Abstract
CDK4/6 inhibitors are FDA-approved drugs for estrogen receptor–positive (ER+) breast cancer and are being evaluated to treat other tumor types, including KRAS-mutant non–small cell lung cancer (NSCLC). However, their clinical utility is often limited by drug resistance. Here, we sought to better understand the resistant mechanisms and help devise potential strategies to overcome this challenge. We show that treatment with CDK4/6 inhibitors in both ER+ breast cancer and KRAS-mutant NSCLC cells induces feedback upregulation of cyclin D1, CDK4, and cyclin E1, mediating drug resistance. We demonstrate that rocaglates, which preferentially target translation of key cell-cycle regulators, effectively suppress this feedback upregulation induced by CDK4/6 inhibition. Consequently, combination treatment of CDK4/6 inhibitor palbociclib with the eukaryotic initiation factor (eIF) 4A inhibitor, CR-1–31-B, is synergistic in suppressing the growth of these cancer cells in vitro and in vivo. Furthermore, ER+ breast cancer and KRAS-mutant NSCLC cells that acquired resistance to palbociclib after chronic drug exposure are also highly sensitive to this combination treatment strategy. Our findings reveal a novel strategy using eIF4A inhibitors to suppress cell-cycle feedback response and to overcome resistance to CDK4/6 inhibition in cancer.
Introduction
CDK4/6 are key cyclin-dependent kinases that promote G1 to S-phase cell-cycle progression. Upon activation by complexing with D-type cyclins, CDK4/6 phosphorylate and inactivate the retinoblastoma protein (RB); this uncouples the inhibitory interaction between RB and E2F transcription factors, which initiate a transcriptional program promoting cell-cycle progression. Given this pivotal role of CDK4/6, their activities are often dysregulated in cancers resulting in aberrant cell proliferation. Thus, CDK4/6 have been key targets of clinical development for cancer therapy (1–3).
Three highly selective CDK4/6 inhibitors, palbociclib (PD-0332991), ribociclib (LEE001), and abemaciclib (LY2835219), are FDA-approved for treating estrogen receptor–positive (ER+) advanced breast cancers, which are often characterized by dysregulated CDK4/6 activation (1–3). These inhibitors, in combination with antiestrogens, effectively target this oncogenic addiction of ER+ breast cancer and have significantly improved patient survival. For example, in the large phase III PALOMA-3 study of women with ER+ metastatic breast cancer, the combination of palbociclib and fulvestrant improved the median overall survival by 6.9 months compared with the fulvestrant alone after 4 years follow up (4). This is important because, of the quarter of a million new cases of breast cancer diagnosed every year in the United States, at least 65% will be ER+, and approximately one-third of these cases will become metastatic at some point and can benefit from CDK4/6 inhibitors. In addition to breast cancer, CDK4/6 inhibitors have shown promising antitumor activities for other cancer types dependent on CDK4/6 activity, such as KRAS-mutant non–small cell lung cancer (NSCLC; refs. 5, 6). It is anticipated that CDK4/6 inhibitors will become standard of care for a variety of malignancies.
Despite these encouraging clinical outcomes, approximately 20% of patients will not respond to CDK4/6 inhibitors and of those initially responding, half will develop drug resistance with progression within 25 months (4, 7). One known resistance mechanism is RB inactivation, as CDK4/6 inhibitors require an active RB pathway to elicit antitumor effects (8, 9). Other possible mechanisms including cyclin E-CDK2 activation (10, 11), CDK6 amplification (12), and enhanced MAPK signaling (13) have been reported. Adaptive response to CDK4/6 inhibition likely contributes to the development of resistance. For example, treatment of CDK4/6 inhibitors in breast cancer cells induces upregulation of cyclin D1 expression, which is dependent on AKT signaling (10). Similarly, CDK4/6 inhibition in pancreatic cancer cells results in upregulation of cyclin D1 and E1, which can be suppressed by PI3K/mTOR inhibitors (14). Furthermore, it has been shown that breast cancer cells can activate autophagy in response to palbociclib and autophagy inhibition sensitizes these cells to CDK4/6 inhibition (11). Thus, effective strategies targeting these adaptive responses may induce durable cell-cycle exit in combination with CDK4/6 inhibitors.
One class of drugs capable of effectively suppressing multiple targets is translation inhibitors. Translation of many oncogenes, including those promoting cell-cycle progression, is likely mainly through a cap-dependent mechanism (15). This is mediated by the eukaryotic initiation factor (eIF) 4F complex, which is composed of the cap binding protein eIF4E, a scaffolding protein eIF4G, and the ATP-dependent RNA helicase eIF4A. Upon binding to the mRNA cap structure, eIF4F remodels the mRNA 5′ leader region and recruits binding of a 40S ribosome (and associated initiation factors); a step that is rate-limiting for translation. Hence, mRNAs must compete for access to eIF4F and one determinant of competitive efficiency is structural barrier (secondary structure, protein:RNA complexes) within 5′ leader regions. Many mRNAs with roles in tumor initiation (e.g., MYC), tumor maintenance (e.g., cyclins, MCL1), and drug response are eIF4F-responsive mRNAs, making eIF4F a potent anticancer target (15–17). In addition, given the large preponderance of human tumors that have activated PI3K/mTOR signaling (18), this is expected to lead to elevated eIF4F levels, making eIF4F a tumor-selective vulnerability.
There are small-molecule compounds that target the regulation of eIF4F assembly (i.e., mTOR inhibitors) or phosphorylation status of eIF4E (i.e., MNK inhibitors; ref. 15). However, these compounds have pleiotropic effects because their target kinases are known to regulate multiple downstream pathways in addition to translation initiation. In contrast, targeting eIF4A can selectively and directly inhibit translation initiation. Several potent eIF4A inhibitors have been identified from the rocaglate family of small molecules, among which are silvestrol and CR-1–31-B (19, 20). Rocaglates are among the more potent eIF4F inhibitors characterized because they elicit two responses: they cause eIF4A to clamp onto polypurine RNA sequences in 5′ leader regions to block initiation (21) and they deplete eIF4F of its eIF4A subunit (22). Silvestrol-responsive mRNAs include oncogenes involved in the inhibition of apoptosis such as MCL1 and BCL2, and in the regulation of cell-cycle progression, notably CCND1, CCND3, CCNE1, and CDK6 (23, 24). Therefore, silvestrol and CR-1–31-B, while also being well tolerated in animals, suppress translation of these oncogenic mRNAs and block proliferation in breast, prostate, lymphoma, and acute lymphoblastic leukemia cancer models (15, 22, 23, 25).
In this study, we employed rocaglates to target cell-cycle feedback response induced by CDK4/6 inhibition in ER+ breast cancer and KRAS-mutant NSCLC cells. We found that targeting eIF4A is synergistic with CDK4/6 inhibitors in suppressing proliferation of these cancer cells and can overcome acquired resistance to CDK4/6 inhibition.
Materials and Methods
Cell culture and viral transduction
MCF-7, T47D, and CAMA-1 were acquired from ATCC and cultured in DMEM with 6% FBS, 1% penicillin/streptomycin antibiotics, and 2 mmol/L l-glutamine, at 37°C and 5% CO2. A549, H358, H2030, and H1944, were acquired from ATCC and cultured in RPMI-1640 media with 6% FBS, 1% penicillin/streptomycin, and 2 mmol/L l-glutamine, at 37°C and 5% CO2. Cell lines were regularly tested for Mycoplasma using the Mycoalert Detection Kit (Lonza). Identity of all cell lines was verified by short tandem repeat (STR) profiling. From thawing, cells recovered for two passages and were passaged maximum 10 times when experiments were performed.
Lentiviral transduction was performed following the guidelines outlined at http://www.broadinstitute.org/rnai/public/resources/protocols. Cells were infected for 30 hours and then selected with puromycin or blasticidin for 2 to 3 days.
Compounds and antibodies
Palbociclib (S1116), abemaciclib (S7158), and ribociclib (S7440) were purchased from Selleck Chemicals. CR-1–31-B and silvestrol were synthesized as described previously (26, 27). Antibodies against HSP90 (H-114), Cyclin D1 (M20), Cyclin D3 (DCS28), Cyclin A2 (BF683), Cyclin E1 (HE12), Cyclin E2 (A-9), CDK2 (D-12), CDK4 (DCS-35), CDK6 (C-21), p16 (C-20), p21 (H164), and p27 (C-19) were purchased from Santa Cruz Biotechnology; p-RB (S795) and Cyclin D2 (D52F9) were purchased from Cell Signaling Technology; RB (554136) was purchased from BD Pharmingen; and eIF4A1 (ab31217) was purchased from Abcam.
Plasmids
Individual shRNA vectors used were obtained from the Mission TRC library (Sigma): shCCND1 #1 (TRCN0000295876), shCCND1 #2 (TRCN0000288598), shCDK4 #1 (TRCN0000000362), shCDK4 #2 (TRCN0000196698), shCCNE1 #1 (TRCN0000045301), and shCCNE1 #2 (TRCN0000045302). Overexpression vectors were obtained from the TRC3 ORF collections from TransOMIC and Sigma: pLX304-GFP, pLX304-CCND1, pLX317-GFP, and pLX317-CDK4. These above plasmids are provided by the Genetic Perturbation Service of Goodman Cancer Research Centre and Biochemistry at McGill University (Montreal, Quebec, Canada).
Cell viability assays
Cells were seeded at a density of 200–2,000 cells per well into 96-well plates and treated with drugs as indicated 24 hours postseeding. Media and drugs were refreshed every 3 days. Cell-Titer-Blue viability assay (Promega) was utilized to measure cell viability and fluorescence (560/590 nm) was recorded in a microplate reader. Cells were grown for 5–8 days depending on cell size, shape, and density.
Colony formation assays
A total of 2–20 × 103 cells were seeded in 6-well plates. For drug assays, 24 hours postseeding, inhibitors were added to the cells. Media and drugs were refreshed every 3 days. Cells were grown for 10–18 days depending on cell size, shape, and density. At end point, cells were fixed with 4% formalin and stained with 0.1% w/v crystal violet before being photographed. All colony formation assays were fixed horizontally.
Drug washout assays
A total of 4–50 × 102 cells were seeded in 6-well plates. Twenty-four hours postseeding, cells were treated with inhibitors for 6 days and refreshed every 3 days. After 6 days of treatment, cells recovered in regular media for 6 additional days until being fixed and stained.
Immunoblots
Twenty-four hours postseeding (6 or 12-well plates), cells were washed with cold PBS, lysed with protein sample buffer, and collected. For drug assays, 24 hours postseeding, the medium was replaced with media containing inhibitors. Cells were collected 24–72 hours posttreatment.
RNA isolation and qRT-PCR
RNA isolation was performed using TRIzol (Invitrogen). Synthesis of cDNAs and qRT-PCR assays were carried out as described previously (28). Relative mRNA levels of each gene shown were normalized to the expression of the housekeeping gene ACTB. The sequences of the primers for assays using SYBR Green Master Mix (Roche) are as follows:
ACTB_Forward: 5′ – GTTGTCGACGACGAGCG – 3′
ACTB_Reverse: 5′ – GCACAGAGCCTCGCCTT – 3′
CCND1_Forward: 5′ – GGCGGATTGGAAATGAACTT – 3′
CCND1_Reverse: 5′ – TCCTCTCCAAAATGCCAGAG – 3′
CCNE1_Forward: 5′ – TCTTTGTCAGGTGTGGGGA – 3′
CCNE1_Reverse: 5′ – GAAATGGCCAAAATCGACAG – 3′
CDK4_Forward: 5′ – GTCGGCTTCAGAGTTTCCAC – 3′
CDK4_Reverse: 5′ – TGCAGTCCACATATGCAAC – 3′
Cell-cycle analysis
Cells were treated with inhibitors for 3 days and then washed with PBS containing 1% FBS and fixed with cold 70% ethanol for 30 minutes on ice. Cells were washed with PBS and treated with 25 μg/mL Ribonuclease A and stained with 50 μg/mL propidium iodide solution for 30 minutes in the dark. A total of 5 × 103 stained cells were then analyzed by Guava easyCyte HT System (Millipore Sigma) to determine proportion of cells in G1, S, or G2 phase of the cell cycle.
Senescence-associated β-galactosidase staining
Cells were seeded at low density (2–40 × 102 cells) and treated with inhibitors for 7 days, refreshed every 3 days. Cells were then washed with PBS and fixed with 0.5% glutaraldehyde solution for 15 minutes at room temperature. Cells were washed with PBS and then with PBS/MgCl2 pH 6.0 solution twice, followed by staining with X-Gal staining solution [0.2 mol/L K3Fe(CN)6, 0.2M K4Fe (CN)6 3H2O, X-Gal stock (40×)] in PBS/MgCl2 in the dark at 37°C for 8 hours. Cells were washed three times with PBS and were photographed. Senescent cells were quantified by counting 100 cells in three different fields for each replicate.
Coimmunoprecipitation assay
Cells were resuspended in ice-cold lysis buffer [50 mmol/L Tris pH 7.5, 150 mmol/L NaCl, 1% NP40, 1 mmol/L dithiothreitol (DTT) and protease/phosphatase inhibitors] and mechanically sheered passing through a needle syringe. After 30-minute incubation on ice, samples were centrifuged at 14,000 × g for 15 minutes at 4°C to collect the supernatant. Three micrograms of IgG or CDK4 (DCS-35, Santa Cruz Biotechnology) antibodies were added to 2 mg of precleared cell lysate in 500 μL of lysis buffer and incubated overnight at 4°C with continuous rocking. Protein immunocomplexes were then incubated with 40 μL protein G sepharose beads (Protein G Sepharose 4 Fast Flow, GE Healthcare) at 4°C for 2 hours. Precipitated proteins were washed three times with lysis buffer and eluted with SDS loading buffer at 95°C for 10 minutes and analyzed by Western blot analysis.
Overlap of TE down genes and Gene Ontology analysis
Two publicly available datasets of cancer cell lines treated with silvestrol were identified and utilized as follows: in the silvestrol-treated KOPT-K1 cells, 281 genes were identified through RNA-seq whose mRNA translation efficiency was decreased, at a cutoff at P < 0.03 (Z-score > 2.5; ref. 23). In the silvestrol-treated MDA-MB-231 cells, 284 genes were identified through RNA-seq whose mRNA translation efficiency was decreased, at a cut-off at Z-score below −1.5 (24). Gene Ontology biological process was performed using the Gene Set Enrichment Analysis (29) provide by Broad Institute on the 33 overlapping TE down genes from these two datasets. The enriched genetic signatures were ranked according to P value, with the top five signatures shown.
In vivo xenografts
All animal procedures (Animal Use Protocol) were approved by the Institutional Animal Care Committee according to guidelines defined by the Canadian Council of Animal Care and were conducted at the Rosalind & Morris Goodman Cancer Centre at McGill University (Montreal, Quebec, Canada). The persons who performed all the tumor measurements and the IHC analysis for the endpoint tumor samples were blinded to the treatment information.
MCF-7 orthotopic xenograft.
For MCF-7 xenograft experiments, estrogen pellets were synthesized (30) and were subcutaneously implanted into 4- to 6-week-old female nude mice (Charles River) prior to injection of cancer cells. A total of 5 × 106 MCF-7 cells were injected at a ratio of 1:1 cells to Matrigel (BD Biosciences) into the fourth right mammary fat pad. Once palpable tumors reached an average volume of 50–100 mm3, tumor-bearing mice were randomized into four groups (n = 4–5) and treated with a combination of either vehicle (5.2% TWEEN 80 and 5.2% polyethylene glycol 400) or 45 mg/kg palbociclib via oral gavage, together with control (50 mmol/L sodium lactate) or 0.35 mg/kg CR-1–31-B via intraperitoneal injection. Mice in the palbociclib alone or palbociclib plus CR-1–31-B group were treated with palbociclib daily for 28 days. Mice in the CR-1–31-B alone or CR-1–31-B plus palbociclib group were treated daily with CR-1–31-B for the first 12 days and recovered for the following 2 days, before continuing daily CR-1–31-B treatment. Tumor volumes were measured with calipers every other day and calculated by the formula [(L × W2)/2]. At the end of treatment, mice were euthanized and tumors were collected for weighing.
H358 subcutaneous xenograft.
A total of 5 × 106 H358 cells were subcutaneously injected at a ratio of 1:1 cells to Matrigel (BD Biosciences) into 6- to 8-week-old male NSG mice (bred in house). Once palpable tumors reached a volume of approximately 200 mm3 (day 5), tumor-bearing mice were randomized into four groups (n = 4–5) and treated daily with a combination of either vehicle (PBS) or 45 mg/kg palbociclib via oral gavage, together with control (50 mmol/L sodium lactate) or 0.2 mg/kg CR-1–31-B via intraperitoneal injection. Mice were treated for a week and recovered for the following week, before continuing daily treatment for 3–4 more weeks. Mice were weighed before measuring tumor volume, and tumor volumes were measured with calipers twice a week and calculated by the formula [(L × W2)/2].
Immunohistochemistry
Isolated xenograft tumors were fixed in 10% formalin before being paraffin embedded, cut into 4-μm–thick sections, and stained using an IntelliPath automated immunostainer (Biocare Medical). Sections were incubated with the primary antibodies: Phospho-Rb Ser807/811 (9308; Cell Signaling Technology), 1/200 dilution; cyclin D1 (SP4; Cell Marque), 1/250 dilution; cyclin E1 (HE12; Abcam), 1/100 dilution; CDK4 (108357; Abcam), 1/500 dilution, and Ki-67 (16667; Abcam), 1/100 dilution. All sections were scanned using an Aperio Scanscope Scanner (Aperio Vista) and images were extracted with Aperio ImageScope.
H-scores of cyclin D1, CDK4, cyclin E1, and pRB staining were calculated using the formula:
(3 × percentage of strongly stained nuclei + 2 × percentage of moderately stained nuclei + 1 × percentage of weakly stained nuclei), giving a H-score range of 0 to 300. For each treatment condition, sections from three independent tumors were stained and from which the average H-score was calculated from four independent fields.
Generation of palbociclib-resistant clones
A total of 2 × 104 MCF-7 or H358 cells were seeded in a 100-mm dish and kept in 300 nmol/L (MCF-7) or 100 nmol/L (H358) of palbociclib for two weeks and then cultured in 900 nmol/L (MCF-7) or 300 nmol/L (H358) of palbociclib for 6 weeks to obtain resistant clones originating from a single cell. Resistant colonies were isolated and continued to be cultured in 900 nmol/L (MCF-7) or 300 nmol/L (H358) of palbociclib.
Combination index calculations
Cells were treated with indicated inhibitors and cell viability was measured by CellTiter-Blue. Drug combination indices were calculated through the Chou–Talalay method using the Compu-Syn software (31). CI of < 1 demonstrates synergism; CI = 1 an additive effect; CI > 1 demonstrates antagonism. The fraction of cells affected was calculated as: 1 – relative cell viability of treated conditions compared with untreated control.
Densitometry
Immunoblot bands were quantified with ImageJ software and then normalized by the loading control HSP90 immunoblot band intensity. Intensity of crystal violet–stained wells from colony formation assays was quantified with ImageJ software. Each well was normalized to the untreated value from each experimental condition.
Statistical analysis
Statistical significance of biological replicates was calculated by two-tailed Student t test and by two-way ANOVA tests, where *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. GraphPad Prism 7 software was used to generate graphs and statistical analyses. All relevant assays were performed independently at least three times.
Results
CDK4/6 inhibition induces feedback upregulation of cyclin D1, CDK4, and cyclin E1, which modulate responses to CDK4/6 inhibitors in ER+ breast cancer
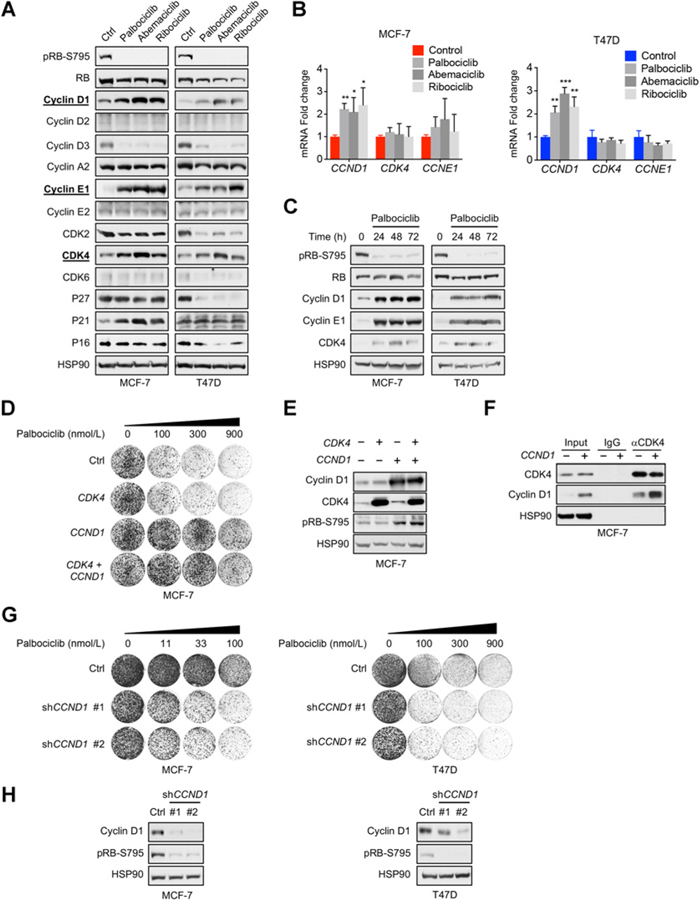
Adaptive response to CDK4/6 inhibition has been suggested to contribute to acquired resistance observed in breast cancer patients. To better characterize this response, we treated two ER+ breast cancer cell lines (MCF-7 and T47D) with three CDK4/6 inhibitors (palbociclib, abemaciclib, and ribociclib) and profiled the expression of key cell-cycle mediators (Fig. 1A; Supplementary Table S1). We focused on factors exhibiting similar regulation in both cell lines across three drug treatments. Among those, we observed downregulation of cyclin D3 and CDK2. This is expected as CCND3 and CDK2 are E2F target genes (32), which would be suppressed due to reactivation of RB upon CDK4/6 inhibition. We also observed consistent upregulation of protein levels of cyclin E1 and CDK4, in addition to previously reported cyclin D1 (10, 33). This is interesting and not expected as both CCNE1 and CCND1 are also known E2F target genes (32). We verified our findings using multiple independent shRNA vectors targeting CCNE1 and CDK4 (Supplementary Fig. S1A and S1B). The upregulated cyclin E1 appears to be an oncogenic low-molecular-weight isoform (34, 35); this isoform, but not the full-length cyclin E1, was shown to confer palbociclib resistance (11). In addition, qRT-PCR analysis revealed that CCND1, but not CDK4 or CCNE1, mRNA was significantly upregulated in both cell lines treated with CDK4/6 inhibitors, indicating posttranscriptional feedback regulation of CDK4 and cyclin E1 (Fig. 1B). Furthermore, this elevation of cyclin D1, cyclin E1, and CDK4 was detected across 72 hours of palbociclib treatment in MCF-7 and T47D cells (Fig. 1C), suggesting that this adaptive response is sustained.
Figure 1.
CDK4/6 inhibition induces feedback upregulation of cyclin D1, CDK4, and cyclin E1, which modulate responses to CDK4/6 inhibitors in ER+ breast cancer. A, Immunoblot profiling of key cell-cycle regulators upregulated after exposure to three CDK4/6 inhibitors. MCF-7 and T47D cells were treated for 24 hours with 300 nmol/L palbociclib, 150 nmol/L abemaciclib, or 1 μmol/L ribociclib. B, qRT-PCR analysis of CCND1, CDK4, and CCNE1 after 24-hour treatment with 300 nmol/L palbociclib, 150 nmol/L abemaciclib, or 1 μmol/L ribociclib. mRNA fold change normalized to mRNA of ACTB housekeeping gene. MCF-7: *, P < 0.05; **, P < 0.01; ***, P < 0.001; by Student t test, unpaired, two-sided; (n = 3). C, Immunoblot showing a time course of sustained upregulation of key cell-cycle regulators. Cells were treated with 300 nmol/L palbociclib for 24, 48, and 72 hours. D, Long-term colony formation assays showing resistance to palbociclib after overexpression of CDK4, CCND1, or their combination in MCF-7 cells. Cells were seeded and treated with palbociclib for 10 to 14 days. Palbociclib was refreshed every 3 days. E, Immunoblot analysis of overexpression of CDK4, CCND1, or their combination in MCF-7 cells. F, Immunoblot detection of CDK4 and cyclin D1 of coimmunoprecipitated samples from MCF-7 cells overexpression a control vector or cyclin D1. Coimmunoprecipitation was performed using an antibody specific against CDK4 or a corresponding IgG control antibody. HSP90 served as a loading control. G, Long-term colony formation assays showing enhanced sensitivity to palbociclib in MCF-7 and T47D after knockdown of CCND1 utilizing two independent shRNAs. Cells were seeded and treated with palbociclib for 10 to 14 days. Drugs were refreshed every 3 days. H, Immunoblot analysis of knockdown of CCND1 utilizing two independent shRNAs in MCF-7 and T47D cells.
Previous studies have implicated cyclin E1 overexpression and amplification in mediating drug resistance to CDK4/6 inhibitors (10, 11). Although plausible, the causal roles of cyclin D1 and CDK4 in mediating drug resistance have not been formally established. We found that exogenous expression of cyclin D1 alone, but not of CDK4, in ER+ breast cancer cells (MCF-7, T47D, and CAMA-1) confers drug resistance to palbociclib in colony-forming assays (Fig. 1D and E; Supplementary Fig. S2A and S2B; Supplementary Table S2). These observations suggest that cyclin D1 is limiting in these cells. Supporting this, ectopic cyclin D1 expression in MCF-7 cells led to a proportional increase of cyclin D1–CDK4 complex and elevated RB phosphorylation (Fig. 1E and F), indicating increased CDK4 kinase activities. In CAMA-1 cells, ectopic expression of cyclin D1 in combination with CDK4 induces slightly further RB phosphorylation and drug resistance, suggesting the additional contribution of exogenous CDK4 expression (Supplementary Fig. S2A and S2B). Consistent with previous reported CCND1 function (10), cyclin D1 knockdown suppressed RB phosphorylation and sensitized MCF-7, T47D, and CAMA-1 to palbociclib in both colony formation (Fig. 1G and H; Supplementary Fig. S2C and S2D) and CellTiter-Blue cell viability assays (Supplementary Fig. S2E). These data casually establish that cyclin D1 is a key determinant of palbociclib responses and that its overexpression can cause drug resistance, which may also be enhanced by CDK4 elevation. Thus, this feedback upregulation of cyclin D1, CDK4, and cyclin E1 induced by CDK4/6 inhibitors may limit the efficiency of these anticancer agents.
eIF4A inhibitors suppress cell-cycle feedback response induced by CDK4/6 inhibition and are synergistic in combination with CDK4/6 inhibitors against ER+ breast cancer cells
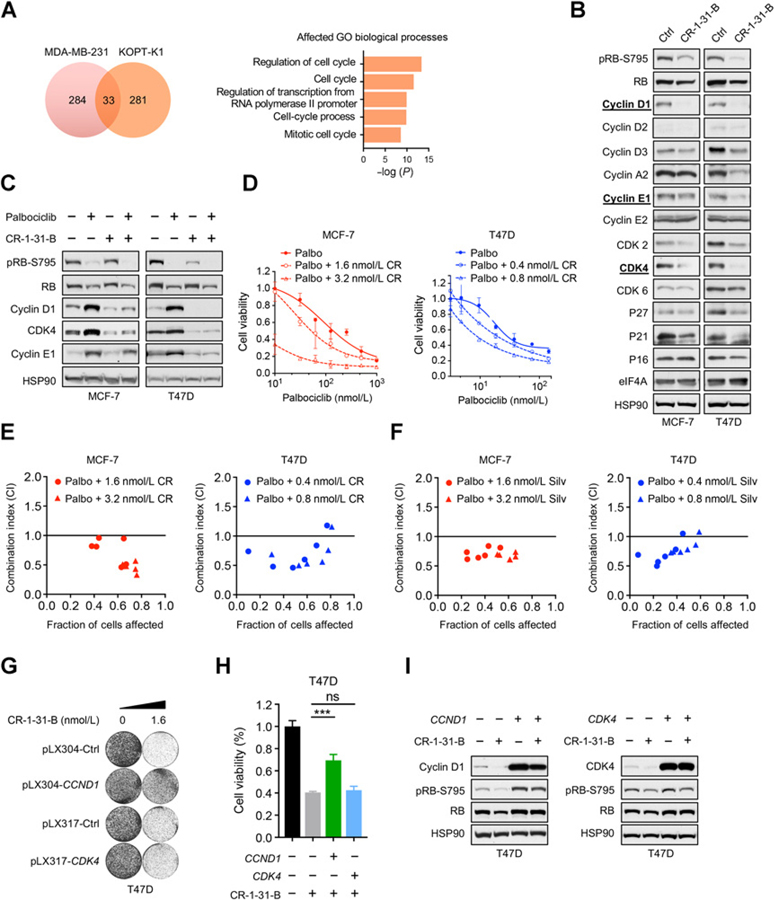
As an approach to simultaneously suppress these cell-cycle mediators induced upon CDK4/6 inhibition, we explored the possibility of targeting translation initiation using rocaglates, silvestrol, and CR-1–31-B. As discussed earlier, these small-molecule compounds selectively and directly target translation initiation by inhibiting eIF4A, a key subunit of eIF4F which is the rate-limiting factor in translation. Many oncogenic mRNAs are known to have more complex mRNA 5′ leader regions and therefore be eIF4F-responsive mRNAs (15–17). In a previous study (23), treatment of KOPT-K1 T-lineage acute lymphoblastic leukemia cells with 25 nmol/L silvestrol resulted in the preferential translational suppression of 281 genes. We compared this publicly available gene set to another study (24), which identified 284 translationally suppressed mRNAs in MDA-MB-231 triple-negative breast cancer treated with silvestrol. This analysis revealed an overlap of 33 genes whose mRNA translation efficiency (TE) was suppressed across both studies (Supplementary Table S3). Gene Ontology analysis of these 33 genes identified the top five biological processes, four of which are cell-cycle related (Fig. 2A). These unbiased findings from KOPT-K1 and MDA-MB-231 cells suggested that eIF4A inhibitors might be effective at suppressing the feedback upregulation of cell-cycle regulators induced by CDK4/6 inhibition.
Figure 2.
eIF4A inhibitors suppress the adaptive response induced by CDK4/6 inhibition and are synergistic in combination with CDK4/6 inhibitors against ER+ breast cancer cells. A, Gene Set Enrichment Analysis of Gene Ontology biological processes for 33 overlapping genes, whose mRNA translation inhibition is suppressed upon treatment with 25 nmol/L silvestrol from two ribosome footprint profiling studies in MDA-MD-231 and KOPT-K1 cells. B, Comprehensive immunoblot profiling of altered protein expression of cell-cycle mediators upon treatment with 3.2 nmol/L CR-1–31-B. MCF-7 and T47D cells were treated for 24 hours. C, Immunoblot analysis showing suppression of cyclin D1, CDK4, and cyclin E1 upon 24-hour treatment with 3.2 nmol/L CR-1–31-B alone or in combination with 300 nmol/L palbociclib in MCF-7 and T47D cells. D, Reduced cell viability upon combination treatment with CR-1–31-B and palbociclib. Cells were treated with indicated concentrations of palbociclib and/or CR-1–31-B for 5 to 8 days, and cell viability was determined using CellTiter-Blue. Error bars represent mean ± SD. CR = CR-1–31-B. E and F, Isobologram synergy analysis at multiple dose combinations of CR-1–31-B (E) or silvestrol (F) with palbociclib. CI < 1.0 suggests synergism, CI > 1 suggests antagonism, CI = 1 suggests an additive effect. Long-term colony formation assays (G) and cell viability assays (H) of T47D cells overexpressing CCND1 or CDK4 and their response to CR-1–31-B treatment. In long-term colony-forming assays, cells were seeded and treated with CR-1–31-B for 10 to 14 days. In cell viability assays, T47D cells were treated for 8 days with 1.6 nmol/L of CR-1–31-B, and cell viability was determined using CellTiter-Blue. Error bars represent mean ± SD (n = 3). ***, P < 0.001 by Student t test, unpaired, two-sided. I, Western blot analysis of T47D cells overexpressing either CCND1 or CDK4 and treated with 3.2 nmol/L of CR-1–31-B for 24 hours. CI, combination index; FA, fraction of cells affected (1 – relative cell viability of treated condition compared with untreated control) ns, not significant.
To confirm the effect of eIF4A inhibitors in ER+ breast cancer cells, we treated MCF-7 and T47D cells with CR-1–31-B and analyzed the expression of a panel of relevant key cell-cycle regulators. We observed consistent suppression of cyclin D1, CDK2, and CDK4 in both cell lines at the low dose of 3.2 nmol/L (Fig. 2B), and suppression of cyclin E1, cyclin A2, and CDK6 at higher concentrations up to 25.6 nmol/L (Supplementary Fig. S3A). We utilized a second eIF4A inhibitor, silvestrol, which exerted a similar degree of growth suppression in both cell lines with half maximal inhibitory concentration (IC50) of approximately 3 nmol/L and approximately 1 nmol/L for MCF-7 and T47D, respectively (Supplementary Fig. S3B), and dose-dependently suppressed cyclin D1, CDK4, and cyclin E1 (Supplementary Fig. S3C). These data indicate that rocaglates are effective in targeting these cell-cycle regulators and suppressing the proliferation of ER+ breast cancer cells.
Next, we investigated the combination of palbociclib and CR-1–31-B in MCF-7 and T47D cells. As expected, addition of 3.2 nmol/L CR-1–31-B reversed the palbociclib-induced upregulation of cyclin D1 and CDK4 to levels lower than basal expression and further inhibited RB phosphorylation in both cell lines (Fig. 2C). In the T47D cell line, which is more sensitive to CR-1–31-B, cyclin E1 feedback was also strongly abrogated at this low dose. Furthermore, the combination of CR-1–31-B and palbociclib further repressed cell viability of MCF-7 and T47D cells (Fig. 2D; Supplementary Table S4). Moreover, this combination of either CR-1–31-B or silvestrol with palbociclib was synergistic as determined by the combination index (31) in both MCF-7 and T47D lines (Fig. 2E and F). Similar synergism was also observed when a second CDK4/6 inhibitor, abemaciclib, was used in combination with CR-1–31-B (Supplementary Fig. S4). Furthermore, we found that CR-1–31-B enhanced G1 cell-cycle arrest induced by CDK4/6 inhibition but had less effect on senescence induction (Supplementary Fig. S5A and S5B). Consistently, drug washout experiments (11) showed that the effect of CR-1–31-B and palbociclib single treatments is partially reversible when drugs are removed; however, the combination can better suppress the regrowth of these cells after drug removal (Supplementary Fig. S5C). These data suggest that the combination of palbociclib and CR-1–31-B can be effective in suppressing breast cancer cells predominantly through mediating cell-cycle arrest.
Given that cyclin D1 and CDK4 are the two common feedback targets being suppressed at low concentrations of CR-1–31-B, we investigated whether they are the determinants of CR-1–31-B responses. As shown in Fig. 2G–I, exogenous expression of cyclin D1 but not CDK4 in T47D cells conferred resistance to the growth inhibition induced by CR-1–31-B in both long-term proliferation and cell viability assays. This is consistent with our findings that cyclin D1 is limiting in these cancer cells (Fig. 1D–H; Supplementary Fig. S2). Although these results do not rule out the contribution of CDK4 suppression (and other targets) to CR-1–31-B–induced growth inhibition, our data suggest that CR-1–31-B exerts its effect, in part, through targeting the dependency of cyclin D1 in these cancer cells. Together, we establish that rocaglates suppress cell-cycle feedback response induced by CDK4/6 inhibition and act synergistically with CDK4/6 inhibitors to target ER+ breast cancer cells.
Drug combination targeting eIF4A and CDK4/6 is synergistic in suppressing ER+ breast cancer growth in vivo
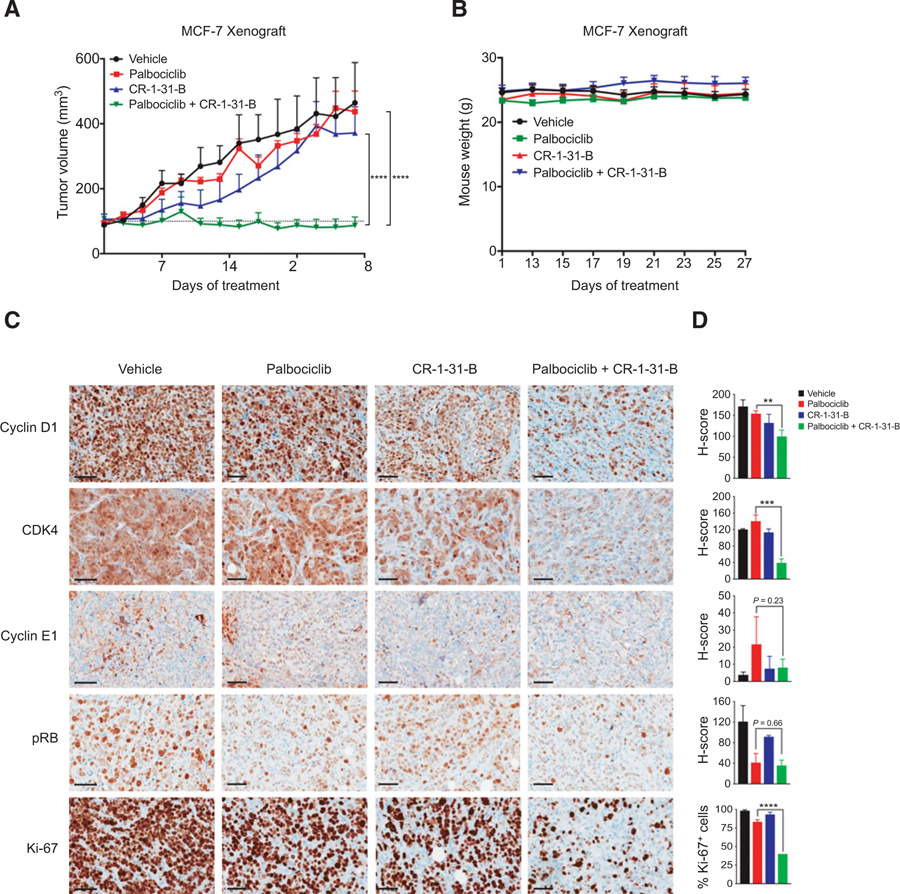
To validate this potential treatment strategy in vivo, we performed orthotopic xenograft experiments by implanting MCF-7 cells into the mammary fat pads of immunodeficient mice. Upon the establishment of tumors, we treated mice with palbociclib or CR-1–31-B at suboptimal doses, or their combination. Although palbociclib or CR-1–31-B treatment alone resulted in marginal growth suppression, their combination elicited a potent growth inhibition of MCF-7 tumors (Fig. 3A). All treatments were well tolerated as no significant weight loss was observed in the animals (Fig. 3B). Using immunohistochemistry (IHC), we analyzed protein expression of the key cell-cycle regulators in tumors isolated at the treatment endpoints (Fig. 3C and D). Palbociclib treatment alone or together with CR-1–31-B led to reduction of RB-phosphorylation, confirming the target modulation by palbociclib. IHC analysis also shows that treatments including CR-1–31-B resulted in decreased expression of cyclin D1, CDK4, and cyclin E1, which is consistent with our in vitro data. While palbociclib or CR-1–31-B treatment alone had minimal effects on expression of the proliferation marker Ki67, their combination resulted in strong suppression of Ki67 expression. Taken together, these results demonstrate that combination treatment of CR-1–31-B and palbociclib is highly effective in suppressing ER+ breast tumor growth in vivo.
Figure 3.
CR-1–31-B in combination with palbociclib is synergistic in suppressing ER+ breast cancer growth in vivo. A, Tumor growth curves of MCF-7 orthotopic xenografts in nude mice treated for 28 days with vehicle (n = 5), palbociclib (45 mg/kg; n = 4), CR-1–31-B (0.35 mg/kg; n = 4), or palbociclib + CR-1–31-B combination (n = 5). Error bars, SEM values. ****, P < 0.0001; by two-way ANOVA. B, Weight of nude mice over xenograft period. Mice were weighed at the beginning of drug treatment and after day 13, every 2 days thereafter. C, IHC staining was performed on formalin-fixed, paraffin-embedded xenografts tumors at endpoint. Representative results are shown. Scale bar for pRB S807/811, cyclin D1, CDK4, cyclin E1, and Ki67: 60 μm. D, H-scores of cyclin D1, CDK4, cyclin E1, and pRB IHC staining of xenograft tumors and percentage of Ki-67-positive cells in each treatment condition. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; by Student t test, unpaired, two-sided.
Combination treatment targeting eIF4A and CDK4/6 synergistically suppresses proliferation of KRAS-mutant NSCLC cells
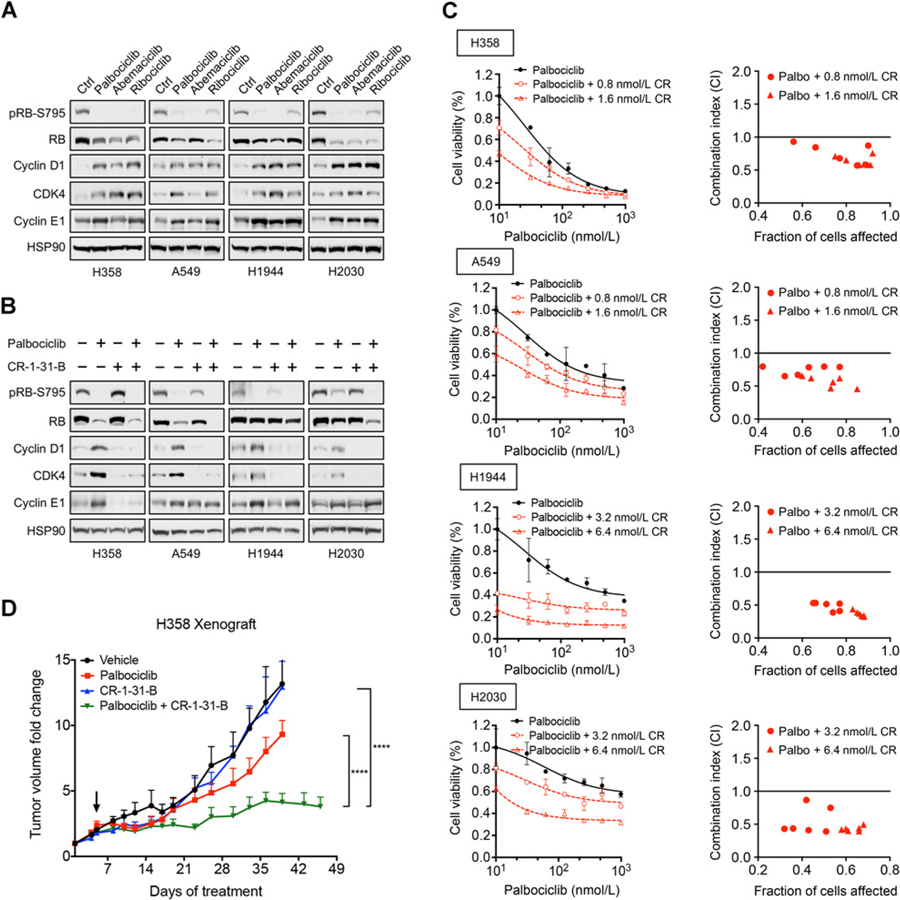
We next aimed to extend our findings to other cancer types where CDK4/6 inhibitors are being evaluated. KRAS-mutant NSCLCs have been shown to respond to CDK/6 inhibitors due to their dependency on CDK4 and these inhibitors are being further evaluated in the clinic (5, 6). KRAS-mutant NSCLC cell lines (H358, A549, H1944, and H2030) have similar sensitivity to palbociclib (Supplementary Fig. S6) as ER+ breast cancer (Fig. 1G). Treatment of all three CDK4/6 inhibitors in these KRAS-mutant NSCLC cells also resulted in upregulation of cyclin D1, CDK4, and cyclin E1 (Fig. 4A), mirroring the feedback response observed in breast cancer cells (Fig. 1A). Furthermore, the addition of CR-1–31-B suppressed this feedback (Fig. 4B) and is synergistic in combination with palbociclib in suppressing proliferation of these NSCLC cells (Fig. 4C; Supplementary Table S4), also predominantly through cell-cycle arrest (Supplementary Fig. S7A–S7C). In support of our MCF-7 ER+ breast cancer xenograft data, combination treatment with suboptimal doses of palbociclib and CR-1–31-B also significantly suppressed tumor proliferation of H358 NSCLC xenografts (Fig. 4D) while also being well-tolerated by animals (Supplementary Fig. S8). Together, our findings demonstrate that eIF4A inhibitors are effective at suppressing the adaptive response induced by CDK4/6 inhibitors and inhibiting the proliferation of KRAS-mutant NSCLC and ER+ breast cancer cells.
Figure 4.
Combination treatment targeting eIF4A and CDK4/6 synergistically suppresses proliferation of KRAS-mutant NSCLC cells. A, Immunoblot analysis showing upregulation of cyclin D1, CDK4, and cyclin E1 of four KRAS-mutant NSCLC cell lines (H358, A549, H1944, and H2030) treated with CDK4/6 inhibitors. Cells were treated for 24 hours with 300 nmol/L palbociclib, 150 nmol/L abemaciclib, or 1 μmol/L ribociclib. B, Immunoblot showing treatment of CR-1–31-B suppresses upregulation of cyclin D1, CDK4, and cyclin E1 induced by palbociclib in KRAS-mutant NSCLC cells. Cells were treated for 48 hours with 300 nmol/L palbociclib, 3.2 nmol/L CR-1–31-B, or their combination. C, Cell viability assays and isobologram synergy analysis of four KRAS-mutant NSCLCs treated with multiple combination doses of CR-1–31-B and palbociclib. Cells were treated with increasing concentrations of palbociclib and/or CR-1–31-B for 5 to 8 days, and cell viability was measured using CellTiter-Blue. Error bars represent mean ± SD. Combination indices were calculated for multiple doses of palbociclib and CR-1–31-B for all four NSCLC cell lines. D, Tumor growth curves of H358 subcutaneous xenografts in NSG mice treated with vehicle (n = 5), palbociclib (45 mg/kg; n = 4), CR-1–31-B (0.2 mg/kg; n = 4), or palbociclib + CR-1–31-B combination (n = 4). Mice treated daily starting on day 5 (arrow) for 1 week and recovered for 1 week. Following recovery, mice were treated daily until endpoint. Error bars indicate SEM values. ****, P < 0.0001; by two-way ANOVA.
eIF4A inhibitors overcome acquired resistance to CDK4/6 inhibition in ER+ breast cancer and NSCLC cells
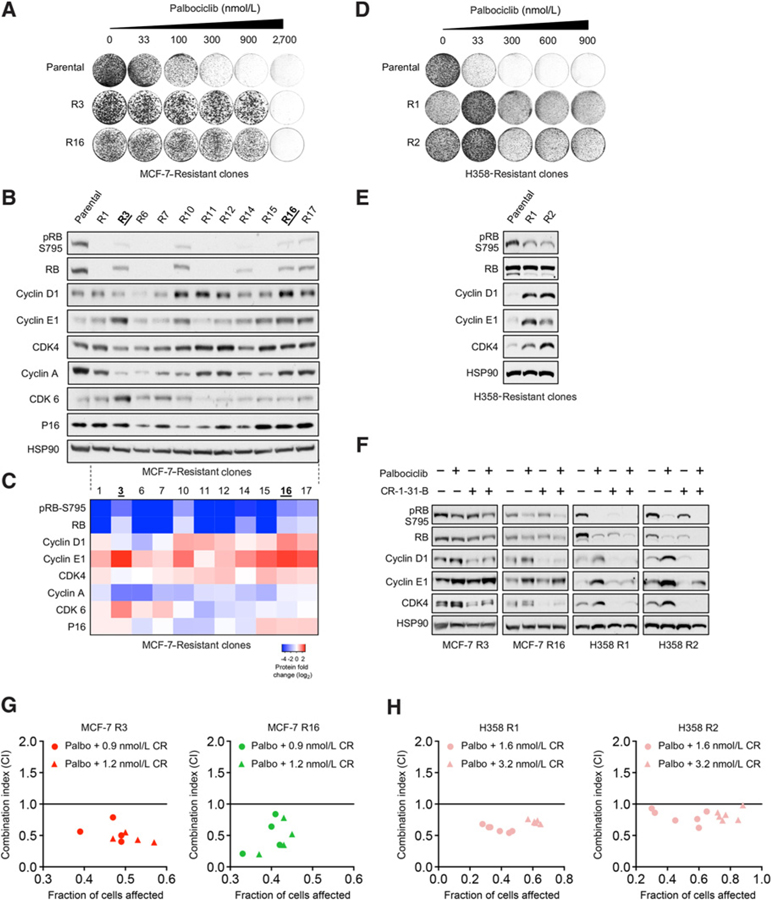
We next investigated whether eIF4A inhibitors are also effective in suppressing breast cancer cells that have acquired resistance to CDK4/6 inhibition. For this purpose, we first generated a panel of spontaneously resistant clones of MCF-7 cells through chronic palbociclib exposure (Fig. 5A; Supplementary Fig. S9). As shown in Fig. 5B and C, one frequent mechanism of acquired resistance was loss of RB expression in 6 of 11 established clones, which is consistent with the established role of RB (1–3, 8). In addition, cyclin E1 was significantly upregulated in 6 clones and cyclin D1 was also upregulated in 5 clones. We also observed the upregulation of CDK6 in 2 clones (R3 and R7) and slight elevation of CDK4 in 3 clones. Thus, the cell-cycle profiles of these resistant clones are in line with previous reports. In a similar manner, we also isolated two stably spontaneously palbociclib-resistant clones of H358 NSCLC cells (Fig. 5D). Indeed, both H358 R1 and R2 clones also had elevated expression of cyclin D1, cyclin E1, and CDK4, and reduced RB phosphorylation (Fig. 5E).
Figure 5.
CR-1–31-B overcomes acquired palbociclib resistance in ER+ breast cancer cells and KRAS-mutant NSCLC cells. A, Colony formation assays of MCF-7 R3 and R16 clones and their resistance to palbociclib compared with the parental line. Cells were seeded and treated with palbociclib for 10 to 14 days at the indicated concentrations. Palbociclib was refreshed every 3 days. B, Immunoblot analysis showing definitive protein expression adaptations mediating acquired resistance to palbociclib in MCF-7 cells. C, Heatmap of protein expression fold change (log2) of key cell-cycle proteins in the MCF-7 palbociclib-resistant clones. Protein expression in B was quantified by densitometry and normalized to the intensity of the HSP90 loading control and then by immunoblot density of the MCF-7 parental line. D, Colony formation assays of two H358 NSCLC clones and their resistance to palbociclib. Cells were seeded and treated with palbociclib for 12 to 16 days at the indicated concentrations. Palbociclib was refreshed every 3 days. E, Immunoblot showing elevated protein expression of cyclin D1, cyclin E1, and CDK4 in H358 R1 and R2 clones relative to the parental line. F, Immunoblot indicating the addition of CR-1–31-B is sufficient to consistently suppress cyclin D1 and CDK4 protein across four MCF-7 and H358 palbociclib-resistant clones. MCF-7 R3, R16, and H358 R1 and R2 cells were treated with either 300 nmol/L palbociclib or 3.2 nmol/L CR-1–31-B alone, or in their combination for 24 hours. G and H, Isobologram synergy analysis of MCF-7 R3 and R16 and H358 R1 and R2 palbociclib-resistant clones treated with multiple combination doses of CR-1–31-B and palbociclib.
To test whether the combination of CR-1–31-B and palbociclib is also effective in targeting the spontaneously palbociclib-resistant cells, we selected MCF-7 clones that were representative of altered signatures–MCF-7 R3 (upregulation of cyclin E1 and CDK6) and MCF-7 R16 (upregulation of cyclin D1 and cyclin E1) and H358 R1 and R2 clones. Indeed, the addition of a low concentration (3.2 nmol/L) of CR-1–31-B was able to suppress the feedback upregulation of cyclin D1 and CDK4 in all palbociclib-resistant clones (Fig. 5F). The inhibition of cyclin E1 upregulation was also pronounced in H358 clones at this low CR-1–31-B concentration, which is consistent with the fact that H358 parental cells are more sensitive to CR-1–31-B compared with MCF-7 parental cells (Supplementary Fig. S10). Furthermore, the drug combination demonstrated synergy in suppressing the growth of resistant populations from both cancer types (Fig. 5G and H). Thus, these results suggest that eIF4A inhibitors can also be viable therapeutic agents treat tumors acquire drug resistance to CDK4/6 inhibitors.
Discussion
In this study, we found that feedback upregulation of cyclin D1, CDK4, and cyclin E1 is a common adaptive response induced by CDK4/6 inhibition in both ER+ breast cancer and KRAS-mutant NSCLC cells. We demonstrate that rocaglates, potent inhibitors of eIF4A, can effectively suppress this feedback response and synergize with CDK4/6 inhibitors targeting these cancer cells and their sub-populations that have acquired resistance to CDK4/6 inhibition. Furthermore, we show that the combination of CR-1–31-B and palbociclib was well tolerated in animals and elicited a potent inhibition to the tumor growth in vivo for both cancer types.
Acquired resistance to CDK4/6 inhibitors is a major challenge that limits their clinical utilities. Cyclin E1 overexpression and amplification have been implicated in mediating drug resistance to CDK4/6 inhibitors (10, 11). Here, we establish a causal role of cyclin D1 in modulating responses to CDK4/6 inhibitors in ER+ breast cancer cells: immunoprecipitation assays indicate that cyclin D1 is limiting in these cells; ectopic expression of cyclin eIF4A Inhibitors Overcome Resistance to CDK4/6 Inhibition D1 led to elevated RB phosphorylation and resistance to CDK4/6 inhibition while cyclin D1 knockdown sensitized these cells to palbociclib. These results are consistent with our previous findings in subtypes of ovarian and lung cancers, where cyclin D1 deficiency limits CDK4/6 activities leading to drug sensitivity to CDK4/6 inhibitors (36, 37).
It is important to note that our results do not rule out other potential contributions of cyclin D1 in mediating drug resistance independent of cell-cycle activities, such as controlling glucose metabolism (38), promoting NF-κB transcriptional regulation (39), and facilitating ER transcriptional activity (40). In addition, this potential role of cyclin D1 in mediating drug resistance remains to be further validated in clinical settings.
Our data suggest that the feedback upregulation of cyclin D1, CDK4, and cyclin E1 induced by CDK4/6 inhibition may contribute to selection of drug-resistant variants after prolonged drug treatment. Indeed, elevation of these cell-cycle regulators is observed in palbociclib-resistant clones of MCF-7 cells and H358 cells generated through chronic drug exposure. In addition, we also observed resistance clones with RB loss, which is consistent with the recent report that RB1 mutations emerged in some tumors of patients treated with palbociclib (9). While RB1 inactivation remains as a challenge, we have identified here a potential strategy to suppress the feedback upregulation of the other cell-cycle regulators contributing to drug resistance.
Our unbiased analysis of previous rocaglate-responsive translatome studies indicated that translation of cell-cycle regulators is particularly sensitive to eIF4A inhibition due to the complex nature of their 5′ leader regions of their mRNAs (23, 24). Indeed, eIF4A inhibitors can effectively suppress the cell-cycle feedback response induced by CDK4/6 inhibition and other key cell-cycle regulators, even at the low nanomolar concentrations used in this study. Thus, this selective feature of eIF4A inhibitors results in a strong synergy in combination with CDK4/6 inhibition against the breast and lung cancer cell lines examined. Consistently, ectopic expression of cyclin D1 from a cDNA lacking the complex endogenous 5′ leaders conferred resistance to CR-1–31-B in breast cancer cells, indicating that the growth-suppressive effect of eIF4A inhibition is in part through targeting cell-cycle pathway. However, translation suppression of other target genes such as c-MYC (22, 41), may also contribute to the potent effect of eIF4A inhibitors and their combination with CDK4/6 inhibitors which require further investigation. In addition, our data do not exclude the possibility of autophagy inhibition induced by eIF4A inhibitors, which may contribute to the synergy in combination with CDK4/6 inhibition—it has been recently shown that targeting autophagy is synergistic with palbociclib (11) and there are emerging connections between protein synthesis, energy homeostasis, and autophagy (42). Nevertheless, our data demonstrate that cotargeting CDK4/6 and eIF4A is an effective combination in better suppressing cancer cells including those that acquired resistance to palbociclib.
Our study highlights the unique ability of eIF4A inhibitors to overcome different modes of resistance to CDK4/6 inhibition, by simultaneously suppressing multiple targets that mediate resistance. Furthermore, given that these targets such as cyclin D1 are often controlled by diverse signaling pathways, targeting their translation is effective regardless the nature of upstream inputs leading to their dysregulation. Supporting this, drug combination targeting eIF4A and BRAF/MEK has also been shown to overcome different resistance mechanisms arising in BRAFV600-mutant cancer models (43). Thus, our data suggest that eIF4A inhibitors, which preferentially target key cell-cycle regulators, could be an effective and novel treatment option to enhance efficiency of CDK4/6 inhibitors and overcome acquired drug resistance.
Supplementary Material
Acknowledgments
We thank M. Biondini and S. Tabaries (McGill University) for the gift of estrogen pellets for xenograft experiments. S. Huang is supported by Canadian Institutes of Health Research (CIHR) grants MOP-130540. ICT-156650J, and PJT-156233; J. Pelletier was supported by a CIHR Foundation grant (FDN-148366); and W.D. Foulkes was supported by a CIHR Foundation Grant (FDN-148390).
Footnotes
Note: Supplementary data for this article are available at Molecular Cancer Therapeutics Online (http://mct.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.O’Leary B, Finn RS, Turner NC. Treating cancer with selective CDK4/6 inhibitors. Nat Rev 2016;13:417–30. [DOI] [PubMed] [Google Scholar]
- 2.Sherr CJ, Beach D, Shapiro GI. Targeting CDK4 and CDK6: from discovery to therapy. Cancer Discov 2016;6:353–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asghar U, Witkiewicz AK, Turner NC, Knudsen ES. The history and future of targeting cyclin-dependent kinases in cancer therapy. Nat Rev Drug Discov 2015;14:130–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turner NC, Slamon DJ, Ro J, Bondarenko I, Im SA, Masuda N, et al. Overall survival with palbociclib and fulvestrant in advanced breast cancer. N Engl J Med 2018;379:1926–36. [DOI] [PubMed] [Google Scholar]
- 5.Patnaik A, Rosen LS, Tolaney SM, Tolcher AW, Goldman JW, Gandhi L, et al. Efficacy and safety of abemaciclib, an inhibitor of CDK4 and CDK6, for patients with breast cancer, non-small cell lung cancer, and other solid tumors. Cancer Discov 2016;6:740–53. [DOI] [PubMed] [Google Scholar]
- 6.Puyol M, Martin A, Dubus P, Mulero F, Pizcueta P, Khan G, et al. A synthetic lethal interaction between K-Ras oncogenes and Cdk4 unveils a therapeutic strategy for non-small cell lung carcinoma. Cancer Cell 2010;18:63–73. [DOI] [PubMed] [Google Scholar]
- 7.Shah M, Nunes MR, Stearns V. CDK4/6 inhibitors: game changers in the management of hormone receptor-positive advanced breast cancer? Oncology (Williston Park) 2018;32:216–22. [PMC free article] [PubMed] [Google Scholar]
- 8.Fry DW, Harvey PJ, Keller PR, Elliott WL, Meade M, Trachet E, et al. Specific inhibition of cyclin-dependent kinase 4/6 by PD 0332991 and associated antitumor activity in human tumor xenografts. Mol Cancer Ther 2004;3: 1427–38. [PubMed] [Google Scholar]
- 9.O’Leary B, Cutts RJ, Liu Y, Hrebien S, Huang X, Fenwick K, et al. The genetic landscape and clonal evolution of breast cancer resistance to palbociclib plus fulvestrant in the PALOMA-3 trial. Cancer Discov 2018;8:1390–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herrera-Abreu MT, Palafox M, Asghar U, Rivas MA, Cutts RJ, Garcia-Murillas I, et al. Early adaptation and acquired resistance to CDK4/6 inhibition in estrogen receptor-positive breast cancer. Cancer Res 2016; 76:2301–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vijayaraghavan S, Karakas C, Doostan I, Chen X, Bui T, Yi M, et al. CDK4/6 and autophagy inhibitors synergistically induce senescence in Rb positive cytoplasmic cyclin E negative cancers. Nat Commun 2017;8:15916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang C, Li Z, Bhatt T, Dickler M, Giri D, Scaltriti M, et al. Acquired CDK6 amplification promotes breast cancer resistance to CDK4/6 inhibitors and loss of ER signaling and dependence. Oncogene 2017;36:2255–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Leeuw R, McNair C, Schiewer MJ, Neupane NP, Brand LJ, Augello MA, et al. MAPK reliance via acquired CDK4/6 inhibitor resistance in cancer. Clin Cancer Res 2018;24:4201–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franco J, Witkiewicz AK, Knudsen ES. CDK4/6 inhibitors have potent activity in combination with pathway selective therapeutic agents in models of pancreatic cancer. Oncotarget 2014;5:6512–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bhat M, Robichaud N, Hulea L, Sonenberg N, Pelletier J, Topisirovic I. Targeting the translation machinery in cancer. Nat Rev Drug Discov 2015; 14:261–78. [DOI] [PubMed] [Google Scholar]
- 16.Ramon YCS, Castellvi J, Hummer S, Peg V, Pelletier J, Sonenberg N. Beyond molecular tumor heterogeneity: protein synthesis takes control. Oncogene 2018;37:2490–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pelletier J, Graff J, Ruggero D, Sonenberg N. Targeting the eIF4F translation initiation complex: a critical nexus for cancer development. Cancer Res 2015;75:250–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y, Kwok-Shing Ng P, Kucherlapati M, Chen F, Liu Y, Tsang YH, et al. A pan-cancer proteogenomic atlas of PI3K/AKT/mTOR pathway alterations. Cancer Cell 2017;31:820–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chu J, Pelletier J. Targeting the eIF4A RNA helicase as an anti-neoplastic approach. Biochim Biophys Acta 2015;1849:781–91. [DOI] [PubMed] [Google Scholar]
- 20.Chu J, Cencic R, Wang W, Porco JA Jr, Pelletier J. Translation inhibition by rocaglates is independent of eIF4E phosphorylation status. Mol Cancer Ther 2016;15:136–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iwasaki S, Floor SN, Ingolia NT. Rocaglates convert DEAD-box protein eIF4A into a sequence-selective translational repressor. Nature 2016;534: 558–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cencic R, Carrier M, Galicia-Vazquez G, Bordeleau ME, Sukarieh R, Bourdeau A, et al. Antitumor activity and mechanism of action of the cyclopenta[b]benzofuran, silvestrol. PLoS One 2009;4:e5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wolfe AL, Singh K, Zhong Y, Drewe P, Rajasekhar VK, Sanghvi VR, et al. RNA G-quadruplexes cause eIF4A-dependent oncogene translation in cancer. Nature 2014;513:65–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rubio CA, Weisburd B, Holderfield M, Arias C, Fang E, DeRisi JL, et al. Transcriptome-wide characterization of the eIF4A signature highlights plasticity in translation regulation. Genome Biol 2014; 15:476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bordeleau ME, Robert F, Gerard B, Lindqvist L, Chen SM, Wendel HG, et al. Therapeutic suppression of translation initiation modulates chemosensitivity in a mouse lymphoma model. J Clin Invest 2008;118: 2651–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodrigo CM, Cencic R, Roche SP, Pelletier J, Porco JA. Synthesis of rocaglamide hydroxamates and related compounds as eukaryotic translation inhibitors: synthetic and biological studies. J Med Chem 2012;55: 558–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gerard B, Cencic R, Pelletier J, Porco JA Jr. Enantioselective synthesis of the complex rocaglate (−)-silvestrol. Angew Chem Int Ed Engl 2007;46: 7831–4. [DOI] [PubMed] [Google Scholar]
- 28.Papadakis AI, Sun C, Knijnenburg TA, Xue Y, Grernrum W, Holzel M, et al. SMARCE1 suppresses EGFR expression and controls responses to MET and ALK inhibitors in lung cancer. Cell Res 2015;25:445–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DeRose YS, Gligorich KM, Wang G, Georgelas A, Bowman P, Courdy SJ, et al. Patient-derived models of human breast cancer: protocols for in vitro and in vivo applications in tumor biology and translational medicine. Curr Protoc Pharmacol 2013;Chapter 14:Unit14.23. [DOI] [PMC free article] [PubMed]
- 31.Chou TC. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res 2010;70: 440–6. [DOI] [PubMed] [Google Scholar]
- 32.Bracken AP, Ciro M, Cocito A, Helin K. E2F target genes: unraveling the biology. Trends Biochem Sci 2004;29:409–17. [DOI] [PubMed] [Google Scholar]
- 33.Jansen VM, Bhola NE, Bauer JA, Formisano L, Lee KM, Hutchinson KE, et al. Kinome-wide RNA interference screen reveals a role for PDK1 in acquired resistance to CDK4/6 inhibition in ER-positive breast cancer. Cancer Res 2017;77:2488–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keyomarsi K, Conte D Jr, Toyofuku W, Fox MP. Deregulation of cyclin E in breast cancer. Oncogene 1995;11:941–50. [PubMed] [Google Scholar]
- 35.Akli S, Zheng PJ, Multani AS, Wingate HF, Pathak S, Zhang N, et al. Tumor-specific low molecular weight forms of cyclin E induce genomic instability and resistance to p21, p27, and antiestrogens in breast cancer. Cancer Res 2004;64:3198–208. [DOI] [PubMed] [Google Scholar]
- 36.Xue Y, Meehan B, Macdonald E, Venneti S, Wang XQD, Witkowski L, et al. CDK4/6 inhibitors target SMARCA4-determined cyclin D1 deficiency in hypercalcemic small cell carcinoma of the ovary. Nat Commun 2019;10:558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xue Y, Meehan B, Fu Z, Wang XQD, Fiset PO, Rieker R, et al. SMARCA4 loss is synthetic lethal with CDK4/6 inhibition in non-small cell lung cancer. Nat Commun 2019;10:557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee Y, Dominy JE, Choi YJ, Jurczak M, Tolliday N, Camporez JP, et al. Cyclin D1-Cdk4 controls glucose metabolism independently of cell cycle progression. Nature 2014;510:547–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Handschick K, Beuerlein K, Jurida L, Bartkuhn M, Muller H, Soelch J, et al. Cyclin-dependent kinase 6 is a chromatin-bound cofactor for NF-kappaB-dependent gene expression. Mol Cell 2014;53:193–208. [DOI] [PubMed] [Google Scholar]
- 40.Zwijsen RM, Wientjens E, Klompmaker R, van der Sman J, Bernards R, Michalides RJ. CDK-independent activation of estrogen receptor by cyclin D1. Cell 1997;88:405–15. [DOI] [PubMed] [Google Scholar]
- 41.Robert F, Roman W, Bramoulle A, Fellmann C, Roulston A, Shustik C, et al. Translation initiation factor eIF4F modifies the dexamethasone response in multiple myeloma. Proc Natl Acad Sci U S A 2014;111:13421–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lindqvist LM, Tandoc K, Topisirovic I, Furic L. Cross-talk between protein synthesis, energy metabolism and autophagy in cancer. Curr Opin Genet Dev 2018;48:104–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boussemart L, Malka-Mahieu H, Girault I, Allard D, Hemmingsson O, Tomasic G, et al. eIF4F is a nexus of resistance to anti-BRAF and anti-MEK cancer therapies. Nature 2014;513:105–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.