellet was collected by anaerobic centrifugation at 3,000 × g, 4 °C for 15 min. Cells were ground using liquid nitrogen in a mortar; detailed information on isolating DNA is described in SI Appendix.
Genome Sequencing.
The obtained C. drakei genome was sequenced using the PacBio system with an average size of 20 kb based on polymerase version P5 and C3 chemistry. Detailed descriptions of genome sequencing, assembly, and annotation are described in SI Appendix. The assembled genome information is deposited in the DNA DataBank of Japan/European Molecular Biology Laboratory/GenBank (DDBJ/EMBL/GenBank) database under accession no. CP020953.
RNA-Seq Library Preparation.
C. drakei was cultivated under heterotrophic and autotrophic conditions, and then sampled at the midexponential phase. A description of extracting total RNA from the collected cells is in SI Appendix. Subsequently, ribosomal RNAs (rRNA) in the obtained RNA were removed using the Ribo-Zero rRNA Removal Kit for Metabacteria (Epicentre) according to the manufacturer’s instruction. The rRNA-depleted RNA was used to construct the RNA libraries; detailed information is described in SI Appendix.
Genome-Scale Model Reconstruction.
C. drakei annotated information was used to construct metabolic networks using COBRA Toolbox v3.0.3 and COBRApy v0.14.1 software. Detailed information on the construction is described in SI Appendix.
Detailed materials and methods conducted in this study are described in SI Appendix. Information on reagents, detailed culture conditions, DNA isolation, genome sequencing, genome annotation, RNA-seq library preparation, RNA-seq analysis, GEM reconstruction, 13C isotope-labeling assay, biochemistry assay, and plasmid construction is described in SI Appendix, Materials and Methods.
Data Access and Availability.
The accession number for the associated data is GSE118519 in the Gene Expression Omnibus. The complete genome sequence of C. drakei SL1T has been deposited in the DDBJ/EMBL/GenBank database under accession no. CP020953.
Supplementary Material
Acknowledgments
This work was supported by the Intelligent Synthetic Biology Centre of the Global Frontier Project (2011-0031957 to B.-K.C.) and the C1 Gas Refinery Program (2018M3D3A1A01055733 to B.-K.C.; 2015M3D3A1A01064882 to E.Y.L.) through the National Research Foundation of Korea funded by the Ministry of Science and Information Communication and Telecommunication (ICT) (MSIT).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. L.T.A. is a guest editor invited by the Editorial Board.
Data deposition: Associated data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE118519). The complete genome sequence of C. drakei SL1T has been deposited in the DNA DataBank of Japan/European Molecular Biology Laboratory/GenBank (DDBJ/EMBL/GenBank) database (accession no. CP020953).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1912289117/-/DCSupplemental.
References
- 1.Fuchs G., Alternative pathways of carbon dioxide fixation: Insights into the early evolution of life? Annu. Rev. Microbiol. 65, 631–658 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Bar-Even A., Noor E., Milo R., A survey of carbon fixation pathways through a quantitative lens. J. Exp. Bot. 63, 2325–2342 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Fast A. G., Papoutsakis E. T., Stoichiometric and energetic analyses of non-photosynthetic CO2-fixation pathways to support synthetic biology strategies for production of fuels and chemicals. Curr. Opin. Chem. Eng. 1, 380–395 (2012). [Google Scholar]
- 4.Dürre P., Eikmanns B. J., C1-carbon sources for chemical and fuel production by microbial gas fermentation. Curr. Opin. Biotechnol. 35, 63–72 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Andreesen J. R., “Acetate via glycine: A different form of acetogenesis” in Acetogenesis, Drake H.L., Ed. (Springer US, Boston, MA, 1994), pp. 568–629. [Google Scholar]
- 6.Poehlein A., et al. , The complete genome sequence of Clostridium aceticum: A missing link between Rnf- and cytochrome-containing autotrophic acetogens. MBio 6, e01168-15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shin J., Song Y., Jeong Y., Cho B. K., Analysis of the core genome and pan-genome of autotrophic acetogenic bacteria. Front. Microbiol. 7, 1531 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song Y., et al. , Determination of the genome and primary transcriptome of syngas fermenting Eubacterium limosum ATCC 8486. Sci. Rep. 7, 13694 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vogels G. D., Van der Drift C., Degradation of purines and pyrimidines by microorganisms. Bacteriol. Rev. 40, 403–468 (1976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Waber L. J., Wood H. G., Mechanism of acetate synthesis from CO2 by Clostridium acidiurici. J. Bacteriol. 140, 468–478 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andreesen J. R., Glycine reductase mechanism. Curr. Opin. Chem. Biol. 8, 454–461 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Figueroa I. A., et al. , Metagenomics-guided analysis of microbial chemolithoautotrophic phosphite oxidation yields evidence of a seventh natural CO2 fixation pathway. Proc. Natl. Acad. Sci. U.S.A. 115, E92–E101 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tashiro Y., Hirano S., Matson M. M., Atsumi S., Kondo A., Electrical-biological hybrid system for CO2 reduction. Metab. Eng. 47, 211–218 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Jeong Y., Song Y., Shin H. S., Cho B. K., Draft genome sequence of acid-tolerant Clostridium drakei SL1T, a potential chemical producer through syngas fermentation. Genome Announc. 2, e00387-14 (2014). Erratum in: Genome Announc.3, e01465-14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ragsdale S. W., Pierce E., Acetogenesis and the Wood-Ljungdahl pathway of CO2 fixation. Biochim. Biophys. Acta 1784, 1873–1898 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bengelsdorf F. R., Straub M., Dürre P., Bacterial synthesis gas (syngas) fermentation. Environ. Technol. 34, 1639–1651 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Wang S., et al. , NADP-specific electron-bifurcating [FeFe]-hydrogenase in a functional complex with formate dehydrogenase in Clostridium autoethanogenum grown on CO. J. Bacteriol. 195, 4373–4386 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schuchmann K., Müller V., Autotrophy at the thermodynamic limit of life: A model for energy conservation in acetogenic bacteria. Nat. Rev. Microbiol. 12, 809–821 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Buckel W., Thauer R. K., Energy conservation via electron bifurcating ferredoxin reduction and proton/Na+ translocating ferredoxin oxidation. Biochim. Biophys. Acta 1827, 94–113 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Okamura-Ikeda K., Ohmura Y., Fujiwara K., Motokawa Y., Cloning and nucleotide sequence of the gcv operon encoding the Escherichia coli glycine-cleavage system. Eur. J. Biochem. 216, 539–548 (1993). [DOI] [PubMed] [Google Scholar]
- 21.Kikuchi G., Motokawa Y., Yoshida T., Hiraga K., Glycine cleavage system: Reaction mechanism, physiological significance, and hyperglycinemia. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 84, 246–263 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fonknechten N., et al. , Clostridium sticklandii, a specialist in amino acid degradation:revisiting its metabolism through its genome sequence. BMC Genom. 11, 555 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caspi R., et al. , The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 44, D471–D480 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.The UniProt Consortium , UniProt: The universal protein knowledgebase. Nucleic Acids Res. 45, D158–D169 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.King Z. A., et al. , BiGG Models: A platform for integrating, standardizing and sharing genome-scale models. Nucleic Acids Res. 44, D515–D522 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanehisa M., Goto S., KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 28, 27–30 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schellenberger J., Palsson B. O., Use of randomized sampling for analysis of metabolic networks. J. Biol. Chem. 284, 5457–5461 (2009). [DOI] [PubMed] [Google Scholar]
- 28.Schellenberger J., et al. , Quantitative prediction of cellular metabolism with constraint-based models: The COBRA Toolbox v2.0. Nat. Protoc. 6, 1290–1307 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Megchelenbrink W., Huynen M., Marchiori E., optGpSampler: An improved tool for uniformly sampling the solution-space of genome-scale metabolic networks. PLoS One 9, e86587 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tatusova T., et al. , Update on RefSeq microbial genomes resources. Nucleic Acids Res. 43, D599–D605 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song Y., et al. , Genome-scale analysis of syngas fermenting acetogenic bacteria reveals the translational regulation for its autotrophic growth. BMC Genom. 19, 837 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu M., et al. , Life in hot carbon monoxide: The complete genome sequence of Carboxydothermus hydrogenoformans Z-2901. PLoS Genet. 1, e65 (2005). Erratum in: PLoS Genet.2, e60 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martin W. F., Hydrogen, metals, bifurcating electrons, and proton gradients: The early evolution of biological energy conservation. FEBS Lett. 586, 485–493 (2012). [DOI] [PubMed] [Google Scholar]
- 34.Marcellin E., et al. , Low carbon fuels and commodity chemicals from waste gases—Systematic approach to understand energy metabolism in a model acetogen. Green Chem. 18, 3020–3028 (2016). [Google Scholar]
- 35.Weghoff M. C., Bertsch J., Müller V., A novel mode of lactate metabolism in strictly anaerobic bacteria. Environ. Microbiol. 17, 670–677 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Jones J. B., Stadtman T. C., Selenium-dependent and selenium-independent formate dehydrogenases of Methanococcus vannielii. Separation of the two forms and characterization of the purified selenium-independent form. J. Biol. Chem. 256, 656–663 (1981). [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.