Graphical abstract
Highlights
-
•
Computational methods identify aggregation-prone regions in protein sequences as a guide for protein engineering.
-
•
Rational methods can reveal genotype-phenotype relationships and to introduce reactive handles for many applications.
-
•
Directed evolution allows the identification of aggregation-resistant proteins in the absence of mechanistic understanding.
-
•
Deep mutational scanning can potentially reveal the effect of every amino acid at every residue on protein aggregation.
Abstract
Protein aggregation occurs through a variety of mechanisms, initiated by the unfolded, non-native, or even the native state itself. Understanding the molecular mechanisms of protein aggregation is challenging, given the array of competing interactions that control solubility, stability, cooperativity and aggregation propensity. An array of methods have been developed to interrogate protein aggregation, spanning computational algorithms able to identify aggregation-prone regions, to deep mutational scanning to define the entire mutational landscape of a protein’s sequence. Here, we review recent advances in this exciting and emerging field, focussing on protein engineering approaches that, together with improved computational methods, hold promise to predict and control protein aggregation linked to human disease, as well as facilitating the manufacture of protein-based therapeutics.
Current Opinion in Structural Biology 2020, 60:157–166
This review comes from a themed issue on Proteins
Edited by Jan Steyaert and Todd O Yeates
For a complete overview see the Issue and the Editorial
Available online 19th February 2020
https://doi.org/10.1016/j.sbi.2020.01.005
0959-440X/© 2020 The Authors. Published by Elsevier Ltd. This is an open access article under the CC BY license (http://creativecommons.org/licenses/by/4.0/).
Introduction
It has been long been recognised that protein aggregation pervades human morbidity and mortality [1] and impinges on our ability to produce life-saving and life-changing protein therapeutics both rapidly and economically [2]. It is now widely understood that as well as adopting soluble, functional structures, many proteins can also self-assemble forming structured aggregates such as amyloid fibrils [3,4], or to undergo liquid-liquid phase-separation [5,6]. The later process drives the formation of membraneless organelles that can be functional (such as in the nucleolus [7]), or causative of cellular dysfunction and disease (such as in virus replication [8] or in protein aggregation disorders [9]) (Figure 1). The ability of proteins to catalyse reactions, to form stable scaffolds, and to bind ligands tightly and with high specificity, has enormous potentials for the use of proteins in industry [10,11]. However, a major challenge in the use of proteins for such applications lies in their instability, conformational dynamics and inherent tendency to aggregate. There is thus an important and currently unmet need to be able to identify protein sequences that may have undesired properties and to engineer their sequences to improve their properties.
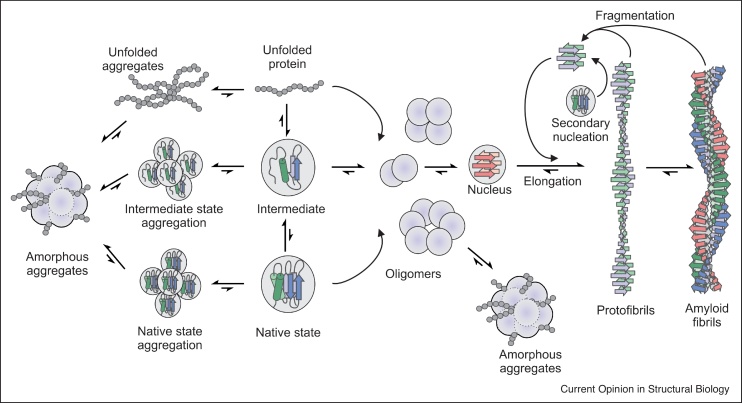
Figure 1.
Schematic illustration of aggregation pathways.
The precursor of aggregation may be the unfolded, partially folded or native state of a protein. During amyloid formation, oligomeric species formed from the initial aggregation-prone monomer, can then assemble further to form higher-order oligomers, one or more of which can form a nucleus, which, by rapidly recruiting other monomers, can nucleate assembly into protofibrils and amyloid fibrils. As fibrils grow, they can fragment, yielding more fibril ends that are capable of elongation by the addition of new aggregation-prone species [86]. Alternatively, amorphous aggregation can occur via one or more aggregation-prone species growing into larger species, by Ostwald ripening or other self-association mechanisms [87].
While aggregation-prone regions (APRs) can be readily identified in short peptide segments using computer algorithms [12, 13, 14, 15], for intrinsically disordered proteins (IDPs) and globular proteins it is still difficult, if not impossible, to identify aggregation-prone and aggregation-resistant sequences under a given set of conditions. This is because aggregation (taken here to be any non-native oligomeric state) can proceed through diverse mechanisms, driven by distinct physico-chemical mechanisms (Figure 1). In addition, the observed aggregation propensity of each protein sequence/structure on each pathway results from a complex convolution of the effects of its sequence on thermodynamic stability, structure, cooperativity and dynamics, which all also depend on the solution conditions (pH, temperature, ionic strength, solvent, nature of surfaces, etc.). For each and all of the pathways traversed, detailed understanding of the molecular mechanisms of the early stages of aggregation remain elusive. By linking changes in sequence to changes in biophysical and cellular behaviour, powerful new approaches in protein engineering are now able to provide a wealth of insight into this process, which can then be used to enhance the performance of computer algorithms so they are better able to predict protein behaviour. Here we discuss how the integration of protein engineering approaches with orthogonal methods including computational and high-throughput phenotypic screening methods, is now set to tackle this difficult problem.
Delineating aggregation mechanisms using rational protein engineering methods
Rational redesign (i.e. the substitution of a small number of residues in a protein sequence with those having the desired physico-chemical or spatial properties) is an attractive approach to modulate protein aggregation when there is prior knowledge of the mechanism of aggregation (Figure 2) (e.g. by altering a protein–protein interface required for aggregation [16, 17, 18]). Approaches such as alanine scanning can also be used to identify or confirm predictions of residues key to the control of aggregation [19,20]. The ability to identify ‘aggregation hotspots’ has been facilitated by the development of at least 40 different algorithms [12, 13, 14, 15]. While differing in their metrics, these programs generally consider three characteristics which control protein aggregation: solubility, thermodynamic stability and aggregation propensity. These computational tools, summarised in Table 1, provide powerful information with which to start any study of protein aggregation by portraying the inherent aggregation propensity of the protein sequence. However, some consider local protein sequences (generally 4-6 residues in length), leaving open the important questions of how this inherent insolubility/aggregation potential is realised in the context of the entire protein sequence, whether disordered (as in the unfolded state or for IDPs) or when ‘hidden’ by the native 3D structure of the protein.
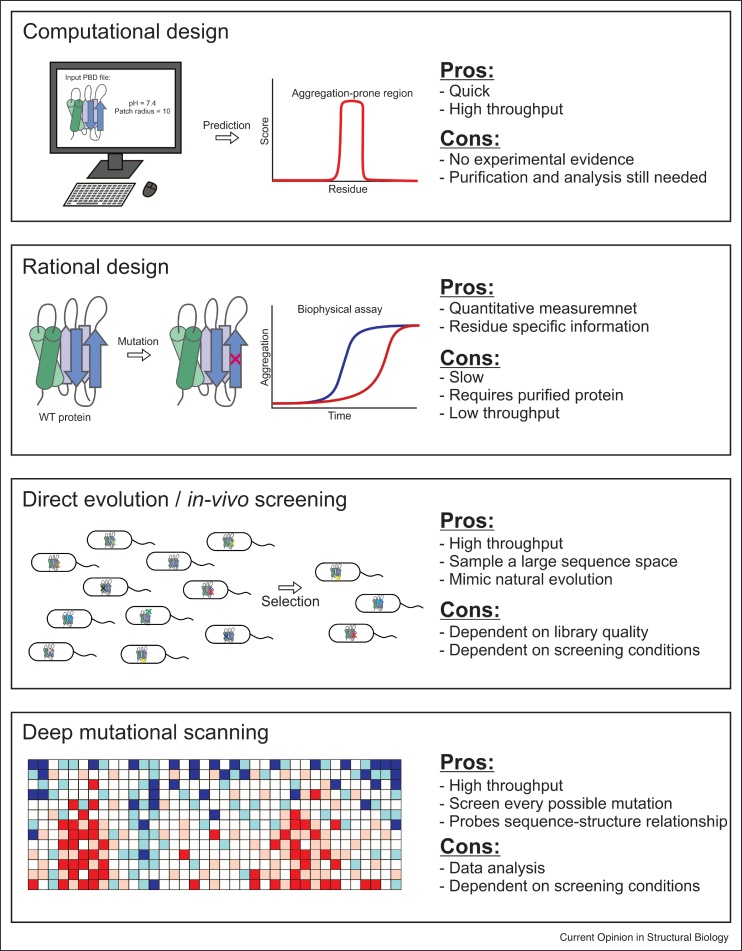
Figure 2.
Summary of different methods for measuring and predicting protein aggregation.
Computational methods can predict aggregation-prone regions using sequence or structure input. Rational design involves introducing specific mutations into a protein and subsequent analysis of the mutational effect in comparison to the behaviour of the wild-type protein. Directed evolution and in vivo screening methods obviate protein purification and large numbers of variants can be screened to identify proteins with enhanced properties. Finally, deep mutational scanning can potentially samples every possible mutation and enables quantification of the effect on protein stability or aggregation to be determined in vivo.
Table 1.
Computational methods to predict and modulate protein aggregation. Methods are grouped by calculated metric and are subdivided into methods that use primary or tertiary sequence data. Algorithms denoted with ‘P’ represent those specific to Prion formation.
| Protein solubility | |
|---|---|
| Sequence | Structure |
| Aggrescan [88] CamSol intrinsic [89] Protein-Sol [90] Proso II [91] |
Aggrescan3D 2.0 [39] CamSol [89] SAP [92] SOLart [93] |
| Aggregation propensity | |
|---|---|
| Sequence | Structure |
| Zyggregator [94] TANGO [22] Pafig [95] SALSA [96] WALTZ-DB 2.0 [97] AmyCoP [98] pWALTZP [99] PrionWP [100] PLAACP [101] pRANKP [102] PAPAP [103] |
PASTA 2.0 [104] Solubis [42] FoldAmyloid [105] NetCSSP [106] BETASCAN [107] STICHER [108] Zipper DB [109] AmyloidMutants [110] AMYLPRED2 [111] |
Detecting aggregation-prone regions in primary sequences
More than 80 % of proteins possess at least one region in their sequence that has a propensity to aggregate (i.e. APRs [21]), calculated based on hydrophobicity, charge patterning, aromatic content and β-sheet propensity [12]. These algorithms use the primary amino acid sequence to predict APRs via empirical training sets or/and calculation based in the known physicochemical properties of the 20 canonical amino acids [12]. One of these algorithms, TANGO [22] (Table 1), identifies APRs by calculating the propensity of penta-peptide sequences to form buried β-sheets, using an algorithm trained on experimental measurement. In an exciting recent application of this algorithm, Khodaparast et al. [23•] identified APRs enriched in the Escherichia coli proteome, and used the resulting information to develop new antibacterial agents by expression of redundant APRs (that were not sequence unique in the genome). Expression of 125 of these sequences resulted in cell death by inducing widespread aggregation of 541 proteins (identified using mass spectrometry) into cross β-structure-enriched inclusion bodies. In marked contrast, overexpression of unique APR sequences within the proteome had no effect. Antimicrobial amyloid-nucleating peptides were bactericidal for a large number of Gram-negative bacteria, suggesting that the approach may have therapeutic potential. Similar approaches from the same groups have also been used as anti-cancer strategies [24], suggesting the general utility of this method to exploit protein aggregation for beneficial purposes.
Effect of ‘order’ in intrinsically disordered proteins
Transient structure formed within IDPs and short peptides can profoundly affect the observed aggregation rate of APRs. For example, the aggregation of Tau, a largely unstructured 441-residue protein which is associated with several neurodegenerative diseases, including Alzheimer’s, Pick and chronic traumatic encephalopathy [25, 26, 27], is thought to be largely driven by the amyloidogenic six-residue peptide sequence 306VQIVYK311 [28••]. Perplexingly, mutations genetically linked to tauopathies such as P301L/S are found outside this sequence. This is similar to the positional relationship between point variants of α-synuclein associated with early onset familial Parkinson’s disease and the non-amyloid component (NAC) region shown to be necessary and sufficient for aggregation [29,30]. Cross-linking studies, together with molecular dynamics (MD) simulations, showed that residues 295-311 of Tau form a β-hairpin, sequestering the APR, and slowing aggregation. Accordingly, destabilising the β-hairpin (by substitution of P301 with a bulky leucine residue) was to found to speed up aggregation, while stabilising it (via adding a Trp-zip motif to the termini of the β-hairpin, in the P301L background) slowed down aggregation [28••]. These elegant protein engineering experiments were thus able to confirm β-hairpin formation as a controlling mechanism of aggregation, in which the aggregation potential of the APR is modulated by specific structure formation in a region that both flanks and overlaps with the APR.
The transient and often promiscuous intra-molecular and inter-molecular interactions that control aggregation of both IDPs and initially structured proteins are challenging to study, but are necessary to understand and map because of their central importance in initiating aggregation. As a consequence of their dynamic and heterogeneous nature, high resolution structural techniques to map these important protein–protein interactions (Figure 1) are difficult, if not impossible to perform. However, these sequences can be engineered to allow site-specific introduction of specific reagents or reporters to gain low resolution information. These include cross-linking reagents, such as diazirines [31•], ruthenium complexes (PICUP) [32] and disuccinimidyl suberate (DSS) [33], which when coupled with mass spectrometric techniques [34], allow identification of pairs of residues that are spatially localised within the dynamic ensemble, even if only transiently populated [35]. Other reagents allow spectroscopic analyses. For example, introduction of spin labels (introduced via unique Cys residues) at single sites across proteins allows identification of transient interactions between sequence-distant residues, or between protein molecules using NMR (using an approach known as Paramagnetic Relaxation Enhancement [36]) or EPR (using Pulsed electron double resonance (PELDOR)/double electron-electron resonance (DEER) EPR spectroscopy [37]. These methods have been applied to α-synuclein, revealing that this IDP makes extensive intra-molecular and inter-molecular contacts which are highly sensitive to environmental conditions [29,37]. Such properties are reminiscent of those described above for Tau, especially as early onset familial missense variants occur outside of the main amyloid core for both proteins, suggestive of similar mechanisms at work that control the aggregation of these IDPs in vitro and possibly also in vivo. Similarly to Tau, formation of a β-hairpin structure (residues 37 to 54) in a region upstream to NAC (residues 61-95) in α-synuclein, induced upon complexation with a β-wrapin engineered binding protein, resulted in inhibition of amyloid formation [38]. These methods often yield low resolution, relatively sparse structural information, but by integrating the outputs from different approaches, remarkably precise molecular mechanisms of aggregation can result, especially when complemented with MD simulations. For example, Bunce et al. [31•] used fluorescence quenching of an extrinsic fluorophore (TAMRA-Ahx) and cross-linking studies to determine how the peptide Aβ16-22 (a fragment of Aβ40/42 associated with Alzheimer’s disease) aggregates and is able to catalyse self-assembly of Aβ40 via secondary surface nucleation.
Effect of ‘disorder’ in the aggregation of globular proteins
Understanding the effect of protein dynamics, sequence and solution conditions is also critically important for determining, and hence predicting, the potential of globular proteins to aggregate. Given that aggregation can occur from the native state, or from partially or globally unfolded species (Figure 1), our ability to predict aggregation requires understanding of the local and global unfolding properties of the protein and how this depends on sequence and solution conditions. Simulation (and quantification) of protein dynamics in silico offers a solution to this problem, but requires greater computational resources and, in some cases, development of force fields able to accurately simulate protein behaviour. Aggrescan3D 2.0 [39] (Table 1) addresses this issue by using CABS-flex [40,41] for rapid simulations (10 nanoseconds length) of near-native dynamics of globular proteins. This ‘dynamic mode’ of Aggrescan3D 2.0 yielded higher aggregation propensity estimates for 80 % of the proteins tested relative to the value obtained from static structures. An alternative approach is to integrate rapid computational methods to predict protein solubility with algorithms able to predict thermodynamic stability. Solubis [42,43] (Table 1), for example, combines TANGO [22] (to identify APRs) with FoldX [44] (to compute the effect of amino acid substitutions on thermodynamic stability). Solubis [42,43] can be used to identify positions in a protein structure able to accommodate gatekeeper residues (i.e. residues with low β-sheet propensity (e.g. Pro) and high solubility such as the charged amino acids (Arg, Lys, Glu and Asp) with minimal changes in protein stability (ΔG°UN). This allows the redesign of proteins to retain stable and native folds, but to reduce aggregation propensity. This approach has been used successfully to decrease the aggregation kinetics of the Protective Antigen protein from B. anthracis [42], a key component in Anthrax vaccines [45], while preserving the native structure and function. This highlights the power of utilising the interdependency of solubility, stability and aggregation propensity to determine and re-engineer a protein’s aggregation potential.
Understanding the diverse effects of electrostatics on protein aggregation
Proteins containing low complexity prion-like domains (PRDs), typically IDPs enriched in glycine and hydrophilic residues, play an important role in the formation of liquid-liquid phase separated membrane-less organelles such as the nucleolus, stress granules and P-bodies [46], and may allow generation of selectable genetic variability akin to that previously reported for prions [47]. Despite the relative depletion of hydrophobic residues in PRDs, reversible amyloid fibril formation can occur upon liquid-liquid phase separation of such sequences, and hence these sequences are known as LARKS (low-complexity aromatic-rich kinked segments) [48]. One such example is hnRNPA1 [49••], an RNA binding protein in which missense mutations are associated with neurodegenerative diseases [50]. Scanning the low complexity (LC) domains for segments containing (Asn)-Asp-(Asn) and (Gly)-Phe/Tyr-(Gly) motifs identified three peptides which formed a hydrogel composed of amyloid fibrils that dissociated upon an increase in temperature [49••]. The structure of the first reversible amyloid core (termed hnRAC1) revealed the cross-β architecture expected for an amyloid fibril, but with notable differences, thought to be important for their function. Firstly, the inter-sheet interface was composed of hydrophilic Asn residues compared with the dry steric zipper typical of amyloid [3,4]. The fibre was further destabilised by the stacking of an aspartic acid (D214) along the exterior face of its parallel in-register β-sheets. Finally, the structure revealed a kink at G211, thought to allow hydrogel formation by sterically facilitating inter-fibrillar cross-linking via π-π stacking of the adjacent Phe and other residues. Accordingly, an hnRAC1 peptide containing G211V, or F210A, or F216A displayed reversible fibril formation, but impaired hydrogel formation. Conversely substituting the destabilising aspartic acid residue (D214V/N) in hnRAC1 resulted in irreversible fibril formation. Interestingly, Asp, Val or Asn substitutions are also found in familial amyotrophic lateral sclerosis (ALS) patients and result in irreversible fibril formation [51]. Taken together, the results provide a structural rationale for ‘maturation’ of irreversible amyloid fibrils within liquid-liquid phase separated low complexity PRDs. The recognition that the aggregation propensity (and liquid-liquid de-mixing) of PRDs is driven by sequences that are chemically and sterically distinct to those involved in amyloid formation [52] has led to the development of AMYCO [53] an algorithm specialised for the prediction of PRDs (Table 1).
Electrostatic interactions are also important drivers and modulators of the aggregation of globular proteins, with pH and ionic strength being important determinants of aggregation both by increasing the probability of proteins unfolding, and by changing the probability of productive protein–protein interactions between transiently exposed APRs in non-native states [54, 55, 56]. The aggregation of natively structured proteins can also be problematic for proteins produced at scale, such as in the biopharmaceutical industry in which proteins are manufactured in high volume and at high concentration [2]. In these cases, aggregation is reversible (at least in the initial stages) and is driven by intermolecular contacts mediated by the presence of hydrophobic or charge-complemented patches on the protein surface, via a mechanism referred to as ‘colloidal aggregation’ [57] (Figure 1). ‘Supercharging’ proteins by introduction of an excess of acidic or basic residues throughout the protein [58, 59, 60] has been shown to reduce aggregation induced by such pathways. Alternatively, introducing defined clusters of specific charged residues that enhance protein stability and reduce protein–protein interactions has been shown to be an effective strategy to reduce aggregation [59,61, 62, 63].
Using directed evolution and in vivo screening to define aggregation landscapes
Directed evolution (DE) methods involve generating diversity in the gene of interest and then isolating variants with improved characteristics from this library using phenotypic selection [64] (Figure 2). DE approaches have been used to develop aggregation-resistant biopharmaceuticals by screening for thermal resistance [65,66], or by utilising three selection methods (temperature, reduction and hydrophobicity) in parallel [67]. A potential disadvantage of optimising a protein’s sequence in this manner is that function is ignored, which can result in proteins with enhanced biophysical properties, but reduced activity, akin to sequence-stability trade-offs [68]. To counter this, Wang et al. described a soluble expression phage assisted continuous evolution method (SE-PACE) [69•]. Here, the protein of interest (POI) is linked in-frame to the N-terminal fragment of a split T7 RNA polymerase (to select for soluble POIs), as well as to the omega subunit of RNA polymerase (RNAP) to select for POIs with high target binding affinity using a bacterial two hybrid approach (using 434 phage cI repressor as the DNA-binding domain). Linking expression of soluble and functional POI to these distinct polymerases allowed both traits to be selected for simultaneously by only allowing expression of the minor coat protein III (pIII) required for progeny phage upon expression and complementation of both the N-terminal and C-terminal fragments of an intein transcribed by RNAP and T7 polymerase, respectively. Using this approach, a fivefold enhancement of expression, but unchanged target affinity was achieved for single-chain antibody fragments (scFvs), as well as enhancement of both expression and activity for the enzyme cytidine deaminase.
If aggregation occurs via a partially folded protein structure, the propensity to aggregate may not always correlate with protein thermal stability. For such proteins, it is necessary to develop alternative screens to create proteins with enhanced solution behaviour. One route to achieve this has recently been developed, in which an E. coli β-lactamase folding reporter links the innate ability of a protein to aggregate to antibiotic sensitivity by fusing the POI between two domains of β-lactamase [70,71]. The system has been shown to be able to differentiate between aggregation-prone and aggregation-resistant variants of diverse protein sequences and structures, including the aggregation-prone peptides Aβ1-42 and amylin, the aggregation-prone protein β2 microglobulin, and single domain antibodies. The system has also been used to screen for small molecule inhibitors of protein aggregation in the periplasm of E. coli [71] and for the selection of excipients able to suppress aggregation [72]. Other screens that link survival to dihydrofolate reductase (DHFR) activity have also been developed and used to identify peptide inhibitors of α-synuclein aggregation [73] and to characterise the phenotypes of 99 % of all the possible single-site substitutions of Aβ1-42 (see below) [74].
Screening peptides in vivo has also been used to gain deeper insight into the pathways by which toxic aggregates are formed [75•]. A combinatorial library of >10 million short cyclic peptides (S/T/C-X1-Xn, where X is any amino-acid and n = 3-5) was produced in bacteria using split intein-mediated circular ligation of peptides and proteins [76]. The peptides were then screened for their ability to reduce aggregation monitored by a reduction in fluorescence of an Aβ1-42-GFP fusion reporter by fluorescence-activated cell sorting [75•]. Biochemical analysis of clones that increased fluorescence revealed penta-peptides that halt Aβ1-42 aggregation by stabilising β-sheet-like structures. These peptides also reduced toxicity measured in primary neuronal cell lines and in vivo.
Developing enhanced understanding of protein behaviour using deep mutational scanning
Deep mutational scanning (DMS) can be used to reveal the effect of thousands of different single amino-acid substitutions on a protein’s properties by quantifying the relative change of abundance of each member of the library under a suitable selective pressure using next generation sequencing methods [77]. This approach is extremely powerful as it combines the strengths of both ‘traditional’ protein engineering methods (quantifiable sequence-phenotype relationships) and DE methods (the ability search vast areas of sequence space without protein purification) (Figure 2). DMS has thus found broad application from structure determination [78,79] to developing a better understanding of the determinants of protein thermodynamic stability [80] and even the utility of alanine scanning [81].
Two studies have recently used DMS to gain a broader understanding of the relationship between sequence and aggregation mechanism. Firstly, to investigate the molecular determinants of Aβ1-42 aggregation Gray et al., [74], used selective growth pressure in yeast cells, by fusing Aβ1-42 to DHFR and growing a library of Aβ1-42 variants using methotrexate as a selective pressure for DHFR function. This screen evaluated 791/798 of all possible single amino acid substitutions of Aβ1-42. Remarkably, 25 % of the variants were more soluble than Aβ1-42, with the others showing unchanged or increased aggregation propensity. Substitutions to Asp and Pro enhanced solubility the most (presumably by increasing charge or decreasing β-strand propensity, respectively), whereas substitutions with Trp or Phe were associated with greater aggregation (presumably by increasing hydrophobicity). This mutational information revealed residues 17-20, 31-32, 34-35, 39 and 41 as ‘hotspots’ important for Aβ1-42 aggregation, which most likely form buried β-strands. Interestingly, these concur with predictions of APRs, for example, using TANGO and Zyggregator [82]. In the second example, Bolognesi et al. exploited the ability to measure the sequence-function relationships of thousands of variants in parallel to understand the relationship between aggregation and toxicity focussing on the PRD of the TAR DNA-binding domain 43 (TDP-43), the aggregation of which is linked to ALS [83••]. Comparison of the relative change in the population of >50 000 variants of yeast cells containing one or two substitutions in TDP-43 before and after induction revealed a 31 residue ‘toxic hotspot’, which correlates with the region of the protein in which mutations occur in ALS patients. Surprisingly, substitutions in this hotspot that increase hydrophobicity decreased toxicity, whereas substitutions that increase charge or polarity increased toxicity. Variants with increased hydrophobicity produced larger, stable aggregates that are less toxic than the small liquid-like loci found at the nuclear periphery for the more toxic variants. Furthermore, epistatic analysis of variants containing two substitutions suggested the presence of secondary structure in this apparently disordered domain. This powerful method can thus be used to identify the structural properties of IDPs in vivo and, further, to interrogate the relationship between the function and toxicity of amyloid versus protein assemblies in liquid-liquid phase separation.
Future perspectives
The synergy between protein engineering and biophysical measurements in vitro with cellular approaches has been integral to developing our understanding of protein aggregation (Figure 2). The diversity of aggregates and aggregation mechanisms, together with the emerging realisation that even IDPs contain transient structure crucial to their function and aggregation potentials, and the finding that native state dynamics are crucial to understanding aggregation propensity, pose enormous current challenges to our ability to predict and modulate aggregation. The ability to rapidly survey the aggregation propensity of large numbers of highly homologous sequences using DMS methods together with statistical and machine learning methods, is now able to guide protein engineering [84,85] and, in the future, is sure to guide the development of new predictive algorithms. These large datasets, when integrated with detailed spectroscopic and cross-linking studies (all made possible by protein engineering approaches), MD simulations and cellular insights, will allow us in the future to define the relationship between sequence, structure, function and aggregation. This will allow genome engineering or the development of small molecules or biomolecules able to control protein aggregation and to develop and manufacture biotherapeutics more rapidly and economically. What is clear is that there is still much to learn, but the powers of modern protein engineering methods, combined with the ability to harness the information that results through machine learning, promises a step change into our ability to understand protein behaviour and to capitalise on the new knowledge to capture the complexity and powers of proteins for the benefits of humankind.
CRediT author statement
JSE and NG prepared the original draft, SER and DJB reviewed and edited the manuscript.
Conflict of interest statement
Nothing declared.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgements
We thank the Brockwell and Radford laboratories especially members of our amyloid and biologics teams for many helpful discussions and comments. We acknowledge funding from the Wellcome Trust (204963) (to S.E.R and N.G) and the Biotechnology and Biological Sciences Research Council (BBSRC) and AstraZeneca for funding J.S.E (BB/M011151/1).
References
- 1.Hartl F.U. Protein misfolding diseases. Annu Rev Biochem. 2017;86:21–26. doi: 10.1146/annurev-biochem-061516-044518. [DOI] [PubMed] [Google Scholar]
- 2.Roberts C.J. Therapeutic protein aggregation: mechanisms, design, and control. Trends Biotechnol. 2014;32:372–380. doi: 10.1016/j.tibtech.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iadanza M.G., Jackson M.P., Hewitt E.W., Ranson N.A., Radford S.E. A new era for understanding amyloid structures and disease. Nat Rev Mol Cell Biol. 2018;19:755–773. doi: 10.1038/s41580-018-0060-8. [DOI] [PubMed] [Google Scholar]
- 4.Gallardo R., Ranson N.A., Radford S.E. Amyloid structures: much more than just a cross-β fold. Curr Opin Struct Biol. 2020;60:7–16. doi: 10.1016/j.sbi.2019.09.001. [DOI] [PubMed] [Google Scholar]
- 5.Hyman A.A., Weber C.A., Ulicher F.J. Liquid-liquid phase separation in biology. Annu Rev Cell Dev Biol. 2014;30:39–58. doi: 10.1146/annurev-cellbio-100913-013325. [DOI] [PubMed] [Google Scholar]
- 6.Boeynaems S., Alberti S., Fawzi N.L., Mittag T., Polymenidou M., Rousseau F., Schymkowitz J., Shorter J., Wolozin B., Van Den Bosch L. Protein phase separation: a new phase in cell biology. Trends Cell Biol. 2018;28:420–435. doi: 10.1016/j.tcb.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frottin F., Schueder F., Tiwary S., Gupta R., Körner R., Schlichthaerle T., Cox J., Jungmann R., Hartl F.U., Hipp M.S. The nucleolus functions as a phase-separated protein quality control compartment. Science. 2019;365:342–347. doi: 10.1126/science.aaw9157. [DOI] [PubMed] [Google Scholar]
- 8.Alenquer M., Vale-Costa S., Etibor T.A., Ferreira F., Sousa A.L., Amorim M.J. Influenza A virus ribonucleoproteins form liquid organelles at endoplasmic reticulum exit sites. Nat Commun. 2019;10:1629. doi: 10.1038/s41467-019-09549-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Babinchak W.M., Haider R., Dumm B.K., Sarkar P., Surewicz K., Choi J.K., Surewicz W.K. The role of liquid-liquid phase separation in aggregation of the TDP-43 low-complexity domain. J Biol Chem. 2019;294:6306–6317. doi: 10.1074/jbc.RA118.007222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Newton M.S., Arcus V.L., Gerth M.L., Patrick W.M. Enzyme evolution: innovation is easy, optimization is complicated. Curr Opin Struct Biol. 2018;48:110–116. doi: 10.1016/j.sbi.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 11.Chiu M.L., Gilliland G.L. Engineering antibody therapeutics. Curr Opin Struct Biol. 2016;38:163–173. doi: 10.1016/j.sbi.2016.07.012. [DOI] [PubMed] [Google Scholar]
- 12.Meric G., Robinson A.S., Roberts C.J. Driving forces for nonnative protein aggregation and approaches to predict aggregation-prone regions. Annu Rev Chem Biomol Eng. 2017;8:139–159. doi: 10.1146/annurev-chembioeng-060816-101404. [DOI] [PubMed] [Google Scholar]
- 13.Buck P.M., Kumar S., Wang X., Agrawal N.J., Trout B.L., Singh S.K. Computational methods to predict therapeutic protein aggregation. Methods Mol Biol. 2012;899:425–451. doi: 10.1007/978-1-61779-921-1_26. [DOI] [PubMed] [Google Scholar]
- 14.Batlle C., Iglesias V., Navarro S., Ventura S. Prion-like proteins and their computational identification in proteomes. Expert Rev Proteomics. 2017;14:335–350. doi: 10.1080/14789450.2017.1304214. [DOI] [PubMed] [Google Scholar]
- 15.Agrawal N.J., Kumar S., Wang X., Helk B., Singh S.K., Trout B.L. Aggregation in protein-based biotherapeutics: computational studies and tools to identify aggregation-prone regions. J Pharm Sci. 2011;100:5081–5095. doi: 10.1002/jps.22705. [DOI] [PubMed] [Google Scholar]
- 16.Dobson C.L., Devine P.W.A., Phillips J.J., Higazi D.R., Lloyd C., Popovic B., Arnold J., Buchanan A., Lewis A., Goodman J. Engineering the surface properties of a human monoclonal antibody prevents self-association and rapid clearance in vivo. Sci Rep. 2016;6 doi: 10.1038/srep38644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karamanos T.K., Kalverda A.P., Thompson G.S., Radford S.E. Visualization of transient protein-protein interactions that promote or inhibit amyloid assembly. Mol Cell. 2014;55:214–226. doi: 10.1016/j.molcel.2014.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rennella E., Morgan G.J., Kelly J.W., Kay L.E. Role of domain interactions in the aggregation of full-length immunoglobulin light chains. Proc Natl Acad Sci U S A. 2019;116:854–863. doi: 10.1073/pnas.1817538116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Williams A.D., Shivaprasad S., Wetzel R. Alanine scanning mutagenesis of Aβ(1-40) amyloid fibril stability. J Mol Biol. 2006;357:1283–1294. doi: 10.1016/j.jmb.2006.01.041. [DOI] [PubMed] [Google Scholar]
- 20.Lupton C.J., Steer D.L., Bottomley S.P., Hughes V.A., Wintrode P.L., Ellisdon A.M. Enhanced molecular mobility of ordinarily structured regions drives polyglutamine disease. J Biol Chem. 2015;290:24190–24200. doi: 10.1074/jbc.M115.659532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prabakaran R., Goel D., Kumar S., Gromiha M.M. Aggregation prone regions in human proteome: Insights from large-scale data analyses. Proteins Struct Funct Bioinformatics. 2017;85:1099–1118. doi: 10.1002/prot.25276. [DOI] [PubMed] [Google Scholar]
- 22.Fernandez-Escamilla A.-M., Rousseau F., Schymkowitz J., Serrano L. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat Biotechnol. 2004;22:1302–1306. doi: 10.1038/nbt1012. [DOI] [PubMed] [Google Scholar]
- 23•.Khodaparast L., Khodaparast L., Gallardo R., Louros N.N., Michiels E., Ramakrishnan R., Ramakers M., Claes F., Young L., Shahrooei M. Aggregating sequences that occur in many proteins constitute weak spots of bacterial proteostasis. Nat Commun. 2018;9 doi: 10.1038/s41467-018-03131-0. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors develop a new antibacterial strategy by targeting redundant APRs in E. coli. In this approach they identify peptides that upon expression trigger widespread and lethal aggregation of bacterial proteins.
- 24.Gallardo R., Ramakers M., De Smet F., Claes F., Khodaparast L., Khodaparast L., Couceiro J.R., Langenberg T., Siemons M., Nyström S. De novo design of a biologically active amyloid. Science. 2016;354 doi: 10.1126/science.aah4949. aah4949. [DOI] [PubMed] [Google Scholar]
- 25.Chong F.P., Ng K.Y., Koh R.Y., Chye S.M. Tau proteins and tauopathies in Alzheimer’s disease. Cell Mol Neurobiol. 2018;38:965–980. doi: 10.1007/s10571-017-0574-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Falcon B., Zhang W., Murzin A.G., Murshudov G., Garringer H.J., Vidal R., Crowther R.A., Ghetti B., Scheres S.H.W., Goedert M. Structures of filaments from Pick’s disease reveal a novel tau protein fold. Nature. 2018;561:137–140. doi: 10.1038/s41586-018-0454-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Falcon B., Zivanov J., Zhang W., Murzin A.G., Garringer H.J., Vidal R., Crowther R.A., Newell K.L., Ghetti B., Goedert M. Novel tau filament fold in chronic traumatic encephalopathy encloses hydrophobic molecules. Nature. 2019;568:420–423. doi: 10.1038/s41586-019-1026-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28••.Chen D., Drombosky K.W., Hou Z., Sari L., Kashmer O.M., Ryder B.D., Perez V.A., Woodard D.R., Lin M.M., Diamond M.I. Tau local structure shields an amyloid-forming motif and controls aggregation propensity. Nat Commun. 2019;10 doi: 10.1038/s41467-019-10355-1. [DOI] [PMC free article] [PubMed] [Google Scholar]; Integration of computational and experimental data shows that disease-related mutations of Tau induce conformational changes that expose an amyloid motif driving aggregation.
- 29.Stephens A.D., Zacharopoulou M., Kaminski Schierle G.S. The cellular environment affects monomeric α-synuclein structure. Trends Biochem Sci. 2019;44:453–466. doi: 10.1016/j.tibs.2018.11.005. [DOI] [PubMed] [Google Scholar]
- 30.Giasson B.I., Murray I.V.J., Trojanowski J.Q., Lee V.M.Y. A hydrophobic stretch of 12 amino acid residues in the middle of α-synuclein is essential for filament assembly. J Biol Chem. 2001;276:2380–2386. doi: 10.1074/jbc.M008919200. [DOI] [PubMed] [Google Scholar]
- 31•.Bunce S.J., Wang Y., Stewart K.L., Ashcroft A.E., Radford S.E., Hall C.K., Wilson A.J. Molecular insights into the surface-catalyzed secondary nucleation of amyloid-β40 (Aβ40) by the peptide fragment Aβ16–22. Sci Adv. 2019;5 doi: 10.1126/sciadv.aav8216. eaav8216. [DOI] [PMC free article] [PubMed] [Google Scholar]; Integration of spectroscopic data with molecular dynamics allowed aggregation to be tracked from the random coil monomer of Aβ16-22 to its secondary nucleation on the surface of Aβ1-40 fibrils.
- 32.Marina G.B., Kirkitadze D., Lomakin A., Vollers S.S., Benedek G.B., Teplow D.B. Amyloid β-protein (Aβ) assembly: Aβ40 and Aβ42 oligomerize through distinct pathways. Proc Natl Acad Sci U S A. 2003;100:330–335. doi: 10.1073/pnas.222681699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bartels T., Choi J.G., Selkoe D.J. α-Synuclein occurs physiologically as a helically folded tetramer that resists aggregation. Nature. 2011;477:107–111. doi: 10.1038/nature10324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schneider M., Belsom A., Rappsilber J. Protein tertiary structure by crosslinking/mass spectrometry. Trends Biochem Sci. 2018;43:157–169. doi: 10.1016/j.tibs.2017.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Preston G.W., Radford S.E., Ashcroft A.E., Wilson A.J. Analysis of amyloid nanostructures using photo-cross-linking: In situ comparison of three widely used photo-cross-linkers. ACS Chem Biol. 2014;9:761–768. doi: 10.1021/cb400731s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karamanos T.K., Kalverda A.P., Thompson G.S., Radford S.E. Mechanisms of amyloid formation revealed by solution NMR. Prog Nucl Magn Reson Spectrosc. 2015;88–89:86–104. doi: 10.1016/j.pnmrs.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen M., Margittai M., Chen J., Langen R. Investigation of α-synuclein fibril structure by site-directed spin labeling. J Biol Chem. 2007;282:24970–24979. doi: 10.1074/jbc.M700368200. [DOI] [PubMed] [Google Scholar]
- 38.Mirecka E.A., Shaykhalishahi H., Gauhar A., Akgül Ş, Lecher J., Willbold D., Stoldt M., Hoyer W. Sequestration of a β-hairpin for control of α-synuclein aggregation. Angew Chemie Int Ed. 2014;53:4227–4230. doi: 10.1002/anie.201309001. [DOI] [PubMed] [Google Scholar]
- 39.Kuriata A., Iglesias V., Pujols J., Kurcinski M., Kmiecik S., Ventura S. Aggrescan3D (A3D) 2.0: prediction and engineering of protein solubility. Nucleic Acids Res. 2019;47:W300–W307. doi: 10.1093/nar/gkz321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuriata A., Gierut A.M., Oleniecki T., Ciemny M.P., Kolinski A., Kurcinski M., Kmiecik S. CABS-flex 2.0: a web server for fast simulations of flexibility of protein structures. Nucleic Acids Res. 2018;46:W338–W343. doi: 10.1093/nar/gky356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kurcinski M., Oleniecki T., Ciemny M.P., Kuriata A., Kolinski A., Kmiecik S. CABS-flex standalone: a simulation environment for fast modeling of protein flexibility. Bioinformatics. 2019;35:694–695. doi: 10.1093/bioinformatics/bty685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van Durme J., De Baets G., Van Der Kant R., Ramakers M., Ganesan A., Wilkinson H., Gallardo R., Rousseau F., Schymkowitz J. Solubis: a webserver to reduce protein aggregation through mutation. Protein Eng Des Sel. 2016;29:285–289. doi: 10.1093/protein/gzw019. [DOI] [PubMed] [Google Scholar]
- 43.van der Kant R., van Durme J., Rousseau F., Schymkowitz J. SolubiS: optimizing protein solubility by minimal point mutations. Methods Mol Biol. 2019;1873:317–333. doi: 10.1007/978-1-4939-8820-4_21. [DOI] [PubMed] [Google Scholar]
- 44.Schymkowitz J., Borg J., Stricher F., Nys R., Rousseau F., Serrano L. The FoldX web server: an online force field. Nucleic Acids Res. 2005;33:W382–W388. doi: 10.1093/nar/gki387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brey R.N. Molecular basis for improved anthrax vaccines. Adv Drug Deliv Rev. 2005;57:1266–1292. doi: 10.1016/j.addr.2005.01.028. [DOI] [PubMed] [Google Scholar]
- 46.Bergeron-Sandoval L.P., Safaee N., Michnick S.W. Mechanisms and consequences of macromolecular phase separation. Cell. 2016;165:1067–1079. doi: 10.1016/j.cell.2016.05.026. [DOI] [PubMed] [Google Scholar]
- 47.Sanchez de Groot N., Torrent Burgas M., Ravarani C.N., Trusina A., Ventura S., Babu M.M. The fitness cost and benefit of phase‐separated protein deposits. Mol Syst Biol. 2019;15:e8075. doi: 10.15252/msb.20178075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hughes M.P., Sawaya M.R., Boyer D.R., Goldschmidt L., Rodriguez J.A., Cascio D., Chong L., Gonen T., Eisenberg D.S. Atomic structures of low-complexity protein segments reveal kinked β sheets that assemble networks. Science. 2018;359:698–701. doi: 10.1126/science.aan6398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49••.Gui X., Luo F., Li Y., Zhou H., Qin Z., Liu Z., Gu J., Xie M., Zhao K., Dai B. Structural basis for reversible amyloids of hnRNPA1 elucidates their role in stress granule assembly. Nat Commun. 2019;10 doi: 10.1038/s41467-019-09902-7. [DOI] [PMC free article] [PubMed] [Google Scholar]; Amyloid fibres within liquid droplets of hnRNPA1 are dynamic intermediates en route to irreversible fibrils. The atomic structure of the reversible amyloid core helps to explain the effect of mutations involved in ALS.
- 50.Kim H.J., Kim N.C., Wang Y.D., Scarborough E.A., Moore J., Diaz Z., MacLea K.S., Freibaum B., Li S., Molliex A. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 2013;495:467–473. doi: 10.1038/nature11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Prasad A., Bharathi V., Sivalingam V., Girdhar A., Patel B.K. Molecular mechanisms of TDP-43 misfolding and pathology in amyotrophic lateral sclerosis. Front Mol Neurosci. 2019;12:25. doi: 10.3389/fnmol.2019.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Toombs J.A., McCarty B.R., Ross E.D. Compositional determinants of prion formation in yeast. Mol Cell Biol. 2010;30:319–332. doi: 10.1128/MCB.01140-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Iglesias V., Conchillo-Sole O., Batlle C., Ventura S. AMYCO: evaluation of mutational impact on prion-like proteins aggregation propensity. BMC Bioinformatics. 2019;20:24. doi: 10.1186/s12859-019-2601-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van der Kant R., Karow-Zwick A.R., Van Durme J., Blech M., Gallardo R., Seeliger D., Aßfalg K., Baatsen P., Compernolle G., Gils A. Prediction and reduction of the aggregation of monoclonal antibodies. J Mol Biol. 2017;429:1244–1261. doi: 10.1016/j.jmb.2017.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu K.P., Baum J. Detection of transient interchain interactions in the intrinsically disordered protein α-synuclein by NMR paramagnetic relaxation enhancement. J Am Chem Soc. 2010;132:5546–5547. doi: 10.1021/ja9105495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goto Y., Adachi M., Muta H., So M. Salt-induced formations of partially folded intermediates and amyloid fibrils suggests a common underlying mechanism. Biophys Rev. 2018;10:493–502. doi: 10.1007/s12551-017-0370-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hofmann M., Gieseler H. Predictive screening tools used in high-concentration protein formulation development. J Pharm Sci. 2018;107:772–777. doi: 10.1016/j.xphs.2017.10.036. [DOI] [PubMed] [Google Scholar]
- 58.Simeonov P., Berger-Hoffmann R., Hoffmann R., Strater N., Zuchner T. Surface supercharged human enteropeptidase light chain shows improved solubility and refolding yield. Protein Eng Des Sel. 2011;24:261–268. doi: 10.1093/protein/gzq104. [DOI] [PubMed] [Google Scholar]
- 59.Lawrence M.S., Phillips K.J., Liu D.R. Supercharging proteins can impart unusual resilience. J Am Chem Soc. 2007;129:10110–10112. doi: 10.1021/ja071641y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Miklos A.E., Kluwe C., Der B.S., Pai S., Sircar A., Hughes R.A., Berrondo M., Xu J., Codrea V., Buckley P.E. Structure-based design of supercharged, highly thermoresistant antibodies. Chem Biol. 2012;19:449–455. doi: 10.1016/j.chembiol.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee C.C., Julian M.C., Tiller K.E., Meng F., DuConge S.E., Akter R., Raleigh D.P., Tessier P.M. Design and optimization of anti-amyloid domain antibodies specific for β-amyloid and islet amyloid polypeptide. J Biol Chem. 2016;291:2858–2873. doi: 10.1074/jbc.M115.682336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Perchiacca J.M., Bhattacharya M., Tessier P.M. Mutational analysis of domain antibodies reveals aggregation hotspots within and near the complementarity determining regions. Proteins Struct Funct Bioinformatics. 2011;79:2637–2647. doi: 10.1002/prot.23085. [DOI] [PubMed] [Google Scholar]
- 63.Austerberry J.I., Dajani R., Panova S., Roberts D., Golovanov A.P., Pluen A., van der Walle C.F., Uddin S., Warwicker J., Derrick J.P. The effect of charge mutations on the stability and aggregation of a human single chain Fv fragment. Eur J Pharm Biopharm. 2017;115:18–30. doi: 10.1016/j.ejpb.2017.01.019. [DOI] [PubMed] [Google Scholar]
- 64.Packer M.S., Liu D.R. Methods for the directed evolution of proteins. Nat Rev Genet. 2015;16:379–394. doi: 10.1038/nrg3927. [DOI] [PubMed] [Google Scholar]
- 65.Jespers L., Schon O., Famm K., Winter G. Aggregation-resistant domain antibodies selected on phage by heat denaturation. Nat Biotechnol. 2004;22:1161–1165. doi: 10.1038/nbt1000. [DOI] [PubMed] [Google Scholar]
- 66.Famm K., Hansen L., Christ D., Winter G. Thermodynamically stable aggregation-resistant antibody domains through directed evolution. J Mol Biol. 2008;376:926–931. doi: 10.1016/j.jmb.2007.10.075. [DOI] [PubMed] [Google Scholar]
- 67.Buchanan A., Ferraro F., Rust S., Sridharan S., Franks R., Dean G., McCourt M., Jermutus L., Minter R. Improved drug-like properties of therapeutic proteins by directed evolution. Protein Eng Des Sel. 2012;25:631–638. doi: 10.1093/protein/gzs054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rabia L.A., Desai A.A., Jhajj H.S., Tessier P.M. Understanding and overcoming trade-offs between antibody affinity, specificity, stability and solubility. Biochem Eng J. 2018;137:365–374. doi: 10.1016/j.bej.2018.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69•.Wang T., Badran A.H., Huang T.P., Liu D.R. Continuous directed evolution of proteins with improved soluble expression. Nat Chem Biol. 2018;14:972–980. doi: 10.1038/s41589-018-0121-5. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors develop a phage-assisted continuous evolution method to simultaneously select for binding affinity and soluble expression.
- 70.Foit L., Morgan G.J., Kern M.J., Steimer L.R., von Hacht A.A., Titchmarsh J., Warriner S.L., Radford S.E., Bardwell J.C.A. Optimizing protein stability in vivo. Mol Cell. 2009;36:861–871. doi: 10.1016/j.molcel.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Saunders J.C., Young L.M., Mahood R.A., Jackson M.P., Revill C.H., Foster R.J., Smith D.A., Ashcroft A.E., Brockwell D.J., Radford S.E. An in vivo platform for identifying inhibitors of protein aggregation. Nat Chem Biol. 2016;12:94–101. doi: 10.1038/nchembio.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hailu T.T., Foit L., Bardwell J.C.A. In vivo detection and quantification of chemicals that enhance protein stability. Anal Biochem. 2013;434:181–186. doi: 10.1016/j.ab.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cheruvara H., Allen-Baume V.L., Kad N.M., Mason J.M. Intracellular screening of a peptide library to derive a potent peptide inhibitor of α-synuclein aggregation. J Biol Chem. 2015;290:7426–7435. doi: 10.1074/jbc.M114.620484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gray V.E., Sitko K., Kameni F.Z.N., Williamson M., Stephany J.J., Hasle N., Fowler D.M. Elucidating the molecular determinants of Aβ aggregation with deep mutational scanning. G3 Genes, Genomes, Genet. 2019;11:3683–3689. doi: 10.1534/g3.119.400535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75•.Matis I., Delivoria D.C., Mavroidi B., Papaevgeniou N., Panoutsou S., Bellou S., Papavasileiou K.D., Linardaki Z.I., Stavropoulou A.V., Vekrellis K. An integrated bacterial system for the discovery of chemical rescuers of disease-associated protein misfolding. Nat Biomed Eng. 2017;1:838–852. doi: 10.1038/s41551-017-0144-3. [DOI] [PubMed] [Google Scholar]; The authors screen a large macrocyclic peptide library in vivo. This method identified cyclic peptides that stabilise β-sheet-like structures in Aβ1-42, slowing amyloid formation.
- 76.Scott C.P., Abel-Santos E., Wall M., Wahnon D.C., Benkovic S.J. Production of cyclic peptides and proteins in vivo. Proc Natl Acad Sci U S A. 1999;96:13638–13643. doi: 10.1073/pnas.96.24.13638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fowler D.M., Fields S. Deep mutational scanning: a new style of protein science. Nat Methods. 2014;11:801–807. doi: 10.1038/nmeth.3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rollins N.J., Brock K.P., Poelwijk F.J., Stiffler M.A., Gauthier N.P., Sander C., Marks D.S. Inferring protein 3D structure from deep mutation scans. Nat Genet. 2019;51:1170–1176. doi: 10.1038/s41588-019-0432-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schmiedel J.M., Lehner B. Determining protein structures using deep mutagenesis. Nat Genet. 2019;51:1177–1186. doi: 10.1038/s41588-019-0431-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nisthal A., Wang C.Y., Ary M.L., Mayo S.L. Protein stability engineering insights revealed by domain-wide comprehensive mutagenesis. Proc Natl Acad Sci U S A. 2019;116:16367–16377. doi: 10.1073/pnas.1903888116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gray V.E., Hause R.J., Fowler D.M. Analysis of large-scale mutagenesis data to assess the impact of single amino acid substitutions. Genetics. 2017;207:53–61. doi: 10.1534/genetics.117.300064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Acharya S., Srivastava K.R., Nagarajan S., Lapidus L.J. Monomer dynamics of Alzheimer peptides and kinetic control of early aggregation in Alzheimer’s disease. Chemphyschem. 2016;17:3470–3479. doi: 10.1002/cphc.201600706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83••.Bolognesi B., Faure A.J., Seuma M., Schmiedel J.M., Tartaglia G.G., Lehner B. The mutational landscape of a prion-like domain. Nat Commun. 2019;10 doi: 10.1038/s41467-019-12101-z. [DOI] [PMC free article] [PubMed] [Google Scholar]; Deep mutational scanning enabled the analysis of >50 000 mutations in TDP-43. Aggregation of TDP-43 is found to be a protective mechanism by decreasing the formation of toxic liquid-like phase foci.
- 84.Yang K.K., Wu Z., Arnold F.H. Machine-learning-guided directed evolution for protein engineering. Nat Methods. 2019;16:687–694. doi: 10.1038/s41592-019-0496-6. [DOI] [PubMed] [Google Scholar]
- 85.Riesselman A.J., Ingraham J.B., Marks D.S. Deep generative models of genetic variation capture the effects of mutations. Nat Methods. 2018;15:816–822. doi: 10.1038/s41592-018-0138-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Meisl G., Kirkegaard J.B., Arosio P., Michaels T.C.T., Vendruscolo M., Dobson C.M., Linse S., Knowles T.P.J. Molecular mechanisms of protein aggregation from global fitting of kinetic models. Nat Protoc. 2016;11:252–272. doi: 10.1038/nprot.2016.010. [DOI] [PubMed] [Google Scholar]
- 87.Roberts C.J. Non-native protein aggregation kinetics. Biotechnol Bioeng. 2007;98:927–938. doi: 10.1002/bit.21627. [DOI] [PubMed] [Google Scholar]
- 88.Conchillo-Solé O., de Groot N.S., Avilés F.X., Vendrell J., Daura X., Ventura S. AGGRESCAN: a server for the prediction and evaluation of “hot spots”; of aggregation in polypeptides. BMC Bioinformatics. 2007;8:65. doi: 10.1186/1471-2105-8-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sormanni P., Aprile F.A., Vendruscolo M. The CamSol method of rational design of protein mutants with enhanced solubility. J Mol Biol. 2015;427:478–490. doi: 10.1016/j.jmb.2014.09.026. [DOI] [PubMed] [Google Scholar]
- 90.Hebditch M., Carballo-Amador M.A., Charonis S., Curtis R., Warwicker J. Protein–Sol: a web tool for predicting protein solubility from sequence. Bioinformatics. 2017;33:3098–3100. doi: 10.1093/bioinformatics/btx345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Smialowski P., Doose G., Torkler P., Kaufmann S., Frishman D. PROSO II--a new method for protein solubility prediction. FEBS J. 2012;279:2192–2200. doi: 10.1111/j.1742-4658.2012.08603.x. [DOI] [PubMed] [Google Scholar]
- 92.Chennamsetty N., Voynov V., Kayser V., Helk B., Trout B.L. Design of therapeutic proteins with enhanced stability. Proc Natl Acad Sci U S A. 2009;106:11937–11942. doi: 10.1073/pnas.0904191106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hou Q., Kwasigroch J.M., Rooman M., Pucci F. SOLart: a structure-based method to predict protein solubility and aggregation. Bioinformatics. 2019 doi: 10.1093/bioinformatics/btz773. [DOI] [PubMed] [Google Scholar]
- 94.Tartaglia G.G., Vendruscolo M. The Zyggregator method for predicting protein aggregation propensities. Chem Soc Rev. 2008;37:1395–1401. doi: 10.1039/b706784b. [DOI] [PubMed] [Google Scholar]
- 95.Tian J., Wu N., Guo J., Fan Y. Prediction of amyloid fibril-forming segments based on a support vector machine. BMC Bioinformatics. 2009;10:S45. doi: 10.1186/1471-2105-10-S1-S45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zibaee S., Makin O.S., Goedert M., Serpell L.C. A simple algorithm locates beta-strands in the amyloid fibril core of alpha-synuclein, Abeta, and tau using the amino acid sequence alone. Protein Sci. 2007;16:906–918. doi: 10.1110/ps.062624507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Louros N., Konstantoulea K., De Vleeschouwer M., Ramakers M., Schymkowitz J., Rousseau F. WALTZ-DB 2.0: an updated database containing structural information of experimentally determined amyloid-forming peptides. Nucleic Acids Res. 2019;48:D389–D393. doi: 10.1093/nar/gkz758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nastou K.C., Nasi G.I., Tsiolaki P.L., Litou Z.I., Iconomidou V.A. AmyCo: the amyloidoses collection. Amyloid. 2019;26:112–117. doi: 10.1080/13506129.2019.1603143. [DOI] [PubMed] [Google Scholar]
- 99.Sabate R., Rousseau F., Schymkowitz J., Ventura S. What makes a protein sequence a prion? PLoS Comput Biol. 2015;11 doi: 10.1371/journal.pcbi.1004013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zambrano R., Conchillo-Sole O., Iglesias V., Illa R., Rousseau F., Schymkowitz J., Sabate R., Daura X., Ventura S. PrionW: a server to identify proteins containing glutamine/asparagine rich prion-like domains and their amyloid cores. Nucleic Acids Res. 2015;43:W331–W337. doi: 10.1093/nar/gkv490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lancaster A.K., Nutter-Upham A., Lindquist S., King O.D. PLAAC: a web and command-line application to identify proteins with prion-like amino acid composition. Bioinformatics. 2014;30:2501–2502. doi: 10.1093/bioinformatics/btu310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Afsar Minhas Ful A., Ross E.D., Ben-Hur A. Amino acid composition predicts prion activity. PLoS Comput Biol. 2017;13 doi: 10.1371/journal.pcbi.1005465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Toombs J.A., Petri M., Paul K.R., Kan G.Y., Ben-Hur A., Rossa E.D. De novo design of synthetic prion domains. Proc Natl Acad Sci U S A. 2012;109:6519–6524. doi: 10.1073/pnas.1119366109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Walsh I., Seno F., Tosatto S.C.E., Trovato A. PASTA 2.0: an improved server for protein aggregation prediction. Nucleic Acids Res. 2014;42:W301–W307. doi: 10.1093/nar/gku399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Garbuzynskiy S.O., Lobanov M.Y., Galzitskaya O.V. FoldAmyloid: a method of prediction of amyloidogenic regions from protein sequence. Bioinformatics. 2010;26:326–332. doi: 10.1093/bioinformatics/btp691. [DOI] [PubMed] [Google Scholar]
- 106.Kim C., Choi J., Lee S.J., Welsh W.J., Yoon S. NetCSSP: web application for predicting chameleon sequences and amyloid fibril formation. Nucleic Acids Res. 2009;37:W469–W473. doi: 10.1093/nar/gkp351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bryan A.W., Menke M., Cowen L.J., Lindquist S.L., Berger B. BETASCAN: probable beta-amyloids identified by pairwise probabilistic analysis. PLoS Comput Biol. 2009;5 doi: 10.1371/journal.pcbi.1000333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bryan A.W., O’Donnell C.W., Menke M., Cowen L.J., Lindquist S., Berger B. STITCHER: dynamic assembly of likely amyloid and prion β-structures from secondary structure predictions. Proteins. 2012;80:410–420. doi: 10.1002/prot.23203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Goldschmidt L., Teng P.K., Riek R., Eisenberg D. Identifying the amylome, proteins capable of forming amyloid-like fibrils. Proc Natl Acad Sci U S A. 2010;107:3487–3492. doi: 10.1073/pnas.0915166107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.O’Donnell C.W., Waldispühl J., Lis M., Halfmann R., Devadas S., Lindquist S., Berger B. A method for probing the mutational landscape of amyloid structure. Bioinformatics. 2011;27:i34–i42. doi: 10.1093/bioinformatics/btr238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tsolis A.C., Papandreou N.C., Iconomidou V.A., Hamodrakas S.J. A consensus method for the prediction of “aggregation-prone” peptides in globular proteins. PLoS One. 2013;8 doi: 10.1371/journal.pone.0054175. [DOI] [PMC free article] [PubMed] [Google Scholar]